- 1Institute for Sustainable Plant Protection of the National Research Council, Unit of Bari, Bari, Italy
- 2Laboratory of Bioinformatics, Mathematical Modelling, and Synthetic Biology, Department of Electrical, Computer and Biomedical Engineering - Centre for Health Technology, University of Pavia, Pavia, Italy
- 3Plant Biotechnology Laboratory, Department of Biology and Biotechnology “L. Spallanzani”, University of Pavia, Pavia, Italy
Introduction: Plant-pathogen interaction is an inexhaustible source of information on how to sustainably control diseases that negatively affect agricultural production. Meloidogyne incognita is a root-knot nematode (RKN), representing a pest for many crops, including tomato (Solanum lycopersicum). RKNs are a global threat to agriculture, especially under climate change, and RNA technologies offer a potential alternative to chemical nematicides. While endogenous microRNAs have been identified in both S. lycopersicum and M. incognita, and their roles have been related to the regulation of developmental changes, no study has investigated the miRNAs cross-kingdom transfer during this interaction.
Methods: Here, we propose a bioinformatics pipeline to highlight potential miRNA-dependent cross-kingdom interactions between tomato and M. incognita.
Results: The obtained data show that nematode miRNAs putatively targeting tomato genes are mostly related to detrimental effects on plant development and defense. Similarly, tomato miRNAs putatively targeting M. incognita biological processes have negative effects on digestion, mobility, and reproduction. To experimentally test this hypothesis, an in vitro feeding assay was carried out using sly-miRNAs selected from the bioinformatics approach. The results show that two tomato miRNAs (sly-miRNA156a, sly-miR169f) soaked by juvenile larvae (J2s) affected their ability to infect plant roots and form galls. This was also coupled with a significant downregulation of predicted target genes (Minc11367, Minc00111), as revealed by a qRT-PCR analysis.
Discussions: Therefore, the current study expands the knowledge related to the cross-kingdom miRNAs involvement in host-parasite interactions and could pave the way for the application of exogenous plant miRNAs as tools to control nematode infection.
1 Introduction
Root-knot nematodes (RKNs) are pathogens that attack many economically important crops, negatively impacting crop yield, quality, and subsequently food security (Kaloshian and Teixeira, 2019). Damage caused by RKNs has been estimated to range between 80 - 157 billion $US per year, although this evaluation may be largely underestimated (Palomares-Rius et al., 2021). Among the RKNs, members of the Meloidogyne genus are the most widespread and have a broad range of hosts. These parasites can penetrate host roots and induce the formation of specialized feeding structures (root galls), which supply the resources required for nematode development. The formation of root galls is highly damaging because they affect the plants’ ability to uptake water and nutrients (Singh et al., 2015). Tomato (Solanum lycopersicum L.), one of the most important and extensively grown horticultural crops in the Mediterranean region (EUROSTAT, 2021), is the preferential host for many Meloidogyne species. Yield losses due to M. incognita RKNs can range between 25-100% (Seid et al., 2015). Control methods generally include the use of chemical fumigants or nematicides, but since the ban of chemicals with a broad action on non-target organisms, emerging RKN populations continue to bypass plant host defenses. Therefore, alternative approaches (e.g., eco-friendly fumigants, bio-control agents) that can stimulate plant defense mechanisms, are needed to control their spread (Leonetti and Molinari, 2020). Currently, much focus is given to the use of RNA interference (RNAi) as an eco-friendly strategy (Banerjee et al., 2018; Banakar et al., 2020; Iqbal et al., 2020), as demonstrated for fungal small RNAs that suppress plant immunity by hijacking host RNAi pathways (Weiberg et al., 2013; Hua et al., 2018). Examples of cross-kingdom RNAi during plant-pathogen interactions are also available. For instance, the Botrytis cinerea sRNA produced by Dicer-like (DCL) proteins can target and silence DCL genes in Arabidopsis with subsequent effects on fungal pathogenicity and growth (Wang et al., 2016). Other examples include the transfer of ds-siRNA and miRNAs from plants to Coleoptera species, with consequences on gene transcription and insect growth (Zhang L. L. et al., 2019; Kleaveland, 2023). Additionally, miRNAs are being investigated as important regulatory actors in the plant-nematode interaction (Jaubert-Possamai et al., 2019), supporting the development of novel tools to advance modern agriculture.
As small, evolutionary conserved, and generally non-coding RNA molecules, microRNAs finely regulate gene expression being involved in developmental and stress responses. In plants, miRNAs achieve their function through perfect or near-perfect complementarity to target mRNAs, while in animals three types of miRNA-target interactions are recognized: (1) partial binding, mainly to the seed region; (2) complete or near-complete binding that enables AGO-mediated endonucleolytic cleavage of the target; and (3) extended/bulged binding to the seed region which specifies target-directed miRNA degradation (Kleaveland, 2023). When addressing host-parasite interactions, the involvement of miRNAs and long non-coding RNAs in the relation between S. lycopersicum and M. incognita, has been investigated (Kaur et al., 2017; Yang et al., 2020, 2022). While Zhang Y. et al. (2016) focused mainly on the high-throughput identification and annotation of miRNAs in M. incognita, Kaur et al. (2017) directed their attention to the identification of tomato miRNAs in susceptible plants during nematode infection. However, no study has investigated the miRNAs cross-species potential during this interaction. Several works have studied miRNAs transfer to other organisms along with possible regulatory functions (LaMonte et al., 2012; Zhang et al., 2012; Cheng et al., 2013; Zhu et al., 2017; Avsar et al., 2020; Cai et al., 2021), making it reasonable to hypothesize that this exchange may be well-represented during plant-parasite interactions, and that this can be exploited as an alternative tool to sustainably fight pathogens (Gualtieri et al., 2020; Rabuma et al., 2022). So far, the miRNA cross-kingdom transfer ability has been demonstrated in several plant-pathogen interactions, like Gossypium hirsutum–Verticillum dahliae (Zhang H. et al., 2016), Triticum aestivum–Puccinia striiformis (Wang et al., 2017), and Arabidopsis thaliana–Plutella xylostella (Zhang L. L. et al., 2019).
In the current study, we propose a bioinformatics pipeline to investigate the putative effects of cross-species miRNAs transfer during the interaction between S. lycopersicum - M. incognita. Specific miRNAs and transcripts from the two species have been retrieved from public databases and used to predict candidate targets in a cross-kingdom manner, based on different miRNA-mRNA hybridization rules. Biological processes of Gene Ontology significantly affected by the target genes were identified and examples of bidirectional cross-targeting predictive miRNAs, with potentially interesting applications to fight disease development, are provided. Additionally, three tomato miRNAs (sly-miRNA 156a, sly-miRNA166b, sly-miRNA169f) were selected for in vitro validation studies on juvenile M. incognita larvae (J2s) infecting the roots of susceptible tomato plants.
2 Materials and methods
The bioinformatics workflow adopted in this work includes the following steps: (1) miRNA and transcript sequence collection from public data, (2) cross-kingdom miRNA target prediction by two different procedures, and (3) analysis of target genes and enriched biological processes resulting from the predicted miRNA-target pairs. Subsequently, experimental methods for the in vitro evaluation of the effect of selected S. lycopersicum miRNAs on M. incognita larvae are provided.
2.1 Datasets
A collection of S. lycopersicum miRNAs (sly-miRNAs) was obtained by merging the entries found in public databases and literature. The miRbase repository (Kozomara et al., 2019) included 147 sly-miRNAs, of which 137 were unique. The sly-miRNA list reported by Kaur et al. (2017) was added, including 136 sequences (56 miRNAs classified as conserved or variants, and 60 as novel miRNAs), which account for 70 unique miRNAs after duplicate removal. Thus, a total of 207 sly-miRNAs (obtained from the two merged lists), with 185 unique sequences, represent the final collection used for tomato miRNAs. The S. lycopersicum transcript dataset (ITAG 2.4.v) was retrieved from psRNATarget (Dai et al., 2018) and included 34,725 transcripts, covering a large part of the tomato annotated genome (37,872 genes) available on NCBI.
Since M. incognita miRNAs (min-miRNAs) are not included in public databases, only literature works were used to define this collection. The list of Zhang Y. et al. (2016) included 144 min-miRNAs (38 classified as conserved and 106 as novel miRNAs), which accounts for 63 unique miRNAs after duplicate removal. The list of Wang et al. (2015) included 102 min-miRNAs, corresponding to 70 unique sequences, of which about 40% is composed of conserved miRNAs. From the 133 min-miRNAs obtained by merging the two lists, 126 were unique and represented the final collection of RKN miRNAs. The M. incognita transcript and CDS datasets were retrieved from the V2 genome assembly (Blanc-Mathieu et al., 2017), available at the INRAE Meloidogyne Genomic Resources website (https://meloidogyne.inrae.fr), which includes 43,718 transcript and CDS sequences. The M. incognita 3’UTRome was obtained by processing the transcripts and CDS lists above with a custom Python (v3.8) script, resulting in 20,201 sequences because not all the full transcripts have an annotated 3’UTR.
2.2 miRNA target prediction
The cross-kingdom search of miRNA targets in S. lycopersicum and M. incognita transcriptome was performed using the psRNATarget (https://www.zhaolab.org/psRNATarget/) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) tools, respectively, previously proposed for miRNA target prediction in plants (Dai et al., 2018) and animals (Kruger and Rehmsmeier, 2006). The collections of M. incognita and S. lycopersicum miRNAs were used as input for both tools, together with the transcriptome of the target organism (tomato and nematode, respectively). The two tools have been validated on intra-kingdom miRNA-target data in their original publications. However, it is worth mentioning that no large-scale validation is available for cross-kingdom predictions because of the very low number of validated miRNA-target pairs. Even though no gold standard tool or set of rules has been defined for cross-kingdom miRNA interactions, we assumed that such regulations follow the rules of the host organism, as it was assumed in other works (Shu et al., 2015; Chin et al., 2016; Zhang H. et al., 2016; Hou et al., 2018; Zhao et al., 2018; Bellato et al., 2019), thus motivating the use of plant- and animal-specific prediction tools. Consistent with previous in silico and in vivo studies (Liu et al., 2017; Li Z. et al., 2018; Lukasik et al., 2018; Mal et al., 2018; Wang et al., 2018), instead of restricting the search to the 3’UTRome, reported to be the preferential target of endogenous miRNA regulation in animals (Bartel, 2004), the full transcript sequences were used to search for targets in the RKN transcriptome. For the reasons above, the miRNA-target pairs found by the two tools represent putative regulations that may occur in nature, even though other herein neglected biological factors could play important roles.
RNAhybrid and psRNATarget were both run by setting a maximum of 50 targets per miRNA as previously done (Zhang H. et al., 2016; Bellato et al., 2019), to obtain a balanced set of miRNA-target pairs to be analyzed downstream. RNAhybrid was run using a Minimum Free Energy (MFE) threshold of -25 kcal/mol, corresponding to the upper bound of many experimentally found miRNA-target pairs (Zhang H. et al., 2016). Putative targets were ranked based on their MFE value (lowest to highest) for further selection of a subset of these genes, as required in the downstream steps (see 2.3). psRNATarget was run with an Expectation value of 3, corresponding to a slightly relaxed threshold in terms of prediction results stringency, as reported by Dai and Zhao (2011). Default values were used for the other psRNATarget parameters, as previously done in other studies (Bellato et al., 2019): Penalty for G:U pair = 0.5, Penalty for other mismatches = 1, Extra weight in seed region = 1.5, Seed region = 2-13 nucleotides, Mismatches allowed in seed region = 0, HSP size = 19.
