
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 March 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1313832
High temperatures affect grape yield and quality. Grapes can develop thermotolerance under extreme temperature stress. However, little is known about the changes in transcription that occur because of high-temperature stress. The heat resistance indices and transcriptome data of five grape cultivars, ‘Xinyu’ (XY), ‘Miguang’ (MG), ‘Summer Black’ (XH), ‘Beihong’ (BH), and ‘Flame seedless’ (FL), were compared in this study to evaluate the similarities and differences between the regulatory genes and to understand the mechanisms of heat stress resistance differences. High temperatures caused varying degrees of damage in five grape cultivars, with substantial changes observed in gene expression patterns and enriched pathway responses between natural environmental conditions (35 °C ± 2 °C) and extreme high temperature stress (40 °C ± 2 °C). Genes belonging to the HSPs, HSFs, WRKYs, MYBs, and NACs transcription factor families, and those involved in auxin (IAA) signaling, abscisic acid (ABA) signaling, starch and sucrose pathways, and protein processing in the endoplasmic reticulum pathway, were found to be differentially regulated and may play important roles in the response of grape plants to high-temperature stress. In conclusion, the comparison of transcriptional changes among the five grape cultivars revealed a significant variability in the activation of key pathways that influence grape response to high temperatures. This enhances our understanding of the molecular mechanisms underlying grape response to high-temperature stress.
Grapes have a long cultivation history and rich germplasm resources. According to latest data from the National Bureau of Statistics, in 2022, China’s grape production is 15.3779 million tons (http://data.stats.gov.cn). Similarly, fresh grapes accounted for approximately 42% of the total. The projected figures indicate that the fresh and dried grape planting area in Xinjiang will remain stable at approximately 0.1 million hectares by 2025. Furthermore, it is anticipated that the yield will exceed 2.5 million tons during this period (Wu et al., 2021). Extreme temperature events are predicted to occur more frequently, intensively, and for longer periods. In several regions, the noon temperature can reach 40°C or higher. High temperatures (HTs) adversely affect the development and composition of grapes, posing a significant threat to their yield and quality. This could undermine the environmental and economic sustainability of grape production (Schultz and Jones, 2010; Fraga et al., 2016). Previous studies on the response and adaptation of grapes to HT have primarily focused on morphological and physiological changes. These include changes in photosynthesis, hormone levels, and antioxidant systems (Wang and Li, 2006; Wang et al., 2009; Luo et al., 2011; Bita and Gerats, 2013). With the availability of the grape genome sequence, more studies have focused on transcriptomic and proteomic changes in response to heat stress. Several transcriptomic studies have explored the effects of heat stress on plant species, such as potatoes, tomatoes, rice, tobacco, and Arabidopsis (Higashi et al., 2015; González-Schain et al., 2016; Keller et al., 2018; Rahmati et al., 2018; Blair et al., 2019; Liu G. et al., 2020; Liu H. et al., 2020; Tang et al., 2020; Bineau et al., 2021; Liu et al., 2021).
Despite the importance of understanding thermotolerance and heat stress responses in grape leaves, evidence of the underlying molecular pathways is limited. However, the mechanism of grape response to extreme temperature stress remains unclear. Heat shock proteins (HSPs) and heat shock transcription factors (HSFs) play significant roles in thermotolerance. The accumulation of HSPs and expression of HSFs has been shown to be related to this process (Larkindale and Huang, 2004; Charng et al., 2006). The role of HSPs in scavenging reactive oxygen species (ROS), maintaining cell membrane integrity, and producing antioxidants and osmolytes is crucial for protecting plants from heat stress. Heat-responsive genes, in combination with transcription factors (TFs), are also induced in response to heat stress. Several transcription factor families, including DREB, MYB, NAC, HSF, and bZIP, play critical roles in heat stress response (Jiang et al., 2020). To better understand the mechanisms of the plant heat stress response, it is important to investigate the roles of different TFs involved in the process.
Transcriptomic analyses have provided comprehensive insights into the mechanisms underlying heat stress responses in grapes (Liu et al., 2012; Rienth et al., 2014; Jiang et al., 2017; Lecourieux et al., 2017; Kim et al., 2018). However, less attention has been paid to heat stress-related metabolic pathways and the molecular signaling networks involved in grapes. One promising avenue of research is examining endoplasmic reticulum (ER) protein processing in plant responses to heat stress (Li et al., 2015; Wang et al., 2017; Jin et al., 2020; Moreno, 2021). Many ER proteins such as HSPs and chaperones, are critical for ER function (Park and Seo, 2015). Accumulation of unfolded or misfolded proteins can lead to ER stress, which activates the unfolded protein response (UPR) to relieve stress. ER-associated degradation (ERAD) is a process that occurs in the ER and involves degradation of misfolded and unfolded proteins through a ubiquitin/proteasome mechanism. Although the above findings suggest a relationship between ERAD, UPR, and heat stress, the specific molecular mechanisms underlying this relationship have not been explored in grapes.
In this study, we selected five representative grape varieties (‘Xinyu,’ ‘Miguang,’ ‘Summer Black,’ ‘Beihong,’ and ‘Flame seedless’) with strong cultivation adaptability and wide planting area in Xinjiang as test materials. We compared the physiological, biochemical, and transcriptome data of five grape varieties after high-temperature treatment by simulating the natural high-temperature environment in a solar greenhouse. The objective of this study was to determine the heat tolerance levels of different grape varieties and identify important genes that may be related to HT resistance, including HSPs and TFs. Additionally, we detected changes in metabolic pathways after exposure to HT stress. This study contributes to a better understanding of transcriptome defense mechanisms associated with heat tolerance in grapes.
This experiment was conducted at the Shihezi University Experiment Station greenhouse. The experimental materials consisted of 2-year-old seedlings of five grape varieties—’Xinyu’ (XY Vitis vinifera L., Originating from Xinjiang, China), ‘Miguang’ (MG V. vinifera L. × Vitis labrusca, originating from Hebei, China), ‘Summer Black’ (XH V. vinifera × V. labrusca, Origin in Japan), ‘Beihong’ (BH V. vinifera × V. amurensis), and ‘Flame seedless’ (FL V. vinifera L, Origin in the United States)—grown in cultivation bags measuring 27 cm (height) × 30 cm (diameter), with a culture medium of pastoral soil and organic matter in a ratio of 2:1. The plant row spacing was set at 80 cm × 100 cm, and soil moisture was maintained between 31% and 35%. Consistent field management practices were implemented for all plants throughout the experiment.
Ten plants with strong growth were randomly selected from each grape cultivar for temperature treatment. The temperature in the solar greenhouse was regulated by an exhaust fan, monitored by a temperature sensor, and maintained at 40 °C ± 2 °C (T: high-temperature treatment). If the temperature rose above 40 °C, the exhaust fan started to work, and the temperature was reduced. The high-temperature period was from 12:00 to 19:30 each day, whereas routine management was conducted during the other periods. The control temperature was set at 35 °C ± 2 °C (CK: control treatment, natural solar greenhouse temperature). In this experiment, a MicroLite-U disk temperature recorder was used to record the temperature once every hour for continuous monitoring of the greenhouse temperature. The temperature data are shown in Figure 1.
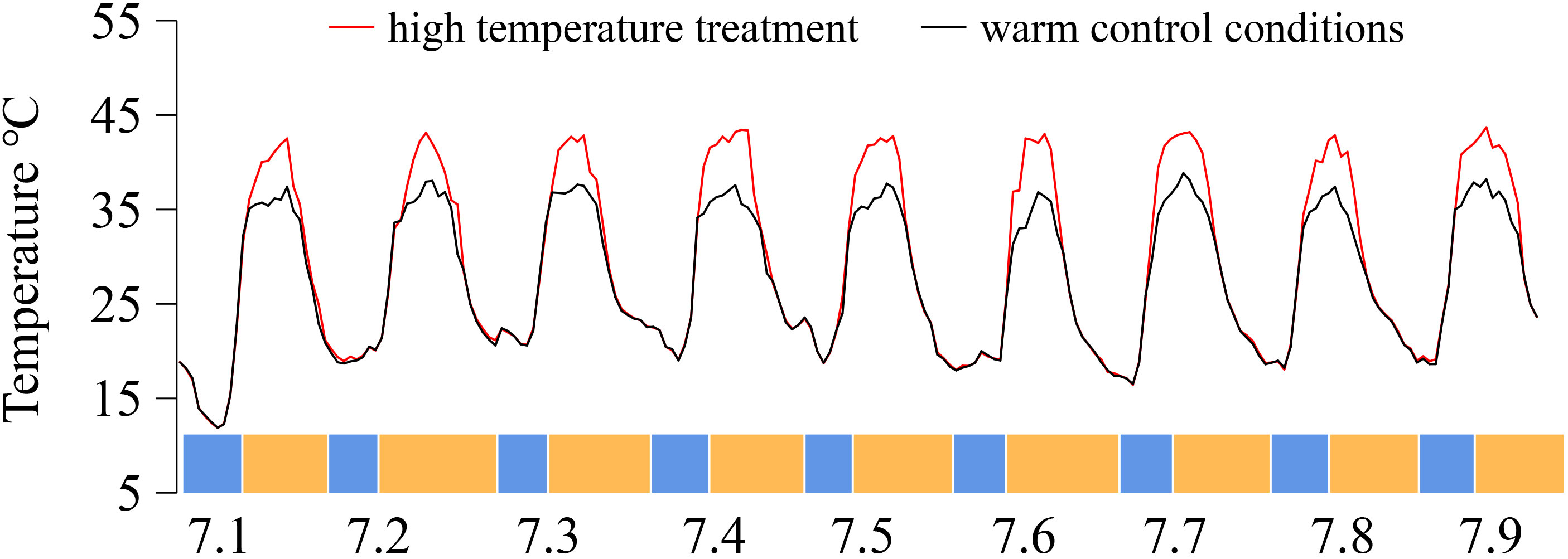
Figure 1 Temperature was recorded of the greenhouse from 1–9 July 2019, China. On the X-axis, blue represents night temperatures, whereas yellow denotes day temperatures.
We conducted high-temperature (HT) treatment from 1 to 7 July 2019 and collected grape leaves on the 7th day of HT treatment. At the same time, the leaves of grapes cultured at 35°C were collected as controls. Each grape variety randomly selected robust young leaves near the functional leaves of sections 9–11 from bottom to top and wrapped in tin foil. They were immediately frozen in liquid nitrogen and stored at −80°C for RNA seq analysis.
Four physiological indexes were measured for each grape cultivar to analyze the physiological changes induced by heat stress. Superoxide dismutase (SOD) activity was determined by the nitrogen blue tetrazolium method, peroxidase (POD) activity was determined using the guaiacol method, and catalase (CAT) activity was detected by ultraviolet spectrophotometry (Cheng et al., 2018). The method described by Liu et al. (2013) was used to determine the production rate of O2−. The malondialdehyde (MDA) and hydrogen peroxide (H2O2) contents were determined using the method described by Li et al. (2015). The method described by Lichtenthaler (1087) was used to determine chlorophylla (Chla) and chlorophyllb (Chlb). A fluorometer (FMS-2, Hansatech, UK) was used to measure chlorophyll fluorescence. The photosynthetic parameters were investigated using a photosynthesizer (Li-6800, LI-COR, US).
On the 7th day of high-temperature stress at 4 p.m., each grape variety randomly selected robust young leaves near the functional leaves of sections 9–11 from bottom to top, immediately frozen with liquid nitrogen, and stored at −80°C for RNA-seq analysis. Total RNA from 30 samples was extracted using TRIzol Reagent (Invitrogen, Canada). The total RNA was quantified using a 2100 Bioanalyzer (Agilent Technologies). These libraries were then sequenced by Shanghai Personal Biotechnology Cp. Ltd. using Next-Generation Sequencing (NGS) technology based on the Illumina sequencing platformAfter on-line sequencing, the samples generated raw Data of FASTQ and were statistically calculated We used the DESeq R package to analyze differences in gene expression (DEGs) and screened for DEGs that met the following conditions: | log2foldchange | ≥1 and p-value ≤0.05.
