
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 July 2024
Sec. Crop and Product Physiology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1393796
This article is part of the Research Topic Physiology and Breeding of Cereals View all 14 articles
The use of wild species as a source of genetic variability is a valued tool in the framework of crop breeding. Hordeum chilense Roem. et Schult is a wild barley species that can be a useful genetic donor for sustainable wheat breeding which carries genes conferring resistance to some diseases or increasing grain quality, among others. Septoria tritici blotch (STB), caused by the Zymoseptoria tritici fungus, is one of the most important wheat diseases worldwide, affecting both bread and durum wheat and having a high economic impact. Resistance to STB has been previously described in H. chilense chromosome 4Hch. In this study, we have developed introgression lines for H. chilense chromosome 4Hch in durum wheat using interspecific crosses, advanced backcrosses, and consecutive selfing strategies. Alien H. chilense chromosome segments have been reduced in size by genetic crosses between H. chilense disomic substitution lines in durum wheat and durum wheat lines carrying the Ph1 deletion. Hordeum chilense genetic introgressions were identified in the wheat background through several plant generations by fluorescence in situ hybridisation (FISH) and simple sequence repeat (SSR) markers. An STB infection analysis has also been developed to assess STB resistance to a specific H. chilense chromosome region. The development of these H. chilense introgression lines with moderate to high resistance to STB represents an important advance in the framework of durum breeding and can be a valuable tool for plant breeders.
Increasing human population and climate change demand greater agricultural productivity. Wheat is a strategic crop, essential for Western countries. It is the most important source of protein, providing 20% of the calories consumed daily by humans, and its consumption is increasing, mainly in developing countries. Although the yield of primary crops, including wheat, in 2022 was double that of 2000, it is estimated that the global demand for wheat production will increase by 60% in 2050 to meet the requirements of a growing population (http://www.fao.org). Europe is the first-world producer of bread wheat and within the European Union; Spain ranks in the fifth position considering cereal production, mainly wheat, only behind France, Germany, Romania, and Poland (http://www.fao.org). Thus, the economic relevance of wheat as a strategic crop in agriculture is clear, but several biotic (diseases) and abiotic (higher temperatures, soil erosion, salinity, water availability, etc.) stresses can compromise the future of this crop. A solid strategy, based on the development of new approaches that allow the adaptation of wheat to climate change and are respectful of the principles of sustainable agriculture, is needed to face the challenge of feeding the expected human population (Hamblin, 1995). Plant breeders are playing a major role in worldwide efforts to understand gene functions and interactions, so the introduction of tolerant genes to both biotic and abiotic stresses can increase the quality and productivity of a key crop such as wheat (Mamrutha et al., 2019; Mondal et al., 2021; Pérez-Méndez et al., 2022).
Durum wheat (Triticum turgidum, 2n = 4x = 28) is a globally relevant species for agriculture and also has an enormous impact in Spain since it is one of the most cultivated cereals together with bread wheat and barley. Despite its high productivity, annual fluctuations can be associated with biotic and abiotic stresses. One of the most important diseases affecting durum wheat is Septoria tritici blotch (STB) caused by the fungus Zymoseptoria tritici (Desm.) (Quaedvlieg et al., 2011), previously known as Septoria tritici (teleomorph Mycospaherella graminicola (Fuckel) J. Schröt.), which triggers severe losses in yield that can reach 50% in susceptible cultivars (Fones and Gurr, 2015). Severe epidemics occur both in areas with frequent rains and in others where rainfall does not exceed 250 mm during the crop cycle. The wide range of temperatures in which the fungus can develop, the use of susceptible varieties, and the rapid evolution and adaptation of Z. tritici to diverse agricultural conditions have allowed a wide geographical distribution of the disease (Fones and Gurr, 2015).
The introgression of resistance genes in the framework of breeding programs is considered the main protection against this disease, and several sources of resistance have yielded adequate protection (Rubiales et al., 2001). Wheat resistance to STB can be qualitative (isolate-specific), depending on major genes with a strong effect according to a gene-for-gene interaction, or quantitative (isolate-non-specific) displaying a partial phenotype controlled by various genes with moderate to small effects (Kema et al., 2000; Brading et al., 2002; Brown et al., 2015). Quantitative resistance is quite significant for its effectiveness against all genotypes of the pathogen and its durability (Brown et al., 2015; Porras et al., 2021). When no sufficient genetic variability or resistant varieties are available, the incorporation of new alleles into the wheat germplasm is considered essential. Thus, genetic crosses between donor species and the crop itself can be a powerful tool to transfer chromosome segments carrying desirable genes from the alien species into wheat.
Breeders have been using related species as genetic donors with the target of widening the genetic basis of this crop obtaining, for example, wheat cultivars better adapted to specific agro-climatic conditions, carrying resistance to pests (Lukaszewski, 2000; Shubing and Honggang, 2005). Indeed, wide-crossing in plant breeding programmes is such an important tool that, sometimes the results are the starting point for new crops (O’mara, 1953; Martin and Sanchez-Mongelaguna, 1982) and marked a milestone in a breeding framework.
Diploid wild barley Hordeum chilense Roem. et Schultz (2n = 2x = 14 HchHch) exhibits valuable agronomic and quality traits for wheat improvement, and it has already been crossed with some species of the tribe Triticeae (grasses), particularly with both durum and bread wheat (Martin et al., 1996; Calderón et al., 2012). This wild species also has enormous potential as a donor of other genetic features such as drought and salt tolerance as well as resistance to pests and diseases (Gallardo and Fereres, 1989; Bothmer et al., 1995; Martin et al., 1996). Particularly, H. chilense contains interesting genes for biotic and abiotic stress resistance, such as resistance to STB, conferred mainly by the 4Hch chromosome (Rubiales et al., 2000). The transfer of H. chilense chromosome 4 in durum wheat has been already developed (Calderón et al., 2012), but, in a plant breeding framework, the size of full introgressed chromosomes from relatives must be reduced to diminish the drag linkage of undesirable genes.
Much of the work aimed to promote interspecific chromosome associations to transfer genetic variability from wild relatives into wheat has relied on the use of the ph1b mutant (Sears and Okamoto, 1958). Homoeologous recombination between chromosomes from related but different genomes can occur in the absence of the Ph1 locus, resulting in the generation of interspecific recombinant chromosomes (Sears and Okamoto, 1958; Othmeni et al., 2019). In the absence of the Ph1 locus, chromosome rearrangements in bread wheat involving wheat and relative H. chilense and H. vulgare chromosomes have already been obtained (Rey et al., 2015a, 2015). In this work, we used the ph1 mutant and the previously developed H. chilense chromosome 4Hch substitution line (Calderón et al., 2012), both in durum wheat background, to carry out genetic crosses to facilitate durum wheat-wild barley interspecific recombination. A set of durum wheat–H. chilense chromosome 4Hch translocation lines were obtained and characterised using molecular markers and fluorescence in situ hybridisation. We also analysed these lines for the incidence of STB under controlled conditions in growth chambers to assess resistance to STB to an H. chilense chromosome 4Hch segment.
Genetic crosses between the 4Hch (4B) substitution line in durum wheat cv. Langdon (T. turgidum, 2n=4x = 28; (Calderón et al., 2012) and durum wheat cv. Capelli carrying the Ph1 deletion (Ceoloni et al., 1992) have been carried out. F1 plants were backcrossed with durum wheat cv. Capelli in the ph1 mutant background. Plants were selected using molecular markers and by in situ hybridisation and then selfed. The procedure is described in Figure 1, and the chromosomal composition of Hordeum chilense chromosome 4Hch introgression lines in durum wheat is summarized in Table 1. Plants carrying H. chilense introgressions were selected for analysis.
