- 1Laboratory of Cryptogamic Biota, Botanical Garden-Institute Far Eastern Branch of the Russian Academy of Sciences (FEB RAS), Vladivostok, Russia
- 2Department of Plant Ecology, Institute of Ecology and Biological Resources, Graduate University of Science and Technology, Vietnam Academy of Science and Technology, Ha Noi, Vietnam
- 3Team of National Ecosystem Survey, National Institute of Ecology, Seocheon, Republic of Korea
- 4Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
An analysis of the phylogeny of Cephaloziellaceae was carried out based on trees constructed for previously and newly obtained sequences of five genes: nuclear ITS1–2 and chloroplast trnL–F, trnG, rbcL, and psbA. Phylogenetic trees inferred from different genes are congruent for the main details; however, the position of several taxa is variable. As a result, a new phylogenetic system of the family was proposed. The narrow genus concept seems to be more appropriate for the family. Cephaloziella spinicaulis is segregated into the new genus Douiniella, the generic status for Prionolobus and Metacephalozia is confirmed, and the dubious generic status of Kymatocalyx is substantiated. The generic independence of Cylindrocolea from Cephaloziella s. str. is confirmed. The small amount of data hinders the description of two more genera from Cephaloziella s.l.
1 Introduction
Cephaloziellaceae plants are the smallest among higher plants. Despite sometimes not reaching a size of 1 mm, they are well developed and have reproductive organs. For a long time, this led to subjective difficulties in comprehending the taxonomy of the family: Cephaloziellaceae are commonly overlooked in field exploration, and when studied in the laboratory, morphological features that delimit species are often difficult to recognize. Despite microscopes now being much better than they were 100 years ago and before, these difficulties, to a certain extent, still challenge researchers who study this group of plants.
Montagne (1856) was the first to describe the genus Dichiton Mont. in the suite of currently recognized Cephaloziellaceae. The genus was forgotten for a long time and remembered when the generic name Cephaloziella (Spruce) Schiffn. became widely known. Moreover, the number of known species was so significant that, despite the earlier appearance, the name Cephaloziella was assigned nom. cons. with priority over Dichiton published 40 years before. The purposeful study of Cephaloziella began with Spruce (1882), who assigned the taxonomic status of this group of plants to Cephalozia subg. Cephaloziella Spruce and subg. Prionolobus Spruce (Spruce, 1882). After 11 years, this subgenus was elevated to the rank of genus by Schiffner (1893): Cephaloziella (Schiffner, 1893). Schiffner also recognized Prionolobus (Spruce) Schiffn., which is actually the evaluated Cephalozia subg. Prionolobus.
A remarkable new step in the knowledge of Cephaloziellaceae was made by Douin, who segregated this family (Douin, 1920) and perfectly described the features differentiating Cephaloziaceae and Cephaloziellaceae. In addition, he also accepted (described or re-evaluated) the following genera within Cephaloziellaceae: Dichiton, Lophoziella Douin (where the type species is the same as that of Dichiton), Prionolobus, Evansia Douin et Schiffn. (illegitimate name, later homonym of Evansia Salisb. ex Decne.), Cephaloziella and Protocephaloziella Douin. The next major contribution to the knowledge of Cephaloziellaceae taxonomy was made by Schuster (several works, with the fundamental one published in 1972) (Schuster, 1972). Schuster described the following genera: Amphicephalozia R.M.Schust., Cephalomitrion R.M.Schust., Cylindrocolea R.M.Schust., and Gymnocoleopsis (R.M.Schust.) R.M.Schust. The latest World Checklist of 2016 includes 171 accepted Cephaloziellaceae species names from 19 genera (Söderström et al., 2016).
All authors of the 19th and 20th centuries regarded Cephaloziella as closely related to Cephalozia (Dumort.) Dumort. However, recent molecular phylogenies (De Roo et al., 2007; Vilnet et al., 2012; Patzak et al., 2016; Bakalin et al., 2021a) have shown that these genera are not related; in contrast, Cephaloziellaceae were found to be genetically more similar to Lophoziaceae Cavers, Scapaniaceae Mig., and Anastrophyllaceae L.Söderstr., commonly showing morphologies unlike the Cephaloziellaceae “archetype” morphology.
Although several robust studies have been carried out, knowledge of intrafamilial taxonomy, ecology, and distribution of Cephaloziellaceae is limited compared with that of many other liverwort families. Moreover, it is noteworthy that the onset of the “molecular era” contributed relatively little to the creation of a clear taxonomic system for the family, and the main reason for this, as best we can infer, is still the same as 100 years ago: the small size of the plants and their common growth in mixtures (often as very scarce admixtures) with other bryophytes make it difficult to collect samples for molecular analysis. The new materials that we obtained during the study of mostly Indochinese liverworts allowed us to shed some light on the taxonomy of Cephaloziellaceae. Moreover, having initially become interested in the study of Cephaloziopsis (Spruce) Schiffn., as a result of our efforts, the system of Cephaloziellaceae was proposed to be revised on the basis of new molecular data as much as possible using the materials and data available to us. The presentation of the results obtained is the goal of this work.
2 Materials and methods
2.1 Taxon sampling
To compile our dataset for molecular phylogenetic analysis, species from the suborder Cephaloziineae Schljakov (Mamontov and Vilnet, 2017) were sampled. Species of Hygrobiella Spruce–Hygrobiella laxifolia (Hook.) Spruce (for rbcL and psbA), Hygrobiella nishimurae N.Kitag., and Hygrobiella squamosa Bakalin et Vilnet (for ITS1–2, trnL–F, and trnG) (Hygrobiellaceae and Jungermanniineae) were selected as an outgroup (Feldberg et al., 2013). In total, 141 species were included in the analysis, one to three species of Hygrobiella (depending on sequences availability) are in outgroups, and others are in ingroups (Supplementary Table 1); sequences of 15 species from the families Cephaloziellaceae and Hygrobiellaceae (5 ITS1–2, 15 trnL–F, 15 trnG, 7 rbcL, and 8 psbA) were obtained by the authors, and nucleotide data for 126 specimens were downloaded from the National Center for Biotechnology Information (NCBI) GenBank. DNA vouchers, including GenBank accession numbers and voucher details, are listed in Supplementary Table 1. The selection of genera involved in the study is identified by the goal of the paper described in the Introduction section (including a study of the taxonomic position of Cephaloziopsis exigua), as well as the presence of data on Cephaloziellaceae s. str. in the GenBank. All Cephaloziellaceae materials available in the GenBank for involved loci were used in this account preparation. Furthermore, in addition to Cephaloziellaceae in the narrow sense, we were involved in the study of related genera belonging to the same superclade Cephaloziellaceae–Scapaniaceae and forming the core of the phylogenetic system of the suborder Cephaloziineae. The involvement of a certain number of genera outside of Cephaloziellaceae s. str. made it possible to more reliably reveal phylogenetic relationships within Cephaloziellaceae and provide a more accurate circumscription of the family in a narrow sense.
2.2 DNA isolation, amplification, and sequencing
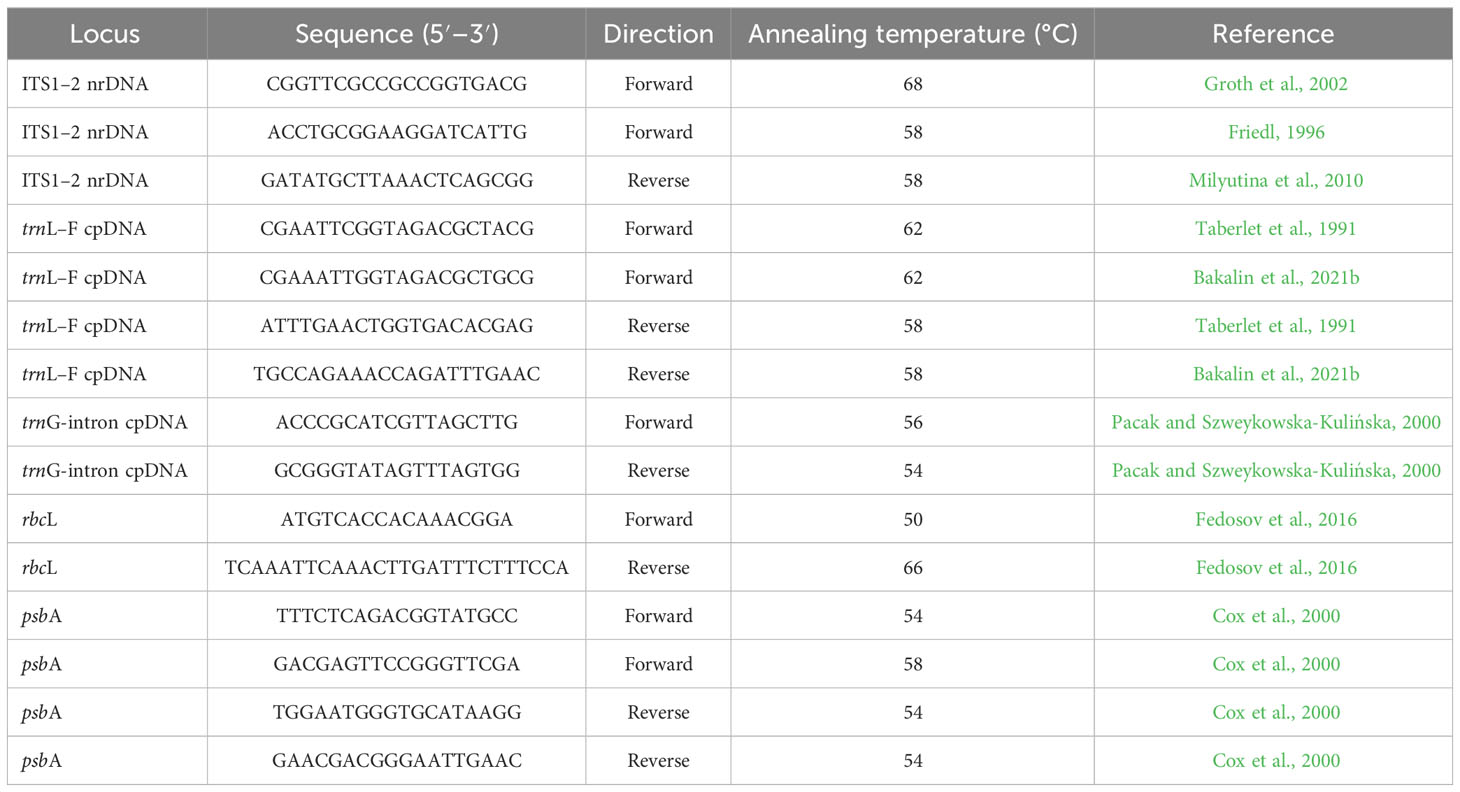
DNA was extracted from dried liverwort tissues using the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) and DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturers’ protocols. Amplification of ITS1–2, trnL–F, trnG-intron, rbcL, and psbA was performed using an Encyclo Plus PCR Kit (Evrogen, Moscow, Russia) with the primers listed in Table 1.
