
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 April 2024
Sec. Plant Systematics and Evolution
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1349358
The genus Argentina Hill belongs to the tribe Potentilleae Sweet and contains approximately 75 species predominantly distributed in the Sino-Himalayan region and the Malesian archipelago. So far we have less knowledge on the phylogenetic relationships within Argentina owing to limited sampling of Argentina taxa or gene fragments in previous studies. Moreover, to date there is no phylogenetic study on Argentina from the perspective of comparative chloroplast (cp) genomics. Here we performed comparative genomic analyses on the cp genomes of 39 accessions representing 18 taxa of Argentina. The Argentina cp genomes presented the typical quadripartite structure, with the sizes ranging from 155 096 bp to 157 166 bp. The 39 Argentina cp genomes contained a set of 112 unique genes, comprising four ribosomal RNA (rRNA) genes, 30 transfer RNA (tRNA) genes, as well as 78 protein-coding genes (PCGs). The cp genome organization, gene content and order in Argentina were highly conserved, but some visible divergences were present in IR/SC boundary regions. Ten regions (trnH-GUG-psbA, trnG-GCC-trnfM-CAU, trnD-GUC-trnY-GUA, rpl32-trnL-UAG, atpH-atpI, rps16-trnQ-UUG, trnS-GCU-trnG-UCC, ndhF-rpl32, trnR-UCU-atpA, and accD-psaI) were identified as excellent candidate DNA markers for future studies on species identification, population genetics and phylogeny of Argentina. Our results indicated that Argentina is monophyletic. In the current sampling, the A. smithiana - A. anserina clade was sister to the remainder of Argentina. Our results corroborated the previous taxonomic treatments to transfer A. phanerophlebia and A. micropetala from the genus Sibbaldia L. to Argentina. Our results showed close relationships among A. stenophylla, A. microphylla, A. taliensis, and A. tatsienluensis, congruent with previous studies based on the morphology of these species. Twenty-six genes (rps3, rps15, rps16, rps19, rpl16, rpl20, rpl22, rpoA, rpoB, rpoC1, rpoC2, atpA, atpF, psbB, psbF, ndhA, ndhB, ndhC, ndhD, ndhF, rbcL, accD, ccsA, matK, ycf1, ycf2) were with sites under positive selection, and adaptive evolution of these genes might have played crucial roles in Argentina species adaptation to the harsh mountain environment. This study will facilitate future work on taxonomy, phylogenetics, and adaptive evolution of Argentina.
In green plants, the chloroplast (cp) is a unique semi-autonomous organelle (Palmer, 1991; Sato et al., 2003), playing a vital role during photosynthesis and synthesis of metabolites (Neuhaus and Emes, 2000; Rodríguez-Ezpeleta et al., 2005). The land plants cp genomes are circular form generally ranging from 115 to 165 kb and containing 120–130 genes (Ravi et al., 2008; Daniell et al., 2016), and those cp genomes are usually quadripartite in structure, with a small single-copy region (SSC) and a large single-copy region (LSC) divided by two inverted repeats (IRs) (Wicke et al., 2011; Daniell et al., 2016). Compared with mitochondrial or nuclear genomes, the cp genomes of land plants are relatively highly conserved in structure, sequence, gene content and order (Palmer, 1991; Wicke et al., 2011; Mower and Vickrey, 2018). Because of advantages of the cp genome such as characterized by usually uniparental inheritance, lack of genetic recombination, and a moderate nucleotide substitution rate (Palmer, 1985; Wolfe et al., 1987; Drouin et al., 2008; Ravi et al., 2008; Wicke et al., 2011; Mower and Vickrey, 2018), cp genomes have been the primary workhorse for researches of phylogenetics, taxonomy, evolution, and species identification in land plants (Zhang et al., 2017; Niu et al., 2018; Fu et al., 2019; Sousa et al., 2020; Daniell et al., 2021; Wu et al., 2021; Du et al., 2022; Wang et al., 2022; Zhang et al., 2022; Hu et al., 2023; Xu et al., 2023; Zhou et al., 2024).
The genus Argentina Hill, belonging to the tribe Potentilleae Sweet (Dobeš and Paule, 2010; Feng et al., 2015; 2017; Li et al., 2024), comprises approximately 75 species predominantly distributed in the Sino-Himalayan region and the Malesian archipelago (Li et al., 2003; Soják, 2010; Kechaykin and Shmakov, 2016). Hill (1756) first separated Argentina from Potentilla L. Rydberg (1898, 1908) supported Hill’s treatment based upon the differences in position of style; Argentina possesses lateral styles, while Potentilla has subterminal ones. Later botanists rarely accepted the genus Argentina and treated it as an infrageneric group of the genus Potentilla (Wolf, 1908; Yü and Li, 1980, 1985; Soják, 1994; Ikeda and Ohba, 1999; Li et al., 2003). Soják (1989, 1994, 2004, 2008) conducted a series of studies on the taxonomic rank and delimitations of Argentina. Until 2010, Soják found that the difference in stipule structure is a main morphological character that distinguishes Argentina from Potentilla. The consistent difference is that Argentina has ventral stipular auricles, but Potentilla s. str. possesses lateral ones (Soják, 2010). Soják (2010, 2012a, b, c) recognized Argentina as a separate genus, according to the difference in stipule structure and results of phylogenetic studies (Eriksson et al., 2003; Dobeš and Paule, 2010). Previous phylogenetic studies (Eriksson et al., 2003; Dobeš and Paule, 2010; Feng et al., 2015, 2017; Li et al., 2024) support that Argentina is distinct from Potentilla. So far, there is no phylogenetic study on Argentina from the perspective of comparative chloroplast genomics. We have little knowledge on the phylogenetic relationships within Argentina because of the limited taxon sampling of Argentina or gene fragments in previous studies, which mainly concentrated on phylogenetics of Rosaceae, Rosoideae, Potentilleae, Fragariinae, Potentilla, or Sibbaldia or report of new Argentina species (Potter et al., 2002, 2007; Eriksson et al., 2003; Lundberg et al., 2009; Dobeš and Paule, 2010; Töpel et al., 2011; Eriksson et al., 2015; Feng et al., 2015, 2017; Persson et al., 2020; Li et al., 2024; Xue et al., 2024). Divergence time estimates indicated that the crown group of Argentina originated in the early Miocene (ca. 18.64 Ma), with relatively old origin compared with other genera within Potentilleae (Li et al., 2024). Argentina species are mostly found in the alpine or subalpine regions, sometimes as the dominant plants (Kalkman, 1989, 1993; Li et al., 2003), and the study of their cp genome adaptation to high-altitude environments is a fascinating question.
Here we conducted comparative genomic analyses on the cp genomes of 39 accessions representing 18 taxa of Argentina by means of bioinformatics. The objectives were to (1) analyze features of Argentina cp genomes, (2) screen divergence hotspots as candidate molecular markers and potential specific barcodes for Argentina, (3) provide insights into the phylogenetic relationships among Argentina species, and (4) explore the adaptive evolution of chloroplast genes of Argentina species. This study will contribute to further studies on species identification, population genetics, phylogenetics, and cp genome evolution of Argentina, and provide a theoretical basis for conservation efforts of Argentina.
In our study, a total of 18 Argentina taxa represented by 39 Argentina accessions were sampled. In addition, seven Potentilla species were chosen as outgroups in the phylogenomic analyses based on previous work (Feng et al., 2017; Zhang et al., 2017; Li et al., 2024). GenBank accession numbers and voucher information of the sampled taxa are presented in Table 1. For fresh or silica-dried leaf samples, genomic DNA was isolated by the CTAB protocol (Doyle and Doyle, 1987). For herbarium leaves sample, genomic DNA was extracted by the SDS method (Dellaporta et al., 1983; Johnson et al., 2023). The extracted DNA was sheared into fragments using sonication. These fragments were used for short-insert library construction with 300 bp insert size by NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®. Finally, the Illumina HiSeq platform in Novogene was used to sequence the pooled libraries.
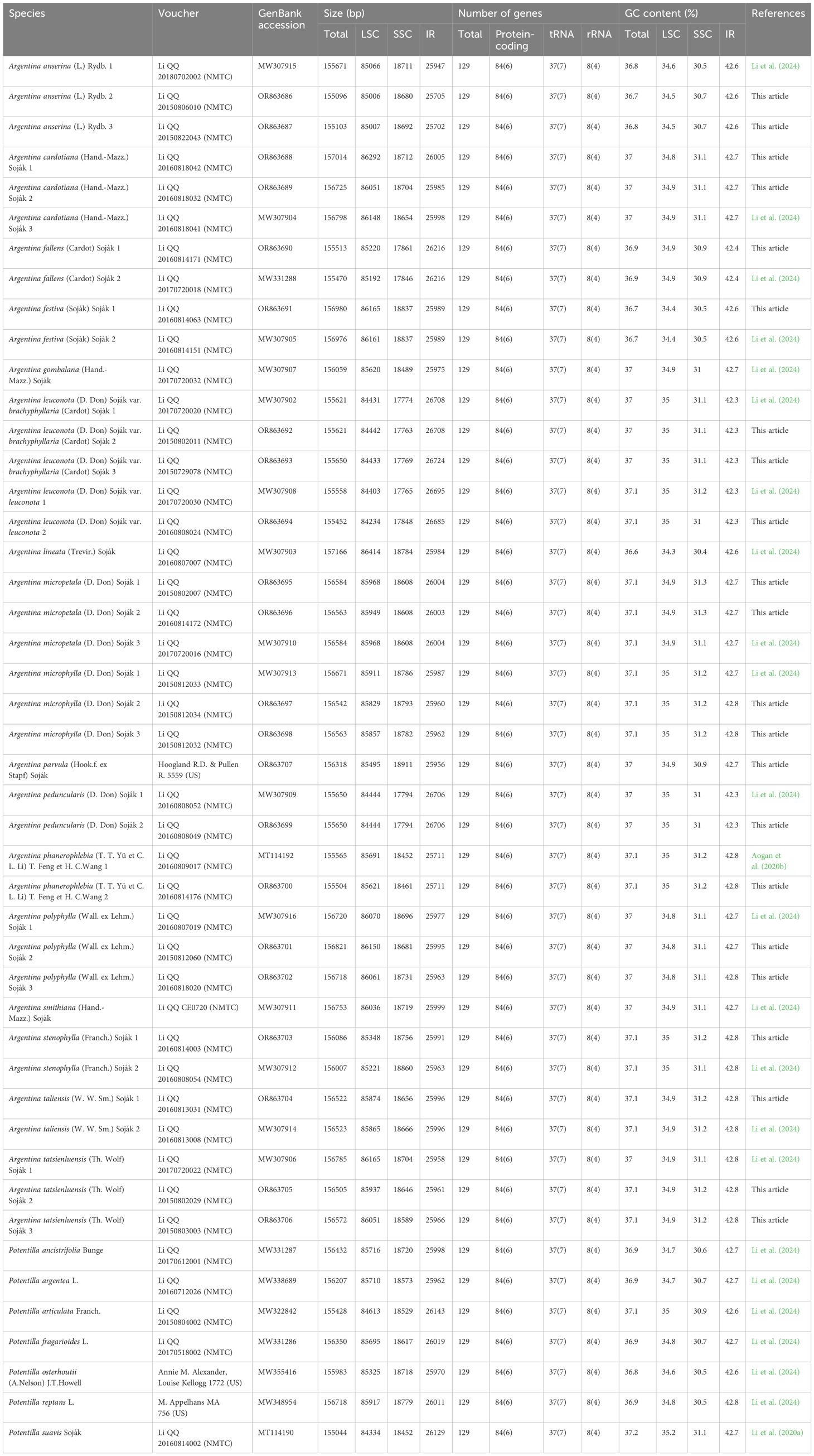
Table 1 Summary of voucher specimens and chloroplast genome characteristics for Argentina species and related outgroups.
