
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 June 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1397289
This article is part of the Research Topic Unraveling Plant Responses to Cadmium Exposure: From Uptake to Stress Resilience View all articles
 Hewan Zhang1
Hewan Zhang1 Lingli Lu1,2*
Lingli Lu1,2*Cadmium (Cd) is a heavy metal highly toxic to living organisms. Cd pollution of soils has become a serious problem worldwide, posing a severe threat to crop production and human health. When plants are poisoned by Cd, their growth and development are inhibited, chloroplasts are severely damaged, and respiration and photosynthesis are negatively affected. Therefore, elucidating the molecular mechanisms that underlie Cd tolerance in plants is important. Transcription factors can bind to specific plant cis-acting genes. Transcription factors are frequently reported to be involved in various signaling pathways involved in plant growth and development. Their role in the resistance to environmental stress factors, particularly Cd, should not be underestimated. The roles of several transcription factor families in the regulation of plant resistance to Cd stress have been widely demonstrated. In this review, we summarize the mechanisms of five major transcription factor families–WRKY, ERF, MYB, bHLH, and bZIP–in plant resistance to Cd stress to provide useful information for using molecular techniques to solve Cd pollution problems in the future.
Cadmium (Cd),a non-essential toxic heavy metal, occurs naturally and is released into the environment through agricultural and industrial activities (Genchi et al., 2020). Resolving Cd pollution presents significant challenges. Cd infiltrates plant organs and tissues via ectoplasmic and symplastic pathways (Song et al., 2017). Under Cd stress, plant growth and cell division slow, while photosynthesis and transpiration rates varyingly decrease (El Rasafi et al., 2022). Furthermore, Cd accumulated in plants can enter the food chain, affecting animals and humans (Okereafor et al., 2020),increasing risks of cardiovascular diseases, and causing kidney dysfunctions and metabolic disorders (Tinkov et al., 2018; Abougabal et al., 2020; Devereux and Navas-Acien, 2013). Recently, heavy metal accumulation rates in the soil of China have notably exceeded those in other countries. Therefore, urgently addressing this issue is crucial (Yuan et al., 2021). Understanding the Cd accumulation mechanisms in plants will enhance their use in bioremediating Cd-contaminated soils.
Cd is transported via trace element pathways that includes iron, manganese, and zinc. It enters plants through the roots and is present as ions in the cytoplasm, vacuoles, and vascular tissues (Sterckeman and Thomine, 2020). Plant respond to Cd stress through three main mechanisms: (I) chelation, (II) compartmentalization, and (III) plant–nutrient interactions. Firstly, although both metallothionein (MT) and phytochelatin (PC) play roles in reducing Cd toxicity, PC is primarily responsible for mitigating heavy metal toxicity in plants (Grennan, 2011). PC, a small cysteine-rich peptide, bind metals via the sulfhydryl (SH) group. Phytochelatin primarily detoxifies Cd, leveraging the high affinity of Cd2+ for sulfur-containing ligands (Seregin and Kozhevnikova, 2023). Studies indicate that Cd stimulates PC biosynthesis in corn, forming PC-Cd complexes that migrate to root vacuoles, thereby reducing Cd toxicity. Additionally, this mechanism prevents root saturation by transferring excess Cd from roots to shoots (Zare et al., 2023). Secondly, to protect vital organs from stress, most plants sequester Cd in vacuoles. For instance, in barley cells, Cd primarily accumulates in vacuoles, facilitated by the stimulating effects of ATP and GSH in the plant (Thomas and Reid, 2021). Finally, appropriate plant nutrition can mitigate Cd damage. Adequate application of Zn fertilizer to plant foliage effectively prevents Cd poisoning in plants and reduces Cd concentrations in wheat seeds harvested from contaminated soils. The protective mechanism likely involves Zn’s crucial role in synthesizing and activating antioxidant enzymes, which diminishes Cd-induced ROS production (Saifullah et al., 2014). Besides the three main mechanisms, plants employ additional strategies to regulate Cd tolerance.
Plant response to Cd stress are closely associated with oxidatives stress and hormone signaling. Cd disrupts the ability of antioxidants in plant to counter oxidative stress due to its high affinity for oxygen, nitrogen, and sulfur atoms. Exogenous NO application enhances antioxidant enzymes activity, limiting Cd absorption and promoting nutrient accumulation in tomato (Ahmad et al., 2018). Together, melatonin and NO enhance Cd tolerance in wheat (Kaya et al., 2019). The interaction between NO and H2S significantly improves plant resistance to Cd by enhancing the antioxidant defenses and essential nutrient absorption (Kaya et al., 2020). This interference reduces catalase (CAT) and superoxide dismutase (SOD) activities, leading to lipid peroxidation and oxidative proteins accumulation (Khanna et al., 2022). Phytohormones enhance plant resistance to heavy metals by activating regulatory and signal transduction molecules. Cd stress suppresses gene expression related to IAA biosynthesis and transport (Chen et al., 2023). Applying abscisic acid (ABA) and salicylic acid (SA) increases Cd transport from roots to stems (Zhu et al., 2020a). In Duckweed Wolffia arrhiza, ABA levels increase while gibberellic acid (GA) levels decrease following Cd treatment (Chmur et al., 2020).
Studying the molecular mechanisms underlying Cd tolerance in plants is essential for further understanding. OsMTP1,a bivalent cation transporter, efficiently translocates Zn, Cd, and other heavy metals, maintaining plant ion homeostasis (Yuan et al., 2012). StMTP9, significantly responds to Cd stress, substantially reducing Cd content in yeast cells (Li et al., 2021a). Overexpression of HaMTP10, an antitransporter of Cd, reduces Cd accumulation (Li et al., 2023). BcNRAMP1 overexpression enhances Cd transport and accumulation (Yue et al., 2021). Overexpression of PcNRAMP1 in Populus x canescens increased Cd net flux by 39–52% (Yu et al., 2022). Arabidopsis reduces Cd toxicity by downregulating genes (AtIRT1, AtHMA2, AtHMA4), enhancing Cd uptake and distal translocation (Lei et al., 2020). ZmHMA3 enhances maize’s tolerance to Cd by facilitating its absorption and transport (Liao et al., 2023). The constitutive expression of NcZNT1 in Noccaea caerulescens roots suggests its role in long-distance metal transport from roots to shoots by aiding Zn/Cd influx into xylem-loading cells (Lin et al., 2016). Upregulation of BcABCC1/2 expression enhances Cd compartmentalization in root vacuoles, reducing its accumulation in the edible parts of pakchoi (Huang et al., 2021). OsABCC9 mediates Cd tolerance and accumulation in rice by compartmentalizing Cd into root vacuoles (Yang et al., 2021). Transcription factors play a crucial role in regulating both biological and abiotic processes in plants. A deeper understanding of transcription factors is essential for elucidating the molecular mechanisms of plant Cd tolerance.
This review summarizes the role of transcription factors in plant Cd tolerance. Transcription factors are crucial for plant transcriptional regulation, affecting growth and adaptation to adverse environments. Consequently, transcription factors are pivotal in regulating plant tolerance to heavy metals (Strader et al., 2022). Plant secondary metabolites, synthesized in specific organs or tissues during distinct developmental stages or in response to environmental stimuli, are crucial for plant-environment interactions. Consequently, these genes are transcriptionally regulated by multiple TFs (Yang et al., 2012). Recent studies confirm the involvement of several transcription factor families in plant Cd tolerance mechanisms. This review aims to summarize how the WRKY, ERF, MYB, bHLH, and bZIP transcription factor families regulate Cd tolerance in plants.
WRKY TFs belong to a large family of transcriptional regulators that are integral to the signaling network regulating various plant processes. WRKY proteins typically function as both repressors and activators, influencing the repression and activation of crucial plant processes (Rushton et al., 2010). Recent studies indicate that WRKY TFs are crucial in regulating plant defense and disease resistance (Eulgem and Somssich, 2007). The expression of several WRKY genes is strongly and quickly induced by drought and high salinity, suggesting their regulatory roles in these signaling pathways (Chen et al., 2012). Additionally, this expression can be triggered by heavy metals. Recently, many studies have focused on how WRKY TFs regulate Cd tolerance in plants. Most WRKY TFs in plants are localized in the nucleus and predominantly expressed in the roots, though they are present in all plant tissues (Table 1).
Cd tolerance mediated by WRKY TFs has been demonstrated across various model plants and food crops. in Arabidopsis thaliana, AtWRKY12 indirectly suppresses genes related to PC synthesis and negatively impacts Cd accumulation and tolerance (Han et al., 2019). Conversely, AtWRKY13 employs distinct mechanisms, enhancing Cd tolerance in plants by influencing H2S synthesis (Sheng et al., 2019; Zhang et al., 2020). Similarly, multiple studies have shown that WRKY transcription factors regulate H2S to affect Cd tolerance. Plants with double or triple mutations in WRKY18, WRKY40, and WRKY60 produced H2S at a higher rate under Cd stress than wild-type and single-mutant plants, suggesting increased Cd resistance. WRKY18, WRKY40, and WRKY60 primarily function as repressors, regulating genes that encode H2S synthase. These findings indicate that WRKY transcription factors regulate the H2S signaling pathway, aiding plant resilience to Cd stress (Liu et al., 2015). Besides H2S, WRKY transcription factors also regulate antioxidant enzyme activity and synthesis; overexpressing TaCOPT3D in wheat decreases ROS levels and enhances antioxidant enzyme activity under Cd stress. Additionally, the transcription factor TaWRKY22 targets the TaCOPT3D promoter, playing a role in its regulation under Cd stress. These findings highlight TaCOPT3D’s crucial role in plant adaptation to Cd stress through its interaction with TaWRKY2 (Liu et al., 2022a). Expression of TaWRKY70 in Arabidopsis enhances growth and biomass production under Cd stress. Moreover, Arabidopsis overexpressing TaWRKY70 exhibit increased SOD, POD, and CAT activities under Cd stress compared with wild-type plants. Thus, the improved growth of transgenic Arabidopsis under Cd stress is due to enhanced antioxidant capacity and reduced oxidative damage (Jia et al., 2021). TaWRKY74 consistently mitigates Cd toxicity in wheat by influencing ASA-GSH synthesis genes and repressing Cd transporter genes (Li et al., 2022). In rice, OsWRKY71 significantly induces Cd stress tolerance in leaves, with transcript levels peaking 24 hours post-stress initiation (Paul and Roychoudhury, 2018).
