
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 January 2024
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1306491
This article is part of the Research TopicPlant-Rhizobia Symbiosis and Nitrogen Fixation in LegumesView all 12 articles
The model legume Medicago truncatula establishes a symbiosis with soil bacteria (rhizobia) that carry out symbiotic nitrogen fixation (SNF) in plant root nodules. SNF requires the exchange of nutrients between the plant and rhizobia in the nodule that occurs across a plant-derived symbiosome membrane. One iron transporter, belonging to the Vacuolar iron Transporter-Like (VTL) family, MtVTL8, has been identified as essential for bacteria survival and therefore SNF. In this work we investigated the spatial expression of MtVTL8 in nodules and addressed whether it could be functionally interchangeable with a similar nodule-expressed iron transporter, MtVTL4. Using a structural model for MtVTL8 and the previously hypothesized mechanism for iron transport in a phylogenetically-related Vacuolar Iron Transporter (VIT), EgVIT1 with known crystal structure, we identified critical amino acids and obtained their mutants. Mutants were tested in planta for complementation of an SNF defective line and in an iron sensitive mutant yeast strain. An extended phylogenetic assessment of VTLs and VITs showed that amino acids critical for function are conserved differently in VTLs vs. VITs. Our studies showed that some amino acids are essential for iron transport leading us to suggest a model for MtVTL8 function, one that is different for other iron transporters (VITs) studied so far. This study extends the understanding of iron transport mechanisms in VTLs as well as those used in SNF.
Symbiotic nitrogen fixation (SNF) in legumes uses energy derived from photosynthesis to reduce N2 gas to ammonia at normal temperature and pressure, and is thus, especially important for sustainable food production. Legumes form a symbiosis with bacteria called rhizobia, which are hosted in specialized root organs called nodules (Hirsch et al., 1992; Oldroyd et al., 2011). These form after a series of interactions between the legume root cells and rhizobia which results in the reprogramming and division of plant cortical cells to form nodule primordia. These primordial cells are colonized by rhizobia in infection threads that deliver the rhizobia to the cells (Oldroyd, 2013; Roy et al., 2020). Rhizobia enter the plant cells in a process resembling endocytosis in which rhizobia are surrounded by a plant-derived symbiosome membrane (SM), through which all plant-microbe nutrient exchange occurs (Udvardi and Poole, 2013). The internalized rhizobia surrounded by the SM and the symbiosome space form symbiosomes, novel organelle-like structures. Rhizobia within symbiosomes, now called bacteroids, grow and divide until tens of thousands fill the infected cells, which increase in ploidy depending on the legume species (Maróti and Kondorosi, 2014). The outer cells of nodules form a gas diffusion barrier resulting in a hypoxic nodule interior (Witty et al., 1987) protecting the rhizobially-encoded oxygen-labile nitrogenase enzyme. Paradoxically, high rates of respiration in nodules are required by rhizobia as well as plant mitochondria during SNF. Respiration is supported by the rapid binding and delivery of oxygen by leghemoglobin, lowering oxygen concentrations further in the nodule interior (Appleby, 1984; Ott et al., 2005).
Many legumes, like the model plant Lotus japonicus and soybean, form round, determinate nodules that only have a transient meristem. Others, like the model Medicago truncatula and pea, form oblong, indeterminate nodules with a persistent meristem. Both nodule types have a central zone of rhizobia infected cells interspersed with uninfected cells (Hirsch, 1992). Indeterminate nodules contain cells in a developmental gradient classified into zones. Zone I (ZI) is the meristem; zone II (ZII) is the invasion zone where rhizobia enter plant cells, divide and differentiate; the interzone (IZ) is where final maturation occurs; and zone III (ZIII) is where SNF takes place. Older nodules also have a senescent zone, zone IV (IV), which likely is involved in nutrient recycling (Vasse et al., 1990). Each zone has characteristic gene expression (Limpens et al., 2013; Roux et al., 2014).
The transition metal iron is a key essential nutrient required for SNF (Clarke et al., 2014; González-Guerrero et al., 2014; Day and Smith, 2021; González-Guerrero et al., 2023) and it accumulates to higher concentrations in nodules than other plant organs (Burton et al., 1998). Low iron levels can hinder SNF in nodules (O'Hara et al., 1988; Tang et al., 1992; Johnston et al., 2001). Iron is a cofactor of multiple metallo-enzymes directly or indirectly implicated in SNF in different stages of the symbiosis process. Rhizobial catalase, containing a catalytic heme iron, has been implicated as having a crucial role early in bacterial infection of nodule primordia (Jamet et al., 2003). Later in nodule development, heme iron is required by rhizobia to sense declining O2 levels (Gilles-Gonzalez et al., 1991), signaling the rhizobia to develop into SNF-capable forms. As rhizobia mature to be able to fix nitrogen, they express nitrogenase comprising the iron-sulfur cluster containing NifH and the iron-molybdenum cofactor-containing NifDK (Hoffman et al., 2014; Einsle and Rees, 2020). Proteins essential for shuttling reducing equivalents to nitrogenase, e.g. FixABCX, contain iron cofactors (Ledbetter et al., 2017). Rhizobial proteins essential for respiration contain iron, either as heme iron (Preisig et al., 1996) or as iron-sulfur clusters. In the plant cell cytosol, leghemoglobin containing heme iron, is abundant, buffering and transporting oxygen to the respiring rhizobia and mitochondria (Appleby, 1984; Ott et al., 2005).
Evidence suggests that iron reaches the nodule via the xylem, chelated to citrate or to nicotianamine, where it is released from the vasculature into the apoplastic space in ZII, the infection and differentiation nodule zone (Rodríguez-Haas et al., 2013). The route from vasculature to infected cells crosses several cell layers, which the iron traverses using symplastic and apoplastic routes (Brear et al., 2013; Rodríguez-Haas et al., 2013; Day and Smith, 2021; González-Guerrero et al., 2023). M. truncatula NRAMP1, a member of the Natural Resistance-Associated Macrophage Protein (NRAMP) family, transports iron from the apoplast into the infected cells’ cytosol (Tejada-Jiménez et al., 2015), potentially assisted by citrate efflux transporter MtMATE67 (Multidrug And Toxic Compound Extrusion) (Kryvoruchko et al., 2018). MtMATE67 transports citrate in an iron-activated manner and appears to enhance iron uptake into infected cells (Kryvoruchko et al., 2018). Once inside the cytoplasm of nodule cells, the iron needs to cross another membrane, the SM, to reach the internalized rhizobia. In M. truncatula, two different transporters have been implicated in the SM transport: MtVTL8 (M. truncatula Vacuolar Iron Transporter (VIT)-Like) (Walton et al., 2020; Cai et al., 2022), the subject of this study, and MtFPN2 (M. truncatula Ferroportin Protein2) (Escudero et al., 2020). Both MtVTL8 and MtFPN2 are ferrous iron efflux transporters. Plants with mutations in either protein’s gene show defects in SNF (Escudero et al., 2020; Walton et al., 2020; Cai et al., 2022).
Rhizobia inside symbiosomes import iron from the symbiosome space. In rhizobia, iron uptake is regulated by the heme-regulated transcription repressor Irr, that represses rirA. RirA is a transcriptional activator of iron-uptake genes (O'Brian, 2015; Sankari et al., 2022). The nodule cysteine-rich peptide (NCR) NCR247 is secreted into symbiosomes and taken up by rhizobia in a bacA-dependent manner (Marlow et al., 2009; Farkas et al., 2014). NCR247 was recently found to bind to and sequester heme, inducing an iron starvation response in rhizobia, resulting in increased iron import (Sankari et al., 2022). Thus, NCR247 enables rhizobia to be better iron sinks.
MtVTL8 is a member of the Cross-Complements Ca2+ phenotype of the csg1/Vacuolar Iron Transporter (CCC1/VIT) family. Both it (encoded by Medtr4094335/MtrunA17Chr4g0050851) and the homolog MtVTL4 (encoded by Medtr4g09325/MtrunA17Chr4g0050811) are expressed in nodules, although their expression profiles are somewhat different temporally and spatially (Roux et al., 2014; Walton et al., 2020; Carrere et al., 2021). MtVTL8 was found to localize on the SM; while MtVTL4 localizes to the plasma membrane and membranes surrounding the infection thread (Walton et al., 2020). Plants with mutations in MtVTL8 have profound defects in nodule development, while those with mutations in MtVTL4 have only minor defects (Walton et al., 2020; Cai et al., 2022). The two Tnt1 mtvtl4 mutants studied were not back-crossed and had similar vegetative growth defects in both low and high nitrogen conditions, with higher numbers of less-developed nodules, suggesting a developmental defect, not a symbiotic defect per se (Walton et al., 2020). MtVTL8 is homologous to L. japonicus LjSEN1 (Stationary Endosymbiont Nodule 1) and to soybean GmVTL1a, which when knocked down or mutated have similar phenotypes to mtvtl8 mutants (Suganuma et al., 2003; Hakoyama et al., 2012; Brear et al., 2020; Liu et al., 2020). MtVTL4, MtVTL8 and GmVTL1a are able to complement the yeast Saccharomyces cerevisiae Δccc1 (ScΔccc1) mutant with a defect in a vacuolar ferrous iron transporter demonstrating their iron efflux activity (Brear et al., 2020; Walton et al., 2020). Two mtvtl8 mutants are available. The first is mtvtl8-1 or 13U, in the A17 genotype, having a large deletion on chromosome 4 that deletes both MtVTL4 and MtVLT8 (Walton et al., 2020). The second is mtvtl8-2 derived from Tnt1 line NF11322 in the R108 background with a homozygous Tnt1 insertion in the exon of MtVTL8 (Cai et al., 2022). Both mtvtl8-1 and mtvtl8-2 form defective Fix- white nodules with Sinorhizobium (Ensifer) meliloti (hereafter referred to as S. meliloti) under limited nitrogen conditions (Walton et al., 2020; Cai et al., 2022).
VIT transporters are predicted to fold into five transmembrane helices (TMH) with a long hydrophilic sequence between TMH2 and TMH3. Similarly, the related VTL transporters contain five TMHs but have a much shorter sequence between TMH2-3. Iron transporters with truncated sequences or VTLs are found among angiosperm plants, both monocots and eudicots (Sorribes-Dauden et al., 2020). Based on phylogenetic analysis of CCC1/VTL sequences, Sorribes-Dauden et al. (2020) proposed a different origin for VITs and VTLs: VITs originated from an ancestral horizontal transfer from bacteria while VTLs were transferred from an archaeal lineage with both transfers dating at the origin of the last common eukaryote ancestor.
The crystal structure of VIT1 from the rose gum Eucalyptus grandis was solved (Kato et al., 2019) and is so far the only solved iron transporter structure from the CCC1/VIT family. The structure confirmed the presence of five TMHs, with the N-terminal end located in the cytoplasm and the C-terminal end in the vacuolar space. In the structure, EgVIT1 is a dimer with TMH1 from each monomer at the center and TMH2-5 arranged clockwise around TMH1 to form the transmembrane domain (TMD). A cavity forms between the monomers and in the crystal structure is open toward the cytoplasm. The cytoplasmic TMH2-3 loop folds into three short α-helices, named H1, H2, and H3. Glutamic residues from H1 and H3 in combination with two glutamic acids from the extended cytoplasmic portion of TMH2 bind three metal ions in each monomer. This region was therefore defined as a metal binding domain (MBD). The dimeric interaction is mediated both by the TMD and the MBD. EgVIT1 as well as another member of the CCC1/VIT family, PfVIT, from the human malaria-causing parasite Plasmodium falciparum, were shown to be Fe2+/H+ antiporters with the exchange driven by the proton electrochemical potential (Labarbuta et al., 2017; Kato et al., 2019). Comparing the EgVIT1 structure with the VTL sequences suggests the absence of an MBD in the subfamily (Sorribes-Dauden et al., 2020).
In this work, we investigated the spatial expression of MtVTL8 in WT and mtvtl8-2 roots during nodulation. We then explored whether altering MtVTL4’s expression using the MtVTL8 cis elements would enable it to functionally complement mtvtl8-2 roots. We used a comparison between the EgVIT1 structure to a structural model of MtVTL8 to identify potentially essential amino acids. We then performed mutagenesis studies to challenge our hypothesis. Our results confirm MtVTL8’s unique role as an iron transporter in nodulation and suggest that it may function differently from previously characterized iron transporters. Models for potential mechanisms of transport are presented.
A structural model for MtVTL8 was obtained from the AlphaFold protein structure database (Jumper et al., 2021; Varadi et al., 2022) using MtVTL8’s UniProt ID: A0A072UNI3. Structural analysis was limited to residues 40-235 due to the low confidence score for the N-terminal amino acids. Structures were visualized by the PyMOL Molecular Graphics System (Schrödinger, LLC). A dimer was generated within PyMOL by aligning the AlphaFold monomer with chains A and B of EgVIT1, pdb 6IU3 (Kato et al., 2019). Structures were positioned in the lipid membrane using the PPM web server (Lomize et al., 2012).
