
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 March 2024
Sec. Plant Cell Biology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1331479
This article is part of the Research Topic Rising Stars: Plant Cell Biology 2022 View all 5 articles
 Yuto Omata1†
Yuto Omata1† Reina Sato1
Reina Sato1 Emi Mishiro-Sato2
Emi Mishiro-Sato2 Keiko Kano2
Keiko Kano2 Haruko Ueda3
Haruko Ueda3 Ikuko Hara-Nishimura3
Ikuko Hara-Nishimura3 Takashi L. Shimada1,4,5,6*
Takashi L. Shimada1,4,5,6*Lipid droplets (LDs) are lipid storage organelles in plant leaves and seeds. Seed LD proteins are well known, and their functions in lipid metabolism have been characterized; however, many leaf LD proteins remain to be identified. We therefore isolated LDs from leaves of the leaf LD–overaccumulating mutant high sterol ester 1 (hise1) of Arabidopsis thaliana by centrifugation or co-immunoprecipitation. We then performed LD proteomics by mass spectrometry and identified 3,206 candidate leaf LD proteins. In this study, we selected 31 candidate proteins for transient expression assays using a construct encoding the candidate protein fused with green fluorescent protein (GFP). Fluorescence microscopy showed that MYOSIN BINDING PROTEIN14 (MYOB14) and two uncharacterized proteins localized to LDs labeled with the LD marker. Subcellular localization analysis of MYOB family members revealed that MYOB1, MYOB2, MYOB3, and MYOB5 localized to LDs. LDs moved along actin filaments together with the endoplasmic reticulum. Co-immunoprecipitation of myosin XIK with MYOB2-GFP or MYOB14-GFP suggested that LD-localized MYOBs are involved in association with the myosin XIK–LDs. The two uncharacterized proteins were highly similar to enzymes for furan fatty acid biosynthesis in the photosynthetic bacterium Cereibacter sphaeroides, suggesting a relationship between LDs and furan fatty acid biosynthesis. Our findings thus reveal potential molecular functions of LDs and provide a valuable resource for further studies of the leaf LD proteome.
Lipids are important as energy sources, membrane components, and signal transducers in plants. Excess lipids are converted to neutral lipids, including triacylglycerols and sterol esters, and are stored in lipid droplets (LDs) (Grimberg et al., 2015; Shimada et al., 2019). LDs are formed on the endoplasmic reticulum (ER). In plants, triacylglycerols are produced by acyl-CoA:diacylglycerol acyltransferase (DGAT) or phosphatidylcholine:diacylglycerol acyltransferase (PDAT), which are localized on the ER (Shockey et al., 2006; Kim et al., 2011). Sterol esters are produced by phospholipid:sterol acyltransferase (PSAT) or acyl-CoA:sterol acyltransferase (ASAT), which are also localized on the ER (Lara et al., 2018; Shimada et al., 2019). In Arabidopsis thaliana, deficiency of DGAT, PDAT, or PSAT inhibits LD formation (Shimada et al., 2019; Bose et al., 2021). LDs are abundant in seed cells of oilseed plants but are also present in leaf cells. Leaf LD formation can be induced by heat stress, cold stress, and phosphate starvation (Shimojima et al., 2015; Gidda et al., 2016). In addition, certain mutants and transformants overaccumulate LDs in leaves (James et al., 2010; Shimada et al., 2019; Vanhercke et al., 2019). For instance, HISE1 down-regulates the content of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR), a rate-limiting enzyme in sterol biosynthesis (Shimada et al., 2019), and leaves of the A. thaliana mutant high sterol ester 1 (hise1) overaccumulate LDs that mainly contain sterol esters (Shimada et al., 2019). Such LD-overaccumulating lines can facilitate the characterization and isolation of LDs.
It was previously assumed that LDs serve only for carbon storage; however, recent studies have revealed LD-localized proteins with unique functions. Members of three protein families, oleosins, caleosins, and steroleosins, have been identified as seed LD proteins (Chapman et al., 2012). Oleosins have an important role in inhibiting LD fusion (Siloto et al., 2006), contributing to freezing tolerance of seeds and facilitating normal germination (Shimada et al., 2008). Leaf LD proteins, including caleosin 3 and α-dioxygenase 1 (α-DOX1), have recently been identified by LD proteomics in A. thaliana leaves (Yang et al., 2012; Shimada et al., 2014; Brocard et al., 2017; Fernandez-Santos et al., 2020; Kretzschmar et al., 2020). Caleosin 3 and α-DOX1 are involved in the production of 2-hydroxy-octadecatrienoic acid, which has antifungal activity against members of the genus Colletotrichum (Shimada et al., 2014), suggesting that leaf LDs contribute to the defense response against fungi. LD-associated proteins (LDAPs), including LDAP3, are highly similar to small rubber particle proteins identified in Hevea brasiliensis and are found in leaf LDs of plants (Gidda et al., 2013; Horn et al., 2013). LDAPs mediate LD size and contribute to drought tolerance (Gidda et al., 2016; Kim et al., 2016). LDAPs of A. thaliana interact with LDAP-INTERACTING PROTEIN (LDIP), which is also a leaf LD protein (Pyc et al., 2017). In addition, some lipases, GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE4 (GPAT4), GPAT8, and CYCLOARTENOL SYNTHASE1 are localized in leaf LDs (Fernandez-Santos et al., 2020). These findings suggest that leaf LDs contain various proteins with important functions; however, few leaf LD proteins have been identified and characterized. To resolve the molecular and physiological functions of leaf LDs, identification of additional leaf LD proteins is necessary.
Previous leaf LD proteomics efforts have used leaf LDs induced by senescence or pathogen infection (Shimada et al., 2014); however, the LD yield from these approaches is low. Here, we used A. thaliana hise1 mutant leaves, which stably accumulate high levels of LDs that are easy to isolate. We performed leaf LD proteomics by mass spectrometry analysis and identified many candidate leaf LD proteins. These included five myosin-binding proteins (MYOBs) and two putative enzymes for furan-containing fatty acid biosynthesis. MYOBs are closely related to myosin and its function, and their localization in LDs represents a breakthrough in elucidating LD movement by myosin. In addition, identification of putative enzymes for furan-containing fatty acid biosynthesis provides insight into plant lipid metabolism by leaf LDs.
A. thaliana accession Columbia (Col-0) was used as the wild type. The A. thaliana T-DNA mutant hise1-2 (Shimada et al., 2019), plants containing Lifeact–Venus driven by the cauliflower mosaic virus 35S promoter (Era et al., 2009), and plants containing myosin XIK fused with yellow fluorescent protein (YFP) driven by the myosin XIK promoter (XIK-YFP plants) (Peremyslov et al., 2012) were in the Col-0 background. Transgenic A. thaliana expressing cytosolic GFP was used as a negative control in the Co-IP experiment (Mano et al., 2002). Seeds were surface sterilized with 70% (v/v) ethanol, dried, and sown on Murashige and Skoog (MS) agar plates (Wako, Tokyo, Japan) containing 1% (w/v) sucrose. The plates were incubated at 23°C under continuous light (100 µE s–1 m–2) for 4 weeks. Seedlings were transplanted to vermiculite and grown at 23°C under continuous light (100 µE s–1 m–2) for 8 weeks.
Vectors were constructed using Gateway Technology (Invitrogen, Carlsbad, CA) with the destination vectors pGWB404m (Nakagawa et al., 2007; Segami et al., 2014), pGWB405m (Nakagawa et al., 2007; Segami et al., 2014), and pGWB406m (Nakagawa et al., 2007; Segami et al., 2014).
The LDAP3 genomic DNA fragment (nucleotides –1,799 to +1,263) was PCR amplified from A. thaliana cDNA (template) using the primer set 5′-CACCAGAAGATGAGTCACTTGAATT-3′ and 5′-ATCAAGTGGATGGAACTCCAA-3′. The PCR product was cloned into pENTR/D-TOPO (Invitrogen) via TOPO cloning to produce the subcloning vector pENTR-proLDAP3:LDAP3. The vector pENTR-proLDAP3:LDAP3 was transferred into the destination vector pGWB404m through an LR recombination reaction (Invitrogen) to create the vector pGWB404m-proLDAP3:LDAP3.
At1g19310 (+1 to +687 bp, where +1 bp refers to the transcription start site) without the stop codon was amplified by PCR from A. thaliana cDNA using the primer set 5′-CACCATGTCTGATGTCCCTTCTTG-3′ and 5′-GCTCAAGAAGAGAGAAAACA-3′. The PCR product was cloned into pENTR/D-TOPO (Invitrogen) via TOPO cloning to produce the subcloning vector pENTR-19310. The pENTR-19310 vector was digested with the restriction enzymes AscI and NotI. The digested DNA fragment (2,555 bp, pENTR-CUT) was purified and used for subcloning.
