
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 May 2024
Sec. Plant Symbiotic Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1394153
Ageratina adenophora can enhance its invasive ability by using beneficial rhizosphere bacteria. Bacillus cereus is able to promote plant growth and provide a positive feedback effect to A. adenophora. However, the interaction between A. adenophora and B. cereus under the influence of native polyphagous insect feeding is still unclear. In this study, Eupatorium lindleyanum, a local species closely related to A. adenophora, was used as a control, aimed to compare the content of B. cereus in the roots of A. adenophora and rhizosphere soil after different densities of Aphis gossypii feeding, and then investigated the variations in the population of A. gossypii and soil characteristics after the addition of B. cereus. The result showed that B. cereus content in the rhizosphere soil and root of A. adenophora increased significantly under A. gossypii feeding compared with local plants, which also led to the change of α-diversity and β-diversity of the bacterial community, as well as the increase in nitrate nitrogen (NO3–N) content. The addition of B.cereus in the soil could also inhibit the population growth of A. gossypii on A. adenophora and increase the content of ammonium nitrogen (NH4+-N) in the soil. Our research demonstrated that B. cereus enhances the ability of A. adenophora to resist natural enemy by increasing soil ammonium nitrogen (NH4+-N) and accumulating other beneficial bacteria, which means that rhizosphere microorganisms help invasive plants defend themselves against local natural enemies by regulating the soil environment.
Plants and soil microbes collaborate to establish a favorable habitat for survival, which can have significant feedback on plant growth and fitness (Bever et al., 2012; Bowen et al., 2017). Rhizosphere microbial communities typically exhibit species-specificity (Singh et al., 2007). In the case of invasive alien plants, soil microorganisms can be harnessed to improve their ability to invade (Zhang et al., 2023). Various hypotheses of invasion mechanism have been proposed to explain this phenomenon, such as the enemy release hypothesis (Keane, 2002) and the mutualist hypothesis (Christian, 2001). Therefore, every element that modifies the interaction between plants and microbes would affect the capacity of nonnative plants to invade. For instance, Batten et al. (2006) discovered that two invasive species, Centaurea solstitialis and Aegilops triuncialis, seem to modify microbial communities in the areas they invade, which promotes their successful invasion. Furthermore, research has demonstrated that the invasive plant species, Impatiens glandulifera, can drastically alter soil microbial characteristics during only two growing seasons, leading to a notable increase in its invasiveness (Stefanowicz et al., 2019).
Ageratina adenophora (Sprengel), originally from Mexico in the Americas, was brought to Europe as a decorative plant and subsequently transferred to Australia and Asia (Darji et al., 2021). In the 1940s, it was introduced from Myanmar to the Chinese province of Yunnan (Wang and Wang, 2006). After 80 years of spreading and dispersing, A. adenophora has extensively colonized throughout south-west China (Du et al., 2022a). Research has also projected that the future range of A. adenophora in China will swiftly grow towards the east and north (Zhao et al., 2019). Moreover, A. adenophora has the ability to inhibit the growth of field crops and can establish a dominating population in the invaded region (Negi et al., 2023). Currently, research has summarized three potential mechanisms that promote the expansion of A. adenophora: phenotypic plasticity, allelopathy, and alteration of soil microbial community structure (Jiao et al., 2021; Khatri et al., 2022; Li et al., 2022).
A. adenophora faces several natural enemies as it spreads in natural habitats (Heystek et al., 2011). Procecidochares utilis Stone (Diptera: Tephritidae) is an obligatory parasitic insect that can induce galls on side branches of A. Adenophora (Gui et al., 2008), but its effectiveness as a biological control agent varies significantly across different regions (Sharma Poudel et al., 2020). For instance, its control effect is not satisfactory in the southwestern region of China (Yuan et al., 2021). Several studies have indicated that native predators can consume invasive plants within the introduced range (Niu et al., 2010; Van Driesche et al., 2010). According to Cheng et al. (2007), it was discovered that cotton aphids (Aphis gossypii Glover, Homoptera: Aphdiate) could complete its life cycle on A. adenophora as a native polyphagous insect, and A. adenophora growth rate was inhibited after A.gossypii feeding (Cheng et al., 2007). Furthermore, with the increase in A.gossypii density and feeding time, the content of secondary metabolites and the activity of antioxidant enzymes changed considerably (Lin et al., 2020).
A. adenophora has implemented many strategies to address external threats. Xia et al. (2023) discovered that the rhizosphere microbiota of A. adenophora harbors a population of plant growth-promoting rhizobacteria (PGPR) that could be utilized as bioinoculants to enhance the growth and productivity of these crops. For instance, invasive plants enhance their ability to invade by regulating soil microorganisms, such as Bacillus, which provide benefits to them, thereby inhibiting the growth of other native plants (Sun et al., 2013). The genus Bacillus can positively impact plant health through three mechanisms: (i) antagonistic activity against pests and pathogens, (ii) enhancement of host nutrition and growth, and (iii) stimulation of plant host defense (Choudhary and Johri, 2009). Such as Bacillus cereus strain D1 (BcD1) induces the production of several secondary metabolites and antioxidant enzymes in plants as a protective mechanism against heat stress and pathogen attack (Tsai et al., 2023). Zhang et al. (2020) identified six bacterial strains that were isolated from the roots of A. adenophora, including B. cereus. Previous research has demonstrated that B. cereus content increased with the invasion process of A. adenophora in the rhizosphere soil (Sun et al., 2021), and the growth of A. adenophora was extensively promoted after adding B.cereus (Du et al., 2023), due to the accumulation of B. cereus could alleviate its autotoxic allelopathy (Wu et al., 2023).
However, the precise contribution that B. cereus plays in facilitating the adaptation and defense against predators in A. adenophora remains uncertain. Consequently, this study aims to examine the response of beneficial bacteria in the rhizosphere of A. adenophora when exposed to A. gossypii feeding. Additionally, it intends to understand the adaptation mechanism of the A. adenophora under the interaction between rhizosphere microorganisms and predators.
The seeds of A. adenophora were obtained from the Xishan District of Yunnan Province, and the Seeds of E. lindleyanum were purchased from Yunxiang Jingpin Flower Seedlings Store in Dali City, Yunnan Province. The seeds of both plants were germinated to the spouting stage and subsequently transplanted to a seedling tray (50 holes/tray). Once the plant reached a height of around 10 cm, it was moved to a plastic cup individually and placed in an artificial climate chamber with parameters set as follows: 25 ± 1°C, 60 ± 5% relative humidity, a photoperiod of 14 h light/10 h dark. The same nutrient soil was used for planting seedlings and transplanting soil, and the experiment was carried out in the greenhouse at Yangzhou University.
Bacillus cereus ATCC14579 was used in the test (concentration maintained at 108 CFU/ml), provided by the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, and stored at a temperature of -20°C. The activated B. cereus was inoculated in 150 mL Luria Bertani (LB) medium and cultured at 28°C, 180 r/min for 24 hours to obtain the B. cereus culture broth.
The number of B.cereus was calculated by dilution broth plate colony counting method. After the serial dilution method was used to dilute the culture broth, the optical density (OD) value of each concentration of culture broth was measured, and 200 μL of culture medium was inoculated into LB ager medium. After 48 hours, the number of colonies in each culture dish was calculated to obtain the number of B. cereus (3 replicates at the same dilution). The calculation method: colony-forming unit (CFU) = the average number of three replicates of the same dilution × dilution multiple × 5, and the unit was CFU/mL.
A.gossypii was obtained from the natural host plant Hibiscus syriacus in the spring of 2021 and reared on A. adenophora and in the insect room (temperature (25 ± 1)°C, humidity (60 ± 5) %, photoperiod (14L:10D)).
E. lindleyanum was employed as a control to determine changes in B. cereus levels after A. gossypii feeding on A. adenophora. Seedlings of both plants were cultivated and then transplanted into plastic cups. 0, 5, 10, and 15 1st-instar A.gossypii nymphs were released on the top of the stem when the plants had grown to 10 cm in length. Until all the A.gossypii grew to the 4th-instar, all the plants were harvested, and the rhizosphere soil from the different treatments was carefully removed, which adhered to the root surface about 1 mm. All samples were sieved through a 40-mesh to determine the content of B.cereus, soil characteristics, and soil bacterial diversity in rhizosphere soil. In addition, plant roots from each treatment were cut for the detection of B. cereus content in five replicates.
Seedlings of two plants were taken from plastic cups (same growth, plant height about 10 cm) in individual, and then 30 mL of sterile water (CK), 200-fold dilution of B.cereus (BC), and 200-fold dilution of Thiessen copper (TC, excluding bacterial treatment) suspension were added to the soil respectively. After that, 4th-instar nymphs with numbers of 0, 5, 10, and 15 were promptly released immediately after the treatment.
After 15 days, the underground part of the plant was harvested, and the rhizosphere soil and non-rhizosphere soil were mixed and filtered using a 40-mesh sieve to determine the physical and chemical properties of the soil.
After the rhizosphere soil was screened by a 40 mesh sieve, the rhizosphere soil was collected and stored at −20°C for DNA extraction by kit (DNeasy ®PowerSoil Pro® Kit, QIAGEN®, Germany). DNA concentration was measured using NanoPhotometer N60 spectrophotometer (IMPLEN, Germany), and the PCR primers for qRT-PCR analysis (Bio-Rad CFX96 touch, Bio-Rad Laboratories Incorporated, USA) were designed based on the sequence of B. cereus strains. The specific determination method referred to the existing research (Sun et al., 2022).
Initially, the plant roots were rinsed multiple times with distilled water, subsequently immersed in anhydrous ethanol, and finally pulverized using liquid nitrogen. DNA was extracted from plant roots using a kit (DNeasy ® PowerPlant ® Kit, QIAGEN ®, Germany). The DNA concentration was determined by NanoPhotometer N60 and quantified by qPCR.
