- 1Department of Grassland Science, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
- 2Guangdong Engineering Research Center for Grassland Science, Guangzhou, China
- 3College of Food Science and Engineering, Foshan University, Foshan, China
- 4Institute for Agricultural Biosciences, Oklahoma State University, Ardmore, OK, United States
- 5College of biology and chemistry, Minzu Normal University of Xingyi, Xingyi, China
Introduction: Members of the plant-specific B3 transcription factor superfamily play crucial roles in various plant growth and developmental processes. Despite numerous valuable studies on B3 genes in other species, little is known about the B3 superfamily in pearl millet.
Methods and results: Here, through comparative genomic analysis, we identified 70 B3 proteins in pearl millet and categorized them into four subfamilies based on phylogenetic affiliations: ARF, RAV, LAV, and REM. We also mapped the chromosomal locations of these proteins and analyzed their gene structures, conserved motifs, and gene duplication events, providing new insights into their potential functional interactions. Using transcriptomic sequencing and real-time quantitative PCR, we determined that most PgB3 genes exhibit upregulated expression under drought and high-temperature stresses, indicating their involvement in stress response regulation. To delve deeper into the abiotic stress roles of the B3 family, we focused on a specific gene within the RAV subfamily, PgRAV-04, cloning it and overexpressing it in tobacco. PgRAV-04 overexpression led to increased drought sensitivity in the transgenic plants due to decreased proline levels and peroxidase activity.
Discussion: This study not only adds to the existing body of knowledge on the B3 family’s characteristics but also advances our functional understanding of the PgB3 genes in pearl millet, reinforcing the significance of these factors in stress adaptation mechanisms.
Introduction
Pearl millet (Pennisetum glaucum), an annual herb native to Africa that grows mainly in arid and semi-arid tropical regions, is the world’s sixth-largest cereal crop (Sun et al., 2020). It has excellent tolerance to environmental stresses, especially high-temperature and drought stresses, and can be grown in areas where other crops cannot survive; it is thus considered an ideal crop for studying plant resilience and has attracted attention since its genome was sequenced (Swaminathan et al., 2008; Varshney et al., 2017; Dudhate et al., 2018; Chanwala et al., 2020; Huang et al., 2021). Transcription factors (TFs) are key regulators that play important roles in the endogenous defense systems of plants in a variety of stress responses. Therefore, the analysis of gene changes in pearl millet under abiotic stress provides a practical basis for cereal crop research for the benefit of modern agriculture.
The B3 family is a class of DNA-binding TFs with a highly conserved domain unique to plants and plays a key role in many aspects of plant development. It is named after its B3 DNA-binding domain; the conserved B3 domain includes approximately 110 amino acid residues for DNA recognition, consisting of seven β-barrels and two short α-helices (Kagaya et al., 1999; Swaminathan et al., 2008). The B3 superfamily is divided into four families: ARF (auxin response factor), LAV (leaf cotyledon2-abscisic acid insensitive3-VAL), RAV (related to ABI3/VP1), and REM (reproductive meristem) (Swaminathan et al., 2008; Sasnauskas et al., 2018). In addition, B3 TFs play important roles in plant adaptation and survival under various abiotic and biotic stresses (Verma and Bhatia, 2019; Ren et al., 2023). Since its discovery, the B3 family has been characterized in many model plants and crops including Arabidopsis, rice, poplar, rapeseed, castor, soybean, tobacco, and grape (Ahmad et al., 2019; Xia et al., 2019; Liu et al., 2020; Wang et al., 2022a; Wei et al., 2023) but not yet in pearl millet.
As a member of the B3 family, RAVs play important roles in developing plants and defense against abiotic stresses. Ectopic expression of soybean GmRAV3 in Arabidopsis can improve the resistance of transgenic lines to high salt and drought conditions and lead to the insensitivity of transgenic plants to exogenous abscisic acid (Zhao et al., 2017). Melon CmRAV1 can be induced by NaCl treatment, and ectopic overexpression of CmRAV1 in Arabidopsis enhances salt tolerance during seed germination and growth (Zhao et al., 2019). According to recent research, ABA and NaCl treatments affect cucumber RAV1, while ectopic expression of CsRAV1 in Arabidopsis enhances its tolerance to NaCl and ABA (Li et al., 2023). Investigations into the function of the wheat TaRAV1 gene revealed that overexpressing TaRAV1 improved the ability of Arabidopsis to tolerate salt (Luo et al., 2022). RAV TFs were also associated with anthocyanin synthesis, further influencing abiotic stress in plants, as demonstrated in strawberries and pears (Zhang et al., 2020b; Liu et al., 2021b).
Pearl millet exhibits excellent tolerance to environmental stresses, particularly to high temperature and drought stress. Among the many regulatory factors in plant growth and development, members of the B3 family play an extremely important role and exhibit unique gene functions among different species. Therefore, research on the B3 family genes in pearl millet can not only deepen the understanding of B3 genes functions but also fill existing knowledge gaps, providing a theoretical basis and practical methods for future research on plant tolerance. We utilized bioinformatics methods to investigate the physicochemical properties, structural characteristics, phylogenetic evolution, chromosomal localization, and gene duplication of the B3 family in pearl millet. We identified a total of 70 B3 family genes that were classified into four subfamilies (ARF, REM, RAV, and LAV) by phylogenetic analysis. These 70 PgB3 genes are distributed across seven chromosomes and were observed to have undergone purifying selection during the evolutionary process. In addition, transcriptomic data and real-time fluorescence quantification analyses revealed that most genes in the B3 family responded to drought and high-temperature stresses. Among these genes, PgRAV-04 expression decreased under drought treatment but increased briefly under high temperature treatment. Therefore, we cloned PgRAV-04 and ectopically overexpressed it in Nicotiana benthamiana to study the function of this gene. Overexpression of the PgRAV-04 gene in tobacco led to an increase in malondialdehyde (MDA) content and decreases in proline content, relative leaf water content, and antioxidant enzyme activity, thereby increasing the plant’s sensitivity to drought. In conclusion, PgRAV-04 acts as a negative regulatory factor in response to drought stress.
Materials and methods
Database research and sequence retrieval
Pearl millet genome and protein sequences were downloaded from the GigaDB database (http://gigadb.org/dataset/100192); the pearl millet protein data set was built using the TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). In addition, the hidden Markov model (HMM) profile of the B3 domain (PF02362) was downloaded from the Pfam database (Pfam; http://pfam.xfam.org/) (Finn et al., 2014) to be utilized for the identification of B3 genes from the local database (E-value < 0.001) using HMMER. All hits were confirmed by Pfam (PF02362) and NCBI conserved domain searches (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/). The SMART database (http://smart.embl-heidelberg.de/smart) and ExPASy-PROSITE database (http://www.expasy.org/prosite) were used to further confirm the presence of the B3 domain in all members of the B3 superfamily. The confirmed B3 protein sequences were aligned using Clustal X (v 2.0 http://www.clustal.org/clustal2/) to remove redundant sequences, then sequences with a confidence level > 97% and an alignment length > 50% were retained. The molecular weights and theoretical isoelectric points of all candidate B3 proteins were calculated using the ExPASy program (https://web.expasy.org/protparam/). The B3 protein sequences of Arabidopsis were downloaded from The Arabidopsis Information Resource (https://www.arabidopsis.org/); rice B3 protein sequences were retrieved from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/).
Phylogenetic tree, motif, and gene structure analyses
A total of 70 B3 protein sequences from pearl millet were aligned using Clustal X (v2.0, http://www.clustal.org/clustal2/). Two neighbor-joining phylogenetic trees were constructed and visualized using the MEGA-X software with 1000 bootstrap replications: one for pearl millet and another for pearl millet, rice, and Arabidopsis. The conserved motifs of the B3 proteins were identified using MEME v5.0.5 (http://meme-suite.org) (Bailey et al., 2009) with the following parameters: the maximum number of motifs was set to 10 for the prediction of subdomains; the motif width was set to between 6 and 100. Motif analysis was carried out using the TBtools visualize domain pattern. The exon–intron substructure map of each B3 gene was generated according to the Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015).
Chromosome location and gene duplication events
The chromosome location images of PgB3 genes were generated using the MapInspect software (http://mapinspect.apponic.com/) (Ma et al., 2013) according to the available information from the genome database. The MCScanX (http://chibba.pgml.uga.edu/mcscan2/) software was used to estimate gene duplication event types in the pearl millet B3 superfamily genes. Tandem gene duplication in pearl millet was defined as paralogous genes physically linked in tandem with less than five genes and/or genes located within 100 kb of each other that belonged to the same family (Zhang et al., 2020a; Liu et al., 2021a). Segmental duplications were those placed on replicated chromosomal blocks from the same genome lineage that isolated more than five genes. Tandem duplications and segmental duplications were visualized using TBtools software. The synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) of duplicated genes were obtained using the online tool kaks_calculator2.0 (https://sourceforge.net/projects/kakscalculator2/) (Gao et al., 2015). The Ka/Ks ratio is commonly used to understand the direction of evolution and can reveal three different situations: Ka/Ks > 1 indicates a positive selection, Ka/Ks < 1 indicates a negative selection, and Ka/Ks = 1 indicates a neutral selection (Li et al., 2009; Wang et al., 2009).
Prediction of the PgB3 protein–protein interaction network
An interaction network of PgB3 proteins was built to retrieve all possible interactions based on orthologous Arabidopsis proteins using the STRING 10 software (https://string-db.org/). Preprocessing of the interaction network included removal of redundancy and self-looping interactions, then target proteins with credibility > 0.7 were selected to construct a regulation network via the OmicStudio tools (https://www.omicstudio.cn/tool); the nodes represent the protein/gene, and the lines indicate the interactions between two proteins/genes.
Plant material and stress treatments
In an artificial climate chamber maintained at 30°C/24° (day/night) with a 16h/8h (day/night) photoperiod, the relative humidity of 50%-75%, and then pearl millet seeds were planted in 1-gallon pots containing a mixture of substrate and vermiculite (4:1, v/v); experiments were repeated in triplicate using a random block design. For stress response analysis, 5-week-old seedlings were treated with high temperature of 42°C/35°C (day/night) with a photoperiod of 16/8h (day/night), the relative humidity of 50%-75%, and drought (non-irrigation) for a specified period, followed by a recovery period. Plant leaves were sampled at different time points under drought (0h, 12h, and 48h, then after 2 days of recovery) and high temperature (0h, 12h, and 148h, then after 2 days of recovery) treatments; these samples were used for transcriptome analysis. Under the same stress treatments, we performed real-time fluorescence quantification experiments for both drought and high temperature at 0h, 12h, and 48h, then after 2 days of recovery. All samples were taken from three replicates and immediately stored at −80° for RNA extraction; the leaves sampled at 0h served as controls for the stress treatments.
