- 1Laboratory of Pollen Biology, Institute for Experimental Botany of the Czech Academy of Sciences, Prague, Czechia
- 2Department of Experimental Plant Biology, Faculty of Science, Charles University, Prague, Czechia
As Arabidopsis flowers mature, specialized cells within the anthers undergo meiosis, leading to the production of haploid microspores that differentiate into mature pollen grains, each containing two sperm cells for double fertilization. During pollination, the pollen grains are dispersed from the anthers to the stigma for subsequent fertilization. Transcriptomic studies have identified a large number of genes expressed over the course of male reproductive development and subsequent functional characterization of some have revealed their involvement in floral meristem establishment, floral organ growth, sporogenesis, meiosis, microsporogenesis, and pollen maturation. These genes encode a plethora of proteins, ranging from transcriptional regulators to enzymes. This review will focus on the regulatory networks that control male reproductive development, starting from flower development and ending with anther dehiscence, with a focus on transcription factors and some of their notable target genes.
1 Introduction
Reproductive development in Arabidopsis occurs within a compact flower, where the developmental stage is correlated with floral bud size (Koltunow et al., 1990), making it convenient to study the different stages of this process. Male reproductive development, specifically, occurs within the anthers of stamen, which hold the pollen sacs where pollen grains develop. Male reproductive development is a very complex process involving a multitude of genetic and molecular cues. These developmental events are precisely timed and choreographed. Transcriptional regulation plays a significant role in influencing various aspects of this process. Our understanding of the role that transcription factors (TFs) play in male reproductive development has been greatly assisted by community resources (e.g., TAIR, ABRC, AGI, etc) and male sterile mutants available for Arabidopsis. Unraveling the identity and functions of these TFs has been a central focus of plant reproductive biology research, as it holds the key to understanding the fundamental mechanisms that govern male fertility and the production of seeds. This review will follow along chronologically from flower development through pollen release, discussing the major transcriptional pathways and players involved. Moreover, to keep the review succinct, details about the TFs, their deduced functions, and mutant phenotypes are provided in Table 1.
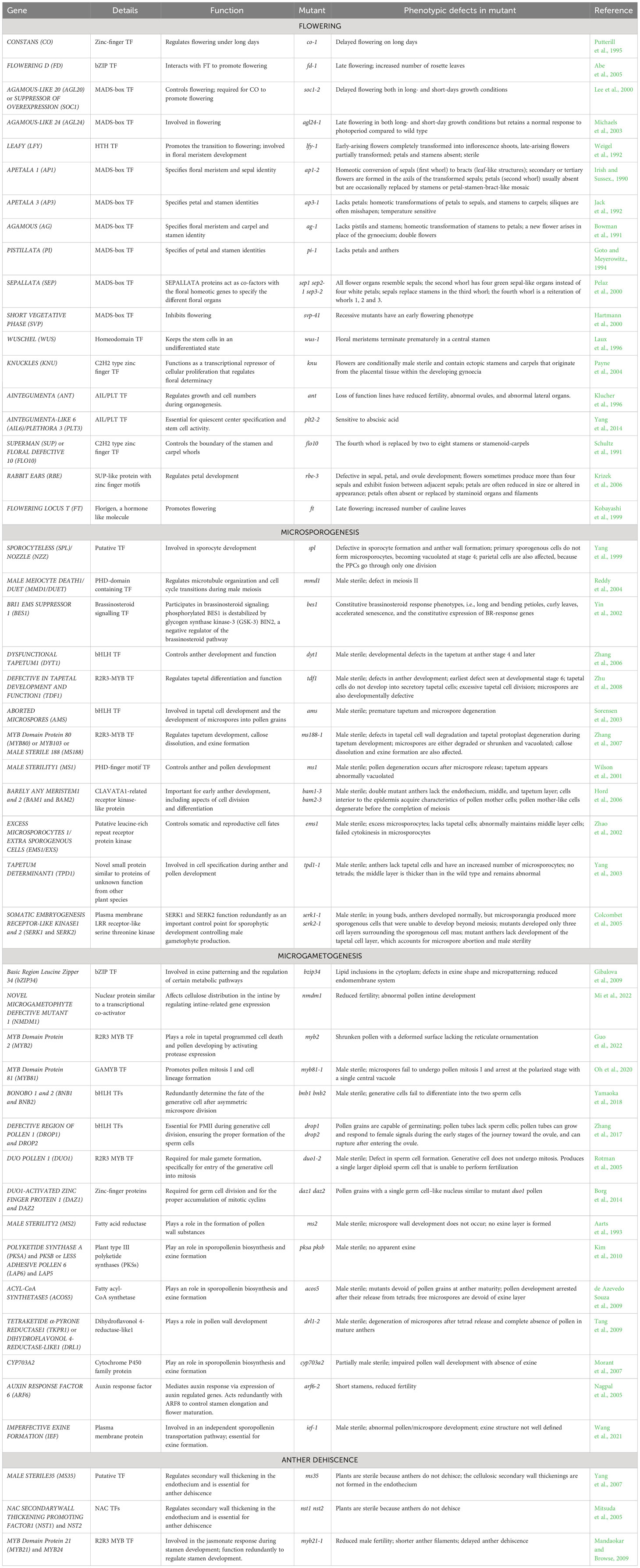
Table 1 The transcription factors and downstream target genes involved in male reproductive development.
2 Floral meristem establishment and floral organ growth
The process of Arabidopsis flower development is a thoroughly researched field with a plethora of studies published on the topic. The following section serves as a brief overview of this process, so as to orient the reader for the sections to come.
Arabidopsis flower development typically follows a precise sequence of events, beginning with the initiation of floral meristems followed by the differentiation of the floral organs (Figure 1). As Arabidopsis enters the reproductive phase, the meristem responsible for vegetative growth transforms into an indeterminate inflorescence meristem that generates floral meristems on its sides. For this to happen, photoreceptors in the leaves determine the length of the day, and when the day length exceeds a critical threshold, flowering is promoted (Srikanth and Schmid, 2011). However, other cues and environmental factors can also promote flowering. Long-day-dependent flowering initiates the photoperiodic pathway, via CONSTANS (CO) (Putterill et al., 1995), which in turn upregulates the expression of FLOWERING LOCUS T (FT) (Yoo et al., 2005), encoding a mobile protein known as florigen (Kardailsky et al., 1999). The latter is transported from the leaves to the shoot apical meristem (SAM) (Corbesier et al., 2007) where it complexes with FLOWERING D (FD) (Abe et al., 2005). This complex in turn activates the expression of AGAMOUS-LIKE 20 (AGL20) in the inflorescence meristem, directing the shift from vegetative growth to reproductive development (Borner et al., 2000). AGL20 then, in combination with AGL24, activates the expression of LEAFY (LFY) (Lee et al., 2008) in the SAM. LFY then initiates a transcriptional cascade of homeotic genes (APETALA 1 [AP1], AP3, AGAMOUS [AG], PISTILLATA [PI], SEPALLATA1–4 [SEP1–4]) that control floral meristem formation and floral organ growth (Kinoshita and Richter, 2020).