Primers and plasmids used for this study are listed in Supplementary Tables S1, S2, respectively. Vectors pMU06 and pMU14 were generous gifts of Drs. Wei Liu and Michael Udvardi. pMU06 contains the pAtUBI-DsRed marker gene for detection of transformed roots using DsRed fluorescence, a site to insert a promoter upstream of GUS, the GUS gene, and the 35S 3’ terminator. pMU14 contains the pAtUBI-DsRed marker gene, the 35S promoter, GFP and 35S 3’ terminator. All constructs were confirmed by sequencing.
For in planta complementation studies, MtVTLs were cloned in the binary vector pMU14 for expression in Medicago as in our previous study of the nitrate transporter MtNPF1.7 (Yu et al., 2021). To express MtVTL8 with its native controlling elements the MtVTL8 promoter (2629 bp), MtVTL8 CDS (708 bp) and MtVTL8 terminator (1083 bp) were amplified from M. truncatula R108 genomic DNA using Q5 High-Fidelity DNA polymerase (New England Biolabs) and primers JYC05-F1/JYC05-R1, JYC05-F2/JYC05-R2, and JYC05-F3/JYC05-R3 respectively. pMU14 was digested with XhoI and HindIII-HF (New England Biolabs). The digested vector, the MtVTL8 promoter, CDS, and terminator PCR fragments were assembled with the Gibson assembly method (NEBuilder HiFi DNA Assembly, New England Biolabs) to form pJYC05 (pMU14/pMtVTL8-MtVTL8-MtVTL8-t). To obtain the MtVTL4 gene driven by pMtVTL8, MtVTL4 was amplified with forward primer JYC24F and reverse primer JYC24R from R108 gDNA. It was assembled with the pJYC05-SpeI/KpnI fragment with the Gibson assembly method to form pJYC24 (pMU14/pMtVTL8-MtVTL4-MtVTL8-t). Plasmids were transformed into electrocompetent Agrobacterium rhizogenes MSU440 by electroporation using Gene Pulser Xcell™ electroporation system (Bio-rad, Hercules, CA, USA) (Valimehr et al., 2014).
The GUS CDS was amplified from the binary vector pMU06 by using primers JYC15-F and JYC15-R. The GUS CDS and JYC05-SpeI-KpnI were assembled with the Gibson assembly method (NEBuilder HiFi DNA Assembly, New England Biolabs) to form pJYC15 (MU06/pMtVTL8-GUS). The construct was introduced into A. rhizogenes as above.
Plant roots transformed with pMtVTL8-GUS were selected by their red fluorescence, demonstrating presence of the transformed vector with the visible marker, at 0, 5 and 15 dpi. Transformed roots were harvested in 0.1 M PBS (Na2HPO4 and NaH2PO4, pH 7.0) and then transferred to GUS solution (44.5 mL of 100 mM PBS-Na pH 7.0, 2 mL of 50 mM K3Fe(CN)6, 2 mL of 50 mM K4Fe(CN)6, 1 mL of 0.5 M EDTA, 0.5 mL of 10% Triton X-100, 50 mg of X-Gluc salt mixed together) followed by vacuum infiltration for 1.5 h. Roots were kept at 37°C overnight. The samples were subsequently washed with 0.1 M PBS (Na2HPO4 and NaH2PO4, pH 7.0) at 4°C. Samples were observed under an Olympus BX50 microscope (Olympus, Tokyo, Japan). GUS-stained nodules were cut and fixed with 4% glutaraldehyde (made in 0.1 M PBS-Na, pH 7.0) under vacuum for 30 min. The samples were kept overnight at 4°C with gentle rotation. Then, the samples were washed three times with 0.1 M PBS-Na and dehydrated with a series of ethanol gradients (30%, 50%, 70%, 90%, 100%), each step was carried out with gentle rotation for 30 min at room temperature. The ethanol was replaced with ethanol: Technovit 7100 (Kulzer Technik, Hanau, Germany) (2:1/v:v) and rotated for one hour at room temperature, followed by ethanol: Technovit 7100 (1:2/v:v) for another hour. The liquid was replaced with 100% Technovit 7100 and rotated gently at room temperature overnight. The next day, the liquid was replaced with fresh Technovit 7100 and rotated at room temperature for 1 h. Resin was prepared by mixing Technovit 7100 with hardener II (15:1/v:v). Resin was added to the mold well and the nodule was placed in the resin. The well was covered with parafilm and left at room temperature for 1 h for polymerization. After polymerization, the parafilm was removed and the block was glued to an adaptor using the Technovit 3040 glue (Kulzer Technik, Hanau, Germany). Five micrometer nodule sections were sliced by a microtome (Leica HistoCore Multicut, Leica) and collected on glass slides. The slides were stained with ruthenium red staining solution (200 mg ruthenium red, 200 mL water) for 5 min followed by rinsing with water until the background was clear. The slides were dried on a hotplate and nodule sections were visualized with an Olympus BX50 microscope.
Mutants of MtVTL8 were constructed in pJYC05, replacing the MtVTL8 cDNA with the mutated gene. Two PCR reactions were carried out resulting in two overlapping fragments, one containing the 5’ end of MtVTL8 to the desired mutation (using primers JYCmut-F and JYC19R2) and the other containing the desired mutation to the 3’ end of MtVTL8 (using primers JYC19F2 and JYCmut-R). The relevant primers for each construct are listed in Supplementary Table S3. Mutated MtVTL8 fragments were subsequently assembled into pJYC05-SpeI/KpnI vector with the Gibson assembly method to form pJYCmut. We obtained the following mutants: MtVTL8_R51A (pJYC16), MtVTL8_D59A (pJYC17), MtVTL8_G88E (pJYC18), MtVTL8_E100A (pJYC19), MtVTL8_E111A (pJYC20), and MtVTL8_K135A (pJYC21). Double mutants MtVTL8_R51E/E100R (pJYC22), and MtVTL8_E111K/K135E (pJYC23) were obtained by the assembly of three PCR fragments with pJYC05-SpeI/KpnI vector.
M. truncatula Tnt1 insertion mutant mtvtl8-2 seeds and control wild type R108 seeds were scarified and germinated as described (Cai et al., 2023). Seedlings of mtvtl8-2 and R108 were transformed with A. rhizogenes MSU440 containing either empty vector (EV) pMU14, pJYC05, or plasmids containing mutated MtVTL8. Transformants were transferred to Fåhraeus medium (Fåhraeus, 1957) containing 5 mg/L nystatin (Millipore-Sigma, Burlington, MA USA) for 5 d in growth chamber with 14 h/10 h (light/dark) at 24°C (Boisson-Dernier et al., 2001). Then, the plants were moved to a 1% MS medium (Millipore-Sigma) plate in between two filter papers covering the roots for 14 d in growth chamber with 14 h/10h (light/dark) at 24°C. Transformed roots were selected based on their expression of the DsRed fluorescent marker, contained in the pMU14 vector. This was done using a Leica MZ10F dissecting microscope (Leica, Deer Park, IL, USA). Non-transformed roots were excised. The plants with transgenic roots were transferred to the aeroponic system with Lullien’s medium (Lullien et al., 1987) without a nitrogen source at 22°C with a 16 h/8 h light/dark photoperiod at 60 μmol m-2s-1 for 5 d (Barker et al., 2006; Cai et al., 2023). Then, the plants were inoculated with S. meliloti Rm41 (Kondorosi et al., 1980). Growth of the transformed plants was checked at 15 dpi and nodules were analyzed and documented using the Leica MZ10F dissecting microscope. Nodules were photographed and their proxy 2-D surface areas were analyzed with Fiji software (Schindelin et al., 2012). For chlorophyll estimation, we followed the control method described (Liang et al., 2017). Briefly, leaves were collected at 28 dpi and frozen in liquid N2. For extraction, 100 mg of leaves were ground under liquid N2, 1.0 mL of 80% acetone was added and the mixture gently agitated for 24 h at room temperature, followed by 15,000g centrifugation for 5 min at 4°C. 100 μL of supernatant was mixed with 400 μL of 80% acetone. Absorbance of the supernatant was measured at wavelength of 645 nm (A645) and 663 nm (A663) with a Bio-Rad SmartSpec Plus spectrophotometer (Hercules, CA, USA) The total chlorophyll content was calculated following the Arnon’s equation (Arnon, 1949): Total chlorophyll (μg/mL) = 20.2 (A645) + 8.02 (A663).
The vector pYES2/CT (ThermoFisher Scientific) was digested with HindIII-HF and BamHI-HF (New England Biolabs). The MtVTL8 gene and its eight different mutant versions were amplified from pJYC16-23 using primers JYC10.FOR and JYC10.REV and assembled with the pYES2/CT-HindIII/BamH1 fragment using the Gibson assembly method to form pJYC10 (MtVTL8-wt) and eight different Mtvtl8-mut versions (pYJYC16-23).
The Saccharomyces cerevisiae wild type strain DY150 and mutant strain Δccc1(ura3, leu2, his3, ade2, can1, CCC1::HIS3) (Li et al., 2001) in the DY150 background were grown in YPD medium at 30°C, 250 rpm for 16 h. Competent cells of DY150 and Δccc1 were produced using the Frozen-EZ Yeast Transformation II kit (Zymo Research). pYES2/CT was transformed into the DY150. The mutant strain Δccc1 was transformed with pYES2/CT, pYES2-AtVTL1 (At1g21140, wild type), pJYC10 or pYJYC16-23. Single colonies, each with a specific construct, were cultured in SC-U+Gal medium at 28°C for 16 h. OD600 was monitored and adjusted to 1 for the spot assay. Ten μL of each culture and their serial dilutions were spotted on SC-U+Gal or SC-U+Gal + 5 mM ammonium ferrous sulfate plates, followed by incubation at 28°C for 3 d. Plates were observed to assess the yeast growth and photographed.
Genes belonging to the CCC1/VIT1 family were identified in the PANTHER18 family library of trees (Thomas et al., 2022) using the keyword PTHR31851. Sequences from one alga and selected plant genomes were retrieved from UniProt Knowledgebase (UniProtKB) (UniProt Consortium, 2023). Sequences for Ceratopteris richardii and Lotus japonicus were obtained on Phytozome v13 (Goodstein et al., 2012). We limited our analysis to the following genomes: Chlamydomonas reinhardtii (green alga); Selaginella moellendorffii (lycophyte); Marchantia polymorpha (liverwort); Physcomitrella patens (bryophyte); Amborella trichopoda (amborella); Ceratopteris richardii (pteridophyte); Brachypodium distachyon, Hordeum vulgare, Musa acuminata, Oryza sativa, Setaria italica, Sorghum bicolor, Triticum aestivum, Tulipa gesneriana, Zea mays, and Zostera marina (monocots); Arabidopsis thaliana, Brassica napus, Capsicum annuum, Citrus sinensis, Cucumis sativus, Erythrante guttata, Eucalyptus grandis, Glycine max, Gossypium hirsutum, Helianthus annuus, Lactuca sativa, Lotus japonicus, Medicago truncatula, Nicotiana tabacum, Populus trichocarpa, Prunus persica, Ricinus communis, Solanum lycopersicum, Solanum tuberosum, Spinacia oleracea, Theobroma cacao, and Vitis vinifera (eudicots). Sequences are listed in Supplementary Table S4. Multiple sequence alignment was obtained using ClustalW. WebLogo 3 was used to create the sequence logos (Crooks et al., 2014). The initial maximum-likelihood phylogenetic tree was calculated by ModelFinder (Kalyaanamoorthy et al., 2017) using the IQ-TREE multicore version 2.1.2 COVID-edition for Linux 64-bit. The best-fit model based on Bayesian Information Criterion was JTT+R6. Branch support for the maximum-likelihood tree was generated with ultrafast bootstrap (Hoang et al., 2018) (1000 replicates) implemented in the IQ-TREE software (Nguyen et al., 2015) Calculations were performed on the CIPRES (CyberInfrastructure for Phylogenetic Research) science gateway platform (Miller et al., 2010). The phylogenetic tree was visualized and annotated with iTOL (Letunic and Bork, 2021) and can be accessed here: https://itol.embl.de/tree/4718713120329291698508806. Predicted protein structures, provided by AlphaFold within UniProtKB, were visually analyzed for each protein in the tree.