Expression profiling of PgB3 genes by RNA-seq and qRT-PCR
To determine the expression profiles of the PgB3 genes under drought and high-temperature stresses, samples from the different stress treatments were subjected to transcriptome sequencing. All samples were stored on dry ice and sent to LC-Bio Technologies (Hangzhou, China) for RNA extraction and sequencing. To identify the differentially expressed genes that respond to drought and high temperature stress, we screened them using the criteria |log2FC| > =1 and q<0.05. Mapped reads were used to measure the fragments per kilobase of transcript per million mapped reads (FPKM) values; heat maps were generated using log2 Z-score values in the OmicStudio tools (https://www.omicstudio.cn/tool) and clustering in heat maps was based on correlation.
For qRT-PCR analyses, total RNA was extracted using TRIzol reagent (TianGen, Beijing, China) and first-strand cDNA was synthesized with a reverse transcription kit (Transgen, Beijing, China). Six gene-specific primer pairs were designed using the Primer Premier5.0 software (Premier Biosoft Intl, California, USA) (Supplementary Table S1). qRT-PCR was performed on the LightCycler 480 Real-Time PCR System using the PerfectStart Green qPCR Supermix reaction kit (Transgen, Beijing, China): 10 µL reactions contained 5 µL 2× PerfectStart Green qPCR Supermix, 0.2 µL each of forward and reverse primer (10.0 µmol/L), 0.5 µL cDNA (5-fold diluted), and 4.1 µL RNase-free water. qRT-PCR amplification was carried out as follows: 94° for 30s, followed by 40 cycles of 94° for 5s and 60° for 30s. The pearl millet EF-1ɑ gene (Shivhare and Lata, 2016) was used as the reference gene to normalize the qRT-PCR data; relative gene expression levels were determined via the 2−ΔΔCT method. Each experiment was performed using three biological replicates.
Gene cloning and transformation of PgRAV-04 into tobacco
The PgRAV-04 gene, involved in the response to drought and high-temperature stresses, was selected to confirm gene functions. PgRAV-04 transgenic lines were generated by cloning the coding sequence of PgRAV-04 into the pUbi-intron-OE vector (preserved in this laboratory) between the Spe I and BamH I restriction sites using the primers listed in Supplementary Table S1; the expression vector Ubi: PgRAV-04 was transformed into Agrobacterium tumefaciens (EHA105), followed by N. benthamiana transformation using the leaf disc method.
Molecular confirmation of transgenic tobacco plants
Genomic DNA was isolated from the leaves of putative transgenic and wild type plants. The presence of PgRAV-04 in the transgenic plants was confirmed by PCR analysis using gene-specific primers (Supplementary Table S1); WT plants were used as negative controls, and plasmids containing fragments of interest were used as positive controls. The amplified products were electrophoresed on a 1% (w/v) agarose gel. PCR-positive plants were further confirmed by qRT-PCR. RNA was isolated and cDNA was synthesized as described above. The tobacco Actin gene was selected as a reference gene for expression in transgenic tobacco (Supplementary Table S1).
Physiological and biochemical indices under stress treatments
For stress response analysis of transgenic tobacco, transgenic plants were planted in 1-gallon pots containing a mixture of substrate and vermiculite (4:1, v/v). The plants were grown in a growth chamber under 16h light (100 μmol m−2 s−1) and 8h dark; humidity was maintained at 60-70% and the temperature was controlled at 28°C/25°C (day/night). Five-week-old PgRAV-04 transgenic tobacco plants were used for drought analysis, it is worth noting that the WT (wild type) material did not undergo transfer of the target gene, nonetheless, the seedlings used in the experiment were obtained through the same tissue culture process that was employed for the transgenic tobacco. In this study, we assessed the drought tolerance of three transgenic tobacco lines compared to the WT plants. Physical indices were determined on days 0d, 9d, and 16d of drought treatment and after 3 days of rehydration; each sample had three biological replicates. Leaf relative water content (RWC), malondialdehyde content (MDA), proline content, superoxide dismutase activity (SOD), peroxidase activity (POD), ascorbate peroxidase activity (APX), and catalase activity (CAT) were determined following previously described protocols (Zhong et al., 2014).
Date statistical analysis
The SPSS software was utilized for significance analysis. Real-time fluorescent quantitative analysis utilized Duncan’s multiple range test while physiological indicators were assessed using independent sample t-tests, with significance levels set at p < 0.05 and p < 0.01 to evaluate the differences among various treatments. The data are presented as mean values with the standard error of the mean for three replicates.
Results
Identification and analysis of pearl millet B3 family genes
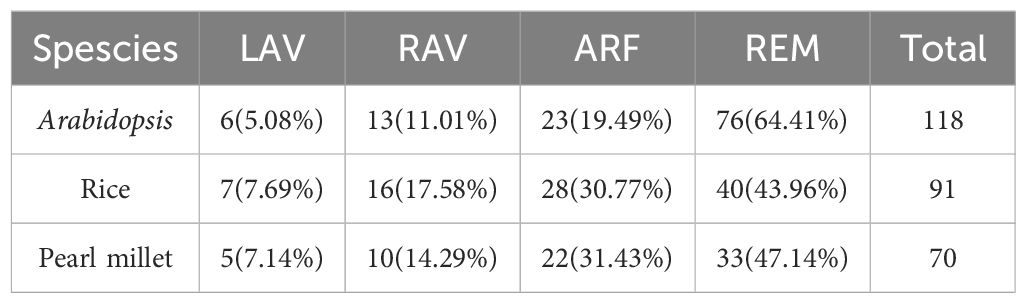
After searching the pearl millet genome, a total of 70 B3 family genes containing the plant-specific B3 domain were identified. Based on other domains in the protein sequences, the PgB3 genes were classified into four subgroups: LAV (5 genes), RAV (10), ARF (22), and REM (33) (Table 1). The gene lengths ranged from 475 bp (PgRAV_03) to 3183 bp (PgARF_17). The amino acid lengths of encoded proteins ranged from 154 to 1039 aa, among which three proteins (PgARF_14, PgARF_17 and PgARF_18) were >1000 aa in length and 32 proteins were >500 aa in length; molecular weights ranged from 17.374 kDa to 140.283 kDa and isoelectric points were between 4.95 and 10.14. In addition, 20 gene members contained more than 10 exons while 4 gene members (PgRAV _ 02, PgRAV _ 03, PgRAV _ 04, and PgRAV _ 06) had only one exon (Supplementary Table S2).
Phylogenetic and structural analyses of pearl millet B3 family genes
To examine the phylogenetic relationships of the B3 gene family in pearl millet and to assist in their classification, we built a neighbor-joining phylogenetic tree. According to the clade support values, the B3 genes were divided into four distinct subfamilies: LAV, RAV, ARF, and REM (Figure 1). The REM subfamily had the most PgB3 genes (33), followed by ARF (22), while the LAV subfamily had the fewest PgB3 genes. In addition, another phylogenetic tree was constructed with the 70 PgB3 genes as well as B3 family genes from other species to confirm the phylogenetic relationships throughout the evolutionary process (Supplementary Figure S1); most results were similar using both approaches except that the rice RAV (Os07g17230) gene was clustered into the REM subfamily. Furthermore, according to the constructed phylogenetic trees and a review of the literature, we were able to pinpoint functionally annotated B3s with their PgB3 homologs (Supplementary Table S3); in all, 34 PgB3 homologous genes were predicted, indicating that the pearl millet B3 superfamily genes were highly conserved during the course of evolution.
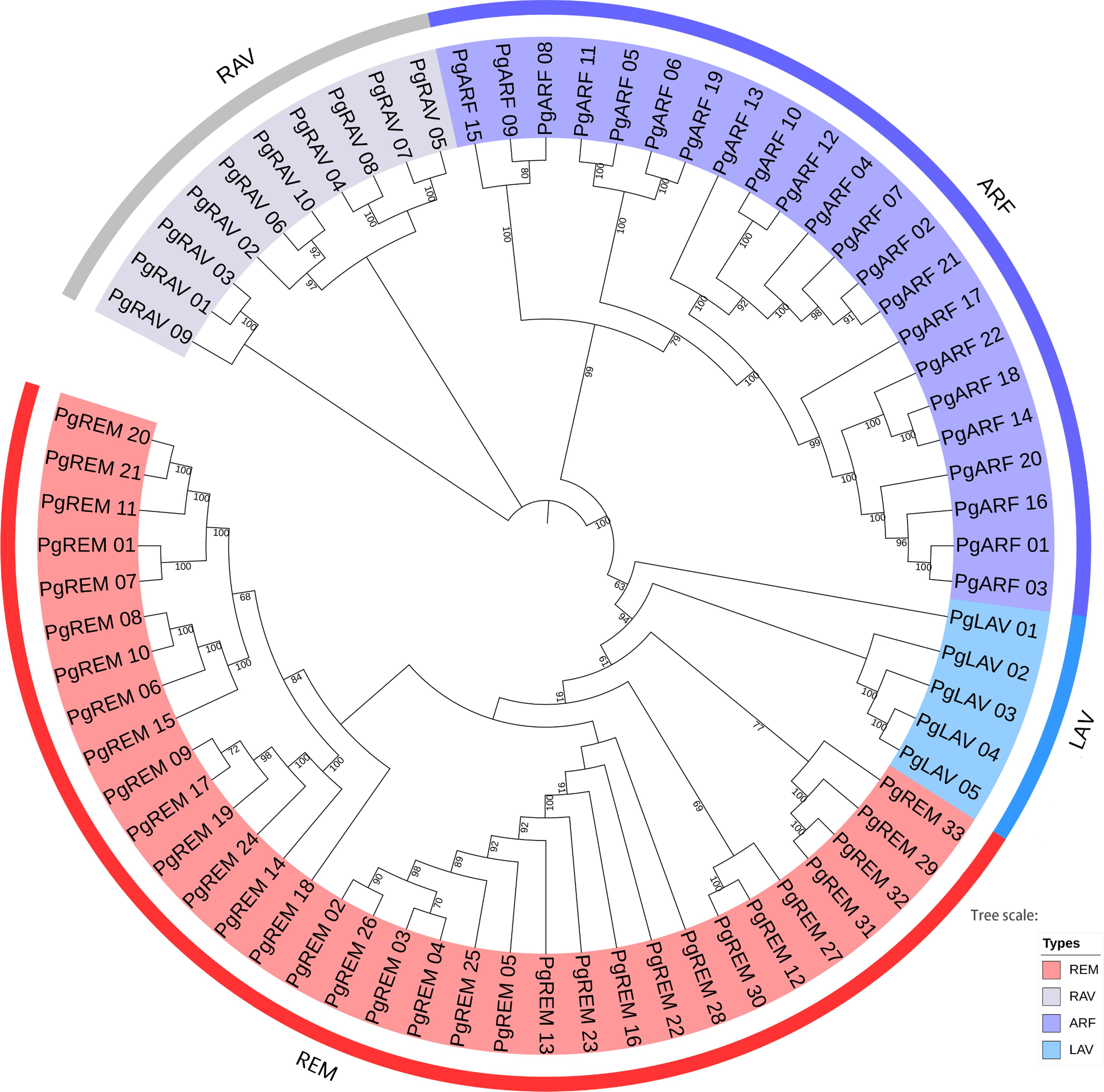
Figure 1 Phylogenetic tree of pearl millet B3 family genes. The neighbor-joining tree is divided into four groups (REM, LAV, ARF, and RAV). The different colored areas indicate different groups of the B3 superfamily.