Polymerase chain reaction was performed in a total volume of 20 µl, including 1 µl of template DNA, 0.4 µl of Encyclo polymerase, 5 µl of Encyclo buffer, 0.4 µl of dNTP mixture (included in the Encyclo Plus PCR Kit), 13.4 µl (for trnL–F, trnG-intron, rbcL, and psbA)/12.4 µl (for ITS1–2) of double-distilled water (Evrogen, Moscow, Russia), 1 µl of dimethylsulfoxide (for ITS1–2), and 0.4 µl of each primer (forward and reverse, at a concentration of 5 pmol/µl).
Polymerase chain reactions were carried out using the following program:

Amplified fragments were visualized after electrophoresis on 1% agarose TAE gels by EthBr staining and purified using the Cleanup Mini Kit (Evrogen, Moscow, Russia). The DNA was sequenced using the ABI PRISM® BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) with further analysis of the reaction products following the standard protocol on an automatic sequencer (3730 DNA Analyzer, Applied Biosystems, USA) at the Genome Center (Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia).
2.3 Phylogenetic analyses
The datasets were produced for the ITS1–2, trnL–F, trnG-intron, rbcL, and psbA loci. All sequences were aligned using MAFFT (Katoh and Standley, 2013) with standard settings and then edited manually in BioEdit ver. 7.2.5 (Hall, 1999). All positions of the final alignments were included in the phylogenetic analyses. Phylogenies were reconstructed under two criteria: maximum likelihood (ML) with IQ-tree ver. 1.6.12 (Nguyen et al., 2015) and Bayesian inference (BI) with MrBayes ver. 3.2.7 (Ronquist et al., 2012).
For the ML analysis with 1,000 bootstrap replicates, the best fitting evolutionary models of nucleotide substitutions according to the BIC value were TIM+F+I+G4 for ITS1–2, HKY+F+G4 for trnL–F, K3Pu+F+G4 for trnG, TN+F+G4 for rbcL, and HKY+F+R2 for psbA as determined by ModelFinder (model-selection method implemented in IQ-tree) (Kalyaanamoorthy et al., 2017). Bootstrap support (BS) percentage values were calculated.
BI was performed by running two parallel analyses using the GTR+I+G model. For all datasets, the analysis consisted of four Markov chains run for five million generations, and trees were sampled every 500th generation. The first 2,500 trees in each run were discarded as burn-in; thereafter, 15,000 trees were sampled from both runs. Bayesian posterior probabilities (PPs) were calculated from the trees sampled after burn-in. The average standard deviation of split frequencies between the two runs was 0.0071 for ITS1–2, 0.0049 for trnL–F, 0.003 for trnG, 0.0047 for rbcL, and 0.003 for psbA.
3 Results
The infraspecific/generic and interspecific/generic variability (Table 2) of all five datasets were quantified as the average pairwise p-distances calculated in Mega XI (Tamura et al., 2021) using the pairwise deletion option for counting gaps.
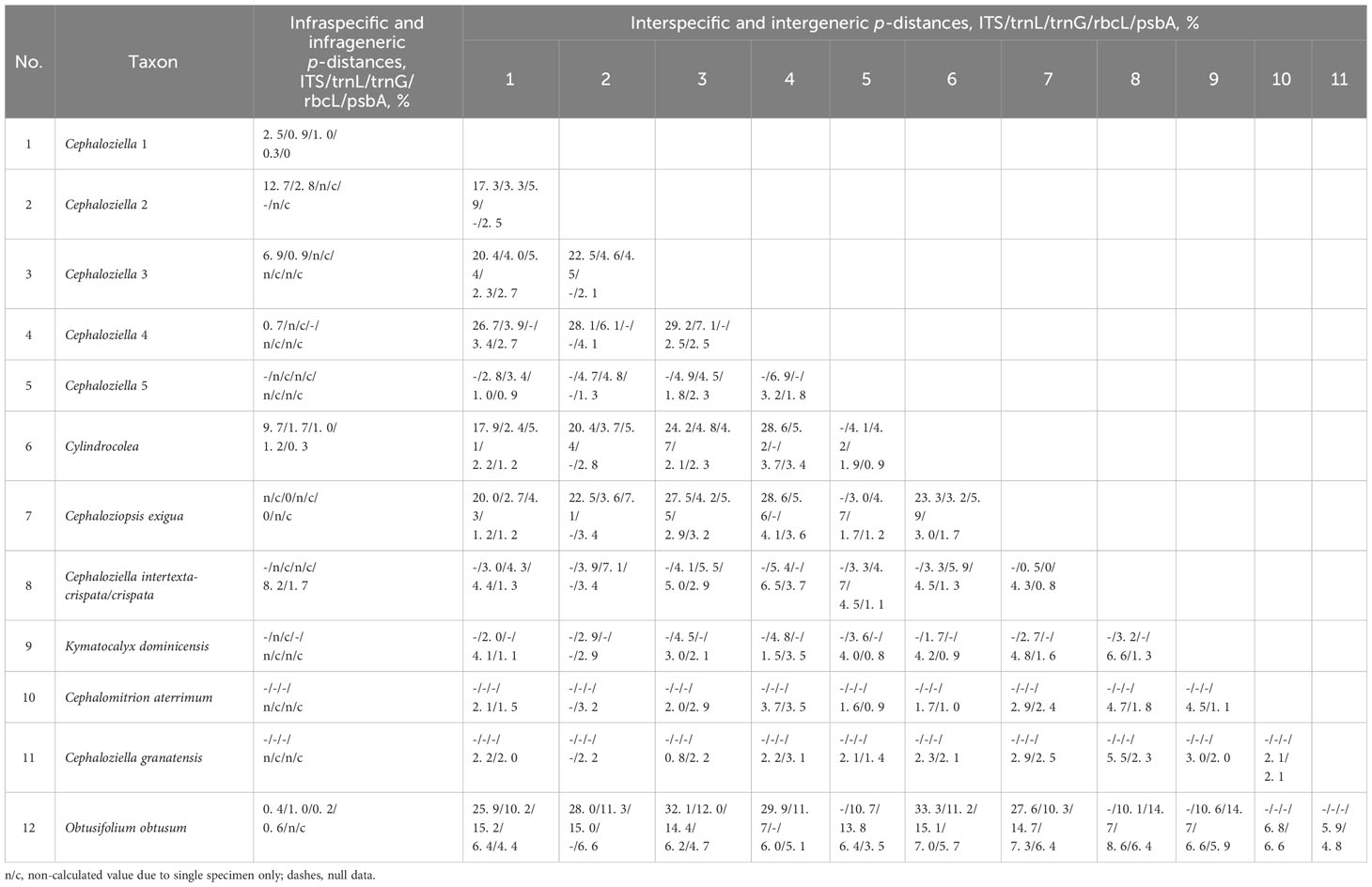
Table 2 The value of infraspecific/generic and interspecific/generic p-distances for the tested nucleotide alignments.
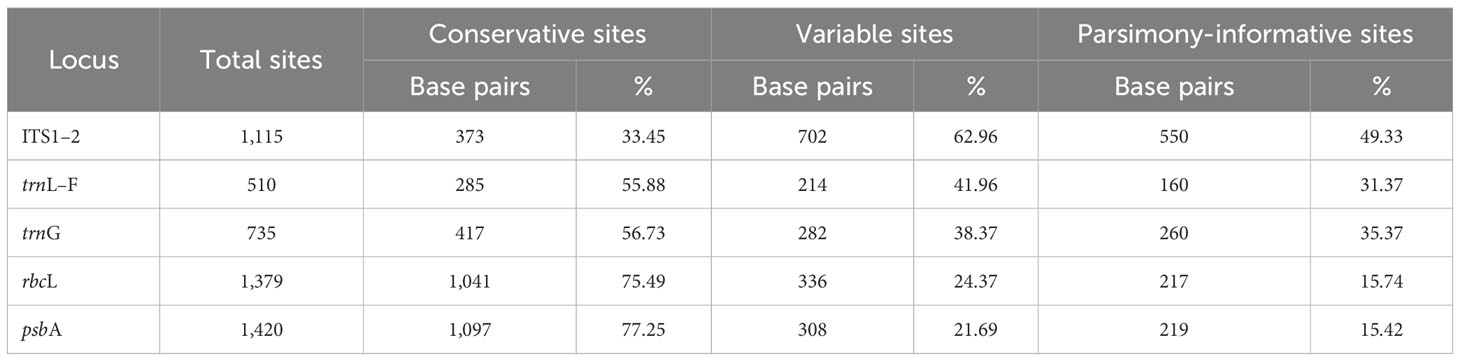
Fifty new sequences from Cephaloziella, Cephaloziopsis, Cylindrocolea, Hygrobiella, and Obtusifolium S.W.Arnell specimens were deposited in the NCBI GenBank (Supplementary Table 1), and the characteristics of the tested alignments are shown in Table 3. The log-likelihood values for the ML analysis and Bayesian analysis of all datasets are presented in Table 4.
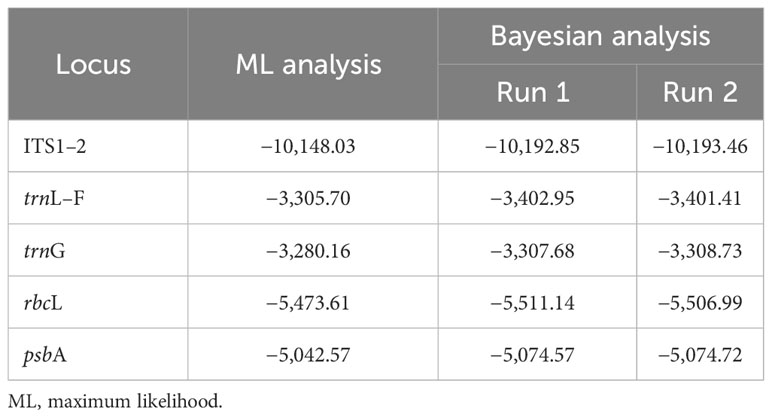
Table 4 The log-likelihood in the ML analysis and in Bayesian analysis (arithmetic mean) of the tested nucleotide sequence alignments.
The tree topologies obtained by the two methods were slightly different, so all of them are presented here. Figures 1–5 show the phylogenetic trees based on the ITS1–2, trnL–F, trnG, rbcL, and psbA datasets retained under ML analysis, along with BS values from the ML analysis and the Bayesian PPs. The differences concern the variable position of some branches of the trees constructed by the two methods. Despite small inconsistencies, the following clades are clearly distinguished on all trees: Cephaloziella 1 (ITS1–2, trnL–F, trnG, rbcL, and psbA), Cephaloziella 2 (ITS1–2, trnL–F, trnG, and psbA), Cephaloziella 3 (ITS1–2, trnL–F, trnG, rbcL, and psbA), Cephaloziella 4 (ITS1–2, rbcL, and psbA), Cephaloziella 5 (trnL–F, trnG, rbcL, and psbA), and Cylindrocolea (ITS1–2, trnL–F, trnG, rbcL, and psbA). Detailed information on these clades is provided in the Discussion section.
