- 1College of Agriculture, Institute of Molecular Agriculture and Bioenergy, Shanxi Agricultural University, Jinzhong, Shanxi, China
- 2Key Laboratory of Coastal Biology and Bio-Resource Utilization, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, Shandon, China
Calcium-dependent protein kinases (CDPKs) serve as calcium ion sensors and play crucial roles in all aspects of plant life cycle. While CDPK gene family has been extensively studied in various plants, there is limited information available for CDPK members in oat, an important cereal crop worldwide. Totally, 60 AsCDPK genes were identified in oat genome and were classified into four subfamilies based on their phylogenetic relationship. The members within each subfamily shared similar gene structure and conserved motifs. Collinearity analysis revealed that AsCDPK gene amplification was attributed to segmental duplication events and underwent strong purifying selection. AsCDPK promoters were predicted to contain cis-acting elements associated with hormones, biotic and abiotic stresses. AsCDPK gene expressions were induced by different salt stresses, exhibiting stress-specific under different salt treatments. Moreover, overexpression of AsCDPK26 gene enhanced salt resistance in C. reinhardtii, a single-cell photoautotrophic model plants. Further analysis revealed a significant correlation between AsCDPK26 and Na+/H+ antiporter 1 (p<0.05), suggesting that AsCDPK26 may interact with ion transporter to modulate salt resistance. These results not only provide valuable insights into AsCDPK genes in response to different salt stresses, but also lay the foundation to mine novel candidates for improving salt tolerance in oat and other crops.
1 Introduction
Plants have developed a series of effective defense mechanisms to cope with complex and variable environmental conditions. Signal transduction pathways are crucial in these processes (Trewavas and Malhó, 1998). Calcium ion (Ca2+), as ubiquitous secondary messengers, play a vital role in signal transduction pathways. Moreover, other molecules also play significant roles in plant stress responses, including small lipid molecules, cAMP (cyclic adenosine monophosphate), and cGMP (cyclic guanosine monophosphate) (Trewavas and Gilroy, 1991; Zhao et al., 2021a). However, free Ca2+ has the widest range of action. When plants encounter abiotic stresses such as temperature, light, salt, and osmotic stress, certain calcium receptors or calcium binding proteins can detect the changes in Ca2+ concentration and recognize the calcium signals. These proteins then transmit signals to downstream components through phosphorylation, leading to the expression of the related genes (Harmon et al., 2001). This mechanism helps plants to cope with environmental stress and enhance their resistance. Plant cells contain a variety of calcium-binding proteins, such as calmodulin (CaM), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinase (CDPKs or CPKs) (Cheng et al., 2002).
CDPK can sense and respond to changes in Ca2+ concentration, thereby regulating various crucial cell signaling processes. Unlike other Ca2+ sensors, CDPK has both Ca2+ binding and kinase activities, allowing it to bind Ca2+ directly and convert the signals into phosphorylation events downstream (Boudsocq and Sheen, 2013). The typical CDPK protein structure consists of four main parts: the N-terminal variable region, the serine/threonine (Ser/Thr) kinase domain, the auto-inhibitory domain (also known as the connection domain), and the calmodulin-like binding domain (Cheng et al., 2002). CDPKs have been identified in several plant species, including Arabidopsis thaliana (Cheng et al., 2002), Triticum aestivum (Liu et al., 2023b), Oryza sativa (Asano et al., 2005), and Zea mays (Kong et al., 2013).
CDPK plays a crucial role in regulating plant growth and responses to various stresses (Kong et al., 2013). Studies have shown that CDPK members within the same plant species exhibit functional diversity, and there are quantitative and functional differences among CDPKs from different plants. For example, in Arabidopsis, the AtCPK1 mutants display sensitivity to salt and drought stress, whereas overexpression of AtCPK1 significantly enhances resistance to these stresses (Huang et al., 2018). Similarly, other CDPK genes in Arabidopsis, such as AtCPK3/4/11, also function as positive regulators in response to salt and drought stress (Mehlmer et al., 2010; Zhu et al., 2007). Conversely, AtCPK23 acts as a negative regulator in plant responses to drought and salt stress. The T-DNA insertion mutant cpk23 exhibits significantly enhanced tolerance to drought and salt stress, whereas the overexpression line is more sensitive to these stresses (Ma and Wu, 2007). In rice, over-expression of OsCDPK7 enhanced induction of some stress-responsive genes in response to salinity/drought, but not to cold stress (Saijo et al., 2000). OsCDPK13 and OsCPK17 are considered as important signaling components in rice response to cold stress (Almadanim et al., 2017; Komatsu et al., 2007). As a direct ‘sensor’ of Ca2+, CDPK can directly or indirectly regulate crucial downstream target proteins, including transcription factors (Yang et al., 2021), ion channels (Latz et al., 2013), or other proteins related to signaling pathways (Zou et al., 2015; Atif et al., 2019). For example, AtCPK8 plays an important role in regulating catalase CAT3 activity via phosphorylation, which is part of ABA-mediated stomatal regulation under drought stress (Zou et al., 2015). In peppers, CaWRKY27b is phosphorylated at Ser137 by CaCDPK29, leading to its translocation from the cytoplasm to the nucleus. This physical interaction enhances the function of CaWRKY40 as a positive regulator of anti-RSI immunity and heat tolerance in peppers (Yang et al., 2021).
Soil salinization is a critical environmental stressor that affects seed germination, crop growth, and productivity, posing a threat to agricultural yields and ecological security globally (Zhou et al., 2024). High sodium concentrations in saline soils limit plant water uptake and nutrient absorption (Zhao et al., 2021b). Water deficiency and nutritional imbalances induce both osmotic stress and ionic stress. These primary stresses can result in oxidative stress, which in turn triggers a cascade of secondary stresses (Zhu, 2002). More than 900 million hectares of the world’s land are affected by soil salinization, and there are no effective practices to control its spread (Fang et al., 2021). It is projected that the global population will reach 9.7 billion by 2050, and the surge in food demand due to this population growth compels us to develop saline areas for cultivation and to breed salt-tolerant crops (Gu et al., 2021). Oat (Avena sativa L.) cultivated extensively in the word is one of the most widely consumed cereal grains, with high nutritional value and potential health benefits for human and animals (Sánchez-Martín et al., 2014). As a traditional grain crop in saline areas, oat has less stringent cultivation soil requirements, exhibiting higher salt tolerance than wheat, rice, and other staple food crops (Han et al., 2014). These attributes render oats a promising crop for the improvement of saline-alkali lands. Therefore, identification and characterization of the salt tolerant genes from oat are urgently needed for breeding of elite oat varieties. As direct sensors of Ca2+, CDPKs play an important role in regulating plant growth and various stress responses. However, the CDPK gene family, especially those associating with salt tolerance, has not been well studied in oat. Understanding the molecular mechanisms of CDPK involvement in salt resistance or tolerance in oat will provide insight on how to improve plant salt stress resistance and is a critical step in improving agricultural productivity and food security. In this study, AsCDPK members in oat were systematically characterized, including their physicochemical properties, chromosome distribution, gene structure and duplication, synteny, and phylogenetic relationship. The expression patterns of AsCDPK genes under different salt stresses were examined using RNA-seq data and further verified by RT-qPCR analysis. Moreover, possible interacting ion transporters were predicted through correlation analysis. Particularly, AsCDPK26 gene was cloned and functionally characterized by heterogenous overexpression in Chlamydomonas reinhardtii (CC849 strain), a single-cell photoautotrophic model plant, followed by phenotypic analysis of the transgenic lines under different salt stresses. And the correlation between AsCDPK26 and ion transporters in Chlamydomonas was also further verified by RT-qPCR analysis. This study provided valuable information for further exploration of the functions of oat AsCDPK genes in responses to salinity and other abiotic stresses, highlighting AsCDPK26 as a suitable target gene in plant biotechnology for improvement of stress resistance in crops.
2 Materials and methods
2.1 Experimental materials and treatments
The oat variety used in this study is CEav5651. This experiment was performed in the greenhouse, and the oat plants underwent hydroponic trials. Equal sized and healthy oat seeds were selected and sterilized using 3% NaClO for 10 min, followed by three rinses with distilled water. The seeds were kept on moist filter paper for germination. Incubation conditions were 16 h of light at 25°C, 8 h of darkness at 20°C, and humidity of 80%. After the seeds germinate fully, the seedlings were transplanted in a 96-hole water boxes. These boxes are filled with 1/2 Hoagland nutrient solution, with 48 seeds per box, cultivated in the greenhouse. The nutrient solution was replaced every 2 days. The greenhouse cultivation conditions consisted of 16 h of light at 25-30°C and 8 h of darkness at 15-18°C. When seedlings grew to two-leaf stage, the plants were treated with salt for 48 h. Saline and/or alkali treatments included three salt conditions (neutral salt stress (NaCl), alkaline salt stress (NaHCO3), and mixed salt-alkali stress (1:1 ratio of NaCl and NaHCO3)), two levels (100 and 200 mmol·L-1), and three replicates. The medium without saline and/or alkali treatment was the control group. Fresh aboveground tissues were immediately frozen in liquid nitrogen and stored at –80°C for transcriptome analysis.
2.2 Identification of AsCDPK gene family and analysis of basic parameters
The amino acid sequences of Arabidopsis AtCDPKs were downloaded from Phytozomev13 (https://phytozome-next.jgi.doe.gov/). The coding sequences (CDS) and the corresponding amino acid sequences were obtained from the oat genome database (https://wheat.pw.usda.gov/GG3/graingenes-downloads/pepsico-oat-oc3098-v2-files-2021). The AtCDPK sequences were used as queries to identify candidate AsCDPK genes by using BLASTP with an E-value less than e-10. Sequences containing both the Pkinase domain and EF-hand motif were filtered using the Interpro tool (http://www.ebi.ac.uk/interpro/). The composition of the identified candidate proteins was further verified in SMART databases (http://smart.embl-heidelberg.de/), while the sequences with errors, shorter length (<100 aa), and containing incomplete Ser/Thr kinase domain were eliminated. Amino acid number (aa), isoelectric point (pI), molecular weight (MW), and other parameters of AsCDPK proteins were predicted using the ExPASy Proteomics Server (https://web.expasy.org/protparam/) (Linghu et al., 2023). WoLF PSORT (https://wolfpsort.hgc.jp) (Yu et al., 2024) predictor was used to predict the subcellular localization of AsCDPK proteins, and the N-terminal myristoylation site was predicted using the GPS-Lipid 1.0 program with default settings and high threshold (Zhao et al., 2021a).
2.3 Multiple sequence alignments and phylogenetic analysis
Multiple alignments of CDPK protein sequences from oat, Arabidopsis, and rice were performed using the Clustal W program, with default parameters implemented in MEGA7.0 software (Chen et al., 2023). The phylogenetic tree was constructed by MEGA7.0 with the neighbor-joining method based on the sequence alignment results. The bootstrap value was set to 1000.
2.4 Chromosome mapping, gene structure and selection pressure analysis
The chromosomal distribution of the AsCDPK genes was visualized using TBtools software (Chen et al., 2020) based on the list of AsCDPK members and the oat genome annotation file. Synteny analysis was performed using the multicollinear scanning tool package (MCScanX) to identify the collinearity pattern of CDPKs among oat (Wang et al., 2023). Additionally, the values of nonsynonymous (Ka) and synonymous (Ks) substitution were calculated for fragment duplication gene pairs. The gene structure was displayed by comparing the coding sequence and the corresponding genomic DNA sequence using the gene structure display server tool (http://gsds.cbi.pku.edu.cn/).
2.5 Conserved motif and functional domain analysis
The conserved motifs were visualized using TBtools software, and each protein’s conserved motifs were analyzed using the MEME program (https://meme-suite.org) (Liu et al., 2023a). The maximum number of motifs was set to 20, and the optimal motif width was set to ranged from 10 to 100 amino acids. The remaining parameters were set to their default values. The Interpro (www.ebi.ac.uk/interpro) and SMART (smart.embl-heidelberg.de) databases were used to identify the functional domains and key sites in AsCDPKs. Subsequently, the TBtools toolkit (https://github.com/CJ-Chen/TBtools) was used to draw the diagram.
2.6 Analysis of cis-acting elements in the promoters of AsCDPK genes
The 2,000 bp promoter sequence upstream of each AsCDPK transcription start point was extracted from the oat genome database. The sequence was analyzed using the PlantCare online software (http://bioinformatics.psb.ugent.be/webtools/PlantCare/html/) to predict potential cis-acting regulatory elements (Cui et al., 2024).
2.7 AsCDPK gene expression profiles and RT- qPCR analysis
The expression patterns of AsCDPK genes were analyzed using a transcriptome database derived from oat seedlings subjected to various salt treatments. The expression levels were measured using the fragments per kilobase of exon model per million mapped fragments (FPKM) method. Total RNA was extracted from oat aboveground parts using Trizol (Simgen, Hangzhou, China). The cDNA was synthesized by using the All-in-One First-Strand cDNA Synthesis Super Mix for qPCR Kit (One-Step gDNA Removal; TransGen, Beijing, China) according to specifications. PCR identification was performed using the gene-specific primers listed in Supplementary Table S1 and the reaction system described in Supplementary Table S2. RT-qPCR detection was conducted using a Takara kit, with each sample repeated three times. The relative expression of genes was calculated using the 2-ΔΔCt method. The PCR amplification conditions were set as follows: (1) 95°C for 10 min; (2) 95°C for 15 s, 60°C for 1 min for 39 cycles.
2.8 Genetic transformation and identification
C. reinhardtii CC849 used as the host in this study was obtained from Hu Zhangli’s research group at Shenzhen University, China. The constitutive expression vector pHR13-AsCDPK26 contained the ORF of AsCDPK26 gene and the hygromycin resistance gene (Hyg). The plasmid pHR13-AsCDPK26 was extracted from the puncture bacteria using the SIMGEN plasmid DNA mini kit (Simgen, Hangzhou, China), and then used for genetic transformation of C. reinhardtii by the glass strain transformation method (Wang et al., 2017a). The algal cells were cultured in 50 mL fresh Tris-Acetate-Phosphate (TAP) medium under white light and selected by 10 mg L−1 hygromycin in TAP agar medium plates. Genomic DNA was extracted from CC849, empty vector (EV) and transgenic strains using the cetyltrimethylammonium bromide (CTAB) method. The positive transgenic algal strains screened by hygromycin were verified using genomic DNA PCR analysis. The primers used were AsCDPK26-F1 and AsCDPK26-R1 (Supplementary Table S1). Total RNA was extracted using Trizol kit (Simgen, Hangzhou, China) and cDNA synthesis performed using the All-in-One First-Strand cDNA Synthesis Super Mix for qPCR Kit (One-Step gDNA Removal; TransGen, Beijing, China). Transgenic algal strains were verified using cDNA PCR analysis with the primers AsCDPK26-F2 and AsCDPK26-R2 (Supplementary Table S1). To quantitatively detect AsCDPK26 gene expression in both wild type and transgenic strains, RT-qPCR was performed with Bio-Rad CFX Connect Optics Module (the primers was listed in Supplementary Table S1). The PCR amplification procedure were as follows: (1) 95°C for 10 min; (2) 95°C for 15 s, 58°C for 1 min for 39 cycles. The relative expression of genes was calculated using the 2-ΔΔCt program. The reaction were conducted with three biological replicates and three technical replicates.
