- College of Life Sciences, Institute for Conservation and Utilization of Agro-Bioresources in Dabie Mountains, Xinyang Normal University, Xinyang, China
In rice cultivation, the traits of semi-dwarfism and glutinous texture are pivotal for optimizing yield potential and grain quality, respectively. Xiangdaowan (XDW) rice, renowned for its exceptional aromatic properties, has faced challenges due to its tall stature and high amylose content, resulting in poor lodging resistance and suboptimal culinary attributes. To address these issues, we employed CRISPR/Cas9 technology to precisely edit the SD1 and Wx genes in XDW rice, leading to the development of stable genetically homozygous lines with desired semi-dwarf and glutinous characteristics. The sd1-wx mutant lines exhibited reduced gibberellin content, plant height, and amylose content, while maintaining hardly changed germination rate and other key agronomic traits. Importantly, our study demonstrated that exogenous GA3 application effectively promoted growth by compensating for the deficiency of endogenous gibberellin. Based on this, a semi-dwarf glutinous elite rice (Oryza sativa L.) Lines was developed without too much effect on most agronomic traits. Furthermore, a comparative transcriptome analysis unveiled that differentially expressed genes (DEGs) were primarily associated with the anchored component of the membrane, hydrogen peroxide catabolic process, peroxidase activity, terpene synthase activity, and apoplast. Additionally, terpene synthase genes involved in catalyzing the biosynthesis of diterpenoids to gibberellins were enriched and significantly down-regulated. This comprehensive study provides an efficient method for simultaneously enhancing rice plant height and quality, paving the way for the development of lodging-resistant and high-quality rice varieties.
Introduction
Rice (Oryza sativa L.) sustains over half of the world’s population as a staple food, making the enhancement of grain quality and yield a global priority (Chandler and Wessler, 2001; Khush, 2005; Zhang, 2007; Demont and Stein, 2013; Qiao et al., 2021). Xiangdaowan (XDW) rice, esteemed for its robust aromatic qualities, confronts a challenge with its traditional varieties exhibiting excessive plant height and poor lodging resistance, adversely impacting rice yield (Singh et al., 2011). Conversely, excessively short plants produce smaller grains and exhibit diminished disease resistance, emphasizing the critical importance of maintaining an optimal plant height for maximizing rice yield (Liu et al., 2018). Gibberellin (GA), a pivotal plant hormone governing stem elongation, holds a key role in regulating plant height (Zhu et al., 2006; Zhang et al., 2014). Numerous genes contribute to GA biosynthesis and signaling, with SD1 (semi dwarf1) standing out as the “Green Revolution” gene. SD1 encodes the crucial enzyme gibberellin 20-oxidase 2 (GA20ox2), essential for gibberellin synthesis (Spielmeyer et al., 2002). Mutations in SD1 result in reduced gibberellin production, leading to a shorter plant stature (Monna et al., 2002; Sasaki et al., 2002). Since the 1930s, the sd1 allele has been used in rice breeding to produce rice varieties with short stature and excellent lodging resistance (Sakamoto and Matsuoka, 2004; Asano et al., 2007). The tall plant phenotype is governed by the SD1 allele, while the recessive sd1 allele induces the semi-dwarf phenotype (Monna et al., 2002; Berry et al., 2004; Magome et al., 2013). Gibberellins, such as GA1 and GA4, composed of diterpene carboxylic acids, play crucial roles in plant growth and development especially in regulating plant height (Thomas and Hedden, 2006; Hedden and Thomas, 2012; Marciniak et al., 2018).
Amylose content (AC) plays a pivotal role in the culinary and physical attributes of rice, exerting a profound influence on its eating and cooking quality (ECQ) (Li et al., 2016; Huang et al., 2021; Zhang et al., 2021a, b). The Waxy gene (Wx), a key player in this saga, encodes granule-bound starch synthase (GBSS) and stands as a major architect dictating amylose synthesis in rice grains. This synthesis, in turn, modulates the AC, gel consistency (GC), and viscosity (Wang et al., 1990; Su et al., 2011; Zhou et al., 2021; Zhang et al., 2021a). Rice varieties are classified into five types based on their AC: high (>25%), intermediate (20-25%), low (10-19%), very low (3-9%), and glutinous (0-2%) (Li and Gilbert, 2018). To date, at least nine allelic variations of Wx, Wxla/Wxmw, Wxlv, Wxa, Wxb, Wxin, Wxop/Wxhp, Wxmp, Wxmq, and wx, have been related to the five AC types found in rice cultivars (Ayres et al., 1997; Cai et al., 1998; Sato et al., 2002; Larkin and Park, 2003; Wanchana et al., 2003; Mikami et al., 2008; Liu et al., 2009; Yang et al., 2013; Zhang et al., 2019; Zhou et al., 2021; Zhang et al., 2021a). In general, rice varieties with higher Wx gene expression exhibit higher GBSS enzyme activity, resulting in a higher AC content and, conversely, lower AC content (Kumar and Khush, 1986; Su et al., 2011). Researchers have explored manipulating the promoter or exon region of the Wx gene to regulated AC, resulting in the development of soft or glutinous rice lines (Han et al., 2018; Yang et al., 2018; Zhang et al., 2018; Pérez et al., 2019; Yunyan et al., 2019; Zeng et al., 2020; Huang et al., 2021; Yang et al., 2022; Tian et al., 2023; Zhang et al., 2023).
Both Wx and SD1 genes play crucial roles in determining rice grain quality and yield potential, respectively. Despite breeding efforts using diverse strategies such as marker-assisted selection, progress has been sluggish, especially in enhancing complex and multiple traits. In recent years, CRISPR/Cas9 genome editing technology has been successfully applied to improve crop traits in rice, maize, wheat, sorghum and soybean, which offers rapid and efficient trait modification (Jiang et al., 2013; Mao et al., 2013; Shan et al., 2013; Liang et al., 2014; Wang et al., 2014; Li et al., 2015). Although some researchers have employed gene editing technology to improve plant height or quality by targeting the SD1 or Wx gene in elite rice landrace or hybrid rice (Zhang et al., 2018; Han et al., 2019; Hu et al., 2019; Yunyan et al., 2019; Nawaz et al., 2020; Zeng et al., 2020; Yang et al., 2022), the simultaneous editing of both genes in the same rice variety remains uncharted.
In this study, we examined Xiangdaowan (XDW) rice, a fragrant, high-quality rice variety characterized by poor lodging resistance and tough rice texture. Despite its robust aromatic qualities, it suffers from a tall stature and elevated AC, resulting in a low harvest index and suboptimal rice quality. Enhancing lodging resistance and refining the overall quality of rice are crucial objectives that can augment yield and the breeding and utilization value by altering plant height and AC. To address these challenges, we utilized CRISPR/Cas9 technology to develop sd1-wx double mutants in Xiangdaowan (XDW) rice. Across various generations, we successfully obtained genetically stable, homozygous mutants. Subsequently, we investigated the phenotype of sd1-wx double mutants, and constructed semi-dwarf glutinous rice varieties with low plant height and AC content without disrupting other essential agronomic characteristics. Further unraveling the intricacies of the SD1 and Wx genes, we conducted comparative RNA-seq analysis on sd1-wx double mutants and the wild type (WT). In summary, our study not only promises increased yield and the provision of high-quality resources for the improvement of new rice varieties but also sheds light on the functional roles of SD1 and Wx genes through advanced RNA-seq analysis.
Materials and methods
Plant material and measurement of agronomic traits
The genetic groundwork for the transgenic plants comprises the esteemed rice landraces Xiangdaowan (Oryza sativa L., ssp. Japonica). All rice cultivation occurred in the natural settings of paddy fields at Xinyang Normal University in Xinyang, China (32°8’30’’N, 114°2’8’’E). Following to rice harvest, a minimum of 10 randomly selected rice plants underwent analysis for agronomic traits, including plant height, tiller number, grain weight, and grain number per panicle.
Construction of CRISPR/Cas9 vectors and plant transformation
The CRISPR/Cas9 vector system tailored for simultaneous targeting of multiple gene sites in monocot plants was procured from Biogle Biotechnology Co., Ltd. The design of gRNA target sequences aligned with the exon sequence of SD1 (LOC_Os01g66100) and Wx (LOC_Os06g04200), as outlined by the Rice Genomics Annotation website (http://rice.plantbiology.msu.edu/). In a nutshell, these target site sequences were incorporated into the sgRNA expression cassette, which included a rice promoter, and subsequently integrated into the Bsa I restriction sites of the pCRISPR/Cas9-BGK032 vector. Detailed primer information for constructing the sg RNA vectors for Wx and SD1 can be found in Supplementary Table S1. Following this, independent introductions of CRISPR/Cas9 constructs were made into Agrobacterium tumefaciens strain EHA105, followed by transformation into the elite rice landraces Xiangdaowan using a method described previously (Nishimura et al., 2006). Hygromycin phosphotransferase (hpt) served as the plant selectable agent during the screening process for positive rice transformants.
RNA-seq analysis
To assess the transcriptome profiling of edited lines resulting from the Wx and SD1 gene modifications, we conducted transcriptomic analysis using both wild-type (WT) and mutant plants. Stem tissues at the heading stage were utilized for RNA extraction. The RNA samples underwent sequencing by APTBIO Co. in Shanghai, China, utilizing the Illumina HiSeq™ 2000 platform. For robustness, three biological replicates were included in the transcriptomic analysis. The analytical methods for transcriptomics were based on previous method (Wang et al., 2023). The resulting clean reads, stored in FASTQ format, were mapped to the reference genome using Bowtie2. Alignment to the Nipponbare reference genome (http://rice.plantbiology.msu.edu/) was achieved with an ideal match using HISAT2 software. The expression level of each gene was normalized to fragments per kilo base per million (FPKM) mapped reads. Differentially expressed genes (DEGs) between mutant and WT plants were identified based on a fold change ≥ 2 and a p-value < 0.05. Mapping of DEGs to GO terms was performed, and the count of genes for each term was determined. Significance of GO term enrichment was assessed using the hypergeometric test, with p-values adjusted. GO terms were considered significantly enriched only when both the p-value and p-adjusted value were < 0.05. Additionally, KEGG analysis was employed to identify significantly enriched signaling and metabolic pathways for each DEG compared with the reference genome. KEGG terms with both a p-value and p-adjusted value of < 0.05 were considered significantly enriched.
