
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 January 2024
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1299461
This article is part of the Research TopicOmics Applications in Agriculture Systems: Unveiling Functionality and PracticalityView all 6 articles
 Philip L. Walker1
Philip L. Walker1 Mark F. Belmonte2
Mark F. Belmonte2 Brent D. McCallum1
Brent D. McCallum1 Curt A. McCartney3
Curt A. McCartney3 Harpinder S. Randhawa4
Harpinder S. Randhawa4 Maria A. Henriquez1,3*
Maria A. Henriquez1,3*Fusarium head blight (FHB) is a devastating fungal disease responsible for significant yield losses in wheat and other cereal crops across the globe. FHB infection of wheat spikes results in grain contamination with mycotoxins, reducing both grain quality and yield. Breeding strategies have resulted in the production of FHB-resistant cultivars, however, the underlying molecular mechanisms of resistance in the majority of these cultivars are still poorly understood. To improve our understanding of FHB-resistance, we performed a transcriptomic analysis of FHB-resistant AC Emerson, FHB-moderately resistant AC Morley, and FHB-susceptible CDC Falcon in response to Fusarium graminearum. Wheat spikelets located directly below the point of inoculation were collected at 7-days post inoculation (dpi), where dual RNA-sequencing was performed to explore differential expression patterns between wheat cultivars in addition to the challenging pathogen. Differential expression analysis revealed distinct defense responses within FHB-resistant cultivars including the enrichment of physical defense through the lignin biosynthesis pathway, and DON detoxification through the activity of UDP-glycosyltransferases. Nucleotide sequence variants were also identified broadly between these cultivars with several variants being identified within differentially expressed putative defense genes. Further, F. graminearum demonstrated differential expression of mycotoxin biosynthesis pathways during infection, leading to the identification of putative pathogenicity factors.
Wheat (Triticum aestivum L.) is a major global cereal crop, accounting for approximately 20% of human consumed calories worldwide (FAOSTAT, 2019). A significant threat to global wheat production is through pathogen attack, which is estimated to be responsible for 21.5% losses in wheat yield globally (Savary et al., 2019). Fusarium graminearum Schwabe is the dominant causative agent of one of the most economically important fungal diseases in wheat, Fusarium head blight (FHB). FHB results in grain contamination with trichothecene mycotoxins, such as the virulence factor deoxynivalenol (DON), which reduces grain quality and yield, while also being hazardous for human and animal consumption (Shin et al., 2014). Economic losses from FHB are expected to increase with rising temperatures and humidity levels due to climate change, likely resulting in more frequent and destructive FHB outbreaks (Shah et al., 2014; Hay et al., 2022). Current preventative measures involve an integrated approach, combining the use of fungicides, cultural practices and FHB-tolerant wheat cultivars to mitigate FHB disease pressure; however, these techniques are currently unable to provide complete FHB resistance (Torres et al., 2019). Further, while FHB-resistant wheat cultivars have been identified, these cultivars display poor agronomic traits, making the generation of elite cultivars challenging (Huang et al., 2018; Yi et al., 2018).
FHB infection initiates on spikelets during wheat anthesis, where the production of mycotoxins and effector molecules contribute to the infection progressing into the rachial node and subsequently progressing up and down the rachis to adjacent spikelets (Dweba et al., 2017; Rocher et al., 2022). While several mycotoxins contribute to FHB virulence, the most influential and well-studied is the trichothecene mycotoxin DON. Prior studies have identified key regulators of DON biosynthesis within the trichothecene cluster, where tri5 and tri6 mutants were unable to produce DON, leading to reduced virulence and host colonization (Audenaert et al., 2014; Wang et al., 2023). As DON plays a critical role in pathogenicity, DON detoxification has been identified in FHB-resistant species and cultivars, both through the activity of glutathione S-transferases like Fhb7 from Thinopyrum elongatum (Guo et al., 2023; Wang et al., 2020), and through UDP-glycosyltransferase activity in Brachypodium distachyon, where DON is conjugated into deoxynivalenol-3-gluscoside (Pasquet et al., 2016; Gatti et al., 2018; He et al., 2020). DON detoxification through UDP-glycosyltransferase activity has also been linked to the Fhb1 quantitative trait locus (QTL), which has shown the strongest and most stable FHB-resistance in wheat and is derived from the most well characterized FHB-resistant cultivar Sumai 3 (Gunupuru et al., 2017; Su et al., 2019). While DON detoxification appears to significantly contribute to a successful host response against FHB, FHB-resistance is a quantitative trait and therefore influenced by several QTLs (Zhang et al., 2021). Research has focused on the identification and confirmation of QTLs in FHB-resistant cultivars using genomic and transcriptomic approaches, with thus far over 500 QTLs associated with FHB-resistance being reported (Wang et al., 2020; Buerstmayr et al., 2021; Nilsen et al., 2021; Xu et al., 2020). Improving our understanding of the genes and enriched host defense pathways associated with and shared between FHB-resistant cultivars and QTLs are critical in improving our understanding FHB-resistance and generating elite FHB-resistant wheat cultivars.
The plant defense response includes the activation of both physical and chemical barriers to deter pathogen progression. Physical defense can occur through cuticle thickening, as well as cell wall lignification and modification which are essential processes in slowing FHB due to the suite of cell wall degrading enzymes released by F. graminearum upon infection (Kikot et al., 2009; Mandalà et al., 2021). Cell wall lignification has been of particular interest to researchers, as multiple FHB-resistance QTLs have identified genes associated with lignin biosynthesis, including the previously mentioned Fhb1 (Soni et al., 2021). Chemical defense responses to pathogen attack include the release of phytoalexins to deter challenging pathogens, as well as reactive oxygen species (ROS) as part of a hypersensitive response at the site of infection to limit pathogen spread throughout the host (Khaledi et al., 2016; Polturak et al., 2022). As these ROS accumulate at the host-pathogen interface, both host and pathogen require antioxidant activity to scavenge ROS and limit oxidative damage through redox homeostasis (Khaledi et al., 2016). This ability to regulate oxidative stress is essential to the host in mounting a successful defense response, as well as to the challenging pathogen throughout infection (Fernando et al., 2019; Matić et al., 2021). Further, priming of this host ROS response through increased antioxidant activity can provide an advantage to the host prior to and upon pathogen attack, leading to reduced disease symptoms (Naguib, 2018; Fujita and Hasanuzzaman, 2022).
The study of gene expression at the host-pathogen interface is essential to understand factors contributing to both pathogenicity and the host defense response. Previous identification of FHB-resistant cultivars provides an opportunity to compare resistant and susceptible cultivars and to explore putative differences at the molecular level (Graf et al., 2013; Pan et al., 2018). Among previously identified resistant winter wheat cultivars are FHB-resistant AC Emerson and FHB-moderately resistant AC Morley (Tamburic-Ilincic et al., 2011; Graf et al., 2013). While AC Emerson and AC Morley demonstrate increased resistance to FHB, the underlying molecular mechanisms leading to this resistance have yet to be explored. Investigation of these mechanisms can not only identify novel defense genes and pathways contributing to FHB-resistance, but further identify shared genes and pathways across previously examined FHB-resistant cultivars and provide additional resources in building resistance to FHB.
To better understand the T. aestivum – F. graminearum interaction, we profiled FHB-resistant AC Emerson, FHB-moderately resistant AC Morley, and FHB-susceptible CDC Falcon winter wheat cultivars in response to F. graminearum at 7-days post inoculation (dpi). Dual RNA-sequencing revealed expression of shared and specific defense gene sets between resistant cultivars prior to, and in response to, F. graminearum infection, while also identifying differentially expressed genes associated with DON biosynthesis and redox homeostasis in F. graminearum. Further, nucleotide sequence variants were identified in differentially expressed genes involved in these pathways and processes. Taken together, these data provide insight into defense genes and pathways that influence FHB-resistance, as well as the pathogenicity factors that may slow FHB disease progression.
A highly virulent 3-acetyldeoxynivalenol producing isolate of F. graminearum (HWW-15-33), obtained from the Henriquez Winter Wheat (HWW) collection of Fusarium isolates at Agriculture and Agri-Food Canada, Morden, Manitoba was used in this study. For conidia production, F. graminearum preserved at -80°C (filter paper Whatman No.1) was plated onto a petri dish (100 mm) containing Spezieller-Nährstoffar Agar (SNA), and incubated at 22°C for 10 days under a combination of fluorescent-UV lights (Nilsen et al., 2021).
Plant materials used in this study included the hard red winter wheat cultivars AC Emerson (Graf et al., 2013), AC Morley (Tamburic-Ilincic et al., 2011) and CDC Falcon (Fowler, 1999). ‘AC Emerson’ is a hard red winter wheat, F1-derived doubled haploid (DH) cultivar produced from the cross McClintock/CDC Osprey, and is resistant to FHB. ‘AC Morley’ is a moderately resistant (MR) cultivar, derived from the cross FR227/17//IP16/20. ‘CDC Falcon’ is susceptible to FHB and derives from the cross Norstar/2/Vona//Abilene. The plant growing conditions were similar to Nilsen et al., 2021. At the growth stage where 50% of the spikelets has anther extrusion, single floret inoculation was performed with 10 µL of F. graminearum inoculum at a concentration of 5 x 104 macroconidia/mL, and water (control) between the lemma and palea at the midpoint of the spike. The inoculated spikelet was marked using a marker pen. This was repeated for five individual wheat plants and three replicates for all cultivars.
Based on significant differences in disease symptoms identified between FHB-resistant and FHB-susceptible cultivars, 7-dpi was the selected time-point to examine gene expression patterns by RNA-sequencing. The spikelet below the point of inoculation was isolated at 7-dpi for F. graminearum and water inoculated treatments for each cultivar. Five spikes were used per replicate, one spike per plant/pot. Each treatment was replicated three times. RNA extraction was performed from 60 sectioned rachis nodes and rachilla samples using Trizol Reagent (Ambion), according to the manufacturer’s instructions. DNase I (Invitrogen) treatment was applied to each sample following the manufacturer’s instructions. RNA yield and quality was assessed using the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano Kit (Agilent). The average RNA Integrity Number (RIN) value for all the samples was 8.4 out of 10. cDNA libraries were prepared by using the NEB rRNA-depleted stranded (plant) kit. Samples were sequenced at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada) using the HiSeq2500 Illumina sequencer with 125-nucleotide paired-end reads.
Raw and processed RNA-sequencing reads were deposited at the Gene Expression Omnibus (GSE233409). Adapter sequences and low-quality reads were removed using Trimmomatic v0.36 (Bolger et al., 2014) (LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36) and surviving reads aligned to the T. aestivum (IWGSC-56) and F. graminearum PH-1 (ASM24013v3) reference assemblies using HISAT2 (Kim et al., 2015). Count data was generated using featureCounts (Liao et al., 2014) and used as input for clustering and differential expression analysis through DESeq2 (Love et al., 2014). Hierarchical clustering (Supplementary Figure 1) and principle component analysis (Supplementary Figure 2) clustering was performed using pvclust (Suzuki and Shimodaira, 2006) and DESeq2 respectively. TMM expression values were calculated using EdgeR (Chen et al., 2016). Differentially expressed genes were identified using an adjusted p-value cut-off of p ≤ 0.01 and log2 FC < -2, > 2 and sorted using Venny 2.1 (Oliveros, 2007). Subsequently, GO enrichment of identified differentially expressed genes (DEGs) was performed using ShinyGO (Ge et al., 2020) and visualized using the conditional formatting function in Excel. DEG lists can be found in Dataset S1. Variants were identified for each sample using BCFtools software (Li, 2011). Low quality SNPs were filtered using the BCFtools filter function based on quality (<20), depth (<10) and variant distance bias (<0.001). GO enrichment was also performed on coding sequences containing nucleotide sequence variants using ShinyGO (Ge et al., 2020).
cDNA was generated using Quantabio qScript cDNA Supermix (Beverly, MA, USA). T. aestivum and F. graminearum RT-qPCR primers were designed using Primer3 and validated using the Primer BLAST tool. Transcript abundance was measured on the Bio-Rad CFX96 Connect Real-Time system using SsoFast EvaGreen Supermix (Bio-rad Laboratories, Hercules, CA, US) in 10 µl reactions according to manufacturer’s protocol using the following conditions: 95°C for 30s, and 45 cycles of: 95°C for 2s and 60°C for 5s. Melt curves were performed at a range of 65 – 95°C with 0.5°C increments to assess nonspecific amplification and potential primer dimers. Relative transcript abundance was calculated using the ΔΔCt method, relative to heterogenous nuclear ribonucleoprotein Q (hnRNP Q) for T. aestivum and β-tubulin (FGRAMPH1_01G26865) (Liang et al., 2023) for F. graminearum using three biological replicates per treatment and three technical replicates per biological replicate. RT-qPCR primers used in this study can be found in Dataset S2. RT-qPCR validation of RNA-sequencing data in both T. aestivum and F. graminearum can be found in Supplementary Figure 3.