Microbial community genomic DNA was extracted from the screened rhizosphere soil samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.). The DNA extract was checked on 1% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA). The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R(5’-GGACTACHVGGGTWTCTAAT-3’) by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). The PCR amplification of 16S rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 10°C. PCR reactions were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using Quantus™ Fluorometer (Promega, USA).
Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited into the CNCB Genome Sequence Archive (GSA) database (GSA Accession Number: CRA016029).
The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 (Chen et al., 2018) and merged by FLASH version 1.2.7 (Magoč and Salzberg, 2011) with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of<20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded, reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of overlap region is 0.2. Reads that could not be assembled were discarded; (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching, 2 nucleotide mismatch in primer matching.
Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed (Stackebrandt and Goebel, 1994; Edgar, 2013). The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 against the 16S rRNA database using confidence threshold of 0.7 (Wang et al., 2007).
Alpha diversity analysis was used to reflect the richness and diversity of bacterial communities in the rhizosphere soil of A. adenophora and E. lindleyanum. The following are the formulas for Shannon’s index (Hshannon) and Simpson’s index (Dsimpson), which are employed to quantify species diversity:
( represents the actual observed OTU; represents the number of sequences contained in the ith OTU; N represents the number of all sequences).
The soil pH was measured according to the glass electrode method by the pH meter (Niu et al., 2007). The drying method was then used to determine the soil moisture content (Samuelsson et al., 2006; Tian et al., 2021). Ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3–N), and available phosphorus (AP) were measured by kits (Ltd. SATD-1-G, SXTD-1-G, ASXL-1-G, Suzhou Keming Biotechnology, China). Flame atomic absorption spectrometry is used to measure available Potassium (AK) (Yushmanov et al., 2007).
Following the inoculation of A.gossypii, a cylindrical sleeve (perforated around) made of a PVC plastic plate covered each plant to prevent the A.gossypii from escaping. Five replicates were set up per treatment (3 A.gossypii densities × 3 soil treatments× 2 plant populations = 18 treatments).
The population of A.gossypii on each plant was recorded at three-day intervals throughout nine days, and the underground parts of the plant were harvested on the tenth day. When the insect’s body turns black and does not respond with a brush tip, it is considered that the insect is dead, and then delicately eliminated the deceased A.gossypii using a brush.
All data were analyzed by DPS 9.01 (Data Processing System v9.01) software. Before Analysis of variance (ANOVA) analysis, all data were evaluated to confirm that they conformed to the normal distribution; the data were the mean ± SE of five replicates per treatment. The effects of varying densities of A.gossypii (the number of A.gossypii was 0,5,10,15) and different plant species (A. adenophora and E. lindleyanum) on the B. cereus content in rhizosphere soil and soil characteristics were analyzed by two-way analysis of variance. The same approach was used to analyze the effects of varying additional treatments (CK\TC\BC) and different plant species on the population growth of A.gossypii and the characteristics of the soil. The difference between the two groups was compared by independent sample t-tests. The significant difference between treatments was based on the LSD test. P< 0.05 was considered statistically significant.
The B. cereus content of the rhizosphere soil changed significantly at different A.gossypii densities within the same plants (F=10.8340, P<0.001, Figure 1A), but it did not differ between plants after A.gossypii feeding. In the treatment group of A. adenophora, the B. cereus content of the A-10 treatment increased by 72.58% compared with the A-0 treatment. Similarly, in the related species E. lindleyanum, the B. cereus content in the L-10 treatment group was 58.65% higher than that in the L-0 treatment group (Figure 1A). This indicated that the A.gossypii infestation promoted the increase of B. cereus content in the rhizosphere soil of A. adenophora and E. lindleyanum.
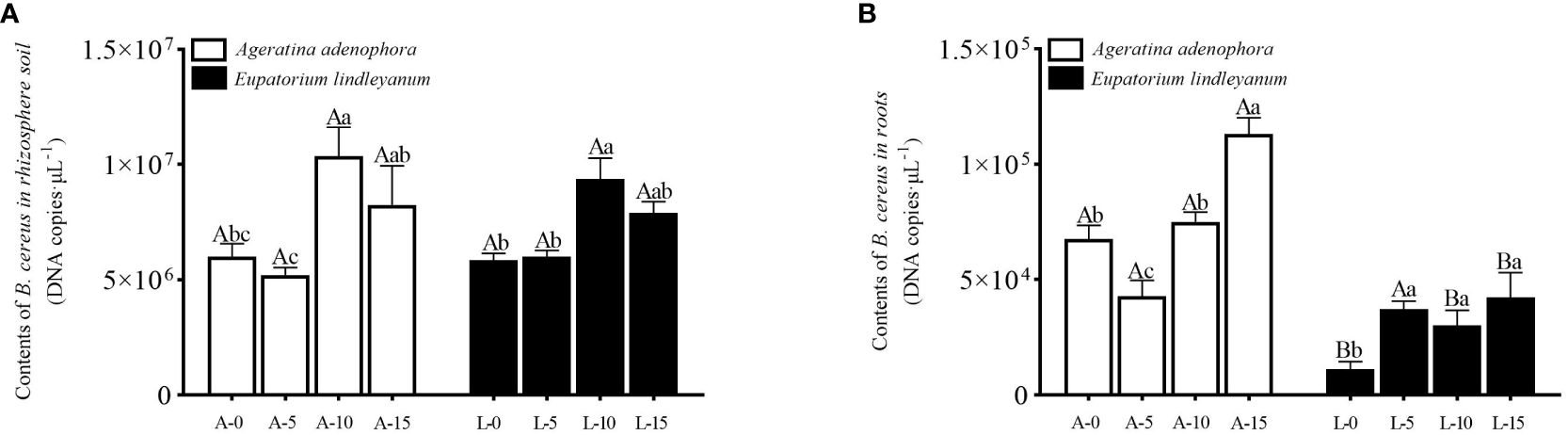
Figure 1 Variation of B. cereus content in root and rhizosphere soil for A. adenophora and E. lindleyanum treated by A.gossypii. (A) B. cereus content in the rhizosphere soil (B) B. cereus content in root; A represents Ageratina adenophora, L represents Eupatorium lindleyanum, 0/5/10/15 represents the number of A.gossypii inoculated, different lowercase letters represent significant difference (LSD’s test, P<0.05) at the same plant, different uppercase letters indicate significant differences between treatments with same A. gossypii infestation (t-test, P<0.05). Error Bars are mean ± SE (n=5).
On the other hand, changes in the B. cereus content in A. adenophora roots differ from those in rhizosphere soil. There are significant differences in different plants (F=86.080, P<0.001, Figure 1B) and different A.gossypii densities (F= 13.619, P<0.001, Figure 1B), as well as interactions between them (F= 8.869, P<0.001, Figure 1B). The A-15 treatment group had significantly higher B. cereus content than the other treatments, and the B. cereus content in the A. gossypii treatment group in E. lindleyanum was higher than in the treatment group without an A. gossypii infestation(Figure 1B). Within the same A.gossypii density group, except for the treatment with inoculation A.gossypii density of 5, the content of B. cereus in the roots of A. adenophora increased by 500.64% compared with that of E. lindleyanum in other treatment groups (Figure 1B), indicating that A.gossypii feeding could significantly increase the B. cereus content in the roots of A. adenophora compared to its local relatives.
The soil’s physical and chemical properties were measured after A.gossypii feeding on two plants. In terms of soil moisture content, the results showed that A.gossypii feeding at varied densities and plants had no significant effect on it (Figure 2A). Nonetheless, the pH of the soil can be greatly impacted by varying A. gossypii feeding densities. Compared with the A-0 treatment, pH decreased by 6.15% in the A-10 treatment, and there was a significant interaction between different plants and different A.gossypii densities (F=7.537, P<0.001, Figure 2B). NH4+-N in rhizosphere soil of A. adenophora did not vary significantly across A.gossypii densities, but it varied widely and was significantly higher in E. lindleyanum compared to A. adenophora at A.gossypii densities of 5 or 15 (Figure 2C). NO3–N content of A. adenophora differed significantly under different treatments (F=5.273, P<0.05, Figure 2D). Specifically, the content of NO3–N in the A-10 treatment increased by 33.72% compared to the A-0 treatment, whereas there was no significant change observed in the control E. lindleyanum (Figure 2D). The content of AP in the rhizosphere soil of the two plants did not significantly differ when exposed to different densities of A.gossypii treatments (Figure 2E). However, in A. adenophora, it decreased significantly by 13.86% in the A-10 treatment as compared to the A-0 treatment (Figure 2E). A.gossypii feeding at different densities (F=38.002, P<0.001, Figure 2F) and plants (F=3.158, P<0.05, Figure 2F) significantly affected soil AK content, and there occurred significant interactions between different plants and A.gossypii densities(F=5.786, P<0.001, Figure 2F). The content of AK decreased to 19.56% and 16.29% in the A-10 and A-15 treatments, respectively, compared to the A-0 treatment (Figure 2F).
