
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 May 2024
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1394213
This article is part of the Research TopicOmics-Driven Crop Improvement for Stress Tolerance, volume IIView all 12 articles
 Hanlin Lai1,2
Hanlin Lai1,2 Yuyang Shen2
Yuyang Shen2 Hong Yang2
Hong Yang2 Dilantha W. G. Fernando3
Dilantha W. G. Fernando3 Chenrong Ren4
Chenrong Ren4 Feifei Deng2
Feifei Deng2 Yi Lu4
Yi Lu4 Na Sun5
Na Sun5 Li Chen2
Li Chen2 Guangkuo Li2
Guangkuo Li2 Huiqing Wang4*
Huiqing Wang4* Haifeng Gao2*
Haifeng Gao2* Yue Li1*
Yue Li1*Background: Stripe rust, caused by the fungus Puccinia striiformis f.sp. tritici (Pst), poses a significant threat to global wheat production.
Objectives: This study aims to analyze the distribution of stripe rust resistance genes, characterize resistance phenotypes at the seedling stage of 137 spring and 149 winter wheat varieties in Xinjiang, China, and discern differences in resistance between spring and winter wheat varieties.
Design: We used various Pst races (CYR23, CYR29, CYR31, CYR32, CYR33, CYR34) to characterize seedling resistance of spring and winter wheat varieties and to correlate resistance to the presence of wheat resistance genes (Yr5, Yr9, Yr10, Yr15, Yr17, Yr18, Yr26, Yr41, Yr80, Yr81) using molecular markers.
Results: Among spring wheat varieties, 62, 60, 42, 26, 51, and 24 varieties exhibited resistance to CYR23, CYR29, CYR31, CYR32, CYR33, and CYR34, respectively, with four varieties resistant to all varieties. Among winter wheat varieties, 66, 32, 69, 26, 83, 40 varieties demonstrated resistance to CYR23, CYR29, CYR31, CYR32, CYR33, and CYR34, respectively, with four varieties resistant to all varieties. Molecular testing revealed that, in spring wheat, 2, 17, 21, 61, 10, 0, 10, 79, and 32 varieties carried Yr9, Yr10, Yr15, Yr17, Yr18, Yr26, Yr41, Yr80, and Yr81 genes, respectively. In winter wheat, 40, 20, 7, 143, 15, 1, 6, 38, and 54 varieties carried Yr9, Yr10, Yr15, Yr17, Yr18, Yr26, Yr41, Yr80, and Yr81 genes, respectively. Notably, winter wheat exhibited a significantly higher resistance frequency than spring wheat, particularly in the incidence of Yr9, Yr10, Yr17, Yr18, and multi-gene combinations.
Conclusion: In summary, this study provides information on seedling stage resistance to stripe rust 286 Xinjiang wheat varieties, elucidates the distribution of resistance genes in this population, and offers a mechanistic basis for breeding durable resistance in wheat. varieties from Xinjiang.
China is the world’s largest producer of wheat, generating 128 million metric tons per annum (Zhao and Kang, 2023). In China, wheat is cultivated across multiple environmentally unique and geographically isolated regions, giving rise to multiple strains of the parasitic fungus, Puccinia striiformis f. sp. tritici (Pst), the causal agent of wheat stripe rust (Zeng and Luo, 2006; Wan et al., 2007; Zhan et al., 2016; Ma et al., 2023). Pst is an ancient airborne pathogen that specifically colonizes wheat during its asexual life cycle (Brown and Hovmoller, 2002; Schwessinger, 2017; Li et al, 2021; Zhao et al., 2013), and Pst urediniospores are dispersed via wind, resulting in specific races localizing to certain regions (Yao et al., 2019a). Virulent strains of Pst cause economically devasting stripe rust outbreaks, which resulted in an average yield loss of 1.54 million metric tons per year from 2000 to 2018 in China alone (Zhang et al., 2022). Wheat stripe rust has been responsible for several pandemics in China, most recently in 2017, which resulted in massive yield losses and significant economic impacts (Shen et al., 2002; Wan et al., 2007; Chen et al., 2009; Ma, 2018).
Local varieties of wheat are often referred to as landrace varieties. These are crop composite populations that have been adapted to specific climatic and geographic conditions of a particular region over a long period of cultivation, and they are relatively genetically stable (Shen et al., 2002). Xinjiang Province is found far northwest of China, bordering Tibet, and is classified as an independent epidemic area of wheat stripe rust (Zeng and Luo, 2006). In China, wheat stripe rust has the characteristics of high epidemic frequency, wide occurrence range, and severe damage to wheat production (Jiang et al., 2022). Xinjiang wheat appears to have originated through natural hybridization between Polish and common wheat, suggesting that local wheat was likely introduced and selectively bred within the region itself rather than through the domestication of wild wheat (Betts et al., 2014). Furthermore, Xinjiang’s distinctive climate allows for the cultivation of both oversummer and overwinter wheat, providing year-round hosts for Pst. Xinjiang is also home to Chinese Berberis, or the barberry plant, the host required for the sexual reproduction phase of the Pst lifecycle, indicating that Xinjiang supports significant diversity in Pst races (Zhuang, 2019). To protect wheat crops within Xinjiang, it is critical to determine the resistance distribution of local wheat cultivars to prevalent races of Pst. Additionally, as wheat varieties expressing a single resistance gene are cultivated over time, selective pressure has driven pathogen evolution to escape gene-for-gene resistance, leading to obsolete resistance genes (Kang et al., 2015; Li et al., 2020). Therefore, Xinjiang’s unique local landraces represent valuable genetic sources for potential resistance genes against Pst that can be bred into major cultivars (Mujeeb-Kazi et al., 2013; Chen et al., 2016, Chen et al., 2018; Yao et al., 2019b; Dai et al., 2020).
Despite the effective use of fungicides to combat recent epidemics, deployment of R-genes against virulent Pst races remains the most economically and environmentally viable approach (Zhao and Kang, 2023). Currently, the dominant Pst races in China, designated with the CYR (Chinese yellow rust) prefix, are CYR32, CYR33, and CYR34 (Zhao and Kang, 2023). Over the last century, the Yr wheat resistance genes Yr9, Yr10, Yr17, and Yr26 have been extensively used in breeding for wheat stripe rust resistance (Han et al., 2010). With the emergence of new virulent races, such as CYR29, CYR31, CYR32, and CYR34, resistance conveyed by Yr9, Yr10, Yr17, and Yr26 has been revealed (Han et al., 2010). Some all-stage resistance (ASR) genes, such as Yr5 and Yr15, are still effective against prevalent races of Pst, but are rarely used in breeding (Zeng et al., 2014). The Yr18 gene confers non-race specific resistance to stripe rust and slows infection time and spore production, resulting in adult plant resistance (APR) (Singh et al., 2000). Presently, the main race types prevalent in Ili prefecture, the largest wheat-producing region of Xinjiang, are CYR34 and Su 11-1 (Chen et al., 2023). It has been shown that Yr5 and Yr15 are highly resistant to stripe rust (Zhou et al., 2023), making the detection of these resistance genes essential for the prevention and control of wheat stripe rust in Xinjiang.
