
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 May 2024
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1415921
 Yanyan Wang1†
Yanyan Wang1† Xinyun Li1†
Xinyun Li1† Yunrong Mo1†
Yunrong Mo1† Caiqian Jiang1
Caiqian Jiang1 Ying Zhou1
Ying Zhou1 Jingyi Hu1
Jingyi Hu1 Youling Zhang1
Youling Zhang1 Junheng Lv1*
Junheng Lv1* Kai Zhao1*
Kai Zhao1* Zhenya Lu2*
Zhenya Lu2*GATA proteins are transcription factors of zinc finger proteins, which play an important role in plant growth development and abiotic stress. However, there have been no identification or systematic studies of the GATA gene family in eggplant. In this study, 28 SmGATA genes were identified in the genome database of eggplant, which could be divided into four subgroups. Plant development, hormones, and stress-related cis-acting elements were identified in promoter regions of the SmGATA gene family. RT-qPCR indicated that 4 SmGATA genes displayed upregulated expressions during fruit developmental stage, whereas 2 SmGATA genes were down-regulated expression patterns. It was also demonstrated that SmGATA genes may be involved in light signals to regulate fruit anthocyanin biosynthesis. Furthermore, the expression patterns of SmGATA genes under ABA, GA and MeJA treatments showed that the SmGATAs were involved in the process of fruit ripening. Notably, SmGATA4 and SmGATA23 were highly correlated with the expression of anthocyanin biosynthesis genes, light-responsive genes, and genes that function in multiple hormone signaling pathways and the proteins they encoded were localized in the nucleus. All these results showed GATA genes likely play a major role in regulating fruit anthocyanin biosynthesis by integrating the light, ABA, GA and MeJA signaling pathways and provided references for further research on fruit quality in eggplant.
Transcription factors (TFs) play a critical role in regulating gene expression in plants. They are specialized proteins capable of interacting with specific DNA sequences, usually within a gene’s promoter region, to regulate its transcription into messenger RNA (mRNA) and subsequently into proteins (Riechmann et al., 2000; Fan et al., 2021). Furthermore, TFs can form complexes with other transcription factors, coactivators, and corepressors to finely regulate gene expression. These complexes allow for the integration of multiple signals and contribute to the precise control of transcriptional activity (Xu, 2020; Lai et al., 2022). Besides their involvement in transcriptional regulation, transcription factors also participate in various physiological and biochemical pathways during the development of higher plants, including plant growth and development (Strader et al., 2022), metabolic networks (Rueda-López et al., 2015), stress responses (Yao et al., 2022), and disease resistance (Li et al., 2017). Within this category, the GATA family has emerged as a prominent group of transcription factors, widely participating in key mechanisms related to plant growth, development, and stress response.
GATA proteins, found across eukaryotes including animals, plants, and fungi, are evolutionarily conserved regulatory factors. They derive their name from their distinctive capability to recognize and bind to the T/AGATAA/G core sequences, exerting regulatory control over various biological processes (Lowry and Atchley, 2000; Behringer and Schwechheimer, 2015). GATA transcription factors typically contain one or two highly conserved type IV zinc finger structural modules, characterized by the sequence signature C-X2-CX17-20-C-X2-C (C, cysteine; X, other residues), followed by a highly basic region (Schwechheimer et al., 2022). The zinc finger structure represents a prevalent DNA-binding domain, stabilizing its three-dimensional conformation through coordination bonds between zinc ions and specific amino acid residues within the protein (Takatsuji, 1998). This structural module in GATA transcription factors enables precise recognition and binding to the promoter regions of target genes, thus regulating their transcriptional activity. The initial discovery of a plant GATA factor, Ntl1 (NIT2-like), from Nicotiana tabacum, is pivotal for understanding nitrogen metabolism (Daniel-Vedele and Caboche, 1993). Presently, GATA TFs have been detected in numerous plant species, such as Arabidopsis thaliana (30 members) (Reyes et al., 2004), Oryza sativa (28 members) (Reyes et al., 2004), Solanum lycopersicum (30 members) (Yuan et al., 2018), Malus domestica (35 members) (Chen et al., 2017), Arachis hypogaea (45 members) (Li et al., 2023), Solanum tuberosum (49 members) (Yu et al., 2021), and Triticum aestivum (79 members) (Feng et al., 2022).
GATA transcription factors, key regulators in plants, have diverse biological functions, such as regulating light responses (Jeong and Shih, 2003), chlorophyll synthesis (An et al., 2014), and responses to environmental stress (Richter et al., 2013). By recognizing and binding to specific DNA sequences, these transcription factors regulate the expression of relevant genes, thereby influencing plant growth, development, and adaptability. Regarding light response regulation, GATA transcription factors modulate plant adaptive responses to changes in light exposure. For instance, GATA2 in Arabidopsis thaliana mediates photomorphogenesis and plays a pivotal role in light signal transduction (Luo et al., 2010). Furthermore, two GATA genes in A. thaliana were strongly light-regulated and expressed in photosynthetic organs. Several GATA genes have shown responsiveness to light, darkness, and circadian rhythm changes (Manfield et al., 2007). Another example is PdGATA19/PdGNC from poplar (Populus deltoides), which contributes to photosynthesis and growth (An et al., 2020). Additionally, GATA genes have been found to be extensively involved in hormone signal transduction. In A. thaliana, AtGATA12 expression is downregulated by gibberellin (GA), promoting seed dormancy (Ravindran et al., 2017), while GATA2 mediates cross-talk between the brassinosteroid and light signaling pathways, regulating plant photomorphogenesis (Luo et al., 2010). PdGNC can be significantly induced by abscisic acid (ABA) and dehydration, thereby improving water use efficiency by reducing stomatal aperture and enhancing water deficit tolerance in poplar (Shen et al., 2021). Proteomic analysis findings indicate that GATAs can interact with JAZ, a key regulator in the jasmonic acid (JA) signaling pathway (Sen et al., 2016).
Eggplant (Solanum melongena L.), a significant economic crop, is cultivated globally for its nutrient-rich fruits. The fruits of purple eggplant have a high content of anthocyanins, which is highly beneficial for health (Bor et al., 2006; García et al., 2008). So far, serval TFs including AP2/ERF and WRKY gene family under abiotic stress conditions from eggplant have been well-characterized (Yang et al., 2020; Li et al., 2021a). However, systematic studies on the eggplant GATA TFs superfamily have not been attempted so far. In the present investigation, we screened 28 GATA genes from the eggplant genome and investigated their phylogenetic correlation, gene structure and motif pattern. We further analyzed their chromosome distribution and putative cis-acting elements on the promoters. More importantly, we studied the expression patterns of SmGATA genes during fruit development. In addition, we analyzed the expression patterns of GATA genes in response to light and multiple hormone signals. The results of this study will provide a platform and cues for further studies related to the functional characterization of eggplant GATAs and identification of pivotal GATA involved in growth and development.
The eggplant cultivar ‘zaohongqie 2’ was used as a material, which were cultivated in a climate chamber at a circadian temperature of 22–25°C with a 16 h photoperiod. Fruit samples were collected at 6 time points during the growing season, including 5 days after flowering (5 d), 10 d, 15 d, 20 d 25 d and 30 d, and which were used to study the expression characteristics of SmGATA genes.
A fruit bagging experiment was conducted at 0 DAF. The bags were removed from eggplant fruits at 25 days after flowering (25 DAF). Samples were taken at 0 d, 2 d, 4 d, 6 d, and 8 d after bag removal. For hormone treatment, Similarly, the eggplant fruit that had been bagged for 25 d will be removed from the bag and then which were treated with water (control), 200 μM ABA, 200 μM GA and 200 μM MeJA. Three replicates were performed in the experiment, with 3 fruits per replicate. The fruits were immediately frozen in liquid nitrogen and stored at -80°C.
The deduced amino acid sequences of 30 GATA genes in Arabidopsis thaliana were used as query sequences for a BlastP search of the eggplant genome database (http://www.eggplant-hq.cn/Eggplant/home/index). The CDD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and the SMART database (http://smart.embl-heidelberg.de) were used to analyze the domains of the candidate GATA proteins. The molecular weights, isoelectric points (pIs), and grand average of hydropathicity (GRAVY) values of the SmGATA proteins were calculated using the ExPASy website (https://web.expasy.org/protparam/).
