- 1School of Life Sciences, Zhengzhou University, Zhengzhou, Henan, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, Henan, China
Weeds present a significant challenge to high crop yield and quality. In our study, we investigated the phytotoxic activity of β-caryophyllene (BCP) and eugenol, which are natural allelopathic chemical compounds, on Arabidopsis seedlings. We found that these compounds inhibited the growth of Arabidopsis thaliana plants. When either BCP or eugenol was applied, it led to decrease in the content of cell wall components such as lignin, cellulose, hemicellulose, and pectin; and increase in the levels of endogenous hormones like ETH, ABA, SA, and JA in the seedlings. Through transcriptome profiling, we identified 7181 differentially expressed genes (DEGs) in the roots and shoots that were induced by BCP or eugenol. The genes involved in the synthesis of lignin, cellulose, hemicellulose, and pectin were down-regulated, whereas genes related to synthesis and signal transduction of ABA, ETH, SA, and JA were up-regulated. However, genes related to IAA synthesis and signal transduction were found to be down-regulated. Furthermore, we characterized 24 hub genes using Weighted Correlation Network Analysis (WGCNA). Among them, the identified 16 genes in response to BCP was primarily associated with hypoxia stress, while 8 genes induced by eugenol were linked to inhibition of cell division. Our results suggested that BCP and eugenol had ability to target multiple genes to inhibit growth and development of Arabidopsis plants. Therefore, they can serve as excellent candidates for natural biological herbicides.
1 Introduction
Weeds pose one of the most significant challenges to crop yields, resulting in significant losses in crop quality (Macias-Rubalcava and Garrido-Santos, 2022). Currently, synthetic herbicides are the primary tools used to control weeds. However, their low biodegradability and high persistence in soil have led to serious environmental risks, including contamination of surface and ground waters. Moreover, the number of herbicide-resistant weeds is continuously increasing. For instance, glyphosate-resistant and synthetic auxin herbicide (SAH) 2,4-D-resistant weed species have become increasingly common (Busi et al., 2018).
Using synthetic herbicides or breeding transgenic crops to control herbicides for weed management can lead to rapid evolution of herbicide-resistant weeds (Duke, 2015). Currently, there are 404 unique cases (species × site of action) of herbicide-resistant weeds globally (Heap, 2014). Thus, relying on synthetic herbicides or transgenic crops for weed management is unsustainable.
Most plants release cytotoxic chemicals into the environment, a process known as allelopathy to compete for resources with their neighbors. This mechanism helps plants to compete with neighboring organisms for limited nutrients (McCoy et al., 2022). Phenolic compounds, terpenoids, and benzoxazinoids are among the most important natural allelochemicals, which can be used as natural herbicides, are helpful in overcoming weed resistance to control weeds in sustainable agricultural systems. (Huang et al., 2010; Macias et al., 2019).
The sesquiterpene β-caryophyllene (BCP) is a major compound found in several plant species, including Psidium guajava L. (Ullah et al., 2021). It is used for remedy in several diseases (Sharma et al., 2016). Additionally, and caryophyllene oxide have been reported as potent anti-inflammatory agents, inducing apoptosis in these cells (Sain et al., 2014). In Arabidopsis thaliana, the miR156-SPL module regulates the formation of BCP during the flowering stage by modulating the expression of the sesquiterpene synthase gene TPS21 (Yu et al., 2015).
Eugenol is a phenolic aromatic ingredient present in clove oil. It not only boosts the antioxidant capacity of cells (Stone, 2017), but only exhibits antimicrobial properties against a range of pathogenic bacteria (Da Camara et al., 2022; Fu et al., 2022). It can easily permeate the lipopolysaccharide layer of cell membranes, penetrate cytoplasmic membrane and cytoplasm of gram-negative bacteria, causing intracellular component leakage (da Silva et al., 2018). Additionally, eugenol has been shown to be highly phytotoxic to several plant species, including amaranth, ryegrass, quinoa, and barnyard grass (Evans et al., 2009), which inhibits the germination and early growth of wild oats, reduces plant photosynthetic efficiency and chlorophyll content, and induces the accumulation of excess ROS (Ahuja et al., 2015).
The application of allelochemicals to control weeds is a preferable method for resistant weeds in sustainable agricultural systems. The molecular mechanism that BCP or eugenol, as major allelochemicals, used to induce death of cells in plants remains unknown. In the present study, the effect of BCP or eugenol on the growth of A. thaliana seedlings was investigated. A combination of transcriptome and WGCN analysis was used to identify key genes, the results extend our knowledge related to the specific action modes of BCP/eugenol on plants.
2 Material and methods
2.1 Plant materials and growing conditions
The A. thaliana seeds were first sterilized by dipping them in 75% ethanol for 1 minute, followed by a 10-minute soak in a 5% NaClO solution. Afterwards, they were washed at least five to ten times with sterilized water. Next, the seeds were placed on Murashige and Skoog (MS) agar medium and stratified at 4° for three days. Subsequently, they were cultured for 15 days under photoperiod at 8 hours light/16 hours dark at 28°C. Following this, uniform seedlings were transplanted into pots containing sterilized perlite and vermiculite and irrigated with a ½ Hoagland solution once every two days for one week. Then the plants were irrigated with 1/2 Hoagland supplemented with 0 (control), 450, 900, and 1800μM of BCP or eugenol once every two days for 7 days. The shoot and root tissues were harvested separately, for analysis.
2.2 Measurement of chlorophyll, malondialdehyde, the relative electrolytic leakage, H2O2, O2-
Chlorophyll extraction was conducted using ethanol as a solvent (Adil et al., 2022). The Malondialdehyde (MDA) content of leaves and roots was estimated using the thiobarbituric (TBA) method, as described by (Xing et al., 2022). The relative electrolytic leakage was measured according to methods of Huang (Huang et al., 2021).
The accumulation of H2O2 was detected by DAB staining at room temperature (Wu et al., 2019). Fresh leaves of different treatment groups were soaked in buffer solution for 24 hours, and then immersed in 100 alcohol: oil (3:1) for 15 minutes. Finally, the leaves were photographed and recorded.
The content of superoxide anion was determined by hydroxylamine oxidation (Kozuleva et al., 2015). 0.2g sample was added to 2 mL 65mM phosphoric acid buffer(PH 7.8), and fully ground, and the supernatant was taken as the extraction solution of superoxide anion separation at 12000 rpm for 10 min. Take 1 mL of extract, add 0.75 ml of phosphoric acid buffer (pH 7.8) and 0.25 mL of 10 mM hydroxylamine hydrochloride. After 25 bath reaction for 30 min, continue to add 2 mL of 7 mM alpha-naphthylamine and 2 mL of 17 mM p-aminobenzenesulfonic acid, 30 OD value was measured at 530 nm after bath reaction for 30 min.
2.3 Determination content of pectin, lignin, cellulose and hemicellulose in shoots and roots
The isolation of cell walls from plants followed the method described by Hu et al. (Hu et al., 2018). First, fresh shoot or root samples were ground in a mortar with pre-cooled PBS (10 mM) buffer to obtain homogenization, which was then vacuum filtered. The resulting filtrate was washed and filtered four times with PBS buffer, followed by washing with acetone until the filtrate became colorless. The obtained filtrate was the cell wall fragments. These fragments were then extracted and dried at 65°C for future use.
Pectin was extracted by boiling 2 mg of cell wall extract in 1 ml of ultrapure water three times, each for 1 hour. After each boiling, the mixture was centrifuged at 4500 g for 3 minutes, and the supernatant was collected into one tube. The uronic acid content was measured using galacturonic acid as a reference, as described by Voragen et al. (Voragen et al., 2009). Specifically, 100 μl of the supernatant was mixed with 500 μl of 98% (v/v) H2SO4 containing 0.0125 M Na2B4O7 and boiled for 5 minutes. After cooling to room temperature, 10 μl of 0.15% (w/v) m-hydroxydiphenyl dissolved in 0.5% (w/v) NaOH was added, and the sample solution was incubated at room temperature for 15 minutes. The absorbance was measured at 520 nm using a spectrophotometer UV-1800PC.
To analyze the content of lignin, 0.1g of cell wall was added to 500 μl of 98% (v/v) H2SO4 and dissolved by adding 2 mL of acetyl bromo-glacial acetic acid (25%). The mixture was heated in a 70° water bath for 30 min, followed by addition of 0.9 mL of NaOH (2M) solution to terminate the reaction. After cooling to room temperature, 2 mL of glacial acetic acid and 0.1 mL of 5 M hydroxylamine hydrochloride were added. The volume was made constant to 7 mL with glacial acetic acid and the mixture was centrifuged at 4500 rpm for 5 min. Then, 200 μL of the supernatant was taken and the absorbance was measured at 280 nm using a spectrophotometer, as described by Hatfield et al. (Hatfield et al., 1999).
To measure the content of cellulose, 0.1g of the cell wall was incubated with 4 mL of cellulase (1 mg/L) for 37 hours under dark conditions. Then, 2 mL of 25% trichloroacetic acid was added and boiled for 10 minutes. The mixture was then centrifuged and the supernatant was collected. Cellulose content in the coarse cell wall was determined by the phenol-sulfuric acid method using glucose as a standard sugar. Briefly, 200 μL of the supernatant was mixed with 50 μL of 2% phenol and 3,500 μL of H2SO4 in a tube and boiled for 15 minutes. After cooling down to room temperature, the absorbance of the supernatant was measured at 490 nm (Hu et al., 2018).
The analysis of hemicellulose was carried out as follows. A 0.1g sample was mixed with 10 ml of PBS and boiled on a heater for 30 minutes. The resultant mixture was subjected to centrifugation and the supernatant was discarded. The pellets were rinsed thrice with distilled water, following which 10 ml of 4M KOH was added to the mixture and stirred at room temperature for 24 hours. The mixture was again centrifuged and the resulting supernatant was collected. The hemicellulose content in coarse cell wall was determined using the phenol-sulfuric acid method with glucose as the standard sugar (Sun et al., 2012).
2.4 Measurement of endogenous hormone in shoots and roots
The content of plant endogenous hormones (IAA, ETH, ABA, SA, JA) in A. thaliana seedlings was measured using ELISA kits. Endogenous hormones were extracted according to a modified method described by Chakraborty et al. (Chakraborty et al., 2013). Specifically, 0.1 g of shoots or roots tissues of each sample were ground with a mortar and pestle at liquid N2 in 0.9 mL of 50 mM PBS buffer. The extract was then centrifuged at 4000 rpm for 15 min and the supernatant was used to detect the concentration of hormones according to the instructions on the ELISA kit. A standard curve was obtained using standard hormones at different known concentrations (3, 6, 12, 24, and 48 pmol/L) and the concentrations of hormones produced were calculated by comparison with the standard curve.
2.5 RNA Extraction, library construction, and illumina sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Prior to library construction, total RNA was treated with RNase-free DNase I (New England Biolabs, USA) to remove any contamination of genomic DNA. After extraction, the RNA concentration was determined using a NanoDrop1000 spectrophotometer. The integrity of RNA was assessed by electrophoresis on a 1% agarose gel. The RNA samples with intact 28S and 18S ribosomal RNA bands were considered to be of good quality and suitable for downstream applications such as reverse transcription and PCR. The mRNA was enriched using oligo (dT) magnetic beads (Qiagen) and fragmented. First-strand cDNA was synthesized using a random hexamer primer, followed by the generation of second-strand cDNA using RNase H and DNA polymerase I. The resulting cDNA was purified, and end reparation and poly (A) addition were performed. Then, sequencing adapters were ligated to the cDNA, and the library was purified through agarose gel electrophoresis and enriched by PCR amplification to generate the final cDNA library. The cDNA libraries were sequenced on the Illumina HiSeq™ 2000 platform using paired-end technology at Yuanshen (Shanghai, China). The resulting clean RNA-seq reads were mapped to the Arabidopsis reference genome.
2.6 Data processing and differential expression
The normalized transcript abundance of the genes was calculated using the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) method. Subsequently, differential expression analysis was conducted using edge R software. The false discovery rate (FDR) was used to determine the threshold of P value in multiple tests to control for false positives. In this study, a threshold of FDR ≤ 0.05 and log2|FC (ratio of BCP or Cary/control)| ≥ 1 were used to determine the significant differences in gene expression between the BCP or eugenol treated samples and the control.
Enrichment analysis of Gene Ontology (GO) terms was performed on significant DEGs using the AgriGO platform (http://geneontology.org/docs/go-enrichment-analysis/). Pathway analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://david.abcc.ncifcrf.gov/, accessed on 15 April 2021) was used to elucidate significantly enriched pathways of the DEGs. In the present study, both GO terms and KEGG pathways with Q-values ≤ 0.05 were considered significantly enriched in DEGs. The heat map was plotted using the OmicShare tools (www.omicshare.com/tools).
