
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 March 2024
Sec. Plant Physiology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1366986
This article is part of the Research Topic Molecular and Genetic Mechanisms of Plant Architecture Regulation View all 8 articles
 Hongming Guo1,2
Hongming Guo1,2 Jianqun Lv1,2
Jianqun Lv1,2 Xiangwen Su1,2
Xiangwen Su1,2 Liang Chen3
Liang Chen3 Juansheng Ren1,2
Juansheng Ren1,2 Liping Liu1
Liping Liu1 Mingxin Ren1
Mingxin Ren1 Song Liu1,2
Song Liu1,2 Mingli Dai1
Mingli Dai1 Guangjun Ren1,2*
Guangjun Ren1,2* Fangyuan Gao1,2*
Fangyuan Gao1,2*The eIF6 proteins are distributed extensively in eukaryotes and play diverse and essential roles. The bona fide eIF6 protein in Arabidopsis, At-eIF6;1, is essential for embryogenesis. However, the role of eIF6 proteins in rice growth and development remains elusive and requires further investigation. Here, we characterized the functions of OseIF6.1, which is homologous to At-eIF6;1. OseIF6.1 encodes an eukaryotic translation initiation factor with a conserved eIF6 domain. The knockdown of OseIF6.1 resulted in a decrease in grain length and pollen sterility, whereas the overexpression of OseIF6.1 displayed opposite phenotypes. Further studies revealed that OseIF6.1 regulates grain shape by influencing cell expansion and proliferation. In addition, OseIF6.1 interacts with OsNMD3, which is a nuclear export adaptor for the 60S ribosomal subunit. The knockdown of OsNMD3 in plants exhibited reduced fertility and seed setting. Therefore, our findings have significantly enriched the current understanding of the role of OseIF6.1 in rice growth and development.
Protein synthesis is a complex process that can be categorized into four distinct stages: initiation, elongation, termination, and ribosome recycling (Sonenberg and Hinnebusch, 2009). Translation initiation represents the crucial phase of protein synthesis, where the ribosome assembles on the mRNA and initiates the synthesis of the polypeptide chain (Merrick and Pavitt, 2018). In eukaryotes, this process is regulated and carried out by a series of protein complexes known as eukaryotic translation initiation factors (eIFs) (Gebauer and Hentze, 2004; Hinnebusch, 2006). These factors orchestrate the assembly of the translation initiation complex, which includes the small ribosomal subunit, initiator tRNA, and mRNA (Merrick and Pavitt, 2018). The eIFs not only facilitate the correct positioning of the ribosome on the mRNA but also regulate the rate and efficiency of translation initiation (Roy and von Arnim, 2013; Raabe et al., 2019). As such, they serve as important regulators of gene expression and can influence various aspects of plant growth and development.
The types of eIFs are diverse and complex, and at least 29 have been identified, including eIF1, eIF1A, eIF2, eIF2α, eIF2B, eIF3a-m, eIF4A1, eIF4A2, eIF4B, eIF4E, eIF4G, eIF4F, eIF5, eIF5A, eIF5B, eIF6.1and eIF6.2. Each exerting distinct roles during the translation initiation process (Jackson et al., 2010; Guo et al., 2011; Aitken and Lorsch, 2012; Hinnebusch and Lorsch, 2012; Raabe et al., 2019; Castellano and Merchante, 2021; Singha et al., 2021; Ma et al., 2022). Among them, eIF6 is an essential protein that possesses a distinctive anti-association activity. As it binds to immature large ribosomal subunits (pre-60S) in the nucleolus, it prevents their premature association with 40S subunits (Miluzio et al., 2009). Subsequently, eIF6 is detected in the nucleoplasm during pre-60S subunit maturation and is exported to the cytosol where it releases the 60S ribosomal subunit. The 60S subunits then join with 40S subunits to form the 80S ribosome complex (Si and Maitra, 1999; Basu et al., 2001; Ceci et al., 2003; Gandin et al., 2008; Miluzio et al., 2009).
Over the past decades, extensive research has been devoted to exploring the function of eIFs in animals and yeast (Kapp and Lorsch, 2004; Sonenberg and Hinnebusch, 2009; Aitken and Lorsch, 2012). Indeed, recent research has suggested that eIFs play a crucial role in regulating cell differentiation, cell cycle progression, and stress responses in plants (Rausell et al., 2003; Thompson et al., 2004; Diédhiou et al., 2008; Singh et al., 2013; Chen et al., 2019; Raabe et al., 2019; Castellano and Merchante, 2021). For example, Fumonisin B1-resistant 12 (FBR12) encodes a putative eIF-5A-2 protein that regulates growth and development of floral organs and sporogenesis by influencing cell division, cell growth, and cell death in Arabidopsis (Feng et al., 2007). In addition, eIF5A played a significant role in the process of cadmium (Cd) accumulation and sensitivity in Arabidopsis. The ateif5a mutant exhibited a higher level of Cd accumulation in both roots and shoots compared to the wild type. Moreover, AteIF5A was found to impact Cd sensitivity by modulating Cd uptake, accumulation, and detoxification (Xu et al., 2015). Furthermore, eIFs are involved in the processes of plant growth and development, such as embryogenesis, flowering, and organogenesis. For example, mutations in the eIF3 subunits eIF3e, eIF3f, and eIF3h in Arabidopsis do not impact pollen formation or maturation. However, they do cause deficiencies in pollen germination and/or pollen tube growth, resulting in reduced efficiency of male gamete transmission (Xia et al., 2010; Roy et al., 2011). Rice eIF3 subunit f has been reported to play an important role in post-meiotic pollen formation, knockdown of OseIF3f showed a large reduction in seed setting and pollen fertility (Li et al., 2016). The expression levels of OseIF3f were significantly higher in unicellular microspores and bicellular pollen than in mature tricellular pollen or germinated pollen, with at least a three-fold increase observed, suggesting that OseIF3f might play a more crucial role in microgametogenesis rather than pollen germination (Wei et al., 2010). Interestingly, OseIF3e interacted with OseIF3f and OseIF6, respectively. OseIF3e-RNAi plants exhibited stunted growth during both the seedling and vegetative stages, and displayed defects in pollen maturation and small grains (Wang et al., 2016).
eIF6 was first discovered and characterized as a wheat protein that associates with the 60S ribosome in crop plants (Russell and Spremulli, 1980). The study on eIF6 in yeast (TIF6) demonstrated its crucial role in ribosome biogenesis. When TIF6 is depleted, it causes abnormal processing of ribosomal RNA (rRNA) precursors and a decrease in the abundance of 60S ribosomal subunits, leading to a lethal phenotype (Wood et al., 1999; Basu et al., 2001). Human eIF6 (p27BBP) interacts with 60S ribosome subunits, preventing the assembly of both 40S and 60S subunits in the cytosol (Sanvito et al., 1999). Embryos lacking eIF6 in mice exhibit a lethal phenotype during the preimplantation stage. Additionally, heterozygous mice display insensitivity to insulin and show reduced hepatic and adipose tissue mass, as well as a decrease in protein synthesis (Gandin et al., 2008). The Arabidopsis at-eif6;1 mutant exhibits an embryonic-lethal phenotype, similar to yeast and mouse. One-third of the pale yellow seeds were observed in heterozygotes mutant siliques (Kato et al., 2010). However, little is known about the function of eIF6 in crop plants, despite these studies indicating a critical role for eIF6 in embryogenesis.
Our previous study has shown that OseIF6.1 interacts with OsLa, and it was impossible to isolate a homozygous oseif6.1 mutant, as the plants displayed abnormal floral organs, leading to a lethal phenotype (Guo et al., 2022). However, the function of OseIF6.1 gene has yet to be characterized. Our present study aimed to validate the roles of OseIF6.1 in the growth and development of rice. In this study, we identified and characterized the functions of OseIF6.1 by using RNA interference (RNAi) approach. We found that OseIF6.1 encodes a protein homologous to At-eIF6;1 in Arabidopsis, and OseIF6.1 acts as a positive regulator in grain size and pollen sterility. In addition, OseIF6.1 physically interacts with OsNMD3, which is a nuclear export adaptor for the 60S ribosomal subunit. The OsNMD3-RNAi lines exhibited a phenotype of reduced fertility. Therefore, these findings help to reveal the function of OseIF6.1 in rice grain size and pollen fertility.
The japonica rice (Oryza sativa) cultivar (Nipponbare, Nip) was used for genetic transformation. Seeds of the wild type (WT), as well as the OseIF6.1 and OsNMD3 transgenic lines, underwent sterilization using a 10% sodium hypochlorite (NaClO) solution for a duration of 30 minutes. After this, they were rinsed thoroughly five times with sterile water. The sterilized seeds were planted in sterile plastic containers half-strength Murashige and Skoog (MS) medium. These containers were subsequently placed in a controlled growth chamber, maintaining a photoperiod of 16 hours of light at an approximate temperature of 28 ± 2°C, followed by 8 hours of darkness at around 25 ± 2°C. The chamber’s relative humidity was kept between 70-85%. After a two-week period of incubation, the seedlings were carefully transplanted to field conditions. All plants were planted in the experimental field of Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China. Each plant was separated by 20 cm within each row, and the rows were also spaced 20 cm apart. The field management was essentially based on standard agricultural practices.
The OseIF6.1 protein homolog sequences were obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). These sequences were aligned to construct phylogenetic trees based on the maximum-likelihood (ML) criterion, employing 1000 bootstraps in MEGA 5. The prediction of their conserved domains was carried out using the NCBI Batch CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and TBtools (Chen et al., 2020).
For the construction of the OseIF6.1 overexpression vector, the complete coding sequence of OseIF6.1 was PCR-amplified from Nipponbare cDNA, followed by its subsequent cloning into the pCXUN-Flag vector using TA cloning method (Chen et al., 2009). For the construction of the OseIF6.1 and OsNMD3 Knockdown vector, the gene-specific sequences of OseIF6.1 and OsNMD3 were cloned into the pH7GWIWGII with the LR Clonase II enzyme (Invitrogen), respectively.
