
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 02 April 2024
Sec. Plant Biotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1377318
Benzylisoquinoline alkaloids (BIAs) produced in opium poppy have been evidenced to heal patients suffering from various diseases. They, therefore, hold an integral position in the herbal drug industry. Despite the adoption of several approaches for the large-scale production of BIAs, opium poppy remains the only platform in this purpose. The only disadvantage associated with producing BIAs in the plant is their small quantity. Thus, recruiting strategies that boost their levels is deemed necessary. All the methods which have been employed so far are just able to enhance a maximum of two BIAs. Thus, if these methods are utilized, a sizable amount of time and budget must be spent on the synthesis of all BIAs. Hence, the exploitation of strategies which increase the content of all BIAs at the same time is more commercially effective and time-saving, avoiding the laborious step of resolving the biosynthetic pathway of each compound. Exposure to biotic and abiotic elicitors, development of a synthetic auto-tetraploid, overexpression of a WRKY transcription factor, formation of an artificial metabolon, and suppression of a gene in the shikimate pathway and miRNA are strategies that turn opium poppy into a versatile bioreactor for the concurrent and massive production of BIAs. The last three strategies have never been applied for BIA biosynthetic pathways.
Opium poppy (Papaver somniferum L.) is widely known as a medicinally significant plant that produces a sizable number of alkaloids, virtually 80 (Hagel and Facchini, 2013). Among these nitrogen-containing compounds are benzylisoquinoline alkaloids (BIAs) which comprise a diverse group of metabolites in terms of structure and function. Morphine, codeine, thebaine, sanguinarine, papaverine, and noscapine are well-established BIAs occurring mainly in opium poppy. Morphine has been administered for sleep disorders, acute pains, and diarrhea (Hamilton and Baskett, 2000). Codeine has the potential to relieve painful muscle tension and to induce sleep as well (Facchini and Park, 2003). Thebaine has been recognized as a sedative agent (Pathan and Williams, 2012). Sanguinarine is a potent drug that remedies inflammation (Dvorák et al., 2006). The healing power of papaverine against muscle spasms and vasoconstriction has been reported (Desgagne´-Penix and Facchini, 2012). Noscapine has been widely prescribed for severe cough and a wide range of cancers (Mahmoudian and Rahimi-Moghaddam, 2009; Rahmanian-Devin et al., 2021). Interestingly, the inhibitory effect of sanguinarine and papaverine on tumorous cells has also been proven (Desgagne´-Penix and Facchini, 2012; Galadari et al., 2017).
If dropping out sanguinarine, the biosynthesis of BIA is strictly associated with differentiation and the development of specific cell types (Facchini and Bird, 1998; Rezaei et al., 2016a; Rezaei et al., 2016b). Thus, with the exception of sanguinarine, cell suspension culture fails to produce BIAs as they lack differentiation. Although offering accelerated and accessible opportunities to synthesize BIA, synthetic approaches are unable to satisfy the long-term advantage. Given that BIAs possess one or more chiral centers and petroleum-based solvents are being used for the production and purification of intermediates and target products (Wu and Chappell, 2008), the chemical synthesis of BIAs is not straightforward, cost-competitive, and environmentally friendly. Carrying differentiated cells and utilizing sunlight and CO2 instead of reactants derived from petroleum, opium poppy is a sole and sustainable option for BIA production. However, their low quantity in poppy plants imposes a barrier to produce them commercially. This prompted us to summarize as well as propose biotechnological strategies beneficial for the concurrent enhancement of the BIA content in opium poppy.
Elicitation has recently emerged as an effective biotechnological approach for stimulating or enhancing plant secondary metabolites. Elicitors are substances of biotic or abiotic origin that trigger plant defense responses and secondary metabolism. This assists the elicited plants to retain their productivity and viability (Jeyasri et al., 2023). Elicitors fall into two categories: abiotic and biotic. Abiotic elicitors can be further divided into physical, chemical, and hormonal stimuli, among which are salinity, drought, UV radiation, heavy metals, nanoparticles, methyl jasmonate (MeJA), and salicylic acid (SA). Signals originated from endophytic fungi or bacteria, pathogenic microorganisms, including lysates and yeast extract, or plants, such as polysaccharides, glycoproteins, and pectin which are counted as biotic elicitors. The biological and non-biological factors have elevated the capacity of opium poppy for producing and accumulating BIAs (Table 1).
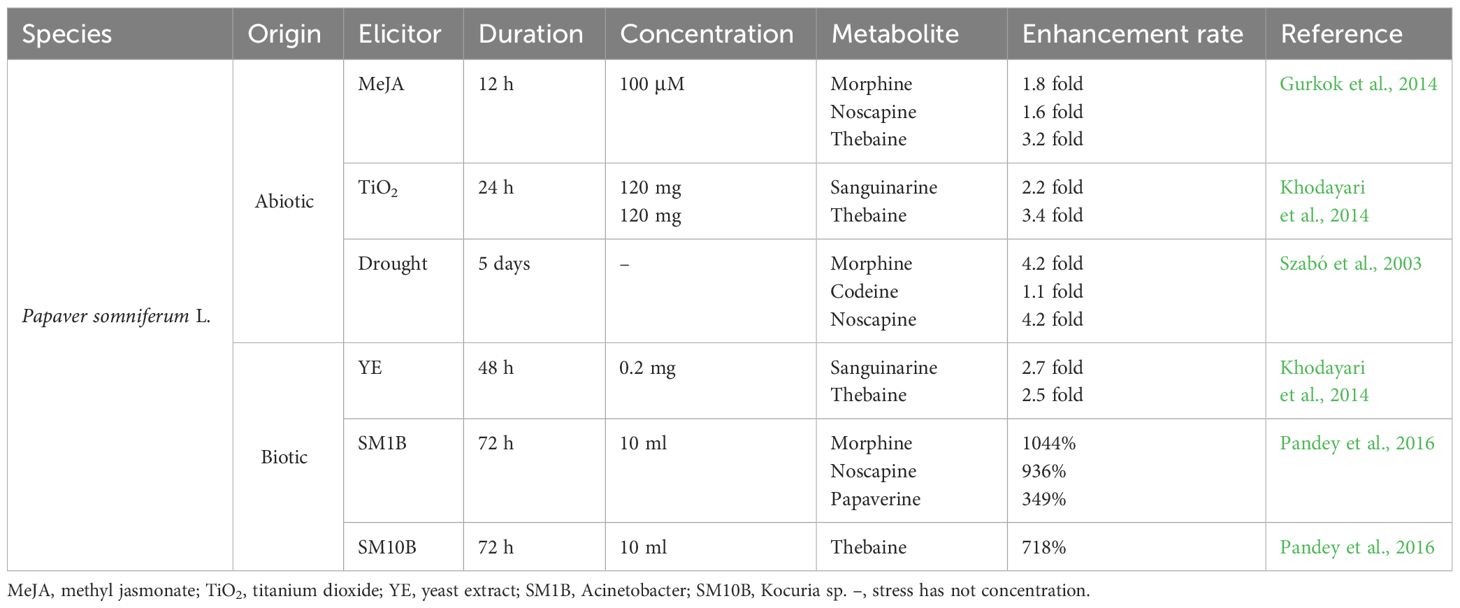
Table 1 Positive impacts of abiotic and biotic elicitors on the benzylisoquinoline alkaloid contents produced in Papaver somniferum L.
