- College of Life Sciences, Shaanxi Normal University, Xi’an, China
All living organisms must develop mechanisms to cope with and adapt to new environments. The transition of plants from aquatic to terrestrial environment provided new opportunities for them to exploit additional resources but made them vulnerable to harsh and ever-changing conditions. As such, the transmembrane receptor-like kinases (RLKs) have been extensively duplicated and expanded in land plants, increasing the number of RLKs in the advanced angiosperms, thus becoming one of the largest protein families in eukaryotes. The basic structure of the RLKs consists of a variable extracellular domain (ECD), a transmembrane domain (TM), and a conserved kinase domain (KD). Their variable ECDs can perceive various kinds of ligands that activate the conserved KD through a series of auto- and trans-phosphorylation events, allowing the KDs to keep the conserved kinase activities as a molecular switch that stabilizes their intracellular signaling cascades, possibly maintaining cellular homeostasis as their advantages in different environmental conditions. The RLK signaling mechanisms may require a coreceptor and other interactors, which ultimately leads to the control of various functions of growth and development, fertilization, and immunity. Therefore, the identification of new signaling mechanisms might offer a unique insight into the regulatory mechanism of RLKs in plant development and adaptations. Here, we give an overview update of recent advances in RLKs and their signaling mechanisms.
1 Introduction
The plant kingdom consists of species with a wide range of morphological diversity ranging from the simple members, such as mosses that have a thickness of the two-cell layer, to the more complex and advanced flowering plants, such as the Brassicaceae family having structures like vascular tissues, stomata, and flowers. This evolutionary journey from simple to complex, finally becoming more adaptive to the ever-changing environment, is challenging due to its sessile nature. Due to these challenges, plants have evolved conserved mechanisms that allow them to utilize different sensory proteins to recognize and perceive various environmental cues and respond appropriately to ensure optimal fitness. Cell-to-cell communication is essential to both plants and animals, as both are multicellular organisms that require this highly regulated mechanism for environmental adaptation and development. Eukaryotic protein kinases (EPKs) are a kinase superfamily that facilitates such cell-to-cell communication and intracellular signal transduction by catalyzing the transfer of ϒ-phosphate from an ATP to a hydroxyl group of serine/threonine or tyrosine residue of the polypeptide. The autophosphorylation of kinases themselves and transphosphorylation of other substrates cause a conformational change, thus activating a series of specific signaling components (Hanks and Hunter, 1995; Lehti-Shiu et al., 2012). There are over 1,000 EPKs in Arabidopsis (The Arabidopsis Genome, 2000), while there are only approximately 500 of them in humans (Manning et al., 2002), suggesting the importance of EPKs in regulating numerous aspects of cellular regulation in eukaryotes, particularly in plants. The first-ever evidence that suggests the process of phosphorylation in plants was found in leaf discs of Chinese cabbage (Ralph et al., 1972). Subsequently, another study based on duckweed showed that phosphorylation takes place at the serine residue in the plant ribosomes (Trewavas, 1973). In 1973, the first-ever EPK was identified and purified from pea plants; however, it was not until 1990 that the first EPK sequences were identified in the rice and pea plants (Keates, 1973; Lawton et al., 1989). Several kinase families, such as calcium-dependent protein kinases (CDPKs), NIMA-related kinases (NEKs), mitogen-activated protein kinases (MAPKs), glycogen synthase kinases (GSKs), and receptor-like kinases (RLKs) are the members of EPK family, with their unique functions and structure characteristics (Shiu and Bleecker, 2003; Lehti-Shiu and Shiu, 2012).
The two most dominant plant receptor families are cell surface-localized receptors RLKs and receptor-like proteins (RLPs) (Couto and Zipfel, 2016; Tang et al., 2017). RLKs are one of the most significant groups of plant cell surface receptors that arose from a common ancestor in green algae, playing a critical role in plant growth and development. In plants, cell-to-cell communication is regulated by their unique and distinctive structure, a hallmark of RLKs. RLK family is the largest gene family of the EPK superfamily with more than 600 members in Arabidopsis, which represent 2.5% of its total genome (Figure 1), and 1,132 members in rice (Shiu et al., 2004). The first RLK was discovered and cloned in maize, and it was found that it has a large putative N-terminal extracellular domain (ECD) together with a transmembrane domain (TM) and a catalytic intracellular kinase domain (KD) (Walker and Zhang, 1990). This discovery led to a distinctive perspective of understanding protein kinases in plants that co-opted a different class of EPKs for function in ligand-based transmembrane signal perception and transduction.
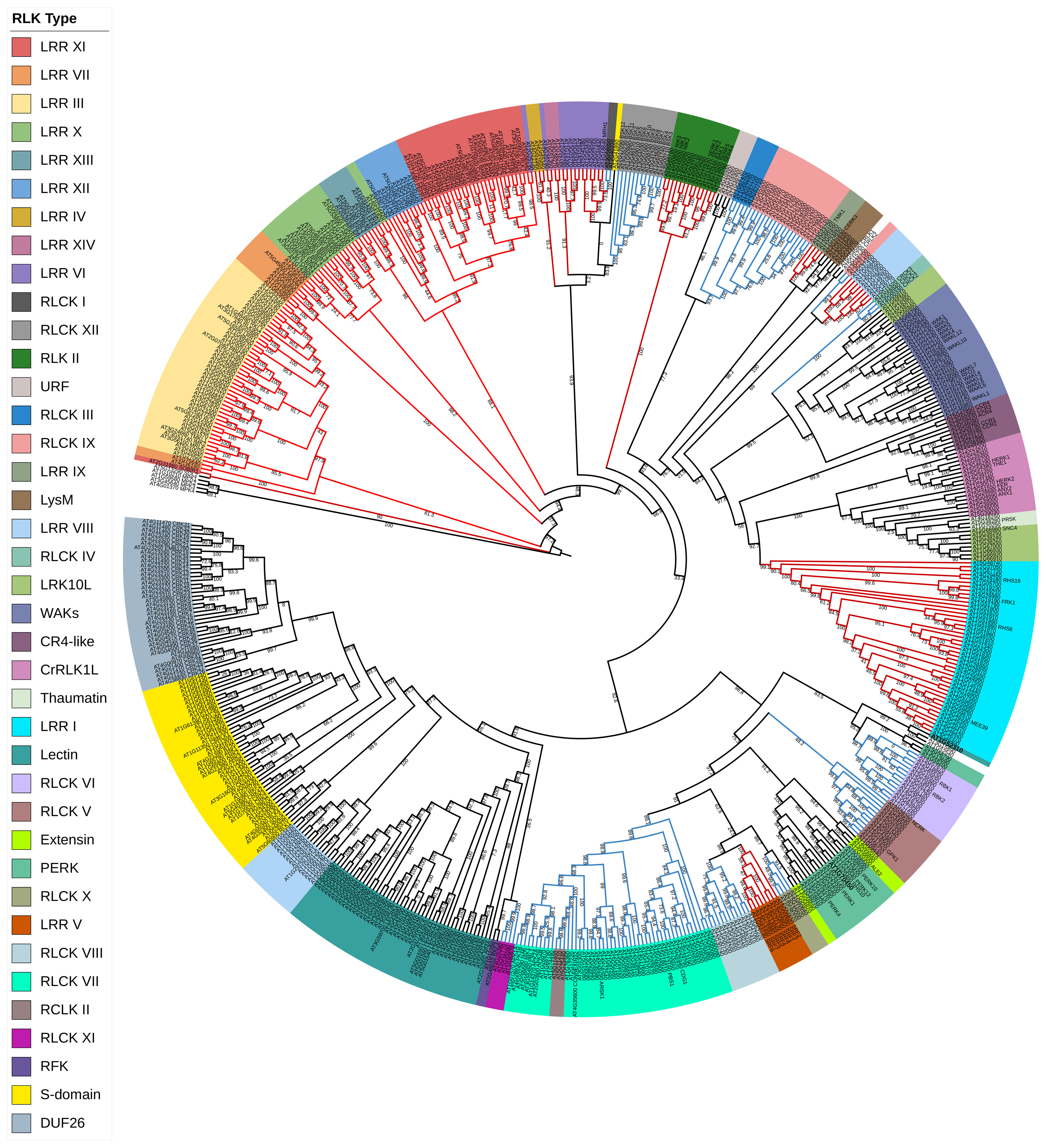
Figure 1 Phylogenetic classification of Arabidopsis thaliana RLKs. Arabidopsis RLK protein sequences were retrieved from the Arabidopsis information resource databases (https://www.arabidopsis.org/). All the retrieved sequences were aligned with MAFFT, and the tree was inferred by the maximum likelihood (ML) method with 1,000 bootstraps using IQ-TREE software (Trifinopoulos et al., 2016). Different taxon colors represent various RLK subgroups. The red color branches denote the LRR-RLK family, while the blue branches represent the RLCK family. RLKs, receptor-like kinases; LRR, leucine-rich repeat.
The plant RLKs have also been identified in animals, collectively called RLK/Pelle family, which forms a monophyletic clade, but no RLKs have been reported in fungi so far (Shiu and Bleecker, 2001b; Shiu and Bleecker, 2003). Pelle kinase is the only member from the RLK/Pelle family in Drosophila, while the human interleukin receptor-associated kinases (HsIRAK-1, HsIRAK-2, HsIRAK-M, and HsIRAK-4), responsible for adaptive and innate immunity, also belong to this family (Shelton and Wasserman, 1993; Janssens and Beyaert, 2003; Lehti-Shiu et al., 2009). The RLKs resemble the animal receptor serine/threonine kinases and receptor tyrosine kinase (RTKs) in their basic structure. The animal RTKs are distinguished from the plant RLKs by their tyrosine kinase specificity, except the transforming growth factor β (TGF-β) receptor in metazoans, which is serine/threonine kinase similar to plant RLKs (Shiu and Bleecker, 2001b; Cock et al., 2002). Some plant RLKs have been reported with both serine/threonine and tyrosine kinase specificity. For example, brassinosteroid insensitive 1 (BRI1), BRI1-associated kinase 1 (BAK1), HAESA (HAE), pollen-expressed kinase 1 (PRK1), and nod factor receptor 1 (NFR1) can phosphorylate at serine/threonine residue as well as at tyrosine residue (Oh et al., 2009; Madsen et al., 2011). In addition to that, both RTKs and RLKs undergo ubiquitination and endocytosis for system desensitization following activation (Martins et al., 2015). In addition, animal RTKs and plant RLK signaling shares similar downstream elements, such as MAPK cascades and reactive oxygen species (ROS) production (Liang and Zhou, 2018).
A large number of RLKs are found that only possess the cytoplasmic KD but lack the ECD. These members of RLKs are known as receptor-like cytoplasmic kinases (RLCKs) and play a vital role in regulating plant cellular activities in response to biotic/abiotic stresses and endogenous extracellular signaling molecules (Shiu and Bleecker, 2001a). The number of RLCKs in Arabidopsis and rice is 149 and 379, respectively (Shiu et al., 2004; Vij et al., 2008), while the reported number of RLCKs in maize is 162 (Fan et al., 2018b). RLPs, on the contrary, lack the KD but possess the ECD and TM or a glycosylphosphatidylinositol anchor. Hence, the RLPs are dependent on RLKs to induce transmembrane signaling (Liang and Zhou, 2018). The number of RLPs in Arabidopsis and rice is approximately 170 and 90, respectively (Fritz-Laylin et al., 2005; Li et al., 2016b).
In this review, we summarize the role of RLKs in crucial diverse processes in plants. Our objectives are to discuss the RLK’s evolution, crystal structure, classification, role in plant growth and development, and immunity. This review also highlights the vital role of RLCKs in RLK signaling.
2 Evolution and expansion of RLKs
During evolution, new species arise when countless tiny variations accumulate in the genome of an existent organism, which are heritable characteristics selected by natural selection. In short, this theory of evolution mainly emphasizes the accumulation of microevolution that is beneficial to the survival of an organism to achieve macroevolution, and eventually, new species emerge (Darwin, 1859; Reznick and Ricklefs, 2009). During this process, a gene duplicates and neofunctionalizes. However, the experimental validation of macroevolution is almost impossible due to the lack of various intermediate species between different lineages. EPKs are a monophyletic clade that controls almost all aspects of the eukaryotic life cycle (Hanks et al., 1988; Shiu and Bleecker, 2001b; Cock et al., 2002). Their sequences and structures are diverse and conserved, which are considered a good model for studying gene duplication and functional diversification (Hanks et al., 1988; Hanks and Hunter, 1995). RLKs are the most diverse class of protein kinases in plants, sensing extracellular signals and triggering the downstream responses, which play a massive role in the process of plant adaptation to the terrestrial environment (Shiu et al., 2004; Gish and Clark, 2011). Therefore, elucidating the functional evolution of plant RLKs is significant. Since all eukaryotic kinases are derived from a common ancestor, their diverse molecular and biological function is achieved through gene duplication and neofunctionalization. Gene duplication is the primary source of evolving new genes and contributes to unique biological traits and even speciation. Therefore, gene duplication and neofunctionalization can be the cornerstones of evolution (Innan and Kondrashov, 2010; Kaessmann, 2010). Most redundant gene copies after gene duplication are likely to be non-functionalized and thus are often deleted from the genome in a few million years (Lynch and Conery, 2000). However, in some cases, a few copies that acquire new functions are retained and diversified (Ohno, 1970; Force et al., 1999; Bergthorsson et al., 2007). That is why a surprising number of protein kinases have emerged during the genome expansion (The Arabidopsis Genome, 2000).
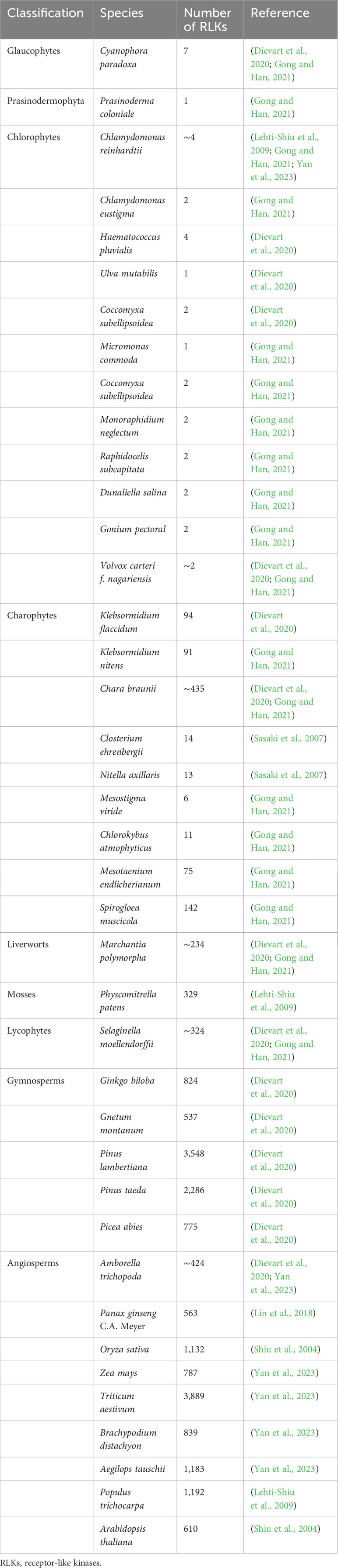
The plant RLKs are more similar to animal RTKs than other plant protein kinases in other groups, such as CDPKs, MAPKs, and GSK3s, and are within a monophyletic protein kinase group. The serine and/or threonine kinase is the ancestor of RLKs, while RTKs evolved from tyrosine kinases (Cock et al., 2002; Gish and Clark, 2011). RLKs dramatically expanded in land plants, especially angiosperms, evident of large family size in plants than its animal counterpart. To understand the evolutionary dynamics of RLKs, several initial comprehensive studies on RLKs’ identification, classification, and evolution and expansion have greatly helped gain further insight into this gene family in plants (Shiu and Bleecker, 2001a; Shiu and Bleecker, 2001b; Shiu et al., 2004; Lehti-Shiu et al., 2009; Lehti-Shiu et al., 2012). Further studies confirmed that RLKs are present in most Charophytes and all land plants, the kinase-associated RLKs first appeared in Chlorophytes, and more than half of analyzed Chlorophytes have RLKs. In 2007, the first cDNA-based report of algae containing RLKs came from Closterium ehrenbergii and Nitella axillaris with 14 and 13 RLKs, respectively, containing an ECD, TM, and a KD (Sasaki et al., 2007). C. braunii, another Charophyte, was initially found to have seven lysine motif kinase (LysM) RLKs in its genome (Nishiyama et al., 2018), which was later identified to have 435 RLKs in total (Dievart et al., 2020), with leucine-rich repeat (LRR), proline-rich, LysM, and malectin as their major domain families. Interestingly, the domain families such as self-incompatibility (S-domain), wall-associated kinases (WAKs), CRINKLY4-like kinase (CR4-like), and lectin (C-Lec) were absent from not only C. braunii but also from Zygnematophyceae and Coleochaetophyceae, suggesting their land plant specificity, as these domain families are found in basal land plants (Dievart et al., 2020; Gong and Han, 2021). Similarly, the first receptor with canonical configuration was found in the earliest Streptophyta, Klebsormidium flaccidum, with 94 RLKs with a TM acquisition, an ECD, and a KD. The LRR, proline-rich, extensin, and C-Lec were the only kinase-associated ECDs found in the RLKs of this genome, suggesting that the first fusion between different domains of a kinase receptor occurred in this lineage (Hori et al., 2014; Dievart et al., 2020). In many other algae members, different independent domains such as KD or conserve motifs of ECD have been found as discrete elements rather than a complete receptor, suggesting that the receptor reshaping had already been set off in green algae just before the divergence of Charophytes into the land plants (Lehti-Shiu et al., 2009). In the earliest-known diverging Charophytes, Mesostigma viride and Chlorokybus atmophyticus from Mesostigmatophyceae and Chlorokybophyceae, respectively, no RLKs were initially identified by using available expressed sequence tag (EST)/cDNA data (Simon et al., 2006; Lemieux et al., 2007). However, a recent study reported 6 and 11 RLKs in M. viride and C. atmophyticus, respectively, suggesting that kinase-associated LRR and C-Lec trace back to the earliest Streptophyte (Table 1) (Gong and Han, 2021).
Several recent attempts have been made to identify RLKs in Chlorophytes. Chlamydomonas reinhardtii was the first reported Chlorophyte to have two RLKs but none in Ostreococcus tauri (Lehti-Shiu et al., 2009). A recent study identified four RLKs in C. reinhardtii with one C-Lec, one L-Lec, and two RLCK members (Yan et al., 2023). Similarly, Gong and Han (2021) studied a total of 18 Chlorophytes, and nine were identified to have RLKs. In addition, RLKs were also present in Prasinodermophyte (Prasinoderma coloniale) and Glaucophyte (Cyanophora paradoxa) but none in Rhodophytes, giving a patchy distribution in Chlorophytes (Table 1) (Gong and Han, 2021). Because of this patchy distribution, the RLKs in Chlorophytes do not form a monophyletic group, which can be explained either by a single origin followed by multiple losses or by multiple horizontal gene transfers to Chlorophytes.
In non-seed land plants, moss (Physcomitrella patens) and lycophyte (Selaginella moellendorffii), the number of identified RLKs is 329 and 324, respectively, accounting for 0.36% and 0.30% of their total protein-coding genes, respectively (Table 1) (Lehti-Shiu et al., 2009; Dievart et al., 2020; Gong and Han, 2021). The liverwort (Marchantia polymorpha) represents all the major domain families except the domain of unknown function 26 (DUF26) that first appeared in S. moellendorffii (Vaattovaara et al., 2019; Gong and Han, 2021). In angiosperms, however, on average, there are approximately 900 RLKs in 53 monocots and 127 dicots species analyzed, which account for 0.67% to 1.39% of their total genome (Shiu et al., 2004; Zulawski et al., 2014). These ratios indicate that the RLK family continued to expand in vascular plants after divergence from the algae and then the lower plant lineage, such as moss, as there are as many as 1.9, 3.3, and 3.6 times RLK members in Arabidopsis, rice, and popular than that of moss, respectively. Moreover, the RLK family is greatly expanded in Triticum aestivum with 3,889 members, which is approximately 6.4 times those in Arabidopsis and 3.4 times in rice (Yan et al., 2023). All these studies suggest that the expansion of RLKs was necessary for the acquisition of their new role in plant survival and adaptation since this expansion is associated with genome complexity rather than genome size (Lehti-Shiu et al., 2009).
3 Conserved crystal structure of RLKs and EPKs
In Eukaryotes, post-translational modification of proteins is the most widespread and conserved process carried out through the common mechanism of phosphorylation. The phosphorylation-catalyzing enzymes are regulated rigorously because phosphorylation controls the activity of a large number of proteins (Olsen and Mann, 2013). In EPKs, the primary regulatory mode is phosphorylation, together with many different ways to regulate EPKs. The cyclic-AMP protein kinase A (PKA) was first identified as a kinase where the C-subunit could phosphorylate the glycogen phosphorylase kinase (Figure 2) (Walsh et al., 1968). Since then, the PKA has been used as a model kinase, which opened up new insights into the concept of kinase signaling. There are many regulatory hotspots that regulate phosphorylation together, but one critical regulatory hotspot is the “activation loop”, where the phosphorylation events occur (Nolen et al., 2004; Beltrao et al., 2012; Bojar et al., 2014). The dynamic assembly of the regulatory spine is induced by the major mechanism of activation loop phosphorylation site activated through phosphorylation (Taylor et al., 2012; Meharena et al., 2013; Taylor et al., 2015). The whole activation process is based on the regulatory spine and is accompanied by structural changes.
