- 1International Research Centre for Environmental Membrane Biology and Department of Horticulture, Foshan University, Foshan, China
- 2Guangdong Key Laboratory for New Technology Research of Vegetables, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 3School of Biological Science, University of Western Australia, Crawley, WA, Australia
Plant respiratory burst oxidase homologs (RBOHs) are key enzymes regulating superoxide production, which is important for plant development and responses to biotic and abiotic stresses. This study aimed to characterize the RBOH gene family in pea (Pisum sativum L.). Seven PsRBOH genes were identified in the pea genome and were phylogenetically clustered into five groups. Collinearity analyses of the RBOHs identified four pairs of orthologs between pea and soybean. The gene structure analysis showed that the number of exons ranged from 6 to 16. Amino acid sequence alignment, conserved domain, and conserved motif analyses showed that all seven PsRBOHs had typical features of plant RBOHs. The expression patterns of PsRBOH genes in different tissues provided suggested their roles in plant growth and organ development. In addition, the expression levels of PsRBOH genes under different abiotic stresses were analyzed via reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The results demonstrated that PsRBOH genes exhibited unique stress-response characteristics, which allowed for functional diversity in response to different abiotic stresses. Furthermore, four PsRBOHs had a high probability of localization in the plasma membrane, and PsRBOH6 was localized to the plasma membrane and endoplasmic reticulum. The results of this study provide valuable information for further functional analysis of pea RBOH genes and their role in plant adaptation to climate-driven environmental constraints.
Introduction
Abiotic stress tolerance of major staple crops was significantly weakened or even lost during their domestication (Palmgren et al., 2015; Rawat et al., 2022). Major abiotic stresses such as drought, salinity, flooding, and extreme temperatures are increasing due to the current climate changes driven by global warming (Liu et al., 2020). Therefore, improving abiotic stress tolerance in crops is critical for future food security.
Despite different plants having different mechanisms for adapting to abiotic stresses, these stresses increase superoxide levels in all plants, and the superoxide is transformed into reactive oxygen species (ROS) via different pathways in different cellular compartments (Sandalio et al., 2021). ROS were initially considered toxic byproducts of aerobic metabolism and deemed harmful to plants; however, their central role in complex adaptive signaling networks has been recently reported (Mittler et al., 2011). While excessive ROS accumulation is toxic, causing DNA backbone damage, protein and lipid oxidation, and inducing apoptosis (Patel et al., 2018; Wang et al., 2020), transient stress-induced ROS spikes are considered central to systemic signaling and adaptation in plants (Bostock et al., 2014; Zhang et al., 2022).
Superoxide can be produced in various cellular compartments, including cytosolic and apoplastic compartments. Respiratory Burst Oxidase Homologues (RBOH) play a key role in apoplastic superoxide production (Suzuki et al., 2011; Zhang et al., 2022). The first identified plant RBOH gene was OsRBOHA in rice (Groom et al., 1996). With the recent increase in whole-genome sequencing of more plant species, RBOH genes have been functionally characterized in various plant species. These plants include Arabidopsis thaliana (Torres et al., 1998; Torres et al., 2002), Lycopersicon esculentum (Sagi et al., 2004), Olea europaea (Jimenez-Quesada et al., 2019), Glycine max (Liu et al., 2019), Brassica rapa (Li et al., 2019), Capsicum annuum (Zhang et al., 2021), Pyropia yezoensis (Gui et al., 2022), and Solanum melongena (Du et al., 2023).
Previous studies showed that many plants have multiple RBOH members, and different members exhibit different expression profiles with distinct functions. For example, there are ten RBOHs in Arabidopsis (AtRBOHA-J) (Sagi and Fluhr, 2006) and nine in rice (OsNox1-9) (Wang et al., 2013). Additional studies showed that AtRBOHD and AtRBOHF were expressed throughout the plant, while AtRBOHA-C, AtRBOHG and AtRBOHI were specifically expressed in the roots. Moreover, AtrbohH and AtrbohJ were specifically expressed in pollens and are thus required for normal pollen tube growth (Sagi and Fluhr, 2006; Kaya et al., 2014). OsNox1-2, OsNox5-6, and OsNox9 were ubiquitously expressed, while OsNox3, OsNox4, OsNox7, and OsNox8 showed tissue-specific expression in rice (Wang et al., 2013). Moreover, OsNox9 were associated with root development (Takeda et al., 2008; Yamauchi et al., 2017).
These studies also provided evidence for the involvement of RBOHs in plant signal transduction during abiotic stress responses and development. Plant RBOHs are involved in several signaling pathways, including Ca2+-dependent protein kinases (CDPKs) (Dubiella et al., 2013), mitogen-activated protein kinases (MAPKs) (Asai et al., 2008), receptor-activated C-kinases (RACKs) (Nakashima et al., 2008), phosphatidylinositol, phospholipase Dα1and phosphatidic acid (Zhang et al., 2009), nitric oxide (NO) (Delledonne et al., 2002), cGMP (Li et al., 2011), extracellular ATP-mediated signaling pathways (Song et al., 2006), and hormonal signaling networks (abscisic acid, salicylic acid, jasmonic acid, and ethylene) (Overmyer et al., 2003). AtRBOHD and AtRBOHF regulate ABA-mediated stomatal closure (Kwak et al., 2003) and salt stress tolerance in Arabidopsis (Chung et al., 2008; Xie et al., 2011).
As a classic model plant and the second most important grain legume, pea (Pisum sativum L., 2n = 14) has been widely used in genetics and developmental biology studies since Mendel (Kreplak et al., 2019; Chen et al., 2022; Chen et al., 2023; Li et al., 2023). Pea has become an important legume crop and green vegetable favored by many people worldwide. Fresh pea is rich in soluble protein, starch, carotenoid, and flavonoids (Yang et al., 2022). However, pea is sensitive to most common abiotic stresses (e.g., drought, cold, high temperatures, and salinity), which cause massive yield losses.
Given the critical role of RBOH in plant adaptive responses to abiotic stresses, in this work we aimed to provide a comprehensive analysis of pea RBOH genes. This study analyzed the chromosomal distribution, developmental evolution, gene structure, conserved domains, subcellular location, and description of cis-acting elements of pea RBOH genes. We also studied the tissue specificity and expression patterns of pea RBOH genes under different abiotic stresses. The reported data provide valuable information for an in-depth understanding of the biological functions of the RBOH gene family in pea, paving the way for improving its abiotic stress tolerance.
Materials and methods
Identification of the PsRBOH genes in pea
The DNA sequences and annotation files of the pea genome were downloaded from the Pea Genome Database (https://www.peagdb.com/). In addition, 17 soybean, 8 tomato, and 10 Arabidopsis RBOH protein sequences were downloaded from the phytozome (https://phytozome-next.jgi.doe.gov/), Sol Genomics Network (https://www.sgn.cornell.edu/) and TAIR (www.arabidopsis.org/) databases, respectively. Arabidopsis is a model plant widely used in various plant studies, including gene family identification studies. Tomato is a horticultural crop that has been widely studied and is one of the most widely eaten vegetables in the world. Its molecular biology research is relatively in-depth. Soybeans, like peas, are important leguminous plants with high protein levels. The domain sequences (HMM model file) of the NADPH oxidase (PF08414), NAD binding (PF08030), FAD binding (PF08022), and Ferric reductase domains (PF01794) were downloaded from the Pfam database (http://pfam.xfam.org/) and used as the seed files to search the RBOH proteins in the pea genome file via a hidden Markov model (HMM) search (e-value 0.01). The conserved domains were verified using CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Analysis of the chromosomal location, phylogenetics, collinearity, gene structure, conserved motifs, and cis-elements
The online software MG2C (http://mg2c.iask.in/mg2c_v2.0/) was used for the chromosome localization analysis. The phylogenetic tree was constructed using the IQ-Tree Wrapper program in TBtools software (Chen et al., 2020) with 1000 bootstrap replicates. The collinearity between pea and soybean sequences was determined using TBtools software (Chen et al., 2020). The Gene Structure Display Server (http://gsds.gao-lab.org) was used to generate the gene structure map, and the online software MEME (http://meme-suite.org/tools/meme) was used to search for conserved motifs with the maximum conserved motif search value set to 10. The 2000-bp sequences upstream of PsRBOH genes were obtained from the pea genome database and used as the promoter, and the cis-regulatory elements of these promoters were identified using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant materials, stress treatments and tissue expression analysis
P. sativum cultivar Zhongwan6 (ZW6) were obtained from the pea seed breeding center in China (Gu’an, Hebei). The seeds were germinated in a liquid culture using a quarter (1/4)-strength modified Hoagland nutrient solution (pH 5.5) in a growth chamber under 25 ± 2°C with a 16 h/8 h light/dark cycle. The four-leaf stage seedlings were treated with 1/4-strength modified Hoagland nutrient solution [Ca (NO3)2·4H2O 236.25 mg/L+KNO3 126.25 mg/L+ NH4 NO3 20 mg/L+ KH2PO4·2H2O 34 mg/L+ MgSO4·7H2O 123.25 mg/L, pH 5.5] supplemented with 100 mM NaCl for the salt-stress treatment, 10% (w/v) polyethylene glycol (PEG) for the drought stress treatment, and 6 μM CdCl2 for the cadmium (Cd) treatment. For the heat and cold stress, the seedlings were treated with 1/4-strength modified Hoagland nutrient solution (pH 5.5) at 38°C and 4°C, respectively. For the low-boron stress treatment, seedlings were initially pretreated with the modified Hoagland nutrient solution containing 0.025 μM boric acid (H3BO3) (B treatment) at the first stage of culture to diminish variations in B concentrations in seeds. Subsequently, the low-boron-treated plants were treated with the modified Hoagland nutrient solution containing 0.025 μM H3BO3, and the controls were treated with the modified Hoagland nutrient solution with 25 μM H3BO3 at the four-leaf-stage seedlings (Li et al., 2018). The treatments were conducted in triplicates with five seedlings per replicate. The leaves of treated seedlings were sampled after 0.5, 3, 6, 12, 24, 36, 48, and 72 h of stress treatment. Pea roots, leaves, stems, flowers, tendrils, one-week-old pea seeds, one-week-old pod, and two-week-old pea seeds were sampled for tissue expression analysis. All the harvested samples were frozen in liquid nitrogen and stored at −80°C until analysis. Each treatment had three independent biological replicates.