Trimmomatic v. 0.33 (Bolger et al., 2014) was utilized in order to clean adapters in raw high-throughput sequencing data. FastQC v. 0.11.8 (Andrews, 2018) was employed to evaluate the quality of the filtered paired-end reads. Then the filtered raw reads of each accession were used for assembly of the cp genome sequence by NOVOPlasty (Dierckxsens et al., 2017), with the parameters of genome range 120000–220000 and K-mer 39. In cp genome sequence assemblies of Argentina species, rbcL gene in Argentina phanerophlebia (GenBank accession no. MT114192) was set as the seed and its cp genome was used as the reference. Using cp genome of Argentina phanerophlebia (MT114192) as the reference, cp genome annotation of Argentina species was performed using Geneious Prime (Kearse et al., 2012) by transferring annotations. The initial annotation results were then manually checked and adjusted in Geneious Prime.
The statistics of genome size, LSC/SSC/IR size, number of genes and GC content were summarized in Geneious Prime. With the purpose of detecting potential rearrangements and inversions, the cp genomes alignment of Argentina species was implemented in MAUVE v. 2.4.0 using the progressiveMauve algorithm (Darling et al., 2004, 2010). The divergence in the LSC/IR/SSC boundaries among 39 cp genomes in Argentina was compared and illustrated to detect the IR expansion/contraction. The mVISTA program (Frazer et al., 2004) was employed to visualize the divergence level among 39 Argentina cp genomes using Shuffle-LAGAN mode and with A. anserina (L.) Rydb. 1 as a reference. To screen divergence hotspots, we extracted the coding and noncoding regions separately in 39 Argentina cp genomes by “extract annotations” in Geneious Prime and furthermore aligned these homologous loci by MAFFT v. 7.450 (Katoh and Standley, 2013). Finally, the nucleotide variability (Pi) of each homologous locus was calculated in DnaSP v. 6.0 (Rozas et al., 2017).
Phylogenetic relationships among the 18 Argentina taxa were inferred by maximum likelihood (ML) and Bayesian inference (BI) methods, with seven Potentilla species as outgroups to root the trees. A total of 46 cp genome sequences were aligned in MAFFT v. 7.450 (Katoh and Standley, 2013) with default parameters. Software trimAL v. 1.4 (Capella-Gutiérrez et al., 2009) was subsequently used to trim the alignment properly. The ML analysis was performed by RAxML v. 8.2.12 (Stamatakis, 2014), under GTRGAMMA model (option “-m GTRGAMMA”) as suggested in the manual, with analysis of rapid bootstrap and search for best-scoring ML tree (option “-f a”) and 1000 replicates bootstrap (option “-N 1000”). MrBayes v. 3.2.7a (Ronquist et al., 2012) was employed to conduct the BI analysis under best-fit model GTR+I+G as recommended by PartitionFinder2 (Lanfear et al., 2017) using the Corrected Akaike Information Criterion (AICc; Sugiura, 1978) according to Posada and Buckley (2004). Four parallel runs were performed, each run with three heated and one cold Markov chain Monte Carlo (MCMC) chains for 6000 000 generations, sampling one tree every 100 generations as well as starting from random trees. The initial 25% of the trees were regarded as burn-in and discarded. A majority-rule consensus tree was generated using the remaining trees. FigTree v. 1.4.4 (Rambaut, 2018) was finally used to visualize the phylogenetic trees.
To identify selection pressures on the Argentina cp genomes, non-synonymous (Ka), synonymous (Ks), and Ka/Ks ratios of 78 protein-coding genes (PCGs) of 39 Argentina accessions were calculated, with Potentilla reptans as the reference. Geneious Prime (Kearse et al., 2012) was employed to extract the 78 PCGs shared among the Argentina species and Potentilla reptans. The amino acids sequences and the relative nucleotide sequences were then aligned and converted into codon alignments by ParaAT v.2.0 (Zhang et al., 2012) with MAFFT as the multiple sequence aligner and with the 11th genetic code (-c 11). The KaKs_Calculator 2.0 program (Wang et al., 2010) was subsequently utilized for the analysis of Ka, Ks, and Ka/Ks ratios, with the 11th genetic code and the default model averaging (MA) method.
We also used site models in CodeML (Yang, 2007) executed in EasyCodeML (Gao et al., 2019) to detect positively selected sites of PCGs in 39 Argentina cp genomes. Firstly, 78 PCGs common to the Argentina species and Potentilla species were extracted in the cp genomes by Geneious Prime (Kearse et al., 2012). Each PCG was aligned according to its codons under MAFFT, followed by manually removing stop codons, and used as input for EasyCodeML. Moreover, these alignments were concatenated into a supermatrix and then the ML tree was established by RAxML v. 8.2.12 (Stamatakis, 2014) as an input tree. Likelihood ratio tests (LRTs) of M7 (beta) vs. M8 (beta and ω >1) and M8a (beta and ω = 1) vs. M8 were performed for detecting positive selection sites. If the LRTs were significant (p-values < 0.05), the Bayes empirical Bayes (BEB) (Yang et al., 2005) analysis was adopted in order to identify positively selected sites with the posterior probabilities threshold of 0.95.
The cp genomes sizes among 39 cp genomes from 18 Argentina taxa ranged from 155,096 bp (A. anserina 2) to 157,166 bp (A. lineata) (Table 1; Figure 1), which was within the cp genome size range in most land plants (Ravi et al., 2008). Argentina cp genomes presented the typical quadripartite structure, which is consistent with that of most other land plants (Wicke et al., 2011; Daniell et al., 2016), including taxa of Rosaceae (Du et al., 2021; Li et al., 2021a; Tang et al., 2022; Wu et al., 2022; Yu et al., 2022; Zhang et al., 2023) (Figure 1). These cp genomes comprised two IRs separating the SSC and LSC region, with lengths of IRs from 25,702 bp (A. anserina 3) to 26,724 bp (A. leuconota var. brachyphyllaria 3), and with lengths of LSC and SSC from 84, 234 bp (A. leuconota var. leuconota 2) to 86,414 bp (A. lineata) and from 17,763 bp (A. leuconota var. brachyphyllaria 2) to 18,911 bp (A. parvula) respectively. The total GC content in Argentina cp genomes was 36.6–37.1% (Table 1), which is roughly comparable to that in other cp genomes of Potentilleae species (e.g., Li et al., 2020b; Rono et al., 2020; Tian et al., 2020; Zhang et al., 2020; Aogan and Li, 2020a; Li et al., 2021b). In addition, GC content in the IR region (42.3–42.8%) was higher compared with that in the SSC and LSC regions (30.4–31.3% and 34.3–35%, respectively), and this phenomenon also exists in cp genomes of other plants (e.g., Liu et al., 2021; Ogoma et al., 2022; Bai et al., 2023; Waswa et al., 2023; Zoclanclounon et al., 2023). The highest GC content in the IR region is ascribed to the existence of rRNA genes (Ravi et al., 2008).
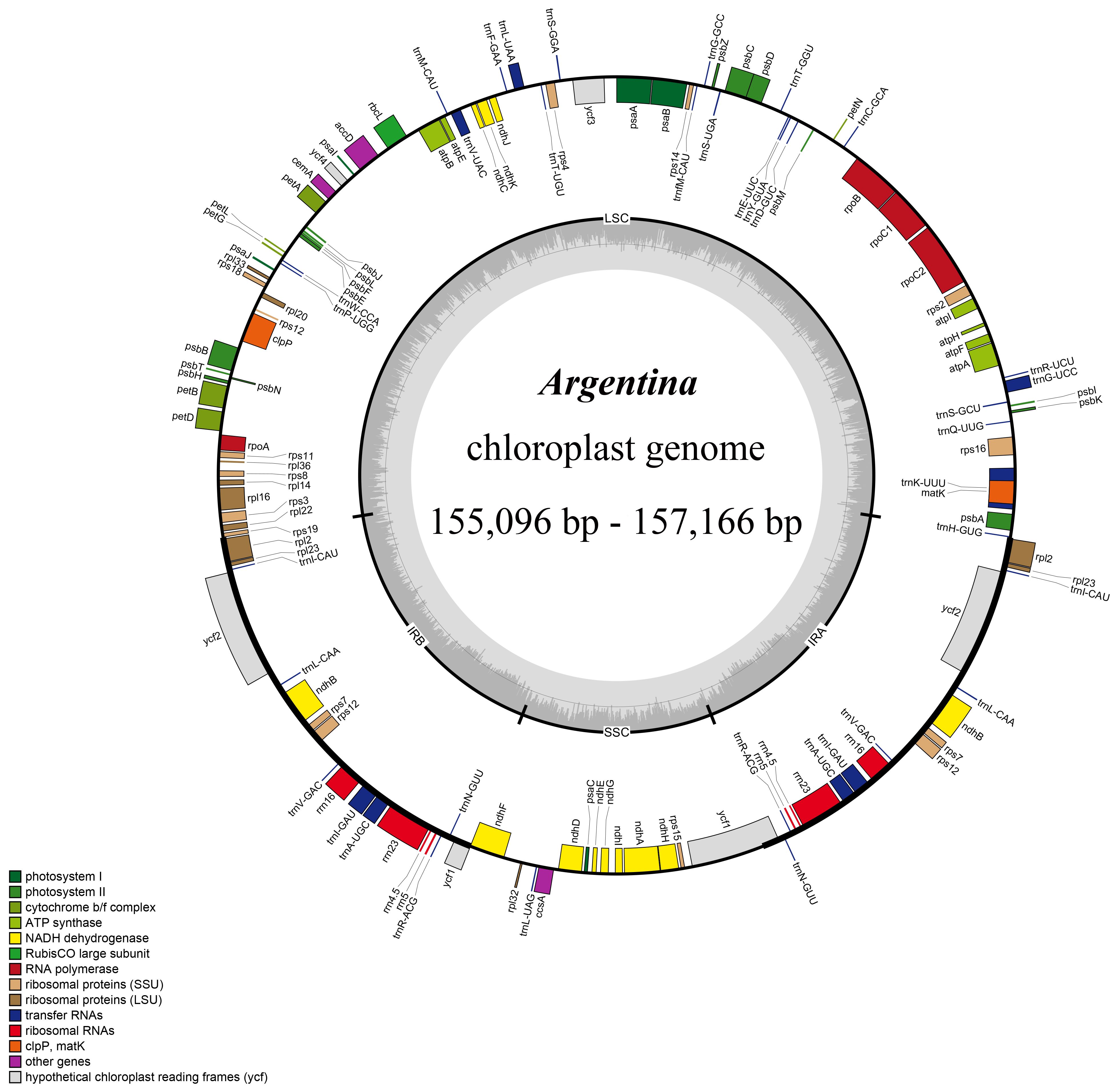
Figure 1 Chloroplast genome map of Argentina species. Genes inside the circle are transcribed clockwise, whereas those outside are transcribed counter-clockwise. The dark and light gray area in the inner circle corresponds to the GC content and AT content, respectively.