Multiple sequence alignments of GATA proteins were analyzed using ClustalW in BioEdit (http://bioedit.software.informer.com), and the phylogenetic tree was constructed with the neighbor-joining algorithm in MEGA 6.0 (Tamura et al., 2013). Bootstrap analysis was carried out with 1000 replicates. WebLogo (http://weblogo.berkeley.edu/logo.cgi) was used to generate sequence logos of the conserved domains. Furthermore, MEME (http://meme-suite.org/) was used tosearch for motifs in all GATA genes. A structure analysis of the SmGATA genes was performed using the Gene Structure Display Server (GSDS) (http://gsds.gao-lab.org/).
The promoter sequences (2 kb upstream from ATG) were extracted from eggplant whole genome, and cis-elements in the promoters were predicted using the PlantCARE online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). TBtools v1.106. https://github.com/CJ-Chen/TBtools/releases was used to map the genes to chromosome duplication events analysis (Chen et al., 2022).
Total RNA was extracted from the samples using a Plant Total RNA Extraction kit (Huayueyang Biotechnology, Beijing, China). The integrity of the RNA was examined by 1.0% agarose gel electrophoresis, and cDNA was synthesized from the RNA using a Reverse Transcription kit (TaKaRa Biotechnology, Dalian, China). All primers used for RT-qPCR are listed in Supplementary Table S1. RT-qPCR was performed using SYBR Premix Ex Taq (Yeasen Biotechnology (Shanghai) Co., Ltd) with the ABI 7500 System. All experiments were carried out with three biological replicates. The relative expression value of each gene was quantified using the 2-△△Ct method (Livak and Schmittgen, 2001).
The differentially expressed genes in the anthocyanin biosynthesis pathway, light signaling pathway, and hormone signaling-related pathways were identified in eggplant fruits with different colors (Supplementary Table S2). Pearson’s correlation tests were performed with SPSS v25 software using the FPKM values of both purple eggplant ‘YN-1’ and yellow eggplant ‘S-12’ samples. Any two genes with an absolute Pearson correlation coefficient of ≥ 0.9 and a p-value of ≤ 0.05 were considered to be significantly co-expressed genes. The co-expression network was visualized using Cytoscape v3.5.1 software.
To examine the subcellular locations of the SmGATA proteins, two full-length SmGATA open reading frames (ORFs) (for SmGATA4 and SmGATA23) without the stop codon were amplified from cDNA from eggplant fruit. Each amplification product was cloned into the pCAMBIA1302 vector with green fluorescence protein (GFP) label under the control of the CaMV35S promoter. The gene-specific primers are listed in Supplementary Table S3.
Nicotiana benthamiana plants were grown in a plant growth chamber at 26°C under a 16-hr light/8-hr dark regimen until they were approximately 15 cm tall and infiltrated with Agrobacterium strain EHA105 harboring the constructs described above. Infiltration was performed as described by (Sparkes et al., 2006). The agroinfiltrated leaves were photographed 2 days after infiltration. GFP fluorescence images were captured using an Olympus laser-scanning confocal microscope.
All data were analyzed using Student’s t-test with the software SPSS 25.0. The values are represented as the means ± SD. P < 0.05 was considered as statistically significant.
To identify and obtain the GATA genes in the eggplant genome, the Arabidopsis GATA proteins were used as a query to search against the eggplant genome via BLAST. Then, the results of the blast search through confirmation of the presence of the GATA domain using SMART. After removing the redundant sequences, 28 full-length GATA homologous sequences were identified in the genomes of eggplant (Supplementary Figure S1). The detailed information of SmGATA genes were listed in Table 1, including gene name, protein length, chromosome location, molecular weight, theoretical isoelectric point, aliphatic index, and GRAVY. The 28 SmGATA proteins had diverse molecular length and weight, ranging from 117 (SmGATA23) to 542 (SmGATA20) in amino acid length. SmGATA23 showed the lowest value of the molecular weight (13.24 kDa), while the highest of the molecular weight (59.98 kDa) was observed in SmGATA22. Theoretical isoelectric points of these SmGATA proteins varied from 5.15 (SmGATA2) to 9.87 (SmGATA3) and the value of the aliphatic index ranged from 41.12 (SmGATA9) to 71.76 (SmGATA3). The GRAVY of all SmGATAs were less than zero, indicating the hydrophilic nature of SmGATA proteins (Table 1).
In order to clarify the characteristics of GATA proteins in eggplant, we first extracted their conserved domain sequences and visualized them (Supplementary Figure S2). Similar to what has been found in other plant species, the conserved domain of GATA proteins in eggplant also contained a consensus sequence of C-X2-C-X18/20-C-X2-C. It is worth noting that besides the highly conserved cysteine residues, there are also many highly conserved amino acid residues in this domain, which may be related to the functions of GATA proteins in eggplant.
To explore the evolutionary relationship and functional divergence of the SmGATA members, a phylogenetic tree was constructed with MEGA6.0 by using the neighbor-joining method according to SmGATAs, AtGATAs and SlGATAs protein sequences (Figure 1), respectively. All SmGATA proteins were clustered into four major subfamilies. Among them, subfamily I contained the largest SmGATA members (14/28), followed by subfamily II (8/28),subfamily III (3/28), and then subfamily IV (3/28) (Figure 2A).
To explore the gene and protein sequence structures of eggplant GATA family members, visualization of their motif compositions and intron/exon structures was performed. Analysis of the motif composition of SmGATAs identified a total of 10 conserved motifs (Figure 2B). Consistent with the expected results, all proteins contained motif 1, annotated as the GATA domain. In subfamily I, most proteins had three motifs (12/14); in subfamily II, the number of motifs was relatively small, with most proteins having two motifs (6/8) and the remaining two proteins only have one motif; in subfamily III, all three SmGATAs had two motifs, and motif 1 was positioned after motif 8 in the amino acid sequence. Furthermore, in subfamily IV, The number of motifs was the highest, with all three SmGATAs had 6 motifs. The differences in the number and variety of conserved motifs in SmGATA proteins reflected the structural diversity of these proteins, while predicting that they have different biological functions.
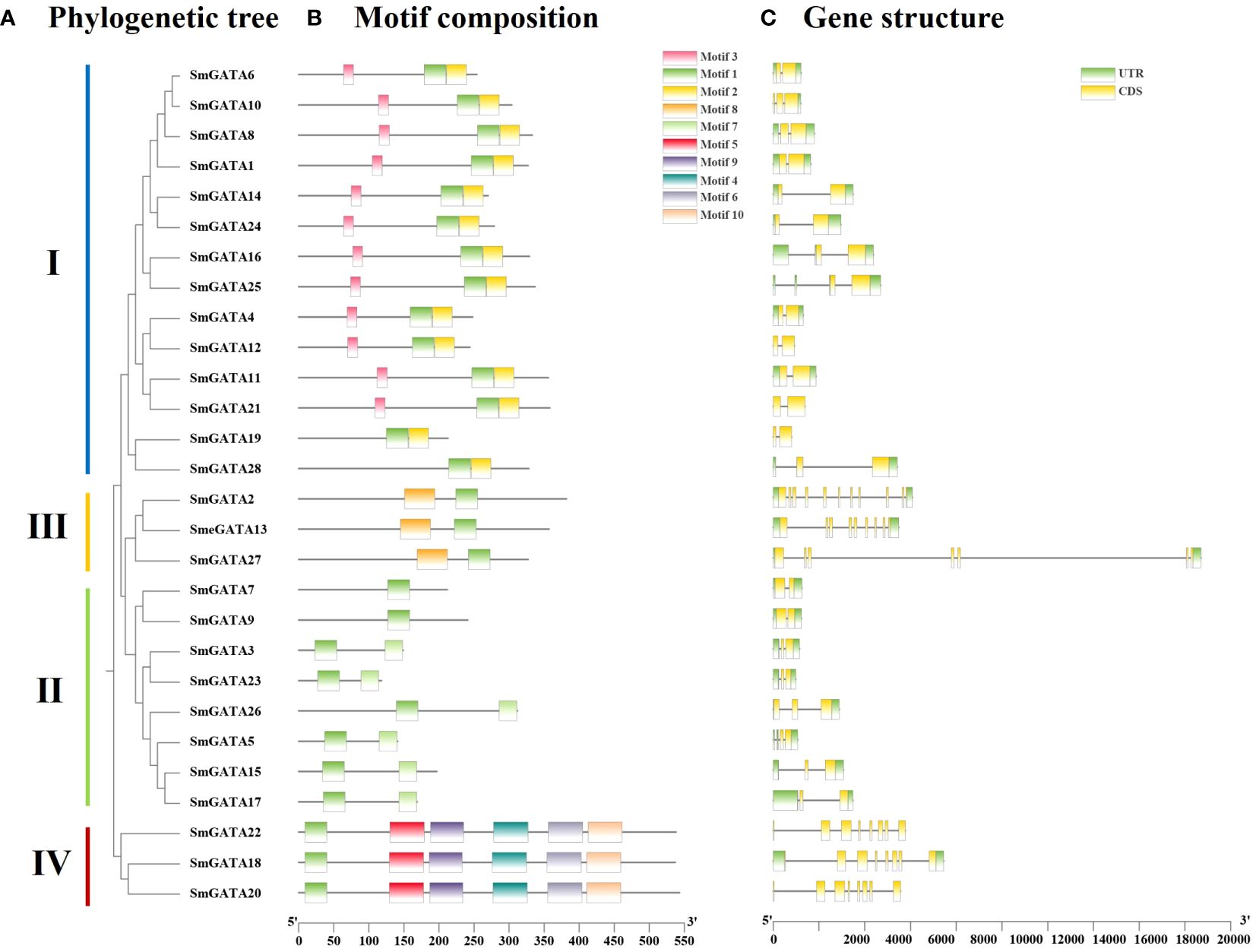
Figure 2 Phylogenetic tree, motif composition and gene structure of GATA genes in eggplant. (A) The phylogenetic tree is constructed by the full-length sequences of eggplant GATA proteins with 1000 replicates on each node. (B) The amino acid motifs (numbered 1-10) in SmGATAs are displayed in ten colored boxes, and black lines indicate protein sequence length. (C) Green rectangles, yellow rectangles lines represent the UTR (untranslated region), CDS (coding sequence or exons), respectively.