Gene structure and conserved motif analyses of pearl millet B3 family genes
To understand the structural diversity of the pearl millet B3 family genes, we analyzed the organization of the exons and introns in the coding regions. The number of introns in PgB3 genes ranged from 0 to 14 (Figure 2A), with ranges of 3-14, 5-10, 1-10, and 0-2 in the ARF, LAV, REM, and RAV groups, respectively. In general, most genes within each B3 subfamily had similar intron numbers, structures, and lengths, supporting the subgroup classification in the phylogenetic tree.
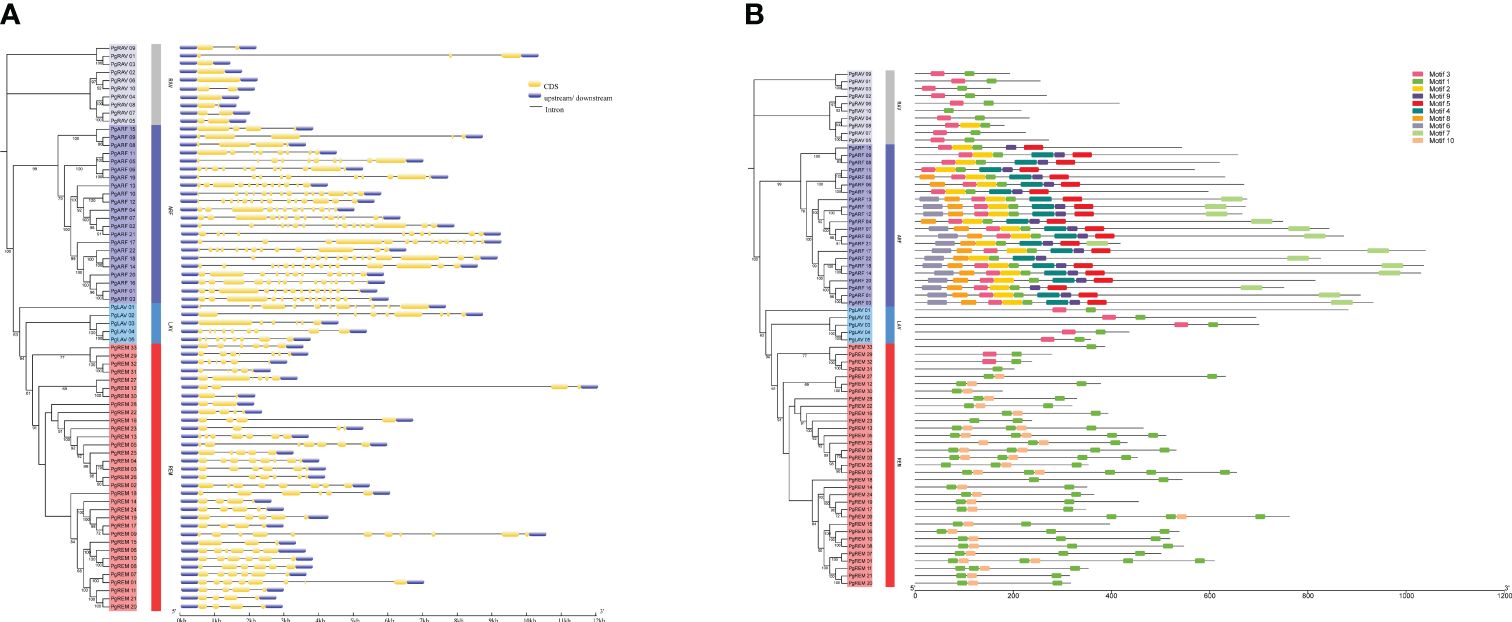
Figure 2 Gene structure and conserved motif analyses of PgB3s. (A) Gene structural analysis was performed according to the phylogenetic tree. The exons, introns, and untranslated regions are represented by yellow boxes, black lines, and blue boxes, respectively. (B) Conserved motif analysis. Each motif is represented by a number of colored boxes; the length of the box indicates the motif length.
We also observed that the distribution of conserved motifs differed among subfamilies. Ten conserved motifs were identified in the PgB3 proteins (Figure 2B): motif 1 was the B3 DNA-binding domain, motifs 4 and 9 corresponded to auxin response factor domains, motif 7 was the Aux/IAA domain (Table 2), and the remaining six were not annotated. Conserved motif 1 was found in almost all members of the pearl millet B3 family; motifs 4 and 9 were found in all members of the ARF subfamily; motif 10 was only present in the REM subfamily; and the RAV, ARF, and LAV subgroups all had motif 3. These results indicate that possession of specific motifs may be associated with gene differentiation and diversity.
Chromosomal location and duplication events of pearl millet B3 family genes
Chromosomal distribution analysis revealed that 69 of the 70 PgB3 genes were randomly distributed across the chromosomes (Figure 3), with the most on Chr3 (15) and the least on Chr7 (2); chromosomes 1, 2, 4, 5, and 6 contained 8, 12, 13, 10, and 9 members, respectively.
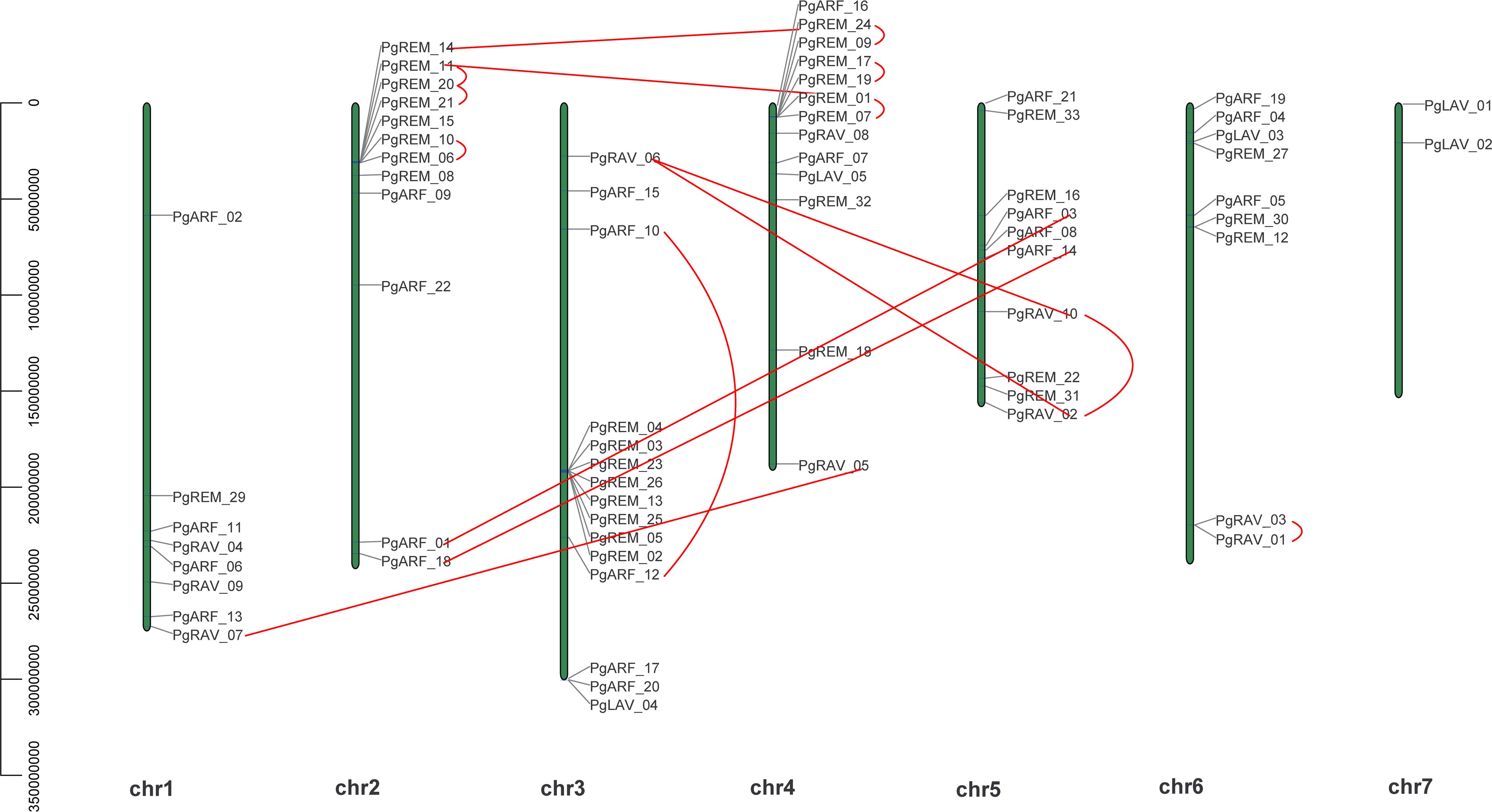
Figure 3 Chromosomal locations of PgB3 genes. The position and full length of PgB3 genes are indicated by the numbers on the left-hand side of the chromosome; the chromosome number is indicated below the chromosome. The segmental duplication is connected by the straight red line, while tandem duplications are connected by curved red lines.