To determine when and where MtVTL8 is expressed in roots and nodules, the coding sequence for the β-glucuronidase (GUS) enzyme (Jefferson et al., 1987) was cloned under the control of the MtVTL8-promoter (pMtVTL8). The pMtVTL8-GUS construct was transferred to an A. rhizogenes strain and expressed via hairy-root transformation in the roots of R108 wild type and mtvtl8-2, a Tnt1 line with a homozygous Tnt1 insertion in the exon of MtVTL8 (Cai et al., 2022). Roots and nodules were examined at 0 dpi, 5 dpi and 15 dpi with S. meliloti Rm41 (Supplementary Figures S1, S2). Blue color was found in nodule primordia and nodules but not in the roots, demonstrating that the MtVTL8 promoter highly and exclusively directs expression in these tissues (Supplementary Figures S1, S2). Longitudinal cross sections of the GUS stained nodules of R108 and mtvtl8-2 at 15 dpi followed by light microscopy imaging showed that pMtVTL8-GUS was expressed from ZII to ZIII in both infected cells (IC) and uninfected cells (UC) with high expression in the infection zone (IZ) (Figure 1; Supplementary Figure S3). Expression was not observed in the interzone (IZ). These results correspond to the RNA-seq data from the Symbimics database (Roux et al., 2014) and reinforce the idea that MtVTL8’s role is to support SNF. Expression of pMtVTL8-GUS was not observed in the vascular system (Figures 1A, E; Supplementary Figures S3A, B). In mtvtl8-2, the MtVTL8 expression was dramatically decreased in the premature senescent zone compared to the zones containing rhizobia (Supplementary Figure S3). However, the expression pattern in R108 was similar to that observed in mtvtl8-2, (Supplementary Figure S3), taking into consideration the developmental defects observed in these nodules due to the absence of MtVTL8 (Walton et al., 2020; Cai et al., 2022). At the 15 dpi time point in R108 nodules, there is not yet a senescent zone; taken together, these results suggest that MtVTL8 may not have a role in reusing Fe2+ during senescence.
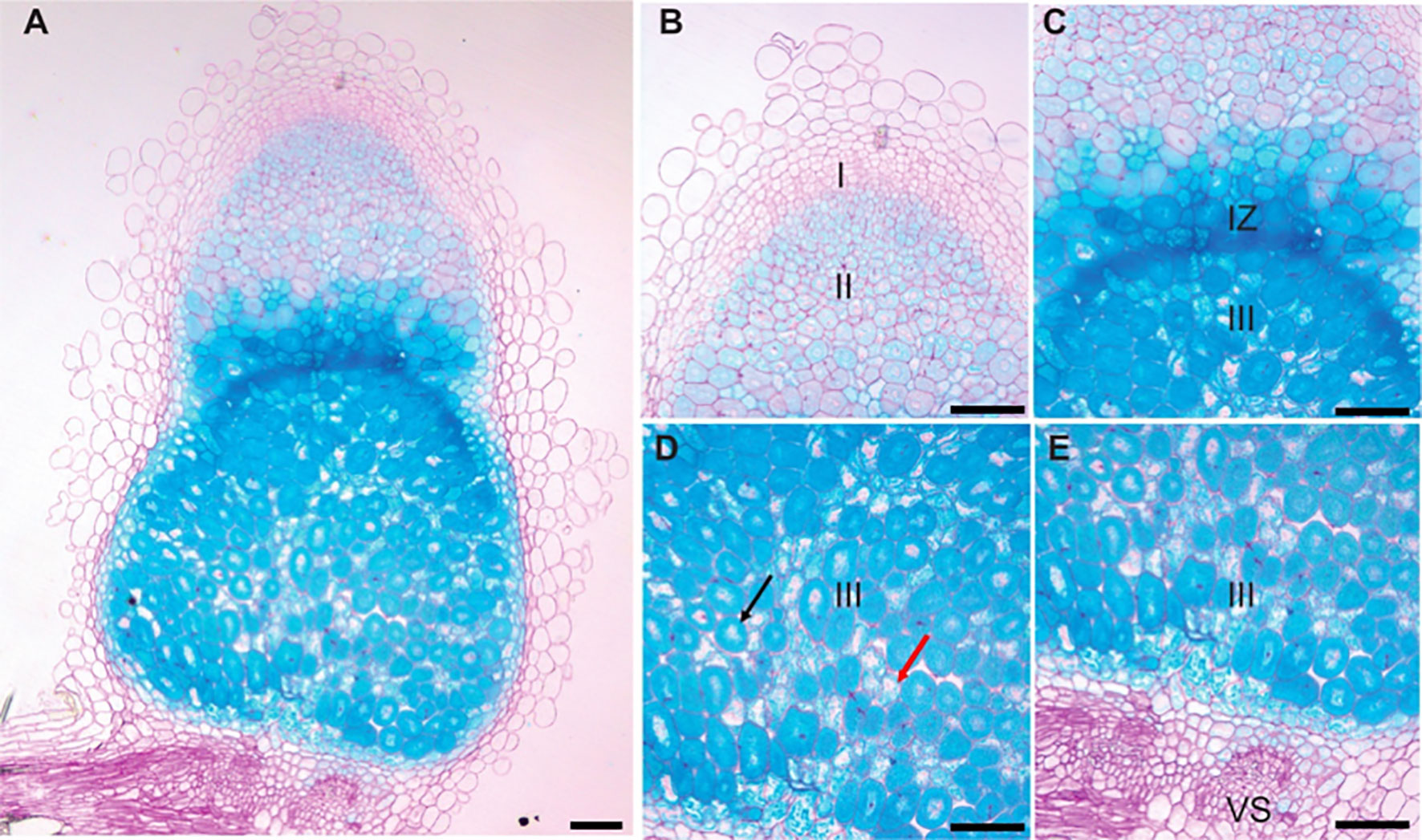
Figure 1 pMtVTL8-GUS is expressed from zone II to zone III in both infected and uninfected cells of Medicago truncatula genotype R108 nodules. Nodules were collected and stained for GUS followed by fixation with glutaraldehyde. Then they were embedded in Technovit 7100 (Kulzer Technik, Hanau, Germany) and sectioned at 5 μm. (A) Expression of pMtVTL8-GUS in the R108 nodule section at 15 dpi with S. meliloti Rm41. (B-E) Details from (A) for expression of pMtVTL8-GUS from Zone I to Zone III. VS, vasculature. I, zone I, meristem zone. II, zone II, infection zone. IZ, interzone, maturation zone. III, zone III, fixation zone. The black arrow indicates an infected cell. The red arrow indicates an uninfected cell. Scale bars represent 0.1 mm.
The yeast Cross-complements Ca2+ phenotype of the Csg1 family (CCC1) (Li et al., 2001) gene encodes an Fe2+/Mn2+ vacuolar transporter. Yeast Δccc1 mutants, containing a deletion of CCC1, show hypersensitivity to high concentrations of external Fe2+ because they fail to sequester the excess Fe2+ in their vacuoles (Li et al., 2001). The ScΔccc1 strain has been used in complementation studies of several plant iron transporters belonging to the CCC1 family including MtVTL4 and MtVTL8 (Walton et al., 2020); AtVIT1 (Kim et al., 2006); AtVTL1, AtVTL2 and AtVTL5 (Gollhofer et al., 2014); TgVIT1 (Momonoi et al., 2009); OsVIT1 and OsVIT2 (Zhang et al., 2012); TaVIT2 (Connorton et al., 2017); EgVIT1 (Kato et al., 2019); GmVTL1a and GmVTL1b (Brear et al., 2020; Liu et al., 2020). Additionally the strain was used to test PfVIT from the P. falciparum parasite (Slavic et al., 2016). All tested transporters were able to complement the ScΔccc1 mutation and restore the growth of the defective strain in high iron conditions.
The S. cerevisiae wild-type DY150 and mutant Δccc1 strains transformed with empty vector (EV) pYES2/CT individually were able to grow on selective medium without Fe2+ (Figures 2A, B, left side). Mutant ScΔccc1 were transformed with pYES2/CT harboring the genes for AtVTL1, a positive control, MtVTL4, or MtVTL8 were able to grow on selective medium without Fe2+ (Figures 2C–E, left side). ScΔccc1 transformed with EV was unable to grow on the selection plates supplied with 5 mM Fe2+ (Figure 2B, right side). In contrast, the growth of the ScΔccc1 strain in the presence of 5 mM Fe2+ was partially restored by heterologous expression of AtVTL1, a positive control, MtVTL4, and MtVTL8 (Figures 2C–E, right side). This confirms that both MtVTL8 and MtVTL4, as well as AtVTL1, are able to transport ferrous ions out of the cytosol either to the vacuole or across the plasma membrane in yeast.

Figure 2 Spot assay of different mutants of MtVTL8 expressed in the S. cerevisiae Δccc1 strain. Spot assay with tenfold dilutions of overnight yeast cultures plated on at 0 or 5 mM ferrous sulfate (left and right panels, respectively) and incubated at 28°C for 3 days. From top to bottom: (A) wild-type strain DY150 transformed with empty vector (EV), (B–M) Δccc1 strain transformed with EV and vector containing MtVTL4, MtVTL8, AtVTL1 or specific MtVTL8 mutant genes under the pGAL1 promoter. Δccc1 expressing AtVTL1, MtVTL4 or MtVTL8 are positive controls. Vector is pYES2/CT.
In addition to the yeast assay, we performed in planta complementation using the SNF defective line mtvtl8-2. In planta complementation is a good platform to investigate plant transporters and has been used in our lab to complement M. truncatula plants with defective nodulation due to a mutated nitrate transporter (Yu et al., 2021). Expression of MtVTL8 driven by its 2.8 kb MtVTL8 native promoter in the mtvtl8-2 root system was accomplished by A. rhizogenes mediated hairy-root transformation (Figures 3). Wildtype MtVTL8 constructs successfully rescued the defective nodule phenotype from small Fix- white nodules with empty vector (Figures 3B, G) to wild-type like (WTL) pink nodules (Figures 3C, H; compare to WT with empty vector, Figures 3A, F). Complemented or control plants’ nodule surface areas were assessed with the aim of determining if MtVTL8- complemented mtvtl8-2 nodules produced statistically similar nodule surface areas to WT R108 nodules, larger than the non-complemented mtvtl8-2 nodules. However, none were found to have statistically different sizes (Supplementary Figure S4). Chlorophyll content of composite MtVTL8- expressing mtvtl8-2 plants was used to assess effective nitrogen fixation. Results showed similar chlorophyll content of plants with MtVTL8- expressing mtvtl8-2 roots was similar to WT R108 plants, which was markedly higher than mtvtl8-2 plants whose roots with an empty vector-transformed (Supplementary Figure S5). This demonstrates that complementation (pink color in nodule and leaf chlorophyll content) can be used to assess MtVTL8 functionality in planta.
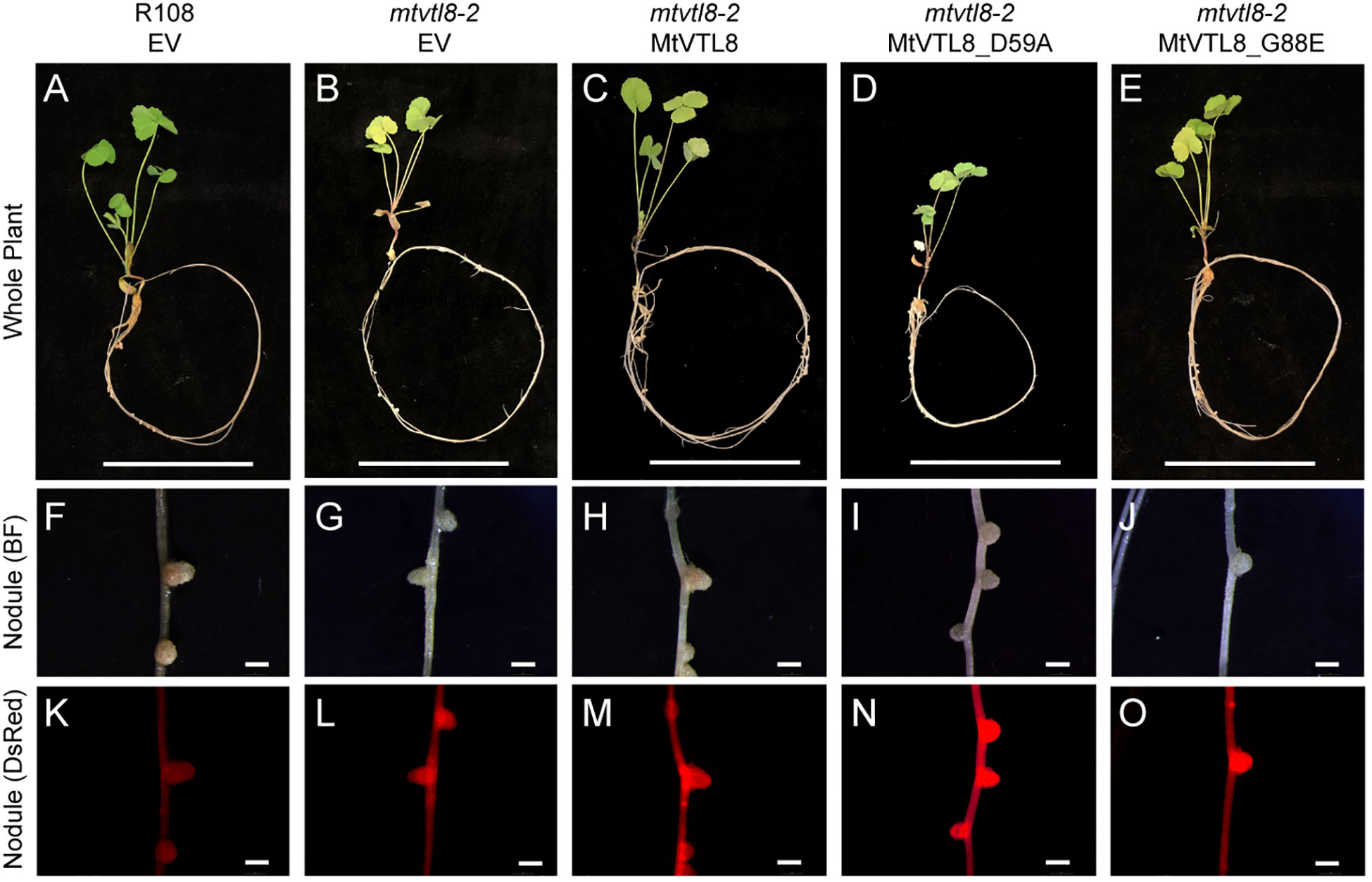
Figure 3 In planta complementation assays of the mtvtl8-2 mutant with the wild-type MtVTL8 and its different mutated versions affected in the transmembrane domain. (A–E) Images of M. truncatula plants transformed with different vectors expressing different mutated versions of MtVTL8. From left to right, (A) R108 plant roots transformed with empty vector (EV). (B–E) Mtvtl8-2 plant roots transformed with (B) EV, (C) MtVTL8 (WT), (D) MtVTL8_D59A, and (E) MtVTL8_G88E expressed under pMtVTL8 promoter. (F–J) Bright field (BF) images of nodules corresponding to (A–E). (K–O) DsRed fluorescence encoded by the DsRed gene under the pAtUBI constitutive promoter in the vector is observed in the transformed nodules corresponding to (F–J). Scale bars represent 5 cm for (A–E) and 1 mm for (F–O).
