
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 March 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1333286
This article is part of the Research TopicCrop Abiotic Stress: Advances in Germplasm/Gene Discovery and UtilizationView all 18 articles
 Mobeen Ali1
Mobeen Ali1 Muhammad Shafiq1*
Muhammad Shafiq1* Muhammad Zeshan Haider2
Muhammad Zeshan Haider2 Adnan Sami2
Adnan Sami2 Pravej Alam3
Pravej Alam3 Thamir Albalawi3
Thamir Albalawi3 Zuha Kamran1
Zuha Kamran1 Saleh Sadiq1
Saleh Sadiq1 Mujahid Hussain4
Mujahid Hussain4 Muhammad Adnan Shahid4
Muhammad Adnan Shahid4 Mouna Jeridi5
Mouna Jeridi5 Ghulam Abbas Ashraf6
Ghulam Abbas Ashraf6 Muhammad Aamir Manzoor7*
Muhammad Aamir Manzoor7* Irfan Ali Sabir8*
Irfan Ali Sabir8*Citrus fruits, revered for their nutritional value, face significant threats from diseases like citrus canker, particularly impacting global citrus cultivation, notably in Pakistan. This study delves into the critical role of NPR1-like genes, the true receptors for salicylic acid (SA), in the defense mechanisms of citrus against Xanthomonas axonopodis pv. citri (Xcc). By conducting a comprehensive genome-wide analysis and phylogenetic study, the evolutionary dynamics of Citrus limon genes across diverse citrus cultivars are elucidated. Structural predictions unveil conserved domains, such as the BTB domain and ankyrin repeat domains, crucial for the defense mechanism. Motif analysis reveals essential conserved patterns, while cis-regulatory elements indicate their involvement in transcription, growth, response to phytohormones, and stress. The predominantly cytoplasmic and nuclear localization of NPR1-like genes underscores their pivotal role in conferring resistance to various citrus species. Analysis of the Ks/Ka ratio indicates a purifying selection of NPR1-like genes, emphasizing their importance in different species. Synteny and chromosomal mapping provide insights into duplication events and orthologous links among citrus species. Notably, Xac infection stimulates the expression of NPR1-like genes, revealing their responsiveness to pathogenic challenges. Interestingly, qRT-PCR profiling post-Xac infection reveals cultivar-specific alterations in expression within susceptible and resistant citrus varieties. Beyond genetic factors, physiological parameters like peroxidase, total soluble protein, and secondary metabolites respond to SA-dependent PR genes, influencing plant characteristics. Examining the impact of defense genes (NPR1) and plant characteristics on disease resistance in citrus, this study marks the inaugural investigation into the correlation between NPR1-associated genes and various plant traits in both susceptible and resistant citrus varieties to citrus bacterial canker.
Globally, citrus fruits stand out as the predominant category of commercially cultivated fruit within the Rutaceae family, offering a wealth of nutritional benefits and playing a crucial role in the world economy (Russo et al., 2021). Unfortunately, the citrus plant faces a significant threat from various diseases such as greening, citrus decline, and gummosis (Malik et al., 2021). Among these is Citrus Canker Disease (CCKD), caused by Xanthomonas axonopodis pv. citri (Xcc), which poses a major risk to citrus crop yield and quality worldwide, with particular severity in Pakistan (Malamud et al., 2011; Sami et al., 2023a; Sami et al., 2023b). The Sargodha region’s citrus orchards contribute over 90% to Pakistan’s national citrus production. Yield losses in Pakistan due to citrus canker can range from 10% to 50%, depending on infection severity and environmental conditions (Sabir et al., 2010; Almas et al., 2023). However, their potential is hampered by the onslaught of canker, evidenced by symptoms like discoloration, abrasions, and water-soaked lesions on fruit, stems, or leaves (Shahbaz et al., 2023). Several commercially important citrus varieties, including grapefruit, sweet oranges [C. sinensis C. sinensis (L.) Osbeck], lemons [C.limon (L.) Burm. F.], and key lime (Citrus aurantifolia Swingle), are susceptible to bacterial canker (Graham et al., 2004) impacting both the quantity and quality of fresh and processed fruit. This, in turn, leads to substantial economic losses for citrus growers (Spreen et al., 2003). The economic repercussions extend beyond the orchards, as limited fruit trade among states or internationally from canker-affected regions takes a severe toll on the overall economy (Duan et al., 2009; Khan et al., 2023).
Several alternative strategies for disease management have been devised with the aim of establishing enduring solutions for disease control in the long term. These methods encompass plant defense mechanisms specifically crafted to counteract various stressors, encompassing attacks from pathogens (Kushalappa et al., 2016; Manzoor et al., 2023). Resistance genes are considered one of the major factors in defense mechanisms against pathogenic attacks. Resistance (R) genes often trigger downstream signaling responses during plant disease resistance (Malamud et al., 2011; Khan et al., 2023). An immune response, SAR (Shahbaz et al., 2022), promotes the exploration of the resistant gene family to an infectious pathogen. SAR is considered a prime factor for activating numerous pathogenesis-related (PR) genes, such as NPR1 gene with its paralogues (NPR3 and NPR4) (Fu and Dong, 2013). Exogenous application of some chemicals, like benzol 1,2,3-thiadiazole-7-carbothermic acid, 2,6-dichloroisonicotinic acid, salicylic acid, and S-methyl ester on plants are considered to be responsible for the stimulation of the SAR system (Durango et al., 2013). Additionally, it is noted that during specific hypersensitive responses, the signal transduction pathway(s) connecting HR to SAR involves the endogenous production of SA (Cao et al., 1997).
Based on structural motifs, transmembrane areas, interleukin-1 receptor domain, leucine rich repeat (LRR) domain, coiled coil (Spoel et al., 2003) domains, and nucleotide-binding sites (NBSs), resistant proteins are grouped into numerous superfamilies. The two most prevalent R genes (NBS-LRR) found in plants (Meyers et al., 2003) are further divided into sub-groups based on N-terminal CC or TIR domain (Li et al., 2018). NPR1 genes and their homologues were revealed to regulate the resistance of disease in the citrus family against various pathogens (Peng et al., 2021). NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1, along with their NPR3 and NPR4 as paralogues) serves as authentic SA receptors, actively participating in both local and systemic immunity by regulating SA-mediated processes (Birkenbihl et al., 2017; Sami et al., 2023c).
The investigation of the resistant gene family across various citrus species is vital in light of the aforementioned findings (Ahmad et al., 2023; Hussain et al., 2023). Thus, this resistant gene family was recognized and analyzed thoroughly in different species of citrus (Citrus sinensis, Citrus reticulata, Citrus fortunella, Citrus maxima, Citrus medica, Citrus ichangenesis, Atlanta buxifolia, and Poncirus trifoliata) that ranged from susceptible to highly resistant against canker. Phylogenic classification, characterization on molecular basis conserved motifs, residues of amino acids, distribution on chromosomes, and composition of protein domains were investigated. Furthermore, the stress-related expression pattern of citrus-resistant genes in different regions was also analyzed using citrus species sequencing of RNA (RNA-seq) datasets (Haider et al., 2023b). The findings underscore the need for substantial support to facilitate applied research and in-depth exploration of resistant genes in citrus species and related plant families. This exploration holds promise for influencing the creation of disease-resistant citrus varieties using various breeding methods such as double haploid technology, hybridization, tissue culture, and backcrossing.
Data pertaining to the amino acid sequences of C. sinensis, C. reticulata, C. fortunella, C. limon, C. maxima, C. ichangenesis, C. maxima, P. trifoliata, C. climentina, A. buxifolia, and C. medica were obtained from Citrus Genome database (https://www.citrusgenomedb.org). To confirm the presence of NPR1-like domains, retrieved amino acid sequences were subjected to searches at the SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2006; Haider et al., 2023a) and NCBI CDD (Conserved Domain Database) (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Marchler-Bauer et al., 2013) with the default parameters.
The NPR reference sequences from A. thaliana were employed to identify homologous genes in the protein database of 11 citrus species via the https://www.citrusgenomedb.org/blast platform, with an E-value threshold of < 1.2 × 10−14. Subsequently, genomic, protein, CDS, and promoter sequences for all NPR genes were retrieved. In-depth analysis included the thorough examination of protein sequences to pinpoint representative sequences within duplicated genes. This process involved the utilization of TBtools and the verification of the existence of distinctive NPR domains, encompassing ANK repeats and N-terminal BTB/POZ domains. Additionally, motif analysis was conducted on these protein sequences using the motif finder tool (https://www.genome.jp/tools-bin/search_motif_lib) (Arabbeigi et al., 2018). The identified motifs were then compared against the canonical pattern found in “AtNPR1”-like proteins.
Multiple sequence alignments of CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs were generated using the MUSCLE Method in the Mega-X program and were visualized (Xu et al., 2020). Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) (Guo et al., 2007) exhibited the exon–intron structures of NPR1-like genes in different citrus species. Moreover, NCBI-Conserved Domain Database (CDD) was used for the identification of the conservative domain compositions of 63 distinct CsNPRs, CrNPRs, CfNPRs, ClNPRs, CcNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs and the results of which were visualized by the TBtools software (https://www.genome.jp/tools/motif/) (Nakasu et al., 2019). The MEME Suite, accessible at https://meme-suite.org/meme/tools/meme, was utilized to identify protein motifs encoded by these genes. Additionally, an online tool was employed to pinpoint the locations of these motifs within the peptide sequences (Haider et al., 2023b).
Furthermore, the protein sequences of A. thaliana were retrieved from the GenBank (Sayers et al., 2022) database (https://www.ncbi.nlm.nih.gov/genbank/) to find out the phylogenetic integration among these NPR1-like proteins and CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs-like proteins. The full-length sequences of Arabidopsis proteins and the protein sequences of NPR1-like genes in 10 different citrus species were aligned using the Clustal W method (Nakasu et al., 2019). As a result, an unrooted phylogenetic tree was generated using the maximum-likelihood (ML) algorithm, employing MEGA-X software, with the specified parameters including the bootstrap method (1,000 replicates), the Jones–Taylor–Thornton model, and the inclusion of all sites (Shafiq et al., 2024).