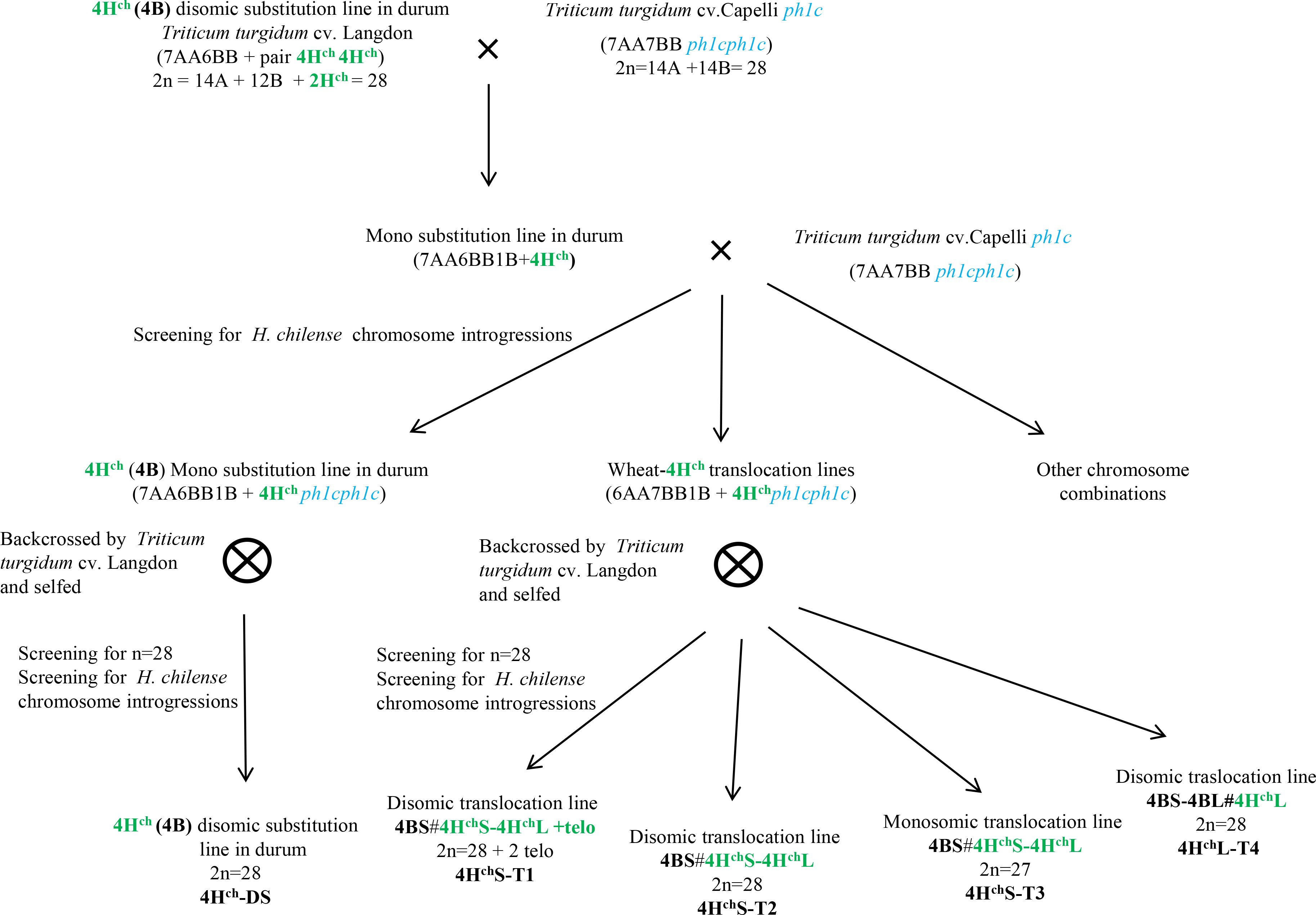
Figure 1 Development of H. chilense introgression lines in tetraploid durum wheat (Triticum turgidum cv. Langdon). Crosses between the 4Hch substitution line in T. turgidum and the ph1c wheat mutant (Triticum turgidum cv. Capelli) were developed to obtain H. chilense genetic introgressions in the absence of the Ph1 locus. Backcrosses by durum wheat (Triticum turgidum cv. Langdon) were developed. Screening for H. chilense genetic introgressions was carried out using molecular markers and in situ hybridization.
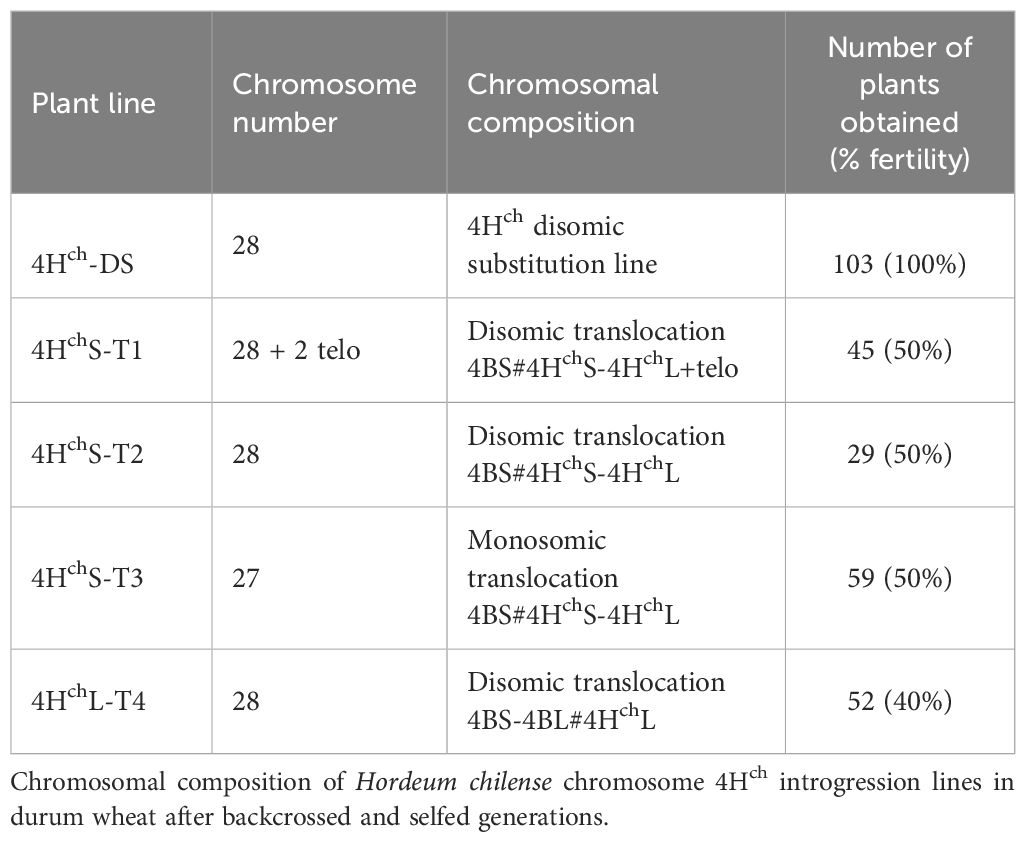
Table 1 Progeny of the crosses between the 4Hch(4B) substitution line in durum wheat (Triticum turgidum) and the durum wheat carrying the Ph1 deletion.
Seeds were incubated in Petri dishes on wet filter paper in the dark for 5 days at 4°C, followed by 24 h of incubation at 24°C to allow germination. Then, roots were cut and incubated for 4 h in 0.05% colchicine solutions at 24°C to accumulate somatic cells in metaphase and finally fixed in ethanol/glacial acetic acid 3:1 (v/v) for 1 month at 4°C.
For identification and characterisation of the H. chilense chromosome 4Hch introgressions and wheat chromosomes involved in translocations, fluorescence in situ hybridisation (FISH) using H. chilense DNA, the GAA-satellite sequence, and the pAs1 probe (Cabrera et al., 1995; Pedersen et al., 1997) was performed. DNA probes were labelled by nick translation with biotin-11 dUTP (Roche Corporate, Basel, Switzerland) and digoxigenin 11-dUTP (Roche Corporate, Basel, Switzerland) and detected with streptavidin-Cy3 (Sigma, St Louis, MO, USA) and anti-digoxigenin–FITC (Roche Corporate Basel, Switzerland), respectively.
