- 1Key Laboratory of Land Resources Evaluation and Monitoring in Southwest, Sichuan Normal University, Ministry of Education, Chengdu, China
- 2College of Life Sciences, Sichuan Normal University, Chengdu, China
- 3Sichuan Environmental Monitoring Center, Chengdu, China
- 4College of Life Sciences, Hunan Normal University, Changsha, China
- 5Sustainable Development Research Center of Resources and Environment of Western Sichuan, Sichuan Normal University, Chengdu, China
Aster L. is an economically and phylogenetically important genus in the tribe Astereae. Here, the complete plastomes of the eight Aster species were assembled and characterized using next-generation sequencing datasets. The results indicated the complete plastomes of Aster had a quadripartite structure. These genomes were 152,045–152,729 bp in length and contained 132–133 genes, including 87 protein-coding genes, 37–38 tRNA genes, and eight rRNA genes. Expansion or contraction of inverted repeat regions and forward, palindromic, complement, and reverse repeats were detected in the eight Aster species. Additionally, our analyses showed the richest type of simple sequence repeats was A/T mononucleotides, and 14 highly variable regions were discovered by analyzing the border regions, sequence divergence, and hotspots. Phylogenetic analyses indicated that 27 species in Astereae were clustered into six clades, i.e., A to D, North American, and outgroup clades, and supported that the genera Heteropappus, Kalimeris, and Heteroplexis are nested within Aster. The results indicated the clades B to D might be considered as genera. Divergence time estimate showed the clades A, B, C, and D diverged at 23.15 Mya, 15.13 Mya, 24.29 Mya, and 21.66 Mya, respectively. These results shed light on the phylogenetic relationships of Aster and provided new information on species identification of Aster and its related genera.
1 Introduction
The tribe Astereaehas has ~222 genera and ~3,100 species, which is the second largest tribe of Asteraceae (Noyes and Rieseberg, 1999; Brouillet et al., 2004; Panero et al., 2004; Panero and Crozier, 2016; Fu et al., 2016). The tribe Senecioneae has over 150 genera and 3,500 species (Nordenstam, 2007), more than the species number of the tribe Astereae. Aster is one of the large genera of Astereae and contains more than 152 species. The majority of Aster species are distributed in Eurasia, with only one species reaching North America (Nesom, 1994a, b; Chen et al., 2011). The species of Aster are mainly perennial herbs and are rarely annual or biennial herbs, subshrubs, or shrubs. The genus is characterized by capitula solitary or arranged in corymbiform or, sometimes, paniculiform capitulescences; white, pink, purple, or blue ray florets; and phyllaries imbricate or arranged in two equal layers.
Traditionally, Aster was defined as a genus encompassing around 300 species distributed in both the New World and the Old World (Jones, 1980; Semple and Brouillet, 1980; Jones and Young, 1983). However, in recent years, studies on the basis of morphology (Nesom, 1994a; Nesom and Robinson, 2007), Restriction Fragment Length Polymorphism (RFLPs) (Xiang and Semple, 1994), or DNA markers (Noyes and Rieseberg, 1999; Selliah and Brouillet, 2008; Li et al., 2012; Jafari et al., 2015; Korolyuk et al., 2015) have shown that the New World Aster species were distinct from the Old World taxa with a considerable genetic divergence. These New World taxa were treated as 13 separate genera (Nesom, 1994a, b), and the generic delimitation of the Old World species remained controversial. Some studies accepted a border Aster s.l., which includes most or all of the species of Aster from the Old World (Merxmüller et al., 1976; Ito et al., 1995; Chen et al., 2011). On the contrary, other studies treated the Old World species into Aster s.s. and 12 segregated genera (e.g., Kalimeris Cass., Heteroplexis C.C.Chang, Heteropappus Less.) (Tamamschjan, 1959; Grierson, 1975; Nesom, 1994b; Nesom and Robinson, 2007; Chen et al., 2011). However, recent molecular phylogenetic analyses have suggested that neither Aster s.l. nor Aster s.s. was monophyletic (Selliah and Brouillet, 2008; Pelser et al., 2010; Li et al., 2012; Jafari et al., 2015; Korolyuk et al., 2015; Fu et al., 2019). On the basis of analyses using Internal Transcribed Spacer (ITS), Enternal Transcribed Spacer (ETS), and trnL-F sequences, Li et al. (Li et al., 2012). showed that Eurasian Aster (referred to as EA Aster hereafter) is polyphyletic and supported that the genera Kalimeris and Heteropappus belonged to Aster. In addition, Li et al. (Li et al., 2012). proposed and suggested that Aster section Alpigenia, Aster ser. Albescentes, and Aster ser. Hersileoides should be elevated to the generic rank. However, the taxonomic position of Aster pycnophyllus Franch. ex Diels. remained unresolved. Another phylogenetic study using ITS and psbA-trnH sequences showed that the genera of Heteropappus and Kalimeris were nested within Aster, supporting the results of Jafari et al. (Jafari et al., 2015). Korolyuk et al. (Korolyuk et al., 2015). divided the Eurasian (EA) Aster into three groups, namely, a typical Eurasian asters group, Heteropappus group, and Asterothamnus group, but the relationships among these three groups were not strongly supported, and, hence, the boundary of Aster remained unclear. Although, the previous studies have indicated that the non-monophyly of Aster, the insufficient sampling of species, and low coverage and inadequate informative sites of molecular markers hampered the resolution of the phylogenetic trees of Aster and its related genera.
The chloroplast genome is one of the three DNA genomes, alongside the nuclear and mitochondrial genomes. In general, it is inherited maternally and possesses a highly conserved circular DNA arrangement, typically ranging from 115 kb to 165 kb in size (Wicke et al., 2011; Daniell et al., 2016). The complete chloroplast genome is a quadripartite structure, consisting of a large single copy (LSC), a small single copy (SSC), and two inverted repeats (IRs) (Daniell et al., 2016). The length differences are mostly due to expansion/contraction of IR regions (Zhu et al., 2016) or gene losses (Magee et al., 2010). In addition, the complete sequences of chloroplast genomes are commonly used for phylogenetic reconstruction at lower taxonomic levels, e.g., within genus, and population genetic analyses in plants. The utilization of complete chloroplast genomes has become widespread as an efficient tool for molecular phylogenetics in Aster (Kumar et al., 2009; Choi and Park, 2015; Zhang et al., 2015; Shen et al., 2017; Wang et al., 2019; Zhang et al., 2019a; Zhang X. et al., 2021; Duan et al., 2022; Palazzesi et al., 2022; Wang and Liu, 2023) and other tribes of Asteraceeae (Vargas et al., 2017; Do et al., 2019; Tyagi et al., 2019; Zhang et al., 2019b; Yu et al., 2022; Liu et al., 2023). Previous studies on Aster classification used one to several molecular markers, such as ITS, ETS, trnL-F, and psbA-trnH sequences (Li et al., 2012; Jafari et al., 2015), and some studies used only ITS sequences (Korolyuk et al., 2015), leaving many phylogenetic and taxonomic questions unresolved. Additionally, the lack of complete chloroplast genome sequences severely hampers the evaluation analyses of the genetic diversity of Aster germplasm resources.
In this study, to explore the genetic variation of Aster and its related genera, we report eight newly sequenced chloroplast genomes in the genus Aster, namely, Aster polius C.K. Schneid., Aster albescens Wall., Aster argyropholis Hand.-Mazz., Aster lavandulifolius Hand.-Mazz., Aster procerus Hemsl., A. pycnophyllus, Aster falcifolius Hand.-Mazz., and Aster yunnanensis Franch. The objectives of this study were to (1) analyze the evolution of chloroplast genomes within Aster using genetic comparative methods, (2) reconstruct the phylogenetic relationships of Aster and its related genera and further determine the phylogenetic backbone of Aster, and (3) estimate the divergence time of Aster and its related genera. This study provides new insights into the phylogenetics and evolution of Aster and its related genera and also shed the lights on the genetic diversity of Aster wild germplasm resources.
2 Materials and methods
2.1 Sampling, extraction, and genome sequencing
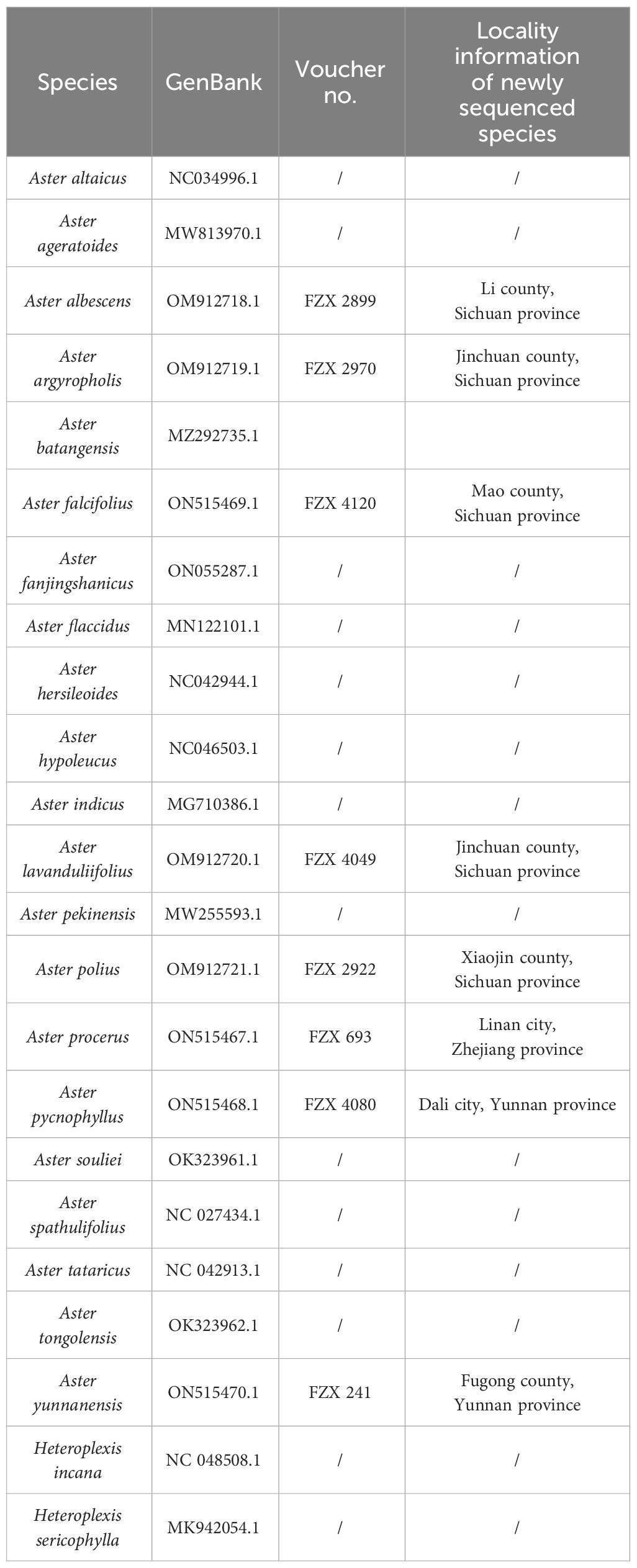
Fresh leaves of the eight Aster species were gathered from the wild (Table 1). The formal identification of the plant material was undertaken by Dr. Zhixi Fu. The voucher specimens were then preserved in the herbarium at Sichuan Normal University in China (SCNU) (contact person: Dr. Zhixi Fu,ZnV6eDIwMTdAc2ljbnUuZWR1LmNu). As these species are not included in List of National Key Protected Wild Plants in China, there was no need to obtain a permit for their collection. Following the CTAB DNA extraction protocol (Allen et al., 2006), genomic DNA was extracted using the Plant Genomic DNA Kit (Tiangen, Beijing, China). The construction of the DNA library was carried out using the Illumina Paired-End DNA Library Kit (Illumina Inc., San Diego, CA, USA), and, subsequently, sequencing was performed on the Illumina Genome Analyzer (Hiseq 2000, Illumina, San Diego, CA, USA). The resulting raw data for each of the eight species consisted of approximately 150-bp paired-end read lengths. The 27 complete chloroplast genomic datasets are available for download on NCBI (Table 2). In the final supermatrix, names of species were checked based on Flora of China (Chen et al., 2011).
