
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 February 2024
Sec. Plant Metabolism and Chemodiversity
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1349009
This article is part of the Research Topic Plant Secondary Metabolite Biosynthesis View all 12 articles
Terpenes are important mediators of plant chemical response to environmental cues. Here, we describe the genome-wide identification and biochemical characterization of TPS-a members in Medicago truncatula, a model legume crop. Genome mining identified thirty-nine full-length terpene synthases with a significant number predicted to produce monoterpenes and sesquiterpenes. Biochemical characterization of the TPS-a subfamily associated with sesquiterpene biosynthesis revealed such compounds, that exhibit substantial biological activity in other plants. Gene expression analysis using qPCR and the Medicago gene atlas illustrated distinct tissue and time-based variation in expression in leaves and roots. Together our work establishes the gene-to-metabolite relationships for sesquiterpene synthases in M. truncatula. Understanding the biosynthetic capacity is a foundational step to defining the ecological roles of this important family of compounds.
Plants produce an array of specialized metabolites in response to environmental stressors. Terpenoids are one such class of natural products with unprecedented chemical diversity. Biosynthesis proceeds from simple core steps which initiate from five-carbon units to make varying lengths of backbones (Figure 1). Terpene synthases (TPS) increase chemical diversity by conversion of precursors into hydrocarbons through complex lysis and allylic diphosphate ester bond cleavage (Christianson, 2017). The resulting compounds are structurally related forming variably sized and species-specific gene families (Pichersky and Raguso, 2018).
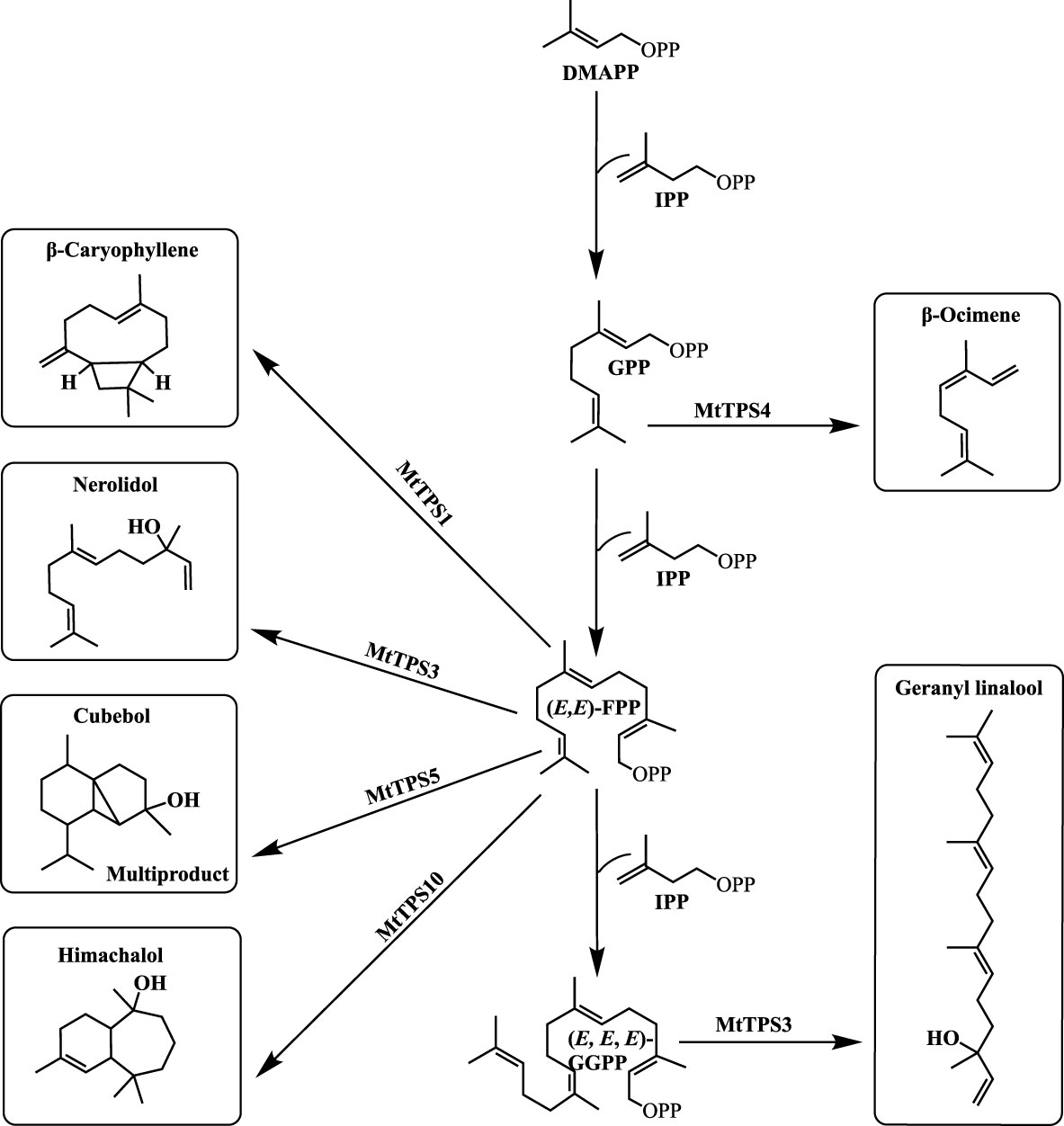
Figure 1 Schematic of terpene biosynthesis. Three major substrates are used in the production of most terpene products by TPSs: GPP (C10), (E, E)-FPP (C15), and (E, E, E)-GGPP (C20). Resultant terpene products of previously characterized MtTPSs: MtTPS1 produces β-caryophyllene, MtTPS3 produces both the sesquiterpene product nerolidol and the diterpene product geranyl linalool, MtTPS4 produces β-ocimene, MtTPS5 produces cubebol as a major product but is a multiproduct enzyme and MtTPS10 produces mainly himachalol.
Phylogenetic analysis of TPSs results in the clustering of genes into seven subfamilies based on sequence homology, gene architecture and functional studies in various plants (Chen et al., 2011). Generally, sesquiterpene synthases (C15) fall primarily into TPS-a, monoterpene synthases (C10) in TPS-b and TPS-g, and diterpene synthases (C20) in TPS-c and TPS-e/f whereas TPS-d and TPS-h are specific to gymnosperms and lycophytes respectively. This vast chemical diversity results in species-specific complex mixtures of terpenes with broadly varying biological activity. Terpenoids mediate interactions between plants and their environment and are critical in coordinating responses to (a)biotic stress (Pichersky and Raguso, 2018). Induced volatile terpene blends composed mainly of lower molecular weight monoterpenes and sesquiterpenes have additional functions in inter and intra-plant communication (He et al., 2022).
