- 1College of Bioengineering, Sichuan University of Science and Engineering, Yibin, China
- 2Institute of Molecular Physiology and Biotechnology of Plants (IMBIO), University of Bonn, Bonn, Germany
- 3Key Laboratory of Horticultural Plant Biology, Ministry of Education, Key Laboratory of Urban Agriculture in Central China, Ministry of Agriculture, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, China
- 4College of Life Science and Technology, Hubei Engineering University, Xiaogan, China
The plant homeodomain (PHD) finger with a conserved Cys4-His-Cys3 motif is a common zinc-binding domain, which is widely present in all eukaryotic genomes. The PHD finger is the “reader” domain of methylation marks in histone H3 and plays a role in the regulation of gene expression patterns. Numerous proteins containing the PHD finger have been found in plants. In this review, we summarize the functional studies on PHD finger proteins in plant growth and development and responses to abiotic stresses in recent years. Some PHD finger proteins, such as VIN3, VILs, and Ehd3, are involved in the regulation of flowering time, while some PHD finger proteins participate in the pollen development, for example, MS, TIP3, and MMD1. Furthermore, other PHD finger proteins regulate the plant tolerance to abiotic stresses, including Alfin1, ALs, and AtSIZ1. Research suggests that PHD finger proteins, as an essential transcription regulator family, play critical roles in various plant biological processes, which is helpful in understanding the molecular mechanisms of novel PHD finger proteins to perform specific function.
Introduction
The plant homeodomain (PHD) finger is a common zinc-binding domain that existed in all eukaryotic genomes (Bienz, 2006). The PHD finger is usually comprised of 50–80 amino acids and typically has a conserved Cys4-His-Cys3 motif containing insertion sequences with various length and composition in the domain (Aasland et al., 1995; Bienz, 2006). The PHD finger exhibits high sequence similarity to the RING finger (Cys3-His-Cys4 type), which also binds to two zinc atoms (Capili et al., 2001). Previously, some RING fingers were mistaken for PHD fingers, leading to the incorrect conclusion that PHD fingers were also involved in E3 ligase activity (Wei et al., 2015). In general, the three-dimensional structure of the PHD finger shows a globular fold, consisting of an alpha-helix and a two-stranded beta-sheet.
Histone acetyltransferases (HATs) catalyze histone acetylation and play an important role in the positive epigenetic regulation of gene expression in eukaryotes (Zhang et al., 2023). Ever since Arabidopsis HAT3.1 was identified as the first PHD finger protein (Schindler et al., 1993), many PHD finger proteins have been found in fungus, animals, and plants (Martin et al., 2006; Baker et al., 2008; Hu et al., 2018). Most of these proteins are localized in the nucleus (Aasland et al., 1995; Gilbert et al., 2014; Shu et al., 2015), while a few are predicted to be localized in the membrane, including the chloroplast thylakoid membrane (Glyma10g05080.1) and mitochondrial inner membrane (Glyma11g11720.1) (Wu and Wang, 2014). A typical PHD finger protein usually contains one or more PHD finger domains. As an independent structure unit, most PHD finger-containing proteins only have PHD finger domain (Wu and Wang, 2014). However, there are other conserved domains in a certain protein concomitant with PHD finger domain, such as DUF3594 (Domain of Unknown Function 3594), BAH (Bromo Adjacent Homology), and DDT (DNA binding homeobox and different transcription factors) domain (Sun et al., 2017). These various domains individually cooperate with PHD finger domain to play a role in a particular biological event (Wu and Wang, 2014). The Alfin1 group belongs to a plant-specific subfamily of PHD finger proteins. Alfin1 from alfalfa (Medicago sativa) is a salt-induced transcription factor and can efficiently bind to the G-rich elements (GNGGTG/GTGGNG) in the promoter region of MsPRP2, a stress-responsive gene (Bastola et al., 1998). Overexpression of Alfin1 in alfalfa increased the transcript of MsPRP2 in roots and enhanced the tolerance of transgenic plants to salt stress (Winicov et al., 2004). So far, an increasing number of Alfin1-like (AL) proteins have been identified and characterized in various plant species, such as Arabidopsis thaliana (Wei et al., 2015), Brassica rapa (Kayum et al., 2015), and Atriplex hortensis (Tao et al., 2018). Except the conserved PHD finger domain in C-terminal, all AL proteins contain the conserved DUF3594 domain in N-terminal with unknown function. AL proteins containing the DUF3594 domain have not been found in animals, fungi, or prokaryotes (Tao et al., 2018).
PHD finger domains are proved to be involved in protein–DNA and protein–protein interaction (Bastola et al., 1998; Wei et al., 2017). The N-terminal tails of nucleosome core histones (H2A, HAB, H3, and H4) are usually modified by methylation or acetylation, which is called post-translational modification in histone proteins (PTM) (Lee et al., 2010; Yang et al., 2013; Sun et al., 2014). The PHD finger is a methyllysine and methylarginine “reader” domain, which can specifically recognize and bind to methylation marks in histone H3 (Sanchez and Zhou, 2011; Milosevich and Hof, 2016; Miura et al., 2020). Lee et al. (2009) first identified that PHD fingers in ING (inhibitor of growth) homologues AtING and AL proteins are able to bind histone H3 at di- or tri-methylated lysine4 (H3K4me2/me3) in Arabidopsis. Additionally, many PHD finger proteins are proved to be involved in chromatin remodeling and have transcriptional regulation activity (Bienz, 2006; Wei et al., 2015; Milosevich and Hof, 2016; Diego-Martin et al., 2022). Chromatin remodeling processes function in the control of gene expression patterns that modulate development in eukaryotic organisms (López-González et al., 2014). Thus, these “reader” proteins are essential for recruiting chromatin remodeling complexes and transcription factors to target loci and regulating their transcriptional status (López-González et al., 2014; Milosevich and Hof, 2016). In this way, the PHD finger proteins play important roles in translating histone modifications into downstream gene expression patterns.
PHD finger proteins function in various biological processes because of high sequence diversity except the eight conserved cysteine/histidine residues (Wei et al., 2015). In plants, diverse functions of PHD finger proteins have been characterized, which are involved in different biological processes, including the regulation of seed dormancy and germination (Ye et al., 2016), vernalization response (Sung and Amasino, 2004; Kim and Sung, 2013), flowering time (Qian et al., 2021), and pollen development (Yang et al., 2019a; Yang et al., 2019b). Furthermore, many genes encoding PHD finger proteins can be induced by environmental stresses and participate in plant abiotic stress responses (Wei et al., 2009; Gao et al., 2018; Alam et al., 2019; Pang et al., 2022). In this review, we aim to summarize and analyze the functions of PHD finger proteins in plants, particularly in plant reproduction development and responses to abiotic stresses. This will provide useful information for studying novel PHD finger proteins and further exploring the molecular mechanisms of these proteins involved in specific biological events.
Roles of PHD finger proteins in plant growth and development
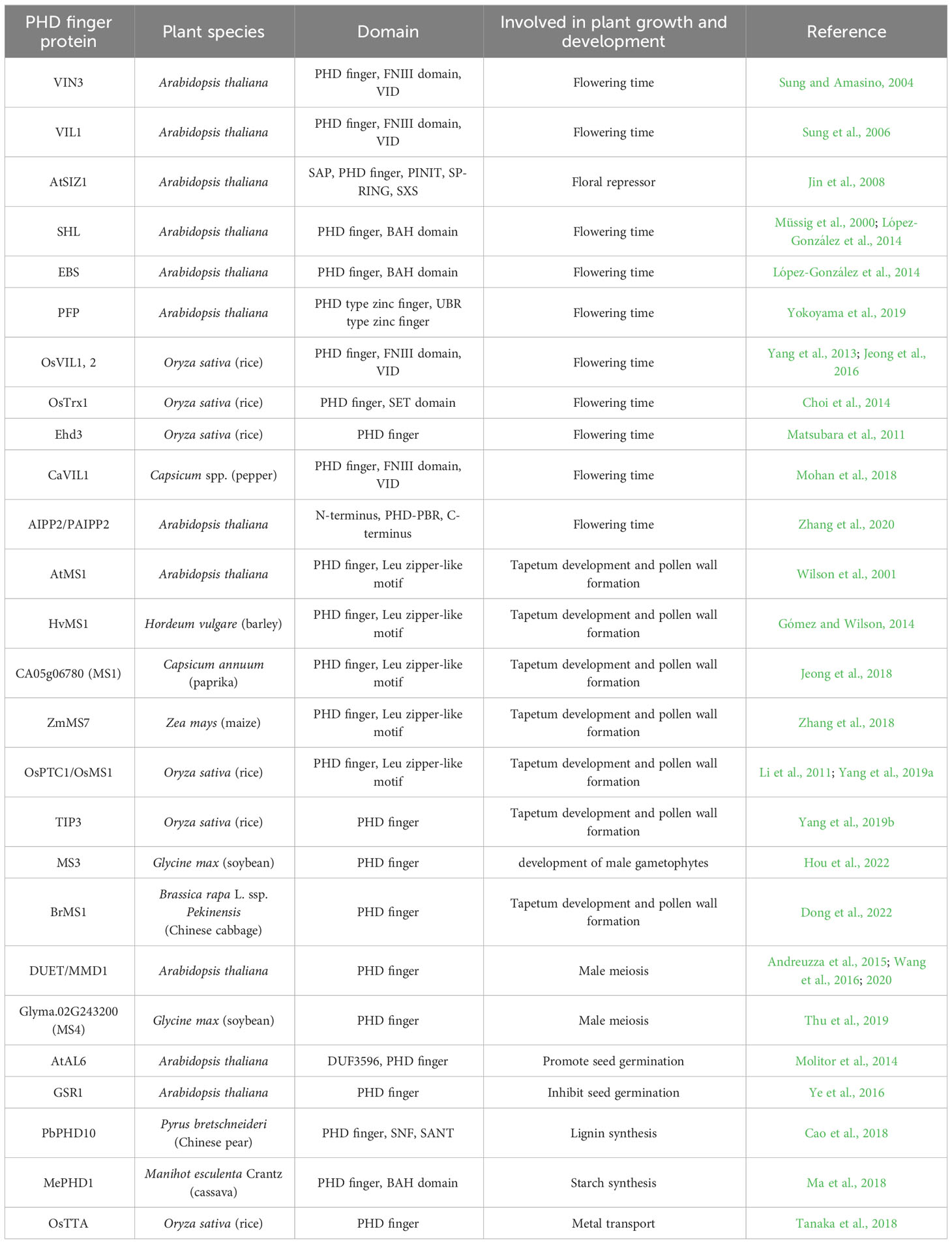
The processes of plant growth and development play vital roles in plant reproduction and the completion of its life cycle. Previous studies showed that lots of PHD finger proteins are involved in these biological events, such as the regulation of flowering time, pollen development, seed germination, metabolite biosynthesis, and metal transport (Table 1).
