- 1Plant Development, The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
- 2Metabolite Traits in Plants, The New Zealand Institute for Plant and Food Research Limited, Palmerston, North, New Zealand
Strigolactones (SLs), a class of carotenoid-derived hormones, play a crucial role in flowering plants by regulating underground communication with symbiotic arbuscular mycorrhizal fungi (AM) and controlling shoot and root architecture. While the functions of core SL genes have been characterized in many plants, their roles in non-tracheophyte plants like liverworts require further investigation. In this study, we employed the model liverwort species Marchantia polymorpha, which lacks detectable SL production and orthologs of key SL biosynthetic genes, including CAROTENOID CLEAVAGE DIOXYGENASE 8 (CCD8) and MORE AXILLARY GROWTH 1 (MAX1). However, it retains some SL pathway components, including DWARF27 (D27) and CCD7. To help elucidate the function of these remaining components in M. polymorpha, knockout mutants were generated for MpD27–1, MpD27–2 and MpCCD7. Phenotypic comparisons of these mutants with the wild-type control revealed a novel role for these genes in regulating the release of gemmae from the gemma cup and the germination and growth of gemmae in the dark. Mpd27–1, Mpd27–2, and Mpccd7 mutants showed lower transcript abundance of genes involved in photosynthesis, such as EARLY LIGHT INDUCED (ELI), and stress responses such as LATE EMBRYOGENESIS ABUNDANT (LEA) but exhibited higher transcript levels of ETHYLENE RESPONSE FACTORS (ERFs) and SL and carotenoid related genes, such as TERPENE SYNTHASE (TS), CCD7 and LECITHIN-RETINAL ACYL TRANSFERASE (LRAT). Furthermore, the mutants of M. polymorpha in the SL pathway exhibited increased contents of carotenoid. This unveils a previously unrecognized role for MpD27–1, MpD27–2 and MpCCD7 in controlling release, germination, and growth of gemmae in response to varying light conditions. These discoveries enhance our comprehension of the regulatory functions of SL biosynthesis genes in non-flowering plants.
Introduction
Carotenoids are a group of isoprenoid metabolites produced by all photosynthetic organisms, including plants, algae, and cyanobacteria, which perform essential roles as accessory and photoprotective pigments (Sun et al., 2022). Carotenoids also play other roles in plants, for example, they provide precursors for strigolactone (SL) and abscisic acid (ABA) synthesis (Sun et al., 2022).
The evolution of mechanisms to communicate with other kingdoms of life has been central to plants adapting to the terrestrial environment (Brundrett, 2002; Bidartondo et al., 2011; Carella and Schornack, 2018; Aquino et al., 2021). One such adaptation to arise after the migration to land is believed to be the symbiotic interaction of plants with arbuscular mycorrhizal (AM) fungi to cope with nutrient starvation (de Saint Germain et al., 2013; Foo and Reid, 2013; Delaux et al., 2015). SLs play a key role in establishing interaction between plant roots and AM fungi (Yeum and Russell, 2002). Beyond their role as rhizosphere signaling molecule, SLs also regulate various aspects of plant growth and development, such as root and shoot architecture, and plant adaptation to stress responses as illustrated in Figure 1A (Yeum and Russell, 2002). In flowering plants, the SL pathway is highly conserved and well characterized, with key biosynthesis and signaling genes having been identified (Gomez-Roldan et al., 2008; Dun et al., 2013; Drummond et al., 2015, 2023). However, in primary land plant lineages such as bryophytes, the SL pathway requires further investigation (Walker et al., 2019).
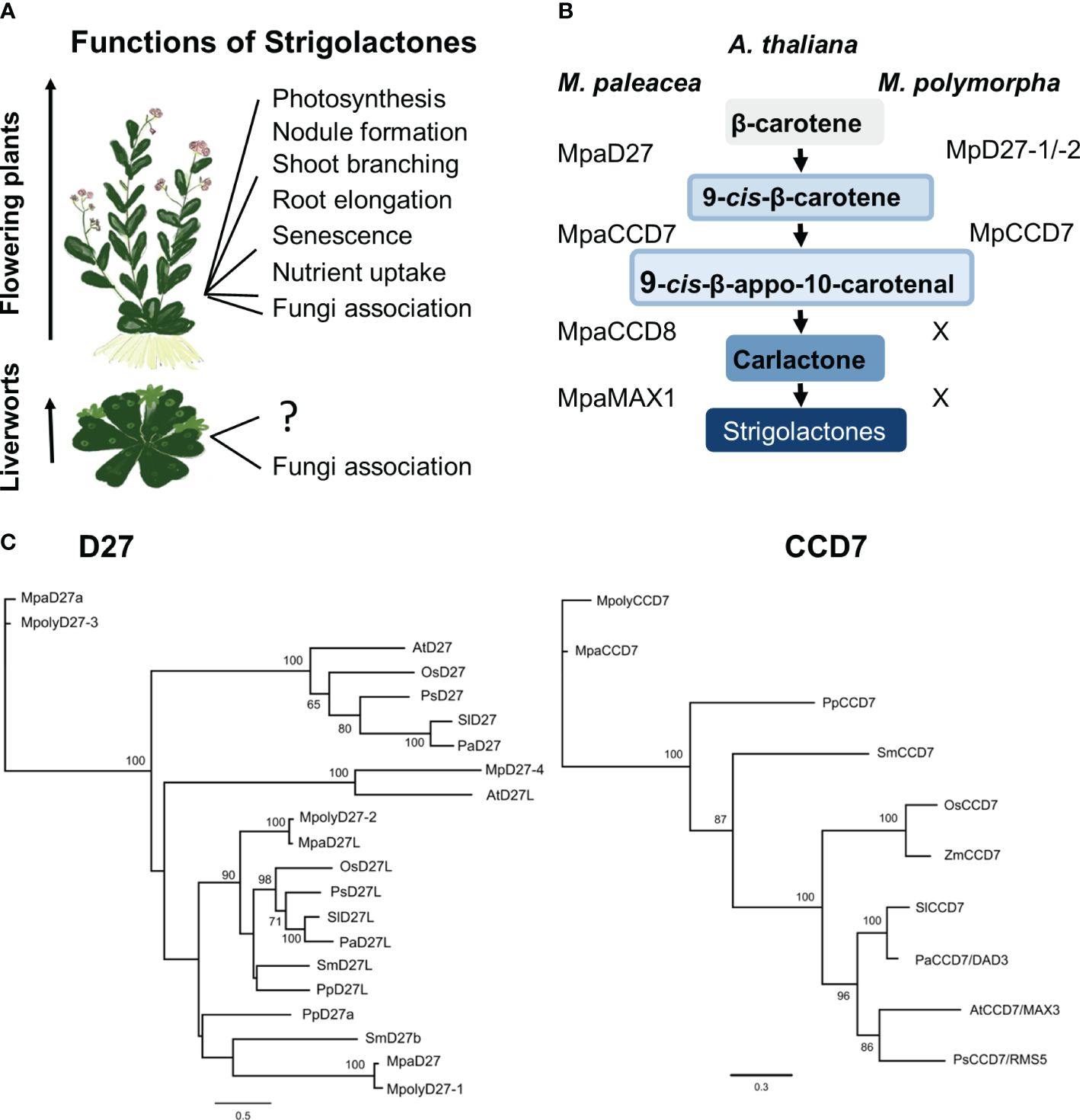
Figure 1 Strigolactones (SL) in flowering plants and liverworts. (A) SL functions in flowering plants and liverworts. (B) Homologs of SL precursor biosynthesis genes in M. polymorpha and M. paleacea. (C) Phylogenetic analysis of D27, D27L and CCD7 proteins across various plant species. Maximum likelihood phylogenetic trees were built by using PhyML with default settings (Guindon and Gascuel, 2003). Alignment was conducted using MUSCLE 3.8.425 (Edgar, 2004) within the Geneious Prime suite of software (https://www.geneious.com). Tree branch labels indicate probability percentages. The horizontal branch length is directly proportional to sequence divergence, as shown by the scale.
Published data suggest that some plant lineages lack a full complement of biosynthetic genes necessary to produce SL (Walker et al., 2019). For example, hornworts lack DWARF 27 (D27), CAROTENOID CLEAVAGE DIOXYGENASE 7 (CCD7), and MORE AXILLARY GROWTH 1 (MAX1) but possess CCD8 and an ancestral form of LATERAL BRANCHING OXIDOREDUCTASE (proto-LBO), the functions of which are unclear. In the model liverwort species M. polymorpha, orthologs for D27 and CCD7 are present, but the key SL biosynthesis genes, CCD8 and MAX1, and canonical SL metabolites have not been reported (Walker et al., 2019) (Figure 1B). In contrast, a close relative, Marchantia paleacea, possesses all the SL biosynthesis enzymes, exhibits growth suppression when treated with synthetic SLs, and interacts with AM fungi to enhance water and nutrient uptake, particularly phosphorous (Humphreys et al., 2010; Costa and Peralta, 2015; Sgroi and Paszkowski, 2020). This raises questions regarding the roles of D27 and CCD7 in M. polymorpha, and whether they contribute to plant adaptation responses.
Bryophytes, including the monophyletic lineages of hornworts, mosses, and liverworts, are thought to be diverged from the common ancestor they share with flowering plants roughly 400 million years ago (Shaw et al., 2011; Morris et al., 2018; Li et al., 2020). M. polymorpha (hereafter referred to as Marchantia) serves as a model plant for the liverworts because it is feasible to cultivate, has a small genome, and the genetic manipulations have been well established (Ishizaki et al., 2016; Poveda, 2020; Kohchi et al., 2021; Bowman et al., 2022). Marchantia produces distinct unisexual individuals, male and female, each with gametophyte and sporophyte generations. The dominant and haploid gametophyte form, contrasts with the dependent diploid sporophyte. Asexual propagation occurs through disc-like vegetative propagules known as gemmae. When conditions become conducive to sexual reproduction, such as long days and an abundance of far-red light, male and female gametophytes produce sex structures known as antheridiophores and archegoniophores, respectively. Liverworts lack true roots but possess root-like structures called rhizoids on the ventral side of the thallus. Rhizoids serve multiple functions, including nutrient and water absorption from soil, facilitating water distribution across the thallus, anchoring the thallus to a substrate, and forming mycorrhizal interactions.
To help establish the function of SL-related genes in Marchantia, we generated knockout mutants for the orthologs of D27–1, D27–2 and CCD7. The mutant lines, along with wild-type plant, were quantitatively assessed for adaptive fitness when experiencing nutrient stress in either standard day/night or dark conditions. Our findings indicate that MpD27–1, MpD27–2 and MpCCD7 have actions in pathways controlling growth of gemmae. Gemmae are formed in the gemma cups, where they remain dormant until dislodged either by physical action, such as impact of rain, or the death of the parental plant. The Mpd27–1, Mpd27–2 and Mpccd7 mutants exhibit increased release of gemmae from the gemma cup and enhanced gemmae germination and growth in the dark compared with wild-type plant. Additionally, the mutants have increased transcript abundance for genes related to ethylene responses and carotenoid metabolism and reduced expression of genes linked to photosynthesis and stress responses. Furthermore, SL mutants in Marchantia exhibit increased amounts of carotenoids. Based on these results, we propose that MpD27 and MpCCD7 regulate the release of gemmae from the gemma cup and their subsequent germination and growth in response to varying light conditions.
Methods
Phylogenetic tree construction
Sequence alignments were performed on amino acid sequences deduced using MUSCLE 3.8.425 (Edgar, 2004) within the Geneious Prime suite of software (https://www.geneious.com). Candidate homologous sequences for Marchantia genes were obtained from Phytozome (https://phytozome-next.jgi.doe.gov/), Sol Genomics Network (https://solgenomics.net/) or NCBI (https://www.ncbi.nlm.nih.gov/), and are provided in Supplementary Data S1-1. Maximum likelihood phylogenetic trees were generated on the alignments using PhyML 2.2.4 (Guindon and Gascuel, 2003) with default settings.
Plant lines and growth conditions
For plant transformation, M. polymorpha L. spores were obtained as described by (Albert et al., 2018). Plant lines were maintained asexually through the propagation of gemmae, either plated directly on 0.5× Gamborg’s B5 medium (Duchefa Biochemie, Haarlem, the Netherlands; 1% (w/v) sucrose, 1% (w/v) agar) or onto a sterile filter paper disc covering the medium. Standard culture conditions were 25°C, 16 h photoperiod and 30 µmol m-2 s-1 light intensity provided by cool white, fluorescent tubes. For carbon and nutrient deprivation experiments, gemmae (four independent transgenic lines and >50 biological replicates per treatment) were plated onto water agar medium solidified using 1% agar and grown for 10 days either in the light/dark or complete darkness.
CRISPR/Cas9 mutagenesis
Three candidate Marchantia genes related to SL pathways were targeted: two genes for DWARF 27, MpD27–1 (Mp6g03970) and MpD27–2 (Mp6g01750), and a CAROTENOID CLEAVAGE DIOXYGENASE 7 (MpCCD7/Mp2g03280). A gene encoding a protein known to regulate gemmae biology, SUPPRESSOR OF MORE AXILLARY GROWTH2-LIKE (MpSMXL/Mp3g06310), was also targeted. CRISPR/Cas9-based genome editing was performed as previously described (Albert et al., 2018). Agrobacterium-mediated transformation was used for transforming spores with a construct containing four guide RNAs per gene (guide sequences are provided in Supplementary Data S1-2). The four 20 bp guides/gene (N17VVR: protospacer adjacent motif) (Doench et al., 2014) were designed using Geneious Prime and synthesized by GenScript Biotech (NJ, USA) as a polycistronic sequence, consisting of 20 bp guide sequences fused to a modified sgRNA scaffold (Dang et al., 2015) separated by glycyl-tRNA sequences. The synthesized sequences were subcloned into pMpU6ENTR and recombined into the binary vector pMpGE010 (Kawamura et al., 2022) using LR Clonase II enzyme mix (Thermo Fisher Scientific). Following transformation, PCR amplification and DNA sequencing were used to find mutations within the target genes (Supplementary Figure S1, Supplementary Data S1-3). At least five independent mutant events were shown for each of the targeted genes. Mutants were found in the initial transgenics (T0) because the main life-stage of Marchantia is gametophytic and, thus, haploid. Mutant lines were propagated through gemmae (G1 generation) and re-sequenced to ensure establishment of non-chimeric lines, as gemmae are derived from single cells. Plants used for experiments were G2 generation or later. For most experiments, four independent lines were used, and all analysis were performed using the parental line as a control.