With the expansion of molecular marker technologies, an increasing number of molecular markers are being used in the breeding of disease-resistant wheat varieties. Currently, the major molecular markers based on genomic DNA molecular polymorphisms include sequence tagged sites (STS), simple sequence repeats (SSR), and kompetitive allele-specific polymerase chain reactions (KASP) (Song et al., 2023). By detecting linked molecular markers, disease resistance genes in wheat can be efficiently detected, either directly or indirectly. Identification of cultivars containing effective resistance genes permits selective breeding for robust resistance and enables the stacking of multiple genes to establish long-lasting disease-resistant varieties (Luo et al., 2023). Presently, 83 stripe rust resistance genes (Yr1-Yr83) have been discovered and conclusively named, and more than 300 genes or QTL have been identified and temporarily named (McIntosh et al., 2009; Li et al., 2020).
A previous study detected a high frequency of Yr9 in wheat cultivars of the Huang-Huai region, along with significant expression of Yr18 in Huang-Huai landraces (Huang et al., 2020). Alternatively, the main wheat varieties in southwest China exhibit a high frequency of polygene polymerization (Xi et al., 2021). Landraces in northwest China display high frequency of Yr9, which has been widely used for breeding, as well as varieties with multiple Yr aggregates (Wang et al., 2018). The ASR genes Yr5 and Yr15 are generally absent from wheat panels in China, where only a few reports identified Yr15 (Li et al., 2016). Regions adjacent to Xinjiang, such as Pakistan and Kazakhstan, have the highest distribution frequency of Yr18 and Yr10, which convey resistance to most races of stripe rust (Sobia et al., 2010; Kokhmetova et al., 2021). The frequencies of Yr26 and Yr10 in Indian wheat are 69.2% and 50%, respectively, with some highly resistant varieties containing stacks of 15 Yr genes (Rani et al., 2019).
Here, we perform a systematic study of phenotypic variation and molecular characterization of stripe rust resistance of winter and spring wheat verities in Xinjiang. We employ a panel of 149 winter wheat varieties and 137 spring wheat varieties in Xinjiang for the characterization of stripe rust resistance against six races at the seedling stage. We further correlate resistance phenotypes to the prevalence of 10 stripe rust resistance genes, as identified with appropriate molecular markers. While the distribution of wheat stripe rust resistance genes in Xinjiang has been recently evaluated, this study is the first to directly assess resistance phenotypes in tandem with the identification of resistance genes (Zhang et al., 2023). This work provides valuable resources for the identification of breeding targets for enhancing wheat stripe rust resistance from the pool of Xinjiang wheat.
A total of 286 wheat varieties, including 137 spring and 149 winter varieties, were tested. The Spring Wheat Breeding Team of the Grain Crops Research Institute of the Xinjiang Academy of Agricultural Sciences provided the 137 spring wheat varieties. The Wheat Breeding Team of the Agricultural Research Institute of the Ili Region contributed 55 YINONG winter wheat varieties. The Winter Wheat Breeding Team of the Grain Crops Research Institute of the Xinjiang Academy of Agricultural Sciences provided 94 wheat varieties of the winter wheat varieties. Associate Professor Zhan Gangming from the College of Plant Protection, North Agriculture and Forestry University of Science and Technology, provided the control materials, including Mingxian 169 and single gene line materials.
Phenotyping of seedling diseases was carried out in climatic chambers at the Institute of Plant Protection, Xinjiang Academy of Agricultural Sciences, Xinjiang, China. In brief, 10–15 seeds from each wheat variety were sown in small pots and cultivated indoors until the one-leaf-one-heart stage. Seedlings were inoculated with Pst races combined with e-fluoridized solution using a pipetting gun set to 5 μL and then maintained in dark conditions for 24 h (10°C). Afterward, seedlings were transferred to climatic chambers (12-hr light/12-hr dark). When symptoms were fully visible in the control group, Mingxian 169, the infection type (IT) was assessed and classed as high resistance (IT: 0–3), moderate resistance (IT: 4–5), or high susceptibility (IT: 6–9).
Appropriate amounts of wheat leaf tissue were collected, and a modified CTAB method was used for the extraction of genomic DNA (Allen et al., 2006). The quality and quantity of the extracted genomic DNA was determined using a NanoDrop 610 spectrophotometer (ThermoScientific, Wilmington, DE, USA), and the DNA was diluted to 100 ng/μL and stored at -20°C. Polymerase chain reaction (PCR) amplification using specific primers for SSR, STS, and SNP molecular markers associated with Yr5, Yr9, Yr10, Yr15, Yr17, Yr18, Yr26, Yr41, Yr80, and Yr81 was performed, followed by agarose gel electrophoresis on 1–2% agarose gel. PCR reactions consisted of 2× Easy Taq PCR mix (12.5 μL), forward primer (F: 1 μL), and reverse primer (R: 1 μL). PCR reactions involved a 4-minute predenaturation at 94°C, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55–65°C for 30 seconds, and extension at 72°C for 30–60 seconds. KASP-SNP molecular marker PCR reactions consisted of 2× KASP mix (170 μL), forward primer 1 (F1: 0.51 μL), forward primer 2 (F2: 0.51 μL), and reverse primer (R: 1.36 μL). Subsequently, 1 μL of DNA template was added to each reaction. Primers were synthesized by Shanghai Bioengineering. Refer to Table 1 for primer sequences.
UpSetR (version 1.4.0) package in the R environment was used for gene combination analysis, and ggplot2 (version 3.4.4) and venn (version 1.11) packages were used for data processing and graph generation, respectively.
The number of varieties of spring wheat resistant to CYR23, CYR29, CYR31, CYR32, CYR33, and CYR34 amounted to 62 (45.26%), 60 (43.8%), 44 (32.12%), 25 (18.25%), 50 (36.60%), and 25 (18.25%), respectively. Notably, Xinchun No. 32, Xinchun No. 51, Liangchun1723, and Liangchun1817 were resistant to all 6 races, accounting for 2.92% of all resistant varieties (Figure 1A; Supplementary Table S1).
