- College of Agronomy, Northwest A&F University, Yangling, Shaanxi, China
Plant-specific VQ proteins have crucial functions in the regulation of plant growth and development, as well as in plant abiotic stress responses. Their roles have been well established in the model plant Arabidopsis thaliana; however, the functions of the potato VQ proteins have not been adequately investigated. The VQ protein core region contains a short FxxhVQxhTG amino acid motif sequence. In this study, the VQ31 protein from potato was cloned and functionally characterized. The complete open reading frame (ORF) size of StVQ31 is 672 bp, encoding 223 amino acids. Subcellular localization analysis revealed that StVQ31 is located in the nucleus. Transgenic Arabidopsis plants overexpressing StVQ31 exhibited enhanced salt tolerance compared to wild-type (WT) plants, as evidenced by increased root length, germination rate, and chlorophyll content under salinity stress. The increased tolerance of transgenic plants was associated with increased osmotic potential (proline and soluble sugars), decreased MDA accumulation, decreased total protein content, and improved membrane integrity. These results implied that StVQ31 overexpression enhanced the osmotic potential of the plants to maintain normal cell growth. Compared to the WT, the transgenic plants exhibited a notable increase in antioxidant enzyme activities, reducing cell membrane damage. Furthermore, the real-time fluorescence quantitative PCR analysis demonstrated that StVQ31 regulated the expression of genes associated with the response to salt stress, including ERD, LEA4-5, At2g38905, and AtNCED3. These findings suggest that StVQ31 significantly impacts osmotic and antioxidant cellular homeostasis, thereby enhancing salt tolerance.
1 Introduction
Salt stress significantly and adversely impacts plant growth and crop yields (Liu et al., 2022; Colin et al., 2023). Plants have evolved numerous strategies to cope with adverse environmental conditions, including but not limited to morphological, physiological, and biochemical adaptations. These mechanisms involve the transcriptional activation of relevant genes, regulating gene expression, and achieving adaptation to salt stress (Sui et al., 2017). Therefore, understanding the molecular mechanisms of plant salt tolerance has scientific importance and practical significance (Liang et al., 2023). To reveal these mechanisms, extensive studies have been conducted to identify and characterize numerous genes and proteins associated with salt tolerance, such as LRX3/4/5 proteins, CCCH-type zinc finger proteins, LncRNA973, AtbZIP17, etc (Zhang et al., 2019; Medina et al., 2020; Zhao et al., 2021; Gong et al., 2022). Among them, transcription factors and their regulatory functions have been demonstrated to have a crucial impact on plant environmental adaptation (Li et al., 2022).
VQ proteins that function as transcriptional regulators were first identified in Arabidopsis thaliana. In terms of protein structure, they possess a core FxxxVQxLTG sequence containing 5 highly conserved amino acids, where x is any amino acid (Shan et al., 2021). Subsequently, VQ genes were discovered in other organisms, such as Arabidopsis, rice, maize, grape, soybean, potato, bamboo, poplar, and tea, with a total of 34, 40, 61, 18, 74, 37, 28, 51, and 25 VQ members identified, respectively (Kim et al., 2013; Wang et al., 2014, 2015, 2015; Chu et al., 2016; Song et al., 2016; Wang et al., 2017; Guo et al., 2018). The VQ family has been extensively studied and, in detail and comprehensively, functionally characterized in Arabidopsis. VQ can interact with different transcription factors, such as WRKY and serine/threonine kinase, to regulate various stress responses in plants (Cheng et al., 2012). Moreover, the plant VQ proteins can regulate gene expression mediated by WRKY by interacting with WRKY transcription factors (Dong et al. and Petcher et al.). This interaction is achieved through the conserved residues V and Q of the FxxhVQxhTG motif (Dong et al., 2003; Pecher et al., 2014). AtCaMBP25 (AtVQ15) plays a role as a suppressor in the regulatory mechanisms of plant seedlings against osmotic stress (Perruc et al., 2004). The interaction between AtVQ9 and WRKY8 reduces WRKY8’s capacity to bind to DNA and mediate salt stress responses, thereby negatively regulating salt tolerance (Hu Y. et al., 2013). PeVQ28 enhances salt tolerance in bamboo via ABA-dependent signaling pathways (Wang et al., 2017). Additionally, salicylic acid and methyl jasmonate strongly upregulated the AtVQ10 gene transcriptional levels in Arabidopsis thaliana (Chen et al., 2018). In maize, ZmVQ19 and ZmVQ54 were highly expressed under drought conditions. In rice, OsVQ2, OsVQ16, and OsVQ20 also exhibited regulatory patterns similar to the maize VQ genes (Kim et al., 2013; Song et al., 2016). Overall, VQ proteins have been shown to play critical roles in many biological processes.
Plant performance under adverse growth conditions can be directly indicated by alterations in total protein content, chlorophyll, soluble sugars, and various other physiological parameters (Li et al., 2022; Hernández-Fernández et al., 2023). For example, when plants are stressed, the content of Pro increases rapidly and acts as a signal to regulate the expression of downstream stress-protective proteins and reduce stress damage to plants (Gu et al., 2019). MDA can seriously damage the protein and enzyme structure and the cell membrane system in plant cells, and changes in MDA content reflect, to a certain extent, the peroxidation degree of plant cell membranes (Chen et al., 2022). The antioxidant enzymes SOD, POD, and CAT can efficiently remove excess peroxide ions within cells when plants are exposed to salt stress, thereby reducing the negative impact of stress on cellular homeostasis (Xu et al., 2018). The antioxidant system’s capacity to scavenge reactive oxygen species and the extent of membrane lipid peroxidation and metabolic disorders in plants can be assessed by measuring H2O2 and O2- (Huang et al., 2022).
In our previous study, we identified 37 VQ proteins in potato, which were classified into 6 subfamilies based on phylogenetic relationships. The qRT-PCR analysis revealed a significant upregulation of StVQ31 in response to abiotic stress, particularly under high salinity stress (Zhai et al., 2022). As a continuation of our previous research, we focused in this paper on the functional characterization of the StVQ31 gene. The differences in related soluble sugar, chlorophyll indexes, total protein, antioxidant enzyme activities (SOD, POD, CAT, MDA, Pro), and ROS (H2O2 and O2-) in transgenic plants were analyzed. Their correlations were also assessed, which will help us further identify the potential underlying molecular mechanisms. Furthermore, the expression of genes related to salt stress was quantitatively analyzed, which revealed that StVQ31 regulated salt tolerance by promoting the expression of specific genes, such as LEA, and AtNCED3. Admittedly, salt stress in potato plants is complex (Wang et al., 2023), but our study adds some insights into how individual genes contribute to salt tolerance. In addition, it offers a foundation for screening candidate genes that control potato salt tolerance.
2 Materials and methods
2.1 Plant materials
The plant material used in this study was the potato cultivar Désirée provided by our laboratory. The tubers were planted in a climate chamber, in a vermiculite substrate mixed with soil, in a 1:1 ratio, v/v, for 4 weeks at a temperature of 23°C ± 1°C, 16h/8h(day/night). The seeds of WT Arabidopsis thaliana (Columbia (Col-0) ecotype) and 3 T3 transgenic lines (L1-L3) were vernalized for 3 days in 1/2MS medium, transferred to an artificial climate chamber at 24°C and a relative humidity of 80% for ten days, and then transplanted to a vermiculite medium with small square pots (7x7cm). Next, plants were grown for 3 weeks (Kim and Hwang, 2015). Samples were taken before and after salt stress treatment and the collected fresh leaves were quickly placed in −80°C for storage (Zhang et al., 2003). At the same time, chlorophyll content, soluble sugar, SOD, MDA, and other physiological indexes and enzyme activities were measured before and after salt stress treatment.
2.2 Generation of overexpression vectors and transgenic Arabidopsis plants
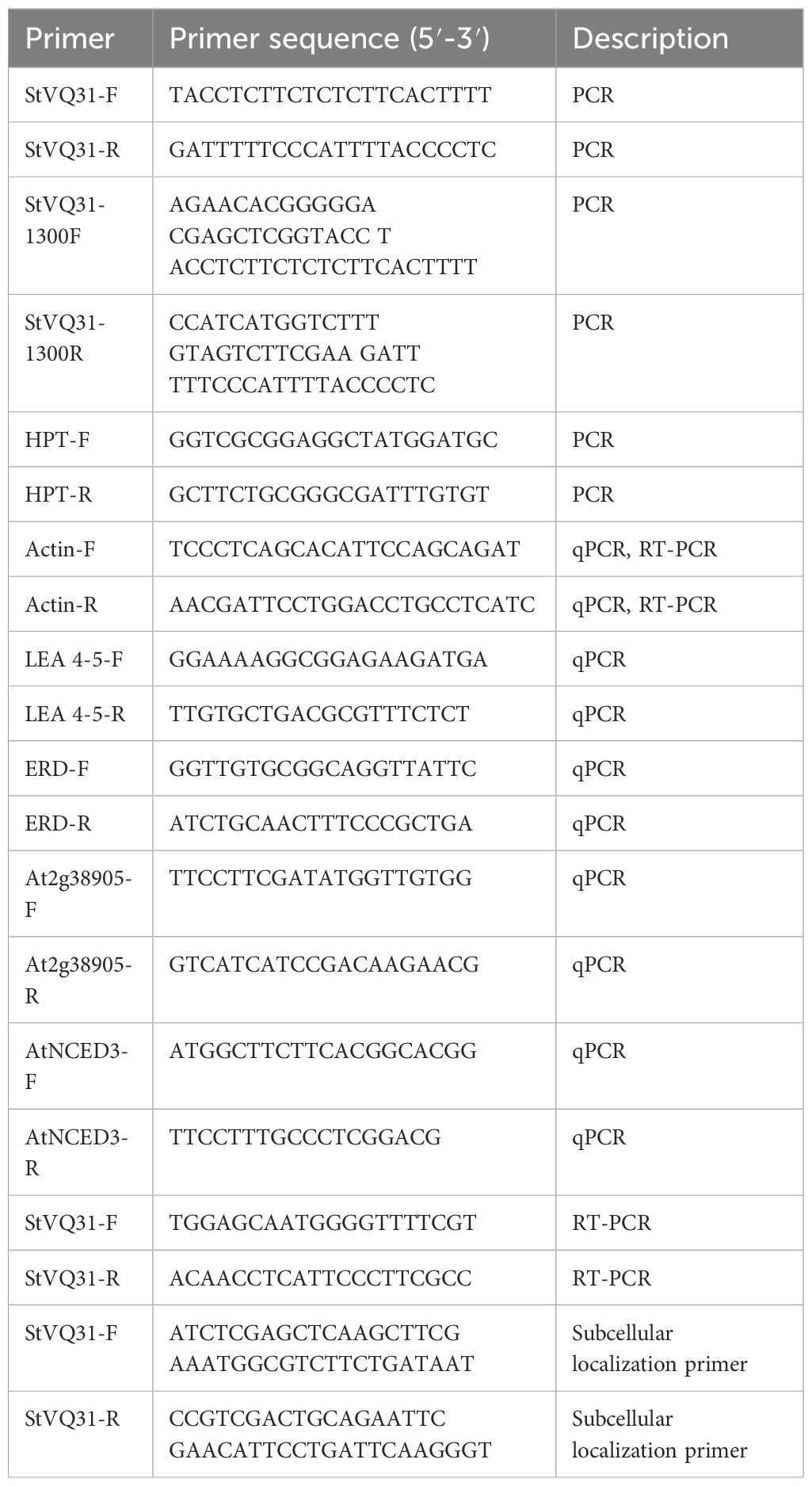
The StVQ31 gene was amplified through PCR using primers specific to the gene (Table 1). The CDS of StVQ31 was ligated to the pCAMBIA1302 overexpression vector within the KpnI and BstbI restriction sites. The recombinant plasmid was verified by sequencing and was then cloned in E. coli DH5α. Subsequently, it was introduced into Agrobacterium GV3101, which was for the genetic transformation of Arabidopsis plants by the floral-dip method (Zhao et al., 2018). When Arabidopsis seeds matured, the T0 generation seeds were harvested. The T0-generation seeds were then sowed on 1/2 MS medium containing 50μg/ml kanamycin, and 16 lines were identified through PCR verification (Figure 1A). The above steps were repeated until T3 transgenic overexpression lines were obtained.
