
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 November 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1293167
This article is part of the Research Topic Physiological and Molecular Mechanisms of Woody Plants Under Abiotic and Biotic Stresses View all 8 articles
Abiotic stresses, such as high salinity, pose a significant threat to plant growth and development, reducing crop yield and quality. Calcineurin B-like (CBL) proteins serve as crucial calcium sensors in plant responses to diverse environmental stresses. However, the CBL family in sweet cherry has not been identified at the genome-wide level, and the regulatory role of CBL proteins in cherry plants’ salt response is unclear. Here, we identified 10 CBL family genes (PavCBLs) from the Prunus avium genome and cloned seven of them. We comprehensively analyzed PavCBL genes for collinearity, phylogenetic relationships, gene structure, and conserved motifs. Expression analysis revealed significant induction of transcription under abiotic stress, with PavCBL4 displaying the most substantial expression change. Additionally, we identified PavCBL4 as a PavSOS2 (Salt Overly Sensitive 2)-interacting protein through Y2H and Split-LUC assays. Subcellular localization analysis indicated that PavCBL4 is present in both the cytoplasm and nucleus. Functional assessment of PavCBL4 in the PavCBL4-overexpressing transgenic ‘Gisela 6’ plants showed its positive role in enhancing salt tolerance in cherry plants. Measurements of Na+ content and antioxidant enzyme activity under salt stress indicated that PavCBL4 functions positively by inhibiting Na+ accumulation and promoting ROS scavenging in response to salt stress. These findings lay the groundwork for a deeper understanding of the molecular mechanisms underlying PavCBL-mediated salt tolerance in sweet cherry.
Plants, immobile by nature, must adapt to environmental changes like pathogens, drought, soil salinity, and extreme temperatures (Dong et al., 2022), significantly affecting global crop growth and productivity. Detecting and interpreting stimuli is crucial for plants to trigger suitable survival responses in challenging settings (Kissoudis et al., 2014). Ca2+ plays a pivotal role in various plant processes: growth, development, and biotic and abiotic stress responses (Harmon, 2003). The Ca2+ signal conveys information about stimuli, activating specific molecular and physiological responses and serving as a crucial specificity layer (Poovaiah and Du, 2018). Additionally, Ca2+-binding proteins and their interactors provide further specificity (Aldon et al., 2018), collectively constituting classical calcium signaling.
In plants, the Ca2+ signals are detected by different types of Ca2+-binding sensors. These include calmodulin (CaM), CaM-like (CML) proteins, calcineurin B-like (CBL) proteins, Ca2+-dependent protein kinases (CDPKs), and calcium/CaM-dependent protein kinase (CCaMK) (Poovaiah and Du, 2018). The CBL family, resembling the calcineurin B subunit of yeast and animal cell protein phosphatases, possesses four elongation factor (EF)-hand motifs that can bind up to four Ca2+ molecules (Kudla et al., 1999; Nagae et al., 2003; Sanchez-Barrena et al., 2005). Studies across plant species highlight CBLs’ significant role in responding to adverse environments. In Arabidopsis, 10 CBL proteins, grouped based on their phylogenetic relationship, subcellular localization, and function, have been identified (Kolukisaoglu et al., 2004). The CBL family’s characterization extends to various plant species, including Populus euphratica (Zhang et al., 2008), Vitis vinifera (Xi et al., 2017), and Solanum melongena (Li et al., 2016).
CBLs, proteins interacting with CIPKs (CBL-interacting protein kinases) (Yadav et al., 2018), translate signals into phosphorylation events, activating plant responses to environmental stresses like high salinity (Albrecht et al., 2001; Xuan et al., 2019). Recent studies have identified many CBL-CIPK signal components that are crucial in abiotic stress signaling pathways (Albrecht et al., 2003; Gao et al., 2020). The CBL-CIPK complex regulates Na+ accumulation and K+ levels, crucial for plant salt tolerance. For instance, CBL1/9-CIPK23 enhances K+ uptake, activating AKT1 during K+ deficiency (Xu et al., 2006; Cheong et al., 2007). This complex also reportedly affects cassava’s low potassium stress response (Yan et al., 2021). Additionally, CBL3-CIPK9 may regulate cellular K+ homeostasis under potassium deficiency by controlling K+ transport across the vacuolar membrane (Amtmann and Armengaud, 2007; Liu et al., 2013). Moreover, CBL1/CBL9-CIPK23 complex regulate nitrate (NO3. −) sensing and uptake via regulating the nitrate sensor and transporter CHL1 (Ho et al., 2009).
Salt stress significantly impacts plant growth and crop productivity (Yang and Guo, 2018). Studies have shown that CBLs regulate plant adaptation to salt stress via a CIPK-dependent salt overly sensitive (SOS) pathway. This pathway facilitates Na+ ion transport out of the cell, which confers plant salt stress tolerance (Liu and Zhu, 1998; Yang and Guo, 2018). In general, SOS3 (CBL4) interacts with SOS2 (CIPK24) in response to salt stress to jointly activate the transmembrane transporter SOS1 (NHX7), thus promoting Na+ efflux and consequently enhances plant salt tolerance (Liu and Zhu, 1998; Qiu et al., 2002). Recent studies showed that the CBL-CIPK complex regulates transport of other ions. For instance, SCaBP3/CBL3 modulates plasma membrane H+-ATPase activity in Arabidopsis (Yang et al., 2019). In Arabidopsis, NRAMP1, a plasma membrane transporter, primarily mediates Mn absorption (Cailliatte et al., 2010). The CBL1/9-CIPK23 complex helps to avoid manganese toxicity by converting the Ca2+ signature into a phosphorylation-mediated regulation of NRAMP1 (Zhang et al., 2023). Furthermore, CPK5 and CBL2/3-CIPK3/9/26 sequentially phosphorylate tonoplast-localized Mn transporter MTP8, providing a regulatory mechanism to fine-tune transporter activation in response to Mn toxicity (Ju et al., 2022).
Sweet cherry (Prunus avium), a Rosaceae family fruit tree common in temperate regions, faces challenges such as high salinity, impacting fruit yield and quality (Esti et al., 2002; Shen et al., 2021). Adverse environmental conditions, including high salinity, severely limit cherry fruit yield and quality. However, the CBL family (PavCBL) in sweet cherry and its role in regulating salinity tolerance remains uncharacterized and limited. This research involved genome-wide identification and cloning of CBL family genes in sweet cherry. Various bioinformatic analyses detailed the identified PavCBLs. Furthermore, the study explored PavCBL gene expression changes under salt stress and interactions between PavCBLs and PavSOS2. Moreover, the role of PavCBL4 in regulating cherry salt tolerance was identified in deatil using transgenic ‘Gisela 6’ plants. The findings elucidated the mechanistic understanding of PavCBL4-mediated salt tolerance, highlighting its role in inhibiting Na+ accumulation and promoting ROS scavenging.
The sweet cherry proteome file was downloaded form the GDR database (https://www.rosaceae.org/; Prunus avium Tieton Genome v2.0) (Wang et al., 2020a). The TAIR database (https://www.org/) provided the protein sequences for 10 Arabidopsis CBLs (AtCBLs). The Pfam database (http://pfam.xfam.org/) was used to retrieve the HMM file EF-hand_7.hmm (PF13499.8). It was then used as a query to search the sweet cherry proteome by HMMER software (version 3.1b2; http://hmmer.org/), along with a E-value threshold of < 0.01. To search for potential PavCBLs, the protein sequences in HMMER screening results and the 10 AtCBLs were used for phylogenetic analysis (Figure S1).
The GDR database provided the GFF file (Prunus_avium_Tieton.annotation.gff3) containing PavCBL genes location data. The collinearity analysis was conducted using the MCScanX software, with visualization using TBtools (Chen et al., 2020). Phylogenetic trees were generated through MEGAX software (version 10.05) employing the NJ (Neighbor-Joining) method. Gene structures’ schematic diagrams were drawn using GSDS software (version 2.0; http://gsds.gao-lab.org/). Conserved motifs within PavCBLs protein sequences were identified via MEME (version 5.5.2; https://meme-suite.org/meme/tools/meme). The PavCBLs and AtCBLs protein sequences’ alignment was performed using DNAMAN software (version 6). For promoter analysis, 2000 bp-long sequences upstream of the start codon of Prunus avium PavCBL genes were collected and analyzed using the online tool PlantCARE (http://bioinformatics.ugent.be/webtools/plantcare/html/). Detailed methods can be found in previous studies (Mao et al., 2017; Mao et al., 2021).
Tissue-cultured ‘Gisela 6’ sweet cherry rootstock seedlings were subjected to abiotic stress treatments to analyze PavCBL gene expression patterns. The seedlings were grown in 1/2 MS rooting medium with 1.0 mg/L IBA, 3% sucrose (w/v), and 0.6% Bacto-agar under long-day conditions (16 h light/8 h dark) at 25 °C. After 35 days, they were transplanted into pots with a 1:1 ratio of nutrient soil and perlite. Following a month, plantlets were chosen and cultured under hydroponic conditions for stress treatments: NaCl (150 mM), cold (4 °C), and PEG6000 (10% w/v). Leaf samples were gathered at specified time points, with 0 h samples acting as the control.