The underlying mechanism whereby an elicitor induces the biosynthesis of BIA in opium poppy is analogous to that of other phytochemical compounds in other plant species. When challenged by abiotic and biotic stimuli, the poppy plants witness a series of intracellular events, ranging from physiological to biochemical (Figure 1): The receptor-like kinases (RLKs) and receptor-like proteins (RLPs) anchoring the plasma membrane sense an external elicitor (Liang and Zhang, 2022), initiating the transfer of the signal to the downstream proteins present in the plasma membrane and cytosol, resulting in their reversible phosphorylation. The receptors are ubiquitous in the plant kingdom. Meanwhile, in opium poppy, the activities of phospholipase A2 (PsPLA2) and lipoxygenase (PsLOX) are increased. PsPLA2 hydrolyzes phospholipids, generates fatty acids (Jablonická et al., 2016), and takes a prominent role in MeJA biosynthesis and signaling (Jeyasri et al., 2023), whereas PsLOX is responsible for the stereospecific deoxygenation of polyunsaturated fatty acids, such as linoleic and linolenic acids (Holková et al., 2010). In addition to that, similar to other plant species, ions are channeled into the inside and outside of the cell, i.e., Ca2+ and proton influx as well as Cl- and K+ efflux, resulting in cytosol acidification as well as apoplast alkalization (Humbal and Pathak, 2023). The deficiency of K+ stimulates the expression of NADPH oxidase which catalyzes the production of reactive oxygen species (ROS), such as O2- and H2O2 (Hajiboland, 2012). Following ROS burst, the synthesis of defense chemicals, including flavonoids, tannins, and phytoalexins, is enhanced. In addition, ROS trigger MeJA and SA signaling. With the MeJA signaling pathway, PsAP2/ERF and PsWRKY transcription factors are upregulated in opium poppy, overexpressing various stress-responsive genes, such as BIAs (Mishra et al., 2013; Mishra et al., 2015). Additionally, the mitogen-activated protein kinase (MAPK) pathway is also activated as the other plant species. Through the MAP kinase cascade, MAP kinase kinases (MAPKKK) phosphorylate MAP kinase kinases (MAPKK) followed by the phosphorylation of MAP kinases (MAPK) by MAPKK. The phosphorylated MAPKs are directed to the nucleus in which they phosphorylate group 1 WRKY transcription factors containing two WRKY domains, WRKYGQK, and a zinc finger motif in the N- and C- terminals, respectively (Ishihama and Yoshioka, 2012). In opium poppy, PsWRKY is characterized as the group 1 WRKY (Agarwal et al., 2016), participating in response to wounding, cold, dehydration, and salt stresses (Mishra et al., 2013). PsWRKY is supposed to regulate the BIA pathway (Mishra et al., 2013) so that its activation by phosphorylation promotes the expression of the genes involved in BIA biosynthetic pathways and ultimately increases BIA concentration.
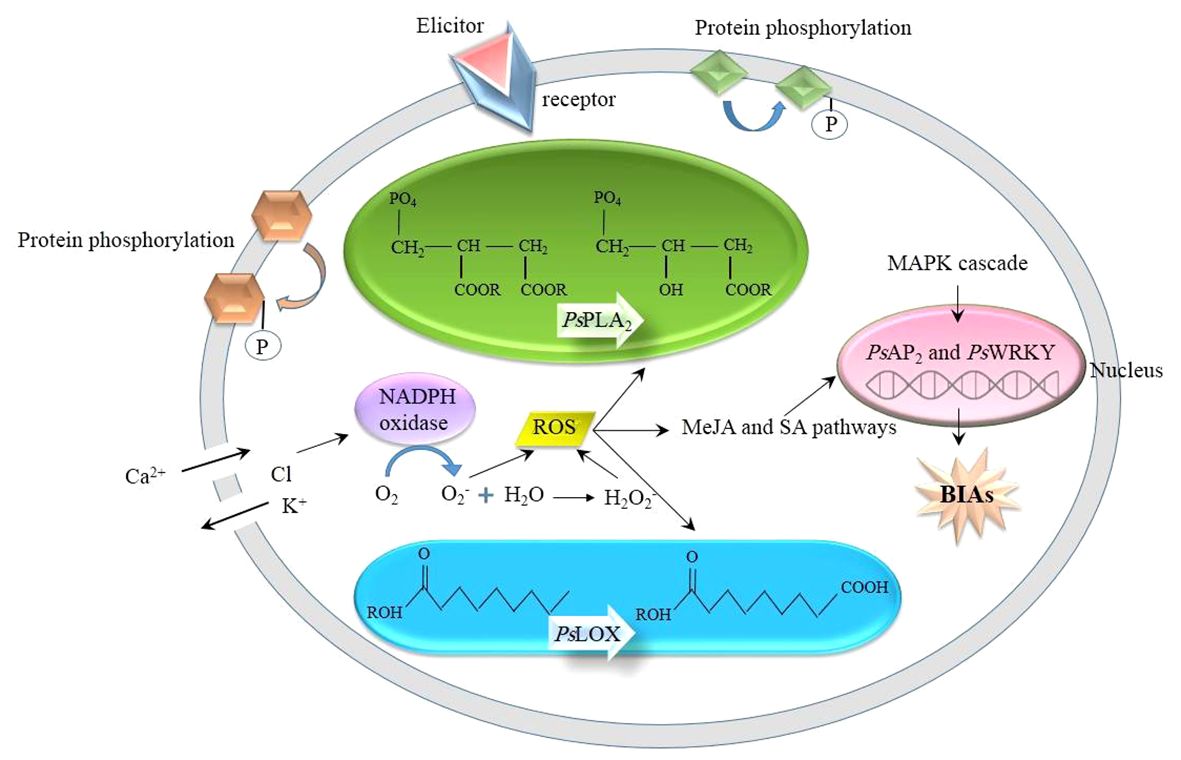
Figure 1 Woven events which are triggered by an external elicitor. BIAs, benzylisoquinoline alkaloids; MAPK; mitogen-activated protein kinase; MeJA, methyl jasmonate; PsLOX, lipoxygenase; PsPLA2, phospholipases A2; ROS, reactive oxygen species; SA, salicylic acid.
Following at least two cycles of whole-genome doubling, polyploid plants are arisen. Genome duplication encourages adaptability, sustainability, tolerance toward biotic and abiotic stresses, growth, and diversity of plants during their historical evolution (Madani et al., 2021). Polyploidization has occurred by either allopolyploidy or autopolyploidy. Allopolyploid organisms arise from hybridization between two genetically divergent species and then the duplication of chromosomes (Yang et al., 2011). On the other hand, autopolyploids are derived from a disturbance in meiotic segregation where chromosomes within a species are multiplied without the subsequent cell division (Figure 2) (Yang et al., 2011). Autopolyploidy can also be induced by the application of chemical and physical agents. The well-known chemicals inhibiting the mitotic cell cycle are colchicine, oryzalin, nitroxide, and dinitroanilines. X-ray, UV radiation, and gamma ray are recognized as physical mutagens (Salma et al., 2017).
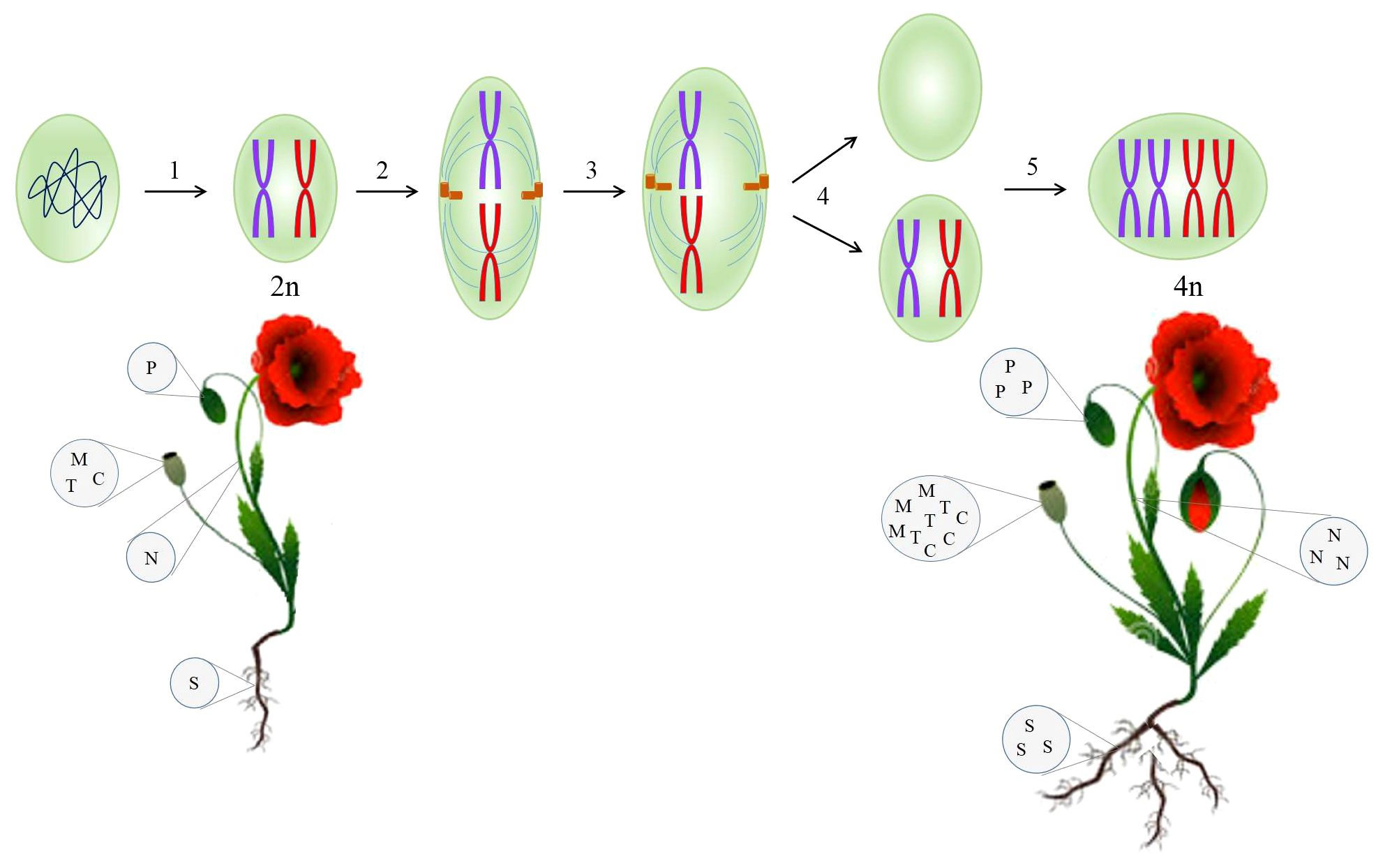
Figure 2 Synthetic autotetraploid developed from a diploid cell through disruption in the mitosis cycle mediated by colchicine, enhancing the biomass and secondary metabolites contents in the resulting plant. 1, prophase; 2, metaphase; 3, anaphase; 4, telophase; 5, S-phase. C, codeine; M, morphine; N, noscapine; P, papaverine; S, sanguinarine; T, thebaine.
