- 1College of Horticulture, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2College of Agricultural, Sichuan Nationalities University, Liangshan Yi autonomous prefecture, Sichuan, China
Sugar is a primary determinant of citrus fruit flavour, but undergoes varied accumulation processes across different citrus varieties owing to high genetic variability. Sucrose phosphate synthase (SPS), a key enzyme in glucose metabolism, plays a crucial role in this context. Despite its significance, there is limited research on sugar component quality and the expression and regulatory prediction of SPS genes during citrus fruit development. Therefore, we analysed the sugar quality formation process in ‘Kiyomi’ and ‘Succosa’, two citrus varieties, and performed a comprehensive genome-wide analysis of citrus CsSPSs. We observed that the accumulation of sugar components significantly differs between the two varieties, with the identification of four CsSPSs in citrus. CsSPS sequences were highly conserved, featuring typical SPS protein domains. Expression analysis revealed a positive correlation between CsSPS expression and sugar accumulation in citrus fruits. However, CsSPS expression displays specificity to different citrus tissues and varieties. Transcriptome co-expression network analysis suggests the involvement of multiple transcription factors in shaping citrus fruit sugar quality through the regulation of CsSPSs. Notably, the expression levels of four CsWRKYs (CsWRKY2, CsWRKY20, CsWRKY28, CsWRKY32), were significantly positively correlated with CsSPSs and CsWRKY20 might can activate sugar accumulation in citrus fruit through CsSPS2. Collectively, we further emphasize the potential importance of CsWRKYs in citrus sugar metabolism, our findings serve as a reference for understanding sugar component formation and predicting CsSPS expression and regulation during citrus fruit development.
Introduction
Citrus, a major global fruit crop, is rich in nutrients such as sugars, organic acids, amino acids, carotenoids, and flavonoids (Guo et al., 2016; Sheng et al., 2017). Sugar metabolism profoundly impacts citrus fruit quality, with sugar content generally increasing during fruit development (Bush, 2020). ‘Kiyomi’ (Citrus unshiu × sinensis) and ‘Succosa’ (Citrus reticulata Blanco cv. Succosa) are important citrus cultivars in Sichuan Province, due to their good fresh-eating quality. However, the sweetness of Kiyomi’s fruit varies greatly before and after ripening, whereas Succosa is stable, relatively. Usually, the sugar amounts in developing fruits significantly vary among citrus cultivars owing to their high genetic variability (Albertini et al., 2006).
Soluble sugar, which is composed of sucrose, fructose, and glucose, constitutes the main sugar in citrus fruits, with sucrose being the principal storage form (Ruan, 2014). Sucrose phosphate synthase (SPS) catalyses the conversion of fructose-6-phosphate (F-6-P) and UDP-glucose (UDP-G) to sucrose 6-phosphate (S-6-P), serving as a rate-limiting enzyme for sucrose synthesis (Winter and Huber, 2000; Coleman et al., 2009). Consequently, SPS plays a crucial role in controlling sucrose synthesis in leaf tissues and sucrose accumulation in fruits (Lunn, 2003).
In recent years, genome-wide data have helped identify SPS genes in plants, forming families with relatively few members. For example, Arabidopsis thaliana has four members, rice (Oryza sativa) has five, and pears have eight (Castleden et al., 2004; Okamura et al., 2011; Koramutla et al., 2019). Despite different SPS numbers in plants, their protein sequences are similar, containing conserved domains for sucrose synthesis, sugar transport-1, and S6PP. Phylogenetic analysis categorises SPS genes into four families (A, B, C, and D), with the D family exclusive to certain monocotyledonous plants (Langenkämper et al., 2002). Tissue-specific expression is observed, with SPS1 and SPS2 preferentially expressed in fruits, whereas SPS3 and SPS4 are expressed in leaves and flowers (Okamura et al., 2011). SPS expression directly influences plant sugar metabolism, with SPS overexpression impacting starch and sucrose proportions in transgenic tomato and Arabidopsis leaves (Worrell et al., 1991; Signora et al., 1998; Anur et al., 2020). Moreover, the SPS expression levels further affect plant morphogenesis by regulating glucose metabolism. For example, overexpression of the spinach SPS gene in cotton can improve cotton fibre quality, and overexpression of SoSPS1 increases plant height and stem number of some transgenic sugarcane strains (Park et al., 2008; Anur et al., 2020).
In addition to factors such as low temperatures, drought, and hormones, transcription factors also regulate SPS expression (Reimholz et al., 2008; Roy Choudhury et al., 2008). For example, the ABA-associated FaMRLK47 regulates sucrose and starch metabolism in strawberry fruit, and silencing the MYB transcription factor FaGAMYB decreases transcription levels of FaSPS3 (Jia et al., 2017). Moreover, FaMYB44.2 can regulate sucrose accumulation by inhibiting the expression of FaSPS3 (Wei et al., 2018).
In this study, we comprehensively investigated the sugar and acid qualities of two citrus varieties (‘Kiyomi’ and ‘Succosa’) and analysed CsSPSs in the citrus genome. Through CsSPSs expression and co-expression analyses, we sought to elucidate their crucial role in sugar metabolism during citrus fruit development, as well as obtain valuable insights into the transcriptional regulation of CsSPSs and advance our understanding of citrus sugar metabolism pathways.
Materials and methods
Plant materials
Mandarin fruits (‘Kiyomi’ and ‘Succosa’) were harvested from a commercial orchard in Liangshan Yi autonomous prefecture, Sichuan province, China, ensuring uniform size and absence of visible injuries. Fruits harvested in September (Sep), October (Oct), November (Nov), and December (Dec)were transported to the laboratory, where pulp samples were frozen, homogenised in liquid nitrogen, and stored at −80°C for subsequent analyses. Three replicates, each consisting of six fruits, were analysed.
Total soluble solid, titratable acidity
‘Kiyomi’ and ‘Succosa’ fruits at each stage were selected to determine the total soluble solid (TSS) and titratable acidity (TA). At each development stage, more than 18 fruits were used for quality assessment, with three replicates. TSS (%) and TA (%) were measured using a digital acidity metre (Pocket PAL-BXIACID1, ATAGO, Tokyo, Japan) following the manufacturer’s instructions.
Determination of glucose, fructose, and sucrose contents
Glucose, fructose, and sucrose levels were determined, as described earlier (Liu et al., 2019). Briefly, 2 g of pulp was homogenised with 10 mL of ddH2O, incubated for 15 min at 80°C, centrifuged, and filtered. The resulting supernatant was analysed on an Agilent 1260 HPLC system (Agilent Technologies) with a refractive index detector using an Innoval NH2 column (4.6 mm × 250 mm, 5 μm, Agela Technologies, Shanghai, China). The mobile phase comprised acetonitrile: water (80:20, v/v) with a flow rate of 1 mL min−1.
Identification and characteristic analysis of the SPS gene family
Citrus SPS genes were identified following established methods (Hu et al., 2015; He et al., 2016). Genomic sequences from various citrus species (Citrus clementina v1.0, Citrus grandis ‘Wanbaiyou’ v1.0, Poncirus trifoliata v1.0, and Citrus sinensis v2.0) were obtained from the Citrus Pan-genome to Breeding Database (CPBD: http://citrus.hzau.edu.cn/index.php). Arabidopsis SPS gene members and their protein sequences were sourced from the Arabidopsis Information Resource (TAIR: https://www.arabidopsis.org/browse/genefamily/index.jsp). HMMER software version 3.0 was utilised to identify C. sinensis SPS (CsSPS) genes. Furthermore, the MapGene2Chrom software (MG2C_v2.1) was employed to generate a chromosome location image of CsSPSs, and TBtools software was used to display the exon/intron structure of all CsSPSs.
Analysis of gene structure and conserved motifs of CsSPSs
Conserved motifs of SPS proteins were identified using the multiple EM for motif elicitation (MEME software) (Brown et al., 2013). The optimal width of each motif ranged from 6–20, with a maximum of six motifs to search and default values for other parameter settings (Bailey et al., 2006). To ensure the inclusion of SPS domains, all candidate SPSs were validated using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CCD) to ensure that they contained the SPS domains. Additionally, the S6PP domain was predicted through multiple sequence alignment using BioEdit.
Phylogenetic relationship analysis of the CsSPS gene family
Multiple sequence alignments of citrus and Arabidopsis thaliana SPS protein sequences were performed using Molecular Evolutionary Genetics Analysis (MEGA) version 6.0, with 1,000 bootstrap replications, pairwise deletion, and Poisson model. Subsequently, neighbour-joining phylogenetic trees were constructed.
RNA-Seq data and qRT-PCR analysis
RNA-Seq data were retrieved from published studies (Terol et al., 2019; Feng et al., 2021; Zhang et al., 2021). The data that support the findings of this study have been deposited in the NCBI BioProject database under accession numbers PRJNA636131, PRJNA517400 and PRJEB12880. RNA-Seq data analysis was performed as described previously (Pertea et al., 2016). Briefly, the analysis included preprocessing for quality using FastQC, trimming low-quality reads (q < 20) and adapters using Trimmomatic, alignment to the C. sinensis genome using HISAT2 with default parameters, and assembly of mapped reads using StringTie. Fragments per kilobase per million mapped fragments (FPKM) was used to represent the gene expression levels. CsSPS expression profiles were extracted from the RNA-Seq data.