Two M. truncatula CCC1/VIT1 genes, MtVTL4 and MtVTL8, are exclusively and highly expressed in nodules. Sequence alignment shows that the two proteins are 55% identical and 72% positives. Expression of MtVTL4 or MtVTL8 was found to successfully rescue the toxicity of iron in yeast ScΔccc1 mutants (Walton et al., 2020) and confirmed in our lab (Figures 2C, D). These results indicate that both transporters can transport ferrous ions out of the cytosol in yeast. While mtvtl8 mutants, either mtvtl8-1 with a large deletion spanning MtVTL8 and MtVTL4 (Walton et al., 2020), or with a homozygous Tnt1 insertion in the MtVTL8 exon (mtvtl8-2) (Cai et al., 2022) display defective white nodules, Walton et al. (2020) found that plants with a Tnt1 homozygous insertion in the MtVTL4 exon display wild-type like nodules, with an apparent developmental delay (Walton et al., 2020). Expression of MtVTL8 under the control of its own promoter successfully rescued the mtvtl8-1 deletion mutant, but MtVTL4 expression under the control of its own promoter did not have the same result (Walton et al., 2020).
Since both MtVTL4 and MtVTL8 transport Fe2+ in yeast, we wondered whether the failure of MtVTL4 expression to complement mtvtl8-1 (Walton et al., 2020) could have been caused by it being expressed at the wrong time and place in maturing nodules. To address this question, we expressed the MtVTL4 gene driven by the constitutive Arabidopsis thaliana translation elongation factor (AtEF1ɑ) promoter (Axelos et al., 1989; Auriac and Timmers, 2007) in the roots of mtvtl8-2. As a control, we expressed MtVTL8 in the same vector. The results showed that expression of neither MtVTL4 nor MtVTL8 driven by the AtEF1ɑ promoter in the roots of mtvtl8-2 rescued the defective nodulation (Supplementary Figures S6D, E, I, J). While these results are not definitive for MtVTL4, they suggest that expression of MtVTL8 in an inappropriate nodule location may be deleterious to nodule development. Alternatively, the AtEF1ɑ promoter may not express well in the area(s) of the nodule where MtVTL8 is needed.
Because MtVTL4 and MtVTL8 have different expression patterns within nodules (Roux et al., 2014; Walton et al., 2020) with MtVTL4 expressed most highly in ZII and MtVTL8 expressed most highly in the IZ and ZIII, we wondered if expressing MtVTL4 under MtVTL8’s promoter might enable MtVTL4 to functionally complement mtvtl8. However, expression of MtVTL4 under the MtVTL8 promoter displayed defective nodulation in mtvtl8-2 (Supplementary Figures S7D, H). Plants with roots transformed with MtVTL4 had leaves containing significantly less chlorophyll compared with the positive controls (Supplementary Figure S5). Thus, our data suggest that MtVTL4’s localization to the plasma membrane and infection thread or other functional differences from MtVTL8 give these two transporters unique capabilities in nodules.
We obtained a structural model for the monomer of MtVTL8 from the AlphaFold protein structure database (Jumper et al., 2021; Varadi et al., 2022). We then produced the dimeric form (Figure 4A) by overlapping the monomer on chains A and B of EgVIT1, pdb 6IU3 (Kato et al., 2019). The dimeric model for MtVTL8 shows the predicted five transmembrane helices (TMHs) from TMH1 to TMH5 (Figure 4B) contributing to form the transmembrane domain (TMD). A cavity forms at the interface between the two monomers and in the model is open toward the cytoplasm, like in the EgVIT1 structure. The transmembrane region does not show remarkable differences between the two transporters (Figure 4C) (RMSD value of 1.23 for 650 atoms). The only exception is TMH2 that in the MtVTL8 model is slightly bent in the direction of TMH1 (Figures 4D, E). This may be due to constraints caused by a shortened sequence between TMH2 and 3. In contrast to the TMD region, structural alignment between the AlphaFold model of MtVTL8 and the crystal structure of EgVIT1 shows intriguing differences in the cytoplasmic region of the two proteins (Figure 4C). Like in EgVIT1, TMH2 in MtVTL8 is longer than the other TMHs and protrudes in the cytoplasm. However, in MtVTL8 the cytoplasmic portion of TMH2 is predicted to be seven residues shorter than TMH2 in EgVIT1 (Supplementary Figure S8A). Also, while in EgVIT1 the extended TMH2 connects with three additional cytoplasmic α-helices to form a metal binding domain (MBD) (Figures 4A, C, D), in MtVTL8 a shorter sequence, MtVTL8(124-138), between TMH2 and 3 is predicted to form one α-helix, H1 (Figures 4B, C, E). In the MtVTL8 model, TMH2 and H1 are almost parallel and are separated by a short loop of seven residues (Figures 4B, C, E; Supplementary Figure S8C). In summary, the overall topology of the transporter is changed from five transmembrane helices plus three cytoplasmic helices in EgVIT1 to five transmembrane helices plus one cytoplasmic helix in MtVTL8 (Supplementary Figures S8B, C).
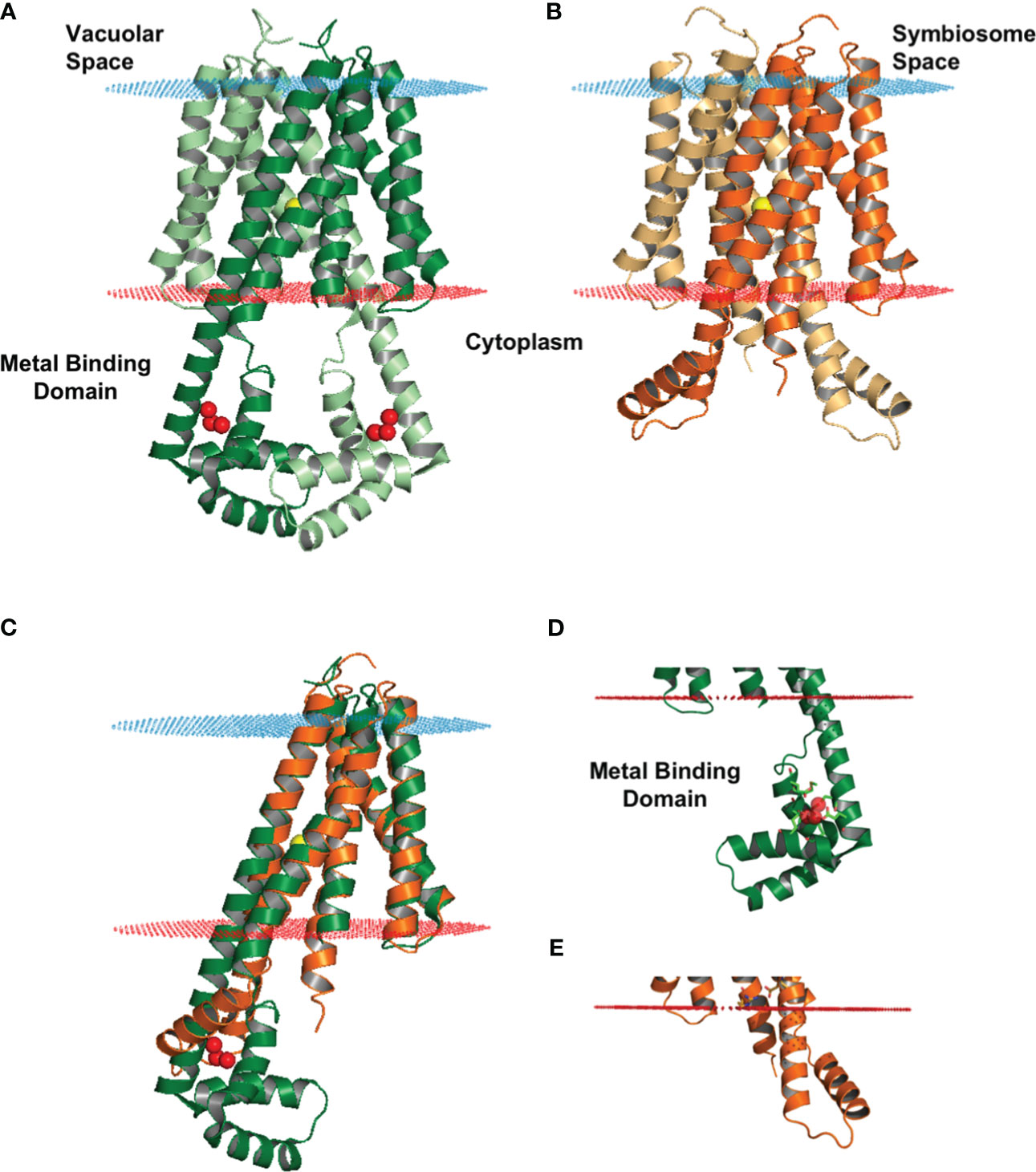
Figure 4 Structural model of MtVTL8 compared to the crystal structure of EgVIT1. (A) Crystal structure of EgVIT1, with monomer A in dark green and monomer B in light green, showing the TMD embedded in the membrane (blue and red dots) and the MBD in the cytoplasm. (B) AlphaFold model of MtVTL8, with monomer A in dark orange and monomer B in light orange, showing a conserved TMD and a reduced cytoplasmic region lacking a MBD. (C) Monomer A of MtVTL8 (dark orange) overlapped on monomer A of EgVIT1 (dark green). (D) Zoom into the MBD of EgVIT1 showing the side chains for the residues that bind the Fe2+ ions. (E) Zoom into the cytoplasmic region of MtVTL8 showing a shorter TMH2 and one α-helix. (A–E) Iron ion in the TMD is a yellow sphere; iron ions in the MBD are red spheres.
In the EgVIT1 structure, five glutamic acid residues, Glu102 (TMH2), Glu105 (TMH2), Glu113 (H1), Glu116 (H1), and Glu153 (H3) and two methionines, Met149 and Met150 (H3), within the MBD are involved in binding and stabilizing transition metal ions like Fe2+ or Mg2+ (Supplementary Figure S8A, red arrowheads). Mutating any of the glutamic acids or the methionines failed to complement the growth inhibition phenotype of Sc△ccc1 and decreased transport activity in liposomes made with the transporter mutant versions (Kato et al., 2019). Additionally, two other residues, Glu32 and Asp36 (Supplementary Figure S8A, red arrowheads), located at the entrance of the ion-translocation pathway on TMH1, may play a role in guiding iron ions from the MBD to the TMD. With the exception of Glu102 on TMH2 (Asp118 in MtVTL8), no amino acid residues with similar biochemical properties in similar locations are found in the cytoplasmic domain of MtVTL8 (Supplementary Figure S8A). This suggests that MtVTL8 is not able to bind metal ions in the cytoplasm.