2.7 Weighted gene co-expression network analysis
A matrix of normalized expression values of DEGs identified by p-value and FDR-based method was analyzed using the weighted gene correlation network analysis (WGCNA) package. Pearson correlations between pairs of DEGs were used to calculate a similarity matrix, and the results were transformed into an adjacency matrix using the following equation: aij = [0.5 ∗ (1 + cor(i, j)]β, where aij represents the connection strength between DEGs. The scale-free topology with R2 cutoff (0.8) was used to choose the soft-thresholding power beta of the co-expression network. Topological Overlap Matrix (TOM) was implemented to cluster DEGs, and a dynamic tree cut algorithm was used to construct gene co-expression modules. Two parameters, including containing at least 50 genes and a cut height higher than 40, were defined to select modules. Genes were clustered into 14 correlated modules.
To further study the gene modules associated with BCP or eugenol treatment, the correlation coefficients between module eigengenes and different samples were calculated. Modules with the highest correlation coefficients were selected as target gene modules, after BCP or eugenol treatment. GO and KEGG analysis is performed for genes in the selected target module. Gene significance (GS) and module correlation degree (MM) of each gene was calculated by the R package and genes with high connectivity tended to be hub genes which may have important functions. Network visualization for target module was performed using the Cytoscape software version 3.9. The gene co-expression network is a scale-free weighted gene network with multiple nodes connected to different nodes via edges. Each node represents a gene, which is connected to a different number of genes. The gene which is connected to a greater number of genes is denoted with a bigger size and is more important for its interaction with a large number of genes.
2.8 qRT-PCR confirmation of the RNA-seq
The RNA-seq data’s reliability was validated by means of qRT-PCR. RNA extraction and purification were undertaken, followed by cDNA synthesis using the RevertAid™ Kit (Fermentas). 16 genes were selected randomly for qRT-PCR analysis. To normalize the cDNA levels in each reaction, A. thaliana Actin gene was employed as endogenous control for relative expression calibration. Supplementary Table 1 contains the primer sequences utilized for the genes selected. Maxima SYBR Green Master Mix (Thermo Scientific) was used for qRT-PCR, which was conducted using the Real-time Quantitative PCR System (iQ5, Bio-Rad, USA), with three repetitions performed. The 2−ΔΔCT method was utilized to analyze the relative expression data.
2.9 Statistical analysis
All experiments were performed in triplicate, and the results were presented as mean ± standard error of the mean (SEM). Statistical analysis of experimental data was performed using GraphPad Prism 8.0 software and SPSS 25.0 software. Data were subjected to analysis of variance using one-way ANOVA, and statistical differences between control and treated groups were determined. P-values less than 0.05 were considered statistically significant.
3 Results
3.1 Effect of both BCP and eugenol on plant physiological parameters
3.1.1 Both BCP and eugenol inhibited growth of A. thaliana plants and damaged their integrity of cells
Our results revealed that BCP or eugenol caused wilting symptoms and obviously inhibited the growth of the seedlings compared to the control group (Supplementary Figures 1A, B). The number of branches and leaves, fresh weight, root length, and plant height were significantly reduced after treatment with BCP or eugenol, except for the lower concentration of BCP or eugenol (100 μM) (Supplementary Figures 1C-F).
Treatment with BCP or eugenol (450, 900, 1800 μM) reduced the content of chlorophyll (Supplementary Figure 2C) in leaves and significantly increased the levels of REC (Supplementary Figure 2A), MDA (Supplementary Figure 2B), O2- (Supplementary Figure 2D), and H2O2 (Supplementary Figure 2E) in both shoots and roots of A. thaliana seedlings compared to the control, except for treatment with concentration at 100 μM BCP or eugenol. These findings indicated that BCP or eugenol caused severe damage to the cell membrane integrity, ultimately affecting the normal growth of seedlings.
3.1.2 BCP and eugenol damaged on components of cell wall in A. thaliana seedlings
After treatment with BCP or eugenol, the content of lignin, pectin, cellulose and hemicellulose, which are the main components of plant cell wall, were decreased in roots and shoots of A. thaliana seedlings compared to control groups (Figure 1). The results suggested that both BCP and eugenol affected the injured the structure and stability of the cell wall, leading to damage to the plant cell.
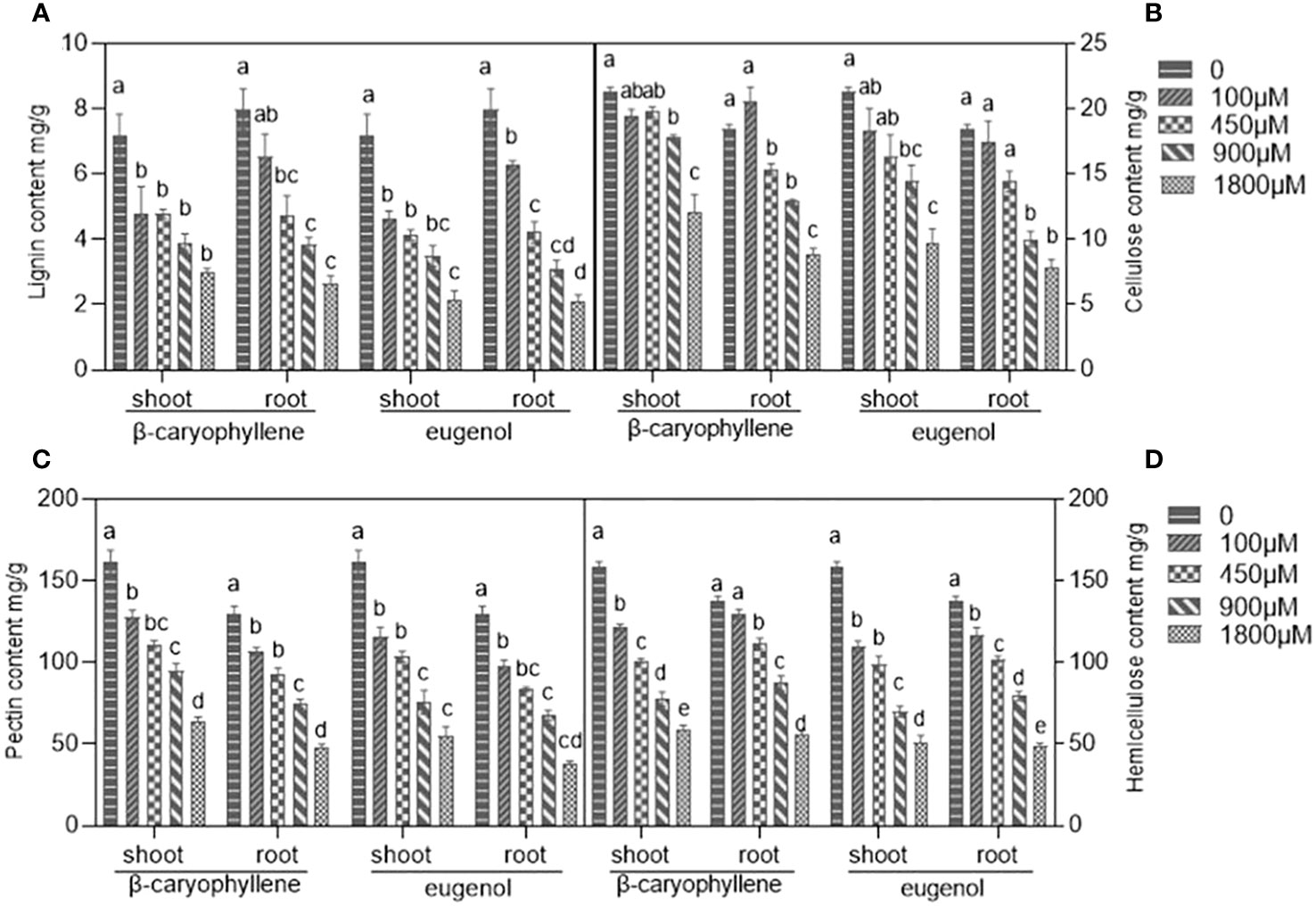
Figure 1 The effects of BCP or eugenol on lignin (A), cellulose (B), pectin (C) and hemicellulose (D) content in Arabidopsis thaliana seedlings. Different lower letters indicate the significant differences of multiple comparisons (P<0.05) using the least significant difference (LSD) method.
3.1.3 Effects of BCP or eugenol on hormone content of A. thaliana seedlings
As shown in Figure 2, the content of phytohormones, including ABA, SA, and ETH in the shoots and roots of seedlings were significantly increased after treatment with BCP or eugenol compared to the control group, except for the IAA, which decreased. The JA content in the shoots and roots of the seedlings increased significantly in response to BCP but decreased in response to eugenol. These results indicated that BCP or eugenol severely disrupted hormone balance of A. thaliana seedlings.
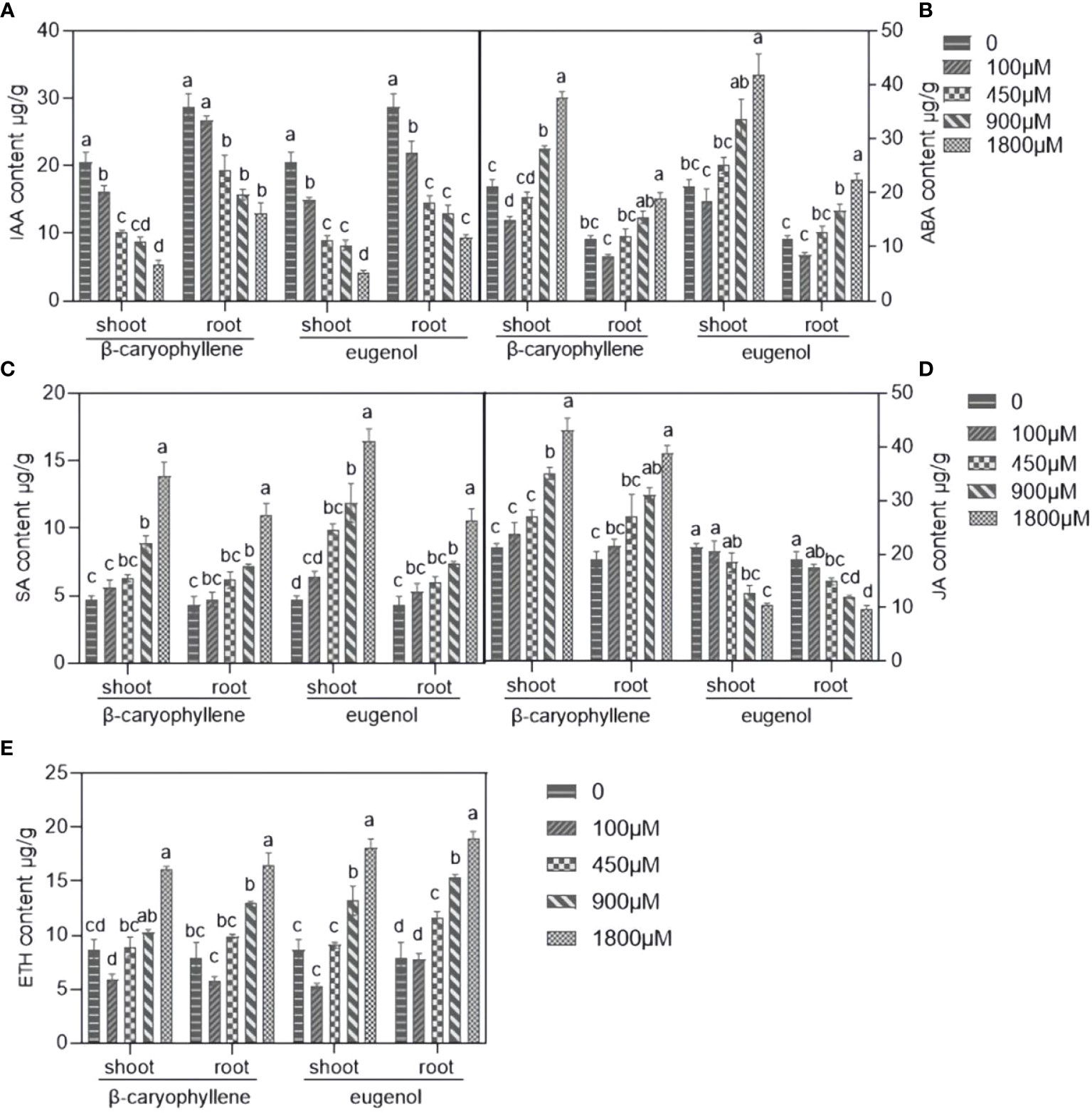
Figure 2 The effects of BCP or eugenol on IAA (A), ABA (B), SA (C), JA (D) and ETH (E) content in Arabidopsis thaliana seedlings. Different lower letters indicate the significant differences of multiple comparisons (P<0.05) using the least significant difference (LSD) method.
3.2 Differential gene expression of A. thaliana in response to BCP or eugenol
We conducted transcriptome analysis of 18 samples of A. thaliana in roots and shoots under treatment with 0 and 900 μM BCP or eugenol using Illumina paired-end sequencing, resulting in a total of 56.93 GB of clean data. Each sample provided 6.48 GB of data with a Q30 base percentage of 93.43% or higher. The alignment efficiency of each sample’s Clean Reads with the defined reference genome ranged from 86.40% to 93.73%.