RNA isolation and RT-qPCR analysis
Total RNA was isolated from the samples using FreeZol Reagent R711 (Nanjing Vazyme Biotech Co., Ltd.), according to the manufacturer’s instructions. Briefly, 50 mg of the ground sample was added into 500 μl of FreeZol Reagent for lysis. The lysate was centrifuged, and the supernatant was collected. A dilution buffer was added to the supernatant, and the mixture was precipitated with isopropanol. After centrifugation, the supernatant was discarded, and the pellet was washed with 75% ethanol and dissolved in RNase-free double distilled water (ddH2O). The quality and concentrations of the isolated RNA samples were determined via 1% agarose gel electrophoresis and a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Reverse transcription PCR was conducted on a QuantStudio™ 6Flex Real-Time PCR System (Applied Biosystems™, Carlsbad, CA, USA) using HiScript® III RT SuperMix for qPCR with (+gDNA wiper) (R323-01) (Vazyme, Nanjin) and ChamQTM Universal SYBR® qPCR Master Mix (Q711) (Vazyme). Three technical replicates were set for each biological sample, and the reaction conditions were as follows: 95°C for 30 s, followed by 40 cycles at 95°C for 10 s and 65°C for 20 s. A melting curve was generated by cooling from 95°C to 65°C then ramping to 95°C, followed by the final cooling to 50°C for 30 s. PsACT (Knopkiewicz and Wojtaszek, 2019) was used as the internal control. The expression level of each PsRBOH gene was calculated using the delta-delta Ct (2−ΔΔCT) method (Livak and Schmittgen, 2001). All analyses were conducted in three biological replicates and three technical replicates. All primer sequences used in this study were designed by Primer Premier 6.0 and are listed in Supplementary Table S1.
Subcellular localization analysis
The coding sequence (CDS) of RBOH6 was amplified from the ZW6 and fused to the N terminus of the green fluorescent protein (GFP) in the pAN580 (GFP) vector. To determine the subcellular localization of RBOH6, the RBHO6-GFP vector was transiently co-expressed with endoplasmic reticulum (ER) marker mCherry-HDEL into Arabidopsis protoplasts (Ishikawa et al., 2018). And, the FM4-64 dye was used to marked plasma membrane. The fluorescent signals were observed using a confocal microscope (Leica SP8, Germany). Fluorescence signals were detected using the following excitation and emission wavelengths: GFP (488 nm/507 nm), FM4-64 (515 nm/640 nm) and mCherry (587 nm/610 nm).
Results
Chromosomal localization analysis of the RBOH gene family in pea
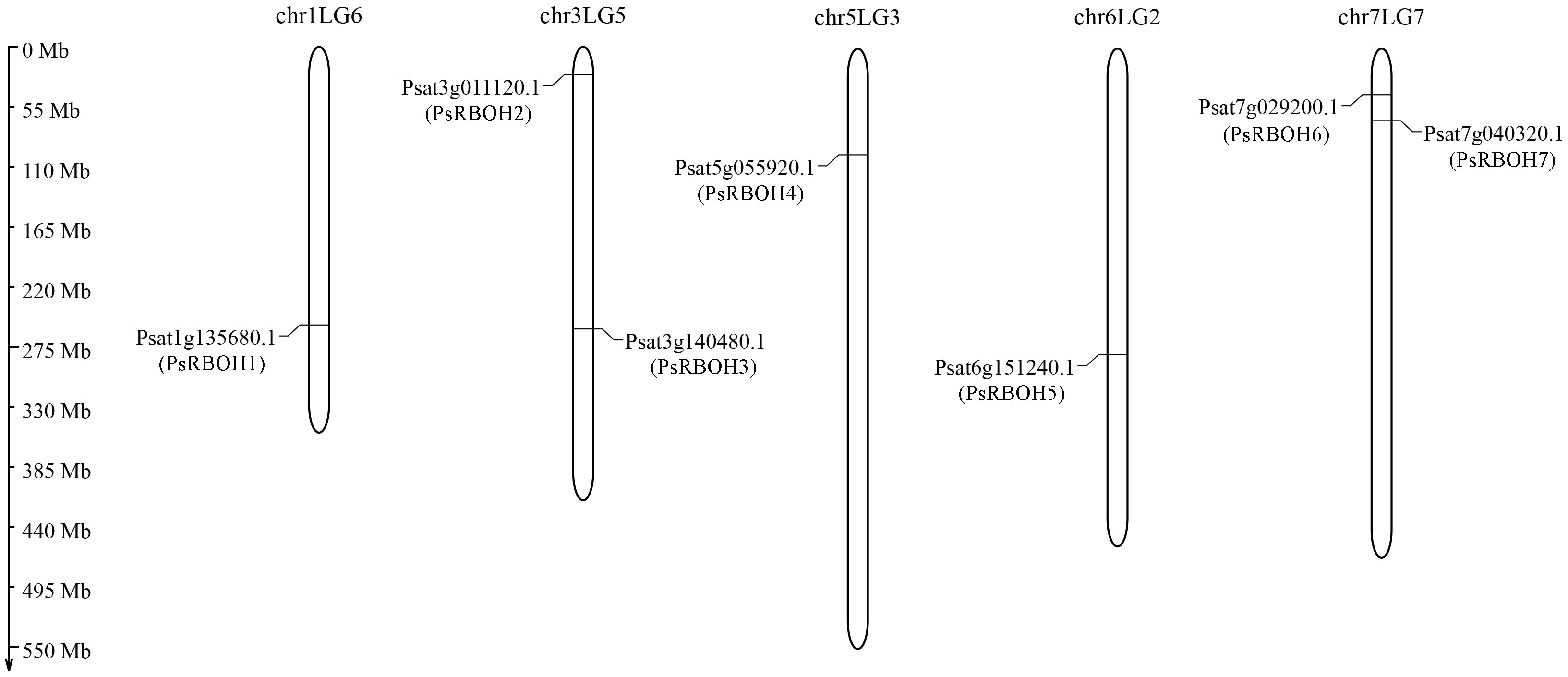
Seven putative PsRBOH genes were retrieved from the pea genome after removing the redundant and repeat sequences. We named these PsRBOH1–7 (Figure 1 and Supplementary Table S2) based on their localization in the P. sativum genome. The distribution of these RBOH genes did not show a certain regularity. There was one RBOH gene on chromosomes chr1LG5 (RBOH1), chr5LG3 (RBOH4), and chr6LG2 (RBOH5) each, while two were on chromosomes chr3LG5 (RBOH2 and 3) and chr7LG2 (RBOH6 and 7) each.
Phylogenetic analysis of the PsRBOHs
To investigate the evolutionary relationships of RBOHs among pea, soybean, tomato, and Arabidopsis, we constructed a neighbor-joining phylogenetic tree using the alignments of seven pea RBOH proteins, 17 soybean RBOH proteins, eight tomato RBOH proteins, and ten Arabidopsis RBOH proteins (Figure 2A). The results showed that 42 RBOHs from pea, soybean, tomato, and Arabidopsis were divided into five groups (I, II, III, IV, and V). Groups I, II, III, IV, and V contained 8, 6, 2, 6, and 20 RBOH proteins, respectively, and 1, 1, 0, 1 and 4 PsRBOHs were clustered in groups I, II, III, IV, and V, respectively.
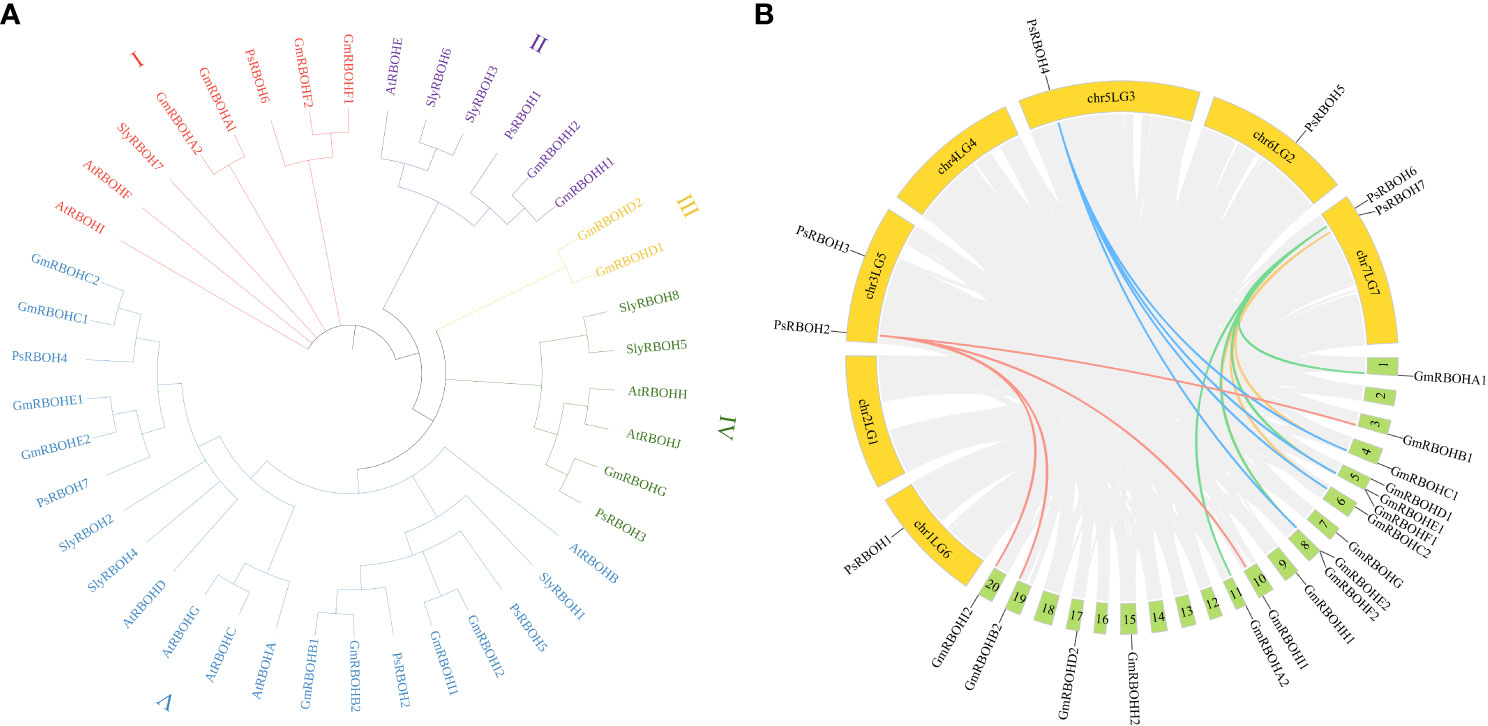
Figure 2 Phylogenetic and collinearity analyses of the PsRBOHs. (A) Phylogenetic analysis of RBOHs of pea, soybean, tomato, and Arabidopsis. (B) Collinearity analysis of RBOHs between pea and soybean. The colored lines represent RBOHs collinearity between species, and the gray lines are collinearity of all gene members between species. Yellow and green boxes represent pea and soybean chromosomes, respectively.