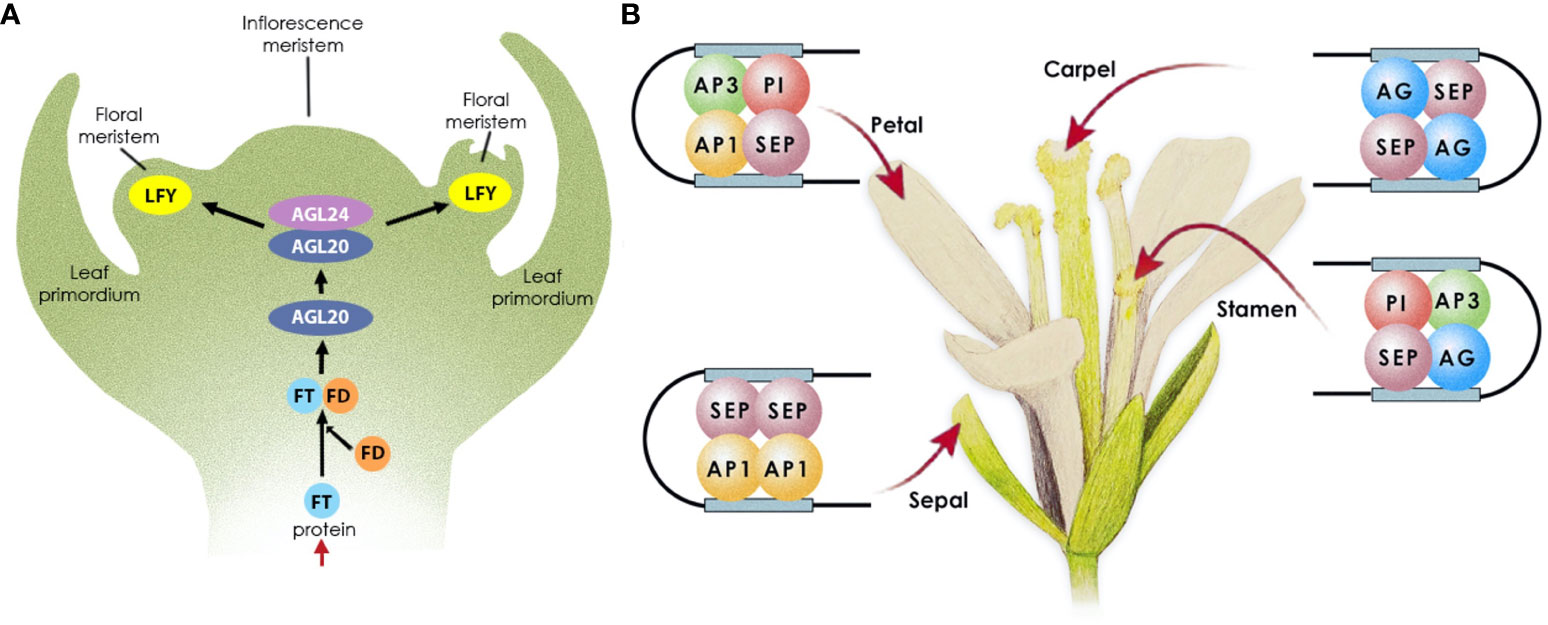
Figure 1 Transcriptional control of flowering in Arabidopsis. (A) Floral meristem establishment. Following FT expression in leaves, the protein is transported through the phloem to the apical meristem, where it complexes with FD, to activate the expression of AGL20; AGL20 then complexes with AGL24 to trigger LFY expression in the floral meristem. (B) Floral organ growth. LFY initiates the expression of the floral homeotic genes, MADS-box TFs, which subsequently form complexes to promote floral organ growth as follows: AP1 and SEP specify sepals in the outermost whorl; AP1, AP3, PI, and SEP specify petal identity in the second whorl; AP3, PI, AG, and SEP specify stamen identity in the third floral whorl; AG and SEP specify carpel identity in the fourth whorl.
LFY and AP1 both function in promoting the expression of the floral homeotic genes required for floral organ development, while concurrently repressing the floral repressor and shoot identity genes, which prevent flowering under unfavourable conditions. Once activated, AP1 and its paralog CAULIFLOWER (CAL) redundantly control the onset of flower development (John L. Bowman et al., 1993; Ferrándiz et al., 2000), by repressing the expression of SHORT VEGETATIVE PHASE (SVP), AGL24, and AGL20 (Yu et al., 2004; Liu et al., 2007; Liu et al., 2008), the protein products of which repress the activity of SEP3. SEP3 and LFY promote flower development by activating the homeotic floral organ identity genes (Honma and Goto, 2001; Castillejo et al., 2005; Immink et al., 2009). Therefore, AP1 activation derepresses the expression of SEP3 to promote flower development. This cascade controls the precise timing of the early events that establish the floral meristem. AG, in turn, promotes floral meristem development by restricting stem cell proliferation (Bowman et al., 1989; Yanofsky et al., 1990). It does so by activating the expression of KNUCKLES (KNU), which negatively regulates the expression of WUSCHEL (WUS) (Liu et al., 2011), effectively terminating the floral meristem (Sun et al., 2009).
Once the floral meristem is established, the different floral organs emerge in whorls around the meristem’s sides, from the outermost to the innermost as follows: four green sepals, four white petals, six stamens, and a single pistil. The homeotic genes activated by LFY specify floral organ identity. Their protein products form tetrameric complexes (Honma and Goto, 2001) that specify the different floral organs (Theißen and Saedler, 2001) as follows: AP1 and SEP specify sepals in the outermost whorl; AP1, AP3, PI, and SEP specify petal identity in the second whorl; AP3, PI, AG, and SEP specify stamen identity in the third floral whorl; AG and SEP specify carpel identity in the fourth whorl.
Recently, a novel set of TFs were identified that regulate aspects of floral organogenesis, namely AINTEGUMENTA (ANT) and AINTEGUMENTA-LIKE 6 (AIL6)/PLETHORA 3 (PLT3) (Krizek et al., 2020). ANT directly influences approximately 200 genes involved in auxin signaling, floral organ identity, polarity establishment, growth regulation, and cell differentiation, including AP1, AP2, and SEP3.
For floral organ development to proceed correctly, boundaries (Breuil-Broyer et al., 2004; Reddy et al., 2004) need to be established to restrict the expression of the homeotic genes to their respective whorls. Several boundary genes have been characterized, including SUPERMAN (SUP) and the closely related RABBIT EARS (RBE), which repress growth at the boundaries between the third and fourth whorls through AP3 and PI repression (Sakai et al., 1995) and the second whorl through AG repression (Krizek et al., 2006), respectively.
3 Microsporogenesis
Following stamen specification in the third whorl, the stamen primordia undergo further differentiation to give rise to the filament and the anther. The differentiation of the anther in Arabidopsis comprises 14 stages divided into two phases (Sanders et al., 1999; Ma, 2005): microsporogenesis (stages 1–7) and microgametogenesis (stages 8–14). Figure 2 outlines the process of microsporogenesis, as discussed below. During stage 1, the anther primordium emerges from the floral meristem in the third whorl, consisting of three germ layers (L1–L3). During stages 2–5, the cells in the L1 layer give rise to the epidermis via anticlinal cell division, the cells in the L3 layer differentiate to form vascular and connective tissues, and the cells in the L2 hypodermal layer undergo periclinal cell division to form four clusters of archesporial cells. During stage 3, the archesporial cells in the L2 layer divide to form the reproductive primary sporogenous cells and the somatic primary parietal layer. The primary parietal layer then divides to give rise to two secondary parietal layers; the outer parietal layer further differentiates during stage 4 into the endothecium and middle layers while the cells of the inner parietal layer develop into the tapetal layer. However, recent evidence suggests that the middle layer originates from both the inner secondary and outer parietal cell layers (Zhao et al., 2002; Xue et al., 2021; Feng and Dickinson 2010).
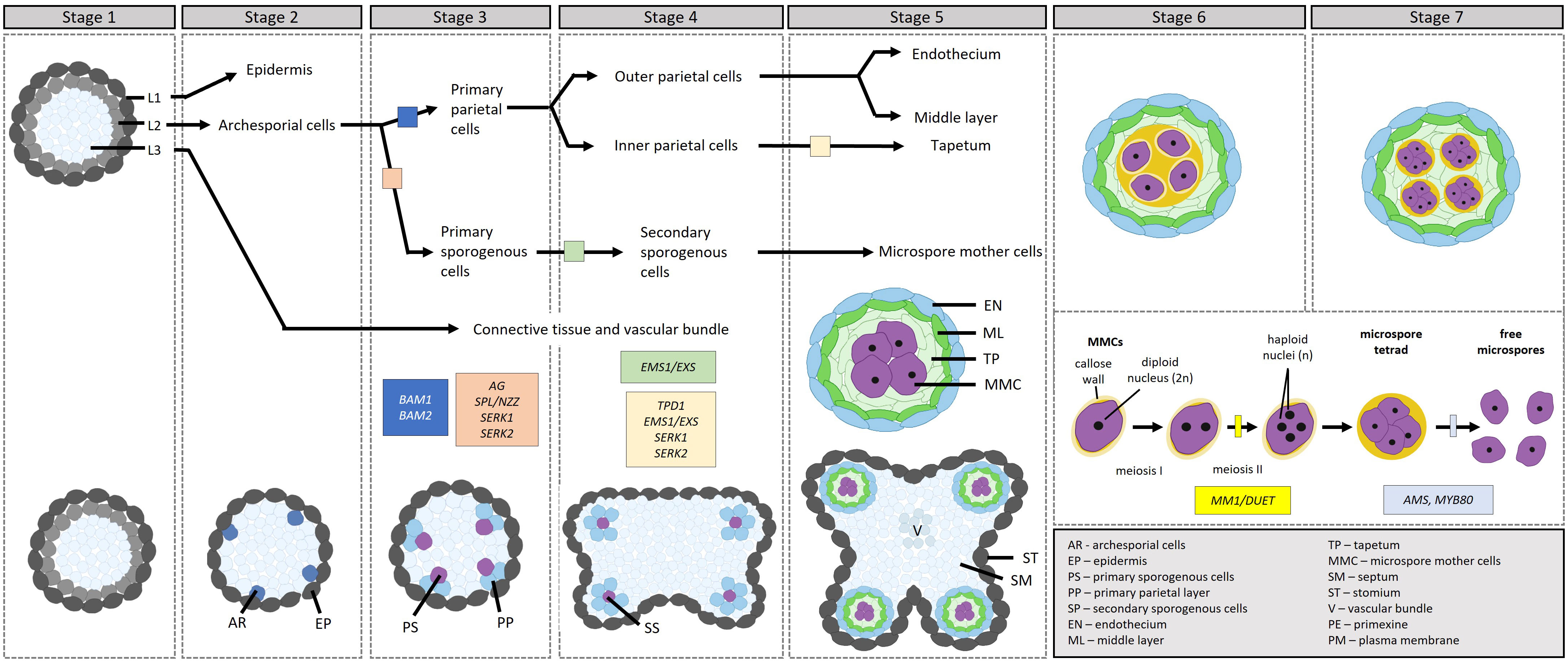
Figure 2 Transcriptional control of microsporogenesis in Arabidopsis, development through stages 1–7. [Stage 1] Stamen primordia differentiate into three cell types, L1, L2 and L3. [Stage 2] The cell types further differentiate into the epidermis, the archesporial cells and vascular and connective tissues. [Stage 3] Archesporial cells divide to form the reproductive primary sporogenous cells and the somatic primary parietal layer. [Stage 4] The primary sporogenous cells differentiate into secondary sporogenous cells through the action of EMS1/EXS, while the primary parietal layer differentiates into the outer and inner secondary parietal layers. [Stage 5] The outer secondary parietal layer differentiates into the endothecium and middle layer, while the inner secondary parietal layer differentiates into the tapetum. In addition, the secondary sporogenous cells differentiate into microspore mother cells. [Stages 6–7] The tapetum differentiates further. Microspore mother cells enter and complete meiosis during stages 6 and 7 to produce four microspores in tetrads surrounded by a callose wall. Before the end of stage 7, callose dissollution releases the microspores from the tetrads. The transcription factors involved in the different steps are indicated in the figure in boxes.
