- 1Department of Crop and Soil Sciences, Washington State University, Pullman, WA, United States
- 2Institute of Biological Chemistry, Washington State University, Pullman, WA, United States
- 3Division of Plant Sciences and Interdisciplinary Plant Group, University of Missouri, Columbia, MO, United States
Lignin is a phenolic heteropolymer found in most terrestrial plants that contributes an essential role in plant growth, abiotic stress tolerance, and biotic stress resistance. Recent research in grass lignin biosynthesis has found differences compared to dicots such as Arabidopsis thaliana. For example, the prolific incorporation of hydroxycinnamic acids into grass secondary cell walls improve the structural integrity of vascular and structural elements via covalent crosslinking. Conversely, fundamental monolignol chemistry conserves the mechanisms of monolignol translocation and polymerization across the plant phylum. Emerging evidence suggests grass lignin compositions contribute to abiotic stress tolerance, and periods of biotic stress often alter cereal lignin compositions to hinder pathogenesis. This same recalcitrance also inhibits industrial valorization of plant biomass, making lignin alterations and reductions a prolific field of research. This review presents an update of grass lignin biosynthesis, translocation, and polymerization, highlights how lignified grass cell walls contribute to plant development and stress responses, and briefly addresses genetic engineering strategies that may benefit industrial applications.
1 Introduction
Given the global dietary and economic importance of cereal grains as well as the UN projected population increase to 9.7 billion people by 2050 (UN, 2022), research is underway to increase cereal yields while maintaining ecosystem services (Rockstrom et al., 2020). Likewise, increasing populations increase energy demands, which drives ongoing research to develop environmentally and economically sustainable biofuels and biochemicals (Chu and Majumdar, 2012). Research into improving cereal yields and sustainable biofuels may converge at the composition of grass cell walls. Research in cereals seeks to improve stress tolerance and sustain yields, which in some cases improves grass cell wall integrity, while much biofuel research seeks to minimize grass cell wall recalcitrance to maximize release of sugars from feedstocks (i.e., saccharification). Both goals hinge on either the improvement or reduction of the second most prolific cell wall polymer on earth: lignin.
Lignin, a primary component of feedstock recalcitrance to saccharification (Zeng et al., 2014), also provides critical structural reinforcement and hydrophobicity to plant tracheary elements (Fan et al., 2006; Chmielewska et al., 2016), rigidity to structural fibers to resist gravity (Li et al., 2022a), and resistance to enzymatic and mechanical pathogen invasion (Nicholson and Hammerschmidt, 1992; Bellincampi et al., 2014; Lee et al., 2019). Grass lignin is a complex hydrophobic hetero-phenolic polymer primarily composed of three phenolic alcohols: p-coumaryl alcohol (H), coniferyl alcohol (G), and sinapyl alcohol (S); two hydroxycinnamic acids: p-coumaric acid (pCA) and ferulic acid (FA); four hydroxycinnamate-phenolic conjugates: γ-p-coumaroylated coniferyl alcohol (pCA-G) and γ-p-coumaroylated sinapyl alcohol (pCA-S), feruloylated coniferyl alcohol (G-FA), and feruloyl sinapyl alcohol (S-FA); and thus far one known flavone: tricin, and many additional conjugated derivatives found in lower concentrations (Ralph, 2010; del Rio et al., 2012; Lan et al., 2015; Karlen et al., 2016; del Río et al., 2022). Grass lignin monomers are synthesized in the phenylpropanoid and flavone pathways, pass through the cell membrane, and are polymerized in secondary cell walls (Ralph et al., 2019; Tobimatsu and Schuetz, 2019). The purpose of this review is threefold: to present the current models of grass lignin synthesis, translocation, and polymerization; to highlight how lignin contributes to grass stress resistance; and to suggest future avenues of research that may benefit both cereal breeders and valorization engineers.
2 Grass lignin biosynthesis
2.1 Phenylpropanoid and flavone enzymes
2.1.1 Substrate promiscuity and subfunctionalization in grass monolignol biosynthesis
Lignin monomers are synthesized in the phenylpropanoid pathway, general models of which have been extensively reviewed (Vanholme et al., 2019; Barros et al., 2019a). However, grass phenylpropanoid biosynthesis research is revealing unique features that diverge from the dicot model, thus leading to a monolignol synthesis model for grasses (Figure 1). For instance, the dicot model suggests phenylalanine ammonia lyase (PAL), which deaminates shikimate-derived phenylalanine into cinnamic acid, to be the singular entry point and rate-limiting step in phenylpropanoid biosynthesis (Barros and Dixon, 2020; Yao et al., 2021). In Brachypodium distachyon, Sorghum bicolor, Triticum aestivum, Bambusa oldhamii, and Zea mays, a bifunctional PAL homolog named phenylalanine-tyrosine ammonia-lyase (PTAL) deaminates both phenylalanine and tyrosine in parallel with PAL (Rosler et al., 1997; Barros et al., 2016; Godin et al., 2016; Jun et al., 2018; Feduraev et al., 2020; Hsieh et al., 2021; Simpson et al., 2021). In addition, PTAL preferentially deaminates tyrosine into pCA, effectively bypassing 4-hydroxylase (C4H) conversion of PAL-derived cinnamic acid to pCA and generating almost half of the total lignin detected (Barros et al., 2016). pCA pools can be converted to p-coumaroyl-CoA via 4-hydroxycinnamate:CoA ligase (4CL) or caffeic acid (CAF) via coumarate 3-hydroxylase (C3H), which has recently been observed in B. distachyon, A. thaliana, and S. bicolor to be an ascorbate peroxidase (Barros et al., 2019a; Zhang et al., 2023b).
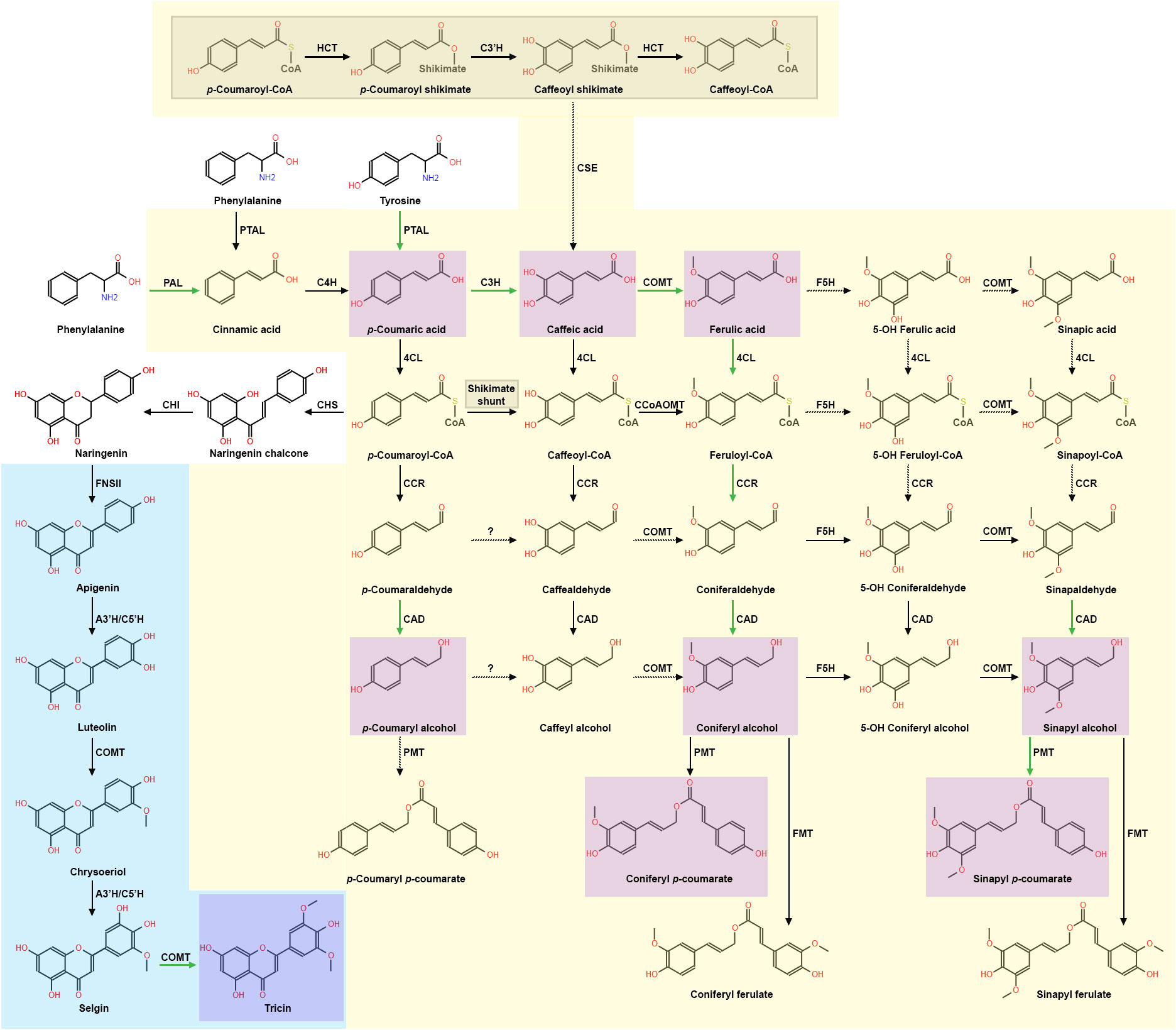
Figure 1 The current model of grass lignin biosynthesis, which includes aspects of the phenylpropanoid pathway and the flavone pathway. Grass preferential carbon flux is shown with green arrows, while dashed arrows represent unverified steps in the pathways. The current grass phenylpropanoid synthesis model is presented with a yellow background. The “shikimate shunt” is expanded on in the grey shaded region and is part of the phenylpropanoid model. The current monocot flavone synthesis model leading to formation of tricin is presented with a blue background. Monocot monolignols and conjugates found in significant concentrations are highlighted in purple. Phenylpropanoid enzymes: PAL, phenylalanine ammonia lyase; PTAL, phenylalanine/tyrosine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxycinnamate:CoA ligase; C3H, 4-coumarate 3-hydroxylase/ascorbate peroxidase; CSE, caffeoyl shikimate esterase; HCT, hydroxycinnamoyl CoA: shikimate/quinate hydroxycinnamoyl transferase; C3´H, 4-coumaroyl shikimate 3´-hydroxylase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; CCR, cinnamoyl CoA reductase; F5H, ferulate/coniferaldehyde 5-hydroxylase; COMT, caffeate/5-hydroxyferulate 3-O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; PMT, p-coumaroyl-CoA: monolignol transferase; FMT, feruloyl-CoA: monolignol transferase. Flavonoid and flavone-related enzymes: CHS, chalcone synthase; CHI, chalcone isomerase; FNSII, flavone synthase II; A3’H/C5’H, apigenin 3′-hydroxylase/chrysoeriol 5′-hydroxylase.
Interestingly, recent evidence suggests grasses may preferentially shunt phenylpropanoid metabolism to hydroxycinnamic acid synthesis (Figure 1). Although grasses use both 4CL and C3H metabolic pathways, high concentrations of PTAL, C3H, and COMT were found in the same fraction derived from B. distachyon stem tissue, which led the authors to postulate that PTAL, C3H, and COMT may form a protein complex that contributes to the observed preferential metabolic flux into hydroxycinnamic acid synthesis (Barros et al., 2019a). 13C flux analysis from the same study suggests B. distachyon caffeate 3-O-methyltransferase (COMT) preferentially methylates CAF, generating significantly more FA compared to the A. thaliana COMT ortholog. Furthermore, analysis of metabolic profiles of mutants in the shikimate intermediate pathway enzymes, hydroxycinnamoyl CoA: shikimate/quinate hydroxycinnamoyl transferase (HCT) and 4-coumaroyl shikimate 3′-hydroxylase (C3’H), resulted in reduced lignin content, but wildtype growth (Barros et al., 2022). In contrast, disruption of A. thaliana hct and c3’h resulted in significantly reduced total lignin and severe dwarfism (Franke et al., 2002a, b; Hoffmann et al., 2004). These data and the previous observation of preferential phenylpropanoid flux into hydroxycinnamic acid metabolism in B. distachyon (Barros et al., 2019a) led to the hypothesis that the phenylpropanoid shikimate intermediate pathway is not essential for B. distachyon development (Barros et al., 2022). In support of this hypothesis, caffeoyl shikimate esterase (CSE) activity is crucial for A. thaliana, Medicago truncatula, and hybrid poplar (Populus tremula × Populus alba) development (Vanholme et al., 2013; Saleme et al., 2017). However, phylogenetic analyses have not identified clear CSE homologs in B. distachyon, O. sativa, Z. mays, Setaria italica, S. bicolor, Oryza minuta, Aegilops tauschii, or Hordeum vulgare (Ha et al., 2016; Serrani-Yarce et al., 2021). A surprising exception is Panicum virgatum, in which both phylogenetic evidence and enzymatic activity of CSE has been detected, leaving the metabolic purpose of CSE in monocots unclear (Ha et al., 2016; Serrani-Yarce et al., 2021). Continued investigation of the functional significance of the shikimate pathway in more grass species will further test the hypothesis of the shikimate pathway being an alternative phenylpropanoid flux route that may provide protective metabolic redundancy (Barros et al., 2022).
Following the action of 4CL, CoA groups are removed from the γ carbon by cinnamoyl-CoA reductase (CCR), resulting in production of cinnamaldehydes. Upstream phenylpropanoid-CoA concentrations likely drive preferential flux through feruloyl-CoA metabolism (Barros et al., 2022) (Figure 1). Multiple CCR isoforms showing differing substrate specificities are found co-expressed in P. virgatum (Escamilla-Trevino et al., 2010), O. sativa (Park et al., 2017), and S. bicolor (Sattler et al., 2017), thus likely providing an additional measure of protective metabolic redundancy. This protective metabolic redundancy was inadvertently demonstrated via single gene disruption of feruloyl-CoA-specific ZmCCR1 in Z. mays (Smith et al., 2017a). Despite the Zmccr1 mutant showing a reported 40% decrease in H monomers, 56% decrease in G monomers, and a 26% decrease in S monomers with WT (1.7:40:59) versus mutant (1.6:27:71) H:G:S ratios, the Zmccr1 mutant showed no significant growth defects. These data support that feruloyl-CoA metabolism is preferential in grasses due to upstream phenylpropanoid pools, and that co-expressed CCR isoforms can compensate for CCR homolog disruption.
Downstream in the phenylpropanoid biosynthesis pathway, S monomers are sequentially generated by the phenylpropanoid enzyme ferulate 5-hydroxylase F5H [also known as coniferaldehyde-5-hydroxylase (CAld5H)] and COMT (Takeda et al., 2017, 2019; Wu et al., 2019). In grasses, F5H primarily converts coniferaldehyde to 5-hydroxyconiferaldehyde, and was observed to be the rate-limiting step of S monomer synthesis in P. virgatum (Wu et al., 2019). However, a f5h null mutant generated in a O. sativa RNAi-comt background unexpectedly produced significant amounts of pCA-S monomers, suggesting that an unidentified enzyme or uncharacterized F5H isoform may generate a separate and discrete pool of 5-hydroxyconiferaldehyde (Takeda et al., 2019). These data led to the hypothesis that a grass-specific lignin pathway may exist to produce pCA-S monomers independently of the current phenylpropanoid biosynthesis model (Takeda et al., 2019; Barros and Dixon, 2020; Shafiei et al., 2023). Lack of F5H-related functional studies in grasses raises new and exciting questions, such as: is the observed separate pool of pCA-S monomers unique to O. sativa or common in all grasses; and is there a biological function of an alternate S monomer biosynthetic pathway?
The final step of the lignin biosynthetic pathway is performed by cinnamyl alcohol dehydrogenase (CAD), which reduces cinnamaldehydes into cinnamyl alcohols: p-coumaraldehyde is reduced to H monomers, coniferaldehyde is reduced to G monomers, and sinapaldehyde is reduced to S monomers (Vanholme et al., 2010; del Río et al., 2022). Predominant CAD isoforms in C4 grasses S. bicolor (Jun et al., 2017) and P. virgatum (Saathoff et al., 2011b, 2012) have reportedly greater substrate specificity for sinapaldehyde; whereas, primary CAD isoforms in C3 grasses T. aestivum (Ma, 2010) and O. sativa (Park et al., 2018) have reportedly greater substrate specificity for coniferaldehyde. Genetic mutations of critical phenylpropanoid genes such as CAD in C4 grasses are easily identifiable due to a conspicuous brown midrib phenotype, observed in S. bicolor (Saballos et al., 2009; Sattler et al., 2009; Li and Huang, 2017), Z. mays (Halpin et al., 1998; Chen et al., 2012b; Liu et al., 2021b), and P. virgatum (Saathoff et al., 2011a). To date, characterized cad mutants show similar phenotypes such as decreased total lignin and increased saccharification efficiency (Zeng et al., 2014; Carmona et al., 2015; Zhong et al., 2021; del Río et al., 2022). However, cad1 mutations in C3 grasses such as B. distachyon, O. sativa, H. vulgare, and T. aestivum do not present a brown midrib, but display increased reddish coloration in various tissues such as the nodes, spike, rachilla, and lemma (d’Yvoire et al., 2013). It is still unclear why the brown midrib phenotype appears in C4 grasses, but not in C3 grasses (Sattler et al., 2010; Eloy et al., 2017). A recent phenolic analysis of zmcad2 in maize showed a significant increase in integration of cinnamaldehydes into lignin and of soluble compounds conjugated with FA, SA, vanillic acid, and hydroxybenzoyl hexose in the mutant (Liu et al., 2021b). These data led to the hypothesis of an uncharacterized metabolic sink that may act as an upper limit on production of FA monomer synthesis, which in turn may moderate diFA-mediated cell wall crosslinking.
2.1.2 Hydroxycinnamic acids and their conjugates
In grasses, both xylan and lignin polymers may harbor substitutions with hydroxycinnamic acids such as ferulic acid (FA) or p-coumaric acid (pCA). Hydroxycinnamic groups are either esterified or etherified to lignin (Hatfield et al., 2017). Feruloylation of xylan facilitates covalent cross linkages of xylan chains or xylan–lignin via diferulate (diFA) bridge formation (Hatfield et al., 2017; Terrett and Dupree, 2019; Feijao et al., 2022). In a recent study, biochemical and histochemical evidence from B. distachyon culms suggest that structural variation in xylan molecules may determine interactions with both lignin and other cell wall components to confer functional specialization (Tryfona et al., 2023).
Recent metabolomic and genetic evidence suggests phenylpropanoid synthesis is preferentially shunted towards hydroxycinnamic acid synthesis in grasses (Ralph, 2010; Karlen et al., 2018; Barros et al., 2022). Thus, it is not surprising that greater amounts of pCA and FA are found in grass cell walls compared to dicots (Figure 1). pCA is a precursor metabolite for most phenylpropanoids and flavonoids (Vogt, 2010). pCA is also abundantly conjugated to S and G monomers via p-coumaroyl-CoA: monolignol acyltransferases (PMT), which are part of the larger “BAHD” acyltransferase protein family (Grabber et al., 1996; Withers et al., 2012; Marita et al., 2014; Petrik et al., 2014; Chandrakanth et al., 2023). The function of pCA-monolignol conjugates is currently unknown (Hatfield et al., 2017), but two hypothesis exist: 1) pCA acts to transfer radicals during S monomer polymerization (Ralph et al., 2004a), and 2) pCA-S monomers terminate polymerization, leading to a more linearized lignin polymer and subsequently change lignin polymer properties (Kishimoto et al., 2005; Hatfield et al., 2017). In addition to conjugating to S monomers, pCA is also acylated to arabinoxylans (AXs) in grass cell walls via BAHD acyltransferases in O. sativa (Bartley et al., 2013), S. viridis (Mota et al., 2021), and P. virgatum (Li et al., 2018a).
FA is also found in grass cell walls at high concentrations relative to dicots (Ralph, 2010). FA is predominantly found esterified to AX sidechains in grass cell walls, but has also been found ether-linked to monolignols in low concentrations (de Oliveira et al., 2015). Recent genetic and metabolomic evidence suggest BAHD acyltransferases may significantly contribute to grass AX feruloylation (de Souza et al., 2018, 2019). Observed AX feruloylation during early stages of secondary cell wall synthesis has led to the hypothesis that FA monomers ester-bound to AX act as nucleation sites for lignin polymerization (Ralph et al., 1995; Grabber et al., 2004; Ralph, 2010; Zhang et al., 2019). DiFA is formed via oxidative radical coupling of FA monomers, facilitated by both laccases and peroxidases (Ralph et al., 1994; Quideau and Ralph, 1997; Carunchio et al., 2001; Ward et al., 2001). DiFAs often link local AX polymers together, resulting in the lignocellulose crosslinking that is characteristic of grass cell walls (Ralph et al., 1995; Grabber et al., 2000; Ralph, 2010; Eugene et al., 2020; del Río et al., 2022). DiFA-mediated cell wall crosslinking has been strongly correlated with greater structural integrity of the cell wall (Jung and Phillips, 2010; Hatfield et al., 2018) and may contribute to both fungal pathogen (Piotrowski et al., 2015) and antinutritional herbivory defense (Barros-Rios et al., 2015). Lastly, recent findings suggest BAHD enzymes with feruloyl-CoA monolignol transferase (FMT) activity may also contribute to FA-monolignol conjugation (Wilkerson et al., 2014; Karlen et al., 2016; Smith et al., 2022).
2.1.3 Promiscuous flavone grass enzymes synthesize the lignin monomer tricin
Tricin was recently determined to be the first non-phenylpropanoid lignin monomer found in both commelinid and non-commelinid monocots (del Rio et al., 2012; Lan et al., 2015; Lan et al., 2016a, b). Tricin has also been detected in dicot legumes Medicago sativa and M. truncatula; synthesis of which is currently hypothesized to result from convergent evolution (Lui et al., 2020). Tricin is found as an external, pendent molecule, coupled via a β-O-4 bond with S and G monolignols. Biomimetic oxidation assays and reaction efficiency assays of acidolysis, thioacidolysis, and derivation followed by reductive cleavage (DFRC) have led to the hypothesis that tricin may serve as a nucleation site for grass lignin polymerization (Lu and Ralph, 1997; Lan et al., 2015; Lan et al., 2016b). Tricin is derived from the phenylpropanoid intermediate pCA-CoA, generated by 4CL during early in the monolignol biosynthesis pathway (Jiang et al., 2016) (Figure 1). pCA-CoA is shunted out of phenylpropanoid metabolism and committed to flavonoid biosynthesis via chalcone synthase (CHS) (Eloy et al., 2017; Lam et al., 2022). Naringenin chalcone is then isomerized to naringenin via chalcone isomerase (CHI) (Tetreault et al., 2021), and flavone synthase (FNS) catalyzes conversion of naringenin to flavone biosynthesis.
Two types of FNS have been characterized: FNS type II (FNSII), which appears to be present in all plants; and FNS type I (FNSI), which has thus far only been found co-expressed with FNSII in O. sativa (Lee et al., 2008; Lam et al., 2014), Z. mays (Righini et al., 2019), and in most of the Apiaceae family (Gebhardt et al., 2005, 2007). Metabolic and histochemical analysis of an osfnsII knockout and osfnsI T-DNA lines suggest that FNSII is the primary FNS involved in grass flavone biosynthesis (Lam et al., 2014, 2017). Tricin synthesis continues with apigenin 3’-hydroxylase/chrysoeriol 5’-hydroxylase (A3’H/C5’H) hydroxylation of the C3 carbon on apigenin, forming luteolin (Lam et al., 2015). Surprisingly, subsequent methylation of the 3’ alcohol of luteolin has been reported to be catalyzed by the phenylpropanoid enzyme COMT in Z. mays (Fornale et al., 2017), S. bicolor (Eudes et al., 2017) and O. sativa (Lam et al., 2019), generating chrysoeriol. Chrysoeriol is then hydroxylated at the C5 carbon by A3’H/C5’H to generate selgin, which is again methylated by COMT, generating tricin (Eudes et al., 2017; Fornale et al., 2017; Lam et al., 2019). Because tricin appears to be present in all tested monocots to date, future grass lignin characterizations should include flavone analysis to investigate how tricin fits into the grass lignin model, such as how its absence may affect saccharification, and how it contributes to overall grass fitness.