Scoring the number of gemmae in a cup
To assay for variation in gemmae numbers, we assigned numbers to the gemma cups on the thallus of 8-week-old wild-type plant. The Marchantia thallus grows outwards from the meristem at the thallus branch tip, showing new gemma cups as it grows. Thus, the oldest cups are at the thallus base and the youngest next to the meristem. Gemma cups at the thallus base were called Gemma Cup 1 (GC1) and successive cups were numbered outwards to the tip of the thallus branch (GC7). For comparisons between transgenic lines and wild-type, GC3 and GC4 were used. To quantify gemmae numbers, gemma cups were removed from the thallus and counted using a magnifying lens.
Scoring the gemmae growth and germination
To quantify gemmae growth during the standard day/night growing conditions, we photographed the growing gemmae and then measured gemmae area by using Image J (Schneider et al., 2012). The germination of gemmae held in the dark was scored by counting the number of elongated gemmae and gemmae that developed rhizoids.
RNA sequencing analysis
The wild-type control and three transgenic lines Mpd27–1#16, Mpd27–2#9 and Mpccd7#17 were grown on half-strength Gamborg’s B5 medium for 5 weeks. For uniform tissue sampling, a 6 mm diameter disc encapsulating GC3 and GC4 was harvested, weighing approximately 160 mg per biological replicate (with three biological replicates per genotype). Total RNA was extracted using the Spectrum™ Plant Total RNA Kit (Sigma Aldrich) following the manufacturer’s instructions. RNA-sequencing library construction and sequencing was performed by Beijing Genomics Institute (Shenzhen, China) using DNBSEQ Eukaryotic Strand-specific mRNA library preparation and the DNBseq platform. The resultant reads were mapped onto M. polymorpha subsp. ruderalis genome version 6.1 (https://marchantia.info/) using HISAT2 (Kim et al., 2019) with default parameters. Read counts were then calculated by Feature Counts (Liao et al., 2014). Feature counts generated were further filtered for genes with lower counts and replicates where genes are not expressed at all using edgeR (filterByExp, with default settings, ensuring removal of genes with lower than 10 counts in all samples). This left a total number of 13,408 genes across all samples. The featurecounts were also processed for K means clustering (top most variable 2000 genes, 6 clusters) and Hierarchical clustering analysis using the iDEP application through the subtraction of each gene’s mean expression level (Ge et al., 2018).
The filtered featurecounts were then processed for pair-wise differential expression (DE) analysis using edgeR (QLFTest) (Robinson et al., 2010). Files for each comparison were then filtered for genes with FDR <0.05 and logFC +-1.5. These files are presented for each comparison and an Upsetplot was performed on all these treatments (https://upsetplot.readthedocs.io/en/stable/formats.html). Raw counts data are described in Supplementary Data S1-4, while differentially expressed genes (DEGs) are summarized in Supplementary Data S1-5.
Ultra-high-performance liquid chromatography (hereafter referred to as UPLC) analysis of SL pathway mutants and wild-type plant
Ten milligrams of powdered freeze-dried thallus tissue from 8 weeks old Marchantia plants was mixed with 1 mL of acetone, homogenized using a vortex mixer for 30 s, and shaken at 4°C for 30 min to allow extraction of the lipophilic compounds. After centrifugation at 10,000 ×g for 10 min, the supernatant was collected. The extraction was repeated on the pellet using the same extraction solvent. Both supernatants were combined, evaporated until dry under a gentle stream of nitrogen, and resuspended in 1 mL of ethanol. The resulting extract was filtered through a 0.45 µm syringe filter and stored at −20°C prior to UPLC analysis. The quantification was carried out using a Waters Acquity UPLC system (Milford, MA, USA) equipped with a Photodiode Array Detector and a Mass Single-quadrupole Detector (QDa, Waters). An aliquot of 2 μL was injected onto an Acquity UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm particle size, Waters) at 40°C, with a flow rate of 0.5 mL min-1. The solvents were: Solvent A: 0.1% formic acid in water, Solvent B: 0.1% formic acid in acetonitrile, and Solvent C: 0.1% formic acid in isopropanol (all v/v). The gradient elution program was: 0 min: 30% A, 66% B, 4% C, 0.5 min: 30% A, 66% B, 4% C, 10 min: 18% A, 72% B, 10% C, 13 min: 2% A, 78% B, 20% C, 17 min: 2% A, 78% B, 20% C, 17.1 min: 30% A, 66% B, 4% C, 20 min: 2% A, 78% B, 20% C. Carotenoids were detected at 420 nm and quantified as lutein or β-carotene equivalents using a six-point calibration curve and lutein and β-carotene calibration standards prepared from commercially available sources. Data acquisition with the QDa mass detector was undertaken in positive mode using the following conditions: total ion current (TIC) between m/z 100–1000 Da, capillary voltage: 1 kV, probe temperature: 600°C, and cone voltage: 5 V. Compounds were confirmed in selected ion recording (SIR) mode.
Transcript quantification by droplet digital PCR
Droplet Digital PCR (ddPCR) was performed to validate the DEGs obtained from the RNA sequencing experiment. We used the BIO-RAD droplet digital PCR system to quantify transcripts of the target genes, with MpACTIN serving as the reference gene. The primer sequences are provided in Supplementary Data S1-3.
Results
Phylogenetic analysis of MpD27–1, MpD27–2, and MpCCD7 to predict gene functions
Gene homologs for two steps of the SL biosynthetic pathway have been identified from Marchantia, DWARF27–1 and DWARF27–2, (MpD27–1/Mp6g03970 and MpD27–2/Mp6g01750) and CAROTENOID CLEAVAGE DIOXYGENASE 7 (MpCCD7/Mp2g03280). However, no gene sequences for two pivotal enzymes responsible for catalysing the final stages of this pathway, MORE AXILLARY GROWTH 1 (MAX1) and CAROTENOID CLEAVAGE DIOXYGENASE 8 (CCD8), have been identified in previous work (Delaux et al., 2012; Walker et al., 2019; Kodama et al., 2022) (Figure 1B). To examine the relationship of the MpD27–1, MpD27–2 and MpCCD7 candidate genes to those of other species, we conducted a phylogenetic analysis of D27 and CCD7 deduced amino acid sequences from representative species of liverworts, lycophytes, gymnosperms, and flowering plants (Figure 1C). Using iterative BLAST searches, a single CCD7 homolog was identified in Marchantia (MpCCD7), along with four homologous D27 genes (MpD27–1, -2, -3 and -4). Of these, MpD27–1 and MpD27–2 were chosen for further analysis because they belong to the same clade. The phylogenetic analysis supported the identification of a single CCD7 ortholog and four closely related D27 homologs in Marchantia (Figure 1C).
Although the function of these genes in M. paleacea has not been characterized yet, it possesses a functional SL pathway (Kodama et al., 2022). Therefore, based on the close sequence identity, MpD27–1, MpD27–2 and MpCCD7 were hypothesized to perform functions related to SL metabolism and chosen for further functional analysis. Based on homology analysis these genes seem to regulate biosynthesis of the SL precursor, 9-cis-10′-apo-beta-carotenal. Considering that ancestors of liverworts had SLs (Kodama et al., 2022), we are referring to these genes in Marchantia as SL precursor biosynthesis genes, despite the absence of detectable SL production in Marchantia. This categorization is based on sequence similarity, probable evolutionary relatedness, and likely functional conservation.
Generation of knockout mutant lines for MpD27–1, MpD27–2 and MpCCD7
To investigate the biological functions of MpD27–1, MpD27–2 and MpCCD7, CRISPR/Cas9 Marchantia knockout mutants were generated for each gene, using Agrobacterium-mediated transformation of spores. The resultant mutations ranged from single nucleotide insertions or deletions to larger modifications, with deletions extending up to 370 bp and insertions reaching 231 bp (Supplementary Figure S1). Most of the genetic changes were predicted to result in truncated proteins. Marchantia is dioicous, so spores for transformation must be generated from a cross of male and female lines, which can generate heterogeneous genetic background among sporelings (Ishizaki et al., 2016). Thus, to isolate the effects of the mutations from genetic background variability, multiple knockout lines were identified for each target gene. Four distinct mutant lines were consequently employed as biological replicates for all experiments (except transcriptomic analysis), in addition to the parental line. Additionally, based on initial analysis of the mutant lines suggesting a phenotypic change in gemmae biology and gemmae gemination on the mother plant, a CRISPR/Cas9 knockout mutant for SUPPRESSOR OF MORE AXILLARY GROWTH2-LIKE (MpSMXL/Mp3g06310) was generated, as this gene regulates karrikin signaling and gemmae numbers in Marchantia (Komatsu et al., 2023) and provided a useful comparative control.
MpD27–1, MpD27–2 and MpCCD7 are required for regulation of gemma release from gemma cups
In tissue culture conditions, the gemmae of wild-type plants typically remain within the gemma cups. However, Mpd27–1, Mpd27–2, and Mpccd7 mutants displayed a higher percentage of gemma cups with gemmae outside of the cups compared with the wild-type control (Figures 2, 3A). Gemmae, upon dispersal from the cup, initiate growth, a phenomenon also observable in SL mutants, as evidenced in Supplementary Figure S2A. Previously, it was reported that mutants of MpSMXL exhibited an increase in gemma number, retarded thallus growth and suppressed gemma dormancy in the dark (Mizuno et al., 2021; Komatsu et al., 2023). Given that karrikins are thought to potentially mimic the bioactivity of SLs, we hypothesized that an increase in the number of gemmae per cup could be a driving factor behind this gemmae escape phenomenon. The Mpsmxl#11 mutant did not exhibit the visible increase in gemmae outside of the cup observed in the SL precursor mutants (Figures 2, 3A). However, to appropriately test the hypothesis, we quantified the gemmae number for both the SL precursor mutants and wild-type. To facilitate this analysis, we first examined how the spatial position of the cups influenced gemmae numbers in wild-type genotype.
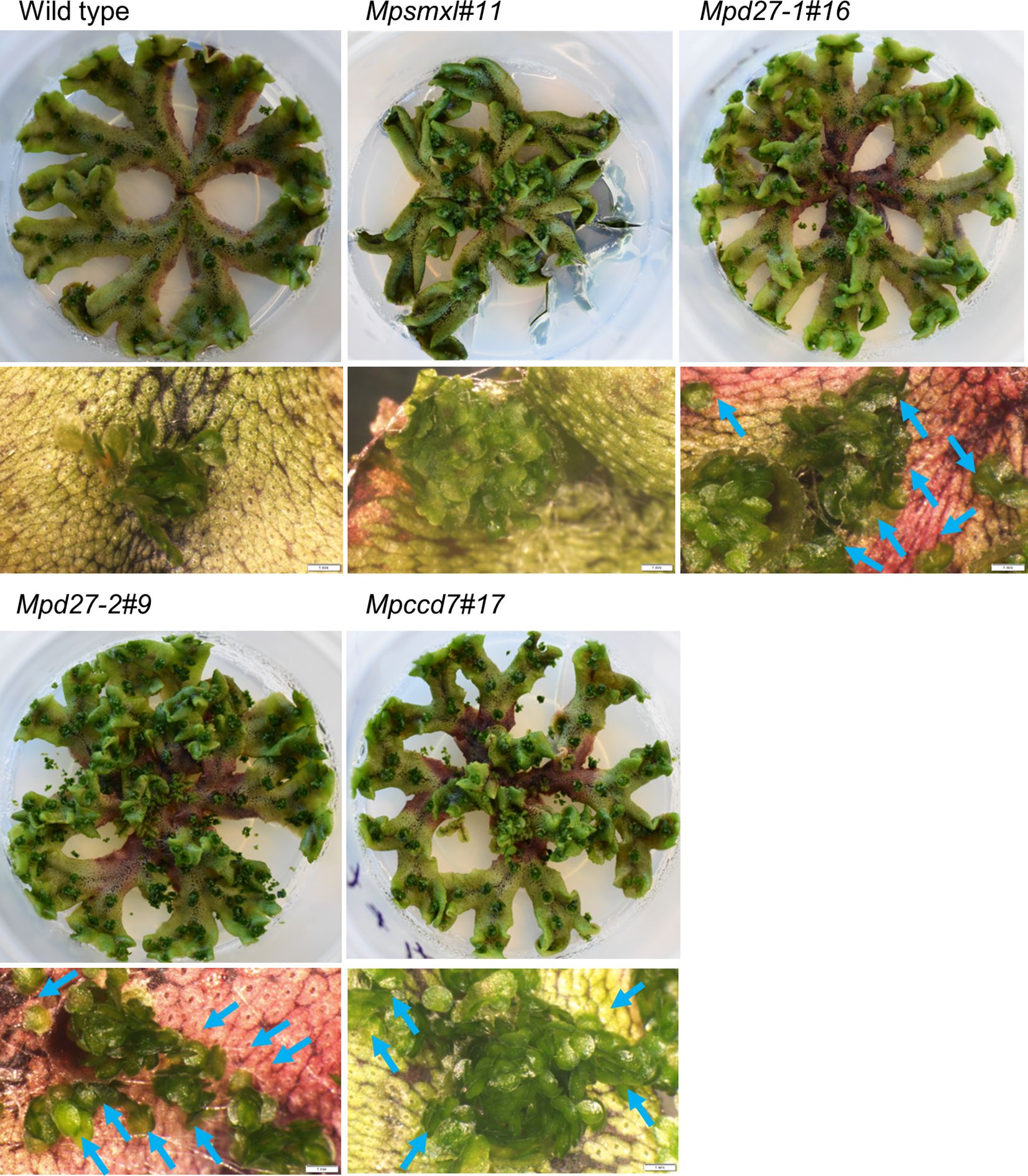
Figure 2 Phenotypes of wild-type and Mpsmxl#11, Mpd27–1#16, Mpd27–2#9 and Mpccd7#17 mutant plants. Plants were grown on complete media (1/2 strength Gamborg’s B5 medium supplemented with 1% sucrose and solidified with 1% agar) for 8 weeks at 25°C, 16-hour photoperiod and a light intensity of 30 mol m-2 s-1, provided by cool fluorescent tubes. Blue arrows indicate the gemmae released from the gemma cup. Scale bars indicate 0.5 cm for the entire plant and 1 mm for the thallus sections imaged with a light microscope.