Naturally, opium poppy underwent two independent genome doubling events, accounting for 7.8 million years ago and a preceding one (Guo et al., 2018). The presence of duplicated genes whose enzymes participate in BIA metabolism proves this evolutionary history in opium poppy (Hu et al., 2018). To take advantage of this natural incident, a synthetic autopolyploid opium poppy was created using exposure to colchicine (Mishra et al., 2010). As a result of doubling chromosome sets, all the genes involved in BIA pathway showed the higher expression contrary to the diploid counterparts (Mishra et al., 2010). It has been documented that there is a positive correlation between polyploidy and an increase in genetic materials indispensable for transcription, such as DNA template and transcription factors. That is why autopolyploidy enhances gene expression (Mishra et al., 2010; Madani et al., 2021) and ultimately the secondary metabolites concentration (Figure 2). A dramatic increase in morphine and noscapine contents (50%) was observed in artificially synthesized autopolyploid opium poppy (Mishra et al., 2010), which can be attributed to the elevated enzymatic and metabolic activities (Dhawan and Lavania, 1996).
WRKY proteins are principal contributors to plant immunity, metabolism, growth, and development. They transcriptionally regulate genes involved in several biological processes, including biotic and abiotic stress responses (Lee et al., 2018), production of elicitor-activating chemicals, phytohormone synthesis (Schluttenhofer and Yuan, 2015), and senescence (Rushton et al., 2010; Cai et al., 2014). Members of the WRKY family are discriminated from the other via one or more copies of a conserved WRKY domain (WRKYGXK) required for binding to DNA at the N-terminal and a zinc finger motif (CX4-8-C-X22-28-H-X1-2-H/C) at the C-terminus (Eulgem et al., 2000). It has been proven that the WRKY transcription factors function as the transcriptional regulator of the genes responsible for alkaloid biosynthesis (Mishra et al., 2013). In terms of BIAs occurring in opium poppy, PsWRKY assumes the responsibility of expressing virtually all genes involved in these pathways (Kawano et al., 2012; Mishra et al., 2013) as the W-box element (TTGACC/T) interacting with the WRKY domain has been recognized in the promoter regions of these genes (Winzer et al., 2012; Agarwal et al., 2016). Moreover, a close association between the gene transcript encoding PsWRKY and tissues synthetizing and accumulating BIAs was reported in opium poppy (Mishra et al., 2013). The other transcription factor isolated from opium poppy is MYB (Kakeshpour et al., 2015). MYB protein is a key transcriptional regulator of genes involved in the monoterpenoid indole alkaloid (MIA) biosynthesis (Labanca et al., 2018). It has been evidenced that MYB motifs are only present in the promoter regions of 10 genes responsible for noscapine synthesis (Winzer et al., 2012). Thus, this TF lacks the capability of comprehensively regulating BIA production, highlighting the fact that the PsWRKY overexpression should be prioritized in this regard.
Transcription factors are known as one of the feasible targets for global manipulation of metabolic pathways. In California poppy, a plant species belonging to the Papaver genus with the BIA production capability, BIA biosynthesis was substantially enhanced as a result of the overexpression of AtWRKY1 isolated from Arabidopsis thaliana (Apuya et al., 2008). Accordingly, given the fact that PsWRKY is a comprehensive activator of the BIA biosynthetic genes, upregulation of the transcription factor is speculated to result in the synchronized accumulation of BIAs in opium poppy through the proposed sequential processes: The PsWRKY protein scans DNA to find the target site (W-box) that shows high affinity for the transcription factor. Following the recognition of the W-box, PsWRKY specifically interacts with it. The other PsWRKYs likewise head the same manner and bind with the other W-boxes. After a while, multiple W-boxes scattered in the promoter regions of the BIA biosynthetic genes (Winzer et al., 2012; Agarwal et al., 2016) are occupied by PsWRKYs. They coordinately initiate the transcription of the genes. Subsequently, several transcripts are produced at the same time. With an enhancement in transcripts, enzymes translated from them are increased as well, resulting in an increase in the number of reactions mediated by these enzymes. Finally, the BIA amount is boosted (Figure 3).
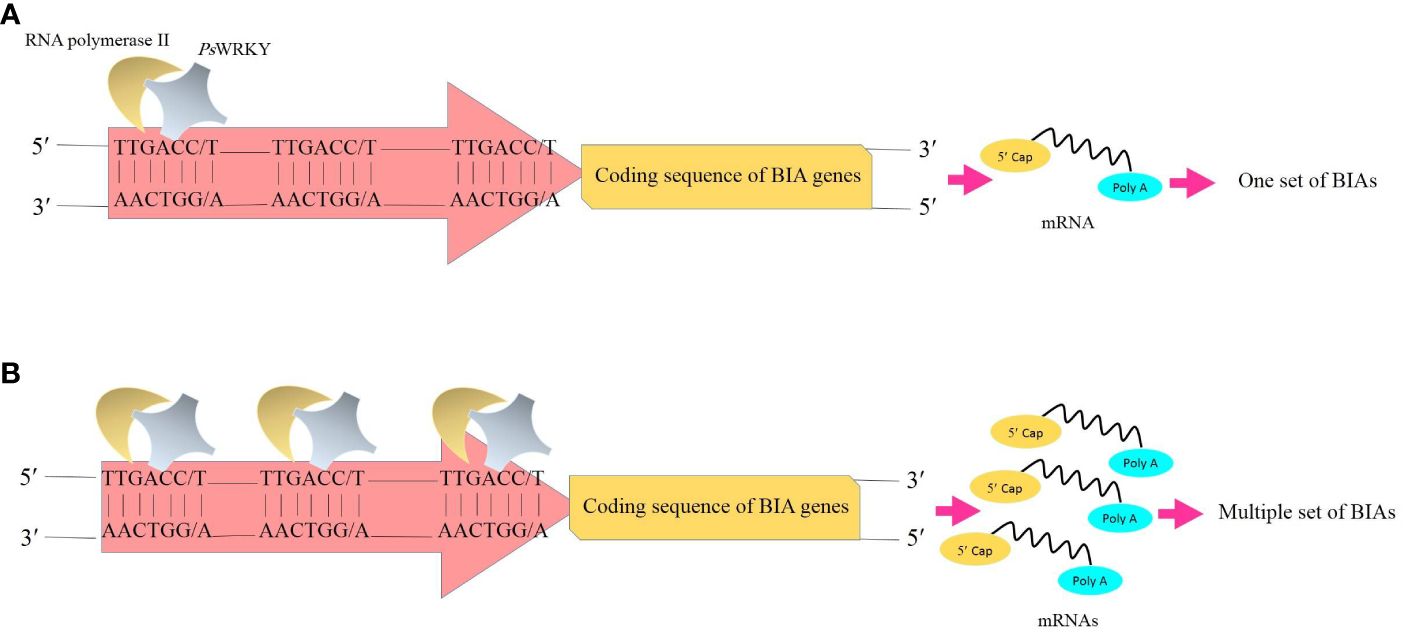
Figure 3 Transcription of the genes whose enzymes are involved in the BIA biosynthetic pathways. (A) Binding of one copy of transcription factor PsWRKY to one W-box (TTGACC/T) in the promoter and subsequently the synthesis of one set of BIAs. (B) Overproduction of BIAs results from the overexpression of PsWRKY and multiple occupancy of W-boxes. BIAs, benzylisoquinoline alkaloids; PsWRKY, Papaver somniferum WRKY transcription factor.
