- 1Department of Plant Pathology, University of Minnesota, Saint Paul, MN, United States
- 2Kenya Agricultural and Livestock Research Organization (KALRO), Food Crops Research Centre, Njoro, Kenya
- 3Department of Agronomy and Plant Genetics, University of Minnesota, Saint Paul, MN, United States
- 4Ethiopian Institute of Agriculture, Debre Zeit Agricultural Research Center, Bishoftu, Ethiopia
- 5International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
- 6Cereal Disease Laboratory, United States Department of Agriculture-Agricultural Research Service, Saint Paul, MN, United States
Wheat stem rust caused by Puccinia graminis f. sp. tritici (Pgt) threatens wheat production worldwide. The objective of this study was to characterize wheat stem rust resistance in ‘Linkert’, a variety with adult plant resistance effective to emerging wheat stem rust pathogen strain Ug99. Two doubled haploid (DH) populations and one recombinant inbred line (RIL) population were developed with ‘Linkert’ as a stem rust resistant parent. Hard red spring wheat variety ‘Forefront’ and genetic stock ‘LMPG’ were used as stem rust susceptible parents of the DH populations. Breeding line ‘MN07098-6’ was used as a susceptible parent of the RIL population. Both DH and RIL populations with their parents were evaluated both at the seedling stage and in the field against Pgt races. Genotyping data of the DH populations were generated using the wheat iSelect 90k SNP assay. The RIL population was genotyped by genotyping-by-sequencing. We found QTL consistently associated with wheat stem rust resistance on chromosome 2BS for the Linkert/Forefront DH population and the Linkert/MN07098-6 RIL population both in Ethiopia and Kenya. Additional reliable QTL were detected on chromosomes 5BL (125.91 cM) and 4AL (Sr7a) for the Linkert/LMPG population in Ethiopia and Kenya. Different QTL identified in the populations reflect the importance of examining the genetics of resistance in populations derived from adapted germplasm (Forefront and MN07098-6) in addition to a genetic stock (LMPG). The associated markers in this study could be used to track and select for the identified QTL in wheat breeding programs.
Introduction
Stem rust of wheat (Triticum aestivum L.), caused by Puccinia graminis f. sp. tritici (Pgt) Erikss. & Henning, is a destructive disease of wheat worldwide (Singh et al., 2011; Savary et al., 2019). The emergence of a race of Pgt known as Ug99 that defeats the widely deployed stem rust resistance gene, Sr31, poses a threat to wheat production (Singh et al., 2015). The use of genetic resistance is a preferred strategy for mitigating losses from crop diseases including stem rust of wheat.
Breeding for disease resistance is one of the primary objectives of wheat improvement programs in the United States and worldwide. Since the early 20th century, there have been international collaborations on genetic resistance breeding for controlling stem rust disease in wheat (Triticum spp.) (Carleton, 1905; Waldron, 1935; Stakman, 1955; Evans, 1980). Consequently, to date 64 stem rust resistance (Sr) genes have been characterized and mapped to chromosome locations in wheat (McIntosh et al., 2020). Out of these, 16 wheat Sr genes have been cloned by either map-based cloning (Sr13, Sr21, Sr33, Sr35, Sr50, Sr55, Sr57, Sr60, and Sr62) or target sequence capture approaches (Sr22, Sr26, Sr27, Sr43, Sr45, Sr46, and Sr61) (Arora et al., 2019; Periyannan et al., 2013; Zhang et al., 2017; Chen et al., 2018; Saintenac et al., 2013; Mago et al., 2015; Moore et al., 2015; Krattinger et al., 2009; Steuernagel et al., 2016; Chen et al., 2020; Zhang et al., 2021; Yu et al., 2022). Since the discovery of Ug99, there has been an intensive search for resistance genes both in domesticated wheat and its wild relatives, and 36 numerically designated stem rust resistance genes that are effective to the Ug99 race group have been characterized (Rouse et al., 2014a; Singh et al., 2015): Sr2, Sr12, Sr13, Sr14, Sr15, Sr21, Sr22, Sr25, Sr26, Sr27, Sr28, Sr29, Sr32, Sr33, Sr35, Sr37, Sr39, Sr40, Sr42, Sr43, Sr44, Sr45, Sr46, Sr47, Sr48, Sr50, Sr51, Sr52, Sr53, Sr55, Sr56, Sr57, Sr59, Sr61, Sr62, and Sr63. However, only few are available in conventional North American spring wheat germplasm including Sr2, Sr12, Sr13, Sr25, Sr55, and Sr57 (Rouse et al., 2014b; Bajgain et al., 2015a; Edae et al., 2018). In addition, many QTL have also been reported using bi-parental populations (Hiebert et al., 2010; Haile et al., 2012; Njau et al., 2013; Zurn et al., 2014; Kumsa et al., 2015; Bajgain et al., 2015b), and association mapping (Yu et al., 2011; Letta et al., 2013; Zhang et al., 2014; Bajgain et al., 2015a; Edae et al., 2018). Despite the availability of numerous genes and QTL, stem rust still remains a threat to wheat production worldwide due to continuous emergence of new Pgt races in different parts of the world and the lack of resistance deployed in conventional wheat varieties effective to the emerging races (Olivera et al., 2015; Newcomb et al., 2016; Olivera et al., 2019, 2022). This implies the importance of persistently carrying out resistance breeding in wheat through the identification and characterization of new resistance genes from both domestic germplasm and its wild relatives.
The development of new sequencing technologies has facilitated the discovery of a large number of SNP markers for many crop species such as hexaploid wheat (Chapman et al., 2015), barley (Mascher et al., 2013), rice (Spindel et al., 2013) and maize (Romay et al., 2013). Genomic/genomic resources such as wheat reference sequences The International Wheat Genome Sequencing Consortium (IWGSC), 2018), wheat 90K iselect SNP chips (Wang et al., 2014) and genotype-by-sequencing (GBS), a reduced representation genotyping platform (Poland et al., 2012), have been used effectively to discover genes/QTL for several traits including disease resistance in wheat.
Genetic resistance to rust pathogens can be generally grouped into two categories: resistance that is effective at all plant growth stages and resistance that is effective at the adult plant stage. All-stage resistance genes often confer major-effects and account for thirty of the thirty-six Ug99-effective genes. Adult plant resistance genes often confer relatively minor effects and include Sr2, Lr67/Sr55/Yr46/Pm46, Sr56, Lr34/Sr57Yr18/Pm38, Lr46/Sr58/Yr29/Pm39, and Sr63 (Mago et al., 2002; Singh et al., 2015). These six designated adult plant resistance genes have not been reported to exhibit race-specificity supporting an expectation (Singh et al., 2011, 2014) that effective adult plant resistance genes are non-race-specific. Though few examples of race-specific adult plant resistance genes have been reported for stripe rust (Yr11, 12, 13, and 14; Milus et al., 2015) and leaf rust (Lr12; Singh and Bowden, 2010), the generally non-race-specific characteristic of adult plant resistance genes justifies an emphasis on adult plant resistance for achieving durable resistance to stem rust in wheat.
Though an abundance of genetic sources of resistance exist to emerging stem rust races, few conventional wheat varieties possess effective resistance. Singh et al., 2015 reported that only 16% of U.S. hard red spring wheat varieties and breeding lines showed resistance to the Ug99 race group. This susceptibility is significant as the U.S. hard red spring wheat growing region is historically vulnerable to major epidemics of wheat stem rust when virulent races are present, resulting in over 50% statewide yield losses in Minnesota and North Dakota during epidemic years. One exception to this widespread vulnerability is hard red spring wheat variety ‘Linkert’, released in 2013 by the University of Minnesota (Anderson et al., 2018). Linkert exhibited consistent adult plant resistance to Ug99 in Kenya (Anderson et al., 2018). Linkert became a successful variety that was grown on 833,900 acres in Minnesota and North Dakota in 2018 and was the most widely grown wheat variety in Minnesota from 2016 to 2020. The objective of this study was to characterize the genetics of stem rust resistance in ‘Linkert’ with particular attention to the adult plant resistance exhibited to Ug99.
Materials and methods
Mapping populations
Two doubled haploid (DH) mapping populations were developed by crossing Linkert with both hard red spring wheat variety ‘Forefront’ (Glover et al., 2013) and genetic stock LMPG-6 (‘Little Club’//3 3 ‘Prelude’/8 3 Marquis’/3/’Gabo’), a highly rust susceptible, day length insensitive line that was developed for the purpose of producing near-isogenic lines for Sr genes (Knott, 2000). ‘Linkert’ is a hard red spring wheat developed and released by the Minnesota Agricultural Experiment Station (MAES) in 2013. Linkert is one of the highest quality hard red winter wheats with yield comparable to other high protein varieties and better disease resistance. Doubled haploid progeny were produced at a service facility at Washington State University through a modified maize pollination method (Niu et al., 2014). A recombinant inbred line (RIL) population was derived through single seed descent to the F6 generation after crossing Linkert and University of Minnesota breeding line ‘MN07098-6’. A total of 107, 190, and 172 lines were derived from the Linkert/Forefront (LK/FF), Linkert/LMPG-6 (LK/LM), and Linkert/MN07098-6 (09X149) populations, respectively.