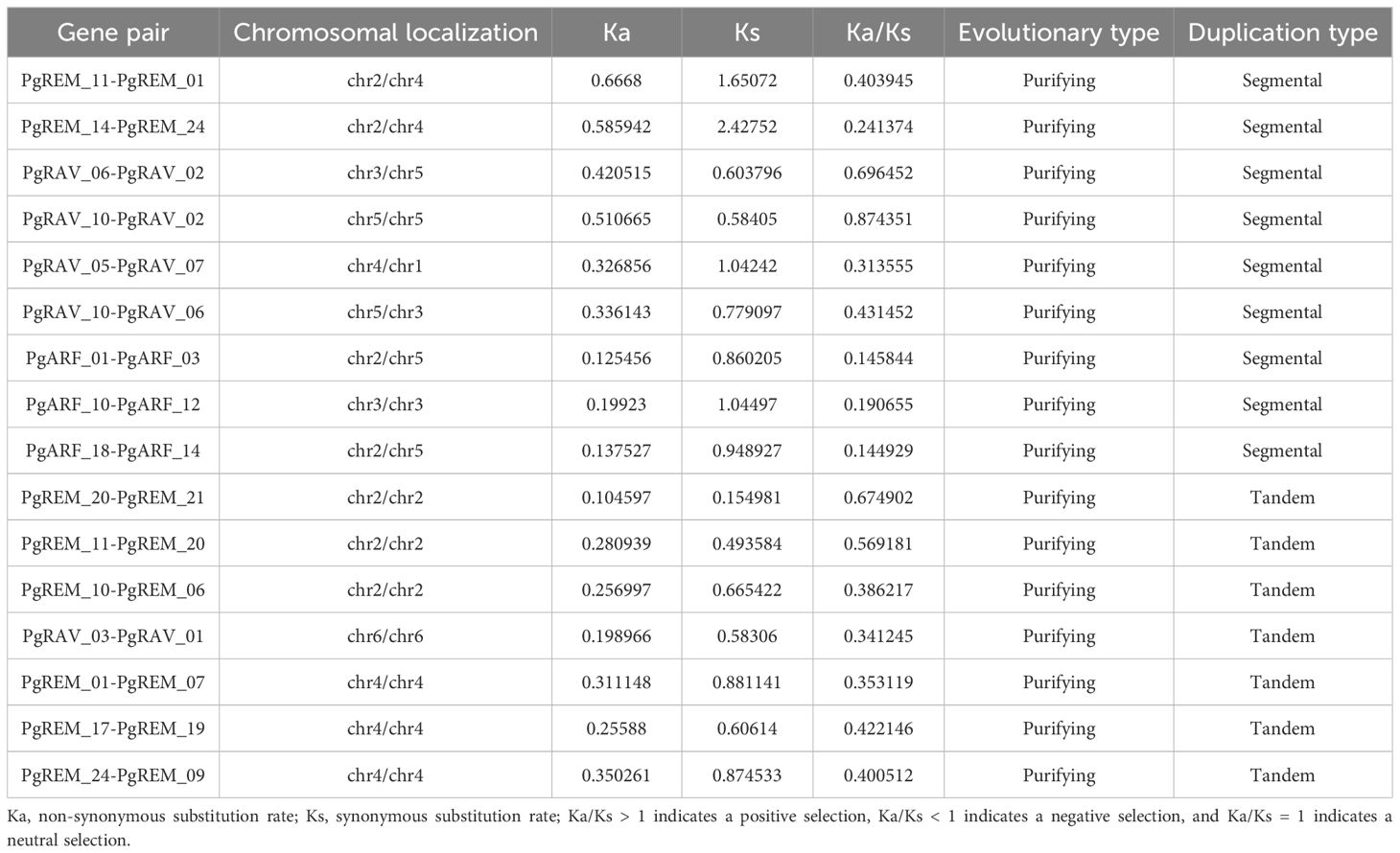
Gene duplications including tandem duplication (in which the duplication segment is contiguous with the original duplication), segmental duplication (the copying of whole blocks of genes from one chromosome to another chromosome) (Leister, 2004), and whole-genome duplication (characterized by an organism’s entire genetic information being copied one or more times) (Lee et al., 2020) may play important roles not only in the process of genome rearrangement and expansion but also in the process of diversification of gene functions and generation of gene families. In this study, a survey of gene duplication analyses indicated that nine gene pairs (17 genes) formed segmental duplication events and seven gene pairs (13 genes) were involved in tandem duplications (Figure 3, Table 3), the number of segmental and tandem duplications was similar, suggesting that both types of gene duplication promote amplification of PgB3 family genes.
Purifying selection preserves the innate adaptability of species, whereas diversifying selection supports evolutionary innovation (beneficial mutations) and species differentiation (Yan et al., 2017). To understand the evolutionary trends of these PgB3 genes, detailed information on Ka, Ks, and Ka/Ks is presented in Table 3. The Ka/Ks values of all tandemly repeated PgB3 sequences were > 1, implying that they were all targets of purifying selection during the evolutionary process.
Analysis of the PgB3 protein interaction network
A network of integrated proteins was built using Arabidopsis homologs to further investigate the relationships between PgB3 proteins (Supplementary Table S4). Among the PgB3 proteins, a total of 25 interacting proteins were identified with high confidence (Figure 4); when combined with the gene annotation results (Supplementary Table S3), a total of 18 of the interacting proteins had predicted functions. The homolog of PgARF_05/06/11/19 is AT2G33860.1, which had the highest number of proteins with direct interactions in the interaction network (six proteins), followed by PgLAV_03/04 and PgARF_14/18/22 interacting with five proteins. Notably, many proteins are involved in stress and disease resistance, for example: PgARF_01/03/06, PgARF_02, PgARF_07, PgARF_14/16, PgARF_14/20, and PgARF_19. In addition, PgARF_17 and PgARF_14/18 were associated with the auxin signaling pathway.
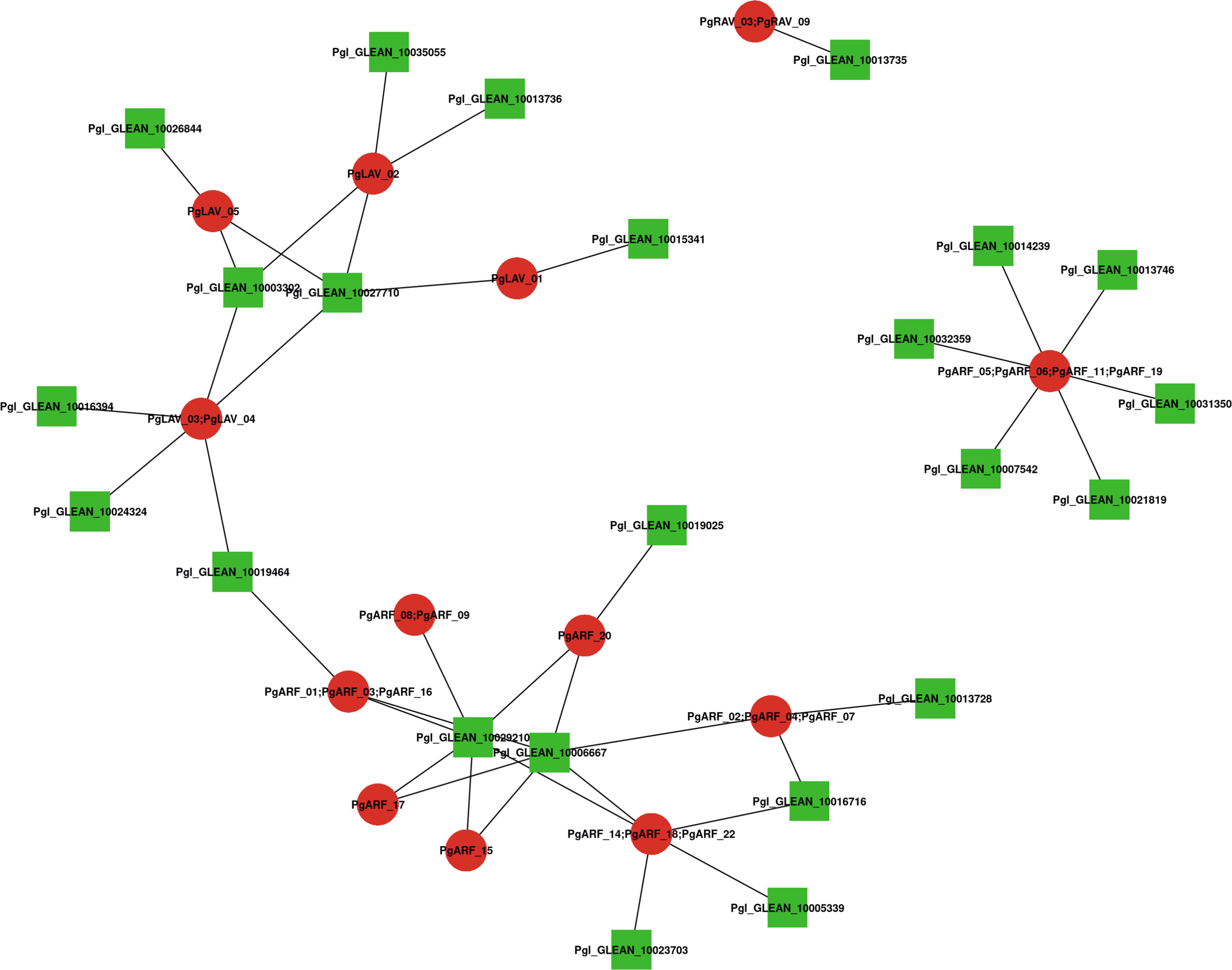
Figure 4 Interaction network analysis of B3 proteins identified in pearl millet according to their orthologs in Arabidopsis. Nodes represent proteins and lines represent interactions between two proteins; red represents PgB3 family proteins, green represents other proteins in pearl millet.
Expression profiles of PgB3 genes under drought and high-temperature stresses
Because the B3 family plays a crucial role in how plants respond to abiotic stresses, we used RNA-seq data from pearl millet under high-temperature and drought treatments to further our understanding of the biological processes involving the PgB3 genes (Figure 5). This analysis demonstrated that 54 PgB3 genes responded to drought stress (Figure 5A; Supplementary Table S5) and 58 PgB3 genes responded to high temperature (Figure 5B; Supplementary Table S6). Several genes were upregulated under both drought and high-temperature stresses, such as PgARF_21, PgARF_05, PgARF_20, PgRAV_10, and PgREM_14. Interestingly, PgRAV_04, PgLAV_02, and PgARF_08 were downregulated by drought stress but upregulated by high-temperature stress. These results suggest that while many PgB3s have roles in drought and high-temperature stress responses, they may be functionally distinct.