Metabolon (or fusion protein) refers to a perpetual or transient complex of enzymes, in which an intermediate produced by one enzyme is directly transferred to the subsequent enzyme without diffusing into the bulk phase of the cell. Aligning active sites of sequential enzymes protects the cell against toxic intermediates, prevents the consumption of intermediates by competitive pathways, and accelerates the reaction rate (Sweetlove and Fernie, 2018). This phenomenon has been evidenced in primary and secondary metabolisms (Obata, 2019). In opium poppy, the biosynthetic pathway of morphine and its derivatives, codeine and thebaine, enjoys a fusion between a cytochrome P450 (CYP) and an oxidoreductase. The CYP enzyme shoulders the conversion of (S)-reticuline to 1,2-dehydroreticuline, while another enzyme, oxidoreductase, is in charge of forming (R)-reticuline from 1,2-dehydroreticuline (Farrow et al., 2015; Winzer et al., 2015).
It is evidenced that all BIAs in opium poppy are originated from a common precursor, (S)-norcoclaurine (Figure 4). (S)-norcoclaurine is yielded by the condensation of dopamine and 4-hydroxyphenylacetaldehyde (4HPAA) as donors of amine and aldehyde moieties, respectively (Beaudoin and Facchini, 2014) (Figure 4). This reaction which is mediated by norcoclaurine synthase (NCS) allows entry into BIA metabolism (Samanani et al., 2004). With the activities of an aminotransferase coupled with an uncharacterized 4-hydroxyphenylpyruvate decarboxylase (4HPPDC), L-tyrosine converts to 4HPAA in the consecutive reactions (Figure 4) (Lee and Facchini, 2011). The L-tyrosine decarboxylation performed by L-tyrosine/L-DOPA decarboxylase (TYDC) generates tyramine which subsequently undergoes 3-hydroxylation, resulting in dopamine synthesis (Facchini and De Luca, 1994). The enzyme catalyzing the hydroxylation reaction has not yet been identified in plants (Figure 4). Dopamine has been found to be synthesized from an alternative route. The hydroxylation of L-tyrosine by polyphenol oxidases generates 3,4-dihydroxy-L-phenylalanine (L-DOPA) which is then accepted by TYDC that removes a carboxyl group from L-DOPA, forming dopamine (Figure 4) (Facchini and De Luca, 1994).
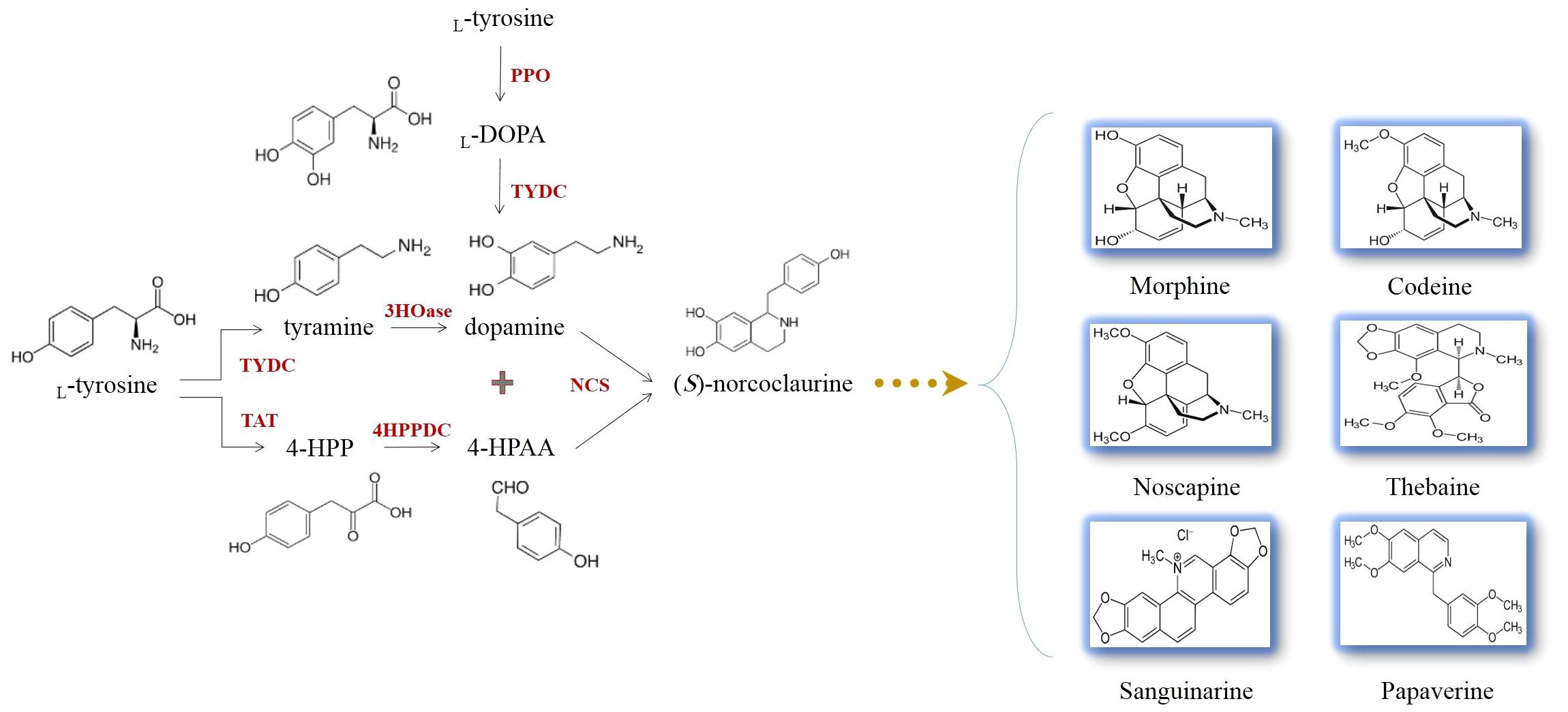
Figure 4 The production of (S)-norcoclaurine allows the biosynthesis of L-tyrosine-derived benzylisoquinoline alkaloids occurring in opium poppy. L-DOPA, 3,4-dihydroxy-L-phenylalanine; 3HOase, L-tyrosine/tyramine 3-hydroxylase; 4-HPAA, 4-hydroxyphenylacetaldehyde; 4-HPP, 4-hydroxyphenylpyruvate; 4HPPDC, 4-hydroxyphenylpuruvate decarboxylase; NCS, norcoclaurine synthase; PPO, polyphenol oxidase; TAT, L-tyrosine aminotransferase; TYDC, L-tyrosine/L-DOPA decarboxylase.
NCS displays cooperation between substrate-binding sites, meaning that the NCS-catalyzing step is rate limiting (Samanani and Facchini, 2001; Samanani and Facchini, 2002). Unfortunately, despite occupying the entry-point location and undertaking the regulatory task in BIA biosynthesis, NCS suffers from catalytic inefficiency. It is calculated that the average catalytic efficiency (kcat/Km) of enzymes is 125 mM, but this parameter for NCS plummets to 1.0 mM (Samanani and Facchini, 2001, 2002). A high Km for both dopamine and 4HPAA highlights the fact that remarkable substrate concentrations should be afforded for NCS to achieve a reasonable turnover (kcat) (Lichman et al., 2015). Among the techniques employed to improve the enzyme kinetic parameters is the formation of a synthetic metabolon. In this regard, since NCS activity is substantially enhanced by an increase in dopamine content (Samanani et al., 2004), an enzyme (i.e., TYDC) that can provide dopamine for NCS is the best partner to fuse with NCS (Figure 5). Interestingly, TYDC shows a strong preference for dopamine than L-tyrosine (Facchini and De Luca, 1994). Thus, NCS–TYDC fusion is expected to supply more available dopamine for NCS and may increase its kcat via confining produced dopamine to a small location and avoiding the entrance of it into competitive pathways, such as phenethylisoquinoline and emetine alkaloids (Figure 5) (Nomura and Kutchan, 2010; Polturak and Aharoni, 2018). This could lead to enhanced (S)-norcoclaurine amount and finally metabolic flux into BIA biosynthesis in opium poppy.