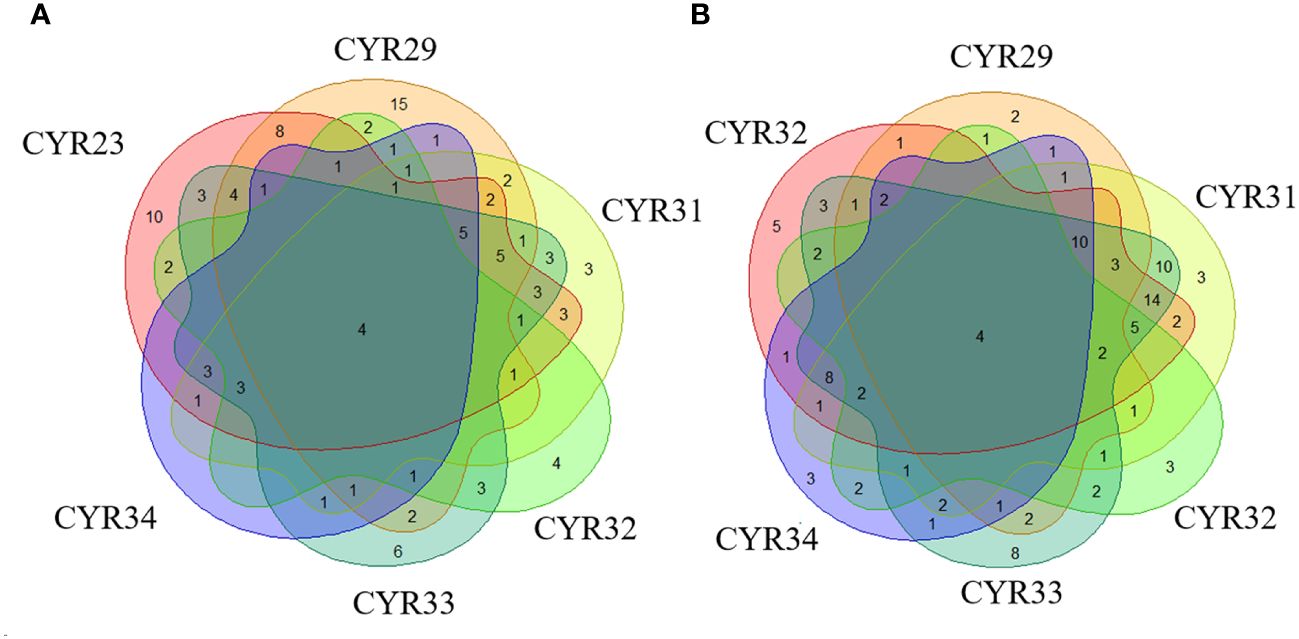
Figure 1 Identification of resistance against six Pst races in spring and winter wheat landraces from Xinjiang. (A) Depicts the number of varieties resistant to different races in spring wheat, and (B) Depicts the number of varieties resistant to different races in winter wheat.
We identified 62 (44.3%), 32 (21.48%), 70 (46.89%), 26 (17.45%), 84 (56.38%), and 40 (26.85%) winter wheat varieties resistant to CYR23, CYR29, CYR31, CYR32, CYR33, and CYR34, respectively. Among these, varieties 2014-132-4-5, 2014-129-13-9, 6239, and 6444 were resistant to all 6 races, accounting for 2.68% of all resistant varieties (Figure 1B; Supplementary Table S2).
Presently, the predominant races in Xinjiang are CYR32, CYR33, and CYR34. A detailed analysis of these races has revealed that 11 spring wheat varieties exhibited resistance to CYR33 and CYR34, including Xinchun No. 12, Xinchun No. 14, and Xinchun No. 16. Four varieties, including Xinchun No. 34, Xinchun No. 39, and Liangchun1934, exhibited resistance to CYR33 and CYR32. Another four varieties, including 2016, Liangchun1758, and Liangchun1832, demonstrated resistance to CYR32 and CYR34. Eight varieties, including Xinchun No. 29, Heli1881, and Liangchun547, exhibited resistance to all three races.
Among the winter wheat varieties, 24 varieties, including Zhaonong147, 2014-132-10-10, and 2014-12-3-1, demonstrated resistance to both CYR33 and CYR34. Twelve varieties exhibited resistance to CYR33 and CYR32, including 2014-129-1-6, Pin I-1, and Pin I-8. Additionally, 2 varieties, namely Jindong008 and Tiandong33, exhibited resistance to both CYR32 and CYR34, whereas 7 varieties, including 2014-132-10-6, 6222, and 6238, demonstrated resistance to all three races. Overall, Xinjiang wheat exhibits the highest frequency of resistance to CYR33 among the Su 11 taxa. Previous surveys have also revealed the highest proportion of Su 11 pathogenic taxa of stripe rust (Chen et al., 2023; Ma et al., 2023). Consequently, Xinjiang wheat possesses resistance to the currently prevalent races.
Our results indicate that the resistance of spring and winter wheat to CYR23 and CYR32 was similar. While the probability of spring wheat possessing resistance to CYR29 was significantly higher than that of winter wheat (Figure 2), the probability of winter wheat possessing resistance to CYR31, CYR33, and CYR34 was significantly higher than that of spring wheat (Figure 2).
Molecular markers closely linked to the Yr5, Yr9, Yr10, Y15, Yr17, Yr18, Yr26, Yr41, Yr80, and Yr81 genes were used to identify potential Yr genes in our spring wheat panel (Figure 3; Supplementary Table S1). Spring wheat varieties Xinchun No. 33 and 2020J/54 were identified as potential carriers of Yr9. The Yr10-linked marker amplified the target band in 17 (12.41%) varieties. We identified 21 (15.32%) varieties potentially carrying Yr15. The VENTRIUP-LN2 marker, which is closely linked to Yr17, amplified the target band in 61 (44.52%) varieties. The KASP marker, which is closely linked to Yr18, amplified the target band in 10 (7.30%) varieties (Figure 3C). BE446068F, which is closely linked to Yr41, was amplified in 10 (7.30%) varieties. The KASP marker, closely linked to Yr80, was amplified in 79 (57.66%) varieties (Figure 3A), while the KASP marker, closely linked to Yr81, was amplified in 32 (23.36%) varieties (Figure 3B). Yr26 and Yr5 was not detected in all spring wheat varieties. Yr17 and Yr80 genes exhibited the highest frequency of distribution in spring wheat.