We introduced recombinant plasmids into Nipponbare callus tissues of rice using the Agrobacterium tumefaciens strain EHA105, employing the previously described transformation protocol (Hiei et al., 1994). All the primers used are listed in Supplementary Table 1.
To determine the subcellular localization of OseIF6.1 protein, its CDS was inserted into the pCXDG vector, creating a fusion with green fluorescent protein (GFP), which was driven by the CaMV35 promoter. The vector was introduced into the A. tumefaciens strain GV3101, followed by the transient transformation into the leaves of Nicotiana benthamiana at the age of four weeks. Confocal microscopy (LSM 780, Carl Zeiss) was used to observe GFP fluorescence signals.
Young panicles and seedlings of WT, and OseIF6.1 transgenic lines were used to isolate total RNA using an Vazyme FastPure Universal Plant Total RNA Isolation Kit. First-strand cDNA was synthesized using Vazyme’s HiScript II Q RT SuperMix for qPCR with gDNA wiper. Quantitative real-time PCR (qRT-PCR) was conducted using a QuantStudio Flex PCR system machine (Thermo Fisher Scientific) with Powerup™ SYBR™ Green Master Mix (Applied Biosystems). The Actin gene of Nip was chosen as the internal reference, and the relative expression levels of the target genes were quantified employing the 2-ΔΔCt method (Livak and Schmittgen, 2001). All the primers used are listed in Supplementary Table 1.
WT and transgenic plants were grown in a rice field and photographed at maturity. Grain traits including plant height, primary branch, secondary branch, panicle length, grain length, grain width, grain thickness, and 1000-grain weight, were measured. Mature grains were dried and sprayed with gold, and then observed using a scanning electron microscope. Cell number and size were determined using Image J software. The sample preparation and TEM observation were carried out according to the previous method for semi-thin section and ultra-thin section assays (Cao et al., 2018).
The OseIF6.1 and OsNMD3 CDS have been cloned into plasmids pGBKT7 and pGADT7, respectively. Ligation-independent cloning (LIC) was used to construct all plasmids (Aslanidis et al., 1994). The Yeast two-hybrid (Y2H) assay was conducted in accordance with the manufacturer’s instructions (Clontech). The pGBKT7-53 plasmid was transformed with pGADT7-T as a positive control, and the pGBKT7-Lam plasmid was transformed with pGADT7-T into the Y2HGold strain as a negative control. All the primers used are listed in Supplementary Table 1.
The CDS of OseIF6.1 was cloned into the N-terminal fragment of the p2YN vector, and the CDS of OsNMD3 was cloned into the C-terminal fragment of the p2YC vector. The LIC method was used to construct all plasmids. For transient expression, N. benthamiana leaves were coinfiltrated with A. tumefaciens strain GV3101 carrying different plasmid combinations and the p19 strain. The confocal microscopy (LSM 780, Carl Zeiss) was used to observe the fluorescent signals of the yellow fluorescent protein (YFP). All the primers used are listed in Supplementary Table S1.
The CDSs of OseIF6.1 and OsNMD3 were cloned into pET-28a (+) and pET-GST vectors, respectively, to generate His-OseIF6.1 and GST-OsNMD3 proteins. All plasmids were constructed using the LIC method, and subsequently transformed into the Escherichia coli BL21 strain. A pull down assay was performed as previously described (Guo et al., 2022). The precipitates were analyzed by western blot using GST and His antibodies (Thermo Fisher Scientific). All the primers used are listed in Supplementary Table 1.
Data are means ± SE from at least three independent experiments. The Statistical analysis was carried out using IBM SPSS software (IBM Corp., Armonk, NY, USA), and means were compared by Student’s t-tests.
First, the eIF6 homologs in eukaryotes were identified by running a BLAST search against the Arabidopsis At-eIF6;1 protein in the NCBI database. A multiple protein sequence alignment of eIF6 protein homologs from Oryza sativa (Genbank accession XP_015647337.1; OseIF6.1), Oryza sativa (XP_015635348.1; OseIF6.2), Arabidopsis thaliana (NP_191121.1; At-eIF6;1), Arabidopsis thaliana (NP_181512.1; At-eIF6;2), Homo sapiens (CAX12724.1; HseIF6), and Saccharomyces cerevisiae (NP_015341.1; TIF6) revealed that the sequences of the six proteins are highly similar (Supplementary Figure 1). An analysis of phylogenetic relationships and structural comparisons between eIF6 proteins from various eukaryotes revealed that they contain a conserved eIF6 domain, as well as OseIF6.1 shares a close relationship with other homologues, and that eIF6 protein is conserved across eukaryotes (Figure 1A).
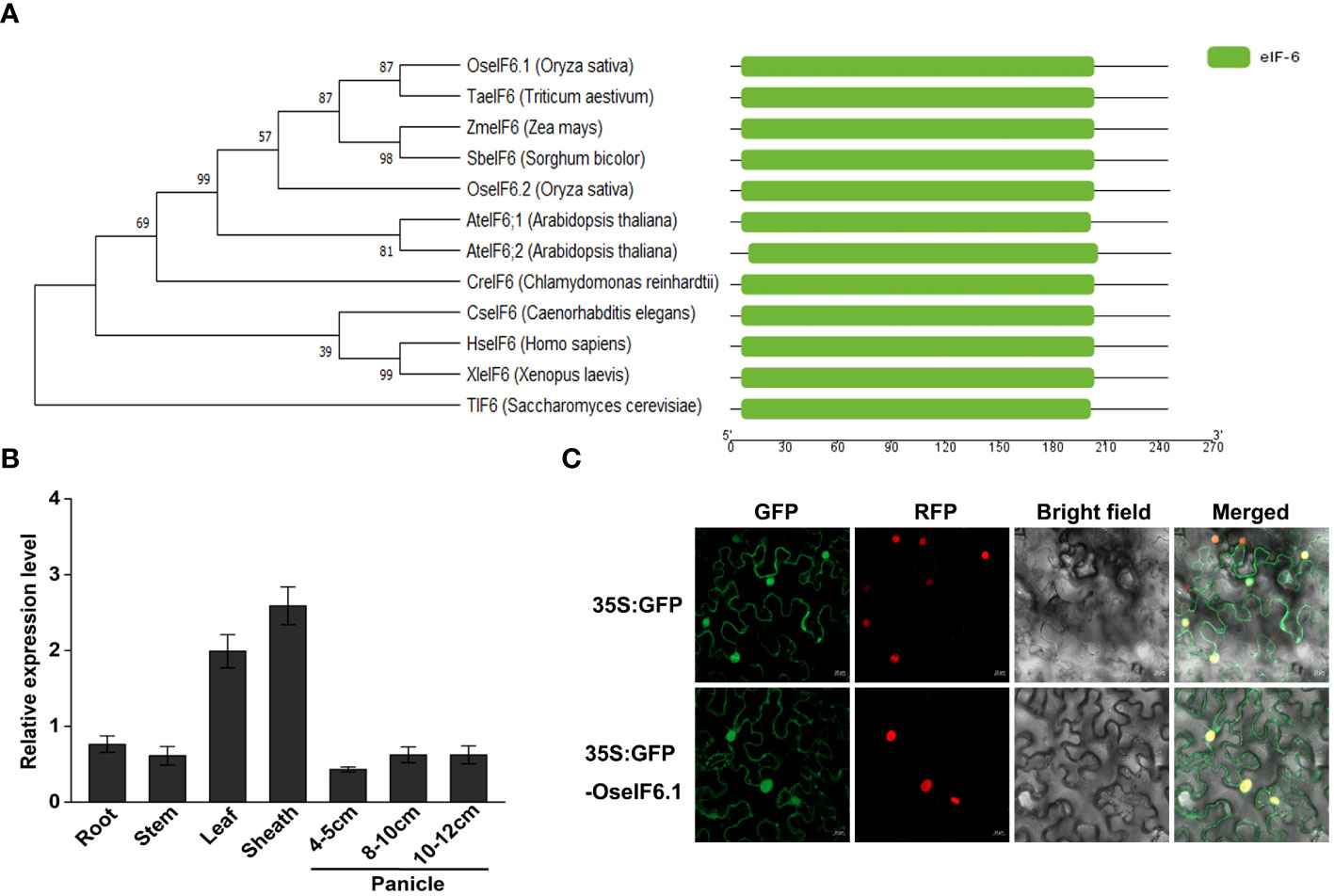
Figure 1 Expression pattern and subcellular localization of OseIF6.1. (A) Comparative analysis of eIF6 protein phylogeny and structure across diverse eukaryotic species. (B) Relative expression of OseIF6.1 in root, stem, leaf, and sheath of young seedlings and developing panicles of 4-5, 8-10, and 10-12 cm. OsActin served as an internal control. The values represent means ± SE derived from at least three independent experiments. (C) Subcellular localization of OseIF6.1 in N. benthamiana leaves. Scale bars = 20 μm.
In order to elucidate the expression profile of OseIF6.1 in rice tissues, we employed qRT-PCR assay. The expression levels of OseIF6.1 transcripts were examined in the root, stem, leaf, and sheath tissues of young seedlings, as well as in developing panicles at three different stages: 4-5 cm, 8-10 cm, and 10-12 cm. qRT-PCR analysis revealed that the expression of OseIF6.1 was detected in all tested tissues, with particularly high expression levels observed in leaves and sheaths (Figure 1B). To investigate the subcellular localization of OseIF6.1, a GFP-OseIF6.1 fusion protein driven by the CaMV 35S promoter was transiently expressed in N. benthamiana leaves. Similar to the GFP signal, confocal images showed that GFP-OseIF6.1 was detected in the cytoplasm, and nucleus (Figure 1C).