For comparing salt treatment-induced phenotypic changes, nine-week-old plants in similar growth stages were irrigated with a 150 mM NaCl solution every five days for 15 days. Each biological replicate comprised at least 20 plants per genotype across three technical replicates.
Sweet cherry leaf total RNA (cultivar ‘Tieton’) was isolated using the Plant RNA Isolation kit from Wolact (Wolact, Vicband Life Sciences Company (Hk) Limited). The cDNAs were obtained using the PrimeScript First-strand cDNA Synthesis kit (TaKaRa, Japan). RT-qPCR analysis followed established methods (Mao et al., 2021), utilizing PavACTIN as the internal reference gene (Sun et al., 2023). Three independent biological replicates, each with four technical replicates, were conducted. The primers used in this study are listed in Table S1.
To explore PavCBLs’ interaction with PavSOS2, their CDSs were amplified with specific primers (PavCBLs-AD-F/R and SOS2-BD-F/R) and inserted PCR products into pGADT7 and pGBKT7 vectors. These constructs were transformed into Y2H-Gold strain with specified combinations. Following growth on SD-Trp/-Leu (DDO) medium for two days, positive transformants were cultured on SD-Trp/-Leu/-His/-Ade (QDO; ± X-α-gal) medium (Yang et al., 2023a).
For split-LUC assays, gene CDSs were cloned into pRI101-nLUC and pRI101-cLUC vectors, then expressed in Nicotiana benthamiana leaves using the Agrobacterium-mediated transient expression method (Mei et al., 2023; Yang et al., 2023a; Yang et al., 2023b). The LUC fluorescence signal was detected using a Lumazone Pylon 2048B imaging system (Roper Scientific, MA, USA).
To determine PavCBL4 protein’s subcellular localization in plant and yeast cells, we used different vectors and transformation methods. PavCBL4-GFP vector (controlled by the 35S promoter) was introduced into Agrobacterium tumefaciens strain GV3101 for expression in tobacco leaves, as previously described. To identify the PavCBL4 localization in yeast cells, PavCBL4 CDS was cloned into the pDR196-GFP vector and subsequently transformed it into yeast cells via the LiAc/PEG method. Fluorescence images were obtained using a confocal laser scanning microscope (Leica TCS-SP8 SR).
For the ‘Gisela 6’ seedlings transformation, PavCBL4 CDS was cloned into pCAMBIA2300-GFP to construct the PavCBL4-OE vector. Agrobacterium rhizogenes C58C1 was used to genetically introduce the PavCBL4-OE vector into ‘Gisela 6’ roots (Sun et al., 2022). Positive transgenic lines were confirmed via genomic PCR analysis of root-extracted total DNA. Additionally, PavCBL4 expression levels in seedling roots were assessed through qRT-PCR.
Relative electrolyte leakage (REL) was calculated as follows: REL = (D1 − D0)/(D2 − D0) × 100 % (Hang et al., 2021; Yang et al., 2021a). Malondialdehyde (MDA) content, superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) enzymatic activities, H2O2 and O2. − content, and root activity were measured using their respective kits (Suzhou Comin Biotechnology Co., Ltd, China) following manufacturer’s instructions. DAB (3,3’-diaminobenzidine) and NBT (nitro blue tetrazolium) histochemical staining procedures followed established methods in prior studies (Zhou et al., 2019; Yang et al., 2021b). Total chlorophyll content was measured as described by Liang et al. (2017). The Open FluorCam FC 800-O assessed the maximal quantum efficiency of photosystem II (PSII; Fv/Fm). Na+ content was determined as described previously (Yang et al., 2021a; Yang et al., 2023b).
Significant differences among means were analyzed using one-way ANOVA and Student’s t-test (p < 0.05) via IBM SPSS software (version 26; IBM, Chicago, Illinois, United States).
To identify PavCBLs in sweet cherry, we employed the HMM file (PF13499.8) as a query to search the Prunus avium proteome using the HMMER software, resulting in 150 putative proteins (Supplementary File 1). These, along with the 10 AtCBLs (Supplementary File 2), were subjected to phylogenetic analysis. The analysis confirmed 10 proteins as sweet cherry CBL family members (Figure S1) grouped into four subgroups (A to D), mirroring the CBL family organization in Arabidopsis (Figure 1A).
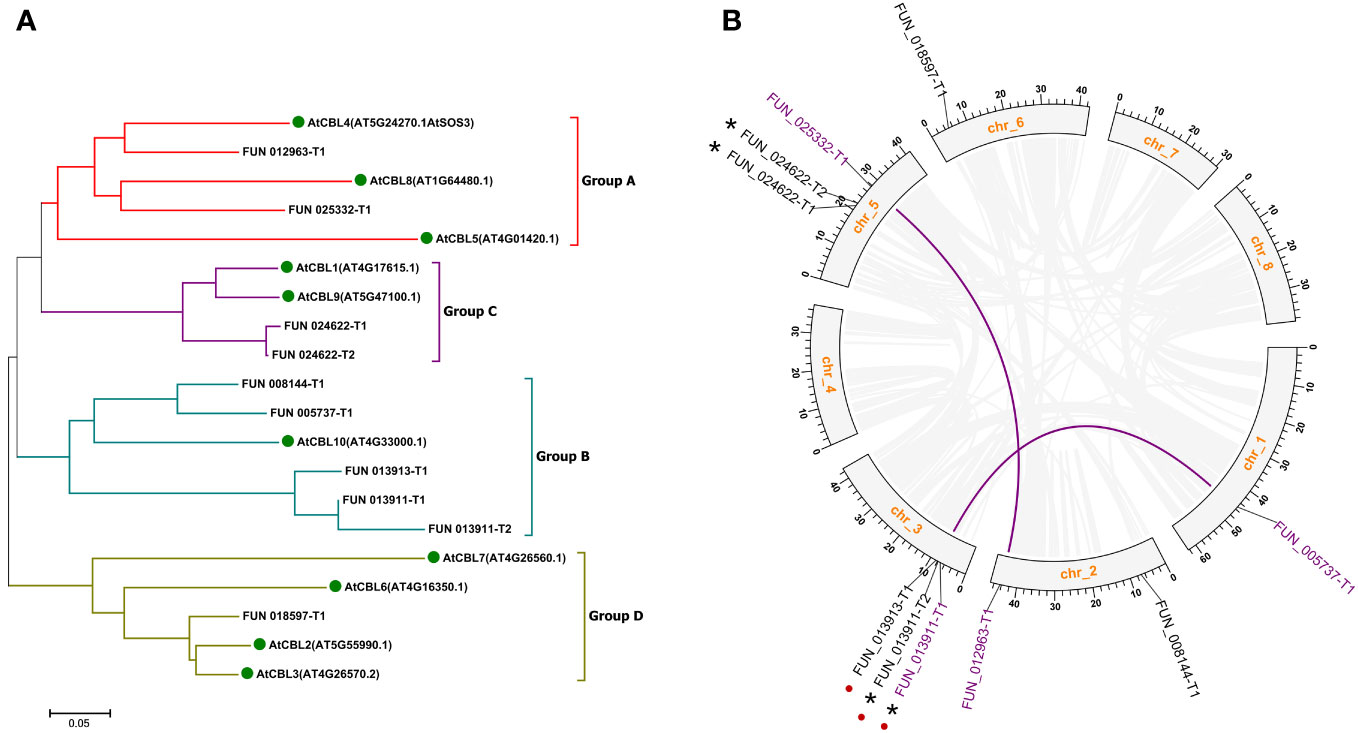
Figure 1 Phylogenic relationships (A) and genome locations (B) of CBL family genes in sweet cherry. Green dots in (A) indicate CBL proteins in Arabidopsis. Grayish lines represent collinear blocks. Purple lines indicate the segmental duplication genes. Red dots indicate the tandem duplication genes. Asterisks indicate the different splicing forms of the same gene loci. Chr, chromosome.
Mapping these 10 PavCBL genes to the sweet cherry genome (Prunus avium Tieton Genome v2.0) revealed their distribution on five of the eight chromosomes (Figure 1B). Plant gene family expansion is predominantly due to segmental and tandem duplications (Leister, 2004). Our collinear analysis uncovered intricate patterns of collinearity between chromosomes, identifying two pairs of segmental duplication genes (FUN_012963-T1 and FUN_025332-T1, FUN_013911-T1 and FUN_005737-T1) and one pair of tandem duplication genes (FUN_013911-T1 and FUN_013913-T1) (Figure 1B; Table 1). FUN_013911-T1/-T2 and FUN_024622-T1/-T2 represent different splicing forms of the gene loci FUN_013911 and FUN_024622 as predicted by the genome (Table 1).
Sequence alignments revealed high conservation of CBL protein sequences in sweet cherry and Arabidopsis, with a large variation primarily in the N-terminal region (Figure S2A). Notably, sequences that were excessively short (FUN_008144-T1, FUN_024622-T1) or long (FUN_013911-T1, FUN_013911-T2) indicated inaccuracies in CDS prediction within the sweet cherry genome, as corroborated by protein sequence alignments (Figure S2; Table 1). To address these inaccuracies, the coding sequence of FUN_013911-T1 was modified based on the protein sequence alignment (Figure S2C). Subsequently, we cloned the remaining 7 out of the 10 PavCBL genes from sweet cherry (cultivar ‘Tieton’) and named them according to their Arabidopsis orthologs (Table 1). The fundamental characteristics of these PavCBLs are also provided in Table 1.