QRT-PCR was performed on nine DEGs related to HT treatment, repeated three times for each sample. QPCR for the relative expression levels of target genes was performed using the CFX Connect Real-time PCR Detection System (BioRad, USA) and SYBR® Premix Ex TaqTM II (TaKaRa). Total RNA was extracted from the grapes using the TRIzol reagent (Invitrogen). This kit was used for gDNA removal (TransGen Biotech, Beijing, China). Actin was used as an internal reference control for data normalization. The expression level of each target gene was calculated using the 2−△△Ct method (Ma et al., 2015).
We built gene co-expression networks based on the DEGs using weighted correlation network analysis (WGCNA) in the R package (Langfelder and Horvath, 2008). DEGs from the five grape varieties (high-temperature and control-treated samples) were used for the co-expression analysis. All physiological indices were examined for correlation with the modules and all genes in each module. Significant high-temperature tolerance-related modules were detected based on the highest correlation values with the physiological indices.
SPSS software (version 19.0) was used for the analysis of variance (P ≤0.05), and Duncan’s method was used for multiple comparisons and significance tests. The results are presented as mean ± standard error of three replicates.
The levels of MDA and H2O2 increased significantly in XH and BH grapes (Figures 2A, B), while O2− content increased in XY and BH grapes (Figure 2C), indicating that oxidative damage occurred in the plants. The activities of SOD, POD, and CAT decreased in all five grape varieties after exposure to HT stress (Figures 2D–F). This reduction in enzyme activity may be due to structural damage and depletion of antioxidant enzymes. Principal component analysis (PCA) showed that the first and second principal components explained 38.6% and 22.2% of the variance, respectively (Figure 2G). Before the high-temperature treatment, the first group of cultivars (BH, XY, and FL grapes) and the second group of cultivars (XH and MG grapes) clustered together. However, after the high-temperature treatment, the five grape varieties showed more significant separation, indicating that HT had different effects on them compared to CK. Supplementary Figure S1 shows the changes in chlorophyll fluorescence and photosynthetic parameters of leaves from different grape cultivars following the HT stress treatment. Under HT conditions, the total chlorophyll content (Chl), intercellular CO2 concentration (Ci), net photosynthetic rate (Pn), transpiration rate (Tr), and stomatal conductance (Gs) decreased. Conversely, the intercellular CO2 concentration (Ci) and initial fluorescence (Fo) increased in all five grape cultivars. Notably, the largest increases in Ci and Fo were observed in BH grapes, suggesting that HT stress had the most significant effect on light energy conversion efficiency in this cultivar.
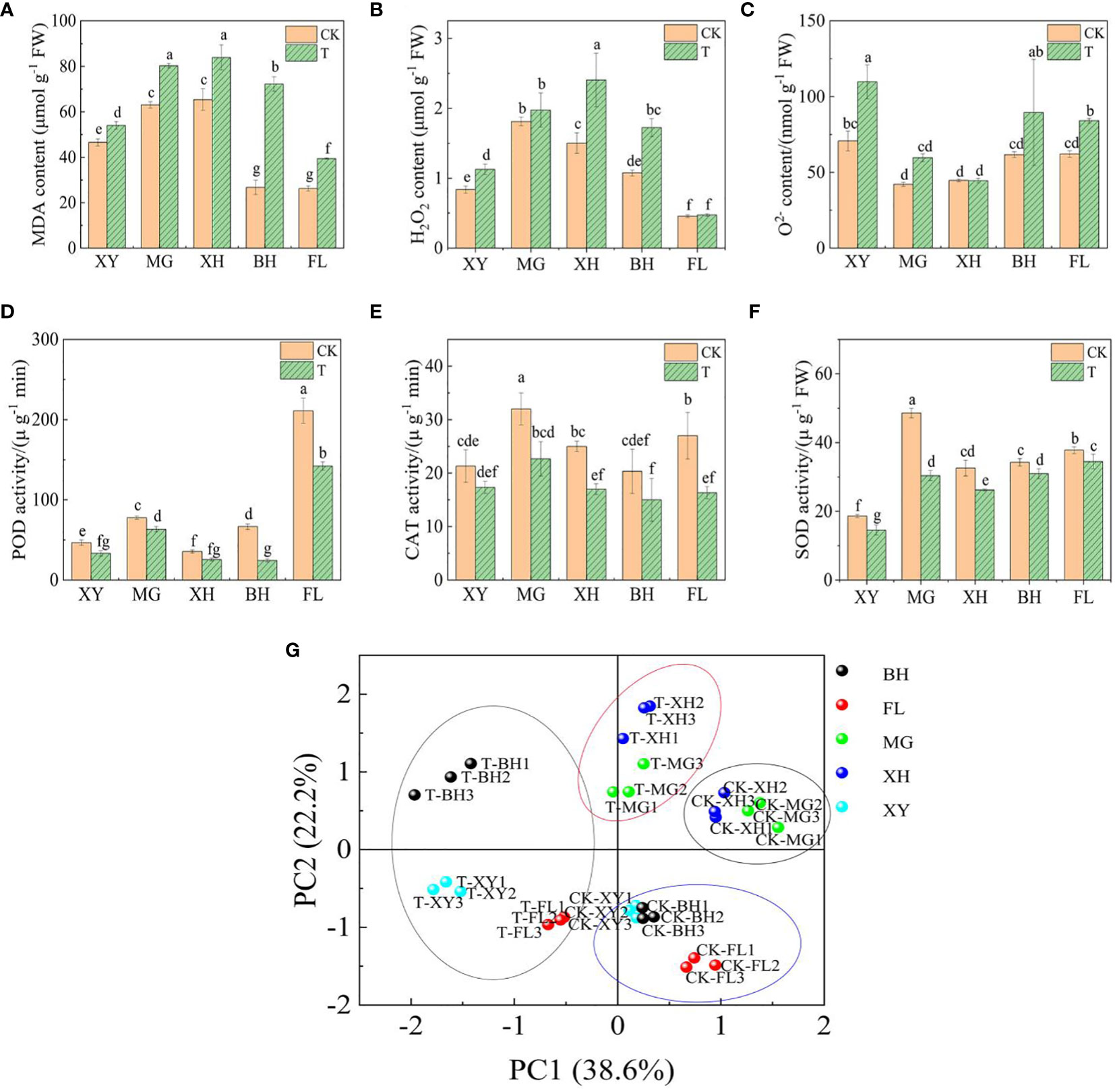
Figure 2 Physiological analysis of five grape varieties under HT stress. The graphs showed the levels of (A) malondialdehyde (MDA) content. (B) hydrogen peroxide (H2O2) content. (C) Superoxide anion (O2−) content. (D) peroxidase (POD) activity. (E) catalase (CAT) activity. (F) Superoxide dismutase (SOD) activity. Values followed by the same letter were not significantly different (p < 0.05) according to Duncan’s assay significant difference test. (G) Principal component analysis (PCA) of the five grape leaves. Small letters indicate significance between the different varieties (P <0.05). In subpanels (A–G), CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively. XY, MG, XH, BH, and FL represent “Xinyu,” “Miguang,” “Summer Black,” “Beihong,” and “Flame seedless” grapes, respectively.
The raw data of 30 samples from XY, MG, XH, BH, and FL grapes were analyzed, and it was found that Q20 (%) >97% and Q30 (%) >93% met the standards for further biological analysis. Transcriptome sequencing data from 30 samples were filtered to remove adapters and low-quality reads to obtain Clean Reads and Clean Data. The ratio of clean reads and clean data obtained from each sample to the original reads was above 92%. The filtered reads were aligned to the reference genome using the upgraded version of HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml) in TopHat2. At least 36,996,094 reads were aligned with the reference genome for each sample, accounting for more than 93.52% of the total clean reads for each sample. Of these, 97.43%–97.75% of the reads were aligned to a unique position in the reference genome, while 2.25% to 2.57% of the reads were aligned to multiple positions in the reference genome (Supplementary Table S1). In general, the RNA-seq data alignment rate was high, indicating that the RNA-Seq data utilization rate was high, which ensured the validity and accuracy of the sequence assembly and post-analysis.
We conducted PCA on the samples and found that the spatial dimensions of the XH and FL samples were similar to those of the control samples after HT treatment; however, the spatial distance was large, indicating that different temperature treatments had a significant impact on the expression of samples (Supplementary Figure S2). Secondly, we analyzed the repeatability of RNA-seq samples using Pearson’s Correlation Coefficient (r) as a parameter of biological repeat correlation. The closer the value of r2 is to 1, the higher the correlation between the two replicate samples, and the closer it is to 0, the weaker the correlation. In this study, the r2 values between the three biological replicates of the same sample were close to 1, indicating that the RNA-Seq samples had good repeatability, and the data were reliable (Supplementary Figure S3).
After analyzing the gene expression of the 10 treatment groups, we compared grape varieties treated at different temperatures with the same grape varieties treated at different temperatures. We used DEGseq to identify genes that were differentially expressed under HT (q-value ≤0.05, |log2 fold-change| ≥1). Hierarchical clustering analysis (HCA) showed that the high-temperature-treated samples and control samples were clustered together, indicating an overlap in responsive genes between these treatments (Supplementary Figure S4).
In this study, we identified 550 (upregulated 314, downregulated 236), 628 (upregulated 414, downregulated 214), 898 (upregulated 487, downregulated 411), 1,013 (upregulated 557, downregulated 456), and 1,062 (upregulated 461, downregulated 601) genes in XY, MG, XH, BH, and FL grapes, respectively. Furthermore, we observed a significant increase in differentially expressed genes (DEGs) under heat stress in different grape varieties. Among these varieties, MG grapes had the least number of DEGs, while XY grapes had the greatest number (Supplementary Table S2A; Supplementary Figure S5). We conducted a comparative analysis of DEGs among grape cultivars exposed to different temperatures. Specifically, we identified the intersection of upregulated and downregulated DEGs across all grape cultivars to determine shared core sets. In addition, we examined the intersection of DEGs among grape varieties under HT treatment to identify the shared core sets. In the control group compared to the HT treatment group, we identified 83 DEGs (Figure 3). We found that HT stress resulted in a greater number of upregulated than downregulated genes in different grape varieties. A detailed list of the shared DEGs grouped by treatment type is presented in Supplementary Table S2B.