Real-time quantitative PCR assays
In order to verify the result of RNA-seq result, RT-qPCR was carried out on selected genes. Trizol reagent was used to isolate total RNA from rice stems tissue by following the manufacturer’s protocol. And then 3 µg RNA samples were synthesized to cDNA using the reverse transcriptase. Quantitative RT-PCR assays were conducted analysis on the CFX96 thermocycler (Bio-rad. USA) PCR System using the SYBR green master. We used three biological and technical replicates. The rice ubiquitin gene was used as an internal control. The expression level of each gene was determined based on previous method (Livak and Schmittgen, 2001). Primers for real-time quantitative PCR are shown in Supplementary Table S1.
Seed germination rate measurement and plant hormone treatments
Following harvest, rice was subjected to a drying process at 37°C for 5 days, after which it was immersed in water, and then placed in a growth chamber maintained at 30 °C in darkness. The assessment of seed dormancy involved visual scoring, specifically when the radicle exceeded 3 mm in length. For H2O and 4 µM GA3 treatments, approximately 50 well-developed seeds were evenly distributed on wet filter paper in culture dishes with a diameter of 90 cm and were subsequently treated with the 8 mL specified reagents (H2O or 4 µM GA3) in incubator at 30°C. It was ensured that the seeds remained moist during this experiment. Germination rates were monitored daily from day 3 to day 5. This process was replicated three times. The resulting germination percentages were averaged for subsequent genetic analysis.
Quantification of the endogenous plant hormone
Guangzhou Hellogene Biotechnology Company conducted the detection of GA compounds using the following procedure: The samples (~1 g) were finely powdered and combined with 4 mL of acetonitrile (v/v) containing 20 ng of each deuterium-labeled internal standard ([2H2]GA3 and [2H6]ABA). This mixture was then placed on a shaker at 300 rpm in the dark at 4 °C for 24 hours. Afterward, the samples underwent centrifugation (12,000 g, 5 minutes), and the resulting supernatant was carefully transferred to a clean tube. To further purify the extract, it was passed through a poroshell 120 SB-C18 column, pre-equilibrated with methanol, followed by an extraction buffer. The column was subsequently rinsed with 500 μL of methanol acidified with 0.1% methanoic acid. The purified extract was then dried using nitrogen gas and reconstituted with 400 μL of 100% methanol at 4 °C in the dark for 24 hours. After centrifugation (12,000 g, 5 minutes), the supernatant was transferred into HPLC vials and subjected to liquid chromatography-mass spectrometry (LC-MS) analysis using a SCIEX6500QTRAP LC/MS/MS system, equipped with an ESI Turbo Ion-Spray interface.
Measurement of grain physicochemical properties
After plant maturation, the harvested rice grains underwent air-drying and were subsequently stored at room temperature for a minimum of three months prior to testing. The flours and starches from brown rice were prepared as previously described (Bao et al., 2006). Measurement of the apparent amylose content (AC) and gel consistency (GC) of brown rice flours was conducted according to the method described by Bao et al (Bao et al., 2006). For scanning and transmission electron microscopy analyses of starch granules, cross sections of brown white rice grains and rice powders were gold-coated under vacuum. Starch granule morphology was then examined using a scanning electron microscope (Regulus-8220, Japan) at an accelerating voltage of 3.0 kV and magnifications of 400×, 2,500×, and 3,000×. The SEM analysis was based on at least three biological replications of mounted specimens, and all procedures strictly adhered to the manufacturer’s protocol.
The GT (gelatinization temperature) values were estimated indirectly by the alkali spreading score (ASS) method. Briefly, 6 grains of whole milled white rice from each strain were placed in a 90 mm petri dish along with 20 ml of 1.7% potassium hydroxide solution. The samples were separated from each other using forceps and incubated at 30°C for 23 h to allow the rice grains to spread. The spreading score of the grains was recorded visually follow previous method. The ASS score of 1-7 was recorded depending on the appearance and degree of dispersion of the endosperm. Unaffected or slightly swollen endosperm was recorded as 1, while completely disappeared was recorded as 7. The GT value was inversely proportional to the ASS score. There are three grades of GT: ASS grades 1 to 2 are high GT means gelatinization temperature >75°C, grades 3 are high-intermediate GT means gelatinization temperature between 70°C-74°C, grades 4 to 5 are intermediate GT means gelatinization temperature between 66°C-69°C, and grades 6 to 7 are low GT means gelatinization temperature between 55°C-65°C.
Statistical analysis
To characterize the samples, we conducted measurements in triplicate, unless stated otherwise. All data are expressed as mean ± standard deviation (SD). Statistical analyses, including one-way ANOVA and Student’s t-test, were performed using the SPSS 16.0 software. P<0.05 was deemed statistically significant.
Results
Editing of OsSD1 and OsWx genes via CRISPR/Cas9 technology
To enhance the plant height and quality of elite rice landrace Xiangdaowan (XDW), we employed CRISPR/Cas9 technology to create single mutants for the sd1 and wx genes, as well as double mutants (sd1-wx). Briefly, we designed and constructed the pCRISPR/Cas9-BGK032-SD1 and pCRISPR/Cas9-BGK032-Wx single target vectors, along with the pCXUN-Wx-SD1 dual target vector (Figure 1A). These vectors were introduced into XDW callus through Agrobacterium-mediated transformation. We obtained 21 XDWsd1 T0 transgenic plants, 5 XDWwx T0 transgenic plants and 20 XDWsd1-wx T0 double mutants transgenic plants, respectively. Subsequently, we utilized PCR to select plants that were free of Cas9 and HPT plants from the transgenic (T1–T2) segregating families. Sanger sequencing of SD1 and Wx target sites identified various insertion/deletion mutations. To produce desirable homozygous and non-transgenic plants, we closely monitored the genotypes of the target sites as well as the segregation of T-DNA from T1 progeny plants to T3 progeny plants. Ultimately, we successfully obtained three lines of homozygous and T-DNA-free editing in the single mutant XDWsd1, one in the single mutant XDWwx, and three in the double mutants XDWsd1-wx. XDWsd1-1 contained eighteen nucleotide deletion in SD1, XDWsd1-2 consisted of one nucleotide deletion (ΔC) in SD1, and XDWsd1-3 contained twenty-three nucleotide deletion in SD1 (Figure 1C). XDWwx-1 contained one nucleotide deletions (ΔC) in Wx (Figure 1D). XDWsd1-wx-1 comprised of three nucleotide deletion (ΔGCC) in SD1 and one nucleotide deletions (ΔC) in Wx (Figure 1B). XDWsd1-wx-2 consisted of one nucleotide deletion (ΔC) in SD1 and Wx (Figure 1B), respectively. XDWsd1-wx-3 contained one nucleotide insertion (ΔG) in SD1 and one nucleotide deletion (ΔC) in Wx (Figure 1B). Most of the mutations were predicted to cause frameshifts, resulting in the translation of non-functional truncated proteins. In conclusion, we employed CRISPR/Cas9 technology to precisely edit the SD1 and Wx genes in XDW rice, leading to the development of stable genetically homozygous lines.
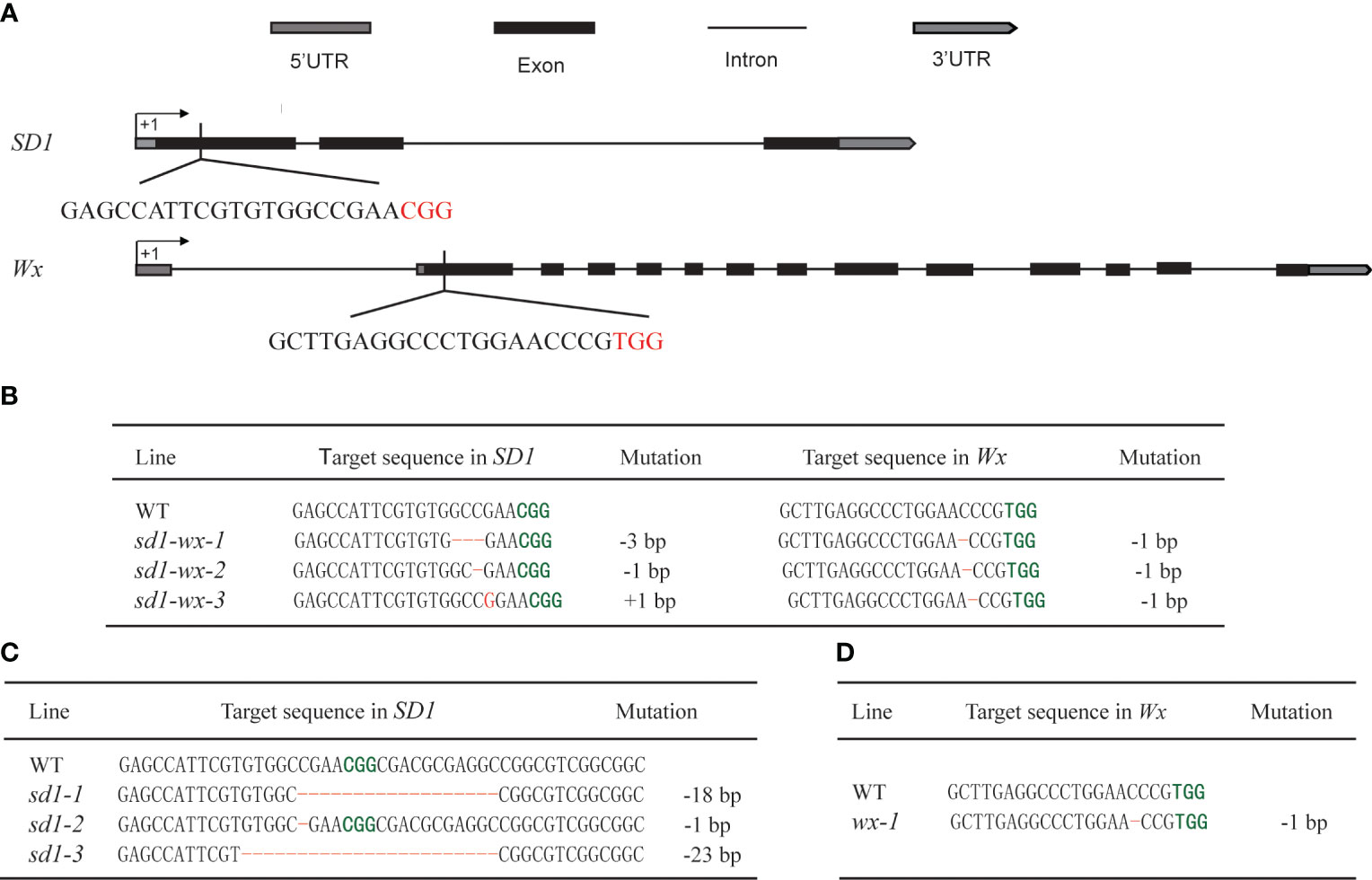
Figure 1 Targeted editing of SD1 and Wx genes based on the CRISPR/Cas9 system. (A) Schematic representation of the gene structure and target sites of SD1 and Wx genes. Introns and exons are indicated by black lines and black rectangles, respectively, and PAM sequences are highlighted in red. (B) The sd1 and wx double mutations in the edited T1 lines. (C) The sd1 single mutation in the edited T1 lines. (D) The wx single mutation in the edited T1 lines. Deletions and insertions are highlighted in red and the PAM sequences are highlighted in green, respectively.
