- 1Department of Botany, University of British Columbia, Vancouver, BC, Canada
- 2Department of Biological Science, University of Toronto Scarborough, Toronto, ON, Canada
- 3Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
High-quality seeds provide valuable nutrients to human society and ensure successful seedling establishment. During maturation, seeds accumulate storage compounds that are required to sustain seedling growth during germination. This review focuses on the epigenetic repression of the embryonic and seed maturation programs in seedlings. We begin with an extensive overview of mutants affecting these processes, illustrating the roles of core proteins and accessory components in the epigenetic machinery by comparing mutants at both phenotypic and molecular levels. We highlight how omics assays help uncover target-specific functional specialization and coordination among various epigenetic mechanisms. Furthermore, we provide an in-depth discussion on the Seed dormancy 4 (Sdr4) transcriptional corepressor family, comparing and contrasting their regulation of seed germination in the dicotyledonous species Arabidopsis and two monocotyledonous crops, rice and wheat. Finally, we compare the similarities in the activation and repression of the embryonic and seed maturation programs through a shared set of cis-regulatory elements and discuss the challenges in applying knowledge largely gained in model species to crops.
Introduction
Seeds contributed greatly to the successful colonization of dry land by spermatophytes (seed plants) by allowing reproduction in the absence of water and dispersal under unfavorable growth conditions. During mid-to-late development, seeds enter the maturation phase, during which the seed accumulates storage compounds (lipids, proteins, or polysaccharides depending on the species) that are needed during germination to sustain post-germinative growth. Seeds can contribute up to 70% of our caloric intake, as food and livestock feed, and therefore play a fundamental role in human nutrition. For these reasons, there is a great interest in understanding the gene regulatory networks (GRNs) controlling seed development and germination to improve qualitative and quantitative traits associated with these processes.
GRN depicts transcriptional regulators and their target genes as nodes and the regulatory relationships as directed edges (Barabási and Oltvai, 2004). GRN analysis often employs one or more of model-, information theory-, and machine learning-based methods (Zhao et al., 2021). Although barebone GRNs can be inferred from transcription profiles of transcription factors (TFs) and their potential target genes based on co-expression in bulk tissues, higher resolution of transcriptome profiles and data examining additional regulatory features will provide valuable information for more accurate inference and a higher understanding of context-specific regulation. However, data availability differs by biological systems. In animals, single-cell (sc) omics data are widely available. These datasets are increasingly paired, which means various types of omics data are collected from the same cells. Equally importantly, a plethora of tools, developed and validated with multi-omics data in animals, are available to infer GRNs (Badia-i-Mompel et al., 2023). By contrast, although a growing amount of plant sc transcriptome and ATAC-seq profiles became available in recent years, sc datasets are still relatively scarce in number and type in plants, and multi-omics assays are usually generated from separate samples, resulting in reduced data resolution, increased noise, and fewer features for multimodal predictions of important biological processes. To date, plant sc transcriptome profiles of vegetative tissues have provided excellent insights into plant development (Shahan et al., 2022), evolution (Guillotin et al., 2023), and response to environmental cues (Wendrich et al., 2020). TF binding sites are widely used together with sc profiles of transcriptome or chromatin accessibility in integrative analysis pipelines such as MINI-EX and MINI-AC to infer cell type-specific GRN (Ferrari et al., 2022; Manosalva Pérez et al., 2024).
There are several challenges in studying transcriptional repression in seeds. First, high-resolution data are limited, especially during late seed maturation. Second, the regulation of gene repression is arguably more complex. For instance, the inference of gene activation is relatively straightforward based on open chromatin, binding (sites) of transcriptional activators, and elevated transcript abundance. By contrast, lack of transcription could result from either active repression or a lack of activation, which may not be distinguishable solely by chromatin accessibility assays. Additionally, both computational analysis (Brooks et al., 2021) and experimental evidence (Zhu et al., 2024) show that TFs may play a dual role of activation and repression depending on their interacting partners, and such characterizations are relatively limited in seeds. In this review, we highlight various omics datasets useful for GRN inference in seeds. We place a special emphasis on the repressive machinery by detailing the genetic and functional genomic characterizations to provide context for their mode of action. Considering complex biological networks are hierarchical and scale-free (Basso et al., 2005), we focus on the master LAFL TFs in the examples because these hubs are likely to capture extensive regulation.
The function of LAFL in seed development and maturation has been extensively studied in Arabidopsis (Lepiniec et al., 2018; Alizadeh et al., 2021; Gazzarrini and Song, 2024). LEC1, FUS3, and LEC2 transcripts accumulate since the zygote or pre-globular stage of embryogenesis, and the expression of ABI3 initiates later at the globular stage. These TFs play a prominent role during seed maturation, where they are required for the accumulation of seed storage compounds, such as triacylglycerols (TAGs), seed storage proteins (SSPs), oleosins (OLEO), and stress proteins (LEA) (Giraudat et al., 1992; Meinke, 1992; Meinke et al., 1994; West et al., 1994; Roscoe et al., 2015). LAFL also promote seed dormancy and suppress precocious germination of immature seeds, known as vivipary or pre-harvest sprouting (PHS) in cereals, by inhibiting cell division, the activity of the shoot and root meristems, and the differentiation of the cotyledon epidermis (trichome development) and vascular system (xylem) (Meinke, 1992; Keith et al., 1994; Nambara et al., 1995; Raz et al., 2001; Bryant et al., 2019). LAFL’s functions in seed development and germination are partly mediated by the hormones auxin, ABA, GA, ethylene, brassinosteroids, and jasmonate (Parcy et al., 1994; Curaba et al., 2004; Gazzarrini et al., 2004; Braybrook et al., 2006; Stone et al., 2008; Lumba et al., 2012; Ryu et al., 2014; Chiu et al., 2016; Pan et al., 2020). The spatiotemporal expression patterns of LAFL are tightly controlled at the transcriptional and epigenetic levels. Epigenetic silencing of LAFL is required to promote post-embryonic development (Lepiniec et al., 2018; Alizadeh et al., 2021; Gazzarrini and Song, 2024).
Seed quality is a holistic term that includes seed viability, moisture and nutrient content, depth of dormancy, longevity, and vigor. Some of these traits, such as seed vigor, are assessed during the transition from seed to seedlings (Finch-Savage and Bassel, 2016). Therefore, we focus on the regulation and performance of seed-to-seedling transition by using this process as a proxy of seed quality in this review. Recent advances made possible by omics tools provide a rich resource to compare and contrast various epigenetic machinery that regulate the transition from seed to seedlings. The information will also provide mechanistic context for GRN inference at this developmental stage. Using a novel transcriptional cofactor, Seed dormancy 4 (Sdr4), as an example, we summarize and discuss the role of this corepressor and its orthologs in Arabidopsis, rice, and wheat. We also discuss cis-regulatory elements (CREs) important for seed maturations as well as a potential regulatory symmetry exerted on these CREs, which together facilitate the activation and repression of the seed maturation program.
Phenotypic and omics resemblance of genetic and epigenetic regulators
Functional characterization of players in seed-to-seedling transition by phenotypic resemblance and genetic evidence
The importance of shutting down the embryonic program and silencing LAFL at the end of seed development is clearly shown by LAFL overexpression/ectopic expression (OE) phenotypes, which include delayed germination, growth and flowering (ABI3, FUS3, and LEC2), development of cotyledon-like organs (LEC1, FUS3, and LEC2), somatic embryos, and callus-like structures (LEC1 and LEC2) (Table 1). Ectopic expression of early-acting genes (LEC1 and LEC2) shows the most dramatic phenotypes, such as somatic embryos and development of callus-like structures. These embryonic structures accumulate seed storage lipids (TAGs), and proteins (2S and 12S), as a result of ectopic expression of the seed maturation program (Table 1). Robust repression of LAFL and the embryonic program during vegetative development is orchestrated by a suite of epigenetic regulators, including Polycomb group (PcG) repressive complex 1 (PRC1) and PRC2, Trithorax group (TrxG), chromatin remodeling factors and other repressive proteins (Figure 1) (Xiao et al., 2017a; Lepiniec et al., 2018; Gazzarrini and Song, 2024). PRC1 catalyzes monoubiquitylation of lysine (K) 121 on histone 2A (H2AK121ub). PRC1 core components include BMI1A, BMI1B, BMI1C, RING1A, and RING1B (Figure 1), all of which have E3 ligase activity (Mozgova and Henning, 2015; Baile et al., 2022). PRC2 promotes trimethylation of H3 on K27 (H3K27me3), catalyzed by the histone methyltransferases SWN, CLF, and MEA. Arabidopsis has three core PRC2 complexes: EMF-PRC2, composed of EMF2, CLF/SWN, FIE, and MSI1; VRN-PRC2, composed of VRN2, CLF/SWN, FIE, and MSI1; and FIS-PRC2, which includes FIS2, MEA, FIE, and MSI1 (Figure 1). These PRC2 complexes have overlapping as well as specific functions throughout development (Bieluszewski et al., 2021; Baile et al., 2022). Several PRC1 and PRC2 accessory proteins that physically interact with the PRC core components to repress transcription, such as VAL1/2, HDAC, LHP1/TFL2, AL6, AL7, EMF1, NDX, and VRN5/VIL1, as well as TrxG factors such as H3K4me3 methyltransferases ATX and ULT1, chromatin remodeling factors such as PKL and PKR2, and corepressors such as TPL, TPR, and AtSDR4L, aid in repression of LAFL and the embryonic program during vegetative development (Gazzarrini and Song, 2024).