MYOB genes, AT3G23510, AT1G30130, AT4G33110, the globular tail domain of myosin XIK (XIKGTD), and other constructs (described in Supplementary Figure S1) were PCR amplified using primer sets (FWD and REV) provided in Supplementary Table S4. To produce entry clones, the PCR products were ligated into pENTR-CUT using an In-Fusion Cloning Kit (Takara, Shiga, Japan). The entry clones were cloned into the destination vector pGWB405m (for C-terminal fusion with mGFP) or pGWB406m (for N-terminal fusion with mGFP) using LR Clonase II (Invitrogen) to create expression vectors for cellular localization.
Transient expression assays were performed as described previously (Shimada et al., 2019). The expression vectors were independently transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101, and the transformed Agrobacterium was infiltrated into the leaves of 3-week-old N. benthamiana plants. The expression vector containing α-DOX1-GFP or α-DOX1-RFP was used as a LD marker (Shimada et al., 2019). One day post-infiltration, leaves were soaked in 3 mM mevalonate solution for 1 day to induce LDs. Two days post-infiltration, the leaves were observed under a fluorescence microscope (BZ-X800; Keyence, Osaka, Japan). GFP fluorescence was examined with a GFP filter (excitation: 450–490 nm; emission: 500–550 nm), and RFP fluorescence was examined with a TRITC filter (excitation: 520–570 nm; emission: 570–640 nm).
Two-week-old Lifeact–Venus plants (wild-type background) were soaked in 3 mM mevalonate solution for 1 day to induce LDs. LDs in the epidermal cells of true leaves were stained with 5 µg/mL BODIPY 493/503 (Thermo Fisher Scientific, Waltham, MA) (Fernandez-Santos et al., 2020) and observed under a fluorescence microscope (BZ-X800). The fluorescent signals of Lifeact–Venus and BODIPY 493/503 were examined with a GFP filter (excitation: 450–490 nm; emission: 500–550 nm). Punctate structures emitting fluorescence were defined as LDs, and filament structures were defined as actin filaments.
Expression vectors containing α-DOX1-RFP (Shimada et al., 2014) and mCherry-HDEL (Nelson et al., 2007) were used as LD and ER markers, respectively. Agrobacterium containing the respective constructs was infiltrated into the leaves of 3-week-old N. benthamiana plants. One day post-infiltration, leaves were soaked in 3 mM mevalonate solution for 1 day to induce LDs. Two days post-infiltration, the epidermal cells of true leaves were observed under a fluorescence microscope (BZ-X800). Fluorescent signals of RFP and mCherry were examined with a TRITC filter (excitation: 520–570 nm; emission: 570–640 nm). Punctate structures emitting fluorescence were defined as LDs, while network structures were defined as ER.
The vector pGWB404m-proLDAP3:LDAP3 was introduced into hise1-2 A. thaliana plants via Agrobacterium-mediated transformation (strain GV3101) using the floral dip method (Clough and Bent, 1998) to express the LDAP3-GFP fusion protein under the control of the LDAP3 promoter (pLDAP3:LDAP3-GFP plants).
Expression vectors containing MYOB2 or MYOB14 in pGWB405m (named pGWB405m-MYOB2 and pGWB405m-MYOB14, respectively) were introduced into wild-type A. thaliana plants via Agrobacterium-mediated transformation (strain GV3101) using the floral dip method (Clough and Bent, 1998) to express the MYOB2-GFP and MYOB14-GFP fusion proteins, respectively. True leaves of 2-week-old plants were treated with 3 mM mevalonate treatment for 1 day, and then the epidermal cells were observed under a fluorescence microscope (BZ-X800).
To isolate LDs from A. thaliana leaves, 3.5 g of leaves from 4-week-old plants (wild type or hise1-2) was homogenized in 5 mL G-20 buffer (100 mM Tris-HCl pH 7.5, 10 mM KCl, 1 mM EDTA, and 20% [w/v] sucrose) using a pestle and mortar. The homogenates were filtered through two layers of cheese cloth and centrifuged for 10 min at 10,000 g at 4°C. Supernatants (1.5 mL) were collected in fresh 2-mL tubes. The resuspensions were covered with 0.2 mL of G-15 buffer (100 mM Tris-HCl pH 7.5, 10 mM KCl, 1 mM EDTA, and 15% [w/v] sucrose) and 0.4 mL of G-5 buffer (100 mM Tris-HCl pH 7.5, 10 mM KCl, 1 mM EDTA, and 5% [w/v] sucrose) and centrifuged for 10 min at 20,000 g at 4°C. Floating lipid pads (0.3 mL) were collected in fresh 2-mL tubes. The resuspensions were covered with 0.4 mL of G-0 buffer (100 mM Tris-HCl pH 7.5, 10 mM KCl, and 1 mM EDTA) and centrifuged for 10 min at 20,000 g at 4°C. Floating lipid pads (0.1 mL) were collected in fresh 2-mL tubes and defined as isolated LDs. The isolated LDs were stained with 5 µg/mL BODIPY 493/503 (Thermo Fisher Scientific). Fluorescence was examined under a fluorescence microscope (BZ-X800) with a GFP filter (excitation: 450–490 nm; emission: 500–550 nm).
The Co-IP LD isolation method was performed using a μMACS GFP-tagged protein isolation kit (Miltenyi Biotec, Gladbach, Germany) following the method of a previous study (Shimada et al., 2014). Four-week-old hise1-2/pLDAP3:LDAP3-GFP plants (0.5 g fresh weight) and wild-type plants expressing cytosolic GFP alone were ground with 1.5 mL extraction buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM CaCl2, and 1 mM MgCl2 using a pestle and mortar. The extracts were centrifuged at 1,000 g for 5 min at 4°C followed by 8,000 g for 10 min at 4°C. The supernatants (1 mL each) were subjected to immunoprecipitation with 50 μL of anti-GFP microbeads (μMACS GFP-tagged protein isolation kit). The samples were incubated at 4°C for 30 min and applied to a μ column (Miltenyi Biotec). The column was washed with 1 mL of the extraction buffer. Pure immunoprecipitates were eluted with 50 μL of sample buffer (100 mM Tris-HCl pH 6.8, 4% [w/v] SDS, 12% [v/v] 2-mercaptoethanol, and 20% [v/v] glycerol) and defined as isolated LDs.
Immunoprecipitated samples were dissolved in sample buffer and resolved (~2 cm) using SDS-PAGE. Each lane was divided into six pieces. In-gel digestion was performed according to a method described previously (Kano et al., 2023). The digested peptides were analyzed by nano-flow reverse-phase LC followed by tandem MS using a Q-Exactive hybrid mass spectrometer (Thermo Fisher Scientific). The capillary reverse-phase HPLC–MS/MS system comprised a Dionex U3000 gradient pump equipped with a VICI CHEMINERT valve. The Q-Exactive mass spectrometer was equipped with a nano-electrospray ionization (NSI) source (AMR, Tokyo, Japan). The desalted peptides were loaded into a separation capillary C18 reverse-phase column (NTCC-360/100–3–125, 125 × 0.1 mm, Nikkyo Technos). Peptide spectra over a mass range of m/z 350–1,800 were recorded using an Xcalibur 3.0.63 system (Thermo Fisher Scientific). MS spectra were subsequently recorded followed by 10 data-dependent high-energy collisional dissociation (HCD) MS/MS spectra generated from the 10 highest intensity precursor ions. MS/MS spectra were interpreted and peak lists were generated using Proteome Discoverer 2.2.0.388 (Thermo Fisher Scientific). Searches were performed using SEQUEST (Thermo Fisher Scientific) against the TAIR10 and cRAP for contaminant databases. Search parameters were set as follows: enzymes selected with a maximum of two missing cleavage sites, a mass tolerance of 10 ppm for peptide tolerance, 0.02 Da for MS/MS tolerance, fixed modification of carbamidomethyl (C), and variable modification of oxidation (M). Peptide identifications were based on significant Xcorr values (high confidence filter). Peptide identification and modification information returned from SEQUEST were manually inspected and filtered to obtain confirmed peptide identification and modification lists from HCD MS/MS.
A. thaliana leaves of 4-week-old hise1-2/pLDAP3:LDAP3-GFP plants were immersed into 2 µM monodansylpentane (MDH; AUTODOT Visualization Dye, Abcepta, San Diego, CA) for 1 min. Fluorescence was examined under a fluorescence microscope (BZ-X800) with a DAPI filter (excitation: 340–380 nm; emission: 435–485 nm). The fluorescent signal from LDAP3-GFP was examined using a GFP filter (excitation: 450–490 nm; emission: 500–550 nm).
The hypocotyls of 4-day-old A. thaliana XIK-YFP plants were soaked in 3 mM mevalonate solution for 1 day to induce LD formation. Fluorescence was examined under a fluorescence microscope (BZ-X800) with a DAPI filter (excitation: 340–380 nm; emission: 435–485 nm). The fluorescent signal from XIK-YFP was examined using a GFP filter (excitation: 450–490 nm; emission: 500–550 nm).
