
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 February 2024
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1325254
This article is part of the Research Topic Molecular Biology and Omic Technologies in the Identification of Plants, Plant Derived-Products, Plant Pathogens, and Plant Microbiomes View all 12 articles
Introduction: Candidatus Liberibacter solanacearum (CLso) is a regulated plant pathogen in European and some Asian countries, associated with severe diseases in economically important Apiaceous and Solanaceous crops, including potato, tomato, and carrot. Eleven haplotypes of CLso have been identified based on the difference in rRNA and conserved genes and host and pathogenicity. Although it is pathogenic to a wide range of plants, the mechanisms of plant response and functional decline of host plants are not well defined. This study aims to describe the underlying mechanism of the functional decline of tomato plants infected by CLso by analyzing the transcriptomic response of tomato plants to CLso haplotypes A and B.
Methods: Next-generation sequencing (NGS) data were generated from total RNA of tomato plants infected by CLso haplotypes A and B, and uninfected tomato plants, while qPCR analysis was used to validate the in-silico expression analysis. Gene Ontology and KEGG pathways were enriched using differentially expressed genes.
Results: Plants infected with CLso haplotype B saw 229 genes upregulated when compared to uninfected plants, while 1,135 were downregulated. Healthy tomato plants and plants infected by haplotype A had similar expression levels, which is consistent with the fact that CLso haplotype A does not show apparent symptoms in tomato plants. Photosynthesis and starch biosynthesis were impaired while starch amylolysis was promoted in plants infected by CLso haplotype B compared with uninfected plants. The changes in pathway gene expression suggest that carbohydrate consumption in infected plants was more extensive than accumulation. In addition, cell-wall-related genes, including steroid biosynthesis pathways, were downregulated in plants infected with CLso haplotype B suggesting a reduction in membrane fluidity, cell signaling, and defense against bacteria. In addition, genes in phenylpropanoid metabolism and DNA replication were generally suppressed by CLso infection, affecting plant growth and defense.
Discussion: This study provides insights into plants’ defense and functional decline due to pathogenic CLso using whole transcriptome sequencing and qPCR validation. Our results show how tomato plants react in metabolic pathways during the deterioration caused by pathogenic CLso. Understanding the underlying mechanisms can enhance disease control and create opportunities for breeding resistant or tolerant varieties.
Candidatus Liberibacter solanacearum (CLso) is a phloem-limited pathogen associated with diseases in many Apiaceous and Solanaceous plants, including potato, tomato, carrot, pepper, eggplant, celery, and leek (Hansen et al., 2008; Liefting et al., 2008; Sumner-Kalkun et al., 2020). This pathogen is associated with foliar dieback in susceptible plants, and causes zebra chip disease of potato (Li et al., 2013). The CLso has a wide range of psyllid vectors, including Bactericera cockerelli, B. trigonica, and Trioza apicalis (Munyaneza et al., 2007b; Sumner-Kalkun et al., 2020). Also, it has been suggested that non-solanaceous psyllids might be the potential vectors of CLso, and therefore more plant hosts might carry the bacteria (Borges et al., 2017; Swisher Grimm and Garczynski, 2019).
CLso is categorized into haplotypes based on single nucleotide polymorphisms (SNPs) in ribosomal RNA and housekeeping genes. Different haplotypes vary in pathogenicity to different host plants (Mendoza-Herrera et al., 2018; Swisher Grimm et al., 2018; Harrison et al., 2019; Harrison et al., 2020; Sumner-Kalkun et al., 2020). Until now, 15 haplotypes of CLso have been identified around the world, including A, B (Wen et al., 2009), C (Munyaneza et al., 2010b), D (Nelson et al., 2013), E (Teresani et al., 2014), F (Swisher Grimm and Garczynski, 2019), G (Mauck et al., 2019), H (Haapalainen et al., 2020), H(Con) (Contreras-Rendon et al., 2020), U (Haapalainen et al., 2018), Cras1, Cras2 (Sumner-Kalkun et al., 2020), Aph1, Aph2, and Aph3 (Grimm et al., 2022). Haplotypes A, B and F are pathogens of ZC in the United States, with haplotypes A and B also occurring in Mexico and New Zealand (Hansen et al., 2008; Liefting et al., 2009; Munyaneza et al., 2009; Antolinez et al., 2017; Haapalainen et al., 2020). Haplotype C was found in carrots in Finland, Sweden and Norway (Nelson et al., 2013). Haplotype D was found in carrots and celeries in Spain, the Canary Islands, Southern Europe and Morocco (Nelson et al., 2013; Alfaro-Fernández et al., 2017). Haplotype E was found in Southern Europe, including Spain and Morocco, in carrot and celery (Alfaro-Fernández et al., 2017; Ben Othmen et al., 2018). Haplotype G was discovered and recovered in herbarium specimens of wild species (Solanum elaeagnifolium, S. americanum, and S. umbelliferum), indicating that CLso has been in South America since at least 1970 (Mauck et al., 2019). Haplotype U was found in Urtica dioica in Finland (Haapalainen et al., 2018). Cras1 and Cras2 were found in psyllid vectors in Scotland (Sumner-Kalkun et al., 2020). Aph1, Aph2 and Aph3 were identified in psyllid vectors collected from yellow sticky cards near potato farms in southern Oregon of the United States; the impact of Aph1 to Aph3 on crops is not known (Grimm et al., 2022)
In general, CLso is injected into the phloem through the psyllids’ saliva. If infected, plants cannot offer an effective immune response, resulting in CLso reproduction in the phloem. This blocks the nutrition transmission in the phloem, inducing erectness and stunting of new foliage, basal cupping of leaves with chlorosis and purpling, upward curving or scorching of all leaves, compressed and enlarged terminal internodes leading to resetting, hypertrophic nodes, axillary branches or tubers on the ground, breach of fruit set, and production of plenty of tiny, misshapen, and bastard fruits (Munyaneza et al., 2007a; Munyaneza et al., 2007b; Liefting et al., 2009; Secor et al., 2009; Crosslin et al., 2010; Munyaneza et al., 2010a; Munyaneza, 2012).
The underground symptoms in infected potatoes include folded stolons, browning of vascular tissue accompanying necrotic flecking of internal tissues, and dark medullary rays, all of which deteriorate throughout the tuber. Upon frying, the tuber-related symptoms are more evident, and chips and crisps present grey blotches and stripes which lead to loss of business value and privation of farmers (Munyaneza et al., 2007a; Munyaneza et al., 2007b; Secor et al., 2009; Crosslin et al., 2010; Miles et al., 2010; Goolsby et al., 2012; Munyaneza, 2012). The term “Zebra Chip disease” generally refers to those symptoms of potato tuber (Munyaneza et al., 2007a; Munyaneza et al., 2007b; Munyaneza, 2012). In carrots with infection of CLso, symptoms include leaf twist and discoloration, stunting of shoots and roots, and proliferation of secondary roots (Munyaneza et al., 2010a; Munyaneza et al., 2010b; Alfaro-Fernandez et al., 2012a; Alfaro-Fernandez et al., 2012b).
Growth experiments show CLso-A-infected tomato plants were stunted after three weeks of infection, but the heights of plants were not significantly less than the negative control group (Harrison et al., 2022). The CLso-B-infected tomato plants’ growth was considerably impaired (Harrison et al., 2022).
Analysis shows that most of the differentially expressed genes were down-regulated in the CLso samples; those genes were generally involved in plant defense against stressors, growth, plant metabolism, transport and signaling and transcription/translation (Levy et al., 2017; Harrison et al., 2022).
Among the genes for enzymes involved in cellulose synthesis in the cell wall, cellulose synthase-like A1, transferase (transferring glycosyl groups), UPA15, glycosyltransferase (CAZy family GT2), and cellulose synthase decreased in transcript abundance (FPKM) in CLso-infected samples comparing to CLso-free samples (Levy et al., 2017). Cell wall modification genes, expansin, XTH3, xyloglucan endotransglycosylase, and xyloglucan endotransglucosylase, were also down-regulated with CLso-infection (Levy et al., 2017).
Although pathogenicity variation is observed, the host response mechanism to the pathogen has not been well studied. This study aims to describe the underlying mechanism of the functional decline of plant hosts infected by CLso by analyzing the transcriptomic response of tomato plants to CLso haplotypes A and B.