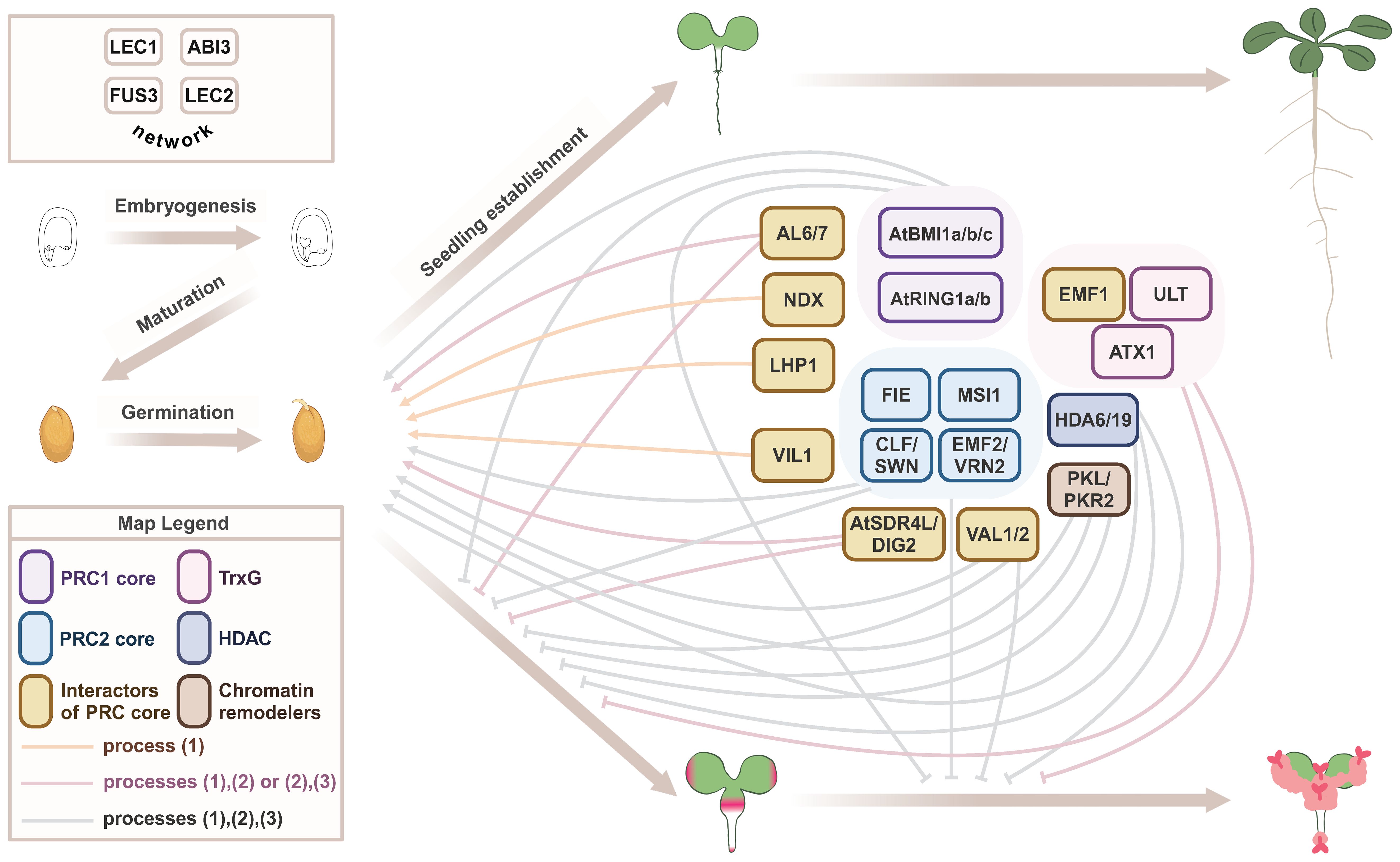
Figure 1 Epigenetic regulators that repress the seed maturation program during seed-to-seedling transition. A proposed model on the regulation of early developmental phase transitions by general and stage-specific epigenetic machinery in Arabidopsis (Table 1). Embryogenesis and maturation phases of seed development are regulated by the LAFL network (top left). The seed-to-seedling phase transition comprises the break of dormancy, germination, and seedling establishment. This normal developmental trajectory requires repression of the seed maturation program by a suite of epigenetic regulators. Loss-of-function mutations in these epigenetic regulators often lead to an alternative developmental trajectory, with mutant seeds exhibiting defects in one or more processes: (1) germination, (2) repression of post-embryonic accumulation of storage compounds, with pink color indicating the accumulation of storage lipids, and (3) repression of subsequent formation of callus and somatic embryos. Severe mutants of general epigenetic machinery usually show all three types of defects, whereas the role of stage-specific regulators may be limited to one or two processes.
Loss-of-function mutants in PRC1 and PRC2 components mimic LAFL OE phenotypes, supporting a role for PRCs in terminating the embryonic program during seed-to-seedling transition. Table 1 and Figure 1 summarize the phenotypes of these mutants and dissect the regulation by the general epigenetic machinery (e.g. core proteins in the PRCs and TrxG) that affects all stages of plant development, as well as developmental stage-specific epigenetic regulators, including the facultative accessory protein of PRCs. Although there is extensive overlap in mutant phenotypes, some epigenetic mutants only show a subset of phenotypes and with variable penetrance. This is because some PRC2 complex subunits are present in all complexes, e.g., FIE and MSI, while others are specific for some developmental stages (e.g., EMF2, VRN2, FIS2, and MEA), and/or their mutant phenotypes are so severe (embryo lethality) that their roles in other developmental stages can be difficult to uncover (FIE, FIS2, and MSI1). For instance, double mutants lacking PRC2 paralogs that are involved in vegetative development and flowering (swn clf and emf2 vrn2), show delayed germination (swn clf) and develop embryonic callus (swn clf and emf2 vrn2). FIE is a single subunit required in all PRC2 complexes and fie mutants are not viable, due to endosperm over-proliferation and embryo arrest at the heart stage. Similar phenotypes are shown by prc2 mutants, fis2, mea, and msi1 (Meinke, 2020). However, evaluation of embryonically-rescued fie plants allowed for the discovery of its role in seed-to-seedling transition. Indeed, embryonically rescued fie lines displayed delayed germination, somatic embryos, and embryonic callus-like structures. Embryonic callus also develops in double mutants lacking PRC1 paralogs (atbmi1a atbmi1b and atring1a atring1b), as well as accessory proteins that promote histone deacetylation (hda6 hda19 RNAi) or recruit PRCs (val1 val2), albeit with different frequencies. The penetrance of the embryonic callus phenotype can be further increased in mutants simultaneously lacking core subunits of PRC1 and PRC2 (atring1a atring1b clf) or in the absence of PRC2 and chromatin remodeling factors (swn clf pkl) (Table 1). Thus, a stable shutdown of LAFL and the embryonic program during vegetative development requires a suite of epigenetic regulators.
Interestingly, mutants in accessory proteins such as al6 al7, lhp1, and vil1 affect germination, dormancy, and sensitivity to ABA and stress, and show repression of late-acting LAFL such as ABI3, suggesting more stage- and context-specific roles for these accessory proteins. While TrxG and PcG play opposite roles in regulating flowering time through FLC (Pien et al., 2008), TrxG’s synergistic role with PRC in transcriptional repression during seed-to-seedling transition does not conform to the norm of TrxG in exerting transcriptional activation. The TrxG mutants atx and ult strongly enhance the phenotype of emf1, with emf1 atx ult showing swollen roots and embryo- and callus-like structures (Xu et al., 2018). Similarly, the TrxG homolog SDG8 acts synergistically with PRC2 EMF2 in repressing seed maturation genes, as shown by the development of somatic embryos in emf2 sdg8 (Table 1). Altogether, LAFL and downstream seed maturation genes serve as excellent models for comprehending the repression of the seed maturation program during the transition from seed to seedling, similar to FLC for the transition from vegetative to reproductive phases (Whittaker and Dean, 2017).
Omics studies facilitate a comprehensive understanding of seed-to-seedling transition
Mutant phenotypes and marker genes are powerful tools to associate regulation with a specific developmental stage. These tools together with omics studies allow for the efficient and comprehensive characterization of biological processes. Transcriptomic and epigenomic datasets confirmed the observation that general epigenetic machinery participates in all major development transitions, and their regulatory specificity is often determined by accessory proteins and interacting transcription factors (Merini et al., 2017; Xiao et al., 2017b). Starting from profiling canonical histone marks such as H3K27me3 (Table 1), our understanding of the epigenetic regulation of plant phase transition has been substantially advanced through the integration of genetic, biochemical, cell biology, and multi-omic results. Here, we discuss some representative examples of how transcriptomic and epigenomic data enable a quantitative assessment of regulatory specificity, suggest interactions between regulators, and uncover crosstalk between regulatory machinery.
Genome-wide comparisons reveal regulatory specificity of PRC components
Both PRC1 and PRC2 contribute to the repression of the embryonic and seed maturation programs (Table 1). Omics studies have strengthened the observation originally made based on marker genes that PRC1 and PRC2 have both shared and unique functions (Wang et al., 2016; Zhou et al., 2017). Furthermore, multi-omic profiling, including chromatin accessibility, H2AK121ub, H3K27me3, and transcriptome of mutants of core and accessory components of PRC1 and PRC2 and wild-type seedlings have shown that PRC1 regulates chromatin accessibility (Yin et al., 2021), and defined PRC1-dependent and -independent repression by H3K27me3 (Kralemann et al., 2020; Yin et al., 2021).
Genome-wide profiles also aid in the characterization of the accessory proteins, which are crucial for general machinery such as PRCs to exert specific roles in plant development and stress responses. The association of LHP1 with PRC2 was supported by extensive overlap between H3K27me3 and genome-wide binding of LHP1 (Turck et al., 2007; Zhang et al., 2007). Additionally, omics assays help to identify or rule out regulators at specific developmental stages. Cross-comparison of differentially expressed (DE) genes and H3K27me3 in the mutants of lhp1 and PRC core components showed that LHP1 regulates vegetative-to-reproductive transition, but lacks a major role in seed-to-seedling transition (Wang et al., 2016). Furthermore, the accumulation of H2AK121ub is similar between lhp1 and wild-type seedlings (Zhou et al., 2017). Collectively, despite the physical association of LHP1 with both PRC1 (Xu and Shen, 2008) and PRC2 (Hecker et al., 2015), the binding of LHP1 to the dormancy promoting loci, ABI3 and DELAY OF GERMINATION 1 (DOG1) (Molitor et al., 2014), and the modest upregulation of DOG1 in lhp1 mutant (Chen et al., 2020) showed a connection to PRC1 and a minor role in germination, with genome-wide evidence indicating that LHP1 mainly functions after the seed-to-seedling transition as a PRC2 accessory protein.
Genome-wide comparisons support functional redundancy and protein–protein interactions
As discussed above, the epigenetic machinery functions through multi-protein complexes. ChIP-seq has been widely used to examine target sites of epigenetic regulators. The sheer number of binding sites across the genome provides numerous data points to assess binding similarities of epigenetic regulators and infer functional redundancy and/or protein–protein interactions (PPIs). In the case of PRC core proteins CLF and SWN, the nearly identical binding patterns (Shu et al., 2019), synergistic phenotype of the clf swn double mutant (Chanvivattana et al., 2004), and the absence of data showing their physical interaction indicate that these two methyltransferases function redundantly in different variants of PRC2 core complexes. By contrast, genomic data can also be used as supporting evidence in functional characterization of VAL proteins. Both VAL1/HSI2 and VAL2/HSL1 bind to the RY motif (CATGCA/TGCATG), and they homo- and heterodimerize via the PHD-L domain (Chen et al., 2020). The physical interaction and association with the same CRE are further supported by the extensive overlap of VAL1 and VAL2 across the genome (Yuan et al., 2021). Similarly, the physical interaction of the VAL proteins with the PRC2 core components SWN and CLF and with the chromatin remodeler PKL was demonstrated by both PPI assays and genome-wide binding similarities (Yuan et al., 2021; Liang et al., 2022).
Genome-wide comparisons suggest crosstalk of epigenetic machinery
The seedling is arguably one of the most vulnerable stages of a plant’s life cycle. Germinating seeds must strike a balance between preserving limited resources to survive uncertain weather patterns in spring and fall and the rapid consumption of storage compounds to establish themselves and outcompete nearby seedlings. To cope with these two seemingly conflicting priorities, master TFs of seed maturation such as ABI3 and FUS3 remain inducible by abiotic stresses during the early stages of germination, while these TFs and the seed maturation program are robustly repressed within a few days after germination if environment is favorable. The robust repression requires coordination of various regulators. For instance, the repression involves a transient increase of histone deacetylase activity soon after germination (Tai et al., 2005), reduced accumulation of active histone marks and increased accumulation of repressive marks at seed maturation and dormancy loci within the first 3 days of germination (Yang et al., 2022a; Pan et al., 2023a), and participation of histone variants (Zhao et al., 2022b). Consistent with the multifaceted regulation, higher-order mutants defective in multiple epigenetic machinery often exhibit more severe phenotypes in germination and seedling establishment (Table 1). The coordination of gene repression is often facilitated by PPI. For instance, VAL1 serves an interaction hub to unite the HDAC and PRC activities (Baile et al., 2021; Mikulski et al., 2022), presumably to reduce the level of active marks such as H3ac, enhance the level of repressive histone marks, and limit chromatin accessibility through H2AK121ub and H3K27me3 (Mikulski et al., 2022). Besides VAL proteins, other TFs that possess an EAR motif can also recruit HDAC and enhance H3K27me3 marking through their physical interaction with TPL or SAP18 (Baile et al., 2021). Crosstalk between repressive machinery has been revealed by omics assays. For instance, LHP1 interacts with ATRX, a chromatin remodeler that deposits histone variant H3.3. The intersection of LHP1 target genes with DE genes in atrx mutant connects repressive histone marks with histone variants (Wang et al., 2018). Another example is the potential crosstalk between PRC and constitutive heterochromatin in the pericentromeric regions. NDX was discovered as a PRC1-associated protein that regulates ABA sensitivity (Zhu et al., 2020). Recently, genome-wide profiling revealed that NDX binds to heterochromatic small RNA loci and affects non-CG DNA methylation (Karányi et al., 2022), suggesting a potential connection between PRC1 with constitutive heterochromatin and chromatin topology.
