
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 December 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1288961
This article is part of the Research Topic Model Organisms in Plant Developmental Biology — their effectiveness and limitations View all 14 articles
The MIXTA family of MYB transcription factors modulate the development of diverse epidermal features in land plants. This study investigates the evolutionary history and function of the MIXTA gene family in the early-diverging eudicot model lineage Thalictrum (Ranunculaceae), with R2R3 SBG9-A MYB transcription factors representative of the pre-core eudicot duplication and thus hereby referred to as “paleoMIXTA” (PMX). Cloning and phylogenetic analysis of Thalictrum paleoMIXTA (ThPMX) orthologs across 23 species reveal a genus-wide duplication coincident with a whole-genome duplication. Expression analysis by qPCR confirmed that the highest expression is found in carpels, while newly revealing high expression in leaves and nuanced differences between paralogs in representative polyploid species. The single-copy ortholog from the diploid species T. thalictroides (TthPMX, previously TtMYBML2), which has petaloid sepals with conical–papillate cells and trichomes on leaves, was functionally characterized by virus-induced gene silencing (VIGS), and its role in leaves was also assessed from heterologous overexpression in tobacco. Another ortholog from a species with conical–papillate cells on stamen filaments, TclPMX, was also targeted for silencing. Overexpression assays in tobacco provide further evidence that the paleoMIXTA lineage has the potential for leaf trichome function in a core eudicot. Transcriptome analysis by RNA-Seq on leaves of VIGS-treated plants suggests that TthPMX modulates leaf trichome development and morphogenesis through microtubule-associated mechanisms and that this may be a conserved pathway for eudicots. These experiments provide evidence for a combined role for paleoMIXTA orthologs in (leaf) trichomes and (floral) conical–papillate cells that, together with data from other systems, makes the functional reconstruction of a eudicot ancestor most likely as also having a combined function.
Plants have evolved diverse epidermal features to adapt to their environment (Javelle et al., 2011), and epidermal cell types are useful micromorphological markers of the different plant organs (Cavallini-Speisser et al., 2021). MYB genes are a prominent land plant gene family that encode transcription factors characterized by having one to four contiguous repeats, with most plants having two repeats, R2 and R3, that function in secondary metabolite production and cell identity (Stracke et al., 2001). The R2R3 MYB subgroup 9A MIXTA/MIXTA-like gene family has specialized roles in epidermal cells (Brockington et al., 2013), including the first characterized function in conical cells (Noda et al., 1994; Baumann et al., 2007; Di Stilio et al., 2009), trichomes (Jakoby et al., 2008; Yan et al., 2018; Qin et al., 2021), and ovule epidermal cells that make cotton fibers (MaChado et al., 2009). Certain MIXTA-like orthologs have evolved multiple functions in epidermal cell patterning, such as Antirrhinum majus AmMYBML1 in regulating trichome branching and conical cells (Pérez-Rodríguez et al., 2005). In addition to the previously reviewed MIXTA family functions (Brockington et al., 2013), recent studies further support roles in glandular trichomes of mint (Qi et al., 2022) and Nepetes (Lamiaceae, Zhou et al., 2022) as well as in trichome patterning and morphogenesis in tomato leaves (Galdon-Armero et al., 2020). A novel function for MIXTA family orthologs in cuticle (Lashbrooke et al., 2015) is further supported in the early land plant model Marchantia (Xu et al., 2020) and in the orchid Phalaenopsis (a monocot, Lu et al., 2022). A MIXTA family ortholog from another orchid, Dendrobium crumenatum, is capable of rescuing the branched trichome phenotype of Arabidopsis thaliana noeck mutants, even though it does not produce trichomes in the orchid (Gilding and Marks, 2010). Since most of the functional information on MIXTA family genes comes from the core eudicots, additional functional studies of MIXTA family genes in early-diverging eudicots are warranted to inform the reconstruction of the ancestral gene function of this important transcription factor family in the eudicots, the most species-rich clade containing three quarters of all angiosperm species (Simpson, 2019).
Trichomes or epidermal hairs represent an emerging model for the study of single-cell differentiation that is structurally simple and easily accessible (Yang and Ye, 2013). Trichomes can be non-glandular or glandular (Feng et al., 2021): the former typically provides protection against UV-B radiation and dehydration, while the latter is a warehouse of defense metabolites against herbivores (Jaime et al., 2013; Yan et al., 2018; Chalvin et al., 2021). On the one hand, differentiation of unicellular trichomes is well understood in A. thaliana, where it is controlled by an activation–inhibition loop (Jakoby et al., 2008). Positive regulators GLABRA3 (GL3), GLABRA1 (GL1), and TRANSPARENT TESTA GLABRA1 (WD40) form a trimeric complex with basic helix–loop–helix proteins (bHLH) known as MYB-bHLH-WD40 that stimulates trichome differentiation in the epidermis (Pesch and Hülskamp, 2009). In the surrounding cells, transcription factors such as TRIPTYCHON (TRY) and TRICHOMELESS1,2 (TCL1 and TCL2) bind and inactivate the bHLH-WD40 complex, thereby suppressing trichome development (Gao et al., 2017). The regulatory network for multicellular trichomes, on the other hand, is less clear, with HD-ZIP IV transcription factors known to interact with MIXTA family orthologs during trichome development in maize, cucumber, and the asterid Artemisia annua (Vernoud et al., 2009; Wang et al., 2016; Yan et al., 2018), while a B-type cyclin plays this role in tomato (Gao et al., 2017). Hence, further investigation of trichome development and the underlying genetic regulators is warranted.
Thalictrum (meadow-rue) are herbaceous perennials in the family Ranunculaceae that lack petals and are pollinated by insects, the wind, or both, with associated floral morphotypes (Martínez-Gómez et al., 2023). Thalictrum MIXTA family orthologs are expressed in the epidermis of floral organs that contain conical–papillate cells (petaloid sepals or showy stamen filaments) that aid in attracting insect pollinators and were previously shown to affect conical–papillate cell elongation when overexpressed in tobacco (Di Stilio et al., 2009). However, the function of these non-core eudicot MIXTA orthologs remains untested in Thalictrum. As predicted then, this gene family represents a lineage that is sister to the core eudicot MIXTA/MIXTA-like duplication (Brockington et al., 2013). We will therefore hereby refer to “paleoMIXTA” (PMX) following the floral MADS box gene nomenclature (paleoAP3; Kramer et al., 1998) and MX shortcut for MIXTA (Brockington et al., 2013) and distinguish this gene lineage from the core eudicot MIXTA/MIXTA-like. Based on this new information, the published gene TthMYBML2 (Di Stilio et al., 2009) is hereby renamed to TthPMX, and the others newly described here will follow this nomenclature scheme.
We chose to compare the gene functions in T. thalictroides, with conical–papillate cells in their petaloid sepals, and T. clavatum, with conical–papillate cells in their showy stamen filaments. The two species are closely related within clade I and represent the ancestral traits for the genus of diploid, hermaphrodite, and insect pollinated (Soza et al., 2012; Soza et al., 2013; Wang et al., 2019). Using virus-induced gene silencing (VIGS) as previously described (Di Stilio et al., 2010), we downregulated the paleoMIXTA ortholog in both species. We show evidence for a new role for this floral MYB transcription factor in leaf trichomes and further show that it is necessary for conical cell formation that contributes to petaloidy in sepals and stamens for these two species of Thalictrum with distinct floral morphologies.
This study aims broadly to provide functional information for members of the paleoMIXTA gene lineage via functional assays in flowers and leaves of early-diverging eudicots to ultimately contribute to an increased understanding of the functional evolution and molecular mechanisms of this important transcription factor family. To that end we (1) reconstruct the evolutionary history of MIXTA family orthologs from the genus Thalictrum, (2) investigate the function of two paleoMIXTA orthologs in the flowers and leaves of two distinct species, and (3) identify candidate genes underlying the MIXTA gene regulatory network (GRN) in leaves by RNA-Seq.
The evidence presented here from functional studies of Thalictrum paleoMIXTA representatives suggests that the function of a putative pre-MIXTA/MIXTA-like core eudicot duplication ancestor likely consisted of a combined role in conical–papillate cells and trichomes. This multifunctional eudicot ancestral gene presumably could have parsed out its distinct roles among paralogs after the core eudicot MIXTA/MIXTA-like duplication as well as within taxa that underwent further lineage-specific duplications. Our finding of a duplication event within Thalictrum resulting in differential patterns of paralogous gene expression provides a potential basis for the evolution of increased epidermal trichomes found on the leaves and flowers of derived wind-pollinated polyploid taxa and thus represents an interesting avenue for future research.
A phylogenetic approach was used to investigate the evolutionary history of the Thalictrum (Ranunculaceae) MYB SBG9-A MIXTA gene family or paleoMIXTA (PMX, Figure 1A inset). To this end, a nucleotide alignment was constructed for 49 MYB SBG9-A Thalictrum sequences isolated from 23 species spanning the two main clades (Soza et al., 2012; Martínez-Gómez et al., 2023) and identified based on the conserved R2R3 and Subgroup 9-A domains (Supplementary Figure S1). The two SBG9-A outgroups chosen were columbine Aqcoe3G346800 (Ranunculaceae, Aquilegia coerulea) and California poppy EscalH2.4G309300 (Papaveraceae, Eschscholzia californica var. Aurantiaca Orange King Plant1.1 v1.1, DOE-JGI, http://phytozome.jgi.doe.gov/info/Ecalifornicavar_AurantiacaOrangeKingPlant1_1HAP2_v1_1).
