- 1National Key Laboratory for Biological Breeding of Tropical Crops, Kunming, Yunnan, China
- 2Sugarcane Research Institute, Yunnan Academy of Agricultural Sciences/Yunnan Key Laboratory of Sugarcane Genetic Improvement, Kaiyuan, Yunnan, China
Under changing climatic scenarios, grassland conservation and development have become imperative to impart functional sustainability to their ecosystem services. These goals could be effectively and efficiently achieved with targeted genetic improvement of native grass species. To the best of our literature search, very scant research findings are available pertaining to gene editing of non-cultivated grass species (switch grass, wild sugarcane, Prairie cordgrass, Bermuda grass, Chinese silver grass, etc.) prevalent in natural and semi-natural grasslands. Thus, to explore this novel research aspect, this study purposes that gene editing techniques employed for improvement of cultivated grasses especially sugarcane might be used for non-cultivated grasses as well. Our hypothesis behind suggesting sugarcane as a model crop for genetic improvement of non-cultivated grasses is the intricacy of gene editing owing to polyploidy and aneuploidy compared to other cultivated grasses (rice, wheat, barley, maize, etc.). Another reason is that genome editing protocols in sugarcane (x = 10–13) have been developed and optimized, taking into consideration the high level of genetic redundancy. Thus, as per our knowledge, this review is the first study that objectively evaluates the concept and functioning of the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technique in sugarcane regarding high versatility, target specificity, efficiency, design simplicity, and multiplexing capacity in order to explore novel research perspectives for gene editing of non-cultivated grasses against biotic and abiotic stresses. Additionally, pronounced challenges confronting sugarcane gene editing have resulted in the development of different variants (Cas9, Cas12a, Cas12b, and SpRY) of the CRISPR tool, whose technicalities have also been critically assessed. Moreover, different limitations of this technique that could emerge during gene editing of non-cultivated grass species have also been highlighted.
1 Introduction
Globally, grasslands are considered the biggest ecosystem and serve as a carbon sink, ecological barriers, a watershed for low riparian regions, feedstock for ruminants, and mineral extraction sites for drilling and mining, and offer numerous associated benefits like wool, herbs for traditional medicines, tourism, and leisure (Wen et al., 2018; Iqbal, 2022). Recently, it has become imperative to conserve grasslands by employing practices that ensure protection and sustainable management of grassland ecosystems by maintaining the biodiversity and ecological integrity for persistent provision of ecosystem services (Abbas et al., 2015; Yu et al., 2019). Contrastingly, different initiatives intended for improving the productivity and sustainability of grasslands for agricultural purposes have been termed as grassland development (Yang et al., 2016; Iqbal et al., 2022; Ijaz et al., 2023). However, grassland conservation and development have remained neglected owing to a multitude of challenges especially climate change (CC). Additionally, overgrazing by livestock has caused serious depletion of grass resources along with adversely affecting the sustainability and health of natural and semi-natural grasslands (Wu et al., 2009; Wen et al., 2018). In addition, soil erosion (a soil quality degradation process that negatively impacts the health of grasses and the entire ecosystem), biodiversity loss, and, more importantly, the invasion of noxious weeds have reduced the productivity of native grass species. The invasive plant species tend to outcompete native grasses, which ultimately alters the grassland’s ecosystem composition and balance (Su et al., 2015; Wen et al., 2018). The underlying reason is that invasive plant species having aggressive growth patterns tend to acquire more growth resources and ultimately disrupt the ecosystem balance by overcoming native grass species (Maqsood et al., 2020; Abbas et al., 2021). Recently, the need for agricultural expansion owing to increasing food demand, rapid urbanization, and numerous abrupt land-use changes has caused grassland conversion into croplands primarily owing to the low productivity of grasses (Liu et al., 2008; Yu et al., 2019). More importantly, global CC has seriously affected grasslands owing to altering temperature and precipitation patterns. Likewise, persistent CC causes periodic fires (planned as well as wild) that are traditionally believed to stimulate the growth of grasses along with controlling the woody vegetation (Abbas et al., 2015). However, fire mismanagement leads to woody plants’ encroachment, which ultimately reduces suitable habitat availability for grass species (Sun et al., 2013; Wang et al., 2016; Yu et al., 2019). Recently, native grasses are exposed to water scarcity owing to changes in precipitation patterns along with other stresses including heat, salinity, and pollution. Figure 1 illustrates the pronounced challenges (environmental, ecological, and anthropogenic) faced by grass species in grasslands. The net result of all these stresses is a significant loss of habitat, which has threatened the survival of grass species; all these stresses have led to a serious decline in ecosystem services provided by grasslands (Di et al., 2014; Dong et al., 2019; Iqbal et al., 2022). Therefore, grassland conservation and development are directly linked to genetic improvement of grasses as CC has posed varying challenges to native grass species. Moreover, different anthropogenic, environmental, ecological, and soil-related challenges are faced by grasses in natural or improved grasslands, which necessitate their genetic improvement in order to impart sustainability to grassland ecosystems.
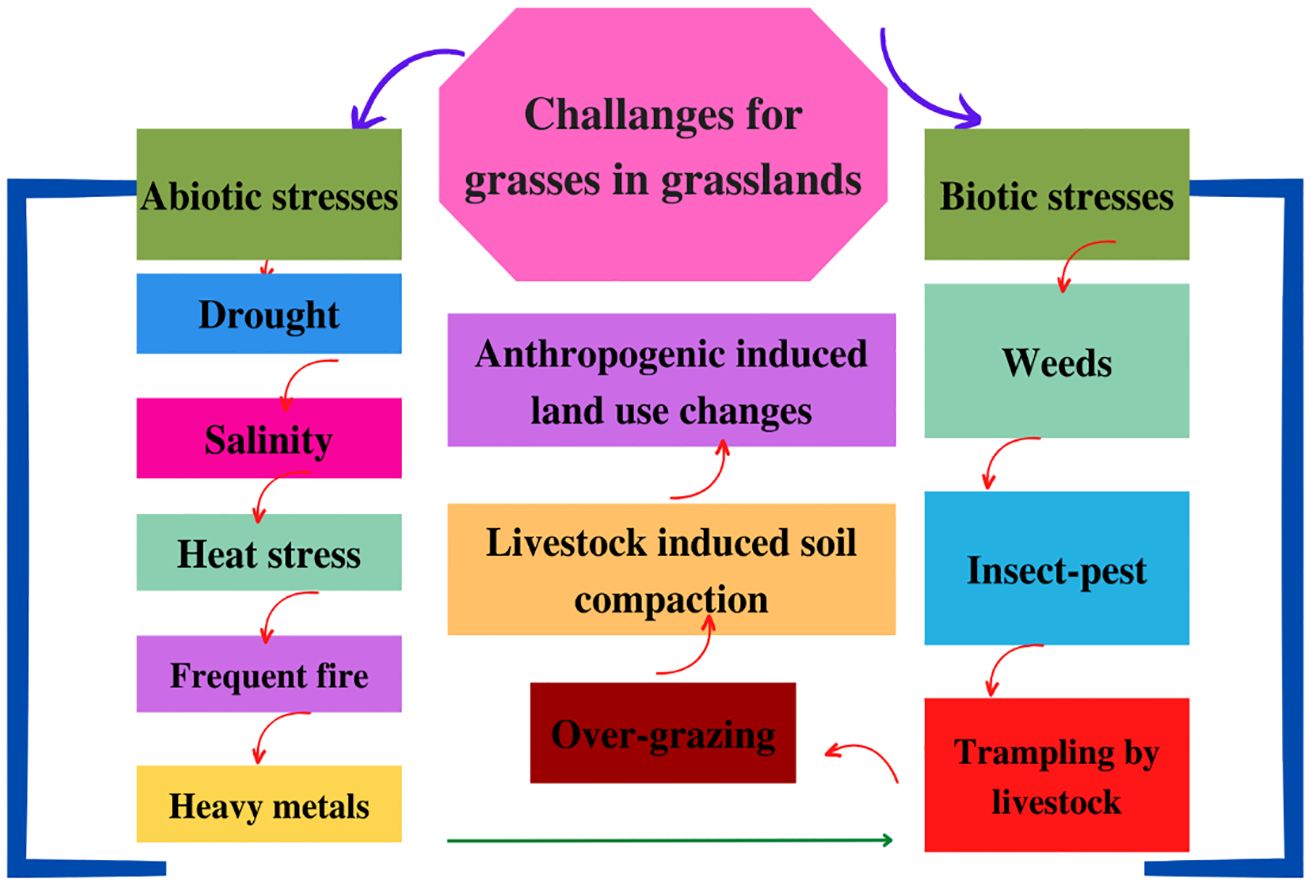
Figure 1 Pronounced abiotic and biotic stresses affect grasses in grasslands, reducing their ecosystem services and necessitating genome editing of grass species for imparting tolerance against biotic and abiotic stresses along with improving their productivity.
The non-cultivated grasses’ genetic improvement has remained neglected owing to the focus on major cultivated grasses like wheat, rice, maize, and sugarcane. In addition, most non-cultivated grasses are polyploidy, which restricts gene editing using traditional approaches. Modern gene editing techniques especially CRISPR (clustered regularly interspaced short palindromic repeats) have effectively inserted and knocked out targeted genes in cultivated grasses for boosting yield attributes under environmental extremes (Xing et al., 2014; Svitashev et al., 2015; Waltz, 2016; Gasparis et al., 2018; Holubová et al., 2018; Brauer et al., 2020). Moreover, this technique has been employed to study gene functions through selective disruption of genes and thereafter observing the resulting effects of altered genes in cultivated grasses (Kim et al., 2022). However, the genome editing of non-cultivated grasses might be initiated by taking sugarcane as a model plant because it is a perennial C-4 grass having exceptional potential for converting solar radiation and farm inputs (nutrients, water, etc.) into chemical harvestable energy (sucrose) (Hoang et al., 2015; Hussin et al., 2022). Furthermore, sugarcane has demonstrated intricacy in its genome editing owing to polyploidy and aneuploidy (Eid et al., 2021). Despite these challenges, genome editing techniques employed in sugarcane have improved yield attributes and plant metabolism, leading to enhanced yield on a sustainable basis (Jung and Altpeter, 2016). More importantly, numerous genetic modifications have been introduced for conferring resistance against diseases that adversely affect sugarcane growth, yield, and sucrose recovery (Viswanathan and Rao, 2011; Ali et al., 2019; Afzal et al., 2020; Wu et al., 2023). Targeted gene editing has assisted in producing sugarcane varieties that are resistant against pests and thus need fewer chemical pesticides. Moreover, these newly developed varieties have the potential to tolerate abiotic stresses (drought, heat, salinity, water logging, etc.) (Ramiro et al., 2016; Hussain et al., 2018; Li et al., 2018; Budeguer et al., 2021).
Thus, to the best of our understanding, this synthesis review is the first study that describes sugarcane as a model crop (because it is a perennial grass and presents high intricacy of gene editing owing to polyploidy), suggesting genetic improvement in non-cultivated grass species. Another reason is genome editing protocols of sugarcane hold bright perspectives for non-cultivated grass improvement because gene editing techniques in sugarcane (x = 10–13) have been developed and subsequently optimized considering the high level of genetic redundancy. Among gene editing techniques, special emphasis has been placed on the basics of CRISPR/Cas9 and its application in sugarcane genome improvement. Last but not least, different potential limitations that might emerge during the deployment of this technique for genetic improvement of non-cultivated grass species have been objectively highlighted.
2 Non-cultivated grasses of economic significance
Grasslands (also known as prairie, savanna, steppe, pampas, etc.) are the areas dominated by grasses (Poaceae family) and different sedges of Cyperaceae family (Iqbal et al., 2022; Ijaz et al., 2023). Recently, grassland conservation has emerged as one of the biggest challenges due to their conversion into croplands (Wen et al., 2018; Yu et al., 2019). Previously, conservation efforts have generally aimed at preventing the loss of grass species, soil degradation, and fragmentation of grasslands for ensuring their long-term sustainability (Dong et al., 2019; Iqbal et al., 2022). Contrastingly, initiatives such as introduction of more efficient grass species and grazing systems along with implementation of sustainable land management techniques are needed for their conservation. In addition, grassland development in a broader economic perspective might involve initiatives to diversify and integrate varying sources or services through the promotion of tourism and affiliated industries (dairy, honey, and medicine) that are compatible with the conservation and sustainable use of grasslands (Saleh and Karwacki, 1996). Therefore, one of the biologically feasible ways of achieving grassland conservation and development could be genetic improvement of native grass species regarding which persistent research efforts are lacking so far. Table 1 presents numerous non-cultivated grasses that hold bright economic perspectives (as biofuel, feed for ruminants, and beverages, and for medicinal use and aesthetic purposes); however, their productivity and nutritional value enhancement through gene editing is still awaited. Therefore, this study proposes to employ modern gene editing techniques of sugarcane especially CRISPR/Cas9 for genome editing of non-cultivated grasses.
