- 1State Key Laboratory of Southwestern Chinese Medicine Resources, School of Pharmacy/College of Modern Chinese Medicine Industry, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 3Departamento de Botánica y Fisiología Vegetal, Universidad de Málaga, Málaga, Spain
- 4Key Laboratory of Plant Resources Conservation and Utilization, College of Biology and Environmental Sciences, Jishou University, Jishou, China
- 5College of Life Science, Sichuan Agricultural University, Ya’an, China
- 6Center for Food Safety and Applied Nutrition, Office of Regulatory Science, U.S. Food and Drug Administration, College Park, MD, United States
Alisma L. is a medicinally important genus of aquatic and wetland plants consisting of c. 10 recognized species. However, largely due to polyploidy and limited taxon and gene sampling, the phylogenomic relationships of Alisma remain challenging. In this study, we sequenced 34 accessions of Alismataceae, including eight of the ten species of Alisma, one species of Echinodorus and one species of Luronium, to perform comparative analyses of plastid genomes and phylogenetic analyses. Comparative analysis of plastid genomes revealed high sequence similarity among species within the genus. Our study analyzed structural changes and variations in the plastomes of Alisma, including IR expansion or contraction, and gene duplication or loss. Phylogenetic results suggest that Alisma is monophyletic, and constitutes four groups: (1) A. lanceolatum and A. canaliculatum; (2) the North American clade of A. subcordatum and A. triviale; (3) A. wahlenbergii and A. gramineum; and (4) A. plantago-aquatica from Eurasia and northern Africa with the eastern Asian A. orientale nested within it. Hence the results challenge the recognition of A. orientale as a distinct species and raise the possibility of treating it as a synonym of the widespread A. plantago-aquatica. The well-known Alismatis Rhizoma (Zexie) in Chinese medicine was likely derived from the morphologically variable Alisma plantago-aquatica throughout its long history of cultivation in Asia. The plastome phylogenetic results also support the tetraploid A. lanceolatum as the likely maternal parent of the hexaploid eastern Asian A. canaliculatum.
1 Introduction
Alisma L. is a medically important genus with about 10 species (Li et al., 2022; Ito and Tanaka, 2023) and is one of the 15 genera of Alismataceae, an ancient aquatic and semi-aquatic group of monocot plants (Les and Tippery, 2013). This genus is widely distributed in temperate and subtropical regions mainly from eastern and South Asia, North America, and Europe to tropical East Africa. Most of the Alisma species may be used as food and medicine. For example, Alisma orientale and A. plantago-aquatica have been utilized in several Asian countries such as China, Japan and Korea (Han et al., 2013) for nearly 2,000 years (Pharmacopoeia of the People’s Republic of China, 2020; The Japanese Pharmacopoeia, 2021). They have been used to treat diseases such as diuresis, oliguria, diabetes, hyperlipidemia, hepatitis and obesity (Liu et al., 2013; Jang et al., 2015). The potential of these two plants’ tubers in cancer treatment had been also explored in one recent research (Jang and Lee, 2021). Furthermore, the extracts of A. canaliculatum have been used to treat gastric cancer (Kwon et al., 2021).
Recent phylogenetic studies on Alismataceae employing plastid, mitochondrial and/or nuclear genes have placed Alisma as closely related to Luronium, Baldellia and Damasonium (Ross et al., 2016; Lehtonen, 2017; Chen et al., 2022). Of particular interest, Chen et al. (2022) have explored the adaptation to the aquatic environments using Alismataceae as a model with extensive nuclear and organelle data. Based on morphology, geographic distribution, ecology, anatomy and cytology, Alisma was divided into two major groups: the gramineum group (A. wahlenbergii; A. gramineum) and the plantago-aquatica group (A. plantago-aquatica, A. orientale, A. triviale) (Hendricks, 1957; Björkqvist, 1968). The groupings are consistent with the taxonomic study on the basis of 38 characters of gross morphology, anatomy and pollen morphology (Zhang and Chen, 1994). However, distinguishing between species within Alisma has been challenging due to the similar morphologies among closely related species (Liu et al., 2020).
Several molecular studies have explored the evolutionary history of Alisma. An earlier molecular phylogenetic study based on nuclear ribosomal internal transcribed spacer (nrITS), plastid DNA (trnL), and RAPD markers (Jacobson and Hedrén, 2007) supported the two major diploid groups (i.e. the plantago-aquatica group and the gramineum group), but the positions of the polyploid Alisma species (A. canaliculatum, A. lanceolatum, A. triviale and A. rariflorum) have remained uncertain. Focusing on the family-level phylogenetic analyses, Chen et al. (2012) sampled only four species of Alisma: A. canaliculatum, A. gramineum, A. nanum and A. plantago-aquatica based on nrITS and plastid DNA (matK, psbA and rbcL) data. Some topological inconsistencies were observed, with A. gramineum being resolved as sister to the other three species in their nrITS tree, but the plastid data supported two clades with the A. gramineum - A. nanum clade being sister to the A. plantago-aquatica - A. canaliculatum clade.
Recently Ito and Tanaka (2023) performed a phylogenetic analysis of Alisma based on several DNA fragments including nuclear (nrITS and phyA) and chloroplast (matK, ndhF, psbA-trnH and rbcL) markers. The study focused on Asian species, sampling six putative species and two varieties particularly from Japan and Europe. The result suggested that A. canaliculatum from eastern Asia and A. rariforum endemic to Japan are sister to each other. A. orientale collected from China, Myanmar and Vietnam was shown to be nested within A. plantago-aquatica. Ito and Tanaka (2023) suggested that A. orientale was likely derived through parapatric speciation in the mountainous regions at the southern edge of the distribution of A. plantago-aquatica.
Due to limited taxon sampling, molecular markers, and small morphological differences between species, as well as the polyploidy of some species, the phylogenetic position of species of Alisma remains uncertain and controversial, such as the low support and ambiguous affinities of the three species of A. plantago-aquatica, A. orientale and A. canaliculatum (Ito and Tanaka, 2023). Therefore, the phylogenetic relationships of Alisma still need further exploration. Compared with plastid gene fragments, plastid genomes (plastomes) contain more informative information. The highly conserved structure of plastid genome is suitable for inferring the evolutionary relationships of higher taxa (Givnish et al., 2018). Plastome sequences have been extensively used to articulate phylogenetic relationships of many plant genera (Lv et al., 2022; Su et al., 2022; Xu et al., 2022; Hu et al., 2023). However, whole plastome sequence data, which could be used to develop novel genetic markers for determining infrageneric relationships, are still lacking for most species of Alisma.
In this study, we sequenced and performed comparative analyses of the plastid genomes of Alisma. The specific aims of this study were to: (1) investigate the structural variation of plastome genomes in Alisma at both the population and species levels, and infer the plastome structural evolution of Alisma; (2) identify the most variable regions of these plastomes as reference genes for future species identification; and (3) uncover the phylogenetic relationships within Alisma. We hope the phylogenetic study of Alisma using whole plastomes will lead to a better phylogenetic framework and improve resource conservation and the downstream application of medicinal resources, which are important for the modern utilization of traditional natural herbs.
2 Materials and methods
2.1 Plant material, DNA extraction and sequencing
A total of 34 accessions, including eight recognized species of Alisma, one species of Echinodorus Rich. and one species of Luronium Raf. of Alismataceae were sampled. All voucher specimens have been deposited in the United States National Herbarium (US), the KHD Herbarium of the Denver Botanic Gardens (KHD), the University of Göttingen Herbarium (GOET), and the Chinese Medicine Herbarium of Chengdu University of Chinese Medicine (CDCM). The voucher information is presented in Supplementary Table 1. Additionally, chloroplast genome data from six accessions, representing Sagittaria (NC_067603, NC_044119, NC_029815) and Caldesia (NC_045925, NC_045926) of Alismataceae, and Butomus (NC_051949) of Butomaceae (Alismatales), were obtained from the NCBI (Luo et al., 2016; Liang et al., 2019; Mwanzia et al., 2019; Yang and Liu, 2019) (Supplementary Table 1).
Total genomic DNAs were isolated from silica-gel dried leaves or herbarium specimens using a modified SDS method (Dellaporta et al., 1983; Johnson et al., 2023). DNA concentration was determined with a Qubit 4.0 fluorometer (Thermo Fisher Scientific) using a high sensitivity dsDNA kit while integrity and fragment size were evaluated with agarose gel electrophoresis (1% agarose SeaKem LE (Lonza Group Ltd. Basal, Switzerland)). DNA libraries were prepared using a KAPA HyperPrep Kit (Hoffmann-La Roche, Basel, Switzerland) and then sequenced with an Illumina Nova-Seq 6000 platform (paired-end, 2x150bp) at Novogene, Sacramento, CA, USA.
2.2 Raw data processing and sequence assembly
The raw reads were trimmed using Trimmomatic v.0.39 (Bolger et al., 2014). We used FastQC v.0.12.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to check the quality of the sequences. For the chloroplast genomes assembly, we employed GetOrganelle v.1.7.7 (Jin et al., 2020) based on trimmed paired-end reads. Circular genomes were generated for 27 taxa. For the seven samples which failed to be assembled by the one step from GetOrganelle due to likely low sequence coverage of some parts, a manual assembly method using closely related references was applied (Zhang et al., 2015; Liu et al., 2019).
For the plastid genomes, de novo assembly graphs were visualized and edited using Bandage v.0.8.1, and a whole or nearly whole circular plastid genome was generated (Wick et al., 2015). Using the plastid genome of A. orientale (OR522698) downloaded from NCBI (National Centre for Biotechnology Information, https://www.ncbi.nlm.nih.gov/) as a reference, all the genes were predicted using CPGAVAS v.2.0 (Liu et al., 2012). Subsequently, the annotation of the new sequences was then manually checked using Geneious v.11.0.18 (Kearse et al., 2012). All the plastid genomes from Alisma, Echinodorus and Luronium specie, were newly obtained in the present study, and submitted to the NCBI after being checked without error. The accession numbers of all samples are shown in Supplementary Table 1. These raw and annotated sequences have also been added to the United States Food and Drug Administration’s GenomeTrakrCP database and the related Bioproject (PRJNA325670, Zhang et al., 2017a). Circular genome visualization was illustrated using the online tool Organellar Genome Draw (OGDRAW) v.1.3.1 (https://chlorobox.mpimpgolm.mpg.de/OGDraw.html) (Greiner et al., 2019).