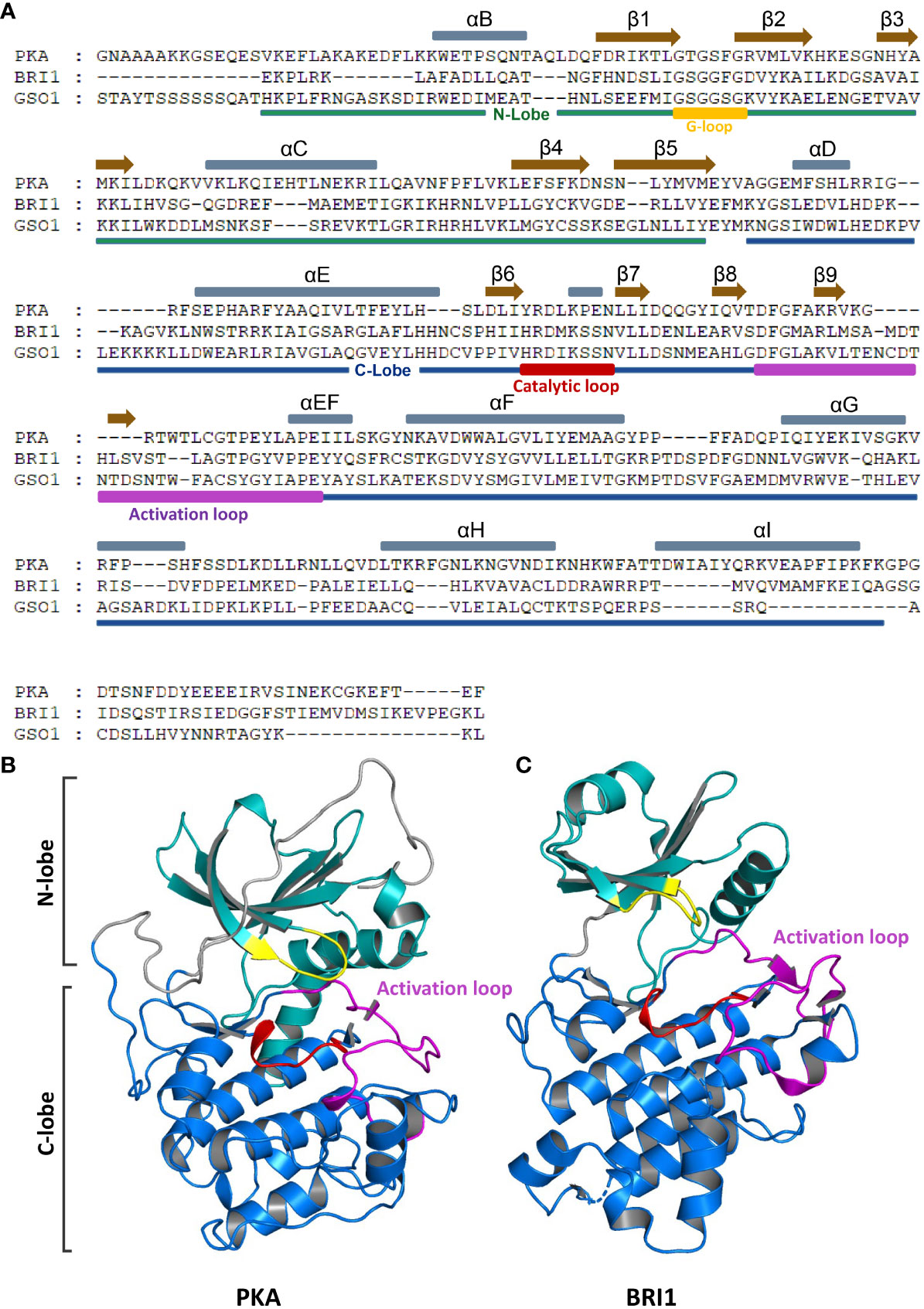
Figure 2 Domain organization and crystal structure of RLKs and EPKs. (A) N-lobe, C-lobe, αC helixes, and β-sheets of RLK (BRI1 and GSO1) and EPK (PKA) are presented in different colors. Gray and brown bars above the sequence represent αC helixes and β-sheets, respectively. The N-lobe and C-lobes are depicted below the sequences in green and blue bars, respectively. Activation loop, G-loop, and catalytic loop are marked in purple, yellow, and red, respectively. (B, C) Crystal structures of PKA (B) and BRI1 (C). Different domains are represented in different colors as shown in panel (A). RLKs, receptor-like kinases; EPKs, eukaryotic protein kinases.
The EPKs are defined by the conserved kinase core that consists of approximately 250 amino acid residues, essential for the activation process. Initially, the kinase core was divided into 12 subdomains based on the conserved residues in the motifs. The crystal structure analysis of EPKs revealed that these motifs could be defined based on their putative functions (Knighton et al., 1991a; Knighton et al., 1991b; Hanks and Hunter, 1995). The main kinase core consists of two lobes, a smaller N-terminal lobe (N-lobe) and a larger C-terminal lobe (C-lobe) (Figures 2A–C). The N-lobe is required for the ATP binding, while the substrate recognition happens at the C-lobe. In protein-activated conformation, a central network of hydrophobic amino acids (spines) connects these lobes non-covalently, resulting in independent folds (Kornev et al., 2008). Each lobe is made up of α-helixes and β-strands (subdomains), and the lobes form a deep cleft that binds adenosine triphosphate (ATP). Phosphoryl transfer occurs at the cleft’s outer edge, where the ϒ-phosphate is positioned. Through opening and closing, the active cleft mediates the catalysis, allowing the transfer of the phosphate group and the release of the nucleotide (Taylor et al., 2012).
The N-lobe consists of five-stranded β-sheets that are antiparallel in configuration involved in the ATP binding. A single conserved helix αC is located between β3 and β4. In addition to αC, there is another shorter helix, αB, which is not conserved in EPKs. A glycine-rich loop (G-loop) links the β1 to the β2, which packs on top of the ATP. In activated form, the N-terminus of αC interacts with the activation loop, while the C-terminus forms part of the active site cleft (Taylor et al., 2012; Beenstock et al., 2016). The αC serves as an allosteric hotspot, and phosphorylation of the activation loop induces the rotation of the αC helix inward to a favorable position, which enables the formation of αC-Glu and β3-Lys salt bridge and a domain encloser between the N-lobe and C-lobe (Beenstock et al., 2016; Kim et al., 2017). On the contrary, the C-lobe mostly consists of α-helixes, αD, αE, αEF, αF, αG, αH, and αI together with four β-strands, β6, β7, β8, and β9. β6 and β7 are connected by a catalytic loop that contains Asp residue, required for the activation of hydroxyl of the phosphoacceptor residue of the substrate (Beenstock et al., 2016; Kim et al., 2017). Between the β8 and β9 is a DFG motif (Asp-Phe-Gly) located right below the C helix and the G-loop between the two lobes, referred to as magnesium positioning loop because the conserved Asp residue binds to the catalytic magnesium ion (Meharena et al., 2013). The DFG motif is the start site of the activation loop, while the APE motif is the end site. The APE motif serves as an anchor for the activation loop (Figure 2A) (Gogl et al., 2019).
The crystal structure-based studies of RLKs allow us to understand the kinase activation mechanism and their transphosphorylation activity in plants. The RLK BRI1 crystal structure and interaction, and kinase activation mechanism with coreceptor BAK1 have been studied extensively. The BRI1 has a dual specificity of serine/threonine and tyrosine kinases (Friedrichsen et al., 2000; Oh et al., 2009). The catalytically competent activation loop becomes active due to the phosphorylation of Thr1039, Ser1042, and 1044. The protein interaction surface is located on the C-lobe of the BRI1 that heterodimerizes with somatic embryogenesis receptor-like kinase (SERK) coreceptor forming catalytically competent rearrangement. This interaction is regulated by the BRI1 inhibitor BKI1 (Figure 2C) (Bojar et al., 2014). For the BRI1 signaling regulation, both BAK1 and BKI1 compete for BRI1 heterodimerization and transphosphorylation (Wang et al., 2014). The island domain (ID) constituted of 70 amino acid residues is required for the BR perception during BR signaling (Li and Chory, 1997). A recent study showed that two conserved amino acids Tyr597 and Tyr 599 are required by BRI1 for BR perception. Mutating these two amino acids in the ID abolished the BR signaling by knocking out or knocking down the phosphorylation activity of BES1 (Li et al., 2022). Previous studies proved that the activation loop, αC helix, and αC-β4 loop are the most critical hotspots for kinase activation, and any abnormality in these regions can lead to the complete eradication of phosphorylation activity and cancer-related diseases in humans (Taylor et al., 2012; Gogl et al., 2019; Yeung et al., 2020). The BRI1 has been shown to allosterically utilize these hotspots together with β4–β5 located in two subdomains to specify its kinase function from GASSHO1 (GSO1). Replacing or deleting either of the subdomains will block the kinase activity in BRI1 and GSO1 (Ali et al., 2021). BAK1-interacting receptor-like kinase 2 (BIR2) is an enzymatic pseudokinase regulator of different signaling pathways that competes for BAK1 (Blaum et al., 2014; Halter et al., 2014). The crystal structure of BIR2 showed that the β1 strand blocks the ATP-binding pocket, and the amino acid variation in the conserved P-loop of kinase core in BIR2 blocks this protein from ATP coordination (Blaum et al., 2014). The structure-based molecular mechanism of GSO1/2 interaction with ligand Casparian strip integrity factors (CIFs) showed that GSO1/2 have evolved unique binding properties in their peptide to control different developmental processes through their signaling mechanism (Okuda et al., 2020). The crystal structural analysis of SOBIR1 displayed an unusually longer β3-αC loop, which together with Thr529 plays a crucial role in SOBIR1-induced Nicotiana benthamiana cell death response (Wei et al., 2022).
4 RLK classification
In Arabidopsis and rice, the RLKs are the largest family of the protein kinase superfamily, which has approximately 610 and 1,132 members, respectively, suggesting the importance of RLKs in plant growth and development (Shiu et al., 2004; Vij et al., 2008). However, not all RLKs are plant receptors; instead, some act as coreceptors or scaffold components of the complex, such as BAK1 and FERONIA (FER), a malectin-like receptor kinase (Li et al., 2002; Chinchilla et al., 2007; Stegmann et al., 2017). Interestingly, in Arabidopsis, out of 610 RLKs, 417 are RLK encoding genes, and 149 are RLCKs. The remaining numbers are RLK entities. Similarly, in rice, there are 379 RLCKs out of 1,132 RLKs (Shiu and Bleecker, 2001b; Shiu et al., 2004; Vij et al., 2008). Based on their versatile ECD, the RLKs can be divided into the following major subgroups.
The RLK classification includes LRR receptor-like kinase (LRR-RLKs), S-domain, lectin (C-Lec, L-Lec), WAKs, CR4-like, extensin, proline-rich extensin-like (PERK), LysM, thaumatin-like, DUF26 (also known as cysteine-rich receptor-like kinases; CRKs), Catharanthus roseus receptor-like kinase 1-like (CrRLK1L), leaf rust kinase-like (LRK), receptor-like kinase in flower (RKF), and kinase with unknown function (Figure 1). The last group of the RLKs has not been reported in Arabidopsis, while the rest are as follows.
4.1 LRR-RLKs
LRR-RLKs represent the largest class of RLKs and have more than 200 RLK encoding genes that are further divided into 15 subclasses (Shiu and Bleecker, 2001a). The first LRR-RLK was reported as a putative transmembrane protein kinase in Arabidopsis, which was later identified as transmembrane kinase 1 (TMK1) (Bleecker, 1991; Chang et al., 1992). Recently, LRR-RLKs have been identified and classified in numerous plant species. For instance, there are total of 379 LRR-type RLK identified in Populus trichocarpa (Zan et al., 2013), 531 in T. aestivum (Shumayla et al., 2016a), 119 and 67 in P. patens and S. moellendorffii, respectively (Liu et al., 2017), 1,641 LRR-RLKs in four Gossypium species (Gossypium arboreum, Gossypium barbadense, Gossypium hirsutum, and Gossypium raimondii) (Sun et al., 2018), 437 LRR-RLKs in Saccharum spontaneum (Cheng et al., 2021), and 444 LRR-RLKs in Brassica napus (Song et al., 2022). The KD is highly conserved among the LRR-RLKs, while ECD is variable with a different number of LRRs, which enables them to recognize a variety of hormonal or peptide-based ligands to control distinct functions (Shiu and Bleecker, 2001b; Gou et al., 2010).
4.2 S-domain
S-domain ECD resembles the ECD of S-locus-specific glycoprotein (SLG) in Brassica with a shared homology of approximately 52%, known to regulate the self-incompatibility of pollen–stigma interaction in mustard (Nasrallah et al., 1988). The S-domain class of RLKs is composed of 40 members in Arabidopsis that have three characteristic subdomains in their ECD: B-Lec (bulb-type lectin), which contains a highly conserved stretch of 40 residues, S-locus, and a 13 conserved cysteine-rich residue cluster called PAN (Nasrallah et al., 1988; Shiu and Bleecker, 2001a).
4.3 WAKs
The WAK family is the third type of RLKs described to have cysteine-containing epidermal growth factor (EGF) motifs in its ECD (Kohorn et al., 1992). WAKs are known to play a role in cell wall integrity and development to protect the plant from injuries during biotic and abiotic stresses. They do so by interacting with pectin or pectin-based molecules in response to changes in cellulose contents during a pathogen attack (Verica and He, 2002; Kohorn, 2015). WAKs have been identified and characterized in several plant species. For instance, Oryza sativa was found to have 125 OsWAKs (Zhang et al., 2005); G. arboreum, G. raimondii, and G. hirsutum genomes consist of 58, 66, and 99 WAKs, respectively (Zhang et al., 2021b); Solanum tuberosum L. has 29 WAKs (Yu et al., 2022); 38 WAKs were characterized in N. benthamiana (Zhong et al., 2023).
4.4 Lec-RLKs
Lec-RLK is the large class of RLKs, only second to LRR-RLKs in plants, playing various roles in plant development and stress tolerance. The Lec-RLK family has three types of lectins, G, L, and C lectin, but only L-Lec and C-Lec are known to exist in Arabidopsis with 47 encoding gene members (Shiu and Bleecker, 2001a; Shiu et al., 2004). They can interact with different kinds of carbohydrate molecules such as glucose-mannose, galactose-GlcNAc, and chitobiose (Sharon and Lis, 1990; Bellande et al., 2017). There is only one C-Lec in Arabidopsis, which is calcium-dependent for their ligand binding, which resembles the mammalian calcium-binding lec motifs (Shiu and Bleecker, 2001a; Shiu and Bleecker, 2001b; Cambi et al., 2005). In addition to Arabidopsis, one C-type, 22 L-type, and 23 G-type Lec-RLKs associated with phytohormones and stress responses were identified in Cucumis sativus L. (Lv et al., 2020). In 2021, 73 putative Lec-RLKs were identified from Vigna radiata L., classified into three subfamilies (Singh et al., 2021). Similarly, the peanut plant (Arachis hypogaea) genome constitutes 1,311 RLKs, among which 274 are Lec-RLKs classified further into three types: C, L, and G (Wang et al., 2021b). The genome-wide analysis of T. aestivum revealed that there are two C-lectin, 84 L-lectin, and 177 Bulb-lectin (B-lectin) in its genome (Shumayla et al., 2016b).
4.5 CR4-like
Maize CRINKLY4 (ACR4) was the first RLK described as crinkly-like (CR-like) RLK. It has a domain of SEVEN tandem repeats of tumor necrosis factor receptor (TNFR) that is 37 amino acids long together with another region of 26 amino acids made up of three cysteine residue tandem repeats similar to TNFR (Becraft et al., 1996; Gifford et al., 2003; Czyzewicz et al., 2016).
4.6 Extensin/proline-rich extensin-like kinases
Extensin is another cell wall development and integrity protein similar to WAKs that constitute Ser-(Hyp)4 repetition in their ECD, while the extensin-like kinases have glycosylated motifs of Ser-(Hyp)3-5 to maintain the integrity and structure of cell wall (Cassab, 1998; Borassi et al., 2016). The PERK1 was first characterized from B. napus and possesses a proline-rich domain in the ECD, similar in sequences to extensin proteins. During a pathogen attack, PERK1 gene is induced rapidly after sensing the cell wall damage, suggesting its role in sensing cell wall modifications (Silva and Goring, 2002). Recently, the PERK family has also been identified in the T. aestivum L. genome with 30 genes classified into eight groups. Their involvement in plant growth processes was suggested by the cis-regulatory elements and expression profile at tissue developmental stages. Furthermore, it was hypothesized that several TaPERK genes were also involved in defense responses by their variable expression under biotic and abiotic stress settings (Shumayla et al., 2022).
4.7 LysM
The lysine motif-containing RLKs are generally known as LysM domain-containing receptor-like kinases (LYKs), which play a major role in defenses and provide fungal resistance, and are involved in chitin signaling. LYKs were identified during the quest for the Nod factor receptors during a legume rhizobial infection (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Recently, Yang et al. found a total of nine LysM-RLKs in Brassica juncea (Yang et al., 2021a). In another three Brassica species (B. napus, Brassica rapa, and Brassica oleracea), the number of the identified LysM is 17, 8, and 8, respectively (Abedi et al., 2022). Shumayla et al. identified 20 TaLysM genes that have tandem duplication with retention of function and 42 TaLysM (Motif) with neofunctionalization and pseudo-functionalization in T. aestivum L. (Shumayla et al., 2021). The ECD of LysM-RLKs is composed of three lysin motifs of approximately 40 amino acids long, which permits LysM-RLKs to perceive chito-oligosaccharide ligands and perform a dual role of immunity against pathogen and rhizobium–legume and mycorrhizal association in plants (Limpens et al., 2015; Buendia et al., 2018).
4.8 Thaumatin-like
Thaumatin-like, also known as PR5K (pathogenesis-related group 5 receptor-like kinase), is another class of RLKs involved in plant resistance against pathogen and chitinase activity. There are three members in Arabidopsis that also go by the name pathogen-related group 5 receptor kinases (PR5K), having 16 conserved residues of cysteine (Wang et al., 1996). In a recent study, thaumatin-like protein kinases were characterized in five cereal crops including two in Brachypodium distachyon and O. sativa, four in Hordeum vulgare and Sorghum bicolor, and 16 in T. aestivum. Their modulated expression in the presence of fungal pathogen, and heat, drought, and salt stress in T. aestivum suggested their roles in stress response (Sharma et al., 2020).
4.9 C. roseus receptor-like kinase 1-like
The first RLK of this class was identified in a higher plant, C. roseus (Schulze-Muth et al., 1996). Arabidopsis possesses 17 members of this family, while T. aestivum was recently identified to encode 15 CrRLK1L family genes that have 43 paralogous copies with three homeologs each, except for paralog -2-D and -7-A, which were absent (Gawande and Sankaranarayanan, 2023). Another study showed that three of 16 O. sativa genes are tandem-duplicated, suggesting possible functional redundancy within this family. However, integrated diurnal expression showed a functional divergence between two of these three genes (Nguyen et al., 2015). Ma et al. (2023) identified 24 CrRLK1L members in Solanum lycopersicum having bacterial and pathogen-associated molecular pattern (PAMP) responses during expression profile (Ma et al., 2023). The ECD of CrRLK1L comprises 450 amino acid residues that possess the carbohydrate-binding domain (malectin-like domain) (Nguyen et al., 2015; Moussu et al., 2018). The members of this class form complexes with other receptors or coreceptors or factors such as glycosylphosphatidylinositol-anchored protein to control plant development or other cell wall sensing responses (Nissen et al., 2016; Ge et al., 2019).
4.10 Leaf rust kinase-like
The first canonical receptor of leaf rust kinase-like (LRK10-like) was receptor-like kinase in flower 3 (RKF3) that has an ECD, a KD, and a TM but lacks distinct motifs in their ECD for signaling peptide (Takahashi et al., 1998). The LRK10 is RLK from wheat leaf rust kinase (WLRK) that is homologous to RLK10-like in sequence and has 14 conserved cysteines in a specific repeated manner of three conserved regions followed by three variable regions in the ECD (Feuillet et al., 1998; Feuillet et al., 2003; Lim et al., 2015). This unique architecture of ECD allows these RLKs to perceive different kinds of ligands, thus playing a diverse role in plant development.
4.11 DUF26
With 44 members in Arabidopsis, DUF26 (CRK) is another cysteine-rich repeat domain kinase receptor that shares similarities with lectins but is different than the S-locus glycoprotein and contains C-X8-C-X2-C motifs (Zeiner et al., 2023). These RLKs containing cysteine-rich repeats were termed cysteine-rich domain-containing receptor-like kinases (CRR RLKs), which play a role in defense and stress responses (Chen, 2001; Vaattovaara et al., 2019). Other cereal crops such as B. distachyon, H. vulgare, O. sativa, S. bicolor, and T. aestivum have also been reported to have 43, 37, 36, 38, and 170 CRKs respectively (Shumayla et al., 2019).
4.12 Receptor-like kinase in flower
The first RLK of this class was found to have an LRR malectin domain in which the LRRs are located upstream of the ECD (Takahashi et al., 1998). The RFK1 receptor has 13 LRRs, and the malectin domain is located between the TM and LRR (Dufayard et al., 2017).
5 Diverse roles of RLKs
The extensive expansion of RLKs in Arabidopsis, rice, and wheat genomes may suggest their role in the perception of a broad range of internal and external signals and stimuli. Although most of the RLK functions have not been identified, still, a large number of RLKs have been studied extensively. Based on their functions, the plant RLKs are divided into two broad categories. The first RLK category is responsible for growth and development, and the second controls plant–microbe interaction, immunity, and stress responses (Shiu and Bleecker, 2001a).