Flowering time
In plant species, the timing of the floral transition is a key developmental switch for the successful propagation. Flowering time is complexly controlled by genetic networks, epigenetic modifications, and other regulatory mechanisms (Khan et al., 2014; Sun et al., 2014). There exist different genetic pathways involved in the induction of flowering in Arabidopsis, such as the vernalizaiton pathway, the photoperiod pathway, and the gibberellin pathway (Khan et al., 2014). The expression of floral integrator genes, including FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), is finely controlled by these floral promoting pathways and floral repressors and then triggers floral initiation under proper conditions (Jarillo and Piñeiro, 2011).
As a floral repressor, FLOWERING LOCUS C (FLC) partly prevents flowering by repressing the expression of floral integrators in the first growing season for biennials and winter-annuals, while vernalization is necessary to promote flowering primarily by repressing FLC expression in the second growing season (Michaels, 2009). In Arabidopsis, VERNALIZATION INSENSITIVE 3 (VIN3) is a chromatin remodeling PHD finger protein and is required to repress FLC by promoting histone H3 deacetylation and increasing H3K9 and H3K27 methylation during vernalization (Sung and Amasino, 2004). VIN3-LIKE (VIL) proteins belong to VIN3 gene family, containing the PHD finger domain, the fibronectin type-III (FNIII) domain, and the VIN3-interacting domain (VID). The PHD finger recognizes histone H3, while VID is responsible for the interaction between VIL proteins (Sung et al., 2006; Jeong et al., 2016). AtVIL1-4 and TmVIL1-3 genes are identified in Arabidopsis and wheat (Triticum monococcum), respectively (Fu et al., 2007). These VIL proteins play a crucial role in the flowering process regulated by vernalization and photoperiod pathways. For example, AtVIL1 cooperates with VIN3 in the chromatin modifications of FLC and FLOWERING LOCUS M (FLM, an FLC-related floral repressor) during vernalization. Prolonged cold treatment induces VIN3 expression; however, the expression of VIL1 is temperature independent and is highly upregulated in short days (SD). Indeed, VIL1 promotes flowering in SD through the VIL1-mediated repression of FLM independent of VIN3. Thus, VIL1 involves in both the vernalization and photoperiod pathways by regulating expression of two floral repressors FLC and FLM (Sung et al., 2006). Wheat VIL genes are upregulated by vernalization and also affected by photoperiod (Fu et al., 2007). However, CaVIL1 is an ortholog of Arabidopsis VIL1 and functions as a flowering promoter in pepper (Capsicum spp.), which is insensitive to vernalization and photoperiod (Mohan et al., 2018). Vernalization is not required for flowering induction in rice (Oryza sativa), which contains four VIL genes (Fu et al., 2007). Among them, OsVIL2 physically associates with EMBRYONIC FLOWER 2b (OsEMF2b), which is a component of Polycomb Repressive Complex 2 (PRC2) with histone methyltransferase (HMTase) activity. The complex of OsVIL2-OsEMF2 induces flowering through epigenetic silencing of the flowering repressor LEAFY COTYLEDON 2 and FUSCA 3-LIKE 1 (OsLFL1) with enriched H3K27me3 under both SD and long days (LD) (Yang et al., 2013). In addition, OsVIL2 interacts with OsVIL1, which is also associated with OsEMF2b to form a PRC2-like complex. Overexpressing of OsVIL1 promotes flowering by reducing the transcripts of the flowering repressor OsLF, a bHLH transcription factor under SD, while it delays flowering by increasing expression of the flowering repressor Grain number, plant height, and heading date 7 (Ghd7) under LD (Jeong et al., 2016).
In addition to VIL gene family, two Arabidopsis paralogs SHORT LIFE (SHL) and EARLY BOLTING IN SHORT DAYS (EBS), belonging to plant-specific transcriptional regulators with a PHD finger domain, function independently in the control of genes modulating flowering. PHD domains presented in SHL and EBS as chromatin effectors recognize H3K4me2/3 and bind to regulatory regions of the floral integrator genes SOC1 and FT, respectively. Moreover, SHL and EBS are necessary to maintain the chromatin of SOC1 and FT in an inactive conformation with low levels of H3 acetylation. These PHD finger proteins are proved to bind HISTONE DEACETYLASE 6 (HDA6) and play important roles in the chromatin-mediated repression of flowering, ensuring the precise control of flowering time (Müssig et al., 2000; López-González et al., 2014). Zhang et al. (2020) reported that the anti-silencing 1 (ASI1)-IMMUNOPRECIPITATED PROTEIN 2 (AIPP2) and PARALOG OF AIPP2 (PAIPP2) could independently interact with BAH domain-containing protein AIPP3 and PolII terminal domain (CTD) phosphatase (CLP2), respectively, through their PHD domain and C-terminus, to form the BAH-PHD-CLP2 (BPC) protein complex. The BPC complex combines the recognition of H3K27me3 and the repression of PolII release to repress the expression of FT in Arabidopsis to delay flowering (Zhang et al., 2020). Furthermore, it was confirmed that six PHD finger proteins in Arabidopsis can enhance the binding of BAH domain-containing transcriptional regulator 1 (BDT1) to the H3K27me3, which is essential for the prevention of early-flowering phenotype (Qian et al., 2021). An Arabidopsis PHD finger protein homolog, PFP (PHD finger domain containing protein), is critical for the flowering repression by upregulating expression of FLC and downregulating FT (Yokoyama et al., 2019). Some proteins only have the PHD finger domain, such as Early heading date 3 (Ehd3) in rice with encoding a nuclear protein containing two PHD finger motifs. As an LD preferential regulator, Ehd3 acts as a repressor upstream of Ghd7 and promotes flowering under LD (Matsubara et al., 2011). It has been reported that Ehd3 associates with Trithorax 1 (OsTrx1), which carries a PHD finger motif and a SET domain with HMTase activity. The suppression of OsTrx1 increases the transcripts of Ghd7 and delays flowering time only under LD conditions (Choi et al., 2014).
Pollen development
Male gametogenesis has important commercial significance for controlling the fertility of crops (Wilson et al., 2001). Microspore mother cells form tetrads after meiosis, and microspores with single nucleus are released from the tetrad. After nuclear fission, the microspores produce mature pollen grains. Acting as the innermost somatic cell layer of the anther locule, the tapetum plays a key role in pollen development (Ito and Shinozaki, 2002). Arabidopsis MALE STERILITY1 (MS1) functions as a transcriptional activator containing Leu zipper-like and PHD finger motifs, which are required for its function (Ito et al., 2007). The MS1 gene is specifically expressed in the sporophytic tapetum for a short time and regulates the development of pollen exine and pollen cytosol and tapetum. The ms1 mutant is male sterile and produces immature pollen with abnormal exine and tapetum (Wilson et al., 2001; Ito and Shinozaki, 2002; Ito et al., 2007).
Based on the information from pollen regulatory gene networks in Arabidopsis, several orthologs of AtMS1 have been identified and functionally characterized in various species, such as PERSISTENT TAPETAL CELL1/OsMS1 (OsPTC1/OsMS1) in rice (Li et al., 2011; Yang et al., 2019a), HvMS1 in barley (Hordeum vulgare) (Gómez and Wilson, 2014), ZmMS7 in maize (Zea mays) (Zhang et al., 2018), and BrMS1 in Chinese cabbage (Brassica rapa L. ssp. pekinensis) (Dong et al., 2022). Using MutMap combined with KASP analysis, Dong et al. (2022) screened out a homologous gene of AtMS1, BrMS1, which plays a transcriptional regulatory role in tapetal programmed cell death (PCD) and pollen wall development. ZmMS7, encoding a PHD finger transcription factor in maize, shows 80.9% and 40.5% amino acid sequence identities with OsPTC1/OsMS1 and AtMS1, respectively (Zhang et al., 2018). Mutation or overexpression of a barley ortholog of AtMS1, HvMS1 results in male sterility. Under control of the native AtMS1 promoter, HvMS1 cDNA successfully complements the Arabidopsis ms1 mutant, which demonstrates the conservation of MS1 function in higher plants (Gómez and Wilson, 2014). Compared to the Arabidopsis ms1 mutant, uncontrolled tapetal proliferation and subsequent necrosis-like tapetal death are uniquely displayed in the rice ptc1 mutant (a single nucleotide insertion in the second exon of LOC_Os09g27620) (Li et al., 2011). Another research reported that the rice osms1 mutant (four nucleotide deletion in the first exon of LOC_Os09g27620) shows significantly reduced transcripts of the genes related to tapetal PCD and pollen wall biosynthesis, including AP25, AP37, EAT1, OsC4, and OsC6. OsMS1 interacts with TDR INTERACTING PROTEIN2 (TIP2), a basic helix–loop–helix (bHLH) transcription factor, and OsMADS15, which are essential for sexual reproduction, through the PHD finger domain to regulate the tapetal PCD and pollen wall formation in rice (Yang et al., 2019a). Subsequently, Yang et al. (2019b) found that TDR INTERACTING PROTEIN3 (TIP3) in rice encodes a PHD finger protein with the transcriptional activation activity. During another development, with the preferential accumulation in tapetum and microspores, TIP3 protein directly interacts with TDR, a bHLH transcription factor, which plays critical roles in the regulation of tapetum development and pollen wall formation. The loss of TIP3 alters the transcript level of genes involved in tapetal PCD, biosynthesis, and transport of sporopollenin precursors, resulting in delayed tapetum degradation and no pollen wall formation in tip3 mutant (Yang et al., 2019b).