2.3 Genome structure and repeat sequence analysis
Geneious was utilized to count the number of genes, types and other information of all samples. The LSC, SSC, and IR regions of each chloroplast genome sequence were also extracted, and the AT and GC contents were counted using BioEdit (Alzohairy, 2011).
REPuter software (Kurtz et al., 2001) was used to identify forward (F), reverse (R), palindrome (P), and complementary (C) repeats in the resultant plastomes with the setting of a minimum repeat size of 30 bp and a maximum of 1000 bp, with sequence identities greater than 90% (Hamming Distance of 3). Simple sequence repeats (SSRs) were predicted using MISA (Beier et al., 2017) by setting the minimum number of repeat units to 10, 5, 4, 3, 3, and 3 for the mono-, di-, tri-, tetra-, penta- and hexa-nucleotides. The maximum length of the sequence between two SSRs to register as a compound SSR was set to 0 bp.
2.4 Comparative genomic analyses
To explore the expansion and contraction of IR regions of Alisma, comparison of boundaries between IRs and single copy regions was performed in CPJSdraw v.1.0.0 (Li et al., 2023). The mVISTA (https://genome.lbl.gov/vista/mvista) (Mayor et al., 2000) was used to assess the similarity among the plastid genomes, and the default parameters were utilized to align the cp genomes in Shuffle-LAGAN mode with A. orientale (OR522698) as the reference. Common sequences of coding regions and intergenic spacer regions (IGS) from 31 samples of Alisma were extracted.
Multiple alignments of sequences were performed using MAFFT v.7.48 with default settings (Katoh et al., 2019). Sliding window analysis of nucleotide variability in the cp genome was conducted using DnaSP v.6.11 (Rozas et al., 2017). The step size was set to 200 bp, with a 600 bp window length.
2.5 Phylogenetic analyses
To explore the phylogenetic relationships in Alisma, we included not only the newly sequenced samples of this genus, but also eight species from other genera within Alismataceae (Caldesia, Echinodorus, Luronium and Sagittaria). Botomus umbellatus (Butomaceae) of Alismatales (NC_051949) downloaded from Genbank was used as an outgroup. The matrix for phylogenomic analyses consisted of complete aligned plastid genomes, and the global alignment was done using MAFFT (Katoh et al., 2019). Poorly aligned regions were then removed using trimAl v1.2 (Capella-Gutiérrez et al., 2009). The final matrix had a total length of 159,457 bp for a total of 40 individuals.
A maximum likelihood (ML) analysis of the complete plastome data was implemented using IQ-TREE v.1.6 (Nguyen et al., 2015) with support values estimated by 10,000 ultrafast bootstrap replicates. The best fit model GTR+I+G model of molecular evolution was used, and the best-fit partitioning schemes and models estimated by ModelFinder (Kalyaanamoorthy et al., 2017). The Bayesian Inference (BI) using MrBayes v.3.2.7 (Ronquist et al., 2012), and the Markov Chain Monte Carlo (MCMC) algorithm was calculated for 1,000,000 generations with a sampling tree every 1000 generations.
3 Results
3.1 Organization of the plastid genomes of species in Alisma
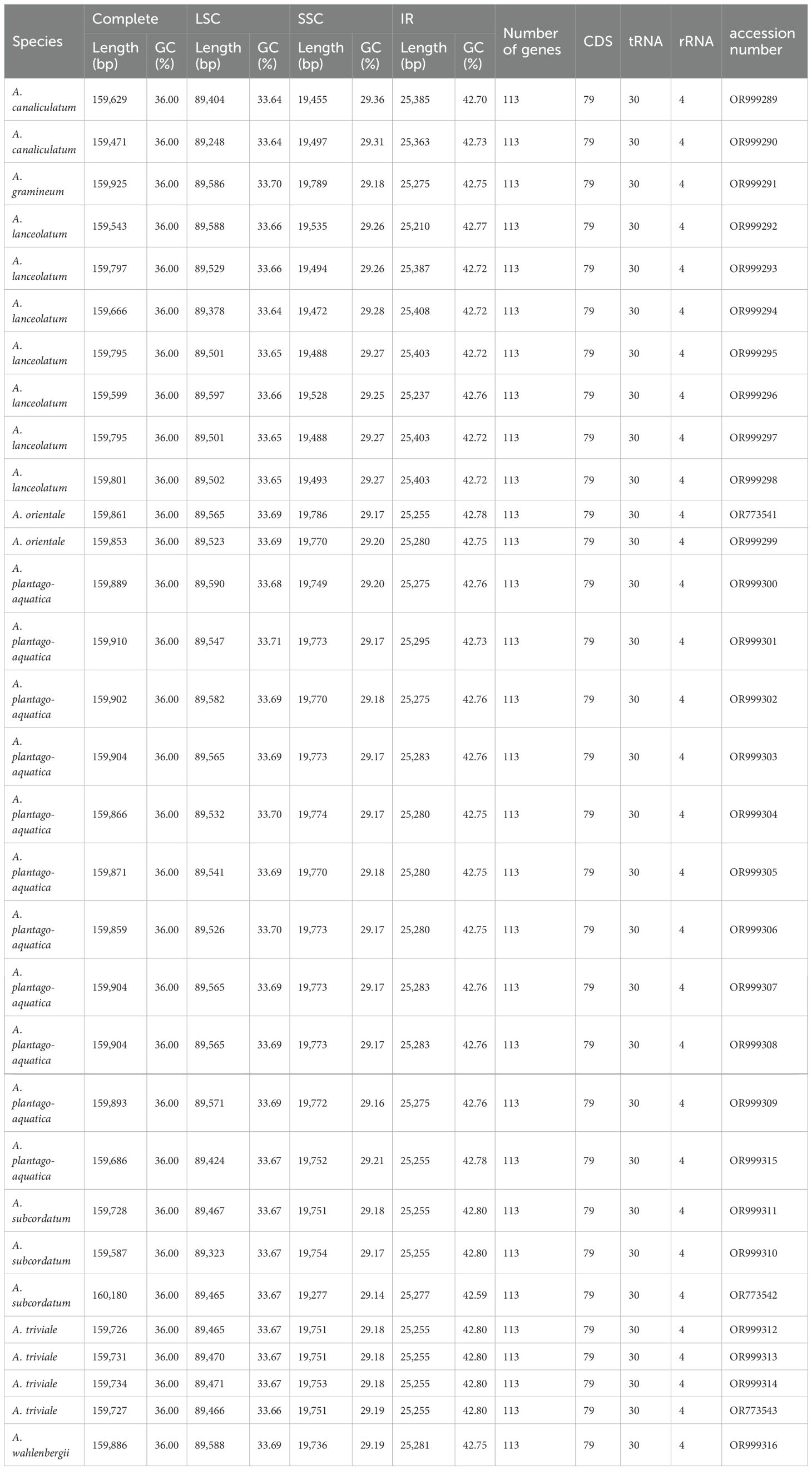
The length of complete plastomes of Alisma ranged from 159,471 bp (A. canaliculatum) to 160,180 bp (A. subcordatum). All of the plastomes displayed a typical quadripartite architecture that contained a large single copy (LSC: 89,248–89,597 bp) and a small single copy (SSC: 19,277–19,789 bp) separated by two copies of an inverted repeat (IR: 25,210–25,408 bp). The total GC content is 36.00%, and the IR regions had the highest GC content (42.70–42.80%) followed by those in the LSC (33.64–33.71%) and SSC (29.14–29.36%) regions (Table 1).
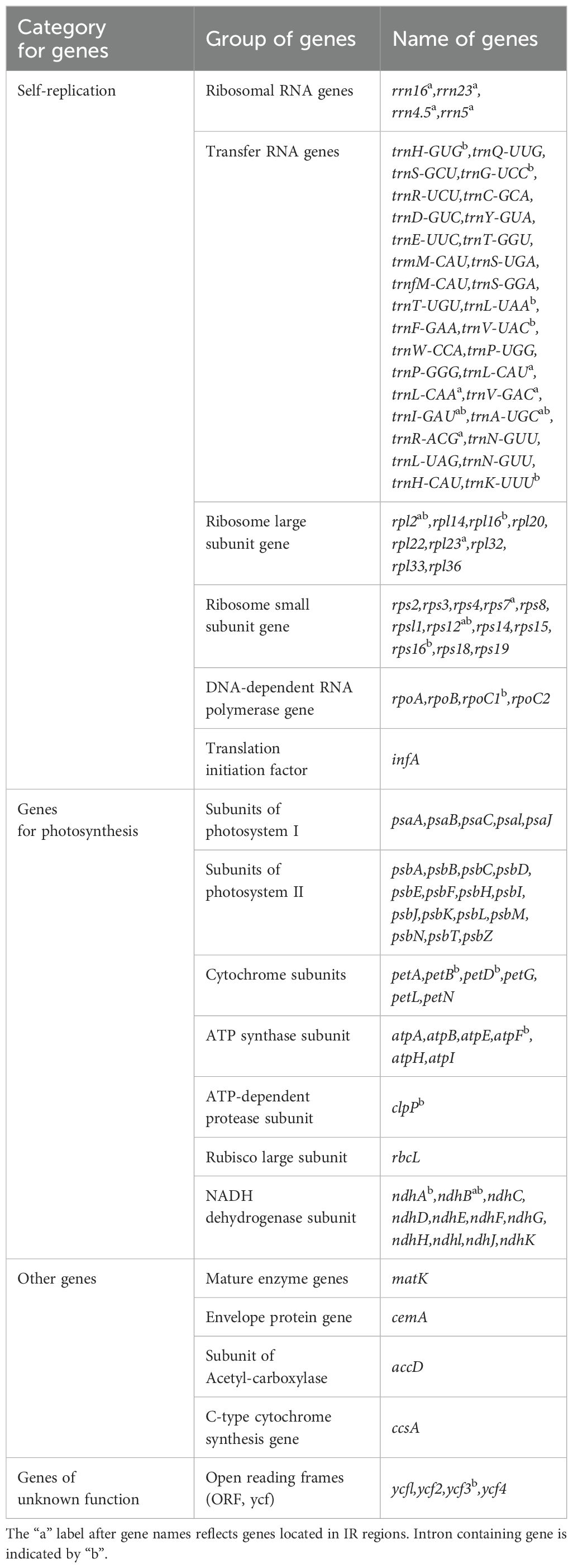
Each plastome encoded 113 unique genes, including 79 protein-coding, 30 tRNA and 4 rRNA genes. Amongst them, 17 genes were duplicated in the IR regions, comprising six protein-coding genes (rps7, rps12, rpl2, rpl23, ndhB and ycf2), four rRNA genes (rrn4.5, rrn23, rrn5 and rrn16), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnV-GAC and trnR-ACG). 17 intron-containing genes were detected, including ten protein-coding genes (atpF, ndhA, ndhB, rpl2, rpl16, rps16, rps12, clpP, rpoC1 and ycf3) and seven tRNA genes (trnH-GUG, trnG-UCC, trnA-UGC, trnI-GAU, trnV-UAC, trnK-UUU and trnL-UAA). Of the 17 genes, three (clpP, rps12 and ycf3) harbored two introns and the other 14 contained only one intron (Table 2).
