
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 April 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1381018
This article is part of the Research TopicHighlights from the 12th Plant Growth-Promoting Rhizobacteria WorkshopView all 12 articles
 Luotao Wang1†
Luotao Wang1† Jianquan Huang2†
Jianquan Huang2† Si Chen1
Si Chen1 Xin Su1
Xin Su1 Xun Zhang3
Xun Zhang3 Lujun Wang4
Lujun Wang4 Wei Zhang4
Wei Zhang4 Zhenshuo Wang1
Zhenshuo Wang1 Qingchao Zeng1
Qingchao Zeng1 Qi Wang1
Qi Wang1 Yan Li1*
Yan Li1*Autolysins are endogenous cell wall degrading enzymes (CWDEs) in bacteria that remodel the peptidoglycan layer of its own cell wall. In the Bacillus subtilis genome, at least 35 autolysin genes have been identified. However, the study of their roles in bacterial physiology has been hampered by their complexity and functional redundancy. B. subtilis GLB191 is an effective biocontrol strain against grape downy mildew disease, the biocontrol effect of which results from both direct effect against the pathogen and stimulation of the plant defense. In this study, we show that the autolysin N-acetylglucosaminidase LytD, a major autolysin of vegetative growth in B. subtilis, plays an important role in its biocontrol activity against grape downy mildew. Disruption of lytD resulted in reduced suppression of the pathogen Plasmopara viticola and stimulation of the plant defense. LytD is also shown to affect the biofilm formation and colonization of B. subtilis on grape leaves. This is the first report that demonstrates the role of an endogenous CWDE in suppressing plant disease infection of a biological control microorganism. These findings not only expand our knowledge on the biological function of autolysins but also provide a new target to promote the biocontrol activity of B. subtilis.
As the demand increases for more environmentally friendly disease control alternatives to the massive use of chemical pesticides, the interest in identifying and developing effective biological control agents of plant diseases has significantly increased in the last decades (Koch et al., 2018; Collinge et al., 2022; Lahlali et al., 2022). Bacteria from genus Bacillus are considered as one of the most important microorganisms in biological control against plant diseases of various crops (Fira et al., 2018; Villarreal-Delgado et al., 2018; Etesami et al., 2023). Besides the high efficacy against phytopathogens, additional advantages of using these bacteria include the ease of culture, storage, and manufacture due to the ability of Bacillus to produce endospores (Biermann and Beutel, 2023). In recent years, commercial formulated Bacillus-based biological pesticides have been increasingly used to control various plant diseases worldwide. To date, B. subtilis is among the most exploited species from the genus (Miljaković et al., 2020).
B. subtilis has been reported to be effective in controlling plant diseases through diverse mechanisms, including both direct and indirect mechanisms. Numerous studies revealed that B. subtilis protects plants against pathogens via a direct antagonistic effect on the pathogens by producing diverse antimicrobial compounds. The three families of Bacillus cyclic lipopeptides (CLPs)—surfactins, iturins, and fengycins—were mostly studied as bioactive compounds due to their antagonistic activity for a wide range of potential phytopathogens, including bacteria, fungi, and oomycetes (Ongena and Jacques, 2008). The antimicrobial activity of B. subtilis could also be due to the production of cell wall degrading enzymes (CWDEs) such as chitinases, glucanases, and proteases, which efficiently hydrolyze the major components of the fungal and bacterial cell walls and limit their growth or activity (Miljaković et al., 2020). The biocontrol activity exhibited by B. subtilis can also be attributed to indirect mechanisms, including biofilm formation, plant growth promotion (PGP), competition for nutrients and colonization sites, and induced systemic resistance (ISR) (Hashem et al., 2019). Besides antimicrobial activity, Bacillus CLPs also influence the colonization and have a key role in the beneficial interaction of B. subtilis with plants by stimulating host defense mechanisms (Ongena and Jacques, 2008; Fan et al., 2017). B. subtilis forms biofilms on the plant, which help to produce lipopeptides and augment their activity (Zeriouh et al., 2014). In addition, B. subtilis could also secrete CWDEs, such as cellulases, hemicellulases, and pectinases, to degrade plant cell wall. The release of cell-wall-derived oligosaccharides with danger-associated molecular pattern (DAMP) capacity could trigger a signaling cascade that leads to the induction of defense responses (Héloir et al., 2019; Miljaković et al., 2020).
One prerequisite of effective biocontrol is the successful colonization of the biocontrol microorganism on plants. Bacteria associated with plant leaves employ a variety of colonization strategies. Steps in these colonization strategies include modification of their environment on leaf, aggregation, ingression, and egression. The formation of bacterial aggregates may enhance the local habitat modification on the surface of leaves (Beattie and Lindow, 1999). In nature, approximately 70% of bacteria live in the form of aggregates on leaves, which confers them a selective advantage for survival and colonization (Monier and Lindow, 2003). Biofilms, which are communities of aggregated cells embedded in a self-produced extracellular polymeric matrix, are critical for Bacillus spp. colonization efficiency and thus the suppression of pathogens (Pandin et al., 2017; Fessia et al., 2022).
The cell wall, which can be found in archaea, bacteria, fungi, plants, and algae, is a complex and selectively permeable layer that surrounds the cell with main functions to provide protection structure and support to the cell (Keegstra, 2010; Free, 2013; Giovannoni et al., 2020). Therefore, the cell wall is resistant to various biotic and abiotic stresses. In this respect, “attacking” organisms evolved various enzymes specialized in cell wall degradation, which are known as exogenous CWDEs. Additionally, organisms contain endogenous CWDEs to remodel their own cell wall structures during development (Giovannoni et al., 2020). These degrading enzymes in bacteria are also collectively referred to as autolysins, which are found in all bacteria that digest the shape-maintaining and stress-bearing peptidoglycan layer of its own cell wall (Smith et al., 2000; Nayyab et al., 2017). Despite extensive researches on CWDEs, much of them focused on exogenous CWDEs, which evolved from organisms to degrade the cell wall of their targets. Few studies, especially in bacteria, have investigated the physiological functions of endogenous CWDEs that remodel their own cell wall structures during development.
Peptidoglycan, the major structure component of bacterial cell wall and the substrate of autolysins, is an alternating heteropolymer of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) with pentapeptide side chains branching from the MurNAc residues. It is a dynamic structure continually being synthesized, modified, and hydrolyzed to allow for cell growth and division (Angeles and Scheffers, 2021). In the B. subtilis genome, at least 35 autolysin genes have been identified. The autolysins can be divided into four classes based on their cleave sites: N-acetylglucosaminidases, muramidases, acetyl-muramyl-L-alanine amidases (amidases), and endopeptidases (Ghuysen et al., 1966; Smith et al., 2000; Nayyab et al., 2017; Angeles and Scheffers, 2021). Among them, N-acetylglucosaminidase LytD (or CwlG) and amidase LytC (or CwlB) are two major autolysins of vegetative growth in B. subtilis, and these two autolysins bear 95% of the autolytic activity of the cells (Smith et al., 2000; Horsburgh et al., 2003; Ren et al., 2022). The autolysins associated with B. subtilis were shown to play a role in several important cellular functions, including differentiation, cell lysis, cell wall growth and turnover, cell separation, competence, and motility (Smith et al., 2000). However, the study of their physiological roles has been hampered by their complexity and functional redundancy.
LytD is a key N-acetylglucosaminidase acting on peptidoglycan in B. subtilis (Smith et al., 2000; Horsburgh et al., 2003; Nayyab et al., 2017). Previous reports showed that inactivation of lytD alone did not affect cell separation, autolysis, cell wall turnover, growth rate, competence, and sporulation (Margot et al., 1994). It is required for the motility function but only in concert with LytC (Rashid et al., 1995). Despite the fact that LytD has been identified for many years, its biological function remains unclear. B. subtilis GLB191 is an efficient biocontrol strain against grape downy mildew disease caused by the pathogen Plasmopara viticola (Zhang et al., 2017). Our previous report showed that the biocontrol effect of B. subtilis GLB191 on grape downy mildew disease is through direct suppression of the pathogen and through stimulating the plant defense, by secreting secondary metabolites fengycin and surfactin (Li et al., 2019). In this study, the role of the N-acetylglucosaminidase LytD in the biological control of B. subtilis GLB191 against grape downy mildew was investigated. We found that LytD affected the direct effect of B. subtilis against the pathogen P. viticola, the stimulation of the plant defenses, colonization on grape leaves, and, consequently, biocontrol activity against grape downy mildew. These findings not only expand our knowledge on the biological function of autolysins but also provide a new target for improving the biocontrol activity of Bacillus and other beneficial microorganisms.
The grapevine cultivar Vitis vinifera L. cv. Red Globe, susceptible to P. viticola, was used in this study. Plant produced from herbaceous cuttings were planted in individual pots at 26 and 18°C (day and night, respectively) with a photoperiod of 16 h of light. The second and third youngest fully expanded leaves were used in the experiments.
