- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Chengdu, Sichuan, China
- 2Maize Research Institute, Sichuan Agricultural University, Chengdu, Sichuan, China
- 3Key Laboratory of Biology and Genetic Improvement of Maize in Southwest Region, Ministry of Agriculture, Chengdu, Sichuan, China
- 4National Key Laboratory of Wheat and Maize Crop Science, College of Agronomy, Henen Agricultural University, Zhengzhou, China
- 5Maize Research Institute, Mianyang Academy of Agricultural Sciences, Mianyang, Sichuan, China
- 6Sichuan Academy of Agricultural Sciences, Biotechnology and Nuclear Technology Research Institute, Chengdu, Sichuan, China
- 7College of Resources, Sichuan Agricultural University, Chengdu, Sichuan, China
- 8Institute of Biotechnology, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
Phosphorus (P) is a crucial macronutrient for plant growth and development, and low-Pi stress poses a significant limitation to maize production. While the role of the SPX domain in encoding proteins involved in phosphate (Pi) homeostasis and signaling transduction has been extensively studied in other model plants, the molecular and functional characteristics of the SPX gene family members in maize remain largely unexplored. In this study, we identified six SPX members, and the phylogenetic analysis of ZmSPXs revealed a close relationship with SPX genes in rice. The promoter regions of ZmSPXs were abundant in biotic and abiotic stress-related elements, particularly associated with various hormone signaling pathways, indicating potential intersections between Pi signaling and hormone signaling pathways. Additionally, ZmSPXs displayed tissue-specific expression patterns, with significant and differential induction in anthers and roots, and were localized to the nucleus and cytoplasm. The interaction between ZmSPXs and ZmPHRs was established via yeast two-hybrid assays. Furthermore, overexpression of ZmSPX1 enhanced root sensitivity to Pi deficiency and high-Pi conditions in Arabidopsis thaliana. Phenotypic identification of the maize transgenic lines demonstrated the negative regulatory effect on the P concentration of stems and leaves as well as yield. Notably, polymorphic sites including 34 single-nucleotide polymorphisms (SNPs) and seven insertions/deletions (InDels) in ZmSPX1 were significantly associated with 16 traits of low-Pi tolerance index. Furthermore, significant sites were classified into five haplotypes, and haplotype5 can enhance biomass production by promoting root development. Taken together, our results suggested that ZmSPX family members possibly play a pivotal role in Pi stress signaling in plants by interacting with ZmPHRs. Significantly, ZmSPX1 was involved in the Pi-deficiency response verified in transgenic Arabidopsis and can affect the Pi concentration of maize tissues and yield. This work lays the groundwork for deeper exploration of the maize SPX family and could inform the development of maize varieties with improved Pi efficiency.
1 Introduction
Maize (Zea mays L.) stands as a pivotal crop globally, not only as a source of food and high-quality feed but also for industrial applications (Feng et al., 2022; Jiao et al., 2023). P is an essential macronutrient for plant growth and development and plays a vital role either directly or indirectly in many physiological and biochemical processes, such as photosynthesis, respiration, signal transduction, and metabolic processes (Luo et al., 2019). Plants primarily acquire P in the inorganic form of Pi from the soil to support their growth and yield (Kumar et al., 2019). The low availability of Pi in soils is a significant limiting factor for plant growth and yield, posing a significant challenge (Balyan et al., 2016). Consequently, the growing need for increased food production and higher crop yields is expected to lead to a rise in the demand for Pi inputs in cropland. The global application of Pi fertilizer on croplands has increased several times from 1961 to 2013 and already surpassed the estimated planetary boundary (Zou et al., 2022). Excessive fertilizer application has led to a myriad of environmental and ecological issues, including water eutrophication.
In response to low-Pi stress, plants have developed a series of adaptive mechanisms, primarily manifested through changes in root morphological structure, physiological and biochemical regulation, and the expression of Pi starvation-inducible genes. Specifically, under low-Pi stress, alterations in root morphology and configuration were characterized by the inhibition of primary roots, accompanied by an increase in the length and quantity of lateral roots and root hairs (Raghothama, 1999; Lynch, 2011). Additionally, the root radius decreases, resulting in a higher root–shoot ratio and a shallower root system. These morphological changes facilitate plants in expanding the contact area between roots and the superficial soil (Raghothama, 1999; Lynch, 2011; Luo et al., 2023). Furthermore, low-Pi stress triggers modifications in plant enzyme system activity, further contributing to the plant’s adaptation to Pi scarcity. For example, Pi deficiency inhibits the activity of ATP synthase, which subsequently impacts ATP and NADPH production, thereby reducing the photosynthetic rate, carbon fixation, and overall plant growth (Carstensen et al., 2018; Garcia et al., 2023; Iqbal et al., 2023; Saengwilai et al., 2023). Enhancing the activities of sulfur lipid and glycolipid synthases, as well as phospholipid hydrolases, contributes to altering the composition of the lipid membrane. Moreover, low-Pi stress can elevate the activity of defense-related enzymes, enabling plants to better adapt to the low-Pi environment (Shimojima, 2011; Liang et al., 2014; Chen et al., 2015). Additionally, the increase in root exudates represents another strategy for plants to cope with low-Pi stress. This includes the secretion of protons, organic acids, and acid phosphatase, which serve to acidify the soil surrounding the roots and release inorganic Pi for plant uptake (Lynch et al., 2005; Shen et al., 2005; Zhang et al., 2010). When encountering Pi deficiency, plants also maintain Pi homeostasis through a series of molecular reactions. For instance, the phosphate starvation response (PSR) pathway involves a MYB transcription factor known as PHR1, which serves as a key regulator in PSR. It governs transcription and takes part in physiological and biochemical adaptations (Zhang et al., 2014). PHR1 is capable of directly binding to the cis-elements P1BS (PHR binding sequence, GNATATNC), which were found in the promoter regions of several PSR genes, including Phosphate 1 (PHO1), phosphate transporter traffic facilitator 1 (PHF1), phosphate transporters (PTs), induced by phosphate starvation (IPS1), miRNA399, and miRNA827 (Rubio et al., 2001; Puga et al., 2017). As Pi sensors, SPX proteins can interact with AtPHR1 or OsPHR2, and under normal conditions, they inhibit their transcriptional activity. This interaction between SPX proteins and PHR transcription factors serves to prevent the toxicity resulting from excessive Pi accumulation (Wang et al., 2014). Notably, the SPX proteins themselves do not directly sense Pi but instead sense soluble inositol polyphosphates (InsPs) with a high affinity (Wild et al., 2016). Recently, the crystal structure of the SPX domain revealed the basic surface for InsP6, and biochemical studies have shown that InsP7 stimulated the interaction between OsSPX4 and OsPHR2 with higher binding affinity than InsP6 (Jung et al., 2018). Among these, InsP8 acts as an intracellular Pi signaling substance, regulating Pi balance by modulating the interaction between AtSPX1 and AtPHR1 (Dong et al., 2019). Consequently, SPX genes play a crucial role in Pi signaling pathways and homeostasis in plants (Li et al., 2021).
The SPX proteins, namely, SYG1 (suppressor of yeast gpa1), Pho81 (CDK inhibitor in the yeast PHO pathway), and XPR1 (xenotropic and polytropic retrovirus receptor), can be classified into four subfamilies based on the presence of structural characteristics: class 1 only contained SPX domain, and the other three have (SPX-MFS, SPX-EXS, and SPX-RING) domains (Chiou and Lin, 2011; Yang et al., 2017; Yue et al., 2017). Recently, two additional classes of SPX proteins, namely, SPX-SLC and SPX-VTC, had been characterized in algae, and they were involved in Pi synthesis and transportation in vacuoles. However, these classes appear to have been lost throughout the evolution of plants, with the type of Pi storage changing from polyphosphates in algae to Pi in the later-diverging streptophytes (Wang et al., 2021a). It seems that the SPX domain has some extra domains that may have been lost during the evolution of SPX proteins and have not been comprehensively identified yet (Kopriva and Chu, 2018; Nezamivand-Chegini et al., 2021). SPX gene family members have been studied for their significant roles in Pi signaling and homeostasis in various plant species, including four SPXs in Arabidopsis thaliana and six SPXs in rice (Secco et al., 2012). In A. thaliana, AtSPX1 and AtSPX3 played a positive role in plant adaptation to Pi starvation. Additionally, AtSPX1 and AtSPX3 demonstrated redundancy, and AtSPX3 may negatively regulate the expression of AtSPX1 (Duan et al., 2008). Moreover, under normal-Pi conditions, AtSPX4 acted as a negative regulator of PSR gene expression in shoots, influencing both PHR1-dependent and PHR1-independent Pi starvation responses. Disruption of the AtSPX4 function led to excessive Pi accumulation in the shoots (Osorio et al., 2019). In rice, OsSPX3/5 negatively regulated the transport of Pi from roots to shoots. OsSPX4 interacted with OsPHR2 in the cytoplasm, inhibiting Pi signaling. Meanwhile, OsSPX6 was upregulated under low-Pi stress, leading to the activation of OsPHR2 (Lv et al., 2014; Zhong et al., 2018). Furthermore, OsSPX4 was involved in the regulation pathway of nitrogen and Pi signals, and ubiquitinated OsSPX4 facilitated the activation of Pi signaling by nitrate, coordinating the utilization of nitrogen and Pi (Hu et al., 2019). Moreover, SPX genes have been reported via bioinformatics analysis in diverse species (Yao et al., 2014b; Du et al., 2017; Kumar et al., 2019; Xiao et al., 2021) because of their possible involvement in a number of physiological and molecular processes. However, despite these advancements, the specific functions of these SPX gene family members have remained unknown in maize. In our previous study, SPX proteins carrying only the SPX domain were predicted under low-Pi conditions in maize (Nie et al., 2021). Therefore, in the present study, we conducted a bioinformatics analysis to investigate the relationship of SPX proteins containing only the SPX domain across various species. Additionally, we explored the expression patterns of these SPX genes in maize, examined their interaction with PHRs, and elucidated the function of ZmSPX1 through transgenic experiments and candidate gene association analysis in maize.
