- College of Traditional Chinese Medicine, Guangzhou University of Chinese Medicine, Guangzhou, China
Cinnamomum cassia (L.) J.Presl, a tropical aromatic evergreen tree belonging to the Lauraceae family, is commonly used in traditional Chinese medicine. It is also a traditional spice used worldwide. However, little is currently known about the extent of the genetic variability and population structure of C. cassia. In this study, 71 individuals were collected from seven populations across two geographical provinces in China. Nine morphological features, three chemical components, and single nucleotide polymorphism (SNP) markers were used in an integrated study of C. cassia germplasm variations. Remarkable genetic variation exists in both phenotypic and chemical compositions, and certain traits, such as leaf length, leaf width, volatile oil content, and geographic distribution, are correlated with each other. One-year-old C. cassia seedling leaf length, leaf width, elevation, and volatile oil content were found to be the main contributors to diversity, according to principal component analysis (PCA). Three major groupings were identified by cluster analysis based on the phenotypic and volatile oil data. This was in line with the findings of related research using 1,387,213 SNP markers; crucially, they all demonstrated a substantial link with geographic origin. However, there was little similarity between the results of the two clusters. Analysis of molecular variance (AMOVA) revealed that the genetic diversity of C. Cassia populations was low, primarily among individuals within populations, accounting for 95.87% of the total. Shannon’s information index (I) varied from 0.418 to 0.513, with a mean of 0.478 (Na=1.860, Ne =1.584, Ho =0.481, He =0.325, and PPB =86.04%). Genetic differentiation across populations was not significant because natural adaptation or extensive exchange of seeds among farmers between environments, thus maintaining the relationship. Following a population structure analysis using the ADMIXTURE software, 71 accessions were found to be clustered into three groups, with 38% of them being of the pure type, a finding that was further supported by PCA. Future breeding strategies and our understanding of the evolutionary relationships within the C. cassia population would benefit greatly from a thorough investigation of phenotypic, chemical, and molecular markers.
1 Introduction
Cinnamomum cassia (L.) J.Presl, a tropical aromatic evergreen tree belonging to the Lauraceae family, is commonly used in traditional Chinese medicine. It is also a traditional spice used worldwide (National Pharmacopoeia Committee, 2020). Cinnamomum cassia Presl is derived from the bark of the tree trunk and is used extensively worldwide owing to its brilliant flavor and smell. It has great economic value and is useful not only as a daily condiment but also as a raw ingredient for pharmaceuticals. More than 160 compounds have been isolated and identified from C. cassia as a result of the numerous investigations that have been conducted on the pharmacology and phytochemicals of this plant. The primary chemical components of its volatile oils possess anti-inflammatory, antibacterial, and anticancer properties (Shen et al., 2021). Numerous studies have demonstrated the broad spectrum of pharmacological effects of C. cassia, including effects against tumors, reduction of inflammation and pain, diabetes mellitus and obesity, prevention of bacteria and viruses, protection of the cardiovascular system, cytoprotection, neuroprotection, and immunoregulatory responses (Kwon et al., 2006; Hong et al., 2012). C. cassia is found in China, India, Vietnam, Indonesia, and other countries.As a major ingredient in traditional Chinese medicine, C. cassia is typically used throughout Asia. More than 500 formulations containing C. cassia are used to treat various illnesses, including inflammatory diseases, chronic gastrointestinal diseases, gynecological disorders, and cardiovascular diseases. C. cassia has been listed in the People’s Republic of China Pharmacopoeia (CH.P) since 1963. In China, Guangdong and Guangxi have the largest cultivated areas of C. cassia, and the C. cassia planting area of Guangdong Province accounts for more than 30% of the country’s planting area. Guangdong’s C. cassia is mainly distributed in the Zhaoqing and Yunfu cities, and the cultivated C. cassia in these two cities is referred to as “Xijianggui.” Guangxi’s C. cassia planting area accounts for more than 50% of the country’s planting area, and Fangchenggang, Dongxing, Yuli, Guiping, and Beiliu cities are the main producing areas of C. cassia. Among them, Fangchenggang and Dongxing predominantly cultivate a variety known as “Fangchenggui”, while Yulin and Guiping mainly cultivate the “Xijianggui” variety (Yang et al., 2020; Li et al., 2023).
A key component of breeding operations is gathering and studying germplasm resources. Although they are currently in danger of becoming extinct, the wild resources of C. cassia are vital to scientific research and applications because they have mostly been subjected to natural selection and have been little affected by artificial selection. Most C. cassia has been sexually propagated, mostly through seeds, and has been grown artificially for many years. Germplasm diversity is the foundation for preventing genetic erosion and gives plant breeders the chance to create new varieties and enhance them with superior features. Previous studies have demonstrated notable variations in the chemical and physical characteristics of various C. cassia germplasm resources. For example, the leaves of C. cassia from Guangdong Province are larger and contain higher amounts of volatile oil, whereas the leaves from Guangxi are noticeably smaller and contain less volatile oil, with the exception of a few C. cassia cultivation areas (Tung et al., 2010; Liang et al., 2016; Li et al., 2021; Narayanankutty et al., 2021; Chen et al., 2022). It is important to analyze genetic diversity and population structure to design breeding methods and research the genetic links of C. cassia plants. Molecular markers have become powerful tools for genetic research on C. cassia populations, including simple sequence repeats (SSR), inter-SSR (ISSR), internal transcribed spacer (ITS), and psbA-trnH (Liang et al., 2016; Wang, 2022). These studies indicate that C. cassia populations are highly genetically diverse. However, the populations of C. cassia used in earlier research had either limited sample sizes or were relatively sparsely distributed geographically, with Guangdong and Guangxi being the two locations that produced C. cassia. It is possible that the current Chinese C. cassia cultivars’ limited genetic base results from the utilization of a small number of parental genotypes. Consequently, to increase the genetic diversity of improved varieties, it is imperative to look for additional breeding materials in China. To the best of our knowledge, no prior research has utilized single nucleotide polymorphism (SNP) markers to investigate the genetic diversity and population structure of C. cassia germplasm. SNP markers are representative of the third generation of molecular marker technology, which generally refers to DNA sequence polymorphisms caused by the mutation of a single base at the genomic level (Belaj et al., 2018). Currently, SNP molecular markers are considered the most promising molecular markers. They have the advantages of abundance, wide distribution, low mutation frequency, and high genetic stability, among others. SNP markers are used to differentiate and identify extensive plant and animal germplasm resources (Maroso et al., 2019; Sun et al., 2020; Perez et al., 2021). Moreover, they are more directly comparable between different genotypes; therefore, they are more suitable for the genetic analysis of complex and diverse traits. They also contribute to the identification of genes that cause population differences (Ren et al., 2013; Maroso et al., 2019; Nantawan et al., 2019). To successfully utilize invaluable germplasm resources in future breeding efforts, both within China and worldwide, it is mandatory to gain insight into the genomic differentiation and variation of C. cassia genotypes.
In this study, we collected 71 germplasm resources of C. Cassia across Guangdong and Guangxi. Morphological, biochemical, and molecular markers were used to study the germplasm variation of C. cassia comprehensively. The objective of this study was to identify and use diverse gene and genotype resources, investigate the genetic differentiation and population structure of C. cassia, clarify the phylogenetic relationships of C. cassia from different locations, search for superior C. cassia accessions, open up new breeding opportunities, and safeguard germplasm resources.
2 Materials and methods
2.1 Plant materials
Seventy-one C. cassia accessions (RG01–RG71) were collected from three populations in Guangdong (Deqing County, Gaoyao County, and Luoding City) and four populations in Guangxi (Guiping, Pingnan, Yulin, and Fangchenggang) (Supplementary Table S1). Fifty-four accessions were from Guangdong Province, whereas the rest originated in Guangxi. Simultaneously, the seeds of each sample were planted in a germplasm resource nursery located in Deqing County, Guangdong Province (Figure 1A). Morphological data of the one-year-old C. cassia seedlings in the resource nursery were recorded later in this study to remove the influence of the ecological environment on the samples. The sample named RG38 was lost, and subsequent analyses, except for the biochemical analysis, did not include RG38. Samples were identified by Professor Ping Ding (Guangzhou University of Chinese Medicine, Guangdong, China).

Figure 1 The phenotypic characters of C. cassia in different populations. (A) The germplasm resource nursery in Deqing county, Guangdong province; (B) The C. cassia form Guangdong province; (C) The C. cassia form Guangxi Zhuang Autonomous Region.
In a previous study, our research group found that the cinnamaldehyde content in different samples of the same batch of cinnamon medicinal materials was considerably different, and that the volatile oil content between different thicknesses from the same tree was irregular. Therefore, we collected another12 cassia bark samples from the Hetai, Gaoliang, and Tanlin Towns in Guangdong Province to determine the content of the volatile oils cinnamaldehyde, cinnamyl alcohol, cinnamic acid, and 2-methoxycinnamaldehyde (Supplementary Table S2). During sampling, cinnamon trees with a growth age of 8 years andheight of approximately 5 m were selected, and the fixed sampling time was October. The tree bark was cut and peeled from 10 cm above the ground, and the bark of each tree trunk was evenly divided into six parts from the bottom to the top (five parts of cinnamon bark in Hetai Town) and dried in the indoor shade.
2.2 Investigation for germplasm resources of C. cassia
First, the distribution and occurrence of C. cassia in Guangdong and Guangxi were obtained by referring to the literature and searching for internet information (Xu et al., 2004; Wei et al., 2006; Lu et al., 2010; Yang et al., 2013). Gaoyao County, Deqing County, Luoding County, Fangchenggang City, Yulin City, and Guiping City were selected as the field investigation sites. The population sites in this survey included 3 districts in Guangdong and 4 districts in Guangxi, and latitudes and longitudes were located between N22°63’–N23°35’ and E108°01’–E112°25’, respectively. The range of annual mean temperature and precipitation in the seven regions in 2022 were 21.5–22.3 °and 1513.0–2690.0 mm, and the range of elevation was 114–493 m, which were well-suited for the growth of C. Cassia (Supplementary Table S1). C. cassia field survey was carried out by investigating nine important datasets: source, germplasm resource type, latitude and longitude, elevation, annual average temperature, annual average precipitation, height of the original tree, trunk girth of the original tree, and tree age of the original tree. Simultaneously, we measured the height, leaf length, leaf width, and stem diameter of the one-year-old C. cassia seedlings. Leaf length and width of the tenth leaf of one-year-old C. cassia seedlings were measured from bottom to top. Ten seedlings were randomly selected from each sample for measurements. The phenotypic characteristics of C. cassia accessions were measured using a Vernier caliper and tapeline, and the latitude and longitude were determined using a global position system navigator. The meteorological data constituted the annual average meteorological data published by the local meteorological bureau.
2.3 Determination of moisture, water-soluble extract, and volatile oil contents
The moisture, water-soluble extract, and volatile oil contents of C. Cassia were determined according to the determination methods (No.0832, No.2201, and No.2204) from the general rules of the CH.P, 2020 edition. The C. cassia samples were ground to a powder and passed through a 40-mesh sieve to obtain a finer powder. Fifty grams of C. cassia powder, which was precisely weighed and 10 times the volume of distilled water, was placed in a round-bottom flask and soaked for 1 h. The water vapor reflux method was used for reflux extraction for 5 h until there were no obvious oil droplets in the effluent. The cooled effluent was dried over anhydrous sodium sulfate to obtain the volatile oil from C. cassia.
Regarding data processing, Spearman correlation analysis using SPSS 28.0 software was performed on seven morphological parameters of cinnamon (height, trunk girth, tree age of the original tree, height, leaf length, leaf width, and stem diameter of the one-year-old C. Cassia seedlings), elevation, latitude and longitude, and volatile oil content.
2.4 Determination of volatile oil and cinnamaldehyde in cassia bark of different thickness
Chromatographic analysis was performed using HPLC (Unimicro Easy SepTM-1020LC, US). The column was C18 (250 mm × 4.6 m, 5 μm, Ecosil, USA). The mobile phase consisted of acetonitrile (A) and -0.1% phosphoric acid aqueous solution (B), and the linear gradient was set as follows: 0–155 min for 32% A to 45% A, 15–21 min for 45% A to 50% A, and 21–26 min for 50% A. Additionally, a volume flow of 1.0 mL·min−1, sample injection volume of 20 μL, room temperature for the column, and detection wavelength of 260 nm were established (Wu et al., 2019).
The standards for volatile oil components, namely cinnamaldehyde, cinnamyl alcohol, cinnamic acid, and 2-methoxycinnamaldehyde were purchased from Shanghai Yuanye Biotechnology Co., LTD (Shanghai, China). The purity of the standards was higher than 98%, and their lot numbers were B21081, B21080, B21082, and B27438, respectively. Following sample crushing, 0.2 g of each sample was soaked in 25 mL of methanol and weighed. After a half-hour ultrasonic extraction, the sample was weighed, and the lost mass was compensated with methanol. After collecting and filtering the upper layer using a 0.22 μm microporous membrane filter, it was transferred for high-performance liquid chromatography (HPLC) analysis (Wu et al., 2019).
2.5 DNA extraction and sequencing
Approximately 100 mg of fresh stem tissue was placed in a mortar and ground with liquid nitrogen. The ground tissue was transferred into a 1.5 mL centrifuge tube for genomic DNA extraction. Genomic DNA was extracted using the FastPure Plant DNA Isolation Mini Kit. Kit Manufacturer: Nanjing Vazyme Biotech Co., Ltd., China; reagent box type: DC104. The extracted genomic DNA was examined and stored at -20°C. DNA integrity was ascertained using an Agilent 2100 Bioanalyzer, and the quality and concentration of the DNA samples were evaluated using a Nanodrop spectrophotometer (Thermo Scientific, USA).
