
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 12 February 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1336892
Flowering represents a crucial stage in the life cycles of plants. Ensuring strong and consistent flowering is vital for maintaining crop production amidst the challenges presented by climate change. In this review, we summarized key recent efforts aimed at unraveling the complexities of plant flowering through genetic, genomic, physiological, and biochemical studies in woody species, with a special focus on the genetic control of floral initiation and activation in woody horticultural species. Key topics covered in the review include major flowering pathway genes in deciduous woody plants, regulation of the phase transition from juvenile to adult stage, the roles of CONSTANS (CO) and CO-like gene and FLOWERING LOCUS T genes in flower induction, the floral regulatory role of GA-DELLA pathway, and the multifunctional roles of MADS-box genes in flowering and dormancy release triggered by chilling. Based on our own research work in blueberries, we highlighted the central roles played by two key flowering pathway genes, FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1, which regulate floral initiation and activation (dormancy release), respectively. Collectively, our survey shows both the conserved and diverse aspects of the flowering pathway in annual and woody plants, providing insights into the potential molecular mechanisms governing woody plants. This paves the way for enhancing the resilience and productivity of fruit-bearing crops in the face of changing climatic conditions, all through the perspective of genetic interventions.
Flowering represents a vital phase in the reproductive developmental of plants, ultimately resulting in the generation of seeds for subsequent generations. In agriculture, a robust flowering process, encompassing floral induction, formation, and developmental programming, stands as a fundamental prerequisite for achieving productive crop cultivation. The persistent trend of global warming, coupled with burgeoning populations and the depletion of natural resources, has presented significant challenges to agricultural production. For staple crops, warming can curtail agricultural output by shifting optimal growth zones and/or diminishing both cropping frequency and yields (Zhu et al., 2022). In the case of woody fruit crops, particularly temperate fruit trees, the impacts of global warming are profound, exerting adverse effects on floral development, dormancy release, and fruit growth (Luedeling et al., 2011). A pertinent instance is the scenario wherein insufficient chilling, triggered by climatic shifts, precipitates decreased bud break and reduced flower quality, leading to a reduction in fruit production (Atkinson et al., 2013).
Annual plants have evolved to respond to seasonal variations, facilitating a seamless transition from vegetative to reproductive phases. Extensive investigations using the model plant Arabidopsis (Arabidopsis thaliana) and cereal crops have yielded a wealth of valuable insights. These studies have unveiled pivotal regulatory nodes governing floral initiation and flowering time, encompassing pathways tied to aging, photoperiod, autonomous/vernalization, and gibberellin stimuli (see reviews by Greenup et al., 2009; Fornara et al., 2010; Andres and Coupland, 2012; Conti, 2017; Kinoshita and Richter, 2020; Izawa, 2021; Liu et al., 2021). At the center of this regulatory matrix stand two major integrators: FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTAN 1 (SOC1). FT exerts a positive influence on SOC1, with this FT-to-SOC1 module occupying a central role in plant flowering, exhibiting evolutionary conservation across diverse plant species (Fornara et al., 2010; Lee and Lee, 2010). In Arabidopsis, FT emerges as a direct downstream target of both CONSTANS (CO) within the photoperiod pathway and FLOWERING LOCUS C (FLC) in the vernalization/autonomous pathway. SOC1, on the other hand, is under the direct sway of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) in the aging pathway, alongside FLC and DELLA proteins within the gibberellin pathway (Wang et al., 2009; Preston and Hileman, 2013; Bao et al., 2020).
The elucidation of the intricate gene networks underpinning each flowering pathway in Arabidopsis has laid a foundational framework, setting the stage for analogous insights into the flowering mechanisms of other plants. In this review, our attention is directed towards the intricacies of flowering mechanisms in woody plants, with a particular focus on fruit-bearing crops. We provide a concise summary of recent advancements and present a gene network that explains possible flowering mechanism in woody plants, specifically focusing on deciduous temperate fruit crops. Within this framework, we underscore the role of FT in floral induction and SOC1’s involvement in floral programming. This gene-centric network not only deepens our understanding of flowering mechanisms in woody plants but also serves as a valuable knowledge resource for orchard management for fruit growers.
The process of flowering in deciduous woody plants is a product of intricate genetic and environmental influences and their interactions in responding to the seasonal changes (Figure 1). The phases of flowering, fruiting, and vegetative growth predominantly unfold mainly during spring and summer, coinciding with the prevalence of elevated average temperatures. Meanwhile new floral buds for many deciduous woody plants are initiated in the summer period and continuously undergo development through fall. In contrast, winter mark the completion of reproductive growth, onset of growth cessation, and the establishment of dormancy in general. During this period, declining temperatures and shortening day period act as key environmental signals to trigger and drive these biological events. It is generally within these colder months that the development of flower buds occurs and the initiation of endodormancy is likely signaled.
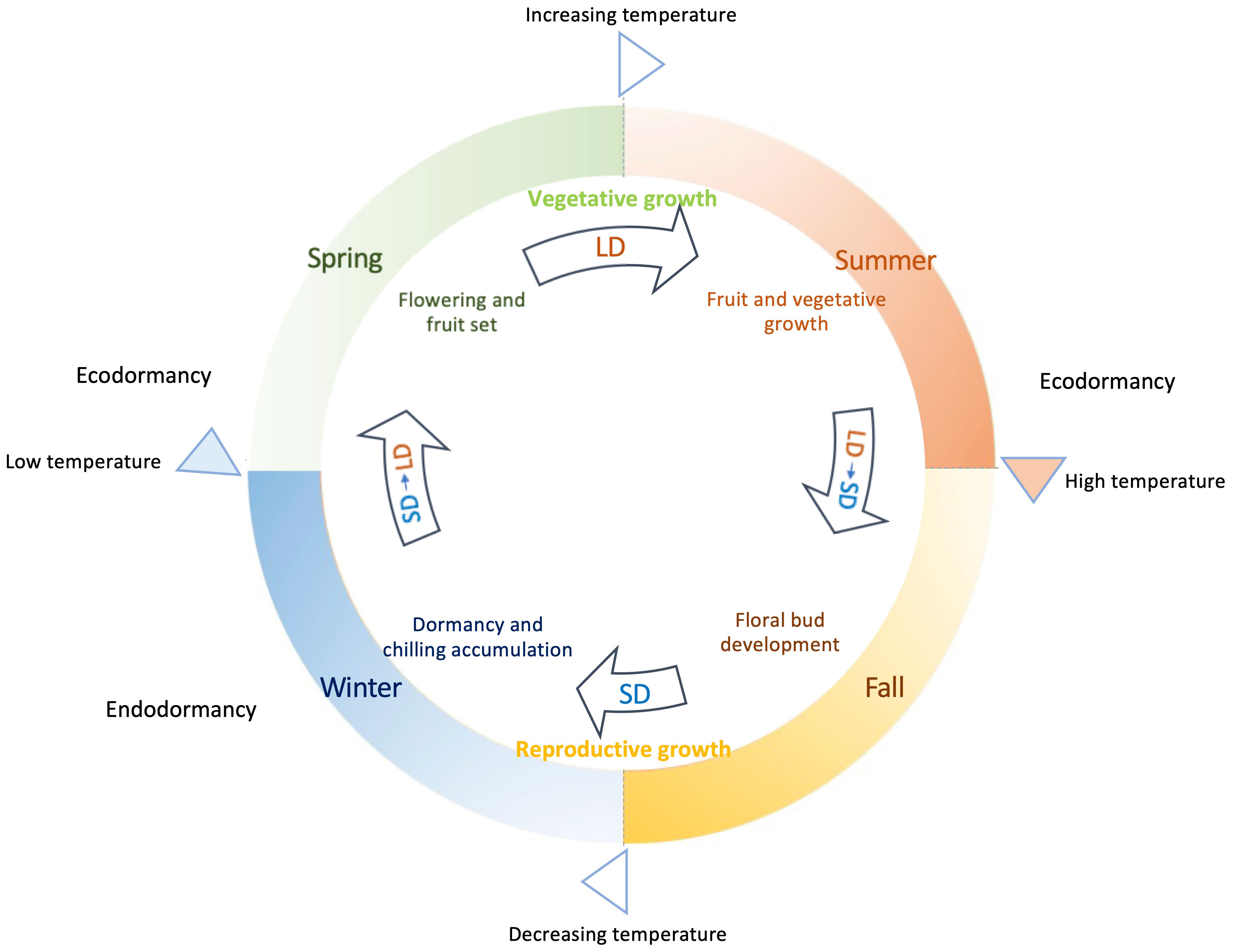
Figure 1 Annual growth cycle of deciduous woody plants guided by seasonal daylight and temperatures. Plant flowering, fruiting, and vegetative growth mainly occur in spring and summer, when it is in long day (LD) conditions with growing daylength associated with increasing/higher average temperatures. Growth cessation and dormancy happen in fall and winter under decreasing temperature and mostly short day (SD) conditions, when flower bud formation is achieved and endodormancy is induced. Enough chilling accumulation is needed before sufficient warm accumulation breaks endodormancy in spring. Low temperatures or insufficient warm accumulation causes ecodormancy in spring for those fully chilled buds. Extreme high temperatures in summer can result in ecodormancy, during which plant growth is temporarily arrested.
A pivotal facet that distinguishes deciduous woody plants from the vernalization/autonomous pathway observed in Arabidopsis lies in the flowering mechanism driven by chilling accumulation, termed chilling requirement (CR). This accumulation of chilling hours occurs before the eventual release of endodormancy, as temperatures start to rise in spring. This CR-mediated process imparts a unique rhythm to the flowering behavior of deciduous woody plants, in contrast to the continuous floral initiation and flowering progression exhibited by vernalized Arabidopsis plants. Deciduous woody plants often have two distinct phases emerge: the initiation of floral buds mostly during the autumn and early winter, followed by the actual blossoming of these buds in spring subsequent to CR fulfillment (Figure 1).
Flower regulation in deciduous fruit trees is distinct from annuals. In annuals, flower initiation begins with the transition from vegetative to inflorescence stage and flower formation and development are typically completed within a single season (see reviews by Baurle and Dean, 2006; Irish, 2010). On the other hand, flowers from seedling-derived fruit trees can only be initiated and developed after the tree reaches adulthood, which can take several years. Even in adult trees, flower initiation and development occur over two growing seasons, not one (Wilkie et al., 2008; Sun et al., 2022). For example, floral bud initiation in apple and peach trees occurs in summer and basic morphological structures such as sepal, petal, stamen, and carpel are developed during the fall before entering a fully dormant state, remaining in a “resting” state during winter and then resumed developmental pace before flowering next spring. Winter chilling is indispensable for driving dormancy out (Arora et al., 2003). It was also found that chilling is essential for driving morphological differentiation within buds during winter (Luna et al., 1990; Luna et al., 1991; Luna et al., 1993; Reinoso et al., 2002a; Reinoso et al., 2002b; Julian et al., 2011). In fact, the formation of specific floral tissue in response to chilling is a morphological indicator that the floral buds are out of dormant state and have entered an ecodormant state capable of responding to warm stimuli, resuming developmental pace and achieving reproduction success (Wang et al., 2004; Wang et al., 2016). Notably, floral initiation, formation and development are regulated by endogenous physiological states and seasonal thermal regimes.
In the genetic realm, the majority of woody fruit crops exhibit a juvenile phase spanning from days to years, during which seedlings remain incapable of flowering even when subjected to suitable environmental triggers (Samach, 2012). In Arabidopsis, the transition from vegetative to inflorescence meristem appears to be regulated by the aging pathway requiring a miR156 (microRNA156) that controls PROMOTER BINDING PROTEIN-LIKE genes (SPLs) (Wang et al., 2009; Wu et al., 2009; Teotia and Tang, 2015; Xu et al., 2016b; Hyun et al., 2017). In this pathway, miR156 negatively regulates the activity of flowering activator SPLs. This family of SPLs, known for their multifunctionality, catalyzes floral transition by augmenting the expression of key genes such as LEAFY (LFY) and MADS box genes SOC1 and AP1 (Albani and Coupland, 2010; Lee and Lee, 2010; Ma et al., 2021). The engagement of the miR156-SPL module in the transition from juvenile to adult phases has been validated in various plants, including apple (Zhang et al., 2015a; Jia et al., 2017; Zheng et al., 2019), the conserved nature of this miR156-SPL module within the aging pathway is acknowledged across annual and perennial plant species. Yet, substantiating this module’s presence in other woody plants other than apple remains a necessity, along with addressing the intriguing query surrounding the divergent durations of juvenile phases observed among different woody plant species (Huijser and Schmid, 2011; Morea et al., 2016) (Figure 2).
