
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 December 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1301084
This article is part of the Research Topic Plant Secondary Metabolite Biosynthesis View all 12 articles
 Ping Wang†
Ping Wang† Yan Yan†
Yan Yan† Min Yan
Min Yan Xiangmin Piao
Xiangmin Piao Yingping Wang
Yingping Wang Xiujuan Lei
Xiujuan Lei He Yang
He Yang Nanqi Zhang
Nanqi Zhang Wanying Li
Wanying Li Peng Di*
Peng Di* Limin Yang*
Limin Yang*Introduction: The BAHD (benzylalcohol O-acetyl transferase, anthocyanin O-hydroxycinnamoyl transferase, N-hydroxycinnamoyl anthranilate benzoyl transferase and deacetylvindoline 4-O-acetyltransferase), has various biological functions in plants, including catalyzing the biosynthesis of terpenes, phenolics and esters, participating in plant stress response, affecting cell stability, and regulating fruit quality.
Methods: Bioinformatics methods, real-time fluorescence quantitative PCR technology, and ultra-high-performance liquid chromatography combined with an Orbitrap mass spectrometer were used to explore the relationship between the BAHD gene family and malonyl ginsenosides in Panax ginseng.
Results: In this study, 103 BAHD genes were identified in P. ginseng, mainly distributed in three major clades. Most PgBAHDs contain cis-acting elements associated with abiotic stress response and plant hormone response. Among the 103 genes, 68 PgBAHDs are WGD (whole-genome duplication) genes. The significance of malonylation in biosynthesis has garnered considerable attention in the study of malonyltransferases. The phylogenetic tree results showed 34 PgBAHDs were clustered with genes that have malonyl characterization. Among them, seven PgBAHDs (PgBAHD4, 45, 65, 74, 90, 97, and 99) showed correlations > 0.9 with crucial enzyme genes involved in ginsenoside biosynthesis and > 0.8 with malonyl ginsenosides. These seven genes were considered potential candidates involved in the biosynthesis of malonyl ginsenosides.
Discussion: These results help elucidate the structure, evolution, and functions of the P. ginseng BAHD gene family, and establish the foundation for further research on the mechanism of BAHD genes in ginsenoside biosynthesis.
The BAHD acyltransferase family mainly uses coenzyme A thioesters as acyl donors and alcohols or amines as acceptors to catalyze acylation reactions to form various acylation products. (St-Pierre and Luca, 2000). A comprehensive comparative analysis of the amino acid sequences of the currently identified BAHD acyltransferase family members reveals that the family proteins’ amino acid sequences all contain two conserved regions, HXXXD and DFGWG (Molina and Kosma, 2015). The HXXXD conserved region located in the active center of the enzyme is also distributed in other acyltransferase families, such as chloramphenicol acetyltransferase type I, II, III (chloramphenicol acetyltransferase,CAT) and choline/carnitine O-acyltransferase. The DFGWG conserved region is located at the C-terminal. In addition, acyltransferases related to anthocyanin/flavonoid biosynthesis also contain YFGNC conserved sequences (Suzuki et al., 2004; Unno et al., 2007; Yu et al., 2009).
Acylation modifications mediated by acyltransferases are prevalent in the structural modification of natural products. They are essential for enriching the structural diversity of plant secondary metabolites, enhancing the stability, lipid solubility, and improving the bioavailability of compounds (Matern et al., 1986; Ardhaoui et al., 2004; Mellou et al., 2006; Taguchi et al., 2010). The BAHD acyltransferase family is a class of proteins unique to plants for acylation modification of secondary metabolites. It is vital in the biosynthesis of a wide range of active acylated natural products (Yu et al., 2009). Researchers have explored the role of BAHD acyltransferase in the biosynthesis of saponin adjuvants from the soapbark tree. Identified one enzyme (encoded by Qs0206480) that generated a product consistent with the addition of an acetyl group, and two enzymes (encoded by Qs0023500 and Qs0213660) that likely corresponded to the addition of L-rhamnose and D-glucose, respectively. The study provides insights into the biosynthetic pathway of saponin adjuvants and highlights the role of BAHD acyltransferases in the modification of the heptasaccharide scaffold (Reed et al., 2023). There is research explored the role of BAHD acyltransferase in the biosynthesis of aescin and aesculin in horse chestnut. AcBAHD3 and AcBAHD6 were able to acetylate the hydroxyl group of aescin precursor, yielding a product called 22-O-acetylprotoaescigenin, and also found that these enzymes could use acetyl-CoA as a donor to catalyze the desacetylation of aescin, resulting in the formation of aesculin. This study provides evidence for the involvement of BAHD acyltransferase in the acylation and deacetylation of triterpenoid compounds in horse chestnut. Highlighted the role of BAHD enzymes in the diversification of triterpenoid compounds in plants (Sun et al., 2023). In the study conducted by GAME36, a BAHD-type acyltransferase, it was found that SGA-acetylation occurs in both cultivated and wild tomatoes. This process involves the conversion of α-tomatine to Esculeoside A, which is non-bitter and less toxic. The researchers successfully elucidated the biosynthesis pathway of core Esculeoside A in ripe tomatoes (Sonawane et al., 2023).
Panax ginseng is a perennial herbaceous plant with a long growth period. Modern pharmacological studies have shown that ginsenosides are the main medicinal components of ginseng. Malonyl ginsenosides are natural ginsenosides that contain a malonyl group attached to a glucose unit of the corresponding neutral ginsenosides (Wang et al., 2016). In ginseng, the proportion of malonyl ginsenosides m-Rb1, m-Rb2, m-Rc, and m-Rd in the total ginsenosides ranged from 35% to 60% (Liu et al., 2012). More than 20 malonyl ginsenosides have been identified by high-resolution mass spectrometry (Sun et al., 2012; Wan et al., 2015). The role of BAHD acyltransferase, a key enzyme in the acylation of secondary metabolites, in the biosynthesis of malonyl ginsenosides has not yet been reported, so the identification and analysis of the ginseng BAHD genes is of great significance.
In this study, a total of 103 BAHD family genes were identified, and the analysis of evolutionary relationships indicated that PgBAHDs could be divided into five evolutionary clades. The gene structure, chromosomal localization, inter-gene, and inter-species collinearity of PgBAHDs were further investigated. In addition, the expression profiles of PgBAHD genes in different tissues and the trend of response to various abiotic treatments were investigated. Co-expression analysis of ginsenoside biosynthesis pathway genes with PgBAHD genes and secondary metabolite malonyl ginsenoside with PgBAHD genes was performed. Eventually eight PgBAHDs were identified as genes that may be involved in malonyl ginsenoside biosynthesis. This study provides a reliable basis for further metabolic regulation of the ginsenoside biosynthetic pathway and for conducting corresponding synthetic biology studies and molecular breeding.
The candidate BAHD genes were initially acquired from the Ginseng Genome Data resource (Wang et al., 2022). The Hidden Markov Models (HMMs) for the conserved BAHD domain (Pfam: PF02458) were extracted from the Pfam database (http://pfam.xfam.org). The HMMER 3.2.1 software was employed to detect the BAHD genes obtained from the ginseng genome, with an E-value threshold set at 10−2. To ensure the presence of the BAHD PF02458 domain, all candidate PgBAHDs were further validated using the SMART data resource (http://smart.embl.de/), NCBI-Conserved Domain Database (CDD), and PlantTFDB (Plant Transcription Factor Database) (http://planttfdb.cbi.pku.edu.cn). Subsequently, their HXXXD and DFGWG motifs were examined.
Furthermore, the online Sequence Manipulation Suite was utilized to predict the theoretical pI and MW of PgBAHD proteins (http://www.detaibio.com/sms2/reference.html) (Stothard, 2000).
Mafft (https://mafft.cbrc.jp/alignment/software/) was used with default parameters to perform multiple alignments of ginseng BAHD sequences and multiple alignments of BAHD, among other species. The ginseng BAHD phylogenetic tree was constructed using the maximum likelihood method IQ-TREE based on the JTTDCMut+F+R4 model (Nguyen et al., 2015), and the nodes were tested 1000 times using bootstrap analysis. Further annotation of the phylogenetic tree results was conducted using Evolview (https://evolgenius.info/).
TBtools 1.053 was employed to demonstrate the gene structure (Chen et al., 2020). Conserved motifs of PgBAHDs were identified using MEME (Multiple expectation maximization for motif elicitation) native software (version 4.12.0) in Linux with a maximum of 10 mismatches and an optimal motif width of 6-100 amino acid residues.
The sequence of 2000 bp upstream of the start codon of PgBAHDs was obtained for promoter analysis. PlantCARE (http://bionformatics.psb.ugent.be/webtools/plantcare/html) was used to predict cis-acting elements in the promoter region, and PlantTFDB software (http://planttfdb.cbi.pku.edu.cn/) was used online to predict the distribution of promoter transcription factor binding sites (p-value ≤ 1e−6).
The MCScanX program was utilized to conduct inter- and intra-species collinearity analysis of proteins, using an E value threshold of 1e-5. Furthermore, the Duplicate Gene Classifier script within the MCScanX program was employed to quantify different types of duplication. The results were visualized using Circos (Krzywinski et al., 2009; Wang et al., 2012).
Using KaKS-Calculator-2.0, this study calculates replicated gene pairs’ Ka and KS. The analysis aims to assess the environmental selection pressure by examining the Ka/KS ratio (Wang et al., 2010).
To analyze gene expression among different tissues and responses to various abiotic treatments, RNA-Seq datasets from 14 different tissues were obtained from NCBI (accession number PRJNA302556) (Wang et al., 2015). Additionally, 15 RNA-Seq datasets for abiotic treatment were retrieved (No.24-38 in the ginseng transcriptome data resource, http://ginsengdb.snu.ac.kr/transcriptome.php) from the Ginseng Genome Data Resource (http://ginsengdb.snu.ac.kr/). The clean reads were aligned to the ginseng genome using Hisat2 software. Assembly and calculation of expression values for each transcript were performed using Hisat2, StringTie, and Ballgown.