During these early stages of anther development, AG initiates the expression of SPOROCYTELESS/NOZZLE (SPL/NZZ), which is expressed in the L2 layer and plays a role in the specification of sporogenous and primary parietal cells; in the spl/nzz mutants, archesporial cells fail to undergo subsequent cell divisions, resulting in sporogenous and primary parietal cells being absent (Schiefthaler et al., 1999; Yang et al., 1999). SPL/NZZ in turn regulates BARELY ANY MERISTEM1 (BAM1) and BAM2, receptor-like kinases (RLKs), which redundantly regulate the formation of the somatic layers (Hord et al., 2006). In the bam1bam2 mutant, the somatic layers are absent while microspore mother cell (MMC)-like cells are present; the latter, however, degenerate before meiosis. SPL/NZZ and BAM1/BAM2 form part of a feedback loop, wherein SPL/NZZ positively regulates BAM1/BAM2, while BAM1/BAM2 restricts SPL/NZZ’s expression to sporogenous cells, ensuring that SPL/NZZ maintains sporogenous activity and BAM1/BAM2 promotes the development of the somatic cell layers. From here, the EXCESS MICROSPOROCYTES 1/EXTRA SPOROGENOUS CELLS (EMS1/EXS), TAPETUM DETERMINANT1 (TPD1), and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 and 2 (SERK1/2) module specifies the tapetal cell layer (Canales et al., 2002; Zhao et al., 2002; Yang S. et al., 2003). The ems1/exs, tpd1, and serk1serk2 mutants are very similar; the L2-derived cell layers are absent, with only the endothecium and middle layers present. In addition, these mutants generate a greater number of MMCs, which can enter meiosis and complete karyokineses, but not cytokinesis, leading to their degeneration. EMS1/EXS and SERK1/2 encode leucine-rich repeat receptor-like kinases (LRR-RLKs) while TPD1 encodes a small protein with a putative signal peptide for secretion. The current model (Ma, 2005) follows: In the differentiating MMCs, the level of EMS1/EXS drops while the level of TPD1 rises; TPD1 is subsequently secreted from the cells and binds to the EMS1/EXS-SERK1/2 complex on the surface of the cells surrounding the MMCs, activating a cascade in the surrounding cells to promote tapetum differentiation. Also downstream of SPL/NZZ, lies the basic region leucine zipper (bZIP) TFs TGA9 and TGA10. These two TFs are expressed in the middle layer and tapetum during stages 5 and 6; mutants show a reduction in tapetum-expressed genes (DYSFUNCTIONAL TAPETUM1 [DYT1] and MALE STERILITY1 [MS1]), affecting microspore and pollen development (Murmu et al., 2010).
By stage 5, all of the somatic cell layers surrounding the developing MMCs are established and the MMCs and tapetal cells are connected via plasmodesmata. Prior to the completion of stage 5, a transient callose (β-1, 3-glucan) layer is synthesized around the MMCs, separating them from each other and the tapetum and breaking the plasmodesmatal connections. Concomitantly, the middle layer thins out. During stage 6, the MMCs enter and complete meiosis to produce four microspores in a tetrad by stage 7, surrounded by a callose wall. Here, MALE MEIOCYTE DEATH1/DUET (MMD1/DUET) is involved in orchestrating the formation of the cell plate, which eventually becomes the cell wall that separates the two daughter cells during meiotic cytokinesis (Yang X. et al., 2003). The accurate positioning and timing of cell plate formation are essential for the successful completion of meiosis.
Whilst encased in a callose layer, following the completion of cytokinesis, the microspores switch from callose to primexine deposition, initiating exine development. The primexine is deposited between the microspore plasma membrane and the callose wall and is largely composed of cellulose, serving as the foundation for sporopollenin precursor accumulation and polymerization on the microspore surface (Fitzgerald and Knox, 1995; Paxson-Sowders et al., 1997; Blackmore et al., 2007). Concurrently, the plasma membrane starts to invaginate, forming undulations, whereon probacula and protectum structures are formed via sporopollenin deposition (Paxson-Sowders et al., 1997).
While the MMCs undergo meiosis, the tapetum further expands and undergoes specialization. Post-specification, the development of the tapetum is controlled by the DYSFUNCTIONAL TAPETUM1 (DYT1), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1), ABORTED MICROSPORES (AMS), MYB80, and MALE STERILITY1 (MS1) module, which regulates genes involved in tapetum programmed cell death (PCD) and degradation and pollen wall development (Yang C. et al., 2007a; Zhu et al., 2008; Phan et al., 2011; Gu et al., 2014; Xu J. et al., 2014; Verma, 2019). It was recently shown that a disruption of fatty acid export-mediated ROS homeostasis suppresses the activation of the DYT1-TDF1-AMS-MYB80-MS1 module, resulting in male sterility (Zhu et al., 2020). In this pathway, the master regulator DYT1 lies downstream of EMS1/EXS-TPD1-SERK1/2 pathway. Briefly, the latter pathway mediates the phosphorylation and nuclear localization of BRI1 EMS SUPPRESSOR 1 (BES1), a TF activated by brassinosteroid (BR) signaling. BRs are known to exert a significant influence on pollen development, with BR-deficient mutants having reduced pollen numbers stemming from decreased microspore mother cells and the early abortion of some microspores (Ye et al., 2010). Once in the nucleus, BES1 activates the expression of DYT1 (Chen et al., 2019); BES1 has also been shown to bind the promoters of SPL/NZZ, TDF1, AMS, MS1, and MS2 (Ye et al., 2010). dyt1 mutants have similar phenotypic defects to that of ems1/exs (arrested meiosis progression with no cytokinesis (Zhang et al., 2006). DYT1 in turn activates the expression of TDF1, which is necessary for tapetal cell differentiation and function (Zhu et al., 2008). TDF1 is necessary for the transition of the tapetum to the secretory type, which provides nutrients (e.g., carbohydrates, lipids, proteins, etc) to developing microspores to support their growth and maturation. The tdf1 mutant shows defective tapetal function and subsequent pollen abortion. TDF1 activates the expression of AMS (Lou et al., 2018), which has a multi-pronged effect on tapetum and pollen development (Ferguson et al., 2016). First, by regulating the expression of TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK (TEK) in the tapetum, encoding an AT-hook nuclear matrix attachment region (MAR) binding protein (Lou et al., 2014), AMS promotes nexine formation in individual microspores of tetrads following meiosis. This takes place through TEK’s positive regulation of Arabinogalactan protein (AGP) genes (i.e., AGP6, AGP11, AGP23, and AGP40), which promote exine formation (Jia et al., 2015). Concomitantly, TEK negatively regulates the expression of CALLOSE SYNTHASE 5 (CalS5) after the tetrad stage, a protein required for callose synthesis (Xiong et al., 2020).
During stage 7, after the secretory tapetum has been established, the tapetum synthesizes and secretes callases (β-1,3-glucanase), enzymes that break down the callose wall surrounding the microspores, releasing them from the tetrad into the anther locule. AMS regulates this process through its target gene MYB80 (Zhang et al., 2006), which plays a role in callose dissolution by regulating the expression of Anther-specific protein 6 (A6), a member of the callase complex. After being expressed and synthesized in the tapetum, UNEVEN PATTERN OF EXINE 1 (UPEX1), encoding an arabinogalactan β-(1,3)-galactosyltransferase, secretes A6 in the anther locule; UPEX1 expression is directly regulated by AMS (Wang et al., 2022). Following callose dissolution, the microspores, covered by the primexine, are released into the anther locule during stage 8, initiating microgametogenesis (Figure 3).