Legumes are important crops providing a significant portion of protein in human and animal diets and play an important role in agricultural sustainability. Early investigations in M. truncatula, have demonstrated that differential terpene-enriched volatile blends are induced in response to herbivores (Leitner et al., 2005; Arimura et al., 2008). Subsequent studies identified biosynthetic genes underlying these observed metabolites summarized in Figure 1 (Gomez et al., 2005; Arimura et al., 2008; Navia-Giné et al., 2009; Garms et al., 2010). MtTPS1 produces a sesquiterpene, (E)-β caryophyllene, which in maize is instrumental in indirect defense by attracting parasitoids of herbivores and entomopathogenic nematodes (Rasmann et al., 2005; Degenhardt et al., 2009). MtTPS3 forms nerolidol and geranyllinalool common to many plants with defense capabilities. In M. truncatula, MtTPS4 which encodes for a monoterpene β-ocimene, was upregulated in response to methyl jasmonate and feeding insects (Navia-Giné et al., 2009). β-ocimene has a priming effect that activates a defense response in Chinese cabbage and cucumber (Navia-Giné et al., 2009; Kang et al., 2018; He et al., 2022). MtTPS5 is a multiproduct TPS, producing up to twenty-seven different terpene products (Garms et al., 2010). More recently, Yadav et al. (2019) showed that MtTPS10, which catalyzes the formation of a sesquiterpene alcohol, himachalol, was upregulated in the root upon infection by an oomycete pathogen, Aphanomyces euteiches (Yadav et al., 2019). The combined literature illustrates that M. truncatula produces a blend of terpene-based compounds upon interactions with biotic stressors as part of its chemical response strategy.
Soybean terpene synthases GmTPS3, GmTPS18 and GmTPS21 have been characterized and shown to produce β-ocimene, geraniol and α-farnesene respectively, each of which have demonstrated protective roles in plants (Liu et al., 2014; Lin et al., 2017; Han et al., 2023). For example, GmTPS21 (which encodes for α-farnesene) provided resistance against soybean cyst nematode belowground and aphids aboveground (Lin et al., 2017; Constantino et al., 2021). In cowpea, terpenes were identified as part of the volatiles produced in response to herbivory whereas biochemical characterization of the genes responsible for floral scent was carried out in sweet pea (Bao et al., 2020; Steinbrenner et al., 2022).
In grasses, such as maize, rice and wheat, terpenes play an important role in the interactions of plants with their environment (Rasmann et al., 2005; Schnee et al., 2006; Schmelz et al., 2014). Although terpenes are widespread there still exist distinct chemotypes in species with new chemistry and biological roles which are still being uncovered (Pichersky and Raguso, 2018; Ding et al., 2020; Murphy and Zerbe, 2020; Zhang et al., 2021). Comparatively less is known about the composition of the terpenome in legumes.
Here, we define the gene-to-metabolite relationships underlying terpene biosynthesis in M. truncatula. Through genome-wide identification of terpene synthases and biochemical characterization of the TPS-a subfamily, we have identified sesquiterpene synthases essential in generating the sesquiterpene-based chemical diversity in M. truncatula. Understanding the terpenoid biosynthetic capacity is a foundational step to defining the ecological roles of this important family of compounds.
Putative terpene synthases (protein models) from the Medicago truncatula cv. Jemalong A17 were collated from the Mt4.0v1 genome in Phytozome (https://phytozome-next.jgi.doe.gov) through an annotation-based search for TPSs. The putative TPSs were further manually curated for the presence of catalytic domains namely the Class I site with characteristic DDxxD and secondary NSE/DTE metal binding motif involved in ionization-dependent cyclization. Class II synthases were identified by the DxDD motif involved in protonation-initiated cyclization characteristic of copalyl diphosphate synthases (CPPs) as shown in Supplementary Figure S1 (Christianson, 2017). For phylogenetic analysis, previously reported and biochemically characterized genes were collated from S. lycopersicum ITAG2.4 and A. thaliana TAIR10 genomes (Zhou and Pichersky, 2020), aligned with M. truncatula TPSs using maximum likelihood analysis with 1000 bootstrap repetitions using CLC software and visualized using iTOL (Letunic and Bork, 2021).
Synthetic MtTPSs codon-optimized for expression in Escherichia coli were ordered through ThermoFisher and subcloned into pET28b using primers listed in Supplementary Table S1. Functional analysis of genes was carried out using a previously described modular metabolic engineering system in E. coli enhanced for sesquiterpene production (Kitaoka et al., 2015). Briefly, TPS-a family genes in pET28b were co-transformed with an (E, E)-FPP synthase from Zea mays (ZmFPPs) in E. coli BL21 DE3-C41 cells (Lucigen). Transformed cultures were grown in 45 mL of Terrific Broth medium to an OD600 of ∼0.6 at 37°C Cultures were cooled to 16°C before induction with 1 mM isopropyl-thiogalactopyranoside and supplemented with 1 mM MgCl2 and 25 mM sodium pyruvate incubation for 72 hours. Dodecane (20 mL) was added to ensure the capture of volatile compounds. In a second flask, enzyme products were extracted with 50 mL of 100% hexane and concentrated under an N2 stream. Samples were resuspended in 1 mL of n-hexane for analysis by gas chromatography–mass spectrometry (GC-MS).
Samples were analyzed on an Agilent 8890B gas chromatograph (GC) coupled to a mass spectrometer (MS) 5977B extractor electron ionization detector at 70 eV. Samples (1 μL) were injected in pulsed spitless mode with the inlet temperature set to 250°C. Separation was achieved on an HP5MS column (30 m, 0.25 mm i.d., 0.25 μm film) using helium as the carrier gas at a flow rate of 1.2 mL/min. The initial oven temperature of 70°C was increased after 1 min to 260°C at a rate of 10°C/min and held for 2 min at 260°C. MS data was collected from 60 to 500 mass-to-charge ratio(m/z). Compounds were identified by comparison of mass spectra and retention times with those of the authentic standards, when available, or with the Wiley, National Institute of Standards and Technology (NIST) mass spectral library.
To obtain sufficient compound for structural elucidation by NMR, cultures were scaled up to 500 mL batches to a total volume of 5 Liters and expressed using the combinatorial expression system described above (Kitaoka et al., 2015). The resultant products were extracted with an equal volume of hexanes. The organic extract was recovered using a separatory funnel and then dried by rotary evaporation. The resulting residue was passed through a silica column and eluted using a hexane: ethyl acetate gradient. Fractions of interest were further purified by HPLC using an Agilent 1260 series instrument equipped with an autosampler, fraction collector and diode array UV detector over a ZORBAX Eclipse XDB C18 (4.6 x 150 mm, 5 mm) at a 0.5 mL/min flow rate using a water and acetonitrile gradient as the mobile phase. Purified compounds were resuspended in CDCl3. Structural analysis was performed using 1D (‘1H, 13C’) and 2D (TOCSY, ROESYPHPR, HMBCGP, DQFCOSY, and 13CHSQC) NMR experiments. Spectra were acquired on a Bruker Avance 500-MHz TopSpin NMR spectrometer.
Germination of M. truncatula cv. Jemalong A17 seeds were carried out by scarification using concentrated anhydrous sulfuric acid for 5 min followed by intensive washing steps with Milli-Q water. Seeds were then placed in water in a 50 ml falcon tube at 4°C for 3 days to break dormancy. Seeds were transferred to moist blotting paper and placed on a petri dish incubated at 26°C. Single seedlings were transferred to pots (9 cm diameter) filled with a perlite and sand mixture (5:1), then grown in a growth chamber with 16 h light/8 h dark for 6 weeks. After 6 weeks of growth, plants were treated by foliar application with solutions containing 1 mM methyl jasmonate (MeJA) in 0.01% Triton-X-100 (Sigma-Aldrich, St Louis, MO, USA) or 1.5 mM salicylic acid solution dissolved in 1% ethanol (SA) (Sigma-Aldrich, St Louis, MO, USA). Plants were treated with 0.01% Triton-X-100 for MeJA control and 1% ethanol for SA. For each plant, 20 ml solution was applied (either MeJA or SA or controls) and at least three biological replicates were conducted for each treatment. The leaf and root tissues were separately collected in time courses from 0h, 2 h, 6 h, 12 h, 24 h, 48 h, 72 h and 96 h and immediately frozen in liquid nitrogen and stored at -80°C for further RNA extraction.