Co-IP assays were performed using a μMACS GFP-tagged protein isolation kit (Miltenyi Biotec). Three-week-old A. thaliana plants (0.5 g fresh weight) expressing MYOB2-GFP or MYOB14-GFP were ground in 1 mL of lysis buffer (150 mM NaCl, 1% [v/v] Ecosurf EH-9, and 50 mM Tris-HCl pH 8.0). The extracts were centrifuged at 1,000 × g for 1 min at 4°C. This was followed by centrifugation of the supernatants at 15,000 × g for 10 min at 4°C. A 1-mL aliquot of each supernatant was subjected to Co-IP with 50 μL of anti-GFP microbeads. Samples were incubated at 4°C for 30 min and applied to a μ column (Miltenyi Biotec). The column was washed with 1 mL of lysis buffer and 0.2 mL of 20 mM Tris-HCl, pH 7.5. Pure immunoprecipitates were eluted with 50 μL of sample buffer (100 mM Tris-HCl pH 6.8, 4% [w/v] SDS, 12% [v/v] 2-mercaptoethanol, and 20% [v/v] glycerol).
Mass spectrometry analysis was outsourced to BGI (Shenzhen, China). To identify proteins in the pure immunoprecipitates from MYOB2-GFP and MYOB14-GFP plants, the samples were separated by gel electrophoresis. Proteins in the samples were digested using trypsin and separated by Thermo UltiMate 3000 UHPLC. The separated peptides were ionized using a nanoESI source and analyzed by a Q-Exactive HF X tandem mass spectrometer (Thermo Fisher Scientific) in data-dependent acquisition mode. The ion source voltage was set to 1.9 kV, MS1 scanning range was 350–1,500 m/z, resolution was set to 60,000, MS2 starting m/z was fixed at 100, and resolution was 15,000. The UniProt protein database (https://www.uniprot.org/) was used for protein profiling.
Immunoprecipitates were subjected to SDS-PAGE on a 5–20% acrylamide-gradient gel (SuperSep Ace, Fuji film, Tokyo, Japan). Silver staining was performed using a Silver Stain MS Kit (Wako, Tokyo, Japan).
Immunoprecipitates were subjected to SDS-PAGE on a 5–20% acrylamide-gradient gel (SuperSep Ace). The proteins were electrophoretically transferred from the gel onto a polyvinylidene difluoride membrane (PVDF; GE Healthcare, Chicago, IL). The membrane was treated with blocking solution (5% [w/v] skim milk, Tris-HCl pH 7.5, and 0.1% [v/v] Triton X) for 30 min and treated with anti-GFP antibody (1:10,000; JL-8, Takara, Shiga, Japan) at 4°C for 24 h. Horseradish peroxidase–conjugated anti-mouse IgG (1:2,000) was used as the secondary antibody. Immunodetection was performed with a chemiluminescent system using ImmunoStar LD (Wako) and a C-DiGit blot scanner (LI-COR, Lincoln, NE).
The plant genes mentioned in this study can be found in the GenBank/EMBL databases under the following accession numbers: LDAP3 (AT3G05500), AtUFAMD1 (AT3G23510), AtUFAO1 (AT1G30130), AtFUFM1/LIME1 (AT4G33110), MYOB1 (AT1G08800), MYOB2 (AT1G70750), MYOB3 (AT5G16720), MYOB4 (AT2G30690), MYOB5 (AT1G18990), MYOB6 (AT1G74830), MYOB7 (AT5G06560), MYOB8 (AT3G11850), MYOB9 (AT3G54740), MYOB10 (AT3G30830), MYOB12 (AT5G57830), MYOB13 (AT4G13630), MYOB14 (AT4G13160), MYOB15 (AT1G04890), MYOB16 (AT1G18265), and myosin XIK (AT5G20490). The proteins of C. sphaeroides mentioned in this study can be found in UniProt (The UniProt, 2017) under the following accession numbers: UfaO (Q3IYV8), FufM (Q6NBA4), UfaM (Q3J4I7), and UfaD (Q3IYV7).
To perform leaf LD proteomics, we isolated LDs from leaves of the A. thaliana hise1-2 mutant via a centrifuge method and a co-immunoprecipitation (Co-IP) method. LDs rise to the top during centrifugation (Shimada et al., 2008), so we centrifuged leaf extracts from wild-type or hise1-2 plants and collected the top fraction. Microscopy observation showed that the top fraction from the hise1-2 mutant contained LDs (Figure 1A). By contrast, we could not collect the top fraction from wild-type leaves (used as a negative control), which have few LDs. We labeled the fraction from the hise1 mutant as isolated leaf LDs. For Co-IP analysis with anti-GFP antibodies, we used a transgenic A. thaliana hise1-2 mutant expressing a LDAP3-GFP fusion under control of the LDAP3 promoter (hise1-2/pLDAP3:LDAP3-GFP plants). Fluorescence microscopy revealed that LDAP3-GFP was localized to LDs in epidermal cells of hise1-2 leaves (Figure 1B). Transformants expressing cytosolic GFP (Mano et al., 2002) were used as a negative control because cytosolic GFP does not fuse with LDs. Because we detected LDAP3-GFP in the co-immunoprecipitants from hise1-2/pLDAP3:LDAP3-GFP plants (Figure 1C), we labeled the co-immunoprecipitants as isolated leaf LDs. Silver staining revealed that the isolated leaf LDs contained a lot of protein (Figures 1D, E). We detected 3,206 proteins in the isolated LDs, but not in the negative controls, using mass spectrometry (Supplementary Table S1). We defined these as candidate leaf LD proteins (Supplementary Table S1, LD candidates = Yes).
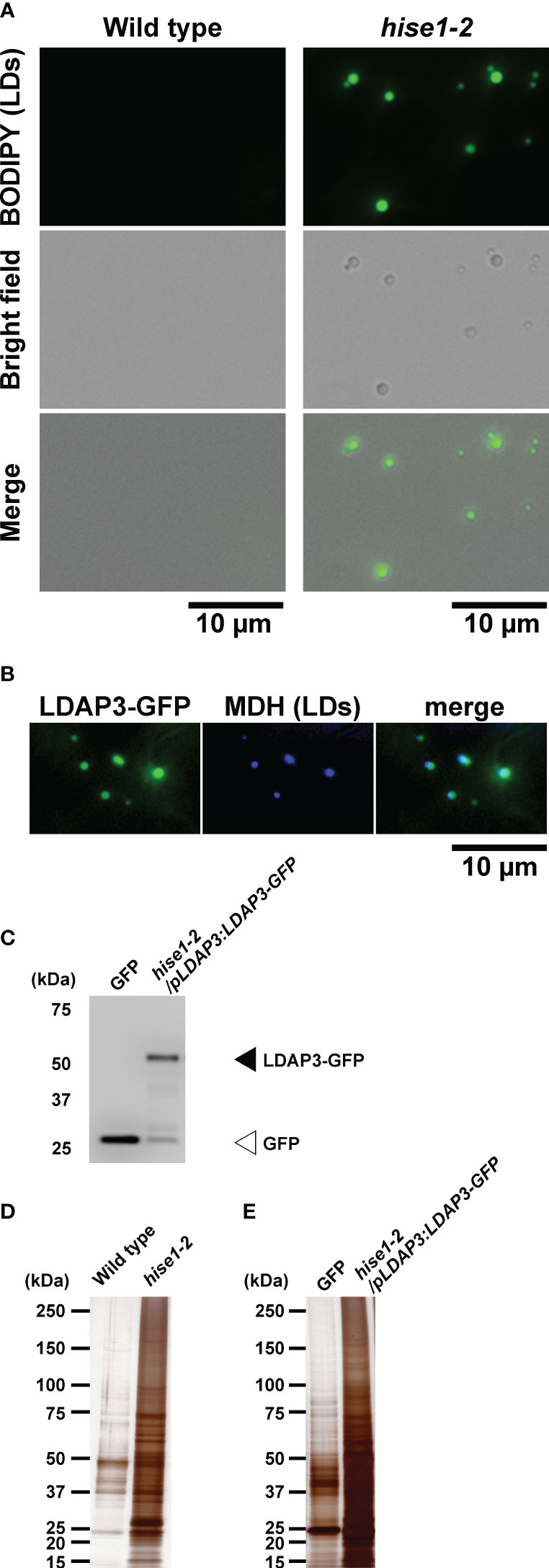
Figure 1 Isolation of leaf LDs from A thaliana leaves using centrifuge and co-immunoprecipitation (Co-IP) methods. (A) BODIPY fluorescence and brightfield images of LDs isolated from the wild type and hise1-2 mutant using a centrifuge method and stained with BODIPY 493/503. (B) Fluorescence images of LDAP3-GFP and MDH (LDs) in epidermal cells of hise1-2/pLDAP3:LDAP3-GFP plants. (C) Immunoblot analysis of the co-immunoprecipitants of hise1-2/pLDAP3:LDAP3-GFP plants, and wild-type plants expressing cytosolic GFP alone, with anti-GFP antibody. (D, E) Silver staining of the co-immunoprecipitants from the centrifuge method (D) and the Co-IP method (E).