Beyond model plants and food crops, the role of WRKY TFs in Cd tolerance has been explored in various other plants. In Sedum alfredii, SaWRKY7 responds to Cd induction, potentially playing a crucial role in Cd accumulation and tolerance (Wang et al., 2019). In Tamarix hispida, ThVHAc1 and ThWRKY7 likely jointly regulate Cd tolerance, with ThWRKY7 potentially control ThVHAc1 to boost abiotic stress tolerance (Yang et al., 2016). Some WRKY TFs increase Zea mays tolerance to Cd by promoting GSH synthesis in vivo (Mei et al., 2017). ZmWRKY64 improves Cd tolerance by upregulating ZmSRG7 transcription, thus maintaining ROS homeostasis (Gu et al., 2024). Cd upregulates of the ZmWRKY4 expression enhances antioxidant enzyme activity (Hong, Cheng et al., 2017). Similarly, a positive feedback loop between H2O2 accumulation and CaWRKY41 upregulation coordinates pepper plants’ responses to Cd exposure. This mechanism might reduce Cd tolerance by enhancing Cd uptake through Zn transporters (Dang et al., 2019). In soybeans, GmWRKY142 directly targets ATCDT1, GmCDT1–1, and GmCDT1–2, reducing plant Cd uptake and enhancing Cd tolerance (Cai et al., 2020). The expression of GmWRKY27 was slightly upregulated under Cd stress compared with that in untreated plants, suggesting that GmWRKY27 may play an important role in Cd tolerance in soybeans (Imran et al., 2022). GmWRKY172 promotes H2O2 accumulation, increases peroxidase (POD) activity, and reduces Cd transport from roots to stems and seeds (Xian et al., 2023). In Kenaf, HcWRKY71 expression decreases as CdCl2 concentrations increase. The most significant change in HcWRKY71 expression occurred in the roots under CdCl2 stress. Thus, HcWRKY71 may enhance Kenaf’s Cd tolerance by regulating root gene expression during stress (Li et al., 2021b). In poplars, overexpression of PyWRKY75 increases chlorophyll content and promotes growth under Cd stress, and correlates with increased tolerance due to the protective effects of antioxidant enzymes (POD, SOD, CAT, APX) and osmoregulatory substances (soluble sugars). Overexpression of PyWRKY75 also increases ASA, GSH, and PCs content in poplars, potentially promoting Cd accumulation. Therefore, PyWRKY75 may serve a positive regulatory role under Cd stress in poplars, suggesting its potential as a candidate gene for plant Cd tolerance (Wu et al., 2022). PyWRKY48 enhances the Cd tolerance by upregulating the expression of PaGRP, PaPER and PaPHOS, which encode antioxidase-related proteins (Wu et al., 2023a). In sugarcane, CdCl2 induces the expression of ScWRKY6, ScWRKY6 likely contributes to sugarcane’s response to Cd stress by promoting stress protein and proline accumulation and enhancing antioxidant enzyme activity (Zhang et al., 2018). StWRKY6 increases CAT activity, enhancing ROS clearance and reducing oxidative stress in photosynthetic tissues (Guandi et al., 2023).
Transcriptome data reveal that the expression level of WRKY TFs changed in different plants in response to Cd stress. For example, In Sedum plumbizincicola, 7 SpWRKYs were significantly upregulated by Cd stress (Wang et al., 2023). The expression level of seven genes (HvWRKY1, 3, 13, 15, 24, 43, and 55) increased by more than two-fold in barley after CdCl2 treatment. The expression levels of HvWRKY3 and HvWRKY24 were 20-fold and 10-fold higher, respectively, than those in controls (Zheng et al., 2021). In particular, the expression of HvWRKY32 and HvWRKY80 was significantly decreased. Conversely, BnaWRKY49 and BnaWRKY38 were upregulated after Cd treatment, and BnaWRKY7, 27, 38, 56, and 60 were identified as key Cd-related genes (Ma et al., 2022). In creeping bentgrass, the expression of WRKY33, WRKY12, WRKY2, WRKY23, WRKY75, and WRKY53 was either upregulated or downregulated after Cd treatment (Yuan et al., 2018). The expression of 11 WRKY genes increased with increasing Cd content in the leaves of golden raintrees (He et al., 2021). In hemp, the expression of CsWRKY genes changed significantly under Cd stress, with 14 of the 40 predicted genes responding to Cd stress. In particular, the expression levels of these three genes increase by more than 10-fold in response to Cd stress (Xin et al., 2016). Finally, the expression of 22 rice WRKY TFs was upregulated under Cd stress (Tan et al., 2017).
In summary, the WRKY TFs modulates Cd tolerance in plants through the redox system, specifically influencing the synthesis and activity of antioxidant enzymes (POD,SOD,CAT and APX) and redox-related substances (ROS,H2S,NO,GSH).Currently, transcriptome analysis. Currently, transcriptome analyses indicate that WRKY factors respond to Cd stress. While specific signaling pathways remain unidentified, these studies guide future research directions.
The ERF TFs regulates stress responses linked to ethylene, a significant stress hormone induced by various abiotic factors (Wang et al., 2022a). ERF TFs were initially identified due to their ability to bind to promoters of stress response genes. Research has shown that besides responding to stress hormones like ethylene, jasmonic acid, and abscisic acid, ERF TFs are also triggered by various biotic and abiotic stresses such as salinity, osmotic stress, drought, hypoxia, temperature fluctuations, and pathogen infections (Licausi et al., 2013). Similarly, ERF TFs are induced by cadmium and play crucial roles in regulating plant Cd tolerance. Most plant ERF TFs are localized in the nucleus and expressed across all tissues, predominantly in the roots. (Table 2).
Cd tolerance mediated by ERF TFs has been demonstrated in various model plants and food crops. For example, in Arabidopsis, the mRNA level of NRT1.8, a nitrate transporter inducible by Cd, are significantly upregulated in the root system following Cd exposure, Specifically, ERF34 and ERF35 bind to the NRT1.8 promoter, regulating Cd tolerance (Xie et al., 2021). Similarly, ERF1B and ERF104 bind directly to the NRT1.8 promoter, mediating its expression under Cd induction, which is crucial for understanding plant responses to Cd stress (Zhang et al., 2014). In wheat, TaMYC8 directly regulates TaERF6 expression and negatively affects Cd tolerance by activating TaERF6 transcription via binding to its promoter (Wang et al., 2022b). TdSHN1, a SHINE-type ERF transcription factor, was significantly upregulated following Cd induction. After TdSHN1 knockout, transgenic tobacco exhibited longer roots, increased biomass, higher chlorophyll content, and reduced ROS production compared to wild-type under heavy metal stress. Additionally, transgenic tobacco displayed increased activity of ROS-scavenging enzymes, SOD and CAT (Djemal and Khoudi, 2022). In barley, the ERF-type transcription factor HvRAF, particularly its C-terminal region when overexpressed, enhances yeast Cd toxicity tolerance (Jung and Kim, 2007). A study on rice’s interaction network revealed that the OsERF141, encoding a transcription factor, is a potential target for specific miRNAs. Interestingly, osa-miR5493 expression negatively correlates with OsERF141, suggesting osa-miR5493’s potential role in regulating Cd stress through OsERF141 expression (Ailing et al., 2020). PvERF15 in soybean (Phaseolus vulgaris) directly binds to the PvMTF-1 promoter and is strongly induced under Cd stress. Knocking down PvERF15 reduces Cd tolerance, positioning PvERF15 as a key element of the Cd stress transcriptional pathway with PvMTF-1 as a downstream target (Lin et al., 2017).
In pepper, CaPF1, an ERF/AP2-type TF, regulates plant tolerance to Cd by regulating the oxidative stress mechanism (Jung Won et al., 2008). In Pinus virginiana Mil, CaPF1 overexpression in transgenic plants resulted in greater Cd tolerance than in the control, presumably by regulating antioxidant enzyme activity and lipid peroxidation under Cd stress (Tang et al., 2005). Furthermore, the relative expression of LrERF061 significantly increased in Glycyrrhiza uralensis Fisch under Cd stress. The activity of antioxidant enzymes (SOD, CAT, and POD) was enhanced in transgenic plants overexpressing LrERF061 at high Cd concentrations (Chahel et al., 2020). In Aquilaria sinensis, the expression level of AsERF1 increased under heavy metal stress, reaching 3.5 times the level of the control group at the highest point. These results indicate that heavy metal stress induces the expression of the AsERF1 encoding gene (Li et al., 2020).
Current transcriptome data show that ERF TFs positively responds to Cd stress. Based on transcriptome analysis, 15 StAP2/ERF genes were randomly selected from 181 potato genes to test their involvement in the plant response to Cd stress. The results showed that most genes were sensitive to Cd stress. Particularly, in the leaves, StAP2/ERF027-, StAP2/ERF140, and StAP2/ERF164 encoding genes were significantly upregulated and showed maximum expression levels 24 h after Cd treatment. The yeast complementation assays showed that StAP2/ERF129 and StAP2/ERF139 (subgroup 1) promoted Cd accumulation (Cd-reduced), whereas StAP2/ERF044 and StAP2/ERF180 (subgroup 2) promoted Cd accumulation and inhibited growth (Cd-accumulated). In turn, the StAP2/ERF075, StAP2/ERF077, and StAP2/ERF126 encoding genes (subgroup 3) promote Cd accumulation and yeast growth (Cd detoxification type) (Tian et al., 2022). In eggplant, seven genes (SmERF9, SmERF30, SmERF59, SmERF61, SmERF85, SmERF86, and SmERF100) showed significantly up-regulated expression levels under Cd treatment and a down-regulation trend under prolonged treatment conditions, whereas four genes (SmERF2, SmERF39, SmERF55, and SmERF57) were consistently down-regulated (Shao et al., 2015). Differentially expressed genes in diploid poplar (Populus simonii × P) were identified experimentally. ERFs and differentially expressed genes exhibited similar expression patterns under salinity (NaCl), chlorine (KCl), heavy metal (CdCl2), and drought stress. ERF76-overexpressing transgenic poplar was superior to the wild-type in terms of morphological and physicochemical traits, indicating that the ERF76 encoding gene played a significant role in stress resistance (Yao et al., 2017). In Lycium chinense, LchERF transgenic plants were more tolerant to Cd stress than non-transgenic plants. Overexpression of LchERF enhanced Cd tolerance in transgenic tobacco, which may be due to the effect of LchERF on tobacco glutathione synthesis-related genes. Under Cd stress, non-transgenic plants consistently accumulate more ROS than those overexpressing LchERF (Guan et al., 2015). Transgenic tobacco overexpressing LcMKK showed greater tolerance to Cd stress with enhanced expression of NtERF and NtGSH1, indicating that LcMKK is associated with enhanced expression levels of ERF synthesis-related genes in tobacco. Evidence suggests that under Cd stress, NtERF may be involved in the accumulation of GSH in the plant body, which in turn reduces the toxic effects of Cd on plants (Guan et al., 2016). Finally, in creeping bentgrass, most genes are upregulated, whereas ERF115 and ERF4 are slightly downregulated under Cd stress (Yuan et al., 2018). Collectively, these results provide a useful reference for further studies on the relationship between members of the ERF TF family and Cd tolerance in plants.
Our findings indicate that ERF TFs actively respond to Cd stress in plants and play a crucial role in regulating Cd tolerance. The mechanisms through which ERF TFs regulate Cd tolerance involve controlling ROS, nitrate, and ethylene synthesis and activity.
The myeloblastosis oncogene (MYB) family, a large group of TF, is prevalent in eukaryotes and widely expressed in most plants (Erpen et al., 2018). Previous research has shown that several MYB TFs regulate plant responses to drought and participate in the ABA signaling network (Cominelli et al., 2005; Lee et al., 2020). Plant MYB proteins are variably regulated by ABA, enhancing abiotic stress tolerance. Additionally, MYB TFs react to Cd stress mechanisms in plants and regulate Cd resistance. Most MYB TFs are localized in the nucleus and expressed in various tissues of the plant, although mainly in the root (Table 3).