The VAHTS Universal DNA Library Prep Kit for Illumina V3 (Nanjing Vazyme Biotech Co., Ltd., China) was used to construct a paired-end sequencing library with fragment size of 350 bp from qualified samples. After the library was constructed, Agilent 2100 Bioanalyzer (Agilent Technologies,USA) were used for quality control. An Illumina NovaSeq 6000 high-throughput sequencing platform (Illumina, USA) was used for DNA library sequencing. The PE150 (pin-end, 150) sequencing strategy was used. Illumina high-throughput sequencing results were initially presented as raw image data files, which were converted into raw reads after base calling using the CASAVA software. High-quality clean reads were obtained from the original sequence using Fastp v0.20.1 software.
2.6 SNP calling
Illumina high-throughput sequencing results were initially presented as image data files, which were converted into raw reads after base calling by CASAVA software (https://www.britannica.com/plant/cassava). High quality clean reads were obtained by using Fastp v0.20.1 software (https://github.com/OpenGene/fastp) with the default parameters for data quality control of the original sequences (Supplementary Table S3).
The BWA software (https://github.com/lh3/bwa) was used to align the clean reads of 71 C. cassia accessions to the reference genome sequence using the default settings (Dobin et al., 2013). To reduce SNP detection errors caused by alignment errors, sequencing fragments that were compared to the SNP region were double-ended and simultaneously aligned to the reference sequence (accession number: CNA0140271).
Based on the comparison results, deepvariant software (version 1.3.0, https://github.com/google/deepvariant) was used to detect the SNPs (Poplin et al., 2018; Eriksson et al., 2022), and samtools-mpileup and Python programs were used to test for SNP genotypes, base sequencing quality, and read comparison quality for the comprehensive identification of SNP polymorphic sites.
2.7 SNP genetic diversity analysis
The Shannon-Weaver (H′) index, minor allele frequency (MAF), observed heterozygosity (Ho), expected heterozygosity (He), observed number of alleles (Na), effective number of alleles (Ne), Shannon’s information index (I), percentage of polymorphic bands (PPB), genetic identity (GI), and genetic diversity (GD) are examples of the parameters used to estimate genetic diversity in populations. These parameters were estimated using PowerMarker 3.25 and Popgene version 1.32 software (Danecek et al., 2011). In this study, VCFtools software (version 0.1.16) was used to convert SNP information into a format recognizable by the PLINK software version v1.90 (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al., 2007). Ho, He, MAF, and polymorphism information content (PIC) were calculated using PIC_CALC 0.6 (Nagy et al., 2012).
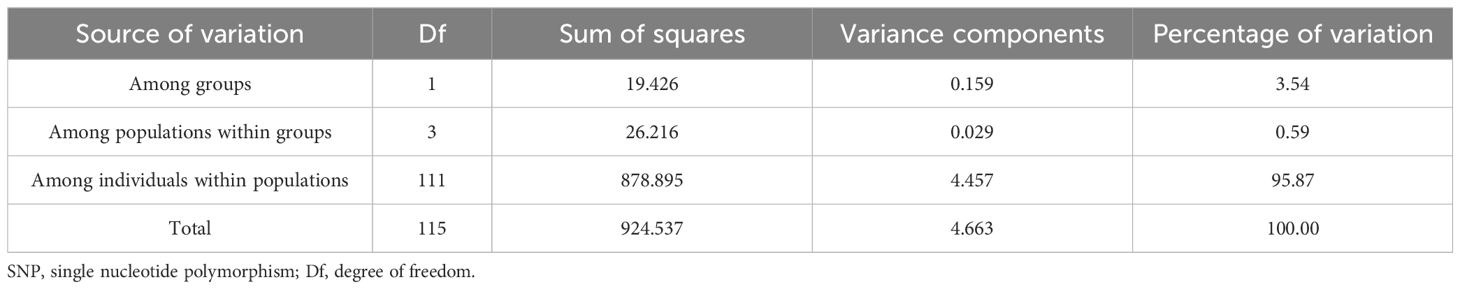
Analysis of molecular variance (AMOVA) using the GeneAlEx 6.502 tool with 1000 permutations was used to characterize the variance components of C. Cassia individuals and population differentiation among the seven postulated subgroups (Peakall and Smouse, 2012).
2.8 Population structure analysis
The ADMIXTURE (Version 1.3.0) software (https://dalexander.github.io/admixture) was used to analyze the population structure (Pritchard et al., 2000). The obtained SNP information is converted into a binary PLINK file that is recognized by ADMIXTURE software. The K value is calculated using ADMIXTURE. Set the K value to 1–24, and theoretically select the K value with the smallest CV error (Cross-validation error) as the best clustering (Perez et al., 2021).
2.9 Phylogenetic and principal component analysis
The ML phylogenetic tree of C. cassia was analyzed using the optimal model GTR+F+R5 implemented in IQ-TREE software (version 2.0.5) (Alexander et al., 2009). The optimal model selection was based on Bayesian Information Criterion (BIC) scores. To provide more evidence for the number and composition of populations of C. cassia accessions, this study also performed principal component analysis (PCA) using the default settings of the Genome-wide Complex-Trait Analysis (GCTA) software (Zheng et al., 2013).
3 Results
3.1 Analysis of morphological traits of C. cassia
The phenotypic characteristics of C. cassia in different populations showed some differences (Figures 1B, C). Table 1 and Supplementary Table S4 show the variations in the primary morphological characteristics of C. cassia from several accessions. In this study, the original C. cassia trees were between 6 and 25 m tall, 35 to 373 cm thick, and 8 to 100 years old; 40 samples were wild and the rest were cultivated. In particular, 37 wild C. cassia samples, which generally had longer growth years and thicker trunks, were sourced from Guangdong, and the other three were sourced from Guangxi. We analyzed the trunk girth and age of the original trees. The average trunk girth and tree age of the original tree from Guangdong were 75.32 cm and 28.32 years, respectively, which were higher than those from Guangxi (40.93 cm and 24.64 years, respectively). The trees in Fangchenggang were older than those in the other areas (Figure 2).
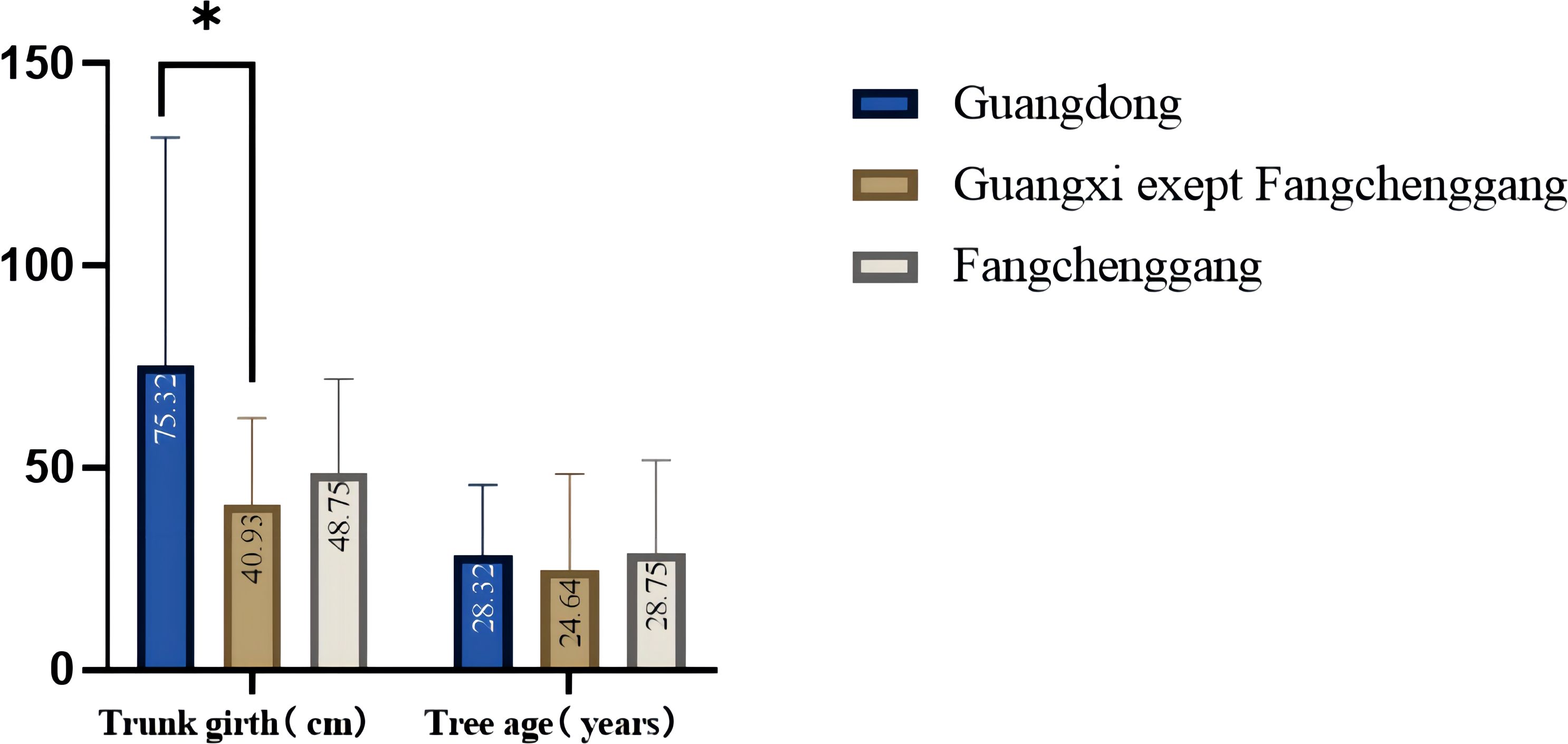
Figure 2 Comparison of trunk girth and age of the original trees in Guangdong and Guangxi (*p<0.05; *p<0.01; ***p<0.001; one-way ANOVA).
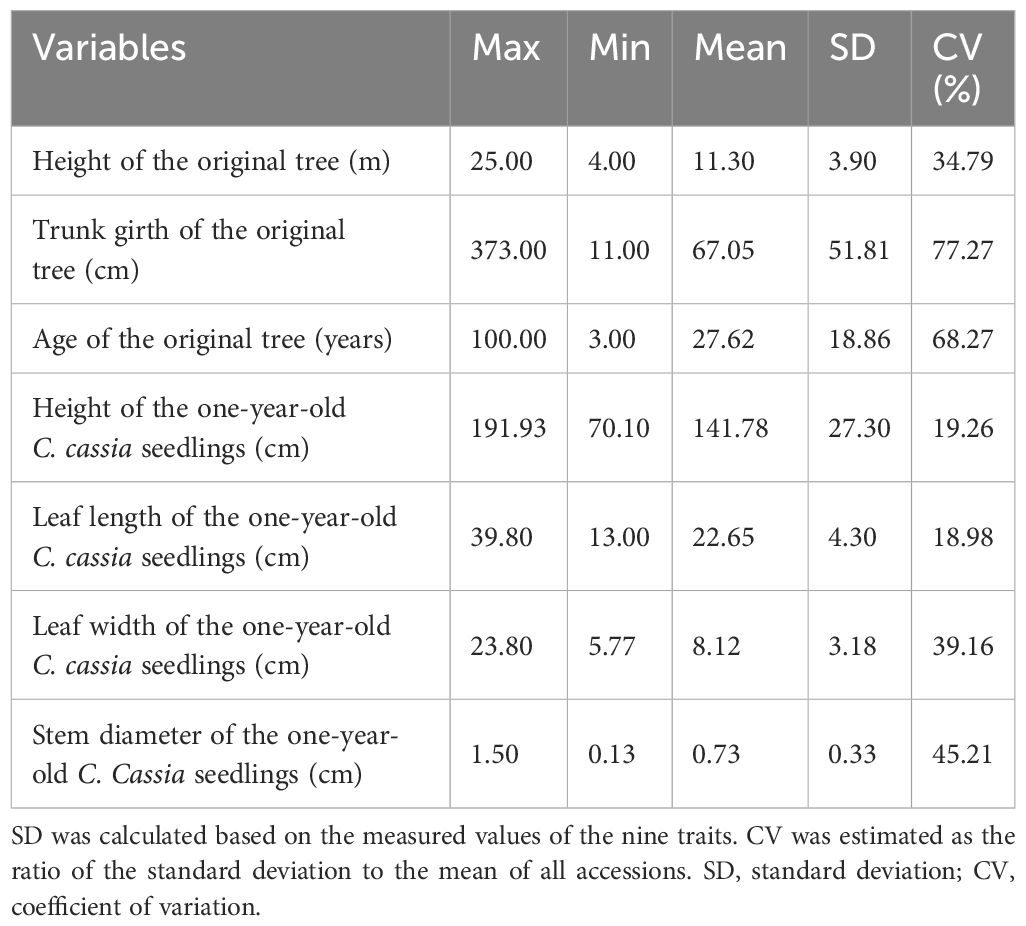
The height of the one-year-old C. cassia seedlings ranged from 70.10 cm to a maximum of 191.93 cm (RG52). The leaf length of the one-year-old C. cassia seedlings ranged from 13.00 cm to a maximum of 39.80 cm (RG70). The leaf width of the one-year-old C. cassia seedlings ranged from 5.77 cm to a maximum of 23.80 cm (RG11). The range of the stem diameter for the one-year-old C. cassia seedlings was 0.13–1.50 cm, with RG68 showing the largest stem diameter (1.50 cm). The average leaf length and width of the one-year-old C. cassia seedlings from Fangchenggang were 32.78 and 13.01 cm, respectively, which were higher than those from Guangdong (21.49 and 7.92 cm, respectively) and other areas of Guangxi (23.47 and 7.04 cm, respectively) (Figure 3). There were no significant differences in leaf length or width between Guangdong and Guangxi, except for Fangchenggang. These findings indicate that the Guangxi germplasm had short development years and fleshy, thick, and relatively wide leaves, particularly Fangchenggang, whereas Guangdong samples generally had lengthy growth years and towering trees with strong trunks.
