
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 February 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1358673
 Miao Zhang1†
Miao Zhang1† Jing-Yun Gao1†
Jing-Yun Gao1† Shi-Chen Dong1
Shi-Chen Dong1 Meng-Han Chang1
Meng-Han Chang1 Jing-Xuan Zhu1
Jing-Xuan Zhu1 Dong-Lin Guo1*
Dong-Lin Guo1* Chang-Hong Guo1*
Chang-Hong Guo1* Ying-Dong Bi2*
Ying-Dong Bi2*Cadmium (Cd) pollution severely affects plant growth and development, posing risks to human health throughout the food chain. Improved iron (Fe) nutrients could mitigate Cd toxicity in plants, but the regulatory network involving Cd and Fe interplay remains unresolved. Here, a transcription factor gene of alfalfa, MsbHLH115 was verified to respond to iron deficiency and Cd stress. Overexpression of MsbHLH115 enhanced tolerance to Cd stress, showing better growth and less ROS accumulation in Arabidopsis thaliana. Overexpression of MsbHLH115 significantly enhanced Fe and Zn accumulation and did not affect Cd, Mn, and Cu concentration in Arabidopsis. Further investigations revealed that MsbHLH115 up-regulated iron homeostasis regulation genes, ROS-related genes, and metal chelation and detoxification genes, contributing to attenuating Cd toxicity. Y1H, EMSA, and LUC assays confirmed the physical interaction between MsbHLH115 and E-box, which is present in the promoter regions of most of the above-mentioned iron homeostasis regulatory genes. The transient expression experiment showed that MsbHLH115 interacted with MsbHLH121pro. The results suggest that MsbHLH115 may directly regulate the iron-deficiency response system and indirectly regulate the metal detoxification response mechanism, thereby enhancing plant Cd tolerance. In summary, enhancing iron accumulation through transcription factor regulation holds promise for improving plant tolerance to Cd toxicity, and MsbHLH115 is a potential candidate for addressing Cd toxicity issues.
Cadmium (Cd) is a biologically non-essential metal and one of the most hazardous environmental pollutants. Cd contamination in crops has become a severe problem due to rapid industrialization and excessive use of pesticides and fertilizers (Palansooriya et al., 2020). Cd injures plant growth and reproduction, disrupts photosynthesis and transpiration, impairs root growth, and reduces biomass production by catalyzing the accumulation of harmful substances (Sandalio et al., 2001; Dias et al., 2012; El Rasafi et al., 2020). Most economic crops are sensitive to Cd toxicity, and high Cd accumulation poses a potential risk to food safety. Therefore, enhancing Cd tolerance and reducing Cd accumulation in edible parts of crops is of great significance.
Iron (Fe) is an essential metal element for plants that plays a role in plant growth and development. Because Fe deficiency hurts plant growth and excess Fe is toxic to plants, a set of transcript factors and transporters tightly regulates Fe homeostasis. Due to the high similarity in ionic hydrated radius of Cd2+ (4.26 am) and Fe2+ (4.28 am) (Nightingale, 1959), Cd can be easily absorbed and transported by competing with Fe (Eide et al., 1996). The crosstalk between Cd and Fe has been found in plants. Cd stress changed the cell wall components and enhanced the binding capacity for Fe, resulting in Fe retention in the apoplast of roots and suppressing Fe translocation from roots to shoots (Xu et al., 2015). The antagonistic interaction between Cd and Fe may contribute to the Cd toxicity symptoms observed in plants (Jian et al., 2019), and Fe deficiency can exacerbate Cd toxicity in plants (Bao et al., 2009; Muneer et al., 2014; Su et al., 2014), while elevating the Fe status reduces the Cd toxicity to plants and alleviate Cd toxicity symptoms, contributing to enhancing the tolerance to Cd stress (Meda et al., 2007; Liu et al., 2020).
Cd and be uptake by identified Fe transporters. The intracellular vesicle membrane protein iron-regulated transporter1 (IRT1) was found to be involved in Cd uptake (Korshunova et al., 1999; Zhang et al., 2020). It has been reported that limiting Fe uptake through the downregulation of Fe acquisition mechanisms confers Si-mediated alleviation of Cd toxicity in Alfalfa (Kabir et al., 2016). Results indicated that argon-stimulated NO production contributes to Cd tolerance by transcriptional reduction in representative target genes involved in heavy metal detoxification, antioxidant defiance, and iron homeostasis (Wang J. et al., 2023). To avoid heavy metal poisoning, plants often up-regulate the expression of genes related to metal chelation, sequestration, and redeployment, including heavy metal-associated domain (HMA), Metal tolerance protein (MTP), Iron regulatory gene (IREG), and other metal transporters (Yao et al., 2018; Sheng et al., 2019; Dang et al., 2022). The conjugation and sequestration of Cd into vacuoles by glutathione and proline are also essential mechanisms promoting Cd tolerance in plants (Xiang et al., 2001; Cai et al., 2011; Koen et al., 2013; Han et al., 2020). Exogenous CH4 mitigates Cd toxicity in alfalfa by inducing miR159 and miR167 to regulate heavy metal transporters, reduce Cd accumulation, and reconstruct glutathione homeostasis (Gu et al., 2018). Although several genes in the synthesis and decomposition of glutathione and proline have been identified, the regulating mechanism of glutathione and proline under Cd stress remains unclear.
Members of TF including WRKY, MYB, NAC, bHLH, and MYC are recognized to enhance Cd tolerance by regulating genes expression in a variety of plants (Sheng et al., 2019; Zhang et al., 2019; Du et al., 2023; Han et al., 2023; Wang B. et al., 2023). Several studies have shown that TFs can regulate genes to improve Fe accumulation and distribution, significantly reducing the toxicity of Cd to plants (Zhang et al., 2019; Shi et al., 2023). bHLH TFs have received increasing attention for their roles in plant development, stress response, and organ communication (Aparicio and Pallás, 2016). Their parts in response networks against heavy metal stress have also been highlighted (Yadav and Mani, 2019). IRT1 is directly controlled by FIT and its interacting partners bHLH38/39 in Fe uptaking (Colangelo and Guerinot, 2004; Schwarz and Bauer, 2020; Riaz and Guerinot, 2021; Liang, 2022). It has been reported that overexpression of FIT with AtbHLH38 or AtbHLH39 enhances Cd tolerance in Arabidopsis by increasing root Cd uptake and improving shoot Fe homeostasis (Wu et al., 2012).
The bHLH IVc genes are widely expressed in plants and have been shown to function as long-distance organ-to-organ signals from shoots to roots, participating in Fe uptake, translocation, and intracellular homeostasis (Grillet et al., 2018). bHLH104 is also responsive to Cd toxicity, and mutant plants lacking bHLH104 are sensitive to Cd stress, while overexpression of bHLH104 increases Cd tolerance by altering Fe translocation (Zhang et al., 2015; Li et al., 2016; Yao et al., 2018; Tissot et al., 2019; Zhou et al., 2019). BTS interacts with bHLH104 and negatively regulates Fe homeostasis at the post-translational level, significantly increasing tolerance to Cd stress and rising Fe and Cd accumulation (Zhu et al., 2020). The ectopic expression of IMA peptides confers Cd tolerance to Arabidopsis by activating Fe-deficiency responses (Meng X. et al., 2022). These studies suggest that bHLH IVc genes are involved in the crosstalk of Fe and Cd. Previously, it was found that the expression of IMAs is regulated by bHLH121, and their expression is highly induced in bhlh121 mutants (Li et al., 2022). Studies suggest that the expression of bHLH IVc may be regulated by bHLH121. Chip assays have demonstrated that bHLH115 regulates downstream bHLH121, FIT, bHLH38/39/100/101, and POPEYE (Li et al., 2016; Liang et al., 2017; Gao et al., 2020). The mechanism by which bHLH115 mediates Cd tolerance in plants remains elusive (Kurt et al., 2019; Lei et al., 2020; Lockhart, 2020). It is unclear whether bHLH115-mediated Fe accumulation can confer Cd tolerance in these overexpressing lines, which could provide new solutions for improving tolerance to Cd toxicity. Activating the Fe-deficiency regulatory system will effectively enhance plant tolerance to Cd (He et al., 2017; Ai et al., 2022). However, the underlying mechanism remains unclear due to a lack of general systemic signaling across species for Fe deficiency regulation and an insufficient understanding of the antagonistic mechanism between Fe and Cd.
Medicago sativa (alfalfa) is an important forage grass (Ndayambaza et al., 2020). Cd adversely affects alfalfa growth(Kabir et al., 2016). In this study, we explored the function of MsbHLH115 in Arabidopsis and determined that MsbHLH115 regulates plant Cd tolerance. Our results suggest that MsbHLH115 may regulate plant Cd tolerance by controlling the expression of iron-regulated transcription factors bHLHs, which increases iron transport. We identified MsbHLH115 as a regulator of Cd tolerance, indicating that MsbHLH115 could be used to design new strategies for crop adaptation to Cd stress.
Alfalfa (Medicago sativa cv. Zhao dong) seeds, provided by the Livestock and Veterinary Research Institute of Heilongjiang Academy of Agricultural Sciences, were germinated in distilled water for 3 days. Seedlings were cultivated in Hoagland’s solution (10 μM MnSO4, 100 μM H3BO3, 0.1 μM CuSO4, 0.1 μM Na2MoO4, 30 μM ZnSO4, 5 μM KI, 0.1 μM CoCl2, 4 mM CaCl2, 1 mM MgSO4, 1 mM KH2PO4, 100 μM Fe(III)-EDTA and 5 mM KNO3, pH 5.8) at 25°C, 16 h light/8 h darkness (Cui et al., 2013). The culture solution was changed every 3 days. Four-week-old alfalfa seedlings were selected for treatment. Alfalfa seedlings were treated with +Cd stress for 3 days to analyze the expression of iron-deficiency response genes. The stress concentration of hydroponics in Hoagland’s solution was −Fe (0 µM Fe-EDTA+0 μM CdCl2), +Cd (100 µM Fe-EDTA+90 μM CdCl2), −Fe+Cd (0 µM Fe-EDTA+90 μM CdCl2), and the control (CK, 100 µM Fe-EDTA+0 μM CdCl2) (Dai et al., 2017; Yang et al., 2021). Alfalfa seedlings were treated with −Fe, +Cd, −Fe+Cd, and CK for 24 h, and the roots, stems, and leaves were collected, respectively, to analyze the tissue-specific expression of MsbHLH115. Alfalfa seedlings were treated with 50 μM, 70 μM, 90 μM, 100 μM, and 200 μM Cd for 24 h to detect the response of MsbHLH115. Alfalfa seedlings were treated with −Fe, +Cd, −Fe+Cd, and CK for 6, 12, 24, 48, and 72 h to detect the temporal properties of MsbHLH115 expression. The above samples were collected, frozen in liquid nitrogen, and stored at −80°C to extract total RNA for Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR).
The cultivation method of Arabidopsis seeds (ecotype Columbia, Col-0) is as follows: surface-sterilized with 10% NaClO for 15 min and then washed six times with distilled water, seeds were plated on 1/2 MS medium (Murashige and Skoog, 1962) with 1% sucrose, 0.8% agar, and 100 µM Fe-EDTA at pH 5.8, cultivated at 22°C, 16 h light/8 h darkness. The Arabidopsis seedlings were treated with −Fe, +Cd, −Fe+Cd, and CK for phenotype observation and physiological detection.
qRT-PCR was performed by the Bio-Rad CFX96 detection system (Bio-Rad, Hercules, CA, USA) using the Top Green qPCR Super Mix (Transgenic Biotechnology Company, Beijing, China). The relative expression levels of genes were calculated using the 2-ΔΔCT comparative method as described by (Livak and Schmittgen, 2001). MsACTIN2 (Wang et al., 2015) and AtACTIN2 (AT3G18780) (Czechowski et al., 2005) were used as internal reference genes. The Arabidopsis genes bHLH115, bHLH104, bHLH121, FIT, bHLH39, FRO2, IRT1, VTL, HMA3, MTP3, NAS4, IREG2, SOD1, POD1, RbohB, RbohD, RbohF, BGLU14, GSTL1/2/4/6, AtP5CS, and AtPRODH were detected in the transgenic Arabidopsis and WT under 90 μM CdCl2 for 24 h. Primers used in this study were designed using Primer 5.0 software (http://www.premierbiosoft.com/primerdesign/) and online NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and listed in Supplementary Table S1.
Total RNA was extracted using the Total RNA Extraction Kit (Omega BioTek, Norcross, GA, USA). Then, RNA was reverse transcribed to cDNA using ReverTra Ace®qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan). Total DNA was extracted using the Total DNA Extraction Kit (OMGAE, USA). Primers are shown in Supplementary Table S1. The PCR products were cloned into the pMD18-T vector (Takara) and sequenced (Sangon, Shanghai, China). Sequence alignment was performed using DANMAN software. The structured domain was obtained through the SMART (HTTP://smart.embl-heidelberg.de/ accessed on 9 June 2022). Phylogenetic analysis was performed using the MEGA 7.0 with neighbor-joining and a bootstrap repeat value of 1000 times. The cis-elements were analyzed using PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 9 June 2022). Finally, TBtools was used to splice.
MsbHLH115 without the stop codon was inserted into the pBWA(V)HS vector (provided by BioRun Biotechnology Co., Ltd, Wuhan, China), fused with the green fluorescent protein (GFP) driven by the 35S promoter. The recombinational pBWA(V)HS-35S::MsbHLH115-Glosgfp plasmid was transformed into Arabidopsis protoplasts through polyethylene glycol treatment (Yoo et al., 2007). nuclear localization of the MsbHLH115-GFP fusion protein. Red fluorescent protein (RFP) is used as a nuclear localization marker. Fluorescence signals were visualized after 12-16 h using a confocal laser-scanning microscope (Olympus FluoView FV1000, Olympus, Tokyo, Japan).