Based on the eggplant genome sequence, a gene structure map of Solanum melongena GATAs was constructed (Figure 2C). 28 SmGATAs all contain no less than 2 CDS regions. Among them, subfamilies I and II both contained 2-4 CDSs, while SmGATA2 in subfamily III had the 10 CDS segments, which was the most abundant. For the untranslated region (UTR), SmGATA12, 19, 20, 21 and 22 had no UTRs, and the remaining GATAs had their UTRs distributed almost at both ends of the gene.
Cis-elements are involved in gene regulation through the interaction with their corresponding trans-regulators and the study of the proposed cis-elements will provide valuable information for the expression of eggplant GATA genes. Therefore, the promoter region of GATA genes in eggplant were obtained and taken to Plantcare database for prediction (Figure 3). The results showed that a total of 26 elements were predicted. among which CAAT-box and TATA-box were present in all SmGATA genes. The remaining elements included light responsive elements, defense response elements and hormone induction elements, such as those for abscisic acid (ABA), auxin (IAA), gibberellin (GA), Salicylic acid (SA) and Methyl jasmonate (MeJA) elements.
In this study, we analyzed gene duplication events of the 28 SmGATA genes to further explore the evolutionary relationships among the GATA genes (Figure 4) and identified 15 pairs of quasi-homologous GATAs in the eggplant genome originated from duplication, of which five pairs belonged to subfamily I. SmGATAs were unevenly distributed among the 12 linked regions (EG) of the eggplant genome, with EG1 containing the largest number of SmGATAs (6/15), followed by EG6 containing three SmGATAs (3/15), while EG7, EG10 and EG11 had no duplicated genes.
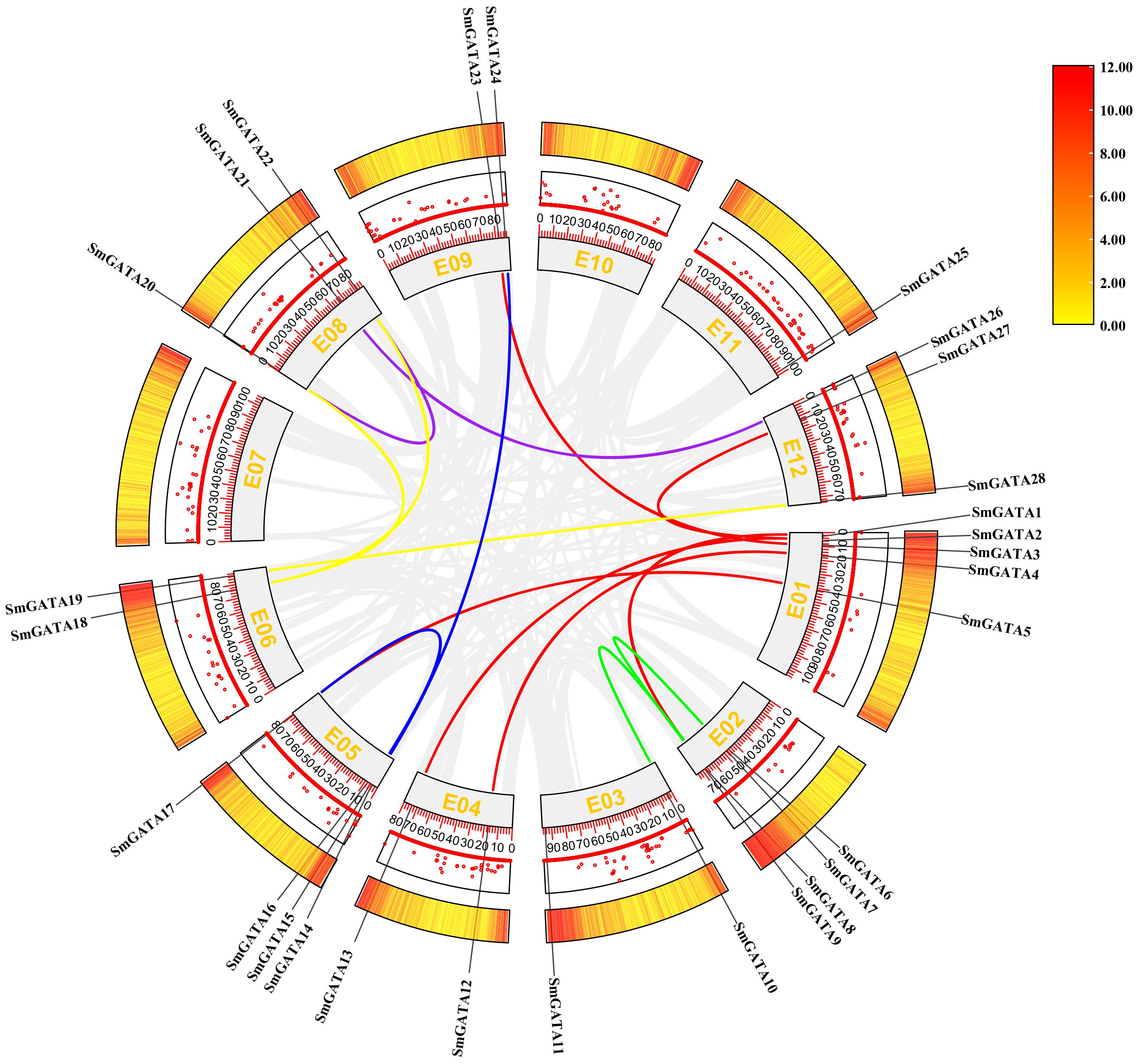
Figure 4 Collinearity of GATA genes in eggplant. Colored lines indicate duplicated GATA gene pairs. The chromosome number is shown inside each chromosome.
GATAs are a class of transcription factors closely related to growth and development (Jeong and Shih, 2003; Richter et al., 2013; An et al., 2014). Therefore, we analyzed the role of SmGATA genes in eggplant fruit development and maturation (Figure 5), the expression patterns of 28 SmGATA genes in five developmental stages of eggplant fruit were studied. Different members of the SmGATA genes showed distinct expression patterns during different eggplant fruit developmental stages. As shown in Figure 5, three genes (SmGATA4, 23 and 24) displayed upregulated expressions during fruit developmental stage and the expression level of SmGATA27 showed a trend of first increasing and then decreasing. Two genes (SmGATA1 and SmGATA26) were down-regulated expression patterns. These results indicated that some SmGATA genes maybe play multiple important roles in eggplant fruit development.