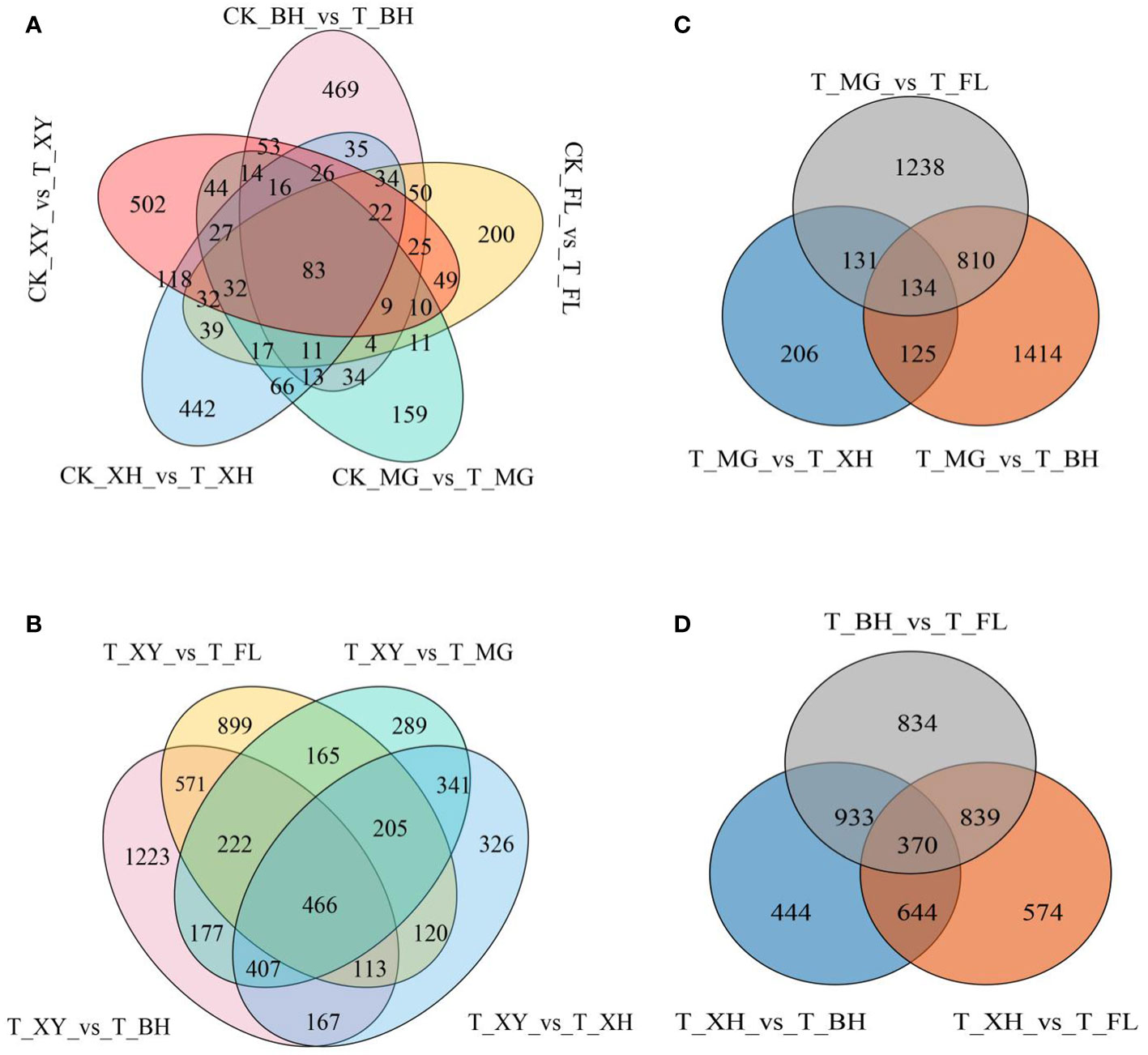
Figure 3 DEG analysis of five grape cultivars after HT treatment. (A) Total number of DEGs and the shared core sets of up and downregulated DEGs in all grape cultivars under different temperature treatments. (B–D) Statistics of DEGs and shared core sets of up and downregulated DEGs in all grape cultivars under HT treatments. In subpanels (A–D), CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively. XY, MG, XH, BH, and FL represented “Xinyu,” “Miguang,” “Summer Black,” “Beihong,” and “Flame seedless” grapes, respectively.
Through GO enrichment analysis, DEGs were enriched into three categories: biological process (BP), cellular component (CC), and molecular function (MF). In the comparison of the same temperature varieties with different varieties, in the BP category, cell communication and defense responses were significantly enriched. ADP binding, adenyl ribonucleotide binding, and adenyl nucleotide binding were significantly enriched in MF category. In the CC category, the cell periphery, membrane, and plasma membrane were significantly enriched (Figures 4A, B). Lists of annotation and enriched GO terms for these DEGs grouped by treatment type can be found (Supplementary Tables S3A, B).
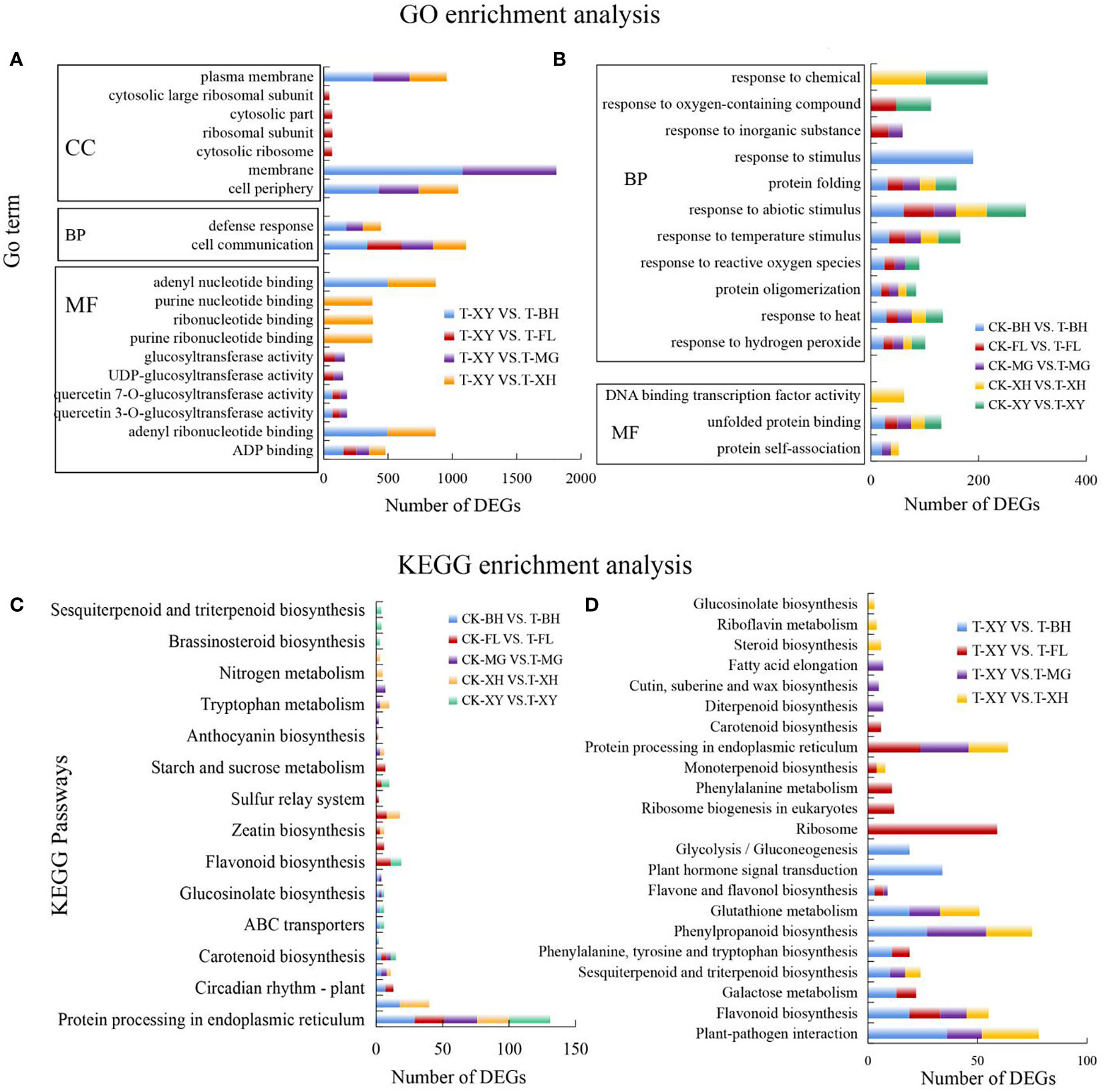
Figure 4 Functional enrichment analysis of differentially expressed genes in the five grape varieties. (A, B) GO enrichment analysis was performed for the DEGs. Y is the term of GO, and X is the number of DEGs. MF, molecular function; CC, cellular component; BP, biological process. Select the top 10 GO term entries with the smallest p-value and the most significant enrichment in each Go classification for display. (C, D) analyzed the significant enrichment of the differential gene KEGG and selected the top 10 pathways with the smallest p-value and the most significant enrichment for display. In subpanels (A–D), CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively. XY, MG, XH, BH, and FL represent “Xinyu,” “Miguang,” “Summer Black,” “Beihong,” and “Flame seedless” grapes, respectively.
To clarify the biochemical metabolism or signal transduction pathways in which different genes may participate in different samples, KEGG pathway enrichment analysis was carried out. In the comparison of five grape varieties under different temperatures, flavonoid biosynthesis, protein processing in the endoplasmic reticulum, plant hormone signal transduction, carotenoid biosynthesis, and starch and sucrose metabolism were significantly enriched (Figure 4C). In the comparison of the control with the five grapes, glutathione metabolism, DNA replication, flavonoid biosynthesis, and phenylpropanoid biosynthesis were significantly enriched (Figure 4D). Lists of annotation and enriched pathways for these DEGs grouped by treatment type can be found (Supplementary Tables S4A, B). These pathways mainly involve were genetic information processing, metabolic pathways, and environmental information processing. HT stress significantly promoted DEGs involved in protein processing in the endoplasmic reticulum pathway, and many HSPs were upregulated. This indicates that grapes can improve their heat tolerance by rapidly accumulating heat shock proteins under HT stress. At the same time, the peroxisome pathway was also activated and upregulated in the five grape varieties, genes involved in photosynthesis and antenna protein were inhibited, and differential genes in the carotenoid biosynthesis pathway were promoted. This may indicate that the growth of the grapes was inhibited. Genes related to glutathione metabolism, proline metabolism, ascorbate and aldate metabolism were upregulated to protect plants.
In this study, 105 genes encoding enzymes were involved in ROS regulation. Two catalase (CATs), two ascorbate peroxidase (APXs), 52 glutathione S-transferase (GSTs), two superoxide dismutase (SODs), two glutathione peroxidase (GPXs), and 49 peroxidase (POXs) genes were differentially expressed after HT treatment compared to the controls (Figures 5A, B). These DEGs showed different expression patterns in different grape cultivars under HT stress. After HT treatment, most DEGs were upregulated in BH, FL, and XH grapes, and downregulated in MG and XY grapes. Among the cultivars, there were 15 genes with differential expression of more than 4.00 times. Lists of these DEGs can be found (Supplementary Table S5). To validate the RNA-Seq data, four DEGs were selected for real-time PCR analysis (Figures 5C–F). The expression patterns of both the qRT-PCR and RNA-Seq data were highly consistent. Therefore, these genes may play an important role in the heat-stress response of grape leaves.
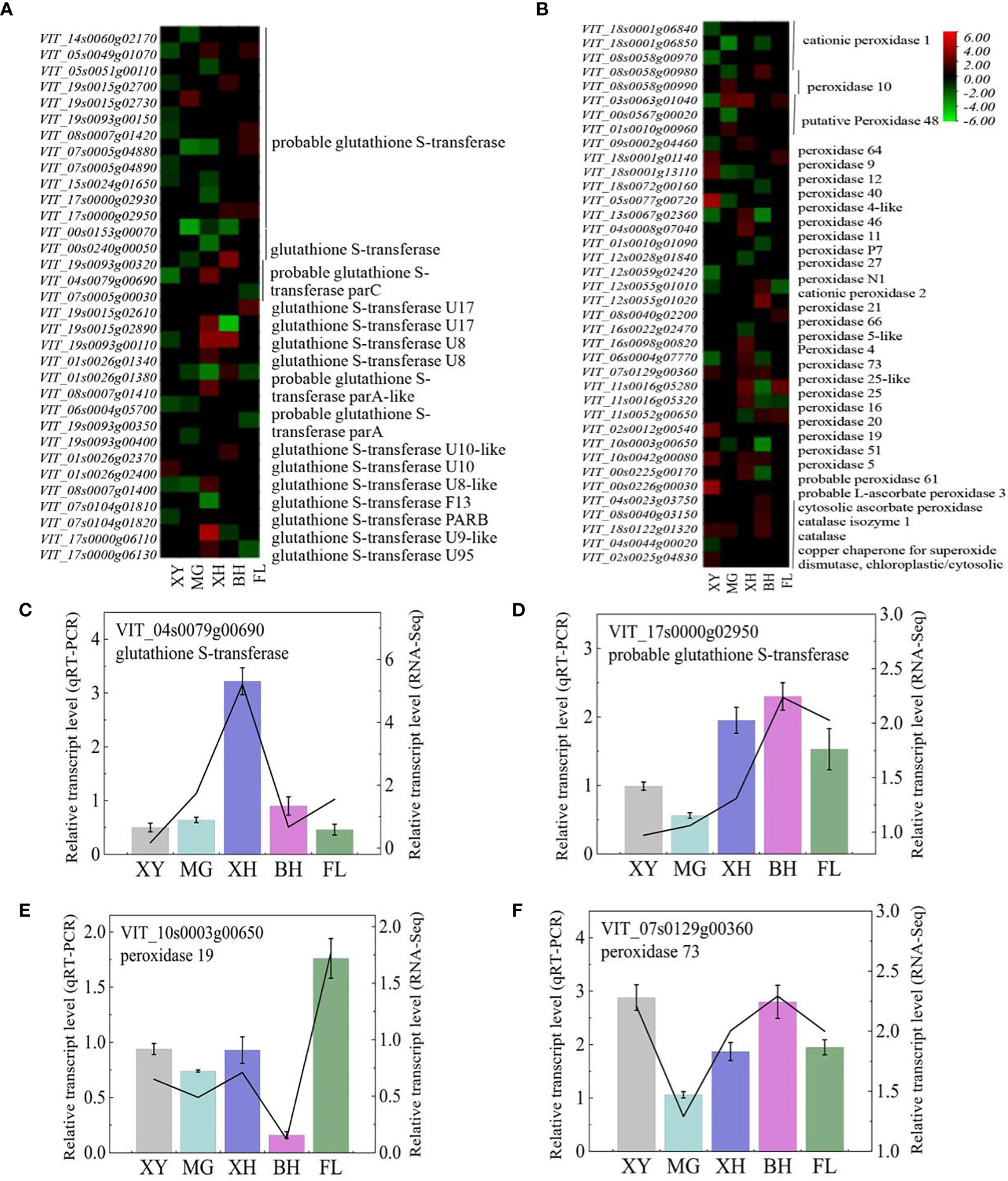
Figure 5 (A, B) Heatmap of the expression patterns of the selected genes involved in ROS scavenging in response to HT stress. The color bar indicates gene fold change, upregulation is indicated in red, and downregulation is indicated in green. (C–F) Expression profiles of the selected DEGs determined using RT-PCR analyses and the line indicates the relative gene transcription level in RNA-Seq. Error bars represent mean ± SD. XY, MG, XH, BH, and FL represent CK_XY_vs_T_XY, CK_MG_vs_T_MG, CK_XH_vs_T_XH, CK_BH_vs_T_BH, and CK_FL_vs_T_FL, respectively. CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively.
