
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 09 February 2024
Sec. Plant Metabolism and Chemodiversity
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1368870
This article is part of the Research TopicProanthocyanidins and IsoflavonoidsView all 12 articles
 Qilin Yang1,2
Qilin Yang1,2 Guodong Wang1,2*
Guodong Wang1,2*Isoflavonoids constitute a well-investigated category of phenylpropanoid-derived specialized metabolites primarily found in leguminous plants. They play a crucial role in legume development and interactions with the environment. Isoflavonoids usually function as phytoalexins, acting against pathogenic microbes in nature. Additionally, they serve as signaling molecules in rhizobial symbiosis. Notably, owing to their molecular structure resembling human estrogen, they are recognized as phytoestrogens, imparting positive effects on human health. This review comprehensively outlines recent advancements in research pertaining to isoflavonoid biosynthesis, transcriptional regulation, transport, and physiological functions, with a particular emphasis on soybean plants. Additionally, we pose several questions to encourage exploration into novel contributors to isoflavonoid metabolism and their potential roles in plant-microbe interactions.
Isoflavonoids, primarily found in legumes, are recognized as phytoestrogens owing to their structural and size resemblance to human estrogens (17β-estradiol and daidzein-derived (S)-equol, structures see Figure 1) (Krizova et al., 2019). Soybeans and soy products are the primary source of these compounds in human diets (Rizzo and Baroni, 2018). Following ingestion, isoflavonoids especially the genistein and daidzein, exhibit the capacity to bind to estrogen receptors, thereby exerting estrogenic or anti-estrogenic effects. (Ariyani et al., 2023). They are thought to confer a protective effect against hormone-related cancers, such as prostate and breast cancer (Boutas et al., 2022; Yu et al., 2023). Besides cancer prevention, isoflavonoids are implicated in averting various diseases, including cardiovascular ailments, Alzheimer’s disease, and possess anti-inflammatory and sterol-lowering properties (Li and Zhang, 2017; Xie et al., 2021; Vina et al., 2022) (Table 1). Isoflavonoids extend beyond human health benefits, crucially influencing plant-microbe interactions. They serve as signals in rhizobia-legume symbiosis, triggering nodulation gene expression in rhizobia and enhancing the nodulation process in legumes (Subramanian et al., 2006; Han et al., 2020). Notably, when investigating isoflavonoids in leguminous plants, it is crucial to consider all specialized metabolites as a whole, including other compounds such as saponins (Sugiyama, 2019; Yuan et al., 2023). This review, however, will focus on isoflavonoid distribution, biosynthesis, regulation, transport, and their pivotal roles in plant ecology.
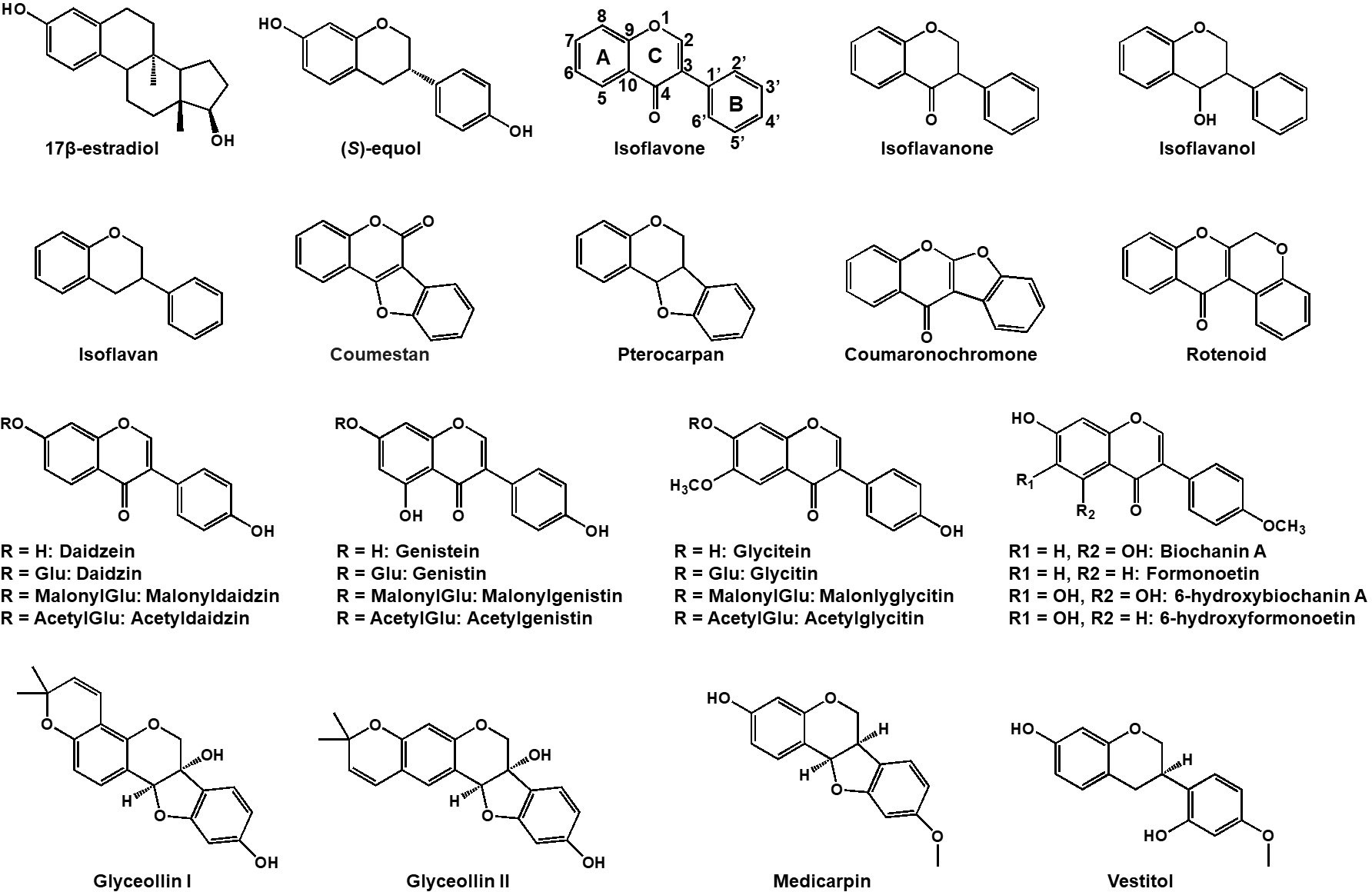
Figure 1 Chemical structures of representative isoflavonoids in leguminous plants. Both 17β-estradiol (estrogen) and daidzein-derived (S)-equol have a high affinity for estrogen receptor in human.