The classification of anthocyanins in eggplants into photosensitive and non-photosensitive types is based on how these pigments respond to light exposure during the growth and development of the eggplant fruit (He et al., 2019; Ohyama et al., 2022), and the signaling pathways for their synthesis are unclear. GATA motif as a light-response promoter element, is usually the binding site of GATA transcription factors (Luo et al., 2010). In order to analyze the potential role of SmGATA gene in photosensitive eggplants in the biosynthesis of anthocyanins, we placed bags on developing eggplant fruits at 0 DAF to block their exposure to light and removed the bags at 25 DAF. The fruits rapidly changed color after bag removal (Figure 6A). We used RT-qPCR to measure SmGATA genes expression in eggplant after bag removal. Different genes showed different expression levels (Figure 6B). As shown in Figure 6C, 14 SmGATA genes were upregulated more than two-fold after light induction. In particular, SmGATA4, 13, 21 and 23 were upregulated approximately 5-fold at 4 d of light induction. These results represented that these GATA genes function in light-induced anthocyanin biosynthesis in eggplant.
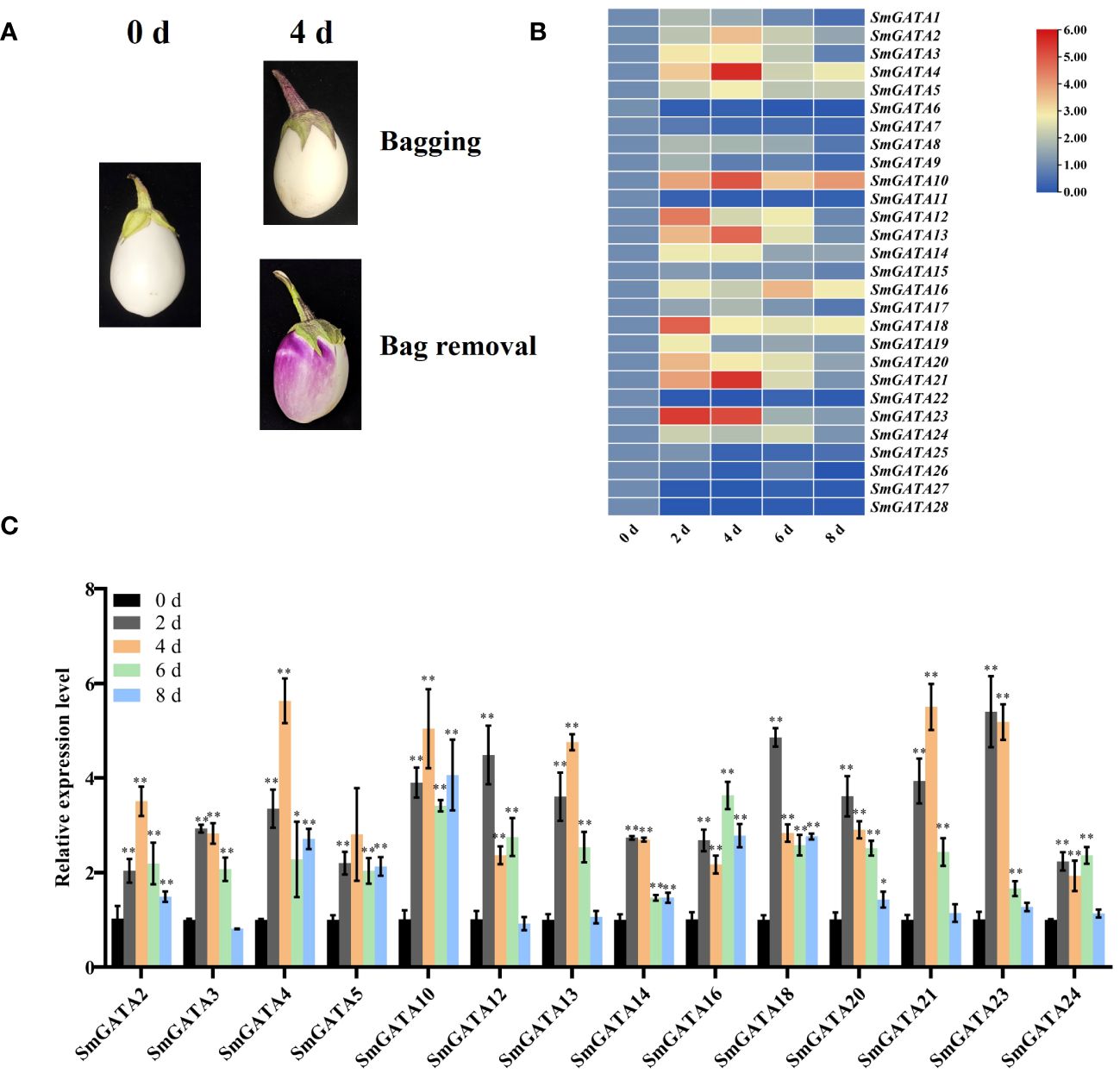
Figure 6 Expression profiles of SmGATA genes in ‘Zaohongqie 2’ fruit following light induction via bag removal. (A) Colour changes of ‘Zaohongqie 2’ fruits after bag removal. (B) Hierarchical clustering of the expression profiles of 28 SmGATA genes after bag removing. (C) RT-qPCR analysis of 14 selected SmGATA genes following light induction. SmActin was used as the internal reference control to normalize template levels. The relative mRNA levels are represented as the mean ± SD (n=3). Statistically significant differences were assessed using Student’s t-test (*P < 0.05, **P < 0.01).
The hormone signals and environmental changes are critical to fruit development and ripening (Santner et al., 2009; Leng et al., 2014; Kuhn et al., 2020; Guo et al., 2021). In our study, the analysis of cis-acting elements in the 2000 bp sequence upstream of the SmGATAs promoter revealed that most members of this family respond to avariety of hormones and stresses, indicating that SmGATAs may be regulated by light, stresses, and phytohormones (Figure 3). In our study, we measured the expression levels of SmGATA genes in response to ABA, GA, and MeJA treatment by RT-qPCR (Figure 7). As expected, the SmGATA genes showed diverse expression patterns during treatment with different hormones. During ABA treatment, 16 SmGATA genes were significantly upregulated, indicating that these genes positively respond to ABA treatment. A majority of SmGATA genes were significantly downregulated under GA treatment, The 28 SmGATA genes responded to MeJA to varying degrees. In general, most SmGATA genes were sensitive to different hormone treatments. It is worth noting that 12 SmGATA (SmGATA3, 4, 6, 7, 8, 9, 10, 12, 18, 19, 21 and 23) exhibit similar expression trends under three treatments. Furthermore, 7 SmGATA genes (SmGATA4, 9, 10, 12, 19, 21 and 23) were significantly upregulated by more than two times under ABA and MeJA treatments, and downregulated by more than two times under GA treatment. Taken together, the response of most SmGATA genes to ABA and MeJA signaling were consistent, and the expression level showed an upward trend. but it exhibited contrary trend of the GA signaling pathway. Notably, three hormones treatment (ABA, GA, and MeJA) had no significant effect on the expression of SmGATA2.