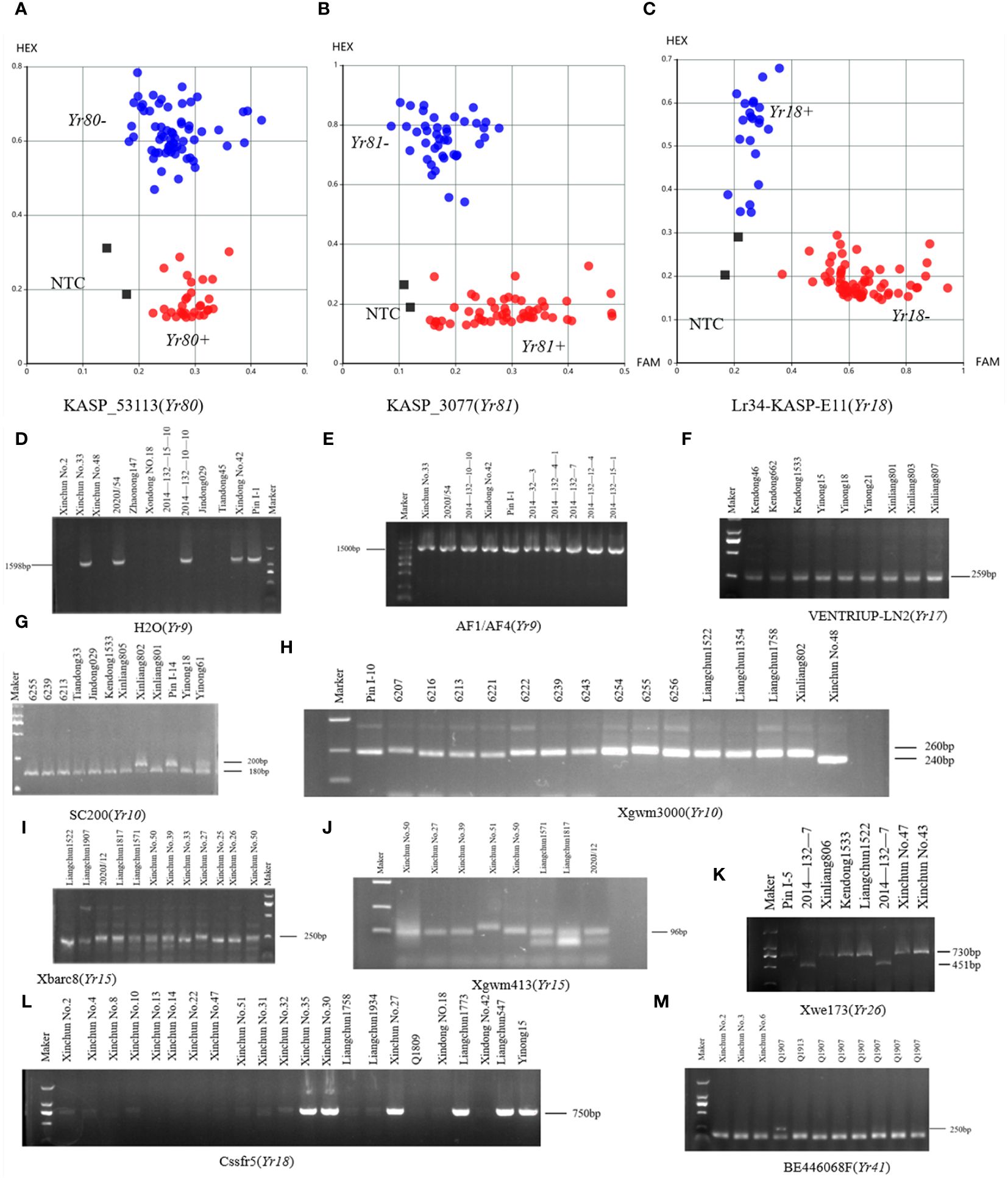
Figure 3 Results of PCR amplification of wheat Yr gene. (A–C) KASP cluster plots of Yr80, Yr81, and Yr18 genotyping. (D–M) Electrophoretograms of the primers for Yr9, Yr10, Yr15, Yr17, Yr18, Yr26, and Yr41 resistance genes.
In the winter wheat panel, 40 (26.85%) varieties were identified as potential carriers of Yr9. The Yr10 marker amplified the target band in 20 (13.42%) varieties. We identified 7 (4.7%) varieties potentially carrying Yr15. The VENTRIUP-LN2 marker amplified the target band in 143 (95.97%) varieties. The KASP marker, which is closely linked to Yr18, amplified the target band in 15 (10.03%) varieties (Figure 3C). Furthermore, 2014-132-7 was found to harbor the Yr26 coding region, where the closely linked WE173 marker amplified the target band. BE446068F was amplified in 6 (4.03%) varieties. The KASP marker, closely linked to Yr80, was amplified in 38 (25.50%) varieties (Figure 3A). Furthermore, 54 (36.24%) varieties amplified the KASP marker, which is closely linked to Yr81 (Figure 3B; Supplementary Table S2). Notably, as observed for spring wheat varieties, Yr17 and Yr80 also showed the highest frequency of distribution in winter wheat.
The results of our Yr gene distribution analysis are summarized in Figure 4. Specifically, we observed that the proportions of winter wheat expressing Yr9, Yr17, Yr18, and Yr81 were 26.85%, 95.97%, 10.07%, and 36.24%, respectively. These genes were significantly more prevalent in winter than in spring wheat, whereas the proportion of winter wheat varieties carrying Yr10 (13.42%) was slightly higher than spring wheat varieties. The proportions of Yr15 (15.32%) and Yr41 (7.3%) in spring wheat were slightly higher than in winter wheat, whereas the proportion of Yr80 (57.66%) in spring wheat was significantly higher than in winter wheat.
To further investigate the distribution of resistance genes in different wheat varieties, we assessed the number of varieties containing aggregates of 2, 3, 4, and 5 Yr genes in spring and winter wheat (Figure 5). The number of multi-gene combined varieties of winter wheat was higher than that of spring wheat. In spring wheat, Yr17+Yr80 was found in 29 of the 137 varieties. However, there were no varieties with 5 or more Yr genes (Figure 5A). Similarly, Yr17+Yr81 was found in 25 of the 149 winter wheat varieties. Notably, we identified an aggregate in Pin I-9 containing Yr17, Yr81, Yr9, Yr80, and Yr18 (Figure 5B).
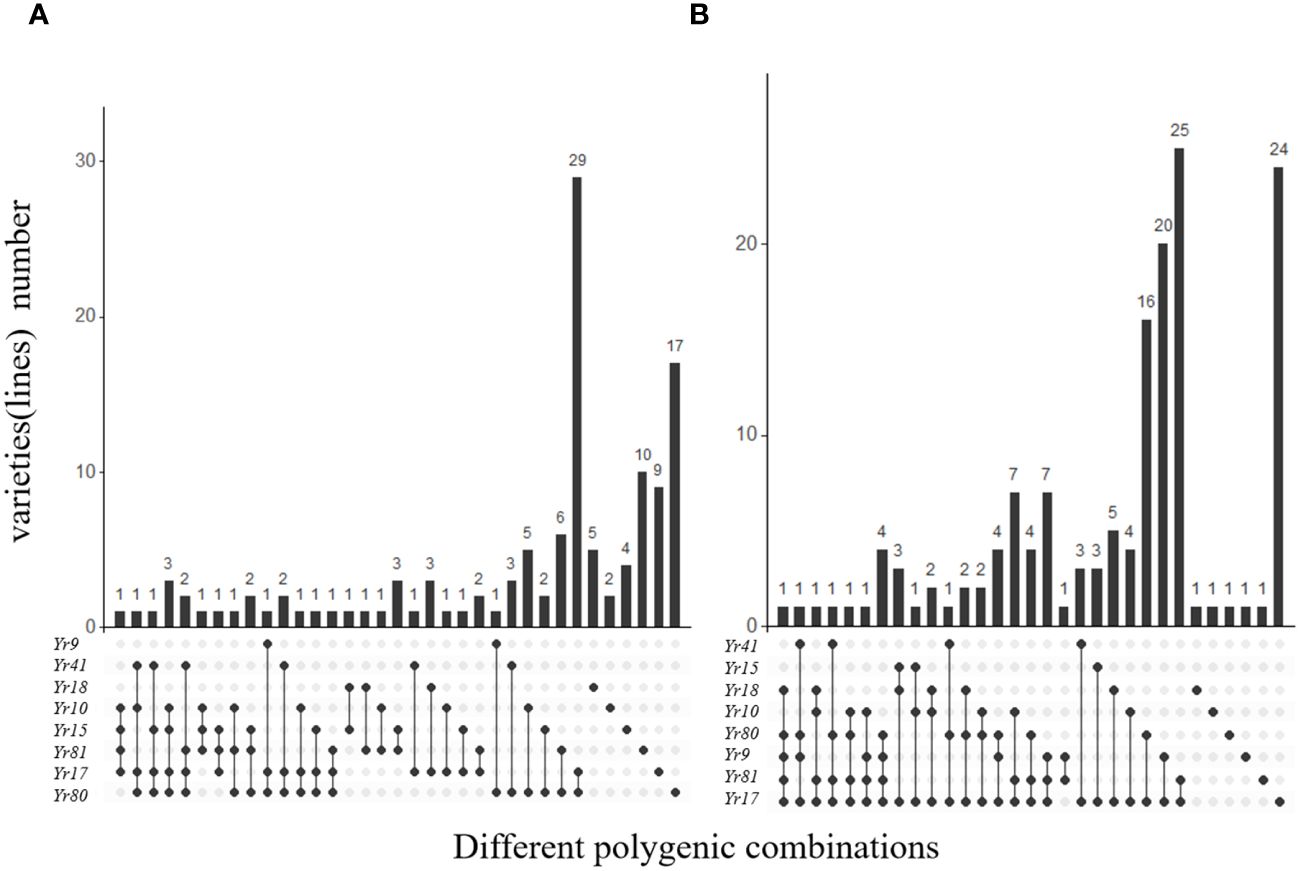
Figure 5 Comparative analysis of Yr genes found in 137 spring and 149 winter wheat varieties from Xinjiang. (A) Gives the combinations of different Yr genes in spring wheat, and (B) Gives the combinations of different Yr genes in winter wheat.