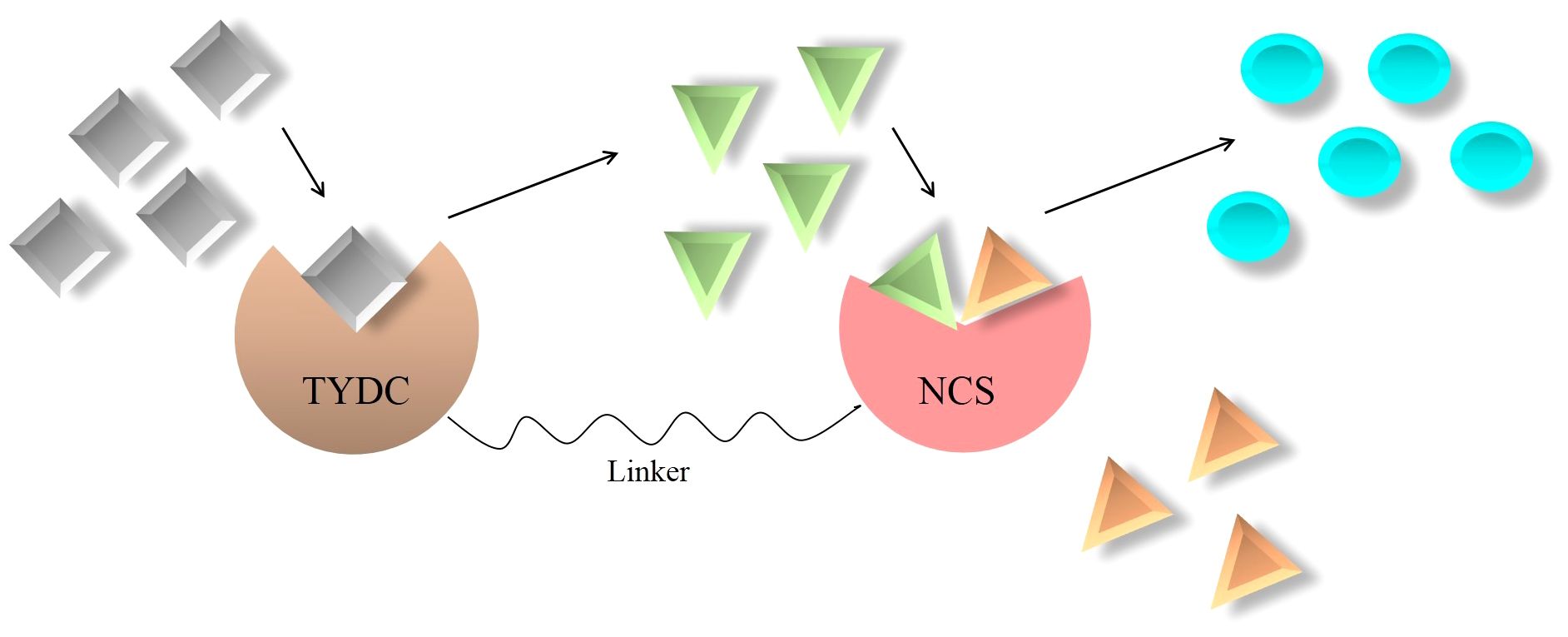
Figure 5 Artificially synthesized metabolon consisting of key enzymes involved in benzylisoquinoline alkaloid biosynthesis. NCS, norcoclaurine synthase; TYDC, L-tyrosine/L-DOPA decarboxylase;  , L-DOPA;
, L-DOPA;  , dopamine;
, dopamine;  , 4-hydroxyphenylacetaldehyde;
, 4-hydroxyphenylacetaldehyde;  , (S)-norcoclaurine.
, (S)-norcoclaurine.
It is noteworthy that the direct fusion of proteins leaves the fusion enzymes crippled. They are unable to fold properly, perform biological activities, and generate sufficient products (Zhang et al., 2009). Therefore, choosing or designing short stretches of amino acids designated as linkers is of great importance. Linkers bring interacting proteins in close proximity, retain their bioactivity, and increase cooperative interactions (Gokhale and Khosla, 2000). Based upon amino acid residues, linkers are put into three groups: flexible, rigid, and cleavable. Flexible linkers usually consist of residues that are small in size and vary in polarity, such as Gly, Ser, and Thr (Chen et al., 2013). The proline-rich linkers possess a rigid structure bringing about hindrance to the mobility of and unfavorable interactions between joined proteins (Chen et al., 2013). Amino acid sequences susceptible to cleavage by enzymes, radiation, or chemicals make up cleavable linkers (Chen et al., 2013). The sequence of residues providing flexibility is among the most preferable linkers. They allow movement, improve conformation, and enhance the performance of the connecting enzymes (Chen et al., 2013). The most widely used flexible linker is (Gly4Ser)n, which can be inserted into the NCS and TYDC fusion (Figure 5). However, the number of repetition “n” must be optimized to acquire enzymes of proper folding and appropriate distance.
Clustered regularly interspaced short palindromic repeats (CRISPR) have been around for approximately two decades in the biotechnology era. This recent methodology functions with the aid of a CRISPR-associated (Cas) endonuclease accompanied by a synthetic single guide RNA (sgRNA) with up to 30 nucleotides. The sgRNA tightly binds with the specific sequence within a gene, marking the desirable site for cleavage. The Cas9 nuclease enzyme recognizes the target location and heads the cleavage of the double-stranded DNA immediately, which is followed by a short sequence named protospacer-adjacent motif (PAM) (Collias and Beisel, 2021). The double-strand break triggers natural DNA repair mechanisms categorized into two distinct groups: non-homologous end joining (NHEJ) and homology-directed (HDR) (Arora and Narula, 2017). The repair mechanisms work well, but sometimes insertion/deletion (InDel) and substitution mutations occur in the target sequence(s). The discriminating attributes of CRISPR in contrast to conventional editing tools are five folds as: robustness, accuracy, sequence specificity, straightforwardness, and commercial effectiveness (Jaganathan et al., 2018). The capability of CRISPR for editing the BIA biosynthetic genes has been proven. The sgRNA was designed according to the 4′OMT2 gene, a regulatory gene acting prior to the divergence of different BIA pathways. The sgRNA-4′OMT2 and Cas9 encoding vectors were delivered into the opium poppy leaves by means of Agrobacterium. The genetically transformed plant carried InDels. Accordingly, BIA production was markedly reduced (Alagoz et al., 2016).
The amino acid which donates the amine group to BIAs is L-tyrosine. L-Tyrosine is generated from the shikimate pathway responsible for the synthesis of myriad primary and secondary metabolites (Maeda and Dudareva, 2012). The steps involved in the shikimate pathway have been fully elucidated. In this pathway, chorismate is yielded via successive reactions. Chorismate is subsequently converted to prephenate by chorismate mutase (CM). The transamination of prephenate results in arogenate synthesis (Maeda and Dudareva, 2012). L-Tyrosine is a product of arogenate dehydration/decarboxylation catalyzed by arogenate dehydrogenase (ADH) (Figure 6). As a branch point intermediate, arogenate is also converted to phenylalanine by the activity of arogenate dehydratase (ADT) (Cho et al., 2007) (Figure 6). A growing body of evidence has indicated that the majority of carbon flux is channeled into phenylalanine biosynthesis (Maeda and Dudareva, 2012; Tohge et al., 2013). Hence, to allocate a high percentage of carbon to L-tyrosine biosynthesis, the suppression of ADT with the CRISPR/Cas9 system may direct more carbon to phenylalanine biosynthesis. If this happens, arogenate is highly likely to be converted to L-tyrosine frequently, culminating in plentiful BIAs (Figure 6).