A new puzzle piece: Sdr4 family in model and crop species
AtSDR4L/ODR1/SFL1 is a nuclear-localized, plant-specific transcriptional corepressor that is devoid of known DNA-binding domains (Moon et al., 2016; Subburaj et al., 2016; Cao et al., 2020; Liu et al., 2020; Wu et al., 2022). Here, we review the functional characterization of AtSDR4L based on the features summarized in Table 1, Figure 1, and omics tools discussed in the previous section.
AtSDR4L and its paralogs are novel corepressors in Arabidopsis seed-to-seedling transition
The role of AtSDR4L is specific to the seed-to-seedling transition, and its expression increases during seed maturation, peaks in dry seeds, and decreases upon imbibition, subsequent to the expression patterns of LAFL that primarily span from embryogenesis to seed maturation (Stone et al., 2001; Baumbusch, 2003; Cao et al., 2020). Loss-of-function mutants of Atsdr4l share many phenotypic and molecular characteristics with mutants listed in Table 1. Mature seeds of Atsdr4l are more dormant (Cao et al., 2020; Liu et al., 2020; Wu et al., 2022; Zheng et al., 2022). Atsdr4l seedlings exhibit stunted growth, with seed storage compounds accumulating to various degrees depending on exogenous sucrose and the duration of after-ripening and cold stratification (Wu et al., 2022; Zheng et al., 2022). A large number of seed maturation genes are upregulated in Atsdr4l seedlings, and AtSDR4L binds to the upstream region of a subset of these, including LEC1 and ABI3 (Wu et al., 2022). Furthermore, AtSDR4L physically interacts with VAL2, and H3K27me3 at a distal regulatory region upstream of ABI3 is decreased in 3-day-old Atsdr4l seedlings (Lu et al., 2024). The lack of strong dedifferentiation phenotypes, such as those shown in LEC1 and LEC2 OE and mutants of PRC core components, indicates that not all genes required for the formation of callus-like structures and somatic embryos are misregulated. Atsdr4l seedlings resemble ML1:FUS3, suggesting that FUS3 could be an indirect target activated by elevated ABI3. Collectively, these data suggest that AtSDR4L functions together with VAL2 to recruit PRC2 to directly or indirectly repress LAFL and other seed maturation genes.
A recent study suggested that AtSDR4L shares partial functional redundancies with its paralogs to form a repressive module in Arabidopsis (Zheng et al., 2022). The paralogs are collectively known as Dynamic Influencer of Gene expression (DIGs) and DIG-like (DILs)/ABA-inducible transcriptional repressors (AITRs)/Seed dormancy Four-Like (SFLs) (Song et al., 2016; Tian et al., 2017; Zheng et al., 2022). Similar to AtSDR4L, DIGs and DILs are nuclear localized (Song et al., 2016; Tian et al., 2017) and physically interact with VAL2 (Lu et al., 2024). An sfl1 sfl4 (Atsdr4l dig2) double mutant shows strongly enhanced seed dormancy and embryonic traits in seedlings compared to Atsdr4l, suggesting synergy between AtSDR4L and its paralogs (Zheng et al., 2022). On the other hand, sfl2 sfl3 sfl4 (aitr2 aitr6 aitr5, dig1 dil1 dig2) triple mutant seeds exhibited reduced dormancy when they were freshly harvested from siliques at 24 days after flowering (DAF), suggesting antagonistic interaction between AtSDR4L and its paralogs at certain developmental stages (Zheng et al., 2022). Additionally, triple mutant seedlings are hyposensitive to ABA and resistant to drought (Tian et al., 2017), while seedlings overexpressing DIG1 or DIG2 are hypersensitive to ABA and salt (Song et al., 2016). These lines of evidence suggest that AtSDR4L and its paralogs may have context-specific functions that require further investigation.
Sdr4 prevents pre-harvest sprouting in rice
Sdr4 is a major quantitative trait locus and a positive regulator for seed dormancy in rice (Sugimoto et al., 2010; Zhao et al., 2022a). Rice Sdr4, herein referred to as OsSdr4, is expressed in the embryo and the protein is localized to the nucleus (Sugimoto et al., 2010). OsSdr4 transcripts begin to accumulate after 7 DAF and increase as seed ripens (Sugimoto et al., 2010). The expression control of OsSdr4 and its orthologs in wheat, a monocot, and Arabidopsis, a dicot, seems well conserved (Figure 2A). Upstream regulatory sequences of OsSdr4 and orthologs contain multiple RY and G-box (CACGTG) motifs, and binding by ABI3/VP1 and bZIP TFs were shown by in vitro or in vivo assays in multiple species (O’Malley et al., 2016; Tian et al., 2020; Chen et al., 2021; Wu et al., 2022; Liu et al., 2024). Knocking down or knocking out OsSdr4 leads to PHS (Sugimoto et al., 2010; Zhao et al., 2022a). In contrast, Atsdr4l mutant seeds harvested at maturity exhibit delayed germination (Cao et al., 2020; Liu et al., 2020; Wu et al., 2022). The seemingly opposite mutant phenotypes of seed germination between rice and Arabidopsis may be attributable to the downstream target genes of Sdr4 (Figure 2B). In accordance with elevated PHS of sdr4 mutants in rice, the expression of a gibberellin biosynthesis gene, OsGA20ox-1, is significantly upregulated and an ABA-responsive gene, OsLEA3, is significantly downregulated compared to wild-type seeds (Sugimoto et al., 2010; Chen et al., 2021; Zhao et al., 2022a). A few other OsLEA genes and a dormancy regulator OsMFT2 are downregulated in the rice sdr4 mutant as opposed to the upregulation of their orthologs in the Atsdr4l mutants (Wu et al., 2022; Zhao et al., 2022a). Interestingly, a recent spatiotemporal single-cell transcriptomic profile of germinating rice embryo revealed that both OsSdr4 and OsMFT2 are expressed in the scutellum, and share a similar temporal expression pattern as the transcript abundance of both genes sharply decreases after imbibition (Yao et al., 2024). While mature Atsdr4l seeds are more dormant, mutant seeds prematurely harvested at 14 DAF germinate better than wild-type seeds (Zheng et al., 2022), suggesting that temporal-specific regulation may also contribute to the phenotypic difference between rice and Arabidopsis mutants. Storage reserve genes were upregulated in Atsdr4l seedlings in Arabidopsis whereas seed storage catabolism genes are upregulated in Ossdr4 seeds in rice (Wu et al., 2022; Zhao et al., 2022a). The major forms of storage reserves in Arabidopsis are lipids and storage proteins that are deposited in cotyledons, and most of the endosperm except the peripheral endosperm layer in Arabidopsis is consumed by the embryo for nutrient uptake during seed maturation (Sreenivasulu and Wobus, 2013; Doll and Ingram, 2022). In contrast, most of the endosperm in Poaceae is retained and accumulates a substantial amount of starch and storage proteins, followed by programmed cell death without full degradation (Sreenivasulu and Wobus, 2013). Thus, the contrasting role of Sdr4 and its annotated orthologs in grasses and dicots may be associated with opposite regulation of key downstream genes, different types of the major forms of storage compounds, and a higher embryo-to-endosperm ratio in Arabidopsis than that in wheat and rice. In summary, Sdr4 homologs in rice and Arabidopsis share similar expression patterns in seeds and the nuclear localization. However, their function in regulating dormancy is species-, developmental stage-, and tissue-dependent.
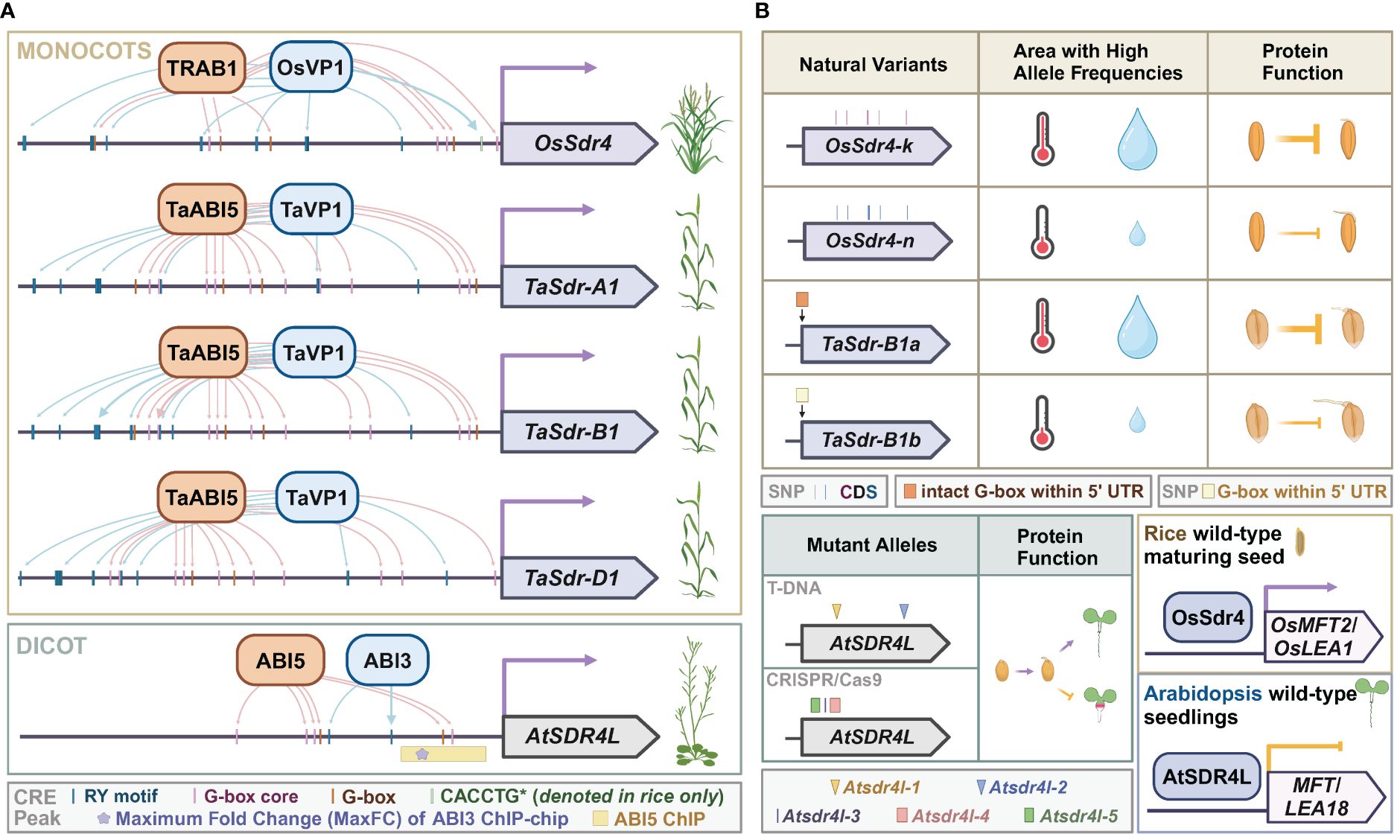
Figure 2 Sdr4 and its orthologs in monocots and dicots. (A) Putative cis-regulatory motifs of OsSdr4 in rice, TaSdrs in wheat, and AtSDR4L in Arabidopsis from 1 bp to 1,500 bp upstream of the corresponding transcription start site. TF binding sites with experimental evidence are indicated by thick arrows protruding from the regulators of SDR4. Data were summarized from O’Malley et al. (2016); Tian et al. (2020); Chen et al. (2021); Wu et al. (2022), and Liu et al. (2024). (B) Top: occurrence of natural variants of OsSdr4 and TaSdr in various climates, and their repression of preharvest sprouting. Bottom left: Mutant alleles of Atsdr4l, and function of wild-type AtSDR4L in promoting germination and preventing fatty acid over-accumulation. Bottom right: the upregulation of the dormancy-related gene, MFT, and the seed reserve-related gene, OsLEA, by Sdr4 and the downregulation of these orthologs by AtSDR4L. Data were summarized from Sugimoto et al. (2010); Zhang et al. (2014); Wu et al. (2022), and Zhao et al. (2022a).