Isoflavonoids commonly harbor a C6-C3-C6 carbon skeleton—comprising two 6-carbon benzene rings A and C and a 3-carbon heterocyclic ring B (Figure 1) (Nabavi et al., 2020). Diverse modifications such as oxidation, glycosylation, acylation, prenylation and others, along with intra-molecular cyclization yield an array of isoflavonoids, including isoflavans, isoflavanones, isoflavanols, coumestans, coumaronochromones, rotenoids, pterocarpans and so on (see Figure 1; Veitch, 2007). Thus far, there are more than 2,000 isoflavonoids were isolated and structurally elucidated in plants (Mackova et al., 2006; Veitch, 2007; Al-Maharik, 2019, 2009; 2013). Legume isoflavonoids are comprised of aglycones and glycosides which include glucosides, malonylglucosides, or acetylglucosides. They prevail and translocate within vacuoles of plant cells (Zhao and Dixon, 2010). Different legume plant species may produce different types of isoflavonoids in response to the growing environment. Soybean produces daizein, genistein, glycitein, and derivatives, with glyceollin synthesized in response to pathogens (Ng et al., 2011), while in Medicago truncatula and Lotus japonicus, the methylated glycones, formononetin and biochanin-A are produced respectively alongside daidzein and genistein. In addition, M. truncatula produces medicarpin, while L. japonicus yields vestitol through further reduction from medicarpin (Naoumkina et al., 2007; Masunaka et al., 2011).
While isoflavonoids are prominently found in legumes, they are also reported to be isolated in numerous non-leguminous plant families (Reynaud et al., 2005). Over 200 isoflavonoids have been identified in 50+ non-leguminous plant families, spanning Bryopsida, Pinopsida, Magnoliopsida, and Liliopsida classes (Lapcík, 2007).
The biosynthetic pathway of isoflavonoids in legumes is currently well investigated, comprising three key phases: the phenylpropanoid pathway, biosynthesis of the isoflavonoid aglycone, and the final production of isoflavonoids (Dixon and Pasinetti, 2010; Sohn et al., 2021). Initially, isoflavonoids originate from the phenylpropanoid pathway, where phenylalanine undergoes a sequential three-step catalytic transformation by phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H), and 4-coumaroyl-CoA ligase (4CL) to produce the flavonoid precursor, p-coumaroyl-CoA. Subsequently, through the catalytic action of chalcone synthase (CHS), p-coumaroyl-CoA condenses with three units of malonyl-CoA to yield naringenin chalcone, a crucial intermediate and the first-rate-limiting step in the flavonoid biosynthetic pathway. Simultaneously, isoliquiritigenin is also formed through the concerted activities of CHS and chalcone reductase (CHR). In soybeans, naringenin chalcone and isoliquiritigenin undergo further enzymatic transformations involving chalcone isomerase (CHI), isoflavone synthase (IFS), and 2-hydroxyisoflavanone dehydratase (HID) to generate three primary isoflavonoid aglycones: genistein, daidzein, and glycitein. However, in other leguminous species such as M. truncatula and L. japonicus, naringenin chalcone and isoliquiritigenin take an alternative route, giving rise to two distinct 4’-methoxyisoflavonoids, biochanin-A and formononetin. This diversion is facilitated by the involvement of a crucial enzyme, 2-hydroxyisoflavanone 4′-O-methyltransferase (HI4’OMT). Subsequently, these isoflavonoid aglycones serve as substrates for various modifying enzymes, including glycosyltransferases and acyltransferases, leading to the formation of diverse isoflavonoid derivatives (Figure 2A). Additionally, further enzymatic reactions contribute to the synthesis of intricate and biologically active isoflavonoids, such as glyceollin, medicarpin, and vestitol. Notably, two tandem P450 enzymes, C4H and IFS, localized to the endoplasmic reticulum (ER), play a pivotal role in anchoring other isoflavonoid enzymes to the ER through protein-protein interactions, thereby establishing an isoflavonoid metabolon on the ER (Dastmalchi et al., 2016).
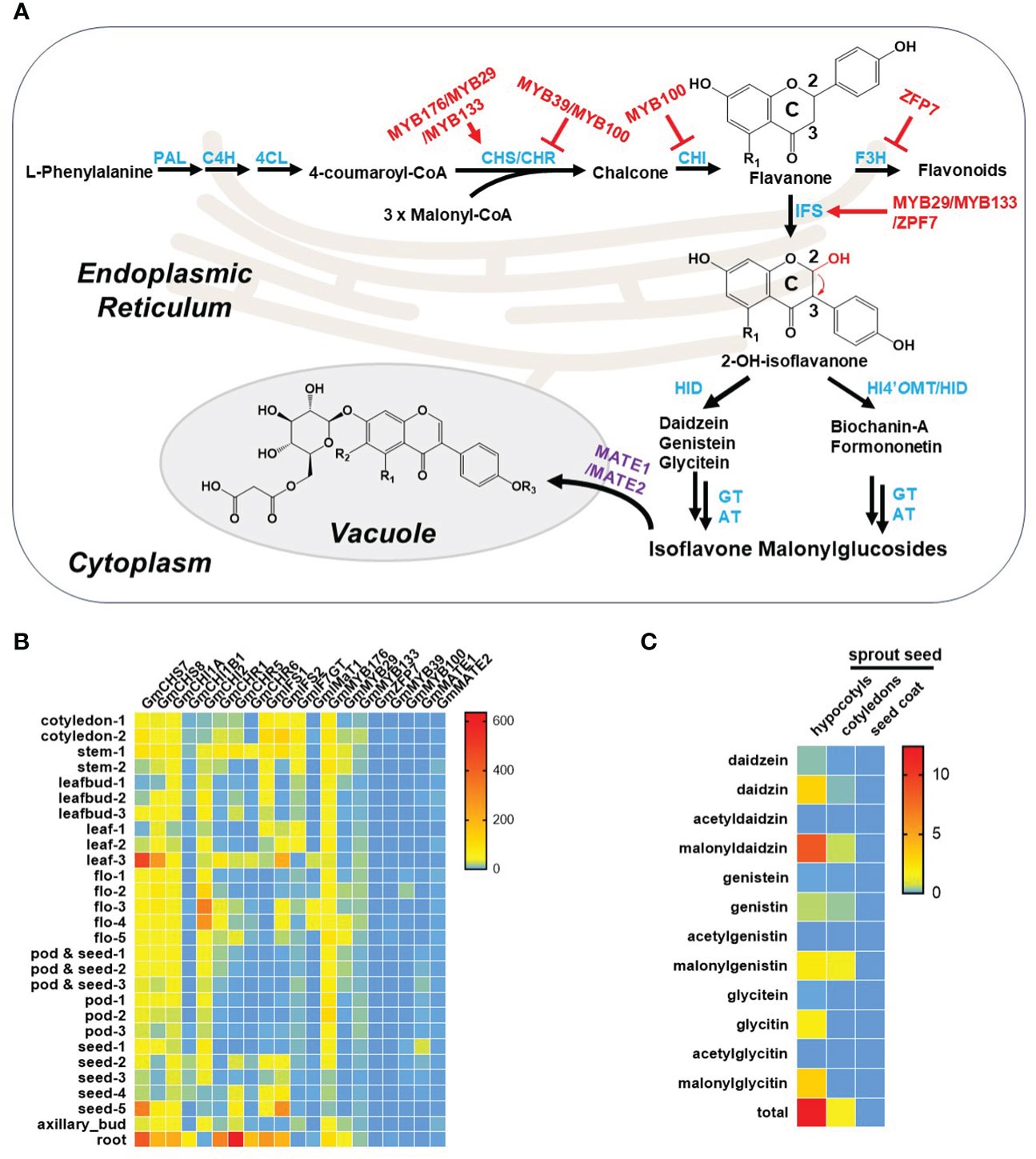
Figure 2 Isoflavonoid metabolism in soybean plants. (A) Summary of isoflavonoid metabolism in soybean plant. Notably, R1 and R2 represent either –H or –OH, and R3 represents either –H or –CH3 in the chemical structures here. The biosynthetic enzymes, transporters, and transcript factors are highlighted in blue, purple, and red, respectively. Abbreviations: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHR, chalcone reductase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; IFS, isoflavone synthase; HID, 2-hydroxyisoflavanone dehydratase; HI4’OMT, 2-hydroxyisoflavanone 4′-O-methyltransferase; GT, glucosyltransferase; AT, acyltransferase. (B) Tissue specificity of isoflavonoid known genes in soybean plants. Transcriptome data are extracted from SoyOmics database (https://ngdc.cncb.ac.cn/soyomics/index). (C) Distribution of isoflavonoids in seeds. Data are extracted from the work by Yuan et al. (Yuan et al., 2009).