Members of the MYB TF family play significant roles in plant Cd tolerance mechanisms. For example, in Arabidopsis, myb49 overexpression results in a significant increase in Cd accumulation, which is significantly reduced by the knockdown of myb49. Further studies have shown that myb49 positively regulates the expression of bHLH38 and bHLH101 by directly binding to their promoters, leading to the activation of iron-regulated TRANSPORTER1, which encodes a metal transporter protein associated with Cd uptake. myb49 also binds to the heavy metal-associated plant heteropropenylated protein (HIPP22) and HIPP44 promoter regions, leading to their upregulation and subsequent Cd accumulation. Therefore, AtMYB49 regulates Cd accumulation by directly regulating the expression of bHLH38, bHLH101, HIPP22, and HIPP44 (Zhang et al., 2019). Furthermore, the germination rate of the Arabidopsis MYB59 knockout mutant was significantly higher than that of the wild type or MYB59 overexpressing mutant in the Cd-containing medium. This suggests that AtMYB59 may be involved in Cd stress resistance in Arabidopsis (Fasani et al., 2019). MYB are crucial for the compartmentalization of Cd.AtMYB75 likely serves as a positive regulator of Cd tolerance by targeting Arabidopsis stress-related genes ACBP2 and ABCC2, which are involved in regulating ATP for Cd compartmentalization in vacuoles (Zheng et al., 2022). They regulate the ATP required for Cd to be compartmentalized by vacuoles. AtMYB4 boosts plant antioxidant activity by engaging in the glutathione pathway, activating and regulating the expression of PCS1 and MT1C in response to Cd stress. This regulation aids in the chelation of Cd by PC and MT, allowing the complexes to be compartmentalized in vacuoles (Agarwal et al., 2020). In rice, OsTAZ4 promotes plant resistance to Cd stress by regulating ROS endostasis; furthermore, OsTAZ4 can promote plant resistance to multiple heavy metals by interacting with the OsMYB34 transcription factor (Shalmani et al., 2021). The reduction in Cd accumulation in the knockout OsMYB36 mutant may be related to the altered abundance of OsNramp5 mRNA or OsNramp5 protein, indirectly suggesting that OsMYB36 plays an important role in Cd tolerance in rice (Wang et al., 2022b). The transgenic rice knockout OsMYB45 mutant was highly sensitive to Cd treatment, and Cd tolerance was greatly reduced, proving that OsMYB45 plays an important role in rice resistance to Cd stress (Hu et al., 2017).
MYB reduces the toxicity of Cd by regulating the redox system. In walnuts, JrVHAG1 acts as a regulator of the Cd stress response by participating in the MYB transcriptional regulatory network by regulating the expression of the JrMYB2 TF, showing that this TF also plays an important role in the regulation of Cd tolerance in walnuts (Xu et al., 2018). In Salicornia brachiate Roxb, the expression of SbMYB15 increased five-fold under Cd stress. In contrast to the wild type, SbMYB15 overexpressing transgenic tobacco showed lower Cd uptake and increased growth and chlorophyll content. Furthermore, transgenic tobacco showed higher scavenging activities of antioxidant enzymes (CAT and SOD) and higher transcript levels of the related antioxidant genes CAT1 and MnSOD (Sapara et al., 2019). In ramie, the expression of BnMYB1 was elevated under Cd treatment and showed a significant and progressive increase with increasing Cd concentrations, suggesting that BnMYB1 may be involved in the mechanism of ramie resistance to Cd tolerance (Zhu et al., 2018). A significant increase in Cd tolerance and accumulation was observed in BnMYB2 overexpressing transgenic Arabidopsis plants. Thus, BnMYB2 regulates Cd tolerance and accumulation (Zhu et al., 2020b). Additionally, BnMYB3 responds positively to Cd stress and is a candidate gene for enhancing Cd tolerance in ramie, with increased expression under Cd stress (Zhu and Shi, 2019). In tomatoes, the GSH-induced enhancement of Cd tolerance is closely associated with the upregulation of the transcription factor MYB1 and some stress-responsive genes (Hasan et al., 2016). In soybean, MYBZ2 regulates Cd tolerance by scavenging the ROS generated by NADPH oxidase (Chmielowska-Bak et al., 2017). Overexpression of PsMYB62 in Potentilla sericea enhances the activities of SOD,POD and CAT, resulting in increased H2O2 content in transgenic plants (Feng et al., 2023). PeRAX2, a MYB TF in Populus euphratica, regulates the transcription of AtANN1, mediates Cd2+ absorption via calcium channels, induces H2O2 synthesis, and enhances CAT, SOD, and POD activities (Yan et al., 2024).
Using transcriptome analysis, the TFs MYB4, MYB10, MYB72, and MYB107 were screened in Arabidopsis after Cd treatment. Owing to their high expression levels, these TFs can be used as candidate genes to study Cd tolerance in this model plant (De Mortel et al., 2008). Similarly, in rice, OsMYB36 expression was significantly upregulated under Cd stress, whereas the genes encoding OsMYB3, OsMYB7, OsMYB15, OsMYB23, OsMYB47, OsMYB76, OsMYB89, and OsMYB93 were significantly downregulated. However, the expression levels of OsMYB2, OsMYB90, and OsMYB99 were not significantly altered (Kang et al., 2022). In hemp, most MYB TF family members respond to Cd stress; specifically, the expression of CsMYB016, CsMYB067, CsMYB098, CsMYB045, and CsMYB024 is downregulated, whereas that of CsMYB028 is upregulated (Yin et al., 2022). In pepper, 10 CaMYB gene members showed a significant increase in expression in response to Cd stress (Xie et al., 2022). Similarly, RNA-seq results have shown that MYB4, MYB39, MYB108, and MYB305 are upregulated in creeping bentgrass under Cd stress (Yuan et al., 2018). Furthermore, the expression of the seven MYB genes gradually decreases with increasing Cd content (He et al., 2021). Meanwhile, in the leaves, the myb-like genes Unigene0019400 and Unigene0027656 were significantly expressed in the absence of Cd stress but were gradually downregulated with increasing Cd concentration. In turn, the expression of most MYB transcription factors in leaves of wild paper mulberry showed a V-shaped expression trend over a period of 120 h of Cd treatment (Xu et al., 2019). In Apocynum venetum, AvMYB4 and AvMYB10 are downregulated under Cd stress, whereas AvMYB8, AvMYB11, AvMYB48, and AvMYB97 are upregulated, indicating that they are candidate genes that respond to Cd stress (Abubakar et al., 2022). In Ipomoea aquatica, the transcript expression levels of IaMYB47, IaMYB86, IaMYB142, IaMYB157, and IaMYB147 were upregulated, whereas those of IaMYB7, IaMYB110, IaMYB121, IaMYB173, and IaMYB176 were downregulated after Cd treatment (Liu et al., 2022b). Finally, in barley, the transcript expression and m6A modification levels of MYB TF family members were significantly upregulated under Cd stress (Su et al., 2022).
These results demonstrate that the MYB transcription factor family plays a crucial role in plant Cd tolerance by regulating the synthesis and activity of antioxidant enzymes and interacting with genes related to Cd compartmentalization.
The basic helix-loop-helix (bHLH) family is a large group of plant transcription factors that function as homodimers or heterodimers to regulate gene expression. Several TFs play unique roles in regulating iron homeostasis in plants (Gao et al., 2019). Recent studies demonstrate that bHLH TFs effectively regulate Cd stress response mechanisms in plants. Subcellular localization analysis shows that most bHLH TFs are located in the nucleus and are predominantly expressed in the root system, though they are found throughout plant tissues (Table 4).
Functional verification has confirmed that bHLH TFs are involved in Cd tolerance mechanisms in both model plants and food crops. The regulation of Cd tolerance by bHLH is related to nicotinamide synthase(NAS).For example, in Arabidopsis, gene expression analysis showed that FIT, AtbHLH38, and AtbHLH39 expression was upregulated in the root systems of Cd-treated plants, and co-expression of all three activated the expression of HMA3, MTP3, IRT2, and IREG2, all of which are associated with heavy metal detoxification. Additionally, co-overexpression enhanced the expression of NAS1 and NAS2, leading to the accumulation of nicotinamide, a key chelator of iron transport and homeostasis. Maintaining high Fe content under Cd stress alleviates Cd toxicity (Wu et al., 2012). In particular, bHLH104 plays an important role in Cd transport from roots to stems. Evidence shows that bHLH104 positively regulates four heavy metal detoxification-related genes, namely IREG2, MTP3, HMA3, and NAS4, which are Cd tolerant genes. This shows that bHLH104 plays an important role in the regulation of Cd-stress plant responses (Yao et al., 2018). Furthermore, bHLH39 and bHLH104 interact functionally with IMA1 and IMA3 to enhance Fe accumulation in plants by inducing the expression of Fe transporter genes, resulting in enhanced Cd tolerance (Meng et al., 2022). The transcription factor bHLH100 is highly expressed under high Cd concentrations and may be a candidate gene for enhancing Cd tolerance (De Morte et al., 2008).In wheat, TabHLH094 mediates Cd tolerance by modulating TaMYC8 transcriptional activity and reducing ethylene production (Du et al., 2023).
Beyond model plants and major food crops, molecular mechanisms in various plants show that bHLH TFs respond to Cd stress. In Broussonetia papyrifera, BpabHLH149 not only responds to Cd stress but also enhances yeast resistance, potentially making it a key candidate gene for stress responses in Caladenia tessellata (He et al., 2022). In soybean, the expression of GmbHLH041 increases very significantly after 24 h of Cd stress and is an important candidate gene for screening Cd stress tolerance (Liu et al., 2018). The bHLH OBP3-responsive gene (GmORG3) was significantly induced by Cd stress in soybean, and GmORG3 overexpressing plants increased the rate of iron translocation from the roots to stems while decreasing that of Cd. This suggests that GmORG3 enhances Cd tolerance by stabilizing iron homeostasis (Xu et al., 2017). In Noccaea caerulescens, genes encoding the TFs bHLH038, bHLH039, bHLH100, and bHLH101 were significantly upregulated under Cd stress, and it is highly likely that they enhance Cd tolerance by maintaining photosynthetic activity (Halimaa et al., 2019).
Current transcriptome data show that bHLH transcription factors positively respond to Cd stress, leading to either up-regulation or down-regulation of their expression. Transcriptome analysis in rice has revealed significant involvement of bHLH TFs in regulating Cd and Zn homeostasis (Adil et al., 2022). Similarly, in Populus simonii × P. nigra cross, 69 bHLH family members responded to Cd stress, of which 29 were upregulated, and 40 were downregulated (Yao et al., 2019). Furthermore, in wild paper mulberry, the expression of most bHLHs was upregulated in wheat leaves under Cd stress (Xu et al., 2019). Consistently, bHLH29, bHLH38, and bHLH47 are upregulated in wheat under Cd stress (Sabella et al., 2021). The expression of bHLHs may be regulated through NA synthesis and maintenance of iron homeostasis in the plant, thereby increasing Cd tolerance (Aprile et al., 2018).