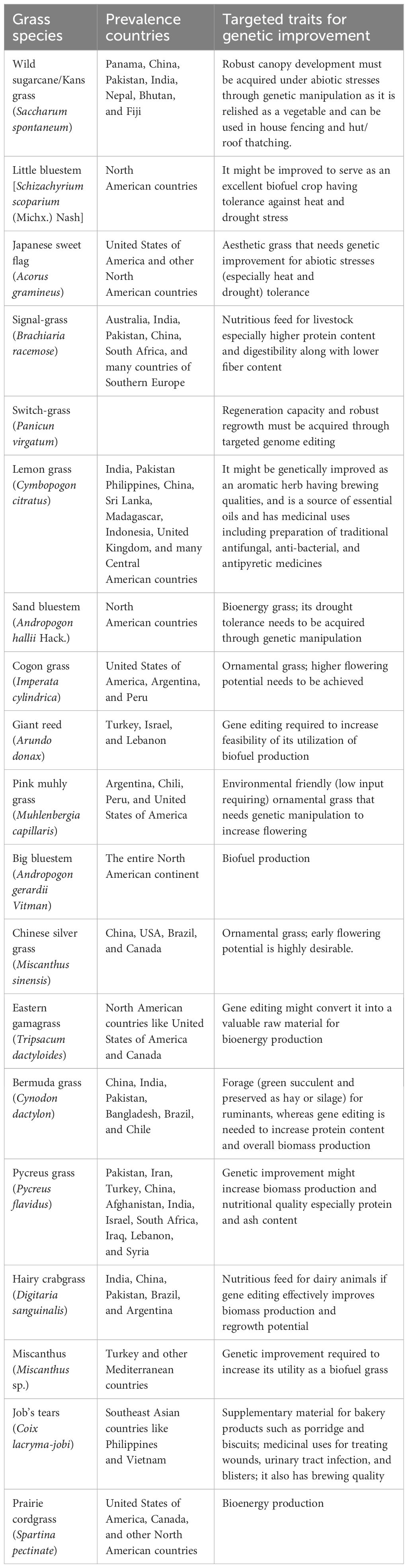
Table 1 Different non-cultivated grass species, their prevalence regions, and prospective uses of economic significance as reported by Iqbal et al. (2022).
3 Sugarcane (a C4 grass) morpho-anatomical features and pertinence
Sugarcane (Saccharum officinarum) belongs to the genus Saccharum that entails many species such as S. robustum, S. officinarum, S. barberi, S. edule, S. sinense, and S. spontaneum (Tew and Cobill, 2008; Taparia et al., 2012; Hussin et al., 2022). These are genetically related to family Poaceae members such as sorghum, Miscanthus, and Erianthus (Saleh and Karwacki, 1996). It is a tropical and subtropical perennial C4 grass (Byrt et al., 2011) that is primarily grown for its high sugar content especially in China, Brazil, India, Thailand, Pakistan, and many other countries of Africa and Americas (Iqbal and Iqbal, 2014; Iqbal and Saleem, 2014; Iqbal et al., 2015). Sugarcane has been classified among the most productive cultivated grasses in modern input-intensive farming systems owing to its superior and unprecedented light, water, and nitrogen use efficiencies (Weeks, 2017). Apart from sugar, this perennial grass also finds its use in the production of ethanol, particularly in countries such as Brazil that promote biofuel production (Iqbal and Iqbal, 2014). Additionally, various by-products such as molasses are produced during sugar-making, which are used for producing ethanol, rum, etc (Iqbal and Saleem, 2014; Ko et al., 2018). Moreover, bagasse (the fibrous residue left after juice extraction) is another useful by-product of sugar production that is used for power and biofuel generation (Tew and Cobill, 2008; Mohan et al., 2020) along with serving as a raw material in paper and board production (Iqbal and Saleem, 2014; Eid et al., 2021).
The frequent occurrence of drought and other CCs have recently imposed pronounced deleterious effects on cane yield of elite cultivars (Andrade et al., 2014; Li et al., 2019). In addition, disruption of rainfall patterns and declining availability of irrigation water are slicing the yield of this higher water requiring cultivated grass (Trujillo et al., 2009; Begcy et al., 2012; Ferreira et al., 2012; Lin et al., 2014; Zhu et al., 2021). Persistent genome editing efforts have been made for improving the agro-botanical traits (enhanced number of leaves and leaf blade area for increasing the rate of photosynthesis, number of nodes and inter-nodal distance, cane diameter, and stronger network of root band to prevent lodging as portrayed in Figure 2) of sugarcane for imparting resilience against weather shifts and shortening of frost-free periods (Enriquez et al., 2000; Mohan, 2016; Nerkar et al., 2018). Figure 2 illustrates prominent morphological and anatomical features of sugarcane plant that have remained the focus of modern breeding and genome editing efforts. The increase in number of nodes (distinct joints on which leaves, buds, and branches emerge) and intermodal distance, improved leaf sheath area, and the higher number of leaves, nodes, and buds per plant resulted in lesser disease attack and herbicide tolerance and in greater light, water, and nutrient absorption, conversion, and use efficiencies in sugarcane (Enríquez-Obregón et al., 1998; Gilbert et al., 2005; Tiwari et al., 2010; Gentile et al., 2015; Mohan, 2017; Oz et al., 2021). Likewise, genetic improvement of anatomical traits especially root primordia (embryonic structures that give rise to roots) and vascular bundles (complex tissues called xylem and phloem, which are the channels for transportation of water, nutrients, and sugars) tends to increase growth, yield attributes, and cane yield. Therefore, it is suggested that these improved traits of sugarcane hold bright perspectives to utilize genome editing techniques for boosting the morphological traits especially higher leaf area and plant height to promote photosynthesis efficiency for producing greater biomass, higher stem diameter, and extended root band to prevent lodging of non-cultivated grasses as well.
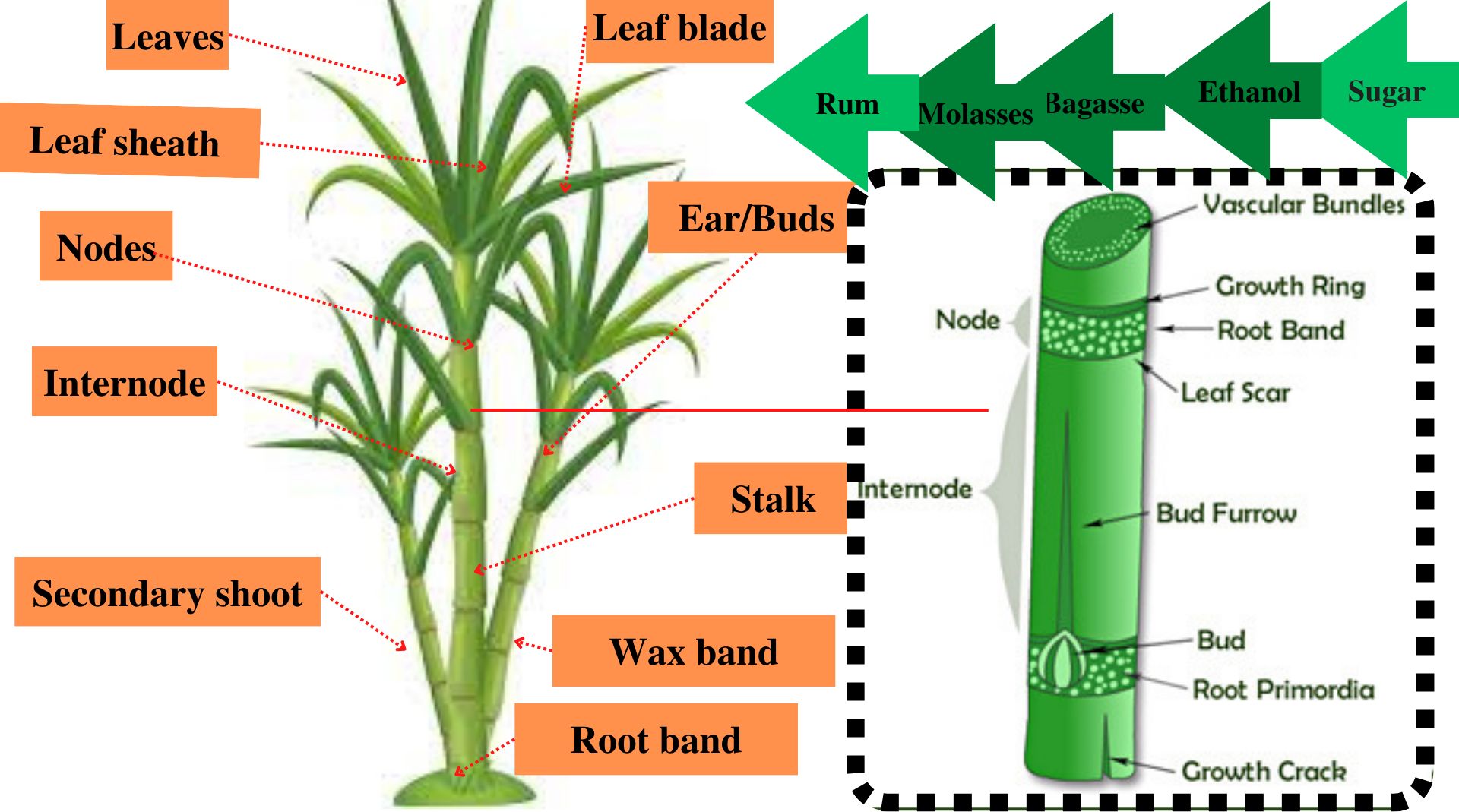
Figure 2 Prominent morphological features (leaf attributes including number of leaves, leaf sheath, and blade thickness along with number of nodes and inter-nodal distance) of the sugarcane plant and the anatomical features of cane/stalk (root primordia, which give rise to the root system, vascular tissues developing in xylem and phloem for transportation of water, nutrients and sugars in a source–sink relationship, leaf scar that serves as a prime feature for cultivar identification in the absence of leaves, etc.) that have been focused on in modern breeding and genetic improvement efforts along with different by-products (molasses, ethanol, bagasse, etc.) prepared directly from sugarcane.
4 Gene editing tools and the CRISPR/CAS9 protocol for genetic improvement of major cultivated grasses
Different gene editing tools such as mitochondrial genome editing, anti-sense transcription, and zinc-finger nuclease techniques have been previously employed to acquire the desired traits in sugarcane (Figure 3). Moreover, other genetic tools such as site-specific recombinase, base editing (Yin et al., 2015; Zong et al., 2017), and transcription activator-like effector nucleases (TALENs) have also been employed for gene’s insertion and/or knocking (Jung and Altpeter, 2016; Kannan et al., 2018) in order to acquire desired morphological traits and improve cane yield, sucrose recovery, etc., but these have demonstrated limited efficacy owing to off-targeting (Peng et al., 2015; Ma et al., 2016; Zaidi et al., 2017). This situation necessitated the development of more advanced genetic tools such as the CRISPR technique (Aitken and McNeil, 2010; Mao et al., 2013; Shan et al., 2018; Hussin et al., 2022) for the gene editing of sugarcane.