A total of 30 ‘Moneymaker’ tomato plants were planted for four weeks, with each plant treated with five male psyllids carrying the same CLso haplotype for 5-7 days in a whole plant cage. The psyllids were vectors of CLso haplotype A or B. Four weeks after inoculation, the DNA of stems and leaves was extracted and purified using the MagneSil KF, Genomic kit according to the manufacturer’s protocol. All tomato plants were tested with CLso PCR primers CLipoF/OI2c (Liefting et al., 2009; Secor et al., 2009) (16S rRNA), and the positive plants were also tested with the 50s rplJ/rplL primers [CL514F/CL514R (Munyaneza et al., 2009)]. PCR gel products of positive samples were extracted using QIAquick Gel Extraction Kit according to the manufacturer’s protocol and sent out for Sanger sequencing at the Ottawa Hospital Research Institute. The raw sequences were trimmed using a 0.01 quality score in CLC Genomics Workbench 20.0.2. Clasnip (the alpha version) was used to confirm CLso haplotypes using the trimmed sequences (Chuan et al., 2023). Grafting was also performed using the branches of tomato plants infected by CLso, which had a probability of producing newly infected plants. The new plants were tested CLso with the same primers as well. Grafted plants were used to maintain the unculturable pathogen, and they were not included in the RNA-Seq samples.
Four weeks after inoculation, total RNA of stem and leaves was extracted from tomato plants infected by CLso haplotype A, tomato plants infected by haplotype B, and healthy tomato plants. NGS sequencing libraries were generated from total RNA using the Illumina TruSeq Library Preparation Kit (300 bp×2) and then sequenced using the Illumina MiSeq Platform. The tomato SL4.0 genome and ITAG4.1 annotation reference were downloaded from the Sol Genomics Network (https://solgenomics.net/). Atria v2.1.0 was used to trim adapter sequences and low-quality reads (Chuan et al., 2021). Salmon v0.12.0 was used for transcript quantification (Patro et al., 2017). The ITAG4.1 coding sequence reference contained hypothetical proteins. However, these proteins lacked experimental evidence to confirm that they could be expressed in vivo. Therefore, coding sequences were removed if they had less than 10x coverage in half of the samples. DESeq2 was used to find differentially expressed genes (DEGs) (Love et al., 2014). DEGs were calculated while controlling for plant and batch variables (Table 1). The ITAG4.1 coding sequences were annotated with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using Blast2GO v1.1.0 (Gotz et al., 2008) and InterPro v70.0 (Paysan-Lafosse et al., 2023). After that, GO and KEGG pathways were enriched using DEGs and visualized with ClusterProfiler v4.2.1 (Wu et al., 2021).
In addition, we used qPCR analysis to validate the gene expression levels estimated from NGS sequencing. We selected 54 (including two internal control) genes to design primers using CLC Genomics Workbench 20.0.2. Those genes were DEGs or enriched in GO or KEGG pathways. Up to three primer sets were designed for each gene. The primers were preliminarily tested with normal PCR using extracted total DNA of tomato samples. Suitable primers were then used to quantify the gene expression levels between three CLso haplotype B infected tomato plants and three healthy tomato plants using SensiMix II Probe Mix and Eva Green dye. Each reaction had two technical replications. A reaction would be re-run if the Ct values of technical replications were not in a reasonable range, such as the Ct difference greater than two. Delta-delta Ct value was used to compute the gene’s expression levels. The expression levels using qPCR and RNA-Seq analyses were tested using Pearson correlation.
A total of 27 samples were successfully sequenced (Table 1). Three samples failed library preparation because of low RNA concentration. The number of clean read pairs ranged from 1.9 to 7.1 million (Table 1). The average read length is around 135 bp (Table 1). The mapping rates of tomato transcriptome varied from 5.3% to 63.3% (Table 1), which might be a result from diverse pathogen to host ratio and rRNA to mRNA ratio. The mean coverage of transcripts ranged from 10.6 to 413.6 (Table 1), indicating all samples had enough sequencing depth for RNA-Seq analysis.
In total, the expression levels of 9,888 genes in tomato plants were quantified in RNA-Seq analysis. The volcano plot illustrated the statistical significance (adjusted P value) versus the magnitude of change (fold change) of DEG results (Figure 1). We selected the differentially expressed genes using adjusted P value (<0.05) and fold change (≥3) (Figures 1–3; Supplementary Table 1). Thus, comparing haplotype B to the negative control, 1,364 DEGs were selected, among which 229 genes were upregulated and 1,135 were downregulated (Figures 2, 3). The Principal Component Analysis (PCA) shows that the principal component 1 (PC1) comprised 68% variance and could be used to differentiate CLso haplotype B and other groups (Figure 2), implying that the expression level of tomato plants infected by haplotype B varied from healthy tomato plants and those infected by CLso haplotype A.
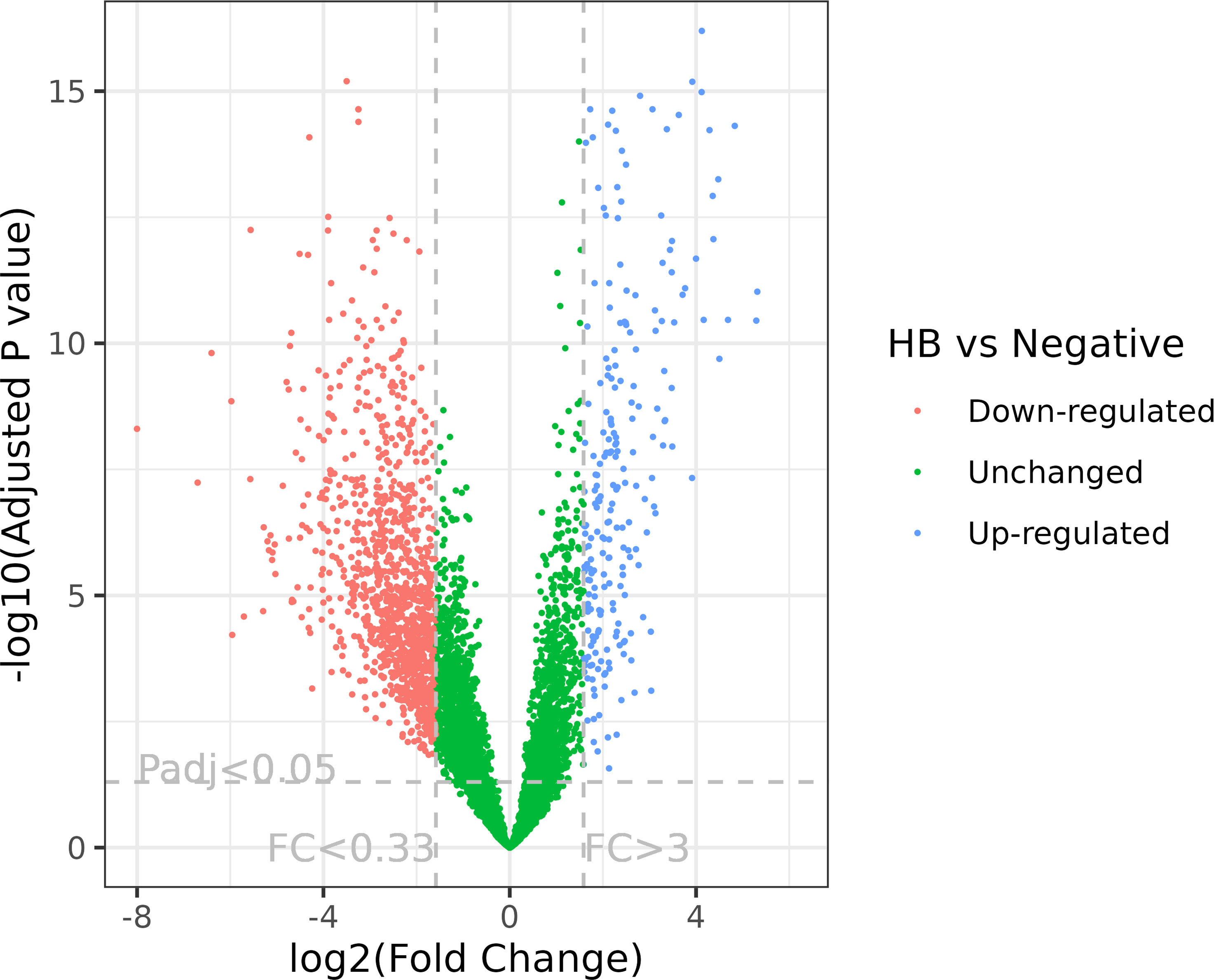
Figure 1 Volcano plot of RNA-Seq expression analysis of CLso haplotype B versus negative plants. FC, fold change; Padj, adjusted P value. The fold change and adjusted P value cutoffs are displayed.