The bacterial strains and plasmids used in this study are listed in Table 1. B. subtilis GLB191 and its derivatives were routinely grown in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, and 1% NaCl) or on LB agar medium supplemented with 1.5% (w/v) agar at 37°C. For experiments, they were grown in handmade potato dextrose broth (PDB, potato broth supplemented with 1.5% dextrose) medium at 37°C with shaking at 180 r min−1 for 48 h. For the biofilm formation assay, B. subtilis was cultivated at 30°C in a minimal salts glutamate glycerol (MSgg) medium that was designed to induce biofilm formation. MSgg is composed of 5 mM potassium phosphate (pH 7.0), 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS, pH 7.0), 2 mM MgCl2, 50 μM MnCl2, 1 μM ZnCl2, 50 μM FeCl3, 700 μM CaCl2, 2 μM thiamine, 0.5% glutamic acid, 50 μg mL−1 phenylalanine, 50 μg mL−1 threonine, 50 μg mL−1 tryptophan, and 0.5% glycerol (Branda et al., 2001). Escherichia coli strains were grown in LB broth at 37°C. When appropriate, antibiotics were added at the following concentrations: 100 μg mL−1 of ampicillin (Amp) for growth of E. coli, 5 μg mL−1 of erythromycin (Em), and 10 μg mL−1 of tetracycline (Tet) for growth of B. subtilis. Primers used for PCR in this report are summarized in Supplementary Table S1.
The markerless lytD deletion mutant (ΔlytD) was constructed by marker exchange mutagenesis using the temperature-sensitive suicide plasmid pMAD as described previously (Li et al., 2019). The flanking regions of lytD (GenBank No. PP434428) were amplified from the genomic DNA of GLB191 by polymerase chain reaction (PCR) using the primer pairs lytD-up-F/lytD-up-R and lytD-dn-F/lytD-dn-R, respectively. The two DNA fragments were joined together by overlap-extension PCR using the primers lytD-up-F and lytD-dn-R (Senanayake and Brian, 1995). The resulting fragment was digested and cloned into pMAD, generating pMAD-lytD in E. coli DH5α. The pMAD-lytD plasmid was purified from E. coli DH5α and mobilized into E. coli EC135 by heat shock and then into GLB191 by electroporation (Zhang et al., 2017). Blue transformants were obtained after incubation at 30°C for 2 days on LB agar plates containing Em and X-Gal (40 μg mL−1), followed by incubation in LB broth containing Em at 42°C with shaking at 180 r min−1 for 8–10 h for the first allelic exchange. Em-resistant and blue transformants were obtained from LB agar plates supplemented with Em and X-Gal and then incubated in LB broth at 25°C with shaking at 180 r min−1 for 24 h for the second allelic exchange. Em-sensitive and white clones were picked and confirmed by PCR amplification and subsequently DNA sequencing to verify the deletion of lytD on the chromosome using primers lytD-L and lytD-D. The primers used are listed in Supplementary Table S1.
To complement the ΔlytD mutation with the wild-type lytD gene, the native promoter and the open reading frame of lytD (GenBank No. PP434428) were amplified using primers lytD-F/lytD-R and then inserted into pHY300PKL, a shuttle plasmid that is replicable in B. subtilis, generating the complementary plasmid pHY-lytD. pHY-lytD was subsequently transformed into ΔlytD by electroporation, resulting in complementary strain ΔlytD (pHY-lytD). pHY300PKL was also transformed into ΔlytD and GLB191, respectively, by electroporation to yield strains ΔlytD(pHY300) and GLB191(pHY300). Transformants were then selected on 10 μg mL−1 Tet-containing LB plates and verified by PCR and DNA sequencing.
The P. viticola isolate used for this study was collected from diseased leaves of Muscat Hamburg in the vineyard in Tianjin Academy of Agricultural Sciences and identified by the analysis of the sequence of internal transcribed spacer regions 1 (ITS) (GenBank No. PP413393) as previously described (Rouxel et al., 2013). The P. viticola isolate was maintained on Red Globe plants in the glasshouse as previously described (Trouvelot et al., 2008). Sporulation was induced after incubation of plants presenting oily spot symptoms in the dark at >95% relative humidity (RH) overnight. Sporangia were then collected from the lower side of leaves using a brush and suspended in distilled water. The concentration was adjusted to 105 sporangia mL−1 using a hemacytometer.
Leaf disc bioassay was carried out to test the biocontrol activity of the bacteria against grape downy mildew. The second and third youngest fully expanded leaves were detached from plants grown in the greenhouse and rinsed with water. Leaf discs of 1 cm diameter were punched from leaves with a cork borer. Discs were soaked in the 48-h culture of GLB191 or its derivatives for 30 min and then plated onto wet paper in Petri dishes with the abaxial side up and air-dried for 2 h. For mock inoculation used as control, the leaf discs were sprayed with PDB medium. One day post-treatment (dpt) with bacterial culture or PDB medium, discs were sprayed with P. viticola sporangia suspension at 105 sporangia mL−1. Petri dishes were placed overnight in a humid chamber (RH > 95%) and then moved back to the growth chamber at 22°C at a day length of 16 h. Nine discs for each replicate and four replicates for each treatment were used. Three independent biological repeats were conducted.
Disease index (DI) was investigated 5–7 days post-inoculation (dpi). The severity was assessed using the following 0–9 scale as previously described: 0, no lesions; 1, 0%–5% of the leaf disc area infected and sporulating; 3, 6%–25%; 5, 26%–50%; 7, 51%–75%; and 9, more than 76% leaf area infected and sporulating (Zhang et al., 2017).
Freshly prepared sporangial suspension of P. viticola was used to detect the direct effect of bacteria on the pathogen in vitro. P. viticola sporangial suspension (500 µL) was incubated with 500 µL of bacterial culture or PDB in 48-well plates in the dark at 18°C until over 50% of sporangia released zoospores for the wild-type strain. Percentage of sporangia released zoospores in the total number of sporangia was visually determined by optical microscopic observation of three different microscopic fields (×400). Three observations for each replicate and three replicates for each treatment were conducted. Counting was repeated on three independent experiments.
Direct effects were also determined on leaves previously treated with bacterial culture as previously described (Li et al., 2019). The lower face of leaves that were treated with bacterial culture or PDB was inoculated with a freshly prepared sporangial suspension (2 × 105 sporangia mL−1) 2 h post-treatment (hpt). Leaves were harvested 24 h post-inoculation (hpi) and discs of 0.5 cm diameter were excised from leaves (four discs per leaf) with a cork borer. The discs were subsequently bleached first with pure methanol at least 2 days until completely decolorized and then with chloral hydrate solution (1.0 g L−1) for 12–24 h until they become completely transparent. Infection sites were detected after 0.05% aniline blue staining. Three representative fields of each disc and four discs per leaf were observed by ultraviolet (UV) epifluorescence microscopy (Nikon Ti-E, Tokyo, Japan). Three replicates for each treatment and three independent biological repeats were conducted.
The lower face of leaves was treated with cell culture or PDB and harvested 3 dpt. Four discs per leaf (0.5 cm in diameter) were punched, and the tissue was cleared first with pure methanol and subsequently with chloral hydrate solution as described above. Callose deposition was revealed after aniline blue staining as previously described (Li et al., 2019). The number of fluorescent spots (callose deposits) was determined by UV epifluorescence microscopy observations. Three replicates for each treatment and three independent biological repeats were conducted.
The abaxial surface of the second youngest expanded leaves of plants was treated with bacterial culture or PDB and harvested 24 hpt. Four leaves from four plants were pooled and ground in liquid nitrogen. Total plant RNA was isolated using the E.Z.N.A. Plant RNA Kit (OMEGA Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. DNA contaminations were removed using the DNA-free™ kit (Invitrogen, Carlsbad, CA, USA). The concentration of RNA extracts was determined by spectrophotometry. The cDNA was synthesized using the M-MLV Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA) through reverse transcription.
The expression of three genes in grape (PR2, PR3, and STS) known to be upregulated by GLB191 treatment was monitored by qRT-PCR in this study. EF1γ encoding the elongation factor 1 gamma was used as the reference gene. The primers are listed in Supplementary Table S1. The qRT-PCR procedure was identical to that of the previous report (Li et al., 2019). Briefly, the SYBR Green PCR Master Mix kit (Takara Bio Inc., Shiga, Japan) was used in this study. Relative gene expression was determined with the formula fold induction = 2−ΔΔCt, where ΔΔCt = ΔCt (treated sample) − ΔCt (control, i.e., PDB-treated sample) and ΔCt = Ct (target gene) − Ct (reference gene) (Livak and Schmittgen, 2001). Three independent biological repeats were included in each treatment.
Bacillus strains were grown in LB broth at 37°C with shaking at 200 r min−1 overnight. Cells were collected after centrifugation at 6,000 × g for 20 min at 4°C and washed three times with distilled water. The cells were re-suspended in water and the final bacterial suspension was adjusted to 1×107 cells mL−1 for use. Grapevine plants with similar growth status were selected. The bacterial inoculum was sprayed on the second youngest leaves of plants until no droplets dripped off. The control treatment was performed on the same plants by using sterilized water. Three plants were used for each replicate and three replicates were used for each treatment. The persistence of bacteria was monitored at 0, 4, 7, 10, 17, 24, and 31 dpt. Leaves were harvested and washed four times with distilled water. Then, the leaves were then dried with sterile filter paper and subsequently ground with 5 mL of distilled water in a sterile mortar with sterile quartz sand being added to improve the tissue disruption. Samples (100 μL) of plant tissue extracts were diluted by 10-fold serial dilutions in sterile water. One hundred microliters of each diluted suspension was plated on LB agar plates supplemented with Tet. After incubation at 37°C for 12 h, the number of bacterial colony-forming units (CFU g−1 fresh leaf weight) was determined. Three independent biological repeats were conducted.