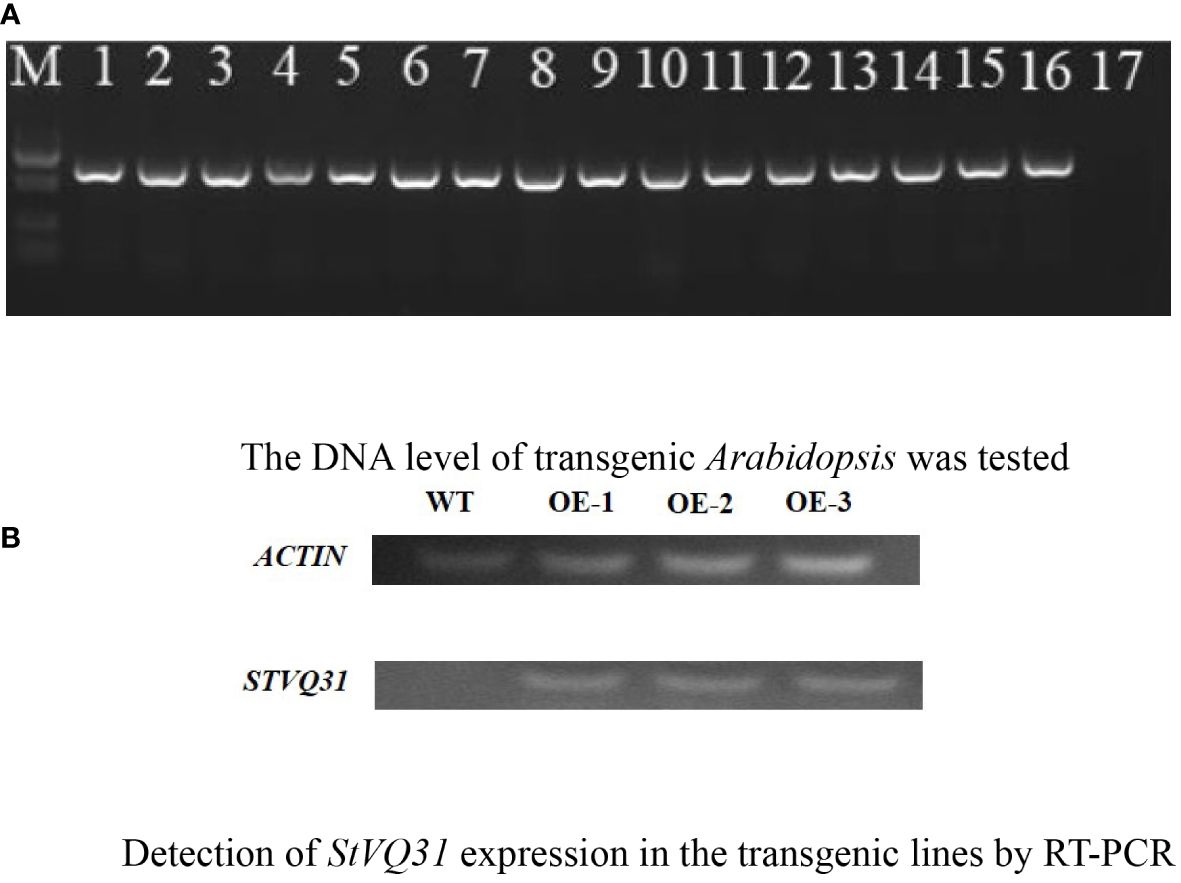
Figure 1 (A) The DNA level of transgenic Arabidopsis was tested; (B) Detection of StVQ31 expression in the transgenic lines by RT-PCR. A:1-16:transgenic Arabidopsis;17: Negative control; M:DL5000 Marker B: Lane WT: wild-type; lanes StVQ31, transgenic plants.
2.3 Prediction of StVQ31 physicochemical properties
We obtained the properties of StVQ31 from the ExPASy website, including its physical and chemical characteristics (http://web.expasy.org/protparam/) (Pedrini et al., 2015). Then, the secondary structure of StVQ31 was predicted using the PHD software (http://www.predictProtein.org/) (Sajid et al., 2018). In addition, the conserved motifs of StVQ31 were determined by MEME (version 4.12.0) (Bailey et al., 2009).
2.4 StVQ31 subcellular localization
The CDS region of StVQ31 (after removal of the stop codon) was ligated to the pCAMBIA1302 vector carrying a green fluorescent protein sequence to produce pCAMBIA1302-StVQ31: GFP. The recombinant plasmid was transferred to Agrobacterium and then infiltrated into tobacco leaves, with the nuclear maker AtWRKY25-mCherry used as a control. All transformed tobacco plants grew in the dark at 22°C for 16 hours and then returned to normal conditions. The GFP and RFP fluorescence signals were observed by IX83-FV1200 confocal laser-scanning microscopy two days after tobacco inoculation.
2.5 Evaluation of seed germination and root length in the presence of salt stress
Seeds from the WT and transgenic plants were cultivated in a growth medium called 1/2 MS, supplemented with 150 mM NaCl. The seeds were incubated at 4°C for 3 days, then transferred to 22°C, with 16 hours of light/8 hours of darkness, and incubated for 10 days. Radicle appearance from the seed coat was used as the criterion for seed germination (Stacey et al., 2016). Meanwhile, seeds of WT and transgenic plants (homozygous lines) were placed in 1/2MS medium for the subsequent experiments. After 7 days, seedlings with uniform appearance and growth were selected, transplanted into 1/2 MS medium containing 150mM NaCl, and grown vertically for 7 days. Subsequently, the changes in root length were recorded. In addition, the salt tolerance of 4-week-old Arabidopsis thaliana transgenic plants was evaluated by irrigation with 150 mM NaCl solution every 2 days for 10 days. The phenotypes of the plants were photographed after 7 days of salt treatment.
2.6 Determination of relevant physiological indicators
Ten physiological indexes in WT and transgenic plants were measured before and after the application of salt treatment (150 mM NaCl). Approximately 0.5 g of leaf tissue, avoiding the midvein of the leaves, was sampled and quickly placed in liquid nitrogen. The samples were fully ground and 9 folds volumes of pH 7.4 PBS buffer were added. After centrifugation at 12,000 ×g at 4°C for 30 min, the supernatant was taken, and the protein concentration was determined using the BCA Protein Assay Kit, following the manufacturer’s instructions (Stanley et al., 2020). Chlorophyll was determined based on the protocol by Guo et al., with slight modifications (Guo et al., 2022). The measurement of soluble sugars was slightly modified based on the Dubois et al. (1951) method, and the content of MDA was determined by the thiobarbituric acid (TBA) method (Basit et al., 2022). The SOD activity was determined using a total SOD assay kit (the wst-8 technique; Beyotime, China) (Chen et al., 2019). Furthermore, the CAT, POD, and Pro antioxidant enzyme activities, as well as the H2O2 and O2- content, were determined using the Solarbio Kit (Beijing, China) following the guidelines provided by the manufacturer (Ren et al., 2021).
2.7 Analysis of the expression of genes related to salt stress
To investigate the regulatory mechanisms of StVQ31, we further examined the expression changes of genes associated with salt stress. Four-week-old transgenic Arabidopsis were moved to MS liquid medium with 150 mM NaCl for 8 hours, while the remainder seedlings were kept in MS liquid medium as a control. Total RNA from the transgenic Arabidopsis leaves was extracted using the RNAprep Pure Plant Total RNA Extraction Kit (DP432), and cDNA was synthesized using the FastKing cDNA First Strand Synthesis Kit (Degenomics)(KR116) (Tiangen, Beijing, China). The template cDNA was then diluted according to experimental requirements. The NCBI primer design suite was used to design gene-specific primers (Table 1) with Actin as the internal reference. qPCR was performed using the ABI QuantStudio7Flex system (Applied Biosystems, USA). The reaction mixture consisted of 2 μl of cDNA, 10 μl of 2× Super Real Color PreMix SYBR (Tiangen, China), 0.6 μl of gene-specific primers, and ddH2O was added up to 20 μl. The reaction conditions were set to: 95 °C 15 min; 95 °C 10 s, 60 °C 32s, 72 °C 32 s, 95 °C 15 s, 60 °C 60 s, 95 °C 15 s for a total of 40 cycles. The expression levels of the evaluated genes were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates were performed for each sample.
2.8 Data analysis
All data were expressed as the mean ± standard errors of three replicates. Prism 7.0 was used for image rendering. The mean differences between groups were statistically analyzed by independent sample t-test, analysis of variance (ANOVA), and post hoc LSD test. SPSS 27.0 was used for correlation analysis.