Disease evaluations
The lines in the DH populations with their parents were evaluated in the field for three years (2016-2018) in Kenya for response to the Ug99 race group (Pgt races including TTKSK, TTKTT), and in Ethiopia for response to Pgt races TKTTF, TTKSK, TRTTF and JRCQC (Kosgey et al., 2021). The RIL population (F6:8 generation) was evaluated in Kenya and Ethiopia for two years (2016 and 2017). The DH populations were also evaluated at Rosemont, MN for three years (2015-2017) with four domestic Pgt races evaluated in single race nurseries inoculated with races QTHJC, QFCSC, TPMKC and RCRSC, according to previously described methods (Edae et al., 2018). Mixtures of spores of Pgt races for field evaluations were inoculated both in Ethiopia and Kenya (Kosgey et al., 2021). Stem rust severity was visually scored in the experimental plots based on the modified Cobb scale of 0-100, where 0 = immunity (no uredinia or any other sign of infection) and 100% = completely susceptible (Peterson et al., 1948). Infection response was rated as resistant (R), small uredinia surrounded by necrosis; moderately resistant (MR), medium-sized uredinia surrounded by necrosis or chlorosis; moderately susceptible (MS), medium-sized uredinia without necrosis; susceptible (S), large uredinia without necrosis; or MRMS, an infection response that included both the MR and MS categories (Roelfs et al., 1992). Coefficient of infection (COI) values were generated by multiplying the stem rust severity value for each line by a constant value for each infection response: 0 = 0, R = 0.2, RMR = 0.3, MR = 0.4, M = 0.6, MS = 0.8, S = 1.0 (Knott, 1989). Average coefficient of infection for the two replicates were determined and used for analyses. Raw mean coefficients of infection values across the two randomized replicates in each environment were used for QTL analyses.
At the seedling stage, the DH populations and parents were evaluated with Pgt races QTHJC (isolate 0069MN399), QFCSC (isolate 95MN1080), TPMKC (isolate 74MN1409), RCRSC (isolate 00MN99C), TRTTF (06YEM34-1), and TKTTF (isolate 13ETH18-1) according to previously described methods (Hundie et al., 2019). All four parents were susceptible as seedlings to race TTKSK (04KEN156/04; Ug99 race group).
DNA extraction and genotyping
Tissues were harvested from approximately two weeks old seedlings of the populations and parents and grown in a greenhouse before placing into 96-well plates. The harvested leaves were dried in a lyophilizer and ground for 2 min using a Genogrinder (SPEX Sample Prep). DNA extraction was carried out following an SDS method: 300 µl extraction buffer (200 mM Tris-HCL pH 8.0, 250 mM NaCl, 25 mM EDTA, 0.5% SDS, ddH20) was added to each well followed by gently shaking the 96-well plates before adding 300 µl of chloroform:isoamyl alcohol (24:1) to each well. The suspension was mixed for 3 minutes on a plate shaker followed by centrifuging for 20 min at 2500 rpm, and the supernatant (200-300 µl) was transferred to separate tubes.
After decanting the supernatant, 300 µl of 70% ethanol was added to each tube, the tubes were centrifuged for 20 min at 2,500 rpm, the ethanol was poured out, and the DNA pellets were air-dried. When the pellets were completely dry, 100 µl of 1xTE was added to each tube to resuspend the pellet. DNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher, Waltham, MA, United States).
Genotyping data of the DH populations were generated using the iSelect 90k SNP assay developed for wheat (Wang et al., 2014). SNPs with minor allele frequency (MAF) less than 25% were removed, all monomorphic markers and markers with missing data points for one or both parental genotypes were also removed. No heterozygote genotypes were retained in the data set. Missing data points were represented by “-”. SNPs with > 10% missing data points were removed, and none of the lines were removed as missing data for all individuals for a given SNP was < 1%. SNPs with segregation distortion p-values of less than 0.5 were removed before creating linkage maps for both DH populations. For the LK/FF population, one SNP was removed due to high missing values in the segregation distortion filtered data set. However, for the LK/LM population, all SNPs had missing values < 6.4% and none of the SNPs were removed. SNPs with lod error (Lincoln and Lander, 1992) values greater than four were removed for the final linkage map construction. After applying the filtering criteria described above, a total of 1,091 and 1,552 high quality SNPs were used for QTL mapping for LK/FF and LK/LM populations, respectively. These SNP markers were used to make linkage groups using JoinMap 4.0 (Upadhyaya, 2006). The Kosambi mapping function was used to estimate the map distance.
Genomic DNA from the Linkert/MN07098-6 population was extracted using the BioSprint 96 DNA Plant Kit (QIAGEN, Valencia, CA) and genotyped using the genotype-by-sequencing approach (Poland et al., 2012). Reads were filtered for phred quality score (Q) of ≥ 30 and de-multiplexed using ‘sabre’ (https://github.com/najoshi/sabre, accessed 05-12-2022). Filtered reads were aligned to the Triticum aestivum v1.0 reference sequence (The International Wheat Genome Sequencing Consortium (IWGSC), 2018) using default parameters in the Burrows-Wheeler Aligner 0.7.5 (Li, 2013). Discovery of genome-wide markers (SNPs) was done using default parameters in SAMtools 1.6 and BCFtools 1.6 (Li et al., 2009; Li, 2011). SNPs with heterozygote level and missing data points greater than 10% and 25%, respectively, were removed. A filtered data set comprised of 1,674 SNPs was used for QTL mapping.
Quantitative trait loci (QTL) analysis was conducted in R package RQTL (Broman et al., 2003) with composite interval mapping (CIM) method using both seedling infection type data and field response data from the populations. Before QTL mapping, categorical field infection responses and seedling infection types were converted to a linear 0-9 scale (Zhang et al., 2014; Gao et al., 2016).
Results
Phenotypic evaluation
Linkert was susceptible at the seedling stage to virulent Pgt races such as TTKSK, TTKTT, and TTRTF, but was resistant to the Ethiopian TKTTF isolate. Forefront was also susceptible to both TTKSK and TTKTT (Table 1). Disease severity values ranged from 1- 47%, 4 - 44% and 4 - 25%, 65 - 70% for Linkert, Forefront, LMPG-6 and MN07098-6, respectively, for response to Pgt races in East Africa (Table 1). Although LMPG-6 had lower average severity percentage than Linkert and Forefront, the average coefficient of infection was similar for all the three parents (16.26- 17.24). For North American Pgt races, Linkert was resistant to all Pgt races (QFCSC, RCRSC, QTHJC and TPMKC) tested at the seedling stage whereas LMPG-6 was susceptible to all races. However, Forefront was resistant to QFCSC and RCRSC but susceptible to QTHJC and TPMKC races at the seedling stage (Table 2). There was strong phenotypic correlation between Ethiopia and Kenya field data based on severity and infection response except for the data from Kenya in 2016 and Ethiopia in 2017. Both severity and infection response data recorded in 2016 in Kenya were poorly correlated with other years in Kenya and Ethiopia (Figure 1; Supplementary Table 1).
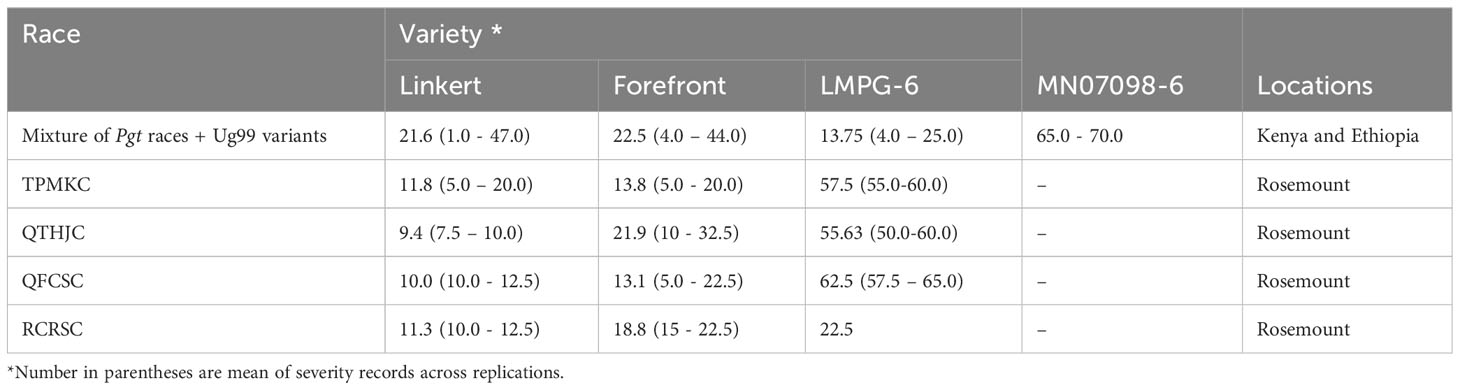
Table 1 Disease severity (%) recorded for Linkert, Forefront, LMPG-6 and MN07098-6 to different stem rust races at different locations (Rosemount, Kenya and Ethiopia).