3.2 SSRs and long repeat sequence
The number of SSRs in each plastome of Alisma ranged from 90 (A. triviale) to 98 (A. lanceolatum). The majority of the SSRs were mononucleotides (49.62%), followed by tetranucleotides (27.41%), dinucleotides (14.66%), trinucleotides (4.78%) and pentanucleotides (3.22%). The smallest number of SSRs were hexanucleotides (0.31%). The mononucleotide SSRs were composed of A/T mofits and most of the dinucleotide ones were composed of AT/TA. SSR was mainly located in the LSC region (76.51%), followed by SSC (17.05%), and then IR (6.44%) (Figure 1).
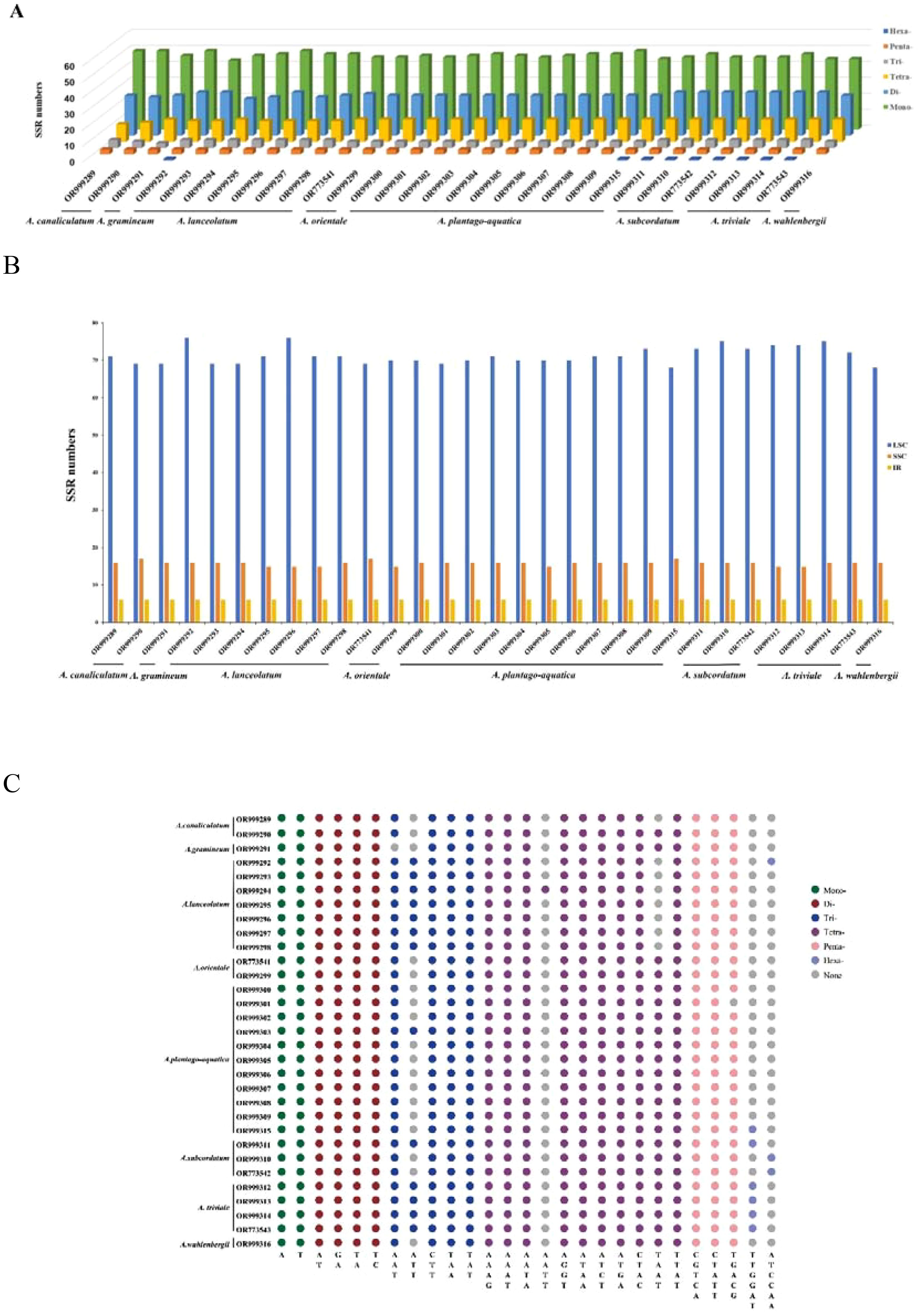
Figure 1. Analysis of simple sequence repeats (SSRs) in the plastid genomes. (A) Number of different SSR types detected in the genomes; (B) number of SSRs identified in LSC, SSC, and IR regions; (C) SSR motifs in different repeat types.
A total of 2157 long repeat sequences with lengths greater than 30 bp, including 871 (40.38%) forward repeats, 939 (43.53%) palindromic repeats, 132 (6.12%) complementary repeats and 215 (9.97%) reverse repeats, were detected in 31 samples from Alisma. The number of repeats for each taxon varied from 46 to 185 and had lengths that ranged from 30 to 55 bp. Most of the long repeats were located in LCS region (62.45%), followed by IR (27.17%) and SSC (10.38%) (Figure 2).
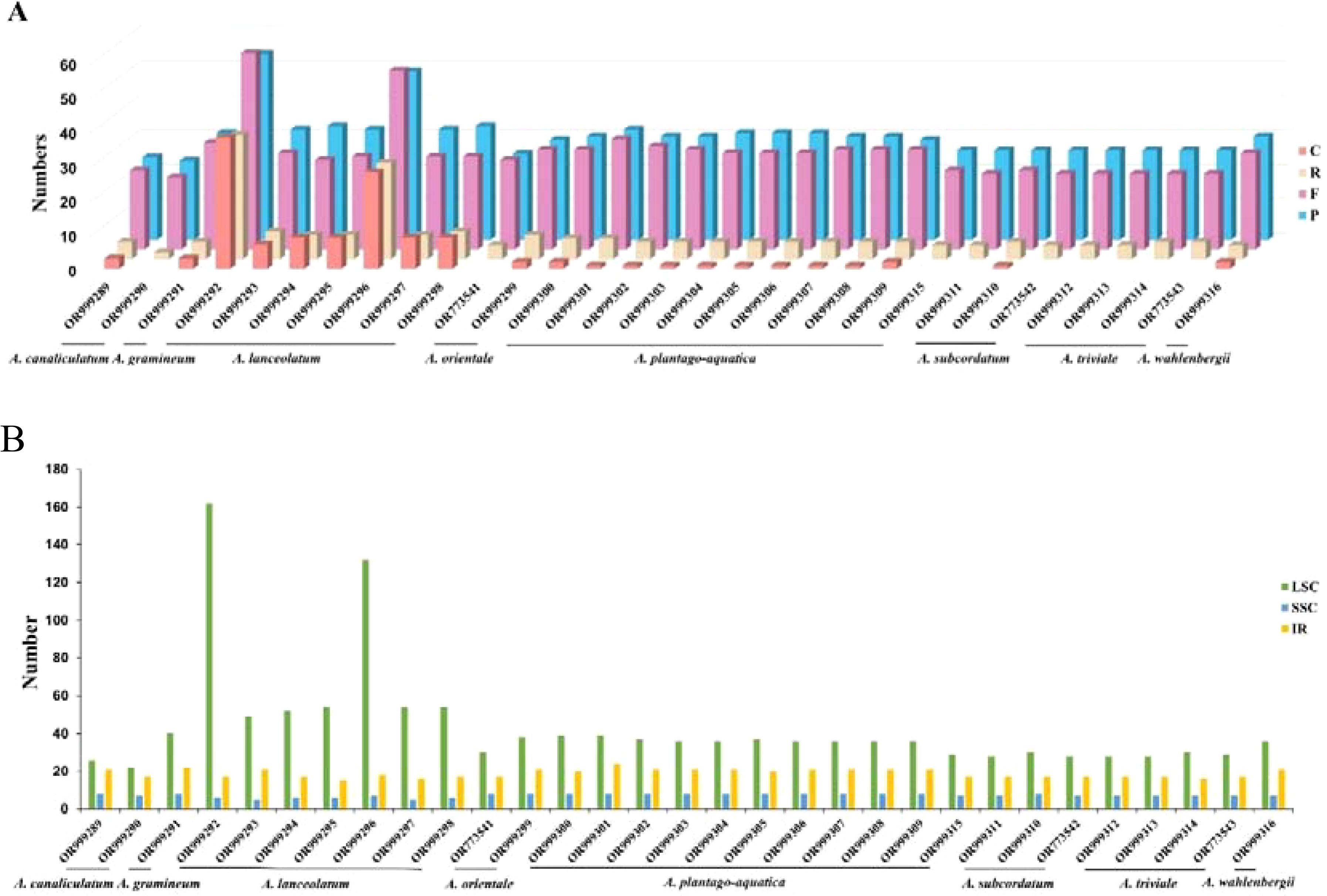
Figure 2. Repeat sequences in the plastid genomes of Alisma. (A) Number of Repeat sequences types detected in the genomes; (B) number of Repeat sequences identified in LSC, SSC, and IR regions.