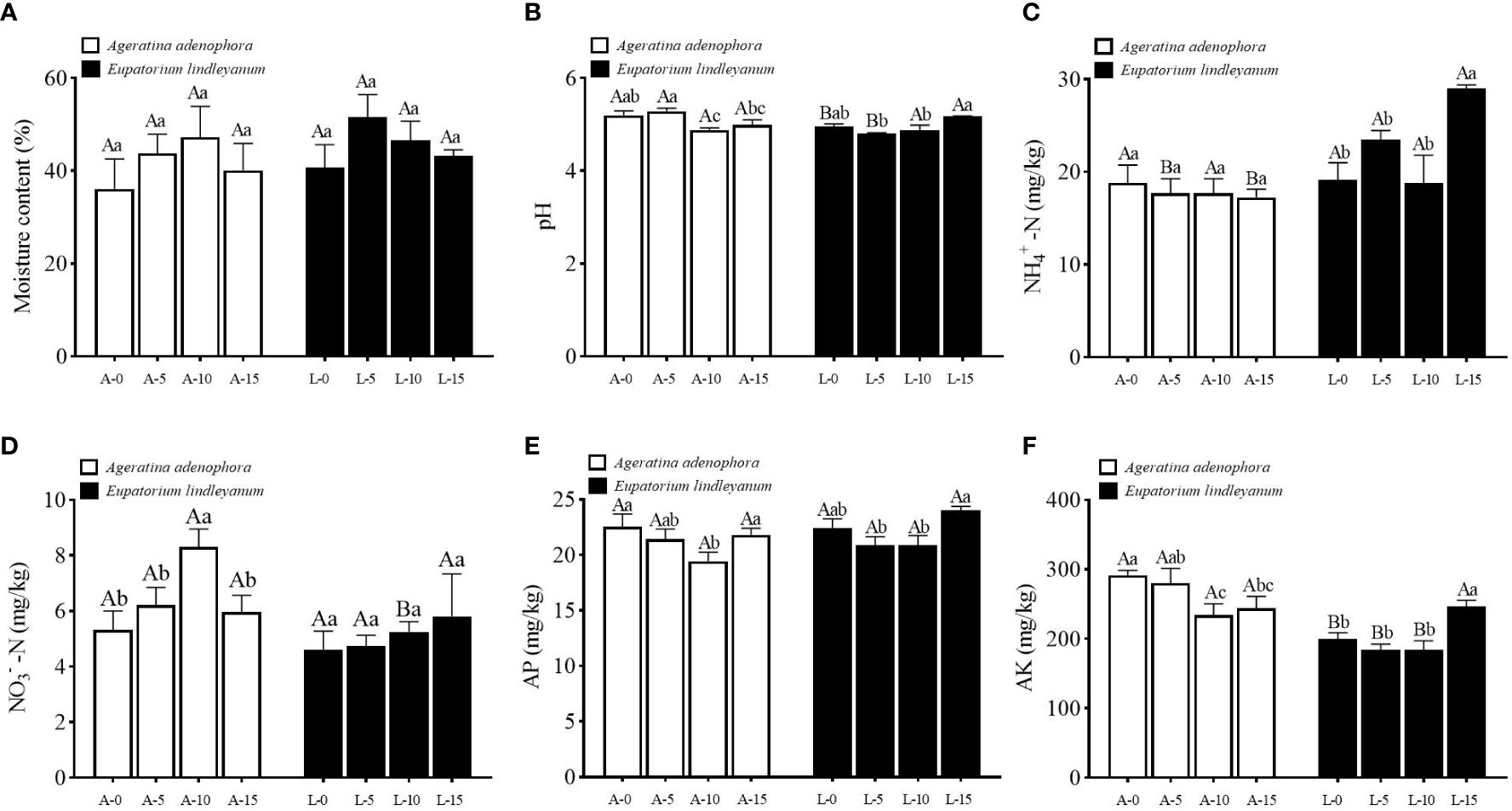
Figure 2 Physicochemical characteristic changes in rhizosphere soil for two kinds of plants treated with A.gossypii. (A) moisture content. (B) pH value. (C) NH4+-N, ammonium nitrogen. (D) NO3–N, nitrate nitrogen. (E) AP, available phosphorus. (F) AK, available potassium. A represents Ageratina adenophora, L represents Eupatorium lindleyanum, 0/5/10/15 represents the number of A.gossypii inoculated, different lowercase letters represent significant difference(LSD’s test, P<0.05) at the same plant, different uppercase letters indicate significant differences between treatments with same A. gossypii infestation (t-test, P<0.05). Error Bars are mean ± SE (n=5).
Different plants and densities of A.gossypii feeding considerably altered both the Shannon and Simpson indexes, with a significant interaction between the two parameters (Shannon index: F= 216.661, P<0.001; F=22.845, P<0.001; F= 23.983, P<0.001; Simpson index: F= 53.609, P<0.001; F= 9.979, P<0.001; F=10.261, P<0.001). However, the ace, chao index could be significantly changed only under different A.gossypii density treatments (ace index: F=24.555, P<0.001; F=22.831, P<0.001; chao index: F=23.160, P<0.001; F=17.770, P<0.001). In A. adenophora, the Shannon index of the rhizosphere soil bacterial community was significantly higher in treatment A-10 along with treatment A-15 than in treatment A-0 together with treatment A-5, and there was no difference in Shannon index was observed among treatment groups in the control plant E. lindleyanum, and there was no difference in Shannon index among treatment groups in the control plant, E. lindleyanum (Figure 3). The composition of the rhizosphere soil bacterial community showed the same trend in Simpson’s index for both plants, with a significant decrease of 74.11% in Simpson’s index for A-10 compared to A-0 (Figure 3). Meanwhile, the ace index and chao index of the A-5 treatment were remarkably lower from other treatments, and a significant difference was observed between the A-5 treatment and the L-5 treatment, and the A-10 treatment and the A-15 treatment had a significantly higher ace index and chao index than the L-10 treatment and the L-15 treatment (Figure 3).
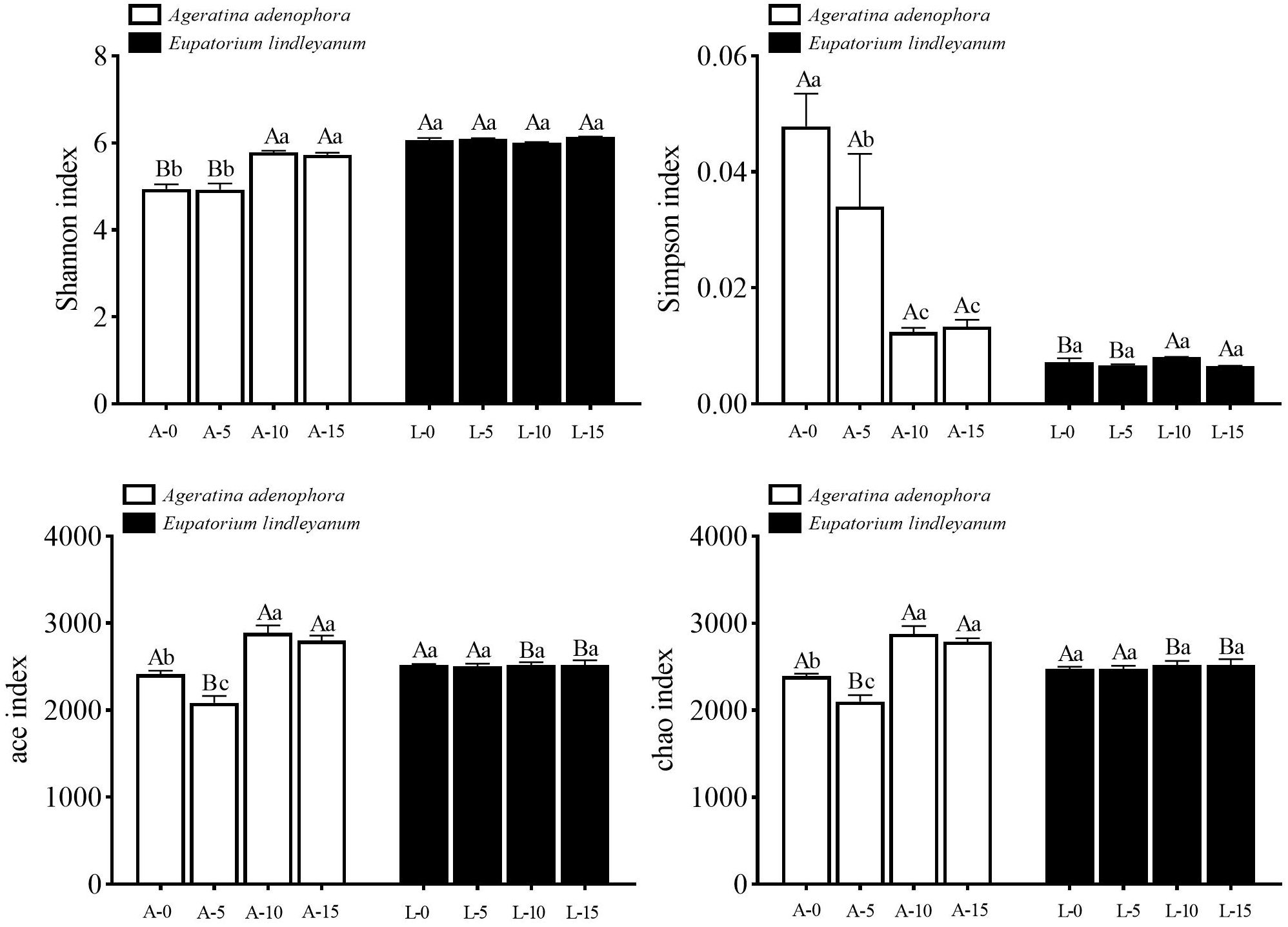
Figure 3 Changes of Alpha diversity indices for bacterial communities in rhizosphere soil for two kinds of plants treated with A.gossypii. A represents Ageratina adenophora, L represents Eupatorium lindleyanum, 0/5/10/15 represents the number of A.gossypii inoculated, different lowercase letters represent significant difference(LSD’s test, P<0.05) at the same plant, different uppercase letters indicate significant differences between treatments with same A. gossypii infestation (t-test, P<0.05). Error Bars are mean ± SE (n=5).
Based on the original OTUs of the plant rhizosphere soils, we carried out non-metric multidimensional scaling (NMDS), which reflects the species composition in each sample on a multidimensional space in the form of points, and the greater the point-to-point distance, the greater the sample differences. As shown in Figure 4, there were significant differences between the soil bacterial communities of the two plants, A. adenophora, and E. lindleyanum, under the same density of A.gossypii feeding (Figure 4). Interestingly, A.gossypii feeding increased the β-diversity of the soil bacterial community in A. adenophora, while there was no increase in β-diversity in the soil bacterial community of E. lindleyanum after A.gossypii inoculation, suggesting that the two plants have different changes in rhizosphere soil microbial communities for the various densities of A.gossypii feeding (Figure 4).