We selected 31 proteins related to lipid metabolism or ER function from the leaf LD protein candidates. To investigate their subcellular localization, we transiently expressed each candidate protein fused with GFP or red fluorescent protein (RFP) in leaves of Nicotiana benthamiana. We induced leaves to form LDs with mevalonate treatment and visualized them using α-DOX1-RFP or BODIPY 493/503 staining. MYOB14-GFP fluorescence was associated with punctate structures (Figure 2A). The punctate structures co-localized with α-DOX1-RFP (Figure 2A), indicating that MYOB14-GFP was localized to LDs. Two uncharacterized proteins (encoded by AT3G23510 and AT1G30130) also co-localized with α-DOX1-RFP, indicating LD localization (Figures 2B, C). A domain search using InterProScan (Paysan-Lafosse et al., 2023) revealed that AT3G23510 encodes a protein classified as cyclopropane-fatty-acyl-phospholipid synthase, and AT1G30130 encodes a protein that contains domain of unknown function 1365 (DUF1365). To our knowledge, this is the first report describing LD localization of these three proteins. The other 28 proteins were not localized to LDs (Supplementary Figure S1).
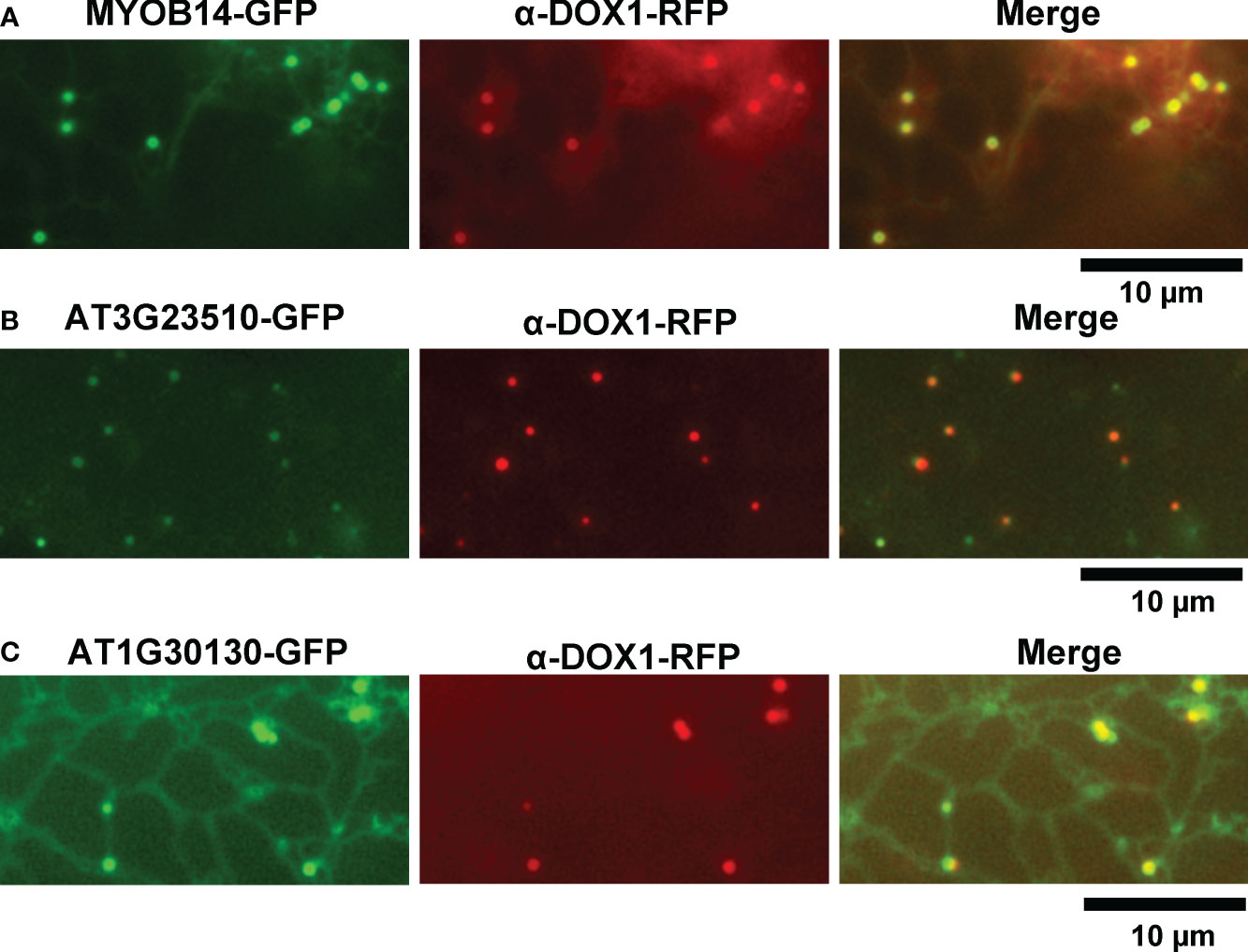
Figure 2 Subcellular localization of MYOB14, AT3G23510, and AT1G30130. (A–C) Fluorescence images of N. benthamiana leaves transiently expressing MYOB14-GFP (A), AT3G23510-GFP (B), or AT1G30130-GFP (C) along with α-DOX1-RFP (LD marker).
MYOBs interact with myosin through the DUF593 domain (Peremyslov et al., 2013). A. thaliana contains 16 MYOBs (Peremyslov et al., 2013; Perico et al., 2021), but their LD localization is unknown. To investigate the LD localization of A. thaliana MYOBs, we fused each of the 14 MYOBs with GFP and transiently expressed them in N. benthamiana leaves. The fluorescence of MYOB1-GFP, MYOB2-GFP, MYOB3-GFP, and MYOB5-GFP co-localized with that of α-DOX1-RFP (Figure 3A), while the other MYOBs did not appear to localize to LDs (Figure 3B). Our results indicate that MYOB1, MYOB2, MYOB3, and MYOB5 also localize to LDs.
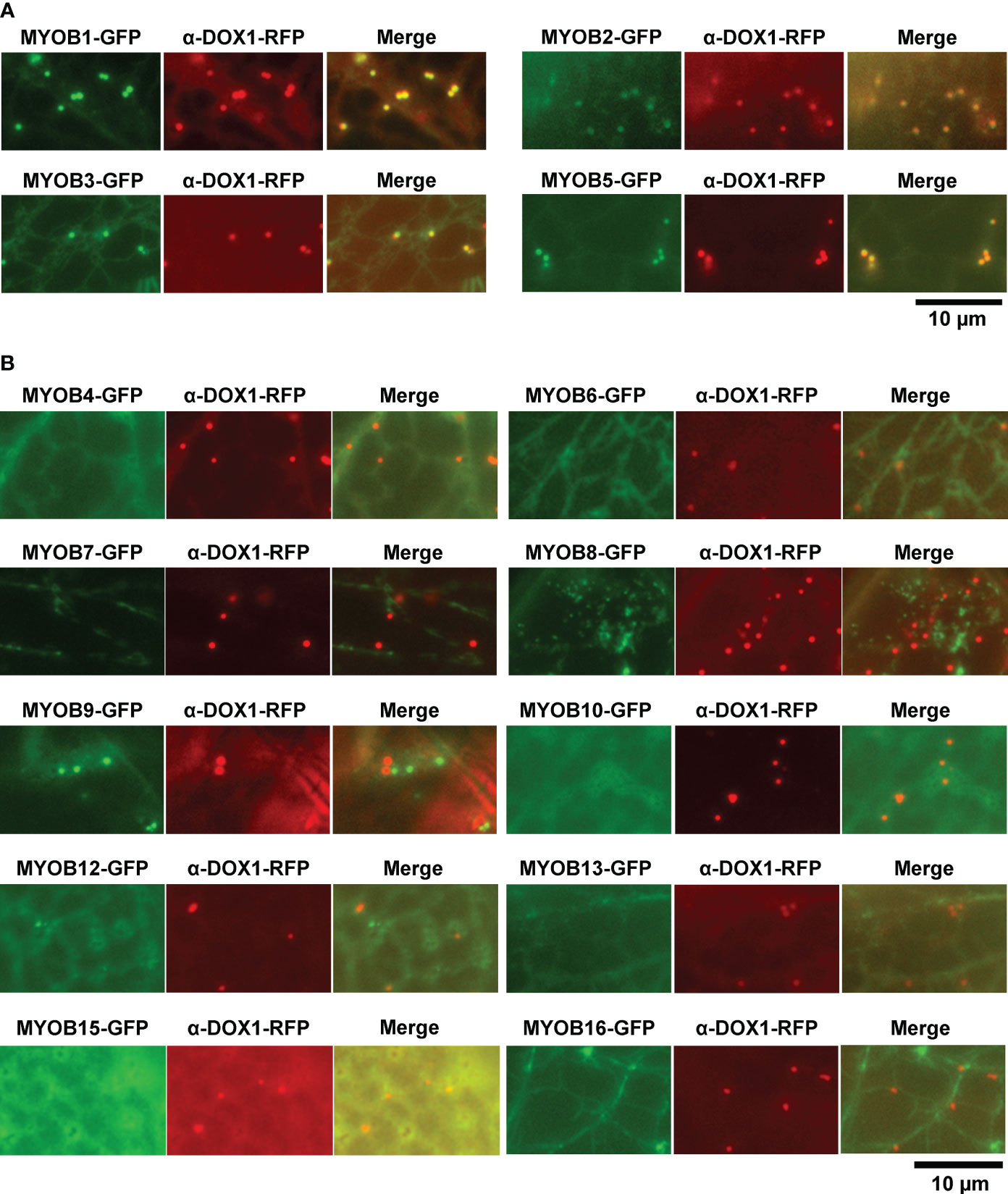
Figure 3 Subcellular localization of MYOB1, MYOB2, MYOB3, and MYOB5. (A) Fluorescence images of N. benthamiana leaves transiently expressing MYOB1-GFP, MYOB2-GFP, MYOB3-GFP, or MYOB5-GFP along with α-DOX1-RFP. (B) Fluorescence images of N. benthamiana leaves transiently co-expressing α-DOX1-RFP and the other MYOB proteins tested in this study.