5.1 RLKs involved in plant growth and development
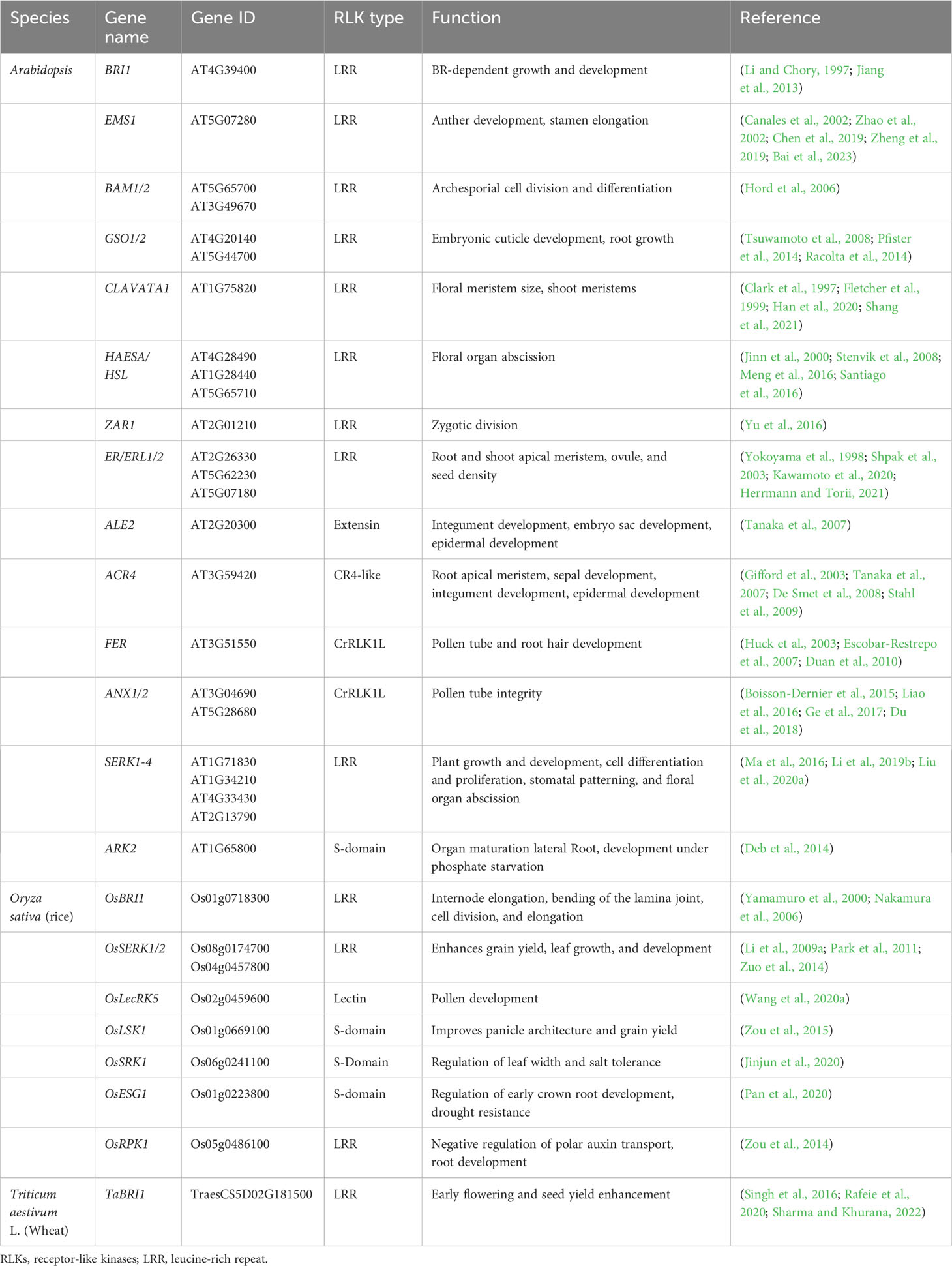
Normal growth and development are defined by the coordinated cell division, differentiation, and morphogenesis of the plant tissues controlled by a complex regulatory mechanism consisting of multiple regulatory genes working together. Several RLKs are known to regulate the development of shoot apical meristem (SAM), which develops into aerial organs in higher plants. The CLAVATA3 (CLV3) is a peptide ligand that forms complexes with CLAVATA1 (CLV1) and CLAVATA2 (CLV2, a receptor-like protein); together, they control the balance between cell differentiation and proliferation (Figure 3; Table 2) (Clark et al., 1997; Fletcher et al., 1999; Jeong et al., 1999). In Arabidopsis, the stem cells in the shoot meristem are regulated by the WUSCHEL (WUS), and SHOOTMERISTEMLESS (STM) interacts with the CLV3 promoter, which enhances the WUS-mediated stem cell activity (Su et al., 2020). Recently, a study reported that CLV1 participates in auxin-dependent meristem maintenance in cool environments. The auxin-dependent floral primordium development requires CLV3 signaling, and both inflorescence fasciation and heat-induced auxin biosynthesis partially mask this role (John et al., 2023). The plant organ shape and inflorescence architecture are monitored by ERECTA (ER), which perceives the epidermal patterning factor (EPF)-like protein (EPFL) as a ligand and expressed in root and shoot apical meristem as well as developing organs (Yokoyama et al., 1998; Herrmann and Torii, 2021). The dwarf phenotype and aberrant flower development of the ER mutants suggest that this family is involved in SAM and flower development, cell proliferation, organ growth, and stomata formation (Table 2) (Shpak et al., 2003; Herrmann and Torii, 2021). Recently, the EPFL family member, EPFL2, has also been reported to play a role in embryogenesis. Its loss-of-function mutants displayed reduced growth in cotyledon (Fujihara et al., 2021). In Arabidopsis, the number of seeds and the growth of pistils and fruits are regulated through coordinated ovule patterning induced by the interaction of EPFL2 and EPFL9 with the ER and ER-like (ERL1/2) receptors (Kawamoto et al., 2020). The ER signaling pathways result from forming complexes with signaling proteins, such as SERK coreceptors and too many mouths (TMM) transmembrane proteins (Lee et al., 2012; Meng et al., 2015). Similarly, another RLK, meristematic receptor-like kinase (MRLK) expressed in root and shoot apical meristem, regulates the floral transition by interacting with the transcription factor AGL24 of the MADS-box family (Fujita et al., 2003). Several RLKs play an equally essential role in root development as the one responsible for the shoot development. One such RLK is CRINKLY4 (ACR4), which is responsible for the normal development of root apical meristem (De Smet et al., 2008; Stahl et al., 2009). ACR4 forms a complex with CLV1 in response to CLE40 peptide association, which regulates the lateral root formation and stem cell differentiation of columella (Stahl et al., 2013). The ACR4 also interacts with Protein Phosphatase 2A (PP2A) to regulate the distal root meristem development. PP2A plays a role in the ACR4 accumulation and plasma membrane localization (Yue et al., 2016). It is also known that the ACR4 regulates the homeostasis of the columella stem cells in the root by forming complexes with CLAVATA3 INSENSITIVE RECEPTOR KINASEs (CIKs) induced by CLV3/EMBRYO SURROUNDING REGION (ESR) 40 (CLE40) (Zhu et al., 2021). During epidermal cell differentiation in leaves, the ACR4 together with ABNORMAL LEAF SHAPE 2 (ALE2) positively regulates protoderm-specific gene expression (Tanaka et al., 2007). The rice OsCR4 is essential for the development of mesophyll tissues and the differentiation of vascular bundles and the epidermis by establishing a local auxin gradient to restrict OsWOX3A (Wang et al., 2020b). The member of the cysteine-rich RLK family, CRK28, is also involved in normal root morphogenesis, and its overexpression phenotype exhibits delayed and reduced lateral root growth, while the crk28 mutant is reported to have longer and branched root (Pelagio-Flores et al., 2019). Two independent studies showed rapid root growth and navigation are regulated through the auxin-induced interaction of TMK1 with H+-ATPas inducing apoplastic acidification and cell expansion (Li et al., 2021b; Lin et al., 2021). S-domain RLK, Arabidopsis receptor kinase2 (ARK2) has been identified to interact with U box/armadillo repeat-containing E3 ligase9 (AtPUB9) mediating lateral root development under phosphate starvation (Deb et al., 2014).
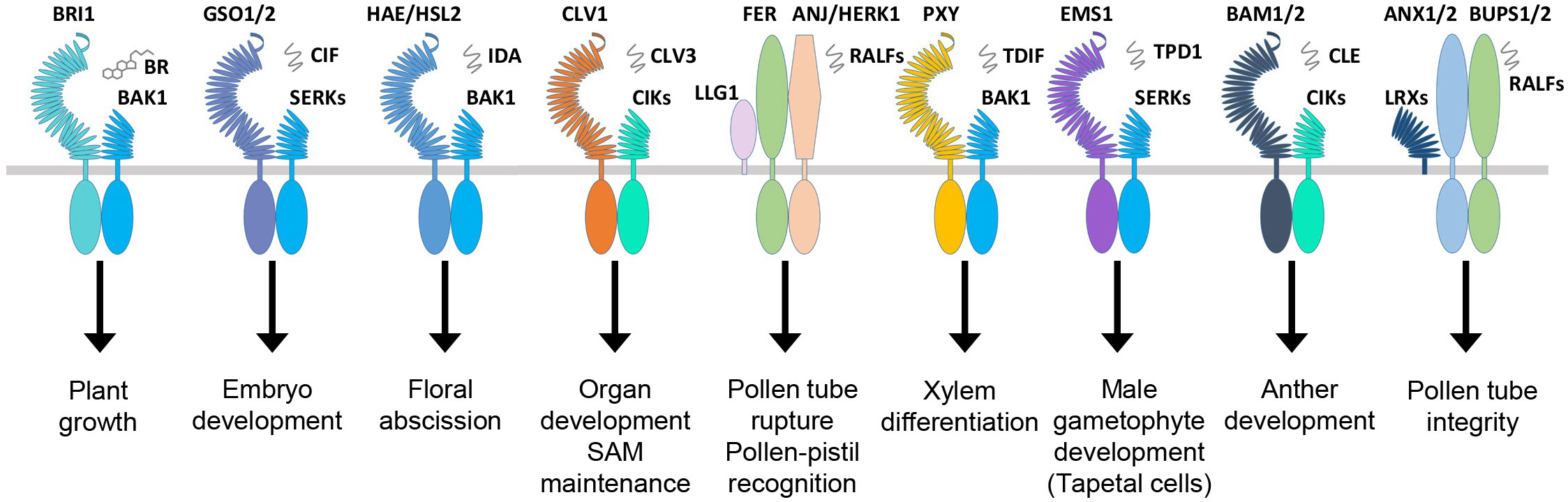
Figure 3 RLKs involved in plant growth and development. The plant hormone BR binds BRI1, and signaling peptides are perceived by the other membrane-localized RLKs. In Arabidopsis, BRI1, GSO1, HAE/HSL, CLV1, FER, PXY, EMS1, BAM1/2, and others make complexes with coreceptors and other scaffold molecules to regulate a variety of growth and developmental processes. RLKs, receptor-like kinases.
HAE is another receptor kinase that expresses in a tissue-specific manner in the areas of abscission zones of floral organs, leaf petiole, and pedicel bases. HAE antisense RNA expression shows delayed abscission in floral organs, suggesting its role in the regulation of the abscission process in floral organs such as stamens, sepals, and petals (Jinn et al., 2000). HAE and HAESA-Like (HSL) interact and bind ligand peptides, inflorescence deficient in abscission (IDA) and IDL, respectively, which leads to the phosphorylation-based activation of MAP kinase signaling. The IDA ligand-based interaction of coreceptor SERK1 with HAE leads to the floral abscission pathway initiating the cell wall hydrolysis at the base of abscission organs (Figure 3; Table 2) (Santiago et al., 2016; Taylor et al., 2016).
In angiosperms, the development of male and female reproductive organs is precisely coordinated to accomplish successful fertilization. There are a number of RLKs involved in the regulation of anther and ovule development. The excess microsporocytes1 (EMS1), receptor-like protein kinases (RPK), barely any meristem (BAM1/2), etc., are known to regulate the pattering and differentiation of cell layers during anther development (Lee et al., 1996; Zhao et al., 2002; Hord et al., 2006). The EMS1 is required for the formation of tapetum during the pollen development inside the anther. Using the pan-brassinosteroid signaling, EMS1 binds the Tapetum Determinant 1 (TPD1) together with coreceptor SERK1/2 to promote the formation of tapetal precursor cells by periclinal division of parietal cells (Jia et al., 2008; Feng and Dickinson, 2010; Zheng et al., 2019; Zheng et al., 2022a). The ems1 and tpd1 mutants have no characteristic tapetum in anthers but produce more microsporocytes than the wild type (Canales et al., 2002; Yang et al., 2003). In another similar study, the EMS1 and BRI1 coordinate stamen elongation in Arabidopsis through the transcription factor BES1/BZR1 (Bai et al., 2023). The RPK2, on the contrary, regulates the anther development through differentiation of the middle layer and tapetum. RPK2 modulates the enzymes involved in lignin biosynthesis and cell wall metabolism by triggering the degradation of tapetum to control their cell fate (Mizuno et al., 2007). BAM1/2, together with RPK2, is required for early anther cell specification. A study of BAM1 and BAM2 mutant bam1bam2 confirmed the lack of endothecium, middle layer, and tapetum in the somatic cell layers, suggesting that both of these genes are responsible for the early-stage regulation of anther differentiation (Table 2) (Hord et al., 2006). In Arabidopsis, coreceptor proteins, CIKs, interact with RPK2 and BAM1/2 to regulate early anther development. As coreceptors of BAM1/2 and RPK2, CIKs control the archesporial cell division and determine the specification of anther parietal cells (Cui et al., 2018). In rice, normal exine development is crucial for pollen grain protection and normal pollination. OsLecRK-S.7 was identified in rice as a vital regulator of pollen development. The kinase of E560K or K418E was found to be vital for the phosphorylation of OsLecRK-S.7 (Peng et al., 2020). Another lectin-type RLK, OsLecRK5, was implicated in anther development by phosphorylating UGP1, enhancing its activity in callose biosynthesis (Wang et al., 2020a). The rice OsERL is a homolog of the Arabidopsis ER gene family implicated in anther lobe development. Mutant erl displayed severe anther development defects and male sterility (Liu et al., 2021). Overexpression of a truncated version of OsLSK1 (including the extracellular and transmembrane domain of OsLSK1 without the intracellular kinase domain) increased plant height and improved yield-related components, including primary branches per panicle and grains per primary branch (Zou et al., 2015).
In plants, after the gamete formation, successful fertilization and reproduction are essential for progeny propagation. It is achieved through a productive pollen and pistil interaction guided by receptors that perceive the ovule-emitted signals in pollen tubes. The RLKs such as PRKs, LURE receptors like Male Discoverer 1 (MDIS1), and MDIS1-interacting receptor-like kinase 1 and 2 (MIK1 and MIK2) sense the ovule secreted peptide, LURE1 (Takeuchi and Higashiyama, 2012; Takeuchi and Higashiyama, 2016; Wang et al., 2016). In Arabidopsis, PERK5 and PERK12, which were recently identified, are necessary for cell wall assembly and ROS homeostasis, guiding proper pollen tube growth. perk5-1 and perk12-1 displayed male gametophytic defect and cell wall of pollen tubes with excessive accumulation of pectins and cellulose (Borassi et al., 2021). Another RLK, FER, expressed in synergids of female gametophyte, comes into play by rupturing the pollen tubes to release the sperm cells (Escobar-Restrepo et al., 2007). The fer mutants display the loss of rupturing ability and overgrowth of pollen tubes (Huck et al., 2003). In rice, CrRLK1L, O. sativa FERONIA-like receptor1 (OsFLR1, also known as DRUS1) and OsFLR2 (DRUS2) are required for maintaining architecture, reproduction, and seed yield (Li et al., 2016a; Pu et al., 2017). Moreover, apple Malus domestica FERONIA-like1 (MdFERL6) and tomato S. lycopersicum FERONIA-like (SlFERL) have been reported in fruit ripening (Jia et al., 2017; Ji et al., 2020). The consistent expression analysis of SlCrRLK1L20 with SlFERL in tomato showed that it might be involved in fruit ripening process similar to SlFERL (Ma et al., 2023). Similarly, the rice-ruptured pollen tube (RUPO) is essential for pollen tube growth and integrity (Liu et al., 2016). The pears (Pyrus bretschneideri), PbrCrRLK1L3, and PbrCrRLK1L26 are also known to regulate the pollen tube rupture process and growth (Kou et al., 2017). The tapetum gene expression and pollen development are regulated by a novel LRR-type PXY-like1 (PXL1) receptor perceiving CLAVATA3/EMBRYO SURROUNDING REGION-RELATED 19 (CLE19) as a ligand (Yu et al., 2023).
Normal zygotic development into an embryo is crucial for plant growth. Several studies have revealed that there are multiple signaling cascades required for embryogenesis. Zygotic arrest 1 (ZAR1) is an RLK that is expressed in the embryo sac starting from the eight-nucleated stage to the mature embryo stage. ZAR1 activates the G protein and calcium-protein signaling, thus regulating the asymmetric division of the zygote and determining the cell fate of daughter cells (Yu et al., 2016). Similarly, GSO1 and GSO2 bind the ligands CIF1 and CIF2 and play an essential role in the development of the normal epidermal surface of the embryo (Figure 3; Table 2) (Nakayama et al., 2017; Okuda et al., 2020). gso1gso2 mutant plants show abnormalities in embryo bending and cotyledon adhesion with highly permeable epidermal structures (Tsuwamoto et al., 2008). A study reported that GSO1 specifies its function from BRI1 by merely two subdomains (Ali et al., 2021).
The development of vascular tissues is a multistep process and requires various RLKs to supervise and regulate the vascular meristem differentiation of the xylem and phloem. For example, the procambial cell division is regulated by phloem intercalated with xylem (PXY), which interacts with the ligand tracheary element differentiation factor (TDIF) to activate the WUSCHEL-related homeobox 4 (WOX4) signaling during vascular development, thus maintaining the cell polarity (Figure 3) (Fisher and Turner, 2007; Hirakawa et al., 2008). The BR receptors BRI1-LIKE 1 (BRL1) and BRI1-LIKE 3 (BRL3) play a role in vascular differentiation (Cano-Delgado, 2004; Zhou et al., 2004; Lozano-Elena and Caño-Delgado, 2019). BRI1-LIKE 2 (BRL2), previously known as vascular highway 1 (VH1), causes abnormal phloem transport in loss-of-function mutants (Clay and Nelson, 2002).
Phytohormone-dependent growth and abiotic stress-related regulations are vital in plant development. Hormone-based RLKs such as BRI1 and receptor dead kinase 1 (RDK1) directly or indirectly perceive the hormone to control various aspects of plant development. BRI1 directly binds to BRs, which activates the downstream signaling cascade, thus activating numerous BR-dependent regulatory genes (Li and Chory, 1997). In this process, upon BR perception, the coreceptor BAK1 associates with the BRI1 receptor, which leads to the transphosphorylation events, thus activating other downstream components of BR signaling (Figure 3) (Li et al., 2002). BRs are also perceived by two other receptors, BRL1 and BRL3, which are involved in the vascular differentiation in Arabidopsis (Cano-Delgado, 2004). A recent study found the BR activity in Ceratopteris richardii for the previously known non-BR receptor, BRL2 (Zheng et al., 2022b). OsBRI1 and its homologs OsBRL1 and OsBRL3 have also been reported in shoot cell elongation and cell division as well as root development (Yamamuro et al., 2000; Nakamura et al., 2006). The wheat TaBRI1 is BR-dependent and responsible for earlier flowering time and higher seed yield in Arabidopsis (Table 2) (Singh et al., 2016; Rafeie et al., 2020; Sharma and Khurana, 2022). Abscisic acid (ABA), however, is mostly reported to be involved in the regulation of abiotic stresses via indirect interactions with RLKs, unlike BRs. For instance, RDK1 interacts indirectly with ABA by recruiting a protein phosphatase, abscisic acid insensitive 1 (ABI1), on the plasma membrane regulating plant responses to abiotic stresses (Kumar et al., 2017). Similarly, the cysteine-rich receptor-like kinase CRK28 and PERK4 are also known to be the indirect regulators of ABA signaling (Bai et al., 2009; Pelagio-Flores et al., 2019). The rice S-receptor protein kinases (OsSRKa) induced by ABA, salt, and polyethylene glycol are vital for leaf width by promoting cell division in the leaf primordium and salt tolerance (Jinjun et al., 2020). Auxin-dependent root growth has been reported in Arabidopsis and rice. In the RHO GTPase (RAC/ROP) signaling pathway, FER as an upstream regulator controls ROS-mediated root hair development (Duan et al., 2010). The auxin- or ABA-induced OsRPK1 is a Ca(2+)-independent Ser/Thr kinase, negatively regulating the adventitious and lateral roots and root apical meristem. Increased plant height, tiller numbers, and growth of transgenic rice plants were reported in the OsRPK1 knockdown mutant (Zou et al., 2014). OsESG1 is an S-domain RLK that regulates early crown root development and drought resistance in rice by utilizing auxin signaling and polar auxin transport (Table 2) (Pan et al., 2020).