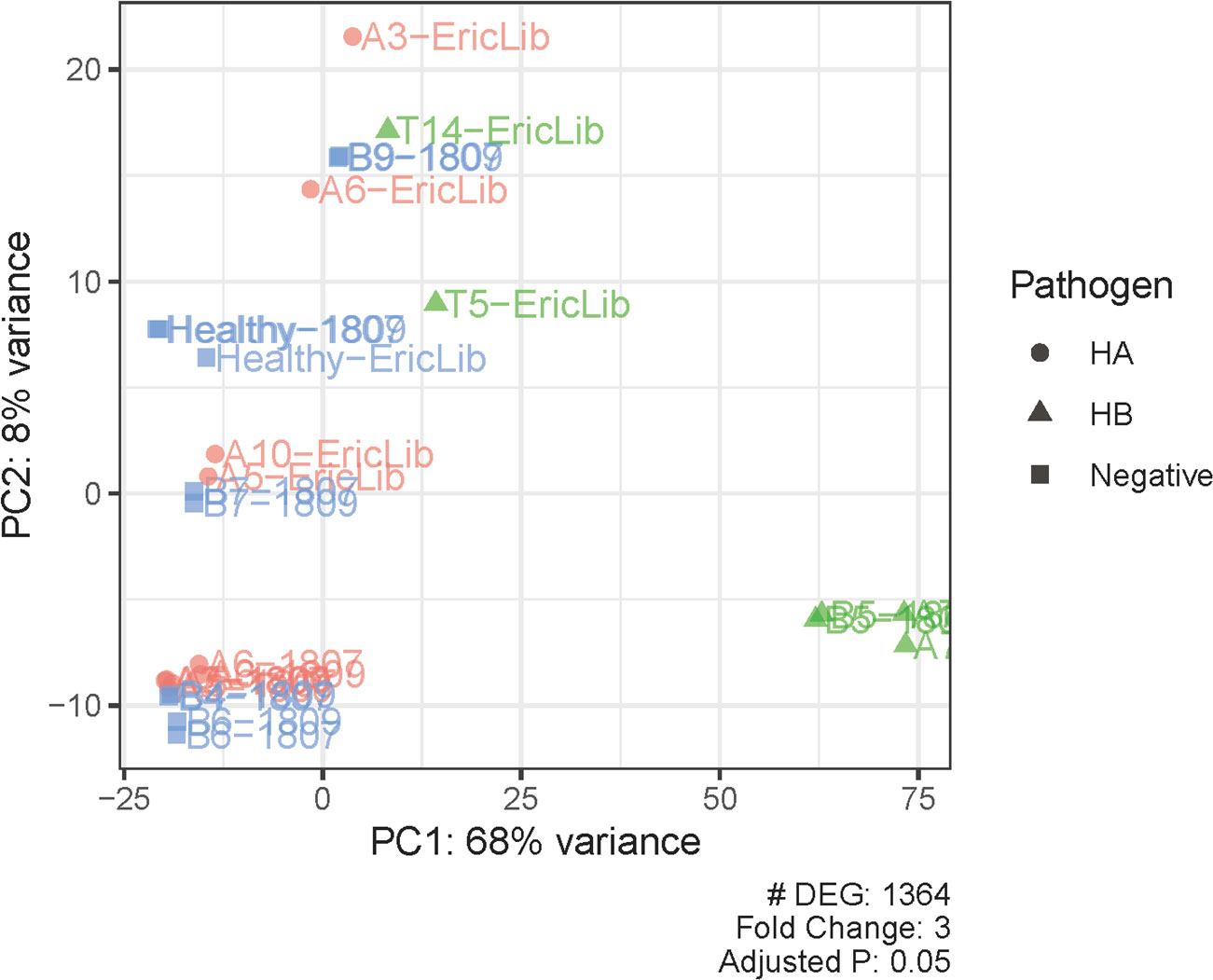
Figure 2 Principal component analysis of differentially expressed genes and samples. PC, principal component. Pathogen statuses are marked with colors and shapes.
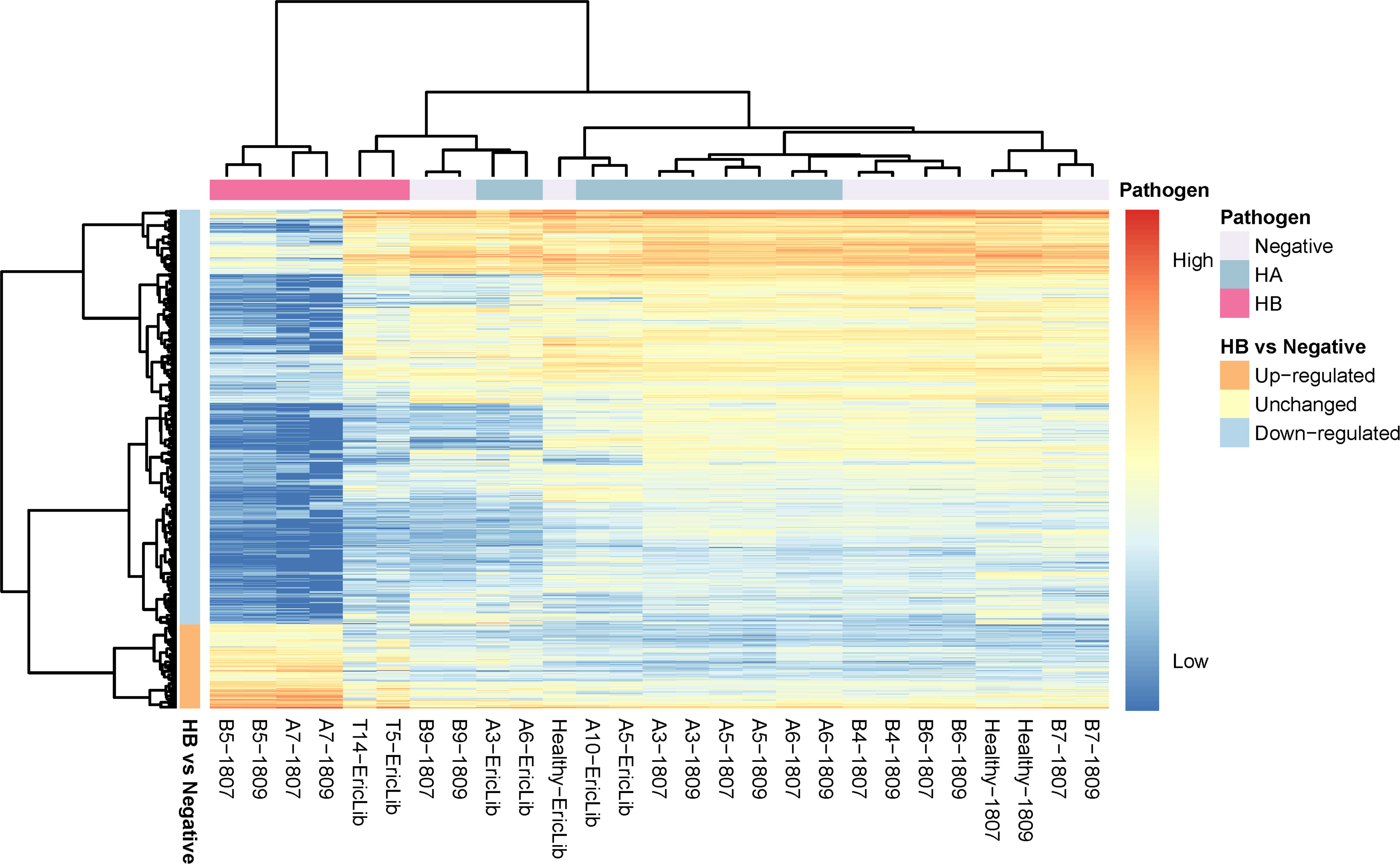
Figure 3 Heatmap of deferentially expressed genes of tomato hosts infected by CLso haplotypes. The column labels are in the sample-batch format, and the rows are deferentially expressed genes (DEGs). The high expression level marks are red, and the low expression level is blue. The pathogen information is labeled at the top. The directions of gene expression (up-regulated, down-regulated) are labeled on the left.