2.2 Grass monolignol translocation and polymerization
2.2.1 Monolignol translocation models are likely conserved among gymnosperms, monocots, and dicots
Once synthesized, grass lignin monomers and conjugates are transported into the apoplast, likely via three mechanisms. Lignin monomer transport models such as passive diffusion (Vermaas et al., 2019b), active transport via ATP-binding cassette transporters (Miao and Liu, 2010; Alejandro et al., 2012), and vesicle storage and transport (Le Roy et al., 2016) have all been proposed to account for the highly variable properties of monolignols and monolignol conjugates and their transport across the cell membrane (del Río et al., 2022) (Figure 2). Although most monolignol transport research has been performed in A. thaliana, chemical properties of monolignols and the proposed biological roles of each transport mechanism suggest that all three models likely function in grasses (Dixon and Barros, 2019).
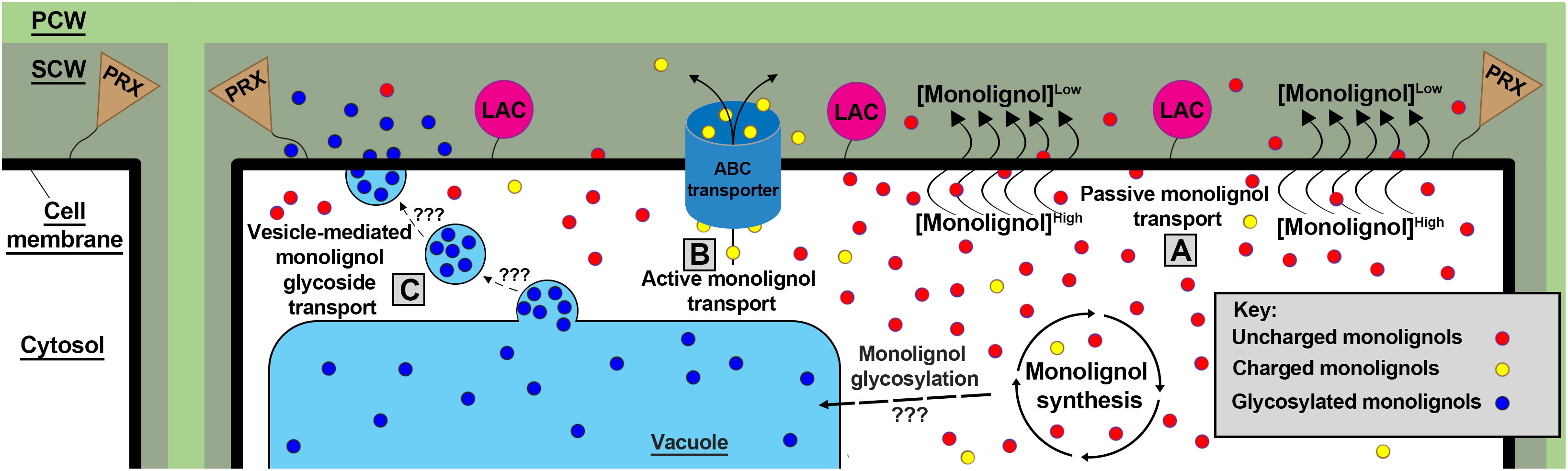
Figure 2 Three proposed models of monolignol transport. (A) Small uncharged monolignols (red) are theorized to passively diffuse through the plasma membrane over a concentration gradient driven by LAC and PRX monolignol polymerization. (B) Charged/conjugated (yellow) monolignols likely require active transport via ABC transporters to translocate across the plasma membrane lipid bilayer. (C) monolignols are glycosylated (blue) by an unknown mechanism and location in grasses. These glycosylated monolignols have been found stored in the vacuole and transported via an unknown mechanism to the plasma membrane via vesicle bodies. Once in the apoplast, membrane-bound LACs and PRXs polymerize monolignols via oxidative radical coupling. PCW, Primary Cell Wall; SCW, Secondary Cell Wall; LACs, laccases; PRXs, peroxidases.
Passive diffusion of monolignols across the plasma membrane is supported by both modeling and experiments. Simulations found that hydrophobic “lignin-related compounds” are capable of diffusing through a homogeneous eukaryotic membrane, Z. mays root cell membranes, and gram-negative bacterial membranes (Vermaas et al., 2019b). Previous modeling corroborates these findings by observing “lignin-like” molecules transgressing a Z. mays cell lipid bilayer (Boija et al., 2007). Furthermore, a recent study performed in A. thaliana observed monolignols accumulating intracellularly when extracellular lignin polymerization was inhibited, suggesting that lignin polymerization generates a concentration gradient that drives passive monolignol diffusion into the apoplast (Perkins et al., 2022). The passive diffusion model is biomechanically simple, and is likely the dominant mechanism of monolignol translocation into the apoplast.
However, modeling experiments suggest some monolignols and glycosylated monolignol conjugates are physically incapable of passively diffusing through lipid bilayers (Vermaas et al., 2019b), and thus require active transport mechanisms to transit into the apoplast. A pair of studies found A. thaliana ATP binding cassette (ABC) transporter genes AtABCG11, AtABCG22, AtABCG29, and ABCG36 were differentially co-expressed with a phenylpropanoid synthesis transcription factor AtMYB58 during cell wall development in stems and roots (Takeuchi et al., 2018a, b). These findings are consistent with an earlier study of A. thaliana AtABCG29, which when expressed in yeast, was observed to actively transport H monomers across yeast cell membranes (Alejandro et al., 2012). The Atabcg29 lines reportedly lack a significant lignin-deficient phenotype but show a decreased root growth phenotype and slight decreases in H, G, S monomers, flavonols, and glucosinolates. The lack of a significant lignin-deficient phenotype in singular lignin transporter mutations and concomitant considerable presence of lignin and glucosinolates in Atabcg29 cell walls suggest that multiple transporters may be involved in actively translocating hydrophilic monolignols and monolignol conjugates, and that gradient-driven hydrophobic monolignol passive diffusion works in parallel with active transport.
Vesicle-mediated monolignol, flavonoid, and other aromatic molecule storage and subsequent deployment may be facilitated glucoside conjugation (Zhao, 2015; Perkins et al., 2019). Monolignol glucosides are synthesized by UDP-glycosyltransferases, and are a putative storage configuration of monolignols (Dima et al., 2015). Glycosylated monolignols are found in vacuoles of A. thaliana leaf cells (Dima et al., 2015), and are transported into the apoplast during cell lignification in a gymnosperm (Vaisanen et al., 2020), dicots (Morikawa et al., 2010; Tsuyama et al., 2013, 2019), and recently in the monocot Phyllostachys pubescens (Shimada et al., 2021). More research is needed to determine if cereals deploy sequestered monolignol glucosides after injury or pathogen detection; a mechanism not yet supported by genetic evidence (Jeon et al., 2022). Furthermore, the magnitude by which grass monolignols and glycosylated monolignol conjugates are actively transported/translocated to the apoplast through passive diffusion, active transport, or vesicle-mediated exocytosis remains uncertain and requires further investigation.
2.2.2 Grass monolignol polymerization is driven by oxidative radical coupling
Once in the apoplast, mobile monolignols undergo polymerization via laccases (LACs) and peroxidases (PRXs) (Zhao et al., 2013; Schuetz et al., 2014; Francoz et al., 2015; Moural et al., 2017; Tobimatsu and Schuetz, 2019). LACs and PRXs facilitate a free radical oxidative coupling reaction, responsible for the combination of lignin monomers and oligomers with greater-order lignin polymer fragments (Ralph et al., 2004b). Radical coupling is capable of generating certain bond configurations of lignin polymers due to the preferential stability of free radical monolignol resonance structures (Ralph, 2010; Ralph et al., 2019). Studies of lignin polymer bonding configurations analyzed in gymnosperms, monocots, and dicots have found the most stable monolignol resonance structures facilitate a high probability of β-O-4, β-β, and β-5 bond formation (Ralph et al., 2004b; Sangha et al., 2012; Kim et al., 2023).
Lignin polymer compositions and purported stereospecificity have been proposed to be templated by dirigent proteins (Davin et al., 1997; Burlat et al., 2001; Davin and Lewis, 2005). However, the role dirigent proteins contribute to lignin polymerization remains somewhat controversial (Hatfield and Vermerris, 2001; Ralph et al., 2008; Paniagua et al., 2017), as lignin polymers are optically inactive (Ralph et al., 1999) and genetic evidence of dirigent proteins orchestrating an optically active lignin polymer is lacking (Ralph et al., 2008). There is evidence that a family of dirigent proteins, such as Enhanced Suberin 1 (ESB1) and dirigent proteins DIR9, DIR16, DIR18, DIR24, and DIR25, which coordinate lignin polymerization during Casparian strip biogenesis (Hosmani et al., 2013; Wang et al., 2019; Wang et al., 2022c; Gao et al., 2023). These findings provide insights into potential connections between lignan and lignin biosynthesis, and support functions of dirigent proteins in cell wall development and in response to stress (Paniagua et al., 2017).
2.2.3 Grass lignin polymerization is tentatively modeled after A. thaliana LAC and PRX functional analysis
The mechanisms that govern lignin polymerization in grasses are largely uncharacterized. However, lignin polymerization studies in A. thaliana suggest that the mechanisms that control A. thaliana lignin deposition may be conserved in grasses. For example, “the good neighbor hypothesis” describes xylem cell lignification as non-cell autonomous (Smith et al., 2013; Smith et al., 2017b). Although the good neighbor hypothesis has thus far only been tested in A. thaliana, apoptotic grass xylem cells cannot autonomously lignify, suggesting that grass xylem cell lignification is also non-cell autonomous.
Apoplastic LACs utilize copper to generate reactive oxygen species (ROS) from molecular oxygen. These ROS are subsequently employed in the combinatorial coupling of monolignols and oligomers with greater-order lignin polymer fragments (Ralph et al., 2004b). LACs are currently thought to significantly contribute to vascular bundle lignification in A. thaliana because a lac4lac11lac17 mutant displayed poor vascular lignification, significantly narrower root diameter, indehiscent anthers, and a severe negative growth phenotype (Zhao et al., 2013). The same study hypothesized that PRX activity is not redundant to LAC activity, as PRX activity was unable to compensate for the triple lac mutant. In a later study, fluorescently labeled AtLAC4 was found immobile once excreted throughout the secondary cell walls of xylem vessels, xylem fibers, and interfascicular fibers, supporting the hypothesis that A. thaliana LAC secretion and activity broadly governs lignification in vascular bundles and interfascicular fibers (Chou et al., 2018). A more recent study of quadruple and quintuple A. thaliana lac mutants found five A. thaliana LAC paralogs to be nonredundant, to lignify specific tracheary and structural cell types, and show different affinities for monolignols (Blaschek et al., 2022).
PRXs generate ROS from H2O2 that also facilitate lignin combinatorial coupling (Ralph et al., 2004b). Observations of LAC and PRX co-expression in lignifying tissues has generated debate over how apoplastic PRXs contribute a non-redundant role to lignin polymerization (Barros et al., 2015). The first substantial clue of PRX nonredundant function was found after Casparian strip (CS) development in A. thaliana was severely delayed due to induced mutation of endodermis-specific prx64 (Lee et al., 2013). Supporting this finding, the A. thaliana lac4lac11lac17 triple mutant (Zhao et al., 2013) was observed to have a lignified and functional CS, suggesting that either a CS-specific LAC homolog was still active, or that LACs do not significantly contribute to CS lignification. Later, fluorescently labeled AtPRX64 was observed to localize in the middle lamella and cell corners of interfascicular fibers, whereas AtLAC4 did not (Chou et al., 2018), suggesting that PRX significantly contributes to cell corner lignification. Recently, PRXs were found to be required and nonredundant for CS lignification after a quintuple A. thaliana prx mutant showed no detectable lignin in the CS, whereas a nonuple lac mutant showed no changes in CS integrity or lignification (Rojas-Murcia et al., 2020). To our knowledge, simultaneous disruption of LAC and PRX genes in grasses has not been studied.
The increasing availability of high-quality genomic databases combined with foundational models established in A. thaliana suggest a promising future to examine grass LAC and PRX function. For instance, LAC genes have been characterized in T. aestivum (Zhong et al., 2023), O. sativa (Liu et al., 2017), and S. bicolor (Wang et al., 2017), whereas PRXs have been identified in B. distachyon (Zhu et al., 2019), P. virgatum (Moural et al., 2017), and Setaria viridis (Simoes et al., 2023). Similar to A. thaliana, both BdLAC5 and BdLAC6 (orthologs to AtLAC17 and AtLAC4, respectively) were observed to localize in the apoplast near tracheary element secondary cell walls (Wang et al., 2015b). In addition, the B. distachyon LAC double mutant (lac5lac8) showed a 20-30% decrease in interfascicular fiber lignin, but only a mild growth defect (Le Bris et al., 2019).
Although these data in grasses are somewhat analogous to LAC studies in A. thaliana, the high concentrations of covalently bound hydroxycinnamic acids and hydroxycinnamic acid conjugates found in grass secondary cell walls may present an additional layer of complexity to the mechanisms that facilitate grass lignin polymerization (Hatfield and Marita, 2010). Recent genetic and metabolomic evidence generated in S. viridis, B. distachyon, and Saccharum spp. suggest that BAHD acyltransferases may significantly contribute to feruloylation of AX (de Souza et al., 2018, 2019). How grass LACs interact with FA is unclear, as the Bdlac5 mutant in B. distachyon displayed a 40% increase in esterified (cell wall-bound) FA and a 15% decrease in ether-bound (protein or lignin bound) FA (Wang et al., 2015b). Likewise, the role of grass PRXs in relation to hydroxycinnamic acid polymerization is largely unknown. A seminal study with Z. mays cell cultures observed that 60% of FA dimerized with dilute H2O2 addition to the cell suspension, suggesting that expression of PRX during early secondary cell wall development may also mediate FA acylation to AX (Grabber et al., 2000; Ralph et al., 2004a). These data, and lack thereof, emphasize there is still much to learn about the specific molecular mechanisms governing grass lignin polymerization, and that not all grass LAC and PRX functions can be simply extrapolated from A. thaliana studies.
3 Biological roles of lignin in grasses
3.1 The role of lignin in stem strength, drought, and heavy metal stress tolerance
3.1.1 diFA-mediated cell wall crosslinking significantly contributes to grass stem mechanical strength
Lignified grass tissue has been reported to facilitate proper development and improve abiotic stress resistance (Table 1). Lignin compositions are variable between and amongst species, tissue types, and even cell wall layers (Vanholme et al., 2010; Zeng et al., 2014; Menard et al., 2022). These lignin compositions are often quantified as a simple ratio measurement of total S monomers to total G monomers, but many past analyses of grass lignocellulose compositions lack quantification of cell wall-bound and ether-linked hydroxycinnamic acids. Future inclusion of this data is significant to grass lignocellulose analysis because recent studies suggest decreased S/G ratios combined with varying cell wall-bound hydroxycinnamic acid content appear to enhance mechanical strength of the load-bearing interfascicular and xylary fibers, tracheary elements, and sclerenchyma cells (Le Bris et al., 2019; Li et al., 2022a).
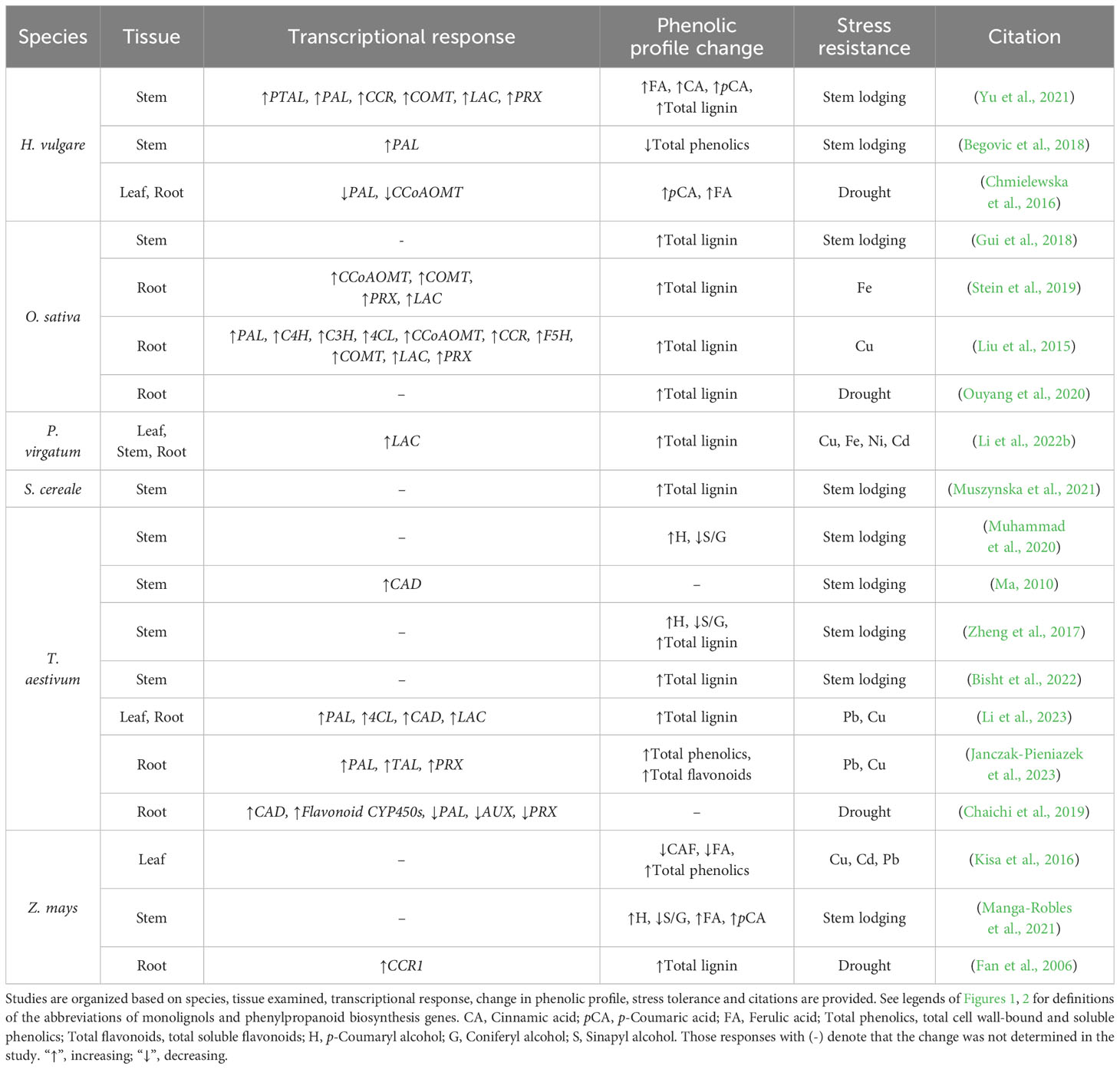
Table 1 A summary of stem lodging, heavy metal, and drought stress response studies that document changes in lignin-related gene expression and lignin content in grasses.
Increased diFA-mediated crosslinking characteristic of monocots significantly contributes to resistance to mechanical stressors (Ralph, 2010; Ralph et al., 2019; Eugene et al., 2020). Consistent with this, decreased S/G ratios and increased diFA content are positively correlated with culm stability and overall plant height (Cesarino, 2019; Xue et al., 2020; Manga-Robles et al., 2021; Li et al., 2022a) (Table 1). Biochemical and phenotypic analysis of lodging resistant Z. mays inbred lines were found to have decreased S/G ratios and significantly increased cell wall FA, diFA, pCA, and AX (Manga-Robles et al., 2021). Similarly, lodging-resistant T. aestivum lines revealed that a 16.9% reduction in S/G ratio, increased size and number of large vascular bundles, and reduced number of small vascular bundles was associated with increased culm strength (Muhammad et al., 2020). Lodging-resistant H. vulgare lines have significantly decreased culm S/G ratio and a significant increase in cell wall-bound FA and pCA compared to lodging susceptible lines (Yu et al., 2021). Finally, a recent analysis of a RNAi knockdown of f5h in H. vulgare found a significant decrease in S/G ratios, cell wall-bound pCA, and cell wall-bound FA, but no change in total lignin content or stem mechanical strength (Shafiei et al., 2023). Taken together, these findings offer evidence to suggest reduced S/G ratios and increased diFA-mediated crosslinking contribute to grass culm mechanical strength and flexibility, and that significant targeted reductions in S monomer content do not significantly compromise grass stem structural integrity.
Although there appears to be a clear correlation between grass cell wall crosslinking and overall stem mechanical strength, the extent to which total lignin deposition contributes to stem strength is still unclear. For instance, studies in O. sativa (Gui et al., 2018), H. vulgare (Begovic et al., 2018), and T. aestivum (Bisht et al., 2022) found total culm lignin content to be positively correlated with increased culm strength. Conversely, no correlation between total stem lignin and stem strength were found in population-scale studies of S. italica (Tian et al., 2015), Avena sativa (Argenta et al., 2022), Z. mays (Manga-Robles et al., 2021), and T. aestivum (Muhammad et al., 2020). These inconsistencies in data may be resolved in future grass lignocellulose analysis by prioritizing more comprehensive cell wall phenolic analyses that include quantification of S/G ratio with cell wall-bound FA, pCA, and tricin. Inclusion of these data with parallel qualitative histochemical assays of structurally significant anatomical features will provide a more holistic analysis of how degrees of cell wall crosslinking, rather than discrete monolignol content, contributes to grass stem strength.
3.1.2 Can drought-induced abscisic acid signaling enhance grass root xylem lignification and improve drought stress resistance?
Drought stress is managed by plants via both avoidance and tolerance strategies. Drought avoidance employs one or more physiologic changes in metabolism or anatomy to mitigate water loss or improve water uptake, and is initiated by drought sensing in roots via a complex series of ROS/Ca2+-mediated signaling pathways that induce changes in abscisic acid (ABA) and auxin production and transport (Zhu, 2016; Kalra et al., 2023). Some of these signals elicit subterranean drought avoidance growth responses such as increased root length, increased root penetration angle, and increased lateral root length—all necessary for plants to access soil moisture at depth (Vadez, 2014; Chaichi et al., 2019; Lamb et al., 2022). On the other hand, less plastic drought tolerance strategies involve innate phenotypes, such as photosynthetic anatomy and leaf shape that contribute to enduring prolonged drought stress. Lignification of secondary cell walls in tracheary elements can be classified as a drought tolerance phenotype, as xylary lignification imbues tracheary elements with hydrophobicity while providing structural reinforcement to resist cavitation caused by negative pressure generated during evapotranspiration (Fan et al., 2006; Chmielewska et al., 2016).
Recent advances in understanding the signaling pathways that govern drought stress suggest that avoidance and tolerance strategies may overlap with root lignification. A previous study proposed to reduce xylem diameter of T. aestivum seminal roots as a means to improve yield (Richards and Passioura, 1989). The authors hypothesized that the reduced seminal root xylem diameter may lower water conductance and slow the depletion of soil moisture, thereby preserving water during anthesis when plants are most sensitive to drought stress (Fageria, 1992). In support of this hypothesis, drought-adapted O. sativa lines showed increased root stele lignification and decreased root diameter after drought treatment (Hazman and Brown, 2018). Drought avoidance root architectural modifications are likely mediated by ABA signaling (Uga et al., 2013; Singh and Laxmi, 2015; Xu et al., 2020; Feng et al., 2022) and partially modulated by endogenous root auxin transport (Uga et al., 2013; Yu et al., 2016) (Figure 3). For example, ABA induces the expression of the grass-specific gene DEEP ROOT 1 (DRO1), which increases root growth angle in the root elongation zone in O. sativa (Arai-Sanoh et al., 2014) and Z. mays (Feng et al., 2022). A positive correlation between increased root angle and drought tolerance has also been reported in Z. mays (Ali et al., 2015) and S. bicolor (Djanaguiraman et al., 2023), suggesting that an uncharacterized ABA-induced activation of DRO1 may also induce lignification in roots.