5.2 RLKs involved in plant defense, immunity, and stress responses
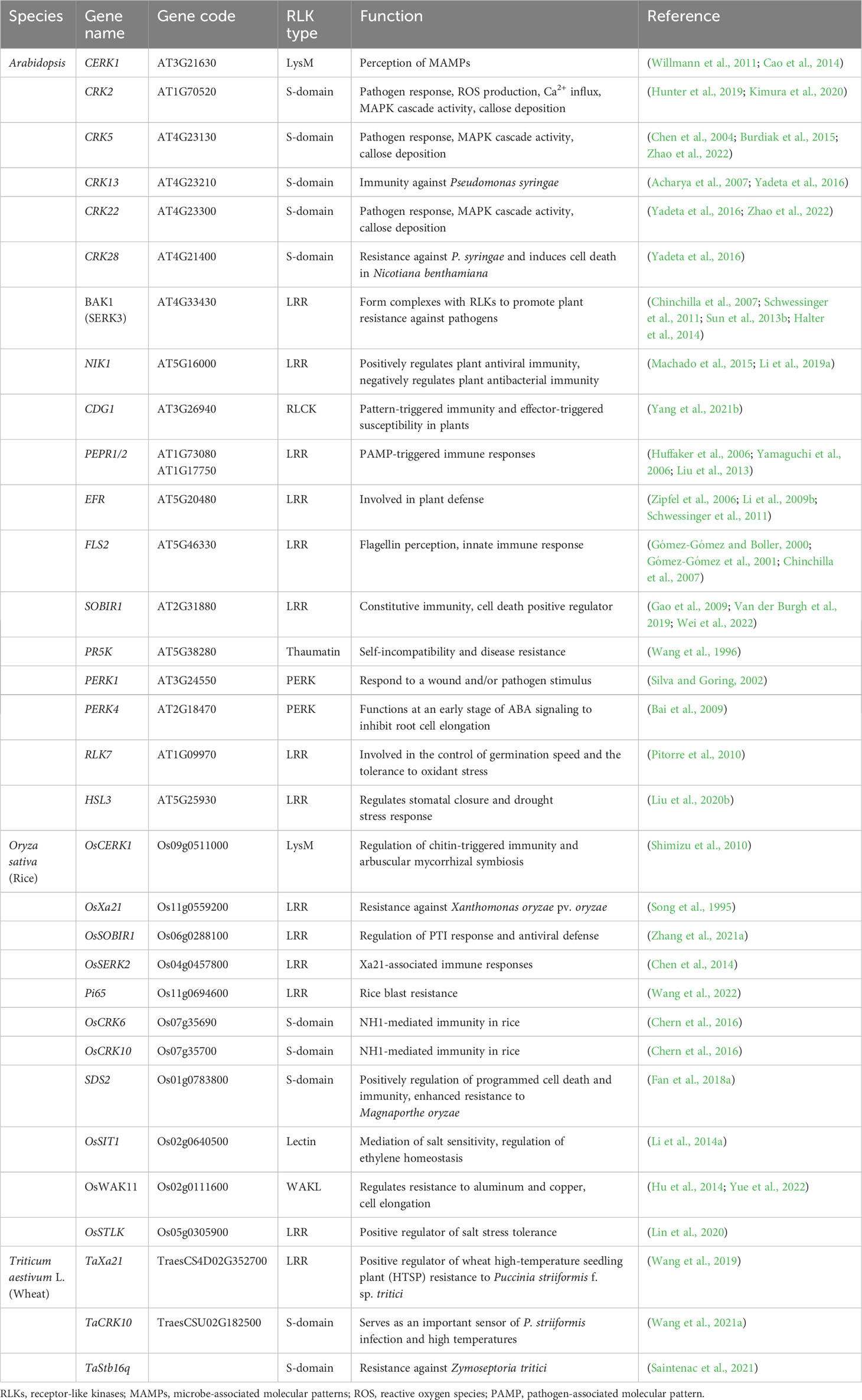
A wide range of RLKs have been identified to provide immunity to the plant against bacterial or fungal pathogens or protein elements of microbial origin. The plants sense these pathogens in the form of host-derived elicitors such as PAMPs, microbe-associated molecular patterns (MAMPs), herbivore-associated molecular patterns (HAMPs), and damage-associated molecular patterns (DAMPs). The responses resulting from these host-derived elicitors are called pathogen-triggered immunity (PTI), while the RLKs involved in this process are known as pattern recognition receptors (PRRs). For instance, the effector-triggered immunity RLKs include tomato Pto and Pto-interacting 1 (Pti1) (Martin et al., 1993; Zhou et al., 1995), Arabidopsis flagellin Insensitive 2 (FLS2), LRK10 (Feuillet et al., 1998), Xa21 in rice (Song et al., 1995), and PBS1 in Arabidopsis (Swiderski and Innes, 2001). Pti1, Pto, and PBS1 are implicated in resistance against bacterial infections. Pti1 itself does not provide immunity against bacteria; instead, it interacts with Pto to confer disease resistance (Zhou et al., 1995). Conversely, Pto gene in tomato confers resistance to races of Pseudomonas syringae pv. tomato, which carries the avirulence gene avrPto (Martin et al., 1993). Similarly, the PBS1 also mediates protection against P. syringae pv. phaseolicola (Swiderski and Innes, 2001).
The FLS2 and Xa21 are closely related in their sequences and belong to the same subgroup of RLKs. The FLS2 recognizes the bacterial epitope flagellin (flg22), which is bacterial elicitor PAMP, and recruits the coreceptor BAK1 or BAK1-like kinase 1 (BKK1), which subsequently activates their transphosphorylation (Figure 4; Table 3) (Gómez-Gómez et al., 1999; Gómez-Gómez and Boller, 2000). This activation leads to the phosphorylation of botrytis-induced kinase 1 (BIK1) and causes its dissociation from the FLS2-BAK1/BKK1 complex. Finally, the PTI responses are regulated through the calcium and ROS as well as the activation of MAPKs or CDPKs (Segonzac and Zipfel, 2011). The FLS2 and BAK1 interaction is negatively regulated by a pseudokinase, BIR2, which competes for the BAK1 interaction (Blaum et al., 2014; Halter et al., 2014). However, the rice OsXa21 provides resistance against Xanthomonas oryzae pv. oryzae, which is a blight pathogen (Song et al., 1995). OsSERK2 associates with OsXa21, OsXa3, and OsFLS2 to positively regulate immune responses in rice (Chen et al., 2014). However, OsSERK1 is involved in rice growth regulation but not reported in immune responses (Zuo et al., 2014). The wheat TaXa21 is highly homologous to OsXa21, cloned from wheat cultivar Xiaoyan 6 (XY 6), and involved in temperature-dependent resistance to strip rust in wheat seedlings. TaXa21 interacts with TaWRKY76 and TaWRKY62 working as a positive regulator under high temperatures, thus providing resistance against Puccinia striiformis (Wang et al., 2019).
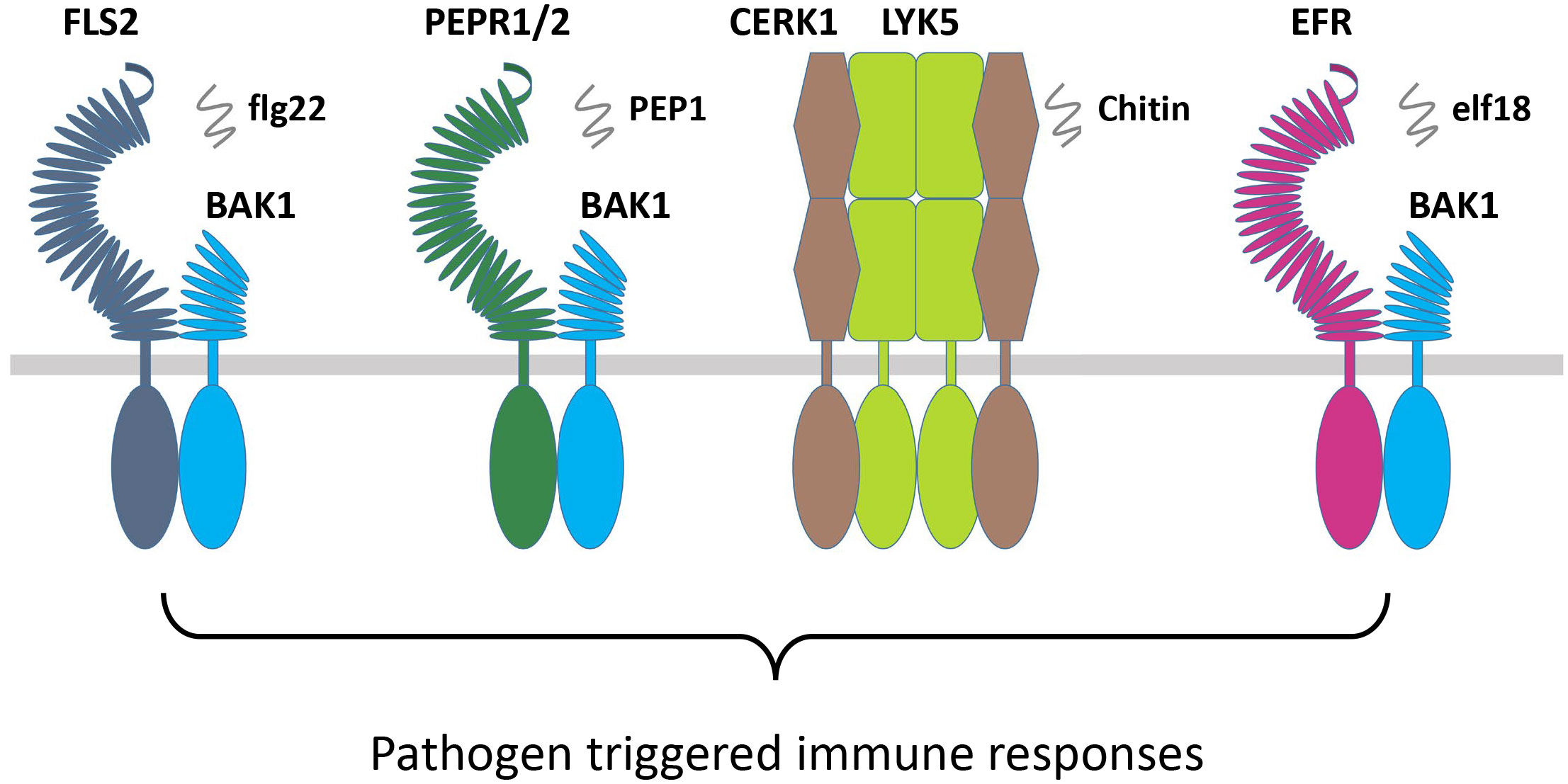
Figure 4 RLKs involved in pathogen immune responses. Pathogen or pathogen elicitor-based RLKs FLS2, PEPR1/2, CERK1/LYK5, and EFR interact with their coreceptors BAK1/SERKs and chitin to induce PTI responses. RLKs, receptor-like kinases; PTI, pathogen-triggered immunity.
Elongation factor (EF-Tu) receptor (EFR) is another PAMP-based RLK that perceives epitopes from bacterial EF-Tu (elf18) to induce immunity against the Agrobacterium transformation (Zipfel et al., 2006). During the process, the PTI pathway is established through the EFR interaction with BAK1 (Schwessinger et al., 2011). In the RLKs, perception of the Arabidopsis danger signal peptides 1 and 2 (PEPR1 and PEPR2) stimulates the plant immunity as a DAMPs, which binds the wound-based perception of damage-associated molecular pattern peptide 1 and 2 (PEP1 and PEP2) (Huffaker et al., 2006; Yamaguchi et al., 2006). The DAMP molecules are produced at the early stages of invasion due to the wounds, microbial infection, or PAMP treatment. Similarly, other RLK complexes that consist of chitin elicitor receptor kinase 1 (CERK1) and LysM receptor-like kinase 1/5 (LYM1/5) recognize the chitin derivative of the fungal cell wall as a MAMP elicitor molecule to regulate the expression of chitin-induced defense genes (Figure 4; Table 3) (Liu et al., 2012; Cao et al., 2014). CERK1 regulates chitin-induced defense gene expression and accumulation of callose by directly interacting and phosphorylating an RLCK PBL27 (Shinya et al., 2014). PBL27 triggers defense responses by phosphorylating MAPKKK5, which activates MPK3/6 and MKK4/5 cascade (Yamada et al., 2016). The BR-signaling component CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) negatively regulates flg22 and chitin-triggered immunity through the interaction with FLS2 and CERK1. CDG1 overexpression causes FLS2 and CERK1 degradation, which reduces flg22 and chitin responses. Depending on its ADP-ribosyl transferase activity, the P. syringae effector AvrRpm1 can cause CDG1 to interact with its host target RPM1-INTERACTING PROTEIN4 (RIN4). CDG1 is capable of phosphorylating RIN4 in vitro at multiple sites, which was diminished in cdg1 null plants, reducing AvrRpm1-induced allergic reaction (Yang et al., 2021b). OsCERK1 is the LysM receptor in O. sativa, which together with CEBiP, regulates chitin elicitor signaling in rice. A significant reduction in defense responses induced by chitin oligosaccharides was observed by knocking down OsCERK1 (Table 3) (Zuo et al., 2014). A novel LRR-type Pi65 was recently cloned from O. sativa japonica that showed resistance against rice blast Magnaporthe oryzae. Decreased resistance was observed in Pi65 knockout gene in the GY129 variety, while its overexpression in the susceptible variety LX1 enhanced resistance to rice blast (Wang et al., 2022). Program cell death is a crucial mechanism for plant immunity but could cause excessive damage to the plant if left unregulated. An S-domain RLK SPL11 cell-death suppressor 2 (SDS2) positively regulates program cell death and immunity in rice. Elevated immune responses and enhanced resistance were observed in the SDS-overexpression lines while showing susceptibility in sds2 mutants against blast fungus M. oryzae (Fan et al., 2018a).
During a pathogen attack, the stress responses are regulated through the activation of cell wall integrity maintenance and immune signaling systems resulting from the cell wall damage. In Arabidopsis, WAK1 and WAK2 are known to maintain cell wall integrity. They do so by binding soluble oligo-galacturonide (OG) pectin and bacterial elicitor activating WAK-dependent, distinct stress-like response pathway to help plants resist a pathogen attack (Kohorn et al., 2016). Recently, several WAKs have been reported in several crop species, playing a role in immune responses against fungal and bacterial pathogens. For instance, individual OsWAKs have been reported to respond differently to rice blast fungus. OsWAK14, OsWAK91, and OsWAK92 positively regulate resistance, while OsWAK112d is a negative regulator of blast resistance. Moreover, enhanced H2O2 production required OsWAK91 to confer resistance during infection (Delteil et al., 2016). The TaWAK2 is responsible for the prevention of Fusarium head blight disease caused by Fusarium graminearum. TaWAK2 produces a more rigid cell wall, limiting the expression of pectin methyl esterase 1 (Gadaleta et al., 2019). Two independent studies identified TaWAK-6D and TaWAK7D in wheat, contributing to resistance against Fusarium pseudograminearum and Rhizoctonia cerealis (Qi et al., 2021a; Qi et al., 2021b). In maize, qHSR1 and ZmWAK-RLK1 (Htn1) have been reported in plant resistance against Sporisorium reilianum and Exserohilum turcicum, respectively (Zuo et al., 2015; Yang et al., 2019).
Cysteine-rich receptor-like kinases (CRKs) have the most significant number of RLKs, which are triggered during defense and oxidative stress mechanisms. CRKs are involved in program cell death and defense responses, employing both PTI and elicitor-triggered immunity (ETI) responses, in which the RLK-mediated signaling upregulates the transcriptional activation of defense (Du and Chen, 2000; Chen, 2001). For example, overexpression of CRK5 and CRK13 upregulates the immunity-related RLKs, such as pathogenesis-related protein1/5 (PR1/5) and isochorismate synthase1 (ICS1), which eventually enhance defenses against P. syringae. Similarly, CRK45 provides similar enhanced immunity against P. syringae, while crk45 mutant displayed more susceptibility to the pathogen (Zhang et al., 2013a). CRK2 is also known to induce full elicitor-induced ROS bursts while regulating plant innate immunity. The mutant crk2 displayed impaired defense responses against P. syringae (Kimura et al., 2020). CRKs, such as CRK4/5 and CRK19/20, are also known to induce cell death hypersensitive-like responses in plants (Chen et al., 2003; Chen et al., 2004; Acharya et al., 2007). CRK28-enhanced expression in Arabidopsis increased disease resistance to P. syringae while inducing cell death in N. benthamiana. CRK28 forms complexes with FLS2 and BAK1 independent of its kinase activity, suggesting that CRK28 kinase activity is not required during this interaction (Yadeta et al., 2016). In rice, the CRK6 and CRK10 are required for NPR1 homolog 1 (NH1)-mediated immunity (Chern et al., 2016). In T. aestivum, TaCRK1 provides protection against R. cerealis infection, while HvCRK1 of H. vulgare contributes to ROS-mediated basal resistance against powdery mildew infection (Rayapuram et al., 2012; Yang et al., 2013). Another wheat CRK Stb16q has been reported to provide resistance against Zymoseptoria tritici, which causes septoria tritici blotch (STB), representing one of the most genetically diverse and devastating wheat pathogens worldwide (Saintenac et al., 2021). Similar responses have also been reported in wheat by studying Stb6, which encodes a WAK-like (WAKL) protein to provide immunity against Z. tritici (Saintenac et al., 2018). A novel wheat TaCRK2 positively regulates resistance against leaf rust, caused by Puccinia triticina in a Ca2+-dependent manner (Gu et al., 2020). Another study found TaCRK3 having two DUF26, providing defense against R. cerealis mycelia (Guo et al., 2021). Similarly, wheat TaCRK10, similar in function to TaXa21, provides resistance against stripe rust under high temperatures. While TaCRK10 and TaXa21 share a similar gene function, their molecular targets differ, suggesting that plants have developed distinct defense mechanisms against the same pathogen. This could work to their benefit if the pathogen suppresses one of the pathways (Table 3) (Wang et al., 2021a). It has been reported that OsRMC, which has DUF26 as an extracellular domain, plays a role in the development of the salt stress response and rice root meander curling (Jiang et al., 2007; Zhang et al., 2009). In Medicago truncatula, the early senescence and defense responses are prevented by SymCRK associated with symbiotic interactions (Berrabah et al., 2014).
In Arabidopsis, SOBIR1 was identified through genetic screening of the suppressors of bir1-1, which showed excessive cell death in mutant form. sobir1 mutant, on the contrary, showed rescued cell death, suggesting that SOBIR1 is a positive regulator of cell death (Gao et al., 2009). Similarly, the Arabidopsis SOBIR1 and kinase-active BAK1 together constitutively activate immune responses through autophosphorylation and transphosphorylation events (Van der Burgh et al., 2019). A recent study reported that the phosphorylation of Thr529 and activation of β3-αC loop are essential for SOBIR1 activation by BAK1 transphosphorylation during SOBIR1-induced cell death response in N. benthamiana (Wei et al., 2022). In rice, immunity against rice black-streaked dwarf virus (RBSDV) infection is mediated by OsSOBIR1 interaction with OsRLP1, triggering PTI responses (Zhang et al., 2021a). The NIK1 is an LRR-type RLK, which was previously found to participate in antiviral immunity by translocation of RLP10 into the nucleus. The RLP10 downregulates the translation machinery by making a complex with LIMYB, resulting in host and viral mRNA inhibition and tolerance against Begomovirus (Machado et al., 2015). Another study reported that NIK1 also acts as a negative regulator of antibacterial immunity. In addition, it also negatively regulates the FLS2-BAK1 complexes. This unique inverse modulation can allow viruses and bacteria to use host immunity against each other (Table 3) (Li et al., 2019a).
During plant growth and development, several environmental and other related factors can lead to the plant’s abiotic stresses. Plants in such conditions use sophisticated mechanisms through utilizing diversified RLKs. In plants, the osmotic imbalance is caused by drought and salinity. It is well recognized that ABA affects osmotic balance and stomatal closure during abiotic stress responses (Zhang et al., 2020). Some CRKs are found to regulate ABA signaling and its biosynthesis. CRK4, CRK5, and CRK19 redundantly confer ABA responses, enhancing drought tolerance (Lu et al., 2016). Similarly, the expression of ABA biosynthesis genes, including abscisic aldehyde oxidase 3 (AAO3), ABA deficient (ABA1 and ABA2), and 9-cis-epoxycarotenoid dioxygenases (NCED3 and NCED5), are positively regulated by cytoplasm-localized CRK45. This increases tolerance to drought and salt stress during germination and post-germination growth (Zhang et al., 2013b). In addition to CRKs, the LRR-type HSL3 also plays a role in tolerance to drought by regulating the stomatal closure. It has been demonstrated that HSL3 can adjust the amount of H2O2 in the guard cells, hence negatively regulating stomatal closure. Dehydration or treatment with ABA or H2O2 greatly upregulated HSL3 (Liu et al., 2020b). Arabidopsis RLK7 is associated with stress and immune responses that interact with phytophthora-inhibited protease 1 (PIP1), activating S-type anion channel SLAC1 to induce stomatal closer (Pitorre et al., 2010; Shen et al., 2020). The plasma membrane-localized PdERECTA reduced stomatal density and limited water-consuming cells, restricting water consumption and enhancing drought resistance in poplar (Li et al., 2021a). A similar study showed that the expression of S. bicolor SbERECTA (SbER) in Arabidopsis and maize confers enhanced drought tolerance (Li et al., 2019c). More recently, it was discovered that BAK1 can activate H+-ATPase isoform 2 (AHA2) by phosphorylating its C-terminal Ser944, which causes guard cells’ cytoplasm to become alkaline, initiating the process of stomatal closure (Pei et al., 2022).
A number of RLKs have been shown to be involved in other stress responses such as salt, metal, and temperature stresses. For instance, WAK1 of the WAK family is implicated in responses against aluminum toxicity in Arabidopsis and rice beans. The AtWAK1 overexpression showed increased tolerance to aluminum stress in Arabidopsis (Sivaguru et al., 2003; Lou et al., 2020). The rice OsWAK11 has been implicated in copper, aluminum, and sodium responses. OsWAK11 detoxified excessive copper and knocked it down, resulting in hypersensitivity to copper toxicity (Hu et al., 2014; Xia et al., 2018). OsSIT1, a lectin-type RLK, is known to negatively regulate resistance to salt stress. OsSIT1 phosphorylates the downstream effectors MPK3 and MPK6, causing the production of ethylene and salt-induced ethylene signaling (Li et al., 2014a). Another rice RLK, OsSTLK, was implicated in salt tolerance with improved salt stress tolerance and reduced salt sensitivity observed in OsSTLK overexpression rice lines (Table 3) (Lin et al., 2020). Similarly, a cold-inducible gene, GsLRPK, from Glycine soja enhances resistance to cold stress and increases the expression of a number of cold-responsive genes when expressed in yeast and Arabidopsis (Yang et al., 2014). Another two cold-tolerance genes, MtCTLK1 and MfCTLK1 from M. truncatula and Medicago falcata, respectively, are also known to regulate cold tolerance (Geng et al., 2021).