Sampling was performed on three biological replicates for both elicitors and controls. The root and leaf tissues were separately ground in liquid nitrogen and 100 mg powder was used for total RNA isolation with an RNeasy Plant Mini Kit (Qiagen), and RNase-Free DNase to remove genomic DNA following the manufacturer’s protocol. RNA purity and yield were checked using a NanoDrop 8000 Spectrophotometer (ThermoFisher Scientific), while RNA integrity and quantification were evaluated with an Agilent 2100 bioanalyzer (Agilent Technologies). First-strand cDNA was synthesized using a Superscript IV Reverse transcriptase kit (ThermoFisher Scientific, USA).
Primers were designed using coding sequences from the genome of M. truncatula (Mt4.0v1) in Phytozome13 (https://phytozome-next.jgi.doe.gov/) (Supplementary Table S2). A quantitative real-time PCR system was carried out using SYBR green with ROX (pre-mixed) as an internal loading standard performed on an Eppendorf Mastercycler. The reaction mixture was 10 µl and comprised of 5 µl of 2× SYBR™ Green qPCR Master Mix (Thermofisher Scientific, USA), 100 nM primers (Thermo Fisher Scientific) and 0.5 µl of (1:10 dilution) cDNA. PCR protocol was followed to initiate polymerase activation: 10 min at 95°C; 40 cycles of 30 s at 95°C, 60 s at 55°C and 30 s at 72°C. Each run of qPCR was followed by a melting curve analysis from 55 to 95 °C. PCR conditions were determined by comparing the threshold values of the RT product (cycle threshold (Ct) value). Relative RNA levels were calibrated and normalized with the level of actin and histone-3 reference genes based on the 2–ΔΔCt method (Rey et al., 2016; Zhang et al., 2020). For each gene, three technical replicates were run for three biological samples. qPCR data were expressed as a fold change with respect to the equivalent time-point in the control groups of MeJA (in 0.01% Triton-X-100 and SA (in 1% ethanol).
We assessed for the genetic potential of M. truncatula cv. Jemalong A17 to produce terpenes using the Mt4.0v1 genome in Phytozome. We identified 54 putative MtTPS gene models using BLAST and subsequent analysis of characteristic motifs (Supplementary Table S3 and Supplementary Figure S1). Further curation through sequence alignments confirmed thirty- nine full-length genes. Twenty-two of the reported fifty-four potential TPSs in M. truncatula were previously assigned numbers by Parker et al., 2014 (MtTPS16 is no longer classified as a putative terpene synthase) (Parker et al., 2014). Supplementary Table S3 is a summary of the previous and newly assigned TPS numbers. A majority of the genes had the Class I DDxxD and secondary NSE/DTE metal binding motif involved in ionization-dependent cyclization (Christianson, 2017). Three Class II synthases were identified by DxDD motifs involved in protonation-initiated cyclization characteristic of copalyl diphosphate synthases (CPPs) involved in diterpene scaffold formation. Phylogenetic analysis of the terpene synthases from M. truncatula and the comprehensively biochemically characterized TPS collections of A. thaliana and S. lycopersicum shows plant species clustering more than enzymes with similar functions (Figure 2) (Chen et al., 2011; Falara et al., 2011; Parker et al., 2014; Zhou and Pichersky, 2020). The genes are distributed into the different clades with fourteen members in TPS-a; five members in TPS-b; three members in TPS-c and four members in TPS e/f. Interestingly there is an expanded TPS-g (thirteen members) subfamily as compared to A. thaliana and S. lycopersicum.
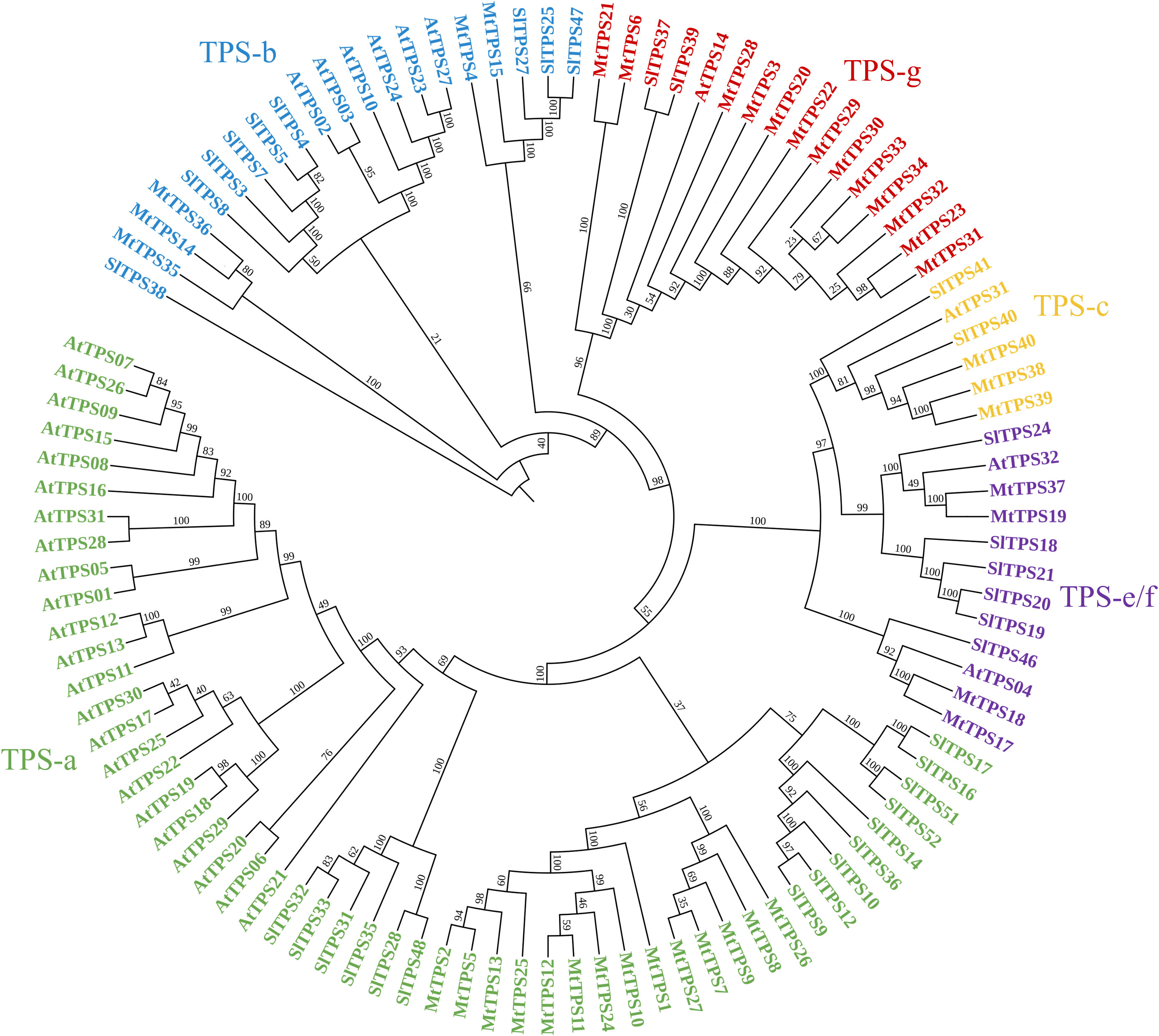
Figure 2 Phylogeny-based analysis of terpene synthases in Medicago truncatula, Arabidopsis thaliana, and Solanum lycopersicum. The sequences were aligned using maximum likelihood analysis with 1000 bootstrap repetitions using CLC software and visualized using iTOLTPS subfamilies are grouped as follows: TPS- a (green), monoterpenes in TPS-b (blue) and TPS-g (red), and diterpenes in TPS-c (orange) and TPS-e/f (purple).
