
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 January 2024
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1320660
Haploid inducers are key components of doubled haploid (DH) technology in maize. Robust agronomic performance and better haploid induction ability of inducers are persistently sought through genetic improvement. We herein developed C1-I inducers enabling large-scale in vivo haploid induction of inducers and discovered superior inducers from the DH progenies. The haploid induction rate (HIR) of C1-I inducers ranged between 5.8% and 12.0%. Overall, the success rate of DH production was 13% on average across the 23 different inducer crosses. The anthesis–silking interval and days to flowering of inducer F1s are significantly correlated with the success rate of DH production (r = −0.48 and 0.47, respectively). Transgressive segregants in DH inducers (DHIs) were found for the traits (days to flowering, HIR, plant height, and total primary branch length). Moreover, the best HIR in DHIs exceeded 23%. Parental genome contributions to DHI progenies ranged between 0.40 and 0.55, respectively, in 25 and 75 percentage quantiles, and the mean and median were 0.48. The allele frequency of the four traits from inducer parents to DHI progenies did not correspond with the phenotypic difference between superior and inferior individuals in the DH populations by genome-wide Fst analysis. This study demonstrated that the recombinant DHIs can be accessed on a large scale and used as materials to facilitate the genetic improvement of maternal haploid inducers by in vivo DH technology.
Doubled haploid (DH) technology has become an established tool in modern plant breeding to rapidly produce homozygous lines. This is accomplished by producing haploid plants with only one set of chromosomes, followed by the doubling of their haploid genomes. DH technology offers numerous benefits, such as a more efficient selection process compared to selection among segregating families, and reducing the time and costs involved in inbred line development by repeated self-pollination. The homozygous nature of DH lines allows for uniform and consistent plant populations, enabling breeders to accurately evaluate the performance of different genetic materials in breeding programs.
Successful implementation of DH technology in maize breeding programs depends on the ability to produce haploids. This is achieved by using pollen from haploid inducers to pollinate the source germplasm from which DH lines will be developed (Chang and Coe, 2009; Chaikam et al., 2019b). The proportion of seeds with haploid embryos detected among the total seeds harvested on the source germplasm is referred to as the haploid induction rate (HIR) (Prigge et al., 2012a). Over the past 15–20 years, more advanced inducers with a high HIR (8%–10%) have been developed based on the original Stock 6 genotype and enabled the production of maize DH lines at a large scale (Geiger and Gordillo, 2009). Improved modern inducers include RWS, RWK-76, UH400, and UH600 (University of Hohenheim, Germany), the PHI series (Procera, Romania), CAU5 (China Agricultural University, China), the BHI series (Iowa State University, USA), and the TAIL series (Tropical Adapted Inducer Lines, CIMMYT) (Geiger and Gordillo, 2009; Rotarenco et al., 2010; Prigge et al., 2012b; Xu et al., 2013; Chaikam et al., 2018; Trentin et al., 2020; Trentin et al., 2023).
Genetic studies of HIRs revealed that it is a quantitative trait controlled by a few major quantitative trait loci (QTLs) along with minor QTLs (Barret et al., 2008; Prigge et al., 2012b). Two major QTLs, qhir1 and qhir8, were shown to be responsible for HIRs in QTL mapping studies (Prigge et al., 2012b; Xu et al., 2013). Later, the gene underlying qhir1 was cloned by three independent research groups simultaneously and called MATRILINEAL, ZmPHOSPHOLIPASE-A1, or NOT LIKE DAD (MTL/ZmPLA1/NLD) (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017). MTL/ZmPLA1/NLD is a pollen-specific gene encoding for a putative phospholipase. A 4-bp insertion mutation in MTL (we will use only this name for simplicity) causes a frameshift and a truncated protein. The presence of a single nucleotide change in the gene ZmDMP in qhir8 has been shown to increase HIRs two- to threefold when MTL is present. However, ZmDMP has a very low HIR (~0.15%) on its own (Zhong et al., 2019). The synergistic effect of mtl and zmdmp mutations suggests that interactions of gene functions after pollination contribute to a high HIR in modern inducers (Jacquier et al., 2020). Furthermore, a loss-of-function mutation of Zea mays PHOSPHOLIPASE D3 (ZmPLD3) induced by the CRISPR-Cas9 machinery showed synergistic effects, increasing HIRs threefold (from 1.19% to 4.13%) in the presence of mtl (Li et al., 2021). Pollen reactive oxygen species (ROS) bursts lead to sperm DNA fragmentation with an impact on haploid induction, as observed in the mtl genotype. In accordance with this finding, mutants of ZmPOD65 discovered as a peroxidase gene in the ROS class induced by CRISPR-Cas9 machinery contributed to the aborted kernel, and the HIRs were between 0.9% and 7.7% (Jiang et al., 2022). Additional genome regions and underlying candidate genes have been identified as putatively controlling HIRs in a genome-wide association study (e.g., Trentin et al., 2023). A novel gene in the Arabidopsis model plant, AtKPl (KOKOPELLI), which is only expressed in male gametophytes, has been discovered to play a role in single fertilization events to trigger maternal haploid induction (Jacquier et al., 2023). Although the biological mechanism of haploid induction in maize remains ambiguous, effective methods for accumulating favorable alleles of other genes affecting HIRs in fixed mtl genetic backgrounds are required for further genetic improvement of inducers and biological studies of haploid induction.
The most cost-effective way to sort haploids from diploids is visual selection at the seed level, followed by the seedling stage (Vanous et al., 2017; Dermail et al., 2023). Anthocyanin biosynthesis structural genes such as A1, A2, C2, Bz1, Bz2, and Pr1 are regulated by the MYB C1 (colored aleurone1)/Pl1 (purple plant1) and bHLH R1 (coloured1)/B1 (plant colour1) gene families. Since each member of these families has a tissue- or development-specific expression, the anthocyanin pigmentation pattern of a maize plant and kernel depends on the allelic constitution at the C1/Pl1 and R1/B1 loci. For example, C1 gene is responsible for anthocyanin development in the aleurone layer of the maize kernel, while Pl1 gene is associated with sun-independent anthocyanin pigmentation in plant tissues and the pericarp of the maize kernel. Likewise, R1 gene activates anthocyanin pigmentation of plant tissues and the aleurone layer in maize kernel, but B1 sun-dependently regulates plant color and is expressed in the pericarp of the maize kernel (Walker et al., 1995; Petroni et al., 2014). The most common haploid kernel identification marker is the purple crown coloration of endosperm and purple coleoptiles of embryos encoded by the dominant mutant allele R1-nj of the red color gene R1 (Nanda and Chase, 1966). Current inducers contain R1-nj gene for identifying donor haploids at the seed stage. In the aleurone, in which cells have one copy of the R1-nj allele, anthocyanin pigments are expressed and allow the identification of contaminations or self-pollinations without coloration. Within the fraction of kernels with a purple kernel crown (usually the majority in controlled induction crosses), haploids are identified by the absence of red coloration of the embryo (Prigge and Melchinger, 2012). The intensity and extent of pigmentation can vary depending on the donor’s and inducer’s genetic backgrounds. For example, in the presence of B1 and Pl1 color genes, R1-nj positively affects the pigmentation of the coleoptile, root color, and plant color. However, if C1-I gene is present in either donor or inducer, the pigmentation of the coleoptile and aleurone layer of maize kernels disappears because C1-I gene inhibits the R1-nj function epistatically in the biosynthesis of anthocyanin in maize kernels. While this is an impediment in regular induction crosses, inhibition of R1-nj by C1-I can be exploited to enable the identification of haploids in genetic backgrounds carrying R1-nj gene, such as inducers. If conventional inducers are crossed with inducers carrying C1-I gene, the resulting F1 (R1-nj × C1-I inducer cross) is expected to have no anthocyanin pigmentation of the embryo unless the embryo has the maternal haploid genotype.