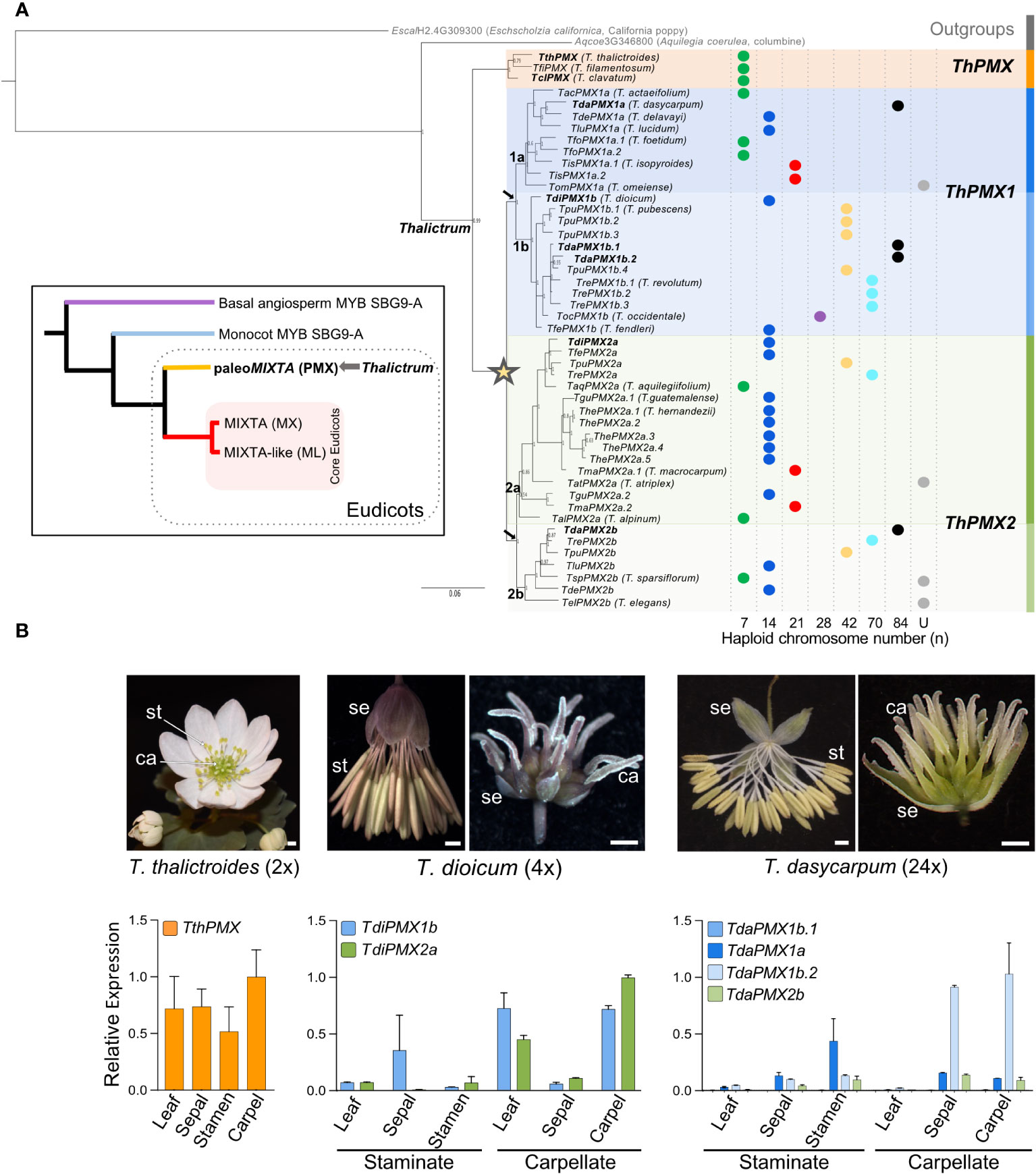
Figure 1 Gene phylogeny of the Thalictrum MIXTA family of MYB transcription factors (R2R3 Subgroup 9A). (A) Bayesian inference phylogeny with Aquilegia coeruleae (Ranunculaceae) and Eschscholzia californica as outgroups showing two major clades: clade I (orange) named Thalictrum paleoMIXTA (ThPMX) and clade II with an inferred gene duplication event (star) leading to subclades ThPMX1 (blue) and ThPMX2 (green). Another duplication in each clade (black arrows) leads to “a” and “b” subclades, and additional species-level duplications are designated with a period followed by a sequential number (e.g., TdaPMX1b.1–TdaPMX1b.4). Posterior probabilities are shown next to the nodes. Branch lengths indicate substitutions per site. Haploid (n) chromosome numbers are shown for each species as colored dots, from n = 7 (diploid) to n = 84 (24-ploid); U = unknown (data from Soza et al., 2013). Genes in bold are the subject of this study. Inset: Simplified tree of the MIXTA gene family in angiosperms [modified from Brockington et al. (2013)] highlighting the ranunculid MIXTA lineage (paleoMIXTA) that includes the Thalictrum orthologs. Organismal coding: purple = ANITA grade; blue = monocots; yellow = early-diverging eudicots; red = core eudicots. (B) Relative expression of Thalictrum paleoMIXTA in leaves and floral tissues of representative species from major clades. From left to right: Representative photos of hermaphrodite flowers of T. thalictroides (diploid) and staminate and carpellate flowers of dioecious T. dioicum (tetraploid) and T. dasycarpum (high-level polyploid). Quantitative RT-PCR showing the average expression level of Thalictrum paleoMIXTA orthologs relative to the average expression of two housekeeping genes ThACTIN and ThEEF1-alpha, normalized to the highest-expressing organ (carpel) and paralog. Color-coding matches the gene lineages shown in (A). Error bars represent ± standard error of the mean. N = 4–7 flowers. Photo credits: Samantha Hartogs (T. thalictroides and T. dasycarpum); Kelsey Galimba (T. dioicum). se, sepals; st, stamens; ca, carpels.
A consensus phylogenetic tree was inferred using the Bayesian optimality criterion implemented in MrBayes (Figure 1A). Thalictrum MIXTA family sequences form two distinct clades with strong support, coincident with the two major clades previously described: clade I Thalictrum paleoMIXTA (ThPMX) containing only diploid species and clade II with subclades paleoMIXTA1 (ThPMX1) and ThPMX2 having both diploid and polyploid species. The gene tree suggests a gene duplication event at the base of the two subclades (Figure 1A, star), with paralogs from polyploid species distributed among the two resulting clades and instances of potential paralog loss (e.g., the 20-ploid T. revolutum has a gene copy in all clades except ThPMX1a). Each of these larger gene clades, in turn, underwent a subsequent duplication, designated as “a” and “b”. Certain polyploid species, particularly those with high ploidy level, like T. dasycarpum (24x) and T. revolutum (20x), had additional species-level single-gene duplications that were consecutively numbered (e.g., TpuPMX1b.1-TpuPMX1b.4). Thus, MIXTA family orthologs only partially follow the species phylogeny, suggesting a reticulated evolutionary history for Thalictrum that could not be previously detected when using chloroplast loci and the transcribed spacers ITS and ETS (Soza et al., 2013; Martínez-Gómez et al., 2023).
To broadly investigate the spatial expression pattern of Thalictrum paleoMIXTA, we performed qPCR on leaves and dissected floral organs (sepals, stamens, and carpels) of recently open flowers, where expression is known to peak in other species (Pérez-Rodríguez et al., 2005; Lau et al., 2015; Lu et al., 2022). We chose diploid T. thalictroides, tetraploid T. dioicum, and high-level polyploid T. dasycarpum (24×) as representative species. Thalictrum thalictroides is in clade I and has the ancestral traits of insect pollination and hermaphrodite flowers, with petaloid sepals that contain conical cells. In contrast, T. dioicum and T. dasycarpum from clade II are wind-pollinated polyploids with small and inconspicuous unisexual flowers that contain long pendulous stamens or plumose stigmas on long styles (Soza et al., 2012; Martínez-Gómez et al., 2023).
There was a trend towards high Thalictrum paleoMIXTA expression for specific paralogs in the carpels of the three species in the leaves of T. dioicum carpellate (female) plants and in the sepals of carpellate T. dasycarpum flowers (Figure 1B). Stamens and sepals from staminate (male) flowers showed a lower combined relative expression across the three species. T. thalictroides had expression in all organs analyzed, likely from a combination of having conical–papillate cells in some organs and trichomes in others, with the highest expression in carpels, followed by sepals, leaves, and stamens. Similarly, both paralogs from T. dioicum showed a high carpel-specific expression, suggesting partial redundancy in function. In T. dasycarpum, the expression was low to undetectable in the leaves, and one paralog (TdaPMX1b.2) had a high expression in carpels while another (TdaPMX1a) had a higher expression in stamens. On closer inspection, the R2 and R3 MYB domains of the lowest-expressing copy (TdaPMX1b.1) had two single amino acid substitutions, K43N and S76N, in the R2 and R3 domain, respectively (Supplementary Figure S1). Single amino acid substitutions in the R2R3 domains can cause loss of promoter–site interactions in other MYB proteins, resulting in the downregulation of expression (Hichri et al., 2011; Dai et al., 2016). Taken together, specific paleoMIXTA paralogs showed a sex-dependent expression pattern in dioecious Thalictrum, with the lower-level polyploid T. dioicum showing an overlapping expression of paralogs and the higher level polyploid T. dasycarpum exhibiting more specialization of paralog expression patterns.