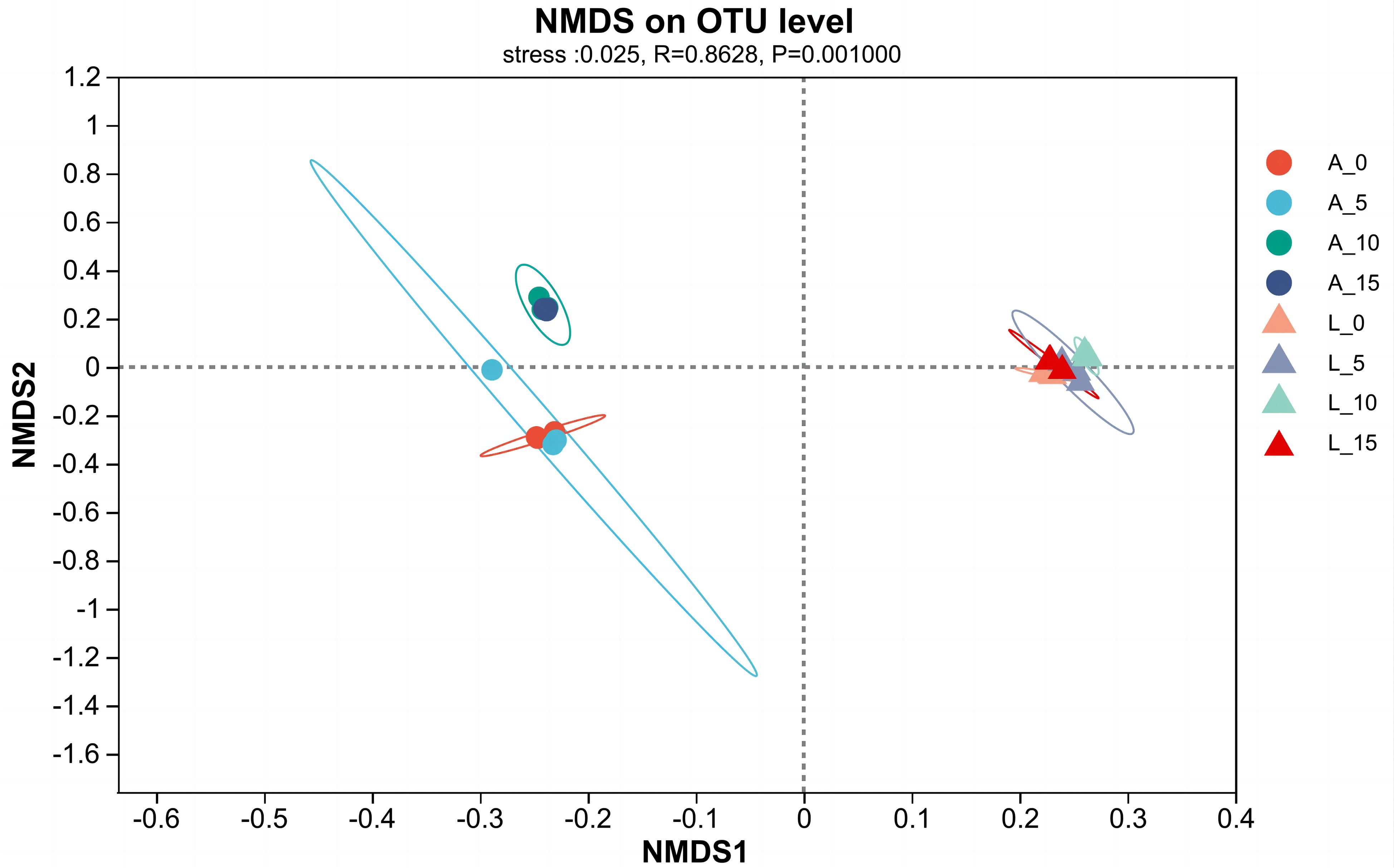
Figure 4 Non-metric multidimensional scaling of β diversity in soil bacterial communities after A.gossypii feeding. A represents Ageratina adenophora, L represents Eupatorium lindleyanum, 0 represents no A.gossypii, 5 represents 5 nymphs, 10 represents 10 nymphs, and 15 represents 15 nymphs.
To further explore the relationship between changes in bacterial communities and A.gossypii feeding, we obtained 11 families of bacteria in the rhizosphere soil of A. adenophora that responded to A.gossypii feeding by sequencing. The number of OTUs represents the abundance of bacteria. As shown in Figure 5, the bacterial abundance of three families in the rhizosphere soil of A. adenophora decreased significantly, namely Chitinophagaceae, Pseudomonadaceae, Micropepsaceae, and compared with the L-10 treatment, the OTU number of Pseudomonadaceae was significantly increased in the A-10 treatment; the OTU number of Micropepsaceae was considerably higher than the control treatment in all treatments (Figure 5).
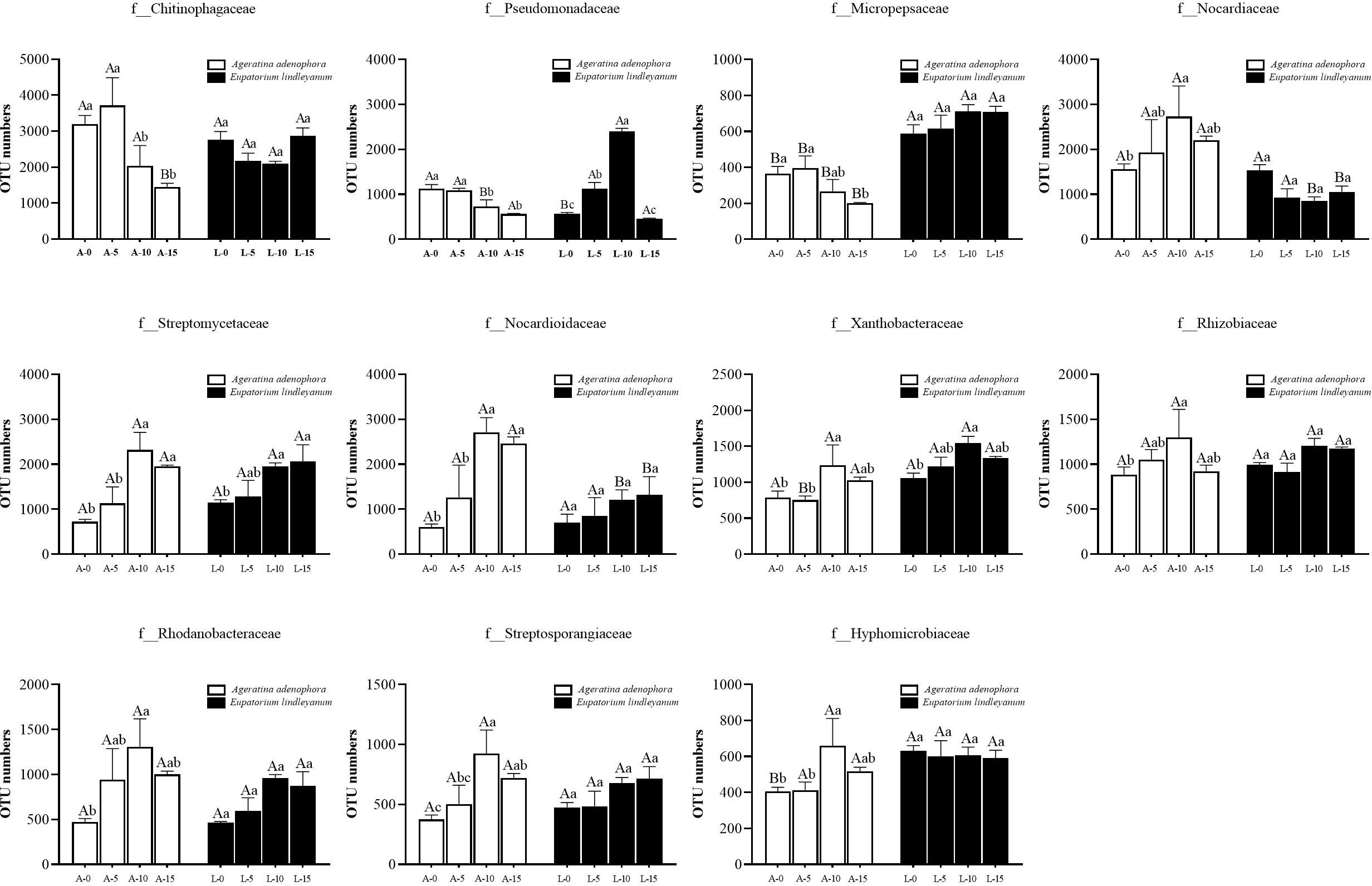
Figure 5 Changes of bacteria in A. adenophora with rhizosphere soil OTU numbers under A. gossypii feeding at the family level. A represents Ageratina adenophora, L represents Eupatorium lindleyanum, 0/5/10/15 represents the number of A.gossypii inoculated, different lowercase letters represent significant difference (LSD’s test, P<0.05) at the same plant, different uppercase letters indicate significant differences between treatments with same A. gossypii infestation (LSD’s test, P<0.05). Error Bars are mean ± SE (n=5).
The abundance of 8 bacterial families has increased: Nocardiaceae, Streptomycetaceae, Nocardioidaceae, Xanthobacteraceae, Rhizobiaceae, Rhodanobacteraceae, Streptosporangiaceae, and Hyphomicrobiaceae (Figure 5). At a feeding density of 10 nymphs, the number of OTU number of Streptomycetaceae, Nocardioidaceae, Xanthobacteraceae, and Streptosporangiaceae were all significantly higher than those of treatment groups with only 5 or no A.gossypii feeding (Figure 5). In addition, the OTU number of Nocardiaceae and Nocardioidaceae in the rhizosphere soil of A. adenophora at the same feeding density (10) was significantly increased by 2.19 and 1.23-folds compared to that of the local relative, E. lindleyanum (Figure 5). Interestingly, the conclusion we emerge from the comparison of the data is that A.gossypii feeding at different densities had no significant effect on the abundance of bacteria in the rhizosphere of E. lindleyanum, except for the three bacteria families, Pseudomonadaceae, Streptomycetaceae, and Xanthobacteraceae (Figure 5).