The objectives of this study were to i) verify that indeed maternal inducer haploids can be produced by in vivo haploid induction using a C1-I inducer, ii) demonstrate that DH inducer (DHI) line development is feasible, iii) determine the relationship between traits of inducer F1s and the success rate of DHI production, iv) understand the relationship between HIRs and agronomic traits in DHI progeny populations, and v) evaluate the ability to accumulate favorable alleles from inducer parents in superior inducers through DH technology.
There are three stages in the DHI production pipeline (Figure 1). First, heterozygous inducer F1s used as donor parents were induced by a C1-I inducer for haploid induction. Second, putative haploid kernels expressing a purple color in the embryo need to be visually sorted. Third, haploid kernels are sown in the greenhouse, and haploid seedlings are injected with colchicine solution for genome doubling at the three-leaf stage (Vanous et al., 2017). Colchicine-treated haploids are subsequently transplanted in the field and self-pollinated to obtain DHI lines. The success rate reflects the percentage of transplanted haploids with seed sets leading to DHI lines.
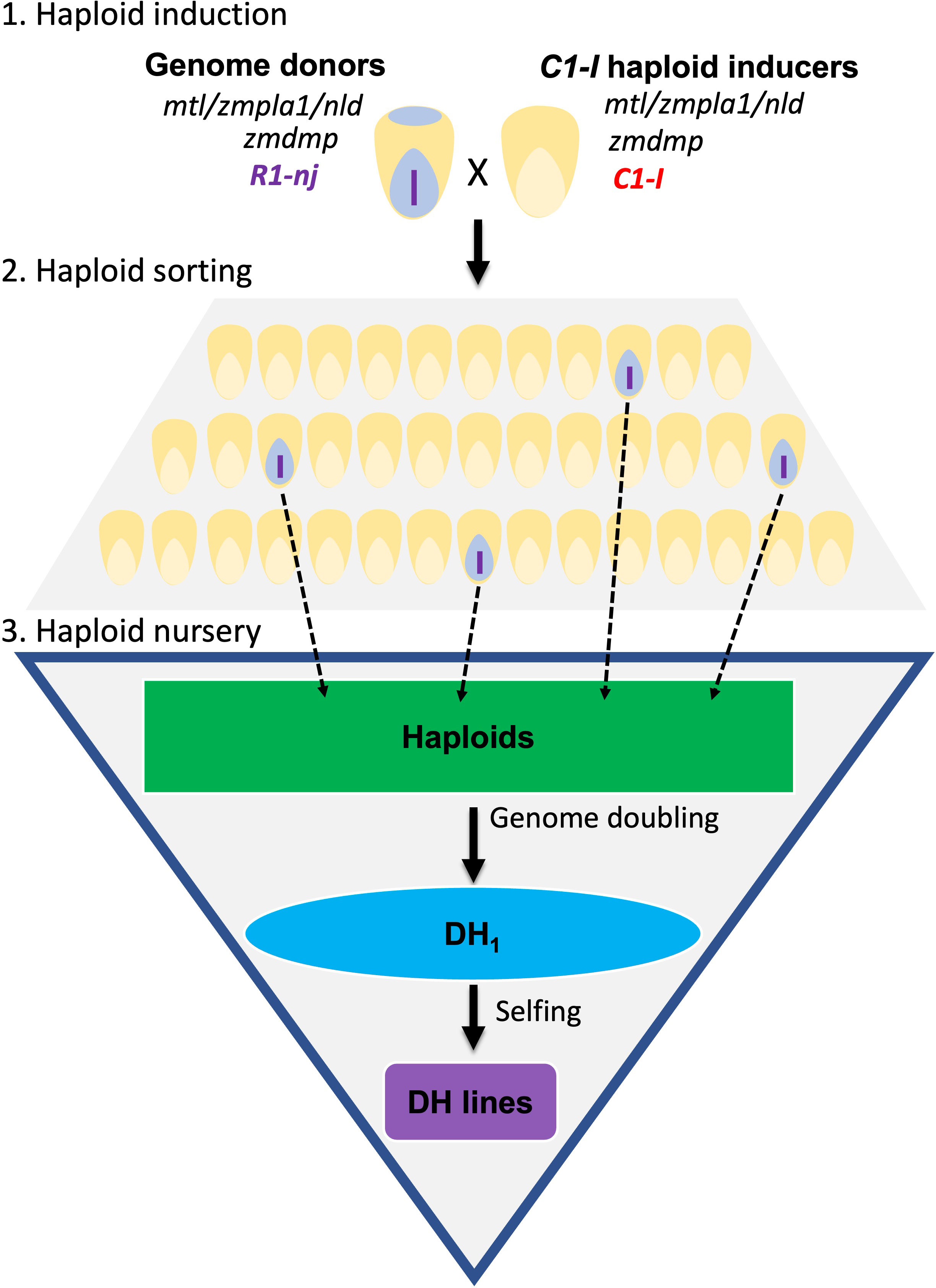
Figure 1 Doubled haploid (DH) technology for DH inducer line development. The diagram illustrates three main steps to obtain DH inducer (DHI) lines. Step 1: C1-I inducers were used as males to cross with regular inducer F1s for the haploid induction. Step 2: sorting the maternal haploids with the purple embryo expressed by R1-nj. Step 3: treating the inducer haploids with colchicine for genome doubling in greenhouse, then transplanting haploid seedlings to a field nursery, and finally obtaining the DHI lines after self-pollination.
The public line Mo47 served as the donor of the C1-I allele. Crosses with three different haploid inducer lines (RWS, BHI306, and BH201) were subsequently performed to combine haploid induction ability with the presence of the C1-I allele. Upon each cross with an inducer line, a generation of self-pollination followed to select ears fixed for the C1-I allele. The resulting families were continued by ear-to-row and selected for their agronomic performance and haploid induction ability in a genetic background fixed for the R1-nj allele. In this study, five F4 generation inducer genotypes were selected for their HIR performance and used for DHI line development.
Eight inducers were used to develop an elite-by-elite DHI population (nos. 1–8 in Table 1). These inducers were selected from the crosses of RWS/RWK76 and exPVP/public maize inbred lines, which were fixed for mtl and zmdmp genes. A total of 28 inducer F1s were created from the eight inducers by a half-diallel mating design in the summer of 2019 and evaluated for haploid induction ability and agronomic performance in the summer of 2020. B73- and PHG83-derived inducers were crossed with MHI (carrying mtl, but not zmdmp) to create elite-by-traditional (HIR< 5%) inducer combinations. The inducer F1s were crossed with C1-I inducers to obtain inducer haploids. Inducer haploids were subjected to the regular protocol of DH technology in maize (Vanous et al., 2017; Aboobucker et al., 2022) to obtain DHI lines. In addition, the HIRs of five C1-I inducers were evaluated by crossing them with nine inducer F1s and BHI306 and B73_R1-nj inbred (Maize Genetic Stock center: X17D), which were used as checks in the summer of 2020.