To enhance comprehension of the correlation amongst PavCBL proteins, we constructed a phylogenetic tree with seven PavCBLs and ten AtCBLs (Figure 2A). Results showed that PavCBLs formed four clusters, akin to observations in Arabidopsis (Figure 2A). Analyzing PavCBL gene structures indicated significantly diverse intron lengths (Figure S3) and noticeable N-terminal variation among distinct groups, yet genes clustered within the same branch shared similar exon-intron patterns (Figures 2A, B). Utilizing MEME software, we also predicted conserved motifs in PavCBLs, with motifs 1, 2, 3, and 7 evident across all proteins (Figures 2C; S4). However, motif 4 was absent in PavCBL10.2, where an analysis revealed a missing fragment in its fifth exon (Figure 2B). Additionally, motifs 6 and 8 exclusively appeared in groups B and D, respectively, while motif 9 was solely present in groups A and C (Figures 2A, C). These outcomes imply the potential utility of these motifs in differentiating CBL proteins among distinct groups.
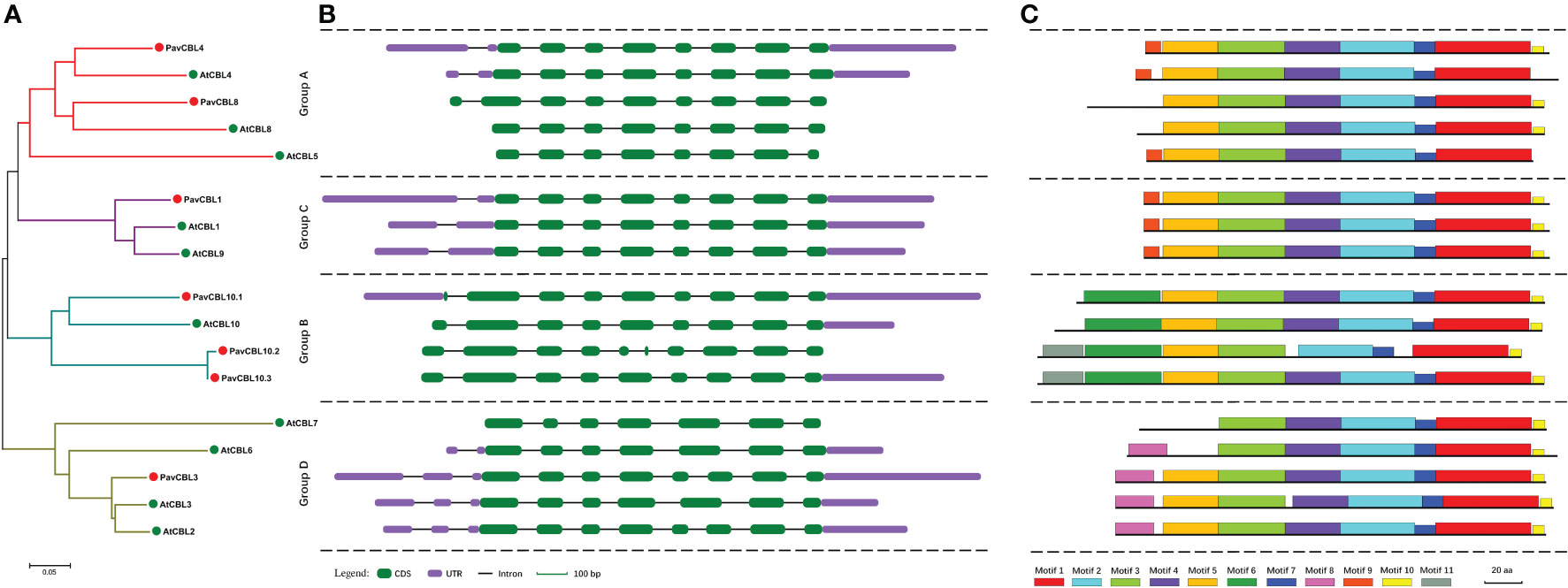
Figure 2 Phylogenic relationships (A), gene structure (B), and conserved motifs (C) for the CBL family proteins in sweet cherry and Arabidopsis. Green and red dots indicate CBL proteins in Arabidopsis (AtCBLs) and sweet cherry (PavCBLs), respectively. Introns are represented by black line segments of the same length to facilitate the comparison of the gene structure composition patterns. Different color boxes represent the 11 conserved motifs.
We obtained the promoter sequences of PavCBL genes from the cherry genome and identified the cis-acting elements contained in them using PlantCARE software. These elements, including those linked to hypoxia, low temperature, water deficit, and plant hormones (SA, ABA, MeJA, GA, and auxin). As expected, we discovered numerous abiotic stress and hormone responsive cis-elements (Figure 3A).
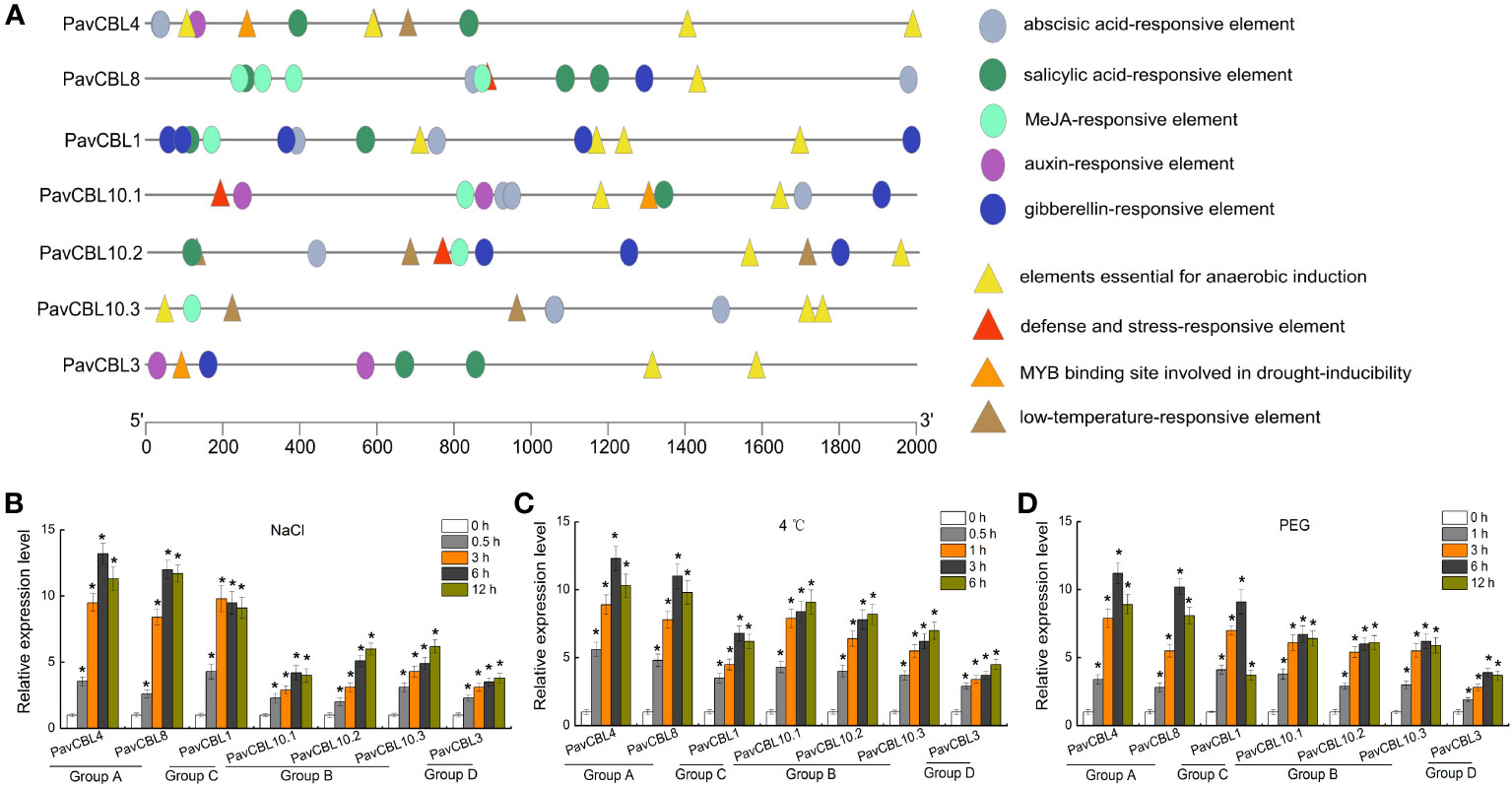
Figure 3 Promoter analysis and expression profile of PavCBL genes. (A) Cis-element analysis of the PavCBL genes promoter regions. Circles and triangles indicate the cis-elements related to hormone response and abiotic stress response, respectively. (B–D) Expression patterns of the PavCBL genes in leaves of ‘Gisela 6’ plants under NaCl (B), cold (C), and PEG (D) treatments. The expression level was calculated with respect to control samples (0 h) with the 2—△△CT method. * in each panel indicates signifcant differences from the control at P < 0.05.