A total of 1,213 transcripts encoding TFs were differentially expressed in the five grape cultivars. Among the differentially expressed TFs, the bHLH, ERF, MYB, WRKY, G2-like, and HSF families were found to be represented by more than 76% of the TF-encoding transcripts in the five grape cultivars (Figure 6A). There were six common transcription factors in CK_BH_vs_T_BH, CK_FL_vs_T_FL, CK_MG_vs_T_MG, CK_XH_vs_T_XH, and CK_XY_vs_T_XY, among which, one HSF (VIT_00s0179g00150, upregulated in five grape cultivars after HT treatment), three ERFs (VIT_11s0016g05340 upregulated in five grape cultivars after HT treatment, VIT_03s0063g00460 and VIT_11s0016g00660 upregulated in BH, FL and XH, but downregulated in MG and XY after HT treatment), one G2-like (VIT_06s0004g05120, downregulated in five grape cultivars after HT treatment) and one bHLH (VIT_13s0047g00450 downregulated in BH, FL, and XH after HT treatment, but upregulated in MG and XY after HT treatment) (Figures 6B, D). In addition, 22 common TFs encoding six ERFs, five bHLHs, four MYBs, four WRKYs, one NF-YB, one MIKC_MADS, and one AP2 in T_XY_vs_T_BH, T_XY_vs_T_FL, T_XY_vs_T_MG, and T_XY_vs_T_XH. Lists of these DEGs can be found (Supplementary Table S6). Among them, 16 DEGs were significantly upregulated after HT treatment and three DEGs were significantly downregulated after HT treatment (Figures 6C, E).

Figure 6 Transcription factor families were differentially expressed in grape cultivars under the HT treatment. (A) Statistics of the differential expression of transcription factors in the five grape varieties under HT stress. (B, C) Venn clustering analysis was conducted. Shared core sets of up and downregulated TFs in all grape cultivars under HT treatment. (D, E) Heat map of core TF expression. Upregulation is indicated in red, and downregulation is indicated in green. In subpanels (A–E), CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively. XY, MG, XH, BH, and FL represented “Xinyu,” “Miguang,” “Summer Black,” “Beihong,” and “Flame seedless” grapes, respectively.
In the HSFs families, five genes were induced during heat treatment. HSFA-6b (VIT_00s0179g00150) was significantly upregulated in all five grape cultivars after HT treatment. HSFA-6b (VIT_05s0020g04090), HSFAB-2b, HSFB-3, and HSFBC-1 were significantly downregulated in XY, BH, and FL grapes after HT treatment, but were not significantly expressed in MG grapes. Interestingly, heat significantly upregulated the bHLHs and WRKYs genes after HT treatment. Several TFs, including ERFs and MYBs, were specifically downregulated after HT treatment. The basic leucine zipper (bZIPs), NACs, C2H2s genes were heat-regulated in grape leaves and showed different expression patterns in five grape cultivars after HT treatment.
In this study, two pathways representing components of ABA, carotenoid biosynthesis, and ABA signal transduction (Figure 7A), were found to be significantly enriched after HT treatment. Four DEGs correlated with ABA biosynthesis and signal transduction were selected for qRT-PCR analysis (Figures 7B–E), and the expression patterns of qRT-PCR and RNA-Seq were highly consistent. Additional information on these genes can be found in Supplementary Table S7. ABA biosynthesis was significantly enriched with two beta-carotene 3-hydroxylase (crtZ) genes and three 9-cis-epoxycarotenoid dioxygenase (NCEDs) genes. Furthermore, beta-carotene hydroxylase was found to be significantly upregulated in XY and MG but downregulated in XH and FL.

Figure 7 ABA biosynthesis and signaling transduction demonstrating a logfold change in DEGs because of the HT treatment. (A) ABA biosynthesis and signaling transduction pathways. Each row represents a significant DEG. The maximum and minimum values of gene expression in the same row are given a corresponding color. Genes without colors were not differentially expressed in this experiment. (B–E) Expression profiles of the selected DEGs determined using RT-PCR analyses and the line indicates the relative gene transcription level in RNA-Seq. Error bars represented the mean ± SD. XY, MG, XH, BH, and FL represent CK_XY_vs_T_XY, CK_MG_vs_T_MG, CK_XH_vs_T_XH, CK_BH_vs_T_BH, and CK_FL_vs_T_FL, respectively. CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively.
The expression of genes associated with the ABA catabolic process, including three (+)-abscisic acid 8’-hydroxylase (CYP707A). Notably, the interaction between ABA and H2O2, ABA promoted H2O2, and CAT gene expression can inhibit H2O2. Except for MG, three ABA receptor-encoding genes PYL and four protein phosphatase 2c (PP2Cs) were significantly expressed in the other four grape cultivars, as were two serine/threonine-protein kinase (SRK2s) genes and one ABA-responsive element binding factor (ABF) gene associated with ABA signaling transduction.
The transcription of the auxin pathway, including auxin synthesis and signal transduction, was very different among the five grape varieties after heat stress (Figure 8A); the information on these genes is shown in Supplementary Table S8. Among the four pathways of ethylene production, the tryptophan–indolepyruvate–IAA pathway enriched the most DEGs, including two L-tryptophan-pyruvate aminotransferase (TAA1) genes and six indole-3-pyruvate monooxygenase (YUCCAs) genes. XH grapes enriched the most DEGs. Of these, four genes were upregulated and two genes were downregulated. BH grape had the least enriched DEGs, with only two genes. It has been speculated that heat stress mainly affects auxin synthesis in grapes through this pathway. Genes associated with IAA signal transduction were the most affected by HT and tended to show complex expression patterns among the five grape varieties. Seven auxin-responsive protein IAA genes, 33 small auxin up RNA (SAUR) family protein genes and five gretchen hagen 3 (GH3) family genes were significantly enriched. Among them, XH enriched the most downregulated genes. BH enriched more upregulated genes and MG enriched the least DEGs. To validate the RNA-Seq data, five DEGs that correlated IAA biosynthesis and signal transduction were selected for qRT-PCR analysis (Figures 8B–F), and the expression patterns of both qRT-PCR and RNA-Seq were highly consistent.
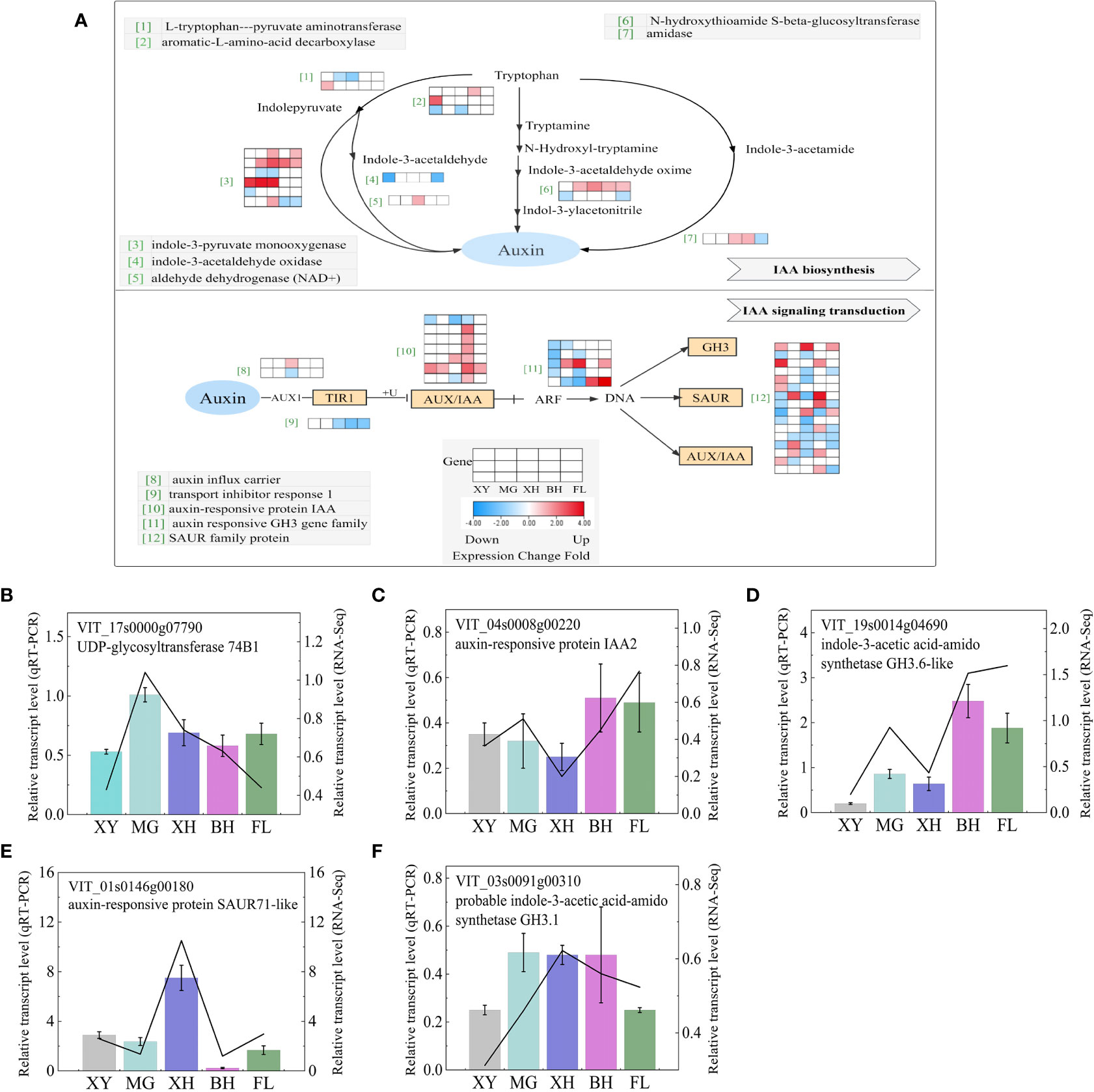
Figure 8 IAA biosynthesis and signaling transduction demonstrating a logfold change in DEGs because of HT treatment. (A) IAA biosynthesis and signaling transduction pathways. Each row represented a significantly DEG. The maximum to minimum values of gene expression in the same row were given a corresponding color. Genes without color were not differentially expressed in this experiment. (B–F) Expression profiles of the selected DEGs determined using RT-PCR analyses and the line indicates the relative gene transcription level in RNA-Seq. Error bars represented the mean ± SD. XY, MG, XH, BH, and FL represent CK_XY_vs_T_XY, CK_MG_vs_T_MG, CK_XH_vs_T_XH, CK_BH_vs_T_BH, and CK_FL_vs_T_FL, respectively. CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively.