2.9 Salt treatment, cell viability determination and pigment measurement
Algal growth study was performed for 96 h in sterile TAP medium after saline and/or alkali treatments. The initial optical density (OD680) was adjusted to approximately 0.3, and the samples were placed in a continuous light incubator at 25°C and 95 μmol·m-2·s-1. The growth characteristics of the algal cells were measured using the 680 nm optical density method. Saline and/or alkali treatments included three salt conditions (neutral salt stress (NaCl), alkaline salt stress (NaHCO3), and mixed salt-alkali stress (1:1 ratio of NaCl and NaHCO3)), three levels (100, 200, and 300 mmol·L-1), and three replicates. The medium without saline and/or alkali treatment was the control group, and culture condition was the same with single strain culture. Manual shaking was performed five times daily to maintain the uniform distribution of culture and medium components. On the final day, a sample of the microalgae was taken and filtered using a 0.45 μm microporous membrane to remove any remaining inorganic salts. The membrane was then dried at 60°C until the weight remained constant to calculate cell mass. All reported data represent the averages of three biological replicates. Total chlorophyll contents in algal cells were measured using spectrophotometry (Hamed et al., 2017). The chlorophyll fluorescence intensity was measured using a Handy-PEA chlorophyll fluorometer (Hansatech Instruments Ltd, UK).
2.10 Statistical analysis
All data were analyzed by Microsoft Excel 2010 and GraphPad Prism 8.0.2 (GraphPad Software, San Diego, USA). Experiments were carried out with biological replicates, and data were presented as the mean with standard deviation (mean ± SD). SPSS26.0 (SPSS, USA) was used for statistical analysis by one-way analysis of variance (ANOVA). For all of data analysis, a p-value<0.01 was considered as highly significant difference while a p-value<0.05 represents statistically significant.
3 Results
3.1 Identification of AsCDPK gene family members and their protein physicochemical properties
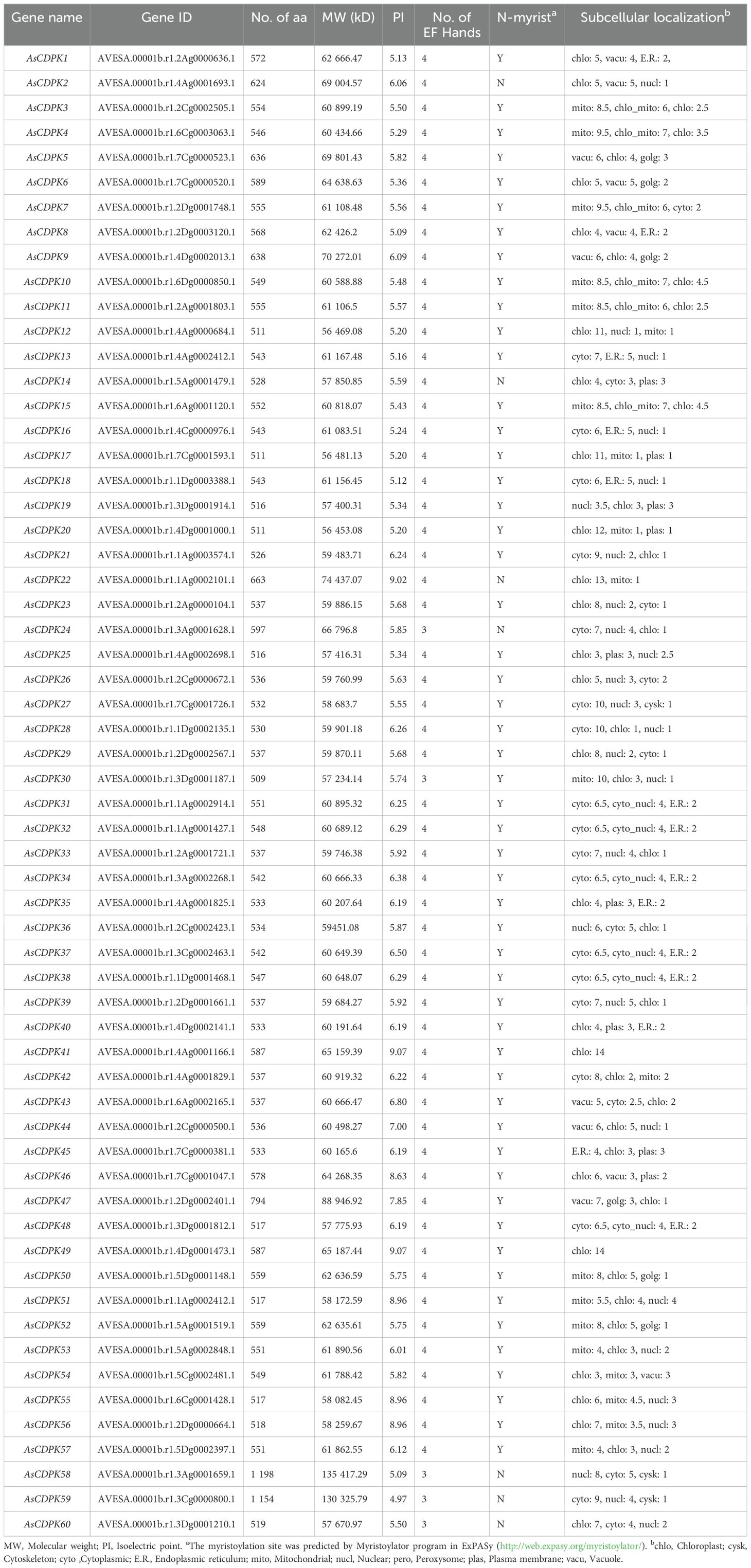
A total of 60 AsCDPK genes were identified in oat genome, using the known Arabidopsis AtCDPK proteins as query sequences. These AsCDPK genes were named AsCDPK1~AsCDPK60 (Table 1), respectively. The encoded amino acid (aa) lengths of these AsCDPK genes ranged from 511 to 1,198 aa, with corresponding molecular weights (MW) varied from 56,453.08 to 135,417.29 bp. The majority of AsCDPK members had isoelectric points (pI) values below 7.0, indicating their acidic nature, except for AsCDPK22/41/46/47/49/51/55/56. Of the 55 AsCDPKs, most contained four EF-hands. However, AsCDPK24/30/58/59/60 had three EF-hands. All AsCDPKs contained the predicted N-terminal myristoylation sites, except for AsCDPK2/14/22/24/58/59/60. In addition, subcellular localization prediction indicated that AsCDPKs were primarily located in the cytoplasm, chloroplasts, and mitochondria, with a smaller numbers found in the nucleus and endoplasmic reticulum.
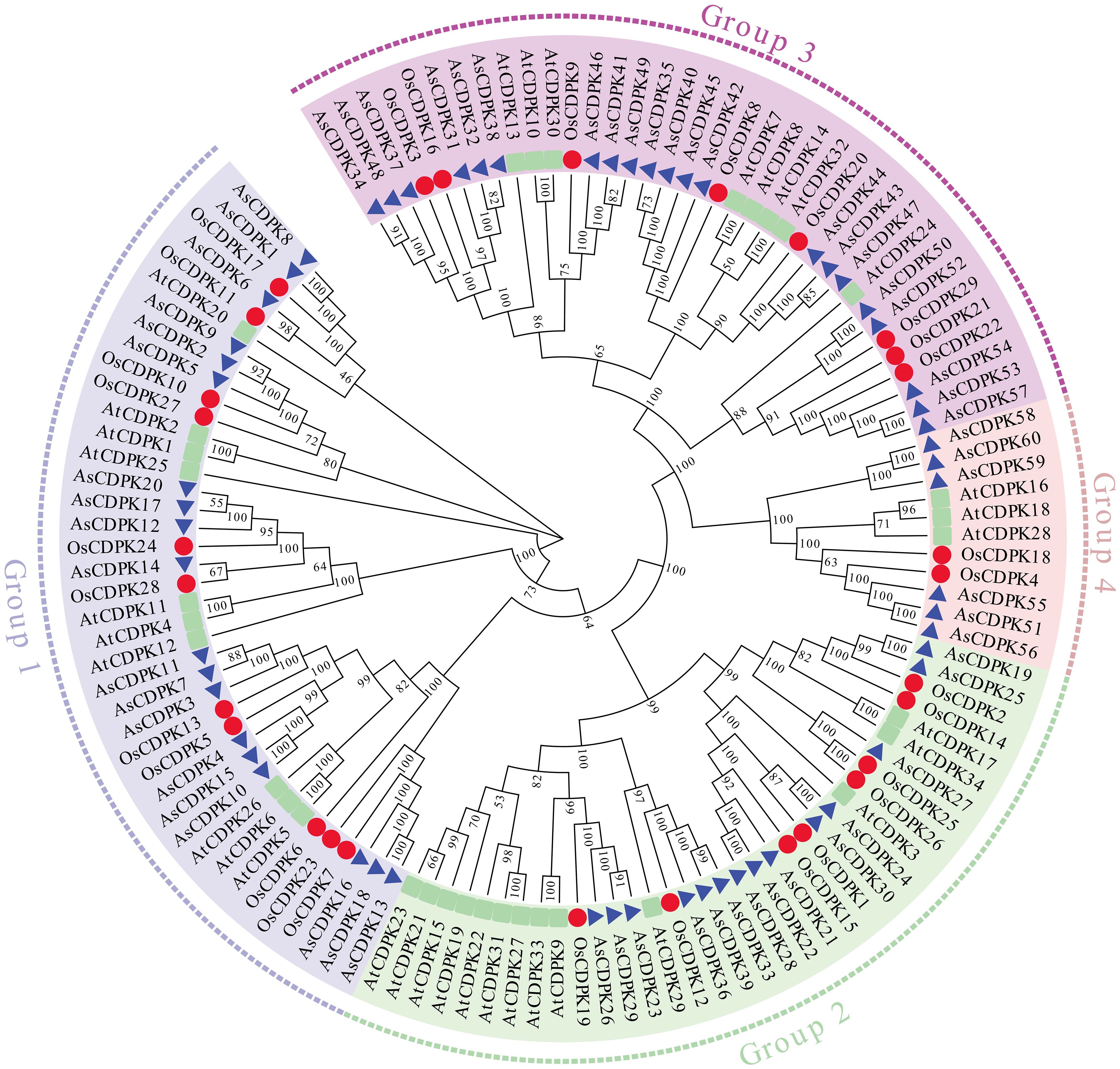
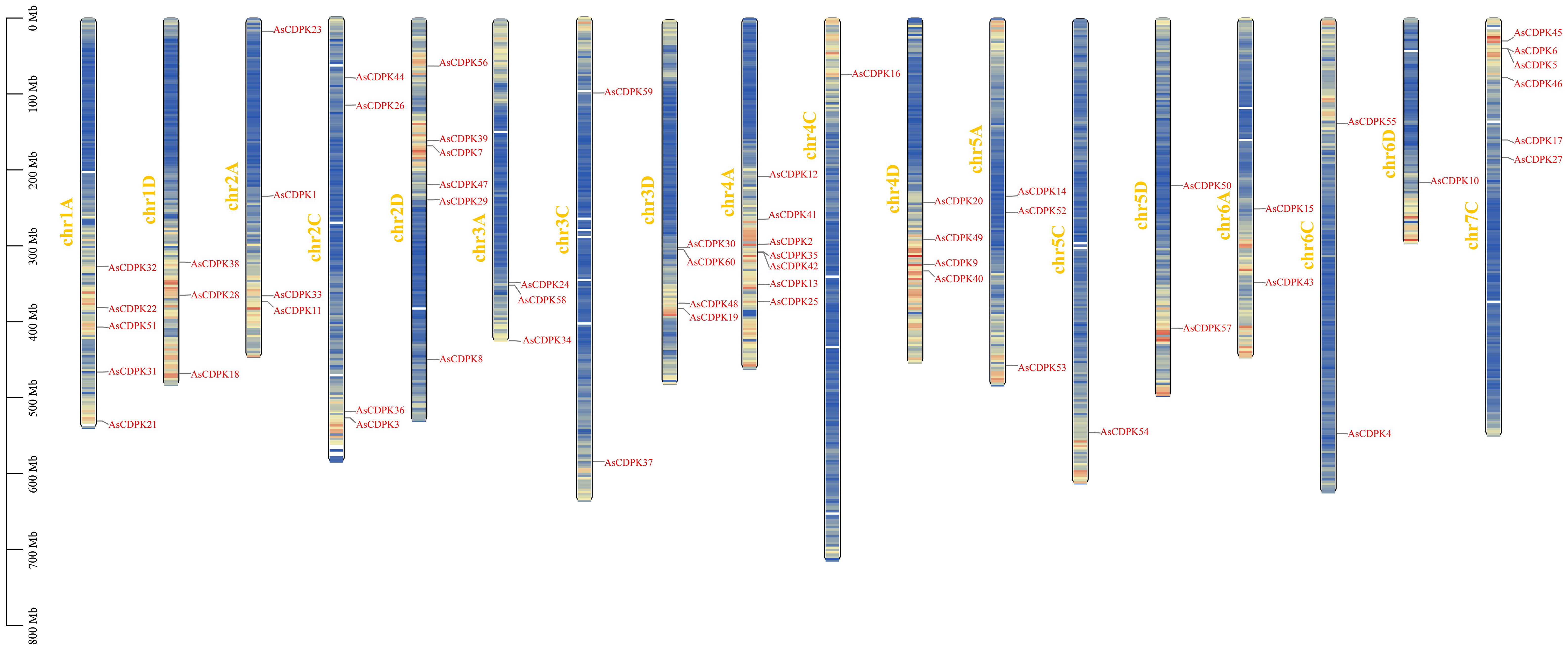
3.2 Phylogenetic analysis and chromosomal distribution of AsCDPK genes
To investigate the evolutionary relationships of CDPK gene family members, we constructed a neighbor-joining (NJ) phylogenic tree using CDPK sequences from Arabidopsis, rice, and oat. A total of 123 CDPK members were classified into four groups (Group 1-4) (Figure 1; Supplementary Table S3), with AsCDPKs distributed across these groups. The chromosomal locations of AsCDPKs were identified using TBtools software, gene annotation data, and gene density analysis, revealing that all AsCDPKs were mapped to 18 chromosomes (Figure 2). Each of the 18 chromosomes contained 1-7 AsCDPK genes, with chromosomes chr4C, chr5C, and chr6D each only having one AsCDPK gene. In addition, collinearity analysis of AsCDPKs was conducted using MCScanX. The results revealed the presence of 78 pairs of segmental duplication genes among AsCDPK gene family, but no tandem duplication event (Figure 3). The selection pressure on gene pairs was evaluated by calculating the nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks), as well as the Ka/Ks ratio, for the identified paralogous AsCDPK gene pairs (Supplementary Table S4). The Ka/Ks value, an indicator for the selection history of these paralogous gene pairs, was below 0.300, indicating that these gene pairs underwent strong purifying selection during the evolutionary process, leading to the function of these gene pairs to be relatively conserved.