All of the 39 Argentina cp genomes contained a set of 112 unique genes, comprising four ribosomal RNA (rRNA) genes, 30 transfer RNA (tRNA) genes, and 78 protein-coding genes (PCGs) (Figure 1; Table 1; Supplementary Table S1). Among the 112 unique genes, four rRNA genes (rrn4.5, rrn5, rrn16, rrn23), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), and six PCGs (ndhB, rpl2, rpl23, rps7, rps12, ycf2) were duplicated. Additionally, three genes (clpP, rps12, ycf3) possessed two introns, while 14 genes (ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) embraced a single intron (Supplementary Table S1).
Mauve alignment analysis of 39 Argentina cp genomes showed no gene rearrangement and inversion and Argentina cp genomes had good collinearity (Supplementary Figure S1). Although the cp genome organization, gene content and order in Argentina were highly conserved, some visible divergences were present in IR/SC boundary regions (Figure 2). The variation of cp genome size across land plants is mainly attributed to expansion and contraction of the IR (Ravi et al., 2008; Mower and Vickrey, 2018). Comparative analysis of IR boundaries indicated that border genes were identical among Argentina cp genomes, but slight differences existed in lengths of these genes (ndhF and ycf1) and relative positions of these genes to the boundaries. In junctions of the LSC/IR and SSC/IR regions, the genes rps19-rpl2-trnH and ycf1-ndhF were located, with rpl2 and ycf1 duplicated or partially duplicated respectively in IR regions. The LSC/IRb junction (JLB) occurred between rps19 gene and one rpl2 gene. Gene rps19 with 279 bp in length occurred entirely in the LSC region with 6–19 bp away from the JLB, while rpl2 with 825 bp in length was placed completely in the IRb region, and the distance between the rpl2 and the JLB were 48–74bp. The IRb/SSC junction (JSB) was located on the truncated ycf1 (ψycf1) and ndhF. The ψycf1 pseudogene with 1071–1842 bp in length spanned the JSB boundary, with a length of 3–30 bp and 1062–1824 bp in the SSC and IRb region, separately. The length of ndhF was 2238–2280 bp, and the overlap between ndhF and ψycf1 existed, in which ndhF expanded into the IRb region for 0–46 bp. The length of ycf1 was 5703–5802 bp and the gene crossed over the SSC/IRa junction (JSA), with a length of 3909–4719 bp and 1062–1824 bp in the SSC and IR region. The IRa/LSC junction (JLA) was located between the other rpl2 and trnH-GUG. Located in the IRa region, rpl2 was separated from the JLA by a spacer varying from 48 bp to 74 bp. Gene trnH-GUG with 74bp in length was located in the LSC region, with 0–11bp away from the JLA. In general, cp genomes boundaries of Argentina species were greatly conserved and no obvious contraction and expansion existed in the IR region, which further supported that IR boundary shifts were relatively minor in closely related species (Zhu et al., 2016; Liu et al., 2018; Ren et al., 2022; Zhang et al., 2022). The similar findings were also observed in other genera in the tribe Potentilleae, such as Alchemilla (Rono et al., 2020), Chamaerhodos (Li et al., 2021a), and Fragaria (Li et al., 2021b). In addition, the boundary features of different populations of the same species were basically stable, although there are some exceptions.
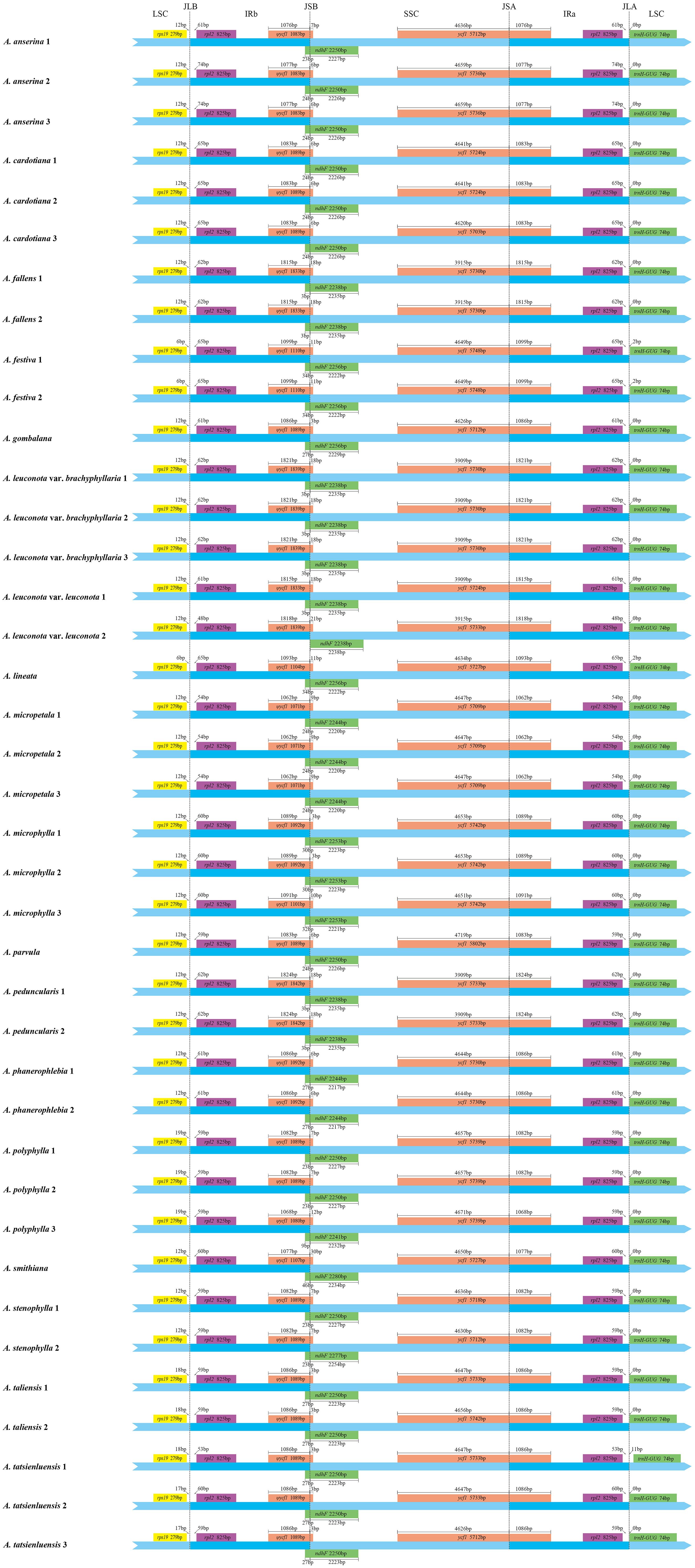
Figure 2 Comparison of the boundaries of the large single-copy (LSC), the small single-copy (SSC) and inverted repeat (IR) regions among 39 Argentina chloroplast genomes. JLB: junction line between LSC and IRb; JSB: junction line between SSC and IRb; JSA: junction line between SSC and IRa; JLA: junction line between LSC and IRa. The figure is not to scale with respect to sequence length and only shows relative changes at or near the IR/SC boundaries.
The mVISTA program (Frazer et al., 2004) was employed to visualize the divergence level among 39 Argentina cp genomes using Shuffle-LAGAN mode and with A. anserina 1 as the reference. The divergence level of the 39 Argentina cp genomes was investigated and plotted using mVISTA (Supplementary Figure S2). Global comparisons of the genomic sequences revealed that at the genome-scale level, Argentina cp genomes were conserved, with a high degree of similarity and synteny. Additionally, the IR regions were more conserved than the SSC and LSC regions, which is a common phenomenon in most angiosperm cp genomes. Overall, compared with the coding regions, the non-coding regions were more divergent, with the highly variable non-coding regions occurred in the intergenic spacers (IGS). This finding was consistent with studies on cp genomes of other angiosperm taxa (e.g., Li et al., 2021a; Zhang et al., 2022; Bai et al., 2023).
In summary, the size, structure, GC contents, gene content and order of the Argentina cp genomes were highly conserved, only with slight differences in the cp genome length, GC content, and IR/SC boundary region for each species.