Metaphase mitotic chromosomes were hybridised with total genomic H. chilense DNA as a probe to identify the chromosome segment introgressed from H. chilense chromosome 4Hch. Afterwards, the preparations were hybridised using the repetitive sequence GAA and the pAs1 probe to characterise both the H. chilense chromosome segment and the wheat chromosome involved in interspecific chromosome translocations. The in situ protocol was performed according to Prieto et al. (2004).
Hybridisation signals were visualised using a Nikon eclipse inflorescence microscopy, and the images were captured with a Nikon CCD camera using the appropriate Nikon 3.0 software (Nikon Instrument Europe BV, Amstelveen, The Netherlands) and processed with Photoshop 4.0 software for brightness and contrast (Adobe Systems Inc., San Jose, CA, USA).
Molecular markers were selected and tested for the specific identification of the short and long arms of H. chilense chromosome 4Hch according to available information (Said and Cabrera, 2009; Sakata et al., 2010).
Genomic DNA was extracted from frozen young leaves using the CTAB method described by Murray and Thompson (1980). We tested 25 and 9 markers to specifically identify both the short arm and the long arm of chromosome 4Hch, respectively. The polymerase chain reaction (PCR) experiments were performed in 20 µL of a mixture containing 4 µL of reaction buffer containing MgCl2 and dNTPs, 5 pmol of selected primers, and 0.4 µL of MyTaq DNA polymerase. Specific primers and PCR conditions to amplify these markers were according to Masaya Sakata et al. (2010) and Said and Cabrera (2009). The PCR products were visualized using SafeView (NBS Biologicals, Cambridgeshire, UK) in an agarose (1%) electrophoresis gel (120 V, 45 min).
We have also used the NCBI’s BLAST (Basic Local Alignment Search Tool) to perform retro-transcription-PCR (RT-PCR) in silico on H. chilense genome and to locate the position of the primers on the H. chilense physical map using the reference genome (txid15565) from the NCBI database (National Center for Biotechnology Information).
The inoculation process was developed as reported by Stewart and McDonald (2014), following minor modifications described by Porras et al. (2021). Briefly, seeds of the 10 durum wheat genotypes, including the positive control, were sown in 8 × 7 × 7 cm pots containing a mix (1:1, v/v) of commercial compost (Suliflor SF1 substrate; Suliflor Lithuania) and sand. Pots were incubated in a growth chamber at 21°C and 70% relative humidity (RH), with 16 h of light. After 16 days, seedlings were fungal inoculated when the second and third leaves emerged [growth stage Z13; (Zadoks et al., 1974)]. Zymoseptoria tritici (isolated from infected durum wheat of Santaella (Cordoba, Spain) and deposited at the NBCI database under the SUB9540116 accession number) inoculum was adjusted to 107 spores mL−1 in distilled water and 0.1% Tween 20. Seedlings were inoculated using a hand sprayer. Once leaves were totally dry, plants were sealed in clear plastic bags to provide 100% RH and maintained for 48 h in a growth chamber at 22/18°C day/night with a 16-h photoperiod. After 48 h, plastic bags were removed, and plants were kept at 75%–80% RH. The amount of infected plants per family was variable, ranging from 6 to 18, based on seed availability. The positive control check, the commercial cultivar ‘SY Leonardo’, was inoculated following the same treatment. Three independent experiments with three replicates each were performed.
At 21 days post-inoculation, the third leaf of each plant was evaluated. The infection process was qualitatively scored using the disease severity (DS) rating scale from 0 to 5 (McCartney et al., 2002), as follows: 0 = immune with no visible symptoms, 1 = highly resistant with hypersensitive flecking, 2 = resistant with small chlorotic or necrotic lesions and no pycnidial development, 3 = moderately resistant, characterised by coalescence of chlorotic and necrotic lesions with slight pycnidial development, 4 = susceptible with moderate pycnidial development and coalesced necrotic lesions, and 5 = very susceptible with large, abundant pycnidia and extensively coalesced necrotic lesions. Porras et al. (2021) showed a digitally infected leaves scale with STB that was used as a reference to evaluate the families of the present study. Reaction types 0–3 were considered resistant, whereas reaction types 4 and 5 were considered susceptible. Although reaction type 3 includes pycnidium development, it is considered resistant because the growth and sporulation of the fungus are quite restricted, and the chlorotic reaction is similar to the chlorotic blotches of reaction type 2.
Crosses between the 4Hch(4B) substitution line in durum wheat cv. Langdon and the durum wheat cv. Capelli carrying the Ph1 deletion were carried out to promote interspecific recombination between chromosome 4Hch and the homoeologous wheat chromosomes in the absence of the Ph1 locus. To obtain the Ph1 deletion in homozygosis, one backcross to durum wheat cv. Capelli carrying the Ph1 deletion was carried out. Wheat plants carrying monosomic and disomic H. chilense introgressions were selected and analysed by multicolour in situ hybridisation and molecular markers. The procedure is summarised in Figure 1. We obtained more than 200 plants having either monosomic or disomic genetic introgressions involving H. chilense chromosome 4Hch in durum wheat (Table 1).
A total of 24 EST-SSR (Expressed Sequence Tags-Simple Single Repeat) specific markers for the short arm of H. chilense chromosome 4Hch and nine barley markers for the long arm of the same chromosome were tested to identify those markers displaying polymorphism between durum wheat and the wild barley (Table 2). Most of them did not show polymorphism between the 4Hch H. chilense and the 4A wheat homoeologous chromosome and were not used for the genetic screening. Only 10 markers revealed polymorphism between 4Hch H. chilense and wheat and were used to identify and characterise the size of the 4Hch H. chilense introgressions (Table 3).

Table 2 Molecular markers used for the screening of Hordeum chilense chromosome introgressions in the durum wheat (DW) background.
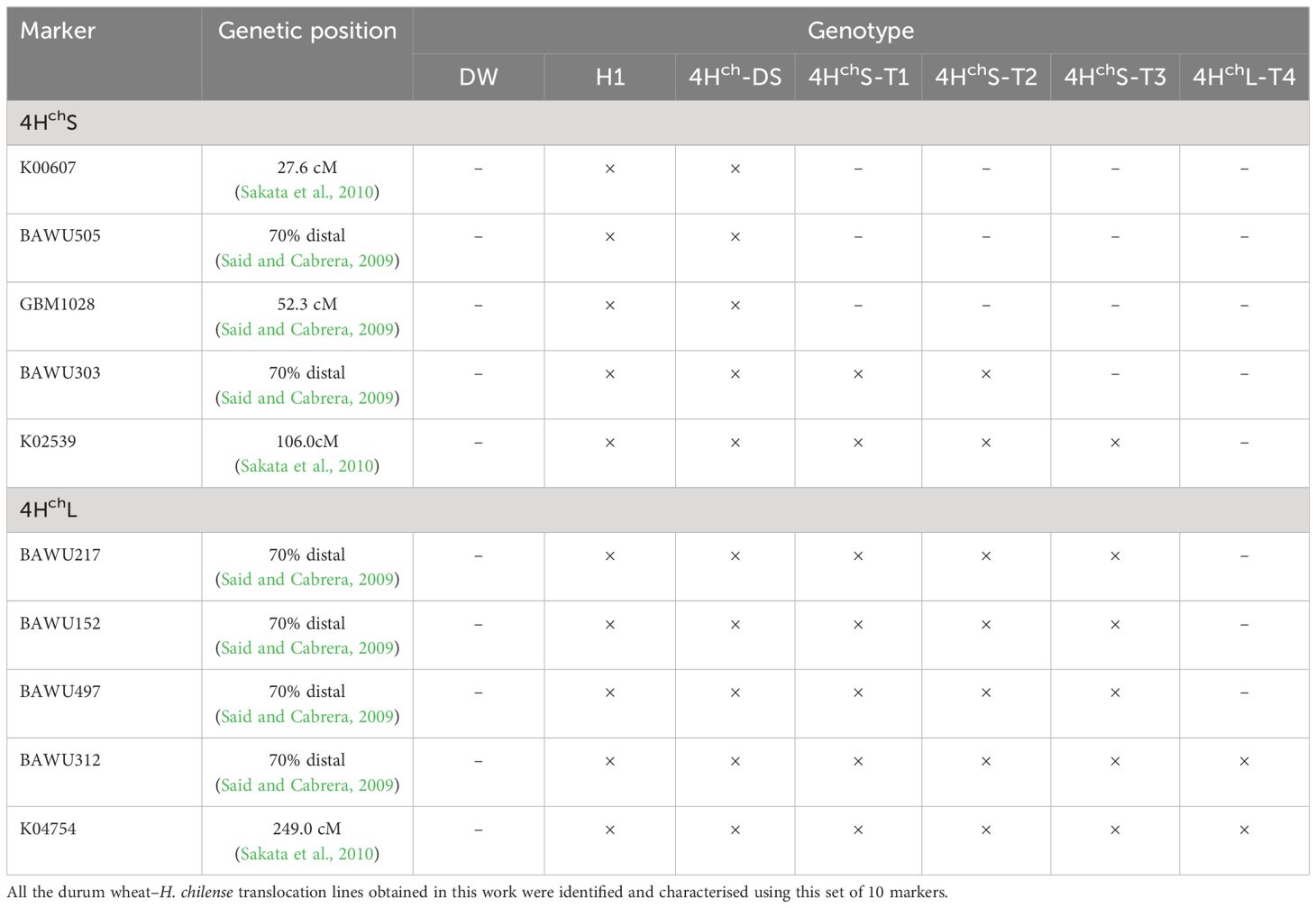
Table 3 Barley SSR markers used for the identification of Hordeum chilense chromosome 4Hch introgressions in the durum wheat (DW) background.