Collinearity analyses of the PsRBOHs
To explore the evolution of RBOH genes, we analyzed the synteny relationship of RBOHs between pea and soybean (Figure 2B). Four ortholog pairs were identified between pea and soybean. PsRBOH2 paired with GmRBOHB1, GmRBOHB2, GmRBOHI1, and GmRBOHI2, while PsRBOH4 paired with GmRBOHC1, GmRBOHC2, GmRBOHE1, and GmRBOHE2. PsRBOH6 paired with GmRBOHA1, GmRBOHA2, GmRBOHF1, and GmRBOHF2, and PsRBOH7 paired with GmRBOHC1, GmRBOHC2, GmRBOHE1, and GmRBOHE2.
Gene structure, conserved motif and conserved domain analysis of the PsRBOHs
A gene structure map of PsRBOHs was constructed based on the pea genome sequence (Figure 3A). The PsRBOHs had varying structures with different numbers of untranslated regions (UTRs) and exons. All PsRBOHs but PsRBOH1 and PsRBOH4 had no UTRs, and the number of exons ranged from six (PsRBOH1) to 14 (PsRBOH6). Ten conserved motifs were identified via the MEME online tool and were used to gain a deeper understanding of various motif compositions of the PsRBOHs. As shown in Figure 3B and Supplementary Figure S1, PsRBOH proteins contained 10 motifs, except PsRBOH1 and PsRBOH4, which had 4 and 9 motifs, respectively. Moreover, all PsRBOH proteins, except PsRBOH1, had four typical conserved domains, including the NADPH-Ox, Ferri-reduction, FAD-binding, and NAD-binding domains (Figure 3C). PsRBOH1 lacked the FAD-binding and NAD-binding domains, and all RBOH proteins contained the EF-hand domain.
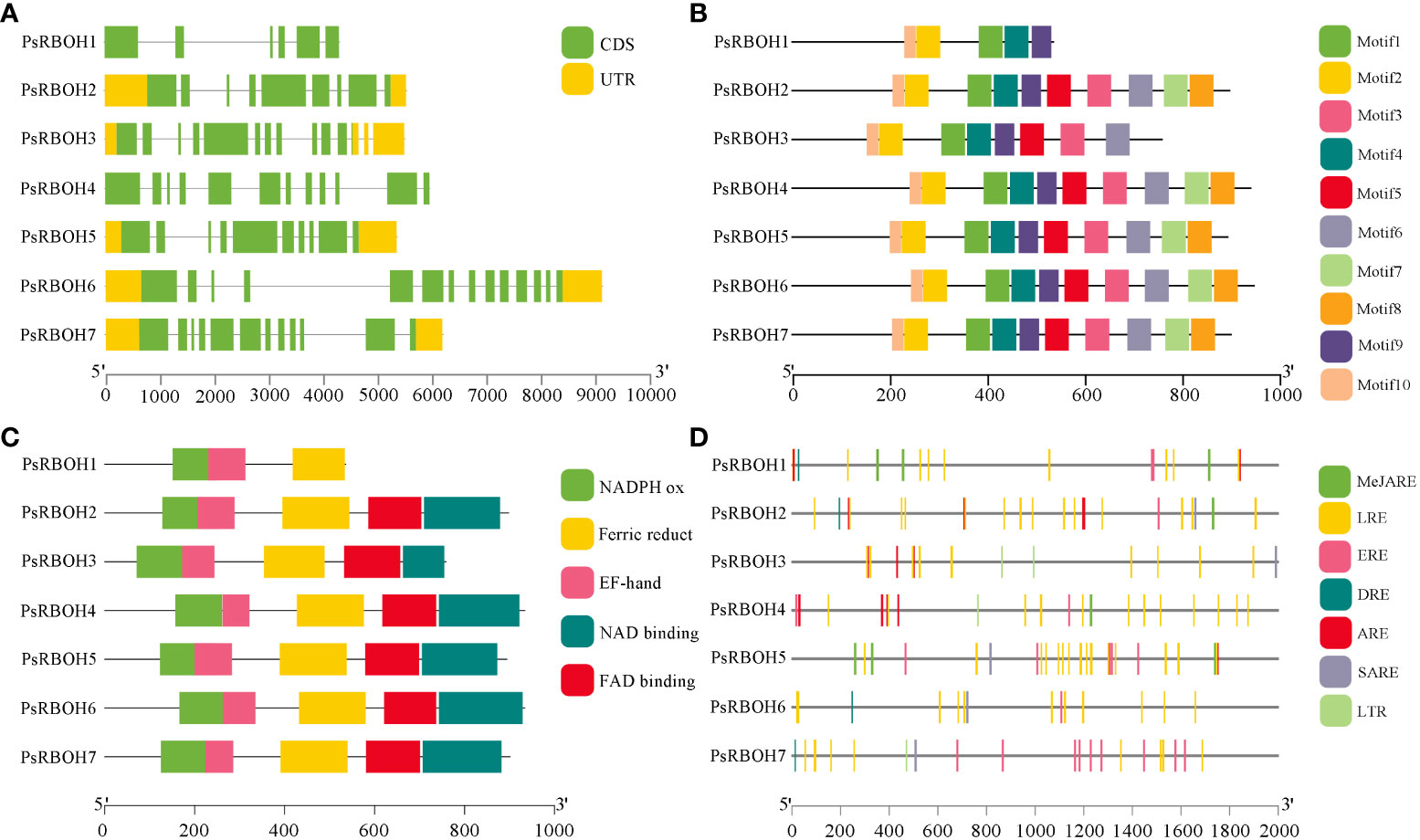
Figure 3 The gene structures, conserved motifs, conserved domain, and cis-elements analysis of putative promoters of PsRBOHs. (A) Gene structure of PsRBOHs. The green block represents the coding sequence (CDS), the yellow block shows the 5’ or 3’ untranslated regions (UTRs), and the black line indicates the intron. The scale bar indicates the length of the DNA sequences. (B) The conserved motifs in PsRBOHs were identified by MEME tools. Each motif is indicated by a rectangular box of a different color and the motifs are numbered from 1 to 10. The black line scale represents the length of amino acids. (C) Conserved domain compositions of PsRBOHs. Only major domains are presented in the figure based on our searches in the Pfam database. The black line scale represents the length of amino acids. (D) Cis-elements analysis of putative promoters of PsRBOHs. The black line scale represents the length of promoter amino acids. The cis-elements are as follows: MeJA-responsive elements (MeJARE), light-responsive elements (LREs), ethylene-responsive elements (EREs), drought-inducible elements (DRE), abscisic acid-responsive element (ARE), salicylic acid-responsive elements (SARE), and low temperature-responsive element (LTR).
Cis-element analysis of the putative PsRBOH promoters
To further explore the function of PsRBOH genes in peas, we predicted the cis-elements of the promoters of these using the PlantCARE database (Figure 3D). The results showed that the light-responsive elements (LREs) were the most abundant (84) among the seven PsRBOH promoter sequences and widely distributed in all promoter sequences, followed by ethylene-responsive elements (EREs) (20), which was also distributed in all promoter sequences (Figure 3D). We also found that the promoter sequences of PsRBOH genes contained three hormone-responsive elements, [abscisic acid-responsive element (ARE, 17), MeJA-responsive element (MeJARE, 16) and salicylic acid-responsive elements (SARE, 5)] and two abiotic response elements [drought-inducible elements (DRE, 4) and low temperature-responsive element (LTR, 4)].
Tissue-specific expression of PsRBOHs in pea
To determine the expression patterns of the PsRBOH genes in different tissues, we studied the tissue-specific transcriptional activity of seven genes in the root, stem, tendril, leaf, flower, pod, one-week-old pea seeds (Pea1W), and two-weeks-old pea seeds (Pea2W) of pea (Figure 4). The results showed that the expressions of the PsRBOH genes had significant differences in different tissues. PsRBOH5 was highly expressed in the roots, while PsRBOH4, PsRBOH6, and PsRBOH7 were highly expressed in the stem. PsRBOH2 was highly expressed in all tissues. PsRBOH6 was highly expressed in the leaves, but PsRBOH3 was not detected in the leaves. Moreover, PsRBOH1 and PsRBOH3 were highly expressed in tendrils, and all seven PsRBOH genes had relatively lower expression levels in the pod and Pea1W. PsRBOH5 and PsRBOH7 were highly expressed in the roots and Pea2W, respectively, but PsRBOH1 was not expressed in flowers and pods.