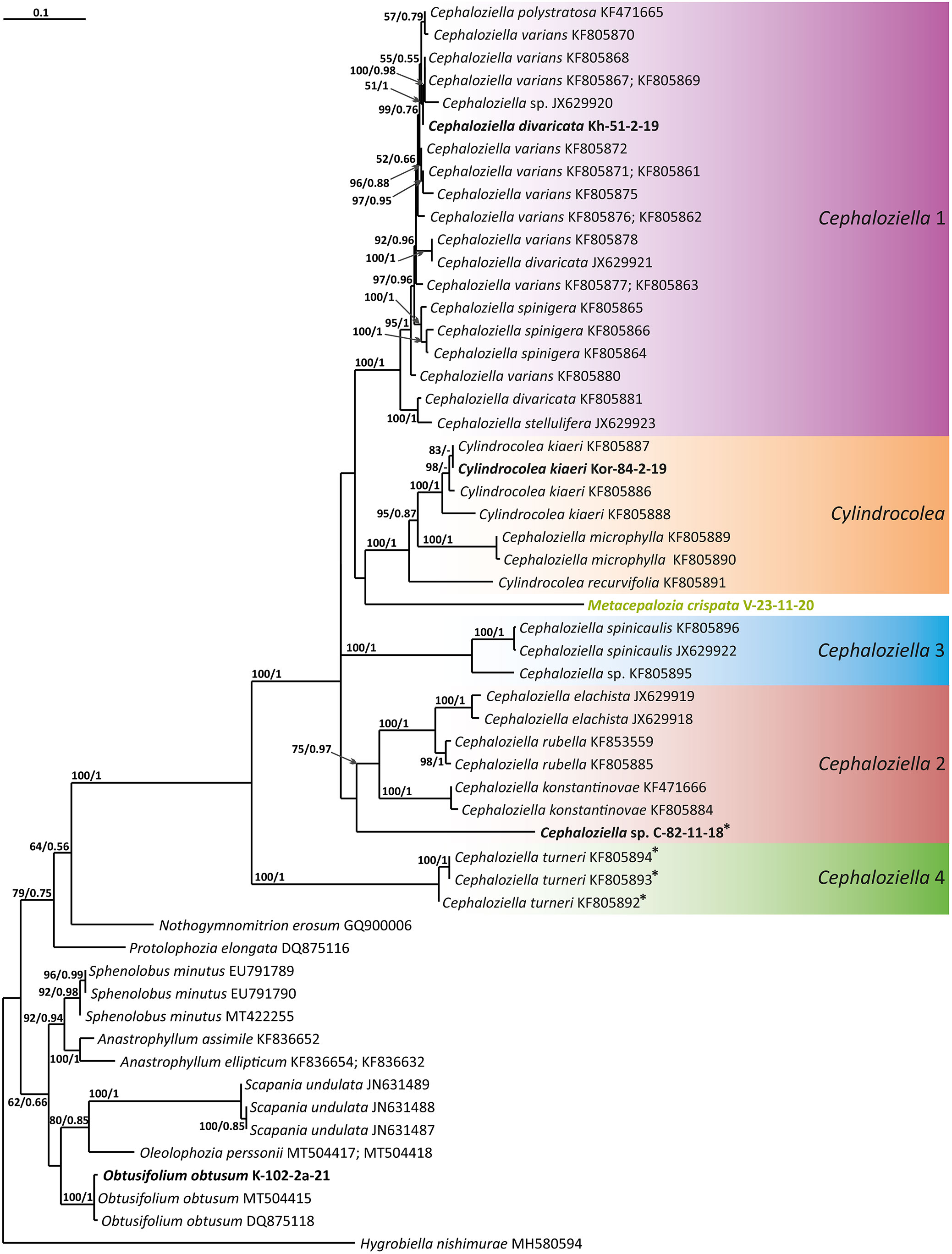
Figure 1 Phylogram obtained from a Bayesian analysis of the listed taxa based on the ITS1–2 dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 50%/0.50 are indicated. Taxon names and GenBank accession numbers or vouchers (for the samples studied by the authors) are provided. Asterisks mark taxa with different positions on ML tree. The bold font indicates the specimens sequenced for the present paper. ML, maximum likelihood.
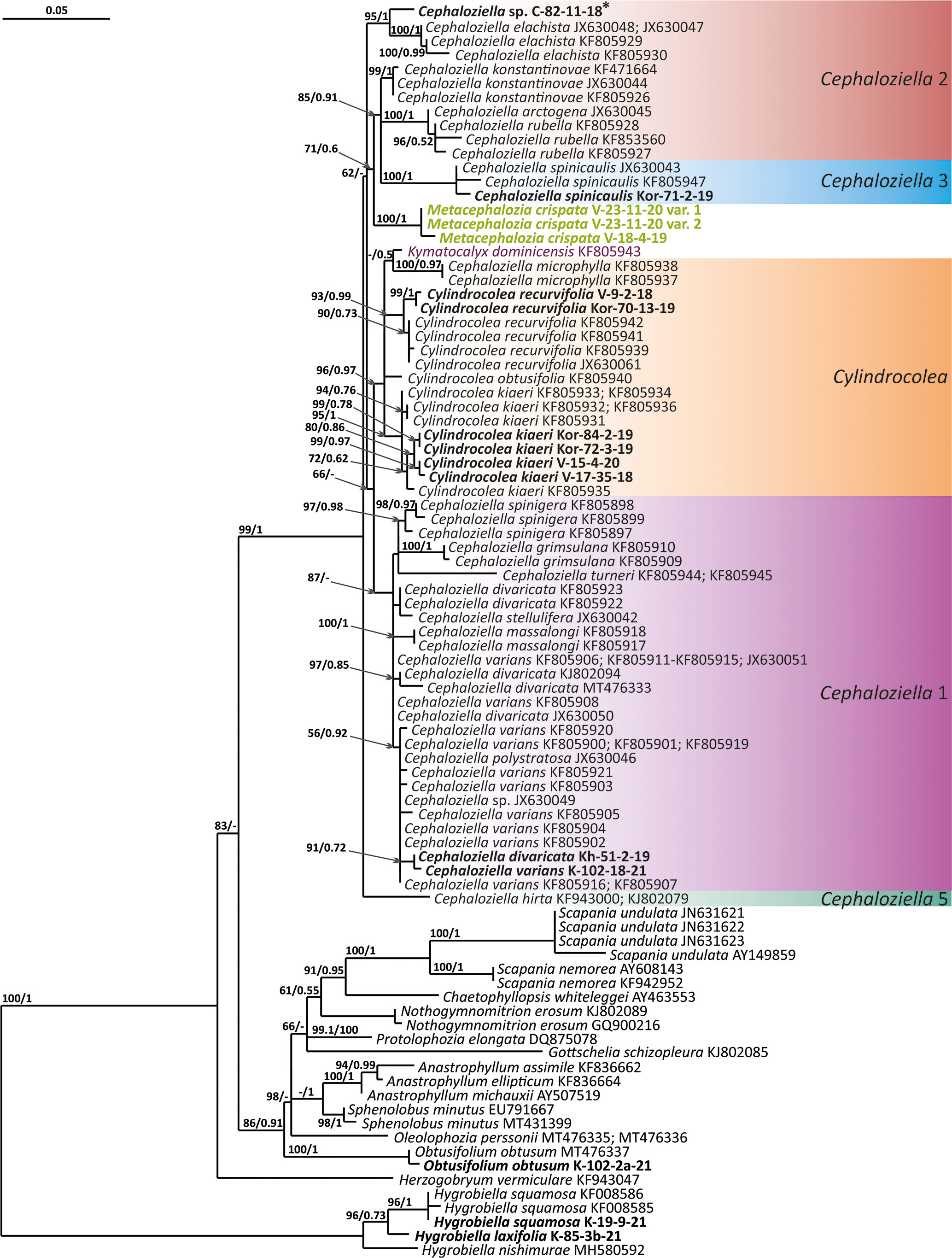
Figure 2 Phylogram obtained from a Bayesian analysis for the listed taxa based on the trnL-F dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 50%/0.50 are indicated. Taxon names and GenBank accession numbers or vouchers (for the samples studied by the authors) are provided. Asterisks mark taxa with different positions on ML tree. The bold font indicates the specimens sequenced for the present paper. ML, maximum likelihood.
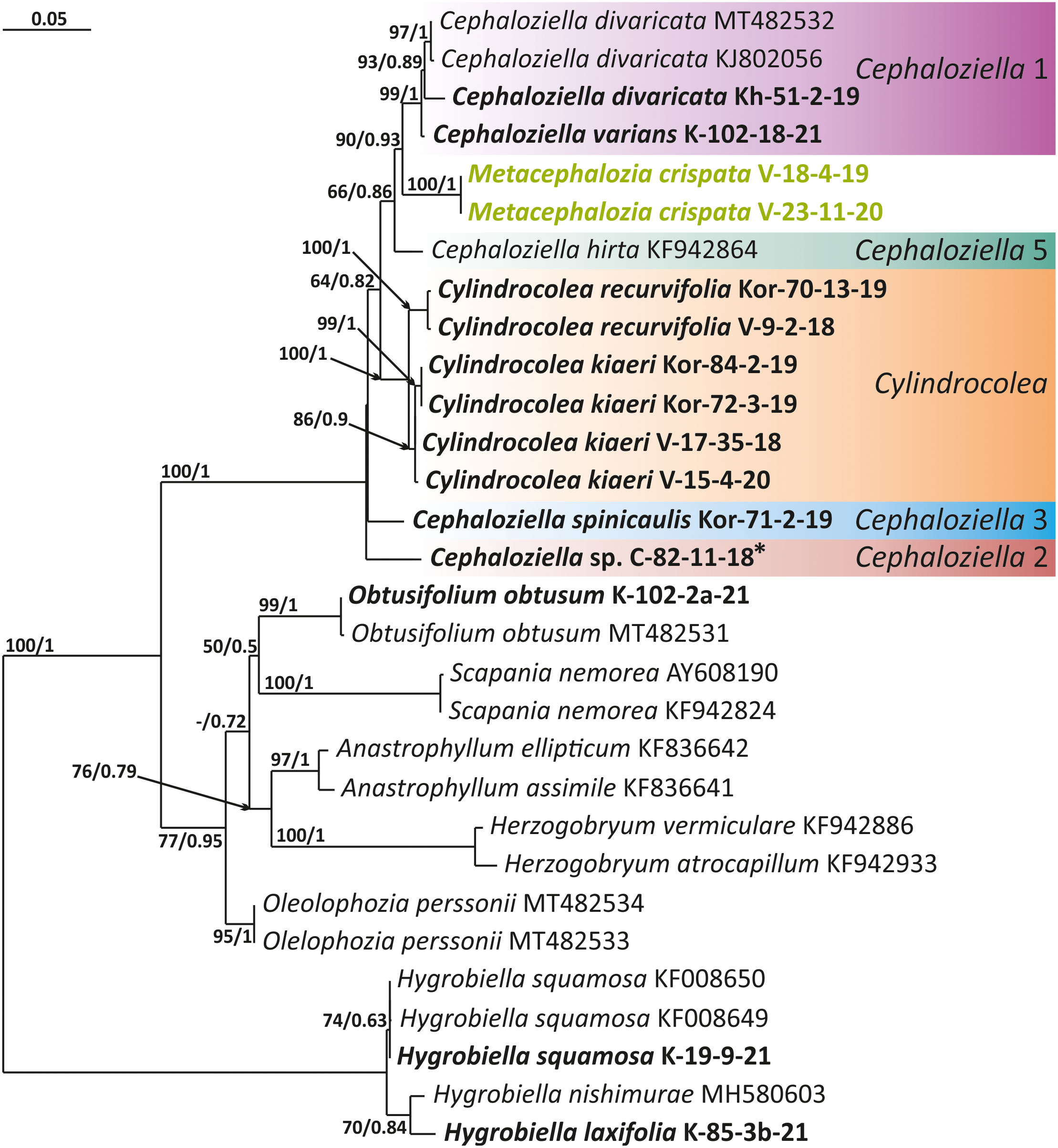
Figure 3 Phylogram obtained from a Bayesian analysis for the listed taxa based on the trnG-intron dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 50%/0.50 are indicated. Taxon names and GenBank accession numbers or vouchers (for the samples studied by the authors) are provided. Asterisks mark taxa with different positions on ML tree. The bold font indicates the specimens sequenced for the present paper. ML, maximum likelihood.