In a previous study, the RG, AR, and CT cDNA libraries were established. These nine cDNA libraries were subsequently sequenced on HiSeq 2500 (Illumina) with the PE125 strategy. The TPM was calculated using the same protocol as the other 16 RNA-Seq datasets. Finally, the heatmap was generated using the R package ‘Heatmap’.
Total RNA was extracted from the samples using an EasyPure Plant RNA Kit (TransGen Biotech), with the inclusion of RNase-free DNase I (TransGen Biotech) to eliminate DNA contamination. The concentration and quality of the RNA samples were assessed using a NanoPhotometer N50 (Implen, GER). Subsequently, the Perfect-Start Uni RT-qPCR Kit (TransGen Biotech) was employed to reverse transcribe RNA into cDNA, followed by two-step quantitative real-time PCR using a Roche Light Cycler 96 (SYBR-GREEN I; No Passive Reference Dye). The β-Actin gene was utilized as the internal control (Hou et al., 2014). Data analyses involved the use of the 2−ΔΔCT method for determining the relative expression of PgBAHD genes (Livak and Schmittgen, 2023). Primers for qRT-PCR were synthesized by Sangon Biotech (ShangHai, China), and their sequences are listed in Supplementary Table S7.
Each dried powdered sample, weighing 0.5 g and with a mesh size of < 40, was accurately weighed. The samples were then sonicated with 10 ml of 80% ethanol for 40 min (100 W, 40 KHz) and centrifugation at 10,000 rpm for 10 min. This process was repeated three times, and the resulting supernatants were combined and transferred into a 10 ml volumetric flask. The volumetric flask was adjusted to a final volume of 10 ml using 80% ethanol and thoroughly mixed. Before injection, the solution was filtered twice through a 0.45 μm organic membrane.
Reference standards of ginsenosides, purchased from Shanghai Yuanye Biotechnology Co., Ltd (Shanghai, China) with purities not less than 98.0%, were used. Approximately 5 mg of each standard was taken into a 5 ml volumetric flask, dissolved with methanol, and diluted to the scale to obtain a reserve solution of each standard with a 1 mg/ml concentration. An appropriate amount of each stock solution was dispensed, diluted with methanol, and adjusted to a final concentration of about 50 μg/ml to create the mixed standard solution.
The sample components were separated using a Thermo Fisher Vanquish liquid chromatography system (Thermo Fisher Scientific, San Jose, CA, USA). The chromatographic column used was a Hypersil Gold Vanquish UHPLC column (100×2.1 mm, 1.9 μm; Thermo). In this experiment, the mobile phase consisted of acetonitrile (A) and 0.1% formic acid in water. The gradient elution program followed the following steps: 0-34.0 min, 15%-55% A; 34-35 min, 55%-98% A; 35-36 min, 98% A; 36-37 min, 98%-15% A; 37-40 min, 15% A. The column temperature was maintained at 35°C, the flow rate was set at 0.30 ml/min, and the injection volume for each sample was 1 μl. During the separation process, the column temperature remained at 35°C, and the flow rate used was 0.30 ml/min, with an injection volume of 1 μl for each sample.
MS spectrometric detection was performed on an Orbitrap Fusion mass spectrometer (FSN10450, Thermo Fisher, USA) equipped with an H-ESI source operating in negative ion modes (Neg Ion Spray Voltage) at 2700 V. Each sample was analyzed separately using an orbitrap full scan in the first stage (full scan, with a resolution of 60,000, RF Lens 50%). For quantitative characterization, MS/MS data were acquired using data-dependent ms2 scans (DDA, resolution 15000, HCD Collision Energy 40%). Ion source conditions were as follows: sheath gas flow, 40 Arb (Arbitrary units); auxiliary gas flow, 5 Arb; ion transfer tube temperature, 320°C; vaporizer temperature, 320°C. Full-scan MS data were collected from 150 to 1500 m/z. Mass data were recorded with Xcalibur 4.0 software.
The TPM (Transcripts Per Million) values of PgBAHDs and ginsenoside biosynthesis pathway genes from AR (adventitiou roots), CT (callus), and RG (fibrous roots) were used to calculate the Pearson’s correlation between these types of genes using the R package ‘Hmisc’. Similarly, the TPM values of PgBAHD genes and malonyl ginsenoside from AR, CT, and RG were used to calculate the Pearson’s correlation between PgBAHD genes and metabolites using the R package ‘Hmisc’.
To identify ginseng BAHD family genes, a search of the ginseng genome using a Hidden Markov Model (HMM) of the transferase family identified 103 genes. All 103 BAHD proteins contained the conserved structural domains HXXXD and DFGWG that characterize the BAHD family. The complete data of these genes, including gene ID, protein length, gene length, molecular weight (MW), and isoelectric point (pI), are shown in Supplementary Table 1. The lengths of these BAHD genes ranged from 741 bp (PgBAHD3 and 80) to 2682 bp (PgBAHD84), while protein lengths ranged from 246 to 893 amino acids (aa). The molecular weight (MW) varied from 27.27 kDa (PgBAHD3) to 98.77 kDa (PgBAHD84). The isoelectric point (pI) is an indicator used to determine the pH, and it varied among PgBAHD proteins from 5.22 (PgBAHD25) to 9.30 (PgBAHD65). Overall, 75 proteins were predicted to have low isoelectric points (pI < 7).
A phylogenetic tree of 103 ginseng BAHDs was constructed to categorize and explore the evolutionary relationships of PgBAHD genes (Figure 1). The analysis included PgBAHD genes and genetically and biochemically characterized BAHD acyltransferases (the information on foreign genes in the phylogenetic tree can be found in Supplementary Table 2) (D’Auria, 2006). The phylogenetic tree results showed that all genes were divided into five clades, of which 103 PgBAHD genes were clustered in three. Clade I contain functionally characterized members that are almost all related to the structural modification of phenolic glycosides, and most of these genes are involved in the acylation of anthocyanins. For example, Dv3MaT is an anthocyanin-like malonyltransferase, and in addition, NtMAT1 catalyzes the malonylation of phenolic glycosides and flavonoid glycosides in Nicotiana tabacum (Suzuki et al., 2002; Taguchi et al., 2005). Twenty-six of the 103 PgBAHD genes were in this clade. Clade III, focuses on acyltransferases related to the biosynthesis of volatile esters in mature fruits and tissues such as flowers and leaves, and 35 of the 103 PgBAHD genes are in this clade (Shalit et al., 2003). In this clade, there are 8 PgBAHD genes closely related to Ss5MaT2 (PgBAHD14, 30, 46, 61, 62, 73, 74 and 88). In clade V, benzyl alcohol/phenylethanol benzoyltransferase (BPBT), methanol acyltransferase (AMAT), and tigloyl transferase (HMT/HLT) were mainly related to the biosynthesis of volatile ester compounds (Wang and Luca, 2005; Okada et al., 2005). There were also genes associated with the biosynthesis of paclitaxel and the biosynthesis of ρ-coumaryl shikimate/quinate esters, and 42 PgBAHD genes were in this clade. In addition, clade II contains only Glossy2 from Zea mays and CER2 from Arabidopsis thaliana (Xia et al., 1997; Costaglioli et al., 2005). Clade IV has only one gene derived from Hordeum vulgare, agmatine coumaroyltransferase (ACT) (Burhenne et al., 2003). This study focuses on 35 PgBAHD genes, including 26 PgBAHDs in cladeI and eight PgBAHDs closely related to Ss5MaT2.
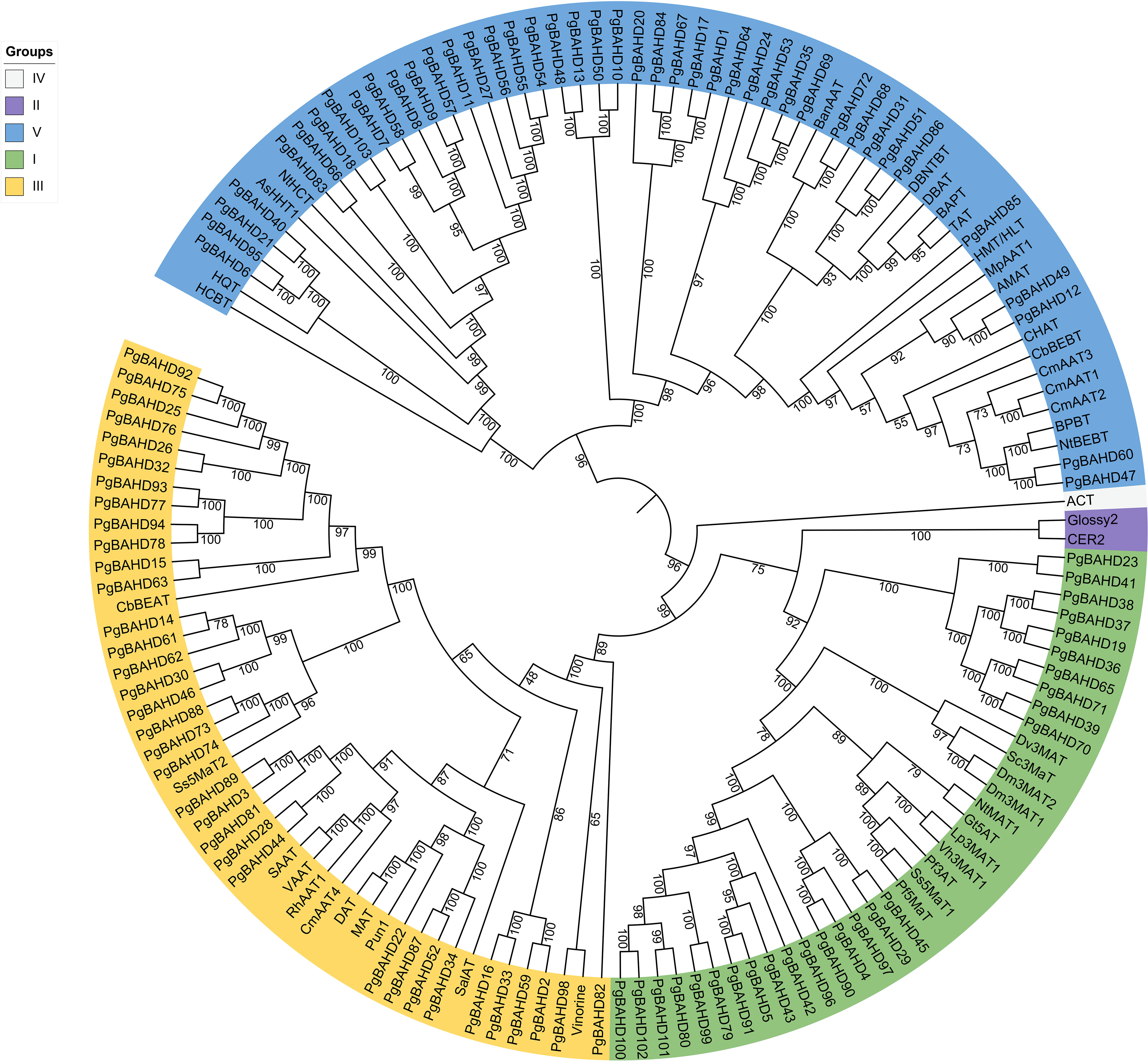
Figure 1 Phylogenetic analysis of PgBAHD genes with genetically or biochemically characterized BAHD acyltransferases.