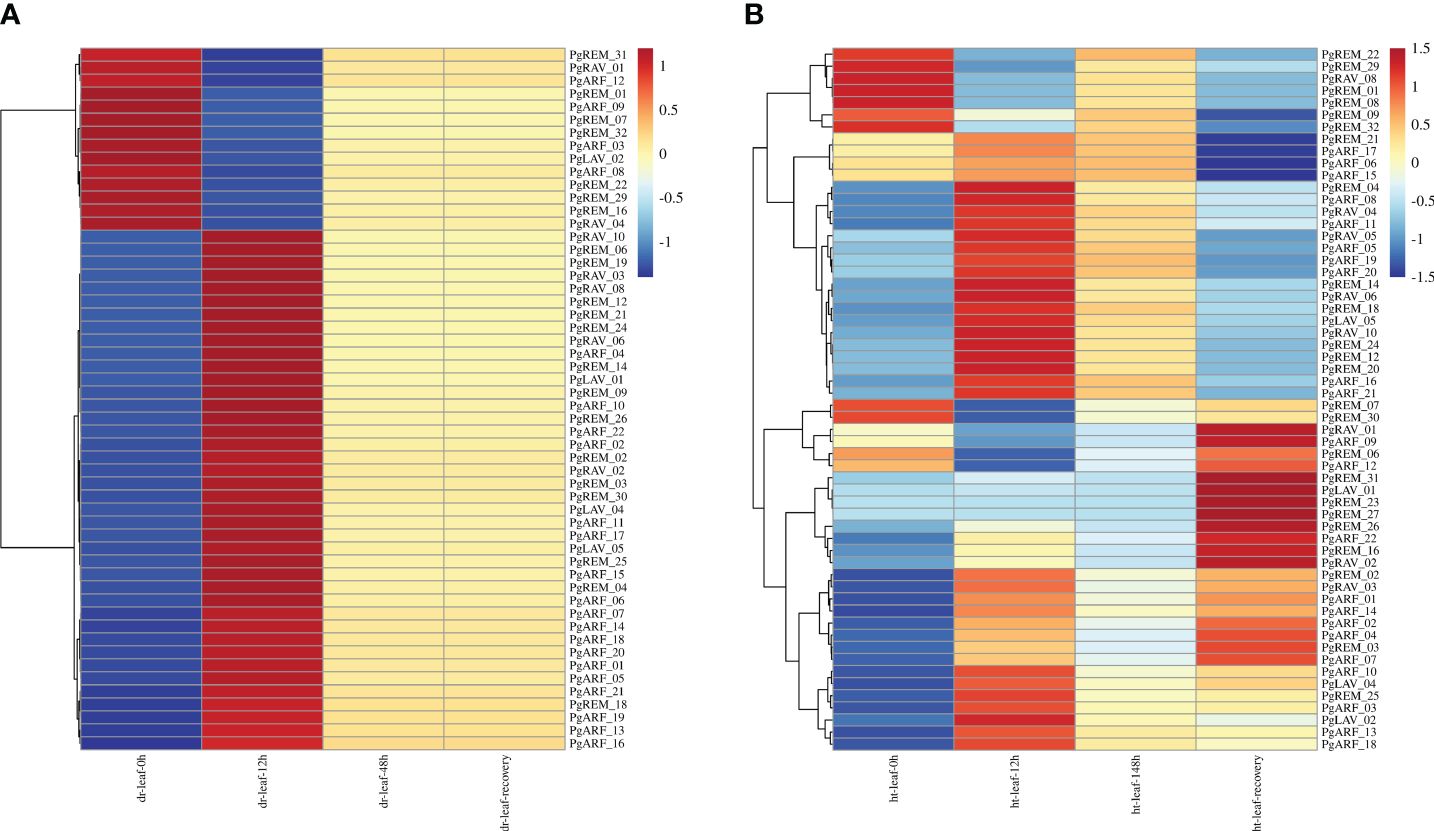
Figure 5 Expression levels of PgB3 gene transcripts in pearl millet in response to abiotic stresses. (A) Drought stress at different treatment times (0h, 12h, 48h and after 2 days of recovery). (B) High-temperature stress at different treatment times (0h, 12h, 148h and after 2 days of recovery). The color scale represents increased (red) and decreased (blue) fold-changes in the levels of PgB3 transcripts at different treatment times when exposed to abiotic stress (dr: drought; ht: high temperature) compared to the respective levels in the controls for each stress. The fold-change is shown on a log2 scale from high to low expression of each PgB3 transcript.
To validate the RNA-seq data under drought and high-temperature stress treatments (Figure 5), we characterized the expression of six PgB3 genes using qRT-PCR. We found that all six genes responded to drought stress: while four genes (PgARF_05, PgARF_19, PgLAV_04, and Pg LAV_05) were upregulated in the early stages of drought, PgRAV-04 and PgRAV-06 were significantly downregulated (Figure 6A). PgLAV_04 and PgLAV_05 were downregulated under high-temperature stress, while PgARF_05, PgARF_19, PgRAV-04, and PgRAV-06 were upregulated (Figure 6B). Notably, the expression levels of PgRAV_04 and PgRAV_06 were significantly low at 8h and 12h of drought treatment but were higher at 8h and 12h of high-temperature treatment. By contrast, expression levels of PgLAV_04 and PgLAV_05 were upregulated under drought stress and downregulated under high-temperature stress; however, PgARF_05 and PgARF_19 were upregulated under both drought and high-temperature stresses (Figures 5, 6). These results support the expression profiles of the transcriptome and demonstrate the validity of the RNA-seq data. To confirm the role of PgRAV_04 in abiotic stress responses, we used the PgRAV_04 OE transgenic line for further analyses.
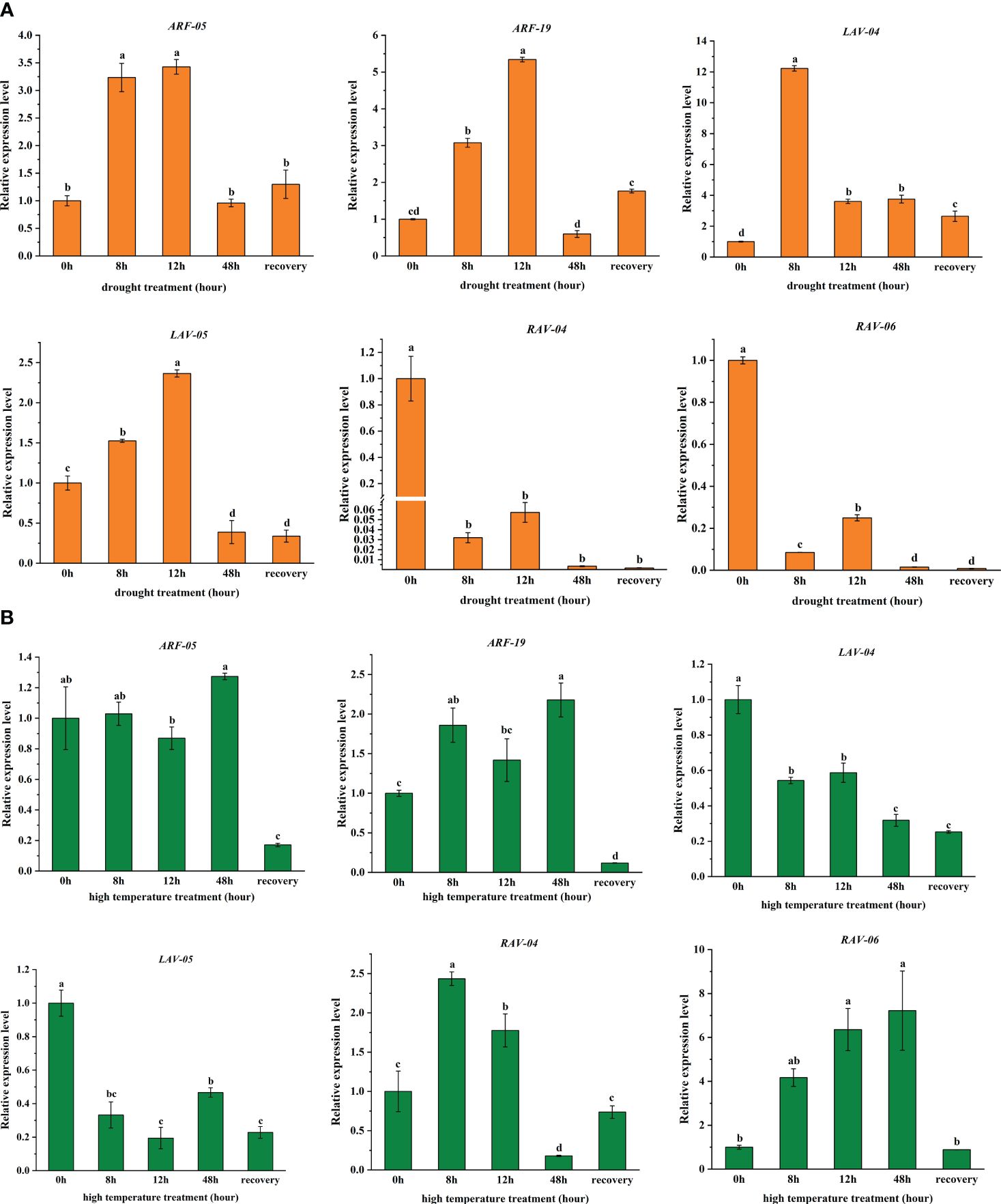
Figure 6 Expression analysis of six selected PgB3 genes based on transcriptome data relating to (A) Drought and (B) High-temperature stresses through qRT-PCR. Relative expression of the PgB3 genes was normalized relative to the reference gene PgEF-1ɑ under different stresses. Significant differences were determined according to Duncan’s multiple tests; data are means (± SDs) of triplicates; bars with different letters were significantly different (P < 0.05).