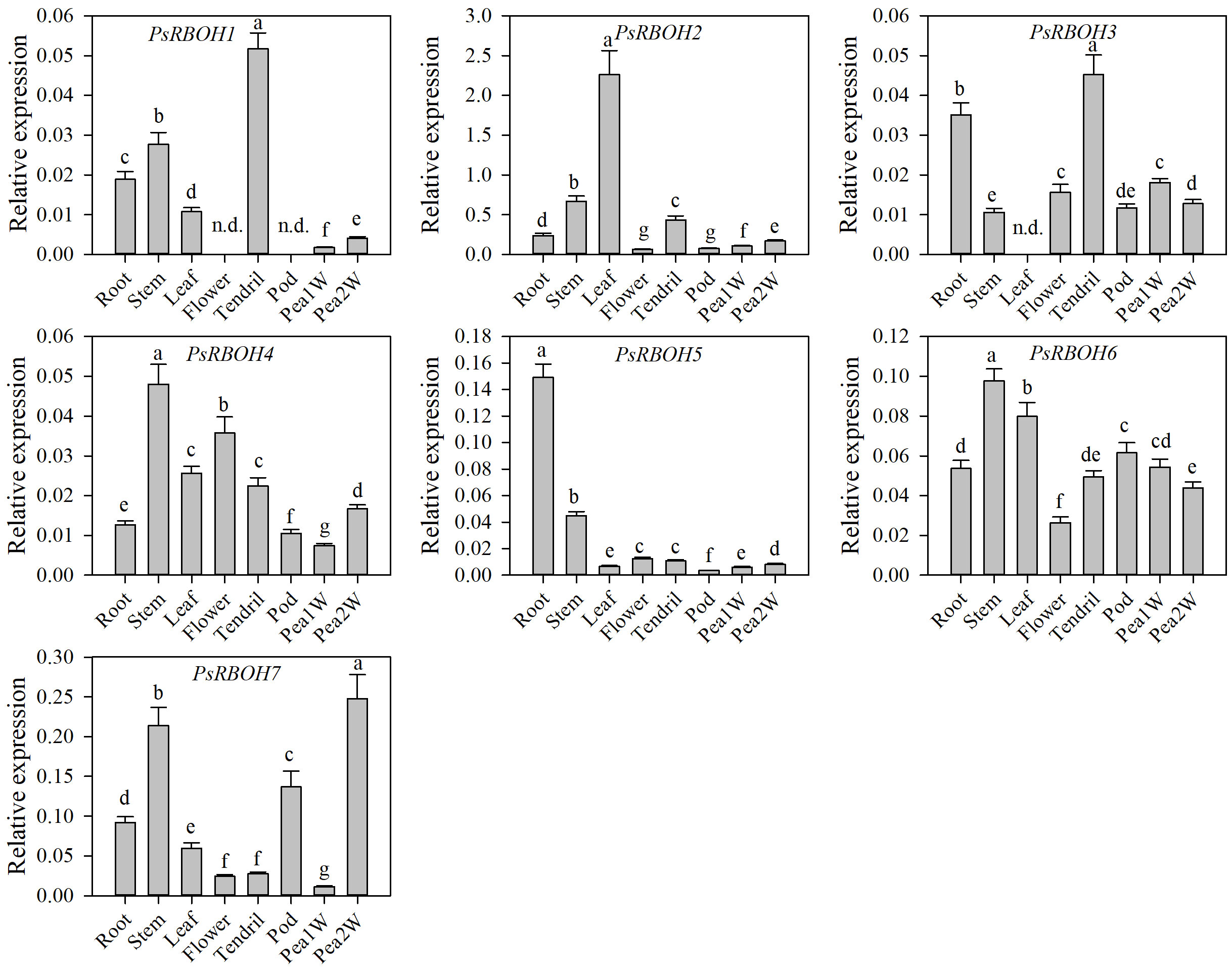
Figure 4 Expression patterns of PsRBOH genes in different tissues. Pea1W: One-week-old pea seeds; Pea2W: Two-week-old pea seeds. Error bars represent standard deviations for three biological replicates. Different letters indicating significant differences among tissues (p<0.05, Duncan’s test). n.d.: Not detected.
Expression of PsRBOHs under different abiotic stresses
To further elucidate the expression patterns of pea RBOH genes under different abiotic stresses, we evaluated the expression profiles of PsRBOHs in the leaves and roots under heat stress, cold stress, salt stress, Cd stress, PEG-induced drought stress (PEG), and low-boron (LB) stress at various time points after the treatment.
The abiotic stresses induced or inhibited the expression of genes in a highly specific manner, with the most significant changes occurring at the early stages of stress treatment (Figure 5). The expression levels of all genes increased in the leaves in all six treatments (Figures 5A–F), except for PsRBOH4 under salt stress (Figure 5C) and PsRBOH5 under Cd stress (Figure 5D). All PsRBOH genes were up-regulated at 6 h under all treatments (Figures 5A–F) except for PsRBOH4 under cold stress (Figure 5B), PsRBOH5 under Cd stress (Figure 5D) and PsRBOH4-6 under salt stress (Figure 5C). PsRBOH1 was up-regulated at 3 h under all six treatments but down-regulated at 24 h under heat (Figure 5A), cold (Figure 5B) and Cd stresses (Figure 5D). PsRBOH3 was not expressed in the leaves under all treatments (Figures 5A–F). Furthermore, PsRBOH4 was down-regulated from 0.5 to 48 h but up-regulated at 72 h under salt stress (Figure 5C). PsRBOH5 was induced at 0.5 h under heat stress (Figure 5A) but down-regulated at 12 h under heat (Figure 5B), salt (Figure 5C), Cd (Figure 5D), and PEG (Figure 5E) stresses. In addition, PsRBOH6 was induced at 6 h under LB stress (Figure 5F) and down-regulated at all time points except 3 h and 6 h under salt stress (Figure 5C).
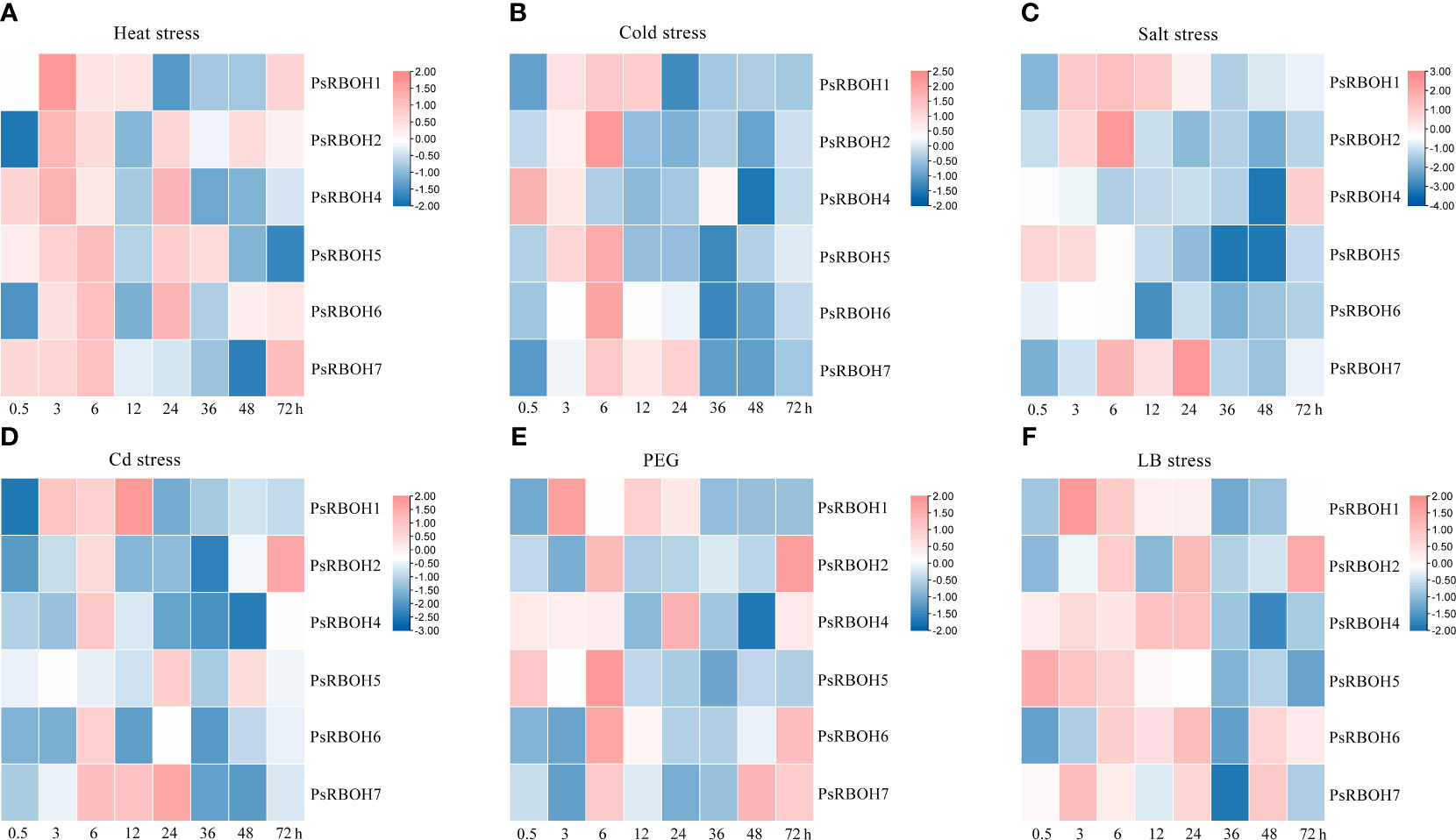
Figure 5 Leaf expression of PsRBOHs under heat stress (A), cold stress (B), salt stress (C), cadmium (Cd) stress (D), polyethylene glycol (PEG)-induced drought stress (PEG) (E), and low boron (LB) stress (F). The log-transformed relative expression levels were used to generate the heatmaps. The color scale is shown on the right.