There were 2620 differentially expressed genes (DEGs) in response to BCP in the A. thaliana seedlings, among which, 921 and 374 genes were up-regulated, 722 and 603 genes were down-regulated in shoots and roots respectively. There were 4561 DEGs to eugenol, among which, 910 and 755 genes were up-regulated, but 2140 and 756 gene down-regulated in shoots and roots, respectively (Figure 3).
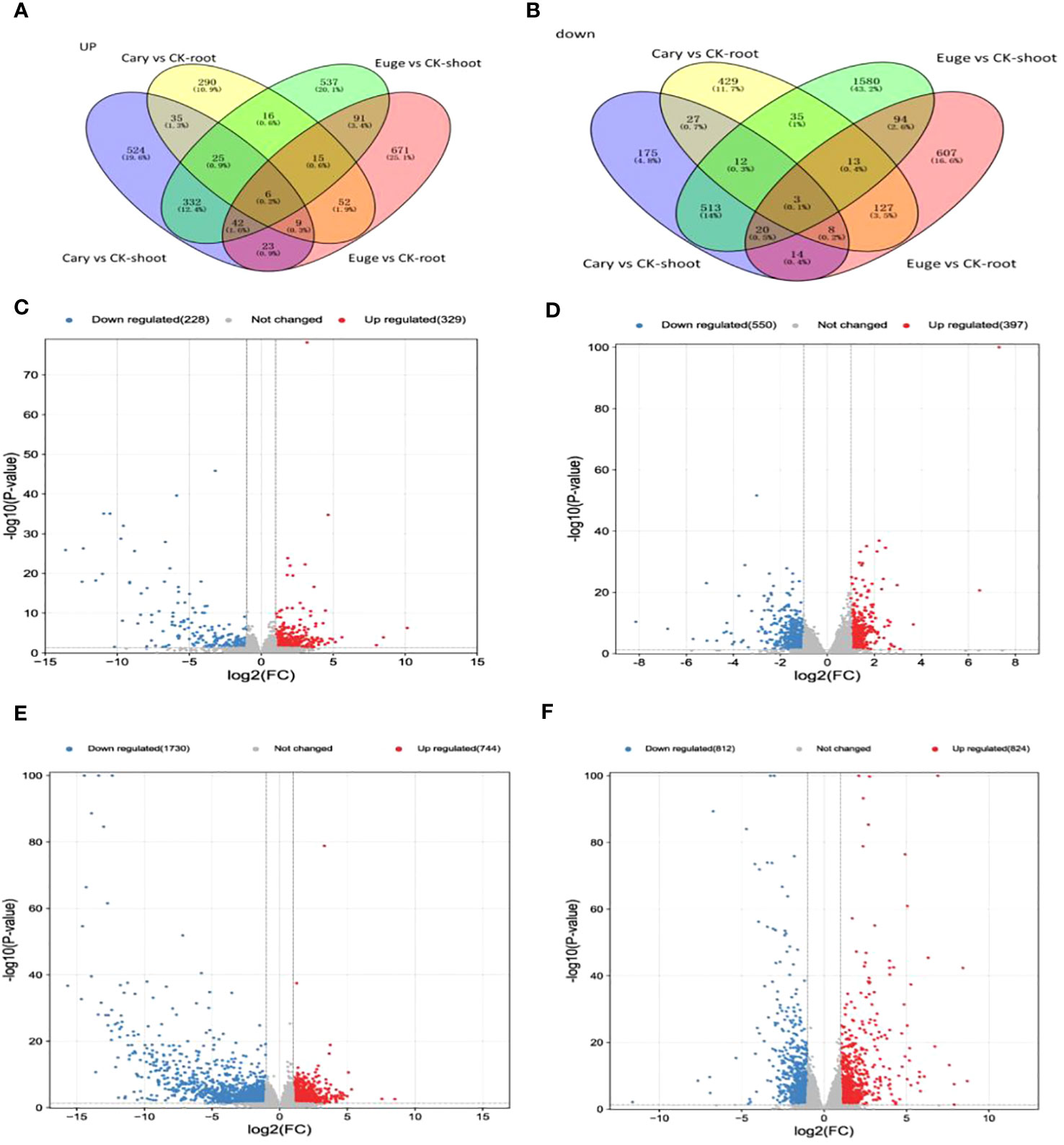
Figure 3 Venn diagram for DEG upregulated (A) and downregulated (B) after BCP treatment or eugenol treatment; Volcanic plot of DEGs in shoot (C) and root (D) after BCP treatment. Volcanic plot of RNA-Seq data using log2 fold change in shoot (E) and root (F) by eugenol treatment. Red and blue dots denote up- and down-regulated genes, respectively, Gray dots denote genes with no significant expression.
3.3 Gene ontology classification and KEGG pathway enrichment of DEGs
The identified DEGs were annotated into the three main GO functional categories, including cellular component (CC), molecular function (MF), and biological process (BP). According to the GO annotations, unigenes were classified into different functional categories.
The GO enrichment analysis showed significant differences in the functional enrichment of genes in response to BCP (Figures 4A-D). DEGs in shoots treated with BCP were categorized into five major sub-categories, including “response to biotic stimulus (GO:0009607),” “response to other organisms (GO:0051707),” “response to external biotic stimulus (GO:0043207), response to hormone (GO:0009725) “ and “defense response (GO:0006952), in the “biological process” category.” Moreover, cellular component category contained terms such as “apoplast (GO:0048046),” “cell wall (GO:0005618),” “external encapsulating structure (GO:0030312),” and “cell periphery (GO:0071944).” In the “molecular function” category, the most abundant sub-categories were “tetrapyrrole binding (GO:0046906),” “oxidoreductase activity (GO:0016491),” and “enzyme inhibitor activity (GO:0004857)” (Figure 4A). Regarding the GO annotation of DEGs in roots, the “biological category” showed that the most DEGs were associated with five subterms: “response to chemical (GO:0042221) “, “response to toxic substance (GO:0009636) “,”response to hormones (GO:0009725),” and “response to endogenous stimulus(GO:0009719), “response to abiotic stimulus GO:0009628”.”The “cell component” category contained three main sub-categories, which were “chloroplast thylakoid membrane protein complex (GO:0098807),” “external encapsulating structure (GO:0030312),” and “extracellular region (GO:0005576).” For the “molecular function” category, the most DEGs were associated with “DNA-binding transcription factor activity (GO:0003700),” “transcription regulator activity (GO:0140110),” “tetrapyrrole binding (GO:0046906),” and “quercetin 3-O-glucosyltransferase activity (GO:0080043)” (Figure 4B).
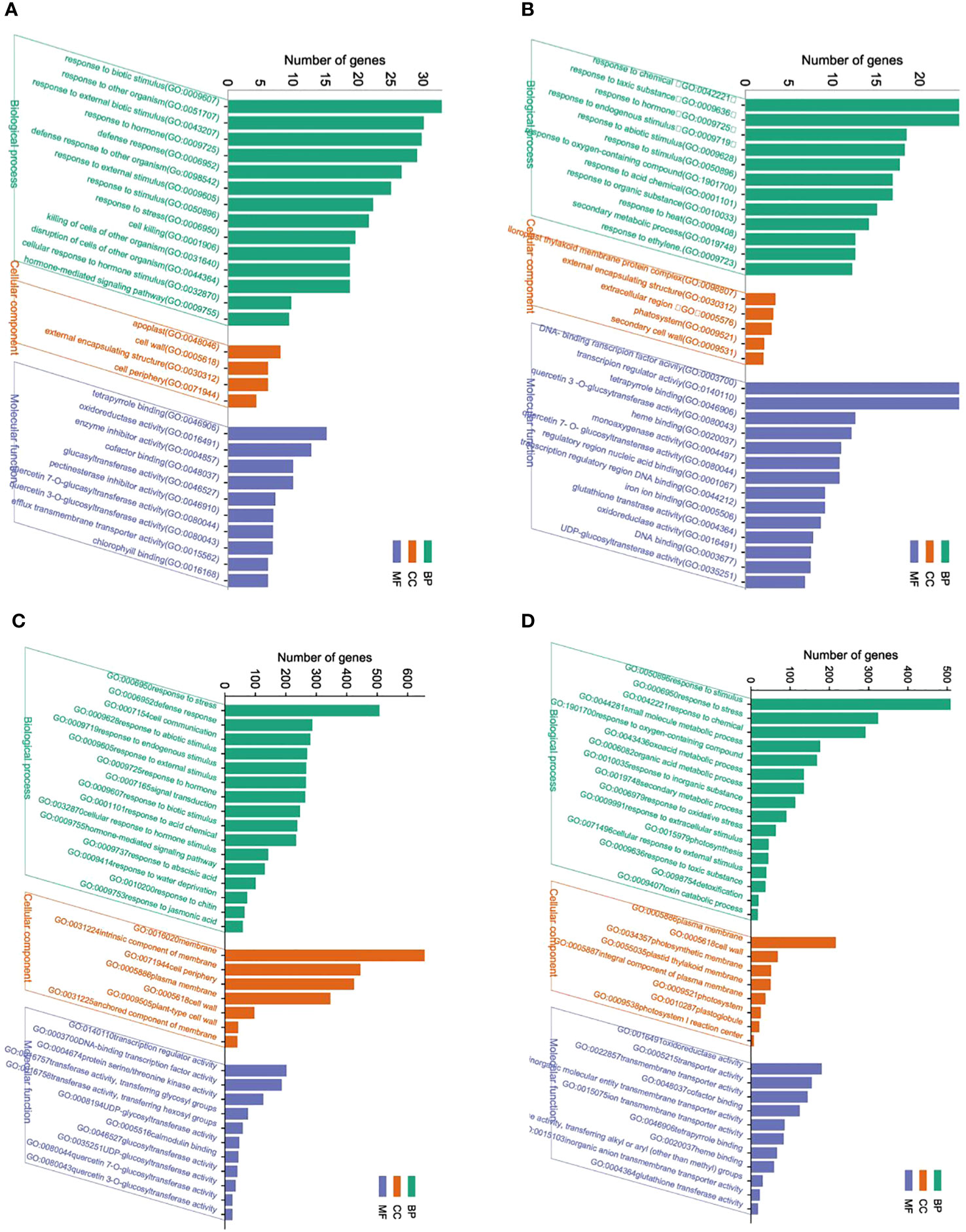
Figure 4 GO functional annotation and classification of DEG of shoot (A) and roots (B) in A. thaliana seedlings after BCP or GO functional annotation and classification of DEG of shoot (C) and roots (D) in A. thaliana seedlings after with eugenol treatment.
Upon treatment with eugenol, DEGs in shoots were associated with three major sub-categories in the biological process category: “response to stress (GO:0006950)”, “defense response (GO:0006952)”, and “cell communication (GO:0007154)”. The most prevalent terms in the cell component category were “membrane (GO:0016020)”, “intrinsic component of membrane (GO:0031224)”, and “cell periphery (GO:0071944)”. In the molecular function category, the three main terms were “transcription regulator activity (GO:0140110)”, “DNA-binding transcription factor activity (GO:0003700)”, and “protein serine/threonine kinase activity (GO:0004674)” (as shown in Figure 4C). For roots, DEGs were predominantly assigned into the biological process category, where the three most abundant terms were “response to stimulus (GO:0050896)”, “response to stress (GO:0006950)”, and “response to chemical (GO:0042221)”. The cell component category primarily involved three sub-categories: “plasma membrane (GO:0005886)”, “cell wall (GO:0005618)”, and “photosynthetic membrane (GO:0034357)”. The most prevalent sub-categories in the molecular function category were “oxidoreductase activity (GO:0016491)”, “transporter activity (GO:0005215)”, and “transmembrane transporter activity (GO:0022857)” (as illustrated in Figure 4D).
The results of KEGG enrichment analysis of shoots and roots after BCP treatment were presented in Figure 5A. The DEGs were distributed across 30 pathways in the shoots. The significant pathways included glutathione metabolism (ko00480), cell cycle (ko04110), cell cycle-yeast (ko04111), plant-pathogen interaction (ko04626), plant hormone signal transduction (ko04075), oxidative phosphorylation (ko00190),biosynthesis of antibiotics (ko01130), microbial metabolism in diverse environments (ko01120), and ribosome(ko03010).Similarly, the detected DEGs in roots were mainly enriched in biosynthesis of amino acids (ko01230), plant hormone signal transduction (ko04075), biosynthesis of antibiotics (ko01130), microbial metabolism in diverse environments (ko01120), Ribosome (ko03010) and plant hormone signal transduction (ko04075) (Figure 5B).