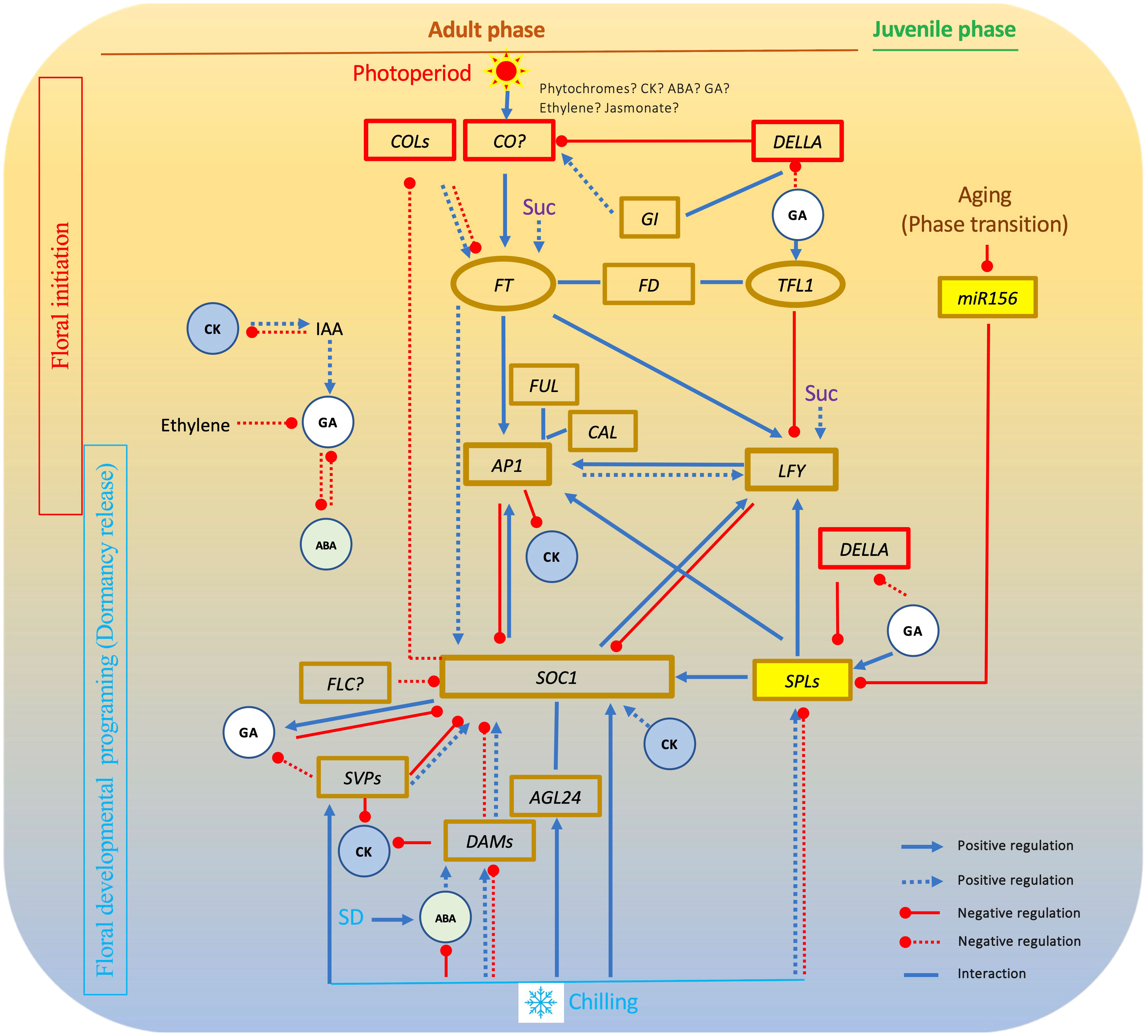
Figure 2 Interactions among key flowering pathway genes in chilling-dependent woody plants. In this gene network, FT plays an essential role in floral initiation and formation. SOC1 has a central role in floral activation by interacting with other MADS box genes. GA affects both floral initiation and floral activation. Other hormones and sucrose have impact on floral initiation or floral activation. Solid lines show relationships revealed in Arabidopsis. Dot lines shows relationships found in woody plants. SD, short day; GA, gibberellin; Suc, sucrose; CK, cytokinin; ABA, abscisic acid; IAA, Indole-3-acetic acid.
Of notable significance is the involvement of the DELLA proteins that act as master regulators that rewire a multitude of transcriptional networks to control diverse biological responses (Briones-Moreno et al., 2023). One of the Arabidopsis DELLA factors exhibits a propensity to interact with distinct SPLs, yielding disparate outcomes. These interactions can either spark the initiation of floral primordia, such as through the activation of AP1 transcription via binding with SPL9, or act to inhibit SPL function, thus serving as a brake on the flowering process. This dual nature confers upon interactions of gibberellin (GA) and DELLA proteins to produce varying effects on the reproductive journey, depending on the developmental stage, all orchestrated by the intricate behavior of DELLA proteins (Yu et al., 2012; Yamaguchi et al., 2014; Bao et al., 2020).
The phase transition in fruit trees is also regulated by TFL1 and its related genes. For example, when the TFL1 gene is knocked out or down in apple plantlets, the flower inhibition imposed by juvenility is erased, allowing for flowering in re-juvenilized shoots as quickly as a few months, instead of the usual 3-6 years (Kotoda et al., 2006; Charrier et al., 2019). However, it is not yet clear how the miR156-SPL pathway interacts with TFL1.
Photoperiod, notably the duration of daylight, constitutes a pivotal environmental cue orchestrating plant flowering. This process elucidates the photoperiod pathway, wherein the CO gene plays a crucial role in transducing light signals to regulate FT expression (Samach et al., 2000; Kinoshita and Richter, 2020). While the CO-FT module has been unequivocally demonstrated in numerous annual plants, suggesting its theoretical conservation across all species (Putterill et al., 1995; Yano et al., 2000; Griffiths et al., 2003; Kim et al., 2008; Samach, 2012; Song et al., 2012; Lymperopoulos et al., 2018; Bao et al., 2020), the centrality of COLs (CO-LIKE genes) in the orchestration of flowering time remains a subject of contention. This is attributed in part to the intricate diversity in characterizing the COL gene family (Wong et al., 2014). Ectopic expression analyses of peach CO in Arabidopsis have indirectly supported the conservation of the CO-FT module in trees, hinting at a broader applicability (Böhlenius et al., 2006; Zhang et al., 2015b). In the case of apple, distinctive expression patterns of apple COLs in comparison to Arabidopsis genes suggest a divergent CO-FT module (Jeong et al., 1999). Expression studies have brought to light varying facets: grape CO linked to flowering initiation and COL1 associated with dormancy (Almada et al., 2009); six pear (Pyrus bretschneideri) COLs, out of a total of 15, showed circadian clock and photoperiod-regulated expression (Wang et al., 2017); and Mango (Mangifera indica L.) CO and COLs implicated in photoperiod-mediated flowering, with CO-FT conservativity warranting further clarification (Liu et al., 2020a; Liu et al., 2022). Similar investigations have extended to bamboo (Phyllostachys violascens), poplar (Populus trichocarpa), and Rabbiteye blueberries (Vaccinium virgatum Aiton), demonstrating their roles in photoperiod-controlled flowering through expression patterns or Arabidopsis ectopic expression (Xiao et al., 2018; Li et al., 2020; Omori et al., 2022).
However, the conclusive validation of the central role of the CO-FT module in woody plants awaits direct evidence from gain-of-function (e.g., overexpression) or loss-of-function (e.g., gene silencing/knockout) studies. The intricate multifunctional roles of the COLs family have added layers of complexity to their functional scrutiny. While the comprehensive understanding remains elusive, the CO-FT involvement in flowering function has yet to be ruled out. Concurrently, the interplay between CO expression and light in Arabidopsis reveals the intricate dance of phytochrome-phytohormone interactions in regulating flowering time and broader aspects of plant growth and development (Fornara et al., 2010; Lymperopoulos et al., 2018). The regulatory role of DELLAs in the GA pathway as a negative regulator of CO expression, along with the indirect influences of abscisic acid (ABA) and Jasmonate, further accentuates the intricate tapestry of these regulatory networks (Davis, 2009; Tiwari et al., 2010; Xu et al., 2016a; Bao et al., 2020; Izawa, 2021; Serrano-Bueno et al., 2021). Flower initiation in apple, peach, and other trees occurs in summer or late summer, suggesting that unlike in Arabidopsis, photoperiod signals may not play a role in flower induction. Instead, other endogenous signals such as physiological state, hormone homeostasis, or nutrient balance (e.g., photosynthetic output) might be influencing flower formation. As a result, the CO-FT module could gain new functions and interact with these endogenous signals in fruit trees to control floral initiation.
In Arabidopsis, phytohormones known as GAs play substantial roles in various aspects of plant growth and development, spanning processes such as seed germination, elongation growth, and the regulation of flowering time (Michniewicz and Lang, 1962; Yamauchi et al., 2004; Willige et al., 2007; Yamaguchi et al., 2014; Ni et al., 2015). This influence is often mediated through intricate interactions with multiple developmental pathways, facilitated by the DELLA domain which functions as a receiver domain for activated GA receptors (Yamauchi et al., 2004; Willige et al., 2007; Yamaguchi et al., 2014; Ni et al., 2015). The impact of GA on flowering time, whether promoting or inhibiting, hinges on the specific plant species and developmental stages at play (Koshita et al., 1999; Goldberg-Moeller et al., 2013; Pearce et al., 2013; Izawa, 2021). Broadly speaking, GAs tend to exert flowering promotion in long-day and biennial plants, while adopting an inhibitory role in other plant categories, encompassing fruit trees such as citrus (Citrus reticulata Blanco × Citrus temple Hort. ex Y. Tanaka) and grape (Boss and Thomas, 2002; Goldberg-Moeller et al., 2013; Li-Mallet et al., 2016; Zhang et al., 2019). Nonetheless, this species- and genotype-dependent function of GA in the intricate relationship of flowering introduces a level of uncertainty and complexity, thereby casting a degree of doubt upon GAs as strong candidates for the role of florigen. This sentiment persists despite findings in the grass species Lolium temulentum, where specific GAs (GA5 and GA6) emerge as an alternative source of florigenic signal distinct from FT, another floral signal originating from leaves (King and Evans, 2003; King et al., 2006).
However, the roles of GAs in regulation of floral formation in perennial flowering species appears to be quite different from that in annual species such as Arabidopsis. GA is thought to be inhibitory to flowering in perennials (Khan et al., 2014), as exemplified by the fact that exogenous applications of GA in citrus, apple and grapevine have been shown to inhibit flower production (Mullins, 1968; Srinivasan and Mullins, 1978; Zhu et al., 2008; Guo et al., 2018). This suggests that GA-generated/stimulated signals are rewired to distinct transcriptional pathways in perennials and annuals, resulting in opposite regulatory outputs. This finding is supported by the gain-of-function mutation of DELLAs, a main target of GA, in annual Arabidopsis and perennial grape, which led to repression and promotion of flowering, respectively (Peng and Harberd, 1997; Dill and Sun, 2001; Silverstone et al., 2001; Fleck and Harberd, 2002; Boss et al., 2006). Thus, DELLAs may be rewiring the same GA signals to transcriptional circuits or modules that have opposing functions, or targeting different factors that regulate flowering, resulting in contrasting flowering phenotypes.
Within Arabidopsis, a collection of seven flowering-promoting genes situated within the autonomous pathway assumes a counteractive role against the MADS box gene FLC, which stands as a pivotal arbiter of flowering time within the vernalization pathway (Michaels and Amasino, 1999; Simpson, 2004; Michaels, 2009). Functioning as a negative regulator, FLC exerts control over flowering by suppressing the expression of FT and SOC1. The phenomenon of vernalization, characterized by prolonged cold exposure, effectively suppresses FLC expression, thus paving the way for the initiation of flowering. The involvement of SHORT VEGETATIVE PHASE (SVP) in this process emerges through its interaction with FLC, mirroring the function of FLC in response to vernalization by curbing GA biosynthesis (Gregis et al., 2006; Andres et al., 2014). Similarly, the vernalization pathway of wheat contains VERNALIZATION 2 (VRN2), a zinc finger protein that parallels FLC in its role within the wheat vernalization pathway. In this context, VRN1 and VRN3 function as counterparts to FT and the MADS box gene AP1, respectively (Gendall et al., 2001; Yan et al., 2003; Yan et al., 2004; Yan et al., 2006; Woods et al., 2016). It is of note that cereals harbor genes akin to FLC, although their functionalities largely remain enigmatic (Kennedy and Geuten, 2020). Collectively, this FLC-regulated vernalization pathway is generally presumed to be conserved in certain plant species, exemplified by Arabidopsis, while displaying evolutionary divergence in others, as seen in the case of cereals (Sheldon et al., 2000; Tadege et al., 2001; Alexandre and Hennig, 2008; Kennedy and Geuten, 2020).
The concept of “Chilling Requirement” (CR), in contrast to vernalization for transition of vegetative to inflorescence meristem in annual plants, elucidates the necessity for an adequate accumulation of chilling hours, pivotal for breaking dormancy and fostering the flowering process within specific woody plant species (Chouard, 1960; Jewaria et al., 2021). In terms of functionality, the CR-mediated flowering pathway in woody plants mirrors the vernalization pathway observed in annual plants (Brunner et al., 2014). The CR in fruit trees and vernalization in annuals and bi-annuals have different impact on flower regulation. Vernalization is required for transition of vegetative to inflorescence meristem while the CR is mostly for regulation of floral bud development rather initiation or formation. Only grape is exception in which conversion of vegetative anagens to inflorescences requires chilling or chilling promotes this conversion. In this sense. chilling acts as a bioregulator and is obligatory for floral development as exemplified by that warm temperature represses the floral development in dormant floral buds but chilling promotes it. As of present, the CR pathways involving FLC or VRN2 have not been definitively substantiated through both forward and reverse genetics methodologies (Table 1). However, insights from transcriptome analyses have unveiled the existence of FLC-like or VRN-like genes across numerous woody plants, including apple, grape, blueberry (Vaccinium corymbosum L.), and kiwifruit (Actinidia chinensis) (Diaz-Riquelme et al., 2009; Diaz-Riquelme et al., 2014; Varkonyi-Gasic et al., 2014; Porto et al., 2015; Kumar et al., 2016; Song and Chen, 2018b). Employing a forward genetics approach, the discovery of six interconnected Dormancy-Associated MADS-Box genes (DAMs) emerged as a hallmark of the CR pathway in the evergrowing mutant of peach. Among these, DAM5 and DAM6 were found to act as repressors of bud break, while DAM4 demonstrated pronounced chilling-induced repressor activity, particularly evident at the level of epigenetic regulation (Bielenberg et al., 2008; Jimenez et al., 2009; Li et al., 2009; Jimenez et al., 2010; Wells et al., 2015; Zhu et al., 2020a; Voogd et al., 2022). These DAMs stand as potential analogs to FLC or VRN2, possibly occupying central roles within the CR pathway (Falavigna et al., 2014; Falavigna et al., 2018; Falavigna et al., 2022). Nonetheless, the deficiency of reverse genetics evidence to confirm the functional role of DAMs in peaches stems from technical challenges arising from the absence of an efficient peach transformation system for conducting functional gene analysis. Moreover, the striking sequence similarities between DAMs, SVPs, and AGAMOUS-LIKE 24 (AGL24) of Arabidopsis, as well as their widespread presence in various woody plants (Table 1), adds to the intrigue and complexity of their roles.
Emerging as pivotal contenders within the CR pathway, both DAMs and FLC-like genes have extensively been studied across various significant woody fruit crops through techniques such as expression analysis, ectopic expression, overexpression, gene silencing, and gene knockout (Table 1). These comprehensive investigations have revealed several key insights:
1) The diversity and prevalence of DAMs and FLC-like genes present in woody plants.
2) Their integral involvement in flowering orchestrated by chilling exposure, while also revealing the nuanced role these genes play, often dependent on the specific species and genotype.
3) Contrary to the well-defined centrality of FLC in Arabidopsis or VRN2 in cereals within their respective vernalization pathways, no singular DAM or FLC-like gene in woody fruit crops appears to assume a universally conserved and central role in the CR pathway.
In fact, for all plant species requiring either vernalization or chilling, the pivotal factor in regulating flowering time is not individual genes like FLC in Arabidopsis, VRN1 in cereals, or DAMs (SVPs or AGL24), but rather the entire MADS-box gene family.