Visual assessments of F. graminearum infected wheat spikes were performed at 7- and 14-dpi and used to score FHB severity in AC Emerson, AC Morley and CDC Falcon cultivars (Figures 1A, B). Here, FHB-susceptible CDC Falcon demonstrated the highest level of FHB severity at both timepoints with an average severity of 20.3% and 74.5% at 7- and 14-dpi respectively, where disease symptoms progressed into adjacent wheat spikelets as early as 7-dpi (Figure 1A)(Dataset S3). FHB-resistant AC Emerson demonstrated the lowest level of FHB severity (7.8% and 24.4%) with infection being limited to the point of inoculation at 7-dpi prior to progression down the rachis internode at 14-dpi (Figures 1A, B)(Dataset S3). AC Morley also demonstrated reduced FHB infection compared to FHB-susceptible CDC Falcon, with infection being limited to the inoculated spikelet at 7-dpi before progressing into adjacent spikelets at 14-dpi (Figures 1A, B). The 7-dpi timepoint was selected for dual RNA-sequencing to represent infected samples, being the earliest timepoint measured demonstrating significant differences in FHB severity between cultivars, while also providing sufficient time for F. graminearum to well colonize the FHB-resistant cultivar AC Emerson.

Figure 1 Fusarium head blight (FHB) disease severity in CDC Falcon, AC Morley and AC Emerson. Wheat spikes at (A) 7- and (B) 14-days post inoculation (dpi) inoculated with F. graminearum. Single florets were inoculated at 50% anther extrusion with 10 µL of 5x104 macroconidia/mL.
Differential gene expression analysis of uninfected control tissues, compared between cultivars, identified both shared and specific significantly up-regulated gene sets in the resistant AC Emerson and moderately resistant AC Morley cultivars relative to the FHB-susceptible CDC Falcon cultivar (Figure 2). Here, 1552 and 829 genes were significantly up-regulated in AC Emerson (R) and AC Morley (MR) cultivars respectively, while 551 genes were significantly up-regulated in both resistant cultivars compared to CDC Falcon (S) (Figure 2B). Gene ontology (GO) term enrichment analysis revealed significant enrichment of oxidoreductase activity (p = 5.8x10-4), cellular redox homeostasis (p = 1.9x10-3), and the immune response (p = 2.0x10-2) in the shared gene set (Figure 2C). Further, we identified 33 putative glutathione S-transferases (GSTs), belonging to the glutathione metabolism (p = 1.4x10-5) enriched GO term, which were up-regulated in either the shared AC Emerson and AC Morley gene set, or the AC Emerson-specific gene set prior to infection (Figure 2D), suggesting priming of redox homeostasis in both resistant cultivars, with GST antioxidant activity playing an important role in AC Emerson. Further, Glutaredoxins (TRAESCS5A02G094700, TRAESCS5A02G094900, TRAESCS5B02G100900, TRAESCS5B02G101100, TRAESCS5D02G107500) and thioredoxins (TRAESCS1B02G462300, TRAESCS6A02G323100) involved in redox homeostasis were also enriched in our resistant cultivars prior to infection (Dataset S1), and like GSTs, have demonstrated enrichment in previous FHB-resistant wheat cultivars in response to FHB, playing an important role in the response to oxidative stress (Li, 2014; Kumar et al., 2020). GO analysis of the AC Emerson-specific gene set also revealed enrichment of cutin biosynthesis (p = 3.0x10-3), as well as chitinase (p = 1.6x10-2) and glucosidase activity (p = 4.2x10-2) (Figure 2C).
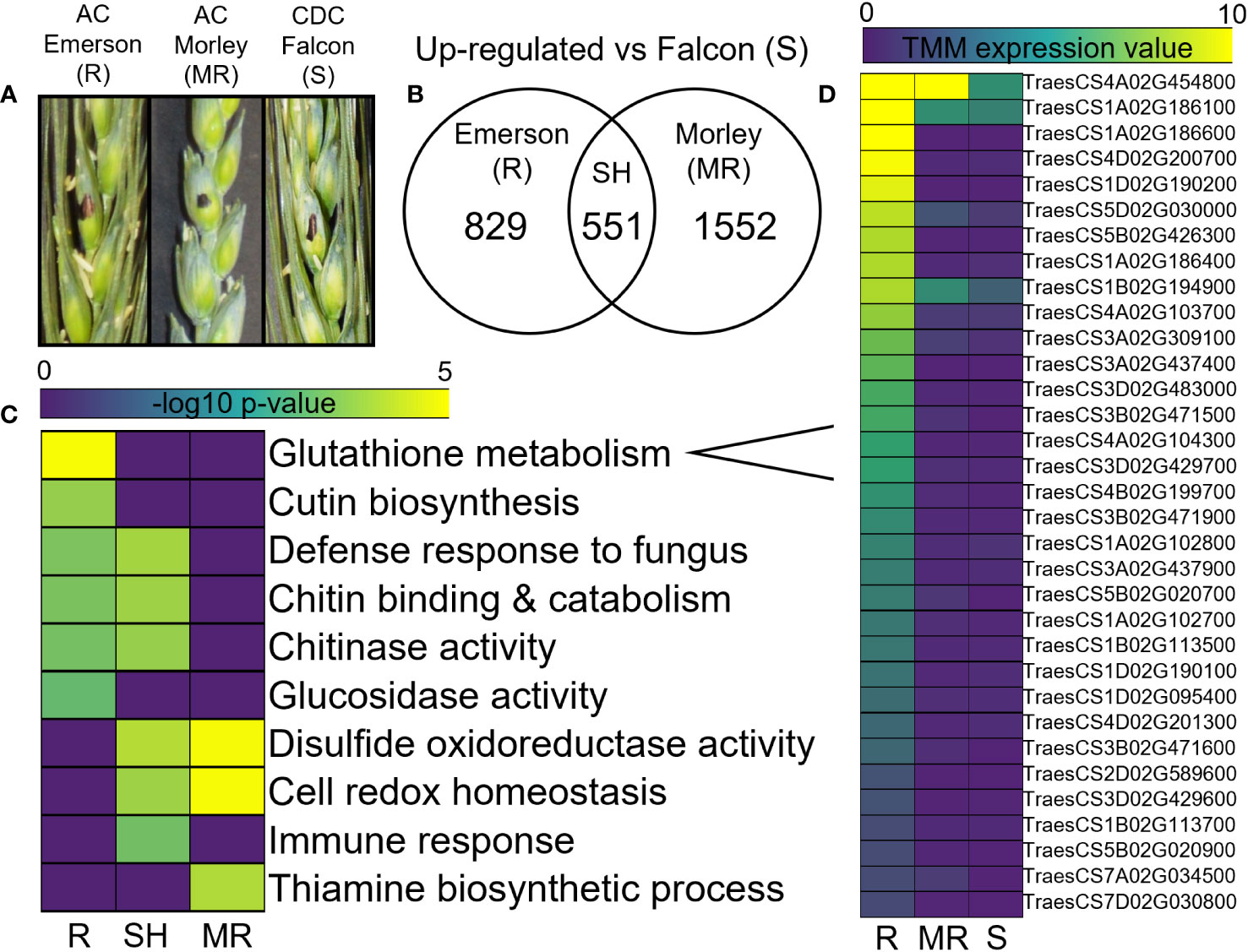
Figure 2 Differential expression analysis of AC Emerson (R), AC Morley (MR) and CDC Falcon (S) T. aestivum. (A) Control water inoculated spikelets for CDC Falcon, AC Morley and AC Emerson T. aestivum cultivars. (B) Significantly up-regulated genes in AC Emerson and AC Morley against FHB-susceptible CDC Falcon detected using DESeq2. (C) Gene ontology (GO) term enrichment of significantly up-regulated gene sets. (D) Trimmed mean of M-values (TMM) gene expression heatmap of glutathione S-transferases in T. aestivum quantified using edgeR. DEGs identified using DESeq2 with a log2 FC < -2, > 2 and an adjusted p-value cut-off of p < 0.01. GO terms considered significantly enriched at p < 0.05 prior to log10 transformation. R = FHB-resistant AC Emerson, MR = FHB-moderately resistant AC Morley, S = FHB-susceptible CDC Falcon, SH = shared. Yellow colour = more statistically enriched, purple colour = less statistically enriched.
Differential gene expression analysis of winter wheat cultivars in response to F. graminearum at 7-dpi compared to respective water inoculated controls identified both shared and specific up-regulated gene sets across AC Emerson, AC Morley and CDC Falcon (Figure 3A). For example, 913, 1436 and 1578 genes were significantly up-regulated specifically in AC Emerson (R), AC Morley (MR) and CDC Falcon (S) cultivars respectively, while 722 genes were up-regulated in both AC Morley and AC Emerson cultivars, and 6522 genes were significantly up-regulated across all three cultivars, compared to the water inoculated controls (Figure 3A). GO enrichment of these gene sets revealed distinct defense responses between cultivars, specifically between FHB-resistant and susceptible cultivars (Figure 3B). For example, enrichment of GO terms associated with physical defense like chorismate (p = 4.1x10-9), cinnamic acid (p = 1.4x10-2) and lignin (p = 4.3x10-2) biosynthesis, as well as DON detoxification through UDP-glycosyltransferase activity (p = 4.8x10-2) in the AC Emerson and AC Morley shared gene set (Figure 3B). To further explore the role of lignin biosynthesis in FHB-resistance, we mapped and plotted the expression of genes within the lignin biosynthesis pathway in response to FHB (Figures 4A, B). Here, we identified enriched mRNA levels of several key regulatory enzymes, including multiple caffeoyl-CoA O-methyltransferases (CCoAOMTs) associated with G-unit monolignol synthesis, shared between AC Emerson and AC Morley cultivars, while also identifying two putative laccases (TRAESCS5B02G149900 (LAC149900) and TRAESCS3B02G37990 (LAC37990)) that were specific to AC Emerson (Figure 4A). Interestingly, we also identified two ferulate 5-hydroxylases (F5H1 and F5H2) involved in S-unit monolignol synthesis that showed down-regulation in resistant cultivars relative to CDC Falcon, suggesting lignin composition may differ between our FHB-resistant and susceptible cultivars (Figure 4A). Fold-change of UDP-glycosyltransferases involved in DON detoxification was also plotted in response to FHB and demonstrated significant enrichment in both AC Emerson and AC Morley cultivars, however, AC Emerson showed a stronger up-regulation of these genes in response to infection (Supplementary Figure 4). Specifically, four UDP-glycosyltransferases demonstrated extremely high levels of up-regulation between both resistant cultivars (TRAESCS2B02G088400 and TRAESCS2A02G028000) or specific to the AC Emerson cultivar (TRAESCS2A02G056400 and TRAESCS2B02G626500) (Supplementary Figure 4). GO analysis further identified enrichment of processes associated with the oxidative stress response, including: glutathione synthase (p = 4.5x10-2), hydrogen peroxide catabolism (p = 3.7x10-2), and increased enrichment of antioxidant (p = 1.8x10-2) and peroxidase activity (p = 1.9x10-2) (Figure 3B). Lastly, several essential defense processes were enriched across all three cultivars including glutathione (p = 6.2x10-75) and ROS (p = 8.4x10-7) metabolism, and chitinase activity (p = 2.1x10-18) (Figure 3B).