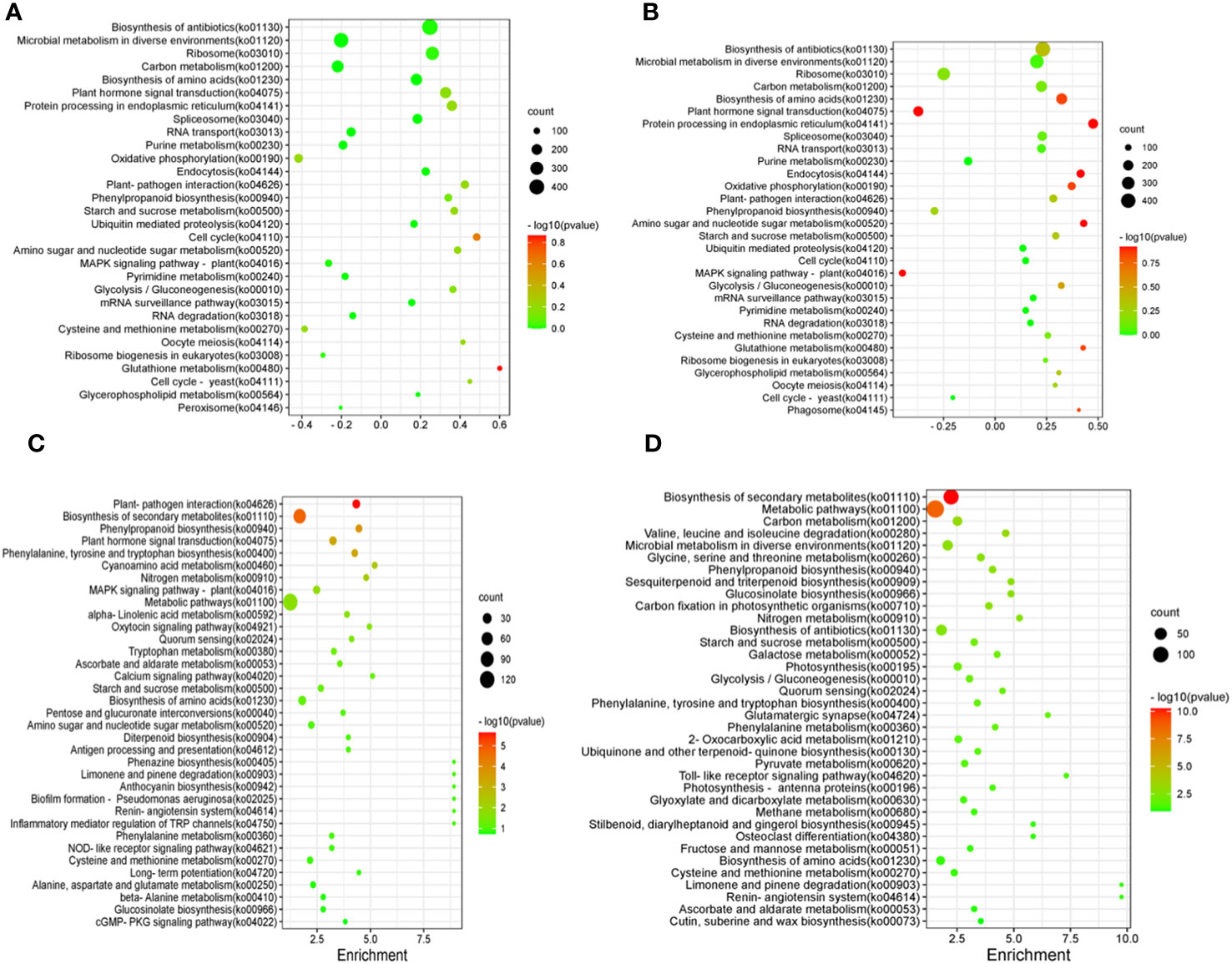
Figure 5 Differentially expressed Gene KEGG pathway enrichment of shoot in A. thaliana seedlings after BCP (A) or eugenol (C) treatment and differentially expressed Gene KEGG pathway enrichment of root in A. thaliana seedlings after BCP (B) or eugenol (D) treatment.
After treatment with eugenol, the KEGG enrichment analysis revealed that DEGs in shoots of A. thaliana seedlings were enriched in 35 pathways. The major pathways included plant-pathogen interaction (ko04626), biosynthesis of secondary metabolites (ko01110), phenylpropanoid biosynthesis (ko00940), plant hormone signal transduction (ko04075), phenylalanine, tyrosine and tryptophan biosynthesis (ko00400), and cyanoamino acid metablism (ko00460) (Figure 5C). DEGs in roots were enriched mainly in biosynthesis of secondary metabolites (ko01110), metabolic pathways (ko01100), carbon metabolism (ko01200), valine, leucine and isoleucine degradation (Figure 5D).
Both BCP and eugenol inhibited weed growth by inhibiting the biosynthesis of cell wall, while eugenol also damaged production of other secondary metabolites. They had diverse effects on the A. thaliana seedlings, affecting multiple pathways and processes.
3.4 DEGs involved in phenylpropanoids-related pathway and their expression pattern
Phenylpropanoids are various natural phenolic compounds in plants, which serve as vital structural components of cell walls and function as phytoalexins in protecting plants from herbivores and pathogens. Phenylpropanoid biosynthesis produces precursors for a wide range of phenolic compounds, including ferulic acid, p-coumaric acid, and caffeic acid (Glagoleva et al., 2022). A total of 47 and 39 DEGs, which associated with phenylpropanoid biosynthesis were identified in response to BCP or eugenol, respectively. These DEGs included genes involved in various process of the phenylpropanoid biosynthesis pathway such as phenylalanine ammonia lyase (PAL), caffeic acid 3-O-methyltransferase (COMT), trans cinnamate 4-monooxygenase (C4H), cinnamol dehydrogenase (CADH), 4-coumaric acid CoA ligase (4CL), laccase (LAC), coumaryl-3-hydroxylase (C3H), peroxidase (POD), cinnamyl-CoA reductase (CCR), and glucosyl transferase (UGT) (De Vries et al., 2021). Most of these DEGs, including C3H, CCR, COMT, and LAC genes, were down-regulated in both the shoots and roots after treatment with BCP or eugenol. However, the PAL, C4H, and 4CL genes were significantly up-regulated in response to BCP, but down-regulated in response to eugenol in both shoots and roots. Additionally, the cinnamyl alcoholdehydrogenase (CAD) gene was down-regulated under BCP treatment, while it was up-regulated in both the shoots and roots with eugenol treatment (Figures 6A, B).
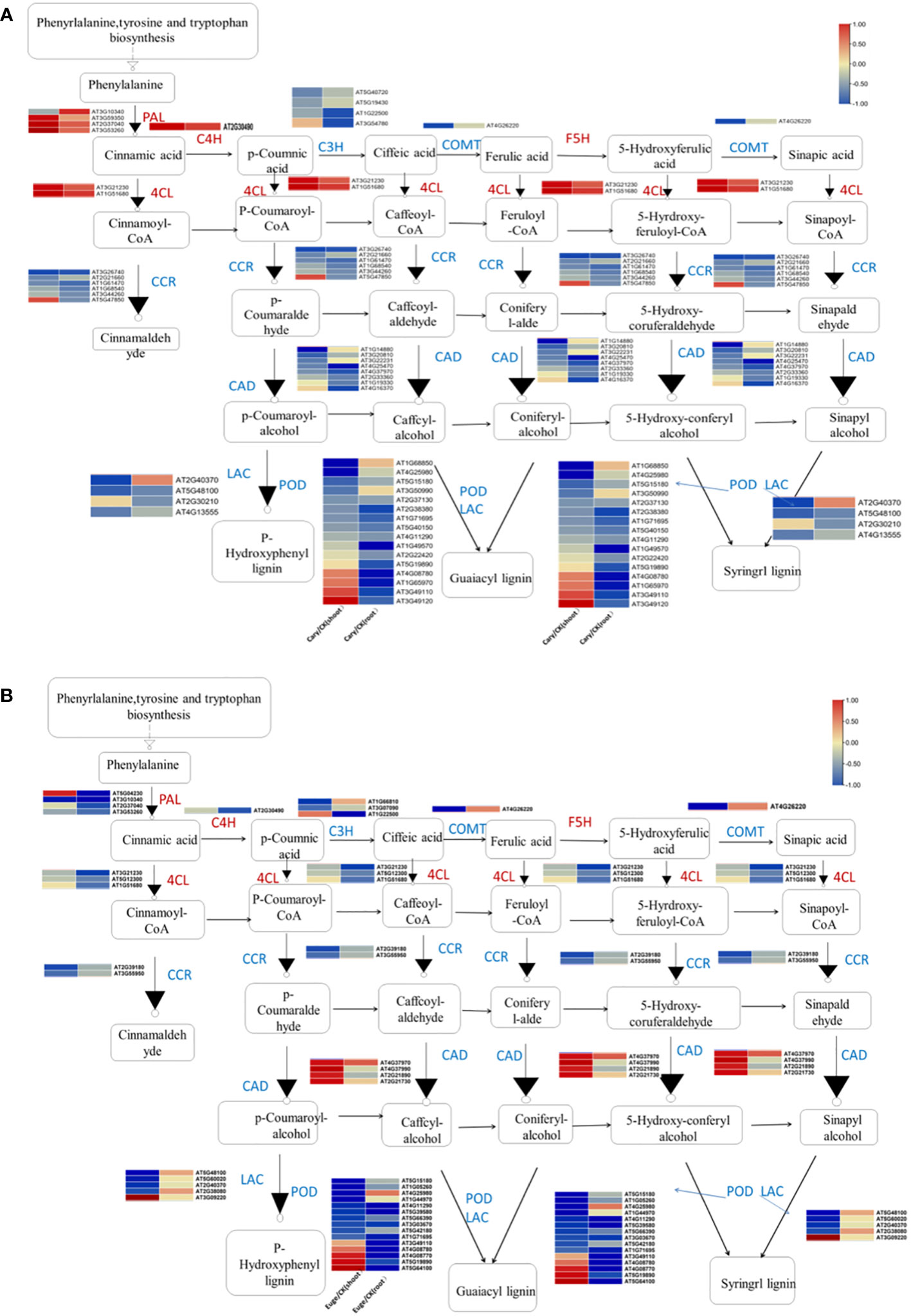
Figure 6 Analysis of genes expression related to phenylpropionic acid biosynthesis in A. thaliana seedlings by BCP (A) or eugenol (B) treatment.
The DEGs involved in the metabolism and biosynthesis of cell wall components were further analyzed. BCP or eugenol induced in the down-regulated expression of most DEGs in the biosynthesis of hemicellulose, pectin, and cellulose in both shoots and roots. Xylose and xyloglucan are the primary components of hemicellulose in dicotyledons. In A. thaliana, genes of the glycosyltransferase 43, 47 (GT43, GT47) family member irregular xylem(IRX), fra8 homolog(F8H), glucuronic acid replacement of xylan (GUX) are associated with xylan biosynthesis. Xyloglucan is mainly synthesized by cellulose synthetase similar to cellulose synthase like-C (CSLC) (Pitaksaringkarn et al., 2014; Sewelam et al., 2021), but xyloglucanendo-transglucosylase/hydrolase (XTH) catalyzes xyloglucan polymers to hydrolyze. BCP or eugenol lead to down-regulated expression of genes including IRX, F8H, GUX, and CSLC for hemicellulose synthesis, while the XTH family genes up-regulated in the shoots (Figures 7A, D).
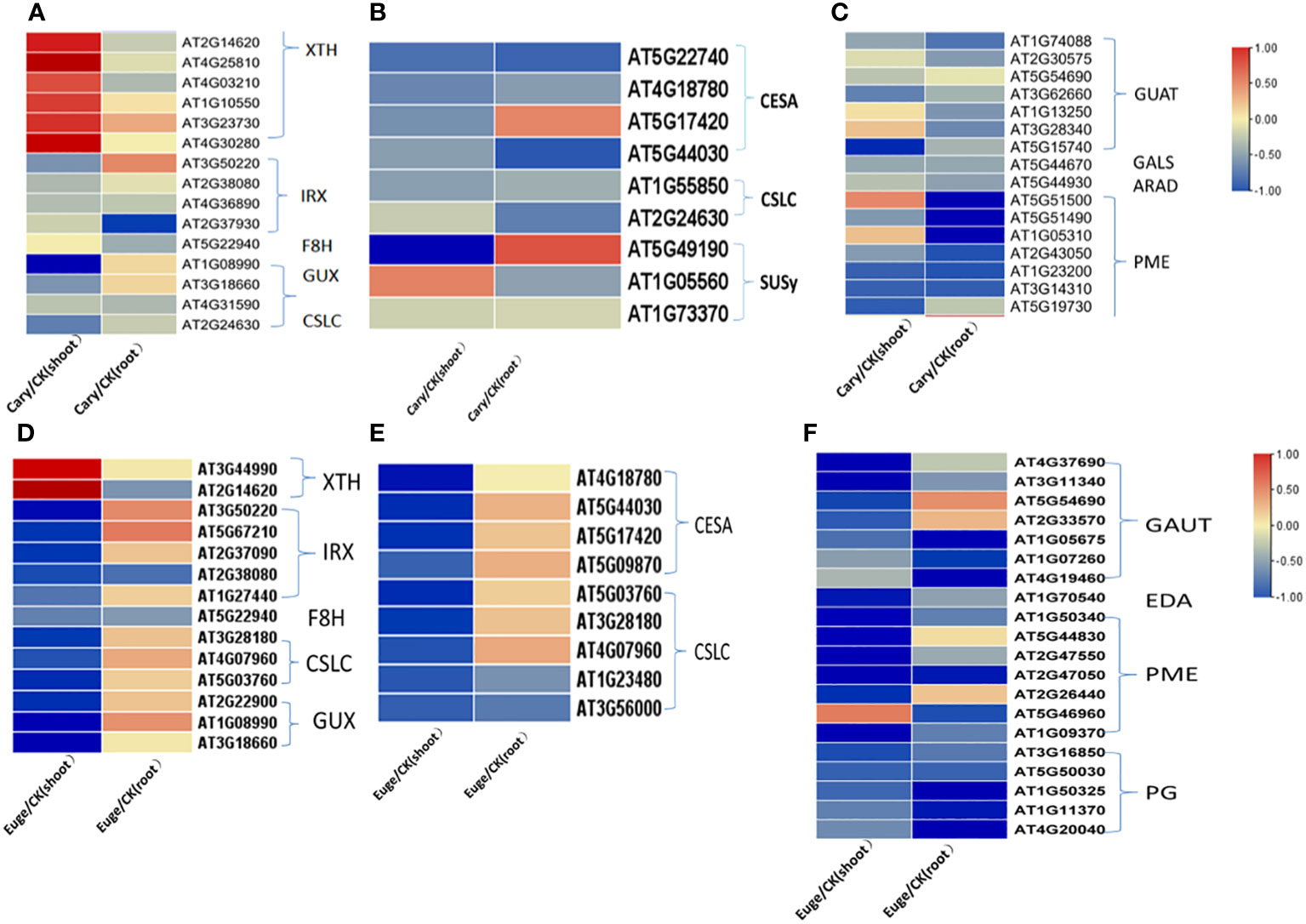
Figure 7 Analysis of gene expression related to the synthesis and metabolism of cell wall components hemicellulose (A), cellulose (B) and pectin (C) of A. thaliana, seedlings by BCP treatment; Eugenol treatment for the expression of hemicellulose (D), cellulose (E), pectin (F) cell wall components hemicellulose (D), cellulose (E), pectin (F) related gene expression of A. thaliana seedlings.