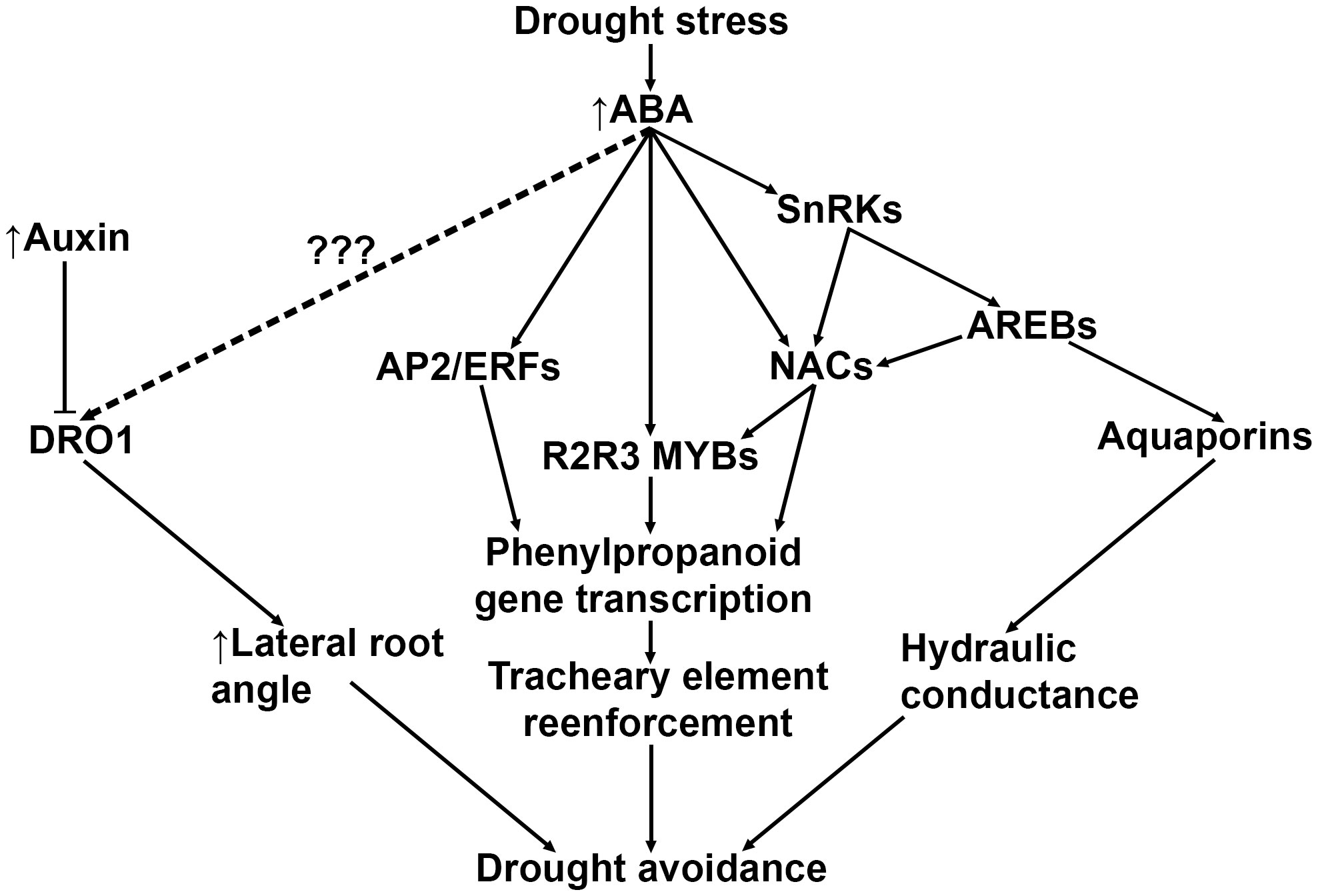
Figure 3 A general model depicting known and predicted drought-induced ABA signaling pathways that result in drought tolerance mechanisms. Drought-induced ABA upregulates DRO1, but the intermediary signaling mechanism is currently unknown. ABA also directly and indirectly upregulates several higher order transcription factors such as AP2/ERFs and NACs, which both directly and indirectly modulate phenylpropanoid gene transcription. Lastly, drought-induced ABA is responsible for upregulating transcription of aquaporins in roots, which appear to be coregulated with genes in the phenylpropanoid biosynthetic pathway. ABA, abscisic acid; DRO1, DEEP ROOT 1; SnRK, Sucrose nonfermenting1-related Kinases; AREBs, Abscisic Acid-Responsive Element Binding proteins; NACs, NAM-ATAF-CUC2 transcription factors; AP2/ERFs, APETALA2/Ethylene-Responsive Factors.
ABA was recently observed to release the constitutive inhibition of Snf1 (Sucrose nonfermenting1)-Related Kinase 2 (SnRK2) in A. thaliana, inducing secondary cell wall remodeling and lignin deposition via phosphorylation of SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1) (Liu et al., 2021a). NST transcription factors are part of the NAM-ATAF-CUC2 (NAC) family of master regulators and form part of a highly conserved multi-tiered transcription factor cascade in A. thaliana that controls activation of the phenylpropanoid biosynthesis pathway (Singh and Laxmi, 2015; Ohtani and Demura, 2019; Miyamoto et al., 2020; Hussey, 2022). NST1 binds to promoters of numerous downstream MYB transcription factors that directly regulate cell wall biogenesis and monolignol enzymes in both A. thaliana (Singh and Laxmi, 2015; Liu et al., 2021a) and grasses (Zhao and Bartley, 2014; Velez-Bermudez et al., 2015; Miyamoto et al., 2020). Both O. sativa and Z. mays NAC transcription factors directly bind to MYB46/83 promoter regions, which modulate secondary cell wall biosynthesis (Zhong et al., 2011). Similarly, overexpression of transcription factor SiMYB56 in O. sativa resulted in increased ABA synthesis, increased lignin deposition in leaves, stems, and roots, and improved overall drought tolerance (Xu et al., 2020).
ABA, jasmonic acid (JA), and salicylic acid (SA) also directly activate NAC promoter elements and modulate lignin biosynthesis independent of SnRK2 signaling (Nakashima et al., 2014; Singh and Laxmi, 2015). For example, ABA, drought, and NaCl treatment and overexpression OsNAC5 were found to upregulate OsCCR10 in drought-resistant O. sativa roots (Bang et al., 2022). In the same study, overexpression of OsNAC5 and OsCCR10 was correlated with improved drought tolerance and significantly increased root lignin content. Similarly, transgenic O. sativa overexpressing OsNAC17 showed increased expression of PAL, CCR, CAD, and PRX genes leading to significant lignin accumulation in leaf and root tissues (Jung et al., 2022).
APETALA2/ethylene-responsive factor (AP2/ERF) transcription factors also mediate drought-induced ABA signaling via activation by DREB transcription factors, resulting in drought tolerance in O. sativa (Liu et al., 2012; Wu et al., 2022; Xie et al., 2022). Treating O. sativa leaves and roots with ABA resulted in upregulation of OsERF83, increase in vascular lignification, enhanced LAC activity, elevated CAD expression, and improved drought tolerance (Jung et al., 2021). Additionally, an ABA-responsive AP2/ERF transcription factor OsERF71 was observed to bind directly to the OsCCR1 promoter (Lee et al., 2016). The same study found overexpression of OsERF71 was correlated with a significant increase in root diameter and a significant increase in PAL, C4H, CCR, and CAD transcription. Taken together, these data suggest that ABA induces multiple drought avoidance responses that modify root architecture, with some of these ABA signaling pathways include upregulation of monolignol synthesis genes and subsequent increased root lignin deposition. How increased root lignin deposition contributes to drought tolerance and/or avoidance and how ABA directly modulates root lignin deposition is still unclear and requires further testing. Elucidation of these ABA signaling pathways in grass roots will likely lead to characterization novel breeding targets that address the need of more drought-tolerant crops.
3.1.3 The lignified and suberized Casparian strip creates an ionic barrier
The transport of both water and ions into the root vasculature is regulated at the CS, the hydrophobic barrier of the root endodermis found in all terrestrial plants (Naseer et al., 2012; Doblas et al., 2017). The hydrophobicity of the CS is attributed to an asymmetric deposition of both lignin and suberin (Doblas et al., 2017; Calvo-Polanco et al., 2021); with low S/G ratio lignin primarily fortifying cell wall junctions, while suberin primarily fortifies the middle lamella (Naseer et al., 2012; Reyt et al., 2021). Recent studies in A. thaliana and O. sativa have observed Casparian Strip Membrane Domain Proteins (CASPs) to initiate CS formation in cell corners, defined by the extracellular protease LORD OF THE RINGS1 excreted by neighboring cells (Kolbeck et al., 2022; Yang et al., 2022; Barbosa et al., 2023). CASPs proceed to direct cell junction lignification via recruitment of the dirigent-domain containing protein Enhanced Suberin1 (ESB1), PRX, and NADPH oxidase F in A. thaliana (Hosmani et al., 2013; Gao et al., 2023), Z. mays (Wang et al., 2022c), and O. sativa (Wang et al., 2019, 2020). A CS hormone/kinase monitoring system characterized in A. thaliana called the Schengen pathway seals and maintains the CS ring utilizing Casparian Strip Integrity Factors (CIFs), GASSHO1 (GSO1)/SCHENGEN3 (SNG3) kinase receptors, PRXs, and a family of dirigent proteins (Doblas et al., 2017; Nakayama et al., 2017; Fujita et al., 2020). Recently, O. sativa CIF1 and SNG3 orthologs were functionally characterized (Zhang et al., 2023a), suggesting grasses possess a similar pathway.
ABA and ethylene antagonistically modulate suberization and lignification of the endodermis to regulate the bidirectional flow of ions and water (Doblas et al., 2017). Surprisingly, two independent studies of A. thaliana Schengen pathway higher order mutants found that disruption of lignification and suberization of the CS did not significantly change hydraulic conductance (Calvo-Polanco et al., 2021; Reyt et al., 2021). Instead, both studies independently concluded that aquaporin expression significantly influences hydraulic conductivity. The A. thaliana Schengen pathway higher order mutants displayed compositionally different lignin in the endodermal cell corners (H: 19%, G: 79%, S: 2%) compared to lignin polymerized in endodermal cell junctions (H:5%, G:87%, S:8%) (Reyt et al., 2021), but the function of this compositional difference is unclear.
Unique metabolic aspects of grass lignin metabolism may be strategically leveraged to quantify grass lignin CS compositions, investigate if lignin compositions in grass CS are a product of selective PRX-mediated polymerization, and test if these compositional differences between cell corners and endodermal junctions contribute to ion permeability. For instance, membrane-bound PRXs have been attributed to the lignification of A. thaliana endodermal cell corners (Rojas-Murcia et al., 2020). Functional characterization of a AtPRX64 ortholog in grasses is first needed to test if CS lignification is also mediated by one or more PRXs. Furthermore, Z. mays PRXs isolated from cell suspension cultures were found to be inefficient at oxidizing S monomers, and were suggested to oxidize S monomers more efficiently through pCA-S via radical transfer (Hatfield et al., 2008). Perturbing PMT function in a grass CS-specific prx mutant background and analyzing its phenolic composition may test if PRX employs pCA-S to facilitate radical transfer and subsequent polymerization of S monomers into the CS (Hatfield and Marita, 2010). Lastly, measuring the lignin composition and changes in CS ion permeability of a grass lac pmt mutant versus a prx pmt mutant may provide insight into the functional significance of differential lignin compositions of cell corners and endodermal junctions, as previously described in A. thaliana Schengen pathway mutants (Reyt et al., 2021).
3.1.4 LAC activity likely contributes to heavy metal ion sequestration in grass secondary cell walls
Research into heavy metal tolerance is increasing as major cereal crops are experiencing significant yield loss due to irrigation containing zinc (Zn), ferrous iron (Fe), nickel (Ni), copper (Cu), cadmium (Cd), chromium (Cr), and lead (Pb) from industrial runoff (Mir et al., 2021; Riyazuddin et al., 2022; Skuza et al., 2022; Li et al., 2022b; Xu et al., 2023). Mounting evidence suggests cell wall thickening of both woody and herbaceous plants contributes a partial but significant exclusion barrier where heavy metals are sequestered via ion exchange and chelation with methyl-esterified pectins, phenolic acids, organic acids, and ROS (Krzeslowska, 2011; Parrotta et al., 2015; Berni et al., 2019; Gao et al., 2021; Sytar et al., 2021) (Table 1). Both the physical barrier created by polysaccharides, callose, and lignin and ROS-mediated chelation of heavy metals into the cell wall are theorized to significantly reduce the rate of heavy metal uptake (Krzeslowska, 2011; Angulo-Bejarano et al., 2021).
LAC activity may significantly contribute to improved heavy metal tolerance by facilitating ROS-mediated chelation of heavy metals into the lignocellulose matrix. High Cu exposure led to significant upregulation of lignin biosynthesis genes, increased lignin deposition and LAC and PRX activity in O. sativa roots (Liu et al., 2015). Exposure to toxic concentrations of Cu, Pb, and Cd triggered a lignin biosynthesis and polymerization response in roots and significantly reduced soluble CAF and FA in Z. mays leaves (Kisa et al., 2016). This same study also demonstrated that stress responses to heavy metal exposure may not be localized only to the treated tissue. Recent studies in P. virgatum (Li et al., 2022b) and T. aestivum (Janczak-Pieniazek et al., 2023; Li et al., 2023) report similar findings of systemic increased monolignol biosynthesis, LAC activity, and lignin deposition in response to heavy metal exposure.
Thus, heavy metal exposure appears to elicit a systemic increase in lignification in grasses, but the reason behind this response is still not yet clear. Furthermore, the significant reduction of soluble CAF and FA reported in Z. mays leaves after Cu, Pb, and Cd root treatments (Kisa et al., 2016) are curious. Perhaps increased monolignol synthesis and LAC activity may lower CAF and FA pools during the stress response. Specific LAC isoforms may also facilitate the hypothesized ROS-mediated chelation of heavy metals, as several non-discrete combinations of O. sativa LACs were found upregulated in response to Cu, Cd, Cr, and Pb exposure (Liu et al., 2017). In support of this postulation, two apoplastic P. virgatum LACs were recently found to be upregulated after Cu, Ni, Fe, and Cd treatments (Li et al., 2022b). Taken together, these studies highlight how model systems and tools developed in basic research can be applied to address modern agricultural problems. However, many questions remain unanswered, such as what role does PRX contribute to heavy metal stress-induced lignification, what compositional changes (if any) arise in lignin in response to heavy metal stress, and how these compositional changes may contribute to decreased heavy metal uptake?
3.2 Lignin and biotic stress resistance in grasses
3.2.1 Do elevated S/G lignin ratios improve resistance of fungal pathogens?
In addition to abiotic stress, lignin has also been linked to biotic stress responses in grasses as summarized in Table 2. Types of fungal pathogenesis can be broadly described in two categories: biotrophs, which develop feeding sites in plant cells and extract nutrients from living plant tissue; and hemibiotrophs/necrotrophs, which secrete digestive cocktails and extract nutrients from the resulting pool of cellular debris (Doehlemann et al., 2017; Rajarammohan, 2021). Elicitation and signal transduction of pathogen triggered immune responses (PTI) and pathogen effector triggered immune responses (ETI) are complex (Yuan et al., 2021), and often lead to hypersensitive response (HR)-mediated cell death (Balint-Kurti, 2019; Guo and Cheng, 2022). One example of a host PTI/ETI response to biotrophic pathogens is a cell autonomous synthesis of papillae (Figure 4), which are hypothesized to slow or prevent biotrophic pathogenesis by reactively encasing the pathogen feeding organ in recalcitrant appositions (Israel et al., 1980; von Ropenack et al., 1998; An et al., 2006; Underwood, 2012; Bellincampi et al., 2014; Wang et al., 2021). Papillae, as seen in biotrophic Blumeria graminis f. sp. Hordei (powdery mildew) resistant H. vulgare, are thought to be generally enriched with increased β-1,3-glucan, callose, “stress lignin,” pectin, and AX (Chowdhury et al., 2014), in addition to antimicrobial peptides, ROS, and other antimicrobial metabolites (An et al., 2006; Underwood, 2012; Bellincampi et al., 2014; Wang et al., 2021). Necrotrophic pathogen PTI/ETI responses differ from biotrophic stress responses. Plants elicit a rapid but localized and controlled HR response at the infection site in response to necrotrophs and surround the affected tissue with “stress lignin” (Malik et al., 2020). Although the efficacy of HR-mediated programmed cell death has been questioned as an efficient defense strategy, an increased HR response combined with “stress lignin” deposition is believed to significantly inhibit pathogen spread and improve plant pathogen resistance (Ride, 1975; Balint-Kurti, 2019; Cesarino, 2019; Lee et al., 2019; Malik et al., 2020).
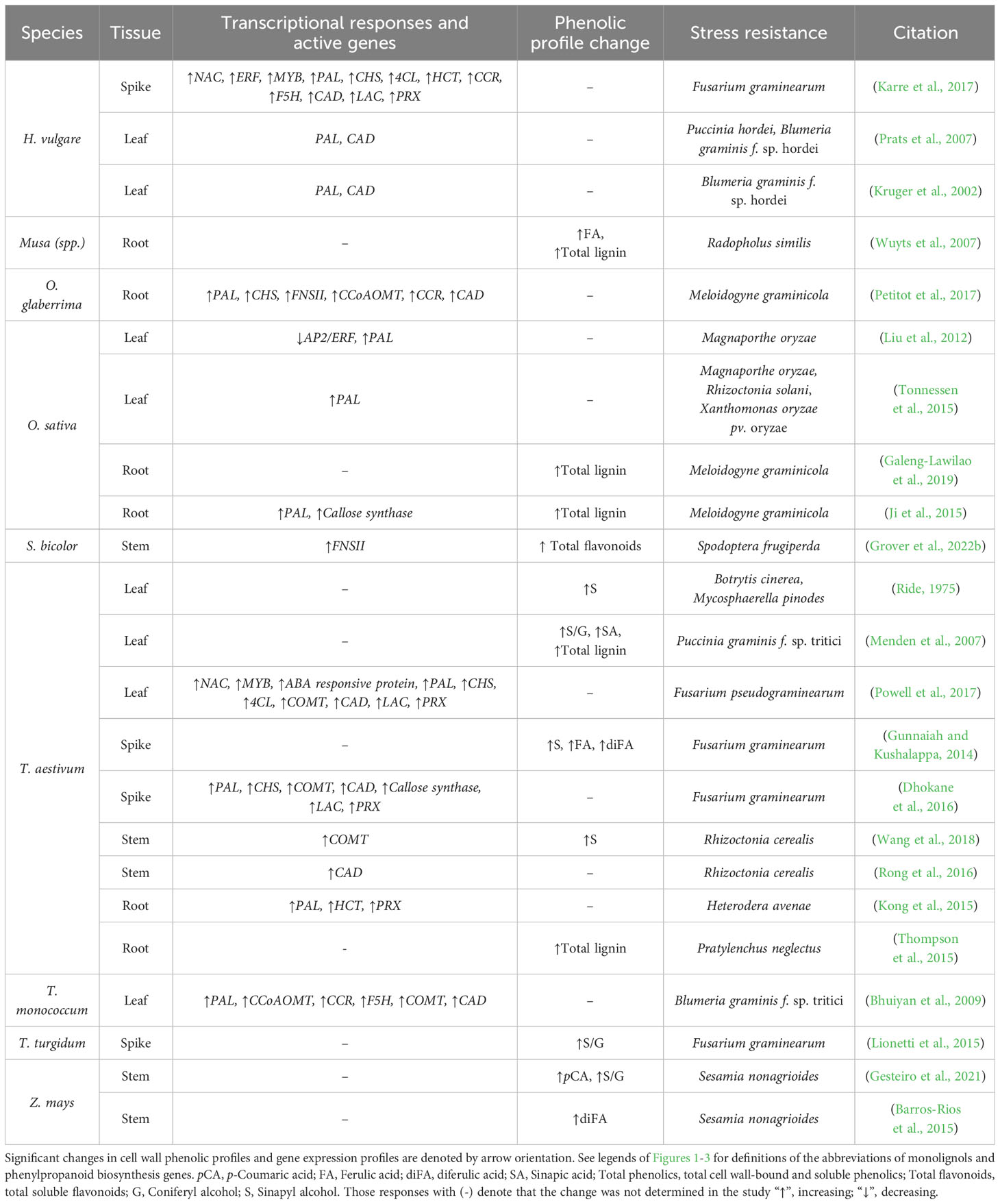
Table 2 A summary of biotic stress studies in monocot crops where differential lignin deposition and/or differential lignin synthesis-related gene expression was reported to significantly correlate with plant pathogen resistance.
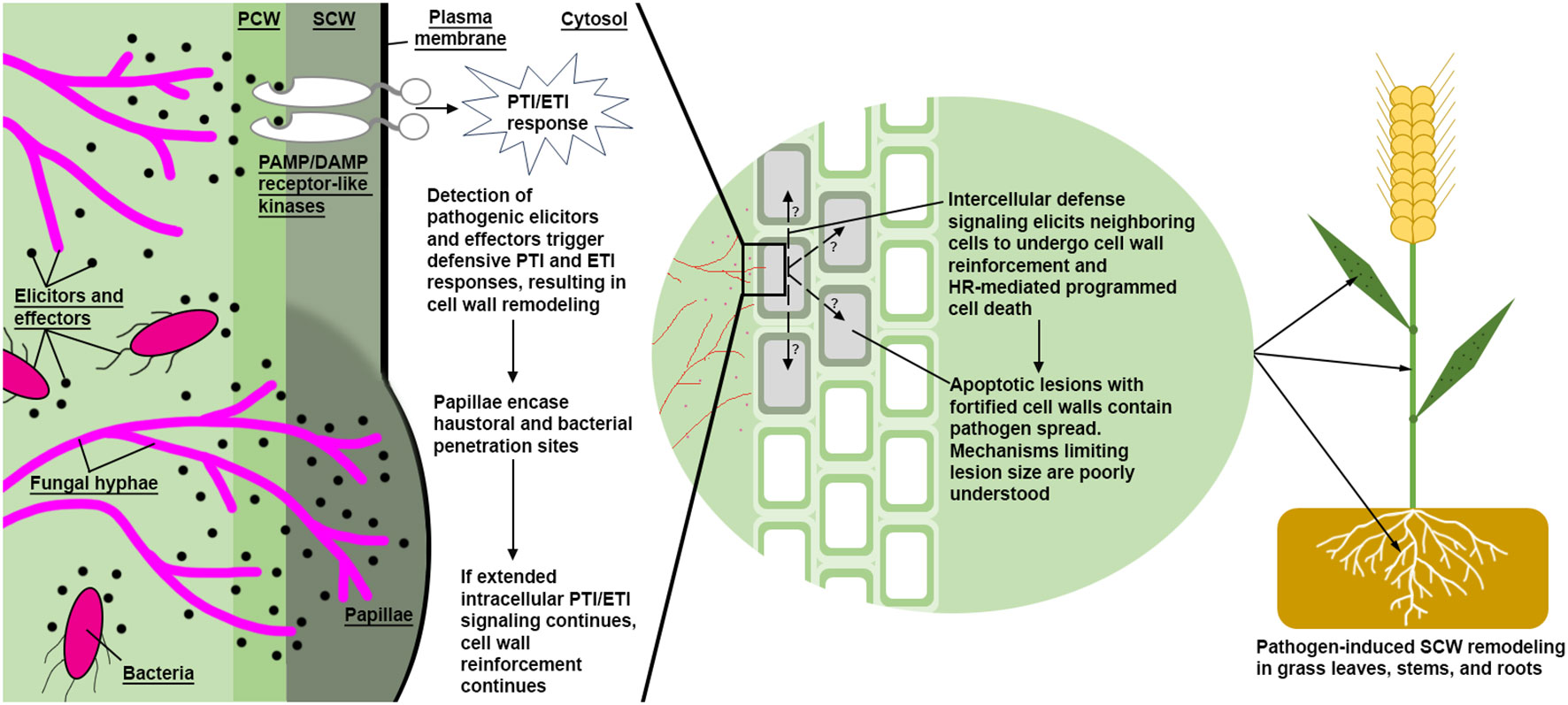
Figure 4 A simplified model of pathogen defense via induced SCW reinforcement. Bacterial and fungal hyphae (magenta colored) both produce PAMPs and DAMPs. Detection of these pathogen elicitors and effectors lead to reactive SCW reinforcement via synthesis and deposition of callose, lignin, arabinoxylans, pectin, ROS, and antimicrobial peptides. A proposed intercellular signal mechanism may transmit to neighboring cells, inducing SCW reinforcement and/or non-cell autonomous hypersensitive response, resulting in apoptotic lesions surrounding infection sites. High S/G ratio stress lignin has been characterized at the margins of lesions, which are thought to physically reinforce affected cells from further pathogen introgression. PCW, Primary Cell Wall; SCW, Secondary Cell Wall; HR, Hypersensitive Response; PTI, Pathogen-Triggered Immune; ETI, Effector-Triggered Immune.