We observed that all HT treatments resulted in the major downregulation of genes leading to the production of d-fructose, d-glucose 6-phosphate, d-glucose, and alpha-trehalose (Figure 9A). Three sucrose synthase (SUSs) genes were upregulated in T_XY_vs_T_FL, and two sucrose-phosphate synthase (SPSs) genes were downregulated in four grapes, but not in MG grapes, and the information on these genes is shown in Supplementary Table S9. Four alpha-amylase (AMYs), three beta-amylase, one glycogen phosphorylase (PYG), and two 4-alpha-glucanotransferase (malQs) genes were downregulated in five grapes after HT treatment (Supplementary Table S9). HT treatment resulted in different expression patterns of genes involved in the conversion of UDP-glucose to d-fructose, d-glucose, and maltose.
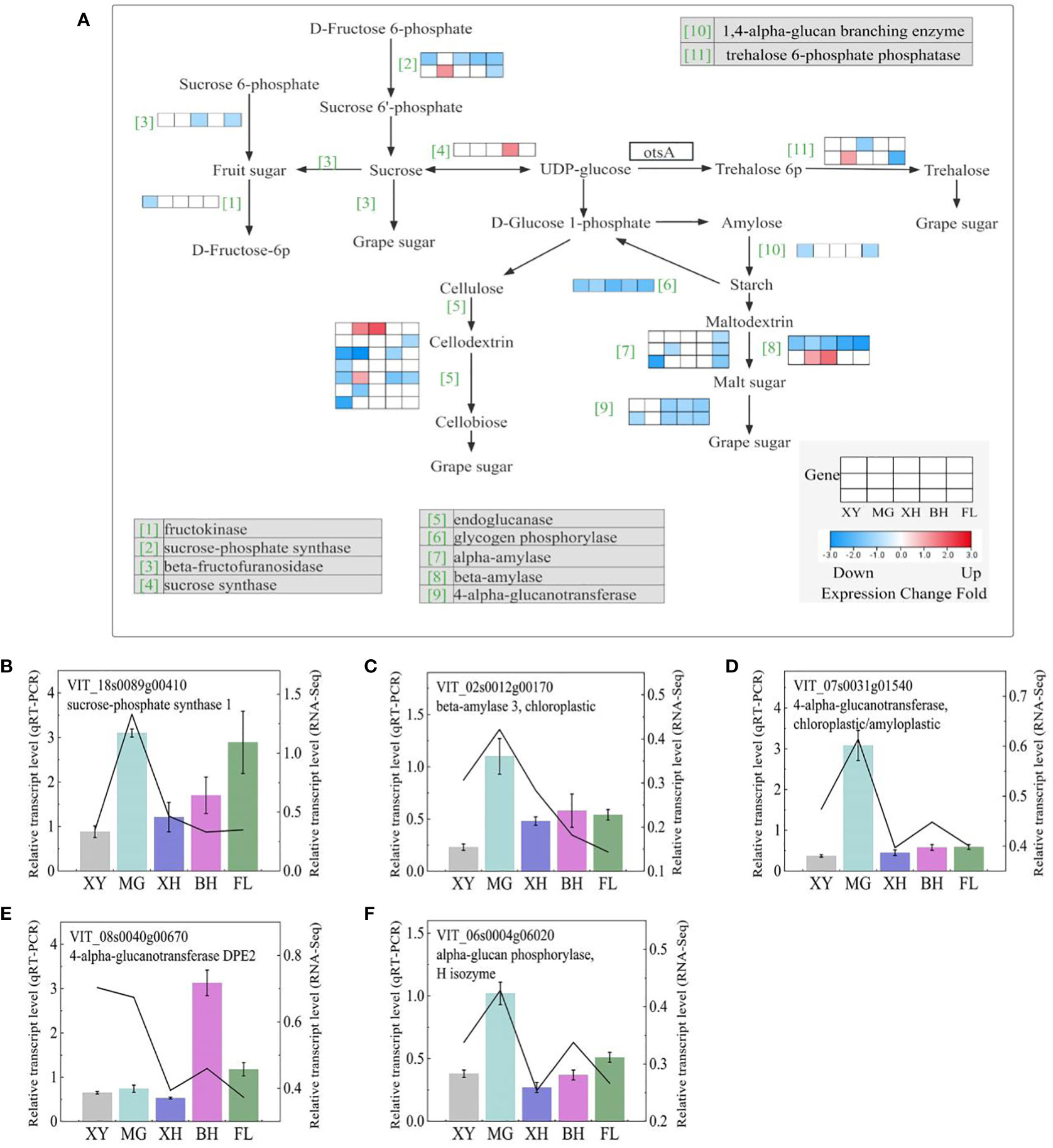
Figure 9 Starch and sucrose metabolism demonstrating major shifts in the expression of metabolic enzymes in response to HT stress in the five grape cultivars. (A) Starch and sucrose metabolism pathways. Each row represents a significant DEGs. The maximum to minimum values of gene expression in the same row were given a corresponding color. Genes without color were not differentially expressed in this experiment. (B–F) Expression profiles of the selected DEGs determined using RT-PCR analyses the line indicated the gene relative transcription level in RNA-Seq. Error bars represent mean ± SD. XY, MG, XH, BH, and FL represent CK_XY_vs_T_XY, CK_MG_vs_T_MG, CK_XH_vs_T_XH, CK_BH_vs_T_BH, and CK_FL_vs_T_FL, respectively. CK and T represent the control treatment (35 °C ± 2 °C) and high-temperature treatment (40 °C ± 2 °C), respectively.
Simultaneously, five DEGs that correlated starch and sucrose metabolism were selected for qRT-PCR analysis (Figures 9B–F), and the expression patterns of both qRT-PCR and RNA-Seq were highly consistent.
A total of 16 WGCNA modules were identified using co-expression network analysis (Figures 10A, B). Among them, the MEpurple module, composed of 167 genes, had the highest correlation with the transpiration rate. The other modules showed lower correlations. There were 17 genes in the MEpurple module (seven HSP20s, three HSPA1s, two HSPE1s, two HSP1s, two HSP90Bs, and one HSPA9) and one heat shock transcription factor (HsfA2). These DEGs were significantly upregulated after high-temperature treatment compared with the controls (Supplementary Figure S6).
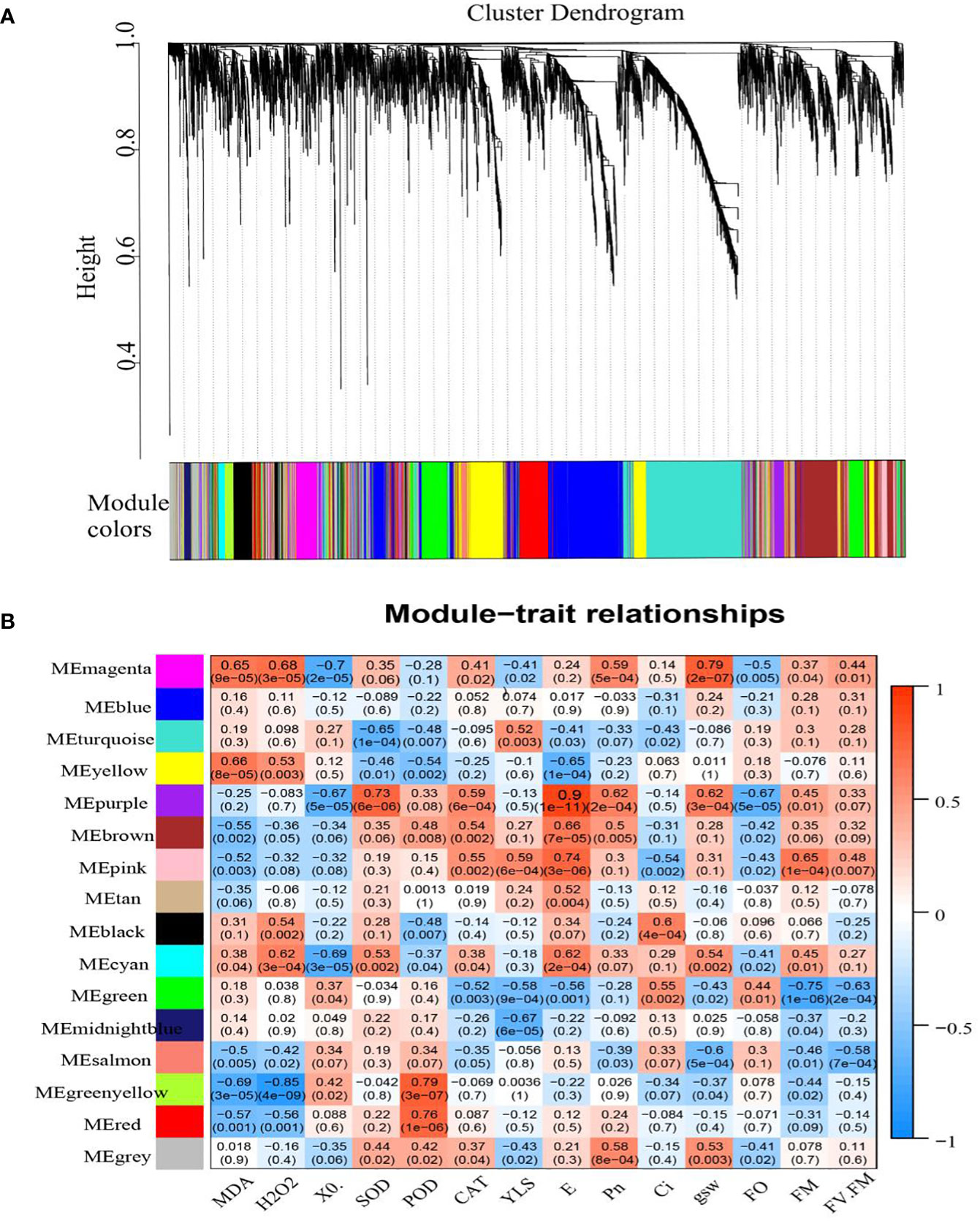
Figure 10 WGCNA of DEGs in grape leaves subjected to HT. (A) Hierarchical clustering of samples. Each color in the figure indicates that each gene in the clustering tree corresponding to one color belongs to the same module. (B) Module—biological character correlations and corresponding p-values. The left panel shows 16 modules. The color scale on the right displays a module-trait correlation from −1 (blue) to 1 (red).
GO enrichment analysis of MEpurple module genes mainly includes “response to temperature stimulus,” “response to heat,” “unfolded protein binding,” “response to abiotic stimulus,” and “response to reactive oxygen species” terms (Supplementary Figure S7A). By comparing the enrichment analysis with KEGG data, the MEpurple module covered pathways related to, “Spliceosome,” “Protein processing in endoplasmic reticulum,” and “Endocytosis” (Q <0.05) (Supplementary Figure S7B).
In addition, we established a MEpurple gene regulation network using WGCNA. The results showed that VIT_06s0004g00240, VIT_05s0051g00340, VIT_01s0011g04990, and VIT_12s0057g00670 were mainly involved in folding, sorting, and degradation (Supplementary Figure S8).
HT stress has been shown to significantly reduce grape yield, leading to a lower economic income. We investigated the physicochemical and transcriptomic changes that occur under these conditions to understand the impact of HT on different heat-sensitive grape varieties.