Allelic variations of Sdr4 correlate with regional weather patterns
Allelic variants of Sdr4 and its orthologs in coding and regulatory sequences are associated with quantitative differences in seed dormancy, germination, and post-germinative growth. In Arabidopsis, developmental defects include inhibited root growth, swollen hypocotyl, and excess buildup of storage lipids. These defects are more severe in the CRISPR/Cas9 frameshift or segmental deletion mutants of Atsdr4l-3, Atsdr4l-4, and Atsdr4l-5 than in the T-DNA insertion mutants of Atsdr4l-1 and Atsdr4l-2, possibly because the mutations reside in the CRISPR/Cas9 lines are in the closer downstream of AtSDR4L start codon (Wu et al., 2022; Lu et al., 2024) (Figure 2B). In rice, a nearly isogenic line of OsSdr4 [NIL(Sdr4)], in which the genomic segment containing OsSdr4 from the Kasalath (indica) group was introgressed into a Nipponbare (japonica) background, had a substantially lower rate of seed germination than that of Nipponbare, demonstrating that the Kasalath allele of Sdr4 (OsSdr4-k) confers deeper dormancy than the Nipponbare allele (OsSdr4-n) (Sugimoto et al., 2010) (Figure 2B). The amino acid sequences of OsSdr4-k and OsSdr4-n alleles differ by approximately 10 amino acid residues, which could potentially affect OsSdr4’s characteristics as a cofactor, thus changing binding behaviors to downstream target loci. The japonica group has only the OsSdr4-n allele, whereas the indica group has both OsSdr4-k and Sdr4-n. Analysis of SNPs flanking the OsSdr4-n locus in the indica cultivars indicated their OsSdr4-n allele was introgressed from the japonica group. A subsequent larger-scale study revealed a correlation between allele frequency and weather patterns (Zhao et al., 2022a). Allele frequency of OsSdr4-k and sequence-similar OsSdr4-k' is higher in regions with high annual temperatures and precipitation, whereas OsSdr4-n is more prevalent in areas with lower annual temperatures and precipitation. Interestingly, different geographic distribution for Sdr4 alleles was also reported in wheat (Zhang et al., 2014). Among three homeologs of wheat Sdr4, namely, TaSdr-A1, TaSdr-B1, and TaSdr-D1, the TaSdr-B1b allele carries an SNP that abolishes a G-box in the 5’ UTR immediately upstream of its start codon and is associated with increased germination compared to that of TaSdr-B1a allele with an intact G-box. The mutation in G-box could affect the binding by bZIP and bHLH TFs t TaSdr-B. Between the two alleles, TaSdr-B1a is likely positively selected for resistance to PHS, since the allele frequency of the TaSdr-B1a is high in areas that are more susceptible to severe PHS and low in areas with reduced rainfall and less damage by PHS. These observations in rice and wheat suggest that selection for Sdr4 alleles best adapted to local climates is a shared feature for the adaptation of staple grains.
Regulatory symmetries of activators and repressors at the same CREs for developmental transitions
Regulatory symmetry via the RY motif
The regulatory symmetry is the activation and repression of genes through the same CRE. The regulatory summary of the seed maturation programs by B3 proteins is well established (Suzuki and McCarty, 2008). The RY motif is enriched in many seed maturation genes and is often bound by the B3 TFs FUS3, LEC2, and ABI3 for transcriptional activation, as well as VAL1/HSI2 and VAL2/HSL1 for transcriptional repression (Reidt et al., 2000; Nakabayashi et al., 2005; Suzuki et al., 2007; Tsukagoshi et al., 2007; Suzuki and McCarty, 2008; Jia et al., 2014; Yuan et al., 2021) (Figure 3). VAL1 and VAL2 can homo- or heterodimerize to target RY motifs in the DOG1 promoter and repress its expression in seedlings (Chen et al., 2020). However, the B3 domains of these TFs exhibit high similarity but differential binding affinity to the target CREs, with the B3 domain of LEC2 (LEC2-B3) and FUS3-B3 having greater affinity than that of VAL1-B3 and ABI3-B3 (Jia et al., 2021), and with VAL1-B3 binding more effectively than VAL2-B3 (Chen et al., 2020). These varying RY-binding efficacies are likely owing to the slightly different structural bases of the B3 domains and the absence or presence of additional domains (Jia et al., 2021). This difference may also explain the more constrained and specific roles of LEC2 and FUS3 in the establishment and maintenance of the embryonic states, as well as lending greater flexibility for ABI3 to incorporate additional cues into more complex target recognition (Jia et al., 2013, 2021; Roscoe et al., 2015).
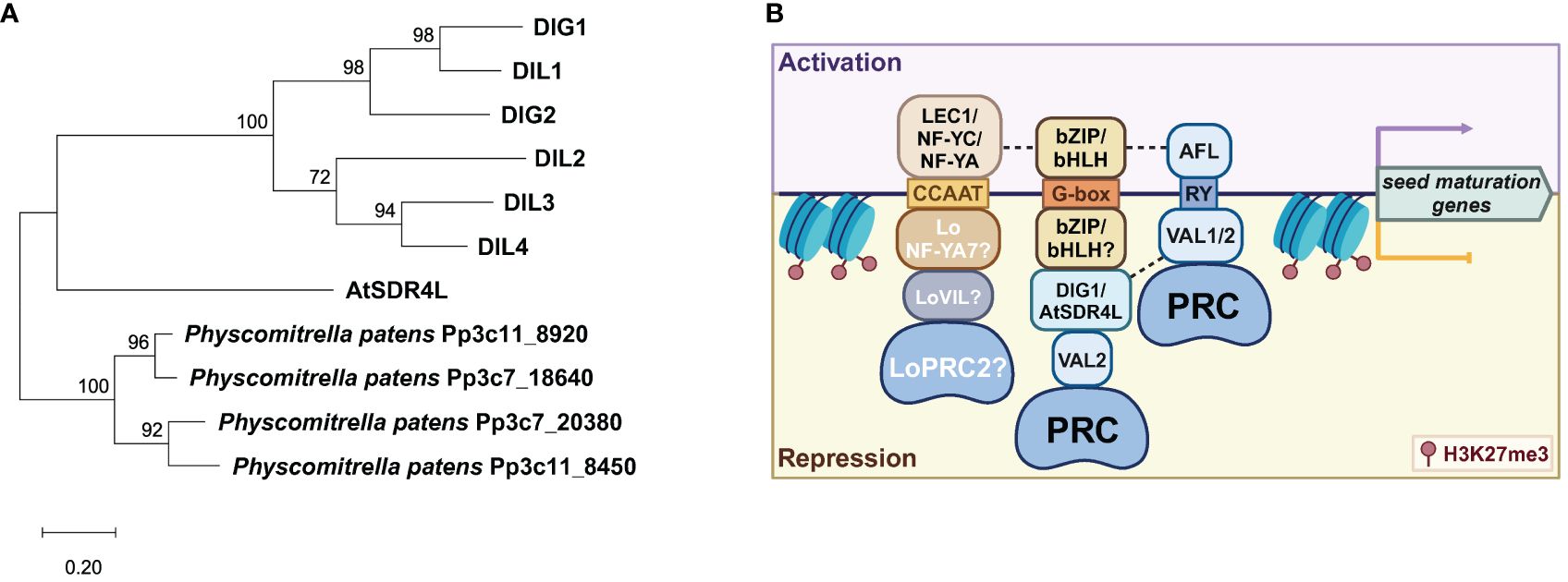
Figure 3 Regulatory symmetry in the activation and repression of seed maturation genes. (A) A phylogenetic tree of the AtSDR4L family in Arabidopsis by the maximum-likelihood method and the JTT-matrix-based model using MEGA11 (Tamura et al., 2021). AtSDR4L’s orthologs in Physcomitrella patens were used as the outgroup (Lang et al., 2018). (B) A working model of the regulatory symmetry between activators and repressors at shared CREs upstream of seed maturation genes. CCAAT is bound by the NF-Y activator complex (top) in Arabidopsis or by the repressor complex of LoNF-YA, LoVIL, and PRC2 in lily (bottom). In Arabidopsis, bZIP/bHLH binding to G-box by itself upregulates the seed maturation genes whereas the G-box binding by bZIP/bHLH coupled with a repressor module of DIG/AtSDR4L-VAL-VIL-PRC2 downregulates these genes. The RY motif can be bound by ABI3/FUS3/LEC1 (AFL) and VAL1/2-PRC, for the activation and repression of the downstream genes, respectively. Dashed lines indicated potentially synergistic effects through protein–protein interaction. Data were summarized from Lumba et al. (2014); Song et al. (2016); Myers and Holt (2018); Bryant et al. (2019); Chen et al. (2020); Liu et al. (2020); Wu et al. (2022), and Pan et al. (2023b).
RY and G-box motifs function coordinately in the activation of seed maturation genes
Transcriptional activation of maturation genes via the RY motif is often coupled with the G-box motif. G-box motifs are preferentially bound by the basic leucine zipper (bZIP) and basic helix-loop-helix (bHLH) TFs, and these CREs are an overrepresented CRE in seed maturation genes (Nakabayashi et al., 2005; Mönke et al., 2012; Mendes et al., 2013; Ezer et al., 2017; Jo et al., 2020). ABI5, a bZIP TF that interacts with ABI3, was found to transactivate the promoter of AtEm6 (Nakamura et al., 2001; Lopez-Molina et al., 2002). G-box-mediated transactivation of gene expression by ABI5 is indicated by the numerous downregulated genes in abi5 dry seed and the enrichment of G-box from these repressed genes (Nakabayashi et al., 2005). Binding to the same RY-containing region upstream of DOG1, AFL TFs may also upregulate DOG1 expression through collaboration with bZIP67 during seed maturation (Bryant et al., 2019). Consistently, G-box and RY motifs are highly enriched in the regulons of ABI3 and FUS3 (Mönke et al., 2012; Wang and Perry, 2013), and ABI3 is believed to induce seed maturation genes via G-box motifs that are in close proximity with RY elements (Suzuki et al., 2003; Jo et al., 2020).
G-box motif and the repression of the seed maturation program
Loci with high occurrence of RY and G-box elements are frequently associated with high occupancy of the repressive histone mark H3K27me3 in the seedlings (Wang et al., 2016; Liu et al., 2019; Baile et al., 2021). Because both AFL and VAL possess the RY-binding B3 domain, AFL may compete with VAL-PRC for the same RY sites in the regulatory regions of seed maturation genes. Emerging evidence suggests that G-box might be recognized by gene repression machinery (Figure 3). Recent studies suggest that AtSDR4L and its paralogs are important for the G-box-dependent transcriptional repression of the seed maturation genes (Song et al., 2016; Liu et al., 2020; Wu et al., 2022). G-box is enriched in AtSDR4L binding regions (Wu et al., 2022). Since AtSDR4L is devoid of known DNA binding domains, it is most likely recruited by bZIP and bHLH family TFs with sequence-specific binding activities to the G-box (Lumba et al., 2014; Liu et al., 2020). It is proposed that AtSDR4L physically interacts with bHLH57 to indirectly inhibit the expression of ABA biosynthetic genes 9-CIS-EPOXYCAROTENOID DIOXYGENASE6 (NCED6) and NCED9, thereby downregulating ABA biosynthesis to counteract seed dormancy (Liu et al., 2020).