The MsbHLH115 promoter fragment was digested with Bgl II and BamH I to construct a pBI121-MsbHLH115pro::GUS and transformed into Agrobacterium rhizogenes K599 by the freeze-thaw method. Soybean (Glycine max cv. Williams 82) hairy roots were induced by needle insertion of K599 (Kereszt et al., 2007). The soybean complex was cultivated in 1/2 Hoagland’s solution for 7 days. The hairy roots were divided into two parts and treated with −Fe, +Cd, −Fe+Cd, and CK for 24 h, respectively (Li et al., 2023). Histochemical staining for GUS was conducted following the method described previously (Li et al., 2023). The photograph was taken using a stereomicroscope (EZ4-HD LEICA, Germany) coupled with a color charge-coupled device (CCD) camera (Zeiss, Germany). The GUS enzyme activity was detected using a plant β-glucuronidase GUS ELISA kit (Coolaber, SL7160) and enzyme-labeler machine (Ferdi Bio, FlexA-200). The absorbance (OD value) was measured at 450 nm wavelength. The GUS enzyme activity was calculated according to the standard curve previously drawn.
The MsbHLH115 gene was digested with Xba I and BamH I and inserted into pBI121. The recombinant vector pBI121-35S:: MsbHLH115 was introduced into Agrobacterium tumefacient GV3101 by freeze-thaw method and then transformed into Arabidopsis by the filter-dipping process (Clough and Bent, 1998). PCR and qRT-PCR confirmed the MsbHLH115 transgenic Arabidopsis. The primers used are listed in Supplementary Table S1. The MsbHLH115 transgenic Arabidopsis produced offspring by self-breeding. T3 seedlings of MsbHLH115 transgenic Arabidopsis and WT were cultivated in 1/2 MS medium for 5 days. The growth of MsbHLH115ox lines and WT plants were treated with different combinations of Fe and Cd treatments for 7 days. The growth of #12-2 plants was compared when different root parts were in the split medium with combinations of Fe and Cd treatments for 7 days. A root-tip elongation experiment was applied on MsbHLH115ox lines and WT. The plants were planted on a split medium and were treated with different combinations of Fe and Cd treatments. The part near the root tip (RT) and the part distal the root tip (RS) were exposed to different treatments for 7 days. Phenotypic photographs were taken, and root length and fresh weight were recorded.
Several previous studies have reported that bHLH115 plays a role in iron homeostasis. In this study, we performed experiments focused on Cd treatment rather than iron deficiency treatment to better explain its role in Cd tolerance, The Arabidopsis leaves were collected and frozen at −80°C for physiological index determination. The total chlorophyll content was measured according to the method described by (Yavari et al., 2021). Leaves from seedlings grown on a medium were collected and ground to powder in liquid nitrogen. The powder was resuspended in 80% (v/v) acetone on ice and centrifuged at 10,000 g at 4°C for 5 min. A664 and A648 were inspected by spectroscopy absorbance measurements, and the chlorophyll a and b contents were calculated according to those described previously. NBT and DAB staining methods refer to (Zhang et al., 2011). For NBT staining, the leaves were soaked in darkness with 1 mg/mL nitro blue tetrazole (pH 7.8) for 40-60 min. For DAB staining, the leaves were washed with 1 mg/mL diaminobenzidine (pH 7.0) in the dark for 8 h. The leaves were then boiled in an ethanol glycerin (3:1) solution for 20 min and photographed. Evans Blue staining was performed as described by (Baker and Mock, 1994). Root damage was evaluated by staining with Evans blue solution (0.25%, w/v), The leaves were then boiled in an ethanol glycerin (3:1) solution for 20 min and photographed. H2O2 content was determined according to Elstner, 1976 and the specific procedures were carried out as per the manufacturer’s instructions. The tissues were homogenized in an ice bath with 0.1% trichloroacetic acid (TCA) and centrifuged at 12,000 ×g for 15 min. The reaction solution contained 0.5 mL of potassium dihydrogen phosphate buffer (pH 7.0), 1 mL of 1 M potassium iodide (KI), and 0.5 mL of supernatant. Following that, the mixture was kept at 25°C for 1 h and its absorbance was measured at the wavelength of 390 nm. The H2O2 content was calculated according to the standard curve obtained using different concentrations of H2O2. O2− content was determined according to Elstner, 1976 and the specific procedures were carried out as per the manufacturer’s instructions. The O2− content was determined by monitoring the absorbance of azo compounds at 530 nm (Willekens et al., 1997). The malondialdehyde (MDA) content of the detailed experimental method refers to (Puckette et al., 2007). Approximately 0.1 g plant tissue was homogenized in 1.5 mL 5% TCA that contained 0.25% TBA, incubated at 100°C for 30 min, cooled to room temperature, and then centrifuged at 15000 ×g for 10 min. The concentration of TBA-reactive substance was then determined by measuring the absorbance of the resulting supernatant at 440, 532, and 600 nm. SOD activity was determined refer to (Giannopolitis et al., 1977) by monitoring the absorbance of blue formazan generated by the reaction of the remaining O2− with nitro-blue tetrazolium at 560 nm. The Catalase (CAT) activity was determined refer to (Soydam Aydin et al., 2013), tested in potassium phosphate buffer (pH 7.8) containing 3 mM H2O2 at 240 nm. The glutathione-s-transferase (GST) activity was determined using a previously described method (Nagalakshmi and Prasad, 2001). The reaction mixture contained 10 μL of 0.1 M 1-chloro-2,4-dinitrobenzene (CDNB), 100 μL of 10 mM GSH, 500 μL of 0.2 M KPO4 buffer, and 390 μL of distilled water. and the increase in absorbance was measured at the wavelength of 340 nm. One unit of GST was defined as the amount of enzyme that increased the absorbance by a unit of 1 per min, at the wavelength of 340 nm. Ferric-chelate reductase activity was determined using a previously described method (Li et al., 2023). The roots were immerged into Fe (III) reductase detection solution (0.1 mM Fe (III)-EDTA and 0.3 mM ferrozine), and 10 intact plants were used in each independent experiment. The reaction solution was placed in darkness at 22°C, and then the supernatant was analyzed spectrophotometrically at 562 nm. Lastly, the FCR activity was measured with the use of a molecular extinction coefficient of 28.6 mM-1cm-1.
The 13-day-old Arabidopsis seedlings were treated with or without 90 μM CdCl2 for 3 days. The roots and shoots were counted and harvested separately, and then washed with ddH2O 3~4 times. After being dried in a conventional oven at 70°C, the samples were digested completely in 65%~68% HNO3 at 120°C. The Fe, Cd, Zn, Mn, and Cu concentrations were determined as described by (Takahashi et al., 2019). Using inductively coupled plasma emission spectrometry ICP-OES8000 (Perkin Elmer, USA). The Fe, Cd, Zn, Mn, and Cu concentrations were calculated according to the method previously described (Wu et al., 2012).
The MsbHLH115 ORF sequence was cloned into the pGADT7 vector to construct the recombinant vector pGADT7-MsbHLH115. The E-box element sequence (CAAATG) and mutated E-box element sequence (ACAATG) were cloned into the pAbAi vector to form pE-box-AbAi and pmE-box-AbAi. The p53-AbAi+pGADT7-p53, pE-box-AbAi+pGADT7-MsbHLH115, and pmE-box-AbAi+pGADT7-MsbHLH115 were transformed into yeast strain Y1H. the yeast was grown in SD/-Leu medium (Takara, Shanghai, China) containing 300 ng/mL AbA. The interaction was studied using a yeast single hybridization system (Clontech, Palo Alto, CA).
The oligonucleotide probe E-box (CAAATG) synthesized the gene promoter and was labeled biotin at the 3’ end by Sangon (Shanghai, China). The MsbHLH115 coding sequence was cloned into the pGEX4T-1 vector (provided by BioRun Biotechnology Co., Ltd, Wuhan, China), and EMSA analysis was performed using the photolytic chemiluminescence EMSA kit (Pierce, Rockford, IL, USA).
The MsbHLH121 promoter was inserted into the pGreenII 0800-LUC (provided by BioRun Biotechnology Co., Ltd, Wuhan, China) to produce a report generator. The MsbHLH115 ORF was inserted into pGreenII 62-SK to produce effectors. The mixture of the above plasmid fusion was injected into the lower epidermis of Nicotiana benthamiana leaves with a 1mL syringe without the needle. The infiltrated tobacco plants were cultured in low light for 2 days. Added 100 μL of freshly prepared Revilla substrate working liquid into the reaction solution, quickly mixed, and detected the Revilla luciferase activity immediately in the fluorescence detector.
pBI121-MsbHLH121pro::GUS and pBI121-3×E-Box::GUS recombinant vectors were constructed and transformed into Agrobacterium GV3101 by freeze-thaw method with pBI121-35S::GUS vector as the control. These vectors infiltrated into 3-week-old tobacco leaves, respectively. GUS activity was detected.
All of the data presented here were mean values for each treatment. There are at least three independent biological replicates in every experiment. SPSS 19.0 software and Origin 2018 software were utilized to perform the statistical analysis and produce the graphs, respectively. Student’s t-test was carried out between WT and transgenic plants in the same treatment, and one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test was carried out between the control and treatments.
To explore the effect of Cd stress on Fe homeostasis, the expression levels of the genes involved in Fe-deficient response under Cd exposure in wild-type alfalfa. Compared with wild-type plants grown on standard medium, the results showed that MsbHLH115, MsbHLH121, MsbHLH25, MsbHLH68, MsFIT, MsWRKY33, MsWRKY40, MsYSL6, and MsNAS were significantly up-regulated; meanwhile, MsFRO2, MsIRT1, and MsIRO3 were considerably down-regulated by Cd stress (p<0.05) (Figure 1A). Interestingly, MsbHLH115 was most strongly induced by Cd at the transcription level, suggesting that MsbHLH115 might be an essential gene involved in the network of Fe and Cd regulation in plants.
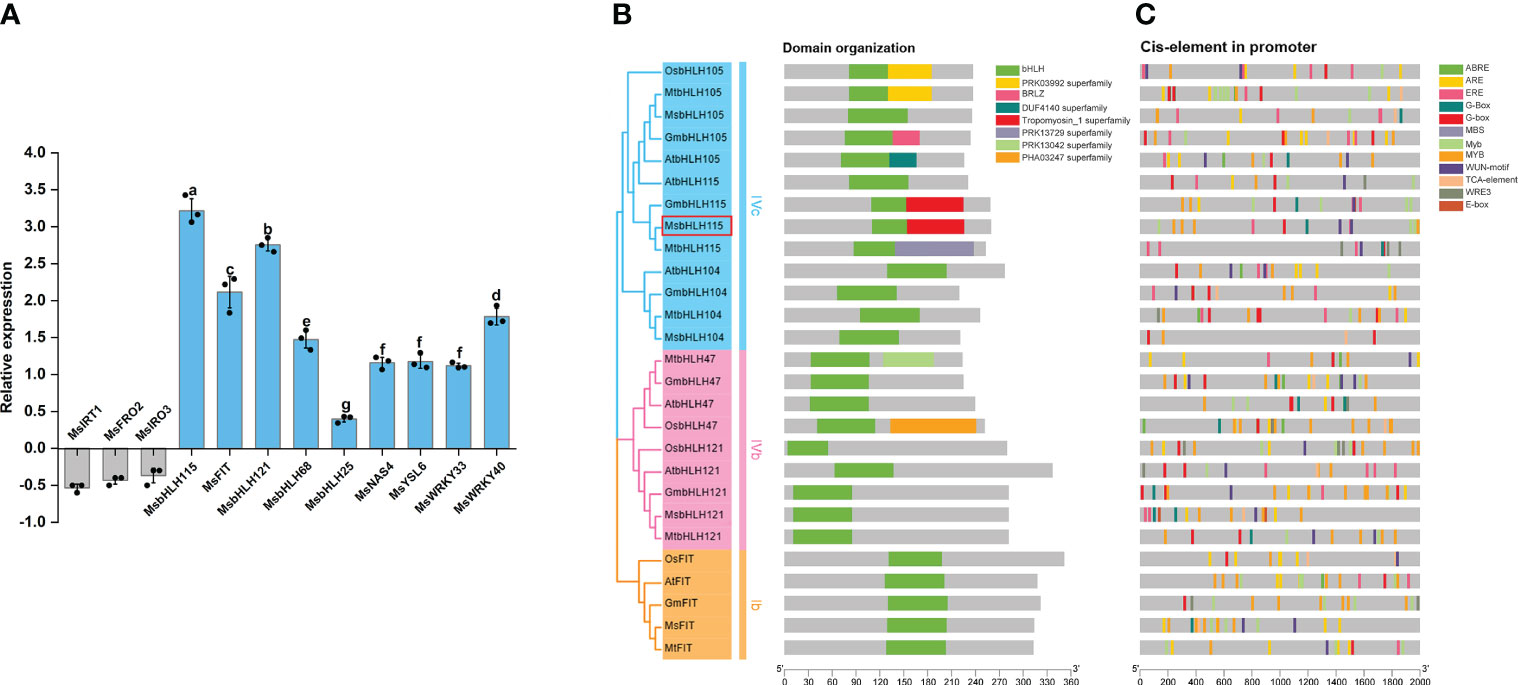
Figure 1 Bioinformatics, Expression level of Fe-deficient responsive genes under Cd stress. (A) Expression level of Fe-deficient responsive genes in the Alfalfa seedlings under Cd stress. Four-week-old alfalfa seedlings were treated with or without 90 μM CdCl2 for 3 days. MsACTIN2 was the normalization control. The y-axis shows gene expression levels normalized to that of the CK. (B) The phylogenetic tree and structure domain of MsbHLH115 and other iron regulation bHLH genes. The MsbHLH115 protein of alfalfa was marked red. (C) Predicted cis-regulatory elements of bHLH promoter. Each type of cis-regulatory element is represented with different color box at the bottom. The five 5′ to 3′ direction represents the orientation of the nucleotide sequence, and the scale at the bottom represents the nucleotide length (bp). One-way ANOVA test. Mean ± standard error, n= 3. Lowercase letters indicate significant at p<0.05.
MsbHLH115 was expressed in roots, leaves, and stems of the alfalfa, mainly in roots. MsbHLH115 was significantly up-regulated by −Fe and +Cd stress and considerably up-regulated by −Fe+Cd stress in roots, while down-regulated by −Fe+Cd stress in the stems and leaves of alfalfa (Figure 2A). The MsbHLH115 expression was significantly increased by −Fe and +Cd at 6 h, 12 h, and 24 h, and was significantly down-regulated at 72 h. The MsbHLH115 expression was significantly up-regulated by −Fe+Cd at 12 h and 24 h and was significantly down-regulated at 48 h and 72 h (Figure 2B). Following the increase of Cd concentration, MsbHLH115 expression was significantly increased, reaching maximum expression at 90 µM, and then decreased (Figure 2C).