6 The role of receptor-like cytoplasmic kinases
RLCKs play a crucial role in RLK-mediated signaling. The N-myristoylation or palmitoylation enables most RLCKs to localize on the plasma membrane (Lin et al., 2013). A combined phylogeny with RLKs conducted in Arabidopsis and rice showed that there are 17 subgroups of RLCKs including subgroups II and IV to XIX (Shiu et al., 2004). Most of the RLCKs consist of Ser/Thr-based KDs, while others possess additional domains such as LRR, EGF, and TM (Vij et al., 2008; Lehti-Shiu et al., 2009; Lin et al., 2013). RLCKs regulate several functions such as hormone signaling, sexual reproduction, shoot and root meristem maintenance, stomatal patterning, petal abscission, differentiation of vascular tissues, innate immunity, and adaptation to abiotic stresses in a coordinated manner with RLKs and RLPs (Lin et al., 2013; Tang et al., 2017; Liang and Zhou, 2018). For example, during the immune responses, the BIK1 and other associated RLCKs directly interact with various immune-related RLKs (Lu et al., 2010; Liu et al., 2013; Kong et al., 2016). Similarly, BR-induced signaling is regulated via direct interaction of RLCK-XII subclass BR signaling kinases (BSKs) with BRI1 to control different subsequent processes (Tang et al., 2008; Sreeramulu et al., 2013). In Arabidopsis, the pollen tube integrity is controlled through the downstream action of RLCK MARIS (MRI) from the two pairs of closely related receptors, buddha’s paper seal (BUPS1) and BUPS2 and ANXUR1 (ANX1) and ANX2 (Figure 3) (Boisson-Dernier et al., 2015; Liao et al., 2016; Ge et al., 2017). In Brassica, the self-incompatibility is regulated through the interaction of an RLCK M-locus protein kinase (MLPK) with S-locus b-lectin receptor kinase (SRK), a female determinant (Kakita et al., 2007). The RLCKs’ primary regulatory mechanism of RLK-mediated signaling is controlled through the transphosphorylation of RLKs and different downstream substrates where the RLCKs activity and stability are firmly regulated (Sreeramulu et al., 2013; Li et al., 2014b; Nolan et al., 2017; Wang et al., 2018).
In RLK-mediated signaling, the RLK KD determines the signal output for specificity. In this case, different kinds of RLCKs are associated with a variety of RLKs. In several cases, different RLKs can associate the same RLCK to regulate distinct functions. For instance, both BRI1 and FLS2 interact with BIK1. The BRI1-BIK1 interaction leads to the negative regulation of root hair, while the FLS2-BIK1 interaction is responsible for the positive regulation of immunity (Veronese et al., 2006; Lu et al., 2010). Similarly, the BSKs can interact with BRI1, EMS1, NILR1, and FLS2 to positively regulate plant growth, tapetum formation, and immunity (Shi et al., 2013; Zheng et al., 2019; Zheng et al., 2022a). In the case of EMS1, BRI1, and NILR1, they all may bind distinct ligands to control their different functions but use the same signaling components, such as BSKs, BKI1, BIN2, and BES1/BZR1, in the same signaling pathway, which opens up a new insight into the regulation of RLK signaling (Figure 5; Table 2) (Zheng et al., 2019; Zheng et al., 2022a). All these studies suggest that the fate of RLCK is determined by ligand-based upstream RLK complexes, but the exact mechanism is still unknown. The RLCKs interpret different signal outputs differently and give rise to distinct ones. BSKs are the direct substrates of BRI1 and are directly phosphorylated by BRI1. Similarly, BSK1 also interacts with and is phosphorylated by NILR1 and FLS2. The phosphorylation of BSK1 by BRI1 takes place in a similar manner to that of FLS2 and NILR1, but this interaction can only induce plant growth but not the immune responses (Shi et al., 2013; Zheng et al., 2022a). It is possible that the BSK1 distinct output signal is caused by the differential phosphorylation. In addition, other signaling components and complexes as well as environmental cues may also play a role in defining the function of RLCKs.
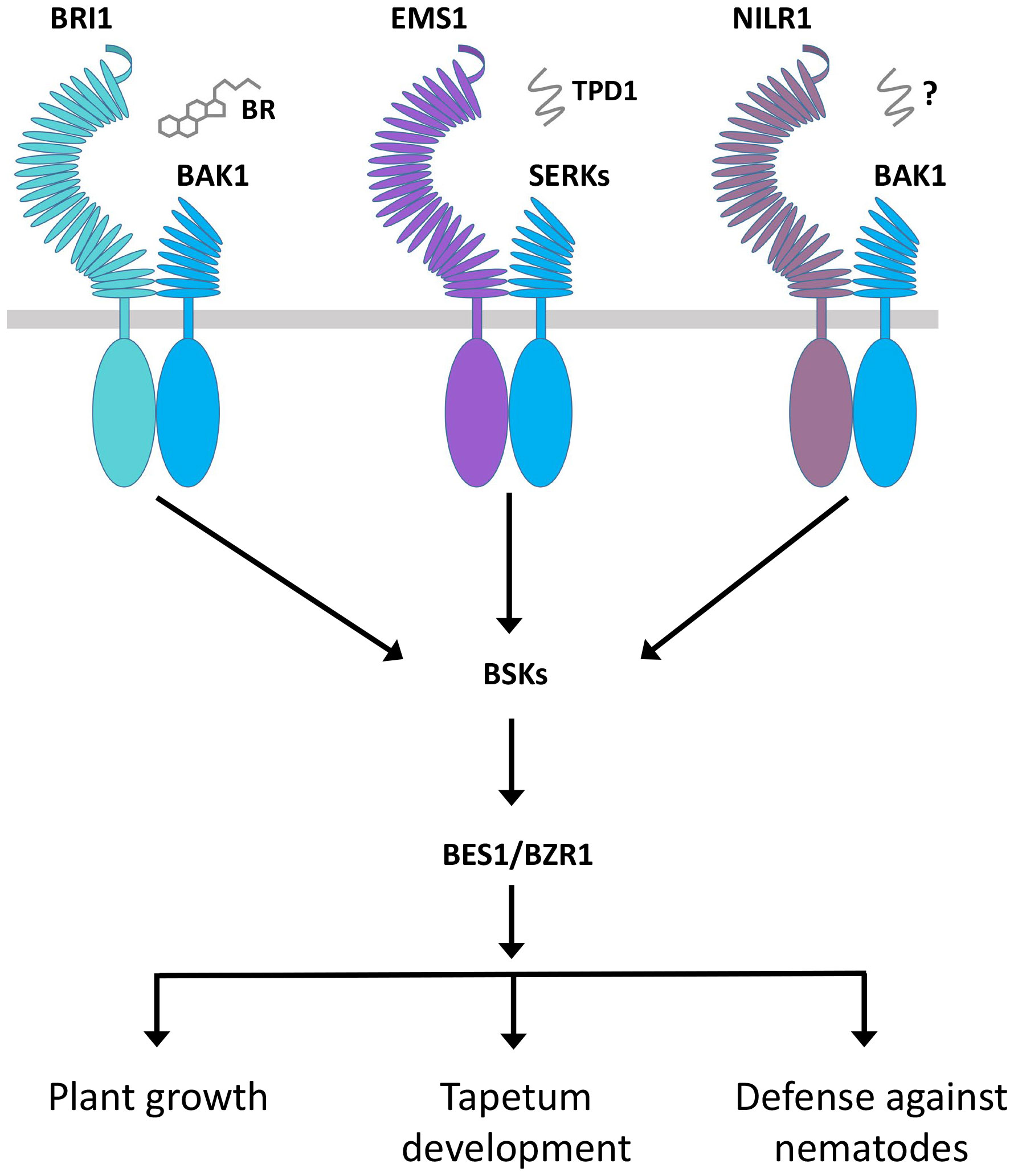
Figure 5 Regulation of diverse functions of BRI1, EMS1, and NILR1 through the same signaling pathway. BRI1 and EMS1 perceive BR hormone and TPD1 peptide ligand, respectively, while NILR1 binds an unknown ligand to collectively initiate a pan-brassinosteroid signaling cascade to control their distinct functions.
7 LRR-RLKs: the biggest class of RLKs
In Arabidopsis, the LRR-RLK subgroup consists of approximately 200 members, making it the largest class of RLKs (Shiu and Bleecker, 2001a). The KD of LRR-RLKs is conserved among the group members, but the ECD that possesses variable numbers and organization of LRRs ranging from 3 to 31, dependent on the kind and the subgroup of LRR, is highly versatile (Pfister et al., 2014; Hohmann et al., 2017). The LRR-RLKs have been studied extensively over the past two decades. Still, due to the large number of RLKs within the group, the number of designated members is approximately 89, and only 60 of them have been characterized functionally, making up 35% of the total LRR-RLKs in Arabidopsis (Wu et al., 2016). Based on experimental studies, these characterized LRR-RLKs can be classified as either plant growth and development regulators or involved in plant defense and immunity (He et al., 2018; Jamieson et al., 2018). Many LRR-RLKs that have been identified were previously misclassified into other gene families (Man et al., 2020). In Arabidopsis, the LRR-RLKs are further divided into 15 subclasses based on their KD conservation (Shiu and Bleecker, 2001a). The LRR is composed of 24 amino acid residues with consensus sequence LxxLxLxxNxLxxGxIPxx, where x represents non-conserved residues (Torii, 2004). In plants, this unique motif of 24 amino acid residues forms the helical horseshoe-like structure characterized by having β-turn or β-sheet acting as a protein–protein interaction site (Kobe and Deisenhofer, 1993; Jones and Jones, 1997). Interestingly, this signature horseshoe-like structure is not common to all LRR-RLKs, but instead, some of the LRRs have a right-handed super-helical structure, such as BRI1, which is different than the solenoid conformation (Hothorn et al., 2011; She et al., 2011). The GxIP sequence configuration forms the helical structure and is common to many LRRs such as FLS2, BRL1, and RPK2 (She et al., 2013; Sun et al., 2013b; Song et al., 2014). There is a comparable size difference among distinct LRR-RLKs, which vary from 19 to 21 LRRs. The super-helical structure is formed by approximately 20–25 LRRs, where the lateral and inner surface of the structure actively achieves its binding with other protein molecules (Sun et al., 2013a; Sun et al., 2013b). Some of the LRR-RLKs with different numbers of LRRs include BRI1 25 LRRs, PSKR1 21 LRRs, GSO1 31 LRRs, FLS2 29 LRRs, HAE 21 LRR, TMK1 13 LRRs, PGIP 10 LRRs, RPK2 22 LRRs, and SERK1 5 LRRs (Pfister et al., 2014; Hohmann et al., 2017). Several studies have suggested that the size of LRR-RLK can be exploited to uncover the function of LRR-RLK as a receptor or coreceptor (Jaillais et al., 2011; Smakowska-Luzan et al., 2018). The small-sized LRRs are generally considered as coreceptor; for example, CIKs and SERKs have been identified with shorter ECD serving as coreceptors for the protein–protein interaction.
In the regular region of LRRs, a spacer ID is identified between LRR21 and LRR22 (corresponding to 584–654 residues), making them the fourth to fifth LRRs from the C-terminus. In BRI1, this 70 amino acid residue segment folds back into the interior of the super-helix, forming a small domain that generates an interaction site for the BR (Li and Chory, 1997; Hothorn et al., 2011). In addition, the crystal structure of RPK2 revealed two ID regions where the ligand binds (Song et al., 2014). Other LRR-RLKs containing an ID include TMK1, PSKR1, and PSY1R (Matsushima and Miyashita, 2012). Some studies have also reported another interesting domain called Cys-pair in the ECD. The Cys-pair is located at the N-terminus near the start codon between the signaling peptide (SP) and the first LRR. In some cases, a second Cys-pair is also present between the TM and the last LRR (Diévart and Clark, 2003). The mutation in Cys-pair does not affect the function in CLV2 (Song et al., 2010), but in some LRR-RLKs, the Cys-pair mutation affects the receptor’s normal function. For example, FLS2 with Cys-pair mutation showed a significant decrease in its activity (Sun et al., 2012). In BRI1, the bri1-5 mutant harboring Cys-pair mutation results in the retention of the bri1 mutant in the endoplasmic reticulum, which suggests that this mutant is not able to pass the endoplasmic reticulum quality control (ERQC) to translocate into the plasma membrane and is eventually degraded by endoplasmic reticulum-associated degradation (ERAD) (Hong et al., 2008). Thus, LRRs, together with other domains in the ECD, are the essential components of the receptor and play a vital role in ligand perception, protein–protein interaction, and signal transduction.
8 Conclusion and perspectives
EPKs control various biological functions through the utilization of ATP, where a γ-phosphate is transferred to the free OH-site of serine/threonine or tyrosine; thus, EPKs are widely considered dynamic molecular switches (Taylor et al., 2012; Zhang et al., 2015). RLKs belong to the EPKs that resemble animal RTKs in their domain organization and play a central role in plant growth and development, reproduction, adaptation to new environmental conditions, and immune responses. The main kinase core of RLKs is highly conserved containing different domains with unique signatures and functions. The evidence of the first RLKs comes from common ancestor green algae, which are vastly duplicated and expanded in angiosperm, implying their significance in plant growth and survival during evolution. Although the exact pattern of RLK evolution is only partially resolved, the sequenced genomes of basal Streptophyte species paved the road to describe the precise evolutionary history. The expansion of RLKs might directly be related to the morphological complexity of the plant species. For example, C. braunii and K. flaccidum are Charophytes with 435 and 94 RLKs, respectively (Table 1), but domain expansion of LyM, CrRLK1L, and S-domain was only specific to C. braunii, suggesting the association of these domains with morphological complexity of a specific lineage (Dievart et al., 2020). Similarly, the ever-changing environments also play a vital role in RLK evolution where their ECD architectures are diversified to enable RLKs to perceive numerous environmental cues and respond accordingly. The intracellular KD remains conserved but evolves enough changes that can recruit different complexes to execute the signals in response to those environmental cues. Gaining an in-depth understanding of RLK families, especially the expansion of the large families, such as LRR, PERK, and Lec, the ancestral and recent lineage-specific duplication, or domain rearrangement events must be taken into account, which makes it a challenging task.
Among the vast number of RLKs, only a handful have been identified and functionally characterized through genetic studies. Plants rely on an RLK‐mediated signal transduction pathway that utilizes intracellular regulatory components to alter gene expression profiles, in response to an extracellular signal. RLKs regulate a vast array of gene functions during plant growth and development. The detailed molecular mechanism of the functionally characterized gene is still not completely defined. Several RLKs regulate multiple developmental processes. For example, HAE and HSL2 play a dual role in regulating cell separation during floral organ abscission and lateral root development (Jinn et al., 2000; Zhu et al., 2019). ACR4 regulates not only root apical meristem but also sepal development, integument development, and epidermal development (Gifford et al., 2003; Tanaka et al., 2007; De Smet et al., 2008). GSO1/2 regulates normal embryo development and root growth development (Tsuwamoto et al., 2008; Racolta et al., 2014). For future studies, it would be interesting to ask how a single RLK can regulate multiple functions or how different RLKs interact with each other to ensure the same functional regulation.
Till recently, traditionally, it was believed that RLKs use unique signaling pathways to control their distinct functions. The first evidence of multiple RLKs utilizing the same signaling cascade to control diverse functions emerged recently, showing that BRI1, EMS1, and NILR1 bind entirely different ligands to control their unique functions; however, they utilize the same pan-brassinosteroid signaling cascade (Figure 5) (Zheng et al., 2019; Zheng et al., 2022a). Studies like this entirely changed the perspective of the RLK signaling mechanism. Techniques such as exchange domain, proteomics-based RLK mining with different genetic backgrounds, single-cell-omic analysis, and cellular and subcellular imaging may help to understand RLK signaling pathways and networks at a higher precision.
One of the most important and challenging tasks regarding the RLK signaling mechanism is the identification of ligands. So far, only a limited number of RLK-binding ligands have been identified, and the majority of RLK-binding ligands are unknown. Novel techniques are expected to be developed, which can be employed together with the existing technologies of proteomic analyses, genetic approaches, structural biology, and photoaffinity crosslinking technology for ligand identification.
As the RLKs are extensively duplicated and expanded in plants, another challenge comes from the functional redundancy of genes, which makes it difficult to identify the functions of RLKs. For example, the BAK1 interacts with BRI1 as a coreceptor to regulate BR-dependent growth promotion. BAK1 also regulates FLS2 and EFR signaling pathways to induce immune responses. Similarly, a double knockout mutant gso1gso2 is required to study the function of GSO1 and GSO2 that interact with two ligand peptides CIF1 and CIF2 (Nakayama et al., 2017). However, advanced techniques, like CRISPR for generating high-order RLK mutants and advanced method phenotypic analysis, will help overcome functional redundancy to understand their roles in the future.
Signaling pathway crosstalk is a common mechanism in plants. One RLK might be associated with multiple pathways apart from the already known ones as in the case of BAK1. The most interesting question about RLKs is how these RLKs define their functions, especially when an opposite output produces from each other if they use the same signaling components or even the same signaling pathways. Technologies such as genetics, structural, and proteomics will play a vital role in defining all these questions. These studies will play a significant role in improving crop yield and more resistant cultivars to biotic and abiotic stresses via bioengineered and plant breeding approaches.
Author contributions
JL: Software, Writing – review & editing. WL: Investigation, Writing – review & editing. GW: Funding acquisition, Validation, Writing – review & editing. KA: Conceptualization, Writing – original draft, Software.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Fundamental Research Fund for Central Universities to AK (GK202304041), JL (2017TS038), and GW (GK201101005 and GK202001010); Chinese Natural Foundation of Science to GW (32070325, 31270324 and 31741014); Grants from the Chinese Ministry of Education (313034) and (20130202110007) to GW.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedi, A., Hajiahmadi, Z., Kordrostami, M., Esmaeel, Q., Jacquard, C. (2022). Analyses of lysin-motif receptor-like kinase (LysM-RLK) gene family in allotetraploid brassica napus L. and its progenitor species: an in silico study. Cells 11 (1), 37. doi: 10.3390/cells11010037
Acharya, B. R., Raina, S., Maqbool, S. B., Jagadeeswaran, G., Mosher, S. L., Appel, H. M., et al. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50 (3), 488–499. doi: 10.1111/j.1365-313X.2007.03064.x
Ali, K., Li, W., Qin, Y., Wang, S., Feng, L., Wei, Q., et al. (2021). Kinase function of brassinosteroid receptor specified by two allosterically regulated subdomains. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.802924
Bai, L., Zhang, G., Zhou, Y., Zhang, Z., Wang, W., Du, Y., et al. (2009). Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 60 (2), 314–327. doi: 10.1111/j.1365-313X.2009.03956.x
Bai, Q., Wang, L., Huang, S., Ali, K., Li, G., Ren, H., et al. (2023). The receptor-like kinase EMS1 and BRI1 coordinately regulate stamen elongation via the transcription factors BES1/BZR1 in Arabidopsis. Plant Sci. 331, 111673. doi: 10.1016/j.plantsci.2023.111673
Becraft, P. W., Stinard, P. S., McCarty, D. R. (1996). CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273 (5280), 1406–1409. doi: 10.1126/science.273.5280.1406
Beenstock, J., Mooshayef, N., Engelberg, D. (2016). How do protein kinases take a selfie (Autophosphorylate)? Trends Biochem. Sci. 41 (11), 938–953. doi: 10.1016/j.tibs.2016.08.006
Bellande, K., Bono, J. J., Savelli, B., Jamet, E., Canut, H. (2017). Plant lectins and lectin receptor-like kinases: how do they sense the outside? Int. J. Mol. Sci. 18 (6), 1164. doi: 10.3390/ijms18061164
Beltrao, P., Albanèse, V., Kenner, L. R., Swaney, D. L., Burlingame, A., Villén, J., et al. (2012). Systematic functional prioritization of protein posttranslational modifications. Cell 150 (2), 413–425. doi: 10.1016/j.cell.2012.05.036
Bergthorsson, U., Andersson, D. I., Roth, J. R. (2007). Ohno's dilemma: Evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U.S.A. 104 (43), 17004–17009. doi: 10.1073/pnas.0707158104
Berrabah, F., Bourcy, M., Eschstruth, A., Cayrel, A., Guefrachi, I., Mergaert, P., et al. (2014). A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol. 203 (4), 1305–1314. doi: 10.1111/nph.12881
Blaum, B. S., Mazzotta, S., Nöldeke, E. R., Halter, T., Madlung, J., Kemmerling, B., et al. (2014). Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 186 (1), 112–121. doi: 10.1016/j.jsb.2014.02.005
Bleecker, A. B. (1991). Genetic analysis of ethylene responses in Arabidopsis thaliana. Symp Soc. Exp. Biol. 45, 149–158.