Under normal conditions, the ROS content in plants is low, which plays an important role in maintaining the stability of the intracellular signaling system (Schneider et al., 2019; Nadarajah, 2020). After HT stress, plants produced a large amount of ROS. This aggravates the degree of membrane lipid peroxidation and leads to cell death, thus inhibiting plant growth (Qi et al., 2010). Important antioxidant enzymes in plant cells include SOD, CAT, and POD. Antioxidant enzymes can remove H2O2 and O2− produced by HT stress to maintain ROS in cells and protect the stability of cell membranes. SOD can convert O2− into H2O2 and O2, whereas POD and CAT can further decompose H2O2 into H2O and O2 (Ruelland and Zachowski, 2010). In this study, the H2O2, O2−, and MDA contents in the five grape leaves increased significantly after HT treatment. BH and XH accumulated higher levels of H2O2 and MDA, which may lead to damage to the cell membrane. However, XY and FL were significantly lower than those of the other cultivars. An increase in MDA and H2O2 content caused by HT stress has also been reported in other plants, such as rice and pepper (Kumar et al., 2012; Li et al., 2015). In this study, the enzyme activities of SOD, CAT, and POD decreased in the five cultivars. This indicated that HT stress inhibited the expression of the enzymes or destroyed their structure of the enzymes, causing a decrease in the activities of the protective enzymes.
We performed mRNA sequencing to reveal differences in gene expression among the five grape varieties. The five grape cultivars exhibited distinct differences in their transcriptome levels in response to heat stress. After HT stress, T_XY_VS_T_BH and CK_XY_VS_T_XY showed the highest numbers of DEGs. A previous study also found that the number of DEGs was far greater in heat-resistant jujube and Pyropia haitanensis strains than in the heat-sensitive jujube and P. haitanensis strains (Wang et al., 2018; Jin et al., 2020). In our study, XY was better able to increase transcriptional regulation in response to HT stress than BH grapes. Through GO and KEGG enrichment analysis, these DEGs were mainly enriched in “response to temperature stimulus,” “response to active oxygen specifications,” “response to heat,” “response to hydrogen peroxide,” “response to abiotic stimulus,” “protein processing in endogenous reticulum,” “plant hormone signal transformation,” and “cartotenoid biosynthesis.” This may reflect their similar responses to heat stress, which is similar to previous studies (Jin et al., 2020; Liu M. et al., 2020). This indicated that similar key regulatory pathways play important roles in the response to high-temperature stress.
HSPs are important cellular response proteins in plants that can be immediately induced by HT (Liu et al., 2012; Wang et al., 2019). HSP70 and HSP90 have been identified as heat response factors in tomatoes, dates, and rice (Frank et al., 2009; Jung et al., 2012; Jin et al., 2020). The expression levels of HSPs (HSP17.6, HSP22, HSP21, and HSP40) are significantly increased in grapevine leaves under heat stress (Wang et al., 2010; Jiang et al., 2017). In this study, a total of 36 HSPs chaperones were significantly up regulated by HT stress, including HSP101, HSP70, HSP83, HSP90, HSP18.1, HSP17.3, HSP17.1, and HSP25.3. These results indicate that upregulated HSP genes play an important role in the heat tolerance of grapes.
TFs are important for regulating plant development and stress responses (Amorim et al., 2017; Ng et al., 2018). Under HT stress, heat stress-induced or heat-suppressed TFs in plants potentially contribute to the differential regulation of downstream genes (Wahid et al., 2007). HSF is closely related to the regulation of plant thermal stress and has been reported in Arabidopsis, rice, tomato, and potato (Song et al., 2016; Ibanez et al., 2017; Singh et al., 2018; Fragkostefanakis et al., 2019). HSFA2 regulates the expression of some HSPs genes, such as HSP101, HSP70, and HSP15.7 (Nishizawa et al., 2006; Jiang et al., 2017). We found 15 HSFs genes, but only four genes were induced during heat treatment. HSFA2 and HSFA-6b were significantly upregulated in five grapes, similar to previous studies (Liu et al., 2012; Jiang et al., 2017). At present, research on WRKY transcription factor under HT stress has mainly focused on Arabidopsis (Li et al., 2011), wheat (He et al., 2016), and pepper (Cai et al., 2015). We found that WRKY33, which could be involved in the heat response of grape leaves, was upregulated in three grape cultivars. Overexpression of MYB genes can enhance the thermotolerance of genetic plants (Amano et al., 2012; Zhao et al., 2017). Wang et al. (2019) found that the expression of some MYB transcription factors changed after heat stimulation in grape leaves. Our results indicated that MYBA6 and MYB108-like are induced by HT. However, MYB114 was strongly repressed in five grapes, indicating that they were involved in the HT stress response. The overexpression of TaNAC2L and NAC019 can improve thermotolerance in Arabidopsis (Guan et al., 2014; Guo et al., 2015). NAC56 was highly expressed in MG grapes, but downregulated in other grapes, suggesting that NAC56 participates in the heat stress response and plays different roles in heat-resistant and heat-sensitive cultivars. bHLH plays an important role in plant stress resistance. However, there are few reports on the response of bHLH cells to HT stress. In our study, many bHLH genes were significantly upregulated after heat treatment, which is consistent with reports on tea (Wang et al., 2019) and wheat (Cui et al., 2018).
Abiotic and biotic stresses are effectively responded to by abscisic acid (ABA) and is called “stress hormone” (Suzuki et al., 2016; Zhu, 2016). Adversity stress leads to a rapid increase in ABA content in plants, which enhances ABA signaling and thus improves stress resistance (Ji et al., 2011). A key rate-limiting enzyme in ABA biosynthesis is 9-cis-epoxycarotenoid dioxygenase (NCED) (Hwang et al., 2018), which has an obvious regulatory effect on abiotic stresses, such as drought (Li et al., 2018) and heat stress (Zhou et al., 2022). In our study, NCED2 was greatly upregulated in three grapes and NCED1 was upregulated in one grape after HT treatment. Drought stress has been reported to be triggered by PYR/PYL/RCAR (hereafter PYLs) proteins. These proteins function as ABA receptors (Pizzio et al., 2013; Liu et al., 2019). Two PYL4s and one PYR1 were upregulated after HT in the four grape cultivars. This indicates that, under HT stress, the VvPYL4 gene is involved in the defense response induced by HT. Protein phosphate 2C (PP2C), a key regulator of the ABA signaling pathway, also has an obvious regulatory effect on abiotic stress (Manabe et al., 2007; Bhaskara et al., 2017; Lenka et al., 2018). In this study, one PP2C and four probable PP2Cs were upregulated and downregulated, respectively, after HT in the four grape cultivars, and there was no differential expression in MG grapes.
In this study, we found that the expression patterns of genes related to IAA synthesis were significantly altered in plants exposed to heat stress. After HT stress, most GH3 genes were downregulated in CK_MG_vs_T_MG, CK_XH_vs_T_XH, and CK_XY_vs_T_XY. This is consistent with the results of previous studies (Du et al., 2013). Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels (Salehin et al., 2019). IAA5, IAA6, and IAA19 are important for drought responses (Shani et al., 2016). Plants overexpressing OsIAA20 showed the opposite phenotype to that of OsIAA20 RNAi transgenic rice (Zhang et al., 2021). After HT treatment, IAA9 was upregulated in the four grape cultivars, and the upregulation in the expression of IAA28 was BH grape-specific.
Many crop species have been shown to be stressed by abiotic stresses related to sugars (Parrotta et al., 2016; Londo et al., 2018; Wang et al., 2020). In higher plants, sucrose synthase (SuS) and sucrose phosphate synthase (SPS) are the key enzymes involved in sucrose metabolism. A recent study found that SUS and SPS might participate in resistance to HT stress (Zhou et al., 2016; Kana et al., 2018; Verma et al., 2018). The HT treatment repressed the expression of SUS7 and SPS1 in the five grape cultivars. When stress was present, SUS and Invertase (INV) levels decreased before ABA levels increased, and stress-induced increases in ABA further inhibited their expression. (Ruan, 2014). HT treatment resulted in the following results: SPS, SUS, and INV-related genes were significantly downregulated in CK_FL_vs_T_FL and CK_XY_vs_T_XY, indicating that HT inhibited sucrose metabolism in XY and FL grapes, whereas XY grapes had a significantly increased sensitivity to HT in their sucrose metabolism.
Alpha-amylase (AMY) and beta-amylase are the main enzymes involved in the catabolism of starch. In our study, the expression of four AMY and two beta-amylase genes encoding enzymes that degrade starch into maltose and dextrin in five grape varieties was downregulated after HT treatment. Two 4-alpha-glucanotransferases (malQ) responsible for the degradation of malt sugar into grape sugar were also downregulated, suggesting that HT stress inhibited starch degradation, thereby repressing the accumulation of grape sugar, especially in FL grapes. The enzyme cellulase hydrolyzes cellulose to glucose, which is then degraded by endoglucanase (EGL) (Payne et al., 2015; Chylenski et al., 2019). AnEGL demonstrated salt tolerance and thermostability in high salinity environments (Cai et al., 2022). Interestingly, six EGLs responsible for the degradation of cellulose into cellodextrin and cellobiose were also downregulated in the five grapes after HT treatment, suggesting that cellobiose may be the terminal response to heat. Compared with other grape cultivars, these EGLs genes were significantly upregulated, indicating that cellulose degradation in XY grapes is more sensitive to HT. Overall, all heat stresses in the five grape cultivars decreased the production of simple soluble sugars used in general metabolism, likely resulting in reduced growth and development.
When plants are exposed to abiotic stresses, the endoplasmic reticulum plays a major role (Liu et al., 2021; Reyes-Impellizzeri and Moreno, 2021; Cao et al., 2022). Arabidopsis antioxidant defenses are modulated by ROS signaling induced by ER stress (Ozgur et al., 2014). Tomato expressing ER-sHSP constitutively displayed improved salinity tolerance (Fu et al., 2016). In our research, 41 HSPs participated in the protein processing in endoplasmic reticulum and significantly up-regulated expression in five grape cultivars after HT treatment, indicating that ER-sHSP actively responds to HT treatment to alleviate the damage of HT to grapes. Lists of these DEGs can be found (Supplementary Table S11). Calreticulin (CRT) and protein disulfide isomerase (PDI) are molecular chaperones that is involved in the pivotal protein folding in ER, they had been reported about abiotic stress (Jia et al., 2008; Xiang et al., 2015; Wang et al., 2017; Feldeverd et al., 2020; Wai et al., 2020; Meng et al., 2021). After HT stress, two CRTs were differentially expressed, of which VIT_14s0060g01290 was upregulated in CK_FL_vs_T_FL, while VIT_07s0005g01390 was differentially expressed among grape cultivars (Supplementary Table S11). One PDI gene (VIT_12s0059g01560) was found to increase expression levels in CK_FL_vs_T_FL and CK_XY_vs_T_XY under HT stress (Supplementary Table S11), this was consistent with previous research results (Wang et al., 2017; Feldeverd et al., 2020). In summary, sHSP, CRT, and PDI participated in the processing of proteins in the endoplasmic reticulum, affecting grape antioxidant defense and leading to changes in heat resistance.
In this study, BH and XH grapes accumulated higher levels of H2O2 and MDA after HT treatment, causing oxidative damage to plants. At the same time, the maximum increases in Ci and Fo were observed in BH grapes. However, XY and FL grapes were significantly lower than the other varieties, indicating that XY and FL grapes were better adapted to this HT environment than BH and XH grapes, followed by MG grapes. Meanwhile, with the help of RNA-Seq analysis, we investigated the underlying mechanisms associated with grape cultivar heat stress responses. We identified 83 shared genes between natural environment control conditions and high-temperature stress treatments across all five grape cultivars. We found that HT treatment resulted in a greater number of upregulated than downregulated genes in different grape varieties. GO and KEGG analyses revealed that DEGs in response to heat stress were enriched in metabolic pathways, protein processing in the endoplasmic reticulum, plant hormone signal transduction, and starch and sucrose metabolism. HT treatment significantly promoted DEGs involved in protein processing in the endoplasmic reticulum pathway, and many HSPs were upregulated. This indicated that grape can improve its heat tolerance by rapidly accumulating heat shock proteins under HT treatment. Our study indicates that examining the HT treatment response of grapevines at 40 °C does not fully elucidate or identify the most important elements of the heat stress response. At 35 °C and 40 °C, we observed very different transcriptional landscapes for key hormones, transcription factors, and sugar pathways under natural temperature and HT conditions. Based on these results, we may gain a better understanding of the molecular mechanisms of the grape heat stress response.
The data presented in the study are deposited in the NCBI SRA database, accession link: https://www.ncbi.nlm.nih.gov/sra/PRJNA1053489.