2.2 Assembly and annotation of chloroplast genome
For the assembly of the chloroplast genome, the software SPAdes 3.15.1-Linux was employed, utilizing the default parameters (Prjibelski et al., 2020). To evaluate the assembly quality, the circular maps were identified using Bandage software (Wick et al., 2015). Subsequently, the resulting assembly was annotated using PGA (Qu et al., 2019), referencing the chloroplast genome sequence of Eschenbachia blinii (H.Lév.) (NC 037605.1). The annotation results were then checked using Geneious R11 (Kearse et al., 2012). The chloroplast genome map was visualized using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Additionally, the tRNA sequences were validated using tRNAscan-SE v2.0 (Chan et al., 2021), available on the Geseq platform (https://chlorobox.mpimp-golm.mpg.de/geseq.html). The annotated chloroplast genomes have been submitted to GenBank (Table 2). Analysis of plastid information was conducted using Geneious R11.
2.3 Comparative genome analysis
To identify potential IR expansion or contraction in eight Aster species, the reference species was used from A. ageratoides. This analysis was conducted using the perl script of Irscope (Amiryousefi et al., 2018). With the A. albescens as reference, the homology of these sequences was visualized using the mVISTA program (Frazer et al., 2004, https://genome.lbl.gov/vista/mvista/submit.shtml) with the LAGAN mode (Brudno et al., 2003).
2.4 Repeat sequences and SSR analysis
In this study, the identification of direct (forward), inverted (palindromic), complement, and reverse repeats elements was identified by REPuter (Kurtz et al., 2001), with maximum computed repeats equal to 50 bp, hamming distance of 3, and minimal repeat size of 30 bp. Furthermore, the detection of simple sequence repeats (SSRs) within the complete chloroplast genomes was performed using Microsatellite (MISA) (Beier et al., 2017). The thresholds for SSR detection were set to 10, 5, 4, 3, 3, and 3, for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively.
The alignment of all sequences from the eight Aster species was performed using the “–auto” strategy of Multiple Alignment using Fast Fourier Transform (MAFFT). Nucleotide diversity was then calculated using a sliding window approach in DnaSP v.6.12.03 (Rozas et al., 2017) with a window length of 600 bp and a step size of 200 bp.
2.5 Codon usage analysis
MEGA v 7.0 was used to analyze the synonymous codon usage and the relative synonymous codon usage (RSCU) of the Aster cp genomes. RSCU values >1 represent frequently used codons than expected, whereas values <1 signify the opposite. Codons having no preference value are set to 1.00.
2.6 Phylogenetic analysis
The phylogenetic analysis of the complete chloroplast genomic dataset, consisting of 27 species of Astereae, was performed using the maximum likelihood (ML) method implemented in RAxML. The species of Nannoglottis ravida and Llerasia caucana from basal group of Astereae were selected as outgroups (Stamatakis et al., 2008). The analysis was performed on the CIPRES platform (Miller et al., 2010) (https://www.phylo.org/portal2/). ModelTest (Posada, 2006) was employed to determine the most suitable model for the dataset. The molecular model GTRCAT was applied for the analysis. For bootstrap support assessment, the fast bootstrap option with 1,000 replicates was utilized in RAxML from CIPRES platform. The morphological identification characteristics of the genus Aster and its related genera have been described more clearly by Chen et al. (2011). Therefore, we define the key to the Aster and related species with reference to the criteria proposed by Chen et al. (2011) in combination with classification of previous studies (Li et al., 2012; Jafari et al., 2015; Korolyuk et al., 2015).
2.7 Divergence time estimations
For divergence time estimation, we used the complete chloroplast sequence dataset. The BEAST v.1.8 (Drummond et al., 2012) was applied to estimate the divergence times with Bayesian uncorrelated lognormal relaxed clock model. The node of Astereae was set at 31.42 Mya according to Zhang C. F. (Zhang C. F. et al. 2021). The tree Yule model was selected. The Markov chain Monte Carlo (MCMC) was run for 10,000,000 generations and sampled every 1,000 generations. TreeAnnotator v. 1.6 (BEAST package) was used to summarize and annotate the tree, with the initial 10% of trees discarded as burn-in. Finally, the tree was visualized in the program Figtree v.1.4.4 (http://tree.bio.ed.ac.uk/) with 95% highest posterior density being shown.
3 Results
3.1 Chloroplast genome structure and feature of Aster
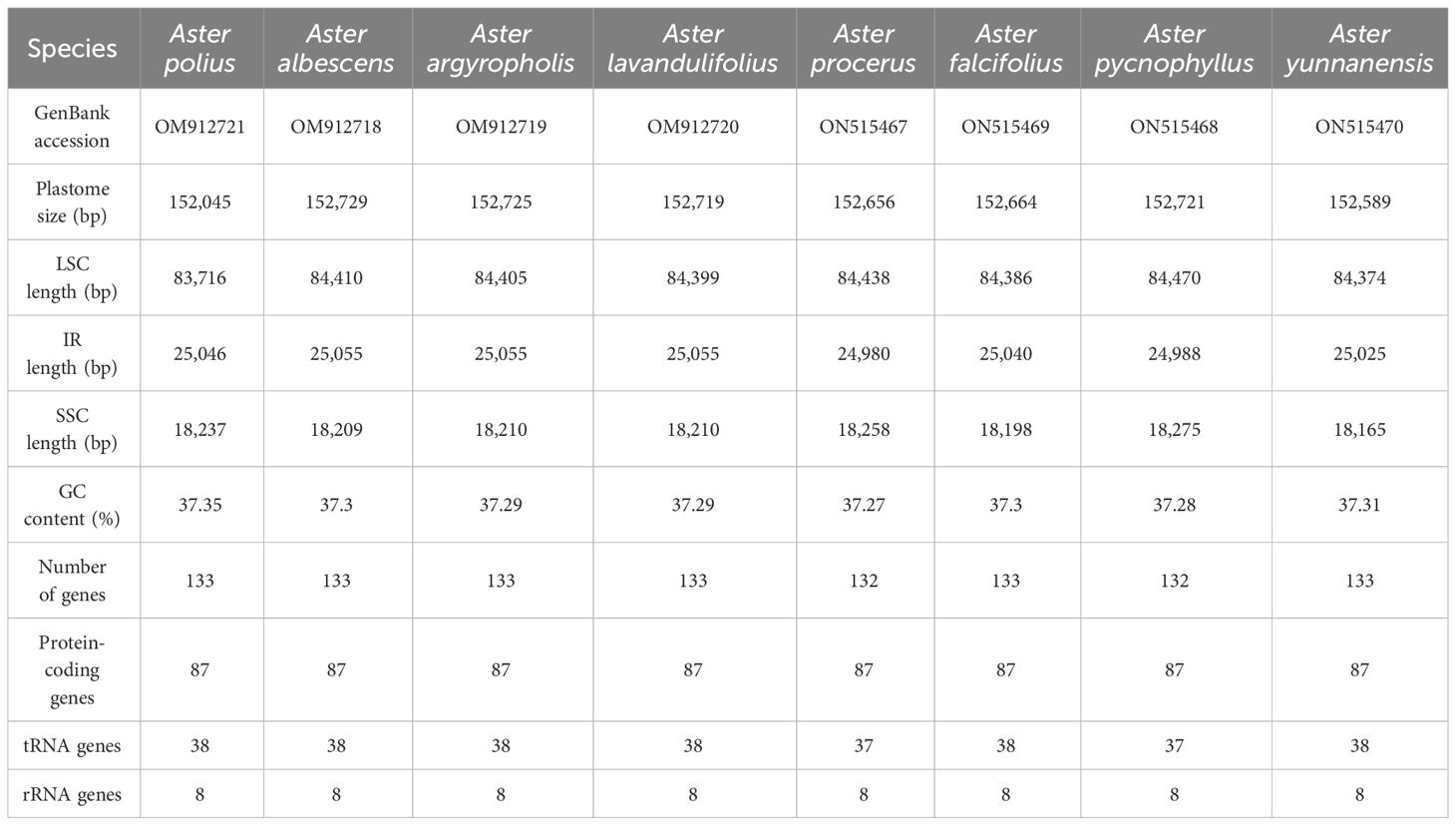
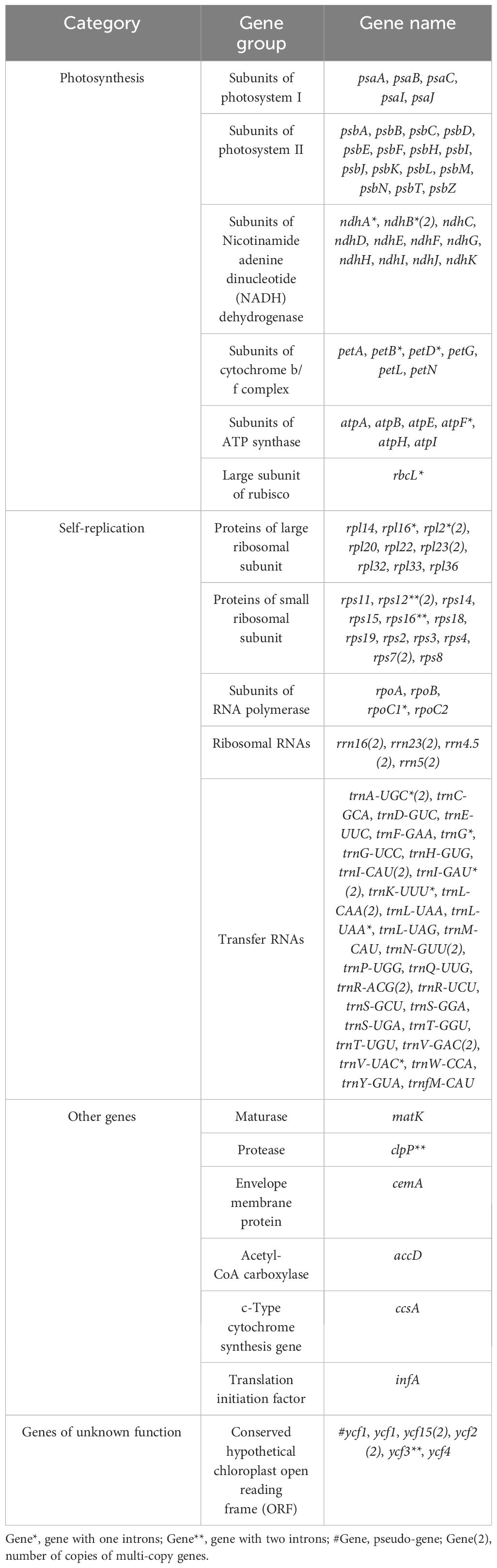
In this study, the complete chloroplast genomes of the eight species of Aster were sequenced and assembled. The results revealed a high degree of conservation in the structures of these genomes (Figure 1). These chloroplast genomes exhibited the standard quadripartite structure, consisting of a LSC region, a SSC region, and a pair of IR regions (IRa and IRb). The size of these genomes varied from 152,045 bp (A. polius) to 152,729 bp (A. albescens) (Table 2). The GC content ranged from 37.27% (A. procerus) to 37.35% (A. polius). Overall, all chloroplast genomes have 133 genes except A. procerus and A. pycnophyllus having 132 genes, including 87 protein-coding genes, 37/38 tRNA genes, and eight rRNA genes. Additionally, 115 of these genes were unique and 18 genes were duplicated in the IR regions (Table 3). The arrangement of these 133 genes in all chloroplast genomes was found to be completely collinear. There were two introns of four genes (rps12, rps12, ycf3, and clpP) and single intron of 16 genes (ndhA, ndhB, petB, petD, atpF, rbcL, rpl16, rpl2, rps16, rpoC1, trnA-UGC, trnG, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC). The gene rps12 was trans-spliced, and the genes ndhD and psbL experienced RNA editing.