The biosynthesis of cellulose is catalyzed by cellulose synthase (CesA), cellulose synthase-like (CSL), and sucrose synthase (SUSy). In response to BCP or eugenol, some genes of CesA, and CSL and SUSy, were significantly down-regulated in the shoots and roots of seedlings (Figures 7B, E). Pectin in the cell wall is composed of homogalacturonan (HG), rhamnogalacturonan-I (RGI), and rhamnogalacturonan-II (RGII). Pectin synthesis involves the participation and regulation of various enzymes, including UDP-GlcA synthesis enzymes, galacturonosyltransferase (GAUT) protein family, and pectin methylesterase (PME), β-1,4galactosyltransferase (GALS), arabinosyltransferase (ARAD) and polygalacturonase (PG). The coordinated action of these enzymes enables the formation of different types of pectin in plant cell walls. Most genes of GAUT, PME and PG in shoots and roots were significantly down-regulated after exposure to BCP or eugenol (Figures 7C, F).
3.5 DEGs related to plant hormone signaling pathway
Previous research indicates that tryptophan, carotenoid, cysteine, and methionine, α-linolenic acid, and phenylalanine serve as the biosynthetic precursors for auxin, abscisic acid, ethylene, jasmonic acid, and salicylic acid, respectively (Zhao, 2018). In this study, our results characterized DEGs, which are enzymes involved in precursors biosynthesis or signaling transduction pathways including IAA, ABA, SA, JA, and BR in Arabdopsis seedlings in the response to BCP or eugenol (Figures 8A-E). The DEGs involved in IAA pathways in plants comprise flavin monooxygenase (YUCCA), tryptophan transaminase (TAA), and nitrilase (NIT) (Yu et al., 2022), which were down-regulated. Additionally, genes such as Auxin response factors (ARF), Auxin/indole acetic acid (AUX/IAA), Gretchen Hagen3 (GH3), and Small auxin up RNA (SAUR), which participate in IAA signal transduction at the earlier stages (Pellizzaro et al., 2020; de Figueiredo et al., 2022), were also down-regulated in the roots and shoots after treatment with BCP or eugenol, especially in roots (Figure 8A).
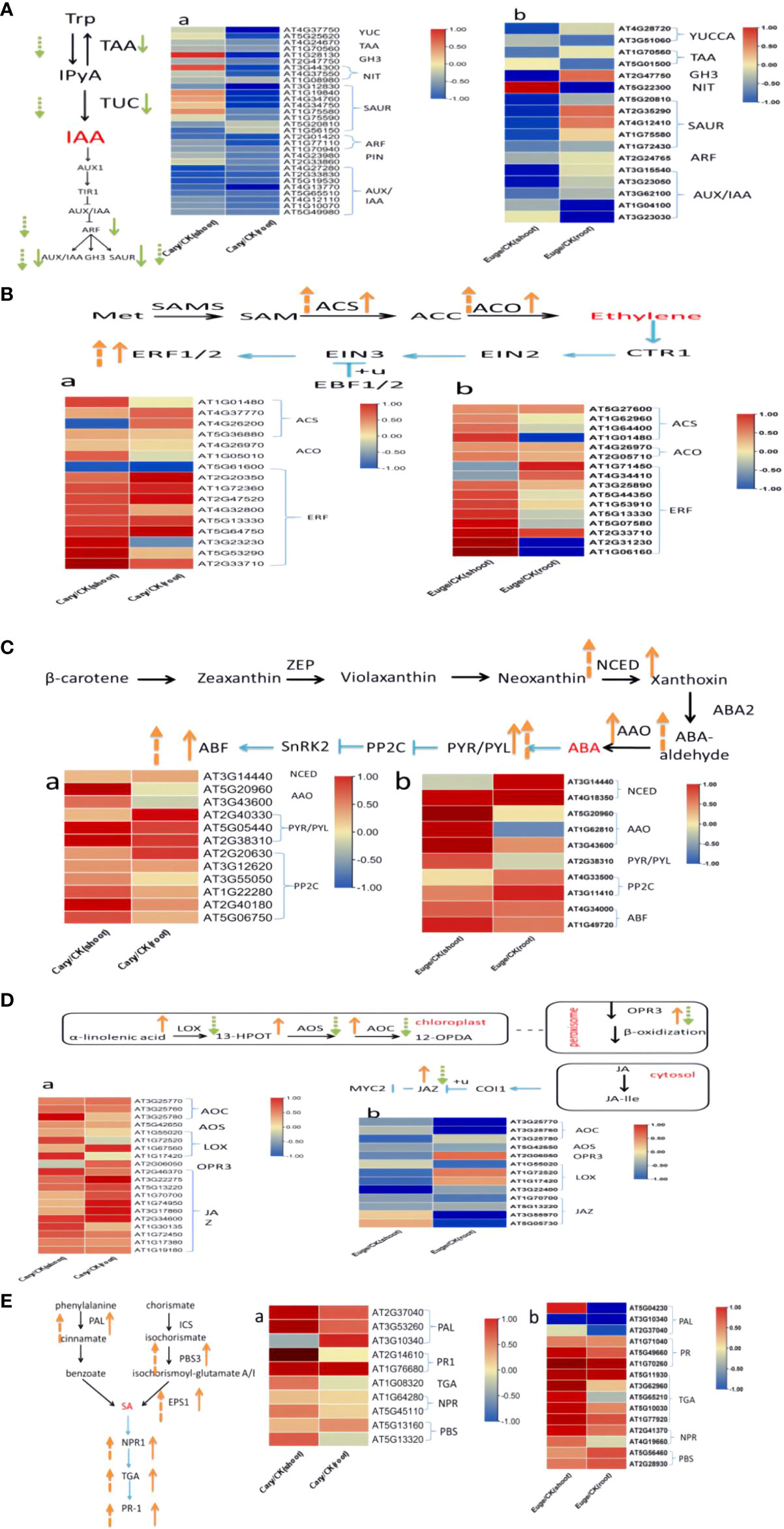
Figure 8 BCP (A) and eugenol (B) treatment for the analysis of DEGs in A. thaliana endogenous hormone auxin (A), ethylene (B), abscisic acid (C), jasmonic acid (D) and salicylic acid (E) synthesis and signal transduction, respectively. The orange arrow in the figure indicates gene up-regulation, and the green arrow indicates down-regulation; the solid arrow represents BCP treatment, and the dotted arrow represents eugenol treatment.
The genes ACC synthase (ACS) and ACC oxidase (ACO) encode enzymes which catalyze the synthesis of ethylene (Li et al., 2020a), these genes were significantly up-regulated in response to BCP or eugenol. Additionally, transcription factors (ERFs) that play a role in the transmission of the ethylene signal were also up-regulated (Figure 8B).
The genes of 9-cis-epoxy dioxygenase (NCED) and acetic aldehyde oxidase (AAO), which are related to ABA synthesis (Fan et al., 2009), were up-regulated in both shoots and roots after treatment with BCP or eugenol. Similarly, ABA receptor gene pyrabactin resistance 1-like (PYL) and protein phosphatase 2C (PP2C) gene were up-regulated as well (Figure 8C). The key genes, in the process of jasmonic acid synthesis including lipoxygenase (LOX), allene oxide cyclase (AOC), 12-oxo plant dienoic acid reductase (OPR3), and acyl coenzyme A oxidase (ACX) (Fidyt et al., 2016), were all up-regulated after treatment with BCP. However, the expression of LOX, AOC, OPR3, and allene oxide synthase (AOS) genes was down-regulated with eugenol. Jasmonate ZIMdomain (JAZ), a protein localized in the cell nucleus that plays a role in jasmonic acid signal transduction, was up-regulated after treatment with BCP but down-regulated with eugenol (Figure 8D). The genes of phenylalanine ammonia lyase (PAL) and aminotransferase (PBS3), which are related to salicylic acid synthesis (Vlot et al., 2009), were significantly up-regulated after treatment with BCP or eugenol. Key genes, involved in disease resistance including pathogenesis-related1 (PR1), TGACG-binding factor (TGA), and nonexpressor of PR genes (NPR) in the SA signal transduction pathway, were also up-regulated (Figure 8E).
3.6 Weighted gene co-expression network construction and module identification
To identify different co-expressed modules under treatment with BCP or eugenol in A. thaliana, we conducted WGCNA analysis to construct the gene co-expression network. We performed gene expression clustering tree and hierarchical clustering tree analyses on all 18 samples to calculate correlation coefficients for each sample’s expression level. The soft threshold β=5 was determined when the fitting curve was close to 0.8 for the first time (Figure 9A). The modules with similar expression were merged by the dynamic cutting tree method.
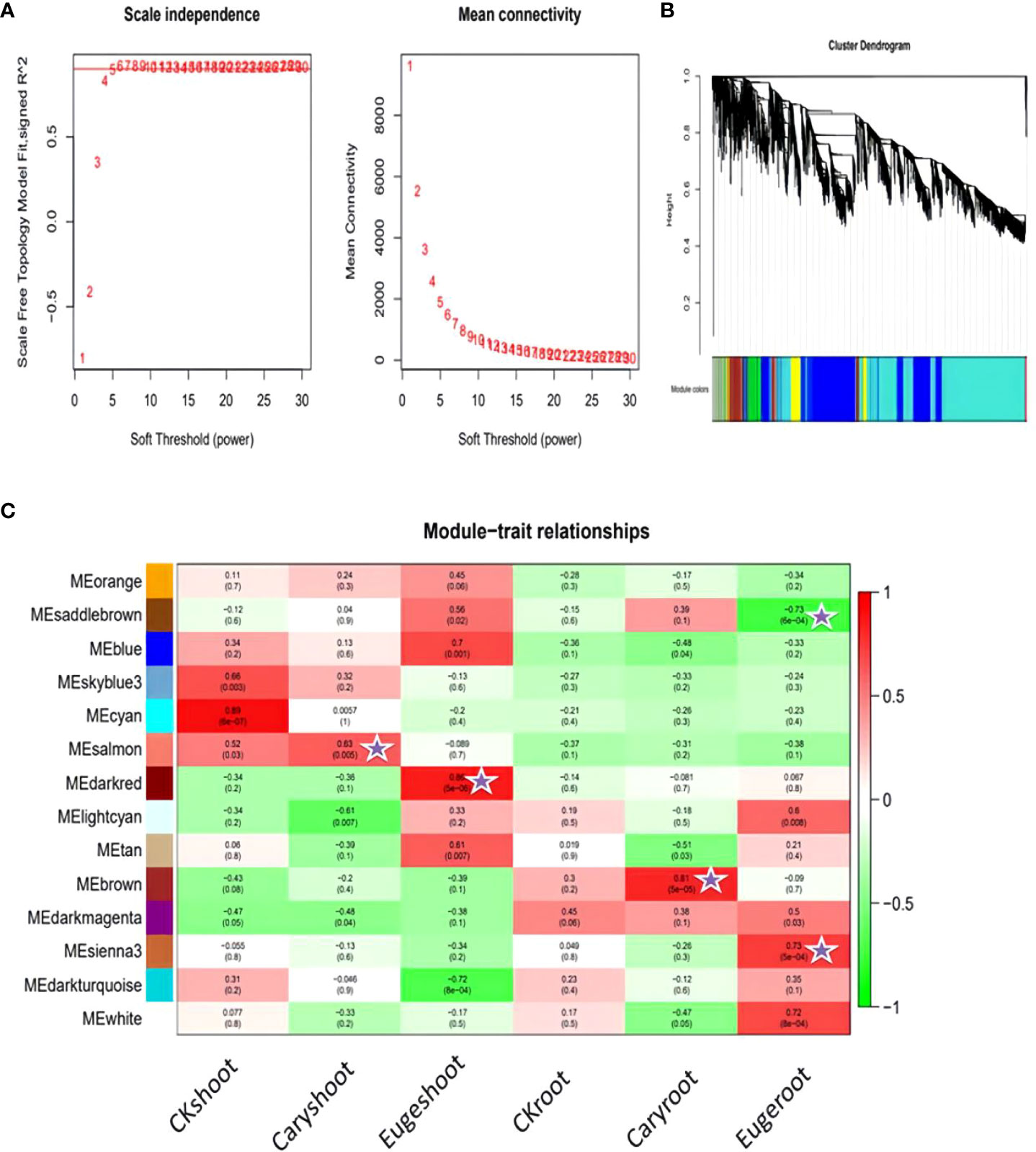
Figure 9 Association analysis of gene co-expression network modules with physiological and biochemical traits: (A) Soft threshold determination (B) Module detection by gene cluster dendrograms (C) Module-trait associations revealed by Pearson correlation coefficient.