3 Results
3.1 Cloning and bioinformatic analysis of StVQ31
A gene-specific amplification primer StVQ31-F/R was designed based on the gene sequence (Table 1), and total RNA was extracted from potato leaves for reverse transcription for the isolation of the complete cDNA of StVQ31 (Supplementary File 1A). DNA Sequencing revealed that the StVQ31 gene fragment amplified by PCR had an open reading frame of 672bp, encoding a protein consisting of 223 amino acids and contained a conserved VQ domain (Supplementary File 1C; Figure 2A). StVQ31 contained only one exon. Based on the ExPASy ProtParam prediction, the theoretical MW of the encoded protein was 23.697 kDa, and the theoretical PI was 8.58. Ser (13%), Gly (10.3%), Val (7.6%), and Thr (7.6%) residues were the most abundant in the StVQ31 protein amino acid sequence. The overall mean hydrophilicity of StVQ31 was -0.295, indicating that the protein was hydrophilic. The secondary structure analysis showed that StVQ31 protein was composed by α helixes covering 19.73%, β fold covering 1.79%, and random coils covering 78.48% of the protein, respectively (Figure 2B). The motif analysis revealed that motif1 depicted in Figure 2C, served as the central conserved region of the protein and encompassed the functional domain (Figure 2A). In addition, the StVQ31 gene promoter contained TGA-element cis-elements, which means it may be involved in the regulation of plant growth and development.
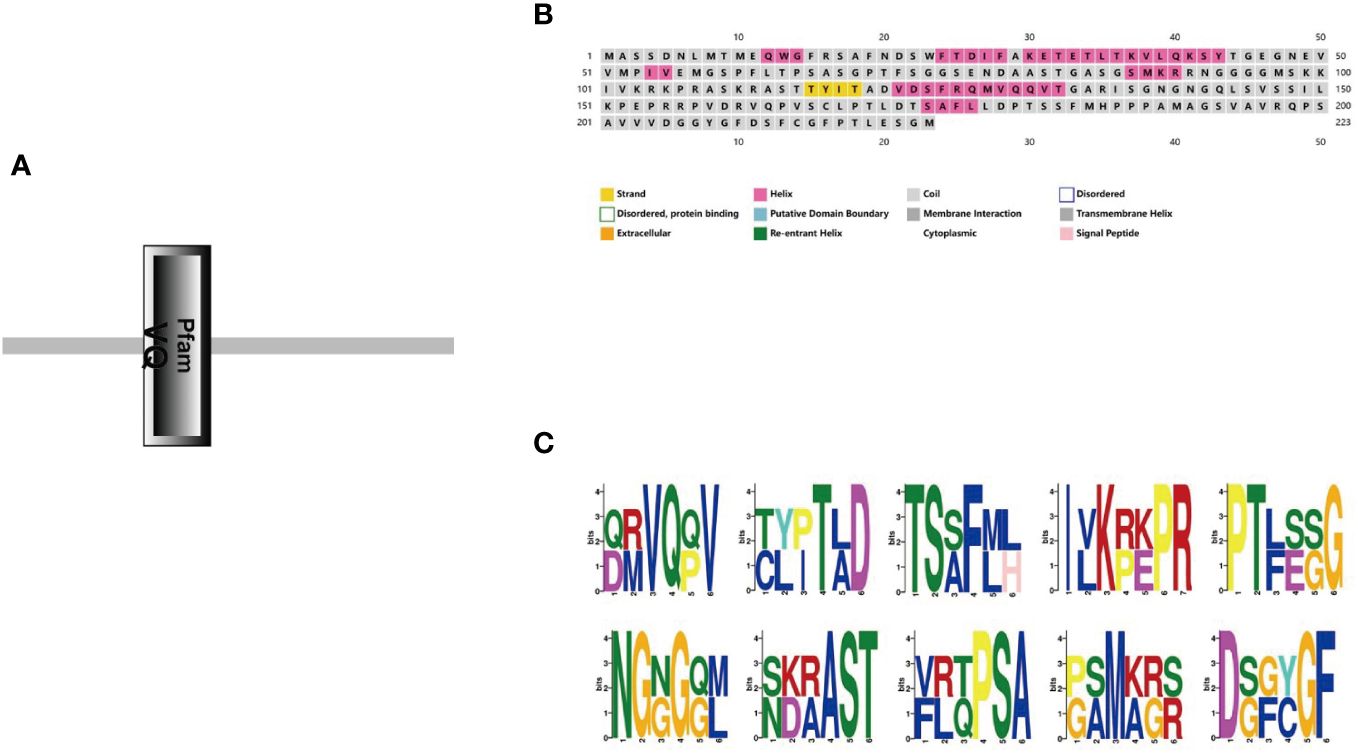
Figure 2 Protein structure of StVQ31 (A) Conservative domain of StVQ31; (B) Secondary structure prediction of StVQ31 Number of amino acids: 223; (C) Conserved motif of StVQ31.
3.2 Subcellular localization of StVQ31
In order to confirm the subcellular location of StVQ31, we removed the termination codon and fused StVQ31 to a GFP vector expressed by the 35s promoter of the tobacco mosaic virus (TMV). Individual cells carrying the control GFP vector exhibited detectable green fluorescence, while the cells expressing STVQ31-GFP emitted a green fluorescence signal from within the nucleus. Thus, the green fluorescence signal emitted by STVQ31-GFP could only be detected in the nucleus, and it overlapped with the red fluorescent nuclear marker AtWRKY25-mCherry (Figure 3), thus indicating that StVQ31 is localized in the nucleus.
Figure 3 (A) A schematic diagram of the subcellular localization vector. (B) The GFP empty vector. (E) The recombinant 35S:StVQ31-GFP vector. (F) The nucleus marker AtWRKY25-mCherry. (C, G) Bright field. (D, H) Merged images. Scale bars: 50 μm.
3.3 Generation and morphological observation of transgenic plants
The transgenic plants were identified by PCR amplification after DNA extraction. Distinct bands were amplified in the 16 transgenic lines identified. In contrast, no corresponding fragment was amplified in the WT plants (Figure 1A). These results were subsequently validated by RT-PCR analysis (Figure 1B), confirming the successful overexpression of StVQ31 in transgenic Arabidopsis lines. In the early growth stages of Arabidopsis plants, we observed that the StVQ31 and wild-type plants exhibited certain differences, with StVQ31 overexpressing plants being bigger than wild-type Arabidopsis (Figure 4B). This discrepancy may be attributed to the elevated expression of StVQ31 protein in the stem, suggesting that it is involved in the regulation of plant development. This result is consistent with the phenotypes exhibited in AtVQ29 expressing plants (Cheng et al., 2012). Furthermore, we conducted a statistical analysis of the phenological stages and flowering dates of transgenic and wild-type Arabidopsis thaliana lines. We found that the flowering in transgenic plants occurred at about 30 days after germination, showing a significant increase in flowering time compared with wild-type Arabidopsis (Figure 4A). The StVQ31 gene not only affected the flowering time of Arabidopsis but also affected its growth and development. This implies that it potentially has pivotal regulatory functions that control Arabidopsis growth. Based on this, we observed the phenotypes of transgenic and wild plants under salt stress. We found that in contrast to the wild type, the leaves of the transgenic plants exhibited a vibrant green color, while the wild type plants had yellow leaves, short plants, and were in a wilted state (Supplementary File 2). These results indicated that the transgenic plants exhibited an increased tolerance to salt stress.

Figure 4 Effect of overexpression of StVQ31 gene on flowering of Arabidopsis thaliana (A); Effects of overexpression of StVQ31 gene on growth and development of Arabidopsis Thaliana (B).
3.4 Germination rate and root length
Seed germination serves as the foundation for plant growth and overall plant vigor. Hence, examining the impact of salt stress on seed germination is critical to assess the resistance of the plants. Under 0mM NaCl treatment, the germination rates of transgenic lines were consistent with that of WT (Figure 5A; Supplementary File 3). After salt treatment, the germination rates of the three transgenic lines evaluated were 89.29%, 78.57%, and 82.14%, respectively (Figure 5D; Supplementary File 3), increased by 66.67%, 46.67%, and 53.33%, respectively, compared to the WT (germination rate of 53.57%). At the same time, the transgenic plants had an increased root length. Specifically, compared to WT plant (3.53cm), the root length of the three transgenic lines was 4.27cm, 4.37cm, and 4.76cm, respectively (Figure 5C; Supplementary File 3), increased by20.96%, 23.80%, and 34.84%, respectively. While under 0mM NaCl treatment, the root length of transgenic lines were consistent with that of WT (Figure 5B; Supplementary File 3).These findings suggest that StVQ31overexpression reduced salinity-mediated inhibition of seed germination, conferring a stronger capacity to the seedlings to reduce salt stress damage and maintain longer root length.