3.3 Expansion and contraction of the IR region
Four boundaries of the plastid genomes, namely JLB (LSC-IRb), JSB (SSC-IRb), JSA (SSC-IRa), and JLA (LSC-IRa) showed remarkable stability among the species in Alisma. The IR/SC junctions of the plastid genomes mainly contained five genes (rpl2, rps19, ycf1, ndhF and trnH). The rps19 gene was completely present within the LSC, at a distance of 29-98 bp to the JLB junction. For the JLA junction, the trnH gene was located in the LSC region 13-64 bp away from the junction, whereas the rpl2 gene was located in the IRa region 55-82 bp away from the junction. We only detected differences in distance to the JLA junction. The SSC/IRa boundary was located within the ycf1 gene and had lengths ranging from 4,557 bp to 5,021 bp in the SSC and of 295 bp or 759 bp in the Ira. In total, expansion of IRa boundaries in A. canaliculatum and A. lanceolatum compared to other species. Genetic variation is somewhat intraspecific conserved and interspecific differences are relatively pronounced in Alisma (Figure 3).
3.4 Hypervariable regions of the plastid genomes
The complete plastid genomes from Alisma were compared using mVISTA with the A. orientale (OR522698) genome as reference. Comparative analysis showed that the plastid genomes were evolutionarily conserved with similar structures and gene orders. IR regions were found to be more conserved than the single copy regions, so were genic regions, coding regions, and exons compared with intergenic regions, non-coding and introns. The regions of 4-12 kb, 46-56 kb, 64-66 kb, 104-106 kb, 114-116 kb, and 124-128 kb have relatively large variations (Figure 4).
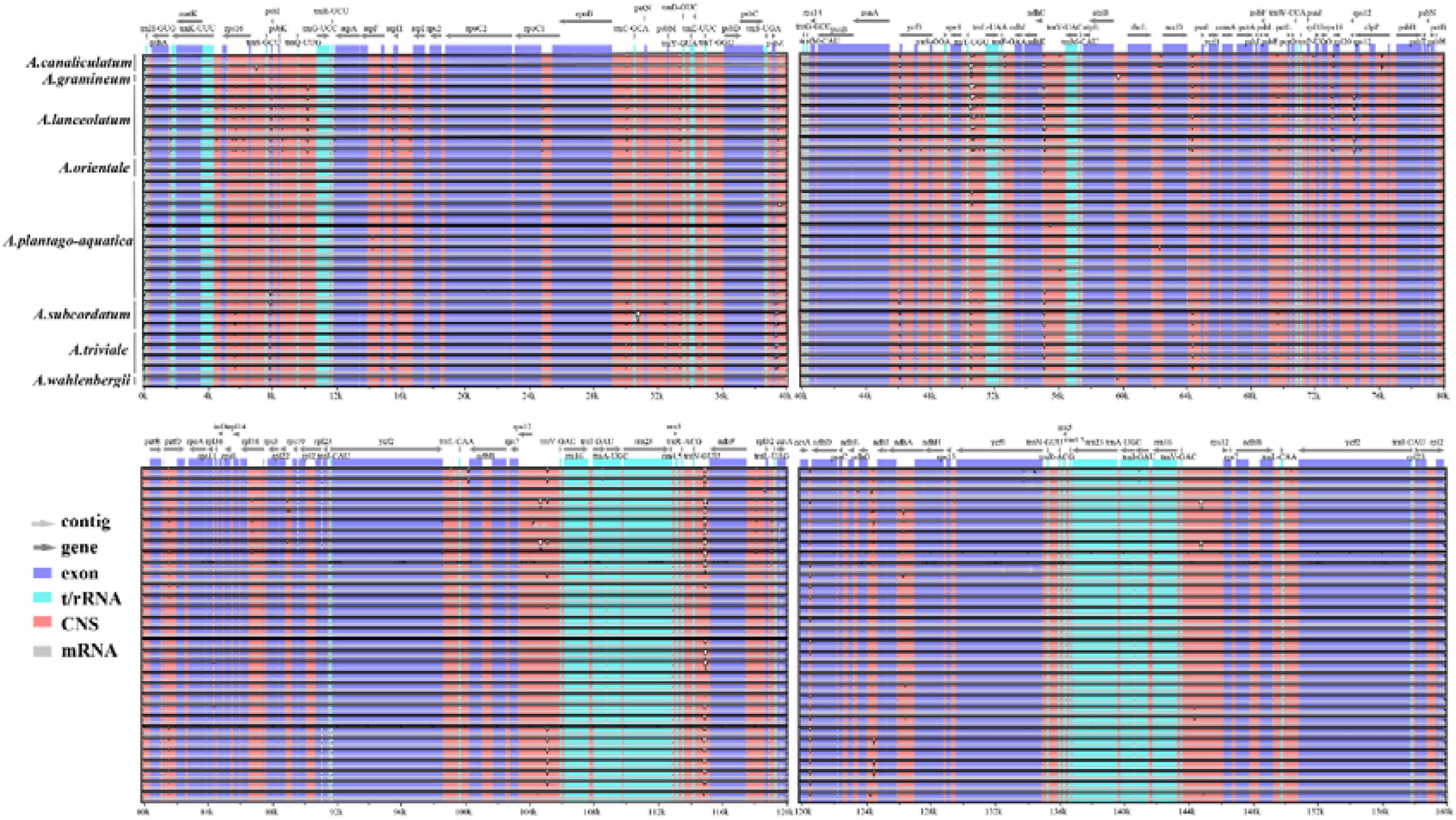
Figure 4. Sequence alignment of the 31 plastid genomes performed using the mVISTA program with Alisma orientale (OR522698) as a reference.
A sliding window analysis was used to estimate the level of variation across regions in coding and non-coding regions. Nucleotide diversity (Pi) values ranged from 0.00000 to 0.01333 for coding regions and 0.00000 to 0.02215 for non-coding regions. All highly divergent sequences were restricted to the single copy (SC) regions, with the highest peak occurring in the SSC region. Among these regions, one coding regions with Pi values exceeding 0.01 and one regions in IGS regions with Pi values surpassing 0.02 were identified as significant hotspot regions, of which one was psbI gene, and the remaining one was intergenic spacer of ccsA-ndhD (Figure 5).
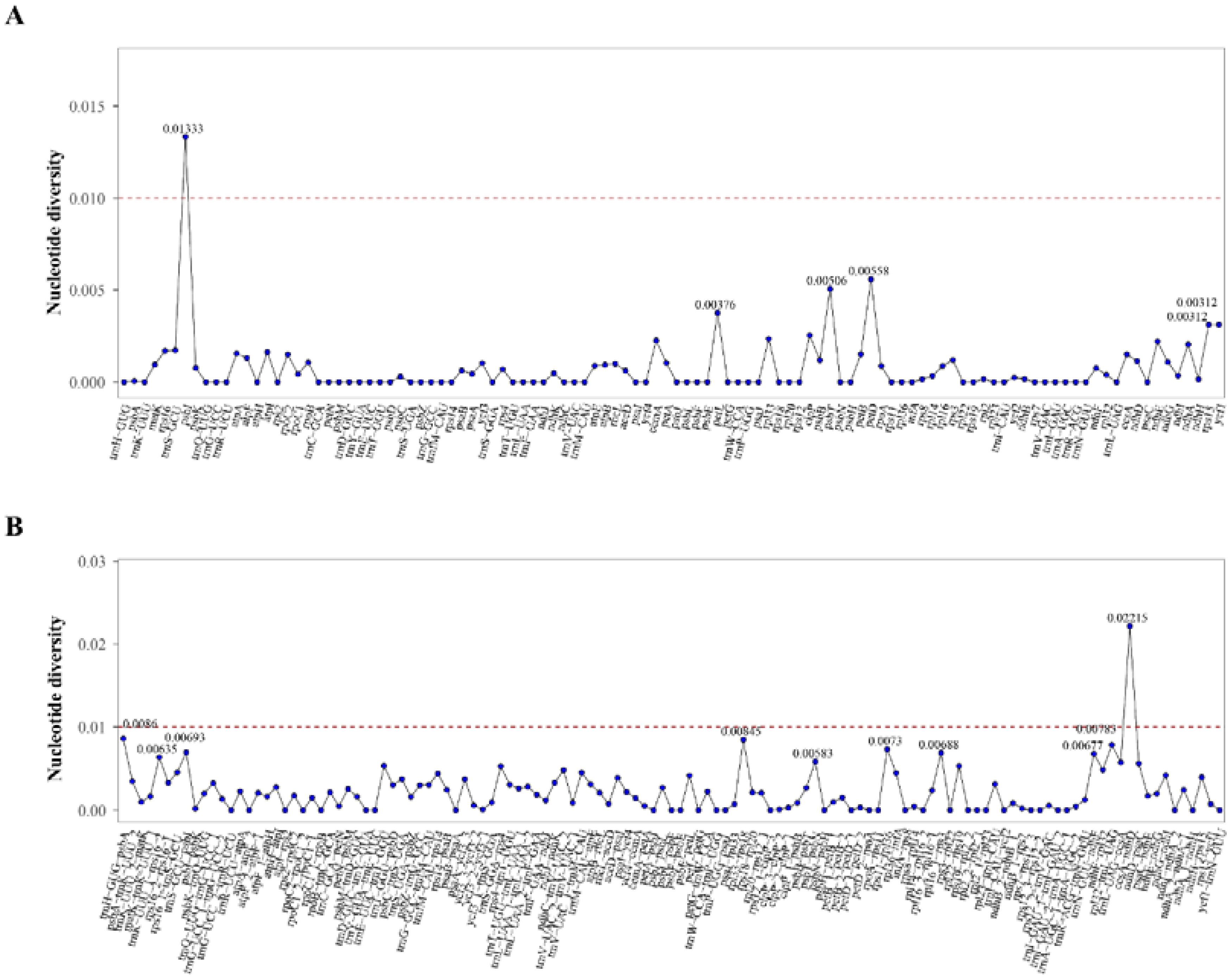
Figure 5. Sliding window analysis of Alisma species. (A) Nucleotide diversity values of common genes within the chloroplast genomes; (B) Nucleotide diversity values of intergenic spacers (IGS) within the chloroplast genomes.