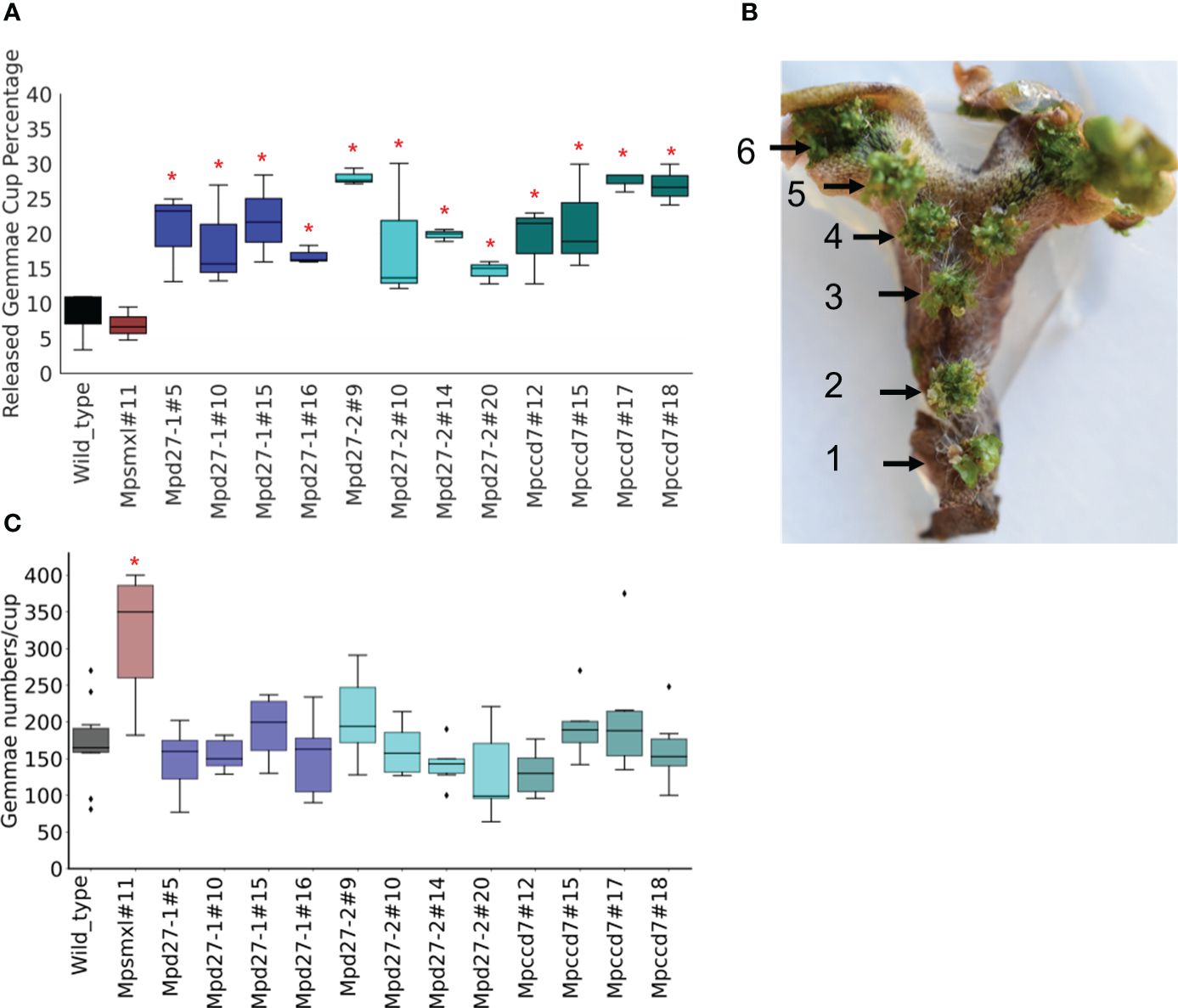
Figure 3 MpD27–1, MpD27–2, and MpCCD7 regulate release of gemmae from gemmae cup. (A) Percentage of gemma cups (GC) was calculated by dividing the number of GC showing gemmae release with the total number of GC. (B) The wild-type thallus branch representing the numbering system used to study gemmae numbers in gemmae cups located at different positions. The numbers and black arrow heads indicate the positions of the gemma cups (GC) on the thallus, representing their location and relative age. Number 1 corresponds to the oldest GC, located closest to the thallus base, whereas number 7 refers to the youngest GC, situated nearest to the apical notch. (C) The combined gemmae numbers for GC3 and GC4 for the wild-type and mutant genotypes, including Mpsmxl#11, Mpd27–1#16, Mpd27–2#9, and Mpccd7#17. The statistical analysis was performed by using Student’s t-test in comparison with wild type and is represented as p < 0.05 (*). The data shown were collected from 8-week-old plants and represent six biological replicates.
A numbering system was used based on the relative age of gemma cups. The oldest cup, situated at the thallus base, was designated as GC1 (Gemma Cup1) and younger cups closer to the apical notch were numbered consecutively from GC1 (reaching GC7), as illustrated in Figure 3B. No significant differences were observed in the number of gemmae within cups positioned at various locations along the thallus (Supplementary Figure S2B). For consistency, subsequent analyses concentrated on GC3 and GC4, with the gemmae from both cups combined for quantification. The Mpd27–1, Mpd27–2, and Mpccd7 mutants had no significant difference in gemmae numbers from wild-type (Figure 3C). This suggested that the observed gemma cup release phenotype in the SL precursor mutants was not due to an increase in gemmae count. Thus, MpD27–1, MpD27–2 and MpCCD7 may have a role in suppressing release of gemmae from the gemma cup, thereby reinforcing gemmae dormancy while on the parent plant.
MpD27–1, MpD27–2 and MpCCD7 regulate gemmae germination and growth in the dark
Given that MpD27–1, MpD27–2 and MpCCD7 influence gemmae dispersal, a process that typically initiates gemmae germination under favorable conditions, we investigated whether the genes also impact gemmae germination and growth. We compared gemmae growth under nutrient- and carbon-starved conditions either in the presence or absence of light. The mutant lines grown in the light did not show any significant differences in gemmae growth compared with wild type, as determined by comparative surface area (Supplementary Figure S3).
The emergence of rhizoids is an indicator of the release of gemmae dormancy: gemma without rhizoids are considered dormant and those with visible rhizoids categorized as non-dormant (Eklund et al., 2015, 2018). Additionally, gemma germination and growth in the dark is typified by elongation of one side of the gemma (Mizuno et al., 2021). To evaluate germination of dark-grown gemmae, elongation and/or the presence of rhizoids were measured in mutants and the wild-type line. This assessment was impractical for light-grown gemmae, as all initially displayed gemmae growth and rhizoid emergence, until development was halted due to nutrient scarcity. Consequently, area-based metrics were exclusively used to gauge growth of light-grown gemmae. In dark conditions, all four lines of Mpd27–1, Mpd27–2 and Mpccd7 exhibited significantly higher numbers of gemmae with elongation compared with wild-type line (Figure 4). Additionally, the majority of the Mpd27–2 mutant lines and all the Mpccd7 mutant lines exhibited an increase in rhizoid production (Supplementary Figure S4). For Mpsmxl#11 gemmae, growth appeared like wild-type line. The experimental evidence shows that MpD27–1, MpD27–2 and MpCCD27 are required to keep gemmae dormant under unfavorable (nutrient depleted and dark) conditions.
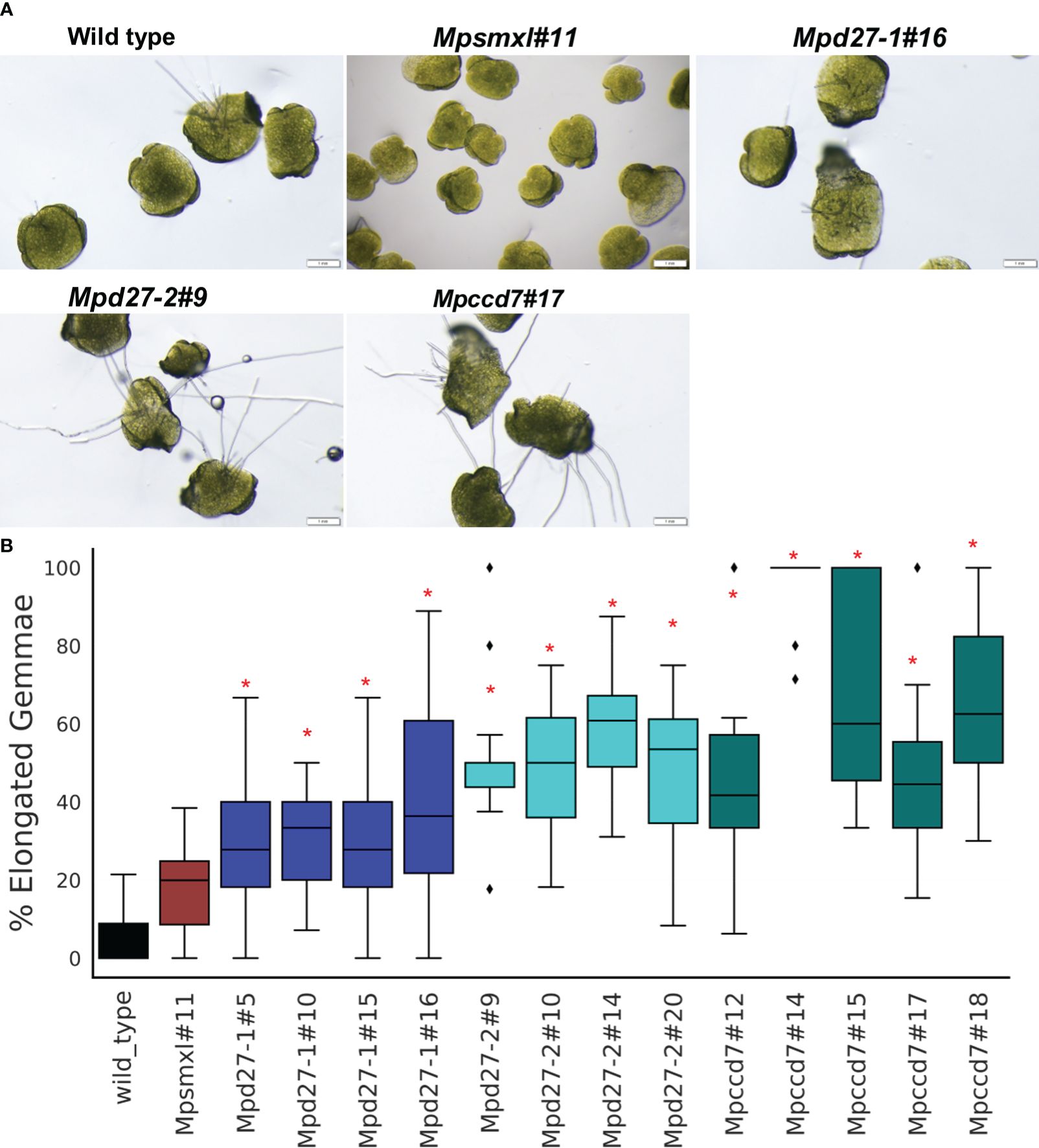
Figure 4 Dormant gemmae of wild-type, Mpsmxl#11, Mpd27–1, Mpd27–2 and Mpccd7 mutant plants grown on carbon- and nutrient-starved medium in darkness for 10 days. (A) Phenotypes of gemmae, with images taken on a light microscope. (B) Percentage of elongated gemmae. Scale bars represent 1 mm. Error bars denote mean ± SD (n = 50). Student’s t-test was employed to compare mutant lines with the wild-type genotype. Statistically significant differences against wild-type are represented as p < 0.05 (*).
Transcriptome analysis of Mpd27–1, Mpd27–2 and Mpccd7 mutants
To find changes in mRNA abundance associated with gemma release, germination, and growth, comparative transcriptome analysis of Mpd27–1, Mpd27–2 and Mpccd7 mutants and wild-type control was conducted, focusing on evaluating the thallus tissue surrounding GC3 and GC4 (Figure 5A). Principal Component Analysis (PCA) demonstrated that biological replicates within each genotype were grouped closely (Figure 5B), whereas distinct groupings were observed among different genotypes, affirming the reliability of the data and the distinctiveness of phenotype between mutant and wild type. The raw counts and results of DEG analysis are given in Supplementary Data S1-4, S1-5.