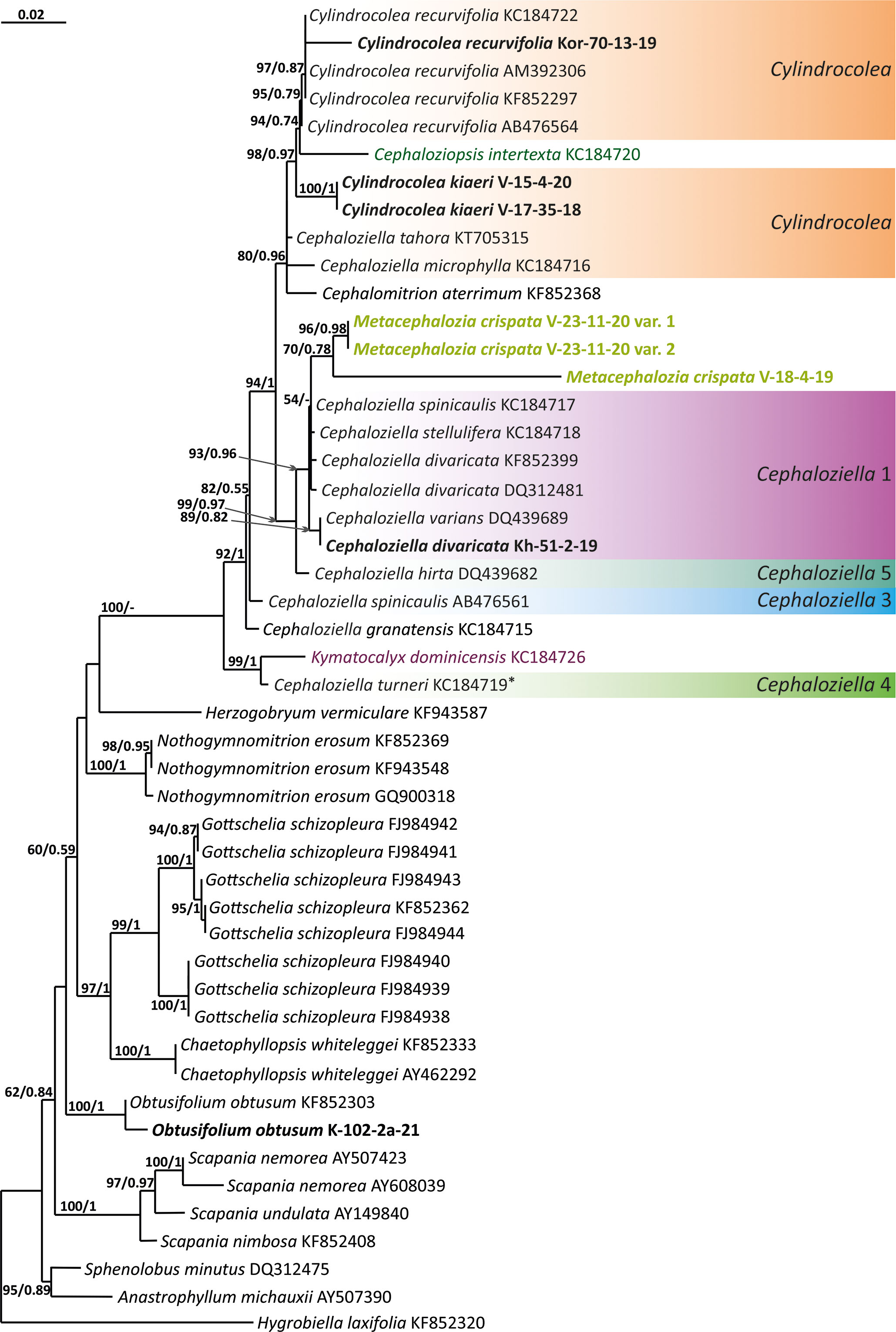
Figure 4 Phylogram obtained from a Bayesian analysis for the listed taxa based on the rbcL dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 50%/0.50 are indicated. Taxon names and GenBank accession numbers or vouchers (for the samples studied by the authors) are provided. Asterisks mark taxa with different positions on ML tree. The bold font indicates the specimens sequenced for the present paper. ML, maximum likelihood.
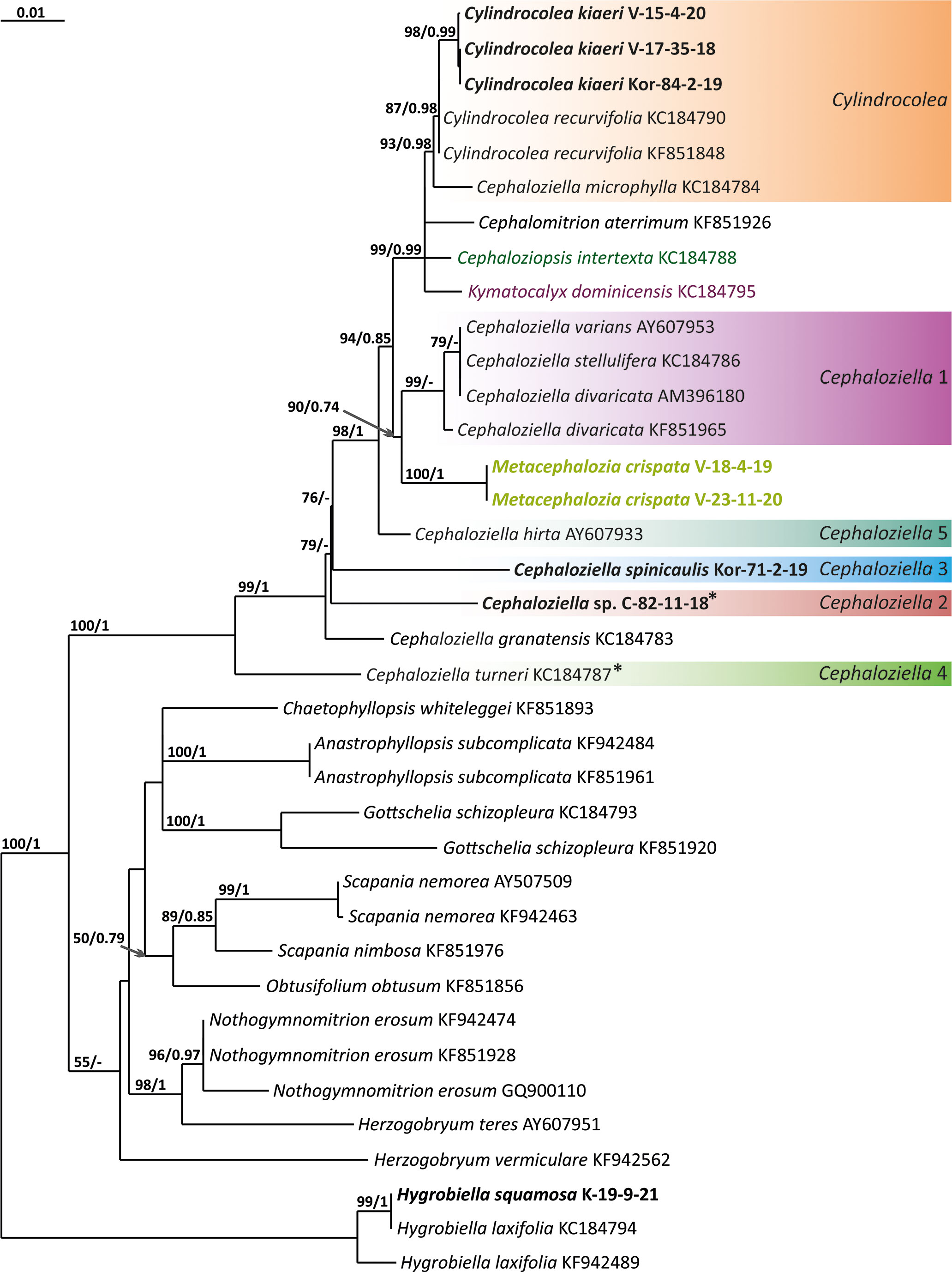
Figure 5 Phylogram obtained from a Bayesian analysis for the listed taxa based on the psbA dataset. The values of bootstrap support from the ML analysis and Bayesian posterior probabilities greater than 50%/0.50 are indicated. Taxon names and GenBank accession numbers or vouchers (for the samples studied by the authors) are provided. Asterisks mark taxa with different positions on ML tree. The bold font indicates the specimens sequenced for the present paper. ML, maximum likelihood.
4 Discussion
The first to demonstrate a sister relationship between Cephaloziellaceae and the Scapaniaceae–Lophoziaceae complex based on two chloroplast genes was De Roo et al. (2007), showing that Cephaloziella is more closely related to Lophozia (Dumort.) Dumort. and Scapania (Dumort.) Dumort. and then to the bulk of taxa later described as Anastrophyllaceae (Söderström et al., 2010). Although Anastrophyllaceae nevertheless seems to be more closely related to Scapaniaceae and Lophoziaceae than Cephaloziellaceae to the last two families, the problem was raised. In the most general approximation, the Cephaloziellaceae phylogeny was reviewed by Vilnet et al. (2012) based on a comparison of the nucleotide sequences of ITS1–2 nuclear and trnL–F chloroplast DNA. The authors emphasized the instability of the position of the family Cephaloziellaceae in trees constructed by different methods. This study is remarkable due to the obtained evidence that the family Cephaloziellaceae is molecularly distinct from the complex of other families and genera with which it was associated in later papers by other authors. Mamontov and Vilnet (2017) described a new species (Cephaloziella konstantinovae Mamontov et Vilnet) and briefly reviewed the Cephaloziellaceae system on the basis of reconstruction from the same two genes, ITS1–2 and trnL–F, using newly obtained materials. Topologies obtained for Cephaloziella s.l. by different methods (Bayesian analysis and maximum parsimony) differ in details, but they clearly show that the genus Cylindrocolea should also be included in Cephaloziella s.l. since it is nested within it. However, the authors did not make any taxonomic decisions in light of the data obtained, except for the recommendation to include Cylindrocolea recurvifolia (Steph.) Inoue within the genus Cephaloziella and treat it as Cephaloziella recurvifolia (Steph.) S. Hatt.