All PgBAHDs contained motif 2and motif 4, annotated as the classic BAHD DNA-binding domain HXXXD and DFGWG, respectively. The 103 PgBAHDs contain three to 10 conserved motifs (Figures 2A; Supplementary Figure 1). Of these, 98 PgBAHDs incorporated six to eight motifs. In addition, the PgBAHD80 has three motifs, the PgBAHD3, 32, and 55 have five motifs, and the PgBAHD93 contains 15 motifs. The PgBAHDs belonging to the same clade have a similar motif composition. For example, in clade I, among the 16 PgBAHD genes, all possess motif 7, and none contain motif 6. Additionally, within this clade, only seven genes have motif 6. Motif 8 is unique to clade V. Motif 9 exclusively occurs in clade III, but PgBAHD44, 77, 82 and 98 in this clade do not contain motif 9.
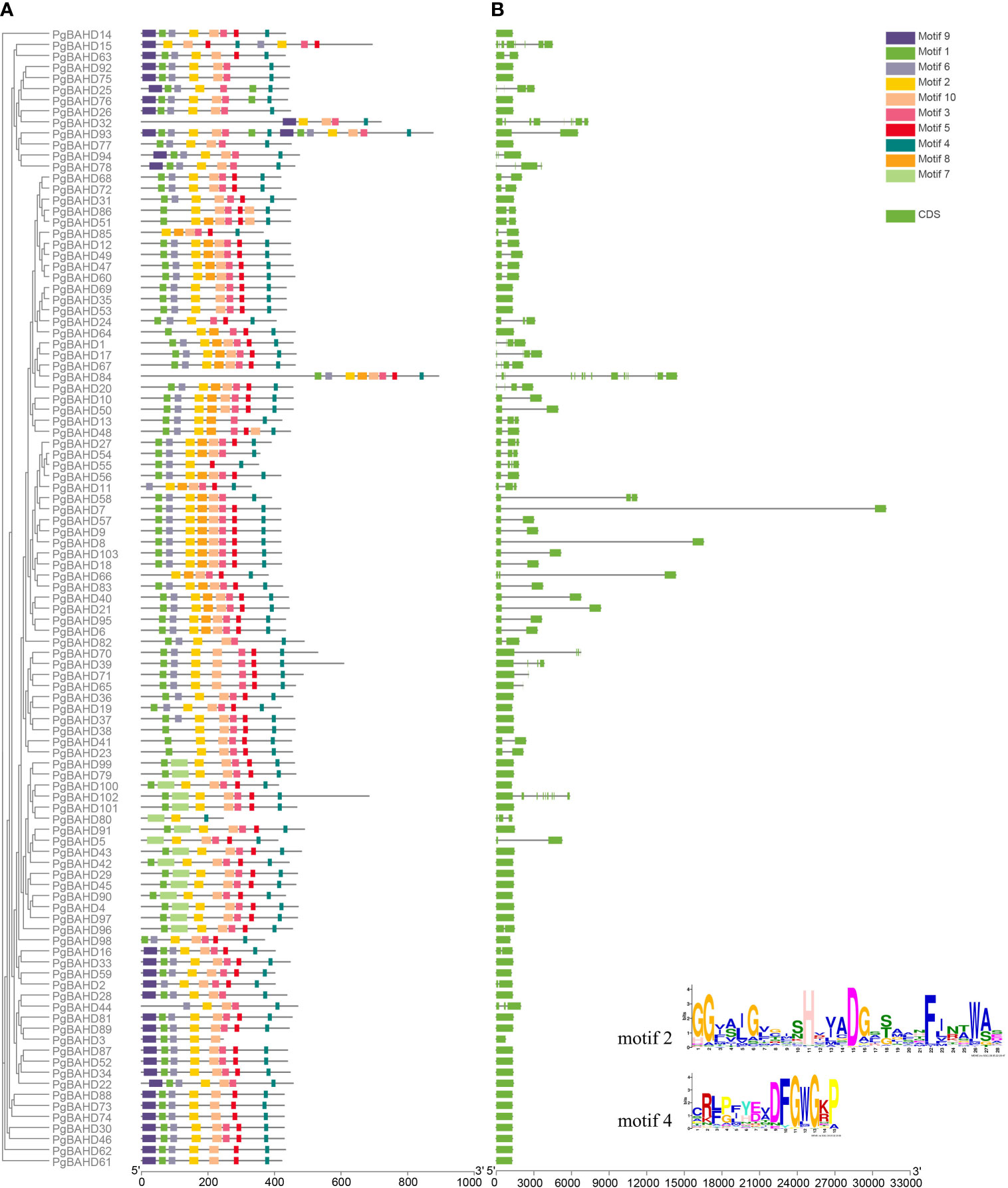
Figure 2 Conserved motif and gene structure analysis of PgBAHDs. (A) Motif distribution of PgBAHDs. (B) Gene structure of 103 PgBAHDs. The Motif composition of PgBAHD was analyzed by the MEME tool. The detailed information of the ten motifs is in Supplementary Figure 1.
To further investigate the gene structure of PgBAHDs, an exon/intron structure diagram was constructed. Of the 103 PgBAHDs, the majority had one to two exons, and 80 PgBAHDs (77.7%) had this structure. A total of 19 PgBAHDs (18.5%) had three to four exons. In addition, PgBAHD15 contained eight exons, PgBAHD32 and 102 contained nine exons, and PgBAHD84 contained 15 exons (Figure 2B). The PgBAHD genes had relatively similar exon numbers.
Several key response elements were identified in the promoter region of PgBAHDs (Supplementary Table 3). The most salient response factors included those related to stress (37%), hormones (27%), light (29%), and growth (7%) (Figure 3A), but no corresponding promoters were identified in PgBAHD37. Several hormone regulatory sites were identified in the study, such as SA (Salicylic acid), IAA (Indole-3-acetic acid), GA3 (Gibberellin), MeJA (Methyl Jasmonate), and ABA (Abscisic acid) (Figure 3B). It was found that the PgBAHD genes might be more guided by ABA and MeJA, depending on the distribution of cis-acting elements in its promoter region. In addition, regulatory elements were identified for various conditions such as dehydration, low temperature, salt stress-responsive, anoxic specific inducibility, wounding responsiveness, and anaerobic induction (Figure 3C).
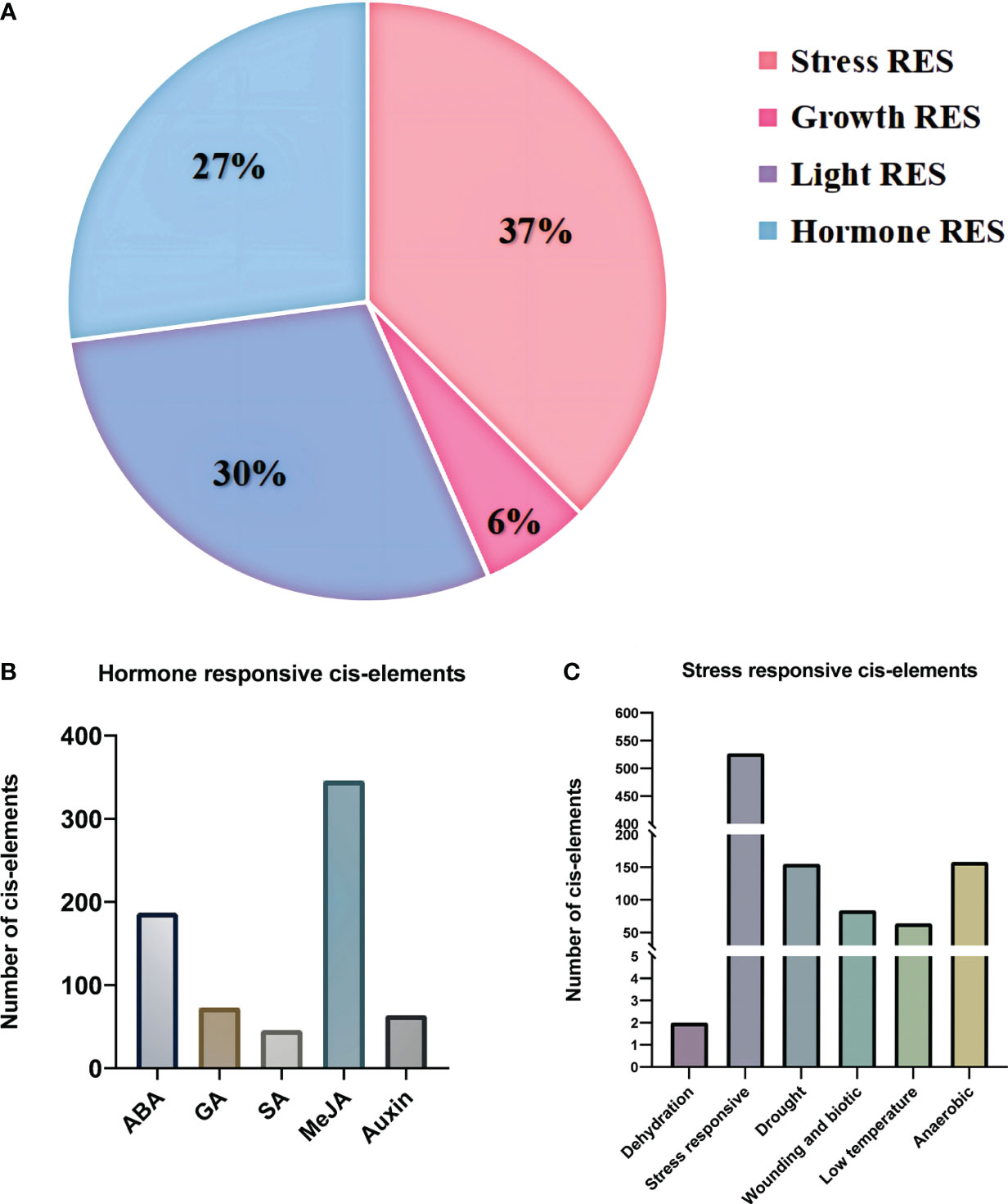
Figure 3 Distribution of cis-regulatory elements into promoter regions of PgBAHD genes. (A) Classification of identified regulatory elements based on function and their response to hormone, light, stress, and growth. (B) Distribution of different types of hormone-related cis-regulatory elements. (C) Cis-regulatory elements are related to various types of stresses.