The biochemical analysis described here focuses on the TPS-a subfamily only, guided by previous studies that showed sesquiterpene compounds are a predominant component of volatile collections of M. truncatula in response to herbivory (Leitner et al., 2005; Arimura et al., 2008). In addition, subsequent biochemical characterization assigned metabolites for three of fourteen sesquiterpene synthases - namely MtTPS1, 5, and 10 - which are in the TPS-a subfamily (Figure 1) (Gomez et al., 2005; Arimura et al., 2008; Garms et al., 2010). MtTPS2 showed increased gene expression upon elicitation and was putatively described as germacrene-D synthase based on phylogenetic analysis but there was no corresponding biochemical confirmation (Gomez et al., 2005). Figure 2 shows little overlap of the members of TPS-a clade amongst plants as they tend to group by species. In S. lycopersicum the fifteen TPSs encode for sesquiterpenes whereas Arabidopsis TPS-a members are not limited to sesquiterpene biosynthetic genes (Zhou and Pichersky, 2020). Combined, these observations guided our investigations to determine the functional landscape of TPS-a subfamily members in M. truncatula through heterologous expression.
To correlate metabolites produced to the identified biosynthetic genes, we assessed for TPS activity and resultant metabolite formation using a modular metabolic engineering system in E. coli (Kitaoka et al., 2015). Briefly, heterologous expression was performed through co-expression of an (E, E)-FPP synthase with uncharacterized full-length TPS-a subfamily genes (MtTPS2, 7, 8, 9, 11, 12, 13, 24, 25, 26 and 27). Metabolite production was analyzed using GC-MS (Figure 3). Compounds were compared to authentic standards when available, the NIST library database, or verified through de novo structural elucidation by NMR.
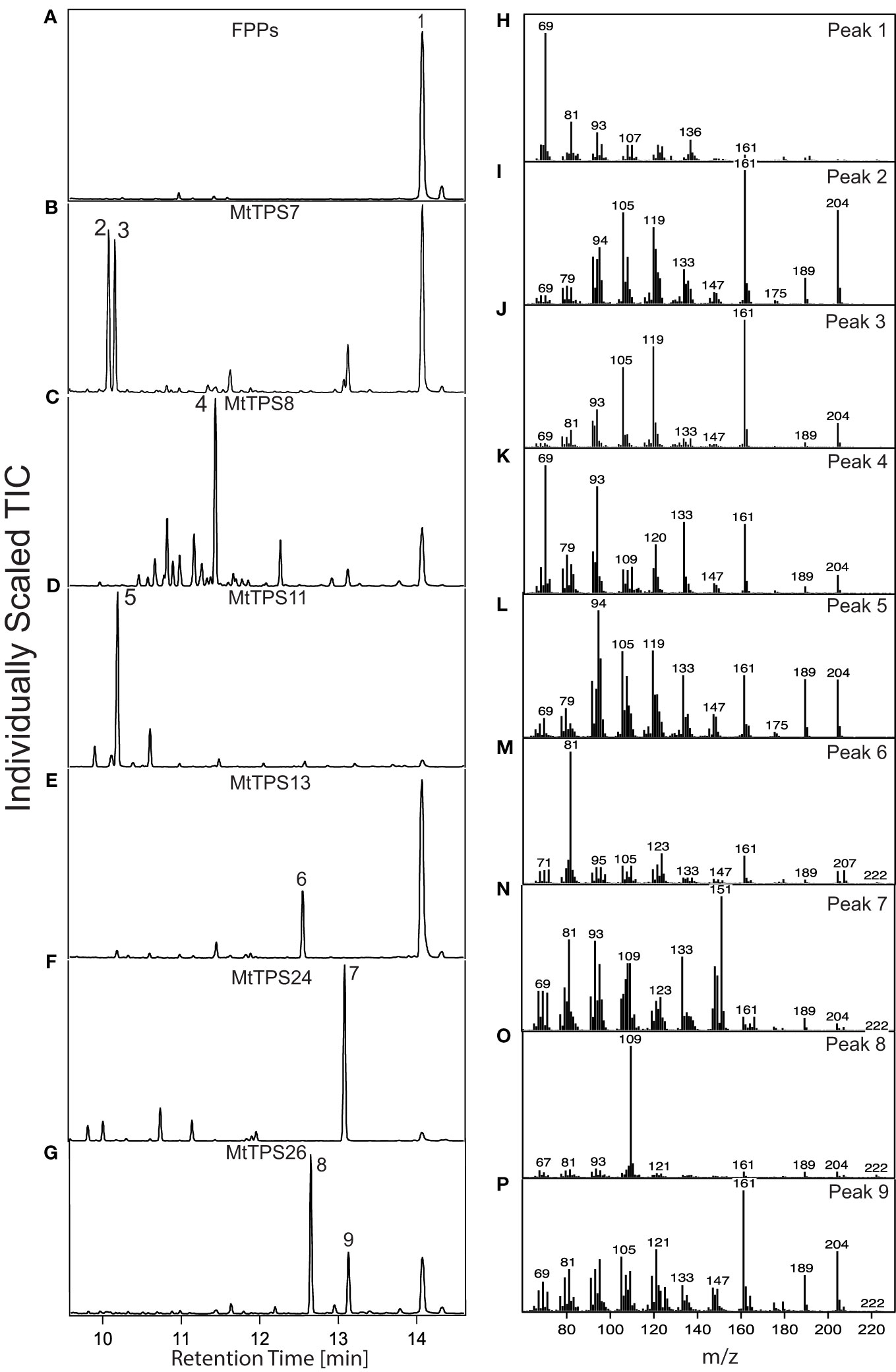
Figure 3 Functional of the TPS-a subfamily in Medicago truncatula. Total ion chromatograms of products resulting from combinatorial expression in E. coli of (E, E)-FPP synthase and corresponding genes from the TPS-a subfamily. The chromatograms focus on the main products. Detailed analysis of the smaller peaks is depicted in Supplementary Figures S3-8. Compound identification was through similarity to library matches (NIST, Wiley) for B-E and NMR verification for G. (A) pET28b containing FPP synthase produces (H) dephosphorylated FPP (farnesol) (1) (B) MtTPS7 produces (I, J) cyclosativene (2), α-copaene (3). (C) MtTPS8 produces (K) farnesene-like product (4). (D) MtTPS11 produces (L) longicyclene (5). (E) MtTPS13 makes a (M) germacrene-D-4-ol product (6), (F) MtTPS24 makes a (N) unknown product (7) and (G) MtTPS26 makes a (O, P) eudesm4(14)-en-6β-ol (8) and a guai-6-en-10β-ol (9).
Analysis of resultant metabolites produced by TPS-a subfamily members reveals the production of an assortment of sesquiterpenes. Figure 3 highlights the most abundant products formed by TPSs and products in smaller quantities are described in the supplementary information (Suppplementary Figures S3-S8). MtTPS7 is a multiproduct enzyme that produces α-copaene (2), cyclosativene (3) and a hydroxylated compound which matches a compound produced by MtTPS26 (Figure 3B; Supplementary Figure S3). MtTPS8 is a multiproduct synthase that produces farnesene (4) as the major compound and exhibits relatively significant turnover to amorphadiene, bergamotene and nerolidol, and production of up to fourteen products in smaller amounts (Figure 3C; Supplementary Figure S4), MtTPS11, produces longicyclene (5) as the main product and a smaller amount of longifolene and longipinene (Figure 3D; Supplementary Figure S5).