Meiosis plays an important role in sexual reproduction, which produces haploid daughter cells essential for maintaining hybrid traits. This process involves two meiotic cell divisions, meiosis I and meiosis II, and each of both is divided into four stages, namely, prophase, metaphase, anaphase, and telophase. During meiosis, a complex series of biological events take place, including chromosome condensation, homologous chromosome recombination and segregation, and sister chromatid separation. The successful completion of meiotic events is necessary to form normal gametes. In Arabidopsis, DUET is also known as MALE MEIOCYTE DEATH1 (MMD1), which encodes a nuclear protein containing a PHD finger and plays important roles in male meiosis. DUET/MMD1 is specifically expressed in male meiocytes, coinciding with the time of meiosis (Reddy et al., 2003; Yang et al., 2003). Yang et al. (2003) showed that the mmd1 mutant displays chromosome fragmentation in meiosis resulting in cell death of male meiocytes. Meanwhile, Reddy et al. (2003) indicated that the loss of DUET negatively affects chromosome condensation and male meiotic progression, leading to the formation of abnormal meiotic products. It was showed that DEUT/MMD1 binds to H3K4me2 in vitro and/or in vivo through the PHD finger domain, which is important for its functions in male meiosis (Andreuzza et al., 2015; Wang et al., 2016). Acting as a transcriptional regulator, DUET is specifically required for the expression of the meiotic gene JASON (JAS) and THREE DIVISION MUTANT 1 (TDM1) critical for spindle organization during meiosis II and cell cycle exit after the second meiosis, respectively. Therefore, DUET functions in the regulation of microtubule organization and cell cycle transitions (Andreuzza et al., 2015). Recently, Liu et al. (2021) found that an Arabidopsis mutant male meiotic restitution 1 (mmr1) is produced by an amino acid change G618D in the PHD finer domain caused by base conversion (C to T) at the third exon of MMD1/DUET gene. The hypomorphic mutant is deficient in spindle organization and mini-phragmoplast formation (Liu et al., 2021).
In addition, MMD1 is necessary to regulate the progression of chromosome condensation during meiotic prophase I. MMD1 PHD finger might directly bind to H3K4me2/3 at the CAP-D3 promoter region to active the expression of CAP-D3, which is a condensin subunit gene belonging to the condensin complex required for chromosome condensation (Wang et al., 2016). Meanwhile, the MMD domain (a conserved domain in plants) of MMD1 interacts with the C-terminal FYR domain of Jumonji C (JmjC)-containing demethylase JMJ16 to broaden the substrate specificity of JMJ16 by binding the H3K9me3 in male meiocytes. Therefore, the interaction between MMD1 and JMJ16 demethylates H3K9 of target genes, for example, CAP-D3, and promotes gene expression, facilitating meiotic chromosome condensation (Wang et al., 2020). In soybean (Glycine max), Glyma.02G243200 is isolated from one male-sterile, female-fertile mutant line (ms4) and named as MS4 protein, which is a homolog of AtMMD1. The Arabidopsis mmd1 mutant with the soybean MS4 gene restores successful tetrad formation and normal stamens and produces fertile pollen and viable seeds (Thu et al., 2019).
Seed germination
Seed germination is a key developmental event that involves the timely transition from stagnant seeds to growing seedlings, which is vital for entering the plant life cycle. During this process, the expression of seed developmental genes needs to be gradually suppressed, such as ABSCISIC ACID INSENSITIVE 3 (ABI3), DELAY OF GERMINATION 1 (DOG1), and CRUCIFERIN 3 (CRU3), to facilitate seedling growth in Arabidopsis. ABI3 is a plant-specific B3 domain transcription factor that regulates the expression of genes involved in seed development. CRU3 is one of 12S seed storage proteins, and its encoding gene is the direct target of ABI3. DOG1 plays a vital role in seed dormancy, and its expression in transcription and protein levels is strictly regulated during seed development (Molitor et al., 2014). The study showed that AtALs directly bind H3K4me3 regions at ABI3 and DOG1 loci through the PHD finger domain and also physically interact with the Polycomb Repressive Complex 1 (PRC1) RING-finger proteins AtRING1a and AtBMI1b through the N-terminal region. AL PHD-PRC1 complexes subsequently recruit PRC2 to establish H3K27me3 accumulation, resulting in a timely conversion from the H3K4me3-marked activation to the H3K27me3-marked repression of seed developmental genes during seed germination (Molitor et al., 2014). Germostatin (GS), a synthetic small molecule, is identified by chemical genetics approaches and can strongly inhibit seed germination through inducing auxin biosynthesis and enhancing auxin responses. GERMOSTATIN RESISTANCE LOCUS 1 (GSR1) with four tandem PHD finger domains binds to non-methylated H3K4 marks and is responsible for GS-induced prevention of seed germination. It physically interacts with AUXIN RESPONSE FACTOR 10/16 (ARF10/16) and IAA17 to form a co-repressor complex involved in auxin-mediated seed germination (Ye et al., 2016).
Metabolite biosynthesis regulation
In cassava (Manihot esculenta Crantz), ADP-glucose pyrophosphorylase small subunit 1a (MeAGPS1a) is a significant catalytic subunit of ADP-glucose pyrophosphorylase (AGPase), which is the first enzyme in starch biosynthesis and determines the efficiency of starch synthesis. MePHD1 can bind to the promoter region of MeAGPS1a and act as a negative transcriptional regulator of MeAGPS1a expression. Many phytohormones (such as ABA, IAA, and GA) and high temperature (42°C) can upregulate the transcript level of MePHD1 (Ma et al., 2018). Another study revealed that 10 PbPHDs are expressed during pear (Pyrus bretschneideri) fruit development. Particularly, the expression of PbPHD10 showed a similar change pattern to the lignin content with the development of the pear fruit, indicating that PbPHD10 is a candidate gene for regulating the lignin synthesis (Cao et al., 2018).
Metal transport
Metal transport from soil to shoots plays an essential role in plant normal growth and is mainly regulated by metal-specific transporters. A PHD finger protein OsTITANIA (OsTTA) is a constitutively expressed transcription factor that can enhance the expression of diverse metal transporter genes in rice. The tta mutant, LOW CADMIUM (LC5), displays decreased growth and lower accumulation of several metals, such as zinc (Zn), copper (Cu), and manganese (Mn), in shoots compared to the wild-type plants (Tanaka et al., 2018).
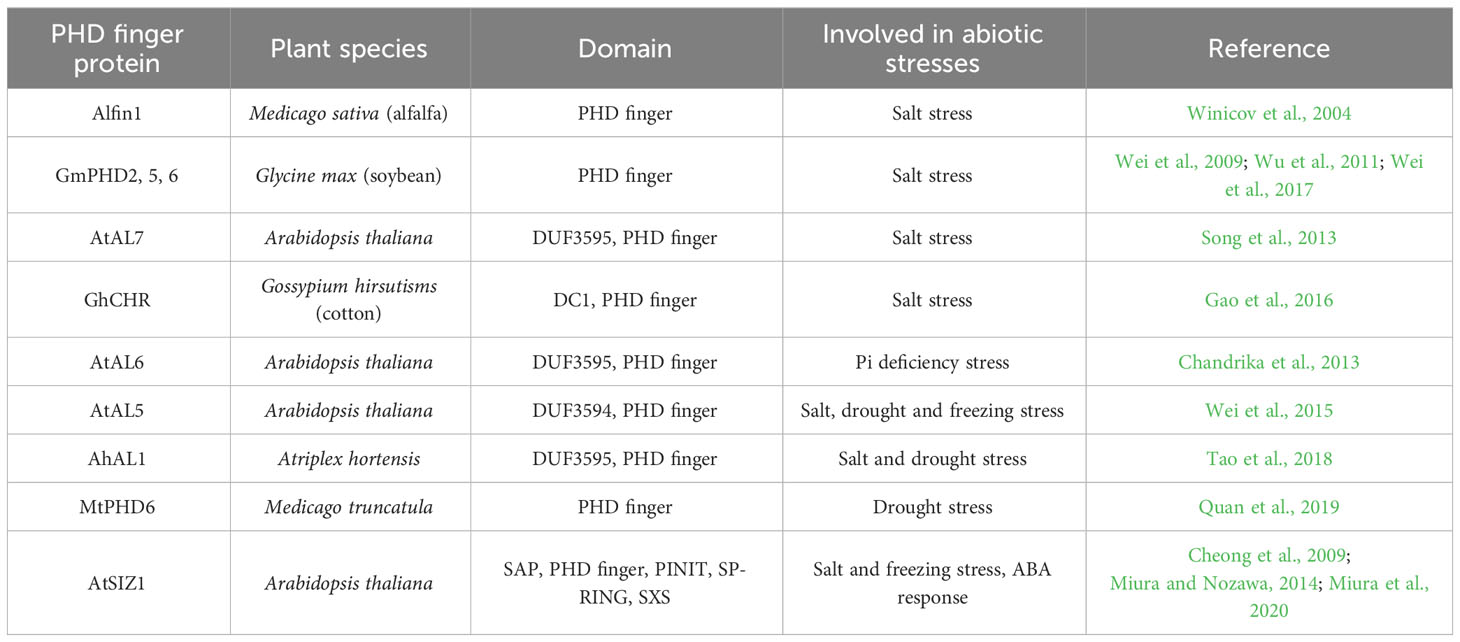
Roles of PHD finger proteins in response to abiotic stresses
Abiotic stresses are major adverse environment factors limiting plant growth, development, and productivity in the whole world (Wang et al., 2019). During the long-time evolution, plants have formed complicated mechanisms to sense, transmit, and respond to these abiotic stress signals in order to survive and reproduce (Gong et al., 2020). The transcriptional regulation of stress-related genes through transcription factors is an important component of plant stress responses (Wang et al., 2019).