The chromosomal location of polymorphic molecular markers on the 4Hch chromosome was performed by in silico RT-PCR on the H. chilense genome, using NCBI’s BLAST for lining up the preexisting primers. Five markers were located on the short arm of chromosome 4Hch (K00607, BAWU505, GBM1028, BAWU303, and K02539) and another five on the 4HchL (BAWU217, BAWU152, BAWU497, BAWU312, and K04754). The molecular analysis of the four translocation lines obtained showed that marker K02539, located near the centromere of 4HchS, amplified in 4HchS-T1, 4HchS-T2, and 4HchS-T3 and was absent in 4HchL-T4, revealing that wheat–H. chilense recombination in 4HchS-T1, 4HchS-T2, and 4HchS-T3 did occur between 4HchS and wheat chromosomes. Only 4HchS-T1 and 4HchS-T2 were positive for the BAWU303 marker. K00607, BAWU505, and GBM1028 markers, located in a distal position on 4HchS, were absent in all translocation lines (Table 3). Thus, three wheat–H. chilense translocation lines involving the short arm of chromosome 4Hch were obtained. Recombination events did occur at least in two different positions on the 4Hch chromosome as 4HchS-T1 and 4HchS-T2 displayed a similar molecular pattern and were different from the 4HchS-T3 translocation line. Related to the molecular analysis of those markers located on the long arm of 4Hch H. chilense chromosome, all translocation lines were positive for distal BAWU312 and K04754 markers. BAWU217, BAWU152, and BAWU497 were positive in all translocation lines except in 4HchL-T4. In fact, the 4HchL-T4 translocation line was only positive for the distal K04754 and BAWU312 markers. Our results displayed that this 4HchL-T4 translocation line only retained a distal 4HchL chromosome segment in the durum wheat background.
Depending on the recombination event between H. chilense and wheat chromosomes, four different types of genetic introgressions for H. chilense chromosome 4Hch were obtained in durum wheat in the ph1 mutant background (Table 1; Figures 2, 3). Most of the interspecific recombination events that we were able to recover occurred between the short 4Hch and 4A chromosome arms. Thus, we obtained three different translocation lines involving the short 4Hch and 4A chromosome arms, named 4HchS-T1, 4HchS-T2, and 4HchS-T3 and one involving the long 4Hch and 4A chromosome arms, named 4HchL-T4.
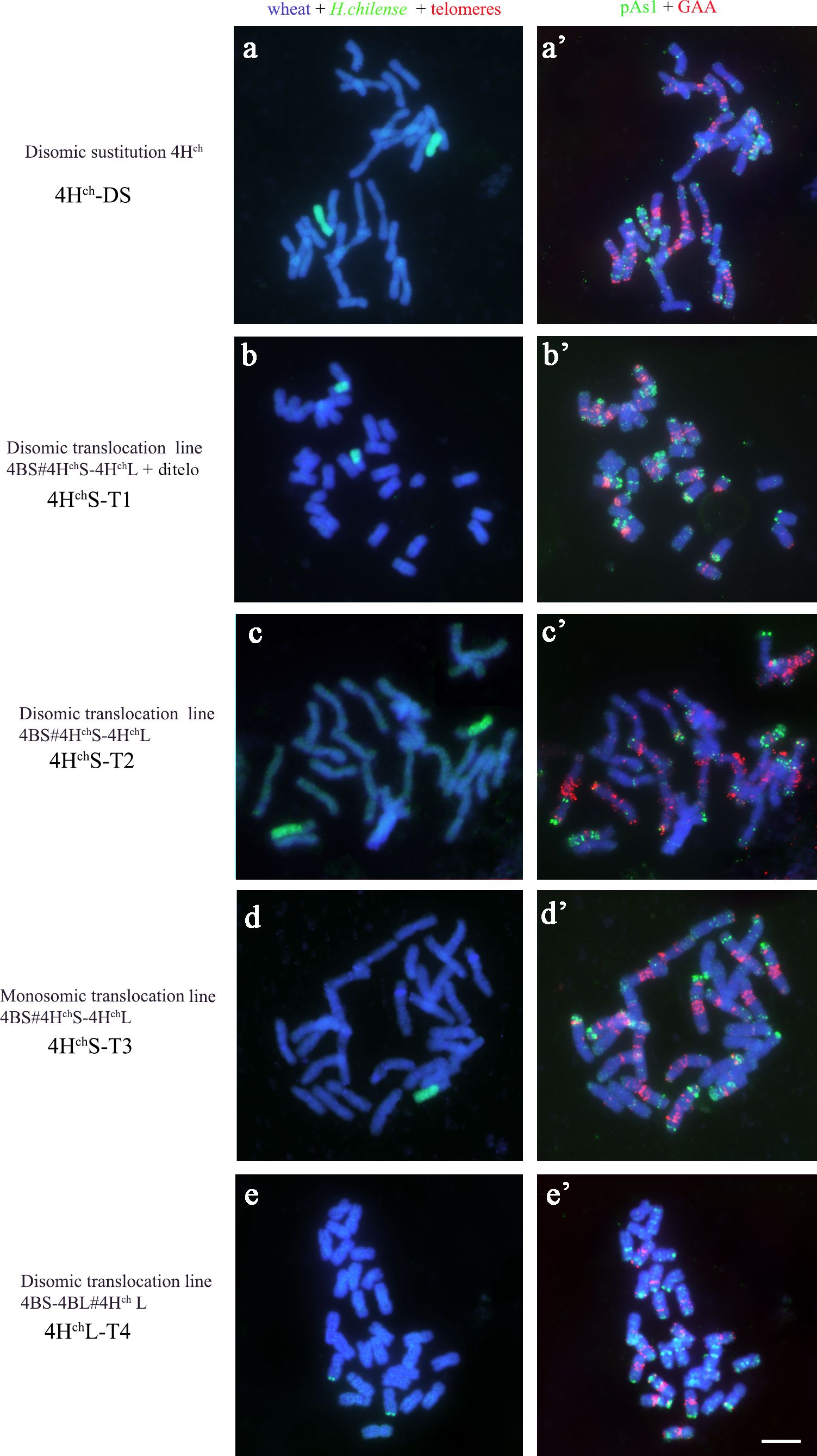
Figure 2 Cytogenetic visualisation of the introgression lines obtained for chromosome 4Hch in Triticum turgidum using fluorescence in situ hybridisation. Hordeum chilense genomic introgressions are shown in green (A-E). Chromosome identification and orientation were confirmed by reprobing with the pAs1 (green) and GAA (red) probes in (A′-E′). DNA was counterstained with DAPI (blue). (A-A′) 4Hch disomic substitution line; (B-B′) 4HchS-T1 translocation line; (C-C′) 4HchS-T2 translocation line, (D-D′) 4HchS-T3 translocation line; (E-E′) 4HchL-T4 translocation line. Scale bar for all panels = 10 µm.