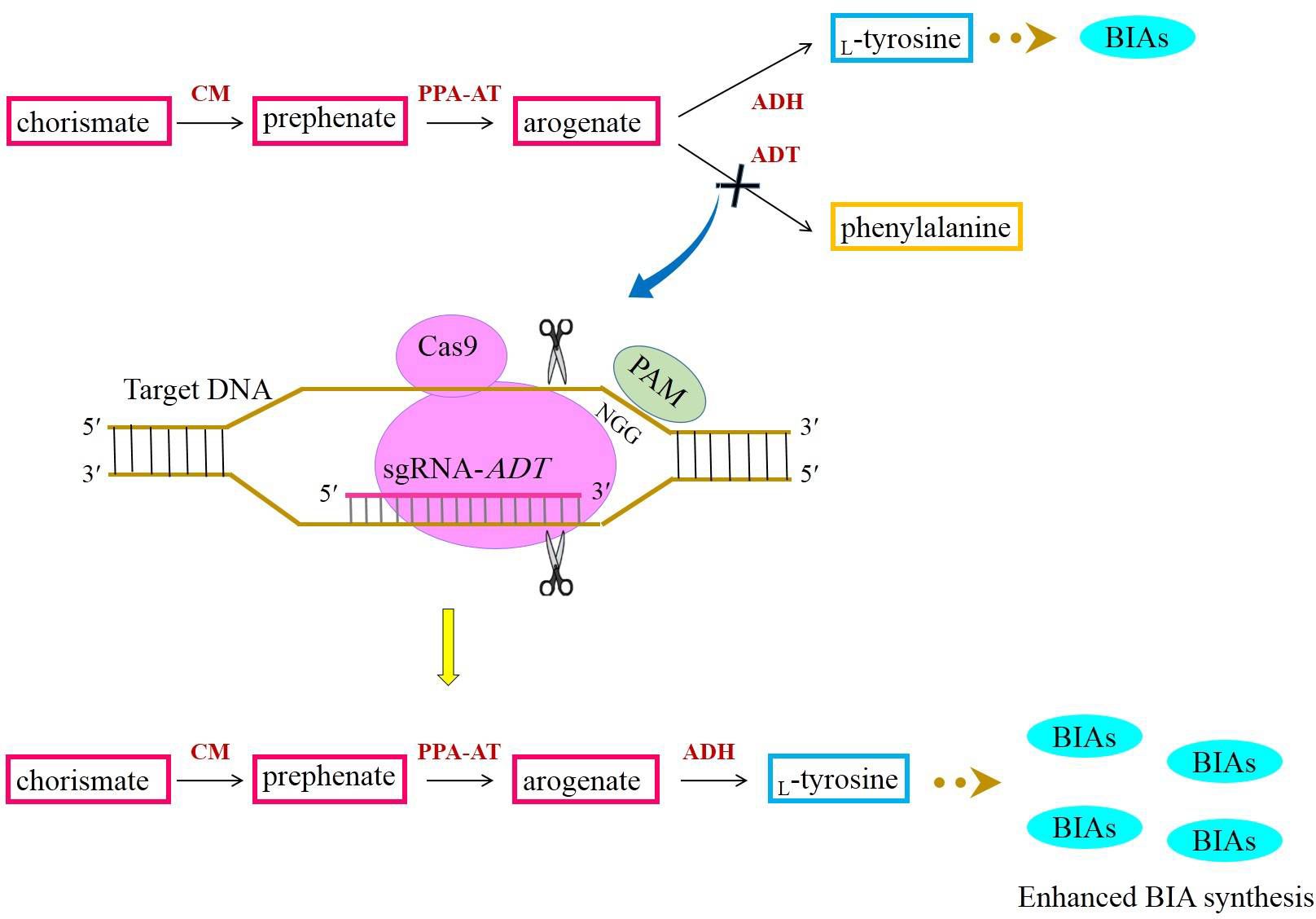
Figure 6 Acquiring higher BIA synthesis through directing more carbon from chorismate to L-tyrosine with the aid of the CRISPR/Cas9-mediated downregulation of ADT. ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; BIAs, benzylisoquinoline alkaloids; CM, chorismate mutase; PAM, protospacer-adjacent motif; PPY-AT, phenylpyruvate aminotransferase.
Phenylalanine is a universal precursor utilized in the biosynthesis of an array of phenolic compounds, such as flavonoids, isoflavones, flavones, phenylpropanoids, anthocyanin, tannins, coumarin, lignans, and lignin (Tohge et al., 2013). Compounds stemmed from phenylalanine encompass 30% of organic matters in plant species (Tohge et al., 2013). It is notable that the plastids are predominantly cellular compartments bearing the phenylalanine biosynthetic route (Maeda and Dudareva, 2012; Lynch, 2022). However, the recent investigations conducted for determining whether the pathway is present in more than one subcellular localization have indicated that the pathway intermediates and enzymes converting chorismate to phenylalanine are localized in the cytosol alike (Lynch, 2022). It means that phenylalanine is also biosynthesized in the cytosol. Thus, if the plastidic ADT is knocked out, the poppy plants will not experience any deleterious effects resulting from the lack of phenylalanine and its derivatives. This is because the cytosolic phenylalanine pathway adequately supplies the plants with phenylalanine.
To globally enhance BIAs, other interesting targets for gene editing using CRISPR are microRNAs (miRNAs). miRNAs are small regulatory RNAs, with nearly 21 nucleotides. They are not capable of coding any proteins but post-transcriptionally regulate the expression of protein-coding genes. In fact, miRNAs connect with complementary sequences present in their target mRNAs, making transcripts vulnerable to cleavage by endonucleases and consequently hampering their translation (Brodersen et al., 2008). miRNAs involved in the growth and development, biotic and abiotic responses, signal transduction, cell differentiation and dedifferentiation, polarity, carbohydrate metabolism, protein conformation, compartmentalization, degradation, and gene transcription have been discovered and functionally characterized in opium poppy (Unver et al., 2010; Boke et al., 2015; Karimi et al., 2017). Furthermore, it has been documented that miRNAs can modulate BIA biosynthesis in poppy plants (Boke et al., 2015; Sabzehzari and Naghavi, 2019). The transcripts of 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT) and 7-O-methyltransferase (7OMT) are subjected to pso-miR2161 and pso-miR13, respectively (Boke et al., 2015). The 4′OMT and 7OMT genes are responsible for the methylation of 3′-hydroxy-N-methylcoclaurine and (S)-reticuline, respectively (Morishige et al., 2000; Ounaroon et al., 2003). pso-miR408 likewise suppresses the genes encoding FAD-binding and BBE domain-containing protein (reticuline oxidase-like protein) (Boke et al., 2015) which form (S)-scoulerine from (S)-reticuline (Facchini et al., 1996). miRNAs targeting the mRNAs of codeinone reductase (COR), salutaridinol 7-O-acetyltransferase (SalAT), and TYDC have also been identified: pso-miR t0047847, pso-miR t0013376, and pso-miR t0000199, respectively (Boke et al., 2015). The two former enzymes yield codeine and thebaine, respectively (Unterlinner et al., 1999; Grothe et al., 2001), while the latter generates tyramine and dopamine (Figure 4) (Facchini and De Luca, 1994). The blocking of pso-miR t0000199 activity using CRISPR may allow the upregulation of TYDC, resulting in higher dopamine production. As depicted in Figure 4, dopamine is a precursor for NCS that synthesizes (S)-norcoclaurine, a distant BIA pathway intermediate. Therefore, by editing the pso-miR t0000199-encoding gene, the poppy plants may witness increased BIA biosynthesis and accumulation.
People of all ages are utilizing plant-derived medicines, in particular, pharmaceutical BIAs, such as morphine and codeine, to relieve their pains. Their expensive chemical synthesis was due largely to their complicated structures coupled with their sparse amounts in plants, which are highly likely to result in the dwindling shortage of herbal remedies. To combat this problem, enhancing the accumulation of such bioactive compounds in the plants that produce them appears to be the most feasible and effective strategy. In this regard, many endeavors have been made with notable successes. However, further studies are still required to identify plants with remarkable concentrations of bioactive compounds. The methods presented here can open a new horizon for further research. If accompanied by omics technologies, bioinformatics, and sequencing, the strategies reviewed and suggested here can broaden our knowledge about regulatory networks and help us to manipulate BIA pathways more precisely, leading to the achievement of the best results.
It is assumed that, in the near future, the techniques mentioned above will be used not only to produce BIAs but also to synthesize other medicinal compounds as they positively affect the metabolic profiles of the plants from which those metabolites originate. They, therefore, would live up to the pharmaceutical industry’s expectations.
ZA: Writing – original draft, Visualization, Investigation, Conceptualization. MN: Writing – review & editing, Supervision. MZ: Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article.