To delve deeper into PavCBLs’ response to abiotic stress, ‘Gisela 6’ seedlings were subjected to hydroponic treatments with NaCl, 4 °C, and PEG6000. qRT-PCR analysis revealed that all PavCBL genes were significantly induced by these treatments (Figures 3B–D). Additionally, genes classified into the same group exhibited similar expression patterns. Among the CBL family, PavCBL4 and PavCBL8 from group A showed the most noticeable changes in expression (Figures 3B–D).
The interaction between CBLs and CIPKs is crucial for plant abiotic stress response. Among these signaling pathways, the SOS pathway has been extensively studied and reportedly positively modulates Arabidopsis salt tolerance (Zhu et al., 1998). The SOS2-SOS3/CBL4 complex is a key component of this pathway (Shi et al., 2000; Qiu et al., 2002). To identify the interaction between PavCBLs and PavSOS2, their CDSs were cloned into pGAD-T7 (AD) and pGBK-T7 (BD) vectors, respectively. Y2H assays in yeast indicated that PavCBL1 and PavCBL4 could interact with PavSOS2, while other PavCBLs could not (Figure 4A).
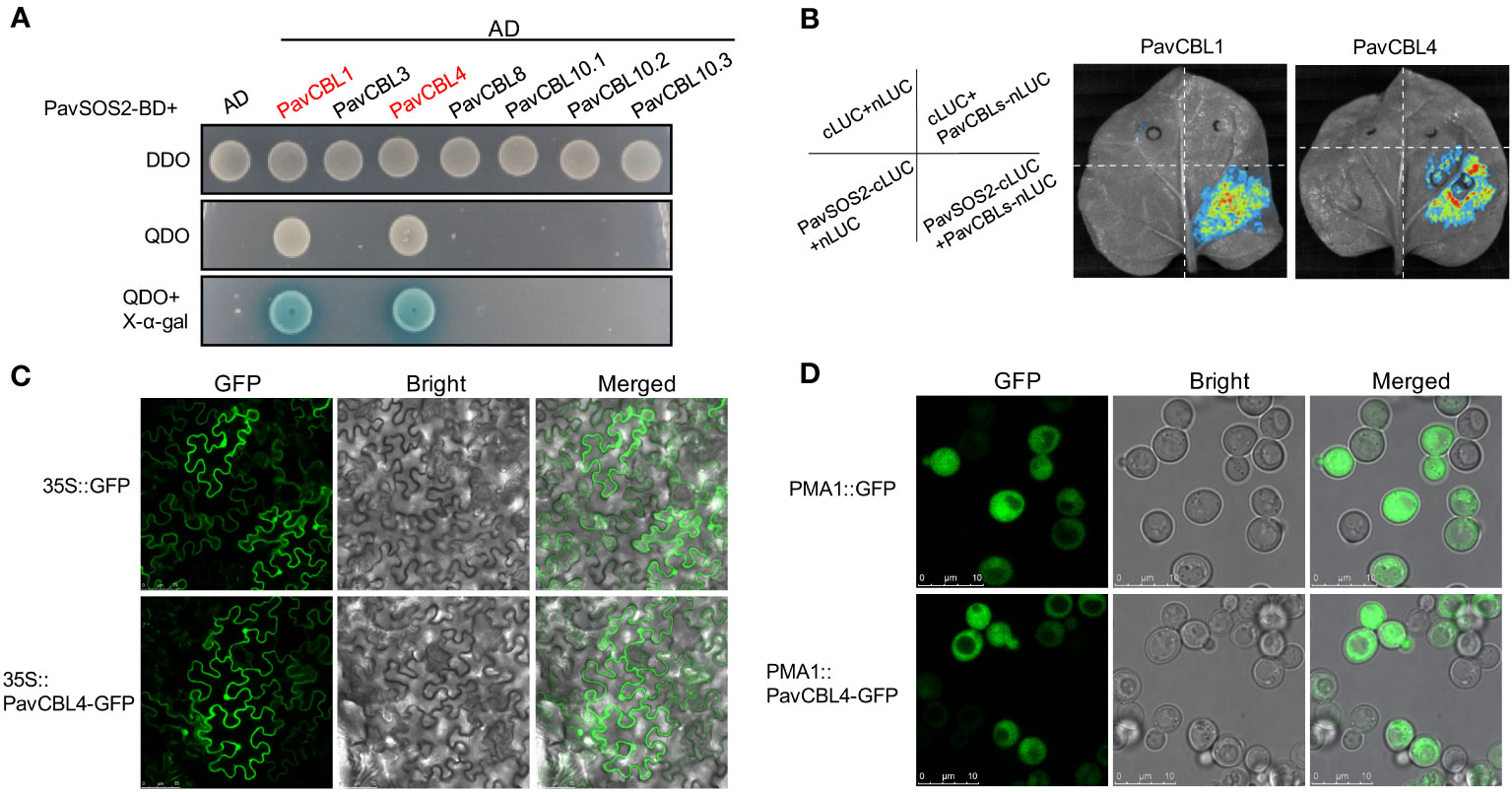
Figure 4 Protein interaction identification between PavCBLs and PavSOS2 and subcellular localization of PavCBL4. (A) Identification of the protein interactions between PavCBLs and PavSOS2 through yeast-two-hybrid (Y2H) assays. AD, pGADT7; BD, pGBKT7. DDO, SD medium without leucine and tryptophan; QDO, SD medium without leucine, tryptophan, histidine, and adenine. (B) Identification of the protein interactions between PavCBL1/PavCBL4 and PavSOS2 through the Split-LUC assays. (C) Subcellular localization of PavCBL4 in epidermal cells of tobacco leaves. Scale bars, 75 μm. (D) Subcellular localization of PavCBL4 in yeast cells. Scale bars, 10 μm.
To identify protein interactions within a living organism, we conducted Split-LUC assays. The CDSs of PavSOS2 and PavCBL1/4 were cloned into c-LUC and n-LUC vectors, respectively. These constructs were then transiently expressed in tobacco leaves in specified combinations (Figure 4B). No fluorescence was detected when PavSOS2 or PavCBL proteins were expressed independently. However, a strong fluorescence signal emerged upon co-expression of PavSOS2 with PavCBL1 (or PavCBL4) (Figure 4B), indicating in vivo interaction between PavSOS2 and PavCBL1 or PavCBL4.
Based on expression analysis and protein interaction identification results, PavCBL4 was selected for further investigation. Certain CBL proteins in Arabidopsis contain a conserved ‘MGCXXSK/T’ motif at their N-terminal, where myristoylation and S-acylation (commonly known as palmitoylation) occur at Gly and Cys residues, respectively. This lipid modification is crucial for membrane localization and function of these CBL proteins (Ishitani et al., 2000; Batistič et al., 2008; Sanyal et al., 2015; Steinhorst et al., 2022). Similar characteristics are observed in other plant CBL proteins like the MdCBL1 in apple (Jiang et al., 2021). Sequence analysis of PavCBL4 showed that it also contained the conserved N-terminal ‘MGCXXSK/T’ motif (Figure S2A), suggesting its potential as a membrane-localized protein. To determine PavCBL4’s location in plant cells, PavCBL4-GFP was expressed in tobacco leaves. Surprisingly, the PavCBL4-GFP fluorescence signal was found ubiquitously throughout the cell, indicating localization not only on the membrane but also in the cytoplasm and nucleus (Figure 4C). Further validation in yeast cells confirmed the widespread distribution of the green fluorescence signal, excluding vacuoles. Previous studies in Arabidopsis have also shown that AtCBL4 is localized to cytoplasm, nucleus, and plasma membrane, excluding the vacuolar membrane (Batistič et al., 2010). Based on these findings, PavCBL4 appears to be widely distributed in the plant cytoplasm, nucleus, and plasma membrane. Numerous studies have also revealed similar findings. For instance, various CBL proteins with the conserved ‘MGCXXSK/T’ motif are not exclusively localized to the membrane, as observed in AtCBL4 and AtCBL5 in Arabidopsis (Batistič et al., 2010; Sanyal et al., 2015) and MdCBL5 in apple (Gu et al., 2015). The diverse subcellular localization patterns of CBL proteins indicate the complex and varied functions of the CBL family in plant stress response (Batistič et al., 2010; Sanyal et al., 2015).
Utilizing A. rhizogenes to create transgenic roots (referred to as ‘composite plants’) provides a convenient method for investigating PavCBL4 function in cherry plants. Through genomic PCR detection and qRT-PCR analysis, at least 20 composite plants overexpressing PavCBL4 (PavCBL4-OE) in roots were chosen for treatment. Plants transformed with the empty vector (EV) were taken as controls. Initially, no significant genotype-based differences were observed prior to NaCl treatment. After a 15-day treatment period, the PavCBL4-OE lines demonstrated superior growth compared to the EV lines (Figure 5A). Histological staining employing DAB and NBT revealed lower H2O2 and O2− levels in the leaves of PavCBL4-OE plants compared to EV lines (Figure 5B). Quantitative measurements of H2O2 and O2. − contents corroborated the staining outcomes (Figure 5C). These results imply that the OE plants accumulated fewer ROS, thus experiencing reduced oxidative stress compared to EV lines.