The expression patterns of PsRBOHs had similarities and differences in the roots and leaves. In the roots, PsRBOH5 was induced at 0.5 h under heat (Figure 6A), salt (Figure 6C), PEG (Figure 6E), and LB (Figure 6F) stresses and maintained the high expression until 72 h under salt (Figure 6C), PEG (Figure 6E) and LB (Figure 6F) stresses. PsRBOH1 was up-regulated at 3 h and maintained a high expression level until 72 h under cold (Figure 6B) and salt stresses (Figure 6C) except at 12 h under cold stress. However, PsRBOH1 was down-regulated at all time points under LB treatment (Figure 6F). PsRBOH6 was induced at 0.5 h under salt (Figure 6C) and Cd (Figure 6D) treatments but down-regulated from 6 to 72 h under Cd (Figure 6D) and LB (Figure 6F) treatments. Additionally, PsRBOH6 had a similar expression pattern with PsRBOH1 under LB stress, except at the 3 h time-point (Figure 6F).
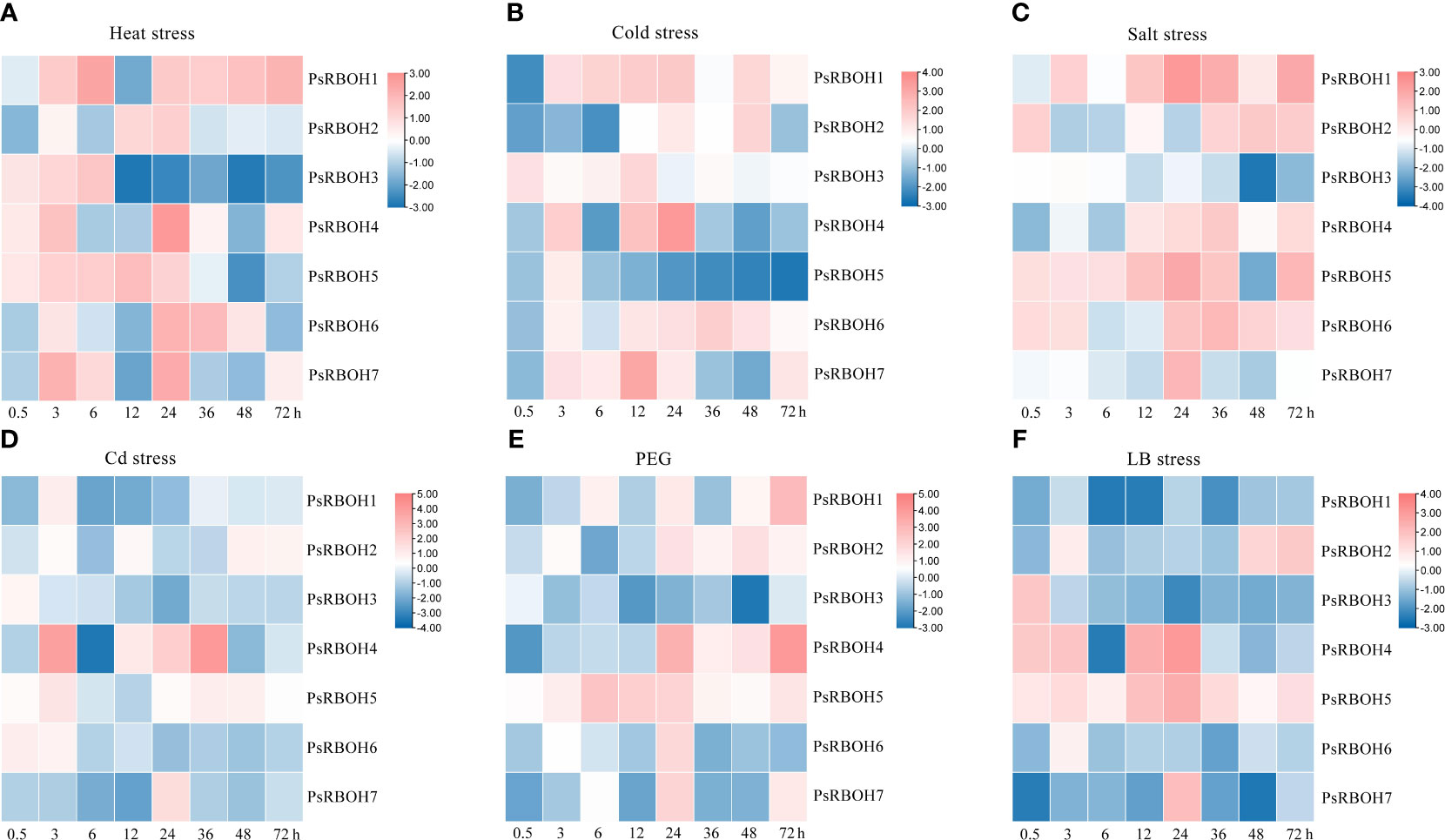
Figure 6 Root expression of PsRBOHs under heat stress (A), cold stress (B), salt stress (C), cadmium (Cd) stress (D), polyethylene glycol (PEG)-induced drought stress (PEG) (E), and low boron (LB) stress (F). The log-transformed relative expression levels were used to generate the heatmaps. The color scale is shown on the right.
Subcellular localization analysis
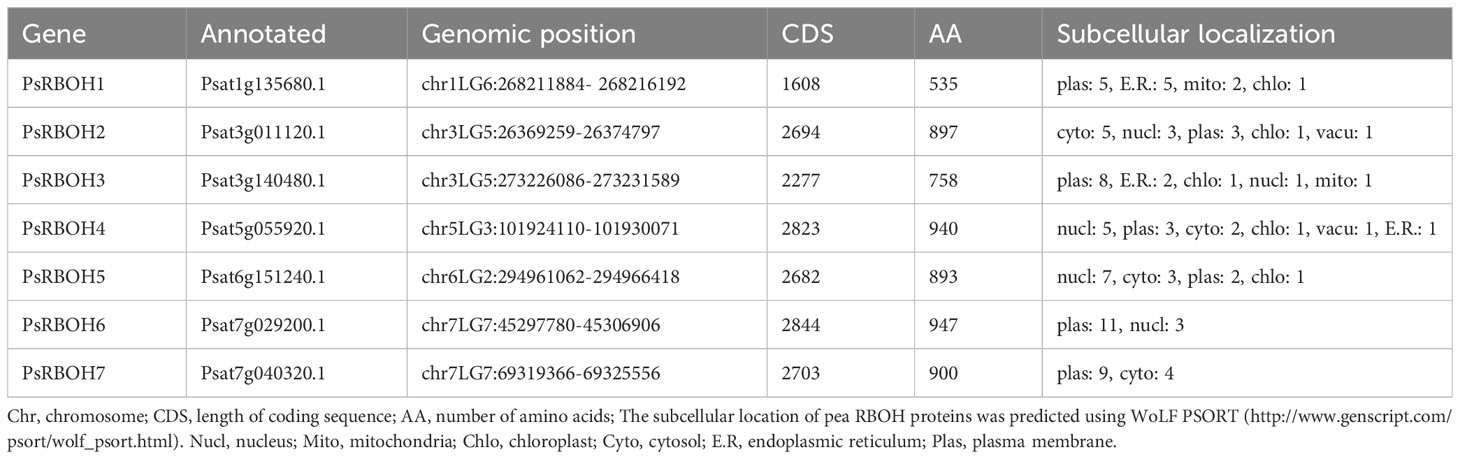
The subcellular localization of pea RBOH proteins was predicted using WoLFPSORT (Table 1). The results indicated that four PsRBOH proteins were highly likely to be located in the plasma membrane. PsRBOH4 and PsRBOH5 were presumably located in the nucleus, and PsRBOH4 in the cytosol. Evolutionary analysis showed that PsRBOH6 was homologous to AtRBOHF and GmRBOHF1/2. It has been shown that multiple calcineurin B-like (CBL) interacting protein kinases (CIPKs) target Arabidopsis RBOHF by directly binding Ca2+ to its EF-hands to fine-tune superoxide production in response to different stimuli (Drerup et al., 2013; Han et al., 2019). Since AtRBOHF localizes in the plasma membrane (Drerup et al., 2013), PsRBOH6 was selected for a transient expression assay using Arabidopsis mesophyll protoplasts (Figure 7). The PsRBOH6-GFP plasmid was transiently co-expressed with the plasma membrane and endoplasmic reticulum markers, FM4-64 (Figure 7A) and mCherry-HDEL (Figure 7B), in Arabidopsis leaf protoplasts, respectively. As shown in Figure 7, GFP-PsRBOH6 fluorescent signals were extensively co-localized with FM4-64 and mCherry-HDEL, suggesting that PsRBOH6 localizes in the plasma membrane and endoplasmic reticulum.