Total RNA was isolated from different tissues, as described previously (Liu et al., 2022). Specific primer pairs for CsSPSs amplification were designed using the Primer Express software (Applied Biosystems, Foster City, CA, USA). The specificity and amplification efficiency of the primers were validated using BLASTN against the sweet orange genome. Relative gene expression values, with the citrus β-actin gene as the internal reference gene, were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The sequences of RT-PCR primers are displayed in Supplementary Table S5. All expression data were processed using the Z-score standardisation method.
Co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) (v1.71) in R was used to construct the co-expression networks (Langfelder and Horvath, 2008). Among 29,138 genes, 16,961 with a sum FPKM < 1 across all samples were removed. The remaining genes were used for the WGCNA. The one-step network construction and module detection function were conducted using an unsigned topological overlap matrix (TOM), a soft-thresholding power b of 14 (R2 > 0.9), a minimal module size of 30, and a branch merge cut height of 0.25. The co-expression network of candidate CsSPSs was visualised using Cytoscape (version 3.6.1) (Shannon et al., 2003).
Dual-luciferase activity assay
The WRKY response cis-acting elements (W-box) in the promoter regions (2000-bp upstream of the initiation codon) of CsSPS1, CsSPS2, CsSPS3 and CsSPS4 using PlantCARE online software (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) were estimated (Supplementary Table S4). Afterwards, CsSPS2 promoter were inserted independently into the pGreen II 0800-LUC double-reporter vector, and the CDSs of CsWRKY2, CsWRKY20, CsWRKY28 and CsWRKY32 were inserted into the constructed pGreen 62-SK vector driven by the 35S promoter as the effector, using the primer sequences listed in Supplementary Table S7. The constructed effector and each reporter plasmid were co-transformed into tobacco leaves using A. tumefaciens strain GV3101(psoup-p19). Plasmids containing CsWRKYs and promoter were combined at a 10:1 ratio (v/v) and then infiltrated into tobacco leaves using needleless syringes. At 2 days after infiltration at 21 ◦C, the LUC and REN activities were measured using a Dual-Luciferase Assay kit (Promega, USA) on a Luminoskan Ascent Microplate Luminometer (Thermo, USA). The results were calculated using the LUC to REN ratio. Six biological repeats were assayed for each combination.
Statistical analysis
All data are presented as the mean (± standard deviation [SD]) of a representative experiment. Significant differences between samples were determined using ANOVA followed by Tukey’s test. The heatmaps were plotted using R studio software using the pheatmap package. The correlation analysis was performed using R studio software. Figures were created using GraphPad Prism (GraphPad Software, CA, USA).
Results
Sugar and organic acid contents in citrus fruit
To comprehend the dynamics of sugar and acid development in citrus, we assessed TSS and TA in two citrus varieties (‘Kiyomi’ and ‘Succosa’). As citrus fruit matured, the pulp colour deepened (Figure 1A), TSS content exhibited an evident upward trajectory (Figure 1B), and TA content significantly decreased (Figure 1C). Notably, the accumulation stages of sugar and acid varied among different citrus varieties. ‘Succosa’ attained higher TSS levels and lower TA content earlier than ‘Kiyomi’, which maintained stability. In contrast, ‘Kiyomi’ demonstrated a faster TSS accumulation rate and significantly higher TA content compared with that of ‘Succosa,’ despite lower TSS levels.
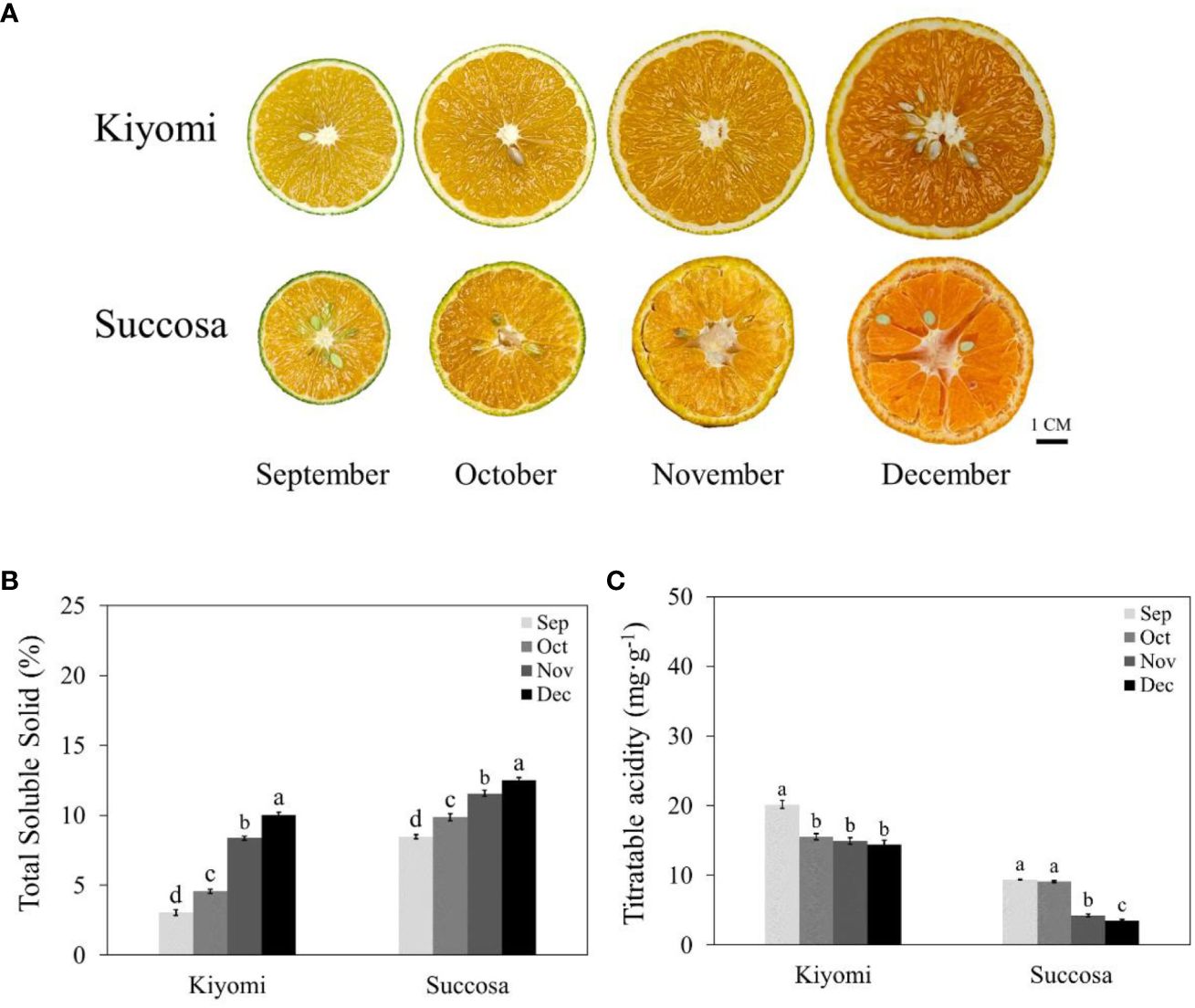
Figure 1 Citrus fruit phenotype. (A) Developmental stages of citrus fruit (‘Kiyomi’ and ‘Succosa’). (B) TSS. (C) TA. TSS, total soluble solid; TA, titratable acid. The different letters represent the significant differences between three groups during storage (P <0.05).
Sucrose, glucose, and fructose contents
As the primary sugars in citrus fruits, the formation of soluble sugars, including sucrose, fructose, and glucose, were analysed for quality assessment during development (Figure 2). We identified sucrose as the predominant component of soluble sugar in citrus pulp, followed by glucose and fructose. All three sugar components increased with citrus fruit development, with sucrose exhibiting the most substantial increase. ‘Succosa’ displayed higher sucrose content and consistent glucose and fructose levels compared with that of ‘Kiyomi.’ However, ‘Kiyomi’ exhibited a faster accumulation rate of sucrose, glucose, and fructose, aligning with the TSS results.