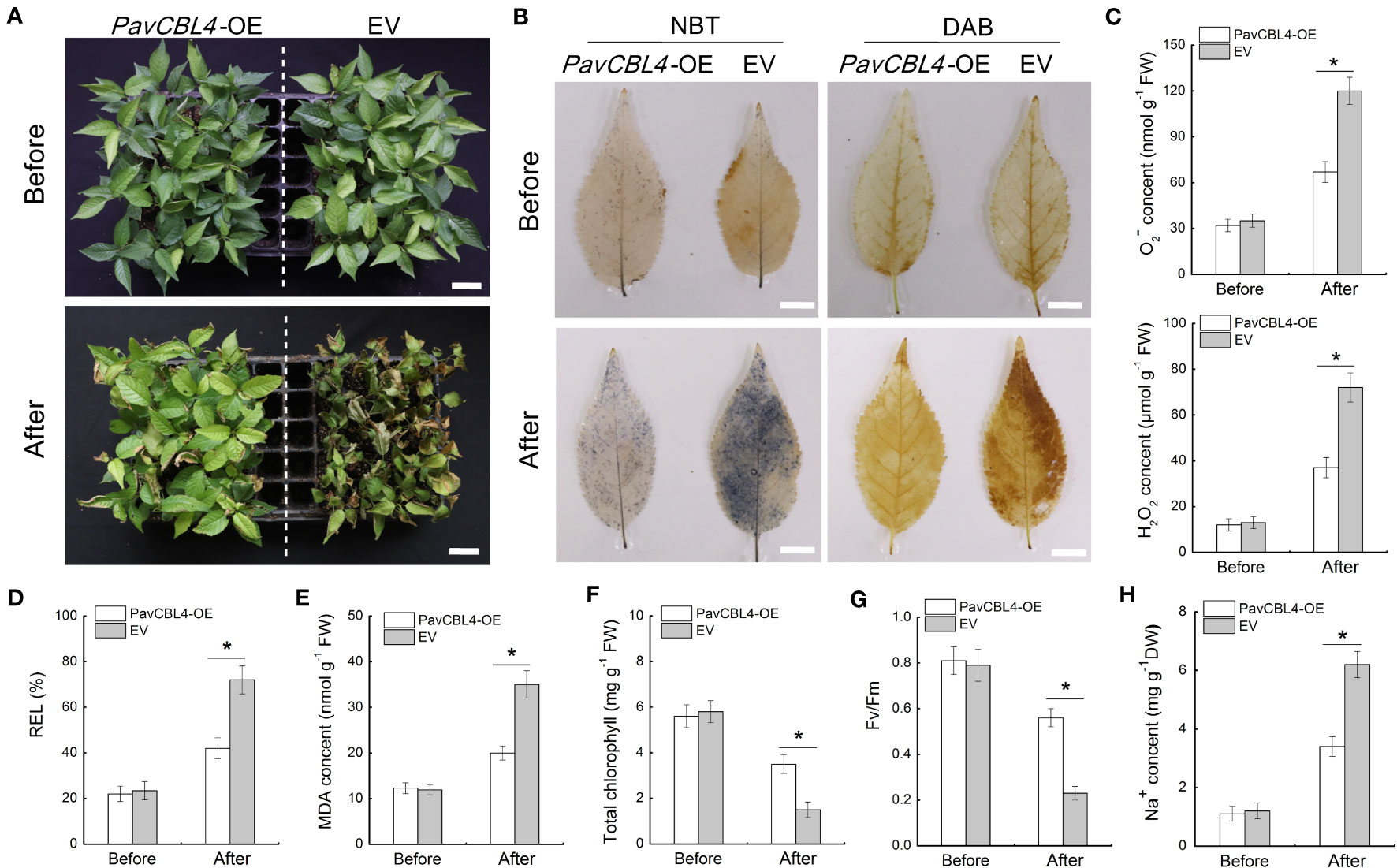
Figure 5 Phenotypic analysis of PavCBL4-overexpressing transgenic ‘Gisela 6’ plants under NaCl treatment. (A) Growth phenotypes of ‘Gisela 6’ plants before and after NaCl treatment. (B) In situ accumulation of H2O2 and O2. - detected by DAB and NBT histochemical staining, respectively. (C) H2O2 and O2. - contents in leaves of ‘Gisela 6’ plants. (D-H) Relative electrolyte leakage (REL) (D), malondialdehyde (MDA) content (E), total chlorophyll content (F), Fv/Fm ratio (G), and Na+ content (H) in leaves of ‘Gisela 6’ plants. * in each panel indicates signifcant differences from the control at P < 0.05.
For further assessment of salt stress-induced damage, various physiological indices related to stress response were evaluated in the leaves of the plants, including relative electrolyte leakage (REL), MDA content, total chlorophyll content, and Fv/Fm (Yang et al., 2021b). No significant differences in these indices were observed between PavCBL4-OE and EV lines prior to salt treatment. Following salt treatment, REL and MDA levels in the PavCBL4-OE leaves markedly decreased compared to the EV lines. Additionally, total chlorophyll levels and Fv/Fm significantly increased (Figures 5D–G). The results showed that PavCBL4 overexpression in roots enhanced the salt stress resistance of ‘Gisela 6’ seedlings. Na+ content determination results revealed a noticeable decrease in Na+ accumulation in PavCBL4-OE leaves compared to the EV lines after salt treatment (Figure 5H). This suggests that PavCBL4 overexpression in roots reduced the transport of Na+ to the aerial parts of the cherry plants.
Since plant roots are the primary part affected by high salinity, the root vitality was measured. No noticeable difference was observed between the root vitalities of PavCBL4-OE and EV lines prior to salt treatment. Post salt stress treatment, PavCBL4-OE plants displayed notably higher root vitality compared to EV lines (Figure 6A), indicating PavCBL4 overexpression mitigated salt-induced damage in roots. Additionally, the PavCBL4-OE lines exhibited reduced Na+ content in roots compared to EV lines under NaCl treatment (Figure 6B), demonstrating that PavCBL4 overexpression suppressed root Na+ accumulation.
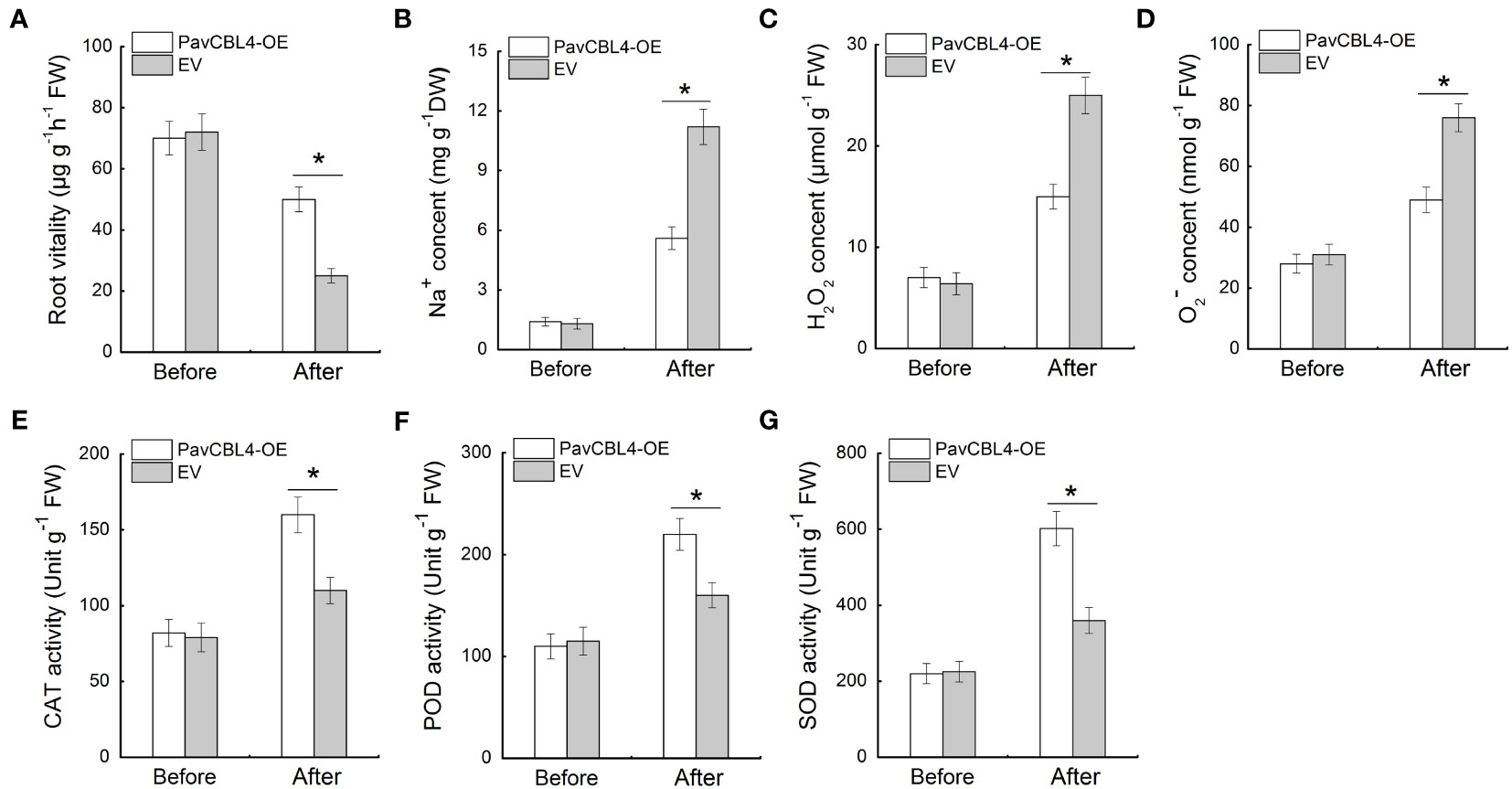
Figure 6 PavCBL4 overexpression inhibits Na+ and ROS accumulation and promotes antioxidant enzyme activities in roots. (A) Root vitality of ‘Gisela 6’ plants. (B) Na+ content in roots. (C, D) H2O2 and O2. - contents in roots of ‘Gisela 6’ plants. (E–G) Enzymatic activity of CAT, POD, and SOD antioxidant enzymes. * in each panel indicates signifcant differences from the control at P < 0.05.