Endogenous GA concentrations and plant heights in sd1 mutants
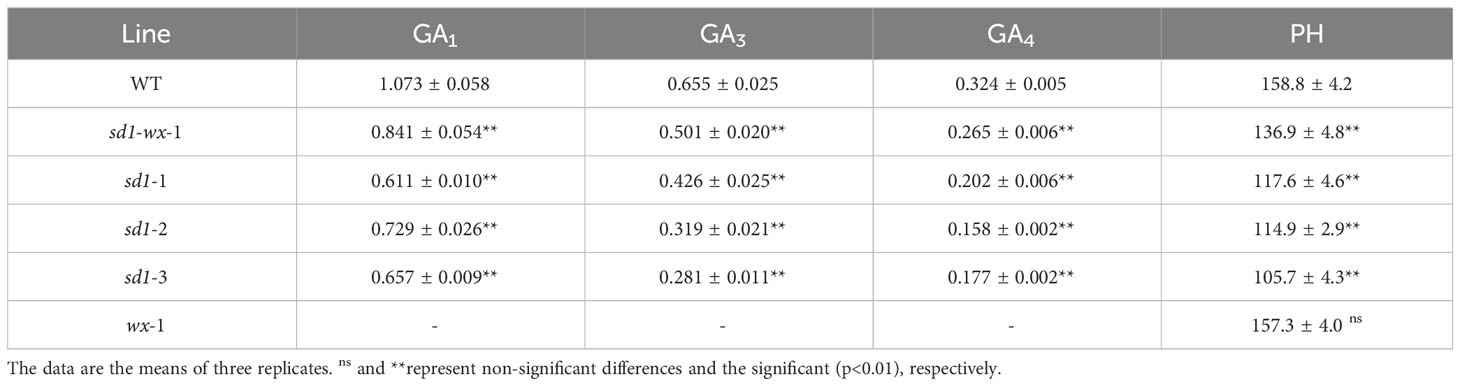
To evaluate the effect of rice plant height in sd1 mutants, we systematically measured the plant height and stem internode length of transgenic plants. The sd1 mutant lines had shorter plant heights and lower GA concentrations compared to WT plants. Specifically, the mutant lines XDWsd1-wx-1 XDWsd1-1, XDWsd1-2, and XDWsd1-3 showed plant heights of 136.9 cm, 117.6 cm, 114.9 cm and 105.7 cm in the T2 generation, respectively, whereas WT had a maximum height of 158.8 cm (Table 1). Analysis of the length of each stem internode showed that the reduction in plant height was caused by a different degree of decline in the length of each stem internode (Figure 2). In order to confirm whether the reduction of mutant lines plant height was due to the reduction of endogenous gibberellin content, we measured the content of gibberellin (GA1, GA3, GA4) through LC-MS analysis. The mutant line XDWsd1-3, with the minimum plant height, had the lowest GA3 content. The mutant lines XDWsd1-1 and XDWsd1-2 had the lowest GA1 and GA4 content (Table 1), respectively. In contrast, the WT with the maximum height had the highest gibberellin content. This suggests that editing the SD1 gene significantly reduces plant height by decreasing endogenous gibberellin levels.
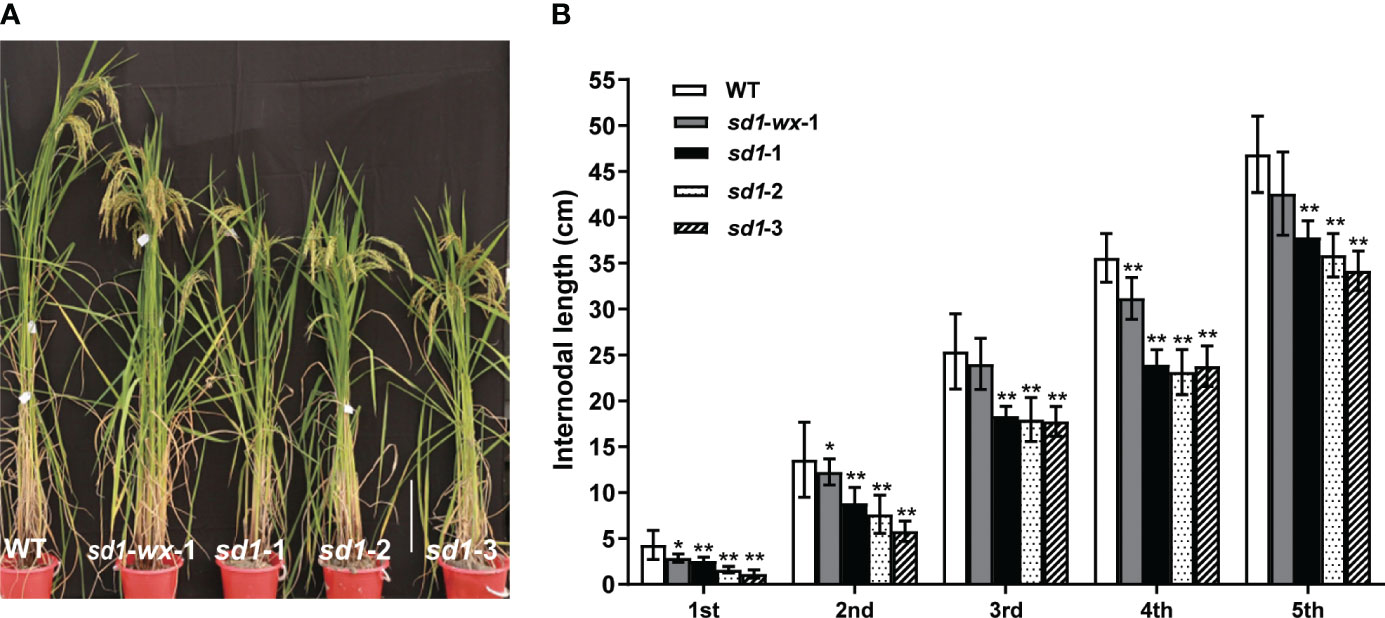
Figure 2 Phenotypes of sd1 and WT lines. (A) The plant height between mutant lines and WT lines at heading stage. Bar = 25 cm. (B) Internode length of mutant and WT lines. **indicates the significant difference (p < 0.01).
Seed germination and effect of exogenous GA3 in sd1 mutants
The concentration of endogenous gibberellin (GA) is crucial for the successful germination of seeds, ensuring the continuous life cycle of seed plants. Quality seed germination is indicative of robust seedling establishment and overall plant production. Herein, we examined the germination rates of XDW, XDWsd1-wx-1 and XDWsd1-3 seeds between H2O and GA3 treatment groups. After imbibition for 5 days, we observed distinct germination responses among WT and mutant seeds. On the third day, the sd1-wx-1 mutant exhibited remarkably lower germination rates compared to WT (Figure 3B), but this difference disappeared by the fifth day (Figures 3C, D). Compared with WT, sd1-3 mutants displayed significantly lower germination rates (Figures 3C, D). Recognizing the pivotal role of GA in seed germination regulation, we explored the impact of exogenous 4 µM GA3 treatment, revealing a significant increase in germination rates. We found that exogenous GA3 treatment can partially compensate lower germination rates in sd1 mutation compared with WT (Figures 3C, D). Further investigation indicated that the sd1-3 mutation somewhat increased seed dormancy at the early stages of seed germination, aligning with previous findings (Ye et al., 2015). Collectively, our results suggest that the sd1 mutation may reduce endogenous GA3 content, leading to decreased germination rates. However, this deficiency can be partially compensated by the application of exogenous GA.