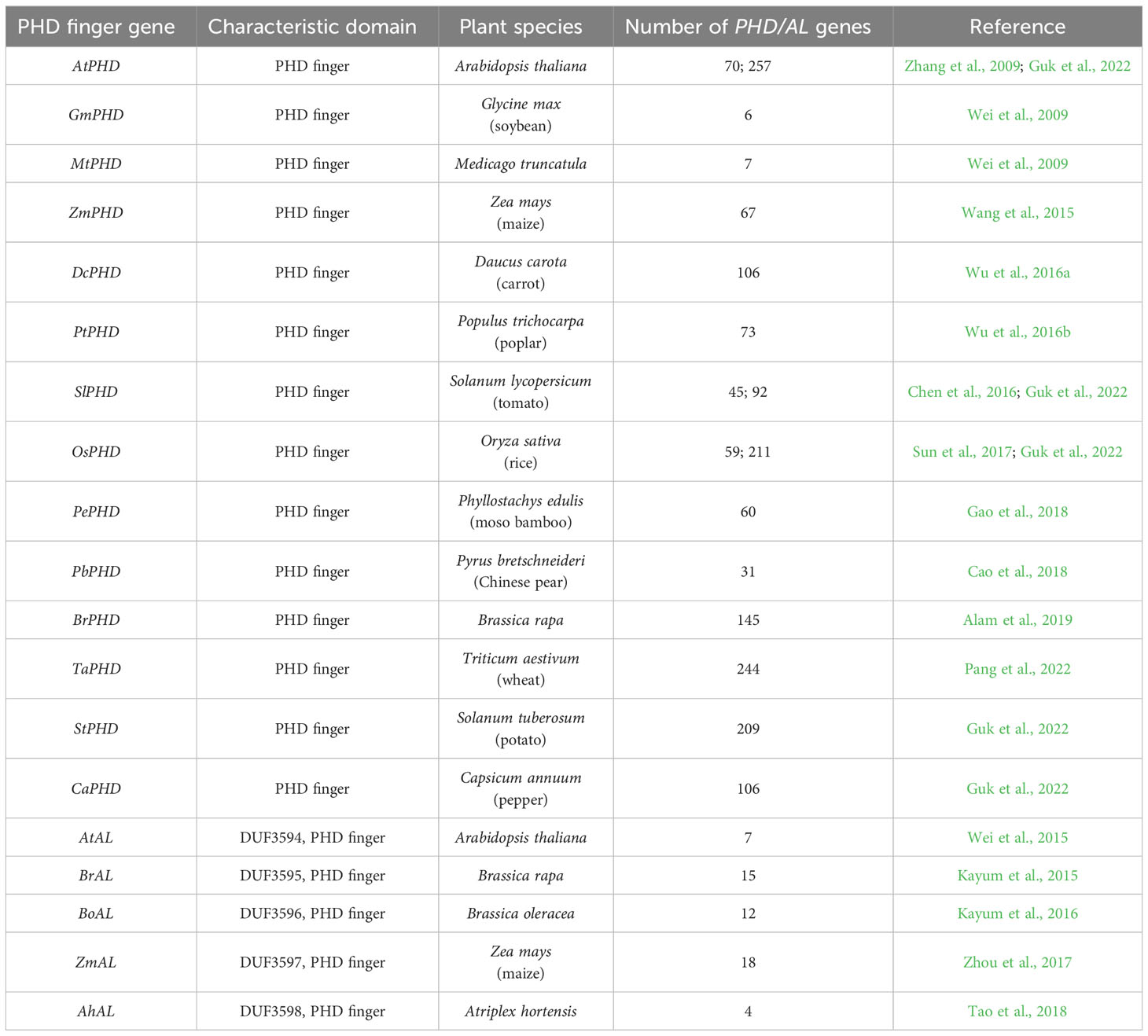
A lot of studies reported that many PHD finger genes are stress responsive and play key roles in plant responses to abiotic stresses, such as salt, drought, and freezing stresses (Table 2). Genome-wide PHD/AL genes are identified and analyzed in various plant species, including Arabidopsis (Zhang et al., 2009; Guk et al., 2022), maize (Wang et al., 2015; Zhou et al., 2017), B. rapa (Kayum et al., 2015; Alam et al., 2019), and wheat (Triticum aestivum L.) (Pang et al., 2022) (Table 3). Wang et al. (2015) reported that 67 PHD finger genes are identified in maize, and 15 ZmPHDs are stress-responsive genes detected by promoter cis-element and expression analysis. When subjected to PEG, NaCl, and ABA treatments, ZmPHD14 and ZmPHD19 are strongly induced or repressed in all stress treatments, while the expression levels of other genes are highly regulated only by one or two treatments. Totally, 73 non-redundant PHD finger genes are isolated from the poplar (Populus trichocarpa) genome. Some paralogous genes have high degrees of sequence homology, such as PtPHD29/PtPHD65, PtPHD35/PtPHD23, and PtPHD45/PtPHD18, suggesting that these genes may have redundant functions. In addition, nine genes, for instance PtPHD68, PtPHD31, and PtPHD65, are strongly regulated under drought, salt, or cold stress (Wu et al., 2016b). In moso bamboo (Phyllostachys edulis), 60 PHD finger genes are classified into 11 subfamilies according to phylogenetic analysis. Among them, 16 PePHDs are stress-responsive genes and differentially induced by drought, low temperature, and NaCl and ABA treatments (Gao et al., 2018). 18 of 145 BrPHD genes from B. rapa are responsive to drought and salt stresses (Alam et al., 2019). As plant genome annotations are updated, a larger number of previously omitted protein-coding genes are re-identified and re-annotated (Cheng et al., 2017; Kim et al., 2020). Through genome-wide re-annotation of PHD finger genes in tomato (Solanum lycopersicum), potato (Solanum tuberosum), pepper (Capsicum annuum), rice, and Arabidopsis, 225 of 875 PHD finger genes were newly identified in the five species, of which 57 is in pepper (Guk et al., 2022). Combining gene expression profiling and GO enrichment analysis showed that many pepper PHD finger differentially expressed genes (DEGs) perform some degree of function in response to salt, heat, or mannitol stress (Guk et al., 2022).
In cotton (Gossypium hirsutum), GhCHR encoding a protein with three PHD finger and two DC1 (Cys5-His) motifs can be induced by salt stress and is the target of miRNVL5. Overexpression of GhCHR in Arabidopsis enhances the tolerance of salt stress with less Na+ accumulation in shoots and better primary root growth, compared with the wild type (Gao et al., 2016). The expression of an M. truncatula gene, MtPHD6, can be induced by osmotic and drought stresses. MtPHD6-overexpressing Arabidopsis plants display lower MDA and ROS contents and higher leaf water content and antioxidant enzyme activities than the wild-type plants under drought stress, leading to the enhanced drought tolerance. Moreover, they found that the transformation of MtPHD6 predominantly upregulates the expression of WRKY, ZINC FINGER, and AP2/EREBP transcription factors (Quan et al., 2019). Plant SIZ proteins encode SUMO (small ubiquitin-related modifier) E3 ligases that play key roles in sumoylation (Jin et al., 2008). AtSIZ1 contains a plant-specific PHD finger domain, while the orthologs in yeast and animals have no PHD finger. The mutation in the PHD finger domain impairs the SUMO conjugate formation and generates the long-hypocotyl phenotype related to sugar and light conditions (Cheong et al., 2009). The PHD finger of AtSIZ1 recognizes H3K4me3, which is important for the suppression of WRKY70 expression and for the interaction with ATX1, a histone lysine methyltransferases. AtSIZ1 without the PHD finger or with mutated PHD finger does not complement the freezing sensitivity and drought tolerance induced by the siz1-2 mutant, whereas AtSIZ1 does, indicating that the PHD finger is necessary for AtSIZ1 function as a transcriptional repressor (Miura et al., 2020). Additionally, the overexpression of AtSIZ1 enhances the tolerance of transgenic plants to freezing and salinity stresses and reduces the inhibition induced by ABA treatment (Miura and Nozawa, 2014).