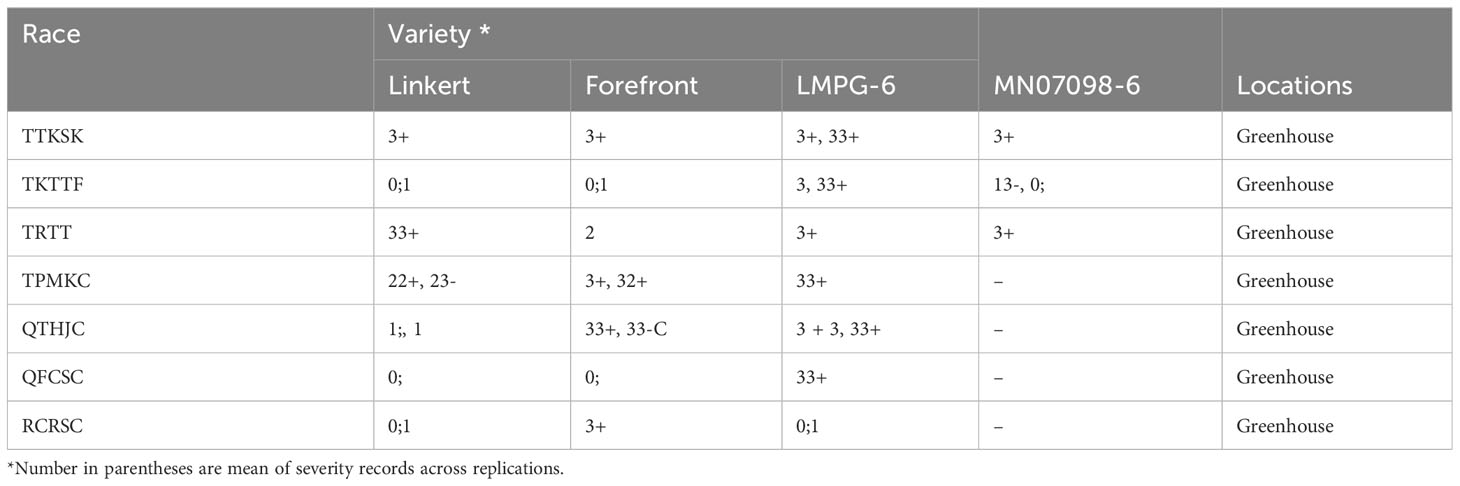
Table 2 Seedling infection types recorded for Linkert, Forefront, LMPG-6 and MN07098-6 to different stem rust race in controlled greenhouse.
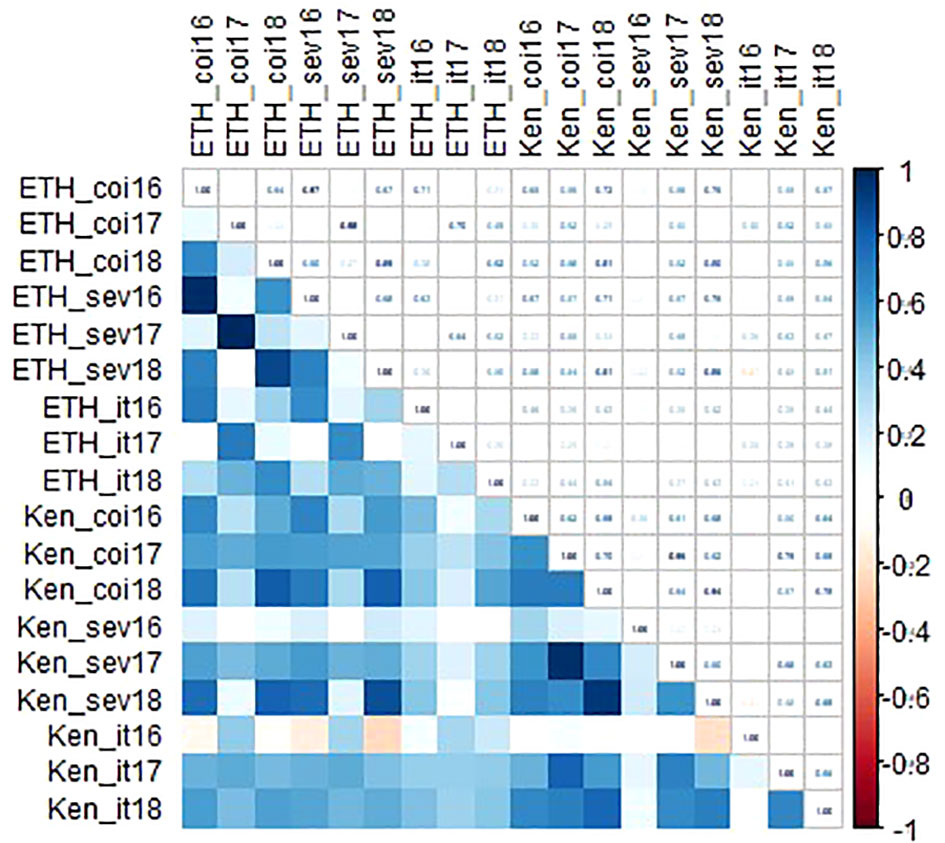
Figure 1 Correlation coefficients among environments for both disease severity and infection response data recorded in Kenya and Ethiopia from 2016-2018.
Seedling infection type (IT) values in linear scale less than six were considered as resistant whereas IT values greater than six were considered as susceptible. All individuals of the Linkert/Forefront population were resistant to Pgt races QFCSC and RCRSC. For all races tested in this study the percentage of resistant lines was greater than 50% except for response to race TPMKC which resulted in resistance in only 35.7% of the Linkert/Forefront and 29.4% of the Linkert/LMPG individuals (Table 3).
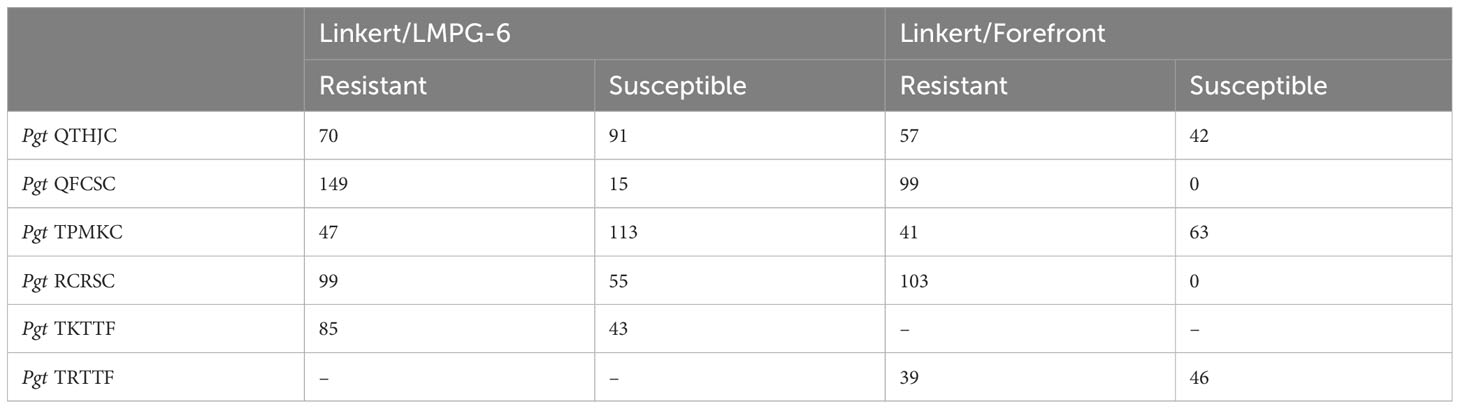
Table 3 Number of resistant and susceptible lines based on seedling reaction following inoculation with six Pgt.