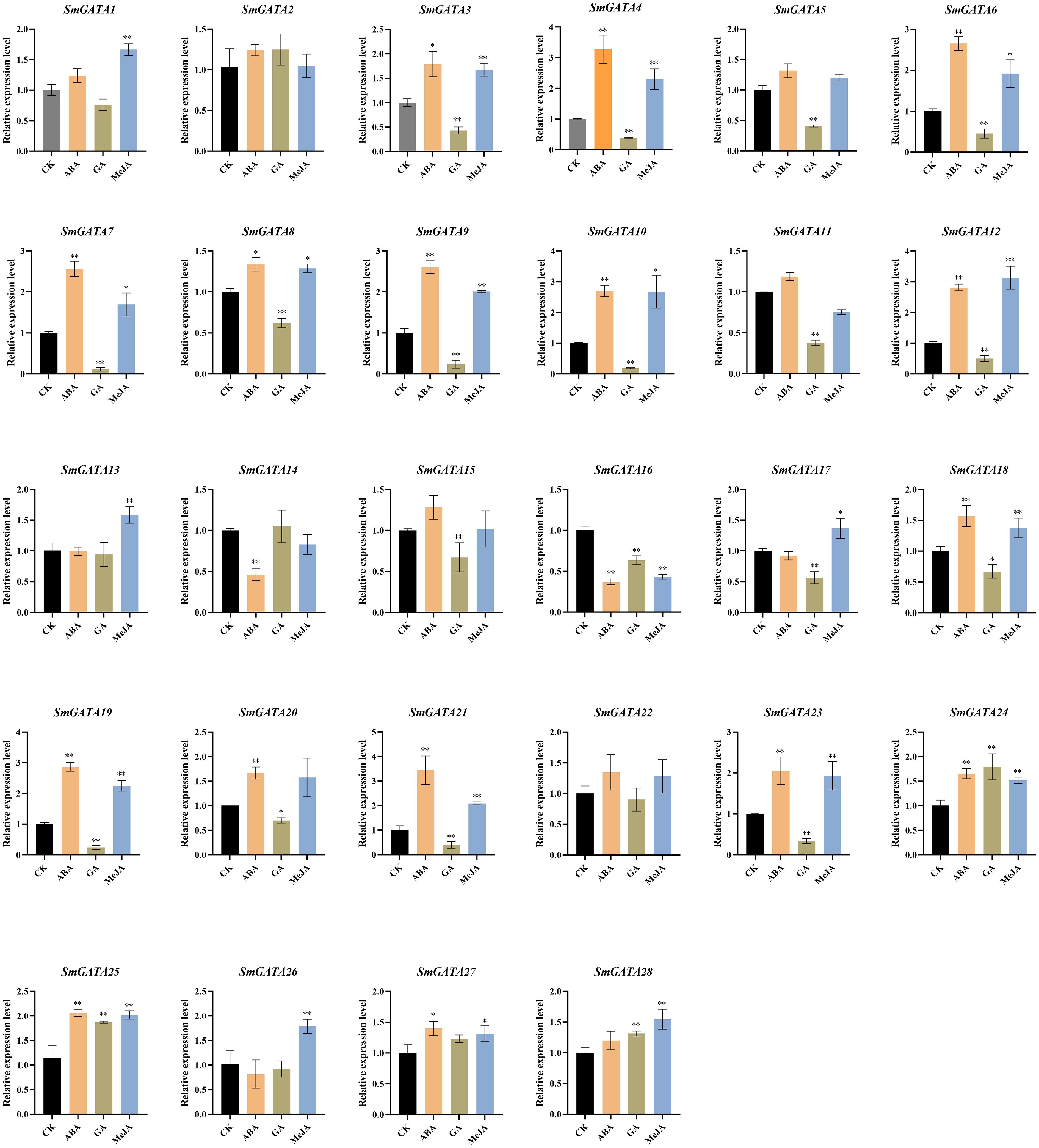
Figure 7 Expression profiles of eggplant GATA genes in response to ABA, GA, and MeJA treatment. SmActin was used as the internal reference control to normalize template levels. Relative mRNA levels are represented as the mean ± SD (n=3). Statistically significant differences were assessed using Student’s t-test (*P < 0.05, **P < 0.01).
We assessed the transcriptome data of eggplant fruits with different colors to examine the interactions between GATA genes, light-responsive genes, multiple hormone signaling genes, and anthocyanin biosynthesis genes in eggplant fruits. Four SmGATA genes (SmGATA4, 10, 23, and 24) according to comprehensive analysis of the expression patterns of SmGATA genes under different developmental stages, as well as under light and hormone treatments were screened. We analyzed the expression of 8 anthocyanin biosynthesis genes (PAL, CHS, CHI, F3H, F3′H, DFR, ANS and UFGT) and 6 light-responsive genes to identify genes that might regulate anthocyanin biosynthesis during fruit development. As shown in Figure 8, genes encoding light signaling components (HY5, PIF3) and anthocyanin biosynthesis genes were co-expressed with ABA, GA and MeJA signaling pathway genes, suggesting that these three hormones might play major roles in anthocyanin biosynthesis in eggplant. Furthermore, the expression patterns of four SmGATA genes (SmGATA4, 10, 23, and 24) were correlated with the expression patterns of anthocyanin biosynthesis genes and light-responsive genes. Notably, GATA4 and GATA23 expression levels were highly correlated with anthocyanin biosynthesis and light-responsive gene expression, and these genes were co-expressed with genes encoding components of the ABA, BR, and MeJA signaling pathways. These results showed that GATA genes likely play major roles in regulating anthocyanin biosynthesis by integrating the light, ABA, GA, and MeJA signaling pathways.
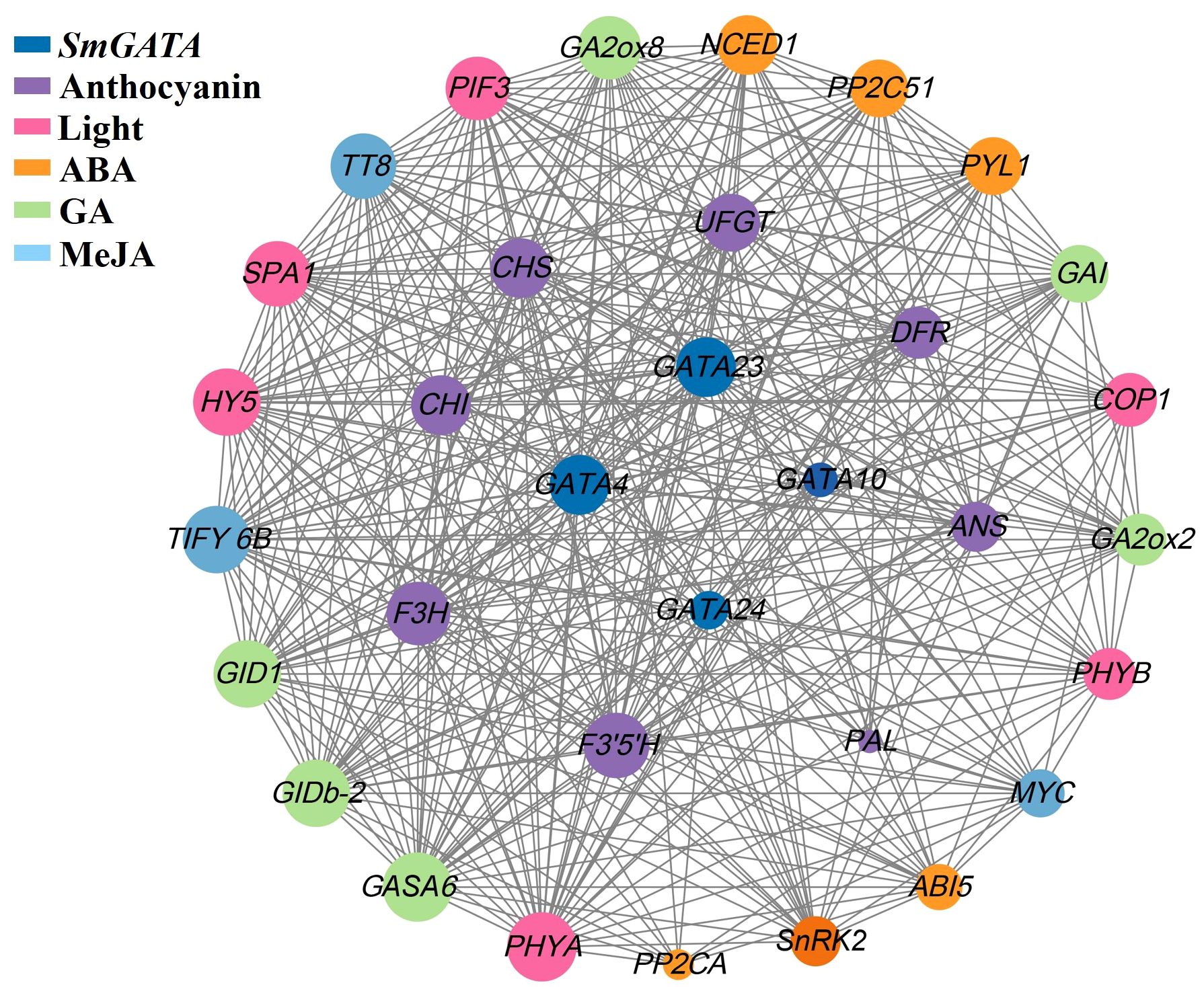
Figure 8 Co-expression network analysis of GATA, anthocyanin biosynthesis, light signaling, and multiple hormone signaling genes in eggplant.