Deposition of distinct types of “stress lignin” in papillae and at the margins of lesions appear to be a common resistance strategy in response to both necrotrophic and biotrophic pathogens. Stress lignin was initially identified as an S-rich lignin through histochemical staining surrounding infection sites of necrotrophic fungus Botrytis cinerea and biotrophic fungus Mycosphaerella pinodes on T. aestivum leaves (Ride, 1975). Later research observed a 4-fold increase in soluble esterified sinapic acid and a S/G ratio increase from 0.55 to 1.43 in T. aestivum leaves seven days after inoculation with the biotroph Puccinia graminis f.sp. tritici (stripe rust) (Menden et al., 2007). Comparison of resistant and susceptible Triticum turgidum lines to Fusarium graminearum (Fusarium head blight) found that increased S/G ratios (0.121 in the resistant line versus 0.031 in the susceptible line) was a significant cell wall resistance factor (Lionetti et al., 2015). In line with this finding, an increase in TaCOMT-3D expression was found in two resistant T. aestivum lines 18 days after inoculation with Rhizoctonia cerealis (sharp eyespot disease) (Wang et al., 2018). The same study overexpressed and suppressed TaCOMT-3D in T. aestivum and found increased TaCOMT-3D expression was positively correlated with increased lignin deposition, particularly enriched with S monomers as determined by Mäule staining, and increased R. cerealis resistance. Taken together, the biosynthesis and localized deposition of chemically distinct stress lignin appears to enhance resistance against both biotrophic and necrotrophic pathogens (Lee et al., 2019).
A question then arises: why would grasses synthesize and polymerize a chemically distinct lignin in response to biotic stress? Recent advances in computational biology have enabled the simulation of small molecule interactions with cell wall carbohydrates, thus providing a method to test if increased S/G ratio lignin can enhance lignocellulose recalcitrance to enzymatic and chemical digestion. One such in silico simulation of lignin monomer, dimer, and polymer binding to a homogeneous cellulose surface found the free energies produced by the aqueous exclusion of the methoxy groups on S monomers and β–O–4 bound dimers likely promote the closest association of the lignin polymer with the cellulose matrix (Vermaas et al., 2019a). These data led to the hypothesis that increased S monomers and β–O–4 bound dimers likely contribute to an enhanced water exclusion mechanism by shielding cell wall carbohydrates from exposure and degradation by pathogen-sourced acids, ROS, and cell wall degrading enzymes. This water-exclusion mechanism of high S monomer lignin may be supported by an earlier study, where high S/G ratio lignin was reported to be more linear (Kishimoto et al., 2005). However, the exact compositions and subsequent chemical functions of a “more linear” high S/G ratio lignin likely differ in dicots versus monocots due to the differences in hydroxycinnamic acid content found in grass cell walls. Increased incorporation of pCA into the lignin polymer appears to reduce lignin recalcitrance, as overexpression of B. distachyon BdPMT1 in a BdF5H overexpression background of poplar (Populus tremula × Populus alba) significantly increased terminal integration of free phenolic groups into the transgenic lignin polymer, improving susceptibility to cold alkali pretreatment (Lapierre et al., 2021). Thus, future in silico simulations of lignin polymer interactions will likely be capable of testing increasingly complex lignocellulose matrices, enabling investigation of how high S/G ratios impact lignin interactions with cell wall carbohydrates in grasses and dicots, how changes in esterified pCA influence diFA crosslinking frequency, or how tricin influences polymerization and lignin association via water exclusion analysis.
3.2.2 PTAL may contribute a significant role in stress lignin synthesis
Biotrophic and necrotrophic resistant cereals have been observed to differentially upregulate lignin biosynthesis genes necessary for stress lignin synthesis, providing genetic support that stress lignin serves a protective function from chemical and enzymatic degradation (Table 2). Triticum monococcum lines resistant to the biotroph Blumeria graminis f. sp. tritici (powdery mildew) showed increased expression of PAL, CCoAOMT, COMT, F5H, and CAD after infection; and increased B. graminis susceptibility was observed after both RNAi-knockdown and chemical inhibition of these upregulated genes (Bhuiyan et al., 2009). A later quantitative trait loci analysis of a F. graminearum-resistant T. aestivum line found upregulation of PAL, CHS, COMT, CAD, two LACs, and one PRX after pathogen inoculation (Dhokane et al., 2016). In F. graminearum- resistant H. vulgare, significant upregulation of PAL, CHS, and HCT in spikelet tissue was observed after infection (Karre et al., 2017). In a subsequent study, similar results from RNAseq of T. aestivum seedlings found significant upregulation of PAL after Fusarium pseudograminearum (Fusarium crown rot) inoculation (Powell et al., 2017). In a recent meta-analysis, all copies of the PAL and C4H genes in Z. mays were consistently upregulated in seven fungal pathogen-resistant Z. mays RNAseq datasets (Wang et al., 2022b). In the same study, 4CL1, 4CL2, and 4CL4 showed consistent downregulation in all datasets, while 4CL3 was upregulated in two of the datasets. This meta-analysis suggests that Z. mays increases hydroxycinnamic acid synthesis in response to fungal pathogen infection, and is in line with recent metabolic phenylpropanoid flux data quantified in B. distachyon (Barros et al., 2022).
A common finding among these grass studies, regardless of host or pathogen species, is the upregulation of PALs and PTALs. Pathogen-induced expression of PAL and PTAL genes has been associated with broad-spectrum pathogen resistance in O. sativa (Zhou et al., 2018; Yadav et al., 2020). Furthermore, analysis of the metabolic consequences of perturbing PAL and PTAL in B. distachyon hinted to the fact that PTAL may contribute to S monolignol synthesis via an uncharacterized pathway (Barros et al., 2022). Specifically, through 13C carbon flux analysis of both BdPAL2i and grass-specific BdPTAL1i in B. distachyon, it was observed that 13C-labeled tyrosine exhibited a preferential incorporation (up to 50%) into hydroxycinnamic acids CAF, pCA, FA, and SA, with significant incorporation into S monomers (Barros et al., 2016). Conversely, 13C-phenylalanine showed a significant preference for incorporation into G monomers. The frequency of PAL/PTAL upregulation in multiple grass species in response to multiple pathogens suggest that PTAL may contribute to a broad-spectrum pathogen resistance by triggering an uncharacterized parallel S monolignol synthesis pathway. Functional studies of PAL/PTAL in S. bicolor (Godin et al., 2016; Jun et al., 2018; Simpson et al., 2021), T. aestivum (Feduraev et al., 2020), and B. oldhamii (Hsieh et al., 2021) could be further leveraged to test how PTAL contributes to pathogen resistance in these and other grass species.
3.2.3 Grass monolignol synthesis enzymes may also function as host-susceptibility factors
Several phenylpropanoid enzymes have been found to function as non-receptor-like susceptibility factors, which play a pivotal role in regulating defense signaling and are often targeted by pathogens to increase their virulence (Saur and Huckelhoven, 2021). Recent studies have found multiple lignin biosynthesis enzymes contribute to pathogen defense-related functions beyond their phenylpropanoid metabolic activity, suggesting that they may be host-susceptibility factors. For example, a meta-analysis of the O. sativa PAL gene family made four strong arguments to suggest that PAL, and possibly PTAL, function as non-receptor-like susceptibility genes by contributing a crucial role in broad-spectrum pathogen resistance: 1) OsPAL genes are localized in pathogen resistant-associated QTLs; 2) increased OsPAL expression is observed in multiple pathogen resistant O. sativa lines; 3) OsPAL regulates SA synthesis in response to multiple pathogens in both aerial and root tissue; and 4) resistance mediated by PAL may be durable due to both PAL expression and subsequent phenylpropanoid metabolites being downstream of pathogen recognition mechanisms (Yadav et al., 2020). Finally, genetic or chemical suppression of PAL in B. distachyon (Cass et al., 2015), O. sativa (Huang et al., 2010; Tonnessen et al., 2015), H. vulgare (Kruger et al., 2002; Prats et al., 2007), and T. aestivum (Bhuiyan et al., 2009) results in increased pathogen susceptibility, further supporting the hypothesis that PAL, and possibly PTAL are host susceptibility factors.
In addition, CCR, HCT, CCoAOMT, and CAD may also be considered host-susceptibility factors. In O. sativa, CCR1 was found to bind to OsRac1, a plant defense-related effector found to upregulate ROS via NADPH oxidases in a GTPase-dependent manner (Kawasaki et al., 2006). In the same study, OsCCR1 was upregulated in response to a sphingolipid elicitor, and constitutive overexpression of OsRac1 led to increased OsCCR1 activity and increased cell wall lignification. In a fascinating series of studies, CCoAOMT and HCT in Z. mays were observed to, both independently and as a complex, modulate activation of the R-gene Rp1-D21 during Puccinia sorghi (maize common rust) infection (Wang et al., 2015a; Wang and Balint-Kurti, 2016). In addition, overexpression of CCoAOMT2 in Z. mays led to broad-spectrum pathogen resistance (Yang et al., 2017). Recently, CCoAOMT in Z. mays was found to be co-expressed with Rp1-D21 at the border of leaf lesions that underwent HR-mediated programmed cell death, and may contribute a role in modulating cell-autonomous HR-mediated programmed cell death (Karre et al., 2021). Z. mays FNSI homologs were found to interact with both the HCT and the Rp-D21 CCD21 binding domain when transiently expressed in Nicotiana benthamiana, suggesting another pathogen defense-related HR attenuation mechanism that is tied to alternative SA synthesis (Zhu et al., 2020). An earlier study of R. cerealis-resistant T. aestivum revealed that TaCAD12 was upregulated in response to the pathogen (Rong et al., 2016). Further functional characterization of TaCAD12 via overexpression increased resistance to R. cerealis and increased co-expression of defense genes (Defensin, PR10, PR17c, and Chitinase1) and other phenylpropanoid genes (TaCAD1, TaCCR, and TaCOMT1). Taken together, phenylpropanoid genes may be an emerging subtype of non-receptor-like susceptibility factors. Continued research on phenylpropanoid enzyme protein-protein interactions may lead to engineering novel broad-spectrum lignin-based herbivory and parasite resistant cereal and grass lines.
3.3 Lignin-based herbivory and parasite resistance in grasses
3.3.1 Constitutive and induced phenolic-based herbivory resistance observed in grass aerial tissues
Grass secondary cell wall compositions contribute a first line of defense against both chewing and invasive herbivore infestation. Unlike pathogenic microorganisms, some herbivores directly and immediately consume the host plant via laceration, chewing, and swallowing of host tissue, and must contend with the immediate composition of consumed lignocellulose, including tissue-specific basal S/G ratios, grass cell wall crosslinking mediated by FA and diFA, and flavonoid-derived phytotoxins (Santiago et al., 2013). Conversely, invasive herbivores physically and enzymatically penetrate plant tissue and induce cell wall modifications via secreted elicitors to feed from host phloem or parasite-induced specialized feeding sites (Waterman et al., 2019; Grover et al., 2022a). Grasses employ two constitutive phenolic-based strategies to resist damage associated with herbivory: toughness, which is the plants pre-digestive anti-nutrition measured via the herbivore’s ability to penetrate or consume the host plant (Lucas et al., 2000; Clissold, 2007), and post-digestive anti-nutrition, which is a measure of a herbivore’s ability to safely and effectively digest the host plant (Clissold, 2007).
Studies into the grass cell wall components that contribute to and enhance pre-digestive herbivory resistance often converge on phenolic compounds that fortify tissue such as hydroxycinnamic acids, lignin, and flavonoids (Hatfield and Marita, 2010; Santiago et al., 2016). For instance, a comparison of cell wall phenolics in Sesamia nonagrioides (Mediterranean corn stalk borer) resistant Z. mays lines found cell wall-bound pCA and S/G ratios in pith tissue to be inversely correlated with stalk penetration (Gesteiro et al., 2021). This study hypothesizes that Z. mays balances the degree of diFA-mediated cell wall crosslinking and pCA incorporation as cell wall FA and diFA concentrations were positively correlated with plant height, blooming time, and grain yield. These data suggest increased diFA-mediated crosslinking is associated with growth and grain production at the expense of tissue toughness, whereas cell wall-bound pCA and increased S/G lignin ratios significantly contribute pre-digestive anti-nutrition herbivory defense at the expense of total plant height and flowering time.
These data also contextualize previous quantifications of Z. mays pith versus rind S/G ratios (1.48 and 1.51), cell wall-bound pCA (2.1% and 2.7%), and cell wall-bound FA (1.4% and 1.5%) (Barros-Rios et al., 2011), suggesting that rind tissue is on average tougher than pith tissue, and said increased toughness contributes to increased pre-digestive anti-nutritional herbivory defense. Observed S. nonagrioides behavior of preferentially infiltrating Z. mays via the base of the internode where the intercalary meristem is located and the cells are the least fortified further support that higher cell wall-bound pCA and S/G ratios correlate with tissue toughness and improved pre-digestive herbivory resistance (Barros et al., 2011; Barros-Rios et al., 2011). However, S. nonagrioides-resistant Z. mays lines with increased diFA in pith tissue reduced stem bore depth by 29%, and recovered fed larvae from the high diFA Z. mays population were 30-40% lower weight than recovered larvae from contrasting low diFA Z. mays population (Barros-Rios et al., 2015), implying that increased diFA-mediated crosslinking is correlated with post-digestive anti-nutritional herbivory defense. In support of these findings, diFA-mediated cell wall crosslinking was negatively correlated with lignocellulose digestibility (Grabber et al., 1998; Grabber, 2005; Hatfield and Marita, 2010).
Grasses are also capable of detecting and reacting to herbivory through inducing phenolic production and cell wall remodeling (Waterman et al., 2019). Chewing herbivores such as Spodoptera frugiperda (fall armyworm) produce saliva containing protein and nonprotein-based herbivore-associated molecular patterns (HAMPs) that have been observed to elicit (Turlings et al., 1993; Chuang et al., 2014; Acevedo et al., 2019; Grover et al., 2022b) and suppress (de Lange et al., 2020; Grover et al., 2022b; Warburton et al., 2023) herbivory defense responses in Z. mays, O. sativa, and S. bicolor. A study of the effects of S. nonagrioides regurgitant on Z. mays leaves showed no significant change in larvae weight or phenolic cell wall composition, suggesting that herbivore-plant interactions are complex and likely require multiple and specific stimuli to elicit HAMP and ETI responses (Santiago et al., 2017). A comparative study of S. frugiperda herbivory on both resistant and susceptible S. bicolor lines observed significantly decreased soluble hydroxycinnamic acids pCA, CAF, FA, and sinapic acid after infestation (Grover et al., 2022b). Furthermore, the resistant S. bicolor line showed increased FNSII transcription and increased concentrations soluble and cell wall-bound flavonoids naringenin, apigenin, luteolin, and quercetin in leaf tissue, suggesting that S. frugiperda ETI response detection may attenuate phenylpropanoid biosynthesis in favor of flavonoid flux to produce defensive phenolic pesticides. Increased traffic and accumulation of vesicle-like bodies containing H2O2 and phenolics at plasma membranes of cells neighboring B. graminis infection in H. vulgare leaves suggest antimicrobials are actively transported into the apoplast (An et al., 2006). The observed ETI-mediated prioritization of flavonoid synthesis may be coupled with a similar intracellular transport and apoplastic excretion mechanism, as recently seen in glycosylated monolignol transport in Physalis pubescens (Shimada et al., 2021), implying grasses may employ a localized phenolic-based defense mechanism at herbivore feed sites (Jeon et al., 2022). Further research is needed to confirm the identity and characteristics of a model for ETI-induced local synthesis and mobilization of phenylpropanoid-derived pesticides at herbivore feeding sites.
3.3.2 Constitutive and induced root lignification may also contribute to endoparasitic nematode resistance
Invasive parasitic herbivores such as endoparasitic nematodes infest grass root systems in a species-dependent manner, and thus may require species-dependent host defense strategies (Okubara et al., 2019). Three types of endoparasitic nematodes that contribute to significant global cereal crop loss are juvenile root knot nematodes (Meloidogyne spp.) (Holbein et al., 2016; Rutter et al., 2022), juvenile cyst nematodes (Heterodera spp. and Globodera spp.) (Holbein et al., 2016), and root lesion nematodes (Pratylenchus spp.) (Fosu-Nyarko and Jones, 2016; Okubara et al., 2019). All three categories of plant endoparasitic nematodes possess a protrusible stylet that aids in physically penetrating plant cell walls. Moreover, all three nematode types secrete a mixture of mostly unidentified effectors and cell wall modifying proteins that facilitate their movement within the host plant, help suppress initial host PTI and subsequent ETI defense responses, and help establish feeding sites that are specific to each nematode species (Wieczorek, 2015; Fosu-Nyarko and Jones, 2016; Holbein et al., 2016).
Emerging research is beginning to suggest that parasitic nematode basal resistance is partially credited to both constitutive and induced root lignification (Okubara et al., 2019). For instance, monocot banana (Musa spp.) cultivars resistant to root lesion nematode Radopholus similis (burrowing nematode) have significantly more cell wall-bound FA in cortical tissue compared to susceptible lines prior to infestation, and significant 1.4-fold increase in root lignin after infestation (Wuyts et al., 2007). In addition, the differential lignification observed extending from the tracheary elements into the pericycle and endodermal cells immediately neighboring R. similis lesion sites were attributed to improved host resistance. Genetic evidence to support endodermal lignification as a significant contributor to nematode-resistance was established after observing A. thaliana CS mutant sgn3-3esb1-1 (no CS lignin) to be significantly more susceptible to Heterodera schachtii (cyst nematode) infestation (Holbein et al., 2019). The same study also demonstrated that disruption of CS lignification and CS suberization via esb1-1CDEF1 (no CS lignin, no CS suberin) significantly increased both H. schachtti and Meloidogyne incognita (root-knot nematode) susceptibility. The Schengen pathway may contribute a significant defense response that increases endodermal cell toughness, thereby limiting the development of root-knot nematode feeding sites (Doblas et al., 2017; Nakayama et al., 2017; Fujita et al., 2020). Furthermore, previously hypothesized PRX-mediated oxidation of S monomers via pCA-S radical transfer (Hatfield et al., 2008) may be key to differentially polymerizing stress lignin at and around root-knot nematode endodermal feeding sites.
Genetic, metabolomic, and histochemical evidence indicate a positive correlation between lignification of roots and nematode resistance. B-aminobutyric acid soil treatment applied to O. sativa roots induced resistance to Meloidogyne graminicola (root knot nematode) infestation and developmental potential by significantly increasing in OsPAL and Callose Synthase expression and increasing lignin and callose deposition around M. graminicola root galls (Ji et al., 2015). In support of these findings, a M. graminicola-resistant variety of O. glaberrima was reported to have upregulated phenylpropanoid (PAL, CCoAOMT, CCR, CAD), flavone (CHS, FNSII), and phytohormone (JA, SA, and ethylene) synthesis genes (Petitot et al., 2017). The same study found chemical inhibition of PAL activity and JA synthesis increased total nematodes and gall number in chemically treated and infested roots (Petitot et al., 2017). O. sativa RILs resistant to M. graminicola were observed to rapidly lignify root epidermal cells after infection, including increased lignification surrounding root galls, resulting in significantly reduced gall numbers (Galeng-Lawilao et al., 2019). In addition, a T. aestivum line resistant to Heterodera avenae (cereal cyst nematode) showed significant upregulation PAL after infestation (Kong et al., 2015). Taken together, grasses seem to employ a localized defensive lignification strategy, reminiscent of pathogen-induced papillae formation (Figure 4), that mitigates some types of nematode infections. Further research focusing on mutants in the grass Schengen pathway may aid in the development of more resistant cereals to root knot and cereal cyst nematodes, whereas resistance to root lesion nematodes may be best achieved by increasing epidermal toughness in the root.
4 Industrial applications of grass lignin
4.1 Advances in grass lignin valorization
4.1.1 Engineering grass lignin qualities to improve saccharification efficiency
Lignin inhibits complete hydrolysis of grass feedstock polysaccharides, and disruption of lignin synthesis improves lignocellulose saccharification efficiencies (Godin et al., 2016; Ralph et al., 2019; Ray et al., 2020; Ha et al., 2021; Ning et al., 2021; Wang et al., 2022a). Molecular simulations have demonstrated that lignin directly and competitively inhibits the recognition mechanism of cellulases (Vermaas et al., 2015). Additionally, in silico simulations of lignin-carbohydrate interfaces found that disruption of phenylpropanoid synthesis likely results in disruption of cell wall crosslinking and decreased noncovalent association of lignin polymers to cell wall carbohydrates, thereby decreasing probability of cellulase inhibition (Carmona et al., 2015; Vermaas et al., 2019a). A recent study tested biomass pretreatment methods on a near-homogeneous lignin polymer composed almost entirely of caffeyl lignin (C monomers) (Li et al., 2018b). C-lignin was initially found in Vanilla planifolia seed coats (Chen et al., 2012a), and was though to arise by the absence of phenylpropanoid O-methyltransferase activity during seed coat lignin biosynthesis (Li et al., 2018b). C-lignin was posited to be an “ideal lignin” because its near homogeneous composition of β-O-4 bonds may depolymerize into a single product by hydrogenolysis if it remained stable during biomass pretreatment conditions. To test this hypothesis, acid pretreatment was applied to V. planifolia seed coats, resulting in near total lignin depolymerization with no detectable undesirable condensation reactions; demonstrating that the theoretical chemistry of an ideal lignin is feasible.