Taken together, our findings suggested the involvement of ZmSPXs in transcription regulations during Pi starvation and found that ZmSPX1 was involved in response to low-Pi conditions and provide a solid foundation for a deeper understanding of the molecular mechanism of genes and improving P use efficiency (PUE) in maize and will promote future studies on this important gene family in plants.
2 Materials and methods
2.1 Plant material and growth conditions
Using the CRISPR/Cas9 (Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) system, the ZmSPX1 gene was knocked out in the maize inbred line KN5585 by the Agrobacterium tumefaciens-mediated transformation method. The mutation site was detected using ZmSPX1-KO-F and R primers (Supplementary Figure S1; Supplementary Table S1). The construction method for maize overexpressing lines utilizes the homologous recombination method to introduce the coding sequence (CDS) of the ZmSPX1 gene into the vector pCAMBIA3301. Subsequently, A. tumefaciens-mediated transformation of the maize recipient line KN5585 was employed to generate transgenic plants. The results for the detection of overexpression and the specific primers used for detection are provided in Supplementary Figure S2 and Supplementary Table S1, respectively. Seeds of 178 (low-Pi tolerant maize inbred line), A. thaliana (Columbia), and tobacco (Nicotiana benthamiana) were provided by the Maize Research Institute of Sichuan Agricultural University. A relatively low-Pi field site in Wenjiang farm (WJ, Chengdu, Sichuan Province, plain region, available Pi 22.8 mg/kg) of Sichuan Agricultural University was selected for the experiments on low-Pi treatment of maize inbred line 178. Each plot received two levels of Pi treatments (low- and normal-Pi conditions), with each treatment replicated thrice. The plots measured 3 m in length with 0.8 m between rows. Prior to reaching the five-leaf stage, plant thinning reduced the number to 14 plants per plot (58,000 plants/ha). In the normal-Pi treatment plots, the following fertilizers (Stanley Agriculture Group Co., Ltd.) were applied: 150 kg of urea, 900 kg of calcium superphosphate, and 350 kg of potassium chloride per hectare before planting; 130 kg of urea per hectare at the six-leaf stage; and 210 kg of urea per hectare as a side dressing prior to tasseling. The low-Pi treatment plots received the same fertilizer combination as the normal-Pi plots, with the exception of the calcium superphosphate. The seeds of A. thaliana were planted in nutrient soil and soil sterilized at 121°C for 20 min and cooled for 30 min. Then, seeds were cultured in nutrient soil at 23°C for 12 h of light and darkness until the A. thaliana mature.
2.2 Bioinformatics identification of ZmSPXs in maize
Based on our previous research (Nie et al., 2021), ZmSPX homologous genes were identified with high similarity sequence: ZmSPX1 (GRMZM2G035579), ZmSPX2 (GRMZM2G171423), ZmSPX3 (GRMZM2G024705), ZmSPX4 (GRMZ2G122108), ZmSPX5 (GRMZM5G828488), ZmSPX6 (GRMZM2G065989), and ZmSPX7 (GRMZM2G083655). The ZmSPX family members were identified in maize, and the reliability of the ZmSPX CDS was predicted using BioXM 2.6 software. The conserved domain of ZmSPXs was verified using the National Center for Biotechnology Information (NCBI) search database (Marchler-Bauer et al., 2015). Amino acid sequences query homologous amino acid sequences of SPXs in maize, wheat, soybean, rape, rice, and Arabidopsis using the BLASTP program from the NCBI (http://www.ncbi.nlm.nih.gov); phylogenetic relationships were aligned using Mega (version 11.0.13) (Kumar et al., 1994). The gene IDs of the SPX gene family in other species were acquired from previous research studies (Secco et al., 2012; Yao et al., 2014b; Du et al., 2017; Kumar et al., 2019). Finally, ZmSPXs were studied for cloning and functional analyses based on protein comparison and transcriptome sequencing, respectively. The promoter region (2,000 bp) of ZmSPXs was utilized to predict the functional region of these genes using the BDGP online database (http://www.fruitfly.org/seq_tools/promoter.html) (Supplementary Table S2), and the cis-acting elements within the promoter were predicted using the plantCARE web server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
2.3 Cloning of ZmSPX family members in maize
Total RNA was extracted from the leaves and roots of 178 inbred lines following the manufacturer’s protocol of Thermo Fisher Scientific, Life Technologies (TRIzol Reagent®; the protocol can be found at https://www.thermofisher.com). The cDNA reverse transcription was performed using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Bio Inc., Otsu, Japan). The CDS of ZmSPXs was then amplified using the cDNA as a template. The polymerase chain reaction (PCR) primers were designed based on the CDS of the B73 reference, targeting the specific sequences of the ZmSPX family members using Primer Premier 5 software. The forward and reverse primers are listed in Supplementary Table S3. The PCR was performed in a 25-µL volume containing phanta max buffer 12.5 μL (Phanta Max Super-Fidelity DNA Polymerase, Nanjing Vazyme Biotech Co., Ltd., Nanjing, China), phanta max super fidelity 0.5 μL, dNTP 4 μL, cDNA 1 μL, 1 μL of each forward and reverse primer, and 7 μL ddH2O. PCR was programmed for 3 min at 95°C followed by 30 cycles of 95°C for 15 s, annealing for 15 s, 72°C for 45 s, extension for 5 min at 72°C, and final 12°C for preservation. Then, PCR products were separated using 1% agarose gel, purified using a DNA purification kit, and cloned into pEASY®-Blunt zero Cloning Vector (TransGen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s protocol. Finally, positive clones were selected for sequencing by TsingKe Biological Technology Co., Ltd. (Beijing, China). The coding sequence of ZmSPXs was compared with that of the B73 reference inbred line using DNAMAN v6.0 software.
2.4 Expression pattern of ZmSPX family members under Pi deficiency in different tissues
Different maize tissues were harvested in the silking stage from normal-Pi and low-Pi treatments. Total RNA was extracted from different maize tissues of 178 inbred lines including leaf, stem, anther, cornsilk, root, ear, and ear bract according to the TRIzol Reagent® (Invitrogen, Carlsbad, CA, USA) manufacturer’s instructions, and reverse transcription was performed using PrimeScript™ II 1st Strand cDNA synthesis kit (Takara Bio Inc., Otsu, Japan). Relative quantitative results were calculated by normalization to the reference gene (GAPDH). At least three independent experiments were performed, and each experiment was performed in technical triplicate. All primers used in the qRT-PCR assay were designed in BEACON DESIGNER 7 and are listed in Supplementary Table S4. qRT-PCR data were analyzed using the 2−ΔΔCT method.
2.5 Subcellular localization of ZmSPX protein
The full-length CDS region was cloned into plant expression vector pCAMBIA2300 for the ZmSPXs. The homologous recombination primers with enzyme cutting sites (SmaI and XbaI) were designed by CEDesignV1.04 software and are shown in Supplementary Table S5. The subcellular localization was assayed in tobacco leaves according to the previously reported transit transformation method (Zhou et al., 2018; Sahito et al., 2020a, b). The transformed tobacco epidermal cells were observed using the A1R-si laser confocal microscope (LSCM, Nikon, Tokyo, Japan) to visualize the green fluorescent protein (GFP) fluorescent signals. The experiment was repeated at least three times to ensure consistency of results.