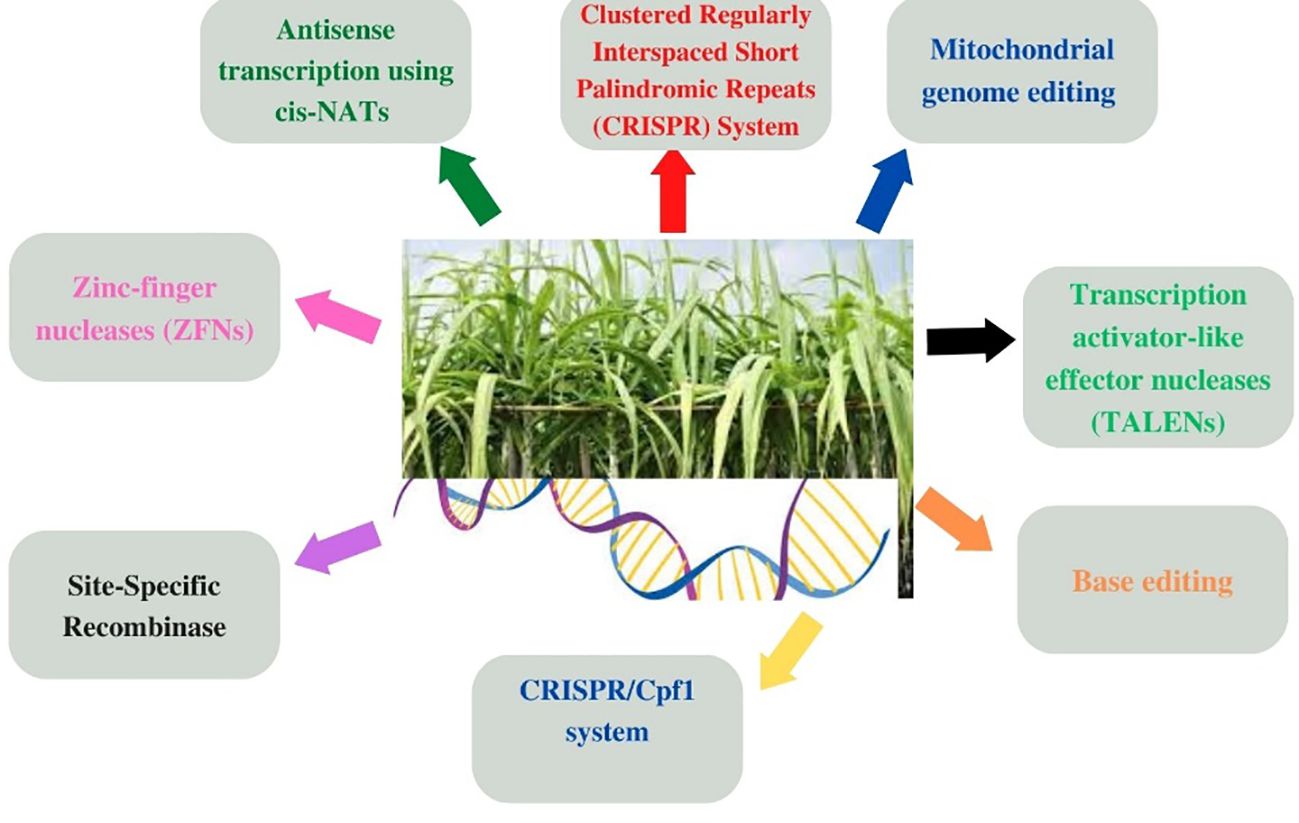
Figure 3 Genetic engineering approaches involving different gene editing techniques used for the genetic improvement of major cultivated grasses (wheat, maize, rice, sugarcane, barley, etc.) by acquiring desired morphological traits as depicted in Figure 2 through gene insertion and/or knocking them out for inducing genetic manipulation and transformation.
Originally, CRISPR/Cas9 was discovered in bacteria and archaea immune systems having a role in detecting and subsequently degrading the invasive DNA from bacteriophages and plasmids (Peng et al., 2015; Ma et al., 2016). Recently, CRISPR has been developed as a revolutionary gene editing technique that performs precise modification of DNA within the host’s genome (Hussin et al., 2022; Riaz et al., 2022; Krishna et al., 2023). The Cas9 part refers to the CRISPR associated protein 9, which serves as molecular scissors. It encompasses two regions, namely, the recognition (REC) lobe and the nuclease (NUC) lobe. Additionally, the REC lobe contains two multi-helix domains that are called REC1 and REC2, which are essential to bind with both guide RNA and target DNA (Xing et al., 2014; Ren et al., 2021). Moreover, REC1 contains α-helical structures of 25 α-helices and 2 β-sheets, while in contrast, REC2 is composed of six-helix structures and gets embedded within the REC1 domain. Similarly, NUC lobe entails three domains called RuvC, HNH, and PAM (protospacer adjacent motif) interacting domains. To cut the DNA’s double strands, firstly, REC lobe triggers sgRNA and DNA binding, whereas the RuvC and HNH domains facilitate to precisely cut target DNA’s complementary as well as non-complementary strands. Another vital component of the system is guide RNA, which is composed of two elements, namely, CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) (Gasparis et al., 2018; Chen et al., 2019). Interestingly, the crRNA is an 18- to 20-base-pair-long sequence that recognizes, specifies, and ensures binding with the target DNA (Barrangou et al., 2007), whereas the tracrRNA (a twisted structure) tends to bind the scaffold for Cas9 nuclease. However, the tracrRNA sequence must be partially complementary with one of the crRNA segment (Ren et al., 2021).
There are different steps involved in the CRISPR/Cas9 working protocol (Mao et al., 2013; Osakabe et al., 2016). The first step involves designing a single guide RNA (sgRNA), which is a synthetic RNA molecule that is compatible with the target DNA sequence. The sgRNA has a vital function as it locates the specific gene or region of interest within the genome of the host organism. The next step is target recognition, whereby sgRNA gets associated with the Cas9 protein (Xing et al., 2014). The resulting complex serves as a pair of molecular scissors that gets triggered for searching the target DNA sequence within the genome of target host. Thereafter, DNA cleavage occurs by the Cas9 protein that induces a break in the DNA at the precise location (Svitashev et al., 2015; Galli et al., 2022). This DNA cleavage tends to trigger the natural repair mechanisms within the cell, which attempts to repair the break with the help of either homology directed repair (HDR) or non-homologous end joining (NHEJ). Interestingly, the NHEJ holds potential to introduce small insertions or deletions, which leads to gene disruption (Ren et al., 2021). In contrast, the HDR provided with a repair template might allow the introduction of specific genetic modifications (Xing et al., 2014; Tang et al., 2017; Miao et al., 2018). For introducing breaks in the double strands, CRISPR needs PAM sequence in the target DNA adjacent to the protospacer complementary sequence, which is a short sequence (2–6 bp) and precedes by the sequence of targeted DNA. This constitutes a serious limitation in its design and has raised the need to develop variants of CRISPR tools having alternative PAM requisites (Xing et al., 2014; Lawrenson et al., 2021). Interestingly, the Cas9 nuclease from the type II CRISPR/Cas9 system of Streptococcus pyogenes is the most frequently used system that requires PAM sequence for DNA targeting and an NGG (N, any nucleotide; G, guanine) component (Jinek et al., 2012). Figure 4 illustrates the schematic working protocol of the CRISPR/Cas9 technique (starting from gene selection and designing of guided RNA and terminates with the growth of transgenic plants) for gene editing of sugarcane. Interestingly, the working efficacy of gene editing depends on two prime components involved in a typically engineered CRISPR/Cas9 system, a Cas (which is an endonuclease protein) and an sgRNA (that is basically a 20-nucleotide sequence) for guiding the Cas enzyme toward the target sequence in order to introduce double-stranded break (DSB) (Xing et al., 2014; Vlcko and Ohnoutkova, 2020).
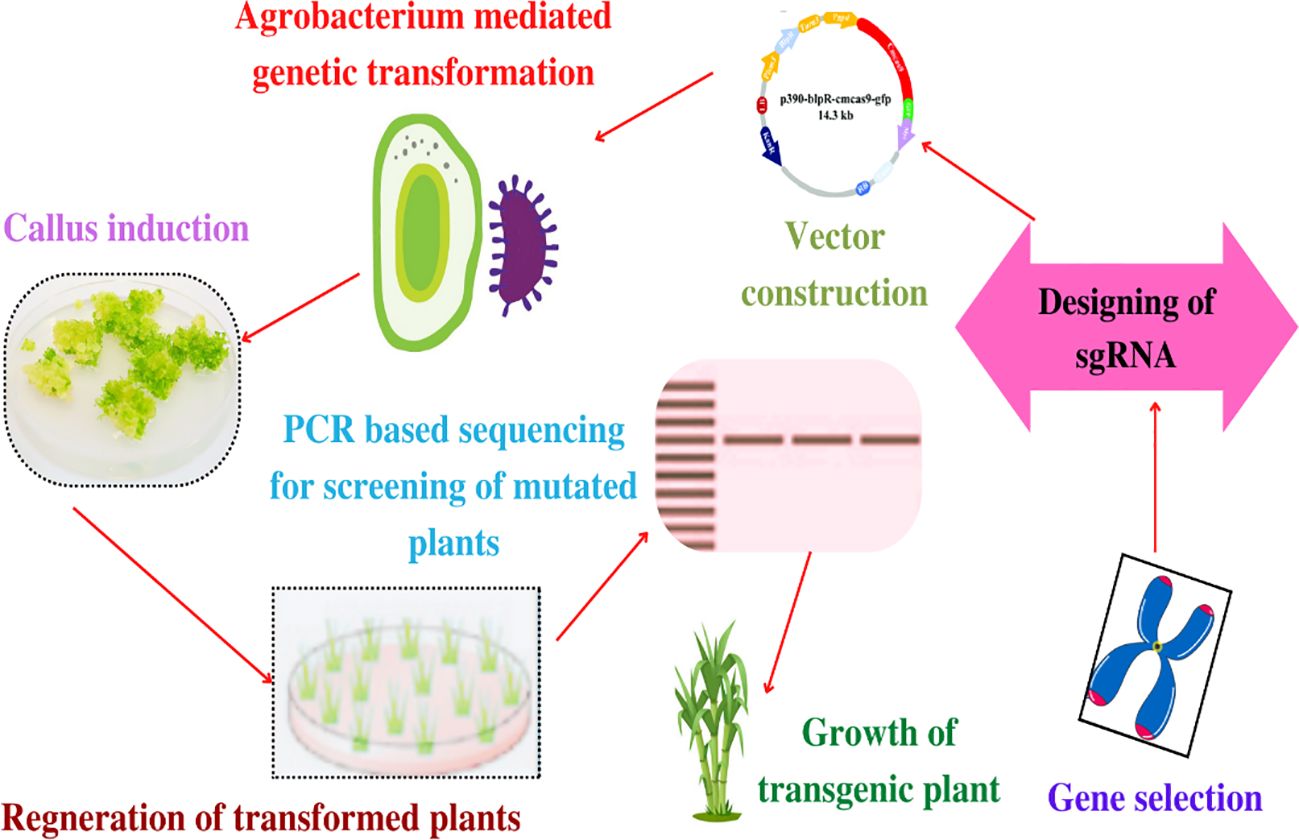
Figure 4 The schematic working protocol of the agrobacterium-mediated CRISPR/Cas9 technique (starting from gene selection and the design of guided RNA and terminating with the growth of transgenic plants after going through agrobacterium-mediated genetic transformation) for producing transgenic elite sugarcane cultivars having the desired agro-botanical and morphological traits.
The components of the CRISPR genetic system could be delivered into the target plant’s genome in the format of DNA, mRNA (in vitro transcripts or IVT), and proteins (Eid et al., 2021). The delivery techniques for CRISPR components include Agrobacterium-mediated infection, agro-infiltration, biolistics (also known as particle bombardment), electroporation, virus-mediated transformation, and PEG-based transformation, which is also referred to as protoplast-based transformation (Liang et al., 2014; Lin et al., 2014, Yin et al., 2015; Malnoy et al., 2016; Zaidi et al., 2017; Bhowmik et al., 2018). The RNP complex direct delivery has been reported to eliminate the risk of foreign DNA introduction into the genome of the host plants (Wolter and Puchta, 2017; 2018). Interestingly, pre-assembled RNP (Cas9-Grna) delivery was precisely attempted in cells (Cho et al., 2013). Later on, Cas9-gRNA RNPs have been successfully delivered into protoplasts by using the PEG-mediated delivery system that was derived from somatic tissues of tobacco, rice, petunia, grapevine, lettuce, apple, and potato Malnoy et al., 2016; Weeks, 2017). Recently, by using the biolistic bombardment protocol, Cas9-gRNA RNPs have also been delivered into maize and wheat embryo cells (Svitashev et al., 2015; Liang et al., 2016).