To reduce the energy and resource input cost generated by the necessary pretreatment of plant biomass, genetic engineering strategies have been pursued to alter lignin compositions (Vanholme et al., 2012; Mottiar et al., 2016; Pazhany and Henry, 2019; Ralph et al., 2019; Liu et al., 2021b). Reducing lignin S/G ratios has been consistently linked to improved saccharification efficiency. However, the specific reasons for this correlation may vary depending on the pretreatment methods used in different studies (Christensen and Rasmussen, 2019). COMT downregulation in grasses is a reliable genetic strategy to lower S/G ratios in P. virgatum (Fu et al., 2011a, b), B. distachyon (Ho-Yue-Kuang et al., 2016), a Saccharum hybrid (Jung and Altpeter, 2016), T. aestivum (Ma and Xu, 2008), Z. mays (Piquemal et al., 2002; Pichon et al., 2006; Fornale et al., 2017), and S. bicolor (Bout and Vermerris, 2003; Oliver et al., 2005; Saluja et al., 2021). Decreasing COMT activity significantly increases saccharification efficiency by increasing concentrations of 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol incorporation into the mutant lignin polymer (Green et al., 2014; Ralph et al., 2019). Interestingly, 5-hydroxyconiferyl alcohol preferentially undergoes β–O–4 coupling, producing surprisingly linear homopolymers that resemble ideal lignin previously observed in C lignin from V. planifolia seed coats (Chen et al., 2012a; Li et al., 2018b; del Río et al., 2022). Reducing COMT activity also likely disrupts FA and tricin synthesis (Barros et al., 2019a) (Figure 1), thus potentially disrupting AX feruloylation and diFA-mediated cell wall crosslinking (Grabber et al., 1998, 2004; de Oliveira et al., 2015). Indirect evidence suggests the availability of these COMT-derived FA pools are positively correlated with grass lignocellulose recalcitrance, as recent perturbation of BAHD acyltransferases resulted in disrupted AX feruloylation and improved lignocellulose saccharification in S. viridis (de Souza et al., 2018) and Saccharum spp (de Souza et al., 2019). Suppression of one or more of these BAHD acyltransferases that mediate AX feruloylation in a comt mutant background may test if COMT activity is a significant source of cell wall-bound FA, while also providing additional evidence grass preferential phenylpropanoid metabolism into hydroxycinnamic acid synthesis.
Lastly, lignin polymers have been genetically engineered to contain more β–O–4 bonds in the lignin backbone (Mottiar et al., 2016). These “Zip-Lignins” are a novel class of exotic “designer lignins” that are susceptible to mild alkaline pretreatment while minimally perturbing plant growth and fitness (Wilkerson et al., 2014). A cell wall modeling system in Z. mays demonstrated successful polymerization of exogenously supplied G-FA into non-lignified cell walls (Grabber et al., 2008). The ester-interunit conjugation of G-FA was found to readily cleave after mild alkaline pretreatment and was positively correlated with improved saccharification of cell wall carbohydrates, independent of pretreatment. A BAHD acyltransferase from Angelica sinensis that generates monolignol ferulate conjugates was characterized as a FMT in poplar (Populus alba × grandidentata) (Wilkerson et al., 2014). In this study, AsFMT conjugated both G and S monomers to FA. Overexpression of FMT led to synthesis of G-FA and S-FA that could be transported, polymerized, and function as Zip-Lignins with no negative growth phenotype. ZmCCR1 substrate specificity for feruloyl-CoA, the ligand for FMT, was successfully leveraged in a genetic engineering strategy to improve lignocellulose digestibility in Z. mays (Smith et al., 2017a). The Zmccr1 mutant showed increased feruloyl-CoA pools, resulting in an increased substrate pool for the native ZmFMT to generate more G-FA and S-FA no significant negative growth phenotype. Recently, grass FMT orthologs have been characterized in P. virgatum and S. bicolor (Smith et al., 2022), demonstrating that a similar genetic engineering strategy may be feasible in other bioenergy grasses.
4.1.2 Prioritizing restrained disruption of monolignol synthesis may mitigate undesirable phenotypes
The adage “too much of a good thing is bad” may apply to genetic engineering of phenylpropanoid metabolism. Altering monolignol biosynthesis in both monocots and dicots can result in unpredictable and sometimes undesirable phenotypes such as dwarfism and sterility (Sattler and Funnell-Harris, 2013; Bonawitz and Chapple, 2013, Ha et al., 2021). For example, comt null mutants brm3 in Z. mays (Vignols et al., 1995) and brm12 in S. bicolor (Bout and Vermerris, 2003; Sattler et al., 2012) show decreased biomass and grain yield. In contrast, Z. mays knockdown comt mutants brm3 (Piquemal et al., 2002; Pichon et al., 2006) and S. bicolor brm12 (Green et al., 2014) show no significant decrease in biomass compared to wild type. Similarly, comt knockdown mutants in P. virgatum (Fu et al., 2011a; Baxter et al., 2014), T. aestivum (Ma and Xu, 2008), H. vulgare (Lee et al., 2021) and B. distachyon (Ho-Yue-Kuang et al., 2016) produce near wild-type biomass. In O. sativa, C3′H-knockout mutants were severely dwarfed and sterile whereas RNAi knockdown mutants reached maturity, set seed, and showed improved saccharification (Takeda et al., 2018). In Z. mays, bm2-bm4 double mutants are dwarfed, show constitutive drought symptoms, and do not reach reproductive maturity, whereas bm1-bm2-bm3 and bm1-bm3-bm4 did reach maturity and were similar to wild-type plants (Vermerris et al., 2010). On the other hand, overexpression of transcription factors that regulate lignification in P. virgatum also led to dwarfing phenotypes (Rao et al., 2019). Taken together, targeting single or even multiple genes to improve saccharification efficiency of lignocellulose should prioritize reducing, rather than completely removing phenylpropanoid enzyme activity from the grass species of interest (Bonawitz and Chapple, 2010, 2013). Application of restrained disruption is can be found in the commercial success following moderate CCoAOMT perturbation of reduced lignin dicot M. sativa “HarvXtra,” specifically marketed to livestock and dairy industries (Barros et al., 2019b).
4.2 Grass lignin as a green source of complex chemical precursors
The structural complexity of lignin polymers (Ralph, 2010; Ralph et al., 2019; del Río et al., 2022) has ignited research into prospecting plant lignin as an environmentally friendly source of industrial chemical precursors (Calvo-Flores and Dobado, 2010; Zakzeski et al., 2010; Banu et al., 2019; Al-Naji et al., 2022). “Lignin first” biorefineries designed to holistically prioritize lignin, cellulose, and hemicellulose valorization follow published methodology with appropriate standards and references to maximize product yields (Renders et al., 2017; Ruiz et al., 2023). Since grass lignin contains relatively high concentrations of pCA, FA, diFA, along with feruloylated and coumaroylated cinnamyl alcohol and carbohydrate conjugates, grass lignin may be a potential source of industrial and pharmaceutical chemical precursors (Islam et al., 2021; Bouyahya et al., 2022). A recent study applied reductive catalytic fractionation (RCF) to P. virgatum, Z. mays stover, and T. aestivum straw to identify the determinants of lignin depolymerization (Chen et al., 2023). The study was able to distinguish core monolignols from pendent phenolate esters conjugated to cell wall carbohydrates. Extraction of chemicals from lignin will likely need to be optimized according to a specific species and tissue type, further stressing the importance of complete cell wall phenolic profiling in future lignin research. Justification for these complex analyses is the potential for discovering natural and renewable sources of pharmaceutical precursors such as the catechol products obtained from V. planifolia seed coat lignin (Sedo et al., 2013; Li et al., 2018b). Although artificial catechol synthesis has been reported (Wu et al., 2017), sourcing naturally occurring phenolic precursors as well as genetically engineering existing sources to favor synthesis of desirable phenolics may be cheaper, cleaner, and more productive alternatives to artificial synthesis (Grossman and Vermerris, 2019). Faster, cleaner, and cheaper pretreatment methods of grass lignin containing high concentrations of pCA were proposed in Z. mays via ozone oxidation of pCA-S monolignols (Danby et al., 2018). Lastly, it could be argued that grasses are preferential sources of bioenergy and industrial feedstocks due to their rapid biomass accumulation. Grasses that harbor natural or engineered phenolic profiles can be quickly grown to produce on-demand precursors, thus acting as renewable and sustainable sources of bio-based chemicals.
5 Conclusions and future directions
This review provides an overview of the recent progress made in understanding the idiosyncrasies of grass monolignol biosynthesis and how grass lignin contributes to abiotic and biotic stress resistance. Differences between dicot and grass monolignol synthesis are reflective of their divergent anatomical characteristics. For instance, grasses appear to preferentially shunt phenylpropanoid flux into hydroxycinnamic acid metabolism, whereas enzymes responsible for the synthesis of phenylpropanoid shikimate intermediates are necessary for dicot lignification. Furthermore, grasses may possess a parallel yet uncharacterized metabolic S monomer synthesis pathway that contribute to synthesis of pCA-S. Grasses may also glycosylate monolignols for storage and transport, but where these conjugation reactions occur, and their exact functions have not been characterized. Moreover, tricin appears to be present in all tested monocots to date, yet the functional role of tricin in grass lignin is currently unknown.
However, the laws of chemistry find common ground between monocot and dicot lignification, such as how small nonpolar monolignols translocate into the apoplast is hypothesized to be facilitated by concentration gradient passive diffusion driven by LAC and PRX polymerization activity, and that monolignols combinatorically polymerize via oxidative radical coupling. Parallel mechanisms that drive Casparian strip development in dicots are quickly being reported in grasses, such as the employment of dirigent proteins that direct the deposition of compositionally different lignin at the Casparian strip compared to surrounding endodermal tissue.
Differences in lignification are also correlated with abiotic stress resistance in both dicots and monocots, but grass studies often do not quantify hydroxycinnamic acid or flavonoid compositions, leaving questions of how these overlooked phenolics may contribute to abiotic stress resistance. Furthermore, the signaling pathways that elicit lignification or changes in lignin in response to abiotic stress in grasses are incidentally reported and are often found not reflective of dicot models. More focus on elucidating grass abiotic stress signaling pathways may lead to discovery of novel quantitative trait loci that can be mapped and selectively bred to improve cereal abiotic stress tolerance. Furthermore, determination of the signaling pathways that govern stress lignin synthesis and deposition may lead to discovery of molecular mechanisms that can be leveraged to artificially prime crops to resist pathogen spread in field conditions. These same mechanisms may also be engineered to favor similar defense responses in flavone signaling that may be tied to the poorly understood mechanisms governing grass monolignol glycosylation, storage, and transport.
Lastly, improving resolution of multi-level omics data and increasingly sophisticated lignocellulose saccharification optimization strategies have led to the development of designer lignins such as Zip Lignins, which reportedly have minimal negative consequences on plant fitness. However, lessons learned from past genetic engineering strategies to improve saccharification suggest future endeavors to employ “restrained disruption” to avoid negative growth phenotypes commonly associated with phenylpropanoid null mutants. Although much has been learned thus far regarding lignification in grasses, future research will most certainly leverage emerging technologies to explore the many uncharacterized mechanisms that drive grass cell wall biology.
Author contributions
LP: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. RP: Writing – review & editing. JB: Writing – review & editing. LB: Supervision, Writing – review & editing. KS: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by USDA/NIFA through Hatch project 1014527 as well as the Washington Grain Commission Grant number 13C_30193687 to KS.
Acknowledgments
We would like to thank the peer reviewers for their critical review and valuable suggestions for improving this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acevedo, F. E., Smith, P., Peiffer, M., Helms, A., Tooker, J., Felton, G. W. (2019). Phytohormones in fall armyworm saliva modulate defense responses in plants. J. Chem. Ecol. 45, 598–609. doi: 10.1007/s10886-019-01079-z.
Alejandro, S., Lee, Y., Tohge, T., Sudre, D., Osorio, S., Park, J., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22, 1207–1212. doi: 10.1016/j.cub.2012.04.064.
Ali, M. L., Luetchens, J., Nascimento, J., Shaver, T. M., Kruger, G. R., Lorenz, A. J. (2015). Genetic variation in seminal and nodal root angle and their association with grain yield of maize under water-stressed field conditions. Plant Soil 397, 213–225. doi: 10.1007/s11104-015-2554-x.
Al-Naji, M., Brandi, F., Driess, M., Rosowski, F. (2022). From lignin to chemicals: an expedition from classical to modern catalytic valorization technologies. Chemie Ingenieur Technik 94, 1611–1627. doi: 10.1002/cite.202200079.
An, Q. L., Huckelhoven, R., Kogel, K. H., Van Bel, A. J. E. (2006). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 8, 1009–1019. doi: 10.1111/j.1462-5822.2006.00683.x.
Angulo-Bejarano, P. I., Puente-Rivera, J., Cruz-Ortega, R. (2021). Metal and metalloid toxicity in plants: an overview on molecular aspects. Plants-Basel 10. doi: 10.3390/plants10040635.
Arai-Sanoh, Y., Takai, T., Yoshinaga, S., Nakano, H., Kojima, M., Sakakibara, H., et al. (2014). Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 4. doi: 10.1038/srep05563
Argenta, J., Pacheco, M. T., Mariath, J. E. D., Federizzi, L. C. (2022). Morphological, anatomical, and chemical characteristics associated with lodging resistance in Avena sativa. Euphytica 218. doi: 10.1007/s10681-022-02971-8.
Balint-Kurti, P. (2019). The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178. doi: 10.1111/mpp.12821.
Bang, S. W., Choi, S., Jin, X. J., Jung, S. E., Choi, J. W., Seo, J. S., et al. (2022). Transcriptional activation of rice CINNAMOYL-CoA REDUCTASE 10 by OsNAC5, contributes to drought tolerance by modulating lignin accumulation in roots. Plant Biotechnol. J. 20, 736–747. doi: 10.1111/pbi.13752.
Banu, J. R., Kavitha, S., Kannah, R. Y., Devi, T. P., Gunasekaran, M., Kim, S. H., et al. (2019). A review on biopolymer production via lignin valorization. Bioresource Technol. 290. doi: 10.1016/j.biortech.2019.121790
Barbosa, I. C. R., De Bellis, D., Flückiger, I., Bellani, E., Grangé-Guerment, M., Hématy, K., et al. (2023). Directed growth and fusion of membrane-wall microdomains requires CASP-mediated inhibition and displacement of secretory foci. Nat. Commun. 14. doi: 10.1038/s41467-023-37265-7.
Barros, J., Dixon, R. A. (2020). Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 25, 66–79. doi: 10.1016/j.tplants.2019.09.011.
Barros, J., Escamilla-Trevino, L., Song, L. H., Rao, X. L., Serrani-Yarce, J. C., Palacios, M. D., et al. (2019a). 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic Ascorbate peroxidase. Nat. Commun. 10, 11. doi: 10.1038/s41467-019-10082-7.
Barros, J., Malvar, R. A., Butron, A., Santiago, R. (2011). Combining abilities in maize for the length of the internode basal ring, the entry point of the Mediterranean corn borer larvae. Plant Breed. 130, 268–270. doi: 10.1111/j.1439-0523.2010.01789.x.
Barros, J., Serk, H., Granlund, I., Pesquet, E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074. doi: 10.1093/aob/mcv046.
Barros, J., Serrani-Yarce, J. C., Chen, F., Baxter, D., Venables, B. J., Dixon, R. A. (2016). Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2. doi: 10.1038/nplants.2016.50.
Barros, J., Shrestha, H. K., Serrani-Yarce, J. C., Engle, N. L., Abraham, P. E., Tschaplinski, T. J., et al. (2022). Proteomic and metabolic disturbances in lignin-modified Brachypodium distachyon. Plant Cell 34, 3339–3363. doi: 10.1093/plcell/koac171.
Barros, J., Templet, S., Dixon, R. A. (2019b). Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 56, 48–54. doi: 10.1016/j.copbio.2018.09.003.
Barros-Rios, J., Malvar, R. A., Jung, H. J. G., Santiago, R. (2011). Cell wall composition as a maize defense mechanism against corn borers. Phytochemistry 72, 365–371. doi: 10.1016/j.phytochem.2011.01.004.
Barros-Rios, J., Santiago, R., Jung, H. J. G., Malvar, R. A. (2015). Covalent Cross-Linking of Cell-Wall Polysaccharides through Esterified Diferulates as a Maize Resistance Mechanism against Corn Borers. J. Agric. Food Chem. 63, 2206–2214. doi: 10.1021/jf505341d.
Bartley, L. E., Peck, M. L., Kim, S. R., Ebert, B., Manisseri, C., Chiniquy, D. M., et al. (2013). Overexpression of a BAHD acyltransferase, osAt10, alters rice cell wall Hydroxycinnamic acid content and Saccharification. Plant Physiol. 161, 1615–1633. doi: 10.1104/pp.112.208694.
Baxter, H. L., Mazarei, M., Labbe, N., Kline, L. M., Cheng, Q. K., Windham, M. T., et al. (2014). Two-year field analysis of reduced recalcitrance transgenic Switchgrass. Plant Biotechnol. J. 12, 914–924. doi: 10.1111/pbi.12195.
Begovic, L., Abicic, I., Lalic, A., Lepedus, H., Cesar, V., Leljak-Levanic, D. (2018). Lignin synthesis and accumulation in barley cultivars differing in their resistance to lodging. Plant Physiol. Biochem. 133, 142–148. doi: 10.1016/j.plaphy.2018.10.036.
Bellincampi, D., Cervone, F., Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00228.
Berni, R., Luyckx, M., Xu, X., Legay, S., Sergeant, K., Hausman, J. F., et al. (2019). Reactive oxygen species and heavy metal stress in plants: Impact on the cell wall and secondary metabolism. Environ. Exp. Bot. 161, 98–106. doi: 10.1016/j.envexpbot.2018.10.017.
Bhuiyan, N. H., Selvaraj, G., Wei, Y. D., King, J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. doi: 10.1093/jxb/ern290.
Bisht, D., Kumar, N., Singh, Y., Malik, R., Djalovic, I., Dhaka, N. S., et al. (2022). Effect of stem structural characteristics and cell wall components related to stem lodging resistance in a newly identified mutant of hexaploid wheat (Triticum aestivum L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1067063.
Blaschek, L., Murozuka, E., Serk, H., Menard, D., Pesquet, E. (2022). Different combinations of laccase paralogs nonredundantly control the amount and composition of lignin in specific cell types and cell wall layers in Arabidopsis. Plant Cell 35 (2), 889–909. doi: 10.1093/plcell/koac344
Boija, E., Lundquist, A., Edwards, K., Johansson, G. (2007). Evaluation of bilayer disks as plant cell membrane models in partition studies. Analytical Biochem. 364, 145–152. doi: 10.1016/j.ab.2007.02.012.
Bonawitz, N. D., Chapple, C. (2010). “The genetics of lignin biosynthesis: connecting genotype to phenotype,” in Annual Review of Genetics, vol. 44. (Annual Reviews, Palo Alto).
Bonawitz, N. D., Chapple, C. (2013). Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24, 336–343. doi: 10.1016/j.copbio.2012.11.004.
Bout, S., Vermerris, W. (2003). A candidate-gene approach to clone the sorghum Brown midrib gene encoding Caffeic acid O-Methyltransferase. Mol. Genet. Genomics 269, 205–214. doi: 10.1007/s00438-003-0824-4.
Bouyahya, A., El Omari, N., Bakrim, S., El Hachlafi, N., Balahbib, A., Wilairatana, P., et al. (2022). Advances in dietary phenolic compounds to improve chemosensitivity of anticancer drugs. Cancers 14. doi: 10.3390/cancers14194573.
Burlat, V., Kwon, M., Davin, L. B., Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. doi: 10.1016/S0031-9422(01)00117-0.
Calvo-Flores, F. G., Dobado, J. A. (2010). Lignin as renewable raw material. Chemsuschem 3, 1227–1235. doi: 10.1002/cssc.201000157.
Calvo-Polanco, M., Ribeyre, Z., Dauzat, M., Reyt, G., Hidalgo-Shrestha, C., Diehl, P., et al. (2021). Physiological roles of Casparian strips and suberin in the transport of water and solutes. New Phytol. 232, 2295–2307. doi: 10.1111/nph.17765.
Carmona, C., Langan, P., Smith, J. C., Petridis, L. (2015). Why genetic modification of lignin leads to low-recalcitrance biomass. Phys. Chem. Chem. Phys. 17, 358–364. doi: 10.1039/C4CP05004E.
Carunchio, F., Crescenzi, C., Girelli, A. M., Messina, A., Tarola, A. M. (2001). Oxidation of ferulic acid by laccase: identification of the products and inhibitory effects of some dipeptides. Talanta 55, 189–200. doi: 10.1016/S0039-9140(01)00417-9.
Cass, C. L., Peraldi, A., Dowd, P. F., Mottiar, Y., Santoro, N., Karlen, S. D., et al. (2015). Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 66, 4317–4335. doi: 10.1093/jxb/erv269.
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012.
Chaichi, M., Sanjarian, F., Razavi, K., Gonzalez-Hernandez, J. L. (2019). Analysis of transcriptional responses in root tissue of bread wheat landrace (Triticum aestivum L.) reveals drought avoidance mechanisms under water scarcity. PloS One 14, 23. doi: 10.1371/journal.pone.0212671
Chandrakanth, N. N., Zhang, C. C., Freeman, J., De Souza, W. R., Bartley, L. E., Mitchell, R. A. C. (2023). Modification of plant cell walls with hydroxycinnamic acids by BAHD acyltransferases. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1088879
Chen, M. J., Li, Y. D., Lu, F. C., Luterbacher, J. S., Ralph, J. (2023). Lignin hydrogenolysis: phenolic monomers from lignin and associated phenolates across plant clades. ACS Sustain. Chem. Eng. 11, 10001–10017. doi: 10.1021/acssuschemeng.3c01320.
Chen, F., Tobimatsu, Y., Havkin-Frenkel, D., Dixon, R. A., Ralph, J. (2012a). A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. United States America 109, 1772–1777. doi: 10.1073/pnas.1120992109.
Chen, W., Vanopdorp, N., Fitzl, D., Tewari, J., Friedemann, P., Greene, T., et al. (2012b). Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol. Biol. 80, 289–297. doi: 10.1007/s11103-012-9948-4.
Chmielewska, K., Rodziewicz, P., Swarcewicz, B., Sawikowska, A., Krajewski, P., Marczak, L., et al. (2016). Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01108
Chou, E. Y., Schuetz, M., Hoffmann, N., Watanabe, Y., Sibout, R., Samuels, A. L. (2018). Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 69, 1849–1859. doi: 10.1093/jxb/ery067.
Chowdhury, J., Henderson, M., Schweizer, P., Burton, R. A., Fincher, G. B., Little, A. (2014). Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp hordei. New Phytol. 204, 650–660. doi: 10.1111/nph.12974.
Christensen, C. S. L., Rasmussen, S. K. (2019). Low lignin mutants and reduction of lignin content in grasses for increased utilisation of lignocellulose. Agronomy-Basel 9, 21. doi: 10.3390/agronomy9050256.
Chu, S., Majumdar, A. (2012). Opportunities and challenges for a sustainable energy future. Nature 488, 294–303. doi: 10.1038/nature11475.
Chuang, W. P., Ray, S., Acevedo, F. E., Peiffer, M., Felton, G. W., Luthe, D. S. (2014). Herbivore cues from the fall armyworm (Spodoptera frugiperda) larvae trigger direct defenses in maize. Mol. Plant-Microbe Interact. 27, 461–470. doi: 10.1094/MPMI-07-13-0193-R.
Clissold, F. J. (2007). The biomechanics of chewing and plant fracture: Mechanisms and implications. Adv. Insect Physiology: Insect Mechanics Control 34, 317–372. doi: 10.1016/S0065-2806(07)34006-X.
d’Yvoire, M. B., Bouchabke-Coussa, O., Voorend, W., Antelme, S., Cezard, L., Legee, F., et al. (2013). Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 73, 496–508. doi: 10.1111/tpj.12053
Danby, A. M., Lundin, M. D., Subramaniam, B. (2018). Valorization of grass lignins: swift and selective recovery of pendant aromatic groups with ozone. ACS Sustain. Chem. Eng. 6, 71–76. doi: 10.1021/acssuschemeng.7b02978.
Davin, L. B., Lewis, N. G. (2005). Lignin primary structures and dirigent sites. Curr. Opin. Biotechnol. 16, 407–415. doi: 10.1016/j.copbio.2005.06.011.