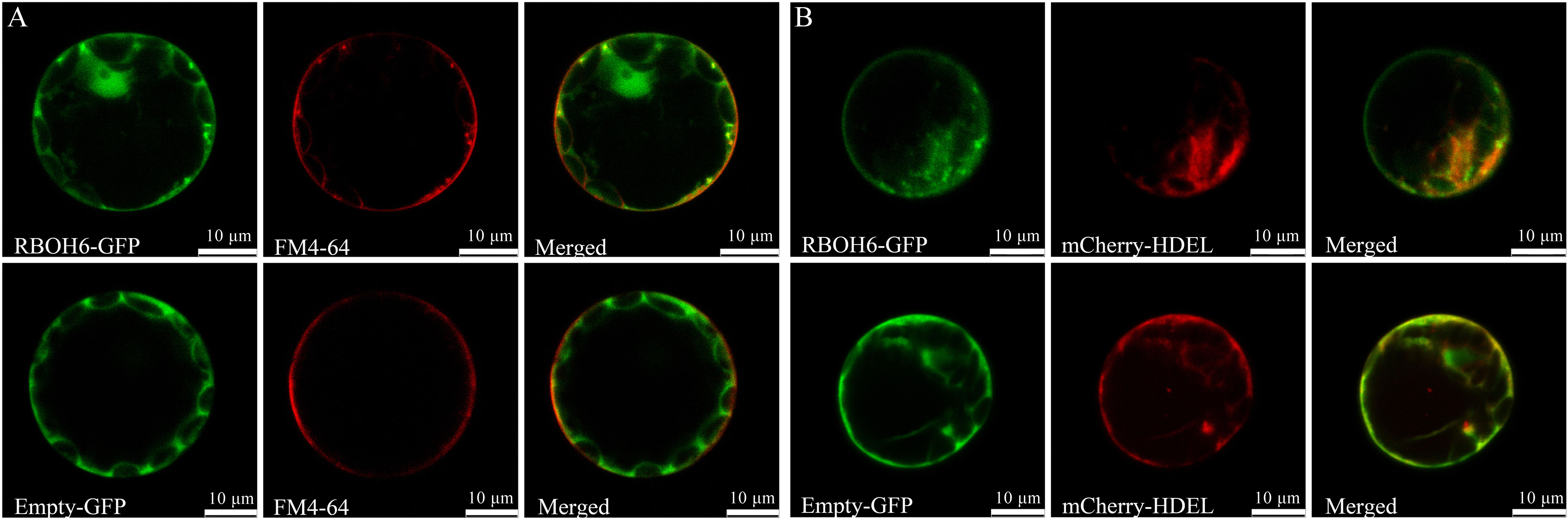
Figure 7 Subcellular localization of PsRBOH6 protein. (A) The plasma membrane was stained by FM4-64. (B) Transient co-expression of PsRBOH6-GFP with endoplasmic reticulum marker mCherry-HDEL. Empty vector was as control. Scale bar = 10 μm. Single optical sections obtained by CLSM.
Discussion
RBOH genes are involved in plant signal transmission, morphogenesis and development, and responses to biotic and abiotic stresses. However, the RBOH gene family has been identified in various plants but not in pea. These studies will be useful for understanding the role of RBOH genes in plant responses to abiotic stresses.
The present study analyzed the RBOH gene family in pea and identified seven RBOH genes were identified in the pea genome. The identified RBOH genes were fewer than those of Arabidopsis (10) (Sagi and Fluhr, 2006), rice (9) (Groom et al., 1996), Pyropia yezoensis (11) (Gui et al., 2022), Solanum melongena (8) (Du et al., 2023), and soybean (17) (Liu et al., 2019), but same those of strawberry (7) (Zhang et al., 2018) and alfalfa (7) (Marino et al., 2011). The chromosomal distribution of RBOH genes did not show a certain regularity, the same as in other plants (Selvi et al., 2020; Gui et al., 2022; Du et al., 2023). Gene structure analysis revealed that the number of PsRBOH exons varied between 6-16 and harbored in 9-12, similar to that of Arabidopsis and rice, which were harbored in 10-14 (Zhang et al., 2018). PsRBOH1 had the lowest number of exons (6), while PsRBOH6 had the most exons (16), indicating that PsRBOHs experienced both conservation and diversification during their evolution.
Determining the phylogenetic relationships among species is fundamental for many biological studies (Kapli et al., 2020). Phylogenetic analysis indicated that members of the RBOH gene families in pea, soybean, tomato, and Arabidopsis could be divided into five groups, indicating that RBOHs are conserved in various species (Chang et al., 2020; Chen and Yang, 2020; Gui et al., 2022; Du et al., 2023).
In soybeans, the ancestral Papilionoideae whole-genome duplication event and Glycine-specific duplication event resulted in nearly 75% of the genes present in multiple copies (Schmutz et al., 2010). While there has no a recent whole-genome duplication but reflects the ancestral Papilionoideae whole-genome duplication event in pea (Kreplak et al., 2019). This also explains why the amount of RBOHs in pea is less than that in soybean and the collinear gene pairs of four PsRBOHs (PsRBOH2, PsRBOH4, PsRBOH6, and PsRBOH7) in soybean.
In Arabidopsis, all ten RBOHs had four typical conserved domains: NADPH-Ox, Ferri-reduction, FAD-binding, and NAD-binding domains (Sagi and Fluhr, 2006). However, in wheat, 4 out of 36 NADPH oxidases had the NADPH_Ox domain but lacked one or two other conserved domains (Hu et al., 2018). In eggplant, 5 out of 8 SmRBOHs lacked the FAD-binding domain, which was substituted by the NOX_Duox_like_FAD_NADP domain (Du et al., 2023). In this study, all PsRBOHs, except PsRBOH1, had the four typical conserved domains. PsRBOH1 lacked the FAD-binding and NAD-binding domains which are crucial for electron transfer and ROS production (Wang et al., 2022). Therefore, the possible function of RBOH1 in the production of superoxide needs further verification.
The tissue-specific expression of genes is crucial for plant growth and development and provides important insights into understanding gene function. The tissue-specific expression patterns of the RBOHs have been reported in many plants; however, the expression patterns are distinct in different plants. Two out of ten, four out of seven, and all seven RBOH genes were expressed throughout Arabidopsis (Sagi and Fluhr, 2006), strawberry (Zhang et al., 2018) and grape (Cheng et al., 2013), respectively. However, only four out of ten AtRBOH genes were specifically expressed in the roots and elongation zone (Sagi and Fluhr, 2006). In this study, five RBOHs (PsRBOH2 and PsRBOH4-7) were expressed in all tissues, while PsRBOH1 was not detected in the flowers and pods, and PsRBOH3 was not expressed in the leaves. In addition, the expression of PsRBOH2 was higher in all tissues, especially in the leaves. In soybeans, GmRBOHB1, which was orthologous with PsRBOH2 and clustered in one cluster, was highly expressed in the leaves (Liu et al., 2019). These results indicated the tissue specificity of the PsRBOH genes, suggesting that these genes may have different functions during plant development.
Cis-acting elements regulate the expression of target genes by binding the trans-acting factors (Yamaguchi-Shinozaki and Shinozaki, 2005). It has been reported that RBOH family members in various plants are induced by different abiotic stress stimuli, such as drought (Duan et al., 2009), salt (Xie et al., 2011), heat (Li et al., 2014; Xia et al., 2014), wounding (Sagi et al., 2004), and cold stress (Zhang et al., 2018). Moreover, RBOHs respond to environmental stimuli through hormonal signaling networks involving abscisic, salicylic, jasmonic acid, and ethylene (Overmyer et al., 2003). In the present study, many hormone-responsive elements (such as ARE, ERE, MeJARE, and SARE) and abiotic response elements (such as DRE and LTR) were found in the promoter of PsRBOHs, indicating their potential roles in pea response to phytohormones and stresses.
The involvement of the RBOH gene family members in abiotic stress responses has been reported in many plants. In grapes, VvRBOHs were significantly increased under salt, drought, powdery mildew, salicylic acid, and abscisic acid treatments (Cheng et al., 2013). It was reported that the expression level of AtRBOHD was significantly increased in Arabidopsis at an early stage in response to hypoxia (Sagi and Fluhr, 2006). Soybean RBOH genes were significantly induced by salt, PEG, cold, and Cu stresses, and GmRBOHD2 was significantly induced by salt, polyethylene glycol, low temperature, and Cu toxicity in the roots (Liu et al., 2019). In the present study, all PsRBOH genes responded to all six stresses. Moreover, PsRBOH3, which had a close evolutionary relationship with GmRBOHD2, was expressed in the roots and not in leaves, and its expression occurred earlier (0.5 - 6 h) after heat, cold, and LB stress treatments. The expression of PsRBOH1 was up-regulated in the leaves 3 h after the abiotic stress treatments and lasted for up to 24 h, after which it was down-regulated under heat, cold, salt and Cd treatments. However, PsRBOH1 was up-regulated in the roots by heat, cold and NaCl treatments but down-regulated by LB treatment. A similar phenomenon was also observed in cotton (Wang et al., 2020). These results indicate that the PsRBOH gene family could be involved in the abiotic stress response, but the regulatory mechanisms are still unclear.
Superoxide can be synthesized in different cellular compartments, such as chloroplasts (Foyer and Hanke, 2022), mitochondria (Postiglione and Muday, 2022), peroxisome (Sandalio et al., 2021), and the plasma membrane oxidoreductase system (Lherminier et al., 2009; Jiménez-Quesada et al., 2022; Miller and Mittler, 2023). In leaves from pea plants grown with 50 µm CdCl2, the accumulation of H2O2 was observed mainly in the plasma membrane of transfer, mesophyll and epidermal cells, as well as in the tonoplast of bundle sheath cells (Romero-Puertas et al., 2004). RBOH is present in the plasma membrane systems of almost all animals and plants (Chen and Yang, 2020). Subcellular location is a key characteristic that determines the function of many proteins, indicating that proteins in different subcellular locations have different functions (Koroleva et al., 2005). All the RBOHs in Arabidopsis (Sagi and Fluhr, 2006), rice (Groom et al., 1996) and wheat (Hu et al., 2018) were predicted to localize to the plasma membrane. Some RBOHs were shown to localize to the chloroplast thylakoid membrane of grapes (Cheng et al., 2013) and strawberry (Zhang et al., 2018). Moreover, in Gossypium barbadense, 71 out of 87 RBOHs were located in the cytoplasm (Chang et al., 2020). Tobacco RBOHD was localized in the plasma membrane and Golgi cisternae (Noirot et al., 2014). In P. vulgaris, PvRBOHA was localized in the plasma membrane of root hair (Arthikala et al., 2017), whereas PvRBOHB was localized in the central apical dome (Montiel et al., 2012). These different subcellular distributions of RBOHs in root hairs resulted in two models that explain root hair development (Arthikala et al., 2017). In pea, four out of the seven RBOHs had a high probability of being localized in the plasma membrane, indicating similar functions. Two of the PsRBOHs were presumably located in the nucleus, while PsRBOH6 was localized in the cell membrane and endoplasmic reticulum, suggesting that they may regulate ROS production at different subcellular locations (Lee et al., 2023).