Genetic linkage maps
For Linkert/Forefront population, a total of 1,088 SNPs from 90k SNP chip mapped to 25 linkage groups, and these 25 linkage groups corresponded to 19 wheat chromosomes. Apart from two wheat chromosomes, 6D and 7D, all wheat chromosomes were represented in the map of Linkert/Forefront population (Figure 2). For both populations, chromosome 7B comprised the highest number of SNPs whereas chromosomes 3D (5 SNPs) and 4D (11 SNPs) had the lowest number of SNPs for LK/FF and LK/LM populations, respectively (Supplementary Tables 2A, B). For the Linkert/LMPG-6 (LK/LM) population a total of 1,552 SNPs from 90k SNP chip mapped to 25 linkage groups. However, the 25 linkage groups were assigned to 17 wheat chromosomes. Five chromosomes (3D, 5D, 6D and 7D) all were from D genome were missing from the linkage (Figure 3; Supplemental Table 2B). Since SNP identification was carried out using wheat genome reference sequence for the validation population, Linkert/MN07098-6, linkage map creation was not attempted because we were able to generate this population specific physical map for the 1,674 GBS SNPs (Figure 3). Unlike 90K SNP chip, all 21 wheat chromosomes were recovered for the GBS platform and chromosome 6B had the highest number of SNPs (2540) whereas only three SNPs passed the filtering criteria for chromosome 4D (Supplementary Table 2C). Relatively GBS SNPs were only evenly distributed on chromosome 6B but for the remaining chromosomes SNPs that met the filtering criteria (see Materials and Methods section) were mainly on the terminal ends of chromosomes. However, unlike GBS, large gap size on linkage maps were not observed for the 90K SNP chip most of the cases.
QTL analyses
In this study, after considering QTL within five centimorgans as redundant QTL, a total of 55 significant QTL distributed on 18 chromosomes were detected for all populations evaluated both under field and seedling stages for resistance to stem rust (Tables 4–7). QTL were detected on all chromosomes except 5D and 7D. Although the phenotypic variance explained was less than 20% in most cases, the QTL detected on chromosomes 1BL and 4DS in the LK/LM population evaluated in the field for race QFCSC and the QTL on chromosome 4AL for race TKTTF at the seedling stage explained substantial amounts of phenotypic variation (38.4-69.0%). In this study, all-stage QTL were detected only on chromosomes 1B (52.0 cM) and 4AL for race RCRSC detected in the Linkert/Forefront and Linkert/LMPG-6 populations, respectively (Tables 4, 5). The remaining detected QTL were expressed only either at seedling or adult plant stages.
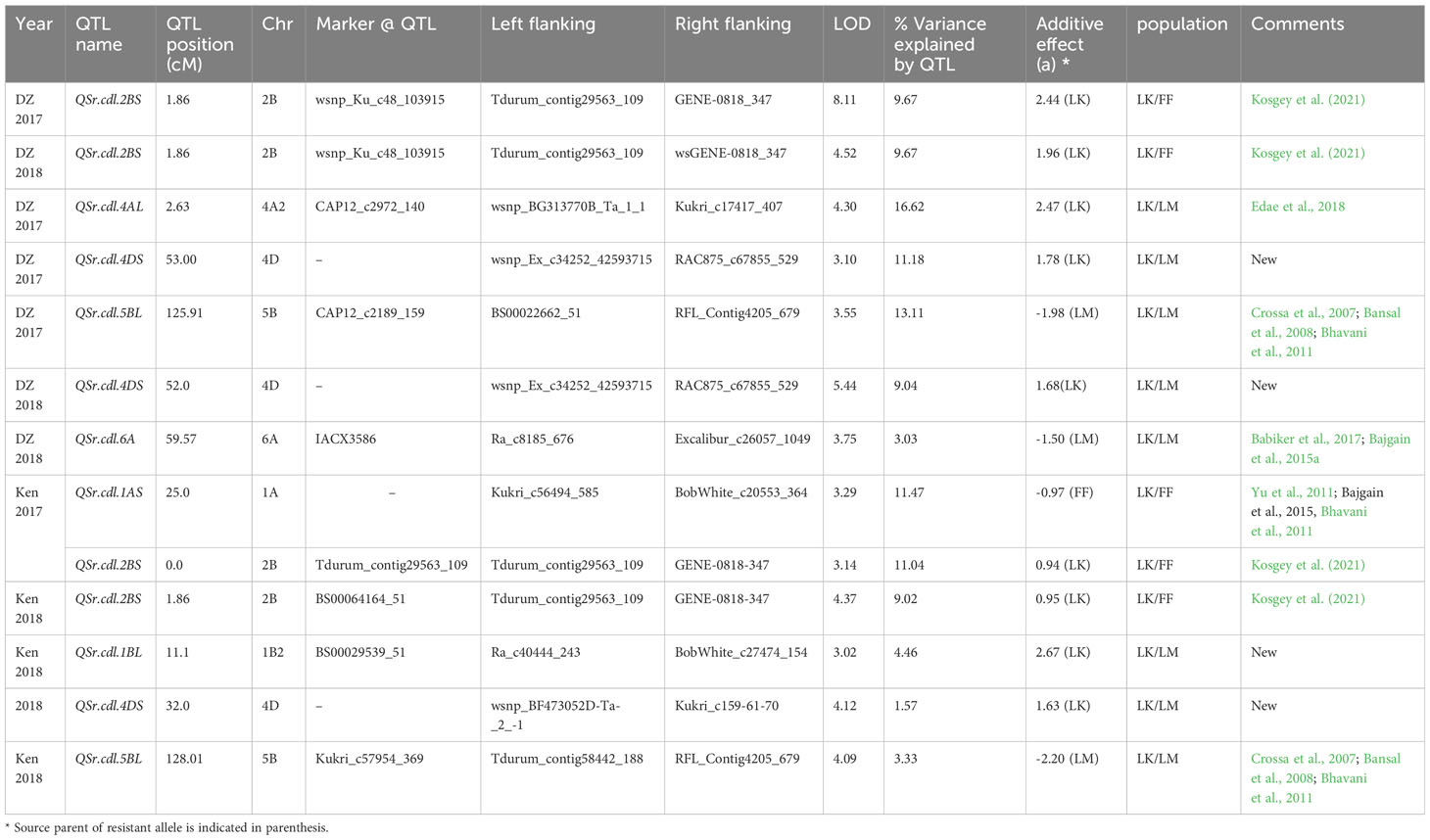
Table 4 Adult-stage stem rust QTL detected for Linkert/Forefront (LK/FF) and Linkert/LMPG-6 (LK/LM) populations in Ethiopia and Kenya using coefficient of infection.
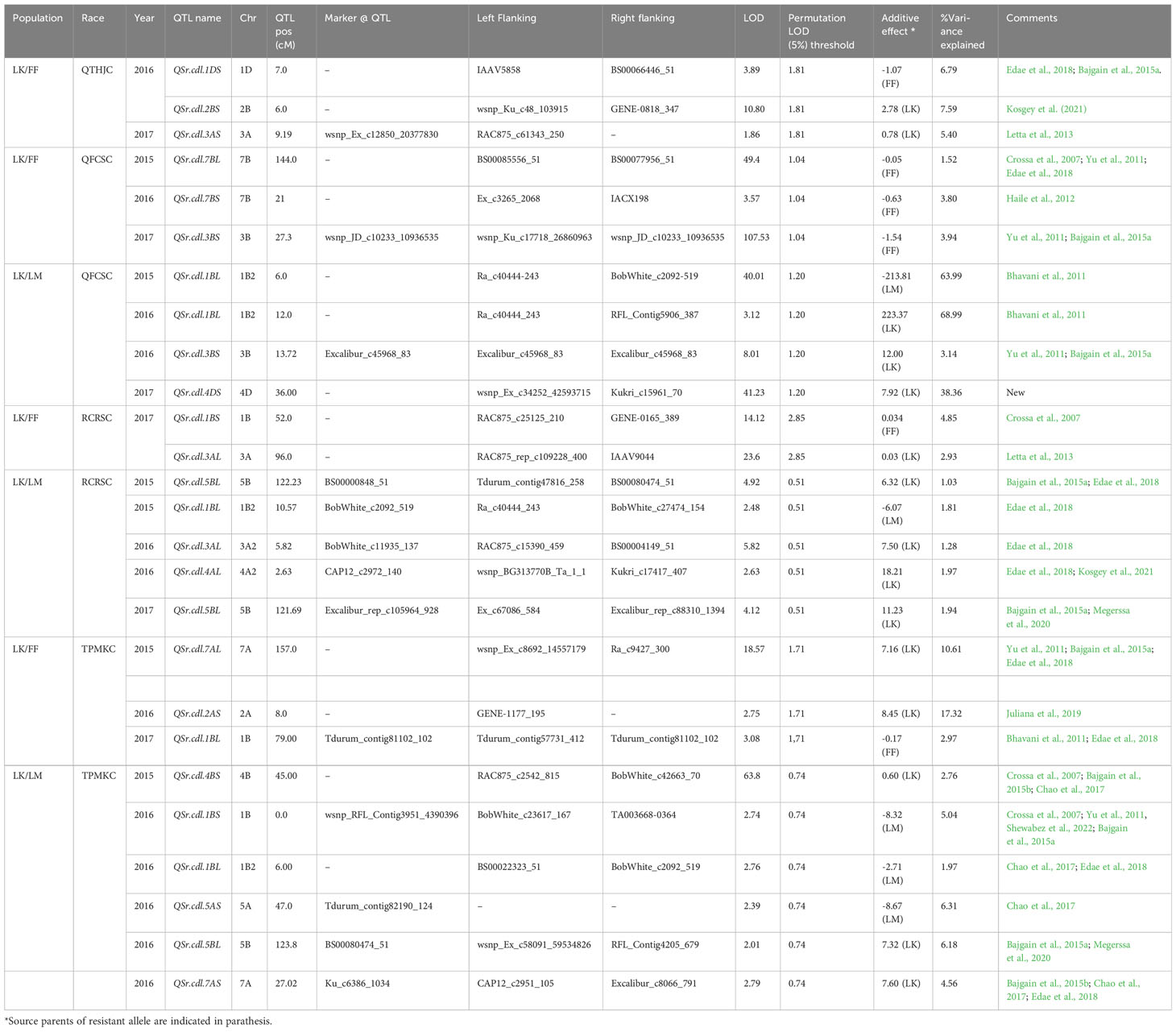
Table 5 QTL identified for North American domestic stem rust races in Linkert/Forefront (LK/FF) and Linkert/LMPG-6 (LK/LM) populations evaluated at Rosemount, MN.