The number of A. gossypii population growth on each treatment was counted on the 10th day after inoculation, and the results of ANOVA showed that the addition of B. cereus significantly changed the number of A. gossypii populations at initial inoculation numbers of 10 and 15 (F=4.784, P<0.05; F=13.070, P<0.001; Figures 6B, C) and that there was a significant interaction between inoculation with B. cereus and different plant sources (F=6.416, P<0.001; F=13.749, P<0.001, Figures 6B, C); and the growth of A. gossypii population was mainly associated with plant source when the initial number of A. gossypii was 5 (F=6.599, P<0.05; Figure 6A).
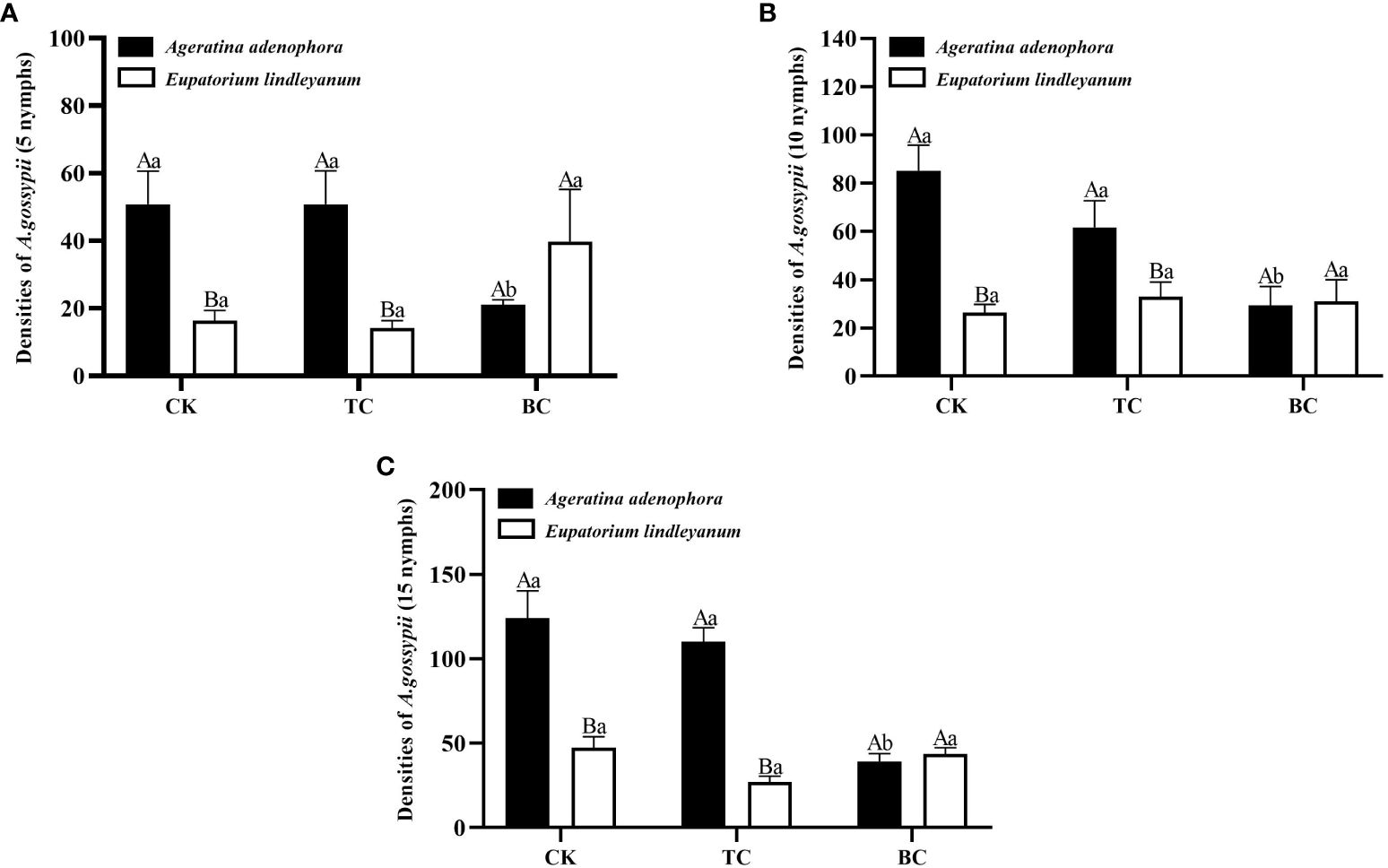
Figure 6 The population growth of A. gossypii on A. adenophora and E. lindleyanum after adding or excluding B. cereus. CK, addition with sterile water; TC, addition with thiosen copper; BC, inoculation with B. cereus. (A) represents infestation with 5 nymphs; (B) represents infestation with 10 nymphs; (C) represents infestation with 15 nymphs. Different lowercase letters above the bars represent significant differences among the three addition treatments (LSD’s test, P<0.05) in the same plant; different uppercase letters indicate significant differences between the same treatments in the different plants (t-test, P<0.05). Error Bars are mean ± SE (n=5).
For A. adenophora, the addition of thiosen copper treatment showed no significant difference in the number of A. gossypii populations with initial inoculation of 5,10, and 15 on the 10th day compared to the sterile water treatment, as well as the addition of B. cereus, which resulted in significant reductions of A. gossypii populations by 58.66% (Figure 6A), 65.49% (Figure 6B), and 68.39% (Figure 6C), respectively, when compared to the sterile water control. For E. lindleyanum, the growth of the A. gossypii population on plants with thiosen copper and with B. cereus was not different from the control under three different A. gossypii feeding density treatments. Meanwhile, the A. gossypii population of A. adenophora was significantly higher than that of E. lindleyanum when no B. cereus was added compared to both plants (Figure 6).
Since the addition of B. cereus at an initial density of 15 A. gossypii inhibited the population growth of A. gossypiis more strongly, the soil physicochemical properties were measured by taking the rhizosphere soil of each plant of this treatment. Except for moisture content and AP, all soil properties varied substantially in soils treated with thiosen copper or B.cereus (Figure 7). The addition of B. cereus significantly increased the pH value of the rhizosphere soil of A. adenophora compared to the control (Figure 7B), and also increased the NH4+-N content by 10% (Figure 7C), while the content of AK decreased significantly by 14.29% (Figure 7F), and both of them was higher than that of the native plant, E. lindleyanum. Further data analysis revealed that after adding thiosen copper, the pH value increased compared to the CK. Additionally, the contents of NH4+-N, NO3–N, and AK significantly decreased by 10.30%, 59.70%, and 55.95%, respectively (Figures 7C, D, F). It was observed that the NH4+-N content in the rhizosphere soil of E. lindleyanum was significantly lower than that of A. adenophora, while the NO3–N and AK content were higher.
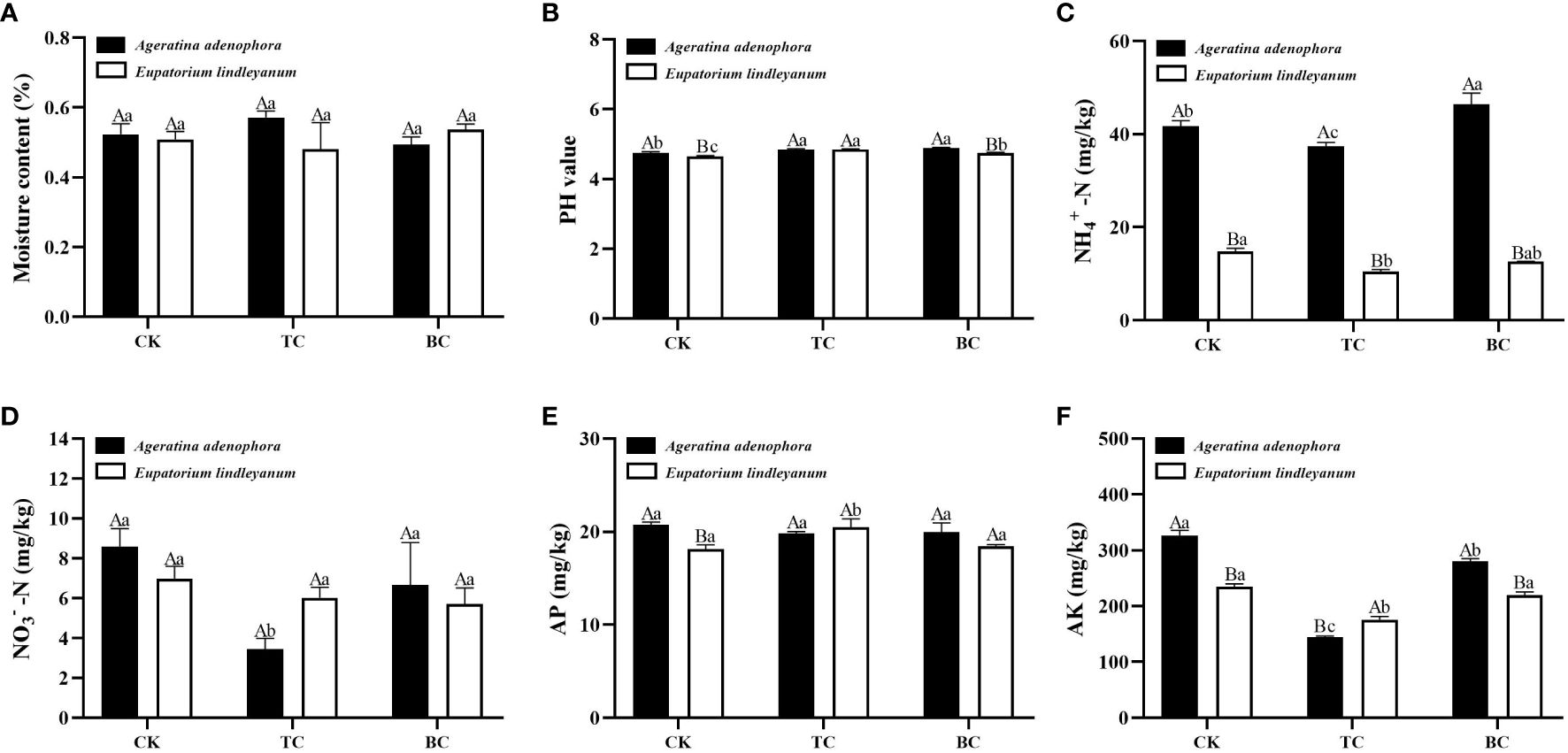
Figure 7 The changes in physicochemical characteristics of rhizosphere soil for two kinds of plants after being fed on by A. gossypii (15 nymphs) with the addition of B.cereus. (A) moisture content. (B) pH value. (C) NH4+-N, ammonium nitrogen. (D) NO3–N, nitrate nitrogen. (E) AP, available phosphorus. (F) AK, available potassium. CK, addition with sterile water; TC, addition with thiosen copper; BC, inoculation with B. cereus. Lowercase letters above the bars represent significant differences among the three addition treatments (LSD’s test, P<0.05) in the same plant, and different uppercase letters indicate significant differences between the same treatments in the different plants (t-test, P<0.05). Error Bars are mean ± SE (n=5).