To investigate the function of OseIF6.1, we generated OseIF6.1 knockdown transgenic lines by RNA interference (RNAi) technology. Additionally, OseIF6.1 was tagged with a Flag under the control of a maize ubiquitin promoter to obtain overexpression lines. These vectors were successfully introduced into Nipponbare through an Agrobacterium tumefaciens-mediated transformation. Further functional analysis was conducted on two OE and two knockdown lines (Figure 2A). The relative expression levels of OseIF6.1 in OseIF6.1-R-1 and OseIF6.1-R-2 were reduced by 0.77-fold and 0.59-fold, respectively, compared to that in WT. In contrast, the expression levels of OseIF6.1 in OseIF6.1-OE-1 and OseIF6.1-OE-2 were 18.9-fold and 31.4-fold higher, respectively, than that of WT (Figure 2A).
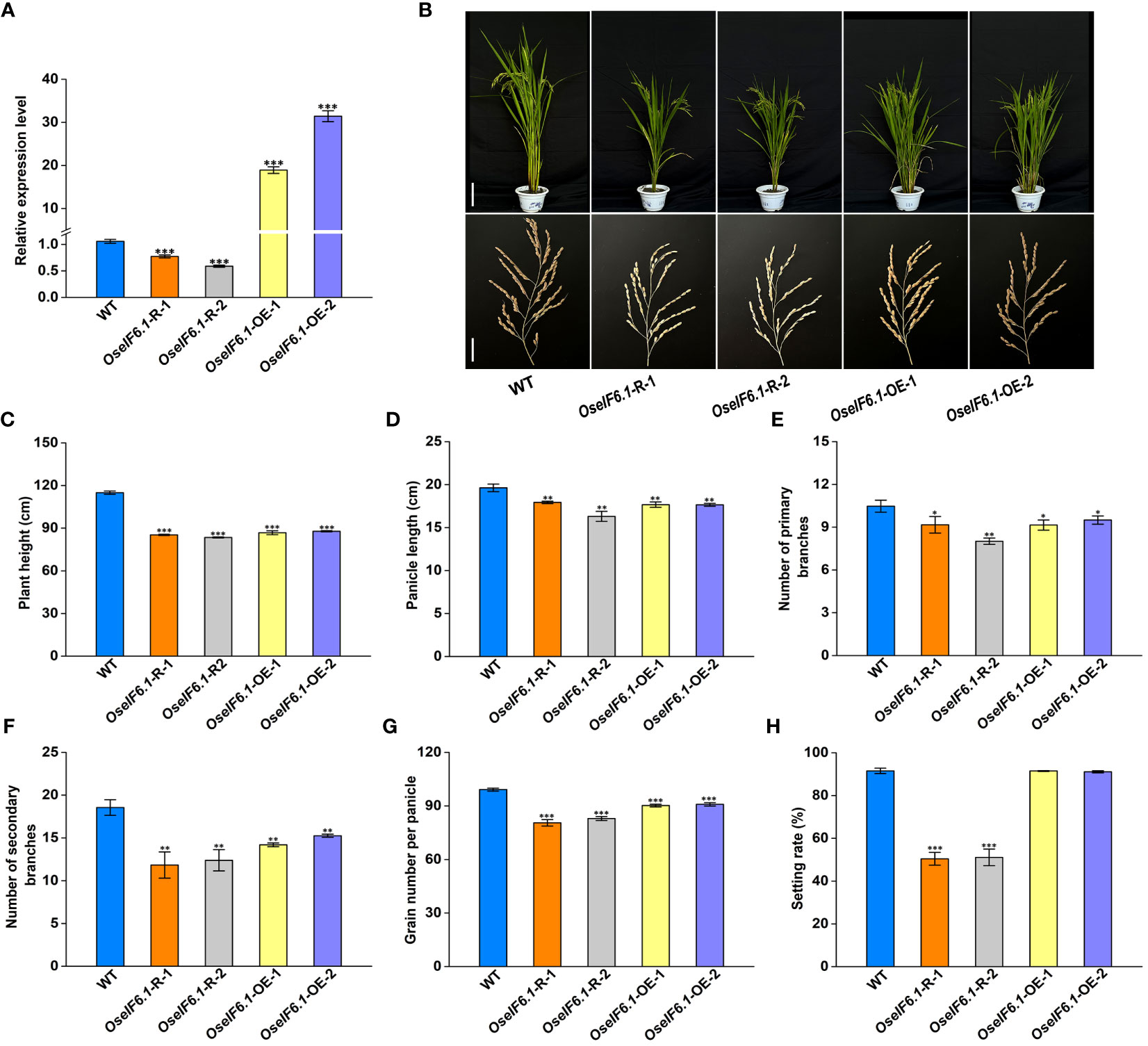
Figure 2 Phenotypic analysis of OseIF6.1-overexpressing and knockdown in rice. (A) Relative expression levels of OseIF6.1 in WT and OseIF6.1 transgenic lines. OsActin served as an internal control. The values represent means ± SE derived from at least three independent experiments. (B) Plants and Panicles of WT and OseIF6.1 transgenic plants at the mature stage. Scale bars = 20 cm for plant height and 3 cm for panicle length. (C) Plant heights of WT and OseIF6.1 transgenic plants. (D) Panicle length of WT and OseIF6.1 transgenic plants. (E) Number of primary branches of WT and OseIF6.1 transgenic panicles. (F) Number of secondary branches of WT and OseIF6.1 transgenic panicles. (G) Grain number per panicle of WT and OseIF6.1 transgenic plants. (H) Setting rate of WT and OseIF6.1 transgenic plants. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
At the mature stage, several agronomic traits between WT and OseIF6.1 transgenic lines were measured (Figure 2B). As shown in Figures 2C, D, the OseIF6.1-R and OseIF6.1-OE plants had reduced plant height and shorter panicles than the WT. In addition, we found that the OseIF6.1-R and OseIF6.1-OE plants had fewer primary and secondary branches, as well as grain number per panicle compared with the WT (Figures 2E–G). Interestingly, the setting rate in OseIF6.1-R plants was dramatically lower when compared with the WT and OseIF6.1-OE lines, while there was no significant difference observed between the WT and OseIF6.1-OE lines (Figure 2H).
Further analysis suggested that OseIF6.1 acts as a positive regulator of grain length. When compared with WT, the OseIF6.1-R plants exhibited shorter and smaller grains (Figures 3A–C). However, the grain lengths in the OseIF6.1-OE1 and OE2 lines were significantly increased by 2.24% and 2.97%, respectively, than those of the WT (Figures 3A, B). Furthermore, both OseIF6.1-R and OseIF6.1-OE grains exhibited a decrease in grain width and thickness compared to WT grains (Figures 3C, D), leading to their lower 1000-grain weights compared to WT (Figure 3E). Therefore, the findings suggest that OsEIF6.1 plays a pivotal role in rice growth and development.
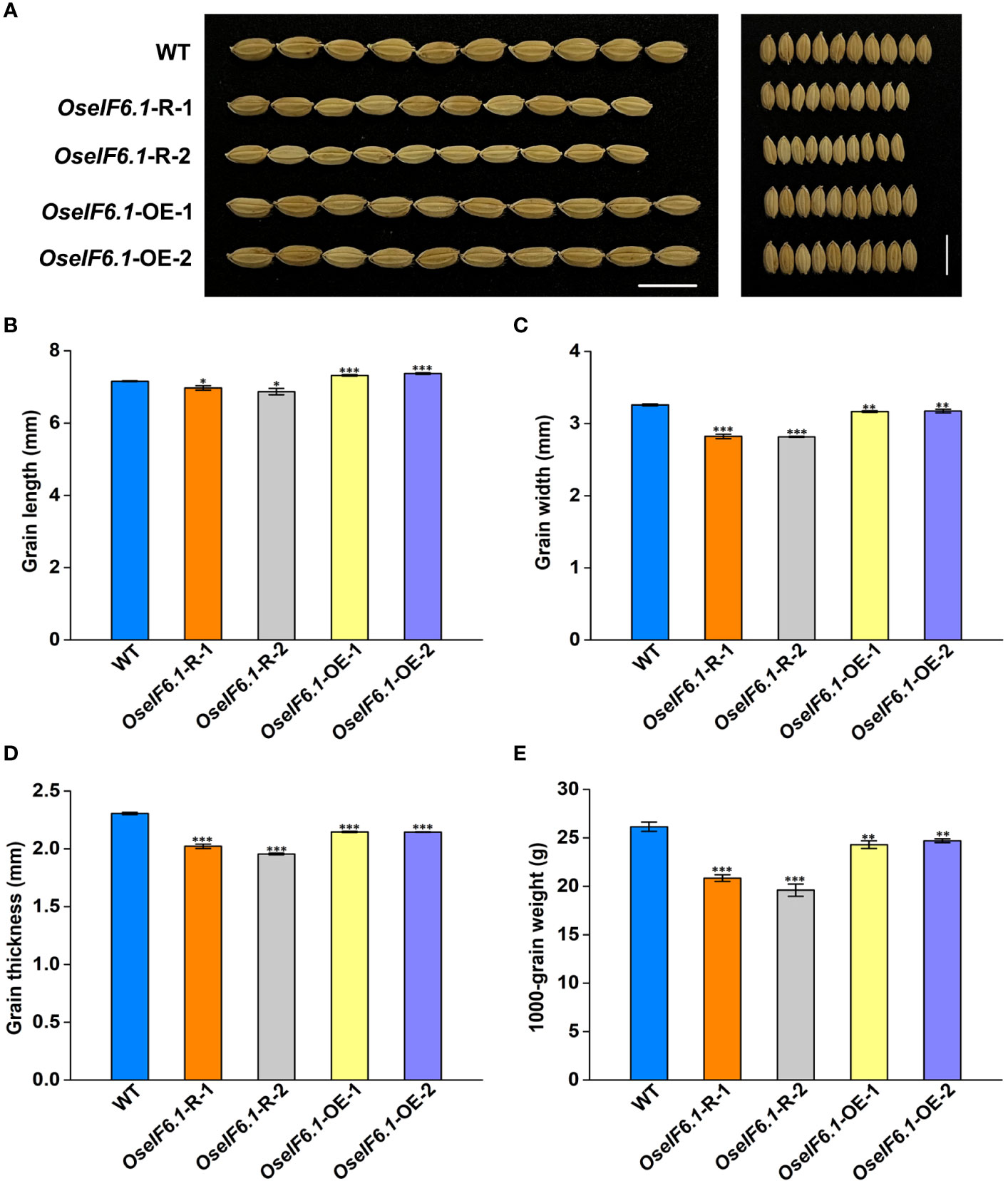
Figure 3 Ectopic expression of OseIF6.1 alters the grain shape. (A) Mature paddy grains of WT and OseIF6.1 transgenic lines. Scale bars =1 cm. (B) Grain length of WT and OseIF6.1 transgenic lines. (C) Grain width of WT and OseIF6.1 transgenic lines. (D) Grain thickness of WT and OseIF6.1 transgenic lines. (E) 1000-grain weight of WT and OseIF6.1 transgenic lines. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
The spikelet hull size is a crucial factor in promoting grain size growth, as it relies on coordinated cell proliferation and expansion (Li and Li, 2016). To comprehensively assess the impact of OseIF6.1 on grain size, we utilized scanning electron microscopy (SEM) to meticulously examine the outer glume of both WT and OseIF6.1 transgenic plants’ spikelet hulls (Figure 4A). The outer epidermal cells in OseIF6.1-R and OseIF6.1-OE lemma were shorter and narrower than those of the WT (Figures 4B, C). In contrast, the number of cells in the outer glume of OseIF6.1-OE was significantly higher than in WT in both the longitudinal and transverse directions (Figures 4D, E). On the other hand, the cell numbers in OseIF6.1-R spikelet hulls were lower than those of the WT (Figures 4D, E). Together, these results suggest that OseIF6.1 plays a positive role in regulating grain size by influencing cell expansion and cell proliferation.