We used the MtVTL8 structural model to identify residues potentially important for transport activity in VTL. After identifying residues that may have a role in metal ion binding or transport, we studied the effect of point mutations on the transport properties of MtVTL8 in complementation studies in the mtvtl8-2 mutated plant line and in ScΔccc1, the iron sensitive mutant yeast strain.
In EgVIT1, two residues from each monomer in the translocation channel, Asp43 and Met80 are predicted to bind metal ions, followed by relaying the metal ions to Glu72 within the central translocation pathway (Kato et al., 2019). The three residues are all essential for EgVIT1 transport activity as demonstrated by yeast spot assay and liposomal essays. Based on sequence alignment between EgVIT1 and MtVTL8, residues Asp43 and Met80 of EgVIT1 correspond to residues Asp59 and Met96 of MtVTL8, respectively (Supplementary Figure S8A). However, Glu72 is not conserved in MtVTL8 as the equivalent position is occupied by a glycine, Gly88 (Supplementary Figure S8A).
To test its impact on the function of MtVTL8, we mutated the polar residue Asp59 to Ala to obtain the MtVTL8_D59A mutant. A. rhizogenes-mediated transformation of mtvtl8-2 mutant roots was used to express constructs of empty vector (EV), MtVTL8, and MtVTL8_D59A (Figures 3A–D, F–I, K–N). Our complementation experiments showed that mtvtl8-2 roots transformed with the MtVTL8_D59A mutant gene displayed defective nodulation with white nodules (Figures 3D, I) compared with the pink nodules from the positive controls, wild type plant R108 transformed with EV (Figures 3A, F) or mtvtl8-2 transformed with MtVTL8 (Figures 3C, H). Mtvtl8-2 plants transformed with MtVTL8_D59A had leaves that contained less chlorophyll compared with positive controls, and no significant difference was observed compared with the negative control (Supplementary Figure S5), suggesting defects in N supply. Taken together, our results indicate that mtvtl8-2 roots transformed with MtVTL8_D59A showed defective SNF and did not complement the mtvtl8-2 phenotype. Similarly, expression of the MtVTL8_D59A mutant failed to complement the yeast ScΔccc1 strain (Figure 2F). Together these results suggest that Asp59 is essential to Fe2+ transport in MtVTL8.
In EgVIT1, Glu72 on TMH2 is an essential residue proposed to translocate the metal ions by displacing its bonded proton along the central ion translocation pathway (Kato et al., 2019). The corresponding residue in MtVTL8 is a glycine, Gly88. We speculated that adding back the glutamic acid in the place of Gly88 in MtVTL8 could have a positive effect on the function. Therefore, we replaced Gly88 with Glu to make the single mutant MtVT8_G88E. Expression of MtVTL8_G88E in mtvtl8-2 hairy roots produced transgenic roots that displayed defective white nodules compared with the pink nodules from the positive controls (Figures 3E, J, O). Mtvtl8-2 plants with roots transformed with MtVTL8_G88E had chlorotic leaves (Supplementary Figure S5). When evaluated in the yeast ScΔccc1 mutant, the MtVTL8_G88E mutant failed to restore iron tolerance (Figure 2G). These data indicate that Gly88 is an essential residue for MtVTL8 function and mutating it to glutamic acid does not restore its putative role as proton transporter proposed for EgVIT1. Interestingly, MtVTL8 does not harbor other negatively chargeable amino acids in the substrate cavity that could potentially fulfill the same role as Glu72. These observations suggest that MtVTL8 may use a different mechanism for iron transport than that hypothesized for EgVIT1. It is also possible that another residue far from MtVTL8’s Gly88 could function in proton transport if MtVTL8 is indeed an iron-proton antiporter.
In silico analysis of the MtVTL8 model indicates the formation of a salt bridge in the TMD between two oppositely charged residues, Arg51 located on TMH1 and Glu100 on TMH2 (Supplementary Figure S9A). Alignment of MtVTL8 with EgVIT1 shows the TMH1-TMH2 salt bridge is only possible in MtVTL8 and not EgVIT1 as the corresponding amino acids in EgVIT1 are Arg35 and Gly84 (Supplementary Figure S8A). Based on our observation that in MtVTL8, Arg51 and Glu100 could form a salt bridge, we constructed mutants with a single mutation of Arg51 or Glu100 into Ala. Expression of MtVTL8_R51A or MtVTL8_E100A in mtvtl8-2 hairy roots produced plants that displayed defective nodulation and symbiotic nitrogen fixation phenotypes (Figures 5; Supplementary Figures S4, S5). Subsequently we tried to restore the putative salt bridge by swapping the charges and we obtained the R51E/E100R double mutant. However, the MtVTL8_R51E/E100R mutant gene failed to rescue the mtvtl8-2 phenotype (Figures 5F, L). Mtvtl8-2 plants transformed with MtVTL8_R51A or MtVTL8_E100A or the double mutant with swapped charges had nodules with no significant difference in size compared with the positive or negative controls (Supplementary Figure S4), whereas the leaves contained significantly less chlorophyll compared with the positive controls (Supplementary Figure S5), indicating a deficiency in SNF. In yeast, neither MtVTL8 proteins with R51A, E100A, nor R51E/E100R were able to rescue the ScΔccc1 iron transport deficiency (Figures 2H–J). These data suggest that these two residues are essential for MtVTL8 function and their location in the protein is important as well, likely contributing to ionic interactions other than a salt bridge between the two residues.
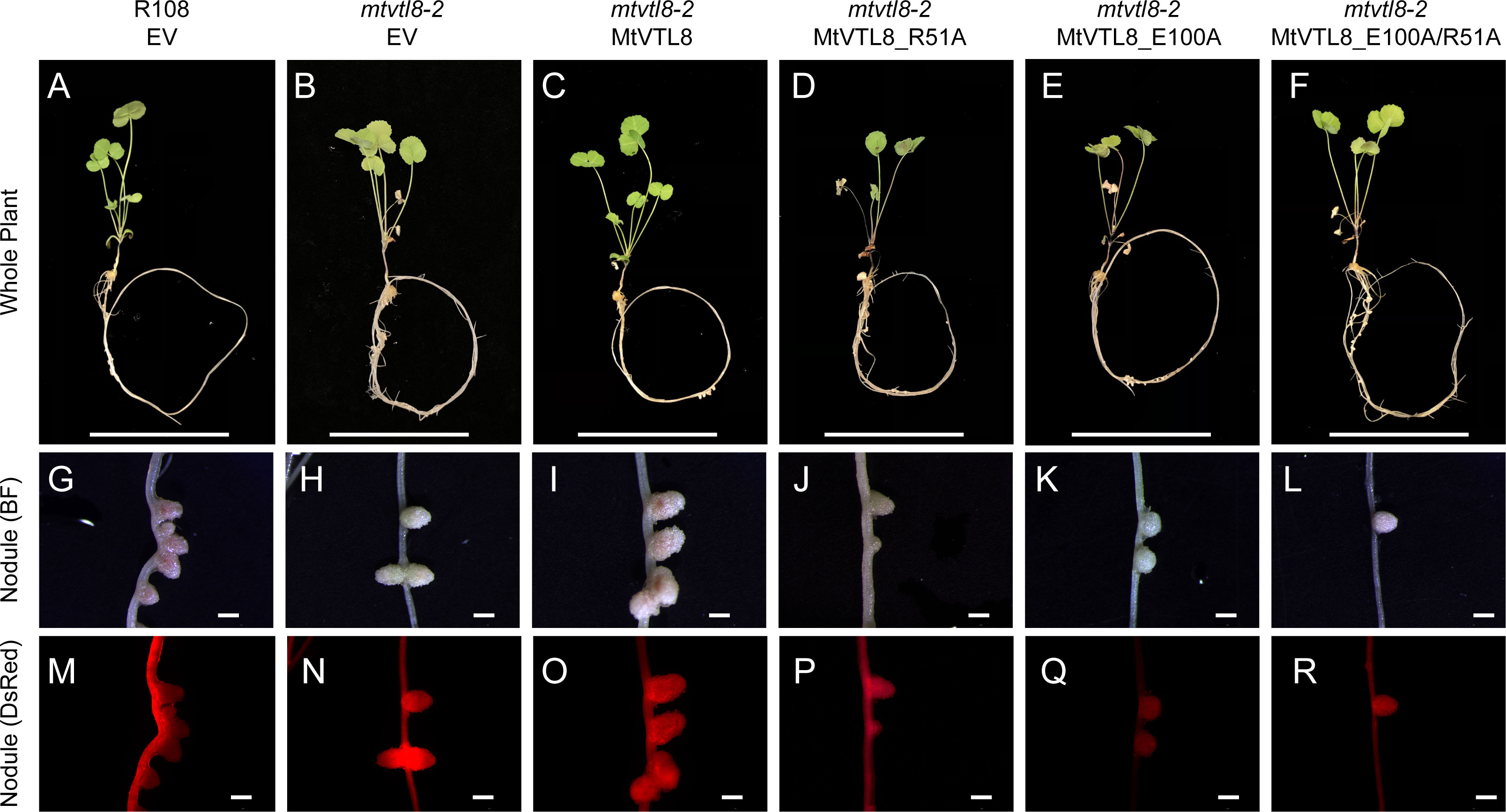
Figure 5 In planta complementation assays of the mtvtl8-2 mutant with wild-type mutations and its different mutated versions affected in the putative TMH1-2 salt bridge. (A–F) Images of M. truncatula plants expressing different mutated versions of MtVTL8. From left to right, (A) R108 plant roots transformed with empty vector (EV); Mtvtl8 plant roots transformed with (B) EV, (C) MtVTL8 under pMtVTL8 (WT), (D) MtVTL8_R51A, (E) MtVTL8_E100A, and (F) MtVTL8_R51E/E100R. (G–L) Bright field (BF) and (M–R) DsRed fluorescence images of transformed nodules corresponding to (A–F). Scale bars represent 5 cm for (A–F) and 1 mm for (G–R).
Our model shows that the cytoplasmic portion of MtVTL8 is dramatically different from that of EgVIT1 with MtVTL8 harboring a lone α-helix MtVTL8(124-138) (H1) instead of the three α-helices as in EgVIT1 (Figure 3; Supplementary Figures S8B, C). In the model, H1 lies almost parallel to the cytoplasmic portion of TMH2. We observed that Lys135 from H1 is at a distance of 2.7 Å from Glu111 (TMH2) which is compatible with the formation of a salt bridge between the two residues (Supplementary Figure S9). In order to assess the role of Glu111 and Lys135, we created the single mutants MtVTL8_E111A and MtVTL8_K135A (Figure 6). The results showed that expression of MtVTL8_E111A in hairy roots of mtvtl8-2 plants failed to rescue the defective nodulation phenotype (Figures 6D, J) whereas expression of MtVTL8_K135A successfully rescued the defective phenotype of mtvtl8-2 (Figures 6E, K). Quantitation of nodule surface areas and leaf chlorophyll of composite plants showed no significant difference with the positive controls (Supplementary Figures S4, S5). To assess the restoration of the putative salt bridge between Glu111 and Lys135, we swapped the charges between the two residues creating the double mutant K111E/E135K. Expression of the double mutant with swapped charges failed to rescue the defective nodulation and nitrogen fixation phenotypes of mtvtl8-2 (Figures 6F, L; Supplementary Figures S4, S5). This data suggests that Glu111 is an essential residue for MtVTL8 function as an Fe2+ transporter in yeast, whereas Lys135 is not in planta. It also suggests that trying to restore the putative salt bridge by swapping the charges in MtVTL8 is insufficient for function in planta. Similarly, when the single or double mutants were expressed in the ScΔccc1 strain they were not able to rescue iron transport deficiency (Figures 2K–M).