In order to investigate the gene function in a paleoMIXTA representative from an early-diverging eudicot, TthpaleoMIXTA (TthPMX) was targeted for downregulation by VIGS in T. thalictroides, on its own or together with PHYTOENE DESATURASE (PDS, as a “reporter” causing visible tissue photobleaching). The infection efficiency was 35%, with 15 out of 45 plants showing a range of photobleaching between 3 to 4 weeks after infiltration. Plants treated with the empty vector (EV = mock control) were indistinguishable from untreated wild-type plants, except for the previously described background effect of small necrotic lesions (Di Stilio et al., 2010) (Supplementary Figure S2). Viral transcripts (TRV1 and TRV2) were detected in treated plants and EV controls (Supplementary Figure S3A).
Targeted gene silencing was validated by qPCR, showing an approximately 11-fold decrease in the average expression of TthPMX in plants treated by VIGS compared to empty vector controls (Figure 2D). Fully photobleached and variegated (partially photobleached) leaves from plants treated with TRV2–ThPDS–ThPMX showed a correlated downregulation of both genes, confirming the utility of ThPDS as a visual indicator of gene silencing (Supplementary Figure S3B).
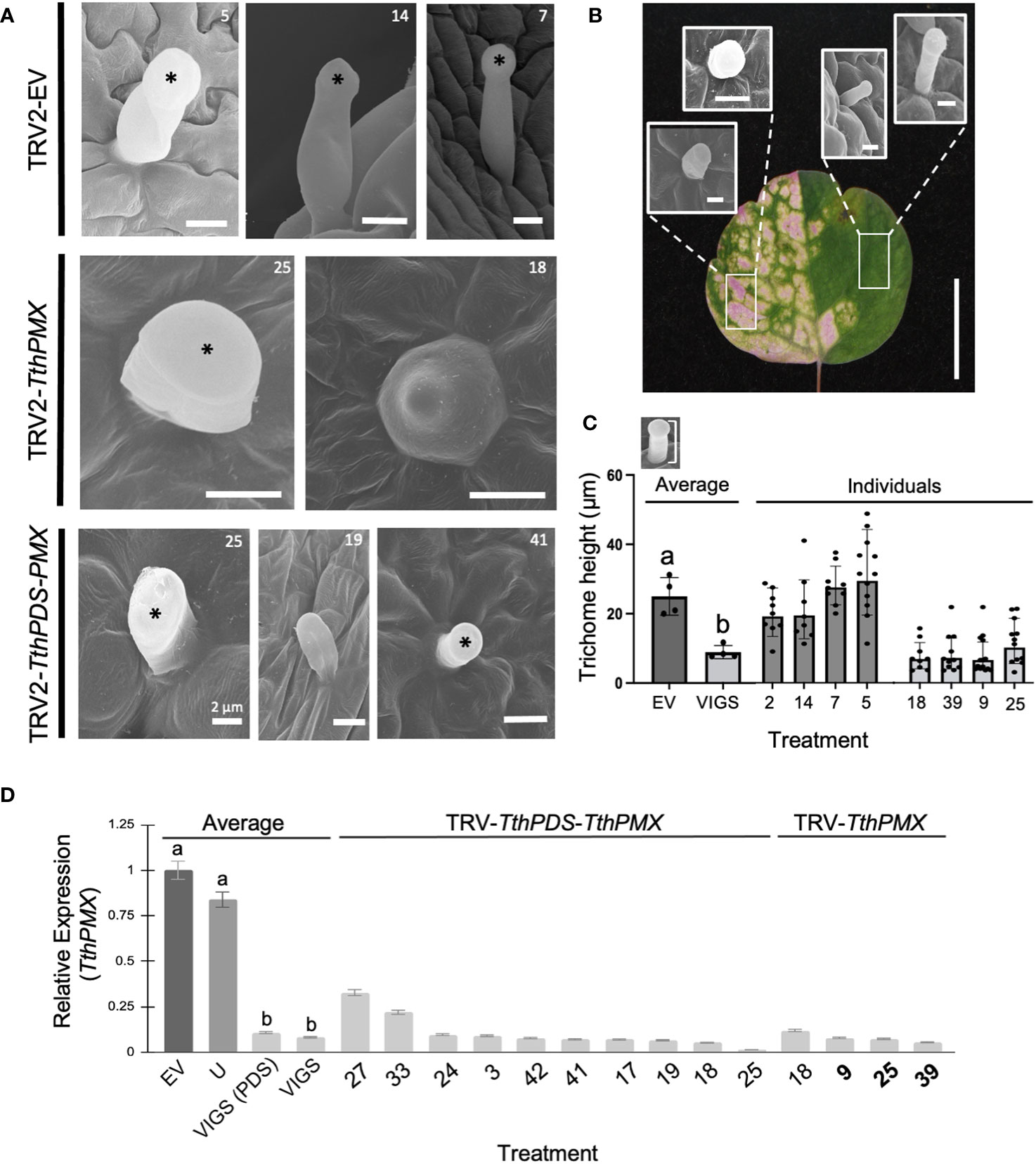
Figure 2 Targeted silencing of a Thalictrum paleoMIXTA ortholog affects leaf trichome morphogenesis. (A) Comparison of trichomes found on the abaxial epidermis of leaves of Thalictrum thalictroides in three representative empty vector control plants (TRV2-EV, top row) and four VIGS-treated plants with the construct TRV2-TthpaleoMIXTA (TthPMX). Asterisks indicate the position of the trichome head. Reduced “stubby” trichomes in plants undergoing targeted gene silencing for TthPMX represent three independent transgenic lines (middle and bottom row, transgenic line numbers shown for cross-reference). Scale bar = 10 μm, except as indicated. (B) Representative trichome phenotypes in green and photobleached sectors within a variegated leaf undergoing partial silencing of TthPDS. Rectangles show areas where the trichomes were sampled. Scale bar = 10 mm. (C) Quantitative difference in trichome cell height, measured as diagrammed in the top inset. Different letters indicate statistical significance between group means in one-way ANOVA with Tukey’s HSD (P ≤ 0.0001, N = 4 independent transgenic plants). (D) Molecular validation of VIGS experiments by qPCR for TthPMX as average expression level relative to TthACTIN and TthEEF1 housekeeping genes, normalized against the empty vector controls. Error bars represent ± standard error of the mean. Different letters indicate statistically significant differences in one-way ANOVA with Tukey’s comparison test (P = 0.008).
To further establish the role of T. thalictroides paleoMIXTA in leaves, we quantified the epidermal features on their adaxial and abaxial epidermis from VIGS-treated plants, including the density and height of trichomes, the stomatal density, and the pavement cell shape. T. thalictroides has sparse trichomes on the abaxial leaf epidermis (Figures 2A, B). Under scanning electron microscopy (SEM), the trichomes appeared unicellular and capitate (enlarged toward the tip), which places them in the glandular category based on morphology (Simpson, 2019; Werker, 2000). The trichomes did not differ between the leaves of empty vector control and untreated plants (Figure 2A, top row). In contrast, the trichomes on the leaves of VIGS-treated plants showed either a “stubby”, short-stalk phenotype (Figure 2A, middle and bottom rows) resembling a conical cell or one that appeared irregular in shape (Figures 2A, B; Supplementary Figure S4).
To further confirm that trichome developmental abnormalities resulted from the downregulation of the target gene, we compared sectors of photobleached and green tissues within variegated leaves (Figure 2B). Green leaf sectors had qualitatively taller trichomes than those found in photobleached sectors (Figure 2B; Supplementary Figure S4). To further quantify this observation and to avoid the potential side effects of PDS downregulation, we measured the trichome height in single-construct VIGS (without PDS); the average trichome was 2.7-fold shorter in VIGS-treated leaves compared to the empty vector controls (Figure 3C, p ≤ 0.0001, N = 41). Taken together, these results suggest that T. thalictroides paleoMIXTA plays a role in the elongation and morphogenesis of leaf trichomes.