In addition to the procedure described above (indicated as cross-kingdom hybridization), we followed a second search method that does not assume any hybridization rule in cross-kingdom interaction. This alternative method (indicated as seed region-based search) was carried out to find sly-miRNA targets in M. incognita transcriptome and included the following steps: (1) intra-kingdom hybridization (i.e., min-miRNAs vs. RKN transcriptome via RNA hybrid); (2) selection of all the sly-miRNAs that share a seed region (nucleotides 2-7 of the miRNA) with the collection of min-miRNAs; (3) association of the selected sly-miRNAs to the targets of the min-miRNAs with the same seed region. Location (CDS or UTRs) of the predicted miRNA binding and sequence similarity outside the seed region were also recorded to support further selection steps.
2.3 Biological process analysis
Statistically over-represented Gene Ontology (GO) terms in the Biological Process (BP) category were computed via enrichment analysis using the predicted miRNA targets obtained above. Duplicate genes in the target list were removed before analysis. The ClueGO application (v2.5.6) (Bindea et al., 2009) of Cytoscape (v3.7.3) (Shannon et al., 2003) was used as a GO analysis tool. A right-sided hypergeometric test with the Benjamini-Hochberg correction for multiple hypothesis testing and a P-value cutoff of 0.05 was used.
For M. incognita enrichment analysis, an ad hoc construction of the GO library was necessary for ClueGO analyses, as this organism was not previously included in this tool. To this aim, the GO biological processes list reported by Somvanshi et al. (2018) was used as a source, converted into a ClueGO-compatible format using a custom Python script, and integrated into ClueGO. In total, 5,508 and 6,098 unique GO terms in the BP category were present for S. lycopersicum and M. incognita, respectively. The BPs presented in this work after enrichment analysis refer to the most relevant term of the functionally grouped term networks provided by ClueGO. The number of input genes in enrichment analysis was set to obtain a comparable number of relevant terms between plant and RKN.
To further analyze the obtained biological processes, individual genes in the miRNA target list associated with the process were considered, and their function was searched in the literature. For S. lycopersicum, gene information was retrieved from the ITAG 2.4 annotations available in Phytozome v.12 (DOE JGI, https://jgi.doe.gov/more-intuitive-phytozome-interface/). For M. incognita, orthologous genes from other nematodes (mainly C. elegans) were obtained using WormBase ParaSite (https://parasite.wormbase.org/index.html) (Howe et al., 2017). This approach was considered because specific information on RKN genes is not directly available in public online resources. Finally, a subset of BP terms from Somvanshi et al. (2018) related to nematode development was selected and used to filter the sly-miRNA targets in the RKN transcriptome.
2.4 In vitro interaction assays of sly-miRNAs soaked by M. incognita juvenile larvae
For the in vitro interaction assay, sly-miR166b, sly-miR169f, and sly-miR156a were selected taking into account the bioinformatics data obtained from the cross-kingdom hybridization and the seed region-based search approach. These microRNAs were obtained from Invitrogen™ Custom Primer Service (BMR Genomics, Padova, Italy) and the relative sequences are given: sly-miR166b (5’-UCGGACCAGGCUUCAUUCCCC-3’, STAR strand GGAAUGUUGUCUGGCUCGAGG), sly-miR156a (5’-UUGACAGAAGAUAGAGAGCAC-3’, STAR strand GCUCUCUAUGCUUCUGUCAU) sly-miR169f (5’-UAGGCGUUGUCUGAGGCUAAC-3’, STAR strand AUCCGUUACUGAGGAACCGAUAG).
Susceptible tomato seedlings (Roma cv.) and axenic cultures of phytoparasitic nematodes were prepared as described by Molinari and Miacola (1997). Freshly hatched M. incognita J2s (infective second-stage juveniles) larvae were used for soaking experiments, following a modified protocol (Danchin et al., 2013; Tan et al., 2013). About 8,000 J2s were soaked for 24 hours in a 40µl final volume of mineral water containing different solutions: (1) a siRNA designed to have no similarity in the M. incognita genome (Dalzell et al., 2010) defined as negative control solution (C-J2); and (2) three different solutions of 0.05 mg/ml sly-miRNAs corresponding to each tested tomato miRNA. J2s from each soaking experiment were washed twice with water by centrifugation at 10,000 g for 3 min, and re-suspended in 100 µl of water. Subsequently, the miRNA-soaked J2s were observed using a Leica M125 stereomicroscope (Leica Microsystem S.r.l, Buccinasco, Milano, Italy) to confirm their vitality and split in two Eppendorf tubes: one used for infection assay and another used for RNA extraction (frozen at -80°C).
For the interaction assay, the sly-miRNAs soaked J2 larvae (approx. 50 J2s/root apex) were loaded in wells placed on agar plates at 0.1 mm from the tomato root. Each plate contained three susceptible tomato seedlings, growth in axenic conditions. The infection was observed for six weeks, during which the penetration in the roots was evaluated in terms of gradual enlargements of the root tip caused by rapid cell division and proliferation. The giant cell induction and galls formation was monitored using a modified LEICA Software image analysis (unpublished), and the following scoring system was used: “+” ranging between 0-30%, “++” ranging between 31-60%, and “+++” ranging between 61-90% (Melillo et al., 2006; Cabrera et al., 2015). The parameters referring to both root enlargements and formation of galls (91-100%), were counted and scored as the number of symptoms normalized to the number of roots in which the phenomena were observed. All experiments were conducted in triplicates coming from two independent trials.
2.5 RNA extraction and qRT-PCR analysis
RNA was extracted using the TRIzol Reagent (Invitrogen, CA, USA) method, as indicated by the manufacturer. For this, 500 J2s soaked for 24 h in the control solution (C-J2) or the respective sly-miRNAs (miR156a-J2, miR169f-J2) solutions, were used. For the reverse transcription, 1 μg of total RNA was used along with the QuantiTect Reverse Transcription Kit (QIAGEN S.r.l, Milano) following the manufacturer’s instructions.
The qRT-PCR reactions were carried out using the StepOnePlus (Applied Biosystems, Life Technologies, Zurich, Switzerland) system and assembled in a reaction with 1.5 μl cDNA, 10 μl SYBR® Select Master Mix (Applied Biosystem, Life Technologies, Zurich, Switzerland), 0.2 μl each of 100 μM of forward and reverse primers, and RNAse free water to 20 μl (total volume). Thermocycling was carried out with one cycle at 95°C for 5 min, followed by 40 cycles of 95°C for 45 s and 58°C for 1 min and 72°C for 45 s. The dissociation curve of the final products was checked to ascertain the presence of a single amplification product. The relative quantification was carried out using the 18S ribosomal RNA gene (NCBI accession HE667742.1, WormBase accession Minc3s09153g42974) as a reference gene. The Minc11367 (WormBase Accession Minc3s00025g01614) gene, was selected and tested as a putative target for sly-miR156a, while Minc00111 (WormBase Accession Minc3s00001g00015) was selected as a putative target of sly-miR169f. Oligonucleotide sequences to amplify the genes of interest were designed with Primer3Plus (https://primer3plus.com) and further validated through the online software Oligo Analyzer (https://eu.idtdna.com/calc/analyzer). All the oligonucleotide sequences are provided in Supplementary Table 1. The ΔΔCt method was used to quantify gene expression. The reactions were performed in triplicate samples of each cDNA and using two independent replicates.
2.6 Statistical analysis
Results of experimental data are shown as mean ± standard deviation (SD) obtained from two independent experiments each with three replicates. Statistical analyses were conducted using the two-way analysis of variance (ANOVA), along with the heteroscedastic Student’s t-test (where *, P ≤ 0.05; **, P ≤ 0.01), available within the Microsoft Excel package.
3 Results and discussion
3.1 Overview of the bioinformatics pipeline
In this work, a bidirectional bioinformatics workflow was carried out to predict miRNAs cross-kingdom potential in the host-parasite interaction between tomato and RKNs. These predictions are based on sequence complementarity between miRNAs and cross-species targets, as well as sequence similarity between tomato and nematode miRNAs. A total of 523 miRNA-target pairs were found in the cross-kingdom hybridization between S. lycopersicum transcripts and min-miRNAs, corresponding to 469 unique genes and 105 unique min-miRNAs (Supplementary Dataset 1). The majority of genes were targeted by a single miRNA, but putative targeting by two miRNAs occurred as well (Figure 1A). All the target genes were adopted for subsequent enrichment analysis that yielded 43 biological processes (BPs), divided into 10 functional groups, as described below in Section 3.2. Conversely, 8,926 transcripts corresponding to 6,428 unique genes were found in M. incognita by cross-kingdom hybridization with the sly-miRNAs (Supplementary Dataset 2). Each gene was putatively targeted by one or more miRNAs (Figure 1B). Based on the MFE of the targets, the 290 top-ranked genes were used for the enrichment analysis that resulted in 86 BPs in the 10 functional groups, described further in Section 3.3.
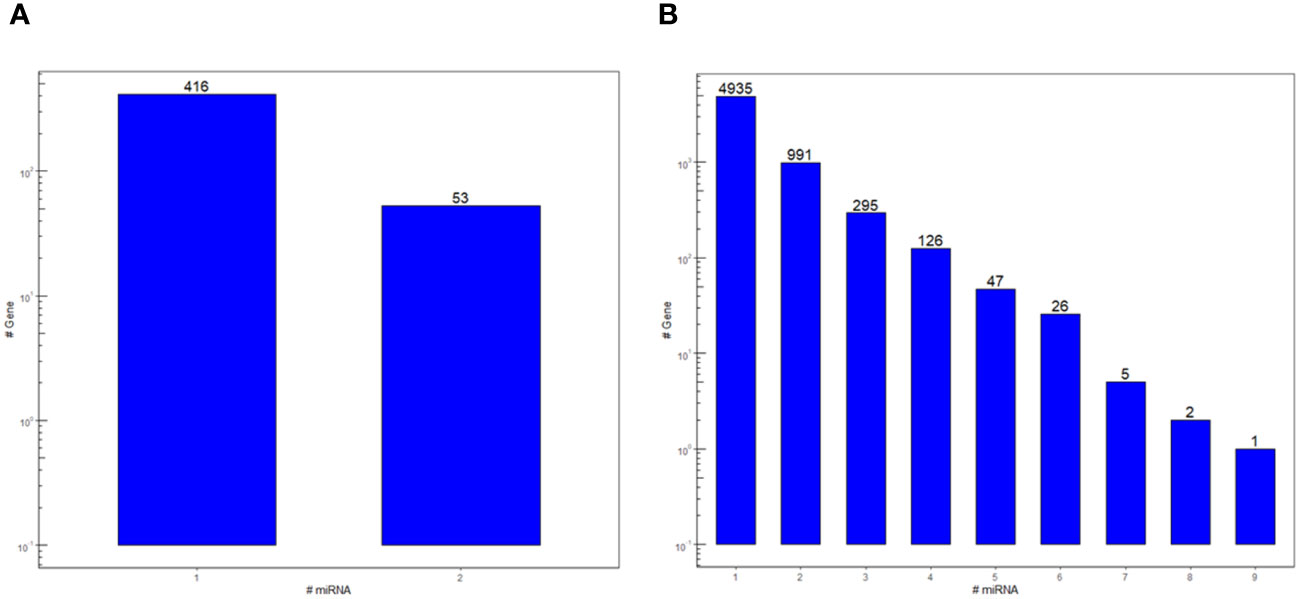
Figure 1 Count of the S. lycopersicum (A) and M. incognita (B) miRNA targets in the cross-kingdom hybridization bioinformatic pipeline run on the two species. Bars represent the number of genes that are putatively targeted by one or more miRNA.
The seed region-based search resulted in the identification of 7 sly-miRNAs having homologies with 9 min-miRNAs; using these sly-miRNAs, a total of 450 putative target genes were identified in M. incognita (Supplementary Dataset 3).