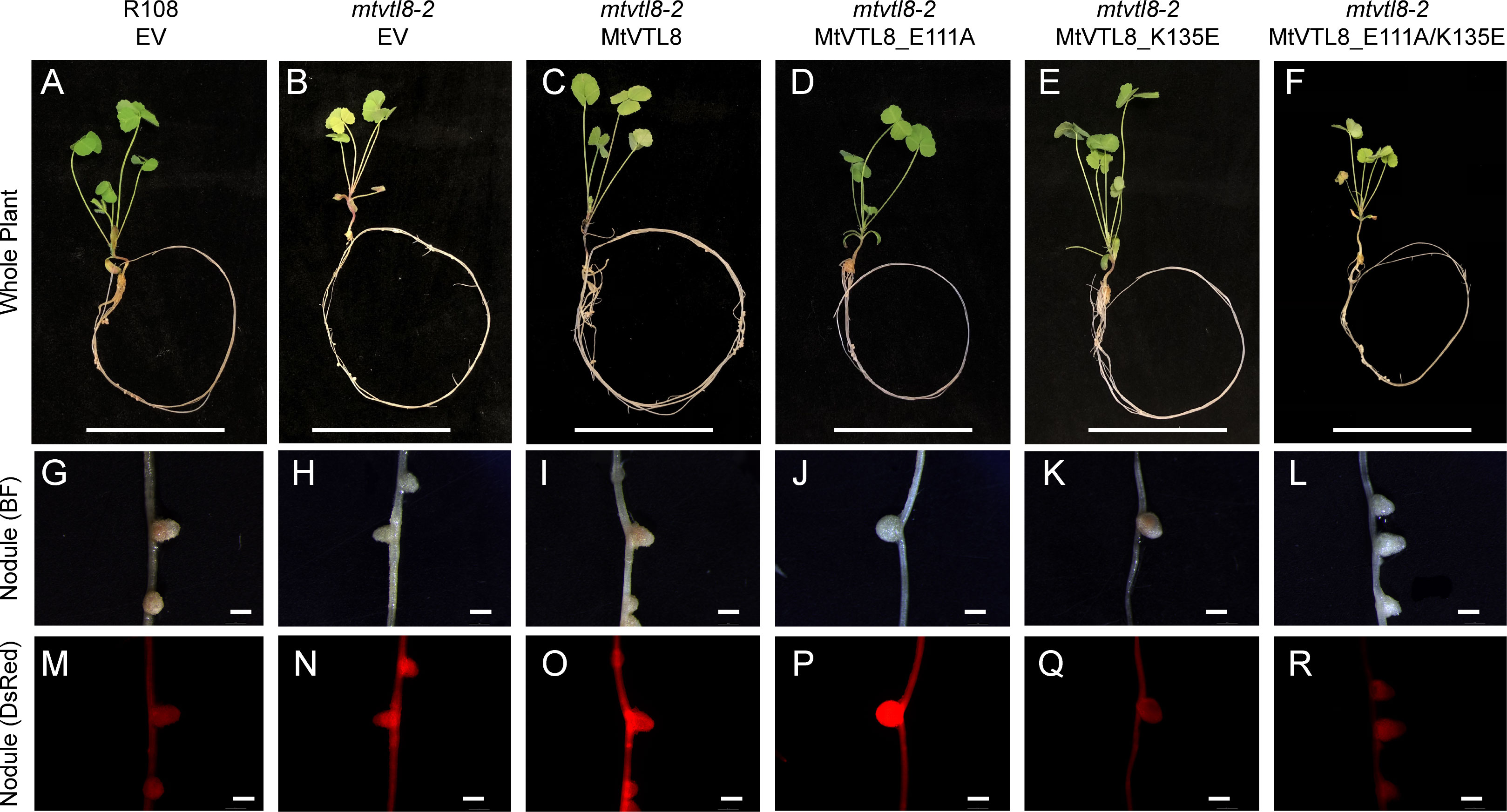
Figure 6 In planta complementation assays of the mtvtl8-2 mutant with wild-type MtVTL8 and its different mutated versions with mutations affected in residues capable of forming a salt bridge on the cytosolic face of the membrane. (A–F) Images of M. truncatula plants expressing different mutated versions of MtVTL8. From left to right, (A) R108 plant roots transformed with empty vector (EV); Mtvtl8-2 plant roots transformed with (B) EV, (C) MtVTL8 under pMtVTL8 (WT), (D) MtVTL8_E111A, (E) MtVTL8_K135A and (F) MtVTL8_E111K/K135E under pVTL8. (G–L) Bright field (BF) and (M–R) DsRed fluorescence images of transformed nodules corresponding to (A–F). Scale bars represent 5 cm for (A–F) and 1 mm for (G–R).
A thorough phylogenetic analysis of plant CCC1/VIT members is not available. Sorribes-Dauden et al. (2020) obtained a phylogenetic tree using 771 sequences from Archaea, bacteria, fungi, and plants showing that CCC1/VIT homologs can be classified in eight different groups. Groups I and II contain only Bacteria or Archaea proteins, respectively. Fungal VITs are clustered in Groups VI, VII and VIII. Plant VITs (69 sequences) belong to Group V, plant VTLs (111 sequences) belong to Group III. Group V also includes several bacterial VITs. Based on their phylogenetic analysis, Sorribes-Dauden et al. (2020) proposed that VITs and VTLs have a different origin: VITs originated from ancestral horizontal transfers from bacteria while VTLs emerged from an archaeal lineage, with both transfers dating at the origin of the last common eukaryote ancestor.
In this paper, we expanded the phylogenetic analysis to include VIT and VTL sequences from 37 plant genomes for a total of 306 sequences (Supplementary Table S4). We also included two sequences from the alga Chlamydomonas reinhardtii. After multiple sequence alignment, we obtained a maximum-likelihood phylogenetic tree (Supplementary Figure S10). The phylogenetic tree supports two main clades: one clade includes transporters that harbor a MBD (purple stars in Supplementary Figure S10) corresponding to VITs (green labels); the other clade contains sequences that lack the MBD corresponding to VITs (orange labels). However, among VTLs, there is an exception with three VTLs that contain a MBD, MpoVTL1, PpaVTL1 and PpaVTL2 (Supplementary Figure S10). In addition, we divided the VITs in two subclades, Group I and II, as they display different structural features as described below.
To learn more about the conservation level of specific amino acids in VTLs and VITs and to validate the results of our mutagenesis experiments, we used our multiple sequence alignment to create separated LOGOs of specific regions for the two phylogenetic groups. We observed that residues corresponding to Asp43 and Met80 in EgVIT1 (Supplementary Figures S11A, C), Asp59 and Met96 in MtVTL8 (Supplementary Figures S11B, D) are strictly conserved in VITs and VTLS, respectively. Intriguingly, we observed that Asp72 which has been proposed to have a role in iron translocation as well as proton movement toward Asp43 (Kato et al., 2019), is only conserved in Group I VITs, while it is a Gln in the 22 VITs belonging to Group II (Supplementary Figure S11C). In VTLs, Asp72 is diverged to a glycine (Gly88 in MtVTL8) or other non-chargeable residue in VTLs (Supplementary Figure S11D).
Residues involved in metal binding in the MBD, Glu102, Glu105, Glu113, Glu116, Met149, Met150, and Glu153 are strictly conserved in Group I VITs (Supplementary Figures S11E–G). However, in Group II VITs, these residues are far less conserved. With the exception of Glu102 that corresponds to Asp118 in MtVTL8, the metal binding residues are absent in VTLs as they lack the MBD region. Note that TMH2 is shorter in MtVTL8 and Asp118 is the last residue of TMH2. On a side note, we should mention that the glutamic acids within the cytoplasmic MBD are conserved in PfVIT but functional expression of mutants with substitutions of glutamic acid residues in ScΔccc1 showed iron tolerance (Sharma et al., 2021). Note that PfVIT was not included in our phylogenetic analysis. Additionally, two residues with a role in transferring iron ions from the MBD to the TMD, Glu32 and Asp36, are highly conserved in VITs, but are instead conserved non-polar residues in VTLs.
We also investigated if residues predicted to form two salt bridges, Arg51-Glu110 (TMH1-TMH2) and Glu111-Lys135 (TMH2-H1), in the MtVTL8’s AlphaFold model are conserved in VTLs and VITs. We obtained different results for the two salt bridges. Arg51 is conserved in both VITs and VTLs (Supplementary Figures S12A, B). However, Glu100 is only conserved in VTLs (Supplementary Figure S12D) and in Group II VITs (Supplementary Figure S12C), while it is a glycine in Group I VITs (Supplementary Figure S12C). Therefore, a salt bridge between TMH1 and TMH2 could only form in VTLs and Group II VITs. Finally, Glu111 (TMH2) is strictly conserved in all VTLs (Supplementary Figure S12D). The quality of alignment for the H1 region among VTLs was not enough to determine the conservation of Lys35 (H1).
These phylogenetic analyses reinforce the conclusions from our complementation studies with mutagenized MtVTL8 proteins. Together, they indicate that MtVTL8 and other VTLs operate by a mechanism that is distinct from that proposed for the VITs. Our analysis also indicates that a subset of VITs may have a different transport mechanism as they lack some of the structural features found in other well-characterized VITs.
Iron is an essential micronutrient for human nutrition and iron-deficiency anemia affects millions of people. Biofortification of crops such as wheat, corn and legumes has been indicated as a sustainable solution to combat malnutrition including iron deficiency (Garg et al., 2018; Ofori et al., 2022). Iron transporters from the CCC1/VIT family have been used to increase iron in the endosperm of wheat and barley by overexpression of TaVIT2 (Connorton et al., 2017) and in cassava roots by overexpression of AtVIT1 (Narayanan et al., 2015; Narayanan et al., 2019). Understanding the mechanism underlying iron transport by iron transporters including VTLs can result in future beneficial application for agronomic biofortification. Equally important is understanding the mechanisms of SNF that contribute to sustainable agriculture.
Our results show the spatial patterns of MtVTL8-promoter directed expression in roots in developing and mature nodules, with expression detected as early as 5 dpi in nodule primordia (Figure 1; Supplementary Figures S1, S2). The results in mature nodules (Figure 1) confirm previous results obtained by laser-capture dissection RNAseq for WT M. truncatula nodules (Roux et al., 2014) showing expression in the proximal areas of ZII, IZ and ZIII. Expression of MtVTL8 was not observed in the vascular system (Figure 1; Supplementary Figures S1A, E, S2A, B, S3A, B). These results are somewhat different from those obtained in a study of the homologous GmVTL1a gene, in which expression was observed in cells surrounding the nodule vasculature and in infected nodule cells (Brear et al., 2020). In mutant mtvtl8-2 nodules, expression was similar to WT (Supplementary Figures S2, S3 compare to Figure 1 and Supplementary Figure S1). It was expressed early in nodulation, in 5 dpi nodule primordia (Supplementary Figures S2H–J) and in the distal zones of the nodule, but much lower in the proximal zones, demarking the nodule zones where MtVTL8 is essential for nodule function (Supplementary Figures S1–3). This suggests that MtVTL8 is dispensable for early nodule development and only becomes essential after the bacteroids are enclosed by symbiosome membranes.
Because both MtVTL4 and MtVTL8 are expressed solely in nodules with different spatial expression patterns as assessed by RNAseq (Roux et al., 2014; Walton et al., 2020), with MtVTL4 only expressed in cells where rhizobia are being released into symbiosomes, we were curious to see if expression of MtVTL4 using MtVTL8 cis elements would complement the defect in mtvtl8-2; i.e., does MtVTL4 have similar functionality if expressed in place of MtVTL8? The p-MtVTL8-MtVTL4 construct did not complement mtvtl8-2 (Supplementary Figure S7), while expression of either gene in the yeast △ccc1 mutant rescued the iron toxicity phenotype (Figures 2C, D), leading us to propose that MtVTL4 and MtVTL8 have distinct functions beyond iron transport per se. We note that our results may be caused by intracellular localization of MtVTL4 vs. MtVTL8 as well as differences in function between the two proteins.
Functional studies for PtVIT and EgVIT1 (Labarbuta et al., 2017; Kato et al., 2019) showed they are Fe2+/H+ antiporters that exchange metal ions and protons on the opposite sides of lipid membranes. The solved crystal structure for EgVIT1, which captured the inside-open conformation, allowed the identification of essential amino acids for the transport of iron/protons (Kato et al., 2019). A model has been proposed in which the proton/iron transport cycle starts on the cytosolic side of the membrane where metal ions are captured by the MBD (Figure 7A). Conserved residues in the MBD and the TMD substrate channel entrance provide a pathway for the iron ions to translocate from the MBD to the TMD. Metal ions are transferred to a metal binding pocket in the TMD that is alternatively exposed to either side of the membrane. Two chargeable amino acids in the TMD, Asp43 and Glu72 are the initial iron and proton binding sides, respectively. After an exchange of position happens, irons are released in the vacuolar space and protons in the cytosol. We note that although most antiporters utilize a so-called single-site alternating-access mechanism, where the transporter can only associate with one substrate at a time, there are some examples of antiporters where ion and substrate binding sites were not overlapping, e.g. in the NorM transporters (Lu et al., 2013; Claxton et al., 2021). Despite the proposed model, many details of the mechanism of transport for EgVIT1 and for VITs in general are not fully understood. For instance, although a cooperative action is expected to take place between the TMD and the auxiliary cytoplasmic MBD, the role of the latter in transport is still not clear. Also, the stoichiometry of transport is not known.