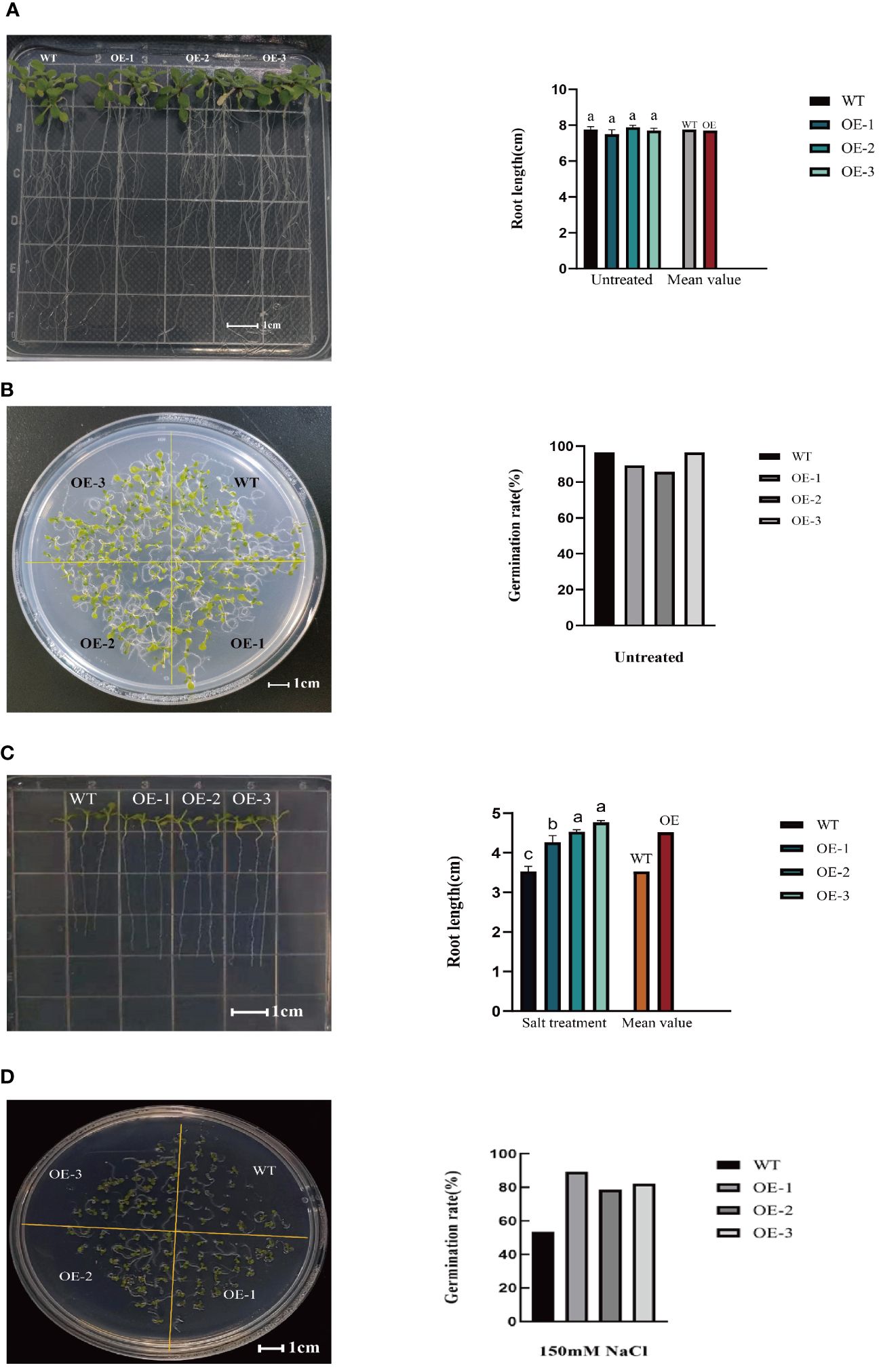
Figure 5 (A) Root length of wild Arabidopsis and transgenic Arabidopsis under 0mM NaCl; (B) Germination experiment of wild Arabidopsis and transgenic Arabidopsis under 0mM NaCl; (C) Root length of wild Arabidopsis and transgenic Arabidopsis after salt stress; (D) Germination experiment of wild Arabidopsis and transgenic Arabidopsis after salt stress. Values are means±SD (n=3), different lowercase letters indicate significant difference from the control (t-test) (P<0.05).
3.5 Effects of StVQ31 overexpression on the antioxidant metabolism under salt stress
Under abiotic stress conditions, considerable amounts of excessive ROS, such as H2O2 and O2− are generated within plant cells. This leads to a rise in lipid peroxidation and subsequent cellular oxidative stress (Figures 6A, B). The contents of H2O2 and O2− in Arabidopsis leaves were determined before and after salt treatment. The H2O2 and O2− contents in WT under salt stress were 1.26 and 1.53 times higher than in the control conditions. The H2O2 and O2−contents increased by 3.45%~5.76% and 17.89%~24.33%, respectively, while the WT increased even more. The increase of transgenic plants was significantly lower than that of WT (Figures 6A, B). The WT plants, overall, did not exhibit any notable variation. Next, we measured the activities of CAT, POD, and SOD. CAT catalyzes H2O2 breakdown, while SOD and POD are responsible for the reduction of stress-induced H2O2 and O2−, respectively. Under salinity stress treatment, the enzyme activities increased in both transgenic plants and WT in different amplitudes. Compared with WT, CAT, SOD, and POD in transgenic plants were 1.66-1.68-fold, 1.37-1.38-fold, and 1.69-1.76-fold higher than WT, respectively (Figures 7A–C). These results are consistent with the H2O2 and O2− contents, suggesting that transgenic plants could reduce cell oxidative damage under stress conditions by increasing ROS clearance capacity.
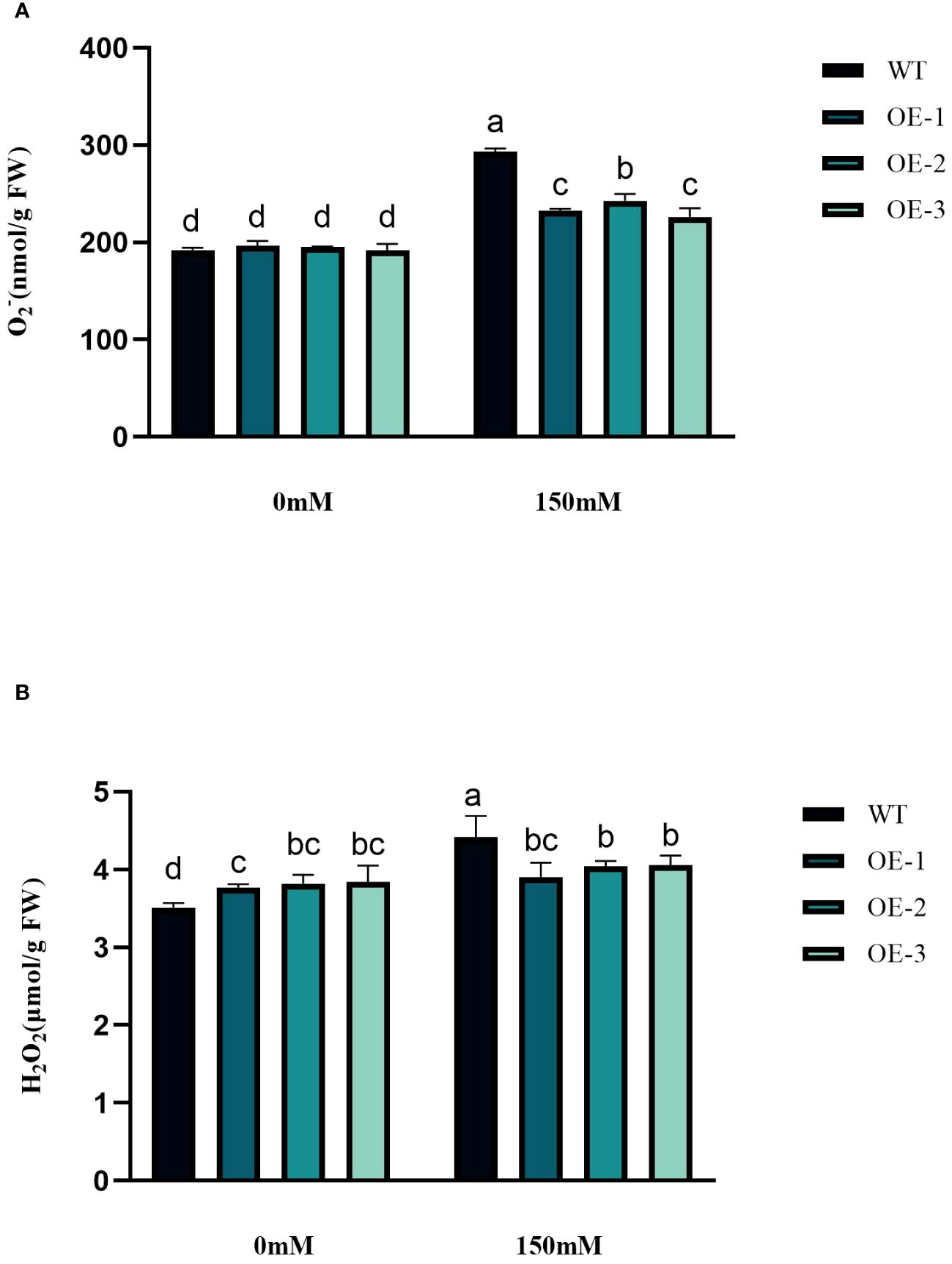
Figure 6 Physiological comparisons of control and StVQ31 overexpressing Arabidopsis plants. (A) O2-; (B) H2O2. All values are presented as mean ± standard error of three replicates. Different letters indicate significant differences in comparisons (P < 0.05).
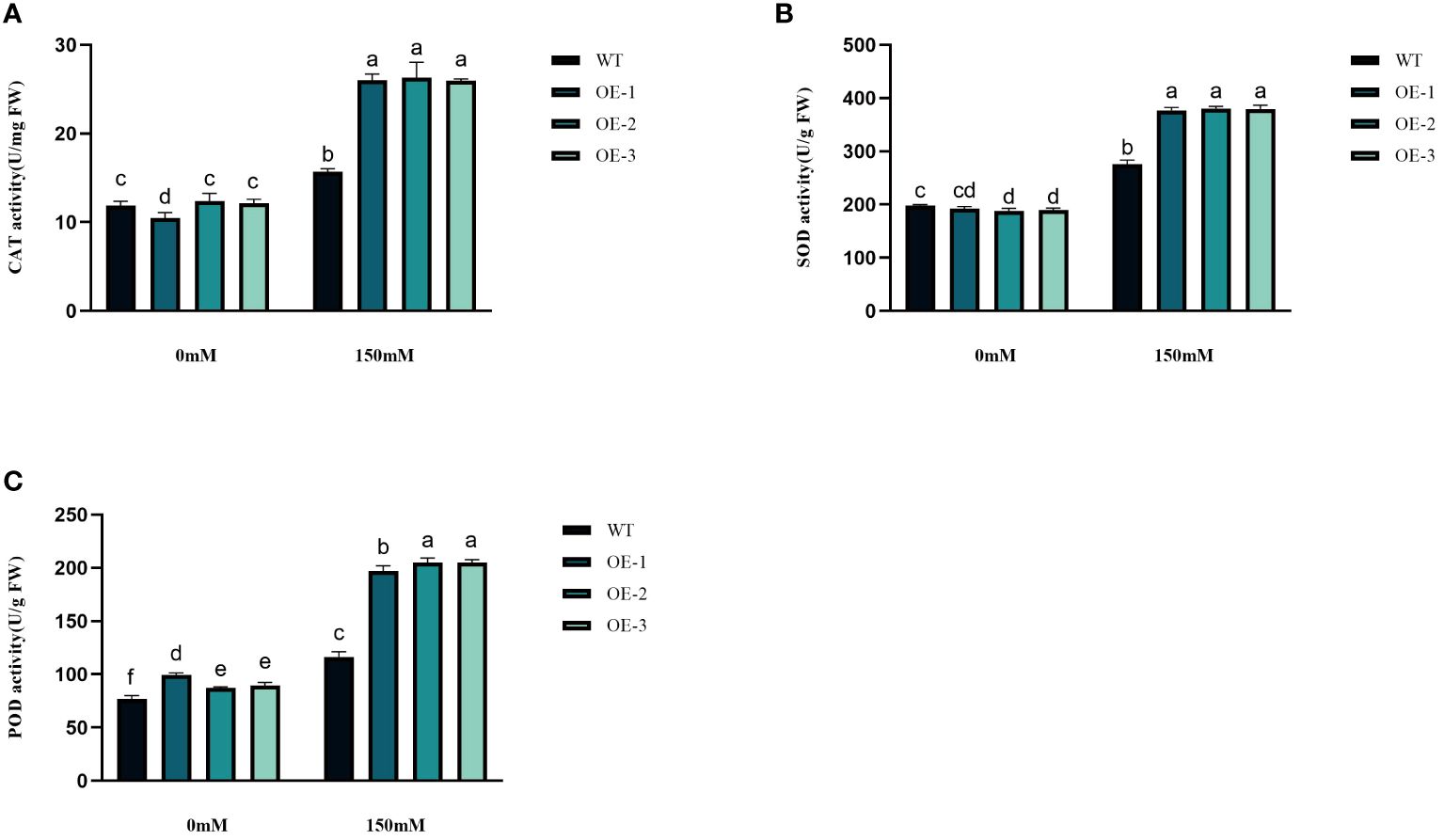
Figure 7 Physiological comparisons of control and StVQ31 overexpressing Arabidopsis plants. (A) CAT activity; (B) SOD activity; (C) POD activity. All values are presented as mean ± standard error of three replicates. Different letters indicate significant differences in comparisons (P < 0.05).