3.2 M. incognita miRNAs predicted to target tomato genes may inhibit plant development
To analyze the obtained datasets, we first looked at the potential effects that min-miRNAs would have on the development of tomato plants providing discussions based on the function of putatively targeted genes. The biological processes found to be enriched among the putative cross-species targets of RKN miRNAs against S. lycopersicum transcripts are reported in Supplementary Dataset 4 and graphically represented in Figure 2. These processes clustered in several major GO terms and the most abundant networks included brassinosteroid-mediated signaling pathway, response to herbivores, and positive regulation of protein catabolic process. Other processes like inositol-lipid mediated signaling, co-translational protein targeting to membrane, non-recombinant repair, rRNA methyl transferase, and cellular response to starvation, were less abundant. Considering the min-miRNAs putatively targeting genes in the tomato dataset, the enriched terms list includes 26 miRNAs and 46 unique targets, with the most interesting ones being summarized in Table 1. Within the brassinosteroid-mediated signaling pathway, different phosphatases, phosphodiesterase, and proteases were predicted to be targeted by NOVEL-18-1, min_miRNA6, and min_miRNA98. Many protease gene families are well-known to be involved in plant immune responses (Balakireva and Zamyatnin, 2018). For instance, aspartyl proteases are specifically linked to systemic acquired resistance (SAR) induced in response to local infections (Breitenbach et al., 2014; Molinari et al., 2014; Molinari and Leonetti, 2019). Other miRNAs, like NOVEL-8, miR-50, and min_miRNA29, are predicted to target genes playing important roles in the response to herbivores. The S-receptor kinase-like genes, aside from being involved in stress responses and host-pathogen defense, have also important functions in cell signaling and development (Takasaki et al., 2000; Becraft, 2002; Pastuglia et al., 2002). Similarly, genes involved in the cuticle and cell wall development and composition (e.g., sterols, mannans) represent important defense lines against many types of pathogens (Wang et al., 2013; Kazan and Gardiner, 2017). Other min-miRNAs (min_miRNA206, NOVEL-18-1) were predicted to target different E3 ubiquitin-protein ligases in tomatoes; these proteins play important roles in the regulation of cell homeostasis and thus they are key regulators of plant growth and stress responses (Liu et al., 2021). Importantly, they are involved in the regulation of plant innate immunity (Duplan and Rivas, 2014).
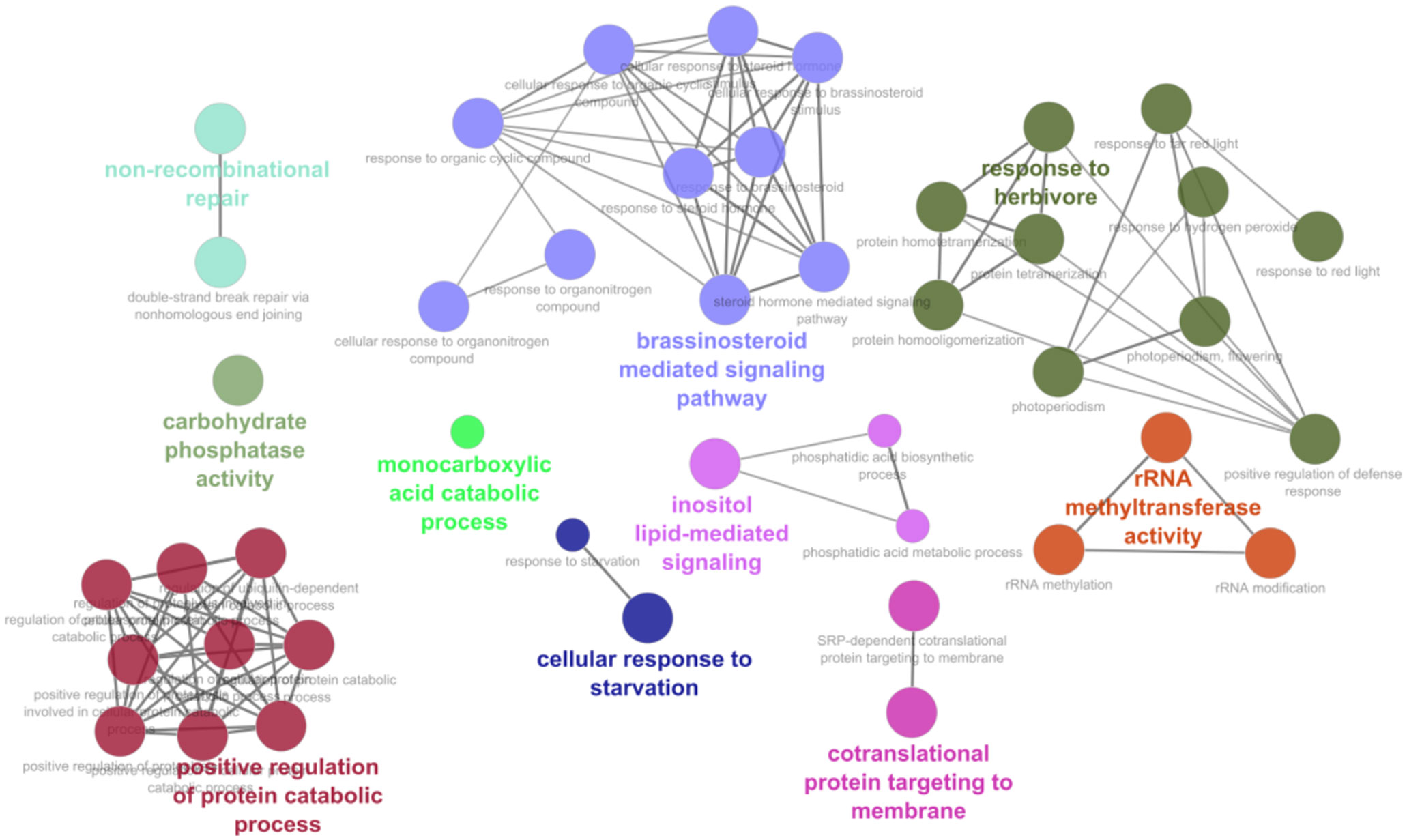
Figure 2 Biological processes and functional network clusters resulting from enrichment analysis carried out using the M. incognita miRNA putatively targeting S. lycopersicum mRNAs. The most relevant terms of the functional groups are highlighted.
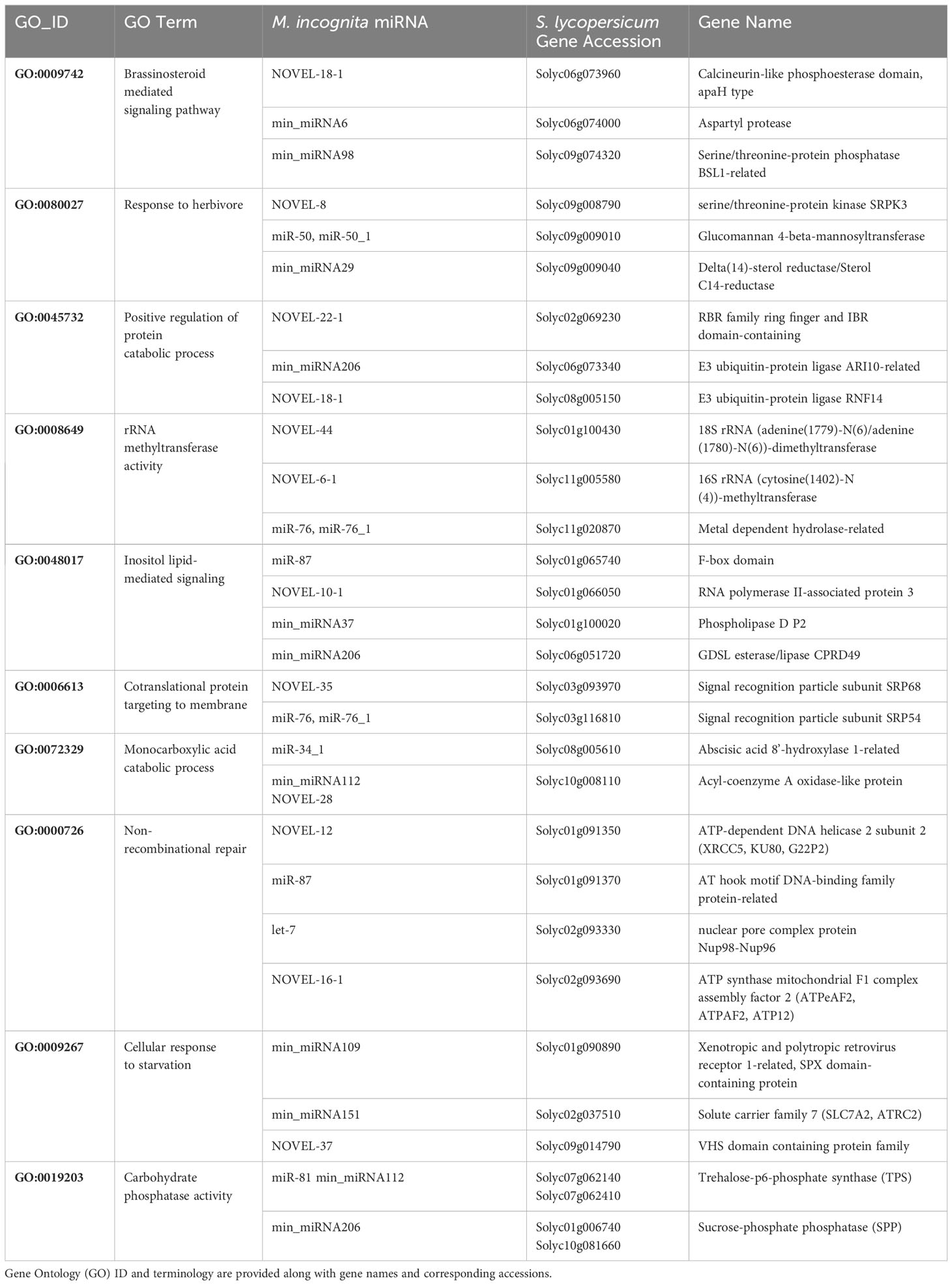
Table 1 List of representative M. incognita miRNAs putatively targeting genes in S. lycopersicum, based on bioinformatics cross-kingdom predictions.
Similarly, miRNAs (e.g., NOVEL-44, NOVEL-6-1, miR-76) affecting translational regulation can play critical roles in the plant defense against pathogen infection. In addition to the essential role of DNA repair in maintaining genome stability, recent works are discussing the involvement of DNA repair proteins in plant-pathogen interactions and SAR (Fu and Dong, 2013; Camborde et al., 2019). Moreover, the miRNA-mediated control over DNA damage responses is starting to gain more interest from both an endogenous (Gualtieri et al., 2021; Macovei et al., 2021) and cross-kingdom fashion (Bellato et al., 2019). An interesting finding in this sense is the case of let-7, one of the most abundant miRNAs found in nematodes, putatively targeting the nuclear pore complex protein Nup98-Nup96 in tomatoes. This complex plays a role in nuclear-cytoplasmic trafficking and mRNA export, being involved in several important biological events such as mitotic checkpoints. Nup98-deficient mutants share pleiotropic phenotypes (decreased root elongation, accelerated floral transition, reduced fertility, and robustness), indicating that it has a critical role in plant development (Parry, 2013).
Other miRNAs worth mentioning are miR-81, min_miRNA112, and min_miRNA206 predicted to putatively target genes involved in sugar metabolism, like sucrose-phosphate phosphatase (SPP) and trehalose-p6-phosphate synthase (TPS). SSP is an important regulator of carbon partitioning and loss-of-function of this gene leads to altered carbohydrate distribution resulting in a reduced growth rate (Chen et al., 2005, 2008). TPS leads to the formation of the trehalose-6-phosphate (T6P), a metabolic intermediate acting as a signaling molecule that regulates sugar metabolism (Paul, 2008), and its silencing in the tomato caused dysfunction in ROS accumulation and decreased expression of genes responsible for defense against pathogenic infections (Suárez et al., 2008).