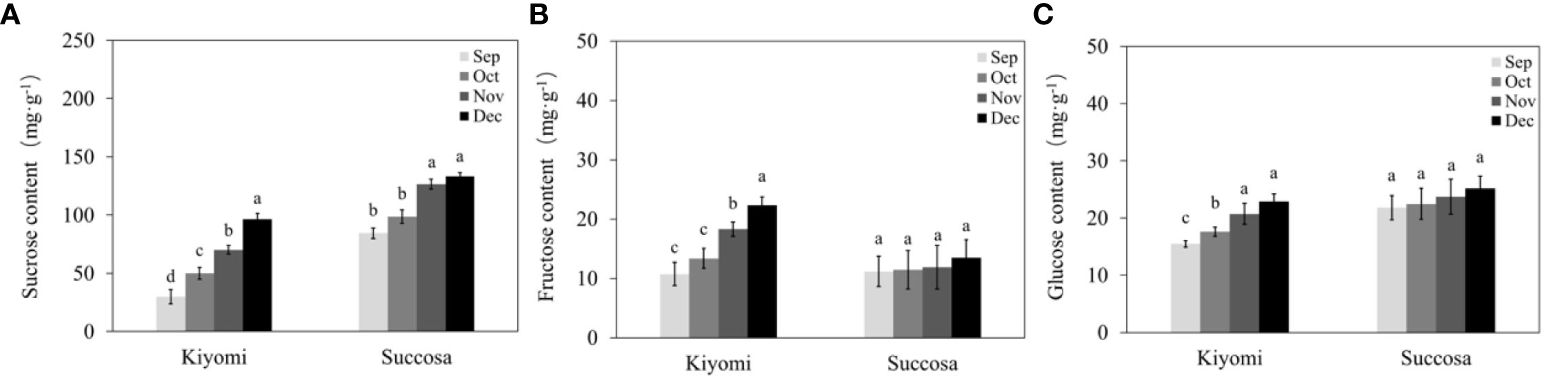
Figure 2 Soluble sugar component in the pulp of citrus fruit. (A) Sucrose. (B) Fructose. (C) Glucose. The different letters represent the significant differences between three groups during storage (P <0.05).
Identification of citrus SPS genes
Sucrose, a vital sugar in citrus fruits, is synthesised by SPS, the rate-limiting enzyme. We analysed the CsSPS family from four representative citrus varieties (Citrus sinensis, Citrus clementina, Citrus grandis, and Pitrus trifoliat) (Supplementary Table S1) and constructed phylogenetic trees to elucidate the evolutionary relationship of SPS genes between citrus and other species (Supplementary Table S2). Citrus SPS genes were categorised into three families, with CsSPS1 (Cs4g_pb004370) and CsSPS2 (Cs4g_pb022560) in family A, CsSPS3 (CsUn_pb042260) in family B, and CsSPS4 (Cs9g_pb011150) in family C (Figure 3A). CsSPS3 demonstrated a closer relationship with Arabidopsis AtSPS3F within the B family, while CsSPS4 also exhibited a closer relationship with Arabidopsis AtSPS4F within the C family. The chromosomal distribution of CsSPSs was analysed based on the physical location data retrieved from the GCA_018104345.1_Cs2.0_genomic database on the NCBI website. Using Dual Synteny Plotter software, we assessed the syntenic relationship between SPS genes in Citrus and Arabidopsis. Our findings revealed the distribution of CsSPSs on three chromosomes: CsSPS1 and CsSPS2 on chromosome 1, CsSPS4 on chromosome 9, and CsSPS3 on an unidentified chromosome (Figure 3B). Synteny analysis between SPS genes in citrus and Arabidopsis identified AtSPS1F-CsSPS1 and AtSPS2F-CsSPS2 as syntenic gene pairs (Figure 3C).
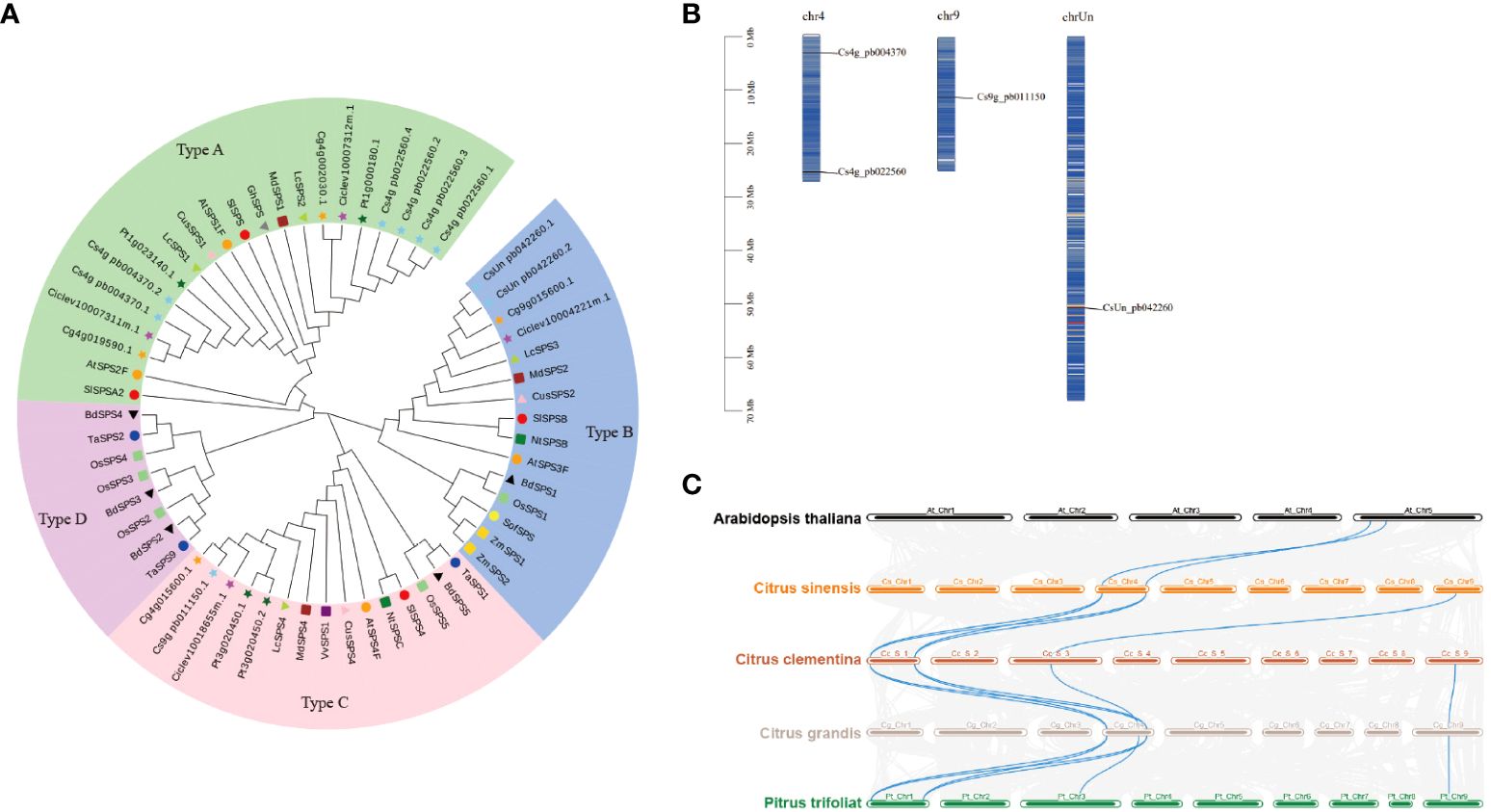
Figure 3 Phylogenetic and genomic analysis of SPS proteins. (A) Phylogenetic relationship of SPS proteins among Citrus and other species. (B) Chromosome distribution of CsSPSs. (C) Collinearity analysis of SPS genes from Arabidopsis and Citrus. Phylogenetic analysis was performed using the neighbour-joining method in MEGA v5.1. SPS proteins in C. sinensis, C. clementina, C. grandis, and P. trifoliata are represented using skyblue star, yellow circles, magenta star, orange star, and dark green star, respectively. Different species and type using different shape and color.
Protein motif, conserved domain, gene sequence, and promoter analysis of CsSPSs
Furthermore, an analysis of conserved motifs and domains was conducted using the MEME web server, revealing a total of six conserved motifs and three conserved domains in citrus SPS proteins (Figure 4A, B). The results indicate that all citrus SPS proteins share the same motifs (Supplementary Table S3) and domains: a Glycos_transf domain, an S6PP domain (C-domain), and a Glycosyltransferase domain (N-domain). Notably, citrus SPS3 possesses a Glycos_transf_4_4 domain that other SPS proteins lack. Concurrently, two transcripts of CsSPS2 (Cs4g_pb022560) lack Motif 6, and one transcript even lacks the Glycos_transf domain, suggesting that these two transcription modes may not be the primary transcription mode of CsSPS2.