The paper by Feldberg et al. (2013), devoted to the estimation of divergence times in liverworts and based on the rbcL and psbA genes, was based on a dataset of Cephaloziellaceae smaller than those in the papers by Vilnet et al. (2012) and Mamontov and Vilnet (2017) cited above. The following clades are distinguished within Cephaloziellaceae: 1) C. recurvifolia, Cephaloziopsis intertexta (Gottsche) R.M.Schust., and Cephaloziella microphylla (Steph.) Douin; 2) Cephaloziella stellulifera (Taylor ex Carrington et Pearson) Croz., Cephaloziella divaricata (Sm.) Schiffn., and Cephaloziella spinicaulis Douin; 3) Cephaloziella granatensis (J.B.Jack ex Steph.) Fulford; and 4) Kymatocalyx dominicensis (Spruce) Váňa and Cephaloziella turneri (Hook.) Müll.Frib. Given that the datasets in the works of Vilnet et al. (2012) and Mamontov and Vilnet (2017), on the one hand, and Feldberg et al. (2013), on the other, are practically not based on the same specimens and provide different topologies, the problems of the Cephaloziellaceae system actually remain unresolved.
Therefore, it was an ambitious goal to seek a broad interpretation of the Cephaloziellaceae in the paper by Váňa et al. (2013), citing the work by Feldberg et al. (2013) as the basis for their solutions, albeit with a remark (Váňa et al., 2013: 1): “The only molecular study dedicated to this group including a fair number of taxa is Feldberg et al. (2013), but as it is a study of divergence time and does not include confidence values, it is difficult to draw too many conclusions from it.” Meanwhile, the authors rightly noted that “As circumscribed here, Cephaloziellaceae is morphologically heterogeneous and is probably best treated as a ‘superfamily’. Morphologically it is difficult to defend such a diverse family and it should probably be separated into further families.” In the list given by Váňa et al. (2013), the genera are conditionally divided into two groups: those whose position is unclear and which belong to Cephaloziellaceae in the broad sense of the interpretation of this family and those that belong to the Cephaloziellaceae “core” group. The last group included eight genera: Amphicephalozia, Cephalojonesia Grolle, Cephaloziella, Cephalomitrion, Cephaloziopsis, Cylindrocolea, Gymnocoleopsis, and Kymatocalyx Herzog. The group belonging to the Cephaloziellaceae in a broad sense includes only 10 genera (as in the cited paper): Allisoniella E.A.Hodgs., Anastrophyllopsis (R.M.Schust.) Váňa et L.Söderstr., Chaetophyllopsis R.M.Schust., Gottschelia Grolle, Herzogobryum Grolle, Lophonardia R.M.Schust., Nothogymnomitrion R.M.Schust., Obtusifolium S.W. Arnell, Oleolophozia L.Söderstr., De Roo et Hedd., and Protolophozia (R.M.Schust.) Schljakov. It is worth mentioning that these two groups are strikingly different in appearance. The Cephaloziellaceae “core” group includes very small plants, with distanced leaves and a seta structure (where known) of 4 + 4, 4 + 8, or similar (corresponding to the reported “archetype” of Cephaloziella of 4 + 4), while another group includes plants much larger and rather “lophozioid” in appearance, with massive setae. This is also confirmed by the fact that genera such as Obtusifolium, Oleolophozia, Protolophozia, and Lophonardia were earlier (in the era of “morphological taxonomy”) commonly referred to Lophozia in a broad interpretation of that genus.
A robust step forward in understanding the phylogenetic relationships of the Cephaloziellaceae was made by Patzak et al. (2016). The authors, based on the analysis of an rps4–rbcL dataset, showed the synonymy of Chonecoleaceae R.M.Schust. ex Grolle and Cephaloziellaceae, although the morphological structure of the seta was not quite typical (eight epidermal and four inner rows, as in Cephalomitrion) for Cephaloziellaceae. However, the Cephaloziellaceae “core” group actually shows deviations and diversity in morphology, even in the already-known species. Patzak et al. (2016) indicated that the structure of the seta within the family (in a narrow sense) varies considerably, namely, (l.c.: 98): “4, rarely (6–)8(–9) epidermal and (3–)4(–14) inner rows”. In addition to clarifying the taxonomic “amount” of the Cephaloziellaceae, based on the same rps4–rbcL dataset, the authors came to the conclusion that the Scapaniaceae family should be interpreted much more widely than is commonly accepted even in broad interpretations of the family (when Lophoziaceae, Anastrophyllaceae, and Scapaniaceae are combined together). The authors moved into the latter group Herzogobryum, Nothogymnomitrion, Anastrophyllopsis, Chaetophyllopsis, Gottschelia, and Obtusifolium, which, 3 years earlier in the concept by Váňa et al. (2013), were considered members of the Cephaloziellaceae superfamily, with the note that (l.c.: 1) “Morphologically it is difficult to defend such a diverse family and it should probably be separated into further families”. Thus, instead, the Cephaloziellaceae superfamily was revised to include Scapaniaceae s.l. (also a superfamily), thus including heterogeneous elements.
In addition to those above, another question remains unresolved: if the topologies obtained by Vilnet et al. (2012) and Mamontov and Vilnet (2017), based on ITS1–2 and trnL–F analyses, differ from the topologies obtained from the rps4–rbcL dataset, can one of these points of view be considered more correct than the other if no cross-comparisons were conducted?
Later, while studying the molecular genetic relationships of Konstantinovia Bakalin et Fedosov (a hitherto monotypic genus) described as new to science, Bakalin et al. (2021a) found it to be most closely related to the genus Obtusifolium, prompting a new attempt to determine the taxonomic position of these two genera. This task was solved on the basis of a comparison of four genes: the nuclear ITS region and the chloroplast rps4 gene, trnL–F region, and trnG-intron. As a result, it was shown that 1) Anastrophyllaceae is clearly distinguished in the system Scapaniaceae s.l. plus Cephaloziellaceae s.l.; 2) Lophoziaceae and Scapaniaceae differ less prominently (actually, Lophoziaceae is nested within Scapaniaceae; however, under certain conditions, these families can be treated separately); 3) Obtusifolium plus Konstantinovia should be segregated into a new family, which is much more closely related to the Scapaniaceae plus Anastrophyllaceae, but not to the Cephaloziellaceae; and 4) Oleolophozia should probably also be placed in a separate family (taxonomic decisions on the last matter have not been made).
Our present study began with the need to determine the position of plants collected in Vietnam, which we identified as C. exigua (Inoue) R.M. Schust. et Inoue, a name that does not appear in the current literature; these plants are commonly known as Cephaloziella crispata N.Kitag. The history of the interpretation of this taxon is rather confusing. It has been described as Metacephalozia exigua Inoue (Inoue, 1973). In the original author’s interpretation, this species (and genus) occupies an intermediate position in terms of morphology between Cephaloziaceae and Cephaloziellaceae (Inoue, 1973). A year later, Inoue and Schuster (1974) published a paper in which, based on a comparison of the morphology of a little-known Neotropical Cephaloziopsis with Metacephalozia Inoue, they came to the conclusion that the latter deserved only the rank of a subgenus within the former (as an older name). The result was also a new combination: C. exigua. At the same time, the authors of the cited work emphasized a serious difference in the distribution of the two subgenera (Neotropics versus warm temperate East Asia), associated with minor morphological differences.
Later, Katagiri and Furuki (2012) stated (in one sentence) that C. crispata, described by Kitagawa in 1969, is synonymous with M. exigua, described by Inoue in 1973. As a result of formal synonymization, this species was placed in the Japanese liverwort checklist under the name C. crispata. The authors (Katagiri and Furuki, 2012) did not express their opinion regarding the status of Metacephalozia or Cephaloziopsis. Interestingly, given the similar morphologies of Cephaloziopsis s. str. and Metacephalozia (which authors of the Japanese checklist did not contest), it would be logical to assume that if one can be declared a synonym of Cephaloziella without much doubt, then it would be better to synonymize the Cephaloziopsis genus with Cephaloziella as well. However, the genus (or subgenus) Metacephalozia has simply disappeared, while Cephaloziopsis “continued to exist”. Cephaloziopsis subg. Cephaloziopsis, as recognized in the World Checklist (Söderström et al., 2016), including one species, C. intertexta. The second species of the genus [following the interpretation by Schuster (1972)] Cephaloziopsis schistophila (Spruce) R.M. Schust., as foreseen by Schuster (l.c.: 182), “I have had one of the two as a subspecies of C. schistophila, and, indeed, subspecific segregation may prove more nearly correct”, was eventually synonymized with the earlier C. intertexta, and the latter name appears on the World Liverwort Checklist (Söderström et al., 2016).
In light of our data on phylogenetic relationships between different groups within Cephaloziellaceae s. str., two options are possible: 1) to consider all genera for which there are nucleotide sequences forming a single unit (including all previously segregated genera, e.g., Cephaloziopsis and Cylindrocolea) as the genus Cephaloziella s.l. and 2) along with the revisiting of existing genera, it is necessary to segregate new genera. Taking into account the general trend and genetic distance values (Table 2) between the identified clades, we chose the second option. Below, we describe clades on phylogenetic trees as harbingers for the possible taxonomic solutions within the Cephaloziellaceae. The morphological background of the obtained clades remains not entirely clear. Probably leaf attachment (oblique or almost transverse) is an important character in the taxonomy of Cephaloziellaceae. Some aberrations in the structure of the sporophyte are also confirmed as significant characters in the taxonomy of Cephaloziellaceae, but there are still very little data on this issue. It seems to us that the most significant criteria are not even morphological, but geographical ones, namely, patterns of distribution, since the distinguished clades (in the most general terms) may differ in the distribution of the species included in them, for example, South American, Paleotropical, mainly East Asian, and North Holarctic. This issue deserves further detailed study.
Clade Cephaloziella 1 is the genus Cephaloziella in the narrow sense. This clade contains the type species of the genus (C. divaricata). In most trees, this clade is quite well isolated from others, and its composition is stable: C. divaricata, Cephaloziella varians (Gottsche) Steph., Cephaloziella spinigera (Lindb.) Jørg., Cephaloziella polystratosa (R.M.Schust. et Damsh.) Konstant., Cephaloziella grimsulana (J.B.Jack ex Gottsche et Rabenh.) Lacout., C. stellulifera, and Cephaloziella massalongi (Spruce) Müll.Frib. In general, this group seems to correspond with the subgenus Cephaloziella of the genus if a broad interpretation is accepted.