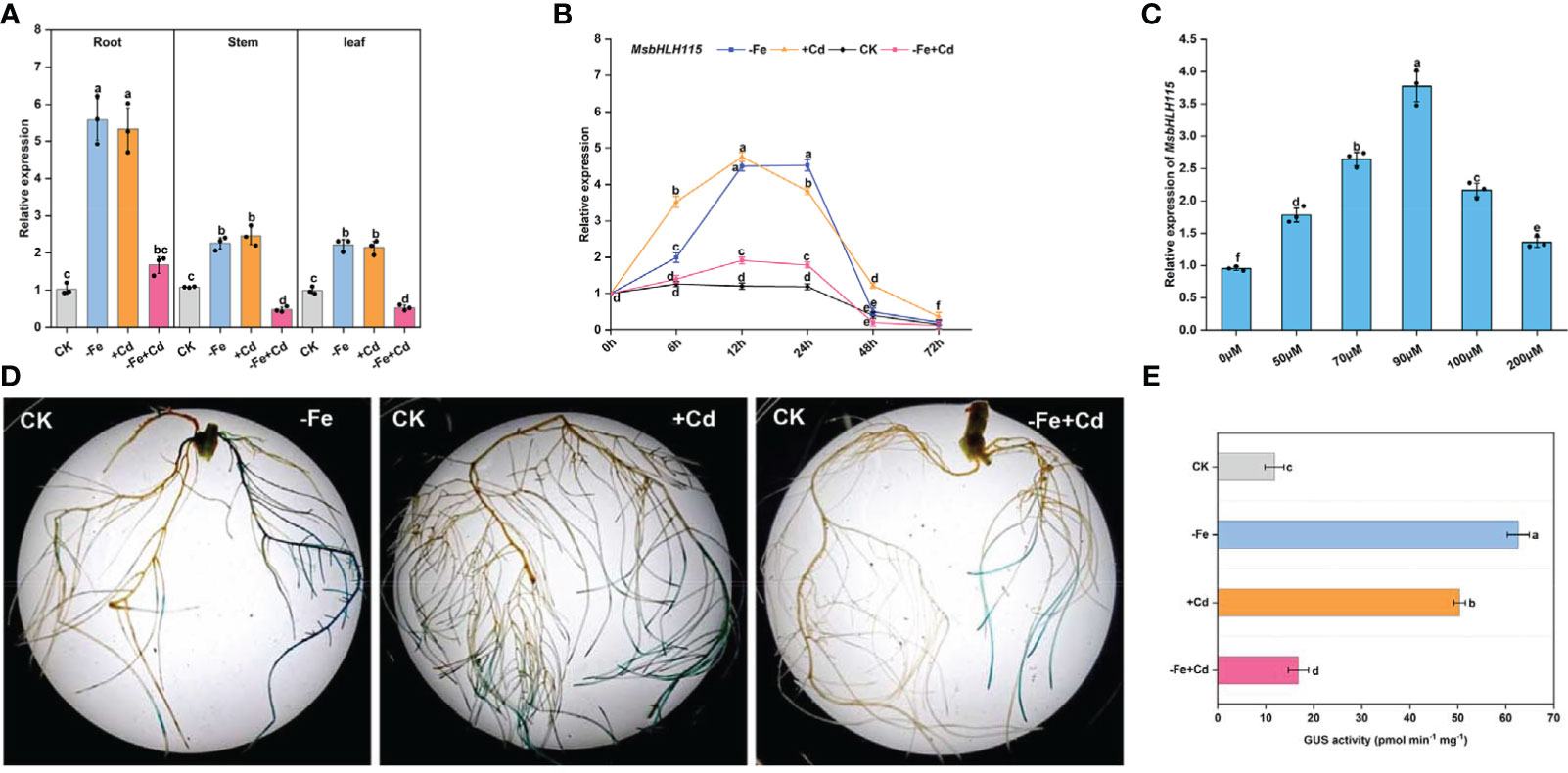
Figure 2 Expression pattern of MsbHLH115. (A) Expression level of MsbHLH115 in different plant tissues. Four-week-old alfalfa seedlings were treated with −Fe, +Cd, and −Fe+Cd stress for 24 h. (B) Time-course expression level of MsbHLH115. Four-week-old alfalfa seedlings were treated with −Fe, +Cd, and −Fe+Cd stress for 6, 12, 24, 48, and 72 h. The expression level at 0 h was set as 1.0. (C) Expression level of MsbHLH115 response to different concentration of Cd. Four-week-old alfalfa seedlings were treated with Cd. MsACTIN2 was the normalization control. (D) Expression pattern of MsbHLH115pro under −Fe, +Cd, −Fe+Cd stress. (E) The MsbHLH115pro GUS activity measurement. One-way ANOVA test. Mean ± standard error, n= 3. Lowercase letters indicate significant at p<0.05.
In the CK, the MsbHLH115pro transformed soybean hairy root showed β-D-glucuronidase (GUS) signals, indicating that the MsbHLH115pro had expression activity. The hairy root tip showed dark GUS staining in all treatments. Under −Fe and +Cd stress, the hairy root was darker blue. Under −Fe+Cd stress, the hairy root staining was darker than that in the CK but shallower than that in −Fe or +Cd treatment (Figure 2D). The value of GUS activity in soybean hairy roots is −Fe>+Cd>−Fe+Cd>CK (p<0.05) (Figure 2E). This result was consistent with the quantification of gene expression, indicating that MsbHLH115 responds to −Fe, +Cd, and −Fe+Cd stress in plants.
A total of 26 bHLHs containing typical bHLH domains were assigned to bHLHIVc, bHLHIVb, and bHLHIb subfamilies. Evolutionary analysis showed that MsbHLH115 belonged to the bHLHIVc subfamily. MsbHLH115 is most closely related to MtbHLH115, GmbHLH115, AtbHLH115, and AtbHLH105 (Figure 1B). There are some cis-regulatory elements such as G-box and E-box in the promoters of bHLH family genes. Several cis-elements involved in stress response and hormone regulation were found in the MsbHLH115 promoter (Figure 1C).
During the transient expression of the fusion protein in Arabidopsis protoplasts, green fluorescence was visible throughout the cytoplasm in protoplasts with the 35S:: GFP control. In protoplasts transiently expressing 35S::MsbHLH115-GFP, nuclear localization of the MsbHLH115-GFP fusion protein was revealed by a nuclear localization marker RFP. The alignment of the predominant red fluorescence in the nucleus with the GFP fluorescence signals confirmed the localization of MsbHLH115 in the nucleus (Figure 3).
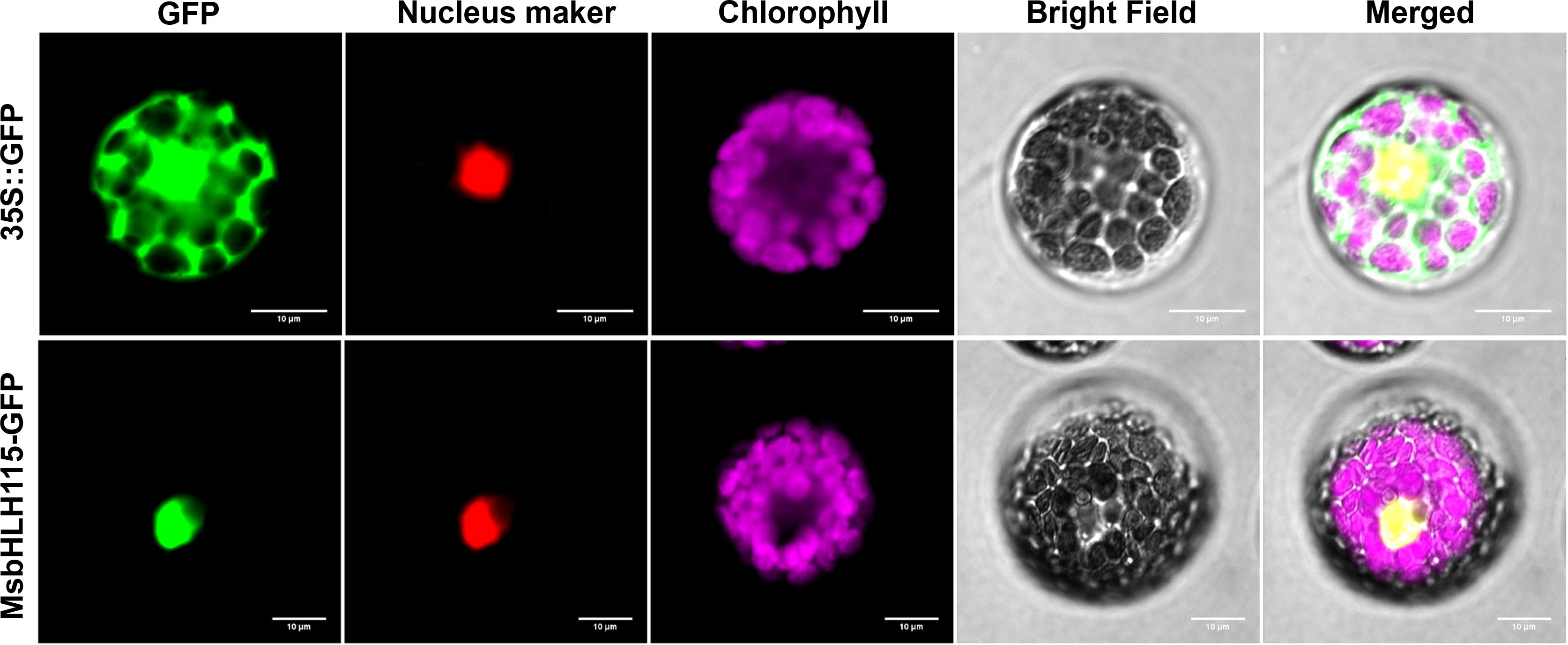
Figure 3 Subcellular localization of MsbHLH115. The 35S::GFP and 35S:: MsbHLH115-GFP plasmid were transformed in Arabidopsis protoplast cells respectively. Red fluorescence signals were used as a nuclear marker. Scale bars: 10 μm.
To investigate the biological function of MsbHLH115, we generated Arabidopsis overexpressed MsbHLH115 lines (MsbHLH115ox). Three transgenic lines with high MsbHLH115 expression levels (ox#2-1, ox#5-1, and ox#12-2) were selected for further analysis (Figure 4B). Under normal conditions(CK), −Fe, +Cd, and −Fe+Cd stress, the MsbHLH115ox lines showed significantly longer roots, higher biomass, and higher chlorophyll content than WT. The degree of impact on plant growth was −Fe+Cd >−Fe>+Cd(p<0.05) (Figures 4A, C–E). The severe growth inhibition caused by −Fe+Cd stress indicated that iron deficiency exacerbated the Cd toxicity in plants.
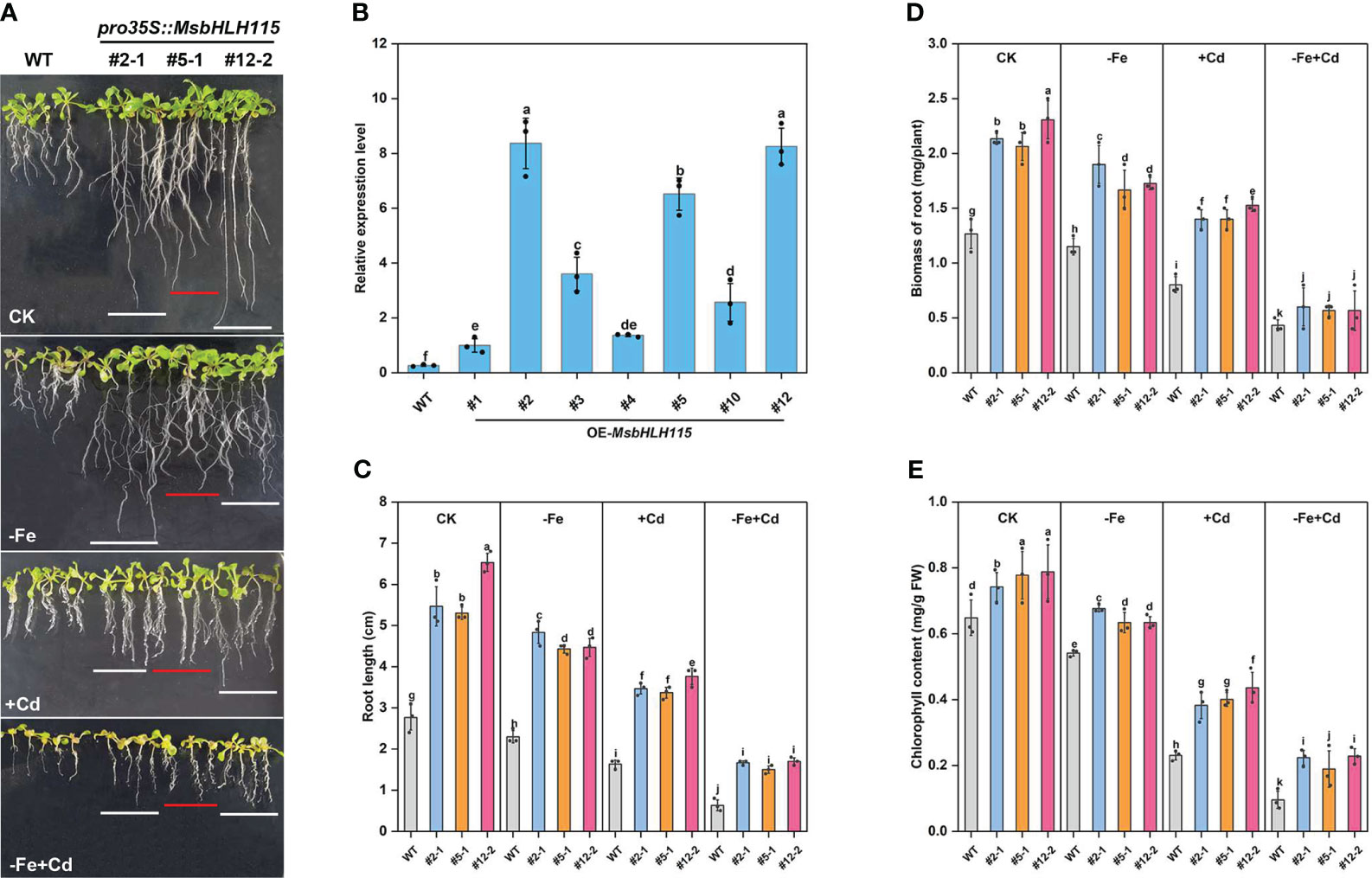
Figure 4 Determination of −Fe and Cd tolerance in overexpressed MsbHLH115ox T3 lines (ox#2-1, ox#5-1, and ox#12-2). (A) Phenotypes of WT and MsbHLH115ox Arabidopsis seedlings under −Fe, +Cd, −Fe+Cd stress and CK. (B) qRT-PCR in overexpressed MsbHLH115ox T3 lines. MsACTIN2 was the normalization control. (C) Root length (D) Biomass of root. (E) The chlorophyll content. The 5-day-old seedlings were cultured on the 1/2 MS medium containing with/without Fe/Cd for 7 days. One-way ANOVA test. Mean ± standard error, n= 3. Lowercase letters indicate significant at p<0.05.