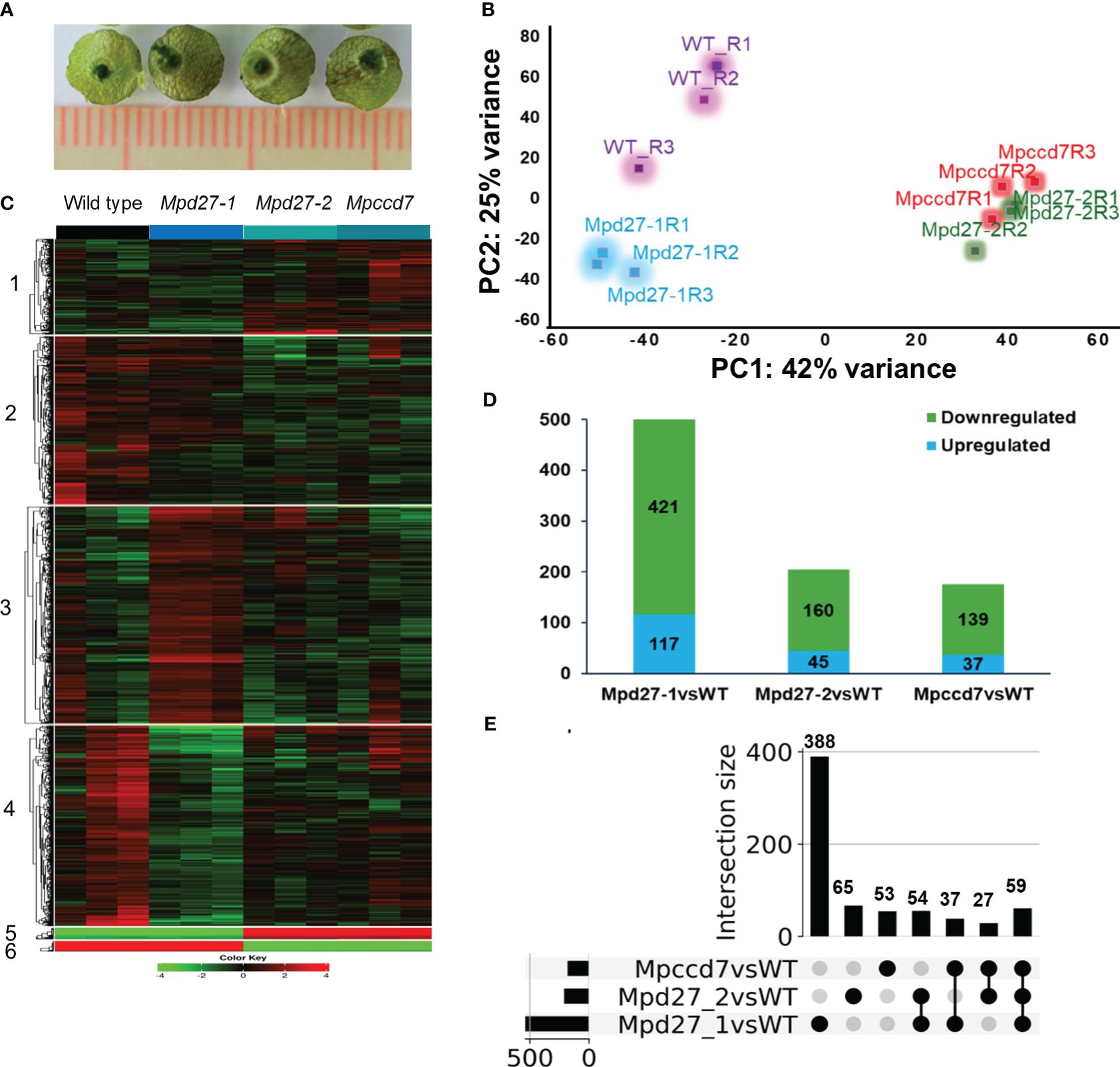
Figure 5 Transcriptome analysis of wild-type and Mpd27–1, Mpd27–2 and Mpccd7 genotypes. (A) Representative samples of the thallus tissue used for RNA sequencing analysis. (B) Principal Component Analysis (PCA) plot displaying variability within the expression dataset. (C) A heatmap of k means clustering applied on normalized (Nr) data through gene standardization. (D) DEGs in Mpd27–1#16, Mpd27–2#9 and Mpccd7#17 mutants compared with wild-type genotype. The p-values were calculated using Fisher’s exact tests. Significance analysis for clustering was conducted on normalized count data via ANOVA, with a false discovery rate (FDR) of < 0.05. (E) UpSet plot of differentially expressed genes from RNA sequencing at a log2 fold change (log2FC) of ±1.5 and FDR at <0.05. The black circle represents the set membership.
Hierarchical clustering of the transcriptomic data grouped the top 2,000 most variable genes into two main clusters (Supplementary Figure S5, Supplementary Data S1-6). Cluster 1 included the transcript data from Mpccd7 and Mpd27–2, while cluster 2 was composed of wild-type and Mpd27–1. This indicated that Mpd27–2 and Mpccd7 exhibited similar gene expression patterns, whereas Mpd27–1 differed from other genotypes. Using K-means clustering yielded the expression patterns into notable six clusters (Figure 5C, Supplementary Data S1-7). Notably, clusters 4 and 2 exhibited the most contrasting profiles based on genotype-based differences. Furthermore, cluster 5 and 6 consisted of genes associated with the genotype’s sex.
To identify the genes that were differentially expressed in a pair-wise fashion between SL mutants and wild type we kept genes that showed log2FC of ± 1.5 at FDR <0.05. Mpd27–1 had 421 downregulated and 117 upregulated genes, Mpd27–2 had 160 downregulated and 45 upregulated genes, and MpCCD7 showed 139 downregulated and 37 upregulated genes (Figure 5D, Supplementary Data S1-7-S1-9).
Employing UpSet plot on this dataset (Figure 5E, Supplementary Data S1-8), identified that 59 genes were differentially regulated across all three mutants. Specifically, 27 genes showed differential expression between Mpccd7 and Mpd27–2, while 37 genes differed between Mpd27–1 and Mpccd7. Among the detected DEGs, with the FDR/fold-change criteria used of <0.05/>1.5-fold, 388 out of 538 were associated only with Mpd27–1. In contrast, far fewer DEGs were found only in the Mpd27–2 and Mpccd7 datasets with these criteria, being 65 out of 205 and 53 out of 176, respectively.
Transcriptomic analysis reveals molecular mechanisms of release, germination, and growth of gemmae
To enhance the clarity of our findings, we concentrated on analyzing DEGs that exhibited consistent changes across all three mutants. Specifically, we focused on genes predicted/known to have biological functions crucial to essential plant developmental processes including photosynthesis, stress adaptation, and terpenoid metabolism (Figure 6, Supplementary Data S1-9). Notably, genes encoding early light induced (ELI) proteins (including Mp4g18580 and Mp4g18590), peroxidases (e.g., Mp5g01640 and Mp3g07970), an α/β hydrolase (including Mp2g15700), and late embryogenesis abundant (LEA) proteins (including Mp4g14840 and Mp4g14870) were predominantly downregulated. Conversely, Mp7g00860/MpERF10, an ethylene response factor, demonstrated increased transcript levels. Genes associated with terpenoid metabolism exhibited variable responses. For example, Mp3g13150/MpTPS6 was significantly upregulated with a log2FC of 3.3, whereas Mp3g21760/MpTPSL18 was downregulated with a log2FC of -5.9 (Figure 6A, Supplementary Data S1-9). Interestingly, the transcript abundance of Mp4g19090/MpTPSL23 was higher in Mpd27–2 and Mpccd7 mutants compared to the Mpd27–1 mutant (Supplementary Data S1-9).
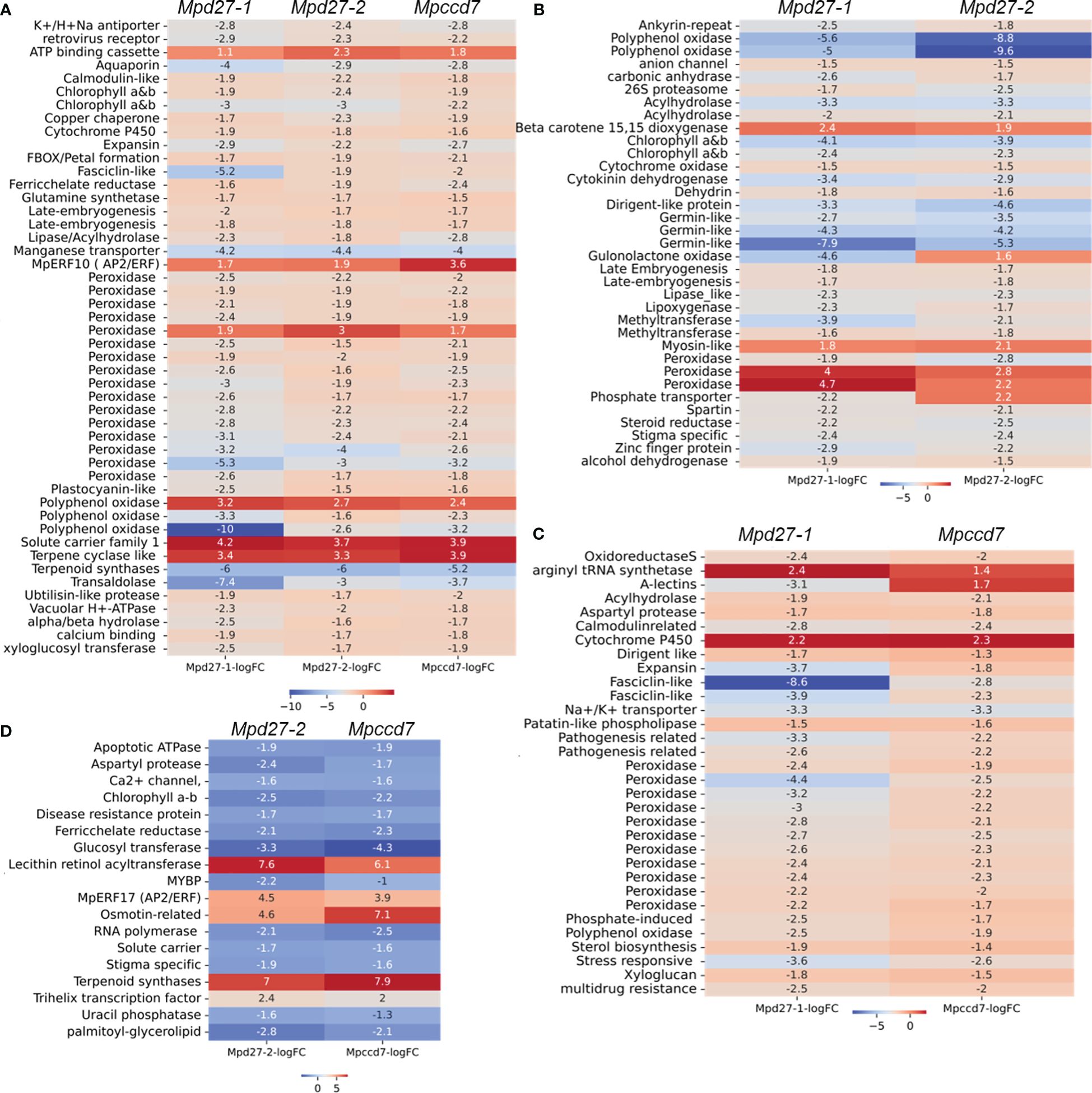
Figure 6 DEG comparisons among Mpd27–1#16vsWT, Mpd27–2#9vsWT, and Mpccd7#17vsWT. The heatmaps illustrate the log2 FC of genes in comparison to wild-type control highlighting genes common across (A) Mpd27–1#16, Mpd27–2#9 and Mpccd7#17, (B) Mpd27–1#16 and Mpd27–2#9, (C) Mpd27–1#16 and Mpccd7#17, and (D) Mpd27–2#9 and Mpccd7#17.
The DEGs associated with photosynthesis, hormones, and stress adaptation were shared between Mpd27–1 and Mpd27–2 mutant RNAseq datasets (Figure 6B, Supplementary Data S1-9). For example, genes including CAB/ELI (e.g., Mp4g18610 and Mp4g18620), GERMIN-LIKE (e.g., Mp5g00880, Mp5g00920, and Mp5g13870), and ABA-responsive e.g., DEHYDRIN (Mp6g15610) and LEA (e.g., Mp7g06630 and Mp4g14880) were downregulated. At the same time BETA-CAROTENE DIOXYGENASE (Mp4g01260) was upregulated in both mutants. Similarly, DEGs between Mpd27–1 and Mpccd7 were mostly associated with stress responses. For example, Mp4g02840 (induced by phosphate), Mp8g18620, (A-type lectin), and Mp5g05460 (known as fasciclin-like) were downregulated, whereas Mp6g18420 (a cytochrome P450), showed an increase in expression (Figure 6C, Supplementary Data S1-9). The downregulated genes common between Mpd27–2 and Mpccd7 included CAB/ELI (e.g., Mp4g18560), Stigma specific (e.g., Mp5g11400) and PALMITOYL GLYCEROLIPID (e.g., Mp6g04480) (Figure 6D, Supplementary Data S1-9). Conversely, genes such as MpERF17/Mp4g22280, TRIHELIX16/Mp3g10500), MpTPSL23/Mp4g19090 and LRAT/Mp3g10470 were upregulated. Taken together, these findings indicate that SL precursor mutants affected many genes regulating essential biological functions such as photosynthesis, ethylene signaling, stress responses, terpenoid and carotenoid metabolism.