In summary, bHLH TFs primarily regulate Cd transport in plants, likely by binding to and regulating the expression of ion channel-related genes. Additionally, some studies indicate that bHLH enhances the accumulation of nicotinamide, which promotes iron transport. Maintaining high iron levels under Cd stress may alleviate Cd toxicity.
Basic (region) leucine zipper (bZIP) TFs are characterized by a basic DNA-binding region and an adjacent leucine zipper that facilitates dimerization (Droge-Laser et al., 2018). bZIP proteins occur in various plants and participate in diverse biological processes, such as seed maturation, flower development, plant senescence, and light signaling. bZIP TFs also exert negative regulation on plant growth and development (Zhang et al., 2011). Subcellular localization analysis indicates that bZIPs are predominantly located in the nuclei of plant cells, with varying expression across different plant tissues. In many plants, bZIP TFs are mainly expressed in the roots, with fewer expressed in the stems or leaves (Table 5).
Cd tolerance mediated by bZIP TFs has been confirmed in various model plants and food crops. For example, in Arabidopsis, AtbZIP30 activates AtGST11 gene expression, enhancing Cd tolerance under stress conditions (Kouno and Ezaki, 2013). AtbZIP44 regulates the expression of AtMAN7, influencing enzyme activity and mannose content in the cell wall, key for compartmentalization (Wu et al., 2023b). Similarly, in ramie, BnbZIP2 overexpressing transgenic Arabidopsis showed higher sensitivity to the heavy metal Cd stress during seed germination, and BnbZIP2 may play a positive regulatory role in the plant response to Cd stress (Huang et al., 2016). In Sedum plumbizincicola, SpbZIP60 may enhance the tolerance of transgenic Arabidopsis plants to Cd by regulating ROS accumulation and protecting photosynthetic organs (Lu et al., 2022). In T. hispida, ThbZIP1 is upregulated under Cd stress, enhancing Cd tolerance through the regulation of SOD activity (Wang et al., 2010). Additionally, the expression of BpbZIP1 can be induced in B. papyrifera by Cd treatment; furthermore, it significantly improves Cd resistance in yeast. These findings indicate that the expression of BpbZIP1 can improve Cd tolerance and is an important gene in the Cd stress response (Chen et al., 2022). Furthermore, the expression of the transcription factor NtbZIP60 in Nicotiana tabacum is upregulated under Cd stress; hence, this gene may be a candidate gene to screen for Cd stress tolerance (Xu et al., 2013). Consistently, overexpression of BjCdR15 (a bZIP-type TF) in Brassica juncea, Arabidopsis, tobacco enhanced Cd tolerance, and BjCdR15 replaced the function of TGA3 and participated in the long-distance transport of Cd from the roots to the stem; moreover, results indicated that the BjCdR15 TF plays a crucial role in regulating Cd uptake by roots and in long-distance root-to-stem transport (Farinati et al., 2010).
Transcriptome analysis revealed that 15 bZIP TF genes responded to Cd stress in ramie, with 14 upregulated and one downregulated (Zheng et al., 2016). In soybeans, genes encoding bZIP44 and bZIP78 are expressed at low levels under Cd stress, indicating that they respond to Cd stress (Chmielowska-Bak et al., 2013). In rice, OsbZIP18 and OsbZIP23 are upregulated by Cd stress (Ailing et al., 2020). Similarly, in Populus simonii × P. nigra, OsbZIP18 and OsbZIP23 are upregulated under Cd stress. In total, 13 genes of the bZIP TF family are upregulated and 19 are downregulated under Cd stress (Yao et al., 2019). Furthermore, in creeping bentgrass, bZIP06, bZIP43, and bZIP19 of the bZIP TFs are upregulated in response to Cd stress (Yuan et al., 2018). Lastly, two genes encoding bZIP (OS06G0614100 and OS02G0194900) were downregulated in response to Cd stress. Hence, these bZIP TF family members may be directly involved in the response of rice plants to Cd stress (Sun et al., 2019).
Current research on bZIP TFs’ role in regulating Cd tolerance lacks detail, but evidence suggests these factors impact Cd’s long-distance transport by influencing compartmentalization and the redox system.
Beyond the five transcription factor families-WRKY, ERF, MYB, bHLH, and bZIP-other factors involved in Cd stress regulation in plants have also gained significant attention. For example, in S. alfredii, SaHsfA4c increases plant resistance to stress by upregulating the activities of ROS-scavenging enzymes and the expression of Hsps (Chen et al., 2020). OsNAC15 reduces Cd toxicity in rice during the vegetative period (Zhan et al., 2022). Overexpression of TaNAC22 in wheat increases the efficiency of the enzymatic antioxidant defense system, consequently boosting Cd tolerance by regulating the expression of stress-responsive genes (Yu and Zhang, 2023). In Arabidopsis,AtNAC102 regulates cell wall pectin metabolism and inhibits its binding to Cd, reducing Cd uptake and accumulation (Han et al., 2023). ZAT6 positively regulates Cd tolerance through a glutathione-dependent pathway (Chen et al., 2016). ZAT10 represses the transcriptional activity of IRT1, which encodes a key metal transporter involved in Cd uptake (Dang et al., 2022). In summary, the extensive role of transcription factors in regulating Cd has been well-documented.
Arabidopsis, as a model plant, has made significant contributions to the study of plant biology. Recently, numerous studies have explored the mechanisms of Cd tolerance in Arabidopsis. However, research into the mechanisms of TF-mediated Cd tolerance and their specific functions in Arabidopsis remains limited. Here, we summarize the relationship between transcription factors and Cd tolerance in Arabidopsis. Transcription factors can regulate downstream genes that are important for Cd tolerance in plants (Figure 1).
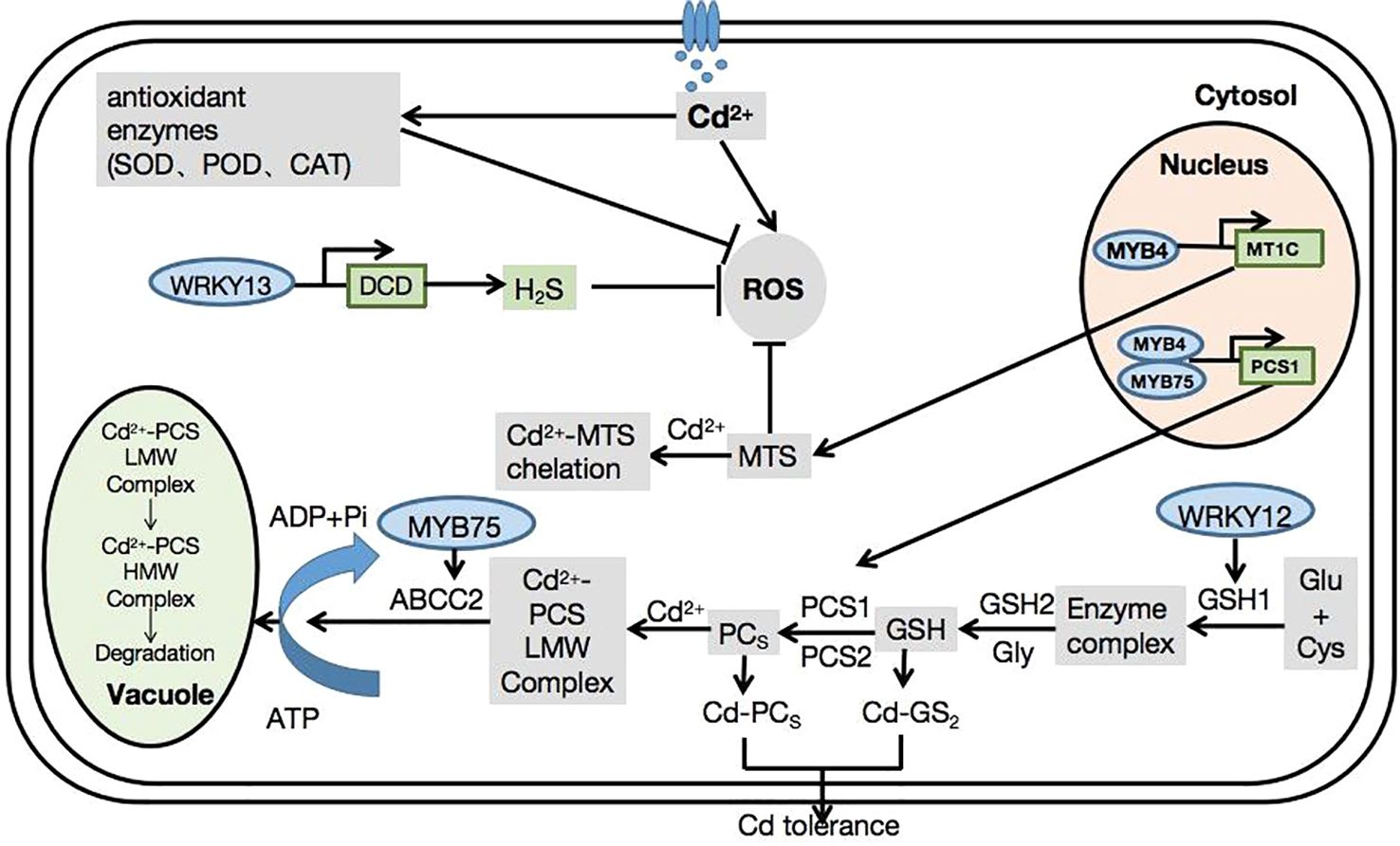
Figure 1 Mechanisms of transcription factor families in Arabidopsis thaliana in response to cadmium stress.
In Arabidopsis, TFs regulate Cd accumulation via several signaling pathways. For example, AtWRKY12 combines with the W-box of the GSH1 promoter to directly target GSH1, inhibit the expression of GSH1, and indirectly inhibit the expression of genes related to PC synthesis (Han et al., 2019). AtWRKY13 activates DCD expression and increases the production of H2S by directly binding to the DCD promoter (Zhang et al., 2020). Furthermore, it binds to the W-box region of the PDR8 promoter, which acts downstream of AtWRKY13, thereby positively regulating Cd tolerance in Arabidopsis through PDR8 (Sheng et al., 2019). Similarly, WRKY18, WRKY40, and WRKY60 mainly act as inhibitors of the transcription of genes encoded by H2S synthases (Liu et al., 2015). ERF34 and ERF35 bind to the promoter of the nitrate transporter NRT1.8 (one of the transporters induced by Cd) to regulate Cd tolerance in plants (Xie et al., 2021). ERF1B and ERF104 directly bind to the NRT1.8 promoter and mediate the expression of the NRT1.8 transporter under the action of Cd. Therefore, the regulation of plant responses to Cd stress is of great significance (Zhang et al., 2014). AtMYB49 positively regulated the expression of bHLH38 and bHLH101 by directly binding to their promoters, leading to the activation of Fe-regulated TRANSPORTER1, which encodes a metal transporter associated with Cd uptake. MYB49 also binds to HIPP22 and the promoter region of HIPP44, leading to the upregulation of their expression (Zhang et al., 2019). AtMYB75 acts as a positive regulator of Cd tolerance by targeting the Arabidopsis Cd stress-related genes ACBP2 and ABCC2 (Zheng et al., 2022). AtMYB4 participates in the glutathione pathway through the transcriptional activation of PCS1 and MT1C, mediating Cd tolerance through enhanced antioxidant activity (Agarwal et al., 2020). bHLH104 positively regulates four Cd tolerance-related genes (IREG2, MTP3, HMA3, and NAS4) involved in heavy metal detoxification (Yao et al., 2018). Furthermore, AtbZIP30 activates AtGST11 gene expression and improves Cd tolerance in plants grown under Cd-stress conditions (Kouno and Ezaki, 2013).