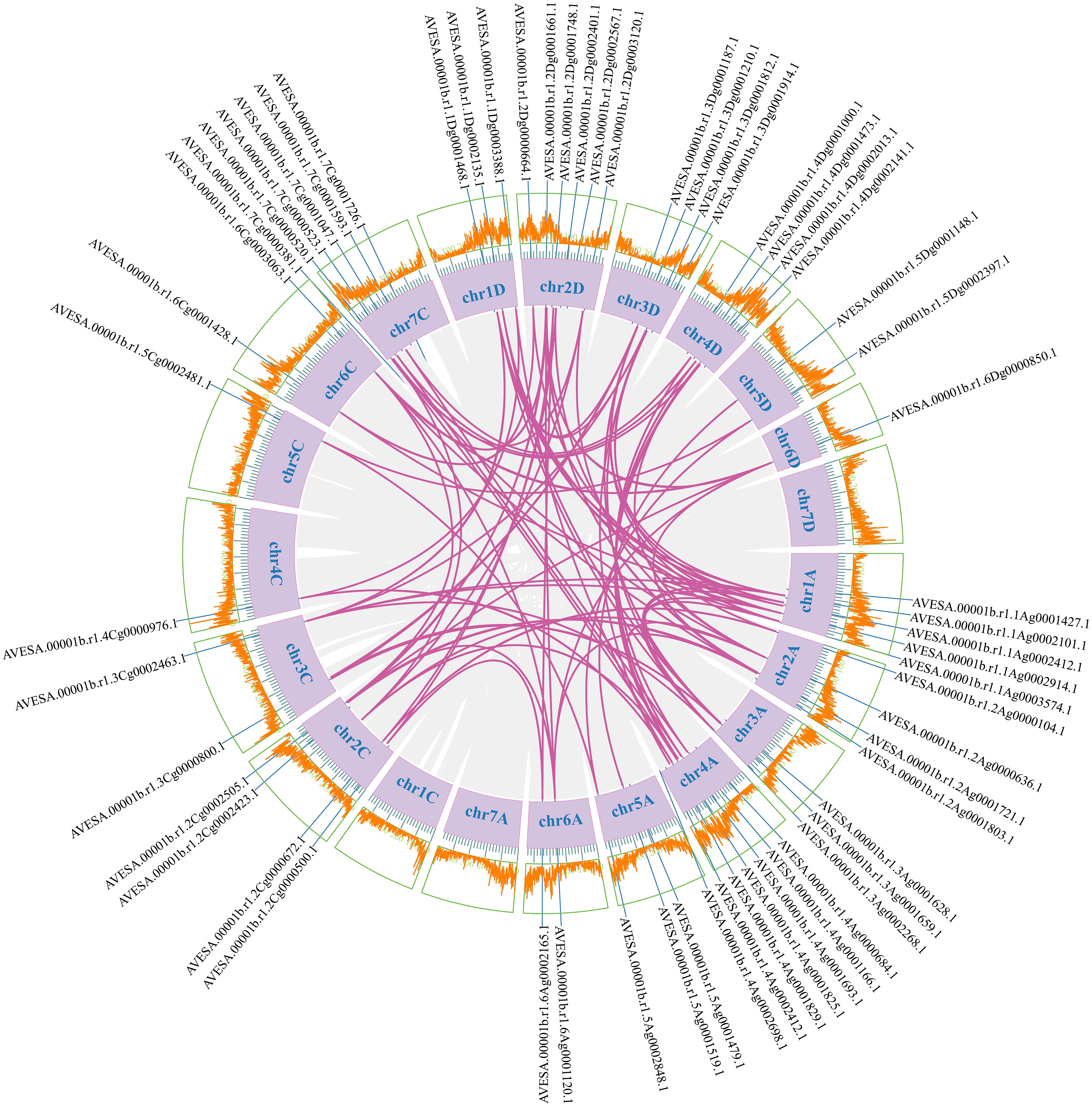
Figure 3. Collinear relationships of AsCDPK genes. The purple lines indicate duplicated CDPK gene pairs. The chromosome number is displayed next to each chromosome.
3.3 Structural analysis of AsCDPK genes
Gene structure analysis provides insights into potential evolutionary relationships among gene families. To better understand the diversity of AsCDPK genes, we constructed an unrooted phylogenetic tree (Figure 1) using the full-length AsCDPK protein sequences and compared this with the gene structure of the corresponding gene sequences (Figure 4). The analysis of the evolutionary relationship aiming these CDPKs showed that most of the AsCDPK members within the same subfamily exhibited similar exon/intron structures. Conserved motifs in homologous proteins may serve functional roles. To further investigate the structural diversity of AsCDPKs, we conducted a comparative analysis of the unrooted phylogenetic tree, focusing on the conserved motifs and domain combinations with the respective gene sequences (Table 1; Figure 5A). Totally, 20 of the most conserved motifs were identified among oat AsCDPKs using online MEME tools (Figure 5B). The unrooted phylogenetic tree revealed that AsCDPKs could be divided into four subgroups. All AsCDPK members contained a protein kinase domain, which was composed of motifs 9, 6, 3, 11, 2, 1, and 7 as illustrated in Figure 5. Motif 9 and 2 were detected as ATP binding site and serine/threonine-protein kinase active site, respectively.
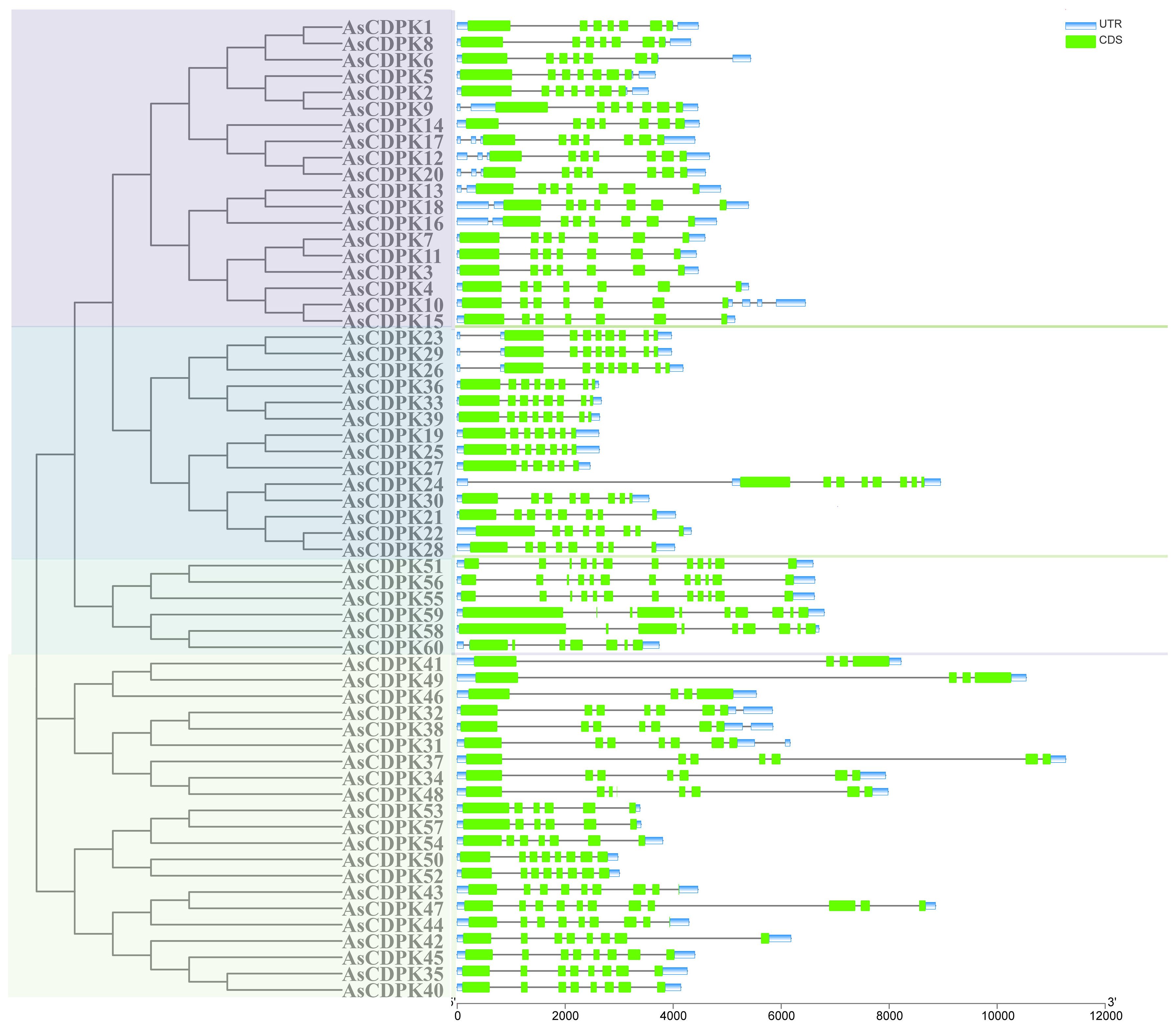
Figure 4. Gene structure analysis of CDPK gene members in oat. Green boxes represent exons, grey lines indicate introns, and blue boxes represent untranslated 5′and 3′ regions.
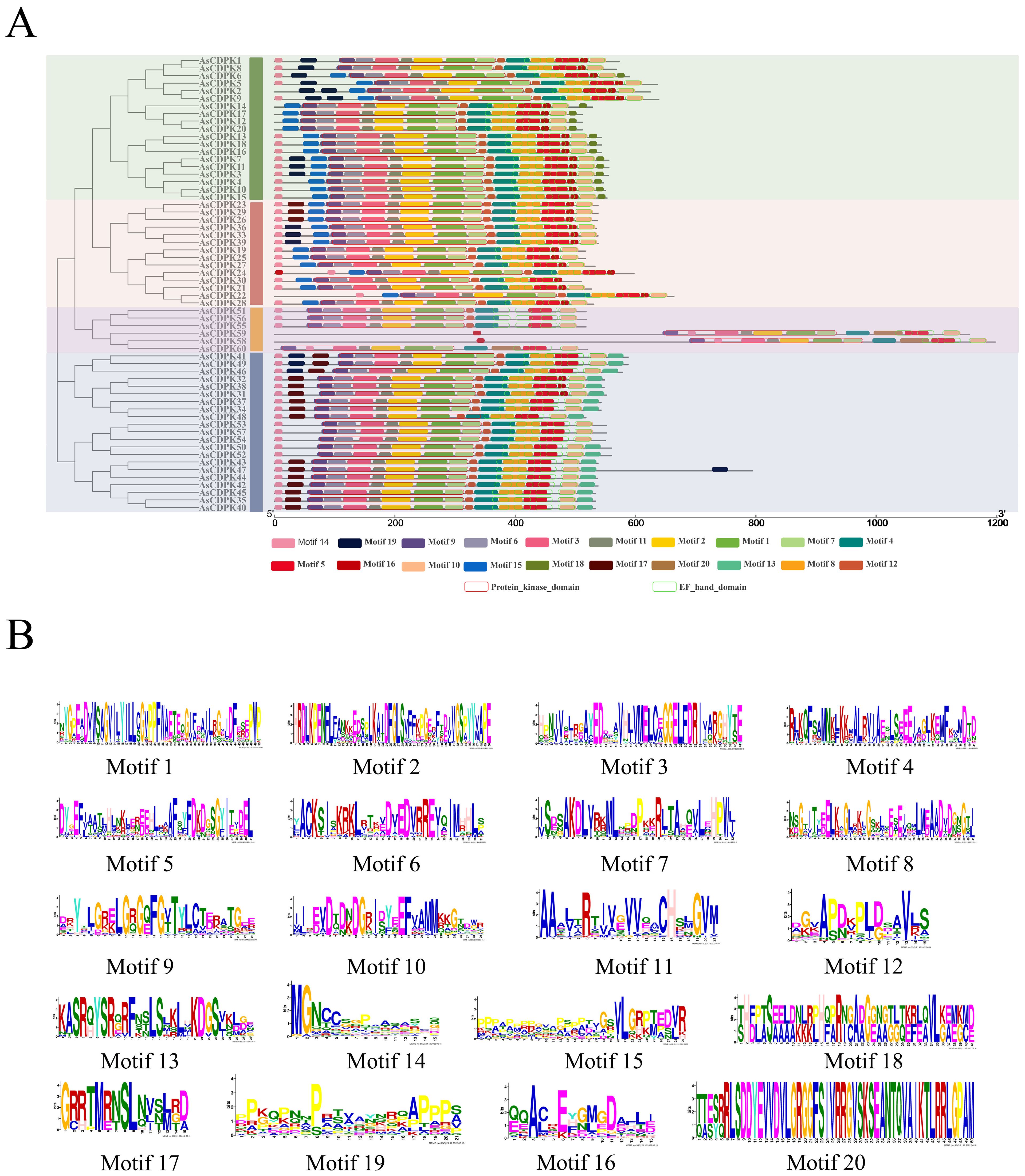
Figure 5. Protein structures of AsCDPKs in oat. (A) Protein structures of AsCDPKs. Different motifs are represented by specific colors. Red hollow box indicted the Serine/Threonine protein kinases domain (SM000220), and green hollow box indicted the EF-hand (SM000054). (B) Conserved motifs of AsCDPKs predicted by MEME.
In oat, 55 CDPKs possessed four Ca2+ binding EF-hand motifs, composed of motifs 4, 8, 5, 16, 10, and 20. Furthermore, the five AsCDPKs (AsCDPK24/30/58/59/60) each contained three EF-hand structures (Figure 5). All AsCDPK members contained motifs 4, 8, 5 and 10, except for the third subfamily without motif 8. Despite variations in motif compositions among different subfamilies, CDPK proteins of the same type typically displayed similar motif components. All AsCDPK members possess ATP-binding sites and serine/threonine-protein kinase active sites within their kinase domains (Figure 6). 95% of AsCDPK members contained three or four Ca2+ binding sites in the EF-hand conserved region, but AsCDPK13/18/16 just contained one Ca2+ binding site. Additionally, AsCDPK59 and AsCDPK58 contained the Kri1-like_C site. In summary, the conserved motif structures within each subgroup supported a close evolutionary relationship between them. However, functional differences may exist among members of different subgroups.