IFS, a cytochrome P450 enzyme, is a pivotal rate-limiting determinant governing the transition of intermediates from the flavonoid pathway to the isoflavonoid pathway. This transformation involves the migration of the B-ring from the C2 to the C3 position of the C-ring, succeeded by hydroxylation at the C2 position of the C-ring (Figure 2A; Dixon and Steele, 1999; Jung et al., 2000). While IFS is prevalent in legumes, it is not exclusive to this plant family, as evidenced by its presence in non-legumes. For example, IFS has been identified in sugarbeet (Beta vulgaris) exhibiting substantial similarity to legume IFS (Jung et al., 2000). In addition to sugarbeet, a wheat-specific IFS gene, TaCYP71F53, has been recently identified in wheat, setting it apart from legume IFSs by utilizing artocarpanone A as a substrate instead of naringenin or liquiritigenin. The wheat IFS (TaCYP71F53) is responsible for aryl migration coupled with C-ring desaturation of artocarpanone A; however, the legume IFSs are usually responsible for aryl migration and 2-hydroxylation of naringenin or liquiritigenin. Phylogenetic analysis further elucidated that legume IFSs belong to the CYP93 family, while the wheat-specific IFS belongs to the CYP71 family (Polturak et al., 2023). This discovery underscores a parallel gain in the capacity for isoflavonoid production in both soybeans and wheat. In essence, the broad distribution of IFS across diverse plant species highlights its evolutionary significance and the adaptability of distinct plants to engage in the synthesis of bioactive isoflavonoids.
Isoflavonoids synthesized in the ER and cytoplasm often undergo subsequent transport processes, with two main possibilities identified: membrane vesicle-mediated transport and membrane transporter-mediated transport (Zhao and Dixon, 2010). Among membrane transporters, multidrug and toxic compound extrusion (MATE) transporters are considered crucial, particularly in the translocation of isoflavonoids from the cytoplasm to the vacuole. Notably, GmMATE1 and GmMATE2, identified as soy isoflavonoid transporters, facilitate the accumulation of isoflavonoids in soybean seeds. However, studies using the yeast system have revealed that these transporters specifically facilitate the transport of aglycones and glycosides but not malonylglycosides (Ng et al., 2021). Similarly, GmMATE4, localized in vacuole-like membranes, plays a role in mediating the transport of isoflavonoids, including daidzein, genistein, glycitein, and glycitin, into the vacuole for storage. The uptake and efflux of isoflavonoids across the tonoplast or plasma membrane involve various transporters. It is suggested that an unknown ATP-binding cassette (ABC) transporter may be involved in the secretion of soy isoflavonoid aglycones. This inference is supported by the ATP-dependent transport of genistein with and without typical ABC transporter inhibitors (Sugiyama et al., 2007). The intricate interplay of these transport mechanisms adds another layer of complexity to the regulation of isoflavonoid compartmentalization and function within plant cells.
Although the isoflavonoid biosynthesis pathway has been well-investigated in plants, little research focuses on the catabolism of isoflavonoids. Recently, Aoki et al. discovered an isoflavone oxidative catabolic gene cluster in legume root bacteria (Variovorax sp.), which belongs to Comamonadaceae. The identified catabolism (ifc) gene cluster includes at least four functional genes, and the final catabolic products of isoflavones eventually enter the tricarboxylic acid cycle in bacteria. The authors further demonstrated that ifc genes are frequently found in bacterial strains isolated from legume plants (Aoki et al., 2023). It can be foreseen that more isoflavone degradation gene clusters will be discovered in near future, especially in plant rhizosphere bacteria.
Isoflavonoid biosynthetic enzymes and transporters definitely play a crucial role in determining the production of isoflavonoids. However, the regulatory network involves transcription factors which can directly/indirectly control the expression of isoflavonoid biosynthetic genes, and consequently altering isoflavonoid content. Numerous studies have identified MYB transcription factors as pivotal regulators in the isoflavonoid biosynthesis. For instance, the R1 MYB transcription factor GmMYB176 was reported to bind with the promoter of CHS8, activating CHS8 expression and enhancing isoflavonoid accumulation (Yi et al., 2010). Similarly, positive regulators like GmMYB29 (R2R3 MYB) and GmMYB133 (CCA1-like MYB) were found to activate the expression of CHS8 and IFS2, leading to increased isoflavonoid content (Chu et al., 2017; Bian et al., 2018). However, not all MYB transcription factors exhibit positive regulation; some, such as GmMYB39 and GmMYB100, act as repressors by inhibiting (iso)flavonoid biosynthesis through the repression of upstream genes such as CHS, CHR, and CHI (Liu et al., 2013; Yan et al., 2015). Moreover, some MYB transcription factors do not act alone. They can interact with other transcription factors to regulate the isoflavonoid biosynthesis. For example, GmMYB176 can interact with GmbZIP5, and the co-overexpression of these two genes was shown to increase isoflavonoid content (Anguraj Vadivel et al., 2021). Additionally, other transcription factors, such as the C2H2-type zinc finger protein GmZFP7, accelerate isoflavonoid synthesis by promoting the expression of GmIFS2 and inhibiting the expression of GmF3H1, thereby increasing the metabolic flux of isoflavonoids and enhancing their levels (Feng et al., 2023). GmNAC42-1 was identified as an essential positive regulator of glyceollin biosynthesis, directly binding to the promoters of IFS2 and G4DT, promoting the expression of these two genes, and increasing glyceollin content (Jahan et al., 2019).