To investigate the ROS accumulation inhibitory function of PavCBL4 in cherry plants, the H2O2 and O2. − levels and antioxidant enzymes’ activity in plant roots were measured. Under salt stress, PavCBL4-OE plant roots accumulated lower H2O2 and O2. − levels compared to EV plants (Figures 6C, D). Additionally, OE lines’ roots showed significantly increased antioxidant enzyme activity (Figures 6E–G). These results indicated that PavCBL4 play a positive role in promoting antioxidant enzyme activity and ROS scavenging, thus mitigating oxidative stress damage caused by salt stress.
Plants, due to their stationary growth nature, encounter diverse environmental stressors. Plants have evolved elaborate mechanisms to perceive surrounding environmental changes, orchestrating growth and stress adaptability (Tang et al., 2012). Calcium serves as a crucial nutrient and functions as a second messenger in responding to diverse environmental stresses like water scarcity, extreme temperatures, and high salinity (Yang and Poovaiah, 2003; Kleist and Luan, 2016; Dong et al., 2022). Calcium sensors like Calcineurin B-like (CBL) proteins are essential in plant adaptation to unfavorable environments (Luan, 2009; Dong et al., 2021). However, most studies of CBLs have focused on model plants, and the information on CBLs in woody plants is limited (Kolukisaoglu et al., 2004). Despite being a globally popular fruit, sweet cherry cultivation faces limitations due to various environmental stresses. Therefore, investigating PavCBLs’ regulatory role in responding to salt stress is imperative for the resistance breeding of cherry plants.
Genome-wide screenings in various plant species aimed to uncover CBL proteins crucial in Ca2+ signaling and stress response, resulting in diverse CBL member counts. Arabidopsis, rice, and Populus revealed 10 CBLs each (Zhang et al., 2008; Kleist et al., 2014; Jiang et al., 2020). Cassava, pepper, and pigeon pea identified nine CBLs each (Mo et al., 2018; Ma et al., 2019; Song et al., 2020). Cotton exhibited 13 CBLs (Sun et al., 2021), while tea plant and eggplant showcased 7 CBLs each (Li et al., 2016; Wang et al., 2020b). Beyond stress response, Ca2+ is crucial for regulating fruit development, quality, and storage (Picchioni et al., 1998; Chardonnet et al., 2003). The CBL family’s exploration extended to fruit crops like grapevine and pineapple (Xi et al., 2017; Aslam et al., 2019). Nevertheless, a detailed investigation of the CBL family in sweet cherry remains absent. Here, 10 PavCBL genes were identified from the sweet cherry genome (Figures 1, S1; Table 1). Collinear analysis unveiled two pairs of segmental duplication genes and one tandem duplication pair (Figure 1B; Table 1), signifying the contribution of both tandem and segmental duplication events to the CBL family’s expansion during the evolution of sweet cherry.
The polygenetic analysis categorized PavCBL proteins into four groups (Figures 1A, 2A), consistent with grouping patterns observed in CBLs of Arabidopsis (Kleist et al., 2014) and various other plant species (Zhang et al., 2008; Jiang et al., 2020; Chen et al., 2021). The phylogenetic tree (Figure 2A) was corroborated by comparing gene structure compositions and conserved motifs of AtCBLs and PavCBLs across distinct groups (Figures 2B, C). In addition, several unique characteristics, such as differing intron counts in the N-terminal region and exclusive conserved motifs in specific groups (Figures 2B, C; Figure S4), were identified among these CBL family members. Similar results have been observed in studies on CBL proteins, exemplified by MdCBLs in apple (Chen et al., 2021). Therefore, these discoveries establish a foundation for discerning CBL proteins within groups and exploring interrelationships among these CBLs in plants.
Environmental stress significantly induced the expression of various CBL genes (Kudla et al., 1999; Aslam et al., 2019; Dong et al., 2021). Stress reportedly induces a transient elevation of intracellular Ca2+ levels, which triggers CBL proteins’ response (Yang and Poovaiah, 2003; Dong et al., 2022). Ca2+ binding by CBLs facilitate their interactions CIPKs (Cheong et al., 2007; Li et al., 2016), notably activating the SOS pathway under salt stress (Qiu et al., 2002; Luan, 2009; Wang et al., 2020b; Yan et al., 2021). In Arabidopsis, the SOS pathway include SOS3 (CBL4) from the CBL family, SOS2 (CIPK24) from the CIPK family, and SOS1 (NHX7) from the NHX (Na+/H+ antiporter) family (Qiu et al., 2002; Sanchez-Barrena et al., 2005). High salinity activates the SOS pathway, which promotes Na+ efflux, thereby mitigating Na+ over-accumulation-induced damage (Qiu et al., 2002; El-Dakak et al., 2021). The positive role of SOS2 in salt stress response and CBLs-SOS2 interactions have been identified in various crops like pepper (Ma et al., 2023), poplar (Tang et al., 2014), and apple (Hu et al., 2012; Hu et al., 2016). This study detected several stress and hormone response-related cis-elements in PavCBLs promoters (Figure 3A). Furthermore, PavCBLs’ transcription levels were obviously up-regulated in response to abiotic stress treatments, especially PavCBL4 and PavCBL8 in group A (Figure 3B). The outcomes, along with the PavCBL4-PavSOS2 interaction (Figure 4), suggested that PavCBL4 may positively regulate sweet cherry salt response by curtailing excessive Na+ accumulation through the SOS pathway. The role of PavCBL4 in modulating sweet cherry salt tolerance was thus investigated using transgenic ‘Gisela 6’ plants, and the Na+ content in roots of was measured. Our findings demonstrate a significant enhancement in salt tolerance in transgenic plants, accompanied by a significant reduction in root Na+ accumulation (Figure 6B), suggesting a positive role of PavCBL4 in salt stress response. The lower Na+ content in leaves may be attributed to decreased root Na+ absorption, leading to lower Na+ transport to the aerial parts (Figure 5H).
Besides the NHX transporters within the SOS pathway, CBL-activated CIPKs interact with and phosphorylate downstream substrates, including ion transporters and enzymes (Xu et al., 2006; Yang et al., 2019; Yan et al., 2021; Ju et al., 2022). This coordination encompasses various plant stress responses, including Na+/K+ balance, osmoregulatory substance accumulation, and antioxidant enzyme activity (Xu et al., 2006; Liu et al., 2013; Ju et al., 2022). Numerous studies indicated that overexpression of SOS2 or CBLs in plants under abiotic stress conditions increases antioxidant enzyme activity, e.g., PtSOS2 in poplar (Zhou et al., 2014), and MdSOS2L1 (Hu et al., 2016) and MdCBL10.1 (Chen et al., 2021) in apple. In PavCBL4-OE transgenic roots, antioxidant enzyme activity was higher than those of controls under salt stress (Figures 6E–G). Furthermore, PavCBL4-OE transgenic roots exhibited significantly lower ROS levels than control roots (Figures 6C, D). These results suggested that, in addition to preventing excessive Na+ accumulation, PavCBL4 enhances cherry plant salt tolerance by improving antioxidant enzyme activity. Overall, this study provides valuable insights for future investigations into the mechanisms underlying PavCBLs-mediated salt stress responses in sweet cherry.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
QF: Writing – original draft. SH: Writing – review & editing. RG: Writing – review & editing. GW: Writing – review & editing. YS: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by innovation projects of Shandong academy of agricultural sciences (CXGC2023G36), Shaanxi Province key research and development plan project (2022ZDLNY03-05), and Tai ‘an agricultural seed improvement project (2022NYLZ04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1293167/full#supplementary-material
File 2 | Protein sequences of the HMMER screening results and the 10 AtCBLs.