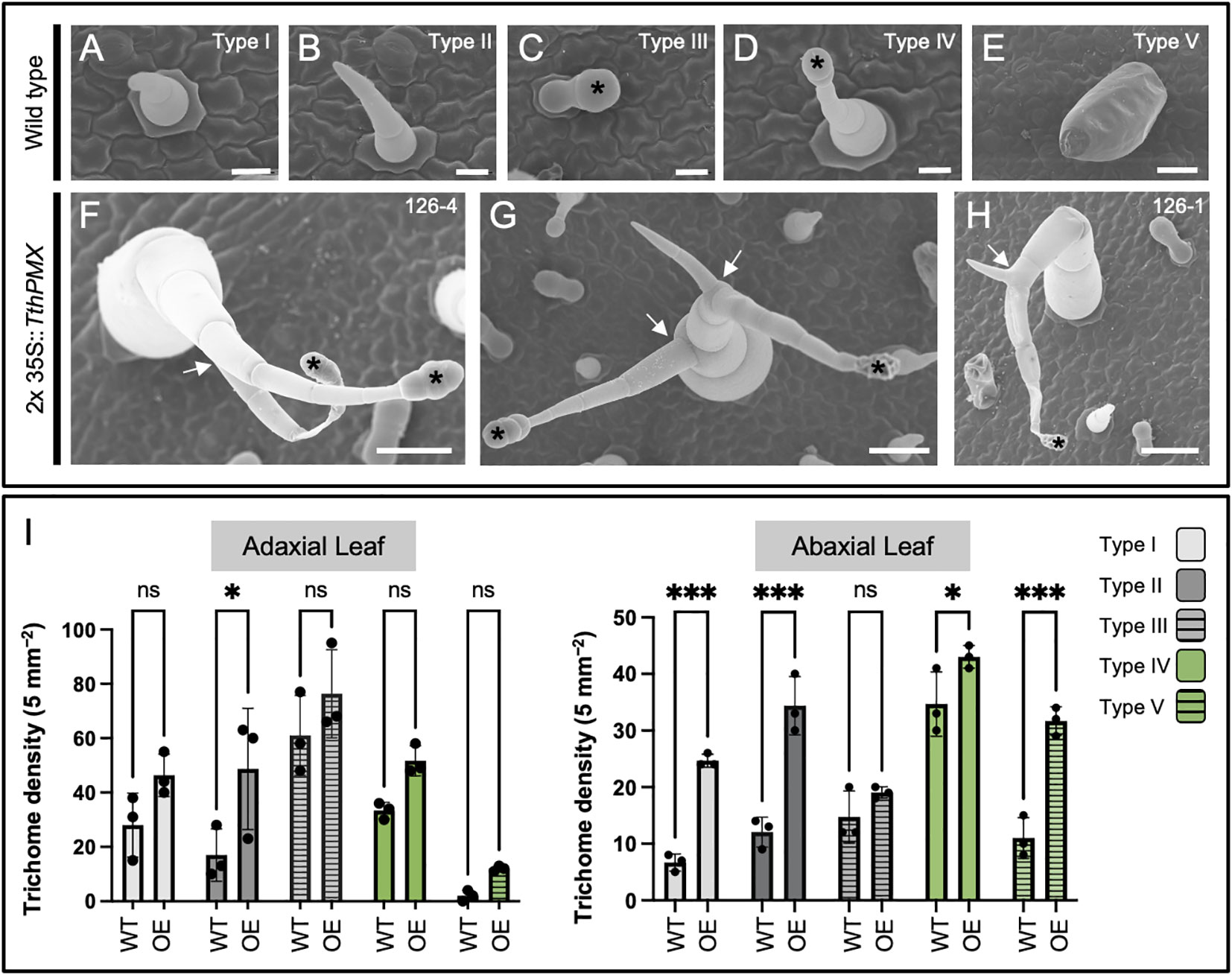
Figure 3 Overexpression of the MIXTA family ortholog TthpaleoMIXTA from Thalictrum thalictroides (Ranunculaceae) in tobacco leaves leads to ectopic and abnormal trichomes. (A–E) Trichome types I–V on the leaves of wild-type tobacco. Black asterisks indicate the position of the glandular head of capitate trichomes. (F–H) Ectopic branched trichomes on transgenic tobacco leaves; white arrows indicate abnormal branching. (I) Average trichome density ± SE by trichome type per 5 mm² on adaxial (left) and abaxial (right) leaf epidermis from three independent transgenic tobacco lines (2x35S::TthPMX) compared to wild type. Asterisks indicate statistically significant differences in trichome counts in a two-way ANOVA with Holm–Šídák’s multiple comparisons. ns, not significant; *P < 0.05; ***P < 0.001. Scale bar = 50 μm.
We investigated additional epidermal features that have been reported to be influenced by other MIXTA family members, such as pavement cell morphology and stomatal density (Brockington et al., 2013). No significant qualitative differences were observed in the morphology of pavement cells between VIGS and control plants (Supplementary Figure S5A). Because stomatal density appeared to vary along the leaf, we sampled three regions on the abaxial side of transgenic plant leaves: top, middle, and base. In spite of some variations in stomatal density (visualized as a heat map, Supplementary Figure S5H), there was no statistically significant difference neither among leaf regions for a given treatment (Supplementary Figures S5I, J) nor between treatments when comparing averages (Supplementary Figure S5G, N = 5 independent transgenic plants per treatment).
Heterologous overexpression was previously pursued using an established bioassay in tobacco to observe changes in ovary wall cell morphology (Di Stilio et al., 2009), and fixed leaves from that experiment were used here for observations of their epidermal features under SEM. In the tobacco family Solanaceae, glandular, non-glandular, and defense trichomes are found (Bar and Shtein, 2019). Two types of non-glandular (types I and II), glandular (III and IV), and one type of defense trichome (type V) were identified on the upper and lower epidermis of wild-type tobacco leaves (Figures 3A–E).
Transgenic plants overexpressing TthPMX exhibited ectopic types II and IV branched trichomes (Figures 3F–H, arrows) and a higher density of type II trichomes adaxially and of types I, II, IV, and V abaxially (Figure 3I). Taken together, these findings suggest that TthPMX is a positive regulator of trichome development, capable of affecting their morphogenesis and patterning on the leaf epidermis of a core eudicot.
To determine the molecular mechanisms underlying the role of paleoMIXTA in trichome development, we compared the transcriptomes of leaves from three mock-treated (empty vector) and three validated TRV2–TthPMX VIGS transgenic lines by RNA-Seq (Figure 4, transgenic line numbers in bold). There were 368,326,658 total reads, with a yield of 110,499 Mb and an average quality score of 35.84 (93.32% bases ≥3) (Supplementary Table S3). To independently validate the RNA-Seq results, we analyzed the expression counts for our target gene (TthPMX) and for genes known to have ubiquitous and stable expression (housekeeping genes). Leaves from VIGS-treated plants exhibited, on average, a fourfold decrease in TthPMX expression compared to controls, while the transcripts of the housekeeping genes TthEEF1-alpha and TthACTIN showed comparable expression levels in both treatments (Figure 4A).

Figure 4 Identification of co-regulated candidate genes in the Thalictrum paleoMIXTA gene regulatory network. RNA-Seq analysis of differentially expressed genes (DEGs) in leaves from plants targeted for TthPMX downregulation (VIGS) vs. empty vector control (EV). (A) RNA-Seq validation with normalized count expression in transcripts per million of TthpaleoMIXTA and the housekeeping genes TthEEF1-alpha and TthACTIN. Means ± SE from three biological replicates; different letters indicate highly statistically significant difference (p < 0.0001) in one-way ANOVA followed by Tukey’s multiple comparisons. (B) Volcano plot depicting log fold change in expression; red indicates upregulated differentially expressed genes (DEGs) and blue downregulated DEGs. Identified candidate genes amongst outliers are shown. (C) Hierarchical clustering and heat map of the top 30 DEGs with the highest or the lowest expression profiles. The color scale indicates the expression levels (log2 fold change) relative to TRV2-EV controls. Genes identified via Diamond BLAST are listed; those of special interest are shown in bold and further discussed in the text.
The RNA-Seq analysis resulted in approximately equal numbers of significantly differentially expressed genes (DEGs) that were either upregulated (49%) or downregulated (50.3%) during the targeted silencing of TthPMX, using a cutoff log2 fold change of ±4, respectively. Differentially expressed genes were mapped onto a volcano plot, where the annotation of outliers identified a Ras-related protein (RABD) and phytochrome B that were significantly upregulated. Downregulated DEGs were identified as coding for DNA Damage Binding1, CULLIN4-associated factor homolog (DCAF), stress and UV damage response protein KIN-17-like, and lysine methyltransferase (Figure 4B). Annotation of the 30 highest- and lowest-expressing DEGs in the heat map uncovered a few more candidate genes upregulated in leaves undergoing VIGS of ThPMX (Figure 4C). These comprised orthologs of microtubule regulating CALMODULIN-BINDING 25-LIKE (CaML-25), kinesin-like protein (KLP), gluconeogenesis-associated phosphoenolpyruvate carboxy-kinase like (PEPCK), and multiple organellar RNA editing factor 2 (MORF2). The downregulated genes included those coding for motor proteins such as KINESIN and 65-kDa microtubule-associated protein (MAP65), defense peptides such as thionins (THI 2.1, THI 2.2, and α-1-purothionin), Rhodanese-like domain, epidermal specific subtilisin-like protease (SDD1; Von Groll et al., 2002), SANT/MYB domain proteins involved in chromatin remodeling, and ETHYLENE RESPONSIVE FACTOR 1 (ETF1) expressed highly in Arabidopsis mature trichomes (Jakoby et al., 2008). Taken together, the comparative transcriptomic analysis suggests that TthpaleoMIXTA influences trichome morphogenesis via the regulation of genes coding for candidate proteins involved in microtubule regulation, epidermal cell patterning, plant defense, and chromatin remodeling.
To further investigate the potential role of ThPMX in the epidermal features of flowers, we conducted functional studies by VIGS in two species of Thalictrum representing distinct flower morphologies, the “petaloid sepal” and “showy stamen” morphotypes (Martínez-Gómez et al., 2023). Untreated T. clavatum flowers are white to light pink and consist of a few small sepals and numerous stamens with showy or “petaloid” filaments (clavate, flattened, and wider at the top, and colored) as previously described (Di Stilio et al., 2010) (Figure 5A). Photobleaching of PDS is detectable in the floral center as a sign of effective gene silencing (Figure 5B). Conical–papillate cells are most prominent in the filaments of outer stamens (Figures 5C, D) and also present adaxially in sepals. In order to test the role of TclPMX in the conical–papillate cells of T. clavatum stamens, we used VIGS and observed the resulting floral cellular phenotypes under SEM. In wild-type plants, this gene is expressed at low levels in sepals and mostly in stamens and carpels, whereas T. thalictroides expresses relatively less in stamens (Figure 5N).