As a plant-specific subfamily of the PHD finger proteins, ALs normally possess transcriptional suppression activity and play crucial roles in biological processes by inhibiting expression of downstream target genes (Wei et al., 2017). Most AL genes are also significantly induced by abiotic stresses, such as cold, salt, drought, and ABA treatment (Wei et al., 2009; Wei et al., 2015; Tao et al., 2018). Of the 12 AL genes from B. oleracea, 10 are abiotic stress responsive (Kayum et al., 2016). In soybean, Wei et al. (2009) identified six Alfin1-type PHD finger proteins, GmPHD1-6, with the ability of binding to the cis-element “GTGGAG.” GmPHDs have transcriptional suppression activity except GmPHD6. Heterologous expression of GmPHD2 in Arabidopsis inhibits the expression of seven negative regulator genes, such as DREB1C, STRS1, and STRS2. Eight genes are upregulated in transgenic plants, including ABI5, WAK5, GLP, MDAR, TPP, and three peroxidase genes. Expression changes in these stress-responsive genes showed that GmPHD2 enhances salt tolerance though affecting stress signaling and by eliminating ROS in transgenic plants (Wei et al., 2009). Under salt stress, GmPHD5 regulates the crosstalk between the methylated H3K4 and the acetylated H3K14 and may recruit chromatin remodeling factors and transcription factors to modulate the transcription of stress-inducible genes, including GmRD22 and GmGST (Wu et al., 2011). GmPHD6 overexpression improves the tolerance to salt stress in soybean through interacting with LHP1 by the PHD finger domain. The GmPHD6 and LHP1 form a transcriptional activation complex to activate expression of downstream stress responsive genes, such as CYP82C4, CYP75B1, and CCD7, suggesting that the GmPHD6-LHP1 complex plays a key role in salt tolerance (Wei et al., 2017). In Arabidopsis, seven AL genes are identified and functionally characterized. AtAL6 is essential for the formation of root hair during phosphate (Pi) deficiency stress. It binds to H3K4me marks at the Myb-type transcription factor ETC1 through its PHD finger domain, which may facilitate the transcription of downstream Pi-responsive genes (Chandrika et al., 2013). The T-DNA insertion mutants of AtAL7 display enhanced salt tolerance with longer root length, indicating that AtAL7 functions as a negative regulator in response to salt stress (Song et al., 2013). Overexpression of AtAL5 improves the tolerance of transgenic plants to salt, drought, and freezing stresses by repressing the transcription of downstream negative regulatory genes, including SHMT7, TAC1, OFE, FAO, and CAX1 (Wei et al., 2015). In addition, four AhAL genes are isolated from A. hortensis and encode nuclear-localized proteins with the transcription repression activities. AhAL1-transgenic Arabidopsis shows the higher survival rate under salt and drought stresses by reducing MDA content and water loss. Through binding to the promoter regions of target genes, AhAL1 represses the expression of negative regulator genes in ABA signaling, such as DREB1C, GRF7, and five group-A protein phosphatase 2C (PP2C)-encoding genes (ABI1, ABI2, AHG3, HAB1, and HAB2), which then induces the activation of some ABA/stress-responsive genes, including DREB1A, DREB2A, and three genes encoding ABA-responsive element (ABRE)-binding factors (ABF2, ABF3, and ABF4) (Tao et al., 2018). In conclusion, the overexpression of PHD finger genes can improve abiotic stress tolerance of transgenic plants with better growth phenotype. PHD finger genes exert the transcriptional regulatory activity by inhibiting or activating the expression of downstream stress-related genes in plant adaptation to adverse environment.
Conclusion and perspectives
Proteins containing the PHD finger domain are widespread in plants and are one of the important transcription regulator families. Recently, many PHD finger proteins are identified in diverse plant species and proved to be involved in various biological processes. In this review, we mainly focused on the roles of PHD finger proteins in plant reproduction development, such as floral transition, tapetum development, and male meiosis (Figure 1), and responses to abiotic stresses, including salinity, drought stress, and low temperature (Figure 2). Moreover, acting as key transcriptional regulators, some PHD finger proteins also function in metal transport and biosynthesis pathways, which are important for the normal growth of plants. In summary, after being exposed to external stimuli, PHD finger proteins can bind to specific regions of downstream target gene promoters through the PHD finger to exert its transcriptional regulatory activity, activate or inhibit the expression of responsive genes related to plant growth and development and stress response, and finally achieve the role of regulating plant development and stress tolerance (Figure 3).
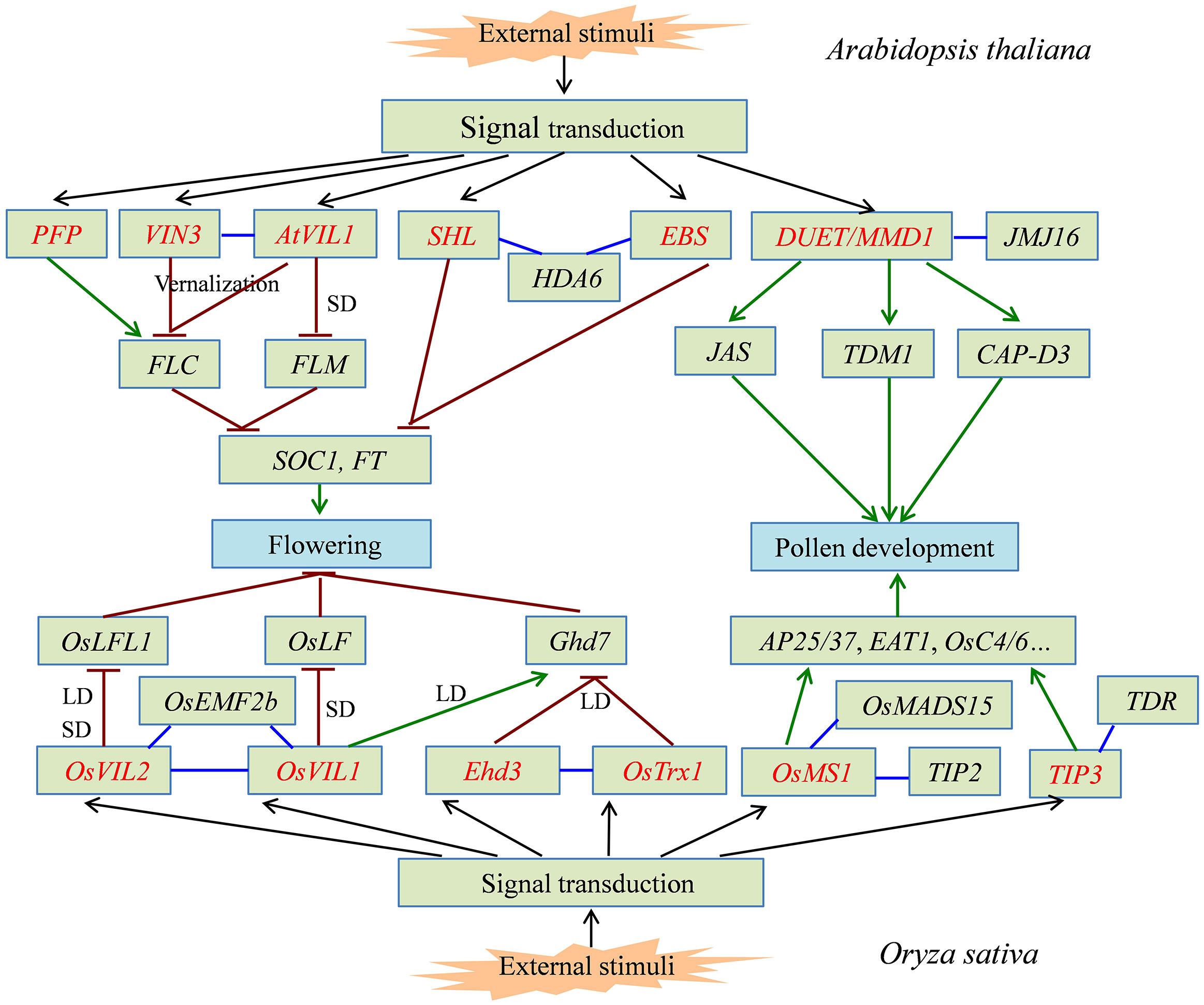
Figure 1 The signal pathway of partial PHD finger proteins involved in plant flowering and pollen development. PHD finger proteins from Arabidopsis thaliana and Oryza sativa directly or indirectly activate (green line) or inhibit (dark red line) the expression of downstream target genes, which can precisely regulate the flowering time and normal pollen development in plants. PHD finger proteins are shown in red font. The interaction between two proteins is shown in blue line.
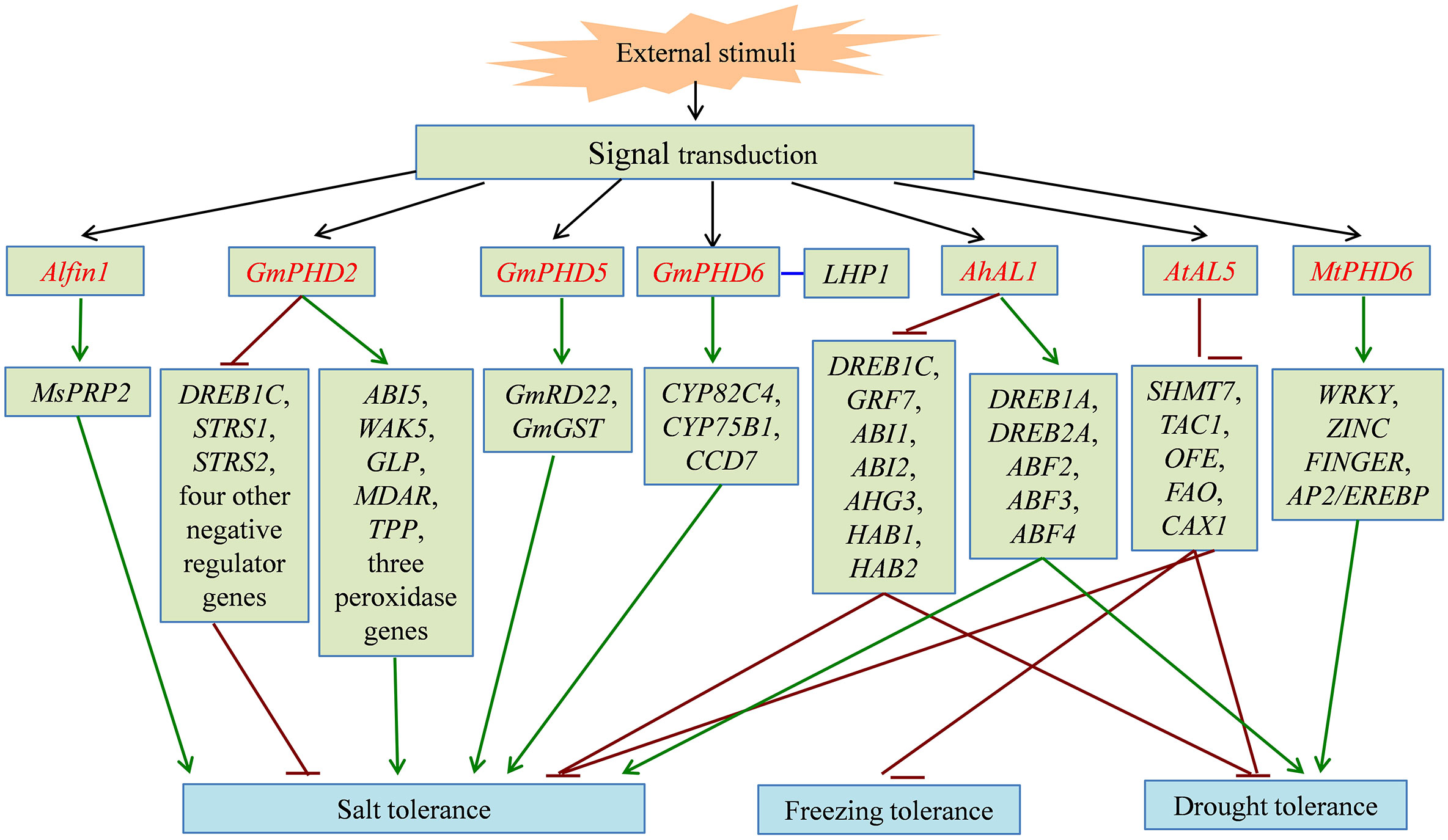
Figure 2 The signal pathway of partial PHD finger proteins involved in plant abiotic stress responses. PHD finger proteins directly or indirectly activate (green line) or inhibit (dark red line) the expression of downstream stress-related genes, and enhance tolerance to salt, freezing, and drought stress in different plant species. PHD finger proteins are shown in red font. The interaction between two proteins is shown in blue line.