In addition to transcription factors, microRNAs might also play a role in mediating the isoflavonoid biosynthetic pathway to regulate isoflavonoid production. Gma-miR26 and Gma-miRNA28, along with their corresponding target genes (Glyma.10G197900 and Glyma.09G127200), were found to be directly related to isoflavonoid content (Gupta et al., 2017). Furthermore, Gma-miR5030 was identified as a mediator of the expression of the target gene GmMYB176, affecting isoflavonoid biosynthesis (Gupta et al., 2019). This intricate regulatory network involving transcription factors and microRNAs adds a layer of complexity to the modulation of isoflavonoid biosynthesis in plants. Notably, the role of microRNAs involved in the regulation of (iso)flavonoid in plants need further experimental validation.
The publicly available transcriptome reveals that genes involved in isoflavone metabolism exhibit higher expression levels in roots and mature seeds compared to other organs, as illustrated in Figure 2B. This heightened expression corresponds to increased accumulation of isoflavonoids in these specific tissues. Notably, seeds play a crucial role as a significant source of soy isoflavones for human consumption through direct intake or soy products. The distribution of soy isoflavones within the mature soy seed varies significantly among its different parts. The highest concentration of all isoflavones is observed in the hypocotyl, followed by the cotyledons, while the seed coat contains almost negligible amounts of isoflavones. Furthermore, within the hypocotyl, daidzein and its derivatives exhibit higher levels compared to the other two isoflavones. In the cotyledons, there is an enrichment of genistein and its derivatives (Yuan et al., 2009) (Figure 2C).
Isoflavonoids play pivotal roles in plant ecology, particularly in plant-microbe interactions, functioning as phytoalexins to combat various diseases caused by nematode, oomycete, fungi, bacteria and virus (Lin et al., 2022), and fostering root-rhizobia symbiosis to regulate nodulation (Hammerschmidt, 1999). Notably, transgenic rice expressing the GmIFS1 gene demonstrated enhanced resistance to the rice blast pathogen, indicating that isoflavonoid biosynthesis, particularly genistein, serves as a phytoalexin in transgenic rice (Pokhrel et al., 2021). In soybeans, glyceollin acts as a crucial phytoalexin induced by cell wall glucan elicitors to resist Phytophthora sojae. The silencing of upstream synthesis genes (IFS and CHR) impairs soybean resistance to P. sojae (Ayers et al., 1976; Graham et al., 2007). Glyceollins also exhibit anti-bacterial, anti-nematode and anti-fungal activities besides anti-oomycetes (P. sojae) (Ng et al., 2011). Also, the soybean plants with 2-fold increased isoflavonoids through simultaneous knockout of three flavanone genes (GmF3H1, GmF3H2 and GmFNSII-1) enhanced the leaf resistance to soya bean mosaic virus (Zhang et al., 2020). In M. truncatula, medicarpin synergizes with SA to combat powdery mildew Erysiphe pisi and Rhizoctonia solani (Gupta et al., 2022).
Isoflavonoids also serve as signals to mediate a symbiotic relationship between legumes and nitrogen-fixing bacteria, primarily Rhizobium, converting atmospheric nitrogen into ammonia for direct plant utilization through the process of biological nitrogen fixation (Yang et al., 2022). Subramanian et al. demonstrated the crucial role of isoflavonoids in nodulation formation, revealing that endogenous production of genistein is sufficient to support nodulation in the soybean hairy root system through RNAi-mediated silencing of the IFS and CHR genes (Subramanian et al., 2006). During nodulation formation, cells transport (iso)flavonoids, mainly aglycones and glycosides, directly into the apoplast (Biala-Leonhard et al., 2021). In the apoplast, isoflavonoid conjugate-hydrolyzing β-glucosidase (ICHG) hydrolyzes isoflavonoid glycosides, forming aglycones (Matsuda et al., 2023). Subsequently, in the soil, legume roots release these (iso)flavonoid aglycones to attract rhizobia to the rhizosphere. Following this, (iso)flavonoids form a complex by binding to the transcriptional activator NodD protein, activating the transcription of rhizobial nod genes (nodA, nodB, and nodC), and inducing rhizobia to synthesize and secrete the Nod factors, like lipo-chitooligosaccharides (Peck et al., 2006; Del Cerro et al., 2019; Rush et al., 2020).
In light of the evident of the beneficial impact of isoflavonoids on human health, numerous endeavors have been undertaken to produce isoflavonoids in various chassis, like Saccharomyces cerevisiae, Escherichia coli and plants (Sajid et al., 2021). Recently, Liu et al. engineered the metabolism of S. cerevisiae for de novo production of isoflavonoid from glucose. The final optimized strain produces up to 85.4 mg L−1 of daidzein and 72.8 mg L−1 puerarin (Liu et al., 2021). The efficient synthesis of isoflavonoids using synthetic biology strategies is expected to be a prominent research focus in this field.
The exploration of customized soybean seeds with enhanced nutrition and the investigation of the physiological functions of isoflavonoids in plant-microbe interactions offer other exciting avenues for future research (Figure 3). Both future researches rely on the comprehensive understanding the biosynthesis and regulation of isoflavonoids and the use of loss-of-function plant materials. Recent years have witnessed the identification of key enzymes in the isoflavonoid biosynthetic pathway, facilitating the utilization of metabolic engineering techniques to expedite the production of higher levels of isoflavonoids for application in the prevention and treatment of associated diseases. Despite the substantial progress, certain isoflavonoid biosynthetic enzymes remain uncharacterized. For instance, although it has been suggested that flavonoid 6-hydroxylase (F6H, a cytochrome P450 monooxygenase belonging to the CYP71D subfamily) may play a role in glycitein synthesis, there is a lack of relevant genetic evidence to substantiate this claim (LatundeDada et al., 2001; Artigot et al., 2013). Continued exploration of these uncharted enzymatic territories holds promise for advancing our understanding and harnessing the full potential of isoflavonoids. Metabolic genome-wide association studies (mGWAS) and quantitative trait locus (QTL), together with co-expression analysis, have emerged as powerful tools in unraveling the complex genetic architecture underlying various biological pathways (Luo, 2015). Given the public availability of soybean-omics databases, these approaches provide a novel and insightful avenue for identifying previously unknown contributors to the isoflavonoid metabolic pathway (Grant et al., 2010; Zheng et al., 2022; Azam et al., 2023; Chu et al., 2023; Liu et al., 2023). The customized soybean seeds thus could be reached by genetic engineering for optimizing isoflavonoid composition and quantity. This process will focus on manipulating key genes involved in the biosynthesis of isoflavonoids and the nutritional knowledge of specific isoflavonoids (Table 2). Finally, investing the potential health benefits of customized soybean seeds enriched with specific isoflavonoids could have impact on human health, such as their antioxidant properties, anti-inflammatory effects, and potential role in preventing chronic diseases. For instance, increasing the metabolic flux to daidzein by reducing glycitein branch might be a good target for customized soybean seeds (Zaheer and Humayoun Akhtar, 2017; Alshehri et al., 2021).