Phenotypes of inducer parents and DHI lines were determined in the summers of 2021 and 2022. However, phenotypes of the C1-I inducers and inducer F1s were only measured in the summer of 2020. A commercial F1 hybrid, Viking 60-01N (released by Albert Lee Seed Company, MN), was used as the donor for testing HIRs of inducers. Plant materials were planted in 3.8-m plots by randomized complete block design in two planting blocks at the Iowa State University Agricultural Engineering and Agronomy Farm in Boone, IA. A total of 537 genotypes were planted in the field for phenotypic measurement in 2021 and 2022. The entries were unbalanced but 42 DHI genotypes, and nine parent founders were common across two years. All trials were sown in loamy soil under rainfed conditions, adopting standard agronomic practices for research at Iowa State University.
Four phenotypic traits were evaluated. Plant height was used as a measure of the agronomic performance of the inducer line and traits influencing immediately the performance of a haploid inducer line: days to flowering, tassel branch length, and HIRs during haploid induction in the field. Days to flowering (DTF) was measured as 50% of plants per plot shedding pollen. Days to silking (DTS) was equivalent to 50% of plants in plots that were silking. The anthesis–silking interval (ASI) was the difference between DF and DS. Plant height was measured as the distance (cm) from the ground to the flag leaf’s base after pollen shedding. The total primary branch length (PBL) was used to represent the tassel size of inducers, which was measured as the cumulative length (cm) of all primary tassel branches. HIRs were calculated using the number of haploid kernels divided by the total number of seeds from at least five donor cobs induced by the inducer genotypes, which were harvested after the R6 stage. Suspicious haploid kernels were cut open using a scalpel to reveal the embryo color underneath the pericarp. Entries with fewer than 800 kernels for HIR evaluation were excluded to ensure high HIR data quality (Chaikam et al., 2018). After phenotypic measurements and data collection, the number of DHIs obtaining phenotypes in this study for DTF, HIRs, PHT, and PBL were 334, 331, 338, and 537, respectively.
Seeds of DHI lines were sown in the agronomy greenhouse at Iowa State University. Leaf tissue from three plants per entry was harvested, the tissue was lyophilized for 24 hours, and leaf samples were shipped to CIMMYT in El Batán, Texcoco, Mexico, for DNA extraction and single-nucleotide polymorphism (SNP) genotyping using DArTseq technology (Jaccoud et al., 2001). A total of 88,421 unimputed SNPs per line were successfully called and reported. Allele sequences were blasted against the B73 reference genome (B73 RefGen_v4) to obtain unique SNP positions to generate the HapMap files. SNP HapMap files were transformed into VCF files and numeric genotype files in TASSEL Version 5.0 (Bradbury et al., 2007). The missing SNP genotype imputation was conducted using Beagle 5.4 software (Browning et al., 2018). Also, minor allele frequencies of SNPs with less than 5% were filtered out. In total, 6636 SNPs across the genome remained and were used for calculating parental genome contribution and fixation index (Fst) in this study. The SNP genotypes of lines were coded as (−1 and 1), where 1 represents homozygosity for the major allele at a given bi-allelic locus and −1 indicates homozygosity for the minor allele.
The least-square mean of the trait performances (responses) in the field trial for genotypes was estimated by the following equation:
where yijk is the response for genotype i in block j nested in year k, μ is the overall mean, genotypei is the fixed effect of genotype, block(year)jk is the fixed effect for block, yeark is the fixed effect for a year, and εijk is the residual, which is independent and identically normal distributed N(0, σε2). The least-square mean of responses of genotypes, which are represented as best linear unbiased estimator (BLUE) values, were estimated using the emmeans package (Searle et al., 1980). To obtain broad-sense heritability (H2) on a plot basis, σg2/(σg2+σε2/r) was calculated, where r is the harmonic mean of replicates of DHIs (Holland et al., 2010). Genotype effects were treated as random to estimate σg2 using the lmer package (Bates et al., 2015).
P1 and P2 are considered the parental lines of the DHI progenies, and (x1, x2, xDHI) represents their SNP genotype matrix. The (m × 1)-dimensional column vector β of effects follows parental identity by descent (IBD) contribution to progeny considering only polymorphic loci between P1 and P2, as follows:
The mean of parental genome contributions in the DHI progeny was computed as
Fst was used to measure the level of genetic differentiation of genome-wide SNPs between two populations in this study, calculated using the gwscaR package (Flanagan and Jones, 2017). In order to understand genetic differentiation after applying DH technology in inducer line development, inducer individuals were classified into groups depending on the pedigree or phenotype. Based on trait performance, three DHI parents were chosen as reference lines. BHI306 was a relatively early flowering and short plant. A PHG83-derived inducer showed a relatively low HIR but had large tassels. A B73-derived inducer was late flowering, with high HIRs and a tall plant but small tassels (Table 1). In terms of the phenotype and genetic source difference of the reference lines, the DHI progenies from their F1 combinations were divided into three nested DHI populations, similar to the nested association mapping (NAM) mating structure. Genome-wide SNP Fst analysis was conducted between the group of DHI parents and their DHI progeny (i.e., based on pedigree) and the group of between phenotypically superior and inferior DHIs for every single trait of interest. The top and bottom two individuals were selected within each family (i.e., based on phenotype).
In the equation above, is the expected heterozygosity of a bi-allelic locus when the two populations are considered as one large meta-population. Moreover, is the average expected heterozygosity for each group within a pedigree-based or phenotype-based population. Fst values range from 0 (no genetic differentiation) to 1 (complete genetic differentiation).
Considering the genetic architecture of the traits of interest were not controlled by an infinitesimal (polygenic) model in this study, the quantile 99% (Q99) was chosen as the threshold to detect outlier Fst values of SNPs. SNPs that have extreme Fst values (Q99 SNPs) in pedigree-based population comparison are the most likely candidates to be affected by any factors and natural selections during the DH technology process. However, the Q99 SNPs in phenotype-based comparison are probably the genome region associated with the phenotypic difference. The sensitivity statistic was used to understand the ratio of the same SNPs detected in the Fst analysis based on both pedigree and phenotype. t-tests were used to test if the sensitivity was greater than zero and the mean differences between groups were equal to zero (null hypothesis) using the rstatix package.
The LS means of HIRs for the five C1-I inducers ranged from 5.8% to 12.0%, and HIRs differed significantly between C1-I inducers (Table 2). The inducer parents used in this study, which have the red root (Pl1) phenotypic marker for haploid sorting, are BHI306, Mo17-derived, A637-derived, and B84-derived inducers (Table 1). The pigmentation of kernels of inducer F1s after haploid induction by C1-I inducers showed that the pigmentation expression depended on the inducer F1 genotypes. Color pigmentation patterns are illustrated by the representative combinations (Figures 2A–E). The kernels of B73-derived/BHI306 and B73-derived/B84-derived inducer F1s after haploid induction by C1-I inducer had similar pigmentation patterns to the check B73_R1-nj inbred, which did not show purple-red endosperm pigmentations of the putative diploid. In contrast, the kernels of Mo17-derived/B84-derived inducer F1 had purple-red pigmentations on all of the kernels after induction, which were like induction outcomes of the check BHI306 inducer inbred. The putative haploids could still be sorted based on anthocyanin pigments in the scutellum. However, the pigmentation of the region around the embryo of the kernel increased the proportion of the suspicious haploids in haploid sorting.