The transcriptome profile unique to Mpd27–1 included many genes linked to stress response and cellular transport (Figure 7A, Supplementary Data S1-9). The ten lectin-related genes, involved in defense and communication responses against biotic and abiotic stress, were suppressed, exhibiting log2FC from -1.6 to -6.4. For example, A-type lectins Mp1g18730 with a log2FC of -2.59 and B-type lectins Mp8g18670 with a log2FC of -6.46 were downregulated. Similarly, transport-related genes involved in transport of iron (including Mp2g25340 with a log2FC of -3.3), phosphate (including Mp4g16570 with a log2FC of -3.46), and nitrate (including Mp4g03070 with a log2FC of -5.6) showed decreased transcript abundance. In contrast, some phosphate transporters like Mp4g11070 were upregulated with a log2FC of 3.3. Furthermore, terpenoid synthase (Mp6g04630/MpMTPSL10) increased by a log2FC of 2, and LOX2 (Mp2g12180) was upregulated with a log2FC of 3.46.
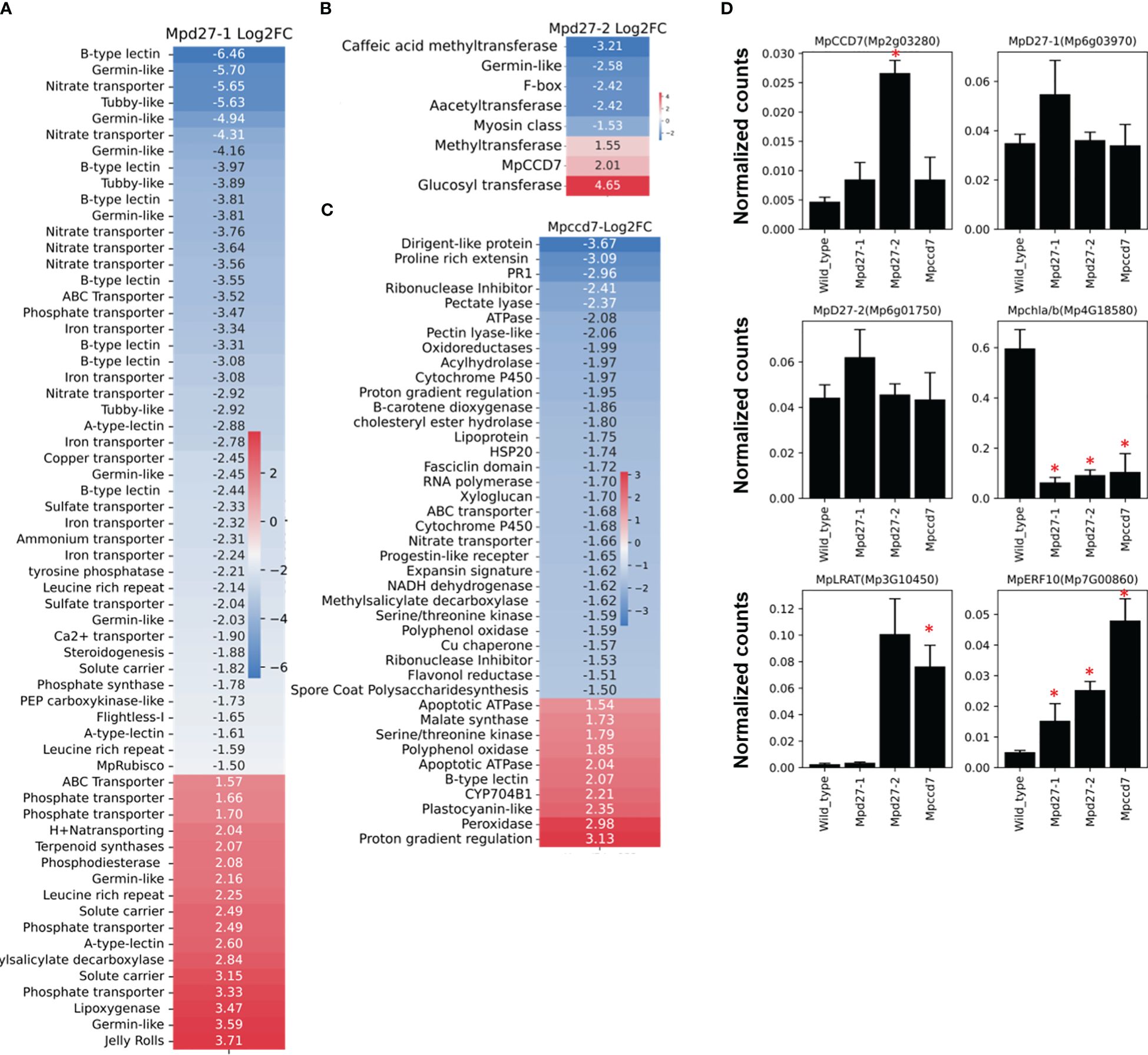
Figure 7 Heatmaps of the log2 FC of genes unique to Mpd27–1#16 (A), Mpd27–2#9 (B), and Mpccd7#17 (C) mutants compared with the wild-type control. (D). Digital Droplet PCR (ddPCR) analysis. The data represent three biological replicates. The y-axis represents the normalized counts, and the x-axis displays the gene names. The expression data were normalized against MpACTIN. The statistical analysis was performed by using Student’s t-test in comparison with wild type and is represented as p < 0.05 (*).
In Mpd27–2, MpCCD7 (Mp2g03280) was upregulated 4-fold, suggesting that the mutant might be responding to a defect in a metabolite biosynthesis process regulated by MpD27–2 (Figure 7B, Supplementary Data S1-9). This change could represent a compensatory mechanism for reduced flux within the pathway or be a result of feedback regulation. These findings suggest a potential link between MpD27–2 and MpCCD7 in the same metabolic pathway.
The Mpccd7 mutant showed 3.6-fold increase in transcript abundance of 9-CIS-EPOXYCAROTENOID DIOXYGENASE (Mp2g07800/MpNCED) (Figure 7C, Supplementary Data S1-9). Other genes with notable reduced expression included SPORE COAT POLYSACCHARIDE (Mp3g15460) with a log2FC of -1.5 and FASCICLIN-LIKE ARABINOGALACTAN (Mp5g05390) with a log2FC of -1.7. In contrast, MALATE SYNTHASE (Mp8g16290), MpCYP704-like8 (Mp3g10820), and MpNBS-LRR5 (Mp3g09150) were upregulated.
Digital droplet PCR confirms the differential gene expression patterns found using RNAseq
Digital Droplet PCR (ddPCR) was used to validate the RNA-Sequencing findings, by examining six genes identified as altered in the mutant lines (Figure 7D). The ddPCR results confirmed the differential expression of the genes compared with the wild-type control. For example, MpCCD7 (Mp2g03280) expression was increased >2-fold in Mpd27–2, while MpERF10 (Mp7G00860) exhibited a more than 4-fold increase in Mpd27–2 and an over 8-fold increase in Mpccd7. In contrast MpCAB (Mp4G18580) was -5-fold downregulated in the Mpd27–1, -3.6-fold in Mpd27–2 and -2.45-fold in Mpccd7 mutants. These results are consistent with the RNAseq data, thereby validating the observed differential gene expression.
The SL mutants showed altered accumulation of carotenoid compounds
Given their orthologous relationship with Arabidopsis thaliana’s D27, MpD27–1 and MpD27–2, are predicted to function as β-carotene isomerases. This is further supported by the observed increase in transcript abundance of carotenoid associated genes in transcriptome analysis of SL precursor mutants. To investigate whether the loss of gene function impacts carotenoid composition, a targeted UHPLC analysis was conducted on wild-type and mutant genotypes (Figure 8A). Additionally, the observed decrease in transcript levels for photosynthesis-related genes, as shown in Figure 6A, prompted the quantification of chlorophyll content in the mutants (Figure 8B).
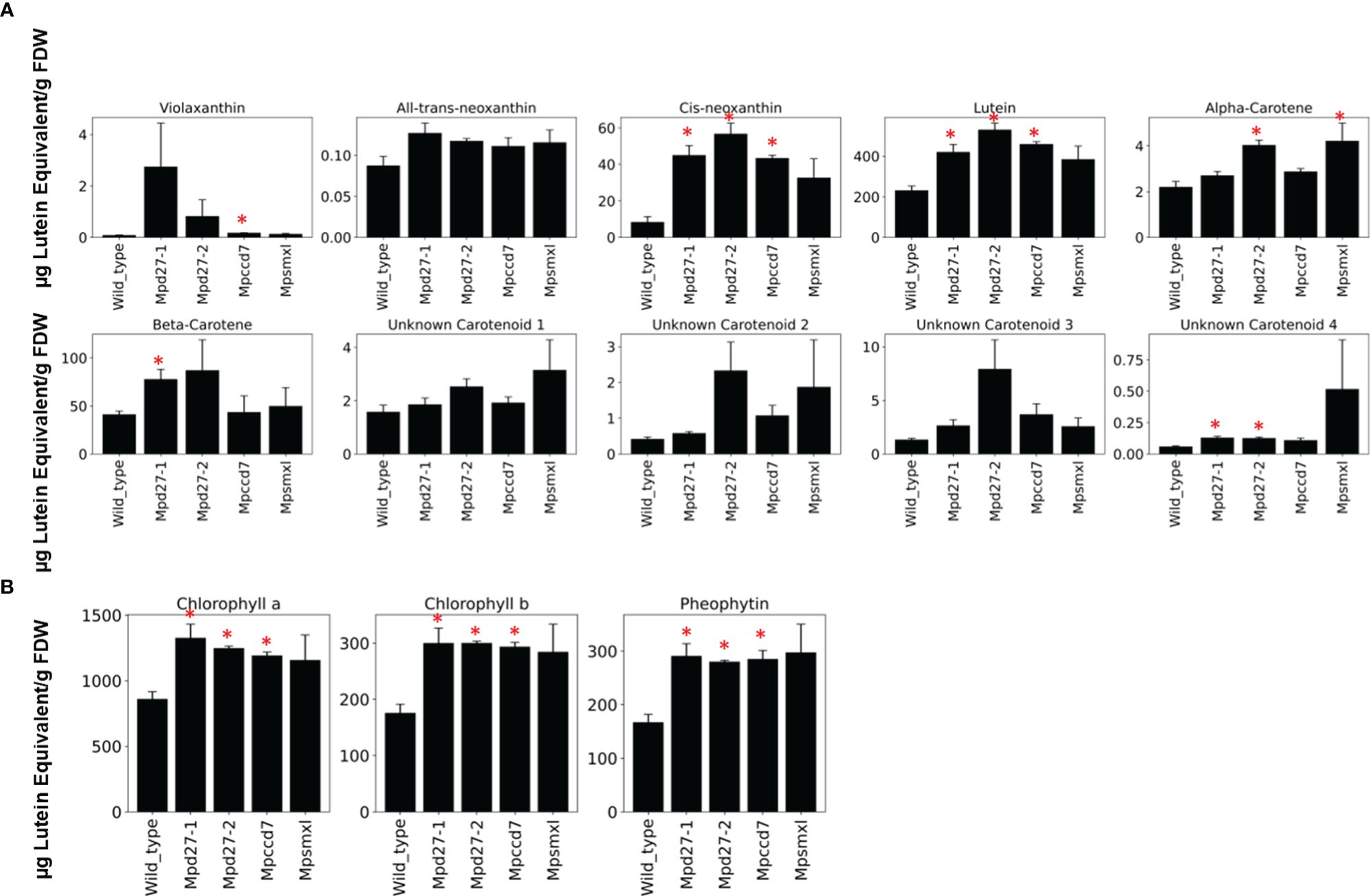
Figure 8 A targeted UHPLC analysis of Mpd27–1#16, Mpd27–2#9, Mpccd7#17, Mpsmxl#11 and wild-type. (A) The amounts of carotenoid-related compounds. (B) The amount of chlorophyll and pheophytin. The data represent three biological replicates. The y-axis represents the concentrations of these compounds as µg Lutein Equivalent gFDW-1, and the x-axis displays the genotype names. Error bars denote mean ± SD (n = 3). Statistically significant differences against the wild-type are represented by Student’s t-test as p < 0.05 (*).
The carotenoids violaxanthin, all-trans- and cis-neoxanthin, lutein, α-carotene and β-carotene have been identified in Marchantia, and several unidentified carotenoids are also present (Takemura et al., 2014). The identification and quantification of carotenoids in the lines studied here showed a significant increase in the levels of cis-neoxanthin and lutein across all three SL mutants. Additionally, violaxanthin levels were higher in Mpccd7, α-carotene was elevated in Mpd27–2 and Mpsmxl, and β-carotene saw an increase in Mpd27–1. An unidentified carotenoid designated as “compound 4”, also exhibited a significant rise in Mpd27–1 and Mpd27–2. Similarly, the chlorophyll data indicated that SL mutants had increased levels of chlorophyll a, b and pheophytin in comparison to the wild-type control (Figure 8B).