As a result, DEGs of different groups were assigned into 14 modules (Figure 9B), with the largest module containing 2693 genes and the smallest module containing 59 genes. We analyzed the correlation between modules and samples, and found that the salmon and brown modules were highly correlated with shoot and root of the seedlings after treatment with BCP, respectively. Similarly, the darked and sienna3 modules were highly correlated with shoot and root of the seedlings after eugenol, respectively (Figure 9C).
3.7 Functional annotation of candidate hub genes
The hub genes in each module with the most significant differential expression pattern across control and treatment were analyzed using Cytoscape. The genes in the target module were sorted using the degree algorithm and GS value, and the top 10 hub genes were selected to construct the core gene network diagram. There were 5, 9, 9, and 10 genes in the Salmon, Brown, Darked, and Sienna3 modules, respectively (Figures 10A-D). A total of 24 genes (Supplementary Table S2) with high connectivity in the four target gene modules were identified as candidate hub genes. The functions of these hub genes were annotated by using the NCBI and TAIR databases.
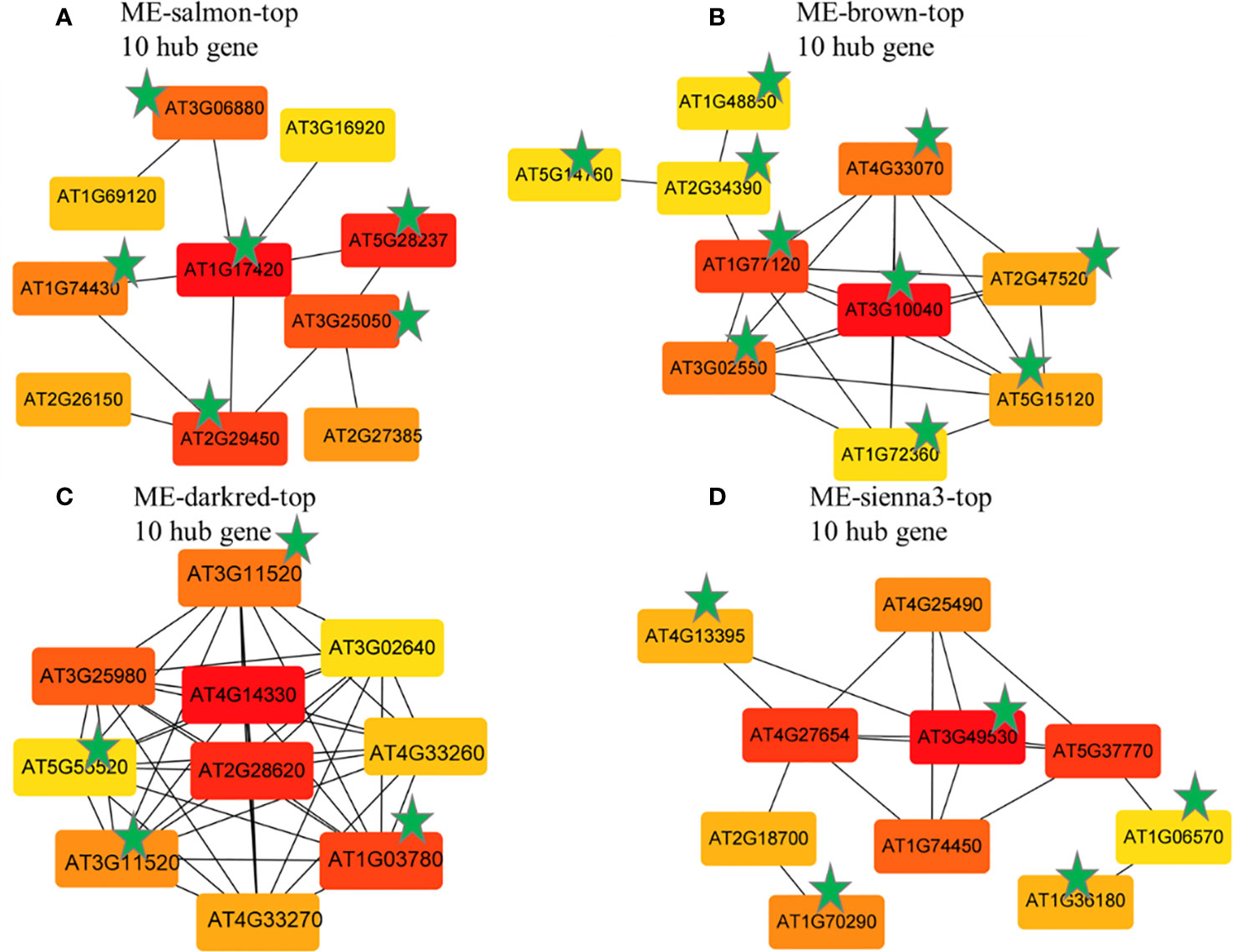
Figure 10 Top hub gene co-expression network of the MEsalmon (A), MEbrown (B), MEdarkred (C) and sienna3 (D) according to the degree value Note: The asterisk indicates the first 10 Hub genes shared by the degree algorithms and GS value.
Among the identified 16 genes in response to BCP, 7 up-regulated genes which encode HRA1 (hypoxia response attenuator1), ADH1 (alcohol dehydrogenase), DUF1637 (ADO, 2-aminoethanethiol dioxygenase), ERF71 (ethylene-responsive transcription factors), ERF73, PDC1 (pyruvate decarboxylase1), and LBD41 (LOB domain-containing protein) respectively, were related to hypoxia. Other up-regulated genes, including aspartate oxidase (AO), xyloglucan endotransglucosylase/hydrolase 3(XTH3), and lipoxygenase 3 (LOX3), were required for the ROS burst, abscission of organs, and cell separation, respectively. The genes down-regulated contained AT3G06880, GSTU5 (glutathione S-transferase class tau 5), AT5G28237 (MYB95), and NIP2.1, all of which played a pivotal role in plant survival. These results suggest that BCP can lead to plant hypoxia stress, which caused excessive ROS, thus brought about lethal effects on plants.
Among the 8 hub genes induced by eugenol, 4 genes were associated with cell division, including CDC20.1 (Cell Division Cycle 20), Cyclin B1-3(CYCB1), TPX2 (Targeting protein for Xklp2), and AT5G55520 (unknown function), which were down-regulated. The other 4 genes, including TPS8 (trehalose-6-P synthesis), PDS1(4-hydroxyphenylpyruvate dioxygenase), NAC062 (NAC domain-containing protein 62) and ACCase (Acetyl- coenzyme A carboxylase), which involved in response to external or internal environmental stress, were up-regulated. These results suggested that eugenol inhibited of cell division to lead to deleterious effects, caused the death of plants.
3.8 Real-time RT-PCR validation of differentially expressed genes
Real-time RT-PCR (qRT-PCR) was performed on 16 DEGs in response to BCP or eugenol. The results from sequencing and qRT-PCR were statistically analyzed, and a high correlation was observed with Pearson correlation coefficient R≈0.97 and R > 0.90 (Supplementary Figure 3). These results indicate that the transcriptome analysis accurately reflected the differential expression of A. thaliana genes under BCP or eugenol treatment, confirmed the reliability of the transcriptome data.
4 Discussion
4.1 The application of BCP and eugenol disrupted the endogenous hormone balance of plants, resulting in dysregulation of gene expression and regulation during growth and development
As natural compounds, both BCP and eugenol, were found to damage plant cell wall components, leading to a decrease in the plant’s ability to maintain cell shape and resist mechanical stress. These findings were consistent with transcriptome and WGCNA analyses, which demonstrated a significant down-regulation of genes involved in cell wall formation pathways, including lignin, cellulose, hemicellulose, and pectin biosynthesis (Hu et al., 2022). In addition, the expression of XTH, responsible for cell wall degradation, was found to be up-regulated. The down-regulation of genes related to cell wall component biosynthesis led to a reduced accumulation of lignin, pectin, cellulose, and hemicellulose, ultimately resulting in a decreased growth rate, developmental abnormalities, or even plant death when treatment with BCP or eugenol.
The growth and development of plant species can be disrupted by natural compounds derived from other plants, which affect their hormone balance. Lee et al. (2015) and Yan et al. (2020) conducted research that revealed how phenolic substances from locust seed extract inhibited the gibberellin pathway and promoted the accumulation of ABA, JA, and SA in plants. Similarly, Zhang et al. (2021) demonstrated that leaf aqueous extract reduced the IAA and GA content in grass seedlings, while increasing the ABA content. Plant stress responses are regulated by hormones such as IAA, ABA, JA, and ethylene (Yang et al., 2008; Li et al., 2020b). The biosynthesis and transport of these hormones are vital for plants to withstand abiotic and biotic stresses (Yang et al., 2008; Chen et al., 2020). However, an excess of hormones can have negative effects. For instance, excessive ABA can result in stomatal closure, reduced photosynthetic rate, and the generation of ROS, ultimately hindering plant growth and triggering senescence (Fan et al., 2009). Similarly, excessive SA can impede electron transport, induce stomatal closure, and inhibit photosynthesis (Ye et al., 2013). Although JA regulates various plant responses, including root growth inhibition, anthocyanin accumulation, and leaf senescence (Mori and Schroeder, 2004), an excessive accumulation of JA can cause stomatal closure, further reducing photosynthesis (Ye et al., 2013). In our study, the application of BCP or eugenol increased ROS levels and significantly altered endogenous hormone levels. The content of ABA, ethylene, JA, and SA increased significantly, while IAA decreased substantially. This disruption of the plant’s endogenous hormone balance inhibited plant growth.
Accordingly, the expression of key enzyme genes involved in the biosynthesis and transduction of hormones such as ethylene, ABA, JA, SA, and IAA in multiple metabolic pathways, was induced in response to BCP and eugenol treatments. This suggested that the application of BCP and eugenol externally disrupted the balance of plant endogenous hormones, leading to disorders in gene expression and regulation during growth and development.
4.2 BCP caused excessive accumulation of ethylene and low oxygen conditions, disrupting metabolic pathways and reducing the plant’s ability to withstand adverse environments while reducing the plant’s ability to detoxify
Characterization of the hub genes provided insight into the molecular mechanism of allelopathy between plants and adjacent organisms. Based on the 16 hub genes that responded to BCP, it can be inferred that BCP leads to excessive ethylene accumulation as well as low oxygen conditions. The expression of ADH1 genes, which is typically regulated by stress-related transcription factors such as ERF71 and ERF73 (Tan and Zwiazek, 2019), was up-regulated. The LBD41 gene has been reported to be associated with hypoxia responses and linked to the ethylene pathway, such as under oil stress in Arabidopsis (Guedes et al., 2018). HRA1 is usually induced by anaerobic conditions, leading to alcohol fermentation in plants (Abdeeva et al., 2018). The expression of PDC1 is strongly up-regulated during flooding in rice (Hess et al., 2011; Zhang et al., 2016). Both cysteine dioxygenase (CDO) and cysteamine dioxygenase (ADO, 2-aminoethanethiol dioxygenase) are essential for taurine biosynthesis, and taurine acts as a biological O2 sensor. In plants, aspartate is oxidized by aspartate oxidase (AO) to produce iminoaspartate, ultimately leading to the synthesis of NAD+, which plays a critical role in maintaining metabolic balance (Saito et al., 2022). AO is also required for RBOHD-dependent stomatal closure, which limits gas exchange between plants and the atmosphere (Macho et al., 2012). Here, the up-regulation of genes LBD41, HRA1, PDC1, ADO, and AO implies that BCP limited gas exchange between the plant and the atmosphere, resulting in low oxygen levels inside the cells.