Cylindrocolea described by Schuster (Schuster, 1969: 666) is treated on the World Checklist as housing 16 species (Söderström et al., 2016). Only a few of these genera were tested by molecular genetic methods; the type of the genus (Cylindrocolea chevalieri (Steph.) R.M. Schust.) was also not tested. In general terms, the genus differs from Cephaloziella s str. (cf. Schuster, 1972) as follows: 1) very obliquely inserted leaves, 2) leaf-free antical sector of the stem, 3) absence of gemmae, and 4) wide or only weakly contracted perianth mouth. Taking into account the available data on the molecular relationships of species, C. recurvifolia can be reliably assigned to this genus. Cylindrocolea kiaeri (Austin) Váňa also belongs to this genus. Moreover, it should be noted that although we call the sequenced specimens C. kiaeri, all sequenced materials (both from us and available from GenBank) originate from East Asia, Indochina, and Malaysia. Meanwhile, the species has been described from Africa. The identity of the Asian and African materials may be questioned, although at the moment we do not have any confirmation of this point of view (a brief discussion on it can be found in Bakalin et al. (2023). The same group clearly includes C. microphylla, previously placed in Cephaloziella subg. Prionolobus (Spruce) Müll. Frib. (type species is C. turneri). C. microphylla clearly differs from other representatives of Cylindrocolea, mainly in the abundance of mammillose protrusions and outgrowths on the abaxial leaf surface. Despite outwardly significant differences, molecular genetic data for four genes (ITS, trnL–F, rbcL, and psbA) agree on the need to refer this species to Cylindrocolea.
There is much less reason to recognize Kymatocalyx as a distinct genus. Four species are known in the genus (type species Kymatocalyx stolonifer Herzog = K. dominicensis). Molecular data are available for the type species only for trnL–F (for which the species is unequivocally included in the genus Cylindrocolea) and rbcL (where Kymatocalyx is found to be related to C. turneri). Given the incongruence found, we refrain from making taxonomic decisions on this matter. However, we note that the phrase of Schuster (Schuster, 1972: 177) “The genus Kymatocalyx is at once distinct from other taxa referred to the Cephaloziellaceae in having quite unlobed leaves and female bracts” can only be taken into account with reservations. For example, C. recurvifolia often has very shallowly emarginate leaves. The same can be said of C. chevalieri. Therefore, if we compare the variability of the leaf shape in Cylindrocolea in the series from C. kiaeri to C. recurvifolia, we can assume that the variants of Kymatocalyx are not sufficiently unique for morphological substantiation of its interpretation as a separate genus. However, due to data limitations, we refrain from formally synonymizing Kymatocalyx with Cylindrocolea.
The Cephaloziella 2 group in the ITS-based phylogenetic tree includes Cephaloziella elachista (J.B.Jack ex Gottsche et Rabenh.) Schiffn., C. konstantinovae, and Cephaloziella rubella (Nees) Warnst. The same species also formed a clade in the ITS1–2 + trnL–F trees in the study by Mamontov and Vilnet (2017). In Figure 1, this group is adjacent with low support to a potentially new species designated Cephaloziella sp. (C-82-11-18). The trnL–F-based tree (Figure 2) shows C. rubella, Cephaloziella arctogena (R.M.Schust.) Konstant., and C. konstantinovae with C. spinicaulis in a clade (BS = 85, PP = 0.91) sister to C. exigua, while C. elachista and C. sp. (C-82-11-18) form a sister branch to all Cephaloziellaceae s. str., except Cephaloziella hirta (Steph.) R.M.Schust.
Cephaloziella sp. (C-82-11-18) forms a sister branch to all Cephaloziellaceae included in the analysis in the trnG-based phylogenetic tree. The sub-basal position is typical for the same species in the psbA-based tree. It is obvious that the complex C. konstantinovae–C. rubella–C. arctogena deserves to be allocated to an independent genus; however, at present, we refrain from nomenclatural combinations due to 1) the ambiguity of phylogenetic relationships with C. elachista and 2) the ambiguity of morphological criteria delimiting the group (this is mainly due to the unstable position of C. elachista).
C. spinicaulis occupies a quite isolated position in the ITS, trnG, rbcL, and psbA trees. This is inconsistent with the morphological expectations of some scholars. Schuster (1980) placed this species in the section Bissaceae R.M. Schust. (=sect. Cephaloziella s. str.), together with the type species of the genus C. divaricata. Douin (1920) had conducted the same even before, placing this species in the “Groupe du C. starkii” (=C. divaricata). However, Schuster (l.c.) indicates that this species has a distanced position in the section due to its rigid texture, rather small leaves relative to the stem diameter, and a main stem having numerous spines that are 1–4 cells long. In addition, Schuster (Schuster, 1980: 107) wrote, “C. spinicaulis appears to be only remotely allied to the sub-Antarctic and Antarctic C. hispidissima R.M. Schust.” (=Cephaloziella verrucosa Steph., cf. Váňa et al., 2014). In the World Checklist (Söderström et al., 2016), the species is placed in the subgenus Cephaloziella, which, however, cannot be accepted according to the available topologies. This species, given the adoption of the generic status for Kymatocalyx and Cylindrocolea, must be allocated to a separate genus. At the same time, the relationship with C. verrucosa needs to be studied, and thus far, we have refrained from formally transferring the latter from one genus to another. We propose naming the new genus that we have identified Douiniella, in honor of Charles Isidore Douin (1858–1944), an outstanding expert on Cephaloziella in the first third of the 20th century. The genus is hitherto monotypic.
Douiniella Bakalin, Maltseva, Troitzk. gen. nov.
Description. Plants dioicous, of rigid texture, with dense and numerous spines covering the stem and adaxial leaf surface.
Type species: Douiniella spinicaulis Bakalin, Maltseva, Troitzk. comb nov. Basionym: C. spinicaulis Douin, Mém. Soc. Bot. France 29: 62, 1920 (Douin, 1920)
In the ITS, rbcL, and psbA trees (Figures 1, 4, 5), C. turneri occupies a sister position to all other Cephaloziella and should also be considered a separate genus. Since the type species for the genus Prionolobus is Prionolobus turneri (Hook.) Schiffn. (cf. Grolle, 1976), it is reasonable to maintain the generic status for the taxon appearing in Söderström et al. (2016) as Cephaloziella subg. Prionolobus. At the same time, however, it must be taken into account that of the eight species referred to this subgenus in the World Checklist (Söderström et al., 2016), at least some definitely cannot be assigned. For example, C. microphylla reliably belongs to Cylindrocolea following the obtained topologies. In addition, in the two available trees, those for rbcL and psbA, C. granatensis does not form a common clade with C. turneri; instead, with the exception of the latter and Kymatocalyx (in the rbcL tree), it is a sister to all other Cephaloziella. No molecular data are available for the remaining Prionolobus species. Therefore, Prionolobus reliably forms a genus, but its content (except for the type species) remains unclear.
Cephaloziella subg. Evansia (Douin et Schiffn.) Müll.Frib., with the type species Cephaloziella dentata (Raddi) Steph., includes (according to the list of Söderström et al. (2016) C. hirta. Apparently, this subgenus should also be treated as a separate genus (that should be supplied with the new generic name since Evansia is the later homonym of the previous Evansia Salisb. ex Decne); however, since we do not have data on the phylogenetic relationships of C. dentata, we refrain from making taxonomic decisions on this matter. However, it is worth mentioning that the morphologies of the two taxa (C. dentata and C. hirta) are very similar. The main difference between C. dentata and C. hirta is in the inflorescence: plants are dioicous in C. dentata but autoicous in C. hirta. In addition, C. dentata is mostly European, but with some reports from China (Hunan, Jiangxi Provinces), Japan, Tasmania, British Columbia, and Mexico (available from https://www.catalogueoflife.org/data/taxon/5XK7F), while C. hirta is Australasian, also with some reports from New Zealand North Island and Juan Fernández Island (available from (https://www.catalogueoflife.org/data/taxon/5XK7G).
Finally, the problem that initiated this work in general terms should be discussed. It was unclear how, in light of the currently accepted treatment (Söderström et al., 2016), to interpret the independence of Cephaloziopsis and Metacephalozia. On the one hand, the authors of the World Checklist (Söderström et al., 2016) agreed that Metacephalozia is a synonym of the genus Cephaloziella and not the same genus but the same named (type) subgenus. On the other hand, no one synonymized monotypic Cephaloziopsis with Cephaloziella. Third, no one has disputed that Metacephalozia and Cephaloziopsis are not the same genus. Thus, a contradiction arose, which we tried to resolve by molecular methods.
First, C. crispata and M. exigua are undoubtedly conspecifics. The first is (according to the type of materials) simply a form of the second arising in shadier habitats. Our studies show that this species is very polymorphic and quite common in the mountains of North Vietnam, where it grows on moist, bare loamy soil (usually without shading or under weak shading). In other words, it tends to prefer habitats favored by many members of Cephaloziella s. str. in the global North. Ecologically determined modifications are genetically almost identical, as the constructed topologies show. The position of M. exigua (=C. crispata) varies from tree to tree. According to the ITS data (Figure 1), the species forms a sister branch to all Cylindrocolea. According to the trnL–F tree (Figure 2), this species is sister to the group C. arctogena-konstantinovae (Cephaloziella 2) + C. spinicaulis (Douiniella, which we propose). According to the trnG, rbcL, and psbA trees (Figures 3–5), this species is sister to Cephaloziella s. str. (C. divaricata and C. varians) (Cephaloziella 1 clade).The position of M. exigua described above differs greatly from the position of C. intertexta (the second species of Cephaloziopsis s.l.), which, according to rbcL, is sister to Cylindrocolea or close to it in the rbcL-based reconstruction. Thus, if the genus Cylindrocolea is distinguished from Cephaloziella s. str. (and even more so if other genera within Cephaloziellaceae s. str. are recognized), then it should be accepted that C. intertexta and M. exigua belong to different genera. Moreover, given the variable position of both genera in phylogenetic trees based on different genes, neither of them can be confidently combined with either the genus Cephaloziella (as it is accepted here, i.e., in the narrow sense) or the genus Cylindrocolea, and at the moment, the best option would be to recognize the independence of the two. At the same time, since C. crispata was described earlier than M. exigua, it is necessary to propose a new combination: Metacephalozia crispata. Taking into account the limited amount of information on this taxon, we provide the corresponding figures for M. crispata (Figures 6–8).