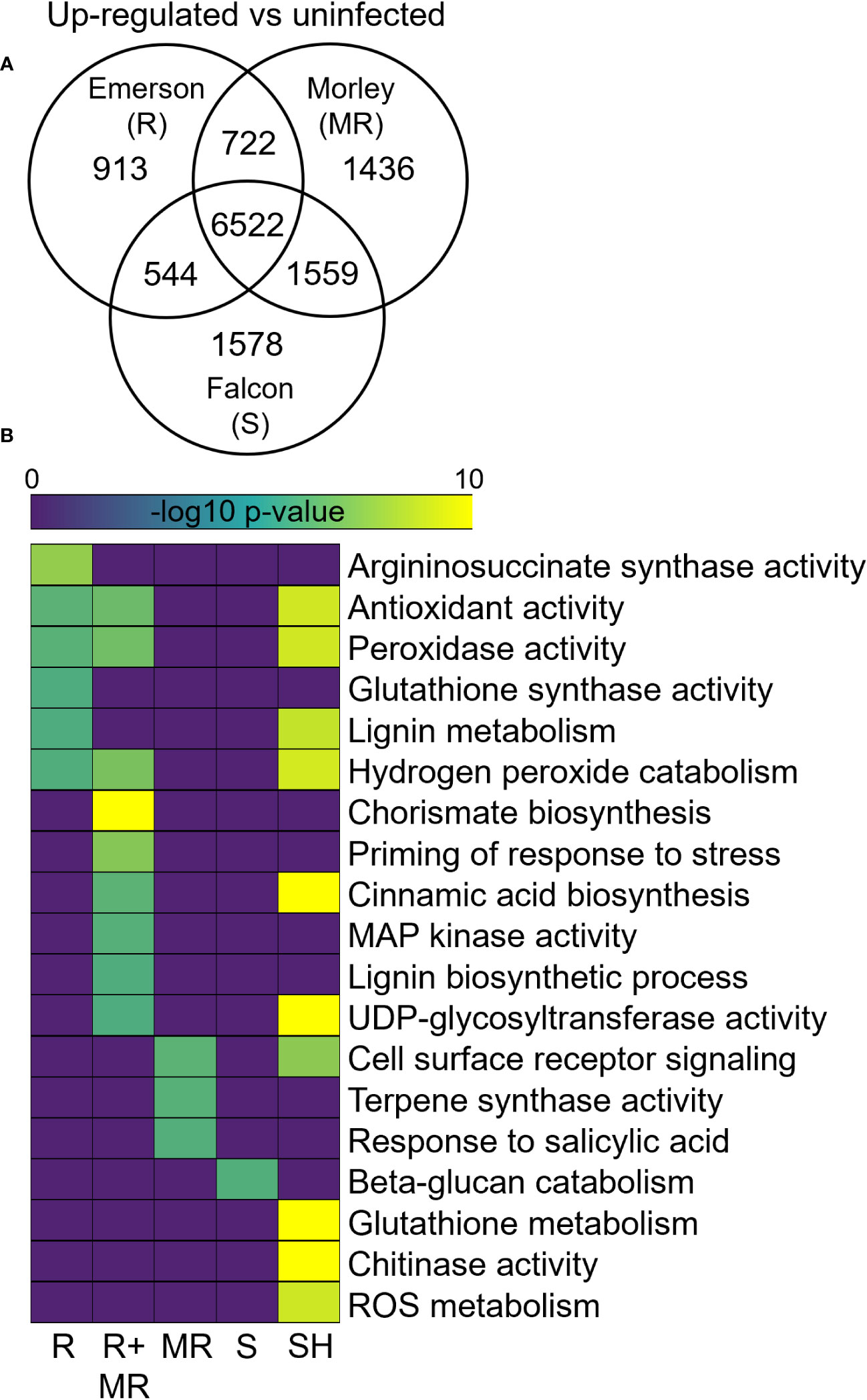
Figure 3 Differential expression analysis of AC Emerson, AC Morley and CDC Falcon T. aestivum at 7-dpi with F. graminearum. Gene ontology enrichment of significantly (A) up-regulated and (B) down-regulated genes in T. aestivum. DEGs identified using DESeq2 with a log2 FC < -2, > 2 and an adjusted p-value cut-off of p < 0.01. GO terms considered significantly enriched at p < 0.05. R = FHB-resistant AC Emerson, MR = FHB-moderately resistant AC Morley, S = FHB-susceptible CDC Falcon, R+MR = AC Emerson and AC Morley, SH = shared. Yellow colour = more statistically enriched, purple colour = less statistically enriched.
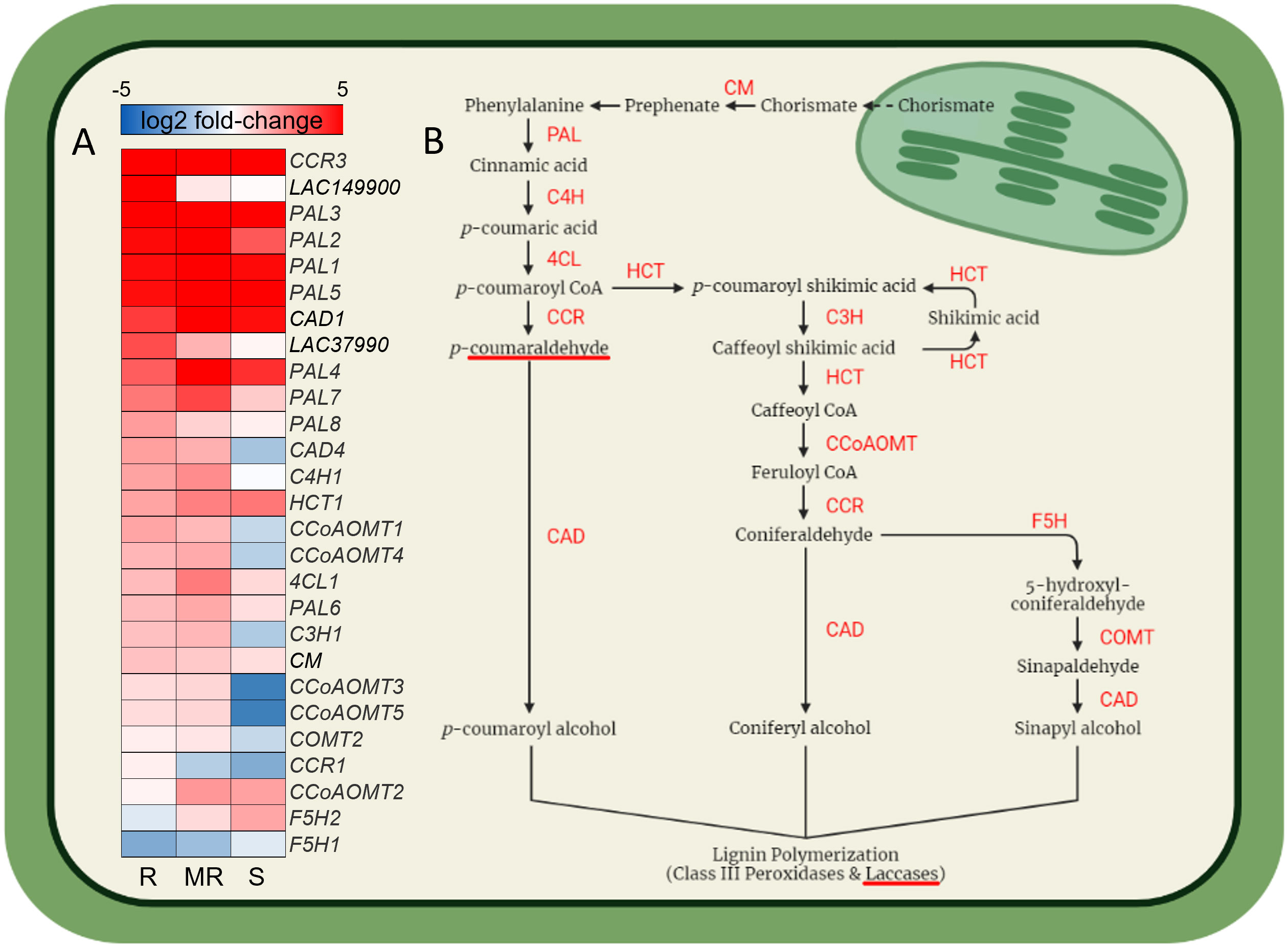
Figure 4 Gene expression heatmap of the lignin biosynthesis pathway. (A) Heatmap represents log2 fold-change between F. graminearum infected and control tissues for AC Emerson (R), AC Morley (MR) and CDC Falcon (S) cultivars. Log2 fold-change quantified using DESeq2. (B) Lignin biosynthesis pathway. CM, chorismite mutase; PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; HCT, p-hydroxycinnamoyl-CoA:qunate/shikimate p-hydroxycinnamoyltransferase; C3H, p-coumarate 3-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid O-methyltransferase. Red colour = up-regulated infected tissue relative to uninfected, blue colour = down-regulation in infected tissue relative to uninfected.
To identify regions of genomic diversity between our FHB-resistant and FHB-susceptible cultivars, we performed a variant analysis comparing AC Emerson and AC Morley against CDC Falcon. Here, we identified 20,401 variants specific to AC Emerson, 16,538 variants specific to AC Morley and 5,848 variants shared between both AC Emerson and AC Morley relative to CDC Falcon (Figure 5A). These variants were then plotted across individual chromosomes, where chromosome 2A, B and D contained the highest rate of variants per million bases in AC Emerson with 3.17, 2.75 and 3.53 variants/Mb respectively, while chromosome 1A, B and D showed the highest rate in AC Morley, with 2.20, 2.99 and 2.02 variants/Mb respectively (Table 1) (Figure 5B). To explore the biological processes and pathways potentially influenced by these variants, we performed a GO enrichment analysis on genes containing these identified variants in both AC Emerson and AC Morley (Figure 5C). Similarly to our differential expression analysis, we identified enrichment of putative FHB-resistance related pathways including, UDP-glycosyltransferase activity (p = 8.66x10-6), lignin biosynthesis through cinnamic acid biosynthesis (p = 4.68x10-5) and phenylalanine ammonia-lyase activity (p = 3.55x10-5), and glutathione S-transferases in glutathione metabolism (p = 1.89x10-8)(Figure 5C). Multiple identified UDP-glycosyltransferases and glutathione S-transferases containing variants also demonstrated significant up-regulation in AC Emerson relative to CDC Falcon (Figure 5D). Further, we plotted variants identified across the 2A, B and D chromosomes in AC Emerson relative to CDC Falcon where we found clusters of sequence diversity located on both ends of the chromosomes, while maintaining low levels of diversity in-between these distal regions (Figure 5E). Here, ~ 90% of variants occurred in the first and last 100 Mb (Figure 5E). We also explored variants within coding sequences of chromosomes 2D and 5A specifically to identify regions of nucleotide diversity within putative defense-related genes that also demonstrate differential expression between FHB-resistant and FHB-susceptible cultivars (Figure 6). Here, we found several differentially expressed variant containing genes encoding glycosyltransferases, glutathione S-transferases and receptor-like kinases in both chromosomes, as well as thaumatin-like proteins (TRAESCS5A02G017900, TRAESCS5A02G01800, TRAESCS5A02G077600) and α-amylase inhibitors (TRAESCS5A02G422900, TRAESCS5A02G554300) in chromosome 5A and agmatine hydroxycinnamoyl transferases (TRAESCS2D02G490900, TRAESCS2D02G491100) in chromosome 2D (Figures 6C, D).
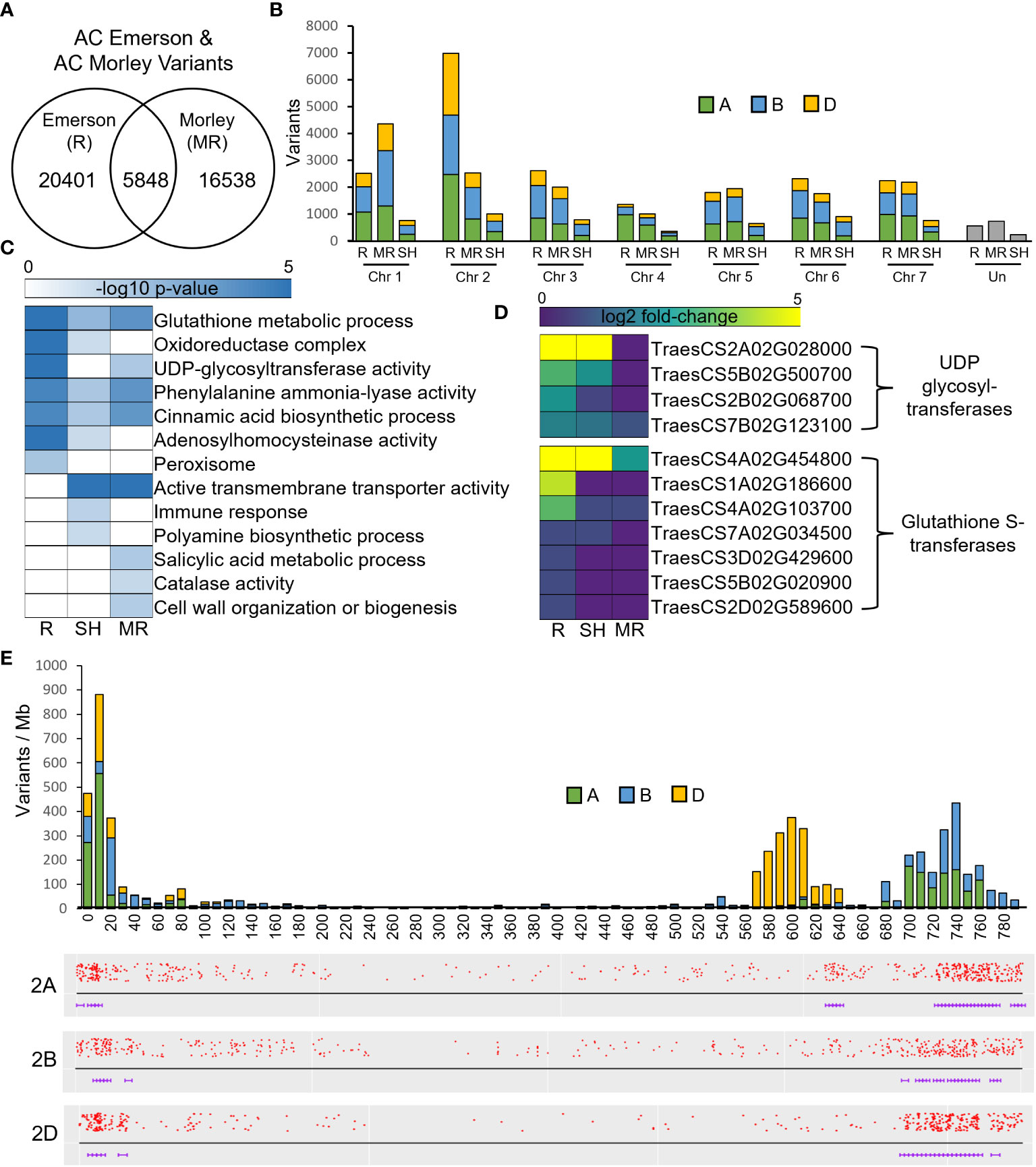
Figure 5 Variant analysis of FHB-resistant AC Emerson (R) and AC Morley (MR) relative to FHB-susceptible CDC Falcon (S). (A) Total variants detected within AC Emerson and AC Morley relative to CDC Falcon, including shared variants between AC Emerson and AC Morley. (B) Chromosomal diversity of total variants detected. (C) GO enrichment of genes containing variants specific to and shared between AC Emerson and AC Morley. (D) Expression of UDP-glycosyltransferases and glutathione S-transferases containing variants in AC Emerson relative to CDC Falcon. (E) Barplots of variants identified between AC Emerson and CDC Falcon along the 2A, 2B and D chromosomes. SH = shared between AC Emerson and AC Morley. Variants detected using BCFtools software.