As elucidated earlier, the five well-established pathways in Arabidopsis—namely age, photoperiod, GA, autonomous, and vernalization—stand as the benchmark for unraveling flowering mechanisms in diverse plant species. The wealth of insights garnered through analyses of flowering pathways in myriad other plants has yielded a plethora of evidence. This evidence aids in discerning both conserved and nonconserved genes and intricate networks governing plant flowering. This accumulation of knowledge paves the way for endeavors aimed at manipulating individual gene(s) to regulate flowering time and enhance yields. Notably, it stands to reason that plants, including woody varieties, have, to varying degrees, evolved distinct flowering pathways. Drawing from the available literature, it becomes evident that at the core of floral induction resides FT-centered processes, while SOC1-centered mechanisms prevail in orchestrating floral activation across a wide spectrum of plants, if not universally so (Figure 2).
FT acts as a critical integrator, assimilating signals for floral transition from approximately 10 activators and 30 repressors largely stemming from photoperiod and vernalization pathways, thus instigating flowering in Arabidopsis (Kobayashi et al., 1999; Wigge et al., 2005; Pin and Nilsson, 2012; Kinoshita and Richter, 2020; Liu et al., 2021). It stands prominently poised as a top contender for the florigen role (Turck et al., 2008; Turnbull, 2011; Pin and Nilsson, 2012). On the converse, TERMINAL FLOWER 1 (TFL1), a homolog of FT, exerts an opposing effect within Arabidopsis (Bradley et al., 1997; Kobayashi et al., 1999). Within the FT/TFL1 gene family, an assemblage of six members comes into view, encompassing FT, TWIN SISTER OF FT, TFL1, BROTHER OF FT AND TFL1 (BFT), MOTHER OF FT AND TFL1 (MFT), and ARABIDOPSIS THALIANA CENTROADIALIS HOMOLOGUE (ATC) (Yoo et al., 2010; Ryu et al., 2011; Liu et al., 2016). FT and TFL1 function through direct interactions with the bZIP transcription factor FD. The constitutive upregulation of FT or its orthologs (hereafter FT-CX) expression aligns with a proclivity for flowering promotion. Conversely, the persistent elevation of TFL1 or its orthologs (hereafter TFL1-CX) tends to elongate the flowering process. This dualistic phenomenon has been empirically validated across a range of plant species, including select woody plants (Table 1). Collectively, mounting evidence gleaned from ectopic expression and overexpression studies substantiates the conservation of FT and its orthologs across diverse plants, underscoring their ubiquitous roles as primary inducers within the flowering transition (Pin and Nilsson, 2012; Kinoshita and Richter, 2020; Liu et al., 2021).
The advancement of flowering in woody plants has been effectively catalyzed through the upregulation of FT-CX, as illustrated in Table 1. Noteworthy examples in deciduous fruit crops include investigations related to a blueberry FT gene, denoted as VcFT (Vc: Vaccinium corymbosum), and a poplar (Populus trichocarpa) FT1 gene (PtFT1) in European plum (Prunus domestica). PtFT1-CX induced continuous flowering as demonstrated by Srinivasan et al. (2012). Rigorous exploration has led to findings wherein the constitutive expression of VcFT (VcFT-CX) leads to a noteworthy shift in the flowering paradigm. Specifically, this alteration is characterized by the partial reversal of chilling requirements and the induction of early flowering within apical shoot meristems. In transgenic blueberry, this phenomenon resulted in the formation of multiple flower buds at each node, diverging from the single bud occurrence observed in their nontransgenic counterparts (Song et al., 2013b; Walworth et al., 2016). It is noteworthy, tough, that while VcFT-CX exerted significant influence, its effects were not fully comprehensive in substituting the requirement for chilling. Under conditions devoid of sufficient chilling hours, nearly 50% of flower buds failed to attain the requisite potential for blooming (Walworth et al., 2016). Therefore, while VcFT-CX did show phenotypic outcomes including expedited flowering and heightened floral bud formation, it remained inadequate in replicating the roles of chilling requirements intrinsic to blueberry flowering. Notably, the trend of expedited flowering attributed to the constitutive expression of FT orthologs and the contrasting delay occasioned by TFL1-CX has been extensively documented across an expanding array of woody plants. Despite this, further investigation is necessary to reveal the intricate involvement of FT in the context of CR-mediated flowering, a domain ripe for exploration (Table 1).
FT exhibits versatile functionality. Evident from previous research, the overexpression of FT orthologs in woody plants such as kiwifruit FT (AcFT) and VcFT-CX resulted in premature flowering within in vitro transformed shoots. However, this effect proved to be potentially overwhelming, hampering the shoots from evolving into viable plants (Moss et al., 2018; Song et al., 2019). The impact of FT-CX at transcript levels becomes readily apparent through comprehensive RNA sequencing analysis. Notably, instances like VcFT-CX provide insight into its extensive impact, significantly elevating VcFT expression in both leaves and flower buds; intriguingly, this upregulation was notably absent in roots. Simultaneously, thousands of differentially expressed genes (DEGs) attributed to VcFT expression varied across different tissues and developmental stages, even within the same tissue (Walworth et al., 2016; Song et al., 2019; Song et al., 2023).
When scrutinizing major blueberry flowering pathway genes including VcSOC1, VcAP1, VcFUL, VcLFY, VcSPLs, and VcSVP across three distinct tissues, intriguing patterns emerge:
1) In the apical shoot meristems where VcFT-CX induces early flowering, its influence extends to the upregulation of VcSOC1, VcAP1, VcFUL, VcLFY, and VcSPLs (Walworth et al., 2016).
2) In the mature leaves along the one-year-old shoot, where VcFT-CX results in the emergence of non-blooming floral buds, VcAP1 and VcFUL experience upregulation, while VcSOC1 and VcSVP are repressed (Walworth et al., 2016).
3) Within nonchilled VcFT-CX buds, VcLFY expression is elevated, whereas VcFUL, VcSOC1, and VcSVP experience repression (Walworth et al., 2016).
4) When considering VcFT-CX influence in roots, VcFUL and VcSPLs encounter increased expression, while VcSOC1 and VcSVP are repressed (Song et al., 2019).
These intricate observations collectively suggest that VcFT-CX induces signals for floral bud formation, at least partly through the upregulation of VcAP1 and VcFUL in leaves. Additionally, the expressions of VcSOC1 and VcSVP appear pivotal in determining the timing of both developing and mature floral bud break. The promotion of flowering by AP1 in various woody plants supports these findings (see Table 1). Counter to the flowering promotion led by VcFT-CX, the functional opposite, VcTFL1, triggers flowering delay (Omori et al., 2020; Omori et al., 2021; Omori et al., 2022). Intriguingly, VcFT-CX resulted in a reduction in VcTFL1 expression within young leaves (Walworth et al., 2016). Drawing parallels from Arabidopsis, where FT competes with TFLs for FD binding (Hanano and Goto, 2011; Zhu et al., 2020b). VcFT-CX led to a surprising decrease in VcFD expression within nonchilled flower buds. This observation underscores the likelihood of an interaction between VcFT and VcFD within floral buds, indicating their interplay.
The hereditary promotion of flowering through FT-CX has been substantiated within both self- and cross-pollinated Eucalyptus seedlings (Klocko et al., 2016). Conversely, in poplar, the constitutive expression of LFY, AP1, and CO resulted in marginal to negligible advancements in early flowering, a contrast to the robust effect observed with FT-CX (Rottmann et al., 2000; Zhang et al., 2010; Klocko et al., 2016). Similarly, the hereditary transmission of VcFT-CX translated to a remarkable reduction in flowering time for cross-pollinated, transgenic blueberry seedlings, swiftly transitioning them to bloom within a few months, in comparison to the 2-3 years characteristic of their nontransgenic counterparts (Our unpublished data). Clearly, FT-CX emerges as a potent factor in accelerating the transition from the juvenile phase.
FT has consistently remained a prominent candidate in the pursuit of identifying the elusive florigen. FT originates within leaves and subsequently moves to the meristems (Turck et al., 2008; Fornara et al., 2010; Krzymuski et al., 2015). Intriguing insights have indicated from transgrafting experiments involving FT-CX materials, underscoring the role of FT-CX in signaling the onset of flowering. In several instances, the FT-CX generated within transgenic leaves, functioning as either a direct or an indirect florigenic signal, exhibited the remarkable capacity to promote flowering in nontransgenic scions through long-distance transportation (Ye et al., 2014; Song et al., 2019; Wu et al., 2022). This phenomenon diverges distinctly from parallel transgrafting studies where FT-CX produced in transgenic roots and stems (in the absence of transgenic leaves) failed to incite flowering in nontransgenic scions (Zhang et al., 2010; Srinivasan et al., 2012; Wenzel et al., 2013; Bull et al., 2017).
In transgrafted blueberry where the transgenic leaves were retained, the influence of VcFT-CX within the transgenic rootstock precipitated floral bud formation within the shoot tips of nontransgenic scions. However, VcFT exhibited negligible alterations, while a cluster of phytohormone genes in nontransgenic scions showed varying expressions (Song et al., 2019). Collectively, the evidence demonstrates the status of FT as a universal catalyst for the initiation of floral bud formation and the hastening flowering process. To gain a more comprehensive understanding of the long-range florigenic signals originating from FT or FT-CX, whether in the form of FT protein, FT mRNA, or other derivatives such as phytohormones, further investigations are needed to unravel this intriguing aspect (Wilkie et al., 2008; Izawa, 2021).
SOC1 stands as a central integrator within the flowering pathway (see review by Lee and Lee, 2010). Evidence across various plant species highlights SOC1’s role as a ubiquitous accelerator of flowering, underscoring its significance (Table 1) (Lee et al., 2004; Lee et al., 2008; Seo et al., 2009; Alter et al., 2016; Han et al., 2021; Song et al., 2021). In Arabidopsis, SOC1 takes on the role of a coordinator, integrating signals from diverse pathways. These connections include the aging pathway, mediated by SPLs, the vernalization/autonomous pathway through FLC, the photoperiod pathway involving FT, and the GA pathway, facilitated by GA (Figure 2). Notably, SOC1’s influence extends to the activation of the LFY gene, a key step in establishing the identity of floral meristems or organs (Lee and Lee, 2010).
SOC1, alongside FLC, AP1, AGL24, SVP, FUL, and CAL, represents a cohort of MADS box genes encoding MIKCc type proteins characterized by four conserved domains: MADS (M-), intervening (I-), Keratin-like (K-), and C-terminal (C-) (Gramzow and Theissen, 2010; Gramzow and Theissen, 2015). This array of MADS box genes assumes dual roles, vital both in the context of the ABC model of floral development and in governing the temporal aspects of flowering (Amasino, 2010; Heijmans et al., 2012; Smaczniak et al., 2012; Su et al., 2018). Conventionally, AP1 and SOC1 have emerged as accelerators of flowering across diverse plant species. Conversely, FLC serves as a repressor, exerting repression on the expression of both FT and SOC1 in Arabidopsis. Within Arabidopsis’ vernalization pathway, the antagonistic action of negative regulators, FLC and SVP, counters the positive regulators SOC1 and AGL24 (Fornara et al., 2010; Lee and Lee, 2010). The investigations on CR-mediated flowering woody plant flowering have identified five groups of MADS box genes, specifically the orthologs of FLC, SOC1, SVP, AGL24, and DAMs (Table 1). Amidst these gene clusters, one consensus emerges: the SOC1 group, a positive regulator, can steer the course of floral initiation and hasten flowering. Yet, the roles of the remaining four gene groups exhibit divergence across diverse plant species. For instance, while FLC-like genes have been identified, the extent of their conserved functions in woody plants remains largely unknown (Table 1). Notably, apple’s FLC-like genes do not mirror FLC’s functions precisely (Porto et al., 2015; Nishiyama et al., 2021). Divergent from expectations, a kiwifruit FLC-like variant expedites flowering, in contrast to FLC’s recognized role in delaying it (Voogd et al., 2022). Similarly, the constitutive expression of an apple FLC3 variant accelerates flowering in blueberry (Zong et al., 2019; Kagaya et al., 2020).
In woody plants, the CR is orchestrated by MADS box genes DAMs, AGL24-like genes, and SVP-like genes (SVLs), which are prominent genes akin to FLC functions albeit the scarcity of reverse genetic substantiation (Zhu et al., 2020a; da Silveira Falavigna et al., 2021; Jewaria et al., 2021). These CR-associated MADS box genes act upstream of SOC1 orthologs and can steer the course of floral development. With chilling accumulation, SOC1 orthologs are activated. For instance, the grape, blueberry, and apple exhibit an increase in expression of SOC1 orthologs in response to the accrual of chilling hours (Hattasch et al., 2008; Song and Chen, 2018b; Kamal et al., 2019). In poplar (Populus tremula × alba), overexpression of a SOC1-like variant leads to bud break (Gómez-Soto et al., 2021; Goralogia et al., 2021). In kiwifruit (Actinidia delicious), SOC1-like genes potentially influence the duration of dormancy, although their role in the transition to flowering remains inconclusive (Voogd et al., 2015). Genetic investigations have shown a linkage between alleles of SOC1 orthologs and the chilling requisites in apricot (Prunus armeniaca L.) and peach genotypes, underlining a pronounced correlation (Trainin et al., 2013; Halasz et al., 2021). Collectively, it is the expression of SOC1 orthologs that governs the poised readiness for floral bud break and activation subsequent to fulfilling the chilling requirement in woody plants.
In Arabidopsis, the decreased expression of SVP during vernalization sets in motion the activation of SOC1 (or SOC1-like gene), thereby initiating the onset of flowering. However, in woody plants, the involvement of SVP homologs and SVLs in the flowering process exhibits a spectrum of variance contingent upon the particular SVP homologs in play. To illustrate, the SVP homologs and SVLs in kiwifruit, trifoliate orange (Poncirus trifoliata L. Raf.), apple, and sweet cherry (Prunus avium L.) play the role of suppressors, effectively suppressing budbreak and the flowering cycle (Gregis et al., 2006; Li et al., 2010; Wu et al., 2017a; Wu et al., 2017b; Wang et al., 2021). Meanwhile, in grapevines, the SVP homologs unveil a degree of inconsistency, alternating between acting as promoters or inhibitors of flowering (Diaz-Riquelme et al., 2012; Li-Mallet et al., 2016; Arro et al., 2019; Kamal et al., 2019; Dong et al., 2022).
The highbush blueberry (2n = 4x = 48), a prominent cultivated member of the Vaccinium fruit crop family, has a rather substantial chilling requirement, typically surpassing 800 chilling units, that must be met to initiate dormancy release during spring (Song et al., 2011; Edger et al., 2022). Over the course of past decades, extensive investigations have been carried out to reveal the flowering mechanism (Song et al., 2023). A summary of these blueberry studies can serve as an illustrative example, providing insights into the network of factors that underlie flowering mechanisms in woody plants.