The Ks and Ka values of genes from CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs were also determined. To achieve this, duplicate gene pairs resembling NPR1-like genes, originating from various duplication mechanisms, were employed to calculate substitution rates for Ka and Ks. TBtools were utilized to compute Ka and Ks values for these duplicated gene pairs and their respective Ka/Ks ratios using the simple Ka/Ks calculator feature. The Ka/Ks ratio was assessed to elucidate the molecular evolutionary rates of each gene pair. Generally, a Ka/Ks value <1 suggests purifying selection in evolution, Ka/Ks equals to 1 indicates neutral selection, while Ka/Ks >1 signifies positive selection (Haider et al., 2023b). Furthermore, the calculation of the divergence time for these gene pairs was conducted using the formula t = Ks/2r, with r (1.5 × 10−8) serving as the representative value for neutral substitution rates (Campos et al., 2023).
Subcellular localization of identified genes expressed was performed by using the WoLF PSORT program (https://wolfpsort.hgc.jp/) (Horton et al., 2006). Similarly, other factors regarding CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs genes and their respective coded proteins like molecular weight (Mw), isoelectric point (pI) value, amino acid (AA) length, length of mRNA, and chromosome number of all CNPR genes were gathered from the online tools like ExPASy-Protparam Tool (https://web.expasy.org/protparam/) (da Silva et al., 2020) and Citrus genome database (https://www.citrusgenomedb.org/).
The chromosomal Id, start and end position of NPR1 genes of CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, CbNPRs, PtNPRs, and CiNPRs were retrieved using “Citrus Genome Database” (https://www.citrusgenomedb.org), and the length of chromosomes was found using “FASTA stats” tool (https://bio.tools/gfastats). Three files, “Chromosome ID and length,” “Chromosome ID, start position/end position,” and “gene pairs”, derived from the Ks/Ka analysis, were driven to the “Gene Location Visualize Advance” window of TBtools, and the graph of chromosomal mapping was retrieved (Islam et al., 2023).
PlantCARE database (http://bioinformatics.pAb.ugent.be/webtools/plantcare/htmL/) (Haider et al., 2023b) was used to identify Cis-regulatory elements. The promoter sequences of NPR1-like genes were extracted from genome sequences of 10 citrus species by TBtools software. Chromosome IDs and coordinates were sourced from the Citrus Genome Database, then Advanced Circos in TBtools was used to create the synteny graph.
The study of ESTs (expressed sequence tags) to find resistance genes in citrus species and characterize their expression levels across multiple citrus plant tissues and organs, leveraging the online NCBI Gene Expression Omnibus (GEO) database (Grechkin et al., 2017). The Blast P search was performed using a default parameter with an e-value of 1e−04. Various genes exhibited expression in distinct segments of the citrus plant, encompassing the flower, root, fruit, leaf, stem, and more, in response to diverse forms of biotic stress and various influencing factors. Heat maps were used to analyze the expression patterns of specific resistance genes across different tissues and organs in different citrus species.
The four citrus cultivars C. sinensis (sweet orange), C. reticulata (mandarin), C. limon (lemon), and C. fortunella (kumquat) were inoculated with citrus bacterial culture (Xanthomonas axonopodis pv. citri) and chosen for gene expression. The experiment was conducted in a greenhouse (day/night temperature, 25°C; light/dark periods, 16/8) using the method of completely randomized design (CRD) and three replicates of each treatment.
mRNA was extracted using the triazole method (Fujino et al., 2016), and first-strand cDNA was synthesized using AB script II cDNA first-strand synthesis kit. SYBR Green (Wiz pure) was used to perform qRT-PCR. Standard curves were created to estimate the proportion of citrus NPR1-like genes, and housekeeping gene quantity in samples was determined using real-time PCR (Illumina) and software (https://biomolecularsystems.com/mic-qpcr/software/). To analyze the dynamics of expression, we conducted three biological and three technical replicates. Significance differences were assessed using Student’s t-tests (p < 0.05).
The data were subjected to statistical analysis using a two-way factorial completely randomized design (CRD). Analysis of variance (ANOVA) was utilized to assess variances, and means were differentiated using the least significant difference (LSD) method, with a significance level set at p < 0.05. The entire statistical analysis was carried out using the Statistix 8.1 software suite.
Approximately 63 non-redundant NPR1-like genes were retrieved from the genome of C. sinensis, C. reticulata, C. fortunella, C. limon, C. maxima, C. ichangenesis, C. maxima, P. trifoliata, C. climentina, A. buxifolia, and C. medica. Utilizing bioinformatics tools like BLAST search, 63 NPR1-like genes were discovered in the genome and named as CsNPRs, CrNPRs, CfNPRs, ClNPRs, CcNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs. Using NCBI CDD Domain analysis, all the proteins that lack N-terminal BTB/POZ domain or ANK repeats in the middle region were excluded. As a result, different species have different number of NPR1-like genes, like nine in C. sinensis; seven in C. reticulata; two in C. fortunella; six in C. limon, A. thaliana, and C. medica; 10 in C. climentina and C. ichangensis; seven in C. maxima and P. trifoliata; and three in A. boxifolia (Figure 1).
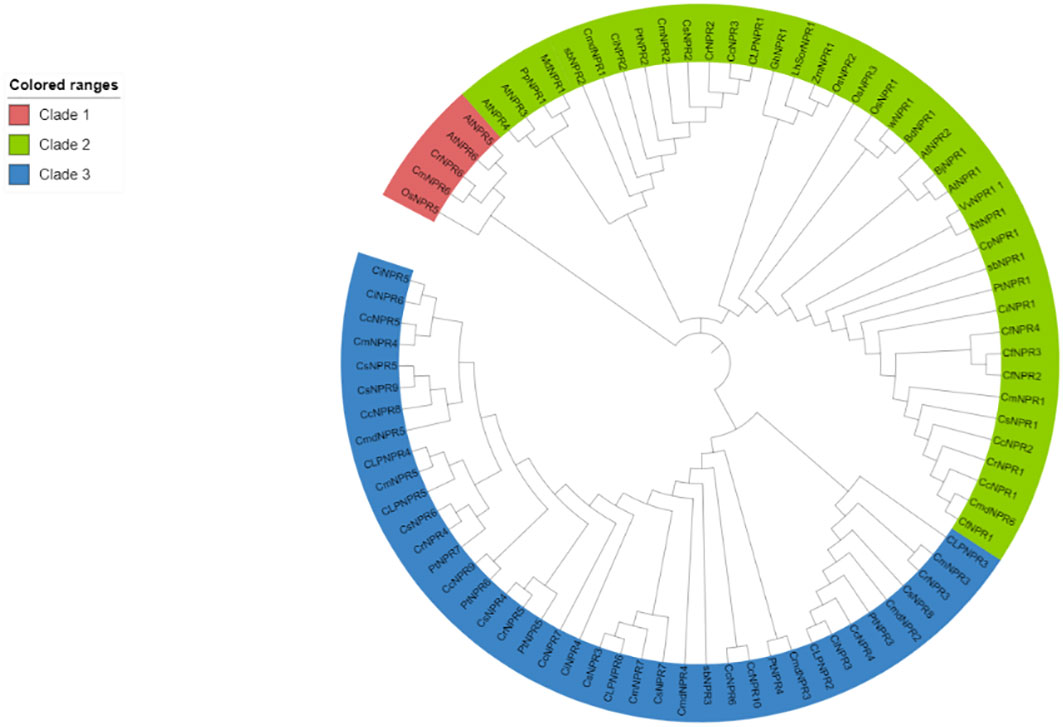
Figure 1 Polygenetic tree of NPR1 like proteins in CsNPRs, CrNPRs, CfNPRs, CcNPRs, CmdNPRs, CmNPRs, sbNPRs, PtNPRs and CiNPRs and its homologous in, A. thaliana (At).
The mRNA lengths of these CsNPRs, CrNPRs, CfNPRs, CcNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs genes have nucleotides that range from 1,119 to 69,428, while the amino acid lengths of their respective proteins vary between 372 and 1,379 amino acids. Additionally, the overall pI values vary from 4.91 to 8.74, and the molecular weight of described genes CsNPRs, CrNPRs, ClNPRs, CfNPRs, CcNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs proteins vary from 118,053.42 to 46,773.06 (Table 1).
The evolutionary tree from the protein sequences of NPR1-like was constructed among the various species of citrus (C. sinensis, C. reticulata, C. fortunella, C. maxima, C. medica, C. ichangenesis, C. limon), and A. buxifolia and P. trifoliata, and references the A. thaliana species as a model (Cao et al., 1997; Spoel et al., 2003; Peng et al., 2021). A 1,000 bootstrap tree was constructed using the UPGMA model in MegaX software. The phylogeny analysis demonstrated the presence of NPR-like proteins categorized into three distinct classes or clades, matching patterns found in other species. Clade I include 41 NPR-related proteins, and clade II comprised 16 NPR-related proteins. Proteins from the same clade appear to be structurally and functionally related (Sami et al., 2024).
To estimate the approximate gene divergence time, the TBtools program was utilized, and it furnished values for Ks, Ka, and Ka/Ks ratios as specified. The value of synonymous substitutions per synonymous site was denoted by Ks, while the value of non-synonymous substitutions per non-synonymous site was quantified by Ka. The balance between synonymous and non-synonymous mutations can be seen in the Ka/Ks ratio. The evolutionary investigation revealed six gene pairs associated with gene duplication within the NPR family. The first pair included ClNPR4/CmNPR5, the second pair CsNPR4/CrNPR5, the third pair CrNPR3/CsNPR8, the fourth pair CmNPR6/CrNPR6, the fifth pair AtNPR6/AtNPR5, and the sixth pair AbNPR1/AtNPR2 (Table 2).