Lipoxygenase (LOX) is responsible for catalyzing the peroxidation of polyunsaturated fatty acids (Suzuki et al., 2015). In Petunia hybrida, PhCS plays a crucial role in the synthesis of chlorophylls, carotenoids, and anthocyanins (Masson et al., 2019). The F-BOX STRESS INDUCED (FBS) proteins have been associated with environmental stress networks, and WD40 repeat-like proteins are known targets of SCFFFBSI complexes (Sepulveda-Garcia et al., 2021). The up-regulation of LOX, CS, and LOX3 genes suggested that BCP induced stress and damaged the metabolic pathway. These results were consistent with the studies of Wang, who have demonstrated that herbicides can regulate LOX3, ADH1, and ERF (Wang et al., 2021).
In plants, MYBs have various roles, such as regulating secondary cell-wall formation and participating in cell communication. At5g28237 (MYB95) encodes tryptophan synthase (Xiao et al., 2021), which is responsible for synthesizing tryptophan, a precursor for IAA. Environmental stresses often cause an increase in H2O2 levels, leading to damage to plant cells, Glutathione S-transferase (GST) utilizes glutathione as a substrate to remove H2O2 (Horváth et al., 2020). In this study, the down-regulation of At5g28237 indicated a decrease in IAA synthesis. Furthermore, the down-regulation of GST suggested that the application of BCP reduced the plant’s ability to withstand adverse environments.
4.3 Eugenol treatment inhibited cell division and decreased the protective effect against osmotic stress and oxidative stress
Eight hub genes were identified under treatment with eugenol, most of which were involved in cell division. Specifically, CYCA, CYCB, and CYCD are known to be involved in controlling the S-phase, G2-to-M-phase, and G1 phase in the cell cycle, respectively (Dante et al., 2014; Niu et al., 2015). CDC20.1 is necessary for the separation of sister chromosomes. TPX2 mediates the assembly and nucleation of microtubules, as well as mitotic spindle assembly in cell division (Tomastikova et al., 2020). Additionally, it is estimated that NAC062 plays a role in regulating the cell cycle, relaying ER stress signaling from the plasma membrane to the nucleus, and regulating UPR downstream gene expression (Yang et al., 2014). Here, the upregulation of CYCB1;3, NAC062, and TPX2 under eugenol treatment indicated that eugenol inhibited cell division and induced ER stress.
ACCase is known to play a pivotal role in fatty acid metabolism (Wang et al., 2018). Transcript changes of ACCase showed that eugenol affected fatty acid metabolism. In Arabidopsis, the enzyme HPPD (also known as PDS1) catalyzes the oxidative decarboxylation and rearrangement of p-hydroxyphenylpyruvate (HPP) to homogentisate (HGA), which is a precursor of tocopherols and plastoquinone, crucial co-factors in photosynthesis. Ahrens et al. (Ahrens, et al., 2013) stated that a mutant of HPPD displays albino and dwarf phenotypes with chlorophyll degradation. Downregulation of PDS1 through eugenol exposure lead to bleaching of Arabidopsis, indicating that the synthesis of vital pigments and co-factors required for plant growth and protection against oxidative stress may be impacted by eugenol.
Exogenous trehalose regulates antioxidant production and osmotic balance to improve cold stress tolerance in rapeseed (Raza et al., 2022). Trehalose biosynthesis involves the conversion of Glc-6-P and UDP-Glc to trehalose-6-P by TPS. The AtTPS8 gene is induced in leaves when sugar starvation occurs during extended night periods (Harthill et al., 2006). Eugenol treatment resulted in the down-regulation of TPS8 genes, leading to dysfunction of sugar signaling at night or reduction in protection from osmotic stress.
4.4 The ability of these BCP and eugenol to target multiple genes through allelopathy allowed them to overcome tolerance to chemical herbicides and could serve as excellent candidates for natural biological herbicides
Natural herbicides derived from the allelopathy of compounds can provide effective alternatives to chemical herbicides for sustainable agricultural practices (Ahuja et al., 2015). In this study, we discovered that BCP and eugenol inhibited the growth of A. thaliana seedlings by affecting the expression of various genes involved in different metabolic pathways. The ability of these natural components to target multiple genes through allelopathy made them capable of overcoming tolerance to chemical herbicides. They induced oxidative stress pathways, disrupted the structure of the cell wall, and affected the plant’s endogenous hormone balance, thereby affecting its morphological phenotype. In terms of gene expression and regulation, BCP treatment primarily resulted in hypoxia stress, while eugenol caused cell division failure. In conclusion, BCP and eugenol have phytotoxic potential, making them excellent candidates for natural biological herbicides.
5 Conclusions
This study firstly provided molecular mechanism of inhibitory action of BCP and eugenol on plants. BCP and eugenol disrupted the endogenous hormone balance of plants, resulting in dysregulation of gene expression and regulation during growth and development. Among the hub genes responsive to BCP, several genes were up-regulated, including HRA1, ADH1, ADO, ERF71 and ERF73, PDC1, and LBD41, all of which are related to hypoxia. Conversely, the down-regulated gene GSTU5 is involved in detoxification, a crucial process for plant survival. Therefore, BCP causes excessive accumulation of ethylene and low oxygen conditions, disrupting metabolic pathways and reducing the plant’s ability to withstand adverse environments. It also impairs the plant’s detoxification abilities, leading to detrimental effects on plants in the long run. Regarding eugenol, four genes associated with cell division, CDC20.1, Cyclin B1-3, TPX2, and AT5G55520, were found to be down-regulated. Eugenol also down-regulated genes of ACCase and PDS1, involved in fatty acid metabolism and photosynthesis, respectively. These findings suggest that eugenol inhibits cell division and decreases the plant’s ability to survive. The continuous use of chemical herbicides has led to weed resistance and poses serious environmental threats. The application of BCP and eugenol could be an environmentally friendly and effective strategy for controlling resistant weeds.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YG: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing, Proofreading. CL: Writing – original draft, Writing – review & editing, Methodology, Software. YZ: Writing – original draft, Writing – review & editing, Methodology. SZ: Formal analysis, Software, Methodology. PC: Formal analysis, Software, Methodology. XW: Writing – review & editing, Methodology. TZ: Methodology, Writing – original draft, Writing – review & editing, Formal analysis, Proofreading.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the scientific and technological projects (Grant Number 231111111200) in Henan province, China.
Acknowledgments
The authors thank the reviews for constructive comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1295779/full#supplementary-material
References
Abdeeva, I. A., Pogorelko, G. V., Maloshenok, L. G., Mokrykova, M. V., Fursova, O. V., bruskin, S. A. (2018). Search for partner proteins of arabidopsis thaliana immunophilins involved in the control of plant immunity. Molecules 23 (4), 953. doi: 10.3390/molecules23040953
Adil, M., Bashir, S., Bashir, S., Aslam, Z., Ahmad, N., Younas, T., et al. (2022). Zinc oxide nanoparticles improved chlorophyll contents, physical parameters, and wheat yield under salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.932861
Ahrens, H., Lange, G., Müller, T., Rosinger, C., Willms, L., van Almsick, A. (2013). 4-Hydroxyphenylpyruvate dioxygenase inhibitors in combination with safeners: solutions for modern and sustainable agriculture. Angew Chem. Int. Ed Engl. 52 (36), 9388–9398. doi: 10.1002/anie.201302365
Ahuja, N., Singh, H. P., Batish, D. R., Kohli, R. K. (2015). Eugenol-inhibited root growth in Avena fatua involves ROS-mediated oxidative damage. Pesticide Biochem. Physiol. 118, 64–70. doi: 10.1016/j.pestbp.2014.11.012
Busi, R., Goggin, D. E., Heap, I. M., Horak, M. J., Jugulam, M., Masters, R. A., et al. (2018). Weed resistance to synthetic auxin herbicides. Pest Manage. Sci. 74 (10), 2265–2276. doi: 10.1002/ps.4823
Chakraborty, U., Chakraborty, B. N., Chakraborty, A. P., Sunar, K., Dey, P. L. (2013). Plant growth promoting rhizobacteria mediated improvement of health status of tea plants. Indian J. Biotechnol. 12(1), 20–31.
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62 (1), 25–54. doi: 10.1111/jipb.12899
Da Camara, C. A. G., Doboszewski, B., De Melo, J. P. R., Nazarenko, A. Y., Dos Santos, R. B., Moraes, M. M. (2022). Novel insecticides from alkylated and acylated derivatives of thymol and eugenol for the control of plutella xylostella (Lepidoptera: plutellidae). J. Braz. Chem. Soc. 33 (2), 196–204. doi: 10.21577/0103-5053.20210137
Dante, R. A., Sabelli, P. A., Nguyen, H. N., Leiva-Neto, J. T., Tao, Y. M., Lowe, K. S., et al. (2014). Cyclin-dependent kinase complexes in developing maize endosperm: evidence for differential expression and functional specialization. Planta 239 (2), 493–509. doi: 10.1007/s00425-013-1990-1
da Silva, F. F. M., Monte, F. J. Q., de Lemos, T. L. G., do Nascimento, P. G. G., Costa, A. K. D., de Paiva, L. M. M. (2018). Eugenol derivatives: synthesis, characterization, and evaluation of antibacterial and antioxidant activities. Chem. Cent. J. 12, 34. doi: 10.1186/s13065-018-0407-4
de Figueiredo, M. R. A., Kupper, A., Malone, J. M., Petrovic, T., de Figueiredo, A., Campagnola, G., et al. (2022). An in-frame deletion mutation in the degron tail of auxin coreceptor IAA2 confers resistance to the herbicide 2,4-D in Sisymbrium orientale. Proc. Natl. Acad. Sci. U S A. 119 (9), e2105819119. doi: 10.1073/pnas.2105819119
De Vries, W., Schulte-Uebbing, L., Kros, H., Voogd, J. C., Louwagie, G. (2021). Spatially explicit boundaries for agricultural nitrogen inputs in the European Union to meet air and water quality targets. Sci. Total Environ. doi: 10.1016/j.scitotenv.2021.147283
Duke, S. O. (2015). Perspectives on transgenic, herbicide-resistant crops in the United States almost 20 years after introduction. Pest Manage. Sci. 71 (5), 652–657. doi: 10.1002/ps.3863
Evans, G. J., Bellinder, R. R., Goffinet, M. C. (2009). Herbicidal Effects of Vinegar and a Clove Oil Product on Redroot Pigweed (Amaranthus retroflexus) and Velvetleaf (Abutilon theophrasti). Weed Technol. 23, 292–299. doi: 10.1614/WT-08-158.1
Fan, J., Hill, L., Crooks, C., Doerner, P., Lamb, C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150 (4), 1750–1761. doi: 10.1104/pp.109.137943
Fidyt, K., Fiedorowicz, A., Strządała, L., Szumny, A. (2016). BCP and BCP oxide-natural compounds of anticancer and analgesic properties. Cancer Med. 5 (10), 3007–3017. doi: 10.1002/cam4.816
Fu, X., Gao, Y., Yan, W. Y., Zhang, Z. L., Sarker, S., Yin, Y. Y., et al. (2022). Preparation of eugenol nanoemulsions for antibacterial activities. Rsc Adv. 12 (6), 3180–3190. doi: 10.1039/d1ra08184e
Glagoleva, A. Y., Vikhorev, A. V., Shmakov, N. A., Morozov, S. V., Chernyak, E. I., Vasiliev, G. V., et al. (2022). Features of activity of the phenylpropanoid biosynthesis pathway in melanin-accumulating barley grains. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.923717
Guedes, F. A. D., Rossetto, P. D., Guimaraes, F., Wilwerth, M. W., Paes, J. E. S., Nicolas, M. F., et al. (2018). Characterization of Laguncularia racemosa transcriptome and molecular response to oil pollution. Aquat. Toxicol. 205, 36–50. doi: 10.1016/j.aquatox.2018.09.001
Harthill, J. E., Meek, S. E. M., Morrice, N., Peggie, M. W., Borch, J., Wong, B. H. C., et al. (2006). Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 47 (2), 211–223. doi: 10.1111/j.1365-313X.2006.02780.x