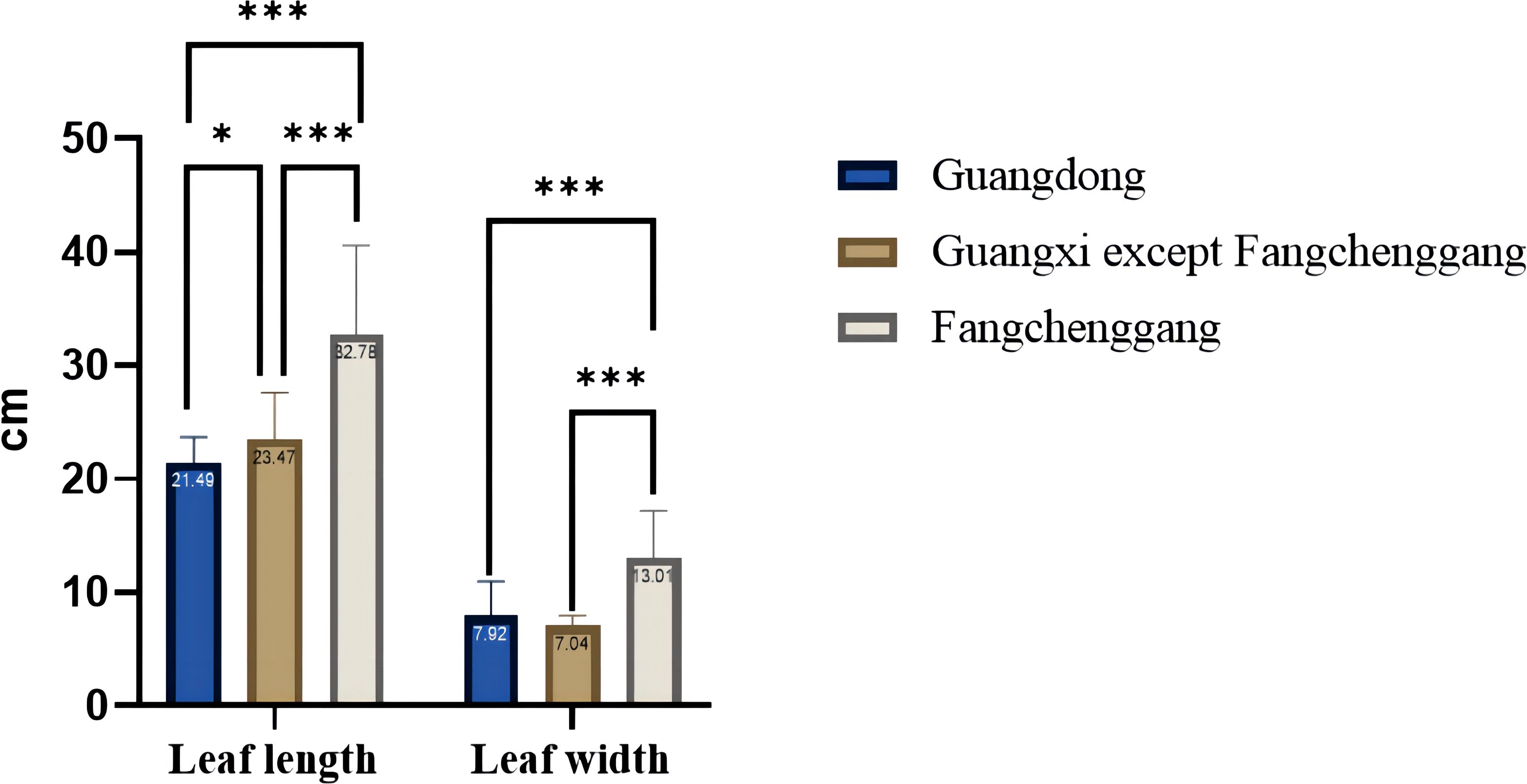
Figure 3 Comparison of leaf length and leaf width from the one-year-old C. cassia seedlings in Guangdong and Guangxi (*p<0.05; *p<0.01; ***p<0.001; one-way ANOVA).
The coefficient of variation (CV) of the phenotypic traits was computed to evaluate the genetic diversity of the C. cassia accessions. With an average of 43.28%, the CVs of the seven attributes ranged from 18.98% to 77.27%. The most variable trait was trunk girth of the original tree (77.27%), followed by tree-age of the original tree (68.27%), stem diameter of the one-year-old C. cassia seedlings (45.21%), and leaf width of the one-year-old C. cassia seedlings (39.16%), while the leaf length of the one-year-old C. cassia seedlings showed the least variation (18.98%), indicating that the tree age varied greatly and the leaf length was almost uniform among different accessions. The majority of the seven phenotypic variables showed CV higher than 20%, indicating clear variability of these features (Table 1).
The morphology of wild species varies greatly across populations. For example, leaves from Guiping and Fangchenggang (Guangxi) are thick, meaty, and somewhat wide. The seeds are black-purple in color and have shiny surfaces. In addition, we found a type of C. cassia with purple volatile oil, strong fragrance, and wider leaves. The local called “Zi You Gui” has larger planting area. In Fangchenggang, we discovered a unique wild plant with extremely large, glabrous leaves and black-purple seeds. The average leaf length and width were 35.00 cm and 15.20 cm, respectively.
3.2 Analysis of moisture, water-soluble extract, and volatile oil contents of C. cassia
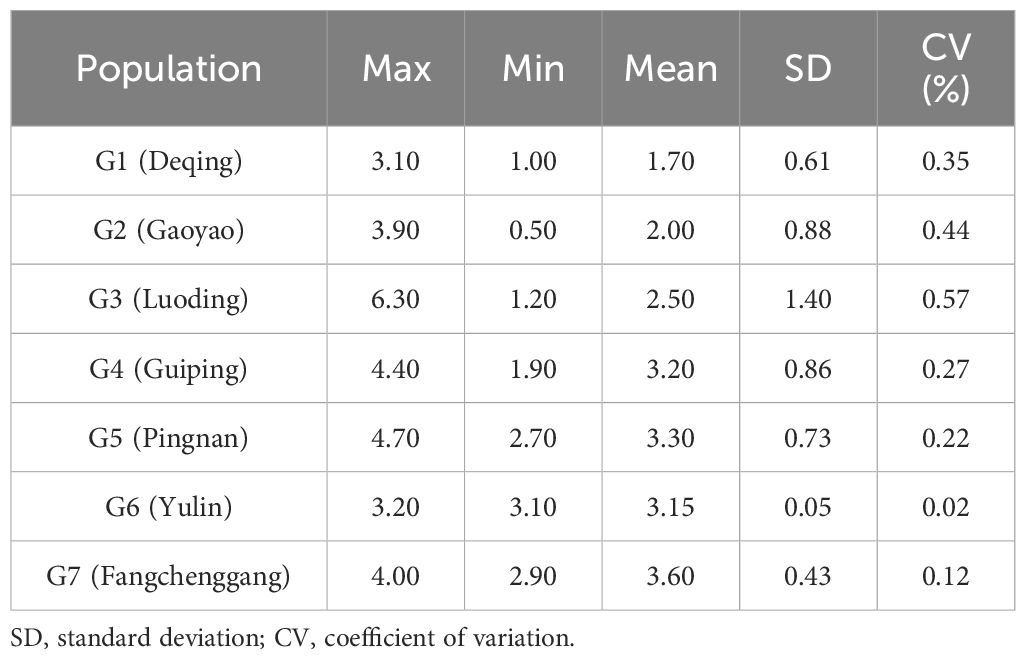
The moisture, water-soluble extract, and volatile oil contents of C. cassia are shown in Supplementary Figure S1 and Supplementary Table S5. The findings demonstrated wide variations in the volatile oil concentrations of C. cassia from various sources. The average amount of volatile oils in each sample was 2.3% and ranged from 0.5% to 6.3%. RG37 contained the highest amount of volatile oils, whereas RG07 contained the lowest. The average volatile oil contents in the seven districts were 1.7%, 2.0%, 2.5%, 3.2%, 3.4%, 3.2%, and 3.6% for Deqing, Gaoyao, Luoding, Guiping, Pingnan, Yulin and Fangchenggang, respectively. The germplasm from Fangchenggang had the highest volatile oil concentration, followed by that from Pingnan, whereas the germplasm from Deqing had the lowest volatile oil concentration. Each sample had a different moisture content ranging from 10.3% to 18.0% on average, with RG52 having the highest moisture content and RG08 having the lowest (Table 2). The average moisture contents in the seven districts were 14.2%, 14.0%, 14.7%,13.5%,15.2%, 14.75%, and 14.3% for Deqing, Gaoyao, Luoding, Guiping, Pingnan, Yulin and Fangchenggang, respectively. The germplasm from Guiping had the lowest moisture content, whereas that from Pingnan had the highest moisture content, followed by that from Yulin. The water-soluble extract content of each sample ranged from 9.9% to 23.0% with an average of 18.0%. RG46 had the highest water-soluble extract concentration, while RG16 had the lowest. The average water-soluble extract contents in the seven districts were 18.1%, 17.4%, 18.9%, 18.5%, 17.8%, 18.4%, and 18.9% for Deqing, Gaoyao, Luoding, Guiping, Pingnan, Yulin and Fangchenggang, respectively. The most water-soluble extract was found in the germplasms of Fangchenggang, whereas the least was found in Yulin and Gaoyao.
To investigate the connection between the phenotypic qualities, chemical components, and geographic location, we performed a Spearman correlation analysis. The results showed that the height of the one-year-old C. cassia seedlings was positively correlated with stem diameter, indicating that the higher the tree, the larger the diameter (Figure 4; Supplementary Table S6). The age of the original tree was also positively correlated with tree height and tree trunk girth, which is consistent with traditional grading standards. Interestingly, we found that three phenotypic characteristics were correlated with volatile oil content, i.e. samples with longer and broader leaves and originating from higher elevations showed higher essential oil content. This provides a scientific foundation for the logical cultivation of C. cassia, suggesting that these traits can be used to create new types with highly active components. However, in contrast to the conventional grading criteria, we discovered a negative association between tree diameter and volatile oil concentration, indicating that samples with a smaller tree diameter had a higher volatile oil content. Furthermore, a negative correlation was observed between latitude and longitude and phenotypic traits such as leaf width, leaf length, tree height, tree diameter, and volatile oil content. These findings indicate that plantations at lower latitudes and longitudes are better suited for the growth and development of C. cassia.
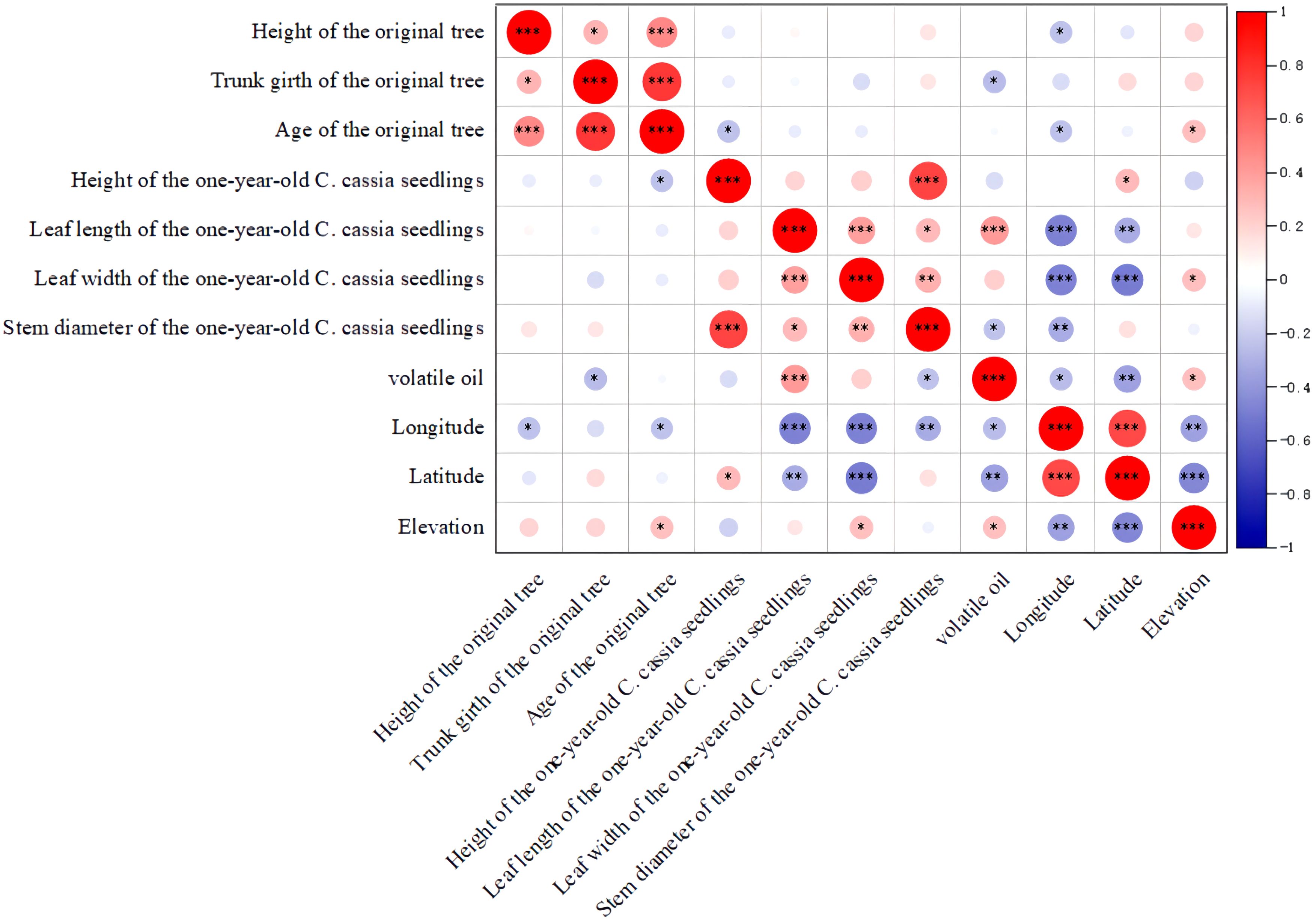
Figure 4 Correlation analysis of morphology and chemical constituents of C. cassia (L.) J.Presl (*p<0.05, **p<0.01, ***p<0.001, Pearson correlation analysis).