Transcription factors play many regulatory roles in plants. The nuclear localization of transcription factors is important for their regulatory roles. Most GATA proteins are located in the nucleus, such as AtGATA2 (Luo et al., 2010), BdGATA13 (Guo et al., 2021). To examine the subcellular locations of GATA proteins in eggplant, we transiently transformed Nicotiana benthamiana epidermal cells with two SmGATA genes (SmGATA4 and SmGATA23) and examined the subcellular localization of the resulting GFP-tagged fusion proteins. As shown in Figure 9, SmGATA4-GFP and SmGATA23-GFP showed green fluorescent signals in the nuclei of N. benthamiana epidermal cells. These results revealed that SmGATA4 and SmGATA23 were nuclear proteins, which was consistent with previous results and their presumed roles as transcription factors (Luo et al., 2010; Guo et al., 2021).
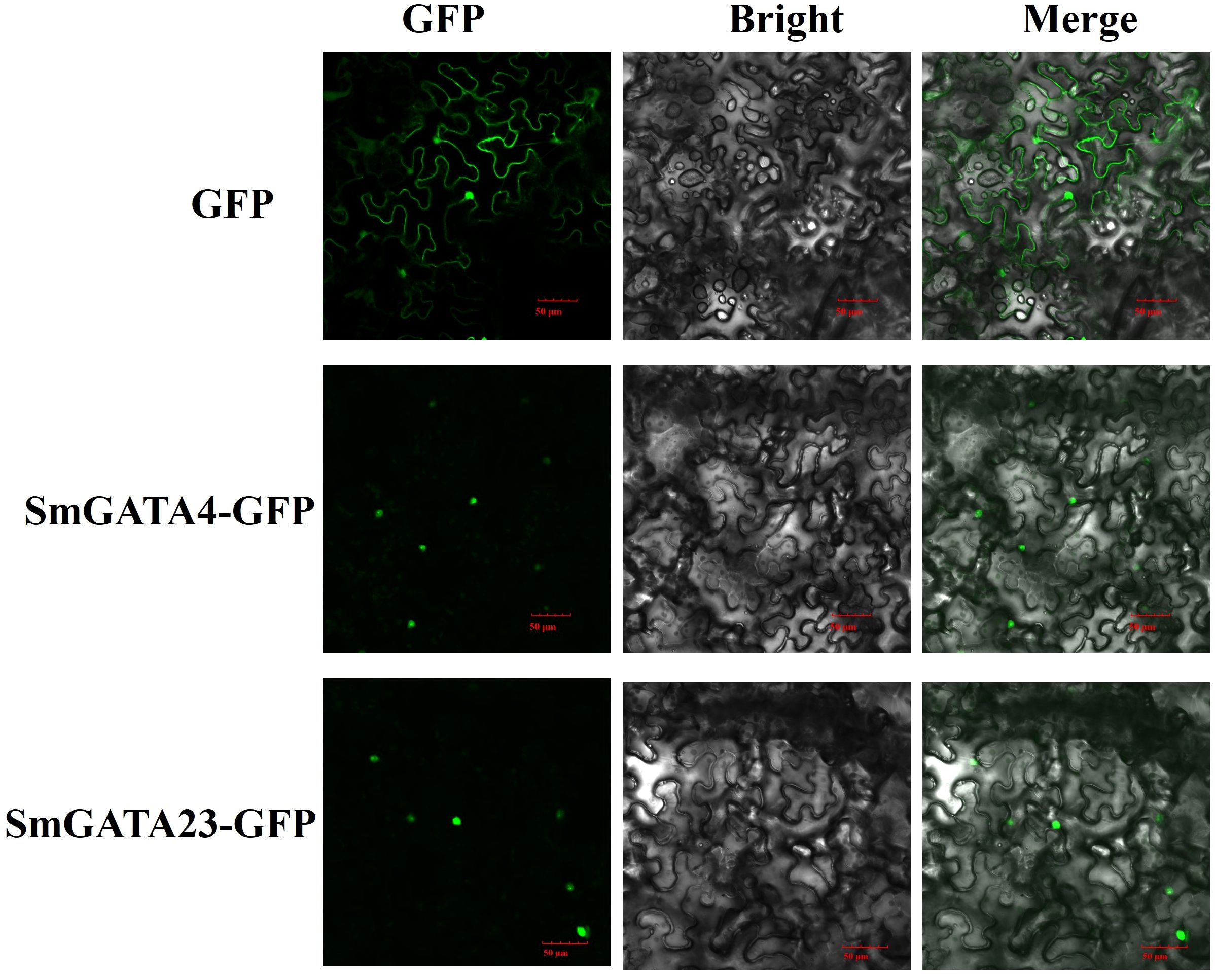
Figure 9 Subcellular localization of two GFP-fused SmGATA proteins. The two SmGATA-GFP fusion proteins (SmGATA4-GFP and SmGATA23-GFP) were transiently expressed in N. benthamiana leaves and observed by fluorescence microscopy 48 h later. Bar = 50 μm.
In the current study, we identified 28 GATA genes in the genome database of eggplant. The number of GATA genes varies among plant species; for example, there are 30 GATA family members in Arabidopsis (Reyes et al., 2004), 28 in rice (Reyes et al., 2004), 30 in tomato (Yuan et al., 2018) and 35 in apple (Chen et al., 2017). These discrepancies may arise from variations in genome size and complexity among these species. The 28 SmGATAs were divided into four subfamilies (I, II, III and IV) (Figure 1), which is consistent with most GATA family studies (Du et al., 2022; Feng et al., 2022), indicating that the GATA family is relatively stable during evolution. We found that the amino acid motifs and gene structures of SmGATA genes are relatively conserved (Figure 2B), the 28 SmGATAs contain 1-10 motifs, with motif 1 being widely distributed, suggesting its role as the Zinc finger GATA motif in this family. At the same time, we found that the SmGATAs genes of subfamily III and IV are rich in CDS (Figure 2C). The distribution of conserved motifs and gene structures is similar among members within the same subfamily, but there are significant differences between subfamilies, which is consistent with prior research findings (Manfield et al., 2007; Yu et al., 2019). Currently, a large number of cis-acting elements and their associated functions or pathways have been deciphered. By analyzing the promoter sequences of SmGATA genes in eggplant, a remarkable number of hormone and stress response elements have been identified ((Figure 3), indicating that they may be involved in multiple biological processes. As shown in Figure 4, the 28 SmGATA genes exhibit uneven distribution across eggplant chromosomes, which may be referred to as the differences in the size and structure of the chromosomes. Notably, 15 pairs of SmGATAs in the eggplant genome were duplicated, suggesting gene duplication as a contributing factor to the expansion of the GATA gene family in eggplant.
The development and ripening of eggplant directly afect the quality of fresh fruit, and which is a complex physiological and biochemical process that is influenced by various transcription factors and regulatory proteins (Li et al., 2021b; Yang et al., 2022; Li et al., 2024). our results revealed that some SmGATA genes were highly expressed in late stage of fruit developments (Figure 5), implying potential roles in development and fruit ripening. There are previously reported examples of GATA genes being involved in these processes in A. thaliana, where GATA proteins have been found to be involved in chlorophyll synthesis and foral development (Bi et al., 2005). In chrysanthemum, CmGATA4 can directly bind to the key gene CmCCD4a-5 for carotenoid degradation, acts as a negative regulator to lower the expression of CmCCD4a-5 resulting in carotenoid accumulation in the mutant (Huang et al., 2022). Additionally, in tomato, SlGATA17 protein interacts with SlHY5, and SlHY5 plays a role upstream of SlGATA17, which inhibits SlGATA17 gene expression by binding to its promter (Wang et al., 2023). Furthermore, SlHY5 regulates fruit ripening both at the transcriptional level by targeting crucial genes involved in anthocyanin biosynthesis and ethylene biosynthesis, which also at the translational level by affecting the protein translation machinery (Wang et al., 2021). Here, the expression level of SmGATA4, SmGATA23 and SmGATA24 are significantly upregulated during fruit development (Figure 5), which revealed that these three genes are involved in the ripening process in eggplant. Finally, two genes (SmGATA1 and SmGATA26) were downregulated during late fruit development, which means that these genes encode negative regulators of fruit ripening in eggplant.