Healthy tomato plants and plants infected by haplotype A had similar expression levels, consistent with observations that CLso haplotype A does not show apparent symptoms in tomato plants (Figures 2, 3). In the hosts infected by CLso haplotype B, the expression levels were generally in a distinct cluster compared to healthy plants and plants infected by CLso haplotype A, despite the fact that T5 and T14 had relatively different patterns in gene expression compared with other haplotype B samples, which was likely the result of batch effect or latent infection (Figures 2, 3).
Figure 4 illustrates the symptoms observed in tomato plants four weeks post-inoculation. The two plants on the left belong to one batch, while the three plants on the right are from a different batch (Figure 4). Notably, the growth patterns of the healthy plants varied between batches (Figure 4). The first healthy plant, despite having larger leaves, was shorter in stature compared to the second healthy plant (Figure 4).

Figure 4 Symptoms of tomato plants infected by CLso. Healthy plants were PCR negative. HA, CLso haplotype A. HB, CLso haplotype B.
The plants with haplotype B also exhibited distinct symptoms (Figure 4). The entirety of the first haplotype B plant’s leaves were curled and displayed yellowing (Figure 4). In contrast, the second haplotype B plant had one stunted stem with scorched leaves, while its other stems appeared healthier, showing no severe curling or yellowing of the foliage (Figure 4).
These observations align with the latent CLso infection previously reported in grafted tomato plants (Li et al., 2013). It’s worth noting that the batch of T5 and T14 differed from the other haplotype B plants, and they could be latent CLso carriers with no severe symptoms at the time of RNA extraction.
Gene set enrichment analysis of gene ontology (GO) identified genes in three categories: cellular component, molecular function, and biological process (Supplementary Table 2). The enrichment map and associations between GO terms are illustrated in Figure 5. The gene ratio within each GO term is shown in Figure 6. The connection between GO terms and genes is depicted using a gene concept network (Figure 7), and the heatmaps of GO terms and detailed gene annotation are plotted (Supplementary Figures 1–8). KEGG pathways were enriched using the over-representation and gene set methods (Supplementary Table 3). The pathways of steroid biosynthesis, phenylpropanoid biosynthesis, flavonoid biosynthesis, DNA replication, and hormone signal transduction are shown and enriched genes are colored with expression levels (Supplementary Figures 10–13).
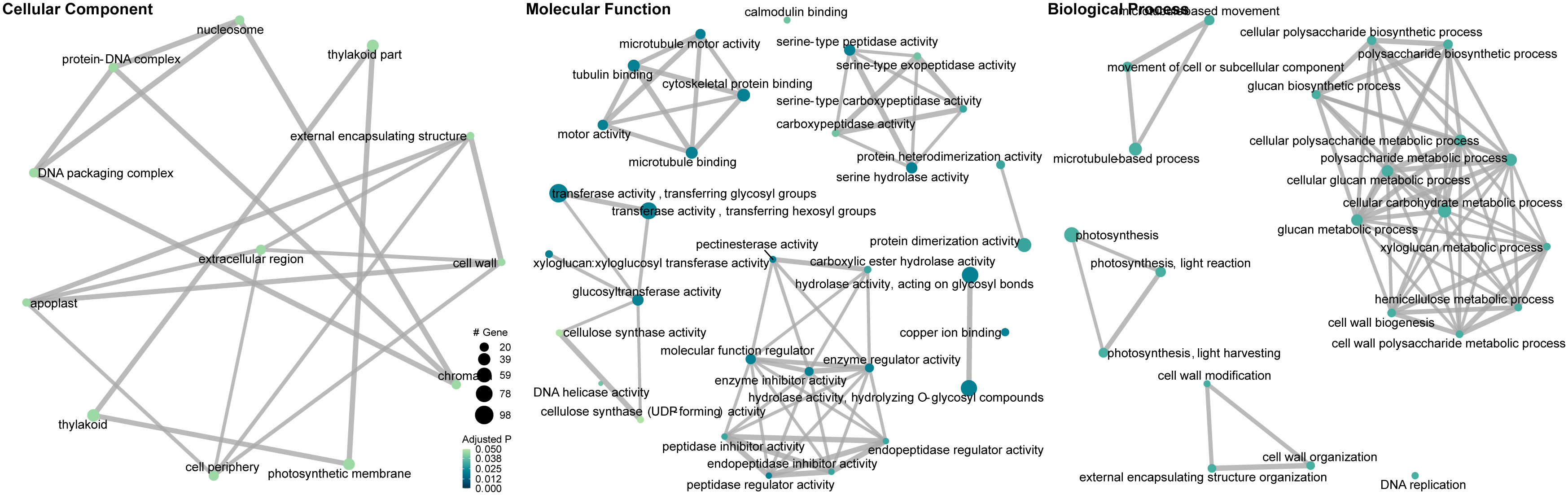
Figure 5 Gene ontology enrichment map and associations between gene ontology terms (CLso haplotype B vs. negative). Dots are GO terms. Grey lines connect adjacent GO terms. Dot size indicates number of DEGs. Darker dot color means lower adjusted P value.
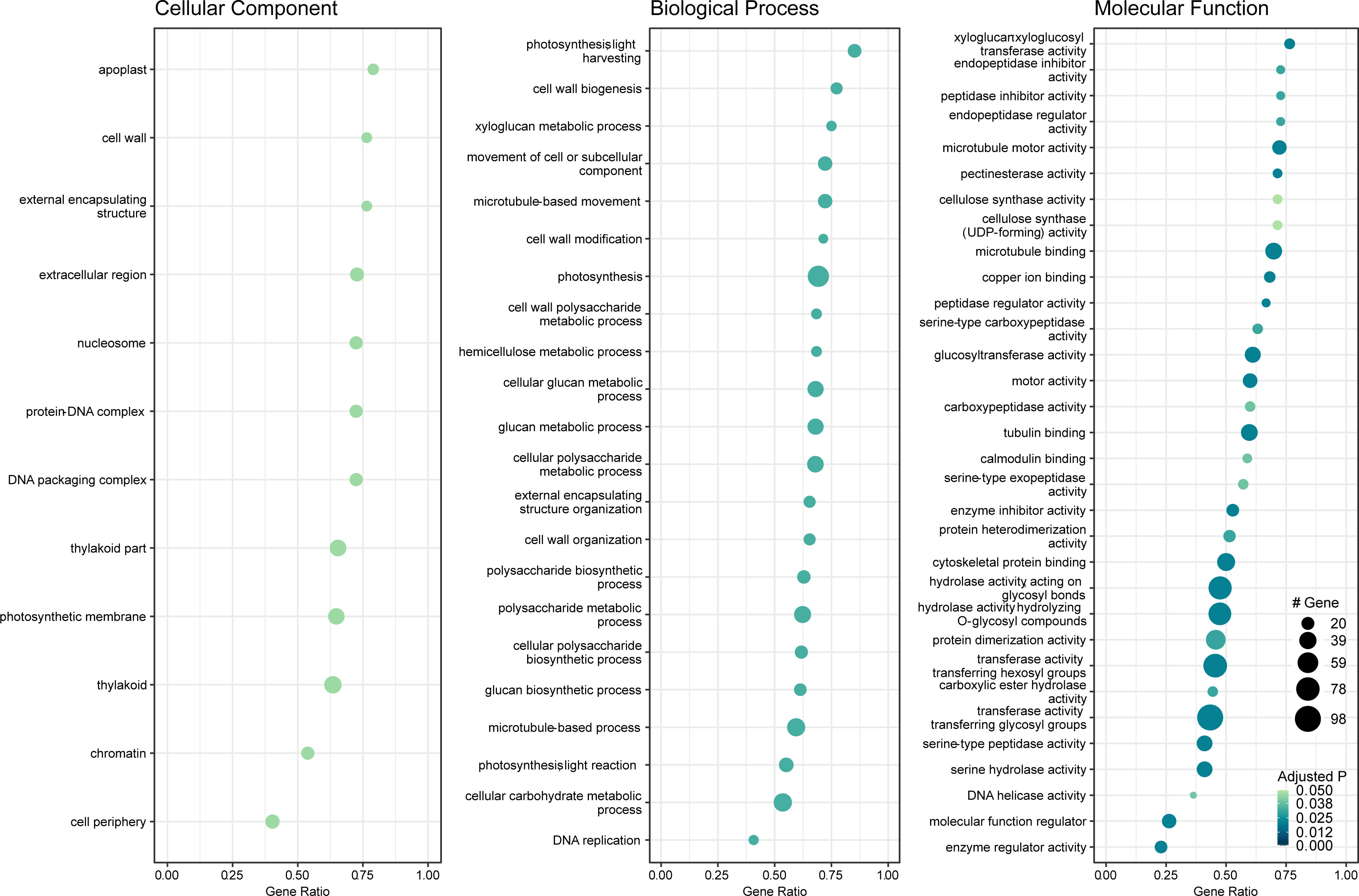
Figure 6 Dot plot of gene set enrichment analysis of gene ontology (GO) (CLso haplotype B vs. negative). X-axis is DEG ratio. Y-axis is the GO terms. Dot size indicates the number of DEGs. Darker dot color means lower adjusted P value.