Additionally, The PCA analysis showed that 75.853% of the diversity was explained by the first four main components (Figure 5A; Supplementary Table S7). With features such as leaf length and leaf leaf width of one-year-old C. cassia seedlings, elevation, and volatile oil, PC1 with an eigenvalue of 3.054 accounted for 27.767% of the total variation. The height, trunk girth, and age of the original tree contributed remarkably to PC2, accounting for 21.194% of the variation. The stem diameter and height of one-year-old C. cassia seedlings were among the variables for which PC3 accounted for 18.812% of the total variation. All 11 variables were slightly differentiated and could be used to discriminate C. cassia accessions. In addition, we also conducted PCA analysis on 71 samples based on these 11 indices, and the results are shown in Figure 5B. The results showed that RG30, RG68, RG69, RG70 and RG71 clustered together, and the rest of the samples clustered together.
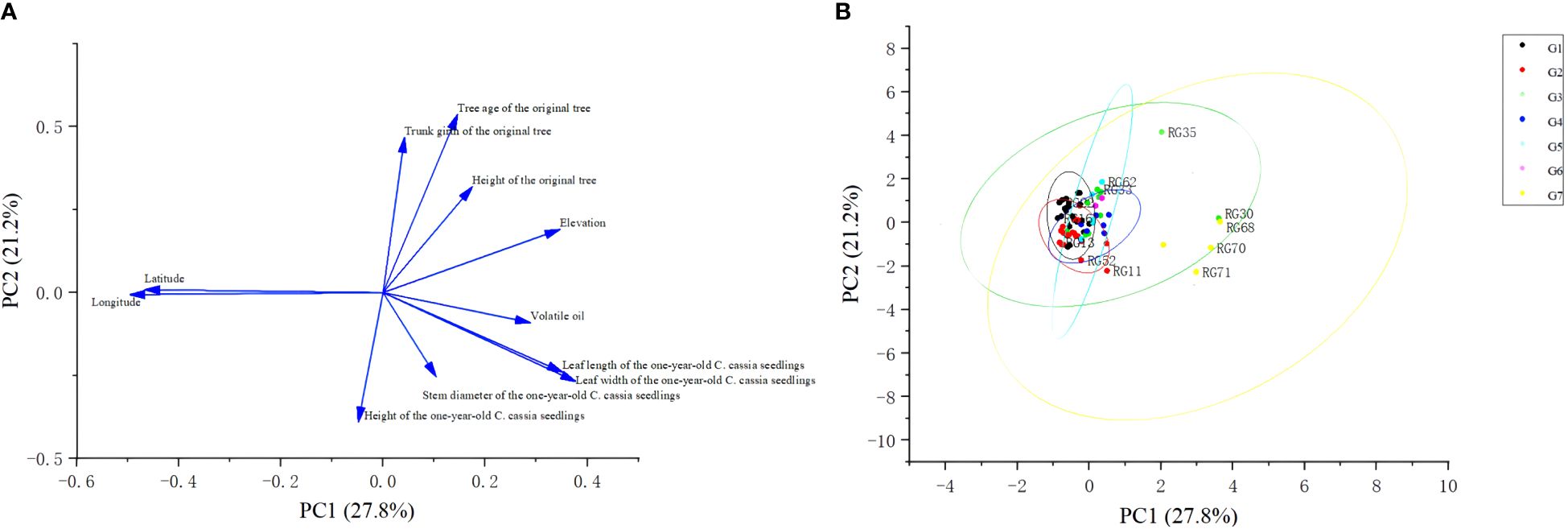
Figure 5 Principal component analysis of variables. (A) Loading plot of 71 C. cassia samples, geographical variables include longitude, latitude and altitude, Chemical composition variable include volatile oil, other variables are morphological indicators. (B) Score plot of 71 C. cassia samples.
3.3 Changes of volatile oil and cinnamaldehyde contents in cassia bark with different thickness
In a previous study, when determining the content of volatile oils in cassia bark, our group found that the cinnamaldehyde content in different samples of cinnamon medicinal materials purchased from the same batch was notably different and that the volatile oil contentbetween different thicknesses from the same tree was irregular, indicating that the quality of the cinnamon medicinal materials was uneven. Although all medicinal compounds of cinnamon are derived from the bark, cinnamon is a tall perennial tree with a long growth period, which is affected by many environmental factors during the growth process, and the distribution of secondary metabolites in the bark may be uneven. Therefore, it is speculated that the content of cinnamaldehyde in the upper, middle, and lower parts of cassia bark may be different.
In this study, HPLC was used to determine the total amount of volatile oil and the difference in the content of four volatile oil components–cinnamaldehyde, cinnamic acid, cinnamyl alcohol, and 2-methoxycinnamaldehyde–in the upper, middle, and lower parts of the C. cassia tree. The results showed that the contents of cinnamaldehyde, cinnamyl alcohol, cinnamic acid, and 2-methoxycinnamaldehyde in the 12 cassia bark samples were 16.10–104.10, 0.16–3.52, 0.18–1.06, and 0–10.24 mg/g, respectively. The mean values were 48.19, 1.08, 0.51 and 1.96 mg/g, respectively. The volatile oil content ranged from 0.85% to 8.05% with an average of 3.70% (Supplementary Table S2). Interestingly, the cinnamaldehyde and cinnamyl alcohol contents first decreased and then increased with increasing cassia bark thickness. Cinnamaldehyde content was highest when the thickness of the cassia bark was 2.60–3.20 mm (up to 44.10 mg/g), while the content of cinnamaldehyde was lowest when the thickness of the cassia bark was 2.20–2.60 mm (up to 36.86 mg/g). The cinnamyl alcohol content was highest when the thickness of the cassia bark was 1.10–1.40 mm (1.35 mg/g), while cinnamaldehyde content was at its lowest (0.83 mg/g) when the thickness of the cassia bark was 1.80–2.20 mm. The cinnamic acid content of the 12 cassia bark samples first increased and then decreased with increasing cassia bark thickness. The content of cinnamic acid was at its highest (0.57 mg/g) when the thickness of the cassia bark was between 1.40 and 1.80 mm. The volatile oil content decreased with an increase in the thickness of the cassia bark, and the content was highest when the thickness of the cassia bark was between 1.10 and 1.40 mm (Figure 6; Supplementary Figure S2). In the same cinnamon tree, the cinnamaldehyde and volatile oil contents were negatively correlated with cross-sectional thickness; that is, the cinnamaldehyde and volatile oil contents increased from the near-ground part to the upper part of each cassia bark sample (Figure 7). The cinnamaldehyde and volatile oil contents in the upper, middle, and lower parts of the cassia bark were markedly different; the difference in cinnamaldehyde content in the same tree was up to two times; and the difference in volatile oil was up to six times, suggesting that this difference might be one of the reasons for the uneven quality of the cassia bark.
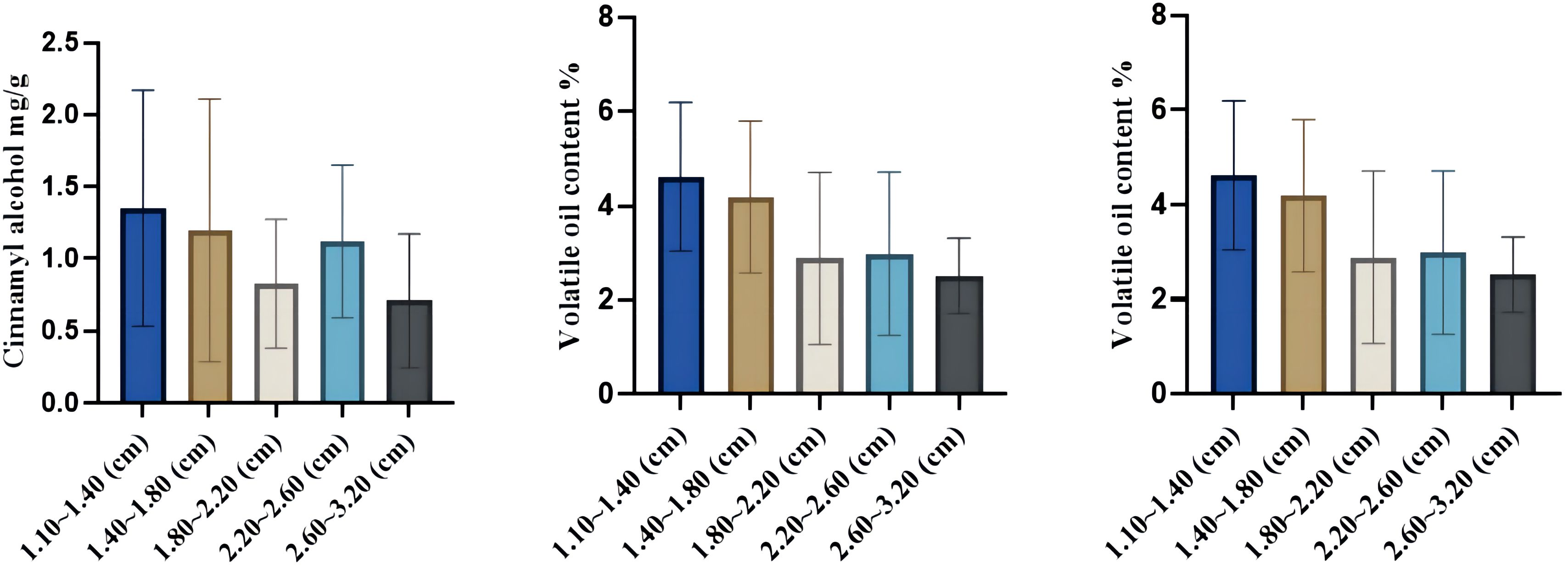
Figure 6 Changes of cinnamaldehyde, cinnamic acid and volatile oil contents incinnamon bark of different thicknesses.
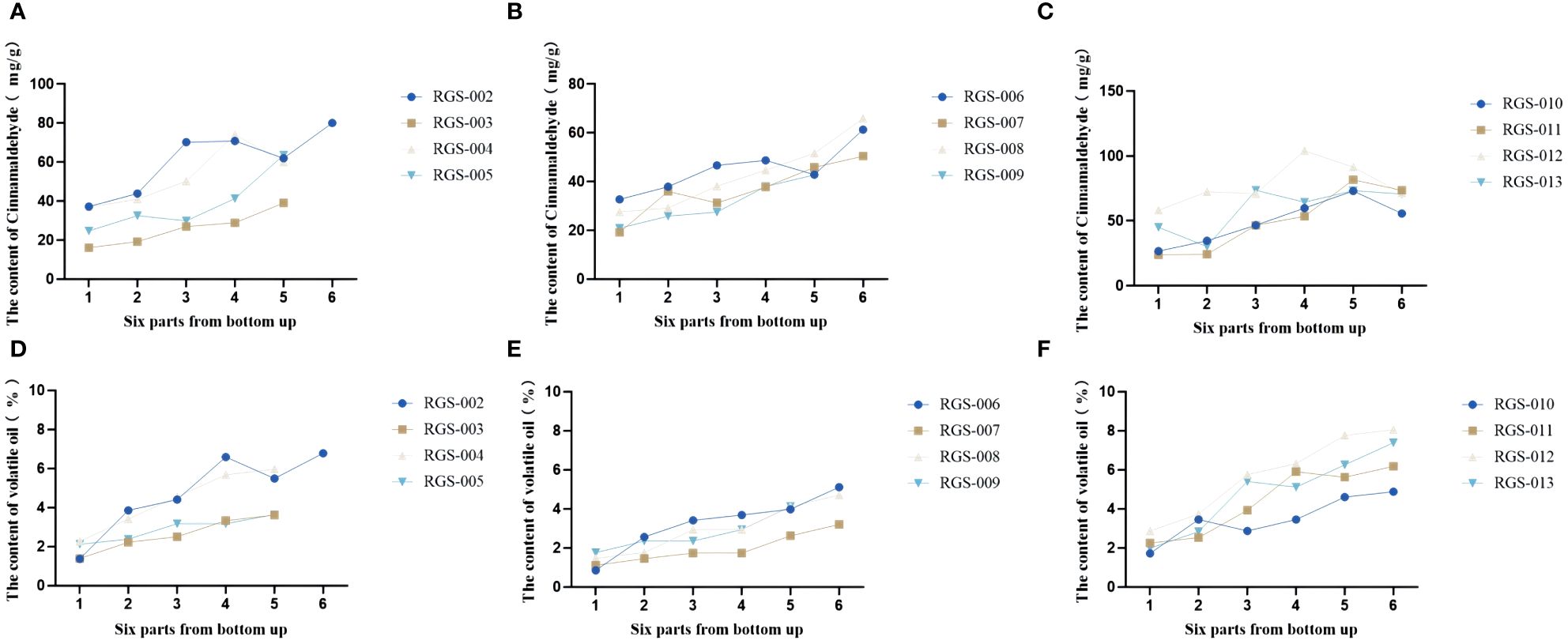
Figure 7 Changes of cinnamaldehyde and volatile oil contents from bottom to top in the bark of the same cinnamon tree. (A–C) The cinnamaldehyde content in cinnamon samples from Hetai Town, Gaoliang Town, Tanbin Town, Guangdong Province. (D–F) The volatile oilcontent in cinnamon samples from Hetai Town, Gaoliang Town, Tanbin Town, Guangdong Province.