Myosin functions in the movement of organelles in association with actin filaments. MYOBs work together with myosin to regulate organelle movement (Peremyslov et al., 2015; Perico et al., 2021). To determine if there is a relationship between LD movement and actin filaments, we used A. thaliana plants (wild-type background) producing Lifeact–Venus, which localizes to actin filaments (Era et al., 2009). After inducing LDs through treatment with mevalonic acid, we co-visualized LDs and actin filaments in the epidermal cells of true leaves stained with BODIPY 493/503. LDs moved along actin filaments and sometimes paused (Figure 4A; Supplementary Movie S1).
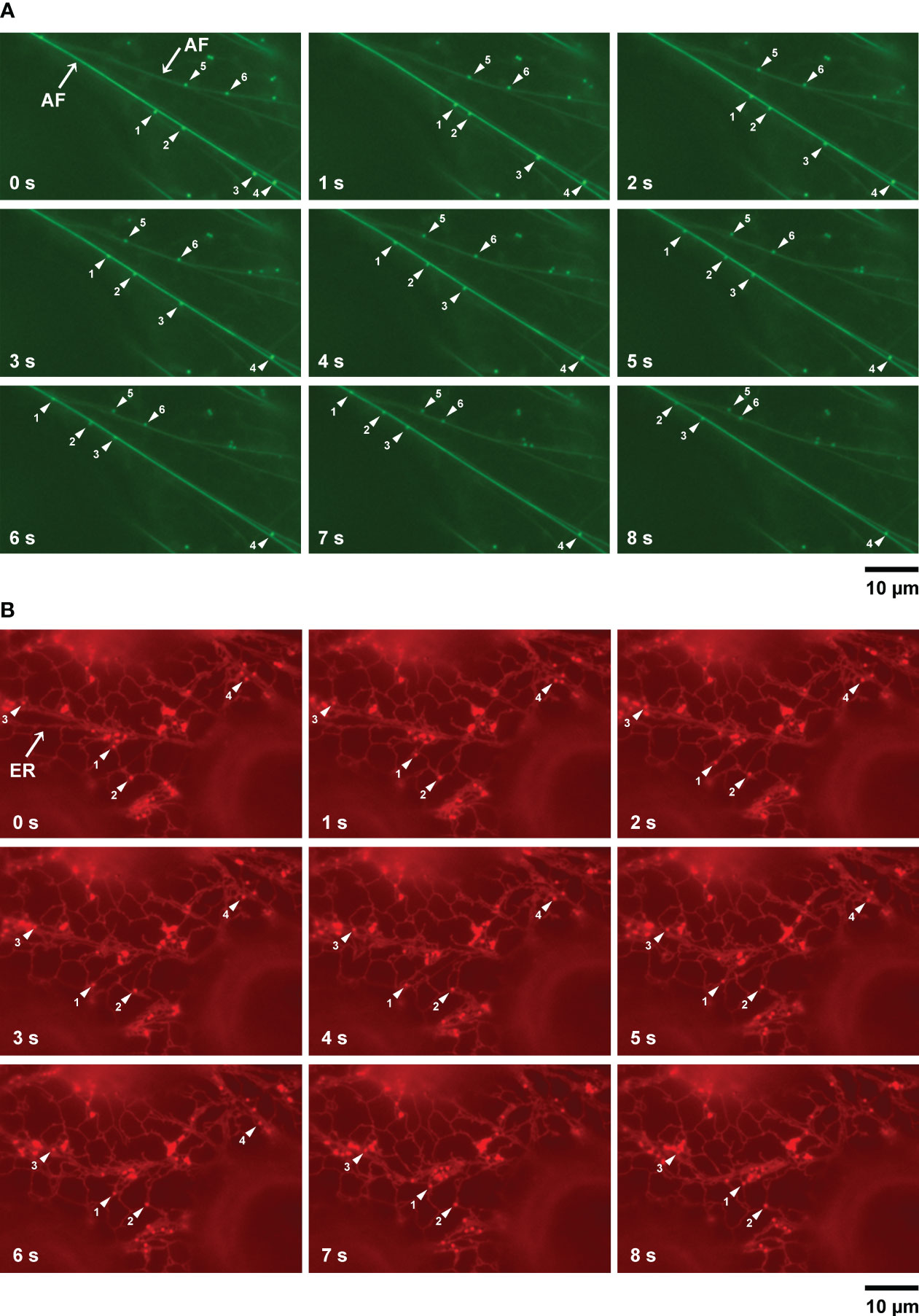
Figure 4 LD movement on actin filaments and the ER captured by time-lapse imaging. (A) Fluorescence images of epidermal cells of true leaves of 2-week-old A thaliana plants expressing Lifeact–Venus and stained with BODIPY 493/503 after inducing LDs with mevalonic acid. Lifeact–Venus, actin filament (filamented structures); BODIPY 493/503, LDs (punctate structures). Arrowheads show LDs (same numbers indicate same LDs). (B) Fluorescence images of epidermal cells of true leaves of N. benthamiana transiently co-expressing α-DOX1-RFP (LDs, punctate structures) and mCherry-HDEL (ER, network structures). Arrowheads show LDs (same numbers indicate same LDs).
ER morphology is involved in actin filament organization (Ueda et al., 2010). To observe the relationship between LD movement and the ER, we co-visualized LDs and the ER in epidermal cells of true leaves of N. benthamiana carrying the LD marker α-DOX1-RFP and the ER marker mCherry-HDEL, after induction of LDs using mevalonic acid. LDs moved together with the ER and sometimes paused, similar to their movement on actin filaments (Figure 4B; Supplementary Movie S2). In addition, we co-visualized LDs and the ER in epidermal cells of true leaves of N. benthamiana carrying the LD marker α-DOX1-GFP and the ER marker mCherry-HDEL, after induction of LDs with mevalonic acid. Almost all LDs co-localized with the ER (Supplementary Figure S2). These results suggest that LDs move along actin filaments together with the ER.
Myosin XIK is a major myosin XI in A. thaliana and interacts with MYOB1, MYOB2, and MYOB3 (Peremyslov et al., 2013; Kurth et al., 2017); the DUF593 domain of MYOBs binds to the globular tail domain of myosin. However, the proteins interacting with MYOB14 are unclear, and there are no interactome data using LD-localized MYOBs. We therefore performed Co-IP using MYOB2 or MYOB14 fused with GFP (MYOB2-GFP and MYOB14-GFP). We generated transgenic A. thaliana plants expressing MYOB2-GFP or MYOB14-GFP (Supplementary Figures S3A, B). Co-immunoprecipitants were prepared from leaves of the transgenic A. thaliana plants using anti-GFP antibodies (Supplementary Figures S3C, D). We performed mass spectrometry analysis of the co-immunoprecipitants and identified candidate proteins that interact with MYOB2 and MYOB14, including myosin XIK (Supplementary Tables S2, S3). In addition, we identified some candidates of interactors with MYOB2 of MYOB14, suggesting that MYOB2 and MYOB14 interact with myosin XIK and other proteins.
To investigate whether myosin XIK localizes to LDs, we co-expressed the GTD domain of myosin XIK (XIKGTD) fused with GFP and the LD marker α-DOX1-RFP in N. benthamiana. We then observed fluorescence of GFP-XIKGTD and α-DOX1-RFP in epidermal cells of true leaves after induction of LDs with mevalonic acid. GFP-XIKGTD fluorescence was observed in the cytosol and did not co-localize with LDs (Supplementary Figure S4A). In addition, we observed hypocotyls of A. thaliana XIK-YFP plants (Peremyslov et al., 2012) following the induction of LD formation with mevalonic acid. This YFP-tagged myosin XIK is previously shown to be a functional myosin (Peremyslov et al., 2012; Okamoto et al., 2015). Fluorescence microscopy showed XIK-YFP signals in a filamentous pattern (Supplementary Figure S4B), as previously observed (Peremyslov et al., 2012). The LDs co-localized with the filamentous XIK-YFP signals. Notably, however, the XIK-YFP fluorescence signals did not resemble LD-like punctate structures (Supplementary Figure S4B), although MYOB1, MYOB2, MYOB3, and MYOB5 localized to LDs (Figures 2A, 3A). These results suggest that binding between myosin XIK and LD-localized MYOBs may occur only at the contact sites between LDs and actin filaments.