Hatfield, R. D., Grabber, J., Ralph, J., Brei, K. (1999). Using the acetyl bromide assay to determine lignin concentrations in herbaceous plants: Some cautionary notes. J. Agric. Food Chem. 47 (2), 628–632. doi: 10.1021/jf9808776
Heap, I. (2014). Global perspective of herbicide-resistant weeds. Pest Manage. Sci. 70 (9), 1306–1315. doi: 10.1002/ps.3696
Hess, N., Klode, M., Anders, M., Sauter, M. (2011). The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiologia Plantarum 143 (1), 41–49. doi: 10.1111/j.1399-3054.2011.01486.x
Horváth, E., Bela, K., Gallé, Á., Riyazuddin, R., Csomor, G., Csenki, D., et al. (2020). Compensation of mutation in Arabidopsis glutathione transferase (AtGSTU) genes under control or salt stress conditions. Int. J. Mol. Sci. 21 (7), 2349. doi: 10.3390/ijms21072349
Hu, S., Kamimura, N., Sakamoto, S., Nagano, S., Takata, N., Liu, S., et al. (2022). Rerouting of the lignin biosynthetic pathway by inhibition of cytosolic shikimate recycling in transgenic hybrid aspen. Plant J. 110 (2), 358–376. doi: 10.1111/tpj.15674
Hu, H., Zhang, R., Tao, Z., Li, X., Li, Y., Huang, J., et al. (2018). Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in arabidopsis. Plant Cell Physiol. 59 (6), 1144–1157. doi: 10.1093/pcp/pcy050
Huang, H., Morgan, C. M., Asolkar, R. N., Koivunen, M. E., Marrone, P. G. (2010). Phytotoxicity of sarmentine isolated from long pepper (Piper longum) fruit. J. Agric. Food Chem. 58 (18), 9994–10000. doi: 10.1021/jf102087c
Huang, D., Wang, Q., Zhang, Z., Jing, G., Ma, M., Ma, F., et al. (2021). Silencing MdGH3-2/12 in apple reduces drought resistance by regulating AM colonization. Horticulture Res. 8 (1), 84. doi: 10.1038/s41438-021-00524-z
Kozuleva, M., Klenina, I., Mysin, I., Kirilyuk, I., Opanasenko, V., Proskuryakov, I., et al. (2015). Quantification of superoxide radical production in thylakoid membrane using cyclic hydroxylamines. Free Radical Biol. Med. 89, 1014–1023. doi: 10.1016/j.freeradbiomed.2015.08.016
Lee, S. M., Radhakrishnan, R., Kang, S. M., Kim, J. H., Lee, I. Y., Moon, B. K., et al. (2015). Phytotoxic mechanisms of bur cucumber seed extracts on lettuce with special reference to analysis of chloroplast proteins, phytohormones, and nutritional elements. Ecotoxicology Environ. Saf. 122, 230–237. doi: 10.1016/j.ecoenv.2015.07.015
Li, Z., Xiong, F. Q., Guo, W. F., Mo, C. M., Wu, H. N., Du, L. (2020b). The root transcriptome analyses of peanut wild species Arachis correntina (Burkart) Krapov. & WC Gregory and cultivated variety Xiaobaisha in response to benzoic acid and p-cumaric acid stress. Genet. Resour. Crop Evol. 67, 9–20. doi: 10.1007/s10722-019-00859-6
Li, Y. Y., Zhao, M., Chen, W. W., Du, H. Y., Xie, X. D., Wang, D. B., et al. (2020a). Comparative transcriptomic analysis reveals that multiple hormone signal transduction and carbohydrate metabolic pathways are affected by Bacillus cereus in Nicotiana tabacum. Genomics 112 (6), 4254–4267. doi: 10.1016/j.ygeno.2020.07.022
Macho, A. P., Boutrot, F., Rathjen, J. P., Zipfel, C. (2012). ASPARTATE OXIDASE plays an important role in arabidopsis stomatal immunity. Plant Physiol. 159 (4), 1845–1856. doi: 10.1104/pp.112.199810
Macias, F. A., Mejias, F. J. R., Molinillo, J. M. G. (2019). Recent advances in allelopathy for weed control: from knowledge to applications. Pest Manage. Sci. 75 (9), 2413–2436. doi: 10.1002/ps.5355
Macias-Rubalcava, M. L., Garrido-Santos, M. Y. (2022). Phytotoxic compounds from endophytic fungi. Appl. Microbiol. Biotechnol. 106 (3), 931–950. doi: 10.1007/s00253-022-11773-w
Masson, N., Keeley, T. P., Giuntoli, B., White, M. D., Puerta, M. L., Perata, P., et al. (2019). Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 365 (6448), 65–69. doi: 10.1126/science.aaw0112
McCoy, R. M., Widhalm, J. R., McNickle, G. G. (2022). Allelopathy as an evolutionary game. Plant Direct 6 (2), e382. doi: 10.1002/pld3.382
Mori, I. C., Schroeder, J. I. (2004). Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol. 135 (2), 702–708. doi: 10.1104/pp.104.042069
Niu, B., Wang, L., Zhang, L., Ren, D., Ren, R., Copenhaver, G. P., et al. (2015). Arabidopsis cell division cycle 20.1 is required for normal meiotic spindle assembly and chromosome segregation. Plant Cell 27 (12), 3367–3382. doi: 10.1105/tpc.15.00834
Pellizzaro, A., Neveu, M., Lalanne, D., Vu, B. L., Kanno, Y., Seo, M., et al. (2020). A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 225 (1), 284–296. doi: 10.1111/nph.16150
Pitaksaringkarn, W., Matsuoka, K., Asahina, M., Miura, K., Sage-Ono, K., Ono, M., et al. (2014). XTH20 and XTH19 regulated by ANAC071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems. Plant J. 80 (4), 604–614. doi: 10.1111/tpj.12654
Raza, A., Su, W., Jia, Z. Q., Luo, D., Zhang, Y., Gao, A., et al. (2022). Mechanistic insights into trehalose-mediated cold stress tolerance in rapeseed (Brassica napus L.) seedlings. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.857980
Sain, S., Naoghare, P. K., Devi, S. S., Daiwile, A., Krishnamurthi, K., Arrigo, P., et al. (2014). Beta caryophyllene and caryophyllene oxide, isolated from Aegle marmelos, as the potent anti-inflammatory agents against lymphoma and neuroblastoma cells. Anti-inflammatory anti-allergy Agents medicinal Chem. 13 (1), 45–55. doi: 10.2174/18715230113129990016
Saito, M., Konishi, M., Miyagi, A., Sakuraba, Y., Kawai-Yamada, M., Yanagisawa, S. (2022). Arabidopsis nitrate-induced aspartate oxidase gene expression is necessary to maintain metabolic balance under nitrogen nutrient fluctuation. Commun. Biol. 5 (1), 432. doi: 10.1038/s42003-022-03399-5
Sepulveda-Garcia, E., Fulton, E. C., Parlan, E. V., O’Connor, L. E., Fleming, A. A., Replogle, A. J., et al. (2021). Unique N-terminal interactions connect F-BOX STRESS INDUCED (FBS) proteins to a WD40 repeat-like protein pathway in arabidopsis. Plants-Basel 10 (10), 2228. doi: 10.3390/plants10102228
Sewelam, N., El-Shetehy, M., Mauch, F., Maurino, V. G. (2021). Combined abiotic stresses repress defense and cell wall metabolic genes and render plants more susceptible to pathogen infection. Plants-Basel 10 (9), 1946. doi: 10.3390/plants10091946
Sharma, C., Al Kaabi, J. M., Nurulain, S. M., Goyal, S. N., Kamal, M. A., Ojha, S. (2016). Polypharmacological properties and therapeutic potential of BCP: A dietary phytocannabinoid of pharmaceutical promise. Curr. Pharm. Des. 22 (21), 3237–3264. doi: 10.2174/1381612822666160311115226
Stone, M. J. (2017). Regulation of chemokine-receptor interactions and functions. Int. J. Mol. Sci. 18 (11), 2415. doi: 10.3390/ijms18112415
Sun, J. P., Tian, C. G., Diamond, S., Glass, N. L. (2012). Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in neurospora crassa. Eukaryot Cell. 11 (4), 482–493. doi: 10.1128/EC.05327-11
Suzuki, Y., Miura, K., Shigemune, A., Sasahara, H., Ohta, H., Uehara, Y., et al. (2015). Marker-assisted breeding of a LOX-3-null rice line with improved storability and resistance to preharvest sprouting. Theor. Appl. Genet. 128 (7), 1421–1430. doi: 10.1007/s00122-015-2516-y
Tan, X., Zwiazek, J. J. (2019). Stable expression of aquaporins and hypoxia-responsive genes in adventitious roots are linked to maintaining hydraulic conductance in tobacco (Nicotiana tabacum) exposed to root hypoxia. PloS One 14 (2), e0212059. doi: 10.1371/journal.pone.0212059
Tomastikova, E. D., Rutten, T., Dvorak, P., Tugai, A., Ptoskova, K., Petrovska, B., et al. (2020). Functional divergence of microtubule-associated TPX2 family members in arabidopsis thaliana. Int. J. Mol. Sci. 21 (6), 2183. doi: 10.3390/ijms21062183
Ullah, H., Di Minno, A., Santarcangelo, C., Khan, H., Daglia, M. (2021). Improvement of oxidative stress and mitochondrial dysfunction by BCP: A focus on the nervous system. Antioxidants (Basel) 10 (4), 546. doi: 10.3390/antiox10040546
Vlot, A. C., Dempsey, D. A., Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Voragen, A. G. J., Coenen, G. J., Verhoef, R. P., Schols, H. A. (2009). Pectin, a versatile polysaccharide present in plant cell walls. Struct. Chem. 20 (2), 263–275. doi: 10.1007/s11224-009-9442-z
Wang, L. Y., Wang, R. L., Lei, W., Wu, J. Y., Li, C. Y., Shi, H. S., et al. (2021). Transcriptome analysis reveals gene responses to herbicide, tribenuron methyl, in Brassica napus L. during seed germination. BMC Genomics 22 (1), 299. doi: 10.1186/s12864-021-07614-1
Wang, J., Xia, H., Zhao, S. Z., Hou, L., Zhao, C. Z., Ma, C. L., et al. (2018). A role of GUNs-Involved retrograde signaling in regulating Acetyl-CoA carboxylase 2 in Arabidopsis. Biochem. Biophys. Res. Commun. 505 (3), 712–719. doi: 10.1016/j.bbrc.2018.09.144
Wu, T. M., Huang, J. Z., Oung, H. M., Hsu, Y. T., Tsai, Y. C., Hong, C. Y. (2019). H2O2-based method for rapid detection of transgene-free rice plants from segregating CRISPR/cas9 genome-edited progenies. Int. J. Mol. Sci. 20 (16), 3885. doi: 10.3390/ijms20163885
Xiao, R., Zhang, C., Guo, X., Li, H., Lu, H. (2021). MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 22 (7), 3560. doi: 10.3390/ijms22073560
Xing, L. J., Zhu, M., Luan, M. D., Zhang, M., Jin, L., Liu, Y. P., et al. (2022). miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 188 (1), 608–623. doi: 10.1093/plphys/kiab498
Yan, Z. Q., Tan, J., Guo, K., Yao, L. G. (2020). Phytotoxic mechanism of allelochemical liquiritin on root growth of lettuce seedlings. Plant Signaling Behav. 15 (10), 1795581. doi: 10.1080/15592324.2020.1795581
Yang, Z. T., Lu, S. J., Wang, M. J., Bi, D. L., Sun, L., Zhou, S. F., et al. (2014). A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 79 (6), 1033–1043. doi: 10.1111/tpj.12604
Yang, G. Q., Wan, F. H., Liu, W. X., Guo, J. Y. (2008). Influence of two allelochemicals from Ageratina adenophora Sprengel on ABA, IAA and ZR contents in roots of upland rice seedlings. Allelopathy J. 21(2), 253–262.
Ye, Z. P., Robakowski, P., Suggett, D. J. (2013). A mechanistic model for the light response of photosynthetic electron transport rate based on light harvesting properties of photosynthetic pigment molecules. Planta 237 (3), 837–847. doi: 10.1007/s00425-012-1790-z
Yu, Z. X., Wang, L. J., Zhao, B., Shan, C. M., Zhang, Y. H., Chen, D. F., et al. (2015). Progressive Regulation of Sesquiterpene Biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-Targeted SPL Transcription Factors. Mol. Plant 8 (1), 98–110. doi: 10.1016/j.molp.2014.11.002
Yu, Z. P., Zhang, F., Friml, J., Ding, Z. J. (2022). Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 64 (2), 371–392. doi: 10.1111/jipb.13225
Zhang, J. Y., Huang, S. N., Wang, G., Xuan, J. P., Guo, Z. R. (2016). Overexpression of Actinidia deliciosa pyruvate decarboxylase 1 gene enhances waterlogging stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 106, 244–252. doi: 10.1016/j.plaphy.2016.05.009
Zhang, X., Wang, Z., Li, H. (2021). Allelopathic effects of Koelreuteria integrifoliola leaf aqueous extracts on Lolium perenne related to mesophyll ultrastructural alterations and endogenous hormone contents. Acta Physiologiae Plantarum 43, 132. doi: 10.1007/s11738-021-03303-4
Keywords: Arabidopsis, β-caryophyllene, eugenol, hormone, cell wall
Citation: Guo Y, Liu C, Zhang Y, Zheng S, Cao P, Wang X and Tian Z (2024) Characterization key genes of Arabidopsis seedlings in response to β-caryophyllene, eugenol using combined transcriptome and WGCN analysis. Front. Plant Sci. 14:1295779. doi: 10.3389/fpls.2023.1295779
Received: 17 September 2023; Accepted: 07 December 2023;
Published: 04 January 2024.
Edited by:
Zhiyong Wang, Hainan University, ChinaReviewed by:
Rongfang Guo, Fujian Agriculture and Forestry University, ChinaXiulan Xie, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Guo, Liu, Zhang, Zheng, Cao, Wang and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengyuan Tian, cherish_lab@163.com
 Yuqi Guo
Yuqi Guo