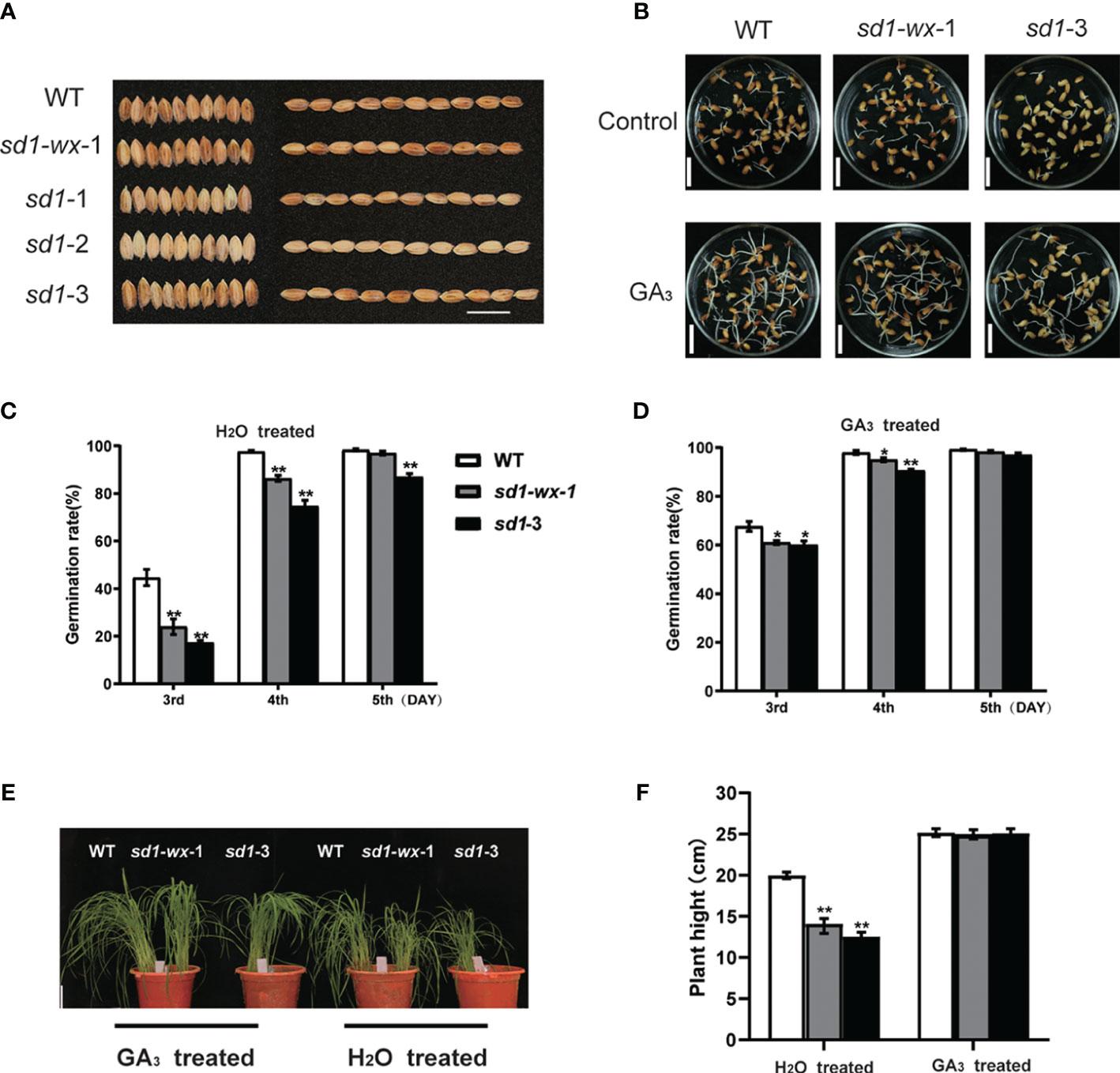
Figure 3 Seed germination and effect of exogenous GA3 in mutant lines and WT. (A) The grain phenotype of mutant line and WT; Bar = 15 mm. (B) Germination phenotypes of WT, sd1-wx-1, and sd1-3. The germination phenotypes are shown at the 3th day after imbibition. Bar = 15 mm. The germination rates of WT, sd1-wx-1, and sd1-3 under 4 µM GA3 treated (C) and H2O treated (D). Seed germination was measured after soaking for 2 days in treatment solution and then imbibing for 3 days. We recorded germination rates from day 3 to day 5. (E) Seedlings phenotype (30 day) of WT and sd1-3 and sd1-wx-1 under 10 µM GA3 treated and H2O treated, respectively. Bar = 4 cm. (F) The plant height of WT and sd1-wx-1 and sd1-3 under 10 µM GA3 treated and H2O treated, ns and ** indicates the non-significant and significant difference at p < 0.01.
To assess the potential of exogenous gibberellin application in addressing endogenous deficiency, we treated XDW plants and the height-reduced mutant line XDWwx-sd1-1 and XDWsd1-3 with 10 µM GA3 at the seedling stage (30 days old) in the greenhouse. Under controlled conditions, XDWwx-sd1-1 and XDWsd1-3 plants initially displayed the lower height (14.11 cm, 12.55 cm) compared to WT XDW plants (19.99 cm) (Figures 3E, F). Following GA3 treatment, the T2 mutant line XDWwx-sd1-1 and XDWsd1-3 exhibited restored plant height (24.95 cm, 25.08 cm), nearly equivalent to that of WT plants (25.17 cm) (Figures 3E, F). These findings demonstrate that the application of exogenous GA3 can enhance growth by supplementing the deficiency of endogenous gibberellin.
Development of glutinous rice with decreased amylose content in grains
To determine the grain quality of mutant lines, amylose content (AC) was investigated. The amylose content of the mutant line XDWsd1-wx-1 and XDWwx-1 were 5.4% and 5.2%, respectively, which showed a sharp decrease compared with the wild type (Figure 4N). All brown rice from the mutant line XDWsd1-wx-1 and XDWwx-1 exhibited an opaque appearance (Figure 4E, I). In contrast, the WT grains had an AC of 22.5% (Figure 4N), and most of the grains displayed a transparent appearance (Figure 4A). Alongside the AC, the gel consistency (GC) in the XDWsd1-wx-1 and XDWwx-1 mutant line showed a significant increase compared to that of WT (Figure 4O). To investigate the microstructural features of mature endosperm, we examined cross sections of the grains using a scanning electron microscope (SEM). The results revealed numerous small cavities in the core of starch granules within the glutinous endosperm of the XDWsd1-wx-1 and XDWwx-1 mutant line (Figures 4F-H, J-L). Interestingly, similar structures were absent in starch granules from transparent WT grains (Figures 4B-D). These small cavities within the starch granules may account for the observed differences in physicochemical properties and transparency between WT and the XDWsd1-wx-1 and XDWwx-1 mutant line. The gelatinization characteristics of starch in urea solution can reflect whether the physicochemical properties of rice have changed. Both WT and wx mutant line starch powder were dissolved in urea solution with different concentrations (0-9 mol/L). Intriguingly, we observed that at 0-3 mol/L urea concentrations, both starch varieties exhibited a certain resistance to dissolution. However, there were notable changes when the urea concentration elevated to the range of 4-9 mol/L. Starch powder of mutant line XDWsd1-wx-1 and XDWwx-1 began to dissolve in a small amount in 4 mol/L urea solution, while the WT was difficult to dissolve (Figure 4M). These findings underscore a shift in the physicochemical attributes of starch subsequent to the editing of the Wx gene. In order to better evaluate the eating and cooking quality (ECQ), we examined the GT (gelatinization temperature) values by the alkali spreading score (ASS). According to the alkali spreading value, the rice can be categorized into four types according to the GT value: high GT (75-79°C), high-intermediate GT (70-74°C), intermediate GT (66-69°C), and low GT (55-65) °C. In our study, the XDWsd1-wx-1 and XDWwx-1 seeds were classified as having high-intermediate GT, while the WT was classified as having low GT (Figures 4P-S). Notable alterations include a marked reduction in AC, conspicuous increases in the GC of the gel and the GT, the emergence of small perforations in the endosperm microstructure, and discernible changes in starch gelatinization dynamics when exposed to urea solutions. These results indicate that the edited mutant line XDWsd1-wx-1 and XDWwx-1 have the characteristics of glutinous rice with good rice quality.
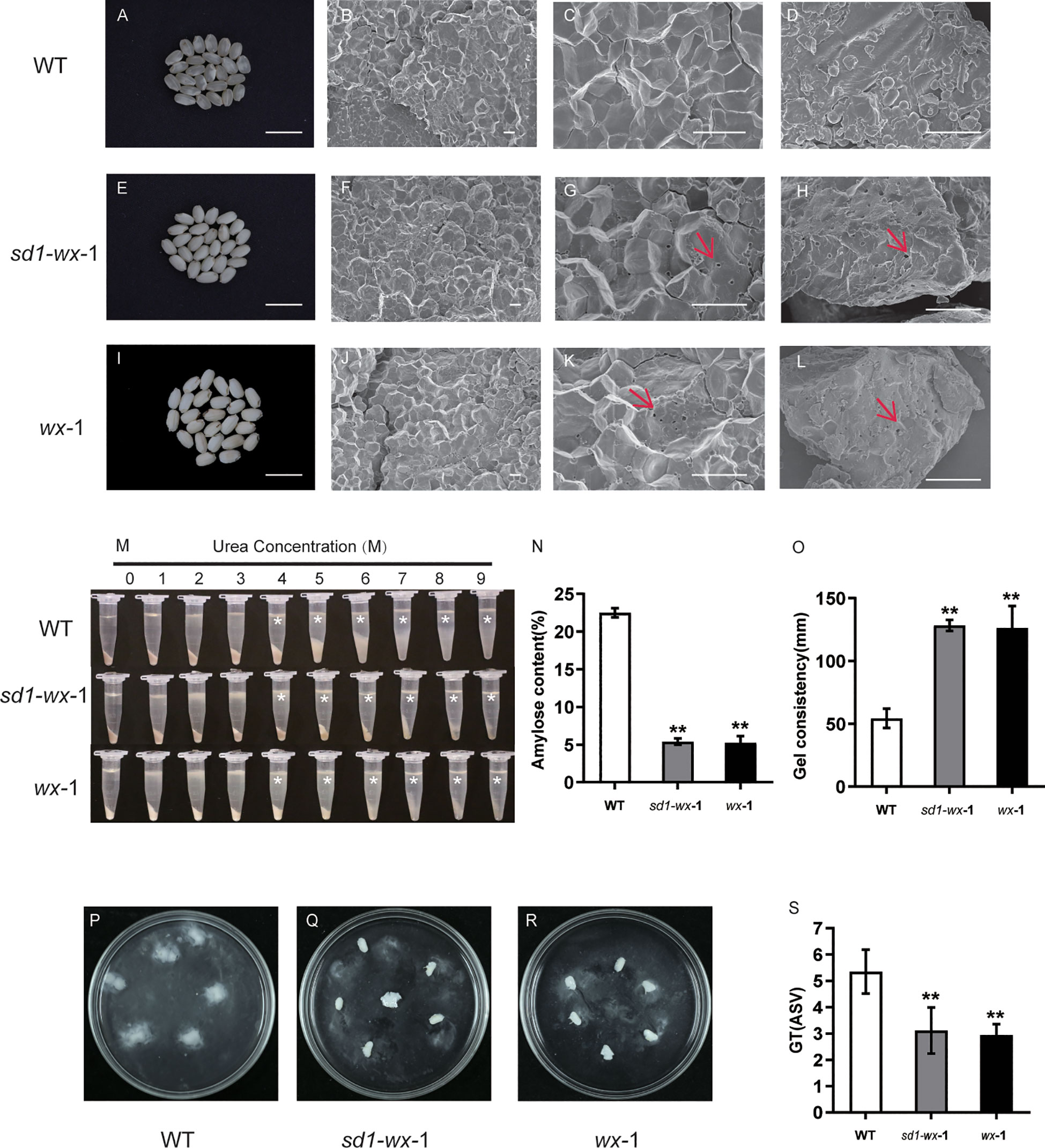
Figure 4 An analysis of starch phenotypic and physicochemical characteristics in WT and wx mutants. Comparative analysis of brown rice appearance between WT (A) and sd1-wx-1 (E) and wx-1 (I); Bar = 1 cm. Scanning electron microscopy images of cross sections of the WT (B, C) and sd1-wx-1 mutant (F, G) grains and wx-1 (J, K). Scale bars, 10 µm and magnifications of 1,000× in (B, F, J); 10 µm and magnifications of 3,000× in (C, G, K). Scanning electron microscope images of WT (D) and sd1-wx-1 mutant (H) and wx-1 (L) brown rice powders. Scale bars, 10 µm and magnifications of 3,000× in (D, H, L). Small cavities are marked by red arrows. (M) Gelatinization properties of starch in urea solutions. Starch powder of WT and sd1-wx-1 and wx-1 was mixed with different concentrations of urea solution. * indicate the starch from the endosperm of sd1-wx-1 and wx-1 were more difficult to gelatinize than that from the endosperm of WT in urea solutions of 4-9 M. (N) The amylose content in endosperm of WT and sd1-wx-1 and wx-1. (O) The gel consistency in endosperm of WT and sd1-wx-1 and wx-1. The seed alkali spreading score (ASS) during WT (P) and sd1-wx-1 (Q) and wx-1 (R). The GT (gelatinization temperature) values were estimated indirectly by the alkali spreading score (ASS) during WT and sd1-wx-1 and wx-1 (S). ** indicate statistical significance between WT and wx mutant lines at p< 0.01.