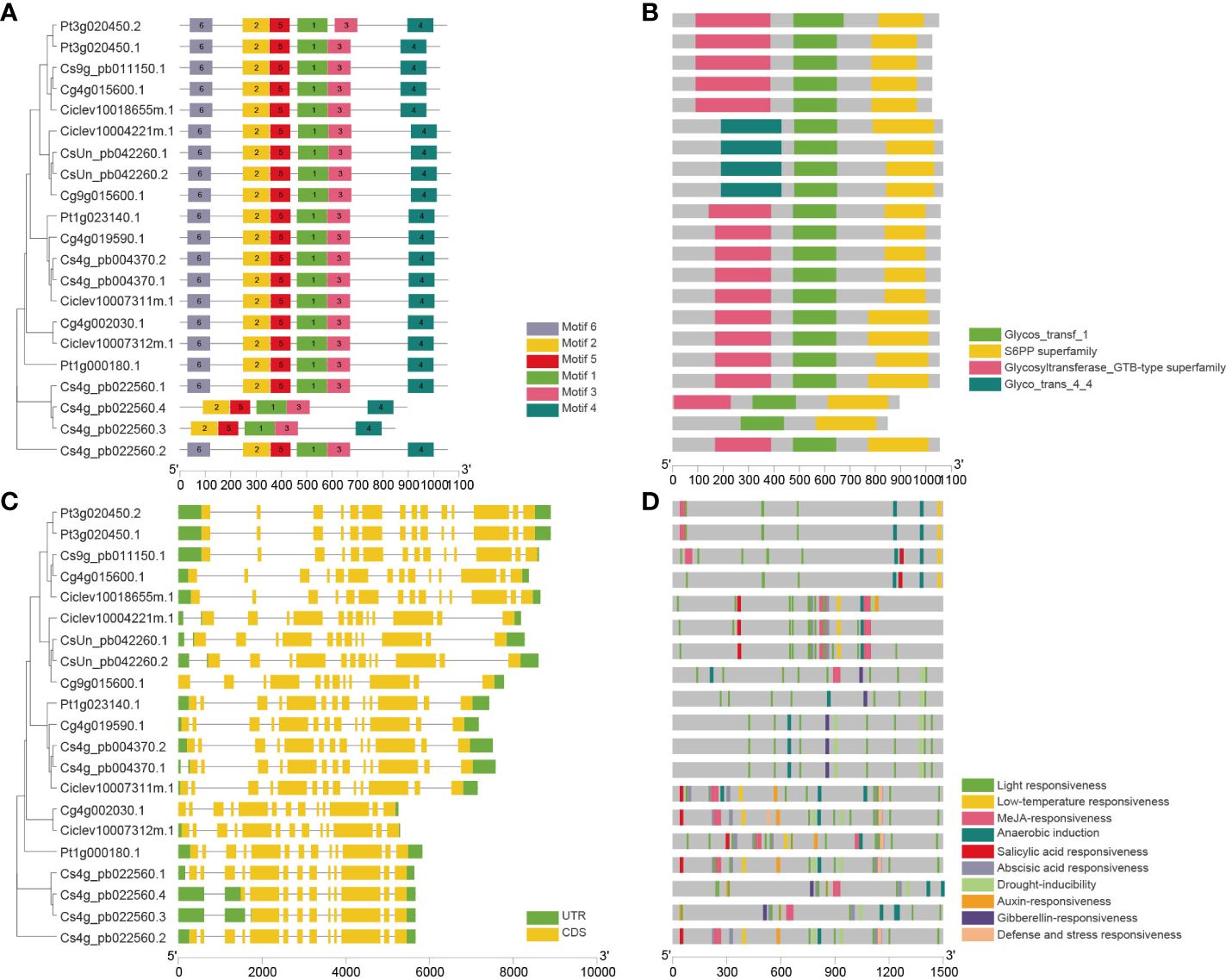
Figure 4 Conserved motifs, domains, gene structure, and predicted cis-elements of CsSPSs. (A) Conserved motifs (numbers 1–6) are highlighted using different coloured boxes. (B) Conserved protein domain of CsSPSs. (C) Exon–intron structure of CsSPSs. Green boxes (5ʹ UTR and 3ʹ UTR), yellow boxes (exons), and grey lines (introns). (D) Predicted cis-elements in CsSPSs promoters are represented using different colours.
The exon–intron structures of CsSPSs were further analysed. Generally, most citrus SPS genes contain twelve to fourteen exons (Figure 4C). Each citrus SPS within the same group shares the same intron–exon structure. For instance, all citrus SPS4 genes contain 14 introns, and every citrus SPS3 gene contains 12 introns. Additionally, each of SPS1 and SPS2 contains 13 introns, except for the two specific transcription modes of CsSPS2.
To gain further insights into the functions of CsSPSs, the 1500 bp promoter regions upstream of the CsSPSs initiation transcription site were analysed using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/) (Figure 4D). The analysis revealed that cis-elements in the promoter region of all identified CsSPSs are primarily involved in responding to light, hormones, and abiotic stress. All CsSPS promoter regions feature photoresponsive elements (Box 4 and G-box), and most CsSPS promoters contain low-temperature, methyl jasmonate (MeJA), and salicylic acid (SA) responsive elements, enabling them to respond to ethylene and abscisic acid (ABA) reactions (Supplementary Table S4). The diversity of cis-acting elements in the upstream promoter region of CsSPSs indicates that their function may involve various reactions, such as hormones, abiotic stress, seed, and endosperm development.
Expression profiles of CsSPSs in citrus
Publicly available transcriptomic data revealed distinct expression patterns of CsSPSs in different citrus tissues. CsSPS1 and CsSPS2 were predominantly expressed in ‘sinensis’ pulp, with CsSPS1 decreasing and CsSPS2 increasing during fruit maturation (Figure 5A). CsSPS3 and CsSPS4 exhibited high expression in leaves, with CsSPS3 also detected in flowers. However, transcriptional expression of CsSPSs in peel and root was low. Across maturing citrus fruits, CsSPSs displayed unique expression characteristics (Figures 5B, C). For example, the transcription level of CsSPSs in ‘Fengjie72-1’ is significantly higher than that in ‘CaraCara’, whereas the transcription level of CsSPSs in Clementina and Hemandina is similar. The transcriptional expression levels of most CsSPSs in pulp were continuously increased. CsSPS1 and CsSPS2 showed an evident upward trend in the four varieties. However, some CsSPSs, such as CsSPS3 in ‘CaraCara’, have decreased expression levels, which reflects the tissue expression specificity of CsSPSs mentioned earlier.
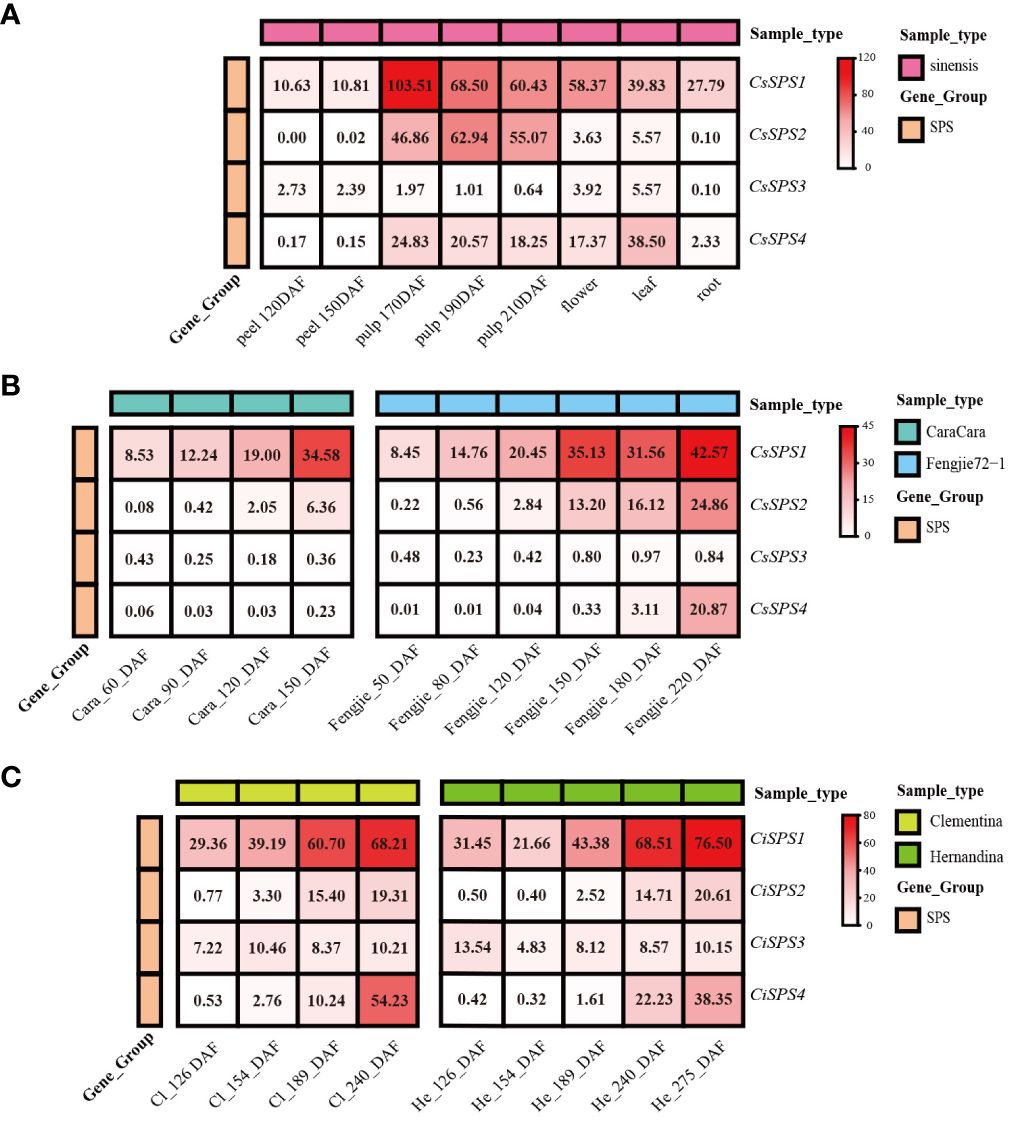
Figure 5 Expression of CsSPSs in citrus tissues and varieties. (A) Transcription levels in peel, pulp, flower, leaf, and root of ‘sinensis’. (B) Transcription levels in the pulp of ‘CaraCara’ and ‘Fengjie 72-1’. (C) Transcription levels in Clementina and ‘Hernandina’.