Discussion
The strigolactone (SL) pathway is only partially present in Marchantia, and the function of the genes involved is unclear (Walker et al., 2019). In this study, we used CRISPR/Cas9 mutagenesis to understand the roles of SL precursor biosynthesis genes, including MpD27–1, MpD27–2 and MpCCD7. The phenotypic comparison of the Marchantia mutants and wild-type plant has elucidated the role of MpD27–1, MpD27–2 and MpCCD7 in the release, growth, and germination of gemmae. Analysis of RNA gene expression patterns in these mutants, compared to wild-type plant, revealed an increased transcript abundance of genes involved in terpenoid (e.g., MpTPS6) and carotenoid metabolism (e.g., MpLRAT, MpCCD7, and BETA-CAROTENE 15,15’-DIOXYGENASE), as well as ETHYLENE RESPONSE FACTORS (e.g., MpERF10 and MpERF17). In contrast, certain genes such as 9-CIS-EPOXYCAROTENOID DIOXYGENASE (MpNCED), EARLY LIGHT INDUCED (ELI), and LATE EMBRYOGENESIS ABUNDANT (LEA) showed decreased transcript abundance. Additionally, mutants in SL precursors displayed elevated levels of carotenoids.
MpD27–1, MpD27–2 and MpCCD7 regulate release of gemmae from the gemmae cup
Bryophytes, which encompass non-vascular plants like liverworts, mosses, and hornworts, can utilize both sexual reproductive organs, such as spores, and vegetative structures like gemmae and some part of the thallus for their propagation (Wyatt, 1982; Chopra and Bhatla, 1990; Buck et al., 2003; Whitaker and Edwards, 2010). Gemmae are small disc-like asexual propagules produced in specialized structures called gemma cups, present on the dorsal side of the thallus (Barnes and Land, 1907, 1908). Raindrops have the potential to expel gemmae from gemma cups (sometimes referred to as ‘splash cups’), allowing plants to grow and establish themselves away from the parental plant (Edwards et al., 2019). The gemmae contained within the gemma cup remain dormant and do not undergo maturation or growth until they are either expelled from the cup or until the maternal plant reaches the end of its life cycle. This suggests a restraint on the liberation and germination of gemmae from the maternal plant. While there is a growing body of information about the genetic factors controlling growth and germination of gemmae (Eklund et al., 2015; Eklund et al., 2018), the genes that regulate the release of gemmae from the cup in liverworts remain elusive.
Here we have shown that defects in functions of MpD27–1, MpD27–2 and MpCCD7 regulate gemma cup release (Figures 2, 3A). These Marchantia mutants, when compared with wild type, displayed elevated levels of transcripts for ETHYLENE REPONSE FACTORS (ERF) orthologs, e.g., Mp7g00860/MpERF10 and Mp4g22280/MpERF17 (Figures 6A, D). The expression of Mp7g00860/MpERF10 was also validated through ddPCR analysis, providing clear evidence of increased MpERF10 levels in all three mutants (Figure 7D). Furthermore, the MBEX expression tool indicated higher expression of MpERF17 in gemmae cups (Kawamura et al., 2022). ERFs, belonging to the family of AP2/ERF transcription factors, regulate many processes in flowering plants, including seed germination and pod shattering (Pirrello et al., 2006; Chung et al., 2010; Müller and Munné-Bosch, 2015; Chandler and Werr, 2020; Hu et al., 2022; Liu et al., 2022; Ali et al., 2023). If we consider gemmae release to be analogous to seed release, we suggest that the SL biosynthesis genes in Marchantia could act to restrain the function of ERFs, thereby inhibiting the release of gemmae.
SL precursor biosynthesis genes positively regulate transcript abundance of ELI
Mutants of SL precursors exhibited a notable decrease in the transcript abundance of EARLY LIGHT INDUCED (ELI) genes (Figure 6A, Supplementary Data S1-9). Specifically, Mp4g18580 and Mp4g18590 showed down regulation across all three mutants, while Mp4g18620 and Mp4g18610 were down regulated in both Mpd27–1 and Mpd27–2, and Mp4g18560 experienced a decrease in Mpd27–2 and Mpccd7. Further independent support for the role of these genes in gemma biology came from gene expression profiles. Results obtained using the MBEX expression tool revealed that all four ELI/CAB genes showed increased expression in the gemma cup (Kawamura et al., 2022). These proteins, while not directly binding chlorophyll (Green et al., 1991), are categorized as part of the CAB family and recognized as early light-induced proteins (ELIPs). ELIPs are triggered by light stress and implicated in pigment biosynthesis and thylakoid membrane assembly, essential for protecting chlorophyll-protein complexes from light damage.
SLs impact the process of senescence by regulating various genetic components involved in photosynthesis (Mayzlish-Gati et al., 2010; Mashiguchi et al., 2021; Li et al., 2022; Schiphorst et al., 2022). For example, the tomato SL biosynthesis Sl-ORT1 mutant showed reduced chlorophyll content and lower levels of light-harvesting genes (e.g., CAB and RUBISCO) (Mayzlish-Gati et al., 2010). Similarly, the Arabidopsis SL signaling max2 mutant exhibited delayed senescence and reduced expression of ELIP1, ELIP2, and HY5 (Shen et al., 2007). The decreased expression of ELI genes in mutants of SL precursors, along with their connection to gemma and gemma cup development, suggests a role for ELI in SL-mediated gemmae development. Furthermore, mutants of SL precursors exhibited higher chlorophyll content compared to the control plants, suggesting that SL regulates senescence in Marchantia (Figure 8B). However, it is important to note that the increase in chlorophyll content in SL precursor mutants may be attributed to an increase in the number of growing gemmae.
SL precursor biosynthesis genes suppress germination and growth of gemmae in the dark
Our data show that MpD27-1, MpD27-2, and MpCCD7 inhibit the germination and growth of gemmae under dark and nutrient-starved conditions (Figure 4, Supplementary Figure S4).
This is consistent with other studies on bryophytes that report enhanced tissues growth in mutants defective in the SL and karrikin pathways. For example, SL-deficient (Ppccd8) and SL-insensitive (Ppkai2Lgjm) moss mutants showed increased extension of moss filaments in the dark (Lopez-Obando et al., 2021). Similarly, karrikin signaling mutants, Mpkai2a and Mpmax2, showed enhanced growth of gemmae under dark conditions (Mizuno et al., 2021).
SL and ABA are known to interact with each other in regulating plant stress responses. For instance, rice mutants defective in SL production (e.g., max4/dwarf10 and max3/dwarf17) and SL perception (e.g., dwarf3) showed higher concentrations of ABA (Haider et al., 2018). Similarly, ABA-deficient tomato mutants exhibited reduced concentrations of SL (López-Ráez et al., 2008, 2010). Furthermore, the external application of the SL analog GR24 in Lotus japonicus led to an increased transcript abundance of ABA biosynthesis and ABA responsive genes (Liu et al., 2015).
In Marchantia, ABA contributes to the induction of gemmae dormancy. For example, ABA-insensitive Marchantia mutants were unable to establish and maintain gemmae dormancy (Eklund et al., 2018). The reduced expression of ABA-biosynthesis genes, e.g., 9-CIS-EPOXYCAROTENOID DIOXYGENASE (MpNCED/Mp2g07800), and ABA-responsive genes, including DEHYDRIN (MpDHN3/Mp6g15610) and LATE EMBRYOGENESIS ABUNDANT (LEA/Mp4g14840, Mp4g14870, Mp7g06630, and Mp4g14880) (Figure 7) in mutants of SL precursors suggests a linkage between SL signaling, ABA production, and the control of gemmae dormancy and germination processes. This conclusion is further supported by evidence from the study by Kawamura et al. (2022), where gene expression profiles generated using the MBEX expression tool indicated that all four MpLEA genes were expressed in the gemma cup and sporeling. Additionally, MpDHN3 and MpNCED exhibited higher expression levels in sporelings. Our analysis revealed that MpNCED expression was notably decreased in the Mpccd7 mutant compared to the Mpd27–1 and -2 mutants (Supplementary Data S1-5). This decrease correlates with enhanced rhizoid production observed in the Mpccd7 mutant lines (Supplementary Figure S4).
MpD27–1 positively regulate the transcript abundance of lectin genes
Mpd7–1 showed reduced transcript abundance of many LECTIN genes, specifically A-type and B-type lectins (Figure 7A). Notably, gene expression profiles generated using the MBEX expression tool indicated that Mp8g18650 was highly expressed in gemmae cups (Kawamura et al., 2022). Lectins play a pivotal in establishing a symbiotic relationship between plants and different type of organism under various stress conditions (De Hoff et al., 2009; Jain et al., 2022). For example, under nitrogen stress, legumes secrete lectins from their roots to facilitate bacterial attachment, leading to nodule formation (Downie, 2010). Similarly, certain lectin genes, specifically induced by mycorrhizal associations, are categorized as AM-induced lectin-like genes (Frenzel et al., 2005, 2006). In lichens, an algal-binding lectin has been found to play a key role in establishing a symbiosis between the mycobiont (fungal component) and the photobiont (photosynthetic component) (Nazem-Bokaee et al., 2021).
SLs are crucial plant hormones that regulate plant architecture and act as signaling molecules exuded into the rhizosphere to foster symbiotic relationships, specifically with AM fungi. Although various species of Marchantia, including M. pappeana and M. paleacea, interact with AM fungi, M. polymorpha stands out as it does not form association with AM fungi. Despite this M. polymorpha possesses the capability to interact with a wide range of microbial communities (Poveda, 2020). For example, M. polymorpha was able to establish interactions with various pathogenic and beneficial endophytic fungi (Nelson et al., 2018; Nelson and Shaw, 2019). Similarly, M. polymorpha and M. paleacea interacted with diverse bacterial genera known for plant-growth promotion, exudate degradation, nitrogen fixation and disease-suppression (Van Damme et al., 2008; Alcaraz et al., 2018).
The observed decrease in lectin genes expression in the SL precursor mutant Mpd27–1 suggests that MpD27–1 might be involved in establishing interactions with microorganisms through lectins during biotic and abiotic stress responses.
SL precursor biosynthesis mutants of Marchantia showed higher transcripts abundance of carotenoid-related genes
In SL precursor biosynthesis mutants, we observed an increased transcript abundance of carotenoid related genes including LRAT (LECITHIN RETINOL ACYLTRANSFERASE) and TS (TERPENE SYNTHASE) (Figure 6, Supplementary Data S1-9). β-carotene is cleaved into two molecules of retinal, which can then be reduced to retinol (vitamin A) (D’Ambrosio et al., 2011). LRAT plays a crucial role in metabolism of vitamin A by catalysing the formation of fatty acid retinyl esters from all-trans-retinol (Sears and Palczewski, 2016). The direct functional counterparts of LRAT are absent in plants, as plants do not require the synthesis of retinol. However, through a sequence-based homology analysis, homologues of LRAT have been identified in plants and are known as the H-Box/NC domain-containing gene family (Kaloudas and Penchovsky, 2018). The roles of LRAT homologs in plants still need investigation. However in Arabidopsis, NC domain containing proteins including AT5G06370, AT3G02700, and AT5G06370 regulate pollen germination and pollen tube growth (Wang et al., 2008). The higher abundance of LRAT homologs in Marchantia SL mutants aligns with their role in enhancing the germination of reproductive structures in Arabidopsis, which, in this study are gemmae. This suggests a conservation of certain functional domains across species, despite the differing metabolic requirements between plants and animals.
Plant terpene synthases (TS) are used to make monoterpenes, sesquiterpenes, and diterpenes that are then utilized in the production of ABA, chlorophyll and carotenoids (Cheng et al., 2007). TS are responsible for converting simple hydrocarbon skeletons into the vast array of terpenes important in plant defense, nodule formation, pollinator attraction, and interplant communication (Huang and Osbourn, 2019). For example, over expression of some terpenoid and terpene synthesis genes from Salvia officinalis in soybean are known to modulate rhizobia interaction and nodulation (Ali et al., 2021). The observed increase in LRAT and TS transcripts in SL precursor biosynthesis mutants may be attributed to either compensation for reduced metabolites controlled by SL precursor biosynthesis genes or because of a feedback mechanism where disrupted SL synthesis affects precursor synthesis. Additionally, Mpd27–2 had significantly higher levels of MpCCD7 (Figure 7B) suggesting that MpD27–2 and MpCCD7 both belong to the same metabolic pathway. When upstream components of a metabolic pathway are compromised then the amount of downstream enzyme could increase due to feedback regulation, compensation mechanisms, regulatory signals, or availability of substrate.
Disruption of SL precursor biosynthesis genes altered carotenoid profiling in Marchantia
Alongside elevated expression of TS and LRAT, Mpd27–1, Mpd27–2 and Mpccd7 mutants exhibited an increased concentration of carotenoid-related metabolites (Figure 8A). Each of the three mutants showed a marked increase in the levels of cis-neoxanthin and lutein. The increased cis-neoxanthin could be attributed to downregulation of MpNCED in the SL mutants (Supplementary Data S1-9). NCED plays a pivotal role in ABA biosynthesis, converting 9-cis-violaxanthin and 9′-cis-neoxanthin into xanthoxin, an ABA precursor (Perreau et al., 2020). Therefore, a decrease in MpNCED abundance could naturally result in the accumulation of neoxanthin, as less of it is converted into xanthoxin. Furthermore, the observed increase in lutein levels may also be attributed to disruptions in the activity of SL precursor biosynthesis enzymes, specifically those involved in cleaving 9-cis-zeaxanthin and 9-cis-lutein. For example, an in vitro study involving CCD7 enzymes sourced from rice, Arabidopsis, and garden pea demonstrated CCD7’s broad specificity. Notably, AtCCD7 and PsCCD7 were shown to effectively catalyze the conversion of 9-cis-lutein into 9-cis-3-OH-ϵ-apo-10′-carotenal (Baz, 2018). This evidence underscores CCD7’s role in carotenoid metabolism and its potential impact on lutein accumulation when the normal function of SL precursor biosynthesis enzymes is disrupted.