Many plants possess the capacity to induce defenses (Rabari et al., 2023). When they are consumed by animals or insects, they increase the synthesis of secondary metabolites or accumulate microorganisms that are beneficial to their growth and development, thus strengthening their defenses (Miller et al., 2005). Our research investigated the beneficial bacteria in the rhizosphere environment of A.adenophora that responded to the action of broad-feeding natural enemies and the changes in the adaptability of A.adenophora under the interaction with them. The results showed that A. gossypii feeding on A.adenophora changed the content of B.cereus in both the rhizosphere soil and the root, and it was significantly increased when the number of A.gossypii reached 15 (Figure 1), which was consistent with the results of previous study on that in the generalist natural enemy, procecidochares utilis (Sun and Yang, 2021). It was found that the parasitism of procecidochares utilis significantly reduced the biomass of A.adenophora, and it counteracted this damage by accumulating the beneficial bacterium B.cereus (Erasmus et al., 1992). It demonstrates that to cope with the feeding of natural enemies, A.adenophora can improve its competitiveness in the environment by gathering B.cereus, which is beneficial to itself. Compared with A.adenophora, the native relative species E.lindleyanum did not have such characteristics.
In terms of soil physicochemical properties, we discovered that pH, AP, and AK contents of the rhizosphere soil of A.adenophora decreased after A. gossypii feeding (Figures 2B, E, F), whereas NO3–N increased significantly (Figure 2D). The metabolism of microorganisms and animals in the rhizosphere of plants directly promotes or inhibits the absorption and growth of nutrients in the roots (Adedayo and Babalola, 2023) and can also affect the transformation of N, P, K, and other substances in the rhizosphere soil (Piton et al., 2023). Relevant studies have shown that the soil characteristics changed after the invasion of A.adenophora, especially the contents of NO3–N, NH4+-N, AK, and AP in the soil increased (Zhao et al., 2019). In this study, it was found that the AK content in the rhizosphere soil of A.adenophora under A.gossypii feeding was higher than that of E.lindleyanum. Furthermore, A.adenophora was found to accumulate B.cereus, which is a kind of growth-promoting bacteria to benefit itself (Chen et al., 2019; Sun et al., 2021). The experimental results revealed the B. cereus mediated defense of A.adenophora against natural enemies.
α diversity refers to the species diversity among individuals within a specific geographic area or ecosystem (Thukral, 2017). Among them, the Chao1 index and ACE index can be used to describe the species abundance of bacteria, while the Shannon index and Simpson index were used to measure the diversity of bacterial community (Xia and Sun, 2023). Compared with the Simpson index, the Shannon index comprehensively takes into account the evenness and richness of the community, and the higher the value is, the greater the diversity is (Kumar et al., 2023). Affected by the species abundance and species evenness in the sample community, under the same species abundance condition, if the species in the community are relatively uniform, the diversity of the community is higher (Kimbro and Grosholz, 2006). Our investigation revealed that there were no significant differences in α-diversity across the treatment groups following the feeding of A.gossypii at various densities on E.lindleyanum.
Nevertheless, The α-diversity of A.adenophora changed significantly. Among the four types of indexes, there were significant differences between the treatment with A.gossypii density of 10, the treatment with A.gossypii density of 15, and the blank control, respectively. There was also a significant difference between the two plants when they were fed by the same A.gossypii density (10 and 15; Figure 3). According to the Shannon index, the diversity of each treatment of A.adenophora was significantly lower than that of E.lindleyanum (Figure 3), which was consistent with the research results that invasive organisms could greatly decrease the α-diversity of soil microbial communities (Stefanowicz et al., 2021). At the same time, the Shannon index of the rhizosphere soil bacterial community of A.adenophora increased significantly under the feeding of A.gossypii, while that of E.lindleyanum did not change significantly (Figure 3), indicating that A.adenophora had a stronger ability to change its bacterial community under the feeding of natural enemies.
β diversity is commonly defined as a consistent change in species between different locations, which provides local-scale biodiversity and a broader regional species pool (Anderson et al., 2011). Non-Metric Multi-Dimensional Scaling (NMDS) is a ranking technique used in β-diversity analysis, specifically in ecological research (Zhao et al., 2020). The results of NMDS analysis revealed that the bacterial structure in the rhizosphere soil of A.adenophora and E.lindleyanum was significantly different when they were not inoculated with A.gossypii (Figure 4). Moreover, the rhizosphere soil bacterial community of A.adenophora with 10 and 15 aphids feeding was significantly changed compared with the blank control and 5 aphids feeding treatments (Figure 4), while that of E.lindleyanum was not, which as previously described that A.adenophora can selectively accumulate bacteria that are advantageous to its own growth (Chen et al., 2019). The bacteria accumulated in the rhizosphere soil showed a specific growth-promoting effect on the plants (Cunha et al., 2024). What’s more. Increases in soil bacterial populations and changes in species structure can provide a competitive advantage for A.adenophora after invasion (Fang et al., 2019). Our results illustrate the conclusion that A.adenophora has a stronger ability to change its bacterial community in response to natural enemy feeding.
Then we continued to analyze the effect of A.gossypii feeding on the number of bacterial OUT in A.adenophora’s rhizosphere soil by sequencing and summarizing the bacterial species with significant changes in abundance at the family level (Figure 5). Among the 11 bacterial families with significant changes, Xanthobacteraceae, Rhizobiaceae, and Hyphomicrobiaceae, which belong to the order Rhizobiales, showed a significant increase in their OTU numbers in the rhizosphere soil of A.adenophora after inoculation with 10 aphids. It was reported that the family Xanthobacteraceae has included many aerobic chemoheterotrophic genera, which contribute to N2 fixation (Ma et al., 2019). Rhizobaceae can provide nitrogen fertilizer for plants and increase the nitrogen content in soil (Sprent et al., 2017), and Hyphomicrobiaceae is a family of photosynthetic bacteria in Rhizobium (Lian et al., 2021). Nitrogen is an indispensable substance for plant growth and is the main source of protein synthesis (Li et al., 2023). Nitrogen-fixing bacteria can provide it for plants through nitrogen fixation, thereby promoting plant growth and development (Masson-Boivin and Sachs, 2018), and the increase in the abundance of these bacteria is positively correlated with the results of our previous study that found an increase in NO3–N content.
Due to the phenotypic plasticity, compensatory or tolerance of invasive species, the negative feedback effect is significantly weakened (Policelli et al., 2018). On the other hand, some rhizosphere-beneficial bacteria have a preferential impact on invasive species (Sun et al., 2022). For instance, mycorrhizal fungi significantly improve the disease resistance of host plants, and the invasive plant Anthemis cotula is protected from the invasion of pests and diseases due to the high infection of arbuscular mycorrhizal fungi (Shah et al., 2008). Our research has revealed that A.gossypii feeding can stimulate the increase of B. cereus content in rhizosphere soil. Therefore, we hypothesize that the addition of B.cereus will affect the growth of the A.gossypii population as well as the soil characteristics. The results showed that the growth rate of the A.gossypii population was significantly slowed down after the addition of B. cereus to A.adenophora (Figure 3C). In the native plant E.lindleyanum, the addition of B. cereus did not impede the growth of A.gossypii population, so it can be speculated that the addition of B.cereus enhanced the resistance of A.adenophora, which is consistent with the previous results of Du et al. (2022b) found that the survival and reproduction of A.gossypii inoculated with arbuscular mycorrhizal fungi on A.adenophora were inhibited. This could be a result of the A.gossypii being poisoned by the leaves’ elevated concentration of insect-resistant chemicals (Zhu et al., 2024).
In the determination of soil physical and chemical properties, we found that even in the case of A.gossypii feeding, the addition of B.cereus significantly increased the content of NH4+-N in the rhizosphere soil of A.adenophora, but this effect was not obvious in E.lindleyanum (Figure 7C). NH4+-N is one of the forms of nitrogen that can be absorbed and utilized by plants (Liu and Von Wirén, 2017). Studies have shown that B. cereus can promote the accumulation of nitrogen in plants (Vessey and Buss, 2002), which is consistent with our experimental results. In addition, the results of this study indicated that the two plants have different nitrogen utilization, and also suggested that the growth-promoting effect or resistance improvement of B.cereus on A.adenophora may be achieved by increasing the ammonium nitrogen in the soil.