Conclusions
This study comprehensively analyzed the RBOH gene family in the pea genome. Seven PsRBOH genes were identified and were divided into five groups, which were distributed on five chromosomes. In addition, we analyzed collinearity, gene structure, conserved domains, conserved motifs, cis-elements, and subcellular distribution of the PsRBOH genes. The PsRBOHs exhibited tissue specificity and functional diversity during plant growth and response to different abiotic stresses. Overall, these results provide valuable information which could be used for further functional analysis of pea RBOH genes in response to climate-driven environmental constraints.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ML: Formal Analysis, Visualization, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Supervision. YZ: Formal Analysis, Visualization, Writing – original draft, Validation. TP: Formal Analysis, Writing – review & editing. YL: Investigation, Writing – review & editing. YH: Investigation, Writing – review & editing. WC: Investigation, Writing – review & editing. YY: Investigation, Writing – review & editing. GZ: Writing – review & editing, Formal Analysis, Visualization, Writing – original draft. SS: Writing – review & editing. MY: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (31902017, 32172672), Guangdong Basic and Applied Basic Research Foundation (2021A1515011020, 2019A1515110070), the Higher Education Department of Guangdong Province (Grant No. 2020KCXTD025), Special fund for scientific innovation strategy-construction of high-level Academy of Agriculture Science (R2022PY-QY002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1321952/full#supplementary-material
Supplementary Figure 1 | Amino acid sequence of predicted motif.
References
Arthikala, M.-K., Montiel, J., Sánchez-López, R., Nava, N., Cárdenas, L., Quinto, C. (2017). Respiratory burst oxidase homolog gene A is crucial for rhizobium infection and nodule maturation and function in common bean. Front. Plant Sci. 8, 2003. doi: 10.3389/fpls.2017.02003
Asai, S., Ohta, K., Yoshioka, H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20 (5), 1390–1406. doi: 10.1105/tpc.107.055855
Bostock, R. M., Pye, M. F., Roubtsova, T. V. (2014). Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Annu. Rev. Phytopathol. 52 (1), 517–549. doi: 10.1146/annurev-phyto-081211-172902
Chang, Y., Li, B., Shi, Q., Geng, R., Geng, S., Liu, J., et al. (2020). Comprehensive analysis of respiratory burst oxidase homologs (Rboh) gene family and function of gbRboh5/18 on verticillium wilt resistance in gossypium barbadense. Front. Genet. 11, 788. doi: 10.3389/fgene.2020.00788
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Humphreys, J. L., Ru, Y., He, Y., Wu, F., Mai, J., et al. (2022). Jasmonate signaling and remodeling of cell wall metabolism induced by boron deficiency in pea shoots. Environ. Exp. Bot. 201, 104947. doi: 10.1016/j.envexpbot.2022.104947
Chen, X., Ru, Y., Takahashi, H., Nakazono, M., Shabala, S., Smith, S. M., et al. (2023). Single-cell transcriptomic analysis of pea shoot development and cell-type-specific responses to boron deficiency. Plant J. doi: 10.1111/tpj.16487
Chen, Q., Yang, G. (2020). Signal function studies of ROS, especially RBOH-dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 39 (1), 157–171. doi: 10.1007/s00344-019-09971-4
Cheng, C., Xu, X., Gao, M., Li, J., Guo, C., Song, J., et al. (2013). Genome-wide analysis of respiratory burst oxidase homologs in grape (Vitis vinifera L.). Int. J. Mol. Sci. 14 (12), 24169–24186. doi: 10.3390/ijms141224169
Chung, J.-S., Zhu, J.-K., Bressan, R. A., Hasegawa, P. M., Shi, H. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53 (3), 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Delledonne, M., Murgia, I., Ederle, D., Sbicego, P. F., Biondani, A., Polverari, A., et al. (2002). Reactive oxygen intermediates modulate nitric oxide signaling in the plant hypersensitive disease-resistance response. Plant Physiol. Bioch. 40 (6), 605–610. doi: 10.1016/S0981-9428(02)01397-9
Drerup, M. M., Schlücking, K., Hashimoto, K., Manishankar, P., Steinhorst, L., Kuchitsu, K., et al. (2013). The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the arabidopsis NADPH oxidase RBOHF. Mol. Plant 6 (2), 559–569. doi: 10.1093/mp/sst009
Du, L., Jiang, Z., Zhou, Y., Shen, L., He, J., Xia, X., et al. (2023). Genome-Wide Identification and Expression Analysis of Respiratory Burst Oxidase Homolog (RBOH) Gene Family in Eggplant (Solanum melongena L.) under Abiotic and Biotic Stress. Genes 14 (9), 1665. doi: 10.3390/genes14091665
Duan, Z.-Q., Bai, L., Zhao, Z.-G., Zhang, G.-P., Cheng, F.-M., Jiang, L.-X., et al. (2009). Drought-stimulated activity of plasma membranenicotinamide adenine dinucleotide phosphate oxidase and its catalytic properties in rice. J. Integr. Plant Biol. 51 (12), 1104–1115. doi: 10.1111/j.1744-7909.2009.00879.x
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C.-P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. P. Natl. Acad. Sci. 110 (21), 8744–8749. doi: 10.1073/pnas.1221294110
Foyer, C. H., Hanke, G. (2022). ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J. 111 (3), 642–661. doi: 10.1111/tpj.15856
Groom, Q. J., Torres, M. A., Fordham-Skelton, A. P., Hammond-Kosack, K. E., Robinson, N. J., Jones, J. D. G. (1996). rbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 10 (3), 515–522. doi: 10.1046/j.1365-313X.1996.10030515.x
Gui, T.-Y., Gao, D.-H., Ding, H.-C., Yan, X.-H. (2022). Identification of respiratory burst oxidase homolog (Rboh) family genes from pyropia yezoensis and their correlation with archeospore release. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.929299
Han, J.-P., Köster, P., Drerup, M. M., Scholz, M., Li, S., Edel, K. H., et al. (2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221 (4), 1935–1949. doi: 10.1111/nph.15543
Hu, C.-H., Wei, X.-Y., Yuan, B., Yao, L.-B., Ma, T.-T., Zhang, P.-P., et al. (2018). Genome-wide identification and functional analysis of NADPH oxidase family genes in wheat during development and environmental stress responses. Front. Plant Sci. 9, 906. doi: 10.3389/fpls.2018.00906
Ishikawa, K., Tamura, K., Ueda, H., Ito, Y., Nakano, A., Hara-Nishimura, I., et al. (2018). Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER tubules. Plant Physiol. 178 (2), 641–653. doi: 10.1104/pp.18.00498
Jiménez-Quesada, M. J., Castro, A. J., Lima-Cabello, E., Alché, J. (2022). Cell localization of DPI-dependent production of superoxide in reproductive tissues of the olive tree (Olea europaea L.). Oxygen 2 (2), 79–90. doi: 10.3390/oxygen2020007
Jimenez-Quesada, M. J., Traverso, J. A., Potocký, M., Žárský, V., Alché, J. (2019). Generation of superoxide by oeRbohH, a NADPH oxidase activity during olive (Olea europaea L.) pollen development and germination. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01149
Kapli, P., Yang, Z., Telford, M. J. (2020). Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 21 (7), 428–444. doi: 10.1038/s41576-020-0233-0
Kaya, H., Nakajima, R., Iwano, M., Kanaoka, M. M., Kimura, S., Takeda, S., et al. (2014). Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 26 (3), 1069–1080. doi: 10.1105/tpc.113.120642
Knopkiewicz, M., Wojtaszek, P. (2019). Validation of reference genes for gene expression analysis using quantitative polymerase chain reaction in pea lines (Pisum sativum) with different lodging susceptibility. Anna Appl. Biol. 174 (1), 86–91. doi: 10.1111/aab.12475
Koroleva, O. A., Tomlinson, M. L., Leader, D., Shaw, P., Doonan, J. H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41 (1), 162–174. doi: 10.1111/j.1365-313X.2004.02281.x
Kreplak, J., Madoui, M.-A., Cápal, P., Novák, P., Labadie, K., Aubert, G., et al. (2019). A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 51 (9), 1411–1422. doi: 10.1038/s41588-019-0480-1
Kwak, J. M., Mori, I. C., Pei, Z.-M., Leonhardt, N., Torres, M. A., Dangl, J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 (11), 2623–2633. doi: 10.1093/emboj/cdg277
Lee, J., Han, M., Shin, Y., Lee, J. M., Heo, G., Lee, Y. (2023). How extracellular reactive oxygen species reach their intracellular targets in plants. Mol. Cells 46 (6), 329–336. doi: 10.14348/molcells.2023.2158
Lherminier, J., Elmayan, T., Fromentin, J., Elaraqui, K. T., Vesa, S., Morel, J., et al. (2009). NADPH oxidase-mediated reactive oxygen species production: subcellular localization and reassessment of its role in plant defense. Mol. Plant 22 (7), 868–881. doi: 10.1094/MPMI-22-7-0868
Li, X., Li, Y., Mai, J., Tao, L., Qu, M., Liu, J., et al. (2018). Boron alleviates aluminum toxicity by promoting root alkalization in transition zone via polar auxin transport. Plant Physiol. 177 (3), 1254–1266. doi: 10.1104/pp.18.00188
Li, H., Liu, S.-S., Yi, C.-Y., Wang, F., Zhou, J., Xia, X.-J., et al. (2014). Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 37 (12), 2768–2780. doi: 10.1111/pce.12360
Li, X., Ou, M., Li, L., Li, Y., Feng, Y., Huang, X., et al. (2023). The wall-associated kinase gene family in pea (Pisum sativum) and its function in response to B deficiency and Al toxicity. J. Plant Physiol. 287, 154045. doi: 10.1016/j.jplph.2023.154045
Li, J., Wang, X., Zhang, Y., Jia, H., Bi, Y. (2011). cGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots. Planta 234 (4), 709–722. doi: 10.1007/s00425-011-1439-3
Li, D., Wu, D., Li, S., Dai, Y., Cao, Y. (2019). Evolutionary and functional analysis of the plant-specific NADPH oxidase gene family in Brassica rapa L. R. Soc Open Sci. 6 (2), 181727. doi: 10.1098/rsos.181727
Liu, J., Lu, H., Wan, Q., Qi, W., Shao, H. (2019). Genome-wide analysis and expression profiling of respiratory burst oxidase homologue gene family in Glycine max. Environ. Exp. Bot. 161, 344–356. doi: 10.1016/j.envexpbot.2018.07.015
Liu, M., Pan, T., Allakhverdiev, S. I., Yu, M., Shabala, S. (2020). Crop halophytism: an environmentally sustainable solution for global food security. Trends Plant Sci. 25 (7), 630–634. doi: 10.1016/j.tplants.2020.04.008
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Marino, D., Andrio, E., Danchin, E. G. J., Oger, E., Gucciardo, S., Lambert, A., et al. (2011). A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189 (2), 580–592. doi: 10.1111/j.1469-8137.2010.03509.x
Miller, G., Mittler, R. (2023). “Plant NADPH oxidases,” in NADPH Oxidases Revisited: From Function to Structure. Ed. Pick, E. (Cham, Switzerland: Springer International Publishing), 445–465.