3.6 Determination of physiological indexes under salt stress
We examined the alterations in proline levels, a significant stress indicator, before and after subjecting plants to salt stress. The results found that The Pro content in WT increased 1.31-fold after salt stress, while the Pro content in transgenic plants was 1.5-1.54-fold increased (Figure 8D). Thus, the increase of Pro content in transgenic plants was considerably higher than that in WT, by 1.19-1.21-fold (Figure 8D). MDA content analysis was conducted to evaluate the oxidative damage caused by salt stress in both WT and transgenic plants. The results showed that all StVQ31 overexpression lines produced significantly less MDA (0.216 to 0.277µmol g−1 FW) than WT plants (0.36µmol g−1 FW) (Figure 8B). These results suggest that overexpression of StVQ31 may contribute to maintaining membrane permeability under salt stress by enhancing their antioxidant metabolism. The accumulation of soluble sugars was similar to that of proline. After being subjected to salinity stress (Figure 8A), the soluble sugar content in WT exhibited a substantial increase of 39.17%. In transgenic plants, soluble sugar content showed an increase ranging from 56.22% to 62.07%. The total protein content decreased in WT decreased under salt stress by 25.82% and in transgenic plants by 3.66%-5.58%. The trend was similar to that of MDA, albeit with a different amplitude of decrease (Figure 8C). Thus, compared with WT, the reduction was lower in transgenic plants. The chlorophyll content is directly related to the light-harvesting process in plants. It can be used as an indicator of plant photosynthesis to determine the plant’s physiological status and salt tolerance. Under salt stress, chlorophyll content decreased, affecting plant growth and development. The contents of chlorophyll a and b and the a/b ratio decreased notably under 150 mM NaCl treatment, but the total content of chlorophyll, chlorophyll a, and chlorophyll b in the transgenic plants was significantly higher than that in the WT plants (Figures 9A–D). These results indicate that the photosynthetic capacity of transgenic plants after salt treatment is even higher than that of WT plants.
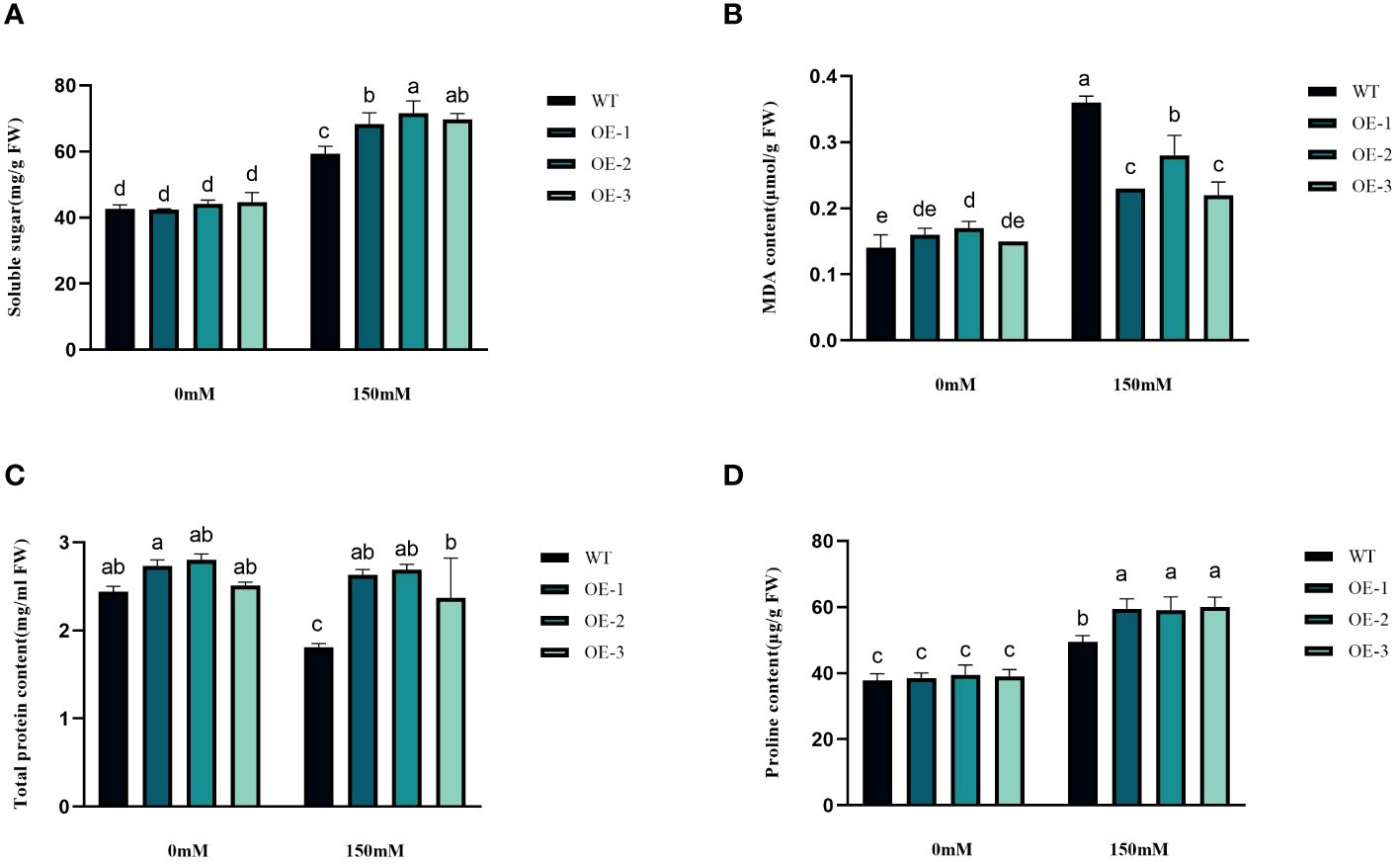
Figure 8 Physiological comparisons of control and StVQ31 overexpressing Arabidopsis plants. (A) Soluble sugar; (B) MDA content; (C) Total protein; (D) Proline All values are presented as mean ± standard error of three replicates. Different letters indicate significant differences in comparisons (P < 0.05).
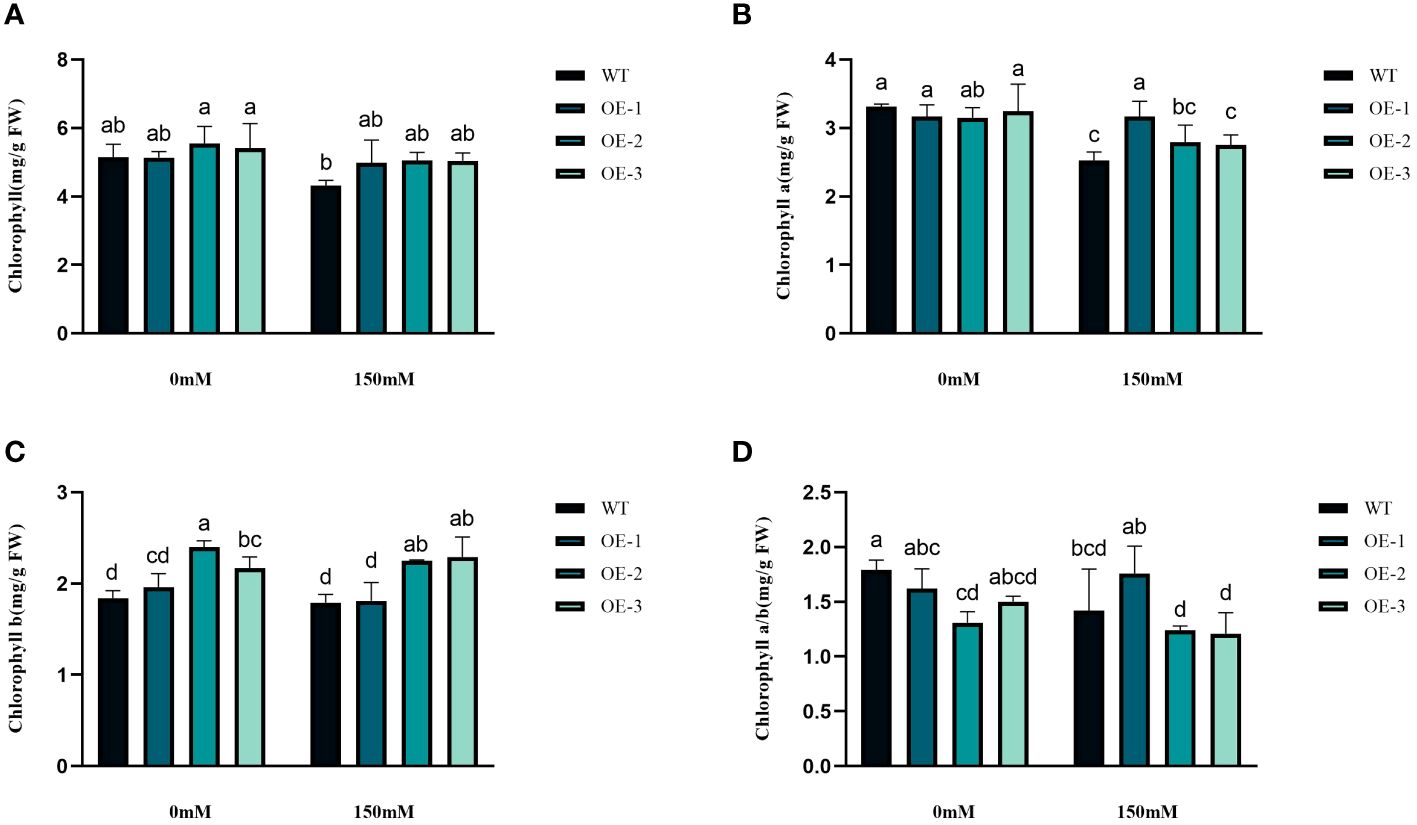
Figure 9 Physiological comparisons of control and StVQ31 overexpressing Arabidopsis plants. (A) Chlorophyll; (B) Chlorophyll a; (C) Chlorophyll b; (D) Chlorophyll a/b. All values are presented as mean ± standard error of three replicates. Different letters indicate significant differences in comparisons (P < 0.05).