There are numerous generalized applications of the CRISPR/Cas9 technique such as gene editing with precise modification of specific genes (addition, deletion, or replacement of DNA sequences) (Westra et al., 2013; Manghwar et al., 2019; Milner et al., 2020). Disease modeling has emerged as another vital application of the CRISPR/Cas9 technique through the creation of model organisms with specific genetic mutations in order to diagnose the potential causes and develop feasible treatments (Kumar et al., 2018; Kim et al., 2022). Another interesting application of this technique is to study gene functions through selective disruption of genes and thereafter observing the resulting effects of altered genes. It is being used to develop genetically modified organisms (GMOs) having desired traits such as pest resistance (Kim et al., 2022). Table 2 illustrates different applications of this technique for the genetic improvement of cultivated grasses (wheat, rice, maize, barley, and sorghum). The increment in yield and quality of different cultivated grasses (wheat, maize, and sorghum) and imparting resistance against biotic and abiotic stresses have been achieved by employing this novel technique (Azevedo et al., 2011; Svitashev et al., 2015; Shimatani et al., 2017; Zong et al., 2017; Bhowmik et al., 2018; Holubová et al., 2018; Li et al., 2019). Such genetic improvements might be attained in non-cultivated grasses as well; however, these might not give desired results for non-cultivated grasses having intricate genetic makeup. Over time, multiple variants of Cas9 and gRNA have been developed (Nishimasu et al., 2018; Chen et al., 2019; Walton et al., 2020), which could hold bright perspectives in genome editing of non-cultivated grasses.
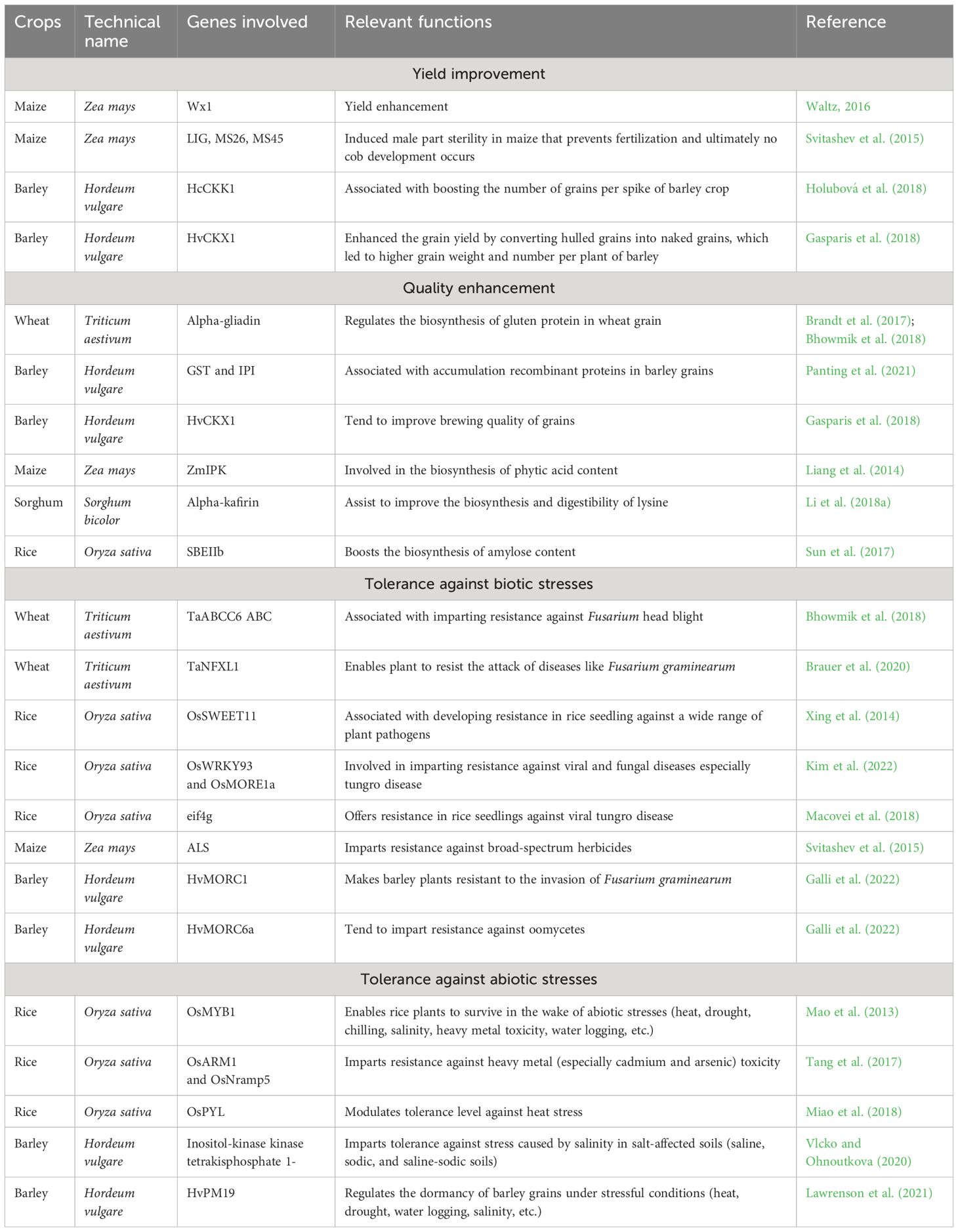
Table 2 Major cultivated grass improvement (yield and quality enhancement along with imparting tolerance against biotic and biotic stresses) using the CRISPR/Cas9 gene editing technique.
5 CRISPR (Cas9, Cas12a, Cas12b, and SpRY) variants
In the CRISPR gene editing system, the guide RNA’s protospacer motif tends to provide target specificity (Liang et al., 2014; Gasparis et al., 2018). However, compatible PAM sequence is a pre-requisite to trigger the cleavage of the targeted DNA region. Additionally, a GC-enriched site is required by PAM prototypical Cas9 derived from S. pyogenes (SpCas9), which reduces flexibility targeting. The PAM presence restrains potential site access, which results in off-targeting. Notwithstanding, Cas enzymes hold potential for target site recognition, which increases the flexibility of target sites (Nishimasu et al., 2018; Miller et al., 2011). Recently, numerous variants of endonuclease enzyme have been developed including Cas12a and Cas12b (Chen et al., 2019; Ming et al., 2020). However, akin to Cas9, these variants are not without PAM requirement and rely on PAM’s T enriched at the 5′-end in the form of TTTV. Recently, Walton et al. (2020) have reported overcoming this limitation through the development of the SpCas9 enzyme variant, which is a structure-guided engineered variant and referred to as SpRY. This newly developed variant holds potential to target the genomic DNA without requiring PAM and might be declared as nearly PAM-less variant. Thereafter, Ren et al. (2021) have reported that SpRY remained equally effective in rice by successfully targeting a large number of NNN PAM sites (NAN/NGN/NCN/NTN). Contrastingly, it was observed that Cas9 was unable to edit a number of relaxed PAM sites and was pronouncedly less efficient in comparison to SpRY for non-canonical PAM sites. Moreover, it was reported that SpRY induced larger deletions (five base pairs at relaxed PAM sites), which was impossible to achieve by using the Cas9 gene editing tool. Interestingly, the PAM requirement elimination induced self-editing in CRISPR-Cas T-DNA, which led to either inactivation or modification of sgRNA (Zong et al., 2017; Miller et al., 2011; Milner et al., 2020).
Likewise, the CRISPR-mediated genome editing tool for single base editing has also been applied in a variety of cultivated grasses. For instance, adenine and cytosine base editing has been effectively optimized in cultivated grasses like rice, wheat, and maize for base editing (Shimatani et al., 2017; Zong et al., 2017; Li et al., 2018). However, those were found inefficient owing to off-targeting effects while more research is needed to enhance the efficiency of base editing tools in monocots. Recently, in rice, SpRY-PmCDA1 (PAM-less C-to-T nucleotide editor) remained effective in converting a C-to-T base (Ren et al., 2021). Thus, it has been inferred that CRISPR-associated SpRY enzyme’s expanded target range might be further harnessed for base editing (nucleotide-level) with high accuracy. This can be achieved by using cytosine base editors at the relaxed PAM (first to sixth base of protospacer) of the SpRY. It was impossible to achieve this using the traditional C-to-T base editors owing to the peculiar distance requirement of editing windows (Manghwar et al., 2019). In contrast, the SpRY-based adenine base editor has demonstrated higher efficiency for A-to-G conversion by using fourth to eighth bases of the protospacer in the editing window (Ren et al., 2021). Hence, it might be inferred that by using SpRY-based editors, a comparatively hefty number of options regarding base edits have become available now. It is worth mentioning that in the CRISPR-based system, PAM tends to differentiate specific Cas enzyme non-self DNA sequences (Westra et al., 2013). The CRISPR tool having PAM-less targeting capacity could limit and restrict self-editing, which could be utilized for secondary off-targeting. The off-target in transgenic rice lines could be prevented by a self-targeting gRNA vector (Ren et al., 2021). These shortcomings compel further investigations pertaining to structural engineering for application in different systems such as single base editing using SpRYABEs (Walton et al., 2020).
6 CRISPR/Cas9 in sugarcane and potential application for non-cultivated grass improvement
The genome size of sugarcane has been estimated to be over 10 Gbp, wherein genes exist in 10–12 allelic forms. Interestingly, depending on a specific cultivar’s ploidy level, monoploid genome size has been estimated to be approximately 800–900 Mb (Zhang et al., 2012; de Setta et al., 2014; Hussin et al., 2022). Because of its high polyploidy (x = 10–13; 2n = 100–130), interspecific, heterozygous, and aneuploidy nature, the genome of sugarcane tends to decelerate the gene editing attempts intended for crop improvement (Le Cunff et al., 2008; de Setta et al., 2014; Oz et al., 2021). Moreover, modern elite cultivars of sugarcane exhibit high level of polyploidy and heterozygosity that necessitate the vegetative propagation of sugarcane in order to prevent allele loss and inhibit detrimental allele accumulation during the process of meiosis (Ali et al., 2019; Krishna et al., 2023). However, most of sugarcane’s parental clones lacking pollen fertility and flowering synchrony have been improved using genetic engineering approaches (Hoang et al., 2015). There have been continuous research efforts to genetically improve sugarcane for boosting cane yield and sucrose recovery (Tew and Cobill, 2008; Hamerli and Birch, 2011; Krishna et al., 2023).
Among major cultivated grasses, taking highly polyploidy sugarcane as a model crop might be a rational approach for genome editing of non-cultivated grasses due to the absence of mutagenesis in diploid grasses (Table 2). The functional redundancy in sugarcane is caused by homeologs and homologs that are present in a large number and restricted genome editing (Taparia et al., 2012; Weeks, 2017; Eid et al., 2021; Oz et al., 2021). However, co-mutated allele numbers are similar to RNAi, offering an unprecedented opportunity to produce a wide range of phenotypes (Eid and Mahfouz, 2016). The CRISPR variants have revolutionized the gene editing process and are being applied in various polyploidy crops including sugarcane for introducing precise genetic modifications with ultimate aims to improve yield, sucrose recovery, biofuel production, disease resistance, and abiotic stress tolerance (Azevedo et al., 2011; Taparia et al., 2012; Hoang et al., 2015; Ali et al., 2019). Figure 5 presents some prominent applications of CRISPR/Cas9 in sugarcane for precision gene editing to acquire the desired morpho-physiological traits. Thus, this technique holds bright perspectives to increase the biomass yield, nutritional quality, and tolerance against biotic and abiotic stresses in non-cultivated grass species through precise screening and targeting of desired genes for acquiring the desired traits.