Figure 2 Pigmentation of kernels induced by C1-I inducers and doubled haploid inducer (DHI) agronomic performance. (A) B73_R1-nj inbred donor (check1). (B) B73-derived inducer/BHI306 inducer F1 donor. (C) B73-derived inducer/B84-derived inducer F1 donor. (D) Mo17-derived inducer/B84-derived inducer F1 donor. (E) BHI306 inbred donor (check2). Light blue arrows point to haploids with clear anthocyanin pigmentation, while orange arrows point to suspicious haploids with vague purple-red pigmentation. The white bar represents 1 cm. (F) Large-scale inducer haploid plant nursery in the field for DHI production. (G) Phenotypic performance of DHI lines in trials.
Applying DH technology for inducers within two seasons allowed us to obtain 100% homozygous DHI populations with uniform agronomic performance (Figure 2G). The HIRs of C1-I inducers (genotypes A, B, and D) exceeded 8%, which enables large-scale DHI production (Figure 2F). Of inducer haploid seedlings treated with colchicine solution for genome doubling, 22% were shedding pollen, with a 13% success rate on average based on initially transplanted inducer haploids to obtain DHI lines in this study. The ASI of inducer F1 genome donors was significantly negatively correlated with the success rate (r = −0.48, p-value = 0.03), and the DTF of inducer F1 genome donors was significantly positively correlated with the success rate (r = 0.47, p-value = 0.03). There was no evidence that HIRs and PBL of inducer F1s were correlated with success rate in DHI production (Figure 3).
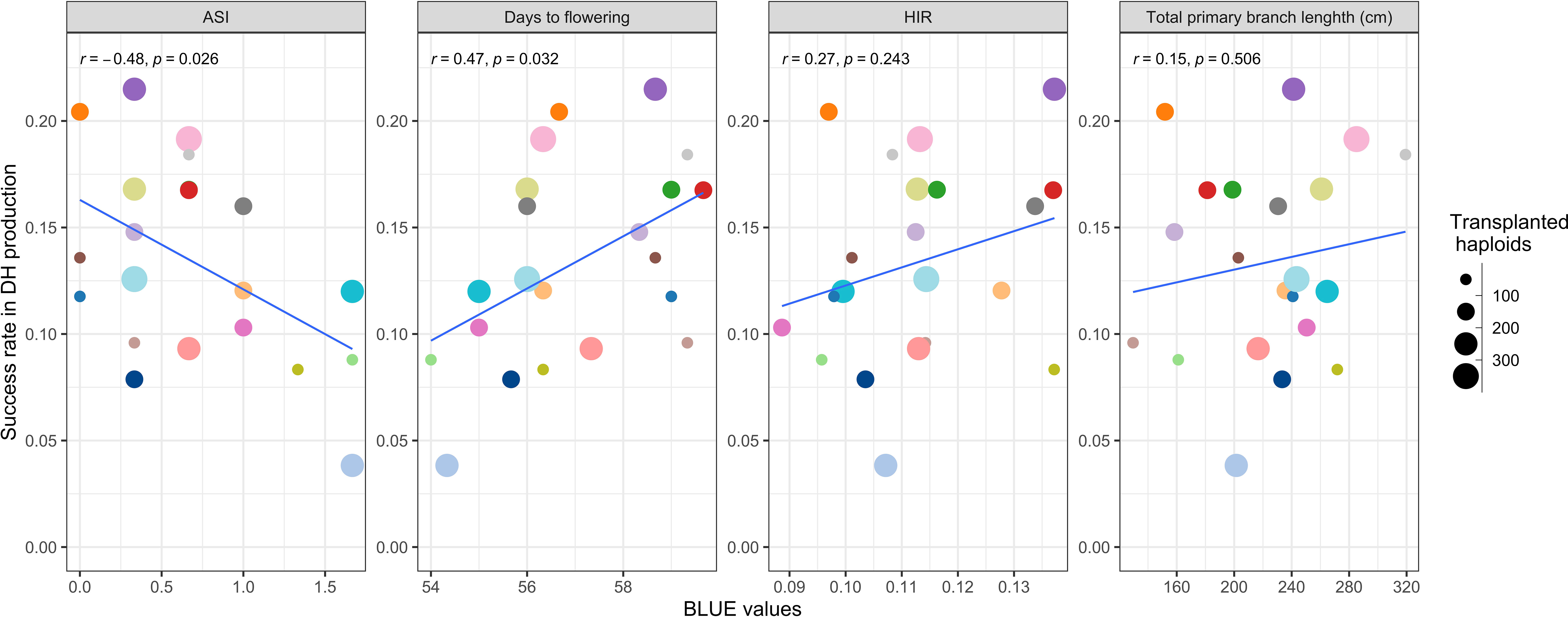
Figure 3 The relationship of the success rate of doubled haploid (DH) production between traits of different inducer F1 genome donors.
The major QTL alleles of haploid induction ability of the eight DHI parents derived from different exPVP/public genetic background lines were contributed by the RWS inducer, while the HIRs of DHI parents all exceeded 12%, and the other three agronomic traits varied among DHI parents (Table 1). The estimated mean of the four trait performances between the DHI parents and the sample of DHI progenies was not significantly different (Table 3). The ranges of maximum and minimum performances of the four traits in DHI progenies were wider than those of DHI parents (Table 3), indicating transgressive segregation (Figure 4). Moreover, the mean differences between inferior and superior groups of DHIs were significant (p< 0.05) for the four traits (Table 3).