Although terpene synthases more commonly form hydrocarbons, the addition of water before terminating deprotonation generates hydroxylated products (Christianson, 2017). M. truncatula produces a set of TPSs capable of producing such hydroxylated compounds as supported by a molecular weight of 222. MtTPS13 produces germacrene-D-4-ol (6) as compared to NIST (Figure 3E; Supplementary Figure S6). The genes, MtTPS24 and MtTPS26 are present in the Mt4.0v1 genome and not identified in earlier genomes (Parker et al., 2014). MtTPS24 produces a compound with a distinct base peak at 151 (7) (Figure 3F; Supplementary Figure S7). However, we were unable to collect NMR data as the compound aggregates in tested deuterated solvents (chloroform, methanol and DMSO) and no further analysis could be performed. Upon expression, MtTPS26 produces two predominant compounds that required structural verification by NMR due to the unavailability of authentic standards. Structural elucidation by NMR and comparison to literature values showed that compound 8 was eudesm4(14)-en-6β-ol (Figure 3G) with chemical shift data closely matching previously reported data for 10-epi-junenol except for the hydroxyl in a 6β-position in 8 (Supplementary Table S5) (F. et al., 1976). Compound 9 was confirmed to be guai-6-en-10β-ol, matching previously reported chemical shifts as shown in Supplementary Table S6 (Lago et al., 2000). No activity was detected with MtTPS2, 9, 12, 25 and 27 upon co-expression with (E, E)-FPP synthase using the combinatorial expression system.
The ability of TPSs to make several products is not uncommon, for example, MtTPS5 was demonstrated to produce up to twenty-seven products (Garms et al., 2010; Vattekkatte et al., 2017). Here, MtTPS8 is the most promiscuous forming up to twenty compounds (Supplementary Figure S4). Multiple products produced in smaller quantities are highlighted for all the genes (Supplementary Figures S3-S8). Similarly, because of the related mechanism of terpene synthases, it is common to observe that some genes will also produce the same compounds in varying amounts in vitro. This is observed for MtTPS7 and MtTPS26 which both produce 9; a similar case is noted for MtTPS11 which produces longicyclene (5) as the main product, but profiles of MtTPS5 & 10 also produce longicyclene in small amounts (Garms et al., 2010; Yadav et al., 2019).
In summary, this biochemical analysis defines gene-to-metabolite relationships in the TPS-a subfamily of M. truncatula. We defined the genetic basis for the biosynthesis of farnesene, nerolidol, cyclosativene, copaene and longicyclene, enabling the association of genes responsible for the biosynthesis of several compounds observed in terpene metabolite response to herbivore feeding (Leitner et al., 2005) and identified a set of sesquiterpene alcohols (Figure 4).
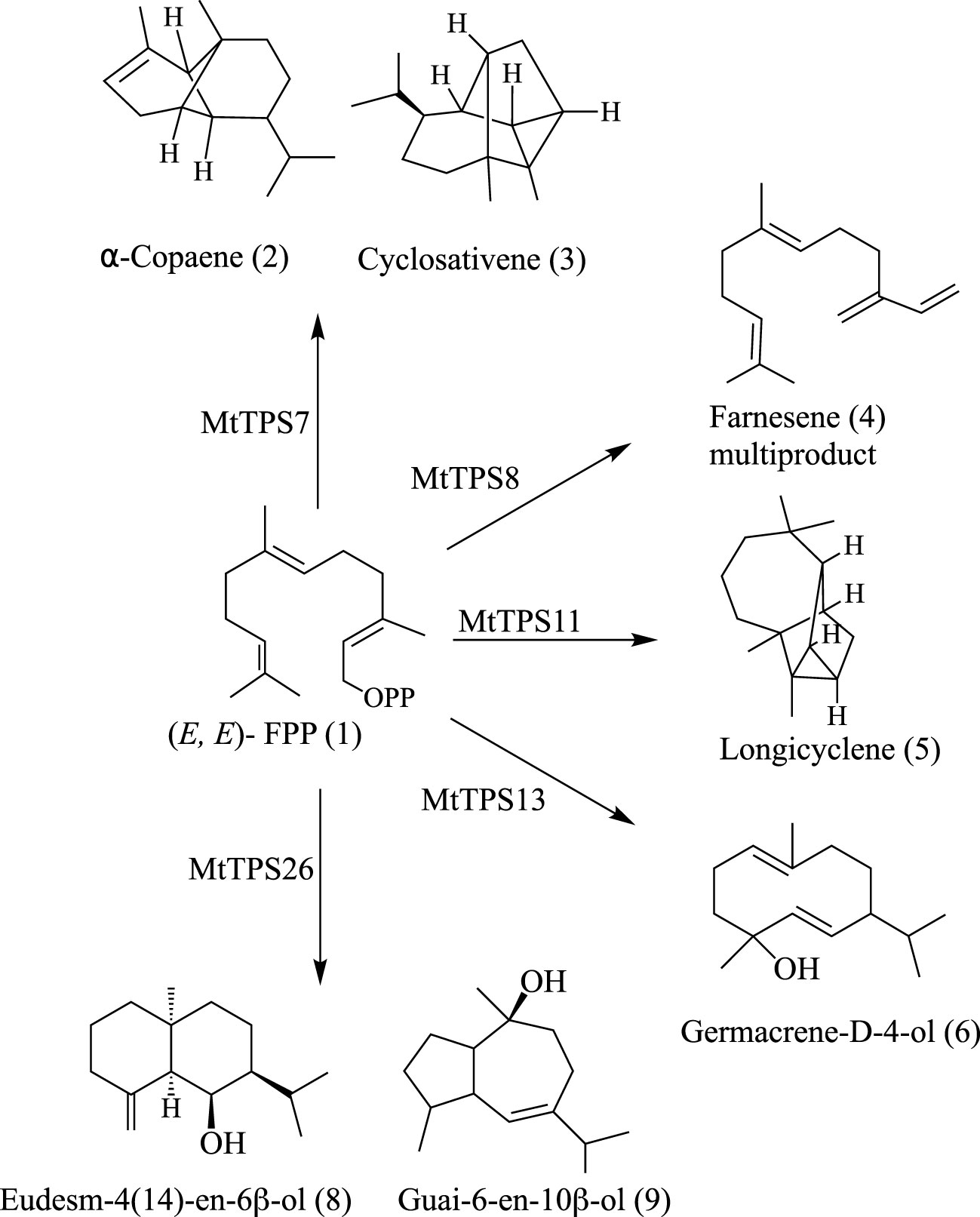
Figure 4 Summary of the main products from biochemical characterization of TPS-a subfamily members in M. truncatula.
Gene expression of the TPS-a subfamily members in response to elicitors was carried out through exogenous application of phytohormones MeJA and SA and analysis by qPCR (Figure 5). Overall, TPS-a genes showed tissue-specific and temporal differences in expression in response to elicitation with MeJA and SA. There was higher expression of TPS-a genes in leaf tissue compared to the root upon elicitation with MeJA. Specifically, our time course evaluations revealed that MeJA application resulted in increased expression (16-20 fold) of MtTPS1 after hours of exposure in leaves whereas expression of MtTPS2 and MtTPS5 was highest after 12 hours. A 4-8 fold increase in expression was observed amongst the MtTPS8, MtTPS10, MtTPS13 and MtTPS26 genes in leaf tissues after 6 and 12 hours of post-treatment of MeJA. MtTPS11, 12, 24, 25 & 27 have low expression in both the leaves and roots. There was a consistently low expression for all the TPS-a members in roots under MeJA elicitation, with MtTPS25 & 27 showing down-regulation.