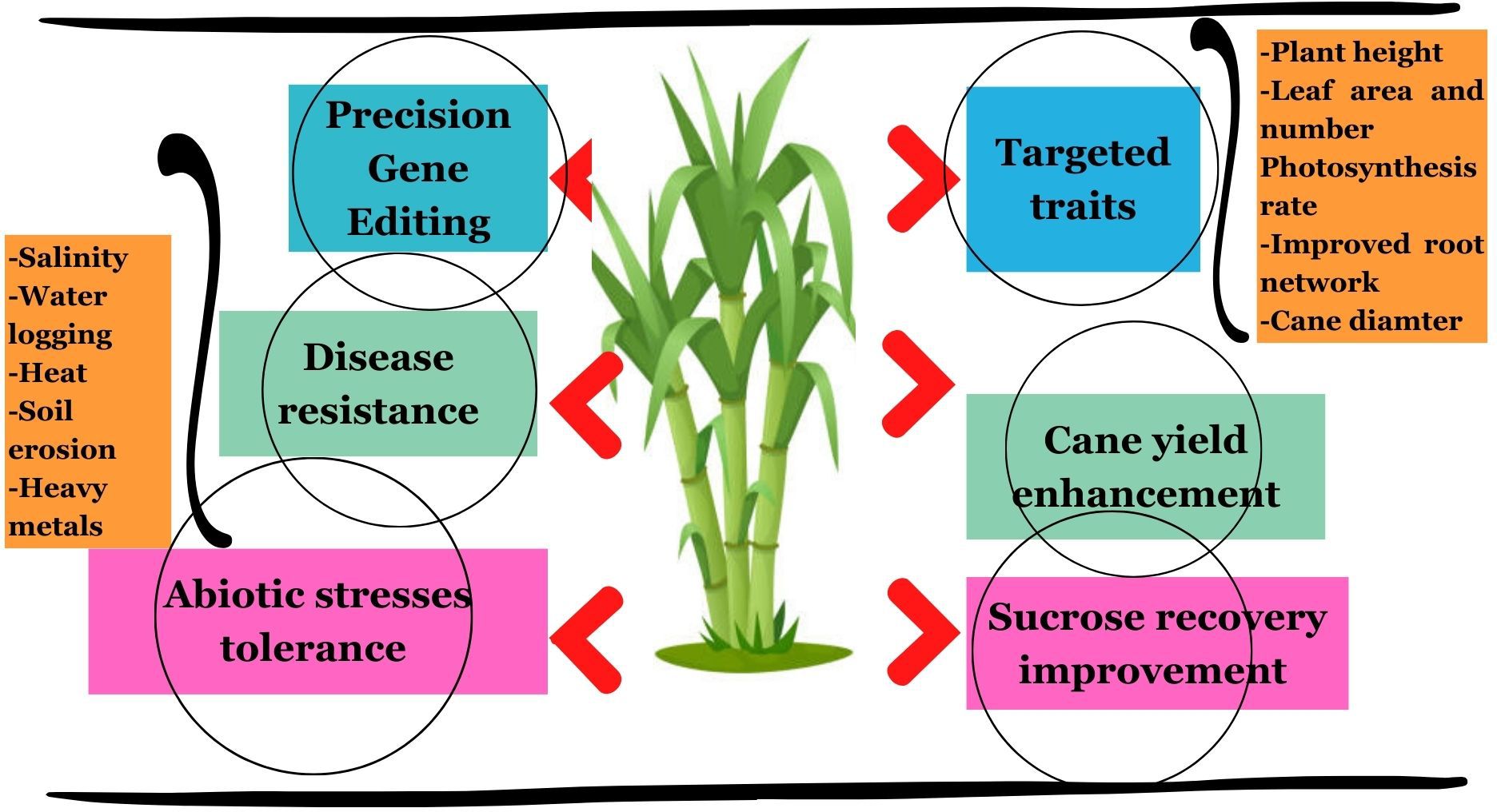
Figure 5 Prominent applications of CRISPR/Cas9 in sugarcane for precise gene editing to acquire the desired traits, especially improvement of morphological attributes (plant height, leaf number per plant, leaf area, cane diameter, etc.), cane yield, sucrose recovery percentage, and tolerance against abiotic stresses (salinity, heat, water logging, soil erosion, and heavy metal toxicity).
Among the specific applications of CRISPR/Cas9 in sugarcane, precise gene modification ranks top by using gRNA to target a specific DNA sequence and the Cas9 enzyme to cut the DNA (Eid and Mahfouz, 2016). In this way, it effectively helps acquire the desired traits such as increased sugar content and resistance against diseases (smut, rust, rot, etc.) (Hoy, 1994; Comstock, 2002; Fitch et al., 2001; Viswanathan and Rao, 2011; Hussain et al., 2018) and abiotic stresses (especially drought, heat, salinity, water logging, heavy metal toxicity, etc.) in an environmentally friendly manner (Gomathi et al., 2015; Tiwari and Lata, 2018; Baig et al., 2020; Rehman et al., 2021; Riyazuddin et al., 2022). Most importantly, this technique in sugarcane has been utilized for developing high-yielding cultivars that require fewer inputs including water, fertilizers, pesticides, etc. that might contribute to impart sustainability to modern intensive sugarcane farming systems. The CRISPR/Cas9 technique deployed in sugarcane as a part of broader efforts in agricultural biotechnology might prove vital in addressing the global challenges of food security, environmental sustainability, and crop resilience (Patade et al., 2008; Sengar et al., 2013; Meena et al., 2020).
Recently, Eid et al. (2021) inferred that the CRISPR/Cas9 technique remained effective in producing a rapidly scorable phenotype in highly polyploid sugarcane through multiallelic, targeted mutagenesis of magnesium chelatase. Likewise, this technique performed precise genome modifications in many elite varieties through bypassing the adverse meiosis in sugarcane (Weeks, 2017) and the same could be repeated in non-cultivated grasses of economic pertinence such as wild sugarcane, Bermuda grass, and Chinese silver grass. Likewise, Oz et al. (2021) reported that efficient and reproducible gene targeting in sugarcane was possible by enabling precise co-editing of multiple alleles via template-mediated and homology-directed repair of DNA double-strand breaks induced by the programmable nuclease CRISPR/Cas9 technique. Ultimately, the co-editing of three acetolactate synthase alleles that could confer herbicide tolerance was confirmed by Sanger sequencing through PCR amplicons. Thus, the CRISPR/Cas9 technique holds potential to precisely target non-cultivated grass species genome for creating tolerance against broad-spectrum herbicides especially in grassland areas adjacent to cultivated lands. It was also inferred that through the comparison of different quantities, delivery of the repair template suggested that exogenously supplied DNA’s excessive quantities might adversely impact the repair process in sugarcane. In addition, Azevedo et al. (2011) opined that CC has asserted extreme pressure on high water-demanding crops like sugarcane, while drought and heat stresses tend to reduce cane yield and sucrose recovery, while CRISPR/Cas9 might be utilized to impart tolerance against terminal heat stress and drought (Meena et al., 2020). This technique could be employed to precisely target the genome of non-cultivated grasses for improving their tolerance against heat and drought stresses. Patade et al. (2008) reported that drought and heat stresses result in salinity owing to higher volatilization from soil surface that causes salt accumulation and, resultantly, sugarcane growth; cane yield and sucrose content were significantly decreased. However, the CRISPR/Cas9 tool holds immense potential to produce elite genotypes of sugarcane having the potential to thrive well on salt-affected soils through precise mutagenesis in sugarcane (Sengar et al., 2013). Soil salinity tolerance in non-cultivated grasses might revolutionize the grassland conservation and development initiatives, leading to ensuring food security and poverty alleviation on a wide scale. Similar results have been reported for sugarcane gene editing for imparting tolerance against other abiotic stresses including water-logging, cold or chilling stress, and heavy metal toxicity using a precise genome editing technique like CRISR/Cas9 (Tiwari et al., 2010; Rehman et al., 2021; Riyazuddin et al., 2022).
Besides abiotic stresses, CRSISPR/Cas9 holds bright perspectives in producing elite genotypes of sugarcane having immense tolerance against biotic stresses. Numerous biotic stresses including weeds, diseases, and a wide range of insects have posed a serious challenge to sugarcane production as per their varietal potential (Hussain et al., 2018). Viswanathan and Rao (2011) reported the precise application of this technique for imparting tolerance against the fungal diseases of sugarcane such as wilt (the causative agent is Fusarium sacchari) and smut (caused by Sporisorium scitamineum) and red rot caused by Colletotrichum falcatum. The same goes for bacterial diseases including ratoon stunting and leaf scald along with sugarcane yellow leaf virus, which cause significant losses in sugarcane (Hoy, 1994; Fitch et al., 2001; Comstock, 2002). Table 3 indicates different candidate genes identified through the CRISPR/Cas9 technique to impart tolerance against biotic and abiotic stresses. These successes might be utilized to initiate genetic improvement of non-cultivated grasses for imparting tolerance against viral, bacterial, and fungal diseases.
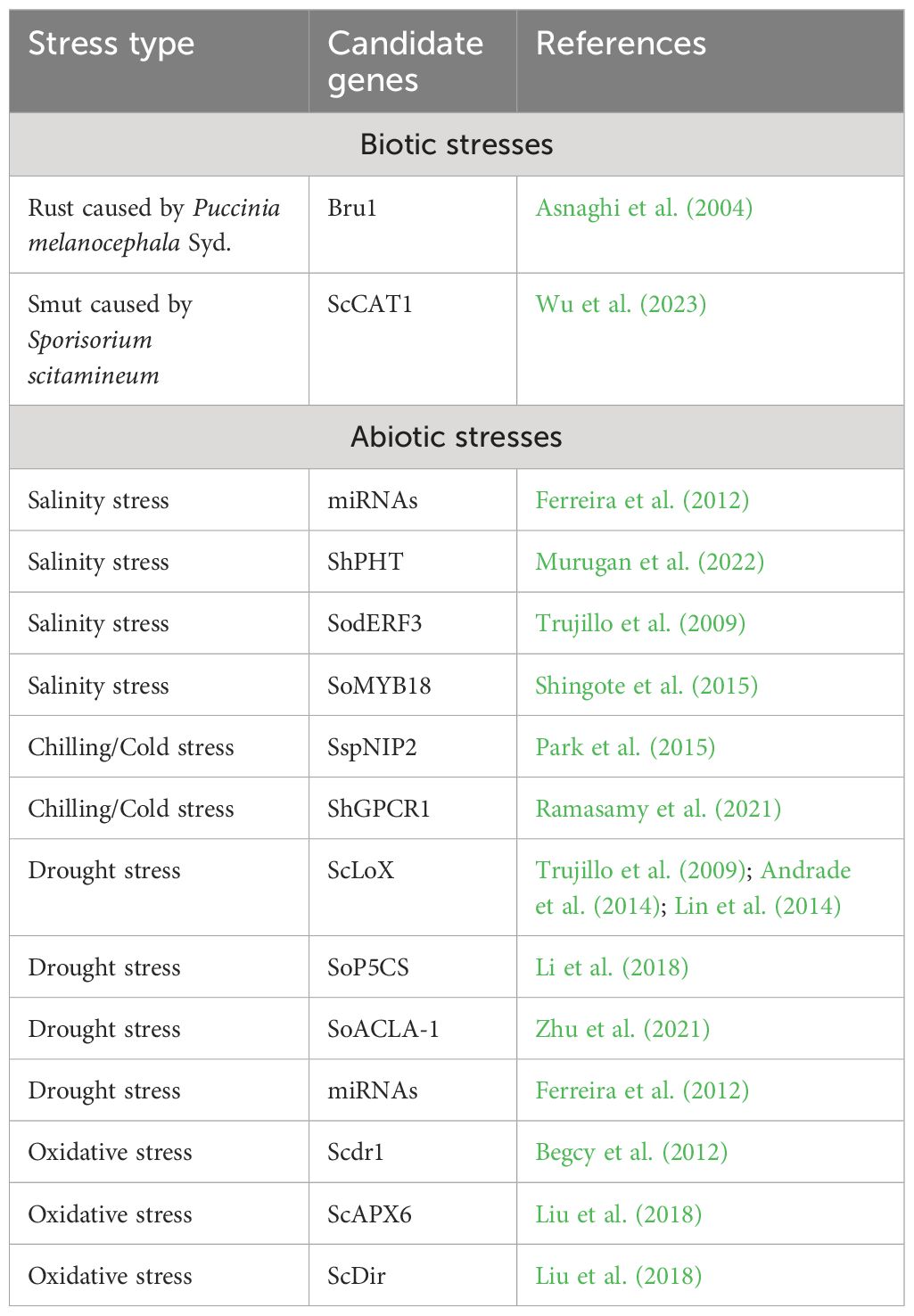
Table 3 Different candidate genes identified for imparting tolerance against rust and smut diseases along with abiotic stresses (drought, salinity, cold, or chilling stress and oxidative stress) in sugarcane through precise genome editing using the CRISPR/Cas9 technique.
7 Limitation of CRISPR/Cas9 for non-cultivated grass improvement and future perspectives
Recently, it has become evident that CRISPR/Cas9 has offered unique efficiency with unmatched precision in gene editing of polyploidy crops like sugarcane (Bhowmik et al., 2018; Hussin et al., 2022); however, its application for non-cultivated grasses’ genetic improvement might raise few technical, ethical, and safety concerns. The key technical limitations of CRISPR/Cas9 might include challenges like achieving 100% precision in gene editing (Ali et al., 2015; Zhang et al., 2019), because few cells could avoid desired genetic modifications leading to low precision in non-cultivated grass species. This could become a serious limitation in cases where high accuracy is crucial and highly desired and the same could be a serious challenge in case of grass mutagenesis. Another limitation might be off-target effects as the Cas9 protein could bind and cleave the target DNA at unintended locations (Wang et al., 2014; Baltes et al., 2015), leading to undesired genetic changes, which, in turn, lead to potentially harmful consequences in terms of biomass production and nutritional value of grass species. For gene editing in polyploidy crops including tetraploid cotton (Gossypium hirsutum), hexaploid wheat (Triticum aestivum), and sugarcane, mutations generally occur in homoeoallele subsets targeted by the same sgRNA (Wang et al., 2017; Zhang et al., 2019). Furthermore, polyploidy due to Mendelian genetics makes transmission and stacking of first-generation mutations harder and even impossible. In addition, one of the prime limitation of the CRISPR/Cas9 technique could be CRISPR/Cas9 component delivery (Hussin et al., 2022) into the target genome of grass species, which would seriously compromise the efficacy and accuracy of the whole gene editing process, while serious research efforts could be required to optimize the delivery system of CRISPR for polyploidy grasses. Likewise, delivery of CRISPR/Cas9 components selectively to specific cell types within a complex genome of host grasses might remain a daunting challenge as that of sugarcane.