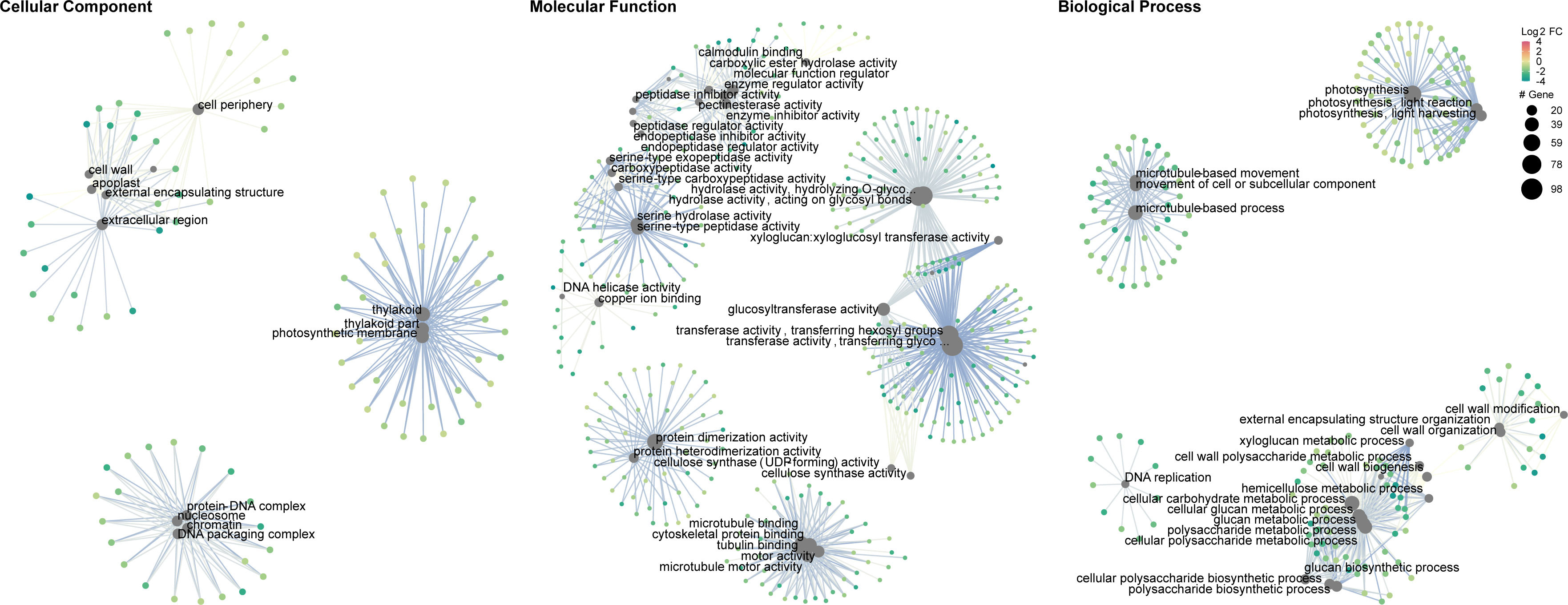
Figure 7 Gene concept network of gene ontology (CLso haplotype B vs. negative). Big grey dots are GO terms. Sizes of grey dots indicate the number of DEGs. Small dots represent genes, colored with fold changes of gene expression levels.
Plants absorb energy from sunlight to synthesize glucose in the chloroplasts, and this process is called photosynthesis. Some glucose is further processed to form starch for energy storage. The energy metabolism of plants can be altered when they are exposed to biotic stress, including bacterial pathogen invasion (Berger et al., 2004).
Photosynthesis, especially in the light reactions, was significantly compromised in CLso haplotype B infected tomato plants (Figures 5–7). The genes in the cellular components of the photosynthetic membrane and thylakoid were downregulated (Figure 7). Photosystems I and II are the two protein complexes with pigments to catalyze primary photosynthetic reactions. Proteins in both photosystem I (PsaE, PsaF, PsaG, PsaH, and PsaL) and photosystem II (PsbO, PsbP, PsbQ, and PsbW) were downregulated in CLso haplotype B infected tomato (Figure 8). Electron transport proteins (PetE, PetF, and PetH) and F-type ATPases were also downregulated (Figure 8).
Concurrently, starch biosynthesis was significantly affected in plants infected by the pathogenic CLso (Figures 7, 8). Core enzymes catalyzing starch biosynthesis from glucose (AGPase [LOC543968] and GBSSI) were downregulated (Figure 8).
Thus, the systematic under expression of both photosynthetic and starch biosynthetic genes influenced the carbon fixation and energy harvesting of the tomato hosts, which would potentially lead to plant wilt and malnutrition.
On the contrary, starch degradation, was upregulated in tomato plants infected by CLso haplotype B (Figure 8). Specifically, alpha-amylase (LOC101257661) and beta-amylase 1 (BAM1) were upregulated (Figure 8). Those alternations indicate that carbohydrate consumption in infected plants was highly active. CLso encodes a glucose/galactose transporter, so it is possible to utilize glucose and galactose from host plants (Lin et al., 2011). This implies that the energy production of infected plants was insufficient for plants to grow and maintain a healthy state. The plants had to use starch to produce disaccharides and monosaccharides, which are uptaken by CLso.
The impact of starch metabolism is more profound in potatoes than in tomatoes. Starch formation and accumulation affect tuber sizes, directly influencing potato yield and profit. Potato tubers infected by CLso caused the conversion of potato starch to water-soluble sugars, which developed discoloration along with the vascular tissue, causing the chips to have unsightly dark blotches, stripes, or streaks after cooking (Pitman et al., 2011; Rashed et al., 2013).
Chromatin, nucleosome, protein-DNA complexes and DNA packing complexes were enriched and clustered in cellular components (Figure 5). Expression variation of those components directly influences the binding factors to DNA, which are critical for gene regulation (Ramirez-Prado et al., 2018; Ahmad et al., 2022). Histones comprise protein complexes to package genomic DNA and form chromatin. Histones H1, H2A, H2B, and H3 were downregulated in CLso haplotype B infected tomato plants (Supplementary Figure 1). The six minichromosome maintenance protein complex (MCM) proteins (MCM2 - MCM7) forming the MCM complexes were downregulated, which affected genomic DNA replication (Supplementary Figure 12) (Maine et al., 1984). This indicates that biotic stress suppressed the expression of histone genes and hindered gene transcription in plants (Yuan et al., 2013).
The cell wall is the outer structure that prevents bacteria from penetrating the host defense mechanisms, and is essential in growth-regulating signal transduction (Rui and Dinneny, 2020). Tomato plants infected by CLso haplotype B expressed lower levels of cell wall modification and biosynthesis proteins, which might fail to prevent the bacteria from initiating direct contact with host cells. Successful adaptation to abiotic stress response in the cell wall is often related to an increased expression of xyloglucan endotransglucosylase/hydrolase (XTH) and expansin proteins (Le Gall et al., 2015). In response to pathogenic CLso, the level of XTH was depressed, implying that plant defense was suppressed by the biotic stress (Supplementary Figures 1, 4).
Besides xyloglucan, pectin metabolism may also play an important role in plant defense response and cell wall integrity (Wang et al., 2023a; Wang et al., 2023b). Pectin is also vital to intercellular communication and signal transduction (Shin et al., 2021). Both pectinesterase and its inhibitors are down-regulated, relating to the antagonistic action of cell-wall breakdown (Supplementary Figure 4).