2.6 Yeast two-hybrid assays of ZmSPXs and ZmPHRs
The full-length CDSs of ZmSPX family members ZmPHR1 (GRMZM2G006477) and ZmPHR2 (GRMZM2G162409) were amplified using gene-specific primers from the cDNA samples. The primers were designed using CE design v1.04 and Primer 5.0 to incorporate (EcorR I, BamH I, and Nde I) restriction sites, as specified in Supplementary Table S6. Phanta Max high-fidelity DNA polymerase (Nanjing Vazyme Biotech Co., Ltd.) was used to amplify the targeted fragments, and then the amplified fragments were inserted into bait (pGBKT7) and prey (pGADT7) vectors. All possible combinations were co-transferred into the Y2HGold yeast-competent cell. pGBKT7-53 and pGBKT7-lam were used as positive and negative controls, respectively. To exclude the possible autoactivation of ZmSPX members, a control experiment was carried out by transformation of loaded bait and prey plasmids with empty prey and bait plasmids, respectively. After screening on solid DDO (−Leu/−Trp) medium for 2–4 days at 28°C, selected monoclonals were inoculated in liquid DDO (−Leu/−Trp) medium until OD600 = 0.3–0.5. These cultures were inoculated on QDO (SD/−Ade/−His/−Trp/−Leu/X-α-Gal) medium after 10-fold dilution. Results were observed after 3–5 days of incubation at 30°C.
2.7 Overexpression of ZmSPX1 in A. thaliana
Cloning of the full open reading frame (ORF) of ZmSPX1 into the vector pCAMBIA3300-35s-PROII MCS-bar and subsequent transformation into wild-type (WT) Arabidopsis produced several transformants through the floral dip method previously described (Clough and Bent, 1998). The seeds of the T0 generation were harvested and sown in the soil to select positive transgenic seedlings. Two-week-old seedlings of T1 plants were screened by spraying with Basta (1/1,000) solution. After the transgenic plants were harvested, DNA was extracted, and PCR was performed to confirm the positive ZmSPX1 transgenic lines.
2.8 Phenotypic characterization of ZmSPX1 transgenic lines under low-Pi stress
The seeds of both WT and ZmSPX1 transgenic lines were sterilized with 75% (v/v) ethanol for 60–90 s and 2% sodium hypochlorite for 8–15 min, followed by several rinses with distilled water. Fifty seeds of both WT and transgenic lines were cultured on 1/2 normal MS agar medium, initially kept at 4°C for 2 days, and subsequently transferred to a greenhouse with a light cycle of 16 hours at 22°C and a dark cycle of 8 hours at 18°C for 7 days. After 7 days, seedlings were transferred to 1/8 normal MS agar medium and subjected to different Pi concentrations: 0 mmol/L (control), low Pi (0.1 mmol/L), normal Pi (1 mmol/L), and high Pi (10 mmol/L). The seedlings were kept in a controlled environment for 2 weeks. Phenotypic observations under different Pi concentrations were recorded after 2 weeks, and their roots were scanned using a WinRHIZO root-scanning method. All Arabidopsis seedlings were dried and digested for the detection of P and nitrogen concentration through a chemical continuous flow analyzer.
2.9 Sequencing and association analysis of ZmSPX1 in maize association population
A total of 211 out of 360 inbred lines were screened from the current Southwest China breeding program (Zhang et al., 2016). These lines were used for association analysis of ZmSPX1 in maize. The genomic DNA was extracted from leaves at the seedling stage according to the cetyltrimethylammonium bromide (CTAB) method (Porebski et al., 1997). The genomic sequence of ZmSPX1 from the B73 inbred line was utilized as the reference sequence and obtained from the Maize Genomic Database (http://www.maizegdb.org). The targeted fragment was amplified using high-fidelity phanta max enzyme polymerase (Vazyme Biotech Co., Ltd.). Amplified fragments were sequenced by TsingKe Biological Technology Co., Ltd. Then, the targeted sequence of ZmSPX1 was compared with the reference genomic sequence of the B73 using DNAMAN v6.0 software, and the sequences of 211 maize inbred lines were trimmed neatly using Bio-Edit 7.1 software (Strable and Scanlon, 2009). Single-nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) were identified in all tested inbred lines with a <0.05 minor allele frequency (MAF), and linkage disequilibrium (LD) between two polymorphic sites was generated using Tassel software v2.1 (Bradbury et al., 2007). Association analysis was carried out between SNPs and InDels and 22 phenotypic traits using Tassel software (Bradbury et al., 2007; Luo et al., 2019). The standard mixed linear model (MLM) including a population structure (Q) and kinship matrix (K) was chosen to detect the significant association of SNPs and InDels as described previously (Zhang et al., 2016; Luo et al., 2019). The association of sites was considered significant at p < 0.05, and the calculated p-values were converted into −log10(p value).
3 Results
3.1 Evolutionary tree analysis of SPXs in plants
The SPX homologous genes ZmSPX1 (GRMZM2G03557), ZmSPX2 (GRMZM2G171423), ZmSPX3 (GRMZM2G024705), ZmSPX4 (GRMZM2G122108), ZmSPX5 (GRMZM5G828488), ZmSPX6 (GRMZM2G065989), and ZmSPX7 (GRMZM2G0836555) were predicted in our previous study, and genes were located on different chromosomes (Supplementary Table S7). However, in the fourth edition of the maize genome, ZmSPX1 and ZmSPX7 were merged into a single gene. Additionally, preliminary transcriptome results indicated that ZmSPX7 (GRMZM2G0836555) did not respond to low-Pi stress (Supplementary Table S7). Hence, in subsequent experiments, the ZmSPX1–6 in the third version of the maize genome were exclusively analyzed. Furthermore, ZmSPX1, ZmSPX2, ZmSPX3, ZmSPX4, ZmSPX5, and ZmSPX6 were amplified in maize according to the targeted CDS by PCR. Specifically, the CDS length of ZmSPX1 was 354 bp, encoding a protein comprising 117 amino acids. Correspondingly, ZmSPX2, ZmSPX3, ZmSPX4, ZmSPX5, and ZmSPX6 have CDS lengths of 846 bp, 687 bp, 996 bp, 765 bp, and 759 bp, encoding proteins of 281, 228, 331, 254, and 252 amino acids, respectively. Simultaneously, SPX genes of wheat, soybean, rape, rice, and Arabidopsis were also searched for in the database (Secco et al., 2012; Yao et al., 2014b; Du et al., 2017; Kumar et al., 2019). Among them, Arabidopsis has four SPX genes (AtSPX1–4), rice has six SPX genes (OsSPX1–6), soybean has nine SPX genes (GmSPX1–9), and rape and wheat have 11 and 15 SPX genes, respectively (Supplementary Table S8). To deepen our understanding of the evolutionary relationships between these SPX proteins, a phylogenetic tree based on the SPX proteins in these plants was constructed (Figure 1). The six SPX genes of maize were distributed across different clades. Notably, ZmSPX5 and ZmSPX6 displayed the closest evolutionary relationship, clustering within the same clade but occupying distinct branch points (Figure 1). Interestingly, the closest evolutionary relationship existed between the SPX genes in maize and rice. Specifically, ZmSPX5 and OsSPX5, ZmSPX3 and OsSPX3, and ZmSPX1 and OsSPX1 were all located at the same branch point (Figure 1). Furthermore, while ZmSPX4 and OsSPX4 were positioned at different branch points on the evolutionary tree, they belonged to the same clade and were situated at adjacent branch points (Figure 1). Remarkably, ZmSPX2 showed a close evolutionary relationship with BnaA3SPX3 (Figure 1).