Under +Cd stress, the Nitro-Blue tetrazolium chloride (NBT), 3,3-diaminobenzidine (DAB), and Evans blue staining in the MsbHLH115ox leaves were lighter (Figure 5A) than WT; the O2-, H2O2, and MDA content in the three MsbHLH115ox lines was lower than WT (Figures 5B–D); the CAT, SOD, and GST activity of the three MsbHLH115ox lines were significantly higher than WT (Figures 5E–G). The result indicated that overexpression of MsbHLH115 enhanced the tolerance of Arabidopsis to Cd stress and reduced the production of reactive oxygen species (ROS) in Arabidopsis under Cd stress.
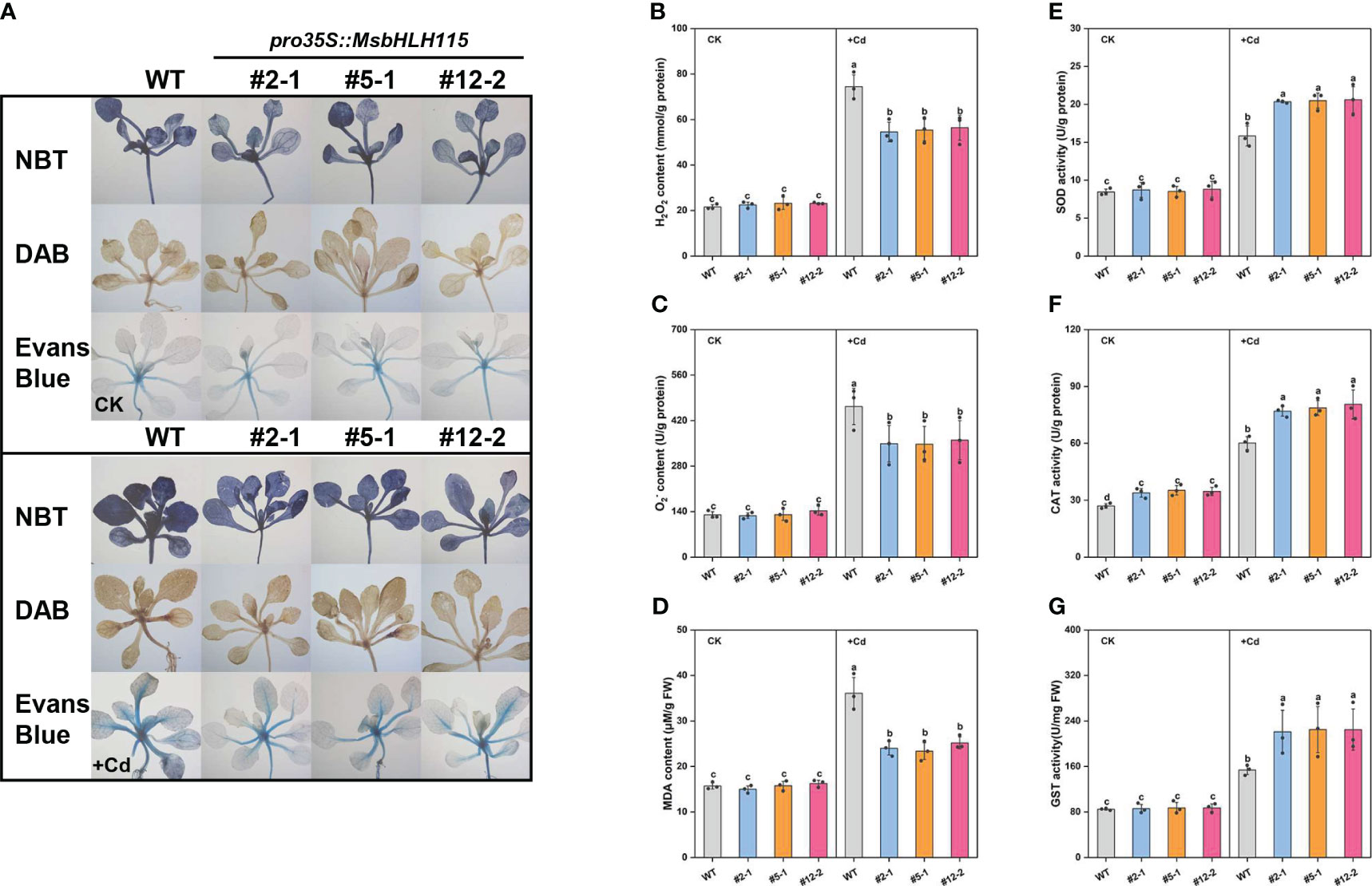
Figure 5 ROS and antioxidant enzyme activity in overexpressed MsbHLH115ox T3 lines (ox#2-1, ox#5-1, and ox#12-2) unde Cd stress. (A) NBT, DAB, and Evans blue staining. (B) H2O2 content. (C) O2- content. (D) MDA content. (E) SOD activity. (F) CAT activity. (G) GST activity. The 5-day-old seedlings were cultured on the 1/2 MS medium containing Cd for 7 days. One-way ANOVA test. Mean ± standard error, n= 3. Lowercase letters indicate significant at p<0.05.
Considering that Fe deprivation visibly elevated the susceptibility of transgenic plants to Cd stress, especially root growth, a split-root experiment was employed to further explore the impact of Fe status on Cd toxicity to plants. We first examined the effects of different Fe and Cd supplies on the growth of MsbHLH115ox line #12-2. In the split medium, the root growth of #12-2 was inhibited by Cd under iron supply conditions (Figure 6A); the Cd inhibition of root growth was more obvious under iron deficiency (Figure 6B); and the Cd inhibition of plant root growth under iron deficiency was more obvious than that under Fe supply (Figure 6C). The result showed that the root elongation of MsbHLH115ox plants was inhibited by Cd, which was exacerbated by iron deficiency (Figures 6A–C, Supplementary Figure S1). We further compared the growth of #12-2 and WT plants when the part near the root tip (RT) and the part distal the root tip (RS) was grown in different conditions. The #12-2 root was longer than WT in the following cases, i. RT or RS was grown in Cd stress and the other part was stressed by iron deficiency (Figures 6D, G); ii. RT was grown in the −Fe+Cd stress and RS was grown in the +Fe−Cd medium (Figures 6E, H); and iii. RT was grown in Cd stress and RS was grown in the +Fe+Cd medium (Figures 6F, I). In other cases, the difference in growth of #12-2 and WT plants was not obvious. The root elongation was affected by the total dose of Cd. In the presence of iron, MsbHLH115 can rescue the root growth inhibition caused by Cd, whether the iron is in RT or RS; but in the absence of iron in the medium, especially in RT, the rescue function of MsbHLH115 was negatively affected. The results showed that the iron in the rhizosphere played a leading role in maintaining root elongation under Cd stress.
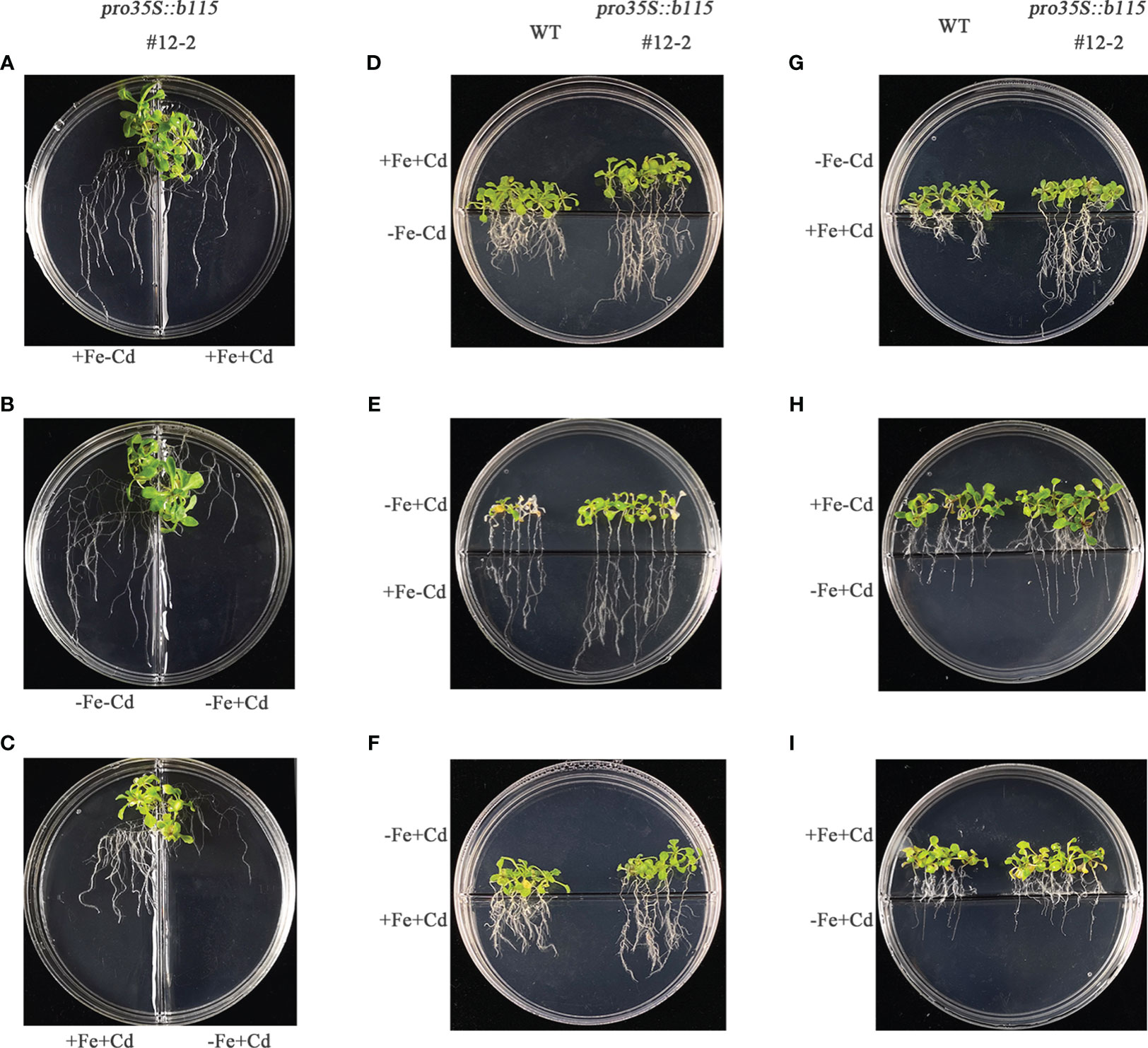
Figure 6 Effect of Fe status on the root elongation in MsbHLH115ox lines unde Cd stress. (A–C) The split-root experiment. The two roots were respectively transferred to different media (e.g., +Fe−Cd and +Fe+Cd, −Fe−Cd and −Fe+Cd or +Fe+Cd and −Fe+Cd) in one petri dish for 7 days when the cut-off-root seedlings were cultured on standard medium for 5 days. (D–I) The root-tip elongation experiment. The root tips and other parts of 5-day-old seedlings were exposed to the different media with/without Fe plus or minus Cd in one petri dish for 7 days.
The Fe concentration in the shoots and roots of three MsbHLH115ox lines was significantly higher than that in WT under both CK and Cd stress. The Fe concentration in shoots and roots of MsbHLH115ox lines and WT significantly decreased by Cd stress (Figures 7A, B). The Cd concentration showed no significant difference in he shoots and roots of MsbHLH115ox lines and WT. The results indicated that overexpression of MsbHLH115 does not affect Cd absorption and translocation (Figure 7C). The Ferric-chelate reductase (FCR) activity in MsbHLH115ox lines was significantly higher than that in WT under CK and Cd stress (Figure 7D). The results indicated that MsbHLH115 positively affects Fe accumulation and translocation in plants. Cd exposure reduced the plant’s ability to absorb Fe and decreased Fe accumulation in plants.