To conclude, the prediction analyses show that the tomato genes putatively targeted by M. incognita miRNAs have essential roles in plant development and stress response, and their silencing can have negative repercussions for the plant. This is in agreement with the parasitic relation between the two organisms, where M. incognita tries to hijack the plant systems to promote its development.
3.3 sly-miRNAs predicted to target M. incognita genes may have detrimental effects on nematode development
When tomato miRNAs were evaluated against the nematode, the enrichment analysis of target genes resulted in BPs related to regulation of Wnt protein secretion, striate muscle contraction, sterol transported activity, ubiquinol-cytochrome-c reductase activity, [2Fe-2S] cluster assembly, glycogen biosynthetic process, defense response to fungus, serine-type exopeptidase, and apoptotic mitochondrial changes (Figure 3; Supplementary Dataset 5).
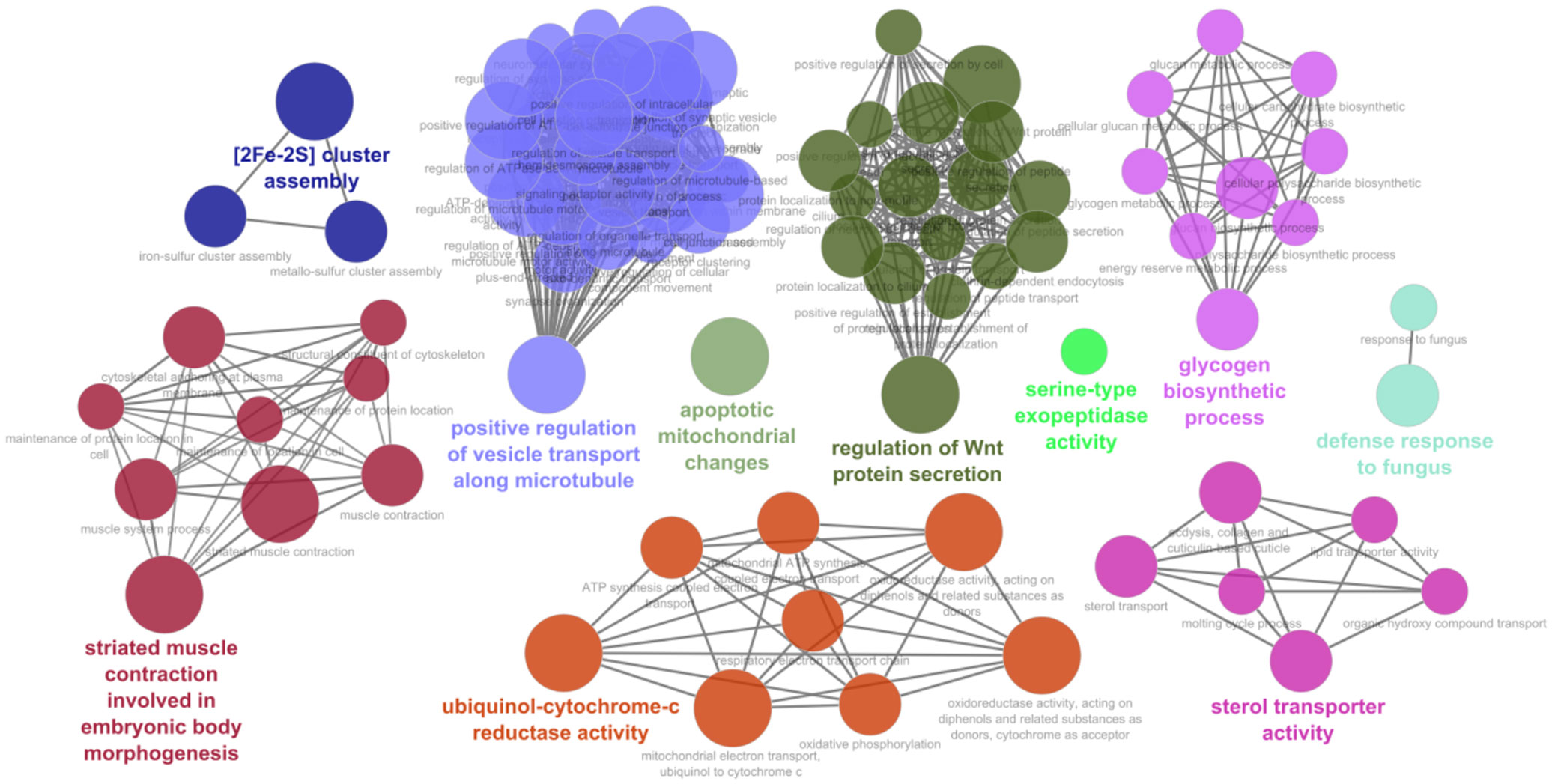
Figure 3 Biological processes and functional network clusters resulting from enrichment analysis carried out using the S. lycopersicum miRNA putatively targeting M. incognita mRNAs. The most relevant terms of the functional groups are highlighted.
Among the most interesting examples investigated are sly-miR156, sly-miR166, and sly-miR319, given that they have been previously identified as highly abundant in roots during M. incognita infection (Kaur et al., 2017). The abundance of these miRNAs may favor their uptake in high amounts during feeding. Table 2 presents the biological processes and putatively cross-kingdom targeted genes in relation to these three miRNAs. The data shows that sly-miR166(b,c) has the most abundant number of putative targets, distributed in GO terms related to response to stress and developmental processes. Although M. incognita genome has been sequenced (Abad et al., 2008), its annotation is not completed. To understand the roles of these putative targets we have looked at orthologues in related species. Among the genes involved in neuromuscular developmental processes, rGCs (guanylyl cyclases) plays a role in sensory processing (Maruyama, 2017), lev-11 (LEVamisole resistant) encodes a conserved CUB-domain containing transmembrane protein (Gally et al., 2004), snt-1 (SyNapTotagmin) functions as a Ca2+ sensor (Li L. et al., 2018), and nep-2 (NEPrilysin metallopeptidase family) is a homolog of the extracellular peptidase neprilysin whose loss-of-function leads to movement anomalies (Soh et al., 2020). Impaired movement can be also caused by dysfunctions in the genes coding for proteins that are part of the collagen extracellular matrix. This may be the case of dpy-17 (DumPY: shorter than wild-type), involved in cuticle development (Lang and Lundquist, 2021), and emb-9 (abnormal EMBroygenesis), an alpha1(IV) collagen gene with embryo lethal effects when mutated (Guo et al., 1991; Li-Leger et al., 2021). Other putative target genes of sly-miR166 are involved in cell cycle regulation, like cdc48 (Franz et al., 2014), rpt-2 (Papaevgeniou and Chondrogianni, 2014), and flub-2 (Haskell and Zinovyeva, 2021), nutrient availability (rpb-7, Collins et al., 2016), and gonad development (gon-1, Blelloch et al., 1999). Regarding sly-miR156a, this was predicted to target dhc1, encoding for the dynein heavy chain protein, and smrc-1, belonging to the Swi/snf chromatin remodeling complex. Dynein is an ATP-powered microtubule-based molecular motor, whose function includes the transport of cargo around the cell, while the loss-of-function of this gene inhibits apoptosis (Harders et al., 2018). The smrc-1 gene is involved in the protection against DNA replication stress and its loss-of-function leads to the accumulation of chronic replication stress (Yang et al., 2019). Finally, sly-miR319a was predicted to target the dlat-1 gene, encoding an enzyme with acetyltransferase activity; its lack of function leads to an early embryonic arrest (Li-Leger et al., 2021).
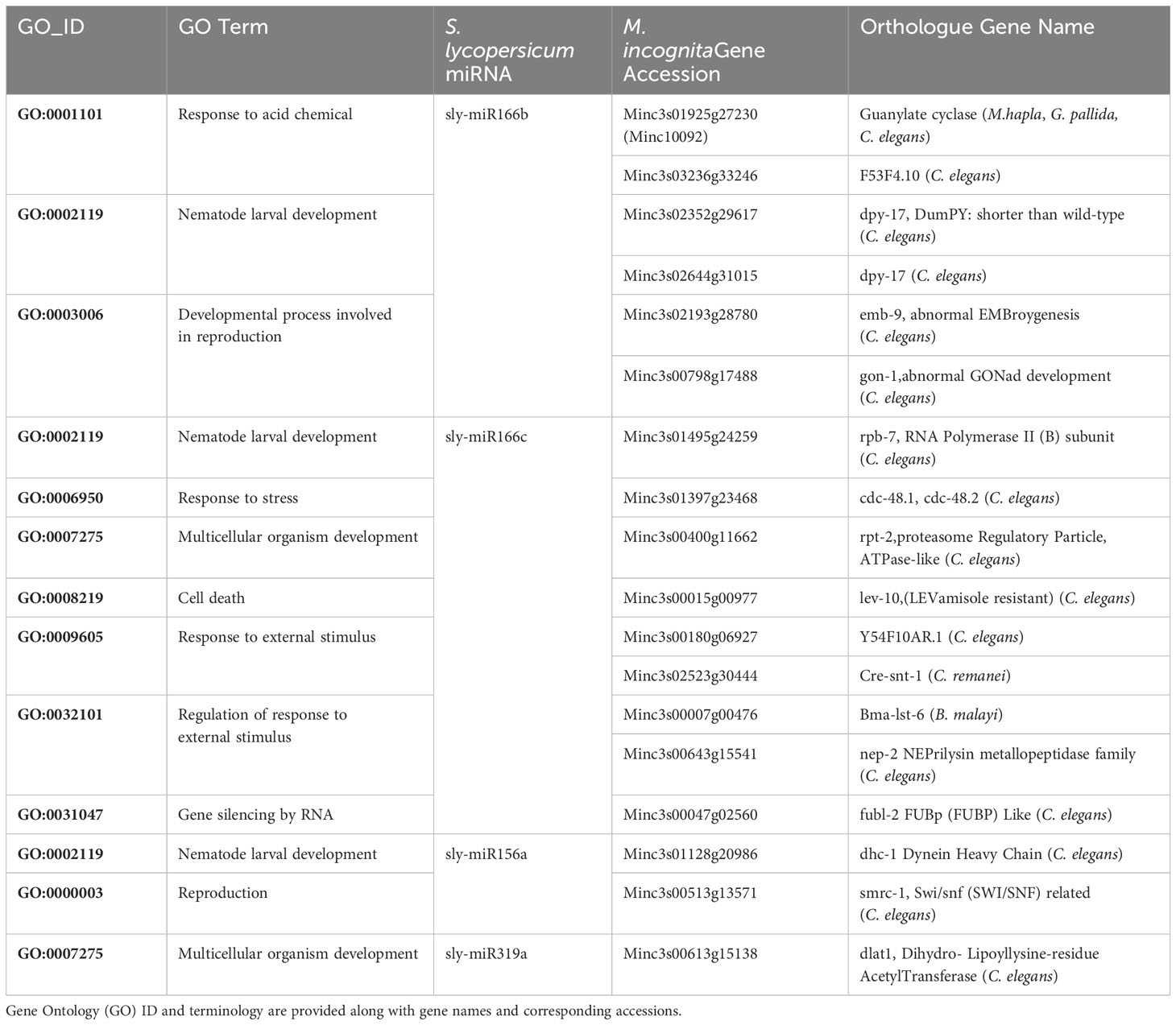
Table 2 List of representative S. lycopersicum miRNAs putatively targeting genes in M. incognita obtained from the cross-kingdom hybridization approach.