Interestingly, the RY motif (CATGCA) is also found in high frequency in AtSDR4L target sites (Lu et al., 2024). Additionally, AtSDR4L and its orthologs also harbor abundant G-box and RY CREs in their own upstream regulatory regions (Figure 2). In Arabidopsis, ABI3 binds to the AtSDR4L promoter and upregulates its expression in the developing seeds (Wu et al., 2022; Zheng et al., 2022). In turn, AtSDR4L represses ABI3 to shut down the seed maturation program in the seedlings, promoting the shift to the vegetative phase (Wu et al., 2022). Intriguingly, AtSDR4L also targets itself, possibly through the G-box and RY motifs in its own promoter (Lu et al., 2024). Similarly, the OsSdr4 promoter contains seven RY and six G-box elements, and one of the RY motifs is in close proximity to a G-box (Sugimoto et al., 2010). The rice ortholog of ABI3, OsVP1, perhaps in collaboration with a rice bZIP TF TRAB1, can induce the expression of OsSdr4 (Sugimoto et al., 2010; Chen et al., 2021). OsSdr4 expression substantially decreases in Osvp1 mutant embryos at the maturation stage. A similar mechanism is conserved in wheat, as TaVP1 binds to the RY motifs, and TaABI5, an ortholog of Arabidopsis bZIP TF ABI5, binds to the G-box in TaSdr promoter to transcriptionally activate TaSdr4 (Liu et al., 2024). These conserved regulatory mechanisms on a key locus of dormancy control in both the model plant and crop species provide further elucidation of the mirrored targeting behaviors by activators and repressors.
Regulatory symmetry in the activation and repression of seed maturation genes via CCAAT and GAGA motifs
Symmetry of activator-repressor binding is not limited to the RY and G-box pairing. The CCAAT motif is a CRE frequently found in the promoters of many genes and specifically targeted by the Nuclear Factor Y (NF-Y) factors for gene regulation (Calvenzani et al., 2012). In Arabidopsis and soybean, the pioneer TF LEC1 (NF-YB factor) can bind to CCAAT box elements as a trimeric complex with the NF-YA and NF-YC subunits to activate the embryonic programs (Yamamoto et al., 2009; Jo et al., 2020) (Figure 3). Conversely, LoNF-YA7 in lily bulbs has been reported to recruit LoVIL1-PRC2 machinery to LoCALS3 locus at CCAAT motif for H3K27me3 deposition, thereby repressing LoCALS3 expression to promote the release of bulb dormancy (Pan et al., 2023b). Similarly, GAGA box-binding BPCs can repress LEC2 during germination (Xiao et al., 2017b) and FUS3 (Wu et al., 2020) in the ovule integuments and endosperm, but activates LEC2 in the embryo (Berger et al., 2011). This is in agreement with GAGA-binding proteins in animals, which function as both activators and repressors (Berger and Dubreucq, 2012). Collectively, these results show that transcriptional activation and repression through the same CREs may be a general mechanism in the activation and repression of the seed maturation program. Regulatory robustness and specificity may be determined by functional coordination of transcriptional regulators that binds to these CREs.
Conclusions and perspectives
Perspectives and challenges to advance basic knowledge
To date, many players involved in the repression of the seed maturation program during the seed-to-seedling transition have been identified. While existing omics data are tremendously useful to understand the general machinery, they often do not fully capture the dynamics of the regulation in seed germination and seedling establishment because many profiling assays were carried out using 10- to 14-day-old seedlings (Table 1). This mismatch with the developmental stages might also miss the identification of stage-specific regulators. For instance, trichostatin A (TSA) treatment to inhibit histone deacetylase activities in 3- and 16-day-old seedlings identified distinct sets of DE genes (Chang and Pikaard, 2005; Tai et al., 2005). Presumably, some regulators of the seed-to-seedling transition might yet to be discovered due to stage-specific regulation and functional redundancy of homologous genes. Additionally, cell biology and biochemical approaches are not always readily applicable to seeds because the seed coat serves as a physical barrier that blocks light and many chemicals. Therefore, tissue- and cell type-specific datasets are often scarce for mature seeds. Previously, transcriptome profiling of dissected developing or germinating seeds (Belmonte et al., 2013; Dekkers et al., 2013) demonstrated tissue- and stage-specific gene expression in Arabidopsis. Single-cell and spatial transcriptomics (Yao et al., 2024) will further advance our understanding of the heterogeneity of gene expression in seeds, allowing a superior statistical power to classify genes based on their expression patterns and designate marker genes to existing and new cell types, thus providing a foundation for understanding cell type-specific GRN. Additional advances may come from integrative analysis of histone modification with omics datasets such as time-course profiles of transcriptome (Narsai et al., 2017), DNA methylation (Bouyer et al., 2017; Kawakatsu et al., 2017), chromatin accessibility and non-coding RNA (Tremblay et al., 2024) during seed-to-seedling transition, and changes in chromatin topology. For instance, histone modifications have been profiled in many epigenetic mutants (Table 1). Combined with other assays such as Hi-C (Huang et al., 2021; Yin et al., 2023) and Hi-ChIP (Huang et al., 2021), these datasets help to reveal how histone marks such as H3K27me3 and H2AK121ub impact spatial genome organization by regulating local and long-range chromatin interactions. Collectively, these data elucidate the unique and shared role of PRC1 and PRC2 in the co-regulation of gene expression, and may contribute to a knowledge framework of multi-loci expression optimization and trait stacking for crop improvement.
Perspectives and challenges for knowledge transfer from model species in the lab to crops in the field
Several PRC-controlled traits, such as dormancy, stress responses, and flowering time, are related to plants’ adaptation to various environments. Regulators specifically targeting these traits are likely to have immediate application value in the field. However, several gaps need to be addressed for knowledge transfer from model species to crop and from controlled laboratory environment to the field. For instance, many important crops and oilseeds are polyploid, which requires additional considerations for homeolog redundancy and subgenome dominance besides optimizing species- and lineage-specific regulation (Ramírez-González et al., 2018; Xiang et al., 2019; Khan et al., 2022). Environmental factors and plant–biotic interactions are prevalent in the field, making trade-offs important considerations to enhance plant performance. For instance, overexpression and mutant phenotypes of master TFs and general epigenetic regulators often reduce fitness, thus requiring more sophisticated engineering of these factors if increased yield under less water and fertilizer usage is the ultimate goal for crop improvement. In summary, research on transcriptional and epigenetic regulation has provided valuable insights into the phase transition from seed to seedlings, and multi-omic studies have revealed many target-specific regulations and crosstalk between regulatory machinery. Further research to identify developmental stage-specific regulators and CREs with minimized fitness trade-off holds strong potential to engineer crops that can adapt to the increasingly stressful environments associated with the increasingly volatile weather patterns from a warming climate.
Author contributions
DG: Conceptualization, Visualization, Writing – original draft. BL: Writing – original draft. MA: Writing – original draft. SG: Conceptualization, Funding acquisition, Writing – review & editing, Writing – original draft. LS: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors thank funding support from the UBC Four Year Doctoral Fellowships to BL and MA, the Natural Sciences and Engineering Research Council of Canada discovery grants (NSERC DG) to SG (NSERC-DG 480529) and LS (RGPIN-2019-05039), and the Canadian Foundation for Innovation (CFI JELF/BCKDF 38187) to LS.
Acknowledgments
The authors thank Dr. Randy Allen for comments and editing, Dr. Shao-shan Carol Huang for discussions, and Jiayi Shan for summarizing some literature. The figures were prepared using BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1416216/full#supplementary-material
Glossary
LEC1, FUS3, LEC2, ABI3 (LAFL):
LEC1: LEAFY COTYLEDON 1
FUS3: FUSCA 3
LEC2: LEAFY COTYLEDON 2
ABI3: ABSCISIC ACID INSENSITIVE 3
Polycomb Repressive Complex 1 (PRC1) Core:
AtRING1A, 1B: Arabidopsis thaliana RING 1A, 1B
AtBMI1A, 1B, 1C: Arabidopsis thaliana BMI1A, 1B, 1C
Polycomb Repressive Complex 2 (PRC2) Core:
CLF: CURLY LEAF
SWN: SWINGER
MEA: MEDEA
EMF2: EMBRYONIC FLOWER 2
VRN2: VERNALIZATION 2
FIS2: FERTILIZATION INDEPENDENT SEED 2
FIE: FERTILIZATION INDEPENDENT ENDOSPERM
MSI1: MULTICOPY SUPPRESSOR OF IRA 1
Proteins that Interact with PRC Core:
AL6, 7: ALFIN-LIKE 6, 7
VAL1/HSI2: VIVIPAROUS1/ABI3-LIKE 1/HIGH-LEVEL EXPRESSION OF SUGAR INDUCIBLE GENE 2
VAL2/HSL1: VIVIPAROUS1/ABI3-LIKE 2/HSI2-LIKE 1
VIL1/VRN5: VERNALIZATION INSENSITIVE 3-LIKE 1/VERNALIZATION 5
LHP1/TFL2: LIKE HETEROCHROMATIN PROTEIN 1/TERMINAL FLOWER 2
EMF1: EMBRYONIC FLOWER 1
BPC1, 2: BASIC PENTACYSTEINE 1, 2
AtSDR4L/SFL1/ODR1: Arabidopsis thaliana SEED DORMANCY FOUR-LIKE 1/SEED DORMANCY FOUR-LIKE 1/REVERSAL OF RDO5 1
DIG1/SFL2/AITR2: DYNAMIC INFLUENCER OF GENE 1/SEED DORMANCY FOUR-LIKE 2/ABA-INDUCED TRANSCRIPTION REPRESSOR 2
DIL1/SFL3/AITR6: DIG-LIKE 1/SEED DORMANCY FOUR-LIKE 3/ABA-INDUCED TRANSCRIPTION REPRESSOR 6
DIG2/SFL4/AITR5: DYNAMIC INFLUENCER OF GENE 2/SEED DORMANCY FOUR-LIKE 4/ABA-INDUCED TRANSCRIPTION REPRESSOR 5
NDX: NODULIN HOMEOBOX
HDA6, 19: HISTONE DEACETYLASE 6, 19
SAP18: SIN3 ASSOCIATED POLYPEPTIDE 18
Other Epigenetic Regulators of Seed-to-Seedling Transition:
ATX1: ARABIDOPSIS TRITHORAX 1
ULT1: ULTRAPETALA 1
PKL: PICKLE
PKR2: PICKLE RELATED 2
SDG8: SET DOMAIN GROUP 8
TPL: TOPLESS
TPR1, 2, 3, 4: TOPLESS-RELATED 1, 2, 3, 4
References
Aichinger, E., Villar, C. B. R., Farrona, S., Reyes, J. C., Hennig, L., Köhler, C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in arabidopsis. PloS Genet. 5, e1000605. doi: 10.1371/journal.pgen.1000605
Alizadeh, M., Hoy, R., Lu, B., Song, L. (2021). Team effort: Combinatorial control of seed maturation by transcription factors. Curr. Opin. Plant Biol. 63, 102091. doi: 10.1016/j.pbi.2021.102091