In Figure 4A, PgBAHD genes are irregularly distributed on all 24 ginseng chromosomes. Subgenome A includes chr2, 3, 4, 5, 6, 7, 8, 9, 11, 13, 18, 23, and subgenome B includes chr1, 10, 12, 14, 15, 16, 17, 19, 20, 21, 22, 24. Among them, chr 5 had the highest density of PgBAHD genes with 10 PgBAHD genes, followed by chr 12 with nine PgBAHD genes. In addition, the vast majority of chromosomes possessed genes ranging from two to six in number. Most genes are distributed primarily at the ends of chromosomes. In addition, to investigate the collinearity of ginseng genes with members of the same family, the genomic collinearity of BAHD was analyzed in P. ginseng and Panax quinquefolium, and P. ginseng and Panax notoginseng (Figure 4B). Seventy-six PgBAHD genes showed collinearity with P. quinquefolium BAHD genes, and 63 PgBAHD genes showed collinearity with P. notoginseng BAHD genes. The results indicated that the PgBAHD genes was more closely related to the P. quinquefolium BAHD genes.
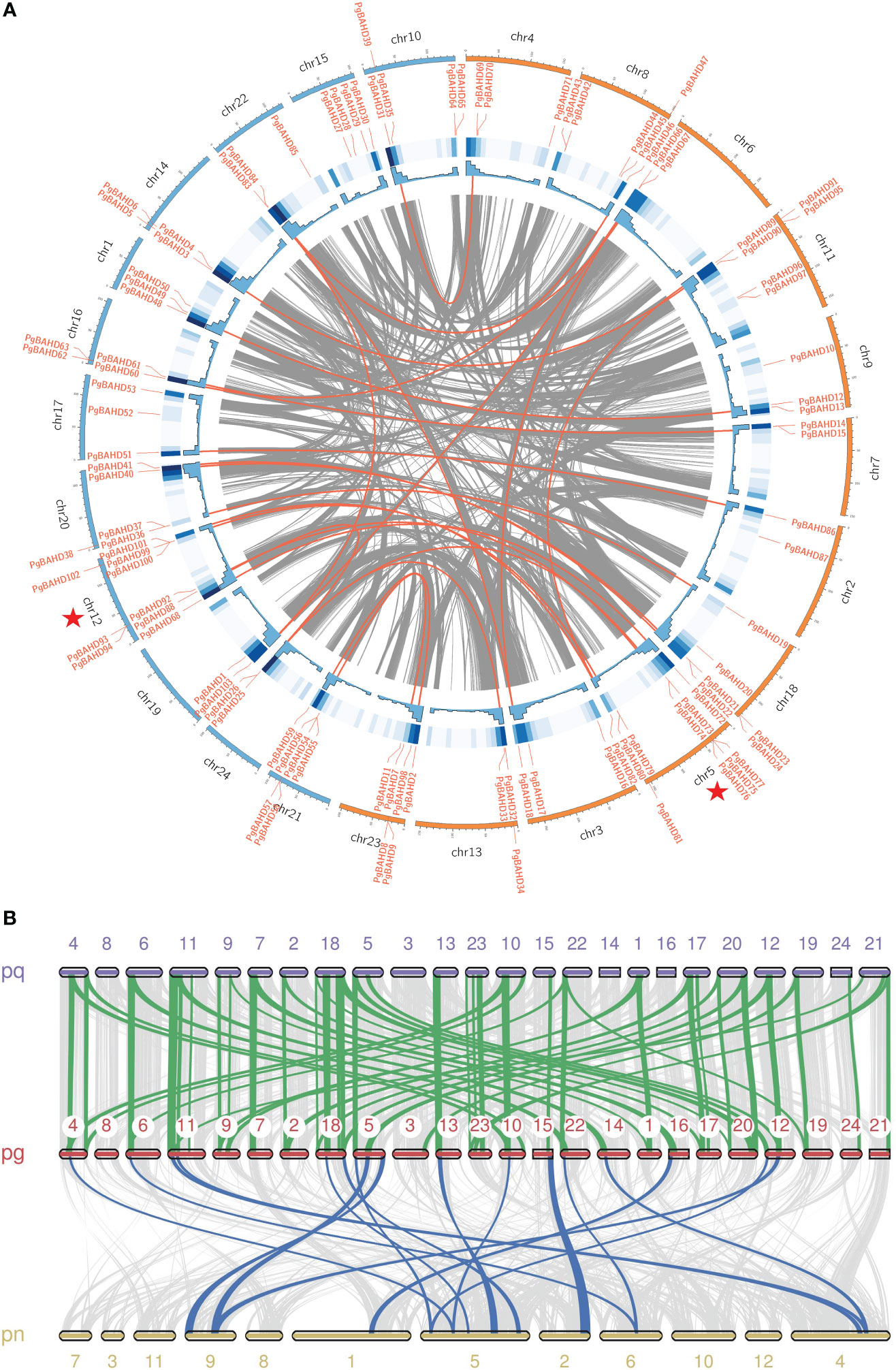
Figure 4 (A) Chromosomal locations and collinearity analysis of the PgBAHD gene family. The red lines indicate probably duplicated PgBAHD gene pairs. (B) Collinearity relationship of BAHD genes among P. ginseng, P. quinquefolium and P. notoginseng.
The 103 PgBAHD in ginseng showed 29 pairs of genes with collinearity. By calculating the non-synonymous substitution rate (Ka) and synonymous substitution rate (KS) of the two protein-coding genes (Supplementary Table 4), the KS value reflects the rate of substitution of background nucleobase during the evolutionary process, and the Ka/KS value determines the selective pressure of these genes during the genetic evolutionary process. The KS values of the PgBAHD gene families ranged from 0.0273 to 0.8226. The Ka/KS values ranged from 0.0692 to 0.8231, indicating that the PgBAHD gene families evolved under purify selection.
Different patterns of gene duplication collectively contribute to the evolution of gene families and are responsible for their functional expansion and diversification. These include whole-genome duplication (WGD) or segmental duplication, tandem duplication (TD), proximal duplication (PD), singleton duplication (SD), and dispersal duplication (DSD). Duplicated BAHD family gene pairs in the ginseng genome were analyzed, and all BAHD gene family members were assigned to WGD, PD, TD, or DSD. Sixty-eight (66.0%) of the 103 ginseng BAHD genes were WGD, 21 (20.4%) were TD, eight were PD, and the remaining six PgBAHDs were DSD. WGD and TD are the duplication patterns that have mainly influenced the evolution of the BAHD superfamily in ginseng.
TPM data from existing transcriptome data (Wang et al., 2015) were used to calculate the expression profiling of the PgBAHD genes in the main root cortex, main root epiderm, leaf blade, leaflet pedicel, fruit peduncle, stem, leaf peduncle, rhizome, leg root, fiber root, arm root, fruit pedicel, fruit flesh, and seed in different tissues. A heat map of BAHD gene expression was produced (Figure 5A; Supplementary Table 5). The results showed that 24 of the 103 PgBAHD genes were not expressed in any tissue (≈ 23.3%; TPM < 1). A total of 56 PgBAHD genes were expressed in at least one tissue (≈ 54.4%; TPM ≥ 1). A total of 23 PgBAHD genes were expressed in all tissues (≈ 22.3%; TPM ≥ 1). The expression patterns of PgBAHDs are low-level, tissue-distinct, and constitutive (Cheng et al., 2019; Liu et al., 2020a). Twelve PgBAHD genes were highly expressed in tissues of the above-ground portion (average TPM > 25), PgBAHD4, 9, 39, 57, 79, 95 and 99 were highly expressed in fruit flesh (TPM > 50). PgBAHD60 was highly expressed in the leaf peduncle (TPM = 91.5). Fruit pedicel had PgBAHD4, 79 and 95 expressions with TPM values greater than 110. In addition, the genes that were highly expressed in the stem were PgBAHD65 (TPM = 72.2), and the genes that were highly expressed in the rhizome were PgBAHD39, 65 and 103 (TPM > 65). While thirteen PgBAHD genes had high expression (TPM > 20) in five tissues in the underground, among them PgBAHD103 was expressed in the leg root, fiber root, and arm root with TPM > 98. Similarly, PgBAHD6 was highly expressed in these three tissues (TPM > 50). PgBAHD18 in the arm root also had high expression (TPM = 91.5). PgBAHD57 was expressed in the main root cortex with TPM = 59.3. PgBAHD6 was low expressed in the main root cortex. PgBAHD family genes expression was mainly in fruit pedicel, fruit flesh, arm root, and fiber root.