Mpd27–1 and Mpd27–2 mutants were observed to have elevated levels of β-carotene and α-carotene, respectively. This rise in carotenoid content within these mutants could be attributed to a decrease in isomerization processes, preventing their usual conversion into other compounds. The D27 enzyme from rice has been demonstrated in vitro to catalyze reactions across a variety of bicyclic carotenoid substrates, each containing at least one unsubstituted β-ionone ring, e.g., β-carotene, α-carotene, and cryptoxanthin (Bruno and Al-Babili, 2016). Similarly, AtD27 was capable of catalyzing the reverse isomerization of all-trans-/9-cis-β-carotene, showcasing enzyme activity that selectively targets specific isomeric forms (Abuauf et al., 2018). Notably, neither the rice nor the Arabidopsis D27 enzyme exhibits isomerization activity with 13-cis- or 15-cis-β-carotene, underscoring a distinct specificity for the C9-C10 double bond in these carotenoids. This specificity may play a critical role in the observed accumulation of β- and α-carotene in the d27 mutants, as it suggests a limited ability of the D27 enzyme to process these carotenoids into other forms, thereby contributing to their increased levels.
Mpd27–1 and Mpd27–2 both exhibited elevated levels of an unidentified carotenoid compound (Figure 8A). This observation suggests that MpD27–1 and MpD27–2 might play roles in the metabolism of carotenoid-like compounds, potentially beyond their established functions in carotene catabolism. Our hypothesis is further supported by a study where the enzymatic action of OsD27 on all-trans-α-carotene might be involved in producing a SL like compound such as heliolactone (Bruno and Al-Babili, 2016).
In this context, we propose that both MpD27–1 and MpD27–2, SL pathway isomerases have specific regulatory functions in the synthesis of SL compounds originating from β-carotene and α-carotene, respectively. This specific role of the two enzymes was evolutionarily favored to produce a diverse range of carotenoid-derived compounds, which might be implicated in controlling plant developmental processes such as regulation of release, germination, and growth of gemmae. Such a mechanism ensures that germination occurs under favorable growth conditions, which include sufficient ambient light and water availability.
Summary
Understanding the role of plant pigments like carotenoids and chlorophylls is vital to gain insights into the remarkable ability of plants to adapt to diverse environmental niches (Boncan et al., 2020). Our study has demonstrated that disruptions in the functioning of SL precursor biosynthesis genes, such as MpD27–1, MpD27–2, and MpCCD7, led to the following outcomes: an increase in gemma release, enhanced germination of gemma when incubated in darkness, reduced transcript levels of genes related to photosynthesis and stress, elevated levels of genes related to ethylene and carotenoids, and an increased abundance of carotenoids. Considering these results, we propose that SL precursor biosynthesis genes have specific regulatory functions in the synthesis of SL compounds. This specificity of SL enzymes was evolutionarily favored to produce a diverse range of carotenoid-derived compounds, which might be implicated in controlling plant developmental processes, such as the regulation of release, germination, and growth of gemmae. Such a mechanism ensures that germination occurs only under favorable growth conditions, which include sufficient ambient light and water availability.
Data availability statement
The data presented in the study are deposited in the BioProject database, NCBI repository, accession numbers PRJNA1123149, SAMN41798178, SAMN41798179, SAMN41798180, SAMN41798181, SAMN41798182, SAMN41798183, SAMN41798184, SAMN41798185, SAMN41798186, SAMN41798187, and SAMN41798188, SAMN41798189.
Author contributions
RJ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. JT: Data curation, Formal Analysis, Software, Writing – review & editing. CA: Methodology, Writing – review & editing. BJ: Methodology, Writing – review & editing. RD: Methodology, Writing – review & editing. NA: Methodology, Writing – review & editing. YZ: Methodology, Writing – review & editing. KD: Conceptualization, Writing – review & editing. KS: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Blue Skies and Growing Futures Funds from The New Zealand Institute for Plant and Food Research Limited.
Acknowledgments
The authors would like to acknowledge Ed Morgan, Janine Johnson, and Cath Kingston for helpful comments on the manuscript.
Conflict of interest
Authors RJ, JT, CA, BJ, RD, NA, YZ, KD and KS were employed by the company The New Zealand Institute for Plant and Food Research Limited.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1358745/full#supplementary-material
Supplementary Figure 1 | CRISPR/Cas9-generated strigolactones mutants of Marchantia. DNA sequence of CRISPR-induced mutations in MpD27–1 (Mp6g03970), MpD27–2 (Mp6g01750), MpCCD7 (Mp2g03280) and MpSMXL (Mp3g06310). The underlined sequences enclosed by large black brackets represent the guides used, the symbol “—” represents the deletion, and the symbol “+ “represents insertions. The purple boxes represent exons, whereas the black lines represent the introns.
Supplementary Figure 2 | (A) Phenotypes of wild-type and Mpsmxl#11, Mpd27–1#16, Mpd27–2#9 and Mpccd7#17 mutant plants. Plants were grown on complete media (1/2 strength Gamborg’s B5 medium supplemented with 1% sucrose and solidified with 1% agar) for 10 weeks at 25°C, 16-hour photoperiod and a light intensity of 30 mol m-2 s-1, provided by cool fluorescent tubes. (B) Total number of gemmae in GC located at different positions across the thallus for the wild-type genotype. The GC positions were numbered as shown in .
Supplementary Figure 3 | Dormant gemmae of Mpsmxl#11, MpD27–1, MpD27–2, MpCCD7 and wild-type were grown on carbon- and nutrient-starved medium under light for 10 days. (A) Phenotypes of gemmae, where images were taken with a light microscope. (B) Area of gemmae, which was measured by image J. Error bars denote mean ± SD (n = 50). The Student’s test was employed to compare mutant lines with wild-type. Statistically significant differences against both wild-types are represented at p < 0.05 (*).
Supplementary Figure 4 | Dormant gemmae of Mpsmxl#11, MpD27–1, MpD27–2, MpCCD7 and wild-type were grown on carbon- and nutrient-starved medium in the complete darkness for 10 days. The graph represents the percentage of gemmae with rhizoids. Scale bars represent 1 mm, images taken with a light microscope. Error bars denote mean ± SD (n = 50). The Student’s t-test was used to compare mutant lines with wild-type. Statistically significant differences against the wild-type are represented as p < 0.05 (*).
Supplementary Figure 5 | Transcriptome analysis of Mpd27–1, Mpd27–2 and Mpccd7 and wild-type genotype. A dendrogram generated by hierarchical clustering illustrating the relationships between the various gene expression profiles. Hierarchical clustering performed on differentially expressed genes defined by ANOVA with a FDR <0.05. Using all replicates per group, top 2000 most variable genes were clustered. The scale bar represents relative expression ±2 SD from the mean.
References
Abuauf, H., Haider, I., Jia, K.-P., Ablazov, A., Mi, J., Blilou, I., et al. (2018). The arabidopsis dwarf27 gene encodes an all-trans-/9-cis-B-carotene isomerase and is induced by auxin, abscisic acid and phosphate deficiency. Plant Sci. 277, 33–42. doi: 10.1016/j.plantsci.2018.06.024
Albert, N. W., Thrimawithana, A. H., Mcghie, T. K., Clayton, W. A., Deroles, S. C., Schwinn, K. E., et al. (2018). Genetic analysis of the liverwort marchantia polymorpha reveals that R2r3 myb activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytol. 218, 554–566. doi: 10.1111/nph.15002
Alcaraz, L. D., Peimbert, M., Barajas, H. R., Dorantes-Acosta, A. E., Bowman, J. L., Arteaga-Vázquez, M. A. (2018). Marchantia liverworts as A proxy to plants’ Basal microbiomes. Sci. Rep. 8, 12712. doi: 10.1038/s41598-018-31168-0
Ali, M., Miao, L., Darwish, D. B., Alrdahe, S. S., Benedito, V. A., Tadege, M., et al. (2021). Overexpression of terpenoid biosynthesis genes from garden sage (Salvia officinalis) modulates rhizobia interaction and nodulation in soybean. Front. In Plant Sci. 12, 783269. doi: 10.3389/fpls.2021.783269
Ali, S., Kucek, L. K., Riday, H., Krom, N., Krogman, S., Cooper, K., et al. (2023). Transcript profiling of hairy vetch (Vicia villosa roth) identified interesting genes for seed dormancy. Plant Genome 16 (2), E20330. doi: 10.1002/tpg2.20330
Aquino, B., Bradley, J. M., Lumba, S. (2021). On the outside looking in: roles of endogenous and exogenous strigolactones. Plant J. 105, 322–334. doi: 10.1111/tpj.15087
Barnes, C. R., Land, W. (1908). Bryological papers. Ii. The origin of the cupule of marchantia. Botanic. Gazette 46, 401–409. doi: 10.1086/329782
Barnes, C. R., Land, W. J. G. (1907). Bryological papers. I. The origin of air chambers. Botanic. Gazette 44, 197–213. doi: 10.1086/329317
Bidartondo, M. I., Read, D. J., Trappe, J. M., Merckx, V., Ligrone, R., Duckett, J. G. (2011). The dawn of symbiosis between plants and fungi. Biol. Lett. 7, 574–577. doi: 10.1098/rsbl.2010.1203
Boncan, D. A. T., Tsang, S. S. K., Li, C., Lee, I. H. T., Lam, H. M., Chan, T. F., et al. (2020). Terpenes and terpenoids in plants: interactions with environment and insects. Int. J. Mol. Sci. 21 (19), 7382. doi: 10.3390/ijms21197382
Bowman, J. L., Arteaga-Vazquez, M., Berger, F., Briginshaw, L. N., Carella, P., Aguilar-Cruz, A., et al. (2022). The renaissance and enlightenment of marchantia as A model system. Plant Cell 34, 3512–3542. doi: 10.1093/plcell/koac219
Brundrett, M. C. (2002). Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304. doi: 10.1046/j.1469-8137.2002.00397.x
Bruno, M., Al-Babili, S. (2016). On the substrate specificity of the rice strigolactone biosynthesis enzyme dwarf27. Planta 243, 1429–1440. doi: 10.1007/s00425-016-2487-5
Buck, W. R., Allen, B., Pursell, R. A. (2003). Recent literature on bryophytes. Bryologist 106 (2), 332–340. doi: 10.1639/0007-2745(2003)106[0332:RLOB]2.0.CO;2
Carella, P., Schornack, S. (2018). Manipulation of bryophyte hosts by pathogenic and symbiotic microbes. Plant And Cell Physiol. 59, 656–665. doi: 10.1093/pcp/pcx182
Chandler, J., Werr, W. (2020). A phylogenetically conserved APETALA2/ethylene response factor, ERF12, regulates arabidopsis floral development. Plant Mol. Biol. 102, 39–54. doi: 10.1007/s11103-019-00936-5
Cheng, A. X., Lou, Y. G., Mao, Y. B., Lu, S., Wang, L. J., Chen, X. Y. (2007). Plant terpenoids: biosynthesis and ecological functions. J. Integr. Plant Biol. 49, 179–186. doi: 10.1111/j.1744-7909.2007.00395.x
Chung, M. Y., Vrebalov, J., Alba, R., Lee, J., Mcquinn, R., Chung, J. D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947. doi: 10.1111/tpj.2010.64.issue-6
Costa, D. P., Peralta, D. F. (2015). Bryophytes diversity in Brazil. Rodriguésia 66, 1063–1071. doi: 10.1590/2175-7860201566409
D’Ambrosio, D. N., Clugston, R. D., Blaner, W. S. (2011). Vitamin A metabolism: an update. Nutrients 3, 63–103. doi: 10.3390/nu3010063
Dang, Y., Jia, G., Choi, J., Ma, H., Anaya, E., Ye, C., et al. (2015). Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 1–10. doi: 10.1186/s13059-015-0846-3
De Hoff, P. L., Brill, L. M., Hirsch, A. M. (2009). Plant lectins: the ties that bind in root symbiosis and plant defense. Mol. Genet. Genomics 282, 1–15. doi: 10.1007/s00438-009-0460-8
Delaux, P.-M., Radhakrishnan, G. V., Jayaraman, D., Cheema, J., Malbreil, M., Volkening, J. D., et al. (2015). Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. 112, 13390–13395. doi: 10.1073/pnas.1515426112
Delaux, P.-M., Xie, X., Timme, R. E., Puech-Pages, V., Dunand, C., Lecompte, E., et al. (2012). Origin of strigolactones in the green lineage. New Phytol. 195, 857–871. doi: 10.1111/j.1469-8137.2012.04209.x
de Saint Germain, A., Bonhomme, S., Boyer, F.-D., Rameau, C. (2013). Novel insights into strigolactone distribution and signalling. Curr. Opin. Plant Biol. 16, 583–589. doi: 10.1016/j.pbi.2013.06.007
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Downie, J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170. doi: 10.1111/j.1574-6976.2009.00205.x