Badia-i-Mompel, P., Wessels, L., Müller-Dott, S., Trimbour, R., Ramirez Flores, R. O., Argelaguet, R., et al. (2023). Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754. doi: 10.1038/s41576-023-00618-5
Baile, F., Gómez-Zambrano, Á., Calonje, M. (2022). Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 3, 100267. doi: 10.1016/j.xplc.2021.100267
Baile, F., Merini, W., Hidalgo, I., Calonje, M. (2021). EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis. Plant Cell 33, 2701–2715. doi: 10.1093/plcell/koab139
Barabási, A.-L., Oltvai, Z. N. (2004). Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113. doi: 10.1038/nrg1272
Basso, K., Margolin, A. A., Stolovitzky, G., Klein, U., Dalla-Favera, R., Califano, A. (2005). Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390. doi: 10.1038/ng1532
Baumbusch, L. O. (2003). LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J. Exp. Bot. 55, 77–87. doi: 10.1093/jxb/erh014
Belmonte, M. F., Kirkbride, R. C., Stone, S. L., Pelletier, J. M., Bui, A. Q., Yeung, E. C., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. 110, E435–E444. doi: 10.1073/pnas.1222061110
Berger, N., Dubreucq, B. (2012). Evolution goes GAGA: GAGA binding proteins across kingdoms. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 1819, 863–868. doi: 10.1016/j.bbagrm.2012.02.022
Berger, N., Dubreucq, B., Roudier, F., Dubos, C., Lepiniec, L. (2011). Transcriptional Regulation of Arabidopsis LEAFY COTYLEDON2 Involves RLE, a cis -Element That Regulates Trimethylation of Histone H3 at Lysine-27. Plant Cell 23, 4065–4078. doi: 10.1105/tpc.111.087866
Bieluszewski, T., Xiao, J., Yang, Y., Wagner, D. (2021). PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 26, 1186–1198. doi: 10.1016/j.tplants.2021.06.006
Bouyer, D., Kramdi, A., Kassam, M., Heese, M., Schnittger, A., Roudier, F., et al. (2017). DNA methylation dynamics during early plant life. Genome Biol. 18, 179. doi: 10.1186/s13059-017-1313-0
Bouyer, D., Roudier, F., Heese, M., Andersen, E. D., Gey, D., Nowack, M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PloS Genet. 7, e1002014. doi: 10.1371/journal.pgen.1002014
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C., Calonje, M. (2010). Keeping cell identity in arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Braybrook, S. A., Stone, S. L., Park, S., Bui, A. Q., Le, B. H., Fischer, R. L., et al. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. 103, 3468–3473. doi: 10.1073/pnas.0511331103
Brooks, M. D., Juang, C.-L., Katari, M. S., Alvarez, J. M., Pasquino, A., Shih, H.-J., et al. (2021). ConnecTF: A platform to integrate transcription factor–gene interactions and validate regulatory networks. Plant Physiol. 185, 49–66. doi: 10.1093/plphys/kiaa012
Bryant, F. M., Hughes, D., Hassani-Pak, K., Eastmond, P. J. (2019). Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in arabidopsis. Plant Cell 31, 1276–1288. doi: 10.1105/tpc.18.00892
Calonje, M., Sanchez, R., Chen, L., Sung, Z. R. (2008). EMBRYONIC FLOWER1 participates in polycomb group–mediated AG gene silencing in arabidopsis. Plant Cell 20, 277–291. doi: 10.1105/tpc.106.049957
Calvenzani, V., Testoni, B., Gusmaroli, G., Lorenzo, M., Gnesutta, N., Petroni, K., et al. (2012). Interactions and CCAAT-binding of arabidopsis thaliana NF-Y subunits. PloS One 7, e42902. doi: 10.1371/journal.pone.0042902
Cao, H., Han, Y., Li, J., Ding, M., Li, Y., Li, X., et al. (2020). Arabidopsis thaliana SEED DORMANCY 4-LIKE regulates dormancy and germination by mediating the gibberellin pathway. J. Exp. Bot. 71, 919–933. doi: 10.1093/jxb/erz471
Carter, B., Bishop, B., Ho, K. K., Huang, R., Jia, W., Zhang, H., et al. (2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in arabidopsis. Plant Cell 30, 1337–1352. doi: 10.1105/tpc.17.00867
Casson, S. A., Lindsey, K. (2006). The turnip mutant of arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 142, 526–541. doi: 10.1104/pp.106.080895
Chang, S., Pikaard, C. S. (2005). Transcript profiling in arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation*[boxs]. J. Biol. Chem. 280, 796–804. doi: 10.1074/jbc.M409053200
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y.-H., Sung, Z. R., et al. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Chen, L., Cheng, J. C., Castle, L., Sung, Z. R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9, 2011–2024. doi: 10.1105/tpc.9.11.2011
Chen, D., Molitor, A., Liu, C., Shen, W.-H. (2010a). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Chen, L.-T., Luo, M., Wang, Y.-Y., Wu, K. (2010b). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, N., Veerappan, V., Abdelmageed, H., Kang, M., Allen, R. D. (2018). HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in arabidopsis. Plant Cell 30, 600–619. doi: 10.1105/tpc.17.00655
Chen, N., Wang, H., Abdelmageed, H., Veerappan, V., Tadege, M., Allen, R. D. (2020). HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 227, 840–856. doi: 10.1111/nph.16559
Chen, W., Wang, W., Lyu, Y., Wu, Y., Huang, P., Hu, S., et al. (2021). OsVP1 activates Sdr4 expression to control rice seed dormancy via the ABA signaling pathway. Crop J. 9, 68–78. doi: 10.1016/j.cj.2020.06.005
Chiu, R. S., Saleh, Y., Gazzarrini, S. (2016). Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signal. Behav. 11, e1247137. doi: 10.1080/15592324.2016.1247137
Curaba, J., Moritz, T., Blervaque, R., Parcy, F., Raz, V., Herzog, M., et al. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in arabidopsis. Plant Physiol. 136, 3660–3669. doi: 10.1104/pp.104.047266
Dekkers, B. J. W., Pearce, S., Van Bolderen-Veldkamp, R. P., Marshall, A., Widera, P., Gilbert, J., et al. (2013). Transcriptional dynamics of two seed compartments with opposing roles in arabidopsis seed germination. Plant Physiol. 163, 205–215. doi: 10.1104/pp.113.223511
Doll, N. M., Ingram, G. C. (2022). Embryo–endosperm interactions. Annu. Rev. Plant Biol. 73, 293–321. doi: 10.1146/annurev-arplant-102820-091838
Ezer, D., Shepherd, S. J. K., Brestovitsky, A., Dickinson, P., Cortijo, S., Charoensawan, V., et al. (2017). The G-box transcriptional regulatory code in arabidopsis. Plant Physiol. 175, 628–640. doi: 10.1104/pp.17.01086
Feng, C., Cai, X.-W., Su, Y.-N., Li, L., Chen, S., He, X.-J. (2021). Arabidopsis RPD3-like histone deacetylases form multiple complexes involved in stress response. J. Genet. Genomics 48, 369–383. doi: 10.1016/j.jgg.2021.04.004
Ferrari, C., Manosalva Pérez, N., Vandepoele, K. (2022). MINI-EX: Integrative inference of single-cell gene regulatory networks in plants. Mol. Plant 15, 1807–1824. doi: 10.1016/j.molp.2022.10.016
Finch-Savage, W. E., Bassel, G. W. (2016). Seed vigour and crop establishment: extending performance beyond adaptation. J. Exp. Bot. 67, 567–591. doi: 10.1093/jxb/erv490
Franco-Echevarría, E., Nielsen, M., Schulten, A., Cheema, J., Morgan, T. E., Bienz, M., et al. (2023). Distinct accessory roles of Arabidopsis VEL proteins in Polycomb silencing. Genes Dev. 37, 801–817. doi: 10.1101/gad.350814.123
Gazzarrini, S., Song, L. (2024). LAFL factors in seed development and phase transitions. Annu. Rev. Plant Biol. 75, 28.1–28.30. doi: 10.1146/annurev-arplant-070623-111458
Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385. doi: 10.1016/j.devcel.2004.06.017
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Guillotin, B., Rahni, R., Passalacqua, M., Mohammed, M. A., Xu, X., Raju, S. K., et al. (2023). A pan-grass transcriptome reveals patterns of cellular divergence in crops. Nature 617, 785–791. doi: 10.1038/s41586-023-06053-0
Hecker, A., Brand, L. H., Peter, S., Simoncello, N., Kilian, J., Harter, K., et al. (2015). The arabidopsis GAGA-binding factor BASIC PENTACYSTEINE6 recruits the POLYCOMB-REPRESSIVE COMPLEX1 component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA motifs. Plant Physiol. 168, 1013–1024. doi: 10.1104/pp.15.00409
Henderson, J. T., Li, H.-C., Rider, S. D., Mordhorst, A. P., Romero-Severson, J., Cheng, J.-C., et al. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Huang, Y., Sicar, S., Ramirez-Prado, J. S., Manza-Mianza, D., Antunez-Sanchez, J., Brik-Chaouche, R., et al. (2021). Polycomb-dependent differential chromatin compartmentalization determines gene coregulation in Arabidopsis. Genome Res. 31, 1230–1244. doi: 10.1101/gr.273771.120
Ikeuchi, M., Iwase, A., Rymen, B., Harashima, H., Shibata, M., Ohnuma, M., et al. (2015). PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1, 15089. doi: 10.1038/nplants.2015.89
Jia, H., McCarty, D. R., Suzuki, M. (2013). Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 163, 1293–1305. doi: 10.1104/pp.113.220988
Jia, H., Suzuki, M., McCarty, D. R. (2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks: Regulation of the seed to seedling developmental phase transition. Wiley Interdiscip. Rev. Dev. Biol. 3, 135–145. doi: 10.1002/wdev.126
Jia, H., Suzuki, M., McCarty, D. R. (2021). Structural variation affecting DNA backbone interactions underlies adaptation of B3 DNA binding domains to constraints imposed by protein architecture. Nucleic Acids Res. 49, 4989–5002. doi: 10.1093/nar/gkab257
Jo, L., Pelletier, J. M., Hsu, S.-W., Baden, R., Goldberg, R. B., Harada, J. J. (2020). Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development. Proc. Natl. Acad. Sci. 117, 1223–1232. doi: 10.1073/pnas.1918441117
Junker, A., Mönke, G., Rutten, T., Keilwagen, J., Seifert, M., Thi, T. M. N., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana: Characterization of the LEC1 regulon. Plant J. 71, 427–442. doi: 10.1111/j.1365-313X.2012.04999.x
Kagaya, Y., Okuda, R., Ban, A., Toyoshima, R., Tsutsumida, K., Usui, H., et al. (2005a). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in arabidopsis. Plant Cell Physiol. 46, 300–311. doi: 10.1093/pcp/pci031
Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., Hattori, T. (2005b). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406. doi: 10.1093/pcp/pci048
Karányi, Z., Mosolygó-L, Á., Feró, O., Horváth, A., Boros-Oláh, B., Nagy, É., et al. (2022). NODULIN HOMEOBOX is required for heterochromatin homeostasis in Arabidopsis. Nat. Commun. 13, 5058. doi: 10.1038/s41467-022-32709-y
Kawakatsu, T., Nery, J. R., Castanon, R., Ecker, J. R. (2017). Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18, 171. doi: 10.1186/s13059-017-1251-x
Keith, K., Kraml, M., Dengler, N. G., McCourt, P. (1994). fusca3: A heterochronic mutation affecting late embryo development in arabidopsis. Plant Cell 6, 589–600. doi: 10.1105/tpc.6.5.589
Khan, D., Ziegler, D. J., Kalichuk, J. L., Hoi, V., Huynh, N., Hajihassani, A., et al. (2022). Gene expression profiling reveals transcription factor networks and subgenome bias during Brassica napus seed development. Plant J. 109, 477–489. doi: 10.1111/tpj.15587