To detect the highly variable regions, the software DnaSP v. 6.0 (Rozas et al., 2017) was utilized to analyze values of the nucleotide variability (Pi) of a total of 263 exons, introns and the IGS regions of Argentina. The range of Pi values was 0–0.08360 and the mean value was 0.01479, which indicated Argentina cp genomes possessed a high level of similarity (Supplementary Table S2; Figure 3). Overall, 40 regions with Pi =0, 97 regions with 0<Pi ≤ 0.01, 51 regions with 0.01<Pi ≤ 0.02, 33 regions with 0.02<Pi ≤ 0.03, 23 regions with 0.03<Pi ≤ 0.04, and 19 regions with Pi>0.04. The results showed that regions located in the IR region had relatively low Pi values, so compared with the SSC and LSC regions, the IR region was less divergent. Moreover, compared to the coding regions, the non-coding regions were relatively more variable. Most of the highly variable regions were presented in the IGS. Nineteen regions (trnH-GUG-psbA, atpA-atpF, trnG-GCC-trnfM-CAU, trnG-UCC-trnR-UCU, rpl33-rps18, rps4-trnT-UGU, trnD-GUC-trnY-GUA, trnL-UAG-ccsA, rpl22-rps19, rpl14-rpl16, rpl32-trnL-UAG, atpH-atpI, ndhI-ndhA, rps16-trnQ-UUG, trnS-GCU-trnG-UCC, ndhF-rpl32, trnR-UCU-atpA, accD-psaI, petD-rpoA) with Pi>0.04 were all intergenic spacer sequences, of which four regions (trnL-UAG-ccsA, rpl32-trnL-UAG, ndhI-ndhA, ndhF-rpl32) are in the SSC region, and the other 15 regions are in the LSC region. Twenty-three regions (5’-rps12-clpP, rps19-rpl2, 5’-trnK-UUU-rps16, psbI-trnS-GCU, rpl36-rps8, petN-psbM, atpF-atpH, ndhC-trnV-UAC, psbK-psbI, psaI-ycf4, ndhE-ndhG, psbC-trnS-UGA, rps8-rpl14, trnT-UGU-trnL-UAA, rps18-rpl20, rps15-ycf1, psaJ-rpl33, ccsA-ndhD, trnP-UGG-psaJ, petA-psbJ, rps3-rpl22, cemA-petA, trnF-GAA-ndhJ) with 0.03<Pi ≤ 0.04 were also intergenic spacer sequences; among these regions, rps19-rpl2 was in the LSC/IR boundary, three regions (ndhE-ndhG, rps15-ycf1, ccsA-ndhD) occurred in the SSC region, and the remaining 19 regions were in the LSC region. The variation range of Pi values of PCGs was 0–0.02264. There were 21 PCGs (ycf1, petL, matK, atpF, ccsA, ndhF, rpl20, rpl33, ndhD, rps19, rpoC2, rps3, ndhI, rpl22, ndhE, rps15, rpoA, accD, ndhG, cemA, ndhH) with Pi>0.01, of which ycf1 was in the SSC/IR boundary, ndhF occurred in the IR/SSC boundary, seven regions (ccsA, ndhD, ndhI, ndhE, rps15, ndhG, ndhH) were present in the SSC region, and the remaining 12 regions were in the LSC region.
Both the sequence variation and the sequence length should be considered when screening hypervariable regions as candidate molecular markers, because if the region length is short, it cannot provide enough informative sites. Among the 19 regions with Pi>0.04, ten regions (trnH-GUG-psbA, trnG-GCC-trnfM-CAU, trnD-GUC-trnY-GUA, rpl32-trnL-UAG, atpH-atpI, rps16-trnQ-UUG, trnS-GCU-trnG-UCC, ndhF-rpl32, trnR-UCU-atpA, accD-psaI) with suitable lengths were identified as candidate DNA markers. Among the 23 regions with 0.03<Pi ≤ 0.04, 12 regions (5’-trnK-UUU-rps16, rpl36-rps8, petN-psbM, atpF-atpH, ndhC-trnV-UAC, psaI-ycf4, trnT-UGU-trnL-UAA, rps15-ycf1, psaJ-rpl33, trnP-UGG-psaJ, petA-psbJ, trnF-GAA-ndhJ) with suitable lengths were selected as useful alternative molecular markers. Although the protein-coding genes were relatively conserved, we proposed that 15 protein-coding genes (ycf1, matK, atpF, ccsA, ndhF, ndhD, rpoC2, rps3, ndhI, rpl22, rpoA, accD, ndhG, cemA, ndhH) with suitable lengths could be utilized as potential DNA markers where there is a lack of information of other excellent molecular markers. Recommended by CBOL Plant Working Group (2009), rbcL is among the core barcodes for land plants, however, its Pi value in our study was only 0.00790. Compared with other above-mentioned molecular markers, rbcL has a relatively low sequence variation, so it is not suitable as a candidate molecular marker of Argentina. In previous studies on Argentina and its related taxa, chloroplast molecular markers matK, ndhF, rbcL, trnC-GCA-ycf6 (trnC-GCA-petN), trnL intron, trnL-UAA-trnF-GAA, trnS-trnG, and trnS-UGA-ycf9 (trnS-UGA-psbZ) were used, among which only matK, ndhF, and trnS-trnG belonged to the candidate molecular markers developed for Argentina. Thus, findings here indicated the necessity to develop exclusive molecular markers for particular groups. In general, the new candidate molecular markers developed in our study will facilitate studies on species identification, population genetics and phylogenetic studies of Argentina.
In recent years, cp genome sequences have been widely applied in plant phylogenetic studies, owning to its own merits such as containing more variable sites than the single fragment or the combination of several fragments (Wu et al., 2021; Zhang et al., 2021). In our present study, phylogenetic relationships among Argentina species were explored based on the cp genome sequences using ML and BI methods. The ML and BI analyses recovered identical topologies, with high maximum likelihood bootstrap support values (ML BS) and posterior probabilities (PP) across most nodes. Therefore, only the ML tree was presented in Figure 4. All the currently sampled Argentina species were clustered together with high support (Figure 4, ML BS = 100%, PP = 1.00). Consistent with former studies (Feng et al., 2015, 2017; Li et al., 2024), our phylogenetic results strongly supported that Argentina is monophyletic. Based on our current sampling, it is possible to get a glimpse into the relationships within Argentina. Within the sampled Argentina species, the clade of A. smithiana (Hand.-Mazz.) Soják and A. anserina (L.) Rydb. was sister to the remainder of the Argentina (Figure 4, ML BS = 100%, PP = 1.00). Argentina phanerophlebia (T.T. Yü et C. L. Li) T. Feng et H. C.Wang and A. micropetala (D. Don) Soják were embedded in the remaining Argentina species, and the result corroborated the previous taxonomic treatments to transferring those two species from the previously accepted genus Sibbaldia L. to Argentina (Soják, 2010; Feng et al., 2015). Argentina cardotiana (Hand.-Mazz.) Soják was sister to a clade including A. gombalana (Hand.-Mazz.) Soják, A. fallens (Cardot) Soják, A. peduncularis (D. Don) Soják, A. leuconota (D. Don) Soják, and A. leuconota var. brachyphyllaria (Cardot) Soják (ML BS = 100%, PP = 1.00). Argentina festiva (Soják) Soják together with A. lineata (Trevir.) Soják was sister to a clade composed of A. parvula (Hook.f. ex Stapf) Soják and A. polyphylla (Wall. ex Lehm.) Soják. It is worth noting that the Malesian archipelago taxon, A.parvula had a close affinity to A. festiva, A. lineata, and A. polyphylla. In terms of distribution, A. festiva and A. lineata are predominantly distributed in the Sino-Himalayan region, while A. polyphylla occurs in the Malesian archipelago and Sino-Himalayan region. A broader sampling of Argentina species from the Malesian archipelago is necessary in order to decipher the phylogenetic relationships between the Sino-Himalayan taxa and the Malesian archipelago taxa in further studies. Argentina stenophylla (Franch.) Soják was sister to a clade comprising A. microphylla (D. Don) Soják, A. taliensis (W. W. Sm.) Soják, and A. tatsienluensis (Th. Wolf) Soják. The close relationship of these species indicated by our molecular phylogenetic analyses was congruent with previous studies based on the morphology of these species (Ikeda and Ohba, 1999; Li et al., 2003).
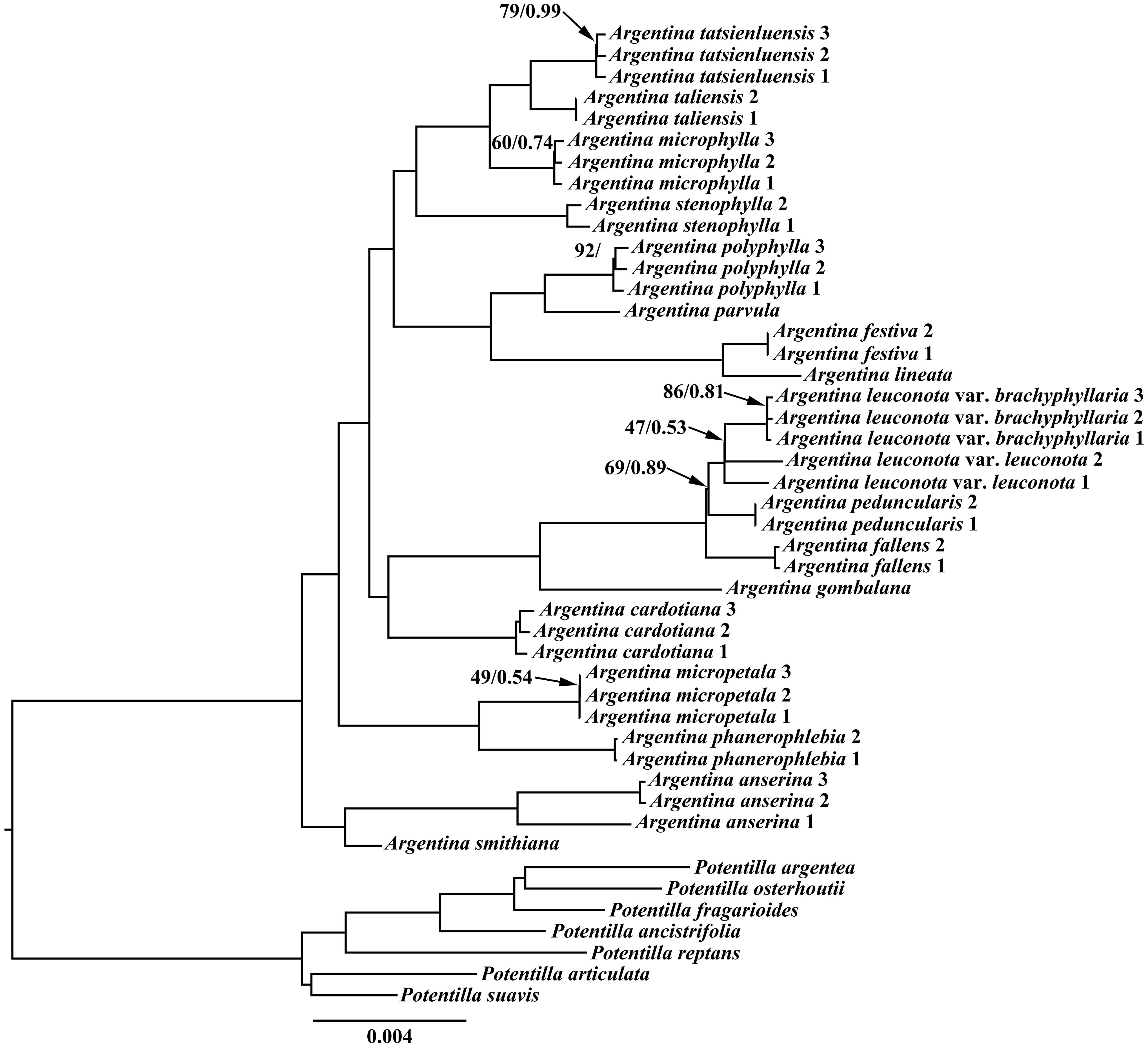
Figure 4 Maximum likelihood (ML) tree based on 39 complete chloroplast genome sequences from Argentina, with seven Potentilla taxa as outgroups. Values along branch represent ML bootstrap percentages (only BS < 100 % are shown) and Bayesian posterior probabilities (only PP < 1.00 are shown), respectively.