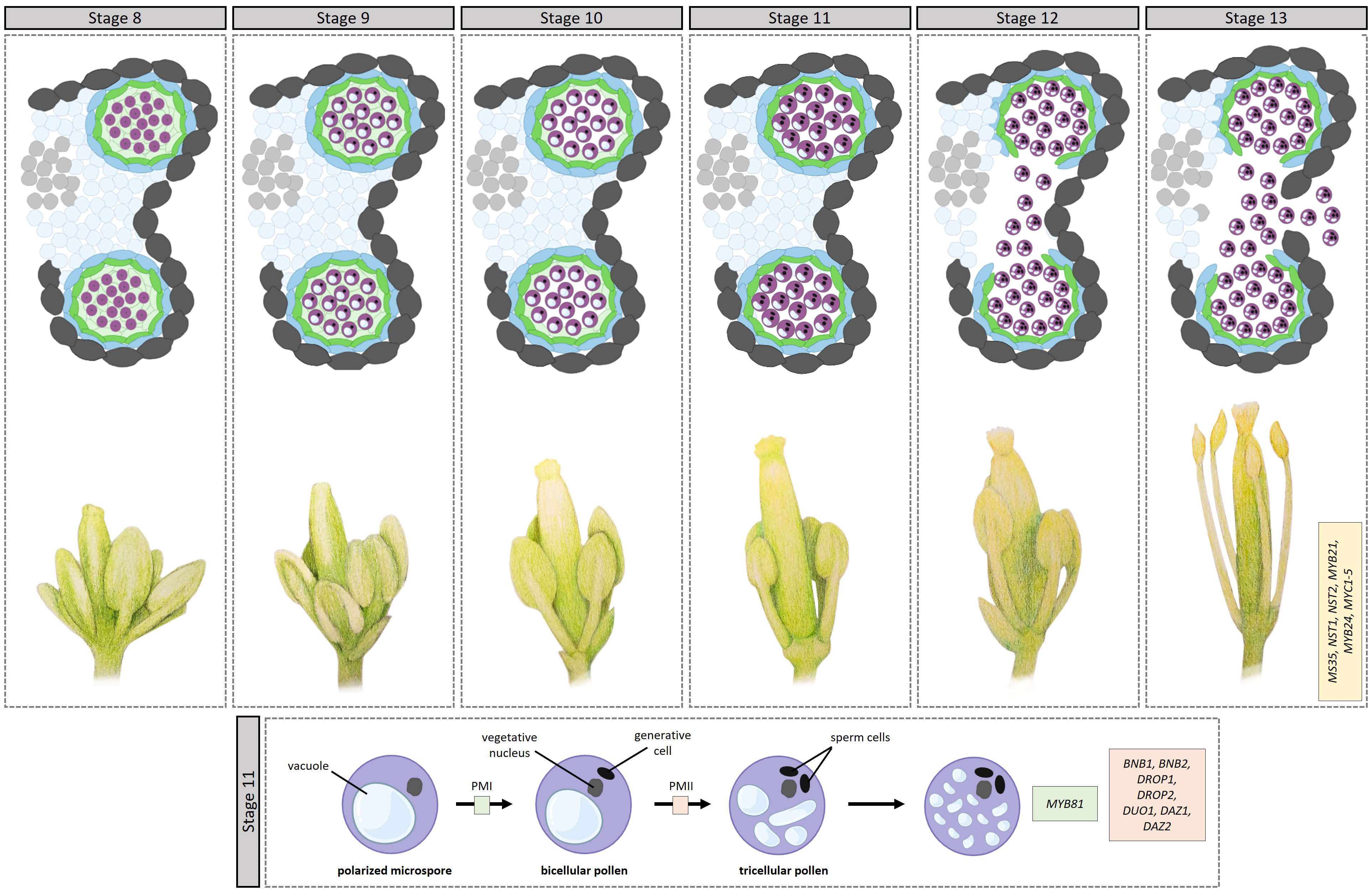
Figure 3 Transcriptional control of microgametogenesis in Arabidopsis. Top half op top panel: development through stages 8–13. [8] Microspores, covered by the primexine, are released into the anther locule. [9] Exine formation starts via sporpollenin deposition on the microspores, with the exine eventually becoming thicker and assuming its basic structure. [10] Intine development starts, appearing between the plasma membrane and the nexine. Additionally, the tapetum starts to degrade. [11] This stage comprises two rounds of pollen mitosis. In addition, the septum starts to degrade. [12] The septum breaks, creating a bilocular anther. Pollen are tricullular. [13] Anther dehiscence leads to the anther breaking open along the stomium, releasing mature pollen grains. Bottom half of top panel: anther positioning during stages 8–13. During stages 11–13, the filament elongates substantially, placing the anther in a position to pollinate the stigma. Bottom panel: During pollen mitosis I (PMI), the polarized microspore undergoes a highly asymmetric cell division giving rise to a large vegetative cell and a small generative cell. The smaller generative cell then divides into two sperm cells through PMII. The transcription factors involved in the different steps are indicated in boxes. bofigure in boxes.
4 Microgametogenesis
Once released into the anther locule, the microspores are surrounded by a fluid containing sugars, amino acids, proteins, and sporopollenin precursors secreted by the tapetum, to aid microspore development and pollen wall formation. During stage 9, exine formation continues via sporopollenin deposition on the microspores, with the exine eventually becoming thicker and assuming its basic structure. During microgametogenesis, the formation of the pollen wall starts in earnest, with the ultimate pollen wall comprising an outer exine and an inner intine; reviewed in (Shi et al., 2015). The intine is a pectocellulosic layer synthesized by the microspore, while the exine is composed mostly of sporopollenin synthesized predominantly in the tapetum. The outer exine is further differentiated into an outer sculpted sexine and an inner nexine. The sexine in turn, comprises columns called baculae that support roof-like structures called tecta; in between, the pollen coat (tryphine, also called the pollenkit) is deposited to fill in the spaces of the exine. The pollen coat protects the pollen from dehydration and aids in pollen-stigma adhesion and communication.
MYB80, through its target genes (Wang et al., 2018), is essential for sporopollenin synthesis in the tapetum and ultimately sexine formation; In the myb80 mutant, the sexine is completely absent (Zhang et al., 2007). Confirmed target genes of MYB80 include: MALE STERILITY 2 (MS2), POLYKETIDE SYNTHASE A (PKSA) and PKSB, and ACYL-CoA SYNTHETASE5 (ACOS5), TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1) and TKPR2, and CYP703A2. The proteins encoded by these genes are responsible for catalyzing specific biochemical reactions that modify fatty acids for sporopollenin biosynthesis (Kim et al., 2010; Shi et al., 2015). For example, ACOS5 catalyzes the activation of fatty acids by attaching a Coenzyme A (CoA) molecule to them (de Azevedo Souza et al., 2009). CYP703A2, a CYTOCHROME P450 family protein, catalyzes the hydroxylation of different long chain fatty acids (Morant et al., 2007), which are then either catalyzed into triketide and tetraketide α-pyrones by PKSA and PKSB, or converted to fatty alcohols by MS2 (Aarts et al., 1993). The tetraketide α-pyrones are then likely reduced to polyhydroxylated tetraketide by TKPR1 and TKPR2 (Grienenberger et al., 2010). Outside of the MYB80 regulon, bHLH010 and bHLH089 redundantly contribute to pollen wall formation by regulating genes implicated in the biosynthesis of glyceryl derivatives and flavonols among others (Lai et al., 2022). In addition, basic region leucine zipper 34 (bZIP34) regulates pollen wall development by either directly or indirectly regulating genes involved in lipid metabolism (Gibalova et al., 2009). Later on, bZIP18 was found to be partially functionally redundant with bZIP34, as evidenced by their similar phenotypes and demonstrated interactions; bzip18 shows pollen abortion, exine formation defects, and the presence of inclusions in the vegetative cell cytoplasm (Gibalová et al., 2017). In addition to these two, a large proportion of other bZIP TFs are expressed in pollen (Honys and Twell, 2004), with interactions among them demonstrated for some (Gibalová et al., 2017). Moreover, bHLH010 and bHLH089 redundantly contribute to pollen wall formation by regulating genes implicated in the biosynthesis of glyceryl derivatives and flavonols among others (Lai et al., 2022).
The sporopollenin precursors are subsequently transported to the anther locule for exine formation by members of the ATP-binding cassette transporter superfamily. AMS regulates the expression of ABCG26, the protein product of which transports lipid precursors and polykedtides (Choi et al., 2011; Quilichini et al., 2014). AMS also regulates the expression of LIPID TRANSFER PROTEIN 12 (LTP12) and IMPERFECTIVE EXINE FORMATION (IEF) (Xu et al., 2010). LTP12 encodes a lipid binding protein that exchanges lipids between membranes while IEF encodes a plasma membrane protein that likely transports materials to the anther locule to reinforce sexine and nexine structures (Wang et al., 2021).