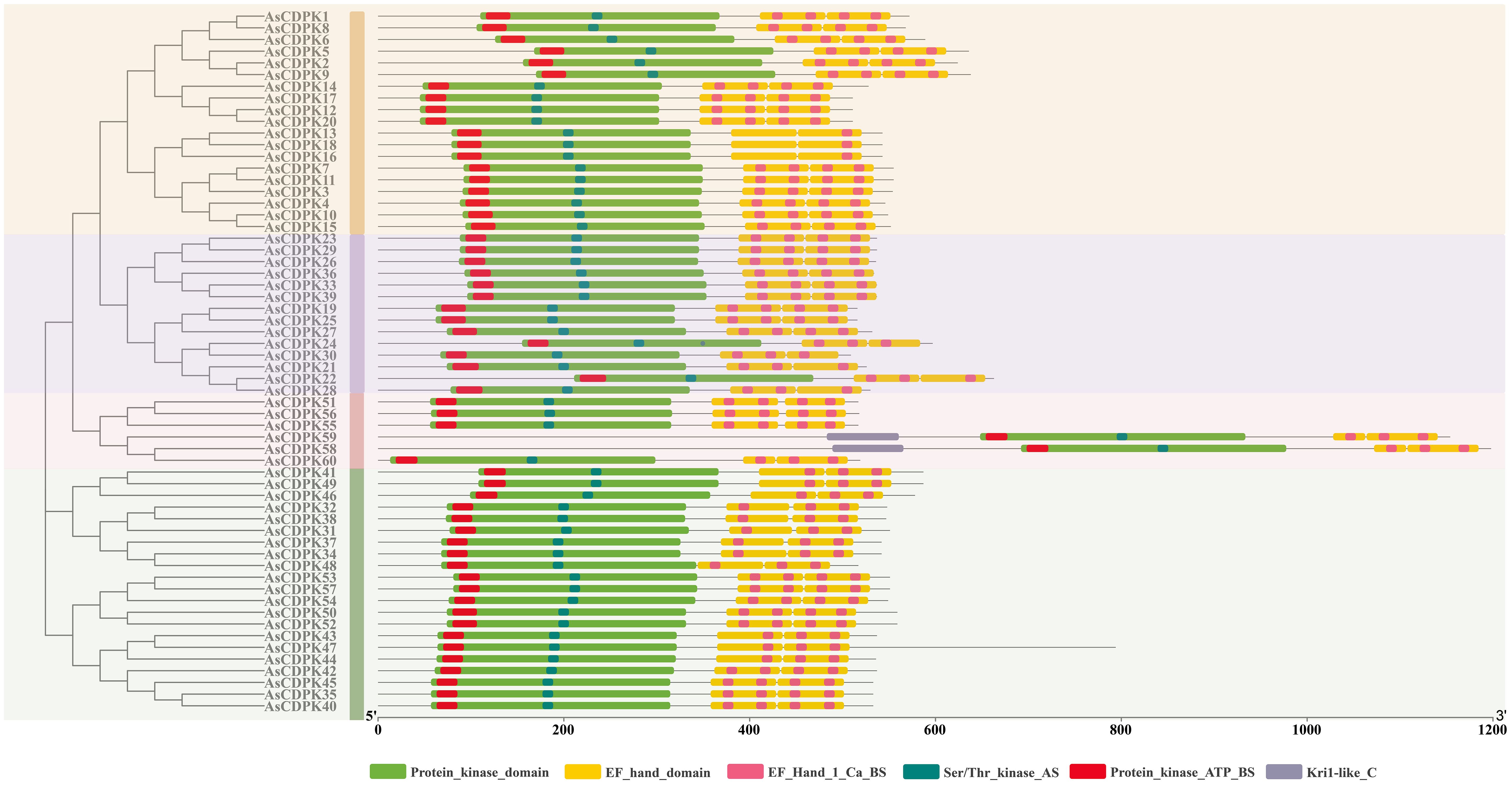
Figure 6. Conserved domain and important sites of AsCDPKs in oat. Different active sites are indicated by different colored boxes.
3.4 Cis-acting elements in the promoters of AsCDPK genes
To further elucidate the functions and regulatory mechanisms of AsCDPK genes in plant development and stress responses, we performed a cis-element analysis of the 2,000 bp promoter region upstream of the start codon for each AsCDPK gene using the PlantCare database. A variety of cis-acting regulatory elements were identified and classified into categories such as phytohormone response, biotic and abiotic stress, and plant growth and development (Figure 7). Transcription factor-associated cis-element (W-box, MBS, MRE and MBSI) were conserved in all AsCDPK genes. AsCDPK gene promoters generally contained cis-elements associated with phytohormone response and biotic and abiotic stress. However, cis-elements related to plant growth and development were only present in a few AsCDPK gene members. For the predicted cis-elements in the promoter, AsCDPK25 just contained the GT1-motif and O2-site elements, while the other members contained the cis-elements associated with phytohormone response (TCA-element, ABRE, TGACG-motif, and CGTCA-motif) and biotic and abiotic stress (ARE and G-box). The auxin response cis-element (AuxRE or TGA-box) was only present in the promoters of AsCDPK4/9/6 genes. For the predicted cis-elements related to plant growth and development, only AsCDPK7/11/3 had endosperm-specific negative expression cis-elements (AACA-motif). Furthermore, AsCDPK36/33/39/52 exclusively possessed regulatory cis-elements related to zein metabolism (O2-site) and meristem-expression element (CAT-box). Approximately 98% of AsCDPKs contained cis-elements responsive to phytohormone and stress, indicating that AsCDPK genes with these cis-elements might be responsive to stress and plant hormones.
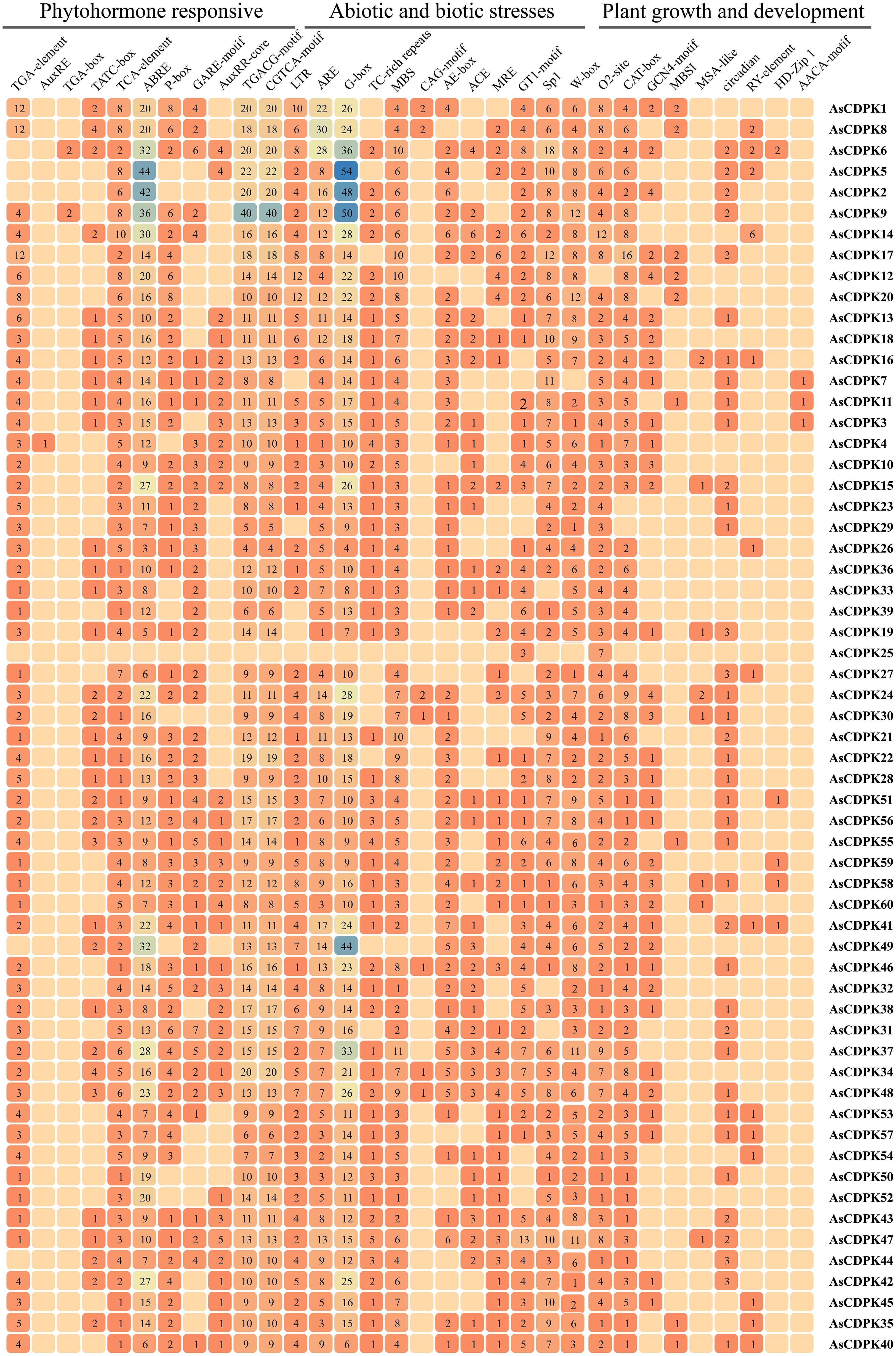
Figure 7. Analysis of cis-acting regulatory elements in the promoter regions of AsCDPK genes. The different colors and numbers of the grid indicate the numbers of different promoter elements in these CDPK genes.
3.5 Expression patterns of AsCDPK genes under different salt stresses
Analysis of gene expression patterns could provide valuable insights into the biological functions of the genes. To explore the roles of the AsCDPK genes in response to salt stress, we examined their expression patterns under different salt stress doses (Figure 8; Supplementary Table S5). The results showed that all 59 AsCDPK genes, with the exception of AsCDPK25, were expressed under the seven treatments (CK, NS100, NS200, AS100, AS200, NAS100, and NAS200). Among these genes, AsCDPK42/54/10/57/32/26/38 exhibited the similar expression patterns. Relative to normal growth conditions, these gene expression levels were upregulated with NS100 treatment and downregulated with AS200 and NAS200 treatments. Furthermore, the expression of these genes decreased as salt dosage increased. Although the number of up-regulated or down-regulated AsCDPK members varied across treatments, 75% of the members showed down-regulated expression under the NAS200 treatment. Cluster analysis further confirmed that the majority of AsCDPK genes displayed the similar expression patterns, despite varying degrees of upregulation or downregulation. Notably, certain AsCDPK gene members showed significant up-regulation or down-regulation in response to saline-alkali treatment, suggesting their roles in the oat’s response process to salt stress. To investigate the expression of AsCDPK genes under salt stress, RT-qPCR analysis was conducted on the five differentially expressed genes identified from the transcriptome in the shoots (Figure 9). The expression profiles of the five AsCDPK genes were found to align with the transcriptome data from salt-stress tissues. Variations in the expression of AsCDPK genes, including up-regulation and down-regulation, were observed under different salt treatment dosages. Specifically, AsCDPK34 and AsCDPK40 exhibited significant differences under NS200 treatment, whereas AsCDPK13/26/40 showed significant differences under NAS100 treatment. Moreover, AsCDPK13 and AsCDPK45 expressions were significantly different under the mixed saline-alkali treatments compared to the normal treatment. These findings highlighted the significant differences in the expression of the AsCDPK genes under various stress conditions, suggesting differential functions of these AsCDPKs in response to diverse stresses.
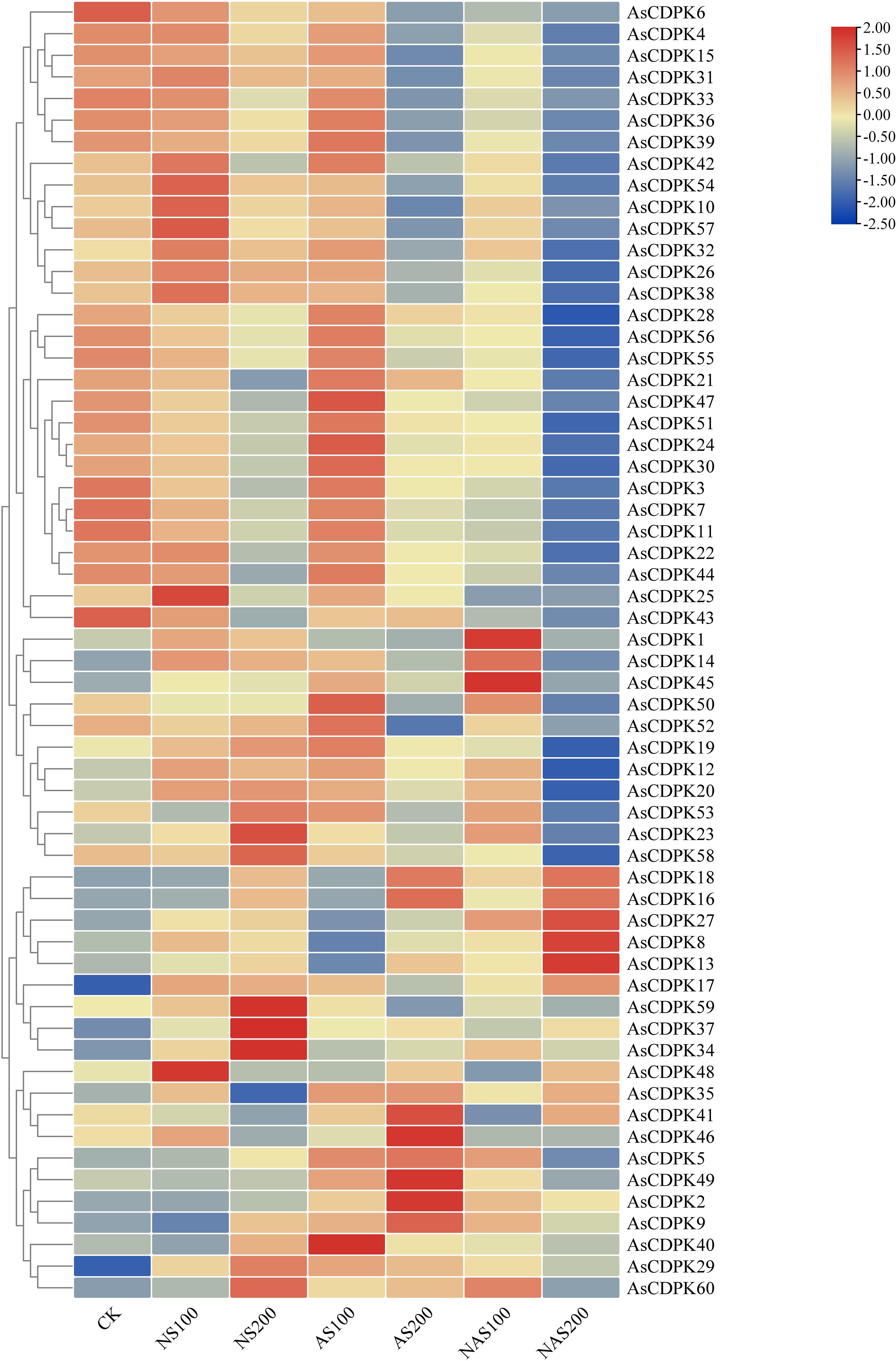
Figure 8. Expression analysis of AsCDPK genes in oat under different salt stresses. CK: normal treatment; NS100: 100 mmol·L-1 neutral salt; NS200: 200 mmol·L-1 neutral salt; AS100: 100 mmol·L-1 alkaline salt; AS200: 200 mmol·L-1 alkaline salt; NAS100: 100 mmol·L-1 mixed salt-alkali; NAS200: 200 mmol·L-1 mixed salt-alkali.