FD: Data curation, Writing – original draft, Writing – review & editing. FP: Methodology, Writing – review & editing. GL: Investigation, Supervision, Writing – review & editing. JZ: Investigation, Supervision, Writing – review & editing. LZ: Data curation, Writing – review & editing. YW: Data curation, Supervision, Validation, Writing – review & editing. HL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program [2018YFD1000304].
The authors acknowledge the specific colleagues, institutions, or agencies that contributed to their efforts.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1313832/full#supplementary-material
Amano, M., Iida, S., Kosuge, K. (2012). Comparative studies of thermotolerance: different modes of heat acclimation between tolerant and intolerant aquatic plants of the genus Potamogeton. Ann. Bot. 109 (2), 443. doi: 10.1093/aob/mcr300
Amorim, L. L. B., dos Santos, R. D., Neto, J. P. B., Guida-Santos, M., Crovella, S., Benko-Iseppon, A. M. (2017). Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18, 335–351. doi: 10.2174/1389203717666160619185308
Bhaskara, G. B., Wen, T., Nguyen, T. T., Verslues, P. E. (2017). Protein phosphatase 2Cs and Microtubule-Associated stress protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 29 (1), 169–191. doi: 10.1105/tpc.16.00847
Bineau, E., Diouf, I., Carretero, Y., Duboscq, R., Bitton, F., Djari, A., et al. (2021). Genetic diversity of tomato response to heat stress at the QTL and transcriptome levels. Plant J. 107 (4), 1213–1227. doi: 10.1111/tpj.15379
Bita, C. E., Gerats (2013). Plant tolerance to HT in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00273
Blair, E. J., Bonnot, T., Hummel, M., Hay, E., Marzolino, J. M., Quijada, I. A., et al. (2019). Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 9 (1), 1–12. doi: 10.1038/s41598-019-41234-w
Cai, H. Y., Yang, S., Yan, Y., Xiao, Z. L., Cheng, J. B., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Cai, L. N., Xu, S. N., Lu, T., Lin, D. Q., Yao, S. J. (2022). Salt-tolerant and thermostable mechanisms of an endoglucanase from marine aspergillus niger. Bioresour. Bioprocess. 9, 44. doi: 10.1186/s40643-022-00533-3
Cao, J., Wang, C., Hao, N., Fujiwara, T., Wu, T. (2022). Endoplasmic reticulum stress and reactive oxygen species in plants. Antioxidants (Basel). 11 (7), 1240. doi: 10.3390/antiox11071240
Charng, Y. Y., Liu, H. C., Liu, N. Y., Hsu, F. C., Ko, S. S. (2006). Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 140 (4), 1297–1305. doi: 10.1104/pp.105.074898
Cheng, Y., Cheng, L., Mi, Y. H., Duan, H. P., Cha, Y. S., Shao, J. L., et al. (2018). Comparative study on various methods for determination of activity of antioxidant enzymes in rice. Jiangxi Agric. J. 30 (02), 108–111. doi: 10.19386/j.cnki.jxnyxb.2018.02.23
Chylenski, P., Bissaro, B., Sørlie, M., Røhr, Å. K., Várnai, A., Horn, S. J., et al. (2019). Lytic polysaccharide monooxygenases in enzymatic processing of lignocellulosic biomass. ACS Catal. 9 (6), 4970–4991. doi: 10.1021/acscatal.9b00246
Cui, X., Wang, Y. X., Liu, Z. W., Wang, W. L., Li, H., Zhuang, J. (2018). Transcriptome-wide identification and expression profile analysis of the bHLH family genes in Camellia sinensis. Funct. Integr. Genomics 18 (5), 489–503. doi: 10.1007/s10142-018-0608-x
Du, H., Liu, H. B., Xiong, L. Z. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00397
Feldeverd, E., Porter, B. W., Yuen, C. Y. L., Iwai, K., Carrillo, R., Smith, T., et al. (2020). The arabidopsis protein disulfide isomerase subfamily m isoform, PDI9, localizes to the endoplasmic reticulum and influences pollen viability and proper formation of the pollen exine during heat stress. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.610052
Fraga, H., García de Cortázar Atauri, I., Malheiro, A. C., Santos, J. A. (2016). Modelling climate change impacts on viticultural yield, phenology and stress conditions in Europe. Global Change Biol. 22 (11), 3774–3788. doi: 10.1111/gcb.13382
Fragkostefanakis, S., Simm, S., El-Shershaby, A., Hu, Y. J., Bublak, D., Mesihovic, A., et al. (2019). The repressor and co-activator HsfB1 regulates the major heat stress transcription factors in tomato. Plant Cell Environ. 42, 874–890. doi: 10.1111/pce.13434
Frank, G., Pressman, E., Ophir, R., Althan, L., Shaked, R., Freedman, M., et al. (2009). Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones,and sugars in the heat stress response. J. Exp. Bot. 60, 3891–3908. doi: 10.1093/jxb/erp234
Fu, C., Liu, X. X., Yang, W. W., Zhao, C. M., Liu, J. (2016). Enhanced salt tolerance in tomato plants constitutively expressing heat-shock protein in the endoplasmic reticulum. Genet. Mol. Res. 15 (2). doi: 10.4238/gmr.15028301
González-Schain, N., Dreni, L., Lawas, L. M. F., Galbiati, M., Colombo, L., Heuer, S., et al. (2016). Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 57 (1), 57–68. doi: 10.1093/pcp/pcv174
Guan, Q. M., Yue, X. L., Zeng, H. T., Zhu, J. H. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26 (1), 438–453. doi: 10.1105/tpc.113.118927
Guo, W. W., Zhang, J. X., Zhang, N., Xin, M. M., Peng, H. R., Hu, Z. R., et al. (2015). The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic arabidopsis. PLoS One 10, 1–11. doi: 10.1371/journal.pone.0135667
He, G. H., Xu, J. Y., Wang, Y. X., Liu, J. M., Li, P. S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16, 116. doi: 10.1186/s12870-016-0806-4
Higashi, Y., Okazaki, Y., Myouga, F., Shinozaki, K., Saito, K. (2015). Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 5 (1), 1–11. doi: 10.1038/srep10533
Hwang, S. G., Lee, C. Y., Tseng, C. S. (2018). Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot. Stud. 59 (1), 1–12. doi: 10.1186/s40529-018-0219-9
Ibanez, C., Poeschl, Y., Peterson, T., Bellstadt, J., Denk, K., Gogol-Doring, A., et al. (2017). Ambient temperature and genotype differentially affect developmental and phenotypic plasticity in Arabidopsis thaliana. BMC Plant Biol. 17, 114. doi: 10.1186/s12870-017-1068-5
Ji, X. M., Dong, B. D., Shiran, B., Talbot, M. J., Edlington, J. E., Hughes, T., et al. (2011). Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156 (2), 647–662. doi: 10.1104/pp.111.176164
Jia, X. Y., Xu, C. Y., Jing, R. L., Li, R. Z., Mao, X. G., Wang, J. P., et al. (2008). Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J. Exp. Bot. 59 (4), 739–751. doi: 10.1093/jxb/erm369
Jiang, H., Du, J., Mao, L., Li, Y., Yue, Y., Lu, J. (2020). Summary of transcription factors in response to HT stress in plants. Mol. Plant Breed. 18 (10), 3251–3258. doi: 10.13271/j.mpb.018.003251
Jiang, J., Liu, X., Liu, C., Liu, G., Li, S., Wang, L. (2017). Integrating omics and alternative splicing reveals insights into grape response to HT. Plant Physiol. 173 (2), 1502–1518. doi: 10.1104/pp.16.01305
Jin, J., Yang, L., Fan, D., Liu, X., Hao, Q. (2020). Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars. PLoS One 15 (9), e0235763. doi: 10.1371/journal.pone.0235763
Jung, K. H., Ko, H. J., Nguyen, M., Kim, S. R., Ronald, P., An, G. (2012). Genome-wide identification and analysis of early heat stress responsive genes in rice. J. Plant Biol. 55, 458–468. doi: 10.1007/s12374-012-0271-z
Kana, T., Kazumasa, M., Takuya, Y., Kohei, Y., Genki, C., Shintaro, K., et al. (2018). Thermo-responsive allele of sucrose synthase 3 (Sus3) provides high-temperature tolerance during the ripening stage in rice (Oryza sativa l.). Breed. Sci. 68 (3), 336–342. doi: 10.1270/jsbbs.18007
Keller, M., Bokszczanin, K., Bostan, H., Bovy, A. G., Simm, S. (2018). The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genomics 19 (1), 447. doi: 10.1186/s12864-018-4824-5
Kim, S. A., Ahn, S. Y., Yun, H. K. (2018). Selection of differentially expressed genes using the transcriptome analysis of ripening grape berries in response to HT. J. Agric. Sci-Sri. Lanka. 13, 15–30. doi: 10.4038/jas.v13i1.8297
Kumar, S., Gupta, D., Nayyar, H. (2012). Comparative response of maize and rice genotypes to heat stress: status of oxidative stress and antioxidants. Acta Physiologiae Plantarum 34 (1), 75–86. doi: 10.1007/s11738-011-0806-9
Langfelder, P., Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559–571. doi: 10.1186/1471-2105-9-559
Larkindale, J., Huang, B. (2004). Changes of lipid composition and saturation level in leaves and roots for heat-stressed and heat-acclimated creeping bentgrass (Agrostis stolonifera). Environ. Exp. Bot. 51 (1), 57–67. doi: 10.1016/S0098-8472(03)00060-1
Lecourieux, F., Kappel, C., Pieri, P., Charon, J., Pillet, J., Hilbert, G., et al. (2017). Dissecting the biochemical and transcriptomic effects of a locally applied heat treatment on developing Cabernet Sauvignon grape berries. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00053
Lenka, S. K., Muthusamy, S. K., Chinnusamy, V., Bansal, K. C. (2018). Ectopic expression of rice PYL3 enhances cold and drought tolerance in Arabidopsis thaliana. Mol. Biotechnol. 60 (5), 350–361. doi: 10.1007/s12033-018-0076-5
Li, S. J., Fu, Q. T., Chen, L. G., Huang, W. D., Yu, D. Q. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, C., Li, C., Wang, B. B., Zhang, R. Q., Fu, K. Y., Gale, W. L., et al. (2018). Programmed cell death in wheat (Triticum aestivum L.) endosperm cells is affected by drought stress. Protoplasma 255 (4), 1039–1052. doi: 10.1007/s00709-018-1203-7
Li, T., Xu, X., Li, Y., Wang, H. M., Li, Z. L., Li, Z. X. (2015). Comparative transcriptome analysis reveals differential transcription in heat-susceptible and heat-tolerant pepper (Capsicum annum L.) cultivars under heat stress. J. Plant Biol. 58 (6), 411–424. doi: 10.1007/s12374-015-0423-z
Liu, M., Ju, Y. L., Min, Z., Fang, Y. L., Meng, J. F. (2020). Transcriptome analysis of grape leaves reveals insights into response to heat acclimation. Scientia Hortic. 272, 109554. doi: 10.1016/j.scienta.2020.109554
Liu, B. L., Kong, L. S., Zhang, Y., Liao, Y. C. (2021). Gene and metabolite integration analysis through transcriptome and metabolome brings new insight into heat stress tolerance in potato (Solanum tuberosum L.). Plants 10 (1), 103. doi: 10.3390/plants10010103
Liu, G. T., Wang, J. F., Cramer, G., Dai, Z. W., Duan, W., Xu, H. G., et al. (2012). Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 12 (1), 174. doi: 10.1186/1471-2229-12-174
Liu, H., Wang, J., Sun, H. M., Han, X. B., Du, B. H. (2020). Transcriptome profiles reveal the growth-promoting mechanisms of Paenibacillus polymyxa YC0136 on tobacco (Nicotiana tabacum L.). Front. Microbiol. 11. doi: 10.3389/fmicb.2020.584174