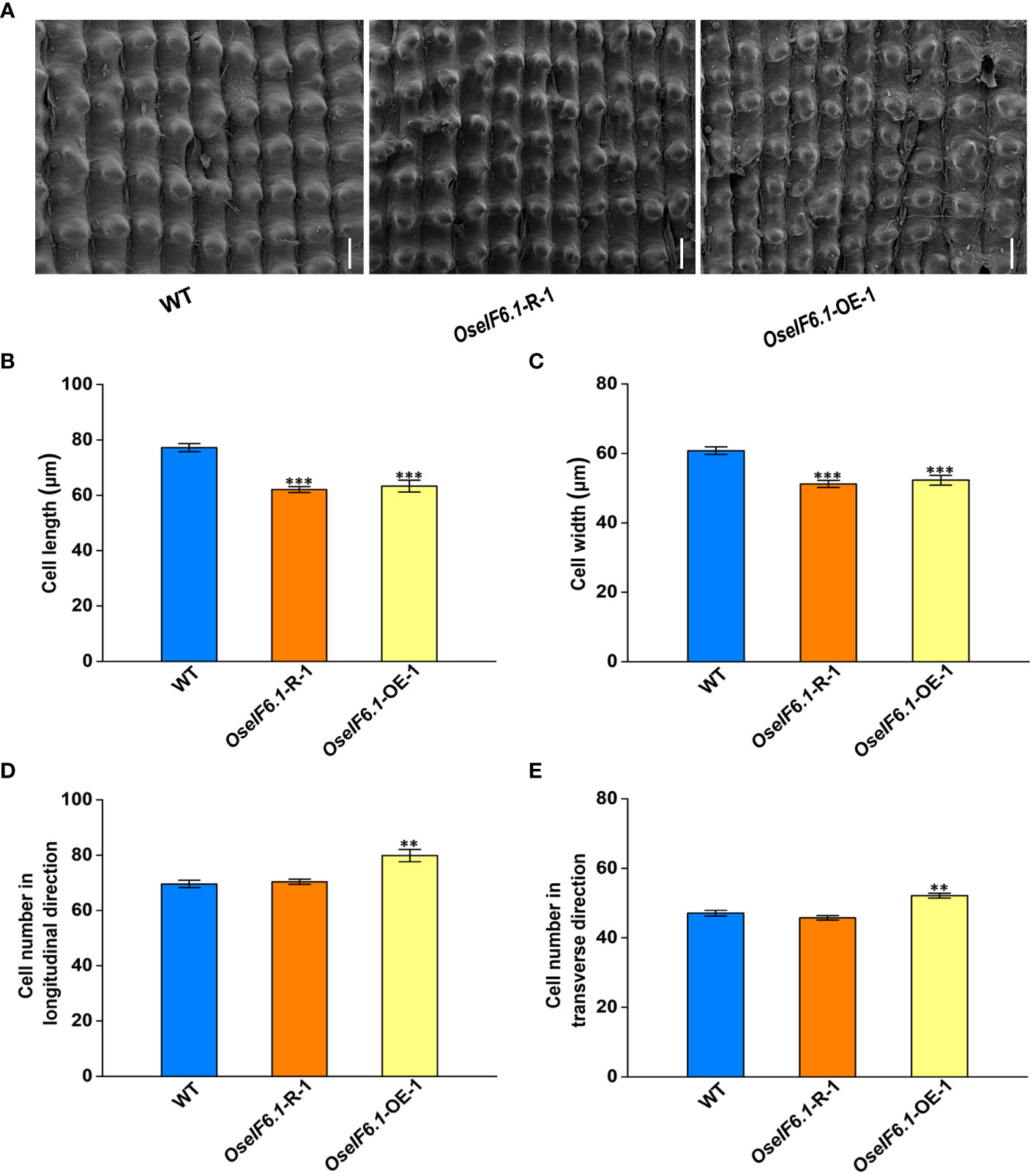
Figure 4 OseIF6.1 regulates grain size by affecting cell expansion and cell proliferation. (A) Scanning electron microscopy images of the glume outer surfaces of WT and OseIF6.1 transgenic lines mature grains. Scale bars = 50 μm. Average length (B) and width (C) of the outer epidermal cells of WT and OseIF6.1 transgenic lines lemmas. (D) Outer epidermal cell number in the longitudinal direction of WT and OseIF6.1 transgenic lines lemmas. (E) Outer epidermal cell number in the transverse direction of WT and OseIF6.1 transgenic lines lemmas. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Previous results have shown that knockdown of OseIF6.1 leads to a reduced seed setting rates. To further investigate this finding, we conducted a pollen viability assay using KI-I2 staining on both WT and OseIF6.1 transgenic plants (Supplementary Figure 2A). The KI-I2 staining rates were 95.7%, 95.6%, and 96.1% in WT and two OseIF6.1-OE anthers, respectively, while the corresponding values were only 48.6% and 50.3% in OseIF6.1-R-1 and OseIF6.1-R-2 anthers, respectively (Supplementary Figure 2B). The lower KI-I2 staining rates in OseIF6.1-R anthers suggest a possible role for OseIF6.1 in the regulation of meiotic progression.
To gain a deeper understanding of the cytological defects in OseIF6.1-R anthers, we conducted a comprehensive analysis of WT and OseIF6.1-R anthers at various stages of development through semithin section assays. During Stage 10, the WT tapetum continues to degrade, with the entire layer of tapetal cells present as a band-like structure. The middle layer is almost completely degraded, and the spherical microspores show multiple small vacuoles (Figure 5A). In contrast, the OseIF6.1-R plants exhibited irregular microspore morphology, with an abnormal distribution of the tapetal cell layer (Figure 5B). Only epidermal cells remained in the WT anthers, the tapetum layer had completely degraded, and the microspores had eventually developed into mature pollen at stage 12 (Figure 5C). However, the OseIF6.1-R anthers exhibit incomplete degradation of the tapetum layer, along with degenerated microspores that show a sickle shape and lack starch accumulation (Figure 5D). To further investigate the defects in anther development of OseIF6.1-R, the transmission electron microscope (TEM) was used to perform ultrathin section assays. As shown in Figures 5E, G, the WT tapetum layer is filled with numerous small vacuoles and organelles, and the surface of the tapetum layer cells is abundant in Ubisch bodies, which play an important role in the transport of substances between the tapetum layer and the microspores. However, the OseIF6.1-R tapetal layer contains fewer intracellular contents and has a reduced number of Ubisch bodies on its surface (Figures 5F, H). Thus, the results obtained in this study demonstrate that the observed pollen defects in OseIF6.1-R anthers are likely linked to tapetal degradation.