Boisson-Dernier, A., Franck, C. M., Lituiev, D. S., Grossniklaus, U. (2015). Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc. Natl. Acad. Sci. U.S.A. 112 (39), 12211–12216. doi: 10.1073/pnas.1512375112
Bojar, D., Martinez, J., Santiago, J., Rybin, V., Bayliss, R., Hothorn, M. (2014). Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78 (1), 31–43. doi: 10.1111/tpj.12445
Borassi, C., Sede, A. R., Mecchia, M. A., Mangano, S., Marzol, E., Denita-Juarez, S. P., et al. (2021). Proline-rich extensin-like receptor kinases PERK5 and PERK12 are involved in pollen tube growth. FEBS Lett. 595 (20), 2593–2607. doi: 10.1002/1873-3468.14185
Borassi, C., Sede, A. R., Mecchia, M. A., Salgado Salter, J. D., Marzol, E., Muschietti, J. P., et al. (2016). An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 67 (2), 477–487. doi: 10.1093/jxb/erv455
Buendia, L., Girardin, A., Wang, T., Cottret, L., Lefebvre, B. (2018). LysM receptor-like kinase and lysM receptor-like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 9 (1531). doi: 10.3389/fpls.2018.01531
Burdiak, P., Rusaczonek, A., Witoń, D., Głów, D., Karpiński, S. (2015). Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J. Exp. Bot. 66 (11), 3325–3337. doi: 10.1093/jxb/erv143
Cambi, A., Koopman, M., Figdor, C. G. (2005). How C-type lectins detect pathogens. Cell Microbiol. 7 (4), 481–488. doi: 10.1111/j.1462-5822.2005.00506.x
Canales, C., Bhatt, A. M., Scott, R., Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12 (20), 1718–1727. doi: 10.1016/s0960-9822(02)01151-x
Cano-Delgado, A. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 (21), 5341–5351. doi: 10.1242/dev.01403
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. doi: 10.7554/eLife.03766
Cassab, G. I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 (1), 281–309. doi: 10.1146/annurev.arplant.49.1.281
Chang, C., Schaller, G. E., Patterson, S. E., Kwok, S. F., Meyerowitz, E. M., Bleecker, A. B. (1992). The TMK1 gene from Arabidopsis codes for a protein with structural and biochemical characteristics of a receptor protein kinase. Plant Cell 4 (10), 1263–1271. doi: 10.1105/tpc.4.10.1263
Chen, Z. (2001). A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 126 (2), 473–476. doi: 10.1104/pp.126.2.473
Chen, K., Du, L., Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53 (1-2), 61–74. doi: 10.1023/b:plan.0000009265.72567.58
Chen, K., Fan, B., Du, L., Chen, Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56 (2), 271–283. doi: 10.1007/s11103-004-3381-2
Chen, W., Lv, M., Wang, Y., Wang, P. A., Cui, Y., Li, M., et al. (2019). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10 (1), 4164. doi: 10.1038/s41467-019-12118-4
Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P. E., Ruan, D., et al. (2014). An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 7 (5), 874–892. doi: 10.1093/mp/ssu003
Cheng, W., Wang, Z., Xu, F., Ahmad, W., Lu, G., Su, Y., et al. (2021). Genome-wide identification of LRR-RLK family in saccharum and expression analysis in response to biotic and abiotic stress. Curr. Issues Mol. Biol. 43 (3), 1632–1651. doi: 10.3390/cimb43030116
Chern, M., Xu, Q., Bart, R. S., Bai, W., Ruan, D., Sze-To, W. H., et al. (2016). A genetic screen identifies a requirement for cysteine-rich–receptor-like kinases in rice NH1 (OsNPR1)-mediated immunity. PloS Genet. 12 (5), e1006049. doi: 10.1371/journal.pgen.1006049
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 (7152), 497–500. doi: 10.1038/nature05999
Clark, S. E., Williams, R. W., Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 (4), 575–585. doi: 10.1016/s0092-8674(00)80239-1
Clay, N. K., Nelson, T. (2002). VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 14 (11), 2707–2722. doi: 10.1105/tpc.005884
Cock, J. M., Vanoosthuyse, V., Gaude, T. (2002). Receptor kinase signalling in plants and animals: distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 14 (2), 230–236. doi: 10.1016/s0955-0674(02)00305-8
Couto, D., Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16 (9), 537–552. doi: 10.1038/nri.2016.77
Cui, Y., Hu, C., Zhu, Y., Cheng, K., Li, X., Wei, Z., et al. (2018). CIK receptor kinases determine cell fate specification during early anther development in arabidopsis. Plant Cell 30 (10), 2383–2401. doi: 10.1105/tpc.17.00586
Czyzewicz, N., Nikonorova, N., Meyer, M. R., Sandal, P., Shah, S., Vu, L. D., et al. (2016). The growing story of (ARABIDOPSIS) CRINKLY 4. J. Exp. Bot. 67 (16), 4835–4847. doi: 10.1093/jxb/erw192
Deb, S., Sankaranarayanan, S., Wewala, G., Widdup, E., Samuel, M. A. (2014). The S-domain receptor kinase arabidopsis receptor kinase2 and the U box/armadillo repeat-containing E3 ubiquitin ligase9 module mediates lateral root development under phosphate starvation in arabidopsis. Plant Physiol. 165 (4), 1647–1656. doi: 10.1104/pp.114.244376
Delteil, A., Gobbato, E., Cayrol, B., Estevan, J., Michel-Romiti, C., Dievart, A., et al. (2016). Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16 (1), 17. doi: 10.1186/s12870-016-0711-x
De Smet, I., Vassileva, V., De Rybel, B., Levesque, M. P., Grunewald, W., Van Damme, D., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322 (5901), 594–597. doi: 10.1126/science.1160158
Diévart, A., Clark, S. E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6 (5), 507–516. doi: 10.1016/s1369-5266(03)00089-x
Dievart, A., Gottin, C., Périn, C., Ranwez, V., Chantret, N. (2020). Origin and diversity of plant receptor-like kinases. Annu. Rev. Plant Biol. 71 (1), 131–156. doi: 10.1146/annurev-arplant-073019-025927
Du, L., Chen, Z. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 24 (6), 837–847. doi: 10.1046/j.1365-313x.2000.00923.x
Du, S., Qu, L.-J., Xiao, J. (2018). Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR2. Protein Sci. 27 (4), 886–892. doi: 10.1002/pro.3381
Duan, Q., Kita, D., Li, C., Cheung, A. Y., Wu, H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107 (41), 17821–17826. doi: 10.1073/pnas.1005366107
Dufayard, J.-F., Bettembourg, M., Fischer, I., Droc, G., Guiderdoni, E., Périn, C., et al. (2017). New insights on leucine-rich repeats receptor-like kinase orthologous relationships in angiosperms. Front. Plant Sci. 8 (381). doi: 10.3389/fpls.2017.00381
Escobar-Restrepo, J. M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W. C., et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317 (5838), 656–660. doi: 10.1126/science.1143562
Fan, J., Bai, P., Ning, Y., Wang, J., Shi, X., Xiong, Y., et al. (2018a). The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 23 (4), 498–510.e495. doi: 10.1016/j.chom.2018.03.003
Fan, M., Ma, W., Liu, C., Zhang, C., Wu, S., Chen, M., et al. (2018b). Evolution and expression characteristics of receptor-like cytoplasmic protein kinases in maize, rice and arabidopsis. Int. J. Mol. Sci. 19 (11), 3680. doi: 10.3390/ijms19113680
Feng, X., Dickinson, H. G. (2010). Tapetal cell fate, lineage and proliferation in the Arabidopsis anther. Development 137 (14), 2409–2416. doi: 10.1242/dev.049320
Feuillet, C., Reuzeau, C., Kjellbom, P., Keller, B. (1998). Molecular characterization of a new type of receptor-like kinase (wlrk) gene family in wheat. Plant Mol. Biol. 37 (6), 943–953. doi: 10.1023/a:1006062016593
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U.S.A. 100 (25), 15253–15258. doi: 10.1073/pnas.2435133100
Fisher, K., Turner, S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17 (12), 1061–1066. doi: 10.1016/j.cub.2007.05.049
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 (5409), 1911–1914. doi: 10.1126/science.283.5409.1911
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 (4), 1531–1545. doi: 10.1093/genetics/151.4.1531
Friedrichsen, D. M., Joazeiro, C. A., Li, J., Hunter, T., Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123 (4), 1247–1256. doi: 10.1104/pp.123.4.1247
Fritz-Laylin, L. K., Krishnamurthy, N., Tor, M., Sjolander, K. V., Jones, J. D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138 (2), 611–623. doi: 10.1104/pp.104.054452
Fujihara, R., Uchida, N., Tameshige, T., Kawamoto, N., Hotokezaka, Y., Higaki, T., et al. (2021). The boundary-expressed EPIDERMAL PATTERNING FACTOR-LIKE2 gene encoding a signaling peptide promotes cotyledon growth during Arabidopsis thaliana embryogenesis. Plant Biotechnol. 38 (3), 317–322. doi: 10.5511/plantbiotechnology.21.0508a
Fujita, H., Takemura, M., Tani, E., Nemoto, K., Yokota, A., Kohchi, T. (2003). An arabidopsis MADS-box protein, AGL24, is specifically bound to and phosphorylated by meristematic receptor-like kinase (MRLK). Plant Cell Physiol. 44 (7), 735–742. doi: 10.1093/pcp/pcg092
Gadaleta, A., Colasuonno, P., Giove, S. L., Blanco, A., Giancaspro, A. (2019). Map-based cloning of QFhb.mgb-2A identifies a WAK2 gene responsible for Fusarium Head Blight resistance in wheat. Sci. Rep. 9 (1), 6929. doi: 10.1038/s41598-019-43334-z
Gao, M., Wang, X., Wang, D., Xu, F., Ding, X., Zhang, Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in arabidopsis. Cell Host Microbe 6 (1), 34–44. doi: 10.1016/j.chom.2009.05.019
Gawande, N. D., Sankaranarayanan, S. (2023). Genome wide characterization and expression analysis of CrRLK1L gene family in wheat unravels their roles in development and stress-specific responses. bioRxiv. doi: 10.1101/2023.05.24.541849
Ge, Z., Bergonci, T., Zhao, Y., Zou, Y., Du, S., Liu, M.-C., et al. (2017). Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358 (6370), 1596–1600. doi: 10.1126/science.aao3642
Ge, Z., Dresselhaus, T., Qu, L. J. (2019). How crRLK1L receptor complexes perceive RALF signals. Trends Plant Sci. 24 (11), 978–981. doi: 10.1016/j.tplants.2019.09.002
Geng, B., Wang, Q., Huang, R., Liu, Y., Guo, Z., Lu, S. (2021). A novel LRR-RLK (CTLK) confers cold tolerance through regulation on the C-repeat-binding factor pathway, antioxidants, and proline accumulation. Plant J. 108 (6), 1679–1689. doi: 10.1111/tpj.15535
Gifford, M. L., Dean, S., Ingram, G. C. (2003). The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130 (18), 4249–4258. doi: 10.1242/dev.00634
Gish, L. A., Clark, S. E. (2011). The RLK/Pelle family of kinases. Plant J. 66 (1), 117–127. doi: 10.1111/j.1365-313X.2011.04518.x
Gogl, G., Kornev, A. P., Remenyi, A., Taylor, S. S. (2019). Disordered protein kinase regions in regulation of kinase domain cores. Trends Biochem. Sci. 44 (4), 300–311. doi: 10.1016/j.tibs.2018.12.002
Gómez-Gómez, L., Bauer, Z., Boller, T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13 (5), 1155–1163.
Gómez-Gómez, L., Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 5 (6), 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gómez-Gómez, L., Felix, G., Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18 (3), 277–284. doi: 10.1046/j.1365-313x.1999.00451.x
Gong, Z., Han, G. Z. (2021). Flourishing in water: the early evolution and diversification of plant receptor-like kinases. Plant J. 106 (1), 174–184. doi: 10.1111/tpj.15157
Gou, X., He, K., Yang, H., Yuan, T., Lin, H., Clouse, S. D., et al. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11 (1), 19. doi: 10.1186/1471-2164-11-19
Gu, J., Sun, J., Liu, N., Sun, X., Liu, C., Wu, L., et al. (2020). A novel cysteine-rich receptor-like kinase gene, TaCRK2, contributes to leaf rust resistance in wheat. Mol. Plant Pathol. 21 (5), 732–746. doi: 10.1111/mpp.12929
Guo, F., Wu, T., Shen, F., Xu, G., Qi, H., Zhang, Z. (2021). The cysteine-rich receptor-like kinase TaCRK3 contributes to defense against Rhizoctonia cerealis in wheat. J. Exp. Bot. 72 (20), 6904–6919. doi: 10.1093/jxb/erab328
Halter, T., Imkampe, J., Blaum, B. S., Stehle, T., Kemmerling, B. (2014). BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal Behav. 9, e28944. doi: 10.4161/psb.28944
Han, H., Liu, X., Zhou, Y. (2020). Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 53, 50–56. doi: 10.1016/j.pbi.2019.10.004
Hanks, S. K., Hunter, T. (1995). The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification1. FASEB J. 9 (8), 576–596. doi: 10.1096/fasebj.9.8.7768349
Hanks, S. K., Quinn, A. M., Hunter, T. (1988). The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241 (4861), 42–52. doi: 10.1126/science.3291115
He, Y., Zhou, J., Shan, L., Meng, X. (2018). Plant cell surface receptor-mediated signaling - a common theme amid diversity. J. Cell Sci. 131 (2), jcs209353. doi: 10.1242/jcs.209353
Herrmann, A., Torii, K. U. (2021). Shouting out loud: signaling modules in the regulation of stomatal development. Plant Physiol. 185 (3), 765–780. doi: 10.1093/plphys/kiaa061
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105 (39), 15208–15213. doi: 10.1073/pnas.0808444105
Hohmann, U., Lau, K., Hothorn, M. (2017). The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137. doi: 10.1146/annurev-arplant-042916-040957
Hong, Z., Jin, H., Tzfira, T., Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 (12), 3418–3429. doi: 10.1105/tpc.108.061879
Hord, C. L., Chen, C., Deyoung, B. J., Clark, S. E., Ma, H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18 (7), 1667–1680. doi: 10.1105/tpc.105.036871
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978–3978. doi: 10.1038/ncomms4978
Hothorn, M., Belkhadir, Y., Dreux, M., Dabi, T., Noel, J. P., Wilson, I. A., et al. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474 (7352), 467–471. doi: 10.1038/nature10153
Hu, W., Lv, Y., Lei, W., Li, X., Chen, Y., Zheng, L., et al. (2014). Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil 384 (1), 335–346. doi: 10.1007/s11104-014-2204-8
Huck, N., Moore, J. M., Federer, M., Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130 (10), 2149–2159. doi: 10.1242/dev.00458
Huffaker, A., Pearce, G., Ryan, C. A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103 (26), 10098–10103. doi: 10.1073/pnas.0603727103
Hunter, K., Kimura, S., Rokka, A., Tran, H. C., Toyota, M., Kukkonen, J. P., et al. (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180 (4), 2004–2021. doi: 10.1104/pp.19.00560
nnan, H., Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11 (2), 97–108. doi: 10.1038/nrg2689
Jaillais, Y., Belkhadir, Y., Balsemão-Pires, E., Dangl, J. L., Chory, J. (2011). Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. U.S.A. 108 (20), 8503–8507. doi: 10.1073/pnas.1103556108
Jamieson, P. A., Shan, L., He, P. (2018). Plant cell surface molecular cypher: Receptor-like proteins and their roles in immunity and development. Plant Sci. 274, 242–251. doi: 10.1016/j.plantsci.2018.05.030
Janssens, S., Beyaert, R. (2003). Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11 (2), 293–302. doi: 10.1016/s1097-2765(03)00053-4
Jeong, S., Trotochaud, A. E., Clark, S. E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 (10), 1925–1934. doi: 10.1105/tpc.11.10.1925
Ji, D., Cui, X., Qin, G., Chen, T., Tian, S. (2020). SlFERL interacts with S-adenosylmethionine synthetase to regulate fruit ripening. Plant Physiol. 184 (4), 2168–2181. doi: 10.1104/pp.20.01203
Jia, M., Du, P., Ding, N., Zhang, Q., Xing, S., Wei, L., et al. (2017). Two FERONIA-like receptor kinases regulate apple fruit ripening by modulating ethylene production. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01406
Jia, G., Liu, X., Owen, H. A., Zhao, D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. U.S.A. 105 (6), 2220–2225. doi: 10.1073/pnas.0708795105
Jiang, J., Li, J., Xu, Y., Han, Y., Bai, Y., Zhou, G., et al. (2007). RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ. 30 (6), 690–699. doi: 10.1111/j.1365-3040.2007.01663.x
Jiang, J., Zhang, C., Wang, X. (2013). Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. J. Integr. Plant Biol. 55 (12), 1198–1211. doi: 10.1111/jipb.12081
Jinjun, Z., Peina, J., Fang, Z., Chongke, Z., Bo, B., Yaping, L., et al. (2020). OsSRK1, an atypical S-receptor-like kinase positively regulates leaf width and salt tolerance in rice. Rice Sci. 27 (2), 133–142. doi: 10.1016/j.rsci.2020.01.004
Jinn, T.-L., Stone, J. M., Walker, J. C. (2000). HAESA,an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. doi: 10.1101/gad.14.1.108
John, A., Smith, E. S., Jones, D. S., Soyars, C. L., Nimchuk, Z. L. (2023). A network of CLAVATA receptors buffers auxin-dependent meristem maintenance. Nat. Plants 9 (8), 1306–1317. doi: 10.1038/s41477-023-01485-y
Jones, D. A., Jones, J. D. G. (1997). “The role of leucine-rich repeat proteins in plant defences,” in Advances in botanical research. Eds. Andrews, J. H., Tommerup, I. C., Callow, J. A. (Cambridge, Massachusetts: Academic Press), 89–167. doi: 10.1016/S0065-2296(08)60072-5
Kaessmann, H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20 (10), 1313–1326. doi: 10.1101/gr.101386.109
Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., et al. (2007). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19 (12), 3961–3973. doi: 10.1105/tpc.106.049999
Kawamoto, N., Del Carpio, D. P., Hofmann, A., Mizuta, Y., Kurihara, D., Higashiyama, T., et al. (2020). A peptide pair coordinates regular ovule initiation patterns with seed number and fruit size. Curr. Biol. 30 (22), 4352–4361.e4354. doi: 10.1016/j.cub.2020.08.050
Keates, R. (1973). Cyclic nucleotide-independent protein kinase from pea shoots. Biochem. Biophys. Res. Commun. 54 2, 655–661. doi: 10.1016/0006-291x(73)91473-3
Kim, J., Ahuja, L. G., Chao, F.-A., Xia, Y., McClendon, C. L., Kornev, A. P., et al. (2017). A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv. 3 (4), e1600663. doi: 10.1126/sciadv.1600663
Kimura, S., Hunter, K., Vaahtera, L., Tran, H. C., Citterico, M., Vaattovaara, A., et al. (2020). CRK2 and C-terminal phosphorylation of NADPH oxidase RBOHD regulate reactive oxygen species production in arabidopsis. Plant Cell 32 (4), 1063–1080. doi: 10.1105/tpc.19.00525
Knighton, D. R., Zheng, J. H., Ten Eyck, L. F., Ashford, V. A., Xuong, N. H., Taylor, S. S., et al. (1991a). Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253 (5018), 407–414. doi: 10.1126/science.1862342
Knighton, D. R., Zheng, J. H., Ten Eyck, L. F., Xuong, N. H., Taylor, S. S., Sowadski, J. M. (1991b). Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253 (5018), 414–420. doi: 10.1126/science.1862343
Kobe, B., Deisenhofer, J. (1993). Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 366 (6457), 751–756. doi: 10.1038/366751a0
Kohorn, B. D. (2015). Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67 (2), 489–494. doi: 10.1093/jxb/erv467
Kohorn, B. D., Hoon, D., Minkoff, B. B., Sussman, M. R., Kohorn, S. L. (2016). Rapid oligo-galacturonide induced changes in protein phosphorylation in arabidopsis. Mol. Cell Proteomics 15 (4), 1351–1359. doi: 10.1074/mcp.M115.055368
Kohorn, B. D., Lane, S., Smith, T. A. (1992). An Arabidopsis serine/threonine kinase homologue with an epidermal growth factor repeat selected in yeast for its specificity for a thylakoid membrane protein. Proc. Natl. Acad. Sci. U.S.A. 89 (22), 10989–10992. doi: 10.1073/pnas.89.22.10989
Kong, Q., Sun, T., Qu, N., Ma, J., Li, M., Cheng, Y.-t., et al. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171 (2), 1344–1354. doi: 10.1104/pp.15.01954
Kornev, A. P., Taylor, S. S., Ten Eyck, L. F. (2008). A helix scaffold for the assembly of active protein kinases. Proc. Natl. Acad. Sci. U.S.A. 105 (38), 14377–14382. doi: 10.1073/pnas.0807988105
Kou, X., Qi, K., Qiao, X., Yin, H., Liu, X., Zhang, S., et al. (2017). Evolution, expression analysis, and functional verification of Catharanthus roseus RLK1-like kinase (CrRLK1L) family proteins in pear (Pyrus bretchneideri). Genomics 109 (3), 290–301. doi: 10.1016/j.ygeno.2017.05.003
Kumar, D., Kumar, R., Baek, D., Hyun, T. K., Chung, W. S., Yun, D. J., et al. (2017). Arabidopsis thaliana RECEPTOR DEAD KINASE1 functions as a positive regulator in plant responses to ABA. Mol. Plant 10 (2), 223–243. doi: 10.1016/j.molp.2016.11.011
Lawton, M. A., Yamamoto, R. T., Hanks, S. K., Lamb, C. J. (1989). Molecular cloning of plant transcripts encoding protein kinase homologs. Proc. Natl. Acad. Sci. U.S.A. 86 (9), 3140–3144. doi: 10.1073/pnas.86.9.3140
Lee, H.-S., Karunanandaa, B., McCubbin, A., Gilroy, S., Kao, T.-h. (1996). PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 9 (5), 613–624. doi: 10.1046/j.1365-313X.1996.9050613.x
Lee, J. S., Kuroha, T., Hnilova, M., Khatayevich, D., Kanaoka, M. M., McAbee, J. M., et al. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26 (2), 126–136. doi: 10.1101/gad.179895.111