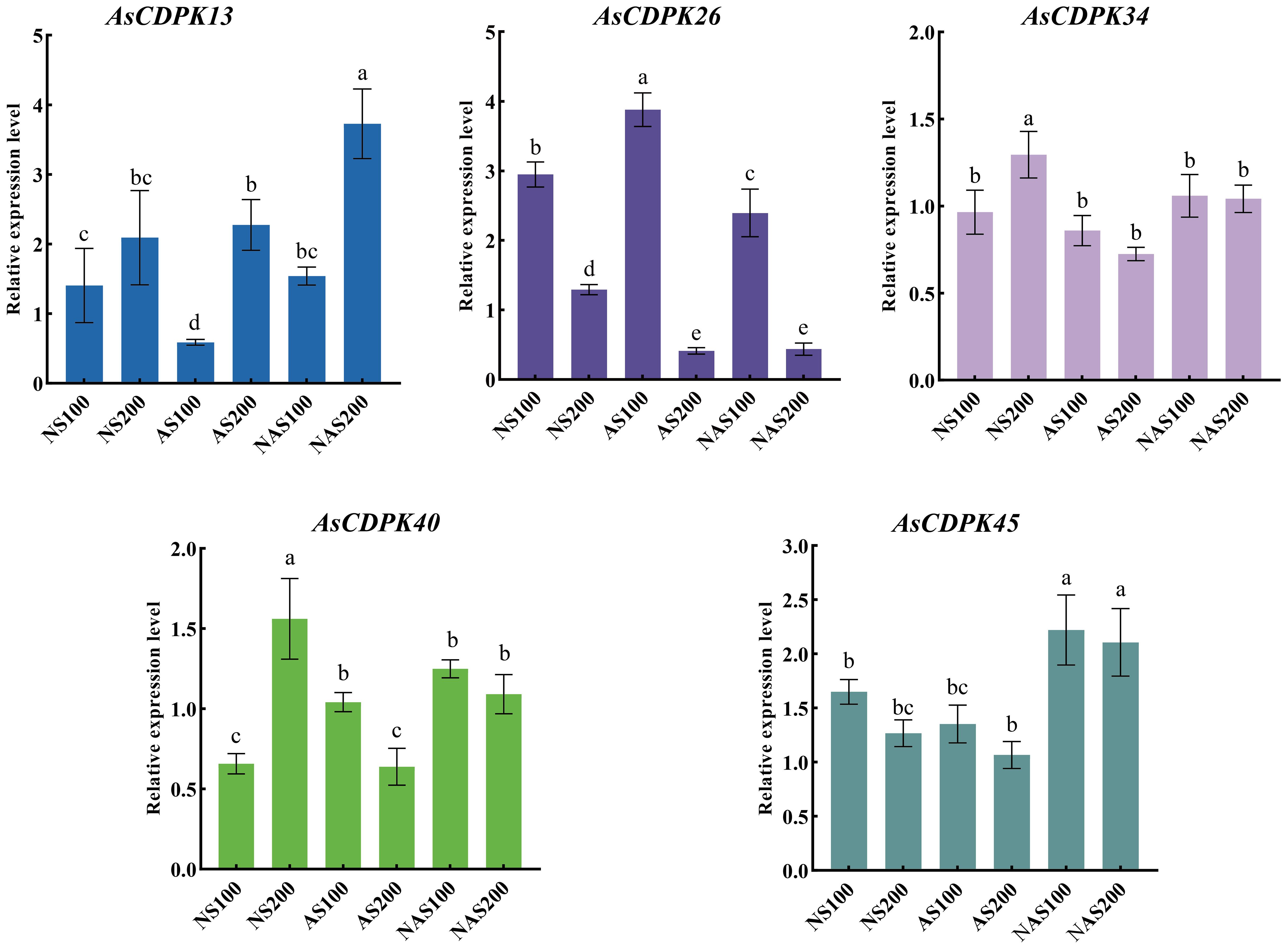
Figure 9. Relative expression levels of AsCDPK genes under different salt stresses. Different lowercase letters indicate significant difference at P<0.05.
3.6 Correlation between AsCDPK and ion transporter gene expression
To investigate the role of AsCDPK’s in saline-alkali stress responses, AsCDPKs and the relevant ion transporters (HKT1, HKT2, HKT3, HKT4, HKT6, HKT7, NHX1, NHX2-1, NHX2-2, NHX3, NHX5, SOS1) were analyzed using oat transcriptome data. Gene expression correlation analysis showed a significant association between AsCDPK genes (excluding AsCDPK5) and ion transporters (P <0.05) (Figure 10A). Notably, 21 AsCDPK genes (AsCDPK 2/3/4/6/7/10/11/13/21/22/26/28/33/36/39/42/44/49/51/55/56) showed an extremely significant correlation (P<0.01) with the ion transporters including AsHKT1, AsHKT2, AsHKT3, AsHKT6, AsNHX1, AsNHX5, and AsSOS1. RT-qPCR analysis validated the correlation between AsCDPK26 and AsHKT1, AsSOS1, and AsNHX1 genes (Figure 10B). A positive correlation was found between AsCDPK26 and both AsHKT1 and AsNHX1 genes, respectively (p<0.05), whereas a negative correlation was observed between AsCDPK26 and the AsSOS1 gene (p<0.05). These findings suggest that the function of AsCDPK26 in response to salt stress may be modulated by the genes AsHKT1, AsSOS1, and AsNHX1.
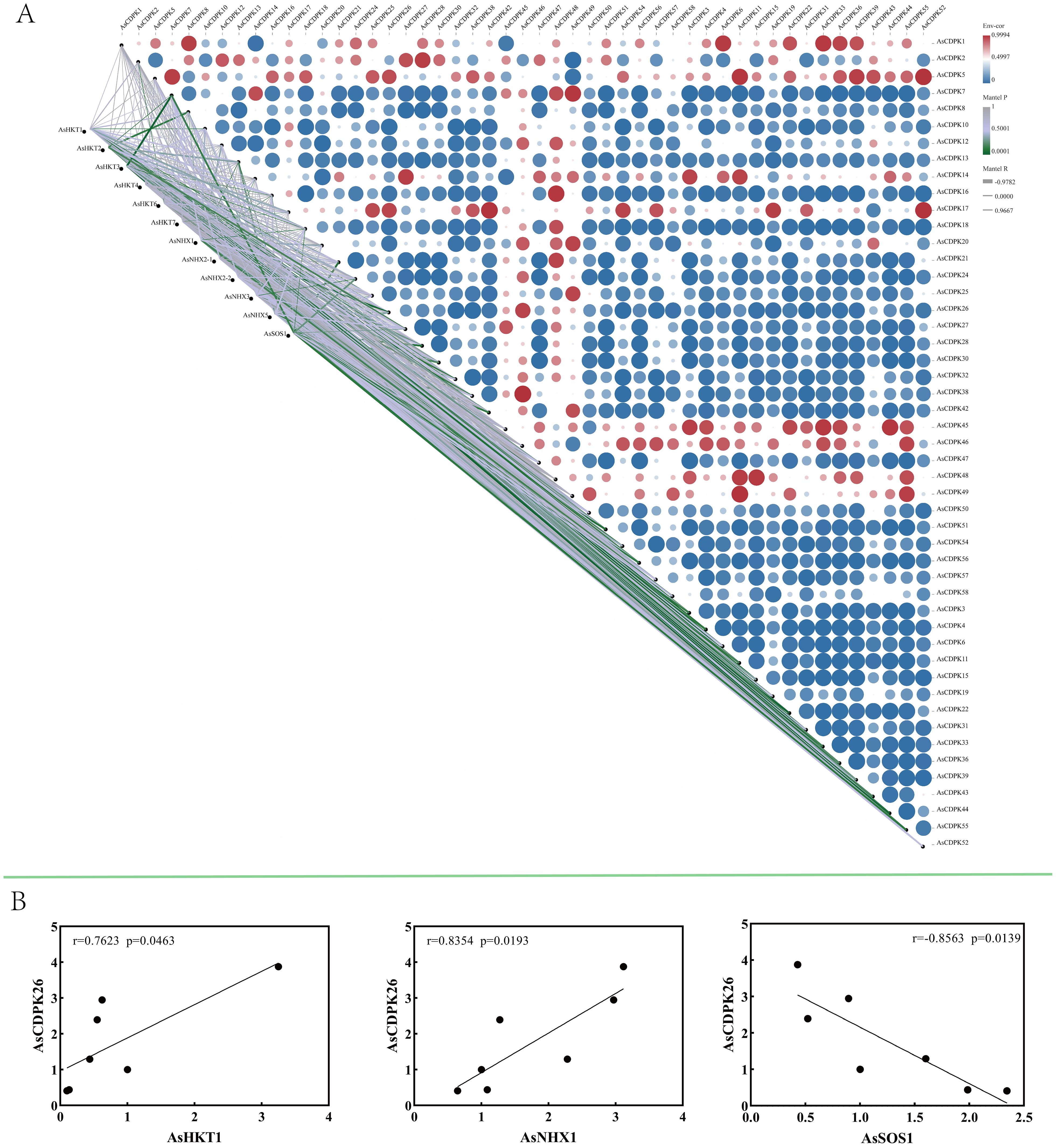
Figure 10. Expression correlation between AsCDPKs and genes related to ion transporters. (A) Correlation analysis was conducted between 49 significantly differentially expressed AsCDPKs and the expression of 12 ion transporter genes. (B) The correlation analysis between the expression levels of AsCDPK26 and AsHKT1, AsNHX1, or AsSOS1 genes were confirmed by RT-qPCR.
3.7 Identification of the transformed algal strains expressing AsCDPK26 gene
The expression analysis above (Figure 8) indicated that a set of AsCDPK genes expressed highly up on different types of salt stresses, including AsCDPK26/34/37/42/2/5/13/16/18/45/21/28/30/40/47/51/55/56. Notably, AsCDPK26 gene was highly expressed under neutral salt, alkaline salt, and mixed salt-alkali stresses (Figure 9) revealed by RT-qPCR. Furthermore, AsCDPK26 was detected to be the most closed with AtCPK27, evidenced by sequence comparative analysis (conserved domain and phylogenetic tree) using AtCPKs and AsCDPKs (Supplementary Figure S1). AtCPK27 has been reported to mediate salt stress tolerance in Arabidopsis by regulating ion and ROS homeostasis (Zhao et al., 2015). It is possible that AsCDPK26 has a similar function to AtCDPK27 in the response to salt stress. Consequently, we selected AsCDPK26 gene for functional analysis in this study. To investigate the function of AsCDPK26 gene, we constructed a novel expression vector for its heterologous expression in the single-cell model organism C. reinhardtii. The constitutive expression vector pHR13-AsCDPK26 contained AsCDPK26 gene and a hygromycin resistance gene (Hyg) (Figure 11A). Wild-type C. reinhardtii CC849 could not grow on plates containing hygromycin (10 mg·L-1) (Figure 11B). Consequently, the transgenic strains and empty vector (EV) lines were selected on TAP solid plates containing 10 mg·L-1 of hygromycin, and the transgenic algal line was cultured in TAP liquid medium (containing hygromycin) for further experiments (Figures 11C–E). The electrophoresis results confirmed that a band matching the size of the plasmid DNA was successfully amplified in the positive transgenic strain, whereas no target band was detected in the negative control and wild type C. reinhardtii CC849. This suggested that the AsCDPK26 gene had been integrated into the C. reinhardtii genome and remained stable genetically (Figure 11F). The total RNA of the wild type C. reinhardtii CC849, EV, and the transformed algal strain was separately extracted, followed by PCR analysis using the primers specific to the target gene. As expected, the specific fragment of AsCDPK26 gene was not detected in the wild type or EV cells, but was successfully amplified in the transgenic strains (Figure 11G). This result demonstrated successful expression of the exogenous AsCDPK26 gene in the algal cells.
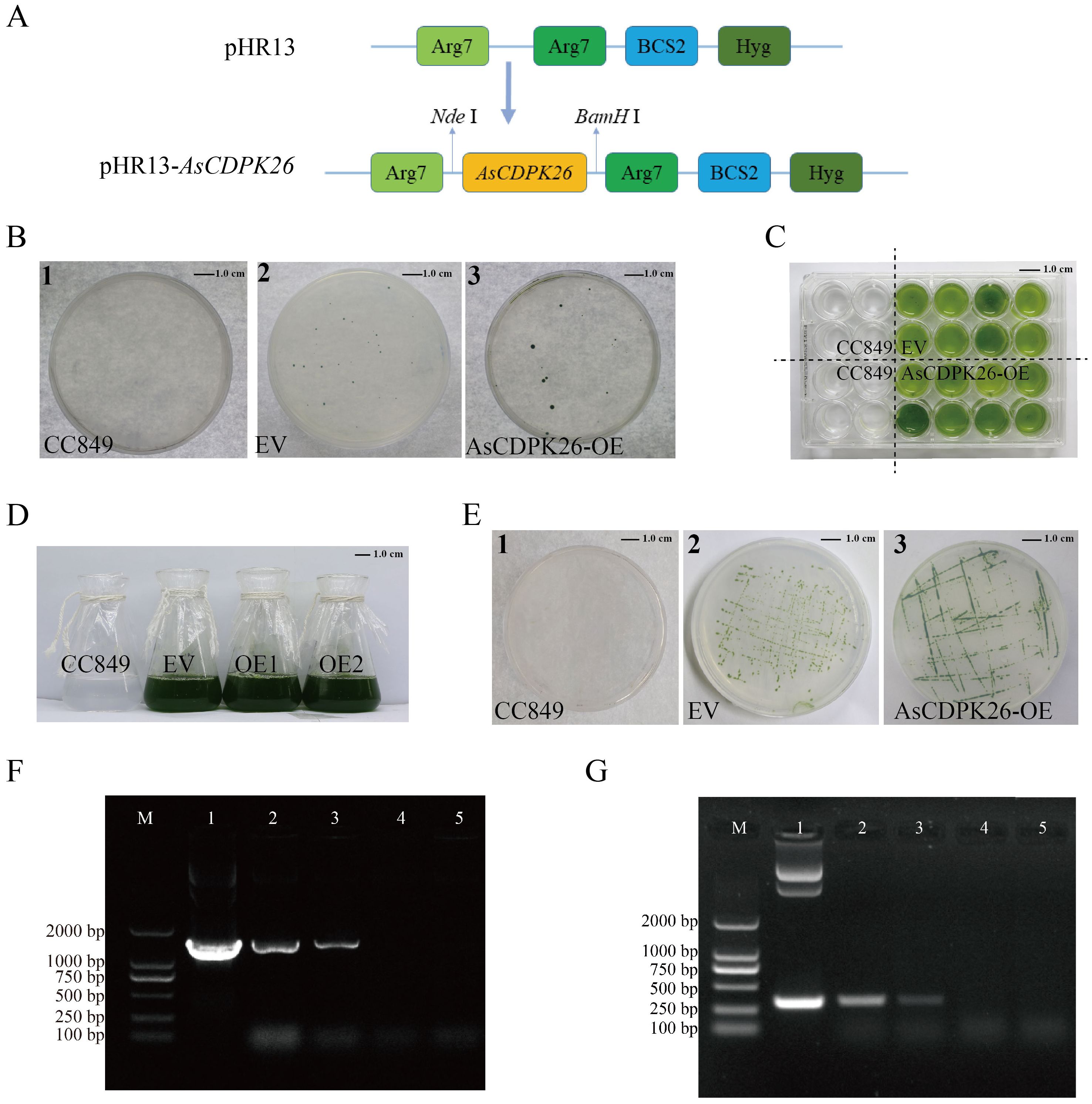
Figure 11. Screening and identification of transgenic algal lines of AsCDPK26 gene. (A) Schematic diagram of expression vector. (B) Transgenic algal lines were screened by TAP agar plate containing 10 mg L-1 hygromycin. (C) 24-well screening plate for the transgenic lines. (D) Liquid culture of wild strain, empty vector (EV) and transformed strains (OE1 and OE2). (E) Subculture plate for the transformed lines. (F) DNA identification of pHR13-AsCDPK26-transgenic C. reinhardtii. M: DNA marker DL2000; 1: Positive control (plasmid); 2-3: Positive transgenic lines; 4: Negative control (wild strain CC849); 5: EV. (G) PCR analysis of pHR13-AsCDPK26-transgenic C. reinhardtii. M: DNA marker DL2000; 1: Positive control (plasmid); 2-3: RT-PCR identification of the transformed lines; 4: Negative control (wild strain CC849); 5: EV.