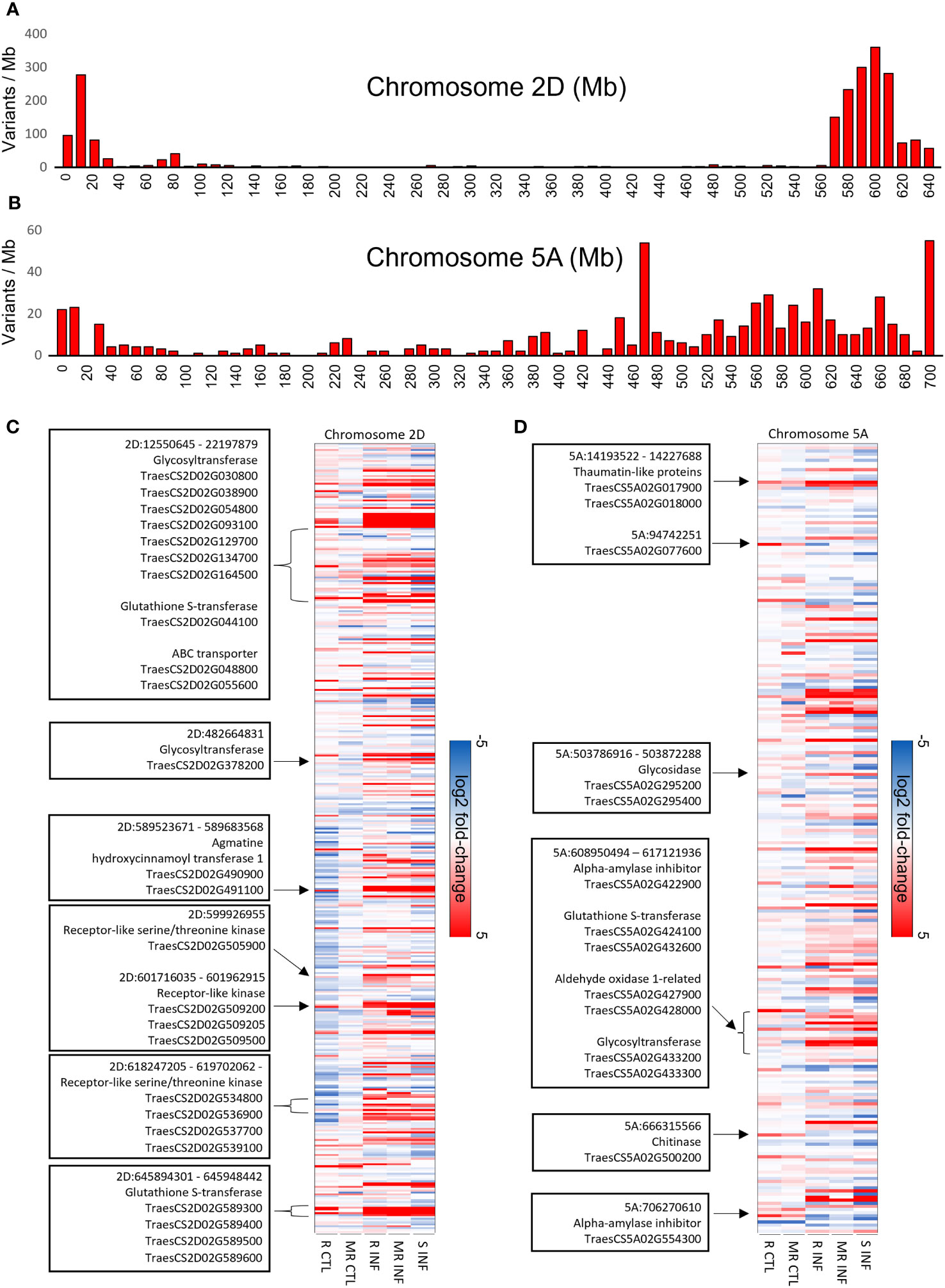
Figure 6 Variant analysis of chromosomes 2D and 5A in FHB-resistant AC Emerson (R) and AC Morley (MR) relative to FHB-susceptible CDC Falcon (S). Barplots of variants identified between AC Emerson and CDC Falcon along the (A) 2D and (B) 5A chromosomes. Gene expression heatmap of variants identified within gene regions in chromosomes (C) 2D and (D) 5A. Highlighted genes/regions represent putative defense genes which demonstrate up-regulation in AC Emerson relative to CDC Falcon. Expression values calculated using log2 fold change between FHB-resistant cultivars and CDC Falcon for control uninfected tissue, and between uninfected and infected tissues for infected sampels. R = AC Emerson, MR AC Morley and S = CDC Falcon. Variants detected using BCFtools software.
Next, we compared the RNA originating from F. graminearum within infected plants to explore differential expression in F. graminearum challenging these resistant and susceptible wheat cultivars to identify genes and processes in the pathogen influenced by distinct defense responses in the host cultivar. Here, we identified 591 and 589 significantly up- and down-regulated genes respectively in F. graminearum RNA infecting AC Morley, and 264 and 134 up- and down-regulated genes respectively in F. graminearum infecting AC Emerson relative to CDC Falcon (Figures 7A, B). Further, we identified 147 and 398 shared up- and down-regulated genes shared between both F. graminearum from AC Morley and AC Emerson (Figures 7A, B). GO enrichment of up-regulated gene sets showed significant enrichment of ABC-type transporters (p = 7.9x10-5) associated with DON export, isoprenoid biosynthesis (p = 3.2x10-2) and phosphopantetheine binding (p = 2.4x10-2) in F. graminearum from AC Emerson (Figure 7A). These enriched GO terms contained several up-regulated polyketide synthases belonging to the trichothecene cluster and DON biosynthesis pathway, including TRI5 and TRI6 which regulate and are necessary for DON biosynthesis (Figures 8A, B)(Cuzick et al., 2008; Wang et al., 2023). Further, several terms associated with the ROS response were enriched in both F. graminearum from AC Morley and AC Emerson, including oxidoreductase activity (p = 3.2x10-12), tetrapyrrole binding (p = 3.4x10-3), antioxidant activity (p = 2.1x10-2) and response to oxidative stress (p = 2.1x10-2) (Figure 7A). Meanwhile, F. graminearum from AC Emerson-specific down-regulated genes showed enrichment of cell wall biogenesis (p = 3.9x10-2), xylan metabolism (p = 1.2x10-2), hemicellulose biosynthesis (p = 1.2x10-2) and GDP-mannose biosynthesis (p = 1.2x10-2) (Figure 7B).
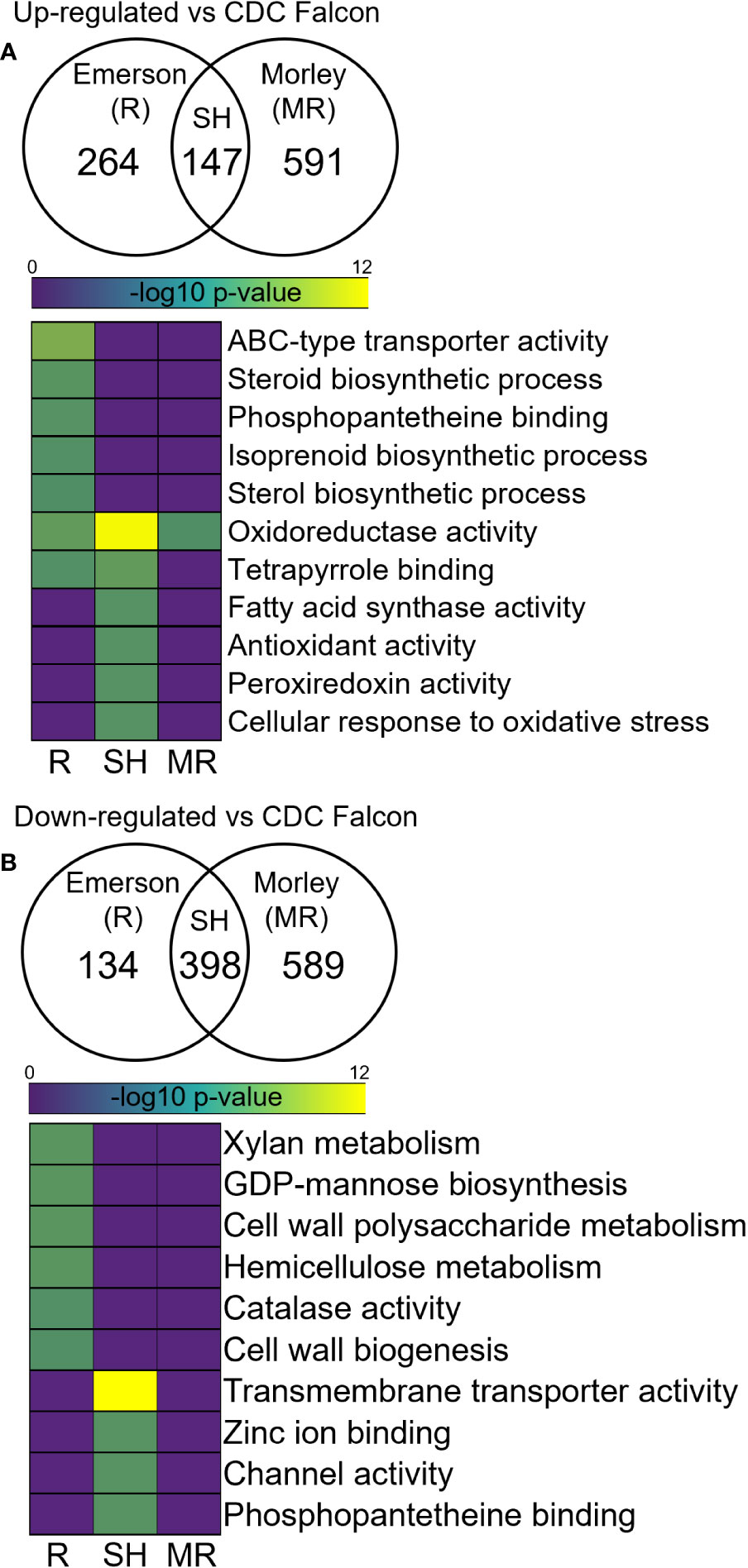
Figure 7 Differential expression analysis in F. graminearum infecting AC Emerson (R) and AC Morley (MR) against CDC Falcon (S). Gene ontology term enrichment of significantly (A) up-regulated and (B) down-regulated genes in F. graminearum. DEGs identified using DESeq2 with an adjusted p-value cut-off of p < 0.05. GO terms considered significantly enriched at p < 0.05 prior to log10 transformation. R = FHB-resistant AC Emerson, MR = FHB-moderately resistant AC Morley, SH = shared. Yellow colour = more statistically enriched, purple colour = less statistically enriched.