During stage 10, intine development also starts, appearing between the plasma membrane and the nexine; here, only the microspores are involved. The NOVEL MICROGAMETOPHYTE DEFECTIVE MUTANT 1 (NMDM1) putative transcription co-activator, regulates intine development by regulating the expression of Arabidogalactin proteins (AGPs) and pectin methylesterases (PMEs) (Mi et al., 2022). Concurrently, the numerous vacuoles in the microspores fuse into a large vacuole, which leads to the migration of the nucleus to one side of the cell, creating a polarized microspore. During stage 10, the tapetum starts degenerating via PCD and pollen coat materials start filling in the sexine cavities. MYB80 plays a role in tapetal PCD through its target genes UNDEAD (A1 aspartic protease) and MS1, the penultimate player of the DYT1-TDF1-AMS-MYB80-MS1 module. UNDEAD hydrolyzes apoptosis-inducing proteins in the tapetal mitochondria (Phan et al., 2011) while MS1 regulates the expression of a number of cysteine proteases (Yang C. et al., 2007a). Outside of MYB80 regulation, MYB2 regulates the expression of CEP1 (Guo et al., 2022), a papain-like cysteine protease that directly participates in tapetal PCD (Zhang et al., 2014).
Stage 11 also comprises two rounds of pollen mitosis. During pollen mitosis I (PMI), the polarized microspore undergoes a highly asymmetric cell division giving rise to a large vegetative cell and a small generative cell. The larger vegetative cell is responsible for forming the pollen tube, which will deliver the male gametes to the female reproductive structures during fertilization, while the smaller generative cell will eventually divide into two sperm cells through PMII, which occurs prior to pollen maturation. One TF essential for PMI is MYB81, a microspore-specific TF; myb81 microspores are unable to undergo PMI, arresting at the polarized microspore stage (Oh et al., 2020). However, the exact mechanisms through which MYB81 promotes microspore progression into PMI remains unclear. Acting later, BONOBO1 (BNB1) and BNB2 redundantly determine the fate of the generative cell after asymmetric microspore division; in the bnb1bnb2 mutant, the generative cells fail to differentiate into the two sperm cells (Yamaoka et al., 2018). Similarly, DEFECTIVE REGION OF POLLEN 1 (DROP1) and DROP2 are essential for PMII during generative cell division, ensuring the proper formation of the sperm cells; drop1drop2 mutants lack sperm cells (Zhang et al., 2017). During PMII, DUO POLLEN1 (DUO1) integrates generative cell mitosis and sperm cell differentiation (Brownfield et al., 2009; Borg and Twell, 2011) via a module including its target genes DUO1-ACTIVATED ZINC FINGER PROTEIN 1 (DAZ1) and DAZ2, encoding transcriptional repressors, and the corepressor TOPLESS (Borg et al., 2014). Following the completion of pollen mitosis I, and before pollen mitosis II, the exine is visually completed.
During stages 11 and 12, the filament elongates substantially, placing the anther in a position to pollinate the stigma. Here, AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8 play a crucial role in filament elongation, by activating the expression of DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1), a gene associated with jasmonic acid (JA) biosynthesis, which is essential for flower opening and anther dehiscence (Tabata et al., 2010). In the arf6/arf8 double mutant, a delay in petal and stamen elongation at anthesis is observed, leading to a protrusion of the stigma in unopened flower buds. Moreover, MYB21, a member of the R2R3-MYB transcription factor family, plays a role in regulating flavonol biosynthesis; flavonols, acting as reactive oxygen species scavengers, contribute to stamen development, particularly in filament elongation (Zhang et al., 2021).
During stage 12, tapetum degeneration is mostly complete, releasing their full complement of materials into the anther locule for pollen wall and coat deposition, including flavonoids, alkanes, lipids, proteins, carotenoids (Hsieh and Huang, 2007). The flavonoids specifically, are deposited onto the sporopollenin wall (Jia et al., 2021; Xue et al., 2023). The eventual pollen coat comprises a sticky material that fills in the exine cavities. The pollen coat plays a crucial role in protecting against desiccation and is essential for adhesion, pollen–stigma recognition, and hydration during interactions with the stigma (Jia et al., 2021). The flavonoids in the pollen wall play a crucial role in protecting haploid pollen and spores against environmental stresses like UV-B light, high temperatures, and water loss (Hsieh and Huang, 2007; Xue et al., 2023). During stage 12, the septa break, creating a bilocular anther.
5 Anther dehiscence
Stages 13 and 14 comprise anther dehiscence, during which, the anther cells switch from a differentiation to a degeneration program. Anther dehiscence involves lignification of the endothecium cells, degradation of the septum cells to create a “bi-locular anther”, and rupture of the stomium to release the pollen grains at anthesis (Goldberg et al., 1993; Sanders et al., 1999). Most research on male reproductive development has focussed on pollen development, with sparse studies available for anther dehiscence. Nevertheless, some studies have identified TFs implicated in this process. MALE STERILE35 (MS35) is involved in the secondary thickening of the endothecium which is necessary for breaking the stomium, by acting upstream of the lignin biosynthetic pathway (Dawson et al., 1999; Steiner-Lange et al., 2001; Yang et al., 2007b). MS35 also regulates the expression of NAC SECONDARYWALL THICKENING PROMOTING FACTOR1 (NST1) and NST2, which act redundantly in regulating secondary cell wall thickening (Mitsuda et al., 2005). The bZIP TFs TGA9 and TGA10 are required for multiple steps in the anther dehiscence program, with mutants showing incomplete dissolution of the middle layer, abnormal lignification of the endothecium, disorganized septums, and nonfunctional stomium-like cells (Murmu et al., 2010). At the intersection of TFs and hormone signaling, MYB21 and MYB24, triggered by jasmonate, play a role in stamen maturation and anther dehiscence (Mandaokar and Browse, 2009). Additional jasmonic acid-regulated TFs include the bHLH TFs MYC1–5, which act redundantly and form transcriptional complexes with MYB21 and MYB24 to control anther dehiscence (Qi et al., 2015). Aside from jasmonic acid, auxin negatively regulates two key events in anther dehiscence, namely endothecium lignification via MYB26 and stomium opening via the control of jasmonic acid (JA) biosynthesis (Cecchetti et al., 2013). Auxin synthesized in anthers, acting through auxin receptors, coordinates anther dehiscence, pollen maturation, and filament elongation; mutants lacking these receptors exhibit premature anther dehiscence (Cecchetti et al., 2008). Moreover, ARF17, is crucial for anther dehiscence by regulating MYB108 expression (Xu et al., 2019), while miR167-mediated downregulation of ARF6 and ARF8 is critical for proper anther growth arrest, enabling anther dehiscence (Zheng et al., 2019).
6 From models to crops – capitalizing on conserved pathways and identifying new players
One of the primary benefits of translational research is the identification and manipulation of key TFs and target genes that regulate pollen development in crop plants. By introducing or modifying these TFs, researchers can potentially enhance pollen quality, pollen viability, and pollen production in crop species. In addition, researchers can engineer male sterility in crop plants for hybrid seed production. This prevents self-pollination, facilitating the controlled cross-breeding of male-sterile and fertile lines to create hybrid seeds with desirable traits (e.g., disease resistance or improved yield). This approach offers a significant advantage over manual emasculation and pollination methods, as it streamlines the breeding process and ensures genetic purity. As discussed in the previous sections, misregulation of several pollen- and tapetum-expressed TFs lead to male sterility. Orthologs of these TFs have been identified in several crop species. For example, orthologs of microgametogenesis TFs in rice and maize include those of DYT1 (OsDYT1, ZmMs32), AMS (OsTDR, ZmbHLH51), MYB80 (OsMYB80, ZmMYB84), and MS1 (OsMS1, ZmMs7) ((Wilson and Zhang, 2009). Orthologs of MYB80 have been also been identified in maize, barley, canola, rapeseed, mustard greens, cabbage, and cotton (Xu Y. et al., 2014). Transcriptomic analyses of fertile and sterile lines have identified additional potential TFs that could be manipulated to bring about male sterility. For example. In rapeseed, ca. 250 TFs were differentially expressed between sterile and fertile lines (Jiang et al., 2022). In addition, transcriptomic analyses of thermosenstitive rapeseed lines showed that MADS, NFY, HSF, MYB/C and WRKY TFs may play a role in male fertility under high temperatures (Tang et al., 2019). Aside from male sterility, breeders can enhance additional traits, such as drought or heat tolerance, disease resistance, or nutrient use efficiency in crops by targeting specific TFs. This targeted breeding approach can accelerate the development of improved crop varieties adapted to changing environmental conditions and increasing agricultural challenges.