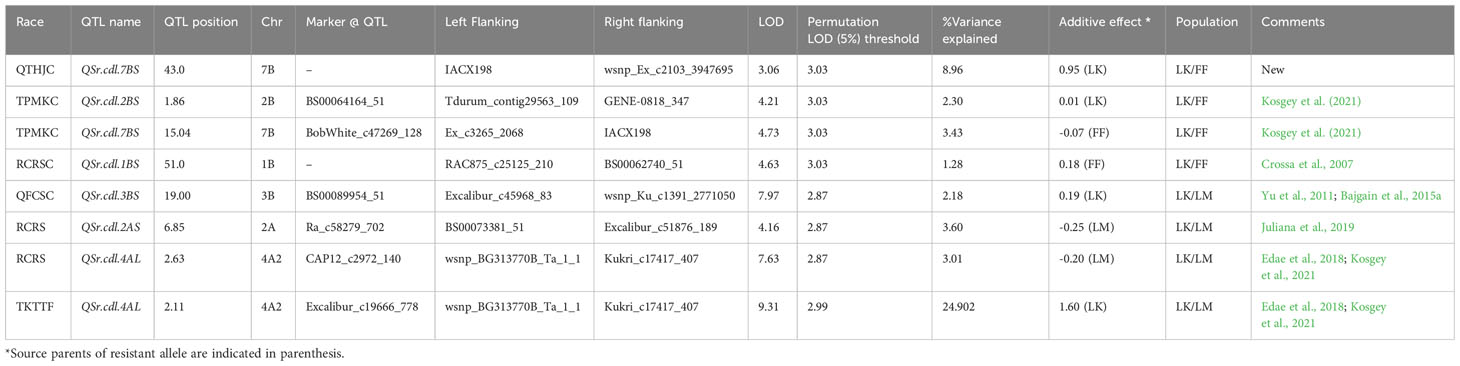
Table 6 Seedling QTL for domestic races and race TKTTF based on infection type (IT) for Linkert/Forefront (LK/FF) and Linkert/LMPG-6 (LK/LM) populations.
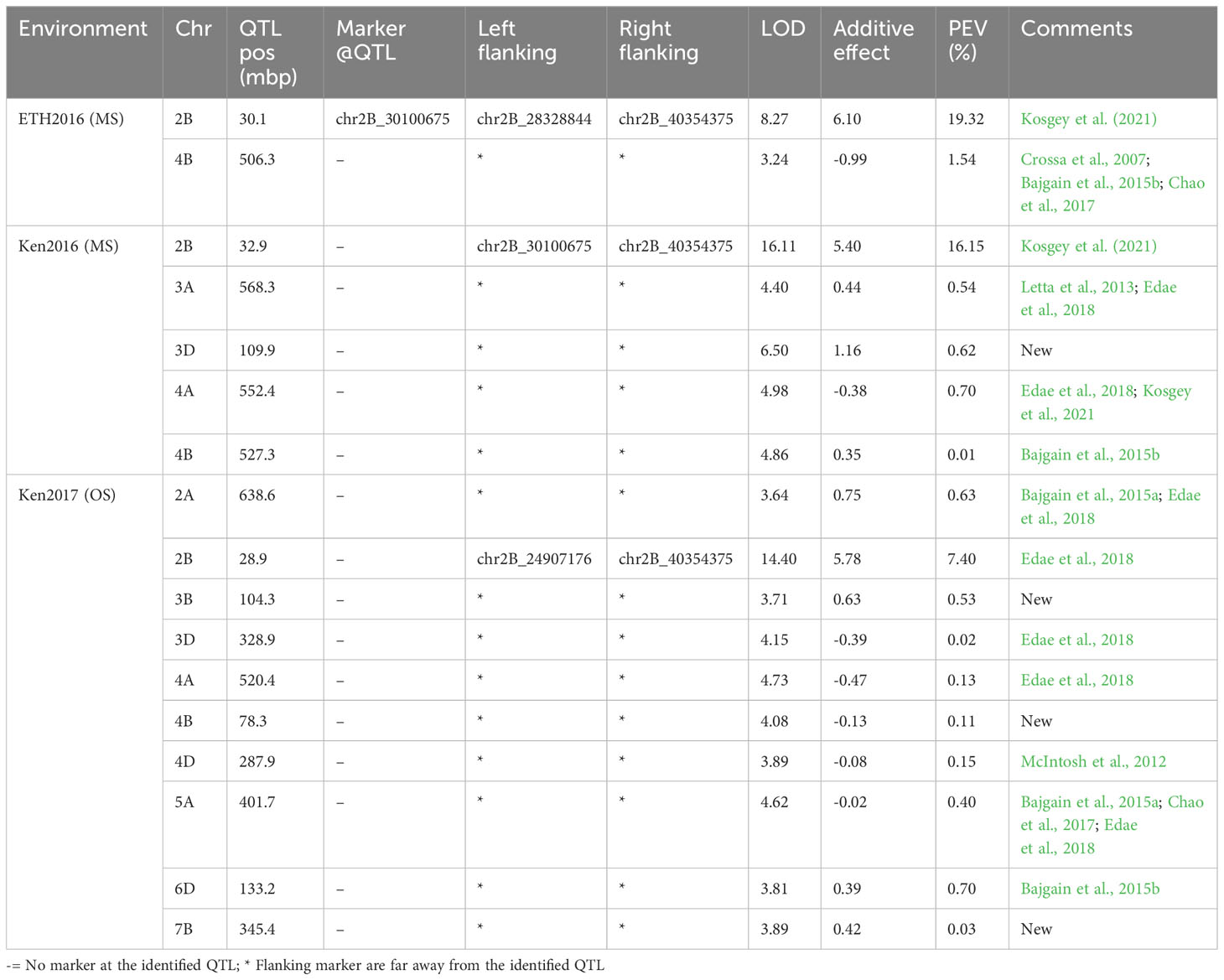
Table 7 QTL identified for Linkert/MN07098-6 population evaluated at Kenya and Ethiopia for two seasons.
A majority of the QTL were population, environment, and race-specific. However, one QTL (marker: CAP12_c2189_159) on chromosome 2BS (1.9 cM) was detected across two populations in multiple environments (Tables 4, 7). It was also detected for field response to race QTHJC and seedling response to race TPMKC (Tables 5, 6). A population-specific stable QTL was detected on chromosome 5BL (125.9 cM) in the LK/LM population across environments in Ethiopia and Kenya (Table 4). It also provided field resistance against races RCRSC and TPMKC (Table 5). The gene Sr7a was detected on 4AL (marker: CAP12QT_c2972_140) in the LK/LM population in Ethiopia and in Rosemont for response to RCRSC in 2016, and seedling response to race TKTTF. In Linkert/MN07098-6, a QTL on chromosome 2BS (30.1 Mbp) was consistently detected and explained up to 19.3% of the phenotypic variation (Table 7). At chromosome level, Linkert/MN07098-6 had a total 11 chromosomes on which QTL were detected and some of the QTL had similar chromosome regions (e.g., 2BS) as detected in other populations.
Discussion
The objective of this study was to characterize the genetics of stem rust resistance in the hard red spring wheat variety Linkert by testing progenies developed by crossing with three stem rust susceptible lines, cultivar Forefront, breeding line MN07098-6, and genetic stock LMPG-6, with different Pgt races in the field and at the seedling stage in a greenhouse.