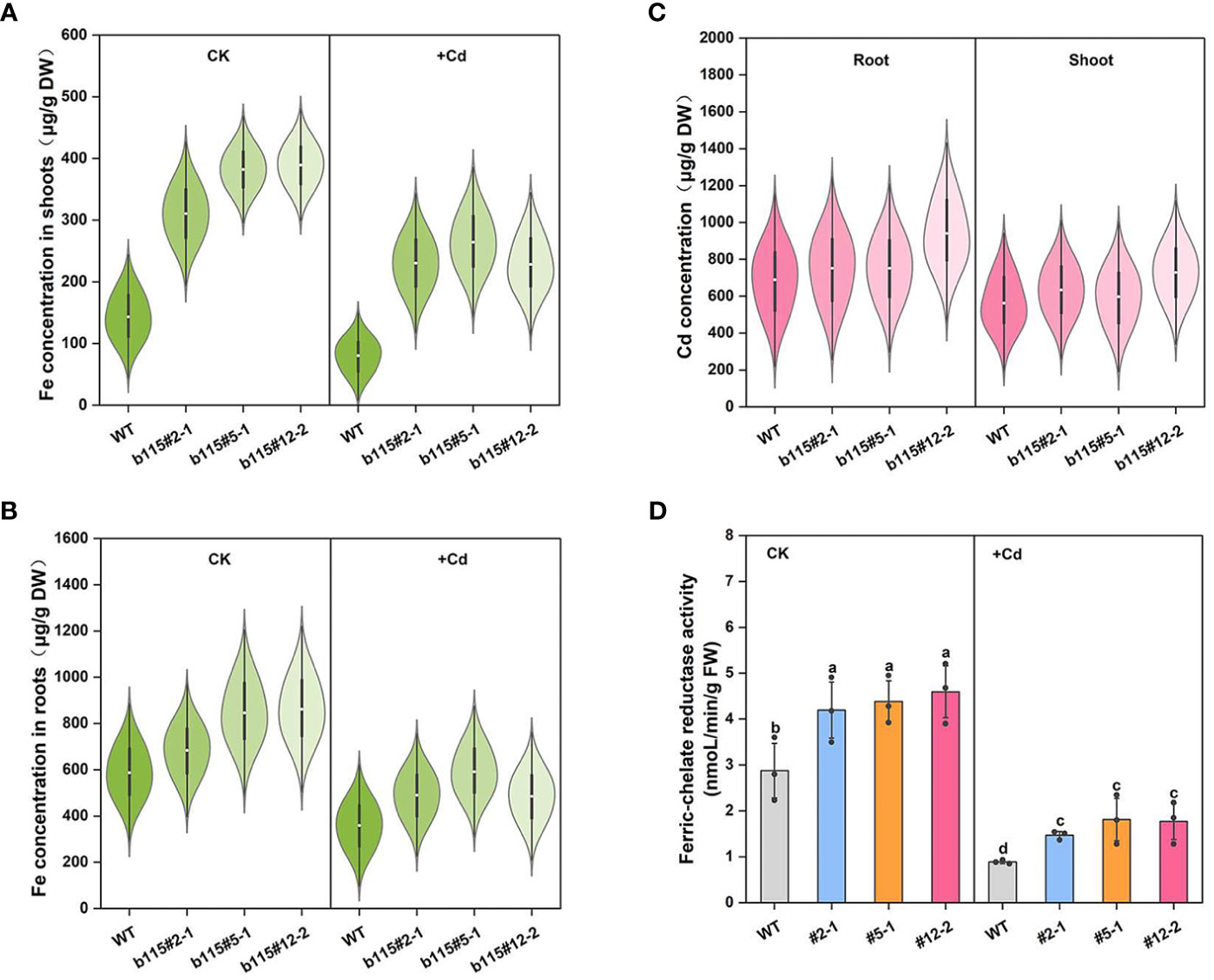
Figure 7 The Cd and Fe concentration and Ferric-chelate reductase activity in MsbHLH115ox lines under Cd stress. (A) Fe concentration in shoots. (B) Fe concentration in roots. (C) Cd concentration in the shoots and roots. (D) Ferric-chelate reductase activity. The 13-day-old seedlings were cultured on the 1/2 MS medium containing 0 or 90 μM CdCl2 for 3 days. Samples were taken for metal concentration determination and Ferric-chelate reductase activity determination. One-way ANOVA test. Mean ± standard error, n=3. Lowercase letters indicate significant at p<0.05.
Zn concentration in MsbHLH115ox was significantly higher than that in WT under Cd treatment or not. Cd stress significantly reduced Zn concentration in MsbHLH115ox lines and WT (Figures 8A, B). The results showed that Cd stress reduced Zn absorption, and led to a decrease in Zn accumulation in plants while MsbHLH115 promotes Zn absorption and transport in plants. Mn and Cu concentration in MsbHLH115ox lines was higher than that in WT under Cd treatment or not, with no significant difference (Figures 8C–F). The results showed that Cd stress reduced Mn and Cu absorption and accumulation and MsbHLH115 had little effect on Mn and Cu absorption and transport.
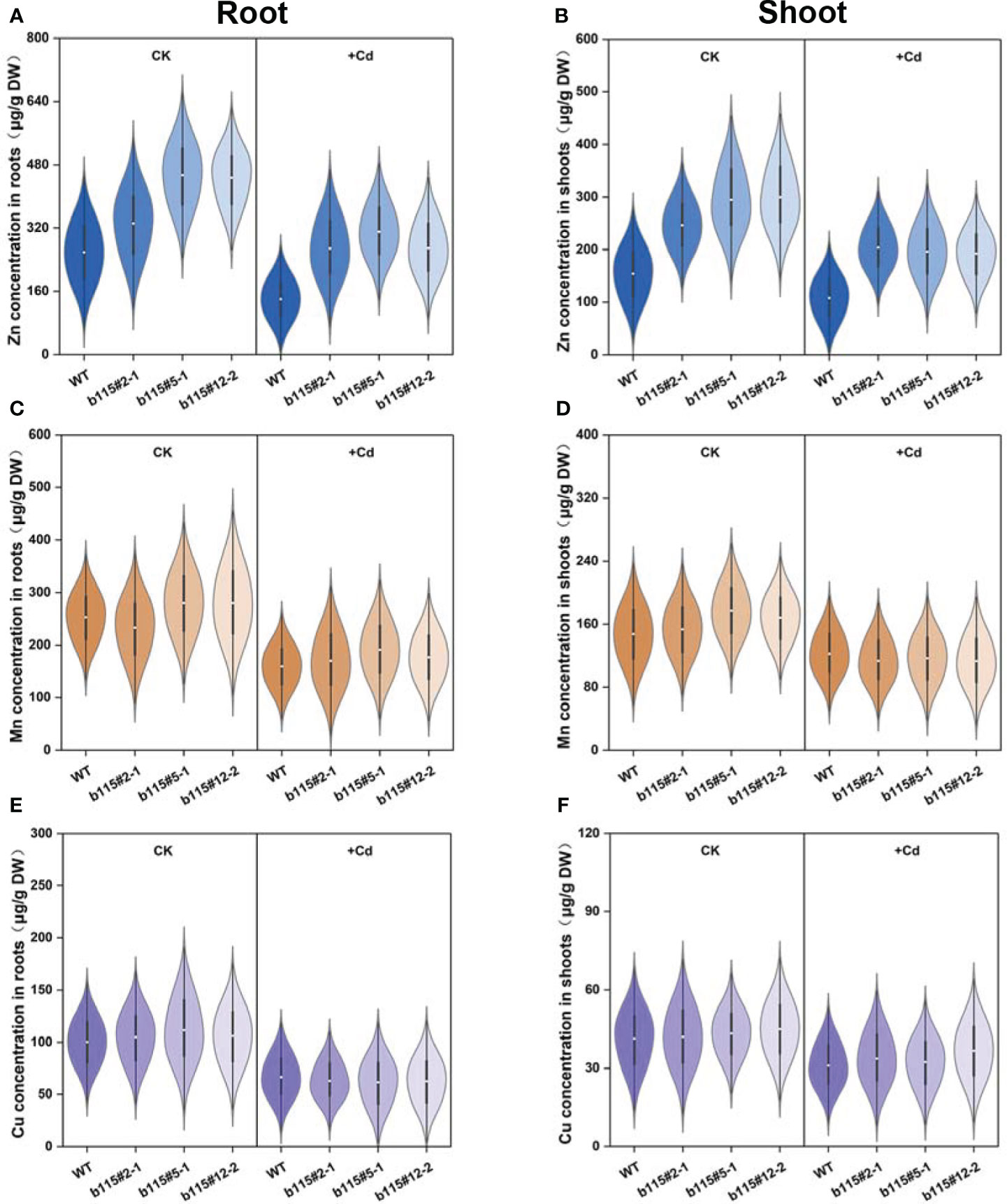
Figure 8 The Zn, Mn and Cu concentration in shoot and root in overexpressed MsbHLH115ox T3 lines (ox#2-1, ox#5-1, and ox#12-2) under Cd stress. (A) Zn concentration in roots. (B) Zn concentration in shoots. (C) Mn concentration in roots. (D) Mn concentration in shoots. (E) Cu concentration in roots. (F) Cu concentration in shoots. The 13-day-old seedlings were cultured on the 1/2 MS medium containing 0 or 90 μM CdCl2 for 3 days. Samples were taken for metal concentration determination. One-way ANOVA test. Mean ± standard error, n= 3.
As shown in Figure 9, the expression level of Fe uptake and translocation genes (IRT1, FRO2, VTL, NAS4, IREG2, HMA3, and MTP3) was higher in the three MsbHLH115ox lines than in the WT, regardless of whether they were under CK or Cd-exposed conditions. Under Cd stress, the expressions of IRT1, FRO2, and VTL expressions decreased, while the expressions of NAS4, HMA3, IREG2, and MTP3 increased in the MsbHLH115ox lines. The terms of these five transcription factors (bHLH115, bHLH39, FIT, bHLH121, and bHLH104) were higher in the MsbHLH115ox lines than in the WT and decreased by Cd stress. The expression of four GSTs was higher in the MsbHLH115ox lines and was enhanced by Cd stress. The expression of RbohB, RbohD, and RbohF was higher in the MsbHLH115ox lines and was enhanced by Cd stress. The expression of SOD1, POD1, and BGLU14, was higher in the MsbHLH115ox lines and was enhanced by Cd stress. The P5CS expression was higher in the MsbHLH115ox lines and was enhanced by Cd stress, while PRODH was higher in the MsbHLH115ox lines but was inhibited by Cd stress (Figure 9A). The promoters of bHLH115, FIT, bHLH121, bHLH104, IRT1, FRO2, VTL, and HMA3 contain E-box elements, while NAS4, IREG2, and MTP3 have no E-box element (Figure 9B).
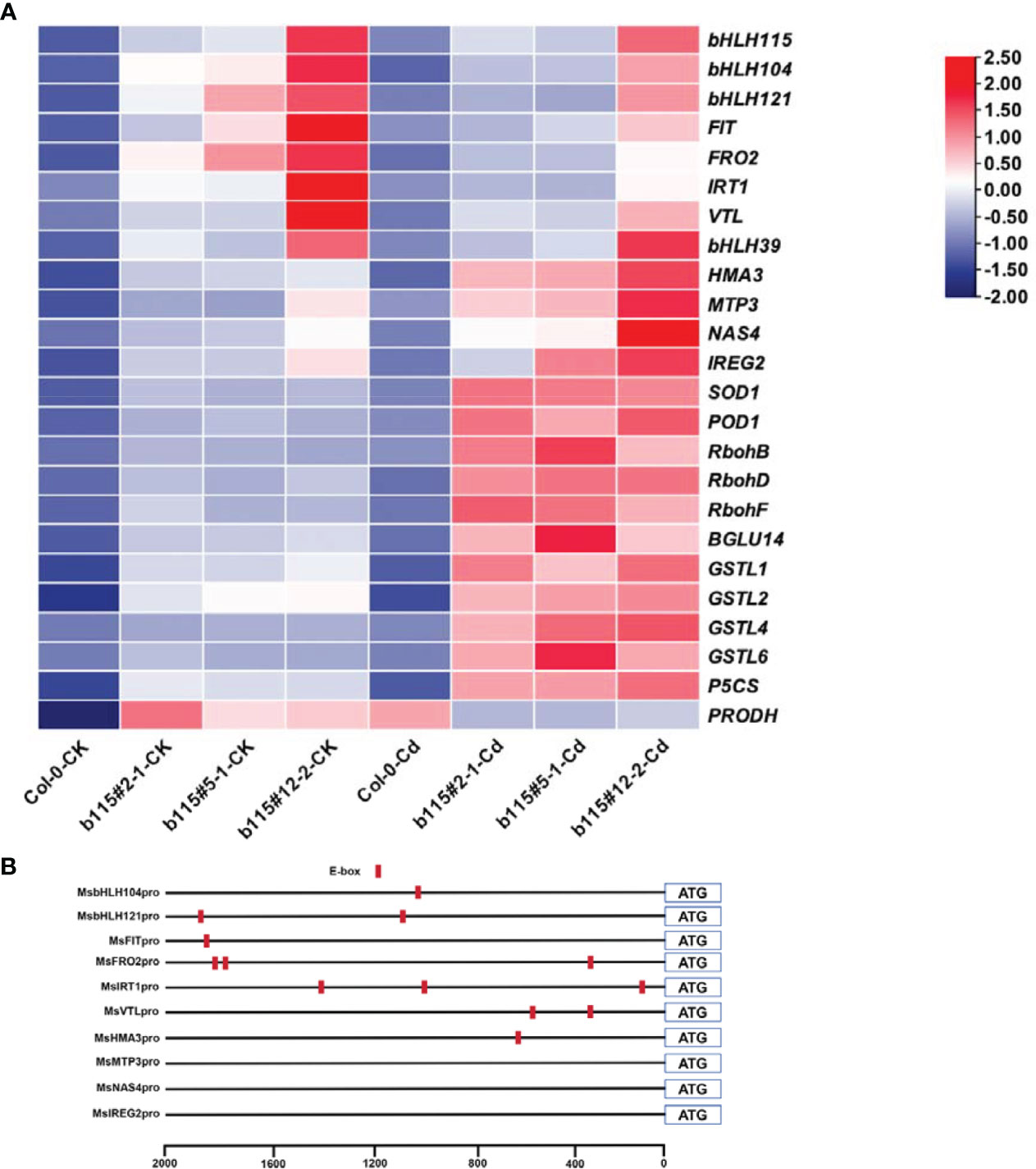
Figure 9 Expression level of various genes regulated by MsbHLH115. (A) Heatmaps showing the fold changes in expression of gene involved in Fe regulate genes (bHLH115, bHLH39, FIT, bHLH121, bHLH104). IRT1, involved in Fe uptake. FRO2 is responsible for ferric reduction. NAS4 is involved in Fe transport in the phloem. HMA3 is involved in Cd transport in vacuoles. IREG2 is involved in iron transport in chloroplasts. MTP3, involved in Fe transport. Glutathione genes (GSTL1, GSTL2, GSTL4, GSTL6). Proline synthesis and decomposition (P5CS, PRODH). Antioxidant genes (SOD1, POD1, and BGLU14). ROS-related genes (RbohB, RbohD, and RbohF). Boxes indicate relative expression level in genes. Log2 (fold changes) are represented by a color scale from white (down-regulated expression) to red (up-regulated expression). The 13-day-old seedlings were cultured on the 1/2 MS medium containing 0 or 90 μM CdCl2 for 24 h. AtACTIN2 was the normalization control. (B) E-box elements of gene promoter. E-box was marked red.