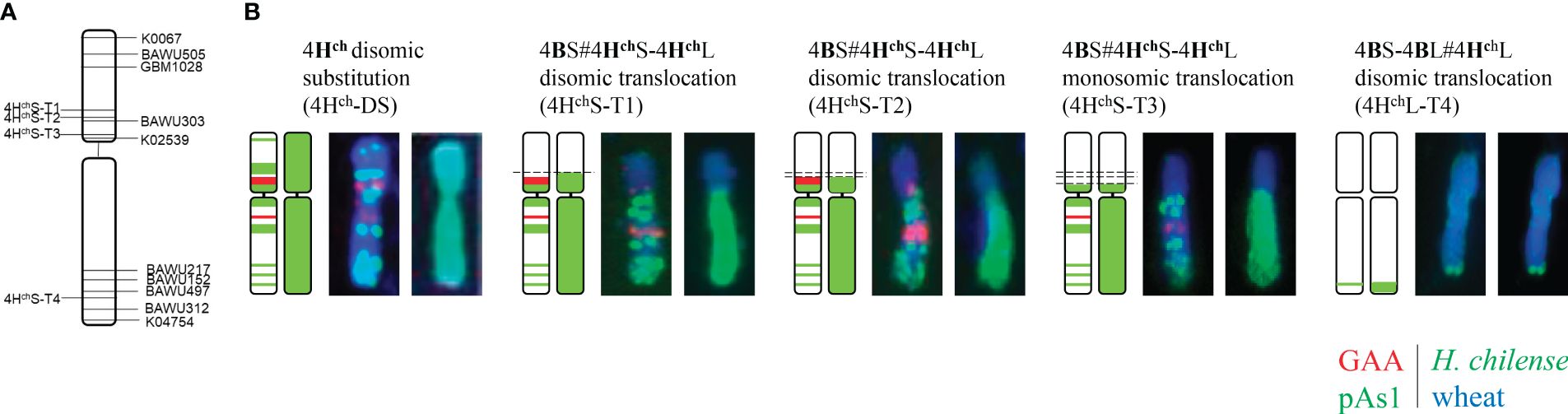
Figure 3 Ideogram of the wheat-H. chilense recombinant chromosomes. (A) Graphical genotyping of the chromosome indicating the recombination breakpoints of the SSR markers. (B) Characterisation of the H. chilense segment introgressed using the pAs1 (in green) and GAA (in red) probes. The whole 4Hch chromosome segment is also shown (green).
Genomic in situ hybridisation contributed to elucidate the exact chromosomal composition of all translocation lines obtained (Figure 2). All the H. chilense–wheat translocation lines obtained were disomic except 4HchS-T3, which was monosomic. The cytogenetic analysis using the psA1 and GAA repeat sequences contributed to characterise the chromosome segment of the H. chilense chromosome 4Hch introgressed in durum wheat by their specific pattern on this chromosome 4Hch (Figure 3B). Thus, 4HchS-T1, 4HchS-T2, and 4HchS-T3 translocation lines contained the full 4HchL chromosome arm and a fragment of the 4HchS chromosome arm, which vary among these translocation lines depending on the position of the recombination event (Figure 3B). The cytogenetic analysis using the psA1 and GAA repeat sequences revealed a similar cytogenetic pattern between 4HchS-T1 and 4HchS-T2 translocation lines (Figure 3B), which agrees with the equivalent result obtained using molecular markers (Table 3). These results suggest that the recombination events occurring between wheat and barley chromosomes in these two 4HchS-T1 and 4HchS-T2 translocation lines were extremely close to each other along the 4HchS chromosome. In addition, a recombination event on the distal region of chromosome 4HchL did occur in the 4HchL-T4 translocation line, retaining only a small 4HchL chromosome segment (Figures 2, 3).
The development of STB in the H. chilense translocation lines obtained in the durum wheat background is shown in Table 4 and Figure 4. Two different experiments were carried out, and differences in Disease Severity (DS) rating scale scores were recorded. DS scores showed differences among the plants studied, ranging from 2 to 4. Most of the studied durum wheat lines exhibited a resistant to moderately resistant reaction, displaying DS ≤ of 3, which indicated a restricted growth and sporulation of the fungus. The 4Hch H. chilense disomic substitution line (4Hch-DS) could be considered the most resistant one, showing a heterogeneous behaviour against the disease but with the lowest DS scores (1-2). This means that some leaves presented reaction types highly resistant with hypersensitive flecking (DS 1), and others showed small necrotic lesions with no pycnidia development (DS 2). No remarkable differences were found between durum wheat–H. chilense translocation lines 4HchS-T1, 4HchS-T2, and 4HchL-T4, showing an average DS score of 3 (moderately resistant), with limited production of pycnidia (DS 3) and small necrosis lesions. In lines 4HchS-T2 and 4HchL-T4, most leaves displayed a mainly moderately resistant response with limited production of pycnidia with chlorotic/necrotic lesions (DS 3), although some leaves allowed higher pycnidia development. Only the family 4HchS-T3 displayed an average DS score higher than 3, showing leaves with two different reaction types, moderately resistant (DS 3) and susceptible (DS 4). This result could be because this translocation line has only one copy of the H. chilense chromosome segment (monosomic translocation line). Lastly, durum wheat cv. Langdon, which is the genetic background of the translocation lines developed in this work, presented an average DS score of 4 (susceptible), which indicates significant fungus-reproduction capability in the form of pycnidia in the necrotic lesions. Strikingly, none of the analysed durum wheat including the Langdon cultivar showed a DS score of 5 (very susceptible with abundant pycnidia and extensively coalesced necrotic lesions). As it was expected, the other control used in this work, cultivar SY Leonardo, was very susceptible, showing most of the screened plants high reproduction of the fungus and extensively coalesced lesions (DS 5).
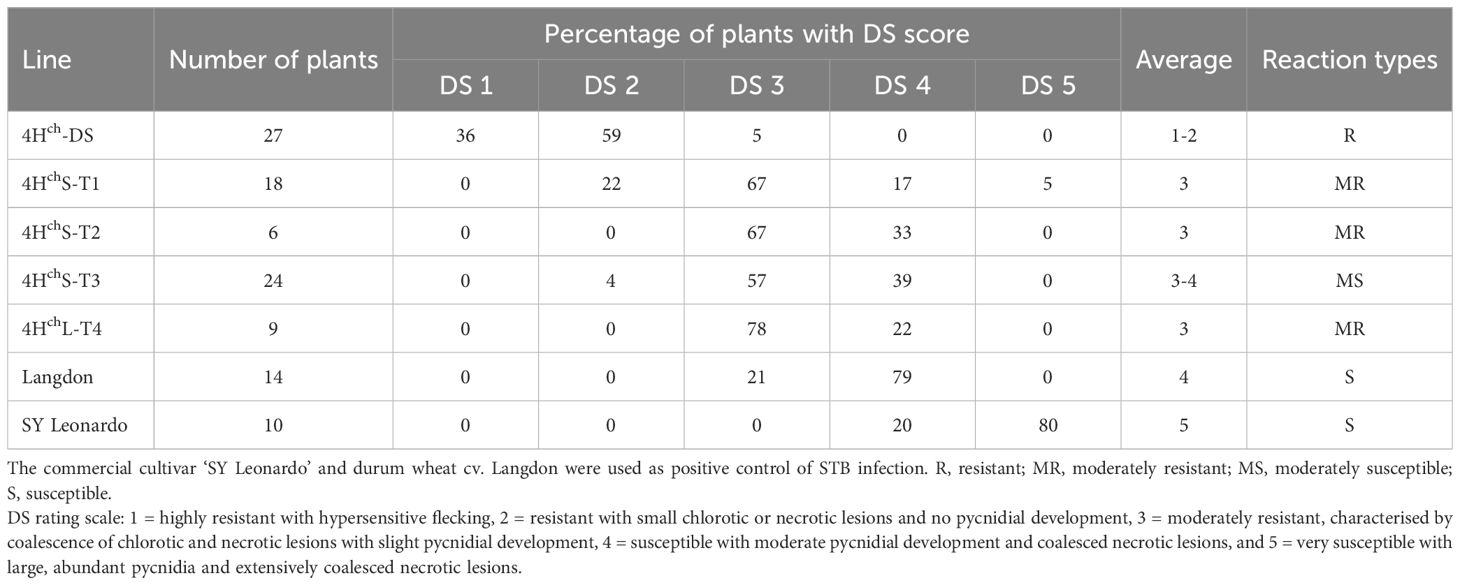
Table 4 Classification of studied families according to their Disease Severity (DS) rating scale (McCartney et al., 2002).