3.8 Overexpression of AsCDPK26 gene enhances salt tolerance of C. reinhardtii
To investigate the function of the exogenous AsCDPK genes in C. reinhardtii, a single-cell photosynthetic model plant, we first examined the growth of the wild type, EV and transgenic strains under normal culture conditions for comparison. Similar growth properties were observed for the wild type, EV and the transgenic strain under normal growth conditions (Figures 12A, B). No significant difference in dry weight at 96 hours (Figure 12C) and total chlorophyll contents (Figure 12D) were observed between the wild type and transgenic strains. Furthermore, the Fv/Fm values were consistent with the change in pigment content for the wild type, EV and transgenic strain (Figure 12E). These findings indicated that the overexpression of the exogenous AsCDPK26 gene did not affect the growth and photosynthesis of the algal cells. After treatment with 100 mM and 200 mM neutral salt for 96 hours, no significant difference in dry weight was observed between the wild type and transgenic strains (Figure 12F). However, when the neutral salt concentration reached 300 mM, the dry weight of the transformed algal lines (AsCDPK26-1 and AsCDPK26-2) was significantly increased by 1.29 and 1.41 times, respectively, compared to the wild strain (Figure 12F). Moreover, RT-qPCR analysis showed that the expression of AsCDPK26 gene in C. reinhardtii was significantly upregulated under neutral salt treatment conditions (Figure 12I). After subjecting the algal cells to 96 hours of alkaline salt treatment at a 100 mM, the transformed strain showed improved growth compared to both the wild strain and the empty vector control (Figure 12A). Furthermore, the dry weight of the transformed strain (AsCDPK26-1 and AsCDPK26-2) was significantly higher, with increases of 1.36 and 1.47 times, respectively, compared to the wild type strain (Figure 12G). Under AS200 treatment, no significant difference in dry weight were observed among the transformed strain, the wild type strain, and the empty vector strain (Figure 12G). However, under AS300 treatment, all algal strains died. The expression of the AsCDPK26 gene was observed to significantly increase under alkaline salt treatment (Figure 12J). Moreover, under high-dose mixed salt-alkali treatments (NAS200 and NAS300), none of the algal strains survived (Figures 12A, H). On the other hand, under low-dose mixed salt-alkali treatments (NAS100), the growth of the transformed strain showed no significant difference compared to that of the wild type strain and the empty vector strain (Figure 12H). However, the expression of the AsCDPK26 gene in the transformed strain was significantly upregulated (Figure 12K). The expression profile of AsCDPK26 in oat showed the similar patterns in C. reinhardtii. In C. reinhardtii, the gene expression of AsCDPK26 increased under all salt treatments, with the most significant upregulation observed under low-dose alkaline salt treatment (AS100). This trend is consistent with the expression patterns of AsCDPK26 in oat. However, under neutral salt treatment, the expression of AsCDPK26 in Chlamydomonas initially increased and then decreased as the salt dose increased, although it remained higher than the control (CK).
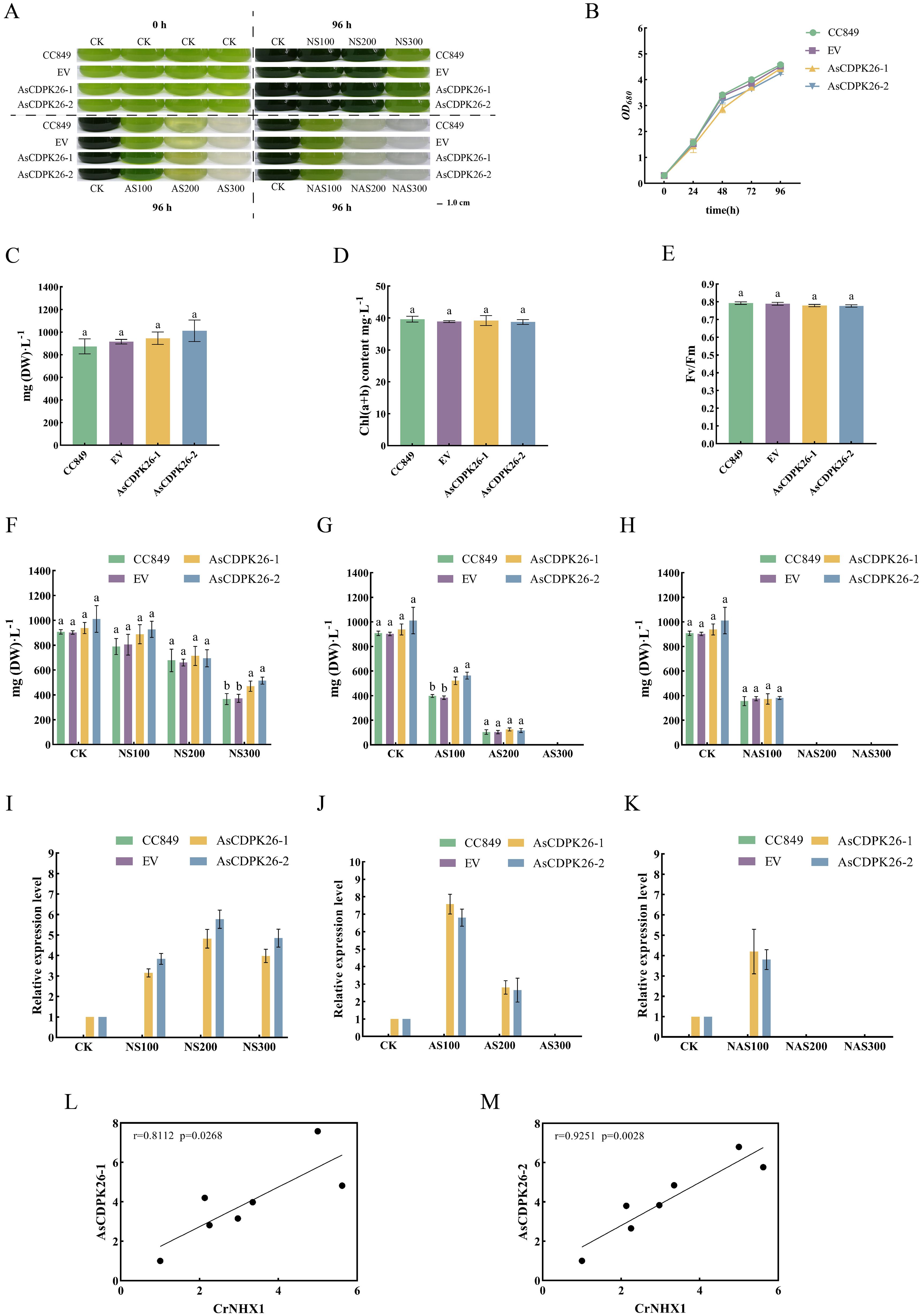
Figure 12. Effect of salt treatment on the algal growth of different genotypes. (A) Growth of C. reinhardtii CC849 and the transgenic algal strains exposed to different doses of salt stresses for 96 h (B) Growth curves of the wild type, EV and transformed algal strains under normal treatment. (C) The biomass, (D) total chlorophyll contents, and (E) Fv/Fm value of the wild type, EV and transgenic algal strains were measured after 96 hours of cultivation under normal conditions. (F-H) The biomass of wild type, EV and transformed algal strains were measured after 96 hours of cultivation under different doses of salt stresses. CK: normal treatment; NS100: 100 mmol·L-1 neutral salt; NS200: 200 mmol·L-1 neutral salt; NS300: 300 mmol·L-1 neutral salt; AS100: 100 mmol·L-1 alkaline salt; AS200: 200 mmol·L-1 alkaline salt; AS300: 300 mmol·L-1 alkaline salt; NAS100: 100 mmol·L-1 mixed salt-alkali; NAS200: 200 mmol·L-1 mixed salt-alkali; NAS300: 300 mmol·L-1 mixed salt-alkali. (I-K) The AsCDPK26 gene expression level in the wild type, EV and transformed algal strains were measured after 96 hours of different salt treatments. (L, M) RT-qPCR verification was conducted to analyze the correlation between the expression levels of AsCDPK26 and NHX1 genes in C. reinhardtii. Different lowercase letters indicate significant difference at P<0.05.
To further elucidate the salt tolerance mechanism in the AsCDPK26-overexpressed algal strain, we performed RT-qPCR analysis to assess the correlation between AsCDPK26 and the ion transporter gene (NHX1) under various salt treatments. The results demonstrated a positive correlation in gene expression levels (p<0.05) (Figures 12L, M), aligning with predictions made in oat. These collective findings suggested that overexpression of AsCDPK26 might enhance salt tolerance in C. reinhardtii by interacting with the NHX1.
4 Discussion
4.1 Characteristics of CDPK genes in oat
CDPK genes present in photosynthetic organisms play a crucial role in regulating various physiological processes, including hormone signaling and stress resistance. They are also critical for plant growth and development (Mehlmer et al., 2010; Boudsocq and Sheen, 2013). CDPK genes have been identified in different plant species, such as A. thaliana (Cheng et al., 2002), O. sativa (Asano et al., 2005), and Solanum. lycopersicum (Rutschmann et al., 2002). In this study, we identified 60 AsCDPK genes based on the oat genome database. By comparing them with CDPK members in Arabidopsis and rice, these AsCDPKs were classified into four groups (Figure 1), like the cases in A. thaliana and Brachypodium distachyon (Cheng et al., 2002; Wen et al., 2020). Phylogenetic analysis revealed that CDPKs, widely present in plants, are relatively conserved. Despite the exon-intron patterns and motifs of AsCDPKs being conserved and similar to those of B. distachyon (Wen et al., 2020), variations were observed among different subfamilies of AsCDPKs in the phylogenetic tree, suggesting that AsCDPKs may serve diverse functions (Figure 4). AsCDPKs primarily underwent segmental duplication, which might be the main driving force for the evolution of AsCDPK gene family. Furthermore, the uneven distribution of AsCDPKs across chromosomes might correlate with species evolution and genetic variation (Figure 2) (Miao et al., 2023). Notably, similar distributional patterns have been observed in other plant species (Miao et al., 2023).
All AsCDPKs identified in our study exhibited the typical characteristic structures of CDPK family, including a variable N-terminal domain, a catalytic Ser/Thr protein kinase domain, an autoinhibitory domain, and an EF-hand domain. Members in the same subfamily exhibited the similar exon-intron structures, biochemical properties, and conserved motif compositions, indicating a close evolutionary relationship among them. The protein kinase domain catalyzes the transfer of the γ-phosphate from ATP to specific amino acids in proteins, resulting in conformational changes and functional alterations (Hanks et al., 1988). AsCDPKs are a type of protein kinase that contain ATP-binding sites (Figure 6). CDPK use ATP as a source of phosphate groups to regulate target substrates in signal transduction pathways through phosphorylation (Wen et al., 2020). The calmodulin-like domain, featuring an EF-hand structure, binds Ca2+, allowing CDPKs to act as Ca2+ sensors. Except for AsCDPK24/30/58/59/60, all AsCDPKs contain four EF-hand structures (Figure 5). This variation in the number of EF-hand structures was also observed in CDPKs from A. thaliana, O. sativa, and Z. mays (Cheng et al., 2002; Asano et al., 2005; Kong et al., 2013). EF hands of CDPKs act as a calcium sensor in calcium-binding affinities (Hu et al., 2016). In agreement with this, AtCPK25 was calcium independent owing to lacking of the functional EF hands (Boudsocq and Sheen, 2013). Therefore, these CDPKs like AsCDPK24/30/58/59/60 might be insensitive to the changes in cellular calcium in plants. Myristoylation is a lipid modification commonly observed in proteins, which enhances their interaction with cell membranes and contributes to protein localization and functional regulation (Wen et al., 2020). Among AsCDPK members, only AsCDPK2/14/22/24/58/59/60 lacked N-myristoylation sequences (Table 1). Furthermore, subcellular localization prediction indicated that AsCDPK proteins were typically located in chloroplasts, cytoplasm, and mitochondria. Previous studies have shown that CDPK has both N-myristoylation and palmitoylation sites, which are crucial for determining subcellular localization (Zhao et al., 2021a). For instance, MtCDPK4/14/16/22 and MtCRK6, which contain both N-myristoylation and palmitoylation sites, are localized to the plasma membrane. MtCDPK7/9/15 without N-acylation site or just having one site are distributed in the cytoplasm and nucleus (Zhao et al., 2021a). Thus, it is likely that the subcellular localization of CDPKs is influenced by other factors that require further investigation.
4.2 AsCDPKs may function importantly in multiple life processes, especially in stress responses
The analysis of cis-acting elements in gene promoter and gene expression patterns can provide valuable insights into the potential functions of the interested genes. Multiple cis-acting elements responsible for phytohormones, stresses, growth and development were observed in the promoter regions of AsCDPKs, indicating the potential role of AsCDPKs in regulating multiple responses to phytohormones, environmental stresses, and development (Figure 7). An increasing evidence has consistently identified that CDPKs play an important role in a variety of abiotic stresses, including drought, salinity, cold, nutrient deficiency, light, hypotonic stress, and so on (Harmon et al., 2001). MtCDPK22 transcript is specifically and strongly expressed under cold stress, and its homologous gene OsCPK17 is involved in cold stress response (Zhao et al., 2021a). SlCDPK5/6, SlCDPK22/27 were highly induced under high temperature in tomato (Hu et al., 2016). Notably, GT1 and W-box elements detected in the AsCDPK promoters were considered to be the cis-acting elements in response to salt stress (Park et al., 2004; Zhou et al., 2008). The cis-acting GT-1 element was also identified to plays a key role in mediating the function of SCaM-4 in inducing salt stress through interaction with GT-3b, a GT-1-like transcription factor (Park et al., 2004). OsWRKY54 physically bound to the promoter sequences of OsHKT1;5 via its W-box motif, promoting expression of the target gene (Huang et al., 2022). These AsCDPK genes (e.g. AsCDPK10/26/34/37 and so on) contain dehydration-responsive elements (GT1 or W-box), were upregulated under neutral salt treatment (Figure 7; Figure 8). In Arabidopsis, some AtCPKs are involved in regulating phytohormone and abiotic stresses signaling when specific cis-acting elements were detected in the promoter regions (Zhang et al., 2020; Li et al., 2016; Huang et al., 2018). Thus, an analysis of similarities and differences between family members based on their expression pattern and promoter can provide candidate genes for further functional analysis at least in part.