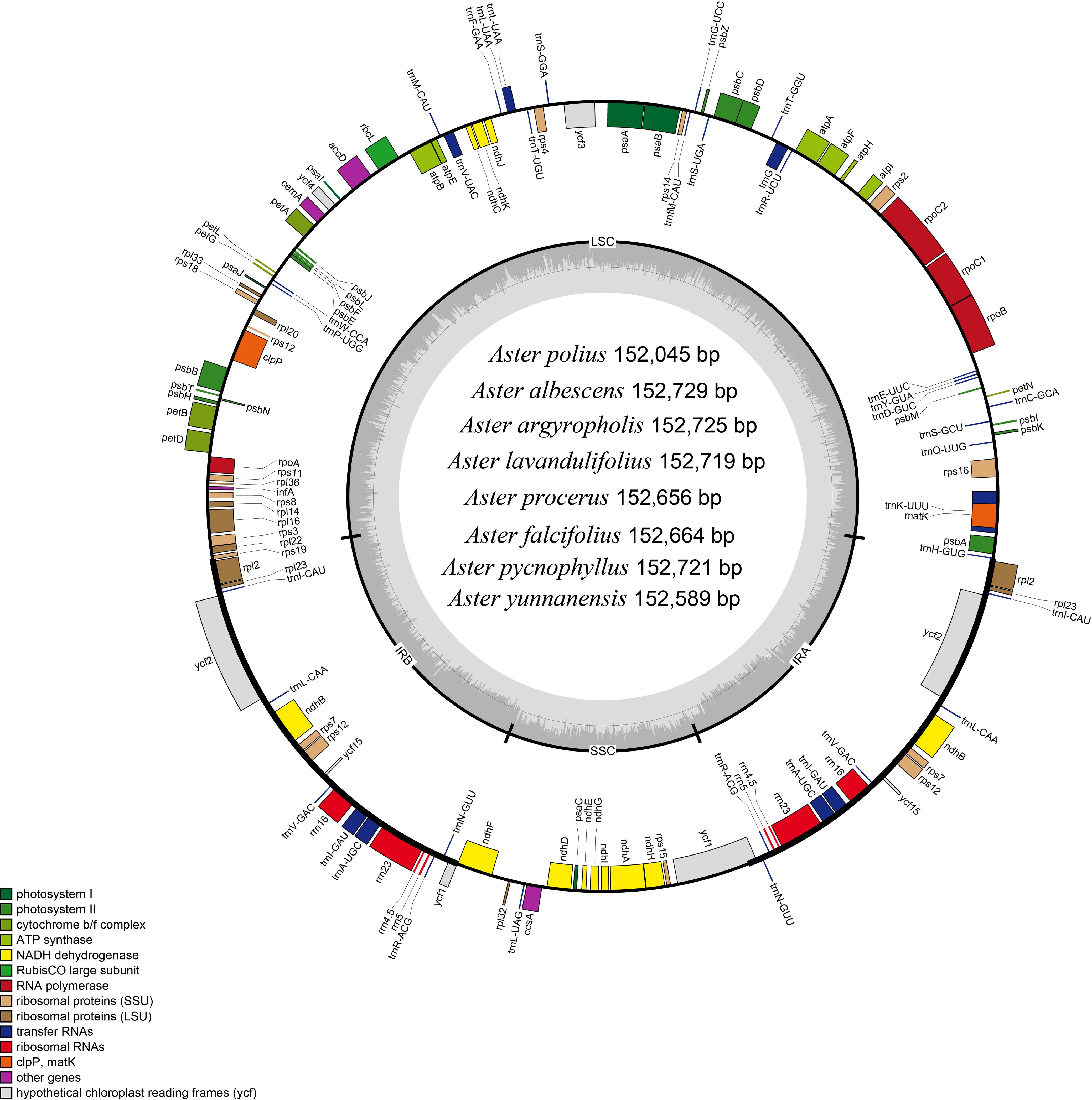
Figure 1 Gene map of the Aster chloroplast genome. Genes shown outside the outer circle are transcribed clockwise, and those insides are transcribed counterclockwise. Genes are color-coded according to different functional groups. The darker gray in the inner circle indicates the GC content, and the lighter gray indicates the AT content. The inner circle also shows that the chloroplast genome contains two copies of inverted repeats (IRA and IRB), a large single-copy (LSC) region, and a small single-copy (SSC) region.
3.2 Expansion and contraction of the border regions
In general, the IR/Single Copy (SC) expansion and contraction might cause the IR/SC junction position change. The IR/SC borders of the eight newly sequenced Aster chloroplast genomes were compared to analyze the expansion and contraction variation in junction regions (Figure 2). Although overall genomic structure including gene order and gene number was well conserved, these genomes exhibited slight differences at four junctions (JLB, JSB, JSA, and JLA). The rps19 gene of all Aster species located the JLBs, with the IRa region including 60 bp to 62 bp, except for A. pycnophyllus (36 bp). Likewise, the JLAs of all Aster species were located between rps12 and trnH. The ndhF gene, related to photosynthesis, was entirely located in the SSC region and the distance to the junction ranged from five to 54 bp. In our newly sequenced genomes, the ycf1 pseudogene was identified in all newly sequenced genomes. The main part of ycf1 gene was in the SSC region, with other 564 bp to 567 bp in the IRa region. The same fragment was also found in the IRb region of the ycf1 pseudogene and extended to SSC region with extension region with 9 bp to 147 bp.

Figure 2 Comparison of IR-SC border positions across plastomes of the eight Aster taxa. Genes are denoted by colored boxes. The gaps between the genes and the boundaries are indicated by the base lengths (bp).
3.3 Repeat sequence analysis
In the SSR analysis of the six species of Astereae, 75 (A. yunnanensis) to 99 (A. pycnophyllus) SSRs were found, showing a similar number of SSRs in Astereae (Figure 3A). In addition, these detected SSRs can be divided into six types, including mononucleotides (38%), dinucleotides (18.4%), trinucleotides (19.7%), tetranucleotides (17.9%), pentanucleotides (5.7%), and hexanucleotides (0.3%) (Figure 3B). The hexanucleotide repeats were only found in the chloroplast genomes of A. falcifolius and A. prorerus. The four dominant motif types of these SSRs were A/T (28–38), AT/AT (15–17), AAT/ATT (11–18), and AAAT/ATTT (7–10) (Figure 3C).
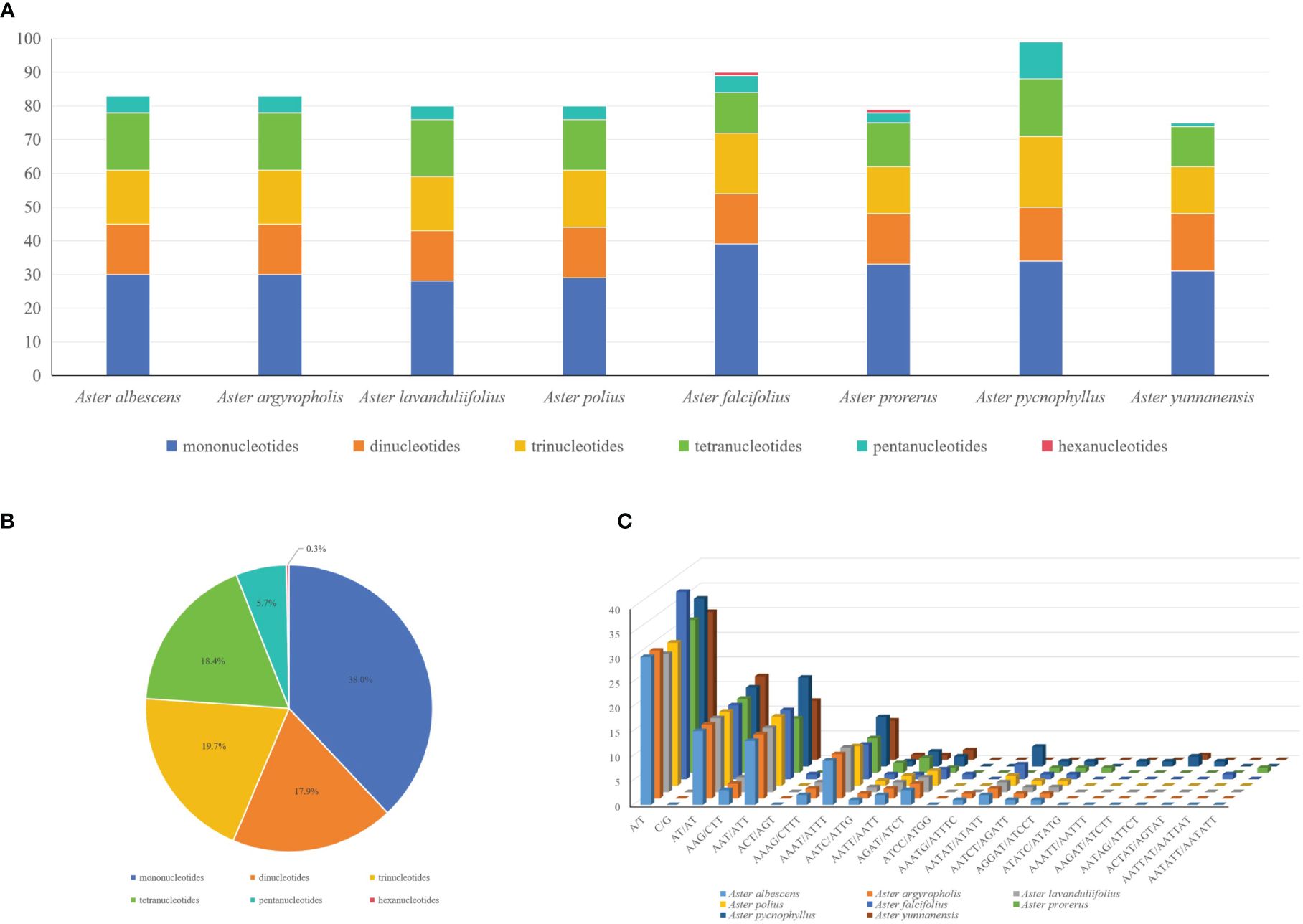
Figure 3 Comparison of simple sequence repeats (SSRs) among eight plastomes. (A) Numbers of SSRs detected in the eight newly sequenced Aster plastomes. (B) Frequencies of identified SSR types in all eight Aster plastomes. (C) Analysis of SSRs in eight Aster plastid genomes species.
The forward, palindromic, complement, and reverse repeats were detected in the eight newly sequenced chloroplast genomes (Figure 4A). A. procerus, A. pycnophyllus, A. yunnanensis, and A. falcifolius had all four type repeats. A. polius, A. albescens, A. argyropholis, and A. lavandulifolius had forward, palindromic, and reverse repeats. On average, 46–49 repeat sequences were identified in these genomes, with 17–23 forward repeats, 19–25 palindromic repeats, and 1–8 reverse repeats. However, complement repeats were only detected in A. procerus, A. pycnophyllus, A. yunnanensis, and A. falcifolius, with number of 1 to 3. Moreover, the repeats with 30 bp to 39 bp in length were the most common type in these genomes (Figure 4B), and none of the repeats with 50 bp to 59 bp in length. These interspersed repeat sequences were mainly present in the intergenic spacers, and several were observed within the coding regions and introns. The ycf15, rps12, ycf2, rrn5, rrn4.5, psbN, trnG, trnT-GGU, ycf4, cemA, trnS-GCU, trnS-UGA, trnS-GGA, psaB, psaA, accD, psal, psbE, petL, ndhD, and psaC genes contained LDRs. The interspersed repeat sequences were also more commonly detected in LSC and IR than SSC regions. The overall distribution of interspersed repeat sequences was similar in both IR regions.