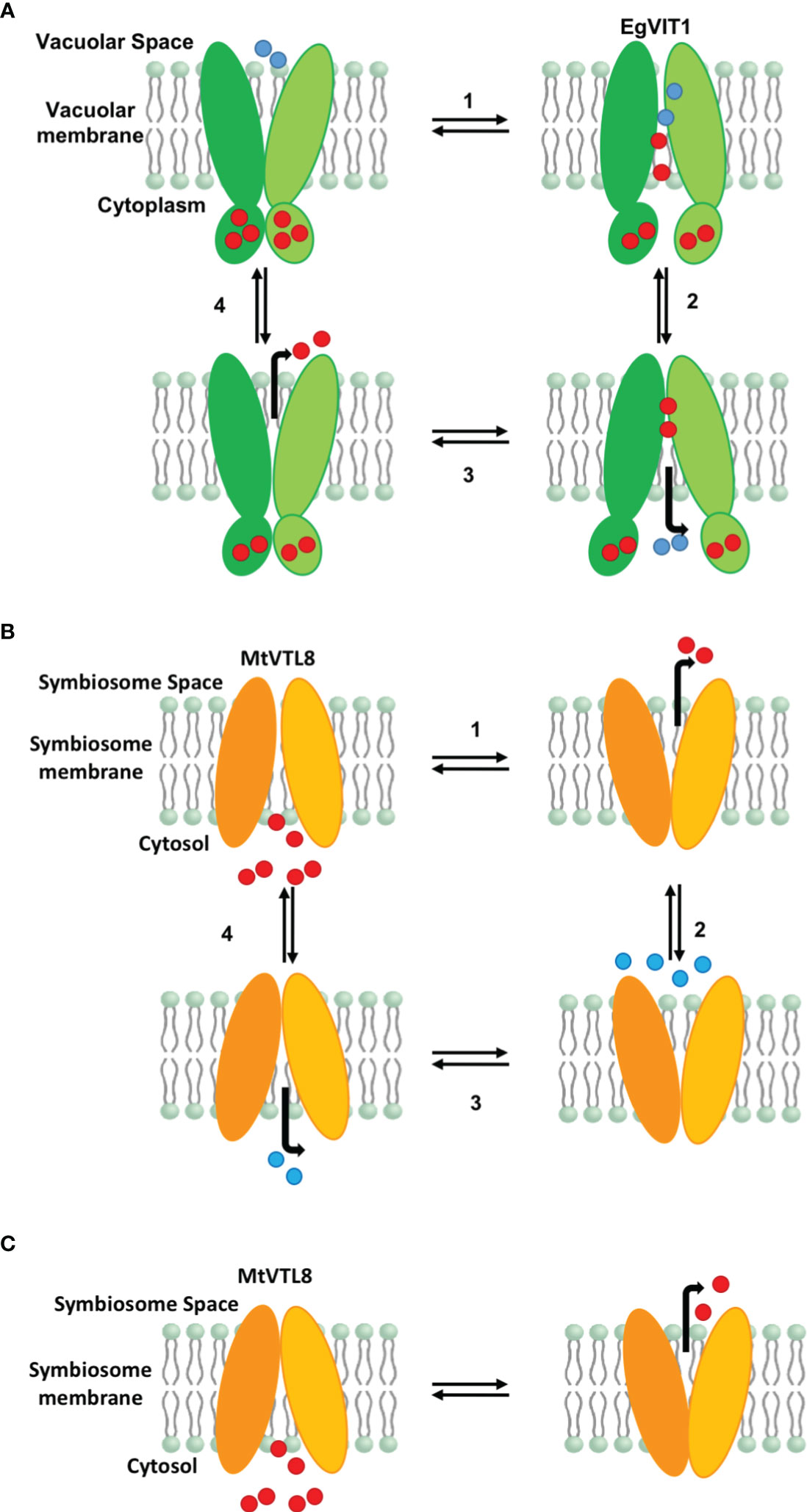
Figure 7 Proposed models for iron transport in plant VITs and VTLs. (A) Proposed model for EgVIT1 as a Fe2+/H+ antiporter that utilizes a multiple-site alternating-access mechanism. Iron ions are captured in the cytosol and stored in the MBD. Iron ions and protons travel in opposite directions through the transporter simultaneously during the alternating access cycle. Protons enter the transport channel with the transporter in the conformation open toward the vacuolar space. Iron ions enter the transport channel in the cytosol open conformation. Iron ions protons are exchanged in the cavity and protons are released in the cytosol. When the transporter opens again toward the vacuolar space, iron ions are released. (B) Proposed model for MtVTL8 as a Fe2+/H+ antiporter that utilizes a single-site alternating-access mechanism. Iron ions and protons do not travel simultaneously in the transport channel. As it lacks a MBD, MtVTL8 does not store iron ions. Instead iron ions enter the transporter channel when the protein is open toward the cytosol and are then released in the symbiosome space in the inward open conformation. Protons enter the transport channel and are released when the transporter opens again toward the cytoplasm. (C) Proposed model for MtVTL8 as a Fe2+ uniporter. In this model MtVTL8 operates as a uniporter and there is no transport of protons. (A, B) Numbers corresponds to the proposed order of events. EgVIT1 in green, MtVTL8 in orange. Iron ions are shown as red spheres, protons as blue spheres.
In the absence of a crystal structure for VTLs, we obtained a model for MtVTL8. Using such model as a guide we identified amino acids potentially involved in iron transport in MtVTL8. We demonstrated that when mutated, the resulting protein cannot restore SNF in plants or iron tolerance in yeast confirming our hypotheses. We proposed that MtVTL8 could rely on a different transport mechanism from that proposed for VITs and suggested two alternative mechanisms. As discussed previously, VTLs, including MtVTL8, lack the cytoplasmic MBD and the amino acids that can capture metal ions in the cytoplasm. Additionally, they lack amino acids that guide the metal ions toward the binding pocket and an essential glutamic acid residue, Glu72, that has been indicated to work as relay for proton-iron exchange in the TMD. In VTLs, Glu72 is substituted with a glycine (Gly88 in MtVTL8) but our studies have shown that the MtVTL8_Gly88Glu mutant is not an active transporter (Figures 2H,3E, J, O). Based on these observations, we have two working hypotheses. Our first hypothesis is that MtVTL8 is a Fe2+/H+ antiporter that uses the symbiosome proton gradient to fuel Fe2+ transport (Figure 7B). In this scenario, in MtVTL8, Asp59, the only membrane-embedded charged residue, is a common binding site for substrate and protons, and the occupancy of this site is mutually exclusive (Figure 7B). This is agreement with what was proposed for other antiporters and called single-site alternating-access mechanism of transport (Yerushalmi and Schuldiner, 2000; Schuldiner, 2014). Another possibility is that MtVTL8 uses an antiporter mechanism similar to that proposed to EgVIT1 but uses an amino acid residue distant from Gly88 as a proton acceptor. Because we lack experimental evidence that MtVTL8 is an Fe2+/H+ antiporter, there exists the less-likely possibility that MtVTL8 and other VTLs may operate as Fe2+ efflux channels instead of being active as active Fe2+/H+ antiporters (Figure 7C). In this model, Fe2+ binds to the conserved Asp59 inside the channel and is transported without an exchange of protons. This hypothetical mechanism spares the pH gradient across the symbiosome membrane required for nitrogen fixation (Udvardi and Day, 1989; Pierre et al., 2013). The gradient is essential for the import of dicarboxylates for SNF and believed to be important for the import of other molecules. In this less-likely scenario, the recently identified sequestration of heme by M. truncatula’s NCR247 that overrides the bacteroids’ negative Fe2+ regulation of Fe2+ import (Sankari et al., 2022) might result in a lowered Fe2+ concentration in the symbiosome space. That lowered Fe2+ concentration might be low enough to provide a sufficient driving force for Fe2+ movement across the symbiosome membrane from the cytosol without requiring an additional source of energy for transport. Additional studies are needed to address these hypotheses including molecular dynamics calculations, transport assays and experimental structures for VTLs.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
JC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. AL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. RD: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partially funded by US National Science Foundation, grant 2139351.
We thank Drs. Wei Liu and Michael Udvardi for the pMU vectors used in this work, Dr. Elinor Lichtenberg for help with statistics, and the reviewers for thoughtful and helpful comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1306491/full#supplementary-material
At, Arabidopsis thaliana; CCC1, Cross-complements Ca2+ phenotype of csg1; Eg, Eucaliptus grandis; Mt, Medicago truncatula; SNF, symbiotic nitrogen fixation; VIT, Vacuolar iron transporter; VTL, Vacuolar iron transporter-like.
Appleby, C. A. (1984). Leghemoglobin and rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478. doi: 10.1146/annurev.pp.35.060184.002303
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts, polyphenolexidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Auriac, M.-C., Timmers, A. C. J. (2007). Nodulation studies in the model legume Medicago truncatula: Advantages of using the constitutive EF1α promoter and limitations in detecting fluorescent reporter proteins in nodule tissues. Mol. Plant Microbe Interact. 20, 1040–1047. doi: 10.1094/MPMI-20-9-1040
Axelos, M., Bardet, C., Liboz, T., Le Van Thai, A., Curie, C., Lescure, B. (1989). The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α: Molecular cloning, characterization and expression. Mol. Gen. Genet. 219, 106–112. doi: 10.1007/BF00261164
Barker, D. G., Pfaff, T., Moreau, D., Groves, E., Ruffel, S., Lepetit, M., et al. (2006). “Growing M. truncatula: choice of substrates and growth conditions,” in The Medicago truncatula handbook. Eds. Mathesius, U., Journet, E.-P., Sumner, L. W. Available at: https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=9ff882f0d91b9e3bbb8217920c8f5eb94d9c3d2c.
Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C., Barker, D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700. doi: 10.1094/MPMI.2001.14.6.695
Brear, E. M., Bedon, F., Gavrin, A., Kryvoruchko, I. S., Torres-Jerez, I., Udvardi, M. K., et al. (2020). GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. New Phytol. 228, 667–681. doi: 10.1111/nph.16734
Brear, E. M., Day, D. A., Smith, P. M. C. (2013). Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci. 13, 359. doi: 10.3389/fpls.2013.00359
Burton, J. W., Harlow, C., Theil, E. C. (1998). Evidence for reutilization of nodule iron in soybean seed development. J. Plant Nutr. 21, 913–927. doi: 10.1080/01904169809365453
Cai, J., Veerappan, V., Arildsen, K., Sullivan, C., Piechowicz, M., Frugoli, J., et al. (2023). A modified aeroponic system for growing small−seeded legumes and other plants to study root systems. Plant Methods 19, 21. doi: 10.1186/s13007-023-01000-6
Cai, J., Veerappan, V., Troiani, T., Mysore, K. S., Wen, J., Dickstein, R. (2022). “Whole genome sequencing identifies a Medicago truncatula Tnt1 insertion mutant in the VTL8 gene that is essential for symbiotic nitrogen fixation,” in The Medicago truncatula Genome. Eds. Sinharoy, S., Kang, Y., Benedito, V. (Switzerland: Springer Nature Switzerland AG).