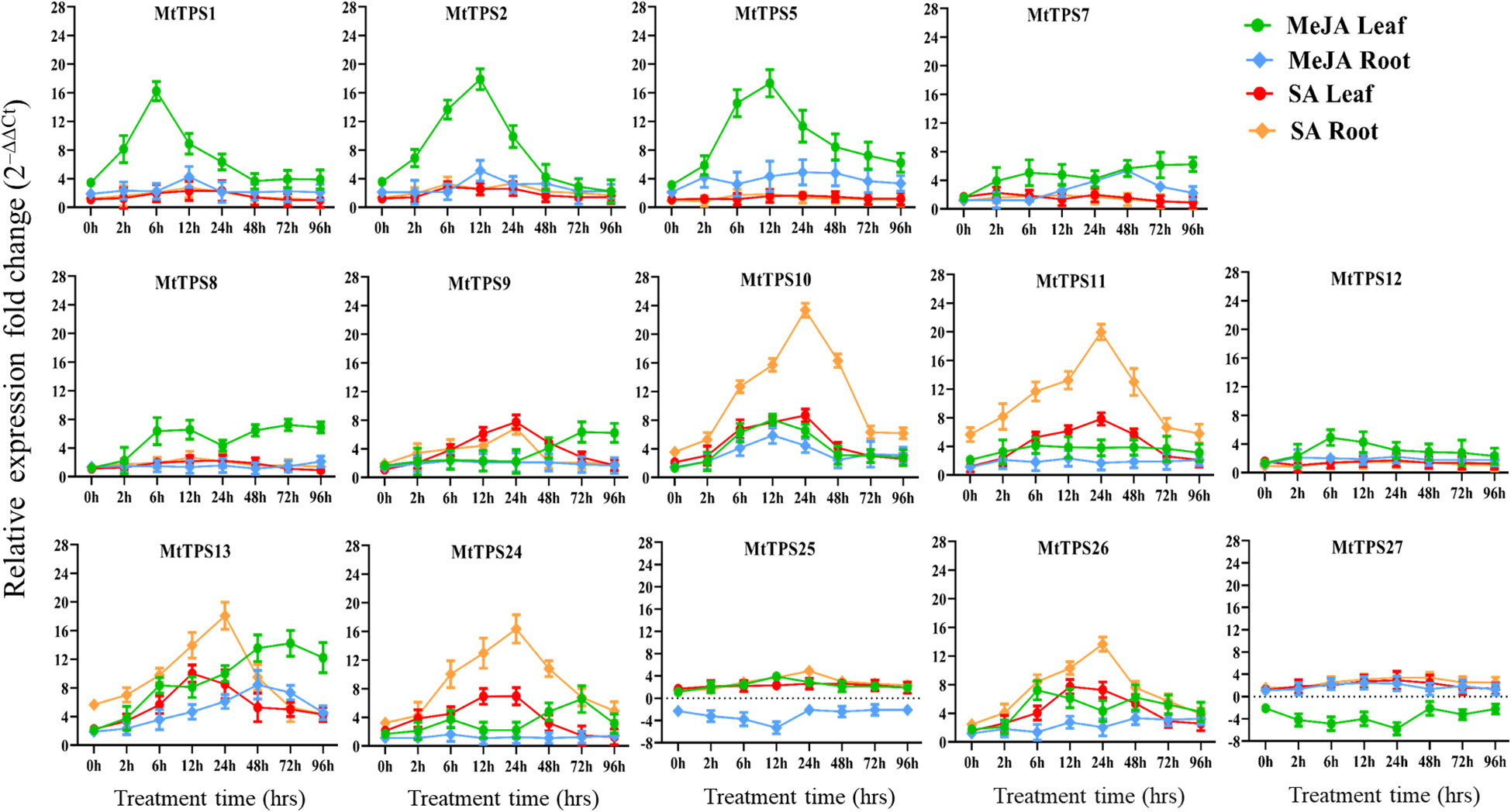
Figure 5 Expression profile of TPS-a terpene synthase genes in leaf and root tissues in response to exogenous application of MeJA and SA in a time course exposure by qRT-PCR. Plants were exposed for 0h, 2 h, 6 h, 12 h, 24 h, 48 h, 72 h and 96 h after foliar application of MeJA (1 mM) or SA (1.5 mM). The data are expressed as the relative level of expression of fold change relative to controls (0.01% Triton-100 for MeJA, or 1% ethanol for SA control) plants, based on the 2−ΔΔct method. Error bars represent the mean ± standard deviation (SD) levels of the relative abundance of three biological replicates.
In contrast, SA induced gene expression occurred mainly in roots. The genes MtTPS10 and MtTPS11 showed the highest expression at 24 hours compared to control with a 20-fold increase whereas a 16-fold increase in expression was observed for genes MtTPS13, 24 & 26 (Figure 5). However, comparatively lower expression was observed in both leaf and root tissue for the remaining eight genes (MtTPS1, 2, 5, 7, 8, 12, 25 & 27). Overall, the TPS-a genes exhibited differential expression to the phytohormones where MeJA largely affected the expression of TPS genes in leaf tissue and SA in root tissue upon foliar treatment.
We used a publicly available expression data set of M. truncatula from the Noble Research Institute legacy Gene Atlas V3 to survey the effect of (a)biotic stress on the TPS-a subfamily (Benedito et al., 2008; Carrere et al., 2021). Mining of the reference data set, which is a collation of gene expression in different tissues, shows that terpene synthases MtTPS1, 2 & 5 which produce volatile compounds exhibited increased expression in the shoots and leaves. Transcript levels of the MtTPS13 were increased during root and nodule development, AM symbiosis, and fungal colonization (Supplementary Figure S23A).
Evaluation of the TPS-a subfamily genes response to abiotic stress highlights increased transcript abundance of MtTPS1, 2, 5 and MtTPS13 under limited nitrogen (N2), ammonium (NH4), nitrate (NO3), drought, and salt stress in shoot and root tissues (Supplementary Figure S23B). An assessment of varied biotic stressors showed that MtTPS1 and 2 had increased expression in leaf tissue after aphid infestation whereas MtTPS13, MtTPS24 and MtTPS25 displayed increased transcript abundance in root tissues under pathogenic fungi, Rhizoctonia solani and Fusarium oxysporum (Supplementary Figure S23C). Notably, datasets obtained from Gene Atlas do not capture the expression of six terpene synthases (MtTPS7,8, 9, 25, 26 & 27). Combined this data supports the variable expression of select terpene synthases dependent on tissue, time and stressor applied.
The catalytic mechanism of sesquiterpenes proceeds via a succession of carbocationic intermediates initiated by dephosphorylation and subsequent deprotonation or water capture (Christianson, 2017). The metabolites of the encoded enzyme of MtTPS1, MtTPS7 and MtTPS8 have characteristic profiles to previously described enzymes, for example, MtTPS8 produces (E)-β-farnesene with bergamotene similar to what is observed for ZmTPS10 (Schnee et al., 2002; Rasmann et al., 2005; Schnee et al., 2006). Notably, MtTPS11 produces longicyclene, a compound commonly found in pine trees (Martin et al., 2004). Previous studies have identified a longifolene synthase with smaller amounts of longicyclene and longipinene - here the enzyme MtTPS11 forms longicyclene as the major product. Olefins (compounds 2-5) produced by M. truncatula are characteristic in volatile blends of several plants upon herbivore feeding and associated interactions (Schnee et al., 2002; Schnee et al., 2006). Here, some of the TPS-a genes quenched the reaction through water capture resulting in sesquiterpene alcohols (compounds 6-9) that have been identified in other plants, but little is known of their biological activity. Our biochemical analysis combined with the previously characterized genes establishes the use of terpenes common to plants generating a species-specific sesquiterpene blend.