On SD/-Leu medium without AbA, p53-AbAi+pGADT7-MsbHLH115, pE-box-AbAi +pGADT7-MsbHLH115, and pmE-box-AbAi+pGADT7-MsbHLH115 transformed yeast strains can grow at diluted concentrations from 10-1 to 10-3. The background expression of pE-box-AbAi(linearized) in yeast was severely inhibited by 300 ng/mL AbA. On SD/-Leu medium supplemented with 300 ng/mL AbA, p53-AbAi+pGADT7-MsbHLH115 and pmE-box-AbAi +pGADT7-MsbHLH115 did not grow, while pE-box-AbAi+pGADT7-MsbHLH115 could grow. The yeast one-hybrid (Y1H) assay showed that MsbHLH115 can bind to E-box cis-element (Figure 10A). The results of EMSA analysis showed that there was a migration band between the E-box probe and the protein GST- MsbHLH115. The 20× competitive examination can compete with part of the migration band of the protein GST- MsbHLH115, and the 100× competitive examination can compete with most of the migration band of the probe and the protein GST- MsbHLH115. No migration band exists between the mutant probe and GST- MsbHLH115, and the E-box interacts with GST- MsbHLH115, further proving that the MsbHLH115 fusion protein can directly bind to the E-box (Figure 10B). LUC test showed that the enzyme activity of double luciferase was significantly higher than that of the control, indicating that MsbHLH115 could directly bind to MsbHLH121 promoter (Figure 10C). The tobacco leaves that co-transformed pBI121-MsbHLH115 and pBI121-MsbHLH121pro::GUS showed more blue spots than those that co-transformed pBI121-MsbHLH115 and pBI121-3×E-box::GUS. However, no blue dots appeared in tobacco leaves of co-transformed pBI121-MsbHLH115 and pBI121-GUS, indicating that MsbHLH115 protein can recognize E-box element and interact with MsbHLH121 promoter (Figure 10D).
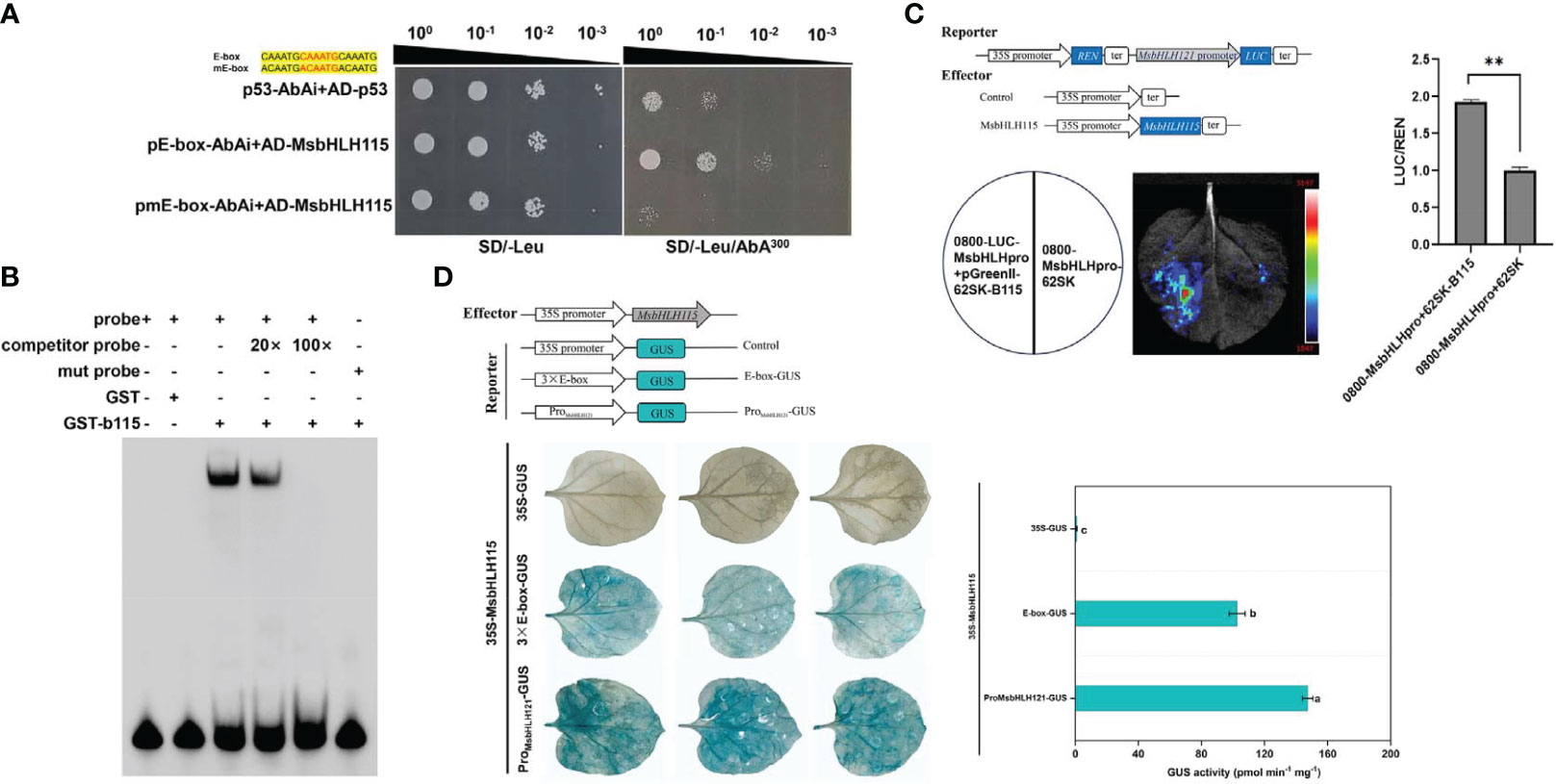
Figure 10 MsbHLH115 interacted with E-box and MsbHLH121pro. (A) Yeast one-hybrid assays of MsbHLH115 binding to the E-box. Constructs pGAD-Rec-p53 and p53-AbAi were used as positive control. Sequences of the MsbHLH115 and corresponding mutant probes used in the electrophoresis mobility shift assay (EMSA). Yellow highlighting denotes the E-box and Mutated nucleotides. (B) EMSA assay of binding between MsbHLH115 and E-box. The competitor probe was added at 20- and 100-fold more than the labeled probes respectively. A negative control was used to validate the EMSA system. (C) LUC assay of binding between MsbHLH115 and MsbHLH121pro.The LUC/REN ratio represents the relative activity of the interaction activation. (D) Transient expression experiment showed that MsbHLH115 interacted with E-box and MsbHLH121pro. GUS staining of representative leaf pieces infiltrated with coinfiltrated with the effector and the reporters. pBI121-MsbHLH115 was used to the effector. pBI121-GUS, pBI121-3×E-box::GUS, pBI121-MsbHLH121pro::GUS was used to the reporters. GUS activity measurements were performed. One-way ANOVA test. Mean ± standard error, n= 3. Lowercase letters indicate significant at p<0.05. ** p<0.01.
The bHLH family members are essential in the regulation of plant iron homeostasis. Alfalfa MsbHLH115 has a typical bHLH structural domain and is a homolog of AtbHLH115, which belongs to the bHLH IVc subfamily. The regulator of iron absorption, bHLH115, is a regulatory gene for FIT and PYE. However, little is known about the role of bHLH115 in plant Cd tolerance. In this study, we carried out a series of experiments on MsbHLH115 to elucidate whether MsbHLH115 is a casual gene at the intersection of Fe and Cd stresses. MsbHLH115 rapidly responded to Fe or Cd deficiency treatment at 6-24 hours, especially in the root of alfalfa (Figure 2). In previous transcriptome studies, 50 days or 42 days Cd treatment induced the expression of FIT, PYE, bHLH38, bHLH39, IRT1, FRO2, NAS, while bHLH115 was not detected (Aprile et al., 2018). We found that MsbHLH115 is an “early” response gene to Cd stress. Interestingly, the expression of MsbHLH115 under −Fe+Cd combined stress was lower than that of −Fe or +Cd alone, indicating that the induced effects of −Fe and +Cd were non-cumulative and mutually inhibitory on MsbHLH115. This non-superposition or even mutual interference between −Fe and +Cd, together with the fact that the two metals Cd and Fe are antagonisms in plants, can be hypothesized that the function of MsbHLH115 may differ in Cd and Fe stress.
Cd is toxic to plants, mainly impairing root growth and decreasing photosynthesis, which can be alleviated by overexpression of Cd resistance genes (Yao et al., 2018). Overexpression of MsbHLH115 in Arabidopsis significantly improved these growth indicators, demonstrating that the MsbHLH115 promoted plant growth and improved the plant tolerance to −Fe and Cd stress. Iron deficiency exacerbated the inhibition of plant growth by Cd, and under the −Fe+Cd complex pressure, although the Cd resistance indicator of the transgenic plants remained higher than WT, the growth advantage of the transgenic plants was significantly deprived (Figure 2). Split-site stress treatments revealed that MsbHLH115 overexpression could reduce the toxic effects of Cd on plants, which is more effective in the presence of iron (Figures 6A–C). Similar to our results, previous studies have identified that the tolerance to Cd toxicity of an iron-regulating factor mutant bts-1 was significantly reduced when iron was removed from the medium (Fan et al., 2020; Zhu et al., 2020). The results imply that MsbHLH115 might enhance plant Cd tolerance by increasing iron uptake capacity. Regardless of Fe supply, Cd had a more significant effect on plant growth when exposure of seedling root tips and Fe supply in the root tips rescued the growth inhibition. The result coincides with the fact that the MsbHLH115 promoter drives the expression of GUS in soybean hairy root tip under Cd stress, suggesting that the root tip is the primary tissue that MsbHLH115 expresses in response to Fe and Cd. Thus, we inferred that MsbHLH115 enhanced Cd tolerance, partly by increasing the iron uptake capacity.
MsbHLH115 increased the Fe concentration, regardless of the presence of Cd, suggesting that MsbHLH115 promotes the uptake and translocation of Fe in plants. Cd treatment induced up-regulation of MxIRT1, MxFRO2-Like, and NtIRT1 (Hodoshima et al., 2007; Gao et al., 2011). These studies concluded that +Cd and −Fe regulate IRT1 expression similarly and suggested that up-regulation of IRT1 could prevent Cd-induced Fe deficiency (Sebastian and Prasad, 2018). Guan et al. concluded that inhibition of IRT1 could reduce Cd uptake by roots (Guan et al., 2019). The up-regulation of FRO2 and IRT1 expression can maintain the high Fe concentration and enhance the tolerance in plants to Cd toxicity(Fan et al., 2020; Zhu et al., 2020). Ferric chelate reductase is required for iron metabolism in plant roots and shoots (Li et al., 2004; Wu et al., 2005). Recently, reduction in Fe reductase activity by Si application to Cd stress in Alfalfa (Kabir et al., 2016). In SlbHLH068 VIGS plants, ferric-chelate reductase reduction in shoots, the expression of LeFRO1, and iron accumulation in leaves and roots were significantly diminished compared with control plants (Du et al., 2015). Two iron uptake genes, IRT1 and FRO2, were significantly up-regulated by the MsbHLH115, which might be the reason for Fe accumulation in transgenic plants. These results suggest that FRO2 and IRT1 might maintain high iron concentrations, thereby reducing Cd toxicity through competition in MsbHLH115 transgenic plants.
Metal translocation in plants is finely regulated by transporter proteins such as VTL, HMA3, MTP3, IREG2, and NAS4. Overexpression of MsbHLH115 induced the expression of these genes. VTL plays an essential role in iron homeostasis in plants, storing excess iron in vesicles for slow release by plants (Ram et al., 2021). In MsbHLH115ox Arabidopsis, Cd stress down-regulates VTL expression might allow iron not to be bound to the vesicle but to be involved in the organism and used in functional organelles. IREG2, NAS4, HMA3, and MTP3 are weakly responsive to Cd, whereas the inducible effects of MsbHLH115 and Cd are cumulative for these genes. NAS4 catalyze the formation of nicotinamide and increases Cd tolerance (Koen et al., 2013). HMA3 and MTP3 are involved in the segregation, retardation, chelation, and detoxification of heavy metals and are commonly used as indicator genes for plant resistance to Cd stress (Morel et al., 2009; Ueno et al., 2010; Qiang et al., 2017; Liu et al., 2018). These genes were positively regulated by Cd tolerant TFs ANAC004, bHLH104, FIT, and AtbHLH38 or AtbHLH39, and ZAT10 TFs (Wu et al., 2012; Dang et al., 2022; Meng Y. et al., 2022), as well as up-regulated by MsbHLH115, suggesting that MsbHLH115, like the above TFs, has a role in enhancing Cd tolerance by regulating the metal transporter genes in plants.
MsbHLH115 up-regulates the expression of TFs, including the endogenous AtbHLH115, bHLH104, and bHLH121 in Arabidopsis, but Cd stress impairs this up-regulatory effect. As a hub gene in the regulatory network, FIT interacts with bHLH38/39 and ZAT10 to up-regulate the expression of IRT1 and FRO2, which helps plants take up iron and also plays a role in Cd tolerance (Wu et al., 2012; Dang et al., 2022). The weakening of the up-regulation of FIT and AtbHLH39 genes in Cd-stressed trans-MsbHLH115 plants may be responsible for the reduced expression of IRT1 and FRO2 and, subsequently, the reduced Fe concentration.