Cd toxicity can be alleviated by maintaining high iron content under Cd stress. Thus, bHLH39 and bHLH104 functionally interact with IMA1 and IMA3 to enhance iron accumulation in plants by inducing the expression of iron transport genes. This leads to an increased Cd tolerance (Meng et al., 2022). Overexpression of FIT, AtbHLH38, and AtbHLH39 activated the expression of HMA3, MTP3, IRT2, and IREG2 in Arabidopsis, all of which are related to heavy metal detoxification. In addition, co-overexpression enhanced the expression of nicotinamine synthase (NAS1) and NAS2, leading to the accumulation of nicotinamine, a key chelating agent for iron transport and homeostasis (Wu et al., 2012).
Food crop safety is closely related to human health. Rice and wheat are common food crops that enter the human body through the daily diet. If rice and wheat are grown in soils contaminated with Cd, it is highly likely that Cd would enter the human body through the food chain. Therefore, studying the mechanism of Cd stress regulation in these plants is highly necessary to provide a solid theoretical basis for further regulation of the mechanism of Cd accumulation in grain crops using molecular genetic engineering.
The TaWRKY22 TF combined with the TaCOPT3D promoter directly targets TaCOPT3D under Cd stress, thus regulating the expression of TaCOPT3D and playing an important role in the regulation of Cd stress in wheat (Liu et al., 2022a). TaWRKY70 transgenic Arabidopsis showed increased SOD, POD, and CAT activities under Cd stress. Therefore, TaWRKY70 regulates Cd stress by improving plant antioxidant capacity (Jia et al., 2021). TaWRKY74 affects the absorption and transport of Cd in wheat by affecting the expression of ASA-GSH synthesis genes and inhibiting the expression of Cd transporter genes (Li et al., 2022). In contrast, TaMYC8 activates TaERF6 transcription by binding to the TaERF6 promoter to directly regulate the expression of TaERF6, thus negatively regulating Cd tolerance (Wang et al., 2022b). TdSHN1 mutant tobacco exhibits higher ROS-scavenging enzyme (SOD and CAT) activity. This might explain why transgenic tobacco plants are more resistant to heavy metals (Djemal and Khoudi, 2022).
In rice, the expression level of osa-miR5493 is negatively correlated with that of OsERF141. This finding implies that osa-miR5493 may also be a key regulator of Cd stress through the regulation of OsERF141 (Ailing et al., 2020). OsTAZ4 promotes plant resistance to Cd stress by regulating ROS homeostasis and can promote plant resistance to various heavy metals by interacting with the OsMYB34 (Shalmani et al., 2021). In the OsMYB36 knockout mutant, Cd accumulation decreased, and the expression of OsNramp5 was significantly altered, indirectly indicating that OsMYB36 may affect rice Cd tolerance by regulating the expression of OsNramp5 (Hu et al., 2017).
The five major transcription factor families in plants-WRKY, ERF, MYB, bHLH, and bZIP-are primarily located in the nucleus and positively respond to Cd stress. Although expressed in all plant tissues, their expression is predominantly in the roots. The WRKY transcription factor family mediates Cd tolerance by regulating the H2S signaling pathway, altering ROS levels, and modulating antioxidant enzyme activities. The ERF transcription factor family regulates Cd tolerance through processes that influence ROS, nitrate, and ethylene synthesis and activity. The MYB transcription factor family regulates the synthesis and activity of antioxidant enzymes, facilitating Cd compartmentalization. The bHLH transcription factor family plays a crucial role in Cd transport from roots to stems and enhances iron accumulation by inducing iron transport gene expression. The bZIP transcription factor family is critical for regulating Cd uptake by roots and its long-distance transport from root to stem. The WRKY, ERF, MYB, bHLH, and bZIP TFs are crucial in regulating Cd tolerance in both model plants and food crops. Therefore, it is essential to study the mechanisms through which these families regulate plant tolerance to Cd.
Although the mechanisms of Cd tolerance in plants are well understood, the specific roles of transcription factors are not thoroughly explored, with limited studies available, Therefore, further investigation of the function of TFs in Cd tolerance in plants, particularly in model plants and food crops, is necessary. A thorough understanding of the molecular mechanisms underlying Cd tolerance in plants can be utilized for genetic engineering. Biotechnology is used to alter the genetic characteristics of organisms from a genetic point of view. This aims to enable the insertion, removal, and replacement of genes of interest, which often play important roles in the regulation of biotic and abiotic stress resistance. Through such purposeful alterations, precise regulation can be achieved to obtain desired plant traits. Therefore, genetic engineering is an effective means to improve biotic and abiotic stress resistance in plants, including that to heavy metals. To appropriately modify plants using genetic engineering, the basic functions of plant genes must be understood.
HZ: Formal analysis, Visualization, Writing – original draft. LL: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China [grant number 22276164]; the Key Research and Development Project of Science and Technology Department of Zhejiang Province [grant number 2023C02020]; and the National Natural Science Foundation of Zhejiang Province [grant number LZ22C150004]. The funding sources had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. We thank Editage for assistance with English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CAT, Catalase; DCD, d-cysteine desulfhydrase; Cd, Cadmium; SOD, superoxide dismutase; MT, Metallothionein; TF, Transcription factor; PC, Phytochelatin; SH, sulfhydryl; IAA, Indole-3-acetic acid; ROS, reactive oxygen species; ABA, Abscisic acid; SA, Salicylic acid; GA, Gibberellic acid.
Abougabal, K., Moselhy, W. A., Korni, F. M. M. (2020). The effect of cadmium toxicity on Oreochromis niloticus and human health. Afr. J. Aquat. Sci. 45, 303–309. doi: 10.2989/16085914.2019.1707429
Abubakar, A. S., Feng, X. K., Gao, G., Yu, C. M., Chen, J. K., Chen, K. M., et al. (2022). Genome wide characterization of R2R3 MYB transcription factor from Apocynum venetum revealed potential stress tolerance and flavonoid biosynthesis genes. Genomics 114. doi: 10.1016/j.ygeno.2022.110275
Adil, M. F., Sehar, S., Chen, S., Lwalaba, J. L. W., Jilani, G., Chen, Z. H., et al. (2022). Stress signaling convergence and nutrient crosstalk determine zinc-mediated amelioration against cadmium toxicity in rice. Ecotoxicol. Environ. Saf. 230, 113128. doi: 10.1016/j.ecoenv.2021.113128
Agarwal, P., Mitra, M., Banerjee, S., Roy, S. (2020). MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 297. doi: 10.1016/j.plantsci.2020.110501
Ahmad, P., Ahanger, M. A., AlYemeni, M. N., Wijaya, L., Alam, P. (2018). Exogenous application of nitric oxide modulates osmolyte metabolism, antioxidants, enzymes of ascorbate-glutathione cycle and promotes growth under cadmium stress in tomato. Protoplasma 255, 79–93. doi: 10.1007/s00709-017-1132-x
Ailing, L., Zhibo, Z., Yake, Y., Guanghui, C. (2020). Transcriptome analysis reveals the roles of stem nodes in cadmium transport to rice grain. BMC Genomics 21, 127. doi: 10.1186/s12864-020-6474-7
Aprile, A., Sabella, E., Vergine, M., Genga, A., Siciliano, M., Nutricati, E., et al. (2018). Activation of a gene network in durum wheat roots exposed to cadmium. BMC Plant Biol. 18. doi: 10.1186/s12870-018-1473-4
Cai, Z. D., Xian, P. Q., Wang, H., Lin, R. B., Lian, T. X., Cheng, Y. B., et al. (2020). Transcription factor gmWRKY142 confers cadmium resistance by up-regulating the cadmium tolerance 1-like genes. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00724
Chahel, A. A., Yousaf, Z., Zeng, S. H., Li, Y. P., Ying, W. (2020). “Growth and physiological alterations related to root-specific gene function of LrERF061-OE in Glycyrrhiza uralensis Fisch. hairy root clones under cadmium stress”. Plant Cell Tissue Organ Cult. 140, 115–127. doi: 10.1007/s11240-019-01715-2
Chen, J., Huang, S. B., Wang, X., Huang, L. Z., Gao, C., Huang, X.-Y., et al. (2023). IAR4 mutation enhances cadmium toxicity by disturbing auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 12. doi: 10.1093/jxb/erad366
Chen, J., Yang, L. B., Yan, X. X., Liu, Y. L., Wang, R., Fan, T. T., et al. (2016). Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in arabidopsis. Plant Physiol. 171, 707–719. doi: 10.1104/pp.15.01882
Chen, L. G., Song, Y., Li, S. J., Zhang, L. P., Zou, C. S., Yu, D. Q. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Et Biophys. Acta-Gene Regul. Mech. 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, S., Xie, M., Cui, M., Li, W., Xu, Z., Jia, C., et al. (2022). “Identification of broussonetia papyrifera transcription factor bpbZIP1 and analysis of its response to cadmium stress”. Bull. Bot. Res. 42, 394–402. doi: 10.7525/j.issn.1673-5102.2022.03.009
Chen, S. S., Yu, M., Li, H., Wang, Y., Lu, Z. C., Zhang, Y. X., et al. (2020). SaHsfA4c from sedum alfredii hance enhances cadmium tolerance by regulating ROS-scavenger activities and heat shock proteins expression. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00142
Chmielowska-Bak, J., Arasimowicz-Jelonek, M., Izbianska, K., Frontasyeva, M., Zinicovscaia, I., Guiance-Varela, C., et al. (2017). NADPH oxidase is involved in regulation of gene expression and ROS overproduction in soybean (Glycine max L.) seedlings exposed to cadmium. Acta Societatis Botanicorum Poloniae 86. doi: 10.5586/asbp.3551
Chmielowska-Bak, J., Lefevre, I., Lutts, S., Deckert, J. (2013). Short term signaling responses in roots of young soybean seedlings exposed to cadmium stress. J. Plant Physiol. 170, 1585–1594. doi: 10.1016/j.jplph.2013.06.019
Chmur, M., Bajguz, A., Piotrowska-Niczyporuk, A. (2020). Effect of cadmium on the level of isoprenoid-derived phytohormones in duckweedWolffia arrhiza. J. Plant Growth Regul. 39, 1518–1530. doi: 10.1007/s00344-020-10154-9