We assessed disease resistance phenotypes against 6 races of Pst in a wheat panel that contained 286 spring and winter wheat varieties found in Xinjiang. Our results show that spring wheat has strong resistance to early popular races CYR23 and CYR29 but weak resistance to currently popular races CYR31, CYR32, CYR33, and CYR34. Only 24 varieties were resistant to CYR34. The resistance of winter wheat to predominant races was generally stronger than that of spring wheat. The number of winter wheat varieties resistant to CYR33 was significantly higher than that of other races, followed by resistance to CYR31 and CYR34, two of the most virulent Pst races (Chen et al., 2023; Ma et al., 2023). As the planting area of winter wheat in Xinjiang has expanded, host plants for Pst are maintained through summer and winter, allowing the pathogen to complete its annual cycle. This makes it particularly important to select winter wheat varieties with high resistance to stripe rust at the seedling stage, which cannot only reduce stripe rust in autumn plantings of winter wheat, but also prevent propagation of the disease in spring wheat.
To identify potential resistance loci in wheat, it is critical to assess resistance at the seedling stage in a climate chamber, followed by field experiments with artificial inoculations. This conventional method is limited by natural conditions and requires multiple generations to produce resistant cultivars. Molecular marker-assisted selection (MAS) is an effective tool to guide breeding for heritable resistance traits in breeding cultivars (Song et al., 2023). Here, we applied MAS to assess the distribution of wheat resistance genes in varieties from Xinjiang, to better understand endogenous resistance, and to identify potential loci that could be bred into agricultural lines for improved resistance.
The resistance genes Yr5 and Yr15 have been reported to convey strong all-stage resistance to the races of stripe rust in China (Zeng et al., 2014). However, Yr5 was not detected in our panel, and Yr15 was identified in only a few varieties (Pin I-6, 1-8-2, Liangchun1817). Among wheat varieties that carried Yr15, only Liangchun1817 showed resistance to all races at the seedling stage, which may be due to false Yr15 positives in the susceptible varieties. Based on a previous study, Yr5 is rarely used in wheat breeding in China, and Yr15 has only been detected in one study (Zeng et al., 2014). However, Kazakhstan, which is adjacent to Xinjiang, has high frequencies of Yr5 and Yr15, suggesting active breeding efforts towards Yr5- and Yr15-positive cultivars (Kokhmetova et al., 2021).
The highly virulent race CYR34 has overcome resistance previously conveyed by Yr26 (Han et al., 2010). Based on a previous study of wheat in Xinjiang, the frequency of Yr5 is approximately 100% in spring and winter wheat, and that of Yr26 is 84.15% (Zhang et al., 2023). This is in contrast to our results, where we did not identify Yr5 in either wheat population, and similarly no Yr26 gene in spring wheat and only one Yr26 gene in a winter wheat line (2014-132-7). This may be due to the use of different molecular markers, and possibly false positives or negatives. We did identify one winter wheat variety with an aggregate containing Yr26, Yr9, and Yr17 that displayed strong resistance to CYR34, indicating that this polygene combination can supplement Yr26-mediated resistance against CYR34. In Indian wheat, Yr26 and Yr10 have high frequencies in the distribution of resistance genes (Rani et al., 2019). While resistance gene distributions in wheat from Xinjiang were similar to those from the rest of China, there were notable differences between Xinjiang and neighboring India and Kazakhstan. This is expected, given the completely different cultivated varieties based on climate and breeder preference. We suggest that a comparative analysis of wheat Yr genes and phenotypes in different regions be performed to reveal the breeding trajectories of different regions.
Yr18 is located on chromosome 7D and codes for an APR gene that slows Pst infections in the adult stage. Additionally, Yr18 conveys resistance to leaf rust, stem rust, and powdery mildew (Singh et al., 2000). While the frequency of Yr18 detection in Chinese wheat landraces is generally high, Yr18 is largely absent from Xinjiang wheat. This could be attributed to the infrequent use of wheat landraces in Xinjiang wheat breeding. Therefore, combination of Yr18 with other ASR and APR genes may significantly improve the stripe rust resistance of Xinjiang wheat varieties.
Yr5, Yr10, Yr15, Yr26, and Yr18 genes exhibit partial resistance to stripe rust and occur infrequently in Xinjiang wheat varieties. Yr26 and Yr10 genes have been overcome by CYR34, yet recent research indicates that the predominant stripe rust population in Xinjiang belongs to the Su 11 pathogenic type (Chen et al., 2023; Ma et al., 2023). Yr26 and Yr10 retain their efficacy in controlling stripe rust in Xinjiang. Wheat varieties in Xinjiang carrying Yr10 and Yr26, such as 2014-129-1-6, Kendong47, and 2014-132-7, can effectively manage the prevalent stripe rust type in the region. Given their strong resistance to Xinjiang stripe rust, these genes can be incorporated into future breeding programs for Xinjiang wheat varieties.