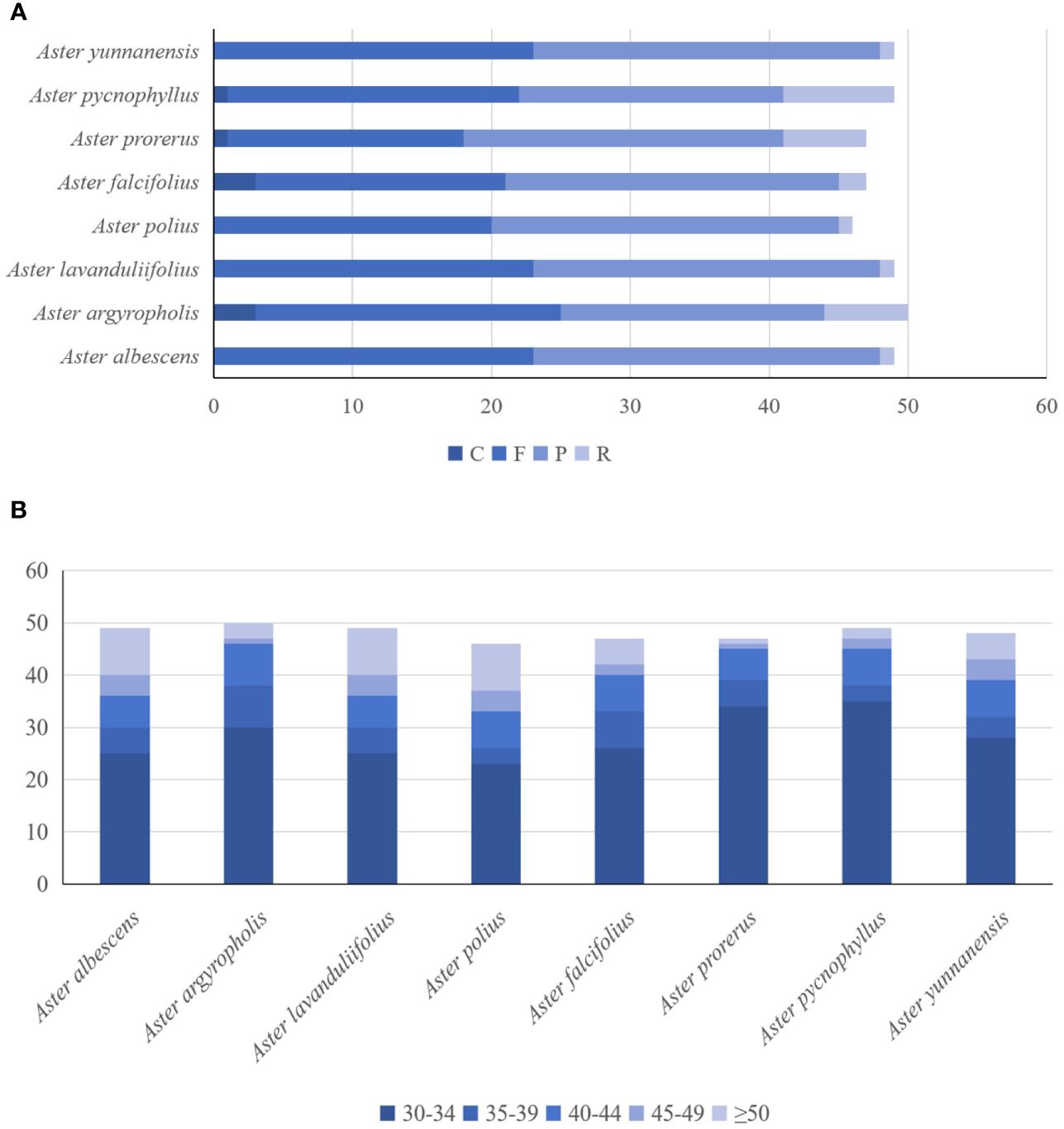
Figure 4 The analysis of the number and length of the long repeats identified from the eight Aster complete chloroplast genomes. The hamming distance of 3, the minimal repeats of 30, and the maximum repeats of 5,000 were applied during the calculating process. (A) Numbers of the type of long repeats contains F, P, R, and C. (B) Numbers of the length of long repeats.
3.4 Sequence divergence and hotspots
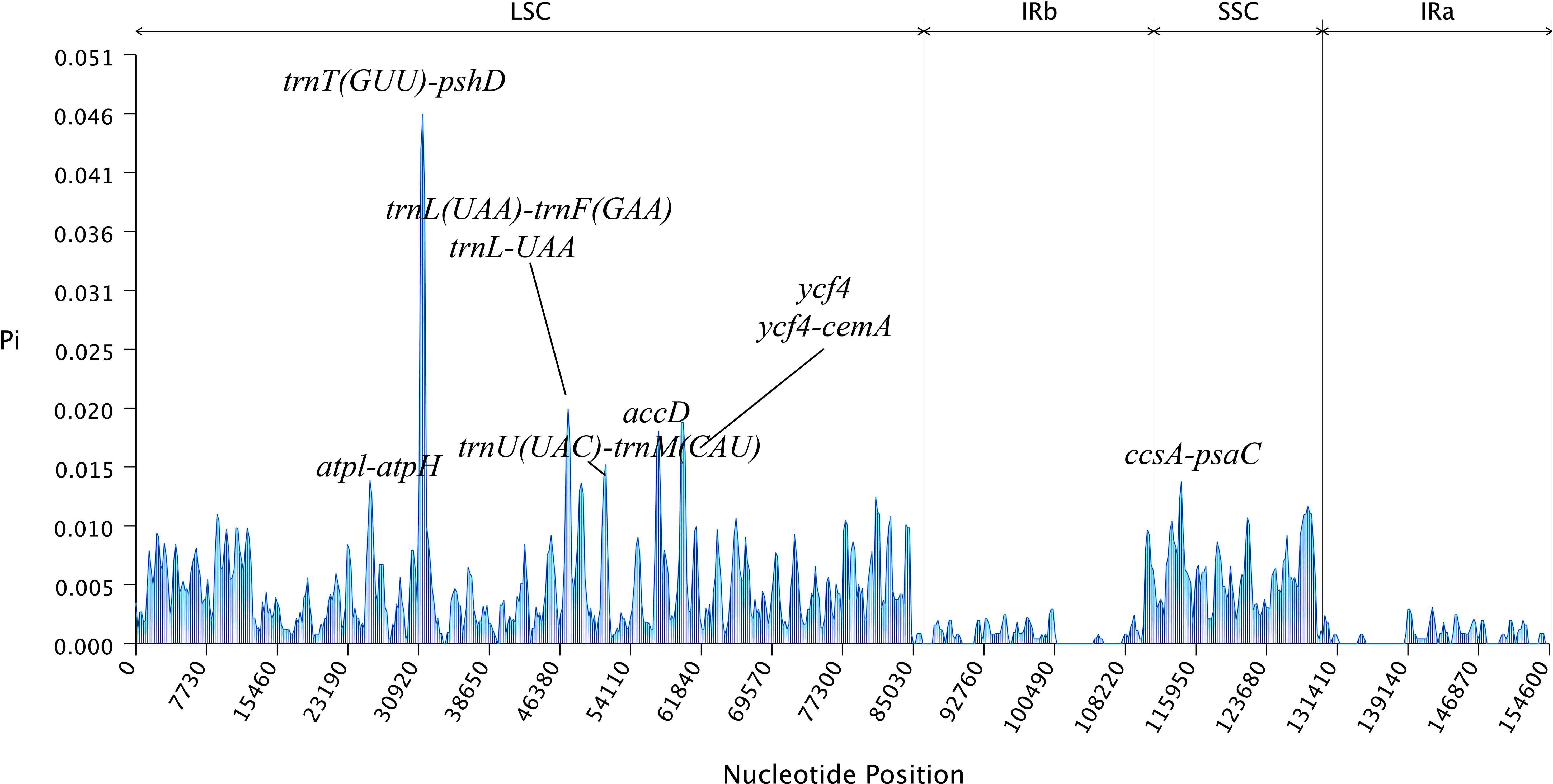
The mVISTA analysis of these eight Aster species indicated the complete chloroplast genome shared high levels of sequence similarity. Genetic variability was more prevalent in the non-coding regions than in the coding regions. The five genes with the highest variation were matK, atpA, rps19, ycf2, and ycf1 (Figure 5). DnaSP analysis revealed nucleotide diversity in single copy genes and intergenic regions with nucleotide diversity (Pi) ranged from 0.00068 to 0.04577. Six mutation hotspots showed significantly high Pi values (π > 0.014) (Figure 6), much higher than the average pi value (π = 0.0038). Among the gene coding regions, the highest Pi values were found in ndhF, followed by ndhC, trnV (UAC), and trnM (CAU). Among intergenic regions, the highest Pi values were detected in the rpl12-ndhF region, followed by ndhC–trnV (UAC), trnV (UAC)–trnM (CAU), and trnM (CAU)–atpE (Figure 6). We analyzed the nucleotide diversity of the complete chloroplast genomes and the LSC, SSC, and IR regions. The nucleotide diversity in the complete chloroplast genome was 0.0038, and higher nucleotide diversity was found in the LSC and SSC regions than the IR region, showing that the IR regions were more conserved than the single-copy regions. We found only two regions with π > 0.02, i.e., trnT (GGU)–psbD and trnL (UAA)–trnF (GAA), and three regions with π>0.015 and <0.02, i.e., trnU (UAC)–trnM (CAU), accD, and ycf4/ycf4-cemA (Figure 6).
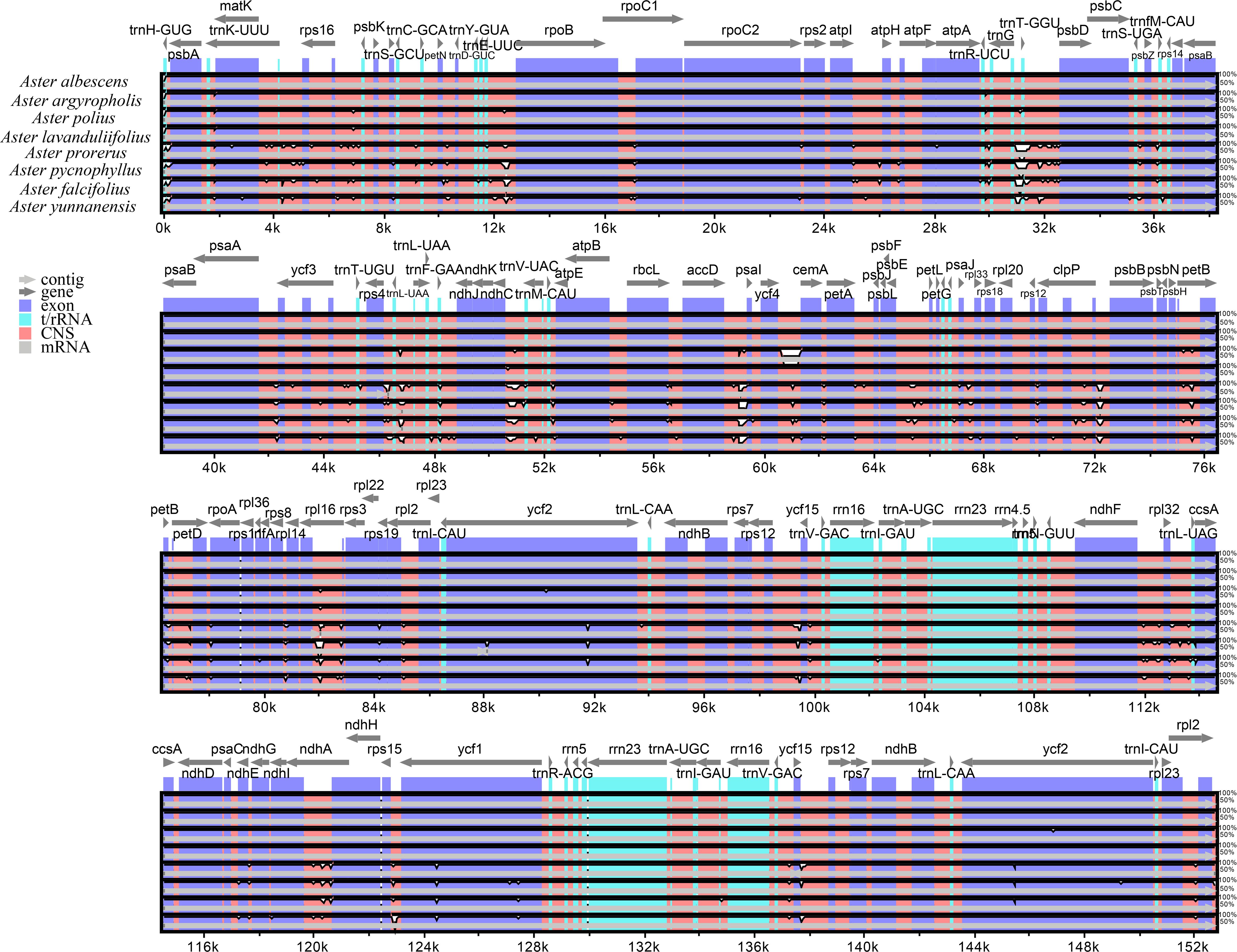
Figure 5 Comparison of the chloroplast genome of the eight Aster newly sequenced species. Dark blue bars represent protein-coding genes, pale blue bars represent rRNA genes, and red bars represent conserved non-coding sequences. The y-scale axis represents the percentage identity (50%–100%). mVISTA was used to perform the comparison.
3.5 Codon usage analysis
The preferences for codon are extremely similar among species. The analyses showed that 87 protein-coding genes were encoded by 64 codons (including three are stop codons: UAA, UGA, and UAG; Figure 7). The most prevalent amino acid was leucine. Leucine was encoded by CUA, CUC, CUG, CUU, UUA, and UUG with 2,420 codons (A. falcifolius) to 2,440 codons (A. yunnanensis). However, the rarest one was cysteine. Cysteine was encoded by UGC and UGU with 251 codons (A. falcifolius) to 252 codons (A. polius). In the complete chloroplast genome of these Aster, only codons tryptophan (encoded by UGG) exhibited no bias with RSCU = 1.00. The common start codon for the protein coding genes was AUG (M), except for the psbL, rps19, and ndhD genes, which have start codons of ACG, GUG, and GUG in all species, respectively.