Davin, L. B., Wang, H. B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., et al. (1997). Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366. doi: 10.1126/science.275.5298.362.
de Lange, E. S., Laplanche, D., Guo, H. J., Xu, W., Vlimant, M., Erb, M., et al. (2020). Spodoptera frugiperda caterpillars suppress herbivore-induced volatile emissions in maize. J. Chem. Ecol. 46, 344–360. doi: 10.1007/s10886-020-01153-x.
del Río, J. C., Rencoret, J., Gutierrez, A., Kim, H., Ralph, J. (2022). Unconventional lignin monomers-Extension of the lignin paradigm. LIGNIN AND HYDROXYCINNAMIC ACIDS: Biosynthesis Buildup Cell Wall 104, 1–39. doi: 10.1016/bs.abr.2022.02.001
del Rio, J. C., Rencoret, J., Prinsen, P., Martinez, A. T., Ralph, J., Gutierrez, A. (2012). Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60, 5922–5935. doi: 10.1021/jf301002n.
de Oliveira, D. M., Finger-Teixeira, A., Mota, T. R., Salvador, V. H., Moreira-Vilar, F. C., Molinari, H. B. C., et al. (2015). Ferulic acid: a key component in grass lignocellulose recalcitrance to hydrolysis. Plant Biotechnol. J. 13, 1224–1232. doi: 10.1111/pbi.12292.
de Souza, W. R., Martins, P. K., Freeman, J., Pellny, T. K., Michaelson, L. V., Sampaio, B. L., et al. (2018). Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytol. 218, 81–93. doi: 10.1111/nph.14970.
de Souza, W. R., Pacheco, T. F., Duarte, K. E., Sampaio, B. L., Molinari, P. A. D., Martins, P. K., et al. (2019). Silencing of a BAHD acyltransferase in sugarcane increases biomass digestibility. Biotechnol. Biofuels 12. doi: 10.1186/s13068-019-1450-7.
Dhokane, D., Karre, S., Kushalappa, A. C., McCartney, C. (2016). Integrated metabolo-transcriptomics reveals Fusarium head blight candidate resistance genes in wheat QTL-Fhb2. PloS One 11. doi: 10.1371/journal.pone.0155851.
Dima, O., Morreel, K., Vanholme, B., Kim, H., Ralph, J., Boerjan, W. (2015). Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27, 695–710. doi: 10.1105/tpc.114.134643.
Dixon, R. A., Barros, J. (2019). Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 9. doi: 10.1098/rsob.190215.
Djanaguiraman, M., Gowsiga, S., Govindaraj, M., Habyarimana, E., Senthil, A., Thavaprakaash, N., et al. (2023). Impact of root architecture and transpiration rate on drought tolerance in stay-green sorghum. Crop Sci. doi: 10.1002/csc2.21108.
Doblas, V. G., Geldner, N., Barberon, M. (2017). The endodermis, a tightly controlled barrier for nutrients. Curr. Opin. Plant Biol. 39, 136–143. doi: 10.1016/j.pbi.2017.06.010.
Doehlemann, G., Okmen, B., Zhu, W. J., Sharon, A. (2017). Plant pathogenic fungi. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.funk-0023-2016
Eloy, N. B., Voorend, W., Lan, W., Saleme, M. D. S., Cesarino, I., Vanholme, R., et al. (2017). Silencing CHALCONE SYNTHASE in maize impedes the incorporation of Tricin into lignin and increases lignin content. Plant Physiol. 173, 998–1016. doi: 10.1104/pp.16.01108.
Escamilla-Trevino, L. L., Shen, H., Uppalapati, S. R., Ray, T., Tang, Y. H., Hernandez, T., et al. (2010). Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 185, 143–155. doi: 10.1111/j.1469-8137.2009.03018.x
Eudes, A., Dutta, T., Deng, K., Jacquet, N., Sinha, A., Benites, V. T., et al. (2017). SbCOMT (Bmr12) is involved in the biosynthesis of Tricin-lignin in sorghum. PloS One 12. doi: 10.1371/journal.pone.0178160.
Eugene, A., Lapierre, C., Ralph, J. (2020). Improved analysis of arabinoxylan-bound hydroxycinnamate conjugates in grass cell walls. Biotechnol. Biofuels 13. doi: 10.1186/s13068-020-01841-6.
Fan, L., Linker, R., Gepstein, S., Tanimoto, E., Yamamoto, R., Neumann, P. M. (2006). Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol. 140, 603–612. doi: 10.1104/pp.105.073130.
Feduraev, P., Skrypnik, L., Riabova, A., Pungin, A., Tokupova, E., Maslennikov, P., et al. (2020). Phenylalanine and Tyrosine as Exogenous Precursors of Wheat (Triticum aestivum L.) Secondary Metabolism through PAL-Associated Pathways. Plants-Basel 9. doi: 10.3390/plants9040476
Feijao, C., Morreel, K., Anders, N., Tryfona, T., Busse-Wicher, M., Kotake, T., et al. (2022). Hydroxycinnamic acid-modified xylan side chains and their cross-linking products in rice cell walls are reduced in the Xylosyl arabinosyl substitution of xylan 1 mutant. Plant J. 109, 1152–1167. doi: 10.1111/tpj.15620.
Feng, X. J., Jia, L., CAI, Y. T., Guan, H. R., Zheng, D., Zhang, W. X., et al. (2022). ABA-inducible DEEPER ROOTING 1 improves adaptation of maize to water deficiency. Plant Biotechnol. J. 20, 2077–2088. doi: 10.1111/pbi.13889.
Fornale, S., Rencoret, J., Garcia-Calvo, L., Encina, A., Rigau, J., Gutierrez, A., et al. (2017). Changes in cell wall polymers and degradability in maize mutants lacking 3 ' -and 5 '-O-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol. 58, 240–255. doi: 10.1093/pcp/pcw198
Fosu-Nyarko, J., Jones, M. G. K. (2016). Advances in understanding the molecular mechanisms of root lesion nematode host interactions. Annu. Rev. Phytopathol. 54, 253–278. doi: 10.1146/annurev-phyto-080615-100257.
Francoz, E., Ranocha, P., Nguyen-Kim, H., Jamet, E., Burlat, V., Dunand, C. (2015). Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21. doi: 10.1016/j.phytochem.2014.07.020.
Franke, R., Hemm, M. R., Denault, J. W., Ruegger, M. O., Humphreys, J. M., Chapple, C. (2002a). Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30, 47–59. doi: 10.1046/j.1365-313X.2002.01267.x.
Franke, R., Humphreys, J. M., Hemm, M. R., Denault, J. W., Ruegger, M. O., Cusumano, J. C., et al. (2002b). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45. doi: 10.1046/j.1365-313X.2002.01266.x.
Fu, C. X., Mielenz, J. R., Xiao, X. R., Ge, Y. X., Hamilton, C. Y., Rodriguez, M., et al. (2011a). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from Switchgrass. Proc. Natl. Acad. Sci. United States America 108, 3803–3808. doi: 10.1073/pnas.1100310108.
Fu, C. X., Xiao, X. R., Xi, Y. J., Ge, Y. X., Chen, F., Bouton, J., et al. (2011b). Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved Saccharification efficiency in Switchgrass. Bioenergy Res. 4, 153–164. doi: 10.1007/s12155-010-9109-z.
Fujita, S., De Bellis, D., Edel, K. H., Koester, P., Andersen, T. G., Schmid-Siegert, E., et al. (2020). SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J. 39. doi: 10.15252/embj.2019103894.
Galeng-Lawilao, J., Kumar, A., Cabasan, M. T. N., De Waele, D. (2019). Comparison of the penetration, development and reproduction of Meloidogyne graminicola, and analysis of lignin and total phenolic content in partially resistant and resistant recombinant inbred lines of Oryza sativa. Trop. Plant Pathol. 44, 171–182. doi: 10.1007/s40858-018-0267-4.
Gao, M. Y., Chen, X. W., Huang, W. X., Wu, L., Yu, Z. S., Xiang, L., et al. (2021). Cell wall modification induced by an Arbuscular mycorrhizal fungus enhanced cadmium fixation in rice root. J. Hazardous Materials 416. doi: 10.1016/j.jhazmat.2021.125894.
Gao, Y.-Q., Huang, J.-Q., Reyt, G., Song, T., Love, A., Tiemessen, D., et al. (2023). A dirigent protein complex directs lignin polymerization and assembly of the root diffusion barrier. Science 382 (6669), 464–471. doi: 10.1126/science.adi5032.
Gebhardt, Y., Witte, S., Forkmann, G., Lukacin, R., Matern, U., Martens, S. (2005). Molecular evolution of flavonoid dioxygenases in the family Apiaceae. Phytochemistry 66, 1273–1284. doi: 10.1016/j.phytochem.2005.03.030.
Gebhardt, Y. H., Witte, S., Steuber, H., Matern, U., Martens, S. (2007). Evolution of flavone synthase I from parsley flavanone 3 beta-hydroxylase by site-directed mutagenesis. Plant Physiol. 144, 1442–1454. doi: 10.1104/pp.107.098392.
Gesteiro, N., Butron, A., Estevez, S., Santiago, R. (2021). Unraveling the role of maize (Zea mays L.) cell-wall phenylpropanoids in stem-borer resistance. Phytochemistry 185. doi: 10.1016/j.phytochem.2021.112683
Godin, B., Nagle, N., Sattler, S., Agneessens, R., Delcarte, J., Wolfrum, E. (2016). Improved sugar yields from biomass sorghum feedstocks: comparing low-lignin mutants and pretreatment chemistries. Biotechnol. Biofuels 9. doi: 10.1186/s13068-016-0667-y.
Grabber, J. H. (2005). How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci. 45, 820–831. doi: 10.2135/cropsci2004.0191.
Grabber, J. H., Hatfield, R. D., Lu, F. C., Ralph, J. (2008). Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9, 2510–2516. doi: 10.1021/bm800528f.
Grabber, J. H., Hatfield, R. D., Ralph, J. (1998). Diferulate cross-links impede the enzymatic degradation of non-lignified maize walls. J. Sci. Food Agric. 77, 193–200. doi: 10.1002/(ISSN)1097-0010.
Grabber, J. H., Quideau, S., Ralph, J. (1996). p-coumaroylated syringyl units in maize lignin: Implications for beta-ether cleavage by Thioacidolysis. Phytochemistry 43, 1189–1194. doi: 10.1016/S0031-9422(96)00431-1.
Grabber, J. H., Ralph, J., Hatfield, R. D. (2000). Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J. Agric. Food Chem. 48, 6106–6113. doi: 10.1021/jf0006978.
Grabber, J. H., Ralph, J., Lapierre, C., Barriere, Y. (2004). Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. Comptes Rendus Biologies 327, 455–465. doi: 10.1016/j.crvi.2004.02.009.
Green, A. R., Lewis, K. M., Barr, J. T., Jones, J. P., Lu, F. C., Ralph, J., et al. (2014). Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations. Plant Physiol. 165, 1440–1456. doi: 10.1104/pp.114.241729.
Grossman, A., Vermerris, W. (2019). Lignin-based polymers and nanomaterials. Curr. Opin. Biotechnol. 56, 112–120. doi: 10.1016/j.copbio.2018.10.009.
Grover, S., Cardona, J. B., Zogli, P., Alvarez, S., Naldrett, M. J., Sattler, S. E., et al. (2022a). Reprogramming of sorghum proteome in response to sugarcane aphid infestation. Plant Sci. 320. doi: 10.1016/j.plantsci.2022.111289.
Grover, S., Shinde, S., Puri, H., Palmer, N., Sarath, G., Sattler, S. E., et al. (2022b). Dynamic regulation of phenylpropanoid pathway metabolites in modulating sorghum defense against fall armyworm. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1019266.
Gui, M. Y., Wang, D., Xiao, H. H., Tu, M., Li, F. L., Li, W. C., et al. (2018). Studies of the relationship between rice stem composition and lodging resistance. J. Agric. Sci. 156, 387–395. doi: 10.1017/S0021859618000369.
Gunnaiah, R., Kushalappa, A. C. (2014). Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against Trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 83, 40–50. doi: 10.1016/j.plaphy.2014.07.002.
Guo, J., Cheng, Y. L. (2022). Advances in fungal elicitor-triggered plant immunity. Int. J. Mol. Sci. 23. doi: 10.3390/ijms231912003.
Ha, C. M., Escamilla-Trevino, L., Yarce, J. C. S., Kim, H., Ralph, J., Chen, F., et al. (2016). An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 86, 363–375. doi: 10.1111/tpj.13177.
Ha, C. M., Rao, X. L., Saxena, G., Dixon, R. A. (2021). Growth-defense trade-offs and yield loss in plants with engineered cell walls. New Phytol. 231, 60–74. doi: 10.1111/nph.17383.
Halpin, C., Holt, K., Chojecki, J., Oliver, D., Chabbert, B., Monties, B., et al. (1998). Brown-midrib maize (bm1) - a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14, 545–553. doi: 10.1046/j.1365-313X.1998.00153.x.
Hatfield, R., Jung, H., Marita, J., Kim, H. (2018). Cell wall characteristics of a maize mutant selected for decreased ferulates. Am. J. Plant Sci. 9 (3), 446–466. doi: 10.4236/ajps.2018.93034.
Hatfield, R. D., Marita, J. M. (2010). Enzymatic processes involved in the incorporation of hydroxycinnamates into grass cell walls. Phytochem. Rev. 9, 35–45. doi: 10.1007/s11101-010-9165-1.
Hatfield, R., Ralph, J., Grabber, J. H. (2008). A potential role for sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta 228, 919–928. doi: 10.1007/s00425-008-0791-4.
Hatfield, R. D., Rancour, D. M., Marita, J. M. (2017). Grass cell walls: A story of cross-linking. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02056.
Hatfield, R., Vermerris, W. (2001). Lignin formation in plants. The dilemma of linkage specificity. Plant Physiol. 126, 1351–1357. doi: 10.1104/pp.126.4.1351.
Hazman, M., Brown, K. M. (2018). Progressive drought alters architectural and anatomical traits of rice roots. Rice 11, 16. doi: 10.1186/s12284-018-0252-z.
Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., et al. (2004). Silencing of hydroxycinnamoy-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16, 1446–1465. doi: 10.1105/tpc.020297.
Holbein, J., Franke, R. B., Marhavy, P., Fujita, S., Gorecka, M., Sobczak, M., et al. (2019). Root endodermal barrier system contributes to defence against plant-parasitic cyst and root-knot nematodes. Plant J. 100, 221–236. doi: 10.1111/tpj.14459.
Holbein, J., Grundler, F. M. W., Siddique, S. (2016). Plant basal resistance to nematodes: an update. J. Exp. Bot. 67, 2049–2061. doi: 10.1093/jxb/erw005.
Hosmani, P. S., Kamiya, T., Danku, J., Naseer, S., Geldner, N., Guerinot, M. L., et al. (2013). Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. United States America 110, 14498–14503. doi: 10.1073/pnas.1308412110.
Ho-Yue-Kuang, S., Alvarado, C., Antelme, S., Bouchet, B., Cezard, L., Le Bris, P., et al. (2016). Mutation in Brachypodium caffeic acid O-methyltransferase 6 alters stem and grain lignins and improves straw saccharification without deteriorating grain quality. J. Exp. Bot. 67, 227–237. doi: 10.1093/jxb/erv446
Hsieh, C. Y., Huang, Y. H., Yeh, H. H., Hong, P. Y., Hsiao, C. J., Hsieh, L. S. (2021). Phenylalanine, Tyrosine, and DOPA Are bona fide Substrates for Bambusa oldhamii BoPAL4. Catalysts 11. doi: 10.3390/catal11111263.
Huang, J. L., Gu, M., Lai, Z. B., Fan, B. F., Shi, K., Zhou, Y. H., et al. (2010). Functional analysis of the arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370.
Hussey, S. G. (2022). Transcriptional regulation of secondary cell wall formation and lignification. LIGNIN AND HYDROXYCINNAMIC ACIDS: Biosynthesis Buildup Cell Wall 104, 317–361. doi: 10.1016/bs.abr.2022.03.007.
Islam, B. U., Suhail, M., Khan, M. K., Zughaibi, T. A., Alserihi, R. F., Zaidi, S. K., et al. (2021). Polyphenols as anticancer agents: Toxicological concern to healthy cells. Phytotherapy Res. 35, 6063–6079. doi: 10.1002/ptr.7216.
Israel, H. W., Wilson, R. G., Aist, J. R., Kunoh, H. (1980). CELL-WALL APPOSITIONS AND PLANT-DISEASE RESISTANCE - ACOUSTIC MICROSCOPY OF PAPILLAE THAT BLOCK FUNGAL INGRESS. Proc. Natl. Acad. Sci. United States America-Biological Sci. 77, 2046–2049. doi: 10.1073/pnas.77.4.2046.
Janczak-Pieniazek, M., Cichonski, J., Michalik, P., Chrzanowski, G. (2023). Effect of heavy metal stress on phenolic compounds accumulation in winter wheat plants. Molecules 28. doi: 10.3390/molecules28010241
Jeon, H. S., Jang, E., Kim, J., Kim, S. H., Lee, M. H., Nam, M. H., et al. (2022). Pathogen-induced autophagy regulates monolignol transport and lignin formation in plant immunity. Autophagy 9 (2), 597–615. doi: 10.1080/15548627.2022.2085496
Ji, H. L., Kyndt, T., He, W., Vanholme, B., Gheysen, G. (2015). beta-aminobutyric acid-induced resistance against root-knot nematodes in rice is based on increased basal defense. Mol. Plant-Microbe Interact. 28, 519–533. doi: 10.1094/MPMI-09-14-0260-R.
Jiang, N., Doseff, A. I., Grotewold, E. (2016). Flavones: from biosynthesis to health benefits. Plants-Basel 5. doi: 10.3390/plants5020027.
Jun, S. Y., Sattler, S. A., Cortez, G. S., Vermerris, W., Sattler, S. E., Kanga, C. H. (2018). Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol. 176, 1452–1468. doi: 10.1104/pp.17.01608.
Jun, S. Y., Walker, A. M., Kim, H., Ralph, J., Vermerris, W., Sattler, S. E., et al. (2017). The enzyme activity and substrate specificity of two major cinnamyl alcohol dehydrogenases in sorghum (Sorghum bicolor), sbCAD2 and sbCAD4. Plant Physiol. 174, 2128–2145. doi: 10.1104/pp.17.00576.
Jung, J. H., Altpeter, F. (2016). TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 92, 131–142. doi: 10.1007/s11103-016-0499-y.
Jung, S. E., Bang, S. W., Kim, S. H., Seo, J. S., Yoon, H. B., Kim, Y. S., et al. (2021). Overexpression of OsERF83, a vascular tissue-specific transcription factor gene, confers drought tolerance in rice. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22147656.
Jung, S. E., Kim, T. H., Shim, J. S., Bang, S. W., Bin Yoon, H., Oh, S. H., et al. (2022). Rice NAC17 transcription factor enhances drought tolerance by modulating lignin accumulation. Plant Sci. 323. doi: 10.1016/j.plantsci.2022.111404.
Jung, H. G., Phillips, R. L. (2010). Putative seedling ferulate ester (sfe) maize mutant: morphology, biomass yield, and stover cell wall composition and rumen degradability. Crop Sci. 50, 403–418. doi: 10.2135/cropsci2009.04.0191.
Kalra, A., Goel, S., Elias, A. A. (2023). Understanding role of roots in plant response to drought: Way forward to climate-resilient crops. Plant Genome e20395. doi: 10.1002/tpg2.20395.
Karlen, S. D., Free, H. C. A., Padmakshan, D., Smith, B. G., Ralph, J., Harris, P. J. (2018). Commelinid monocotyledon Lignins are acylated by p-coumarate. Plant Physiol. 177, 513–521. doi: 10.1104/pp.18.00298.
Karlen, S. D., Zhang, C. C., Peck, M. L., Smith, R. A., Padmakshan, D., Helmich, K. E., et al. (2016). Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2. doi: 10.1104/pp.18.00298
Karre, S., Kim, S. B., Kim, B. S., Khangura, R. S., Sermons, S. M., Dilkes, B., et al. (2021). Maize plants chimeric for an autoactive resistance gene display a cell-autonomous hypersensitive response but non-cell autonomous defense signaling. Mol. Plant-Microbe Interact. 34, 606–616. doi: 10.1094/MPMI-04-20-0091-R.
Karre, S., Kumar, A., Dhokane, D., Kushalappa, A. C. (2017). Metabolo-transcriptome profiling of barley reveals induction of chitin elicitor receptor kinase gene (HvCERK1) conferring resistance against Fusarium graminearum. Plant Mol. Biol. 93, 247–267. doi: 10.1007/s11103-016-0559-3.
Kawasaki, T., Koita, H., Nakatsubo, T., Hasegawa, K., Wakabayashi, K., Takahashi, H., et al. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. United States America 103, 230–235. doi: 10.1073/pnas.0509875103.
Kim, H., Rencoret, J., Elder, T. J., Del Rio, J. C., Ralph, J. (2023). Biomimetic oxidative copolymerization of hydroxystilbenes and monolignols. Sci. Adv. 9. doi: 10.1126/sciadv.ade5519
Kisa, D., Elmastas, M., Ozturk, L., Kayir, O. (2016). Responses of the phenolic compounds of Zea mays under heavy metal stress. Appl. Biol. Chem. 59, 813–820. doi: 10.1007/s13765-016-0229-9.
Kishimoto, T., Uraki, Y., Ubukata, M. (2005). Easy synthesis of beta-O-4 type lignin related polymers. Organic Biomolecular Chem. 3, 1067–1073. doi: 10.1039/B416699J.
Kolbeck, A., Marhavy, P., De Bellis, D., Li, B. H., Kamiya, T., Fujiwara, T., et al. (2022). CASP microdomain formation requires cross cell wall stabilization of domains and non-cell autonomous action of LOTR1. Elife 11. doi: 10.7554/eLife.69602.sa2.
Kong, L. A., Wu, D. Q., Huang, W. K., Peng, H., Wang, G. F., Cui, J. K., et al. (2015). Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics 16. doi: 10.1186/s12864-015-2037-8.
Kruger, W. M., Carver, T. L. W., Zeyen, R. J. (2002). Effects of inhibiting phenolic biosynthesis on penetration resistance of barley isolines containing seven powdery mildew resistance genes or alleles. Physiol. Mol. Plant Pathol. 61, 41–51. doi: 10.1016/S0885-5765(02)90415-7.
Krzeslowska, M. (2011). The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiologiae Plantarum 33, 35–51. doi: 10.1007/s11738-010-0581-z.
Lam, P. Y., Liu, H. J., Lo, C. (2015). Completion of Tricin biosynthesis pathway in rice: cytochrome P450 75B4 is a unique Chrysoeriol 5 '-hydroxylase. Plant Physiol. 168, 1527–U760. doi: 10.1104/pp.15.00566.
Lam, P. Y., Tobimatsu, Y., Matsumoto, N., Suzuki, S., Lan, W., Takeda, Y., et al. (2019). OsCAldOMT1 is a bifunctional O-methyltransferase involved in the biosynthesis of tricin-lignins in rice cell walls. Sci. Rep. 9. doi: 10.1038/s41598-019-47957-0
Lam, P. Y., Tobimatsu, Y., Takeda, Y., Suzuki, S., Yamamura, M., Umezawa, T., et al. (2017). Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 174, 972–985. doi: 10.1104/pp.16.01973.
Lam, P. Y., Wang, L. X., Lui, A. C. W., Liu, H. J., Takeda-Kimura, Y., Chen, M. X., et al. (2022). Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol. 188, 1993–2011. doi: 10.1093/plphys/kiab606.
Lam, P. Y., Zhu, F. Y., Chan, W. L., Liu, H. J., Lo, C. (2014). Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of Tricin O-linked conjugates in rice. Plant Physiol. 165, 1315–1327. doi: 10.1104/pp.114.239723.