The bacterial colonization on grape leaves was also observed at 31 dpt using scanning electron microscopy (SEM) as described by Sun et al. (2021). Briefly, samples of approximately 1 cm2 of leaf blades were fixed with 2.5% dialdehyde in Na-K-phosphate buffer (0.1 M, pH 7. 2) for 2 h at room temperature. The samples were then postfixed in 1% (v/v) OsO4 in the PBS (pH 7.2) at room temperature for 2 h and washed three times with PBS (pH 7.2). Subsequently, the samples were dehydrated through an ethanol series (30%, 50%, 70%, 80%, 90%, and 100% for 15 min each). The samples were then critical point dried using a LEICA EM CPD300 (Leica, Mannheim, Germany) and coated with a gold layer using EIKO IB-3 (EIKO, Tokyo, Japan). The bacterial colonization was observed using a HITACHI S-3400N scanning electron microscope (Hitachi, Tokyo, Japan).
The bacterial cell culture was grown in 5 mL of LB liquid medium supplemented with Tet overnight and then transferred to 5 mL of fresh LB medium to grow to mid-log phase (OD600 0.8–1.0). Bacterial cell culture (4 µL) was added to 4 mL of MSgg medium in a 12-well microtiter plate and incubated at 30°C for 48–72 h. Images were taken using a Nikon BM-7 digital camera (Nikon Corporation, Tokyo, Japan). Four replicates were set for each treatment. Three independent biological repeats were conducted.
Analysis of variance (ANOVA) was performed with the statistical program SPSS software 21.0 using the Duncan test or Student’s t-test to assess significant differences between treatments (p < 0.05).
To test whether N-acetylglucosaminidase LytD is involved in the biocontrol efficacy of B. subtilis GLB191 (hereafter called GLB191) against grape downy mildew, lytD deletion mutant (ΔlytD) was constructed and its biocontrol activity was compared with the wild-type strain. Foliar treatment of an in vitro culture of the wild-type GLB191 significantly suppressed the DI of downy mildew on grapevine compared to the medium-only treatment. However, the biocontrol activity of GLB191 was significantly compromised upon deletion of lytD (Figure 1), suggesting that LytD is important for the biocontrol activity of B. subtilis GLB191 against P. viticola. The reduced biocontrol activity of ΔlytD can be complemented by pHY-lytD (Figure 1), which confirms our conclusion. These results suggest that N-acetylglucosaminidase LytD plays an important role in the biocontrol of GLB191 against P. viticola.
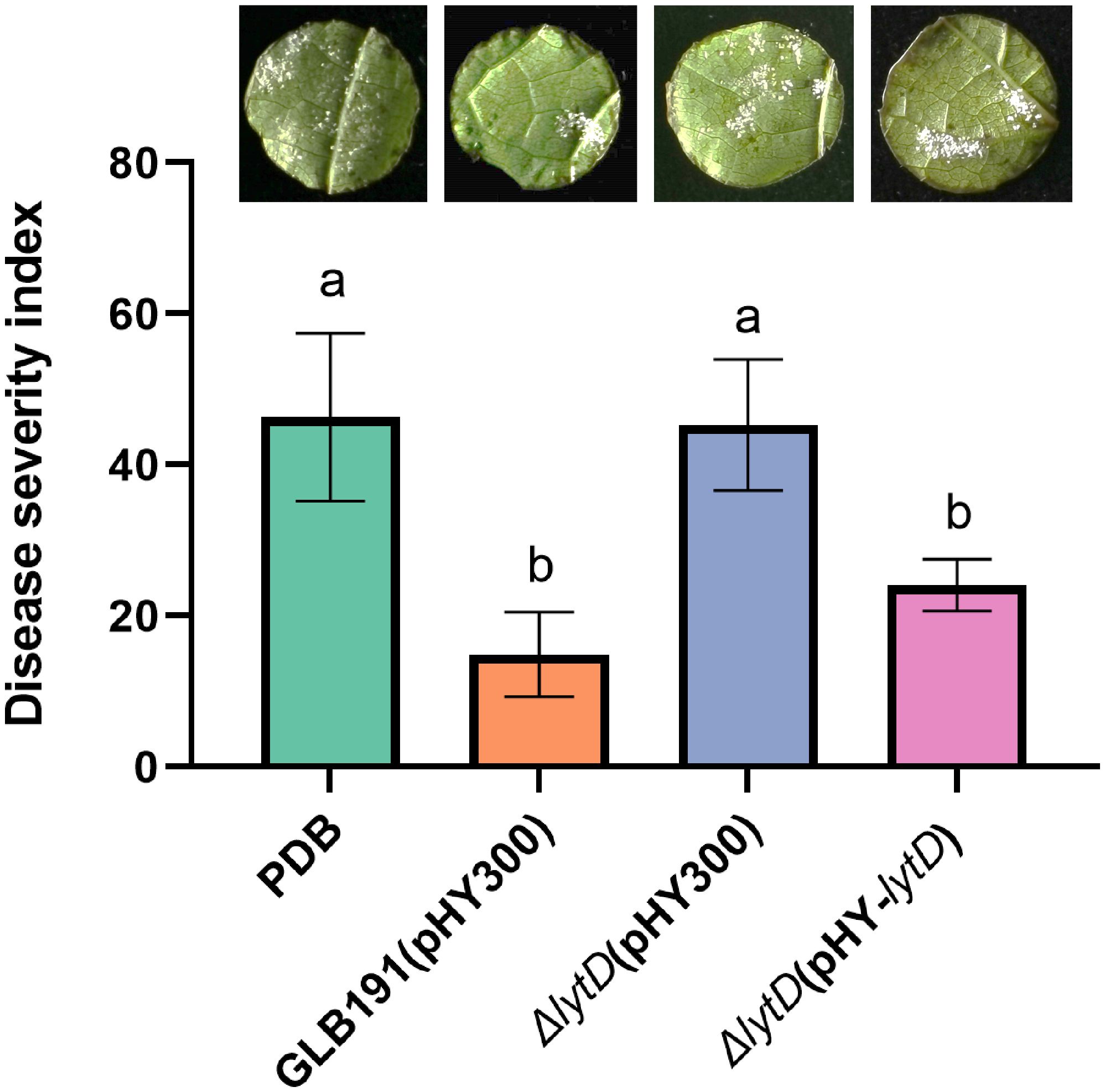
Figure 1 Effect of lytD on the protection of grapevine leaves against grape downy mildew induced by B. subtilis GLB191. Disease severity was assessed with the grape downy mildew on susceptible cultivar cv. Red Globe treated with PDB medium (PDB) or the 48-h culture of the wild-type B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD). Values are the means ± standard deviations (SDs) obtained from 36 discs of four replicates. The data are representative of three independent experiments. Different letters on the column indicate significant differences according to the Duncan test (p < 0.05).
The biocontrol activity of GLB191 against downy mildew disease results from both direct suppression of the pathogen P. viticola and stimulation of the plant defense (Li et al., 2019). In order to further unravel the mechanisms of LytD in biocontrol of GLB191, we first examined the impact of lytD deletion on P. viticola growth under in vitro conditions. Percentage of sporangia released zoospores of P. viticola was determined in the presence and absence of the GLB191 strains. Wild-type GLB191 treatment significantly reduced the percentage of sporangia released zoospores compared to the medium-only treatment (Figure 2), indicating that GLB191 inhibits the viability of sporangia. Comparable percentage of sporangia released zoospores was observed in ΔlytD and ΔlytD(pHY-lytD) as compared to the wild-type GLB191. These results suggest that LytD did not impact the effect of B. subtilis on the sporangia viability under in vitro conditions.

Figure 2 Direct effect of B. subtilis GLB191 and its derivatives on P. viticola sporangia releasing zoospores in vitro. B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD) were cultured in PDB broth for 48 h at 37°C. P. viticola sporangial suspension (500 µL) was incubated with 500 µL of bacterial culture or PDB for 1–3 (h) The percentage of sporangia released zoospores was determined by optical microscopic observations. Data are means ± SD from nine observations of three replicates. Different letters on the column indicate significant differences according to the Duncan test (p < 0.05).
The anti-oomycete effect of GLB191 and ΔlytD was further compared under in vivo conditions. The number of infection sites represented by the number of stomata with encysted zoospores of P. viticola was determined at 24 hpi by UV epifluorescence observations after aniline blue staining of the pathogen. The highest number of infection sites (6.0 ± 1.6 per observation field) was observed for the PDB media-only treatment (Figure 3). Consistent with previous results, almost no infection sites (0.5± 0.2 per observation field) could be observed on leaves treated with wild-type GLB191 (Figure 3), suggesting that GLB191 has a significant direct anti-oomycete effect on zoospores. Interestingly, deletion of lytD resulted in a significantly higher number of infection sites (3.7 ± 0.6 per observation field) compared to the wild type. Complementing the ΔlytD strain with an in trans lytD gene partially restored this phenotype to a comparable level of the wild-type GLB191 (Figure 3). The results above indicate that N-acetylglucosaminidase LytD of B. subtilis GLB191 is required for its anti-oomycete effect against P. viticola zoospores infection although it does not affect its antagonism of sporangia viability.