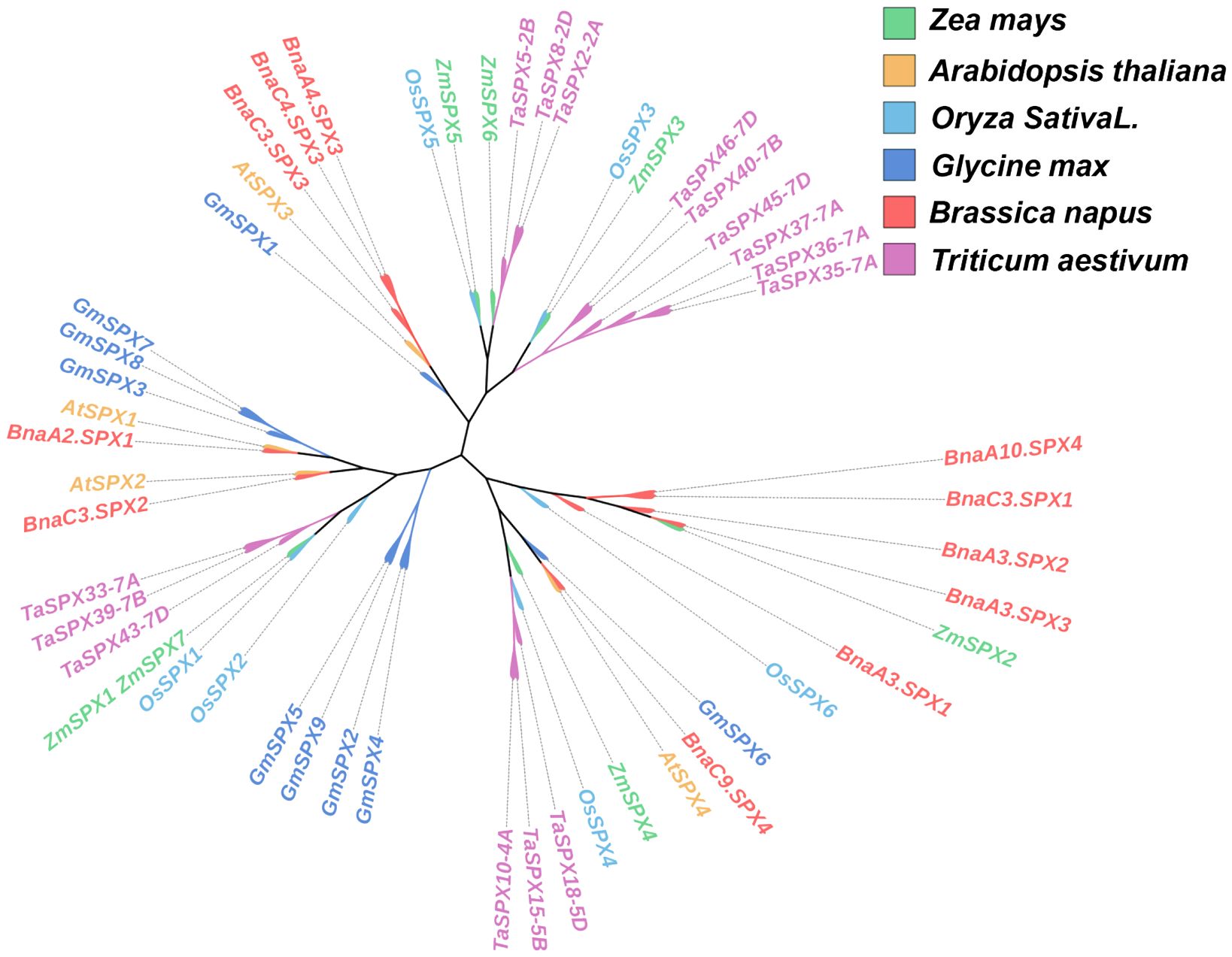
Figure 1 Phylogenetic tree of SPXs in plants. The phylogenetic tree was created in the Mega 11 program. In plants, proteins harboring the SPX domain are classified into four families based on the presence of additional domains in their structure, namely, the SPX, SPX-EXS, SPX-MFS, and SPX-RING families. The SPX proteins of this phylogenetic tree carry only the SPX domain.
3.2 cis-Acting element analysis of promoter region of ZmSPXs
Generally, the results showed that ZmSPX gene family members were associated with abscisic acid response, light reaction, methyl jasmonate (MeJA) reaction, low-temperature response, auxin response, and regulation of zein metabolism cis-acting elements (Supplementary Table S9). Specifically, ZmSPX gene family members also possess unique cis-acting regulatory elements. For instance, ZmSPX1 contains response elements for gibberellin and salicylic acid. The ZmSPX2 gene responds to light and is also involved in defense and stress responses. ZmSPX3 is involved in the light responsiveness. ZmSPX4 contains a conserved DNA module (ATCT-motif) involved in photoreaction. ZmSPX5 is involved in anaerobic induction. ZmSPX6 contains DNA-binding protein binding sites and responds to light. It also contains MYB binding sites involved in the regulation of flavonoid biosynthesis genes. Previous studies have demonstrated that SPXs can interact with the MYB domain and the CC domain of PHRs (Jia et al., 2023). Additionally, our results also indicated that ZmSPX gene family members also contain MYB binding sites or MYB recognition sites (Supplementary Table S9). All of these findings suggested that ZmSPXs may play a role in the regulation of ZmPHRs in association with certain hormones.
3.3 Expression pattern of ZmSPXs under low- and normal-Pi conditions
Expression patterns of ZmSPXs were analyzed under low- and normal-Pi treatments in different tissues of 178 maize inbred lines, including ear leaf, stem, anther, cornsilk, root, ear, and ear bract (Figure 2). Notably, the ZmSPXs exhibited higher expression levels in anthers compared to other tissues but were significantly inhibited under low-Pi conditions. Meanwhile, ZmSPX1–5 were induced by low-Pi stress in roots (Figure 2). Collectively, these results showed that the ZmSPX family members were involved in response to the low-Pi stress and play a specific function in different tissues of maize.
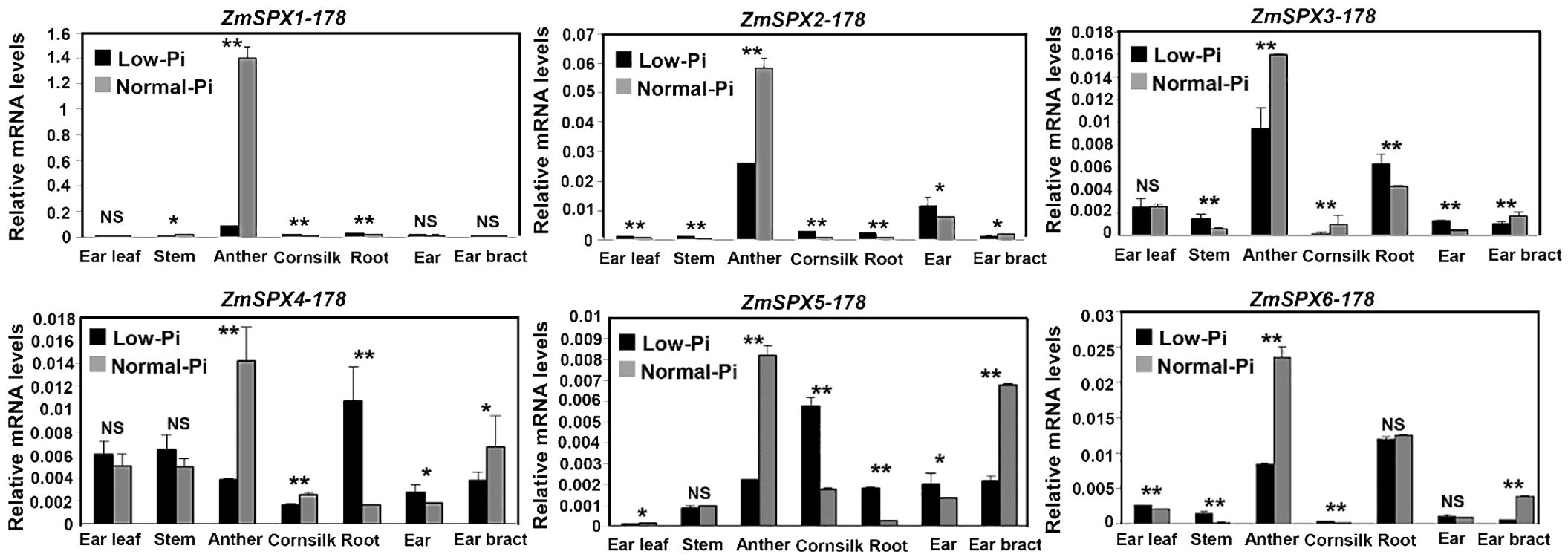
Figure 2 Expression pattern of ZmSPX family members in different tissues of 178 inbred lines under low- and normal-Pi conditions. Significant differences are indicated by Student’s t-test: *p < 0.05, **p < 0.01. NS, No significance.
3.4 Subcellular localization of ZmSPX proteins
ZmSPX1, ZmSPX3, ZmSPX4, ZmSPX5, and ZmSPX6 were found to be localized in the nucleus and cytoplasm (Supplementary Figure S3). ZmSPX2 was mainly localized in the nucleus (Supplementary Figure S3). In a previous study (Xiao et al., 2021), subcellular localization studies of ZmSPXs in maize protoplasts revealed that ZmSPX1 (equivalent to ZmSPX5 in this study) and ZmSPX3 (equivalent to ZmSPX4 in this study) were localized in both the nucleus and cytoplasm. ZmSPX5 (equivalent to ZmSPX2 in this study) was exclusively localized in the nucleus. Moreover, ZmSPX4 (equivalent to ZmSPX6 in this study) and ZmSPX5 (equivalent to ZmSPX2 in this study) were detected in both the nucleus and cytoplasm, while ZmSPX6 (equivalent to ZmSPX1 in this study) was found in the chloroplast. The results showed partial consistency in the subcellular localization between tobacco tissue and maize protoplasts. However, some discrepancies were observed, presumably attributed to variations in cell types across different species.
3.5 Interaction between ZmSPXs and ZmPHRs in maize
In this study, a yeast two-hybrid assay was conducted to identify the interaction between ZmSPX and ZmPHR proteins. Initially, the self-activation of ZmSPX and ZmPHR genes was assessed. Our findings showed that ZmPHR1 and ZmPHR2 demonstrated normal growth on SD/−Ade/−His/−Trp-deficient medium, while the ZmSPXs transformed with yeast cells did not grow on the selective medium (Figure 3A), indicating that the ZmSPXs had no self-activation effect. Therefore, SPX protein was chosen as bait and PHR protein as prey to verify the interaction. Furthermore, protein interaction was examined in various combinations of ZmSPXs, ZmPHR1, and ZmPHR2. Results revealed that combinations such as ZmSPX1 and ZmPHR2, ZmSPX2 and ZmPHR1, ZmSPX3 and ZmPHR1, ZmSPX5 and ZmPHR1, and ZmSPX6 and ZmPHR1 all exhibited robust growth on SD/−Ade/−His/−Leu/−Trp/+X-α-gal medium, indicating their protein interactions (Figure 3B). It is suggested that ZmSPXs and ZmPHRs may modulate the maize response to low-Pi stress at the post-transcriptional level.