The authors gratefully acknowledge the University of Tehran for providing the research facilities. Moreover, this study was supported by the RUDN University Strategic Academia Leadership Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agarwal, P., Pathak, S., Lakhwani, D., Gupta, P., Asif, M. H., Trivedi, P. K. (2016). Comparative analysis of transcription factor gene families from Papaver somniferum: identification of regulatory factors involved in benzylisoquinoline alkaloid biosynthesis. Protoplasma 253, 857–871. doi: 10.1007/s00709-015-0848-8
Alagoz, Y., Gurkok, T., Zhang, B., Unver, T. (2016). Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 6, 309–310. doi: 10.1038/srep30910
Apuya, N. R., Park, J. H., Zhang, L., Ahyow, M., Davidow, P., Van Fleet, J., et al. (2008). Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol. J. 6, 160–175. doi: 10.1111/j.1467-7652.2007.00302.x
Arora, L., Narula, A. (2017). Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01932
Beaudoin, G. A., Facchini, P. J. (2014). Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240, 19–32. doi: 10.1007/s00425-014-2056-8
Boke, H., Ozhuner, E., Turktas, M., Parmaksiz, I., Ozcan, S., Unver, T. (2015). Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol. J. 13, 409–420. doi: 10.1111/pbi.12346
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
Cai, Y., Chen, X., Xie, K., Xing, Q., Wu, Y., Li, J., et al. (2014). Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PloS One 9, 1–13. doi: 10.1371/journal.pone.0102529
Chen, X., Zaro, J. L., Shen, W. C. (2013). Fusion protein linkers: property, design and functionality. Adv. Drug Deliv Rev. 65, 1357–1369. doi: 10.1016/j.addr.2012.09.039
Cho, M. H., Corea, O. R. A., Yang, H., Bedgar, D. L., Laskar, D. D., Anterola, A. M., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana—identification and characterization of arogenate dehydratases. J. Biol. Chem. 282, 30827–30835. doi: 10.1074/jbc.M702662200
Collias, D., Beisel, C. L. (2021). CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 12, 555. doi: 10.1038/s41467-020-20633-y
Desgagne´-Penix, I., Facchini, P. J. (2012). Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72, 331–344. doi: 10.1111/j.1365-313X.2012.05084.x
Dhawan, O. E., Lavania, U. C. (1996). Enhancing the productivity of secondary metabolites via induced polyploidy: a review. Euphytica 87, 81–89. doi: 10.1007/BF00021879
Dvorák, Z., Vrzal, R., Maurel, P., Ulrichová, J. (2006). Differential effects of selected natural compounds with anti-inflammatory activity on the glucocorticoid receptor and NF-kappaB in HeLa cells. Chem. Biol. Interact. 159, 117–128. doi: 10.1016/j.cbi.2005.10.105
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Facchini, P. J., Bird, D. A. (1998). Developmental regulation of benzylisoquinoline alkaloids biosynthesis in opium poppy plants and tissue cultures. Vitro Cell Dev. Biol.–Plant 34, 69–79. doi: 10.1007/BF02823126
Facchini, P. J., De Luca, V. (1994). Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J. Biol. Chem. 269, 26684–26690. doi: 10.1016/S0021-9258(18)47073-1
Facchini, P. J., Park, S. U. (2003). Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochem 64, 177–186. doi: 10.1016/S0031-9422(03)00292-9
Facchini, P. J., Penzes, C., Johnson, A., Bull, D. (1996). Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 112, 1669–1677. doi: 10.1104/pp.112.4.1669
Farrow, S. C., Hagel, J. M., Beaudoin, G. A., Burns, D. C., Facchini, P. J. (2015). Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 11, 728–732. doi: 10.1038/nchembio.1879
Galadari, S., Rahman, A., Pallichankandy, S., Thayyullathil, F. (2017). Molecular targets and anticancer potential of sanguinarine–a benzophenanthridine alkaloid. Phytomed 4, 143–153. doi: 10.1016/j.phymed.2017.08.006
Gokhale, R. S., Khosla, C. (2000). Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol. 4, 22–27. doi: 10.1016/S1367-5931(99)00046-0
Grothe, T., Lenz, R., Kutchan, T. M. (2001). Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy P. Somniferum. J. Biol. Chem. 276, 30717–30723. doi: 10.1074/jbc.M102688200
Guo, L., Winzer, T., Yang, X., Li, Y., Ning, Z., He, Z., et al. (2018). The opium poppy genome and morphinan production. Science 362, 343–347. doi: 10.1126/science.aat4096
Gurkok, T., Turktas, M., Parmaksiz, I., Unver, T. (2014). Transcriptome profiling of alkaloid biosynthesis in elicitor induced opium poppy. Plant Mol. Biol. Rep. 33, 673–688. doi: 10.1007/s11105-014-0772-7
Hagel, J. M., Facchini, P. J. (2013). Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672. doi: 10.1093/pcp/pct020
Hajiboland, R. (2012). “Effect of micronutrient deficiencies on plants stress responses,” in Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability. Eds. Ahmad, P., Prasad, M. N. V. (Springer press, New York), 283–329.
Hamilton, G. R., Baskett, T. F. (2000). In the arms of morpheus the development of morphine for postoperative pain relief. Can. J. Anaesth. 47, 367–374. doi: 10.1007/BF03020955
Holková, I., Bezáková, L., Bilka, F. S., Balazová, A., Vanko, M., Blanáriková, V. (2010). Involvement of lipoxygenase in elicitor-stimulated sanguinarine accumulation in Papaver somniferum suspension cultures. Plant Physiol. Biochem. 48, 887–892. doi: 10.1016/j.plaphy.2010.08.004
Hu, Y., Zhao, R., Xu, P., Jiao, Y. (2018). The genome of opium poppy reveals evolutionary history of morphinan pathway. Genom. Proteom. Bioinform. 16, 460–462. doi: 10.1016/j.gpb.2018.09.002
Humbal, A., Pathak, B. (2023). Influence of exogenous elicitors on the production of secondary metabolite in plants: A review (VSI: secondary metabolites). Plant Stress 8, 1–13. doi: 10.1016/j.stress.2023.100166
Ishihama, N., Yoshioka, H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437. doi: 10.1016/j.pbi.2012.02.003
Jablonická, V., Mansfeld, J., Heilmann, I., zinský, M. O., Heilmann, M. (2016). Identification of a secretory phospholipase A2 from Papaver somniferum L. that transforms membrane phospholipids. Phytochem 129, 4–13. doi: 10.1016/j.phytochem.2016.07.010
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., Venkataraman, G. (2018). CRISPR for crop improvement: An update review. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00985
Jeyasri, R., Muthuramalingam, P., Karthick, K., Shin, H., Choi, S. H., Ramesh, M. (2023). Methyl jasmonate and salicylic acid as powerful elicitors for enhancing the production of secondary metabolites in medicinal plants: an updated review. Plant Cell Tiss. Organ Cult. 153, 447–458. doi: 10.1007/s11240-023-02485-8
Kakeshpour, T., Nayebi, S., Rashidi Monfared, S., Moieni, A., Karimzadeh, G. (2015). Identification and expression analyses of MYB and WRKY transcription factor genes in Papaver somniferum L. Physiol. Mol. Biol. Plants 21, 465–478. doi: 10.1007/s12298-015-0325-z
Karimi, A. A., Naghavi, M. R., Nasiri, J. (2017). Identification of miRNAs and their related target genes in Papaver somniferum. Iran. J. Field Crop Sci. 48, 1161–1170. doi: 10.22059/ijfcs.2017.230802.654301
Kawano, N., Kiuchi, F., Kawahara, N., Yoshimatsu, K. (2012). Genetic and phenotypic analyses of a Papaver somniferum T-DNA insertional mutant with altered alkaloid composition. Pharm. (Basel) 5, 133–154. doi: 10.3390/ph5020133
Khodayari, M., Omidi, M., Shahnejat Boushahri, A. K., Yazdani, D., Naghavi, M. R., Kadkhoda, Z. (2014). Effect of biotic elicitor and nanoelicitor on the increase in the production of some alkaloids in opium poppy (Papaver somniferum). Iran. J. Hortic. Sci. 45, 287–295. doi: 10.22059/ijhs.2014.52877
Labanca, F., Ovesnà, J., Milella, L. (2018). Papaver somniferum L. taxonomy, uses and new insight in poppy alkaloid pathways. Phytochem. Rev. 17, 853–871. doi: 10.1007/s11101-018-9563-3