7 Directives for future research
Despite significant progress in the field, much remains to be discovered or understood. For example:
● The regulatory networks governing early anther development and the specification of archesporial cells are not fully elucidated.
● Pollen development is regulated by multiple hormones (e.g., gibberellins, auxins, cytokinins, and jasmonic acid), however, the crosstalk and integration of these hormonal signals with transcriptional regulation remain largely unknown.
● Many TFs undergo post-translational modifications (e.g., phosphorylation, ubiquitination, and acetylation) which can significantly impact their activity and stability. The functional significance of these modifications in pollen development and their precise regulatory roles are still not fully elucidated.
● Pollen development is influenced by various environmental factors (e.g., temperature, light, and nutrient availability), however, how these environmental cues are perceived and transduced into specific transcriptional responses during pollen development requires further investigation.
● Several TFs involved in pollen development belong to gene families, leading to functional redundancy and potential compensation among family members. Understanding the extent of this redundancy and how individual factors contribute to specific aspects of pollen development remains challenging.
● TFs often function by forming complexes and interacting with co-regulators to modulate gene expression. The nature and dynamics of these protein-protein interactions during pollen development are yet to be fully unraveled.
Addressing these open questions will require the integration of various -omics approaches along with functional studies and advanced genetic tools.
8 Conclusions
In conclusion, Arabidopsis has emerged as a powerful model system for studying the intricate molecular and genetic mechanisms underlying male reproductive development. Throughout this literature review, we have explored the roles of various TFs in the different stages of anther and pollen development, shedding light on their functions in specifying cell fate, regulating pollen wall formation, and ensuring pollen maturation and germination. We have also discussed how the knowledge gained from Arabidopsis research on TFs has significant implications for crop improvement. As the field continues to advance, addressing current gaps and open questions will further enrich our understanding of male reproductive development, contributing to the broader goal of sustainable agriculture and global food security.
Author contributions
AW: Conceptualization, Writing – original draft, Writing – review & editing. ET: Writing – original draft, Writing – review & editing. DH: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Czech Science Foundation (Grantová agentura České republiky; Grant no. 23-07000S.
Acknowledgments
We would like to thank Jan Fíla and Said Hafidh for their critical reading of the manuscript and Suné Horn for the pencil illustrations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aarts, M., Dirkse, W., Stiekema, W., Pereira, A. (1993). Transposon tagging of a male sterility gene in Arabidopsis. Nature 363 (6431), 715–717. doi: 10.1038/363715a0
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science 309 (5737), 1052–1056. doi: 10.1126/science.1115983
Blackmore, S., Wortley, A. H., Skvarla, J. J., Rowley, J. R. (2007). Pollen wall development in flowering plants. New Phytol. 174 (3), 483–498. doi: 10.1111/j.1469-8137.2007.02060.x
Borg, M., Rutley, N., Kagale, S., Hamamura, Y., Gherghinoiu, M., Kumar, S., et al. (2014). An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell 26 (5), 2098–2113. doi: 10.1105/tpc.114.124743
Borg, M., Twell, D. (2011). “Pollen: structure and development,” in eLS (American Cancer Society). doi: 10.1002/9780470015902.a0002039.pub2
Borner, R., Kampmann, G., Chandler, J., Gleißner, R., Wisman, E., Apel, K., et al. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24 (5), 591–599. doi: 10.1046/j.1365-313x.2000.00906.x
Bowman, J., Alvarez, J., Weigel, D., Meyerowitz, E., Smyth., D. (1993). “Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119 (3), 721–743. doi: 10.1242/dev.119.3.721
Bowman, J., Smyth, D., Meyerowitz., E. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 (1), 37–52. doi: 10.1105/tpc.1.1.37
Bowman, J., Smyth, D., Meyerowitz, E. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112 (1), 1–20. doi: 10.1242/dev.112.1.1
Breuil-Broyer, S., Morel, P., de Almeida-Engler, J., Coustham, V., Negrutiu, I., Trehin, C. (2004). High-Resolution Boundary Analysis during Arabidopsis thaliana Flower Development. Plant J. For. Cell Mol. Biol. 38 (1), 182–192. doi: 10.1111/j.1365-313X.2004.02026.x
Brownfield, L., Hafidh, S., Borg, M., Sidorova, A., Mori, T., Twell, D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PloS Genet. 5 (3), e1000430. doi: 10.1371/journal.pgen.1000430
Canales, C., Bhatt, A. M., Scott, R., Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12 (20), 1718–1727. doi: 10.1016/s0960-9822(02)01151-x
Castillejo, C., Romera-Branchat, M., Pelaz, S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43 (4), 586–596. doi: 10.1111/j.1365-313X.2005.02476.x
Cecchetti, V., Altamura, M. M., Brunetti, P., Petrocelli, V., Falasca, G., Ljung, K., et al. (2013). Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. For. Cell Mol. Biol. 74 (3), 411–422. doi: 10.1111/tpj.12130
Cecchetti, V., Altamura, M. M., Falasca, G., Costantino, P., Cardarelli, M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20 (7), 1760–1774. doi: 10.1105/tpc.107.057570
Chen, W., Lv, M., Wang, Y., Wang, P.-A., Cui, Y., Li, M., et al. (2019). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10 (1), 4164. doi: 10.1038/s41467-019-12118-4
Choi, H., Jin, J.-Y., Choi, S., Hwang, J.-U., Kim, Y.-Y., Suh, M. C., et al. (2011). An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant Journal: For Cell Mol. Biol. 65 (2), 181–193. doi: 10.1111/j.1365-313X.2010.04412.x
Colcombet, J., Boisson-Dernier, A., Ros-Palau, R., Vera, C. E., Schroeder, J. I. (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17 (12), 3350–3361. doi: 10.1105/tpc.105.036731
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 (5827), 1030–1033. doi: 10.1126/science.1141752
Dawson, J., Sözen, E., Vizir, I., Van Waeyenberge, S., Wilson, Z. A., Mulligan., B. J. (1999). Characterization and genetic mapping of a mutation (Ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144 (2), 213–222. doi: 10.1046/j.1469-8137.1999.00507.x
de Azevedo Souza, C., Soo Kim, S., Koch, S., Kienow, L., Schneider, K., McKim, S. M., et al. (2009). A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21 (2), 507–525. doi: 10.1105/tpc.108.062513
Ferguson, A. C., Pearce, S., Band, L. R., Yang, C., Ferjentsikova, I., King, J., et al. (2016). Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 14, 778–790. doi: 10.1111/nph.14200
Ferrándiz, C., Gu, Q., Martienssen, R., Yanofsky., M. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 (4), 725–734. doi: 10.1242/dev.127.4.725
Fitzgerald, M., Knox, B. (1995). Initiation of primexine in freeze-substituted microspores of brassica campestris. Sexual Plant Reprod. 8 (2), 99–104. doi: 10.1007/BF00230896
Gibalova, A., Chab, D., Twell, D., Honys., D. (2009). AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol. Biol. 21, 581–601. doi: 10.1007/s11103-009-9493-y
Gibalová, A., Steinbachová, L., Hafidh, S., Bláhová, V., Gadiou, Z., Michailidis, C., et al. (2017). Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod. 30, 1–17. doi: 10.1007/s00497-016-0295-5
Goldberg, R., Beals, T., Sanders, P. (1993). Anther development: basic principles and practical applications. Plant Cell 5 (10), 1217–1229. doi: 10.1105/tpc.5.10.1217
Goto, K., Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 (13), 1548–1560. doi: 10.1101/gad.8.13.1548
Grienenberger, E., Kim, S. S., Lallemand, B., Geoffroy, P., Heintz, D., Souza, C. d. A., et al. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE Function in Arabidopsis thaliana Reveals a Previously Unknown, but Conserved, Biochemical Pathway in Sporopollenin Monomer Biosynthesis. Plant Cell 22 (12), 4067–4083. doi: 10.1105/tpc.110.080036
Gu, J.-N., Zhu, J., Yu, Y., Teng, X.-D., Lou, Y., Xu, X.-F., et al. (2014). DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 80 (6), 1005–1013. doi: 10.1111/tpj.12694