Steroids are essential components of cell membranes, altering membrane fluidity and functioning as signaling molecules. The steroid biosynthesis pathway was significantly impaired in tomato plants infected by CLso haplotype B (Supplementary Figure 9). Key proteins in the steroid biosynthesis pathway were all downregulated, such as sterol side chain reductase (SISSR1), sterol C14-demethylase (CPY51), and delta14-sterol reductase (FK), resulting insufficient production of cholesterol and other phytosterols (Supplementary Figure 9). A fungal and maize study also reports steroids are essential components of network regulation of plant immunity (Agostini et al., 2019).
Phenylpropanoids are one of the largest classes of secondary metabolites, including flavonoids, anthocyanins, monolignols, and tannins, functioning in photosynthesis, growth regulation, nutrient process, and stress response (Pratyusha and Sarada, 2022). Phenylpropanoids are required for plant immune response to biotic and abiotic stresses (Bauters et al., 2021). In response to CLso haplotype B, the gene expression levels of the tomato plants were generally suppressed, such as cinnamoyl-CoA reductase 2 (CCR2), peroxidase (CEVI-1) and hydroxycinnamoyl CoA quinate transferase (HQT) (Supplementary Figure 10). Carrying the pathogen, the host cannot produce sufficient secondary metabolites for plant development and defense, including scopoline, coumarin, lignin, coniferin, and syringin (Supplementary Figure 10).
The flavonoid biosynthesis pathway was also widely suppressed, such as chalcone synthase 1 (CHS1) and chalcone isomerase 1 (CHI1), flavanone 3-dioxygenase (F3H) and hydroxycinnamoyl CoA quinate transferase (HQT) (Supplementary Figure 11). The function of flavonoids spans from plant development and pigmentation to defense and signaling between plants and microorganisms (Mathesius, 2018).
Plant hormones regulate downstream signaling components through a core pathway or independently from pathways (Bunsick et al., 2021), and are essential in plant growth, development, and stress responses (Jaillais and Chory, 2010; Ku et al., 2018).
KEGG analysis shows that disease resistance and stomatal closure pathways were promoted, and cell elongation, enlargement, and division signals were suppressed in the tomato plants infected by CLso haplotype B (Supplementary Figure 13). Specifically, in disease resistance, NPR1 was down-regulated, while its downstream genes TGA and PR-1 were upregulated (Supplementary Figure 13). NPR1 is the key regulator of salicylic acid in the systemic acquired resistance pathways (Fan and Dong, 2002). In the nucleus, NPR1 interacts with TGA transcription factors to promote the expression of PR genes (Chen et al., 2019). The proteins encoded by those PR genes triggered broad-spectrum resistance to pathogens (Fan and Dong, 2002; Kesarwani et al., 2007; Ding et al., 2020). However, CLso can counteract expression of NPR1 by producing salicylate hydroxylase to degrade salicylic acid (Wang et al., 2021; Levy et al., 2023). This explains why NPR1 was down-regulated in the CLso-B infected tomato plants.
ABF and snRK2 were promoted for stomatal closure and seed dormancy (Supplementary Figure 13), which play a crucial role in plant responses to environmental stresses (Nakashima and Yamaguchi-Shinozaki, 2013; Rehman et al., 2021). The signal transduction genes of cell enlargement and plant growth were generally suppressed, such as AUX1, TIR1, IAA21, ARF, GH3, and SAUR (Supplementary Figure 13). Cell elongation and division genes BZR1, BZR2, TCH4 and CYCD3 were impaired (Supplementary Figure 13). A-AAR and B-AAR, all of which are related to cell division and shoot initiation, were suppressed (Supplementary Figure 13). Those suppressed genes align with the plant’s mechanism against pathogens, which can be seen as a trade-off between growth and defense, where the plant reallocates its resources from growth to defense in response to pathogen attack (Zhang et al., 2022).
In silico RNA-Seq expression was validated using qPCR analysis. We successfully designed 150 primer sets for the 54 selected genes (Supplementary Table 4). In the preliminary primer validation, 52 primer sets were selected for 52 genes, except that two genes, Solyc05g052280.3.1 (PSEP7) and Solyc01g107590.3.1 (CAD1), had no valid primer set (Supplementary Table 4). The expression levels of 47 genes were successfully generated from qPCR analysis, and five genes failed (Supplementary Tables 4, 5). Among the five failed genes, two [Solyc07g052510.4.1 (PSE3) and Solyc02g083490.3.1 (PSE64)] had no ΔCt value for positive samples, Solyc04g077970.5.1 (APRT) was an unused internal control, and two [Solyc02g082930.3.1 (CHI17) and Solyc02g030170.4.1 (SISSR1)] were failed because of primer design or low concentration (Supplementary Table 5). The Pearson correlation coefficient between the expression levels of qPCR and Bioinformatics was 0.62 (P value = 2.75×10-5) (Figure 9), indicating the expression levels of qPCR and RNA-Seq were consistent, and the differential expression analysis of RNA-Seq was reliable.
This study provided insights into plants’ defense and functional decline to pathogenic CLso, using whole transcriptome sequencing and qPCR validation. Our results showed how tomato plants react in metabolic pathways during the deterioration caused by pathogenic CLso. Understanding the underlying mechanisms can enhance disease control and create opportunities for breeding resistant or tolerant varieties.
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author/s.
JC: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JN: Methodology, Writing – review & editing. WRC: Methodology, Resources, Writing – review & editing. WC: Data curation, Formal analysis, Resources, Software, Writing – review & editing. LH: Project administration, Resources, Supervision, Writing – review & editing. XL: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was partially funded by the Interdepartmental funding of Genomics Research and Development Initiatives (GRDI) # CHA-P-3058 (N000469), Canada to XL. The financial support of CFIA and University of Prince Edward Island to JC is greatly appreciated.
The advice, encouragement, and support of Drs. Christian Lacroix and Stevan Springer to JC are greatly appreciated. Copy Right © His Majesty the King in Right of Canada, 2024.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor GB declared a shared affiliation with the author XL, JC, and JN at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1325254/full#supplementary-material
Agostini, R. B., Postigo, A., Rius, S. P., Rech, G. E., Campos-Bermudez, V. A., Vargas, W. A. (2019). Long-lasting primed state in maize plants: salicylic acid and steroid signaling pathways as key players in the early activation of immune responses in silks. Mol. Plant-Microbe Interact. 32, 95–106. doi: 10.1094/MPMI-07-18-0208-R
Ahmad, K., Henikoff, S., Ramachandran, S. (2022). Managing the steady state chromatin landscape by nucleosome dynamics. Annu. Rev. Biochem. 91, 183–195. doi: 10.1146/annurev-biochem-032620-104508
Alfaro-Fernandez, A., Cebrian, M. C., Villaescusa, F. J., de Mendoza, A. H., Ferrandiz, J. C., Sanjuan, S., et al. (2012b). First report of 'Candidatus Liberibacter solanacearum' in carrot in mainland Spain. Plant Dis. 96, 582. doi: 10.1094/PDIS-11-11-0918-PDN
Alfaro-Fernández, A., Hernández-Llopis, D., Font, M. I. (2017). Haplotypes of ‘Candidatus Liberibacter solanacearum’ identified in Umbeliferous crops in Spain. Eur. J. Plant Pathol. 149, 127–131. doi: 10.1007/s10658-017-1172-2