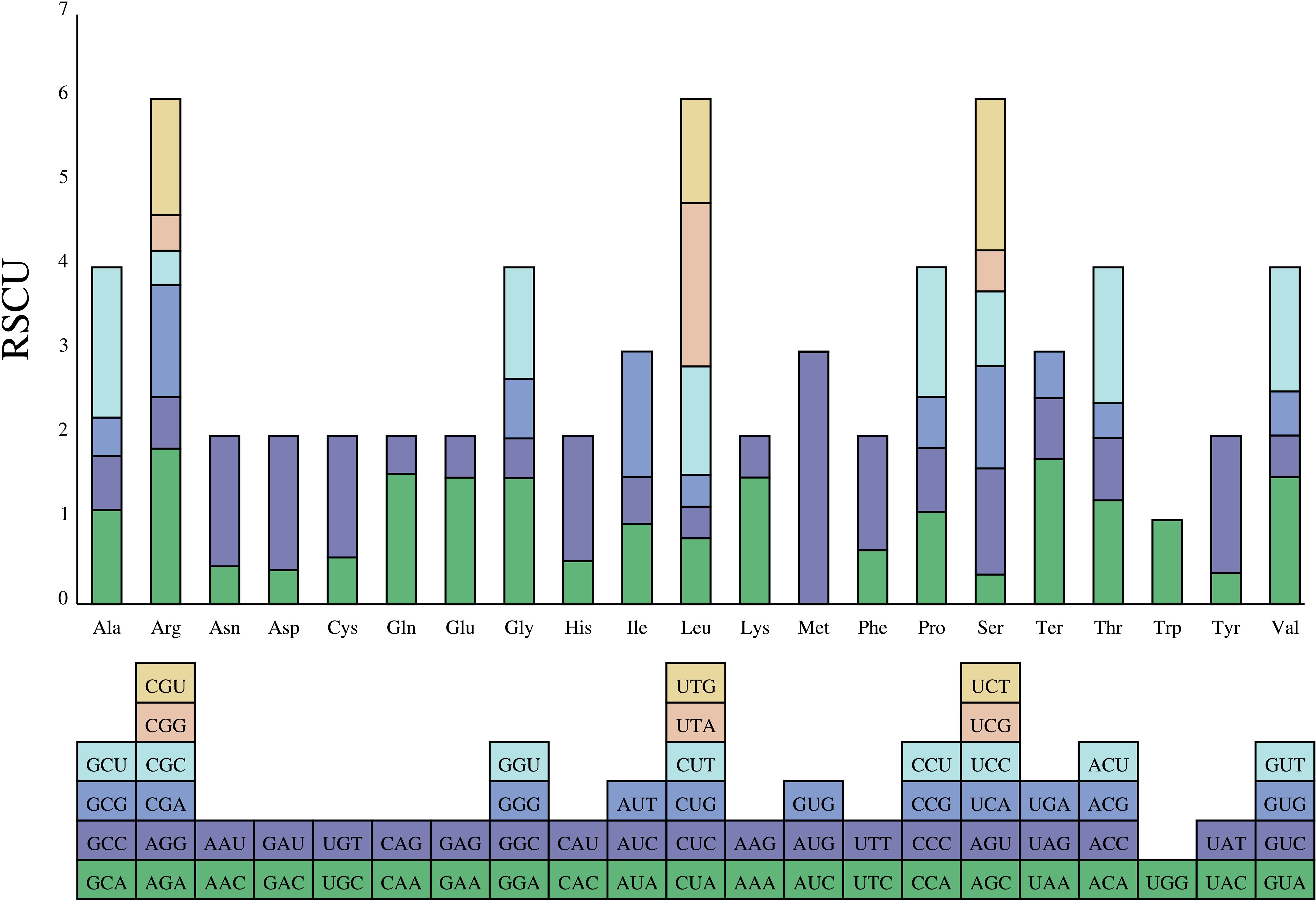
Figure 7 The RSCU values of the 20 amino acids of the complete chloroplast genome of the eight Aster taxa and their different codon usages.
3.6 Phylogenetic analysis
In this study, the complete chloroplast genomes of 27 Astereae species were used to perform phylogenetic reconstruction, with Nannoglottis ravida (C.Winkl.) Y.L.Chen and Llerasia caucana (S.F.Blake) Cuatrec used as outgroup. Phylogenetic analyses of the supermatrix of 25 taxa (not include outgroups) using the ML methods yielded a topology (Figure 8) with in-group fell into five clades: clade A, clade B, clade C, clade D, and North American clade (Figure 8). Clade A was the largest clade and was strongly supported (Bootstrap value (BP) = 97/100), containing 12 Aster species and two Heteroplexis species. The newly sequenced species, namely, A. falcifolius, A. pycnophyllus, and A. procerus, were nested in clade A. Other four newly sequenced species, namely, A. argyropholis, A. albescens, A. lavandulifolius, and A. polius, together with Aster hypoleucus Hand.-Mazz. formed the strongly supported clade B. A. hersileoides lonely formed clade C, as the sister group of clade B with a moderate support (BS = 68). Symphyotrichum subulatum (Michx.) G.L. Nesom and Erigeron canadensis L. formed the North American clade, and it was a sister group of clades A, B, and C with a high support (BS = 100). The newly sequenced A. yunnanensis were placed together with A. flaccidus and A. batangensis in clade D with strong support (BS = 100).

Figure 8 The best maximum likelihood (ML) phylogram inferred from 27 chloroplast genomes (bootstrap value are indicated on the branches). The circled species are the newly sequenced species in this study.
3.7 Divergence time estimations
On the basis of the newly reconstructed phylogeny, the origin and divergence times of lineages within the genus Aster were estimated (Figure 9). Divergent time estimate showed that the divergent time of clade A was dated back to 23.15 Mya. clades B, C, and D were divergent from 15.13 Mya, 24.29 Mya, and 21.66 Mya, respectively.
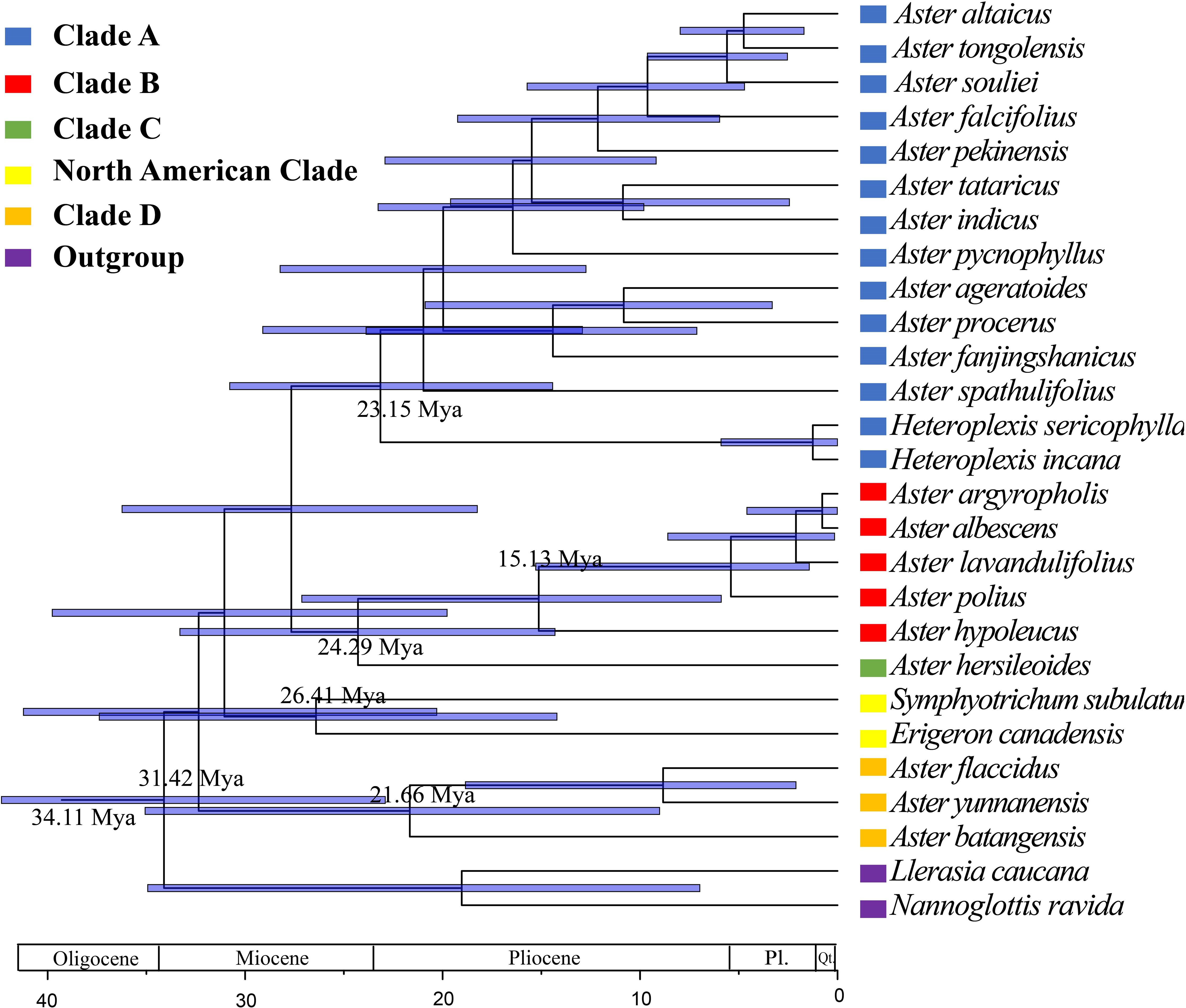
Figure 9 Divergence time estimates of Aster based on complete cp genomes, based on BEAST analysis using the complete chloroplast genomes dataset. Blue bars indicate 95% highest posterior density intervals.
4 Discussion
4.1 Plastome structure and characteristics analysis
The structure, gene position, size, orientation, and gene content of the plastid genomes of the eight Aster species were highly conserved (Doorduin et al., 2011; Curci et al., 2015; Cheon et al., 2017; Chen et al., 2018). These genomes of Aster have a standard quadripartite structure, including a LSC, a SSC, and a pair of IRs (IRa and IRb), which was the same as that reported for most other Aster (Shen et al., 2018; Wang et al., 2019; Zhang X. et al., 2021). The sizes of plastomes of the eight Aster species are between 152,045 bp and 152,729 bp (Table 2). In addition, these plastomes did not have any loss of introns. Additionally, the GC contents, which are a crucial factor in genome organization and stability, of the chloroplast genomes were low (37.3%) and were similar to that of other Asteraceae species, such as Aster spathulifolius Maxim. (37.28%) and A. hypoleucus (37.3%) (Ravi et al., 2008; Tyagi et al., 2019; Wang et al., 2019). We detected losses of the trnT (GGU) gene in A. procerus and A. pycnophyllus. In previous study, the loss of the tRNA was detected in some Asteraceae species (Lee et al., 2017).
4.2 Expansion and contraction of the border regions
The IR regions are known to be highly conserved in the genome of chloroplasts. During evolution, the expansion and contraction of the IR, LSC, and SSC regions are common, which leads to variability in genome length (Kim and Lee, 2004). In this study, the examination of chloroplast genome variation (Figure 2) showed that great expansion or contraction of the chloroplast IR region was not detected. However, ycf1 and ndhF genes located at the SSC/IR border had the slight variation in position and length in the eight Aster chloroplast genomes, suggesting boundary contraction and expansion between the SSC/IR regions in Aster (Liu et al., 2018).
4.3 Repeat sequence analysis
The SSRs are effective molecular markers, and they are often used for species identification and population genetic analyses (Thiel et al., 2003). In the eight Aster species analyzed here, A/T repeats, AT/AT repeats, AAT/ATT repeats, and AAAT/ATTT repeats were commonly detected (Figure 3C). This phenomenon may be related to that the AT preference pattern is widely reported in many plant plastids (Somaratne et al., 2019). In the rearrangement of the complete chloroplast genomes and sequence divergence, larger and more complex repeat sequences may play an important role (Weng et al., 2014). The interspersed repeat sequences were more prevalent in the non-coding regions than the coding regions (Kim et al., 2015). In our study, the ycf2 gene includes rich repeats, which contained many repeats: forward and palindromic. This result was consistent with the previous analysis that showed the gene has already been shown to be associated with many evolutionary events (Huang et al., 2010).