The impact of VcFT-CX is evident across different plant species. In tobacco (Nicotiana tabacum) and petunia (Petunia x hybrid), VcFT-CX not only induced early flowering but also led to plant dwarfing (Song et al., 2013b). Similarly, under nonchilling conditions, the northern highbush blueberry cultivar Aurora exhibited precocious flowering as a result of VcFT-CX (Song et al., 2013b). At the transcript level, VcFT-CX triggered a substantial increase in VcFT expression in leaves and nonchilled floral buds, while its effect on young roots was not significant (Walworth et al., 2016; Song et al., 2019; Song et al., 2023). Notably, VcFT-CX displayed distinct effects on various tissues and developmental stages (Walworth et al., 2016; Song et al., 2019; Song et al., 2023). In young leaves, it upregulated the expressions of VcAP1/VcFUL, blueberry SEPALLATA (VcSEP), VcLFY, VcSOC1, and VcTFL1, with no significant changes in VcSVP and VcFD (Figure 3A) (Walworth et al., 2016). In mature leaves, VcFT-CX enhanced VcAP1/VcFUL and VcSEP expressions, while repressing VcSOC1 and VcSVP, and it had minimal impact on VcLFY, VcFD, and VcTFL1 (Figure 3B) (Song et al., 2023). In nonchilled flower buds, VcFT-CX upregulated VcLFY expression, downregulated VcSEP, VcSOC1, VcSVP, VcFD, and VcTFL1, while VcAP1/VcFUL expression remained relatively unaffected (Figure 3C) (Song et al., 2023). In young roots, VcAP1/VcFUL expression increased, while VcSOC1 and VcSVP decreased; VcSEP, VcLFY, VcFD, and VcTFL1 showed no significant changes (Figure 3D) (Song et al., 2019). Key takeaways from the analysis of VcFT-CX tissues include: 1) Varied responses of major flowering pathway genes (e.g., VcSEP3, VcSOC1, and VcSVP) to VcFT-CX across tissues and developmental stages, with a consistent promotion of VcLFY and VcAP1/VcFUL expression; 2) Enhanced expressions of VcAP1/VcFUL and VcSEP in leaf tissues due to VcFT-CX, indicating the potential role of these genes in floral initiation, while VcFD and VcTFL1 seem less involved in promoting floral initiation; 3) Repression of VcFD and VcTFL1 expressions in nonchilled floral buds by VcFT-CX; and 4) Likely pivotal roles of VcSOC1 and VcSVP in the activation of floral buds under chilling conditions.
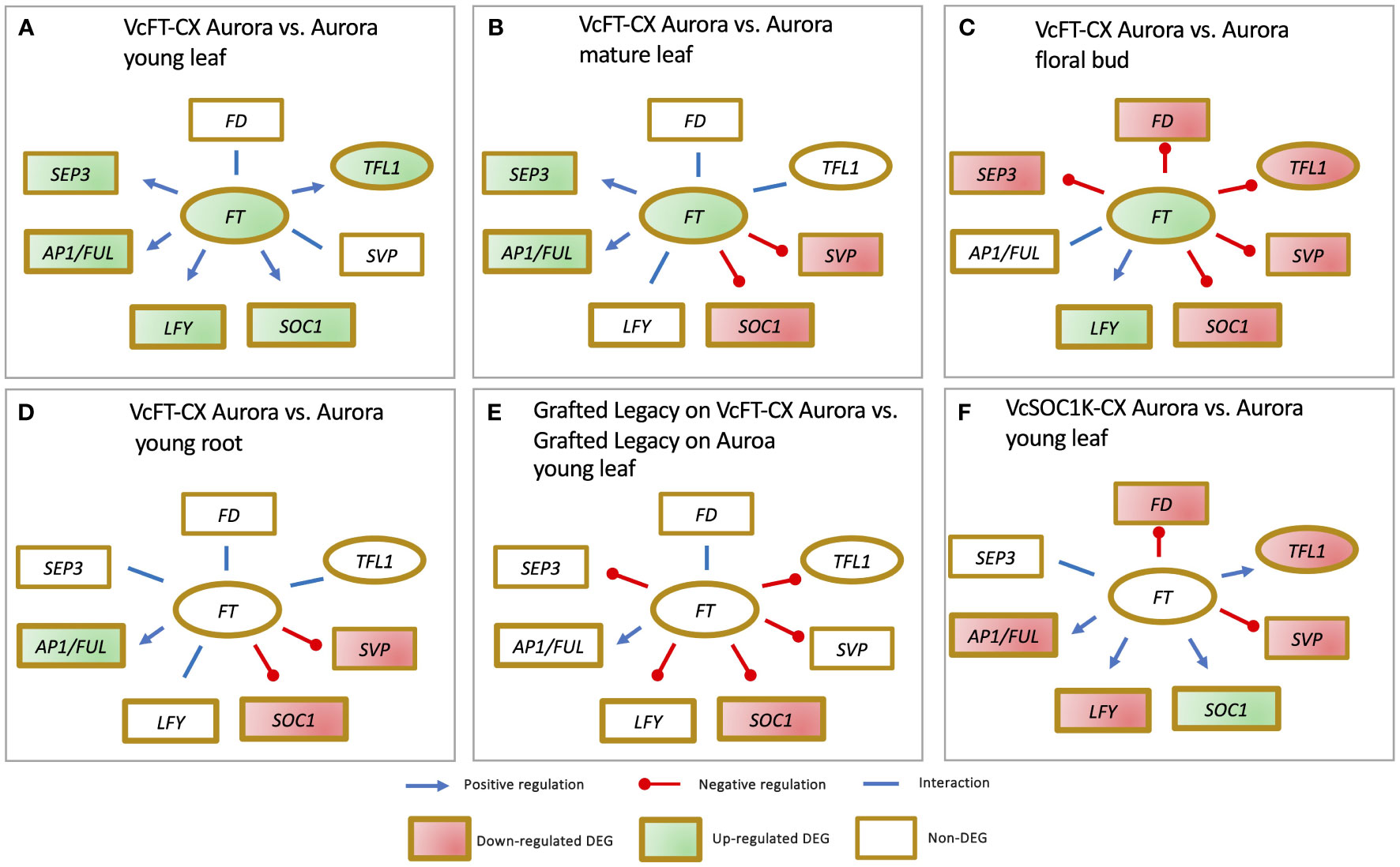
Figure 3 RNA-sequencing data reveals impact of VcFT-CX (A-E) and VcSOC1K-CX (F) on major flowering pathway genes in different tissues.
Moreover, as rootstocks, VcFT-CX produced signals in leaves that were effectively conveyed through transgrafting to nontransgenic scions (cv. Legacy), yielding a distinctive enhancement in floral bud formation (Song et al., 2019). Intriguingly, at the transcript level, VcFT-CX in rootstocks did not trigger differential expression of VcFT, VcFD, VcTFL1, VcAP1/VcFUL, VcLFY, VcSVP, and VcSEP3 in the grafted nontransgenic scions. Interestingly, expression of VcSOC1 was significantly downregulated (Figure 3E). Notably, there is an instance where none of the identified major flowering pathway genes (e.g., VcFT, VcAP1/VcFUL, VcSOC1, and VcLFY) appear to solely account for the promoted floral bud formation, indicating that an escalated VcFT expression is not always the sole requirement for initiating flowering (Song et al., 2019). As for the potential long-distance florigenic signals inducing from VcFT-CX, their precise nature remains to be discerned from candidates like VcFT protein/mRNA, cytokinin, or other hormonal factors (Gao et al., 2016; Walworth et al., 2016; Song et al., 2019).
Comparative analyses of floral buds have been conducted for four genotypes, including a nontransgenic northern highbush variety Aurora, a VcFT-CX transgenic ‘Aurora’, a nontransgenic southern highbush variety Legacy, and a transgenic Legacy mutant (Mu1-Legacy) (Figure 4) (Song and Chen, 2018b; Song and Walworth, 2018; Song et al., 2023). Among these four comparisons: 1) VcFT expression demonstrated either negligible differential expression in nontransgenic cultivars or downregulation in the two transgenic genotypes, suggesting that VcFT may not a primary target of chilling accumulation for bud break; 2) Expression of VcLFY, VcTFL1, and VcFD remained either suppressed or constant post full chilling; 3) VcAP1/VcFUL expression increased in three genotypes and decreased in VcFT-CX ‘Aurora’; and 4) VcSOC1 expression was upregulated in three genotypes and exhibited no significant differential expression in VcFT-CX ‘Aurora’, while VcSVP expression was elevated in all four genotypes (Figure 4). Collectively, VcSOC1 and VcSVP played key roles in chilling requirement-mediated floral activation. Interestingly, in transcriptomic comparisons between late pink buds and fully chilled stages for two genotypes, expressions of VcFT, VcFD, VcTFL1, VcAP1/VcFUL, VcLFY, and VcSOC1 were uniformly repressed in late pink buds (Figure 4) (Song and Chen, 2018b).
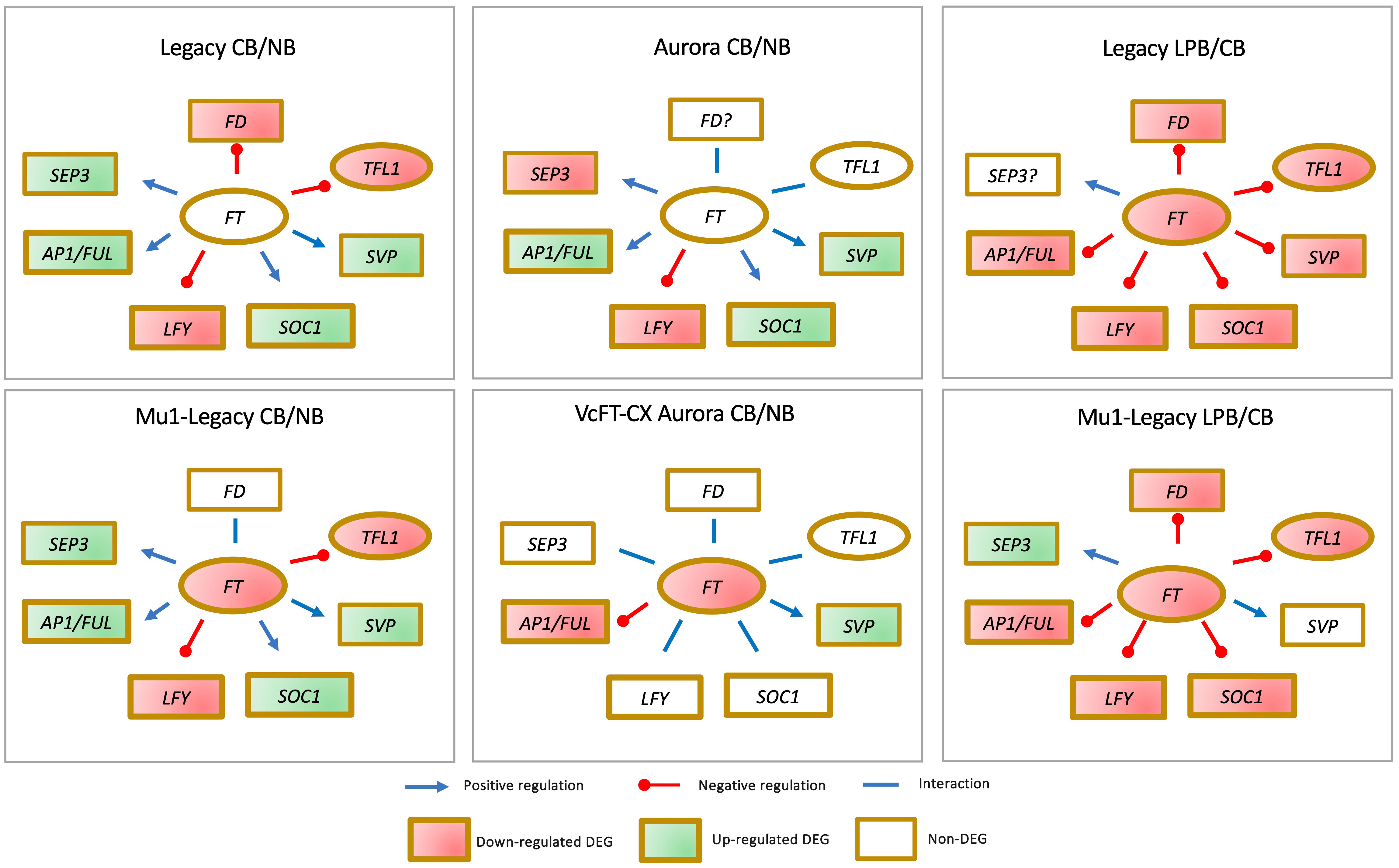
Figure 4 Distinctive gene expression patterns observed in the comparisons between fully chilled floral buds (CB) and nonchilled floral buds (NB), as well as late pink bud (LPB) versus CB, across different genotypes.
Further evidence supporting VcSOC1 as a significant floral activator is the fact that the constitutive expression of the K domain of VcSOC1 (VcSOC1K-CX) led to the flowering of transgenic ‘Aurora’ plants under nonchilling conditions, a condition where nontransgenic ‘Aurora’ plants remained unable to flower (Song and Chen, 2018a). SOC1 is classified as a type-II plant-specific MIKC protein, characterized by its conserved MADS (M-), intervening (I), keratin-like (K-), and C-terminal (C-) domains (Theissen et al., 1996). The K domain is instrumental in facilitating interactions among various MADS box genes. Remarkably, VcSOC1K-CX has also demonstrated the ability to accelerate flowering in tobacco and maize (Song et al., 2013a; Song and Han, 2021). In blueberry, the promotion of flowering through VcSOC1K-CX was associated by the increased expression of VcSOC1, which in turn led to the repression of VcFT, VcFD, VcTFL1, VcAP1/VcFUL, VcLFY, and VcSVP, offering another piece of evidence that the elevation of VcFT expression is not always a prerequisite for flowering promotion in ‘Aurora’ (Figure 3F) (Song and Chen, 2018a).
While the expression of VcSOC1 is indeed crucial for CR-mediated floral activation in blueberry, it is important to note that VcSOC1 does not always play an obligatory role in floral bud activation. An intriguing instance is presented by the Mu1-Legacy genotype, which carries an overexpressed blueberry DWARF AND DELAYING FLOWERING 1 gene (VcDDF1), allowing it to flower under nonchilling conditions, a feat nontransgenic ‘Legacy’ plants could not achieve (Song and Walworth, 2018). Remarkably, in this scenario, none of the major genes—VcSOC1, VcFT, VcAP1/VcFUL, VcLFY, VcSVP, and VcSEP3—displayed discernible differential expression in both young leaves and floral buds (Figure 5) (Song and Walworth, 2018; Lin et al., 2019).