Ka/Ks ratios of PCGs in Argentina cp genomes were less than 1 (Supplementary Table S3), indicating that these genes probably experienced purifying selection (Nei and Kumar, 2000). Adaptive evolution typically occurs at only a few amino acid sites, so averaging rates across all sites can result in low ability to detect positive selection (Anisimova et al., 2001). Considering that KaKs_Calculator 2.0 program (Wang et al., 2010) explores selection pressures by MA method, CodeML (Yang, 2007) implemented in EasyCodeML (Gao et al., 2019) was used to identify positively selected sites of PCGs of 39 Argentina cp genomes. The LRTs of M7 vs. M8 and M8a vs. M8 for PCGs of Argentina cp genomes were significant (p-values < 0.05), indicating that M8 (model of positive selection) should be accepted (Table 2). Based on BEB analysis, a total of 26 genes with sites under positive selection were detected (Table 3). The number of positively selected sites among these genes was 1–66: 11 genes (atpA, rpoC1, rpoB, ndhC, psbF, rpl20, psbB, rpl16, rps19, ndhB, rps15) possessing one site, three genes (rpoA, rps3, ndhA) containing two sites, five genes (rps16, atpF, accD, rpl22, ycf2) having three sites, two genes (ccsA, ndhD) harboring six sites, rbcL with 10 sites, rpoC2 with 12 sites, two genes (matK, ndhF) with 13 sites, and ycf1 containing the largest number of sites. These 26 genes included four small subunit of ribosome genes (rps3, rps15, rps16, rps19), three large subunit of ribosome genes (rpl16, rpl20, rpl22), four DNA dependent RNA polymerase genes (rpoA, rpoB, rpoC1, rpoC2), two subunits of ATP synthase genes (atpA, atpF), two subunits of photosystem II genes (psbB, psbF), five subunits of NADH-dehydrogenase genes (ndhA, ndhB, ndhC, ndhD, ndhF), subunit of Rubisco gene (rbcL), subunit of Acetyl-CoA-carboxylase (ACCase) gene (accD), c-type cytochrom synthesis gene (ccsA), maturase gene (matK), and ycf1 and ycf2.
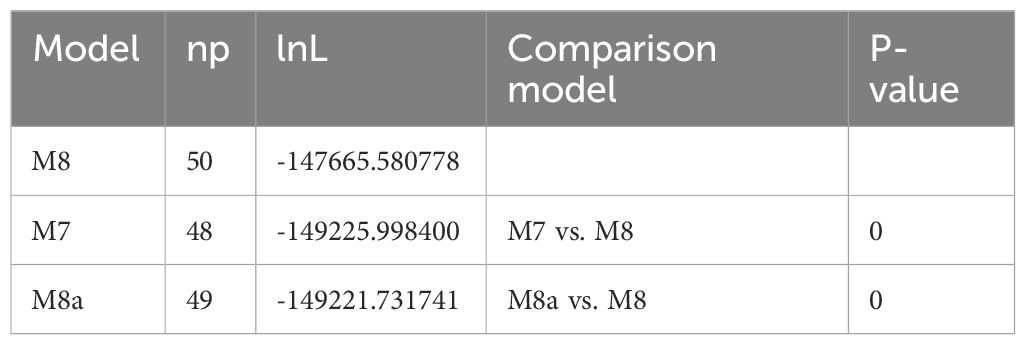
Table 2 Likelihood ratio tests (LRTs) statistics for detecting positive selection sites of protein-coding genes (PCGs) in Argentina chloroplast genomes based on site models.
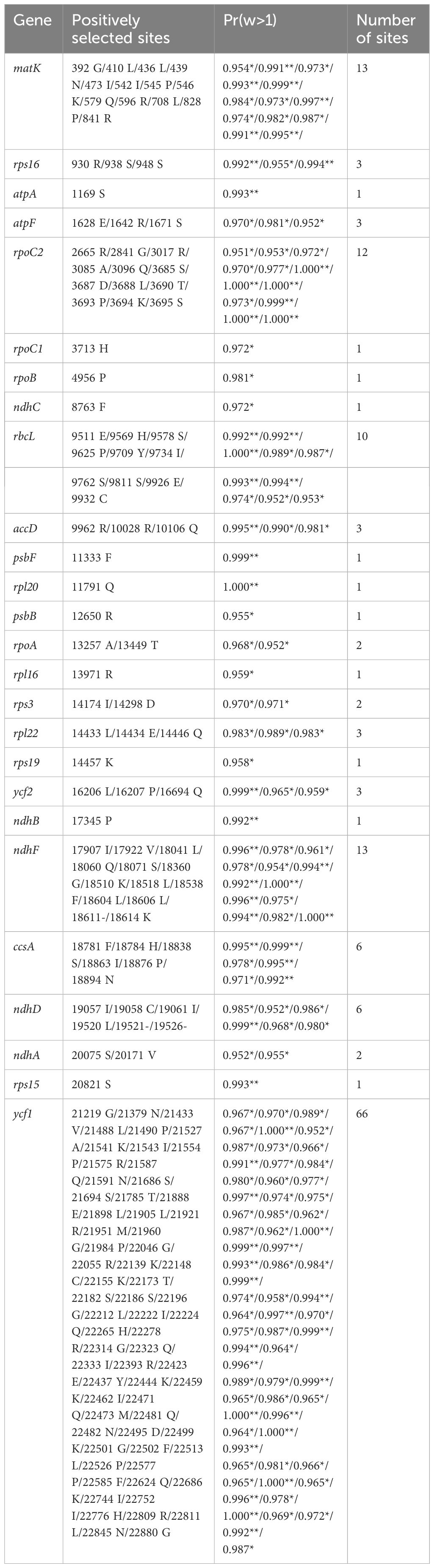
Table 3 Positively selected sites (*: P>95%; **: P>99%) identified in the chloroplast genomes of Argentina. Amino acids refer to sequence of A. gombalana.
The majority of Argentina species occurs at (sub)alpine areas above 3000 meters altitude (Kalkman, 1989, 1993; Ikeda and Ohba, 1999; Li et al., 2003; Kechaykin and Shmakov, 2016). High-altitude mountains habitats are characterized by low water availability, intense ultraviolet radiation, strong wind and/or abrasion, low air density, extreme temperatures or large diurnal/seasonal thermal fluctuations (Körner, 2003; Körner et al., 2011). The occupation of the high-altitude mountain environments indicates that Argentina species inhabiting these areas have adapted to conditions of high-altitude habitats. Species of Argentina show several morphological adaptations to high-altitude mountains habitats, such as small and compact rosettes, pinnate leaves with small leaflets, plant surface covered with hairs (Kalkman, 1989, 1993; Ikeda and Ohba, 1999; Li et al., 2003). Here possible evidence of positive selection in chloroplast coding genes was detected to reveal the adaptation of Argentina species to high-altitude mountains habitats at the molecular level. A total of 26 genes with sites under positive selection were detected and adaptive evolution of these genes might have helped Argentina species to adapt to the harsh mountain environment. As important constituents of protein synthesis machinery, cp ribosomal proteins participate in various processes of plant growth, development as well as reaction to unfavorable conditions (Schippers and Mueller-Roeber, 2010; Fleischmann et al., 2011; Tiller et al., 2012; Tiller and Bock, 2014; Zhang et al., 2016; Robles and Quesada, 2022). Adaptive evolution of these seven subunits of ribosome genes (rps3, rps15, rps16, rps19, rpl16, rpl20, rpl22) may be helpful for the normal growth and development of Argentina species under the extreme environments. Previous studies revealed that 30S ribosomal protein S15 is essential for the maintenance of high cp translational capacity under the cold stress (Fleischmann et al., 2011). Four enzymatic subunits α, β, β’ and β’’ encoded by rpoA, rpoB, rpoC1 and rpoC2 respectively, constitute catalytic core of the plastid-encoded plastid RNA polymerase (PEP) (Zhelyazkova et al., 2012). Genes of photosystems I and II are only transcribed by PEP promoters, and PEP represents the main transcription machinery of mature chloroplasts (Hajdukiewicz et al., 1997; Zhelyazkova et al., 2012; Kindgren and Strand, 2015). Genes rpoA, rpoB, rpoC1, rpoC2 were all under positive selection, which may be conducive to the transcription of photosynthetic genes of Argentina species in the harsh environments. Generating ATP from ADP using the proton gradient across the membrane (Hahn et al., 2018), the cp ATP synthase is essential for photosynthesis and plant growth (Yamamoto et al., 2023). Six ATP synthase subunit genes (atpA, atpB, atpE, atpF, atpH, and atpI) are encoded by the cp (Wicke et al., 2011), and two out of six genes (atpA and atpF) were subjected to positive selection in the present study. The 47 kDa chlorophyll α-binding protein (CP47) encoded by psbB, acts as the light-capturing antenna for the core complex of photosystem II (PSII) (Hird et al., 1991; Swiatek et al., 2003). The beta subunit of Cytochrome (cyt) b-559 protein encoded by psbF, is essential for proper assembly and activity of PSII reaction center (Pakrasi et al., 1990, 1991; Swiatek et al., 2003; Nakamura et al., 2019). The positive selection pressure on psbB and psbF may reflect adaptation of PSII to the alpine environments such as strong light. The ndh genes (ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK) in cp encode eleven NADH-dehydrogenase subunits of the Ndh1-complex bound to the thylakoid membrane (Ifuku et al., 2011). The thylakoid Ndh1-complex is involved in cyclic electron transfer around photosystem I (PSI) and in chlororespiration (Suorsa et al., 2009), which seems to be important in adaptation to stress conditions, such as high light and low temperature (Endo et al., 1999; Rumeau et al., 2007; Yamori et al., 2011). Positive selection of five ndh genes (ndhA, ndhB, ndhC, ndhD, ndhF) in Argentina may reflect the adaptation to alpine environmental stress, such as low temperature and intense light. The rbcL gene encodes the Rubisco large subunit (Wicke et al., 2011). Rubisco catalyzes the assimilation of atmospheric CO2 during photosynthesis (Wilson and Hayer-Hartl, 2018; Whitney and Sharwood, 2021). In land plants, positive selection of rbcL is quite common (Kapralov and Filatov, 2007). Positive selection of rbcL in Argentina is related to adaptation to low CO2 concentrations in the alpine environments. Gene accD encodes the beta carboxyl transferase subunit of ACCase, and ACCase is an essential enzyme that catalyzes de novo fatty acid biosynthesis (Rawsthorne, 2002; Kode et al., 2005). The accD gene could affect several biological processes such as cp division, leaf development, and seed development and storage compound metabolism (Madoka et al., 2002; Kode et al., 2005; Caroca et al., 2021). The gene ccsA encodes cytochrome c biogenesis protein, which is essential during c-type cytochromes biogenesis in the heme attachment step (Xie and Merchant, 1996). Maturase K protein encoded by the gene matK, is the only putative group II intron maturase of the cp (Neuhaus and Link, 1987). The maturase matK is required for splicing its own and other additional group II introns (Zoschke et al., 2010) and functions in photosynthesis and plant development (Barthet and Hilu, 2007). Essential genes ycf1 and ycf2 in higher plant cp genomes encode products that are indispensable for cell survival (Drescher et al., 2000). In all, the 26 genes with sites under positive selection are associated with biological processes such as self-replication, photosynthesis and biosynthesis, which may have played crucial roles in Argentina adaptation to the harsh mountain environment.