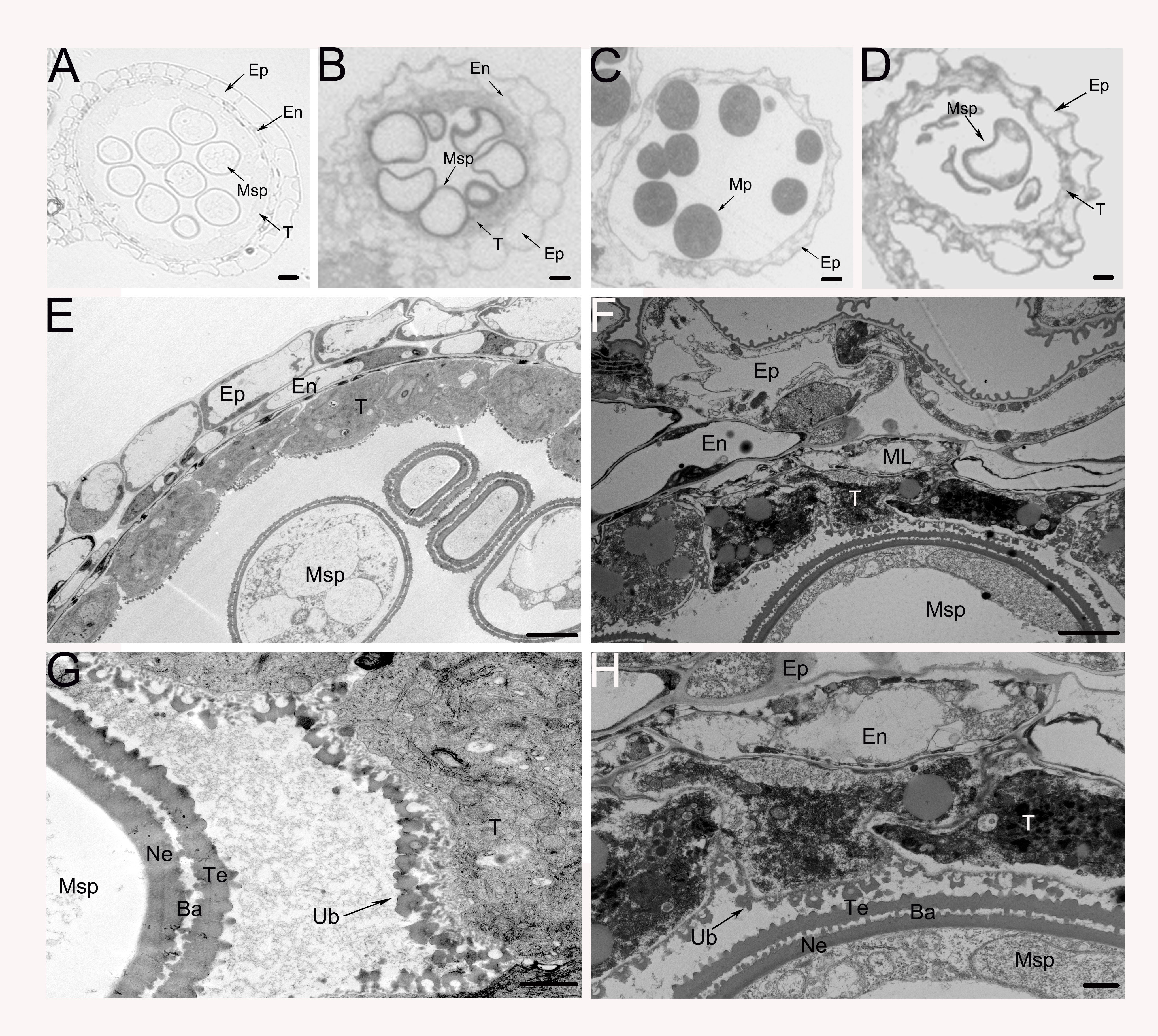
Figure 5 Transverse section and comparison between WT and OseIF6.1 knockdown transgenic anthers. Cross sections of WT (A, C) and OseIF6.1 knockdown transgenic lines (B, D) anthers at stage 10 (A, B) and stage 12 (C, D). Scale bars = 50 μm. Transmission electron microscopy images of WT (E, G) and OseIF6.1 knockdown transgenic lines (F, H) anthers at stage 10. Scale bars = 5 μm in (E) and (F). Scale bars = 1 μm in (G) and (H). Ba, bacula; En, endothecium; Ep, epidermis; ML, middle layer; Msp, microspores; MP, mature pollen; Ne, nexine; T, tapetum; Te, tectum; Ub, Ubisch body.
To test this hypothesis, we conducted an analysis of the expression of genes related to anther development in both WT and OseIF6.1-R young panicles. The expression of PTC2, TDR, and EAT1, which play a crucial role in tapetum programmed cell death (PCD) and pollen wall formation (Zhang et al., 2008; Niu et al., 2013; Uzair et al., 2020), was found to be downregulated in OseIF6.1-R plants compared to WT (Supplementary Figure 3). In addition, similar expression tendency of DPW and CYP703A3 was also observed in OseIF6.1-R plants (Supplementary Figure 3), which regulates sporopollenin precursor biosynthesis (Shi et al., 2011; Yang et al., 2014). Taken together, these findings suggest that OseIF6.1 positively regulates seed setting and pollen viability, and may also affect anther development by altering the expression of related genes.
To further investigate the potential function of OseIF6.1 in rice growth and development, we utilized the publicly accessible rice interactome network to identify proteins that interact with OseIF6.1 (https://bar.utoronto.ca/eplant_rice/ and http://bioinfo.sibs.ac.cn/plant-regulomics/index.php/ Waese et al., 2017; Ran et al., 2020). In the list of proteins predicted to interact with OseIF6.1, OsNMD3 was identified as a potential candidate, which encodes a nuclear export adaptor for the 60S ribosomal subunit. As expected, the yeast two-hybrid (Y2H) results showed that OseIF6.1 interacted with OsNMD3 (Figure 6A). The interaction between OseIF6.1 and OsNMD3 was further confirmed by bimolecular fluorescence complementation (BiFC) assays. OseIF6.1 and OsNMD3 were fused to the N-terminal and C-terminal of YFP, respectively. By co-expressing the OseIF6.1-nYFP and OsNMD3-cYFP constructs in N. benthamiana cells, the reconstitution of YFP fluorescence indicated the direct binding of these two proteins in vivo (Figure 6B). Furthermore, we conducted a pull-down assay to investigate whether OseIF6.1 can engage in direct interaction with OsNMD3 in vitro. We expressed and purified His-tagged OseIF6.1 (OseIF6.1-His) and glutathione S-transferase (GST) tag-fused OsNMD3 (OsNMD3-GST) in Escherichia coli, respectively. As shown in Figure 6C, OseIF6.1-His bound to OsNMD3-GST, but not to the negative control. Thus, these findings suggest that OseIF6.1 can interact with OsNMD3.
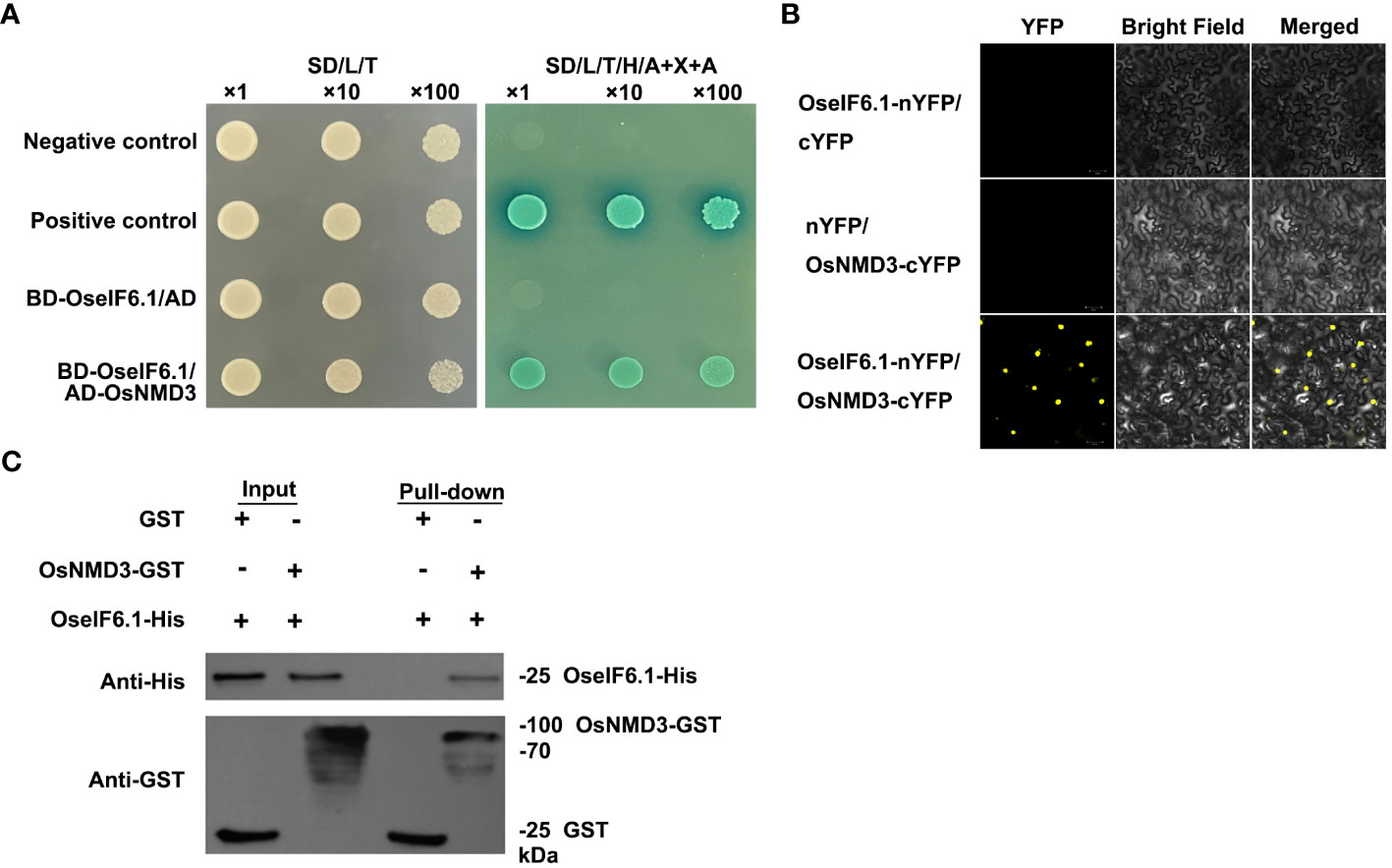
Figure 6 OseIF6.1 physically interacts with OsNMD3. (A) OseIF6.1 interacts with OsNMD3 in yeast cells. Transformed cells were cultured on DDO or QDO/X/A media. (B) Bimolecular fluorescence complementation assays verifies the interaction between OseIF6.1 and OsNMD3 in N. benthamiana. OseIF6.1-nYFP was coexpressed with OsNMD3-cYFP in cells of N. benthamiana. Scale bars = 50 μm. (C) Pull-down assay indicates that OseIF6.1 binds OsNMD3 in vitro. OseIF6.1-His was incubated with OsNMD3-GST and pulled down by OsNMD3-GST.