3.5 Phylogenetic inference
The plastid genome topologies of Alisma based on ML and BI analyses were consistent (Figure 6). The monophyly of Alisma was strongly supported (100%ML BS, 1.0 PP). The whole plastid genome data provided strong support for the position of A. canaliculatum (100% ML BS, 1 PP), A. lanceolatum (100% ML BS, 1 PP), A. wahlenbergii (86% ML BS, 1 PP) and A. gramineum (86% ML BS, 1 PP). The eight Alisma species sampled were grouped into four clades. Group I consists of A. canaliculatum and A. lanceolatum, which diverged first. Group II is composed of two species, A. subcordatum and A. triviale, both from North America. Groups III and IV were sister to each other, and each clade is composed of two species. Group III includes A. gramineum and A. wahlenbergii, which are very similar in morphology. It should be noted that the two species A. plantago-aquatica and A. orientale in Group IV are largely distributed in Eurasia and they cannot be clearly delineated. Furthermore, the two North American species, A. subcordatum and A. triviale in Group II are intermixed, and each did not form a clade (Figure 6).
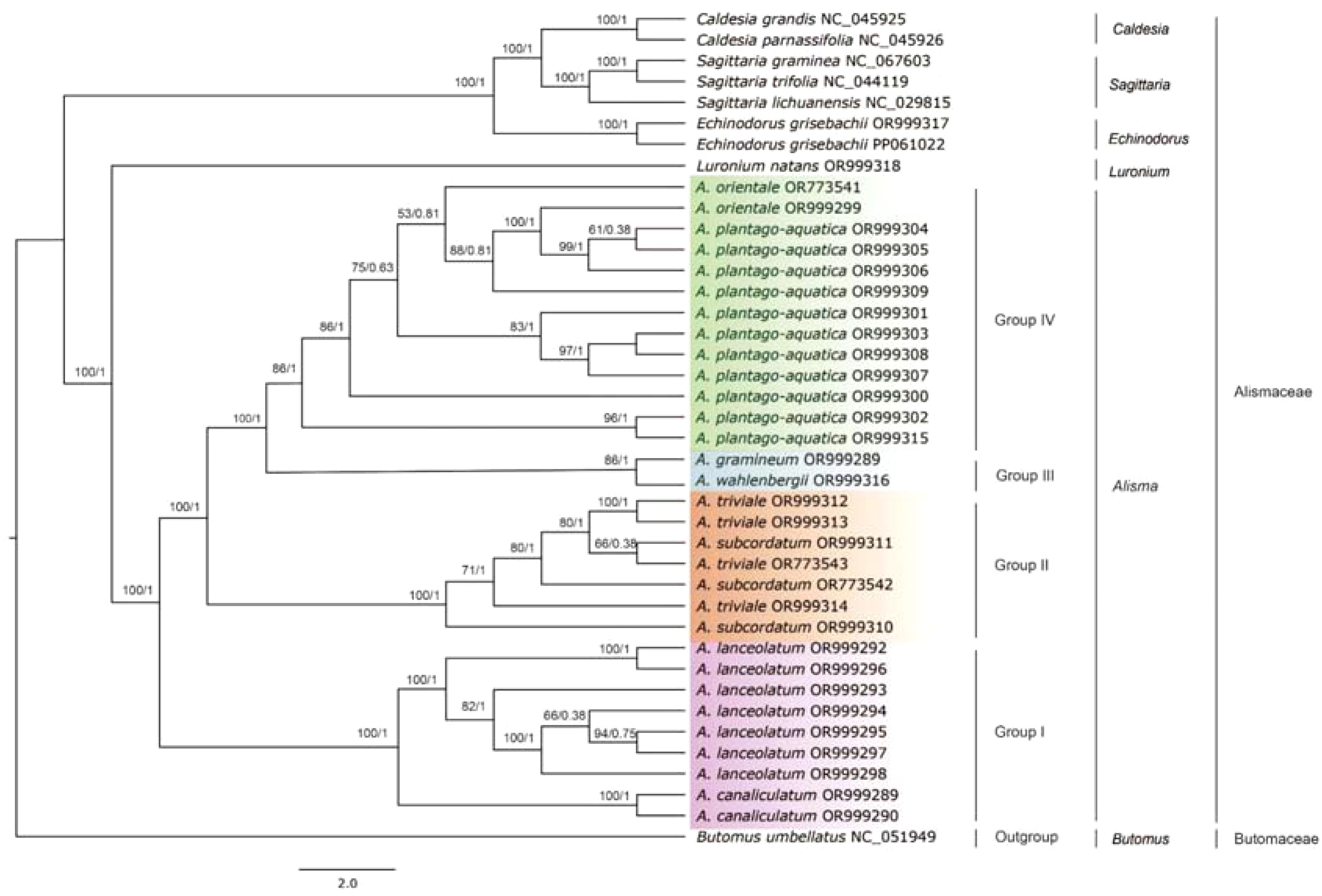
Figure 6. Plastid phylogeny of Alisma using Maximum Likelihood (ML) and Bayesian Inference (BI) methods based on the whole plastome sequences. Likelihood bootstrap support values (BS) and Bayesian posterior probability values (PP) are shown next to the nodes.
4 Discussion
4.1 Gene content and structural variations of Alisma plastomes
Plastome structure and gene content are generally considered to be conserved in flowering plants, but vary in length from 120 kb to 160 kb and consist of 100-120 unique genes (Bai et al., 2021). In this study, 113 genes were annotated across the genomes of the species in Alisma and one specie in Luronium, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes that showed good conservatism across species in this genus. The length of complete plastid genomes of Alisma ranged from 159,471 bp in A. canaliculatum to 160,180 bp in A. subcordatum, which are clearly shorter than Sagittaria and Caldesia in Alismataceae (Liang et al., 2019; Mwanzia et al., 2019). However, the length of the plastid sequences of the Alisma species is similar to that of Hydrocharitaceae and Butomaceae, which are close relatives of Alismataceae in Alismatales. In addition to the obvious difference in length, the genera of Alismataceae also differed greatly in the type, number and length of ycf genes. For example, Caldesia parnassifolia (NC_045926) did not contain ycf1 gene; Sagittaria graminea (NC_067603) contained two ycf1 genes; and the 32 samples from Alisma and Luronium sequenced in the present study contained only one ycf1 gene. In addition to the Alismataceae, the Alismatales contain other species with deletions of ycf1 genes, including the Hydrocharitaceae, Butomaceae, and Tofieldiaceae. In general, although ycf1 is a highly variable gene that is often used as a DNA barcode in phylogenetic analyses (Dong et al., 2015; Zhao et al., 2020), ycf1 remains highly conserved among species within Alisma as this study shows.
SSRs have the lowest number of hexanucleotides, with only one hexanucleotide found in nine samples from three Alisma species, including A. triviale (OR999314, OR773543, OR999312, OR999313), A. subcordatum (OR999311, OR773542, OR999310), A. lanceolatum (OR999292) and A. plantago-aquatica (OR999315). No hexanucleotides were found in the other samples. Notably, A. triviale and A. subcordatum are two species from North America. They had slightly more SSRs than other species of the genus, and both species contained hexanucleotides SSRs. Meanwhile, most of the SSRs are located in the LSC and SSC, revealing conservatism of the IR regions, which was also supported by the distributions of the hypervariable regions (HVR). As SSRs are highly reliable, reproducible, and highly polymorphic, analysis of the SSR loci in the plastid genomes of Alisma has the potential for future population genetic studies, genetic diversity and species identification (Raza et al., 2019; Tang et al., 2021).
Among the four long repetitive sequences, forward repeats, palindromic repeats, complementary repeats, and reverse repeats, we identified the lowest number of complementary repeats. In A. triviale (OR999314, OR773543, OR999312, OR999313), A. subcordatum (OR999311, OR773542), A. plantago-aquatica (OR999315), A. orientale (OR773541) and A. canaliculatum (OR999290) in which no complementary repeat sequences were found. However, 38 and 28 complementary repeats were respectively found in two samples of A. lanceolatum (OR999292, OR999296). The number of complementary repeats found in other samples of A. lanceolatum was also higher than that of other species. Long repetitive sequences may promote variation and rearrangement in the plastid genomes, thus they play an important role in plant evolution (Wang et al., 2008). The plastome variation in A. lanceolatum may be further explored with a broader populational sampling.
Although it is generally accepted that the plastid genome is quite conserved in terms of structural organization and gene arrangement, expansion and contraction of the IR region at the boundary with the LSC and SSC regions are extremely common in chloroplast genomes (Wang et al., 2018; Ye et al., 2018). These changes are closely related to the differences in chloroplast genome length between taxa at the taxonomic hierarchy above the genus level (Stamatakis, 2014). In this study, by comparing the boundary regions of 31 Alisma plastid genomes, the IR boundaries of both A. canaliculatum and A. lanceolatum were found to be somewhat expanded, but the size and distribution of genes in the boundary region of the taxon remained conservative. It is also evident that the differences among the four boundary regions combined could not clearly and accurately reflect the inter- and intraspecific differences in Alisma. This further suggests that the developmental process of Alisma also relies on genetic changes in other regions.