4.4 Sequence divergence and hotspots
DNA barcoding technology has been widely used in the species identification, phylogeny, and evolution (Doorduin et al., 2011; Palazzesi et al., 2022). In mVISTA analysis, the matK, atpA, rps19, ycf2, and ycf1 genes had large differences and were putative markers for population genetic and barcoding analyses (Figure 5). Among these genes, the matK and ycf1 genes have been used in previous plant phylogenetic and DNA barcoding analyses for land plants (Dong et al., 2015). Some regions of the plastomes of the eight Aster species showed high sequence divergence and might be used for phylogenetic reconstruction. However, these regions are different from the phylogenetic markers previously reported for Asteraceae (Do et al., 2019). Therefore, the complete chloroplast genome sequences and molecular markers might provide fundamental data for further studies on genus of Aster and related species in tribe Astereae.
4.5 Codon usage analysis
Thirty-one codons with RSCU value >1 were found, indicating that these codons are preferentially used in coding amino acids. An identical trend was discovered among the eight species (Figure 7). Leucine was the most abundant amino acid, whereas the cysteine was the least abundant amino acid, which is consistent with other Asteraceae species (Salih et al., 2017; Shen et al., 2017). In addition, most of A/U-ending codons had RSCU values >1; meanwhile, most of G/C-ending codons had RSCU values <1, indicating that amino acids tended to using A/U-ending codons, similar to a previous study (Zhao et al., 2021).
4.6 Phylogenetic analysis
In the tribe Astereae, there are numerous morphologically similar but distantly related taxa, such as some species of Aster (Noyes and Rieseberg, 1999). Whether some taxa should remain as genera or be merged into a single genus remains to be determined, such as genus Kalimeris, Heteropappus, and Heteroplexis, as wells as Aster series Albescentes, Aster Ser. Hersileoides, and Aster section Alpigenia (Nesom, 1994b; Noyes and Rieseberg, 1999; Li et al., 2012; Jafari et al., 2015; Korolyuk et al., 2015; Nesom, 2020). Nesom (1994b) proposed that Aster series Albescentes should be removed from Aster. In the Flora of China, Aster series Albescentes species A. nitidus and A. hersileoides were treated as the unplaced Aster group. The molecular phylogeny of Li et al. (2012) suggested that Aster section Alpigenia should be elevated to the new genera, series Albescentes is considered to be more closely related to section Alpigenia, and the Aster series Hersileoides is a well-supported monophyletic group. Therefore, according to the results of previous studies and the phylogenetic tree of this study, we classified the 25 species (not including outgroups) into five clades: clade A (core Aster), clade B (Aster series Albescentes), clade C (Aster Ser. Hersileoides), clade D (Alpine Aster, Aster section Alpigenia), and North American clade. Besides, the phylogenetic analysis of complete chloroplast genomes provided strong supports for these five clades (clade A, BS = 100; clade B, BS = 100; clade C, BS = 100; North American Clade, BS = 100; and clade D, BS = 100).
4.6.1 Clade A (core Aster)
The eight species of Aster formed clade A with high support (Figure 8): Aster tongolensis Franch., Aster souliei Franch., A. falcifolius, Aster tataricus L.f., A. pycnophyllus, Aster ageratoides Turcz., Aster fanjingshanicus Y.L.Chen & D.J.Liu, and A. spathulifolius. Additionally, six species of the closely related genera Kalimeris, Heteroplexis, and Heteropappus were also included within clade A, supporting the placement of Kalimeris, Heteroplexis, and Heteropappus within Aster. The general characteristics of Aster are as follows: large herbs, leaves cauline, basal leaves, and proximal leaves withered at anthesis usually, stem leaves well developed, nearly as long as basal leaves, capitula many, much branched, in corymbiform, terminal solitary rarely, involucres herbaceous or membranous, involucres 3-numerous, unequal, imbricate, 2-3(7)–ribbed, and secretory cavity few.
In the previous studies, Heteropappus was considered for generic rank based on the heteromorphic pappus of ray and disc flowers (Jones, 1980; Noyes and Rieseberg, 1999; Chen et al., 2011; Jafari et al., 2015). Based on RFLPs and gene sequences, it is suggested that Heteropappus altaicus should be classified within Aster (Ito et al., 1998; Li et al., 2012). Our study showed that Aster altaicus Willd. (=Heteropappus altaicus) belong to clade A (BS = 100), supporting the previous results. Li et al. (2012) proposed A. pycnophyllus should be kept separate as it was found to be nested within a clade with Myriactis Less. and distantly related to Aster. In our study, the result shown that A. pycnophyllus was nested within clade A, with a strong support (BS = 97).
The genus Kalimeris is defined by the compressed obovoid-oblong of achenes and short lobe only comprising K. indica (Ito et al., 1995, Ito et al., 1998; Chen et al., 2011). These traits have also been analyzed in previous studies. However, many species of Aster also exhibit similar characteristics such as Aster smithianus Hand.-Mazz., Aster souliei Franch., and Aster hunanensis Hand.-Mazz. Hybridization between Aster and Kalimeris was also observed frequently (Tara, 1972; Gu and Hoch, 1997; Li et al., 2012). Gu and Hoch (1997) revised the genus Kalimeris based on morphological and cytological evidence showing a close phylogenetic relationship between Kalimeris and Heteropappus. Using RFLPs and DNA molecular markers, Kalimeris was shown to be not an independent genus and embedded within the genus Aster (Ito et al., 1995; Li et al., 2012). In our study (Figure 8), Aster indicus L. (=Kalimeris indicus), Aster pekinensis (Homce) F.H.Chen (=Kalimeris pekinensis), and Aster procerus (=Kalimeris procerus) fall within clade A (BS = 100), supporting the including of Kalimeris in Aster.
The genus Heteroplexis, comprising five species, is an herb endemic to Guangxi, China (Chen et al., 2011). In the Flora of China, the genus Heteroplexis shares similarities in morphology and inflorescence with the genus Aster, but they could be distinguished by its bilaterally symmetrical corolla and climbing or erect herb (Chen et al., 2011). Therefore, it is placed within the subtribe Asterinae as close allies of Aster. Based on the number of outer flowers over the number of bisexual flowers, Zhang and Bremer (1993) treated Heteroplexis in Erigeron-Conyza group. According to some characters, e.g., disciform capitula, oblong-obovoid achenes, and long corolla lobes, Nesom (1994a) treated Heteroplexis as a member of Baccharidinae. In recent study, it is the unplaced Aster group (Nesom and Robinson, 2007). Our results suggested that Heteroplexis should be included with Aster and treated as a synonym of Aster.
4.6.2 Clade B (Aster series Albescentes)
The species of clade B exhibit a shrubby growth habit and are classified within the Aster series Albescentes (Li et al., 2012; Nesom, 2020). Based on the characters of muti-layers involucre and muti-ribs achene, Nesom (1994b) proposed that Aster series Albescentes has a distinct position in Aster. Based on the character of pappus, Nesom (1994b) noted that A. series Albescentes is sister to the NA Doellingeria Nees. In the Flora of China, the species of Aster ser. Albescentes were considered as the unplaced Aster group (Chen et al., 2011). Li et al. (2012) showed that ser. Albescentes is a monophyletic taxon with high support in a polytomy with Myriactis and other segregates of Aster s.s., implying that series Albescentes may belong to the Australasian lineages, in disagreement with the study of Nesom (1994b). In this study, A. albescens, A. argyropholis, A. lavandulifolius, A. polius, and A. hypoleucus formed a strong supported clade B (BS = 88/100) as sister of clade C (Aster Ser. Hersileoides) (Figure 8). It demonstrates that series Albescentes is a well-supported monophyletic genus. The newly defined taxon possesses the following distinct characteristics: shrubs, leaves cauline, basal leaves and proximal leaves withered at anthesis usually, non-rosulate, stem leaves well developed, nearly as long as basal leaves, capitula many, much branched, in corymbiform, terminal solitary rarely, involucres herbaceous or membranous, involucres 3-5, imbricate, margin membranous, irregularly lobed. margin membranous, irregularly lobed, and 4-5(8)–ribbed.
4.6.3 Clade C (Aster Ser. Hersileoides)
Aster series Hersileoides consists of two species, Aster hersileoides C.K.Schneid. and Aster nitidus C.C.Chang (Yin et al., 2010; Chen et al., 2011). Chen et al. (2011) treated the A. hersileoides within the unplaced status in Aster. Based on molecular phylogenetic studies, Li et al. (2012) strongly suggested that Aster ser. Hersileoides should be removed from Aster and considered as a separate genus. Our results supported that the series (represented by A. hersileoides) should be kept separately from Aster. They are characterized by shrubs, leaves cauline, non-rosulate, leaf oblanceolate and glabrous, capitula many, terminal solitary, involucres 3-5, imbricate, 2 inner involucres equaling, and 3-ribbed. It is reasonable to propose the elevation of the Aster Series Hersileoides to a generic rank, considering its unique traits and the phylogenetic results here. Further investigations and comprehensive molecular analyses will be essential in demonstrating the full taxonomic status and evolutionary relationships of this clade.
4.6.4 Clade D (Alpine Aster)
In our study, clade D contained Aster batangensis Bureau & Franch., Aster flaccidus Bunge, and A. yunnanensis (BS = 97). In previous study, these species have been placed in the genus Aster (Nesom, 1994b; Nesom and Robinson, 2007; Chen et al., 2011). Li et al. (2012) recognized that A. batangensis is closely allied with Aster senecioides Franch. and Aster fuscescens Bureau & Franch. from Aster section Alpigenia. However, based on morphologic differences, the study of Li et al. (2012) supported that A. batangensis might represent a monotypic genus. In our study, A. batangensis also has a distinct position in clade D. The clade has some distinctive characteristics: herbs dwarf, leaves rosulate, basal leaves at anthesis, cauline leaves reduced, significantly shorter than basal leaves, capitula solitary few, scapose, rarely branched, involucres herbaceous 2-3, subequal, non-imbricate, 3-4(6), secretory cavity, and secretory cavity few. Our molecular findings strongly support that clade D is an independent group.
In conclusion, the previously classification and definition of Aster is not monophyletic. Clade A (core Aster) includes most Aster taxa, Heteropappus, Kalimeris, and Heteroplexis. Additionally, clade B (Aster series Albescentes), clade C (Aster Ser. Hersileoides), and clade D (Alpine Aster) are identified as independent groups. Furthermore, it is estimated that the genus Aster comprise more than 152 species. However, this study only encompassed 25 species and two outgroup species. Therefore, a more comprehensive and extensive sampling of chloroplast genome and more data are necessary to conduct a thorough and comprehensive phylogenetic study of the genus Aster and its related genera. Based on the results of both general morphological and molecular phylogenetic analysis, the identification key was presented as following.
Key to the Aster and related species (clades A to D):
1. herbs, achenes 2-3(7)–ribbed, phyllaries 2-numerous-layers
2. herbs large or occasionally dwarf, achenes 2-3(7)–ribbed, phyllaries 3-numerous-layers……………………………………………………………….Clade A (core genus Aster)
2. herbs dwarf or occasionally large few, achenes 3-4(6)–ribbed, phyllaries 2-3-layers…………………………………………………………………………………………………………………………Clade D (alpine Aster)
1. shrubs, achenes (3)5-7-ribbed, phyllaries 3-numerous-layers
3. achenes 4-5(8)–ribbed………Clade B (Aster ser. Albescentes)
3. achenes 3 ribbed……….Clade C (Aster ser. Hersileoides)
4.7 Divergence time estimations
The divergence time estimation of Aster relied on secondary calibration because of the lack of fossil record for most Aster taxa. Most species of Aster and its related genera are distributed in East Asia (Nesom, 1994b; Brouillet et al., 2009; Chen et al., 2011). The result of Brouillet et al. (2009) indicated that Aster originated from a clade with a dispersal from Australasia into East Asia. The result of our molecular dating suggests that clades A and C began to diversify in the late Oligocene (23.15 Mya and 24.29Mya, respectively). Clades B and D originated in the Early Miocene (15.13 Mya and 21.66Mya, respectively). The rapid radiation may be related to collisions between geological plates (Audley-Charles, 1987; Liu et al., 2002). Geologic uplift events (first of which began at about 50 Ma) have taken place in the Tibetan Plateau during at least four different periods since the early Miocene, i.e., 22 Mya, 15–13 Mya, 8–7 Mya, and 3.5–1.6 Mya (Shi et al., 1998; Guo et al., 2002; Spicer et al., 2003). The origin of the four clades (Clade A-D) likely occurred independently at first two stages of the uplift and formation of the Tibetan Plateau. Geological evidence suggests that the strong uplift of the Tibetan Plateau, coupled with favorable oceanic and continental environments, produced a strong Asian monsoon dominated by the Summer Monsoon (Shi et al., 1998; Ding et al., 2020). During this uplift movement, the original Planetary Wind System in East Asia was changed and the arid zone retreated to the northwest (Shi et al., 1998; Ding et al., 2020). Eastern China was gradually covered by tropical or subtropical forests. This scenario also correlates with the current habitat preferences of the studied taxa (clade A, understorey vegetation; clades B and C, dry slopes and scree regions; and clade D, cold and dry alpine meadows). The similar rapid radiation has also been found in other groups of Asteraceae in the Tibetan Plateau, such as Saussurea (Liu et al., 2002) and the Dolomiaea-Diplazoptilon-Xanthopappus group (Wang et al., 2007).