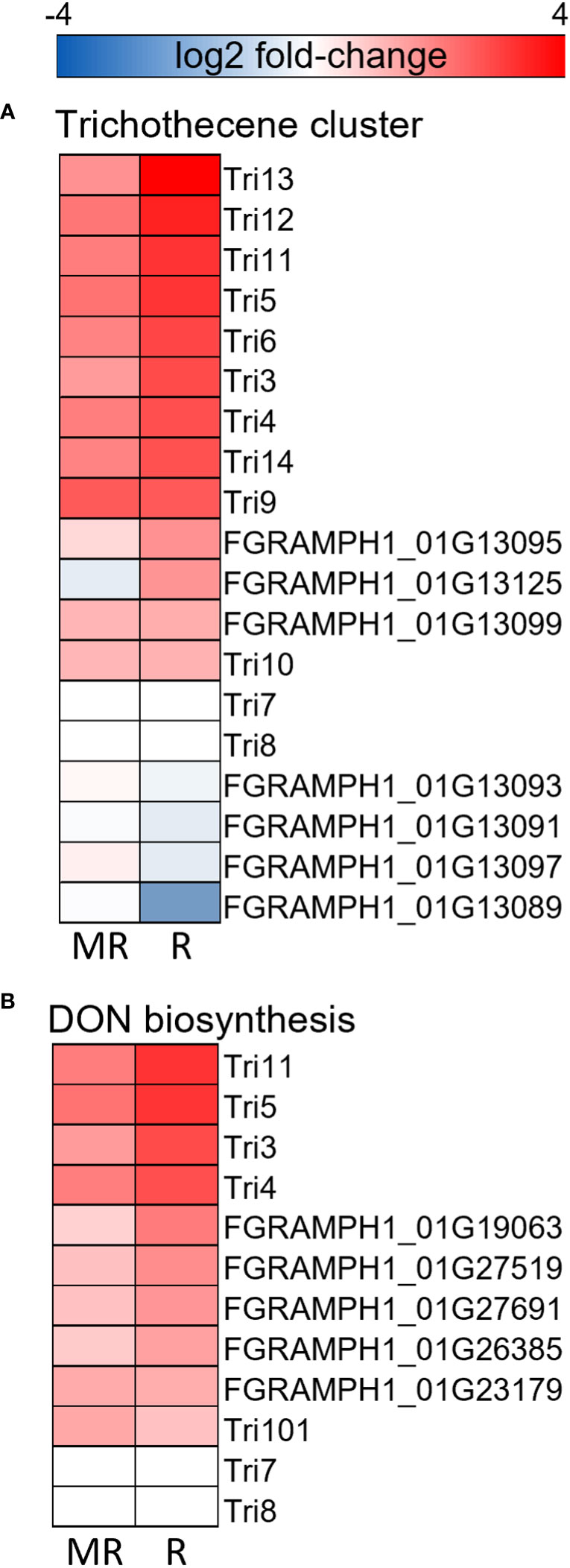
Figure 8 Gene expression heatmaps of the (A) trichothecene cluster, (B) DON biosynthesis pathway. Heatmaps represent log2 fold-change between AC Emerson (R) and AC Morley (MR) against CDC Falcon (S). Red colour = up-regulated in R, MR relative to S, blue colour = down-regulation in R, MR relative to S.
As our three winter wheat cultivars demonstrated distinct levels of FHB-resistance (Figure 1), we performed a transcriptomic analysis of both uninfected and infected tissues to identify genes and biological processes underlying these different responses. When comparing the three cultivars, differential gene expression and GO analysis of uninfected control tissues identified enriched defense processes in AC Morley and AC Emerson relative to FHB-susceptible CDC Falcon. Enrichment of oxidoreductase activity and cellular redox homeostasis in the shared gene set suggests a potential priming of redox activity prior to infection in these resistant cultivars (Figure 2C). This may lead to a more immediate and enhanced response to oxidative stress, which is typically triggered in response to pathogen attack, as ROS are produced and accumulate at the host-pathogen interface upon infection, requiring antioxidant activity to prevent cellular damage (Khaledi et al., 2016). ROS production can be further elicited throughout infection by trichothecenes produced by F. graminearum, ultimately resulting in host programmed cell death (Waśkiewicz et al., 2014; Khaledi et al., 2016). The most common antioxidant induced in response to ROS accumulation in wheat are glutathione S-transferases (GSTs) (Buerstmayr et al., 2021). Increased activity of these GSTs has been previously associated with FHB-resistant cultivars in response to infection, with the GST encoded by Fhb7 conferring resistance to FHB through trichothecene detoxification (Wang et al., 2020; Kumar et al., 2020; Buerstmayr et al., 2021). As we identified 33 putative GSTs enriched in our uninfected tissues, these data suggest a potential priming of GST activity prior to infection that may provide an increased ability to detoxify trichothecenes which are produced by F. graminearum during early stages of FHB infection (24 – 96 hpi) (Gunupuru et al., 2017).
AC Emerson-specific enrichment of cutin biosynthesis, as well as chitinase and glucosidase activity were also identified through GO analysis of uninfected control tissues (Figure 2). Chitinases and β-glucosidases are commonly up-regulated by hosts immediately upon pathogen recognition to degrade the invading pathogen, where upon secretion, these enzymes can breakdown fungal cell wall components, subsequently acting as signalling molecules to activate host immunity pathways (Wan et al., 2008). In a study by Anand et al., 2003, co-expression of two pathogenesis-related (PR) genes encoding a chitinase and β-1,3-glucosidase led to increased resistance in transgenic spring wheat compared to background FHB-susceptible lines. Further, increased chitinase and glucosidase activity was identified in the FHB-resistant cultivar Sumai 3 (Kosaka et al., 2015). Specific chitinase and glucosidase genes have also been identified in resistance against rust diseases, with the chitinase CHT20.2 (TRAESCS5B02G403700) conferring resistance to wheat stripe rust (Bai et al., 2021) and the β-1,3-glucosidase TRAESCS3A02G483000 being significantly up-regulated in the yellow stripe rust resistant cultivar Gannong 2 (Zhao et al., 2022). Our data identified both CHT20.2 and TRAESCS3A02G483000 as significantly up-regulated in uninfected AC Emerson tissue relative to CDC Falcon (Dataset S1). Here, activity of these enzymes, prior to infection, may lead to a more immediate and enhanced pathogen recognition, breakdown and therefore immune signalling response.
Further, cutin is the major component of the outer waxy cuticle which serves as a physical barrier to abiotic and biotic stressors, acting as the first point of contact to pathogen attack (Serrano et al., 2014). Genes responsible for cuticle reinforcement were found to be enriched in FHB-resistant barley, with knockdown of the regulatory transcription factor of these genes (HvWIN1) in RNAi transgenic lines leading to reduced resistance (Kumar et al., 2016). As our data indicates significant enrichment of cutin biosynthesis in FHB-resistant AC Emerson, cuticle density and composition at the point of pathogen contact may be associated with increased tolerance to FHB through slowing pathogen attack and allowing the host sufficient time to mount a successful defense response.
Taken together, these data suggest a priming of the defense response through enhanced antioxidant activity and redox homeostasis shared between resistant cultivars, while AC Emerson further demonstrates amplified enrichment of cellular detoxification through increased glutathione S-transferase activity, as well as enrichment of chitinases and glucosidases capable of degrading fungal cell walls upon infection. These processes, as well as bolstered physical barriers, have historically been essential during early stages of pathogen attack (Kosaka et al., 2015; Wang et al., 2020), which may provide insight into why priming of these responses are associated with FHB-resistance in AC Emerson. Lastly, further transcriptomic investigation of earlier infection stages could help determine whether priming of these host defense pathways results in improved antioxidant activity and pathogen degradation at the onset of FHB infection in AC Emerson and AC Morley cultivars.
Differential expression analyses between uninfected and F. graminearum infected tissues was subsequently performed to provide insight into the specific genes and pathways that may be critical to FHB resistance during infection. GO analysis of the up-regulated shared AC Morley and AC Emerson gene set in response to infection revealed enrichment of cell wall lignification through chorismate, cinnamic acid and lignin biosynthesis (Figure 3B). Cell wall lignification is a common defense response to pathogen attack which bolsters physical defense by filling gaps present between the cellulose, hemicellulose and pectin matrix of plant cell walls (Wang et al., 2013; Ziv et al., 2018). Prior studies have reported increased lignin content as well as increased activation of lignin biosynthesis regulators in FHB-resistant cultivars upon infection relative to susceptible lines (Kang and Buchenauer, 2000; Soni et al., 2021), with genes belonging to the caffeoyl-coenzyme A O-methyltransferase (CCoAOMT) family being identified as key regulators of lignin biosynthesis and therefore potentially FHB-resistance (Yang et al., 2021). Dehydrogenative polymerization of p-coumaryl, sinapyl and coniferyl alcohols via laccases and class III peroxidases give rise to the p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) units of lignin respectively (Nguyen et al., 2016; Yang et al., 2021). Distinct enzymatic families contribute to the synthesis of these specific monolignol units, with CCoAOMTs being associated with G-unit monolignol synthesis and ferulate-5-hydroxylase (F5Hs) being associated with S-unit synthesis (Cao et al., 2022). Lignin composition, specifically the S/G unit ratio, has been primarily explored for its affect on biomass recalcitrance, however, studies have also demonstrated an influence on disease resistance in plants (Bhuiyan et al., 2009; Cao et al., 2022). A recent study by Cao et al. (2022) demonstrated that silencing of F5H, which is necessary for guaiacyl (G) conversion to the syringyl (S) monolignol unit, leads to reduced S/G ratios and increased S. sclerotiorum resistance in f5h Brassica napus knockout mutants. Interestingly, our data suggests a similar pattern, with FHB-resistant cultivars AC Emerson and AC Morley demonstrating reduced expression of F5H genes relative to FHB-susceptible CDC Falcon (Figure 4A). Further, we also find increased expression of CCOAOMT genes associated with G-unit synthesis in AC Emerson and AC Morley relative to CDC Falcon (Figure 4A). These data suggest that lignin composition and the S/G unit ratio of lignin polymers may contribute to resistance against FHB in wheat through the activity of key monolignol synthesis enzymes. Further, the final step of lignin biosynthesis is monolignol polymerization through oxidation of p-coumaryl, sinapyl and coniferyl alcohols via activity of laccases and class III peroxidases (Nguyen et al., 2016; Rossini et al., 2022). Differential expression analysis identified two plant laccases (TRAESCS5B02G149900 (LAC149900) and TRAESCS3B02G37990 (LAC37990)) to be significantly up-regulated in AC Emerson in response to FHB (Figure 4A). Identification of specific laccases and characterization of their role in lignification in response to FHB has yet to be explored in wheat, partly due to the large number of predictive laccases identified in the genome (Hoffmann et al., 2020; Sun et al., 2022). Here, identification of LAC149900 and LAC37990 could serve as ideal candidates for further characterization in cell wall lignification in response to FHB.
While laccases are known to play a role in physical defense through their role in lignin polymerization, a study by Sun et al. (2022) also suggested that these laccases can interact with DON, potentially preventing DON from entering host cells through direct interaction or oxidization. Data on laccases and their ability to interact with DON are currently incomplete, however, DON detoxification has primarily been demonstrated through UDP-glycosylation, where DON is conjugated into its detoxified product deoxynivalenol-3-glucoside (Pasquet et al., 2016; Tian et al., 2016). This DON glycosylation via UDP-glycosyltransferases has been previously demonstrated to increase resistance against FHB in wheat (Li et al., 2017; He et al., 2020), and within our shared AC Emerson and AC Morley up-regulated gene set, we found significant enrichment of UDP-glycosyltransferase activity (Figure 3B). While identified UDP-glycosyltransferases showed significant up-regulation in both AC Morley and AC Emerson cultivars, many genes showed a stronger or specific (TRAESCS2A02G056400 and TRAESCS2B02G626500) up-regulation in AC Emerson relative to CDC Falcon (Supplementary Figure 4). As our dataset identified a broader pattern of increased UDP-glycosyltransferase enrichment correlating to increased levels of FHB-resistance, these specific highly up-regulated UDP-glycosyltransferases in resistant cultivars relative to susceptible CDC Falcon may serve as ideal candidate genes for further characterization for their role in DON detoxification and FHB-resistance.
AC Emerson also demonstrated an enrichment of processes associated with oxidative stress, including: hydrogen peroxide catabolism, peroxidase activity, antioxidant activity and glutathione synthase activity (Figure 3B). As previously mentioned, the ability to maintain oxidation levels and reduce oxidative stress is critical for stress tolerance in response to both abiotic and biotic stressors (Matić et al., 2021), as ROS can accumulate at high levels requiring both enzymatic and non-enzymatic antioxidants to scavenge the abundant free radicals (Kapoor et al., 2019). Peroxidases are multi-functional enzymes, demonstrating antioxidant activity through hydrogen peroxide catabolism in response to stress (Tyagi et al., 2018), while also contributing to lignin polymerization in cell wall reinforcement through class III peroxidases (Hoffmann et al., 2020). ROS can also be detoxified through non-enzymatic antioxidants like glutathione, whose synthesis is catalyzed via glutathione synthase (Hasanuzzaman et al., 2017). Increased levels of glutathione during early infection stages has been previously associated with increased levels of tolerance across multiple plant pathosystems (Dubreuil-Maurizi and Poinssot, 2012; Zechmann, 2020). Further, glutathione and ROS metabolism were also enriched across all three cultivars in response to FHB, along with increased chitinase activity (Figure 3B). Interestingly, while enrichment of these processes were shared across all three cultivars in response to infection, they were also enriched specifically within uninfected AC Emerson tissue compared uninfected AC Morley and CDC Falcon, further suggesting a potential priming of these key defense processes that are critical in response to FHB.