Figure 3 Yeast two-hybrid (Y2H) assay of ZmSPXs and ZmPHRs. (A) Self-activation analysis of ZmSPXs, ZmPHR1, and ZmPHR2. The medium is SD/−Leu-Trp-His-Ade (the medium lacking leucine, tryptophan, histidine, and adenine). (B) Y2H assay of ZmSPXs and ZmPHRs. PGADT7 and PGBKT7 vectors were used for positive and negative control, respectively. Yeast dilution ratio was 1, 1:10, 1:100, 1:1,000, and 1:10,000. The medium is SD/−Ade/−His/−Leu/−Trp/+X-α-Gal.
3.6 Overexpressing and knocking out ZmSPX1 in maize revealed its correlation with Pi absorption and its effect on yield and yield-related traits
To investigate the function of ZmSPXs in response to low-Pi stress, overexpression lines of ZmSPXs were created in Arabidopsis. However, only the overexpression of ZmSPX1 served to enhance root sensitivity to Pi deficiency and high-Pi conditions in A. thaliana (Supplementary Figures S4, S5). In order to further explore the function and role of ZmSPX1, we generated knockout and overexpressing maize transgenic lines for this gene. Our experimental results demonstrated a significant reduction in the hundred-grain weight and grain weight per year in the overexpressing lines compared to the wild type, while the knockout lines exhibited a marked increase (Figures 4A, B, D, E). Moreover, analysis of the P concentration in the grains of the knockout and overexpressing lines revealed no significant difference compared to the wild type (Figures 4C, F). We also assessed the P concentration in the internode, sheath, and leaf of the transgenic plants in the field trials. The findings indicated a higher P concentration in the knockout lines in these tissues than in the wild-type lines, whereas the overexpressing lines showed lower P concentrations compared to the wild-type lines (Figures 4G–I). These results suggested that ZmSPX1 affected the P concentration in maize stems and leaves and exerted a certain impact on the yield of maize, but not on the grains.
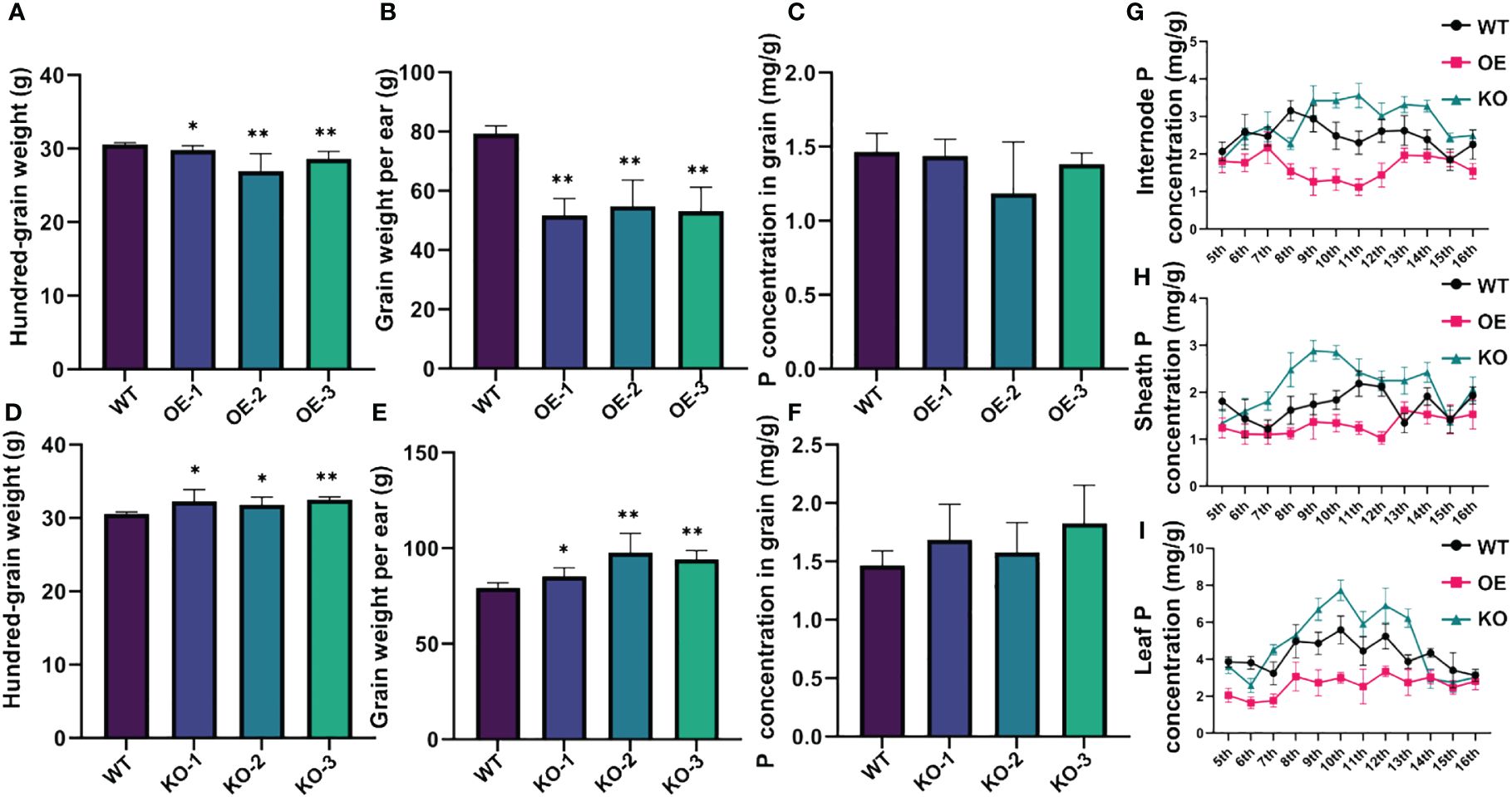
Figure 4 Phenotypic identification of ZmSPX1 transgenic maize materials. (A–C) Statistical analysis of differences in hundred-grain weight, grain weight per ear, and P concentration in grain between ZmSPX1 overexpressing lines and wild-type lines. (D–F) Statistical analysis of differences in hundred-grain weight, grain weight per ear, and P concentration in grain between ZmSPX1 knockout lines and wild-type lines. (G–I) The internode P concentration, sheath P concentration, and leaf P concentration in ZmSPX1 overexpressing lines, knockout lines, and wild-type lines. Significant differences are indicated by Student’s t-test: *p < 0.05, **p < 0.01.
3.7 Sequence variation and association analysis of ZmSPX1
To identify significant variations associated with phenotypic traits, we performed genome sequence amplification on 211 inbred lines and conducted multiple sequence alignments. We obtained a total of 1,390 bp of sequences, comprising 780 bp upstream of the initiation codon, 354 bp of the coding region, and 256 bp downstream of the initiation codon (Supplementary Table S10). We identified a total of 41 variants, which included 34 SNPs and seven InDels. The average distances between SNPs and InDels were 40.89 bp and 198.57 bp, respectively. The sequence variation frequencies varied across regions, with the highest frequency observed in the upstream sequence of the initiation codon (0.038) and the lowest in the coding region (0.006) (Supplementary Table S10). By utilizing a 200-bp sliding window with a 50-bp step size, we analyzed the nucleotide diversity (π × 1,000) of the ZmSPX1. We observed the overall nucleotide diversity to be 0.009, with the upstream sequence of the initiation codon exhibiting the highest diversity (0.013) among all regions and the coding region displaying the lowest diversity (0.002). Additionally, we investigated the selection pressure of ZmSPX1 using Tajima’s D, Fu and Li’s D*, and F* tests (Supplementary Table S10). The results showed that the upstream sequences displayed positive values, indicating a mode of balanced selection in their sequence evolution, with both Tajima’s D and Fu and Li’s F* tests being significant (Supplementary Table S10). Conversely, the coding region and downstream showed negative values, suggesting that these regions have experienced either negative selection or population expansion (Supplementary Table S10).