Overexpression of the PgRAV-04 gene resulted in drought-sensitive transgenic tobacco
We confirmed the successful transformation of PgRAV-04 in transgenic plants by PCR and qRT-PCR (Supplementary Figures S2, S3). To investigate the function of the PgRAV_04 gene in plant growth and development under drought stress, we subjected 5-week-old transgenic and WT plants to drought treatment (Figure 7). The transgenic plants showed severe wilting symptoms under severe stress (16 days of drought treatment) compared to WT plants (Figure 7A). After 16 days without irrigation, the MDA content in the three overexpression lines were higher than in the WT plants. Among them, the MDA content of OE2 is particularly notable, being 1.51 times that of the WT plants (Figure 7B; Supplementary Table S7). The relative water content of the three transgenic lines were generally lower than the WT plants at all stages of the drought treatment (Figure 7C; Supplementary Table S7). The proline content of the three transgenic tobaccos were significantly lower than the WT at 0 day, suggesting that our transgenic materials were inherently intolerant to osmotic stress; meanwhile, the proline content of the three transgenic plants were similarly lower than in the WT plants as drought increased (Figure 7D; Supplementary Table S7), further indicating that the PgRAV-04 overexpression plants were sensitive to drought. In summary, the transgenic plants had higher MDA levels, lower RWC and proline levels, and a more pronounced wilt phenotype, suggesting that the transgenic plants were more sensitive to drought.
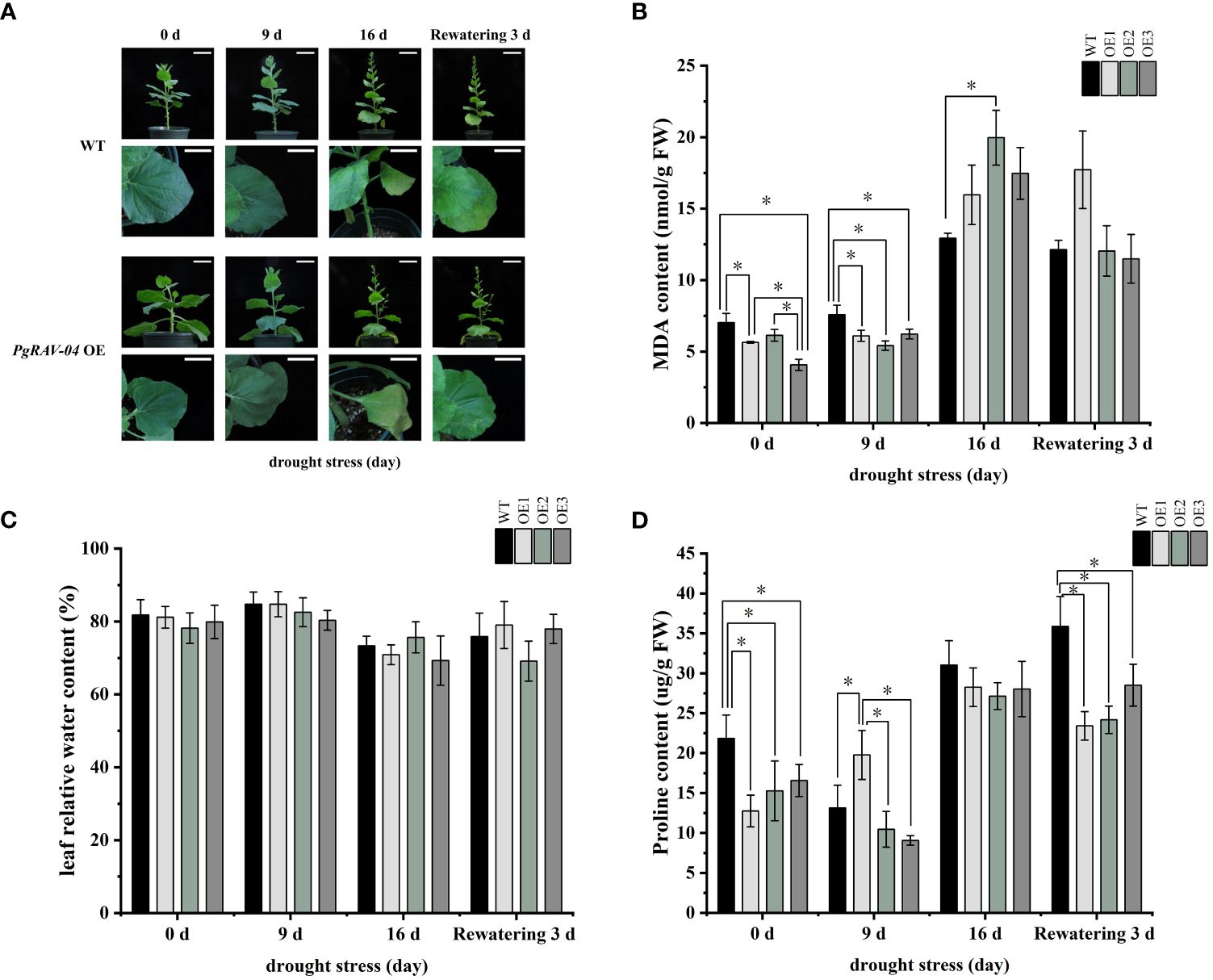
Figure 7 Effect of PgRAV-04 overexpression (OEs) on tobacco drought tolerance during different periods of drought treatment. (A) Phenotypes of wild-type (WT) and the three PgRAV-04 OE lines during drought treatment and after 3 days of recovery. (B) Malondialdehyde content (MDA), (C) Leaf relative water content (RWC), and (D) Proline content in WT and the three PgRAV-04 OE lines during drought stress and after 3 days of recovery. Data of the three transgenic lines are presented as means (± SDs) of triplicates, while the WT is represented by the average value of 9 repetitions. Significant differences between WT and transgenic lines were determined using independent samples t-tests. *P < 0.05.
Because the MDA content was higher in the transgenic plants, which would imply membrane lipid peroxidation, we measured peroxidase activity, which quenches toxic free radicals. APX activities were initially higher in the three transgenic plants than in the WT plants but were significantly lower in the three transgenic plants compared to the WT after 16 days of drought (Figure 8A; Supplementary Table S7). At the same time, after 16 days of drought treatment, CAT, SOD, and POD activities were lower in the transgenic plants than in the WT plants (Figures 8B–D; Supplementary Table S7). In a word, these antioxidant enzyme activity results further suggest that the transgenic plants were more sensitive to drought. Altogether, we conclude that PgRVA-04 strongly responds to drought but acts as a negative regulator under drought stress.
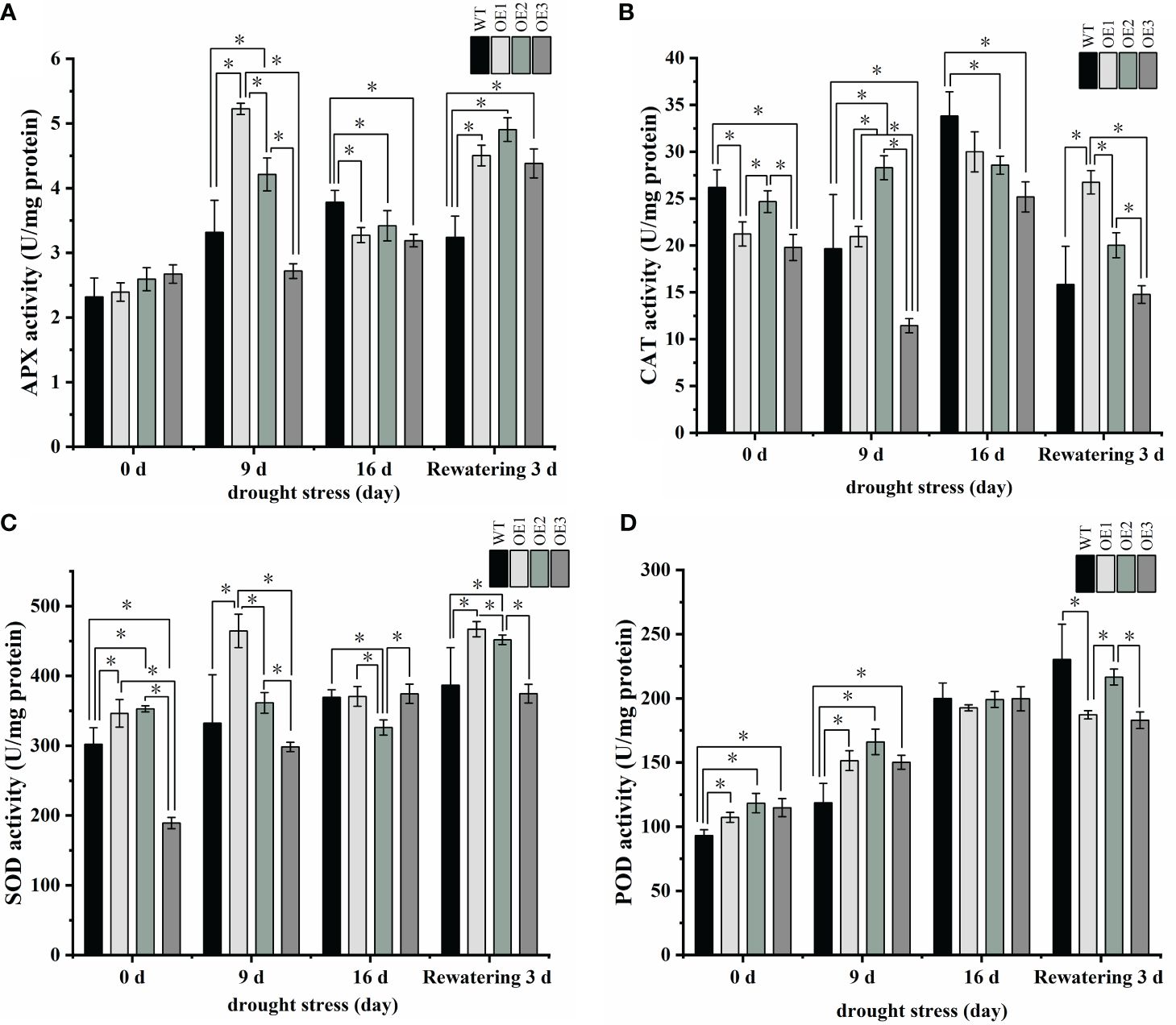
Figure 8 Antioxidant enzyme activity in WT and the three PgRAV-04 OE lines. (A) Ascorbate peroxidase (APX), (B) Catalase (CAT), (C) Superoxide dismutase (SOD), and (D) Peroxidase (POD) activities in the leaves of WT and the three PgRAV-04 OE lines under drought stress and after 3 days of recovery. Data of the three transgenic lines are presented as means (± SDs) of triplicates, while the WT is represented by the average value of 9 repetitions. Significant differences between WT and transgenic lines were determined using independent samples t-tests. *P < 0.05.