Liu, S. L., Yang, R. J., Pan, Y. Z., Ding, J. J., He, Y., Wang, L. (2013). Effects of exogenous nitric oxide on lipid peroxidation and ATPase activity in plasma membrane and photosynthetic characteristics of catharanthus roseus under cadmium stress. J. Agro-Environment Sci. 32 (12), 2360–2368. doi: 10.11564/jaes.2013.12.008
Liu, G., Zha, Z. P., Cai, H. Y., Qin, D. D., Jiao, C. H. (2020). Dynamic transcriptome analysis of anther response to heat stress during anthesis in thermotolerant rice (Oryza sativa L.). Int. J. Mol. Sci. 21 (3), 1155. doi: 10.3390/ijms21031155
Liu, J., Zhao, F. L., Guo, Y., Fan, X. C., Wang, Y. J., Wen, Y. Q. (2019). The ABA receptor-like gene VyPYL9 from drought-resistance wild grapevine confers drought tolerance and ABA hypersensitivity in Arabidopsis. Plant Cell Tissue Organ Cult. 138 (3), 543–558. doi: 10.1007/s11240-019-01650-2
Londo, J. P., Kovaleski, A. P., Lillis, J. A. (2018). Divergence in the transcriptional landscape between low temperature and freeze shock in cultivated grapevine (Vitis vinifera). Hortic. Res. 5, 10. doi: 10.1038/s41438-018-0020-7
Luo, H. B., Ling, M., Xi, H. F., Duan, W., Li, S. H., Wayne, L., et al. (2011). Photosynthetic responses to heat treatments at different temperatures and following recovery in grapevine (vitis amurensis L.) leaves. PLoS One 6 (8), e23033. doi: 10.1371/journal.pone.0023033
Ma, C., Burd, S., Lers, A. (2015). miR408 is involved in abiotic stress responses in arabidopsis. Plant J. 84 (1), 169–187. doi: 10.1111/tpj.12999
Manabe, Y., Bressan, R. A., Wang, T., Li, F., Koiwa, H., Sokolchik, I., et al. (2007). The Arabidopsis kinase-associated protein phosphatase regulates adaptation to Na+ stress. Plant Physiol. 146 (2), 612–622. doi: 10.1104/pp.107.109009
Meng, Z., Zhao, Y., Liu, L., Du, X. (2021). Genome-wide characterization of the PDI gene family in medicago truncatula and their roles in response to endoplasmic reticulum stress. Genome 64 (6), 599–614. doi: 10.1139/gen-2020-0064
Moreno, A. A. (2021). The endoplasmic reticulum role in the plant response to abiotic stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.755447
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21 (15), 5208. doi: 10.3390/ijms21155208
Ng, D. W. K., Abeysinghe, J. K., Kamali, M. (2018). Regulating the regulators: the control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19, 3737. doi: 10.3390/ijms19123737
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Ozgur, R., Turkan, I., Uzilday, B., Sekmen, A. H. (2014). Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of arabidopsis thaliana. J. Exp. Bot. 65, 1377–1390. doi: 10.1093/jxb/eru034
Park, C. J., Seo, Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31 (4), 323. doi: 10.5423/PPJ.RW.08.2015.0150
Parrotta, L., Faleri, C., Cresti, M., Cai, G. (2016). Heat stress affects the cytoskeleton and the delivery of sucrose synthase in tobacco pollen tubes. Planta 243 (1), 43–63. doi: 10.1007/s00425-015-2394-1
Payne, C. M., Knott, B. C., Mayes, H. B., Hansson, H., Himmel, M. E., Sandgren, M., et al. (2015). Fungal cellulases. Chem. Rev. 115 (3), 1308–1448. doi: 10.1021/cr500351c
Pizzio, G. A., Rodriguez, L., Antoni, R., Gonzalez-Guzman, M., Yunta, C., Merilo, E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163 (1), 441–455. doi: 10.1104/pp.113.224162
Qi, Y. C., Wang, H. J., Zou, Y., Liu, C., Liu, Y. Q., Wang, Y., et al. (2010). Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett. 585 (1), 231–239. doi: 10.1016/j.febslet.2010.11.051
Rahmati, I. M., Brown, E., Weigand, C., Tillett, R. L., Schlauch, K. A., Miller, G., et al. (2018). A comparison of heat-stress transcriptome changes between wild-type Arabidopsis pollen and a heat-sensitive mutant harboring a knockout of cyclic nucleotide-gated cation channel 16 (cngc16). BMC Genomics 19 (1), 1–19. doi: 10.1186/s12864-018-4930-4
Reyes-Impellizzeri, S., Moreno, A. A. (2021). The endoplasmic reticulum role in the plant response to abiotic stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.755447
Rienth, M., Torregrosa, L., Luchaire, N., Chatbanyong, R., Lecourieux, D., Kelly, M. T., et al. (2014). Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 14 (1), 1–18. doi: 10.1186/1471-2229-14-108
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65 (1), 33–67. doi: 10.1146/annurev-arplant-050213-040251
Ruelland, E., Zachowski, A. (2010). How plants sense temperature. Environ. Exp. Bot. 69 (3), 225–232. doi: 10.1016/j.envexpbot.2010.05.011
Salehin, M., Li, B., Tang, M., Katz, E., Song, L., Ecker, J. R., et al. (2019). Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 10 (1), 4021. doi: 10.1038/s41467-019-12002-1
Schneider, J. R., Caverzan, A., Chavarria, G. (2019). Water deficit stress, ROS involvement, and plant performance. Arch. Agron. Soil Sci. 65 (8), 1160–1181. doi: 10.1080/03650340.2018.1556789
Schultz, H. R., Jones, G. V. (2010). Climate induced historic and future changes in viticulture. J. Wine Res. 21 (2-3), 137–145. doi: 10.1080/09571264.2010.530098
Shani, E., Salehin, M., Zhang, Y. Q., Sanchez, S. E., Doherty, C., Wang, R. H., et al. (2016). Plant stress tolerance requires Auxin-sensitive Aux/IAA transcriptional repressors. Curr. Biol. 27 (3), 437–444. doi: 10.1016/j.cub.2016.12.016
Singh, G., Sarkar, N. K., Grover, A. (2018). Mapping of domains of heat stress transcription factor OsHsfA6a responsible for its transacti vation activity. Plant Sci. 274, 80–90. doi: 10.1016/j.plantsci.2018.05.010
Song, C., Chung, W. S., Lim, C. O. (2016). Overexpression of heat shock factor gene HsfA3 Increases galactinol levels and oxidative stress tolerance in arabidopsis. Mol. Cells 39, 477–483. doi: 10.14348/molcells.2016.0027
Suzuki, N., Bassil, E., Hamilton, J. S., Inupakutika, M. A., Zandalinas, S. I., Tripathy, D., et al. (2016). ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One 11 (1), e0147625. doi: 10.1371/journal.pone.0147625
Tang, R. M., Gupta, S. K., Niu, S. Y., Li, X. Q., Yang, Q., Chen, G. S., et al. (2020). Transcriptome analysis of heat stress response genes in potato leaves. Mol. Biol. Rep. 47 (6), 4311–4321. doi: 10.1007/s11033-020-05485-5
Verma, E., Sharma, B., Singal, H. R., Munjal, R. (2018). Purification of sucrose synthase from thermotolerant wheat grains and its characterization. J. Environ. Biol. 39 (4), 459–466. doi: 10.22438/jeb/39/4/MRN-503
Wahid, A., Gelani, S., Ashraf, M., Foolad, M. R. (2007). Heat tolerance in plants: An overview. Environ. Exp. Bot. 61 (3), 199–223. doi: 10.1016/j.envexpbot.2007.05.011
Wai, A. H., Waseem, M., Khan, A.B.M.M.M., Nath, U. K., Lee, D. J., Kim, S. T., et al. (2020). Genome-wide identification and expression profiling of the PDI gene family reveals their probable involvement in abiotic stress tolerance in tomato (Solanum lycopersicum l.). Genes (Basel) 12 (1), 23. doi: 10.3390/genes12010023
Wang, L. J., Fan, L., Loescher, W., Duan, W., Liu, G. J., Cheng, J. S., et al. (2010). Salicylic acid alleviates decreases in photosynthesis under heat stress and accelerates recovery in grapevine leaves. BMC Plant Biol. 10, 34. doi: 10.1186/1471-2229-10-34
Wang, L. J., Li, S. H. (2006). Salicylic acid-induced heat or cold tolerance in relation to Ca2+ homeostasis and antioxidant systems in young grape plants. Plant Sci. 170 (4), 685–694. doi: 10.1016/j.plantsci.2005.09.005
Wang, W. L., Lin, Y. H., Teng, F., Ji, D. H., Xu, Y., Chen, C. S., et al. (2018). Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to HT stress. Algal Res. 29, 104–112. doi: 10.1016/j.algal.2017.11.026
Wang, L. J., Loescher, W., Duan, W., Li, W. D., Yang, S. H., Li, S. H. (2009). Heat acclimation induced acquired heat tolerance and cross adaptation in different grape cultivars: relationships to photosynthetic energy partitioning. Funct. Plant Biol. 36, 516–526. doi: 10.1071/FP09008
Wang, H., Niu, H., Zhai, Y., Lu, M. (2017). Characterization of BiP genes from pepper (Capsicum annuum l.) and the role of CaBiP1 in response to endoplasmic reticulum and multiple abiotic stresses. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01122
Wang, L., Xiang, L., Hong, J., Xie, Z., Li, B. (2019). Genome-wide analysis of bHLH transcription factor family reveals their involvement in biotic and abiotic stress responses in wheat (Triticum aestivum L.). 3 Biotech. 9 (6), 236. doi: 10.1007/s13205-019-1742-4
Wang, Y. L., Zhang, Y. K., Shi, Q. H., Chen, H. Z., Xiang, J., Hu, G. H., et al. (2020). Decrement of sugar consumption in rice young panicle under HT aggravates spikelet number reduction. Rice Sci. 27 (01), 44–55. doi: 10.1016/j.rsci.2019.12.005
Wu, J. Y., Liu, L. Y., Xu, G. X., Jiang, J. F., Lian, W. J., Zhou, H., et al. (2021). Grape planting situation and regional spatial analysis in Xinjiang, China. IOP Conf. Ser.: Earth Environ. Sci. 705, 12028. doi: 10.1088/1755-1315/705/1/012028
Xiang, Y., Lu, Y. H., Song, M., Wang, Y., Xu, W. Q., Wu, L. T., et al. (2015). Overexpression of a triticum aestivum calreticulin gene (TaCRT1) improves salinity tolerance in tobacco. PloS One 10 (10), e0140591. doi: 10.1371/journal.pone.0140591
Zhang, A. Y., Yang, X., Lu, J., Song, F. Y., Sun, J. H., Wang, C., et al. (2021). OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 308, 110903. doi: 10.1016/j.plantsci.2022.111403
Zhao, Y., Tian, X., Wang, F., Zhang, L., Xin, M., Hu, Z., et al. (2017). Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 17 (1), 208. doi: 10.1186/s12870-017-1158-4
Zhou, H., Wang, Y. F., Zhang, Y. J., Xiao, Y. H., Liu, X., Deng, H. B., et al. (2022). Comparative analysis of heat-tolerant and heat-susceptible rice highlights the role of OsNCED1 gene in heat stress tolerance. Plants (Basel) 11 (8), 1062. doi: 10.3390/plants11081062
Zhou, Z. P., Yuan, Y. Z., Zhou, W., Zhang, C. F. (2016). Effects of exogenously supplied sucrose on OsSUTs and OsSPSs transcript abundances and rice root ammonium assimilation. Acta Physiol. Plant 38, 274. doi: 10.1007/s11738-016-2285-5
Keywords: grape, high temperature stress, transcriptome, heat shock protein, plant physiology
Citation: Dou F, Phillip FO, Liu G, Zhu J, Zhang L, Wang Y and Liu H (2024) Transcriptomic and physiological analyses reveal different grape varieties response to high temperature stress. Front. Plant Sci. 15:1313832. doi: 10.3389/fpls.2024.1313832
Received: 10 October 2023; Accepted: 17 January 2024;
Published: 08 March 2024.
Edited by:
Ildiko Matusikova, University of St. Cyril and Methodius, SlovakiaReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences (JIRCAS), JapanCopyright © 2024 Dou, Phillip, Liu, Zhu, Zhang, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaifeng Liu, bGhmX2FnckBzaHp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.