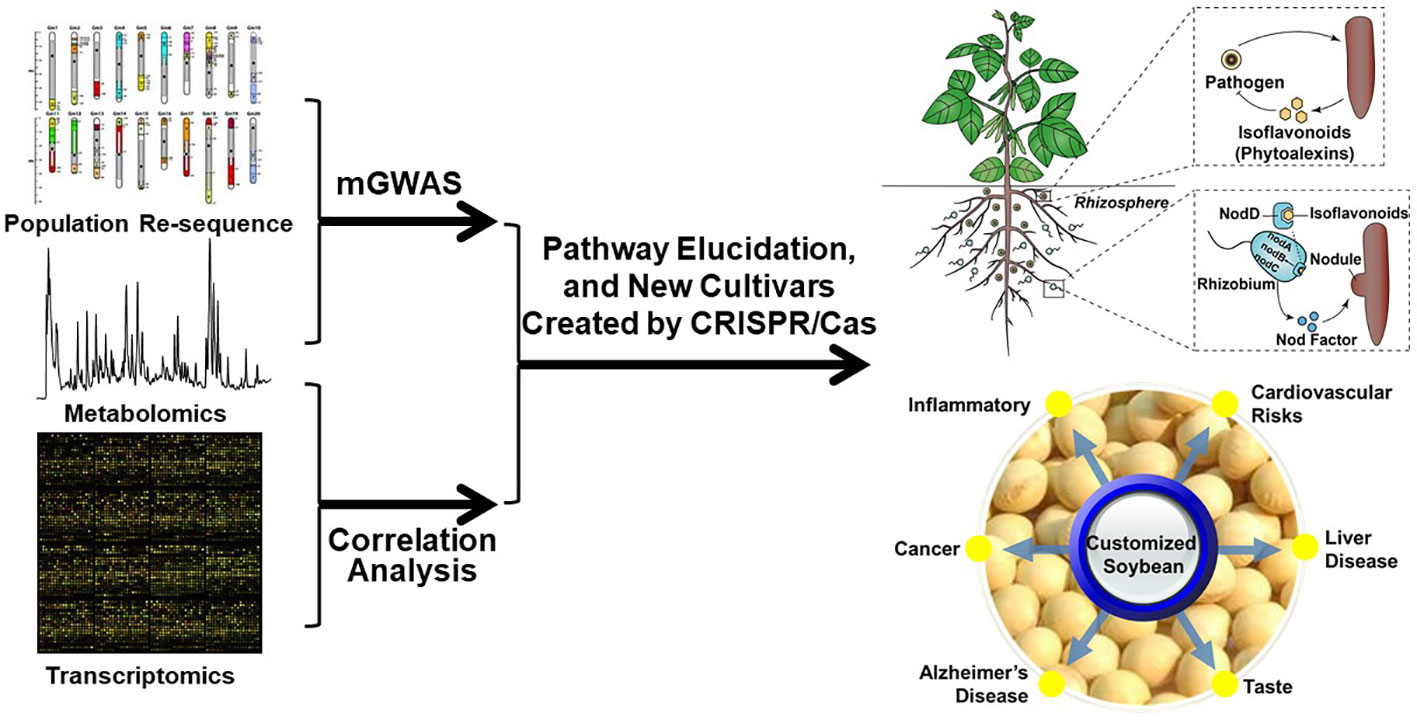
Figure 3 Workflow for isoflavonoid pathway elucidation and their applications in either rhizosphere or seeds.
As above-mentioned, isoflavonoids play a crucial role as signals mediating plant-microbe symbiosis, particularly in the context of rhizobial interactions. This symbiotic relationship is essential for the ability of legumes to thrive in nitrogen-poor soils, offering a distinct advantage over non-leguminous plants that rely on chemical fertilizers for normal development in low-nitrogen environments. Modifying key enzymes involved in isoflavonoid synthesis in non-leguminous plants holds the potential to promote the establishment of this symbiosis, thereby reducing the dependence on nitrogen fertilizers. The overexpression of IFS in transgenic rice has been shown to induce nod gene expression in rhizobia, opening up the possibility of symbiosis (Sreevidya et al., 2006). Furthermore, isoflavonoids can influence the bacterial community composition in the soil. For instance, the bacterial communities in daidzein-treated soils more closely resemble those in the soybean rhizosphere (Okutani et al., 2020). The addition of daidzein enriches the abundance of bacteria in Comamonadaceae, a predominant bacterial family in soybean roots. This suggests that manipulating isoflavonoid and other type of plant specialized metabolites could be a strategic approach to change the soil environment, potentially improving crop yields in non-leguminous plants through methods such as crop rotations in nitrogen-deficient conditions (Chen et al., 2019; Sugiyama, 2019; Wang et al., 2023). Understanding the regulation of isoflavonoids in the context of legume-rhizobial symbiosis allows for the development of crops with improved nitrogen fixation capabilities, which is essential for sustainable agriculture by reducing the need for synthetic fertilizers.