3.4 SNP markers quality and diversity
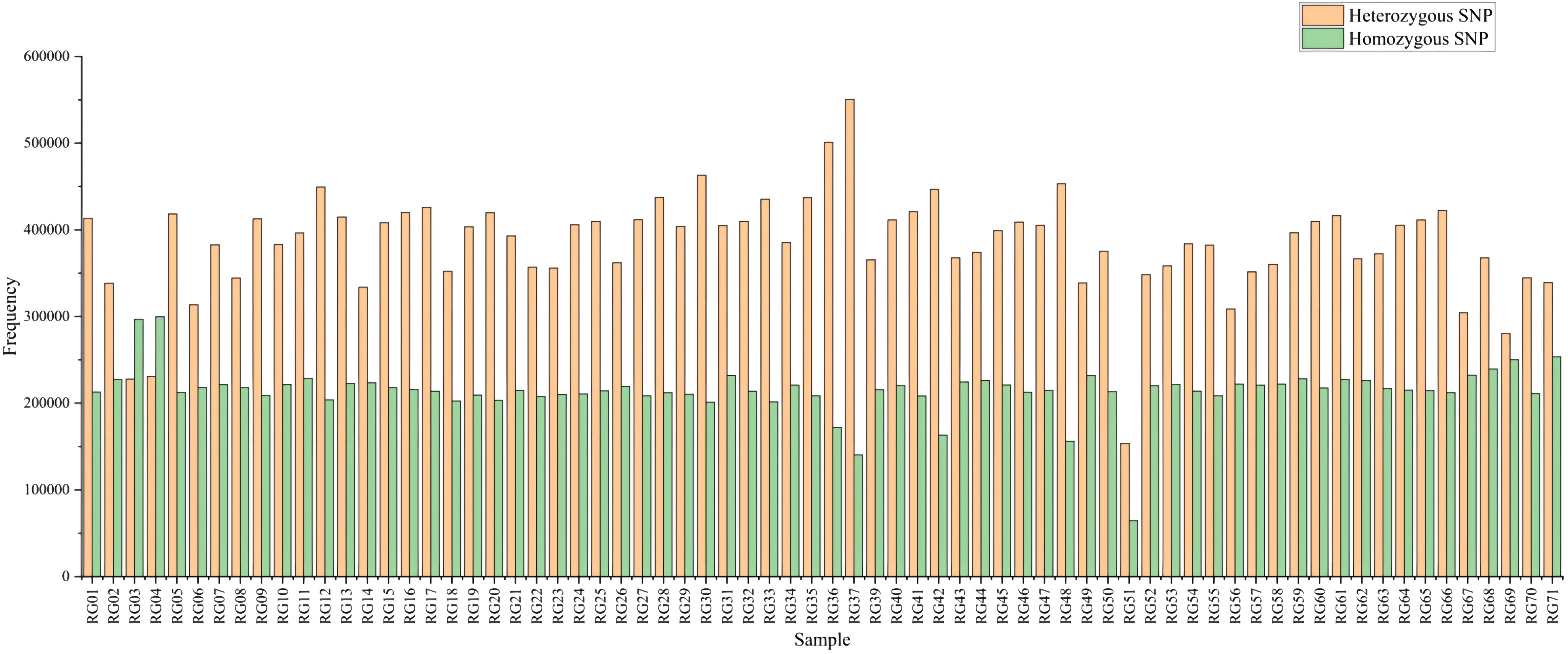
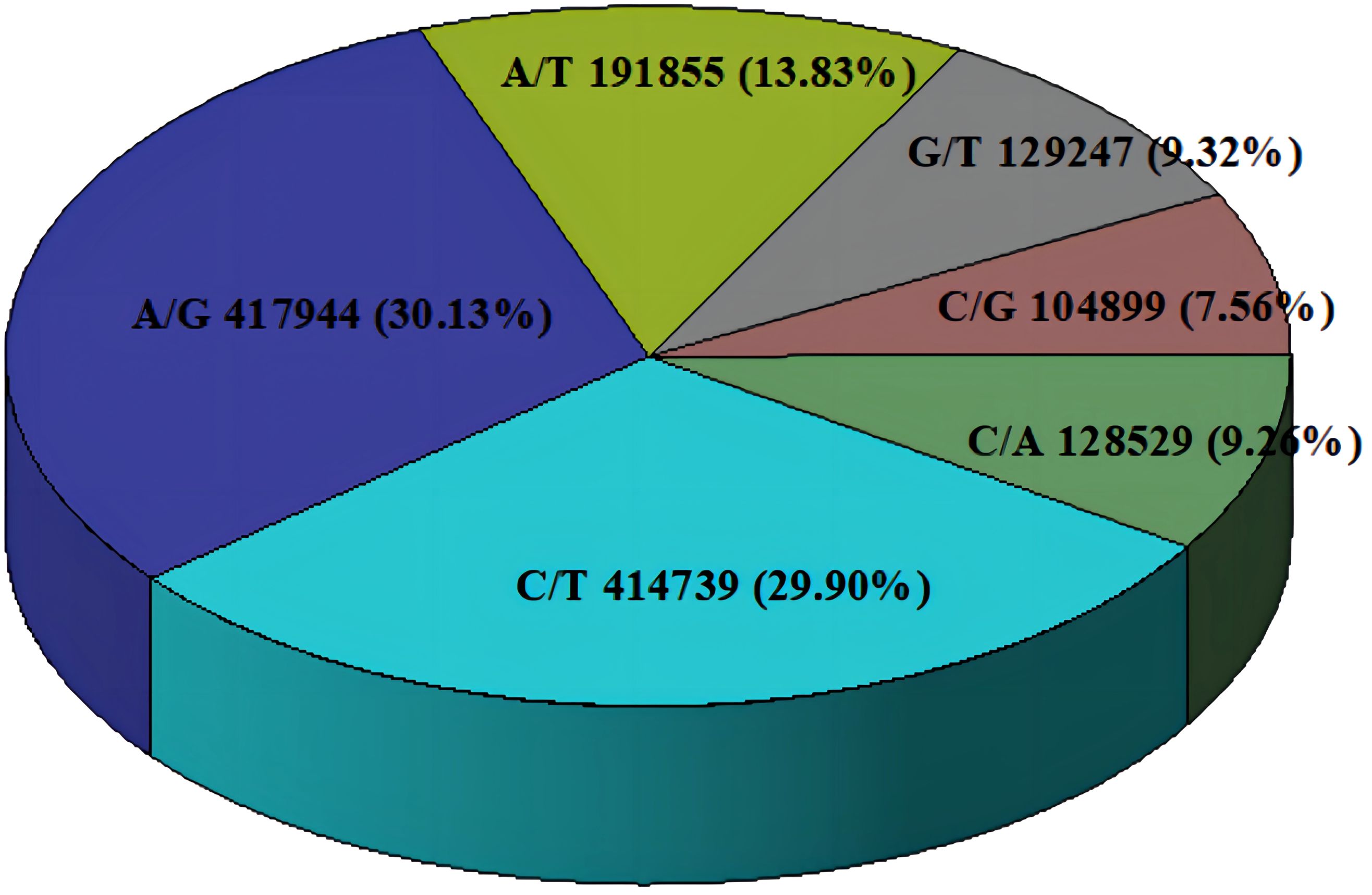
A total of 1,387,213 SNP markers were used in subsequent analyses when the identified SNPs of all 71 C. cassia samples were combined. SNP markers included across all libraries were screened for non-biallelic sites, sites with MAF < 0.05, and sites with deletion rates greater than 20%. In contrast, 1,387,213 SNP markers were sufficient to estimate the genetic diversity and population structure of C. cassia. The average number of SNPs per sample was 5598,397, with a range of 218,134–6690,682. The sample with the highest number of SNPs was RG37, whereas the sample with the lowest number was RG51. All 71 accessions had 15,014,708 homozygous and 26,863,047 heterozygous SNPs, representing 35.87% and 64.13% of all the SNPs, respectively. There was no discernible difference between the wild (64.15%) and cultivated (63.31%) samples, with an average heterozygosity rate of 63.93% (Figure 8; Supplementary Table S8). The two most common types of substitutions in the SNP dataset were transversions (C/A, 9.26%; G/T, 9.32%; C/G, 7.56%; A/T, 13.83%) and transitions (C/T, 29.90%; A/G, 30.13%). These substitutions included 832,683 (60.01%) and 554,530 (39.99%) SNPs, respectively (Figure 9; Supplementary Table S9). Heterozygosity is considered the best metric for assessing the genetic diversity of a population because it can represent the genetic variance of the population at several loci. A population’s genetic diversity can be measured using Ho; the higher the Ho, the more diverse the population’s genetic makeup. With an average of 0.32 and 0.49, respectively, Ho and He ranged from 0.14 to 0.44 and 0.18 to 0.92, respectively, indicating that C. Cassia populations were less impacted by inbreeding, artificial selection, and other factors and were in a state of genetic balance.
The Luoding population had the greatest Na at 1.919, while the Yulin population (1.655) had the lowest value. In the Yulin population, the Ne varied from 1.545 to 1.610 in the Gaoyao population. With a mean of 0.478, the Shannon’s information index (I) ranged from 0.418 (Yulin) to 0.513 (Gaoyao), indicating a comparatively high level of community diversity in Gaoyao. The PPB percentage varied between Yulin (65.52%) and Deqing (98.88%) with an average of 86.94%. In contrast, the Ho and He ranged from 0.2 to 0.4 and 0.4 to 0.5, indicating the largest number of SNP loci (464,901 and 611,493, respectively), followed by 0 to 0.2 and 0.1 to 0.2 (Table 3). As seen in Supplementary Figure S3, the MAF distribution was examined. The highest number of SNP loci (approximately 320,000) in MAF ranged from 0.05 to 0.10, followed by 0.10 to 0.15, suggesting that the C. cassia populations under investigation had a low level of genetic diversity. The polymorphism height of the molecular markers was quantified using the PIC metric. It is generally believed that 0.25 < PIC < 0.5 represents a moderate polymorphism site, PIC > 0.5 represents a high polymorphism site, and PIC < 0.25 represents a low polymorphism site (Wang et al., 2019). The PIC varied from 0.075 to 0.375, and the highest number of SNPs (approximately 550,000) was between 0.300–0.375 and 0.225–0.300, respectively. These results suggest that the C. cassia populations under investigation had a moderate degree of genetic variation (Supplementary Figure S4).
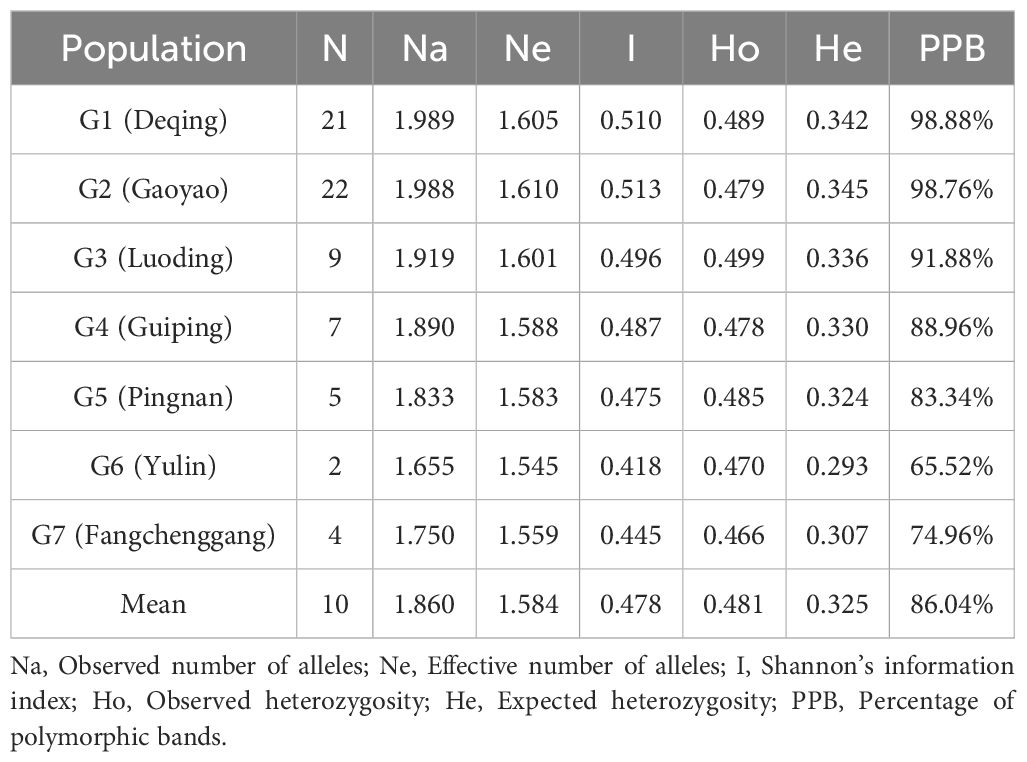
Table 3 Summary statistics of molecular diversity revealed by SNP markers in Seven C. cassia populations from two provinces in China.