In summary, comparative analyses were conducted on 39 Argentina cp genomes, which revealed that the size, structure, GC contents, gene content and order of Argentina cp genomes were highly conserved. Twenty-two regions (trnH-GUG-psbA, trnG-GCC-trnfM-CAU, trnD-GUC-trnY-GUA, rpl32-trnL-UAG, atpH-atpI, rps16-trnQ-UUG, trnS-GCU-trnG-UCC, ndhF-rpl32, trnR-UCU-atpA, accD-psaI, 5’-trnK-UUU-rps16, rpl36-rps8, petN-psbM, atpF-atpH, ndhC-trnV-UAC, psaI-ycf4, trnT-UGU-trnL-UAA, rps15-ycf1, psaJ-rpl33, trnP-UGG-psaJ, petA-psbJ, trnF-GAA-ndhJ) were identified as candidate molecular markers for species identification, population genetics as well as phylogenetic researches of Argentina. The phylogenetic relationships among Argentina species were explored using the cp genome sequences, which were helpful for deciphering the evolutionary relationships of Argentina species. Twenty-six genes were with sites under positive selection and adaptive evolution of these genes might have helped Argentina species to adapt to the harsh mountain environment. Our findings provide insights into species identification, cp genome evolution and phylogeny in Argentina, and adaptation of Argentina species to high-altitude mountain habitats. Expanding species sampling and incorporating single-copy nuclear genes in further studies will contribute to deeper understanding about Argentina taxonomy, phylogeny, and adaptive evolution.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank of NCBI (https://www.ncbi.nlm.nih.gov/genbank/), MT114190, MT114192, MW307902-MW307916, MW322842, MW331286-MW331288, MW338689, MW348954, MW355416, and OR863686-OR863707.
Q-QL: Conceptualization, Formal analysis, Investigation, Writing – original draft. Z-PZ: Formal analysis, Writing – original draft. A: Investigation, Writing – original draft. JW: Conceptualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 32260053, 31460051), the China Scholarship Council (No. 201808155039), Inner Mongolia Autonomous Region “Grassland Talents” Project Youth Innovation and Entrepreneurship Talent Training Program (No. Q2022096), and the Fundamental Research Funds for Inner Mongolia Normal University (No. 2022JBTD010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1349358/full#supplementary-material
Andrews, S. (2018) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed March 25, 2020).
Anisimova, M., Bielawski, J. P., Yang., Z. (2001). Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18, 1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945
Aogan, Khasbagan, Li, Q. Q. (2020a). The complete chloroplast genome of Farinopsis salesoviana (Rosaceae: Potentilleae). Mitochondrial DNA Part B 5, 1363–1364. doi: 10.1080/23802359.2020.1735275
Aogan, Khasbagan, Li, Q. Q. (2020b). The complete chloroplast genome of Argentina phanerophlebia (Rosaceae: Potentilleae). Mitochondrial DNA Part B 5, 1763–1764. doi: 10.1080/23802359.2020.1748549
Bai, X., Wang, G., Ren, Y., Su, Y., Han, J. (2023). Insights into taxonomy and phylogenetic relationships of eleven Aristolochia species based on chloroplast genome. Front. Plant Sci. 14, 1119041. doi: 10.3389/fpls.2023.1119041
Barthet, M. M., Hilu, K. W. (2007). Expression of matK: functional and evolutionary implications. Am. J. Bot. 94, 1402–1412. doi: 10.3732/ajb.94.8.1402
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Caroca, R., Howell, K. A., Malinova, I., Burgos, A., Tiller, N., Pellizzer, T., et al. (2021). Knockdown of the plastid-encoded acetyl-CoA carboxylase gene uncovers functions in metabolism and development. Plant Physiol. 185, 1091–1110. doi: 10.1093/plphys/kiaa106
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Daniell, H., Jin, S., Zhu, X. G., Gitzendanner, M. A., Soltis, D. E., Soltis, P. S. (2021). Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol. J. 19, 430–447. doi: 10.1111/pbi.13556
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Darling, A. C. E., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Darling, A. E., Mau, B., Perna, N. T. (2010). ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/journal.pone.0011147
Dellaporta, S. L., Wood, J., Hicks, J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/BF02712670
Dierckxsens, N., Mardulyn, P., Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Dobeš, C., Paule, J. (2010). A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol. Phylogenet. Evol. 56, 156–175. doi: 10.1016/j.ympev.2010.03.005
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Drescher, A., Ruf, S., Calsa, T., Jr., Carrer, H., Bock, R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22, 97–104. doi: 10.1046/j.1365-313x.2000.00722.x
Drouin, G., Daoud, H., Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Du, X. Y., Kuo, L. Y., Zuo, Z. Y., Li, D. Z., Lu, J. M. (2022). Structural variation of plastomes provides key insight into the deep phylogeny of ferns. Front. Plant Sci. 13, 862772. doi: 10.3389/fpls.2022.862772
Du, Z., Lu, K., Zhang, K., He, Y., Wang, H., Chai, G., et al. (2021). The chloroplast genome of Amygdalus L. (Rosaceae) reveals the phylogenetic relationship and divergence time. BMC Genomics 22, 645. doi: 10.1186/s12864-021-07968-6
Endo, T., Shikanai, T., Takabayashi, A., Asada, K., Sato, F. (1999). The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457, 5–8. doi: 10.1016/S0014-5793(99)00989-8
Eriksson, T., Hibbs, M. S., Yoder, A. D., Delwiche, C. F., Donoghue, M. J. (2003). The phylogeny of Rosoideae (Rosaceae) based on sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA and the trnL/F region of chloroplast DNA. Int. J. Plant Sci. 164, 197–211. doi: 10.1086/346163
Eriksson, T., Lundberg, M., Töpel, M., Östensson, P., Smedmark, J. E. E. (2015). Sibbaldia: a molecular phylogenetic study of a remarkably polyphyletic genus in Rosaceae. Plant Syst. Evol. 301, 171–184. doi: 10.1007/s00606-014-1063-3
Feng, T., Moore, M. J., Sun, Y. X., Meng, A. P., Chu, H. J., Li, J. Q., et al. (2015). A new species of Argentina (Rosaceae, Potentilleae) from Southeast Tibet, with reference to the taxonomic status of the genus. Plant Syst. Evol. 301, 911–921. doi: 10.1007/s00606-014-1125-6
Feng, T., Moore, M. J., Yan, M. H., Sun, Y. X., Zhang, H. J., Meng, A. P., et al. (2017). Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J. Syst. Evol. 55, 177–191. doi: 10.1111/jse.12243
Fleischmann, T. T., Scharff, L. B., Alkatib, S., Hasdorf, S., Schottler, M. A., Bock, R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155. doi: 10.1105/tpc.111.088906
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Fu, C. N., Mo, Z. Q., Yang, J. B., Ge, X. J., Li, D. Z., Xiang, Q. Y., et al. (2019). Plastid phylogenomics and biogeographic analysis support a trans-Tethyan origin and rapid early radiation of Cornales in the Mid-Cretaceous. Mol. Phylogenet. Evol. 140, 106601. doi: 10.1016/j.ympev.2019.106601
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., Ho, S. Y. W. (2019). EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Hahn, A., Vonck, J., Mills, D. J., Meier, T., Kühlbrandt, W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318. doi: 10.1126/science.aat4318
Hajdukiewicz, P. T., Allison, L. A., Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. doi: 10.1093/emboj/16.13.4041
Hill, J. (1756). The British herbal: an history of plants and trees, natives of Britain, cultivated for use, or raised for beauty (London: Printed for Osborne, T., Shipton, J., Hodges, J., Newbery, J., Collins, B., Crowder, S., and Woodgate, H). doi: 10.5962/bhl.title.51133
Hird, S. M., Webber, A. N., Wilson, R. J., Dyer, T. A., Gray, J. C. (1991). Differential expression of the psbB and psbH genes encoding the 47 kDa chlorophyll a-protein and the 10 kDa phosphoprotein of photosystem II during chloroplast development in wheat. Curr. Genet. 19, 199–206. doi: 10.1007/BF00336487
Hu, H. S., Mao, J. Y., Wang, X., Liang, Y. Z., Jiang, B., Zhang, D. Q. (2023). Plastid phylogenomics and species discrimination in the “Chinese” clade of Roscoea (Zingiberaceae). Plant Divers 45, 523–534. doi: 10.1016/j.pld.2023.03.012
Ifuku, K., Endo, T., Shikanai, T., Aro, E. M. (2011). Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52, 1560–1568. doi: 10.1093/pcp/pcr098
Ikeda, H., Ohba, H. (1999). “A systematic revision of Potentilla L. section Leptostylae (Rosaceae) in the Himalaya and adjacent regions,” in The Himalayan Plants, vol. 3 . Ed. Ohba, H. (University of Tokyo Press, Tokyo), 31–117.