Lamb, A., Weers, B., Mckinley, B., Rooney, W., Morgan, C., Marshall-Colon, A., et al. (2022). Bioenergy sorghum's deep roots: A key to sustainable biomass production on annual cropland. Global Change Biol. Bioenergy 14, 132–156. doi: 10.1111/gcbb.12907.
Lan, W., Lu, F. C., Regner, M., Zhu, Y. M., Rencoret, J., Ralph, S. A., et al. (2015). Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 167, 1284–U265. doi: 10.1104/pp.114.253757.
Lan, W., Morreel, K., Lu, F. C., Rencoret, J., Del Rio, J. C., Voorend, W., et al. (2016a). Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol. 171, 810–820. doi: 10.1104/pp.16.02012.
Lan, W., Rencoret, J., Lu, F. C., Karlen, S. D., Smith, B. G., Harris, P. J., et al. (2016b). Tricin-lignins: occurrence and quantitation of Tricin in relation to phylogeny. Plant J. 88, 1046–1057. doi: 10.1111/tpj.13315.
Lapierre, C., Sibout, R., Laurans, F., Lesage-Descauses, M. C., Déjardin, A., Pilate, G. (2021). p-Coumaroylation of poplar lignins impacts lignin structure and improves wood saccharification. Plant Physiol. 187, 1374–1386. doi: 10.1093/plphys/kiab359.
Le Bris, P., Wang, Y., Barbereau, C., Antelme, S., Cezard, L., Legee, F., et al. (2019). Inactivation of LACCASE8 and LACCASE5 genes in Brachypodium distachyon leads to severe decrease in lignin content and high increase in saccharification yield without impacting plant integrity. Biotechnol. Biofuels 12, 11. doi: 10.1186/s13068-019-1525-5.
Lee, M. H., Jeon, H. S., Kim, S. H., Chung, J. H., Roppolo, D., Lee, H. J., et al. (2019). Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 38, 17. doi: 10.15252/embj.2019101948.
Lee, D. K., Jung, H., Jang, G., Jeong, J. S., Kim, Y. S., Ha, S. H., et al. (2016). Overexpression of the osERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 172, 575–588. doi: 10.1104/pp.16.00379.
Lee, Y. J., Kim, J. H., Kim, B. G., Lim, Y., Ahn, J. H. (2008). Characterization of flavone synthase I from rice. Bmb Rep. 41, 68–71. doi: 10.5483/BMBRep.2008.41.1.068.
Lee, Y., Rubio, M. C., Alassimone, J., Geldner, N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412. doi: 10.1016/j.cell.2013.02.045.
Lee, J. H., Won, H. J., Tran, P. H. N., Lee, S. M., Kim, H. Y., Jung, J. H. (2021). Improving lignocellulosic biofuel production by CRISPR/Cas9-mediated lignin modification in barley. Global Change Biol. Bioenergy 13, 742–752. doi: 10.1111/gcbb.12808.
Le Roy, J., Huss, B., Creach, A., Hawkins, S., Neutelings, G. (2016). Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00735
Li, Q., Fu, C. F., Liang, C. L., Ni, X. J., Zhao, X. H., Chen, M., et al. (2022a). Crop lodging and the roles of lignin, cellulose, and hemicellulose in lodging resistance. Agronomy-Basel 12. doi: 10.3390/agronomy12081795.
Li, H., Huang, Y. H. (2017). Expression of brown-midrib in a spontaneous sorghum mutant is linked to a 5 '-UTR deletion in lignin biosynthesis gene SbCAD2. Sci. Rep. 7. doi: 10.1038/s41598-017-10119-1
Li, G. T., Jones, K. C., Eudes, A., Pidatala, V. R., Sun, J., Xu, F., et al. (2018a). Overexpression of a rice BAHD acyltransferase gene in switchgrass (Panicum virgatum L.) enhances saccharification. BMC Biotechnol. 18. doi: 10.1186/s12896-018-0464-8.
Li, Y. D., Li, S., Kim, H., Motagamwala, A. H., Mobley, J. K., Yue, F., et al. (2018b). An "ideal lignin" facilitates full biomass utilization. Sci. Adv. 4. doi: 10.1126/sciadv.aau2968.
Li, Y. P., Shi, S. Q., Zhang, Y., Zhang, A. M., Wang, Z. F., Yang, Y. L. (2023). Copper stress-induced phytotoxicity associated with photosynthetic characteristics and lignin metabolism in wheat seedlings. Ecotoxicology Environ. Saf. 254. doi: 10.1016/j.ecoenv.2023.114739.
Li, R., Zhao, Y., Sun, Z., Wu, Z. Y., Wang, H. L., Fu, C. X., et al. (2022b). Genome-wide identification of Switchgrass laccases involved in lignin biosynthesis and heavy-metal responses. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23126530.
Lionetti, V., Giancaspro, A., Fabri, E., Giove, S. L., Reem, N., Zabotina, O. A., et al. (2015). Cell wall traits as potential resources to improve resistance of durum wheat against Fusarium graminearum. BMC Plant Biol. 15. doi: 10.1186/s12870-014-0369-1.
Liu, D. F., Chen, X. J., Liu, J. Q., Ye, J. C., Guo, Z. J. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079.
Liu, Q. Q., Luo, L., Wang, X. X., Shen, Z. G., Zheng, L. Q. (2017). Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of osLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 18. doi: 10.3390/ijms18020209.
Liu, X. Y., van Acker, R., Voorend, W., Pallidis, A., Goeminne, G., Pollier, J., et al. (2021b). Rewired phenolic metabolism and improved saccharification efficiency of a Zea mays cinnamyl alcohol dehydrogenase 2 (zmcad2) mutant. Plant J. 105, 1240–1257. doi: 10.1111/tpj.15108.
Liu, C., Yu, H. S., Rao, X. L., Li, L. G., Dixon, R. A. (2021a). Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. United States America 118. doi: 10.1073/pnas.2010911118.
Liu, Q. Q., Zheng, L., He, F., Zhao, F. J., Shen, Z. G., Zheng, L. Q. (2015). Transcriptional and physiological analyses identify a regulatory role for hydrogen peroxide in the lignin biosynthesis of copper-stressed rice roots. Plant Soil 387, 323–336. doi: 10.1007/s11104-014-2290-7.
Lu, F. C., Ralph, J. (1997). DFRC method for lignin analysys.: 1.: New method for βaryl ether cleavage:: Lignin model studies. J. Agric. Food Chem. 45, 4655–4660. doi: 10.1021/jf970539p
Lucas, P. W., Turner, I. M., Dominy, N. J., Yamashita, N. (2000). Mechanical defences to herbivory. Ann. Bot. 86, 913–920. doi: 10.1006/anbo.2000.1261.
Lui, A. C. W., Lam, P. Y., Chan, K. H., Wang, L. X., Tobimatsu, Y., Lo, C. V. (2020). Convergent recruitment of 5 '-hydroxylase activities by CYP75B flavonoid B-ring hydroxylases for Tricin biosynthesis in Medicago legumes. New Phytol. 228, 269–284. doi: 10.1111/nph.16498.
Ma, Q. H. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 61, 2735–2744. doi: 10.1093/jxb/erq107.
Ma, Q. H., Xu, Y. (2008). Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie 90, 515–524. doi: 10.1016/j.biochi.2007.09.016.
Malik, N. A. A., Kumar, I. S., Nadarajah, K. (2020). Elicitor and receptor molecules: orchestrators of plant defense and immunity. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21030963
Manga-Robles, A., Santiago, R., Malvar, R. A., Moreno-Gonzalez, V., Fornale, S., Lopez, I., et al. (2021). Elucidating compositional factors of maize cell walls contributing to stalk strength and lodging resistance. Plant Sci. 307. doi: 10.1016/j.plantsci.2021.110882.
Marita, J. M., Hatfield, R. D., Rancour, D. M., Frost, K. E. (2014). Identification and suppression of the p-coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea Mays L. Plant J. 78, 850–864. doi: 10.1111/tpj.12510.
Menard, D., Blaschek, L., Kriechbaum, K., Lee, C. C., Serk, H., Zhu, C. T., et al. (2022). Plant biomechanics and resilience to environmental changes are controlled by specific lignin chemistries in each vascular cell type and morphotype. Plant Cell 34, 4877–4896. doi: 10.1093/plcell/koac284.
Menden, B., Kohlhoff, M., Moerschbacher, B. M. (2007). Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 68, 513–520. doi: 10.1016/j.phytochem.2006.11.011.
Miao, Y. C., Liu, C. J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. United States America 107, 22728–22733. doi: 10.1073/pnas.1007747108
Mir, A. R., Pichtel, J., Hayat, S. (2021). Copper: uptake, toxicity and tolerance in plants and management of Cu-contaminated soil. Biometals 34, 737–759. doi: 10.1007/s10534-021-00306-z.
Miyamoto, T., Tobimatsu, Y., Umezawa, T. (2020). MYB-mediated regulation of lignin biosynthesis in grasses. Curr. Plant Biol. 24. doi: 10.1016/j.cpb.2020.100174.
Morikawa, Y., Yoshinaga, A., Kamitakahara, H., Wada, M., Takabe, K. (2010). Cellular distribution of coniferin in differentiating xylem of Chamaecyparis obtusa as revealed by Raman microscopy. Holzforschung 64, 61–67. doi: 10.1515/hf.2010.015.
Mota, T. R., De Souza, W. R., Oliveira, D. M., Martins, P. K., Sampaio, B. L., Vinecky, F., et al. (2021). Suppression of a BAHD acyltransferase decreases p-coumaroyl on arabinoxylan and improves biomass digestibility in the model grass Setaria viridis. Plant J. 105, 136–150. doi: 10.1111/tpj.15046.
Mottiar, Y., Vanholme, R., Boerjan, W., Ralph, J., Mansfield, S. D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200. doi: 10.1016/j.copbio.2015.10.009.
Moural, T. W., Lewis, K. M., Barnaba, C., Zhu, F., Palmer, N. A., Sarath, G., et al. (2017). Characterization of class III peroxidases from Switchgrass. Plant Physiol. 173, 417–433. doi: 10.1104/pp.16.01426.
Muhammad, A., Hao, H. H., Xue, Y. L., Alam, A., Bai, S. M., Hu, W. C., et al. (2020). Survey of wheat straw stem characteristics for enhanced resistance to lodging. Cellulose 27, 2469–2484. doi: 10.1007/s10570-020-02972-7.
Muszynska, A., Guendel, A., Melzer, M., Moya, Y. A. T., Roder, M. S., Rolletschek, H., et al. (2021). A mechanistic view on lodging resistance in rye and wheat: a multiscale comparative study. Plant Biotechnol. J. 19, 2646–2661. doi: 10.1111/pbi.13689.
Nakashima, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00170.
Nakayama, T., Shinohara, H., Tanaka, M., Baba, K., Ogawa-Ohnishi, M., Matsubayashi, Y. (2017). A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355, 284–286. doi: 10.1126/science.aai9057.
Naseer, S., Lee, Y., Lapierre, C., Franke, R., Nawrath, C., Geldner, N. (2012). Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. United States America 109, 10101–10106. doi: 10.1073/pnas.1205726109
Nicholson, R. L., Hammerschmidt, R. (1992). PHENOLIC-COMPOUNDS AND THEIR ROLE IN DISEASE RESISTANCE. Annu. Rev. Phytopathol. 30, 369–389. doi: 10.1146/annurev.py.30.090192.002101.
Ning, P., Yang, G. F., Hu, L. H., Sun, J. X., Shi, L. N., Zhou, Y. H., et al. (2021). Recent advances in the valorization of plant biomass. Biotechnol. Biofuels 14. doi: 10.1186/s13068-021-01949-3.
Ohtani, M., Demura, T. (2019). The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 56, 82–87. doi: 10.1016/j.copbio.2018.10.002.
Okubara, P. A., Peetz, A. B., Sharpe, R. M. (2019). Cereal root interactions with soilborne pathogensFrom trait to gene and back. Agronomy-Basel 9. doi: 10.3390/agronomy9040188.
Oliver, A. L., Pedersen, J. F., Grant, R. J., Klopfenstein, T. J. (2005). Comparative effects of the sorghum bmr-6 and bmr-12 genes: I. Forage sorghum yield and quality. Crop Sci. 45, 2234–2239. doi: 10.2135/cropsci2004.0644.
Ouyang, W. J., Yin, X. Y., Yang, J. C., Struik, P. C. (2020). Comparisons with wheat reveal root anatomical and histochemical constraints of rice under water-deficit stress. Plant Soil 452, 547–568. doi: 10.1007/s11104-020-04581-6.
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301. doi: 10.1093/jxb/erx141.
Park, H. L., Bhoo, S. H., Kwon, M., Lee, S. W., Cho, M. H. (2017). Biochemical and expression analyses of the rice cinnamoyl-coA reductase gene family. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02099.
Park, H. L., Kim, T. L., Bhoo, S. H., Lee, T. H., Lee, S. W., Cho, M. H. (2018). Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family. Molecules 23, 17. doi: 10.3390/molecules23102659.
Parrotta, L., Guerriero, G., Sergeant, K., Cal, G., Hausman, J. F. (2015). Target or barrier? The cell wall of early- and later-diverging plants vs cadmium toxicity: differences in the response mechanisms. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00133.
Pazhany, A. S., Henry, R. J. (2019). Genetic modification of biomass to alter lignin content and structure. Ind. Eng. Chem. Res. 58, 16190–16203. doi: 10.1021/acs.iecr.9b01163.
Perkins, M. L., Schuetz, M., Unda, F., Chen, K. T., Bally, M. B., Kulkarni, J. A., et al. (2022). Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell 34, 2080–2095. doi: 10.1093/plcell/koac051.
Perkins, M., Smith, R. A., Samuels, L. (2019). The transport of monomers during lignification in plants: anything goes but how? Curr. Opin. Biotechnol. 56, 69–74. doi: 10.1016/j.copbio.2018.09.011.
Petitot, A. S., Kyndt, T., Haidar, R., Dereeper, A., Collin, M., Engler, J. D., et al. (2017). Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 119, 885–899. doi: 10.1093/aob/mcw256.
Petrik, D. L., Karlen, S. D., Cass, C. L., Padmakshan, D., Lu, F. C., Liu, S., et al. (2014). p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J. 77, 713–726. doi: 10.1111/tpj.12420.
Pichon, M., Deswartes, C., Gerentes, D., Guillaumie, S., Lapierre, C., Toppan, A., et al. (2006). Variation in lignin and cell wall digestibility in caffeic acid O-methyltransferase down-regulated maize half-sib progenies in field experiments. Mol. Breed. 18, 253–261. doi: 10.1007/s11032-006-9033-2.
Piotrowski, J. S., Okada, H., Lu, F. C., Li, S. C., Hinchman, L., Ranjan, A., et al. (2015). Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc. Natl. Acad. Sci. United States America 112, E1490–E1497. doi: 10.1073/pnas.1410400112.
Piquemal, J., Chamayou, S., Nadaud, I., Beckert, M., Barriere, Y., Mila, I., et al. (2002). Down-regulation of caffeic acid O-methyltransf erase in maize revisited using a transgenic approach. Plant Physiol. 130, 1675–1685. doi: 10.1104/pp.012237.
Powell, J. J., Carere, J., Fitzgerald, T. L., Stiller, J., Covarelli, L., Xu, Q., et al. (2017). The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Ann. Bot. 119, 853–867. doi: 10.1093/aob/mcw207
Prats, E., Martinez, F., Rojas-Molina, M. M., Rubiales, D. (2007). Differential effects of phenylalanine ammonia lyase, cinnamyl alcohol dehydrogenase, and energetic metabolism inhibition on resistance of appropriate host and nonhost cereal-rust interactions. Phytopathology 97, 1578–1583. doi: 10.1094/PHYTO-97-12-1578.
Quideau, S., Ralph, J. (1997). Lignin-ferulate cross-links in grasses .4. Incorporation of 5-5-coupled dehydrodiferulate into synthetic lignin. J. Chem. Society-Perkin Trans. 1, 2351–2358. doi: 10.1039/a701808h.
Rajarammohan, S. (2021). Redefining plant-necrotroph interactions: the thin line between hemibiotrophs and necrotrophs. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.673518.
Ralph, J. (2010). Hydroxycinnamates in lignification. Phytochem. Rev. 9, 65–83. doi: 10.1007/s11101-009-9141-9.
Ralph, J., Brunow, G., Harris, P. J., Dixon, R. A., Schatz, P. F., Boerjan, W. (2008). Lignification: are Lignins Biosynthesized via simple Combinatorial Chemistry or via Proteinaceous Control and Template Replication? Recent Adv. Polyphenol Research Vol 1 1, 36–3+. doi: 10.1002/9781444302400.ch2
Ralph, J., Bunzel, M., Marita, J. M., Hatfield, R. D., Lu, F., Kim, H., et al. (2004a). Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem. Rev. 3, 79–96. doi: 10.1023/B:PHYT.0000047811.13837.fb.
Ralph, J., Grabber, J. H., Hatfield, R. D. (1995). LIGNIN-FERULATE CROSS-LINKS IN GRASSES - ACTIVE INCORPORATION OF FERULATE POLYSACCHARIDE ESTERS INTO RYEGRASS LIGNINS. Carbohydr. Res. 275, 167–178. doi: 10.1016/0008-6215(95)00237-N.
Ralph, J., Lapierre, C., Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi: 10.1016/j.copbio.2019.02.019.
Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P. F., et al. (2004b). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, 29–60. doi: 10.1023/B:PHYT.0000047809.65444.a4.
Ralph, J., Peng, J. P., Lu, F. C., Hatfield, R. D., Helm, R. F. (1999). Are lignins optically active? J. Agric. Food Chem. 47, 2991–2996. doi: 10.1021/jf9901136.
Ralph, J., Quideau, S., Grabber, J. H., Hatfield, R. D. (1994). IDENTIFICATION AND SYNTHESIS OF NEW FERULIC ACID DEHYDRODIMERS PRESENT IN GRASS CELL-WALLS. J. Chem. Society-Perkin Trans. 116 (21), 9448–9456. doi: 10.1021/ja00100a006.
Rao, X. L., Chen, X., Shen, H., Ma, Q., Li, G. F., Tang, Y. H., et al. (2019). Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum). Plant Biotechnol. J. 17, 580–593. doi: 10.1111/pbi.13000.
Ray, A. E., Williams, C. L., Hoover, A. N., Li, C. L., Sale, K. L., Emerson, R. M., et al. (2020). Multiscale characterization of lignocellulosic biomass variability and its implications to preprocessing and conversion: a case study for corn stover. ACS Sustain. Chem. Eng. 8, 3218–3230. doi: 10.1021/acssuschemeng.9b06763.
Renders, T., van den Bosch, S., Koelewijn, Schutyser, W., Sels, B. F. (2017). Lignin-first biomass fractionation: the advent of active stabilisation strategies. Energy Environ. Sci. 10, 1551–1557. doi: 10.1039/C7EE01298E.
Reyt, G., Ramakrishna, P., Salas-Gonzalez, I., Fujita, S., Love, A., Tiemessen, D., et al. (2021). Two chemically distinct root lignin barriers control solute and water balance. Nat. Commun. 12. doi: 10.1038/s41467-021-22550-0.
Richards, R. A., Passioura, J. B. (1989). A BREEDING PROGRAM TO REDUCE THE DIAMETER OF THE MAJOR XYLEM VESSEL IN THE SEMINAL ROOTS OF WHEAT AND ITS EFFECT ON GRAIN-YIELD IN RAIN-FED ENVIRONMENTS. Aust. J. Agric. Res. 40, 943–950. doi: 10.1071/AR9890943.
Ride, J. P. (1975). LIGNIFICATION IN WOUNDED WHEAT LEAVES IN RESPONSE TO FUNGI AND ITS POSSIBLE ROLE IN RESISTANCE. Physiol. Plant Pathol. 5, 125–134. doi: 10.1016/0048-4059(75)90016-8.
Righini, S., Rodriguez, E. J., Berosich, C., Grotewold, E., Casati, P., Ferreyra, M. L. F. (2019). Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant Cell Environ. 42, 495–508. doi: 10.1111/pce.13428.
Riyazuddin, R., Nisha, N., Ejaz, B., Khan, M. I. R., Kumar, M., Ramteke, P. W. W., et al. (2022). A comprehensive review on the heavy metal toxicity and sequestration in plants. Biomolecules 12. doi: 10.3390/biom12010043
Rockstrom, J., Edenhofer, O., Gaertner, J., Declerck, F. (2020). Planet-proofing the global food system. Nat. Food 1, 3–5. doi: 10.1038/s43016-019-0010-4.
Rojas-Murcia, N., Hematy, K., Lee, Y., Emonet, A., Ursache, R., Fujita, S., et al. (2020). High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc. Natl. Acad. Sci. United States America 117, 29166–29177. doi: 10.1073/pnas.2012728117
Rong, W., Luo, M. Y., Shan, T. L., Wei, X. N., Du, L. P., Xu, H. J., et al. (2016). A wheat cinnamyl alcohol dehydrogenase taCAD12 contributes to host resistance to the sharp eyespot disease. Front. Plant Sci. 7, 15. doi: 10.3389/fpls.2016.01723.
Rosler, J., Krekel, F., Amrhein, N., Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179. doi: 10.1104/pp.113.1.175.
Ruiz, H. A., Sganzerla, W. G., Larnaudie, V., Veersma, R. J., van Erven, G., Shiva, Ríos-González, L. J., et al. (2023). Advances in process design, techno-economic assessment and environmental aspects for hydrothermal pretreatment in the fractionation of biomass under biorefinery concept. Bioresource Technol. 369. doi: 10.1016/j.biortech.2022.128469.
Rutter, W. B., Franco, J., Gleason, C. (2022). Rooting out the mechanisms of root-knot nematode-plant interactions. Annu. Rev. Phytopathol. 60, 43–76. doi: 10.1146/annurev-phyto-021621-120943.
Saathoff, A. J., Hargrove, M. S., Haas, E. J., Tobias, C. M., Twigg, P., Sattler, S., et al. (2012). Switchgrass pviCAD1: understanding residues important for substrate preferences and activity. Appl. Biochem. Biotechnol. 168, 1086–1100. doi: 10.1007/s12010-012-9843-0.
Saathoff, A. J., Sarath, G., Chow, E. K., Dien, B. S., Tobias, C. M. (2011a). Downregulation of cinnamyl-alcohol dehydrogenase in Switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PloS One 6, 10. doi: 10.1371/journal.pone.0016416.
Saathoff, A. J., Tobias, C. M., Sattler, S. E., Haas, E. J., Twigg, P., Sarath, G. (2011b). Switchgrass contains two cinnamyl alcohol dehydrogenases involved in lignin formation. Bioenergy Res. 4, 120–133. doi: 10.1007/s12155-010-9106-2.
Saballos, A., Ejeta, G., Sanchez, E., Kang, C., Vermerris, W. (2009). A Genomewide Analysis of the Cinnamyl Alcohol Dehydrogenase Family in Sorghum Sorghum bicolor (L.) Moench Identifies SbCAD2 as the Brown midrib6 Gene. Genetics 181, 783–795. doi: 10.1534/genetics.108.098996.
Saleme, M. D. S., Cesarino, I., Vargas, L., Kim, H., Vanholme, R., Goeminne, G., et al. (2017). Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves Saccharification in poplar. Plant Physiol. 175, 1040–1057. doi: 10.1104/pp.17.00920.
Saluja, M., Zhu, F. Y., Yu, H. F., Walia, H., Sattler, S. E. (2021). Loss of COMT activity reduces lateral root formation and alters the response to water limitation in sorghum brown midrib (bmr) 12 mutant. New Phytol. 229, 2780–2794. doi: 10.1111/nph.17051.
Sangha, A. K., Parks, J. M., Standaert, R. F., Ziebell, A., Davis, M., Smith, J. C. (2012). Radical coupling reactions in lignin synthesis: A density functional theory study. J. Phys. Chem. B 116, 4760–4768. doi: 10.1021/jp2122449.