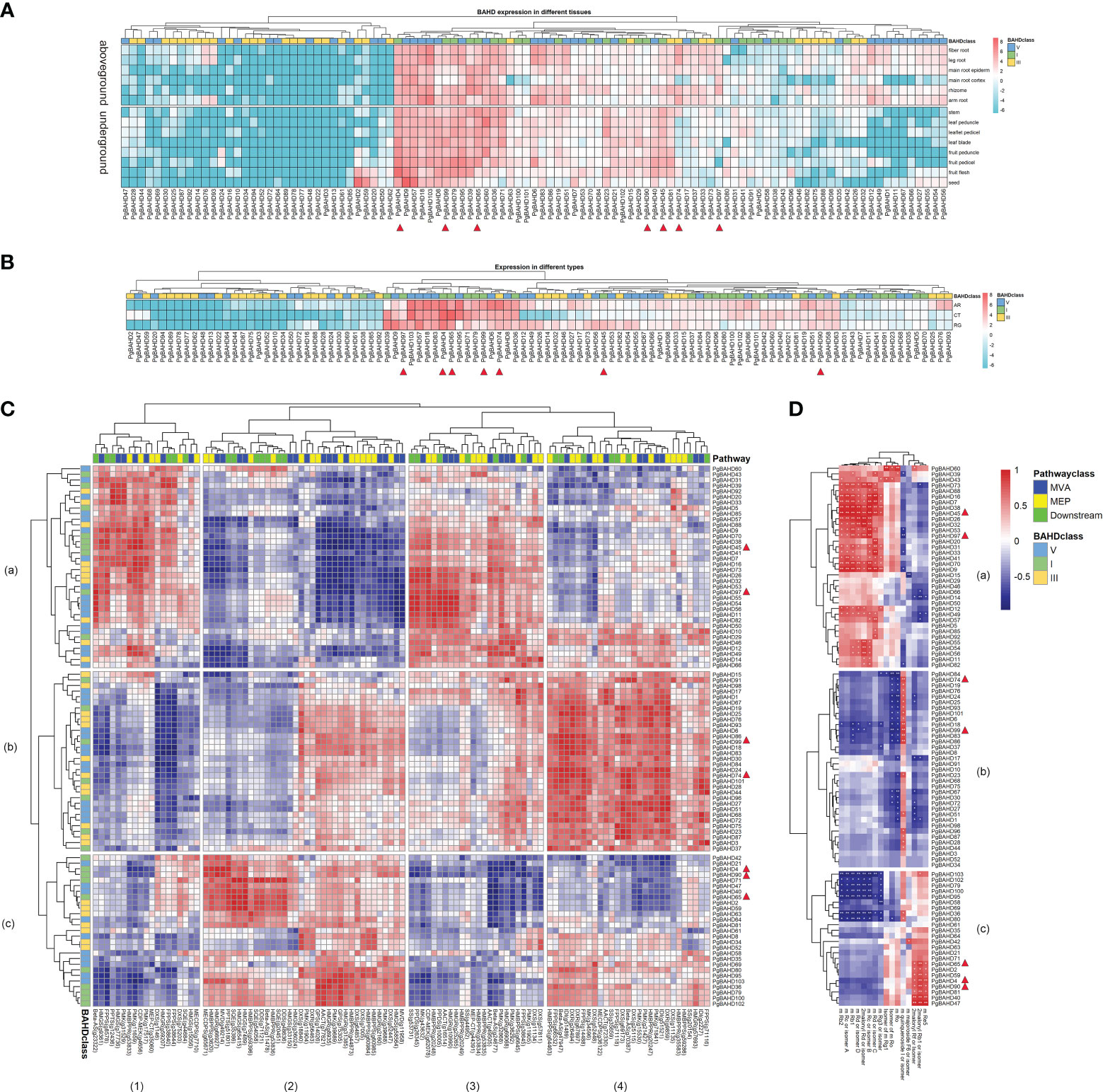
Figure 5 (A) Expression heat map of PgBAHD genes in different ginseng tissues. 14 samples were used for expression analysis, including main root cortex, main root epiderm, leaf blade, leaflet pedicel, fruit peduncle, stem, leaf peduncle, rhizome, leg root, fiber root, arm root, fruit pedicel, fruit flesh, and seed. Expression values from RNA-seq data were log2-transformed and are displayed as filled blocks in blue to red. (B) Heatmap of PgBAHD genes in different morphology samples of AR, CT and RG based on transcriptome data. Three biological replicates were set up in this samples. Note: Blue: Low expression level; Red: High expression level; The gene expression values present as log2-transformed normalized TPM values. (C) Coexpression between PgBAHD genes and ginsenoside biosynthesis pathway genes. Red indicates high correlation, and blue indicates low correlation. (D) Correlation between PgBAHD genes and malonyl ginsenosides. Red indicates high correlation, and blue indicates low correlation (*p < 0.05, **p < 0.01).
Among the three different morphology samples (fibrous roots, RG; adventitiou roots, AR; and callus, CT) of ginseng samples (Figure 5B; Supplementary Table 6), A total of 27 of 103 PgBAHD genes were expressed in at least one sample (TPM ≥ 1). Forty-four PgBAHD genes were expressed in all samples (≈ 33.9%; TPM ≥ 1). There were 65 PgBAHD genes expressed in AR, 61 in RG, and 50 in CT (TPM ≥ 1). In addition, only 32 genes were not expressed (TPM < 1) in all three samples. The highest expression of PgBAHD4 was found in AR (TPM = 150.9), and this gene also had an enriched expression in RG (TPM = 131.7). The gene with the highest expression in CT was PgBAHD65, with an TPM value of 324.1. However, the expression level of this gene is relatively low in the other two tissues. Overall, most genes showed high expression levels in AR and CT samples. Among the 35 key PgBAHD genes mentioned in result 3.1, a total of 13 genes (PgBAHD4, 19, 36, 39, 45, 65, 71, 74, 79, 90, 97, 99 and 102) in this section had a TPM > 20 in at least one tissue, and a TPM > 5 in at least one type of ginseng sample. Further qRT PCR analysis was performed on the candidate PgBAHD genes. The results showed that the expression patterns of seven candidate genes (PgBAHD4, 45, 65, 74, 90, 97, and 99) were basically consistent with RNA seq data (Supplementary Figure 2).
Different abiotic treatments of ginseng, such as cold, heat, drought, and salt, were applied in a previous study to investigate the response of PgBAHD genes to abiotic stresses. These treatments offer valuable insights for further studies in this area (Supplementary Figure 33; Supplementary Table 8). The results showed that the expression of seven PgBAHDs (TPM = 0; Fold change = 0) remained unchanged under four different stress conditions, and four of these genes (PgBAHD13, 22, 48, and 77) did not have any expression in the 14 ginseng tissues and in the different morphology samples described in the previous two section as well. In the previous section, we focused on 13 PgBAHD genes that were significantly upregulated under three weeks of high temperature and drought stress. Among them, PgBAHD39, 45, 65, and 71 were also relatively upregulated under low temperature and salt stress. However, one week of high temperature stress seemed to have no effect on these genes.
About 43 (≈ 41.8%) genes were up-regulated in expression under cold treatment compared to the control, of which 14 genes showed more significant changes (fold change > 2), and six genes showed more than five-fold changes. In addition, an 11.4-fold change was observed in the PgBAHD43 gene, while PgBAHD35 and 69 only showed relatively high expression (fold change > 6) under cold treatment. About 36 (≈ 35.0%) PgBAHDs had up-regulated gene changes under drought treatment compared to control, of which about 19 genes had more than two-fold changes and ten genes showed about five-fold changes after drought treatment. There were also five genes with more than ten-fold change in expression, including PgBAHD20 with 85.4-fold change. Salt treatment resulted in the up-regulation of the expression of 33 (≈ 32.0%) PgBAHD genes compared to the control, of which eight PgBAHDs were up-regulated (fold change >2), four PgBAHDs were significantly up-regulated (fold change >5), and in addition, the expression of the gene PgBAHD39 was very significantly up-regulated (fold change >15). While the other PgBAHDs were slightly changed. The findings indicate that the expression patterns of PgBAHDs significantly changed under drought treatments, while they only showed slight changes in response to cold and salt treatments.
PgBAHDs exhibited varying response patterns to different treatments. Among the heat treatments, eight PgBAHDs (fold change >2) displayed significant changes between the control and one-week heat treatments. Notably, PgBAHD43 exhibited a remarkable change of more than 23-fold. In the three-week heat-treated group, about 27 genes showed significant up-regulation of expression (fold change >2) compared to the control group, of which 19 PgBAHDs had more than four-fold changes, with PgBAHD39, 43, 65, and 79 changes most especially above 20-fold. A total of 20 PgBAHDs were found to be changed at both one and three weeks of treatment, and 32 genes with a significantly higher amount of gene change (fold change > 2) were found in the three-week heat treatment compared with the one-week heat treatment. The expression trend analysis indicates that several PgBAHDs might play a role in the response to heat treatment after three weeks. Expression profiles also confirmed that PgBAHDs have a variety of functional and physicochemical properties.
To investigate the relationship between PgBAHD genes and ginsenoside biosynthesis, co-expression analysis of PgBAHDs and ginsenoside biosynthesis pathway genes was performed. This pathway includes the upstream MEP and MVA pathways and the downstream pathway (Figure 5C; Supplementary Table 9). This section focuses on the 13 key PgBAHD genes mentioned earlier. Among them, a total of 12 genes have a correlation greater than 0.9 with at least one pathway gene. Among these, PgBAHD39, 45, and 97 were predominantly localized within modules a1 and a3, PgBAHD19, 74, and 99 exhibit primary concentration within module b4, whereas PgBAHD4, 36, 65, 79, and 90 were primarily situated in module c2. PgBAHD45 was mainly highly correlated with CDP-MEK (g4656) and PMK (g11659), while PgBAHD97 was highly correlated with Beta-AS (g23322) and CDP-MEK (g62078). In c2 module, PgBAHD4 and 90 were highly correlated with MVD(g49214) and HMGR(g66927), and PgBAHD65 with HMGS(g65415), HMGS(g7489), SQE(g35386) and PPTS(g51801) were highly correlation. PgBAHD74 and 99 were highly correlated with Beta-AS (g47947), DXR (g26494), and IDI (g51489). In addition, PgBAHD74 is also highly correlated with Beta-AS (g70387), DXS (g69173), FPPS (g14488), and DXR (g9859).