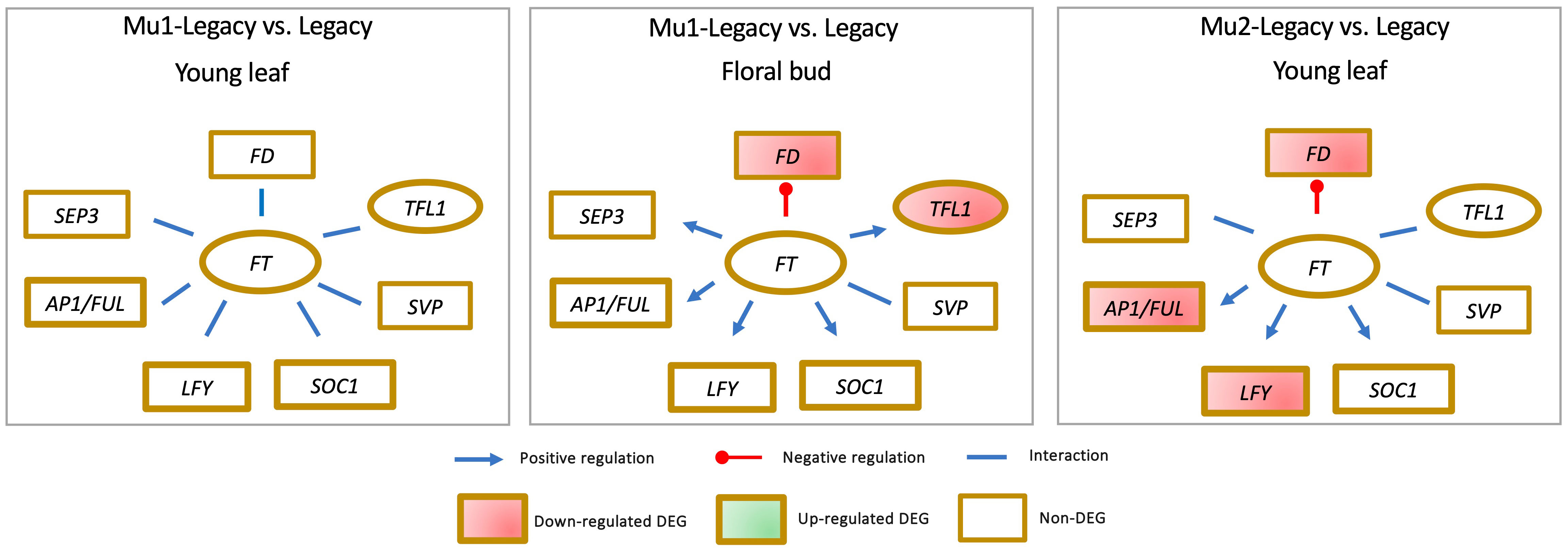
Figure 5 The identification of differential expression of genes (DEGs) in blueberry mutants indicates that elevated VcFT expression is not always a prerequisite for achieving precocious or early flowering.
As of now, functional FLC-like candidates have not been definitively identified, despite the presence of orthologues of many other vernalization pathway genes from Arabidopsis in blueberry (Walworth et al., 2016). Notably, the intriguing case of an apple FLC3-like gene stands out, as its constitutive expression surprisingly promotes flowering instead of causing the expected delay (Zong et al., 2019). Although there have been studies examining the effects of chilling accumulation on the expression of flowering pathway genes, the impact of warm accumulation on the activation of fully chilled floral buds remains an area yet to be thoroughly explored. In light of the available literature, it is apparent that among the genes within the flowering pathway, VcSOC1 plays a pivotal role as a major floral activator.
In general, the expression of FT within leaves is significantly influenced by light conditions, whereas SOC1 expression is modulated in response to temperature changes. Notably, VcFT attains its peak expression in floral buds, while VcSOC1 reaches its highest expression level in leaves (Walworth et al., 2016). Recently, a regulatory framework centered on the ratio of FT-to-SOC1 expression (VcFT/VcSOC1) has been proposed, providing a valuable lens through which to comprehend the processes of floral initiation and activation. According to this model, an elevated VcFT/VcSOC1 ratio in leaves serves to stimulate floral initiation, while heightened VcSOC1 expression can lead to early flowering. Within flower buds, the VcFT/VcSOC1 ratios often decline in chilled buds due to the increasing VcSOC1 expression during chilling accumulation, whereas emerging flower buds exhibit rising VcFT/VcSOC1 ratios due to the more rapid decline of VcSOC1 expression relative to VcFT (Song et al., 2023). This principle is further bolstered by observations of reduced FT/SOC1 ratios in polar buds during chilling accumulation, marked by an upsurge in SOC1 expression alongside neutral FT levels (Gómez-Soto et al., 2021). Nonetheless, it’s important to acknowledge that this VcFT/VcSOC1 ratio might not be universally applicable to all tested blueberry genotypes; for instance, the altered flowering pattern in the Legacy-mutant1 was not attributable to major flowering pathway genes (Song and Walworth, 2018). Collectively, this FT/SOC1 ratio emerges as a potential determinant of both leaf-based floral initiation and bud-based floral activation, given that these genes hold pivotal roles as integrators within the flowering pathway.
In addition to the well-defined flowering pathway genes, there exists a range of genes from other pathways that exert influence over floral initiation and activation. A notable instance, as discussed earlier, is the altered floral initiation and activation process observed in Mu1-Legacy, wherein neither VcFT nor VcSOC1 played a role (Figure 5). Upon scrutinizing the transcriptomic analysis of blueberry flowering pathway genes, it becomes evident that numerous genes from hormone and sugar pathways are intricately linked to floral bud initiation or activation, whether through direct or indirect means (Gao et al., 2016; Lin et al., 2019; Song et al., 2019). This underlies the fact that hormones and sugar pathway genes, in conjunction with the flowering pathway genes, likely participate in a coordinated manner to regulate the process of flowering (Izawa, 2021).
The well-established genetic framework governing flowering pathways in Arabidopsis has served as a cornerstone for unraveling the intricate mechanisms operating in other plants. Woody plants, however, have developed notably complex flowering pathways compared to Arabidopsis, although some key flowering pathway genes maintain largely conserved roles (Table 1, Figure 2). The CO-FT module within the photoperiod pathway, which is crucial in Arabidopsis, appears to be considerably conserved in woody plants, although the functions of CO or COL orthologues in this context warrant further investigation. The miR156-SPL module of the age pathway exhibits conservation across all plant species, notwithstanding the varied roles of SPLs in woody plants. In the GA pathway, the interactions involving GA and DELLA factors demand deeper exploration in both Arabidopsis and woody plants due to their extensive influence on both floral initiation and activation. The FLC-mediated vernalization pathway, a central mechanism in Arabidopsis, exhibits the least conservation in woody plants, where effective chilling is requisite to initiate flowering. Nonetheless, it’s noteworthy that MADS-box genes play significant roles in floral activation. In essence, among the individual flowering pathway genes, FT and its orthologues serve as pivotal floral initiators, while SOC1 and its orthologs stand as the principal floral activators. Remarkably, this pattern remains highly conserved across plant species.
G-qS: Conceptualization, Writing – original draft, Writing – review & editing. ZL: Writing – review & editing. G-yZ: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Albani, M. C., Coupland, G. (2010). Comparative analysis of flowering in annual and perennial plants. Curr. Top. Dev. Biol. 91, 323–348. doi: 10.1016/s0070-2153(10)91011-9
Alexandre, C. M., Hennig, L. (2008). FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59 (6), 1127–1135. doi: 10.1093/jxb/ern070
Almada, R., Cabrera, N., Casaretto, J. A., Ruiz-Lara, S., Gonzalez Villanueva, E. (2009). VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28 (8), 1193–1203. doi: 10.1007/s00299-009-0720-4
Alter, P., Bircheneder, S., Zhou, L. Z., Schluter, U., Gahrtz, M., Sonnewald, U., et al. (2016). Flowering time-regulated genes in maize include the transcription factor zmMADS1. Plant Physiol. 172 (1), 389–404. doi: 10.1104/pp.16.00285
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61 (6), 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Andres, F., Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13 (9), 627–639. doi: 10.1038/nrg3291
Andres, F., Porri, A., Torti, S., Mateos, J., Romera-Branchat, M., Garcia-Martinez, J. L., et al. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. United. States America 111 (26), E2760–E2769. doi: 10.1073/pnas.1409567111
Arora, R., Rowland, L. J., Tanino, K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38, 11. doi: 10.21273/HORTSCI.38.5.911
Arro, J., Yang, Y., Song, G.-Q., Zhong, G.-Y. (2019). RNA-Seq reveals new DELLA targets and regulation in transgenic GA-insensitive grapevines. BMC Plant Biol. 19 (1), 80. doi: 10.1186/s12870-019-1675-4
Atkinson, C. J., Brennan, R. M., Jones, H. G. (2013). Declining chilling and its impact on temperate perennial crops. Environ. Exp. Bot. 91, 48–62. doi: 10.1016/j.envexpbot.2013.02.004
Bai, S., Tuan, P. A., Saito, T., Ito, A., Ubi, B. E., Ban, Y., et al. (2017). Repression of TERMINAL FLOWER1 primarily mediates floral induction in pear (Pyrus pyrifolia Nakai) concomitant with change in gene expression of plant hormone-related genes and transcription factors. J. Exp. Bot. 68 (17), 4899–4914. doi: 10.1093/jxb/erx296
Bao, S. J., Hua, C. M., Shen, L. S., Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62 (1), 118–131. doi: 10.1111/jipb.12892
Baurle, I., Dean, C. (2006). The timing of developmental transitions in plants. Cell 125 (4), 655–664. doi: 10.1016/j.cell.2006.05.005
Bielenberg, D. G., Wang, Y., Li, Z. G., Zhebentyayeva, T., Fan, S. H., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 (3), 495–507. doi: 10.1007/s11295-007-0126-9
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 (5776), 1040–1043. doi: 10.1126/science.1126038
Boss, P. K., Sreekantan, L., Thomas, M. R. (2006). A grapevine TFL1 homologue can delay flowering and alter floral development when overexpressed in heterologous species. Funct. Plant Biol. 33 (1), 31–41. doi: 10.1071/fp05191
Boss, P. K., Thomas, M. R. (2002). Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416 (6883), 847–850. doi: 10.1038/416847a
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275 (5296), 80–83. doi: 10.1126/science.275.5296.80
Branchereau, C., Quero-Garcia, J., Zaracho-Echague, N. H., Lambelin, L., Fouche, M., Wenden, B., et al. (2022). New insights into flowering date in Prunus: fine mapping of a major QTL in sweet cherry. Horticult. Res. 9, uhac042. doi: 10.1093/hr/uhac042
Briones-Moreno, A., Hernández-García, J., Vargas-Chávez, C., Blanco-Touriñán, N., Phokas, A., Úrbez, C., et al. (2023). DELLA functions evolved by rewiring of associated transcriptional networks. Nat. Plants 9 (4), 535–543. doi: 10.1038/s41477-023-01372-6
Brunner, A. M., Evans, L. M., Hsu, C. Y., Sheng, X. (2014). Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00732
Bull, S. E., Alder, A., Barsan, C., Kohler, M., Hennig, L., Gruissem, W., et al. (2017). FLOWERING LOCUS T triggers early and fertile flowering in glasshouse cassava (Manihot esculenta crantz). Plants (Basel) 6 (2), 22. doi: 10.3390/plants6020022
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-cas9 system. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00040
Chen, Y. H., Jiang, P., Thammannagowda, S., Liang, H. Y., Wilde, H. D. (2013). Characterization of peach TFL1 and comparison with FT/TFL1 gene families of the rosaceae. J. Am. Soc. Hortic. Sci. 138 (1), 12–17. doi: 10.21273/Jashs.138.1.12
Chouard, P. (1960). Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. Plant Mol. Biol. 11, 191–238. doi: 10.1146/annurev.pp.11.060160.001203
Conti, L. (2017). Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 430 (2), 288–301. doi: 10.1016/j.ydbio.2017.03.024
da Silveira Falavigna, V., Severing, E., Lai, X., Estevan, J., Farrera, I., Hugouvieux, V., et al. (2021). Unraveling the role of MADS transcription factor complexes in apple tree dormancy. New Phytol. 232 (5), 2071–2088. doi: 10.1111/nph.17710
Davis, S. J. (2009). Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 32 (9), 1201–1210. doi: 10.1111/j.1365-3040.2009.01968.x
Diaz-Riquelme, J., Grimplet, J., Martinez-Zapater, J. M., Carmona, M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 12, 181. doi: 10.1186/1471-2229-12-181
Diaz-Riquelme, J., Lijavetzky, D., Martinez-Zapater, J. M., Carmona, M. J. (2009). Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 149 (1), 354–369. doi: 10.1104/pp.108.131052
Diaz-Riquelme, J., Martinez-Zapater, J. M., Carmona, M. J. (2014). Transcriptional analysis of tendril and inflorescence development in grapevine (Vitis vinifera L.). PloS One 9 (3), e92339. doi: 10.1371/journal.pone.0092339
Dill, A., Sun, T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 (2), 777–785. doi: 10.1093/genetics/159.2.777
Do, V. G., Lee, Y., Kim, S., Kweon, H., Do, G. (2022). Antisense expression of apple TFL1-like gene (MdTFL1) promotes early flowering and causes phenotypic changes in tobacco. Int. J. Mol. Sci. 23 (11), 6006. doi: 10.3390/ijms23116006
Dong, Y., Khalil-Ur-Rehman, M., Liu, X., Wang, X., Yang, L., Tao, J., et al. (2022). Functional characterisation of five SVP genes in grape bud dormancy and flowering. Plant Growth Regul. 97 (3), 511–522. doi: 10.1007/s10725-022-00817-w
Edger, P. P., Iorizzo, M., Bassil, N. V., Benevenuto, J., Ferrão, L. F. V., Giongo, L., et al. (2022). There and back again; historical perspective and future directions for Vaccinium breeding and research studies. Horticult. Res. 9, uhac083. doi: 10.1093/hr/uhac083
Falavigna, V. D. S., Guitton, B., Costes, E., Andres, F. (2018). I want to (Bud) break free: the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01990
Falavigna, V. D., Porto, D. D., Buffon, V., Margis-Pinheiro, M., Pasquali, G., Revers, L. F. (2014). Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol. Biol. Rep. 32 (4), 796–813. doi: 10.1007/s11105-013-0690-0
Falavigna, V. S., Severing, E., Estevan, J., Farrera, I., Hugouvieux, V., Revers, L. F., et al. (2022). Potential role of apple SOC1-like transcription factors in gene regulatory networks involved in bud dormancy. Acta Hortic. 1342), 8. doi: 10.17660/ActaHortic.2022.1342.6
Fang, Z. Z., Kui, L. W., Dai, H., Zhou, D. R., Jiang, C. C., Espley, R. V., et al. (2022). The genome of low-chill Chinese plum “Sanyueli” (Prunus salicina Lindl.) provides insights into the regulation of the chilling requirement of flower buds. Mol. Ecol. Resour. 22 (5), 1919–1938. doi: 10.1111/1755-0998.13585
Flachowsky, H., Hattasch, C., Hofer, M., Peil, A., Hanke, M. V. (2010). Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta 231 (2), 251–263. doi: 10.1007/s00425-009-1041-0
Flachowsky, H., Szankowski, I., Waidmann, S., Peil, A., Trankner, C., Hanke, M. V. (2012). The MdTFL1 gene of apple (Malus x domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 32 (10), 1288–1301. doi: 10.1093/treephys/tps080
Fleck, B., Harberd, N. P. (2002). Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J. 32 (6), 935–947. doi: 10.1046/j.1365-313X.2002.01478.x
Fornara, F., de Montaigu, A., Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. Cell 141 (3), 550–550e2. doi: 10.1016/j.cell.2010.04.024
Freiman, A., Golobovitch, S., Yablovitz, Z., Belausov, E., Dahan, Y., Peer, R., et al. (2015). Expression of flowering locus T2 transgene from Pyrus communis L. delays dormancy and leaf senescence in Malus x domestica Borkh, and causes early flowering in tobacco. Plant Sci. 241, 164–176. doi: 10.1016/j.plantsci.2015.09.012