3.7 Expression of salt-stress related genes
The involvement of the StVQ31 gene in the adaptive response of plants to salt stress was determined by analyzing the expression of four genes that function as stress markers. Under high salinity conditions, the expression of the genes encoding LEA 4-5, NCED-3, ERD, and At2g38905 was notably upregulated in transgenic plants, as shown in Figures 10A–D. While under non-stress conditions, their expression in transgenic plants and wild-type plants remained unaltered. Taken together, these findings suggest that the enhanced salt tolerance in plants that overexpress StVQ31 genes is also associated with increased expression levels of LEA 4-5, NCED-3, ERD, and At2g38905genes.
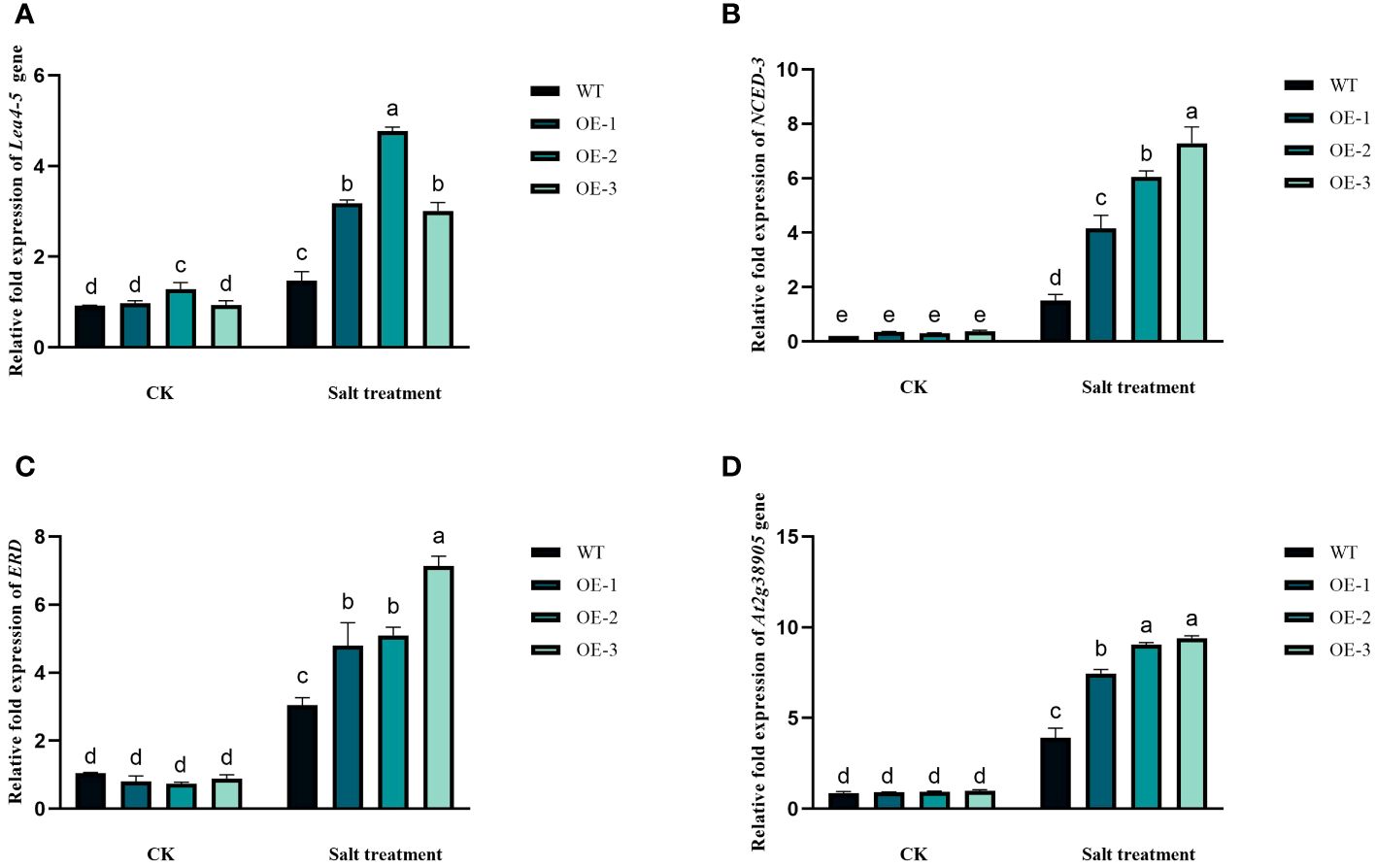
Figure 10 Expression analysis of abiotic stress marker genes in StVQ31 transgenic Arabidopsis lines by real-time RT-PCR. The transcript levels of (A) ERD; (B) salt responsive gene (At2g38905); (C) LEA 4-5, and (D) NCED-3 in WT and transgenic Arabidopsis plants were analyzed under both control and salt treatment conditions. Actin was used as housekeeping genes for normalization. Values are means ± SD (n=3), different lowercase letters indicate significant difference from the control (t-test) (P<0.05).
3.8 Correlation analysis
Based on the results, 61.99% of the measured indicators were correlated, of which 47.95% were strongly correlated and 39.77% were weakly correlated (Figure 11). Root length was found to have a positive correlation with SOD and POD activities (P<0.01), while the levels of superoxide anion and MDA were significantly negatively correlated. This suggests that reactive oxygen species play a role in regulating root growth. Similarly, the germination rate showed a positive correlation with SOD and CAT, and a negative correlation with superoxide anion and MDA levels. These findings indicate that superoxide anion and MDA have varying degrees of negative regulatory effects on both germination rate and root length. Moreover, total protein, SOD, POD, CAT, and Pro showed positive correlations with other indexes, whereas MDA and H2O2 were predominantly negatively correlated with other indexes. Specifically, CAT was highly correlated with SOD and POD. Root length was highly correlated with the expression of At2g38905 genes and NCED-3. Moreover, certain indexes, such as H2O2 and Chlorohyll b, were not correlated. The findings suggest a possible correlation between the indexes, which could collectively impact the salt tolerance of the StVQ31 overexpressing plants.
4 Discussion
VQ protein is a plant-specific protein (Wang et al., 2010). Growing evidence indicates that VQ regulates plant response to various abiotic stresses, including salinity, drought, cold and high temperature (Kim et al., 2013; Song et al., 2016; Wang et al., 2017; Chen et al., 2018). In Arabidopsis thaliana, AtVQ9 and AtVQ15 genes significantly affect its salt stress tolerance (Perruc et al., 2004; Hu et al., 2013). In our previous study, we identified 37 VQ genes in potato, which were classified into 6 subfamilies based on evolutionary tree relationships. Based on the phylogenetic tree, StVQ31 is the closest to AtVQ24 in phylogeny and is also phylogenetically close to AtVQ15 (Zhai et al., 2022). Based on similar evolutionary functions of the same subfamily (Wang et al., 2015), StVQ31 may have similar functions to AtVQ15. However, the function of the StVQ gene has not been previously investigated. Combined with previous studies and the literature, we selected StVQ31, a gene potentially involved in abiotic stress adaptation, for further functional analysis. StVQ31 has a typical VQ conserved domain, and based on its gene structure, it contains only one exon. In addition, the StVQ31 protein is also hydrophilic, with an average hydrophilic coefficient of -0.295. Subcellular localization revealed that StVQ31 is a nuclear-localized protein (Figure 3).
In this study, we cloned the StVQ31 gene and transformed it in Arabidopsis. Based on the morphological analysis results, transgenic plants exhibited a relatively improved growth compared to WT under high salt conditions (Supplementary File 2). These observations indicated that StVQ31 may be involved in plant salt stress adaptation and tolerance. Our findings are similar to the results of Kim et al (Kim et al., 2013). Seed germination is considered to be greatly affected by stress and is regarded as a crucial phase in the growth cycle of crops (Chen et al., 2020). A higher germination rate reflects a better growth capacity of the seedlings, which is essential for achieving a high yield during maturity (Wilson et al., 2014). Based on the germination rate experiment results, the survival rate of transgenic plants was higher, indicating that transgenic plants had greater salt tolerance. In addition, plant roots are important organs that absorb water and nutrients, and vigorous root growth directly affects the capacity of plants to adapt to environmental stress (Zhou et al., 2013). The taproot length of all transgenic strains was significantly longer than that of WT. Thus, overexpression of the VQ31 gene could promote the root growth of Arabidopsis thaliana under salt stress, increase the root absorption area, and improve the salt tolerance of the plants. Therefore, we speculate that the StVQ31 gene is involved in morphogenesis, growth, and development in Arabidopsis thaliana.