Correlation analysis between sugar and CsSPSs in citrus fruit
To understand the expression of CsSPSs in citrus pulp, we investigated the transcriptional expression of CsSPSs in ‘Kiyomi’ and ‘Succosa’ (Figure 6A). Consistent with the transcriptome results, the transcriptional expression of the four CsSPSs in citrus pulp increased with fruit development, and the types of CsSPSs expression patterns differed in the pulp of different citrus varieties. For example, The transcription expression of CsSPS2 and CsSPS3 is dominant in ‘Kiyomi’, while the transcription expression of CsSPS3 is dominant in ‘Succosa’. Unexpectedly, CsSPS1 is considered to be the most important SPS gene in the pulp of other citrus varieties, but in the pulp development of ‘Kiyomi’ and ‘Succosa’, the relative transcriptional expression of CsSPS1 is lower than that of other CsSPSs. Simultaneously, the stable, high transcription level of CsSPS3 and the rapid increase in CsSPS4 expression levels in the two materials indicated the importance of CsSPS3 and CsSPS4 in the development of citrus pulp.
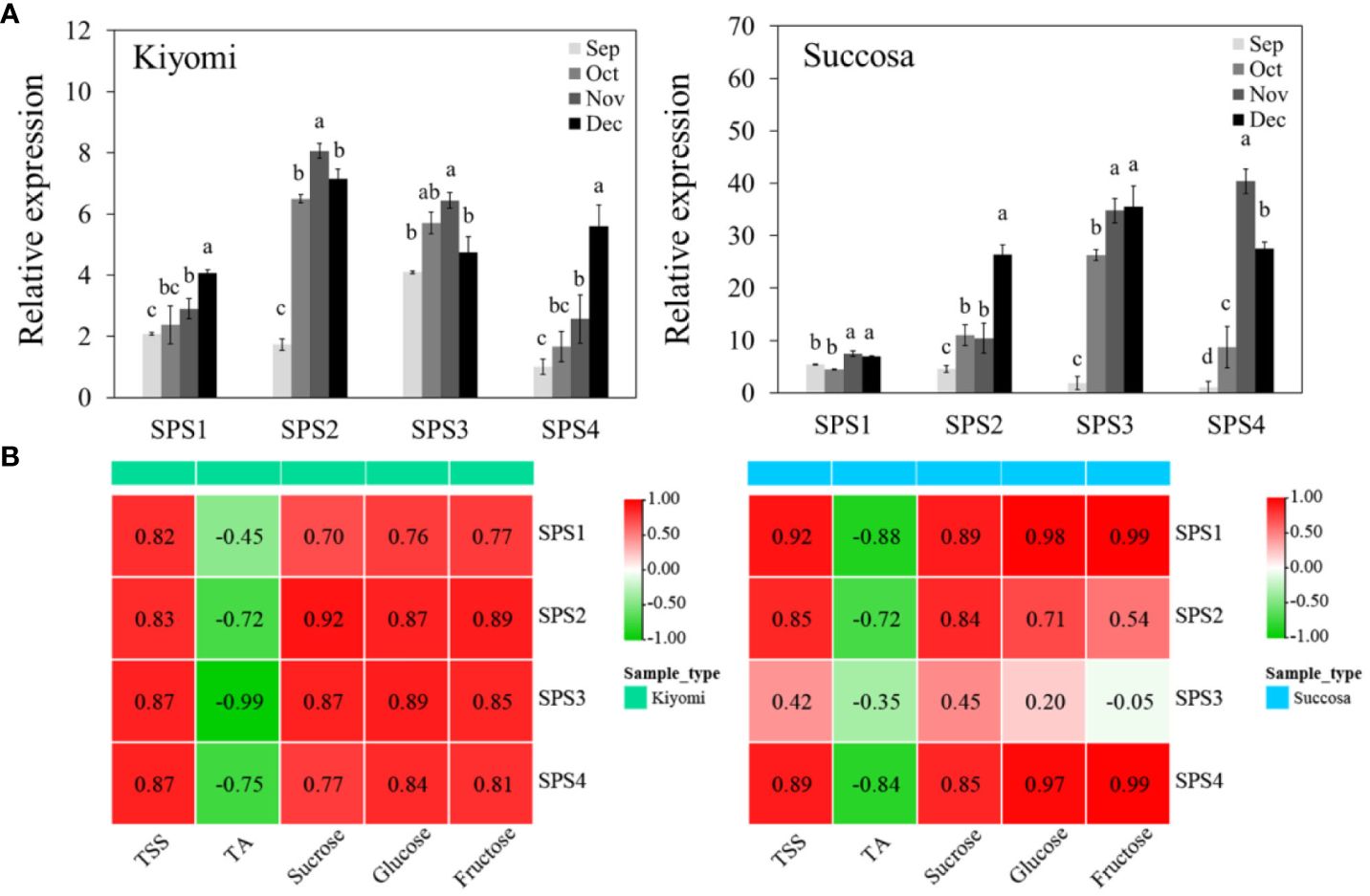
Figure 6 Expression of CsSPSs in ‘Kiyomi’ and ‘Succosa’. (A) Expression levels. (B) Correlation analysis with TSS, TA, sucrose, glucose, and fructose.
To comprehensively highlight the significance of CsSPSs in citrus sugar accumulation, we analysed Pearson’s correlation coefficients between CsSPSs expression and sugar content (Figure 6B). As expected, most CsSPSs were significantly and positively correlated with sugar content and its components. However, in ‘Succosa’, CsSPS1 with low transcriptional expression level and CsSPS4 with late rapid elevation demonstrated the most significant positive correlation with sugar content. Counterintuitively, despite having the highest transcriptional expression level, CsSPS3 had a low correlation with sugar content.
Co-expression network analysis of candidate CsSPSs during fruit development
To further understand the possible regulatory pathways of citrus sugar metabolism, we analysed the co-expression of CsSPSs based on our previous study (Zhang et al., 2023a). The results showed that all four CsSPSs were in the blue module (4,871 genes), which was positively correlated with the total sugar content, and the expression levels of most genes in the blue module increased with fruit development (Figure 7A). Importantly, in the blue module, we observed 196 transcription factors (Supplementary Table S6) that have a significant co-expression relationship with CsSPSs (weight > 0.15), including bHLH (15), ERF (12), bZIP (7), MADS (10), NAC (17) and MYB (18) (Figure 7B). It is worth noting that we found that eight WRKY transcription factors (CsWRKY20, CsWRKY47, CsWRKY32, CsWRKY2, CsWRKY65, CsWRKY3, CsWRKY28, CsWRKY74) have a co-expression relationship with CsSPSs. A positive relationship between CsWRKY47, CsWRKY3, CsWRKY28, CsWRKY74 and sugar was found in our previous study, indicating that there may be a regulatory relationship between CsWRKYs and CsSPSs.