There are currently no reports on the functions of the two proteins encoded by AT3G23510 and AT1G30130. To characterize their functions, we performed homology searches using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI). Our similarity searches revealed that amino acid sequences of the two proteins were highly similar to those of enzymes involved in furan-containing fatty acid (FU-FA) biosynthesis (Figure 5A) in the photosynthetic bacterium Cereibacter sphaeroides (Lemke et al., 2020). The amino acid sequence identity of AT3G23510 and UfaM was 34.26%, that of AT3G23510 and UfaD was 38.50%, and that of AT1G30130 and UfaO was 26.42% using UniProt (The Uniprot, 2017) (Figures 5B–D). Additionally, our similarity searches revealed that the amino acid sequence of FufM was highly similar to that of AT4G33110, with 37.87% amino acid sequence identity (The Uniprot, 2017) (Figure 5E). AT4G33110 (named LD METHYLTRANSFERASE1, LIME1) is classified as an S-adenosyl-L-methionine-dependent methyltransferase superfamily protein and localizes to LDs (Kretzschmar et al., 2020). We confirmed that LIME1-GFP localized to LDs (Supplementary Figure S5). These results suggest that the enzymes for FU-FA biosynthesis exist in A. thaliana and localize to LDs. We named AT3G23510, AT1G30130, and AT4G33110 as AtUFAMD1, AtUFAO1, and AtFUFM1/LIME1, respectively.
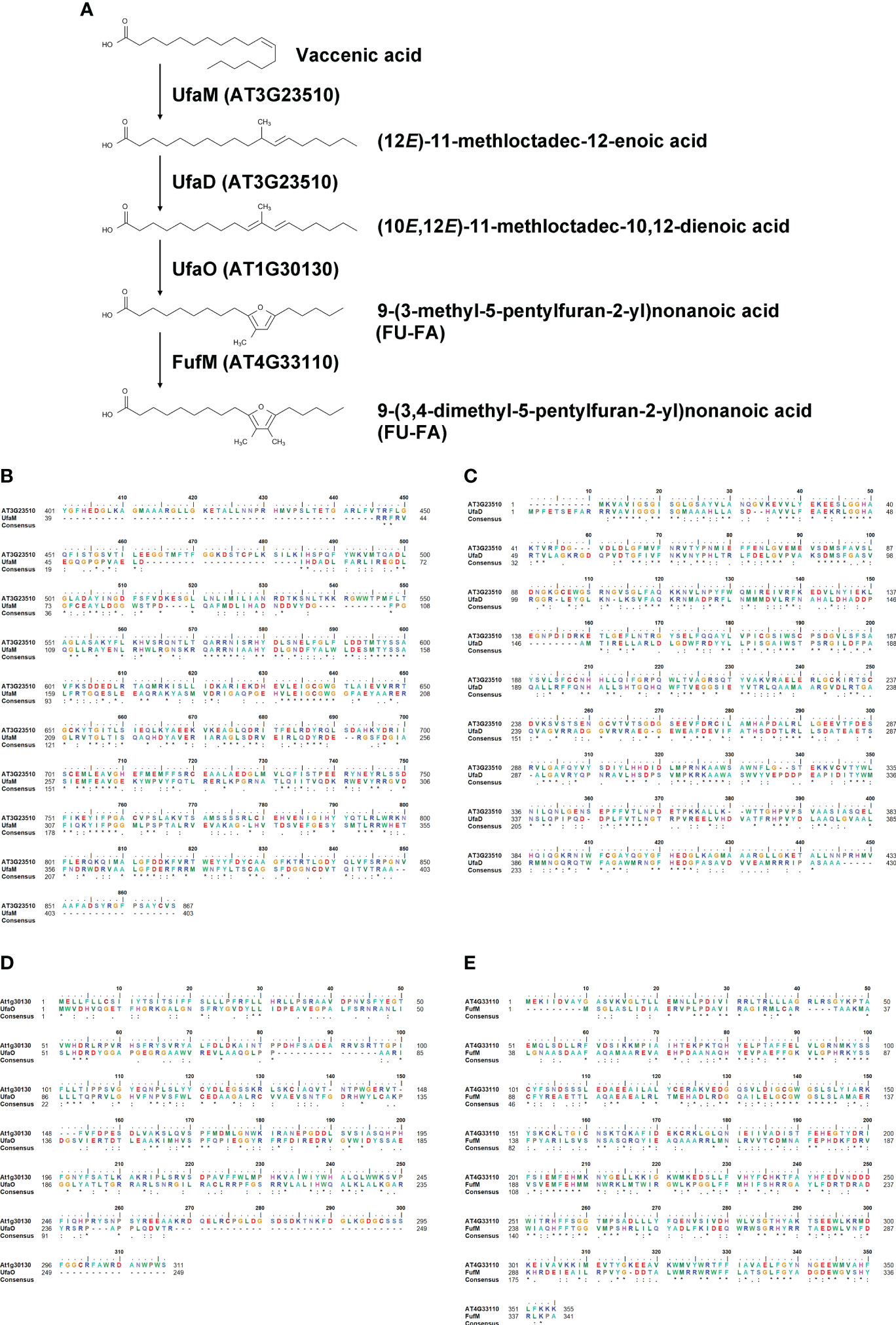
Figure 5 Homology search of LD-associated proteins AT3G23510, AT1G30130, and AT4G33110. (A) A furan-containing fatty acid biosynthesis pathway. (B–E) Amino acid sequence alignments between A. thaliana and C. sphaeroides: AT3G23510 and UfaM (B); AT3G23510 and UfaD (C); AT1G30130 and UfaO (D); and AT4G33110 and FufM (E).
In this study, we performed LD proteomics to identify leaf LD proteins. Among 3,206 candidate leaf LD proteins, we identified MYOB14, AT3G23510, and AT1G30130, which localize to LDs. Since LD localization of MYOB14, AT3G23510, and AT1G30130 has not been reported previously, our proteome data may include many unidentified LD-associated proteins. Our LD proteomics method and the resulting data are therefore useful for investigating leaf LD proteins.
LD movement is mediated by myosin in animals and plants (Pfisterer et al., 2017; Veerabagu et al., 2020; Han et al., 2023). MYOBs have a plant-specific DUF593 (Holding et al., 2007) and are involved in the movement of organelles in plant cells (Peremyslov et al., 2013; Peremyslov et al., 2015; Perico et al., 2021). However, the LD localization of MYOBs is unknown. MYOB1 and MYOB2 were previously reported to localize to motile vesicles (Peremyslov et al., 2013), and MYOB14 was reported to localize to uncharacterized compartments (Kurth et al., 2017). Here, we determined that MYOB1, MYOB2, MYOB3, MYOB5, and MYOB14 localize to LDs in A. thaliana, indicating that these proteins may be involved in LD movement. Our microscopy observations revealed that LDs move along actin filaments, suggesting that this movement is mediated by myosin and MYOBs (Figure 6). In addition, we observed that LDs move together with the ER. Organelle dynamics of the ER are closely related to those of actin filaments (Ueda et al., 2010). Our data suggest that LD movement along actin filaments is mediated by myosin XI and LD-localized MYOBs.