Additionally, insertion of large DNA sequences continues to remain one of the pronounced challenges (Liang et al., 2016; Gao et al., 2020; Eid et al., 2021; Oz et al., 2021), which decreases its ability to add large regulatory elements into the targeted genome of non-cultivated grasses. Moreover, one of the limitations of the CRISPR/Cas9 technique is mosaicism (Yin et al., 2015) whereby this technique could induce non-uniform genetic modification in different cells of non-cultivated grasses; thus, before employing this technique for grass species, optimization of the CRISPR system might be required. Although not in sugarcane, but the immune system of many hosts has responded negatively to CRISPR/Cas9 components (Baltes et al., 2015), limiting the efficacy of the gene editing process, and the same could be happen in the case of a few non-cultivated grass species. Presently, gene editing by using CRSPR/Cas9 has assisted cultivated grass improvement by facilitating precise knock-in, triggering accurate knockout and desired replacement, and promoting planned point mutations and gene fine-tuning (Svitashev et al., 2015; Tang et al., 2017; Li et al., 2018; Miao et al., 2018; Gao et al., 2020). For non-cultivated grass improvement, the potential development of the CRISPR method to facilitate on-target editing and circumvent the vector self-editing that reduces off-targeting may be further explored. Although Cas9 is incapable to act in a PAM-less editing mode, its accuracy and efficiency have remained far better than SpRY, which highlights Cas9’s unterminated pertinence for genetic modification of grass species (Ren et al., 2021). Recently, SpRY has been developed as a more precise choice for exploring the genome of crop plants especially its application in rice as proved by its efficacy in terms of unconstrained targeting using PAM-less editing (Tang et al., 2017; Macovei et al., 2018). The application of Cas9 and SpRY is bound to inspire numerous exciting investigations including in vivo directed evolution for acquiring desired characteristics that bolster plant establishment against biotic and abiotic stresses under changing climate scenario.
Furthermore, there remain few ethical and social concerns regarding the potential application of the CRISPR/Cas9 technique in creating GMO crops (Malnoy et al., 2016; Zaidi et al., 2017). Previously, CRISPR/Cas9 use in sugarcane has also raised ethical and regulatory considerations (Ostengo et al., 2022), and the same could be expected for non-cultivated grass species as well. Different countries may not have regulations regarding the genetically modified grass species, which could delay the initiation and execution of genome editing programs. Furthermore, application of this technique could prompt the need to devise robust regulatory frameworks to ensure its responsible use and to avoid the potential unintended consequences of genome editing non-cultivated grasses. Despite unprecedented opportunities offered by the CRISPR/Cas9 technique regarding the precise genome editing, these limitations must be given due consideration before considering this technique for genetic improvement of non-cultivated grasses.
8 Conclusions
Owing to CC, global warming, rapidly increasing human population, and decreasing agricultural land area, it is about time to initiate out-of-the-box conservation strategies for grasslands. This goal could be effectively achieved through genetic improvement of native grass species in order to diversify and multiply their ecosystem services. Gene editing techniques might be utilized to genetically improve native grasses based on the pattern of cultivated grasses like sugarcane. Among the recent genetic techniques employed in sugarcane, CRISPR/Cas9 has emerged with an immense potential to precisely modify the specific genes in the target host’s genome with unprecedented accuracy and efficiency. This technique has produced marvelous results in sugarcane gene editing for acquiring desired traits like higher cane yield, sucrose recovery, and tolerance against biotic and abiotic stresses, and the same might be utilized for grass species of grasslands. Future research must strive to attain abiotic stress tolerance in non-cultivated grass species using the CRISPR/Cas9 technique and other desired characteristics including higher biomass productivity, regrowth capacity, nutritional quality (especially higher protein and digestibility and lower fiber content) of grasses for consumption as forage for ruminants, biofuel production potential, and flowering capacity. After genetic improvement, one of the vital aspects would be the introduction of new seeds into the grasslands that can be economically achieved through over-seeding. However, unlike cultivated grasses, future genome editing research has to face novel challenges like gene delivery issue, off-targeting, and limited efficacy of gene editing procedures, but such research struggles are bound to open new frontiers of genome editing of non-cultivated grasses, which might contribute to ensuring food security in the future.
Author contributions
CL: Conceptualization, Writing – original draft, Writing – review & editing, Methodology, Project administration. MAI: Conceptualization, Writing – original draft, Writing – review & editing, Investigation, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Key R&D Program of China (2022YFD2301100), Yunnan Science and Technology Talent and Platform Program (202205AM070001), and Yunnan Intelligence Union Program (202103AM140033).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, R. N., Iqbal, A., Iqbal, M. A., Ali, O. M., Ahmed, R., Ijaz, R., et al. (2021). Weed-Free durations and fertilization regimes boost nutrient uptake and paddy yield of direct-seeded fine rice (Oryza sativa L.). Agronomy 11, 2448. doi: 10.3390/agronomy11122448
Abbas, S., Qamer, F. M., Murthy, M. S. R., Tripathi, N. K., Ning, W., Sharma, E., et al. (2015). Grassland growth in response to climate variability in the upper Indus Basin, Pakistan. Climate 3, 697–714. doi: 10.3390/cli3030697
Afzal, S., Sirohi, P., Singh, N. K. (2020). A review of crispr associated genome engineering: application, advances and future prospects of genome targeting tool for crop improvement. Biotechnol. Lett. 42, 1611–1632. doi: 10.1007/s10529-020-02950-w
Aitken, K., McNeil, M. (2010). Diversity analysis. In Genetics, Genomics and Breeding of Sugarcane (Boca Raton, FL, USA: CRC Press), 19–42.
Ali, A., Khan, M., Sharif, R., Mujtaba, M., Gao, S.-J. (2019). Sugarcane omics: an update on the current status of research and crop improvement. Plants 8, 344. doi: 10.3390/plants8090344
Ali, Z., Abul-faraj, A., Li, L., Ghosh, N., Piatek, M., Mahjoub, A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/cas9 system. Mol. Plant 8, 1288–1291. doi: 10.1016/j.molp.2015.02.011
Andrade, L. M., Benatti, T. R., Nobile, P. M., Goldman, M. H., Figueira, A., Marin, A. L. A., et al. (2014). Characterization, isolation and cloning of sugarcane genes related to drought stress. BMC Proc. BioMed. Cent. 8, 110. doi: 10.1186/1753-6561-8-S4-P110
Asnaghi, C., Roques, D., Ruffel, S., Kaye, C., Hoarau, J.-Y., Telismart, H., et al. (2004). Targeted mapping of a sugarcane rust resistance gene (Bru 1) using bulked segregant analysis and AFLP markers. Theor. Appl. Genet. 108, 759–764. doi: 10.1007/s00122-003-1487-6
Azevedo, R. A., Carvalho, R. F., Cia, M. C., Gratão, P. L. (2011). Sugarcane under pressure: An overview of biochemical and physiological studies of abiotic stress. Trop. Plant Biol. 4, 42–51. doi: 10.1007/s12042-011-9067-4
Baig, M. A., Qamar, S., Ali, A. A., Ahmad, J., Qureshi, M. I. (2020). “Heavy metal toxicity and tolerance in crop plants,” in Contaminants in Agriculture: Sources, Impacts and Management. Eds. Naeem, M., Ansari, A. A., Gill, S. S. (Springer, Cham, Switzerland), 201–216.
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1, 15145. doi: 10.1038/nplants.2015.145
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). Crispr provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Begcy, K., Mariano, E. D., Gentile, A., Lembke, C. G., Zingaretti, S. M., Souza, G. M., et al. (2012). A novel stress-induced sugarcane gene confers tolerance to drought, salt and oxidative stress in transgenic tobacco plants. PloS One 7, e44697. doi: 10.1371/journal.pone.0044697
Bhowmik, P., Ellison, E., Polley, B., Bollina, V., Kulkarni, M., Ghanbarnia, K., et al. (2018). Targeted mutagenesis in wheat microspores using CRISPR/cas9. Sci. Rep. 8, 6502. doi: 10.1038/s41598-018-24690-8
Brandt, K. M., Gunn, H., Buschke, B. L., Heesacker, A., Moretti, N., Karasev, A., et al. (2017). Testing Non-transgenic Crispr Technology for Wheat Improvement. Austria: Presentation. 13th IWGS–Tulln. Oregon State University Press, USA.
Brauer, E. K., Balcerzak, M., Rocheleau, H., Leung, W., Schernthaner, J., Subramaniam, R., et al. (2020). Genome editing of a deoxynivalenol- induced transcription factor confers resistance to fusarium graminearum in wheat. Mpmi 33, 553–560. doi: 10.1094/MPMI-11-19-0332-R
Budeguer, F., Enrique, R., Perera, M. F., Racedo, J., Castagnaro, A. P., Noguera, A. S., et al. (2021). Genetic transformation of sugarcane, current status and future prospects. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.768609
Byrt, C. S., Grof, C. P., Furbank, R. T. (2011). C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 53, 120–135. doi: 10.1111/j.1744-7909.2010.01023.x
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). Crispr/cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Cho, S. W., Lee, J., Carroll, D., Kim, J. S., Lee, J. (2013). Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics 195, 1177–1180. doi: 10.1534/genetics.113.155853
de Setta, N., Monteiro-Vitorello, C. B., Metcalfe, C. J., Cruz, G. M. Q., Del Bem, L. E., Vicentini, R., et al. (2014). Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15, 540. doi: 10.1186/1471-2164-15-540
Di, H. J., Cameron, K. C., Podolyan, A., Robinson, A. (2014). Effect of soil moisture status and a nitrification inhibitor, dicyandiamide, on ammonia oxidizer and denitrifier growth and nitrous oxide emissions in a grassland soil. Soil Biol. Biochem. 73, 59–68. doi: 10.1016/j.soilbio.2014.02.011
Dong, S. K., Zhang, J., Li, Y. Y., Liu, S. L., Dong, Q. M., Zhou, H. K., et al. (2019). Effect of grassland degradation on aggregate-associated soil organic carbon of alpine grassland ecosystems in Qinghai-Tibetan Plateau. Eur. J. Soil Sci. 71 (1). doi: 10.1111/ejss.12835
Eid, A., Mahfouz, M. M. (2016). Genome editing: the road of CRISPR/Cas9 from bench to clinic. Exp. Mol. Med. 48, e265. doi: 10.1038/emm.2016.111
Eid, A., Mohan, C., Sanchez, S., Wang, D., Altpeter, F. (2021). Multiallelic, targeted mutagenesis of magnesium chelatase with CRISPR/cas9 provides a rapidly scorable phenotype in highly polyploid sugarcane. Front. Genome Ed. 3. doi: 10.3389/fgeed.2021.654996
Enriquez, G. A., Trujillo, L. E., Menendez, C., Vazquez, R. I., Tiel, K., Dafhnis, F., et al. (2000). “Sugarcane (Saccharun hybrid) genetic transformation mediated by Agrobacterium tumefaciens: production of transgenic plants expressing proteins with agronomic and industrial value,” in Plant Genetic Engineering: Towards the Third Millenium. Ed. Arencibia, A. D. (Elsevier, Amsterdam), 76–81.
Enríquez-Obregón, G. A., Vázquez-Padrón, R. I., Prieto-Samsonov, D. L., Gustavo, A., Selman-Housein, G. (1998). Herbicide-resistant sugarcane (Saccharum officinarum L.) plants by Agrobacterium-mediated transformation. Planta 206, 20–27.