To research the metabolites associated with malonyl ginsenoside biosynthesis, this study used ultra-high-performance liquid chromatography combined with an Orbitrap mass spectrometer (UHPLC-Orbitrap MS) for non-targeted detection of ginsenosides in ginseng tissues of different types (fibrous roots, RG; adventitious root, AR; and callus, CT). A total of 16 metabolites (malonyl ginsenosides) were identified in nine samples (Supplementary Table 10). To further validate the relationship between PgBAHD genes and malonyl ginsenoside biosynthesis, coexpression analysis was performed (Figure 5D; Supplementary Table 11). Selected PgBAHD4, 45, 65, 74, 90, 97, and 99 as candidate genes, which have a correlation greater than 0.8 with at least one malonyl ginsenoside. The two genes PgBAHD45 and 97 screened in the a1 and a3 modules mentioned above were mainly distributed in the a module in the analysis of their correlation with saponins (Figure 5D). They were highly correlated with m-Rc or isomer-B, m-Rb, and 2malonyl-Rd or isomer. Among them, 45 was also highly correlated with m-Rd or isomer-C, m-Rc or isomer-A, m-Rb2, and m-Rd or isomer-D. The PgBAHD74 and 99 screened by the b4 module are highly correlated only with m-quinquenode I or isolmer, and they are distributed in module b in Figure 5D. The genes in module c in Figure 5D correspond to module c2 in 5C, where PgBAHD4, 65, and 90 are highly correlated with 2malonyl-Rb1 or isomer, and PgBAHD4 is also highly correlated with m-Re5. The results suggest that PgBAHDs are related to the metabolism of malonyl ginsenosides, and these PgBAHDs may promote the biosynthesis of malonyl ginsenosides in ginseng.
BAHD acyltransferase plays a widespread role in acylation modification (Kusano et al., 2019; Luo et al., 2007; Grienenberger et al., 2009; Li et al., 2018). It is important for the biosynthesis of various active acylated natural products (Liu et al., 2017). Acylation products include lignin monomers, anthocyanins, terpenes, and esters. It also involves plant growth and development, environmental stress response, and fruit ripening (D’Auria, 2006; Zheng et al., 2009). Therefore, there is a need to analyze the possible roles of BAHD gene families in ginseng systematically. In this study, 103 BAHD genes were identified from ginseng. The number of genes in gene families also varied by species (Abdullah et al., 2021). In previous studies, 52 BAHD genes were identified in A. thaliana with a genome size of 0.125 Gb, Prunus avium (125; 0.125 Gb), Rubus mesogaeus (69, 0.24 Gb), Brachypodium distachyon (15; 0.26 Gb), Musa acuminata (46; 0.43 Gb), P. ginseng (103; 2.98 Gb), Hordeum vulgare (116; 5.1 Gb). Therefore, The number of BAHD genes did not seem to be correlated with genome size, which is in line with the results of the previous studies (Xu et al., 2021).
The BAHD family genes of ginseng were clustered into five evolutionary clades (I, II, III, IV, and V), and the results were consistent with the previously reported evolutionary clades of the plant BAHD acyltransferase family (D’Auria, 2006). Based on the results of sequence alignment of the encoded proteins, D’Auria J C performed a phylogenetic analysis of the relationships of 46 BAHD acyltransferase genes that have been functionally characterized (D’Auria, 2006). The clades were distinguished from each other based on differences in substrate and enzyme activities. This distinction revealed the evolutionary progression of BAHD family members’ functions and aided in predicting the activities of enzymes with unknown functions. NtMAT1 catalyzes the malonylation of phenolic and flavonoid glycosides in N. tabacum and belongs to the clade I. In addition, based on a conserved sequence, YFGNC (motif 3), is shared by all the clade members. The motif is an acyltransferase associated with the biosynthesis of anthocyanins/flavonoid compounds (Unno et al., 2007; Yu et al., 2008). Successfully cloned a new enzyme Dv3MaT from Ahlia variabilis flowers, which is a cDNA coding 3-glucoside-specific malonyltransferase for anthocyanins (Suzuki et al., 2002). The hydroxycinnamoylation at positions three and five of the glycan chain of anthocyanins deepens the color, while malonylation increases stability (D’Auria et al., 2007; Luo et al., 2007). Nicotiana benthamiana BAHD family malonyltransferase NbMaT1 displays significant substrate tolerance to a wide range of natural products with different glycosyl substitutions at different positions in the flavonoid, coumarin, and phenylethylchromone skeletons (Liu et al., 2017). There are 26 ginseng BAHD genes in clade I, so it is assumed that these PgBAHD genes are closely related to the malonylation of ginsenosides. Malonyltransferase Ss5MaT2, although also associated with acyl modification, is classified in clade III because it does not contain conserved sequences common to clade I. Therefore, attention will also be paid to PgBAHD14, 30, 46, 61, 62, 73, 74 and 88. Based on sequence structure, physicochemical properties, function, and distribution on chromosomes, the identified PgBAHDs showed a high degree of diversity, which is consistent with previous reports on BAHD gene families (Moglia et al., 2016; Zhang et al., 2019; Ahmad et al., 2020; Liu et al., 2020b).
Gene sequences and molecular weights differed significantly, but the characteristic structural domains and constituent motifs were relatively conserved. The exon/intron composition analysis showed that most 103 PgBAHDs had one to four exons. The exon numbers of PgBAHD genes within the same group were relatively similar, which is a phenomenon that most plants have(D’Auria, 2006; Tuominen et al., 2011; Liu et al., 2020b). Conservative motif and gene structure analyses showed that the identification and grouping of PgBAHDs was reliable. The presence of cis-acting elements related to stress responses in the promoter region of the PgBAHD gene indicates that this gene family may be activated by transcription factors associated with stress stimuli, as observed in previous studies (Ahmadizadeh and Heidari, 2014; Heidari et al., 2019). At the promoter site, a high frequency of the cis-acting elements related to responsive hormones such as ABA and MeJA were detected, indicating that stress-related hormonal signals primarily induce the PgBAHD gene.
Gene duplication patterns can be classified into five distinct types (Maher et al., 2006; Qiao et al., 2015). Each gene duplication pattern contributes differently to the expansion of gene families (Freeling, 2009). WGD, TD, and DSD are considered significant features of eukaryotic genome evolution, primarily propelling the emergence of novel functionalities within genomes and genetically evolved systems (Friedman and Hughes, 2001; Moore and Purugganan, 2003). Studies have estimated that WGD account for approximately 90% of the genetic expansion observed in the Arabidopsis lineage (Maere et al., 2005). The Hydroxycinnamoyltransferase and SWEET gene families have primarily expanded through WGD and DSD (Li et al., 2017; Ma et al., 2017).TD is the primary driving force behind the expansion of the AP2/ERF gene family (Du et al., 2013; Guo et al., 2014). Research results indicate that WGD and TD are the primary expansion modes for the ginseng BAHD gene family. Ginseng possibly experienced two WGD events between 2.2 million and 28 million years ago (Kim et al., 2018; Liu et al., 2022). TD and SD contribute to the domestication, survival, and resistance to both biotic and abiotic stresses in plants. These duplications lead to the creation of structural and functional diversity within genes (Schilling et al., 2020; Zan et al., 2020; Liu et al., 2021). The Ka, KS results show that the KS values of the PgBAHD gene pairs are all less than one. Among them, 17 gene pairs have relatively low KS values (< 0.1), suggesting that these genes have undergone fewer mutations in a short period and may possess stable functions during the evolutionary process. On the other hand, 12 gene pairs have relatively high KS values (> 0.1), indicating that these genes have undergone significant evolution over a more extended period, potentially facilitating functional evolution and family expansion. The Ka/KS ratio is less than one, suggesting that purifying selection pressure has been acting on the ginseng BAHD gene family, leading to relatively stable expression. This implies that the gene family plays an important role in the growth and development of ginseng.
The molecular weight of BAHD proteins ranges from 48-55 kDa. They utilize CoA thioesters as acyl donors to transfer acetyl, malonyl, tigloyl, benzoyl, and hydroxycinnamoyl moieties, thereby regulating the structure and content of compounds in secondary metabolic pathways, ultimately influencing and modifying their properties (Kuate et al., 2008). Malonyl transferase is a class of acyltransferases that transfer malonyl molecules to sugar moieties, forming a wide range of biologically active natural acylated glycosidic constituents. Some malonyl ginsenosides have been isolated in ginseng in studies. For example, reports indicate the presence of malonyl ginsenosides Rb1, Rb2, Rc, and Rd in both P. ginseng and P. quinquefolius (Nakai and Yamamoto, 1984). Malonyl notoginsenoside R4 and malonyl-ginsenoside Ra3 have been isolated from the fresh roots of P. ginseng, respectively (Ruan et al., 2010). The malonyl ginsenosides isolated above are all derived from protopanaxadiol (PPT) type ginsenosides (Wan et al., 2015). Wang et al. isolated the PPT-type M-Re (malonyl-ginsenoside Re) from the flower buds of P. ginseng for the first time (Wang et al., 2016). The malonylation of plants primarily serves several purposes: stabilizing unstable structures, increasing the solubility of target compounds in water, and facilitating the transfer of target compounds into the vacuoles (Suzuki et al., 2002; Luo et al., 2007; Zhao et al., 2011; Matern et al., 1983). During ginsenoside biosynthesis, malonylation may also be associated with the transfer of ginsenosides into the vacuoles, thus affecting the accumulation of total ginsenosides in ginseng. Therefore, the specific functions and modes of action of different PgBAHDs in ginseng need to be further investigated.