A summary of the known gene-to-metabolite relationships in the TPS-a subfamily of M. truncatula shows functional divergence from the TPS profiles of the two comprehensively biochemically characterized eudicots (Figure 4). Previously characterized genes from M. truncatula mainly fall in the TPS-a clade (Figures 1 & 2). Comparison of the currently defined M. truncatula TPS-a subfamily members compared to those of Arabidopsis and tomato show a difference in metabolite profiles. In tomato, the fifteen TPSs encode for sesquiterpenes whereas the product outcome of the TPS-a subfamily in A. thaliana is not limited to sesquiterpenes (Supplementary Table S4) (Zhou and Pichersky, 2020). Biochemical analysis here revealed that a subset of the metabolites synthesized by M. truncatula sesquiterpene synthases was specific to the species, revealing functional divergence.
In this study, we elicited TPS expression by exogenous application of phytohormones. The genes that produce volatile compounds had increased expression on the early onset of foliar application of MeJA (Figures 1, 3). Genes corresponding to the production of volatile compounds β-caryophyllene (MtTPS1), putative germacrene-D (MtTPS2) and MtTPS5 a multiproduct synthase with the demonstrated antiherbivore response were upregulated by 16 to 20-fold (Figure 5). These observations correlate in part to a study by Leitner et al. (2005), which showed that herbivore interactions and artificial wounding induced the emission of up to 23 compounds (Leitner et al., 2005; Arimura et al., 2008). This observation is supported by data from gene atlas that also showed increased expression of the volatile terpenes predominantly in the leaves under various stresses (Supplementary Figure S23). However, ten out of fourteen TPS-a subfamily members exhibited moderate upregulation by 4-6-fold using MeJA.
In contrast, elicitation using SA increased the transcript abundance in root tissue of select genes; MtTPS10, MtTPS11, MtTPS13, MtTPS24 and MtTPS26 (Figure 5). In general, SA mediates response to biotrophic and hemibiotrophic pathogens and triggers systemic acquired resistance (Anand et al., 2008). Yadav et al. (2019) demonstrated that MtTPS10, which produces himalachol, reduced susceptibility to the oomycete pathogen Aphanomyces euteiches (Yadav et al., 2019). The terpene scaffolds may be elaborated by downstream enzymes resulting in further oxygenated non-volatile sesquiterpenoids. The resultant compounds may have various bioactivity similar to previously identified antifungals and insect deterrents such as germacrene-A based costunolides in lettuce; costic acids derived from selinene and β-macrocarpene based zealexins produced in maize under pathogen attack (Nguyen et al., 2010; Ding et al., 2017; Ding et al., 2019).
In conclusion, the results reported here establish gene-metabolite relationships of the TPS-a subfamily that encodes for sesquiterpene synthases in M. truncatula. Biochemical analysis reveals a blend of sesquiterpenes that highlight the evolution of species-specific chemotypes. Gene expression analysis shows a temporal tissue-specific and stress induced variation. While determining the function of these compounds is beyond the scope of this work, our study further defines the sesquiterpene biosynthetic capacity and provides a foundation to understand the biological roles of the compounds.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
HH: Investigation, Methodology, Writing – original draft, Writing – review & editing. MI: Investigation, Writing – original draft. GW: Formal analysis, Writing – review & editing. SM: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support was provided by the Department of Biochemistry and Molecular Biology at the University of Massachusetts – Amherst; Richard and Susan Smith Family Foundation and National Science Award 2300060 to SM. HH was supported by the National Research Service Award T32 GM135096 from the National Institutes of Health.
Mass spectral data were obtained at the University of Massachusetts Mass Spectrometry Core Facility, RRID: SCR_019063. NMR data were obtained at Rensselaer Polytechnic Institute (RPI) NMR Core Faculties: NIST Award # 60NANB22D167.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1349009/full#supplementary-material
Anand, A., Uppalapati, S. R., Ryu, C. M., Allen, S. N., Kang, L., Tang, Y., et al. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146 (2), 703–715. doi: 10.1104/pp.107.111302
Arimura, G., Garms, S., Maffei, M., Bossi, S., Schulze, B., Leitner, M., et al. (2008). Herbivore-induced terpenoid emission in Medicago truncatula: concerted action of jasmonate, ethylene and calcium signaling. Planta 227 (2), 453–464. doi: 10.1007/s00425-007-0631-y
Bao, T., Shadrack, K., Yang, S., Xue, X., Li, S., Wang, N., et al. (2020). Functional characterization of terpene synthases accounting for the volatilized-terpene heterogeneity in Lathyrus odoratus cultivar flowers. Plant Cell Physiol. 61 (10), 1733–1749. doi: 10.1093/pcp/pcaa100
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55 (3), 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Carrere, S., Verdier, J., Gamas, P. (2021). MtExpress, a comprehensive and curated RNAseq-based gene expression atlas for the model legume Medicago truncatula. Plant Cell Physiol. 62 (9), 1494–1500. doi: 10.1093/pcp/pcab110
Chen, F., Tholl, D., Bohlmann, J., Pichersky, E.. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66 (1), 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Christianson, D. W. (2017). Structural and chemical biology of terpenoid cyclases. Chem. Rev. 117 (17), 11570–11648. doi: 10.1021/acs.chemrev.7b00287
Constantino, N., Oh, Y., Şennik, E., Andersen, B., Warden, M., Oralkan, Ö, et al. (2021). Soybean cyst nematodes influence aboveground plant volatile signals prior to symptom development. Front. Plant Sci. 12, 749014. doi: 10.3389/fpls.2021.749014
Degenhardt, J., Hiltpold, I., Köllner, T. G., Frey, M., Gierl, A., Gershenzon, J., et al. (2009). Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. U.S.A. 106 (32), 13213–13218. doi: 10.1073/pnas.0906365106
Ding, Y., Huffaker, A., Köllner, T. G., Weckwerth, P., Robert, C.A.M., Spencer, J.L., et al. (2017). Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol. 175 (3), 1455–1468. doi: 10.1104/pp.17.00879
Ding, Y., Murphy, K. M., Poretsky, E., Mafu, S., Yang, B., Char, S. N., et al. (2019). Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants 5 (10), 1043–1056. doi: 10.1038/s41477-019-0509-6
Ding, Y., Weckwerth, P. R., Poretsky, E., Murphy, K. M., Sims, J., Saldivar, E., et al. (2020). Genetic elucidation of interconnected antibiotic pathways mediating maize innate immunity. Nat. Plants 6 (11), 1375–1388. doi: 10.1038/s41477-020-00787-9
F., Thomas, Ozainne, M., Decorzant, R., Näf, F. (1976). 10-epijunenol, a new cis-eudesmane sesquiterpenoid. Tetrahedron 32, 2261–2264. doi: 10.1016/0040-4020(76)85143-5
Falara, V., Akhtar, T. A., Nguyen, T. T., Spyropoulou, E. A., Bleeker, P. M., Schauvinhold, I., et al. (2011). The tomato terpene synthase gene family. Plant Physiol. 157 (2), 770–789. doi: 10.1104/pp.111.179648
Garms, S., Köllner, T. G., Boland, W. (2010). A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J. Org. Chem. 75 (16), 5590–5600. doi: 10.1021/jo100917c
Gomez, S. K., Cox, M. M., Bede, J. C., Inoue, K., Alborn, H. T., Tumlinson, J. H., et al. (2005). Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula. Arch. Insect Biochem. Physiol. 58 (2), 114–127. doi: 10.1002/arch.20037
Han, T., Y., Shao, R., Gao, J., Gao, Y., Jiang, Y., Yang, Y., et al. (2023). Functional Characterization of a (E)-B ocimene synthase gene contributing to the defense against Spordoptera litura. Int. J. Mol. Sci. 24 (8), 7182. doi: 10.3390/ijms24087182.