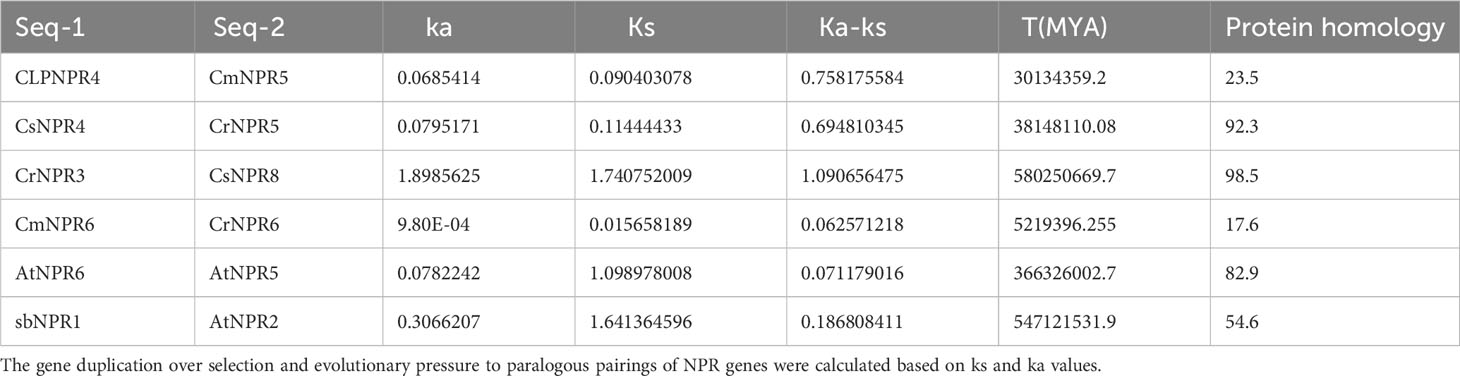
Table 2 The expression ka/ks represents the ratio of mutations involving synonymous substitutions (ks) to mutations involving non-synonymous substitutions (ka).
A measure that evaluates the selection pressure affecting amino acid substitutions was the Ka/Ks ratio. When the Ka/Ks ratio declines below 1, it denotes an evolutionary process of purifying selection (Parmley and Hurst, 2007). If this ratio was exceeding value 1, it may indicate the probability of positive selection (Biswas and Akey, 2006). The findings showed that the Ka/Ks ratios varied significantly across NPR groups. The Ka/Ks ratios for ClNPR4/CmNPR5, CsNPR4/CrNPR5, CmNPR6/CrNPR6, AtNPR6/AtNPR5, and AbNPR1/AtNPR2 indicate purifying selection, with values below 1, while the Ka/Ks ratio for CrNPR3/CsNPR8 suggests positive selection, as it exceeds 1, indicating environmental influence (Sami et al., 2023d).
To gain deeper insights into the functions of CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs genes, structural features and sequence composition analysis was conducted using online tools GSDS and NCBI-CDD. Intron/exon profiles of NPR genes were generated using GSDS 2.0 (Figure 2). Notably, the majority of NPRs, including CsNPR6, CcNPR9, CmdNPR5, CcNPR8, CcNPR5, CsNPR5, CsNPR9, CiNPR5, CiNPR6, CiNPR7, CiNPR4, CsNPR3, CsNPR7, CmdNPR4, AbNPR3, CcNPR6, CcNPR10, CmdNPR3, CsNPR8, CmdNPR2, CcNPR4, CiNPR3, AbNPR2, CiNPR2, CmdNPR1, CsNPR2, CcNPR3, AbNPR1, CiNPR1, CsNPR1, CiNPR2, and CmdNPR6, exhibited lack of introns. In contrast, the four Arabidopsis sequences, AtNPR1, AtNPR2, AtNPR3, and AtNPR4, contained two introns, while AtNPR5 and AtNPR6 had only one intron each. ClNPR4 had seven introns; CmNPR4 had six introns; CmNPR5, CrNPR4, and ClNPR3 had four introns; and ClNPR5, PtNPR6, CmNPR7, CmNPR3, PtNPR3, ClNPR3, AtNPR3, AtNPR4, PtNPR2, CmNPR2, and CrNPR1 had three introns. CrNPR6, CmNPR1, and CrNPR3 had two introns, while PtNPR5, CmdNPR5, PtNPR4, and CmNPR6 possessed a single intron only. The number of introns per gene ranged from 0 to 7. Interestingly, no results were obtained for CcNPR1, CfNPR1, CfNPR2, and CcNPR6. These findings highlight significant variations in the number of introns–exons among CsNPRs, CrNPRs, CfNPRs, ClNPRs, CcNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, CiNPRs, and A. thaliana genes.
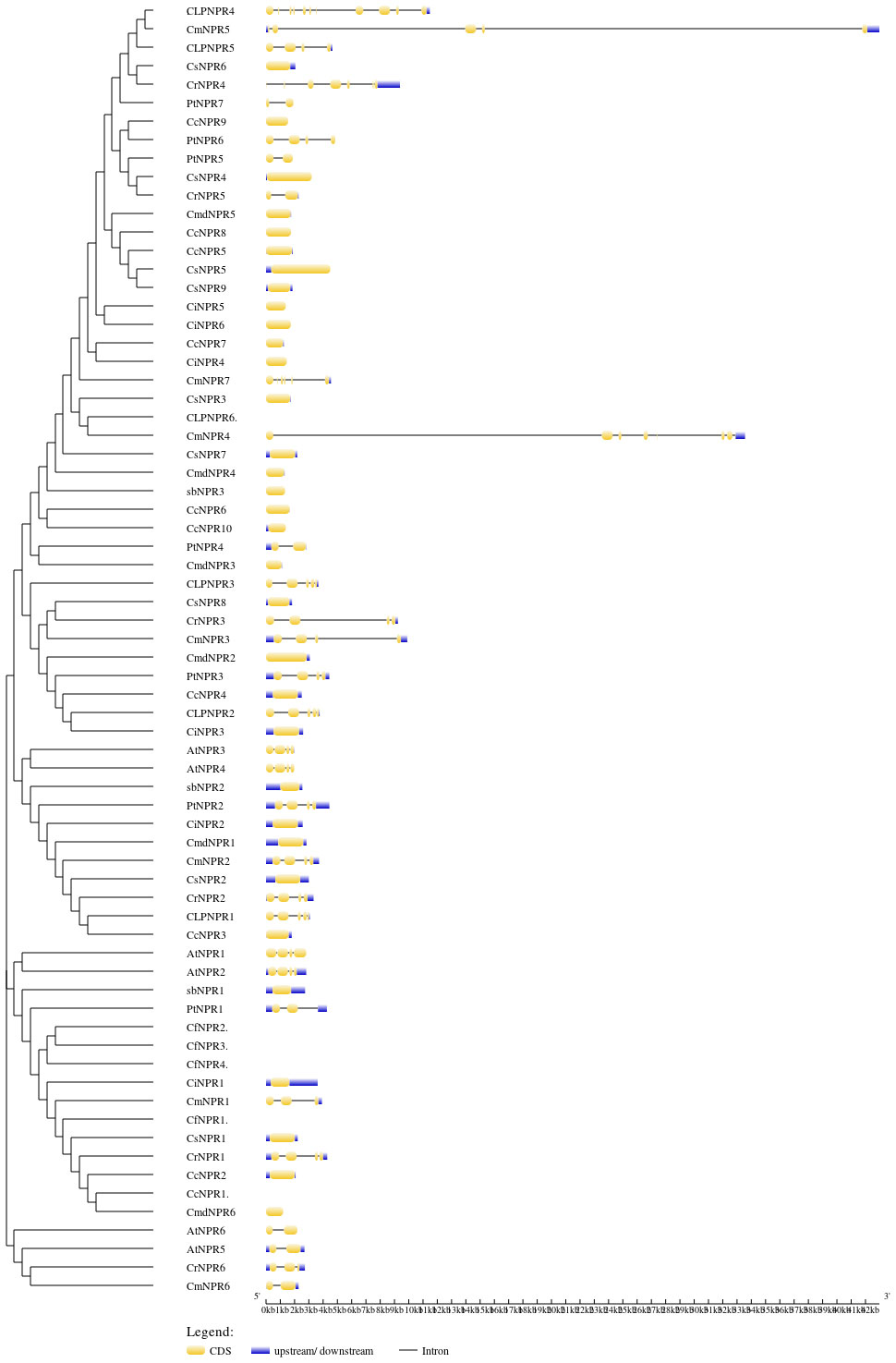
Figure 2 Phylogenetic relationship and gene structure of NPR1 genes from CsNPRs, CrNPRs, CfNPRs, CcNPRs, CmdNPRs, CmNPRs, sbNPRs, PtNPRs and CiNPRs. The phylogenetic tree was constructed using full length sequences of CsNPRs, CrNPRs, CfNPRs, CcNPRs, CmdNPRs, CmNPRs, sbNPRs, PtNPRs and CiNPRs-like genes. Yellow boxes indicate exons; and black lines indicate introns.
Conserved domain analysis was employed to filter out sequences exclusively featuring the BTB domain. Following that, motif analysis, along with conserved domain analysis, was carried out using TBtools. The analysis revealed that several genes, including ClNPR4, CmNPR5, ClNPR5, CsNPR6, CcNPR9, PtNPR6, CcNPR8, CcNPR5, CmNPR2, CsNPR2, CrNPR2, CLNPR1, AtNPR1, AtNPR2, AbNPR1, PtNPR1, CfNPR2, and CrNPR4 possessed motifs in the order of 8, 7, 2, 9, 5, 6, 1, 4, 3, and 10. However, PtNPR5 shows all these motifs except motif 5, while CrNPR5 featured all of them except for motifs 8 and 10, and CsNPR4 included motifs 7, 2, 9, 5, 6, 1, 4, 3, and 10, in addition to others. CsNPR5 and CsNPR9 displayed motifs 8, 9, 7, 2, 5, 6, 1, 4, 3, and 10, followed by motifs 9, 5, 6, 7, 4, 3, and 10. Some sequences had minor variations, such as ClNPR6 lacking motifs 2 and 3; CcNPR6 lacking motif 5; CmdNPR3 and CmdNPR2 featuring motifs 7, 2, 9, 5, 6, 1, and 4 in addition; and CrNPR5 missing motifs 8 and 10, along with motifs 6, 1, 4, 3, and 10. CsNPR7 possessed an additional motif 10; CmdNPR6 lacked motifs 8, 7, 2, 1, and 6; CsNPR8 lacked motifs 4, 1, 3, and 10; ClNPR2 had an additional motif 3; CmNPR4 had motifs 8, 9, 1, 4, 3, and 10 in addition; CmdNPR3 lacked motifs 5, 6, and 10; AtNPR3 and AtNPR4 were lacking motif 3; and CcNPR3 lacked motif 1. PtNPR7 exhibited motifs 7, 2, 9, 5, 6, and 1, while CiNPR5 lacked motif 6, as did CiNPR6 and CcNPR7. CmNPR7 only possessed motifs 8, 7, 2, 9, 4, and 10. CiNPR1, CmNPR1, CfNPR1, CsNPR1, CrNPR1, CcNPR2, and CcNPR1 did not have motif 6, whereas AtNPr5, AtNPR6, CrNPR6, and CmNPR6 had an additional motif 6 (Figure 3).