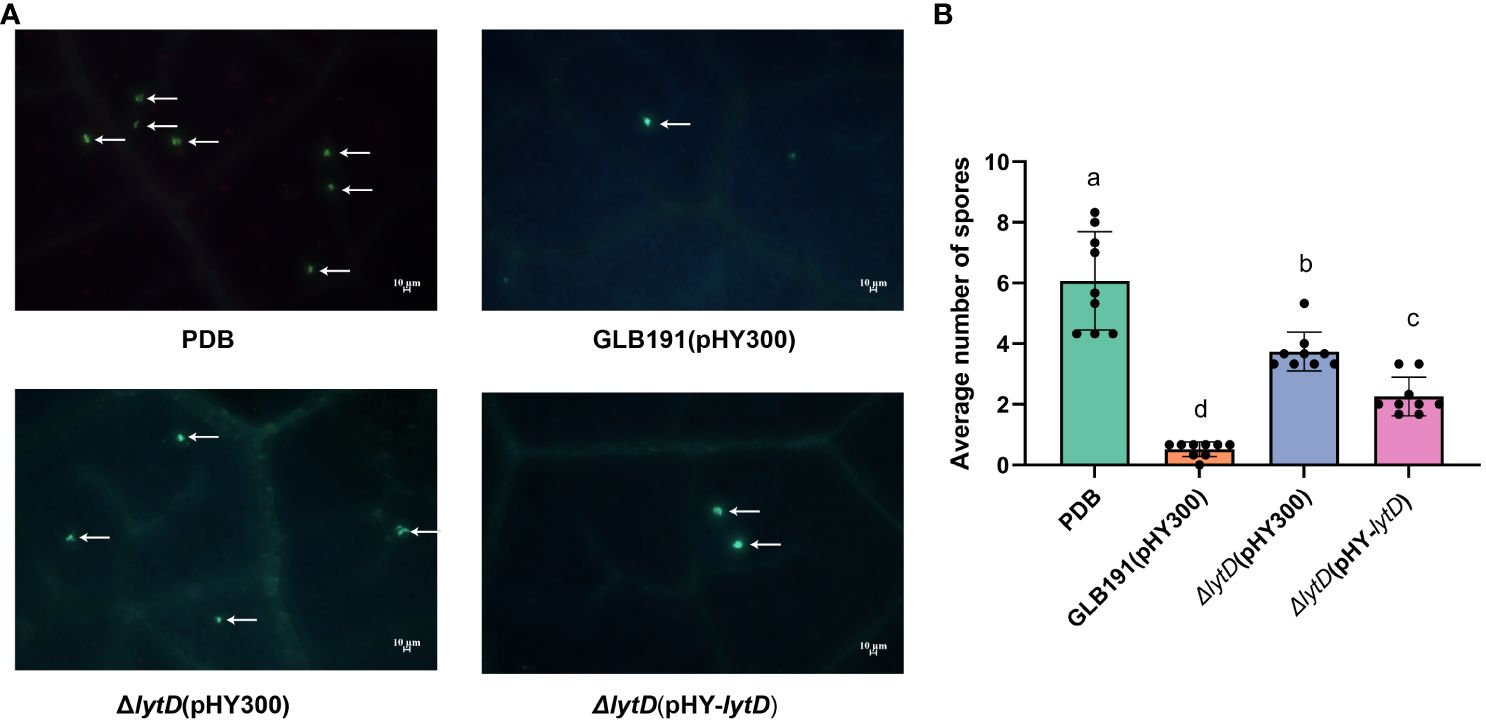
Figure 3 Direct effect of B. subtilis GLB191 and its derivatives on P. viticola zoospores on grapevine leaves. B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD) were cultured in PDB medium for 48 h at 37°C. Grapevine (V. vinifera cv. Red Globe) leaves were treated with PDB medium (PDB) or the culture of GLB191(pHY300), ΔlytD(pHY300), and ΔlytD(pHY-lytD), and inoculated with P. viticola sporangia 2 h post-treatment. Leaf discs were then punched out from leaves 24 h post-inoculation, and the number of infection sites (i.e., stomata with encysted zoospores of P. viticola) was determined by UV epifluorescence observations after aniline blue staining of the pathogen. (A) Representative fields of fluorescence microscopy observations. Arrows indicate the infection sites (encysted zoospores). Scale bars represent 10 μm. (B) Values are the mean number of infection sites (means ± SD) for 9 discs from three replicates. Different letters on the column indicate significant different at p < 0.05 according to the Duncan test. The data are representative of three independent experiments.
In order to further determine whether lytD is involved in inducing defense responses of grapevine leaves, callose production and defense gene expression measured by qRT-PCR were chosen as two representative phenotypical markers of plant defense to be evaluated upon treatment of wild-type GLB191 and ΔlytD (Li et al., 2019). The in vitro culture of Bacillus strains was sprayed on grape leaves and callose production was monitored at 3 dpi by UV epifluorescence after aniline blue staining. As anticipated, the number of fluorescent spots was significantly increased after treatment by GLB191 (6.4 ± 0.9 per observation field) as compared to the media-only treatment (0.9 ± 0.2 per observation field) (Figure 4), indicating a strong induction of callose production by GLB191. In contrast, disruption of lytD resulted in a marked decrease in the number of spots (3.6 ± 0.6 per observation field) (Figure 4). This phenotype can be complemented by ΔlytD(pHY-lytD) (5.5 ± 0.5 per observation field) (Figure 4), thus confirming the important role of lytD in inducing callose deposition in grapevine leaves.
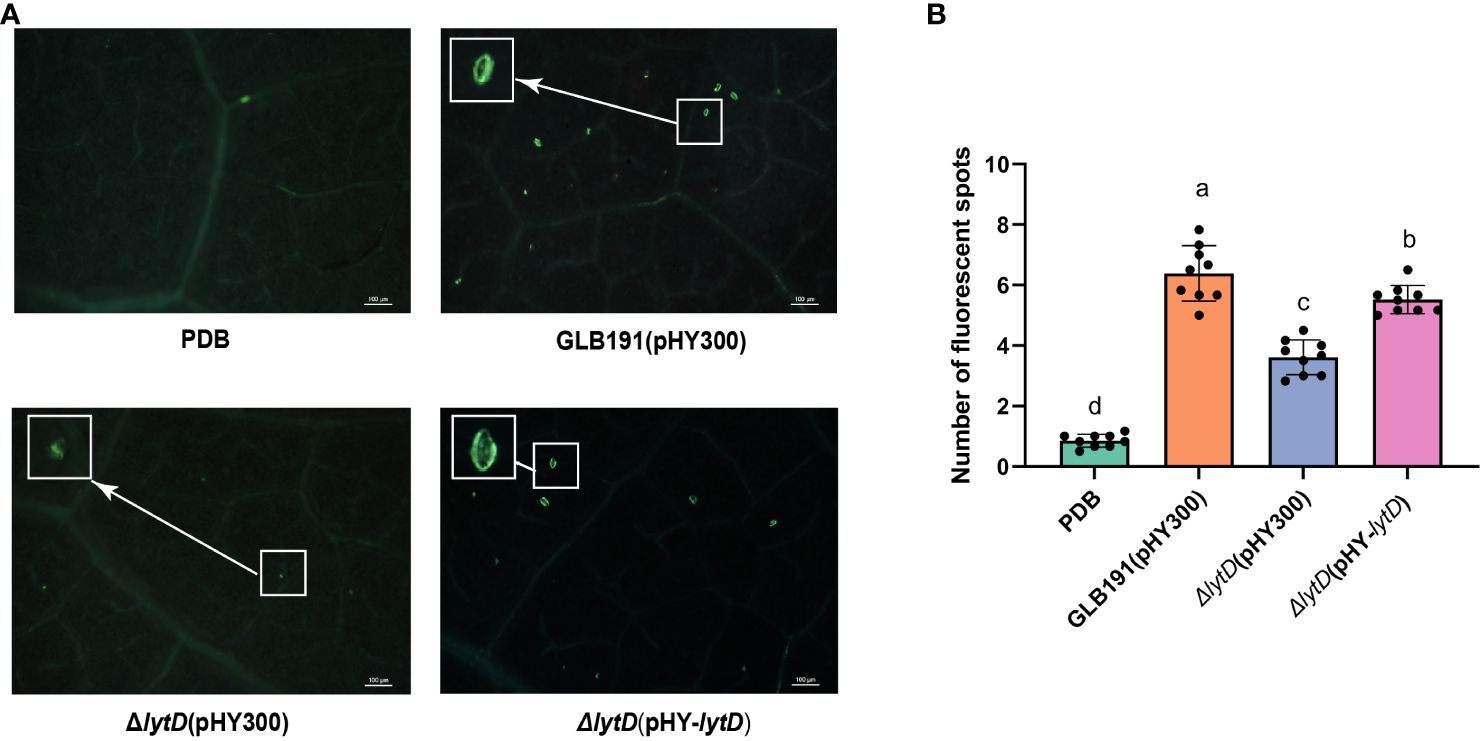
Figure 4 Callose deposition induced by B. subtilis GLB191 and its derivatives on grapevine leaves. Grapevine (V. vinifera cv. Red Globe) leaves were treated with PDB medium (PDB) or the culture of the wild-type B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD). Callose production (fluorescent spots) was observed 3 days post-treatment by epifluorescence observations after aniline blue staining. (A) Representative fields observed using a fluorescence microscope. Scale bars represent 100 μm. (B) Values are the mean number of fluorescent spots (means ± SD) for 9 discs from three replicates. Different letters on the column indicate significant difference at p < 0.05 according to the Duncan test. The data are representative of three independent experiments.