Performance of agronomic and quality traits
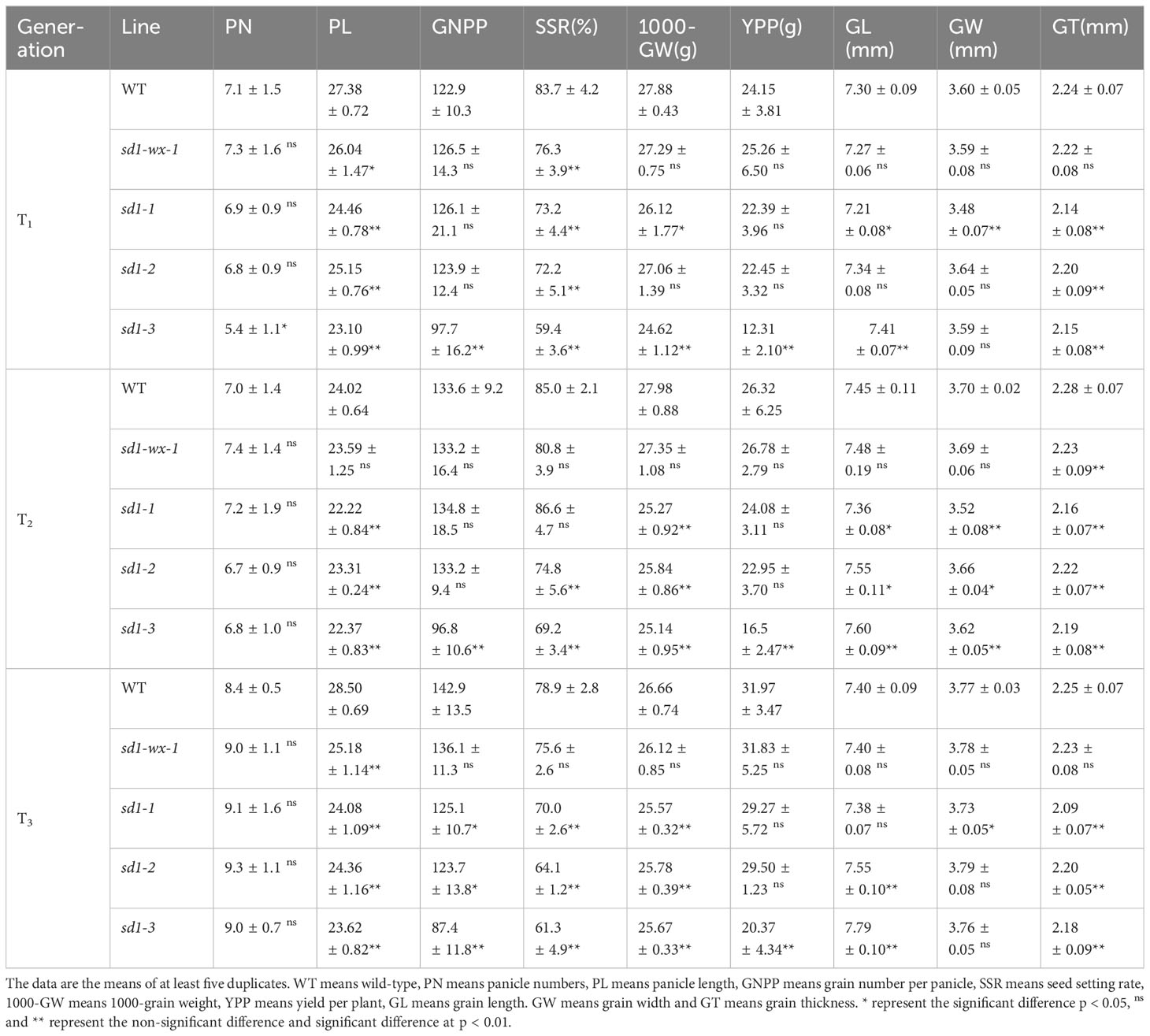
To evaluate the impact of the sd1-wx double mutant lines on yield, we conducted a comprehensive analysis of major agronomic traits across multiple generations, ranging from T1 to T3 (Table 2). Consistently, the results demonstrated discernible effects of the mutations persisting through subsequent generations. Panicle numbers (PN) exhibited no significant difference between the WT plants and mutant lines, indicating a degree of stability in this trait. Significant variations emerged in certain semi-dwarf lines, with shorter panicle length (PL) and lower seed setting rates (SSR). Grain number per panicle (GNPP) displayed no marked distinctions in most mutants when compared to the WT, although specific sd1-3 mutant lines exhibited diverse effects. Notably, the XDWsd1-3 mutant line, characterized by minimal plant height, exhibited substantial reductions in PL, GNPP, SSR, 1000-grain weight (GW), and yield per plant (YPP) compared to the WT. Intriguingly, it displayed longer grain length (GL) (Figure 3A). Of particular interest is the semi-dwarf XDWsd1-wx-1 double mutant line, which demonstrated a comparable yield per plant to the WT. Across T1 to T3 generations, the YPP for WT and XDWsd1-wx-1 double mutant line exhibited changes from 24.15 g to 25.26 g, 26.32 g to 26.78 g, and 31.97 g to 31.83 g, respectively. Besides, other agronomic traits between WT and XDWsd1-wx-1 double mutant line showed no significant differences. This suggests that XDWsd1-wx-1 double mutant line holds promise as a potential breeding material. The application of CRISPR/Cas9-based mutagenesis targeting Wx and SD1 in generating semi-dwarf glutinous elite rice lines could contribute to enhanced genetic diversity, ultimately benefiting rice yield and quality.
Transcriptomic analysis in sd1-wx double mutants
To evaluate the impact of the sd1-wx knockout double mutant line on the rice transcriptome, we employed RNA-seq analysis on both sd1-wx double mutant and WT plants at the heading stage. Comparison of the transcriptomes of WT and sd1-wx mutant stems during the heading stage revealed 1665 DEGs, with 787 downregulated and 878 upregulated genes (Figures 5A, B; Supplementary Table S3). Employing GOseq30 for functional categorization, the DEGs were classified into 76 subclasses (Supplementary Table S5). The enriched GO terms are predominantly related to molecular functions and biological processes. Notably, the DEGs were significantly involved in biological processes and cellular components, with 54 secondary items exhibiting substantial enrichment (p value < 0.01 and p adjust value < 0.01). Among these, the top five items were apoplast (gene ratio 3.96%, GO:0048046), anchored component of membrane (gene ratio 3.14%, GO: 0031225), hydrogen peroxide catabolic process (gene ratio 2.72%, GO:0042744), peroxidase activity (gene ratio 2.72%, GO:0004601), and terpene synthase activity (gene ratio 1.90%, GO:0010333).