With the source variation of stripe rust and the emergence of new virulence factors, many single resistance genes have been overcome. In a previous analysis of wheat resistance genes, polymerization or stacking of multiple genes has been shown to effectively improve resistance (Zhang et al., 2023). In this study, we found that most of the wheat varieties in Xinjiang contained two or more Yr genes. We found high prevalence of Yr80, Yr81 and all-stage resistant Yr17 in both spring and winter wheat, mainly as components of polygene doublets of Yr17+Yr80 and Yr17+Yr81, as well as some other combinations. The results of wheat resistance to stripe rust in Sichuan, the Huang-Huai-Hai region, and Yunnan were similar to ours, indicating that polygene polymerization, specifically Yr17+Yr81, can improve the resistance to wheat stripe rust (Li et al., 2016; Wang et al., 2018; Guan et al., 2020; Huang et al., 2020; Xi et al., 2021). Yr17 from the 2NS/2AL translocation line of Aegilops has all-stage resistance and the distribution frequency in spring and winter wheat, however, all the prevalent Pst races have overcome Yr17-mediated resistance. Since the emergence of CYR29, only wheat varieties carrying Yr9 alone have lost their resistance. In this study, we found that wheat varieties with Yr9+Yr17 showed strong resistance to wheat stripe rust races. Notably, we identified two Yr9+Yr17 winter wheat lines, 2014-132-4-5 and 2014-129-13-9, that displayed seedling resistance to all 6 races, indicating that the combination of Yr9 and Yr17 can effectively defend against all prevalent Pst races.
Our comprehensive analysis of 286 varieties of spring and winter wheat in Xinjiang provides valuable insights into the dynamics of stripe rust resistance. Spring wheat demonstrates robust resistance to early races CYR23 and CYR29 but shows weak resistance against contemporary races CYR31, CYR32, CYR33, and CYR34, with only 24 varieties displaying resistance to CYR34. Conversely, winter wheat exhibits stronger resistance, particularly against CYR33, CYR31, and CYR34, highlighting the importance of selecting resistant winter wheat varieties to curtail stripe rust propagation into spring wheat.
Utilizing MAS, we identify the distribution of key resistance genes, revealing variations in the prevalence of genes such as Yr5, Yr15, Yr26, and Yr18 compared to previous studies. The absence of Yr5 and limited detection of Yr15 suggest regional disparities in breeding priorities, as seen in neighboring Kazakhstan. The emergence of highly virulent CYR34, overcoming Yr26-mediated resistance, underscores the evolving nature of stripe rust and the need for adaptable resistance strategies.
Our findings emphasize the significance of polygene combinations in enhancing resistance. Notably, the combination of Yr17 and Yr81 demonstrates effectiveness in resisting stripe rust, as observed in the prevalence of Yr17+Yr80 and Yr17+Yr81 combinations. Furthermore, the synergy between Yr9 and Yr17 proves highly effective against all prevalent Pst races, offering promising avenues for breeding resilient wheat varieties.
The absence of Yr18 in Xinjiang wheat, despite its widespread presence in Chinese landraces, underscores the potential for improved resistance through strategic gene combinations. We propose that combining Yr18 with other APR genes could significantly enhance stripe rust resistance in Xinjiang wheat. In conclusion, our study contributes crucial data for breeding durable and resilient wheat varieties in Xinjiang, offering insights into regional resistance dynamics and highlighting the efficacy of polygene combinations in mitigating the impact of evolving stripe rust races. These findings can guide future breeding strategies and foster international collaboration for a more comprehensive understanding of wheat resistance across diverse regions.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
HL: Writing – original draft, Conceptualization, Data curation, Formal analysis. YS: Conceptualization, Writing – original draft. HY: Formal analysis, Writing – original draft. DF: Data curation, Formal analysis, Writing – original draft. CR: Conceptualization, Data curation, Writing – original draft. FD: Data curation, Methodology, Writing – review & editing. YiL: Formal analysis, Funding acquisition, Writing – review & editing. NS: Project administration, Validation, Writing – review & editing. LC: Investigation, Methodology, Writing – review & editing. GL: Funding acquisition, Resources, Writing – review & editing. HW: Funding acquisition, Writing – review & editing. HG: Funding acquisition, Writing – review & editing. YuL: Methodology, Project administration, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by the Science and Technology Assistance Project of Xinjiang Uygur Autonomous Region (2022E02070); Regional Coordinated Innovation Project, Shanghai Cooperation Organization Science and Technology Partnership Program (2023E01015); Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01A135); Key Laboratory of Integrated Pest Management on Crop in Northwestern Oasis (KFJJ202104); Third Tianshan Talent Plan of Xinjiang Uygur Autonomous Region (2022TSYCCX0081); Earmarked Fund for XJARS-01; Science and Technology Major Project of Xinjiang Uygur Autonomous Region (2023A02009); Urumqi Integrated Experimental Station of China Agriculture Research System for Wheat (CARS-03-88); and Project of Fund for Stable Support to Agricultural Sci-Tech Renovation (XJNKYWDZC-2023004-1).
We would like to thank A&L Scientific Editing (www.alpublish.com) for the linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1394213/full#supplementary-material
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Betts, A., Jia, P. W., Dodson, J. (2014). The origins of wheat in China and potential pathways for its introduction: as review. Quatern Int. 348, 158–168. doi: 10.1016/j.quaint.2013.07.044
Brown, J. K., Hovmoller, M. S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 297, 537–541. doi: 10.1126/science.1072678