Santiago, R., Barros-RIOS, J., Malvar, R. A. (2013). Impact of cell wall composition on maize resistance to pests and diseases. Int. J. Mol. Sci. 14, 6960–6980. doi: 10.3390/ijms14046960.
Santiago, R., Cao, A., Butron, A., Lopez-Malvar, A., Rodriguez, V. M., Sandoya, G. V., et al. (2017). Defensive changes in maize leaves induced by feeding of Mediterranean corn borer larvae. BMC Plant Biol. 17. doi: 10.1186/s12870-017-0991-9.
Santiago, R., Malvar, R. A., Barros-Rios, J., Samayoa, L. F., Butron, A. (2016). Hydroxycinnamate synthesis and association with Mediterranean corn borer resistance. J. Agric. Food Chem. 64, 539–551. doi: 10.1021/acs.jafc.5b04862.
Sattler, S. E., Funnell-Harris, D. L. (2013). Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00070.
Sattler, S. E., Funnell-Harris, D. L., Pedersen, J. F. (2010). Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci. 178, 229–238. doi: 10.1016/j.plantsci.2010.01.001.
Sattler, S. E., Palmer, N. A., Saballos, A., Greene, A. M., Xin, Z. G., sarath, G., et al. (2012). Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Res. 5, 855–865. doi: 10.1007/s12155-012-9197-z.
Sattler, S. E., Saathoff, A. J., Haas, E. J., Palmer, N. A., Funnell-Harris, D. L., Sarath, G., et al. (2009). A Nonsense Mutation in a Cinnamyl Alcohol Dehydrogenase Gene Is Responsible for the Sorghum brown midrib6 Phenotype. Plant Physiol. 150, 584–595. doi: 10.1104/pp.109.136408.
Sattler, S. A., Walker, A. M., Vermerris, W., Sattler, S. E., Kang, C. (2017). Structural and biochemical characterization of cinnamoyl-coA reductases. Plant Physiol. 173, 1031–1044. doi: 10.1104/pp.16.01671.
Saur, I. M. L., Huckelhoven, R. (2021). Recognition and defence of plant-infecting fungal pathogens. J. Plant Physiol. 256. doi: 10.1016/j.jplph.2020.153324.
Schuetz, M., Benske, A., Smith, R. A., Watanabe, Y., Tobimatsu, Y., Ralph, J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–U489. doi: 10.1104/pp.114.245597.
Sedo, J., Saiz-Poseu, J., Busque, F., Ruiz-Molina, D. (2013). Catechol-based biomimetic functional materials. Advanced Materials 25, 653–701. doi: 10.1002/adma.201202343
Serrani-Yarce, J. C., Escamilla-Trevino, L., Barros, J., Gallego-Giraldo, L., Pu, Y. Q., Ragauskas, A., et al. (2021). Targeting hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase for lignin modification in Brachypodium distachyon. Biotechnol. Biofuels 14. doi: 10.1186/s13068-021-01905-1.
Shafiei, R., Hooper, M., McClellan, C., Oakey, H., Stephens, J., Lapierre, C., et al. (2023). Downregulation of barley ferulate 5-hydroxylase dramatically alters straw lignin structure without impact on mechanical properties. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1125003.
Shimada, N., Munekata, N., Tsuyama, T., Matsushita, Y., Fukushima, K., Kijidani, Y., et al. (2021). Active transport of lignin precursors into membrane vesicles from lignifying tissues of bamboo. Plants-Basel 10. doi: 10.3390/plants10112237.
Simoes, M. S., Carvalho, G. G., Ferreira, S. S., Cesarino, I. (2023). Toward the identification of class III peroxidases potentially involved in lignification in the model C4 grass Setaria viridis. Theor. Exp. Plant Physiol. 35, 111–131. doi: 10.1007/s40626-023-00273-5.
Simpson, J. P., Olson, J., Dilkes, B., Chapple, C. (2021). Identification of the tyrosine- and phenylalanine-derived soluble metabolomes of sorghum. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.714164.
Singh, D., Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00895.
Skuza, L., Szucko-Kociuba, I., Filip, E., Bozek, I. (2022). Natural molecular mechanisms of plant hyperaccumulation and hypertolerance towards heavy metals. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23169335.
Smith, R. A., Beebe, E. T., Bingman, C. A., Vander Meulen, K., Eugene, A., Steiner, A. J., et al. (2022). Identification and characterization of a set of monocot BAHD monolignol transferases. Plant Physiol. 189, 37–48. doi: 10.1093/plphys/kiac035.
Smith, R. A., Cass, C. L., Mazaheri, M., Sekhon, R. S., Heckwolf, M., Kaeppler, H., et al. (2017a). Suppression of CINNAMOYL-CoA REDUCTASE increases the level of monolignol ferulates incorporated into maize lignins. Biotechnol. Biofuels 10, 10. doi: 10.1186/s13068-017-0793-1.
Smith, R. A., Schuetz, M., Karlen, S. D., Bird, D., Tokunaga, N., Sato, Y., et al. (2017b). Defining the diverse cell populations contributing to lignification in Arabidopsis stems. Plant Physiol. 174, 1028–1036. doi: 10.1104/pp.17.00434.
Smith, R. A., Schuetz, M., Roach, M., Mansfield, S. D., Ellis, B., Samuels, L. (2013). Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25, 3988–3999. doi: 10.1105/tpc.113.117176.
Stein, R. J., Duarte, G. L., Scheunemann, L., Spohr, M. G., De Araujo, A. T., Ricachenevsky, F. K., et al. (2019). Genotype variation in rice (Oryza sativa L.) tolerance to fe toxicity might be linked to root cell wall lignification. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00746
Sytar, O., Ghosh, S., Malinska, H., Zivcak, M., Brestic, M. (2021). Physiological and molecular mechanisms of metal accumulation in hyperaccumulator plants. Physiologia Plantarum 173, 148–166. doi: 10.1111/ppl.13285
Takeda, Y., Koshiba, T., Tobimatsu, Y., Suzuki, S., Murakami, S., Yamamura, M., et al. (2017). Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 246, 337–349. doi: 10.1007/s00425-017-2692-x.
Takeda, Y., Suzuki, S., Tobimatsu, Y., Osakabe, K., Osakabe, Y., Ragamustari, S. K., et al. (2019). Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J. 97, 543–554. doi: 10.1111/tpj.14141.
Takeda, Y., Tobimatsu, Y., Karlen, S. D., Koshiba, T., Suzuki, S., Yamamura, M., et al. (2018). Downregulation of p-COUMAROYL ESTER 3-HYDROXYLASE in rice leads to altered cell wall structures and improves biomass saccharification. Plant J. 95, 796–811. doi: 10.1111/tpj.13988.
Takeuchi, M., Kegasa, T., Watanabe, A., Tamura, M., Tsutsumi, Y. (2018a). Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during Tracheary element differentiation. J. Plant Res. 131, 297–305. doi: 10.1007/s10265-017-0979-4.
Takeuchi, M., Watanabe, A., Tamura, M., Tsutsumi, Y. (2018b). The gene expression analysis of Arabidopsis thaliana ABC transporters by real-time PCR for screening monolignol-transporter candidates. J. Wood Sci. 64, 477–484. doi: 10.1007/s10086-018-1733-9.
Terrett, O. M., Dupree, P. (2019). Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 56, 97–104. doi: 10.1016/j.copbio.2018.10.010.
Tetreault, H. M., Gries, T., Liu, S. R., Toy, J., Xin, Z. G., Vermerris, W., et al. (2021). The sorghum (Sorghum bicolor) brown midrib 30 gene encodes a chalcone isomerase required for cell wall lignification. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.732307.
Thompson, A. L., Smiley, R. W., Garland-Campbell, K. (2015). Registration of the LouAu (Louise/IWA8608077) wheat recombinant inbred line mapping population. J. Plant Registrations 9, 424–429. doi: 10.3198/jpr2015.01.0002crmp.
Tian, C., Jiang, Q., Wang, F., Wang, G. L., Xu, Z. S., Xiong, A. S. (2015). Selection of Suitable Reference Genes for qPCR Normalization under Abiotic Stresses and Hormone Stimuli in Carrot Leaves. PloS One 10, 16. doi: 10.1371/journal.pone.0117569.
Tobimatsu, Y., Schuetz, M. (2019). Lignin polymerization: how do plants manage the chemistry so well? Curr. Opin. Biotechnol. 56, 75–81. doi: 10.1016/j.copbio.2018.10.001.
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9.
Tryfona, T., Bourdon, M., Marques, R. D., Busse-Wicher, M., Vilaplana, F., Stott, K., et al. (2023). Grass xylan structural variation suggests functional specialization and distinctive interaction with cellulose and lignin. Plant J. 113, 1004–1020. doi: 10.1111/tpj.16096.
Tsuyama, T., Kawai, R., Shitan, N., Matoh, T., Sugiyama, J., Yoshinaga, A., et al. (2013). Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 162, 918–926. doi: 10.1104/pp.113.214957.
Tsuyama, T., Matsushita, Y., Fukushima, K., Takabe, K., Yazaki, K., Kamei, I. (2019). Proton gradient-dependent transport of p-glucocoumaryl alcohol in differentiating xylem of woody plants. Sci. Rep. 9. doi: 10.1038/s41598-019-45394-7
Turlings, T. C. J., Mccall, P. J., Alborn, H. T., Tumlinson, J. H. (1993). AN ELICITOR IN CATERPILLAR ORAL SECRETIONS THAT INDUCES CORN SEEDLINGS TO EMIT CHEMICAL SIGNALS ATTRACTIVE TO PARASITIC WASPS. J. Chem. Ecol. 19, 411–425. doi: 10.1007/BF00994314.
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–109+. doi: 10.1038/ng.2725.
UN. (2022). World population prospects 2022: summary of results (Population Division of the United Nations Department of Economic and Social Affairs). Available at: https://www.un.org/development/desa/pd/sites/www.un.org.development.desa.pd/files/undesa_pd_2022_wpp_key-messages.pdf.
Underwood, W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3, 6. doi: 10.3389/fpls.2012.00085.
Vadez, V. (2014). Root hydraulics: The forgotten side of roots in drought adaptation. Field Crops Res. 165, 15–24. doi: 10.1016/j.fcr.2014.03.017.
Vaisanen, E., Takahashi, J., Obudulu, O., Bygdell, J., Karhunen, P., Blokhina, O., et al. (2020). Hunting monolignol transporters: membrane proteomics and biochemical transport assays with membrane vesicles of Norway spruce. J. Exp. Bot. 71, 6379–6395. doi: 10.1093/jxb/eraa368.
Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y. G., Sundin, L., Goeminne, G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602.
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119.
Vanholme, R., De Meester, B., Ralph, J., Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018.
Vanholme, R., Morreel, K., Darrah, C., Oyarce, P., Grabber, J. H., Ralph, J., et al. (2012). Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000. doi: 10.1111/j.1469-8137.2012.04337.x.
Velez-Bermudez, I. C., Salazar-Henao, J. E., Fornale, S., Lopez-Vidriero, I., Franco-Zorrilla, J. M., Grotewold, E., et al. (2015). A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell 27, 3245–3259. doi: 10.1105/tpc.15.00545.
Vermaas, J. V., Crowley, M. F., Beckham, G. T. (2019a). A quantitative molecular atlas for interactions between lignin and cellulose. ACS Sustain. Chem. Eng. 7, 19570–19583. doi: 10.1021/acssuschemeng.9b04648.
Vermaas, J. V., Dixon, R. A., Chen, F., Mansfield, S. D., Boerjan, W., Ralph, J., et al. (2019b). Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. United States America 116, 23117–23123. doi: 10.1073/pnas.1904643116.
Vermaas, J. V., Petridis, L., Qi, X. H., Schulz, R., Lindner, B., Smith, J. C. (2015). Mechanism of lignin inhibition of enzymatic biomass deconstruction. Biotechnol. Biofuels 8. doi: 10.1186/s13068-015-0379-8.
Vermerris, W., Sherman, D. M., Mcintyre, L. M. (2010). Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J. Exp. Bot. 61, 2479–2490. doi: 10.1093/jxb/erq093.
Vignols, F., Rigau, J., Torres, M. A., Capellades, M., Puigdomenech, P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid o-methyltransferase. Plant Cell 7, 407–416. doi: 10.1105/tpc.7.4.407
von Ropenack, E., Parr, A., Schulze-Lefert, P. (1998). Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022. doi: 10.1074/jbc.273.15.9013.
Wang, G. F., Balint-Kurti, P. J. (2016). Maize homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR rp1 protein to modulate the defense response. Plant Physiol. 171, 2166–2177. doi: 10.1104/pp.16.00224.
Wang, Y., Bouchabke-Coussa, O., Lebris, P., Antelme, S., Soulhat, C., Gineau, E., et al. (2015b). LACCASE5 is required for lignification of the brachypodium distachyon culm. Plant Physiol. 168, 192–U951. doi: 10.1104/pp.114.255489.
Wang, Y. Y., Cao, Y. B., Liang, X. Y., Zhuang, J. H., Wang, X. F., Qin, F., et al. (2022c). A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 13. doi: 10.1038/s41467-022-29809-0.
Wang, J. H., Feng, J. J., Jia, W. T., Fan, P. X., Bao, H., Li, S. Z., et al. (2017). Genome-wide identification of sorghum bicolor laccases reveals potential targets for lignin modification. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00714.
Wang, Y. L., Gui, C. J., Wu, J. Y., Gao, X., Huang, T., Cui, F. J., et al. (2022a). Spatio-temporal modification of lignin biosynthesis in plants: A promising strategy for lignocellulose improvement and lignin valorization. Front. Bioengineering Biotechnol. 10. doi: 10.3389/fbioe.2022.917459.
Wang, G. F., He, Y. J., Strauch, R., Olukolu, B. A., Nielsen, D., Li, X., et al. (2015a). Maize homologs of hydroxycinnamoyltransferase, A key enzyme in lignin biosynthesis, bind the nucleotide binding leucine-rich repeat Rp1 proteins to modulate the defense response. Plant Physiol. 169, 2230–2243. doi: 10.1104/pp.15.00703
Wang, Y., Li, X. F., Fan, B. F., Zhu, C., Chen, Z. X. (2021). Regulation and function of defense-related callose deposition in plants. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22052393.
Wang, Y. P., Li, T., Sun, Z. D., Huang, X. J., Yu, N. B., Tai, H. H., et al. (2022b). Comparative transcriptome meta-analysis reveals a set of genes involved in the responses to multiple pathogens in maize. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.971371.
Wang, Z. G., Shi, M. X., Wei, Q. X., Chen, Z. W., Huang, J. J., Xia, J. X. (2020). OsCASP1 forms complexes with itself and OsCASP2 in rice. Plant Signaling Behav. 15. doi: 10.1080/15592324.2019.1706025.
Wang, Z. G., Yamaji, N. K., Huang, S., Zhang, X., Shi, M. X., Fu, S., et al. (2019). OsCASP1 is required for Casparian strip formation at endodermal cells of rice roots for selective uptake of mineral elements. Plant Cell 31, 2636–2648. doi: 10.1105/tpc.19.00296.
Wang, M. X., Zhu, X. L., Wang, K., Lu, C. G., Luo, M. Y., Shan, T. L., et al. (2018). A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 8, 14. doi: 10.1038/s41598-018-24884-0.
Warburton, M. L., Woolfolk, S. W., Smith, J. S., Hawkins, L. K., Castano-Duque, L., Lebar, M. D, et al. (2023). Genes and genetic mechanisms contributing to fall armyworm resistance in maize. Plant Genome 16 (2). doi: 10.1002/tpg2.20311.
Ward, G., Hadar, Y., Bilkis, I., Konstantinovsky, L., Dosoretz, C. G. (2001). Initial steps of ferulic acid polymerization by lignin peroxidase. J. Biol. Chem. 276, 18734–18741. doi: 10.1074/jbc.M009785200.
Waterman, J. M., Cazzonelli, C. I., Hartley, S. E., Johnson, S. N. (2019). Simulated herbivory: the key to disentangling plant defence responses. Trends Ecol. Evol. 34, 447–458. doi: 10.1016/j.tree.2019.01.008.
Wieczorek, K. (2015). Cell wall alterations in nematode-infected roots. Plant Nematode Interactions: View Compatible Interrelationships 73, 61–90. doi: 10.1016/bs.abr.2014.12.002.
Wilkerson, C. G., Mansfield, S. D., Lu, F., Withers, S., Park, J. Y., Karlen, S. D., et al. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344, 90–93. doi: 10.1126/science.1250161.
Withers, S., Lu, F. C., Kim, H., Zhu, Y. M., Ralph, J., Wilkerson, C. G. (2012). Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 287, 8347–8355. doi: 10.1074/jbc.M111.284497.
Wu, Y., Li, X., Zhang, J. N., Zhao, H. Q., Tan, S. L., Xu, W. H., et al. (2022). ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1042084.
Wu, Z. Y., Wang, N. F., Hisano, H., Cao, Y. P., Wu, F. Y., Liu, W. W., et al. (2019). Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alters effects of COMT suppression on syringyl lignin biosynthesis. Plant Biotechnol. J. 17, 836–845. doi: 10.1111/pbi.13019.
Wu, Q., Yan, D. Y., Chen, Y., Wang, T., Xiong, F., Wei, W., et al. (2017). A redox-neutral catechol synthesis. Nat. Commun. 8. doi: 10.1038/ncomms14227.
Wuyts, N., Lognay, G., Verscheure, M., Marlier, M., De Waele, D., Swennen, R. (2007). Potential physical and chemical barriers to infection by the burrowing nematode Radopholus similis in roots of susceptible and resistant banana (Musa spp.). Plant Pathol. 56, 878–890. doi: 10.1111/j.1365-3059.2007.01607.x.
Xie, W., Ding, C. Q., Hu, H. T., Dong, G. J., Zhang, G. H., Qian, Q., et al. (2022). Molecular events of rice AP2/ERF transcription factors. Int. J. Mol. Sci. 23. doi: 10.3390/ijms231912013.
Xu, J., Li, Y. Y., Wang, S. L., Long, S., Wu, Y. N., Chen, Z. M. (2023). Sources, transfers and the fate of heavy metals in soil-wheat systems: The case of lead (Pb)/zinc (Zn) smelting region. J. Hazardous Materials 441. doi: 10.1016/j.jhazmat.2022.129863.
Xu, W. Y., Tang, W. S., Wang, C. X., Ge, L. H., Sun, J. C., Qi, X., et al. (2020). SiMYB56Confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00785
Xue, J., Gao, S., Fan, Y. H., Li, L. L., Ming, B., Wang, K. R., et al. (2020). Traits of plant morphology, stalk mechanical strength, and biomass accumulation in the selection of lodging-resistant maize cultivars. Eur. J. Agron. 117. doi: 10.1016/j.eja.2020.126073.
Yadav, V., Wang, Z. Y., Wei, C. H., Amo, A., Ahmed, B., Yang, X. Z., et al. (2020). Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens 9. doi: 10.3390/pathogens9040312.
Yang, Q., He, Y. J., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., et al. (2017). A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–136+. doi: 10.1038/ng.3919.
Yang, X. F., Xie, H. F., Weng, Q. Q., Liang, K. J., Zheng, X. J., Guo, Y. C., et al. (2022). Rice OsCASP1 orchestrates Casparian strip formation and suberin deposition in small lateral roots to maintain nutrient homeostasis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1007300.
Yao, T., Feng, K., Xie, M., Barros, J., Tschaplinski, T. J., Tuskan, G. A., et al. (2021). Phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.704697.
Yu, P., Gutjahr, C., Li, C. J., Hochholdinger, F. (2016). Genetic control of lateral root formation in cereals. Trends Plant Sci. 21, 951–961. doi: 10.1016/j.tplants.2016.07.011.
Yu, M. Z., Wang, M., Gyalpo, T., Basang, Y. Z. (2021). Stem lodging resistance in hulless barley: Transcriptome and metabolome analysis of lignin biosynthesis pathways in contrasting genotypes. Genomics 113, 935–943. doi: 10.1016/j.ygeno.2020.10.027.
Yuan, M. H., Ngou, B. P. M., Ding, P. T., Xiu-Fan, X. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62. doi: 10.1016/j.pbi.2021.102030.
Zakzeski, J., Bruijnincx, P. C. A., Jongerius, A. L., Weckhuysen, B. M. (2010). The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 110, 3552–3599. doi: 10.1021/cr900354u.
Zeng, Y. N., Zhao, S., Yang, S. H., Ding, S. Y. (2014). Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 27, 38–45. doi: 10.1016/j.copbio.2013.09.008.
Zhang, Y., Legland, D., El Hage, F., Devaux, M. F., Guillon, F., Reymond, M., et al. (2019). Changes in cell walls lignification, feruloylation and p-coumaroylation throughout maize internode development. PloS One 14. doi: 10.1371/journal.pone.0219923.
Zhang, B. X., Lewis, J. A., Vermerris, W., Sattler, S. E., Kang, C. L. (2023b). A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids. Plant Physiol. 192, 102–118. doi: 10.1093/plphys/kiac604.
Zhang, B., Xin, B., Sun, X., Chao, D., Zheng, H., Peng, L., et al. (2023a). Small peptide signaling via OsCIF1/2 mediates Casparian strip formation at the root endodermal and nonendodermal cell layers in rice. Plant Cell: Oxford Acad. 36 (2), 383–403. doi: 10.1093/plcell/koad269.
Zhao, J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007.
Zhao, K. M., Bartley, L. E. (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol. 14. doi: 10.1186/1471-2229-14-135.
Zhao, Q., Nakashima, J., Chen, F., Yin, Y. B., Fu, C. X., Yun, J. F., et al. (2013). LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770.
Zheng, M. J., Chen, J., Shi, Y. H., Li, Y. X., Yin, Y. P., Yang, D. Q., et al. (2017). Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci. Rep. 7, 12. doi: 10.1038/srep41805.
Zhong, R. Q., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123.
Zhong, X., Li, M. J., Zhang, M. M., Feng, Y., Zhang, H., Tian, H. (2023). Genome-wide analysis of the laccase gene family in wheat and relationship with arbuscular mycorrhizal colonization. Planta 257. doi: 10.1007/s00425-022-04048-1.
Zhong, H. M., Zhou, J. Y., Abdelrahman, M., Xu, H., Wu, Z., Cui, L. C., et al. (2021). The effect of lignin composition on ruminal fiber fractions degradation from different roughage sources in water buffalo (Bubalus bubalis). Agriculture-Basel 11. doi: 10.3390/agriculture11101015.
Zhou, X. G., Liao, H. C., Chern, M. S., Yin, J. J., Chen, Y. F., Wang, J. P., et al. (2018). Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. United States America 115, 3174–3179. doi: 10.1073/pnas.1705927115
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029.
Zhu, Y. X., Ge, C. X., Ma, S. J., Liu, X. Y., Liu, M. J., Sun, Y., et al. (2020). Maize zmFNSI homologs interact with an NLR protein to modulate hypersensitive response. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21072529.
Keywords: lignin, hydroxycinnamic acids, grass, cereals, abiotic stress, biotic stress, monolignol transport, monolignol polymerization
Citation: Peracchi LM, Panahabadi R, Barros-Rios J, Bartley LE and Sanguinet KA (2024) Grass lignin: biosynthesis, biological roles, and industrial applications. Front. Plant Sci. 15:1343097. doi: 10.3389/fpls.2024.1343097
Received: 22 November 2023; Accepted: 06 February 2024;
Published: 23 February 2024.
Edited by:
Olga A. Zabotina, Iowa State University, United StatesReviewed by:
Jan Lyczakowski, Jagiellonian University, PolandWout Boerjan, Flanders Institute for Biotechnology, Belgium
Copyright © 2024 Peracchi, Panahabadi, Barros-Rios, Bartley and Sanguinet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen A. Sanguinet, karen.sanguinet@wsu.edu
 Luigi M. Peracchi
Luigi M. Peracchi