Next, the expression of three plant defense-related genes, PR2 (encoding the PR protein 2 β-1,3-glucanase), PR3 (encoding the PR protein 3 chitinase 4c), and STS (encoding stilbene synthase, downstream of PAL and responsible for the synthesis of resveratrol, the main phytoalexin produced by grapevine in response to biotic or abiotic stresses), previously known to be induced by GLB191 (Li et al., 2019) was evaluated upon deletion of lytD. As expected, PR2, STS, and PR3 were highly upregulated upon GLB191 treatment by 8.5-, 20.2-, and 3.8-fold, respectively (Figure 5). However, the transcript accumulation of these defense genes was only 2.0-, 1.5-, and 1.4-fold relative to PDB in response to ΔlytD treatment (Figure 3), suggesting that LytD contributes to the induction of defense gene expression after GLB191 treatment. Taken together, the results above indicate that LytD is involved in the induction of plant defense response by B. subtilis GLB191.
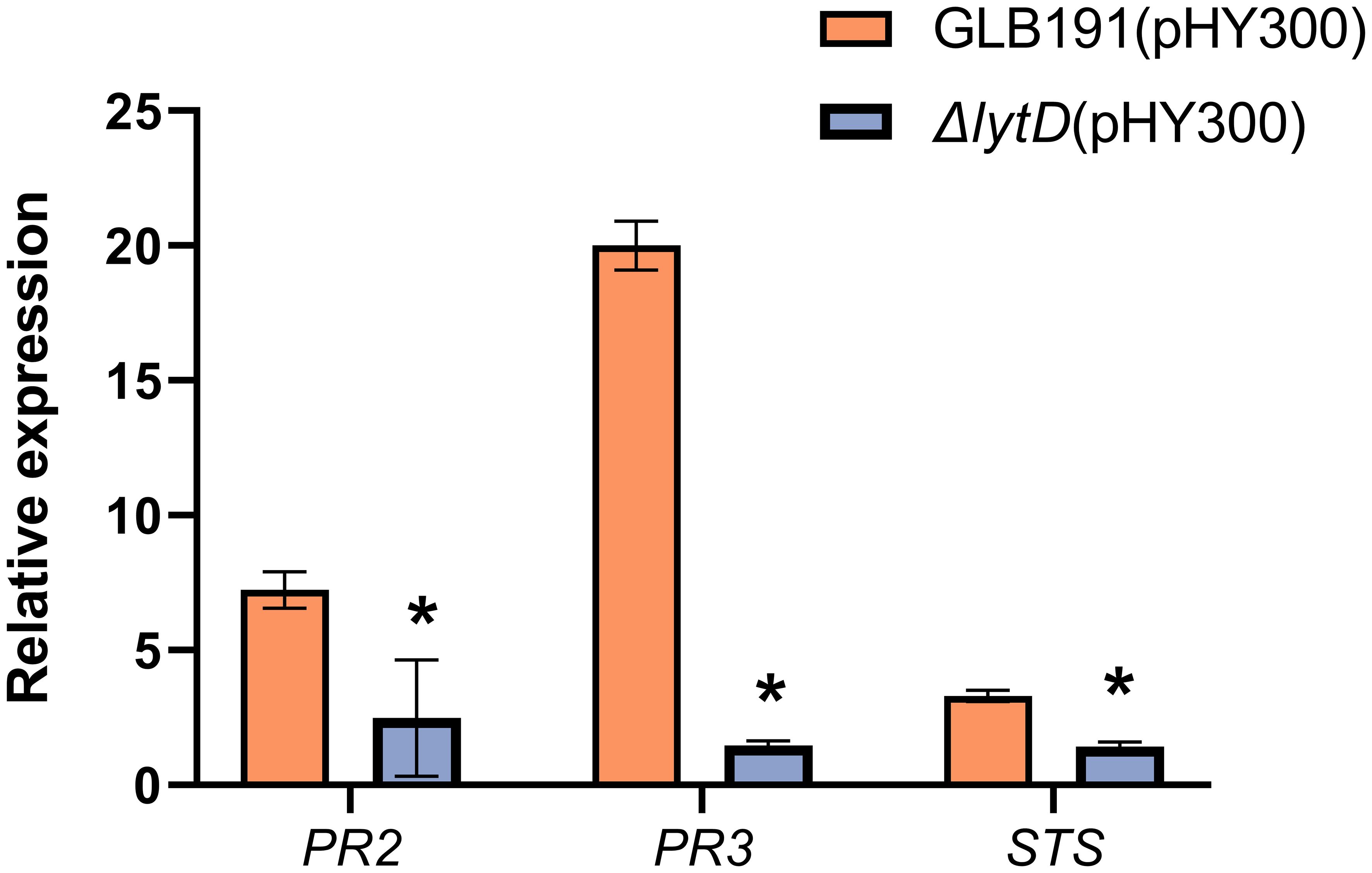
Figure 5 Defense-related gene expression in grapevine leaves treated with B. subtilis GLB191 and its derivatives. Grapevine (V. vinifera cv. Red Globe) leaves were treated with PDB medium (PDB) or the culture of the wild-type B. subtilis GLB191(pHY300) [GLB191(pHY300)] and ΔlytD(pHY300) and harvested 24 h post-treatment. The relative transcript accumulation of defense genes PR2 (encoding the pathogenesis-related protein 2 β-1,3-glucanase), PR3 (encoding the pathogenesis-related protein 3 chitinase 4c), and STS (encoding stilbene synthase) was determined by qRT-PCR. Values are the fold increase in transcript level compared to the PDB-treated leaves. Data represent mean ± SD of triplicate measurements from a representative experiment. Asterisks above the column indicate significant difference in transcript level in leaves treated with ΔlytD(pHY300) compared to that of GLB191(pHY300)-treated ones at p < 0.05 according to Student’s t-test.
Sufficient colonization of biocontrol microorganism on plants is as an important first step required for the subsequent biocontrol activities (Sachdev and Singh, 2018). We next determined whether LytD affects the growth of B. subtilis GLB191 under in vitro conditions and, subsequently, its colonization on grape leaves. Cultured in LB medium, the growth of GLB191(pHY300), ΔlytD(pHY300), and ΔlytD(pHY-lytD) strains was similar to each other within the test period of 19 h (Supplementary Figure S1). Interestingly, the population of B. subtilis GLB191 determined by plating on LB medium supplemented with the antibiotic Tet revealed a reduction in B. subtilis colonization on grape leaves upon deletion of ΔlytD. Compared to wild-type B. subtilis GLB191 that showed an initial decrease (4 dpi), followed by a significant increase (17 dpi) and then stability (24 dpi) (Figure 6A), ΔlytD showed a dramatic reduction in its population except on 0 dpi. The reduced plant colonization phenotype could be restored by the complementary strain ΔlytD(pHY-lytD) (Figure 6A). No bacteria were recovered on the LB containing tetracycline from the uninoculated plants throughout the experiments (data not shown). These results suggest that the LytD is important for grape leaf colonization although this is not due to the growth impediment.
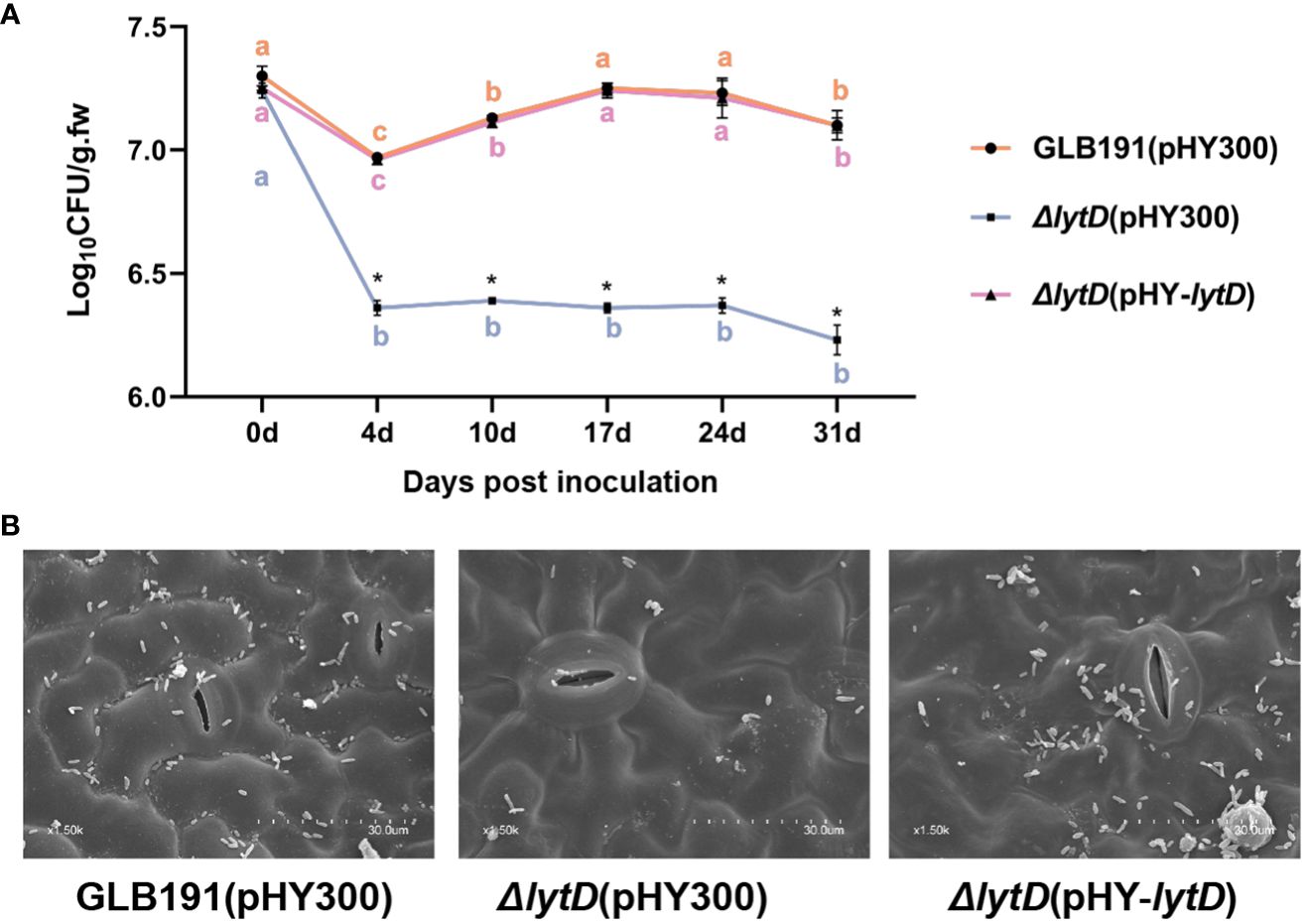
Figure 6 Colonization of B. subtilis GLB191 and its derivatives on grapevine leaves. Grapevine (V. vinifera cv. Red Globe) leaves were treated with the cells of the wild-type B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD). (A) Bacterial population was isolated from grapevine leaves at 0, 4, 7, 10, 17, 24, and 31 days post-treatment and determined through plate CFU counts. Values correspond to the mean bacterial population (CFU g−1 fresh weight) (mean ± SD) from three grapevine leaves. The data are representative of three independent experiments. Different letters in the same color on the curve indicate significant differences between different time points for the same strain according to the Duncan test (p < 0.05). Asterisks above the curve indicate significant difference in bacterial population at the same time point between different strains according to Student’s t-test (p < 0.05). (B) Bacterial colonization on grapevine leaves observed at 31 days post-treatment through scanning electron microscopy.