In the subsequent analysis, tomato miRNAs that share sequence homology with M. incognita miRNAs were analyzed via the seed region-based approach. Annotated orthologues of some predicted targets are collected in Table 3. Among the targeted biological processes, neurological development is much represented by putative targets such as tgs-1 (trimethyl guanosine synthase), gcy-9 (receptor-type guanylate cyclase), mig-6 (abnormal cell MIGration, papilin), kal-1 (human KALlmann syndrome homolog), as well as chemosensory genes like Mi-odr-1 (Minc3s00015g01026, Minc3s00056g02910). A recent study indicated that Mi-odr-1 is present in two copies in M. incognita, and was found to be expressed in the cell bodies of amphidal neurons and phasmids (Shivakumara et al., 2019). Silencing the Mi-odr and Mi-gpa genes could affect the nematode perception and infestation of the tomato root system (Li et al., 2022). Loss of tgs-1 function in C. elegans leads to neurological phenotypes similar to those caused by the survival motor neuron (SMN) deficiency (Chen et al., 2022) whereas gcy-9 mutants have different physiology in relation to adaptation and plasticity (Rossillo and Ringstad, 2020). The C. elegans kal-1 gene affects epidermal morphogenesis by regulating the development of the substrate neuroblasts, and kal-1 mutants show delayed migration of the ventral neuroblasts (Hudson et al., 2006). Loss-of-function mutants for the extracellular matrix molecule mig-6 result in defects in dendrite formation (Ramirez-Suarez et al., 2019). Other important genes, like unc-52 and cbp-1,2,3, are involved in larval development (Merz et al., 2003) and embryogenesis (Shi and Mello, 1998). Inhibition of these genes results in defects in myofilament assembly, larval movement deficiencies, or developmental arrest (Rogalski et al., 1995; Shi and Mello, 1998).
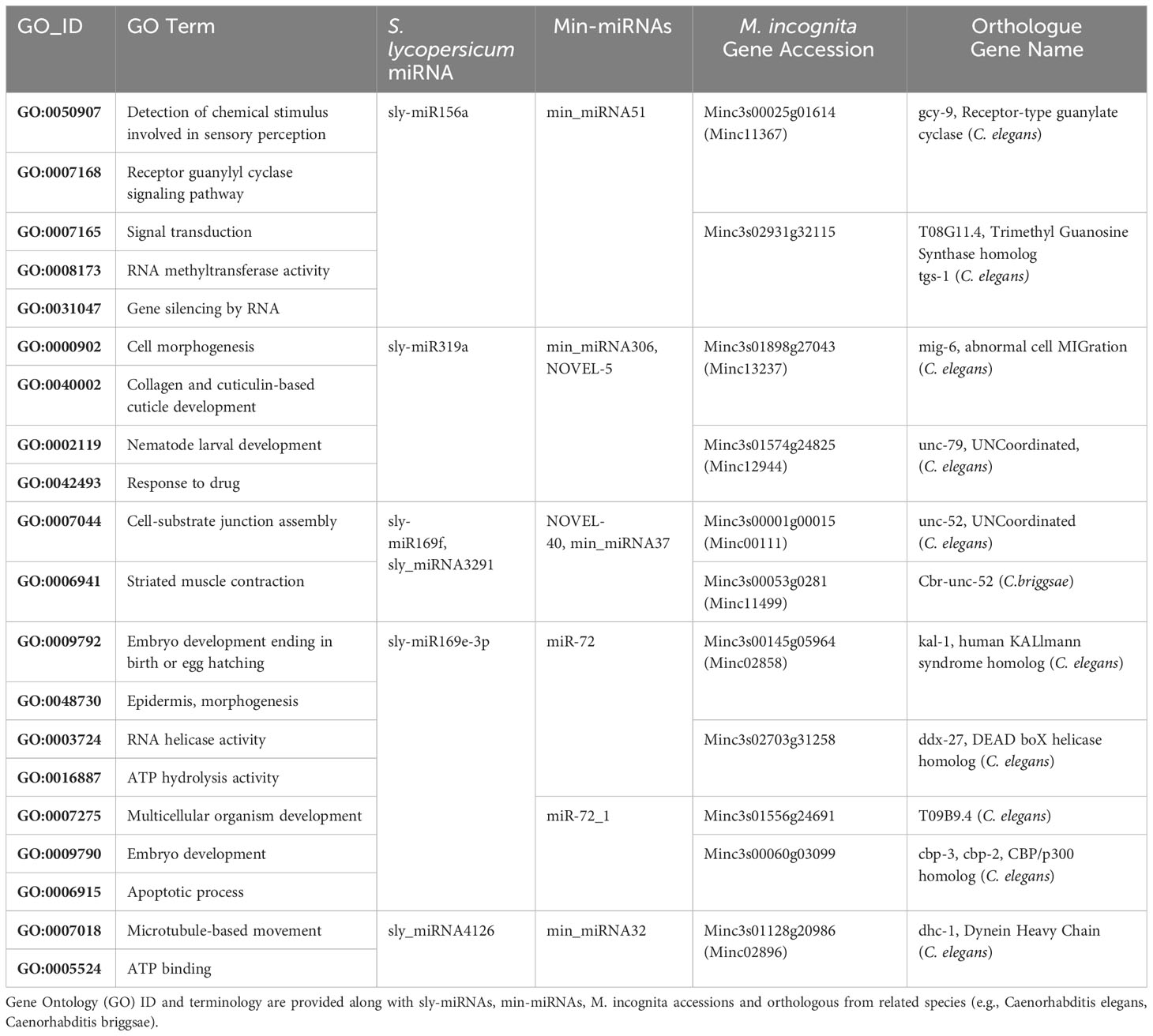
Table 3 List of some representative S. lycopersicum miRNAs putatively targeting genes in M. incognita obtained from the seed region-based search approach.
Hence, most M. incognita genes putatively targeted by sly-miRNAs have important roles in nematode development, leading to adverse effects on digestion, mobility, and reproduction, often with lethal outcomes. This finding is of utmost importance for the agricultural sector in view of developing plant miRNA-based technologies to control nematode diffusion.
When considering the use of different computational methods to predict miRNA targets in a cross-kingdom manner, such as the ones used in this work, it is important to underline that these rely on different assumptions on targeting rules and are still necessary to face our currently limited knowledge in miRNA trans-species interactions. Experimental confirmations are therefore needed to understand the actual targeting roles of the illustrated miRNAs. The availability of these computational methods is however useful to guide researchers in the selection of miRNA candidates for further investigation, despite the target prediction algorithm outcomes could be different due to the different underlying assumptions.
3.4 In vitro experimental validation of tomato miRNAs influencing M. incognita infection
To investigate the hypothesized cross-kingdom miRNAs transfer along with its potential to control the RKN infection, an in vitro experimental system was designed based on soaking assay, nematode larvae phenotyping, and gene expression profiling. To this purpose, J2 larvae were fed with solutions containing tomato miRNAs (sly-miR166b, sly-miR156a, sly-miR169f) selected from the bioinformatics data. The sly-miR166b - Minc3s01925g27230 (formerly named Minc10092) pair was chosen from the cross-kingdom hybridization approach (Table 2). No C. elegans homolog was found for this gene but a protein orthologue was identified as guanylate cyclase (UniProtKB/TrEMBL accession A0A1I8B0V8_MELHA) in M. hapla. The sly-miRNA156a - Minc3s00025g01614 (formerly named Minc11367) pair was selected from the seed-based approach (Table 3). This miRNA has a complete seed-region homology with min_miRNA51 targeting the Minc11367 gene, as shown in Supplementary Figure 1. In plants, it is well known that miR156 targets the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor, controlling genes involved in the regulation of reactive oxygen species (ROS) (Yin et al., 2019). In the RKN trans-kingdom approach, this sly-miRNA putative target was predicted as the C. elegans homologous gcy-9 (guanylyl cyclase, WBGene00001536). A homologue of this gene was also identified in the parasitic nematode Haemonchus contortus, a close relative of M. incognita (Winter et al., 2012). Lastly, sly-miR169f was predicted to target Minc3s00001g00015 (formerly named Minc00111, Supplementary Figure 2), an orthologue of the C. elegans unc-52 (WBGene00006787) gene. In the GO analysis, this accession was related to functions connected to cell junction organization or muscle system.
Following the selection of tomato miRNAs to be tested in the nematode system, an in vitro soaking assay was performed as described in Section 2.4. The motility and vitality of the J2 larvae were observed and compared with those of larvae kept in the control solution. Subsequently, these larvae were used to infect tomato seedlings. Figure 4 shows the effects of the infection symptoms classified as “+”, “++”, “+++”, along with galls formation on the roots. The values represent the percentage of symptoms normalized to the number of roots where the phenomena were observed. When the effects of the control solution and sly-miRNA156a soaked J2 larvae were compared, the best statistically significant reduction was detected in terms of gall formation. The amount of “+/roots”, “++/roots” and “+++/roots” was also reduced up to 1/3, while the number of “galls/roots” was reduced up to 1/4. A significant reduction of the same parameters was observed also for the J2s soaked with sly-miR169f, where root enlargements (“+/roots”, “++/roots”, “+++/roots”) were reduced up to 60% compared to control, and the number of “galls/roots” was halved. In the case of sly-miR166b, the observed reduction was less prominent, ranging from 15% (“+/roots”) to 25% (“galls/roots”).
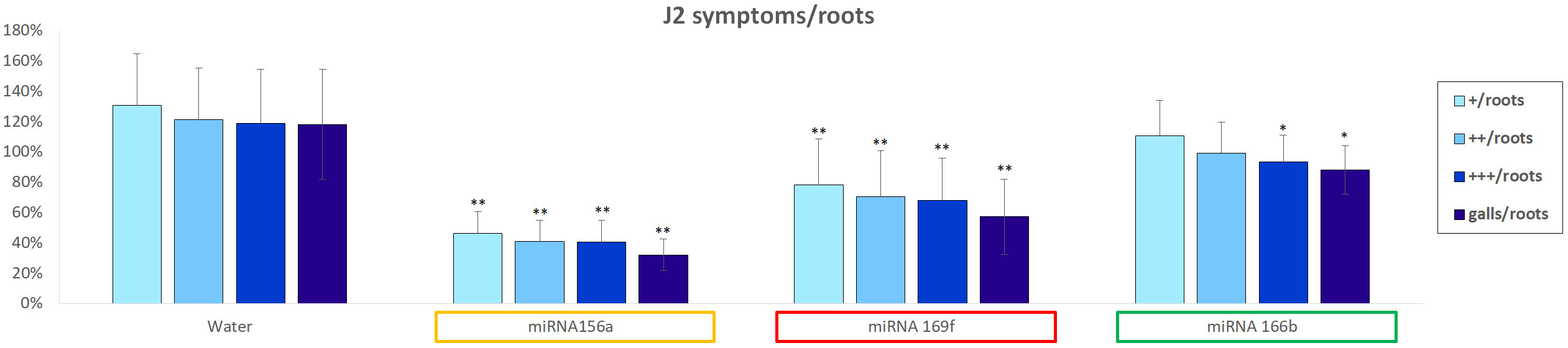
Figure 4 In vitro assay regarding soaked sly-miRNAs J2 larvae and susceptibility of the tomato root infections. The symptoms of infection (normalized with respect to the number of roots) with increasing apex enlargement, are categorized as “+”, “++”, “+++” and “galls”. Data represents percentages expressed compared to the soaked J2s control solution, expressed as mean ± SD of three independent experiments for each plant/miRNA. Statistical differences, in terms of Student’s t-test, are given (*P ≤ 0.05; **P ≤ 0.01). FDR, False Discovery Rate.
Because the most prominent reduction of the galling process was observed for the sly-miRNA156a and sly-miRNA169f soaked larvae, these were selected for further molecular analysis. Quantitative RealTime-PCR was employed to measure the relative expression of the Minc11367 (Minc3s00025g01614) and Minc00111(Minc3s00001g00015) genes, putatively targeted by these miRNAs. The obtained data show a downregulation of 54% in the miR156a-J2 and 29% in the miR169f-J2 treated larvae compared to the control (Figure 5). This result indirectly indicates that, in the in vitro experimental setup applied in this study, sly-miRNA156a and sly-miRNA169f have a cross-kingdom influence on the expression of Minc11367 and Minc00111 nematode genes.