Chen, L., Awais, M., Yang, H., Shen, Y. Y., Li, G. K., Gao, H. F., et al. (2023). Races CYR34 and Suwon11-1 of Puccinia striiformis f. sp. tritici Played an Important Role in Causing the Stripe Rust Epidemic in Winter Wheat in Yili, Xinjiang, China. J. Fungi. 9, 436. doi: 10.3390/jof9040436
Chen, Q., Song, J., Du, W. P., Xu, L. Y., Jiang, Y., Zhang, J., et al. (2018). Phylogenetic analyses of four Chinese endemic wheat landraces based on two single copy genes. Cereal Res. Commun. 46, 191–200. doi: 10.1556/0806.46.2018.01
Chen, Q., Song, J., Du, W. P., Xu, L. Y., Yu, G. R. (2016). Possible origin of Triticum petropavlovskyi based on cytological analyses of crosses between T. petropavlovskyi and tetraploid, hexaploid, and synthetic hexaploid (SHW-DPW) wheat accessions. Span J. Agric. Res. 14, e0713–e0713. doi: 10.5424/sjar/2016144-8476
Chen, X., Soria, M. A., Yan, G., Sun, J., Dubcovsky, J. (2003). Development of sequence tagged site and cleaved amplified polymorphic sequence markers for wheat stripe rust resistance gene Yr5. Crop Sci. 43, 2058–2064. doi: 10.2135/cropsci2003.2058
Chen, W. Q., Wu, L. R., Liu, T. G., Xu, S. C., Jin, S. L., Peng, Y. L., et al. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93, 1093–1101. doi: 10.1094/PDIS-93-11-1093
Dai, S. F., Xu, D. Y., Yan, Y. L., Wen, Z. J., Zhang, J. B., Chen, H. X., et al. (2020). Characterization of high-and low-molecular-weight glutenin subunits from Chinese Xinjiang wheat landraces and historical varieties. J. Food Sci. Technol. 57, 3823–3835. doi: 10.1007/s13197-020-04414-5
Fang, T. L., Lei, L., Li, G. Q., Powers, C., Hunger, R. M., Carver, B. F., et al. (2020). Development and deployment of KASP markers for multiple alleles of Lr34 in wheat. Theor. Appl. Genet. 133, 2183–2195. doi: 10.1007/s00122-020-03589-x
Francis, H. A., Leitch, A. R., Koebner, R. M. D. (1995). Conversion of a RAPD-generated PCR product, containing a novel dispersed repetitive element, into a fast and robust assay for the presence of rye chromatin in wheat. Theor. Appl. Genet. 90, 636–642. doi: 10.1007/BF00222127
Gessese, M., Bariana, H., Wong, D., Hayden, M., Bansal, U. (2019). Molecular mapping of stripe rust resistance gene Yr81 in a common wheat landrace Aus27430. Plant Dis. 103, 1166–1171. doi: 10.1094/PDIS-06-18-1055-RE
Guan, F. N., Long, L., Yao, F. J., Wang, Y. Q., Jiang, Q. T., Kang, H. Y., et al. (2020). Evaluation of resistance to stripe rust and molecular detection of important known Yr gene (s) of 152 Chinese wheat landraces from the Huang-huai-hai. Scientia Agricultura Sin. 53, 3629–3637. doi: 10.3864/J.issn.0578-1752.2020.18.001
Han, D. J., Wang, Q., Zhang, L., Wei, G., Zeng, Q., Zhao, J., et al. (2010). Evaluation of resistance of current wheat cultivars to stripe rust in northwest China, north China and the middle and lower reaches of Changjiang river epidemic area. Scientia Agricultura Sinica. 43, 2889–2896. doi: 10.3864/j.issn.0578-1752.2010.14.007
Huang, L., Xiao, X. Z., Liu, B., Gao, L., Gong, G. S., Chen, W. Q., et al. (2020). Identification of stripe rust resistance genes in common wheat cultivars from the Huang-Huai-Hai region of China. Plant Dis. 104, 1763–1770. doi: 10.1094/PDIS-10-19-2119-RE
Jia, J. Q., Lie, M. P., Liu, C., Li, G. R., Yang, Z. J. (2010). Exploitation and application of a new SCAR marker linked to strip rust resistance gene Yr17 in wheat. J. Triticeae Crops. 30, 11–16doi: 10.09-1041(2010)01-0011-06
Jiang, Q., Wang, H. L., Wang, H. G. (2022). Two new methods for severity assessment of wheat stripe rust caused by Puccinia striiformis f. sp. tritici. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1002627
Kang, Z. S., Wang, X. J., Zhao, J., Tang, C. L., Huang, L. L. (2015). Advances in research of pathogenicity and virulence variation of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Sci. Agric. Sin. 48, 3439–3453. doi: 10.3864/j.issn.0578-1752.2015.17.011
Kokhmetova, A., Rsaliyev, A., Malysheva, A., Atishova, M., Kumarbayeva, M., Keishilov, Z. (2021). Identification of stripe rust resistance genes in common wheat cultivars and breeding lines from Kazakhstan. Plants. 10, 2303. doi: 10.3390/plants10112303
Lagudah, E. S., Krattinger, S. G., Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., Spielmeyer, W., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 119, 889–898. doi: 10.1007/s00122-009-1097-z
Li, J. B., Dundas, I., Dong, C. M., Li, G. R., Trethowan, R., Yang, Z. J., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, Q., Wang, B. T., Chao, K. X., Guo, J., Song, J. R., Yue, W. Y., et al. (2016). Molecular detection of stripe rust resistance gene (s) in 115 wheat cultivars (lines) from the Yellow and Huai River valley wheat region. J. Phytopathol. 164, 946–958. doi: 10.1111/jph.12515
Li, S. N., Wen, C., Tian, X. X., Yao, L., Huang, L. L., Kang, Z. S., et al. (2021). Identification of eight Berberis species from the Yunnan-Guizhou plateau as aecial hosts for Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen. J. Integr. Agr. 20, 1563–1569. doi: 10.1016/S2095-3119(20)63327-5
Liu, C., Yang, Z. J., Li, G. R., Zeng, Z. X., Zhang, Y., Zhou, J. P., et al. (2008). Isolation of a new repetitive DNA sequence from Secale africanum enables targeting of Secale chromatin in wheat background. Euphytica. 159, 249–258. doi: 10.1007/s10681-007-9484-5
Luo, K., He, D. J., Guo, J., Li, G. W., Li, B. L., Chen, X. L. (2023). Molecular advances in breeding for durable resistance against pests and diseases in wheat: opportunities and challenges. Agronomy. 13, 628. doi: 10.3390/agronomy13030628
Ma, Z. H. (2018). Researches and control of wheat stripe rust in China. J. Plant protection. 45, 1–6. doi: 10.13802/j.cnki.zwbhxb.2018.2018900
Ma, J. B., Awais, M., Chen, L., Yang, H., Lai, H. L., Shen, Y., et al. (2023). Identification of Puccinia striiformis races from the spring wheat crop in Xinjiang, China. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1273306
Marchal, C., Zhang, J. P., Zhang, P., Fenwick, P., Steuernagel, B., Adamski, N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. plants. 4, 662–668. doi: 10.1038/s41477-018-0236-4
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., Appels, R., Xia, X. C., et al. (2009). Catalogue of gene symbols for wheat: 2009 supplement. Annu. wheat newsletter. 55, 256–278.