AMOVA, which can yield important information, was performed using a model-based analysis to assess the population’s genetic constitution based on its consistency and reliability (Table 4). The findings showed that individuals within populations accounted for the majority of the genetic variation (95.87%; Df = 111; sum squares = 878.895), whereas populations within groups accounted for 0.59% of the variation (Df = 3; sum squares = 26.216), and the remaining variation was found among groups. AMOVA demonstrated that individuals accounted for the majority of the genetic diversity in the C. cassia germplasm. In addition, research has revealed that there is little genetic variation among the seven populations, owing to gene crossovers. The genetic distances (GD) and genetic identities (GI) between seven populations were computed based on the SNP locus data. The findings indicated that the GD between Fangchenggang and the other populations were greater, ranging from 0.049 to 0.082, with an average of 0.059, whereas the GD between Deqing and Gaoyao were lowest (0.008). Furthermore, the differences between the other populations were comparatively small, averaging 0.030 and ranging from 0.014 to 0.057 (Supplementary Table S10). Except for Fangchenggang, there was a high degree of genetic relatedness among the C. cassia subpopulations, which is in line with the findings of the study of plant morphological characteristics. Fangchenggang and Yulin showed the greatest GD (0.082), whereas Deqing and Gaoyao showed the shortest GD (0.049). A mean GI of 0.965 was obtained, ranging from 0.921 to 0.992, A trend with which the GD was at odds.
3.5 Phylogenetic analysis of the C. cassia population
Based on genetic distance, a phylogenetic tree was created using the Neighbour Joining (NJ), and the results showed that the 71 C. cassia accessions were mostly grouped into two clusters (Figure 10). Three accessions from Fangchenggang City, which included a sample of wild C. cassia (RG68), were included in cluster I. The genetic difference between RG68 and RG69 in this cluster was the highest at GD = 0.210, whereas the genetic distance between GX68 and GX71 was the lowest at GD = 0.197. Despite their genetic diversity, all these individuals could be clearly separated from cultivars based on their phenotypic features. Because the Fangchenggang accessions were closely linked to one another, it was possible to use them to increase the genetic background. Cluster II contained 68 accessions, most of which developed in Guangdong and Guangxi, with the exception of Fangchenggang. It contained all wild-type samples, except for RG68. The GD in this clade ranged from 0.150 to 0.306, and some accessions such as RG26 and RG27 had very little genetic distance, making them closely related (Supplementary Table S11). Overall, there was a strong correlation between the genetic distances between C. cassia accessions and their geographical origins. This indicates that most C. cassia germplasms could be gathered from the same or comparable origin, with a small amount of test materials mixed with other groups. Except for Fangchenggang, the accessions from Guangdong and Guangxi were closely related. In 71 samples, it was not possible to distinguish between wild and cultivated types, which is consistent with the results of the cluster analysis based on phenotypic characteristics.
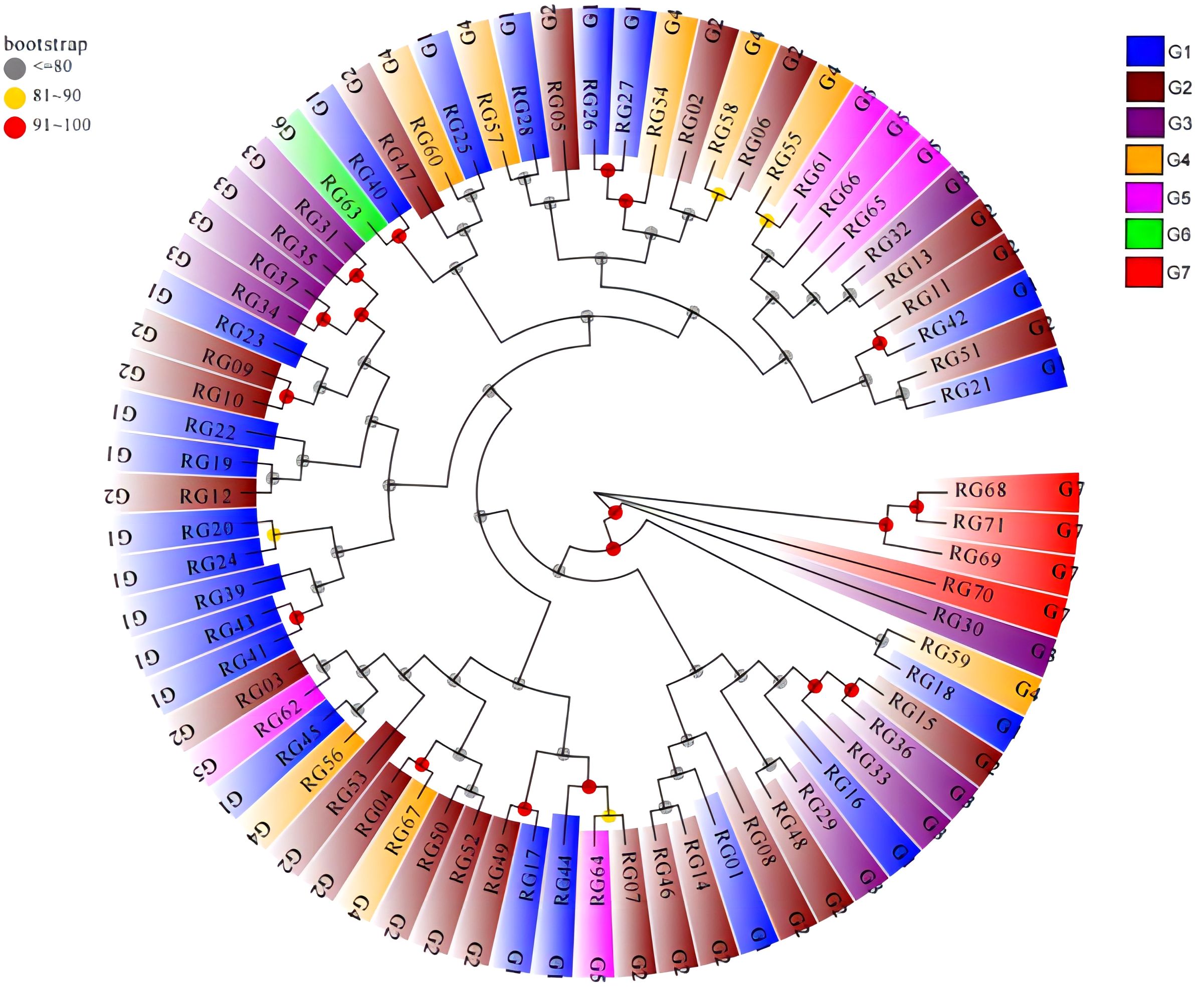
Figure 10 Phylogenetic tree of the 71 C. cassia accessions based on SNP marker. G1: Deqing, G2: Gaoyao, G3: Luoding, G4: Guiping, G5: Pingnan, G6: Yulin, G7: Fangchenggang.
3.6 Structure analysis of the C. cassia population
To identify the population groups, structure analysis based on a mixed Bayesian clustering model was applied. The findings indicated that K (the number of random mating subgroups) was equal to three as the optimal number of groups (Supplementary Figure S5; Supplementary Table S12), indicating that the 71 accessions could be divided into three subpopulations (I, II, and III) (Figure 11). Based on shared genomic areas, the genotypes of the various populations were separated into pure and mixed types. Specifically, genotypes scoring ≥0.80 were classified as pure and split into appropriate subgroups, while genotypes scoring <0.80 were termed admixed (Luo et al., 2022). In total, 47 (or about 62%) of the 71 accessions were admixed, and the remaining 27 (approximately 38%) were pure. The admixtures showed that 75% of the genotypes were from Guangdong and 25% were from Guangxi, including three wild-types. This suggests that the genotypes from Guangxi and Guangdong have a more complex genetic background with mixed genes from multiple populations and a higher level of genetic diversity, whereas the pure genotypes from Gaoliang Town, Guangdong, and Fangchenggang City (Guangxi) have a narrower genetic background. These results are consistent with those of the phylogenetic analysis. RG70 and RG71 were pure types that originated from Fangchenggang. This suggests that they may have originated in Vietnam and might have been utilized in the breeding of C. cassia.
3.7 Principal component analysis of the C.cassia population
The results of the PCA, which revealed three separate clusters, agreed with those of the population structure analysis. The data shown in Figure 12 indicate that the Guangxi accessions were more dispersed than the Guangdong accessions. Except for RG30, RG68, RG69, RG70, and RG71, the Guangdong and Guangxi accessions were clustered together. This is consistent with PCA analysis results of 11 indicators. This suggests that there was low genetic variation among the germplasm lines and that there was only a small genetic divergence of genotypes from Guangdong and Guangxi. Interestingly, accessions from Fangchenggang in Guangxi were unique. One possible explanation for this could be the introduction of dispersed germplasms from other areas.
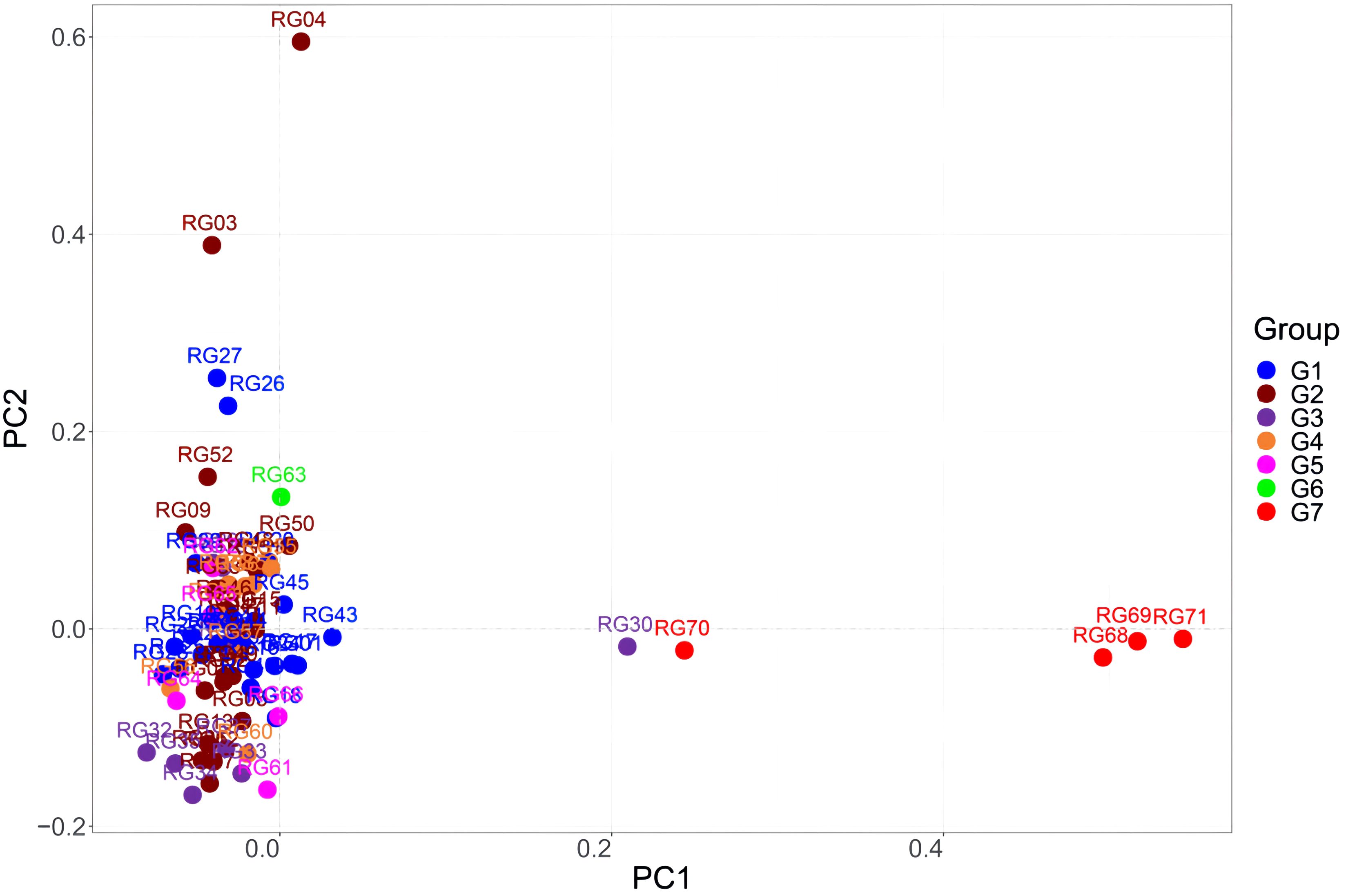
Figure 12 Principal component analysis (PCA) of the 71 C. cassia accessions. G1: Deqing, G2: Gaoyao, G3: Luoding, G4: Guiping, G5: Pingnan, G6: Yulin, G7: Fangchenggang.
4 Discussion
The total genetic variety among individuals within a species or population is referred to as genetic diversity (Pyne et al., 2018; Verma et al., 2019). The study of genetic variation can be used to improve breeding plans for medicinal plants by choosing parents with superior genotypes and providing information on the degree of genetic organization within populations (Yang et al., 2018; Niu et al., 2019). Based on phenotypic and chemical composition and SNP markers, we examined the genetic diversity of C. cassia samples in this study. These findings demonstrated that populations of C. cassia exhibited a low degree of genetic variation.