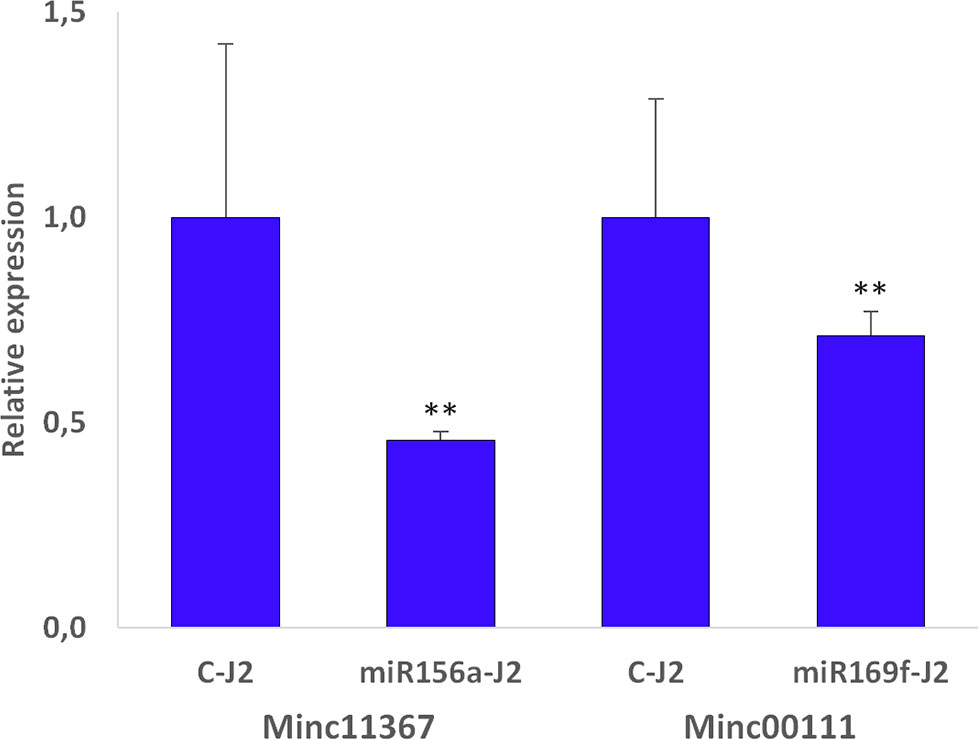
Figure 5 Relative expression data in miRNA156a and miRNA169f soaked J2s relative to control (C-J2). Transcript levels for Minc11367 (Minc3s00025g01614) and Minc00111 (Minc3s00001g00015) were measured by qRT-PCR after soaking treatment and compared to their transcript level in control samples. Data are shown as mean ± SD from two independent replicates. Statistical differences, in terms of Student’s t-test, are given (**P ≤ 0.01).
To summarize these findings, Figure 6 shows a schematic representation of how the in vitro sly-miRNAs soaking experiments performed on J2 larvae led to a decrease in the M. incognita infection symptoms along with the downregulation of putative cross-kingdom targets. To our knowledge, this is the first evidence showing that tomato miRNAs can be used to alter the infection ability of M. incognita larvae. Other studies related to the trans-kingdom transfer of tomato miRNAs to pathogens focused mainly on the Botrytis cinerea fungal infection. For instance, sly-miR1001 was shown to inhibit fungal virulence and conidiospore germination by targeting genes encoding for an ATP-dependent metallopeptidase and a cysteine-type endopeptidase (Meng et al., 2020). More recently, a genome-wide study identified multiple sRNAs and miRNAs with antifungal properties (Wu et al., 2023). The same study demonstrated that exogenous application of sly-miR396a was able to suppress the virulence of B. cinerea. Therefore, the data provided in our study together with other experimental evidence form the literature, evidence that tomato miRNAs can be effectively used to limit plant pathogen infection. However, given that most studies provide in vitro evidence to support this fact, in the future it would be required to further investigate how this transfer occurs and how is it conditioned by concentration and bioavailability in an in vivo system.
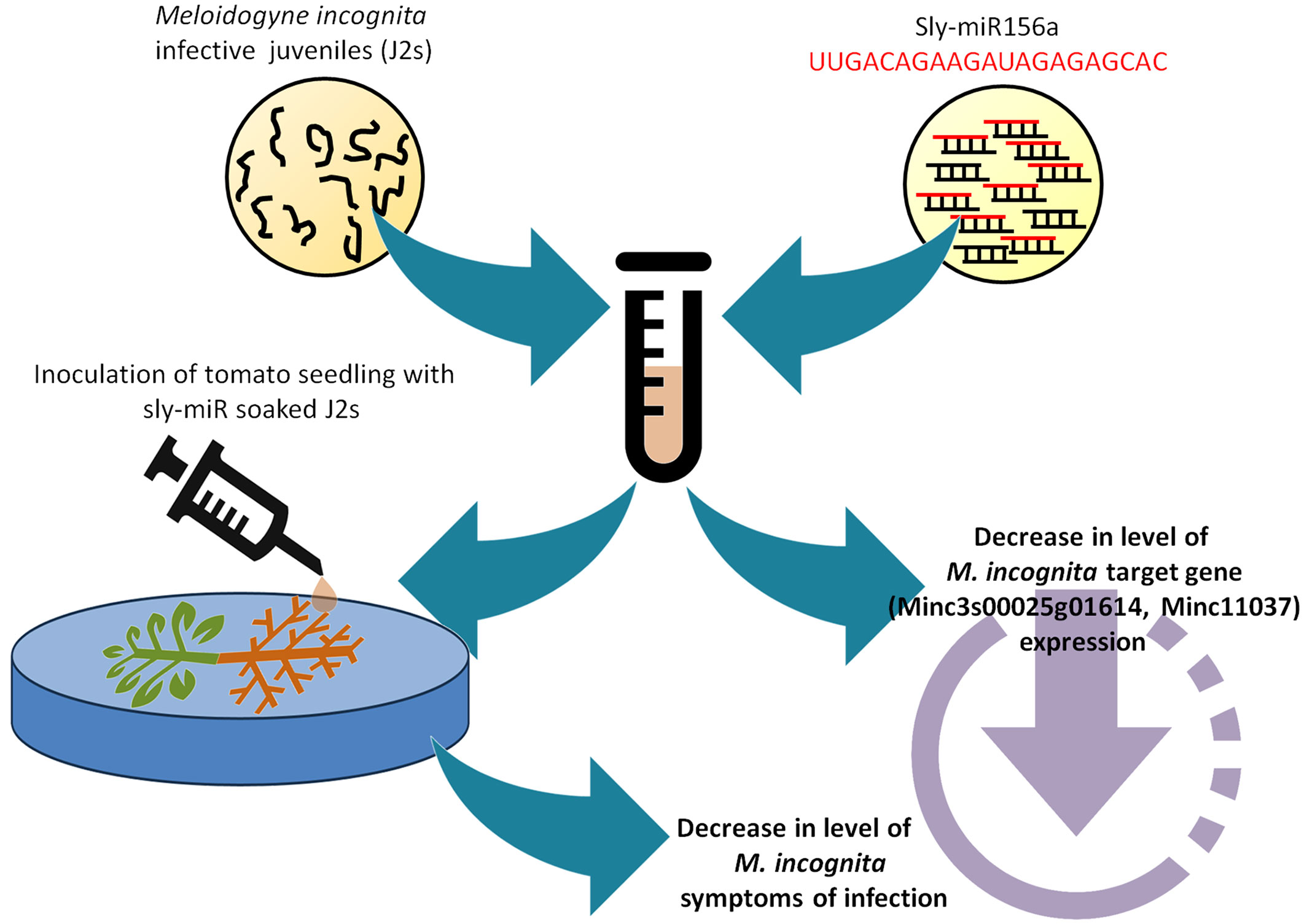
Figure 6 Schematic representation of the in vitro feeding assay showing the Sly-miRNAs soaking experiments performed on J2 larvae. The inoculation of tomato seedlings with sly-miR caused a decrease in the M. incognita infection symptoms and downregulation of putative cross-kingdom targets.
4 Conclusions
Following the increasing availability of omics data, bioinformatic studies are providing powerful tools to aid in setting up pertinent experimental designs based on preliminary predictions. This applies to miRNA-target interactions (both intra- and inter-specific) since different tools are available for target predictability based on sequence complementarity or hybridization energy. In this work, bidirectional bioinformatics analyses were conducted to uncover the potential miRNA-dependent cross-kingdom interactions between tomato and the phytoparasitic nematode M. incognita in terms of putative repressed genes and related biological processes. The obtained results are compatible with the host-parasite interactions between tomato and RKNs, suggesting that exogenous miRNAs may play a role in such processes. Although bioinformatics provides solid grounds to formulate hypotheses, some limitations are present considering the use of different computational methods, target prediction algorithms, and the lack of general guidelines applicable specifically for cross-kingdom predictions. Therefore, an in vitro experimental system was developed to support the potential cross-kingdom miRNAs effect during the S. lycopersicum - M. incognita interaction. The presented data evidence that the administration of sly-miR156a and sly-miR169f to J2s larvae was able to significantly lower root infection. This was also coupled with the downregulation of predicted cross-kingdom targets, Minc11367 and Minc00111. Thus, the current study expands the knowledge on the host-parasite interactions between tomato and RKNs paving the way for future application of exogenous miRNAs as tools to control M. incognita infection. However, this is still preliminary work and further analyses should be taken into account to understand the highly complex in vivo mechanism of plant miRNA-mediated gene control in the context of nematode research.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
PL: Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Data curation. DD: Writing – review & editing, Software. DM: Writing – review & editing, Software. PC: Writing – review & editing, Software. LP: Writing – original draft, Writing – review & editing, Conceptualization, Data curation. AM: Writing – original draft, Writing – review & editing, Conceptualization, Data curation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PL acknowledges the support from the Project NUTR-AGE (FOE-2021) DSBAD005.225.
Acknowledgments
PL would like to thank Campanale Antonia for the technical assistance during the in vitro experiments and Vitantonio Pantaleo for critical reading and discussions of the study. LP kindly thanks Gabriela Bindea (INSERM, Laboratory of Integrative Cancer Immunology, Université Paris Descartes, France) for the help provided for the ClueGO analysis on RKNs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1383986/full#supplementary-material
Supplementary Data Sheet 1 | Complete dataset regarding cross-kingdom prediction based on M. incognita miRNAs putatively targeting genes in S. lycopersicum. Information regarding miRNA and target sequence, perfect seed complementarity (no GU), seed complementarity (with GU), percentage (%) of the match without seed and GU, % match without seed but with GU, and multiplicity, are provided.
Supplementary Data Sheet 2 | Complete dataset generated using the cross-kingdom hybridization approach regarding S. lycopersicum miRNAs putatively targeting genes in M. incognita. Information regarding Minimum Free Energy (MFE), miRNA and target sequence, perfect seed complementarity (no GU), seed complementarity (with GU), percentage (%) of the match without seed and GU, % match without seed but with GU, multiplicity, and 3’ÚTR targeting, are provided.
Supplementary Data Sheet 3 | Complete dataset using the seed region-based search approach regarding S. lycopersicum miRNAs putatively targeting genes in M. incognita. Information regarding Minimum Free Energy (MFE), miRNA and target sequence, perfect seed complementarity (no GU), seed complementarity (with GU), percentage (%) of the match without seed and GU, % match without seed but with GU, multiplicity, and 3’ÚTR targeting, are provided.
Supplementary Data Sheet 4 | List of enriched S. lycopersicum Biological Process GO terms, with the relevant terms of the functionally grouped term networks provided by ClueGO shown in bold. Genes relative to each term and the miRNAs targeting each gene are shown.
Supplementary Data Sheet 5 | List of enriched M. incognita Biological Process GO terms, with the relevant terms of the functionally grouped term networks provided by ClueGO shown in bold. Genes relative to each term and the miRNAs targeting each gene are given.