Guo, X., Li, L., Liu, X., Zhang, C., Yao, X., Xun, Z., et al. (2022). MYB2 is important for tapetal PCD and pollen development by directly activating protease expression in Arabidopsis. Int. J. Mol. Sci. 23 (7), 3563. doi: 10.3390/ijms23073563
Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., Huijser., P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant Journal: For Cell Mol. Biol. 21 (4), 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Honma, T., Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 (6819), 525–529. doi: 10.1038/35054083
Honys, D., Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5. doi: 10.1186/gb-2004-5-11-r85
Hord, C., Chen, C., DeYoung, B., Clark, S., Ma., H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18 (7), 1667–1680. doi: 10.1105/tpc.105.036871
Hsieh, K., Huang, A. H. C. (2007). Tapetosomes in brassica tapetum accumulate endoplasmic reticulum–derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19 (2), 582–596. doi: 10.1105/tpc.106.049049
Immink, R., Tonaco, I., de Folter, S., Shchennikova, A., Dijk, A. v., Busscher-Lange, J., et al. (2009). SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10 (2), R24. doi: 10.1186/gb-2009-10-2-r24
Irish, V., Sussex, I. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2 (8), 741–753. doi: 10.2307/3869173
Jack, T., Brockman, L., Meyerowitz., E. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 (4), 683–697. doi: 10.1016/0092-8674(92)90144-2
Jia, X.-L., Xue, J.-S., Zhang, F., Yao, C., Shen, S.-Y., Sui, C.-X., et al. (2021). A dye combination for the staining of pollen coat and pollen wall. Plant Reprod. 34 (2), 91–101. doi: 10.1007/s00497-021-00412-5
Jia, Q.-S., Zhu, J., Xu, X.-F., Lou, Y., Zhang, Z.-L., Zhang, Z.-P., et al. (2015). Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for nexine formation. Mol. Plant 8 (2), 251–260. doi: 10.1016/j.molp.2014.10.001
Jiang, J., Xu, P., Zhang, J., Li, Y., Zhou, X., Jiang, M., et al. (2022). Global transcriptome analysis reveals potential genes associated with genic male sterility of rapeseed (Brassica napus L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1004781
Kardailsky, I., Shukla, V., Ahn, J. H., Dagenais, N., Christensen, S., Nguyen, J., et al. (1999). Activation tagging of the floral inducer FT. Science 286 (5446), 1962–1965. doi: 10.1126/science.286.5446.1962
Kim, S. S., Grienenberger, E., Lallemand, B., Colpitts, C., Kim, S. Y., de Azevedo Souza, C., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22 (12), 4045–4066. doi: 10.1105/tpc.110.080028
Kinoshita, A., Richter, R. (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71 (9), 2490–2504. doi: 10.1093/jxb/eraa057
Klucher, K., Helen, C., Leonore, R., Robert., F. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8 (2), 137–153. doi: 10.1105/tpc.8.2.137
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki., T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 (5446), 1960–1962. doi: 10.1126/science.286.5446.1960
Koltunow, A., Truettner, J., Cox, K., Wallroth, M., Goldberg., R. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 (12), 1201–1224. doi: 10.1105/tpc.2.12.1201
Krizek, B., Ivory, B., Yen-Yi, H., Nowlan, F., Ann., L. (2020). The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. 103 (2), 752–768. doi: 10.1111/tpj.14769
Krizek, B., Lewis, M., Fletcher., J. (2006). RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant Journal: For Cell Mol. Biol. 45 (3), 369–383. doi: 10.1111/j.1365-313X.2005.02633.x
Lai, Z., Wang, J., Peng, S.-Q., Chang, F. (2022). bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana. Int. J. Mol. Sci. 23 (19), 11683. doi: 10.3390/ijms231911683
Laux, T., Mayer, K., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 (1), 87–96. doi: 10.1242/dev.122.1.87
Lee, J., Oh, M., Park, H., Lee., I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant Journal: For Cell Mol. Biol. 55 (5), 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J. H., Kim, S.-G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14 (18), 2366–2376. doi: 10.1101/gad.813600
Liu, C., Chen, H., Er, H. L., Soo, H. M., Kumar, P., Han, J.-H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135 (8), 1481–1491. doi: 10.1242/dev.020255
Liu, X., Kim, Y. J., Müller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell 23 (10), 3654–3670. doi: 10.1105/tpc.111.091538
Liu, C., Zhou, J., Bracha-Drori, K., Yalovsky, S., Ito, T., Yu., H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134 (10), 1901–1910. doi: 10.1242/dev.003103
Lou, Y., Xu, X.-F., Zhu, J., Gu, J.-N., Blackmore, S., Yang, Z.-N. (2014). The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 5 (1), 3855. doi: 10.1038/ncomms4855
Lou, Y., Zhou, H.-S., Han, Y., Zeng, Q.-Y., Zhu, J., Yang, Z.-N. (2018). Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 217 (1), 378–391. doi: 10.1111/nph.14790
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Mandaokar, A., Browse, J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149 (2), 851–862. doi: 10.1104/pp.108.132597
Mi, L., Mo, A., Yang, J., Liu, H., Ren, D., Che, W., et al. (2022). Arabidopsis novel microgametophyte defective mutant 1 is required for pollen viability via influencing intine development in Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.814870
Michaels, S., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., Amasino, R. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant Journal: For Cell Mol. Biol. 33 (5), 867–874. doi: 10.1046/j.1365-313x.2003.01671.x
Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi., M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17 (11), 2993–3006. doi: 10.1105/tpc.105.036004
Morant, M., Jørgensen, K., Schaller, H., Pinot, F., Møller, B. L., Werck-Reichhart, D., et al. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19 (5), 1473–1487. doi: 10.1105/tpc.106.045948
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154 (3), 1492–1504. doi: 10.1104/pp.110.159111
Nagpal, P., Christine, E., Hans, W., Sara, P., Lana, B., Thomas, G., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 (18), 4107–4118. doi: 10.1242/dev.01955
Oh, S.-A., Hoai, T. N. T., Park, H.-J., Zhao, M., Twell, D., Honys, D., et al. (2020). MYB81, a microspore-specific GAMYB transcription factor, promotes pollen mitosis I and cell lineage formation in Arabidopsis. Plant J. 101 (3), 590–603. doi: 10.1111/tpj.14564
Paxson-Sowders, D., Owen, H., Makaroff., C. (1997). A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma 198 (1), 53–65. doi: 10.1007/BF01282131
Payne, T., Johnson, S., Koltunow., A. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131 (15), 3737–3749. doi: 10.1242/dev.01216
Pelaz, S., Ditta, G., Baumann, E., Wisman, E., Yanofsky., M. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 (6783), 200–203. doi: 10.1038/35012103
Phan, H. A., Iacuone, S., Li, S., Parish, R. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23 (6), 2209–2224. doi: 10.1105/tpc.110.082651
Putterill, J., Robson, F., Lee, K., Simon, R., Coupland., G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 (6), 847–857. doi: 10.1016/0092-8674(95)90288-0
Qi, T., Huang, H., Song, S., Xie, D. (2015). Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27 (6), 1620–1633. doi: 10.1105/tpc.15.00116
Quilichini, T., Samuels, L., Douglas, C. (2014). ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 26 (11), 4483–4498. doi: 10.1105/tpc.114.130484
Reddy, V., Heisler, M., Ehrhardt, D., Meyerowitz., E. (2004). Real-Time Lineage Analysis Reveals Oriented Cell Divisions Associated with Morphogenesis at the Shoot Apex of Arabidopsis thaliana. Development 131 (17), 4225–4237. doi: 10.1242/dev.01261
Rotman, N., Durbarry, A., Wardle, A., Yang, W. C., Chaboud, A., Faure, J.-E., et al. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15 (3), 244–248. doi: 10.1016/j.cub.2005.01.013
Sakai, H., Medrano, L., Meyerowitz, E. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378 (6553), 199–203. doi: 10.1038/378199a0
Sanders, P., Bui, A., Weterings, K., McIntire, K. N., Hsu, Y.-C., Lee, P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reprod. 11 (6), 297–322. doi: 10.1007/s004970050158
Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., Schneitz., K. (1999). Molecular Analysis of NOZZLE, a Gene Involved in Pattern Formation and Early Sporogenesis during Sex Organ Development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. United States America 96 (20), 11664–11669. doi: 10.1073/pnas.96.20.11664
Schultz, E., Pickett, B., Haughn., G. (1991). The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3 (11), 1221–1237. doi: 10.2307/3869229