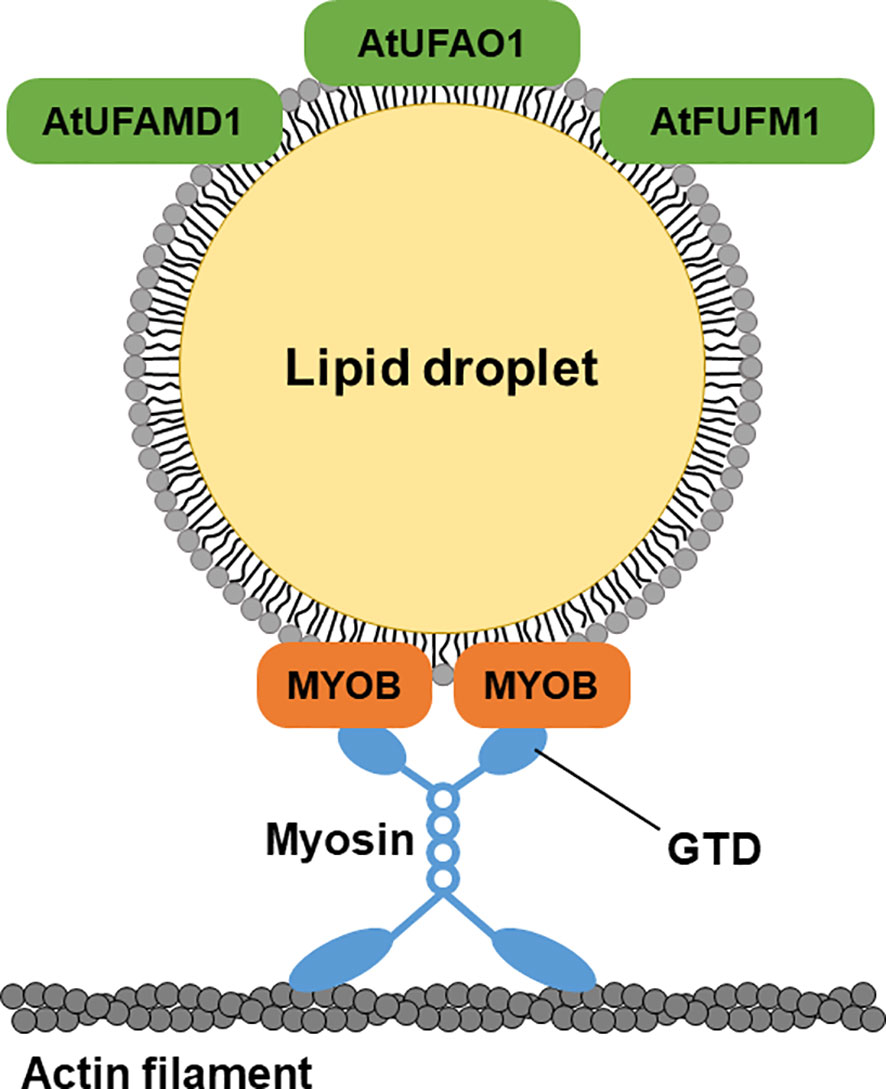
Figure 6 A hypothetical model of LD-localized proteins. LD-localized MYOBs for LD movement and LD-localized enzymes for furan-containing fatty acid biosynthesis.
A. thaliana has 16 MYOBs (Peremyslov et al., 2013; Perico et al., 2021), and we determined that 5 of these localize to LDs, suggesting that there might be various LD-localized MYOBs in plant cells. LD localization of MYOBs may play important roles in plant physiology. MYOBs are characterized into three groups (Peremyslov et al., 2013): MYOB1, MYOB2, MYOB3, and MYOB5 are in group I, and MYOB14 is in group III. Myosin XIK interacts with MYOB1, MYOB2, MYOB3, MYOB5, and MYOB14 in A. thaliana (Peremyslov et al., 2013; Kurth et al., 2017). We detected myosin XIK in our Co-IP assays using MYOB2-GFP (Supplementary Table S2) and MYOB14-GFP (Supplementary Table S3) as well as in our LD proteomics data (Supplementary Table S1). Thus, LD-localized MYOBs may function with myosin XIK in LD movement.
UfaM, UfaD, UfaO, and FufM are enzymes involved in FU-FA biosynthesis in the photosynthetic bacterium C. sphaeroides (Lemke et al., 2014; Lemke et al., 2020; Yu and Shanklin, 2020). A. thaliana contains three proteins whose amino acid sequences are highly similar to those of the bacterial enzymes for FU-FA biosynthesis: AT3G23510 (AtUFAMD1), AT1G30130 (AtUFAO1), and AT4G33110 (AtFUFM1/LIME1). These proteins localize to LDs (Figure 6). FU-FAs have been detected in Hevea brasiliensis latex (Liengprayoon et al., 2011) and in soybean (Glycine max) and soy products (Müller et al., 2020); however, FU-FA biosynthesis in plants is largely unknown. Our findings indicate that FU-FA biosynthesis may occur in LDs. It will be interesting to investigate whether AtUFAMD1, AtUFAO1, and AtFUFM1/LIME1 directly function in FU-FA biosynthesis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YO: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Visualization, Writing – original draft. RS: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. EM-S: Data curation, Formal analysis, Methodology, Writing – original draft. KK: Data curation, Formal analysis, Methodology, Writing – original draft. HU: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Writing – original draft. IH-N: Conceptualization, Data curation, Formal analysis, Project administration, Supervision, Writing – original draft. TS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) to TS (nos. 19K05809 and 23K17986), HU (nos. 19K06732 and 18H05496), and IH-N (no. 15H05776); Kato Memorial Bioscience Foundation (to TS); a SUNBOR GRANT from the Suntory Foundation for Life Sciences (to TS); Takeda Science Foundation (to TS); a research grant from The Yanmar Environmental Sustainability Support Association to TS; Inamori Foundation (to TS); JGC-S Scholarship Foundation (to TS); the Leading Initiative for Excellent Young Researchers (LEADER) from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT) to TS (no. J16HJ00026); the Phytochemical Plant Molecular Science of Strategic Priority Research Promotion Program from Chiba University (to TS and KS); and the Hirao Taro Foundation of KONAN GAKUEN for Academic Research (to HU and IH-N).
We thank Shoji Mano (National Institute for Basic Biology), Shoji Segami (National Institute for Basic Biology), Tsuyoshi Nakagawa (Shimane University), and Sumie Ishiguro (Nagoya University) for donating the vectors; Takashi Ueda (National Institute for Basic Biology) for donating the Lifeact–Venus plants; Andreas Nebenführ (University of Tennessee) for donating mCherry-HDEL; Valerian V. Dolja (Oregon State University) for donating XIK-YFP plants; Shoji Mano for donating GFP-expressing A. thaliana; and the Arabidopsis Biological Resource Center for providing the seeds of the A. thaliana T-DNA insertion mutant.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1331479/full#supplementary-material
Supplementary Figure 1 | Fluorescence images of N. benthamiana leaves transiently co-expressing α-DOX1-RFP and the GFP-fused candidate proteins.
Supplementary Figure 2 | Fluorescence images of epidermal cells of true leaves of N. benthamiana transiently co-expressing α-DOX1-GFP (LDs, punctate structures) and mCherry-HDEL (ER, network structures).
Supplementary Figure 3 | Co-immunoprecipitation using transgenic A. thaliana expressing MYOB2-GFP or MYOB14-GFP. (A, B) Fluorescence images of MYOB2-GFP (A) and MYOB14-GFP (B) in epidermal cells of true leaves of 2-week-old A. thaliana plants. Arrowheads indicate LDs. (C, D) Immunoblot analysis of the co-immunoprecipitates of MYOB2-GFP (C) and MYOB14-GFP (D) plants using anti-GFP antibody.
Supplementary Figure 4 | Subcellular localization of myosin XIK and LDs. (A) Fluorescence images of N. benthamiana leaves transiently expressing GFP-XIKGTD along with α-DOX1-RFP. (B) Fluorescence images of myosin XIK-YFP and MDH (LDs) in the hypocotyls of XIK-YFP plants following the induction of LD formation with mevalonic acid.
Supplementary Figure 5 | Subcellular localization of AT4G33110. Fluorescence images of N. benthamiana leaves transiently expressing AT4G33110-GFP along with α-DOX1-RFP.
Supplemental Movie S1 | Fluorescence in Lifeact–Venus plants (for visualizing actin filaments) stained with BODIPY 493/503 for visualizing LDs.
Supplemental Movie S2 | Fluorescence in N. benthamiana leaves transiently co-expressing α-DOX1-RFP for LD visualizing and mCherry-HDEL for ER visualizing.