Additionally, to further investigate the relationship between sequence variations of ZmSPX1 and the phenotype in the maize seedling stage, we employed an MLM for candidate gene association analysis. We identified, under normal-Pi treatment, 37 markers (30 SNPs and seven InDels) as significantly associated with 17 traits, explaining 2.6% to 8.6% of the phenotypic variation (Supplementary Table S11). We found, under low-Pi treatment, a total of 10 markers to be significantly correlated with 13 traits, with R2 ranging from 2.8% to 5.4% (Supplementary Table S12). Moreover, 37 markers were significantly correlated with 16 traits of low-Pi tolerance index, explaining 2.8% to 20.2% of the phenotypic variation (Figures 5A, B; Supplementary Table S13). As the low-Pi tolerance index integrated traits under low- and normal-Pi conditions, we utilized all sequence variant sites significantly associated with the low-Pi tolerance index for haplotype division. First, LD analysis revealed the presence of multiple LD blocks with high linkage relationships (Figure 5C). Then, we categorized all variant sites into five haplotypes (MAF > 0.05) and conducted comparisons among the different haplotypes (Figure 5D; Supplementary Table S14). The findings indicated that in the low-Pi tolerance index of root traits, the fresh weight of the crown root index of Hap5 was significantly higher than that of Hap1, Hap2, and Hap4, while the fresh weight of the seminal root index was higher than that of Hap3 and Hap1. Additionally, the root volume of Hap5 was significantly higher than that of Hap1. Furthermore, we observed that the dry weight of the whole plant index of Hap5 was significantly higher than that of other haplotypes. Based on these results, it can be inferred that Hap5 enhances biomass production by promoting root development.
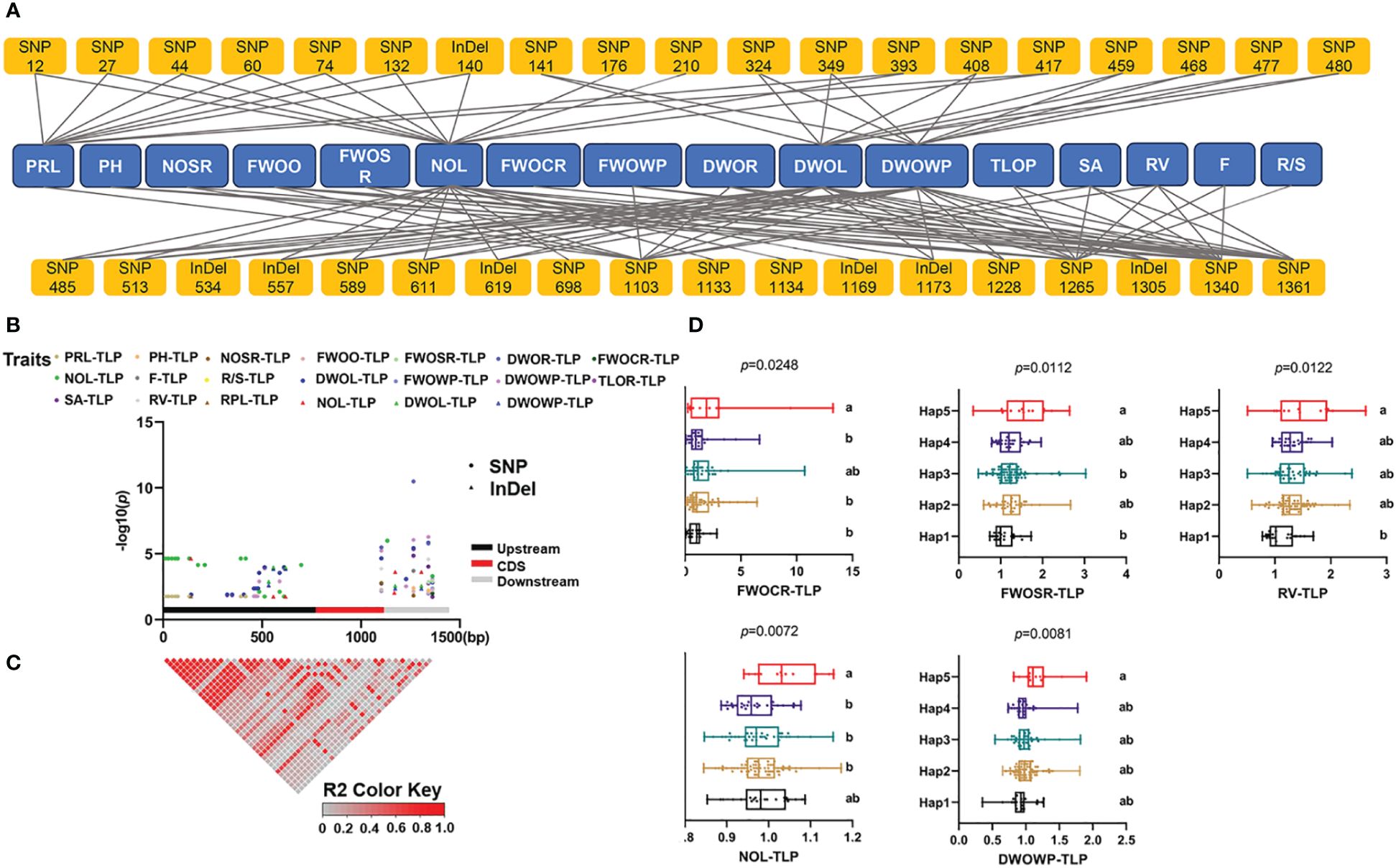
Figure 5 Candidate gene association analysis of ZmSPX1 with traits of low-Pi tolerance index. (A, B) The results of the candidate gene association analysis. (C) Linkage disequilibrium (LD) heatmap of ZmSPX1. (D) Phenotypic difference between different haplotypes. The abbreviation for each trait: primary root length (PRL), plant height (PH), number of seminal roots (NOSR), fresh weight of overground (FWOO), fresh weight of seminal root (FWOSR), dry weight of root (DWOR), fresh weight of crown root (FWOCR), number of leaves (NOL), number of root forks (F), root/shoot ratio (R/S), dry weight of leaves (DWOL), fresh weight of whole plant (FWOWP), dry weight of whole plant (DWOWP), total length of root (TLOR), root surface area (SA), and root volume (RV). TLP means the low-Pi tolerance index (Low-Pi/Normal-Pi). Means with the same letter in (D) are not significantly different at p < 0.05 according to one-way ANOVA followed by Tukey’s multiple comparison test.
4 Discussion
4.1 The maize transgenic lines of ZmSPX1 exhibited significant impacts on P concentration and yield
Pi homeostasis is crucial for plant growth and development, with ZmSPX1 playing a pivotal role in Pi uptake by plants from both the source and reservoir. Consequently, we first analyzed the phenotypes in ZmSPX1 overexpression transgenic A. thaliana and WT under different Pi concentration treatments. Under both Pi deficiency and high-Pi conditions, we observed a more robust root system development in the WT plants, whereas the growth of the root system was inhibited in the overexpressing (OE) lines. These results indicated that overexpression of the ZmSPX1 gene in A. thaliana enhanced the sensitivity of roots to both Pi deficiency and high-Pi conditions. Therefore, we proceeded to construct transgenetic lines for ZmSPX1 in maize. The knockout ZmSPX1 in maize led to an increase in P content in the stems and leaves. Additionally, a significant rise in hundred-grain weight and grain yield per ear was also observed in the knockout maize lines. Conversely, overexpression of ZmSPX1 in maize demonstrated contrasting phenotypes. Studies in dicotyledonous model plants have demonstrated that SPXs, functioning as negative regulators, can impede the central regulator PHR1 from acting on downstream low-Pi response genes (Jia et al., 2023). Conversely, research reports in monocotyledonous model plants such as A. thaliana and soybean suggest that SPXs serve as positive regulators. Overexpression of SPX1 induced the expression of low-Pi response genes and thus increased plant P concentration (Duan et al., 2008; Yao et al., 2014a). These results indicated that the expression patterns and functions of the SPX1 gene in maize and A. thaliana are different, which may also explain why the P concentration in A. thaliana overexpressing lines of SPX was higher than that in wild-type plants under normal-Pi conditions.
Additionally, a previous study demonstrated that OsPHO1;2 and ZmPHO1;2 play a crucial role in Pi allocation during grain filling. The activity of ADP-glucose pyrophosphorylase (AGPase) was inhibited in the knockout mutants of OsPHO1;2 and ZmPHO1;2, resulting in a defect in grain filling and a reduction in hundred-grain weight (Ma et al., 2021). Regarding the plant Pi signaling pathway (Wang et al., 2021b), the E2 ubiquitin-binding enzyme PHO2 can degrade PHO1 through its N-terminal SPX domain. The mRNA level of PHO2 was regulated by the cleavage of miR399. PHR1 can bind to the promoter region of miRNA399 to modulate its expression. Therefore, the relationship between SPX proteins and PHO1 was established through this network. However, further research is needed to investigate whether the impact of ZmSPX1 on maize grain yield, as observed in this study, is related to ZmPHO1.
4.2 Association analysis of the ZmSPX1 gene: implications for low-Pi signaling pathway and marker development for efficient Pi utilization in maize
To further investigate the relationship between the ZmSPX1 gene and low-Pi stress response-related traits, we conducted an association analysis utilizing sequence variations of the gene in the population and integrated the corresponding phenotypes. The results revealed that variant sites in ZmSPX1 are significantly associated with numerous low-Pi-related traits. This finding complements the findings from the phenotypic identification experiments in Arabidopsis involving the overexpression of the ZmSPX1 gene, providing strong evidence for the pivotal role of the ZmSPX1 gene in the low-Pi stress response pathway in maize. Additionally, we identified beneficial haplotypes based on 37 markers significantly associated with low-Pi tolerance indexes. Among these, Hap5 manifested a notable low-Pi tolerance phenotype, presenting an opportunity for the development of low-Pi-tolerant maize materials based on this outcome, and the identification of novel markers for screening purposes.