Lee, E. J., Facchini, P. J. (2011). Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol. 157, 1067–1078. doi: 10.1104/pp.111.185512
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11, 1–12. doi: 10.1186/s12284-018-0199-0
Liang, X., Zhang, J. (2022). Regulation of plant responses to biotic and abiotic stress by receptor-like cytoplasmic kinases. Stress Biol. 2, 25. doi: 10.1007/s44154-022-00045-2
Lichman, B. R., Gershater, M. C., Lamming, E. D., Pesnot, T., Sula, A., Keep, N. H., et al. (2015). Dopamine-first' mechanism enables the rational engineering of the norcoclaurine synthase aldehyde activity profile. FEBS J. 282, 1137–1151. doi: 10.1111/febs.13208
Lynch, J. H. (2022). Revisiting the dual pathway hypothesis of chorismate production in plants. Hortic. Res. 9, uhac052. doi: 10.1093/hr/uhac052
Madani, H., Escrich, A., Hosseini, B., Sanchez-Muñoz, R., Khojasteh, A., Palazon, J. (2021). Effect of polyploidy induction on natural metabolite production in medicinal plants. Biomolecules 11, 1–14. doi: 10.3390/biom11060899
Maeda, H., Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Mahmoudian, M., Rahimi-Moghaddam, P. (2009). The Anti-cancer activity of noscapine: A review. Recent Pat. Anti-Cancer Drug Discovery 4, 92–97. doi: 10.2174/157489209787002524
Mishra, B. K., Pathak, S., Sharma, A., Trivedi, P. K., Shukla, S. (2010). Modulated gene expression in newly synthesized auto-tetraploid of Papaver somniferum L. S. Afr. J. Bot. 76, 447–452. doi: 10.1016/j.sajb.2010.02.090
Mishra, S., Phukan, U. J., Tripathi, V., Singh, D. K., Luqman, S., Shukla, R. K. (2015). PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 89, 173–186. doi: 10.1007/s11103-015-0361-7
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M. M., Shanker, K., et al. (2013). Wound induced transcriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PloS One 8, 1–5. doi: 10.1371/journal.pone.0052784
Morishige, T., Tsujita, T., Yamada, Y., Sato, F. (2000). Molecular characterization of the S-adenosyl-l-methionine: 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in C. Japonica. J. Biol. Chem. 275, 23398–23405. doi: 10.1074/jbc.M002439200
Nomura, T., Kutchan, T. M. (2010). Three new O-methyltransferases are sufficient for all O-methylation reactions of ipecac alkaloid biosynthesis in root culture of Psychotria ipecacuanha. J. Biol. Chem. 285, 7722–7738. doi: 10.1074/jbc.M109.086157
Obata, T. (2019). Metabolons in plant primary and secondary metabolism. Phytochem. Rev. 18, 1483–1507. doi: 10.1007/s11101-019-09619-x
Ounaroon, A., Decker, G., Schmidt, J., Lottspeich, F., Kutchan, T. M. (2003). (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum—-cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 36, 808–819. doi: 10.1046/j.1365-313X.2003.01928.x
Pandey, S. S., Singh, S., Babu, C. S. V., Shanker, K., Shrivastava, N. K., Kalra, A. (2016). Endophytes of opium poppy differentially modulate host plant productivity and genes for the biosynthetic pathways of benzylisoquinoline alkaloids. Planta 243, 1097–1114. doi: 10.1007/s00425-016-2467-9
Pathan, H., Williams, J. (2012). Basic opioid pharmacology: an update. Br. J. Pain. 6, 11–16. doi: 10.1177/2049463712438493
Polturak, G., Aharoni, A. (2018). ‘‘La Vie en Rose’’: biosynthesis, sources, and applications of betalain pigments. Mol. Plant 11, 7–22. doi: 10.1016/j.molp.2017.10.008
Rahmanian-Devin, P., Baradaran Rahimi, V., Jaafari, M. R., Golmohammadzadeh, S., Sanei-Far, Z., Askari, V. R. (2021). Noscapine, an emerging medication for different diseases: A mechanistic review. Evid. Based Complement Alternat. Med. 2021, 8402517. doi: 10.1155/2021/8402517
Rezaei, M., Naghavi, M. R., Hoseinzade, A. H., Abbasi, A. (2016a). Developmental accumulation of thebaine and some gene transcripts in different organs of Papaver bracteatum. Ind. Crops Prod. 80, 262–268. doi: 10.1016/j.indcrop.2015.11.009
Rezaei, M., Naghavi, M. R., Hoseinzade, A. H., Abbasi, A. (2016b). Measurement of some benzylisoquinoline alkaloids in different organs of Persian poppy during ontogenetical stages. Chem. Biodivers. 13, 539–543. doi: 10.1002/cbdv.201500172
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sabzehzari, M., Naghavi, M. R. (2019). Phyto-miRNAs-based regulation of metabolites biosynthesis in medicinal plants. Gene 682, 13–24. doi: 10.1016/j.gene.2018.09.049
Salma, U., Kundu, S., Mandal, N. (2017). Artificial polyploidy in medicinal plants: Advancement in the last two decades and impending prospects. J. Crop Sci. Biotechnol. 20, 9–19. doi: 10.1007/s12892-016-0080-1
Samanani, N., Facchini, P. J. (2001). Isolation and partial characterization of norcoclaurine synthase, the first committed step in benzylisoquinoline alkaloid biosynthesis, from opium poppy. Planta 213, 898–906. doi: 10.1007/s004250100581
Samanani, N., Facchini, P. J. (2002). Purification and characterization of norcoclaurine synthase. The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J. Biol. Chem. 277, 33878–33883. doi: 10.1074/jbc.M203051200
Samanani, N., Liscombe, D. K., Facchini, P. J. (2004). Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40, 302–313. doi: 10.1111/j.1365-313X.2004.02210.x
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Sweetlove, L. J., Fernie, A. R. (2018). The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 9, 1–12. doi: 10.1038/s41467-018-04543-8
Szabó, B., Tyihák, E., Szabó, G., Botz, L. (2003). Mycotoxin and drought stress induced change of alkaloid content of Papaver somniferum plantlets. Acta Bot. Hung. 45, 409–417. doi: 10.1556/ABot.45.2003.3-4.15
Tohge, T., Watanabe, M., Hoefgen, R., Fernie, A. R. (2013). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00062
Unterlinner, B., Lenz, R., Kutchan, T. M. (1999). Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy P. Somniferum. Plant J. 18, 465–745. doi: 10.1046/j.1365-313X.1999.00470.x
Unver, T., Parmaksız, I., Du¨ndar, E. (2010). Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Rep. 29, 757–769. doi: 10.1007/s00299-010-0862-4
Winzer, T., Gazda, V., He, Z., Kaminski, F., Kern, M., Larson, T. R., et al. (2012). A Papaver somniferum 10- gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708. doi: 10.1126/science.1220757
Winzer, T., Kern, M., King, A. J., Larson, T. R., Teodor, R. I., Donninger, S. L., et al. (2015). Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349, 309–312. doi: 10.1126/science.aab1852
Wu, S., Chappell, J. (2008). Metabolic engineering of natural products in plants; tools of the trade and challenges for the future. Curr. Opin. Biotechnol. 19, 145–152. doi: 10.1016/j.copbio.2008.02.007
Yang, X., Ye, C. Y., Cheng, Z. M., Tschaplinski, T. J., Wullschleger, S. D., Yin, W., et al. (2011). Genomic aspects of research involving polyploid plants. Plant Cell Tiss. Organ Cult. 104, 387–397. doi: 10.1007/s11240-010-9826-1
Keywords: autopolyploidy, CRISPR/Cas9, elicitor, opium poppy, PsWRKY, synthetic metabolon
Citation: Aghaali Z, Naghavi MR and Zargar M (2024) Promising approaches for simultaneous enhancement of medicinally significant benzylisoquinoline alkaloids in opium poppy. Front. Plant Sci. 15:1377318. doi: 10.3389/fpls.2024.1377318
Received: 27 January 2024; Accepted: 14 March 2024;
Published: 02 April 2024.
Edited by:
Wusirika Ramakrishna, Central University of Punjab, IndiaReviewed by:
Konstantinos Koudounas, Aristotle University of Thessaloniki, GreeceCopyright © 2024 Aghaali, Naghavi and Zargar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Reza Naghavi, bW5hZ2hhdmlAdXQuYWMuaXI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.