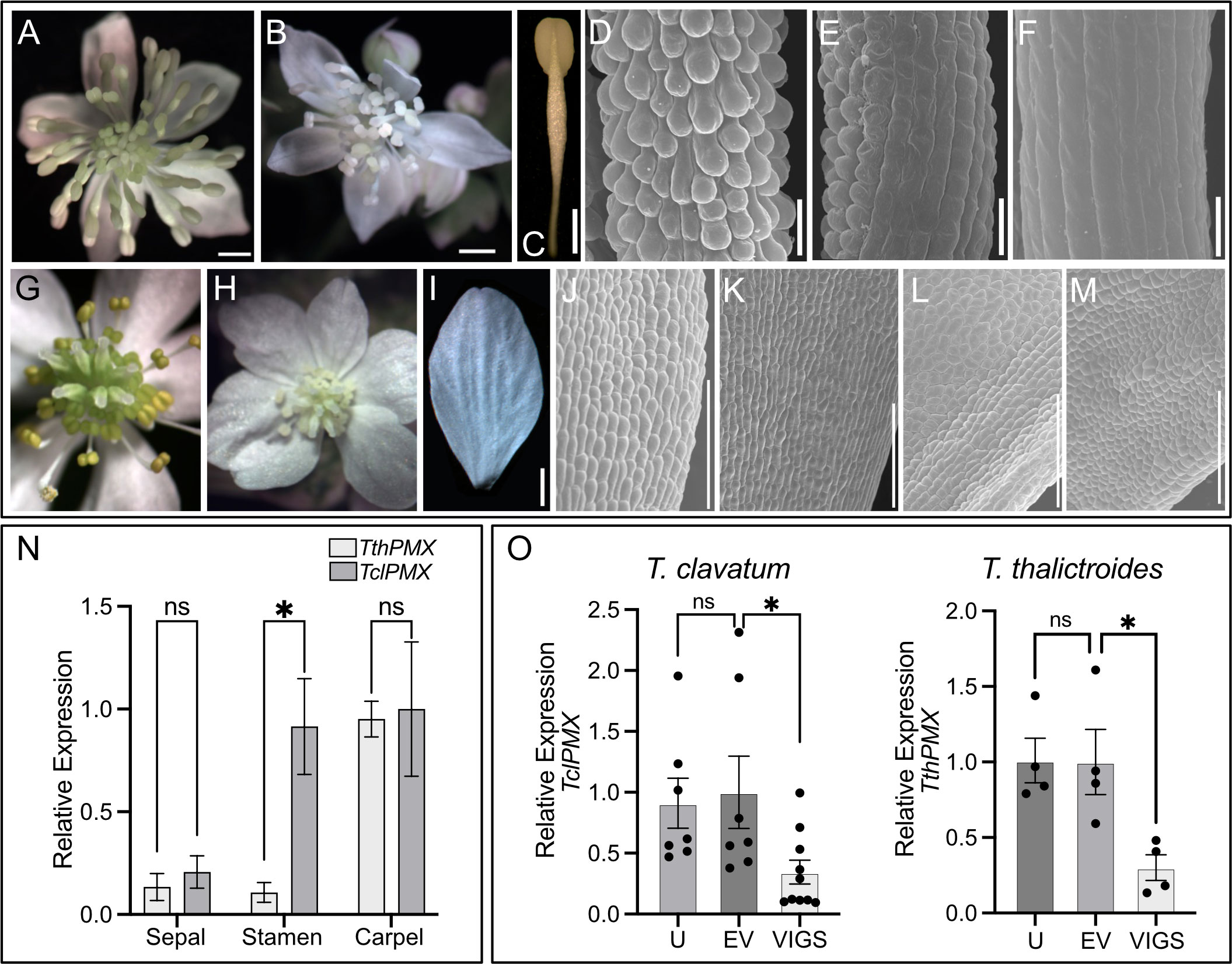
Figure 5 Thalictrum paleoMIXTA orthologs promote conical–papillate cells on the epidermis of flower organs. Virus-induced gene silencing (VIGS) of ThPMX in Thalictrum clavatum (A–F) and T. thalictroides (G–M). (A) T. clavatum whole untreated flower. (B) VIGS-treated flower showing signs of photobleaching in the central region. (C) Control (untreated) outer stamen, whole view. (D) Detail of stamen filament in (C), with protruding conical–papillate cells. (E) Stamen filament detail of flower treated with TRV2–PDS–TclPMX, showing signs of incomplete gene silencing; note the conical–papillate cells on the left half compared to the flatter cells on the right half. (F) Fully silenced stamen filament with flat epidermal cells. All filaments depicted are from fully mature outer stamens (before anther dehiscence). (G) T. thalictroides whole untreated flower. (H) VIGS-treated flower showing signs of photobleaching in the central region (immature stamens and carpels). (I) Control (untreated) outer sepal, whole view. (J, K) Detail of the adaxial epidermis of two untreated sepals with slightly protruding conical–papillate cells. (L, M) Detail of sepal adaxial epidermis undergoing VIGS in flowers from two independent plants treated with TRV2–TthPDS–TthPMX, with flatter epidermal cells. (N) Wild-type relative expression by qPCR of TthPMX and TclPMX in dissected floral organs of T. thalictroides and T. clavatum, respectively. (O) Molecular validation of VIGS experiments in T. clavatum and T. thalictroides. U, untreated; EV, empty vector control; VIGS, virus-induced gene silencing. The values shown in (N, O) are average expression levels relative to TthACTIN and TthEEF1 housekeeping genes, normalized to the highest expressing organ in (N) (carpel) or to EV control in (O). The asterisk denotes statistical significance at P < 0.05 by Dunnett’s multiple-comparisons test, ns: not significant. The scale bar is 1 mm in (A–C, G–I) and 100 µm in (D–F, J–M).
As in the leaf experiments, plants were visually assessed for photobleaching in leaves and flowers post-infiltration. T. clavatum began to show signs of photobleaching in leaves 13 days after infiltration, and the first buds opened 4 days later. The photobleached flowers tested positive for the TRV1 and TRV2 viruses as expected in all 10 plants that displayed the phenotype. The mock-treated plants displayed an identical TRV1 as well as a lower-molecular-weight TRV2 band, confirming the absence of insert (Supplementary Figure S6). The untreated plants did not show signs of TRV1 or TRV2 (Supplementary Figure S6A). Target gene silencing was validated by qPCR, showing a significant downregulation of approximately threefold in VIGS treatments compared to controls (P = 0.04). The SEM of stamens showed epidermal cells that looked flatter in VIGS treatments, either partially (Figure 5E) or fully along the filament (Figure 5F), and these observations were confirmed in the stamens of at least three independent transgenic plants.
Untreated T. thalictroides flowers have white petaloid sepals, stamens that are green to yellow, and carpels that are green toward the base (Figures 5G, I), therefore allowing for the detection of photobleaching in the flower center (Figure 5H). The conical–papillate cells are subtle in this species but can be observed adaxially at the base of the sepals under SEM (Figures 5J, K). Flowers showing signs of photobleaching similarly contained TRV virus (Supplementary Figure S6B), and the qPCR of dissected sepals showed a significant downregulation of TthPMX by approximately threefold (P = 0.02, N = 4 independent transgenic plants) (Figure 5O). Their sepal epidermis had flatter cells adaxially than the untreated controls (compare Figures 5J–M).
Thus, these results confirm a role for TthPMX in conical–papillate cells on the perianth in the non-core eudicot Thalictrum, previously suggested via heterologous expression in tobacco (Di Stilio et al., 2009). Taken together, the T. clavatum experiments suggest a more general function for ThPMX in epidermal protrusions beyond the perianth and into the reproductive organs.
Within the early-diverging eudicots, Thalictrum is in the order Ranunculales, sister to the rest of the eudicots. Since MIXTA and MIXTA-like are found exclusively within the core eudicots, we propose here that Thalictrum orthologs represent a “paleo” MIXTA lineage, analogous to how the APETALA3 subfamily of floral MADS-box genes has a paleoAP3 lineage in early-diverging eudicots sister to the euAP3/TM6 core eudicot gene duplication (Kramer et al., 1998). Therefore, Thalictrum paleoMIXTA is informative of the ancestral function of MIXTA family orthologs.
The phylogenetic inference of the MIXTA-like family in Thalictrum uncovered a lineage-specific duplication that coincides with a WGD event in the genus. This result suggests that, early in the history of the genus, the evolution of the MIXTA-like gene family was impacted by polyploidy, which is ubiquitous in Thalictrum.
The MYB family R2R3 SBG9-A gene TthPMX (previously TthMYBML2) from T. thalictroides had been shown to increase the height of ovary epidermal cells when overexpressed in tobacco (Di Stilio et al., 2009). This study provides evidence for the role of TthPMX in conical–papillate cells in the floral organs of Thalictrum while uncovering a novel function as a positive regulator of leaf trichome development. Most subgroup 9-A MIXTA family genes such as Antirrhinum majus (snapdragon) AmMYBML1 (Pérez-Rodríguez et al., 2005) and A. annua AaMIXTA1 (Shi et al., 2018) are positive regulators of trichome development. In tomato, the ectopic expression of SIMIXTA increases type I trichomes, whereas RNA silencing reduces them (Ewas et al., 2016; Schuurink and Tissier, 2020). However, certain MIXTA-like genes have developed inhibitory roles in trichome initiation, such as Mimulus guttatus MgMYBML8 (Scoville et al., 2011) and Arabidopsis MYB106 (Gilding and Marks, 2009). Hence, our finding that Thalictrum paleoMIXTA positively affects trichome development suggests that the negative role of MIXTA-like genes in trichome development could be a derived function in certain core eudicots.