We employed RNAi approach to generate OsNMD3 knockdown transgenic lines, as we were concerned that gene editing of OsNMD3 may result in a lethal phenotype similar to that of the oseif6.1 mutant. Two OsNMD3-R lines were chosen for further analysis. The expression levels of OsNMD3 were reduced by 0.52 and 0.69 fold in OsNMD3-R-1 and OsNMD3-R-2, respectively, compared to that in WT (Supplementary Figure 4). Subsequently, we conducted an assessment of agronomic traits in mature WT and OsNMD3-R plants (Figure 7A). The OsNMD3-R plants exhibited shorter plant height and smaller panicles compared to the WT (Figures 7B, C). Furthermore, the OsNMD3-R plants had fewer primary and secondary branches than the WT, resulting in a decrease in grain number per panicle (Figures 7D–F). Interestingly, the setting rate in OsNMD3-R plants was significantly lower compared to the WT (Figure 7G). We then performed pollen viability assays on WT and OsNMD3-R anthers using KI-I2 staining. The results showed that the pollen viability of OsNMD3-R-1 and OsNMD3-R-2 anthers was 31.8% and 34.2%, respectively, which was markedly lower compared to the WT anthers (Supplementary Figure 5). Taken together, these results suggest that the knockdown of OsNMD3 has negative impacts on plant architecture and pollen viability.
Originally purified from wheat germ, eukaryotic initiation factor 6 (eIF6) was discovered to function as a ribosome dissociation factor (Russell and Spremulli, 1980). By binding to the 60S ribosome subunit, eIF6 prevents its association with the 40S ribosome subunit (Russell and Spremulli, 1980). Homologous proteins of wheat eIF6 were subsequently isolated and purified from various sources, including rabbit (Raychaudhuri et al., 1984), calf, human (Si et al., 1997), yeast (Si and Maitra, 1999), and Arabidopsis (Kato et al., 2010). The phylogenetic analysis indicates that the eIF6 protein is conserved among eukaryotes (Figure 1A). Structural comparisons of eIF6 proteins have further revealed that OseIF6.1 shares a close relationship with other homologues (Supplementary Figure 1). Several studies on yeast and mammals have demonstrated the crucial role of eIF6 in growth regulation. Depletion of eIF6 is lethal in both yeast and mouse models (Si et al., 1997; Sanvito et al., 1999; Wood et al., 1999; Gandin et al., 2008). Like in yeast and mouse, the null allele of the At-eIF6;1 in Arabidopsis results in an embryonic-lethal phenotype, underscoring the essential role of eIF6 in plant embryogenesis (Kato et al., 2010). Although numerous studies have examined the role of eIF6 proteins in eukaryotes, our previous study demonstrated that oseif6.1 mutant was unable to harvest seeds, which resulted in a lethal phenotype (Guo et al., 2022). Therefore, the functional characterization of the OseIF6.1 gene remains to be fully understood.
Given that the mutant of OseIF6.1 results in embryonic lethality, we conducted an investigation into the function of OseIF6.1 in rice by either knocking down or overexpressing its expression in Nipponbare. In this study, we found that OseIF6.1 plays an important role in the development of plant architecture and grain shape. Both the knockdown and overexpression of OseIF6.1 resulted in a dwarfing phenotype (Figure 2A). It has been widely reported that gibberellins (GA) are the primary regulators of plant height, influencing the growth and development of stems and determining the overall stature of the plant (Salas Fernandez et al., 2009). The expression of the genes related to GA biosynthesis in stems of WT and OseIF6.1 transgenic plants was examined. We found that the expression of OsGA20ox1 (Oikawa et al., 2004), OsGA20ox2 (Sasaki et al., 2002; Su et al., 2021), and OsKO2 (Itoh et al., 2004) was suppressed in OseIF6.1-R and OseIF6.1-OE stems (Supplementary Figure 6). These results suggest that altered eIF6.1 expression affects the expression of genes related to GA biosynthesis and ultimately plant height. Furthermore, the knockdown of OseIF6.1 resulted in short, narrow, and thin grains, whereas the overexpression of OseIF6.1 resulted in long grains (Figures 3A–D), indicating that OseIF6.1 acts as a positive regulator of grain shape. The conclusion was reinforced through scanning electron microscopy observations, which disclosed that OseIF6.1 has a positive impact on grain size by regulating cell elongation and proliferation (Figure 4). Therefore, OseIF6.1 seems to be a key determinant of plant structure and rice grain morphology. These lines exhibit no difference in seed germination compared to the WT. However, the OseIF6.1-R and OE lines both exhibit a reduced 1000-grain weight compared to the WT (Figure 3E), potentially due to changes in OseIF6.1 expression that affect the translation efficiency of mRNAs related to grain development. Despite the OseIF6.1-OE lines having increased translation efficiency, there is a decrease in both grain width and thickness. This could potentially be attributed to compensatory mechanisms or feedback regulation, resulting in a similar 1000-grain weight phenotype to that observed in the OseIF6.1-R lines.
The At-eIF6;1 heterozygous plants exhibit a 1:3 ratio of growth defects in their silique seeds, which display a pale yellow coloration (Kato et al., 2010). Interestingly, we have noted that plants with OseIF6.1-R show decreased seed-setting rates alongside diminished pollen fertility (Figure 2H; Supplementary Figure 2). Subsequent analysis using transmission electron microscopy revealed that the abortion of OseIF6.1-R pollens is associated with the degradation of the tapetum layer (Figure 5). Numerous investigations have identified a range of genes that play a pivotal role in tapetum PCD and the formation of pollen walls. These genes include Udt1 (Jung et al., 2005), Wda1 (Jung et al., 2006), TDR (Zhang et al., 2008), DPW (Shi et al., 2011), EAT1 (Niu et al., 2013), CYP703A3 (Yang et al., 2014), and PTC2 (Uzair et al., 2020). We observed that the expression of CYP703A3, PTC2, TDR, EAT1 and DPW genes was downregulated in OseIF6.1-R young panicles (Supplementary Figure 3). These findings suggest that OseIF6.1 is essential for anther development. Consistent with this, the depletion of eIF6 in yeast causes a reduction in the availability of free 60S ribosomal subunits, resulting in a decrease in translational activity, which is eventually lethal (Wood et al., 1999; Basu et al., 2001). In addition, it has been reported that eIF6-null embryos are lethal at the preimplantation stage in mice, and the quality of hepatic and adipose tissue in the heterozygous eIF6 mice is reduced due to the reduction in cell number and perturbation of the G1/S cell cycle process (Gandin et al., 2008). Together with our results, these data suggest that OseIF6.1 plays a vital role in embryonic development across eukaryotes.
The biogenesis of ribosomes is a complex process that involves various trans-acting factors. eIF6 can bind to immature large ribosomal subunits as well as other trans-acting factors in the nucleolus (Miluzio et al., 2009). NMD3 is a conserved transcriptional factor that encodes the nuclear export adaptor for the 60S ribosomal subunit. This protein is characterized by its N-terminus, which contains Cx2C repeats and a nuclear localization sequence. The C-terminus of NMD3 features a nuclear export sequence, which is essential for maintaining the efficiency of normal protein synthesis (Shi et al., 2014). Overexpression of OsNMD3ΔNLS, which lacks a nuclear localization site, caused abnormal plant growth and development in rice, including dwarfism and reduced grain size (Shi et al., 2014). In the Arabidopsis AtNMD3ΔNES OE line, pleiotropic phenotypes were observed, such as reduced plant height and stamen size, as well as an obvious curly shape of the rosette leaves (Chen et al., 2012). NMD3 may interact with specific ribosomal proteins, especially those associated with the 60S ribosomal subunit, facilitating their proper assembly and export from the nucleus to the cytoplasm, given the roles of its homologs in other species (Hedges et al., 2006; Sengupta et al., 2010; Chen et al., 2012). It has been reported that OsNMD3 interacts with the 60S subunit through OsRPL10Ac1 (Shi et al., 2014). In this study, we have discovered that OseIF6.1 interacts with OsNMD3 (Figure 6). Interestingly, the OsNMD3-R plants exhibit similar phenotypic traits to those of OseIF6.1-R plants. The OsNMD3-R plants also exhibit reduced plant height and pollen fertility (Figure 7; Supplementary Figure 5). The expression of the GA biosynthesis-related genes, including OsGA20ox1, OsGA20ox2, and OsKO2 were decreased in stems of OsNMD3-R plants (Supplementary Figure 7). Similar dwarfism and sterility phenomena have also been observed in other eIFs RNAi plants. For example, it has been reported that OseIF3e protein interacts with OseIF6. Notably, RNAi targeting OseIF3e causes significant plant height and pollen maturation defects (Wang et al., 2016). The mutation of OseIF3h has been found to cause abnormal development in rice, particularly affecting plant height and pollen fertility (Huang et al., 2021). Similarly, the T-DNA mutants of AteIF3e exhibited diminished pollen fertility, along with various floral and reproductive defects. Overexpressing AteIF3e led to dwarfism and affected seed formation (Yahalom et al., 2008). The differences in seed setting between the OseIF6.1-R and OsNMD3-R lines suggest that they may regulate rice growth via distinct or partially overlapping pathways. Higher seed setting rates in OseIF6.1-R lines imply that its role might be partly independent of OsNMD3, indicating the existence of additional pathways or partners for OseIF6.1. The more significant effect observed with OsNMD3 reduction could indicate that OsNMD3 operates downstream of OseIF6.1 or in a parallel pathway that is crucial for rice fertility. In future studies, genetic epistasis analysis can elucidate their relationships in rice development.