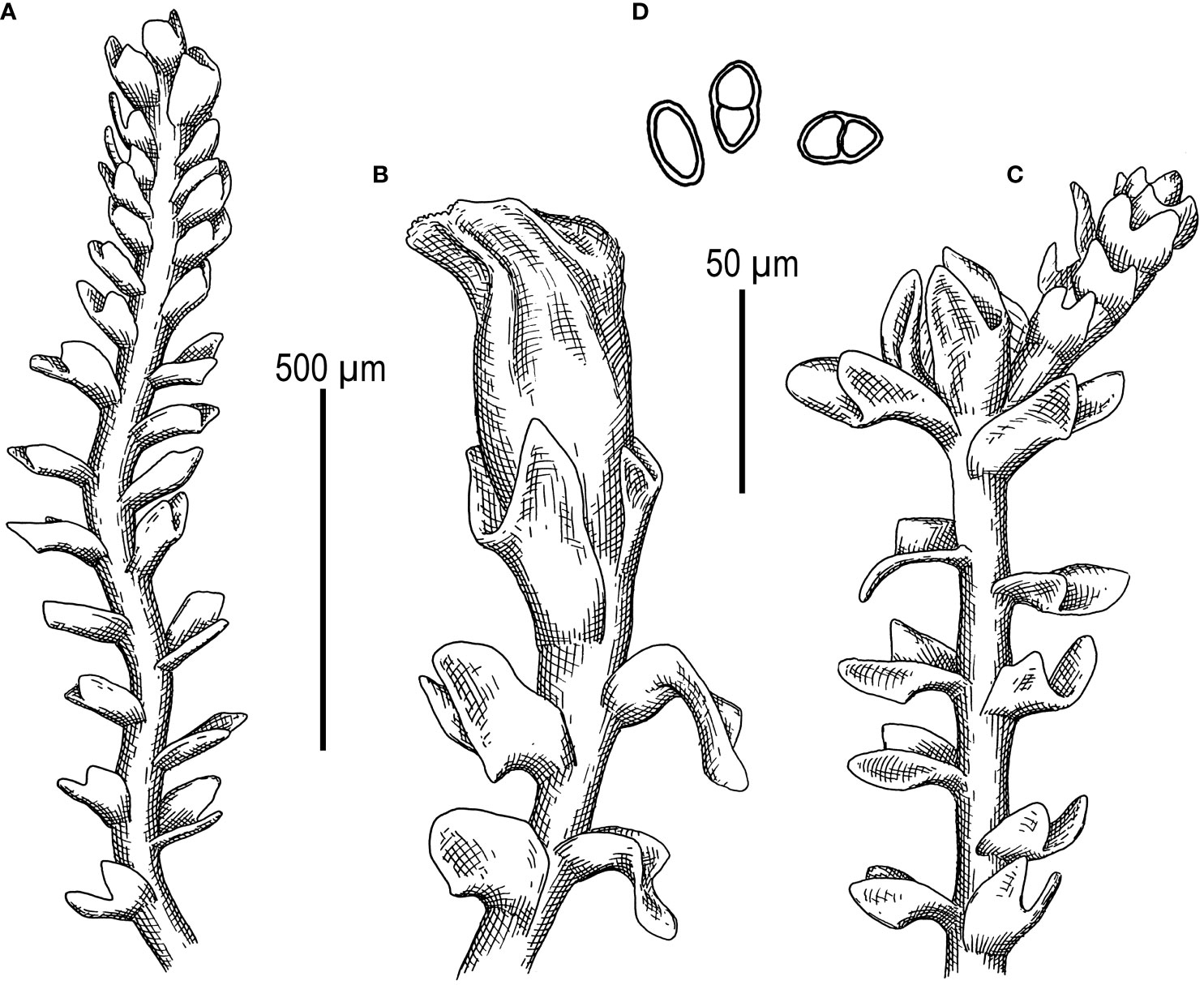
Figure 6 Metacephalozia crispata (N.Kitag.) Bakalin, Maltseva, Troitzk. (A) Shoot, fragment, dorsal view. (B) Perianthous shoot, fragment, dorsal view. (C) Shoot with lateral branch, fragment, dorsal view. (D) Gemmae. Scales: 500 µm for panels (A–C) and 50 µm for (D) All from V-23-11-20 (VBGI).

Figure 7 Metacephalozia crispata (N.Kitag.) Bakalin, Maltseva, Troitzk. (A) Mat. (B, C) Gemmiparous shoot, fragment, ventral view. (D) Oil bodies in leaf cells near shoot apex. (E) Plagiotropic shoot, fragment. (F–H) Leaves with oil bodies (mostly destructed). (A, F–H) From V-18-4-19. (C, B) From V-23-11-20. (D, E) From V-23-21-22. All from VBGI.
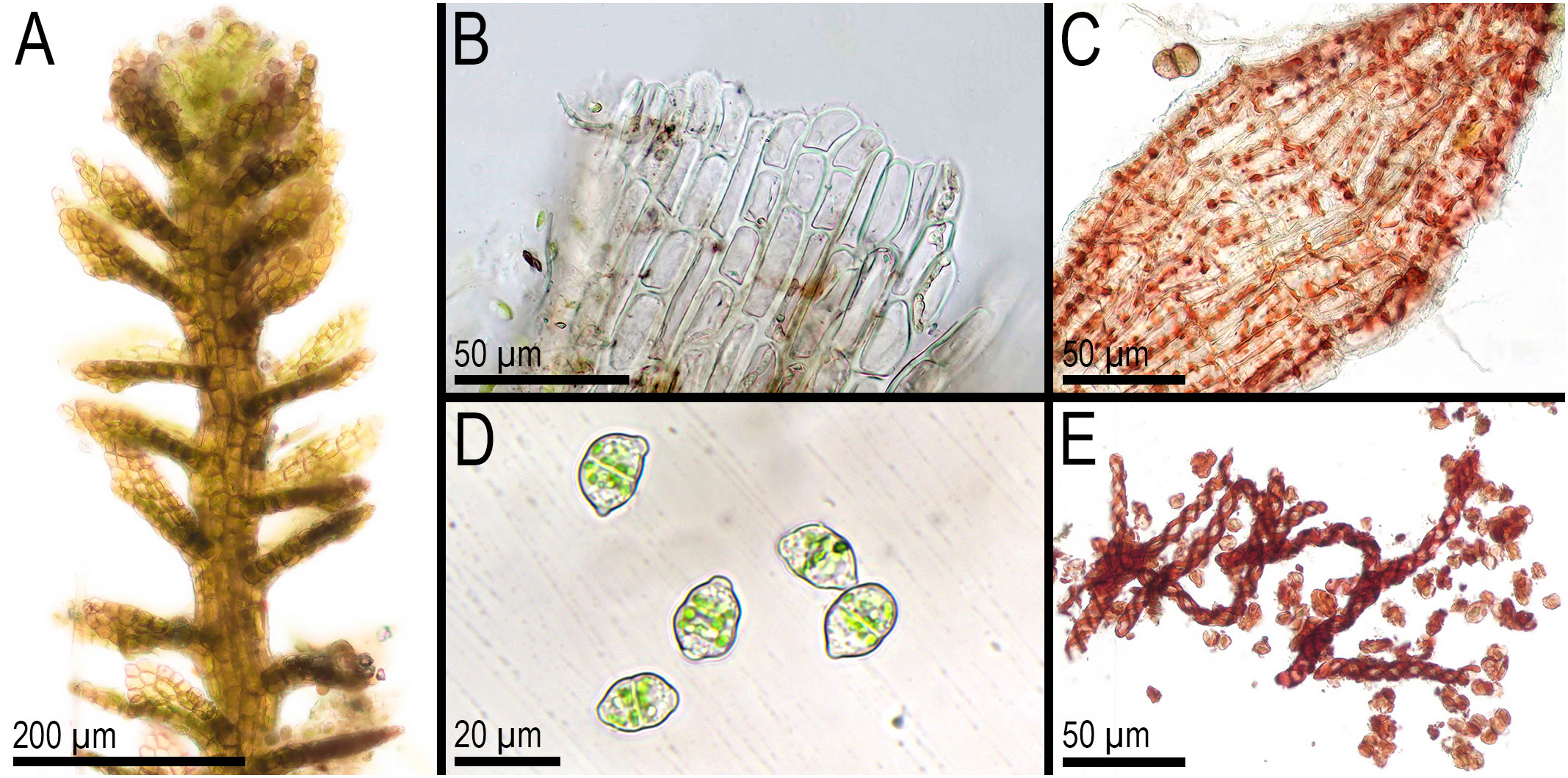
Figure 8 Metacephalozia crispata (N.Kitag.) Bakalin, Maltseva, Troitzk. (A) Shoot, fragment, dorsal view. (B) Perianth mouth. (C) Outer cells of capsule lobe. (D) Gemmae. (E) Spores and elaters (spores are distinctly collapsed). (A–C, E) From V-23-11-20. (D) From V-6-20-22. All from VBGI.
M. crispata (N.Kitag.) Bakalin, Maltseva, Troitzk. comb. nov.
Basionym: C. crispata N.Kitag., J. Hattori Bot. Lab. 32: 301 1969 (Kitagawa 1969c). = C. exigua (Inoue) R.M. Schust. & Inoue Bull. Natl. Sci. Mus. Tokyo, n.s. 17: 162 1974; = M. exigua Inoue J. Hattori Bot. Lab. 37: 287 1973.
At last, we provide the generic list of Cephaloziellaceae s. str. as it is accepted in the present paper. The asterisk indicates that the genus was not studied by molecular genetic methods and that its status, as well as its scope, is not definitely known. The exclamation mark indicates that at least one species of the genus was involved in the present study and that its position within Cephaloziellaceae s. str. is now confirmed.
*Amphicephalozia R.M.Schust., Nova Hedwigia 22: 131, 1971 [1972]. The genus includes three species, and none of them are tested genetically. Type: Amphicephalozia amplexicaulis R.M. Schust. Nova Hedwigia 22(1–2): 133. 1971[1972].
*Cephalojonesia Grolle, Rev. Bryol. Lichénol. 37: 763 1971. The genus is monospecific. Type: Cephalojonesia incuba Grolle & Vanden Berghen Rev. Bryol. Lichénol. 37: 764, pl. 1–2 1970 [1971].
!Cephaloziella (Spruce) Schiffn., Hepat. (Engl.-Prantl): 98 1893. Basionym: Cephalozia subgen. Cephaloziella Spruce, Cephalozia: 62 1882. The exact number of species is unknown due to results obtained in the present paper; this genus should be split into several genera. Type: C. divaricata (Sm.) Schiffn., Hepat. (Engl.-Prantl): 99 1893.
!Cephalomitrion R.M.Schust., Nova Hedwigia 61: 550 1995. The genus is monospecific. Type: Cephalomitrion aterrimum (Steph.) R.M. Schust., Nova Hedwigia 61 (3/4): 554 1995.
!Cephaloziopsis (Spruce) Schiffn., Hepat. (Engl.-Prantl): 85 1893. Basionym: Jungermannia sect. Cephaloziopsis Spruce, Trans. & Proc. Bot. Soc. Edinburgh 15: 511, 1885. The genus is monospecific. Type: C. intertexta (Gottsche) R.M.Schust., Nova Hedwigia 22: 183 1971 [1972].
!Cylindrocolea R.M.Schust., Bull. Natl. Sci. Mus. Tokyo 12: 664 1969. The genus includes 18 recognized taxa, and only a few of them were tested genetically. Type: C. chevalieri (Steph.) R.M.Schust., Bull. Natl. Sci. Mus. Tokyo (n.ser.) 12 (3): 666 1969.
!Douiniella Bakalin, Maltseva, Troitzk. (the present paper). The genus is confirmed as monospecific (may be bi-specific, but it requires additional studies). Type: D. spinicaulis Bakalin, Maltseva, Troitzk. (the present paper).
*Gymnocoleopsis (R.M.Schust.) R.M.Schust., Phytologia 39: 243, 1978. Basionym: Gymnocolea subgen. Gymnocoleopsis R.M.Schust., Bryologist 70: 111, 1967. The genus includes two recognized taxa, but none of them were tested genetically. Type: Gymnocoleopsis multiflora (Steph.) R.M. Schust. Phytologia 39: 243. 1978. (=Gymnocoleopsis cylindriformis (Mitt.) R.M. Schust., J. Hattori Bot. Lab. 78: 126, 1995).