QTLs associated with FHB-resistance have predominately been identified in spring wheat cultivars, with the most well established and repeatable QTLs occurring on chromosomes 3B (Fhb1; Qfhs.ndsu-3BS), 5A (Fhb5; Qfhs.ifa-5A) and 6B (Fhb2). However, a study by Clinesmith et al., 2019 identified four QTLs conferring resistance to FHB in winter wheat cultivars, with two of the four being located on chromosome 2D. Further, one of these QTLs (Qfhb.ksu-2D.1) was identified at 580.12 Mb, a region where we identified high levels of variation in AC Emerson (Figure 5G). Two FHB disease index marker trait associations (MTAs) were also identified on chromosome 2A and 2B in winter wheat, where similarly to Qfhb.ksu-2D.1, these MTAs were located towards the end of the long arm of chromosomes 2A and 2B, occurring at the 722 and 725 Mb positions respectively (Zhang et al., 2022). These chromosomal regions also demonstrate high levels of sequence diversity in AC Emerson (Figures 5E, F), suggesting these regions may be contributing to FHB resistance across multiple winter wheat cultivars. The majority of variants identified in both AC Emerson and AC Morley occurred at the distal regions of chromosomes (Dataset S4) (Figures 5E-G). In a study by Nilsen et al. (2021) exploring variants present in chromosome 5A, a similar phenomena was discovered, with a significant number of variant clusters occurring near the end of chromosome 5A. This disproportionate clustering of variants is likely due to the increased recombination rates associated with the distal regions of chromosomes in crop species with large genomes, including wheat, maize and barley (Ganal et al., 2011; Mayer et al., 2012; Choulet et al., 2014). Further, while these distal chromosomal regions have demonstrated greater gene density across several grass genomes (Schnable et al., 2009; Choulet et al., 2014), GO analysis has also described an enrichment of genes associated host stress responses in these distal genes (Choulet et al., 2014). These data suggest distal chromosomal regions may be of particular interest in the discovery of FHB and other resistance associated QTLs. GO enrichment of variants within annotated genes in our data identified high levels of enrichment for similar processes and pathways as identified in our differential expression analysis, including UDP-glycosyltransferase activity, glutathione S-transferase activity and lignin biosynthesis through phenylalanine ammonia-lyase activity and cinnamic acid biosynthesis (Figure 5C). These data suggest that sequence variation in genes associated with these processes may contribute to their differential expression across AC Emerson and AC Morley cultivars relative to CDC Falcon.
To further explore these findings we mapped expression of all gene variants identified in chromosomes 2D and 5A, as multiple important FHB-related QTLs have been previously identified in these regions (McCartney et al., 2016; Clinesmith et al., 2019; Steiner et al., 2019). Here, we discovered two distinct clusters of differentially expressed receptor-like kinases (RLKs), known for their role in defense signalling, within chromosome 2D located at the 601 and 618 Mb regions (Figure 6C). A previous study by Hu et al., 2019 identified four receptor-like kinases as DEG candidates genes associated with the FHB 2DL-QTL, as these RLKs showed significant up-regulation in FHB-resistant near isogenic lines. While several of the RLKs identified in our study were induced in response to F. graminearum, multiple were also upregulated in uninfected AC Emerson relative to CDC Falcon coupled with a stronger induction in response to infection (Figures 6C, D). This increased activity prior to infection may result in a more immediate signalling response and subsequent down-stream defense gene activation upon infection. We also identified variants within two agmatine hydroxycinnamoyl transferases (TRAESCS2D02G490900, TRAESCS2D02G491100) that demonstrated up-regulation in response to F. graminearum in AC Emerson. Hydroxycinnamoyl transferases are involved in lignin biosynthesis through the synthesis of hydroxycinnamic acid amides (HCAAs), where silencing of these transferases has been shown to inhibit lignin polymerization in N. benthamiana (Hoffmann et al., 2004). The agmatine coumaroyl transferase, TaACT, has previously been described in the FHB QTL-2DL and is a rate limiting step in the synthesis of HCAAs, where silencing of TaACT leads to reduced HCAA accumulation and increased FHB susceptibility (Kage et al., 2017). As hydroxycinnamoyl transferases also play a vital role in HCAA production, TRAESCS2D02G490900 and TRAESCS2D02G491100 may also be associated with FHB resistance on chromosome 2D through HCAA biosynthesis. Further, several differentially expressed glycosyltransferases and glutathione S-transferases were once again identified across both chromosomes 2D and 5A, however, on chromosome 5A we also found thaumatin-like proteins (TLPs) and α-amylase inhibitors which were significantly up-regulated in AC Emerson relative to CDC Falcon prior to infection (Figures 6C, D). Thaumatin-like proteins belong to the pathogenesis-related (PR) protein 5 family and are commonly induced in response to fungal pathogen attack (Liu et al., 2010; Ren et al., 2022). TLPs have demonstrated the ability to inhibit pathogen growth across several plant species as well as in vitro (Ren et al., 2022). A previous genome wide characterization of thaumatin-like proteins in response to FHB infection identified a disproportionate distribution of these genes across the genome, with the highest number of TLPs (22) being found on chromosome 5A (Ren et al., 2022). A further analysis of differential expression between F. graminearum inoculated and mock inoculated near isogenic lines expressing Fhb1 and Qfhs.ifa-5A FHB-resistance QTLs showed TRAESCS5A02G017900 to be one of four TLPs to be significantly up-regulated across all timepoints tested (24hpi – 48 hpi) (Ren et al., 2022). Our data indicates not only do we find induction of this TLP in response to F. graminearum at 7 dpi across AC Emerson and AC Morley, we also find increased activity in AC Emerson relative to CDC Falcon prior to infection (Figure 6D). Further, variants in two α-amylase inhibitors were identified on chromosome 5A that also demonstrate significant up-regulation in AC Emerson relative to CDC Falcon prior to infection (Figure 6D). These α-amylase inhibitors have been shown to accumulate at significantly higher levels in wheat kernels of FHB-resistant cultivars relative to FHB-susceptible lines (Perlikowski et al., 2014). α-amylases are necessary for starch hydrolysis in Fusarium graminearum, where reducing α-amylase activity can lead to reduced trichothecene production and ultimately reduced pathogenicity in wheat (Oh et al., 2016) Further, these α-amylase inhibitors have been shown to inhibit pathogen growth in vitro and reduce mycotoxin production by the pathogen (Mendes et al., 2015).
Differential expression analysis of F. graminearum infecting AC Emerson, AC Morley and CDC Falcon revealed significant up-regulation of genes in mycotoxin clusters and biosynthesis pathways in FHB-resistant cultivars (Figure 8). Trichothecene mycotoxins produced by Fusarium species have been demonstrated to increase pathogenicity in multiple hosts, acting through inhibition of protein synthesis and the generation of free radicals leading to oxidative stress (Wu et al., 2017; Villafana et al., 2019). The C2H2 transcription factor TRI6 has been described as a positive regulator of trichothecene gene expression necessary for DON biosynthesis, with tri6 mutants demonstrating reduced expression of all TRI cluster genes, with the exception of TRI10 which is also hypothesized to be a positive regulator of TRI expression (Wang et al., 2023). Further, TRI5 activity has also been described as essential for DON biosynthesis, with tri5 mutants not being able to produce DON and demonstrating significantly reduced pathogenicity (Cuzick et al., 2008). All three positive regulators of TRI gene cluster activity and DON biosynthesis (TRI6, TRI10 and TRI5) showed significant up-regulation when F. graminearum was analyzed from both AC Morley and AC Emerson cultivars, along with several other members of the trichothecene cluster and DON biosynthesis pathway (Figures 8A, B). Enrichment of ATP-binding cassette (ABC)-type transporters in F. graminearum infected FHB-resistant AC Emerson was also identified (Figure 7). ABC-type transporters and major facilitator superfamily (MFS) transporters have been hypothesized to be responsible for the export of DON and other mycotoxins produced by F. graminearum during pathogenesis (O’Mara et al., 2021). Prior studies have demonstrated that silencing of ABC1, ABC3 and ABC6 each result in significantly reduced DON accumulation (O’Mara et al., 2021), while the MFS transporter, TRI12, also results in reduced DON accumulation when silenced (Menke et al., 2012). Our data identified ABC1, ABC3, and ABC6 to be significantly up-regulated in F. graminearum infecting AC Emerson, along with several other ABC-type transporters, while TRI12 was significantly up-regulated in F. graminearum infecting both AC Morley and AC Emerson (Supplementary Figure 5; Dataset S1). Prior studies have hypothesized that ABC6 and MFS1 may be regulated by TRI6 and play a role in DON export only when TRI12 is absent, as these transporters were highly up-regulated with TRI6 in tri12 mutants during periods of DON production (O’Mara et al., 2021). While our data supported this hypothesis for MFS1, which showed no activity in F. graminearum infecting AC Emerson or AC Morley cultivars when TRI12 was up-regulated, ABC6 did show up-regulation in F. graminearum infecting AC Emerson, suggesting these two transporters may not be redundant in function (Dataset S1). As export of DON and other mycotoxins has yet to be well characterized, the ABC-type and MFS transporters identified in this study may serve as ideal candidates for further characterization for their role in F. graminearum pathogenesis (Supplementary Figure 5). Enrichment of these genes involved in the trichothecene cluster, as well as DON biosynthesis and export, could ultimately be due to the suppression at earlier stages of infection in resistant cultivars, leading to a need to up-regulate these key pathogenicity factors. Alternatively, as DON biosynthesis genes and the trichothecene cluster has previously demonstrated increased activity during early stages of infection (48h – 96h) and decreased activity at later stages, these findings could be attributed to the progression of FHB in CDC Falcon and the inability of infection to progress in resistant AC Emerson and AC Morley cultivars beyond the early infection stages where activity of these genes is highest (Lysøe et al., 2011).
Further, oxidative stress has been demonstrated to activate toxin production, specifically the trichothecene cluster where increased H2O2 leads to increased TRI gene expression in F. graminearum (Montibus et al., 2013). This correlates with our data, which showed enrichment of the trichothecene cluster and ROS response in F. graminearum infecting AC Emerson (Figures 7A, 8). Further, the ROS-degrading enzymes, catalases, have been identified as pathogenicity factors that can neutralize host-derived ROS present at the host-pathogen interface, however, CAT1 and CAT3 which have been previously identified as essential in maintaining redox homeostasis, were down-regulated in F. graminearum infecting our resistant cultivars relative to infecting the FHB-susceptible cultivar CDC Falcon (Montibus et al., 2016). These data further support a study by Ponts et al. (2007), where catalase treated F. graminearum resulted in reduced TRI gene expression and toxin accumulation, which is what our data indicates in F. graminearum infecting susceptible CDC Falcon where CAT1 and CAT3 are expressed. Conversely, down-regulated genes in F. graminearum challenging our resistant cultivars demonstrated increased cell wall modification and biogenesis (Figure 7). This enrichment suggests reduced fungal growth in resistant cultivars compared to F. graminearum in CDC Falcon which is demonstrating consistent growth as indicated by the significantly greater spread of infection and FHB-severity in CDC Falcon.
Taken together, we identified several enriched defense processes prior to and in response to F. graminearum infection associated with FHB-resistance in AC Emerson and AC Morley winter wheat cultivars, including: redox homeostasis, lignin biosynthesis and DON detoxification. Within these defense processes we have identified novel putative defense regulators as well as defense genes previously associated with FHB-resistance in other FHB-resistant cultivars and species. Finally, dual RNA-sequencing identified novel putative pathogenicity factors in F. graminearum which were differentially expressed when infecting these FHB-resistant cultivars compared to CDC Falcon. These data provide insight into identifying new sources of FHB-resistance in winter wheat and improving our understanding of this important pathosystem.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. MB: Supervision, Writing – review & editing. BM: Resources, Writing – review & editing. CM: Resources, Writing – review & editing. HR: Resources, Writing – review & editing. MH: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by Agriculture and Agri-Food Canada’s Abase project ID: J-001386.