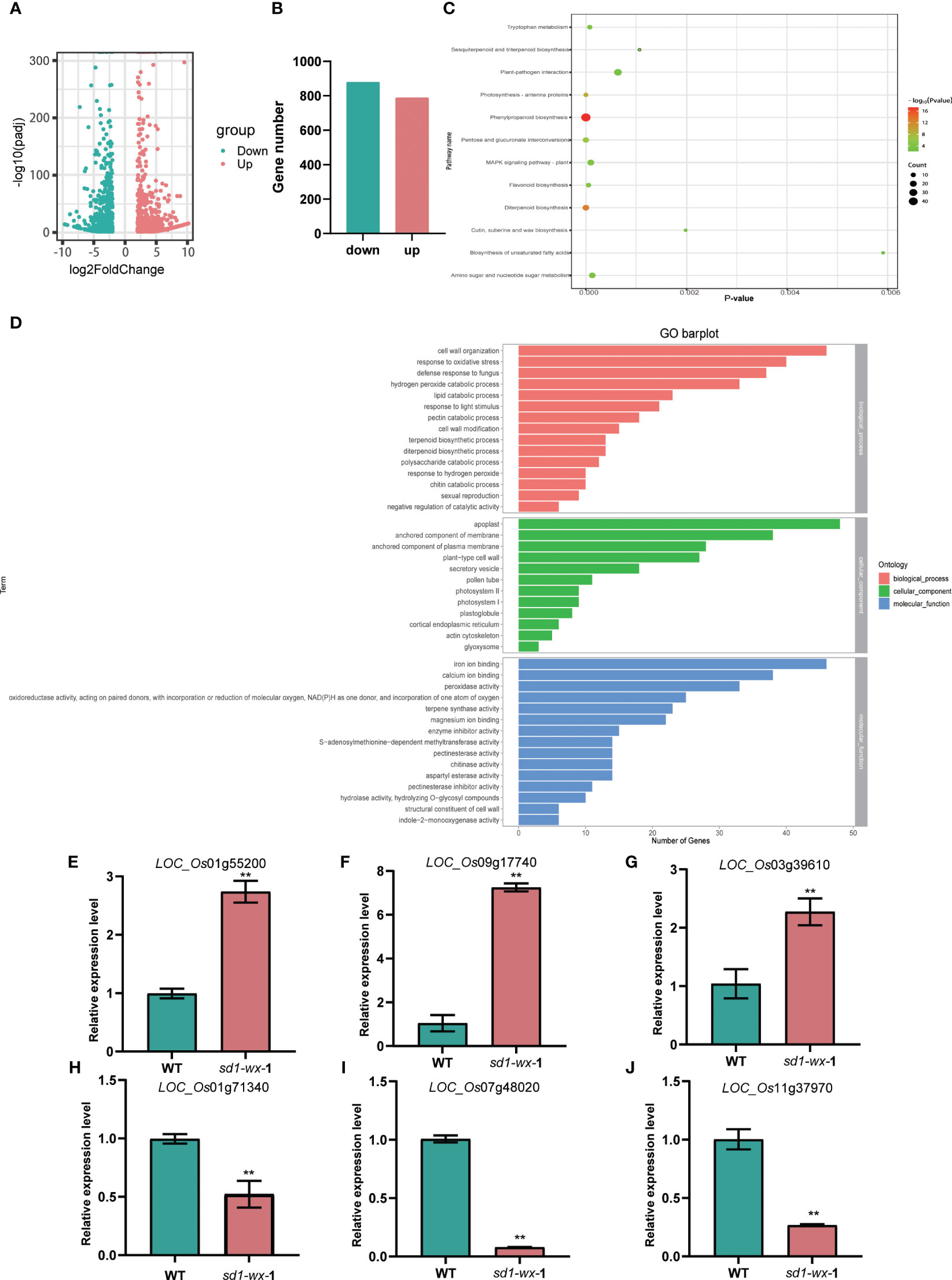
Figure 5 RNA-seq analysis between the sd1-wx and WT. (A) Volcano plot displaying DEGs between the sd1-wx and WT. (B) The number of DEGs in sd1-wx double mutant lines. (C) KEGG classifications of DEGs. Enrichment 12 significant pathways (both p value and p adjust value <0.05). (D) GO analysis of the DEGs. Enriched significant different GO terms with both p value and p adjust value<0.05. (E-J) RT-qPCR to confirm the transcript level of down-regulated genes LOC_Os01g71340 (PR2), LOC_Os07g48020 (POX22.3), and LOC_Os11g37970 (PR4a) and up-regulated genes LOC_Os01g55200 (KAT1), LOC_Os09g17740 (LHCB1.3), and LOC_Os03g39610 (LHCB2) at the stem.
To further understand the biological function, we delving into biological pathways via KEGG pathways. 194 out of the 1665 DEGs were implicated in 12 pathways (Supplementary Table S4). The top two enriched pathways were phenylpropanoid biosynthesis (ko00940, gene ratio 10.50%) and diterpenoid biosynthesis (ko00904, gene ratio 4.11%), aligning with the results obtained from GO analysis. These pathways included annotations in peroxidase activity and terpene synthase activity. Additionally, other pathways such as photosynthesis-antenna proteins, pentose and glucuronate interconversions, flavonoid biosynthesis, tryptophan metabolism, MAPK signaling pathway-plant, amino sugar and nucleotide sugar metabolism, plant-pathogen interaction, sesquiterpenoid and triterpenoid biosynthesis, cutin, suberine and wax biosynthesis, and biosynthesis of unsaturated fatty acids were identified (Figure 5C; Supplementary Table S4). Furthermore, we found that most of the DEGs related to diterpenoid biosynthesis were down-regulated in sd1-wx double mutants. There were 18 genes associated with diterpenoid biosynthesis, including two gene encoding ent-copalyl diphosphate synthase (OsCPS2, OsCPS4), which is the first key enzyme catalyzes gibberellin biosynthesis pathway, five genes encoding terpene synthase/ent-kaurene synthase (OsKS3, OsKS4, OsKS5, OsKSL8, OsKSL10), which is the second enzyme in the gibberellin synthesis step, one gene encoding ent-kaurene oxidase (OsKO1), six genes encoding cytochrome P450, two genes encoding short-chainalcohol dehydrogenase (OsSDR110C-MS1, OsSDR110C-MS2), two gene encoding gibberellin biosynthesis gene (OsGA20ox3, OsGA3ox2) (Figure 6; Supplementary Table S2). This comprehensive RNA-seq analysis at the stem level unequivocally confirms the influence of sd1 knockout on the expression of gibberellin biosynthesis-related genes.
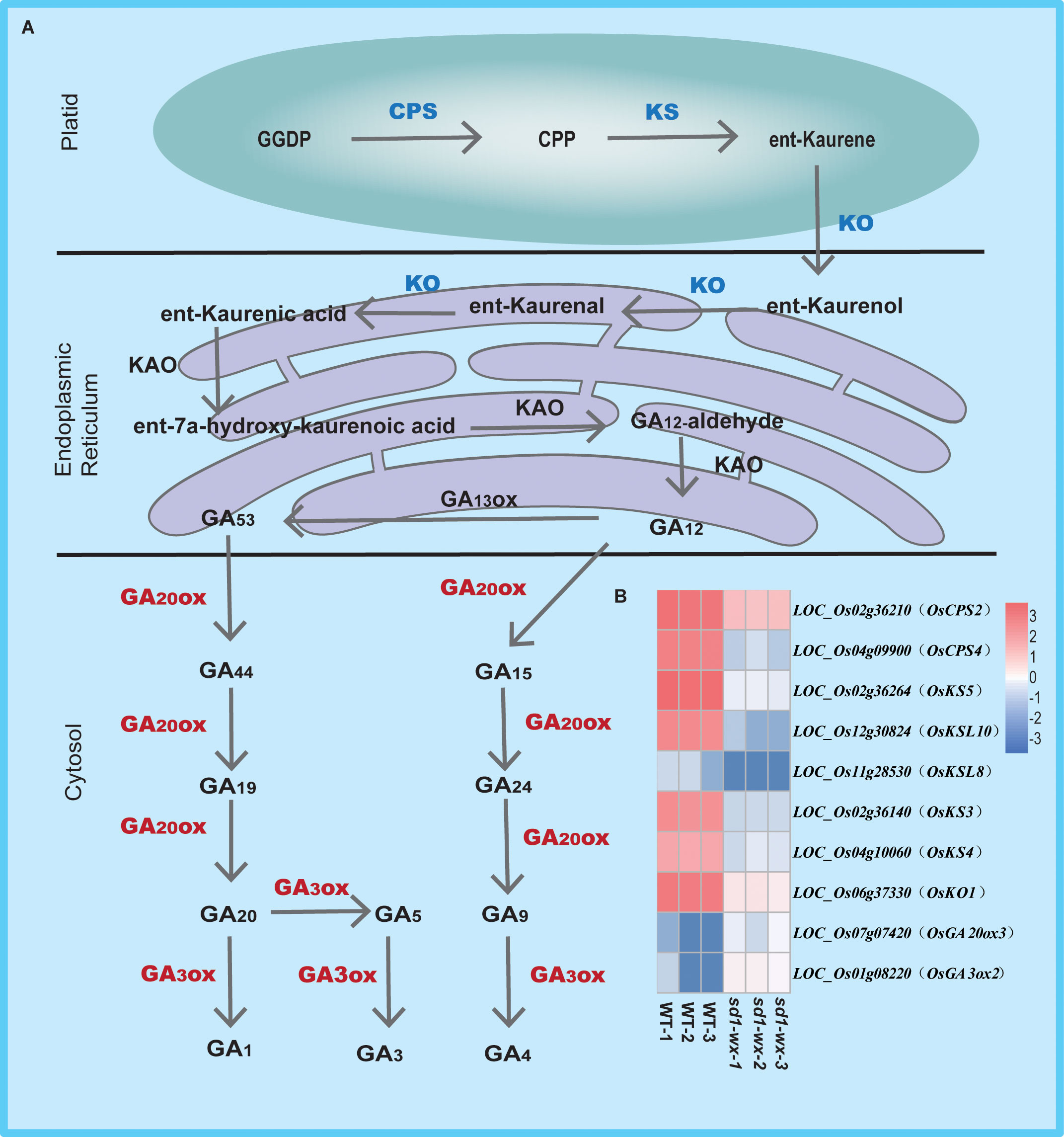
Figure 6 Pathway of gibberellin biosynthesis in WT and sd1 knockout plants. (A) Pathway of gibberellin biosynthesis. (B) Heatmap analysis of the expression of 10 DEGs associated with gibberellin biosynthesis. Note: red indicates up-regulated genes, blue indicates down-regulated genes.
To validate the DEGs identified from the RNA-seq analysis, we used RT-qPCR to determine the expression levels of the DEGs. We randomly selected 6 genes that were significantly up-regulated or down-regulated compared to WT. The results showed that the expression levels of three randomly selected up-regulated DEGs, LOC_Os01g55200 (KAT1), LOC_Os09g17740 (LHCB1.3), and LOC_Os03g39610 (LHCB2) RT-qPCR results were greatly reduced, while three randomly selected down-regulated DEGs, LOC_Os01g71340 (PR2), LOC_Os07g48020 (POX22.3), and LOC_Os11g37970 (PR4a) RT-qPCR results were greatly increased (Figures 5E-J). The RT-qPCR results were in good agreement with the RNA-seq analysis.