5 Conclusion
In conclusion, our study delineated the evolutionary relationship of maize SPXs with counterparts in other plants, particularly revealing a close relationship and collinearity with rice SPX genes. The presence of hormone-responsive elements in ZmSPXs promoters suggests their involvement at the nexus of hormone and Pi signaling. Expression pattern analyses indicated that ZmSPXs genes were upregulated under low-Pi stress, with pronounced expression in anthers and roots, and were localized to the nucleus and cytoplasm. The interaction of ZmSPXs with PHR proteins underscored the significance of SPXs in maize’s low-Pi stress signaling pathways. Specifically, overexpression of ZmSPX1 can enhance root sensitivity to Pi deficiency and high-Pi conditions in A. thaliana, emphasizing the pivotal role of the SPX gene family in the Pi stress response in maize. Finally, ZmSPX1 transgenic maize lines exhibited changes in P concentration in different tissues and yield, demonstrating that ZmSPX1 plays an important role in regulating the transport and distribution of P in maize and ultimately influencing yield. These findings will provide valuable information for further investigations into the mechanisms of the SPX-mediated Pi signaling pathway.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
BL: Conceptualization, Formal analysis, Funding acquisition, Methodology, Visualization, Writing – original draft, Writing – review & editing. JS: Conceptualization, Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. HZ: Formal analysis, Visualization, Writing – original draft. JZ: Writing – review & editing. GY: Writing – review & editing. WW: Writing – review & editing. JG: Writing – review & editing. SZ: Writing – review & editing. PM: Writing – review & editing. ZN: Writing – review & editing. XZ: Writing – review & editing. DL: Project administration, Resources, Writing – review & editing. LW: Project administration, Resources, Writing – review & editing. DG: Project administration, Resources, Writing – review & editing. SQG: Project administration, Resources, Writing – review & editing. SS: Project administration, Resources, Writing – review & editing. ZG: Investigation, Writing – review & editing. SBG: Writing – review & editing, Conceptualization, Funding acquisition, Methodology, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (grant no. 32101655), the China Postdoctoral Science Foundation (grant no. 2022M712292), Sichuan Science and Technology Support Project (grant nos. 2023YFH0038, 2021YFYZ0027, 2021YFYZ0020, and 2021YFFZ0017), and the earmarked fund for China Agriculture Research System (grant no. CARS-02).
Acknowledgments
The bioinformatics analyses in this study were supported by the High-Performance Computing Platform of Sichuan Agricultural University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1385977/full#supplementary-material
Supplementary Figure 1 | CRISPR/Cas9-induced mutation in the ZmSPX1 gene.
Supplementary Figure 2 | Detection of expression level of ZmSPX1 gene in overexpressing plants. Significant differences are indicated by Student’s t-test: *P < 0.05, **P < 0.01.
Supplementary Figure 3 | Subcellular localization of ZmSPXs in tobacco epidermal. GFP, green fluorescence protein.
Supplementary Figure 4 | Detection of overexpression of the ZmSPX1 in Arabidopsis thaliana. M: DNA marker BM2000.
Supplementary Figure 5 | Characterization of Arabidopsis thaliana ZmSPX1-overexpression plants in response to low-Pi conditions. (A) The WT and OE seedlings were subjected to treatments with P concentrations of 0 mM, 0.1 mM, 1 mM, and 10 mM for 14 days. (B-C) The expression of AtPHR1 and ZmSPX1 in WT and OE. The numbers 1 and 2 on the abscissa represent different independent replicates. (D-F) Measurement of root traits. Significant differences are indicated by Student’s t-test: *P < 0.05, **P < 0.01. Under Pi-free and high-Pi conditions, only the ZmSPX1 overexpression lines exhibited negative regulation of root growth when compared with the WT. Phenotypic of OE and WT was analyzed under Pi deficiency (0 mmol/L), low-Pi (0.1 mmol/L), normal-Pi (1 mmol/L) and high-Pi (10 mmol/L) treatments. The results indicated that, except under low-Pi conditions, the total root length, total root projected area, and total root surface area of the OE were significantly lower than those of the WT. Furthermore, the expression patterns of the AtPHR1 and ZmSPX1 genes were analyzed in the WT and OE plants. The results demonstrated significant downregulation of the AtPHR1 in the OE plants and substantial upregulation in the WT plants. In contrast, ZmSPX1 was upregulated in the OE plants and downregulated in the WT plants. Concurrently, we also examined the nitrogen and P concentration of the OE and WT (Supplementary Figure S6). Under low-Pi conditions, the nitrogen concentration in the OE was notably lower than that in the WT. However, under 1mM Pi concentration treatment, the nitrogen concentration in the OE was significantly higher than in the WT, while at 10mM Pi concentration conditions, the difference in nitrogen concentration between OE and WT was not statistically significant. Under Pi deficiency and low-Pi conditions, there was no significant difference in P concentration between the OE and WT. Nevertheless, at Pi concentrations of 1mM and 10mM, the P concentration in the OE was markedly elevated compared to that in the WT. Owing to the exceptionally low biomass of Arabidopsis tissues, there was a potential for deviations in the measurement of nutrient concentrations, and no consistent results were observed with regard to root traits. As a result, this study advanced to develop overexpression and knockout lines for ZmSPX1 in maize to investigate the gene’s impact on P concentration in maize tissues.
Supplementary Figure 6 | The nitrogen and P concentrations of ZmSPX1 overexpression Arabidopsis lines under varying Pi levels. (A) Nitrogen concentrations. (B) P concentrations.
References
Balyan, H. S., Gahlaut, V., Kumar, A., Jaiswal, V., Dhariwal, R., Tyagi, S., et al. (2016). Nitrogen and phosphorus use efficiencies in wheat: physiology, phenotyping, genetics, and breeding. Plant Breed. Rev. 40, 167–234. doi: 10.1002/9781119279723.ch4
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Carstensen, A., Herdean, A., Schmidt, S. B., Sharma, A., Spetea, C., Pribil, M., et al. (2018). The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant Physiol. 177, 271–284. doi: 10.1104/pp.17.01624
Chen, S., Ding, G., Wang, Z., Cai, H., Xu, F. (2015). Proteomic and comparative genomic analysis reveals adaptability of Brassica napus to phosphorus-deficient stress. J. Proteomics 117, 106–119. doi: 10.1016/j.jprot.2015.01.012
Chiou, T.-J., Lin, S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dong, J., Ma, G., Sui, L., Wei, M., Satheesh, V., Zhang, R., et al. (2019). Inositol pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant 12, 1463–1473. doi: 10.1016/j.molp.2019.08.002
Du, H., Yang, C., Ding, G., Shi, L., Xu, F. (2017). Genome-wide identification and characterization of SPX domain-containing members and their responses to phosphate deficiency in Brassica napus. Front. Plant Sci. 8, 35. doi: 10.3389/fpls.2017.00035
Duan, K., Yi, K., Dang, L., Huang, H., Wu, W., Wu, P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. doi: 10.1111/j.1365-313X.2008.03460.x
Feng, X., Jia, L., Cai, Y., Guan, H., Zheng, D., Zhang, W., et al. (2022). ABA-inducible DEEPER ROOTING 1 improves adaptation of maize to water deficiency. Plant Biotechnol. J. 20, 2077–2088. doi: 10.1111/pbi.13889
Garcia, A., Gaju, O., Bowerman, A. F., Buck, S. A., Evans, J. R., Furbank, R. T., et al. (2023). Enhancing crop yields through improvements in the efficiency of photosynthesis and respiration. New Phytol. 237, 60–77. doi: 10.1111/nph.18545
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate–NRT1. 1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Iqbal, A., Qiang, D., Xiangru, W., Huiping, G., Hengheng, Z., Xiling, Z., et al. (2023). Genotypic variation in cotton genotypes for low phosphorus tolerance and efficiency under different growth conditions. Gesunde Pflanzen 75, 1975–1993. doi: 10.1007/s10343-022-00823-y
Jia, X., Wang, L., Nussaume, L., Yi, K. (2023). Cracking the code of plant central phosphate signaling. Trends Plant Science 28, 267–270. doi: 10.1016/j.tplants.2022.12.008