Lehti-Shiu, M. D., Shiu, S. H. (2012). Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R Soc. Lond B Biol. Sci. 367 (1602), 2619–2639. doi: 10.1098/rstb.2012.0003
Lehti-Shiu, M. D., Zou, C., Hanada, K., Shiu, S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150 (1), 12–26. doi: 10.1104/pp.108.134353
Lehti-Shiu, D. M., Zou, C., Shiu, S.-H. (2012). Origin, diversity, expansion history and functional evolution of the plant receptor-like kinase/pelle family. Receptor-like Kinases Plants 13, 1–22. doi: 10.1007/978-3-642-23044-8_1
Lemieux, C., Otis, C., Turmel, M. (2007). A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol. 5 (1), 2. doi: 10.1186/1741-7007-5-2
Li, H., Cai, Z., Wang, X., Li, M., Cui, Y., Cui, N., et al. (2019b). SERK receptor-like kinases control division patterns of vascular precursors and ground tissue stem cells during embryo development in arabidopsis. Mol. Plant 12 (7), 984–1002. doi: 10.1016/j.molp.2019.04.011
Li, J., Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/s0092-8674(00)80357-8
Li, B., Ferreira, M. A., Huang, M., Camargos, L. F., Yu, X., Teixeira, R. M., et al. (2019a). The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 10 (1), 4996. doi: 10.1038/s41467-019-12847-6
Li, H., Han, X., Liu, X., Zhou, M., Ren, W., Zhao, B., et al. (2019c). A leucine-rich repeat-receptor-like kinase gene SbER2–1 from sorghum (Sorghum bicolor L.) confers drought tolerance in maize. BMC Genomics 20 (1), 737. doi: 10.1186/s12864-019-6143-x
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014b). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15 (3), 329–338. doi: 10.1016/j.chom.2014.02.009
Li, L., Verstraeten, I., Roosjen, M., Takahashi, K., Rodriguez, L., Merrin, J., et al. (2021b). Cell surface and intracellular auxin signalling for H+ fluxes in root growth. Nature 599 (7884), 273–277. doi: 10.1038/s41586-021-04037-6
Li, C., Wang, L., Cui, Y., He, L., Qi, Y., Zhang, J., et al. (2016a). Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Mol. Breed. 36 (11), 151. doi: 10.1007/s11032-016-0580-x
Li, D., Wang, L., Wang, M., Xu, Y. Y., Luo, W., Liu, Y. J., et al. (2009a). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7 (8), 791–806. doi: 10.1111/j.1467-7652.2009.00444.x
Li, C. H., Wang, G., Zhao, J. L., Zhang, L. Q., Ai, L. F., Han, Y. F., et al. (2014a). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26 (6), 2538–2553. doi: 10.1105/tpc.114.125187
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 (2), 213–222. doi: 10.1016/s0092-8674(02)00812-7
Li, H., Yang, Y., Wang, H., Liu, S., Jia, F., Su, Y., et al. (2021a). The receptor-like kinase ERECTA confers improved water use efficiency and drought tolerance to poplar via modulating stomatal density. Int. J. Mol. Sci. 22 (14), 7245. doi: 10.3390/ijms22147245
Li, L., Yu, Y., Zhou, Z., Zhou, J.-M. (2016b). Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 59 (9), 878–888. doi: 10.1007/s11427-016-0115-2
Li, W., Zhang, J., Tian, X., Liu, H., Ali, K. (2022). Two conserved amino acids characterized in the island domain are essential for the biological functions of brassinolide receptors. Int. J. Mol. Sci. 23 (19), 11454. doi: 10.3390/ijms231911454
Li, J., Zhao-Hui, C., Batoux, M., Nekrasov, V., Roux, M., Chinchilla, D., et al. (2009b). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106 (37), 15973–15978. doi: 10.1073/pnas.0905532106
Liang, X., Zhou, J. M. (2018). Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 69, 267–299. doi: 10.1146/annurev-arplant-042817-040540
Liao, H. Z., Zhu, M. M., Cui, H. H., Du, X. Y., Tang, Y., Chen, L. Q., et al. (2016). MARIS plays important roles in Arabidopsis pollen tube and root hair growth. J. Integr. Plant Biol. 58 (11), 927–940. doi: 10.1111/jipb.12484
Lim, C. W., Yang, S. H., Shin, K. H., Lee, S. C., Kim, S. H. (2015). The AtLRK10L1.2, Arabidopsis ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 34 (3), 447–455. doi: 10.1007/s00299-014-1724-2
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 (5645), 630–633. doi: 10.1126/science.1090074
Limpens, E., van Zeijl, A., Geurts, R. (2015). Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu. Rev. Phytopathol. 53, 311–334. doi: 10.1146/annurev-phyto-080614-120149
Lin, F., Li, S., Wang, K., Tian, H., Gao, J., Zhao, Q., et al. (2020). A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 296, 110465. doi: 10.1016/j.plantsci.2020.110465
Lin, W., Ma, X., Shan, L., He, P. (2013). Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 55 (12), 1188–1197. doi: 10.1111/jipb.12071
Lin, Y., Wang, K., Li, X., Sun, C., Yin, R., Wang, Y., et al. (2018). Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng C.A. Meyer. Mol. Genet. Genomics 293 (4), 845–859. doi: 10.1007/s00438-018-1425-6
Lin, W., Zhou, X., Tang, W., Takahashi, K., Pan, X., Dai, J., et al. (2021). TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 599 (7884), 278–282. doi: 10.1038/s41586-021-03976-4
Liu, P.-L., Du, L., Huang, Y., Gao, S.-M., Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17 (1), 47. doi: 10.1186/s12862-017-0891-5
Liu, T., Jiang, G. Q., Yao, X. F., Liu, C. M. (2021). The leucine-rich repeat receptor-like kinase OsERL plays a critical role in anther lobe formation in rice. Biochem. Biophys. Res. Commun. 563, 85–91. doi: 10.1016/j.bbrc.2021.05.059
Liu, J., Li, J., Shan, L. (2020a). SERKs. Curr. Biol. 30 (7), R293–R294. doi: 10.1016/j.cub.2020.01.043
Liu, X. S., Liang, C. C., Hou, S. G., Wang, X., Chen, D. H., Shen, J. L., et al. (2020b). The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.548034
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336 (6085), 1160–1164. doi: 10.1126/science.1218867
Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q., et al. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. U.S.A. 110 (15), 6205–6210. doi: 10.1073/pnas.1215543110
Liu, L., Zheng, C., Kuang, B., Wei, L., Yan, L., Wang, T. (2016). Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PloS Genet. 12 (7), e1006085. doi: 10.1371/journal.pgen.1006085
Lou, H. Q., Fan, W., Jin, J. F., Xu, J. M., Chen, W. W., Yang, J. L., et al. (2020). A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 43 (2), 463–478. doi: 10.1111/pce.13676
Lozano-Elena, F., Caño-Delgado, A. I. (2019). Emerging roles of vascular brassinosteroid receptors of the BRI1-like family. Curr. Opin. Plant Biol. 51, 105–113. doi: 10.1016/j.pbi.2019.06.006
Lu, K., Liang, S., Wu, Z., Bi, C., Yu, Y. T., Wang, X. F., et al. (2016). Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 67 (17), 5009–5027. doi: 10.1093/jxb/erw266
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107 (1), 496–501. doi: 10.1073/pnas.0909705107
Lv, D., Wang, G., Xiong, L.-R., Sun, J.-X., Chen, Y., Guo, C.-L., et al. (2020). Genome-wide identification and characterization of lectin receptor-like kinase gene family in cucumber and expression profiling analysis under different treatments. Genes 11 (9), 1032. doi: 10.3390/genes11091032
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 (5494), 1151–1155. doi: 10.1126/science.290.5494.1151
Ma, W., Liu, X., Chen, K., Yu, X., Ji, D. (2023). Genome-wide re-identification and analysis of crRLK1Ls in tomato. Int. J. Mol. Sci. 24 (4), 3142. doi: 10.3390/ijms24043142
Ma, X., Xu, G., He, P., Shan, L. (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21 (12), 1017–1033. doi: 10.1016/j.tplants.2016.08.014
Machado, J. P., Brustolini, O. J., Mendes, G. C., Santos, A. A., Fontes, E. P. (2015). NIK1, a host factor specialized in antiviral defense or a novel general regulator of plant immunity? Bioessays 37 (11), 1236–1242. doi: 10.1002/bies.201500066
Madsen, E. B., Antolín-Llovera, M., Grossmann, C., Ye, J., Vieweg, S., Broghammer, A., et al. (2011). Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 65 (3), 404–417. doi: 10.1111/j.1365-313X.2010.04431.x
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 (6958), 637–640. doi: 10.1038/nature02045
Man, J., Gallagher, J. P., Bartlett, M. (2020). Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 226 (5), 1492–1505. doi: 10.1111/nph.16455
Manning, G., Whyte, D. B., Martinez, R., Hunter, T., Sudarsanam, S. (2002). The protein kinase complement of the human genome. Science 298 (5600), 1912–1934. doi: 10.1126/science.1075762
Martin, G. B., Brommonschenkel, S. H., Chunwongse, J., Frary, A., Ganal, M. W., Spivey, R., et al. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 (5138), 1432–1436. doi: 10.1126/science.7902614
Martins, S., Dohmann, E. M. N., Cayrel, A., Johnson, A., Fischer, W., Pojer, F., et al. (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6 (1), 6151. doi: 10.1038/ncomms7151
Matsushima, N., Miyashita, H. (2012). Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules 2 (2), 288–311. doi: 10.3390/biom2020288
Meharena, H. S., Chang, P., Keshwani, M. M., Oruganty, K., Nene, A. K., Kannan, N., et al. (2013). Deciphering the structural basis of eukaryotic protein kinase regulation. PloS Biol. 11 (10), e1001680. doi: 10.1371/journal.pbio.1001680
Meng, X., Chen, X., Mang, H., Liu, C., Yu, X., Gao, X., et al. (2015). Differential function of arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25 (18), 2361–2372. doi: 10.1016/j.cub.2015.07.068
Meng, X., Zhou, J., Tang, J., Li, B., de Oliveira, M. V. V., Chai, J., et al. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in arabidopsis. Cell Rep. 14 (6), 1330–1338. doi: 10.1016/j.celrep.2016.01.023
Mizuno, S., Osakabe, Y., Maruyama, K., Ito, T., Osakabe, K., Sato, T., et al. (2007). Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50 (5), 751–766. doi: 10.1111/j.1365-313X.2007.03083.x
Moussu, S., Augustin, S., Roman, A. O., Broyart, C., Santiago, J. (2018). Crystal structures of two tandem malectin-like receptor kinases involved in plant reproduction. Acta Crystallogr. D Struct. Biol. 74 (Pt 7), 671–680. doi: 10.1107/s205979831800774x
Nakamura, A., Fujioka, S., Sunohara, H., Kamiya, N., Hong, Z., Inukai, Y., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140 (2), 580–590. doi: 10.1104/pp.105.072330
Nakayama, T., Shinohara, H., Tanaka, M., Baba, K., Ogawa-Ohnishi, M., Matsubayashi, Y. (2017). A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355 (6322), 284–286. doi: 10.1126/science.aai9057
Nasrallah, J. B., Yu, S. M., Nasrallah, M. E. (1988). Self-incompatibility genes of Brassica oleracea: Expression, isolation, and structure. Proc. Natl. Acad. Sci. U.S.A. 85 (15), 5551–5555. doi: 10.1073/pnas.85.15.5551
Nguyen, Q. N., Lee, Y. S., Cho, L. H., Jeong, H. J., An, G., Jung, K. H. (2015). Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 241 (3), 603–613. doi: 10.1007/s00425-014-2203-2
Nishiyama, T., Sakayama, H., de Vries, J., Buschmann, H., Saint-Marcoux, D., Ullrich, K. K., et al. (2018). The chara genome: secondary complexity and implications for plant terrestrialization. Cell 174 (2), 448–464.e424. doi: 10.1016/j.cell.2018.06.033
Nissen, K. S., Willats, W. G. T., Malinovsky, F. G. (2016). Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci. 21 (6), 516–527. doi: 10.1016/j.tplants.2015.12.004
Nolan, T., Chen, J., Yin, Y. (2017). Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474 (16), 2641–2661. doi: 10.1042/bcj20160633
Nolen, B., Taylor, S., Ghosh, G. (2004). Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15 (5), 661–675. doi: 10.1016/j.molcel.2004.08.024
Oh, M. H., Wang, X., Kota, U., Goshe, M. B., Clouse, S. D., Huber, S. C. (2009). Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 (2), 658–663. doi: 10.1073/pnas.0810249106
Okuda, S., Fujita, S., Moretti, A., Hohmann, U., Doblas, V. G., Ma, Y., et al. (2020). Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2. Proc. Natl. Acad. Sci. U.S.A. 117 (5), 2693–2703. doi: 10.1073/pnas.1911553117
Olsen, J. V., Mann, M. (2013). Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteomics 12 (12), 3444–3452. doi: 10.1074/mcp.O113.034181
Pan, J., Li, Z., Wang, Q., Yang, L., Yao, F., Liu, W. (2020). An S-domain receptor-like kinase, OsESG1, regulates early crown root development and drought resistance in rice. Plant Sci. 290, 110318. doi: 10.1016/j.plantsci.2019.110318
Park, H. S., Ryu, H. Y., Kim, B. H., Kim, S. Y., Yoon, I. S., Nam, K. H. (2011). A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Molecules Cells 32 (6), 561–569. doi: 10.1007/s10059-011-0178-4
Pei, D., Hua, D., Deng, J., Wang, Z., Song, C., Wang, Y., et al. (2022). Phosphorylation of the plasma membrane H+-ATPase AHA2 by BAK1 is required for ABA-induced stomatal closure in Arabidopsis. Plant Cell 34 (7), 2708–2729. doi: 10.1093/plcell/koac106
Pelagio-Flores, R., Muñoz-Parra, E., Barrera-Ortiz, S., Ortiz-Castro, R., Saenz-Mata, J., Ortega-Amaro, M. A., et al. (2019). The cysteine-rich receptor-like protein kinase CRK28 modulates Arabidopsis growth and development and influences abscisic acid responses. Planta 251 (1), 2. doi: 10.1007/s00425-019-03296-y
Peng, X., Wang, M., Li, Y., Yan, W., Chang, Z., Chen, Z., et al. (2020). Lectin receptor kinase OsLecRK-S.7 is required for pollen development and male fertility. J. Integr. Plant Biol. 62 (8), 1227–1245. doi: 10.1111/jipb.12897
Pfister, A., Barberon, M., Alassimone, J., Kalmbach, L., Lee, Y., Vermeer, J. E. M., et al. (2014). A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3, e03115. doi: 10.7554/eLife.03115
Pitorre, D., Llauro, C., Jobet, E., Guilleminot, J., Brizard, J. P., Delseny, M., et al. (2010). RLK7, a leucine-rich repeat receptor-like kinase, is required for proper germination speed and tolerance to oxidative stress in Arabidopsis thaliana. Planta 232 (6), 1339–1353. doi: 10.1007/s00425-010-1260-4
Pu, C.-X., Han, Y.-F., Zhu, S., Song, F.-Y., Zhao, Y., Wang, C.-Y., et al. (2017). The rice receptor-like kinases DWARF AND RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell 29 (1), 70–89. doi: 10.1105/tpc.16.00218
Qi, H., Guo, F., Lv, L., Zhu, X., Zhang, L., Yu, J., et al. (2021a). The Wheat Wall-Associated Receptor-Like Kinase TaWAK-6D Mediates Broad Resistance to Two Fungal Pathogens Fusarium pseudograminearum and Rhizoctonia cerealis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.758196
Qi, H., Zhu, X., Guo, F., Lv, L., Zhang, Z. (2021b). The wall-associated receptor-like kinase TaWAK7D is required for defense responses to rhizoctonia cerealis in wheat. Int. J. Mol. Sci. 22 (11), 5629. doi: 10.3390/ijms22115629
Racolta, A., Bryan, A. C., Tax, F. E. (2014). The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev. Dyn 243 (2), 257–278. doi: 10.1002/dvdy.24066
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 (6958), 585–592. doi: 10.1038/nature02039
Rafeie, M., Amerian, M. R., Sorkhi, B., Heidari, P., Asghari, H. R. (2020). Effect of Exogenous Brassinosteroid Application on Grain Yield, some Physiological Traits and Expression of Genes Related to This Hormone Signaling Pathway in Wheat under Drought Stress. Plant Genet. Res. 6 (2), 157–172. doi: 10.29252/pgr.6.2.157
Ralph, R. K., McCombs, P. J., Tener, G., Wojcik, S. J. (1972). Evidence for modification of protein phosphorylation by cytokinins. Biochem. J. 130 (4), 901–911. doi: 10.1042/bj1300901a
Rayapuram, C., Jensen, M. K., Maiser, F., Shanir, J. V., Hornshøj, H., Rung, J. H., et al. (2012). Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol. Plant Pathol. 13 (2), 135–147. doi: 10.1111/j.1364-3703.2011.00736.x
Reznick, D. N., Ricklefs, R. E. (2009). Darwin's bridge between microevolution and macroevolution. Nature 457 (7231), 837–842. doi: 10.1038/nature07894
Saintenac, C., Cambon, F., Aouini, L., Verstappen, E., Ghaffary, S. M. T., Poucet, T., et al. (2021). A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against Septoria tritici blotch. Nat. Commun. 12 (1), 433. doi: 10.1038/s41467-020-20685-0
Saintenac, C., Lee, W.-S., Cambon, F., Rudd, J. J., King, R. C., Marande, W., et al. (2018). Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50 (3), 368–374. doi: 10.1038/s41588-018-0051-x
Santiago, J., Brandt, B., Wildhagen, M., Hohmann, U., Hothorn, L. A., Butenko, M. A., et al. (2016). Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5, e15075. doi: 10.7554/eLife.15075
Sasaki, G., Katoh, K., Hirose, N., Suga, H., Kuma, K., Miyata, T., et al. (2007). Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: Extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene 401 (1-2), 135–144. doi: 10.1016/j.gene.2007.07.009
Schulze-Muth, P., Irmler, S., Schröder, G., Schröder, J. (1996). Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus). cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. J. Biol. Chem. 271 (43), 26684–26689. doi: 10.1074/jbc.271.43.26684
Schwessinger, B., Roux, M., Kadota, Y., Ntoukakis, V., Sklenar, J., Jones, A., et al. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PloS Genet. 7 (4), e1002046. doi: 10.1371/journal.pgen.1002046
Segonzac, C., Zipfel, C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14 (1), 54–61. doi: 10.1016/j.mib.2010.12.005
Shang, E., Wang, X., Li, T., Guo, F., Ito, T., Sun, B. (2021). Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc. Natl. Acad. Sci. U.S.A. 118 (36), e2102826118. doi: 10.1073/pnas.2102826118
Sharma, N., Khurana, P. (2022). Genome-wide identification, characterization and expression analysis of the BRI1 gene family in Triticum aestivum L. Plant Biotechnol. Rep. 16 (6), 777–791. doi: 10.1007/s11816-022-00762-0
Sharma, A., Shumayla, Tyagi, S., Alok, A., Singh, K., Upadhyay, S. K. (2020). Thaumatin-like protein kinases: Molecular characterization and transcriptional profiling in five cereal crops. Plant Sci. 290, 110317. doi: 10.1016/j.plantsci.2019.110317
Sharon, N., Lis, H. (1990). Legume lectins–a large family of homologous proteins. FASEB J. 4 (14), 3198–3208. doi: 10.1096/fasebj.4.14.2227211
She, J., Han, Z., Kim, T.-W., Wang, J., Cheng, W., Chang, J., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474 (7352), 472–476. doi: 10.1038/nature10178
She, J., Han, Z., Zhou, B., Chai, J. (2013). Structural basis for differential recognition of brassinolide by its receptors. Protein Cell 4 (6), 475–482. doi: 10.1007/s13238-013-3027-8
Shelton, C. A., Wasserman, S. A. (1993). pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell 72 (4), 515–525. doi: 10.1016/0092-8674(93)90071-w
Shen, J., Diao, W., Zhang, L., Acharya, B. R., Wang, M., Zhao, X., et al. (2020). Secreted peptide PIP1 induces stomatal closure by activation of guard cell anion channels in arabidopsis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01029
Shi, H., Shen, Q., Qi, Y., Yan, H., Nie, H., Chen, Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in arabidopsis. Plant Cell 25 (3), 1143–1157. doi: 10.1105/tpc.112.107904
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64 (2), 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shinya, T., Yamaguchi, K., Desaki, Y., Yamada, K., Narisawa, T., Kobayashi, Y., et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79 (1), 56–66. doi: 10.1111/tpj.12535
Shiu, S.-H., Bleecker, A. B. (2001a). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE, re22. doi: 10.1126/stke.2001.113.re22
Shiu, S. H., Bleecker, A. B. (2001b). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98 (19), 10763–10768. doi: 10.1073/pnas.181141598
Shiu, S. H., Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132 (2), 530–543. doi: 10.1104/pp.103.021964
Shiu, S.-H., Karlowski, W. M., Pan, R., Tzeng, Y.-H., Mayer, K. F. X., Li, W.-H. (2004). Comparative analysis of the receptor-like kinase family in arabidopsis and rice. Plant Cell 16 (5), 1220–1234. doi: 10.1105/tpc.020834
Shpak, E. D., Lakeman, M. B., Torii, K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15 (5), 1095–1110. doi: 10.1105/tpc.010413
Shumayla, Madhu, Singh, K., Upadhyay, S. K. (2021). LysM domain-containing proteins modulate stress response and signalling in Triticum aestivum L. Environ. Exp. Bot. 189, 104558. doi: 10.1016/j.envexpbot.2021.104558
Shumayla, Mendu, V., Singh, K., Upadhyay, S. K. (2022). Insight into the Roles of Proline-Rich Extensin-like Receptor Protein Kinases of Bread Wheat (Triticum aestivum L.). Life 12 (7), 941. doi: 10.3390/life12070941