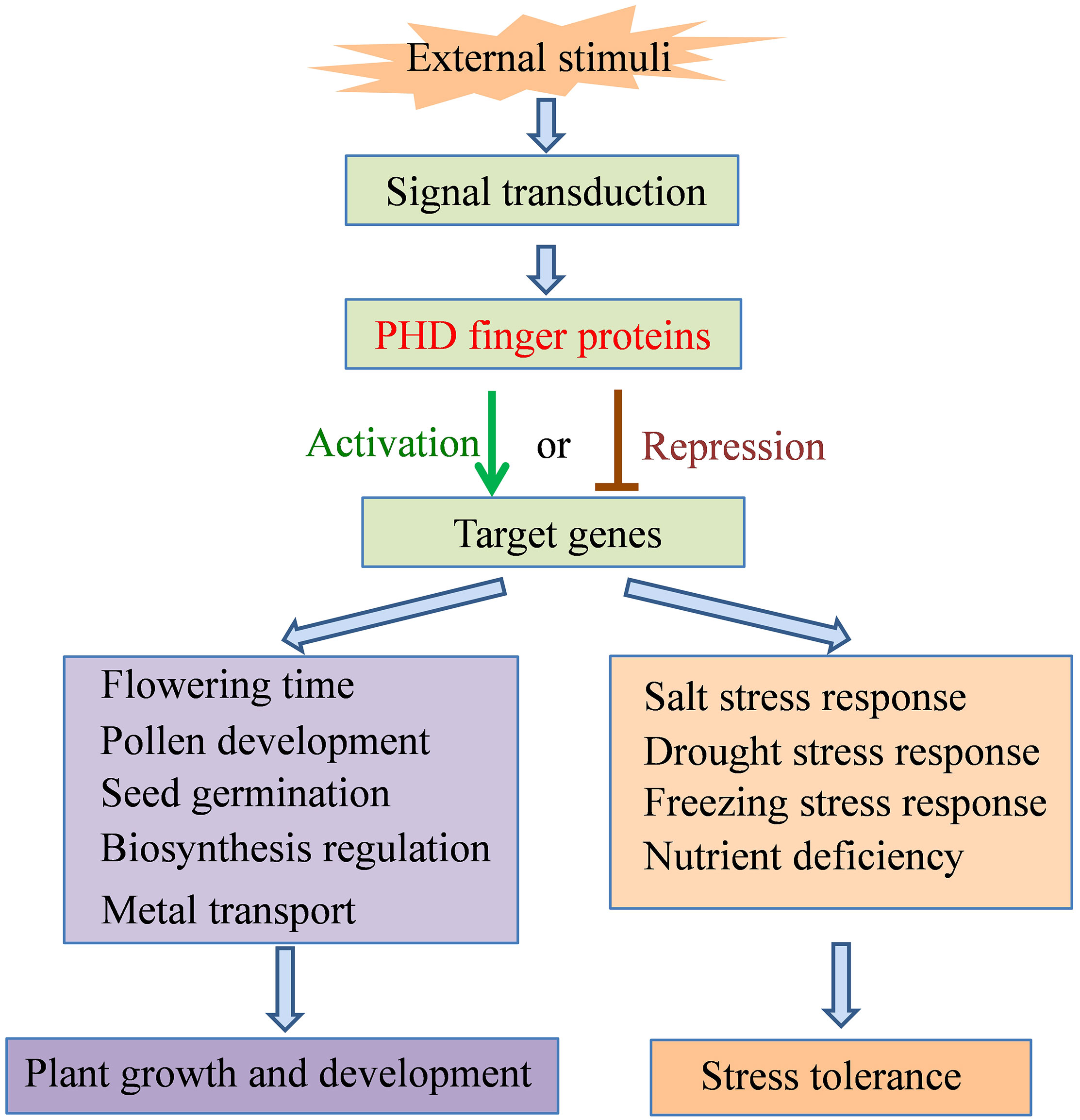
Figure 3 Summary framework of the roles of PHD finger proteins in plant growth and development and stress tolerance.
The PHD finger is the “reader” domain of epigenetic modification and directly binds to the methylated histone H3, which plays key roles in chromatin remodeling and transcription regulation of target genes (Sung and Amasino, 2004; Wu et al., 2011; López-González et al., 2014; Wang et al., 2016; Miura et al., 2020). Furthermore, PHD finger proteins can interact with other proteins through the PHD finger domain to regulate specific biological processes. For example, the interaction of OsMS1 with TIP2 and OsMADS15 is necessary for the tapetum development and pollen wall formation, both of which play crucial roles in the production pf mature pollen grains (Yang et al., 2019a). In order to verify the functional importance of the PHD finger, the target gene with or without the PHD finger domain is transferred into the corresponding mutant plants to observe whether the abnormal phenotypes can be restored. For instance, under the control of the AtSIZ1 promoter, the recombinant vector containing AtSIZ1 gene with or without the PHD finger is transferred to the siz1-2 mutant. The expression of ProSIZ1::SIZ1:GFP could be able to complement the defective phenotypes of the siz1-2 mutant, while ProSIZ1::SIZ1(ΔPHD):GFP does not, demonstrating that the PHD finger is critical for AtSIZ1 in plant response to cold stress, drought stress, and ABA treatment (Miura et al., 2020).
With the development of transcriptome-wide sequencing and the updating of genome-wide annotation, more and more PHD finger genes will be excavated or be re-annotated in plant species; however, further studies still need to focus on the exact functions of PHD finger proteins. For instance, many PHD finger genes are proved to be stress responsive, but their biological functions in plant responses to abiotic stresses remain to be confirmed. By using PHD finger gene-specific overexpression and mutant lines is helpful for analyzing their functions. In addition, it is worth noting that a PHD finger protein may have multiple roles participated in different biological events, such as AtAL6 (Chandrika et al., 2013; Molitor et al., 2014) and AtSIZ1 (Jin et al., 2008; Miura et al., 2020). Last but not the least, the signaling transduction pathways by which many PHD finger proteins perform specific biological functions are still unclear. At present, the research on some PHD finger proteins is limited to the identification of biological functions. The molecular mechanism of its specific biological function has not been studied deeply. Many studies have screened differentially expressed genes through transcriptome sequencing, so as to obtain downstream functional genes modulated by a PHD finger protein. The upstream regulators of this protein expression and their functions have been less studied. Identifying the upstream and downstream interaction factors of PHD finger proteins is essential to better understand their molecular mechanisms in a biological process. The yeast two-hybrid assay and other protein interaction methods are useful for analyzing and verifying proteins interacted with PHD finger proteins. To sum up, the in-depth research on the biological functions of PHD finger proteins and the construction of their molecular regulatory networks will enrich and improve our understanding of the roles of PHD finger proteins in various biological events. Meanwhile, this will also provide a valuable scientific basis for the study of new PHD finger proteins. In addition, the PHD finger protein encoding genes may be used as novel candidate genes to modify the plant genome for enhancing the tolerance of transgenic plants to adverse environment or improving their growth and development, ultimately leading to the plant biomass increase.
Author contributions
WQ: Writing – original draft. ZC: Writing – review & editing. PW: Writing – review & editing. YM: Writing – review & editing. DB: Writing – review & editing. XL: Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. WQ was supported by the National Natural Science Foundation of China (No. 32202727). XL was funded by the Natural Science Foundation of Sichuan Province (No. 2022NSFSC1782) and Sichuan Province Scientific Research Foundation for the Returned Overseas Chinese Scholars (2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aasland, R., Gibson, T. J., Stewart, A. F. (1995). The PHD finger: implications for chromatin mediated transcriptional regulation. Trends Biochem. Sci. 20 (2), 56–59. doi: 10.1016/S0968-0004(00)88957-4
Alam, I., Liu, C. C., Ge, H. L., Batool, K., Yang, Y. Q., Lu, Y. H. (2019). Genome wide survey, evolution and expression analysis of PHD finger genes reveal their diverse roles during the development and abiotic stress responses in Brassica rapa L. BMC Genomics 20 (1), 773. doi: 10.1186/s12864-019-6080-8
Andreuzza, S., Nishal, B., Singh, A., Siddiqi, I. (2015). The chromatin protein DUET/MMD1 controls expression of the meiotic gene TDM1 during male meiosis in Arabidopsis. PloS Gene 11 (9), e1005396. doi: 10.1371/journal.pgen.1005396
Baker, L. A., Allis, C. D., Wang, G. G. (2008). PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat. Res-Fund. Mol. M. 647 (1-2), 3–12. doi: 10.1016/j.mrfmmm.2008.07.004
Bastola, D. R., Pethe, V. V., Winicov, I. (1998). Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol. 38, 1123–1135. doi: 10.1023/A:1006081926699
Bienz, M. (2006). The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31 (1), 35–40. doi: 10.1016/j.tibs.2005.11.001
Cao, Y., Han, Y., Meng, D., Abdullah, M., Li, D., Jin, Q., et al. (2018). Systematic analysis and comparison of the PHD-Finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct. Integr. Genomics 18, 519–531. doi: 10.1007/s10142-018-0609-9
Capili, A. D., Schultz, D. C., RauscherIII, F. J., Borden, K. L. (2001). Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20 (1-2), 165–177. doi: 10.1093/emboj/20.1.165
Chandrika, N. N. P., Sundaravelpandian, K., Schmidt, W. (2013). A PHD in histone language: On the role of histone methylation in plant responses to phosphate deficiency. Plant Signal. Behav. 8 (6), e24381. doi: 10.4161/psb.24381
Chen, X., Hayat, S., Cheng, Z. (2016). Genome-wide identification and analysis of genes encoding PHD-finger protein in Tomato. Pak. J. Bot. 48, 155–165. Available at: https://www.researchgate.net/publication/295185906.