Discussion
Evolutionary diversity in the pearl millet B3 superfamily
The B3 superfamily has been extensively studied in various plants due to its diverse roles in vegetative and reproductive development. However, this study is the first report of a genome-wide analysis of the B3 superfamily in pearl millet, identifying a total of 70 B3 superfamily genes. This number is lower than those found in Arabidopsis (118), rice (91) (Swaminathan et al., 2008), and soybean (143) (Ren et al., 2023), suggesting that different B3 family genes have undergone different gene duplication events. Based on phylogenetic analyses, the 70 PgB3 genes were categorized into four subfamilies (Figure 1; Supplementary Table S2), consistent with the classification of the B3 superfamily in citrus (Liu et al., 2020), chickpea (Verma and Bhatia, 2019), tobacco (Xia et al., 2019), and Medicago truncatula (Gao et al., 2023). Structure and motif analyses revealed similar gene structures and motifs within each B3 gene subfamily (Figure 2), supporting the reliability and accuracy of the subfamily classification. Gene families are founded by a single ancestor; through duplications and deletions, the size of the family fluctuates over time, with the possibility of the family disappearing from the genome (Cannon et al., 2004). Analysis of gene duplication events and the Ka/Ks ratios of duplicated pairs (Figure 3, Table 3) indicated that the evolutionary type of all PgB3 gene pairs was purifying selection, which further explains why pearl millet has fewer B3 family genes than other species.
PgB3s play an important role in the stress response
Previous studies have identified important roles for B3 superfamily members in plant responses to a range of biotic and abiotic stresses. For example, oil palm ARF subfamily genes play an important role in abiotic stress (Jin et al., 2022); the wheat REM gene responds to cold stress (Badawi et al., 2019); overexpression of TaRAV1 improved salt tolerance in Arabidopsis and upregulated the expression of SOS genes and other stress response genes (Luo et al., 2022). However, in this study, we found that 54 genes of the pearl millet B3 family were responsive to drought; the majority of these genes were upregulated under drought stress but a few such as PgARF-03, PgREM-16, PgLAV-02, and PgRAV-04 were downregulated (Figures 5A, 6A), suggesting their potential roles in the plant drought stress response. Similarly, 58 genes also responded to high-temperature stress, especially after 12h of heat treatment; many genes such as PgLAV-02, PgRAV-04, and PgREM-20 were upregulated, indicating their involvement in regulating the plant’s response to heat stress (Figures 5B, 6B). Taken together with previous findings, the variability in the functions of B3 superfamily genes under abiotic stresses is critical to plant stress responses.
PgRAV-04 is a negative regulator of drought stress in transgenic tobacco
Drought is a major environmental stressor that constrains plant growth and development. Plants primarily adopt two strategies to cope with drought stress: drought avoidance and enhancement of drought resistance (Basu et al., 2016). In this study, we systematically investigated the function of PgRAV-04 in response to drought stress. Compared to the WT, the transgenic tobacco plants had a reduced ability to cope with drought stress. This conclusion was supported by the positive MDA content, the decreases in proline content and RWC, as well as the observed wilting in the leaves of transgenic plants after 16 days of water deficit. Plants defend against drought by synthesizing and accumulating osmoprotectants (including glycine, betaine, myo-inositol, and proline) to stabilize cellular substructures and maintain internal equilibrium under drought stress (Zulfiqar et al., 2020; Wang et al., 2022b). Our findings corroborate this understanding: the PgRAV-04 overexpression plants had significantly lower proline levels than the WT (Figure 7D), even at 0h of drought, suggesting that the transgenic tobacco had poor drought resistance. As the drought stress intensified, the proline content remained significantly lower than the WT at 16h of drought treatment (Figure 7D) and the RWC was also lower (Figure 7C). At the same time, we observed a significant increase in MDA after 16h of drought treatment (Figure 7B), indicating severe cell damage under drought stress. Therefore, based on these observations, plants overexpressing PgRAV-04 were more sensitive to drought.
In addition to combating drought through the accumulation of osmolytes, plants can also mitigate stress-induced harm through the activity of peroxidases (Minibayeva et al., 2015; Kidwai et al., 2020; Nadarajah, 2020). In this study, the activities of APX and CAT in plants overexpressing PgRAV-04 were significantly lower than in the WT after 16 days of drought (Figures 8A, B); similarly, in plants overexpressing PgRAV-04, the activities of SOD and POD were also lower than those in the WT under severe stress conditions (Figures 8C, D). This further suggests that under severe drought stress, the ability of peroxidases to rescue plants is compromised. It has been reported that when stress levels increase to the point where the accumulation of reactive oxygen species exceeds the threshold of the protective enzyme system, the antioxidant capacity of plants gradually decreases (Hussain et al., 2020); this was well corroborated in our study.
Drought is a pervasive and extreme environmental challenge that poses a significant threat to plant survival and development (Chaves et al., 2003). Under conditions of water shortage, plants undergo a series of physiological and biochemical changes that not only affect their growth rate and yield (Gupta et al., 2020) but can also lead to metabolic imbalances and cellular structural damage (Joshi et al., 2016; Hussain et al., 2018). To maintain hydration and cellular homeostasis, plants initiate complex coping mechanisms that include regulating the opening and closing of stomata to reduce water loss (Basu et al., 2016), accumulating osmotic adjustment substances such as proline to maintain cell sap concentration (Hayat et al., 2012; Ghosh et al., 2022), enhancing root depth and density to improve water absorption efficiency (Vadez et al., 2013), and neutralizing reactive oxygen species produced due to water deficiency by bolstering the antioxidant defense system (Shao et al., 2007; Zhang et al., 2017; Hasanuzzaman et al., 2020; Nadarajah, 2020). In our study, overexpression of PgRAV-04 not only diminished the drought resistance of transgenic plants through a reduction in osmolytes (proline content) (Figure 7D) but also decreased peroxidase activity (Figure 8). Among the plethora of mechanisms for stress response in plants, these two pathways in our study both contributed to the reduced drought tolerance of the transgenic plants. Therefore, we conclude that PgRAV-04 is a negative regulator in the drought stress response.
Conclusion
This is the first comprehensive analysis of the pearl millet B3 superfamily. We identified a total of 70 PgB3 genes, categorized into four subfamilies. Detailed analyses were conducted on their phylogenetic relationships, exon-intron structures, chromosomal localization, gene duplication events, protein interaction network, and expression patterns under conditions of drought and high-temperature stresses. Moreover, we cloned a member of the RAV subfamily, PgRAV-04 from pearl millet, for ectopic overexpression in tobacco and evaluated its function during drought stress. The overexpression of PgRAV-04 increased the MDA content while decreasing proline levels and peroxidase activity, thereby enhancing the sensitivity of the transgenic plants to drought conditions. This research sets the stage for further exploration of the functional roles of the PgB3 genes in pearl millet.
Data availability statement
The data presented in this study were deposited in the NCBI repository, accession number PP239383. Further inquiries can be directed to the corresponding author.
Author contributions
Y-HW: Formal analysis, Methodology, Writing – original draft, Validation. XY: Formal analysis, Methodology, Validation, Writing – original draft. B-YZ: Data curation, Investigation, Software, Writing – review & editing. W-JW: Data curation, Investigation, Software, Writing – review & editing. Z-FZ: Data curation, Investigation, Software, Writing – review & editing. X-QZ: Data curation, Formal analysis, Writing – review & editing. JD: Data curation, Formal analysis, Writing – review & editing. J-LS: Data curation, Formal analysis, Writing – review & editing. X-LH: Data curation, Writing – review & editing. K-XO: Data curation, Writing – review & editing. T-XZ: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. F-XL: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Project No.31802116, No. 32071823).