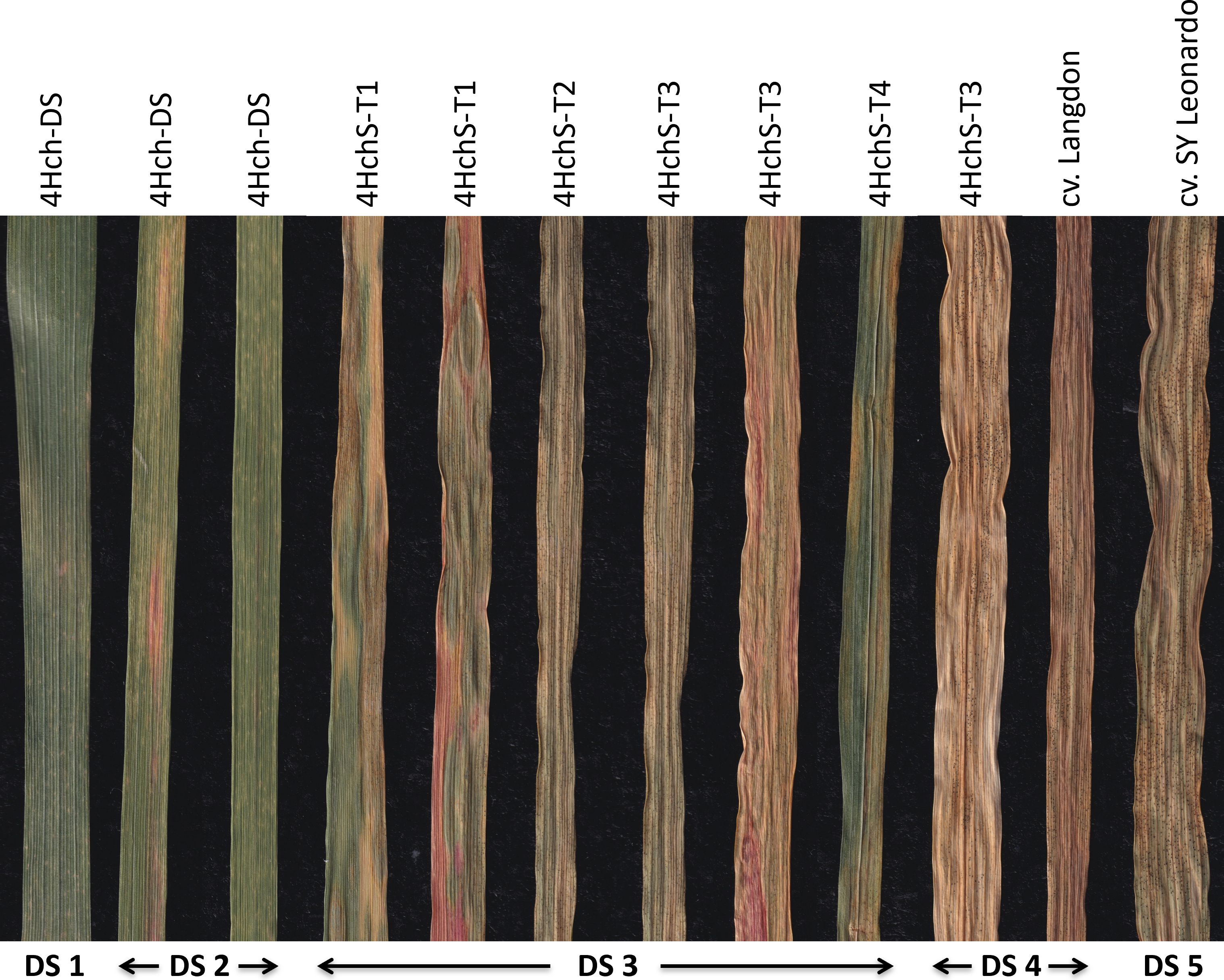
Figure 4 Examples of infected leaves with Septoria tritici blotch (STB) showing diverse disease severity rating scale (DS) scores. Leaves from the studied genotypes with different average DS scores. The 4Hch disomic substitution, 4Hch-DS, line displayed DS1-2. All the translocation lines 4HchS-T1, 4HchS-T2, 4HchS-T3, and 4HchL-T4 displayed DS 3, although some leaves from 4HchS-T3 also showed DS4 as the durum wheat cultivar Langdon. The most sensitive line (DS5) was the control cultivar SY Leonardo.
The effective introgression of genes from alien species like H. chilense into cultivated species such as durum wheat entails that target genes or the chromosome segment carrying them must be incorporated into wheat chromosomes as recombinant segments or translocations. Examples of an important source of chromosome introgressions are centric-break fusion events found several decades ago in bread wheat–rye translocations (e.g., 1BL-lRS and lAL-lRS), which have importantly contributed to global wheat production (Pena et al., 1990). Genetic introgressions from H. chilense chromosome 4Hch, which contains resistance to STB (Rubiales et al., 2000), have been previously developed in the durum wheat background (Calderón et al., 2012). In this work, genetic crosses between the durum wheat line carrying a disomic 4Hch(4B) chromosome substitution and durum wheat carrying the Ph1 deletion were developed to promote interspecific recombination between wheat and H. chilense. Recombination events between bread wheat and H. chilense lines in the absence of the Ph1 locus have been previously promoted for both fundamental and applied purposes, which is to shed light into the mechanism of this Ph1 locus or in a plant breeding framework to specifically introgress H. chilense chromosome 7Hch to increase the carotenoid content in bread wheat (Rey et al., 2015a). To the best of our knowledge, it is the first time that the ph1 mutant has been used in genetic crosses between H. chilense and wheat in the tetraploid durum background. The use of the ph1 mutants in this work has enabled the possibility of promoting recombination events between 4A wheat and 4Hch H. chilense homoeologous chromosomes, providing the opportunity to introgress part of 4Hch chromosome in durum wheat and the development of a series of genetic introgressions for this H. chilense chromosome for durum breeding purposes. The durum wheat–H. chilense introgression lines for chromosome 4Hch developed in this work might facilitate the possibility of transferring resistance to STB in durum wheat as the 4Hch chromosome was previously targeted as a potential source to introduce this type of resistance in wheat (Rubiales et al., 2000).
The interaction between Z. tritici and durum wheat has been poorly studied, and no Stb genes have been identified in durum wheat so far (Somai-Jemmali et al., 2017). This lack of identified and characterised Stb resistance genes is a challenge for plant breeders to find durable sources of resistance to STB in durum wheat. Resistance to STB is generally expressed through the reduction of the foliar area covered with pycnidia and necrosis (Porras et al., 2021). In this work, we have obtained several durum wheat–H. chilense translocation lines including chromosome 4Hch that present a moderate resistance to STB. The reduction of the diseased in the 4Hch H. chilense introgression lines in durum wheat is attributed to the contribution of chromosome 4Hch compared with both control lines, the cultivar Langdon, which is the genetic background of the H. chilense introgressions developed, and the cultivar SY Leonardo, which is routinely used as a susceptible control to STB. Both durum cultivars displayed elevated susceptibility with abundant development of the fungus and extensively coalesced lesions (DS 4 and 5, respectively) in contrast to the DS scores 1 and 2 expressed by the 4Hch disomic substitution line, 4hch-DS, or the average DS score of 3 displayed by the translocation lines 4HchS-T1, 4HchS-T2, and 4HchL-T4. Thus, our results demonstrate the contribution of chromosome 4Hch for introgressing resistance to STB in durum wheat.