The result of the global visualization showed that the Alisma plastid genomes had considerable similarity, little interspecific variation, and are developmentally conserved, with coding regions showing stronger conservation than non-coding regions, and the IR region being more conserved than the two single-copy regions. A. canaliculatum and A. lanceolatum had more variable sites compared to the reference sequence (A. orientale OR522698). In addition, A. subcordatum and A. triviale from North America also showed greater variation than others. Combined with the analysis of nucleotide polymorphisms, the degree of variation in the non-coding regions of the chloroplast genome is generally higher than that in the coding regions, which is consistent with the results of most plant studies (Zhao et al., 2020; Su et al., 2022). Despite the typically stable nucleotide content and highly conserved gene structure of chloroplast genomes, mutation hotspots can still occur. Among the protein-coding genes, the psbI gene (pi = 0.01333) had a relatively high level of polymorphism, the intergenic spacer of ccsA-ndhD had the highest polymorphism (pi = 0.02215). They can be considered as a molecular marker for Alisma systematic, genealogical and biogeographic studies. However, compared to other genera, such as Gaultheria (Xu et al., 2022), Alisma has fewer loci with significant nucleotide variants and low nucleotide diversity values. Alismataceae is one of the oldest lineages within monocots (Chen et al., 2012). However, the low genetic diversity in Alisma suggests that while it has an ancient origin, it diverged relatively recently. In addition, the high hybridization affinity among species of Alisma (Jacobson and Hedrén, 2007) may also be responsible for the relatively weak reproductive isolation among Alisma species.
4.2 Plastid phylogenomics of Alisma
According to our ML and BI trees using the complete plastomes (Figure 6), Alisma is divided into four groups (I, II, III, and IV) (100% ML BS, 1 PP). Group I, including A. lanceolatum and A. canaliculatum, is sister to all other taxa of the genus; Group II consists of two New World species (A. subcordatum and A. triviale) and is sister to the clade of Group III (A. wahlenbergii and A. gramineum) and Group IV (A. plantago-aquatica and A. orientale).
Group I: Two Eurasian species (A. lanceolatum and A. canaliculatum): Our plastome phylogeny showed that two polyploid species A. lanceolatum (2n = 28) and A. canaliculatum (2n = 42) constitute one clade (100% ML BS, 1PP). However, the phylogenetic analyses result based on the nuclear ITS and RAPD data (Jacobson and Hedrén, 2007) did not place these two species together. A. lanceolatum from temperate Europe, western Asia and North Africa was unresolved in the RAPD tree, while it was sister to the clade of the remaining Alisma species in the ITS tree. Jacobson and Hedrén (2007) suggested an ancient allotetraploid origin of Alisma lanceolatum. The placement of the hexaploid eastern Asian A. canaliculatum as the sister to the widespread A. lanceolatum suggests that A. lanceolatum may have served as the maternal parental species for A. canaliculatum. Our result of the IR boundary analysis and sequence variation analysis also showed that A. canaliculatum and A. lanceolatum differed to a greater extent from other species within the genus and to a lesser extent between the two species.
Björkqvist (1968) found that A. canaliculatum and A. lanceolatum were both very uniform and well distinguished in many characteristics from other taxa of the genus, and are most similar to each other vegetatively. Yet the leaf shape of the two species was usually different. The two species have aerial leaves that are lanceolate to broadly lanceolate and have cuneate leaf bases. But A. lanceolatum has thin and translucent lateral pericarps, substraight styles, and anthers 1-1.2 mm, whereas A. canaliculatum has thickish and opaque lateral pericarp, recurved styles, and anthers 0.5~0.8 mm long.
Our analysis showed that the A. canaliculatum - A. lanceolatum clade was the first diverged clade in Alisma (Figure 6). Jacobson and Hedrén (2007) also showed A. lanceolatum as sister to other Alisma species, and they argued that the allopolyploid origin of A. lanceolatum may have led to the hybrid ITS sequence produced by concerted evolution.
Group II: the North American clade of A. subcordatum and A. triviale: The two North American species A. subcordatum and A. triviale formed a clade with strong support (100% ML BS, 1 PP). But the two species are each unresolved. According to Björkqvist (1968), A. triviale and A. subcordatum both seem to be isolated from all other taxa of the genus with absolute sterility barriers, and both are distributed in New World (North America). Alisma subcordatum has a more southeastern distribution, whereas A. triviale has a more northern and western distribution in North America (Björkqvist, 1968).
With the two North American species each not forming a clade, it is worth mentioning that these two taxa are morphologically highly similar (Björkqvist, 1968). Haynes and Hellquist (2000) indicated that A. triviale and A. subcordatum were occasionally treated as varieties of a widespread A. plantago-aquatica. But they differ in ploidy levels with A. triviale as a polyploid (2n = 28) and A. subcordatum as a diploid (2n = 14) (Björkqvist, 1968). We thus speculate that A. triviale may be of allopolyploid origin in North America. We must test this hypothesis by employing nuclear genome data and conducting phylogenetic network analysis (Ferreira de Carvalho et al., 2019; Mendez-Reneau et al., 2023).
Group III: the clade of A. gramineum and A. wahlenbergii: Samuelsson (1931) first suggested that A. wahlenbergii was a relict endemic in the Baltic Sea area, but later regarded it as a newly formed taxon that had evolved as an ecotype of the morphologically similar A. gramineum at the shores of the Baltic Sea (Samuelsson, 1934). The latter hypothesis was also supported by Björkqvist (1968). Jacobson and Hedrén (2007) reported small genetic differences between A. gramineum and A. wahlenbergii, supporting the newly evolved hypothesis of A. wahlenbergii. Our result of comparative plastid genome and phylogenetic analyses is consistent with this hypothesis.
Alisma gramineum and A. wahlenbergii formed a clade with 100% support. The hypothesis that the Baltic endemic A. wahlenbergii was probably originated relatively recently from A. gramineum (Jacobson and Hedrén, 2007) may be supported by two aspects of evidence. First, both species live under water and their habitats obviously differ from those of the other congeneric species. Alisma gramineum distributes widely in Europe, Asia, and North America, and it is more adapted to submerged conditions than most other taxa of the genus. Alisma wahlenbergii has a more restricted distribution in Europe from Sweden, along the Finish coast of the Gulf of Bothnia and the Gulf of Finland, to the western part of Russia. Second, ecological environment determines that the morphology characters of the two species are very similar. Alisma wahlenbergii is similar to A. gramineum in many ways, including leaf shape, petal shape, anther shapes and size, style shape and length, achene shape. However, compared A. gramineum, A. wahlenbergii is more slender and smaller in most characteristics, both vegetatively and florally (Björkqvist, 1968).
Group IV: the clade of A. plantago-aquatica and A. orientale: A. plantago-aquatica from Eurasia and northern Africa and the eastern Asian A. orientale form a clade, yet the latter is nested within the widespread A. plantago-aquatica. The result hence supports treating A. orientale as a synonym of A. plantago-aquatica. But we will further test this hypothesis with the nuclear data.
The systematic relationship between the two taxa has been controversial. Based on the evidence of morphology, anatomy, pollen morphology and geographic distribution, Björkqvist (1968) and Zhang and Chen (1994) found that A. orientale from East and Central Asia was very uniform throughout the range and vegetatively similar to A. plantago-aquatica, but can be distinguished in floral characteristics, e.g. petal size, anther and style shape, stigma regions, shape of achenes, indicated that although there were interspecific and intraspecific variation in the two species, but most of these morphological variations were continuous and many were overlapping. The interspecific boundary between the two species seems blurred. The long-term cultivation and domestication of the well-known traditional medicine Alismatis Rhizoma in different areas in China may have also led to the formation of two different populations of “the A. plantago-aquatica complex”.
Rhizoma Alismatis is widely used in China, Japan, Malaysia and South Korea with over two thousand years of medicinal history. As the original plants of Alismatis Rhizoma, A. plantago-aquatica and A. orientale were listed in the latest edition of Chinese Pharmacopoeia (Pharmacopoeia of the People’s Republic of China, 2020), respectively. Although it had been cultivated for thousands of years in China, the botanical origins of the two species have caused much controversy due to the very little morphological distinctions. According to the Flora of China treatment (Wang et al., 2010), the distinguishing features between A. plantago-aquatica and A. orientale are few, primarily related to the length of styles, the margin of petals, and the regularity/irregularity of carpel arrangement. Additionally, these two species have significant geographic overlap in China (Wang and Li, 2002; Wu et al., 2008; Chen et al., 2009; Ma et al., 2015; Zhang et al., 2016). Based on morphological comparisons of the flowers and fruits, Liu et al. (2020) suggested that the differences between the two are very minimal. The identification results from DNA barcoding also reflected their similarities, with a single-base difference at position 247 out of 534 bases, and their homology exceeding 99.8%. Based on nuclear DNA (nrITS and phyA) and chloroplast DNA (matK, ndhF, psbA-trnH and rbcL) sequence data, Ito and Tanaka (2023) showed that the A. planago-aquatica complex including A. orientale was resolved to be monophyletic. They further suggest that A. orientale endemic to the Southeast Asian Massif may have originated via parapatric speciation from the progenitor species A. planago-aquatica, but the interspecific phylogenetic relationship was scarcely resolved.
Our results in this study challenge the species status of A. orientale, as it is nested within the widespread A. planago-aquatica. This result may also be related to the medicinal utilization history of nearly two thousand years and the very rare status of both species in the wild. Therefore, further analyses are needed to understand the phylogeographic pattern and species delimitation in this group by expanding the taxon sampling including both wild and cultivated in Asia especially in southeastern China and using nuclear genome analysis to further explore the phylogeny and delimitation of the two species. Such analyses will help reveal the origin, differentiation and domestication process of the important medicine “Alismatis Rhizoma” and provide an important reference for solving the taxonomic disputes for the two taxa.
Compared with traditional molecular phylogenetic studies based on several loci, the whole plastomes provide more informative sites for phylogenetic studies, which substantially improves the resolution of phylogenetic trees of some taxa at different taxonomic levels (Zhang et al., 2017b; Zhao et al., 2018). Indeed, compared with the previous molecular phylogenetic trees constructed from a few gene fragments (Jacobson and Hedrén, 2007; Ito and Tanaka, 2023), the support of the phylogenetic tree constructed based on the complete plastid genome data of Alisma in this study was much improved. But the plastomes still cannot fully resolve the phylogeny and species differentiation of Alisma due to allopolyploidy in the genus (Jacobson and Hedrén, 2007; Zhao et al., 2017). Therefore, future phylogenetic studies of Alisma need to be performed using nuclear genes, such as the Hyb-Seq approaches (Ma et al., 2021; Liu et al., 2023; Talavera et al., 2023) to unravel the allopolyploid speciation history of several polyploid species.