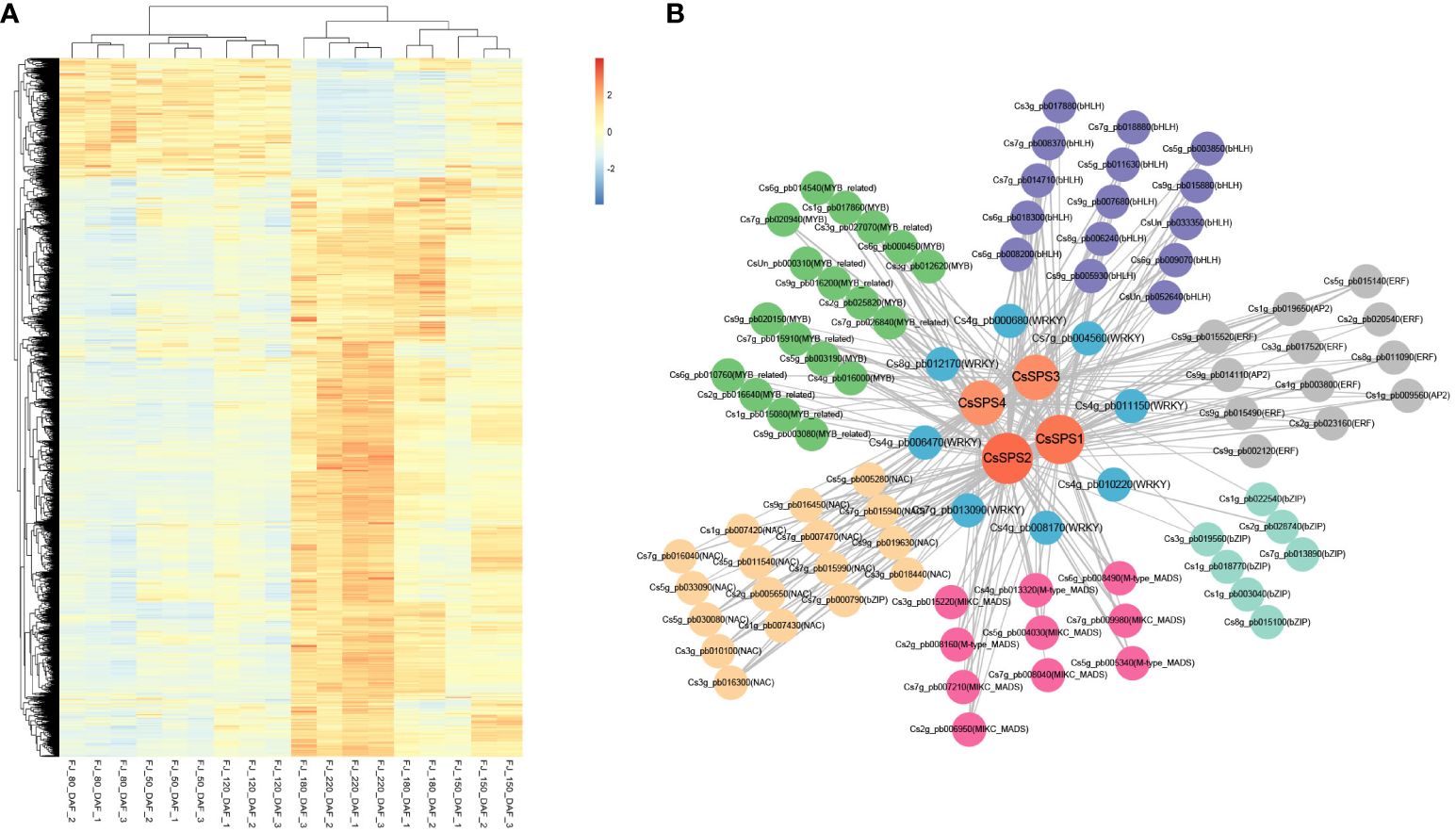
Figure 7 Weighted gene co-expression network analysis of CsSPSs. (A) Expression of genes in the blue module. (B) Co-expression network analysis of CsSPSs. (CsSPSs. Orange circles, WRKYs. Blue circles, MYBs. Green circles, bHLHs. Purple circles, ERFs. Grey circles, bZIPs. Turquoise circles, MADS. Pink circles, NACs. Yellow circles).
Expression analysis of CsWRKYs and dual-luciferase activity assay
To elaborate on the potential involvement of CsWRKYs in the sugar metabolism of citrus fruits through CsSPSs, we examined the expression of co-expressed WRKY genes in Kiyomi’ and ‘Succosa’. The results revealed a continuous increase in the transcriptional expressions of CsWRKY2, CsWRKY20, CsWRKY28, and CsWRKY32, and CsWRKY47 with the maturation of citrus pulp (Figure 8A).
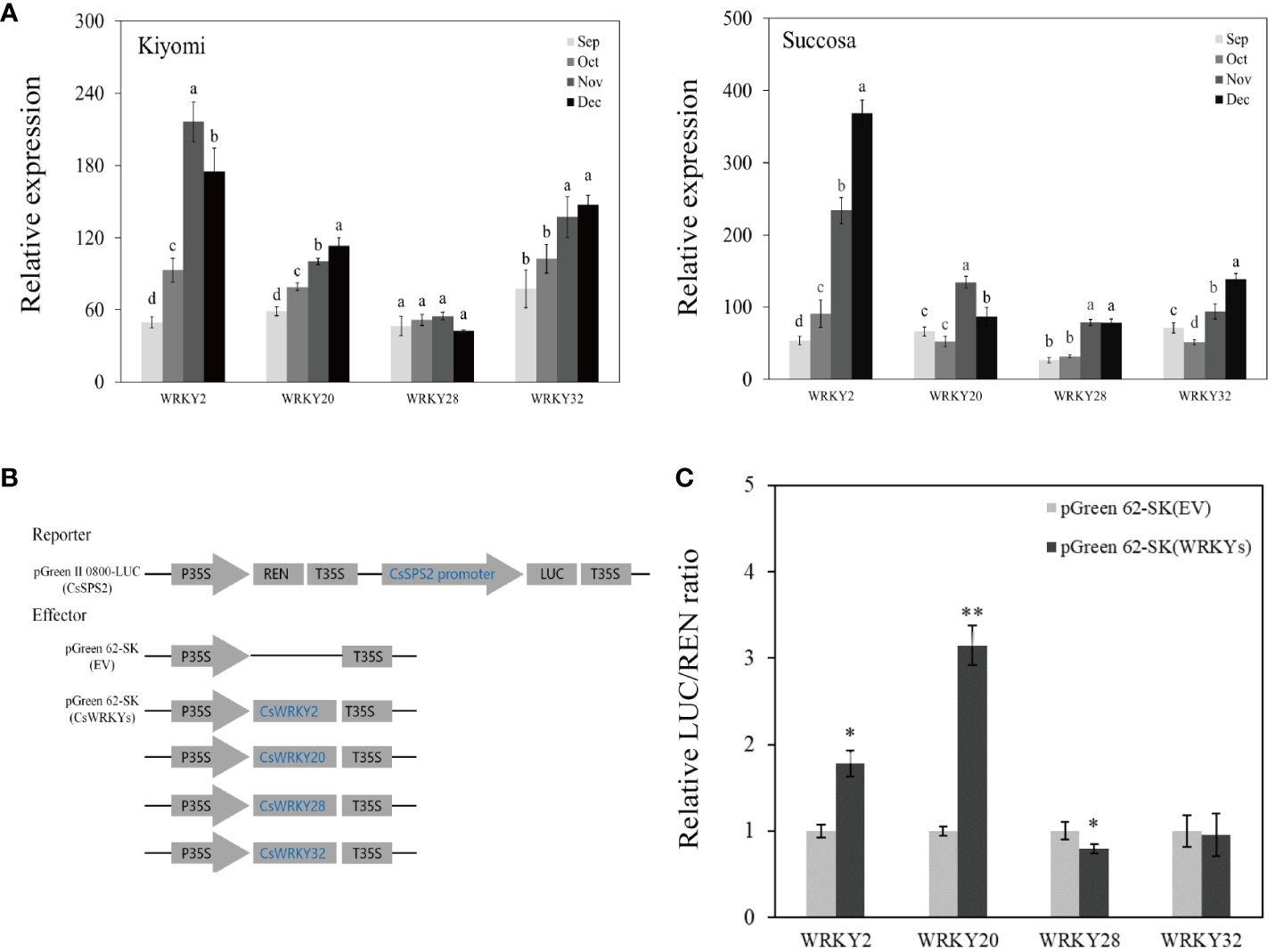
Figure 8 Expression analysis of CsWRKYs, Dual-luciferase activity assay of CsWRKYs and CsSPS2 promoter. (A) Expression of CsWRKYs in ‘Kiyomi’ and ‘Succosa’. (B) Schematic diagrams of vectors used for the dual-luciferase activity assay. (C) The relative LUC/REN ratio of pGreen II 0800-LUC (CsSPS2) × pGreen 62-SK (EV, empty vector) as the control and pGreen II 0800-LUC (CsSPS2) ×pGreen 62-SK(CsWRKYs). Asterisks indicate significant differences as determined by Student–Newman–Keuls multiple range test at P < 0.01 level.
Notably, W-box cis-elements was found in CsSPS2 promoter, but not in other CsSPSs (Supplementary Table S4). The dual-LUC activity assay between CsWRKYs and CsSPS2 indicated that the co-expression of CsWRKY2, CsWRKY20 with the CsSPS2 promoter significantly enhanced the LUC to REN ratio, and co-expression of CsWRKY28 with the CsSPS2 promoter significantly reduced the LUC to REN ratio, whereas no significant LUC to REN ratios were observed in the independent co-expression of CsWRKY32 with the promoters of CsSPS2. This indicated that CsWRKY2, CsWRKY20 might act as an activator in citrus fruit of sugar accumulation through CsSPS2, while CsWRKY28 inhibits sugar accumulation through CsSPS2 (Figure 8C).