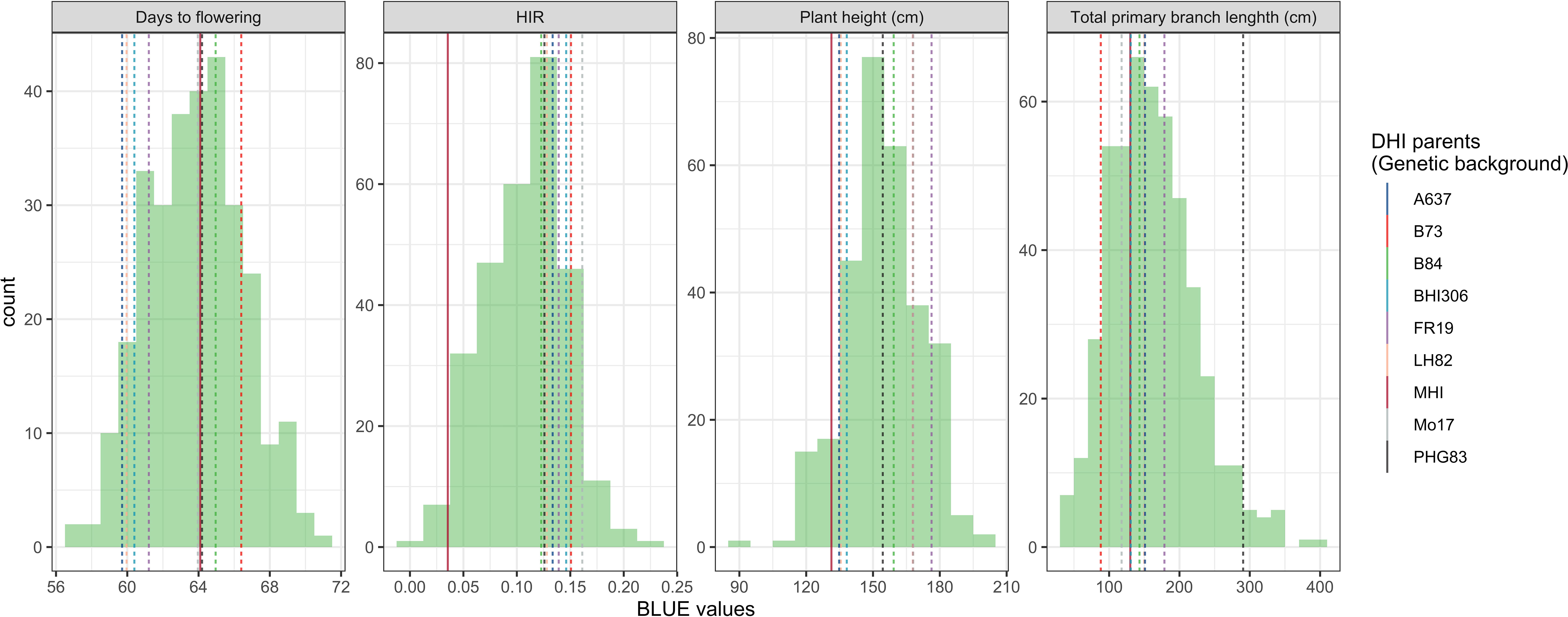
Figure 4 Phenotypic distribution of four traits in the doubled haploid inducer (DHI) sample population. The dashed lines represent the performances of the eight elite inducer parents, which had more than 10% haploid induction rate (HIR). The solid line represents the performances of traditional inducer (MHI).
The majority of DHIs obtained approximately equal genome contributions from both parents (Figure 5). The first quantile, mean/median, and third quantile of parental genome contribution were 0.40, 0.48, and 0.55, respectively. Most of the SNP outliers, which exceeded the 99% quantile (Q99 SNPs), were dependent on the population (Figure 6). This means that extraordinary individuals from the three NAM populations had different SNP genotypes associated with phenotypic differences. Comparing Q99 SNPs detected in both pedigree and phenotype groups, the mean sensitivity of Q99 SNPs was below 0.05 for the four traits (Table 3). There is, therefore, no evidence that DHI production was affected by a change in allele frequencies when considering the four traits between superior and inferior DHI lines (Figure 6).
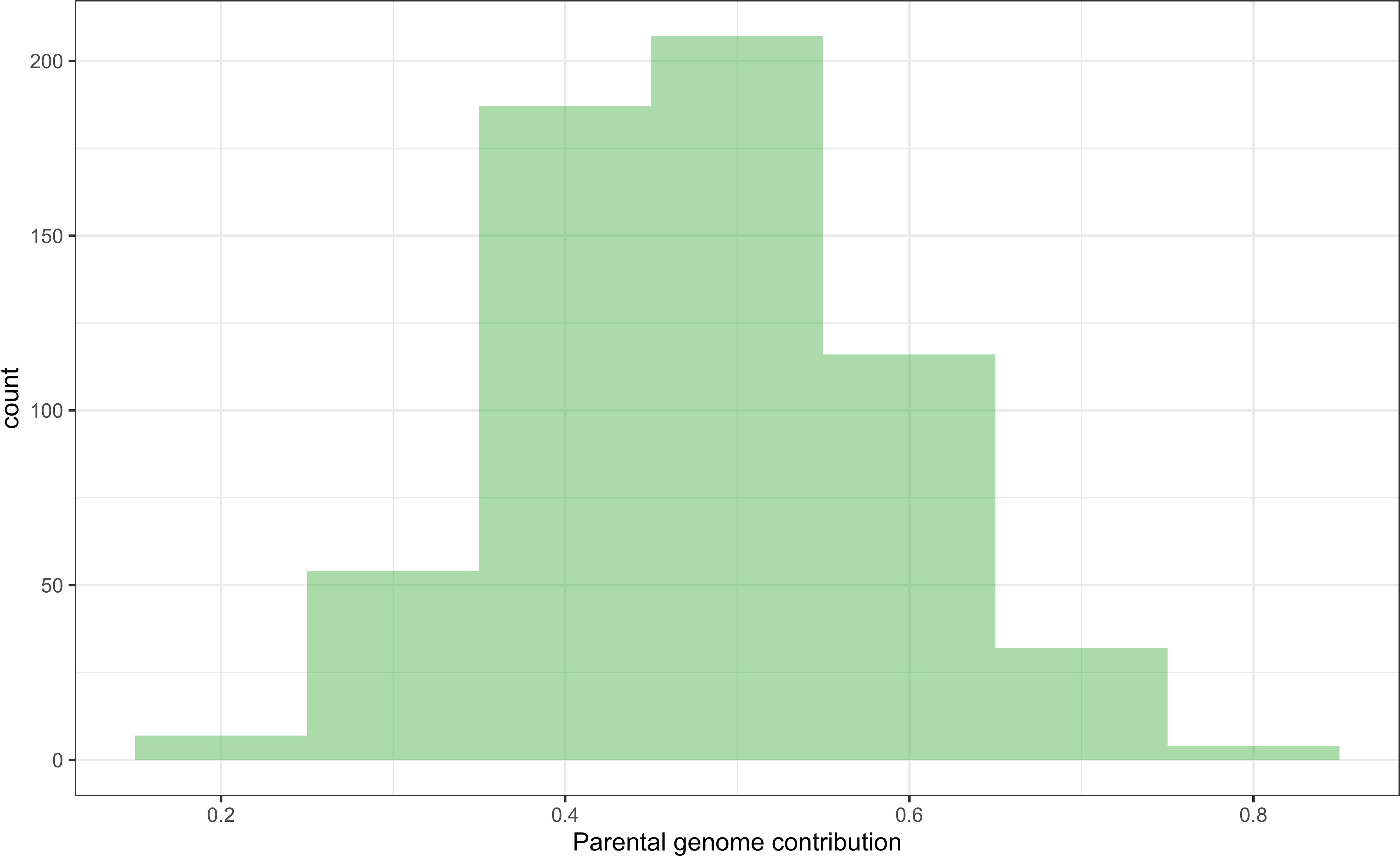
Figure 5 The distribution of parental genome contribution to doubled haploid inducers (DHIs) derived from the 23 biparental crosses.