5 Conclusions
In this study, we sequenced and assembled the plastid genomes of eight species of the medicinally important genus Alisma, plus one species of Echinodorus and one species of Luronium of Alismataceae. The reported plastid genomes in Alisma are conserved, showing high levels of consistency and similarity in terms of gene content, order and structure. The plastid phylogenomic analyses have improved the phylogenetic resolution, and supported four main clades within Alisma. Our results support the tetraploid A. lanceolatum as the likely maternal parent of the hexaploid eastern Asian A. canaliculatum. However, plastid genomes still cannot resolve the complicated phylogenetic relationships and species discrimination especially within the North American clade and among polyploid species. Further studies are needed to test the extent of hybridization and allopolyploidy during the evolutionary diversification of Alisma using nuclear sequence data and a broader taxon sampling.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number OR773541-OR773543, OR999289-OR999318, PP061022.
Author contributions
Z-QL: Writing – original draft, Writing – review & editing, Funding acquisition, Supervision. WZ: Writing – original draft, Formal analysis, Writing – review & editing. AT: Formal analysis, Writing – review & editing. Z-LN: Formal analysis, Writing – review & editing. JL: Formal analysis, Writing – review & editing. GJ: Writing – review & editing, Formal analysis. X-MY: Writing – review & editing, Data curation. W-QZ: Resources, Writing – review & editing. Z-YZ: Resources, Writing – review & editing. SH: Writing – review & editing. JW: Writing – original draft, Writing – review & editing, Funding acquisition, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Smithsonian Barcode Network, the China Scholarship Council under Grant (202008515111) and Project of Xinglin Scholar Program for Discipline Talents and Research Enhancement at Chengdu University of Traditional Chinese Medicine (XCZX2022006). AT was supported by a “Margarita Salas” postdoctoral grant funded by the Ministerio de Universidades of Spain and the European Union “NextGenerationEU” and a grant from the Smithsonian National Museum of Natural History ADCS Core Grant Program (PI, JW).
Acknowledgments
The study was supported by the Laboratories of Analytical Biology of the Smithsonian National Museum of Natural History. The authors acknowledge the Smithsonian High Performance Cluster (SI/HPC) (https://doi.org/10.25572/SIHPC) and Chengdu University of Traditional Chinese Medicine for providing computational resources. The authors thank Dr. Marc Appelhans at GOET, Dr. Jennifer Ackerfield at KHD, Ceming Tan at Jiangxi Jiujiang Forest Herbarium, JuanYu at Herbarium of Inner Mongolia Medical University, Shigui Mo from Fujian Chengtian Pharmaceutical Co., Ltd, Xiaoqing Ning and Liuping Wang from Guangxi University of Chinese Medicine, Wei Qiu at Herbarium of Jiangxi University of Chinese Medicine, Ziwei Li at Herbarium of Chongqing Academy of Chinese Materia Medica for access to specimens for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1415253/full#supplementary-material
References
Alzohairy, A. M. (2011). BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Bai, H. R., Oyebanji, O., Zhang, R., Yi, T. S. (2021). Plastid phylogenomic insights into the evolution of subfamily Dialioideae (Leguminosae). Plant Div. 43, 27–34. doi: 10.1016/j.pld.2020.06.008
Beier, S., Thiel, T., Münch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Björkqvist, I. (1968). Studies in Alisma L. 2. Chromosome studies, crossing experiments and taxonomy. Opera Bot. 19, 1–138.
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, J. Y., Su, H. L., Huang, Y. J., Ge, P. S., Lv, Z. Q. (2009). Comparative studies on genetic resource of characteristics of. Alisma. China J. Chin. Materia Medica. 34, 2713–2717. doi: 10.3321/j.issn:1001-5302.2009.21.005
Chen, L. Y., Chen, J. M., Gituru, R. W., Temam, T. D., Wang, Q. F. (2012). Generic phylogeny and historical biogeography of Alismataceae, inferred from multiple DNA sequences. Mol. Phylogenet. Evol. 63, 407–416. doi: 10.1016/j.ympev.2012.01.016
Chen, L. Y., Lu, B., Morales-Briones, D. F., Moody, M. L., Liu, F., Hu, G. W., et al. (2022). Phylogenomic analyses of Alismatales shed light into adaptations to aquatic environments. Mol. Biol. Evol. 39, msac079. doi: 10.1093/molbev/msac079
Chinese Pharmacopoeia Commission (2020). Pharmacopoeia of the People’s Republic of China (Beijing: China Medical Science Press).
Dellaporta, S. L., Wood, J., Hicks, J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/BF02712670
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5, 8348. doi: 10.1038/srep08348
Ferreira de Carvalho, J., Lucas, J., Deniot, G., Falentin, C., Filangi, O., Gilet, M., et al. (2019). Cytonuclear interactions remain stable during allopolyploid evolution despite repeated whole-genome duplications in Brassica. Plant J. 98, 434–447. doi: 10.1111/tpj.14228
Givnish, T. J., Zuluaga, A., Spalink, D., Soto Gomez, M., Lam, V. K., Saarela, J. M., et al. (2018). Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Amer. J. Bot. 105, 1888–1910. doi: 10.1002/ajb2.1178
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nuc. Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Han, C. W., Kwun, M. J., Kim, K. H., Choi, J. Y., Oh, S. R., Ahn, K. S., et al. (2013). Ethanol extract of Alismatis Rhizoma reduces acute lung inflammation by suppressing NF-κB and activating Nrf2. J. Ethnopharm. 146, 402–410. doi: 10.1016/j.jep.2013.01.010
Hendricks, A. J. (1957). A revision of the genus alisma (Dill.) L. Amer. Midl. Nat. 58, 470–493. doi: 10.2307/2422629
Hu, H. S., Mao, J. Y., Wang, X., Liang, Y. Z., Jiang, B., Zhang, D. Q. (2023). Plastid phylogenomics and species discrimination in the “Chinese” clade of Roscoea (Zingiberaceae). Plant Divers. 45, 523–534. doi: 10.1016/j.pld.2023.03.012
Ito, Y., Tanaka, N. (2023). Phylogeny of Alisma (Alismataceae) revisited: implications for polyploid evolution and species delimitation. J. Plant Res. 136, 613–629. doi: 10.1007/s10265-023-01477-1
Jacobson, A., Hedrén, M. (2007). Phylogenetic relationships in Alisma (Alismataceae) based on RAPDs, and sequence data from ITS and trn L. Plant Syst. Evol. 265, 27–44. doi: 10.1007/s00606-006-0514-x
Jang, E., Lee, J. H. (2021). Promising Anticancer Activities of Alismatis rhizome and Its Triterpenes via p38 and PI3K/Akt/mTOR Signaling Pathways. Nutrients 13, 2455. doi: 10.3390/nu13072455
Jang, M. K., Han, Y. R., Nam, J. S., Han, C. W., Kim, B. J., Jeong, H. S., et al. (2015). Protective effects of Alisma orientale extract against hepatic steatosis via inhibition of endoplasmic reticulum stress. Int. J. Mol. Sci. 16, 26151–26165. doi: 10.3390/ijms161125944
Japanese Pharmacopoeia Commission (2021). The Japanese Pharmacopoeia (Tokyo: Japan Ministry of Health and Welfare Press).