QY: Writing – original draft, Conceptualization. GW: Conceptualization, Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Key Research and Development Projects (2018YFA0900603 to GW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Maharik, N. (2019). Isolation of naturally occurring novel isoflavonoids: an update. Nat. Prod. Rep. 36 (8), 1156–1195. doi: 10.1039/c8np00069g
Alshehri, M. M., Sharifi-Rad, J., Herrera-Bravo, J., Jara, E. L., Salazar, L. A., Kregiel, D., et al. (2021). Therapeutic potential of isoflavones with an emphasis on daidzein. Oxid. Med. Cell Longev. 2021, 6331630. doi: 10.1155/2021/6331630
Anguraj Vadivel, A. K., McDowell, T., Renaud, J. B., Dhaubhadel, S. (2021). A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 4 (1), 356. doi: 10.1038/s42003-021-01889-6
Aoki, N., Shimasaki, T., Yazaki, W., Sato, T., Nakayasu, M., Ando, A., et al. (2023). Discovery of an isoflavone oxidative catabolic pathway in legume root microbiota. bioRxiv. doi: 10.1101/2023.08.07.552369
Ariyani, W., Amano, I., Koibuchi, N. (2023). Isoflavones mediate dendritogenesis mainly through estrogen receptor α. Int. J. Mol. Sci. 24 (10), 9011. doi: 10.3390/ijms24109011
Artigot, M. P., Baes, M., Dayde, J., Berger, M. (2013). Expression of flavonoid 6-hydroxylase candidate genes in normal and mutant soybean genotypes for glycitein content. Mol. Biol. Rep. 40 (7), 4361–4369. doi: 10.1007/s11033-013-2526-2
Ayers, A. R., Ebel, J., Valent, B., Albersheim, P. (1976). Host-pathogen interactions: X. fractionation and biological activity of an elicitor isolated from the mycelial walls of Phytophthora megasperma var. sojae. Plant Physiol. 57 (5), 760–765. doi: 10.1104/pp.57.5.760
Azam, M., Zhang, S., Li, J., Ahsan, M., Agyenim-Boateng, K. G., Qi, J., et al. (2023). Identification of hub genes regulating isoflavone accumulation in soybean seeds via GWAS and WGCNA approaches. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1120498
Babapour, M., Khorvash, F., Rouhani, M. H., Ghavami, A., Ghasemi-Tehrani, H., Heidari, Z., et al. (2022). Effect of soy isoflavones supplementation on migraine characteristics, mental status and calcitonin gene-related peptide (CGRP) levels in women with migraine: results of randomised controlled trial. Nutr. J. 21 (1), 50. doi: 10.1186/s12937-022-00802-z
Biala-Leonhard, W., Zanin, L., Gottardi, S., de Brito Francisco, R., Venuti, S., Valentinuzzi, F., et al. (2021). Identification of an isoflavonoid transporter required for the nodule establishment of the Rhizobium-Fabaceae symbiotic interaction. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.758213
Bian, S., Li, R., Xia, S., Liu, Y., Jin, D., Xie, X., et al. (2018). Soybean CCA1-like MYB transcription factor GmMYB133 modulates isoflavonoid biosynthesis. Biochem. Biophys. Res. Commun. 507 (1-4), 324–329. doi: 10.1016/j.bbrc.2018.11.033
Bilir, B., Sharma, N. V., Lee, J., Hammarstrom, B., Svindland, A., Kucuk, O., et al. (2017). Effects of genistein supplementation on genome-wide DNA methylation and gene expression in patients with localized prostate cancer. Int. J. Oncol. 51 (1), 223–234. doi: 10.3892/ijo.2017.4017
Boutas, I., Kontogeorgi, A., Dimitrakakis, C., Kalantaridou, S. N. (2022). Soy isoflavones and breast cancer risk: a meta-analysis. Vivo 36 (2), 556–562. doi: 10.21873/invivo.12737
Cai, Z., Cheng, Y., Ma, Z., Liu, X., Ma, Q., Xia, Q., et al. (2018). Fine-mapping of QTLs for individual and total isoflavone content in soybean (Glycine max L.) using a high-density genetic map. Theor. Appl. Genet. 131 (3), 555–568. doi: 10.1007/s00122-017-3018-x
Chen, Q., Jiang, T., Liu, Y. X., Liu, H., Zhao, T., Liu, Z., et al. (2019). Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci 62 (7), 947–958. doi: 10.1007/s11427-019-9521-2
Chu, D., Zhang, Z., Hu, Y., Fang, C., Xu, X., Yuan, J., et al. (2023). Genome-wide scan for oil quality reveals a coregulation mechanism of tocopherols and fatty acids in soybean seeds. Plant Commun. 4 (5), 100598. doi: 10.1016/j.xplc.2023.100598
Chu, S., Wang, J., Zhu, Y., Liu, S., Zhou, X., Zhang, H., et al. (2017). An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PloS Genet. 13 (5), e1006770. doi: 10.1371/journal.pgen.1006770
Dastmalchi, M., Bernards, M. A., Dhaubhadel, S. (2016). Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 85 (6), 689–706. doi: 10.1111/tpj.13137
Del Cerro, P., Megias, M., Lopez-Baena, F. J., Gil-Serrano, A., Perez-Montano, F., Ollero, F. J. (2019). Osmotic stress activates nif and fix genes and induces the Rhizobium tropici CIAT 899 Nod factor production via NodD2 by up-regulation of the nodA2 operon and the nodA3 gene. PloS One 14 (3), e0213298. doi: 10.1371/journal.pone.0213298
Dixon, R. A., Pasinetti, G. M. (2010). Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol. 154 (2), 453–457. doi: 10.1104/pp.110.161430
Dixon, R. A., Steele, C. L. (1999). Flavonoids and isoflavonoids - a gold mine for metabolic engineering. Trends Plant Sci. 4 (10), 394–400. doi: 10.1016/s1360-1385(99)01471-5
Feng, Y., Zhang, S., Li, J., Pei, R., Tian, L., Qi, J., et al. (2023). Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean. New Phytol. 237 (5), 1794–1809. doi: 10.1111/nph.18610
Graham, T. L., Graham, M. Y., Subramanian, S., Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144 (2), 728–740. doi: 10.1104/pp.107.097865
Grant, D., Nelson, R. T., Cannon, S. B., Shoemaker, R. C. (2010). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38 (Database issue), D843–D846. doi: 10.1093/nar/gkp798
Gupta, A., Awasthi, P., Sharma, N., Parveen, S., Vats, R. P., Singh, N., et al. (2022). Medicarpin confers powdery mildew resistance in Medicago truncatula and activates the salicylic acid signalling pathway. Mol. Plant Pathol. 23 (7), 966–983. doi: 10.1111/mpp.13202
Gupta, O. P., Dahuja, A., Sachdev, A., Kumari, S., Jain, P. K., Vinutha, T., et al. (2019). Conserved miRNAs modulate the expression of potential transcription factors of isoflavonoid biosynthetic pathway in soybean seeds. Mol. Biol. Rep. 46 (4), 3713–3730. doi: 10.1007/s11033-019-04814-7
Gupta, O. P., Nigam, D., Dahuja, A., Kumar, S., Vinutha, T., Sachdev, A., et al. (2017). Regulation of isoflavone biosynthesis by miRNAs in two contrasting soybean genotypes at different seed developmental stages. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00567