Shi, J., Cui, M., Yang, L., Kim, Y.-J., Zhang, D. (2015). Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20 (11), 741–753. doi: 10.1016/j.tplants.2015.07.010
Sorensen, A.-M., Kröber, S., Unte, U., Huijser, P., Dekker, K., Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33 (2), 413–423. doi: 10.1046/j.1365-313X.2003.01644.x
Srikanth, A., Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sciences: CMLS 68 (12), 2013–2037. doi: 10.1007/s00018-011-0673-y
Steiner-Lange, S., Gremse, M., Kuckenberg, M., Nissing, E., Schächtele, D., Spenrath, N., et al. (2001). Efficient identification of Arabidopsis knock-out mutants using DNA-arrays of transposon flanking sequences. Plant Biol. 3 (4), 391–397. doi: 10.1055/s-2001-16468
Sun, B., Xu, Y., Ng, K.-H., Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23 (15), 1791–1804. doi: 10.1101/gad.1800409
Tabata, R., Ikezaki, M., Fujibe, T., Aida, M., Tian, C.-e., Ueno, Y., et al. (2010). Arabidopsis AUXIN RESPONSE FACTOR6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51 (1), 164–175. doi: 10.1093/pcp/pcp176
Tang, L. K., Chu, H., Yip, W. K., Yeung, E., Lo., C. (2009). An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 181 (3), 576–587. doi: 10.1111/j.1469-8137.2008.02692.x
Tang, X., Hao, Y.-J., Lu, J.-X., Lu, G., Zhang., T. (2019). Transcriptomic Analysis Reveals the Mechanism of Thermosensitive Genic Male Sterility (TGMS) of Brassica napus under the High Temperature Inducement. BMC Genomics 20, 644. doi: 10.1186/s12864-019-6008-3
Verma, N. (2019). Transcriptional regulation of anther development in Arabidopsis. Gene 689, 202–209. doi: 10.1016/j.gene.2018.12.022
Wang, K., Guo, Z.-L., Zhou, W.-T., Zhang, C., Zhang, Z.-Y., Lou, Y., et al. (2018). The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 178 (1), 283–294. doi: 10.1104/pp.18.00219
Wang, K.-Q., Yu, Y.-H., Jia, X.-L., Zhou, S.-D., Zhang, F., Zhao, X., et al. (2022). Delayed callose degradation restores the fertility of multiple P/TGMS lines in arabidopsis. J. Integr. Plant Biol. 64 (3), 717–730. doi: 10.1111/jipb.13205
Wang, K., Zhao, X., Pang, C., Zhou, S., Qian, X., Tang, N., et al. (2021). IMPERFECTIVE EXINE FORMATION (IEF) is required for exine formation and male fertility in Arabidopsis. Plant Mol. Biol. 105 (6), 625–635. doi: 10.1007/s11103-020-01114-8
Weigel, D., Alvarez, J., Smyth, D., Yanofsky, M., Meyerowitz., E. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 (5), 843–859. doi: 10.1016/0092-8674(92)90295-N
Wilson, Z., Morroll, S., Dawson, J., Swarup, R., Tighe, P. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant Journal: For Cell Mol. Biol. 28 (1), 27–39. doi: 10.1046/j.1365-313x.2001.01125.x
Wilson, Z., Zhang, D.-B. (2009). From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60 (5), 1479–1492. doi: 10.1093/jxb/erp095
Xiong, S.-X., Zeng, Q.-Y., Hou, J.-Q., Hou, L.-L., Zhu, J., Yang, M., et al. (2020). The temporal regulation of TEK contributes to pollen wall exine patterning. PloS Genet. 16 (5), e1008807. doi: 10.1371/journal.pgen.1008807
Xu, Y., Iacuone, S., Li, S. F., Parish, R. W. (2014). MYB80 homologues in Arabidopsis, cotton and brassica: regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 14, 271. doi: 10.1186/s12870-014-0278-3
Xu, X.-F., Wang, B., Feng, Y.-F., Xue, J.-S., Qian, X.-X., Liu, S.-Q., et al. (2019). AUXIN RESPONSE FACTOR17 directly regulates MYB108 for anther dehiscence. Plant Physiol. 181 (2), 645–655. doi: 10.1104/pp.19.00576
Xu, J., Yang, C., Yuan, Z., Zhang, D., Gondwe, M., Ding, Z., et al. (2010). The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22 (1), 91–107. doi: 10.1105/tpc.109.071803
Xu, J., Zhiwen, D., Gema, V.-B., Jianxin, S., Wanqi, L., Zheng, Y., et al. (2014). ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. . Plant Cell 26 (4), 1544–1556. doi: 10.1105/tpc.114.122986
Xue, J.-S., Qiu, S., Jia, X.-L., Shen, S.-Y., Shen, C.-W., Wang, S., et al. (2023). Stepwise changes in flavonoids in spores/pollen contributed to terrestrial adaptation of plants. Plant Physiol. 193 (1), 627–642. doi: 10.1093/plphys/kiad313
Xue, J.-S., Yao, C., Xu, Q.-L., Sui, C.-X., Jia, X.-L., Hu, W.-J., et al. (2021). Development of the middle layer in the anther of Arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.634114
Yamaoka, S., Nishihama, R., Yoshitake, Y., Ishida, S., Inoue, K., Saito, M., et al. (2018). Generative cell specification requires transcription factors evolutionarily conserved in land plants. Curr. Biol. 28 (3), 479–486.e5. doi: 10.1016/j.cub.2017.12.053
Yang, L., Jing, Z., Junna, H., Yingying, Q., Deping, H., Ying, D., et al. (2014). ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PloS Genet. 10 (12), e1004791. doi: 10.1371/journal.pgen.1004791
Yang, X., Makaroff, C., Ma., H. (2003). The arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15 (6), 1281–1295. doi: 10.1105/tpc.010447
Yang, C., Vizcay-Barrena, G., Conner, K., Wilson., Z. A. (2007a). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19 (11), 3530–3548. doi: 10.1105/tpc.107.054981
Yang, S.-L., Xie, L.-F., Mao, H.-Z., Puah, C. S., Yang, W.-C., Jiang, L., et al. (2003). TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15 (12), 2792–2804. doi: 10.1105/tpc.016618
Yang, C., Xu, Z., Song, J., Conner, K., Barrena, G. V., Wilson, Z. (2007b). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19 (2), 534–548. doi: 10.1105/tpc.106.046391
Yang, W.-C., Ye, D., Xu, J., Sundaresan., V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 (16), 2108–2117. doi: 10.1101/gad.13.16.2108
Yanofsky, M., Ma, H., Bowman, J., Drews, G., Feldmann, K., Meyerowitz, E. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 (6279), 35–39. doi: 10.1038/346035a0
Ye, Q., Wenjiao, Z., Lei, L., Shanshan, Z., Yanhai, Y., Hong, M., et al. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. United States America 107 (13), 6100–6105. doi: 10.1073/pnas.0912333107
Yin, Y., Zhi-Yong, W., Santiago, M.-G., Jianming, L., Shigeo, Y., Tadao, A., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 (2), 181–191. doi: 10.1016/S0092-8674(02)00721-3
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139 (2), 770–778. doi: 10.1104/pp.105.066928
Yu, H., Ito, T., Wellmer, F., Meyerowitz, E. M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36 (2), 157–161. doi: 10.1038/ng1286
Zhang, X., He, Y., Li, L., Liu, H., Hong, G. (2021). Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J. Exp. Bot. 72 (12), 4319–4332. doi: 10.1093/jxb/erab156
Zhang, J., Huang, Q., Zhong, S., Bleckmann, A., Huang, J., Guo, X., et al. (2017). Sperm cells are passive cargo of the pollen tube in plant fertilization. Nat. Plants 3 (6), 17079. doi: 10.1038/nplants.2017.79
Zhang, D., Liu, D., Lv, X., Wang, Y., Xun, Z., Liu, Z., et al. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26 (7), 2939–2961. doi: 10.1105/tpc.114.127282
Zhang, W., Yujin, S., Ljudmilla, T., Changbin, C., Ueli, G., Hong, M. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133 (16), 3085–3095. doi: 10.1242/dev.02463
Zhang, Z.-B., Zhu, J., Gao, J.-F., Wang, C., Li, H., Li, H., et al. (2007). Transcription factor atMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis: molecular cloning and functional analysis of atMYB103. Plant J. 52 (3), 528–538. doi: 10.1111/j.1365-313X.2007.03254.x
Zhao, D.-Z., Wang, G.-F., Speal, B., Ma., H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 (15), 2021–2031. doi: 10.1101/gad.997902
Zheng, L., Nagpal, P., Villarino, G., Trinidad, B., Bird, L., Huang, Y., et al. (2019). miR167 limits anther growth to potentiate anther dehiscence. Development 146 (14), dev174375. doi: 10.1242/dev.174375
Zhu, J., Chen, H., Li, H., Gao, J.-F., Jiang, H., Wang, C., et al. (2008). Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55 (2), 266–277. doi: 10.1111/j.1365-313X.2008.03500.x
Keywords: flower, anther, pollen, reproductive development, transcription factors, pathways
Citation: Wiese AJ, Torutaeva E and Honys D (2024) The transcription factors and pathways underpinning male reproductive development in Arabidopsis. Front. Plant Sci. 15:1354418. doi: 10.3389/fpls.2024.1354418
Received: 12 December 2023; Accepted: 15 January 2024;
Published: 08 February 2024.
Edited by:
Helena Fernández, University of Oviedo, SpainReviewed by:
Jingshi Xue, Shanghai Normal University, ChinaSubramanian Sankaranarayanan, Indian Institute of Technology Gandhinagar, India
Copyright © 2024 Wiese, Torutaeva and Honys. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Honys, aG9ueXNAdWViLmNhcy5jeg==
 Anna Johanna Wiese
Anna Johanna Wiese Elnura Torutaeva1,2
Elnura Torutaeva1,2 David Honys
David Honys