Discussion
Similar to most citrus fruits, the sugar accumulation in citrus fruits (‘Kiyomi’ and ‘Succosa’) increases during fruit development, with sucrose being the predominant soluble sugar component (Ren et al., 2023). However, distinct citrus varieties exhibit varying rates of sucrose accumulation (Figure 1B). The rapid increase in glucose and fructose content, particularly in ‘Kiyomi’, contributes to the swift sucrose accumulation, whereas the stable substrate content in ‘Succosa’ leads to a slower rate of sucrose accumulation (Figure 2).
SPS genes identified in various plant species, and investigated in this study, revealing four SPS genes in representative citrus species (Citrus sinensis, Citrus clementina, Citrus grandis, Pitrus trifoliat) (Figure 3A). CsSPS1 and CsSPS2, belonging to subfamily A, exhibit high homology with Arabidopsis AtSPS1 and AtSPS2 (Figure 3C). CsSPS3 and CsSPS4 belong to subfamilies B and C, respectively. All CsSPSs share typical SPS family domains and similar intron–exon structures, indicating functional similarity among citrus SPSs (Figures 4A-C). The promoters of CsSPSs exhibit responsiveness to light, hormones, and abiotic stress, suggesting their pivotal role in citrus sugar signal responses to external abiotic cues (Figure 4D).
As a rate-limiting enzyme in sucrose metabolism, the transcriptional expression of SPS directly influences sucrose synthesis (Ruan, 2014; Wan et al., 2018). CsSPSs, akin to other species, generally exhibit increased transcriptional expression levels with fruit ripening, showing a significant positive correlation with fruit sugar content (Figure 6). However, CsSPSs display expression specificity in different tissues and varieties. For example, CsSPS1 and CsSPS2 are predominantly expressed in citrus fruits, whereas CsSPS3 and CsSPS4 are expressed in leaves and flowers (Figure 5). However, CsSPS3 is the dominant CsSPS expression type in ‘Kiyomi’ and ‘Succosa’ fruits. In addition, the expression level of CsSPS2 in ‘Kiyomi’ was significantly higher than ‘Succosa’, which may be one of the reasons why the sucrose content of ‘Kiyomi’ increased faster than ‘Succosa’. Remarkably, the expression levels of CsSPS4 increase rapidly during late fruit development in several citrus varieties. Therefore, we believe that CsSPS1 and CsSPS2 play a major role in sugar accumulation in citrus fruits, while CsSPS4 plays a vital role in the late development of citrus fruit. To sum up, the expression types and patterns of CsSPSs in different citrus varieties and tissues are different.
Sugar signals play an important role in plant development and resistance to external stress (Gill and Tuteja, 2010; Sperdouli and Moustakas, 2012; Proels and Hückelhoven, 2014). But the synthesis of sugar are subject to regulation by various transcription factors. Silencing strawberry FaGAMYB and FaMYB10 has been shown to reduce the transcriptional expression of FaSPS3 and FaSPS1, respectively (Vallarino et al., 2015). FaMYB44.2 can directly bind to the FaSPS3 promoter to inhibit sucrose accumulation (Wei et al., 2018). However, WRKY transcription factors can also be involved in the regulation of sugar metabolism. In the present study, we found that CsWRKY2, CsWRKY20, CsWRKY28 and CsWRKY32 had a significant positively correlated with CsSPSs (Figure 7). In previous research, WRKY2 was found to regulate circadian expression, mediated seed germination, regulation of pollen development, and play an important role in regulating tolerance to abiotic stresses such as cold damage and drought (Jiang and Yu, 2009; Niu et al., 2012; Lei et al., 2017; Wang et al., 2022); Rice OsWRKY47 and soybean GsWRKY20 mainly respond to drought stress (Luo et al., 2013; Ning et al., 2017; Li et al., 2023); WRKY28 is thought to be involved in plant salt tolerance, element absorption and morphogenesis (Yang et al., 2022; Zhang et al., 2023b, Zhang et al., 2023c); Tomato SlWRKY32 can regulate ethylene signal and affect tomato fruit colouring (Zhao et al., 2021); Nonetheless, There is no doubt that the promoter activity of CsSPS2 can be activated by CsWRKY2 and CsWRKY2, restrained by CsWRKY28. Therefore, we suggest that CsWRKY2, CsWRKY20 and CsWRKY28 may affect the sugar quality of citrus fruit by regulating CsSPS2.
Conclusion
In conclusion, we investigated the sugar composition in ‘Kiyomi’ and ‘Succosa’ and elucidated the complexities of sugar quality formation in citrus fruits. Simultaneously, we conducted a comprehensive identification of SPS genes in the citrus genome, revealing four CsSPSs belonging to three types with conserved sucrose phosphate synthase domains. Our expression analysis demonstrated that most CsSPSs exhibit increased expression levels with citrus fruit maturity, tightly correlated with sucrose content. CsSPSs manifest specific expression patterns in different tissues and citrus varieties, with CsSPS4 emerging as a key player in late fruit development. These findings highlight the pivotal role of CsSPSs in citrus fruit sugar metabolism. Notably, building upon our previous studies, we suggest that the identified CsWRKYs, particularly CsWRKY2, CsWRKY20, CsWRKY28 and CsWRKY32, may participate in regulating CsSPS expression. CsWRKY20 might act as an activator in citrus fruit of sugar accumulation through CsSPS2, and CsWRKY28 might can inhibits sugar accumulation through CsSPS2. Our results not only elucidate the process of sugar quality formation in citrus fruits but also provide novel insights into the mechanisms through which WRKY transcription factors modulate sugar metabolism.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
WL: Investigation, Resources, Formal Analysis, Writing – original draft. WH: Investigation, Resources, Writing – original draft. KL: Investigation, Resources, Writing – original draft. JL: Investigation, Writing – original draft. CY: Investigation, Writing – original draft. YS: Investigation, Writing – original draft. ZH: Investigation, Writing – original draft. BP: Resources, Supervision, Writing – review & editing. HL: Resources, Supervision, Writing – review & editing. BX: Resources, Supervision, Writing – review & editing. LL: Resources, Supervision, Writing – review & editing. JH: Resources, Supervision, Writing – review & editing. MZ: Conceptualization, Funding acquisition, Investigation, Resources, Validation, Writing – review & editing. XW: Conceptualization, Writing – review & editing. ZW: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Sichuan Province Science and Technology Department Project (2023NSFSC1248).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1372809/full#supplementary-material
References
Albertini, M. V., Carcouet, E., Pailly, O., Gambotti, C., Luro, F., Berti, L. (2006). Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. J. Agric. Food Chem. 54, 8335–8339. doi: 10.1021/jf061648j
Anur, R. M., Mufithah, N., Sawitri, W. D., Sakakibara, H., Sugiharto, B. (2020). Overexpression of sucrose phosphate synthase enhanced sucrose content and biomass production in transgenic sugarcane. Plants (Basel Switzerland) 9. doi: 10.3390/plants9020200
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Brown, P., Baxter, L., Hickman, R., Beynon, J., Moore, J. D., Ott, S. (2013). MEME-LaB: motif analysis in clusters. Bioinformatics 29, 1696–1697. doi: 10.1093/bioinformatics/btt248
Bush, D. R. (2020). Identifying the pathways that control resource allocation in higher plants. Proc. Natl. Acad. Sci. U. S. A. 117, 8669–8671. doi: 10.1073/pnas.2002581117
Castleden, C. K., Aoki, N., Gillespie, V. J., MacRae, E. A., Quick, W. P., Buchner, P., et al. (2004). Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol. 135, 1753–1764. doi: 10.1104/pp.104.042457
Coleman, H. D., Yan, J., Mansfield, S. D. (2009). Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. U. S. A. 106, 13118–13123. doi: 10.1073/pnas.0900188106
Feng, G., Wu, J., Xu, Y., Lu, L., Yi, H. (2021). High-spatiotemporal-resolution transcriptomes provide insights into fruit development and ripening in Citrus sinensis. Plant Biotechnol. J. 19, 1337–1353. doi: 10.1111/pbi.13549
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guo, L. X., Shi, C. Y., Liu, X., Ning, D. Y., Jing, L. F., Yang, H., et al. (2016). Citrate accumulation-related gene expression and/or enzyme activity analysis combined with metabolomics provide a novel insight for an orange mutant. Sci. Rep. 6, 29343. doi: 10.1038/srep29343
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in brassica napus. PloS One 11, e0157558. doi: 10.1371/journal.pone.0157558