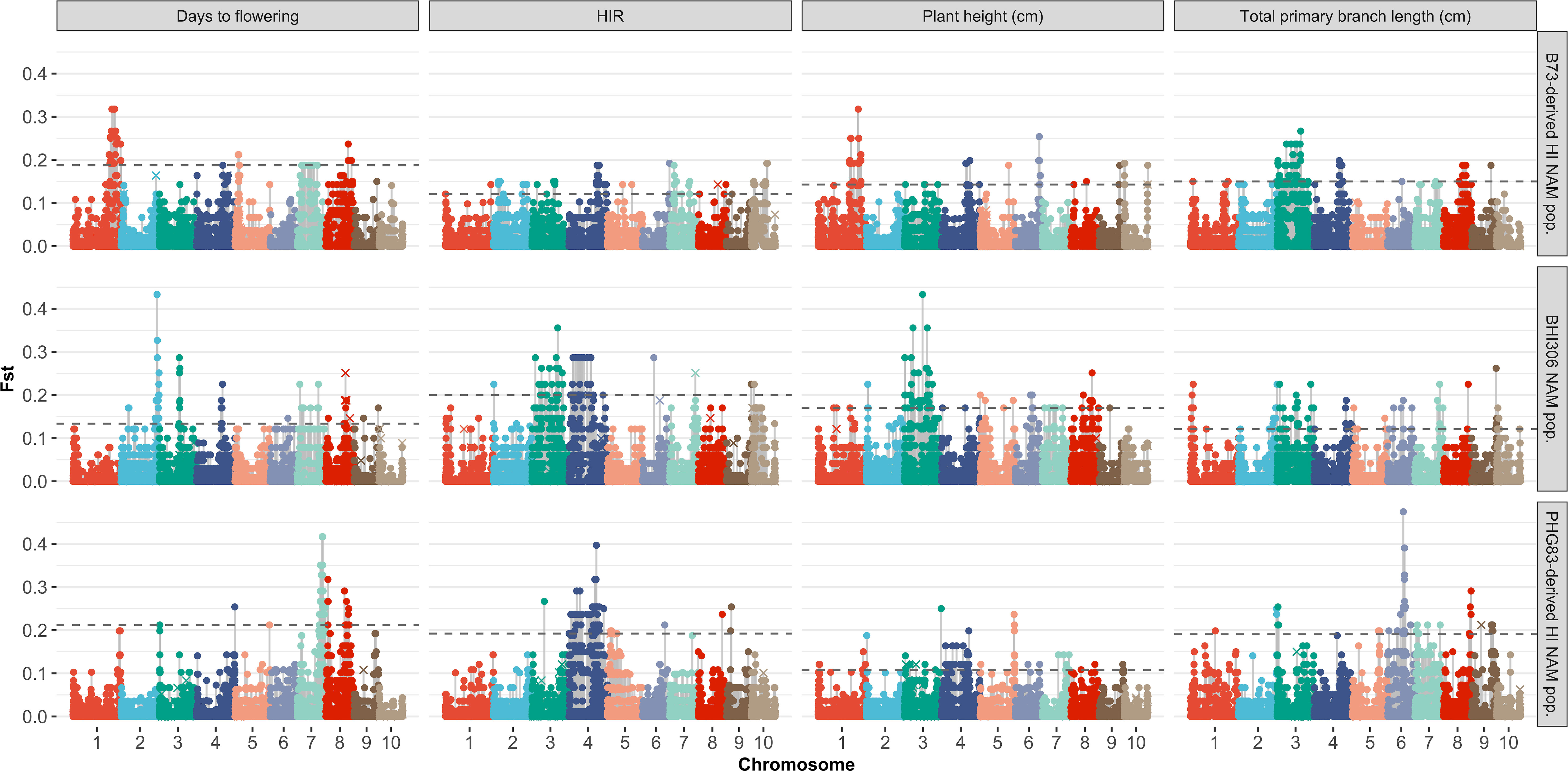
Figure 6 Genome-wide SNP Fst analysis of the traits of superior versus inferior doubled haploid inducers (DHIs) from three samples of inducer-derived nested association mapping (NAM) populations. The horizontal dash line was the Q99 threshold used to detect the outlier Fst SNPs. The × symbol represents Q99 outlier Fst SNPs detected from the groups of parents and their respective derived DHI progenies. The three lines chosen as the reference lines in the NAM samples were BHI306, which had relatively early flowering and short plant; PHG83-derived inducer, which had relatively low haploid induction rate (HIR) but big tassels; and B73-derived inducer, which had late flowering, relatively high HIR, and tall plant but small tassels.
Chaikam et al. (2018) observed that crosses of haploid inducer lines with agronomical superior and environmentally well-adapted non-inducer lines not only improved the agronomic performance of the existing inducer but also had a positive effect on HIRs. The DHI parents in this study were derived from crosses between the haploid inducer line RWS and several ex-PVP/public non-inducer inbreds. Transgressive segregants were detected for all four quantitative traits including HIRs, when samples of more than 50 inducer haploid plants were transplanted per cross (Figures 3, 4). Moreover, the best DHI progenies from the parental crosses (PHG83-derived/BHI306 and PHG83-derived/Mo17-derived) with an HIR > 13.9% on average outperformed their parental lines with induction rates exceeding 20% (Figure 4). The majority of parental genome contributions to DHIs ranged from 0.40 to 0.55. In conclusion, elite × elite inducer crosses appear promising to derive even better performing DHIs.
The DHI lines in this study were obtained from maternal haploids, which had the mtl and zmdmp alleles fixed. Thus, other genes must be responsible for increasing haploid induction ability in our DHIs. Those could be the more recently identified HIR genes (Li et al., 2021; Jiang et al., 2022; Jacquier et al., 2023). However, Trentin et al. (2023) and others have shown that there is additional genetic variation available for HIRs, in addition to the four identified genes to date. Thus, it appears promising to combine DHI technology with genomic selection, while accumulating known HIR genes, to develop superior maternal haploid inducers in the future, with HIRs >20% (observed in this study) or even >25%.
DHIs in this study were obtained from the induction of inducer F1s (Figure 1). While only a single recombination happens in the F1 generation, it still resulted in significant phenotypic mean differences between superior and inferior DH line families (Table 3). This suggests that obtaining DHIs from inducer F1 generation is sufficient to create substantial phenotypic variation for selection. This is consistent with Sleper and Bernardo (2016), who reported that superior DHs can be obtained from inducing F1 rather than F2 generations.
There is a continuous need for further improved haploid inducers to enhance the efficiency of DH technology in maize overall and to adapt haploid inducers to different environments (Trentin et al., 2020; Dermail et al., 2021). Applying DH technology not only shortens the time needed for inducer line development but also allows for more efficient genotypic selection for qualitative traits using haploid plants or DHIs, rather than segregating inducer families (Lübberstedt and Frei, 2012). For example, color appearance traits controlled by B1 and Pl1 anthocyanin regulatory genes, common rust, and leaf blight major resistance genes (Prigge et al., 2012a) are straightforward to be observed and selected for in the haploid nursery (Figure 2).
The overall success rate for DH production ranges from 10% to 22% for Dent Corn using artificial genome doubling by colchicine (Melchinger et al., 2016). The success rate of DH production in the haploid inducer nursery ranged from 3.8% to 21.5%, with 13.3% on average (Figure 3). Thus, the efficiency of colchicine application for artificial genome doubling in Dent Corn and inducers was similar. A negative correlation between the success rate of DH production and ASI of inducer F1s was observed. ASI of lines is an important indicator to assess the ability of flowering and nicking in water-deficit environments (Ribaut et al., 1996; Sah et al., 2020). Selecting closer ASIs of inducer F1s has the potential to increase the success rate of DH production and obtain DHI progenies with better drought tolerance.