The reduced plant colonization was confirmed by SEM. Heavy colonization of GLB101 was observed at 31 dpi on the surface of the GLB191-treated leaves. In contrast, markedly less bacteria were observed on the surface of the ΔlytD-treated leaves. When introducing a complementary plasmid pHY-lytD into ΔlytD, the bacteria amount was almost restored to the wild-type level (Figure 6B). These results confirm that LytD is important for B. subtilis GLB191 colonization on grape leaves.
It is widely recognized that biofilm formation is critical for the colonization of bacteria on plant surfaces, especially for Bacillus species (Chen et al., 2013; Arnaouteli et al., 2021; Gong et al., 2022). To determine whether the impaired plant colonization of ΔlytD is due to a reduced biofilm formation, we measured the pellicle biofilm formation of the wild type and ΔlytD of GLB191 in MSgg medium. Compared to wild-type GLB191 that formed a strong biofilm in MSgg medium, the disruption of lytD led to reduced biofilm formation. The biofilm formation of the complementary strain ΔlytD(pHY-lytD) was restored to the wild-type level (Figure 7). These results suggest that LytD is involved in B. subtilis GLB191 biofilm formation.
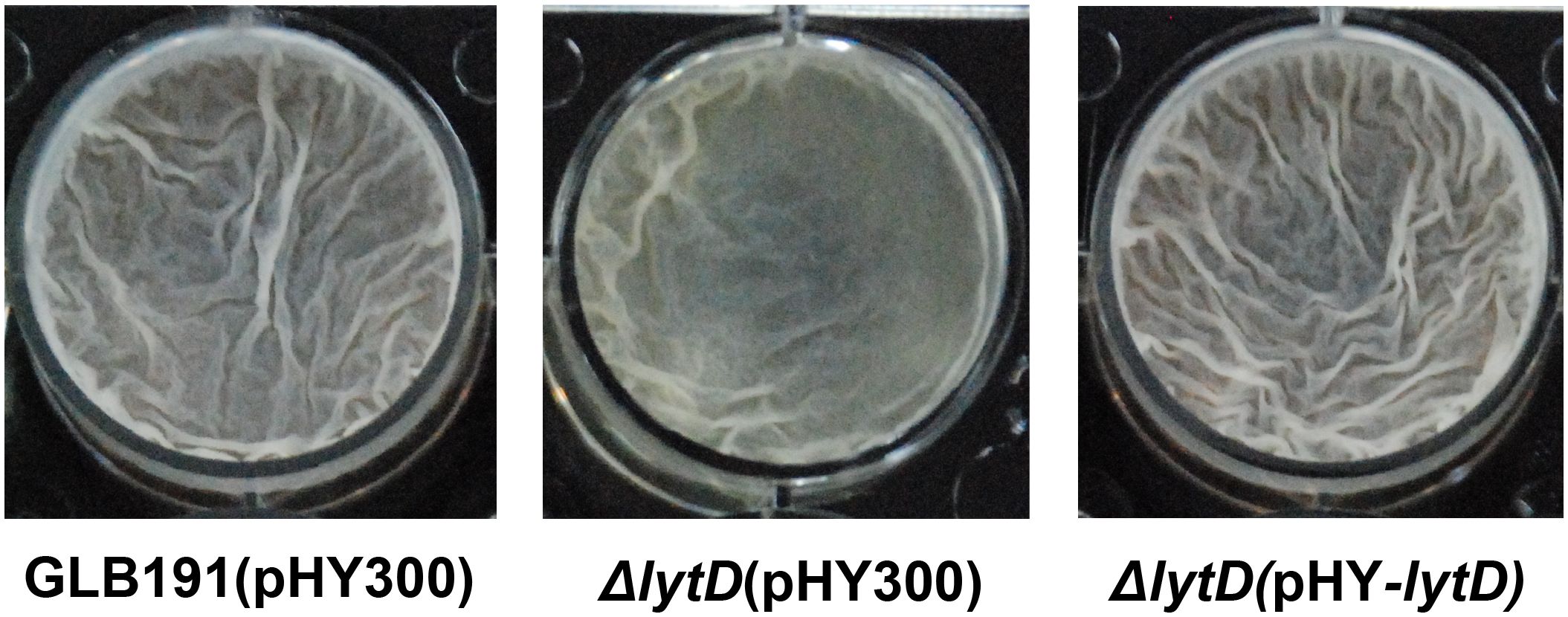
Figure 7 Biofilm formation of B. subtilis GLB191 and its derivatives. The wild-type B. subtilis GLB191(pHY300) [GLB191(pHY300)], ΔlytD(pHY300), and ΔlytD(pHY-lytD) were grown in LB at 37°C overnight and then transferred to fresh LB to grow to mid-log phase (OD600 0.8–1.0). Cultures (4 μL) were added into 4 mL of MSgg medium in a 12-well microtiter plate and incubated at 30°C. Images were taken using a Nikon BM-7 digital camera after 48–72 h.
In summary, the roles of LytD in biological control of B. subtilis are depicted in Figure 8. LytD positively impacts bacterial biofilm formation and thereby colonization on grape leaves, which determine its efficiency to stimulate the plant defenses and direct effect on the pathogen, and finally impact its biocontrol activity.
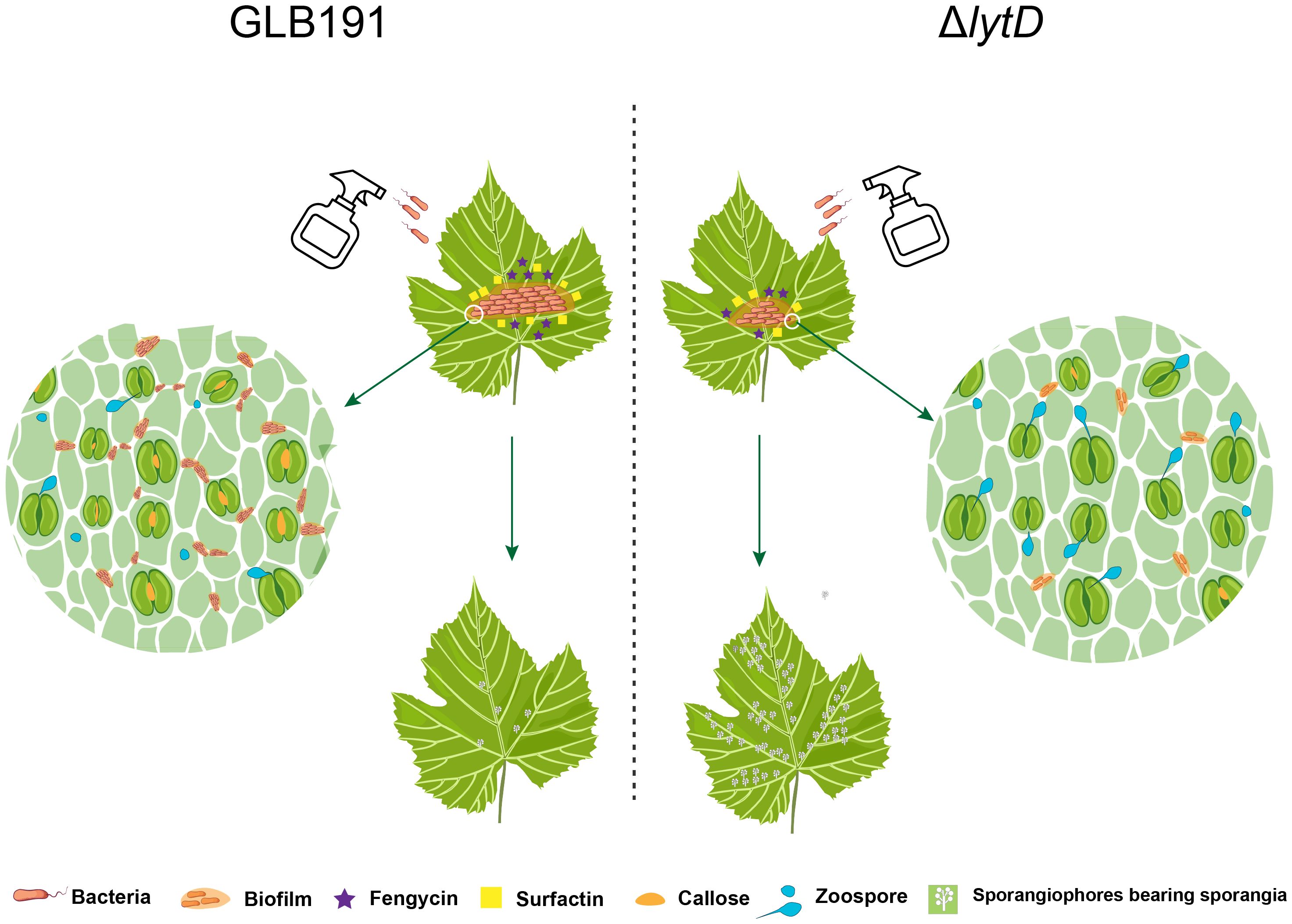
Figure 8 Model of LytD regulation on the biocontrol efficacy of B. subtilis GLB191 against grape downy mildew. Upon landing on a grapevine leaf, vegetative cells of B. subtilis GLB191 (GLB191) produce the antimicrobial lipopeptide surfactin (Srf) to antagonize the pathogen P. viticola and to activate biofilm formation and further colonize grapevine leaf. Besides surfactin, it also produces fengycin (Fen). Both surfactin and fengycin contribute to inhibit the infection (germination) of P. viticola and the induction of defense resistance, which was associated with high callose deposition. Inactivation of lytD (ΔlytD) resulted in decreased biofilm formation and the consequent colonization on grape leaves, which determine its efficiency to direct effect on the pathogen and induction of defense responses, thus reducing biocontrol activity against grape downy mildew.