Light is an important factor that influences plant growth and development (Jaakola, 2013). Response promoter elements, G-box and GATA motif, is critical for promoter activation in response to the signals from multiple photoreceptors (Chattopadhyay et al., 1998). In fungi, such as Neurospora, two GATA-type factors bind to GATA element and regulate gene expression in response to light signal (Scazzocchio, 2000). Additionally, In A. thaliana, AtGATA2 was reported as a rositive regulator of photomorphogenesis and GATA21 and GATA22 are induced by red light in a PIF3-dependent manner (Monte et al., 2004; Luo et al., 2010). Furthermore, anthocyanin accumulation is highly dependent on light in photosensitive eggplants. Based on our present results, three SmGATA genes are upregulated and two SmGATA genes are downregulated during light exposure, proposed that these differentially expressed genes might function in light-induced anthocyanin biosynthesis in eggplant (Figure 6).
Exogenous hormone treatment can promote fruit ripening (Forlani et al., 2019). As a non-climacteric fruit, mutiple hormones play an important role in regulating the ripening process of eggplants, including MeJA (Li et al., 2024). Additionally, the roles of SmGATA proteins in hormone signaling pathways are currently unclear. Several reports document the roles of GATA genes in hormonal pathways in other plants (Wei et al., 2023). TaGATA1–TaELF6-A1–TaABI5, which contributes to seed dormancy through the ABA signaling pathway in Triticum aestivum (Wei et al., 2023). The GATA-type transcription factors GNC and GNL/CGA1, is a negative regulatory factor of GA signaling, functions in downstream from DELLA proteins (Richter et al., 2010). Other example suggested GATAs can interact with JAZ, a key regulator in the jasmonic acid (JA) signaling pathway (Sen et al., 2016). In our study, RT-qPCR revealed that the SmGATA genes were responsive to numerous hormonal treatments (Figure 7). More than half of the SmGATA genes were regulated by more than one hormone treatment, providing that these genes may be involved in the interactions of different hormone signals at the physiological level. Furthermore, the expression patterns of GATA genes were highly correlated with those of anthocyanin biosynthesis genes, light-responsive genes, and ABA, GA and MeJA signaling pathway genes (Figure 8). In addition, SmGATA4 and SmGATA23 responded to three hormone signals and light treatment, indicating that these genes encode proteins that integrate light and hormone signals to regulate anthocyanin biosynthesis.
In this study, 28 SmGATA genes were identified in the eggplant genome database, and their gene structures and expression patterns were comprehensively investigated. The results illustrated that the SmGATA genes significantly contributed to fruit development and ripening, primarily manifested in the expression patterns of the SmGATA gene under light exposure and changes in hormone levels. Especially by the correlation between several SmGATA genes and the expression of light-responsive genes, various hormone signaling genes, and anthocyanin biosynthesis genes. Particularly noteworthy was the upregulation of several SmGATA genes in response to hormone treatments such as ABA, GA and MeJA, indicating their role in integrating light, ABA, GA and MeJA signaling pathways to regulate anthocyanin biosynthesis. In conclusion, our comprehensive analysis of the SmGATA family across the genome provides a basis for further investigations into the biological roles of these genes in fruit development and ripening.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YW: Writing – original draft, Writing – review & editing. XL: Investigation, Writing – original draft. YM: Software, Writing – original draft. CJ: Investigation, Writing – original draft. YZ: Data curation, Writing – original draft. JH: Methodology, Writing – original draft. YLZ: Methodology, Writing – original draft. JL: Conceptualization, Methodology, Software, Writing – review & editing. KZ: Conceptualization, Funding acquisition, Writing – review & editing. ZL: Conceptualization, Software, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (32160715); Yunnan Fundamental Research Projects (202301AS070078); Major science and technology project of Yunnan Province (202402AE090012), Yunnan Province “Xing Dian talent support plan” project (XDYC-QNRC-2022-0227) and Science Research Fund Project of Yunnan Provincial Department of Education (2024J0484), the Joint Project of Basic Agricultural Research in Yunnan Province (202401BD070001-047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1415921/full#supplementary-material
ABA, Abscisic acid; MeJA, Methyl jasmonate; GA, Gibberellic acid; AA, amino acid residues; MW, molecular weight; pI, theoretical isoelectric point; GRAVY, grand average of hydropathicity; FPKM, Fragments per kilobase of exon per million mapped fragments; ORF, Open reading frame; GFP, Green fluorescent protein
An, Y., Han, X., Tang, S., Xia, X. L., Yin, W. L. (2014). Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 119, 313–327. doi: 10.1007/s11240-014-0536-y
An, Y., Zhou, Y. Y., Han, X., Shen, C., Wang, S., Liu, C., et al. (2020). The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 71, 1969–1984. doi: 10.1093/jxb/erz564
Behringer, C., Schwechheimer, C. (2015). B-GATA transcription factors—Insights into their structure, regulation, and role in plant development. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00090
Bi, Y. M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T., Rothstein, S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44, 680–692. doi: 10.1111/j.1365-313X.2005.02568.x
Bor, J. Y., Chen, H. Y., Yen, G. C. (2006). Evaluation of antioxidant activity and inhibitory effect on nitric oxide production of some common vegetables. J. Agric. Food Chem. 54, 1680–1686. doi: 10.1021/jf0527448
Chattopadhyay, S., Puente, P., Deng, X. W., Wei, N. (1998). Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 15, 69–77. doi: 10.1046/j.1365-313X.1998.00180.x
Chen, C. J., Wu, Y., Xia, R. (2022). A painless way to customize circos plot: From data preparation to visualization using TBtools. iMeta 1, e35. doi: 10.1002/imt2.35
Chen, H. F., Shao, H. X., Li, K., Zhang, D., Fan, S., Li, Y., et al. (2017). Genome-wide identifcation, evolution, and expression analysis of GATA transcription factors in apple (Malus x domestica Borkh.). Gene 627, 460–472. doi: 10.1016/j.gene.2017.06.049
Daniel-Vedele, F., Caboche, M. (1993). A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Genet. Genom. 240, 365–373. doi: 10.1007/BF00280388
Du, K., Xia, Y. F., Zhan, D. J., Xu, T. T., Lu, T., Yang, J., et al. (2022). Genome-wide identification of the Eucalyptus urophylla GATA gene family and its diverse roles in chlorophyll biosynthesis. Int. J. Mol. Sci. 23, 5251. doi: 10.3390/ijms23095251
Fan, Y., Wei, X. B., Lai, D. L., Yang, H., Feng, L., Li, L., et al. (2021). Genome-wide investigation of the GRAS transcription factor family in foxtail millet (Setaria italica l.). BMC Plant Biol. 21, 1–19. doi: 10.1186/s12870-021-03277-y
Feng, X., Yu, Q., Zeng, J. B., He, X. Y., Liu, W. X. (2022). Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 22, 372. doi: 10.1186/s12870-022-03733-3
Forlani, S., Masiero, S., Mizzotti, C. (2019). Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 70, 2993–3006. doi: 10.1093/jxb/erz112
García, R., Aguilera, A., Contreras-Esquivel, J. C., Rodríguez, R., Aguilar, C. N. (2008). Extraction of condensed tannins from Mexican plant sources. Z. Naturforsch. C J. Biosci. 63, 17–20. doi: 10.1515/znc-2008-1-204
Guo, J., Bai, X. H., Dai, K. L., Yuan, X. Y., Guo, P. Y., Zhou, M. X., et al. (2021). Identification of GATA transcription factors in Brachypodium distachyon and functional characterization of BdGATA13 in drought tolerance and response to gibberellins. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.763665
He, Y. J., Chen, H., Zhou, L., Liu, Y., Chen, H. Y. (2019). Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genomics 20, 678. doi: 10.1186/s12864-019-6023-4