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Dang, F. F., Li, Y. J., Wang, Y. F., Lin, J. H., Du, S. X., Liao, X. Y. (2022). ZAT10 plays dual roles in cadmium uptake and detoxification in Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994100
Dang, F. F., Lin, J. H., Chen, Y. P., Li, G. X., Guan, D. Y., Zheng, S. J., et al. (2019). A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J. Exp. Bot. 70, 1581–1595. doi: 10.1093/jxb/erz006
De Mortel, J. E. V., Schat, H., Moerland, P. D., Van Themaat, E. V. L., van der Ent, S., Blankestijn, H., et al. (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 31, 301–324. doi: 10.1111/j.1365-3040.2007.01764.x
Devereux, R. B., Navas-Acien, A. (2013). Cadmium exposure and incident cardiovascular disease. Epidemiology 24, 421–429. doi: 10.1097/EDE.0b013e31828b0631
Djemal, R., Khoudi, H. (2022). The ethylene-responsive transcription factor of durum wheat, TdSHN1, confers cadmium, copper, and zinc tolerance to yeast and transgenic tobacco plants. Protoplasma 259, 19–31. doi: 10.1007/s00709-021-01635-z
Droge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The Arabidopsis bZIP transcription factor family - an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Du, X. Y., Fang, L. H., Li, J. X., Chen, T. J., Cheng, Z., Zhu, B., et al. (2023). The TabHLH094-TaMYC8 complex mediates the cadmium response in wheat. Mol. Breed. 43. doi: 10.1007/s11032-023-01404-1
El Rasafi, T., Oukarroum, A., Haddioui, A., Song, H., Kwon, E. E., Bolan, N., et al. (2022). Cadmium stress in plants: A critical review of the effects, mechanisms, and tolerance strategies. Crit. Rev. Environ. Sci. Technol. 52, 675–726. doi: 10.1080/10643389.2020.1835435
Erpen, L., Devi, H. S., Grosser, J. W., Dutt, M. (2018). Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 132, 1–25. doi: 10.1007/s11240-017-1320-6
Eulgem, T., Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Farinati, S., DalCorso, G., Varotto, S., Furini, A. (2010). The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol. 185, 964–978. doi: 10.1111/j.1469-8137.2009.03132.x
Fasani, E., DalCorso, G., Costa, A., Zenoni, S., Furini, A. (2019). The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol. Biol. 99, 517–534. doi: 10.1007/s11103-019-00833-x
Feng, Z. H., Gao, B., Gao, Y., Wu, J. H. (2023). Cloning of PsMYB62 and analysis of cadmium resistant in Potentilla sericea. Biocell 47, 1571–1582. doi: 10.32604/biocell.2023.029106
Gao, F., Robe, K., Gaymard, F., Izquierdo, E., Dubos, C. (2019). The transcriptional control of iron homeostasis in plants: A tale of bHLH transcription factors? Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00006
Genchi, G., Sinicropi, M. S., Lauria, G., Carocci, A., Catalano, A. (2020). The effects of cadmium toxicity. Int. J. Environ. Res. Public Health 17. doi: 10.3390/ijerph17113782
Grennan, A. K. (2011). Metallothioneins, a diverse protein family. Plant Physiol. 155, 1750–1751. doi: 10.1104/pp.111.900407
Gu, L., Hou, Y. Y., Sun, Y. Y., Chen, X. X., Wang, G. Y., Wang, H. C., et al. (2024). The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep. 43, 44. doi: 10.1007/s00299-023-03112-8
Guan, C. F., Ji, J., Li, X. Z., Jin, C., Wang, G. (2016). LcMKK, a MAPK kinase from Lycium chinense, confers cadmium tolerance in transgenic tobacco by transcriptional upregulation of ethylene responsive transcription factor gene. J. Genet. 95, 875–885. doi: 10.1007/s12041-016-0710-6
Guan, C. F., Ji, J., Wu, D. Y., Li, X. Z., Jin, C., Guan, W. Z., et al. (2015). The glutathione synthesis may be regulated by cadmium-induced endogenous ethylene in Lycium chinense, and overexpression of an ethylene responsive transcription factor gene enhances tolerance to cadmium stress in tobacco. Mol. Breed. 35, 123. doi: 10.1007/s11032-015-0313-6
Guandi, H., Muhammad, S., Tingfei, D., Zhuoyan, Z., Tengbing, H., Jiahai, W. (2023). Unraveling the mechanism of stWRKY6 in potato (Solanum tuberosum)s cadmium tolerance for ensuring food safety. Foods 12, 2303. doi: 10.3390/foods12122303
Halimaa, P., Blande, D., Baltzi, E., Aarts, M. G. M., Granlund, L., Keinanen, M., et al. (2019). Transcriptional effects of cadmium on iron homeostasis differ in calamine accessions of Noccaea caerulescens. Plant J. 97, 306–320. doi: 10.1111/tpj.14121
Han, G. H., Huang, R. N., Hong, L. H., Xu, J. X., Hong, Y. G., Wu, Y. H., et al. (2023). The transcription factor NAC102 confers cadmium tolerance by regulating WAKL11 expression and cell wall pectin metabolism in Arabidopsis. J. Integr. Plant Biol. 65, 2262–2278. doi: 10.1111/jipb.13557
Han, Y. Y., Fan, T. T., Zhu, X. Y., Wu, X., Ouyang, J., Jiang, L., et al. (2019). WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol. Biol. 99, 149–159. doi: 10.1007/s11103-018-0809-7
Hasan, M. K., Liu, C. C., Wang, F. N., Ahammed, G. J., Zhou, J., Xu, M. X., et al. (2016). Glutathione-mediated regulation of nitric oxide, S-nitrosothiol and redox homeostasis confers cadmium tolerance by inducing transcription factors and stress response genes in tomato. Chemosphere 161, 536–545. doi: 10.1016/j.chemosphere.2016.07.053
He, Y., Zhang, X., Xie, M., Chen, X., Zhao, T., Chen, Y., et al. (2022). Cd stress response function analysis of bpbHLH149 transcription factor from broussonetia papyifera in yeast. J. North-East Forest. Univ. 50, 21–26.
He, Q. H., Zhou, T., Sun, J. K., Wang, P., Yang, C. P., Bai, L., et al. (2021). Transcriptome profiles of leaves and roots of goldenrain tree (Koelreuteria paniculata laxm.) in response to cadmium stress. Int. J. Environ. Res. Public Health 18, 12046. doi: 10.3390/ijerph182212046
Hong, C. Y., Cheng, D., Zhang, G. Q., Zhu, D. D., Chen, Y. H., Tan, M. P. (2017). The role of ZmWRKY4 in regulating maize antioxidant defense under cadmium stress. Biochem. Biophys. Res. Commun. 482, 1504–1510. doi: 10.1016/j.bbrc.2016.12.064
Hu, S. B., Yu, Y., Chen, Q. H., Mu, G. M., Shen, Z. G., Zheng, L. Q. (2017). OsMYB45 plays an important role in rice resistance to cadmium stress. Plant Sci. 264, 1–8. doi: 10.1016/j.plantsci.2017.08.002
Huang, Y. F., Chen, J. H., Zhang, D. R., Fang, B., YangJin, T., Zou, J. W., et al. (2021). Enhanced vacuole compartmentalization of cadmium in root cells contributes to glutathione-induced reduction of cadmium translocation from roots to shoots in pakchoi (Brassica chinensis L.). Ecotoxicol. Environ. Saf. 208. doi: 10.1016/j.ecoenv.2020.111616
Huang, C. J., Zhou, J. H., Jie, Y. C., Xing, H. C., Zhong, Y. L., Yu, W. L., et al. (2016). A ramie bZIP transcription factor bnbZIP2 is involved in drought, salt, and heavy metal stress response. DNA Cell Biol. 35, 776–786. doi: 10.1089/dna.2016.3251
Imran, M., Khan, A. L., Mun, B. G., Bilal, S., Shaffique, S., Kwon, E. H., et al. (2022). Melatonin and nitric oxide: Dual players inhibiting hazardous metal toxicity in soybean plants via molecular and antioxidant signaling cascades. Chemosphere 308, 3. doi: 10.1016/j.chemosphere.2022.136575
Jia, Z. Z., Li, M. Z., Wang, H. C., Zhu, B., Gu, L., Du, X. Y., et al. (2021). TaWRKY70 positively regulates TaCAT5 enhanced Cd tolerance in transgenic Arabidopsis. Environ. Exp. Bot. 190. doi: 10.1016/j.envexpbot.2021.104591
Jung, J., Kim, M. (2007). Overexpression of the acidic C-terminus region of HvRAF from barley confers enhanced cadmium tolerance in yeast. J. Plant Biol. 50, 383–384. doi: 10.1007/BF03030672
Jung Won, Y., Jae Heung, J., Doil, C., So Young, Y., Hyouk, J., Hyun Soon, K. (2008). Ectopic expression of pepper CaPF1 in potato enhances multiple stresses tolerance and delays initiation of in vitro tuberization. Planta 228, 701–708. doi: 10.1007/s00425-008-0782-5
Kang, L. H., Teng, Y. Y., Cen, Q. W., Fang, Y. X., Tian, Q. X., Zhang, X. Q., et al. (2022). Genome-wide identification of R2R3-MYB transcription factor and expression analysis under abiotic stress in rice. Plants-Basel 11, 15. doi: 10.3390/plants11151928
Kaya, C., Ashraf, M., AlYemeni, M. N., Ahmad, P. (2020). Responses of nitric oxide and hydrogen sulfide in regulating oxidative defence system in wheat plants grown under cadmium stress. Physiol. Plant. 168, 345–360. doi: 10.1111/ppl.13012
Kaya, C., Okant, M., Ugurlar, F., AlYemeni, M. N., Ashraf, M., Ahmad, P. (2019). Melatonin-mediated nitric oxide improves tolerance to cadmium toxicity by reducing oxidative stress in wheat plants. Chemosphere 225, 627–638. doi: 10.1016/j.chemosphere.2019.03.026
Khanna, K., Kohli, S. K., Ohri, P., Bhardwaj, R., Ahmad, P. (2022). Agroecotoxicological aspect of cd in soil-plant system: uptake, translocation and amelioration strategies. Environ. Sci. pollut. Res. 29, 30908–30934. doi: 10.1007/s11356-021-18232-5
Kouno, T., Ezaki, B. (2013). Multiple regulation of Arabidopsis AtGST11 gene expression by four transcription factors under abiotic stresses. Physiol. Plant. 148, 97–104. doi: 10.1111/j.1399-3054.2012.01699.x
Lee, H.-O., Hong, Y., Etlioglu, H. E., Cho, Y. B., Pomelle, V., Van den Bosch, B., et al. (2020). Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 52, 594. doi: 10.1038/s41588-020-0636-z
Lei, G. J., Sun, L., Sun, Y., Zhu, X. F., Li, G. X., Zheng, S. J. (2020). Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 62, 218–227. doi: 10.1111/jipb.12801
Li, J., Abbas, M., Desoky, E. M., Zafar, S., Soaud, S. A., Hussain, S. S., et al. (2023). Analysis of metal tolerance protein (MTP) family in sunflower (Helianthus annus L.) and role of HaMTP10 as Cadmium antiporter under moringa seed extract. Ind. Crops Prod. 202, 117023. doi: 10.1016/j.indcrop.2023.117023
Li, D. D., He, G. D., Tian, W. J., Saleem, M., Huang, Y., Meng, L. L., et al. (2021a). Comparative and systematic omics revealed low cd accumulation of potato stMTP9 in yeast: suggesting a new mechanism for heavy metal detoxification. Int. J. Mol. Sci. 22, 19. doi: 10.3390/ijms221910478
Li, H., Li, D., Deng, Y., Pan, G., Chen, A., Zhao, L., et al. (2021b). Expression analysis of abiotic stress response gene HcWRKY71 in kenaf and transformation of Arabidopsis. Acta Agronom. Sin. 47, 1090–1099.