References
Abad, P., Gouzy, J., Aury, J.-M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Avsar, B., Zhao, Y., Li, W., Lukiw, W. J. (2020). Atropa belladonna expresses a microRNA (aba-miRNA-9497) highly homologous to Homo sapiens miRNA-378 (hsa-miRNA-378); both miRNAs target the 3’-untranslated region (3’-UTR) of the mRNA encoding the neurologically relevant, zinc-finger transcription factor ZNF-691. Cell Mol. Neurobiol. 40, 179–188. doi: 10.1007/s10571-019-00729-w
Balakireva, A. V., Zamyatnin, A. A. (2018). Indispensable role of proteases in plant innate immunity. Int. J. Mol. Sci. 19, 629. doi: 10.3390/ijms19020629
Banakar, P., Hada, A., Papolu, P. K., Rao, U. (2020). Simultaneous RNAi knockdown of three FMRFamide-like peptide genes, Mi-flp1, Mi-flp12, and Mi-flp18 provides resistance to root-knot nematode, Meloidogyne incognita. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.573916
Banerjee, S., Gill, S. S., Gawade, B. H., Jain, P. K., Subramaniam, K., Sirohi, A. (2018). Host delivered RNAi of two cuticle collagen genes, Mi-col-1 and Lemmi-5 hampers structure and fecundity in Meloidogyne incognita. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02266
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Becraft, P. W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18, 163–192. doi: 10.1146/annurev.cellbio.18.012502.083431
Bellato, M., De Marchi, D., Gualtieri, C., Sauta, E., Magni, P., Macovei, A., et al. (2019). A bioinformatics approach to explore microRNAs as tools to bridge pathways between plants and animals. Is DNA Damage Response (DDR) a potential target process? Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01535
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally groupedgene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Blanc-Mathieu, R., Perfus-Barbeoch, L., Aury, J. M., Da Rocha, M., Gouzy, J., Sallet, E., et al. (2017). Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PloS Genet. 13, e1006777. doi: 10.1371/journal.pgen.1006777
Blelloch, R., Anna-Arriola, S. S., Gao, D., Li, Y., Hodgkin, J., Kimble, J. (1999). The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 216, 382–393. doi: 10.1006/dbio.1999.9491
Breitenbach, H. H., Wenig, M., Wittek, F., Jordá, L., Maldonado-Alconada, A. M., Sarioglu, H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol. 165, 791–809. doi: 10.1104/pp.114.239665
Cabrera, J., Diaz-Manzano, F. E., Barcala, M., Arganda-Carreras, I., de Almeida-Engler, J., Engler, G., et al. (2015). Phenotyping nematode feeding sites: Three-dimensional reconstruction and volumetric measurements of giant cells induced by root-knot nematodes in Arabidopsis. New Phytol. 206, 868–880. doi: 10.1111/nph.13249
Cai, C., Li, C., Sun, R., Zhang, B., Nichols, R. L., Hake, K. D., et al. (2021). Small RNA and degradome deep sequencing reveals important roles of microRNAs in cotton (Gossypium hirsutum L.) response to root-knot nematode Meloidogyne incognita infection. Genomics 113, 1146–1156. doi: 10.1016/j.ygeno.2021.02.018
Camborde, L., Raynaud, C., Dumas, B., Gaulin, E. (2019). DNA-damaging effectors: New players in the effector arena. Trends Plant Sci. 24, 1094–1101. doi: 10.1016/j.tplants.2019.09.012
Chen, L., Roake, C. M., Maccallini, P., Bavasso, F., Dehghannasiri, R., Santonicola, P., et al. (2022). TGS1 controls snRNA 3’ end processing, prevents neurodegeneration and ameliorates SMN-dependent neurological phenotypes in vivo. Nucleic Acids Res. 28, 12400–12424. doi: 10.1101/2020.10.27.356782
Chen, S., Hajirezaei, M., Peisker, M., Tschiersch, H., Sonnewald, U., Börnke., F. (2005). Decreased sucrose-6-phosphate phosphatase level in transgenic tobacco inhibits photosynthesis, alters carbohydrate partitioning, and reduces growth. Planta 221, 479–492. doi: 10.1007/s00425-004-1458-4
Chen, S., Hajirezaei, M. R., Zanor, M.-I., Hornyik, C., Debast, S., Lacomme, C., et al. (2008). RNA interference-mediated repression of sucrose-phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose-to-sucrose ratio upon cold storage with only minor effects on total soluble carbohydrate accumulation. Plant Cell Environ. 31, 165–176. doi: 10.1111/j.1365-3040.2007.01747.x
Cheng, G., Luo, R., Hu, C., Cao, J., Jin, Y. (2013). Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology 140, 1751–1761. doi: 10.1017/S0031182013000917
Chin, A. R., Fong, M. Y., Somlo, G., Wu, J., Swiderski, P., Wu, X., et al. (2016). Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 26, 217–228. doi: 10.1038/cr.2016.13
Collins, K. M., Bode, A. R., Fernandez, R. W., Tanis, J. E., Brewer, J. C., Creamer, M. S., et al. (2016). Activity of the C. elegans egg-laying behavior circuit is controlled by competing activation and feedback inhibition. eLife 5, e21126. doi: 10.7554/eLife.21126
Dai, X., Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Dalzell, J. J., McMaster, S., Fleming, C. C., Maule, A. G. (2010). Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol. 40, 91–100. doi: 10.1016/j.ijpara.2009.07.003
Danchin, E., Arguel, M., Campan-Fournier, A., Perfus-Barbeoch, L., Magliano, M., Rosso, M., et al. (2013). Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PloS Pathog., e1003745. doi: 10.1371/journal.ppat.1003745
Duplan, V., Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00042
EUROSTAT (2021). Available online at: https://ec.europa.eu/info/sites/default/files/food-farming-fisheries/farming/documents/tomatoes-production_en.pdf.
Franz, A., Ackermann, L., Hoppe, T. (2014). Create and preserve: proteostasis in development and aging is governed by Cdc48/p97/VCP. Biochim. Biophys. Acta 1843, 205–215. doi: 10.1016/j.bbamcr.2013.03.031
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gally, C., Eimer, S., Richmond, J. E., Bessereau, J. L. (2004). A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431, 578–582. doi: 10.1038/nature02893
Gualtieri, C., Gianella, M., Pagano, A., Cadeddu, T., Araújo, S., Balestrazzi, A., et al. (2021). Exploring microRNA signatures of DNA Damage Response using an innovative system of genotoxic stress in Medicago truncatula seedlings. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.645323
Gualtieri, C., Leonetti, P., Macovei, A. (2020). Plant miRNA cross-kingdom transfer targeting parasitic and mutualistic organisms as a tool to advance modern agriculture. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00930
Guo, X. D., Johnson, J. J., Kramer, J. M. (1991). Embryonic lethality caused by mutations in basement membrane collagen of C. elegans. Nature 349, 707–709. doi: 10.1038/349707a0
Harders, R. H., Morthorst, T. H., Lande, A. D., Hesselager, M. O., Mandrup, O. A., Bendixen, E., et al. (2018). Dynein links engulfment and execution of apoptosis via CED-4/Apaf1 in C. elegans. Cell Death Dis. 9, 1012. doi: 10.1038/s41419-018-1067-y
Haskell, D., Zinovyeva, A. (2021). KH domain containing RNA-binding proteins coordinate with microRNAs to regulate Caenorhabditis elegans development. G3 (Bethesda) 11, jkab013. doi: 10.1093/g3journal/jkab013
Hou, D., He, F., Ma, L., Cao, M., Zhou, Z., Wei, Z., et al. (2018). The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J. Nutr. Biochem. 57, 197–205. doi: 10.1016/j.jnutbio.2018.03.026
Howe, K. L., Bolt, B. J., Shafie, M., Kersey, P., Berriman, M. (2017). WormBase ParaSite - a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2–10. doi: 10.1016/j.molbiopara.2016.11.005
Hua, C., Zhao, J. H., Guo, H. S. (2018). Trans-kingdom RNA silencing in plant-fungal pathogen interactions. Mol. Plant 11, 235–244. doi: 10.1016/j.molp.2017.12.001
Hudson, M. L., Kinnunen, T., Cinar, H. N., Chisholm, A. D. (2006). C. elegans Kallmann syndrome protein KAL-1 interacts with syndecan and glypican to regulate neuronal cell migrations. Dev. Biol. 294, 352–365. doi: 10.1016/j.ydbio.2006.02.036
Iqbal, S., Fosu-Nyarko, J., Jones, M. G. K. (2020). Attempt to silence genes of the RNAi pathways of the root-knot nematode, Meloidogyne incognita results in diverse responses including increase and no change in expression of some genes. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00328
Jaubert-Possamai, S., Noureddine, Y., Favery, B. (2019). MicroRNAs, new players in the plant-nematode interaction. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01180
Kaloshian, I., Teixeira, M. (2019). Advances in plant-nematode interactions with emphasis on the notorious nematode genus Meloidogyne. Phytopathology 109, 1988–1996. doi: 10.1094/PHYTO-05-19-0163-IA
Kaur, P., Shukla, N., Joshi, G., VijayaKumar, C., Jagannath, A., Agarwal, M., et al. (2017). Genome-wide identification and characterization of miRNAome from tomato (Solanum lycopersicum) roots and root-knot nematode (Meloidogyne incognita) during susceptible interaction. PloS One 12, e0175178. doi: 10.1371/journal.pone.0175178
Kazan, K., Gardiner, D. M. (2017). Targeting pathogen sterols: Defence and counterdefence? PloS Pathog. 13, e1006297. doi: 10.1371/journal.ppat.1006297
Kleaveland, B. (2023). SnapShot: Target-directed miRNA degradation. Cell 25, 5674–5674. doi: 10.1016/j.cell.2023.11.020
Kozomara, A., Birgaoanu, M., Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kruger, J., Rehmsmeier, M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454. doi: 10.1093/nar/gkl243
LaMonte, G., Philip, N., Reardon, J., Lacsina, J. R., Majoros, W., Chapman, L., et al. (2012). Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12, 187–199. doi: 10.1016/j.chom.2012.06.007
Lang, A. E., Lundquist, E. A. (2021). The collagens DPY-17 and SQT-3 direct anterior-posterior migration of the Q neuroblasts in C. elegans. J. Dev. Biol. 9, 7. doi: 10.3390/jdb9010007
Leonetti, P., Molinari, S. (2020). Epigenetic and metabolic changes in root-knot nematode-plant interactions. Int. J. Mol. Sci. 21, 7759. doi: 10.3390/ijms21207759
Li, L., Liu, H., Wang, W., Chandra, M., Collins, B. M., Hu, Z. (2018). SNT-1 functions as the Ca2+ sensor for tonic and evoked neurotransmitter release in Caenorhabditis Elegans. J. Neurosci. 38, 5313–5324. doi: 10.1523/JNEUROSCI.3097-17.2018
Li, Y., Ren, Q., Bo, T., Mo, M., Liu, Y. (2022). AWA and ASH homologous sensing genes of Meloidogyne incognita contribute to the tomato infection process. Pathogens 11, 1322. doi: 10.3390/pathogens11111322
Li, Z., Xu, R., Li, N. (2018). MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. (Lond). 15, 68. doi: 10.1186/s12986-018-0305-8
Li-Leger, E., Feichtinger, R., Flibotte, S., Holzkamp, H., Schnabel, R., Moerman, D. G. (2021). Identification of essential genes in Caenorhabditis elegans through whole-genome sequencing of legacy mutant collections. G3 (Bethesda) 11, jkab328. doi: 10.1093/g3journal/jkab328
Liu, Y. C., Chen, W. L., Kung, W. H., Huang, H. D. (2017). Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genomics 18, 112. doi: 10.1186/s12864-017-3502-3
Liu, R., Xia, R., Xie, Q., Wu, Y. (2021). Endoplasmic reticulum-related E3 ubiquitin ligases: Key regulators of plant growth and stress responses. Plant Commun. 2, 100186. doi: 10.1016/j.xplc.2021.100186
Lukasik, A., Brzozowska, I., Zielenkiewicz, U., Zielenkiewicz, P. (2018). Detection of plant miRNAs abundance in human breast milk. Int. J. Mol. Sci. 19, E37. doi: 10.3390/ijms19010037
Macovei, A., Rubio-Somoza, I., Paiva, J. A. P., Araújo, S., Donà, M. (2021). Editorial: MicroRNA signatures in plant genome stability and genotoxic stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.683302
Mal, C., Aftabuddin, M., Kundu, S. (2018). IIKmTA: inter and intra kingdom miRNA-target analyzer. Interdiscip. Sci. 10, 538–543. doi: 10.1007/s12539-018-0291-6
Maruyama, I. N. (2017). Receptor guanylyl cyclases in sensory processing. Front. Endocrinol. (Lausanne) 7. doi: 10.3389/fendo.2016.00173
Melillo, M. T., Leonetti, P., Bongiovanni, M., Castagnone-Sereno, P., Bleve-Zacheo, T. (2006). Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. New Phytol. 170, 501–512. doi: 10.1111/j.1469-8137.2006.01724.x
Meng, X., Jin, W., Wu, F. (2020). Novel tomato miRNA miR1001 initiates cross-species regulation to suppress the conidiospore germination and infection virulence of Botrytis cinerea in vitro. Gene 759, 145002. doi: 10.1016/j.gene.2020.145002
Merz, D. C., Alves, G., Kawano, T., Zheng, H., Culotti, J. G. (2003). UNC-52/perlecan affects gonadal leader cell migrations in C. elegans hermaphrodites through alterations in growth factor signaling. Dev. Biol. 256, 173–186. doi: 10.1016/S0012-1606(03)00014-9
Molinari, S., Miacola, C. (1997). Antioxidant enzymes in phytoparasitic nematodes. J. Nematol., 153–159.