Plants have evolved various mechanisms to adapt to environmental changes (Yang et al., 2020). When plants are subjected to abiotic stress, large amounts of reactive oxygen species are produced. The plant antioxidant mechanisms can eliminate the damage caused by excessive ROS accumulation by regulating CAT, SOD, POD, and other antioxidant enzyme activities (Yang et al., 2017). The accumulation of osmoregulatory substances is an important indicator of plant stress tolerance. The accumulation of proline, soluble sugars, and total proteins can protect against oxidative stress by scavenging free radicals (Qiu et al., 2020). After salt stress, the increase of Pro content in transgenic plants was significantly higher (by 1.19-1.21-fold) than that in WT. Increased Pro content can account for elevated osmotic pressure, reducing the intracellular water potential and enhancing plant water conservation (Li et al., 2013). The change in soluble sugar content was similar to that of Pro, while the total protein content decreased to different degrees. It is speculated that these osmoregulatory compounds also help maintain the integrity of the cellular membranes (Kok et al., 2021; He et al., 2022). Malondialdehyde production and accumulation is also a common indicator of reactive oxygen species accumulation and oxidative stress in plant cells. Under salt stress, the MDA content of WT plants was higher than that of StVQ31 transgenic plants, indicating that overexpression of StVQ31 may lead to increased resistance to salt stress-induced oxidative stress (Figure 8B). In addition, the maintenance of chlorophyll and its reduced breakdown significantly affects the photosynthetic efficiency under stress (Ahammed et al., 2013). Based on our results, the chlorophyll content of transgenic lines was significantly higher than that of WT under high salt conditions. Therefore, overexpression of StVQ31 in Arabidopsis may enhance its photosynthetic capacity by the maintenance of chlorophyll content, thus ensuring normal plant growth and development under stress. This is consistent with the phenotypic observations of the transgenic plants, which exhibited robust growth under salt stress. Enhanced antioxidant enzyme activities increased the capacity of transgenic plants to eliminate peroxy ions, reduced the peroxidation of cells’ plasma membrane, and improved the survival ability of plants under high salinity conditions. Furthermore, the H2O2 and O2− levels in transgenic plants were remarkably lower than those of WT, this is consistent with the results of enzyme activity determination (Song et al., 2016).
Expression analysis of salt stress response genes could, in part, explain the molecular mechanism of salt tolerance derived from the StVQ31 gene overexpression (Shkolnik et al., 2019). AtNCED3, a crucial gene for ABA production in Arabidopsis thaliana, plays a definite role in regulating stomatal closure and enhancing the plant’s salt stress tolerance through the synthesis and accumulation of ABA in the leaves (Lv et al., 2011). Based on our results, the expression of NCED3 in transgenic plants was 2.65-4.48-fold higher than in WT. These results indicated that StVQ31 could promote the expression of salt-stress-related genes and positively regulate ABA biosynthesis and signal transduction pathways, thereby improving plant salt tolerance. In addition, the high expression level of AT2g38905 (salt-responsive protein family) in transgenic plants is consistent with the increase in the concentration of osmoprotectants under salinity conditions (Mishra et al., 2021), which leads to improved plant salt tolerance. The upregulation of LEA 4-5 indicates that protein and membrane integrity preservation and the sequestration of ions are crucial in salt stress adaptation and tolerance (Huang et al., 2018). Erd-encoding proteins can protect macromolecules and membranes under stress (Kasuga et al., 2004), consistent with our findings of higher expression levels of ERD in transgenic plants under salt stress compared to WT plants.
A high salinity environment can lead to increased oxidative stress in plants, affecting normal growth and metabolism (Hasanuzzaman et al., 2021). The overexpression of the StVQ31 gene may promote and enhance the activity of the antioxidant system by reducing the content of MDA, reducing the damage by oxidative stress, and reducing the osmotic stress damage caused by high salinity by increasing the content of osmoprotectant and osmoregulatory compounds. In addition, the Arabidopsis’s tolerance to salt stress can be improved by regulating the expression of stress-responsive genes. However, more work is needed to elucidate the StVQ31 gene’s precise mechanism of action. Based on the findings of our study, StVQ31 can be exploited to enhance salt tolerance in other plants via genetic engineering.
5 Conclusion
In this study, a VQ gene, StVQ31, was cloned from potato, and it was functionally characterized by overexpression in transgenic Arabidopsis thaliana plants. StVQ31 overexpression significantly enhanced Arabidopsis tolerance to salt stress in terms of growth and its overall physiological status. Salt stress significantly enhanced the activity of the antioxidant enzyme system and maintained the stability of membrane lipids and reactive oxygen species homeostasis. At the same time, it also resulted in changes in the germination capacity and flowering of Arabidopsis thaliana seedlings under salt stress. In addition, overexpression of StVQ31 induced the expression of genes involved in salt stress adaptation. The results showed that the StVQ31 gene plays a critical role in plant responses to salt stress. However, due to the complexity of StVQ31’s function, its precise mechanism of action still needs to be further explored.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
MZ: Conceptualization, Formal analysis, Software, Visualization, Writing – original draft, Writing – review & editing. ZA: Formal analysis, Software, Writing – review & editing. HQ: Formal analysis, Software, Writing – review & editing. DG: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. MZ, DG: Designed and carried out the experiments. MZ, ZA: Analyzed the experimental results. MZ, HQ, ZA: Analyzed the data, developed the analysis tools, and wrote the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1347861/full#supplementary-material
Supplementary file 1 | Determination of total RNA in potato leaves by agarose gel electrophoresis: Agarose gel electrophoresis of StVQ31 cDNA amplification and PLB- StVQ31positive identification A:M: DL2000 DNA Marker, 1-5: RNA; B: 1-2: blank control, 3-4: the PCR detection result of StVQ31-1300 bacterial solution, M: DL 2000 DNA Marker C: cDNA amplification of StVQ31 gene; D: positive identification of two single colonies of PLB- StVQ31; M: DL2000 DNA Marker, 1-2: StVQ31.
Supplementary file 2 | Morphology of wild Arabidopsis and transgenic Arabidopsis before and after salt stress (150mM NaCl).
Supplementary file 3 | Root length and germination rate of wild Arabidopsis and transgenic Arabidopsis before(0 mM NaCl) and after salt stress (150mM NaCl).
Abbreviations
LEA 4–5, late embryogenesis abundant proteins; At2g38905, salt-sensitive protein family gene; ERD, monosaccharides encodes ESL1; NCED, 9-cis-epoxycarotenoid dioxygenase; Chl; chlorophyll; Chla, chlorophyll a; Chlb, chlorophyll b; SS, soluble sugar; Pro, proline; Tp, total protein; H2O2, hydrogen peroxide; O2-, superoxide anion; SOD, superoxide dismutase; POD, peroxidase; CAT, catalase; MDA, malondialdehyde; ROS, reactive oxygen species; PCR, polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction.
References
Ahammed, G. J., Choudhary, S. P., Chen, S., Xia, X., Shi, K., Zhou, Y., et al. (2013). Role of brassinosteroids in alleviation of phenanthrene-cadmium co-contamination-induced photosynthetic inhibition and oxidative stress in tomato. J. Exp. Bot. 64, 199–213. doi: 10.1093/jxb/ers323
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Basit, F., Bhat, J. A., Ulhassan, Z., Noman, M., Zhao, B., Zhou, W., et al. (2022). Seed priming with spermine mitigates chromium stress in rice by modifying the ion homeostasis, cellular ultrastructure and phytohormones balance. Antioxid. (Basel Switzerland) 11 (9), 1704. doi: 10.3390/antiox11091704
Chen, J., Wang, H., Li, Y., Pan, J., Hu, Y., Yu, D. (2018). Arabidopsis VQ10 interacts with WRKY8 to modulate basal defense against Botrytis cinerea. J. Integr. Plant Biol. 60, 956–969. doi: 10.1111/jipb.12664
Chen, Q. W., Dong, K., Qin, H. X., Yang, Y. K., He, J. L., Li, J., et al. (2019). Direct and indirect inhibition effects of resveratrol against toxoplasma gondii tachyzoites in vitro. Antimicrobial Agents Chemother. 63, null. doi: 10.1128/AAC.01233-18
Chen, F., Zhou, W., Yin, H., Luo, X., Chen, W., Liu, X., et al. (2020). Shading of the mother plant during seed development promotes subsequent seed germination in soybean. J. Exp. Bot. 71, 2072–2084. doi: 10.1093/jxb/erz553
Chen, Y. B., Zhang, Y. B., Wang, Y. L., Kaur, P., Yang, B. G., Zhu, Y., et al. (2022). A novel inhalable quercetin-alginate nanogel as a promising therapy for acute lung injury. J. Nanobiotechnol. 20, 272. doi: 10.1186/s12951-022-01452-3
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J., Chen, J. Y., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Chu, W., Liu, B., Wang, Y., Pan, F., Chen, Z., Yan, H., et al. (2016). Genome-wide analysis of poplar VQ gene family and expression profiling under PEG, NaCl, and SA treatments. Tree Genet. Genomes 12, 124. doi: 10.1007/s11295-016-1082-z
Colin, L., Ruhnow, F., Zhu, J. K., Zhao, C., Zhao, Y., Persson, S. (2023). The cell biology of primary cell walls during salt stress. Plant Cell 35, 201–217. doi: 10.1093/plcell/koac292
Dong, J., Chen, C., Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., Smith, F. A. J. N. (1951). A colorimetric method for the determination of sugars. Nature 168, 167. doi: 10.1038/168167a0
Gong, Y., Liu, X., Chen, S., Li, H., Duanmu, H. (2022). Genome-wide identification and salt stress response analysis of the bZIP transcription factor family in sugar beet. Int. J. Mol. Sci. 23 (19), 11573. doi: 10.3390/ijms231911573