5 Conclusions
The complete chloroplast genomes of the eight Aster species were sequenced in this study. The results revealed that cp genome size, structure, gene content, as well as compositional organization were highly conserved among these species. The chloroplast genomes of all species exhibited the standard quadripartite structure, and the size of these species of Aster varied from 152,045 bp to 152,729 bp. They include 87 protein-coding genes, 37/38 tRNA genes, and eight rRNA genes. They have three/four types of repeats, and the number of SSRs ranged from 75 to 99. Genes located at the junctions were well conserved among the Aster species. Furthermore, the genic and IR regions were more conserved than the intergenic and SC regions, respectively. In addition, the plastid genome structure of Aster exhibited high consistency and was obviously different in some regions, such as rps19, ycf1, and ndhf. Furthermore, the preferences for codon use in our study are all similar. The most prevalent amino acid was leucine, whereas the rarest one was cysteine. Moreover, we detected six hotspots that could be used as candidate DNA barcodes. The analysis of complete chloroplast genomes and combined datasets provided clear evidence supporting the moderate to strong differentiation of clades (clades A, B, C, and D and North American clade). The phylogenetic results showed that the traditionally defined Aster was not monophyly. For the delimitation of the genus Aster, Kalimeris, Heteropappus, and Heteroplexis, the closed allied genera of Aster were revealed to be nested within the Aster clade and should be included in Aster. Additionally, we suggest that the clade B (Aster series Albescentes), clade C (Aster Ser. Hersileoides), and clade D (Alpine Aster) should be treated as separated genera and taxonomic treatment. Divergent time estimate showed that the divergent time of clade A was dated back to 23.15Mya. Clades B, C, and D were divergent from 15.13 Mya, 24.29 Mya, and 21.66Mya, respectively. Our analyses suggested that the divergence of the genus Aster is closely related to the uplift of the Qinghai-Tibet Plateau. This study sequenced eight plastid genomes of Aster, provided a well resolved phylogenetic tree of Aster and related genera, and selected putative markers for further barcoding analysis. This study is important for us to understand the phylogeny and evolution of Aster and the further phylogenetic, population genetic, and related studies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OM912721; https://www.ncbi.nlm.nih.gov/, ON515470; https://www.ncbi.nlm.nih.gov/, ON515468; https://www.ncbi.nlm.nih.gov/, ON515469; https://www.ncbi.nlm.nih.gov/, ON515467; https://www.ncbi.nlm.nih.gov/, OM912720; https://www.ncbi.nlm.nih.gov/, OM912719; https://www.ncbi.nlm.nih.gov/, OM912718.
Author contributions
HC: Writing – original draft, Conceptualization. TL: Writing – original draft, Software, Methodology, Formal analysis. XC: Writing – original draft, Conceptualization. TQ: Writing – review & editing, Software, Methodology, Formal analysis. XZ: Writing – review & editing, Software, Methodology, Formal analysis. JL: Writing – original draft, Conceptualization. BL: Writing – review & editing, Supervision. GZ: Writing – review & editing, Supervision. ZF: Writing – review & editing, Supervision, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Natural Science Foundation of China (No. 32000158); the National Science and Technology Fundamental Resources Investigation Program of China (No. 2021XJKK0702); the Foundation of Sustainable Development Research Center of Resources and Environment of Western Sichuan, Sichuan Normal University (No. 2020CXZYHJZX03); Key Laboratory of Chemistry in Ethnic Medicinal Resources (Yunnan Minzu University); State Ethnic Affairs Commission and Ministry of Education (No. MZY2301); and Laboratory equipment research projects, Sichuan Normal University (No. SYJS2022014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer QM declared a past co-authorship with the author ZF to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, G. C., Flores-Vergara, M. A., Krasnyanski, S., Kumar, S., Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Audley-Charles, M. G. (1987). “Dispersal of Gondwanaland: relevance to evolution of the Angiosperms,” in Biogeographical Evolution of the Malay Archipelago. Ed. Whitemore, T. C. (Clarendon Press, Oxford).
Beier, S., Thiel, T., Munch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Brouillet, L., Lowrey, T. K., Urbatsch, L., Karaman-Castro, V., Sancho, G., Wagstaff, S., et al. (2009). “Astereae,” in Systematics, evolution and biogeography of the Compositae, vol. 37 . Eds. Funk, V. A., Susanna, A., Stuessy, T., Bayer, R. (IAPT, Vienna), 449–490.
Brouillet, L., Urbatsch, L., Roberts, R. P. (2004). Tonestus kingii and T. Aberrans are related to Eurybia and the Machaerantherinae (Asteraceae: Astereae) based on nrDNA (ITS and ETS) data: reinstatement of Herrickia and a new genue, Triniteurybia. Sida 21, 889–900.
Brudno, M., Do, C. B., Cooper, G. M., Kim, M. F., Davydov, E., Green, E. D., et al. (2003). LAGAN and multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13, 721–731. doi: 10.1101/gr.926603
Chan, P. P., Lin, B. Y., Mak, A. J., Lowe, T. M. (2021). tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096. doi: 10.1093/nar/gkab688
Chen, X. L., Zhou, J. G., Cui, Y. X., Wang, Y., Duan, B. Z., Yao, H. (2018). Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front. Pharmacol. 9. doi: 10.3389/fphar.2018.00695
Chen, Y. L., Brouillet, L., Semple, J. C. (2011). “Aster,” in Flora of China, vol. 20/21 . Eds. Wu, Z. Y., Raven, P. H. (Science Press, Beijing), 574–632.
Cheon, K. S., Kim, H. J., Han, J. S., Kim, K. A., Yoo, K. O. (2017). The complete chloroplast genome sequence of Saussurea chabyoungsanica (Asteraceae), an endemic to Korea. Conserv. Genet. Resour. 9, 51–53. doi: 10.1007/s12686-016-0617-9
Choi, K. S., Park, S. (2015). The complete chloroplast genome sequence of Aster spathuhfolius (Asteraceae), genomic features and relationship with Asteraceae. Gene 572, 214–221. doi: 10.1016/j.gene.2015.07.020
Curci, P. L., De, P. D., Danzi, D., Vendramin, G. G., Sonnante, G. (2015). Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PloS One 10, e0120589. doi: 10.1371/journal.pone.0120589
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Ding, W. N., Ree, R. H., Spicer, R. A., Xing, Y. W. (2020). Ancient orogenicand monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578–581. doi: 10.1126/science.abb4484
Do, H. D. K., Jung, J., Hyun, J., Yoon, S. J., Lim, C., Park, K., et al. (2019). The newly developed single nucleotide polymorphism (SNP) markers for a potentially medicinal plant, Crepidiastrum denticulatum (Asteraceae), inferred from complete chloroplast genome data. Mol. Biol. Rep. 46, 3287–3297. doi: 10.1007/s11033-019-04789-5
Dong, W. P., Xu, C., Li, C. H., Sun, J. H., Zuo, Y. J., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5, 8348. doi: 10.1038/srep08348
Doorduin, L., Gravendeel, B., Lammers, Y., Ariyurek, Y., Chin-A-Woeng, T., Vrieling, K. (2011). The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18, 93–105. doi: 10.1093/dnares/dsr002
Drummond, A. J., Suchard, M. A., Xie, D., Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Duan, N., Deng, L. L., Zhang, Y., Shi, Y. C., Liu, B. B. (2022). Comparative and phylogenetic analysis based on chloroplast genome of Heteroplexis (Compositae), a protected rare genus. BMC Plant Biol. 22, 1–10. doi: 10.1186/s12870-022-04000-1
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Fu, Z. X., Jiao, B. H., Nie, B., Zhang, G. J., Gao, T. G. (2016). A comprehensive generic-level phylogeny of the sunflower family: Implications for the systematics of Chinese Asteraceae. J. Syst. Evol. 54, 416–437. doi: 10.1111/jse.12216
Fu, Z. X., Zhang, G. J., Shao, Y. Z., Liu, Y. Y., Yang, X. C., Gao, T. G., et al. (2019). Rediscovery of Aster polius (Astereae: Asteraceae), a rare and endemic species from China, after one century. Phytotaxa 423, 247–258. doi: 10.11646/phytotaxa.423.4.3
Grierson, A. J. C. (1975). “Aster,” in Flora of Turkey and the East Aegean Islands. Ed. Davis, P. H. (Edinburgh University Press, Britain), 1–124.
Gu, H., Hoch, P. C. (1997). Systematics of Kalimeris (Asteraceae: astereae). Ann. Mo. Bot. Gard. 84, 762–814. doi: 10.2307/2992027
Guo, Z. T., Ruddiman, W. F., Hao, Q. Z., Wu, H. B., Qiao, Y. S., Zhu, R. X., et al. (2002). Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416, 159–163. doi: 10.1038/416159a
Huang, J. L., Sun, G. L., Zhang, D. M. (2010). Molecular evolution and phylogeny of the angiosperm ycf2 gene. J. Syst. Evol. 48, 240–248. doi: 10.1111/jse.2010.48.issue-4
Ito, M., Soejima, A., Hasebe, M., Watanabe, K. (1995). A chloroplast-DNA phylogeny of Kalimeris and Aster, with reference to the generic circumscription. J. Plant Res. 108, 93–96. doi: 10.1007/BF02344311
Ito, M., Soejima, A., Watanabe, K. (1998). Phylogenetic relationships of Japanese Aster (Asteraceae, Astereae) sensu lato based on chloroplast-DNA restriction site mutations. J. Plant Res. 111, 217–223. doi: 10.1007/BF02512173
Jafari, F., Osaloo, S. K., Mozffarian, V. (2015). Molecular phylogeny of the tribe Astereae (Asteraceae) in SW Asia based on nrDNA ITS and cpDNA psbA-trnH sequences. Willdenowia 45, 77–92. doi: 10.3372/wi.45.45108
Jones, A. G. (1980). A classification of the New world species of Aster (Asteraceae). Brittonia 32, 230–239. doi: 10.2307/2806795
Jones, A. G., Young, D. A. (1983). Generic concepts of Aster (Asteraceae): a comparison of cladistics, phenetic and cytological approaches. Syst. Bot. 8, 71–84. doi: 10.2307/2418564
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, K. J., Lee, H. L. (2004). Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11, 247–261. doi: 10.1093/dnares/11.4.247
Kim, K., Lee, S. C., Lee, J., Yu, Y., Yang, K., Choi, B. S., et al. (2015). Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci. Rep. 5, 15655. doi: 10.1038/srep15655
Korolyuk, E., Makunin, A., Matveeva, T. (2015). Relationships and generic delimitation of Eurasian genera of the subtribe Asterinae (Astereae, Asteraceae) using molecular phylogeny of ITS. Turk. J. Bot. 39, 808–824. doi: 10.3906/bot-1410-12
Kumar, S., Hahn, F. M., McMahan, C. M., Cornish, K., Whalen, M. C. (2009). Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 9, 131. doi: 10.1186/1471-2229-9-131
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Lee, D. H., Cho, W. B., Choi, B. H., Lee, J. H. (2017). Characterization of two complete chloroplast genomes in the tribe Gnaphalieae (Asteraceae): gene loss or pseudogenization of trnT-GGU and implications for phylogenetic relationships. Hortic. Sci. Technol. 35, 769–783. doi: 10.12972/kjhst.20170081
Li, W. P., Yang, F. S., Jivkova, T., Yin, G. S. (2012). Phylogenetic relationships and generic delimitation of Eurasian Aster (Asteraceae: Astereae) inferred from ITS, ETS and trnL-F sequence data. Ann. Bot. 109, 1341–1357. doi: 10.1093/aob/mcs054
Liu, J. Q., Gao, T. G., Chen, Z. D., Lu, A. M. (2002). Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae). Mol. Phylogenet. Evol. 23, 307–325. doi: 10.1016/S1055-7903(02)00039-8
Liu, X. F., Luo, J. J., Zhang, M. K., Wang, Q., Liu, J., Wu, D., et al. (2023). Phylogenomic analysis of two species of Parasenecio and comparative analysis within tribe Senecioneae (Asteraceae). Diversity 15, 563. doi: 10.3390/d15040563
Liu, X., Zhou, B., Yang, H., Li, Y., Yang, Q., Lu, Y., et al. (2018). Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable. Molecules 23, 1358. doi: 10.3390/molecules23061358
Magee, A. M., Aspinall, S., Rice, D. W., Cusack, B. P., Sémon, M., Perry, A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710. doi: 10.1101/gr.111955.110
Merxmüller, H., Schreiber, A., Yeo, P. F. (1976). “Aster,” in Flora Europaea, vol. 4 . Eds. Tutin, T. G., Heywood, V. H., Burges, N. A. (Cambridge University Press, Cambridge), 112–116.