Kralemann, L. E. M., Liu, S., Trejo-Arellano, M. S., Muñoz-Viana, R., Köhler, C., Hennig, L. (2020). Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable Polycomb-mediated gene repression in Arabidopsis. Genome Biol. 21, 144. doi: 10.1186/s13059-020-02062-8
Lang, D., Ullrich, K. K., Murat, F., Fuchs, J., Jenkins, J., Haas, F. B., et al. (2018). The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533. doi: 10.1111/tpj.13801
Lepiniec, L., Devic, M., Roscoe, T. J., Bouyer, D., Zhou, D.-X., Boulard, C., et al. (2018). Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 31, 291–307. doi: 10.1007/s00497-018-0337-2
Liang, Z., Yuan, L., Xiong, X., Hao, Y., Song, X., Zhu, T., et al. (2022). The transcriptional repressors VAL1 and VAL2 mediate genome-wide recruitment of the CHD3 chromatin remodeler PICKLE in Arabidopsis. Plant Cell 34, 3915–3935. doi: 10.1093/plcell/koac217
Liu, C., Cheng, J., Zhuang, Y., Ye, L., Li, Z., Wang, Y., et al. (2019). Polycomb repressive complex 2 attenuates ABA -induced senescence in Arabidopsis. Plant J. 97, 368–377. doi: 10.1111/tpj.14125
Liu, J., Deng, S., Wang, H., Ye, J., Wu, H.-W., Sun, H.-X., et al. (2016). CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis. Plant Physiol. 171, 424–436. doi: 10.1104/pp.15.01335
Liu, S., Li, L., Wang, W., Xia, G., Liu, S. (2024). TaSRO1 interacts with TaVP1 to modulate seed dormancy and pre-harvest sprouting resistance in wheat. J. Integr. Plant Biol. 66, 36–53. doi: 10.1111/jipb.13600
Liu, F., Zhang, H., Ding, L., Soppe, W. J. J., Xiang, Y. (2020). REVERSAL OF RDO5 1, a homolog of rice seed dormancy4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in arabidopsis. Plant Cell 32, 1933–1948. doi: 10.1105/tpc.20.00026
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., Chua, N.-H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Lotan, T., Ohto, M., Yee, K. M., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lu, B., Alizadeh, M., Hoy, R., Zheng, R., Go, D., Song, L. (2024). Co-repressors AtSDR4L and DIG1 interact with transcription factor VAL2 and promote Arabidopsis seed-to-seedling transition. Plant Physiol., kiae225. doi: 10.1093/plphys/kiae225
Lumba, S., Toh, S., Handfield, L.-F., Swan, M., Liu, R., Youn, J.-Y., et al. (2014). A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in arabidopsis. Dev. Cell 29, 360–372. doi: 10.1016/j.devcel.2014.04.004
Lumba, S., Tsuchiya, Y., Delmas, F., Hezky, J., Provart, N. J., Shi Lu, Q., et al. (2012). The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol. 10, 8. doi: 10.1186/1741-7007-10-8
Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., et al. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7, 947–952. doi: 10.1038/sj.embor.7400760
Manosalva Pérez, N., Ferrari, C., Engelhorn, J., Depuydt, T., Nelissen, H., Hartwig, T., et al. (2024). MINI-AC : inference of plant gene regulatory networks using bulk or single-cell accessible chromatin profiles. Plant J. 117, 280–301. doi: 10.1111/tpj.16483
Meinke, D. W. (1992). A homoeotic mutant of arabidopsis thaliana with leafy cotyledons. Science 258, 1647–1650. doi: 10.1126/science.258.5088.1647
Meinke, D. W. (2020). Genome-wide identification of EMBRYO - DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 226, 306–325. doi: 10.1111/nph.16071
Meinke, D. W., Franzmann, L. H., Nickle, T., Yeung, E. (1994). Leafy cotyledon mutants of arabidopsis. Plant Cell 6, 1049–1064. doi: 10.1105/tpc.6.8.1049
Mendes, A., Kelly, A. A., van Erp, H., Shaw, E., Powers, S. J., Kurup, S., et al. (2013). bZIP67 regulates the omega-3 fatty acid content of arabidopsis seed oil by activating FATTY ACID DESATURASE3. Plant Cell 25, 3104–3116. doi: 10.1105/tpc.113.116343
Merini, W., Romero-Campero, F. J., Gomez-Zambrano, A., Zhou, Y., Turck, F., Calonje, M. (2017). The arabidopsis polycomb repressive complex 1 (PRC1) components atBMI1A, B, and C impact gene networks throughout all stages of plant development. Plant Physiol. 173, 627–641. doi: 10.1104/pp.16.01259
Mikulski, P., Wolff, P., Lu, T., Nielsen, M., Echevarria, E. F., Zhu, D., et al. (2022). VAL1 acts as an assembly platform co-ordinating co-transcriptional repression and chromatin regulation at Arabidopsis FLC. Nat. Commun. 13, 5542. doi: 10.1038/s41467-022-32897-7
Molitor, A. M., Bu, Z., Yu, Y., Shen, W.-H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10, e1004091. doi: 10.1371/journal.pgen.1004091
Monfared, M. M., Simon, M. K., Meister, R. J., Roig-Villanova, I., Kooiker, M., Colombo, L., et al. (2011). Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis: Roles of BASIC PENTACYSTEINE genes. Plant J. 66, 1020–1031. doi: 10.1111/j.1365-313X.2011.04562.x
Mönke, G., Seifert, M., Keilwagen, J., Mohr, M., Grosse, I., Hähnel, U., et al. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254. doi: 10.1093/nar/gks594
Moon, H.-D., Lee, M.-S., Kim, S.-H., Jeong, W.-J., Choi, D.-W. (2016). Identification of a drought responsive gene encoding a nuclear protein involved in drought and freezing stress tolerance in Arabidopsis. Biol. Plant 60, 105–112. doi: 10.1007/s10535-015-0567-1
Mozgova, I., Hennig, L. (2015). The Polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Mu, Y., Zou, M., Sun, X., He, B., Xu, X., Liu, Y., et al. (2017). Basic pentacysteine proteins repress abscisic acid insensitive4 expression via direct recruitment of the polycomb-repressive complex 2 in arabidopsis root development. Plant Cell Physiol. 58, 607–621. doi: 10.1093/pcp/pcx006
Myers, Z. A., Holt, B. F. (2018). NUCLEAR FACTOR-Y: still complex after all these years? Curr. Opin. Plant Biol. 45, 96–102. doi: 10.1016/j.pbi.2018.05.015
Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed: Molecular profiling in Arabidopsis seed. Plant J. 41, 697–709. doi: 10.1111/j.1365-313X.2005.02337.x
Nakamura, S., Lynch, T. J., Finkelstein, R. R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. Cell Mol. Biol. 26, 627–635. doi: 10.1046/j.1365-313x.2001.01069.x
Nambara, E., Keith, K., McCourt, P., Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636. doi: 10.1242/dev.121.3.629
Narsai, R., Gouil, Q., Secco, D., Srivastava, A., Karpievitch, Y. V., Liew, L. C., et al. (2017). Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 18, 172. doi: 10.1186/s13059-017-1302-3
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Ogas, J., Cheng, J.-C., Sung, Z. R., Somerville, C. (1997). Cellular Differentiation Regulated by Gibberellin in the Arabidopsis thaliana pickle Mutant. Science 277, 91–94. doi: 10.1126/science.277.5322.91
Pan, J., Hu, Y., Wang, H., Guo, Q., Chen, Y., Howe, G. A., et al. (2020). Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in arabidopsis. Plant Cell 32, 3846–3865. doi: 10.1105/tpc.19.00838
Pan, W., Li, J., Du, Y., Zhao, Y., Xin, Y., Wang, S., et al. (2023b). Epigenetic silencing of callose synthase by VIL1 promotes bud-growth transition in lily bulbs. Nat. Plants 9, 1451–1467. doi: 10.1038/s41477-023-01492-z
Pan, J., Zhang, H., Zhan, Z., Zhao, T., Jiang, D. (2023a). A REF6-dependent H3K27me3-depleted state facilitates gene activation during germination in Arabidopsis. J. Genet. Genomics 50, 178–191. doi: 10.1016/j.jgg.2022.09.001
Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. doi: 10.1105/tpc.6.11.1567
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. 114, E6710–E6719. doi: 10.1073/pnas.1707957114
Perruc, E., Kinoshita, N., Lopez-Molina, L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52, 927–936. doi: 10.1111/j.1365-313X.2007.03288.x
Pien, S., Fleury, D., Mylne, J. S., Crevillen, P., Inzé, D., Avramova, Z., et al. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20, 580–588. doi: 10.1105/tpc.108.058172
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089. doi: 10.1126/science.aar6089
Ramirez-Prado, J. S., Latrasse, D., Rodriguez-Granados, N. Y., Huang, Y., Manza-Mianza, D., Brik-Chaouche, R., et al. (2019). The Polycomb protein LHP 1 regulates Arabidopsis thaliana stress responses through the repression of the MYC 2-dependent branch of immunity. Plant J. 100, 1118–1131. doi: 10.1111/tpj.14502
Raz, V., Bergervoet, J. H. W., Koornneef, M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128, 243–252. doi: 10.1242/dev.128.2.243
Reidt, W., Wohlfarth, T., Ellerström, M., Czihal, A., Tewes, A., Ezcurra, I., et al. (2000). Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 21, 401–408. doi: 10.1046/j.1365-313x.2000.00686.x
Roscoe, T. T., Guilleminot, J., Bessoule, J.-J., Berger, F., Devic, M. (2015). Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in arabidopsis. Plant Cell Physiol. 56, 1215–1228. doi: 10.1093/pcp/pcv049
Ryu, H., Cho, H., Bae, W., Hwang, I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5, 4138. doi: 10.1038/ncomms5138
Santos Mendoza, M., Dubreucq, B., Miquel, M., Caboche, M., Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Letters 579, 4666–4670. doi: 10.1016/j.febslet.2005.07.037
Schneider, A., Aghamirzaie, D., Elmarakeby, H., Poudel, A. N., Koo, A. J., Heath, L. S., et al. (2016). Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 85, 305–319. doi: 10.1111/tpj.13106
Schubert, D., Clarenz, O., Goodrich, J. (2005). Epigenetic control of plant development by Polycomb-group proteins. Curr. Opin. Plant Biol. 8, 553–561. doi: 10.1016/j.pbi.2005.07.005
Shahan, R., Hsu, C.-W., Nolan, T. M., Cole, B. J., Taylor, I. W., Greenstreet, L., et al. (2022). A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Dev. Cell 57, 543–560.e9. doi: 10.1016/j.devcel.2022.01.008
Shen, Y., Devic, M., Lepiniec, L., Zhou, D.-X. (2015). Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 13, 811–820. doi: 10.1111/pbi.12315
Shu, J., Chen, C., Thapa, R. K., Bian, S., Nguyen, V., Yu, K., et al. (2019). Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 3, e00100. doi: 10.1002/pld3.100
Song, L., Huang, S.-S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550–aag1550. doi: 10.1126/science.aag1550
Sreenivasulu, N., Wobus, U. (2013). Seed-development programs: A systems biology–based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64, 189–217. doi: 10.1146/annurev-arplant-050312-120215
Stone, S. L., Braybrook, S. A., Paula, S. L., Kwong, L. W., Meuser, J., Pelletier, J., et al. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3151–3156. doi: 10.1073/pnas.0712364105
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. 98, 11806–11811. doi: 10.1073/pnas.201413498
Subburaj, S., Cao, S., Xia, X., He, Z. (2016). Phylogenetic Analysis, Lineage-Specific Expansion and Functional Divergence of seed dormancy 4-Like Genes in Plants. PloS One 11, e0153717. doi: 10.1371/journal.pone.0153717