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16 (6), 300–309. doi: 10.1016/j.tplants.2011.03.007
Montiel, J., Nava, N., Cárdenas, L., Sánchez-López, R., Arthikala, M.-K., Santana, O., et al. (2012). A phaseolus vulgaris NADPH oxidase gene is required for root infection by rhizobia. Plant Cell Physiol. 53 (10), 1751–1767. doi: 10.1093/pcp/pcs120
Nakashima, A., Chen, L., Thao, N. P., Fujiwara, M., Wong, H. L., Kuwano, M., et al. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20 (8), 2265–2279. doi: 10.1105/tpc.107.054395
Noirot, E., Der, C., Lherminier, J., Robert, F., Moricova, P., Kiêu, K., et al. (2014). Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 65 (17), 5011–5022. doi: 10.1093/jxb/eru265
Overmyer, K., Brosché, M., Kangasjärvi, J. (2003). Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8 (7), 335–342. doi: 10.1016/S1360-1385(03)00135-3
Palmgren, M. G., Edenbrandt, A. K., Vedel, S. E., Andersen, M. M., Landes, X., Østerberg, J. T., et al. (2015). Are we ready for back-to-nature crop breeding? Trends Plant Sci. 20 (3), 155–164. doi: 10.1016/j.tplants.2014.11.003
Patel, R., Rinker, L., Peng, J., Chilian, W. M. (2018). “Reactive oxygen species: The good and the bad,” in Reactive oxygen species (ROS) in living cells. Ed. Albu, C. (IntechOpen, Rijeka, London), 216.
Postiglione, A. E., Muday, G. K. (2022). Abscisic acid increases hydrogen peroxide in mitochondria to facilitate stomatal closure. Plant Physiol. 192 (1), 469–487. doi: 10.1101/2022.01.11.475946
Rawat, N., Wungrampha, S., Singla-Pareek, S. L., Yu, M., Shabala, S., Pareek, A. (2022). Rewilding staple crops for the lost halophytism: Toward sustainability and profitability of agricultural production systems. Mol. Plant 15 (1), 45–64. doi: 10.1016/j.molp.2021.12.003
Romero-Puertas, M. C., Rodriquez-Serrano, M., Corpas, F. J., Gomez, M., Rio, D., L.A.andSandalio, L. M. (2004). Cadmium-induced subcellular accumulation of O2·– and H2O2 in pea leaves. Plant Cell Environ. 27 (9), 1122–1134. doi: 10.1111/j.1365-3040.2004.01217.x
Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J. W., et al. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16 (3), 616–628. doi: 10.1105/tpc.019398
Sagi, M., Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141 (2), 336–340. doi: 10.1104/pp.106.078089
Sandalio, L. M., Peláez-Vico, M. A., Molina-Moya, E., Romero-Puertas, M. C. (2021). Peroxisomes as redox-signaling nodes in intracellular communication and stress responses. Plant Physiol. 186 (1), 22–35. doi: 10.1093/plphys/kiab060
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 (7278), 178–183. doi: 10.1038/nature08670
Selvi, A., Devi, K., Manimekalai, R., Prathima, P. T. (2020). Comparative analysis of drought-responsive transcriptomes of sugarcane genotypes with differential tolerance to drought. 3 Biotech. 10 (6), 236. doi: 10.1007/s13205-020-02226-0
Song, C. J., Steinebrunner, I., Wang, X., Stout, S. C., Roux, S. J. (2006). Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 140 (4), 1222–1232. doi: 10.1104/pp.105.073072
Suzuki, N., Miller, G., Morales, J., Shulaev, V., Torres, M. A., Mittler, R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14 (6), 691–699. doi: 10.1016/j.pbi.2011.07.014
Takeda, S., Gapper, C., Kaya, H., Bell, E., Kuchitsu, K., Dolan, L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319 (5867), 1241–1244. doi: 10.1126/science.1152505
Torres, M. A., Dangl, J. L., Jones, J. D. G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. P. Natl. Acad. Sci. 99 (1), 517–522. doi: 10.1073/pnas.012452499
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., Jones, J. D. G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14 (3), 365–370. doi: 10.1046/j.1365-313X.1998.00136.x
Wang, W., Chen, D., Liu, D., Cheng, Y., Zhang, X., Song, L., et al. (2020). Comprehensive analysis of the Gossypium hirsutum L. respiratory burst oxidase homolog (Ghrboh) gene family. BMC Genomics 21 (1), 91. doi: 10.1186/s12864-020-6503-6
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2022). The conserved domain database in 2023. Nucleic Acids Res. 51 (D1), D384–D388. doi: 10.1093/nar/gkac1096
Wang, G.-F., Li, W.-Q., Li, W.-Y., Wu, G.-L., Zhou, C.-Y., Chen, K.-M. (2013). Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 14 (5), 9440–9458. doi: 10.3390/ijms14059440
Xia, X.-J., Gao, C.-J., Song, L.-X., Zhou, Y.-H., Shi, K., Yu, J.-Q. (2014). Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ. 37 (9), 2036–2050. doi: 10.1111/pce.12275
Xie, Y.-J., Xu, S., Han, B., Wu, M.-Z., Yuan, X.-X., Han, Y., et al. (2011). Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J. 66 (2), 280–292. doi: 10.1111/j.1365-313X.2011.04488.x
Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10 (2), 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamauchi, T., Yoshioka, M., Fukazawa, A., Mori, H., Nishizawa, N. K., Tsutsumi, N., et al. (2017). An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29 (4), 775–790. doi: 10.1105/tpc.16.00976
Yang, Y., Yuan, Z., Ning, C., Zhao, B., Wang, R., Zheng, X., et al. (2022). The pea R2R3-MYB gene family and its role in anthocyanin biosynthesis in flowers. Front. Genet. 13, 936051. doi: 10.3389/fgene.2022.936051
Zhang, Y., Li, Y., He, Y., Hu, W., Zhang, Y., Wang, X., et al. (2018). Identification of NADPH oxidase family members associated with cold stress in strawberry. FEBS Open Bio. 8 (4), 593–605. doi: 10.1002/2211-5463.12393
Zhang, J., Xie, Y., Ali, B., Ahmed, W., Tang, Y., Li, H. (2021). Genome-wide identification, classification, evolutionary expansion and expression of Rboh family genes in pepper (Capsicum annuum L.). Trop. Plant Biol. 14 (3), 251–266. doi: 10.1007/s12042-021-09286-3
Zhang, H., Zhu, J., Gong, Z., Zhu, J.-K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23 (2), 104–119. doi: 10.1038/s41576-021-00413-0
Keywords: reactive oxygen species, abiotic stress, plasma membrane, drought, salinity, cadmium, boron
Citation: Liu M, Zhang Y, Pan T, Li Y, Hong Y, Chen W, Yang Y, Zhao G, Shabala S and Yu M (2023) Genome-wide analysis of respiratory burst oxidase homolog gene family in pea (Pisum sativum L.). Front. Plant Sci. 14:1321952. doi: 10.3389/fpls.2023.1321952
Received: 15 October 2023; Accepted: 27 November 2023;
Published: 12 December 2023.
Edited by:
Dongmei Li, Shandong Agricultural University, ChinaReviewed by:
Yuan Huang, Huazhong Agricultural University, ChinaJuan de Dios Alché, Spanish National Research Council (CSIC), Spain
Copyright © 2023 Liu, Zhang, Pan, Li, Hong, Chen, Yang, Zhao, Shabala and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Yu, eXVtaW5AZm9zdS5lZHUuY24=; Sergey Shabala, c2VyZ2V5LnNoYWJhbGFAdXdhLmVkdS5hdQ==; Gangjun Zhao, emhhb2dhbmdqdW5AZ2RhYXMuY24=
 Minmin Liu1
Minmin Liu1 Yu Zhang
Yu Zhang Ting Pan
Ting Pan Gangjun Zhao
Gangjun Zhao Min Yu
Min Yu