Arabidopsis has become a model to study the morphogenesis and anisotropic development of epidermal cells (Yang et al., 2019). Microtubule binding proteins are important components of the plant cytoskeleton that have been implicated in the development of both conical cells and branched trichomes (Reddy et al., 2004). Previously, trichomes with short stalks were observed in Arabidopsis loss-of-function mutants for kinesin-like calmodulin-binding protein (KCBP) (Oppenheimer et al., 1997). However, the mechanisms of trichome morphogenesis in species outside of this derived model plant are poorly understood.
Our RNA-Seq data shows the downregulation of genes coding for the microtubule motor protein KINESIN, DCAF, KIN-17-like, and MAP65 and the upregulation of microtubule-regulating CaML-25 and KLP. In Arabidopsis, calmodulin-like proteins interact with kinesin microtubule motor proteins to regulate trichome morphogenesis (Oppenheimer et al., 1997; Reddy et al., 2004). In addition, there was downregulation of subtilisin-like proteases such as SDD1 that play a role in cell signaling and spacing of stomatal cells (Von Groll et al., 2002); however, their role in the spacing of other epidermal cells is unknown. While our qualitative SEM analysis of VIGS-treated plants did not uncover statistically significant differences in the morphology of pavement cells or the stomatal density, there was visible variability in stomatal densities among individual transgenic plants across the top, middle, and bottom leaf abaxial regions (Supplementary Figure S5H), thus warranting further investigation.
The top candidates included DEGS that are also highly expressed in mature Arabidopsis trichomes (top 5%), such as PEPCK, ETF1, KINASE, and RABD. These results together suggest that MIXTA family orthologs from phylogenetically distant plant species may have converged in regulating cell elongation in trichomes through similar microtubule-mediated mechanisms. Comparable genetic components point to the potential conservation of the genetic machinery for the development of epidermal extensions in eudicots.
Thalictrum species are often cultivated for their wealth of biochemical compounds such as alkaloids, found in roots and leaves (Hao et al., 2021). It is currently unknown whether Thalictrum trichomes have the ability to synthesize or accumulate bio-compounds. Our study provides evidence that targeted silencing of TthPMX affects trichome morphology, and modulates genes related to plant stress and defense. Basic secretory protein (BSP), Rhodanese-like domain, multiple enzymes and genes encoding antimicrobial thionins were co-repressed in transgenic plants undergoing gene silencing. Glandular trichomes of Artemisia annua secrete Rhodanese-like domain protein for defense and detoxification (Wu et al., 2012), while leaf specific-thionins are small metabolites with roles in plant defense against pathogens and herbivores (Escudero-Martinez et al., 2017). Therefore, it appears likely that Thalictrum trichomes play a role in defense against herbivores. Moreover, the downregulation of secondary metabolites such as secretory proteins, sugar transport proteins and enzymes like methyltransferase, amidase, and protease in VIGS-treated plants supports a glandular role for Thalictrum trichomes that is consistent with their morphology (Figure 2).
It has been speculated that timing, developmental stage, or localization may determine the specialized cell forms that develop from the expression of MIXTA-like genes (Pérez-Rodríguez et al., 2005). Interestingly, our comparative transcriptomic approach revealed that TthPMX positively regulates a SANT/MYB domain-containing protein involved in local chromatin remodeling (Boyer et al., 2004), suggesting that paleoMIXTA genes could potentially reprogram epidermal cell fate via the regulation of chromatin remodeling complexes.
Functional studies of MIXTA family orthologs in the monocot and core eudicot clades suggest that they are sufficient to drive the development of multiple epidermal features. For instance, in the orchid P. aphrodite, both PaMYB9A1/2 paralogs can drive the differentiation of conical cells and cuticle biosynthesis in petals (Lu et al., 2022). Similarly, snapdragon AmMYBML1 can modulate the differentiation of trichomes in leaves and of conical cells in petals (Pérez-Rodríguez et al., 2005). In the liverwort Marchantia polymorpha (a bryophyte, or non-vascular plant), MIXTA family homologs play a role in cuticle in addition to papillate cells, representing a more ancestral function potentially common to all land plants (Xu et al., 2020).
Here, we show that Thalictrum paleoMIXTA from the early-diverging eudicots can modulate the development of leaf trichomes and floral conical–papillate cells. Hence, if the trichome and conical cell roles found here were extended to other early-diverging eudicots, it may be possible to reconstruct a combined function back to the last common ancestor of eudicots. In closing, our findings in Thalictrum join those in other plant systems to suggest a deeply conserved role for MIXTA family transcription factors in trichomes and conical–papillate, readily producing a diversity of epidermal features.
T. thalictroides bare root plants were purchased from Sundquist nursery (Poulsbo, WA) and mature T. clavatum plants from Gardens of the Blue Ridge Inc (Pineola, NC). Both sets of plants were vernalized at 4°C before infiltration. Voucher specimens are listed in Supplementary Table S1.
MIXTA family homologs were recovered from 23 species representative of all major clades in the Thalictrum phylogeny by PCR and cloning and/or direct sequencing (vouchers listed in Supplementary Table S1). Genomic DNA was extracted using the MP Bio101 FastDNA Kit, and complete or near-complete paleoMIXTA loci were amplified by PCR with locus-specific primers (Supplementary Table S2) and cloned with pCRII TOPO TA (Invitrogen, Carlsbad, CA, USA). Positive clones were purified using a 5 Prime FastPlasmid Miniprep kit and sequenced (Genewiz, South Plainfield, NJ, USA). At least eight colonies per diploid species or 15 colonies per polyploid species were sequenced to encompass all paralogs. The sequences were edited in Sequencher version 4.9 (Gene Codes Corporation, Ann Arbor, MI, USA) and aligned using MUSCLE (Edgar, 2004). INDELS were coded as gaps and missing data was coded as “?”. The models of evolution for each data set were determined using jModelTest version 2.1 (Posada, 2008). The GTR+I+Γ model was selected based on the Akaike Information Criterion (Akaike, 1974). Bayesian inference analysis was conducted using MrBayes v.3.2 (Ronquist et al., 2012) with a parallel MCMC analysis of 50 million generations, sampling every 1,000 generations. Convergence was checked using the average standard deviation of split frequencies (<0.01) and the effective sample size (ESS) values (>200). The first 25% of trees were discarded as burn-in. The remaining trees were pooled to construct a 50% majority rule consensus tree and visualized using FigTree v1.4.4 (Rambaut et al., 2018).
To determine paralog-specific expression, real-time quantitative PCR (qPCR) was carried out on a Bio-Rad CFX qPCR system, and the results were analyzed in BioRad CFX Manager3.0 software (Bio-Rad laboratories, Hercules, CA, USA). Locus-specific primers were designed in NCBI’s Primer3 (Supplementary Table S2), and primer specificity was tested via melting curve analysis, resulting in a single peak per primer set. Quantification of expression for TthPMX, TthPDS, TclPMX, TdiPM1b, TdiPMX2a, TdaPMX1a, TdaPMX1b.1, TdaPMX1b.2, and TdaPMX2b was performed as previously described (Galimba et al., 2018). Briefly, each 10-μL reaction contained 5 μL of SYBR Green PCR Master Mix (Bio-Rad, Hercules, CA, USA), 0.5 μL (10 μM) of locus-specific primers, 1 μL of template cDNA, and 3 μL of water. The samples were amplified for 40 cycles in duplicate, including a no-template control. The reactions were normalized to the Thalictrum orthologs of two housekeeping genes, ACTIN and EEF1 (EUKARYOTIC ELONGATION FACTOR 1), using the 2-ΔΔCT relative quantification method (Livak and Schmittgen, 2001). The standard deviation of Ct values of reference genes was calculated, and the average of three technical replicates was used to ensure minimal variation in gene expression. Average values and standard errors were graphed and compared statistically by single-factor ANOVA followed by Tukey’s comparisons or Dunnett’s multiple-comparisons test.
T. clavatum and T. thalictroides share 99.8% nucleotide identity for PDS, allowing us to use the same PDS sequence across the two species. TthPDS had been previously cloned into TRV2 (Di Stilio et al., 2010) and was used here as a marker in a double construct with the target gene, also previously cloned (Di Stilio et al., 2009). Briefly, the TRV2–TthPDS construct was double-digested with XbaI and BamHI and then ligated to a TclPMX or TthPMX 400-bp fragment, half of which comprised the end of the C terminal and the other half the 3′UTR. The target sequences were obtained by PCR amplification from plasmid DNA containing the whole coding region of TthPMX (previously TthMYBML2, Di Stilio et al., 2009) using primers with added restriction sites (Supplementary Table S2).
The infiltration of T. thalictroides and T. clavatum was carried out as previously described (Di Stilio et al., 2010) with minor modifications. Briefly, TRV1, TRV2–TthPMX/TclPMX, and TRV2–ThPDS–ThPMX starter cultures were grown overnight with selective antibodies and subsequently used to inoculate 500-mL cultures. Agrobacterium was centrifuged at 4°C and 4,000g for 15 min before being resuspended in an infiltration medium (10 mM MES, 20 μM acetosyringone, and 10 mM MgCl2). Each culture was resuspended to a final OD600 of 2.0 and incubated for 3 h at room temperature. Islet L-77 (Lehle Seeds, Round Rock, TX, USA) was added at 100 uL/L as a surfactant.