Sugimoto, K., Takeuchi, Y., Ebana, K., Miyao, A., Hirochika, H., Hara, N., et al. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. 107, 5792–5797. doi: 10.1073/pnas.0911965107
Sung, Z. R., Belachew, A., Shunong, B., Bertrand-Garcia, R. (1992). EMF, an arabidopsis gene required for vegetative shoot development. Science 258, 1645–1647. doi: 10.1126/science.258.5088.1645
Suzuki, M., Ketterling, M. G., Li, Q.-B., McCarty, D. R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132, 1664–1677. doi: 10.1104/pp.103.022475
Suzuki, M., McCarty, D. R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11, 548–553. doi: 10.1016/j.pbi.2008.06.015
Suzuki, M., Wang, H. H.-Y., McCarty, D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3 - LIKE B3 genes. Plant Physiol. 143, 902–911. doi: 10.1104/pp.106.092320
Tai, H. H., Tai, G. C. C., Beardmore, T. (2005). Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol. Biol. 59, 909–925. doi: 10.1007/s11103-005-2081-x
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tanaka, M., Kikuchi, A., Kamada, H. (2008). The arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. doi: 10.1104/pp.107.111674
Tang, X., Lim, M.-H., Pelletier, J., Tang, M., Nguyen, V., Keller, W. A., et al. (2012). Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J. Exp. Bot. 63, 1391–1404. doi: 10.1093/jxb/err383
Tian, H., Chen, S., Yang, W., Wang, T., Zheng, K., Wang, Y., et al. (2017). A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 40, 2958–2971. doi: 10.1111/pce.13058
Tian, R., Wang, F., Zheng, Q., Niza, V. M. A. G. E., Downie, A. B., Perry, S. E. (2020). Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. Cell Mol. Biol. 103, 1679–1694. doi: 10.1111/tpj.14854
Tremblay, B. J. M., Santini, C. P., Cheng, Y., Zhang, X., Rosa, S., Qüesta, J. I. (2024). Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition. Nat. Commun. 15, 1724. doi: 10.1038/s41467-024-46082-5
Tsai, A. Y.-L., Gazzarrini, S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis: Interaction between FUSCA3 and AKIN10. Plant J. 69, 809–821. doi: 10.1111/j.1365-313X.2011.04832.x
Tsukagoshi, H., Morikami, A., Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. 104, 2543–2547. doi: 10.1073/pnas.0607940104
Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M.-L., Guillaume, E., Buisine, N., et al. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PloS Genet. 3, e86. doi: 10.1371/journal.pgen.0030086
Veerappan, V., Chen, N., Reichert, A. I., Allen, R. D. (2014). HSI2/VAL1 PHD-like domain promotes H3K27 trimethylation to repress the expression of seed maturation genes and complex transgenes in Arabidopsis seedlings. BMC Plant Biol. 14, 293. doi: 10.1186/s12870-014-0293-4
Veerappan, V., Wang, J., Kang, M., Lee, J., Tang, Y., Jha, A. K., et al. (2012). A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta 236, 1–17. doi: 10.1007/s00425-012-1630-1
Veluchamy, A., Jégu, T., Ariel, F., Latrasse, D., Mariappan, K. G., Kim, S.-K., et al. (2016). LHP1 regulates H3K27me3 spreading and shapes the three-dimensional conformation of the arabidopsis genome. PloS One 11, e0158936. doi: 10.1371/journal.pone.0158936
Wang, H., Jiang, D., Axelsson, E., Lorković, Z. J., Montgomery, S., Holec, S., et al. (2018). LHP1 Interacts with ATRX through Plant-Specific Domains at Specific Loci Targeted by PRC2. Mol. Plant 11, 1038–1052. doi: 10.1016/j.molp.2018.05.004
Wang, H., Liu, C., Cheng, J., Liu, J., Zhang, L., He, C., et al. (2016). Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PloS Genet. 12, e1005771. doi: 10.1371/journal.pgen.1005771
Wang, F., Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi: 10.1104/pp.112.212282
Wendrich, J. R., Yang, B., Vandamme, N., Verstaen, K., Smet, W., Van De Velde, C., et al. (2020). Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370, eaay4970. doi: 10.1126/science.aay4970
West, M., Yee, K. M., Danao, J., Zimmerman, J. L., Fischer, R. L., Goldberg, R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in arabidopsis. Plant Cell 6, 1731–1745. doi: 10.1105/tpc.6.12.1731
Whittaker, C., Dean, C. (2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Wu, T., Alizadeh, M., Lu, B., Cheng, J., Hoy, R., Bu, M., et al. (2022). The transcriptional co-repressor SEED DORMANCY 4-LIKE (AtSDR4L) promotes the embryonic-to-vegetative transition in Arabidopsis thaliana. J. Integr. Plant Biol. 64, 2075–2096. doi: 10.1111/jipb.13360
Wu, J., Mohamed, D., Dowhanik, S., Petrella, R., Gregis, V., Li, J., et al. (2020). Spatiotemporal restriction of FUSCA3 expression by class I BPCs promotes ovule development and coordinates embryo and endosperm growth. Plant Cell 32, 1886–1904. doi: 10.1105/tpc.19.00764
Xiang, D., Quilichini, T. D., Liu, Z., Gao, P., Pan, Y., Li, Q., et al. (2019). The transcriptional landscape of polyploid wheats and their diploid ancestors during embryogenesis and grain development. Plant Cell 31, 2888–2911. doi: 10.1105/tpc.19.00397
Xiao, J., Jin, R., Wagner, D. (2017a). Developmental transitions: integrating environmental cues with hormonal signaling in the chromatin landscape in plants. Genome Biol. 18, 88. doi: 10.1186/s13059-017-1228-9
Xiao, J., Jin, R., Yu, X., Shen, M., Wagner, J. D., Pai, A., et al. (2017b). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552. doi: 10.1038/ng.3937
Xu, F., Kuo, T., Rosli, Y., Liu, M.-S., Wu, L., Chen, L.-F. O., et al. (2018). Trithorax group proteins act together with a polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in arabidopsis. Mol. Plant 11, 659–677. doi: 10.1016/j.molp.2018.01.010
Xu, L., Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Yamamoto, A., Kagaya, Y., Toyoshima, R., Kagaya, M., Takeda, S., Hattori, T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856. doi: 10.1111/j.1365-313X.2009.03817.x
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., Calonje, M. (2013). VAL- and atBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yang, Z., Liu, X., Wang, K., Li, Z., Jia, Q., Zhao, C., et al. (2022b). ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 73, 2077–2092. doi: 10.1093/jxb/erab524
Yang, D., Zhao, F., Zhu, D., Chen, X., Kong, X., Wu, Y., et al. (2022a). Progressive chromatin silencing of ABA biosynthesis genes permits seed germination in Arabidopsis. Plant Cell 34, 2871–2891. doi: 10.1093/plcell/koac134
Yao, J., Chu, Q., Guo, X., Shao, W., Shang, N., Luo, K., et al. (2024). Spatiotemporal transcriptomic landscape of rice embryonic cells during seed germination. Dev. Cell. doi: 10.1016/j.devcel.2024.05.016
Yin, X., Romero-Campero, F. J., de Los Reyes, P., Yan, P., Yang, J., Tian, G., et al. (2021). H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots. Nat. Commun. 12, 315. doi: 10.1038/s41467-020-20614-1
Yin, X., Romero-Campero, F. J., Yang, M., Baile, F., Cao, Y., Shu, J., et al. (2023). Binding by the Polycomb complex component BMI1 and H2A monoubiquitination shape local and long-range interactions in the Arabidopsis genome. Plant Cell 35, 2484–2503. doi: 10.1093/plcell/koad112
Yuan, L., Song, X., Zhang, L., Yu, Y., Liang, Z., Lei, Y., et al. (2021). The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 49, 98–113. doi: 10.1093/nar/gkaa1129
Zhang, M., Cao, X., Jia, Q., Ohlrogge, J. (2016). FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 88, 95–107. doi: 10.1111/tpj.13233
Zhang, X., Garreton, V., Chua, N.-H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543. doi: 10.1101/gad.1318705
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., Jacobsen, S. E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhang, Y., Miao, X., Xia, X., He, Z. (2014). Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor. Appl. Genet. 127, 855–866. doi: 10.1007/s00122-014-2262-6
Zhao, M., He, W., Tang, J., Zou, Q., Guo, F. (2021). A comprehensive overview and critical evaluation of gene regulatory network inference technologies. Brief. Bioinform. 22, bbab009. doi: 10.1093/bib/bbab009
Zhao, T., Lu, J., Zhang, H., Xue, M., Pan, J., Ma, L., et al. (2022b). Histone H3.3 deposition in seed is essential for the post-embryonic developmental competence in Arabidopsis. Nat. Commun. 13, 7728. doi: 10.1038/s41467-022-35509-6
Zhao, B., Zhang, H., Chen, T., Ding, L., Zhang, L., Ding, X., et al. (2022a). Sdr4 dominates pre-harvest sprouting and facilitates adaptation to local climatic condition in Asian cultivated rice. J. Integr. Plant Biol. 64, 1246–1263. doi: 10.1111/jipb.13266
Zheng, L., Otani, M., Kanno, Y., Seo, M., Yoshitake, Y., Yoshimoto, K., et al. (2022). Seed dormancy 4 like1 of Arabidopsis is a key regulator of phase transition from embryo to vegetative development. Plant J. 112, 460–475. doi: 10.1111/tpj.15959
Zhou, Y., Romero-Campero, F. J., Gómez-Zambrano, Á., Turck, F., Calonje, M. (2017). H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18, 69. doi: 10.1186/s13059-017-1197-z
Zhou, Y., Tan, B., Luo, M., Li, Y., Liu, C., Chen, C., et al. (2013). HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in arabidopsis seedlings. Plant Cell 25, 134–148. doi: 10.1105/tpc.112.096313
Zhou, Y., Yang, P., Zhang, F., Luo, X., Xie, J. (2020). Histone deacetylase HDA19 interacts with histone methyltransferase SUVH5 to regulate seed dormancy in Arabidopsis. Plant Biol. 22, 1062–1071. doi: 10.1111/plb.13158
Zhu, Y., Hu, X., Duan, Y., Li, S., Wang, Y., Rehman, A. U., et al. (2020). The arabidopsis nodulin homeobox factor atNDX interacts with atRING1A/B and negatively regulates abscisic acid signaling. Plant Cell 32, 703–721. doi: 10.1105/tpc.19.00604
Zhu, T., Wei, C., Yu, Y., Zhang, Z., Zhu, J., Liang, Z., et al. (2024). The BAS chromatin remodeler determines brassinosteroid-induced transcriptional activation and plant growth in Arabidopsis. Dev. Cell 59, 924–939.e6. doi: 10.1016/j.devcel.2024.01.021
Keywords: climate, crop adaptation, epigenetic regulation, HDAC, LAFL, PKL, PRC, Sdr4
Citation: Go D, Lu B, Alizadeh M, Gazzarrini S and Song L (2024) Voice from both sides: a molecular dialogue between transcriptional activators and repressors in seed-to-seedling transition and crop adaptation. Front. Plant Sci. 15:1416216. doi: 10.3389/fpls.2024.1416216
Received: 12 April 2024; Accepted: 20 June 2024;
Published: 06 August 2024.
Edited by:
Raju Datla, Global Institute for Food Security (GIFS), CanadaReviewed by:
Sarah J. Whitcomb, Agricultural Research Service (USDA), United StatesZhihua Hua, Ohio University, United States
Copyright © 2024 Go, Lu, Alizadeh, Gazzarrini and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Song, bGlhbmcuc29uZ0Bib3RhbnkudWJjLmNh
 Dongeun Go
Dongeun Go Bailan Lu
Bailan Lu Milad Alizadeh1
Milad Alizadeh1 Sonia Gazzarrini
Sonia Gazzarrini Liang Song
Liang Song