The DHI line development in this study depends on the selection of R1-nj expression on the kernels of female parents (Figure 1). When using non-inducer inbreds as genetic sources to improve the inducer breeding population, mtl, zmdmp, and R1-nj genes need to be fixed in the source germplasm. Moreover, inducers with purple-red kernels used as females in F1 crosses tend to negatively affect haploid identification after induction (Figure 2) because B1 Pl1 genes induce anthocyanin synthesis in the pericarp of purple corn (Petroni et al., 2014). In contrast, inducers with white/yellow kernels used as the female parent in F1 crosses enabled clear haploid identification (Figure 2), reducing efforts in haploid sorting and rogueing false-positive haploid plants in the haploid nursery.
The ability and efficiency to obtain DH lines from haploids influenced the utilization of DH technology. One major constraint is the proportion of haploid plants with restored male fertility after successful artificial genome doubling. QTL-enhancing haploid spontaneous genome doubling (SHGD) ability has been discovered (Chaikam et al., 2019a; Ren et al., 2020; Trampe et al., 2020; Verzegnazzi et al., 2021). Introgressing the desirable SHGD allele into inducer backgrounds could improve the proportion of haploid male fertility for obtaining more DH lines. Moreover, the high HIR performance of inducers increases the probability of haploids by self-induction, which would have a negative effect on the available pollen for haploid induction and line maintenance. Therefore, introgressing the desirable alleles from QTLs associated with SHGD into inducers is a target to further improve DHI line development by DH technology. Moreover, haploid inducibility of donors has been found to influence haploid induction rates (De La Fuente et al., 2018; Ren et al., 2022; Trampe et al., 2022; Trentin et al., 2022). Inadvertent selection likely increases haploid inducibility gradually when applying in vivo haploid induction for DHI line development over multiple selection cycles.
The data for this study can be found online: https://doi.org/10.25380/iastate.24527653.v1. The haploid inducers in this study could be accessed from Doubled Haploid Facility at Iowa State University.
YC: Conceptualization, Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft. TL: Funding acquisition, Project administration, Supervision, Writing – review & editing. UF: Conceptualization, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by USDA’s National Institute of Food and Agriculture (grant numbers: IOW04714, IOW05520; IOW05510; IOW05656; NIFA award 2018-51181-28419 and 2020-51300-32180), US. Department of Agriculture, Agricultural Research Service, the Iowa State University Plant Sciences Institute, Iowa State University Crop Bioengineering Center, R.F. Baker Center for Plant Breeding, and the K.J. Frey Chair in Agronomy at Iowa State University.
We thank Dr. Jianming Yu and Dr. William Beavis for helping us improve the content of the manuscript. We also thank Elizabeth Bovenmyer, Iris Ma, Blessing Ngara, Smit Muttha, Abil Dermail, Rakumar Zunjare, Alice Santana, and Alison Uberti for their assist work in haploid induction, sorting, and nursery.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aboobucker, S. I., Jubery, T. Z., Frei, U. K., Chen, Y. R., Foster, T., Ganapathysubramanian, B., et al. (2022). Protocols for in vivo doubled haploid (DH) technology in maize breeding: from haploid inducer development to haploid genome doubling. Methods Mol. Biol. 2484, 213–235. doi: 10.1007/978-1-0716-2253-7_16
Barret, P., Brinkmann, M., Beckert, M. (2008). A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 117, 581–594. doi: 10.1007/S00122-008-0803-6/FIGURES/6
Bates, D., Mächler, M., Bolker, B. M., Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw 67, 1–48. doi: 10.18637/JSS.V067.I01
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/BIOINFORMATICS/BTM308
Browning, B. L., Zhou, Y., Browning, S. R (2018). A one-penny imputed genome from next generation reference panels. Am. J. Hum. Genet 103 (3), 338–348. doi: 10.1016/j.ajhg.2018.07.015
Chaikam, V., Gowda, M., Nair, S. K., Melchinger, A. E., Boddupalli, P. M. (2019a). Genome-wide association study to identify genomic regions influencing spontaneous fertility in maize haploids. Euphytica 215, 1–14. doi: 10.1007/s10681-019-2459-5
Chaikam, V., Molenaar, W., Melchinger, A. E., Boddupalli, P. M. (2019b). Doubled haploid technology for line development in maize: technical advances and prospects. Theor. Appl. Genet. 132, 3227–3243. doi: 10.1007/S00122-019-03433-X
Chaikam, V., Nair, S. K., Martinez, L., Lopez, L. A., Utz, H. F., Melchinger, A. E., et al. (2018). Marker-assisted breeding of improved maternal haploid inducers in maize for the tropical/subtropical regions. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01527
Chang, M. T., Coe, E. H. (2009). Doubled haploids. Biotechnol. Agric. Forestry 63, 127–142. doi: 10.1007/978-3-540-68922-5_10/COVER
De La Fuente, G. N., Frei, U. K., Trampe, B., Nettleton, D., Zhang, W., Lübberstedt, T. (2018). A diallel analysis of a maize donor population response to in vivo maternal haploid induction: I. inducibility. Crop Sci. 58, 1830–1837. doi: 10.2135/cropsci2017.05.0285
Dermail, A., Chankaew, S., Lertrat, K., Lübberstedt, T., Suriharn, K. (2021). Selection gain of maize haploid inducers for the tropical savanna environments. Plants 10, 2812. doi: 10.3390/PLANTS10122812/S1
Dermail, A., Lübberstedt, T., Suwarno, W. B., Chankaew, S., Lertrat, K., Ruanjaichon, V., et al (2023). Combining ability of tropical× temperate maize inducers for haploid induction rate, R1-nj seed set, and agronomic traits. Front. in Plant Sci. 14, 1154905. doi: 10.3389/fpls.2023.1154905
Flanagan, S. P., Jones, A. G. (2017). Genome-wide selection components analysis in a fish with male pregnancy. Evol. (N Y) 71, 1096–1105. doi: 10.1111/EVO.13173
Geiger, H. H., Gordillo, G. A. (2009). Doubled haploids in hybrid maize breeding. Maydica 54, 485–499.