Ferreira, T. H., Gentile, A., Vilela, R. D., Costa, G. G. L., Dias, L. I., Endres, L., et al. (2012). microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp. ). PloS One 7, e46703. doi: 10.1371/journal.pone.0046703
Fitch, M., Lehrer, A. T., Komor, E., Moore, P. H. (2001). Elimination of Sugarcane yellow leaf virus from infected sugarcane plants by meristem tip culture visualized by tissue blot immunoassay. Plant Pathol. 50, 676–680. doi: 10.1046/j.1365-3059.2001.00639.x
Galli, M., Martiny, E., Imani, J., Kumar, N., Koch, A., Steinbrenner, J., et al. (2022). CRISPR/sp cas9-mediated double knockout of barley microrchidia MORC1 and MORC6a reveals their strong involvement in plant immunity, transcriptional gene silencing and plant growth. Plant Biotechnol. J. 20, 89–102. doi: 10.1111/pbi.13697
Gao, H., Mutti, J., Young, J. K., Yang, M., Schroder, M., Lenderts, B., et al. (2020). Complex trait loci in maize enabled by CRISPR-cas9 mediated gene insertion. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00535
Gasparis, S., Kała, M., Przyborowski, M., Łyżnik, L. A., Orczyk, W., Nadolska- Orczyk, A. (2018). A simple and efficient crispr/cas9 platform for induction of single and multiple, heritable mutations in barley (Hordeum vulgare L.). Plant Methods 14, 111. doi: 10.1186/s13007-018-0382-8
Gentile, A., Dias, L. I., Mattos, R. S., Ferreira, T. H., Menossi, M. (2015). MicroRNAs and drought responses in sugarcane. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00058
Gilbert, R. A., Gallo-Meagher, M., Comstock, J. C., Miller, J. D., Jain, M., Abouzid, A. (2005). Agronomic evaluation of sugarcane lines transformed for resistance to sugarcane mosaic virus strain E. Crop Sci. 45, 2060–2067. doi: 10.2135/cropsci2004.0771
Gomathi, R., Gururaja Rao, P., Chandran, K., Selvi, A. (2015). Adaptive responses of sugarcane to waterlogging stress: An over view. Sugar Tech 17, 325–338. doi: 10.1007/s12355-014-0319-0
Hamerli, D., Birch, R. G. (2011). Transgenic expression of trehalulose synthase results in high concentrations of the sucrose isomer trehalulose in mature stems of field-grown sugarcane. Plant Biotechnol. J. 9, 32–37. doi: 10.1111/j.1467-7652.2010.00528.x
Hoang, N. V., Furtado, A., Botha, F. C., Simmons, B. A., Henry, R. J. (2015). Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front. Bioeng. Biotechnol. 3. doi: 10.3389/fbioe.2015.00182
Holubová, K., Hensel, G., Vojta, P., Tarkowski, P., Bergougnoux, V., Galuszka, P. (2018). Modification of barley plant productivity through regulation of cytokinin content by reverse-genetics approaches. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01676
Hoy, J. (1994). Sugarcane leaf scald distribution, symptomatology. Plant Dis. 78, 1083–1087. doi: 10.1094/PD-78-1083
Hussain, S., Khaliq, A., Mehmood, U., Qadir, T., Saqib, M., Iqbal, M. A., et al. (2018). “Sugarcane production under changing climate: Effects of environmental vulnerabilities on sugarcane diseases, insects and weeds,” in Sugarcane Production-Agronomic, Scientific and Industrial Perspectives (IntechOpen, Rijeka, Croatia), 1–17.
Hussin, S. H., Liu, X., Li, C., Diaby, M., Jatoi, G. H., Ahmed, R., et al. (2022). An Updated Overview on Insights into Sugarcane Genome Editing via CRISPR/Cas9 for Sustainable Production. Sustainability 14, 12285. doi: 10.3390/su141912285
Ijaz, R., Harun, N., Iqbal, M. (2023). “Grasslands Development for Ecotourism: Aesthetic Perspectives,” in Grasslands-Conservation and Development. Ed. Iqbal, M. A. (Intech open Ltd, London). doi: 10.5772/intechopen.112588
Iqbal, M. A. (2022). “Grasslands Development: Green Ecological Economy and ecosystem services Perspectives,” in Grasses and Grasslands: New Perspectives. Ed. Iqbal, M. A. (Intech Open, UK).
Iqbal, M. A., Iqbal, A. (2014). Sugarcane production, economics and industry in Pakistan. American-Eurasian J. Agric. Environ. Sci. 14, 1470–1477. doi: 10.5829/idosi.aejaes.2014.14.12.12479
Iqbal, M. A., Iqbal, A., Ali, K., Ali, H., Khan, R. D., Ahmad, B., et al. (2015). Integration of forage sorghum and by-products of sugarcane and sugar beet industries for ruminant nutrition: A review. Glob. Vet. 14, 752–760.
Iqbal, M. A., Khalid, S., Raees, A., Khan, M. Z., Nagina, R., Raina, I., et al. (2022). “Underutilized Grasses Production: New Evolving Perspectives,” in Grasses and grassland: New perspectives. Ed. Iqbal, M. A. (Intech open Ltd, London), 1–19. doi: 10.5772/intechopen
Iqbal, M. A., Saleem, A. M. (2014). Sugar beet potential to beat sugarcane as a sugar crop in Pakistan. American-Eurasian J. Agric. Environ. Sci. 15, 36–44. doi: 10.5829/idosi.aejaes.2015.15.1.12480
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-rna-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jung, J. H., Altpeter, F. (2016). TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 92, 131–142. doi: 10.1007/s11103-016-0499-y
Kannan, B., Jung, J. H., Moxley, G. W., Lee, S. M., Altpeter, F. (2018). TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16, 856–866. doi: 10.1111/pbi.12833
Kim, C. Y., Park, J. Y., Choi, G., Kim, S., Vo, K. T. X., Jeon, J. S., et al. (2022). A rice gene encoding glycosyl hydrolase plays contrasting roles in immunity depending on the type of pathogens. Mol. Plant Pathol. 23, 400–416. doi: 10.1111/mpp.13167
Ko, J. K., Jung, J. H., Altpeter, F., Kannan, B., Kim, H. E., Kim, K. H., et al. (2018). Largely enhanced bioethanol production through the combined use of ligninmodified sugarcane and xylose fermenting yeast strain. Bioresour. Technol. 256, 312–320. doi: 10.1016/j.biortech.2018.01.123
Krishna, S. S., Harish Chandar, S. R., Ravi, M., Valarmathi, R., Lakshmi, K., Prathima, P. T., et al. (2023). Transgene-free genome editing for biotic and abiotic stress resistance in sugarcane: prospects and challenges. Agronomy 13, 1000. doi: 10.3390/agronomy13041000
Kumar, N., Galli, M., Ordon, J., Stuttmann, J., Kogel, K. H., Imani, J. (2018). Further analysis of barley morc1 using a highly efficient rna-guided cas9 gene-editing system. Plant Biotechnol. J. 16, 1892–1903. doi: 10.1111/pbi.12924
Lawrenson, T., Hinchliffe, A., Clarke, M., Morgan, Y., Harwood, W. (2021). In-planta gene targeting in barley using cas9 with and without geminiviral replicons. Front. Genome Ed. 3. doi: 10.3389/fgeed.2021.663380
Le Cunff, L., Garsmeur, O., Raboin, L. M., Pauquet, J., Telismart, H., Selvi, A., et al. (2008). Diploid/polyploid syntenic shuttle mapping and haplotype-specific chromosome walking toward a rust resistance gene (Bru1) in highly polyploid sugarcane (2 N ~ 12 X ~ 115). Genetics 180, 649–660. doi: 10.1534/genetics.108.091355
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018a). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438. doi: 10.1104/pp.18.00200
Li, J., Li, Y., Ma, L. G. (2019). Crispr/cas9-based genome editing and its applications for functional genomic analyses in plants. Small Methods 3, 1800473. doi: 10.1002/smtd.201800473
Li, J., Phan, T.-T., Li, Y.-R., Xing, Y.-X., Yang, L.-T. (2018). Isolation, transformation and overexpression of sugarcane SoP5CS gene for drought tolerance improvement. Sugar Tech 20, 464–473. doi: 10.1007/s12355-017-0568-9
Liang, Z., Zhang, K., Chen, K., Gao, C. (2014). Targeted mutagenesis in zea mays using talens and the crispr/cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Liang, G., Zhang, H., Lou, D., Yu, D. (2016). Selection of highly efficient sgRNAs for CRISPR/Cas9 based plant genome editing. Sci. Rep. 6, 21451. doi: 10.1038/srep21451
Lin, S., Chen, T., Qin, X., Wu, H., Khan, M. A., Lin, W. (2014). Identification of microrna families expressed in sugarcane leaves subjected to drought stress and the targets thereof. Pak. J. Agric. Sci. 51, 925–934.
Liu, F., Huang, N., Wang, L., Ling, H., Sun, T., Ahmad, W., et al. (2018). A novel L-ascorbate peroxidase 6 gene, ScAPX6, plays an important role in the regulation of response to biotic and abiotic stresses in sugarcane. Front. Plant Sci. 8, 2262. doi: 10.3389/fpls.2017.02262
Liu, J., Xu, X., Shao, Q. (2008). The spatial and temporal characteristics of grassland degradation in the Three-River Headwaters region in Qinghai Province. Acta Geogr. Sin. 63, 364–377.
Ma, X., Zhu, Q., Chen, Y., Liu, Y. G. (2016). CRISPR/cas9 platforms for genome editing in plants: developments and applications. Mol. Plant 9, 961–974. doi: 10.1016/j.molp.2016.04.009
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/cas9-targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi: 10.3389/fpls.2016.01904
Manghwar, H., Lindsey, K., Zhang, X., Jin, S. (2019). Crispr/cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., Zhu, J. K. (2013). Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi: 10.1093/mp/sst121
Maqsood, Q., Abbas, R. N., Iqbal, M. A., Aydemir, S. K., Iqbal, A., El Sabagh, A. (2020). Overviewing of weed management practices to reduce weed seed bank and to increase maize yield. Planta Daninha 38, e020199716. doi: 10.1590/s0100-83582020380100075
Meena, M. R., Kumar, R., Chinnaswamy, A., Karuppaiyan, R., Kulshreshtha, N., Ram, B. (2020). Current breeding and genomic approaches to enhance the cane and sugar productivity under abiotic stress conditions. 3 Biotech. 10, 440. doi: 10.1007/s13205-020-02416-w
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. U.S.A. 115, 6058–6063. doi: 10.1073/pnas.1804774115
Miller, J. C., Tan, S., Qiao, G., Barlow, K. A., Wang, J., Xia, D. F., et al. (2011). A tale nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148. doi: 10.1038/nbt.1755
Milner, M. J., Craze, M., Hope, M. S., Wallington, E. J. (2020). Turning up the temperature on CRISPR: increased temperature can improve the editing efficiency of wheat using CRISPR/Cas9. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.583374
Ming, M., Ren, Q., Pan, C., He, Y., Zhang, Y., Liu, S., et al. (2020). Crispr-cas12b enables efficient plant genome engineering. Nat. Plants 6, 202–208. doi: 10.1038/s41477-020-0614-6
Mohan, C. (2016). Genomeeditinginsugarcane:challenges ahead. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01542
Mohan, C. (2017). Sugarcane Biotechnology: Challenges and Prospects (Cham: Springer), 1–176. doi: 10.1007/978-3-319-58946-6
Mohan, C., Narayan, J. A., Esterling, M., Yau, Y. Y. (2020). “Current transformation methods for genome–editing applications in energy crop sugarcane,” in Climate Change, Photosynthesis and Advanced Biofuels. Eds. Kumar, A., Yau, Y. Y., Ogita, S., Scheibe, R. (Springer, Singapore), 369–388.