A reliable stem rust QTL was identified on chromosome 2BS in the Linkert/Forefront and Linkert/MN07098-6 populations. Interestingly, in addition to conferring adult plant resistance to the Ug99 race group in African field environments, it was detected at the seedling stage for response to race TPMKC. In addition to relatively consistent expression under different environments, the phenotypic variation explained by the QTL was also substantial (9.02-19.32%). Seven Sr genes have been reported on 2BS, and out of these Sr36, Sr39, and Sr40 (Wu et al., 2009; Niu et al., 2011; Rouse et al., 2012) are effective against the Ug99 race TTKSK. However, the QTL detected in this study is only effective to the Ug99 race group at the adult growth stage, excluding the previously described major effect all-stage resistance genes on chromosome arm 2BS. The coincidence of this QTL with seedling response to race TPMKC warrants further study. Kosgey et al. (2021) also detected a QTL (QSr.cdl-2BS.2) on 2BS using field-tested recombinant inbred lines derived from a cross made between CI 14275 (resistant parent) and LMPG-6 (susceptible parent), and the KASP marker that was developed from SNP Excalibur_c7963_1722_C1 was also associated with reduced stem rust severity. Thus, the resistance in Linkert and CI 14275 is most likely similar. Previous QTL mapping studies have also shown that there were at least eight QTL regions on the short arm of chromosome 2B (from 0-160 mbp range) detected in both GWAS and bi-parental populations (Crossa et al., 2007; Bajgain et al., 2015a; Bajgain et al., 2015b; Bajgain et al., 2016; Edae et al., 2018), and the associated SNPs were IWB2369 (pos: 48.5 cM; 31.50 mbp), IWB69830 (pos: 46.76 cM; 31.30 mbp), IWB24614 (104. 80 mbp), IWB32327 (104.80 mbp), IWB23439 (48.04 cM; 55.04 mbp), and BS00073426-51 (160.00 mbp). Gene Sr23 mapped on distal end of 2BS and was described as completely linked with Lr16 (McIntosh and Luig, 1973b) and expressed under conditions of high temperature and high light intensity (Luig, 1983). All cultivars that possess Lr16 also were reported to possess Sr23 (Kassa et al., 2017). Cultivars such as Exchange, Selkirk, Warden and Etoile de choisy are sources of Sr23. A recent report also indicated that Lr16 is present in North American cultivars such as ‘AC Domain’, ‘AC Karma’, ‘AC Majestic’, ‘AC Splendor’, ‘Columbus’, and ‘Grandin’ (Kassa et al., 2017). Although major gene Lr16/Sr23 was mapped on chromosome 2BS, it is unlikely that the QTL detected in the current study is Lr16/Sr23 because the QTL is mapped 21 cM away from that of the Lr16/Lr23 position.
The Linkert/LMPG-6 population possessed a relatively consistent QTL on chromosome 4AL (SNP: CAP12_c2972_140) that provided resistance against different Pgt races both at seedling and adult stages implying it can provide all-stage resistance against stem rust races RCRSC and TKTTF. Edae et al., 2018 reported a strong association signal in the region of major gene Sr7a for resistance against race RCRSC. Similarly, in this study the QTL on chromosome 4AL was detected at adult and seedling stages for Pgt race RCRSC, indicating Linkert possesses gene Sr7a, as previously postulated (Anderson et al., 2018).
The QTL detected on chromosome 5BL in Linkert/LMPG-6 was relatively stable. This region of 122-128 cM on the long arm of chromosome 5B harbored a QTL that provided field resistance against virulent races in East Africa (Ethiopia and Kenya) and North American Pgt races (RCRSC and TPMKC). This QTL was detected in the Linkert/LMPG-6 population but not in the Linkert/Forefront or Linkert/MN07098-6 populations. Stem rust resistance gene Sr56 that confers adult plant resistance (APR) was mapped on chromosome 5BL (McIntosh et al., 2012). Several QTL have been reported using both bi-parental populations and GWAS including adult plant resistance gene Sr56 (Crossa et al., 2007; Bansal et al., 2008; Bhavani et al., 2011). In previous studies the 5BL QTL representing Sr56 explained 10-13% phenotypic variation and contributed an important component of the “Sr2 complex” (Bhavani et al., 2011). The QTL detected in the current study explained up to 13% of phenotypic variation, and the QTL, QSr.Sun-5BL, reported by Bansal et al., 2008 also explained 11 to 12% of the phenotypic variation in adult plant stem rust response and was responsible for a 12-15% reduction in stem rust severity. Since Sr56 was first reported from European ‘Arina’ winter wheat, a pedigree link between Sr56 and Linkert is not known. Further studies are warranted to determine if the 5BL QTL is Sr56. It is possible that both Forefront and MN07098-6 also possess the resistance allele of the 5BL QTL, which would explain why it was not detected in the corresponding populations.
Besides stem rust QTL on 2BS, 5BL, and 4AL, numerous environment and population-specific QTL were found that corresponded with previously reported QTL. Three QTL were detected on chromosome 7A. Two of them provided resistance against Pgt race TPMKC (pos: 157.0 cM and 27.0 cM). Several QTL have been reported on 7AL (Bajgain et al., 2015a; Edae et al., 2018) for stem rust resistance, and the major genes Sr15 and Sr22 are located on 7AL (Sears and Briggle, 1969; The and McIntosh, 1975). Similarly, QTL were detected on chromosome 7B for stem rust resistance. Two QTL that provided field resistance against races QTHJC (pos: 126.0 cM) and QFCSC (pos: 144.0 cM) were detected on 7BL. Edae et al., 2018 also reported QTL for Pgt race QFCSC on 7BL (pos: 109.0 cM) using association mapping.
The seedling and single-race field experiments in Rosemount, MN facilitated a precise evaluation of resistance QTL to the four races evaluated. As expected, all-stage resistance gene Sr7a was consistently detected in both seedling and field studies in response to avirulent race RCRSC. However, the other QTL were detected only in seedling or field environments. This seems to contrast with the expectation that QTL detected at the seedling stage would be effective in the field consistent with “all-stage” resistance. This may be explained by the relatively low effectiveness of the seedling QTL detected other than Sr7a. Not detecting these weakly effective QTL in the field may be the result of (1) true ineffectiveness of these QTL in the field or (2) the masking of weakly effective all-stage QTL by the presence of adult plant resistance loci. Our results are similar with the findings of an association mapping study conducted by Edae et al., 2018 where conventional North American spring wheat lines possesses largely seedling- or field-effective QTL. Only the most strongly effective QTL were detected in both seedling and field studies.
In conclusion, despite the large number of environment and race-specific QTL identified in the current study, we found Linkert derived QTL that were consistently associated with adult plant resistance to Ug99 stem rust on chromosome 2BS in the Linkert/Forefront and Linkert/MN07098-6 populations. Additional QTL were detected consistently on chromosomes 5BL (125.9 cM) and 4AL (Sr7a) with the Linkert/LMPG population. Combined with the detection of the QTL on 2BS in previous studies, the identification of the 2BS QTL in the two populations derived from conventional germplasm demonstrates the importance of this QTL in contributing towards stem rust adult plant resistance in United States hard red spring wheat.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author contributions
EE: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ZK: Writing – review & editing. PB: Data curation, Writing – review & editing. KN: Conceptualization, Data curation, Investigation, Writing – review & editing. AG: Data curation, Methodology, Writing – review & editing. SB: Resources, Writing – review & editing. JA: Funding acquisition, Project administration, Resources, Writing – review & editing. MR: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support was obtained from the USDA-ARS National Plant Disease Recovery System.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1343148/full#supplementary-material
References
Anderson, J. A., Wiersma, J. J., Linkert, G. L., Reynolds, S. K., Kolmer, J. A., Jin, Y., et al. (2018). Registration of 'Linkert' spring wheat with good straw strength and adult plant resistance to the Ug99 family of stem rust race. J. Plant Registration 12, 208–214.
Arora, A., Steuernagel, B., Gaurav, K., Chandramohan, S., Long, Y., Matny, O., et al. (2019). Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. doi: 10.1038/s41587-018-0007-9
Bajgain, P., Rouse, M. N., Bhavani, S., Anderson, J. A. (2015b). QTL mapping of adult plant resistance to Ug99 stem rust in the spring wheat population RB07/MN06113-8. Mol. Breed. 35, 170. doi: 10.1007/s11032-015-0362-x
Bajgain, P., Rouse, M. N., Bulli, P., Bhavani, S., Gordon, T., Wanyera, R., et al. (2015a). Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biol. 15, 249. doi: 10.1186/s12870-015-0628-9
Bajgain, P., Rouse, M. N., Tsilo, T. J., Macharia, G. K., Bhavani, S., Jin, Y., et al. (2016). Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PloS One 11, e0155760. doi: 10.1371/journal.pone.0155760
Babiker, E. M., Gordon, v., Bonman, J. M. (2017). Genetic Loci conditioning adult plant resistance to the Ug99 race group and seedling resistance to races TRTTF abd TTTTF of the stem rust pathogen in wheat landrace Cltr 15026. Plant Dis. 101, 496–501.