Mujeeb-Kazi, A., Kazi, A. G., Dundas, I., Rasheed, A., Ogbonnaya, F., Kishii, M., et al. (2013). Genetic diversity for wheat improvement as a conduit to food security. Adv. Agron. 122, 179–257. doi: 10.1016/B978-0-12-417187-9.00004-8
Nsabiyera, V., Bariana, H. S., Qureshi, N., Wong, D., Hayden, M. J., Bansal, U. K. (2018). Characterisation and mapping of adult plant stripe rust resistance in wheat accession Aus27284. Theor. Appl. Genet. 131, 1459–1467. doi: 10.1007/s00122-018-3090-x
Peng, J. H., Fahima, T., Röder, M. S., Huang, Q. Y., Dahan, A., Li, Y. C., et al. (2000). High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica. 109, 199–210. doi: 10.1023/A:1017573726512
Rani, R., Singh, R., Yadav, N. R. (2019). Evaluating stripe rust resistance in Indian wheat genotypes and breeding lines using molecular markers. Cr Biol. 342, 154–174. doi: 10.1016/j.crvi.2019.04.002
Schwessinger, B. (2017). Fundamental wheat stripe rust research in the 21st century. New Phytol. 213, 1625–1631. doi: 10.1111/nph.14159
Shao, Y. T., Niu, Y. C., Zhu, L. H., Zhai, W. X., Xu, S. C., Wu, L. R. (2001). Identification of an AFLP marker linked to the stripe rust resistance gene Yr10 in wheat. Chin. Sci. Bull. 46, 1466–1468. doi: 10.1007/BF03187033
Shen, Y. H., Wang, H. Q., Yang, T. Y., Zhang, H. G., Huang, X. G. (2002). Evolvement of genetic diversity of spring wheat varieties in Gansu and Qinghai Provinces. Acta Botanica Boreali-occidentalia Sinica. 22, 731–740. doi: 10.1023/A:1017573726512
Singh, R. P., Nelson, J. C., Sorrells, M. E. (2000). Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155. doi: 10.2135/cropsci2000.4041148x
Sobia, T., Muhammad, A., Chen, X. M. (2010). Evaluation of Pakistan wheat germplasms for stripe rust resistance using molecular markers. Sci. China Life Sci. 53, 1123–1134. doi: 10.1007/s11427-010-4052-y
Song, L. Q., Wang, R. H., Yang, X. J., Zhang, A. M., Liu, D. C. (2023). Molecular markers and their applications in marker-assisted selection (MAS) in bread wheat (Triticum aestivum L.). Agriculture. 13, 642. doi: 10.3390/agriculture13030642
Wan, A. M., Chen, X. M., He, Z. H. (2007). Wheat stripe rust in China. Aust. J. Agr Res. 58, 605–619. doi: 10.1071/AR06142
Wang, S. H., Gong, K. Y., Chu, B. Y., Sun, Q. Y., Luo, Y., Ma, Z. H. (2018). Molecular detection of stripe rust resistance gene (s) in 100 wheat cultivars (lines) from Sichuan Province in China. Acta Phytopathologica Sinica. 48, 195–206. doi: 10.13926/j.cnki.apps.000163
Wang, L. F., Ma, J. X., Zhou, R. H., Wang, X. M., Jia, J. Z. (2002). Molecular tagging of the yellow rust resistance gene Yr10 in common wheat, PI 178383 (Triticum aestivum L.). Euphytica. 124, 71–73. doi: 10.1023/A:1015689817857
Wang, C. M., Zhang, Y. P., Han, D. J., Kang, Z. S., Li, G. P., Cao, A. Z., et al. (2008). SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica. 159, 359–366. doi: 10.1007/s10681-007-9524-1
Xi, L., Wang, Y., Yang, X., Zhu, W., Chen, G. Y., Wang, Y., et al. (2021). Evaluation of resistance to stripe rust and molecular detection of resistance gene(s) in 243 common wheat landraces from the Yunnan Province. Scientia Agricultura Sinica. 54, 684–695. doi: 10.3864/j.issn.0578-1752.2021.04.002
Yao, Q., Huang, S., Yan, J., Hou, L., Guo, Q., Zhao, J., et al. (2019a). Investigation and identification of barberry as alternate hosts for Puccinia striiformis f. sp. tritici in eastern Qinghai. Acta Phytopathologica Sinica. 49, 370–378. doi: 10.13926/j.cnki.apps.000290
Yao, F. J., Zhang, X. M., Ye, X. L., Li, J., Long, L., Yu, C., et al. (2019b). Characterization of molecular diversity and genome-wide association study of stripe rust resistance at the adult plant stage in Northern Chinese wheat landraces. BMC Genet. 20, 1–16. doi: 10.1186/s12863-019-0736-x
Zeng, Q. D., Han, D. J., Wang, Q. L., Yuan, F. P., Wu, J. H., Zhang, L., et al. (2014). Stripe rust resistance and genes in Chinese wheat cultivars and breeding lines. Euphytica. 196, 271–284. doi: 10.1007/s10681-013-1030-z
Zeng, S., Luo, Y. (2006). Long-distance spread and interregional epidemics of wheat stripe rust in China. Plant Dis. 90, 980–988. doi: 10.1094/PD-90-0980
Zhan, G. M., Wang, F. P., Wan, C. P., Han, Q. M., Huang, L. L., Kang, Z. S., et al. (2016). Virulence and molecular diversity of the Puccinia striiformis f. sp. tritici population in Xinjiang in relation to other regions of western China. Plant Dis. 100, 99–107. doi: 10.1094/PDIS-11-14-1142-RE
Zhang, Z. J. (2016). Fine mapping of a stripe rust resistance gene Yr41 and the function of two genes induced by Pstriiformis W. @ in Wheat. PHD Dissertation (Yaan, Chain: Sichuan Agricultural University).
Zhang, Q. Q., Men, X. Y., Hui, C., Ge, F., Ouyang, F. (2022). Wheat yield losses from pests and pathogens in China. Agr Ecosyst. Environ. 326, 107821. doi: 10.1016/j.agee.2021.107821
Zhang, M. H., Saimi, A., Liu, Q., Ma, Z. Y., Chen, J. (2023). The detection of yr genes in xinjiang wheat cultivars using different molecular markers. Int. J. Mol. Sci. 24, 13372. doi: 10.3390/ijms241713372
Zhao, J., Kang, Z. S. (2023). Fighting wheat rusts in China: a look back and into the future. Phytopathol. Res. 5, 6. doi: 10.1186/s42483-023-00159-z
Zhao, J., Wang, L., Wang, Z. Y., Chen, X. M., Zhang, H. C., Yao, J. N., et al. (2013). Identification of eighteen Berberis species as alternate hosts of Puccinia striiformis f. sp. tritici and virulence variation in the pathogen isolates from natural infection of barberry plants in China. Phytopathology. 103, 927–934. doi: 10.1094/PHYTO-09-12-0249-R
Zhou, A. H., Wang, J., Chen, X. M., Xia, M. H., Feng, Y. X., Ji, F., et al. (2023). Virulence Characterization of Puccinia striiformis f. sp. tritici in China Using the Chinese and Yr single-gene Differentials. Plant Dis. 108, 671–683. doi: 10.1094/PDIS-08-23-1524-RE
Keywords: wheat variety, disease resistance, molecular detection, Puccinia striiformis f. sp. tritici, Xinjiang
Citation: Lai H, Shen Y, Yang H, Fernando DWG, Ren C, Deng F, Lu Y, Sun N, Chen L, Li G, Wang H, Gao H and Li Y (2024) Comparative analysis of stripe rust resistance in seedling stage and Yr gene incidence in spring and winter wheat from Xinjiang, China. Front. Plant Sci. 15:1394213. doi: 10.3389/fpls.2024.1394213
Received: 01 March 2024; Accepted: 18 April 2024;
Published: 01 May 2024.
Edited by:
Weicong Qi, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaReviewed by:
Chao Zhang, Hebei Agricultural University, ChinaCopyright © 2024 Lai, Shen, Yang, Fernando, Ren, Deng, Lu, Sun, Chen, Li, Wang, Gao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiqing Wang, eGp3aHExQDE2My5jb20=; Haifeng Gao, Z2hmMjAwNDQ2NjZAMTYzLmNvbQ==; Yue Li, bGl5dWU2OTA1QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.