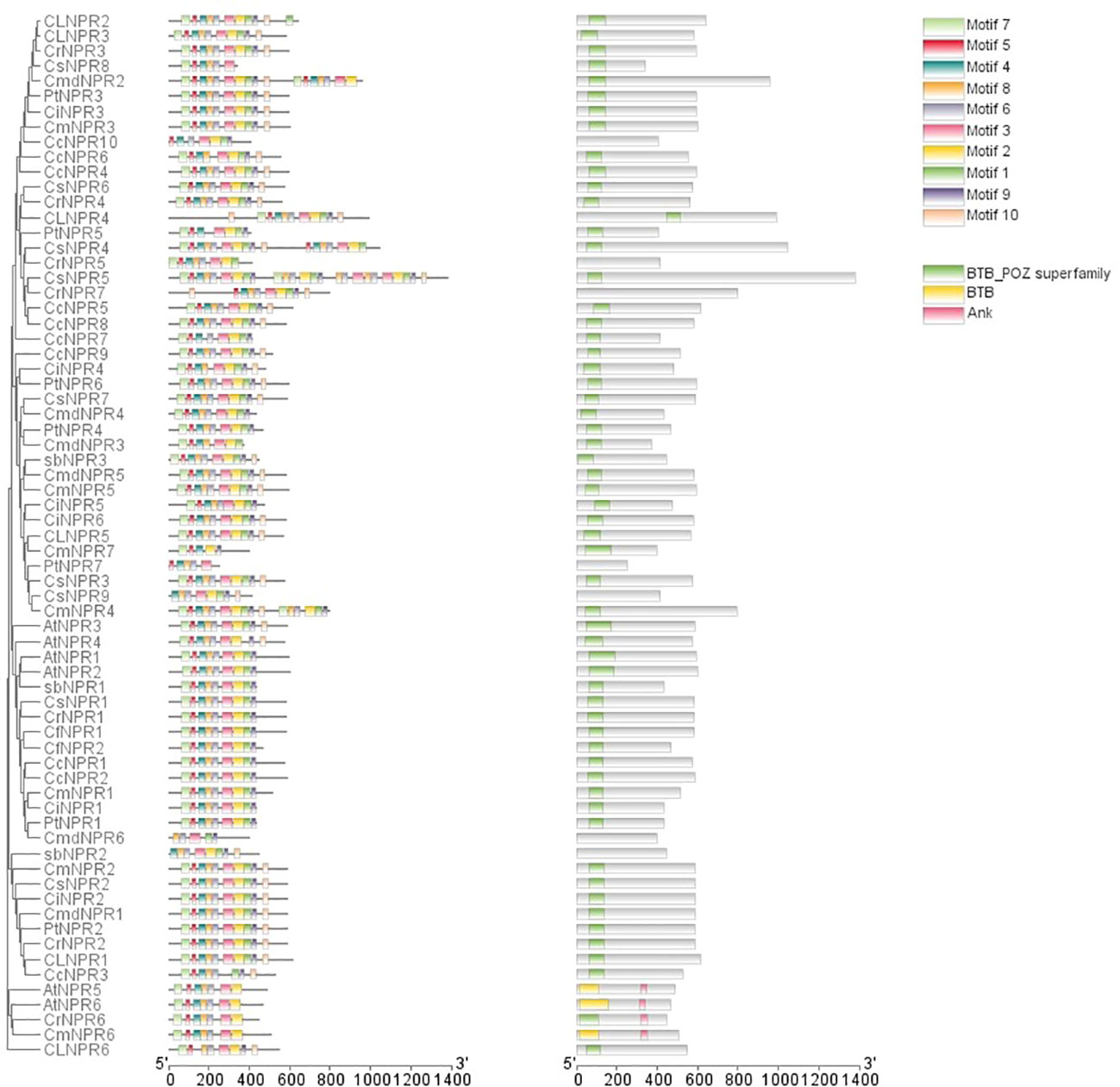
Figure 3 The domain arrangements of putative NPR1-like genes in C. sinensis, C. reticulata, C. fortunella, C. limon, C. medica, C. maxima, C. clementina, C. ichangensis, P. trifoliate, A. buxifolia, and A. thaliana connected with the phylogenetic tree.
The subcellular localization analysis indicated that cytoplasm was the predominant location for CsNPR1, CrNPR1, CcNPR2, PtNPR7, CiNPR1, and PtNPR1. Chloroplasts and plastids showed lower quantities of CsNPR9, CsNPR7, CLNPR5, CcNPR7, CcNPR10, PtNPR4, CcNPR5, CsNPR3, CLNPR5, CmNPR4, CfNPR2, CmNPR1, CcNPR1, PtNPR2, AbNPR1, CfNPR1, CmdNPR6, CmNPR7, CsNPR5, CsNPR9, CcNPR6, PtNPR6, CiNPR4, CmdNPR3, and CmdNPR4. Chloroplasts had lower levels of ClNPR1. Cyto-nucleus contained reduced amounts of ClNPR1, CiNPR3, CcNPR4, PtNPR3, CrNPR3, CcNPR3, and CmdNPR2. Peroxisomes contained minimal quantities of CiNPR2, CmdNPR1, CsNPR2, CrNPR2, and CcNPR3. The nucleus was the primary site for ClNPR3, ClNPR1, CiNPR3, CcNPR4, PtNPR3, CrNPR3, and CmNPR3, with CmNPR6, CrNPR5, and PtNPR7, absent from the nucleus. Other proteins were present in lower amounts in the nucleus. ER exhibited limited concentrations of CcNPR7, CcNPR10, PtNPR4, CsNPR4, CmNPR6, CrNPR6, CsNPR6, CmdNPR5, and CcNPR8. Vacuoles contained CcNPR6, CmNPR4, CLNPR6, CcNPR5, CLNPR5, CsNPR7, CCNPR9, CrNPR6, CmNPR6, CsNPR4, CLNPR2, and AbNPR3 in small quantities. Mitochondria housed the highest quantity of CmNPR5, while CcNPR7, CcNPR10, PtNPR4, CrNPR4, PtNPR5, CrNPR6, and CmNPR6 were present in smaller amounts. R-vacuoles contained small numbers of CmdNPR6, CfNPR1, PtNPR1, CiNPR1, and CmdNPR1. Golgi apparatus had ClNPR2, CLNPR1, and PtNPR4 in small quantities. ER plastids contained CcNPR10 and CcNPR7 in lower numbers, and cyto-ER contained only CmdNPr6 in moderate quantities (Figure 4).
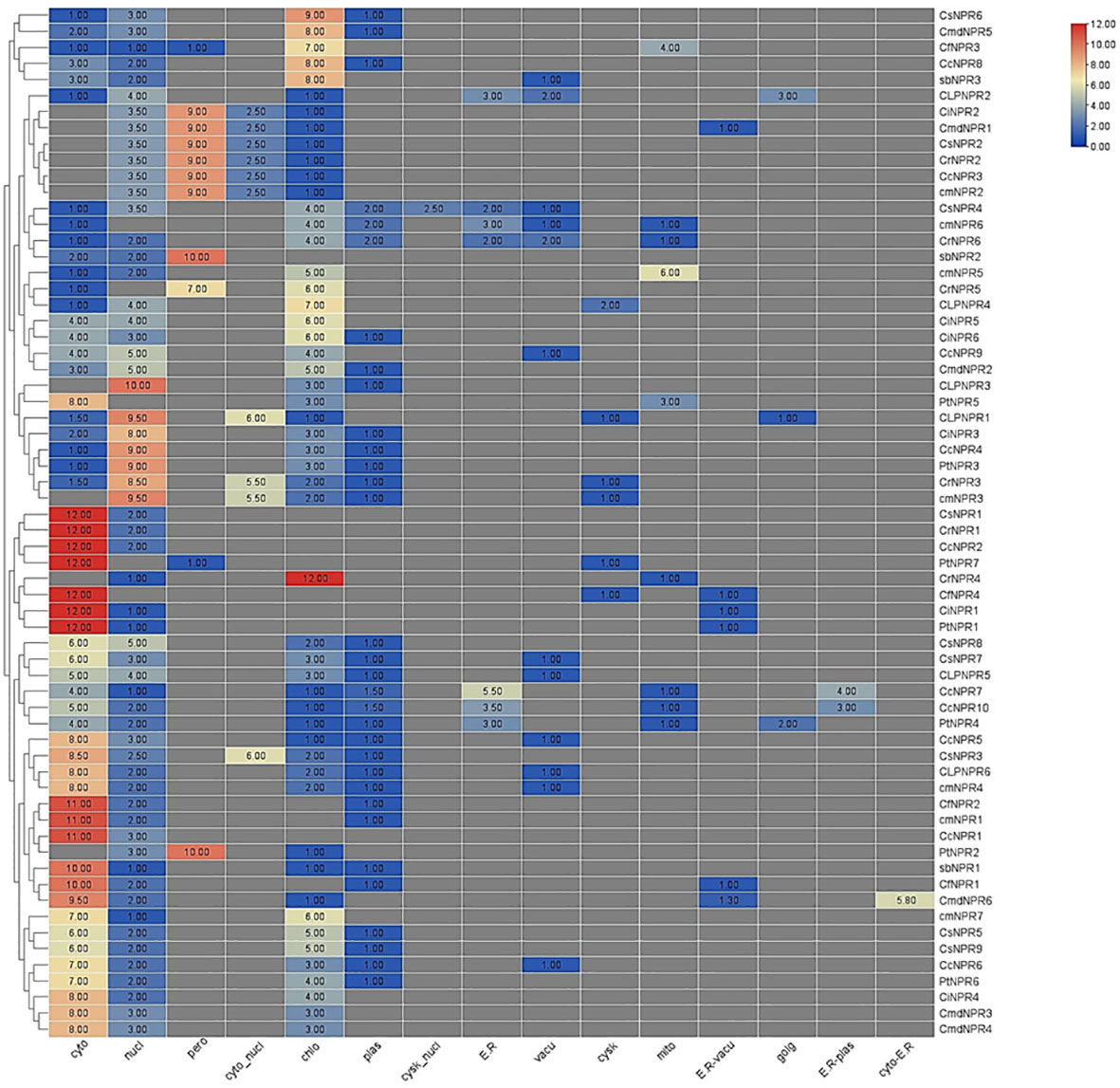
Figure 4 An abundance of putative NPR1-like genes in various species of citrus, connected to a phylogeny tree, enables a better understanding of gene function and analogy. Cytoplasm exhibited the highest abundance of CsNPR1, CrNPR1, CcNPR2, PtNPR7, CiNPR1, and PtNPR1, while various organelles had distinct but lower quantities of these proteins, indicating diverse subcellular distributions.