Bansal, U. K., Bossolini, E., Miah, H., Keller, B., Park, R. F., Bariana, H. S. (2008). Genetic mapping of seedling and adult plant stem rust resistance in two European winter wheat cultivars. Euphytica 164, 821–828. doi: 10.1007/s10681-008-9736-z
Bhavani, S., Singh, R. P., Argillier, O., Huerta-Espino, J., Singh, S., Njau, P., et al. (2011). “Mapping durable adult plant stem rust resistance to the race Ug99 group in six CIMMYT wheats,” in 2011 BGRI technical workshop Borlaug Global Rust Initiative, St. Paul, MN, pp 43–pp 53.
Broman, K. W., Wu, H., Sen, Ś., Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Carleton, M. A. (1905). Lessons from the grain-rust epidemic of 1904. Farmers’ Bulletin No. 219, 1–24.
Chao, S., Rouse, M. N., Acevedo, M., Szabo-Hever, A., Bockelman, H., Bonman, M. J., et al. (2017). Evaluation of genetic diversity and host resistance to stem rust in USDA NSGC durum wheat accessions. Plant Genome 10, (2). doi: 10.3835/plantgenome2016.07.0071
Chapman, J., Mascher, M., Buluç, A., Barry, K., Georganas, E., Session, A., et al. (2015). A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol. 16, 26. doi: 10.1186/s13059-015-0582-8
Chen, S., Rouse, M. N., Zhang, W., Zhang, X., Guo, Y., Briggs, J., et al. (2020). Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol 225, 948-959. doi: 10.1111/nph.16169
Chen, S., Zhang, W., Bolus, S., Rouse, M. N., Dubcovsky, J. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PloS Genet. 14, e1007287. doi: 10.1371/journal.pgen.1007287
Crossa, J., Burgueno, J., Dreisigacker, S., Vargas, M., Herrera-Foesse, S. A., Lillemo, M., et al. (2007). Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177, 1889–1913. doi: 10.1534/genetics.107.078659
Edae, A. E., Pumphrey, M. O., Rouse, M. N. (2018). A genome wide association study of resistance to North American stem rust races in a hexaploid spring wheat (Triticum aestivum L.) breeding germplasm. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00052
Evans, L. T. (1980). Responses to challenge: William Farrer and the making of wheat. J. Aust. Inst. Agric. Sci. 46, 3–13.
Gao, L., Turner, M. K., Chao, S., Kolmer, J., Anderson, J. A. (2016). Genome wide association study of seedling and adult plant leaf rust resistance in elite spring wheat breeding lines. PloS One 11, e0148671. doi: 10.1371/journal.pone.0148671
Glover, K. D., Rudd, J. C., Devkota, R. N., Hall, R. G., Jin, Y., Osborne, L. E., et al. (2013). Registration of “Forefront” wheat. J. Plant Registrat. 7, 184–190. doi: 10.3198/jpr2012.07.0007crc
Haile, J. K., Nachit, M. M., Hammer, K., Badebo, A., Roder, M. S. (2012). QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. Tritici in durum wheat (Triticum durum Desf.). Mol. Breed. 30, 1479–1493. doi: 10.1007/s11032-012-9734-7
Hiebert, C. W., Fetch, T. G., Zegeye, T. (2010). Genetics and mapping of stem rust resistance to Ug99 in the wheat cultivar Webster. Theor. Appl. Genet. 121, 65–69. doi: 10.1007/s00122-010-1291-z
Hundie, B., Girma, B., Tadesse, Z., Edae, E., Olivera, P., Hailu, A. E., et al. (2019). Characterization of ethiopian wheat germplasm for resistance to four puccinia graminis f. sp. tritici races facilitated by single-race nurseries. Plant Dis. 103, 2359–2366. doi: 10.1094/PDIS-07-18-1243-RE
Juliana, P., Poland, J., Huerta-Espino, J., Shrestha, S., Crossa, J., Crespo-Herrera, L., et al. (2019). Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat. Genet. 51, 1530–1539. doi: 10.1038/s41588-019-0496-6
Krattinger, S., Lagudah, E., Spielmeyer, W., Singh, R., Huerta-Espino, J., Mcfadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323 (5919), 1360–1363.
Kassa, M., You, F., Hiebert, C., Pozniak, C., Fobert, P., Sharpe, A., et al. (2017). Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16. BMC Plant Biol. 17, 45. doi: 10.1186/s12870-017-0993-7
Knott, D. R. (2000). Inheritance of resistance to stem rust in ‘Triumph 64’ Winter wheat. Crop Sci. 40, 1237–1124. doi: 10.2135/cropsci2000.4051237x
Kosgey, Z. C., Edae, E. A., Dill-Macky, R., Jin, Y., Bulbula, W. D., Gemechu, A., et al. (2021). Mapping and validation of stem rust resistance loci in spring wheat line CI 14275. Front. Plant Sci. 11, 609659. doi: 10.3389/fpls.2020.609659
Kumsa, T. T., Baenziger, P. S., Rouse, M. N., Guttieri, M., Dweikat, I., Brown-Guedira, G., et al. (2015). Characterization of stem rust resistance in wheat cultivar Gage. Crop Sci. 55, 229–239. doi: 10.2135/cropsci2014.05.0348
Lincoln, S. E., Lander, E. S. (1992). Systematic detection of errors in genetic linkage data. Genomics 14, 604–610. doi: 10.1016/S0888-7543(05)80158-2
Letta, T., Maccaferri, M., Badebo, A., Ammar, K., Ricci, A., Crossa, J., et al. (2013). Searching for novel sources of field resistance to Ug99 and Ethiopian stem rust races in durum wheat via association mapping. Theor. Appl. Genet. 126, 1237–1256. doi: 10.1007/s00122-013-2050-8
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM (Cambridge, MA 02142: Oxford University Press), 1–3. Available at: https://arxiv.org/abs/1303.3997.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinf. (Oxford England) 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Luig, N. H. (1983). A survey of virulence genes in wheat stem rust, Puccinia graminis f. sp. tritici. Fortschr. der Pflanzenzuechtung 11. 198PP.
Mago, R., Spielmeyer, W., Lawrence, G., Lagudah, E., Ellis, J., Pryor, A. (2002). Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor. Appl. Genet. 104, 1317–1324. doi: 10.1007/s00122-002-0879-3
Mago, R., Zhang, P., Vautrin, S., Šimková, H., Bansal, U., Luo, M. C., et al. (2015). The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 1, 15186. doi: 10.1038/nplants.2015.186
Mascher, M., Muehlbauer, G. D., Rokhsar, D., Chapman, J., Schmutz, J., Barry, K., et al. (2013). Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J. 76, 718–727. doi: 10.1111/tpj.12319
Megerssa, S., Ammar, K., Acevedo, M., Brown-Guedira, G., Warrd, B., et al. (2020). Multiple-race stem rust resistance loci identified in durum wheat using genome-wide association mapping. Front. Plant Sc. 11. doi: 10.3389/fpls/.2020.598509
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C. F., Appels, R., Xia, X. C. (2012). Catalogue of gene symbols for wheat: supplement. Ann. Wheat Newsl 58, 259.
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., Raupp, W.J. (2020). Catalogue of gene symbols for wheat: supplement. Ann. Wheat Newsl 66, 109-128.
McIntosh, R. A., Luig, N. H. (1973). Linkage of genes for reaction to Puccinia graminis f. sp. tritici and P. recondita in Selkirk wheat and related cultivars. Aust. J. Biol. Sci. 26, 1145–1152. doi: 10.1071/BI9731145
Milus, E., Moon, D., Lee, K. D., Maton, E. (2015). Race-specific adult plant resistance in winter wheat to stripe rust and characterization of pathogen virulence patterns. Phytopathology 105, 1114–1122. doi: 10.1094/PHYTO-11-14-0305-R
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498.
Newcomb, M., Olivera, P. D., Rouse, M. N., Szabo, L. J., Johnson, J., Gale, S., et al. (2016). Kenyan isolates of Puccinia graminis f. sp. tritici from 2008 to 2014: Virulence to SrTmp in the Ug99 race group and implications for breeding programs. Phytopathology 106 (7), 729–736 8.
Niu, Z., Jiang, A., Abu Hammad, W., Oladzadabbasabadi, A., Xu, S. S., Mergoum, M., et al. (2014). Review of doubled haploid production in durum and common wheat through wheat × maize hybridization. Plant Breed. 133, 313–320. doi: 10.1111/pbr.12162
Niu, Z., Kloindworth, D. I., Friesen, T. I., Chao, S., Jin, Y., Cai, X., et al. (2011). Target introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi: 10.1534/genetics.110.123588
Njau, P. N., Bhavani, S., Huerta-Espino, J., Keller, B., Singh, R. P. (2013). Identification of QTL associated with durable adult plant resistance to stem rust race Ug99 in wheat cultivar ‘Pavon 76’. Euphytica 190, 33–44. doi: 10.1007/s10681-012-0763-4
Olivera, P., Newcomb, M., Szabo, L. J., Rouse, M., Johnson, J., Gale, S., et al. (2015). Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013-14. Phytopathology 105 (7),917–928.