Albrecht, V., Ritz, O., Linder, S., Harter, K., Kudla, J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20 (5), 1051–1063. doi: 10.1093/emboj/20.5.1051
Albrecht, V., Weinl, S., Blazevic, D., D'Angelo, C., Batistic, O., Kolukisaoglu, U., et al. (2003). The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 36 (4), 457–470. doi: 10.1046/j.1365-313X.2003.01892.x
Aldon, D., Mbengue, M., Mazars, C., Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19 (3), 665. doi: 10.3390/Ijms19030665
Amtmann, A., Armengaud, P. (2007). The role of calcium sensor-interacting protein kinases in plant adaptation to potassium-deficiency: new answers to old questions. Cell Res. 17 (6), 483–485. doi: 10.1038/cr.2007.49
Aslam, M., Fakher, B., Jakada, B. H., Zhao, L. H., Cao, S. J., Cheng, Y., et al. (2019). Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple (Ananas comosus) and the role of acCBL1 in abiotic and biotic stress response. Biomolecules 9 (7), 293. doi: 10.3390/Biom9070293
Batistič, O., Sorek, N., Schultke, S., Yalovsky, S., Kudla, J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20, 1346–1362. doi: 10.1105/tpc.108.058123
Batistič, O., Waadt, R., Steinhorst, L., Held, K., Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Cailliatte, R., Schikora, A., Briat, J. F., Mari, S., Curie, C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22 (3), 904–917. doi: 10.1105/tpc.109.073023
Chardonnet, C. O., Charron, C. S., Sams, C. E., Conway, W. S. (2003). Chemical changes in the cortical tissue and cell walls of calcium-infiltrated 'Golden Delicious' apples during storage. Postharvest Biol. Technol. 28 (1), 97–111. doi: 10.1016/S0925-5214(02)00139-4
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, P. H., Yang, J., Mei, Q. L., Liu, H. Y., Cheng, Y. P., Ma, F. W., et al. (2021). Genome-wide analysis of the apple CBL family reveals that mdcbl10.1 functions positively in modulating apple salt tolerance. Int. J. Mol. Sci. 22 (22), 12430. doi: 10.3390/Ijms222212430
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52 (2), 223–239. doi: 10.1111/j.1365-313X.2007.03236.x
Dong, Q. Y., Bai, B. W., Almutairi, B. O., Kudla, J. (2021). Emerging roles of the CBL-CIPK calcium signaling network as key regulatory hub in plant nutrition. J. Plant Physiol. 257, 153335. doi: 10.1016/J.Jplph.2020.153335
Dong, Q., Wallrad, L., Almutairi, B. O., Kudla, J. (2022). Ca(2+) signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 64 (2), 287–300. doi: 10.1111/jipb.13228
El-Dakak, R., El-Aggan, W., Badr, G., Helaly, A., Tammam, A. (2021). Positive Salt Tolerance Modulation via Vermicompost Regulation of SOS1 Gene Expression and Antioxidant Homeostasis in Vicia faba Plant. Plants-Basel 10 (11), 2477. doi: 10.3390/Plants10112477
Esti, M., Cinquanta, L., Sinesio, F., Moneta, E., Di Matteo, M. (2002). Physicochemical and sensory fruit characteristics of two sweet cherry cultivars after cool storage. Food Chem. 76 (4), 399–405. doi: 10.1016/S0308-8146(01)00231-X
Gao, H., Wang, C., Li, L., Fu, D., Zhang, Y., Yang, P., et al. (2020). A novel role of the calcium sensor CBL1 in response to phosphate deficiency in Arabidopsis thaliana. J. Plant Physiol. 253, 153266. doi: 10.1016/j.jplph.2020.153266
Gu, Z. Y., Meng, D., Yang, Q., Yuan, H., Wang, A. D., Li, W., et al. (2015). A CBL gene, MdCBL5, controls the calcium signal and influences pollen tube growth in apple. Tree Genet. Genomes 11, 27. doi: 10.1007/s11295-015-0853-2
Hang, N., Shi, T. R., Liu, Y. R., Ye, W. X., Taier, G., Sun, Y., et al. (2021). Overexpression of Os-microRNA408 enhances drought tolerance in perennial ryegrass. Physiologia Plantarum 172 (2), 733–747. doi: 10.1111/ppl.13276
Harmon, A. C. (2003). Calcium-regulated protein kinases of plants. Gravit Space Biol. Bull. 16 (2), 83–90.
Ho, C. H., Lin, S. H., Hu, H. C., Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 (6), 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hu, D. G., Li, M., Luo, H., Dong, Q. L., Yao, Y. X., You, C. X., et al. (2012). Molecular cloning and functional characterization of MdSOS2 reveals its involvement in salt tolerance in apple callus and Arabidopsis. Plant Cell Rep. 31 (4), 713–722. doi: 10.1007/s00299-011-1189-5
Hu, D. G., Ma, Q. J., Sun, C. H., Sun, M. H., You, C. X., Hao, Y. J. (2016). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiologia Plantarum 156 (2), 201–214. doi: 10.1111/ppl.12354
Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W., Zhu, J. K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12, 1667–1678. doi: 10.2307/3871181
Jiang, H., Ma, Q. J., Zhong, M. S., Gao, H. N., Li, Y. Y., Hao, Y. J. (2021). The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1–MdCIPK13–MdSUT2.2 pathway. Plant J. 106, 689–705. doi: 10.1111/tpj.15191
Jiang, M., Zhao, C. L., Zhao, M. F., Li, Y. Z., Wen, G. S. (2020). Phylogeny and evolution of calcineurin B-like (CBL) gene family in grass and functional analyses of rice CBLs. J. Plant Biol. 63 (2), 117–130. doi: 10.1007/s12374-020-09240-y
Ju, C., Zhang, Z., Deng, J., Miao, C., Wang, Z., Wallrad, L., et al. (2022). Ca(2+)-dependent successive phosphorylation of vacuolar transporter MTP8 by CBL2/3-CIPK3/9/26 and CPK5 is critical for manganese homeostasis in Arabidopsis. Mol. Plant 15 (3), 419–437. doi: 10.1016/j.molp.2021.11.012
Kissoudis, C., van de Wiel, C., Visser, R. G. F., van der Linden, G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 5. doi: 10.3389/Fpls.2014.00207
Kleist, T. J., Luan, S. (2016). Constant change: dynamic regulation of membrane transport by calcium signalling networks keeps plants in tune with their environment. Plant Cell And Environ. 39 (3), 467–481. doi: 10.1111/pce.12599
Kleist, T. J., Spencley, A. L., Luan, S. (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 5. doi: 10.3389/Fpls.2014.00187
Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., Kudla, J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134 (1), 43–58. doi: 10.1104/pp.103.033068
Kudla, J., Xu, Q., Harter, K., Gruissem, W., Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. U. S. A. 96 (8), 4718–4723. doi: 10.1073/pnas.96.8.4718
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends In Genet. 20 (3), 116–122. doi: 10.1016/j.tig.2004.01.007
Li, J., Jiang, M. M., Ren, L., Liu, Y., Chen, H. Y. (2016). Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 291 (4), 1769–1781. doi: 10.1007/s00438-016-1218-8
Liang, B. W., Li, C. Y., Ma, C. Q., Wei, Z. W., Wang, Q., Huang, D., et al. (2017). Dopamine alleviates nutrient deficiency-induced stress in Malus hupehensis. Plant Physiol. Biochem. 119, 346–359. doi: 10.1016/j.plaphy.2017.09.012
Liu, L. L., Ren, H. M., Chen, L. Q., Wang, Y., Wu, W. H. (2013). A protein kinase, calcineurin B-like protein-interacting protein kinase9, interacts with calcium sensor calcineurin B-like protein3 and regulates potassium homeostasis under low-potassium stress in arabidopsis. Plant Physiol. 161 (1), 266–277. doi: 10.1104/pp.112.206896
Liu, J. P., Zhu, J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 (5371), 1943–1945. doi: 10.1126/science.280.5371.1943
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends In Plant Sci. 14 (1), 37–42. doi: 10.1016/j.tplants.2008.10.005
Ma, X., Gai, W. X., Qiao, Y. M., Ali, M., Wei, A. M., Luo, D. X., et al. (2019). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics 20 (1), 775. doi: 10.1186/S12864-019-6125-Z
Ma, X., Yu, Y. N., Li, Y., Gong, Z. H. (2023). The CBL-interacting protein kinase CaCIPK7 enhances drought resistance in pepper. Scientia Hortic. 310, 111726. doi: 10.1016/j.scienta.2022.111726
Mao, K., Dong, Q. L., Li, C., Liu, C. H., Ma, F. W. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8. doi: 10.3389/Fpls.2017.00480
Mao, K., Yang, J., Wang, M., Liu, H. Y., Guo, X., Zhao, S., et al. (2021). Genome-wide analysis of the apple CaCA superfamily reveals that MdCAX proteins are involved in the abiotic stress response as calcium transporters. BMC Plant Biol. 21 (1), 81. doi: 10.1186/S12870-021-02866-1