Drummond, R. S., Janssen, B. J., Luo, Z., Oplaat, C., Ledger, S. E., Wohlers, M. W., et al. (2015). Environmental control of branching in petunia. Plant Physiol. 168, 735–751. doi: 10.1104/pp.15.00486
Drummond, R. S., Lee, H. W., Luo, Z., Janssen, B. J., Snowden, K. C. (2023). Varying the expression pattern of the strigolactone receptor gene DAD2 results in phenotypes distinct from both wild type and knockout mutants. Front. Plant Sci. 14, 1277617. doi: 10.3389/fpls.2023.1277617
Dun, E. A., De Saint Germain, A., Rameau, C., Beveridge, C. A. (2013). Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol. Plant 6, 128–140. doi: 10.1093/mp/sss131
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edwards, J., Laskowski, M., Baskin, T., Mitchell, N., Demeo, B. (2019). The role of water in fast plant movements. Integr. Comp. Biol. 59, 1525–1534. doi: 10.1093/icb/icz081
Eklund, D. M., Ishizaki, K., Flores-Sandoval, E., Kikuchi, S., Takebayashi, Y., Tsukamoto, S., et al. (2015). Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27, 1650–1669. doi: 10.1105/tpc.15.00065
Eklund, D. M., Kanei, M., Flores-Sandoval, E., Ishizaki, K., Nishihama, R., Kohchi, T., et al. (2018). An evolutionarily conserved abscisic acid signaling pathway regulates dormancy in the liverwort Marchantia polymorpha. Curr. Biol. 28, 3691–3699. e3. doi: 10.1016/j.cub.2018.10.018
Foo, E., Reid, J. B. (2013). Strigolactones: new physiological roles for an ancient signal. J. Plant Growth Regul. 32, 429–442. doi: 10.1007/s00344-012-9304-6
Frenzel, A., Manthey, K., Perlick, A. M., Meyer, F., Pühler, A., Küster, H., et al. (2005). Combined transcriptome profiling reveals a novel family of arbuscular mycorrhizal-specific Medicago truncatula lectin genes. Mol. Plant-Microbe Interact. 18, 771–782. doi: 10.1094/MPMI-18-0771
Frenzel, A., Tiller, N., Hause, B., Krajinski, F. (2006). The conserved arbuscular mycorrhiza-specific transcription of the secretory lectin mtlec 5 is mediated by A short upstream sequence containing specific protein binding sites. Planta 224, 792–800. doi: 10.1007/s00425-006-0262-8
Ge, S. X., Son, E. W., Yao, R. (2018). Idep: an integrated web application for differential expression and pathway analysis of rna-seq data. BMC Bioinf. 19, 1–24. doi: 10.1186/s12859-018-2486-6
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A., Pillot, J.-P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi: 10.1038/nature07271
Green, B. R., Pichersky, E., Kloppstech, K. (1991). Chlorophyll A/B-binding proteins: an extended family. Trends In Biochem. Sci. 16, 181–186. doi: 10.1016/0968-0004(91)90072-4
Guindon, S., Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. System. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Haider, I., Andreo-Jimenez, B., Bruno, M., Bimbo, A., Floková, K., Abuauf, H., et al. (2018). The interaction of strigolactones with abscisic acid during the drought response in rice. J. Exp. Bot. 69, 2403–2414. doi: 10.1093/jxb/ery089
Hu, Y., Han, Z., Wang, T., Li, H., Li, Q., Wang, S., et al. (2022). Ethylene response factor MdERF4 and histone deacetylase MdHDA19 suppress apple fruit ripening through histone deacetylation of ripening-related genes. Plant Physiol. 188, 2166–2181. doi: 10.1093/plphys/kiac016
Huang, A. C., Osbourn, A. (2019). Plant terpenes that mediate below-ground interactions: prospects for bioengineering terpenoids for plant protection. Pest Manage. Sci. 75, 2368–2377. doi: 10.1002/ps.5410
Humphreys, C. P., Franks, P. J., Rees, M., Bidartondo, M. I., Leake, J. R., Beerling, D. J. (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 1, 103. doi: 10.1038/ncomms1105
Ishizaki, K., Nishihama, R., Yamato, K. T., Kohchi, T. (2016). Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–270. doi: 10.1093/pcp/pcv097
Jain, M., Amera, G. M., Muthukumaran, J., Singh, A. K. (2022). Insights into biological role of plant defense proteins: A review. Biocatal. Agric. Biotechnol. 40, 102293. doi: 10.1016/j.bcab.2022.102293
Kaloudas, D., Penchovsky, R. (2018). Arabidopsis homologues to the LRAT a possible substrate for new plant-based anti-cancer drug development. Int. J. Biomed. Clin. Eng. (IJBCE) 7, 40–52. doi: 10.4018/IJBCE
Kawamura, S., Romani, F., Yagura, M., Mochizuki, T., Sakamoto, M., Yamaoka, S., et al (2022). MarpolBase expression: A web-based, comprehensive platform for visualization and analysis of transcriptomes in the liverwort Marchantia polymorpha. Plant Cell Physiol. 63, 1745–1755.
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kodama, K., Rich, M. K., Yoda, A., Shimazaki, S., Xie, X., Akiyama, K., et al. (2022). An ancestral function of strigolactones as symbiotic rhizosphere signals. Nat. Commun. 13, 3974. doi: 10.1038/s41467-022-31708-3
Kohchi, T., Yamato, K. T., Ishizaki, K., Yamaoka, S., Nishihama, R. (2021). Development and molecular genetics of Marchantia polymorpha. Annu. Rev. Plant Biol. 72, 677–702. doi: 10.1146/annurev-arplant-082520-094256
Komatsu, A., Kodama, K., Mizuno, Y., Fujibayashi, M., Naramoto, S., Kyozuka, J. (2023). Control of vegetative reproduction in Marchantia polymorpha by the KAI2-ligand signaling pathway. Curr. Biol. 33, 1196–1210. e4. doi: 10.1016/j.cub.2023.02.022
Li, F.-W., Nishiyama, T., Waller, M., Frangedakis, E., Keller, J., Li, Z., et al. (2020). Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 6, 259–272. doi: 10.1038/s41477-020-0618-2
Li, Z., Pi, Y., Zhai, C., Xu, D., Ma, W., Chen, H., et al. (2022). The strigolactone receptor SlDWARF14 plays a role in photosynthetic pigment accumulation and photosynthesis in tomato. Plant Cell Rep. 41, 2089–2105. doi: 10.1007/s00299-022-02908-4
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, J. W., He, H. Z., Vitali, M., Visentin, I., Charnikhova, T., Haider, I., et al. (2015). Osmotic stress represses strigolactone biosynthesis in roots: exploring the interaction between strigolactones and ABA under abiotic stress. Planta 241, 1435–1451. doi: 10.1007/s00425-015-2266-8
Liu, J., Zhang, Y., Jiang, Y., Sun, H., Duan, R., Qu, J., et al. (2022). Formation mechanism and occurrence law of pod shattering in soybean: A review. Phyton 91. doi: 10.32604/phyton.2022.019870
Lopez-Obando, M., Guillory, A., Boyer, F.-D., Cornu, D., Hoffmann, B., Le Bris, P., et al. (2021). The Physcomitrium (Physcomitrella) patens PpKAI2L receptors for strigolactones and related compounds function via MAX2-dependent and-independent pathways. Plant Cell 33, 3487–3512. doi: 10.1093/plcell/koab217
López-Ráez, J. A., Charnikhova, T., Gómez-Roldán, V., Matusova, R., Kohlen, W., De Vos, R., et al. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178, 863–874. doi: 10.1111/j.1469-8137.2008.02406.x
López-Ráez, J. A., Kohlen, W., Charnikhova, T., Mulder, P., Undas, A. K., Sergeant, M. J., et al. (2010). Does abscisic acid affect strigolactone biosynthesis? New Phytol. 187, 343–354. doi: 10.1111/j.1469-8137.2010.03291.x
Mashiguchi, K., Seto, Y., Yamaguchi, S. (2021). Strigolactone biosynthesis, transport and perception. Plant J. 105, 335–350. doi: 10.1111/tpj.15059
Mayzlish-Gati, E., Lekkala, S. P., Resnick, N., Wininger, S., Bhattacharya, C., Lemcoff, J. H., et al. (2010). Strigolactones are positive regulators of light-harvesting genes in tomato. J. Exp. Bot. 61, 3129–3136. doi: 10.1093/jxb/erq138
Mizuno, Y., Komatsu, A., Shimazaki, S., Naramoto, S., Inoue, K., Xie, X., et al. (2021). Major components of the KARRIKIN INSENSITIVE2-dependent signaling pathway are conserved in the liverwort Marchantia polymorpha. Plant Cell 33, 2395–2411. doi: 10.1093/plcell/koab106
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. 115, E2274–E2283. doi: 10.1073/pnas.1719588115
Müller, M., Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nazem-Bokaee, H., Hom, E. F., Warden, A. C., Mathews, S., Gueidan, C. (2021). Towards a systems biology approach to understanding the lichen symbiosis: opportunities and challenges of implementing network modelling. Front. Microbiol. 12, 667864. doi: 10.3389/fmicb.2021.667864
Nelson, J. M., Hauser, D. A., Hinson, R., Shaw, A. J. (2018). A novel experimental system using the liverwort Marchantia polymorpha and its fungal endophytes reveals diverse and context-dependent effects. New Phytol. 218, 1217–1232. doi: 10.1111/nph.15012
Nelson, J., Shaw, A. J. (2019). Exploring the natural microbiome of the model liverwort: fungal endophyte diversity in Marchantia polymorpha L. Symbiosis 78, 45–59. doi: 10.1007/s13199-019-00597-4
Perreau, F., Frey, A., Effroy-Cuzzi, D., Savane, P., Berger, A., Gissot, L., et al. (2020). ABSCISIC ACID-DEFICIENT4 has an essential function in both cis-violaxanthin and cis-neoxanthin synthesis. Plant Physiol. 184, 1303–1316. doi: 10.1104/pp.20.00947
Pirrello, J., Jaimes-Miranda, F., Sanchez-Ballesta, M. T., Tournier, B., Khalil-Ahmad, Q., Regad, F., et al. (2006). Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 47, 1195–1205. doi: 10.1093/pcp/pcj084
Poveda, J. (2020). Marchantia polymorpha as a model plant in the evolutionary study of plant-microorganism interactions. Curr. Plant Biol. 23, 100152. doi: 10.1016/j.cpb.2020.100152
Robinson, M. D., Mccarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schiphorst, C., Achterberg, L., Gómez, R., Koehorst, R., Bassi, R., Van Amerongen, H., et al. (2022). The role of light-harvesting complex I in excitation energy transfer from LHCII to photosystem I in Arabidopsis. Plant Physiol. 188, 2241–2252. doi: 10.1093/plphys/kiab579
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sears, A. E., Palczewski, K. (2016). Lecithin: retinol acyltransferase: a key enzyme involved in the retinoid (visual) cycle. Biochemistry 55, 3082–3091. doi: 10.1021/acs.biochem.6b00319
Sgroi, M., Paszkowski, U. (2020). Transcriptional responses to arbuscular mycorrhizal symbiosis development are conserved in the early divergent Marchantia paleacea. BioRxiv, 422721. doi: 10.1101/2020.12.14.422721
Shaw, A. J., Szövényi, P., Shaw, B. (2011). Bryophyte diversity and evolution: windows into the early evolution of land plants. Am. J. Bot. 98, 352–369. doi: 10.3732/ajb.1000316
Shen, H., Luong, P., Huq, E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145, 1471–1483. doi: 10.1104/pp.107.107227
Sun, T., Rao, S., Zhou, X., Li, L. (2022). Plant carotenoids: recent advances and future perspectives. Mol. Horticult. 2, 3. doi: 10.1186/s43897-022-00023-2
Takemura, M., Maoka, T., Misawa, N. (2014). Carotenoid analysis of a liverwort Marchantia polymorpha and functional identification of its lycopene β-and ϵ-cyclase genes. Plant Cell Physiol. 55, 194–200. doi: 10.1093/pcp/pct170
Van Damme, E. J., Lannoo, N., Peumans, W. J. (2008). “Plant lectins,” in Advances in botanical research (Elsevier). doi: 10.1016/S0065-2296(08)00403-5
Walker, C. H., Siu-Ting, K., Taylor, A., O’connell, M. J., Bennett, T. (2019). Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol. 17, 1–19. doi: 10.1186/s12915-019-0689-6
Wang, Y., Zhang, W.-Z., Song, L.-F., Zou, J.-J., Su, Z., Wu, W.-H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211. doi: 10.1104/pp.108.126375
Whitaker, D. L., Edwards, J. (2010). Sphagnum moss disperses spores with vortex rings. Science 329, 406–406. doi: 10.1126/science.1190179
Wyatt, R. (1982). Population ecology of bryophytes. J. Hattori Botanic. Lab. 52, 179–198. doi: 10.18968/jhbl.52.0_179
Keywords: Marchantia polymorpha, strigolactones, DWARF27, Carotenoid Cleavage Dioxygenase 7, gemma, gemma cup, ethylene
Citation: Jibran R, Tahir J, Andre CM, Janssen BJ, Drummond RSM, Albert NW, Zhou Y, Davies KM and Snowden KC (2024) DWARF27 and CAROTENOID CLEAVAGE DIOXYGENASE 7 genes regulate release, germination and growth of gemma in Marchantia polymorpha. Front. Plant Sci. 15:1358745. doi: 10.3389/fpls.2024.1358745
Received: 20 December 2023; Accepted: 07 June 2024;
Published: 25 June 2024.
Edited by:
Jorge Poveda, University of Valladolid, SpainReviewed by:
Claudia Renate Stange, University of Chile, ChileJean-David Rochaix, University of Geneva, Switzerland
Copyright © 2024 Jibran, Tahir, Andre, Janssen, Drummond, Albert, Zhou, Davies and Snowden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rubina Jibran, cnViaW5hLmppYnJhbkBwbGFudGFuZGZvb2QuY28ubno=
 Rubina Jibran
Rubina Jibran Jibran Tahir
Jibran Tahir Christelle M. Andre
Christelle M. Andre Bart J. Janssen
Bart J. Janssen Revel S. M. Drummond
Revel S. M. Drummond Nick W. Albert
Nick W. Albert Yanfei Zhou
Yanfei Zhou Kevin M. Davies
Kevin M. Davies Kimberley C. Snowden
Kimberley C. Snowden