In summary, compared to local closely related species, A. gossypii feeding significantly increased the content of B. cereus in the rhizosphere and in the roots of A. adenophora, induced remarkable changes in the α-diversity and β-diversity of rhizosphere bacterial communities, along with an increase in NO3–N content in the rhizosphere soil, which confirms that A. adenophora can exert a biased impact on the rhizosphere soil environment under the influence of a generalist predator. The B. cereus addition/exclusion test revealed that an elevated concentration of B. cereus content in the soil could inhibit the population growth of A. gossypii on A. adenophora. Apart from that, the NH4+-N content in the rhizosphere soil also enhanced, suggesting that the increase of B. cereus content provided positive assistance in helping A. adenophora resist insect feeding. We investigated the role of B. cereus in the adaptation of A. gossypii to a wide range of predatory natural enemies from the perspective of the bidirectional role of natural enemies and inter-root beneficial bacteria, and the further research will explore how B. cereus activates the JA and SA pathways in A. adenophora from the viewpoint of triggering induced systemic resistance (ISR) by beneficial rhizosphere bacteria, and analyze the molecular mechanisms by which beneficial bacteria enhance the adaptability of A. adenophora to predators, laying the foundation for evaluating the biological control effect of A. gossypii on A. adenophora.
The original contributions presented in the study are included in the article/Supplementary Material, and https://bigd.big.ac.cn/gsa/browse/CRA016029, accession number: CRA016029. further inquiries can be directed to the corresponding author/s.
The manuscript presents research on animals that do not require ethical approval for their study.
YY: Data curation, Writing – original draft, Writing – review & editing, Formal Analysis, Investigation. ZY: Data curation, Investigation, Writing – original draft, Formal Analysis, Methodology. MH: Data curation, Investigation, Writing – original draft. SS: Conceptualization, Methodology, Writing – review & editing. GX: Conceptualization, Writing – review & editing, Methodology. GY: Conceptualization, Funding acquisition, Writing – review & editing, Methodology.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Nature Science Foundation of China (NSFC) (31772229).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1394153/full#supplementary-material
Adedayo, A. A., Babalola, O. O. (2023). Fungi that promote plant growth in the rhizosphere boost crop growth. J. Fungi 9, 239. doi: 10.3390/jof9020239
Anderson, M. J., Crist, T. O., Chase, J. M., Vellend, M., Inouye, B. D., Freestone, A. L., et al. (2011). Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist: Roadmap for beta diversity. Ecol. Letters. 14, 19–28. doi: 10.1111/j.1461-0248.2010.01552.x
Batten, K. M., Scow, K. M., Davies, K. F., Harrison, S. P. (2006). Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol. Invasions. 8, 217–230. doi: 10.1007/s10530-004-3856-8
Bever, J. D., Platt, T. G., Morton, E. R. (2012). Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 66, 265–283. doi: 10.1146/annurev-micro-092611-150107
Bowen, J. L., Kearns, P. J., Byrnes, J. E. K., Wigginton, S., Allen, W. J., Greenwood, M., et al. (2017). Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun. 8, 433. doi: 10.1038/s41467-017-00626-0
Chen, L., Fang, K., Zhou, J., Yang, Z. P., Dong, X. F., Dai, G. H., et al. (2019). Enrichment of soil rare bacteria in root by an invasive plant Ageratina adenophora. Sci. Total Environment. 683, 202–209. doi: 10.1016/j.scitotenv.2019.05.220
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng, L., Ren, Q., Liu, X., Guo, C., Teng, Z., Zhang, Q. (2007). Behavioral responses of Aphis gossypii and Coccinella septempunctata to volatiles from Eupatorium adenophorum and an analysis of chemical components of the volatiles. Acta Entomol Sin. 50, 1194–1199. doi: 10.16380/j.kcxb.2007.11.006
Choudhary, D. K., Johri, B. N. (2009). Interactions of Bacillus spp. and plants – With special reference to induced systemic resistance (ISR). Microbiological Res. 164, 493–513. doi: 10.1016/j.micres.2008.08.007
Christian, C. E. (2001). Consequences of a biological invasion reveal the importance of mutualism for plant communities. Nature. 413, 635–639. doi: 10.1038/35098093
Cunha, I., de, C. M. da., Silva, A. V. R. da., Boleta, E. H. M., Pellegrinetti, T. A., Zagatto, L. F. G., et al. (2024). The interplay between the inoculation of plant growth-promoting rhizobacteria and the rhizosphere microbiome and their impact on plant phenotype. Microbiological Res. 283, 127706. doi: 10.1016/j.micres.2024.127706
Darji, T. B., Adhikari, B., Pathak, S., Neupane, S., Thapa, L. B., Bhatt, T. D., et al. (2021). Phytotoxic effects of invasive Ageratina adenophora on two native subtropical shrubs in Nepal. Sci. Rep. 11, 13663. doi: 10.1038/s41598-021-92791-y
Du, E., Chen, Y., Li, Y., Sun, Z., Gui, F. (2022a). Rhizospheric bacillus-facilitated effects on the growth and competitive ability of the invasive plant ageratina adenophora. Front. Plant Science. 13. doi: 10.3389/fpls.2022.882255
Du, E., Chen, Y., Li, Y., Zhang, F., Sun, Z., Hao, R., et al. (2022b). Effect of arbuscular mycorrhizal fungi on the responses of Ageratina adenophora to Aphis gossypii herbivory. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1015947
Du, X., Ge, Y., Zhang, Y., Hu, H., Zhang, Y., Yang, Z., et al. (2023). Responses of soil carbon cycling microbial functional genes to nitrogen and phosphorus addition in saline-sodic soils. Plant Soil. 490, 261–277. doi: 10.1007/s11104-023-06070-y
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Erasmus, D. J., Bennett, P. H., Staden, J. V. (1992). The effect of galls induced by the gall fly Procecidochares utilis on vegetative growth and reproductive potential of crofton weed, Ageratina adenophora. Ann. Appl. Biol. 120, 173–181. doi: 10.1111/j.1744-7348.1992.tb03414.x
Fang, K., Chen, L., Zhou, J., Yang, Z. P., Dong, X. F., Zhang, H. B. (2019). Plant–soil–foliage feedbacks on seed germination and seedling growth of the invasive plant Ageratina adenophora. Proc. R. Soc. B: Biol. Sci. 286, 20191520. doi: 10.1098/rspb.2019.1520
Gui, F. R., Wan, F. H., Guo, J. Y. (2008). Population genetics of Ageratina adenophora using inter-simple sequence repeat (ISSR) molecular markers in China. Plant Biosystems-An Int. J. Dealing all Aspects Plant Biol. 142, 255–263. doi: 10.1080/11263500802150399
Heystek, F., Wood, A. R., Neser, S., Kistensamy, Y. (2011). Biological control of two ageratina species (Asteraceae: eupatorieae) in South Africa. Afr. Entomology. 19, 208–216. doi: 10.4001/003.019.0208
Jiao, Y., Li, Y., Yuan, L., Huang, J. (2021). Allelopathy of uncomposted and composted invasive aster (Ageratina adenophora) on ryegrass. J. Hazardous Materials. 402, 123727. doi: 10.1016/j.jhazmat.2020.123727
Keane, R. (2002). Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evolution. 17, 164–170. doi: 10.1016/S0169-5347(02)02499-0
Khatri, K., Negi, B., Bargali, K., Bargali, S. S. (2022). Effects of elevation and habitat on leaf and reproductive traits of Ageratina adenophora (Sprengel) King & Robinson. South Afr. J. Botany. 147, 859–870. doi: 10.1016/j.sajb.2022.03.033
Kimbro, D. L., Grosholz, E. D. (2006). Disturbance influences oyster community richness and evenness, but not diversity. Ecology. 87, 2378–2388. doi: 10.1890/0012-9658(2006)87[2378:DIOCRA]2.0.CO;2
Kumar, V., Thukral, A. K., Sharma, A., Bhardwaj, R. (2023). Extending the concept of entropy-negentropy for the assessment of ecological dominance and diversity at alpha, beta and gamma levels. Geology Ecology Landscapes. 7, 27–39. doi: 10.1080/24749508.2021.1923270
Li, M., Gao, L., White, J. C., Haynes, C. L., O’Keefe, T. L., Rui, Y., et al. (2023). Nano-enabled strategies to enhance biological nitrogen fixation. Nat. Nanotechnol 18, 688–691. doi: 10.1038/s41565-023-01392-5
Li, W., Li, Y., Lv, J., He, X., Wang, J., Teng, D., et al. (2022). Rhizosphere effect alters the soil microbiome composition and C, N transformation in an arid ecosystem. Appl. Soil Ecology. 170, 104296. doi: 10.1016/j.apsoil.2021.104296
Lian, J., Schimmel, P., Sanchez Garcia, S., Wijffels, R. H., Smidt, H., Sipkema, D. (2021). Different co-occurring bacteria enhance or decrease the growth of the microalga Nannochloropsis sp. CCAP211/78. Microbial Biotechnol. 14, 1159–1170. doi: 10.1111/1751-7915.13784
Lin, Y., He, S. Q., Lu, Z. H., Gao, Y. L., Duan, Y. R., Li, Z. Y., et al. (2020). Influence of Aphis gossypii feeding on defense strategy of native and introduced populations of Ageratina adenophora. Arthropod-Plant Interactions. 14, 345–356. doi: 10.1007/s11829-020-09748-7
Liu, Y., Von Wirén, N. (2017). Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Botany. 68, 2581–2592. doi: 10.1093/jxb/erx086
Ma, J., Zhu, D., Sheng, G. D., O'Connor, P., Zhu, Y. G. (2019). Soil oxytetracycline exposure alters the microbial community and enhances the abundance of antibiotic resistance genes in the gut of Enchytraeus crypticus. Sci. Total Environment. 673, 357–366. doi: 10.1016/j.scitotenv.2019.04.103