Johnson, G., Canty, S. W. J., Lichter-Marck, I. H., Wagner, W., Wen, J. (2023). Ethanol preservation and pretreatments facilitate quality DNA extractions in recalcitrant plant species. Appl. Plant Sci. 11, e11519. doi: 10.1002/aps3.11519
Kalkman, C. (1989). Potentilla (Rosaceae) in New Guinea: Census, key, and some new taxa. Blumea 34, 143–160.
Kalkman, C. (1993). “Rosaceae,” in Flora Malesiana, vol. 11 . Ed. Van Steenis, C. G. G. J. (Noordhoff-Kolff, Djakarta), 227–351.
Kapralov, M. V., Filatov, D. A. (2007). Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 7, 73. doi: 10.1186/1471-2148-7-73
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kechaykin, A. A., Shmakov, A. I. (2016). A system of subtribe Potentillinae J. Presl (Rosaceae Juss.). Turczaninowia 19, 114–128. doi: 10.14258/turczaninowia.19.4.16
Kindgren, P., Strand, A. (2015). Chloroplast transcription, untangling the Gordian Knot. New Phytol. 206, 889–891. doi: 10.1111/nph.13388
Körner, C. (2003). Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems. 2nd ed (Berlin: Springer-Verlag). doi: 10.1007/978-3-642-18970-8
Körner, C., Paulsen, J., Spehn, E. M. (2011). A definition of mountains and their bioclimatic belts for global comparisons of biodiversity data. Alp. Bot. 121, 73–78. doi: 10.1007/s00035-011-0094-4
Kode, V., Mudd, E. A., Iamtham, S., Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., Calcott, B. (2017). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Li, C., Cai, C., Tao, Y., Sun, Z., Jiang, M., Chen, L., et al. (2021b). Variation and evolution of the whole chloroplast genomes of Fragaria spp. (Rosaceae). Front. Plant Sci. 12, 754209. doi: 10.3389/fpls.2021.754209
Li, C. L., Ikeda, H., Ohba, H. (2003). “Potentilla L.,” in Flora of China, vol. 9 . Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Science Press/Missouri Botanical Garden Press, Beijing/St. Louis), 291–327.
Li, Y. K., Khasbagan, Li, Q. Q. (2020b). The complete chloroplast genome of Drymocallis saviczii (Rosaceae: Potentilleae). Mitochondrial DNA Part B 5, 2036–2037. doi: 10.1080/23802359.2020.1756966
Li, Q. Q., Khasbagan, Zhang, Z. P., Wen, J., Yu, Y. (2024). Plastid phylogenomics of the tribe Potentilleae (Rosaceae). Mol. Phylogenet. Evol. 190, 107961. doi: 10.1016/j.ympev.2023.107961
Li, Q. Q., Yu, Y., Zhang, Z. P., Wen, J. (2021a). Comparison among the chloroplast genomes of five species of Chamaerhodos (Rosaceae: Potentilleae): phylogenetic implications. Nord. J. Bot. 39, e03121. doi: 10.1111/njb.03121
Li, Q. Q., Zhang, Z. P., Khasbagan (2020a). The complete chloroplast genome of Potentilla suavis (Rosaceae: Potentilleae). J. Inner Mongolia Norm. Univ. (Nat. Sci. Edn) 49, 471–474.
Liu, C., Chen, H. H., Tang, L. Z., Khine, P. K., Han, L. H., Song, Y., et al. (2021). Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae). Plant Divers. 44, 377–388. doi: 10.1016/j.pld.2021.11.009
Liu, L., Wang, Y., He, P., Li, P., Lee, J., Soltis, D. E., et al. (2018). Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics 19, 235. doi: 10.1186/s12864-018-4633-x
Lundberg, M., Töpel, M., Eriksen, B., Nylander, J. A. A., Eriksson, T. (2009). Allopolyploidy in Fragariinae (Rosaceae): comparing four DNA sequence regions, with comments on classification. Mol. Phylogenet. Evol. 51, 269–280. doi: 10.1016/j.ympev.2009.02.020
Madoka, Y., Tomizawa, K. I., Mizoi, J., Nishida, I., Nagano, Y., Sasaki, Y. (2002). Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 43, 1518–1525. doi: 10.1093/pcp/pcf172
Mower, J. P., Vickrey, T. L. (2018). Structural diversity among plastid genomes of land plants. Adv. Bot. Res. 85, 263–292. doi: 10.1016/bs.abr.2017.11.013
Nakamura, M., Boussac, A., Sugiura, M. (2019). Consequences of structural modifications in cytochrome b559 on the electron acceptor side of Photosystem II. Photosynth. Res. 139, 475–486. doi: 10.1007/s11120-018-0521-0
Nei, M., Kumar, S. (2000). Molecular Evolution and Phylogenetics (Oxford: Oxford university press). doi: 10.1093/oso/9780195135848.001.0001
Neuhaus, H. E., Emes, M. J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140. doi: 10.1146/annurev.arplant.51.1.111
Neuhaus, H., Link, G. (1987). The chloroplast tRNALys (UUU) gene from mustard (Sinapsis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr. Genet. 11, 251–257. doi: 10.1007/BF00355398
Niu, Y. T., Jabbour, F., Barrett, R. L., Ye, J. F., Zhang, Z. Z., Lu, K. Q., et al. (2018). Combining complete chloroplast genome sequences with target loci data and morphology to resolve species limits in Triplostegia (Caprifoliaceae). Mol. Phylogenet. Evol. 129, 15–26. doi: 10.1016/j.ympev.2018.07.013
Ogoma, C. A., Liu, J., Stull, G. W., Wambulwa, M. C., Oyebanji, O., Milne, R. I., et al. (2022). Deep insights into the plastome evolution and phylogenetic relationships of the tribe Urticeae (family Urticaceae). Front. Plant Sci. 13, 870949. doi: 10.3389/fpls.2022.870949
Pakrasi, H. B., De Ciechi, P., Whitmarsh, J. (1991). Site directed mutagenesis of the heme axial ligands of cytochrome b559 affects the stability of the photosystem II complex. EMBO J. 10, 1619–1627. doi: 10.1002/j.1460-2075.1991.tb07684.x
Pakrasi, H. B., Nyhus, K. J., Granok, H. (1990). Targeted deletion mutagenesis of the beta subunit of cytochrome b559 protein destabilizes the reaction center of photosystem II. Z. Naturforsch. C J. Biosci. 45, 423–429. doi: 10.1515/znc-1990-0519
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Palmer, J. D. (1991). “Plastid chromosomes: structure and evolution,” in The molecular biology of plastids, vol. 7A . Eds. Bogorad, L., Vasil, I. K. (Academic Press, San Diego), 5–53.
Persson, N. L., Toresen, I., Andersen, H. L., Smedmark, J. E. E., Eriksson, T. (2020). Detecting destabilizing species in the phylogenetic backbone of Potentilla (Rosaceae) using low-copy nuclear markers. AoB Plants 12, plaa017. doi: 10.1093/aobpla/plaa017
Posada, D., Buckley, T. R. (2004). Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808. doi: 10.1080/10635150490522304
Potter, D., Eriksson, T., Evans, R. C., Oh, S., Smedmark, J. E. E., Morgan, D. R., et al. (2007). Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266, 5–43. doi: 10.1007/s00606-007-0539-9
Potter, D., Gao, F., Bortiri, P. E., Oh, S. H., Baggett, S. (2002). Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data. Plant Syst. Evol. 231, 77–89. doi: 10.1007/s006060200012
Rambaut, A. (2018) FigTree ver. 1.4.4. Available online at: http://tree.bio.ed.ac.uk/software/figtree (Accessed March 20, 2020).
Ravi, V., Khurana, J. P., Tyagi, A. K., Khurana, P. (2008). An update on chloroplast genomes. Plant Syst. Evol. 271, 101–122. doi: 10.1007/s00606-007-0608-0
Rawsthorne, S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41, 182–196. doi: 10.1016/S0163-7827(01)00023-6
Ren, J., Tian, J., Jiang, H., Zhu, X. X., Mutie, F. M., Wanga, V. O., et al. (2022). Comparative and phylogenetic analysis based on the chloroplast genome of Coleanthus subtilis (Tratt.) Seidel, a protected rare species of monotypic genus. Front. Plant Sci. 13, 828467. doi: 10.3389/fpls.2022.828467
Robles, P., Quesada, V. (2022). Unveiling the functions of plastid ribosomal proteins in plant development and abiotic stress tolerance. Plant Physiol. Biochem. 189, 35–45. doi: 10.1016/j.plaphy.2022.07.029
Rodríguez-Ezpeleta, N., Brinkmann, H., Burey, S. C., Roure, B., Burger, G., Löffelhardt., W., et al. (2005). Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330. doi: 10.1016/j.cub.2005.06.040
Rono, P. C., Dong, X., Yang, J. X., Mutie, F. M., Oulo, M. A., Malombe, I., et al. (2020). Initial complete chloroplast genomes of Alchemilla (Rosaceae): comparative analysis and phylogenetic relationships. Front. Genet. 11, 560368. doi: 10.3389/fgene.2020.560368
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Rumeau, D., Peltier, G., Cournac, L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30, 1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x
Rydberg, P. A. (1898). A monograph of the North American Potentilleae, Memoirs from the Department of Botany of Columbia University Vol. 2 (Lancaster: Press of the New Era Printing Company).
Sato, N., Terasawa, K., Miyajima, K., Kabeya, Y. (2003). Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 232, 217–262. doi: 10.1016/S0074-7696(03)32006-6
Schippers, J. H. M., Mueller-Roeber, B. (2010). Ribosomal composition and control of leaf development. Plant Sci. 179, 307–315. doi: 10.1016/j.plantsci.2010.06.012
Soják, J. (1989). Die generische Problematik von Potentilla s.l. Čas. Nár. Muz., Řada Přír 154, 117–118.
Soják, J. (1994). Notes on Potentilla (Rosaceae) X–XII. X. The section Dumosae. XI. The P. microphylla and P. stenophylla groups (sect. Pentaphylloides). XII. Key to the taxa of P. sect. Pentaphylloides (Anserina). Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 116, 11–81.