Gilles, L. M., Khaled, A., Laffaire, J.-B., Chaignon, S., Gendrot, G., Laplaige, J., et al. (2017). Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717. doi: 10.15252/EMBJ.201796603
Holland, J. B., Nyquist, W. E., Cervantes-Martínez, C. T. (2010). Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev. 9–112. doi: 10.1002/9780470650202.CH2
Jaccoud, D., Peng, K., Feinstein, D., Kilian, A. (2001). Diversity Arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 29, e25–e25. doi: 10.1093/NAR/29.4.E25
Jacquier, N. M. A., Calhau, A. R. M., Fierlej, Y., Martinant, J.-P., Rogowsky, P. M., Gilles, L. M., et al. (2023). In planta haploid induction by kokopelli mutants. Plant Physiol. 193, 182–185. doi: 10.1093/PLPHYS/KIAD328
Jacquier, N. M. A., Gilles, L. M., Pyott, D. E., Martinant, J.-P., Rogowsky, P. M., Widiez, T. (2020). Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 6, 610–619. doi: 10.1038/s41477-020-0664-9
Jiang, C., Sun, J., Li, R., Yan, S., Chen, W., Guo, L., et al. (2022). A reactive oxygen species burst causes haploid induction in maize. Mol. Plant 15, 943–955. doi: 10.1016/J.MOLP.2022.04.001
Kelliher, T., Starr, D., Richbourg, L., Chintamanani, S., Delzer, B., Nuccio, M. L., et al. (2017). MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542 (7639), 105–109. doi: 10.1038/nature20827
Li, Y., Lin, Z., Yue, Y., Zhao, H., Fei, X., Lizhu, E., et al. (2021). Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 7 (12), 1579–1588. doi: 10.1038/s41477-021-01037-2
Liu, C., Li, X., Meng, D., Zhong, Y., Chen, C., Dong, X., et al. (2017). A 4-bp insertion at zmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10, 520–522. doi: 10.1016/J.MOLP.2017.01.011
Lübberstedt, T., Frei, U. K. (2012). Application of doubled haploids for target gene fixation in backcross programmes of maize. Plant Breed. 131, 449–452. doi: 10.1111/J.1439-0523.2011.01948.X
Melchinger, A. E., Molenaar, W. S., Mirdita, V., Schipprack, W. (2016). Colchicine alternatives for chromosome doubling in maize haploids for doubled-haploid production. Crop Sci. 56, 559–569. doi: 10.2135/CROPSCI2015.06.0383
Nanda, D. K., Chase, S. S. (1966). An embryo marker for detecting monoploids of maize (Zea mays L.)1. Crop Sci. 6, 213–215. doi: 10.2135/CROPSCI1966.0011183X000600020036X
Petroni, K., Pilu, R., Tonelli, C. (2014). Anthocyanins in corn: a wealth of genes for human health. Planta 240, 901–911. doi: 10.1007/S00425-014-2131-1/FIGURES/4
Prigge, V., Melchinger, A. E. (2012). Production of haploids and doubled haploids in maize. Methods Mol. Biol. 877, 161–172. doi: 10.1007/978-1-61779-818-4_13
Prigge, V., Schipprack, W., Mahuku, G., Atlin, G. N., Melchinger, A. E. (2012a). Development of in vivo haploid inducers for tropical maize breeding programs. Euphytica 185, 481–490. doi: 10.1007/s10681-012-0657-5
Prigge, V., Xu, X., Li, L., Babu, R., Chen, S., Atlin, G. N., et al. (2012b). New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 190, 781–793. doi: 10.1534/GENETICS.111.133066
Ren, J., Boerman, A., Liu, R., Wu, P., Trampe, B., Vanous, K., et al. (2020). Mapping of QTL and identification of candidate genes conferring spontaneous haploid genome doubling in maize (Zea mays L.). Plant Sci. 293, 110337. doi: 10.1016/j.plantsci.2019.110337
Ren, J., Zhang, X., Li, Z., Wu, P. (2022). Genetic analysis of maternal haploid inducibility for in vivo haploid induction in maize. Agriculture 12, 845. doi: 10.3390/AGRICULTURE12060845
Ribaut, J. M., Hoisington, D. A., Deutsch, J. A., Jiang, C., Gonzalez-De-Leon, D. (1996). Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis-silking interval. Theor. Appl. Genet. 92, 905–914. doi: 10.1007/BF00221905/METRICS
Rotarenco, V., Dicu, G., State, D., Fuia, S. (2010). New inducers of maternal haploids in maize. Maize Genet. Cooperation Newslett. 84, 21–22. Available at: https://mnl.maizegdb.org/84/HTML/15rotarenco.htm.
Sah, R. P., Chakraborty, M., Prasad, K., Pandit, M., Tudu, V. K., Chakravarty, M. K., et al. (2020). Impact of water deficit stress in maize: Phenology and yield components. Sci. Rep. 10 (1), 1–15. doi: 10.1038/s41598-020-59689-7
Searle, S. R., Speed, F. M., Milliken, G. A. (1980). Population marginal means in the linear model: An alternative to least squares means. Am. Statistician 34, 216–221. doi: 10.1080/00031305.1980.10483031
Sleper, J. A., Bernardo, R. (2016). Recombination and genetic variance among maize doubled haploids induced from F1 and F2 plants. Theor. Appl. Genet. 129, 2429–2436. doi: 10.1007/S00122-016-2781-4/FIGURES/1
Trampe, B., Batîru, G., Pereira da Silva, A., Frei, U. K., Lübberstedt, T. (2022). QTL mapping for haploid inducibility using genotyping by sequencing in maize. Plants 11, 878. doi: 10.3390/PLANTS11070878
Trampe, B., dos Santos, I. G., Frei, U. K., Ren, J., Chen, S., Lübberstedt, T. (2020). QTL mapping of spontaneous haploid genome doubling using genotyping-by-sequencing in maize (Zea mays L.). Theor. Appl. Genet. 133, 2131–2140. doi: 10.1007/s00122-020-03585-1
Trentin, H. U., Batîru, G., Frei, U. K., Dutta, S., Lübberstedt, T. (2022). Investigating the effect of the interaction of maize inducer and donor backgrounds on haploid induction rates. Plants 11, 1527. doi: 10.3390/PLANTS11121527/S1
Trentin, H. U., Dalsente Krause, M., Zunjare, R. U., Almeida, V. C., Peterlini, E., Rotarenco, V., et al. (2023). Genetic basis of maize maternal haploid induction beyond MATRILINEAL and ZmDMP. Front. Plant Sci. 14. doi: 10.3389/FPLS.2023.1218042
Trentin, H. U., Frei, U. K., Lübberstedt, T. (2020). Breeding maize maternal haploid inducers. Plants 9, 614. doi: 10.3390/PLANTS9050614
Vanous, K., Vanous, A., Frei, U. K., Lübberstedt, T. (2017). Generation of Maize (Zea mays) Doubled Haploids via Traditional Methods. Curr. Protoc. Plant Biol. 2, 147–157. doi: 10.1002/CPPB.20050
Verzegnazzi, A. L., dos Santos, I. G., Krause, M. D., Hufford, M., Frei, U. K., Campbell, J., et al. (2021). Major locus for spontaneous haploid genome doubling detected by a case–control GWAS in exotic maize germplasm. Theor. Appl. Genet. 134, 1423–1434. doi: 10.1007/S00122-021-03780-8/FIGURES/7
Walker, E. L., Robbins, T. P., Bureau, T. E., Kermicle, J., Dellaporta, S. L. (1995). Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 14, 2350–2363. doi: 10.1002/J.1460-2075.1995.TB07230.X
Xu, X., Li, L., Dong, X., Jin, W., Melchinger, A. E., Chen, S. (2013). Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J. Exp. Bot. 64, 1083–1096. doi: 10.1093/JXB/ERS393
Keywords: doubled haploids, haploid inducers, in vivo induction, line development, transgressive segregation
Citation: Chen Y-R, Lübberstedt T and Frei UK (2024) Development of doubled haploid inducer lines facilitates selection of superior haploid inducers in maize. Front. Plant Sci. 14:1320660. doi: 10.3389/fpls.2023.1320660
Received: 12 October 2023; Accepted: 11 December 2023;
Published: 05 January 2024.
Edited by:
Xiwen Cai, United States Department of Agriculture, United StatesReviewed by:
Vignesh Muthusamy, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2024 Chen, Lübberstedt and Frei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ursula K Frei, dWZyZWlAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.