This study subjected the BAHD gene family in ginseng to identification, phylogenetic construction, gene structure analysis, chromosomal localization, expression pattern analysis, and co-expression analysis. The results showed a high correlation between PgBAHDs and the critical enzyme genes of the ginsenoside biosynthesis pathway and with malonyl ginsenosides. This study provides a reliable basis for further metabolic regulation of the ginsenoside biosynthesis pathway, synthetic biology research, and molecular breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PW: Writing – original draft, Conceptualization, Data curation. YY: Conceptualization, Writing – original draft. MY: Data curation, Writing – review & editing. XP: Methodology, Writing – review & editing. YW: Data curation, Writing – review & editing. XL: Data curation, Writing – review & editing. HY: Conceptualization, Writing – review & editing. NZ: Methodology, Writing – review & editing. WL: Data curation, Writing – review & editing. PD: Conceptualization, Writing – review & editing. LY: Conceptualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Jilin Province Science and Technology Development Project (20210101190JC), Key Research and Development Project of Jilin Province Science and Technology Development Program (20230204001YY), National Key Research and Development Program of China (2021YFD1600900) and Natural Science Foundation of China (U21A20405).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1301084/full#supplementary-material
HMMs, Hidden Markov Models; CDD, Conserved Domain Database; MW, Molecular weight; pI, Isoelectric point; aa, amino acid; kDa, Kilodaltons; CDS, Coding sequences; MEME, Multiple expectation maximization for motif elicitation; TPM, Transcripts Per Million; HMM, Hidden Markov model; MEGA, Molecular evolutionary genetics analysis; MYA, Million years ago; AR, adventitiou roots; CT, callus; RG, fibrous roots; SA, Salicylic acid; IAA, Indole-3-acetic acid; GA3, Gibberellin; MeJA, Methyl Jasmonate; ABA, Abscisic acid; chr, Chromosome; Ka, Non-synonymous; KS, Synonymous; WGD, Whole-genome duplication; TD, tandem duplication; PD, proximal duplication; SD, singleton duplication; DSD, dispersal duplication; RNA-Seq, RNA sequencing; qRT-PCR, Quantitative real-time PCR; H-ESI, High Energy Spark-Induced Breakdown Ionization.
Abdullah, F. S., Heidari, P., Poczai, P. (2021). The BAHD gene family in cacao (Theobroma cacao, malvaceae): genome-wide identification and expression analysis. Front. Ecol. Evol. 9, 707708. doi: 10.3389/fevo.2021.707708
Ahmad, M. Z., Zeng, X., Dong, Q., Manan, S., Jin, H., Li, P., et al. (2020). Global dissection of the BAHD acyltransferase gene family in soybean: Expression profiling, metabolic functions, and evolution. Res. Square doi: 10.21203/rs.2.21482/v2
Ahmadizadeh, M., Heidari, P. (2014). Bioinformatics study of transcription factors involved in cold stress. Biharean Biol. 8, 83–86.
Ardhaoui, M., Falcimaigne, A., Ognier, S., Engasser, J. M., Moussou, P., Pauly, G., et al. (2004). Effect of acyl donor chain length and substitutions pattern on the enzymatic acylation of flavonoids. J. Biotechnol. 110, 265–272. doi: 10.1016/j.jbiotec.2004.03.003
Burhenne, K., Kristensen, B. K., Rasmussen, S. K. (2003). A new class of N-hydroxycinnamoyltransferases: Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J. Biol. Chem. 278, 13919–13927. doi: 10.1074/jbc.M213041200
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, Y., Ahammed, G. J., Yao, Z., Ye, Q., Ruan, M., Wang, R., et al. (2019). Comparative genomic analysis reveals extensive genetic variations of WRKYs in Solanaceae and functional variations of CaWRKys in pepper. Front. Genet. 10, 492. doi: 10.3389/fgene.2019.00492
Costaglioli, P., Joubès, J., Garcia, C., Stef, M., Arveiler, B., Lessire, R., et al. (2005). Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1734, 247–258. doi: 10.1016/j.bbalip.2005.04.002
D’Auria, J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340. doi: 10.1016/j.pbi.2006.03.016
D’Auria, J. C., Reichelt, M., Luck, K., Svatoš, A., Gershenzon, J. (2007). Identification and characterization of the BAHD acyltransferase malonyl CoA: Anthocyanidin 5-O-glucoside-6″-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett. 581, 872–878. doi: 10.1016/j.febslet.2007.01.060
Du, D., Hao, R., Cheng, T., Pan, H., Yang, W., Wang, J., et al. (2013). Genome-wide analysis of the AP2/ERF gene family in prunus mume. Plant Mol. Biol. Rep. 31, 741–750. doi: 10.1007/s11105-012-0531-6
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Friedman, R., Hughes, A. L. (2001). Pattern and timing of gene duplication in animal genomes. Genome Res. 11, 1842–1847. doi: 10.1101/gr.200601
Grienenberger, E., Besseau, S., Geoffroy, P., Debayle, D., Heintz, D., Lapierre, C., et al. (2009). A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J. 58, 246–259. doi: 10.1111/j.1365-313X.2008.03773.x
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Heidari, P., Ahmadizadeh, M., Izanlo, F., Nussbaumer, T. (2019). In silico study of the CESA and CSL gene family in Arabidopsis thaliana and Oryza sativa: Focus on post-translation modifications. Plant Gene 19, 100189. doi: 10.1016/j.plgene.2019.100189
Hou, S. L., Han, M., Liu, C. J., Yang, L. M. (2014). Development of real-time fluorescence quantitative RT-PCR assay for β-actin gene of Panax ginseng. Chin. Tradit. Herb. Drugs 45, 2530–2533. doi: 10.7501/j.issn.0253-2670.2014.17.020
Kim, N. H., Jayakodi, M., Lee, S. C., Choi, B. S., Jang, W., Lee, J., et al. (2018). Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 16, 1904–1917. doi: 10.1111/pbi.12926
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kuate, S. P., Pádua, R. M., Eisenbeiss, W. F., Kreis, W. (2008). Purification and characterization of malonyl-coenzyme A: 21-hydroxypregnane 21-O-malonyltransferase (Dp21MaT) from leaves of Digitalis purpurea L. Phytochemistry 69, 619–626. doi: 10.1016/j.phytochem.2007.08.025
Kusano, H., Li, H., Minami, H., Kato, Y., Tabata, H., Yazaki, K. (2019). Evolutionary developments in plant specialized metabolism, exemplified by two transferase families. Front. Plant Sci. 10, 794. doi: 10.3389/fpls.2019.00794
Li, G., Jones, K. C., Eudes, A., Pidatala, V. R., Sun, J., Xu, F., et al. (2018). Overexpression of a rice BAHD acyltransferase gene in switchgrass (Panicum virgatum L.) enhances saccharification. BMC Biotechnol. 18, 1–10. doi: 10.1186/s12896-018-0464-8
Li, J., Qin, M., Qiao, X., Cheng, Y., Li, X., Zhang, H., et al. (2017). A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (pyrus bretschneideri). Plant Cell Physiol. 58, 839–850. doi: 10.1093/pcp/pcx025
Liu, A., Liu, C., Lei, H., Wang, Z., Zhang, M., Yan, X., et al. (2020a). Phylogenetic analysis and transcriptional profiling of WRKY genes in sunflower (Helianthus annuus L.): Genetic diversity and their responses to different biotic and abiotic stresses. Ind. Crops Prod. 148, 112268. doi: 10.1016/j.indcrop.2020.112268
Liu, C., Qiao, X., Li, Q., Zeng, W., Wei, S., Wang, X., et al. (2020b). Genome-wide comparative analysis of the BAHD superfamily in seven Rosaceae species and expression analysis in pear (Pyrus bretschneideri). BMC Plant Biol. 20, 14. doi: 10.1186/s12870-019-2230-z
Liu, C., Wu, Y., Liu, Y., Yang, L., Dong, R., Jiang, L., et al. (2021). Genome-wide analysis of tandem duplicated genes and their contribution to stress resistance in pigeonpea (Cajanus cajan). Genomics 113, 728–735. doi: 10.1016/j.ygeno.2020.10.003
Liu, L., Li, X., Li, B., Sun, M.y., Li, S.x. (2022). Genome-wide analysis of the GRF gene family their expression profiling in peach (Prunus persica). J. Plant Interact. 17, 437–449. doi: 10.1080/17429145.2022.2045370
Liu, Y., Wang, X., Mo, T., Yan, Y., Song, Y., Zhao, Y., et al. (2017). Identification and functional application of a new malonyltransferase NbMaT1 towards diverse aromatic glycosides from Nicotiana benthamiana. RSC Adv. 7, 21028–21035. doi: 10.1039/C7RA01940H
Liu, Z., Li, Y., Li, X., Ruan, C. C., Wang, L. J., Sun, G. Z. (2012). The effects of dynamic changes of malonyl ginsenosides on evaluation and quality control of Panax ginseng C.A. Meyer. J. Pharm. Biomed. Anal. 64–65, 56–63. doi: 10.1016/j.jpba.2012.02.005
Livak, K. J., Schmittgen, T. D. (2023). Analysis of relative gene expression data using real- time quantitative PCR and the 2 Ϫ⌬⌬ C T method. Methods 408, 402–408. doi: 10.1006/meth.2001.1262
Luo, J., Nishiyama, Y., Fuell, C., Taguchi, G., Elliott, K., Hill, L., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50, 678–695. doi: 10.1111/j.1365-313X.2007.03079.x
Ma, C., Zhang, H., Li, J., Tao, S., Qiao, X., Korban, S. S., et al. (2017). Genome-wide analysis and characterization of molecular evolution of the HCT gene family in pear (Pyrus bretschneideri). Plant Syst. Evol. 303, 71–90. doi: 10.1007/s00606-016-1353-z
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Maher, C., Stein, L., Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16, 510–519. doi: 10.1101/gr.4680506