B. subtilis is one of the most widely studied biocontrol agents (BCAs). Many of the factors, genes, and conditions needed by this organism to protect plants are well studied. However, many aspects of the regulation of the protection are not fully elucidated. In this study, the autolysin N-acetylglucosaminidase LytD, an endogenous CWDE of B. subtilis acting on the peptidoglycan layer of its own cell wall, was found to play an important role in the biocontrol activity of B. subtilis against grape downy mildew. Evidence also showed that N-acetylglucosaminidase LytD of B. subtilis regulates its biocontrol activity through biofilm formation and the consequent colonization on grape leaves, which determine its direct effect on the pathogen and induction of defense responses. This is the first study to report on the role of an endogenous CWDE in biological control.
Competent colonization is the most important prerequisite towards efficient biocontrol (Sachdev and Singh, 2018). An efficient BCA must be able to establish itself and survive on plant. Therefore, the first step towards an efficient biocontrol is to demonstrate the successful colonization on the plant (Maurer et al., 2013). There is ongoing research to identify genes or factors needed for colonization. Among these studies, biofilm is at the top, which plays essential roles in the colonization of the BCA after inoculation (Fessia et al., 2022). GLB191 has a strong protective activity against grape downy mildew in both leaf disk assays and the field (Zhang et al., 2017). As expected, it also showed strong biofilm formation and exceptionally high and stable cell densities on grapevine leaves (Figures 6, 7), indicating competent colonization on grapevine leaves, which makes it a promising candidate for BCA.
The cell wall is the first line of defense against pathogen attacks. BCAs produce an array of CWDEs to degrade the cell wall of phytopathogenic fungi. For example, B. subtilis produces chitinases and glucanases, which are important in the biological control of phytopathogenic fungi (Manjula and Podile, 2005; Shrestha et al., 2015). Additionally, CWDEs can act as elicitors of host defense reaction (Kikot et al., 2009; Brunner et al., 2013; Kubicek et al., 2014; Tayi et al., 2016; Bacete et al., 2018; Giovannoni et al., 2020). Moreover, CWDEs also contribute to the colonization of bacteria on plants. For example, endo-β-1,4-glucanase of B. amyloliquefaciens is needed for its optimal endophytic colonization (Fan et al., 2016). In this study, we found that autolysin N-acetylglucosaminidase LytD of B. subtilis plays an important role in the suppression of the pathogen P. viticola, stimulation of the plant defense, biofilm formation and colonization, and the biocontrol activity against grape downy mildew (Figures 1–7). These results are in accordance with previous reports, indicating that endogenous CWDEs also play important roles in multiple aspects besides cell development. These findings expand our knowledge on the biological function of autolysins and provide a new target to promote the biocontrol activity of B. subtilis.
Even though the direct effect on the pathogen and the stimulation of the defense, which are the main mechanisms of B. subtilis GLB191 to protect grapevine against downy mildew (Li et al., 2019), reduced the suppression of the pathogen P. viticola and the stimulation of the plant defense by the inactivation of LytD, they are probably not the direct reasons for its reduced biocontrol efficacy since colonization is a prerequisite towards efficient biocontrol among all mechanisms. Given that the bacterial growth rate under in vitro conditions and the percentage of sporangia released zoospores in vitro were not significantly affected by disruption of lytD (Figure 2; Supplementary Figure S1), we speculate that the reduction of biofilm and, therefore, colonization are the direct reasons for its reduced biocontrol efficacy in ΔlytD. Therefore, we propose that N-acetylglucosaminidase LytD of B. subtilis regulates its biocontrol efficacy against grape downy mildew through colonization. However, the effect of LytD on the production of surfactin and fengycin, which are the main active compounds for its biocontrol efficacy against grape downy mildew, still needs to be determined. In addition, it is worth determining whether lytD was induced when B. subtilis interacts with the grapevine leaves.
The genome of B. subtilis harbors at least 35 autolysin genes. Although autolysins associated with B. subtilis have been known for many years, the study of their physiological roles has been hampered by their great number and functional redundancy (Smith et al., 2000). Therefore, analysis of multiply inactivated mutants provides the possibility to define the roles played by individual autolysins in a number of important cellular processes. LytC and LytD are two major autolysins expressed during the vegetative phase of growth in B. subtilis 168. Both enzymes are dispensable for growth but have mutually compensatory roles in cell wall turnover, motility, and cell separation (Rashid et al., 1995; Ren et al., 2022). Inconsistent with B. subtilis 168, single deletion of lytD of B. subtilis GLB191 showed significant phenotype, probably due to the different strain. Therefore, it will be important to identify and analyze the total complement of autolysins to determine their individual and combined roles in B. subtilis GLB191. It will provide a better understanding of the complex roles in biological control played by the autolysins of B. subtilis GLB191.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
LTW: Formal analysis, Investigation, Methodology, Software, Writing – original draft. JQH: Methodology, Resources, Supervision, Writing – review & editing. SC: Methodology, Resources, Review & editing. XS: Methodology, Software, Writing – editing. XZ: Resources, Writing – review & editing. LJW: Resources, Review – editing. WZ: Writing – review & editing. ZSW: Methodology, Review – editing. QCZ: Methodology, Review – editing. QW: Resources, Supervision, Review – editing. LY: Conceptualization, Methodology, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31972982 and 32272613), the National Key R&D Program of China (2023YFE0104800 and 2023YFD1401400), and the earmarked fund for China Agricultural Research System (No. CARS-29-bc-3).