Comparative chromosomal mapping of several citrus species, including C. sinensis, C. climentina, C. medica, C. maxima, C. ichangenesis, C. limon, C. fortunella, A. buxifolia, and P. trifoliata, about A. thaliana, revealed the presence of five pairs of orthologous genes among these citrus species. Specifically, Scaffold 86 of C. reticulata contained CrNPR6, which had an ortholog, CsNPR4, on chromosome 7 of C. sinensis. Additionally, CrNPR3 on Scaffold 86 of C. reticulata exhibited orthology with CsNPR7 on chromosome 7 of C. sinensis, while CrNPR5 on Scaffold 86 of C. reticulata shared orthology with CmNPR6 on chromosome 9 of C. maxima. Furthermore, AbNPR1 on Scaffold 15601 of A. buxifolia was found to be orthologous to AtNPR2 on chromosome 4 of A. thaliana, and AtNPR5 on Chromosome 2 of A. thaliana displayed orthologous relationships with AtNPR6 on chromosome 3 of A. thaliana (Figure 5).
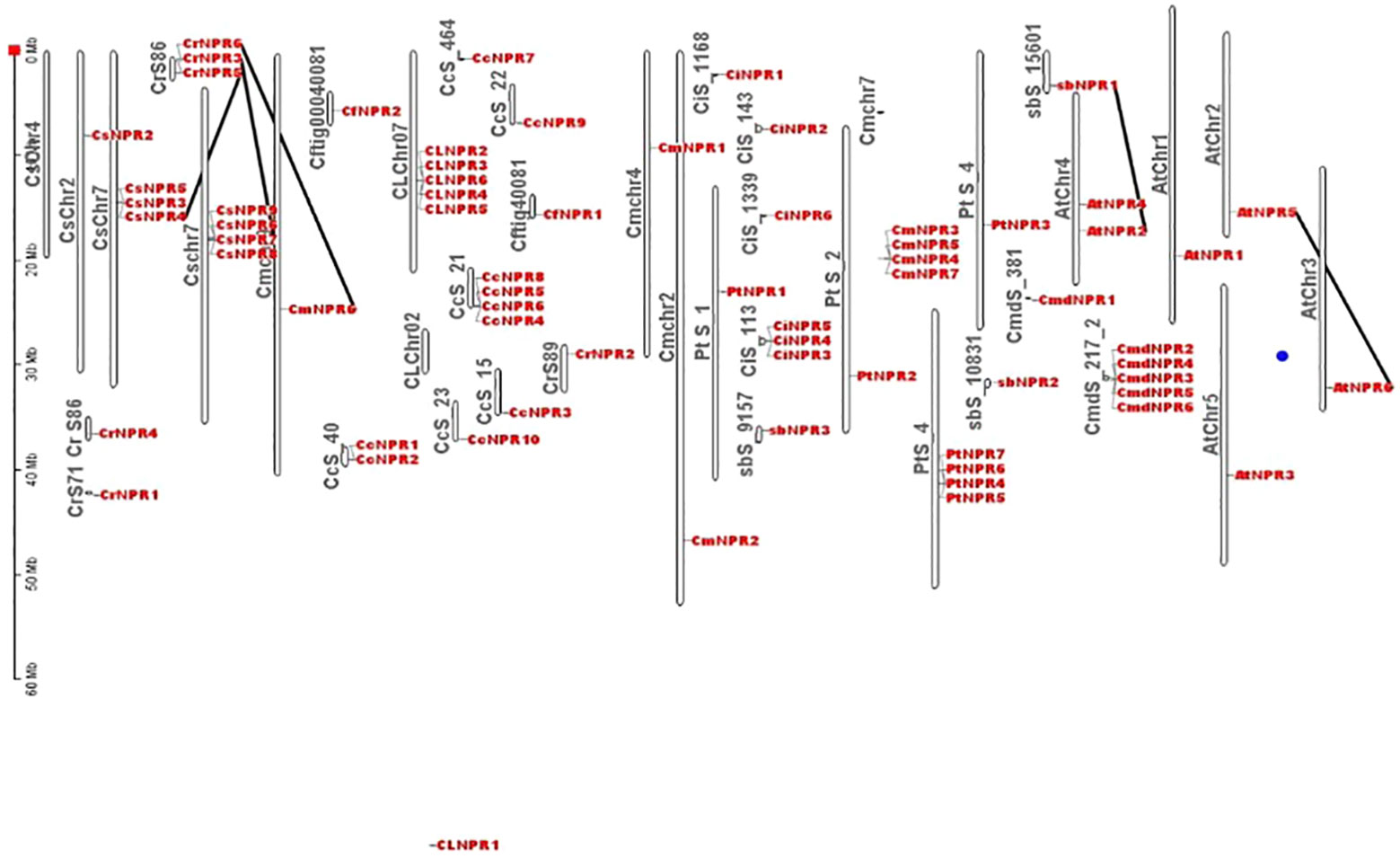
Figure 5 Different citrus species chromosomes location and duplication occurrences to NPR1-like genes revealed 5 orthologous pairs, e.g., CrNPR6 in C. reticulata aligns with CsNPR4 in C. sinensis. Additionally, AbNPR1 in A. buxifolia corresponds to AtNPR2 in A. thaliana, while AtNPR5 aligns with AtNPR6 within A. thaliana.
Numerous cis-regulatory elements were identified within the 55 promoter regions of CsNPRs, CrNPRs, CfNPRs, CcNPRs, ClNPRs, CmdNPRs, CmNPRs, AbNPRs, PtNPRs, and CiNPRs, including widely observed CAAT and TATA boxes. Additionally, phytohormone-responsive elements such as ABRE, TGACG-motif, TCA-element, P-box, GARE-motif, TATC-box, TGA-box, and AuxRR-core were detected across various genes and are involved in the response to auxin, GA, SA, abscisic acid, etc., prominently in CsNPRs, CrNPRs, CcNPRs, CfNPRs, CiNPRs, CmdNPRs, CmNPRs, PtNPRs, AbNPRs, and AtNPRs. Light-responsive elements such as Box 4, CAG-motif, G-Box, GT1-motif, ACE, AT1-motif, TCT-motif, I-box, TCCC-motif, chs-CMA2a, AE-box, ATCT-motif, GATA-motif, Gap-box, LAMP-element, GA-motif, Pc-CMA2c, and Sp1 were notably present in various genes. Furthermore, specific cis-regulatory elements like LRR, ACII, and ref2f-1 were discovered in selected genes, implicated in regulating circadian rhythm, zein metabolism, anaerobic induction, and drought stress among other processes (Figure 6).
To determine the collinearity relationships among orthologous genes of C. sinensis, C. fortunella, C. reticulata, C. medica, C. maxima, C. limon, A. buxifolia, P. trifoliata, C. ichangenesis, and C. climentina, a synteny analysis was conducted. Notably, scaffolds 1, 2, and 4 of P. trifoliata did not exhibit orthologous gene counterparts. In A. buxifolia, AbNPR1 genes located on scaffold 10 shared orthologs with chromosome 4 of A. thaliana, which also houses AtNPR4 and AtNPR3. Similarly, in A. thaliana, the gene AtNPR5 found on chromosome 2 exhibited orthologs on chromosome 3, including AtNPR6. Moreover, genes located on scaffold 86 of C. reticulata—specifically, CrNPR3, CrNPR4, CrNPR5, and CrNPR6 showed orthology with CmNPR6 on chromosome 9 of C. maxima and with genes CsNPR3 through CsNPR9 on chromosome 7 of C. sinensis (Figure 7).