Cd accumulation leads to oxidative damage in plants. This study reduced ROS accumulation in MsbHLH115ox plants under Cd stress, similar to the performance of AtbHLH104 and AtILNF-YC6 overexpression plants. Increasing SOD, POD, and CAT enzyme activity helped to scavenge oxygen radicals in MsbHLH115ox plants. ROS-related genes (RbohB, RbohD, RbohF), SOD1, and BGLU14 were up-regulated at the transcriptional level, suggesting that MsbHLH115 participates in Cd toxicity tolerance. In addition, GSH scavenges H2O2, thereby reducing Cd-induced oxidative stress in cells. Previous studies have found that PyWRKY48 can promote Cd detoxification in plants by regulating GSH synthesis and GST expression (Wu et al., 2023). In this study, GST activity increased, and GSTs up-regulated in MsbHLH115ox plants under Cd stress, suggesting that MsbHLH115 is involved in Cd detoxification by inducing GST expression and up-regulating GSH synthesis (Sebastian and Prasad, 2018). Proline is one of the plants’ most critical abiotic osmolytes (Kim and Nam, 2013). P5CS gene was up-regulated, and PRODH was down-regulated in MsbHLH115ox plants under Cd stress, suggesting that MsbHLH115 promotes proline synthesis and inhibits its catabolism, resulting in the accumulation of proline in the plant (Cecchini et al., 2011; Funck et al., 2012). Under Fe-deficient conditions, Cd-induced oxidative damage in MsbHLH115ox plants was still lower than in WT. Combined with the gene expression results, we hypothesized that MsbHLH115 could also increase Cd tolerance in plants by regulating the expression of oxidase and detoxification genes in a Fe-independent manner. Heavy metals can affect photosynthesis and water metabolism in plants, resulting in wilting of leaves and slow growth (Xuan et al., 2016; Ma et al., 2017; Bashir et al., 2020). Cd may interfere with the plant’s water absorption and transpiration, causing the plant to be unable to absorb and utilize water effectively, thereby triggering drought stress (Xia et al., 2015). Several studies mentioned that plant biomass decreased due to water stress and Cd toxicity (Bashir et al., 2019; Khan et al., 2019). MsbHLH115 might also alleviate drought stress caused by Cd by regulating antioxidant genes, thereby improving plant growth, although not confirmed by experiment in this study.
We found most MsbHLHs promoters contained E-box element, which could be bind to MsbHLH115. The instantaneous expression analysis and EMSA provide strong evidence to support the direct and specific interaction between the MsbHLH115 and MsbHLH121 promoter. These data lead us to speculate that MsbHLH121 is the target gene of MsbHLH115, suggesting that MsbHLH121 may be assigned to bHLH to participate in Cd regulation. bHLH121 controls the expression of many iron-deficiency response genes (Kim et al., 2019; Gao et al., 2020). Recent studies have shown that IMAs increase iron content and Cd tolerance in plants by activating iron absorption regulatory networks (Lei et al., 2020). MsbHLH115 may also regulate other genes such as FIT and other bHLHs which we will examine in future work to construct the molecular regulatory network of cadmium tolerance. We will further elucidate the function of MsbHLH115 by homologous transformation or inhibition of expression in alfalfa. Interestingly, the expression of the antioxidant genes was increased when MsbHLH115 overexpressed in plants under Cd stress, which is also worth studying. There is also a strong possibility that some MsbHLH115 controlled transporter genes, different from IRT1, transport Fe but not Cd. These candidate genes are looking forward to obtaining for the improvement of the Cd tolerance of plants.
In summary, we have identified a novel MsbHLH115 transcription factor, which regulates Cd tolerance in Arabidopsis. Overexpression of MsbHLH115 leads to iron accumulation increasing and downstream gene activation in Arabidopsis. Furthermore, MsbHLH115 interacts with the E-box element. We also demonstrated direct and specific interactions between MsbHLH115 and the promoter of MsbHLH121. Taken together, our findings suggest that MsbHLH115 might regulate Cd tolerance by controlling the expression of the iron-regulate transcript factor bHLHs which increases iron transport in plants.
The original contributions presented in the study are included in the article/Supplementary Material. further inquiries can be directed to the corresponding authors.
MZ: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. J-YG: Conceptualization, Supervision, Writing – review & editing. S-CD: Conceptualization, Supervision, Writing – review & editing. M-HC: Conceptualization, Supervision, Writing – review & editing. J-XZ: Conceptualization, Supervision, Writing – review & editing. D-LG: Conceptualization, Resources, Writing – review & editing, Funding acquisition. C-HG: Funding acquisition, Resources, Writing – review & editing. Y-DB: Funding acquisition, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Key Research and Development Project of Heilongjiang Province (No.2022ZX02B05), the National Natural Science Foundation of China (No. U21a20182, 31972507), the National Key R&D Program of China (2022YFE0203300) the National Natural Science Foundation of China general project (No. 31771823).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1358673/full#supplementary-material
Ai, H., Wu, D., Li, C., Hou, M. (2022). Advances in molecular mechanisms underlying cadmium uptake and translocation in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1003953
Aparicio, F., Pallás, V. (2016). The coat protein of the Alfalfa mosaic virus interacts and interferes with the transcriptional activity of the bHLH transcription factor ILR3, promoting a salicylic acid-dependent defense signaling response. Mol. Plant Pathol. 18 (2), 173–186. doi: 10.1111/mpp.12388
Aprile, A., Sabella, E., Vergine, M., Genga, A., Siciliano, M., Nutricati, E., et al. (2018). Activation of a gene network in durum wheat roots exposed to cadmium. BMC Plant Biol. 18 (1), 238. doi: 10.1186/s12870-018-1473-4
Baker, C. J., Mock, N. M. (1994). An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 39, 7–12. doi: 10.1007/BF00037585
Bao, T., Sun, L., Sun, T., Zhang, P., Niu, Z. (2009). Iron deficiency induces cadmium uptake and accumulation in Solanum nigrum L. Bull. Environ. contamination Toxicol. 82 (3), 338–342. doi: 10.1007/s00128-008-9573-8
Bashir, W., Anwar, S., Zhao, Q., Hussain, I., Xie, F. (2019). Interactive effect of drought and cadmium stress on soybean root morphology and gene expression. Ecotoxicology Environ. Saf. 175, 90–101. doi: 10.1016/j.ecoenv.2019.03.042
Bashir, A., Rizwan, M., Ali, S., Adrees, M., Rehman, M. Z. U., Qayyum, M. F. (2020). Effect of composted organic amendments and zinc oxide nanoparticles on growth and cadmium accumulation by wheat; a life cycle study. Environ. Sci. pollut. Res. Int. 27 (19), 23926–23936. doi: 10.1007/s11356-020-08739-8
Cai, Y., Cao, F., Wei, K., Zhang, G., Wu, F. (2011). Genotypic dependent effect of exogenous glutathione on Cd-induced changes in proteins, ultrastructure and antioxidant defense enzymes in rice seedlings. J. Hazardous Materials 192 (3), 1056–1066. doi: 10.1016/j.jhazmat.2011.06.011
Cecchini, N. M., Monteoliva, M. I., Alvarez, M. E. (2011). Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol. 155 (4), 1947–1959. doi: 10.1104/pp.110.167163
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colangelo, E. P., Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 (12), 3400–3412. doi: 10.1105/tpc.104.024315
Cui, W., Gao, C., Fang, P., Lin, G., Shen, W. (2013). Alleviation of cadmium toxicity in Medicago sativa by hydrogen-rich water. J. Hazard Mater 260, 715–724. doi: 10.1016/j.jhazmat.2013.06.032
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 (1), 5–17. doi: 10.1104/pp.105.063743
Dai, C., Cui, W., Pan, J., Xie, Y., Wang, J., Shen, W. (2017). Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J. Proteomics 152, 109–120. doi: 10.1016/j.jprot.2016.10.013
Dang, F., Li, Y., Wang, Y., Lin, J., Du, S., Liao, X. (2022). ZAT10 plays dual roles in cadmium uptake and detoxification in Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994100
Dias, M. C., Monteiro, C., Moutinho-Pereira, J., Correia, C., Gonçalves, B., Santos, C. (2012). Cadmium toxicity affects photosynthesis and plant growth at different levels. Acta Physiologiae Plantarum 35 (4), 1281–1289. doi: 10.1007/s11738-012-1167-8
Du, J., Huang, Z., Wang, B., Sun, H., Chen, C., Ling, H. Q., et al. (2015). SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato. Ann. Bot. 116 (1), 23–34. doi: 10.1093/aob/mcv058
Du, X., Zhou, L., Zhu, B., Gu, L., Yin, H., Wang, H. (2023). The TabHLH35-TaWAK20-TaSPL5 pathway positively regulates Cd stress in wheat. Theor. Appl. Genet. 136 (7), 153. doi: 10.1007/s00122-023-04400-3
Eide, D., Broderius, M., Fett, J., Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. United States America 93 (11), 5624–5628. doi: 10.1073/pnas.93.11.5624
El Rasafi, T., Oukarroum, A., Haddioui, A., Song, H., Kwon, E. E., Bolan, N., et al. (2020). Cadmium stress in plants: A critical review of the effects, mechanisms, and tolerance strategies. Crit. Rev. Environ. Sci. Technol. 52, 675–726. doi: 10.1080/10643389.2020.1835435
Elstner, A. (1976). Formation of hydrogen peroxide by isolated cell walls from horseradish (Armoracia lapathifolia Gilib). Planta 130 (2), 175–180. doi: 10.1007/BF00384416
Fan, S. K., Ye, J. Y., Zhang, L. L., Chen, H. S., Zhang, H. H., Zhu, Y. X., et al. (2020). Inhibition of DNA demethylation enhances plant tolerance to cadmium toxicity by improving iron nutrition. Plant Cell Environ. 43 (1), 275–291. doi: 10.1111/pce.13670
Funck, D., Winter, G., Baumgarten, L., Forlani, G. (2012). Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 12, 191. doi: 10.1186/1471-2229-12-191
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., et al. (2020). The Transcription Factor bHLH121 Interacts with bHLH105 (ILR3) and Its Closest Homologs to Regulate Iron Homeostasis in Arabidopsis. Plant Cell 32 (2), 508–524. doi: 10.1105/tpc.19.00541
Gao, C., Wang, Y., Xiao, D. S., Qiu, C. P., Han, D. G., Zhang, X. Z., et al. (2011). Comparison of cadmium-induced iron-deficiency responses and genuine iron-deficiency responses in Malus xiaojinensis. Plant Sci. 181 (3), 269–274. doi: 10.1016/j.plantsci.2011.05.014
Giannopolitis, C. N., Ries, S. K. (1977). Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 59 (2), 309–314. doi: 10.1104/pp.59.2.309
Grillet, L., Lan, P., Li, W., Mokkapati, G., Schmidt, W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 4 (11), 953–963. doi: 10.1038/s41477-018-0266-y
Gu, Q., Chen, Z., Cui, W., Zhang, Y., Hu, H., Yu, X., et al. (2018). Methane alleviates alfalfa cadmium toxicity via decreasing cadmium accumulation and reestablishing glutathione homeostasis. Ecotoxicology Environ. Saf. 147, 861–871. doi: 10.1016/j.ecoenv.2017.09.054
Guan, M. Y., Zhu, Y. X., Liu, X. X., Jin, C. W. (2019). Induction of S-nitrosoglutathione reductase reduces root cadmium uptake by inhibiting Iron-regulated transporter 1. Plant Soil 438 (1-2), 251–262. doi: 10.1007/s11104-019-04014-z
Han, G. H., Huang, R. N., Hong, L. H., Xu, J. X., Hong, Y. G., Wu, Y. H., et al. (2023). The transcription factor NAC102 confers cadmium tolerance by regulating WAKL11 expression and cell wall pectin metabolism in Arabidopsis. J. Integr. Plant Biol. 65 (10), 2262–2278. doi: 10.1111/jipb.13557
Han, X., Zhang, Y., Yu, M., Zhang, J., Xu, D., Lu, Z., et al. (2020). Transporters and ascorbate-glutathione metabolism for differential cadmium accumulation and tolerance in two contrasting willow genotypes. Tree Physiol. 40 (8), 1126–1142. doi: 10.1093/treephys/tpaa029
He, X. L., Fan, S. K., Zhu, J., Guan, M. Y., Liu, X. X., Zhang, Y. S., et al. (2017). Iron supply prevents Cd uptake in Arabidopsis by inhibiting IRT1 expression and favoring competition between Fe and Cd uptake. Plant Soil 416 (1-2), 453–462. doi: 10.1007/s11104-017-3232-y
Hodoshima, H., Enomoto, Y., Shoji, K., Shimada, H., Goto, F., Yoshihara, T. (2007). Differential regulation of cadmium-inducible expression of iron-deficiency-responsive genes in tobacco and barley. Physiologia Plantarum 129 (3), 622–634. doi: 10.1111/j.1399-3054.2006.00825.x
Jian, L., Jingchun, L., Chongling, Y., Daolin, D., Haoliang, L. (2019). The alleviation effect of iron on cadmium phytotoxicity in mangrove A. marina. Alleviation effect of iron on cadmium phytotoxicity in mangrove Avicennia marina (Forsk.) Vierh. Chemosphere 226, 413–420. doi: 10.1016/j.chemosphere.2019.03.172
Kabir, A. H., Hossain, M. M., Khatun, M. A., Mandal, A., Haider, S. A. (2016). Role of silicon counteracting cadmium toxicity in alfalfa (Medicago sativa L.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01117
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2 (4), 948–952. doi: 10.1038/nprot.2007.141
Khan, M. R., Adam, V., Rizvi, T. F., Zhang, B., Ahamad, F., Jośko, I., et al. (2019). Nanoparticle-plant interactions: two-way traffic. Small (Weinheim an der Bergstrasse Germany) 15 (37), e1901794. doi: 10.1002/smll.201901794
Kim, S. A., LaCroix, I. S., Gerber, S. A., Guerinot, M. L. (2019). The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. U.S.A. 116 (50), 24933–24942. doi: 10.1073/pnas.1916892116