Carrere, S., Verdier, J., Gamas, P. (2021). MtExpress, a comprehensive and curated RNAseq-based gene expression atlas for the model legume Medicago truncatula. Plant Cell Physiol. 62, 1494–1500. doi: 10.1093/pcp/pcab110
Clarke, V. C., Loughlin, P. C., Day, D. A., Smith, P. M. C. (2014). Transport processes of the legume symbiosome membrane. Front. Plant Sci. 5, 699. doi: 10.3389/fpls.2014.00699
Claxton, D. P., Jagessar, K. L., Mchaourab, H. S. (2021). Principles of alternating access in multidrug and toxin extrusion (MATE) transporters. J. Mol. Biol. 6, 166959. doi: 10.1016/j.jmb.2021.166959
Connorton, J. M., Jones, E. R., Rodríguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., Balk, J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
Crooks, G. E., Hon, G., Chandonia, J.-M., Brenner, S. E. (2014). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Day, D. A., Smith, P. M. C. (2021). Iron transport across symbiotic membranes of nitrogen-fixing legumes. Int. J. Mol. Sci. 22, 432. doi: 10.3390/ijms22010432
Einsle, O., Rees, D. C. (2020). Structural enzymology of nitrogenase enzymes. Chem. Rev. 120, 4969–5004. doi: 10.1021/acs.chemrev.0c00067
Escudero, V., Abreu, I., Tejada-Jiménez, M., Rosa-Núñez, E., Quintana, J., Prieto, R. I., et al. (2020). Medicago truncatula Ferroportin2 mediates iron import into nodule symbiosomes. New Phytol. 228, 194–209. doi: 10.1111/nph.16642
Fåhraeus, G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16, 374–381. doi: 10.1099/00221287-16-2-374
Farkas, A., Maróti, G., Dürgő, H., Györgypál, Z., Lima, R. M., Medzihradszky, K. F., et al. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 5183–5188. doi: 10.1073/pnas.1404169111
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 433, 166959. doi: 10.3389/fnut.2018.00012
Gilles-Gonzalez, M. A., Ditta, G. S., Helinski, D. R. (1991). A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350, 170–172. doi: 10.1038/350170a0
Gollhofer, J., Timofeev, R., Lan, P., Schmidt, W., Buckhout, T. J. (2014). Vacuolar-Iron-Transporter1-Like proteins mediate iron homeostasis in Arabidopsis. PloS One 9, e110468. doi: 10.1371/journal.pone.0110468
González-Guerrero, M., Matthiadis, A., Sáez, Á., Long, T. A. (2014). Fixating on metals: new insights into the role of metals in nodulation and symbiotic nitrogen fixation. Front. Plant Sci. 5, 45. doi: 10.3389/fpls.2014.00045
González-Guerrero, M., Navarro-Gómez, C., Rosa-Núñez, E., Echávarri-Erasun, C., Imperial, J., Escudero, V. (2023). Forging a symbiosis: transition metal delivery in symbiotic nitrogen fixation. New Phytol. 239, 2113–2125. doi: 10.1111/nph.19098
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hakoyama, T., Niimi, K., Yamamoto, T., Isobe, S., Sato, S., Nakamura, Y., et al. (2012). The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 53, 225–236. doi: 10.1093/pcp/pcr167
Hirsch, A. M. (1992). Developmental biology of legume nodulation. New Phytol. 122, 211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hoffman, B. M., Lukoyanov, D., Yang, Z.-Y., Dean, D. R., Seefeldt, L. C. (2014). Mechanism of nitrogen fixation by nitrogenase: The next stage. Chem. Rev. 114, 4041–4062. doi: 10.1021/cr400641x
Jamet, A., Sigaud, S., Van de Sype, G., Puppo, A., Hérouart, D. (2003). Expression of the bacterial catalase genes during Sinorhizobium meliloti–Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant Microbe Interact. 16, 217–225. doi: 10.1094/MPMI.2003.16.3.217
Jefferson, R., Kavanagh, T., Bevan, M. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Johnston, A. W., Yeoman, K. H., Wexler, M. (2001). Metals and the rhizobial-legume symbiosis–uptake, utilization and signalling. Adv. Microbial. Physiol. 45, 113–156. doi: 10.1016/S0065-2911(01)45003-X
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kato, T., Kumazaki, K., Wada, M., Taniguchi, R., Nakane, T., Yamashita, K., et al. (2019). Crystal structure of plant vacuolar iron transporter VIT1. Nat. Plants 5, 308–315. doi: 10.1038/s41477-019-0367-2
Kim, S. A., Punshon, T., Lanzirotti, A., Li, L., Alonso, J. M., Ecker, J. R., et al. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314, 1295–1298. doi: 10.1126/science.1132563
Kondorosi, A., Vincze, E., Johnston, A. W. B., Beringer, J. E. (1980). A comparison of three Rhizobium linkage maps. Mol. Gen. Genet. 178, 403–408. doi: 10.1007/BF00270491
Kryvoruchko, I. S., Routray, P., Sinharoy, S., Torres-Jerez, I., Tejada-Jiménez, M., Finney, L. A., et al. (2018). An iron-activated citrate transporter, MtMATE67, is required for symbiotic nitrogen fixation. Plant Physiol. 176, 2315–2329. doi: 10.1104/pp.17.01538
Labarbuta, P., Duckett, K., Botting, C. H., Chahrour, O., Malone, J., Dalton, J. P., et al. (2017). Recombinant vacuolar iron transporter family homologue PfVIT from human malaria-causing Plasmodium falciparum is a Fe2+/H+exchanger. Sci. Rep. 7, 42850. doi: 10.41038/srep42850
Ledbetter, R. N., Garcia Costas, A. M., Lubner, C. E., Mulder, D. W., Tokmina-Lukaszewska, M., Artz, J. H., et al. (2017). The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: Generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56, 4177–4190. doi: 10.1021/acs.biochem.7b00389
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, L., Chen, O. S., Ward, D. M., Kaplan, J. (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276, 29515–29519. doi: 10.1074/jbc.M103944200
Liang, Y., Urano, D., Liao, K.-L., Hedrick, T. L., Gao, Y., Jones, A. M. (2017). A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 13, 26. doi: 10.1186/s13007-017-0174-6
Limpens, E., Moling, S., Hooiveld, G., Pereira, P. A., Bisseling, T., Becker, J. D., et al. (2013). Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PloS One 8, e64377. doi: 10.1371/journal.pone.0064377
Liu, S., Liao, L. L., Nie, M. M., Peng, W. T., Zhang, M. S., Lei, J. N., et al. (2020). A VIT-like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. New Phytol. 225, 1413–1428. doi: 10.1111/nph.16506
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I., Lomize, A. L., et al. (2012). OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370-376. doi: 10.1093/nar/gkr703
Lu, M., Symersky, J., Radchenko, M., Koide, A., Guo, Y., Nie, R., et al. (2013). Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U.S.A. 110, 2099–2104. doi: 10.1073/pnas.1219901110
Lullien, V., Barker, D. G., de Lajudie, P., Huguet, T. (1987). Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol. Biol. 9, 469–478. doi: 10.1007/BF00015878
Marlow, V. L., Haag, A. F., Kobayashi, H., Fletcher, V., Scocchi, M., Walker, G. C., et al. (2009). Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191, 1519–1527. doi: 10.1128/JB.01661-08
Maróti, G., Kondorosi, É. (2014). Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Microbiol. 5, 326. doi: 10.3389/fmicb.2014.00326
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees (New Orleans, LA, USA: 2010 Gateway Computing Environments Workshop (GCE), 1–8.
Momonoi, K., Yoshida, K., Mano, S., Takahashi, H., Nakamori, C., Shoji, K., et al. (2009). A vacuolar iron transporter in tulip, TgVit1, is responsible for blue coloration in petal cells through iron accumulation. Plant J. 59, 437–447. doi: 10.1111/j.1365-313X.2009.03879.x
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitán-Solís, E., Gehan, J., Butts, P., et al. (2019). Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 37, 144–151. doi: 10.1038/s41587-018-0002-1
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitán-Solis, E., Grusak, M. A., Taylor, N., et al. (2015). Overexpression of Arabidopsis VIT1 increases accumulation of iron in cassava roots and stems. Plant Sci. 240, 170–181. doi: 10.1016/j.plantsci.2015.09.007
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
O'Brian, M. R. (2015). Perception and momeostatic control of iron in the rhizobia and related bacteria. Annu. Rev. Microbiol. 69, 229–245. doi: 10.1146/annurev-micro-091014-104432
O'Hara, G. W., Boonkerd, N., Dilworth, M. J. (1988). Mineral constraints to nitrogen fixation. Plant Soil 108, 93–110. doi: 10.1007/BF02370104
Ofori, K. F., Antoniello, S., English, M. M., Aryee, A. N. A. (2022). Improving nutrition through biofortification–A systematic review. Front. Nutr. 9, 1043655. doi: 10.3389/fnut.2022.1043655
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E. D., Murray, J. D., Poole, P. S., Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Ott, T., van Dongen, J. T., Gunther, C., Krusell, L., Desbrosses, G., Vigeolas, H., et al. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15, 531–535. doi: 10.1016/j.cub.2005.01.042
Pierre, O., Engler, G., Hopkins, J., Brau, F., Boncompagni, E., Hérouart, D. (2013). Peribacteroid space acidification: a marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 36, 2059–2070. doi: 10.1111/pce.12116
Preisig, O., Zufferey, R., Thöny-Meyer, L., Appleby, C. A., Hennecke, H. (1996). A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178, 1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996
Rodríguez-Haas, B., Finney, L., Vogt, S., González-Melendi, P., Imperial, J., González-Guerrero, M. (2013). Iron distribution through the developmental stages of Medicago truncatula nodules. Metallomics 5, 1247–1253. doi: 10.1039/c3mt00060e
Roux, B., Rodde, N., Jardinaud, M.-F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. doi: 10.1111/tpj.12442
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Sankari, S., Babu, V. M. P., Bian, K., Alhhazmi, A., Andorfer, M. C., Avalos, D. M., et al. (2022). A haem-sequestering plant peptide promotes iron uptake in symbiotic bacteria. Nat. Microbiol. 7, 1453–1465. doi: 10.1038/s41564-022-01192-y
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schuldiner, S. (2014). Competition as a way of life for H(+)-coupled antiporters. J. Mol. Biol. 426, 2539–2546. doi: 10.1016/j.jmb.2014.05.020
Sharma, P., Tóth, V., Hyland, E. M., Law, C. J. (2021). Characterization of the substrate binding site of an iron detoxifying membrane transporter from Plasmodium falciparum. Malaria J. 20, 295. doi: 10.1186/s12936-021-03827-7
Slavic, K., Krishna, S., Lahree, A., Bouyer, G., Hanson, K. K., Vera, I., et al. (2016). A vacuolar iron-transporter homologue acts as a detoxifier in Plasmodium. Nat. Commun. 7, 10403. doi: 10.1038/ncomms10403
Sorribes-Dauden, R., Peris, D., Martínez-Pastor, M. T., Puig, S. (2020). Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi. Comput. Struct. Biotechnol. J. 18, 3712–3722. doi: 10.1016/j.csbj.2020.10.044
Suganuma, N., Nakamura, Y., Yamamoto, M., Ohta, T., Koiwa, H., Akao, S., et al. (2003). The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol. Genet. Genomics 269, 312–320. doi: 10.1007/s00438-003-0840-4
Tang, C., Robson, A. D., Dilworth, M. J. (1992). The role of iron in the (brady) Rhizobium legume symbiosis. J. Plant Nutr. 15, 2235–2252. doi: 10.1080/01904169209364471
Tejada-Jiménez, M., Castro-Rodríguez, R., Kryvoruchko, I., Lucas, M. M., Udvardi, M., Imperial, J., et al. (2015). Medicago truncatula Natural Resistance-Associated Macrophage Protein1 is required for iron uptake by rhizobia-infected nodule cells. Plant Physiol. 168, 258–272. doi: 10.1104/pp.114.254672
Thomas, P. D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, L.-P., Mi, H. (2022). PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22. doi: 10.1002/pro.4218
Udvardi, M. K., Day, D. A. (1989). Electrogenic ATPase activity on the peribacteroid membrane of soybean (Glycine max L.) root nodules. Plant Physiol. 90, 982–987. doi: 10.1104/pp.90.3.982
Udvardi, M., Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
UniProt Consortium (2023). UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531. doi: 10.1093/nar/gkac1052
Valimehr, S., Sanjarian, F., Sohi, H. H., Sharafi, A., Sabouni, F. (2014). A reliable and efficient protocol for inducing genetically transformed roots in medicinal plant Nepeta pogonosperma. Physiol. Mol. Biol. Plants 20, 351–356. doi: 10.1007/s12298-014-0235-5
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Vasse, J., de Billy, F., Camut, S., Truchet, G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172, 4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990
Walton, J. H., Kontra-Kováts, G., Green, R. T., Domonkos, Á., Horváth, B., Brear, E. M., et al. (2020). The Medicago truncatula Vacuolar iron Transporter-Like proteins VTL4 and VTL8 deliver iron to symbiotic bacteria at different stages of the infection process. New Phytol. 228, 651–666. doi: 10.1111/nph.16735
Witty, J. F., Skøt, L., Revsbech, N. P. (1987). Direct evidence for changes in the resistance of legume root nodules to O2 diffusion. J. Exp. Bot. 38, 1129–1140. doi: 10.1093/jxb/38.7.1129
Yerushalmi, H., Schuldiner, S. (2000). A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett. 476, 1873–3468. doi: 10.1016/S0014-5793(00)01677-X
Yu, Y.-C., Dickstein, R., Longo, A. (2021). Structural modeling and in planta complementation studies link mutated residues of the Medicago truncatula nitrate transporter NPF1.7 to functionality in root nodules. Front. Plant Sci. 12, 685334. doi: 10.3389/fpls.2021.685334
Keywords: Medicago truncatula, symbiotic nitrogen fixation, iron transporter, site-directed mutagenesis, protein models, transporter function
Citation: Cai J, Longo A and Dickstein R (2024) Expression and mutagenesis studies in the Medicago truncatula iron transporter MtVTL8 confirm its role in symbiotic nitrogen fixation and reveal amino acids essential for transport. Front. Plant Sci. 14:1306491. doi: 10.3389/fpls.2023.1306491
Received: 03 October 2023; Accepted: 28 November 2023;
Published: 04 January 2024.
Edited by:
Chang Fu Tian, China Agricultural University, ChinaReviewed by:
Igor Kryvoruchko, United Arab Emirates University, United Arab EmiratesCopyright © 2024 Cai, Longo and Dickstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca Dickstein, YmVjY2FkQHVudC5lZHU=
†These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.