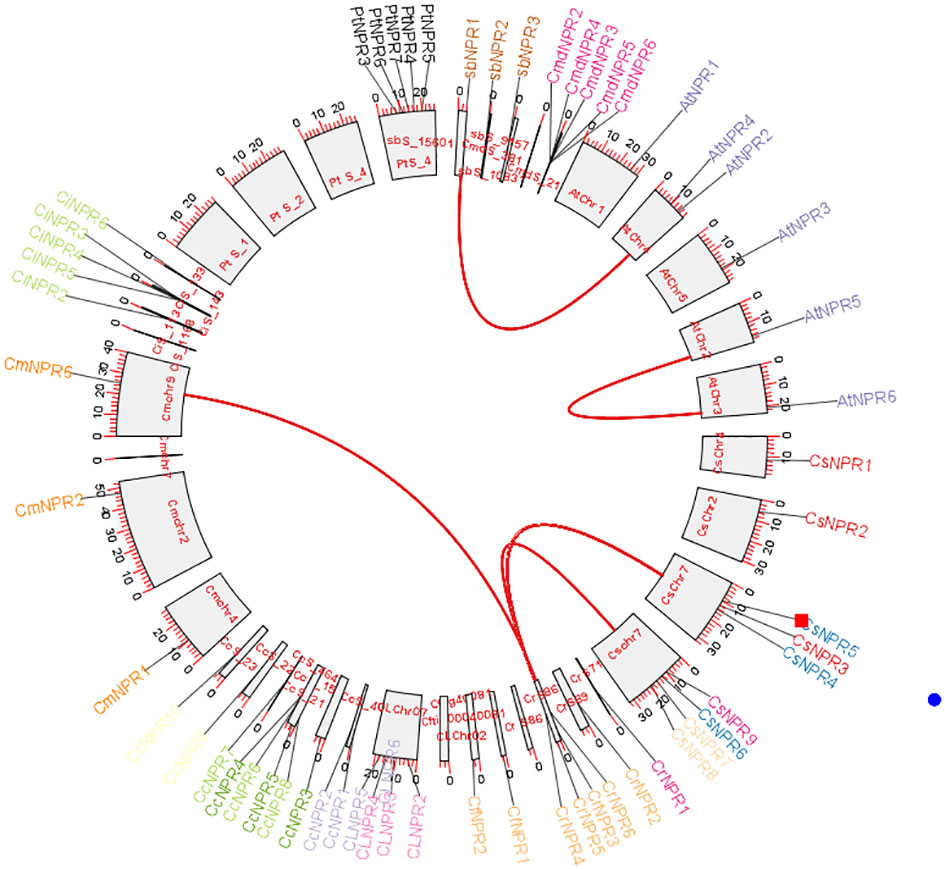
Figure 7 A Circos plot depicting the NPR1-like gene family in citrus species. The red connections indicate instances of gene duplications occurring both within and between various species. Synteny analysis revealed collinearity relationships among orthologs from various citrus species and identified specific gene matches, such as AbNPR1 in A. buxifolia, AtNPR5 in A. thaliana, and CrNPR3, CrNPR4, CrNPR5, CrNPR6 in C. reticulata with their respective orthologs in related species.
CmNPR1 and CmNPR5 were derived from previously generated in silico data that focused on NPR1 expression during the flavonoid biosynthesis process in both red and white cultivars of C. maxima. The results did not indicate any substantial upregulation or downregulation of CmNPR1 or CmNPR5, implying that NPR1 might not have a significant influence on flavonoid biosynthesis (Zhao et al., 2020) within C. maxima.
The role of CsNPR1 in the developmental phases of C. sinensis was scrutinized, revealing its active involvement across various stages. Notably, the expression of CsNPR1 remained consistent across all four treatments, suggesting a stable pattern without significant fluctuations in up- or downregulation. This constancy in expression implies that CsNPR1 maintains a steady involvement throughout the developmental processes, indicating a potential regulatory role that remains relatively unaltered across the conditions tested. The observed consistency in expression underscores the significance of CsNPR1 in the intricate orchestration of developmental events within C. sinensis.
In the face of diverse low pH conditions ranging from 2 to 6, tested at intervals of 1 h, 2 h, 3 h, 4 h, and 10 h, CsNPR1 displayed a remarkable resilience. Importantly, throughout these varying acidic stress conditions, there was no discernible alteration in the expression of CsNPR1. This steadfastness in expression levels strongly suggests that CsNPR1 remains unaffected by acidic stress within the tested pH range and exposure durations. The consistent expression of CsNPR1 despite the challenging acidic conditions underscores its robust nature and resilience to environmental stressors, highlighting its potential role as a key player in mitigating the impact of low pH stress on the biological processes it governs NPR1 (Zhang et al., 2022).
CsNPR1 expression was identified in the regulation of cuticle formation during two distinct developmental stages, a finding consistent across two separate replicates. Importantly, there were no significant fluctuations observed in the expression levels of CsNPR1 during these stages. This stability in expression implies that CsNPR1 consistently plays a role in the control of cuticle formation at these specific developmental points. The lack of notable changes in CsNPR1 expression levels underscores its reliability as a regulator in the intricate processes involved in cuticle development, emphasizing its potential importance in maintaining the integrity and protective function of the cuticle across these specific developmental milestones.
The quantitative analysis using qRT-PCR revealed the presence of NPR1 genes expression in C. sinensis following inoculation with Xanthomonas axonopodis pv. citri. Out of the nine CsNPR1 genes, statistical analysis showed that three genes (CsNPR1, CsNPR4, and CsNPR5) did not exhibit significant differences (Figure 8). Conversely, the remaining six genes (CsNPR2, CsNPR3, CsNPR6, CsNPR7, CsNPR8, and CsNPR9) displayed statistically significant responses. Notably, all samples that were treated with infected citrus bacterial canker exhibited higher relative expression of the NPR1 gene compared to the control group. Among them, CsNPR7- and CsNPR9-treated samples demonstrated the highest gene expression levels. However, it is important to note that control samples displayed higher expression levels than the other control samples.
The results obtained from qRT-PCR analysis indicated the expression of NPR1 genes across all treatments in C. reticulata. Notably, all NPR1 genes in C. reticulata displayed statistically significant expression, except for CrNPR1, which exhibited varying results. Among these genes, CrNPR3, CrNPR4, and CrNPR6 demonstrated the highest levels of expression in the treated samples compared to the control group, while CrNPR2 and CrNPR5 exhibited higher expression levels in the control samples than in other genes (Figure 9).
In the case of C. fortunella, the real-time qPCR findings showed significant expression of NPR1 genes, particularly CfNPR1 and CfNPR2. Interestingly, CfNPR2 displayed increased expression in the control samples compared to the treated (infected with citrus bacterial canker) samples (Figure 10).
Similarly, NPR1 genes were found to be expressed in all treated samples of C. limon compared to the control. Among these, ClNPR1, ClNPR3, and ClNPR6 yielded statistically non-significant results, while ClNPR2, ClNPR4, and ClNPR5 exhibited statistically significant responses. Notably, ClNPR2 and ClNPR4 upregulated the expression of the NPR1 gene in the treated samples, whereas ClNPR5 showed higher expression in the control samples compared to the treated ones (Figure 11).
This study marks the inaugural genome-wide exploration of NPR1-like gene families and their corresponding expression patterns in reaction to Xanthomonas axonopodis pv. citri infection within four distinct citrus species (Cao et al., 1997; Spoel et al., 2003; Li et al., 2018; Peng et al., 2021).
NPR1 (Non-expressor of pathogenesis related genes 1) serves as the authentic receptor for salicylic acid, playing a crucial role in plant immunity (Cao et al., 1997; Spoel et al., 2003; Li et al., 2018; Peng et al., 2021). NPR1 plays a pivotal role in the signaling pathway of salicylic acid and systemic acquired resistance (SAR) in plants. Additionally, other members of the NPR1 gene family are associated with various reactions to both biotic and abiotic stresses (Cao et al., 1997; Peng et al., 2021). The basic significance of studies on NPR1 is due to its extensive role in plant defense (Cao et al., 1997; Peng et al., 2021). It is generally postulated that salicylic acid-based defense shields plants against host- or semi-host-dependent pathogens, while jasmonic acid-based defense protects against necrotrophic pathogens (Wally et al., 2009). This study represents the first comprehensive examination of the NPR1-like gene family in various citrus species, identifying a total of 63 NPR1-like genes across 10 citrus species (Irfan et al., 2023).
The phylogenetic tree illustrates three distinct clades of NPR genes, with members within the same clade grouping together based on their presumed similar functions. Gene structure predictions using intron–exon displays and motif analysis revealed that most NPR1 sequences lacked introns, although some had between 1 and 7. In motif analysis, it was observed that motifs 8, 2, 7, 9, 5, 1, 4, 3, and 10 were commonly found in the majority of sequences. Nevertheless, variations, including the presence or absence of specific motifs, were evident, even among sequences within the same clade. These variations suggest that the NPR1 gene has undergone structural mutations, including insertions and deletions, over time. The orthologous relationships inferred from gene mapping and syntenic analysis support the notion of a shared evolutionary history among NPR sequences. For NPR1 to upregulate plant resistance genes, it must localize in the nucleus (Kinkema et al., 2000). Changes in redox potentials influence its nuclear-cytoplasmic localization (Bassene et al., 2011). Typically, NPR1 exists as a cytoplasmic oligomer under normal conditions. However, upon detecting pathogen infection and salicylic acid accumulation, which alters cellular redox potential, NPR1 translocates to the nucleus as an active monomer, where it interacts with bZIP transcription factors from the TGA family (Després et al., 2000). The results of subcellular localization confirm that NPR1 is primarily located in both the cytoplasm and the nucleus, thereby substantiating its involvement in conferring resistance among various citrus species (Spoel et al., 2003; Wally et al., 2009; Després et al., 2000).
Examining selective pressure unveils the benefits of particular amino acid sequence modifications within proteins, offering insights into functional residues and alterations in protein function. The analysis of synteny and chromosomal mapping of NPR1-like genes reveals evidence of tandem and segmental duplications, along with the identification of orthologous relationships among different citrus species. This information provides valuable insights into gene duplications, rearrangements, and the evolutionary history of these species. C. sinensis and C. reticulata share a common ancestor, supported by the presence of various orthologs on chromosome 7 of C. sinensis and scaffold 86 of C. reticulata. A duplication event occurred in scaffold 86 of C. reticulata and chromosome 9 of C. maxima, A. buxifolia, and A. thaliana diverged from a common ancestor, sharing an ortholog.