Conflict of interest
The reviewer KS declared a shared affiliation with the authors Y-HW, XY, B-YZ, W-JW, Z-FZ, X-LH, K-XO, T-XZ and F-XL to the handling editor at the time of review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1400301/full#supplementary-material
References
Ahmad, B., Zhang, S. L., Yao, J., Rahman, M. U., Hanif, M., Zhu, Y. X., et al. (2019). Genomic organization of the B3-Domain transcription factor family in grapevine (Vitis vinifera L.) and expression during seed development in seedless and seeded cultivars. Int. J. Mol. Sci. 20, 4453. doi: 10.3390/ijms20184553
Badawi, M. A., Agharbaoui, Z., Zayed, M., Li, Q., Byrns, B., Zou, J. T., et al. (2019). Genome-wide identification and characterization of the wheat remorin (TaREM) family during cold acclimation. Plant Genome 12 (2), 180040. doi: 10.3835/plantgenome2018.06.0040
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Basu, S., Ramegowda, V., Kumar, A., Pereira, A. (2016). Plant adaptation to drought stress. F1000Res 5, 1554. doi: 10.12688/f1000research.7678.1
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chanwala, J., Satpati, S., Dixit, A., Parida, A., Giri, M. K., Dey, N. (2020). Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genomics 21, 231. doi: 10.1186/s12864-020-6622-0
Chaves, M. M., Maroco, J. P., Pereira, J. S. (2003). Understanding plant responses to drought - from genes to the whole plant. Funct. Plant Biol. 30, 239–264. doi: 10.1071/FP02076
Dudhate, A., Shinde, H., Tsugama, D., Liu, S. K., Takano, T. (2018). Transcriptomic analysis reveals the differentially expressed genes and pathways involved in drought tolerance in pearl millet [Pennisetum glaucum (L.) R. Br]. PloS One 13, 14. doi: 10.1371/journal.pone.0195908
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Gao, Y. N., Gao, S. H., Xiong, C., Yu, G., Chang, J., Ye, Z. B., et al. (2015). Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 96, 141–153. doi: 10.1016/j.plaphy.2015.07.025
Gao, J., Ma, G., Chen, J., Gichovi, B., Cao, L., Liu, Z., et al. (2023). The B3 gene family in medicago truncatula: genome-wide identification and the response to salt stress. Plant Physiol. Biochem. 206, 108260. doi: 10.1016/j.plaphy.2023.108260
Ghosh, U. K., Islam, M. N., Siddiqui, M. N., Cao, X., Khan, M. A. R. (2022). Proline, a multifaceted signalling molecule in plant responses to abiotic stress: understanding the physiological mechanisms. Plant Biol. 24, 227–239. doi: 10.1111/plb.13363
Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science. 368, 266–269. doi: 10.1126/science.aaz7614
Hasanuzzaman, M., Bhuyan, M., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants. 9, 681. doi: 10.3390/antiox9080681
Hayat, S., Hayat, Q., AlYemeni, M. N., Wani, A. S., Pichtel, J., Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, D. J., Sun, M., Zhang, A. L., Chen, J. S., Zhang, J., Lin, C., et al. (2021). Transcriptional changes in pearl millet leaves under heat stress. Genes. 12, 1716. doi: 10.3390/genes12111716
Hussain, H. A., Hussain, S., Khaliq, A., Ashraf, U., Anjum, S. A., Men, S. N., et al. (2018). Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00393
Hussain, H. A., Men, S. N., Hussain, S., Zhang, Q. W., Ashraf, U., Anjum, S. A., et al. (2020). Maize tolerance against drought and chilling stresses varied with root morphology and antioxidative defense system. Plants. 9, 720. doi: 10.3390/plants9060720
Jin, L. F., Yarra, R., Zhou, L. X., Cao, H. X. (2022). The auxin response factor (ARF) gene family in oil palm (Elaeis guineensis Jacq.): Genome-wide identification and their expression profiling under abiotic stresses. Protoplasma 259, 47–60. doi: 10.1007/s00709-021-01639-9
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01029
Kagaya, Y., Ohmiya, K., Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Kidwai, M., Ahmad, I. Z., Chakrabarty, D. (2020). Class III peroxidase: an indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 39, 1381–1393. doi: 10.1007/s00299-020-02588-y
Lee, S., Choi, S., Jeon, D., Kang, Y., Kim, C. (2020). Evolutionary impact of whole genome duplication in poaceae family. J. Crop Sci. Biotechnol. 23, 413–425. doi: 10.1007/s12892-020-00049-2
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi: 10.1016/j.tig.2004.01.007
Li, J. L., Song, C. Y., Li, H. M., Wang, S. Q., Hu, L. Y., Yin, Y. L., et al. (2023). Comprehensive analysis of cucumber RAV family genes and functional characterization of CsRAV1 in salt and ABA tolerance in cucumber. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1115874
Li, J., Zhang, Z., Vang, S. R., Yu, J., Wong, G. K. S., Wang, J. (2009). Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 68, 414–423. doi: 10.1007/s00239-009-9222-9
Liu, J. L., Deng, Z. W., Liang, C. L., Sun, H. W., Li, D. L., Song, J. K., et al. (2021b). Genome-wide analysis of RAV transcription factors and functional characterization of anthocyanin-biosynthesis-related RAV genes in Pear. Int. J. Mol. Sci. 22, 5567. doi: 10.3390/ijms22115567
Liu, Z., Ge, X. X., Wu, X. M., Xu, Q., Atkinson, R. G., Guo, W. W. (2020). Genome-wide analysis of the citrus B3 superfamily and their association with somatic embryogenesis. BMC Genomics 21, 305. doi: 10.1186/s12864-020-6715-9
Liu, C., Wu, Y. H., Liu, Y. X., Yang, L. Y., Dong, R. S., Jiang, L. Y., et al. (2021a). Genome-wide analysis of tandem duplicated genes and their contribution to stress resistance in pigeonpea (Cajanus cajan). Genomics. 113, 728–735. doi: 10.1016/j.ygeno.2020.10.003
Luo, Y. X., Chen, S. K., Wang, P. D., Peng, D., Zhang, X., Li, H. F., et al. (2022). Genome-wide analysis of the RAV gene family in wheat and functional identification of TaRAV1 in salt stress. Int. J. Mol. Sci. 23, 8834. doi: 10.3390/ijms23168834
Ma, L. C., Wang, Y. R., Liu, W. X., Liu, Z. P. (2013). Expression analysis of seed-specific genes in four angiosperm species with an emphasis on the unconserved expression patterns of homologous genes. Seed Sci. Res. 23, 223–231. doi: 10.1017/S0960258513000305
Minibayeva, F., Beckett, R. P., Kranner, I. (2015). Roles of apoplastic peroxidases in plant response to wounding. Phytochemistry. 112, 122–129. doi: 10.1016/j.phytochem.2014.06.008
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21, 5208. doi: 10.3390/ijms21155208
Ren, C. Y., Wang, H. M., Zhou, Z. H., Jia, J. R., Zhang, Q., Liang, C. Z., et al. (2023). Genome-wide identification of the B3 gene family in soybean and the response to melatonin under cold stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1091907
Sasnauskas, G., Manakova, E., Lapenas, K., Kauneckaite, K., Siksnys, V. (2018). DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. FEBS. J. 285, 4041–4059. doi: 10.1111/febs.14649
Shao, H. B., Chu, L. Y., Lu, Z. H., Kang, C. M. (2007). Primary antioxidant free radical scavenging and redox signaling pathways in higher plant cells. Int. J. Biol.Sci. 4, 8–14. doi: 10.7150/ijbs.4.8
Shivhare, R., Lata, C. (2016). Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6, 23036. doi: 10.1038/srep23036
Sun, M., Huang, D. J., Zhang, A. L., Khan, I., Yan, H. D., Wang, X. S., et al. (2020). Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol. 20, 323. doi: 10.1186/s12870-020-02530-0
Swaminathan, K., Peterson, K., Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 13, 647–655. doi: 10.1016/j.tplants.2008.09.006
Vadez, V., Rao, J. S., Bhatnagar-Mathur, P., Sharma, K. K. (2013). DREB1A promotes root development in deep soil layers and increases water extraction under water stress in groundnut. Plant Biol. 15, 45–52. doi: 10.1111/j.1438-8677.2012.00588.x
Varshney, R. K., Shi, C. C., Thudi, M., Mariac, C., Wallace, J., Qi, P., et al. (2017). Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat.Biotechnol. 35, 969–976. doi: 10.1038/nbt.3943
Verma, S., Bhatia, S. (2019). A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum L.). 3 Biotech. 9, 346. doi: 10.1007/s13205-019-1875-5
Wang, W. B., Ao, T., Zhang, Y. Y., Wu, D., Xu, W., Han, B., et al. (2022a). Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 44, 201–212. doi: 10.1016/j.pld.2021.06.008
Wang, D. P., Wan, H. L., Zhang, S., Yu, J. (2009). Gamma-MYN: a new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct. 4, 20. doi: 10.1186/1745-6150-4-20
Wang, Z. Y., Yang, Y. C., Yadav, V., Zhao, W., He, Y. P., Zhang, X., et al. (2022b). Drought-induced proline is mainly synthesized in leaves and to roots in watermelon under water deficit. Hortic. Plant J. 8, 615–626. doi: 10.1016/j.hpj.2022.06.009
Wei, M. K., Li, H., Wang, Q., Liu, R., Yang, L. X., Li, Q. Z. (2023). Genome-wide identification and expression profiling of B3 transcription factor genes in Populus alba × Populus glandulosa. Front. Plant Sci. 14. doi: 10.3389/fpls.2023
Xia, F., Sun, T. T., Yang, S. J., Wang, X., Chao, J. T., Li, X. X., et al. (2019). Insight into the B3 transcription factor superfamily and expression profiling of B3 genes in axillary buds after topping in tobacco (Nicotiana tabacum L.). Genes. 10, 164. doi: 10.3390/genes10020164
Yan, H. D., Zhang, A. L., Ye, Y. T., Xu, B., Chen, J., He, X. Y., et al. (2017). Genome-wide survey of switchgrass NACs family provides new insights into motif and structure arrangements and reveals stress-related and tissue-specific NACs. Sci. Rep. 7, 3056. doi: 10.1038/s41598-017-03435-z
Zhang, J., Fu, X. X., Li, R. Q., Zhao, X., Liu, Y., Li, M. H., et al. (2020a). The hornwort genome and early land plant evolution. Nat. Plants. 6, 107–118. doi: 10.1038/s41477-019-0588-4
Zhang, Z. Y., Shi, Y. N., Ma, Y. C., Yang, X. F., Yin, X. R., Zhang, Y. Y., et al. (2020b). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhang, G. Q., Zhang, M., Zhao, Z. X., Ren, Y. Q., Li, Q. X., Wang, W. (2017). Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci. Rep. 7, 7549. doi: 10.1038/s41598-017-08181-w
Zhao, S. P., Xu, Z. S., Zheng, W. J., Zhao, W., Wang., Y. X., Yu, T. F., et al. (2017). Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00905
Zhao, L. N., Zhang, F. R., Liu, B., Yang, S. L., Xiong, X., Hassani, D., et al. (2019). CmRAV1 shows differential expression in two melon (Cucumis melo L.) cultivars and enhances salt tolerance in transgenic Arabidopsis plants. Acta Biochim. Biophys. Sin. 51, 1123–1133. doi: 10.1093/abbs/gmz107
Zhong, T., Zhang, L., Sun, S., Zeng, H., Han, L. (2014). Effect of localized reduction of gibberellins in different tobacco organs on drought stress tolerance and recovery. Plant Biotechnol. Rep. 8, 399–408. doi: 10.1007/s11816-014-0330-7
Keywords: B3 superfamily, drought tolerance, gene expression, pearl millet, PgRAV-04
Citation: Wang Y-H, Ye X, Zhao B-Y, Wang W-J, Zhou Z-F, Zhang X-Q, Du J, Song J-L, Huang X-L, Ouyang K-X, Zhong T-X and Liao F-X (2024) Comprehensive analysis of B3 family genes in pearl millet (Pennisetum glaucum) and the negative regulator role of PgRAV-04 in drought tolerance. Front. Plant Sci. 15:1400301. doi: 10.3389/fpls.2024.1400301
Received: 13 March 2024; Accepted: 08 July 2024;
Published: 29 July 2024.
Edited by:
Hamidreza Sharifan, The University of Texas at El Paso, United StatesReviewed by:
Lihu Wang, Hebei University of Engineering, ChinaLida Zhang, Shanghai Jiao Tong University, China
Arash Fazeli, University of Ilam, Iran
Kamran Shah, South China Agricultural University, China
Copyright © 2024 Wang, Ye, Zhao, Wang, Zhou, Zhang, Du, Song, Huang, Ouyang, Zhong and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Xiu Zhong, emhvbmd4aW5iaUAxNjMuY29t; Fei-Xiong Liao, ZnhsaWFvQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
 Yin-Hua Wang
Yin-Hua Wang Xing Ye1,2†
Xing Ye1,2† Wen-Jing Wang
Wen-Jing Wang Juan Du
Juan Du Jian-Ling Song
Jian-Ling Song Kun-Xi Ouyang
Kun-Xi Ouyang Tian-Xiu Zhong
Tian-Xiu Zhong