Shumayla, Sharma, S., Kumar, R., Mendu, V., Singh, K., Upadhyay, S. K. (2016a). Genomic dissection and expression profiling revealed functional divergence in triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01374
Shumayla, Sharma, S., Pandey, A. K., Singh, K., Upadhyay, S. K. (2016b). Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PloS One 11 (4), e0153925. doi: 10.1371/journal.pone.0153925
Shumayla, Tyagi, S., Sharma, A., Singh, K., Upadhyay, S. K. (2019). Genomic dissection and transcriptional profiling of Cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int. J. Biol. Macromol 134, 316–329. doi: 10.1016/j.ijbiomac.2019.05.016
Silva, N. F., Goring, D. R. (2002). The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 50 (4), 667–685. doi: 10.1023/A:1019951120788
Simon, A., Glöckner, G., Felder, M., Melkonian, M., Becker, B. (2006). EST analysis of the scaly green flagellate Mesostigma viride (Streptophyta): Implications for the evolution of green plants (Viridiplantae). BMC Plant Biol. 6 (1), 2. doi: 10.1186/1471-2229-6-2
Singh, A., Breja, P., Khurana, J. P., Khurana, P. (2016). Wheat brassinosteroid-insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in arabidopsis. PloS One 11 (6), e0153273. doi: 10.1371/journal.pone.0153273
Singh, P., Mishra, A. K., Singh, C. M. (2021). Genome-wide identification and characterization of Lectin receptor-like kinase (LecRLK) genes in mungbean (Vigna radiata L. Wilczek). J. Appl. Genet. 62 (2), 223–234. doi: 10.1007/s13353-021-00613-8
Sivaguru, M., Ezaki, B., He, Z.-H., Tong, H., Osawa, H., Baluška, F., et al. (2003). Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in arabidopsis. Plant Physiol. 132 (4), 2256–2266. doi: 10.1104/pp.103.022129
Smakowska-Luzan, E., Mott, G. A., Parys, K., Stegmann, M., Howton, T. C., Layeghifard, M., et al. (2018). An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553 (7688), 342–346. doi: 10.1038/nature25184
Song, X., Guo, P., Li, C., Liu, C.-M. (2010). The cysteine pairs in CLV2 are not necessary for sensing the CLV3 peptide in shoot and root meristems. J. Integr. Plant Biol. 52 9, 774–781. doi: 10.1111/j.1744-7909.2010.00978.x
Song, W., Han, Z., Sun, Y., Chai, J. (2014). Crystal structure of a plant leucine rich repeat protein with two island domains. Sci. China Life Sci. 57 (1), 137–144. doi: 10.1007/s11427-013-4586-x
Song, M., Linghu, B., Huang, S., Li, F., An, R., Xie, C., et al. (2022). Genome-wide survey of leucine-rich repeat receptor-like protein kinase genes and CRISPR/cas9-targeted mutagenesis BnBRI1 in brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.865132
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 (5243), 1804–1806. doi: 10.1126/science.270.5243.1804
Sreeramulu, S., Mostizky, Y., Sunitha, S., Shani, E., Nahum, H., Salomon, D., et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74 (6), 905–919. doi: 10.1111/tpj.12175
Stahl, Y., Grabowski, S., Bleckmann, A., Kühnemuth, R., Weidtkamp-Peters, S., Pinto, K. G., et al. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23 (5), 362–371. doi: 10.1016/j.cub.2013.01.045
Stahl, Y., Wink, R. H., Ingram, G. C., Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19 (11), 909–914. doi: 10.1016/j.cub.2009.03.060
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355 (6322), 287–289. doi: 10.1126/science.aal2541
Stenvik, G. E., Tandstad, N. M., Guo, Y., Shi, C. L., Kristiansen, W., Holmgren, A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20 (7), 1805–1817. doi: 10.1105/tpc.108.059139
Su, Y. H., Zhou, C., Li, Y. J., Yu, Y., Tang, L. P., Zhang, W. J., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 117 (36), 22561–22571. doi: 10.1073/pnas.2015248117
Sun, W., Cao, Y., Jansen Labby, K., Bittel, P., Boller, T., Bent, A. F. (2012). Probing the arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24 (3), 1096–1113. doi: 10.1105/tpc.112.095919
Sun, Y., Han, Z., Tang, J., Hu, Z., Chai, C., Zhou, B., et al. (2013a). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23 (11), 1326–1329. doi: 10.1038/cr.2013.131
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013b). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342 (6158), 624–628. doi: 10.1126/science.1243825
Sun, R., Wang, S., Ma, D., Liu, C. (2018). Genome-wide analysis of LRR-RLK gene family in four gossypium species and expression analysis during cotton development and stress responses. Genes 9 (12), 592. doi: 10.3390/genes9120592
Swiderski, M. R., Innes, R. W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26 (1), 101–112. doi: 10.1046/j.1365-313x.2001.01014.x
Takahashi, T., Mu, J. H., Gasch, A., Chua, N. H. (1998). Identification by PCR of receptor-like protein kinases from Arabidopsis flowers. Plant Mol. Biol. 37 (4), 587–596. doi: 10.1023/a:1005924817190
Takeuchi, H., Higashiyama, T. (2012). A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PloS Biol. 10 (12), e1001449. doi: 10.1371/journal.pbio.1001449
Takeuchi, H., Higashiyama, T. (2016). Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531 (7593), 245–248. doi: 10.1038/nature17413
Tanaka, H., Watanabe, M., Sasabe, M., Hiroe, T., Tanaka, T., Tsukaya, H., et al. (2007). Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134 (9), 1643–1652. doi: 10.1242/dev.003533
Tang, W., Kim, T. W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321 (5888), 557–560. doi: 10.1126/science.1156973
Tang, D., Wang, G., Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29 (4), 618–637. doi: 10.1105/tpc.16.00891
Taylor, S. S., Keshwani, M. M., Steichen, J. M., Kornev, A. P. (2012). Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos. Trans. R Soc. Lond B Biol. Sci. 367 (1602), 2517–2528. doi: 10.1098/rstb.2012.0054
Taylor, S. S., Shaw, A. S., Kannan, N., Kornev, A. P. (2015). Integration of signaling in the kinome: Architecture and regulation of the alphaC Helix. Biochim. Biophys. Acta 1854 (10 Pt B), 1567–1574. doi: 10.1016/j.bbapap.2015.04.007
Taylor, I., Wang, Y., Seitz, K., Baer, J., Bennewitz, S., Mooney, B. P., et al. (2016). Analysis of phosphorylation of the receptor-like protein kinase HAESA during arabidopsis floral abscission. PloS One 11 (1), e0147203. doi: 10.1371/journal.pone.0147203
The Arabidopsis Genome, I. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 (6814), 796–815. doi: 10.1038/35048692
Torii, K. U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol 234, 1–46. doi: 10.1016/s0074-7696(04)34001-5
Trewavas, A. (1973). The phosphorylation of ribosomal protein in lemna minor. Plant Physiol. 51 (4), 760–767. doi: 10.1104/pp.51.4.760
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44 (W1), W232–W235. doi: 10.1093/nar/gkw256
Tsuwamoto, R., Fukuoka, H., Takahata, Y. (2008). GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 54 (1), 30–42. doi: 10.1111/j.1365-313X.2007.03395.x
Vaattovaara, A., Brandt, B., Rajaraman, S., Safronov, O., Veidenberg, A., Luklová, M., et al. (2019). Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2 (1), 56. doi: 10.1038/s42003-019-0306-9
Van der Burgh, A. M., Postma, J., Robatzek, S., Joosten, M. (2019). Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20 (3), 410–422. doi: 10.1111/mpp.12767
Verica, J. A., He, Z.-H. (2002). The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129 (2), 455–459. doi: 10.1104/pp.011028
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 (1), 257–273. doi: 10.1105/tpc.105.035576
Vij, S., Giri, J., Dansana, P. K., Kapoor, S., Tyagi, A. K. (2008). The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 1 (5), 732–750. doi: 10.1093/mp/ssn047
Walker, J. C., Zhang, R. (1990). Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345 (6277), 743–746. doi: 10.1038/345743a0
Walsh, D. A., Perkins, J. P., Krebs, E. G. (1968). An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243 (13), 3763–3765. doi: 10.1016/S0021-9258(19)34204-8
Wang, B., Fang, R., Zhang, J., Han, J., Chen, F., He, F., et al. (2020a). Rice LecRK5 phosphorylates a UGPase to regulate callose biosynthesis during pollen development. J. Exp. Bot. 71 (14), 4033–4041. doi: 10.1093/jxb/eraa180
Wang, J., Grubb, L. E., Wang, J., Liang, X., Li, L., Gao, C., et al. (2018). A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69 (3), 493–504.e496. doi: 10.1016/j.molcel.2017.12.026
Wang, J., Jiang, J., Wang, J., Chen, L., Fan, S. L., Wu, J. W., et al. (2014). Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 24 (11), 1328–1341. doi: 10.1038/cr.2014.132
Wang, T., Liang, L., Xue, Y., Jia, P. F., Chen, W., Zhang, M. X., et al. (2016). A receptor heteromer mediates the male perception of female attractants in plants. Nature 531 (7593), 241–244. doi: 10.1038/nature16975
Wang, L., Ma, Z., Kang, H., Gu, S., Mukhina, Z., Wang, C., et al. (2022). Cloning and functional analysis of the novel rice blast resistance gene Pi65 in japonica rice. Theor. Appl. Genet. 135 (1), 173–183. doi: 10.1007/s00122-021-03957-1
Wang, J., Wang, J., Li, J., Shang, H., Chen, X., Hu, X. (2021a). The RLK protein TaCRK10 activates wheat high-temperature seedling-plant resistance to stripe rust through interacting with TaH2A.1. Plant J. 108 (5), 1241–1255. doi: 10.1111/tpj.15513
Wang, J., Wang, J., Shang, H., Chen, X., Xu, X., Hu, X. (2019). TaXa21, a Leucine-Rich Repeat Receptor–Like Kinase Gene Associated with TaWRKY76 and TaWRKY62, Plays Positive Roles in Wheat High-Temperature Seedling Plant Resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 32 (11), 1526–1535. doi: 10.1094/mpmi-05-19-0137-r
Wang, X., Wu, M.-H., Xiao, D., Huang, R.-L., Zhan, J., Wang, A.-Q., et al. (2021b). Genome-wide identification and evolutionary analysis of RLKs involved in the response to aluminium stress in peanut. BMC Plant Biol. 21 (1), 281. doi: 10.1186/s12870-021-03031-4
Wang, J., Yan, L.-L., Yue, Z.-L., Li, H.-Y., Ji, X.-J., Pu, C.-X., et al. (2020b). Receptor-like kinase OsCR4 controls leaf morphogenesis and embryogenesis by fixing the distribution of auxin in rice. J. Genet. Genomics 47 (9), 577–589. doi: 10.1016/j.jgg.2020.08.002
Wang, X., Zafian, P., Choudhary, M., Lawton, M. (1996). The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc. Natl. Acad. Sci. U.S.A. 93 (6), 2598–2602. doi: 10.1073/pnas.93.6.2598
Wei, X., Wang, Y., Zhang, S., Gu, T., Steinmetz, G., Yu, H., et al. (2022). Structural analysis of receptor-like kinase SOBIR1 reveals mechanisms that regulate its phosphorylation-dependent activation. Plant Commun. 3 (2), 100301. doi: 10.1016/j.xplc.2022.100301
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M. A., Kolb, D., Tsuda, K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108 (49), 19824–19829. doi: 10.1073/pnas.1112862108
Wu, Y., Xun, Q., Guo, Y., Zhang, J., Cheng, K., Shi, T., et al. (2016). Genome-wide expression pattern analyses of the arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 9 (2), 289–300. doi: 10.1016/j.molp.2015.12.011
Xia, Y., Yin, S., Zhang, K., Shi, X., Lian, C., Zhang, H., et al. (2018). OsWAK11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of Cu in cell walls. Environ. Exp. Bot. 150, 99–105. doi: 10.1016/j.envexpbot.2018.03.005
Yadeta, K. A., Elmore, J. M., Creer, A. Y., Feng, B., Franco, J. Y., Rufian, J. S., et al. (2016). A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 173 (1), 771–787. doi: 10.1104/pp.16.01404
Yamada, K., Yamaguchi, K., Shirakawa, T., Nakagami, H., Mine, A., Ishikawa, K., et al. (2016). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35 (22), 2468–2483. doi: 10.15252/embj.201694248
Yamaguchi, Y., Pearce, G., Ryan, C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 103 (26), 10104–10109. doi: 10.1073/pnas.0603729103
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 (9), 1591–1606. doi: 10.1105/tpc.12.9.1591
Yan, J., Su, P., Meng, X., Liu, P. (2023). Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genomics 24 (1), 224. doi: 10.1186/s12864-023-09303-7
Yang, H., Bayer, P. E., Tirnaz, S., Edwards, D., Batley, J. (2021a). Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea. Biology 10 (1), 17. doi: 10.3390/biology10010017
Yang, Q., Guo, J., Zeng, H., Xu, L., Xue, J., Xiao, S., et al. (2021b). The receptor-like cytoplasmic kinase CDG1 negatively regulates Arabidopsis pattern-triggered immunity and is involved in AvrRpm1-induced RIN4 phosphorylation. Plant Cell 33 (4), 1341–1360. doi: 10.1093/plcell/koab033
Yang, P., Praz, C., Li, B., Singla, J., Robert, C. A. M., Kessel, B., et al. (2019). Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 221 (2), 976–987. doi: 10.1111/nph.15419
Yang, K., Rong, W., Qi, L., Li, J., Wei, X., Zhang, Z. (2013). Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis. Sci. Rep. 3 (1), 3021. doi: 10.1038/srep03021
Yang, L., Wu, K., Gao, P., Liu, X., Li, G., Wu, Z. (2014). GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 215-216, 19–28. doi: 10.1016/j.plantsci.2013.10.009
Yang, S. L., Xie, L. F., Mao, H. Z., Puah, C. S., Yang, W. C., Jiang, L., et al. (2003). Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15 (12), 2792–2804. doi: 10.1105/tpc.016618
Yeung, W., Ruan, Z., Kannan, N. (2020). Emerging roles of the alphaC-beta4 loop in protein kinase structure, function, evolution, and disease. IUBMB Life 72 (6), 1189–1202. doi: 10.1002/iub.2253
Yokoyama, R., Takahashi, T., Kato, A., Torii, K., Komeda, Y. (1998). The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 15 (3), 301–310. doi: 10.1046/j.1365-313X.1998.00203.x
Yu, T. Y., Shi, D. Q., Jia, P. F., Tang, J., Li, H. J., Liu, J., et al. (2016). The arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PloS Genet. 12 (3), e1005933. doi: 10.1371/journal.pgen.1005933
Yu, Y., Song, W., Zhai, N., Zhang, S., Wang, J., Wang, S., et al. (2023). PXL1 and SERKs act as receptor–coreceptor complexes for the CLE19 peptide to regulate pollen development. Nat. Commun. 14 (1), 3307. doi: 10.1038/s41467-023-39074-4
Yu, H., Zhang, W., Kang, Y., Fan, Y., Yang, X., Shi, M., et al. (2022). Genome-wide identification and expression analysis of wall-associated kinase (WAK) gene family in potato (Solanum tuberosum L.). Plant Biotechnol. Rep. 16 (3), 317–331. doi: 10.1007/s11816-021-00739-5
Yue, Z. L., Liu, N., Deng, Z. P., Zhang, Y., Wu, Z. M., Zhao, J. L., et al. (2022). The receptor kinase OsWAK11 monitors cell wall pectin changes to fine-tune brassinosteroid signaling and regulate cell elongation in rice. Curr. Biol. 32 (11), 2454–2466.e2457. doi: 10.1016/j.cub.2022.04.028
Yue, K., Sandal, P., Williams, E. L., Murphy, E., Stes, E., Nikonorova, N., et al. (2016). PP2A-3 interacts with ACR4 and regulates formative cell division in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 113 (5), 1447–1452. doi: 10.1073/pnas.1525122113
Zan, Y., Ji, Y., Zhang, Y., Yang, S., Song, Y., Wang, J. (2013). Genomewide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genomics 14, 318. doi: 10.1186/1471-2164-14-318
Zeiner, A., Colina, F. J., Citterico, M., Wrzaczek, M. (2023). CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASES - their evolution, structure and roles in stress response and development. J. Exp. Bot. 74 (17), 4910–4927. doi: 10.1093/jxb/erad236
Zhang, S., Chen, C., Li, L., Meng, L., Singh, J., Jiang, N., et al. (2005). Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139 (3), 1107–1124. doi: 10.1104/pp.105.069005
Zhang, H., Chen, C., Li, L., Tan, X., Wei, Z., Li, Y., et al. (2021a). A rice LRR receptor-like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol. J. 19 (11), 2319–2332. doi: 10.1111/pbi.13663
Zhang, X., Han, X., Shi, R., Yang, G., Qi, L., Wang, R., et al. (2013a). Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 73, 383–391. doi: 10.1016/j.plaphy.2013.10.024
Zhang, P., Kornev, A. P., Wu, J., Taylor, S. S. (2015). Discovery of allostery in PKA signaling. Biophys. Rev. 7 (2), 227–238. doi: 10.1007/s12551-015-0170-x
Zhang, Z., Ma, W., Ren, Z., Wang, X., Zhao, J., Pei, X., et al. (2021b). Characterization and expression analysis of wall-associated kinase (WAK) and WAK-like family in cotton. Int. J. Biol. Macromol 187, 867–879. doi: 10.1016/j.ijbiomac.2021.07.163
Zhang, L., Tian, L. H., Zhao, J. F., Song, Y., Zhang, C. J., Guo, Y. (2009). Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 149 (2), 916–928. doi: 10.1104/pp.108.131144
Zhang, X., Yang, G., Shi, R., Han, X., Qi, L., Wang, R., et al. (2013b). Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol. Biochem. 67, 189–198. doi: 10.1016/j.plaphy.2013.03.013
Zhang, H., Zhao, Y., Zhu, J.-K. (2020). Thriving under stress: how plants balance growth and the stress response. Dev. Cell 55 (5), 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhao, J., Sun, Y., Li, X., Li, Y. (2022). CYSTEINE-RICH RECEPTOR-LIKE KINASE5 (CRK5) and CRK22 regulate the response to Verticillium dahliae toxins. Plant Physiol. 190 (1), 714–731. doi: 10.1093/plphys/kiac277
Zhao, D.-Z., Wang, G.-F., Speal, B., Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 (15), 2021–2031. doi: 10.1101/gad.997902
Zheng, B., Bai, Q., Li, C., Wang, L., Wei, Q., Ali, K., et al. (2022a). Pan-brassinosteroid signaling revealed by functional analysis of NILR1 in land plants. New Phytol. 235 (4), 1455–1469. doi: 10.1111/nph.18228
Zheng, B., Bai, Q., Wu, L., Liu, H., Liu, Y., Xu, W., et al. (2019). EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 10 (1), 4165. doi: 10.1038/s41467-019-12112-w
Zheng, B., Xing, K., Zhang, J., Liu, H., Ali, K., Li, W., et al. (2022b). Evolutionary analysis and functional identification of ancient brassinosteroid receptors in ceratopteris richardii. Int. J. Mol. Sci. 23 (12), 6795. doi: 10.3390/ijms23126795
Zhong, X., Li, J., Yang, L., Wu, X., Xu, H., Hu, T., et al. (2023). Genome-wide identification and expression analysis of wall-associated kinase (WAK) and WAK-like kinase gene family in response to tomato yellow leaf curl virus infection in Nicotiana benthamiana. BMC Plant Biol. 23 (1), 146. doi: 10.1186/s12870-023-04112-2
Zhou, J., Loh, Y. T., Bressan, R. A., Martin, G. B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83 (6), 925–935. doi: 10.1016/0092-8674(95)90208-2
Zhou, A., Wang, H., Walker, J. C., Li, J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 40 (3), 399–409. doi: 10.1111/j.1365-313X.2004.02214.x
Zhu, Y., Hu, C., Cui, Y., Zeng, L., Li, S., Zhu, M., et al. (2021). Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis. Mol. Plant 14 (7), 1119–1134. doi: 10.1016/j.molp.2021.04.001
Zhu, Q., Shao, Y., Ge, S., Zhang, M., Zhang, T., Hu, X., et al. (2019). A MAPK cascade downstream of IDA–HAE/HSL2 ligand–receptor pair in lateral root emergence. Nat. Plants 5 (4), 414–423. doi: 10.1038/s41477-019-0396-x
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 (4), 749–760. doi: 10.1016/j.cell.2006.03.037
Zou, Y., Liu, X., Wang, Q., Chen, Y., Liu, C., Qiu, Y., et al. (2014). OsRPK1, a novel leucine-rich repeat receptor-like kinase, negatively regulates polar auxin transport and root development in rice. Biochim. Biophys. Acta 1840 (6), 1676–1685. doi: 10.1016/j.bbagen.2014.01.003
Zou, X., Qin, Z., Zhang, C., Liu, B., Liu, J., Zhang, C., et al. (2015). Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J. Exp. Bot. 66 (22), 7197–7209. doi: 10.1093/jxb/erv417
Zulawski, M., Schulze, G., Braginets, R., Hartmann, S., Schulze, W. X. (2014). The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genomics 15 (1), 548. doi: 10.1186/1471-2164-15-548
Zuo, W., Chao, Q., Zhang, N., Ye, J., Tan, G., Li, B., et al. (2015). A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47 (2), 151–157. doi: 10.1038/ng.3170
Keywords: receptor-like kinases, evolution, phosphorylation, signaling, ligand, development
Citation: Liu J, Li W, Wu G and Ali K (2024) An update on evolutionary, structural, and functional studies of receptor-like kinases in plants. Front. Plant Sci. 15:1305599. doi: 10.3389/fpls.2024.1305599
Received: 03 October 2023; Accepted: 03 January 2024;
Published: 31 January 2024.
Edited by:
Shumayla, Texas Tech University, United StatesReviewed by:
Anil Kumar, National Institute of Plant Genome Research (NIPGR), IndiaGabriela Cabrales Orona, Texas Tech University, United States
Nasir Khan, Texas Tech University, United States
Copyright © 2024 Liu, Li, Wu and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khawar Ali, YWFsaUBzbm51LmVkdS5jbg==; Guang Wu, Z3d1M0Bzbm51LmVkdS5jbg==
 Jing Liu
Jing Liu Wenjuan Li
Wenjuan Li Guang Wu
Guang Wu Khawar Ali
Khawar Ali