Hammerschmidt, R. (1999). Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306. doi: 10.1146/annurev.phyto.37.1.285
Han, F., He, X., Chen, W., Gai, H., Bai, X., He, Y., et al. (2020). Involvement of a novel tetR-like regulator (BdtR) of Bradyrhizobium diazoefficiens in the efflux of isoflavonoid genistein. Mol. Plant Microbe Interact. 33 (12), 1411–1423. doi: 10.1094/MPMI-08-20-0243-R
Jahan, M. A., Harris, B., Lowery, M., Coburn, K., Infante, A. M., Percifield, R. J., et al. (2019). The NAC family transcription factor GmNAC42-1 regulates biosynthesis of the anticancer and neuroprotective glyceollins in soybean. BMC Genomics 20 (1), 149. doi: 10.1186/s12864-019-5524-5
Jung, W., Yu, O., Lau, S. M., O'Keefe, D. P., Odell, J., Fader, G., et al. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18 (2), 208–212. doi: 10.1038/72671
Kim, J. M., Lyu, J. I., Kim, D., Hung, N. N., Seo, J. S., Ahn, J., et al. (2022). Genome wide association study to detect genetic regions related to isoflavone content in a mutant soybean population derived from radiation breeding. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.968466
Krizova, L., Dadakova, K., Kasparovska, J., Kasparovsky, T. (2019). Isoflavones. Molecules 24 (6), 1076. doi: 10.3390/molecules24061076
Lapcík, O. (2007). Isoflavonoids in non-leguminous taxa: a rarity or a rule? Phytochemistry 68 (22-24), 2909–2916. doi: 10.1016/j.phytochem.2007.08.006
LatundeDada, A. O., CabelloHurtado, F., Czittrich, N., Didierjean, L., Schopfer, C., Hertkorn, N., et al. (2001). Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. J. Biol. Chem. 276 (3), 1688–1695. doi: 10.1074/jbc.M006277200
Li, Y., Zhang, H. (2017). Soybean isoflavones ameliorate ischemic cardiomyopathy by activating Nrf2-mediated antioxidant responses. Food Funct. 8 (8), 2935–2944. doi: 10.1039/c7fo00342k
Lin, F., Chhapekar, S. S., Vieira, C. C., Da Silva, M. P., Rojas, A., Lee, D., et al. (2022). Breeding for disease resistance in soybean: a global perspective. Theor. Appl. Genet. 135 (11), 3773–3872. doi: 10.1007/s00122-022-04101-3
Liu, Q., Liu, Y., Li, G., Savolainen, O., Chen, Y., Nielsen, J. (2021). De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories. Nat. Commun. 12 (1), 6085. doi: 10.1038/s41467-021-26361-1
Liu, X. Q., Yuan, L. L., Xu, L., Xu, Z. L., Huang, Y. H., He, X. L., et al. (2013). Over-expression of GmMYB39 leads to an inhibition of the isoflavonoid biosynthesis in soybean (Glycine max. L). Plant Biotechnol. Rep. 7 (4), 445–455. doi: 10.1007/s11816-013-0283-2
Liu, Y., Zhang, Y., Liu, X., Shen, Y., Tian, D., Yang, X., et al. (2023). SoyOmics: A deeply integrated database on soybean multi-omics. Mol. Plant 16 (5), 794–797. doi: 10.1016/j.molp.2023.03.011
Lu, L. W., Chen, N., Brunder, D. G., Nayeem, F., Nagamani, M., Nishino, T. K., et al. (2022). Soy isoflavones decrease fibroglandular breast tissue measured by magnetic resonance imaging in premenopausal women: a 2-year randomized double-blind placebo controlled clinical trial. Clin. Nutr. ESPEN 52, 158–168. doi: 10.1016/j.clnesp.2022.10.007
Luo, J. (2015). Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 24, 31–38. doi: 10.1016/j.pbi.2015.01.006
Mackova, Z., Koblovska, R., Lapcik, O. (2006). Distribution of isoflavonoids in non-leguminous taxa - an update. Phytochemistry 67 (9), 849–855. doi: 10.1016/j.phytochem.2006.01.020
Masunaka, A., Hyakumachi, M., Takenaka, S. (2011). Plant growth-promoting fungus, Trichoderma koningi suppresses isoflavonoid phytoalexin vestitol production for colonization on/in the roots of Lotus japonicus. Microbes Environments 26 (2), 128–134. doi: 10.1264/jsme2.me10176
Matsuda, H., Yamazaki, Y., Moriyoshi, E., Nakayasu, M., Yamazaki, S., Aoki, Y., et al. (2023). Apoplast-localized beta-glucosidase elevates isoflavone accumulation in the soybean rhizosphere. Plant Cell Physiol. 64 (5), 486–500. doi: 10.1093/pcp/pcad012
Nabavi, S. M., Samec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 38, 107316. doi: 10.1016/j.bioteChadv.2018.11.005
Naoumkina, M., Farag, M. A., Sumner, L. W., Tang, Y., Liu, C. J., Dixon, R. A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 104 (46), 17909–17915. doi: 10.1073/pnas.0708697104
Ng, M. S., Ku, Y. S., Yung, W. S., Cheng, S. S., Man, C. K., Yang, L., et al. (2021). MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 22 (21), 12017. doi: 10.3390/ijms222112017
Ng, T. B., Ye, X. J., Wong, J. H., Fang, E. F., Chan, Y. S., Pan, W., et al. (2011). Glyceollin, a soybean phytoalexin with medicinal properties. Appl. Microbiol. Biotechnol. 90 (1), 59–68. doi: 10.1007/s00253-011-3169-7
Okutani, F., Hamamoto, S., Aoki, Y., Nakayasu, M., Nihei, N., Nishimura, T., et al. (2020). Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ. 43 (4), 1036–1046. doi: 10.1111/pce.13708
Pawlowski, J. W., Martin, B. R., McCabe, G. P., McCabe, L., Jackson, G. S., Peacock, M., et al. (2015). Impact of equol-producing capacity and soy-isoflavone profiles of supplements on bone calcium retention in postmenopausal women: a randomized crossover trial. Am. J. Clin. Nutr. 102 (3), 695–703. doi: 10.3945/ajcn.114.093906
Peck, M. C., Fisher, R. F., Long, S. R. (2006). Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J. Bacteriol. 188 (15), 5417–5427. doi: 10.1128/JB.00376-06
Pei, R., Zhang, J., Tian, L., Zhang, S., Han, F., Yan, S., et al. (2018). Identification of novel QTL associated with soybean isoflavone content. Crop J. 6 (3), 244–252. doi: 10.1016/j.cj.2017.10.004
Pokhrel, S., Ponniah, S. K., Jia, Y. L., Yu, O., Manoharan, M. (2021). Transgenic rice expressing isoflavone synthase gene from soybean shows resistance against blast fungus (Magnaporthe oryzae). Plant Dis. 105 (10), 3141–3146. doi: 10.1094/Pdis-08-20-1777-Re
Polturak, G., Misra, R. C., El-Demerdash, A., Owen, C., Steed, A., McDonald, H. P., et al. (2023). Discovery of isoflavone phytoalexins in wheat reveals an alternative route to isoflavonoid biosynthesis. Nat. Commun. 14 (1), 6977. doi: 10.1038/s41467-023-42464-3
Reynaud, J., Guilet, D., Terreux, R., Lussignol, M., Walchshofer, N. (2005). Isoflavonoids in non-leguminous families: an update. Nat. Prod. Rep. 22 (4), 504–515. doi: 10.1039/b416248j
Rizzo, G., Baroni, L. (2018). Soy, soy foods and their role in vegetarian diets. Nutrients 10 (1), 43. doi: 10.3390/nu10010043