Analyzing gene expression patterns of NPR1-like genes using existing data reveals variations in their expression levels under different stresses (biotic and abiotic).The expression of CsNPR1 exhibits a remarkable consistency and stability across diverse conditions, reflecting its pivotal role in C.sinensis development and stress response. In developmental phases, CsNPR1 maintains a steady presence, suggesting its regulatory involvement throughout these processes. Its resilience to varying low pH conditions, tested over different time intervals, underscores its robust nature in withstanding acidic stress. Notably, CsNPR1 expression remains unaffected, implying a key role in mitigating the impact of low pH stress on biological processes. Furthermore, in the regulation of cuticle formation, CsNPR1 demonstrates reliability with no significant fluctuations in expression levels across distinct developmental stages. Collectively, these findings emphasize the versatility and importance of CsNPR1, positioning it as a central player in orchestrating developmental events, responding to environmental stressors, and regulating key processes like cuticle formation in C. sinensis. A joint transcriptome analysis was conducted to indicate changes in gene expression of CmNPR1 and CmNPR5 during flavonoid biosynthesis, although the differences were not significant (Chen et al., 2022). When C. sinensis is subjected to acidic stress for a long time, the expression of CsNPR1 modulates in the leaves, although this variation is minimal. Similar variations in CsNPR1 expression were detected during cuticle development in Newhall navel orange fruits (Liu et al., 2022). In C. sinensis, all NPR1-like genes exhibited upregulated expression in response to Xac infection compared to the control. CsNPR3, CsNPR7, CsNPR8, and CsNPR9 lacked significant expression in the control group. Except for CrNPR2 and CrNPR5, which exhibited no significant expression in Xac-infected samples, all NPR1-like genes in C. reticulata showed higher expression. Conversely, NPR1-like genes in C. fortunella exhibited downregulated expression in response to Xac treatment compared to control samples, while in C. limon, NPR1-like genes were downregulated except for ClNPR4 and ClNPR6.
Citrus bacterial canker (Xanthomonas axonopodis pv. citri) stress was studied in terms of NPR1-like genes and their associated expression. Citrus species were picked based on their reported resistance to citrus bacterial canker. Two resilient species, C. sinensis and C. limon, and two vulnerable species, C. sinensis and C. limon, were picked for a complete comparative study. C. sinensis and C. limon are vulnerable to citrus bacterial canker, although C. reticulata and C. fortunella are resistant (Licciardello et al., 2022). The first identified and characterized citrus bacterial canker through cultural, biochemical, and molecular analysis, confirming the presence of Xanthomonas axonopodis pv. citri, a Gram-negative bacterium (Chaudhari et al., 2022). PCR results using Xanthomonas axonopodis pv. citri-specific primers XACF (5′-CGTCGCAATACGATTGGAAC-3′) and reverse primer XACR (5′−CGGAGGCATTGTCGAAGGAA-3′) yielded a 581-bp product.
Previous investigations into NPR1-like gene families in model organism A. thaliana had already established the importance of these families of genes in enhancing tolerance to both biotic and abiotic stress factors (Cao et al., 1997; Després et al., 2000; Kinkema et al., 2000; Spoel et al., 2003; Wally et al., 2009; Peng et al., 2021). This current study unveils alterations in the expression analysis of NPR1-like genes in response to an attack by Xac bacteria, with varying responses observed across different citrus species. Notably, in C. limon and C. sinensis, there was an upregulation in the expression of NPR1-like genes. Conversely, in C. fortunella, a lower expression of NPR1-like genes was observed under Xac attack, rendering it more resistant to the pathogen. C. reticulata exhibited varying NPR1-like gene expression, indicating a moderate level of resistance to Xac attack. These findings were further corroborated by physiological and biochemical analyses in C. limon, which showed decreased levels of chlorophyll a and b, alongside an increase in flavonoid and carotenoid compounds, known for their roles in defense mechanisms against biotic stress. However, the production of secondary metabolites like phenols and soluble proteins was suppressed. Additionally, the activity of vital enzymes, such as catalase and peroxidase, was found to decrease, subsequently impeding the plant’s growth and development. This groundbreaking research addresses a critical gap by conducting the inaugural analysis of the correlation between NPR1-associated genes and diverse physiological plant parameters in both citrus cultivars susceptible and resistant to citrus bacterial canker. The findings offer potential implications for the development of disease-resistant citrus varieties through employing multiple breeding techniques, including double haploid technology, hybridization, tissue culture, and backcrossing.
In conclusion, the comprehensive genome-wide analysis of NPR1-like gene families across diverse citrus species provides compelling evidence of their pivotal role in citrus plants resistance mechanisms against biotic stress factors. This exploration not only enhances our understanding of the evolutionary relationships within these gene families but also offers promising avenues for addressing challenges in citrus production and elevating fruit quality. Our hypothesis proposes the targeted modification of the NPR1 gene through CRISPR Cas9 technology, with the potential to yield a resilient C. limon variant resistant to Xac infection. Moreover, a thorough investigation into the roles played by SA and NPR1 during biotic stress emerges as a promising direction for future research. By integrating modern natural breeding techniques—such as double haploids, hybridization, tissue culture, and backcrossing—we aim to fortify susceptibility-prone varieties, bolstering their resilience against both biotic and abiotic stresses. In this way, our holistic approach not only advances citrus resilience but also contributes to a broader understanding of plant resistance mechanisms, marking a significant step forward in the pursuit of sustainable and robust citrus cultivation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
MA: Conceptualization, Methodology, Writing – original draft. MS: Conceptualization, Data curation, Writing – review & editing. MZH: Conceptualization, Data curation, Visualization, Writing – review & editing. AS: Formal analysis, Investigation, Methodology, Writing – review & editing. PA: Project administration, Validation, Writing – review & editing. TA: Funding acquisition, Supervision, Validation, Writing – review & editing. ZK: Conceptualization, Formal analysis, Resources, Writing – review & editing. SS: Conceptualization, Data curation, Software, Writing – review & editing. MH: Investigation, Methodology, Writing – review & editing. MAS: Funding acquisition, Supervision, Visualization, Writing – review & editing. MJ: Funding acquisition, Visualization, Writing – review & editing. GA: Formal analysis, Funding acquisition, Investigation, Visualization, Writing – review & editing. MM: Data curation, Investigation, Methodology, Writing – review & editing. IS: Funding acquisition, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia for funding this work through Large Groups Project under Grant Number (RGP.2/176/44).
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia for funding this work through Large Groups Project under Grant Number (RGP.2/176/44). The authors express their gratitude to Khalid Ali Khan, for his unwavering support and encouragement throughout the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, B., Mahmood, A., Sami, A., Haider, M. Z. (2023). Impact of climate change on fruits and crops production in south punjab: farmer’s perspective. Biol. Agric. Sci. Res. J. 2023, 22. doi: 10.54112/basrj.v2023i1.22
Almas, M. H., Sami, A., Shafiq, M., Bhatti, M. H. T., Haider, M. Z., Hashmi, M., et al. (2023). Sale price comparison of saggian flower market: a case study. Bull. Biol. Allied Sci. Res. 2023, 39. doi: 10.54112/bbasr.v2023i1.39
Arabbeigi, M., Arzani, A., Majidi, M. M., Sayed-Tabatabaei, B. E., Saha, P. (2018). Expression pattern of salt tolerance-related genes in Aegilops cylindrica. Physiol. Mol. Biol. Plants 24, 61–73. doi: 10.1007/s12298-017-0483-2
Bassene, J.-B., Froelicher, Y., Navarro, L., Ollitrault, P., Ancillo, G. (2011). Influence of mitochondria on gene expression in a citrus cybrid. Plant Cell Rep. 30, 1077–1085. doi: 10.1007/s00299-011-1014-1
Birkenbihl, R. P., Liu, S., Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Biswas, S., Akey, J. M. (2006). Genomic insights into positive selection. Trends Genet. 22, 437–446. doi: 10.1016/j.tig.2006.06.005
Campos, P. E., Pruvost, O., Boyer, K., Chiroleu, F., Cao, T. T., Gaudeul, M., et al. (2023). Herbarium specimen sequencing allows precise dating of Xanthomonas citri pv. citri diversification history. Nat. Commun. 14, 4306. doi: 10.1038/s41467-023-39950-z
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Chaudhari, K., Kolase, S., Shete, M., Chandanshive, A., Patil, M. (2022). Cultural and bio-chemical characterization of bacterial canker disease caused by Xanthomonas axonopodis pv. Citri Acid lime. doi: 10.22271/tpi
Chen, J., Shi, Y., Zhong, Y., Sun, Z., Niu, J., Wang, Y., et al. (2022). Transcriptome Analysis and HPLC Profiling of Flavonoid Biosynthesis in Citrus aurantium L. during Its Key Developmental Stages. Biology 11, 1078. doi: 10.3390/biology11071078
da Silva, E. S., Huber, S., Alcantara-Neves, N. M., Asam, C., Silveira, E. F., de Andrade Belitardo, E. M. M., et al. (2020). N-terminal peptide deletion influences immunological and structural features of Blo t 5. Allergy 75, 1503–1507. doi: 10.1111/all.14176
Després, C., DeLong, C., Glaze, S., Liu, E., Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. doi: 10.1105/tpc.12.2.279
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus huanglongbing bacterium,’Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant-Microbe Interact. 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Durango, D., Pulgarin, N., Echeverri, F., Escobar, G., Quiñones, W. (2013). Effect of salicylic acid and structurally related compounds in the accumulation of phytoalexins in cotyledons of common bean (Phaseolus vulgaris L.) cultivars. Molecules 18, 10609–10628. doi: 10.3390/molecules180910609
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fujino, T., Suzuki, T., Okada, K., Kogashi, K., Yasumoto, K.-I., Sogawa, K., et al. (2016). Chimeric RNA oligonucleotides incorporating triazole-linked trinucleotides: synthesis and function as mRNA in cell-free translation reactions. J. Organic Chem. 81, 8967–8976. doi: 10.1021/acs.joc.6b01618
Graham, J. H., Gottwald, T. R., Cubero, J., Achor, D. S. (2004). Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. doi: 10.1046/j.1364-3703.2004.00197.x
Grechkin, M., Poon, H., Howe, B. (2017). Wide-Open: Accelerating public data release by automating detection of overdue datasets. PLOS Biol. 15. doi: 10.1371/journal.pbio.2002477
Guo, A.-Y., Zhu, Q.-H., Chen, X., Luo, J.-C. (2007). GSDS: a gene structure display server. Yi chuan= Hereditas 29, 1023–1026. doi: 10.1360/yc-007-1023
Haider, M. Z., Sami, A., Mazhar, H., Akram, J., Nisa, B. U., Umar, M., et al. (2023a). Exploring morphological traits variation in gomphrena globosa: a multivariate analysis. Biol. Agric. Sci. Res. J. 2023, 21. doi: 10.54112/basrj.v2023i1.21
Haider, M. Z., Sami, A., Shafiq, M., Anwar, W., Ali, S., Ali, Q., et al. (2023b). Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1269995
Horton, P., Park, K.-J., Obayashi, T., Nakai, K. (2006). “Protein subcellular localization prediction with WoLF PSORT,” in Proceedings of the Proceedings of the 4th Asia-Pacific bioinformatics conference. 3:39–48.