Mei, C., Yang, J., Mei, Q., Jia, D., Yan, P., Feng, B., et al. (2023). MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 21 (10), 2057–2073. doi: 10.1111/pbi.14112
Mo, C. Y., Wan, S. M., Xia, Y. Q., Ren, N., Zhou, Y., Jiang, X. Y. (2018). Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 9. doi: 10.3389/Fpls.2018.00269
Nagae, M., Nozawa, A., Koizumi, N., Sano, H., Hashimoto, H., Sato, M., et al. (2003). The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J. Biol. Chem. 278 (43), 42240–42246. doi: 10.1074/jbc.M303630200
Picchioni, G. A., Watada, A. E., Conway, W. S., Whitaker, B. D., Sams, C. E. (1998). Postharvest calcium infiltration delays membrane lipid catabolism in apple fruit. J. Agric. Food Chem. 46 (7), 2452–2457. doi: 10.1021/Jf971083e
Poovaiah, B. W., Du, L. Q. (2018). Calcium signaling: decoding mechanism of calcium signatures. New Phytol. 217 (4), 1394–1396. doi: 10.1111/nph.15003
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U. S. A. 99 (12), 8436–8441. doi: 10.1073/pnas.122224699
Sanchez-Barrena, M. J., Martinez-Ripoll, M., Zhu, J. K., Albert, A. (2005). The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 345 (5), 1253–1264. doi: 10.1016/j.jmb.2004.11.025
Sanyal, S. K., Pandey, A., Pandey, G. K. (2015). The CBL–CIPK signaling module in plants: a mechanistic perspective. Physiologia Plantarum 155, 89–108. doi: 10.1111/ppl.12344
Shen, T. J., Wen, X. P., Wen, Z., Qiu, Z. L., Hou, Q. D., Li, Z. C., et al. (2021). Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.). Scientia Hortic. 279, 109905. doi: 10.1016/j.scienta.2021.109905
Shi, H. Z., Ishitani, M., Kim, C. S., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6896–6901. doi: 10.1073/pnas.120170197
Song, Z. H., Dong, B. Y., Yang, Q., Niu, L. L., Li, H. H., Cao, H. Y., et al. (2020). Screening of CBL genes in pigeon pea with focus on the functional analysis of CBL4 in abiotic stress tolerance and flavonoid biosynthesis. Environ. Exp. Bot. 177, 104102. doi: 10.1016/j.envexpbot.2020.104102
Steinhorst, L., He, G. F., Moore, L. K., Schultke, S., Schmitz-Thom, I., Cao, Y. B., et al. (2022). A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev. Cell 57, 2081–2094. doi: 10.1016/j.devcel.2022.08.001
Sun, Y. T., Wang, Y. Y., Xiao, Y. Q., Zhang, X., Du, B. Y., Turupu, M., et al. (2023). Genome-wide identification of the SQUAMOSA promoter-binding protein-like (SPL) transcription factor family in sweet cherry fruit. Int. J. Mol. Sci. 24 (3), 2880. doi: 10.3390/Ijms24032880
Sun, W. N., Zhang, B., Deng, J. W., Chen, L., Ullah, A., Yang, X. Y. (2021). Genome-wide analysis of CBL and CIPK family genes in cotton: conserved structures with divergent interactions and expression. Physiol. Mol. Biol. Plants 27 (2), 359–368. doi: 10.1007/s12298-021-00943-1
Sun, Y., Zhao, X. H., Gao, Y. H., Jiao, J., Sun, Y. D., Zhu, D. Z., et al. (2022). Genome-wide analysis of lectin receptor-like kinases (LecRLKs) in sweet cherry (Prunus avium) and reveals PaLectinL16 enhances sweet cherry resistance with salt stress. Environ. Exp. Bot. 194, 104751. doi: 10.1016/j.envexpbot.2021.104751
Tang, R. J., Liu, H., Yang, Y., Yang, L., Gao, X. S., Garcia, V. J., et al. (2012). Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 22 (12), 1650–1665. doi: 10.1038/cr.2012.161
Tang, R. J., Yang, Y., Yang, L., Liu, H., Wang, C. T., Yu, M. M., et al. (2014). Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 37 (3), 573–588. doi: 10.1111/pce.12178
Wang, L., Feng, X., Yao, L. N., Ding, C. Q., Lei, L., Hao, X. Y., et al. (2020b). Characterization of CBL-CIPK signaling complexes and their involvement in cold response in tea plant. Plant Physiol. Biochem. 154, 195–203. doi: 10.1016/j.plaphy.2020.06.005
Wang, J. W., Liu, W. Z., Zhu, D. Z., Hong, P., Zhang, S. Z., Xiao, S. J., et al. (2020a). Chromosome-scale genome assembly of sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read and Hi-C sequencing. Hortic. Res. 7 (1), 122. doi: 10.1038/s41438-020-00343-8
Xi, Y., Liu, J. Y., Dong, C., Cheng, Z. M. (2017). The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 8. doi: 10.3389/Fpls.2017.00978
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 (7), 1347–1360. doi: 10.1016/j.cell.2006.06.011
Xuan, Y. H., Kumar, V., Han, X., Kim, S. H., Jeong, J. H., Kim, C. M., et al. (2019). CBL-INTERACTING PROTEIN KINASE 9 regulates ammonium-dependent root growth downstream of IDD10 in rice (Oryza sativa). Ann. Bot. 124 (6), 947–960. doi: 10.1093/aob/mcy242
Yadav, A. K., Jha, S. K., Sanyal, S. K., Luan, S., Pandey, G. K. (2018). Arabidopsis calcineurin B-like proteins differentially regulate phosphorylation activity of CBL-interacting protein kinase 9. Biochem. J. 475, 2621–2636. doi: 10.1042/Bcj20180372
Yan, Y., He, M., Guo, J. R., Zeng, H. Q., Wei, Y. X., Liu, G. Y., et al. (2021). The CBL1/9-CIPK23-AKT1 complex is essential for low potassium response in cassava. Plant Physiol. Biochem. 167, 430–437. doi: 10.1016/j.plaphy.2021.08.026
Yang, Y. Q., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523–539. doi: 10.1111/nph.14920
Yang, J., Guo, X., Li, W. H., Chen, P. H., Cheng, Y. P., Ma, F. W., et al. (2021a). MdCCX2 of apple functions positively in modulation of salt tolerance. Environ. Exp. Bot. 192, 104663. doi: 10.1016/j.envexpbot.2021.104663
Yang, J., Guo, X., Mei, Q., Qiu, L., Chen, P., Li, W., et al. (2023a). MdbHLH4 negatively regulates apple cold tolerance by inhibiting MdCBF1/3 expression and promoting MdCAX3L-2 expression. Plant Physiol. 191 (1), 789–806. doi: 10.1093/plphys/kiac512
Yang, J., Li, W. H., Guo, X., Chen, P. H., Cheng, Y. P., Mao, K., et al. (2021b). Cation/ca2+ Exchanger 1 (MdCCX1), a plasma membrane-localized na+ Transporter, enhances plant salt tolerance by inhibiting excessive accumulation of na+ and reactive oxygen species. Front. Plant Sci. 12. doi: 10.3389/Fpls.2021.746189
Yang, T. B., Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends In Plant Sci. 8 (10), 505–512. doi: 10.1016/j.tplants.2003.09.004
Yang, J., Qiu, L., Mei, Q., Sun, Y., Li, N., Gong, X., et al. (2023b). MdHB7-like positively modulates apple salt tolerance by promoting autophagic activity and Na(+) efflux. Plant J. doi: 10.1111/tpj.16395
Yang, Y., Wu, Y., Ma, L., Yang, Z., Dong, Q., Li, Q., et al. (2019). The ca(2+) sensor SCaBP3/CBL7 modulates plasma membrane H(+)-ATPase activity and promotes alkali tolerance in Arabidopsis. Plant Cell 31 (6), 1367–1384. doi: 10.1105/tpc.18.00568
Zhang, Z., Fu, D., Xie, D., Wang, Z., Zhao, Y., Ma, X., et al. (2023). CBL1/9-CIPK23-NRAMP1 axis regulates manganese toxicity. New Phytol. 239 (2), 660–672. doi: 10.1111/nph.18968
Zhang, H. C., Yin, W. L., Xia, X. L. (2008). Calcineurin B-Like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 56 (2), 129–140. doi: 10.1007/s10725-008-9293-4
Zhou, K., Hu, L., Li, Y., Chen, X., Zhang, Z., Liu, B., et al. (2019). MdUGT88F1-mediated phloridzin biosynthesis regulates apple development and valsa canker resistance. Plant Physiol. 180 (4), 2290–2305. doi: 10.1104/pp.19.00494
Zhou, J., Wang, J. J., Bi, Y. F., Wang, L. K., Tang, L. Z., Yu, X., et al. (2014). Overexpression of ptSOS2 enhances salt tolerance in transgenic poplars. Plant Mol. Biol. Rep. 32 (1), 185–197. doi: 10.1007/s11105-013-0640-x
Keywords: sweet cherry (Prunus avium), calcineurin B-like proteins (CBLs), salt stress, PavCBL4, Na+ accumulation, antioxidant enzyme
Citation: Fu Q, Hou S, Gao R, Wei G and Sun Y (2023) Functional identification of the calcineurin B-like protein PavCBL4 in modulating salt tolerance in sweet cherry. Front. Plant Sci. 14:1293167. doi: 10.3389/fpls.2023.1293167
Received: 12 September 2023; Accepted: 03 November 2023;
Published: 22 November 2023.
Edited by:
Qinglong Dong, Hebei Agricultural University, ChinaReviewed by:
Ke Mao, Northwest A&F University, ChinaCopyright © 2023 Fu, Hou, Gao, Wei and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqin Wei, Z3VvcWludzE5ODNAMTI2LmNvbQ==; Yugang Sun, c2RzMTI5QDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.