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). "Creating the CIPRES science gateway for inference of large phylogenetic trees," in Gateway Computing Environments Workshop, ed. Proceedings of a meeting held New Orleans (New York: IEEE), 1–8.
Nesom, G. L. (1994a). Subtribal classification of the astereae (Asteraceae). Phytologia 76, 193–274. doi: 10.5962/bhl.part.4090
Nesom, G. L. (1994b). Review of the taxonomy of Aster sensu lato (Asteraceae: Astereae), emphasizing the New World species. Phytologia 77, 141–297.
Nesom, G. L., Robinson, H. (2007). “Astereae,” in The Families and Genera of Vascular Plants, vol. 8 . Eds. Kadereit, J. W., Jeffrey, C. (Springer, Berlin), 284–342.
Nordenstam, B. (2007). “Tribe senecioneae,” in The Families and Genera of Vascular Plants, vol. 8 . Eds. Kadereit, J. W., Jeffrey, C. (Springer, Berlin), 208–241.
Noyes, R. D., Rieseberg, L. H. (1999). ITS sequence data support a single origin for North American Astereae (Asteraceae) and reflect deep geographic divisions in Aster s.l. Am. J. Bot. 86, 398–412. doi: 10.2307/2656761
Palazzesi, L., Pellicer, J., Barreda, V. D., Loeuille, B., Mandel, J. R., Pokorny, L., et al. (2022). Asteraceae as a model system for evolutionary studies: from fossils to genomes. Bot. J. Linn. Soc 200, 143–164. doi: 10.1093/botlinnean/boac032
Panero, J. L., Crozier, B. S. (2016). Macroevolutionary dynamics in the early diversification of Asteraceae. Mol. Phylogenet. Evol. 99, 116–132. doi: 10.1016/j.ympev.2016.03.007
Panero, J. L., Freire, S. E., Espinar, L. A., Crozier, B. S., Barboza, G. E., Cantero, J. J. (2004). Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Mol. Phylogenet. Evol. 80, 43–53. doi: 10.1016/j.ympev.2014.07.012
Pelser, P. B., Kennedy, A. H., Tepe, E. J., Shidler, J. B., Nordenstam, B., Kadereit, J. W., et al. (2010). Patterns and causes of incongruence between plastid and nuclear senecioneae (Asteraceae) phylogenies. Am. J. Bot. 97, 856–873. doi: 10.3732/ajb.0900287
Posada, D. (2006). ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic. Acids Res. 34, W700–W703. doi: 10.1093/nar/gkl042
Prjibelski, A., Antipov, D., Meleshko, D. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinformatics 70, 1–29. doi: 10.1002/cpbi.102
Qu, X. J., Moore, M. J., Li, D. Z., Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Ravi, V., Khurana, J. P., Tyagi, A. K., Khurana, P. (2008). An update on chloroplast genomes. Plant Syst. Evol. 271, 101–122. doi: 10.1007/s00606-007-0608-0
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Salih, R. H. M., Majesky, L., Schwarzacher, T., Gornall, R., Heslop-Harrison, P. (2017). Complete chloroplast genomes from apomictic Taraxacum (Asteraceae): identity and variation between three microspecies. PloS One 12, e0168008. doi: 10.1371/journal.pone.0168008
Selliah, S., Brouillet, L. (2008). Molecular phylogeny of the North American eurybioid asters (Asteraceae, Astereae) based on the nuclear ribosomal internal and external transcribed spacers. Botany 86, 901–915. doi: 10.1139/B08-070
Semple, J. C., Brouillet, L. (1980). A synopsis of North American Asters: the subgenera, sections and subsections of Aster and Lasallea. Am. J. Bot. 67, 1010–1026. doi: 10.1002/j.1537-2197.1980.tb07733.x
Shen, X. F., Guo, S., Yin, Y., Zhang, J. J., Yin, X. M., Liang, C. L., et al. (2018). Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules 23, 2426. doi: 10.3390/molecules23102426
Shen, X. F., Wu, M. L., Liao, B. S., Liu, Z. X., Bai, R., Xiao, S. M., et al. (2017). Complete Chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 22, 1330. doi: 10.3390/molecules22081330
Shi, Y. F., Li, J. J., Li, B. Y. (1998). Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic (Guangzhou: Guangdong Science and Technology Press).
Somaratne, Y., Guan, D. L., Wang, W. Q., Zhao, L., Xu, S. Q. (2019). Complete chloroplast genome sequence of Xanthium sibiricum provides useful DNA barcodes for future species identification and phylogeny. Plant Syst. Evol. 305, 949–960. doi: 10.1007/s00606-019-01614-1
Spicer, R. A., Harris, N. B., Widdowson, W. M., Herman, A. B., Guo, S., Valdes, P. J., et al. (2003). Constant elevation of southern Tibet over the past 15 million years. Nature 421, 622–624. doi: 10.1038/nature01356
Stamatakis, A., Hoover, P., Rougemont, J. A. (2008). Rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Tamamschjan, S. G. (1959). “Astereae,” in Flora of the USSR, vol. 25 . Ed. Schischkin, B. K. (Akademiya Nauk SSSR Publishers, Moscow and Leningrad), 24–290.
Tara, M. (1972). Cytogenetic studies on natural intergeneric hybridization in Aster alliances. Bot. Mag. 85, 219–240. doi: 10.1007/BF02489214
Thiel, T., Michalek, W., Varshney, R., Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Tyagi, S., Jung, J. A., Kim, J. S., Won, S. Y. (2019). Comparative analysis of the complete chloroplast genome of mainland Aster spathulifolius and other Aster species. Plants 9, 568. doi: 10.3390/plants9050568
Vargas, O. M., Ortiz, E. M., Simpson, B. B. (2017). Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 214, 1736–1750. doi: 10.1111/nph.14530
Wang, X. Y., Liu, Y. (2023). The complete chloroplast genome of Aster altaicus Willd. (Asteraceae: Aster) and phylogenetic analysis. Mitochondrial DNA Part B. 8, 819–822. doi: 10.1080/23802359.2023.2238358
Wang, Y. J., Liu, J. Q., Miehe, G. (2007). Phylogenetic origins of the Himalayan endemic Dolomiaea, Diplazoptilon and Xanthopappus (Asteraceae: Cardueae) based on three DNA regions. Ann. Bot. 99, 311–322. doi: 10.1093/aob/mcl259
Wang, Q., Wang, X. H., Yang, X. C., Fu, L. F., Gong, Q. J., Fu, Z. X. (2019). The complete chloroplast genome of Aster hypoleucus (Asteraceae: Astereae): an endemic species from China. Mitochondrial DNA Part B. 4, 2647–2648. doi: 10.1080/23802359.2019.1644219
Weng, M. L., Blazier, J. C., Govindu, M., Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Wick, R. R., Schultz, M. B., Justin, Z., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 20, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Müller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Xiang, C., Semple, J. D. (1994). “Molecular systematic study of Aster sensu lato and related genera (Asteraceae: Astereae) based on chloroplast DNA restriction site analyses and mainly North American taxa,” in Compositae: systematics, Proceedings of the International Compositae Conference, vol. 1 . Eds. Hind, D. J. N., Beentje, H. J. (Royal Botanic Gardens, Kew), 393–423.
Yin, G. S., Li, W. P., Chen, S. M., Liu, S. X. (2010). A karyotypic study on Aster Series Hersileoides ling (Asteraceae). J. Wuhan Botanical Res. 28, 406–409. doi: 10.3724/SP.J.1142.2010.40406
Yu, S. H., Yang, X. C., Tian, X. Y., Liu, X. F., Lu, X., Huang, C. P., et al. (2022). The complete chloroplast genome sequence of the monotypic and enigmatic genus Cavea (tribe Gymnarrheneae) and a comparison with other species in Asteraceae. J. Genet. 101, 20. doi: 10.1007/s12041-022-01360-3
Zhang, X. P., Bremer, K. (1993). A cladistic analysis of the tribe Astereae (Asteraceae) with notes on their evolution and subtribal classification. Plant Syst. Evol. 184, 259–283.
Zhang, X., Deng, T., Moore, M. J., Ji, Y. H., Lin, N., Zhang, H. J., et al. (2019b). Plastome phylogenomics of Saussurea (Asteraceae: cardueae). BMC Plant Biol. 19, 290. doi: 10.1186/s12870-019-1896-6
Zhang, G. J., Hu, H. H., Gao, T. G., Gilbert, M. G., Jin, X. F. (2019a). Convergent origin of the narrowly lanceolate leaf in the genus Aster-with special reference to an unexpected discovery of a new Aster species from East China. Peer J. 7, e6288. doi: 10.7717/peerj.6288
Zhang, G. J., Hu, H. H., Zhang, C. F., Tian, X. J., Peng, H., Gao, T. G. (2015). Inaccessible biodiversity on limestone cliffs: aster tianmenshanensis (Asteraceae), a new critically endangered species from China. PloS One 10, e0134895. doi: 10.1371/journal.pone.0134895
Zhang, C. F., Huang, C. H., Liu, M., Hu, Y., Panero, J. L., Luebert, F., et al. (2021). Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation. J. Integr. Plant Bio. 63, 1273–1293. doi: 10.1111/jipb.13078
Zhang, X., Jiang, P. P., Fan, S. J. (2021). Characterization of the complete plastome of Aster pekinensis (Asteraceae), a perennial herb. Mitochondrial DNA Part B. 6, 1064–1065. doi: 10.1080/23802359.2021.1899081
Zhao, C., Xu, W. F., Huang, Y., Sun, Q. W., Wang, B., Chen, C. L., et al. (2021). Chloroplast genome characteristics and phylogenetic analysis of the medicinal plant Blumea balsamifera (L.) DC. Genet. Mol. Biol. 44, e20210095. doi: 10.1590/1678-4685-gmb-2021-0095
Keywords: Aster, chloroplast genome, comparative analysis, Astereae, phylogenetic relationship, divergence time
Citation: Chen H, Li T, Chen X, Qu T, Zheng X, Luo J, Li B, Zhang G and Fu Z (2024) Insights into comparative genomics, structural features, and phylogenetic relationship of species from Eurasian Aster and its related genera (Asteraceae: Astereae) based on complete chloroplast genome. Front. Plant Sci. 15:1367132. doi: 10.3389/fpls.2024.1367132
Received: 08 January 2024; Accepted: 25 March 2024;
Published: 26 April 2024.
Edited by:
Enrico Vito Perrino, International Centre for Advanced Mediterranean Agronomic Studies, ItalyReviewed by:
Robert Philipp Wagensommer, Free University of Bozen-Bolzano, ItalyQing Ma, Zhejiang Shuren University, China
Copyright © 2024 Chen, Li, Chen, Qu, Zheng, Luo, Li, Zhang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojin Zhang, Z3VvamluX3poYW5nQDE2My5jb20=; Zhixi Fu, ZnV6eDIwMTdAc2ljbnUuZWR1LmNu
 Hui Chen1,2
Hui Chen1,2 Xinyu Chen
Xinyu Chen Xinyi Zheng
Xinyi Zheng Zhixi Fu
Zhixi Fu