Cheng, C. Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., Town, C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89 (4), 789–804. doi: 10.1111/tpj.13415
Cheong, M. S., Park, H. C., Hong, M. J., Lee, J., Choi, W., Jin, J. B., et al. (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 151 (4), 1930–1942. doi: 10.1104/pp.109.143719
Choi, S. C., Lee, S., Kim, S. R., Lee, Y. S., Liu, C., Cao, X., et al. (2014). Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol. 164 (3), 1326–1337. doi: 10.1104/pp.113.228049
Diego-Martin, B., Pérez-Alemany, J., Candela-Ferre, J., Corbalán-Acedo, A., Pereyra, J., Alabadí, D., et al. (2022). The TRIPLE PHD FINGERS proteins are required for SWI/SNF complex-mediated +1 nucleosome positioning and transcription start site determination in Arabidopsis. Nucleic Acids Res. 50 (18), 10399–10417. doi: 10.1093/nar/gkac826
Dong, S., Zou, J., Fang, B., Zhao, Y., Shi, F., Song, G., et al. (2022). Defect in BrMS1, a PHD-finger transcription factor, induces male sterility in ethyl methane sulfonate-mutagenized Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.992391
Fu, D., Dunbar, M., Dubcovsky, J. (2007). Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Mol. Genet. Genomics 277, 301–313. doi: 10.1007/s00438-006-0189-6
Gao, S., Yang, L., Zeng, H. Q., Zhou, Z. S., Yang, Z. M., Li, H., et al. (2016). A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 6 (1), 19736. doi: 10.1038/srep19736
Gao, Y., Liu, H., Wang, Y., Li, F., Xiang, Y. (2018). Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 123, 378–391. doi: 10.1016/j.plaphy.2017.12.034
Gilbert, T. M., McDaniel, S. L., Byrum, S. D., Cades, J. A., Dancy, B. C. R., Wade, H., et al. (2014). A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell. Proteomics 13 (11), 2883–2895. doi: 10.1074/mcp.M114.038224
Gómez, J. F., Wilson, Z. A. (2014). A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12 (6), 765–777. doi: 10.1111/pbi.12181
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Guk, J. Y., Jang, M. J., Kim, S. (2022). Identification of novel PHD−finger genes in pepper by genomic re−annotation and comparative analyses. BMC Plant Biol. 22 (1), 206. doi: 10.1186/s12870-022-03580-2
Hou, J., Fan, W., Ma, R., Li, B., Yuan, Z., Huang, W., et al. (2022). MALE STERILITY 3 encodes a plant homeodomain-finger protein for male fertility in soybean. J. Integr. Plant Biol. 64 (5), 1076–1086. doi: 10.1111/jipb.13242
Hu, Y., Yang, G., Zhang, D., Liu, Y., Li, Y., Lin, G., et al. (2018). The PHD transcription factor Rum1 regulates morphogenesis and aflatoxin biosynthesis in Aspergillus flavus. Toxins 10 (7), 301. doi: 10.3390/toxins10070301
Ito, T., Nagata, N., Yoshiba, Y., Ohme-Takagi, M., Ma, H., Shinozaki, K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19 (11), 3549–3562. doi: 10.1105/tpc.107.054536
Ito, T., Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger Motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43 (11), 1285–1292. doi: 10.1093/pcp/pcf154
Jarillo, J. A., Piñeiro, M. (2011). Timing is everything in plant development. The central role of floral repressors. Plant Sci. 181 (4), 364–378. doi: 10.1016/j.plantsci.2011.06.011
Jeong, K., Choi, D., Lee, J. (2018). Fine mapping of the genic male−sterile ms1 gene in Capsicum annuum L. Theor. Appl. Genet. 131, 183–191. doi: 10.1007/s00122-017-2995-0
Jeong, H. J., Yang, J., Cho, L. H., An, G. (2016). OsVIL1 controls flowering time in rice by suppressing OsLF under short days and by inducing Ghd7 under long days. Plant Cell Rep. 35, 905–920. doi: 10.1007/s00299-015-1931-5
Jin, J. B., Jin, Y. H., Lee, J., Miura, K., Yoo, C. Y., Kim, W. Y., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53 (3), 530–540. doi: 10.1111/j.1365-313X.2007.03359.x
Kayum, M. A., Park, J. I., Ahmed, N. U., Jung, H. J., Saha, G., Kang, J. G., et al. (2015). Characterization and stress-induced expression analysis of Alfin-like transcription factors in Brassica rapa. Mol. Genet. Genomics 290, 1299–1311. doi: 10.1007/s00438-015-0993-y
Kayum, M. A., Park, J. I., Ahmed, N. U., Saha, G., Chung, M. Y., Kang, J. G., et al. (2016). Alfin-like transcription factor family: characterization and expression profiling against stresses in Brassica oleracea. Acta Physiol. Plant 38, 127. doi: 10.1007/s11738-016-2139-1
Khan, M. R. G., Ai, X. Y., Zhang, J. Z. (2014). Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip. Rev. RNA 5 (3), 347–359. doi: 10.1002/wrna.1215
Kim, S., Cheong, K., Park, J., Kim, M., Kim, J., Seo, M., et al. (2020). TGFam-Finder: a novel solution for target-gene family annotation in plants. New Phytol. 227 (5), 1568–1581. doi: 10.1111/nph.16645
Kim, D. H., Sung, S. (2013). Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25 (2), 454–469. doi: 10.1105/tpc.112.104760
Lee, W. Y., Lee, D., Chung, W. I., Kwon, C. S. (2009). Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58 (3), 511–524. doi: 10.1111/j.1365-313X.2009.03795.x
Lee, J. S., Smith, E., Shilatifard, A. (2010). The language of histone crosstalk. Cell 142 (5), 682–685. doi: 10.1016/j.cell.2010.08.011
Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C., Liang, W., Zong, J., et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156 (2), 615–630. doi: 10.1104/pp.111.175760
Liu, B., Jin, C., De Storme, N., Schotte, S., Schindfessel, C., De Meyer, T., et al. (2021). A hypomorphic mutant of PHD domain protein Male Meiocytes Death 1. Genes 12 (4), 516. doi: 10.3390/genes12040516
López-González, L., Mouriz, A., Narro-Diego, L., Bustos, R., Martínez-Zapater, J. M., Jarillo, J. A., et al. (2014). Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 26 (10), 3922–3938. doi: 10.1105/tpc.114.130781
Ma, P., Chen, X., Liu, C., Xia, Z., Song, Y., Zeng, C., et al. (20182831). MePHD1 as a PHD-finger protein negatively regulates ADP-glucose pyrophosphorylase small subunit1a gene in cassava. Int. J. Mol. Sci. 19 (9). doi: 10.3390/ijms19092831
Martin, D. G. E., Baetz, K., Shi, X., Walter, K. L., MacDonald, V. E., Wlodarski, M. J., et al. (2006). The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26 (21), 7871–7879. doi: 10.1128/MCB.00573-06
Matsubara, K., Yamanouchi, U., Nonoue, Y., Sugimoto, K., Wang, Z., Minobe, Y., et al. (2011). Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 66 (4), 603–612. doi: 10.1111/j.1365-313X.2011.04517.x
Michaels, S. D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12 (1), 75–80. doi: 10.1016/j.pbi.2008.09.005
Milosevich, N., Hof, F. (2016). Chemical inhibitors of epigenetic methyllysine reader proteins. Biochemistry 55 (11), 1570–1583. doi: 10.1021/acs.biochem.5b01073
Miura, K., Nozawa, R. (2014). Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana. Plant Biotechnol. (Tokyo) 31 (2), 167–172. doi: 10.5511/plantbiotechnology.14.0109a
Miura, K., Renhu, N., Suzaki, T. (2020). The PHD finger of Arabidopsis SIZ1 recognizes trimethylated histone H3K4 mediating SIZ1 function and abiotic stress response. Commun. Biol. 3 (1), 23. doi: 10.1038/s42003-019-0746-2
Mohan, V., Borovsky, Y., Kamara, I., Zemach, H., Paran, I. (2018). CaVIL1, a plant homeodomain gene that promotes flowering in pepper. Theor. Appl. Genet. 131, 2639–2649. doi: 10.1007/s00122-018-3179-2
Molitor, A. M., Bu, Z., Yu, Y., Shen, W. H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10 (1), e1004091. doi: 10.1371/journal.pgen.1004091
Müssig, C., Kauschmann, A., Clouse, S. D., Altmann, T. (2000). The Arabidopsis PHD-finger protein SHL is required for proper development and fertility. Mol. Gen. Genet. 264, 363–370. doi: 10.1007/s004380000313
Pang, F., Niu, J., Solanki, M. K., Nosheen, S., Liu, Z., Wang, Z. (2022). PHD-finger family genes in wheat (Triticum aestivum L.): Evolutionary conservatism, functional diversification, and active expression in abiotic stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1016831
Qian, F., Zhao, Q. Y., Zhang, T. N., Li, Y. L., Su, Y. N., Li, L., et al. (2021). A histone H3K27me3 reader cooperates with a family of PHD finger-containing proteins to regulate flowering time in Arabidopsis. J. Integr. Plant Biol. 63 (4), 787–802. doi: 10.1111/jipb.13067