Discussion
The quality of rice and its resistance to lodging are pivotal factors influencing both grain quality and yield potential. These traits are primarily governed by the major genes Wx and SD1, respectively. Using CRISPR/Cas9 genome editing technology for crop improvement has demonstrated remarkable efficiency. In the present study, we not only employed this technology to generate Wx gene or SD1 gene knockout single mutant, but also utilized it to produce sd1-wx double mutants. Ultimately, this approach led to the development of elite rice lines with desirable traits, characterized by semi-dwarfism and glutinous properties. In addition, our investigation extended beyond genetic modifications to encompass a comprehensive analysis of various agronomic traits, quality attributes, physiological characteristics and physical properties of the modified plants. Furthermore, a comparative RNA-seq analysis was performed on both sd1-wx double mutants and WT plants. The integrative analysis of transcriptomic and physiological data of sd1-wx double mutants knockout lines could deepen our understanding of the development of plant height and amylose metabolism in rice.
In our current investigation, we have successfully generated an exceptional breeding line, denoted as XDWsd1-wx-1, featuring double mutant plants. Notably, this edited line exhibited a reduction in endogenous gibberellin content, leading to a decrease in plant height, although it does not represent the absolute minimum height within the mutant lines, and a concomitant decline in amylose content. Interestingly, despite these alterations, agronomic traits, yield parameters, and seed germination rates demonstrated no statistically significant differences. Our findings align with previous reports highlighting the efficacy of CRISPR/Cas9 genome editing technology in manipulating the SD1 gene to achieve lower plant height and enhanced yield potential (Han et al., 2019; Hu et al., 2019; Nawaz et al., 2020). Some researchers have suggested that although plant height decreases, seed yield remains unaffected due to the selective expression of GA20ox-2 in young tissues, while its homologous gene, GA20ox-1, is preferentially expressed in unopened flowers (Sasaki et al., 2002). Our exploration further uncovered intriguing characteristics in the XDWsd1-3 mutant line, characterized by the lowest endogenous gibberellin content and the most diminutive plant height among the mutants. Despite a lower seed setting rate, this mutant line exhibited an elongated grain length. The observed decrease in seed setting rate may be attributed to the absence of endogenous gibberellin content, potentially disrupting pollen development, as suggested by prior studies (Sakata et al., 2014).
Maintaining an optimal plant height is paramount for enhancing rice lodging resistance and overall yield. In our study, we engineered various alleles of the SD1 gene, resulting in distinct plant height phenotypes. Specifically, the transgenic plants XDWsd1-2 and XDWsd1-3 harbored 1-bp and 23-bp nucleotide deletions in SD1, respectively. These deletions induced frame-shift mutations, predicting the translation of non-functional truncated proteins that disrupted gibberellin function. Consequently, these mutants exhibited the shortest plant height compared to the WT plants. Our observations extended to XDWsd1-wx-1 and XDWsd1-1 transgenic plants, carrying 3-bp and 18-bp nucleotide deletions at the first exon of the SD1 gene. These deletions caused either one or six amino acid deletions in the SD1 protein, respectively. Intriguingly, we also found that XDWsd1-wx-1 and XDWsd1-1 transgenic plants displayed varying degrees of endogenous gibberellin content and plant height reduction compared to the WT plants. The SD1 gene encodes gibberellin 20 oxidase 2, a pivotal enzyme in gibberellin biosynthesis. However, despite its crucial role, the SD1 gene did not emerge as differentially expressed in the comparative transcriptome analysis between sd1-wx-1 and WT plants. This anomaly suggests that the deletion of one or six amino acids might disrupt the catalytic activity of gibberellin 20 oxidase 2, thereby impairing the gibberellin metabolic pathway.
Gibberellins, a class of diterpenoids, are widely distributed in plants and are primarily synthesized in the young apical parts of plants (Hedden, 2020). The synthesis of gibberellins begins with the precursor geranylgeranyl diphosphate (GGPP), which undergoes sequential catalysis by ent-copalyl diphosphate synthase (CPS), ent-kaurenoic acid oxidase (KAO), ent-kaurene oxidase (KO), and ent-kaurene synthase (KS) to culminate in the formation of GA12, an integral intermediate in the gibberellin biosynthetic pathway (Hedden and Thomas, 2012; Hedden and Sponsel, 2015; Salazar-Cerezo et al., 2018; Hedden, 2020). Our study, utilizing comparative transcriptome analysis, unveiled a significant enrichment and down-regulation of differentially expressed genes associated with diterpenoid biosynthesis. Particularly noteworthy were the findings that two genes encoding CPS, five genes encoding KS, and one gene encoding KO were substantially down-regulated in sd1-wx double mutants. This observation raises the possibility that the compromised catalytic activity of gibberellin 20 oxidase 2 restricts the efficient conversion of GGPP to GA12 (Figure 6). Interestingly, we also found that two genes encoding gibberellin oxidase (OsGA20ox3, OsGA3ox2) exhibited remarkable up-regulated expression patterns in sd1-wx double mutants. This increased expression of gibberellin oxidase genes suggests a potential compensatory mechanism in response to the compromised function of gibberellin 20 oxidase 2. The up-regulation of these genes might serve to augment the production of downstream gibberellin compounds, offsetting the limitations imposed by the impairment in GA12 biosynthesis.
In our transcriptome analysis, we unearthed compelling insights into the regulatory landscape of gene expression. Phenylpropanoid biosynthesis emerged as the most significantly enriched pathway in the KEGG analysis, leading to 46 DEGs. Furthermore, the hydrogen peroxide catabolic process (enriched with 33 genes) and peroxidase activity (also enriched with 33 genes) could act as the most prominently enriched terms in the GO analysis (Figure 5D; Supplementary Table S5). These findings underscore a pronounced modulation of the phenylpropanoid metabolism machinery in sd1-wx double mutants. Noteworthy is the absence of down-regulation of the Wx gene expression in sd1-wx double mutants compared to the WT plants, as revealed by our transcriptome analysis. Equally intriguing is the lack of enrichment for genes related to the starch metabolism pathway. This discrepancy can be attributed to the specific high expression of Wx genes in the endosperm, contrasting with their low expression in the stem, a dynamic that underscores the intricacies of gene expression patterns in different plant tissues.
In conclusion, our study harnessed the precision of the CRISPR/Cas9 gene editing system to modify both the SD1 and Wx genes in Xiangdaowan (XDW) rice, an elite landrace known for its robust aromatic properties. Across various generations, we successfully generated genetically stable, semi-dwarf glutinous rice lines, ensuring homozygosity. These edited lines exhibited a spectrum of alterations, including reduced gibberellin content, diminished plant height, lower AC, elevated GC, modified starch gelatinization characteristics in urea solutions, and the development of small holes in the endosperm microstructure. Notably, the germination rate and other key agronomic traits remained unaffected. To compensate for the growth defects associated with endogenous gibberellin deficiency, we applied exogenous GA3, revealing its potential to mitigate the observed growth-related issues. Our comparative transcriptome analysis uncovered that the compromised catalytic activity of gibberellin 20 oxidase 2 played a pivotal role in limiting the metabolism of diterpenoid biosynthesis. Taken together, our results highlight the efficacy of CRISPR/Cas9 technology in enhancing both plant height and grain quality in rice. By strategically manipulating key genes, we achieved improvements that not only offer agronomic benefits but also hold the potential to increase overall yield and enhance the quality of the crop.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
QW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. HG: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. KLi: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. HW: Data curation, Formal analysis, Methodology, Software, Writing – review & editing. FZ: Data curation, Formal analysis, Investigation, Software, Writing – review & editing. LW: Formal analysis, Investigation, Methodology, Software, Writing – review & editing. KLu: Formal analysis, Investigation, Software, Writing – review & editing. ML: Formal analysis, Software, Writing – review & editing. YS: Formal analysis, Software, Writing – review & editing. JZ: Funding acquisition, Validation, Writing – review & editing. WZ: Funding acquisition, Validation, Writing – review & editing. BP: Funding acquisition, Validation, Writing – review & editing. HY: Conceptualization, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research work was supported by grant (31801332, 31901533, U2004141) of the National Natural Science Foundation of China, Scientific and Technological Frontiers in Project of Henan Province (222102110115, 212102110249), Key Scientific Research Projects of Universities in Henan Province (24B210016), and Nanhu Scholars Program for Young Scholars of XYNU.