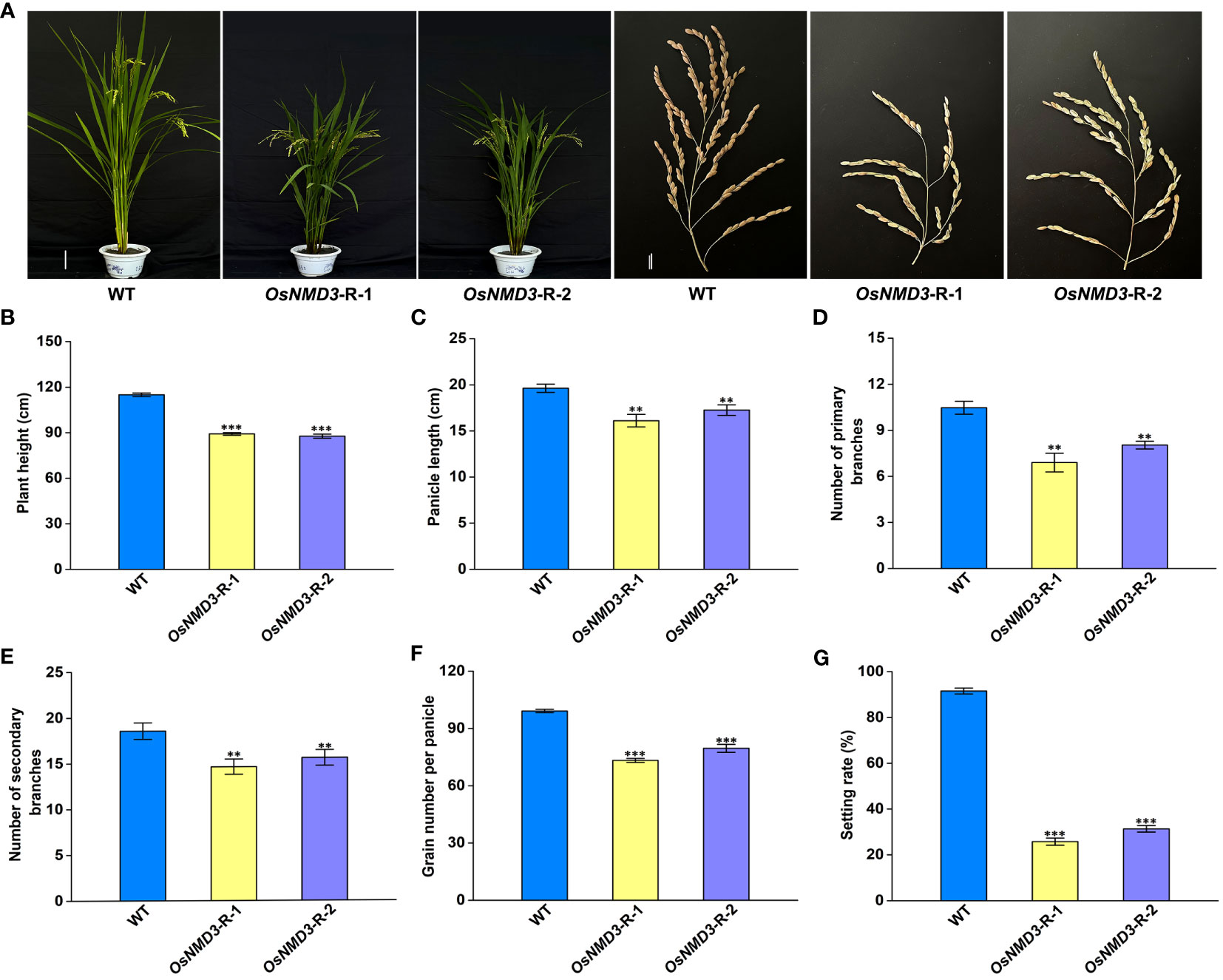
Figure 7 Knockdown of OsNMD3 affects multiple agronomic traits. (A) Plants and Panicles of WT and OsNMD3 knockdown transgenic plants at the mature stage. Scale bars = 20 cm for plant height and 3 cm for panicle length. (B) Plant heights of WT and OsNMD3 knockdown transgenic plants. (C) Panicle length of WT and OsNMD3 knockdown transgenic plants. (D) Number of primary branches of WT and OsNMD3 knockdown transgenic panicles. (E) Number of secondary branches of WT and OsNMD3 knockdown transgenic panicles. (F) Grain number per panicle of WT and OsNMD3 knockdown transgenic plants. (G) Setting rate of WT and OsNMD3 knockdown transgenic plants. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
In conclusion, our findings indicate that OseIF6.1 plays a regulatory role in the expression of sporopollenin precursor biosynthesis genes, as well as genes involved in tapetal PCD, thereby influencing tapetal development and the formation of pollen. Additionally, it modulates the expression of GA biosynthesis-related genes, thus regulating plant height. Furthermore, OseIF6.1 interacts with OsNMD3, which also plays a crucial role in regulating pollen fertility and plant height. By identifying and characterizing OseIF6.1, the research provides insights into the genetic factors that influence rice growth and development. Understanding these genetic components is crucial for breeding and genetic engineering efforts aimed at crop improvement. The specific roles of OseIF6.1 in regulating grain length and fertility highlight it as a potential target for genetic engineering.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HG: Investigation, Writing – original draft, Writing – review & editing, Data curation. JL: Data curation, Writing – original draft. XS: Investigation, Writing – original draft. LC: Data curation, Investigation, Writing – original draft. JR: Data curation, Writing – original draft. LL: Data curation, Writing – original draft. MR: Writing – original draft. SL: Writing – original draft. MD: Writing – original draft. GR: Writing – original draft, Writing – review & editing. FG: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Natural Science Foundation of Sichuan Province (2023NSFSC1277), the earmarked fund for China Agriculture Research System (CARS-01), Independent Innovation Transformation Project of Crop Research Institute of Sichuan Academy of Agricultural Sciences (QNCX2301), the Independent Innovation Project of Sichuan Finance (2022ZZCX001), and the Original Innovation Program of Sichuan Academy of Agricultural Sciences (YSCX2035-010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1366986/full#supplementary-material
Supplementary Figure 1 | Alignment of eIF6 homologues from Oryza sativa, Arabidopsis thaliana, Homo sapiens, and Saccharomyces cerevisiae.
Supplementary Figure 2 | Fertility analysis of WT and OseIF6.1 transgenic plants. (A) KI-I2 staining of WT, OseIF6.1-R-1, and OseIF6.1-OE-1 pollen. Normal pollen appears dark colored. Scale bars = 500 μm. (B) KI-I2 staining rate of WT and OseIF6.1 transgenic plants pollen. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 3 | Expression levels of genes involved in anther development in WT and OseIF6.1 knockdown transgenic plants young panicles. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 4 | OsNMD3 expression in WT and OsNMD3 knockdown transgenic lines. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 5 | Fertility analysis of WT and OsNMD3 knockdown transgenic plants. (A) KI-I2 staining of WT and OsNMD3 knockdown transgenic lines pollen. Normal pollen appears dark colored. Scale bars = 1 mm. (B) KI-I2 staining rate of WT and OsNMD3 knockdown transgenic lines pollen. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 6 | Expression levels of genes involved in gibberellin biosynthesis in stems of WT and OseIF6.1 transgenic plants. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 7 | Expression levels of genes involved in gibberellin biosynthesis in stems of WT and OsNMD3 knockdown transgenic plants. The values represent means ± SE derived from at least three independent experiments. Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Figure 8 | A working model for the role of OseIF6.1 in rice growth and development.