4.3 AsCDPK gene exhibited specific expression patterns under various salt stresses
In response to salt stress, plants can resist damage by maintaining balance of ion, osmotic potential, and reactive oxygen species (ROS) (Zhao et al., 2020). CDPK regulate the activity of transcription factors, ion channels, transporters, and other proteins involved in signaling pathways through phosphorylation, thereby controlling gene expression, ion balance, and ROS homeostasis (Yang et al., 2021). For example, in Arabidopsis, AtCPK3 can phosphorylate the vacuolar potassium channel TPK1 to regulate intracellular K+/Na+ balance in response to salt stress (Latz et al., 2013). Conversely, overexpression of CPK23 renders plants more sensitive to drought and salt stress, whereas the T-DNA insertion mutant of AtCPK23, cpk23, exhibits an increased tolerance to these stresses compared to the wild type. This process may negatively regulate drought and salt resistance by inhibiting K+ uptake (Ma and Wu, 2007). Previous studies indicated that CDPKs may enhance plant tolerance to natural salt stress. However, the role of CDPKs in plant responses to alkaline salt and mixed salt-alkali stresses remains unclear. In this study, we identified the specific expression patterns of AsCDPKs under different salt treatments (Figure 8). The expressions of certain AsCDPK genes (AsCDPK17/29/34/37) were found to be enhanced under all salt treatments, indicating their potential regulatory functions. Similar gene-specific expression patterns have also been observed in Prunus mume and B. distachyons (Miao et al., 2023; Wen et al., 2020). The expression level of CsCDPK6 was significantly up-regulation under NaCl treatment of cucumber seedlings (Zhu et al., 2021). Similarly, the gene expression of AsCDPK10/25/26/32/38/42/48/54/57 in oat was also increased under NS100 stress (Figure 8). After 48 hours of neutral salt stress, CgCDPK expression decreased initially before increasing with the rise in NaCl dosage (Wang et al., 2017b). More likely, the gene expression of AsCDPK1/8/10/14/19/25/26/32/38/42/46/48/54/57 in oat was also increased while AsCDPK16/18/23/29/34/37/58/59/60 genes showed an increased expression under higher dosage of NaCl stress. This observation could be attributed to the variations in sequences and functional diversity present among CDPK members (Figure 8). Although the role of CDPKs in neutral salt stress has been extensively studied, the literature still lacks information on their role in alkaline salt and mixed salt-alkali stresses. Salinity stress is a widespread environmental problem (Zhou et al., 2024). Despite considerable efforts to address this issue, two crucial aspects have been neglected, i.e. salt–alkali stress and complex salt stress (Shi and Sheng, 2005). In fact, soil salinization from NaHCO3 and Na2CO3 may be more severe than that caused by neutral salts like NaCl and Na2SO4 (Shi and Sheng, 2005). In this study, as alkalinity levels increased, oat plants suffered greater damage, which corresponded to decreased expression levels of most AsCDPKs. Interestingly, a small subset of AsCDPK genes actively responded to these stresses, showing a significant increase in expression levels. Specifically, AsCDPK23/58/59/37/34 genes were involved in response to neutral salt stress while AsCDPK18/16/41/46/49/2 genes responded to alkaline salt stress. In contrast, AsCDPK18/16/27/8/13 genes strongly responded to mixed salt-alkali stresses (Figure 8). RT-qPCR analysis further validated the expression pattern of AsCDPK13/26/34/40/45 genes, indicating their potential role in mitigating plant stress damage and enhancing plant salt tolerance (Figure 9).
In addition, the AsCDPK26 gene was found to be positively correlated with the transcription levels of AsHKT1 and AsNHX1 genes (p<0.05), but negatively correlated with that of AsSOS1 (p<0.05) (Figure 10). Biotic and abiotic stresses, as well as certain intracellular stimuli, can alter cytosolic Ca2+ concentrations. As a calcium ion sensor, CDPK can recognize calcium signals and act directly or indirectly on downstream interacting proteins, thereby triggering a series of physiological responses (Pirayesh et al., 2021). Previous studies have demonstrated that exposure to salt stress leads to an elevation in the concentration of calcium ions within the cell, which is detected by the calcineurin B-like protein CBL4 (also known as SOS3). This protein subsequently interacts with the CBL-interacting protein kinase CIPK24 (SOS2). The resulting SOS3/SOS2 complex then localizes to the plasma membrane and triggers the activation of the membrane-bound Na+/H+ antiporter (SOS1) via phosphorylation (Qiu et al., 2003; Shi et al., 2002). In Gossypium barbadense, the protein-protein prediction study revealed that GbNHX7 is involved in the CBL-CIPK protein interaction pathway (Akram et al., 2020). The HKT gene in barley is regulated by Ca2+ signals. Specifically, HvCaM1 plays a crucial role in regulating Na+ transport by preferentially controlling the transcription of HvHKT1s in response to salt stress. This regulatory process leads to the downregulation of HvHKT1;5 and the upregulation of HvHKT1;1 through the interaction between HvCaM1 and CAMTA4 (calmodulin-binding transcription activator). Consequently, the Na+ transport in the roots is effectively regulated, enabling barley to achieve higher salt tolerance (Shen et al., 2020). Additionally, CaM protein can also regulate the expression of AtHKT1;1, MtHKT1;1, and MtHKT1;2 (Galon et al., 2010; Shkolnik et al., 2019; Zhang et al., 2019). Thereby preventing the transportation of Na+ is not disrupted by external stress. Thus, it is hypothesized that in response to salt stress, AsCDPK26 may interact directly or indirectly with AsHKT1, AsNHX1, and AsSOS1 proteins to regulate Na+ transport, thereby reducing the damage from ion toxicity in oat (Figure 10).
4.4 AsCDPK26 may be the valuable target gene in biotechnology to enhance host tolerance to salt stress
To investigate the function of the CDPK gene, we performed an overexpression experiment of the AsCDPK26 gene in C. reinhardtii CC849, a unicellular model plant. The algae strains were treated with three types of salt stress, and subsequently, the growth features of the transgenic algal strains were measured. Compared to other plants, C. reinhardtii grows quickly and requires minimal cultivation costs. Especially, it is easily genetically modified and is referred as the “green yeast” (Rochaix, 1995). Thus, this algal species has been widely used to study various metabolic regulations in plants, including photosynthesis, storage substance biosynthesis and stress responses. For example, the overexpression of LHCSR and PsbS resulted in the improvement of light tolerance in C. reinhardtii (Wilson et al., 2023). Additionally, the overexpression of native ORANGE (OR) gene and OR mutant in C. reinhardtii enhanced carotenoid and ABA accumulation, leading to the increased resistance to abiotic stresses (Yazdani et al., 2021). Another study showed that the overexpression of phosphoribosyl pyrophosphate synthase promotes the resistance of Chlamydomonas to ionizing radiation (Jung et al., 2021).
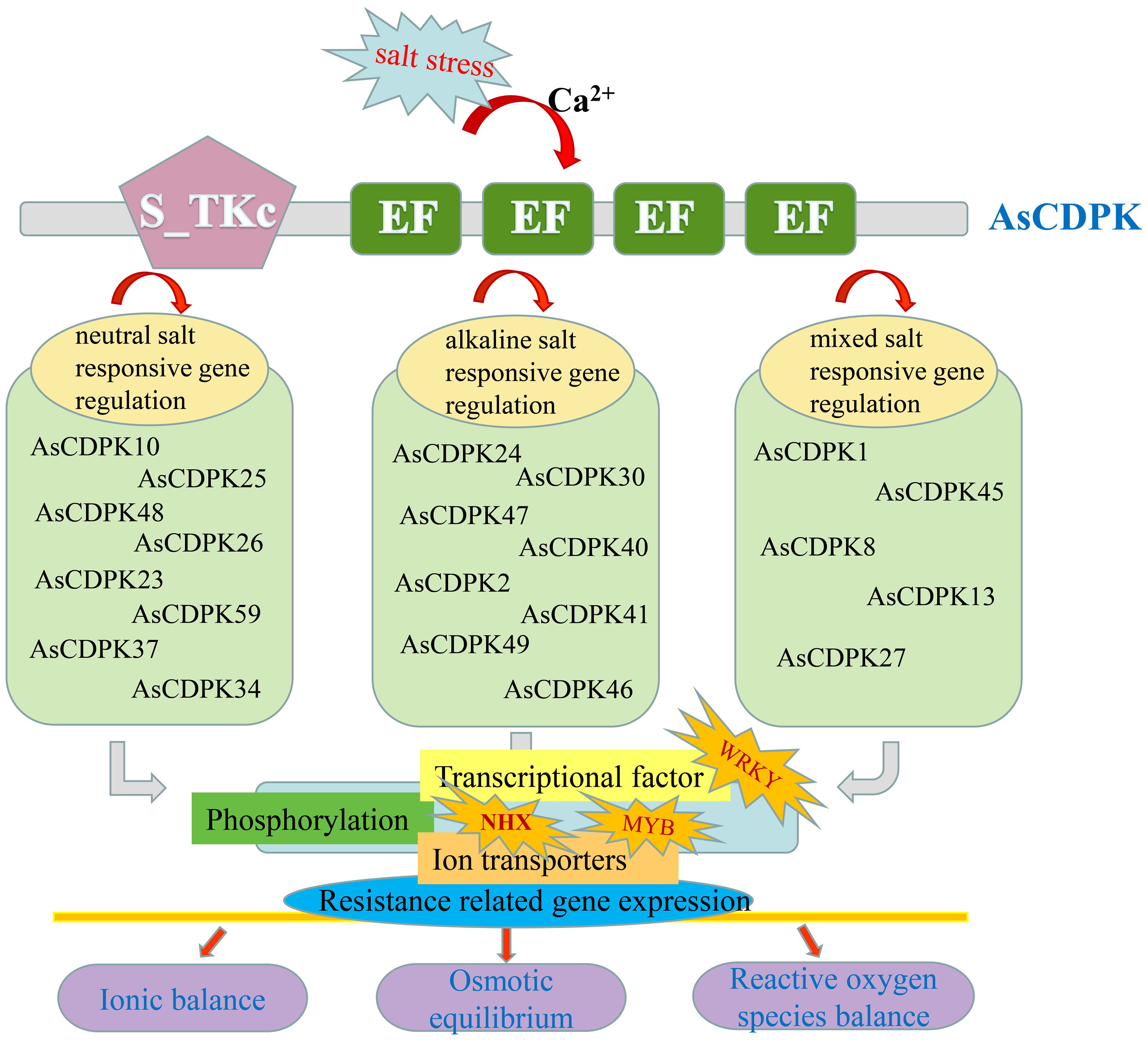
In this study, overexpression of AsCDPK26 gene improved C. reinhardtii tolerance to various types of salt stresses. For example, overexpressing AsCDPK26 improved the growth of Chlamydomonas in response to high-dose neutral salt treatment (Figure 12). Additionally, AsCDPK26 overexpression increased Chlamydomonas’ survival rate under AS100 treatment. AsCDPK26 gene expression was significantly up-regulation, indicating that the transgenic cells have a stronger salt tolerance (Figures 12I-K). However, the algal strains did not survive when subjected to AS300, NAS200, and NAS300 treatments, respectively. Possibly, alkaline salt and mixed saline-alkali are more detrimental to plants than neutral salts (Shi and Sheng, 2005). In addition, OsCPK4 overexpression plants accumulate less Na+ in their roots compared with control plants (Campo et al., 2014). The upregulation of ion transporter genes in roots of OsCPK4 rice plants could be, at least in part, responsible for the observed lower level of Na+ accumulation in roots of OsCPK4 rice plants (Campo et al., 2014). In our study, overexpressing the AsCDPK26 gene in C. reinhardtii showed a significant correlation with the expression of the NHX1 gene (p<0.05) (Figures 12L, M). This suggests that NHX1 plays an important role in improving the salt tolerance of Chlamydomonas by overexpressing the AsCDPK26 gene. Previous studies have shown that CDPK in plants plays an important role in responses to biotic or abiotic stresses by phosphorylating specific substrate proteins or transcription factors (Yang et al., 2021; Latz et al., 2013; Zou et al., 2015; Atif et al., 2019). Therefore, we hypothesize that AsCDPK26 might interact with the ion transporter protein (NHX1) to regulate Na+ balance, thus enhancing salt tolerance in the transgenic strains. However, this hypothesis needed to be tested by yeast two-hybrid system. In summary, we hypothesized that a set of AsCDPK members might play key roles in oat response to salt stress by regulating the activity of a specific substrate protein (ion transporter) or transcription factor through phosphorylation (Figure 13). However, further experimental validation is necessary to elucidate the specific molecular mechanisms underlying this response.
5 Conclusions
In this study, a total of 60 AsCDPK gene members were identified in oat and classified into four subfamilies, followed by comprehensively clarifying their molecular characteristics, including gene structures, chromosomal locations, phylogenetic relationships, motif composition, domain distribution, synthesis analysis and cis-acting elements. Moreover, transcriptome analysis plus PCR verification revealed that a set of AsCDPKs (e.g. AsCDPK13/26/34/40/45 and so on) might function importantly in responses to different salt stresses. The 49 differentially expressed AsCDPKs showed a significant positive correlation with the transcription levels of ion transporter genes NHX and HKT (p<0.05), yet a negative correlation with the transcription levels of the SOS1 gene (p<0.05). This suggests that AsCDPKs may interact directly or indirectly with the ion transporters NHX, HKT, or SOS1, mediating the response of oats to salt stress. Importantly, overexpression of AsCDPK26 gene in C. reinhardtii enhanced the host salt tolerance, highlighting the crucial role of AsCDPK26 in the salt-stress signaling pathway. Further correlation analysis demonstrated a positive correlation between AsCDPK26 and the transcription level of NHX1 gene (p<0.05) in the transgenic Chlamydomonas. These results not only provide the first knowledge of function and structure of AsCDPK members in oat, particularly in responses to different salt stresses, but also establish a foundation for further exploration of the molecular mechanism underlying the abiotic stress responses in oat and other crops despite diverse functions of AsCDPKs needed to be proved experimentally in future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Y-NL: Data curation, Methodology, Visualization, Writing – original draft, Formal analysis. CL: Methodology, Writing – review & editing. QY: Methodology, Writing – review & editing. XY: Investigation, Writing – review & editing. SL: Investigation, Writing – review & editing. YS: Methodology, Supervision, Writing – review & editing. CJ: Methodology, Supervision, Writing – review & editing. CZ: Methodology, Supervision, Writing – review & editing. J-AX: Conceptualization, Resources, Supervision, Writing – review & editing. HC: Conceptualization, Resources, Supervision, Writing – review & editing. RL: Resources, Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2021YFD1901105), State Key Laboratory of Integrative Sustainable Dryland Agriculture (in preparation), Shanxi Agricultural University (202105D121008-3-6), Six New Project of Agriculture Department of Shanxi Province (unnumbered), National Natural Science Foundation of China (31902394), Key Research and Development Planning Project of Shanxi Province (201803D31063), Science and Technology Innovation Planning Project of Shanxi Agricultural University (2018YJ16), Outstanding Doctor to Work in Shanxi Province Research Project (SXYBKY2019036), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (2021L119).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1395696/full#supplementary-material
Supplementary Figure 1 | Phylogenic tree of AsCDPKs and AtCPK27.