Li, G. Z., Zheng, Y. X., Liu, H. T., Liu, J., Kang, G. Z. (2022). WRKY74 regulates cadmium tolerance through glutathione-dependent pathway in wheat. Environ. Sci. pollut. Res. 29, 68191–68201. doi: 10.1007/s11356-022-20672-6
Li, T.-z., Zheng, Y.-z., Rong, Y.-q., Wei, S.-l., Wang, X.-h., Tu, P.-f. (2020). Gene cloning, subcellular localization and expression analysis of the AsERF1 gene from Aquilaria sinensis. Yaoxue Xuebao 55, 1957–1964. doi: 10.16438/j.0513-4870.2020-0176
Liao, C. J., Li, Y. Q., Wu, X. H., Wu, W. M., Zhang, Y., Zhan, P. L., et al. (2023). ZmHMA3, a member of the heavy-metal-transporting ATPase family, regulates cd and zn tolerance in maize. Int. J. Mol. Sci. 24, 13496. doi: 10.3390/ijms241713496
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lin, Y. F., Hassan, Z., Talukdar, S., Schat, H., Aarts, M. G. M. (2016). Expression of the ZNT1 Zinc Transporter from the Metal Hyperaccumulator Noccaea caerulescens Confers Enhanced Zinc and Cadmium Tolerance and Accumulation to Arabidopsis thaliana. PloS One 11, 3. doi: 10.1371/journal.pone.0149750
Lin, T. T., Yang, W. N., Lu, W., Wang, Y., Qi, X. T. (2017). Transcription factors pvERF15 and pvMTF-1 form a cadmium stress transcriptional pathway. Plant Physiol. 173, 1565–1573. doi: 10.1104/pp.16.01729
Liu, X., Chen, H., Zhang, H., Yuan, X., Cui, X., Chen, X. (2018). Bioinformatics analysis of gmbHLH041 of soybean and its response to cadmium stress. Acta Agricult. Boreali-Sinica 33, 14–18.
Liu, Z. Q., Fang, H. H., Pei, Y. X., Jin, Z. P., Zhang, L. P., Liu, D. M. (2015). WRKY transcription factors down-regulate the expression of H2S-generating genes, LCD and DES in Arabidopsis thaliana. Sci. Bull. 60, 995–1001. doi: 10.1007/s11434-015-0787-y
Liu, X. J., Wang, H. C., He, F., Du, X. Y., Ren, M. J., Bao, Y. G. (2022a). The taWRKY22-taCOPT3D pathway governs cadmium uptake in wheat. Int. J. Mol. Sci. 23, 18. doi: 10.3390/ijms231810379
Liu, Z., Zhang, Y. X., Altaf, M. A., Hao, Y. Y., Zhou, G. Z., Li, X. Y., et al. (2022b). Genome-wide identification of myeloblastosis gene family and its response to cadmium stress in Ipomoea aquatica. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.979988
Lu, Z., Qiu, W., Jin, K., Yu, M., Han, X., He, X., et al. (2022). Identification and Analysis of bZIP Family Genes in Sedum plumbizincicola and Their Potential Roles in Response to Cadmium Stress. Front. Plant Sci. 13, 859386. doi: 10.3389/fpls.2022.859386
Ma, S. J., Zheng, L. W., Liu, X. H., Zhang, K. Y., Hu, L. L., Hua, Y. P., et al. (2022). Genome-wide identification of brassicaceae hormone-related transcription factors and their roles in stress adaptation and plant height regulation in allotetraploid rapeseed. Int. J. Mol. Sci. 23, 15. doi: 10.3390/ijms23158762
Mei, L., Pengfei, H., Fangbin, C. (2017). Glutathione-induced alleviation of cadmium toxicity in Zea mays. Plant Physiol. Biochem. 119, 240–249. doi: 10.1016/j.plaphy.2017.09.005
Meng, X. X., Li, W. F., Shen, R. F., Lan, P. (2022). Ectopic expression of IMA small peptide genes confers tolerance to cadmium stress in Arabidopsis through activating the iron deficiency response. J. Hazard. Mater. 422. doi: 10.1016/j.jhazmat.2021.126913
Okereafor, U., Makhatha, M., Mekuto, L., Uche-Okereafor, N., Sebola, T., Mavumengwana, V. (2020). Toxic metal implications on agricultural soils, plants, animals, aquatic life and human health. Int. J. Environ. Res. Public Health 17, 2204. doi: 10.3390/ijerph17072204
Paul, S., Roychoudhury, A. (2018). Transcriptome profiling of abiotic stress-responsive genes during cadmium chloride-mediated stress in two indica rice varieties. J. Plant Growth Regul. 37, 657–667. doi: 10.1007/s00344-017-9762-y
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. X. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sabella, E., Luvisi, A., Genga, A., De Bellis, L., Aprile, A. (2021). Molecular responses to cadmium exposure in two contrasting durum wheat genotypes. Int. J. Mol. Sci. 22, 7343. doi: 10.3390/ijms22147343
Saifullah, Sarwar, N., Bibi, S., Ahmad, M., Ok, Y. S. (2014). Effectiveness of zinc application to minimize cadmium toxicity and accumulation in wheat (Triticum aestivum L.). Environ. Earth Sci. 71, 1663–1672. doi: 10.1007/s12665-013-2570-1
Sapara, K. K., Khedia, J., Agarwal, P., Gangapur, D. R., Agarwal, P. K. (2019). SbMYB15 transcription factor mitigates cadmium and nickel stress in transgenic tobacco by limiting uptake and modulating antioxidative defence system. Funct. Plant Biol. 46, 702–714. doi: 10.1071/FP18234
Seregin, I. V., Kozhevnikova, A. D. (2023). Phytochelatins: sulfur-containing metal(loid)-chelating ligands in plants. 24, 2430. doi: 10.3390/ijms24032430
Shalmani, A., Ullah, U., Muhammad, I., Zhang, D., Sharif, R., Jia, P., et al. (2021). The TAZ domain-containing proteins play important role in the heavy metals stress biology in plants. Environ. Res. 197. doi: 10.1016/j.envres.2021.111030
Shao, X., Li, T., Li, Z., Yu, L., Li, Z., Xu, X. (2015). Genome-wide identification and expression analysis in oxidative stress of AP2/ERF gene family in eggplant (Solanum melongena). Plant Physiol. J. 51, 1901–1918.
Sheng, Y. B., Yan, X. X., Huang, Y., Han, Y. Y., Zhang, C., Ren, Y. B., et al. (2019). The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 42, 891–903. doi: 10.1111/pce.13457
Song, Y., Jin, L., Wang, X. J. (2017). Cadmium absorption and transportation pathways in plants. Int. J. Phytoremed. 19, 133–141. doi: 10.1080/15226514.2016.1207598
Sterckeman, T., Thomine, S. (2020). Mechanisms of cadmium accumulation in plants. Crit. Rev. Plant Sci. 39, 322–359. doi: 10.1080/07352689.2020.1792179
Strader, L., Weijers, D., Wagner, D. (2022). Plant transcription factors-being in the right place with the right company. Curr. Opin. Plant Biol. 65. doi: 10.1016/j.pbi.2021.102136
Su, T. T., Fu, L. B., Kuang, L. H., Chen, D. Y., Zhang, G. P., Shen, Q. F., et al. (2022). Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J. Hazard. Mater. 423, 127140. doi: 10.1016/j.jhazmat.2021.127140
Sun, L. J., Wang, J., Song, K., Sun, Y. F., Qin, Q., Xue, Y. (2019). Transcriptome analysis of rice (Oryza sativa L.) shoots responsive to cadmium stress. Sci. Rep. 9, 10177. doi: 10.1038/s41598-019-46684-w
Tan, M. P., Cheng, D., Yang, Y. N., Zhang, G. Q., Qin, M. J., Chen, J., et al. (2017). Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 17, 194. doi: 10.1186/s12870-017-1143-y
Tang, W., Charles, T. M., Newton, R. J. (2005). Overexpression of the pepper transcription factor CaPF1 in transgenic virginia pine (Pinus virginiana mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 59, 603–617. doi: 10.1007/s11103-005-0451-z
Thomas, M., Reid, R. (2021). Vacuolar compartmentalisation and efflux of cadmium in barley. Botany 99, 1–8. doi: 10.1139/cjb-2020-0080
Tian, W. J., Huang, Y., Li, D. D., Meng, L. L., He, T. B., He, G. D. (2022). Identification of StAP2/ERF genes of potato (Solanum tuberosum) and their multiple functions in detoxification and accumulation of cadmium in yest: Implication for Genetic-based phytoremediation. Sci. Total Environ. 810. doi: 10.1016/j.scitotenv.2021.152322
Tinkov, A. A., Filippini, T., Ajsuvakova, O. P., Skalnaya, M. G., Aaseth, J., Bjorklund, G., et al. (2018). Cadmium and atherosclerosis: A review of toxicological mechanisms and a meta-analysis of epidemiologic studies. Environ. Res. 162, 240–260. doi: 10.1016/j.envres.2018.01.008
Wang, Y. C., Gao, C. Q., Liang, Y. N., Wang, C., Yang, C. P., Liu, G. F. (2010). A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol. 167, 222–230. doi: 10.1016/j.jplph.2009.09.008
Wang, J., Qiu, W., Jin, K., Lu, Z., Han, X., Zhuo, R., et al. (2023). Comprehensive analysis of WRKY gene family in Sedum plumbizincicola responding to cadmium stress. J. Nanjing Forest. University. Natural Sci. Edit. 47, 49–60.
Wang, Y., Qiu, W., Li, H., He, X., Liu, M., Han, X., et al. (2019). Research on the response of SaWRKY7 gene to cadmium stress in Sedum alfredii Hance. J. Nanjing Forest. University. Natural Sci. Edit. 43, 59–66.