4.1 Variations based on morphological traits
In this study, we analyzed 11 indicators of cultivated and wild C. cassia samples, including leaf length, leaf width, tree height, tree age, elevation, longitude, and latitude. These findings demonstrate that the C. cassia population’s phenotypic features exhibited a high degree of genetic variation. Variations in C. cassia differed greatly in phenotypic traits. For example, Fangchenggang, a variety of Guangxi, has long and broad leaves, whereas Guangdong has very narrow leaves. We found that Guangdong samples generally had long growth years and tall trees with strong trunks, whereas Guangxi germplasm had short growth years and fleshy, thick, and relatively wide leaves, especially in Fangchenggang. In our previous study, we found that the C. cassia planting area in Guangdong Province accounted for more than 30% of the country’s total C. cassia planting area. The Xijiang River Basin in Zhaoqing City, Guangdong Province, is suitable for C. cassia growth under the geographical environment, soil, soil quality, water quality, climate, and light conditions, which are crucial factors for C. cassia -producing areas in China. The main cultivated C. cassia is Xijiang C. cassia, also known as “Xijianggui,” which has the characteristics of thin skin, thick meat, rich oil, moist color, and fragrant moderately sweet and spicy taste. Guangxi C. cassia accounts for more than 50% of the country’s C. cassia planting area. Fangchenggang, Dongxing, Yulin, Guiping, and Beiliu cities are the main producing areas of C. cassia. Among them, the C. cassia cultivated in Fangchenggang and Dongxing is mainly “Fangchenggui,” whereas that cultivated in Yulin and Guiping is mainly “Xijianggui.” Therefore, based on the morphological data analysis of 71 C. cassia samples, we inferred that all the samples were “Xijianggui,” except for 4 samples obtained from Fangchenggang (Wei et al., 2006; Chen et al., 2014; Lin et al., 2016). Among the C. cassia varieties, there is also a variety from Vietnam, which is named Qinghuagui (C. cassia Bl forma macrophylla), famous for its large leaf shape, thick skin, and high volatile oil content. Since the morphological characteristics of the four C. cassia samples from Fangchenggang were similar to those of Qinghuagui, and Fangchenggang is near Vietnam, we speculated that these four samples might be from Qinghuagui. Liang et al., 2016 reported that the leaf length, leaf width, fresh leaf weight, and leaf area of the Qinghuagui family were markedly higher than those of the “Xijianggui” family by 37.03%, 26.80%, 174.88%, and 41.32%, respectively, thereby confirming our results. Genetic divergence due to genetic drift, local adaptation, and gene flow limitation may result in the phenotypic differentiation of traits (Agre et al., 2019; Arab et al., 2019).
4.2 Variations based on chemical components
Modern pharmacological studies have revealed that cinnamaldehyde, the main component of volatile oils, has a wide range of pharmacological effects, such as vascular dilation, anti-gastric ulcer, bacteriostasis, and anti-oxidation. Therefore, in this study, the volatile oil, moisture, and water-soluble extract contents of 71 C. cassia accessions were analyzed. The results showed that the volatile oil content of C. cassia samples from Guangxi was generally higher than those from other places except Luoding, up to 4.7%, whereas the volatile oil content of C. cassia samples from Luoding was the highest in Guangdong Province, up to 6.3%. The reasons for this may be related to planting history, cultivation years, or the ecological environment. Luoding, the hometown of C. cassia in China, has a long history of cultivation, and its ecological environment is suitable for its cultivation of C. cassia. The volatile oil content of C. cassia samples from Guangxi was generally higher than that from Guangdong Province, which may be due to the short planting period of C. cassia in Guangdong Province.
In a previous study by our group, we found that the planting years of Guangdong’s C. cassia samples were mostly 7–8 years, which might be the main reason why the volatile oil content in C. cassia produced in Guangdong was lower than that in Guangxi (Wei et al., 2017). Guangxi is the traditional planting base for C. cassia medicinal materials, and the planting years for these medicinal materials are mostly 10 years or more, mainly for clinical medicine. If the quality standards of C. cassia medicinal materials are to be improved, it is recommended that Guangdong’s C. cassia planting base increase the planting years of C. cassia, with 10 to 15 years deemed optimal (Xu et al., 2004). Wu et al. (2019) determined the content of cinnamaldehyde and four other volatile oil components in C. cassia from Guangdong and Guangxi and found that the cinnamaldehyde content in Guangxi’s C. cassia was generally higher than that in Guangdong’s C. cassia, which was also confirmed in our study (Wei et al., 2019). Furthermore, our study found that the average content of volatile oil of C. cassia was highest in Fangchenggang, where “Ziyougui” was also found. We speculate that perhaps because Fangchenggang is adjacent to Vietnam, some of the C. cassia in this area may represent the “Qinghuagui” introduced by Vietnam. Zhang. (2019) measured 45 batches of cinnamon samples from different origins using HPLC, and the results showed that the volatile oil content of cinnamon in Fangchenggang was as high as 5.2%. Qian et al. (2020) determined the cinnamaldehyde content in 11 cinnamon samples from Guangdong and Guangxi using HPLC. The results showed that the cinnamaldehyde content in Fangchenggang was 33.52 mg/g, whereas that in Luoding was only 16.23 mg/g. Wang showed that the volatile oil content of Qinghuagui (Cinnamomum cassia var. macrophyllum Chu) was considerably higher than that in Chinese cassia (Wang, 2011).
In a previous study, our research group found that the cinnamaldehyde content in different samples of the same batch of cinnamon medicinal materials was remarkably different and that the content difference between different thicknesses was irregular. Therefore, we studied the changes in cinnamaldehyde and volatile oil contents in the upper, middle, and lower parts of the bark of the same tree. The results showed that the contents of cinnamaldehyde and volatile oil varied greatly in the upper, middle, and lower parts of the cassia bark. The content of cinnamaldehyde in the same tree was up to two times, and the total volatile oil was up to six times, suggesting that this difference might be one of the reasons for the uneven quality of the cassia bark. In the same cinnamon tree, the cinnamaldehyde and volatile oil contents were negatively correlated with cross-sectional thickness; that is, the cinnamaldehyde and volatile oil contents increased from the near-ground part to the upper part of each cassia bark sample. The studies (Hou et al., 2013; Wang et al., 2022) have shown that the cinnamaldehyde and cinnamic acid content of Ramulus Cinnamomi and cinnamonic fruit is higher than that of cassia bark. Ramulus Cinnamomi and cinnamonic fruit are known in China as “Guizhi” and “Guiding”. “Guizhi” are branches of the cinnamon tree and “Guiding” is the tender stem of cinnamon fruit. They’re both at the top of the cinnamon tree. This may confirm our research. Therefore, it is necessary to further study the distribution of cinnamaldehyde and volatile oils in cassia bark to provide an experimental basis for the formulation of extraction technology and specification grade of cassia bark and to lay a foundation for the improvement of quality control standards.
4.3 Genetic diversity based on SNP
Current research indicates that domestication has less of an effect on genetic diversity reduction than breeding practices, which leads to cultivated varieties having less genetic diversity than wild counterparts (Guan et al., 2019; Wang et al., 2019; Wolfe et al., 2019). In this study, we discovered that C. cassia from Fangchenggang differed greatly from the others in terms of genotype, chemical composition, and morphological characteristics. This is most likely because Fangchenggang harbors ancient landraces that descended naturally from the early landraces and influenced the genetic diversification of C. cassia populations. However, we did not find a significant difference between the wild and cultivated types in our samples, which may be due to the fact that the wild C. Cassia, we were looking for were only artificially planted older trees rather than true wild C. cassia trees. Therefore, we need to explore wild C. cassia that inhabits less explored areas for further study. Wild types may be less susceptible to genetic influence from other C. cassia variants owing to their comparatively isolated ecological setting. The restricted genetic diversity of cultivars not only hinders the breeding process of C. cassia, but also elevates the likelihood of natural disasters (Luo et al., 2019; Mwale et al., 2023). Both landraces and wild-type plants may offer invaluable genetic resources for breeding initiatives, and new C. cassia. cultivars may be bred to diversify their genetic makeup.
Compared to other species, including rice (Peringottillam et al., 2022), wheat (Ouaja et al., 2021), corn (Rivas et al., 2022), C. cassia (L.), J.Presl has a substantially higher heterozygosity rate (average heterozygosity rate = 63.93%). This is most likely due to the fact that these crops have long been subjected to manual selection, which has decreased their genetic diversity. In contrast, C. cassia has a complex genetic basis because it has been propagated for many years using a variety of techniques, including seed, layering, and cutting propagation (Li and Jiang, 2022). The low genetic diversity within the population obtained from AMOVA could be a result of natural adaptation or extensive exchange of seeds among farmers between environments or because of the common origin of the population, which might have led to C. Cassia growers using the same seed continuously, without new introductions. Furthermore, we found that heterozygosity in the wild was higher than that in the cultivar types, which is consistent with findings from other species (Uba et al., 2021; Yao et al., 2012). However, the difference is not substantial. This finding may explain why C. cassia is propagated from one generation to another, and variation gradually accumulates, whereas wild species live in a relatively isolated setting with a lower genetic variety over extended periods of time. Therefore, they gradually become similar. Owing to the limitations of the wild individuals evaluated in this study, these are only our first conclusions about genetic variance among wild populations, and additional wild samples from various populations must be gathered for additional examination. Phylogenetic and population structure analyses demonstrated a strong positive correlation between geographical distance and gene exchange, displaying an isolated pattern within the distribution of C. cassia. This finding further supports the notion that there is minimal gene flow between populations, which promotes local adaptability. The clustering of C. cassia accessions in the current study not clearly separated the accessions based on their geographic origin, as determined by genetic distance or model simulation. The tested accessions clustered closely, both within and between neighboring locations. Using the ADMIXTURE program, all C. cassia accessions were assigned to three populations representing various ecogeographic regions. Cluster I was a unique and distinct Fangchenggang population, cluster II contained RG30 and RG70 from Guangdong and Guangxi, respectively, and cluster III represented populations from Deqing, Gaoyao, Luoding, Pingnan, Yulin, and Guiping. Guangdong and Guangxi were determined to have lower levels of genetic variety than Fangchenggang based on genetic differentiation in these inferred populations. The closest GD was found between Gaoyao and Deqing, while the greatest GD was found between Fangchenggang and Yulin. This is related to the distance between these species. With the exception of Fangchenggang, the GD between C. cassia accessions from Guangdong and Guangxi was reasonably small. In 71 samples, it was not possible to discriminate between the cultivated and wild types of the species, which is consistent with the findings of cluster analysis based on phenotypic features. According to our conjecture, the evolutionary process may have involved the ancestral population of the Guangxi area being more suited for the growth of C. cassia. because of the local climate and habitat, which allowed for relatively quick local breeding before spreading to Guangdong.
In summary, SNP markers provide a clear picture of the population structure and genetic diversity of C. cassia. This provided insightful conclusions regarding the collection, preservation, and use of Chinese C. cassia germplasms. The ecogeographic distribution of genetic diversity may provide information about the DNA-level spread of C. cassia from its genesis center to other regions of China. Our research verified that the geographic origin of C. cassia germplasms was linked to their population structure and that wild C. cassia has a more complicated genetic structure than local landraces. Furthermore, greater variation was found within the population than between populations, which directed us to gather more individuals within the same population to preserve a sufficient number of representative varieties of local C. cassia germplasms, especially for wild populations and landraces. In the meantime, in order to increase genetic diversity, we should gather and conserve genetic resources that come from various ecogeographic groups. From the comparatively high diversity and simple population of Fangchenggang, we were able to infer how to efficiently identify and utilize beneficial genes in local landraces and wild C. cassia to breed exceptional cultivars and expand their genetic base.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Bioproject accession number: PRJNA1074496.
Author contributions
PH: Writing – original draft. JC: Writing – review & editing. ZC: Writing – review & editing. XC: Writing – review & editing. ZP: Writing – review & editing. PD: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Technology Innovation and Promotion Project (No. 2023KJ142), the Guangdong Provincial Rural Revitalization Strategy Special Project (No.2021KJ268), and the Guangzhou Key R&D Project (No.202206010010).
Acknowledgments
We would like to thank Ruifa Li (Deqing County Dexin Agricultural Development Co., LTD, Zhaoqing, China) for providing us with the plant materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1374648/full#supplementary-material
References
Agre, P., Asibe, F., Darkwa, K., Edemodu, A., Bauchet, G., Asiedu, R., et al. (2019). Phenotypic and molecular assessment of genetic structure and diversity in a panel of winged yam (Dioscorea alata) clones and cultivars. Sci. Rep. 9, 18221. doi: 10.1038/s41598-019-54761-3
Alexander, D. H., Novembre, J., Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Arab, M. M., Marrano, A., Abdollahi-Arpanahi, R., Leslie, C. A., Askari, H., Neale, D. B., et al. (2019). Genome-wide patterns of population structure and association mapping of nut-related traits in Persian walnut populations from Iran using the Axiom J. regia 700K SNP array. Sci. Rep. 9, 6376. doi: 10.1038/s41598-019-42940-1
Belaj, A., Raul, R., Lorite, I. J., Mariotti, R., Cultrera, N. G. M., Beuzón, C. R., et al. (2018). Usefulness of a new large set of high throughput EST-SNP markers as a tool for olive germplasm collection management. Front. Plant Sci. 9, 1320. doi: 10.3389/fpls.2018.01320
Chen, D. Y., Zhao, L., Wu, Z. Q. (2014). Biological characteristics and scientific planting techniques of Cinnamomum cassia Presl. Agric. Technol. Service 31, 15–16.
Chen, R. Z., Wang, X. P., Gao, W. C., Li, Z. (2022). A comparative analysis of quality of Rougui from different origins. Clin. J. Chin. Med. 14, 41–44.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Eriksson, P., Marzouka, N. A., Sjödahl, G., Bernardo, C., Liedberg, F., Höglund, M. (2022). A comparison of rule-based and centroid single-sample multiclass predictors for transcriptomic classification. Bioinformatics 38, 1022–1029. doi: 10.1093/bioinformatics/btab763
Guan, C., Liu, S., Wang, M., Ji, H., Ruan, X., Wang, R., et al. (2019). Comparative transcriptomic analysis reveals genetic divergence and domestication genes in Diospyros. BMC Plant Biol. 19, 227. doi: 10.1186/s12870-019-1839-2
Hong, J. W., Yang, G. E., Kim, Y. B., Eom, S. H., Lew, J. H., Kang, H. (2012). Anti-inflammatory activity of cinnamon water extract in vivo and in vitro LPS-induced models. BMC Complement Altern. Med. 12, 237. doi: 10.1186/1472-6882-12-237
Hou, W. X., Wu, C., Zhou, Y. T., Deng, X. J., Yin, X. Y., Xie, Z. Y., et al. (2013). “Study on contents and distribution of four active components in different parts of Cinnamomum cassia Presl,” in Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology, vol. 15. , 254–259.