!Kymatocalyx Herzog, Memoranda Soc. Fauna Fl. Fenn. 25: 55, 1950. The genus includes four recognized taxa, and one of them was tested genetically. Type: K. stolonifer Herzog Memoranda Soc. Fauna Fl. Fenn. 25: 55 1950. (=K. dominicensis (Spruce) Váňa, Österr. Bot. Z. 118 (5): 575 1970).
!Metacephalozia Inoue J. Hattori Bot. Lab. 37: 287 1973. The genus is monospecific: M. crispata (N.Kitag.) Bakalin, Maltseva, Troitzk. (the present paper). The generic status is confirmed in the present paper.
!Prionolobus (Spruce) Schiffn. Hepaticae … aus Engler-Prantl 98 1893. The genus is confirmed as monospecific (actually it may include more taxa, but it requires additional studies). Type: P. turneri (Hook.) Schiffn. Hepaticae … aus Engler-Prantl 98 1893.
5 Conclusions
In general, the attempt to construct Cephaloziellaceae trees based on five genes revealed how little information is available on this family. A number of genera have not yet been studied genetically. For Cephalomitrion (characterized by the peculiar structure of seta 4 + 8), the data do not give an unambiguous answer to the question of whether it should be included in Cylindrocolea or considered as a separate genus. The resulting tree topologies highlight the need for critical studies of other genera within Cephaloziellaceae s. str. In contrast, it may be assumed that the reduction of Dichiton to the synonyms of Cephaloziella (such as Cephaloziella subg. Dichiton (Mont.) Müll.Frib.) may be quite premature, given the large genetic distances between subgenera that have already been studied.
As shown by the present study, the Cephaloziellaceae system recognized in the World Liverwort Checklist (Söderström et al., 2016) does not perfectly reflect phylogenetic relationships and should be corrected. Some of the widely recognized genera may not deserve independence (Kymatocalyx, Cephalomitrion). At the same time, some species groups of the large genus Cephaloziella should be segregated into distinct genera. Some of them are simply genera described a long time ago and undeservedly forgotten. Given that molecular data are available for only less than 15% of the recognized species within Cephaloziellaceae, a large number of new “surprises” can be expected in further studies of this family of the smallest higher plants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
VB: Funding acquisition, Investigation, Writing – review & editing, Data curation, Project administration, Supervision, Validation, Writing – original draft. YM: Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft. KK: Investigation, Resources, Visualization, Writing – review & editing. VN: Formal Analysis, Investigation, Resources, Writing – review & editing. SC: Funding acquisition, Investigation, Resources, Writing – review & editing. AT: Data curation, Formal Analysis, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work of VB, YM, and KK is within the frame of the institutional research project “Cryptogamic Biota of Pacific Asia” (no. 122040800088-5). The work of VN was supported by the Vietnam Academy of Science and Technology (NVCC09.01/23-24). The work of SC was partially supported by a grant from the National Ecosystem Survey of the National Institute of Ecology (NIE-A-2023-01) of Korea. The work of AT is within the frame of the institutional research project # AAAA-A17-117120540067-0 “Genosystematics of Eukaryotes”.
Acknowledgments
The authors are sincerely grateful to Matvei Bakalin for line-art figure preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1326810/full#supplementary-material
References
Bakalin, V. A., Choi, S. S., Park, S. J. (2023). Liverwort diversity in Cambodia: new records show there is still much to learn. Diversity 15, 241. doi: 10.3390/d15020241
Bakalin, V. A., Fedosov, V. E., Fedorova, A. V., Ma, W. Z. (2021a). Obtusifoliaceae, a new family of leafy liverworts to accommodate Konstantinovia, newly described from the Hengduan Mts. (South China) and Obtusifolium (Cephaloziineae, Marchantiophyta). Plant Syst. Evol. 307, 62. doi: 10.1007/s00606-021-01779-8
Bakalin, V., Maltseva, Y., Vilnet, A., Choi, S. S. (2021b). The transfer of Tritomaria koreana to Lophozia has led to recircumscription of the genus and shown convergence in Lophoziaceae (Hepaticae). Phytotaxa 512, 41–56. doi: 10.11646/phytotaxa.512.1.3
Cox, C. J., Goffinet, B., Newton, A. E., Shaw, A. J., Hedderson, T. A. J. (2000). Phylogenetic relationships among the diplolepideous-alternate mosses (Bryidae) inferred from nuclear and chloroplast DNA sequences. Bryologist 103, 224–241. doi: 10.1639/0007-2745(2000)103[0224:PRATDA]2.0.CO;2
De Roo, R. T., Hedderson, T. A., Söderström, L. (2007). Molecular insights into the phylogeny of the leafy liverwort family Lophoziaceae cavers. Taxon 56, 301–314. doi: 10.1002/tax.562005
Fedosov, V. E., Fedorova, A. V., Fedosov, A. E., Ignatov, M. S. (2016). Phylogenetic inference and peristome evolution in haplolepideous mosses, focusing on Pseudoditrichaceae and Ditrichaceae s.l. Bot. J. Linn. Soc 181, 139–155. doi: 10.1111/boj.12408
Feldberg, K., Heinrichs, J., Schmidt, A. R., Váňa, J., Schneider, H. (2013). Exploring the impact of fossil constraints on the divergence time estimates of derived liverworts. Plant Syst. Evol. 299, 585–601. doi: 10.1007/s00606-012-0745-y
Friedl, T. (1996). Evolution of the polyphyletic genus Pleurastrum (Chlorophyta): inferences from nuclear-encoded ribosomal DNA sequences and motile cell ultrastructure. Phycologia 35, 456–469. doi: 10.2216/i0031-8884-35-5-456.1
Grolle, R. (1976). Verzeichnis der lebermoose Europas und benachbarter gebiete. Feddes Repert. 87, 171–279. doi: 10.1002/fedr.19760870303
Groth, H., Helms, G., Heinrichs, J. (2002). The systematic status of Plagiochila Sects. Bidentes Carl and Caducilobae Inoue (Hepaticae) inferred from NrDNA ITS sequences. Taxon 51, 675–684. doi: 10.2307/3647332
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Inoue, H., Schuster, R. M. (1974). The taxonomic status of the genus Metacephalozia Inoue. Bull. Natl. Sci. Mus 17, 161–162.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katagiri, T., Furuki, T. (2012). Checklist of Japanese liverworts and hornworts. Bryological Res. 10, 193–210.
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Mamontov, Y. S., Vilnet, A. A. (2017). Cephaloziella konstantinovae (Cephaloziellaceae, Marchantiophyta), a new leafy liverwort species from Russia and Mongolia identified by integrative taxonomy. Pol. Bot. J. 62, 1–19. doi: 10.1515/pbj-2017-0001
Milyutina, I. A., Goryunov, D. V., Ignatov, M. S., Ignatova, E. A., Troitsky, A. V. (2010). The phylogeny of Schistidium (Bryophyta, Grimmiaceae) based on the primary and secondary structure of nuclear RDNA internal transcribed spacers. Mol. Biol. 44, 883–897. doi: 10.1134/S0026893310060051
Montagne, J. F. C. (1856). Sylloge generum specierumque Cryptogamarum: quas in variis operibus descriptas iconibusque illustratas, nunc ad diagnosim reductas, nonnullasque novas interjectas, ordine systematico disposuit (Parisiis: J.-B. Baillière).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pacak, A., Szweykowska-Kulińska, Z. (2000). Molecular data concerning alloploid character and the origin of chloroplast and mitochondrial genomes in the liverwort Pellia borealis. Plant Biotechnol. J. 2, 101–108.
Patzak, S. D. F., Schäfer-Verwimp, A., Váňa, J., Renner, M. A. M., Peralta, D. F., Heinrichs, J. (2016). Chonecoleaceae (Lophocoleineae) is a synonym of Cephaloziellaceae (Cephaloziineae) and Rivulariella (Jungermanniineae) belongs to Scapaniaceae s.l. (Cephaloziineae) Phytotaxa 267, 91–102. doi: 10.11646/phytotaxa.267.2.1
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schuster, R. M. (1969). Studies on hepaticae XLVI-XLVII. On Alobiella (Spr.) schiffn. and Alobiellopsis schust. Bull. Natn. Sci. Mus. Tokyo New Ser. 12, 659–683.
Schuster, R. M. (1980). The Hepaticae and Anthocerotae of North America East of the Hundredth Meridian. Vol. IV (New York: Columbia University Press).
Söderström, L., De Roo, R., Hedderson, T. (2010). Taxonomic novelties resulting from recent reclassification of the Lophoziaceae/Scapaniaceae clade. Phytotaxa 3, 47–53. doi: 10.11646/phytotaxa.3.1.7
Söderström, L., Hagborg, A., von Konrat, M., Bartholomew-Began, S., Bell, D., Briscoe, L., et al. (2016). World checklist of hornworts and liverworts. PhytoKeys 59, 1–828. doi: 10.1093/sysbio/sys029
Spruce, R. (1882). On Cephalozia, its subgenera and some allied genera (Malton: J. W. Slater). doi: 10.5962/bhl.title.46289
Taberlet, P., Gielly, L., Pautou, G., Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/BF00037152
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Váňa, J., Söderström, L., Hagborg, A., Von Konrat, M. (2013). Notes on early land plants today. 40. Notes on Cephaloziellaceae (Marchantiophyta). Phytotaxa 112, 1–6. doi: 10.11646/phytotaxa.112.1.1
Váňa, J., Söderström, L., Hagborg, A., Von Konrat, M. (2014). Notes on early land plants today. 61. New synonyms and new combinations in Cephaloziaceae and Cephaloziellaceae (Marchantiophyta). Phytotaxa 183, 290–292. doi: 10.11646/phytotaxa.183.4.10
Keywords: Cephaloziella, Cephaloziopsis, ITS1–2, Metacephalozia, psbA, rbcL, trnG, trnL–F
Citation: Bakalin VA, Maltseva YD, Klimova KG, Nguyen V S, Choi SS and Troitsky AV (2024) New insight into the taxonomy of Cephaloziellaceae (Marchantiophyta): the family of the smallest higher plants on Earth. Front. Plant Sci. 15:1326810. doi: 10.3389/fpls.2024.1326810
Received: 23 October 2023; Accepted: 06 February 2024;
Published: 29 February 2024.
Edited by:
Gabriele Casazza, University of Genoa, ItalyReviewed by:
Llorenç Saez, Autonomous University of Barcelona, SpainYing Yu, Huangshan University, China
Copyright © 2024 Bakalin, Maltseva, Klimova, Nguyen, Choi and Troitsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vadim A. Bakalin, dmFiYWthbGluQGdtYWlsLmNvbQ==; Seung Se Choi, aGVwYXRpY2FlQGpibnUuYWMua3I=
 Vadim A. Bakalin
Vadim A. Bakalin Yulia D. Maltseva1
Yulia D. Maltseva1 Seung Se Choi
Seung Se Choi Aleksey V. Troitsky
Aleksey V. Troitsky