Rush, T. A., Puech-Pages, V., Bascaules, A., Jargeat, P., Maillet, F., Haouy, A., et al. (2020). Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 11 (1), 3897. doi: 10.1038/s41467-020-17615-5
Sajid, M., Stone, S. R., Kaur, P. (2021). Recent advances in heterologous synthesis paving way for future green-modular bioindustries: a review with special reference to isoflavonoids. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.673270
Sathyapalan, T., Aye, M., Rigby, A. S., Thatcher, N. J., Dargham, S. R., Kilpatrick, E. S., et al. (2018). Soy isoflavones improve cardiovascular disease risk markers in women during the early menopause. Nutr. Metab. Cardiovasc. Dis. 28 (7), 691–697. doi: 10.1016/j.numecd.2018.03.007
Sohn, S. I., Pandian, S., Oh, Y. J., Kang, H. J., Cho, W. S., Cho, Y. S. (2021). Metabolic engineering of isoflavones: an updated overview. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.670103
Squadrito, F., Imbalzano, E., Rottura, M., Arcoraci, V., Pallio, G., Catalano, A., et al. (2023). Effects of genistein aglycone in glucocorticoid induced osteoporosis: a randomized clinical trial in comparison with alendronate. BioMed. Pharmacother. 163, 114821. doi: 10.1016/j.biopha.2023.114821
Sreevidya, V. S., Srinivasa Rao, C., Sullia, S. B., Ladha, J. K., Reddy, P. M. (2006). Metabolic engineering of rice with soybean isoflavone synthase for promoting nodulation gene expression in rhizobia. J. Exp. Bot. 57 (9), 1957–1969. doi: 10.1093/jxb/erj143
Subramanian, S., Stacey, G., Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48 (2), 261–273. doi: 10.1111/j.1365-313X.2006.02874.x
Sugiyama, A. (2019). The soybean rhizosphere: Metabolites, microbes, and beyond-A review. J. Adv. Res. 19, 67–73. doi: 10.1016/j.jare.2019.03.005
Sugiyama, A., Shitan, N., Yazaki, K. (2007). Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol. 144 (4), 2000–2008. doi: 10.1104/pp.107.096727
Tiscione, D., Gallelli, L., Tamanini, I., Luciani, L. G., Verze, P., Palmieri, A., et al. (2017). Daidzein plus isolase associated with zinc improves clinical symptoms and quality of life in patients with LUTS due to benign prostatic hyperplasia: results from a phase I-II study. Arch. Ital Urol. Androl. 89 (1), 12–16. doi: 10.4081/aiua.2017.1.12
Veitch, N. C. (2007). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 24 (2), 417–464. doi: 10.1039/b511238a
Veitch, N. C. (2009). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 26 (6), 776–802. doi: 10.1039/b616809b
Veitch, N. C. (2013). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 30 (7), 988–1027. doi: 10.1039/c3np70024k
Vina, J., Escudero, J., Baquero, M., Cebrian, M., Carbonell-Asins, J. A., Munoz, J. E., et al. (2022). Genistein effect on cognition in prodromal Alzheimer's disease patients. Genial Clin. Trial Alzheimers Res. Ther. 14 (1), 164. doi: 10.1186/s13195-022-01097-2
Wang, X., Zhang, J., Lu, X., Bai, Y., Wang, G. (2023). Two diversities meet in the rhizosphere: root specialized metabolites and microbiome. J. Genet. Genomics. doi: 10.1016/j.jgg.2023.10.004
Watanabe, S., Yamada, R., Kanetake, H., Kaga, A., Anai, T. (2019). Identification and characterization of a major QTL underlying soybean isoflavone malonylglycitin content. Breed Sci. 69 (4), 564–572. doi: 10.1270/jsbbs.19027
Wu, D., Li, D., Zhao, X., Zhan, Y., Teng, W., Qiu, L., et al. (2020). Identification of a candidate gene associated with isoflavone content in soybean seeds using genome-wide association and linkage mapping. Plant J. 104 (4), 950–963. doi: 10.1111/tpj.14972
Xie, C. L., Park, K. H., Kang, S. S., Cho, K. M., Lee, D. H. (2021). Isoflavone-enriched soybean leaves attenuate ovariectomy-induced osteoporosis in rats by anti-inflammatory activity. J. Sci. Food Agric. 101 (4), 1499–1506. doi: 10.1002/jsfa.10763
Yan, J. H., Wang, B., Zhong, Y. P., Yao, L. M., Cheng, L. J., Wu, T. L. (2015). The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89 (1-2), 35–48. doi: 10.1007/s11103-015-0349-3
Yang, J., Lan, L., Jin, Y., Yu, N., Wang, D., Wang, E. (2022). Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 64 (2), 244–267. doi: 10.1111/jipb.13207
Yi, J., Derynck, M. R., Li, X., Telmer, P., Marsolais, F., Dhaubhadel, S. (2010). A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62 (6), 1019–1034. doi: 10.1111/j.1365-313X.2010.04214.x
Yu, X., Yan, J., Li, Y., Cheng, J., Zheng, L., Fu, T., et al. (2023). Inhibition of castration-resistant prostate cancer growth by genistein through suppression of AKR1C3. Food Nutr. Res. 67, 9024. doi: 10.29219/fnr.v67.9024
Yuan, J., Ma, L., Wang, Y., Xu, X., Zhang, R., Wang, C., et al. (2023). A recently evolved BAHD acetyltransferase, responsible for bitter soyasaponin A production, is indispensable for soybean seed germination. J. Integr. Plant Biol. 65 (11), 2490–2504. doi: 10.1111/jipb.13553
Yuan, J. P., Liu, Y. B., Peng, J., Wang, J. H., Liu, X. (2009). Changes of isoflavone profile in the hypocotyls and cotyledons of soybeans during dry heating and germination. J. Agric. Food Chem. 57 (19), 9002–9010. doi: 10.1021/jf902248b
Zaheer, K., Humayoun Akhtar, M. (2017). An updated review of dietary isoflavones: Nutrition, processing, bioavailability and impacts on human health. Crit. Rev. Food Sci. Nutr. 57 (6), 1280–1293. doi: 10.1080/10408398.2014.989958
Zhang, P., Du, H., Wang, J., Pu, Y., Yang, C., Yan, R., et al. (2020). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18 (6), 1384–1395. doi: 10.1111/pbi.13302
Zhang, H., Gordon, R., Li, W., Yang, X., Pattanayak, A., Fowler, G., et al. (2019). Genistein treatment duration effects biomarkers of cell motility in human prostate. PloS One 14 (3), e0214078. doi: 10.1371/journal.pone.0214078
Zhao, J., Dixon, R. A. (2010). The 'ins' and 'outs' of flavonoid transport. Trends Plant Sci. 15 (2), 72–80. doi: 10.1016/j.tplants.2009.11.006
Keywords: isoflavonoids, metabolism, human health, nodulation, plant-microbe interactions
Citation: Yang Q and Wang G (2024) Isoflavonoid metabolism in leguminous plants: an update and perspectives. Front. Plant Sci. 15:1368870. doi: 10.3389/fpls.2024.1368870
Received: 11 January 2024; Accepted: 29 January 2024;
Published: 09 February 2024.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Xianzhi He, North Carolina State University, United StatesCopyright © 2024 Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, Z2R3YW5nQGdlbmV0aWNzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.