Wang, Z. G., Zhang, B. L., Chen, Z. W., Wu, M. J., Chao, D., Wei, Q. X., et al. (2022b). Three OsMYB36 members redundantly regulate Casparian strip formation at the root endodermis. Plant Cell 34, 2948–2968. doi: 10.1093/plcell/koac140
Wang, H. C., Zuo, D., Zhu, B., Du, X. Y., Gu, L. (2022a). TaMYC8 regulates TaERF6 and inhibits ethylene synthesis to confer Cd tolerance in wheat. Environ. Exp. Bot. 198. doi: 10.1016/j.envexpbot.2022.104854
Wu, X. L., Chen, Q., Chen, L. L., Tian, F. F., Chen, X. X., Han, C. Y., et al. (2022). A WRKY transcription factor, PyWRKY75, enhanced cadmium accumulation and tolerance in poplar. Ecotoxicol. Environ. Saf. 239. doi: 10.1016/j.ecoenv.2022.113630
Wu, H. L., Chen, C. L., Du, J., Liu, H., Cui, Y., Zhang, Y., et al. (2012). Co-overexpression FIT with atbHLH38 or atbHLH39 in arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158, 790–800. doi: 10.1104/pp.111.190983
Wu, X. L., Chen, L. L., Lin, X. Y., Chen, X. X., Han, C. Y., Tian, F. F., et al. (2023a). Integrating physiological and transcriptome analyses clarified the molecular regulation mechanism of PyWRKY48 in poplar under cadmium stress. Int. J. Biol. Macromol. 238. doi: 10.1016/j.ijbiomac.2023.124072
Wu, Q., Meng, Y. T., Feng, Z. H., Shen, R. F., Zhu, X. F. (2023b). The endo-beta mannase MAN7 contributes to cadmium tolerance by modulating root cell wall binding capacity in Arabidopsis thaliana. J. Integr. Plant Biol. 65, 1670–1686. doi: 10.1111/jipb.13487
Xian, P. Q., Yang, Y., Xiong, C. W., Guo, Z. B., Alam, I., He, Z. H., et al. (2023). Overexpression of GmWRKY172 enhances cadmium tolerance in plants and reduces cadmium accumulation in soybean seeds. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1133892
Xie, Q. Q., Yu, Q., Jobe, T. O., Pham, A., Ge, C. N., Guo, Q. Q., et al. (2021). An amiRNA screen uncovers redundant CBF and ERF34/35 transcription factors that differentially regulate arsenite and cadmium responses. Plant Cell Environ. 44, 1692–1706. doi: 10.1111/pce.14023
Xie, Y. F., Zhang, R. X., Qin, L. J., Song, L. L., Zhao, D. G., Xia, Z. M. (2022). Genome-wide identification and genetic characterization of the CaMYB family and its response to five types of heavy metal stress in hot pepper (Capsicum annuum cv. CM334). Plant Physiol Biochem. 170, 98–109. doi: 10.1016/j.plaphy.2021.11.024
Xin, P. F., Gao, C. S., Cheng, C. H., Tang, Q., Dong, Z. X., Zhao, L. N., et al. (2016). Identification and characterization of hemp WRKY transcription factors in response to abiotic stresses. Biol. Plant. 60, 489–495. doi: 10.1007/s10535-016-0621-7
Xu, Z. G., Dong, M., Peng, X. Y., Ku, W. Z., Zhao, Y. L., Yang, G. Y. (2019). New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotoxicol. Environ. Saf. 171, 301–312. doi: 10.1016/j.ecoenv.2018.12.084
Xu, Z. G., Ge, Y., Zhang, W., Zhao, Y. L., Yang, G. Y. (2018). The walnut JrVHAG1 gene is involved in cadmium stress response through ABA-signal pathway and MYB transcription regulation. BMC Plant Biol. 18, 19. doi: 10.1186/s12870-018-1231-7
Xu, Z. L., Liu, X. Q., He, X. L., Xu, L., Huang, Y. H., Shao, H. B., et al. (2017). The soybean basic helix-loop-helix transcription factor ORG3-like enhances cadmium tolerance via increased iron and reduced cadmium uptake and transport from roots to shoots. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01098
Xu, H., Xu, W. Z., Xi, H. M., Ma, W. W., He, Z. Y., Ma, M. (2013). The ER luminal binding protein (BiP) alleviates Cd2+-induced programmed cell death through endoplasmic reticulum stress-cell death signaling pathway in tobacco cells. J. Plant Physiol. 170, 1434–1441. doi: 10.1016/j.jplph.2013.05.017
Yan, C., Feng, B., Zhao, Z., Zhang, Y., Yin, K., Liu, Y., et al. (2024). Populus euphratica R2R3-MYB transcription factor RAX2 binds ANN1 promoter to increase cadmium enrichment in Arabidopsis. Plant sci.: an Int. J. Exp. Plant Biol. 344, 112082. doi: 10.1016/j.plantsci.2024.112082
Yang, C. Q., Fang, X., Wu, X. M., Mao, Y. B., Wang, L. J., Chen, X. Y. (2012). Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 54, 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yang, G. Z., Fu, S., Huang, J. J., Li, L. Y., Long, Y., Wei, Q. X., et al. (2021). The tonoplast-localized transporter OsABCC9 is involved in cadmium tolerance and accumulation in rice. Plant Sci. 307, 110894. doi: 10.1016/j.plantsci.2021.110894
Yang, G. Y., Wang, C., Wang, Y. C., Guo, Y. C., Zhao, Y. L., Yang, C. P., et al. (2016). Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of Cadmium stress. Sci. Rep. 6, 18752. doi: 10.1038/srep18752
Yao, X. N., Cai, Y. R., Yu, D. Q., Liang, G. (2018). bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 60, 691–702. doi: 10.1111/jipb.12658
Yao, W. J., Zhang, X. M., Zhou, B. R., Zhao, K., Li, R. H., Jiang, T. B. (2017). Expression Pattern of ERF Gene Family under Multiple Abiotic Stresses in Populus simonii x P-nigra. Front. Plant Sci. 8, 181. doi: 10.3389/fpls.2017.00181
Yao, W. J., Zhou, B. R., Zhang, X. M., Zhao, K., Cheng, Z. H., Jiang, T. B. (2019). Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii x P.nigra. Gene 707, 189–197. doi: 10.1016/j.gene.2019.04.071
Yin, M., Pan, G., Tao, J., Doblin, M. S., Zeng, W., Pan, L. L., et al. (2022). Identification of MYB genes reveals their potential functions in cadmium stress response and the regulation of cannabinoid biosynthesis in hemp. Ind. Crops Prod. 180, 114607. doi: 10.1016/j.indcrop.2022.114607
Yu, W. J., Deng, S. R., Chen, X., Cheng, Y., Li, Z. R., Wu, J. T., et al. (2022). PcNRAMP1 Enhances Cadmium Uptake and Accumulation in Populus x canescens. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23147593
Yu, Y. A., Zhang, L. (2023). The wheat NAC transcription factor TaNAC22 enhances cadmium stress tolerance in wheat. Cereal Res. Commun. 51, 867–877. doi: 10.1007/s42976-023-00354-w
Yuan, J. B., Bai, Y. Q., Chao, Y. H., Sun, X. B., He, C. Y., Liang, X. H., et al. (2018). Genome-wide analysis reveals four key transcription factors associated with cadmium stress in creeping bentgrass (Agrostis stolonifera L.). Peerj 6, 15. doi: 10.7717/peerj.5191
Yuan, X. H., Xue, N. D., Han, Z. G. (2021). A meta-analysis of heavy metals pollution in farmland and urban soils in China over the past 20 years. J. Environ. Sci. 101, 217–226. doi: 10.1016/j.jes.2020.08.013
Yuan, L. Y., Yang, S. G., Liu, B. X., Zhang, M., Wu, K. Q. (2012). Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 31, 67–79. doi: 10.1007/s00299-011-1140-9
Yue, X. M., Song, J. X., Fang, B., Wang, L., Zou, J. W., Su, N. N., et al. (2021). BcNRAMP1 promotes the absorption of cadmium and manganese in Arabidopsis. Chemosphere 283, 131113. doi: 10.1016/j.chemosphere.2021.131113
Zare, L., Ronaghi, A., Ghasemi, R., Zarei, M., Sepehri, M. (2023). Alterations in glutathione, phytochelatin and micronutrients of corn plants exposed to cadmium stress at different time periods. Commun. Soil Sci. Plant Anal. 54, 1185–1197. doi: 10.1080/00103624.2022.2138909
Zhan, J. H., Zou, W. L., Li, S. Y. Y., Tang, J. C., Lu, X., Meng, L. J., et al. (2022). OsNAC15 regulates tolerance to zinc deficiency and cadmium by binding to osZIP7 and osZIP10 in rice. Int. J. Mol. Sci. 23, 11771. doi: 10.3390/ijms231911771
Zhang, Q., Cai, W., Ji, T. T., Ye, L., Lu, Y. T., Yuan, T. T. (2020). WRKY13 enhances cadmium tolerance by promoting D-CYSTEINE DESULFHYDRASE and hydrogen sulfide production(1). Plant Physiol. 183, 345–357. doi: 10.1104/pp.19.01504
Zhang, X., Ling, H., Liu, F., Huang, N., Wang, L., Mao, H., et al. (2018). Cloning and expression analysis of a IId sub-group WRKY transcription factor gene from sugarcane. Sci. Agricult. Sin. 51, 4409–4423.
Zhang, J.-y., Qu, S.-c., Guo, Z.-r., Du, X.-l., Du, B.-b., Zhang, Z. (2011). Biology function of bZIP transcription factors in plants. Acta Botanica Boreali-Occidentalia Sin. 31, 1066–1075.
Zhang, P., Wang, R. L., Ju, Q., Li, W. Q., Tran, L. S. P., Xu, J. (2019). The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 180, 529–542. doi: 10.1104/pp.18.01380
Zhang, G. B., Yi, H. Y., Gong, J. M. (2014). The arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26, 3984–3998. doi: 10.1105/tpc.114.129296
Zheng, T., Lu, X. B., Yang, F., Zhang, D. W. (2022). Synergetic modulation of plant cadmium tolerance via MYB75-mediated ROS homeostasis and transcriptional regulation. Plant Cell Rep. 41, 1515–1530. doi: 10.1007/s00299-022-02871-0
Zheng, J. J., Zhang, Z. L., Tong, T., Fang, Y. X., Zhang, X., Niu, C. Y., et al. (2021). Genome-wide identification of WRKY gene family and expression analysis under abiotic stress in barley. Agronomy-Basel 11, 521. doi: 10.3390/agronomy11030521
Zheng, X., Zhu, S. Y., Tang, S. W., Liu, T. M. (2016). Identification of drought, cadmium and root-lesion nematode infection stress-responsive transcription factors in ramie. Open Life Sci. 11, 191–199. doi: 10.1515/biol-2016-0025
Zhu, H. H., Chen, L., Xing, W., Ran, S. M., Wei, Z. H., Amee, M., et al. (2020a). Phytohormones-induced senescence efficiently promotes the transport of cadmium from roots into shoots of plants: A novel strategy for strengthening of phytoremediation. J. Hazard. Mater. 388, 122080. doi: 10.1016/j.jhazmat.2020.122080
Zhu, S., Shi, W. (2019). Cloning and expression pattern analysis of bnMYB3 transcription factor gene in ramie. Acta Botanica Boreali-Occidentalia Sin. 39, 422–429. doi: 10.7606/j.issn.1000-4025.2019.03.0422
Zhu, S., Shi, W., Jie, Y., Zhou, Q. (2018). Cloning and expression analysis of cadmium-responsive transcription factor gene bnMYB1 from ramie (Boehmeria nivea). Journal Agric. Biotechnol. 26, 774–783.
Keywords: cadmium, transcription factor, WRKY, ERF, MYB, bHLH
Citation: Zhang H and Lu L (2024) Transcription factors involved in plant responses to cadmium-induced oxidative stress. Front. Plant Sci. 15:1397289. doi: 10.3389/fpls.2024.1397289
Received: 07 March 2024; Accepted: 15 May 2024;
Published: 13 June 2024.
Edited by:
Luis E. Hernandez, Autonomous University of Madrid, SpainReviewed by:
Shikha Verma, Ben-Gurion University of the Negev, IsraelCopyright © 2024 Zhang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingli Lu, bHVsaW5nbGlAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.