Huang, H. F., Gao, X. K., Gao, X., Zhang, S. Q., Zheng, Y., Zhang, N., et al. (2022). Flower color mutation, pink to orange, through CmGATA4-CCD4a-5 module regulates carotenoids degradation in chrysanthemum. Plant Sci. 322, 111290. doi: 10.1016/j.plantsci.2022.111290
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jeong, M. J., Shih, M. C. (2003). Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 300, 555–562. doi: 10.1016/S0006-291X(02)02892-9
Kuhn, N., Ponce, C., Arellano, M., Time, A., Sagredo, B., Donoso, J. M., et al. (2020). Gibberellic acid modifies the transcript abundance of ABA pathway orthologs and modulates sweet cherry (Prunus avium) fruit ripening in early and mid-season varieties. Plants (Basel) 9, 1796. doi: 10.3390/plants9121796
Lai, D. L., Fan, Y., Xue, G. X., He, A. L., Yang, H., He, C. L., et al. (2022). Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). BMC Genomics 23, 389. doi: 10.1186/s12864-022-08633-2
Leng, P., Yuan, B., Guo, Y. D. (2014). The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 65, 4577–4588. doi: 10.1093/jxb/eru204
Li, X. J., Deng, X. X., Han, S. Y., Zhang, X. Y., Dai, T. B. (2023). Genome-wide identification and expression analysis of GATA Gene family under different nitrogen levels in Arachis hypogaea L. Agronomy 13, 215. doi: 10.3390/agronomy13010215
Li, S. H., Dong, Y. X., Li, D. L., Shi, S. L., Zhao, N., Liao, J. L., et al. (2024). Eggplant transcription factor SmMYB5 integrates jasmonate and light signaling during anthocyanin biosynthesis. Plant Physiol. 194, 1139–1165. doi: 10.1093/plphys/kiad531
Li, L. Z., He, Y. J., Ge, H. Y., Liu, Y., Chen, H. Y. (2021b). Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 302, 110696. doi: 10.1016/j.plantsci.2020.110696
Li, D. L., He, Y. J., Li, S. H., Shi, S. L., Li, L. Z., Liu, Y., et al. (2021a). Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 167, 492–503. doi: 10.1016/j.plaphy.2021.08.006
Li, W. T., Zhu, Z. W., Chern, M., Yin, J. J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126. doi: 10.1016/j.cell.2017.06.008
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- △△Ct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowry, J. A., Atchley, W. R. (2000). Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 50, 103–115. doi: 10.1007/s002399910012
Luo, X. M., Lin, W. H., Zhu, S., Zhu, J. Y., Sun, Y., Fan, X. Y., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19, 872–883. doi: 10.1016/j.devcel.2010.10.023
Manfield, I. W., Devlin, P. F., Jen, C. H., Westhead, D. R., Gilmartin, P. M. (2007). Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 143, 941–958. doi: 10.1104/pp.106.090761
Monte, E., Tepperman, J. M., Al-Sady, B., Kaczorowski, K. A., Alonso, J. M., Ecker, J. R., et al. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U. S. A. 101, 16091–16098. doi: 10.1073/pnas.0407107101
Ohyama, A., Yamaguchi, H., Miyatake, K., Negoro, S., Nunome, T., Saito, T., et al. (2022). Genetic mapping of simply inherited categorical traits, including anthocyanin accumulation profiles and fruit appearance, in eggplant (Solanum melongena). Mol. Biol. Rep. 49, 9147–9157. doi: 10.1007/s11033-022-07737-y
Ravindran, P., Verma, V., Stamm, P., Kumar, P. P. (2017). A novel RGL2–DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10, 1307–1320. doi: 10.1016/j.molp.2017.09.004
Reyes, J. C., Muro-Pastor, M. I., Florencio, F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. doi: 10.1104/pp.103.037788
Richter, R., Bastakis, E., Schwechheimer, C. (2013). Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering Time in Arabidopsis. Plant Physiol. 162, 1992–2004. doi: 10.1104/pp.113.219238
Richter, R., Behringer, C., Müller, I. K., Schwechheimer, C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24, 2093–2104. doi: 10.1101/gad.594910
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rueda-López, M., Cañas, R. A., Canales, J., Cánovas, F. M., Á vila, C. (2015). The overexpression of the pine transcription factor PpDof5 in Arabidopsis leads to increased lignin content and affects carbon and nitrogen metabolism. Physiol. Plant 155, 369–383. doi: 10.1111/ppl.12381
Santner, A., Calderon-Villalobos, L. I., Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Scazzocchio, C. (2000). The fungal GATA factors. Curr. Opin. Microbiol. 3, 126–131. doi: 10.1016/S1369-5274(00)00063-1
Schwechheimer, C., Schröder, P. M., Blaby-Haas, C. E. (2022). Plant GATA factors: their biology, phylogeny, and phylogenomics. Annu. Rev. Plant Biol. 73, 123–148. doi: 10.1146/annurev-arplant-072221-092913
Sen, S., Kundu, S., Dutta, S. K. (2016). Proteomic analysis of JAZ interacting proteins under methyl jasmonate treatment in finger millet. Plant Physiol. Biochem. 108, 79–89. doi: 10.1016/j.plaphy.2016.05.033
Shen, C., Zhang, Y., Li, Q., Liu, S. J., He, F., An, Y., et al. (2021). PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus. New Phytol. 230, 1868–1882. doi: 10.1111/nph.17301
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Strader, L., Weijers, D., Wagner, D. (2022). Plant transcription factors - being in the right place with the right company. Curr. Opin. Plant Biol. 65, 102136. doi: 10.1016/j.pbi.2021.102136
Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell Mol. Life Sci. 54, 582–596. doi: 10.1007/s000180050186
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wang, Y. Q., Cao, X. Y., Zhang, D. K., Li, Y. Q., Wang, Q. Q., Ma, F., et al. (2023). SlGATA17, A tomato GATA protein, interacts with SlHY5 to modulate salinity tolerance and germination. Environ. Exp. Bot. 206, 105191. doi: 10.1016/j.envexpbot.2022.105191
Wang, W. H., Wang, P. W., Li, X. J., Wang, Y. Y., Tian, S. P., Qin, G. Z. (2021). The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hortic. Res. 8, 83. doi: 10.1038/s41438-021-00523-0
Wei, X. N., Li, Y. Y., Zhu, X. L., Liu, X., Ye, X. G., Zhou, M. P., et al. (2023). The GATA transcription factor TaGATA1 recruits demethylase TaELF6-A1 and enhances seed dormancy in wheat by directly regulating TaABI5. J. Integr. Plant Biol. 65, 1262–1276. doi: 10.1111/jipb.13437
Xu, D. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Yang, G. B., Li, L. J., Wei, M., Li, J., Yang, F. J. (2022). SmMYB113 is a key transcription factor responsible for compositional variation of anthocyanin and color diversity among eggplant peels. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.843996
Yang, Y., Liu, J., Zhou, X. H., Liu, S. Y., Zhuang, Y. (2020). Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. Peer J. 8, e8777. doi: 10.7717/peerj.8777
Yao, X., Zhou, M. L., Ruan, J. J., Peng, Y., Ma, C., Wu, W. J., et al. (2022). Physiological and biochemical regulation mechanism of exogenous hydrogen peroxide in alleviating NaCl stress toxicity in tartary buckwheat (Fagopyrum tataricum (L.) gaertn). Int. J. Mol. Sci. 23, 10698. doi: 10.3390/ijms231810698
Yu, R. M., Chang, Y. N., Chen, H. Z., Feng, J. L., Wang, H. J., Tian, T., et al. (2021). Genome-wide identifcation of the GATA gene family in potato (Solanum tuberosum L.) and expression analysis. J. Plant Biochem. Biot. 31, 37–48. doi: 10.1007/s13562-021-00652-6
Yu, M. N., Yu, J. J., Cao, H. J., Yong, M. L., Liu, Y. F. (2019). Genome-wide identification and analysis of the GATA transcription factor gene family in Ustilaginoidea virens. Genome 62, 807–816. doi: 10.1139/gen-2018-0190
Keywords: Solanum melongena L., GATA, gene family, light, hormones
Citation: Wang Y, Li X, Mo Y, Jiang C, Zhou Y, Hu J, Zhang Y, Lv J, Zhao K and Lu Z (2024) Identification and expression profiling of SmGATA genes family involved in response to light and phytohormones in eggplant. Front. Plant Sci. 15:1415921. doi: 10.3389/fpls.2024.1415921
Received: 11 April 2024; Accepted: 14 May 2024;
Published: 28 May 2024.
Edited by:
Zhichao Wu, National Cancer Institute Bethesda, United StatesReviewed by:
Kamran Shah, South China Agricultural University, ChinaCopyright © 2024 Wang, Li, Mo, Jiang, Zhou, Hu, Zhang, Lv, Zhao and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junheng Lv, MjAyMDA2MUB5bmF1LmVkdS5jbg==; Kai Zhao, MjAxMzAzNUB5bmF1LmVkdS5jbg==; Zhenya Lu, emhlbnlhbHVAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.