Jiao, P., Liu, T., Zhao, C., Fei, J., Guan, S., Ma, Y. (2023). ZmTCP14, a TCP transcription factor, modulates drought stress response in Zea mays L. Environ. Exp. Bot. 208, 105232. doi: 10.1016/j.envexpbot.2023.105232
Jung, J.-Y., Ried, M. K., Hothorn, M., Poirier, Y. (2018). Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 49, 156–162. doi: 10.1016/j.copbio.2017.08.012
Kopriva, S., Chu, C. (2018). Are we ready to improve phosphorus homeostasis in rice? J. Exp. Bot. 69, 3515–3522. doi: 10.1093/jxb/ery163
Kumar, A., Sharma, M., Gahlaut, V., Nagaraju, M., Chaudhary, S., Kumar, A., et al. (2019). Genome-wide identification, characterization, and expression profiling of SPX gene family in wheat. Int. J. Biol. macromolecules 140, 17–32. doi: 10.1016/j.ijbiomac.2019.08.105
Kumar, S., Tamura, K., Nei, M. (1994). MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 10, 189–191. doi: 10.1093/bioinformatics/10.2.189
Li, C., You, Q., Zhao, P. (2021). Genome-wide identification and characterization of SPX-domain-containing protein gene family in Solanum lycopersicum. PeerJ 9, e12689. doi: 10.7717/peerj.12689
Liang, C., Wang, J., Zhao, J., Tian, J., Liao, H. (2014). Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 21, 59–66. doi: 10.1016/j.pbi.2014.06.009
Luo, B., Ma, P., Nie, Z., Zhang, X., He, X., Ding, X., et al. (2019). Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 97, 947–969. doi: 10.1111/tpj.14160
Luo, B., Zhang, Z., Li, B., Zhang, H., Ma, J., Li, J., et al. (2023). Chromatin remodeling analysis reveals the RdDM pathway responds to low-phosphorus stress in maize. Plant J. 117, 33–52. doi: 10.1111/tpj.16468
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. doi: 10.1105/tpc.114.123208
Lynch, J. P. (2011). Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156, 1041–1049. doi: 10.1104/pp.111.175414
Lynch, J. P., Ho, M. D., Phosphorus, L. (2005). Rhizoeconomics: Carbon costs of phosphorus acquisition. Plant Soil 269, 45–56. doi: 10.1007/s11104-004-1096-4
Ma, B., Zhang, L., Gao, Q., Wang, J., Li, X., Wang, H., et al. (2021). A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 53, 906–915. doi: 10.1038/s41588-021-00855-6
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Nezamivand-Chegini, M., Ebrahimie, E., Tahmasebi, A., Moghadam, A., Eshghi, S., Mohammadi-Dehchesmeh, M., et al. (2021). New insights into the evolution of SPX gene family from algae to legumes; a focus on soybean. BMC Genomics 22, 915. doi: 10.1186/s12864-021-08242-5
Nie, Z., Luo, B., Zhang, X., Wu, L., Liu, D., Guo, J., et al. (2021). Combined transcriptome and proteome analysis of maize (Zea mays l.) reveals a complementary profile in response to phosphate deficiency. Curr. Issues Mol. Biol. 43, 1142–1155. doi: 10.3390/cimb43020081
Osorio, M. B., Ng, S., Berkowitz, O., De Clercq, I., Mao, C., Shou, H., et al. (2019). SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 181, 332–352. doi: 10.1104/pp.18.00594
Porebski, S., Bailey, L. G., Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108
Puga, M. I., Rojas-Triana, M., De Lorenzo, L., Leyva, A., Rubio, V., Paz-Ares, J. (2017). Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr. Opin. Plant Biol. 39, 40–49. doi: 10.1016/j.pbi.2017.05.007
Raghothama, K. G. (1999). PHOSPHATE ACQUISITION. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Saengwilai, P. J., Bootti, P., Klinnawee, L. (2023). Responses of rubber tree seedlings (Hevea brasiliensis) to phosphorus deficient soils. Soil Sci. Plant Nutr. 69, 78–87. doi: 10.1080/00380768.2022.2164675
Sahito, J. H., Zhang, X., Zhong, H., He, X., Zhen, C., Ma, P., et al. (2020a). Identification, association of natural variation and expression analysis of ZmNAC9 gene response to low phosphorus in maize seedling stage. Plants 9, 1447. doi: 10.3390/plants9111447
Sahito, J. H., Zheng, F., Tang, H., He, X., Luo, B., Zhang, X., et al. (2020b). Identification, association, and expression analysis of ZmNAC134 gene response to phosphorus deficiency tolerance traits in maize at seedling stage. Euphytica 216, 1–19. doi: 10.1007/s10681-020-02634-6
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Shen, J., Li, H., Neumann, G., Zhang, F. (2005). Nutrient uptake, cluster root formation and exudation of protons and citrate in Lupinus albus as affected by localized supply of phosphorus in a split-root system. Plant Sci. 168, 837–845. doi: 10.1016/j.plantsci.2004.10.017
Shimojima, M. (2011). Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 50, 234–239. doi: 10.1016/j.plipres.2011.02.003
Strable, J., Scanlon, M. J. (2009). Maize (Zea mays): a model organism for basic and applied research in plant biology. Cold Spring Harbor Protoc. 2009, pdb. emo132. doi: 10.1101/pdb.emo132
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wang, L., Jia, X., Zhang, Y., Xu, L., Menand, B., Zhao, H., et al. (2021a). Loss of two families of SPX domain-containing proteins required for vacuolar polyphosphate accumulation coincides with the transition to phosphate storage in green plants. Mol. Plant 14, 838–846. doi: 10.1016/j.molp.2021.01.015
Wang, Y., Chen, Y. F., Wu, W. H. (2021b). Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52. doi: 10.1111/jipb.13053
Wild, R., Gerasimaite, R., Jung, J.-Y., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Xiao, J., Xie, X., Li, C., Xing, G., Cheng, K., Li, H., et al. (2021). Identification of SPX family genes in the maize genome and their expression under different phosphate regimes. Plant Physiol. Biochem. 168, 211–220. doi: 10.1016/j.plaphy.2021.09.045
Yang, J., Wang, L., Mao, C., Lin, H. (2017). Characterization of the rice NLA family reveals a key role for OsNLA1 in phosphate homeostasis. Rice 10, 1–6. doi: 10.1186/s12284-017-0193-y
Yao, Z., Liang, C., Zhang, Q., Chen, Z., Xiao, B., Tian, J., et al. (2014a). SPX1 is an important component in the phosphorus signalling network of common bean regulating root growth and phosphorus homeostasis. J. Exp. Bot. 65, 3299–3310. doi: 10.1093/jxb/eru183
Yao, Z., Tian, J., Liao, H. (2014b). Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann. Bot. 114, 477–488. doi: 10.1093/aob/mcu147
Yue, W., Ying, Y., Wang, C., Zhao, Y., Dong, C., Whelan, J., et al. (2017). Os NLA 1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 90, 1040–1051. doi: 10.1111/tpj.13516
Zhang, H., Huang, Y., Ye, X., Xu, F. (2010). Analysis of the contribution of acid phosphatase to P efficiency in Brassica napus under low phosphorus conditions. Sci. China Life Sci. 53, 709–717. doi: 10.1007/s11427-010-4008-2
Zhang, Z., Liao, H., Lucas, W. J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56, 192–220. doi: 10.1111/jipb.12163
Zhang, X., Zhang, H., Li, L., Lan, H., Ren, Z., Liu, D., et al. (2016). Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genomics 17, 697. doi: 10.1186/s12864-016-3041-3
Zhong, Y., Wang, Y., Guo, J., Zhu, X., Shi, J., He, Q., et al. (2018). Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 219, 135–148. doi: 10.1111/nph.15155
Zhou, L., Zhou, J., Xiong, Y., Liu, C., Wang, J., Wang, G., et al. (2018). Overexpression of a maize plasma membrane intrinsic protein ZmPIP1; 1 confers drought and salt tolerance in Arabidopsis. PLoS One 13, e0198639. doi: 10.1371/journal.pone.0198639
Keywords: maize, low-Pi stress, SPX gene family, PHRS, candidate gene association analysis
Citation: Luo B, Sahito JH, Zhang H, Zhao J, Yang G, Wang W, Guo J, Zhang S, Ma P, Nie Z, Zhang X, Liu D, Wu L, Gao D, Gao S, Su S, Gishkori ZGN and Gao S (2024) SPX family response to low phosphorus stress and the involvement of ZmSPX1 in phosphorus homeostasis in maize. Front. Plant Sci. 15:1385977. doi: 10.3389/fpls.2024.1385977
Received: 14 February 2024; Accepted: 17 June 2024;
Published: 08 July 2024.
Edited by:
Md Sazzad Hossain, University of Kiel, GermanyReviewed by:
Hongmei Cai, Huazhong Agricultural University, ChinaGuillaume Pilot, Virginia Tech, United States
Copyright © 2024 Luo, Sahito, Zhang, Zhao, Yang, Wang, Guo, Zhang, Ma, Nie, Zhang, Liu, Wu, Gao, Gao, Su, Gishkori and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shibin Gao, shibingao@163.com
†These authors have contributed equally to this work and share first authorship
 Bowen Luo1,2,3†
Bowen Luo1,2,3† Javed Hussain Sahito
Javed Hussain Sahito Haiying Zhang
Haiying Zhang Peng Ma
Peng Ma Shibin Gao
Shibin Gao