Gao, X., Walworth, A. E., Mackie, C., Song, G. Q. (2016). Overexpression of blueberry FLOWERING LOCUS T is associated with changes in the expression of phytohormone-related genes in blueberry plants. Hortic. Res. 3, 16053. doi: 10.1038/hortres.2016.53
Gao, Y. H., Yang, Q. S., Yan, X. H., Wu, X. Y., Yang, F., Li, J. Z., et al. (2021). High-quality genome assembly of ‘Cuiguan’ pear (Pyrus pyrifolia) as a reference genome for identifying regulatory genes and epigenetic modifications responsible for bud dormancy. Horticult. Res. 8 (1), 197. doi: 10.1038/s41438-021-00632-w
Gendall, A. R., Levy, Y. Y., Wilson, A., Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 (4), 525–535. doi: 10.1016/S0092-8674(01)00573-6
Goldberg-Moeller, R., Shalom, L., Shlizerman, L., Samuels, S., Zur, N., Ophir, R., et al. (2013). Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci. 198, 46–57. doi: 10.1016/j.plantsci.2012.09.012
Gómez-Soto, D., Ramos-Sánchez, J. M., Alique, D., Conde, D., Triozzi, P. M., Perales, M., et al. (2021). Overexpression of a SOC1-related gene promotes bud break in ecodormant poplars. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.670497
Goralogia, G. S., Howe, G. T., Brunner, A. M., Helliwell, E., Nagle, M. F., Ma, C., et al. (2021). Overexpression of SHORT VEGETATIVE PHASE-LIKE (SVL) in Populus delays onset and reduces abundance of flowering in field-grown trees. Hortic. Res. 8 (1), 167. doi: 10.1038/s41438-021-00600-4
Gramzow, L., Theissen, G. (2010). A hitchhiker’s guide to the MADS world of plants. Genome Biol. 11 (6), 214. doi: 10.1186/gb-2010-11-6-214
Gramzow, L., Theissen, G. (2015). Phylogenomics reveals surprising sets of essential and dispensable clades of MIKC(c)-group MADS-box genes in flowering plants. J. Exp. Zool. B. Mol. Dev. Evol. 324 (4), 353–362. doi: 10.1002/jez.b.22598
Greenup, A., Peacock, W. J., Dennis, E. S., Trevaskis, B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103 (8), 1165–1172. doi: 10.1093/aob/mcp063
Gregis, V., Sessa, A., Colombo, L., Kater, M. M. (2006). AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18 (6), 1373–1382. doi: 10.1105/tpc.106.041798
Griffiths, S., Dunford, R. P., Coupland, G., Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131 (4), 1855–1867. doi: 10.1104/pp.102.016188
Guo, R., Wang, B., Lin, L., Cheng, G., Zhou, S., Xie, S., et al. (2018). Evolutionary, interaction and expression analysis of floral meristem identity genes in inflorescence induction of the second crop in two-crop-a-year grape culture system. J. Genet. 97 (2), 439–451. doi: 10.1007/s12041-018-0929-5
Halasz, J., Hegedus, A., Karsai, I., Tosaki, A., Szalay, L. (2021). Correspondence between SOC1 genotypes and time of endodormancy break in peach (Prunus persica L. Batsch) cultivars. Agronomy-Basel 11 (7), 1298. doi: 10.3390/agronomy11071298
Han, Y., Tang, A. Y., Yu, J. Y., Cheng, T. R., Wang, J., Yang, W. R., et al. (2019). RcAP1, a homolog of APETALA1, is associated with flower bud differentiation and floral organ morphogenesis in rosa chinensis. Int. J. Mol. Sci. 20 (14), 3557. doi: 10.3390/ijms20143557
Han, X., Wang, D. C., Song, G. Q. (2021). Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 11 (1), 12758. doi: 10.1038/s41598-021-92215-x
Hanano, S., Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23 (9), 3172–3184. doi: 10.1105/tpc.111.088641
Hattasch, C., Flachowsky, H., Kapturska, D., Hanke, M. V. (2008). Isolation of flowering genes and seasonal changes in their transcript levels related to flower induction and initiation in apple (Malus domestica). Tree Physiol. 28 (10), 1459–1466. doi: 10.1093/treephys/28.10.1459
Heijmans, K., Morel, P., Vandenbussche, M. (2012). MADS-box genes and floral development: the dark side. J. Exp. Bot. 63 (15), 5397–5404. doi: 10.1093/jxb/ers233
Huijser, P., Schmid, M. (2011). The control of developmental phase transitions in plants. Development 138 (19), 4117–4129. doi: 10.1242/dev.063511
Hyun, Y., Richter, R., Coupland, G. (2017). Competence to flower: age-controlled sensitivity to environmental cues. Plant Physiol. 173 (1), 36–46. doi: 10.1104/pp.16.01523
Irish, V. F. (2010). The flowering of Arabidopsis flower development. Plant J. 61 (6), 1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x
Izawa, T. (2021). What is going on with the hormonal control of flowering in plants? Plant J. 105 (2), 431–445. doi: 10.1111/tpj.15036
Jeong, D.-H., Sung, S.-K., An, G. (1999). Molecular cloning and characterization of constans-like cDNA clones of the fuji apple. J. Plant Biol. 42 (1), 23–31. doi: 10.1007/BF03031143
Jewaria, P. K., Hanninen, H., Li, X., Bhalerao, R. P., Zhang, R. (2021). A hundred years after: endodormancy and the chilling requirement in subtropical trees. New Phytol. 231 (2), 565–570. doi: 10.1111/nph.17382
Jia, X. L., Chen, Y. K., Xu, X. Z., Shen, F., Zheng, Q. B., Du, Z., et al. (2017). miR156 switches on vegetative phase change under the regulation of redox signals in apple seedlings. Sci. Rep. 7 (1), 14223. doi: 10.1038/s41598-017-14671-8
Jiang, Y., Peng, J., Wang, M., Su, W., Gan, X., Jing, Y., et al. (2019a). The role of ejSPL3, ejSPL4, ejSPL5, and ejSPL9 in regulating flowering in loquat (Eriobotrya japonica lindl.). Int. J. Mol. Sci. 21 (1), 248. doi: 10.3390/ijms21010248
Jiang, Y., Peng, J., Zhang, Z., Lin, S., Lin, S., Yang, X. (2019b). The role of ejSVPs in flower initiation in eriobotrya japonica. Int. J. Mol. Sci. 20 (23), 5933. doi: 10.3390/ijms20235933
Jiang, Y., Peng, J., Zhu, Y., Su, W., Zhang, L., Jing, Y., et al. (2019c). The role of ejSOC1s in flower initiation in eriobotrya japonica. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00253
Jiang, Y., Zhu, Y., Zhang, L., Su, W., Peng, J., Yang, X., et al. (2020). EjTFL1 genes promote growth but inhibit flower bud differentiation in loquat. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00576
Jimenez, S., Lawton-Rauh, A. L., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9, 81. doi: 10.1186/1471-2229-9-81
Jimenez, S., Reighard, G. L., Bielenberg, D. G. (2010). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73 (1-2), 157–167. doi: 10.1007/s11103-010-9608-5
Julian, C., Rodrigo, J., Herrero, M. (2011). Stamen development and winter dormancy in apricot (Prunus Armeniaca). Ann. Bot. 108 (4), 617–625. doi: 10.1093/aob/mcr056
Kagaya, H., Ito, N., Shibuya, T., Komori, S., Kato, K., Kanayama, Y. (2020). Characterization of FLOWERING LOCUS C homologs in apple as a model for fruit trees. Int. J. Mol. Sci. 21 (12). doi: 10.3390/ijms21124562
Kamal, N., Ochßner, I., Schwandner, A., Viehöver, P., Hausmann, L., Töpfer, R., et al. (2019). Characterization of genes and alleles involved in the control of flowering time in grapevine. PloS One 14 (7), e0214703. doi: 10.1371/journal.pone.0214703
Kennedy, A., Geuten, K. (2020). The role of FLOWERING LOCUS C relatives in cereals. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.617340
Khan, M. R., Ai, X. Y., Zhang, J. Z. (2014). Genetic regulation of flowering time in annual and perennial plants. Wiley. Interdiscip. Rev. RNA 5 (3), 347–359. doi: 10.1002/wrna.1215
Kim, S. Y., Yu, X., Michaels, S. D. (2008). Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiol. 148 (1), 269–279. doi: 10.1104/pp.108.122606
King, R. W., Evans, L. T. (2003). Gibberellins and flowering of grasses and cereals: prizing open the lid of the “florigen” black box. Annu. Rev. Plant Biol. 54, 307–328. doi: 10.1146/annurev.arplant.54.031902.135029
King, R. W., Moritz, T., Evans, L. T., Martin, J., Andersen, C. H., Blundell, C., et al. (2006). Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol. 141 (2), 498–507. doi: 10.1104/pp.106.076760
Kinoshita, A., Richter, R. (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71 (9), 2490–2504. doi: 10.1093/jxb/eraa057
Klocko, A. L., Goddard, A. L., Jacobson, J. R., Magnuson, A. C., Strauss, S. H. (2021). RNAi suppression of LEAFY gives stable floral sterility, and reduced growth rate and leaf size, in field-grown poplars. Plants (Basel) 10 (8), 1594. doi: 10.3390/plants10081594
Klocko, A. L., Ma, C., Robertson, S., Esfandiari, E., Nilsson, O., Strauss, S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14 (2), 808–819. doi: 10.1111/pbi.12431
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 (5446), 1960–1962. doi: 10.1126/science.286.5446.1960
Koshita, Y., Takahara, T., Ogata, T., Goto, A. (1999). Involvement of endogenous plant hormones (IAA, ABA, GAs) in leaves and flower bud formation of satsuma mandarin (Citrus unshiu Marc.). Sci. Hortic. 79 (3-4), 185–194. doi: 10.1016/S0304-4238(98)00209-X
Kotoda, N., Hayashi, H., Suzuki, M., Igarashi, M., Hatsuyama, Y., Kidou, S., et al. (2010). Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus x domestica Borkh.). Plant Cell Physiol. 51 (4), 561–575. doi: 10.1093/pcp/pcq021
Kotoda, N., Iwanami, H., Takahashi, S., Abe, K. (2006). Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc. Hortic. Sci. 131 (1), 74–81. doi: 10.21273/JASHS.131.1.74
Kotoda, N., Wada, M., Kusaba, S., Kano-Murakami, Y., Masuda, T., Soejima, J. (2002). Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci. 162 (5), 679–687. doi: 10.1016/S0168-9452(02)00024-9
Krzymuski, M., Andres, F., Cagnola, J. I., Jang, S., Yanovsky, M. J., Coupland, G., et al. (2015). The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J. 83 (6), 952–961. doi: 10.1111/tpj.12938
Kumar, G., Arya, P., Gupta, K., Randhawa, V., Acharya, V., Singh, A. K. (2016). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 6, 20695. doi: 10.1038/srep20695
Lee, S., Kim, J., Han, J. J., Han, M. J., An, G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38 (5), 754–764. doi: 10.1111/j.1365-313X.2004.02082.x
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61 (9), 2247–2254. doi: 10.1093/jxb/erq098
Lee, J., Oh, M., Park, H., Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55 (5), 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Li, J., Gao, K., Yang, X., Khan, W. U., Guo, B., Guo, T., et al. (2020). Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus. Int. J. Biol. Macromol. 161, 999–1010. doi: 10.1016/j.ijbiomac.2020.06.056
Li, Z., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 (12), 3521–3530. doi: 10.1093/jxb/erp195
Li, Z.-M., Zhang, J.-Z., Mei, L., Deng, X.-X., Hu, C.-G., Yao, J.-L. (2010). PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol. Biol. 74 (1), 129–142. doi: 10.1007/s11103-010-9660-1
Li-Mallet, A., Rabot, A., Geny, L. (2016). Factors controlling inflorescence primordia formation of grapevine: their role in latent bud fruitfulness? A review. Botany 94 (3), 247–163. doi: 10.1139/cjb-2015-0108
Lin, T. Y., Walworth, A., Zong, X. J., Danial, G. H., Tomaszewski, E. M., Callow, P., et al. (2019). VcRR2 regulates chilling-mediated flowering through expression of hormone genes in a transgenic blueberry mutant. Horticult. Res. 6, 96. doi: 10.1038/s41438-019-0180-0
Liu, Y. X., Hu, G. B., Lin, S. Q., Han, Z. H. (2011). Ectopic expression of LFY and AP1 homologues of loquat. Iii. Int. Symposium. Loquat. 887, 227–232. doi: 10.17660/ActaHortic.2011.887.37
Liu, Y. X., Kong, J., Li, T. Z., Wang, Y., Wang, A. D., Han, Z. H. (2013a). Isolation and characterization of an APETALA1-like gene from pear (Pyrus pyrifolia). Plant Mol. Biol. Rep. 31 (4), 1031–1039. doi: 10.1007/s11105-012-0540-5
Liu, Y., Luo, C., Liang, R., Lan, M., Yu, H., Guo, Y., et al. (2022). Genome-wide identification of the mango CONSTANS (CO) family and functional analysis of two MiCOL9 genes in transgenic Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1028987
Liu, Y., Luo, C., Zhang, X.-J., Lu, X.-X., Yu, H.-X., Xie, X.-J., et al. (2020a). Overexpression of the mango MiCO gene delayed flowering time in transgenic Arabidopsis. Plant Cell Tissue Organ Culture (PCTOC) 143 (1), 219–228. doi: 10.1007/s11240-020-01894-3
Liu, Y. X., Song, H. W., Liu, Z. L., Hu, G. B., Lin, S. Q. (2013b). Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 70 (3), 287–296. doi: 10.1007/s10725-013-9800-0
Liu, Z., Wu, X., Cheng, M., Xie, Z., Xiong, C., Zhang, S., et al. (2020b). Identification and functional characterization of SOC1-like genes in Pyrus bretschneideri. Genomics 112 (2), 1622–1632. doi: 10.1016/j.ygeno.2019.09.011
Liu, L., Xuan, L. J., Jiang, Y. P., Yu, H. (2021). Regulation by FLOWERING LOCUS T and TERMINAL FLOWER 1 in flowering time and plant architecture. Small. Structures 2 (4), 2000125. doi: 10.1002/sstr.202000125
Liu, Y. Y., Yang, K. Z., Wei, X. X., Wang, X. Q. (2016). Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 212 (3), 730–744. doi: 10.1111/nph.14066
Luedeling, E., Girvetz, E. H., Semenov, M. A., Brown, P. H. (2011). Climate change affects winter chill for temperate fruit and nut trees. PloS One 6 (5), e20155. doi: 10.1371/journal.pone.0020155
Luna, V., Lorenzo, E., Reinoso, H., Tordable, M. C., Abdala, G., Pharis, R. P., et al. (1990). Dormancy in Peach (Prunus persica L.) Flower Buds: I. Floral Morphogenesis and Endogenous Gibberellins at the End of the Dormancy Period. Plant Physiol. 93 (1), 20–25. doi: 10.1104/pp.93.1.20
Luna, V., Reinoso, H., Lorenzo, E., Bottini, R., Abdala, G. (1991). Dormancy in peach (Prunus persica L.) flower buds. Trees 5 (4), 244–246. doi: 10.1007/BF00227532
Luna, V., Soriano, M., Bottini, R., Sheng, C., Pharis, R. (1993). Dormancy in peach (Prunus persica L.) flower buds. III. Levels of endogenous gibberellins, abscisic acid, indole-3- acetic acid, and naringenin during dormancy of peach flower buds. Acta Hortic. 329, 4.