Bose, D., Ngo, A. H., Nguyen, V. C., Nakamura, Y. (2021). Non-specific phospholipases C2 and C6 redundantly function in pollen tube growth via triacylglycerol production in Arabidopsis. Plant J. 106, 409–418. doi: 10.1111/tpj.15172
Brocard, L., Immel, F., Coulon, D., Esnay, N., Tuphile, K., Pascal, S., et al. (2017). Proteomic analysis of lipid droplets from arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 8, 894. doi: 10.3389/fpls.2017.00894
Chapman, K. D., Dyer, J. M., Mullen, R. T. (2012). Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J. Lipid Res. 53, 215–226. doi: 10.1194/jlr.R021436
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Era, A., Tominaga, M., Ebine, K., Awai, C., Saito, C., Ishizaki, K., et al. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50, 1041–1048. doi: 10.1093/pcp/pcp055
Fernandez-Santos, R., Izquierdo, Y., Lopez, A., Muniz, L., Martinez, M., Cascon, T., et al. (2020). Protein Profiles of Lipid Droplets during the Hypersensitive Defense Response of Arabidopsis against Pseudomonas Infection. Plant Cell Physiol. 61, 1144–1157. doi: 10.1093/pcp/pcaa041
Gidda, S. K., Park, S., Pyc, M., Yurchenko, O., Cai, Y., Wu, P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. doi: 10.1104/pp.15.01977
Gidda, S. K., Watt, S., Collins-Silva, J., Kilaru, A., Arondel, V., Yurchenko, O., et al. (2013). Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal Behav. 8, e27141. doi: 10.4161/psb.27141
Grimberg, A., Carlsson, A. S., Marttila, S., Bhalerao, R., Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15, 192. doi: 10.1186/s12870-015-0579-1
Han, L., Huang, D., Wu, S., Liu, S., Wang, C., Sheng, Y., et al. (2023). Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. Nat. Cell Biol. 25, 1033–1046. doi: 10.1038/s41556-023-01162-4
Holding, D. R., Otegui, M. S., Li, B., Meeley, R. B., Dam, T., Hunter, B. G., et al. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19, 2569–2582. doi: 10.1105/tpc.107.053538
Horn, P. J., James, C. N., Gidda, S. K., Kilaru, A., Dyer, J. M., Mullen, R. T., et al. (2013). Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162, 1926–1936. doi: 10.1104/pp.113.222455
James, C. N., Horn, P. J., Case, C. R., Gidda, S. K., Zhang, D., Mullen, R. T., et al. (2010). Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. U.S.A. 107, 17833–17838. doi: 10.1073/pnas.0911359107
Kano, K., Noda, S., Sato, S., Kuwata, K., Mishiro-Sato, E. (2023). An efficient in-gel digestion method on small amounts of protein sample from large intact gel pieces. Separation Sci. Plus 6. doi: 10.1002/sscp.202200121
Kim, H. U., Lee, K. R., Go, Y. S., Jung, J. H., Suh, M. C., Kim, J. B. (2011). Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol. 52, 983–993. doi: 10.1093/pcp/pcr051
Kim, E. Y., Park, K. Y., Seo, Y. S., Kim, W. T. (2016). Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170, 2494–2510. doi: 10.1104/pp.16.00165
Kretzschmar, F. K., Doner, N. M., Krawczyk, H. E., Scholz, P., Schmitt, K., Valerius, O., et al. (2020). Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol. 182, 1326–1345. doi: 10.1104/pp.19.01255
Kurth, E. G., Peremyslov, V. V., Turner, H. L., Makarova, K. S., Iranzo, J., Mekhedov, S. L., et al. (2017). Myosin-driven transport network in plants. Proc. Natl. Acad. Sci. U.S.A. 114, E1385–E1394. doi: 10.1073/pnas.1620577114
Lara, J. A., Burciaga-Monge, A., Chavez, A., Reves, M., Lavilla, R., Arro, M., et al. (2018). Identification and characterization of sterol acyltransferases responsible for steryl ester biosynthesis in tomato. Front. Plant Sci. 9, 588. doi: 10.3389/fpls.2018.00588
Lemke, R., Olson, S. M., Morse, K., Karlen, S. D., Higbee, A., Beebe, E. T., et al. (2020). A bacterial biosynthetic pathway for methylated furan fatty acids. J. Biol. Chem. 295, 9786–9801. doi: 10.1074/jbc.RA120.013697
Lemke, R. A., Peterson, A. C., Ziegelhoffer, E. C., Westphall, M. S., Tjellstrom, H., Coon, J. J., et al. (2014). Synthesis and scavenging role of furan fatty acids. Proc. Natl. Acad. Sci. U.S.A. 111, E3450–E3457. doi: 10.1073/pnas.1405520111
Liengprayoon, S., Sriroth, K., Dubreucq, E., Vaysse, L. (2011). Glycolipid composition of Hevea brasiliensis latex. Phytochemistry 72, 1902–1913. doi: 10.1016/j.phytochem.2011.04.023
Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M., Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43, 331–341. doi: 10.1093/pcp/pcf037
Müller, F., Hogg, M., Vetter, W. (2020). Valuable furan fatty acids in soybeans and soy products. Eur. Food Res. Technol. 246, 1383–1392. doi: 10.1007/s00217-020-03497-w
Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. doi: 10.1271/bbb.70216
Nelson, B. K., Cai, X., Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Okamoto, K., Ueda, H., Shimada, T., Tamura, K., Kato, T., Tasaka, M., et al. (2015). Regulation of organ straightening and plant posture by an actin-myosin XI cytoskeleton. Nat. Plants 1, 15031. doi: 10.1038/nplants.2015.31
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. doi: 10.1093/nar/gkac993
Peremyslov, V. V., Cole, R. A., Fowler, J. E., Dolja, V. V. (2015). Myosin-powered membrane compartment drives cytoplasmic streaming, cell expansion and plant development. PloS One 10, e0139331. doi: 10.1371/journal.pone.0139331
Peremyslov, V. V., Klocko, A. L., Fowler, J. E., Dolja, V. V. (2012). Arabidopsis myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Front. Plant Sci. 3, 184. doi: 10.3389/fpls.2012.00184
Peremyslov, V. V., Morgun, E. A., Kurth, E. G., Makarova, K. S., Koonin, E. V., Dolja, V. V. (2013). Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25, 3022–3038. doi: 10.1105/tpc.113.113704
Perico, C., Gao, H., Heesom, K. J., Botchway, S. W., Sparkes, I. A. (2021). Arabidopsis thaliana myosin XIK is recruited to the Golgi through interaction with a MyoB receptor. Commun. Biol. 4, 1182. doi: 10.1038/s42003-021-02700-2
Pfisterer, S. G., Gateva, G., Horvath, P., Pirhonen, J., Salo, V. T., Karhinen, L., et al. (2017). Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat. Commun. 8, 14858. doi: 10.1038/ncomms14858
Pyc, M., Cai, Y., Gidda, S. K., Yurchenko, O., Park, S., Kretzschmar, F. K., et al. (2017). Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92, 1182–1201. doi: 10.1111/tpj.13754
Segami, S., Makino, S., Miyake, A., Asaoka, M., Maeshima, M. (2014). Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26, 3416–3434. doi: 10.1105/tpc.114.127571
Shimada, T. L., Shimada, T., Okazaki, Y., Higashi, Y., Saito, K., Kuwata, K., et al. (2019). HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat. Plants 5, 1154–1166. doi: 10.1038/s41477-019-0537-2
Shimada, T. L., Shimada, T., Takahashi, H., Fukao, Y., Hara-Nishimura, I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55, 798–809. doi: 10.1111/j.1365-313X.2008.03553.x
Shimada, T. L., Takano, Y., Shimada, T., Fujiwara, M., Fukao, Y., Mori, M., et al. (2014). Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 164, 105–118. doi: 10.1104/pp.113.230185
Shimojima, M., Madoka, Y., Fujiwara, R., Murakawa, M., Yoshitake, Y., Ikeda, K., et al. (2015). An engineered lipid remodeling system using a galactolipid synthase promoter during phosphate starvation enhances oil accumulation in plants. Front. Plant Sci. 6, 664. doi: 10.3389/fpls.2015.00664
Shockey, J. M., Gidda, S. K., Chapital, D. C., Kuan, J. C., Dhanoa, P. K., Bland, J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. doi: 10.1105/tpc.106.043695
Siloto, R. M., Findlay, K., Lopez-Villalobos, A., Yeung, E. C., Nykiforuk, C. L., Moloney, M. M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18, 1961–1974. doi: 10.1105/tpc.106.041269
The Uniprot, C. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Ueda, H., Yokota, E., Kutsuna, N., Shimada, T., Tamura, K., Shimmen, T., et al. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6894–6899. doi: 10.1073/pnas.0911482107
Vanhercke, T., Belide, S., Taylor, M. C., El Tahchy, A., Okada, S., Rolland, V., et al. (2019). Up-regulation of lipid biosynthesis increases the oil content in leaves of Sorghum bicolor. Plant Biotechnol. J. 17, 220–232. doi: 10.1111/pbi.12959
Veerabagu, M., K Paul, L., Rinne, P. L., van der Schoot, C. (2020). Plant lipid bodies traffic on actin to plasmodesmata motorized by myosin XIs. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21041422
Yang, L., Ding, Y., Chen, Y., Zhang, S., Huo, C., Wang, Y., et al. (2012). The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53, 1245–1253. doi: 10.1194/jlr.R024117
Keywords: lipid droplet proteomics, myosin-binding protein, enzymes for furan-containing fatty acid biosynthesis, actin filament, endoplasmic reticulum, Arabidopsis thaliana
Citation: Omata Y, Sato R, Mishiro-Sato E, Kano K, Ueda H, Hara-Nishimura I and Shimada TL (2024) Lipid droplets in Arabidopsis thaliana leaves contain myosin-binding proteins and enzymes associated with furan-containing fatty acid biosynthesis. Front. Plant Sci. 15:1331479. doi: 10.3389/fpls.2024.1331479
Received: 01 November 2023; Accepted: 13 February 2024;
Published: 01 March 2024.
Edited by:
Andreas Nebenführ, The University of Tennessee, Knoxville, United StatesCopyright © 2024 Omata, Sato, Mishiro-Sato, Kano, Ueda, Hara-Nishimura and Shimada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi L. Shimada, dGxzaGltYWRhQGNoaWJhLXUuanA=
†Present address: Yuto Omata, Department of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.