Molinari, S., Fanelli, E., Leonetti, P. (2014). Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 15, 255–264. doi: 10.1111/mpp.12085
Molinari, S., Leonetti, P. (2019). Bio-control agents activate plant immune response and prime susceptible tomato against root-knot nematodes. PloS One 14, e0213230. doi: 10.1371/journal.pone.0213230
Palomares-Rius, J. E., Hasegawa, K., Siddique, S., Vicente, C. S. L. (2021). Editorial: Protecting our crops - Approaches for plant parasitic nematode control. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.726057
Papaevgeniou, N., Chondrogianni, N. (2014). The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol. 2, 333–347. doi: 10.1016/j.redox.2014.01.007
Parry, G. (2013). Assessing the function of the plant nuclear pore complex and the search for specificity. J. Exp. Bot. 64, 833–845. doi: 10.1093/jxb/ers289
Pastuglia, M., Swarup, R., Rocher, A., Saindrenan, P., Roby, D., Dumas, C., et al. (2002). Comparison of the expression patterns of two small gene families of S gene family receptor kinase genes during the defence response in Brassica oleracea and Arabidopsis thaliana. Gene 282, 215–225. doi: 10.1016/S0378-1119(01)00821-6
Paul, M. J. (2008). Trehalose 6-phosphate: a signal of sucrose status. Biochem. J. 412, e1–e2. doi: 10.1042/BJ20080598
Rabuma, T., Gupta, O. P., Chhokar, V. (2022). Recent advances and potential applications of cross-kingdom movement of miRNAs in modulating plant’s disease response. RNA Biol. 19, 519–532. doi: 10.1080/15476286.2022.2062172
Ramirez-Suarez, N. J., Belalcazar, H. M., Salazar, C. J., Beyaz, B., Raja, B., Nguyen, K. C. Q., et al. (2019). Axon-dependent patterning and maintenance of somatosensory dendritic arbors. Dev. Cell. 48, 229–244.e4. doi: 10.1016/j.devcel.2018.12.015
Rogalski, T. M., Gilchrist, E. J., Mullen, G. P., Moerman, D. G. (1995). Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics 139, 159–169. doi: 10.1093/genetics/139.1.159
Rossillo, M., Ringstad, N. (2020). Development of specialized sensory neurons engages a nuclear receptor required for functional plasticity. Genes Dev. 34, 1666–1679. doi: 10.1101/gad.342212.120
Seid, A., Fininsa, C., Mekete, T., Decraemer, W., Wesemael, W. M. L. (2015). Tomato (Solanum lycopersicum) and root-knot nematodes (Meloidogyne spp.) – a century-old battle. Nematology 17, 995–1009. doi: 10.1163/15685411-00002935
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, Y., Mello, C. (1998). A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12, 943–955. doi: 10.1101/gad.12.7.943
Shivakumara, T. N., Dutta, T. K., Chaudhary, S., von Reuss, S. H., Williamson, V. M., Rao, U. (2019). Homologs of Caenorhabditis elegans chemosensory genes have roles in behavior and chemotaxis in the root-knot nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 32, 876–887. doi: 10.1094/MPMI-08-18-0226-R
Shu, J., Chiang, K., Zempleni, J., Cui, J. (2015). Computational characterization of exogenous microRNAs that can be transferred into human circulation. PloS One 10, e0140587. doi: 10.1371/journal.pone.0140587
Singh, S., Singh, B., Singh, A. P. (2015). Nematodes: A threat to sustainability of agriculture. Proc. Environ. Sci. 29, 215–216. doi: 10.1016/j.proenv.2015.07.270
Soh, M. S., Cheng, X., Vijayaraghavan, T., Vernon, A., Liu, J., Neumann, B. (2020). Disruption of genes associated with Charcot-Marie-Tooth type 2 lead to common behavioural, cellular and molecular defects in Caenorhabditis elegans. PloS One 15, e0231600. doi: 10.1371/journal.pone.0231600
Somvanshi, V. S., Ghosh, O., Budhwar, R., Dubay, B., Shukla, R. N., Rao, U. (2018). A comprehensive annotation for the root-knot nematode Meloidogyne incognita proteome data. Data Brief. 19, 1073–1079. doi: 10.1016/j.dib.2018.05.131
Suárez, R., Wong, A., Ramírez, M., Barraza, A., Del Carmen Orozco, M., Cevallos, M. A., et al. (2008). Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in Rhizobia. Mol. Plant Microbe Interact. J. 21, 958–966. doi: 10.1094/MPMI-21-7-0958
Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A., Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916. doi: 10.1038/35002628
Tan, J. A., Jones, M. G., Fosu-Nyarko, J. (2013). Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 133, 166–178. doi: 10.1016/j.exppara.2012.11.011
Wang, W., Liu, D., Zhang, X., Chen, D., Cheng, Y., Shen, F. (2018). Plant microRNAs in cross-kingdom regulation of gene expression. Int. J. Mol. Sci. 19, E2007. doi: 10.3390/ijms19072007
Wang, Y., Mao, Z., Yan, J., Cheng, X., Liu, F., Xiao, L., et al. (2015). Identification of microRNAs in Meloidogyne incognita using deep sequencing. PloS One 10, e0133491. doi: 10.1371/journal.pone.0133491
Wang, Y., Mortimer, J. C., Davis, J., Dupree, P., Keegstra, K. (2013). Identification of an additional protein involved in mannan biosynthesis. Plant J. 73, 105–117. doi: 10.1111/tpj.12019
Wang, B., Sun, Y., Song, N., Zhao, M., Liu, R., Feng, H., et al. (2017). Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 215, 338–350. doi: 10.1111/nph.14577
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants. 2, 16151. doi: 10.1038/nplants.2016.151
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Winter, A. D., Weir, W., Hunt, M., Berriman, M., Gilleard, J. S., Devaney, E., et al. (2012). Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genomics 13, 4. doi: 10.1186/1471-2164-13-4
Wu, F., Huang, Y., Jiang, W., Jin, W. (2023). Genome-wide identification and validation of tomato-encoded sRNA as the cross-species antifungal factors targeting the virulence genes of Botrytis cinerea. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1072181
Yang, F., Ding, L., Zhao, D., Fan, H., Zhu, X., Wang, Y., et al. (2022). Identification and functional analysis of tomato microRNAs in the biocontrol Bacterium pseudomonas putida induced plant resistance to Meloidogyne incognita. Phytopathology 112, 2372–2382. doi: 10.1094/PHYTO-03-21-0101-R
Yang, B., Xu, X., Russell, L., Sullenberger, M. T., Yanowitz, J. L., Maine, E. M. (2019). A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line. PloS Genet. 15, e1007992. doi: 10.1371/journal.pgen.1007992
Yang, F., Zhao, D., Fan, H., Zhu, X., Wang, Y., Liu, X., et al. (2020). Functional analysis of long non-coding RNAs reveal their novel roles in biocontrol of bacteria-induced tomato resistance to Meloidogyne incognita. Int. J. Mol. Sci. 21, 911. doi: 10.3390/ijms21030911
Yin, H., Hong, G., Li, L., Zhang, X., Kong, Y., Sun, Z., et al. (2019). miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 109, 632–642. doi: 10.1094/PHYTO-08-18-0306-R
Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126. doi: 10.1038/cr.2011.158
Zhang, L. L., Jing, X. D., Chen, W., Wang, Y., Lin, J., Zheng, L., et al. (2019). Host plant-derived miRNAs potentially modulate the development of a cosmopolitan insect pest, Plutella xylostella. Biomolecules 9, 602. doi: 10.3390/biom9100602
Zhang, H., Li, Y., Liu, Y., Liu, H., Wang, H., Jin, W., et al. (2016). Role of plant microRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 10, 60. doi: 10.1186/s12918-016-0292-1
Zhang, B., Li, W., Zhang, J., Wang, L., Wu, J. (2019). Roles of small RNAs in virus-plant interactions. Viruses 11, 827. doi: 10.3390/v11090827
Zhang, Y., Wang, Y., Xie, F., et al. (2016). Identification and characterization of microRNAs in the plant-parasitic root-knot nematode Meloidogyne incognita using deep sequencing. Funct. Integr. Genomics 16, 127–142. doi: 10.1007/s10142-015-0472-x
Zhao, Q., Mao, Q., Zhao, Z., Dou, T., Wang, Z., Cui, X., et al. (2018). Prediction of plant-derived xenomiRs from plant miRNA sequences using random forest and one-dimensional convolutional neural network models. BMC Genomics 19, 839. doi: 10.1186/s12864-018-5227-3
Keywords: bioinformatics pipeline, miRNAs, cross-species target prediction, plant-pathogen interaction, tomato, root-knot nematode
Citation: Leonetti P, Dallera D, De Marchi D, Candito P, Pasotti L and Macovei A (2024) Exploring the putative microRNAs cross-kingdom transfer in Solanum lycopersicum-Meloidogyne incognita interactions. Front. Plant Sci. 15:1383986. doi: 10.3389/fpls.2024.1383986
Received: 08 February 2024; Accepted: 22 April 2024;
Published: 08 May 2024.
Edited by:
Andressa Machado, Agronema, BrazilReviewed by:
Raghavendra Aminedi, Indian Council of Agricultural Research (ICAR), IndiaAbdelfattah A. Dababat, International Maize and Wheat Improvement Center, Mexico
Copyright © 2024 Leonetti, Dallera, De Marchi, Candito, Pasotti and Macovei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Leonetti, cGFvbGEubGVvbmV0dGlAY25yLml0; Lorenzo Pasotti, bG9yZW56by5wYXNvdHRpQHVuaXB2Lml0; Anca Macovei, YW5jYS5tYWNvdmVpQHVuaXB2Lml0
 Paola Leonetti
Paola Leonetti Debora Dallera
Debora Dallera Davide De Marchi
Davide De Marchi Pamela Candito2
Pamela Candito2 Lorenzo Pasotti
Lorenzo Pasotti Anca Macovei
Anca Macovei