The combination of cytogenetic tools with a molecular analysis offers a reliable method of selection of H. chilense introgressions in the durum wheat background during a plant breeding programme. These combined approaches provide the possibility to detect and analyse a high number of plant lines carrying chromosome modifications in a suitable frame of time, and for this, they have been extensively used for the identification and characterisation of plant material for decades (Nalam et al., 2006; Faris et al., 2008; Wang et al., 2009; Calderón et al., 2012). In this work, genomic in situ hybridisation contributed to elucidate the exact chromosomal composition of all translocation lines obtained. All the H. chilense–wheat translocations obtained were disomic except 4HchS-T3, which was monosomic. Disomic 4HchS-T1, 4HchS-T2, and 4HchL-T4 translocation lines showed moderate resistance to STB, with limited production of pycnidia and small necrosis lesions, although the monosomic 4HchS-T3 line displayed moderately susceptible reaction with limited pycnidia development (DS 3-4). In contrast, the disomic line 4Hch-DS showed a resistant behaviour to STB displaying hypersensitive flecking (DS1) and necrotic lesions with no presence of pycnidia (DS 2). These results confirmed an apparent dosage effect on STB resistance as it was previously described in Agropyron–wheat derivates (Rillo et al., 1970) and agree with the presence of different Stb genes along the H. chilense chromosome 4Hch.
The similar pattern obtained by the cytogenetic analysis and molecular markers in 4HchS-T1 and 4HchS-T2 translocation lines suggested that the recombination events occurring in each of these translocation lines were extremely close to each other along the 4HchS chromosome. The fact of being the same recombination event is discarded as both 4HchS-T1 and 4HchS-T2 translocation lines were obtained from different genetic crosses between the original 4Hchdisomic line and the ph1 mutants.
All the durum wheat–H. chilense translocation lines obtained in this work retained the distal region of H. chilense chromosome 4HchL, which might contain some homology to bread wheat chromosome 4AL, where QTL7 and Stb7 and Stb12 resistance genes are located (Brown et al., 2015). All the durum wheat–H. chilense translocation lines developed in this work (4HchS-T1, 4HchS-T2, 4HchS-T3, and 4HchL-T4) displayed moderate resistance to STB compared with the control lines. However, some other gene(s) might also be located on the distal region of chromosome 4HchS, as the 4Hch H. chilense disomic substitution (4HchDS) line could be considered the most resistant one, showing a heterogeneous behaviour against the disease but with the lowest DS scores (1–2). In addition, no remarkable differences in the STB resistance were found between 4HchS-T1 and 4HchS-T2 translocation lines, indicating that putative resistance genes might not be located in the small segment of chromosome 4HchS in which these two durum wheat–H. chilense translocation lines might be different. Thus, that putative additional gene(s) should be included in the most distal 4HchS chromosome region. Consequently, our results suggest that H. chilense chromosome 4HchL might contain genes conferring resistance to STB on the distal 4HchL chromosome arm. These genes could be homoeologous to those Stb7 and Stb12 resistance genes or STB7 QTL described previously in the 4A bread wheat chromosome (Brown et al., 2015), but, interestingly enough, additional gene(s) conferring stronger resistance to STB might also be included in the distal region of the H. chilense chromosome 4HchS. To sum up, this present study sheds light on the interaction of Z. tritici and durum wheat, giving some clues to identify possible Stb genes in durum wheat. Therefore, the plant material obtained in this work constitutes a valuable germplasm to introduce resistance to STB in durum wheat.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors. Zymoseptoria tritici isolated is deposited in the public NBCI database under the MZ026796 accession number.
ZC: Writing – original draft, Investigation. M-CC: Investigation, Writing – review & editing. CM-R: Investigation, Writing – review & editing. JS: Investigation, Writing – review & editing. PP: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work has been supported by P20_00971 grant and the Qualifica Project QUAL21_023 IAS both from Consejería de Transformación Económica, Industria, Conocimiento y Universidades/Cofinanciación FEDER 80%—Programa Operativo FEDER de Andalucía 2014-2020.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bothmer, R., Jacobsen, N., Baden, C., Jørgensen, R., Linde-Laursen, I. (1995). An ecogeographical study of the genus Hordeum. 2nd edition (Rome: IPGRI). pp. 129.
Brading, P. A., Verstappen, E. C. P., Kema, G. H. J., Brown, J. K. M. (2002). A gene-for-gene relationship between wheat and mycosphaerella graminicola, the septoria tritici blotch pathogen. Phytopatology 92, 439–445. doi: 10.1094/PHYTO.2002.92.4.439
Brown, J. K. M., Chartrain, L., Lasserre-Zuber, P., Saintenac, C. (2015). Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 79, 33–41. doi: 10.1016/j.fgb.2015.04.017
Cabrera, A., Friebe, B., Jiang, J., Gill, B. S. (1995). Characterization of Hordeum Chilense chromosomes by C-banding and in situ hybridization using highly repeated DNA probes. Genome 38, 435–442. doi: 10.1139/g95-057
Calderón, M. C., Ramírez, M. C., Martín, A., Prieto, P. (2012). Development of Hordeum Chilense 4Hch introgression lines in durum wheat: A tool for breeders and complex trait analysis. Plant Breed. 131, 733–738. doi: 10.1111/j.1439-0523.2012.02010.x
Ceoloni, C., Signore, G., Ercoli, L., Donini, P. (1992). Locating the alien chromatin segment in common wheat- Aegilops longissima mildew resistant transfers. Hereditas 116, 239–245. doi: 10.1111/j.1601-5223.1992.tb00148.x
Faris, J. D., Xu, S. S., Cai, X., Friesen, T. L., Jin, Y. (2008). Molecular and cytogenetic characterization of a durum wheat- Aegilops speltoides chromosome translocation conferring resistance to stem rust. Chromosom. Res. 16, 1097–1105. doi: 10.1007/s10577-008-1261-3
Fones, H., Gurr, S. (2015). The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genet. Biol. 79, 3–7. doi: 10.1016/j.fgb.2015.04.004
Gallardo, M., Fereres, E. (1989). Drought resistance in tritordeum ( Hordeum Chilense x Triticum turgidum) in relation to wheat, barley and triticale. Invest. Agrar. Prod. Prot. Veg. 4, 361–375.
Hamblin, A. (1995). The concept of agricultural sustainability. Ed. Andrews, J. H. (Tommerup: Academic Press). I. C. B. T.-A. @ in P. P. doi: 10.1016/S0736-4539(06)80003-6
Kema, G. H. J., Verstappen, E. C. P., Waalwijk, C. (2000). Avirulence in the wheat Septoria tritici leaf blotch fungus Mycosphaerella graminicola is controlled by a single locus. Mol. Plant-Microbe Interact. 13, 1375–1379. doi: 10.1094/MPMI.2000.13.12.1375
Lukaszewski, A. J. (2000). Manipulation of the 1RS.1BL translocation in wheat by induced homoeologous recombination. Crop Sci. 40, 216–225. doi: 10.2135/cropsci2000.401216x
Mamrutha, H. M., Singh, R., Sharma, D., Venkatesh, K., Pandey, G. C., Kumar, R., et al. (2019). Physiological and Molecular Basis of Abiotic Stress Tolerance in Wheat BT - Genetic Enhancement of Crops for Tolerance to Abiotic Stress: Mechanisms and Approaches, Vol. I. Eds. Rajpal, V. R., Sehgal, D., Kumar, A., Raina, S. N. (Cham: Springer International Publishing), 99–124. doi: 10.1007/978-3-319-91956-0_5
Martin, A., Martínez-Araque, C., Rubiales, D., Ballesteros, J. (1996). “Tritordeum: Triticale´s New Brother Cereal,” in Triticale: Today and Tomorrow. Developments in Plant Breeding, vol. 5 . Eds. Guedes-Pinto, H., Darvey, N., Carnide, V. P. (Springer, Dordrecht), 57–72.