He, J., Verstappen, F., Jiao, A., Dicke, M., Bouwmeester, H. J., Kappers, I. F. (2022). Terpene synthases in cucumber (Cucumis sativus) and their contribution to herbivore-induced volatile terpenoid emission. New Phytol. 233 (2), 862–877. doi: 10.1111/nph.17814
Kang, Z. W., Liu, F.H., Zhang, Z. F., Tian, H.G., Liu, T.X. (2018). Volatile β-ocimene can regulate developmental performance of peach aphid. Front. Plant Sci. 9, 708. doi: 10.3389/fpls.2018.00708
Kitaoka, N., Wu, Y., Xu, M., Peters, R. J. (2015). Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl. Microbiol. Biotechnol. 99 (18), 7549–7558. doi: 10.1007/s00253-015-6496-2
Lago, J. H., Brochini, C. B., Roque, N. F. (2000). Terpenes from leaves of Guarea macrophylla (Meliaceae). Phytochemistry 55 (7), 727–731. doi: 10.1016/S0031-9422(00)00302-2
Leitner, M., Boland, W., Mithöfer, A. (2005). Direct and indirect defences induced by piercing-sucking and chewing herbivores in Medicago truncatula. New Phytol. 167 (2), 597–606. doi: 10.1111/j.1469-8137.2005.01426.x
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Lin, J., Wang, D., Chen, X., Köllner, T. G., Mazarei, M., Guo, H., et al. (2017). An (E,E)-α-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 15 (4), 510–519. doi: 10.1111/pbi.12649
Liu, J., Huang, F., Wang, X., Zhang, M., Zheng, R., Wang, J., et al. (2014). Genome-wide analysis of terpene synthases in soybean: functional characterization of GmTPS3. Gene 544 (1), 83–92. doi: 10.1016/j.gene.2014.04.046
Martin, D. M., Fäldt, J., Bohlmann, J. (2004). Functional characterization of nine Norway Spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 135 (4), 1908–1927. doi: 10.1104/pp.104.042028
Murphy, K. M., Zerbe, P. (2020). Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 172, 112289. doi: 10.1016/j.phytochem.2020.112289
Navia-Giné, W. G., Yuan, J. S., Mauromoustakos, A., Murphy, J. B., Chen, F., Korth, K.L. (2009). Medicago truncatula (E)-beta-ocimene synthase is induced by insect herbivory with corresponding increases in emission of volatile ocimene. Plant Physiol. Biochem. 47 (5), 416–425. doi: 10.1016/j.plaphy.2009.01.008
Nguyen, D. T., Göpfert, J.C., Ikezawa, N., Macnevin, G., Kathiresan, M., Conrad, J., et al. (2010). Biochemical conservation and evolution of germacrene A oxidase in asteraceae. J. Biol. Chem. 285 (22), 16588–16598. doi: 10.1074/jbc.M110.111757
Parker, M. T., Zhong, Y., Dai, X., Wang, S., Zhao, P.. (2014). Comparative genomic and transcriptomic analysis of terpene synthases in Arabidopsis and Medicago. IET Syst. Biol. 8 (4), 146–153. doi: 10.1049/iet-syb.2013.0032
Pichersky, E., Raguso, R. A. (2018). Why do plants produce so many terpenoid compounds? New Phytol. 220 (3), 692–702. doi: 10.1111/nph.14178
Rasmann, S., Köllner, T. G., Degenhardt, J., Hiltpold, I., Toepfer, S., Kuhlmann, U., et al. (2005). Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434 (7034), 732–737. doi: 10.1038/nature03451
Rey, T., Laporte, P., Bonhomme, M., Jardinaud, M. F., Huguet, S., Balzergue, S., et al. (2016). A central transcriptional regulator of symbiotic nodule development, is also a determinant of. Front. Plant Sci. 7, 1837. doi: 10.3389/fpls.2016.01837
Schmelz, E. A., Huffaker, A., Sims, J.W., Christensen, S.A., Lu, X., Okada, K., et al. (2014). Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 79 (4), 659–678. doi: 10.1111/tpj.12436
Schnee, C., Köllner, T. G., Gershenzon, J., Degenhardt, J. (2002). The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-beta-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 130 (4), 2049–2060. doi: 10.1104/pp.008326
Schnee, C., Köllner, T. G., Held, M., Turlings, T. C., Gershenzon, J., Degenhardt, J. (2006). The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. U.S.A. 103 (4), 1129–1134. doi: 10.1073/pnas.0508027103
Steinbrenner, A. D., Saldivar, E., Hodges, N., Guayazán-Palacios, N., Chaparro, A. F., Schmelz, E. A. (2022). Signatures of plant defense response specificity mediated by herbivore-associated molecular patterns in legumes. Plant J. 110 (5), 1255–1270. doi: 10.1111/tpj.15732
Vattekkatte, A., Garms, S., Boland, W. (2017). Alternate cyclization cascade initiated by substrate isomer in multiproduct Terpene synthase from Medicago truncatula. J. Org. Chem. 82 (6), 2855–2861. doi: 10.1021/acs.joc.6b02696
Yadav, H., Dreher, D., Athmer, B., Porzel, A., Gavrin, A., Baldermann, S., et al. (2019). Medicago TERPENE SYNTHASE 10 is involved in defense against an oomycete root pathogen. Plant Physiol. 180 (3), 1598–1613. doi: 10.1104/pp.19.00278
Zhang, H., Cao, Y., Xu, Y., Zhou, C., Liu, W., Zhu, R., et al. (2020). Efficient generation of CRISPR/Cas9-mediated homozygous/Biallelic. Front. Plant Sci. 11, 294. doi: 10.3389/fpls.2020.00294
Zhang, J. R., Li, M., Xu, R. I., Hoffmann, Y., Zhang, B., Liu, M., et al. (2021). A (conditional) role for labdane-related diterpenoid natural products in rice stomatal closure. New Phytol. 230 (2), 698–709. doi: 10.1111/nph.17196
Keywords: Medicago truncatula, terpene synthase, sesquiterpenes, biochemical analysis, chemical diversity
Citation: Hendrickson H, Islam M, Wabo GF and Mafu S (2024) Biochemical analysis of the TPS-a subfamily in Medicago truncatula. Front. Plant Sci. 15:1349009. doi: 10.3389/fpls.2024.1349009
Received: 04 December 2023; Accepted: 19 January 2024;
Published: 15 February 2024.
Edited by:
Naoki Kitaoka, Hokkaido University, JapanReviewed by:
Naoki Ube, Toyama Prefectural University, JapanCopyright © 2024 Hendrickson, Islam, Wabo and Mafu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sibongile Mafu, c21hZnVAdW1hc3MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.