We thank Quan Zeng at the Connecticut Agricultural Experiment Station in the United States for helpful comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1381018/full#supplementary-material
Angeles, D. M., Scheffers, D. J. (2021). The cell wall of Bacillus subtilis. Curr. Issues Mol. Biol. 41, 539–596. doi: 10.21775/cimb.041.539
Arnaouteli, S., Bamford, N. C., Stanley-Wall, N. R., Kovacs, A. T. (2021). Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 19, 600–614. doi: 10.1038/s41579-021-00540-9
Arnaud, M., Chastanet, A., Débarbouillé, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004
Bacete, L., Melida, H., Miedes, E., Molina, A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. doi: 10.1111/tpj.13807
Beattie, G. A., Lindow, S. E. (1999). Bacterial colonization of leaves: A spectrum of strategies. Phytopathology 89, 353–359. doi: 10.1094/PHYTO.1999.89.5.353
Biermann, R., Beutel, S. (2023). Endospore production of Bacillus spp. for industrial use. Eng. Life Sci. 23, e2300013. doi: 10.1002/elsc.202300013
Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R., Kolter, R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. United States America 98, 11621–11626. doi: 10.1073/pnas.191384198
Brunner, P. C., Torriani, S. F. F., Croll, D., Stukenbrock, E. H., McDonald, B. A. (2013). Coevolution and life cycle specialization of plant cell wall degrading enzymes in a hemibiotrophic pathogen. Mol. Biol. Evol. 30, 1337–1347. doi: 10.1093/molbev/mst041
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Collinge, D. B., Jensen, D. F., Rabiey, M., Sarrocco, S., Shaw, M. W., Shaw, R. H. (2022). Biological control of plant diseases - What has been achieved and what is the direction? Plant Pathol. 71, 1024–1047. doi: 10.1111/ppa.13555
Etesami, H., Jeong, B. R., Glick, B. R. (2023). Biocontrol of plant diseases by Bacillus spp. Physiol. Mol. Plant Pathol. 126, 102048. doi: 10.1016/j.pmpp.2023.102048
Fan, H. Y., Zhang, Z. W., Li, Y., Zhang, X., Duan, Y. M., Wang, Q. (2017). Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01973
Fan, X., Yang, R., Qiu, S., Cai, X., Zou, H., Hu, F. (2016). The endo-β-1, 4-glucanase of Bacillus amyloliquefaciens is required for optimum endophytic colonization of plants. J. Microbiol. Biotechnol. 26, 946–952. doi: 10.4014/jmb.1512.12055
Fessia, A., Barra, P., Barros, G., Nesci, A. (2022). Could Bacillus biofilms enhance the effectivity of biocontrol strategies in the phyllosphere? J. Appl. Microbiol. 133, 2148–2166. doi: 10.1111/jam.15596
Fira, D., Dimkic, I., Beric, T., Lozo, J., Stankovic, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Free, S. J. (2013). Fungal cell wall organization and biosynthesis. Adv. Genet. 81, 33–82. doi: 10.1016/B978-0-12-407677-8.00002-6
Ghuysen, J.-M., Tipper, D. J., Strominger., J. L. (1966). “Enzymes that degrade bacterial cell walls,” in Methods in Enzymology, (Academic Press), 685–699. doi: 10.1016/0076-6879(66)08124-2
Giovannoni, M., Gramegna, G., Benedetti, M., Mattei, B. (2020). Industrial use of cell wall degrading enzymes: The fine line between production strategy and economic feasibility. Front. Bioengineering Biotechnol. 8. doi: 10.3389/fbioe.2020.00356
Gong, H. L., Jiang, W. X., Yang, Y., Zhang, Y., Chen, X. F., Li, W., et al. (2022). Cyclic di-GMP regulates bacterial colonization and further biocontrol efficacy of Bacillus velezensis against apple ring rot disease via its potential receptor YdaK. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1034168
Hashem, A., Tabassum, B., Abd_Allah, E. F. (2019). Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 26, 1291–1297. doi: 10.1016/j.sjbs.2019.05.004
Héloir, M.-C., Adrian, M., Brulé, D., Claverie, J., Cordelier, S., Daire, X., et al. (2019). Recognition of elicitors in grapevine: From MAMP and DAMP perception to induced resistance. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01117
Horsburgh, G. J., Atrih, A., Williamson, M. P., Foster, S. J. (2003). LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: The major active glucosaminidase. Biochemistry 42, 257–264. doi: 10.1021/bi020498c
Ishiwa, H., Shibahara-Sone, H. (1986). New shuttle vectors for Escherichia coli and Bacillus subtilis. IV. The nucleotide sequence of pHY300PLK and some properties in relation to transformation. Japanese J. Genet. 61, 515–528. doi: 10.1266/jjg.61.515
Kikot, G. E., Hours, R. A., Alconada, T. M. (2009). Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: a review. J. Basic Microbiol. 49, 231–241. doi: 10.1002/jobm.200800231
Koch, E., Ole Becker, J., Berg, G., Hauschild, R., Jehle, J., Köhl, J., et al. (2018). Biocontrol of plant diseases is not an unsafe technology. J. Plant Dis. Prot. 125, 121–125. doi: 10.1007/s41348-018-0158-4
Kubicek, C. P., Starr, T. L., Glass, N. L. (2014). Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Lahlali, R., Ezrari, S., Radouane, N., Kenfaoui, J., Esmaeel, Q., El Hamss, H., et al. (2022). Biological control of plant pathogens: A global perspective. Microorganisms 10, 596. doi: 10.3390/microorganisms10030596
Li, Y., Heloir, M. C., Zhang, X., Geissler, M., Trouvelot, S., Jacquens, L., et al. (2019). Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 20, 1037–1050. doi: 10.1111/mpp.12809
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Manjula, K., Podile, A. (2005). Production of fungal cell wall degrading enzymes by a biocontrol strain of Bacillus subtilis AF 1. Indian J. Exp. Biol. 43, 892–896. http://repository.ias.ac.in/104085/
Margot, P., Mauel, C., Karamata, D. (1994). The gene of the N-Acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase not involved in vegetative cell autolysis. Mol. Microbiol. 12, 535–545. doi: 10.1111/j.1365-2958.1994.tb01040.x
Maurer, K. A., Zachow, C., Seefelder, S., Berg, G. (2013). Initial steps towards biocontrol in hops: successful colonization and plant growth promotion by four bacterial biocontrol agents. Agronomy 3, 583–594. doi: 10.3390/agronomy3040583
Miljaković, D., Marinković, J., Balešević-Tubić, S. (2020). The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 8, 1037. doi: 10.3390/microorganisms8071037
Monier, J.-M., Lindow, S. E. (2003). Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. 100, 15977–15982. doi: 10.1073/pnas.2436560100
Nayyab, S., O’Connor, M., Brewster, J., Gravier, J., Jamieson, M., Magno, E., et al. (2017). Diamide inhibitors of the Bacillus subtilis N-acetylglucosaminidase LytG that exhibit antibacterial activity. ACS Infect. Dis. 3, 421–427. doi: 10.1021/acsinfecdis.7b00005
Ongena, M., Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Pandin, C., Le Coq, D., Canette, A., Aymerich, S., Briandet, R. (2017). Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microbial Biotechnol. 10, 719–734. doi: 10.1111/1751-7915.12693
Rashid, M. H., Mori, M., Sekiguchi, J. (1995). Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology-Sgm 141, 2391–2404. doi: 10.1099/13500872-141-10-2391
Ren, K. X., Wang, Q., Hu, M. K., Chen, Y., Xing, R. F., You, J. J., et al. (2022). Research progress on the effect of autolysis to Bacillus subtilis fermentation bioprocess. Fermentation 8, 685. doi: 10.3390/fermentation8120685
Rouxel, M., Mestre, P., Comont, G., Lehman, B. L., Schilder, A., Delmotte, F. (2013). Phylogenetic and experimental evidence for host-specialized cryptic species in a biotrophic oomycete. New Phytologist 197, 251–263. doi: 10.1111/nph.12016
Sachdev, S., Singh, R. (2018). Root colonization: imperative mechanism for efficient plant protection and growth. MOJ Ecol. Environ. Sci. 3, 240–242. doi: 10.15406/MOJES.2018.03.00094
Senanayake, S. D., Brian, D. A. (1995). Precise large deletions by the PCR-based overlap extension method. Mol. Biotechnol. 4, 13–15. doi: 10.1007/BF02907467
Shrestha, A., Sultana, R., Chae, J.-C., Kim, K., Lee, K.-J. (2015). Bacillus thuringiensis C25 which is rich in cell wall degrading enzymes efficiently controls lettuce drop caused by Sclerotinia minor. Eur. J. Plant Pathol. 142, 577–589. doi: 10.1007/s10658-015-0636-5
Smith, T. J., Blackman, S. A., Foster, S. J. (2000). Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology-Uk 146, 249–262. doi: 10.1099/00221287-146-2-249
Sun, W., Ma, N., Huang, H., Wei, J., Ma, S., Liu, H., et al. (2021). Photosynthetic contribution and characteristics of cucumber stems and petioles. BMC Plant Biology 21, 454. doi: 10.1186/s12870-021-03233-w
Tayi, L., Maku, R., Patel, H. K., Sonti, R. V. (2016). Action of multiple cell wall-degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol. Plant-Microbe Interact. 29, 599–608. doi: 10.1094/MPMI-02-16-0039-R
Trouvelot, S., Varnier, A.-L., Allègre, M., Mercier, L., Baillieul, F., Arnould, C., et al. (2008). A β-1, 3 glucan sulfate induces resistance in grapevine against Plasmopara viticola through priming of defense responses, including HR-like cell death. Mol. Plant-Microbe Interact. 21, 232–243. doi: 10.1094/MPMI-21-2-0232
Villarreal-Delgado, M. F., Villa-Rodríguez, E. D., Cira-Chávez, L. A., Estrada-Alvarado, M. I., Parra-Cota, F. I., Santos-Villalobos, S. (2018). The genus Bacillus as a biological control agent and its implications in the agricultural biosecurity. Rev. mexicana fitopatología 36, 95–130. doi: 10.18781/R.MEX.FIT.1706-5
Zeriouh, H., de Vicente, A., Pérez-García, A., Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16, 2196–2211. doi: 10.1111/1462-2920.12271
Zhang, G., Wang, W., Deng, A., Sun, Z., Zhang, Y., Liang, Y., et al. (2012). A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PloS Genet. 8, e1002987. doi: 10.1371/journal.pgen.1002987
Keywords: autolysin, peptidoglycan, cell wall degrading enzyme, colonization, biofilm, induced resistance, biological control
Citation: Wang L, Huang J, Chen S, Su X, Zhang X, Wang L, Zhang W, Wang Z, Zeng Q, Wang Q and Li Y (2024) Endogenous cell wall degrading enzyme LytD is important for the biocontrol activity of Bacillus subtilis. Front. Plant Sci. 15:1381018. doi: 10.3389/fpls.2024.1381018
Received: 02 February 2024; Accepted: 14 March 2024;
Published: 10 April 2024.
Edited by:
Choong-Min Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Republic of KoreaReviewed by:
Lifang Zou, Shanghai Jiao Tong University, ChinaCopyright © 2024 Wang, Huang, Chen, Su, Zhang, Wang, Zhang, Wang, Zeng, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, bGl5YW5jYXVAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.