Soják, J. (2004). Potentilla L. (Rosaceae) and related genera in the former USSR (identification key, checklist and figures). Notes on Potentilla XVI. Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 125, 253–340. doi: 10.1127/0006-8152/2004/0125-0253
Soják, J. (2008). Notes on Potentilla XXI. A new division of the tribe Potentilleae (Rosaceae) and notes on generic delimitations. Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 127, 349–358. doi: 10.1127/0006-8152/2008/0127-0349
Soják, J. (2010). Argentina Hill, a genus distinct from Potentilla (Rosaceae). Thaiszia J. Bot. 20, 91–97.
Soják, J. (2012a). Argentina recognita (Rosaceae, Potentilleae), a new species from New Guinea, with a key to the species known from the island. Willdenowia 42, 89–93. doi: 10.3372/wi.42.42111
Soják, J. (2012b). Argentina adulterina (Rosaceae-Potentilleae), a new species from New Guinea. Feddes Repert. 123, 51–54. doi: 10.1002/fedr.201200006
Soják, J. (2012c). Potentilla L. (Rosaceae) and related genera in Asia (excluding the former USSR), Africa and New Guinea. Notes on Potentilla XXVIII. Plant Div. Evol. 130, 7–157. doi: 10.1127/1869-6155/2012/0130-0060
Sousa, F., Civáň, P., Foster, P. G., Cox, C. J. (2020). The chloroplast land plant phylogeny: analyses employing better-fitting tree- and site-heterogeneous composition models. Front. Plant Sci. 11, 1062. doi: 10.3389/fpls.2020.01062
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sugiura, N. (1978). Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun. Stat. Theory Methods A7, 13–26. doi: 10.1080/03610927808827599
Suorsa, M., Sirpiö, S., Aro, E. M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2, 1127–1140. doi: 10.1093/mp/ssp052
Swiatek, M., Regel, R. E., Meurer, J., Wanner, G., Pakrasi, H. B., Ohad, I., et al. (2003). Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: the psbEFLJ operon in Nicotiana tabacum. Mol. Genet. Genomics 268, 699–710. doi: 10.1007/s00438-002-0791-1
Tang, C., Chen, X., Deng, Y., Geng, L., Ma, J., Wei, X. (2022). Complete chloroplast genomes of Sorbus sensu stricto (Rosaceae): comparative analyses and phylogenetic relationships. BMC Plant Biol. 22, 495. doi: 10.1186/s12870-022-03858-5
Tian, W. J., Khasbagan, Li, Q. Q. (2020). The complete chloroplast genome of Sibbaldianthe adpressa (Rosaceae: Potentilleae). Mitochondrial DNA Part B 5, 1563–1564. doi: 10.1080/23802359.2020.1742601
Tiller, N., Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Tiller, N., Weingartner, M., Thiele, W., Maximova, E., Schottler, M. A., Bock, R. (2012). The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 69, 302–316. doi: 10.1111/j.1365-313X.2011.04791.x
Töpel, M., Lundberg, M., Eriksson, T., Eriksen, B. (2011). Molecular data and ploidal levels indicate several putative allopolyploidization events in the genus Potentilla (Rosaceae). PloS Curr. 3, RRN1237. doi: 10.1371/currents.RRN1237
Wang, J., Fu, C. N., Mo, Z. Q., Möller, M., Yang, J. B., Zhang, Z. R., et al. (2022). Testing the complete plastome for species discrimination, cryptic species discovery and phylogenetic resolution in Cephalotaxus (Cephalotaxaceae). Front. Plant Sci. 13, 768810. doi: 10.3389/fpls.2022.768810
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Waswa, E. N., Mkala, E. M., Odago, W. O., Amenu, S. G., Mutinda, E. S., Muthui, S. W., et al. (2023). Comparative chloroplast genome analysis of Sambucus L. (Viburnaceae): inference for phylogenetic relationships among the closely related Sambucus adnata Wall. ex DC Sambucus javanica Blume. Front. Plant Sci. 14, 1179510. doi: 10.3389/fpls.2023.1179510
Whitney, S. M., Sharwood, R. E. (2021). Rubisco engineering by plastid transformation and protocols for assessing expression. Methods Mol. Biol. 2317, 195–214. doi: 10.1007/978-1-0716-1472-3_10
Wicke, S., Schneeweiss, G. M., depamphilis, C. W., Müller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wilson, R. H., Hayer-Hartl, M. (2018). Complex chaperone dependence of Rubisco biogenesis. Biochemistry 57, 3210–3216. doi: 10.1021/acs.biochem.8b00132
Wolfe, K. H., Li, W. H., Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, X., Luo, D., Zhang, Y., Yang, C., Crabbe, M. J. C., Zhang, T., et al. (2022). Comparative genomic and phylogenetic analysis of chloroplast genomes of hawthorn (Crataegus spp.) in Southwest China. Front. Genet. 13, 900357. doi: 10.3389/fgene.2022.900357
Wu, L., Wu, M., Cui, N., Xiang, L., Li, Y., Li, X., et al. (2021). Plant super-barcode: a case study on genome-based identification for closely related species of Fritillaria. Chin. Med. 16, 52. doi: 10.1186/s13020-021-00460-z
Xie, Z., Merchant, S. (1996). The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 271, 4632–4639. doi: 10.1074/jbc.271.9.4632
Xu, X. M., Liu, D. H., Zhu, S. X., Wang, Z. L., Wei, Z., Liu, Q. R. (2023). Phylogeny of Trigonotis in China—with a special reference to its nutlet morphology and plastid genome. Plant Divers. 45, 409–421. doi: 10.1016/j.pld.2023.03.004
Xue, T. T., Janssens, S. B., Liu, B. B., Yu, S. X. (2024). Phylogenomic conflict analyses of the plastid and mitochondrial genomes via deep genome skimming highlight their independent evolutionary histories: a case study in the cinquefoil genus Potentilla sensu lato (Potentilleae, Rosaceae). Mol. Phylogenet. Evol. 190, 107956. doi: 10.1016/j.ympev.2023.107956
Yamamoto, H., Cheuk, A., Shearman, J., Nixon, P. J., Meier, T., Shikanai, T. (2023). Impact of engineering the ATP synthase rotor ring on photosynthesis in tobacco chloroplasts. Plant Physiol. 192, 1221–1233. doi: 10.1093/plphys/kiad043
Yamori, W., Sakata, N., Suzuki, Y., Shikanai, T., Makino, A. (2011). Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 68, 966–976. doi: 10.1111/j.1365-313X.2011.04747.x
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S. W., Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yu, J., Fu, J., Fang, Y., Xiang, J., Dong, H. (2022). Complete chloroplast genomes of Rubus species (Rosaceae) and comparative analysis within the genus. BMC Genomics 23, 32. doi: 10.1186/s12864-021-08225-6
Yü, T. T., Li, C. L. (1980). A study on the genus Potentilla of China. Acta Phytotax. Sin. 18, 1–14.
Yü, T. T., Li, C. L. (1985). “Potentilla L.,” in Flora Reipublicae Popularis Sinicae, vol. 37 . Ed. Yü, T. T. (Science Press, Beijing), 233–331.
Zhang, Y. M., Han, L. J., Yang, C. W., Yin, Z. L., Tian, X., Qian, Z. G., et al. (2021). Comparative chloroplast genome analysis of medicinally important Veratrum (Melanthiaceae) in China: insights into genomic characterization and phylogenetic relationships. Plant Divers. 44, 70–82. doi: 10.1016/j.pld.2021.05.004
Zhang, S. D., Jin, J. J., Chen, S. Y., Chase, M. W., Soltis, D. E., Li, H. T., et al. (2017). Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367. doi: 10.1111/nph.14461
Zhang, X. H., Khasbagan, Li, Q. Q. (2020). The complete chloroplast genome of Sibbaldia aphanopetala (Rosaceae: Potentilleae). Mitochondrial DNA Part B 5, 2026–2027. doi: 10.1080/23802359.2020.1756945
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419, 779–781. doi: 10.1016/j.bbrc.2012.02.101
Zhang, S. D., Yan, K., Ling, L. Z. (2023). Characterization and phylogenetic analyses of ten complete plastomes of Spiraea species. BMC Genomics 24, 137. doi: 10.1186/s12864-023-09242-3
Zhang, J., Yuan, H., Yang, Y., Fish, T., Lyi, S. M., Thannhauser, T. W., et al. (2016). Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 67, 2731–2744. doi: 10.1093/jxb/erw106
Zhang, G. J., Zhang, Z. P., Li, Q. Q. (2022). Comparative analysis of chloroplast genomes of Sanguisorba species and insights into phylogenetic implications and molecular dating. Nord. J. Bot. 2022, e03719. doi: 10.1111/njb.03719
Zhelyazkova, P., Sharma, C. M., Forstner, K. U., Liere, K., Vogel, J., Borner, T. (2012). The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24, 123–136. doi: 10.1105/tpc.111.089441
Zhou, N., Miao, K., Liu, C., Jia, L., Hu, J., Huang, Y., et al. (2024). Historical biogeography and evolutionary diversification of Lilium (Liliaceae): New insights from plastome phylogenomics. Plant Divers 46, 219–228. doi: 10.1016/j.pld.2023.07.009
Zhu, A., Guo, W., Gupta, S., Fan, W., Mower, J. P. (2016). Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209, 1747–1756. doi: 10.1111/nph.13743
Zoclanclounon, Y. A. B., Thamilarasan, S. K., Mo, Y., Ahn, B. O., Kim, J. G., Lee, K. (2023). Insights into chloroplast genome structure and phylogenetic relationships within the Sesamum species complex (Pedaliaceae). Front. Genet. 14, 1207306. doi: 10.3389/fgene.2023.1207306
Keywords: Argentina, Potentilleae, adaptive evolution, chloroplast genome, comparative analyses, phylogeny
Citation: Li Q-Q, Zhang Z-P, Aogan and Wen J (2024) Comparative chloroplast genomes of Argentina species: genome evolution and phylogenomic implications. Front. Plant Sci. 15:1349358. doi: 10.3389/fpls.2024.1349358
Received: 04 December 2023; Accepted: 25 March 2024;
Published: 30 April 2024.
Edited by:
Francesco Sunseri, Mediterranean University of Reggio Calabria, ItalyReviewed by:
Zhechen Qi, Zhejiang Sci-Tech University, ChinaCopyright © 2024 Li, Zhang, Aogan and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Wen, d2VuakBzaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.