Aitken, C. E., Lorsch, J. R. (2012). A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576. doi: 10.1038/nsmb.2303
Aslanidis, C., de Jong, P. J., Schmitz, G. (1994). Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl. 4, 172–177. doi: 10.1101/gr.4.3.172
Basu, U., Si, K., Warner, J. R., Maitra, U. (2001). The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell Biol. 21, 1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001
Cao, J. B., Cheng, K., Yuan, M. (2018). Observation of rice tissues with semi-thin section. Bio 101, e1010142. doi: 10.21769/BioProtoc.1010142
Castellano, M. M., Merchante, C. (2021). Peculiarities of the regulation of translation initiation in plants. Curr. Opin. Plant Biol. 63, 102073. doi: 10.1016/j.pbi.2021.102073
Ceci, M., Gaviraghi, C., Gorrini, C., Sala, L. A., Offenhäuser, N., Marchisio, P. C., et al. (2003). Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584. doi: 10.1038/nature02160
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, K., Guo, T., Li, X. M., Zhang, Y. M., Yang, Y. B., Ye, W. W., et al. (2019). Translational regulation of plant response to high temperature by a dual-function tRNAHis guanylyltransferase in rice. Mol. Plant 12, 1123–1142. doi: 10.1016/j.molp.2019.04.012
Chen, M. Q., Zhang, A. H., Zhang, Q., Zhang, B. C., Nan, J., Li, X., et al. (2012). Arabidopsis NMD3 is required for nuclear export of 60S ribosomal subunits and affects secondary cell wall thickening. PloS One 7, e35904. doi: 10.1371/journal.pone.0035904
Chen, S., Songkumarn, P., Liu, J., Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Diédhiou, C. J., Popova, O. V., Dietz, K. J., Golldack, D. (2008). The SUI-homologous translation initiation factor eIF-1 is involved in regulation of ion homeostasis in rice. Plant Biol. 10, 298–309. doi: 10.1111/j.1438-8677.2008.00037.x
Feng, H., Chen, Q., Feng, J., Zhang, J., Yang, X., Zuo, J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144, 1531–1545. doi: 10.1104/pp.107.098079
Gandin, V., Miluzio, A., Barbieri, A. M., Beugnet, A., Kiyokawa, H., Marchisio, P. C., et al. (2008). Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455, 684–688. doi: 10.1038/nature07267
Gebauer, F., Hentze, M. W. (2004). Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835. doi: 10.1038/nrm1488
Guo, H., Cui, Y., Huang, L., Ge, L., Xu, X., Xue, D., et al. (2022). The RNA binding protein OsLa influences grain and anther development in rice. Plant J. 110, 1397–1414. doi: 10.1111/tpj.15746
Guo, J., Jin, Z., Yang, X., Li, J., Chen, J. (2011). Eukaryotic initiation factor 6, an evolutionarily conserved regulator of ribosome biogenesis and protein translation. Plant Signal Behav. 6, 766–771. doi: 10.4161/psb.6.5.15438
Hedges, J., Chen, Y., West, M., Bussiere, C., Johnson, A. W. (2006). Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 281, 36579–36587. doi: 10.1074/jbc.M606798200
Hiei, Y., Ohta, S., Komari, T., Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hinnebusch, A. G. (2006). eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562. doi: 10.1016/j.tibs.2006.08.005
Hinnebusch, A. G., Lorsch, J. R. (2012). The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544. doi: 10.1101/cshperspect.a011544
Huang, Y., Zheng, P., Liu, X., Chen, H., Tu, J. (2021). OseIF3h Regulates Plant Growth and Pollen Development at Translational Level Presumably through Interaction with OsMTA2. Plants (Basel). 10, 1101. doi: 10.3390/plants10061101
Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., et al. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54, 533–547. doi: 10.1023/B:PLAN.0000038261.21060.47
Jackson, R. J., Hellen, C. U., Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi: 10.1038/nrm2838
Jung, K. H., Han, M. J., Lee, Y. S., Kim, Y. W., Hwang, I., Kim, M. J., et al. (2005). Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 17, 2705–2722. doi: 10.1105/tpc.105.034090
Jung, K. H., Han, M. J., Lee, D. Y., Lee, Y. S., Schreiber, L., Franke, R., et al. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. 18, 3015–3032. doi: 10.1105/tpc.106.042044
Kapp, L. D., Lorsch, J. R. (2004). The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657–704. doi: 10.1146/annurev.biochem.73.030403.080419
Kato, Y., Konishi, M., Shigyo, M., Yoneyama, T., Yanagisawa, S. (2010). Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: the essential role in embryogenesis and their differential expression in Arabidopsis and rice. Biochem. Biophys. Res. Commun. 397, 673–678. doi: 10.1016/j.bbrc.2010.06.001
Li, Q., Deng, Z., Gong, C., Wang, T. (2016). The rice eukaryotic translation initiation factor 3 subunit f (OseIF3f) is involved in microgametogenesis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00532
Li, N., Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Yang, Y., Wang, Y., Cheng, K., Zhou, X., Li, J., et al. (2022). SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato. Plant Cell. 34, 2747–2764. doi: 10.1093/plcell/koac104
Merrick, W. C., Pavitt, G. D. (2018). Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 3, a033092. doi: 10.1101/cshperspect.a033092
Miluzio, A., Beugnet, A., Volta, V., Biffo, S. (2009). Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 10, 459–465. doi: 10.1038/embor.2009.70
Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A., Hu, J., et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4, 1445. doi: 10.1038/ncomms2396
Oikawa, T., Koshioka, M., Kojima, K., Yoshida, H., Kawata, M. (2004). A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 55, 687–700. doi: 10.1007/s11103-004-1692-y
Raabe, K., Honys, D., Michailidis, C. (2019). The role of eukaryotic initiation factor 3 in plant translation regulation. Plant Physiol. Biochem. 145, 75–83. doi: 10.1016/j.plaphy.2019.10.015
Ran, X., Zhao, F., Wang, Y., Liu, J., Zhuang, Y., Ye, L., et al. (2020). Plant Regulomics: a data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant J. 101, 237–248. doi: 10.1111/tpj.14526
Rausell, A., Kanhonou, R., Yenush, L., Serrano, R., Ros, R. (2003). The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 34, 257–267. doi: 10.1046/j.1365-313X.2003.01719.x
Raychaudhuri, P., Stringer, E. A., Valenzuela, D. M., Maitra, U. (1984). Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr = 25,000). J. Biol. Chem. 259, 11930–11935. doi: 10.1016/S0021-9258(20)71300-1
Roy, B., Copenhaver, G. P., Von Arnim, A. G. (2011). Fluorescence-tagged transgenic lines reveal genetic defects in pollen growth-application to the eIF3 complex. PloS One 6, e17640. doi: 10.1371/journal.pone.0017640
Roy, B., von Arnim, A. G. (2013). Translational regulation of cytoplasmic mRNAs. Arabidopsis Book. 11, e0165. doi: 10.1199/tab.0165
Russell, D. W., Spremulli, L. L. (1980). Mechanism of action of the wheat germ ribosome dissociation factor: interaction with the 60 S subunit. Arch. Biochem. Biophys. 201, 518–526. doi: 10.1016/0003-9861(80)90540-8
Salas Fernandez, M. G., Becraft, P. W., Yin, Y., Lübberstedt, T. (2009). From dwarves to giants? plant height manipulation for biomass yield. Trends Plant Sci. 14, 454–461. doi: 10.1016/j.tplants.2009.06.005
Sanvito, F., Piatti, S., Villa, A., Bossi, M., Lucchini, G., Marchisio, P. C., et al. (1999). The β4 integrin interactor p27BBP/eIF6 is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 144, 823–837. doi: 10.1083/jcb.144.5.823
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Sengupta, J., Bussiere, C., Pallesen, J., West, M., Johnson, A., Frank, J. (2010). Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 189, 1079–1086. doi: 10.1083/jcb.201001124
Shi, Y., Liu, X., Li, R., Gao, Y., Xu, Z., Zhang, B., et al. (2014). Retention of OsNMD3 in the cytoplasm disturbs protein synthesis efficiency and affects plant development in rice. J. Exp. Bot. 65, 3055–3069. doi: 10.1093/jxb/eru150
Shi, J., Tan, H., Yu, X. H., Liu, Y., Liang, W., Ranathunge, K., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell. 23, 2225–2246. doi: 10.1105/tpc.111.087528
Si, K., Chaudhuri, J., Chevesich, J., Maitra, U. (1997). Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc. Natl. Acad. Sci. U.S.A. 194, 14285–14290. doi: 10.1073/pnas.94.26.14285
Si, K., Maitra, U. (1999). The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell Biol. 19, 1416–1426. doi: 10.1128/MCB.19.2.1416
Singh, B., Chauhan, H., Khurana, J. P., Khurana, P., Singh, P. (2013). Evidence for the role of wheat eukaryotic translation initiation factor 3 subunit g (TaeIF3g) in abiotic stress tolerance. Gene 532, 177–185. doi: 10.1016/j.gene.2013.09.078
Singha, D. L., Maharana, J., Panda, D., Dehury, B., Modi, M. K., Singh, S. (2021). Understanding the thermal response of rice eukaryotic transcription factor eIF4A1 towards dynamic temperature stress: insights from expression profiling and molecular dynamics simulation. J. Biomol Struct. Dyn. 39, 2575–2584. doi: 10.1080/07391102.2020.1751295
Sonenberg, N., Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. doi: 10.1016/j.cell.2009.01.042
Su, S., Hong, J., Chen, X., Zhang, C., Chen, M., Luo, Z., et al. (2021). Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 19, 2304–2318. doi: 10.1111/pbi.13661
Thompson, J. E., Hopkins, M. T., Taylor, C., Wang, T. W. (2004). Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 9, 174–179. doi: 10.1016/j.tplants.2004.02.008
Uzair, M., Xu, D., Schreiber, L., Shi, J., Liang, W., Jung, K. H., et al. (2020). PERSISTENT TAPETAL CELL2 is required for normal tapetal programmed cell death and pollen wall patterning. Plant Physiol. 182, 962–976. doi: 10.1104/pp.19.00688
Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., et al. (2017). ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell. 29, 1806–1821. doi: 10.1105/tpc.17.00073
Wang, W., Xu, M., Liu, X., Tu, J. (2016). The rice eukaryotic translation initiation factor 3 subunit e (OseIF3e) influences organ size and pollen maturation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01399
Wei, L. Q., Xu, W. Y., Deng, Z. Y., Su, Z., Xue, Y., Wang, T. (2010). Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11, 338. doi: 10.1186/1471-2164-11-338
Wood, L. C., Ashby, M. N., Grunfeld, C., Feingold, K. R. (1999). Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J. Biol. Chem. 274, 11653–11659. doi: 10.1074/jbc.274.17.11653
Xia, C., Wang, Y. J., Li, W. Q., Chen, Y. R., Deng, Y., Zhang, X. Q., et al. (2010). The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 63, 189–202. doi: 10.1111/j.1365-313X.2010.04237.x
Xu, X. Y., Ding, Z. J., Chen, L., Yan, J. Y., Li, G. X., Zheng, S. J. (2015). An eukaryotic translation initiation factor, AteIF5A-2, affects cadmium accumulation and sensitivity in Arabidopsis. J. Integr. Plant Biol. 57, 848–858. doi: 10.1111/jipb.12329
Yahalom, A., Kim, T. H., Roy, B., Singer, R., von Arnim, A. G., Chamovitz, D. A. (2008). Arabidopsis eIF3e is regulated by the COP9 signalosome and has an impact on development and protein translation. Plant J. 53, 300–311. doi: 10.1111/j.1365-313X.2007.03347.x
Yang, X., Wu, D., Shi, J., He, Y., Pinot, F., Grausem, B., et al. (2014). Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56, 979–994. doi: 10.1111/jipb.12212
Keywords: OseIF6.1, eukaryotic translation initiation factor, grain shape, pollen sterility, seed setting
Citation: Guo H, Lv J, Su X, Chen L, Ren J, Liu L, Ren M, Liu S, Dai M, Ren G and Gao F (2024) Rice OseIF6.1 encodes a eukaryotic translation initiation factor and is essential for the development of grain and anther. Front. Plant Sci. 15:1366986. doi: 10.3389/fpls.2024.1366986
Received: 08 January 2024; Accepted: 11 March 2024;
Published: 21 March 2024.
Edited by:
Muhammad Jamil, Kohat University of Science and Technology, PakistanReviewed by:
Kamran Iqbal Shinwari, Nanjing Agricultural University, ChinaCopyright © 2024 Guo, Lv, Su, Chen, Ren, Liu, Ren, Liu, Dai, Ren and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangjun Ren, Z3VhbmdqdW42MUBzaW5hLmNvbQ==; Fangyuan Gao, Z2Z5MjQ2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.