Lymperopoulos, P., Msanne, J., Rabara, R. (2018). Phytochrome and phytohormones: working in tandem for plant growth and development. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01037
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P. T., et al. (2021). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19 (2), 311–323. doi: 10.1111/pbi.13464
Michaels, S. D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12 (1), 75–80. doi: 10.1016/j.pbi.2008.09.005
Michaels, S. D., Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 (5), 949–956. doi: 10.1105/tpc.11.5.949
Michniewicz, M., Lang, A. (1962). Effect of nine different gibberellins on stem elongation and flower formation in cold-requiring and photoperiodic plants grown under non-inductive conditions. Planta 58 (5), 549–563. doi: 10.1007/bf01928367
Mimida, N., Kotoda, N., Ueda, T., Igarashi, M., Hatsuyama, Y., Iwanami, H., et al. (2009). Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus x domestica Borkh.). Plant Cell Physiol. 50 (2), 394–412. doi: 10.1093/pcp/pcp001
Mimida, N., Saito, T., Moriguchi, T., Suzuki, A., Komori, S., Wada, M. (2015). Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol. Plantarum. 59 (2), 237–244. doi: 10.1007/s10535-015-0503-4
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62 (4), 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Morea, E. G., da Silva, E. M., e Silva, G. F., Valente, G. T., Barrera Rojas, C. H., Vincentz, M., et al. (2016). Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 16, 40. doi: 10.1186/s12870-016-0716-5
Moss, S. M. A., Wang, T. C., Voogd, C., Brian, L. A., Wu, R. M., Hellens, R. P., et al. (2018). AcFT promotes kiwifruit in vitro flowering when overexpressed and Arabidopsis flowering when expressed in the vasculature under its own promoter. Plant Direct. 2 (7), 68. doi: 10.1002/pld3.68
Mullins, M. G. (1968). Regulation of inflorescence growth in cuttings of the grape vine (Vitis vinifera L.). J. Exp. Bot. 19 (3), 532–543. doi: 10.1093/jxb/19.3.532
Ni, J., Gao, C., Chen, M. S., Pan, B. Z., Ye, K., Xu, Z. F. (2015). Gibberellin promotes shoot branching in the perennial woody plant jatropha curcas. Plant Cell Physiol. 56 (8), 1655–1666. doi: 10.1093/pcp/pcv089
Nishiyama, S., Matsushita, M. C., Yamane, H., Honda, C., Okada, K., Tamada, Y., et al. (2021). Functional and expressional analyses of apple FLC-like in relation to dormancy progress and flower bud development. Tree Physiol. 41 (4), 562–570. doi: 10.1093/treephys/tpz111
Omori, M., Cheng, C.-C., Hsu, F.-C., Chen, S.-J., Yamane, H., Tao, R., et al. (2022). Off-season flowering and expression of flowering-related genes during floral bud differentiation of rabbiteye blueberry in a subtropical climate. Sci. Hortic. 306, 111458. doi: 10.1016/j.scienta.2022.111458
Omori, M., Yamane, H., Li, K.-T., Matsuzaki, R., Ebihara, S., Li, T.-S., et al. (2020). Expressional analysis of FT and CEN genes in a continuously flowering highbush blueberry ‘Blue Muffin’. Acta Hortic. 1280), 6. doi: 10.17660/ActaHortic.2020.1280.27
Omori, M., Yamane, H., Osakabe, K., Osakabe, Y., Tao, R. (2021). Targeted mutagenesis of CENTRORADIALIS using CRISPR/Cas9 system through the improvement of genetic transformation efficiency of tetraploid highbush blueberry. J. Hortic. Sci. Biotechnol. 96 (2), 153–161. doi: 10.1080/14620316.2020.1822760
Pearce, S., Vanzetti, L. S., Dubcovsky, J. (2013). Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 163 (3), 1433–1445. doi: 10.1104/pp.113.225854
Pena, L., Martin-Trillo, M., Juarez, J., Pina, J. A., Navarro, L., Martinez-Zapater, J. M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19 (3), 263–267. doi: 10.1038/85719
Peng, J., Harberd, N. P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 113 (4), 1051–1058. doi: 10.1104/pp.113.4.1051
Pillitteri, L. J., Lovatt, C. J., Walling, L. L. (2004). Isolation and characterization of LEAFY and APETALA1 homologues from Citrus sinensis L. Osbeck ‘Washington’. J. Am. Soc. Hortic. Sci. 129 (6), 846–856. doi: 10.21273/Jashs.129.6.0846
Pin, P. A., Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35 (10), 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Porto, D. D., Bruneau, M., Perini, P., Anzanello, R., Renou, J. P., dos Santos, H. P., et al. (2015). Transcription profiling of the chilling requirement for bud break in apples: a putative role for FLC-like genes. J. Exp. Bot. 66 (9), 2659–2672. doi: 10.1093/jxb/erv061
Preston, J. C., Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00080
Putterill, J., Robson, F., Lee, K., Simon, R., Coupland, G. (1995). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 (6), 847–857. doi: 10.1016/0092-8674(95)90288-0
Quesada-Traver, C., Guerrero, B. I., Badenes, M. L., Rodrigo, J., Rios, G., Lloret, A. (2020). Structure and expression of bud dormancy-associated MADS-box genes (DAM) in european plum. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01288
Randoux, M., Daviere, J. M., Jeauffre, J., Thouroude, T., Pierre, S., Toualbia, Y., et al. (2014). RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol. 202 (1), 161–173. doi: 10.1111/nph.12625
Reinoso, H., Luna, V., Dauriía, C., Pharis, R., Bottini, R. (2002a). Dormancy in peach (Prunus persica) flower buds. VI. Effects of gibberellins and an acylcyclohexanedione (trinexapac-ethyl) on bud morphogenesis in field experiments with orchard trees and on cuttings. Can. J. Bot. 80, 11. doi: 10.1139/b02-051
Reinoso, H., Luna, V., Pharis, R., Bottini, R. (2002b). Dormancy in peach (Prunus persica) flower buds. V. Anatomy of bud development in relation to phenological stage. Can. J. Bot. 80, 8. doi: 10.1139/b02-052
Rothkegel, K., Sanchez, E., Montes, C., Greve, M., Tapia, S., Bravo, S., et al. (2017). DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.). Tree Physiol. 37 (12), 1739–1751. doi: 10.1093/treephys/tpx055
Rottmann, W. H., Meilan, R., Sheppard, L. A., Brunner, A. M., Skinner, J. S., Ma, C., et al. (2000). Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22 (3), 235–245. doi: 10.1046/j.1365-313x.2000.00734.x
Ryu, J. Y., Park, C. M., Seo, P. J. (2011). The floral repressor BROTHER OF FT AND TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Mol. Cells 32 (3), 295–303. doi: 10.1007/s10059-011-0112-9
Samach, A. (2012). “25 - control of flowering,” in Plant biotechnology and agriculture. Eds. Altman, A., Hasegawa, P. M. (San Diego: Academic Press), 387–404.
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 (5471), 1613–1616. doi: 10.1126/science.288.5471.1613
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157 (1), 485–497. doi: 10.1104/pp.111.181982
Seo, E., Lee, H., Jeon, J., Park, H., Kim, J., Noh, Y. S., et al. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21 (10), 3185–3197. doi: 10.1105/tpc.108.063883
Serrano-Bueno, G., Sanchez de Medina Hernandez, V., Valverde, F. (2021). Photoperiodic signaling and senescence, an ancient solution to a modern problem? Front. Plant Sci. 12. doi: 10.3389/fpls.2021.634393
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97 (7), 3753–3758. doi: 10.1073/pnas.060023597
Silverstone, A. L., Jung, H. S., Dill, A., Kawaide, H., Kamiya, Y., Sun, T. P. (2001). Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 (7), 1555–1566. doi: 10.1105/tpc.010047
Simpson, G. G. (2004). The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7 (5), 570–574. doi: 10.1016/j.pbi.2004.07.002
Sinn, J. P., Held, J. B., Vosburg, C., Klee, S. M., Orbovic, V., Taylor, E. L., et al. (2021). Flowering Locus T chimeric protein induces floral precocity in edible citrus. Plant Biotechnol. J. 19 (2), 215–217. doi: 10.1111/pbi.13463
Smaczniak, C., Immink, R. G., Angenent, G. C., Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139 (17), 3081–3098. doi: 10.1242/dev.074674
Soares, J. M., Weber, K. C., Qiu, W., Stanton, D., Mahmoud, L. M., Wu, H., et al. (2020). The vascular targeted citrus FLOWERING LOCUS T3 gene promotes non-inductive early flowering in transgenic Carrizo rootstocks and grafted juvenile scions. Sci. Rep. 10 (1), 21404. doi: 10.1038/s41598-020-78417-9
Song, G. Q., Carter, B. B., Zhong, G. Y. (2023). Multiple transcriptome comparisons reveal the essential roles of FLOWERING LOCUS T in floral initiation and SOC1 and SVP in floral activation in blueberry. Front. Genet. 14. doi: 10.3389/fgene.2023.1105519
Song, G.-Q., Chen, Q. (2018a). Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 276, 10. doi: 10.1016/j.plantsci.2018.07.018
Song, G. Q., Chen, Q. (2018b). Comparative transcriptome analysis of nonchilled, chilled, and late-pink bud reveals flowering pathway genes involved in chilling-mediated flowering in blueberry. BMC Plant Biol. 18 (1), 98. doi: 10.1186/s12870-018-1311-8
Song, G.-q., Han, X. (2021). K-domain technology: constitutive expression of a blueberry keratin-like domain mimics expression of multiple MADS-box genes in enhancing maize grain yield. Front. Plant Sci. 12 (844). doi: 10.3389/fpls.2021.664983
Song, G. Q., Han, X., Ryner, J. T., Thompson, A., Wang, K. (2021). Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize. Plant Cell Rep. 40 (9), 1679–1693. doi: 10.1007/s00299-021-02722-4
Song, G.-Q., Hancock, J. F., Kole, C. (2011). “Vaccinium,” in Wild Crop Relatives: Genomic and Breeding Resources: Temperate Fruits (SpringerVerlag Berlin Heidelberg: Springer), 197–221. doi: 10.1007/978-3-642-16057-8_10
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336 (6084), 1045–1049. doi: 10.1126/science.1219644
Song, G. Q., Walworth, A. (2018). An invaluable transgenic blueberry for studying chilling-induced flowering in woody plants. BMC Plant Biol. 18 (1), 265. doi: 10.1186/s12870-018-1494-z
Song, G. Q., Walworth, A., Lin, T., Chen, Q., Han, X., Irina Zaharia, L., et al. (2019). VcFT-induced mobile florigenic signals in transgenic and transgrafted blueberries. Hortic. Res. 6, 105. doi: 10.1038/s41438-019-0188-5
Song, G. Q., Walworth, A., Zhao, D. Y., Hildebrandt, B., Leasia, M. (2013a). Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 32 (11), 1819–1826. doi: 10.1007/S00299-013-1495-1
Song, G. Q., Walworth, A., Zhao, D. Y., Jiang, N., Hancock, J. F. (2013b). The Vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): a flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Rep. 32 (11), 1759–1769. doi: 10.1007/S00299-013-1489-Z
Sreekantan, L., Thomas, M. R. (2006). VvFT and VvMADS8, the grapevine homologues of the floral integrators FT and SOC1, have unique expression patterns in grapevine and hasten flowering in Arabidopsis. Funct. Plant Biol. 33 (12), 1129–1139. doi: 10.1071/Fp06144
Srinivasan, C., Dardick, C., Callahan, A., Scorza, R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PloS One 7 (7), e40715. doi: 10.1371/journal.pone.0040715
Srinivasan, C., Mullins, M. G. (1978). Control of flowering in the grapevine (Vitis vinifera L.): formation of inflorescences in vitro by isolated tendrils. Plant Physiol. 61 (1), 127–130. doi: 10.1104/pp.61.1.127
Su, M., Wang, N., Jiang, S., Fang, H., Xu, H., Wang, Y., et al. (2018). Molecular characterization and expression analysis of the critical floral gene MdAGL24-like in red-fleshed apple. Plant Sci. 276, 189–198. doi: 10.1016/j.plantsci.2018.08.021
Sun, L., Nie, T., Chen, Y., Yin, Z. (2022). From floral induction to blooming: the molecular mysteries of flowering in woody plants. Int. J. Mol. Sci. 23 (18), 10959. doi: 10.3390/ijms231810959
Tadege, M., Sheldon, C. C., Helliwell, C. A., Stoutjesdijk, P., Dennis, E. S., Peacock, W. J. (2001). Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28 (5), 545–553. doi: 10.1046/j.1365-313X.2001.01182.x
Tan, F. C., Swain, S. M. (2007). Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol. Plant 131 (3), 481–495. doi: 10.1111/j.1399-3054.2007.00971.x
Teotia, S., Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant 8 (3), 359–377. doi: 10.1016/j.molp.2014.12.018
Theissen, G., Kim, J. T., Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43 (5), 484–516. doi: 10.1007/BF02337521
Tiwari, S. B., Shen, Y., Chang, H. C., Hou, Y. L., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187 (1), 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Tomes, S., Gunaseelan, K., Dragulescu, M., Wang, Y.-Y., Guo, L., Schaffer, R. J., et al. (2023). A MADS-box gene-induced early flowering pear (Pyrus communis L.) for accelerated pear breeding. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1235963
Trainin, T., Bar-Ya’akov, I., Holland, D. (2013). ParSOC1, a MADS-box gene closely related to Arabidopsis AGL20/SOC1, is expressed in apricot leaves in a diurnal manner and is linked with chilling requirements for dormancy break. Tree Genet. Genomes 9 (3), 753–766. doi: 10.1007/s11295-012-0590-8
Trankner, C., Lehmann, S., Hoenicka, H., Hanke, M. V., Fladung, M., Lenhardt, D., et al. (2010). Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta 232 (6), 1309–1324. doi: 10.1007/s00425-010-1254-2
Tuan, P. A., Bai, S., Saito, T., Ito, A., Moriguchi, T. (2017). Dormancy-associated MADS-box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 58 (8), 1378–1390. doi: 10.1093/pcp/pcx074
Turck, F., Fornara, F., Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59 (1), 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Turnbull, C. (2011). Long-distance regulation of flowering time. J. Exp. Bot. 62 (13), 4399–4413. doi: 10.1093/jxb/err191
Ubi, B. E., Sakamoto, D., Ban, Y., Shimada, T., Ito, A., Nakajima, I., et al. (2010). Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J. Am. Soc. Hortic. Sci. 135 (2), 174–182. doi: 10.21273/JASHS.135.2.174
Varkonyi-Gasic, E., Wang, T., Karunairetnam, S., Nain, B., Wu, R., Hellens, R. P. (2014). Analysis of kiwifruit MADS box genes with potential roles in bud dormancy and flower development. Ii. Int. Symposium. Biotechnol. Fruit Species. 1048, 107–112.