We thank Debbie Miranda and Otto Gruenke for their technical support maintaining plants, preparing inoculum and processing RNA extractions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1299461/full#supplementary-material
Supplementary Figure 1 | Hierarchical clustering analysis of CDC Falcon, AC Morley and AC Emerson T. aestivum control and F. graminearum infected (7-dpi) samples. Count data was generated using featureCounts and used as input. Dendrogram generated using the pvclust package in r. CTL, control water inoculated samples; INF, F. graminearum infected samples at 7-dpi. A = bio-replicate #1, B = bio-replicate #2, C = bio-replicate #3.
Supplementary Figure 2 | Principal component analysis of (A) T. aestivum (CDC Falcon (S), AC Morley (MR) and AC Emerson (R)) and (B) F. graminearum infecting CDC Falcon (S), AC Morley (MR) and AC Emerson (R) at 7-dpi. Count data generated using featureCounts was used as input and PCA plot generated using DESeq2 in r. CTL = control water inoculated samples, INF = F. graminearum infected samples at 7-dpi.
Supplementary Figure 3 | Target T. aestivum and F. graminearum gene expression correlation of RNA sequencing and qPCR data. Log2 fold-change of AC Emerson and AC Morley cultivars compared to CDC Falcon and deltaCT qPCR expression data. T. aestivum expression data comparisons of (A) AC Morley vs CDC Falcon and (B) AC Emerson vs CDC Falcon. F. graminearum expression data comparisons of (C) AC Morley vs CDC Falcon and (D) AC Emerson vs CDC Falcon. RNA sequencing data represented as bars and qPCR data represented with diamond marked line plot.
Supplementary Figure 4 | Gene expression heatmap of UDP-glycosyltransferases. Heatmaps represent log2 fold-change between infected and control tissues for AC Emerson (R), AC Morley (MR) and CDC Falcon (S) cultivars. Red colour = up-regulated infected tissue relative to uninfected, blue colour = down-regulation in infected tissue relative to uninfected.
Supplementary Figure 5 | Gene expression heatmap of ABC-type and MFS transporters. Heatmaps represent log2 fold-change between AC Emerson (R) and AC Morley (MR) against CDC Falcon (S). Red colour = up-regulated in R, MR relative to S, blue colour = down-regulation in R, MR relative to S.
Anand, A., Zhou, T., Trick, H. N., Gill, B. S., Bockus, W. W., Muthukrishnan, S. (2003). Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 54 (384), 1101–1111. doi: 10.1093/jxb/erg110
Audenaert, K., Vanheule, A., Höfte, M., Haesaert, G. (2014). Deoxynivalenol: A major player in the multifaceted response of Fusarium to its environment. Toxins (Basel) 6 (1), 1–19. doi: 10.3390/toxins6010001
Bai, X., Zhan, G., Tian, S., Peng, H., Cui, X., Islam, M. A., et al. (2021). Transcription factor BZR2 activates chitinase Cht20.2 transcription to confer resistance to wheat stripe rust. Plant Physiol. 187 (4), 2749–2762. doi: 10.1093/plphys/kiab383
Bhuiyan, N. H., Selvaraj, G., Wei, Y., King, J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60 (2), 509–521. doi: 10.1093/jxb/ern290
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Buerstmayr, M., Wagner, C., Nosenko, T., Omony, J., Steiner, B., Nussbaumer, T., et al. (2021). Fusarium head blight resistance in European winter wheat: insights from genome-wide transcriptome analysis. BMC Genomics 22 (1), 470. doi: 10.1186/s12864-021-07800-1
Cao, Y., Yan, X., Ran, S., Ralph, J., Smith, R. A., Chen, X., et al. (2022). Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ. 45 (1), 248–261. doi: 10.1111/pce.14208
Chen, Y., Lun, A. A. T., Smyth, G. K. (2016). From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 5, 1438. doi: 10.12688/f1000research.8987.2
Choulet, F., Alberti, A., Theil, S., Glover, N., Barbe, V., Daron, J., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Sci. (New York N.Y.) 345 (6194), 1249721. doi: 10.1126/science.1249721
Clinesmith, M. A., Fritz, A. K., Lemes da Silva, C., Bockus, W. W., Poland, J. A., Dowell, F. E., et al. (2019). QTL mapping of fusarium head blight resistance in winter wheat cultivars ‘Art’ and ‘Everest’. Crop Sci. 59, 911–924. doi: 10.2135/cropsci2018.04.0276
Cuzick, A., Urban, M., Hammond-Kosack, K. (2008). Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. New Phytol. 177 (4), 990–1000. doi: 10.1111/j.1469-8137.2007.02333.x
Dubreuil-Maurizi, C., Poinssot, B. (2012). Role of glutathione in plant signaling under biotic stress. Plant Signaling Behav. 7 (2), 210–212. doi: 10.4161/psb.18831
Dweba, C. C., Figlan, S., Shimelis, H. A., Motaung, T. E., Sydenham, S., Mwadzingeni, L., et al. (2017). Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 91, 114–122. doi: 10.1016/j.cropro.2016.10.002
Fernando, U., Chatur, S., Joshi, M., Bonner, C. T., Fan, T., Hubbard, K., et al. (2019). Redox signalling from NADPH oxidase targets metabolic enzymes and developmental proteins in Fusarium graminearum. Mol. Plant Pathol. 20 (1), 92–106. doi: 10.1111/mpp.12742
Food and Agriculture Organization of the United Nations, FAOSTAT (2019). Available at: https://www.fao.org/faostat/en/#data/QC (Accessed 1 May 2023).
Fowler, D. B. (1999). CDC Falcon winter wheat. Can. J. Plant Sci. 79, 599–5601. doi: 10.4141/P99-024
Fujita, M., Hasanuzzaman, M. (2022). Approaches to enhancing antioxidant defense in plants. Antioxid. (Basel Switzerland) 11 (5), 925. doi: 10.3390/antiox11050925
Ganal, M. W., Durstewitz, G., Polley, A., Bérard, A., Buckler, E. S., Charcosset, A., et al. (2011). A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PloS One 6 (12), e28334. doi: 10.1371/journal.pone.0028334
Gatti, M., Choulet, F., Macadré, C., Guérard, F., Seng, J. M., Langin, T., et al. (2018). Identification, molecular cloning, and functional characterization of a wheat UDP-glucosyltransferase involved in resistance to Fusarium Head Blight and to mycotoxin accumulation. Front. Plant Sci. 13. doi: 10.3389/fpls.2018.01853
Ge, S. X., Jung, D., Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36 (8), 2628–2629. doi: 10.1093/bioinformatics/btz931
Graf, R. J., Beres, B. L., Laroche, A., Gaudet, D. A., Eudes, F., Pandeya, R. S., et al. (2013). Emerson hard red winter wheat. Can. J. Plant Sci. 93 (4), 741–748. doi: 10.4141/cjps2012-262
Gunupuru, L. R., Perochon, A., Doohan, F. M. (2017). Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 42, 175–183. doi: 10.1007/s40858-017-0147-3
Guo, X., Shi, Q., Wang, M., Yuan, J., Zhang, J., Wang, J. (2023). Functional analysis of the glutathione S-transferases from Thinopyrum and its derivatives on wheat Fusarium head blight resistance. Plant Biotechnol. J 21 (6), 1091–1093. doi: 10.1111/pbi.14021
Hasanuzzaman, M., Nahar, K., Anee, T. I., Fujita, M. (2017). Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol Mol Biol Plants 23 (2), 249–268. doi: 10.1007/s12298-017-0422-2
Hay, W. T., Anderson, J. A., McCormick, S. P., Hojilla-Evangelista, M. P., Selling, G. W., Utt, K. D., et al. (2022). Fusarium head blight resistance exacerbates nutritional loss of wheat grain at elevated CO2. Sci. Rep. 12, 15. doi: 10.1038/s41598-021-03890-9
He, Y., Wu, L., Liu, X., Jiang, P., Yu, L., Qiu, J., et al. (2020). TaUGT6, a novel UDP-glycosyltransferase gene enhances the resistance to FHB and DON accumulation in wheat. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.574775
Hoffmann, N., Benske, A., Betz, H., Schuetz, M., Samuels, A. L. (2020). Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 184 (2), 806–822. doi: 10.1104/pp.20.00473
Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., et al. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16 (6), 1446–1465. doi: 10.1105/tpc.020297
Hu, X., Rocheleau, H., McCartney, C., Biselli, C., Bagnaresi, P., Balcerzak, M., et al. (2019). Identification and mapping of expressed genes associated with the 2DL QTL for fusarium head blight resistance in the wheat line Wuhan 1. BMC Genet. 20 (1), 47. doi: 10.1186/s12863-019-0748-6
Huang, Y., Haas, M., Heinen, S., Steffenson, B. J., Smith, K. P., Muehlbauer, G. J. (2018). QTL mapping of Fusarium Head Blight and correlated agromorphological traits in an elite barley cultivar Rasmusson. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01260
Kage, U., Karre, S., Kushalappa, A. C., McCartney, C. (2017). Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol. J. 15 (4), 447–457. doi: 10.1111/pbi.12641
Kang, Z., Buchenauer, H. (2000). Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiol. Mol. Plant Pathol. 57 (6), 255–268. doi: 10.1006/pmpp.2000.0305
Kapoor, D., Singh, S., Kumar, V., Romero, R., Prasad, R., Singh, J. (2019). Antioxidant enzymes regulation in plants in reference to reactive oxygen species (ROS) and reactive nitrogen species (RNS). Plant Gene 19, 100182. doi: 10.1016/j.plgene.2019.100182
Khaledi, N., Taheri, P., Falahati-Rastegar, M. (2016). Reactive oxygen species and antioxidant system responses in wheat cultivars during interaction with Fusarium species. Australas. Plant Pathol. 45, 653–670. doi: 10.1007/s13313-016-0455-y
Kikot, G. E., Hours, R. A., Alconada, T. M. (2009). Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: a review. J. Basic Microbiol. 49 (3), 231–241. doi: 10.1002/jobm.200800231
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Kosaka, A., Manickavelu, A., Kajihara, D., Nakagawa, H., Ban, T. (2015). Altered gene expression profiles of wheat genotypes against Fusarium head blight. Toxins (Basel) 7 (2), 604–620. doi: 10.3390/toxins7020604
Kumar, J., Rai, K. M., Pirseyedi, S., Elias, E. M., Xu, S., Dill-Macky, R., et al. (2020). Epigenetic regulation of gene expression improves Fusarium head blight resistance in durum wheat. Sci. Rep. 10, 17610. doi: 10.1038/s41598-020-73521-2
Kumar, A., Yogendra, K. N., Karre, S., Kushalappa, A. C., Dion, Y., Choo, T. M. (2016). WAX INDUCER1 (HvWIN1) transcription factor regulates free fatty acid biosynthetic genes to reinforce cuticle to resist Fusarium head blight in barley spikelets. J. Exp. Bot. 67 (14), 4127–4139. doi: 10.1093/jxb/erw187
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinf. (Oxford England) 27 (21), 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, S. (2014). Redox modulation matters: emerging functions for glutaredoxins in plant development and stress responses. Plants (Basel) 3 (4), 559–582. doi: 10.3390/plants3040559
Li, X., Michlmayr, H., Schweiger, W., Malachova, A., Shin, S., Huang, Y., et al. (2017). UDP-glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 68 (9), 2187–2197. doi: 10.1093/jxb/erx109
Liang, N., Charron, J. B., Jabaji, S. (2023). Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis E68 against Fusarium graminearum DAOMC 180378, the causal agent of Fusarium head blight. PloS One 18 (1), e0277983. doi: 10.1371/journal.pone.0277983
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi: 10.1093/bioinformatics/btt656
Liu, J. J., Sturrock, R., Ekramoddoullah, A. K. (2010). The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 29 (5), 419–436. doi: 10.1007/s00299-010-0826-8
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. doi: 10.1186/s13059-014-0550-8
Lysøe, E., Seong, K. Y., Kistler, H. C. (2011). The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact. 24 (9), 995–1000. doi: 10.1094/MPMI-02-11-0038
Mandalà, G., Ceoloni, C., Busato, I., Favaron, F., Tundo, S. (2021). Transgene pyramiding in wheat: Combination of deoxynivalenol detoxification with inhibition of cell wall degrading enzymes to contrast Fusarium Head Blight and Crown Rot. Plant Sci. 313, 111059. doi: 10.1016/j.plantsci.2021.111059
Matić, M., Vuković, R., Vrandečić, K., Štolfa Čamagajevac, I., Ćosić, J., Vuković, A. (2021). Oxidative status and antioxidative response to Fusarium attack and different nitrogen levels in winter wheat varieties. Plants (Basel) 10 (4), 611. doi: 10.3390/plants10040611