Magoč, T., Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Masson-Boivin, C., Sachs, J. L. (2018). Symbiotic nitrogen fixation by rhizobia-the roots of a success story. Current Opinion in Plant Biology. 44, 7–15. doi: 10.1016/j.pbi.2017.12.001
Miller, B., Madilao, L. L., Ralph, S., Bohlmann, J. (2005). Insect-induced conifer defense. White pine weevil and methyl jasmonate induce traumatic resinosis, de novo formed volatile emissions, and accumulation of terpenoid synthase and putative octadecanoid pathway transcripts in sitka spruce. Plant Physiol. 137, 369–382. doi: 10.1104/pp.104.050187
Negi, B., Khatri, K., Bargali, S. S., Bargali, K. (2023). Invasive ageratina adenophora (Asteraceae) in agroecosystems of Kumaun Himalaya, India: A threat to plant diversity and sustainable crop yield. Sustainability. 15, 10748. doi: 10.3390/su151410748
Niu, H.b., Liu, W. X., Wan, F. H., Liu, B. (2007). An invasive aster (Ageratina adenophora) invades and dominates forest understories in China: altered soil microbial communities facilitate the invader and inhibit natives. Plant Soil. 294, 73–85. doi: 10.1007/s11104-007-9230-8
Niu, Y., Feng, Y., Xie, J., Luo, F. (2010). Noxious invasive Eupatorium adenophorum may be a moving target: Implications of the finding of a native natural enemy, Dorylus orientalis. Chin. Sci. Bulletin. 55, 3743–3745. doi: 10.1007/s11434-010-4117-0
Piton, G., Allison, S. D., Bahram, M., Hildebrand, F., Martiny, J. B. H., Treseder, K. K., et al. (2023). Life history strategies of soil bacterial communities across global terrestrial biomes. Nat. Microbiol. 8, 2093–2102. doi: 10.1038/s41564-023-01465-0
Policelli, N., Chiuffo, M. C., Moyano, J., Torres, A., Rodriguez-Cabal, M. A., Nuñez, M. A. (2018). Pathogen accumulation cannot undo the impact of invasive species. Biol. Invasions. 20, 1–4. doi: 10.1007/s10530-017-1439-8
Rabari, A., Ruparelia, J., Jha, C. K., Sayyed, R. Z., Mitra, D., Priyadarshini, A., et al. (2023). Articulating beneficial rhizobacteria-mediated plant defenses through induced systemic resistance: A review. Pedosphere. 33, 556–566. doi: 10.1016/j.pedsph.2022.10.003
Samuelsson, R., Burvall, J., Jirjis, R. (2006). Comparison of different methods for the determination of moisture content in biomass. Biomass Bioenergy. 30, 929–934. doi: 10.1016/j.biombioe.2006.06.004
Shah, M. A., Reshi, Z., Rashid, I. (2008). Mycorrhizal source and neighbour identity differently influence Anthemis cotula L. invasion in the Kashmir Himalaya, India. Appl. Soil Ecology. 40, 330–337. doi: 10.1016/j.apsoil.2008.06.002
Sharma Poudel, A., Babu Shrestha, B., Kumar Jha, P., Bahadur Baniya, C., Muniappan, R. (2020). Stem galling of Ageratina adenophora (Asterales: Asteraceae) by a biocontrol agent Procecidochares utilis (Diptera: Tephritidae) is elevation dependent in central Nepal. Biocontrol Sci. Technology. 30, 611–627. doi: 10.1080/09583157.2020.1749991
Singh, B. K., Munro, S., Potts, J. M., Millard, P. (2007). Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecology. 36, 147–155. doi: 10.1016/j.apsoil.2007.01.004
Sprent, J. I., Ardley, J., James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytologist. 215, 40–56. doi: 10.1111/nph.14474
Stackebrandt, E., Goebel, B. M. (1994). Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Systematic Bacteriology. 44, 846–849. doi: 10.1099/00207713-44-4-846
Stefanowicz, A. M., Kapusta, P., Stanek, M., Frąc, M., Oszust, K., Woch, M. W., et al. (2021). Invasive plant Reynoutria japonica produces large amounts of phenolic compounds and reduces the biomass but not activity of soil microbial communities. Sci. Total Environment. 767, 145439. doi: 10.1016/j.scitotenv.2021.145439
Stefanowicz, A. M., Stanek, M., Majewska, M. L., Nobis, M., Zubek, S. (2019). Invasive plant species identity affects soil microbial communities in a mesocosm experiment. Appl. Soil Ecology. 136, 168–177. doi: 10.1016/j.apsoil.2019.01.004
Sun, X., Gao, C., Guo, L. (2013). Changes in soil microbial community and enzyme activity along an exotic plant Eupatorium adenophorum invasion in a Chinese secondary forest. Chin. Sci. Bulletin. 58, 4101–4108. doi: 10.1007/s11434-013-5955-3
Sun, C., Li, Q., Han, L., Chen, X., Zhang, F. (2022). The effects of allelochemicals from root exudates of Flaveria bidentis on two Bacillus species. Front. Plant Science. 13. doi: 10.3389/fpls.2022.1001208
Sun, Y. Y., Yang, G. Q. (2021). Ecological adaptation mechanism of Ageratina adenophpra and its specialist herbivore Procecidochares utilis mediated by Bacillus cereus. Yangzhou university, Yangzhou.
Sun, Y. Y., Zhang, Q. X., Zhao, Y. P., Diao, Y. H., Gui, F. R., Yang, G. Q. (2021). Beneficial rhizobacterium provides positive plant–soil feedback effects to Ageratina adenophora. J. Integr. Agriculture. 20, 1327–1335. doi: 10.1016/S2095-3119(20)63234-8
Thukral, A. (2017). A review on measurement of Alpha diversity in biology. Agric. Res. J. 54, 1. doi: 10.5958/2395-146X.2017.00001.1
Tian, J., Yue, J., Philpot, W. D., Dong, X., Tian, Q. (2021). Soil moisture content estimate with drying process segmentation using shortwave infrared bands. Remote Sens. Environment. 263, 112552. doi: 10.1016/j.rse.2021.112552
Tsai, S. H., Hsiao, Y. C., Chang, P. E., Kuo, C. E., Lai, M. C., Chuang, H. (2023). Exploring the biologically active metabolites produced by bacillus cereus for plant growth promotion, heat stress tolerance, and resistance to bacterial soft rot in arabidopsis. Metabolites. 13, 676. doi: 10.3390/metabo13050676
Van Driesche, R. G., Carruthers, R. I., Center, T., Hoddle, M. S., Hough-Goldstein, J., Morin, L., et al. (2010). Classical biological control for the protection of natural ecosystems. Biol. Control. 54, S2–S33. doi: 10.1016/j.biocontrol.2010.03.003
Vessey, J. K., Buss, T. J. (2002). Bacillus cereus UW85 inoculation effects on growth, nodulation, and N accumulation in grain legumes: Controlled-environment studies. Can. J. Plant Science. 82, 282–290. doi: 10.4141/P01-047
Wang, Q., Garrity, G. M., Tiedje, J. M., Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 3, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, R., Wang, Y. Z. (2006). Invasion dynamics and potential spread of the invasive alien plant species Ageratina adenophora (Asteraceae) in China. Diversity Distributions. 12, 397–408. doi: 10.1111/j.1366-9516.2006.00250.x
Wu, A. P., Bai, Z. X., Li, J., Liu, H., Chen, F. L., Zhang, M. Y., et al. (2023). Invasive Ageratina adenophora can maintain its ecological advantages over time through releasing its autotoxicity by accumulating a bacterium Bacillus cereus. Heliyon. 9, e12757. doi: 10.1016/j.heliyon.2022.e12757
Xia, Y., Sun, J. (2023). “Alpha Diversity,” in Bioinformatic and Statistical Analysis of Microbiome Data (Springer, Cham), 289–333. doi: 10.1007/978-3-031-21391-5_9
Xia, Y., Zhang, H., Zhang, Y., Zhang, Y., Liu, J., Seviour, R., et al. (2023). Screening plant growth-promoting bacteria from the rhizosphere of invasive weed Ageratina adenophora for crop growth. PeerJ 11, e15064. doi: 10.7717/peerj.15064
Yuan, C., Wang, Q., Chen, Y., Zhang, L. D., Tan, L., Fu, R. H., et al. (2021). Impacts of a biocontrol agent on invasive Ageratina adenophora in Southwest China: Friend or foe? Biol. Control. 152, 104471. doi: 10.1016/j.biocontrol.2020.104471
Yushmanov, V. E., Kharlamov, A., Boada, F. E., Jones, S. C. (2007). Monitoring of brain potassium with rubidium flame photometry and MRI. Magnetic Resonance Med. 57, 494–500. doi: 10.1002/mrm.21155
Zhang, X., Van Kleunen, M., Chang, C., Liu, Y. (2023). Soil microbes mediate the effects of resource variability on plant invasion. Ecology. 104, e4154. doi: 10.1002/ecy.4154
Zhang, M., Wang, H., Hussain, M., Qi, J., Ma, C., Lan, M., et al. (2020). Identification and functional assessment of endophytic bacterial diversity in Ageratina adenophora (Sprengel) and their interactions with the host plant. South Afr. J. Botany. 134, 99–108. doi: 10.1016/j.sajb.2020.07.038
Zhao, Q., Bai, J., Gao, Y., Zhao, H., Zhang, G., Cui, B. (2020). Shifts in the soil bacterial community along a salinity gradient in the Yellow River Delta. Land Degradation Dev. 31, 2255–2267. doi: 10.1002/ldr.3594
Zhao, M., Lu, X., Zhao, H., Yang, Y., Hale, L., Gao, Q., et al. (2019). Ageratina adenophora invasions are associated with microbially mediated differences in biogeochemical cycles. Sci. Total Environment. 677, 47–56. doi: 10.1016/j.scitotenv.2019.04.330
Keywords: Ageratina adenophora, Bacillus cereus, Aphis gossypii, soil physicochemical properties, native polyphagous insect
Citation: Yu Y, Yang Z, Han M, Sun S, Xu G and Yang G (2024) Beneficial rhizosphere bacteria provides active assistance in resisting Aphis gossypiis in Ageratina adenophora. Front. Plant Sci. 15:1394153. doi: 10.3389/fpls.2024.1394153
Received: 01 March 2024; Accepted: 29 April 2024;
Published: 15 May 2024.
Edited by:
Roberta Pastorelli, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Ugur Azizoglu, Kayseri University, TürkiyeCopyright © 2024 Yu, Yang, Han, Sun, Xu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqing Yang, Z3F5YW5nQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.