Dormant tubers of both species that had been kept at 4°C for 3 weeks were removed from soil and rinsed, and a small razor blade incision was made away from the meristem before being submerged in an infiltration medium containing a 1:1 ratio of TRV1 and TRV2 cultures. T. thalictroides tubers were treated with a construct with the gene of interest alone or with both the target gene and the marker gene phytoene desaturase (ThPDS), whose downregulation induces photobleaching. T. clavatum tubers were treated with the double construct only. In total, 40 tubers were vacuum-infiltrated per species and then potted in 2.5″ Deepots™ (Stuewe & Sons, Tangent, OR, USA) using Sunshine Mix 4 soil (Sun Gro, Bellevue, WA, USA). The plants were kept in growth chambers under a 16-h light–dark cycle at 22°C/18°C. An additional 15–20 plants per species were infiltrated with empty TRV2 (mock). This control was used to test for background viral effects. A total of 15 untreated plants were grown under the same conditions as an additional control.
Fully expanded mature leaves and open flowers before anther dehiscence were collected from plants at 3 to 4 weeks post-infiltration, flash-frozen with liquid nitrogen, and stored at -80°C. Total RNA was extracted using the Spectrum Plant Total RNA Kit or Trizell reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The samples were treated with DNAase I (Thermo Scientific™, Waltham, MA, USA), and first-strand cDNA was synthesized using iScript cDNA synthesis kit (Sigma, Burlington, MA, USA). cDNA was synthesized using TRV1- and TRV2 -specific primers (for viral detection by RT-PCR) or the manufacturer’s mix of random and polyT primers (for qPCR) and amplified using a 51 × 30 PCR cycle and locus-specific primers (Supplementary Table S2). The RT-PCR products were run on 1.2% agarose gel and photographed on GelDoc Image lab software package (Bio-Rad, Hercules, CA, USA).
To determine if target genes had been successfully downregulated, qPCR was conducted as explained above with ThPDS- and ThPMX-specific primers (Supplementary Table S2) on leaves or flowers (whole or dissected) at 4 to 10 samples each of untreated, mock (empty vector), and treatment. Average values and standard deviations for each biological replicate were graphed relative to the highest-expressing tissue.
Plants of both species were photographed using a Nikon D3400 hand-held camera and a dissecting microscope (Nikon SMZ800, Nikon Instruments Inc., Melville, NY, USA) equipped with a QImaging MicroPublisher 3.3 RTV digital camera (Surrey, BC, Canada).
Fully expanded mature leaves and open flowers were collected whole or dissected and fixed in 10% formaldehyde, 5% acetic acid, and 50% alcohol (FAA) for 1 h at RT and overnight at 4°C, dehydrated through an alcohol series, critical point-dried, mounted on stubs, and sputter-coated with gold. Observations were made on a JEOL NeoScope JCM-7000 (University of Washington Microscopy Facility). The leaves fixed in FAA from a previously published tobacco bioassay overexpressing TthMYBML2 (here renamed to TthPMX) under a strong constitutive promoter (Di Stilio et al., 2009) were similarly prepared for SEM. The images were assembled in Affinity Publisher (Serif (Europe) Ltd) or Microsoft Powerpoint.
T. thalictroides leaves and dissected floral organs were photographed under SEM. For leaf trichomes, a 1-mm2 surface adjacent to the mid vein was sampled adaxially and abaxially in three regions: top, middle, and base. Trichomes were found only on the abaxial side and were measured from the base to the tip with Image J (Schneider et al., 2012) using either a straight or segmented line from four independent transgenic plants (at eight to 14 trichomes each).
Tobacco leaf trichomes were sampled in a 5-mm2 area at the base adjacent to the mid vein both adaxially and abaxially. The trichome type was identified, and the number of trichomes found for each type (I–V) was averaged per treatment (three independent transgenic plant lines per treatment). Statistical significance was determined using a two-factor ANOVA followed by Holm–Šídák’s test for multiple comparisons. Chimeric trichomes such as those with two or more types represented within one individual trichome were counted as a separate category and removed from the count data to avoid redundancy.
Four biological samples of T. thalictroides leaves were collected and validated from mock-treated and VIGS-treated lines (TRV2–TthPMX), and their total RNA was extracted for the RNA-Seq analysis. Sequencing was outsourced to Azenta-Genewiz (Burlington, MA, USA), where sample quality was evaluated in an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., CA, USA) before library preparation. mRNA sequencing was conducted using polyA selection on a HiSeq 4000, and the adapters were removed from the sequence reads with Trimmomatic v.0.36 (Bolger et al., 2014). The resulting reads were mapped to a de novo Thalictrum thalictroides transcriptome, assembled with Trinity v2.5 (Haas et al., 2013). Open reading frames were identified with EMBOSS GetOrf, and the transcriptome assembly was annotated using Diamond BLASTx.
To determine changes in transcript expression between controls and plants targeted for gene silencing, gene hit counts were used for downstream differential expression analysis (DEGs). Briefly, the standard bioinformatic analysis package (Azenta-Genewiz, South Plainfield, NJ, USA) consisted of DeSeq2 to generate normalized hit counts, Wald test to generate p-values for statistical significance (Wald, 1943) and log2 fold changes to quantify the expression change between groups. The Benjamini–Hochberg test (Benjamini and Hochberg, 1995) was used to generate adjusted p-values. Genes with adjusted p-values <0.05 and absolute log2 fold change >1 or ≤1 were regarded as differentially expressed. Significant DEGs were functionally annotated, and transcripts of interest, such as TthPMX and orthologs of the reference genes EEF1-alpha and ACTIN, were identified in the assemblies using BLAST. Transcript normalized counts (transcripts per million) were used to graph their relative expression as a validation step. The average expression for three bio-replicates was normalized to mock-treated controls (empty vector, EV), and the statistical significance was calculated using single-factor ANOVA and Tukey’s test for multiple comparison.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA data: PRJNA1030785, NCBI SRA PRJNA1030785 and DRYAD dataset https://doi.org/10.5061/dryad.tdz08kq5k.
SZ: Writing – review & editing, Formal Analysis, Investigation, Visualization, Writing – original draft. AS: Investigation, Writing – review & editing. VD: Writing – review & editing, Conceptualization, Funding acquisition, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by National Science Foundation (USA) Division of Environmental Biology (DEB), Evolutionary Processes, Mary Gates Research Scholarship and Frye-Hotson-Rigg award to AS, grant number 1911539 to VD; Hall International Fellowship and Lawrence Giles and Kruckeberg-Walker Awards to SZ (University of Washington, Department of Biology).
Cathie Martin (John Innes center, Norwich, UK) sent the transformed tobacco seeds. Kacie McCarty, Alessandra (Oddone) Sullivan, and Janet Solano ran the preliminary VIGS experiments. Jens Johnson and Valerie Soza contributed to an early version of the gene phylogeny. Patricia Salles and Christina Owen contributed to the cloning of MIXTA orthologs. Anthony García provided advice on RNA-seq analysis and assistance in locating the samples. We thank Wai Pang Chang (University of Washington microscopy facility) for help in the preparation and observation of SEM samples and the Student Technology Fee (STF) for funding the SEM. We thank Stephanie Ickert-Bond (U. of Alaska) for collecting T. sparsiflorum specimens and sharing seeds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1288961/full#supplementary-material
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723. doi: 10.1109/TAC.1974.1100705
Bar, M., Shtein, I. (2019). Plant trichomes and the biomechanics of defense in various systems, with Solanaceae as a model. Botany 97, 651–660. doi: 10.1139/cjb-2019-0144
Baumann, K., Perez-Rodriguez, M., Bradley, D., Venail, J., Bailey, P., Jin, H., et al. (2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134, 1691–1701. doi: 10.1242/dev.02836
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B. (Methodological). 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyer, L. A., Latek, R. R., Peterson, C. L. (2004). The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163. doi: 10.1038/nrm1314
Brockington, S. F., Alvarez-Fernandez, R., Landis, J. B., Alcorn, K., Walker, R. H., Thomas, M. M., et al. (2013). Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol. Biol. Evol. 30, 526–540. doi: 10.1093/molbev/mss260
Cavallini-Speisser, Q., Morel, P., Monniaux, M. (2021). Petal cellular identities. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.745507
Chalvin, C., Drevensek, S., Gilard, F., Mauve, C., Chollet, C., Morin, H., et al. (2021). Sclareol and linalyl acetate are produced by glandular trichomes through the MEP pathway. Horticult. Res. 8, 206. doi: 10.1038/s41438-021-00640-w
Dai, X., Zhou, L., Zhang, W., Cai, L., Guo, H., Tian, H., et al. (2016). A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3: Ser92 in GL1 is critical for its function. Plant. Cell Environ. 39, 897–907. doi: 10.1111/pce.12695
Di Stilio, V. S., Kumar, R. A., Oddone, A. M., Tolkin, T. R., Salles, P., McCarty, K. (2010). Virus-induced gene silencing as a tool for comparative functional studies in thalictrum. PloS One 5, e12064. doi: 10.1371/journal.pone.0012064
Di Stilio, V. S., Martin, C., Schulfer, A. F., Connelly, C. F. (2009). An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol. 183, 718–728. doi: 10.1111/j.1469-8137.2009.02945.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. doi: 10.1093/nar/gkh340
Escudero-Martinez, C. M., Morris, J. A., Hedley, P. E., Bos, J. I. B. (2017). Barley transcriptome analyses upon interaction with different aphid species identify thionins contributing to resistance: Barley transcriptional responses to aphids. Plant. Cell Environ. 40, 2628–2643. doi: 10.1111/pce.12979
Ewas, M., Gao, Y., Wang, S., Liu, X., Zhang, H., Nishawy, E. M. E., et al. (2016). Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci. Bull. 61, 1413–1418. doi: 10.1007/s11434-016-1108-9
Feng, Z., Bartholomew, E. S., Liu, Z., Cui, Y., Dong, Y., Li, S., et al. (2021). Glandular trichomes: new focus on horticultural crops. Hortic. Res. 8, 158. doi: 10.1038/s41438-021-00592-1
Galdon-Armero, J., Arce-Rodriguez, L., Downie, M., Li, J., Martin, C. (2020). A scanning electron micrograph-based resource for identification of loci involved in epidermal development in tomato: elucidation of a new function for the mixta-like transcription factor in leaves[OPEN]. Plant Cell 32, 1414–1433. doi: 10.1105/tpc.20.00127
Galimba, K. D., Martínez-Gómez, J., Di Stilio, V. S., S, V. (2018). Gene duplication and transference of function in the paleoAP3 lineage of floral organ identity genes. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00334
Gao, S., Gao, Y., Xiong, C., Yu, G., Chang, J., Yang, Q., et al. (2017). The tomato B-type cyclin gene, SlCycB2 , plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 262, 103–114. doi: 10.1016/j.plantsci.2017.05.006
Gilding, E. K., Marks, M. D. (2009). NOECK, a gene related to MIXTA of snapdragon, controls cell size and shape in Arabidopsis trichomes in unexpected ways. Plant Biol. (Rockville). 2009, 181.