Mayer, K. F., Waugh, R., Brown, J. W., Schulman, A., Langridge, P., Platzer, M., et al. (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491 (7426), 711–716. doi: 10.1038/nature11543
McCartney, C. A., Brûlé-Babel, A. L., Fedak, G., Martin, R. A., McCallum, B. D., Gilbert, J., et al. (2016). Fusarium head blight resistance QTL in the spring wheat cross Kenyon/86ISMN 2137. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01542
Mendes, G. R. L., Alves, C. L., Lopes Cavalheiro, P., Bretanha, C. C., Pagnussatt, F. A., Badiale-Furlong, E. (2015). α-amylase inhibitors from wheat against development and toxigenic potential of Fusarium verticillioides. CCHEM 92, 611–616. doi: 10.1094/CCHEM-11-14-0227-R
Menke, J., Dong, Y., Kistler, H. C. (2012). Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol. Plant Microbe Interact. 25 (11), 1408–1418. doi: 10.1094/MPMI-04-12-0081-R
Montibus, M., Ducos, C., Bonnin-Verdal, M. N., Bormann, J., Ponts, N., Richard-Forget, F., et al. (2013). The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum. PloS One 8 (12), e83377. doi: 10.1371/journal.pone.0083377
Montibus, M., Khosravi, C., Zehraoui, E., Verdal-Bonnin, M. N., Richard-Forget, F., Barreau, C. (2016). Is the Fgap1 mediated response to oxidative stress chemotype dependent in Fusarium graminearum? FEMS Microbiol. Lett. 363 (2), fnv232. doi: 10.1093/femsle/fnv232
Naguib, D. M. (2018). Control of Fusarium wilt in wheat seedlings by grain priming with defensin-like protein. Egypt J. Biol. Pest Control. 28, 68. doi: 10.1186/s41938-018-0073-9
Nguyen, T. N., Son, S., Jordan, M. C., Levin, D. B., Ayele, B. T. (2016). Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biol. 16, 28. doi: 10.1186/s12870-016-0717-4
Nilsen, K. T., Walkowiak, S., Kumar, S., Molina, O. I., Randhawa, H. S., Dhariwal, R., et al. (2021). Histology and RNA sequencing provide insights into Fusarium Head Blight resistance in AAC Tenacious. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.570418
Oh, M., Son, H., Choi, G. J., Lee, C., Kim, J. C., Kim, H., et al. (2016). Transcription factor ART1 mediates starch hydrolysis and mycotoxin production in Fusarium graminearum and F. verticillioides. Mol. Plant Pathol. 17 (5), 755–768. doi: 10.1111/mpp.12328
Oliveros, J. C. (2007). Venny. An interactive tool for comparing lists with venn’s diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
O’Mara, S. P., Broz, K., Dong, Y. H., Kistler, C. (2023). The Fusarium graminearum transporters Abc1 and ABC6 are important for xenobiotic resistance, trichothecene accumulation, and virulence to wheat. Phytopathol. 113 (10), 1916–1923. doi: 10.1101/2021.06.15.448535
Pan, Y., Liu, Z., Rocheleau, H., Fauteux, F., Wang, Y., McCartney, C., et al. (2018). Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics 19, 642. doi: 10.1186/s12864-018-5012-3
Pasquet, J. C., Changenet, V., Macadré, C., Boex-Fontvieille, E., Soulhat, C., Bouchabké-Coussa, O., et al. (2016). UDP-glycosyltransferase confers root tolerance to deoxynivalenol and resistance to Fusarium infection. Plant Physiol. 172 (1), 559–574. doi: 10.1104/pp.16.00371
Perlikowski, D., Wiśniewska, H., Góral, T., Kwiatek, M., Majka, M., Kosmala, A. (2014). Identification of kernel proteins associated with the resistance to fusarium head blight in winter wheat (Triticum aestivum L.). PloS One 9 (10), e110822. doi: 10.1371/journal.pone.0110822
Polturak, G., Dippe, M., Stephenson, M. J., Misra, C. R., Owen, C., Ramirez-Gonzalez, R. H., et al. (2022). Pathogen-induced biosynthetic pathways encode defense-related molecules in bread wheat. Proc. Natl. Acad. Sci. 119 (16), e2123299119. doi: 10.1073/pnas.2123299119
Ponts, N., Pinson-Gadais, L., Barreau, C., Richard-Forget, F., Ouellet, T. (2007). Exogenous H(2)O(2) and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 581 (3), 443–447. doi: 10.1016/j.febslet.2007.01.003
Ren, R., Zhou, X., Zhang, X., Li, X., Zhang, P., He, Y. (2022). Genome-wide identification and characterization of thaumatin-like protein family genes in wheat and analysis of their responses to Fusarium head blight infection. Food Prod. Process Nutr. 4, 24. doi: 10.1186/s43014-022-00105-0
Rocher, F., Alouane, T., Philippe, G., Martin, M. L., Label, P., Langin, T., et al. (2022). Fusarium graminearum infection strategy in wheat involves a highly conserved genetic program that controls the expression of a core effectome. Int. J. Mol. Sci. 23 (3), 1914. doi: 10.3390/ijms23031914
Rossini, B. C., Bouvet, J. M., Frouin, J., Guerrini, I. A., de Freitas, T. C. M., da Silva, M. R., et al. (2022). Differential gene expression in Eucalyptus clones in response to nutrient deficiency. Tree Genet. Genomes 18, 14. doi: 10.1007/s11295-022-01546-x
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Sci. (New York N.Y.) 326 (5956), 1112–1115. doi: 10.1126/science.1178534
Serrano, M., Coluccia, F., Torres, M., L’Haridon, F., Métraux, J. P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00274
Shah, D. A., De Wolf, E. D., Paul, P. A., Madden, L. V. (2014). Predicting Fusarium head blight epidemics with boosted regression trees. Phytopathology 104 (7), 702–714. doi: 10.1094/PHYTO-10-13-0273-R
Shin, S., Kim, K. H., Kang, C. S., Cho, K. M., Park, C. S., Okagaki, R., et al. (2014). A simple method for the assessment of Fusarium Head Blight resistance in Korean wheat seedlings inoculated with Fusarium graminearum. Plant Pathol. J. 30 (1), 25–32. doi: 10.5423/PPJ.OA.06.2013.0059
Soni, N., Altartouri, B., Hegde, N., Duggavathi, R., Nazarian-Firouzabadi, F., Kushalappa, A. C. (2021). TaNAC032 transcription factor regulates lignin-biosynthetic genes to combat Fusarium head blight in wheat. Plant Sci. 304, 110820. doi: 10.1016/j.plantsci.2021.110820
Steiner, B., Buerstmayr, M., Wagner, C., Danler, A., Eshonkulov, B., Ehn, M., et al. (2019). Fine-mapping of the Fusarium head blight resistance QTL Qfhs.ifa-5A identifies two resistance QTL associated with anther extrusion. TAG. Theor. Appl. Genet. Theoretische und angewandte Genetik 132 (7), 2039–2053. doi: 10.1007/s00122-019-03336-x
Su, Z., Bernardo, A., Tian, B., Chen, H., Wang, S., Ma, H., et al. (2019). A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51 (7), 1099–1105. doi: 10.1038/s41588-019-0425-8
Sun, Z., Zhou, Y., Hu, Y., Jiang, N., Hu, S., Li, L., et al. (2022). Identification of wheat LACCASEs in response to Fusarium graminearum as potential Deoxynivalenol trappers. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.832800
Suzuki, R., Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22 (12), 1540–1542. doi: 10.1093/bioinformatics/btl117
Tamburic-Ilincic, L., Falk, D., Schaafsma, A. (2011). Fusarium ratings in Ontario winter wheat performance trial (OWWPT) using an index that combines Fusarium head blight symptoms and deoxynivalenol levels. Czech J. Genet. Plant Breed. 47, 115–122. doi: 10.17221/3265-CJGPB
Tian, Y., Tan, Y., Liu, N., Yan, Z., Liao, Y., Chen, J., et al. (2016). Detoxification of deoxynivalenol via glycosylation represents novel insights on antagonistic activities of Trichoderma when confronted with Fusarium graminearum. Toxins (Basel) 8 (11), 335. doi: 10.3390/toxins8110335
Torres, A. M., Palacios, S. A., Yerkovich, N., Palazzini, J. M., Battilani, P., Leslie, J. F. (2019). Fusarium head blight and mycotoxins in wheat: prevention and control strategies across the food chain. World Mycotoxin J. 12, 4, 333–4, 355. doi: 10.3920/WMJ2019.2438
Tyagi, S., Himani, Sembi, J. K., Upadhyay, S. K. (2018). Gene architecture and expression analyses provide insights into the role of glutathione peroxidases (GPXs) in bread wheat (Triticum aestivum L.). J. Plant Physiol. 223, 19–31. doi: 10.1016/j.jplph.2018.02.006
Villafana, R. T., Ramdass, A. C., Rampersad, S. N. (2019). Selection of Fusarium trichothecene toxin genes for molecular detection depends on tri gene cluster organization and gene function. Toxins 11 (1), 36. doi: 10.3390/toxins11010036
Waśkiewicz, A., Morkunas, I., Bednarski, W., Mai, V. C., Formela, M., Beszterda, M., et al. (2014). Deoxynivalenol and oxidative stress indicators in winter wheat inoculated with Fusarium graminearum. Toxins (Basel) 6 (2), 575–591. doi: 10.3390/toxins6020575
Wan, J., Zhang, X. C., Stacey, G. (2008). Chitin signaling and plant disease resistance. Plant Signaling Behav. 3 (10), 831–833. doi: 10.4161/psb.3.10.5916
Wang, Y., Chantreau, M., Sibout, R., Hawkins, S. (2013). Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00220
Wang, W., Liang, X., Li, Y., Wang, P., Keller, N. P. (2023). Genetic regulation of mycotoxin biosynthesis. J Fungi (Basel) 9 (1), 21. doi: 10.3390/jof9010021
Wang, H., Sun, S., Ge, W., Zhao, L., Hou, B., Wang, K., et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368 (6493), eaba5435. doi: 10.1126/science.aba5435
Wu, Q., Wang, X., Nepovimova, E., Wang, Y., Yang, H., Li, L., et al. (2017). Antioxidant agents against trichothecenes: new hints for oxidative stress treatment. Oncotarget 8 (66), 110708–110726. doi: 10.18632/oncotarget.22800
Xu, Q., Xu, F., Qin, D., Li, M., Fedak, G., Cao, W., et al. (2020). Molecular mapping of QTLs conferring Fusarium Head Blight resistance in chinese wheat cultivar Jingzhou 66. Plants (Basel) 9 (8), 1021. doi: 10.3390/plants9081021
Yang, G., Pan, W., Zhang, R., Pan, Y., Guo, Q., Song, W., et al. (2021). Genome-wide identification and characterization of caffeoyl-coenzyme A O-methyltransferase genes related to the Fusarium head blight response in wheat. BMC Genomics 22, 504. doi: 10.1186/s12864-021-07849-y
Yi, X., Cheng, J., Jiang, Z., Hu, W., Bie, T., Gao, D., et al. (2018). Genetic analysis of Fusarium Head Blight resistance in CIMMYT bread wheat line C615 using traditional and conditional QTL mapping. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00573
Zechmann, B. (2020). Subcellular roles of glutathione in mediating plant defense during biotic stress. Plants (Basel Switzerland) 9 (9), 1067. doi: 10.3390/plants9091067
Zhang, J., Gill, H. S., Halder, J., Brar, N. K., Ali, S., Bernardo, A., et al. (2022). Multi-locus genome-wide association studies to characterize fusarium head blight (FHB) resistance in hard winter wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946700
Zhang, Y., Yang, Z., Ma, H., Huang, L., Ding, F., Du, Y., et al. (2021). Pyramiding of Fusarium Head Blight resistance quantitative trait loci, Fhb1, Fhb4, and Fhb5, in modern chinese wheat cultivars. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.694023
Zhao, F., Niu, K., Tian, X., Du, W. (2022). Triticale improvement: mining of genes related to yellow rust resistance in triticale based on transcriptome sequencing. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.883147
Keywords: AC Emerson, AC Morley, dual RNA-sequencing, F. graminearum, FHB, T. aestivum, transcriptomics, resistance
Citation: Walker PL, Belmonte MF, McCallum BD, McCartney CA, Randhawa HS and Henriquez MA (2024) Dual RNA-sequencing of Fusarium head blight resistance in winter wheat. Front. Plant Sci. 14:1299461. doi: 10.3389/fpls.2023.1299461
Received: 22 September 2023; Accepted: 29 November 2023;
Published: 04 January 2024.
Edited by:
Thiruvarangan Ramaraj, DePaul University, United StatesReviewed by:
Ajay Kumar Thakur, Central Potato Research Institute (ICAR), IndiaCopyright © 2024 Mark F. Belmonte, Curt A. McCartney and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Philip L. Walker, Brent D. McCallum, Harpinder S. Randhawa and Maria A. Henriquez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria A. Henriquez, TWFyaWFBbnRvbmlhLkhlbnJpcXVlekBhZ3IuZ2MuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.