Kwon, K. B., Kim, E. K., Jeong, E. S., Lee, Y. H., Lee, Y. R., Park, J. W., et al. (2006). Cortex cinnamomi extract prevents streptozotocin-and cytokine-induced beta-cell damage by inhibiting NF-kappaB. World J. Gastroenterol. 12, 4331–4337. doi: 10.3748/wjg.v12.i27.4331
Li, C., Luo, Y., Zhang, W., Cai, Q., Wu, X., Tan, Z., et al. (2021). A comparative study on chemical compositions and biological activities of four essential oils: Cymbopogon citratus (DC.) Stapf, Cinnamomum cassia (L.) Presl, Salvia japonica Thunb. and Rosa rugosa Thunb. J. ethnopharmacology 280, 114472. doi: 10.1016/j.jep.2021.114472
Li, K. G., Jiang, X. J. (2022). Breeding methods and planting techniques of cinnamom. Special Economic Anim. Plants 25, 99–100.
Li, L., Wu, G. F., Luo, Y., Li, L. L., Ling, L., Ma, S. C. (2023). DNA molecular identification of genuine medicinal material Cinnamomum cassia and Cinnamomum burmanni in Guangxi. Food Fermentation Industries 50 (4), 191–196+203.
Liang, X. J., Li, K. X., Liang, W. H., Huang, K. S., Li, B. C., Liang, J. K., et al. (2016). Analysis on leaf phenotypic traits of different Cinnamomum cassia species. Guangxi Forestry Sci. 45 (01), 40–44. doi: 10.19692/j.cnki.gfs.2016.01.008
Liang, W. H., Liu, K., Huang, K. S., Liang, X. J., Li, B. C., Li, K. X. (2016). Genetic background analysis of Cinnamomum cassia genealogies by ISSR. Guangxi Forestry Sci. 45 (01), 35–39. doi: 10.19692/j.cnki.gfs.2016.01.007
Lin, X. J., Zhou, H. S., Wu, H. S., Zhao, J. P., Hao, C. Y. (2016). Investigation report of cinnamom industry in Guangdong province. Chin. J. Trop. Agric. 36, 80–84.
Lu, Z. J., Chen, D. Y., Cao, M. H. (2010). MAnalysis of cinnamom's planting and climatic conditions in Gaoyao City. Guangdong Agric. Sci. 37 (05), 37–38. doi: 10.16768/j.issn.1004-874x.2010.05.040
Luo, Z., Brock, J., Dyer, J. M., Kutchan, T., Schachtman, D., Augustin, M., et al. (2019). Genetic diversity and population structure of a Camelina sativa spring panel. Front. Plant Sci. 10, 184. doi: 10.3389/fpls.2019.00184
Luo, Z., Chen, Z., Liu, M., Yang, L., Zhao, Z., Yang, D., et al. (2022). Phenotypic, chemical component and molecular assessment of genetic diversity and population structure of Morinda officinalis germplasm. BMC Genomics 23, 605. doi: 10.1186/s12864-022-08817-w
Maroso, F., Gracia, C. P., Iglesias, D., Cao, A., Díaz, S., Villalba, A., et al. (2019). A Useful SNP Panel to Distinguish Two Cockle Species, Cerastoderma edule and C. glaucum, Co-Occurring in Some European Beds, and Their Putative Hybrids. Genes 10 (10), 760. doi: 10.3390/genes10100760
Mwale, S. E., Shimelis, H., Abincha, W., Nkhata, W., Sefasi, A., Mashilo, J. (2023). Genetic differentiation of a southern Africa tepary bean (Phaseolus acutifolius A Gray) germplasm collection using high-density DArTseq SNP markers. PloS One 18, e0295773. doi: 10.1371/journal.pone.0295773
Nagy, S., Poczai, P., Cernák, I., Gorji, A. M., Hegedűs, G., Taller, J. (2012). PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 50, 670–672. doi: 10.1007/s10528-012-9509-1
Nantawan, U., Kanchana-Udomkan, C., Bar, I., Ford, R. (2019). Linkage mapping and quantitative trait loci analysis of sweetness and other fruit quality traits in papaya. BMC Plant Biol. 19, 449. doi: 10.1186/s12870-019-2043-0
Narayanankutty, A., Kunnath, K., Alfarhan, A., Rajagopal, R., Ramesh, V. (2021). Chemical composition of Cinnamomum verum leaf and flower essential oils and analysis of their antibacterial, insecticidal, and larvicidal properties. Molecules (Basel Switzerland) 26, 6303. doi: 10.3390/molecules26206303
National Pharmacopoeia Committee (2020). Chinese pharmacopoeia (Beijing: China Medical Science and Technology Press), 142.
Niu, S., Song, Q., Koiwa, H., Qiao, D., Zhao, D., Chen, Z., et al. (2019). Genetic diversity, linkage disequilibrium, and population structure analysis of the tea plant (Camellia sinensis) from an origin center, Guizhou plateau, using genome-wide SNPs developed by genotyping-by-sequencing. BMC Plant Biol. 19, 328. doi: 10.1186/s12870-019-1917-5
Ouaja, M., Bahri, B. A., Aouini, L., Ferjaoui, S., Medini, M., Marcel, T. C., et al. (2021). Morphological characterization and genetic diversity analysis of Tunisian durum wheat (Triticum turgidum var. durum) accessions. BMC Genom Data 22, 3. doi: 10.1186/s12863-021-00958-3
Peakall, R., Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Perez, G. A., Tongyoo, P., Chunwongse, J., Jong, H., Wongpraneekul, A., Sinsathapornpong, W., et al. (2021). Genetic diversity and population structure of ridge gourd (Luffa acutangula) accessions in a Thailand collection using SNP markers. Sci. Rep. 11, 15311. doi: 10.1038/s41598-021-94802-4
Peringottillam, M., Kunhiraman Vasumathy, S., Selvakumar, H. K. K., Alagu, M. (2022). Genetic diversity and population structure of rice (Oryza sativa L.) landraces from Kerala, India analyzed through genotyping-by-sequencing. Mol. Genet. Genomics 297, 169–182. doi: 10.1007/s00438-021-01844-4
Poplin, R., Chang, P. C., Alexander, D., Schwartz, S., Colthurst, T., Ku, A., et al. (2018). A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987. doi: 10.1038/nbt.4235
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Pyne, R. M., Honig, J. A., Vaiciunas, J., Wyenandt, C. A., Simon, J. E. (2018). Population structure, genetic diversity and downy mildew resistance among Ocimum species germplasm. BMC Plant Biol. 18, 69. doi: 10.1186/s12870-018-1284-7
Qian, J. P., Luo, B., Zhong, L., Lv, J., Li, H. Q., Zhang, Y. Q. (2020). Effects of different producing area, tree age, cutting processing on the contents of active compounds in Connamomi Cortex. J. Chin. Medicinal Materials 43 (12), 2887–2892. doi: 10.13863/j.issn1001-4454.2020.12.007
Ren, J., Sun, D., Chen, L., You, F. M., Wang, J., Peng, Y., et al. (2013). Genetic diversity revealed by single nucleotide polymorphism markers in a worldwide germplasm collection of durum wheat. Int. J. Mol. Sci. 14, 7061–7088. doi: 10.3390/ijms14047061
Rivas, J. G., Gutierrez, A. V., Defacio, R. A., Schimpf, J., Vicario, A. L., Hopp, H. E., et al. (2022). Morphological and genetic diversity of maize landraces along an altitudinal gradient in the Southern Andes. PloS One 17, e0271424. doi: 10.1371/journal.pone.0271424
Shen, M. T., Bai, D. N., Wang, Q. W., Ping, Y., Zhao, H., Wang, L. H., et al. (2021). Research progress on anti-inflammatory mechanism of Cinnamomum cassia and its active components. Chin. Traditional Herbal Drugs 53 (10), 3218–3225.
Sun, C., Dong, Z., Zhao, L., Ren, Y., Zhang, N., Chen, F. (2020). The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 18, 1354–1360. doi: 10.1111/pbi.13361
Tung, Y. T., Yen, P. L., Lin, C. Y., Chang, S. T. (2010). Anti-inflammatory activities of essential oils and their constituents from different provenances of indigenous cinnamon (Cinnamomum osmophloeum) leaves. Pharm. Biol. 48, 1130–1136. doi: 10.3109/13880200903527728
Uba, C. U., Oselebe, H. O., Tesfaye, A. A., Abtew, W. G. (2021). Genetic diversity and population structure analysis of bambara groundnut (Vigna subterrenea L) landraces using DArT SNP markers. PloS One 16, e0253600. doi: 10.1371/journal.pone.0253600
Verma, H., Borah, J. L., Sarma, R. N. (2019). Variability assessment for root and drought tolerance traits and genetic diversity analysis of rice germplasm using SSR markers. Sci. Rep. 9, 16513. doi: 10.1038/s41598-019-52884-1
Wang, S. W. (2011). The study on essential oil components and comparison of identification in Chinese cassia and Cinnamomum cassia var. macrophyllum Chu. Chin. Arch. Traditional Chin. 29, 1401–1402.
Wang, R. Y. (2022). Authenticity Identification of Five Plant Spices using DNA Molecular Markers (China (IL: Yan Tai University).
Wang, J., Li, X., Do Kim, K., Scanlon, M. J., Jackson, S. A., Springer, N. M., et al. (2019). Genome-wide nucleotide patterns and potential mechanisms of genome divergence following domestication in maize and soybean. Genome Biol. 20 (1), 74. doi: 10.1186/s13059-019-1683-6
Wang, Y. X., Liao, Y. Y., Q, A. Q., Li, H. J., Xu, H. W., Yang, J. T., et al. (2019). Correlation analysis between LHX3 gene polymorphism and growth traits in three sheep breeds. Biotechnol. Bull. 35, 162–168. doi: 10.1186/s13059-019-1683-6
Wang, X. J., Sun, S. N., Gao, H. Y. (2022). Components and sensory quality comparison of essential oil from different parts of Guangxi Cinnamomum cassia. Flavour Fragrance Cosmetics 04), 8–12+36.
Wei, R. P., Huang, Y. F., Hu, D. H., Zheng, Y. G. (2006). Status and trends of researches on Cinnamomum cassia Presl. Non-wood For. Res. 03), 65–70.
Wei, C. H., Su, M., Li, Q., Ding, P. (2017). Investigation on Cinnamomum cassia Presl resource in Guangdong and Guangxi. Res. Pract. Chin. Medicines 31, 14–17+21.
Wolfe, M. D., Bauchet, G. J., Chan, A. W., Lozano, R., Ramu, P., Egesi, C., et al. (2019). Historical introgressions from a wild relative of modern cassava improved important traits and may be under balancing selection. Genetics 213, 1237–1253. doi: 10.1534/genetics.119.302757
Wu, C. H., Feng, C., Yang, L., Ding, P. (2019). Determination of four essential oils in Cinnamomum cassia by quantitative analysis of multi-components by single marker. Chin. Pharm. J. 54, 400–406.
Xu, Y., Cheng, B. Q., Ding, J. K., Yu, Z., Chen, Z. H., Zeng, J. N. (2004). Investigation on cinnamon resource, growth and yield of oil in Guangxi and Yunnan. Trop. Agric. Sci. Technol. 03), 4–7+26.
Yang, J. B., Dong, Y. R., Wong, K. M., Gu, Z. J., Yang, H. Q., Li, Z. (2018). Genetic structure and differentiation in Dendrocalamus sinicus (Poaceae: Bambusoideae) populations provide insight into evolutionary history and speciation of woody bamboos. Sci. Rep. 8, 16933. doi: 10.1038/s41598-018-35269-8
Yang, M., Li, L. J., Lan, J. X., Chen, L. J., Ye, S. M. (2013). Biomass and productivity of Strified Mixed Stands of Pinus massoniana and Cinnamomum cassia. Acta Botanica Boreali-Occidentalia Sin. 33, 585–591.
Yang, Y. L., Luo, B., Zhang, H., Zheng, W. J., Wu, M. L. (2020). Advances in quality research of Cinnamomum cassia. China J. Chin. Materia Med. 45, 2792–2799.
Yao, M. Z., Ma, C. L., Qiao, T. T., Jin, J. Q., Chen, L. (2012). Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genomes 8, 205–220. doi: 10.1007/s11295-011-0433-z
Zhang, Y. (2019). Study on Quantity Evaluation of Cinnamomi Cortex (China (IL: Shanghai University of Traditional Chinese Medicine).
Keywords: Cinnamomum cassia (L.) J.Presl, cinnamon, volatile oil, single-nucleotide polymorphism, genetic diversity, population structure, germplasm resources of traditional Chinese medicine
Citation: Han P, Chen J, Chen Z, Che X, Peng Z and Ding P (2024) Exploring genetic diversity and population structure in Cinnamomum cassia (L.) J.Presl germplasm in China through phenotypic, chemical component, and molecular marker analyses. Front. Plant Sci. 15:1374648. doi: 10.3389/fpls.2024.1374648
Received: 22 January 2024; Accepted: 14 June 2024;
Published: 03 July 2024.
Edited by:
Jim Leebens-Mack, University of Georgia, United StatesReviewed by:
Manuela Bog, University of Greifswald, GermanyLaura Emma Maria Morello, National Research Council (CNR), Italy
Copyright © 2024 Han, Chen, Chen, Che, Peng and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Ding, ZGluZ3BpbmdAZ3p1Y20uZWR1LmNu
 Panpan Han
Panpan Han Jinfang Chen
Jinfang Chen