Murugan, N., Palanisamy, V., Channappa, M., Ramanathan, V., Ramaswamy, M., Govindakurup, H., et al. (2022). Genome- wide in silico identification, structural analysis, promoter analysis, and expression profiling of PHT gene family in sugarcane root under salinity stress. Sustainability 14, 15893. doi: 10.3390/su142315893
Nerkar, G., Thorat, A., Sheelavantmath, S., Kassa, H. B., Devarumath, R. (2018). “Genetic transformation of sugarcane and field performance of transgenic sugarcane,” in Biotechnologies of Crop Improvement, vol. 2 . Eds. Gosal, S., Wani, S. (Springer, Cham). doi: 10.1007/978-3-319-90650-8_9
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., et al. (2018). Engineered crispr-cas9 nuclease with expanded targeting space. Science 361, 1259–1262. doi: 10.1126/science.aas9129
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of crispr/cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6, 26685. doi: 10.1038/srep26685
Ostengo, S., Serino, G., Perera, M. F., Racedo, J., Mamaní Gonzales, S., Yáñez Cornejo, F., et al. (2022). Sugarcane Breeding, Germplasm Development and Supporting Genetic Research in Argentina. Sugar Tech 24, 166–180. doi: 10.1007/s12355021-00999-z
Oz, M. T., Altpeter, A., Karan, R., Merotto, A., Altpeter, F. (2021). CRISPR/cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance. Front. Genome Ed. 3. doi: 10.3389/fgeed.2021.673566
Panting, M., Holme, I. B., Björnsson, J. M., Zhong, Y., Brinch-Pedersen, H. (2021). Crispr/cas9 and transgene verification of gene involvement in unfolded protein response and recombinant protein production in barley grain. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.755788
Park, J.-W., Benatti, T. R., Marconi, T., Yu, Q., Solis-Gracia, N., Mora, V., et al. (2015). Cold responsive gene expression profiling of sugarcane and Saccharum spontaneum with functional analysis of a cold inducible Saccharum homolog of NOD26-like intrinsic protein to salt and water stress. PloS One 10, e0125810. doi: 10.1371/journal.pone.0125810
Patade, V. Y., Suprasanna, P., Bapat, V. A. (2008). Effects of salt stress in relation to osmotic adjustment on sugarcane (Saccharum officinarum L. ) callus cultures. Plant Growth Regul. 55, 169–173. doi: 10.1007/s10725-008-9270-y
Peng, R., Lin, G., Li, J. (2015). Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 283, 1218–1231. doi: 10.1111/febs.13586
Ramasamy, M., Damaj, M. B., Vargas-Bautista, C., Mora, V., Liu, J., Padilla, C. S., et al. (2021). A sugarcane G-protein-coupled receptor, ShGPCR1, confers tolerance to multiple abiotic stresses. Front. Plant Sci. 12, 745891. doi: 10.3389/fpls.2021.745891
Ramiro, D. A., Melotto-Passarin, D. M., Barbosa, M., Dos Santos, F., Gomez, S. G. P., Massola Júnior, N. S., et al. (2016). Expression of Arabidopsis Bax Inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol. J. 14, 1826–1837. doi: 10.1111/pbi.12540
Rehman, S., Muhammad, K., Novaes, E., Que, Y., Din, A., Islam, M., et al. (2021). Expression analysis of transcription factors in sugarcane during cold stress. Braz. J. Biol. 83, e242603. doi: 10.1590/1519-6984.242603
Ren, Q., Sretenovic, S., Liu, S., Tang, X., Huang, L., He, Y., et al. (2021). Pam-less plant genome editing using a crispr-spry toolbox. Nat. Plants 7, 25–33. doi: 10.1038/s41477-020-00827-4
Riaz, A., Kanwal, F., Ahmad, I., Ahmad, S., Farooq, A., Madsen, C. K., et al. (2022). New hope for genome editing in cultivated grasses: CRISPR variants and application. Front. Genet. 13. doi: 10.3389/fgene.2022.866121
Riyazuddin, R., Nisha, N., Ejaz, B., Khan, M. I. R., Kumar, M., Ramteke, P. W., et al. (2022). A comprehensive review on the heavy metal toxicity and sequestration in plants. Biomolecules 12, 43. doi: 10.3390/biom12010043
Saleh, F., Karwacki, J. (1996). Revisiting the ecotourist: The case of grasslands national park. J. Sustain. Tourism 4, 61–80. doi: 10.1080/09669589608667259
Sengar, K., Sengar, R., Singh, A. (2013). Biotechnological and genomic analysis for salinity tolerance in sugarcane. Int. J. Biotechnol. Bioeng. Res. 4, 407–414.
Shan, S., Mavrodiev, E. V., Li, R., Zhang, Z., Hauser, B. A., Soltis, P. S., et al. (2018). Application of CRISPR/Cas9 to Tragopogon (Asteraceae), an evolutionary model for the study of polyploidy. Mol. Ecol. Resour. 18, 1427–1443. doi: 10.1111/1755-0998.12935
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a crispr-cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. doi: 10.1038/nbt.3833
Shingote, P. R., Kawar, P. G., Pagariya, M. C., Kuhikar, R. S., Thorat, A. S., Babu, K. (2015). SoMYB18, a sugarcane MYB transcription factor improves salt and dehydration tolerance in tobacco. Acta Physiol. Plant 37, 217. doi: 10.1007/s11738-015-1961-1
Su, X. K., Wu, Y., Dong, S. K., Wen, L., Li, Y. Y., Wang, X. X. (2015). Effects of grassland degradation and re-vegetation on carbon and nitrogen storage in the soils of the Headwater Area Nature Reserve on the Qinghai-Tibetan Plateau, China. J. Mt. Sci. 12, 582–591. doi: 10.1007/s11629-014-3043-z
Sun, J., Cheng, G. W., Li, W. P. (2013). Meta-analysis of relationships between environmental factors and aboveground biomass in the alpine grassland on the Tibetan Plateau. Biogeosciences 10, 1707–1715. doi: 10.5194/bg-10-1707-2013
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high- amylose rice through crispr/cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00298
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using cas9 and guide rna. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of osnramp5 using the crispr/cas9 system produces low cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 14438. doi: 10.1038/s41598-017-14832-9
Taparia, Y., Gallo, M., Altpeter, F. (2012). Comparison of direct and indirect embryogenesis protocols, biolistic gene transfer and selection parameters for efficient genetic transformation of sugarcane. Plant Cell Tissue Organ. Cult. 111, 131–141. doi: 10.1007/s11240-012-0177-y
Tew, T. L., Cobill, R. M. (2008). “Genetic improvement of sugarcane (Saccharum spp.) as an energy crop,” in Genetic Improvement of Bioenergy Crops. Ed. Vermerris, W. (Springer New York, New York, NY), 273–294. doi: 10.1007/978-0-387-70805-8_9
Tiwari, A. K., Bharti, Y. P., Tripathi, S., Mishra, N., Rao, M., Sharma, G. P., et al. (2010). Biotechnological approaches to improve sugarcane crop with special reference to disease resistance. Acta Phytopathol. Entomol. Hung. 45, 235–249. doi: 10.1556/APhyt.45.2010.2.1
Tiwari, S., Lata, C. (2018). Heavy metal stress, signaling, and tolerance due to plant-associated microbes: An overview. Front. Plant Sci. 9, 452. doi: 10.3389/fpls.2018.00452
Trujillo, L. E., Menéndez, C., Ochogavía, M. E., Hernández, I., Borrás, O., Rodríguez, R., et al. (2009). Engineering drought and salt tolerance in plants using SodERF3, a novel sugarcane ethylene responsive factor. Biotecnol. Apl. 26, 168–171.
Viswanathan, R., Rao, G. (2011). Disease scenario and management of major sugarcane diseases in India. Sugar Tech 13, 336–353. doi: 10.1007/s12355-011-0102-4
Vlcko, T., Ohnoutkova, L. (2020). Allelic variants of crispr/cas9 induced mutation in an inositol trisphosphate 5/6 kinase gene manifest different phenotypes in barley. Plants (Basel) 9, 195. doi: 10.3390/plants9020195
Walton, R. T., Christie, K. A., Whittaker, M. N., Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-pamless engineered crispr-cas9 variants. Science 368, 290–296. doi: 10.1126/science.aba8853
Waltz, E. (2016). CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34, 582. doi: 10.1038/nbt0616-582
Wang, F. Z., Chen, M. X., Yu, L. J., Xie, L. J., Yuan, L. B., Qi, H., et al. (2017). Osarm1, an R2r3 myb transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01868
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Z., Zhang, Y., Yang, Y., Zhou, W., Gang, C., Zhang, Y., et al. (2016). Quantitative assess the driving forces on the grassland degradation in the Qinghai–Tibet Plateau. China. Ecol. Inform. 33, 32–44. doi: 10.1016/j.ecoinf.2016.03.006
Weeks, D. P. (2017). “Chapter Four - Gene editing in polyploid crops: wheat, camelina, canola, potato, cotton, peanut, sugar cane, and citrus,” in Progress in Molecular Biology and Translational Science, vol. 149 . Eds. Weeks, D. P., Yang, B. (Academic Press, San Diego, CA), 65–80. doi: 10.1016/bs.pmbts.2017.05.002
Wen, L., Jinlan, W., Xiaojiao, Z., Shangli, S., Wenxia, C. (2018). Effect of degradation and rebuilding of artificial grasslands on soil respiration and carbon and nitrogen pools on an alpine meadow of the Qinghai-Tibetan Plateau. Ecol. Eng. 111, 134–142. doi: 10.1016/j.ecoleng.2017.10.013
Westra, E. R., Semenova, E., Datsenko, K. A., Jackson, R. N., Wiedenheft, B., Severinov, K., et al. (2013). Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PloS Genet. 9, e1003742. doi: 10.1371/journal.pgen.1003742
Wolter, F., Puchta, H. (2017). Knocking out consumer concerns and regulator’s rules: Efficient use of CRISPR/Cas ribonucleo- protein complexes for genome editing in cereals. Genome Biol. 18, 43. doi: 10.1186/s13059-017-1179-1
Wolter, F., Puchta, H. (2018). The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 94, 767–775. doi: 10.1111/tpj.13899
Wu, Q., Chen, Y., Zou, W., Pan, Y.-B., Lin, P., Xu, L., et al. (2023). Genome-wide characterization of sugarcane catalase gene family identifies a ScCAT1 gene associated disease resistance. Int. J. Biol. Macromol. 232, 123398. doi: 10.1016/j.ijbiomac.2023.123398
Wu, G. L., Du, G. Z., Liu, Z. H., Thirgood, S. (2009). Effect of fencing and grazing on a Kobresia-dominated meadow in the Qinghai-Tibetan plateau. Plant Soil 319, 115–126. doi: 10.1007/s11104-008-9854-3
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A crispr/cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. doi: 10.1186/s12870-014-0327-y
Yang, Y., Wang, Z., Li, J., Gang, C., Zhang, Y., Zhang, Y., et al. (2016). Comparative assessment of grassland degradation dynamics in response to climate variation and human activities in China, Mongolia, Pakistan and Uzbekistan from 2000 to 2013. J. Arid. Environ. 135, 164–172. doi: 10.1016/j.jaridenv.2016.09.004
Yin, K., Han, T., Liu, G., Chen, T., Wang, Y., Yu, A. Y., et al. (2015). A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 5, 14926. doi: 10.1038/srep14926
Yu, L., Chen, Y., Sun, W., Huang, Y. (2019). Effects of grazing exclusion on soil carbon dynamics in alpine grasslands of the Tibetan Plateau. Geoderma 353, 133–143. doi: 10.1016/j.geoderma.2019.06.036
Zaidi, S. S., Mahfouz, M. M., Mansoor, S. (2017). CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci. 22, 550–553. doi: 10.1016/j.tplants.2017.05.001
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via crispr/cas9-targeted mutagenesis of the osrr22 gene. Mol. Breed. 39, 47. doi: 10.1007/s11032-019-0954-y
Zhang, J., Nagai, C., Yu, Q., Pan, Y.-B., Ayala-Silva, T., Schnell, R. J., et al. (2012). Genome size variation in three Saccharum species. Euphytica 185, 2336. doi: 10.1007/s10681-012-0664-6
Zhu, K., Huang, C., Phan, T.-T., Yang, L.-T., Zhang, B.-Q., Xing, Y.-X., et al. (2021). Overexpression of SoACLA-1 gene confers drought tolerance improvement in sugarcane. Plant Mol. Biol. Rep. 39, 489–500. doi: 10.1007/s11105-020-01263-6
Keywords: agrobacterium, genetic transformation, polyploidy, steppe and pampas, transgenic plants, lemon grass, wild sugarcane
Citation: Li C and Iqbal MA (2024) Leveraging the sugarcane CRISPR/Cas9 technique for genetic improvement of non-cultivated grasses. Front. Plant Sci. 15:1369416. doi: 10.3389/fpls.2024.1369416
Received: 12 January 2024; Accepted: 11 March 2024;
Published: 27 March 2024.
Edited by:
Avinash Mishra, Central Salt & Marine Chemicals Research Institute (CSIR), IndiaReviewed by:
Vijay Sheri, East Carolina University, United StatesPhetole Mangena, University of Limpopo, South Africa
Copyright © 2024 Li and Iqbal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Aamir Iqbal, YWFtaXIxODAxQHlhaG9vLmNvbQ==; Chunjia Li, bGNqQHlhYXMub3JnLmNu
†These authors have contributed equally to this work and share first authorship
 Chunjia Li1,2*†
Chunjia Li1,2*† Muhammad Aamir Iqbal
Muhammad Aamir Iqbal