Alfaro-Fernandez, A., Siverio, F., Cebrian, M. C., Villaescusa, F. J., Font, M. I. (2012a). 'Candidatus Liberibacter solanacearum' associated with Bactericera trigonica-affected carrots in the Canary Islands. Plant Dis. 96, 581. doi: 10.1094/PDIS-10-11-0878-PDN
Antolinez, C. A., Fereres, A., Moreno, A. (2017). Risk assessment of 'Candidatus Liberibacter solanacearum' transmission by the psyllids Bactericera trigonica and B-tremblayi from Apiaceae crops to potato. Sci. Rep. 7, 45534. doi: 10.1038/srep45534
Bauters, L., Stojilkovic, B., Gheysen, G. (2021). Pathogens pulling the strings: effectors manipulating salicylic acid and phenylpropanoid biosynthesis in plants. Mol. Plant Pathol. 22, 1436–1448. doi: 10.1111/mpp.13123
Ben Othmen, S., Morán, F. E., Navarro, I., Barbé, S., Martínez, C., Marco-Noales, E., et al. (2018). ‘Candidatus Liberibacter solanacearum’ haplotypes D and E in carrot plants and seeds in Tunisia. J. Plant Pathol. 100, 197–207. doi: 10.1007/s42161-018-0045-7
Berger, S., Papadopoulos, M., Schreiber, U., Kaiser, W., Roitsch, T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant 122, 419–428. doi: 10.1111/j.1399-3054.2004.00433.x
Borges, K. M., Cooper, W. R., Garczynski, S. F., Thinakaran, J., Jensen, A. S., Horton, D. R., et al. (2017). "Candidatus Liberibacter solanacearum" associated with the psyllid, Bactericera maculipennis (Hemiptera: Triozidae). Environ. Entomology 46, 210–216. doi: 10.1093/ee/nvw174
Bunsick, M., McCullough, R., McCourt, P., Lumba, S. (2021). Plant hormone signaling: is upside down right side up? Curr. Opin. Plant Biol. 63, 102070. doi: 10.1016/j.pbi.2021.102070
Chen, J., Mohan, R., Zhang, Y., Li, M., Chen, H., Palmer, I. A., et al. (2019). NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Chuan, J., Xu, H., Hammill, D. L., Hale, L. R., Chen, W., Li, X. (2023). Clasnip: a web-based intraspecies classifier and multi-locus sequence typing for pathogenic microorganisms using fragmented sequences. PeerJ 11, e14490. doi: 10.7717/peerj.14490
Chuan, J., Zhou, A., Hale, L. R., He, M., Li, X. (2021). Atria: an ultra-fast and accurate trimmer for adapter and quality trimming. Gigabyte 2021, 1–18. doi: 10.46471/gigabyte.31
Contreras-Rendon, A., Sanchez-Pale, J. R., Fuentes-Aragon, D., Alanis-Martinez, I., Silva-Rojas, H. V. (2020). Conventional and qPCR reveals the presence of 'Candidatus Liberibacter solanacearum' haplotypes A, and B in Physalis philadelphica plant, seed, and Betaactericera cockerelli psyllids, with the assignment of a new haplotype H in Convolvulaceae. Antonie Van Leeuwenhoek 113, 533–551. doi: 10.1007/s10482-019-01362-9
Crosslin, J. M., Munyaneza, J. E., Brown, J. K., Liefting, L. W. (2010). Potato zebra chip disease: a phytopathological tale. Plant Health Prog. 11, 33. doi: 10.1094/PHP-2010-0317-01-RV
Ding, Y., Dommel, M. R., Wang, C., Li, Q., Zhao, Q., Zhang, X., et al. (2020). Differential quantitative requirements for NPR1 between basal immunity and systemic acquired resistance in Arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.570422
Fan, W., Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Goolsby, J. A., Adamczyk, J. J., Crosslin, J. M., Troxclair, N. N., Anciso, J. R., Bester, G. G., et al. (2012). Seasonal population dynamics of the potato psyllid (Hemiptera: Triozidae) and its associated pathogen "Candidatus Liberibacter solanacearum" in potatoes in the Southern Great Plains of North America. J. Econ Entomol 105, 1268–1276. doi: 10.1603/EC11435
Gotz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grimm, K. D. S., Horton, D. R., Lewis, T. M., Garczynski, S. F., Jensen, A. S., Charlton, B. A. (2022). Identification of three new 'Candidatus Liberibacter solanacearum' haplotypes in four psyllid species (Hemiptera: Psylloidea). Sci. Rep. 12, 20618. doi: 10.1038/s41598-022-24032-9
Haapalainen, M., Latvala, S., Wickström, A., Wang, J., Pirhonen, M., Nissinen, A. I. (2020). A novel haplotype of ‘Candidatus Liberibacter solanacearum’ found in Apiaceae and Polygonaceae family plants. Eur. J. Plant Pathol. 156, 413–423. doi: 10.1007/s10658-019-01890-0
Haapalainen, M., Wang, J., Latvala, S., Lehtonen, M. T., Pirhonen, M., Nissinen, A. I. (2018). Genetic variation of ‘Candidatus Liberibacter solanacearum’ haplotype C and identification of a novel haplotype from Trioza urticae and Stinging nettle. Phytopathology 108, 925–934. doi: 10.1094/PHYTO-12-17-0410-R
Hansen, A. K., Trumble, J. T., Stouthamer, R., Paine, T. D. (2008). A new huanglongbing species, "Candidatus liberibacter psyllaurous," found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Appl. Environ. Microbiol. 74, 5862–5865. doi: 10.1128/AEM.01268-08
Harrison, K., Levy, J. G., Tamborindeguy, C. (2022). Effects of 'Candidatus Liberibacter solanacearum' haplotypes A and B on tomato gene expression and geotropism. BMC Plant Biol. 22, 156. doi: 10.1186/s12870-022-03505-z
Harrison, K., Tamborindeguy, C., Rondon, S. I., Levy, J. G. (2020). Effects of 'Candidatus Liberibacter Solanacearum' haplotype on Atlantic potato tuber emergence rate in South Texas. Am. J. Potato Res. 97, 489–496. doi: 10.1007/s12230-020-09796-y
Harrison, K., Tamborindeguy, C., Scheuring, D. C., Herrera, A. M., Silva, A., Badillo-Vargas, I. E., et al. (2019). Differences in zebra chip severity between "Candidatus Liberibacter solanacearum" haplotypes in Texas. Am. J. Potato Res. 96, 86–93. doi: 10.1007/s12230-018-9692-7
Jaillais, Y., Chory, J. (2010). Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 17, 642–645. doi: 10.1038/nsmb0610-642
Kesarwani, M., Yoo, J., Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Ku, Y. S., Sintaha, M., Cheung, M. Y., Lam, H. M. (2018). Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 19, 3206. doi: 10.3390/ijms19103206
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Levy, J. G., Mendoza, A., Miller, Jr. J.C., Tamborindeguy, C., Pierson, E. A. (2017). Global gene expression in two potato cultivars in response to 'Candidatus Liberibacter solanacearum' infection. BMC Genomics 18, 960. doi: 10.1186/s12864-017-4313-2
Levy, J. G., Mendoza-Herrera, A., Merchant, N., Berg-Falloure, K. M., Kolomiets, M. V., Tamborindeguy, C. (2023). Evaluation of the effect of ‘Candidatus Liberibacter solanacearum’ haplotypes in tobacco infection. Agronomy 13, 569. doi: 10.3390/agronomy13020569
Li, X., Nie, J., Arsenault, H., Hammill, D. L., Xu, H., De Boer, S. H. (2013). Tomato can be a latent carrier of ‘CandidatusLiberibacter solanacearum’, the causal agent of potato zebra chip disease. EPPO Bull. 43, 229–237. doi: 10.1111/epp.12032
Liefting, L. W., Perez-Egusquiza, Z. C., Clover, G. R. G., Anderson, J. A. D. (2008). A new 'Candidatus Liberibacter' species in Solanum tuberosum in New Zealand. Plant Dis. 92, 1474. doi: 10.1094/PDIS-92-10-1474A
Liefting, L. W., Weir, B. S., Pennycook, S. R., Clover, G. R. (2009). 'Candidatus Liberibacter solanacearum', associated with plants in the family Solanaceae. Int. J. Syst. Evol. Microbiol. 59, 2274–2276. doi: 10.1099/ijs.0.007377-0
Lin, H., Lou, B., Glynn, J. M., Doddapaneni, H., Civerolo, E. L., Chen, C., et al. (2011). The complete genome sequence of 'Candidatus Liberibacter solanacearum', the bacterium associated with potato zebra chip disease. PloS One 6, e19135. doi: 10.1371/journal.pone.0019135
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Maine, G. T., Sinha, P., Tye, B. K. (1984). Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106, 365–385. doi: 10.1093/genetics/106.3.365
Mathesius, U. (2018). Flavonoid functions in plants and their interactions with other organisms. Plants 7, 30. doi: 10.3390/plants7020030
Mauck, K. E., Sun, P., Meduri, V. R., Hansen, A. K. (2019). New Ca. Liberibacter psyllaurous haplotype resurrected from a 49-year-old specimen of Solanum umbelliferum: A native host of the psyllid vector. Sci. Rep. 9, 1–13.