Martin, A., Sanchez-Mongelaguna, E. (1982). Cytology and morphology of the amphiploid Hordeum Chilense × Triticum turgidum conv. Durum. Euphytica 31, 261–267. doi: 10.1007/BF00028329
McCartney, C. A., Brûlé-Babel, A. L., Lamari, L. (2002). Inheritance of race-specific resistance to Mycosphaerella graminicola in wheat. Phytopathology 92, 138–144. doi: 10.1094/PHYTO.2002.92.2.138
Mondal, S., Sallam, A., Sehgal, D., Sukumaran, S., Farhad, M., Navaneetha Krishnan, J., et al. (2021). “Advances in Breeding for Abiotic Stress Tolerance in Wheat BT,” in Genomic Designing for Abiotic Stress Resistant Cereal Crops. Ed. Kole, C. (Springer International Publishing, Cham), 71–103. doi: 10.1007/978-3-030-75875-2_2
Murray, M. G., Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nalam, V. J., Vales, M. I., Watson, C. J. W., Kianian, S. F., Riera-Lizarazu, O. (2006). Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat ( Triticum turgidum L.). Theor. Appl. Genet. 112, 373–381. doi: 10.1007/s00122-005-0140-y
Othmeni, M., Grewal, S., Hubbart-Edwards, S., Yang, C., Scholefield, D., Ashling, S., et al. (2019). The use of pentaploid crosses for the introgression of amblyopyrum muticum and D-genome chromosome segments into durum wheat. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01110
Pedersen, C., Zimny, J., Becker, D., Jähne-Gärtner, A., Lörz, H. (1997). Localization of introduced genes on the chromosomes of transgenic barley, wheat and triticale by fluorescence in situ hybridization. Theor. Appl. Genet. 94, 749–757. doi: 10.1007/s001220050474
Pena, R. J., Amaya, A., Rajaram, S., Mujeeb-Kazi, A. (1990). Variation in quality characteristics associated with some spring 1B/1R translocation wheats. J. Cereal Sci. 12, 105–112. doi: 10.1016/S0733-5210(09)80092-1
Pérez-Méndez, N., Miguel-Rojas, C., Jimenez-Berni, J. A., Gomez-Candon, D., Pérez-De-luque, A., Fereres, E., et al. (2022). Plant breeding and management strategies to minimize the impact of water scarcity and biotic stress in cereal crops under mediterranean conditions. Agronomy 12 (1), 75. doi: 10.3390/agronomy12010075
Porras, R., Pérez-De-luque, A., Sillero, J. C., Miguel-Rojas, C. (2021). Behavior of spanish durum wheat genotypes against Zymoseptoria tritici : Resistance and susceptibility. Spanish J. Agric. Res. 19 (3), e1002. doi: 10.5424/sjar/2021193-17953
Prieto, P., Martín, A., Cabrera, A. (2004). Chromosomal distribution of telomeric and telomeric-associated sequences in Hordeum Chilense by in situ hybridization. Hereditas 141, 122–127. doi: 10.1111/j.1601-5223.2004.01825.x
Quaedvlieg, W., Kema, G. H. J., Groenewald, J. Z., Verkley, G. J. M., Seifbarghi, S., Razavi, M., et al. (2011). Zymoseptoria gen. nov.: A new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia Mol. Phyl. Evol. Fungi 26, 57–69. doi: 10.3767/003158511X571841
Rey, M.-D., Calderón, M. C., Prieto, P. (2015b). The use of the ph1b mutant to induce recombination between the chromosomes of wheat and barley. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00160
Rey, M.-D., Calderón, M.-C., Rodrigo, M. J., Zacarías, L., Alós, E., Prieto, P. (2015a). Novel bread wheat lines enriched in carotenoids carrying Hordeum Chilense chromosome arms in the ph1b background. PloS One 10 (8), e0134598. doi: 10.1371/journal.pone.0134598
Rillo, A. O., Caldwell, R. M., Glover, D. V. (1970). Cytogenetics of resistance to wheat leaf blotch (Septoria tritici) in backcross derivatives of an agrotricum line. Crop Sci. 10, 223–227. doi: 10.2135/cropsci1970.0011183X001000030004x
Rubiales, D., Moral, A., Martín, A. (2001). Chromosome location of resistance to septoria leaf blotch and common bunt in wheat-barley addition lines. Euphytica 122, 369–372. doi: 10.1023/A:1012952819255
Rubiales, D., Reader, S. M., Martín, A. (2000). Chromosomal location of resistance to Septoria tritici in Hordeum Chilense determined by the study of chromosomal addition and substitution lines in “Chinese Spring” wheat. Euphytica 115, 221–224. doi: 10.1023/A:1004097830103
Said, M., Cabrera, A. (2009). A physical map of chromosome 4Hch from Hordeum Chilense containing SSR, STS and EST-SSR molecular markers. Euphytica 167, 253–259. doi: 10.1007/s10681-009-9895-6
Sakata, M., Nasuda, S., Endo, T. R. (2010). Dissection of barley chromosome 4H in common wheat by the gametocidal system and cytological mapping of chromosome 4H with EST markers. Genes Genet. Syst. 85, 19–29. doi: 10.1266/ggs.85.19
Sears, E. R., Okamoto, M. (1958). “Intergenomic chromosome relationships in hexaploid wheat,” in Paper Presented at the Proceedings of the 10th International Congress of Genetics. (Montreal) 258–259.
Shubing, L., Honggang, W. (2005). Characterization of a wheat- Thinopyron intermedium substitution line with resistance to powdery mildew. Euphytica 143, 229–233. doi: 10.1007/s10681-005-3862-7
Somai-Jemmali, L., Siah, A., Harbaoui, K., Fergaoui, S., Randoux, B., Magnin-Robert, M., et al. (2017). Correlation of fungal penetration, CWDE activities and defense-related genes with resistance of durum wheat cultivars to Zymoseptoria tritici. Physiol. Mol. Plant Pathol. 100, 117–125. doi: 10.1016/j.pmpp.2017.08.003
Stewart, E. L., McDonald, B. A. (2014). Measuring quantitative virulence in the wheat pathogen zymoseptoria tritici using high-throughput automated image analysis. Phytopathology 104, 985–992. doi: 10.1094/PHYTO-11-13-0328-R
Wang, B., Ding, Z., Liu, W., Pan, J., Li, C., Ge, S., et al. (2009). Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol. Biol. 9, 1–13. doi: 10.1186/1471-2148-9-250
Keywords: Septoria leaf blotch, plant breeding, Ph1 locus, foliar disease, chromosome manipulation
Citation: Cifuentes Z, Calderón M-C, Miguel-Rojas C, Sillero JC and Prieto P (2024) Development and characterisation of novel durum wheat–H. chilense 4Hch chromosome lines as a source for resistance to Septoria tritici blotch. Front. Plant Sci. 15:1393796. doi: 10.3389/fpls.2024.1393796
Received: 29 February 2024; Accepted: 29 June 2024;
Published: 23 July 2024.
Edited by:
Ignacio Romagosa, Agrotecnio Center, SpainReviewed by:
Paula Martins-Lopes, University of Trás-os-Montes and Alto Douro, PortugalCopyright © 2024 Cifuentes, Calderón, Miguel-Rojas, Sillero and Prieto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pilar Prieto, cGlsYXIucHJpZXRvQGlhcy5jc2ljLmVz
†These authors share first authorship
‡ORCID: Pilar Prieto, orcid.org/0000-0002-8160-808X
Josefina C. Sillero, orcid.org/0000-0002-5878-9054
Cristina Miguel-Rojas, orcid.org/0000-0002-7245-9342
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.