Olivera, P. D., Sikharulidze, Z., Dumbadze, R., Szabo, L. J., Newcomb, M., Natsarishvili, K., et al. (2019). Presence of a Sexual Population of Puccinia graminis f. Sp. Tritici in Georgia Provides a Hotspot for Genotypic and Phenotypic Diversity. Phytopathology 109 (12), 2152–2160.
Olivera, P. D., Villegas, D., Cantero-Martínez, C., Szabo, L. J., Rouse, M. N., Luster, D. G., et al. (2022). A unique race of the wheat stem rust pathogen with virulence on Sr31 identified in Spain and reaction of wheat and durum cultivars to this race. Plant pathology 71 (4),873–889 17. doi: 10.1111/ppa.13530
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 80). 341, 786–788. doi: 10.1126/science.1239028
Peterson, R. F., Cambell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can. J. Res. Sect. C. 26, 496–500. doi: 10.1139/cjr48c-033
Poland, J. A., Brown, P. J., Sorrells, M. E., Jannink, J. L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PloS One 7, e32253. doi: 10.1371/journal.pone.0032253
Roelfs, A. P., Singh, R. P., Saari, E. E. (1992). Rust disease of wheat: Concepts and Methods of Disease Management (Mexico: CIMMYT).
Romay, M., Millard, M., Glaubitz, J., Peiffer, J., Swarts, K., Casstevens, T. M., et al. (2013). Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14, R55. doi: 10.1186/gb-2013-14-6-r55
Rouse, M. N., Nava, I. C., Chao, S., Anderson, J. A., Jin, Y. (2012). Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor. Appl. Genet. 125, 877–885. doi: 10.1007/s00122-012-1879-6
Rouse, M. N., Nirmala, J., Jin, Y., Chao, S., Fetch, T. G., Pretorius, Z. A., et al. (2014a). Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor. Appl. Genet. 127, 1681–1688. doi: 10.1007/s00122-014-2330-y
Rouse, M. N., Talbert, L. E., Singh, D., Sherman, J. D. (2014b). Complementary epistasis involving Sr12 explains adult plant resistance to stem rust in Thatcher wheat (Triticum aestivum L.). Theor. Appl. Genet. 127, 1549–1559.
Saintenac, N., Zhang, W., Salcedo, A., Rouse, M. N., Trick, H. N., Akhunov, E., et al. (2013). Identification of wheat gene Sr35 that Confers resistance to Ug99 stem rust race group. Science 341 (16), 783–786.
Savary, S., Willocquet, L., Pethybridge , S. J., Esker, P., McRobertsm, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Sears, E. R., Briggle, L. W. (1969). Mapping gene Pm1 for resistance to Erysiphe graminis f. sp. tritici on chromosome 7A of wheat. Crop Sci. 9, 96. doi: 10.2135/cropsci1969.0011183X000900010033x
Shewabez, E., Bekele, E., Alemu, A., Mugnai, L., Tadesse, W. (2022). Genetic characterization and genome-wide association mapping for stem rust resistance in spring bread wheat. BMC Genomic Data 23, 11. doi: 10.1186/s12863-022-01030-4
Singh, S., Bowden, R. L. (2010). Molecular mapping of adult-plant race-specific leaf rust resistance gene Lr12 in bread wheat. Mol. Breed 28, 137-142. doi: 10.1007/s11032-010-9467-4
Singh, R., Herrera-Foessel, S., Huerta-Espino, J., Singh, S., Bhavani, S., Lan, C., et al. (2014). Progress towards genetics and breeding for minor genes-based resistance to Ug99 and other rusts in CIMMYT high-yielding spring wheat. J. Integr. Agric. 13, 255–261. doi: 10.1016/S2095-3119(13)60649-8
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Singh, R. P., Huerta-Espino, J., Bhavani, S., Herrerra-Foessel, S., Singh, D., Singh, P., et al. (2011). Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179, 175–186. doi: 10.1007/s10681-010-0322-9
Spindel, J., Wright, M., Chen, C., Cobb, J., Gage, J., Harrington, S., et al. (2013). Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor. Appl. Genet. 126, 2699–2716. doi: 10.1007/s00122-013-2166-x
Stakman, E. C. (1955). Progress and problems in plant pathology. Ann. Appl. Biol. 42, 22–33. doi: 10.1111/j.1744-7348.1955.tb02407.x
Steuernagel, B., Periyannan, S. K., Hernández-Pinzón, I. H., Witek, K., Rouse, M. N., Yu, G., et al. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. doi: 10.1038/nbt.3543
The, T. T., McIntosh, R. A. (1975). Cytogenetical studies in wheat VIII. Telocentric mapping and linkage studies involving Sr22 and other genes in chromosome 7AL. Aust. J. Biol. Sci. 28, 531–538. doi: 10.1071/BI9750531
The International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Upadhyaya, N. M., Mago, R., Panwar, V., Hewitt , T., Luo, M., Chen , J., et al. (2006). JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Ed. Kyazma, B. V. (Wageningen, Netherlands).
Waldron, L. R. (1935). Stem rust epidemics and wheat breeding (North Dakota Agricultural College: Agricultural Experiment Station).
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wu, S., Pumphary, M., Bai, G. (2009). Molecular mapping of stem-rust resistance gene Sr40 in wheat. Crop Sci. 49, 1681–1686.
Yu, L. X., Lorenz., A., Rutkoski, J., Singh, R. P., Bhavani, S., HuertaEspino, J., et al. (2011). Association mapping and gene-gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor. Appl. Genet. 123, 1257–1268. doi: 10.1007/s00122-011-1664-y
Yu, G., Matny, O., Champouret, N., Steuernagel, B., Moscou., M. J., Hernández-Pinzón, I., et al. (2022). Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nat. Commun. 13, 1607. doi: 10.1038/s41467-022-29132-8
Yu, G., Matny, O., Gourdoupis, S., Nayapuram, N., Aljedaani, F. R., Wang, Y. L., et al. (2023). The wheat stem rust resistance gene Sr43 encodes an unusual protein kinase. Nat. Genet. 55, 921–926. doi: 10.1038/s41588-023-01402-1
Zhang, D., Bowden, R. L., Jin, Y., Carver, B. F., Bai, G. (2014). Association analysis of stem rust resistance in U.S. winter wheat. PloS One 9, e103747. doi: 10.1371/journal.pone.0103747
Zhang, W., Chena, S., Abatea, Z., Nirmala, J., Rouseb, M., Dubcovsky, J. (2017). Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. U.S.A. 114, E9483–E9492. doi: 10.1073/pnas.1706277114
Zhang, J., Hewitt, T. C., Boshoff, W. H. P., Dundas, I., Upadhyaya, N., Li, J., et al. (2021). A recombined Sr26 and Sr61 disease resistance gene stack in wheat encodes unrelated NLR genes. Nat. Commun. 12, 3378. doi: 10.1038/s41467-021-23738-0
Keywords: QTL, stem rust, wheat, disease resistance, 90K iSelect
Citation: Edae EA, Kosgey Z, Bajgain P, Ndung'u KC, Gemechu A, Bhavani S, Anderson JA and Rouse MN (2024) The genetics of Ug99 stem rust resistance in spring wheat variety ‘Linkert‘. Front. Plant Sci. 15:1343148. doi: 10.3389/fpls.2024.1343148
Received: 23 November 2023; Accepted: 12 February 2024;
Published: 07 March 2024.
Edited by:
Dilip R. Panthee, North Carolina State University, United StatesReviewed by:
Jingzhong Xie, Chinese Academy of Sciences (CAS), ChinaDragan Perovic, Julius Kühn-Institut, Germany
Copyright © 2024 Edae, Kosgey, Bajgain, Ndung'u, Gemechu, Bhavani, Anderson and Rouse. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erena A. Edae, ZWRhZXgwMDFAdW1uLmVkdQ==; Matthew N. Rouse, bWF0dGhldy5yb3VzZUB1c2RhLmdvdg==
 Erena A. Edae
Erena A. Edae Zennah Kosgey
Zennah Kosgey Prabin Bajgain
Prabin Bajgain Kimani C. Ndung'u
Kimani C. Ndung'u Ashenafi Gemechu
Ashenafi Gemechu Sridhar Bhavani
Sridhar Bhavani James A. Anderson
James A. Anderson Matthew N. Rouse1,6*
Matthew N. Rouse1,6*