Gu, H., Yang, Y., Xing, M., Yue, C., Wei, F., Zhang, Y., et al. (2019). Physiological and transcriptome analyses of Opisthopappus taihangensis in response to drought stress. Cell Biosci. 9, 56. doi: 10.1186/s13578-019-0318-7
Guo, J., Chen, J., Yang, J., Yu, Y., Yang, Y., Wang, W. (2018). Identification, characterization and expression analysis of the VQ motif-containing gene family in tea plant (Camellia sinensis). BMC Genomics 19, 710. doi: 10.1186/s12864-018-5107-x
Guo, C., Li, X., Zhang, Z., Wang, Q., Zhang, Z., Wen, L., et al. (2022). The INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE6 peptide functions as a positive modulator of leaf senescence in arabidopsis thaliana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.909378
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22, 9326. doi: 10.3390/ijms22179326
He, X., Long, F., Li, Y., Xu, Y., Hu, L., Yao, T., et al. (2022). Comparative transcriptome analysis revealing the potential mechanism of low-temperature stress in Machilus microcarpa. Front. Plant Sci. 13, 900870. doi: 10.3389/fpls.2022.900870
Hernández-Fernández., L., Vázquez., J. G., Hernández, L., Pérez-Bonachea., L., Campbell, R., Martínez, J., et al. (2023). Soluble phenolics, chlorophylls, and malondialdehyde are the best indicators of salt stress in Eichornia crassipes. Vegetos 0, 0–0. doi: 10.1007/s42535-023-00669-3
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J.: Cell Mol. Biol. 74, 730–745. doi: 10.1111/tpj.12159
Huang, K. C., Lin, W. C., Cheng, W. H. (2018). Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 18, 40. doi: 10.1186/s12870-018-1255-z
Huang, Z., Liu, L., Jian, L., Xu, W., Wang, J., Li, Y., et al. (2022). Heterologous expression of mfWRKY7 of resurrection plant myrothamnus flabellifolia enhances salt and drought tolerance in arabidopsis. Int. J. Mol. Sci. 23 (14), 7890. doi: 10.3390/ijms23147890
Kasuga, M., Miura, S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Kim, N. H., Hwang, B. K. (2015). Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol. 167, 307–322. doi: 10.1104/pp.114.253898
Kim, D. Y., Kwon, S. I., Choi, C., Lee, H., Ahn, I., Park, S. R., et al. (2013). Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 529, 208–214. doi: 10.1016/j.gene.2013.08.023
Kok, A. D., Mohd Yusoff, N. F., Sekeli, R., Wee, C. Y., Lamasudin, D. U., Ong-Abdullah, J., et al. (2021). Pluronic F-68 improves callus proliferation of recalcitrant rice cultivar via enhanced carbon and nitrogen metabolism and nutrients uptake. Front. Plant Sci. 12, 667434. doi: 10.3389/fpls.2021.667434
Li, J., Besseau, S., Törönen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, D., Li, J., Xiang, S., Pan, A. (2022). PSegNet: simultaneous semantic and instance segmentation for point clouds of plants. Plant Phenomics (Washington D.C.) 2022, 9787643. doi: 10.34133/2022/9787643
Liang, X., Li, J., Yang, Y., Jiang, C., Guo, Y. (2023). Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol 66 (3), 303–329. doi: 10.1111/jipb.13599
Liu, Y., Liu, X., Dong, X., Yan, J., Xie, Z., Luo, Y. (2022). The effect of Azorhizobium caulinodans ORS571 and γ-aminobutyric acid on salt tolerance of Sesbania rostrata. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.926850
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego Calif.) 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, W. T., Lin, B., Zhang, M., Hua, X. J. (2011). Proline accumulation is inhibitory to Arabidopsis seedlings during heat stress. Plant Physiol. 156, 1921–1933. doi: 10.1104/pp.111.175810
Medina, C. A., Hawkins, C., Liu, X. P., Peel, M., Yu, L. X. (2020). Genome-wide association and prediction of traits related to salt tolerance in autotetraploid alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 21 (9), 3361. doi: 10.3390/ijms21093361
Mishra, M. K., Tiwari, S., Misra, P. (2021). Overexpression of WssgtL3.1 gene from Withania somnifera confers salt stress tolerance in Arabidopsis. Plant Cell Rep. 40, 2191–2204. doi: 10.1007/s00299-021-02666-9
Pecher, P., Eschen-Lippold, L., Herklotz, S., Kuhle, K., Naumann, K., Bethke, G., et al. (2014). The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of 'VQ-motif'-containing proteins to regulate immune responses. New Phytol. 203, 592–606. doi: 10.1111/nph.12817
Pedrini, N., Ortiz-Urquiza, A., Huarte-Bonnet, C., Fan, Y., Juárez, M. P., Keyhani, N. O. (2015). Tenebrionid secretions and a fungal benzoquinone oxidoreductase form competing components of an arms race between a host and pathogen. Proc. Natl. Acad. Sci. United States America 112, E3651–E3660. doi: 10.1073/pnas.1504552112
Perruc, E., Charpenteau, M., Ramirez, B. C., Jauneau, A., Galaud, J. P., Ranjeva, R., et al. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J.: Cell Mol. Biol. 38, 410–420. doi: 10.1111/j.1365-313X.2004.02062.x
Qiu, J. R., Huang, Z., Xiang, X. Y., Xu, W. X., Wang, J. T., Chen, J., et al. (2020). MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 20, 542. doi: 10.1186/s12870-020-02732-6
Ren, Y., Che, X., Liang, J., Wang, S., Han, L., Liu, Z., et al. (2021). Brassinosteroids benefit plants performance by augmenting arbuscular mycorrhizal symbiosis. Microbiol. Spectr. 9, e0164521. doi: 10.1128/spectrum.01645-21
Sajid, M., Geng, C., Li, M., Wang, Y., Liu, H., Zheng, J., et al. (2018). Whole-genome analysis of bacillus thuringiensis revealing partial genes as a source of novel cry toxins. Appl. Environ. Microbiol. 84 (14), e00277-18. doi: 10.1128/AEM.00277-18
Shan, N., Xiang, Z., Sun, J., Zhu, Q., Xiao, Y., Wang, P., et al. (2021). Genome-wide analysis of valine-glutamine motif-containing proteins related to abiotic stress response in cucumber (Cucumis sativus L.). BMC Plant Biol. 21, 492. doi: 10.1186/s12870-021-03242-9
Shkolnik, D., Finkler, A., Pasmanik-Chor, M., Fromm, H. (2019). CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A key regulator of NA+ Homeostasis during germination. Plant Physiol. 180, 1101–1118. doi: 10.1104/pp.19.00119
Song, W., Zhao, H., Zhang, X., Lei, L., Lai, J. (2016). Genome-wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in maize. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01177
Stacey, M. G., Cahoon, R. E., Nguyen, H. T., Cui, Y., Sato, S., Nguyen, C. T., et al. (2016). Identification of homogentisate dioxygenase as a target for vitamin E biofortification in oilseeds. Plant Physiol. 172, 1506–1518. doi: 10.1104/pp.16.00941
Stanley, L. E., Ding, B., Sun, W., Mou, F., Hill, C., Chen, S., et al. (2020). A tetratricopeptide repeat protein regulates carotenoid biosynthesis and chromoplast development in monkeyflowers (Mimulus). Plant Cell 32, 1536–1555. doi: 10.1105/tpc.19.00755
Sui, N., Tian, S., Wang, W., Wang, M., Fan, H. (2017). Overexpression of glycerol-3-phosphate acyltransferase from suaeda salsa improves salt tolerance in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01337
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J.: Cell Mol. Biol. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Wang, H., Li, J., Liu, H., Chen, S., Zaman, Q. U., Rehman, M., et al. (2023). Variability in morpho-biochemical, photosynthetic pigmentation, enzymatic and quality attributes of potato for salinity stress tolerance. Plant Physiol. Biochem. PPB 203, 108036. doi: 10.1016/j.plaphy.2023.108036
Wang, Y., Liu, H., Zhu, D., Gao, Y., Yan, H., Xiang, Y. (2017). Genome-wide analysis of VQ motif-containing proteins in Moso bamboo (Phyllostachys edulis). Planta 246, 165–181. doi: 10.1007/s00425-017-2693-9
Wang, M., Vannozzi, A., Wang, G., Zhong, Y., Corso, M., Cavallini, E., et al. (2015). A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00417
Wang, X., Zhang, H., Sun, G., Jin, Y., Qiu, L. (2014). Identification of active VQ motif-containing genes and the expression patterns under low nitrogen treatment in soybean. Gene 543, 237–243. doi: 10.1016/j.gene.2014.04.012
Wilson, R. L., Kim, H., Bakshi, A., Binder, B. M. (2014). The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of arabidopsis during salt stress. Plant Physiol. 165, 1353–1366. doi: 10.1104/pp.114.241695
Xu, M., Chen, C., Cai, H., Wu, L. (2018). Overexpression of peHKT1;1 improves salt tolerance in populus. Genes 9, 475. doi: 10.3390/genes9100475
Yang, L., Chen, X., Wang, Z., Sun, Q., Hong, A., Zhang, A., et al. (2020). HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 226, 507–522. doi: 10.1111/nph.16380
Yang, Q., Li, Y., Li, C., Zhang, H., Jiang, Z., Zhang, X., et al. (2017). Antioxidative enzymes and substances involve in the activity of improving the oxidative tolerance of Pichia caribbica by ascorbic acid. Biol. Control 108, 83–88. doi: 10.1016/j.biocontrol.2017.02.013
Zhai, M., Liu, N., Xu, R., Li, H., Liu, B., Wang, K., et al. (2022). Identification and expression analysis of VQ gene family in solanum tuberosum. J. Agric. Biotechnol. 30, 25–37. doi: 10.3969/j.issn.1674-7968.2022.01.003
Zhang, X., Dong, J., Deng, F., Wang, W., Cheng, Y., Song, L., et al. (2019). The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 19, 459. doi: 10.1186/s12870-019-2088-0
Zhang, Z., Quick, M. K., Kanelakis, K. C., Gijzen, M., Krishna, P. (2003). Characterization of a plant homolog of hop, a cochaperone of hsp90. Plant Physiol. 131, 525–535. doi: 10.1104/pp.011940
Zhao, C., Jiang, W., Zayed, O., Liu, X., Tang, K., Nie, W., et al. (2021). The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 8, nwaa149. doi: 10.1093/nsr/nwaa149
Zhao, Y., Luo, L., Xu, J., Xin, P., Guo, H., Wu, J., et al. (2018). Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 28, 448–461. doi: 10.1038/s41422-018-0024-8
Keywords: StVQ31, high salt stress, functional characterization, gene expression, physiology and biochemistry
Citation: Zhai M, Ao Z, Qu H and Guo D (2024) Overexpression of the potato VQ31 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 15:1347861. doi: 10.3389/fpls.2024.1347861
Received: 01 December 2023; Accepted: 18 March 2024;
Published: 05 April 2024.
Edited by:
Sachin Teotia, Sharda University, IndiaReviewed by:
Walid Zorrig, Center of Biotechnology of Borj Cedria (CBBC), TunisiaJuli Jing, Cornell University, United States
Copyright © 2024 Zhai, Ao, Qu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongwei Guo, Z2R3ZWlAbndzdWFmLmVkdS5jbg==
 Mingming Zhai
Mingming Zhai Zhengxiong Ao
Zhengxiong Ao Dongwei Guo
Dongwei Guo