Varkonyi-Gasic, E., Wang, T., Voogd, C., Jeon, S., Drummond, R. S. M., Gleave, A. P., et al. (2019). Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 17 (5), 869–880. doi: 10.1111/pbi.13021
Vergara, R., Noriega, X., Pérez, F. J. (2021). VvDAM-SVPs genes are regulated by FLOWERING LOCUS T (VvFT) and not by ABA/low temperature-induced VvCBFs transcription factors in grapevine buds. Planta 253 (2), 31. doi: 10.1007/s00425-020-03561-5
Voogd, C., Brian, L. A., Wu, R. M., Wang, T. C., Allan, A. C., Varkonyi-Gasic, E. (2022). A MADS-box gene with similarity to FLC is induced by cold and correlated with epigenetic changes to control budbreak in kiwifruit. New Phytol. 233 (5), 2111–2126. doi: 10.1111/nph.17916
Voogd, C., Wang, T., Varkonyi-Gasic, E. (2015). Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 66 (15), 4699–4710. doi: 10.1093/jxb/erv234
Wada, M., Cao, Q. F., Kotoda, N., Soejima, J., Masuda, T. (2002). Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol. Biol. 49 (6), 567–577. doi: 10.1023/A:1015544207121
Walworth, A. E., Chai, B., Song, G. Q. (2016). Transcript profile of flowering regulatory genes in vcFT-overexpressing blueberry plants. PloS One 11 (6), e0156993. doi: 10.1371/journal.pone.0156993
Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in arabidopsis thaliana. Cell 138 (4), 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, J., Gao, Z., Li, H., Jiu, S., Qu, Y., Wang, L., et al. (2020). Dormancy-associated MADS-box (DAM) genes influence chilling requirement of sweet cherries and co-regulate flower development with SOC1 gene. Int. J. Mol. Sci. 21 (3), 921. doi: 10.3390/ijms21030921
Wang, J., Jiu, S., Xu, Y., Sabir, I. A., Wang, L., Ma, C., et al. (2021). SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium L.). Plant Physiol. Biochem. 159, 277–284. doi: 10.1016/j.plaphy.2020.12.013
Wang, P., Liu, Z., Cao, P., Liu, X. Y., Wu, X. P., Qi, K. J., et al. (2017). PbCOL8 is a clock-regulated flowering time repressor in pear. Tree Genet. Genomes 13 (5), 107. doi: 10.1007/s11295-017-1188-y
Wang, Y., Pijut, P. M. (2013). Isolation and characterization of a TERMINAL FLOWER 1 homolog from Prunus serotina Ehrh. Tree Physiol. 33 (8), 855–865. doi: 10.1093/treephys/tpt051
Wang, S., Yuan, C., Dai, Y., Shu, H., Yang, T., Zhang, C. (2004). Development of flower organs in sweet cherry in shanghai area. Acta Hortic. Sin. 31 (3), 357–359.
Wang, L., Zhang, L., Ma, C., Xu, W.-p., Liu, Z.-r., Zhang, C.-x., et al. (2016). Impact of chilling accumulation and hydrogen cyanamide on floral organ development of sweet cherry in a warm region. J. Integr. Agric. 15 (11), 2529–2538. doi: 10.1016/S2095-3119(16)61341-2
Wang, J., Zhang, X. M., Yan, G. H., Zhou, Y., Zhang, K. C. (2013). Over-expression of the PaAP1 gene from sweet cherry (Prunus avium L.) causes early flowering in Arabidopsis thaliana. J. Plant Physiol. 170 (3), 315–320. doi: 10.1016/j.jplph.2012.09.015
Wells, C. E., Vendramin, E., Jimenez Tarodo, S., Verde, I., Bielenberg, D. G. (2015). A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 15, 41. doi: 10.1186/s12870-015-0436-2
Wenzel, S., Flachowsky, H., Hanke, M.-V. (2013). The Fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell. Tissue Organ Culture. (PCTOC). 115 (2), 127–137. doi: 10.1007/s11240-013-0346-7
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 (5737), 1056–1059. doi: 10.1126/science.1114358
Wilkie, J. D., Sedgley, M., Olesen, T. (2008). Regulation of floral initiation in horticultural trees. J. Exp. Bot. 59 (12), 3215–3228. doi: 10.1093/jxb/ern188
Willige, B. C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E. M., Maier, A., et al. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 (4), 1209–1220. doi: 10.1105/tpc.107.051441
Wong, A. C., Hecht, V. F., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00486
Woods, D. P., McKeown, M. A., Dong, Y., Preston, J. C., Amasino, R. M. (2016). Evolution of VRN2/ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170 (4), 2124–2135. doi: 10.1104/pp.15.01279
Wu, R., Cooney, J., Tomes, S., Rebstock, R., Karunairetnam, S., Allan, A. C., et al. (2021). RNAi-mediated repression of dormancy-related genes results in evergrowing apple trees. Tree Physiol. 41 (8), 1510–1523. doi: 10.1093/treephys/tpab007
Wu, Y. M., Ma, Y. J., Wang, M., Zhou, H., Gan, Z. M., Zeng, R. F., et al. (2022). Mobility of FLOWERING LOCUS T protein as a systemic signal in trifoliate orange and its low accumulation in grafted juvenile scions. Horticult. Res. 9, uhac056. doi: 10.1093/hr/uhac056
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 (4), 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017a). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00477
Wu, R., Wang, T., McGie, T., Voogd, C., Allan, A. C., Hellens, R. P., et al. (2014). Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 65 (17), 4985–4995. doi: 10.1093/jxb/eru264
Wu, R., Wang, T., Warren, B. A. W., Allan, A. C., Macknight, R. C., Varkonyi-Gasic, E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68 (5), 1071–1082. doi: 10.1093/jxb/erx014
Xiao, G., Li, B., Chen, H., Chen, W., Wang, Z., Mao, B., et al. (2018). Overexpression of PvCO1, a bamboo CONSTANS-LIKE gene, delays flowering by reducing expression of the FT gene in transgenic Arabidopsis. BMC Plant Biol. 18 (1), 232. doi: 10.1186/s12870-018-1469-0
Xu, M. L., Hu, T. Q., Zhao, J. F., Park, M. Y., Earley, K. W., Wu, G., et al. (2016b). Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PloS Genet. 12 (8). doi: 10.1371/journal.pgen.1006263
Xu, F., Li, T., Xu, P. B., Li, L., Du, S. S., Lian, H. L., et al. (2016a). DELLA protein e1006263s physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590 (4), 541–549. doi: 10.1002/1873-3468.12076
Yamaguchi, N., Winter, C. M., Wu, M. F., Kanno, Y., Yamaguchi, A., Seo, M., et al. (2014). Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344 (6184), 638–641. doi: 10.1126/science.1250498
Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 (2), 367–378. doi: 10.1105/tpc.018143
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103 (51), 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L. L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 (5664), 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100 (10), 6263–6268. doi: 10.1073/pnas.0937399100
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12 (12), 2473–2483. doi: 10.1105/tpc.12.12.2473
Yarur, A., Soto, E., Len, G., Almeida, A. M. (2016). The sweet cherry (Prunus avium) FLOWERING LOCUS T gene is expressed during floral bud determination and can promote flowering in a winter-annual Arabidopsis accession. Plant Reprod. 29 (4), 311–322. doi: 10.1007/s00497-016-0296-4
Ye, J., Geng, Y., Zhang, B., Mao, H., Qu, J., Chua, N. H. (2014). The Jatropha FT ortholog is a systemic signal regulating growth and flowering time. Biotechnol. Biofuels 71, 10. doi: 10.1186/1754-6834-7-91
Yoo, S. J., Chung, K. S., Jung, S. H., Yoo, S. Y., Lee, J. S., Ahn, J. H. (2010). BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 63 (2), 241–253. doi: 10.1111/j.1365-313X.2010.04234.x
Yu, S., Galvão, V. C., Zhang, Y.-C., Horrer, D., Zhang, T.-Q., Hao, Y.-H., et al. (2012). Gibberellin Regulates the Arabidopsis Floral Transition through miR156-Targeted SQUAMOSA PROMOTER BINDING–LIKE Transcription Factors. Plant Cell 24 (8), 3320–3332. doi: 10.1105/tpc.112.101014
Zhang, X., An, L., Nguyen, T. H., Liang, H., Wang, R., Liu, X., et al. (2015b). The cloning and functional characterization of peach CONSTANS and FLOWERING LOCUS T homologous genes ppCO and ppFT. PloS One 10 (4), e0124108. doi: 10.1371/journal.pone.0124108
Zhang, S., Gottschalk, C., van Nocker, S. (2019). Genetic mechanisms in the repression of flowering by gibberellins in apple (Malus x domestica Borkh.). BMC Genomics 20 (1), 747. doi: 10.1186/s12864-019-6090-6
Zhang, H., Harry, D. E., Ma, C., Yuceer, C., Hsu, C. Y., Vikram, V., et al. (2010). Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. J. Exp. Bot. 61 (10), 2549–2560. doi: 10.1093/jxb/erq092
Zhang, L., Hu, Y. B., Wang, H. S., Feng, S. J., Zhang, Y. T. (2015a). Involvement of miR156 in the regulation of vegetative phase change in plants. J. Am. Soc. Hortic. Sci. 140 (5), 387–395. doi: 10.21273/Jashs.140.5.387
Zheng, J., Ma, Y., Zhang, M., Lyu, M., Yuan, Y., Wu, B. (2019). Expression Pattern of FT/TFL1 and miR156-Targeted SPL Genes Associated with Developmental Stages in Dendrobium catenatum. Int. J. Mol. Sci. 20 (11), 2725. doi: 10.3390/ijms20112725
Zhu, P., Burney, J., Chang, J. F., Jin, Z. N., Mueller, N. D., Xin, Q. C., et al. (2022). Warming reduces global agricultural production by decreasing cropping frequency and yields. Nat. Climate Change 12 (11), 1016–101+. doi: 10.1038/s41558-022-01492-5
Zhu, H., Chen, P. Y., Zhong, S., Dardick, C., Callahan, A., An, Y. Q., et al. (2020a). Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Hortic. Res. 7 (1), 114. doi: 10.1038/s41438-020-0336-y
Zhu, Y., Klasfeld, S., Jeong, C. W., Jin, R., Goto, K., Yamaguchi, N., et al. (2020b). TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 11 (1), 5118. doi: 10.1038/s41467-020-18782-1
Zhu, L. H., Li, X. Y., Welander, M. (2008). Overexpression of the Arabidopsis gai gene in apple significantly reduces plant size. Plant Cell Rep. 27 (2), 289–296. doi: 10.1007/s00299-007-0462-0
Zong, X., Zhang, Y., Walworth, A., Tomaszewski, E. M., Callow, P., Zhong, G. Y., et al. (2019). Constitutive expression of an apple FLC3-like gene promotes flowering in transgenic blueberry under nonchilling conditions. Int. J. Mol. Sci. 20 (11), 2775. doi: 10.3390/ijms20112775
Keywords: chilling requirement, deciduous plant, florigen, floral activation, floral initiation, flowering mechanism, woody plant
Citation: Song G-q, Liu Z and Zhong G-y (2024) Regulatory frameworks involved in the floral induction, formation and developmental programming of woody horticultural plants: a case study on blueberries. Front. Plant Sci. 15:1336892. doi: 10.3389/fpls.2024.1336892
Received: 11 November 2023; Accepted: 26 January 2024;
Published: 12 February 2024.
Edited by:
Bo Sun, Nanjing University, ChinaReviewed by:
Maria Manuela Ribeiro Costa, University of Minho, PortugalCopyright © 2024 Song, Liu and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-qing Song, c29uZ2dAbXN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.