Hu, W., Hou, X., Huang, C., Yan, Y., Tie, W., Ding, Z., et al. (2015). Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 16, 19728–19751. doi: 10.3390/ijms160819728
Jia, M., Ding, N., Zhang, Q., Xing, S., Wei, L., Zhao, Y., et al. (2017). A FERONIA-Like receptor kinase regulates strawberry (fragaria × ananassa) fruit ripening and quality formation. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01099
Jiang, W., Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9, 96. doi: 10.1186/1471-2229-9-96
Koramutla, M. K., Ram, C., Bhatt, D., Annamalai, M., Bhattacharya, R. (2019). Genome-wide identification and expression analysis of sucrose synthase genes in allotetraploid Brassica juncea. Gene 707, 126–135. doi: 10.1016/j.gene.2019.04.059
Langenkämper, G., Fung, R. W., Newcomb, R. D., Atkinson, R. G., Gardner, R. C., MacRae, E. A. (2002). Sucrose phosphate synthase genes in plants belong to three different families. J. Mol. Evol. 54, 322–332. doi: 10.1007/s00239-001-0047-4
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Lei, R., Li, X., Ma, Z., Lv, Y., Hu, Y., Yu, D. (2017). Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 91, 962–976. doi: 10.1111/tpj.13619
Li, D., Gu, B., Huang, C., Shen, J., Wang, X., Guo, J., et al. (2023). Functional study of amorpha fruticosa WRKY20 gene in response to drought stress. Int. J. Mol. Sci. 24. doi: 10.3390/ijms241512231
Liu, G., Zhang, D., Zhao, T., Yang, H., Jiang, J., Li, J., et al. (2022). Genome-wide analysis of the WRKY gene family unveil evolutionary history and expression characteristics in tomato and its wild relatives. Front. Genet. 13. doi: 10.3389/fgene.2022.962975
Liu, J., Yue, R., Si, M., Wu, M., Cong, L., Zhai, R., et al. (2019). Effects of exogenous application of melatonin on quality and sugar metabolism in ‘Zaosu’ Pear Fruit. J. Plant Growth Regul. 38, 1161–1169. doi: 10.1007/s00344-019-09921-0
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lunn, J. E. (2003). Sucrose-phosphatase gene families in plants. Gene 303, 187–196. doi: 10.1016/S0378-1119(02)01177-0
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Ning, W., Zhai, H., Yu, J., Liang, S., Yang, X., Xing, X., et al. (2017). Overexpression of glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 37. doi: 10.1007/s11032-016-0614-4
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Okamura, M., Aoki, N., Hirose, T., Yonekura, M., Ohto, C., Ohsugi, R. (2011). Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 181, 159–166. doi: 10.1016/j.plantsci.2011.04.019
Park, J. Y., Canam, T., Kang, K. Y., Ellis, D. D., Mansfield, S. D. (2008). Over-expression of an arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 17, 181–192. doi: 10.1007/s11248-007-9090-2
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Proels, R. K., Hückelhoven, R. (2014). Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 15, 858–864. doi: 10.1111/mpp.12139
Reimholz, R., Geiger, M., Haake, V., Deiting, U., Krause, K. P., Sonnewald, U., et al. (2008). Potato plants contain multiple forms of sucrose phosphate synthase, which differ in their tissue distributions, their levels during development, and their responses to low temperature. Plant Cell Environ. 20, 291–305. doi: 10.1046/j.1365-3040.1997.d01-83.x
Ren, Y., Liao, S., Xu, Y. (2023). An update on sugar allocation and accumulation in fruits. Plant Physiol. 193, 888–899. doi: 10.1093/plphys/kiad294
Roy Choudhury, S., Roy, S., Das, R., Sengupta, D. N. (2008). Differential transcriptional regulation of banana sucrose phosphate synthase gene in response to ethylene, auxin, wounding, low temperature and different photoperiods during fruit ripening and functional analysis of banana SPS gene promoter. Planta 229, 207–223. doi: 10.1007/s00425-008-0821-2
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sheng, L., Shen, D., Luo, Y., Sun, X., Wang, J., Luo, T., et al. (2017). Exogenous γ-aminobutyric acid treatment affects citrate and amino acid accumulation to improve fruit quality and storage performance of postharvest citrus fruit. Food Chem. 216, 138–145. doi: 10.1016/j.foodchem.2016.08.024
Signora, L., Galtier, N., Skøt, L., Lucas, H., Foyer, C. H. (1998). Over-expression of sucrose phosphate synthase in Arabidopsis thaliana results in increased foliar sucrose/starch ratios and favours decreased foliar carbohydrate accumulation in plants after prolonged growth with CO2 enrichment. J. Exp. Bot. 49, 669–680. doi: 10.1093/jxb/49.321.669%J
Sperdouli, I., Moustakas, M. (2012). Interaction of proline, sugars, and anthocyanins during photosynthetic acclimation of Arabidopsis thaliana to drought stress. J. Plant Physiol. 169, 577–585. doi: 10.1016/j.jplph.2011.12.015
Terol, J., Nueda, M. J., Ventimilla, D., Tadeo, F., Talon, M. (2019). Transcriptomic analysis of Citrus clementina mandarin fruits maturation reveals a MADS-box transcription factor that might be involved in the regulation of earliness. BMC Plant Biol. 19, 47. doi: 10.1186/s12870-019-1651-z
Vallarino, J. G., Osorio, S., Bombarely, A., Casañal, A., Cruz-Rus, E., Sánchez-Sevilla, J. F., et al. (2015). Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 208, 482–496. doi: 10.1111/nph.13463
Wan, H., Wu, L., Yang, Y., Zhou, G., Ruan, Y. L. (2018). Evolution of sucrose metabolism: the dichotomy of invertases and beyond. Trends Plant Sci. 23, 163–177. doi: 10.1016/j.tplants.2017.11.001
Wang, S., Sun, Q., Zhang, M., Yin, C., Ni, M. (2022). WRKY2 and WRKY10 regulate the circadian expression of PIF4 during the day through interactions with CCA1/LHY and phyB. Plant Commun. 3, 100265. doi: 10.1016/j.xplc.2021.100265
Wei, L., Mao, W., Jia, M., Xing, S., Ali, U., Zhao, Y., et al. (2018). FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J. Exp. Bot. 69, 4805–4820. doi: 10.1093/jxb/ery249
Winter, H., Huber, S. C. (2000). Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit. Rev. Biochem. Mol. Biol. 35, 253–289. doi: 10.1080/10409230008984165
Worrell, A. C., Bruneau, J. M., Summerfelt, K., Boersig, M., Voelker, T. A. (1991). Expression of a maize sucrose phosphate synthase in tomato alters leaf carbohydrate partitioning. Plant Cell 3, 1121–1130. doi: 10.1105/tpc.3.10.1121
Yang, H., Zhu, Z., Zhang, M., Li, X., Xu, R., Zhu, F., et al. (2022). CitWRKY28 and CitNAC029 promote the synthesis of cuticular wax by activating CitKCS gene expression in citrus fruit. Plant Cell Rep. 41, 905–920. doi: 10.1007/s00299-021-02826-x
Zhang, H., Chen, J., Peng, Z., Shi, M., Liu, X., Wen, H., et al. (2021). Integrated transcriptomic and metabolomic analysis reveals a transcriptional regulation network for the biosynthesis of carotenoids and flavonoids in 'Cara cara' navel Orange. BMC Plant Biol. 21, 29. doi: 10.1186/s12870-020-02808-3
Zhang, Z., Chen, L., Yu, J. (2023c). Maize WRKY28 interacts with the DELLA protein D8 to affect skotomorphogenesis and participates in the regulation of shade avoidance and plant architecture. J. Exp. Bot. 74, 3122–3141. doi: 10.1093/jxb/erad094
Zhang, M., Lu, W., Yang, X., Li, Q., Lin, X., Liu, K., et al. (2023a). Comprehensive analyses of the citrus WRKY gene family involved in the metabolism of fruit sugars and organic acids. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1264283
Zhang, M., Zhao, R., Wang, H., Ren, S., Shi, L., Huang, S., et al. (2023b). OsWRKY28 positively regulates salinity tolerance by directly activating OsDREB1B expression in rice. Plant Cell Rep. 42, 223–234. doi: 10.1007/s00299-022-02950-2
Keywords: citrus, sugar metabolism, sucrose phosphate synthase, fruit quality, expression network
Citation: Lu W, Hao W, Liu K, Liu J, Yin C, Su Y, Hang Z, Peng B, Liu H, Xiong B, Liao L, He J, Zhang M, Wang X and Wang Z (2024) Analysis of sugar components and identification of SPS genes in citrus fruit development. Front. Plant Sci. 15:1372809. doi: 10.3389/fpls.2024.1372809
Received: 18 January 2024; Accepted: 20 March 2024;
Published: 28 March 2024.
Edited by:
Heping Cao, United States Department of Agriculture (USDA), United StatesReviewed by:
Yuanyue Shen, Beijing University of Agriculture, ChinaChen Zhu, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Lu, Hao, Liu, Liu, Yin, Su, Hang, Peng, Liu, Xiong, Liao, He, Zhang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Wang, d2FuZ3poaWh1aTMxOEBzaWNhdS5lZHUuY24=; Xun Wang, d3gwMTA0QHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Wen Lu1†
Wen Lu1† Bo Xiong
Bo Xiong Mingfei Zhang
Mingfei Zhang Zhihui Wang
Zhihui Wang