Mendoza-Herrera, A., Levy, J., Harrison, K., Yao, J., Ibanez, F., Tamborindeguy, C. (2018). 'Infection by Candidatus Liberibacter solanacearum’ haplotypes A and B in Solanum lycopersicum ‘Moneymaker’. Plant Dis. 102, 2009–2015. doi: 10.1094/PDIS-12-17-1982-RE
Miles, G. P., Samuel, M. A., Chen, J. C., Civerolo, E. L., Munyaneza, J. E. (2010). Evidence that cell death is associated with zebra chip disease in potato tubers. Am. J. Potato Res. 87, 337–349. doi: 10.1007/s12230-010-9140-9
Munyaneza, J. E. (2012). Zebra chip disease of potato: biology, epidemiology, and management. Am. J. Potato Res. 89, 329–350. doi: 10.1007/s12230-012-9262-3
Munyaneza, J. E., Crosslin, J. M., Upton, J. E. (2007b). Association of Bactericera cockerelli (Homoptera : Psyllidae) with "zebra chip," a new potato disease in southwestern United States and Mexico. J. Economic Entomology 100, 656–663. doi: 10.1093/jee/100.3.656
Munyaneza, J. E., Fisher, T. W., Sengoda, V. G., Garczynski, S. F., Nissinen, A., Lemmetty, A. (2010a). Association of "Candidatus Liberibacter solanacearum" with the psyllid, Trioza apicalis (Hemiptera: Triozidae) in Europe. J. Econ Entomol 103, 1060–1070. doi: 10.1603/EC10027
Munyaneza, J. E., Fisher, T. W., Sengoda, V. G., Garczynski, S. F., Nissinen, A., Lemmetty, A. (2010b). First report of "Candidatus Liberibacter solanacearum" associated with psyllid-affected carrots in Europe. Plant Dis. 94, 639–639. doi: 10.1094/PDIS-94-5-0639A
Munyaneza, J. E., Goolsby, J. A., Crosslin, J. M., Upton, J. E. (2007a). Further evidence that zebra chip potato disease in the lower Rio Grande Valley of Texas is associated with Bactericera cockerelli. Subtrop Plant Sci. 59, 30–37.
Munyaneza, J. E., Sengoda, V. G., Crosslin, J. M., Garzon-Tiznado, J. A., Cardenas-Valenzuela, O. G. (2009). First report of “Candidatus Liberibacter solanacearum” in tomato plants in Mexico. Plant Dis. 93, 1076–1076.
Nakashima, K., Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nelson, W. R., Sengoda, V. G., Alfaro-Fernandez, A. O., Font, M. I., Crosslin, J. M., Munyaneza, J. E. (2013). A new haplotype of "Candidatus Liberibacter solanacearum" identified in the Mediterranean region. Eur. J. Plant Pathol. 135, 633–639. doi: 10.1007/s10658-012-0121-3
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–DD27. doi: 10.1093/nar/gkac993
Pitman, A. R., Drayton, G. M., Kraberger, S. J., Genet, R. A., Scott, I. A. W. (2011). Tuber transmission of 'Candidatus Liberibacter solanacearum' and its association with zebra chip on potato in New Zealand. Eur. J. Plant Pathol. 129, 389–398. doi: 10.1007/s10658-010-9702-1
Pratyusha, D. S., Sarada, D. V. L. (2022). MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 41, 2245–2260. doi: 10.1007/s00299-022-02927-1
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., Benhamed, M. (2018). Modify the histone to win the battle: chromatin dynamics in plant-pathogen interactions. Front. Plant Sci. 9, 355. doi: 10.3389/fpls.2018.00355
Rashed, A., Wallis, C. M., Paetzold, L., Workneh, F., Rush, C. M. (2013). Zebra chip disease and potato biochemistry: tuber physiological changes in response to 'Candidatus Liberibacter solanacearum' infection over time. Phytopathology 103, 419–426. doi: 10.1094/PHYTO-09-12-0244-R
Rehman, A., Azhar, M. T., Hinze, L., Qayyum, A., Li, H., Peng, Z., et al. (2021). Insight into abscisic acid perception and signaling to increase plant tolerance to abiotic stress. J. Plant Interact. 16, 222–237. doi: 10.1080/17429145.2021.1925759
Rui, Y., Dinneny, J. R. (2020). A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.16166
Secor, G. A., Rivera, V. V., Abad, J. A., Lee, I. M., Clover, G. R. G., Liefting, L. W., et al. (2009). Association of 'Candidatus Liberibacter solanacearum' with zebra chip disease of potato established by graft and psyllid transmission, electron microscopy, and PCR. Plant Dis. 93, 574–583. doi: 10.1094/PDIS-93-6-0574
Shin, Y., Chane, A., Jung, M., Lee, Y. (2021). Recent advances in understanding the roles of pectin as an active participant in plant signaling networks. Plants 10, 1712. doi: 10.3390/plants10081712
Sumner-Kalkun, J. C., Highet, F., Arnsdorf, Y. M., Back, E., Carnegie, M., Madden, S., et al. (2020). 'Candidatus Liberibacter solanacearum' distribution and diversity in Scotland and the characterisation of novel haplotypes from Craspedolepta spp. (Psyllidae: Aphalaridae). Sci. Rep. 10, 16567.
Swisher Grimm, K. D., Garczynski, S. F. (2019). Identification of a new haplotype of ‘Candidatus Liberibacter solanacearum’ in Solanum tuberosum. Plant Dis. 103, 468–474. doi: 10.1094/PDIS-06-18-0937-RE
Swisher Grimm, K. D., Mustafa, T., Cooper, W. R., Munyaneza, J. E. (2018). Role of 'Candidatus Liberibacter solanacearum' and Bactericera cockerelli haplotypes in zebra chip incidence and symptom severity. Am. J. Potato Res. 95, 709–719. doi: 10.1007/s12230-018-9678-5
Teresani, G. R., Bertolini, E., Alfaro-Fernandez, A., Martinez, C., Tanaka, F. A., Kitajima, E. W., et al. (2014). Association of 'Candidatus Liberibacter solanacearum' with a vegetative disorder of celery in Spain and development of a real-time PCR method for its detection. Phytopathology 104, 804–811. doi: 10.1094/PHYTO-07-13-0182-R
Wang, J., Haapalainen, M., Nissinen, A. I., Pirhonen, M. (2021). Dual transcriptional profiling of carrot and ‘Candidatus Liberibacter solanacearum’ at different stages of infection suggests complex host-pathogen interaction. Mol. Plant-Microbe Interact. 34, 1281–1297. doi: 10.1094/MPMI-10-20-0274-R
Wang, D., Kanyuka, K., Papp-Rupar, M. (2023b). Pectin: a critical component in cell-wall-mediated immunity. Trends Plant Sci. 28, 10–13. doi: 10.1016/j.tplants.2022.09.003
Wang, D. D., Lu, Q. H., Jin, S. D., Fan, X. Y., Ling, H. (2023a). Pectin, lignin and disease resistance in Brassica napus L.: an update. Horticulturae 9, 112. doi: 10.3390/horticulturae9010112
Wen, A., Mallik, I., Alvarado, V. Y., Pasche, J. S., Wang, X., Li, W., et al. (2009). Detection, distribution, and genetic variability of 'Candidatus Liberibacter' species associated with zebra complex disease of potato in North America. Plant Dis. 93, 1102–1115. doi: 10.1094/PDIS-93-11-1102
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141.
Yuan, L., Liu, X., Luo, M., Yang, S., Wu, K. (2013). Involvement of histone modifications in plant abiotic stress responses'. J. Integr. Plant Biol. 55, 892–901. doi: 10.1111/jipb.12060
Keywords: Candidatus Liberibacter solanacearum, zebra chip, transcriptomics, gene expression, bioinformatics
Citation: Chuan J, Nie J, Cooper WR, Chen W, Hale L and Li X (2024) The functional decline of tomato plants infected by Candidatus Liberbacter solanacearum: an RNA-seq transcriptomic analysis. Front. Plant Sci. 15:1325254. doi: 10.3389/fpls.2024.1325254
Received: 20 October 2023; Accepted: 15 January 2024;
Published: 01 February 2024.
Edited by:
Guillaume J. Bilodeau, Canadian Food Inspection Agency (CFIA), CanadaReviewed by:
Tianrun Li, University of California, Davis, United StatesCopyright © 2024 Chuan, Nie, Cooper, Chen, Hale and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Li, c2Vhbi5saUBpbnNwZWN0aW9uLmdjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.