Quan, W., Liu, X., Wang, L., Yin, M., Yang, L., Chan, Z. (2019). Ectopic expression of Medicago truncatula homeodomain finger protein, MtPHD6, enhances drought tolerance in Arabidopsis. BMC Genomics 20 (1), 982. doi: 10.1186/s12864-019-6350-5
Reddy, T. V., Kaur, J., Agashe, B., Sundaresan, V., Siddiqi, I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130 (24), 5975–5987. doi: 10.1242/dev.00827
Sanchez, R., Zhou, M. M. (2011). The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36 (7), 364–372. doi: 10.1016/j.tibs.2011.03.005
Schindler, U., Beckmann, H., Cashmore, A. R. (1993). HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4 (1), 137–150. doi: 10.1046/j.1365-313X.1993.04010137.x
Shu, Y., Tao, Y., Wang, S., Huang, L., Yu, X., Wang, Z., et al. (2015). GmSBH1, a homeobox transcription factor gene, relates to growth and development and involves in response to high temperature and humidity stress in soybean. Plant Cell Rep. 34, 1927–1937. doi: 10.1007/s00299-015-1840-7
Song, Y., Gao, J., Yang, F., Kua, C. S., Liu, J., Cannon, C. H. (2013). Molecular Evolutionary Analysis of the Alfin-Like Protein Family in Arabidopsis lyrata, Arabidopsis thaliana, and Thellungiella halophile. PloS One 8 (7), e66838. doi: 10.1371/journal.pone.0066838
Sun, C., Chen, D., Fang, J., Wang, P., Deng, X., Chu, C. (2014). Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 5 (12), 889–898. doi: 10.1007/s13238-014-0068-6
Sun, M., Jia, B., Yang, J., Cui, N., Zhu, Y., Sun, X. (2017). Genome-wide identification of the PHD-finger family genes and their responses to environmental stresses in Oryza sativa L. Int. J. Mol. Sci. 18 (9), 2005. doi: 10.3390/ijms18092005
Sung, S., Amasino, R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 (6970), 159–164. doi: 10.1038/nature02195
Sung, S., Schrmitz, R. J., Amasino, R. M. (2006). A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 20 (23), 3244–3248. doi: 10.1101/gad.1493306
Tanaka, N., Uraguchi, S., Kajikawa, M., Saito, A., Ohmori, Y., Fujiwara, T. (2018). A rice PHD-finger protein OsTITANIA, is a growth regulator that functions through elevating expression of transporter genes for multiple metals. Plant J. 96 (5), 997–1006. doi: 10.1111/tpj.14085
Tao, J. J., Wei, W., Pan, W. J., Lu, L., Li, Q. T., Ma, J. B., et al. (20182707). An Alfin-like gene from Atriplex hortensis enhances salt and drought tolerance and abscisic acid response in transgenic Arabidopsis. Sci. Rep. 8 (1). doi: 10.1038/s41598-018-21148-9
Thu, S. W., Rai, K. M., Sandhu, D., Rajangam, A., Balasubramanian, V. K., Palmer, R. G., et al. (2019). Mutation in a PHD-finger protein MS4 causes male sterility in soybean. BMC Plant Biol. 19 (1), 378. doi: 10.1186/s12870-019-1979-4
Wang, K., Ding, Y., Cai, C., Chen, Z., Zhu, C. (2019). The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plant 165 (4), 690–700. doi: 10.1111/ppl.12728
Wang, Q., Liu, J., Wang, Y., Zhao, Y., Jiang, H., Cheng, B. (2015). Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. Int. J. Mol. Sci. 16 (10), 23517–23544. doi: 10.3390/ijms161023517
Wang, J., Niu, B., Huang, J., Wang, H., Yang, X., Dong, A., et al. (2016). The PHD finger protein MMD1/DUET ensures the progression of male meiotic chromosome condensation and directly regulates the expression of the condensing gene CAP-D3. Plant Cell 28 (8), 1894–1909. doi: 10.1105/tpc.16.00040
Wang, J., Yu, C., Zhang, S., Ye, J., Dai, H., Wang, H., et al. (2020). Cell-type-dependent histone demethylase specificity promotes meiotic chromosome condensation in Arabidopsis. Nat. Plants 6 (7), 823–837. doi: 10.1038/s41477-020-0697-0
Wei, W., Huang, J., Hao, Y. J., Zou, H. F., Wang, H. W., Zhao, J. Y., et al. (2009). Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PloS One 4, e7209. doi: 10.1371/journal.pone.0007209
Wei, W., Tao, J. J., Chen, H. W., Li, Q. T., Zhang, W. K., Ma, B., et al. (2017). A histone code reader and a transcriptional activator interact to regulate genes for salt tolerance. Plant Physiol. 175 (3), 1304–1320. doi: 10.1104/pp.16.01764
Wei, W., Zhang, Y. Q., Tao, J. J., Chen, H. W., Li, Q. T., Zhang, W. K., et al. (2015). The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J. 81 (6), 871–883. doi: 10.1111/tpj.12773
Wilson, Z. A., Morroll, S. M., Dawson, J., Swarup, R., Tighe, P. J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28 (1), 27–39. doi: 10.1046/j.1365-313x.2001.01125.x
Winicov, I., Valliyodan, B., Xue, L., Hoober, J. K. (2004). The MsPRP2 promoter enables strong heterologous gene expression in a rootspecific manner and is enhanced by overexpression of Alfin1. Planta 219, 925–935. doi: 10.1007/s00425-004-1296-4
Wu, X. J., Li, M. Y., Que, F., Wang, F., Xu, Z. S., Xiong, A. S. (2016a). Genome-wide analysis of PHD family transcription factors in carrot (Daucus carota L.) reveals evolution and response to abiotic stress. Acta Physiol. Plant 38, 67. doi: 10.1007/s11738-016-2086-x
Wu, T., Pi, E. X., Tsai, S. N., Lam, H. M., Sun, S. M., Kwan, Y. W., et al. (2011). GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 11, 178. doi: 10.1186/1471-2229-11-178
Wu, Q., Wang, C. T. (2014). Genome wide analysis of PHD finger family in Soybean (Glycine max). Adv. Mat. Res. 864, 2503–2508. doi: 10.4028/www.scientific.net/AMR.864-867.2503
Wu, S., Wu, M., Dong, Q., Jiang, H., Cai, R., Xiang, Y. (2016b). Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene 575 (1), 75–89. doi: 10.1016/j.gene.2015.08.042
Yang, J., Lee, S., Hang, R., Kim, S. R., Lee, Y. S., Cao, X., et al. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73 (4), 566–578. doi: 10.1111/tpj.12057
Yang, Z., Liu, L., Sun, L., Yu, P., Zhang, P., Abbas, A., et al. (2019a). OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99, 175–191. doi: 10.1007/s11103-018-0811-0
Yang, X., Makaroff, C. A., Ma, H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15 (6), 1281–1295. doi: 10.1105/tpc.15.010447
Yang, Z. F., Sun, L., Zhang, P., Zhang, Y., Yu, P., Liu, L., et al. (2019b). TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice. Plant J. 99 (5), 844–861. doi: 10.1111/tpj.14365
Ye, Y., Gong, Z., Lu, X., Miao, D., Shi, J., Lu, J., et al. (2016). Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant J. 85 (1), 3–15. doi: 10.1111/tpj.13086
Yokoyama, Y., Kobayashi, S., Kidou, S. I. (2019). PHD type zinc finger protein PFP represses flowering by modulating FLC expression in Arabidopsis thaliana. Plant Growth Regul. 88, 49–59. doi: 10.1007/s10725-019-00487-1
Zhang, Z., Qian, H., Wang, Z., Pang, Y., Guan, X., Poetsch, A., et al. (2023). Characterization of histone acetyltransferases and deacetylases and their roles in response to dehydration stress in Pyropia yezoensis (Rhodophyta). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1133021
Zhang, D., Wu, S., An, X., Xie, K., Dong, Z., Zhou, Y., et al. (2018). Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 16 (2), 459–471. doi: 10.1111/pbi.12786
Zhang, F., Yang, Z. N., Zhang, S. (2009). Genome-wide analysis of PHD-finger protein family in Arabidopsis thaliana. Acta Bot. Yunnan. 31 (3), 227–238. doi: 10.3724/SP.J.1143.2009.09006
Zhang, Y. Z., Yuan, J., Zhang, L., Chen, C., Wang, Y., Zhang, G., et al. (20206212). Coupling of H3K27me3 recognition with transcriptional repression through the BAH-PHD-CPL2 complex in Arabidopsis. Nat. Commun. 11. doi: 10.1038/s41467-020-20089-0
Keywords: PHD finger, methylation, transcription regulator, plant development, abiotic stress, molecular mechanism
Citation: Quan W, Chan Z, Wei P, Mao Y, Bartels D and Liu X (2023) PHD finger proteins function in plant development and abiotic stress responses: an overview. Front. Plant Sci. 14:1297607. doi: 10.3389/fpls.2023.1297607
Received: 20 September 2023; Accepted: 30 October 2023;
Published: 17 November 2023.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Rashmi Kaul, International Centre for Genetic Engineering and Biotechnology, IndiaSridhar Malkaram, West Virginia State University, United States
Copyright © 2023 Quan, Chan, Wei, Mao, Bartels and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Liu, eHVubGl1MDEyM0Bob3RtYWlsLmNvbQ==; Dorothea Bartels, ZGJhcnRlbHNAdW5pLWJvbm4uZGU=
 Wenli Quan
Wenli Quan Zhulong Chan
Zhulong Chan Piwei Wei1
Piwei Wei1 Dorothea Bartels
Dorothea Bartels Xun Liu
Xun Liu