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Gen. Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Johnson, G., Canty, S. W. J., Lichter-Marck, I. H., Wagner, W., Wen, J. (2023). Ethanol preservation and pretreatments facilitate quality DNA extractions in recalcitrant plant species. Appl. Plant Sci. 11, e11519. doi: 10.1002/aps3.11519
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Meth. 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinf. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucl. Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Kwon, M. J., Kim, J. N., Park, J., Kim, Y. T., Lee, M. J., Kim, B. J. (2021). Alisma canaliculatum extract affects AGS gastric cancer cells by inducing apoptosis. Int. J. Med. Sci. 18, 2155–2161. doi: 10.7150/ijms.55212
Lehtonen, S. (2017). Splitting caldesia in favour of albidella (Alismataceae). Aust. Syst. Bot. 30, 64–69. doi: 10.1071/SB16050
Les, D. H., Tippery, N. P. (2013). “In time and with water … the systematics of alismatid monocotyledons,” in Early events in monocot evolution, vol. 83 . Eds. Wilkin, P., Mayo, S. J. (Cambridge Univ. Press, Cambridge), 118–164. The Systematics Association Special. doi: 10.1016/j.jep.2013.01.010
Li, H., Guo, Q., Xu, L., Gao, H., Liu, L., Zhou, X. (2023). CPJSdraw: analysis and visualization of junction sites of chloroplast genomes. PeerJ 11, e15326. doi: 10.7717/peerj.15326
Li, Z. Z., Lehtonen, S., Martins, K., Wang, Q. F., Chen, J. M. (2022). Complete genus-level plastid phylogenomics of Alismataceae with revisited historical biogeography. Mol. Phylogenet. Evol. 166, 107334. doi: 10.1016/j.ympev.2021.107334
Liang, J., Ma, Q., Yang, Z. (2019). The first complete chloroplast genomes of two Alismataceae species, and the phylogenetic relationship under order Alismatales. Mitochondr. DNA B Resour. 4, 122–123. doi: 10.1080/23802359.2018.1536486
Liu, B. B., Hong, D. Y., Zhou, S. L., Xu, C., Dong, W. P., Johnson, G., et al. (2019). Phylogenomic analyses of the Photinia complex support the recognition of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Rosaceae). J. Syst. Evol. 57, 678–694. doi: 10.1111/jse.12542
Liu, C., Shi, L., Zhu, Y., Chen, H., Zhang, J., Lin, X., et al. (2012). CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 13, 1–7. doi: 10.1186/1471-2164-13-715
Liu, J., Nie, Z. L., Ren, C., Su, C., Wen, J. (2023). Phylogenomics of Aralia sect. Aralia (Araliaceae): Signals of hybridization and insights into its species delimitations and intercontinental biogeography. Mol. Phylogenet. Evol. 181, 107727. doi: 10.1016/j.ympev.2023.107727
Liu, S. S., Wei, S. H., Zhu, J. J., Zhang, W. W., Jiao, S., Guo, J., et al. (2020). Herbal textural research, morphologic characteristics and DNA barcoding of botanical origins of Alismatis Rhizoma. China J. Chin. Materia Medica. 45, 1536–1544. doi: 10.19540/j.cnki.cjcmm.20200318.201
Liu, Z. L., Xie, L. Z., Zhu, J., Li, G. Q., Grant, S. J., Liu, J. P. (2013). Herbal medicines for fatty liver diseases. Cochrane Database Syst. Rev. 8, CD009059. doi: 10.1002/14651858.CD009059.pub2
Luo, Y., Ma, P. F., Li, H. T., Yang, J. B., Wang, H., Li, D. Z. (2016). Plastid phylogenomic analyses resolve tofieldiaceae as the root of the early diverging monocot order alismatales. Genome Biol. Evol. 8, 932–945. doi: 10.1093/gbe/evv260
Lv, S. Y., Ye, X. Y., Li, Z. H., Ma, P. F., Li, D. Z. (2022). Testing complete plastomes and nuclear ribosomal DNA sequences for species identification in a taxonomically difficult bamboo genus. Fargesia. Plant Divers. 45, 147–155. doi: 10.1016/j.pld.2022.04.002
Ma, X. C., Yao, H., Xin, T. Y., Chen, X. C., Song, J. Y. (2015). SNP identification of Alisma orientale, Alisma plantago-aquatica and Alismatis Rhizoma in the market based on DNA barcoding. Chin. Pharmaceut. J. 50, 1474–1478. doi: 10.11669/cpj.2015.17.003
Ma, Z. Y., Nie, Z. L., Ren, C., Liu, X. Q., Zimmer, E. A., Wen, J. (2021). Phylogenomic relationships and character evolution of the grape family (Vitaceae). Mol. Phylogenet. Evol. 154, 106948. doi: 10.1016/j.ympev.2020.106948
Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., et al. (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047. doi: 10.1093/bioinformatics/16.11.1046
Mendez-Reneau, J., Gordon Burleigh, J., Sigel, E. M. (2023). Target capture methods offer insight into the evolution of rapidly diverged taxa and resolve allopolyploid homeologs in the fern genus Polypodium ss. Syst. Bot. 48, 96–109. doi: 10.1600/036364423X16758873924135
Mwanzia, V. M., Nzei, J. M., Yan, D. Y., Kamau, P. W., Chen, J. M., Li, Z. Z. (2019). The complete chloroplast genomes of two species in threatened monocot genus Caldesia in China. Genetica 147, 381–390. doi: 10.1007/s10709-019-00079-x
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Raza, A., Saher, M. S., Farwa, A., Ahmad, K. R. S. (2019). Genetic diversity analysis of Brassica species using PCR-based SSR markers. Gesunde Pflanzen. 71, 1–7. doi: 10.1007/s10343-018-0435-y
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ross, T. G., Barrett, C. F., Soto Gomez, M., Lam, V. K. Y., Henriquez, C. L., Les, D. H., et al. (2016). Plastid phylogenomics and molecular evolution of Alismatales. Cladistics 32, 160–178. doi: 10.1111/cla.12133
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Samuelsson, G. (1934). Die Verbreitung der höheren Wasserpflanzen in Nordeuropa. Acta Phytogeogr. Suec. 6, 1–200.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Su, T., Geng, Y. F., Xiang, C. L., Zhao, F., Wang, M., Gu, L., et al. (2022). Chloroplast genome of salvia sect. Drymosphace: comparative and phylogenetic analysis. Divers 14, 324. doi: 10.3390/D14050324
Talavera, A., Nie, Z. L., Ma, Z. Y., Johnson, G., Ickert-Bond, S. M., Zimmer, E. A., et al. (2023). Phylogenomic analyses using a new 1013-gene Vitaceae bait-set support major groups of North American Vitis. Mol. Phylogenet. Evol. 186, 107866. doi: 10.1016/j.ympev.2023.107866
Tang, H., Tang, L., Shao, S., Peng, Y., Li, L., Luo, Y. (2021). Chloroplast genomic diversity in Bulbophyllum section Macrocaulia (Orchidaceae, Epidendroideae, Malaxideae): Insights into species divergence and adaptive evolution. Plant Divers. 43, 350–361. doi: 10.1016/j.pld.2021.01.003
Wang, Q. F., Haynes, R. R., Hellquist, C. B. (2010). “Alismataceae,” in Flora of China, vol. 23 . Eds. Wu, C. Y., Hong, D. Y., Raven, P. H. (Missouri Botanical Garden Press, St. Louis & Science Press, Beijing), 84–89.
Wang, R. J., Cheng, C. L., Chang, C. C., Wu, C. L., Su, T. M., Chaw, S. M. (2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 8, 1–14. doi: 10.1186/1471-2148-8-36
Wang, S. L., Li, Y. J. (2002). Studies on standard operating procedure (SOP) for Alisma plantago-aquatica. Chin. Trad. Herb. Drugs 33, 64–67. doi: 10.3321/j.issn:0253-2670.2002.04.032
Wang, Y. H., Wicke, S., Wang, H., Jin, J. J., Chen, S. Y., Zhang, S. D., et al. (2018). Plastid genome evolution in the early-diverging legume subfamily cercidoideae (Fabaceae). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00138
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wu, S. S., Chen, J. Y., Huang, Z. H., Fan, S. M. (2008). “Distinguishing Alisma orientale and Alisma plantago-aquatica from the point of view of plant taxonomy and molecular biology,” in The 8th National Symposium on Natural Medicinal Resources Proceedings and Abstracts, Vol. 5. 99–103.
Xu, Y. L., Shen, H. H., Du, X. Y., Lu, L. (2022). Plastome characteristics and species identification of Chinese medicinal wintergreens (Gaultheria, Ericaceae). Plant Divers. 44, 519–529. doi: 10.1016/j.pld.2022.06.002
Yang, Z., Liu, L. (2019). The complete chloroplast genome of Butomus umbellatus L. and its phylogenetic position. Mitochondr. DNA B Resour. 4, 3700–3701. doi: 10.1080/23802359.2019.1678435
Ye, W. Q., Yap, Z. Y., Li, P., Comes, H. P., Qiu, Y. X. (2018). Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae). Mol. Phylogenet. Evol. 127, 978–987. doi: 10.1016/j.ympev.2018.07.001
Zhang, M. L., Chen, J. K. (1994). Quantitative and taxonomic studies on Alisma domestica–Annotation on the phenological relationships among Alisma species in the world. Plant Sci. J. 12, 18–24.
Zhang, N., Ramachandran, P., Wen, J., Duke, J. A., Metzman, H., McLaughlin, W., et al. (2017a). Development of a reference standard library of chloroplast genome sequences, genomeTrakrCP. Planta Med. 83, 1420–1430. doi: 10.1055/s-0043-113449
Zhang, N., Wen, J., Zimmer, E. A. (2015). Congruent deep relationships in the grape family (Vitaceae) based on sequences of chloroplast genomes and mitochondrial genes via genome skimming. PloS One 10, e0144701. doi: 10.1371/journal.pone.0144701
Zhang, N. N., Xin, T. Y., Jin, Y. (2016). A study on the identification of zeolite seeds based on the DNA bar coding system of Chinese herbal medicine. Modern. Tradit. Chin. Med. Materia Medica-World Sci. Technol. 18, 18–23. doi: 10.11842/wst.2016.01.003
Zhang, X., Zhou, T., Kanwal, N., Zhao, Y., Bai, G., Zhao, G. (2017b). Completion of eight gynostemma BL. (Cucurbitaceae) chloroplast genomes: characterization, comparative analysis, and phylogenetic relationships. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01583
Zhao, F., Drew, B. T., Chen, Y. P., Hu, G. X., Li, B., Zhang, C. L. (2020). The chloroplast genome of Salvia: genomic characterization and phylogenetic analysis. Int. J. Plant Sci. 181, 812–830. doi: 10.1371/journal.pone.0196473
Zhao, J. L., Zhong, J., Fan, Y. L., Xia, Y. M., Li, Q. J. (2017). A preliminary species-level phylogeny of the alpine ginger Roscoea: Implications for speciation. J. Syst. Evol. 55, 215–224. doi: 10.1111/jse.12247
Keywords: Alisma, phylogenomics, plastid genome, structural variation, medicinal plant
Citation: Lan Z-Q, Zheng W, Talavera A, Nie Z-L, Liu J, Johnson G, Yin X-M, Zhao W-Q, Zhao Z-Y, Handy SM and Wen J (2024) Comparative and phylogenetic analyses of plastid genomes of the medicinally important genus Alisma (Alismataceae). Front. Plant Sci. 15:1415253. doi: 10.3389/fpls.2024.1415253
Received: 10 April 2024; Accepted: 31 July 2024;
Published: 20 August 2024.
Edited by:
Linchun Shi, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Jing Li, Integrated DNA Technologies (United States), United StatesZhechen Qi, Zhejiang Sci-Tech University, China
Copyright © 2024 Lan, Zheng, Talavera, Nie, Liu, Johnson, Yin, Zhao, Zhao, Handy and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Qiong Lan, bGFubGFuMTk3OTUxMkAxMjYuY29t; Jun Wen, d2VuakBzaS5lZHU=
†These authors have contributed equally to this work and share first authorship
 Zhi-Qiong Lan
Zhi-Qiong Lan Wen Zheng1†
Wen Zheng1† Alicia Talavera
Alicia Talavera Ze-Long Nie
Ze-Long Nie Gabriel Johnson
Gabriel Johnson Sara M. Handy
Sara M. Handy Jun Wen
Jun Wen