Kim, G. B., Nam, Y. W. (2013). A novel Delta(1)-pyrroline-5-carboxylate synthetase gene of Medicago truncatula plays a predominant role in stress-induced proline accumulation during symbiotic nitrogen fixation. J. Plant Physiol. 170 (3), 291–302. doi: 10.1016/j.jplph.2012.10.004
Koen, E., Besson-Bard, A., Duc, C., Astier, J., Gravot, A., Richaud, P., et al. (2013). Arabidopsis thaliana nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant Sci. 209, 1–11. doi: 10.1016/j.plantsci.2013.04.006
Korshunova, Y. O., Eide, D., Clark, W. G., Guerinot, M. L., Pakrasi, H. B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40 (1), 37–44. doi: 10.1023/a:1026438615520
Kurt, F., Filiz, E., Kurt, B. (2019). A key gene bHLH115 in iron homeostasis: comprehensive bioinformatics analyses in Arabidopsis, tomato, rice, and maize. Biometals 32 (4), 641–656. doi: 10.1007/s10534-019-00199-z
Lei, R., Li, Y., Cai, Y., Li, C., Pu, M., Lu, C., et al. (2020). bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining fe homeostasis in arabidopsis. Mol. Plant 13 (4), 634–649. doi: 10.1016/j.molp.2020.01.006
Li, L., Cheng, X., Ling, H. Q. (2004). Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol. Biol. 54 (1), 125–136. doi: 10.1023/B:PLAN.0000028774.82782.16
Li, Y., Lei, R., Pu, M., Cai, Y., Lu, C., Li, Z., et al. (2022). bHLH11 inhibits bHLH IVc proteins by recruiting the TOPLESS/TOPLESS-RELATED corepressors. Plant Physiol. 188 (2), 1335–1349. doi: 10.1101/2020.04.09.035097
Li, X., Zhang, H., Ai, Q., Liang, G., Yu, D. (2016). Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 170 (4), 2478–2493. doi: 10.1104/pp.15.01827
Li, T., Zhang, X. M., Gao, J. L., Wang, L., Si, L., Shu, Y. J., et al. (2023). Soybean gmVIT1 gene confers plant tolerance to excess Fe/Mn stress. Agronomy 13 (2), 384. doi: 10.3390/agronomy13020384
Liang, G. (2022). Iron uptake, signaling, and sensing in plants. Plant Commun. 3 (5), 1–13. doi: 10.1016/j.xplc.2022.100349
Liang, G., Zhang, H., Li, X., Ai, Q., Yu, D. (2017). bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68 (7), 1743–1755. doi: 10.1093/jxb/erx043
Liu, Z.-Q., Li, H.-L., Zeng, X.-J., Lu, C., Fu, J.-Y., Guo, L.-J., et al. (2020). Coupling phytoremediation of cadmium-contaminated soil with safe crop production based on a sorghum farming system. J. Cleaner Production 275. doi: 10.1016/j.jclepro.2020.123002
Liu, S. J., Liu, Y. G., Tan, X. F., Zeng, G. M., Zhou, Y. H., Liu, S. B., et al. (2018). The effect of several activated biochars on Cd immobilization and microbial community composition during in-situ remediation of heavy metal contaminated sediment. Chemosphere 208, 655–664. doi: 10.1016/j.chemosphere.2018.06.023
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lockhart, J. (2020). Personal trainer: bHLH121 functions upstream of a transcriptional network of heavy lifters involved in balancing iron levels. Plant Cell 32 (2), 293–294. doi: 10.1105/tpc.19.00918
Ma, D., Sun, D., Wang, C., Ding, H., Qin, H., Hou, J., et al. (2017). Physiological responses and yield of wheat plants in zinc-mediated alleviation of drought stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00860
Meda, A. R., Scheuermann, E. B., Prechsl, U. E., Erenoglu, B., Schaaf, G., Hayen, H., et al. (2007). Iron acquisition by phytosiderophores contributes to cadmium tolerance. Plant Physiol. 143 (4), 1761–1773. doi: 10.1104/pp.106.094474
Meng, X., Li, W., Shen, R., Lan, P. (2022). Ectopic expression of IMA small peptide genes confers tolerance to cadmium stress in Arabidopsis through activating the iron deficiency response. J. Hazard Mater 422, 126913. doi: 10.1016/j.jhazmat.2021.126913
Meng, Y. T., Zhang, X. L., Wu, Q., Shen, R. F., Zhu, X. F. (2022). Transcription factor ANAC004 enhances Cd tolerance in Arabidopsis thaliana by regulating cell wall fixation, translocation and vacuolar detoxification of Cd, ABA accumulation and antioxidant capacity. J. Hazard Mater 436, 129121. doi: 10.1016/j.jhazmat.2022.129121
Morel, M., Crouzet, J., Gravot, A., Auroy, P., Leonhardt, N., Vavasseur, A., et al. (2009). AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149 (2), 894–904. doi: 10.1104/pp.108.130294
Muneer, S., Jeong, B. R., Kim, T. H., Lee, J. H., Soundararajan, P. (2014). Transcriptional and physiological changes in relation to Fe uptake under conditions of Fe-deficiency and Cd-toxicity in roots of Vigna radiata L. J. Plant Res. 127 (6), 731–742. doi: 10.1007/s10265-014-0660-0
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15 (3), 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nagalakshmi, N., Prasad, M. N. (2001). Responses of glutathione cycle enzymes and glutathione metabolism to copper stress in Scenedesmus bijugatus. Plant Sci. an Int. J. Exp. Plant Biol. 160 (2), 291–299. doi: 10.1016/s0168-9452(00)00392-7
Ndayambaza, B., Jin, X., Min, X., Lin, X., Yin, X., Liu, W. (2020). Genome-Wide Identification and Expression Analysis of the Barrel Medic (Medicago truncatula) and Alfalfa (Medicago sativa L.) Basic Helix-Loop-Helix Transcription Factor Family Under Salt and Drought Stresses. J. Plant Growth Regul. 40 (5), 2058–2078. doi: 10.1007/s00344-020-10252-8
Nightingale, E. R. (1959). Phenomenological theory of ion solvation-effective radii of hydrated ions. J. Phys. Chem. 63, 1381–1387. doi: 10.1021/j150579a011
Palansooriya, K. N., Shaheen, S. M., Chen, S. S., Tsang, D. C. W., Hashimoto, Y., Hou, D., et al. (2020). Soil amendments for immobilization of potentially toxic elements in contaminated soils: A critical review. Environ. Int. 134, 105046. doi: 10.1016/j.envint.2019.105046
Puckette, M. C., Weng, H., Mahalingam, R. (2007). Physiological and biochemical responses to acute ozone-induced oxidative stress in Medicago truncatula. Plant Physiol. Biochem. 45 (1), 70–79. doi: 10.1016/j.plaphy.2006.12.004
Qiang, J., Tao, Y. F., He, J., Xu, P., Bao, J. W., Sun, Y. L. (2017). miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat Toxicol. 182, 39–48. doi: 10.1016/j.aquatox.2016.11.009
Ram, H., Sardar, S., Gandass, N. (2021). Vacuolar Iron Transporter (Like) proteins: Regulators of cellular iron accumulation in plants. Physiol. Plant 171 (4), 823–832. doi: 10.1111/ppl.13363
Riaz, N., Guerinot, M. L. (2021). All together now: regulation of the iron deficiency response. J. Exp. Bot. 72 (6), 2045–2055. doi: 10.1093/jxb/erab003
Sandalio, L. M., Dalurzo, H. C., Gómez, M., Romero-Puertas, M. C., del Río, L. A. (2001). Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J. Exp. Bot. 52 (364), 2115–2126. doi: 10.1093/jexbot/52.364.2115
Schwarz, B., Bauer, P. (2020). FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 71 (5), 1694–1705. doi: 10.1093/jxb/eraa012
Sebastian, A., Prasad, M. N. V. (2018). Exogenous citrate and malate alleviate cadmium stress in Oryza sativa L.: Probing role of cadmium localization and iron nutrition. Ecotoxicol Environ. Saf. 166, 215–222. doi: 10.1016/j.ecoenv.2018.09.084
Sheng, Y., Yan, X., Huang, Y., Han, Y., Zhang, C., Ren, Y., et al. (2019). The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 42 (3), 891–903. doi: 10.1111/pce.13457
Shi, Y. J., Niu, M.-X., Feng, C.-H., Li, J.-L., Lin, T.-T., Wang, T., et al. (2023). Overexpression of PtrAREB3 improved cadmium enrichment and tolerance in poplar. Environ. Exp. Bot. 210, 43–56. doi: 10.1016/j.envexpbot.2023.105343
Soydam Aydin, S., Büyük, I., Aras, S. (2013). Relationships among lipid peroxidation, SOD enzyme activity, and SOD gene expression profile in Lycopersicum esculentum L. exposed to cold stress. Genet. Mol. research: GMR 12 (3), 3220–3229. doi: 10.4238/2013.August.29.6
Su, Y., Liu, J., Lu, Z., Wang, X., Zhang, Z., Shi, G. (2014). Effects of iron deficiency on subcellular distribution and chemical forms of cadmium in peanut roots in relation to its translocation. Environ. Exp. Bot. 97, 40–48. doi: 10.1016/j.envexpbot.2013.10.001
Takahashi, F., Hanada, K., Kondo, T., Shinozaki, K. (2019). Hormone-like peptides and small coding genes in plant stress signaling and development. Curr. Opin. Plant Biol. 51, 88–95. doi: 10.1016/j.pbi.2019.05.011
Tissot, N., Robe, K., Gao, F., Grant-Grant, S., Boucherez, J., Bellegarde, F., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 223 (3), 1433–1446. doi: 10.1111/nph.15753
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. U.S.A. 107 (38), 16500–16505. doi: 10.1073/pnas.1005396107
Wang, X., Fu, Y., Ban, L., Wang, Z., Feng, G., Li, J., et al. (2015). Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet. Syst. 90 (3), 175–180. doi: 10.1266/ggs.90.175
Wang, J., Liu, B., Jin, Z., Li, L., Shen, W. (2023). Argon-stimulated nitric oxide production and its function in alfalfa cadmium tolerance. Environ. pollut. (Barking Essex 1987) 333, 122009. doi: 10.1016/j.envpol.2023.122009
Wang, B., Wang, Y., Yuan, X., Jiang, Y., Zhu, Y., Kang, X., et al. (2023). Comparative transcriptomic analysis provides key genetic resources in clove basil (Ocimum gratissimum) under cadmium stress. Front. Genet. 14. doi: 10.3389/fgene.2023.1224140
Willekens, H., Chamnongpol, S., Davey, M., Schraudner, M., Langebartels, C., Van Montagu, M., et al. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16 (16), 4806–4816. doi: 10.1093/emboj/16.16.4806
Wu, H., Chen, C., Du, J., Liu, H., Cui, Y., Zhang, Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158 (2), 790–800. doi: 10.1104/pp.111.190983
Wu, X., Chen, L., Lin, X., Chen, X., Han, C., Tian, F., et al. (2023). Integrating physiological and transcriptome analyses clarified the molecular regulation mechanism of PyWRKY48 in poplar under cadmium stress. Int. J. Biol. Macromol 238, 124072. doi: 10.1016/j.ijbiomac.2023.124072
Wu, H., Li, L., Du, J., Yuan, Y., Cheng, X., Ling, H. Q. (2005). Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 46 (9), 1505–1514. doi: 10.1093/pcp/pci163
Xia, S., Wang, X., Su, G., Shi, G. (2015). Effects of drought on cadmium accumulation in peanuts grown in a contaminated calcareous soil. Environ. Sci. pollut. Res. Int. 22 (23), 18707–18717. doi: 10.1007/s11356-015-5063-9
Xiang, C., Werner, B. L., Christensen, E. M., Oliver, D. J. (2001). The biological functions of glutathione revisited in arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 126 (2), 564–574. doi: 10.1104/pp.126.2.564
Xu, L., Wang, Y., Liu, W., Wang, J., Zhu, X., Zhang, K., et al. (2015). De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant science: an Int. J. Exp. Plant Biol. 236, 313–323. doi: 10.1016/j.plantsci.2015.04.015
Xuan, Y., Zhou, Z. S., Li, H. B., Yang, Z. M. (2016). Identification of a group of XTHs genes responding to heavy metal mercury, salinity and drought stresses in Medicago truncatula. Ecotoxicology Environ. Saf. 132, 153–163. doi: 10.1016/j.ecoenv.2016.06.007
Yadav, B. S., Mani, A. (2019). Analysis of bHLH coding genes of Cicer arietinum during heavy metal stress using biological network. Physiol. Mol. Biol. Plants 25 (1), 113–121. doi: 10.1007/s12298-018-0625-1
Yang, X., Kong, L., Wang, Y., Su, J., Shen, W. (2021). Methane control of cadmium tolerance in alfalfa roots requires hydrogen sulfide. Environ. pollut. 284, 117123. doi: 10.1016/j.envpol.2021.117123
Yao, X., Cai, Y., Yu, D., Liang, G. (2018). bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 60 (8), 691–702. doi: 10.1111/jipb.12658
Yavari, N., Tripathi, R., Wu, B. S., MacPherson, S., Singh, J., Lefsrud, M. (2021). The effect of light quality on plant physiology, photosynthetic, and stress response in Arabidopsis thaliana leaves. PloS One 16 (3), e0247380. doi: 10.1371/journal.pone.0247380
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 (7), 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, X., Li, X., Tang, L., Peng, Y., Qian, M., Guo, Y., et al. (2020). The root iron transporter 1 governs cadmium uptake in Vicia sativa roots. J. Hazard Mater 398, 122873. doi: 10.1016/j.jhazmat.2020.122873
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27 (3), 787–805. doi: 10.1105/tpc.114.132704
Zhang, P., Wang, R., Ju, Q., Li, W., Tran, L. P., Xu, J. (2019). The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 180 (1), 529–542. doi: 10.1104/pp.18.01380
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75 (4-5), 365–378. doi: 10.1007/s11103-011-9732-x
Zhou, L. J., Zhang, C. L., Zhang, R. F., Wang, G. L., Li, Y. Y., Hao, Y. J. (2019). The SUMO E3 ligase mdSIZ1 targets mdbHLH104 to regulate plasma membrane H(+)-ATPase activity and iron homeostasis. Plant Physiol. 179 (1), 88–106. doi: 10.1104/pp.18.00289
Keywords: cadmium, iron nutrient, MsbHLH115, transcription factors, alfalfa
Citation: Zhang M, Gao J-Y, Dong S-C, Chang M-H, Zhu J-X, Guo D-L, Guo C-H and Bi Y-D (2024) Alfalfa MsbHLH115 confers tolerance to cadmium stress through activating the iron deficiency response in Arabidopsis thaliana. Front. Plant Sci. 15:1358673. doi: 10.3389/fpls.2024.1358673
Received: 20 December 2023; Accepted: 29 January 2024;
Published: 12 February 2024.
Edited by:
Paul Larsen, University of California, Riverside, United StatesReviewed by:
Sheliang Wang, Huazhong Agricultural University, ChinaCopyright © 2024 Zhang, Gao, Dong, Chang, Zhu, Guo, Guo and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Lin Guo, Z3VvZG9uZ2xpbkBocmJudS5lZHUuY24=; Chang-Hong Guo, a2FrdTMwMDhAaHJibnUuZWR1LmNu; Ying-Dong Bi, eWluZ2RvbmdiaUBoYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.