References
Akram, U., Song, Y., Liang, C., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX gene family under salinity stress in Gossypium barbadense and its comparison with Gossypium hirsutum. Genes 11, 803. doi: 10.3390/genes11070803
Almadanim, M. C., Alexandre, B. M., Rosa, M. T. G., Sapeta, H., Leitao, A. E., Ramalho, J. C., et al. (2017). Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 40, 1197–1213. doi: 10.1111/pce.12916
Asano, T., Tanaka, N., Yang, G., Hayashi, N., Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1111/j.1365-313X.2011.04766
Atif, R. M., Shahid, L., Waqas, M., Ali, B., Rashid, M. A. R., Azeem, F., et al. (2019). Insights on calcium-dependent protein kinases (CPKs) signaling for abiotic stress tolerance in plants. Int. J. Mol. Sci. 20, 5298. doi: 10.3390/ijms20215298
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Song, H., Xin, J., Dong, G., Xu, F., Su, Y., et al. (2023). Comprehensive genome-wide identification and functional characterization of MAPK cascade gene families in Nelumbo. Int. J. Biol. Macromol. 233, 123543. doi: 10.1016/j.ijbiomac.2023.123543
Cheng, S. H., Willmann, M. R., Chen, H. C., Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Cui, X., Gu, J., Liu, P., Fu, H., Wang, F., Qi, M., et al. (2024). Genome-wide identification and expression analysis of the UPF0016 family in tomato (Solanum lycopersicum) under drought stress. Environ. Exp. Bot. 219, 105607. doi: 10.1016/j.envexpbot.2023.105607
Fang, S., Hou, X., Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.667458
Galon, Y., Finkler, A., Fromm, H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3, 653–669. doi: 10.1093/mp/ssq019
Gu, D., Andreev, K., Dupre, M. E. (2021). Major trends in population growth around the world. China CDC. Weekly. 3, 604–613. doi: 10.46234/ccdcw2021.160
Hamed, S. M., Selim, S., Klöck, G., AbdElgawad, H. (2017). Sensitivity of two green microalgae to copper stress: growth, oxidative and antioxidants analyses. Ecotoxicol. Environ. Saf. 144, 19–25. doi: 10.1016/j.ecoenv.2017.05.048
Han, L., Eneji, A. E., Steinberger, Y., Wang, W., Yu, S., Liu, H., et al. (2014). Comparative biomass production of six oat varieties in a saline soil ecology. Commun. Soil Sci. Plan. 45, 2552–2564. doi: 10.1080/00103624.2014.912299
Hanks, S. K., Quinn, A. M., Hunter, T. (1988). The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52. doi: 10.1126/science.3291115
Harmon, A. C., Gribskov, M., Gubrium, E., Harper, J. F. (2001). The CDPK superfamily of protein kinases. New Phytol. 151, 175–183. doi: 10.1046/j.1469-8137.2001.00171
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00469
Huang, J., Liu, F., Chao, D., Xin, B., Liu, K., Cao, S., et al. (2022). The WRKY transcription factor OsWRKY54 is involved in salt tolerance in rice. Int. J. Mol. Sci. 23, 11999. doi: 10.3390/ijms231911999
Huang, K., Peng, L., Liu, Y., Yao, R., Liu, Z., Li, X., et al. (2018). Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 498, 92–98. doi: 10.1016/j.bbrc.2017.11.175
Jung, S., Koo, K. M., Ryu, J., Baek, I., Kwon, S. J., Kim, J. B., et al. (2021). Overexpression of phosphoribosyl pyrophosphate synthase enhances resistance of Chlamydomonas to ionizing radiation. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.719846
Komatsu, S., Yang, G., Khan, M., Onodera, H., Toki, S., Yamaguchi, M. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723. doi: 10.1007/s00438-007-0220-6
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 433. doi: 10.1186/1471-2164-14-433
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Li, C. L., Wang, M., Wu, X. M., Chen, D. H., Lv, H. J., Shen, J. L., et al. (2016). THI1, a thiamine thiazole synthase, interacts with Ca2+-dependent protein kinase CPK33 and modulates the S-Type anion channels and stomatal closure in Arabidopsis. Plant Physiol. 170, 1090–1104. doi: 10.1104/pp.15.01649
Linghu, B., Xu, Z., Chu, Y., Yan, Y., Nie, X., Weining, S. (2023). Genome-wide analysis of calcium-dependent protein kinase (CDPK) family and functional characterization of TaCDPK25-U in response to drought stress in wheat. Environ. Exp. Bot. 209, 105277. doi: 10.1016/j.envexpbot.2023.105277
Liu, H., Wang, K., Mei, Q., Wang, X., Yang, J., Ma, F., et al. (2023a). Genome-wide analysis of the Actinidia chinensis NHX family and characterization of the roles of AcNHX3 and AcNHX7 in regulating salt tolerance in Arabidopsis. Environ. Exp. Bot. 214, 105477. doi: 10.1016/j.envexpbot.2023.105477
Liu, M., Wang, C., Xu, Q., Pan, Y., Jiang, B., Zhang, L., et al. (2023b). Genome-wide identification of the CPK gene family in wheat (Triticum aestivum L.) and characterization of TaCPK40 associated with seed dormancy and germination. Plant Physiol. Biochem. 196, 608–623. doi: 10.1016/j.plaphy.2023.02.014
Ma, S. Y., Wu, W. H. (2007). AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 65, 511–518. doi: 10.1007/s11103-007-9187-2
Mehlmer, N., Wurzinger, B., Stael, S., Hofmann-Rodrigues, D., Csaszar, E., Pfister, B., et al. (2010). The Ca2+ -dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 63, 484–498. doi: 10.1111/j.1365-313X.2010.04257.x
Miao, R., Li, M., Wen, Z., Meng, J., Liu, X., Fan, D., et al. (2023). Whole-genome identification of regulatory function of cdpk gene families in cold stress response for Prunus mume and Prunus mume var. Tortuosa. Plants (Basel). 12, 2548. doi: 10.3390/plants12132548
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135, 2150–2161. doi: 10.1104/pp.104.041442
Pirayesh, N., Giridhar, M., Ben Khedher, A., Vothknecht, U. C., Chigri, F. (2021). Organellar calcium signaling in plants: An update. Biochim. Biophys. Acta Mol. Cell Res. 1868, 118948. doi: 10.1016/j.bbamcr.2021.118948
Qiu, Q. S., Barkla, B. J., Vera-Estrella, R., Zhu, J. K., Schumaker, K. S. (2003). Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 132, 1041–1052. doi: 10.1104/pp.102.010421
Rochaix, J. D. (1995). Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 29, 209–230. doi: 10.1146/annurev.ge.29.120195.001233
Rutschmann, F., Stalder, U., Piotrowski, M., Oecking, C., Schaller, A. (2002). LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129, 156–168. doi: 10.1104/pp.000869
Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. doi: 10.1046/j.1365-313x.2000.00787.x
Sánchez-Martín, J., Rubiales, D., Flores, F., Emeran, A. A., Shtaya, M. J. Y., Sillero, J. C., et al. (2014). Adaptation of oat (Avena sativa) cultivars to autumn sowings in Mediterranean environments. Field Crops Res. 156, 111–122. doi: 10.1016/j.fcr.2013.10.018
Shen, Q., Fu, L., Su, T., Ye, L., Huang, L., Kuang, L., et al. (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 183, 1650–1662. doi: 10.1104/pp.20.00196
Shi, H., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Shi, D., Sheng, Y. (2005). Effect of various salt–alkaline mixed stress conditions on sunflower seedlings and analysis of their stress factors. Environ. Exp. Bot. 54, 8–21. doi: 10.1016/j.envexpbot.2004.05.003
Shkolnik, D., Finkler, A., Pasmanik-Chor, M., Fromm, H. (2019). CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A key regulator of Na+ homeostasis during germination. Plant Physiol. 180, 1101–1118. doi: 10.1104/pp.19.00119
Trewavas, A., Gilroy, S. (1991). Signal transduction in plant cells. Trends Genet. 7, 356–361. doi: 10.1016/0168-9525(91)90255-O
Trewavas, A. J., Malhó, R. (1998). Ca2+ signalling in plant cells: the big network! Curr. Opin. Plant Biol. 1, 428–433. doi: 10.1016/S1369-5266(98)80268-9
Wang, C., Chen, X., Li, H., Wang, J., Hu, Z. (2017a). Artificial miRNA inhibition of phosphoenolpyruvate carboxylase increases fatty acid production in a green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels. 10, 91. doi: 10.1186/s13068-017-0779-z
Wang, J., Cheng, G., Wang, C., He, Z., Lan, X., Zhang, S., et al. (2017b). The bHLH transcription factor CgbHLH001 is a potential interaction partner of CDPK in halophyte Chenopodium glaucum. Sci. Rep. 7, 8441. doi: 10.1038/s41598-017-06706-x
Wang, Z., Zhang, Z., Wang, P., Qin, C., He, L., Kong, L., et al. (2023). Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int. J. Biol. Macromol. 240, 124436. doi: 10.1016/j.ijbiomac.2023.124436
Wen, F., Ye, F., Xiao, Z., Liao, L., Li, T., Jia, M., et al. (2020). Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass Brachypodium distachyon. BMC Genom. 21, 53. doi: 10.1186/s12864-020-6475-6
Wilson, S., Kim, E., Ishii, A., Ruban, A. V., Minagawa, J. (2023). Overexpression of LHCSR and PsbS enhance light tolerance in Chlamydomonas reinhardtii. J. Photochem. Photobiol. B. 244, 112718. doi: 10.1016/j.jphotobiol.2023.112718
Yang, S., Cai, W., Shen, L., Cao, J., Liu, C., Hu, J., et al. (2021). A CaCDPK29–CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 233, 1843–1863. doi: 10.1111/nph.17891
Yazdani, M., Croen, M. G., Fish, T. L., Thannhauser, T. W., Ahner, B. A. (2021). Overexpression of native ORANGE (OR) and OR mutant protein in Chlamydomonas reinhardtii enhances carotenoid and ABA accumulation and increases resistance to abiotic stress. Metab. Eng. 68, 94–105. doi: 10.1016/j.ymben.2021.09.006
Yu, H., Li, J., Chang, X., Dong, N., Chen, B., Wang, J., et al. (2024). Genome-wide identification and expression profiling of the WRKY gene family reveals abiotic stress response mechanisms in Platycodon grandiflorus. Int. J. Biol. Macromol. 257, 128617. doi: 10.1016/j.ijbiomac.2023.128617
Zhang, H. F., Liu, D. Y., Yang, B., Liu, W. Z., Mu, B. B., Song, H. X., et al. (2020). Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 71, 188–203. doi: 10.1093/jxb/erz432
Zhang, X., Wang, T., Liu, M., Sun, W., Zhang, W. H. (2019). Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ. Exp. Bot. 157, 79–90. doi: 10.1016/j.envexpbot.2018.09.022
Zhao, P., Liu, Y., Kong, W., Ji, J., Cai, T., Guo, Z. (2021a). Genome-wide identification and characterization of calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in Medicago truncatula. Int. J. Mol. Sci. 22, 1044. doi: 10.3390/ijms22031044
Zhao, R., Sun, H., Zhao, N., Jing, X., Shen, X., Chen, S. (2015). The Arabidopsis Ca²+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 563, 203–214. doi: 10.1016/j.gene.2015.03.024
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, P. (2021b). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22, 4609. doi: 10.3390/ijms22094609
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb). 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhou, H., Shi, H., Yang, Y., Feng, X., Chen, X., Xiao, F., et al. (2024). Insights into plant salt stress signaling and tolerance. J. Genet. Genomics 51, 16–34. doi: 10.1016/j.jgg.2023.08.007
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, H., He, M., Jahan, M. S., Wu, J., Gu, Q., Shu, S., et al. (2021). CsCDPK6, a CsSAMS1-interacting protein, affects polyamine/ethylene biosynthesis in cucumber and enhances salt tolerance by overexpression in Tobacco. Int. J. Mol. Sci. 22, 11133. doi: 10.3390/ijms222011133
Zhu, S. Y., Yu, X. C., Wang, X. J., Zhao, R., Li, Y., Fan, R. C., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036. doi: 10.1105/tpc.107.050666
Zou, J. J., Li, X. D., Ratnasekera, D., Wang, C., Liu, W. X., Song, L. F., et al. (2015). Arabidopsis calcium-dependent protein kinase8 and catalase3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Keywords: oat (Avena sativa L.), saline-alkali stresses, calcium-dependent protein kinase (CDPK), Chlamydomonas reinhardtii, genetic transformation, Na+/ H+ antiporter 1 (NHX1)
Citation: Li Y-n, Lei C, Yang Q, Yu X, Li S, Sun Y, Ji C, Zhang C, Xue J-a, Cui H and Li R (2024) Identification and expression analysis of calcium-dependent protein kinase family in oat (Avena sativa L.) and their functions in response to saline-alkali stresses. Front. Plant Sci. 15:1395696. doi: 10.3389/fpls.2024.1395696
Received: 04 March 2024; Accepted: 18 September 2024;
Published: 10 October 2024.
Edited by:
David Wm Leung, University of Canterbury, New ZealandReviewed by:
Anna Kulik, Polish Academy of Sciences, PolandAoxue Wang, Northeast Agricultural University, China
Copyright © 2024 Li, Lei, Yang, Yu, Li, Sun, Ji, Zhang, Xue, Cui and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-ai Xue, MzA2MjE0ODAzQHFxLmNvbQ==; Hongli Cui, Y3VpaG9uZ2xpQHN4YXUuZWR1LmNu; Runzhi Li, cmxpMjAwMUAxMjYuY29t
 Ya-nan Li1
Ya-nan Li1 Xiao Yu
Xiao Yu Yan Sun
Yan Sun Chunli Ji
Chunli Ji Jin-ai Xue
Jin-ai Xue Hongli Cui
Hongli Cui Runzhi Li
Runzhi Li