Matern, U., Heller, W., Himmelspach, K. (1983). Conformational changes of apigenin 7-O-(6-O-malonylglucoside), a vacuolar pigment from parsley, with solvent composition and proton concentration. Eur. J. Biochem. 133, 439–448. doi: 10.1111/j.1432-1033.1983.tb07483.x
Matern, U., Reichenbach, C., Heller, W. (1986). Efficient uptake of flavonoids into parsley (Petroselinum hortense) vacuoles requires acylated glycosides. Planta 167, 183–189. doi: 10.1007/BF00391413
Mellou, F., Loutrari, H., Stamatis, H., Roussos, C., Kolisis, F. N. (2006). Enzymatic esterification of flavonoids with unsaturated fatty acids: Effect of the novel esters on vascular endothelial growth factor release from K562 cells. Process Biochem. 41, 2029–2034. doi: 10.1016/j.procbio.2006.05.002
Moglia, A., Acquadro, A., Eljounaidi, K., Milani, A. M., Cagliero, C., Rubiolo, P., et al. (2016). Genome-wide identification of bahd acyltransferases and in vivo characterization of HQT-like enzymes involved in caffeoylquinic acid synthesis in globe artichoke. Front. Plant Sci. 7, 1424. doi: 10.3389/fpls.2016.01424
Molina, I., Kosma, D. (2015). Role of HXXXD-motif/BAHD acyltransferases in the biosynthesis of extracellular lipids. Plant Cell Rep. 34, 587–601. doi: 10.1007/s00299-014-1721-5
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U. S. A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nakai, Y., Yamamoto (1984). NII-electronic library service. Chem. Pharm. Bull. 32, 685–691. doi: 10.1248/cpb.32.685
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Okada, T., Hirai, M. Y., Suzuki, H., Yamazaki, M., Saito, K. (2005). Molecular characterization of a novel quinolizidine alkaloid O-tigloyltransferase: cDNA cloning, catalytic activity of recombinant protein and expression analysis in Lupinus plants. Plant Cell Physiol. 46, 233–244. doi: 10.1093/pcp/pci021
Qiao, X., Li, M., Li, L., Yin, H., Wu, J., Zhang, S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 15, 1–16. doi: 10.1186/s12870-014-0401-5
Reed, J., Orme, A., El-Demerdash, A., Owen, C., Martin, L. B. B., Misra, R. C., et al. (2023). Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Sci. 379, 1252–1264. doi: 10.1126/science.adf3727
Ruan, C. C., Liu, Z., Li, X., Liu, X., Wang, L. J., Pan, H. Y., et al. (2010). Isolation and characterization of a new ginsenoside from the fresh root of panax ginseng. Molecules 15, 2319–2325. doi: 10.3390/molecules15042319
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S., Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1111/nph.16122
Shalit, M., Guterman, I., Volpin, H., Bar, E., Tamari, T., Menda, N., et al. (2003). Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/citronellol acetyltransferase in developing rose petals. Plant Physiol. 131, 1868–1876. doi: 10.1104/pp.102.018572
Sonawane, P. D., Gharat, S. A., Jozwiak, A., Barbole, R., Heinicke, S., Almekias-Siegl, E., et al. (2023). A BAHD-type acyltransferase concludes the biosynthetic pathway of non-bitter glycoalkaloids in ripe tomato fruit. Nat. Commun. 14, 4540. doi: 10.1038/s41467-023-40092-5
Stothard, P. (2000). Internet on-ramp internet on-ramp. Biotechniques 28, 1102–1104. doi: 10.2144/00286ir01
St-Pierre, B., Luca, V. (2000). Chapter Nine Evolution of acyltransferase genes: Origin and diversification fo the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv. Phytochem. 34, 285–315. doi: 10.1016/S0079-9920(00)80010-6
Sun, B. S., Xu, M. Y., Li, Z., Wang, Y. B., Sung, C. K. (2012). UPLC-Q-TOF-MS/MS analysis for steaming times-dependent profiling of steamed Panax quinquefolius and its ginsenosides transformations induced by repetitious steaming. J. Ginseng Res. 36, 277–290. doi: 10.5142/jgr.2012.36.3.277
Sun, W., Yin, Q., Wan, H., Gao, R., Xiong, C., Xie, C., et al. (2023). Characterization of the horse chestnut genome reveals the evolution of aescin and aesculin biosynthesis. Nat. Commun. 14, 1–15. doi: 10.1038/s41467-023-42253-y
Suzuki, H., Nakayama, T., Yonekura-Sakakibara, K., Fukui, Y., Nakamura, N., Yamaguchi, M. A., et al. (2002). cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme A:anthocyanidin 3-O-glucoside-6′-O-malonyltransferase from dahlia flowers. Plant Physiol. 130, 2142–2151. doi: 10.1104/pp.010447
Suzuki, H., Sawada, S., Watanabe, K., Nagae, S., Yamaguchi, M. A., Nakayama, T., et al. (2004). Identification and characterization of a novel anthocyanin malonyltransferase from scarlet sage (Salvia splendens) flowers: An enzyme that is phylogenetically separated from other anthocyanin acyltransferases. Plant J. 38, 994–1003. doi: 10.1111/j.1365-313X.2004.02101.x
Taguchi, G., Shitchi, Y., Shirasawa, S., Yamamoto, H., Hayashida, N. (2005). Molecular cloning, characterization, and downregulation of an acyltransferase that catalyzes the malonylation of flavonoid and naphthol glucosides in tobacco cells. Plant J. 42, 481–491. doi: 10.1111/j.1365-313X.2005.02387.x
Taguchi, G., Ubukata, T., Nozue, H., Kobayashi, Y., Takahi, M., Yamamoto, H., et al. (2010). Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63, 1031–1041. doi: 10.1111/j.1365-313X.2010.04298.x
Tuominen, L. K., Johnson, V. E., Tsai, C. J. (2011). Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12, 236. doi: 10.1186/1471-2164-12-236
Unno, H., Ichimaida, F., Suzuki, H., Takahashi, S., Tanaka, Y., Saito, A., et al. (2007). Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J. Biol. Chem. 282, 15812–15822. doi: 10.1074/jbc.M700638200
Wan, J. Y., Fan, Y., Yu, Q. T., Ge, Y. Z., Yan, C. P., Alolga, R. N., et al. (2015). Integrated evaluation of malonyl ginsenosides, amino acids and polysaccharides in fresh and processed ginseng. J. Pharm. Biomed. Anal. 107, 89–97. doi: 10.1016/j.jpba.2014.11.014
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinforma. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, J., Luca, V. (2005). The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including “foxy” methylanthranilate. Plant J. 44, 606–619. doi: 10.1111/j.1365-313X.2005.02552.x
Wang, K., Jiang, S., Sun, C., Lin, Y., Yin, R., Wang, Y., et al. (2015). The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C. A. Meyer. Sci. Rep. 5, 1–12. doi: 10.1038/srep18283
Wang, Y. S., Jin, Y. P., Gao, W., Xiao, S. Y., Zhang, Y. W., Zheng, P. H., et al. (2016). Complete1H-NMR and13C-NMR spectral assignment of five malonyl ginsenosides from the fresh flower buds of Panax ginseng. J. Ginseng Res. 40, 245–250. doi: 10.1016/j.jgr.2015.08.003
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, 1–14. doi: 10.1093/nar/gkr1293
Wang, Z.-H., Wang, X.-F., Lu, T., Li, M.-R., Jiang, P., Zhao, J., et al. (2022). Reshuffling of the ancestral core-eudicot genome shaped chromatin topology and epigenetic modification in Panax. Nat. Commun. 13, 1–12. doi: 10.1038/s41467-022-29561-5
Xia, Y., Nikolau, B. J., Schnable, P. S. (1997). Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 115, 925–937. doi: 10.1104/pp.115.3.925
Xu, Y., Tie, W., Yan, Y., Xu, B., Liu, J., Li, M., et al. (2021). Identification and expression of the BAHD family during development, ripening, and stress response in banana. Mol. Biol. Rep. 48, 1127–1138. doi: 10.1007/s11033-020-06132-9
Yu, X. H., Chen, M. H., Liu, C. J. (2008). Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J. 55, 382–396. doi: 10.1111/j.1365-313X.2008.03509.x
Yu, X.-H., Gou, J.-Y., Liu, C.-J. (2009). BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol. Biol. 70, 421–442. doi: 10.1007/s11103-009-9482-1
Zan, T., Li, L., Xie, T., Zhang, L., Li, X. (2020). Genome-wide identification and abiotic stress response patterns of abscisic acid stress ripening protein family members in Triticum aestivum L. Genomics 112, 3794–3802. doi: 10.1016/j.ygeno.2020.04.007
Zhang, T., Huo, T., Ding, A., Hao, R., Wang, J., Cheng, T., et al. (2019). Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume. PloS One 14, 1–18. doi: 10.1371/journal.pone.0223974
Zhao, J., Huhman, D., Shadle, G., He, X. Z., Sumner, L. W., Tang, Y., et al. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23, 1536–1555. doi: 10.1105/tpc.110.080804
Keywords: Panax ginseng, BAHD gene family, malonyltransferase, malonyl ginsenoside, biosynthesis
Citation: Wang P, Yan Y, Yan M, Piao X, Wang Y, Lei X, Yang H, Zhang N, Li W, Di P and Yang L (2023) Identification and analysis of BAHD superfamily related to malonyl ginsenoside biosynthesis in Panax ginseng. Front. Plant Sci. 14:1301084. doi: 10.3389/fpls.2023.1301084
Received: 24 September 2023; Accepted: 30 November 2023;
Published: 14 December 2023.
Edited by:
Zhichao Wu, National Institutes of Health (NIH), United StatesReviewed by:
Yang Chu, China Academy of Chinese Medical Sciences, ChinaCopyright © 2023 Wang, Yan, Yan, Piao, Wang, Lei, Yang, Zhang, Li, Di and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Di, ZGlAamxhdS5lZHUuY24=; Limin Yang, eWFuZ2xpbWluQGpsYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.