Acknowledgments
We thank Xinyang Academy of Agricultural Sciences for providing rice varieties.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1333191/full#supplementary-material
References
Asano, K., Takashi, T., Miura, K., Qian, Q., Kitano, H., Matsuoka, M., et al. (2007). Genetic and molecular analysis of utility of sdl alleles in rice breeding. Breed. Sci. 57, 53–58. doi: 10.1270/jsbbs.57.53
Ayres, N., Mcclung, A., Larkin, P., Bligh, H., Jones, C., Park, W. (1997). Microsatellites and a single-nucleotide polymorphism differentiate apparentamylose classes in an extended pedigree of US rice germ plasm. Theor. Appl. Genet. 94, 773–781. doi: 10.1007/s001220050477
Bao, J., Shen, S., Sun, M., Corke, H. (2006). Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch-Stärke 58, 259–267. doi: 10.1002/star.200500469
Berry, P., Sterling, M., Spink, J., Baker, C., Sylvester-Bradley, R., Mooney, S., et al. (2004). Understanding and reducing lodging in cereals. Adv. Agron. 84, 215–269. doi: 10.1016/S0065-2113(04)84005-7
Cai, X. L., Wang, Z. Y., Xing, Y. Y., Zhang, J. L., Hong, M. M. (1998). Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. doi: 10.1046/j.1365-313X.1998.00126.x
Chandler, V. L., Wessler, S. (2001). Grasses. A collective model genetic system. Plant Physiol. 125, 1540. doi: 10.2307/4279780
Demont, M., Stein, A. J. (2013). Global value of GM rice: a review of expected agronomic and consumer benefits. New Biotechnol. 30, 426–436. doi: 10.1016/j.nbt.2013.04.004
Han, Y., Luo, D., Usman, B., Nawaz, G., Zhao, N., Liu, F., et al. (2018). Development of high yielding glutinous cytoplasmic male sterile rice (Oryza sativa L.) lines through CRISPR/Cas9 based mutagenesis of Wx and TGW6 and proteomic analysis of anther. Agronomy 8, 290. doi: 10.3390/agronomy8120290
Han, Y., Teng, K., Nawaz, G. (2019). Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech. 9, 387. doi: 10.1007/s13205-019-1919-x
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hedden, P., Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Hedden, P., Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hu, X., Cui, Y., Dong, G., Feng, A., Wang, D., Zhao, C., et al. (2019). Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci. Rep. 9, 19096. doi: 10.1038/s41598-019-55757-9
Huang, L., Gu, Z., Chen, Z., Yu, J., Chu, R., Tan, H., et al. (2021). Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 106, 419–432. doi: 10.1007/s11103-021-01162-8
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. doi: 10.1093/nar/gkt780
Khush, G. S. (2005). What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 59, 1–6. doi: 10.1007/S11103-005-2159-5
Kumar, I., Khush, G. (1986). Gene dosage effects of amylose content in rice endosperm. Japan. J. Genet. 61, 559–568. doi: 10.1266/jjg.61.559
Larkin, P. D., Park, W. D. (2003). Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol. Breed. 12, 335–339. doi: 10.1023/B:MOLB.0000006797.51786.92
Li, H., Gilbert, R. G. (2018). Starch molecular structure: The basis for an improved understanding of cooked rice texture. Carbohydr. Polymers 195, 9–17. doi: 10.1016/j.carbpol.2018.04.065
Li, Z., Liu, Z.-B., Xing, A., Moon, B. P., Koellhoffer, J. P., Huang, L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. doi: 10.1104/pp.15.00783
Li, H., Prakash, S., Nicholson, T. M., Fitzgerald, M. A., Gilbert, R. G. (2016). The importance of amylose and amylopectin fine structure for textural properties of cooked rice grains. Food Chem. 196, 702–711. doi: 10.1016/j.foodchem.2015.09.112
Liang, Z., Zhang, K., Chen, K., Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Liu, L., Ma, X., Liu, S., Zhu, C., Jiang, L., Wang, Y., et al. (2009). Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71, 609–626. doi: 10.1007/s11103-009-9544-4
Liu, F., Wang, P., Zhang, X., Li, X., Yan, X., Fu, D., et al. (2018). The genetic and molecular basis of crop height based on a rice model. Planta 247, 1–26. doi: 10.1007/s00425-017-279
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Magome, H., Nomura, T., Hanada, A., Takeda-Kamiya, N., Ohnishi, T., Shinma, Y., et al. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. 110, 1947–1952. doi: 10.1073/pnas.1215788110
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., Zhu, J.-K. (2013). Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi: 10.1093/mp/sst121
Marciniak, K., Kućko, A., Wilmowicz, E., Świdziński, M., Kęsy, J., Kopcewicz, J. (2018). Photoperiodic flower induction in Ipomoea nil is accompanied by decreasing content of gibberellins. Plant Growth Regul. 84, 395–400. doi: 10.1093/mp/sst121
Mikami, I., Uwatoko, N., Ikeda, Y., Yamaguchi, J., Hirano, H.-Y., Suzuki, Y., et al. (2008). Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 116, 979–989. doi: 10.1007/s00122-008-0729-z
Monna, L., Kitazawa, N., Yoshino, R., Suzuki, J., Masuda, H., Maehara, Y., et al. (2002). Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9, 11–17. doi: 10.1093/dnares/9.1.11
Nawaz, G., Usman, B., Zhao, N., Han, Y., Li, Z., Wang, X., et al. (2020). CRISPR/Cas9 directed mutagenesis of OsGA20ox2 in high yielding basmati rice (Oryza sativa L.) line and comparative proteome profiling of unveiled changes triggered by mutations. Int. J. Mol. Sci. 21, 6170. doi: 10.3390/ijms21176170
Nishimura, A., Aichi, I., Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Pérez, L., Soto, E., Farré, G., Juanos, J., Villorbina, G., Bassie, L., et al. (2019). CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep. 38, 417–433. doi: 10.1007/s00299-019-02388-z
Qiao, J., Jiang, H., Lin, Y., Shang, L., Wang, M., Li, D., et al. (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 14, 1683–1698. doi: 10.1016/j.molp.2021.06.023
Sakamoto, T., Matsuoka, M. (2004). Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 15, 144–147. doi: 10.1016/j.copbio.2004.02.003
Sakata, T., Oda, S., Tsunaga, Y., Shomura, H., Kawagishi-Kobayashi, M., Aya, K., et al. (2014). Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164, 2011–2019. doi: 10.1104/pp.113.234401
Salazar-Cerezo, S., Martínez-Montiel, N., García-Sánchez, J., Pérez-Y-Terrón, R., Martínez-Contreras, R. D. (2018). Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 208, 85–98. doi: 10.1016/j.micres.2018.01.010
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Sato, H., Suzuki, Y., Sakai, M., Imbe, T. (2002). Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.). Breed. Sci. 52, 131–135. doi: 10.1270/jsbbs.52.131
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Singh, A., Gopalakrishnan, S., Singh, V., Prabhu, K., Mohapatra, T., Singh, N., et al. (2011). Marker assisted selection: a paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 71, 120. doi: 10.1002/pca.1274
Spielmeyer, W., Ellis, M. H., Chandler, P. M. (2002). Semidwarf (sd-1),”green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. 99, 9043–9048. doi: 10.1073/pnas.132266399
Su, Y., Rao, Y., Hu, S., Yang, Y., Gao, Z., Zhang, G., et al. (2011). Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by. Waxy in rice (Oryza sativa L.). Theor. Appl. Genet. 123, 859. doi: 10.1007/s00122-011-1632-6
Thomas, S. G., Hedden, P. (2006). Gibberellin metabolism and signal transduction. Annu. Plant Rev. Vol. 24: Plant Hormone Signaling 24, 147–184. doi: 10.1002/9780470988800.ch6
Tian, Y., Zhou, Y., Gao, G., Zhang, Q., Li, Y., Lou, G., et al. (2023). Creation of two-line fragrant glutinous hybrid rice by editing the Wx and OsBADH2 genes via the CRISPR/Cas9 system. Int. J. Mol. Sci. 24, 849. doi: 10.1007/s11032-023-01368-2
Wanchana, S., Toojinda, T., Tragoonrung, S., Vanavichit, A. (2003). Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 165, 1193–1199. doi: 10.1016/S0168-9452(03)00326-1
Wang, Q., Chen, P., Wang, H., Chao, S., Guo, W., Zhang, Y., et al. (2023). Physiological and transcriptomic analysis of OsLHCB3 knockdown lines in rice. Mol. Breed. 43, 38. doi: 10.1007/s11032-023-01387-z
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Z.-y., Wu, Z., Xing, Y., Zheng, F., Guo, X., Zhang, W., et al. (1990). Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 18, 5898. doi: 10.1093/nar/18.19.5898
Yang, J., Guo, X., Wang, X., Fang, Y., Liu, F., Qin, B., et al. (2022). Development of soft rice lines by regulating amylose content via editing the 5′ UTR of the wx gene. Int. J. Mol. Sci. 23, 10517. doi: 10.3390/ijms231810517
Yang, J., Wang, J., Fan, F. J., Zhu, J. Y., Chen, T., Wang, C. L., et al. (2013). Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in Milky P rincess and its application in japonica soft rice (Oryza sativa L.) breeding. Plant Breed. 132, 595–603. doi: 10.1111/pbr.12088
Yang, B., Xu, S., Xu, L., You, H., Xiang, X. (2018). Effects of Wx and its interaction with SSIII-2 on rice eating and cooking qualities. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00456
Ye, H., Feng, J., Zhang, L., Zhang, J., Mispan, M. S., Cao, Z., et al. (2015). Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169, 2152–2165. doi: 10.1104/pp.15.01202
Yunyan, F., Jie, Y., Fangquan, W., Fangjun, F., Wenqi, L., Jun, W., et al. (2019). Production of two elite glutinous rice varieties by editing Wx gene. Rice Sci. 26, 118–124. doi: 10.1016/j.rsci.2018.04.007
Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., et al. (2020). Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18, 2385. doi: 10.1111/pbi.13427
Zhang, Q. (2007). Strategies for developing green super rice. Proc. Natl. Acad. Sci. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Zhang, J., Liu, X., Li, S., Cheng, Z., Li, C. (2014). The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PloS One 9, e88068. doi: 10.1371/journal.pone.0088068
Zhang, C., Yang, Y., Chen, S., Liu, X., Zhu, J., Zhou, L., et al. (2021a). A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 63, 889–901. doi: 10.1111/jipb.13010
Zhang, C., Yun, P., Xia, J., Zhou, K., Wang, L., Zhang, J., et al. (2023). CRISPR/Cas9-mediated editing of Wx and BADH2 genes created glutinous and aromatic two-line hybrid rice. Mol. Breed. 43, 24. doi: 10.1007/s11032-023-01368-2
Zhang, J., Zhang, H., Botella, J. R., Zhu, J. K. (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 60, 369–375. doi: 10.1111/jipb.12620
Zhang, L., Zhang, C., Yan, Y., Hu, Z., Wang, K., Zhou, J., et al. (2021b). Influence of starch fine structure and storage proteins on the eating quality of rice varieties with similar amylose contents. J. Sci. Food Agric. 101, 3811–3818. doi: 10.1002/jsfa.11014
Zhang, C., Zhu, J., Chen, S., Fan, X., Li, Q., Lu, Y., et al. (2019). Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhou, H., Xia, D., Zhao, D., Li, Y., Li, P., Wu, B., et al. (2021). The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 63, 878–888. doi: 10.1111/jipb.13011
Keywords: rice, Sd1, Wx, gibberellin, amylose, transcriptome
Citation: Wang Q, Gao H, Liu K, Wang H, Zhang F, Wei L, Lu K, Li M, Shi Y, Zhao J, Zhou W, Peng B and Yuan H (2024) CRISPR/Cas9-mediated enhancement of semi-dwarf glutinous traits in elite Xiangdaowan rice (Oryza sativa L.): targeting SD1 and Wx genes for yield and quality improvement. Front. Plant Sci. 15:1333191. doi: 10.3389/fpls.2024.1333191
Received: 04 November 2023; Accepted: 02 February 2024;
Published: 16 February 2024.
Edited by:
Ignacio Romagosa, Agrotecnio Center, SpainCopyright © 2024 Wang, Gao, Liu, Wang, Zhang, Wei, Lu, Li, Shi, Zhao, Zhou, Peng and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Yuan, eWhvbmd5dTkyQDE2My5jb20=
 Quanxiu Wang
Quanxiu Wang Haolin Gao
Haolin Gao Bo Peng
Bo Peng