Hussain, Z., Muzamil, M., Saeed, M. R., Naheed, K., Kareem, M., Munir, A., et al. (2023). Trait correlations and implications for yield potential in cotton: a comprehensive study. Biol. Agric. Sci. Res. J. 2023, 24. doi: 10.54112/basrj.v2023i1.24
Irfan, U., Haider, M., Shafiq, M., Sami, A., Ali, Q. (2023). Genome editing for early and late flowering in plants. Bull. Biol. Allied Sci. Res. 2023, 45–45. doi: 10.54112/bbasr.v2023i1.45
Islam, M. A. U., Nupur, J. A., Shafiq, M., Ali, Q., Sami, A., Shahid, M. A. (2023). In silico and computational analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes in chilli (Capsicum annuum L). BMC Genomics 24, 603. doi: 10.1186/s12864-023-09682-x
Khan, F., Shafiq, M., Haider, M. Z., Sami, A., Arshad, A., Anees, M. M. (2023). Impact of various concentrations of nacl on morphological attributes of different citrus rootstocks under field conditions. Bull. Biol. Allied Sci. Res. 2023, 50. doi: 10.54112/bbasr.v2023i1.50
Kinkema, M., Fan, W., Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Kushalappa, A. C., Yogendra, K. N., Karre, S. (2016). Plant innate immune response: qualitative and quantitative resistance. Crit. Rev. Plant Sci. 35, 38–55. doi: 10.1080/07352689.2016.1148980
Letunic, I., Copley, R. R., Pils, B., Pinkert, S., Schultz, J., Bork, P. (2006). SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. doi: 10.1093/nar/gkj079
Li, M., Chen, H., Chen, J., Chang, M., Palmer, I. A., Gassmann, W., et al. (2018). TCP transcription factors interact with NPR1 and contribute redundantly to systemic acquired resistance. Front. Plant Sci. 9, 1153. doi: 10.3389/fpls.2018.01153
Licciardello, G., Caruso, P., Bella, P., Boyer, C., Smith, M. W., Pruvost, O., et al. (2022). Pathotyping citrus ornamental relatives with Xanthomonas citri pv. citri and X. citri pv. aurantifolii refines our understanding of their susceptibility to these pathogens. Microorganisms 10, 986.
Liu, D., Guo, W., Guo, X., Yang, L., Hu, W., Kuang, L., et al. (2022). Ectopic overexpression of CsECR from navel orange increases cuticular wax accumulation in tomato and enhances its tolerance to drought stress. Front. Plant Sci. 13, 924552. doi: 10.3389/fpls.2022.924552
Malamud, F., Torres, P. S., Roeschlin, R., Rigano, L. A., Enrique, R., Bonomi, H. R., et al. (2011). The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 157, 819–829. doi: 10.1099/mic.0.044255-0
Malik, A. U., Hasan, M. U., Khalid, S., Mazhar, M. S., Shafique, M., Khalid, M. N. K., et al. (2021). Biotic and abiotic factors causing rind blemishes in citrus and management strategies to improve the cosmetic quality of fruits. Int. J. Agric. Biol. 25, 298–318. doi: 10.17957/IJAB
Manzoor, M., Hamza, A., Javaid, A., Anees, M., Tariq, M. R., Firdosi, M. F. H., et al. (2023). Bioefficacy of Some Botanical Extracts against Brinjal Fruit and Shoot Borer [Leucinodes orbonalis (Guenee); Lepidoptera, Pyrallidae. Plant Prot. 7, 263–272. doi: 10.33804/pp.007.02.4728
Marchler-Bauer, A., Zheng, C., Chitsaz, F., Derbyshire, M. K., Geer, L. Y., Geer, R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352. doi: 10.1093/nar/gks1243
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Nakasu, E. Y., Hedil, M., Nagata, T., Michereff-Filho, M., Lucena, V. S., Inoue-Nagata, A. K. (2019). Complete genome sequence and phylogenetic analysis of a novel dicistrovirus associated with the whitefly Bemisia tabaci. Virus Res. 260, 49–52. doi: 10.1016/j.virusres.2018.11.008
Parmley, J. L., Hurst, L. D. (2007). How common are intragene windows with KA> KS owing to purifying selection on synonymous mutations? J. Mol. Evol. 64, 646–655. doi: 10.1007/s00239-006-0207-7
Peng, A., Zou, X., He, Y., Chen, S., Liu, X., Zhang, J., et al. (2021). Overexpressing a NPR1-like gene from Citrus paradisi enhanced Huanglongbing resistance in C. sinensis. Plant Cell Rep. 40, 529–541. doi: 10.1007/s00299-020-02648-3
Russo, C., Maugeri, A., Lombardo, G. E., Musumeci, L., Barreca, D., Rapisarda, A., et al. (2021). The second life of Citrus fruit waste: A valuable source of bioactive compounds. Molecules 26, 5991. doi: 10.3390/molecules26195991
Sabir, H. M., Khan, M. B., Hussain, Z. (2010). Marketing margin of mandarin: A case study of Sargodha region, Pakistan. Pakistan J. Soc. Sci. 30, 275–291.
Sami, A., Haider, M. Z., Imran, M., Abbas, A., Javed, M. M. (2023a). Synergizing food safety, quality and genetic improvement: the intersection of food microbiology and processing. Bull. Biol. Allied Sci. Res. 2023, 44. doi: 10.54112/bbasr.v2023i1.44
Sami, A., Haider, M. Z., Iqbal, M., Bhatti, M. H. T., Ahmad, S., Khalid, M. N. (2023b). Deterrence effect of colored diversion sheets on the population density of melon fruit flies bactrocera cucurbitae (coquillett) and yield parameters of bitter gourd (momordica charantia l.). Biol. Agric. Sci. Res. J. 2023, 17. doi: 10.54112/basrj.v2023i1.17
Sami, A., Haider, M. Z., Meeran, M. W., Ali, M. H., Abbas, A., Ali, Q., et al. (2023c). Exploring morphological traits variation in chenopodium murale: a comprehensive multivariate analysis. Bull. Biol. Allied Sci. Res. 2023, 43. doi: 10.54112/bbasr.v2023i1.43
Sami, A., Haider, M. Z., Shafiq, M. (2024). “Microbial nanoenzymes: Features and applications,” in Fungal secondary metabolites (Elsevier), 353–367. doi: 10.1016/B978-0-323-95241-5.00015-0
Sami, A., Haider, M. Z., Shafiq, M., Sadiq, S., Ahmad, F. (2023d). Genome-wide identification and in-silico expression analysis of CCO gene family in sunflower (Helianthus annnus). Plant Mol. Biol. doi: 10.21203/rs.3.rs-3344879/v1
Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Schoch, C. L., Sherry, S. T., et al. (2022). GenBank. Nucleic Acids Res. 50, D161–D164. doi: 10.1093/nar/gkab1135
Shafiq, M., Manzoor, M., Bilal, M., Manzoor, T., Anees, M. M., Rizwan, M., et al. (2024). Genome-wide analysis of plant specific YABBY transcription factor gene family in watermelon (Citrullus lanatus) and arabidopsis. J. Appl. Res. Plant Sci. 5, 63–78.
Shahbaz, E., Ali, M., Shafiq, M., Atiq, M., Hussain, M., Balal, R. M., et al. (2022). Citrus canker pathogen, its mechanism of infection, eradication, and impacts. Plants 12, 123. doi: 10.3390/plants12010123
Shahbaz, E., Ali, M., Shafiq, M., Atiq, M., Hussain, M., Balal, R. M., et al. (2023). Citrus canker pathogen, its mechanism of infection, eradication, and impacts. Plants 12, 123.
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate-and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Spreen, T. H., Zansler, M. L., Muraro, R. P. (2003). “The costs and value loss associated with Florida citrus groves exposed to citrus canker,” in Proceedings of the Proceedings of the Florida State horticultural society, vol. pp. , 289–294.
Wally, O., Jayaraj, J., Punja, Z. K. (2009). Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta 231, 131–141. doi: 10.1007/s00425-009-1031-2
Xu, S., Yao, S., Huang, R., Tan, Y., Huang, D. (2020). Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in Gynostemma pentaphyllum. Plant Physiol. Biochem. 154, 238–247. doi: 10.1016/j.plaphy.2020.05.040
Zhang, J., Chen, X.-F., Huang, W.-T., Chen, H.-H., Lai, N.-W., Yang, L.-T., et al. (2022). Mechanisms for increased pH-mediated amelioration of copper toxicity in Citrus sinensis leaves using physiology, transcriptomics and metabolomics. Environ. Exp. Bot. 196, 104812. doi: 10.1016/j.envexpbot.2022.104812
Keywords: citrus, expression profile of NPR1, phylogenetic analysis, salicylic acid, biotic stress
Citation: Ali M, Shafiq M, Haider MZ, Sami A, Alam P, Albalawi T, Kamran Z, Sadiq S, Hussain M, Shahid MA, Jeridi M, Ashraf GA, Manzoor MA and Sabir IA (2024) Genome-wide analysis of NPR1-like genes in citrus species and expression analysis in response to citrus canker (Xanthomonas axonopodis pv. citri). Front. Plant Sci. 15:1333286. doi: 10.3389/fpls.2024.1333286
Received: 05 November 2023; Accepted: 08 February 2024;
Published: 25 March 2024.
Edited by:
Hui Song, Qingdao Agricultural University, ChinaReviewed by:
Md Mahfuzur Rob, Sylhet International University, BangladeshCopyright © 2024 Ali, Shafiq, Haider, Sami, Alam, Albalawi, Kamran, Sadiq, Hussain, Shahid, Jeridi, Ashraf, Manzoor and Sabir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irfan Ali Sabir, aXJmYW5hbGlzYWJpckBzanR1LmVkdS5jbg==; Muhammad Shafiq, c2hhZmlxLmlhZ3NAcHUuZWR1LnBr; Muhammad Aamir Manzoor, YWFtaXJtYW56b29yMUBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.