Gilding, E. K., Marks, M. D. (2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64, 304–317. doi: 10.1111/j.1365-313X.2010.04329.x
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hao, D.-C., Li, P., Xiao, P.-G., He, C.-N. (2021). Dissection of full-length transcriptome and metabolome of Dichocarpum (Ranunculaceae): implications in evolution of specialized metabolism of Ranunculales medicinal plants. PeerJ 9, e12428. doi: 10.7717/peerj.12428
Hichri, I., Deluc, L., Barrieu, F., Bogs, J., Mahjoub, A., Regad, F., et al. (2011). A single amino acid change within the R2 domain of the VvMYB5b transcription factor modulates affinity for protein partners and target promoters selectivity. BMC Plant Biol. 11, 117. doi: 10.1186/1471-2229-11-117
Jaime, R., Rey, P. J., Alcántara, J. M., Bastida, J. M. (2013). Glandular trichomes as an inflorescence defence mechanism against insect herbivores in Iberian columbines. Oecologia 172, 1051–1060. doi: 10.1007/s00442-012-2553-z
Jakoby, M. J., Falkenhan, D., Mader, M. T., Brininstool, G., Wischnitzki, E., Platz, N., et al. (2008). Transcriptional profiling of mature arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 148, 1583–1602. doi: 10.1104/pp.108.126979
Javelle, M., Vernoud, V., Rogowsky, P. M., Ingram, G. C. (2011). Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39. doi: 10.1111/j.1469-8137.2010.03514.x
Kramer, E. M., Dorit, R. L., Irish, V. F. (1998). Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783. doi: 10.1093/genetics/149.2.765
Lashbrooke, J., Adato, A., Lotan, O., Alkan, N., Tsimbalist, T., Rechav, K., et al. (2015). The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169, 2553–2571. doi: 10.1104/pp.15.01145
Lau, S.-E., Schwarzacher, T., Othman, R. Y., Harikrishna, J. A. (2015). dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 15, 194. doi: 10.1186/s12870-015-0577-3
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H.-C., Lam, S.-H., Zhang, D., Hsiao, Y.-Y., Li, B.-J., Niu, S.-C., et al. (2022). R2R3-MYB genes coordinate conical cell development and cuticular wax biosynthesis in Phalaenopsis aphrodite. Plant Physiol. 188, 318–331. doi: 10.1093/plphys/kiab422
MaChado, A., Wu, Y., Yang, Y., Llewellyn, D. J., Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Martínez-Gómez, J., Park, S., Hartogs, S. R., Soza, V. L., Park, S., Di Stilio, V. S. (2023). Flower morphology as a predictor of pollination mode in a biotic to abiotic pollination continuum. Ann. Bot. 132, 61–76. doi: 10.1093/aob/mcad069
Noda, K., Glover, B. J., Linstead, P., Martin, C. (1994). Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369, 661–664. doi: 10.1038/369661a0
Oppenheimer, D. G., Pollock, M. A., Vacik, J., Szymanski, D. B., Ericson, B., Feldmann, K., et al. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 94, 6261–6266. doi: 10.1073/pnas.94.12.6261
Pérez-Rodríguez, M., Jaffe, F. W., Butelli, E., Glover, B. J., Martin, C. (2005). Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132, 359–370. doi: 10.1242/dev.01584
Pesch, M., Hülskamp, M. (2009). One, two, three…models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Qi, X., Chen, Z., Yu, X., Li, L., Bai, Y., Fang, H., et al. (2022). Characterisation of the Mentha canadensis R2R3-MYB transcription factor gene McMIXTA and its involvement in peltate glandular trichome development. BMC Plant Biol. 22, 1–11. doi: 10.1186/s12870-022-03614-9
Qin, W., Xie, L., Li, Y., Liu, H., Fu, X., Chen, T., et al. (2021). An R2R3-MYB transcription factor positively regulates the glandular secretory trichome initiation in artemisia annua L. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.657156
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reddy, V. S., Day, I. S., Thomas, T., Reddy, A. S. N. (2004). KIC, a novel calcium binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16, 185–200. doi: 10.1105/tpc.016600
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schuurink, R., Tissier, A. (2020). Glandular trichomes: micro-organs with model status? New Phytol. 225, 2251–2266. doi: 10.1111/nph.16283
Scoville, A. G., Barnett, L. L., Bodbyl-Roels, S., Kelly, J. K., Hileman, L. C. (2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191, 251–263. doi: 10.1111/j.1469-8137.2011.03656.x
Shi, P., Fu, X., Shen, Q., Liu, M., Pan, Q., Tang, Y., et al. (2018). The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. Burlington, MA 217, 261–276. doi: 10.1111/nph.14789
Simpson, M. G. (2019). Plant systematics (Elsevier-Academic Press), 774. doi: 10.1016/B978-0-12-812628-8.50006-7
Soza, V. L., Brunet, J., Liston, A., Salles Smith, P., Di Stilio, V. S. (2012). Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae). Mol. Phylogenet. Evol. 63, 180–192. doi: 10.1016/j.ympev.2012.01.009
Soza, V. L., Haworth, K. L., Stilio, V. S. D. (2013). Timing and consequences of recurrent polyploidy in meadow-rues (Thalictrum, ranunculaceae). Mol. Biol. Evol. 30, 1940–1954. doi: 10.1093/molbev/mst101
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Vernoud, V., Laigle, G., Rozier, F., Meeley, R. B., Perez, P., Rogowsky, P. M. (2009). The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 59, 883–894. doi: 10.1111/j.1365-313X.2009.03916.x
Von Groll, U., Berger, D., Altmann, T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during arabidopsis stomatal development. Plant Cell 14, 1527–1539. doi: 10.1105/tpc.001016
Wald, A. (1943). Tests of statistical hypotheses concerning several parameters when the number of observations is large. Trans. Am. Math. Soc. 54, 426–482. doi: 10.1090/S0002-9947-1943-0012401-3
Wang, T. N., Clifford, M. R., Martínez-Gómez, J., Johnson, J. C., Riffell, J. A., Di Stilio, V. S. (2019). Scent matters: differential contribution of scent to insect response in flowers with insect vs. wind pollination traits. Ann. Bot. 123, 289–301. doi: 10.1093/aob/mcy131
Wang, Y.-L., Nie, J., Chen, H.-M., Guo, C., Pan, J., He, H.-L., et al. (2016). Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 129, 305–316. doi: 10.1007/s00122-015-2628-4
Werker, E. (2000). “Trichome diversity and development,” in Advances in botanical research (Academic Press), 1–35. doi: 10.1016/S0065-2296(00)31005-9
Wu, T., Wang, Y., Guo, D. (2012). Investigation of glandular trichome proteins in artemisia annua L. Using comparative proteomics. PloS One 7, e41822. doi: 10.1371/journal.pone.0041822
Xu, B., Taylor, L., Pucker, B., Feng, T., Glover, B. J., Brockington, S. F. (2020). The land plant-specific MIXTA-MYB lineage is implicated in the early evolution of the plant cuticle and the colonization of land. New Phytol 229, 2324–2338. doi: 10.1111/nph.16997
Yan, T., Li, L., Xie, L., Chen, M., Shen, Q., Pan, Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218, 567–578. doi: 10.1111/nph.15005
Yang, Y., Huang, W., Wu, E., Lin, C., Chen, B., Lin, D. (2019). Cortical microtubule organization during petal morphogenesis in arabidopsis. IJMS 20, 4913. doi: 10.3390/ijms20194913
Yang, C., Ye, Z. (2013). Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Keywords: epidermis, MIXTA family, non-core eudicots, polyploidy, trichomes, conical cells, ranunculid, R2R3 MYB
Citation: Zahid S, Schulfer AF and Di Stilio VS (2023) A eudicot MIXTA family ancestor likely functioned in both conical cells and trichomes. Front. Plant Sci. 14:1288961. doi: 10.3389/fpls.2023.1288961
Received: 05 September 2023; Accepted: 27 November 2023;
Published: 18 December 2023.
Edited by:
Dianella G. Howarth, St. John’s University, United StatesReviewed by:
Harold Suárez-Baron, Pontificia Universidad Javeriana Cali, ColombiaCopyright © 2023 Zahid, Schulfer and Di Stilio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Verónica S. Di Stilio, ZGlzdGlsaW9AdXcuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.