- 1Plant Breeding, Wageningen University & Research, Wageningen, Netherlands
- 2Wageningen Food & Biobased Research, Wageningen University & Research, Wageningen, Netherlands
The growing interest in plant protein sources, such as pulses, is driven by the necessity for sustainable food production and climate change mitigation strategies. Faba bean (Vicia faba L.) is a promising protein crop for temperate climates, owing to its remarkable yield potential (up to 8 tonnes ha−1 in favourable growing conditions) and high protein content (~29% dry matter basis). Nevertheless, the adoption of faba bean protein in plant-based products that aim to resemble animal-derived counterparts is hindered by its distinctive taste and aroma, regarded as “off-flavors”. In this review, we propose to introduce off-flavor as a trait in breeding programs by identifying molecules involved in sensory perception and defining key breeding targets. We discuss the role of lipid oxidation in producing volatile and non-volatile compounds responsible for the beany aroma and bitter taste, respectively. We further investigate the contribution of saponin, tannin, and other polyphenols to bitterness and astringency. To develop faba bean varieties with diminished off-flavors, we suggest targeting genes to reduce lipid oxidation, such as lipoxygenases (lox) and fatty acid desaturases (fad), and genes involved in phenylpropanoid and saponin biosynthesis, such as zero-tannin (zt), chalcone isomerase (chi), chalcone synthase (chs), β-amyrin (bas1). Additionally, we address potential challenges, including the need for high-throughput phenotyping and possible limitations that could arise during the genetic improvement process. The breeding approach can facilitate the use of faba bean protein in plant-based food such as meat and dairy analogues more extensively, fostering a transition toward more sustainable and climate-resilient diets.
1 Introduction
Natural resources depletion and climate change exacerbate the challenge to meet the future food demand in view of anticipated population growth by 2050 (Calicioglu et al., 2019). In the past decades, rising incomes and urbanization have driven the increased consumption of animal proteins. Animal-based products contribute significantly to environmental pressures, including land and water use, greenhouse gas (GHG) emissions, acidification and eutrophication potential. In the depicted scenario, the transition from meat-intensive diets towards plant protein-based diets (“protein transition”) is crucial to meet the climate change mitigation targets and ensure food security (D’Odorico et al., 2018).
The European food market has already witnessed a burgeoning trajectory of plant-based food retail sales, especially meat and dairy analogues (Aschemann-Witzel et al., 2021). Also known as replacers or alternatives, analogues are plant-based processed foods designed to resemble animal products in appearance, texture and taste (Fresán et al., 2019; Kyriakopoulou et al., 2019). At present, soybean, wheat, and pea-based products dominate the market for meat analogues, while soybean and almond-based products do for dairy analogues (Nawaz et al., 2022). A more recent area of development involves the use of soy and wheat protein in fish analogues (Nowacka et al., 2023). Environmental concerns around soy production in South America (e.g., deforestation, land-use change, transport-associated emissions) and increased nitrogen emissions have led to consumer and policy-maker interest in alternative pulse-based proteins (Tsolakis et al., 2019). Pulses are the dried seeds of certain legumes (e.g., pea, chickpea, lentil, lupin, faba bean, and diverse other dry beans), thus excluding those that are used as vegetables (e.g., fresh peas, or green beans) or for oil extraction (e.g., soybean and peanut) (Iriti and Varoni, 2017). Among pulses, the use of protein-rich ingredients from faba bean (Vicia faba L.) (i.e., flours, concentrates, isolates) is still minor (Multari et al., 2015).
Faba bean is in the spotlight for its high yield potential. The yield of superior cultivars adapted to specific environments and grown with appropriate husbandry can reach up to 8 to 9 tonnes ha−1 in favourable European climates (Metayer, 2004). However, less than half of the production potential is usually achieved due to biotic and abiotic stress, as well as inadequate agronomic management (Metayer, 2004). In Europe, the difference in actual yield between countries is remarkable. Yield ranges from an average of 3.9 t ha−1 in Germany and UK to 1.4 t ha−1 in the Mediterranean-type environments, such as Spain (Mínguez and Rubiales, 2021). Faba bean ranks behind other pulses such as lentil, cowpea, field pea, chickpea, and common bean in global harvested area and production, being superior only to lupin (FAOSTAT, 2021) (Figure 1). However, the high yield and protein content makes faba bean superior to most other pulses in terms of protein yield per unit of land area (protein ha−1).
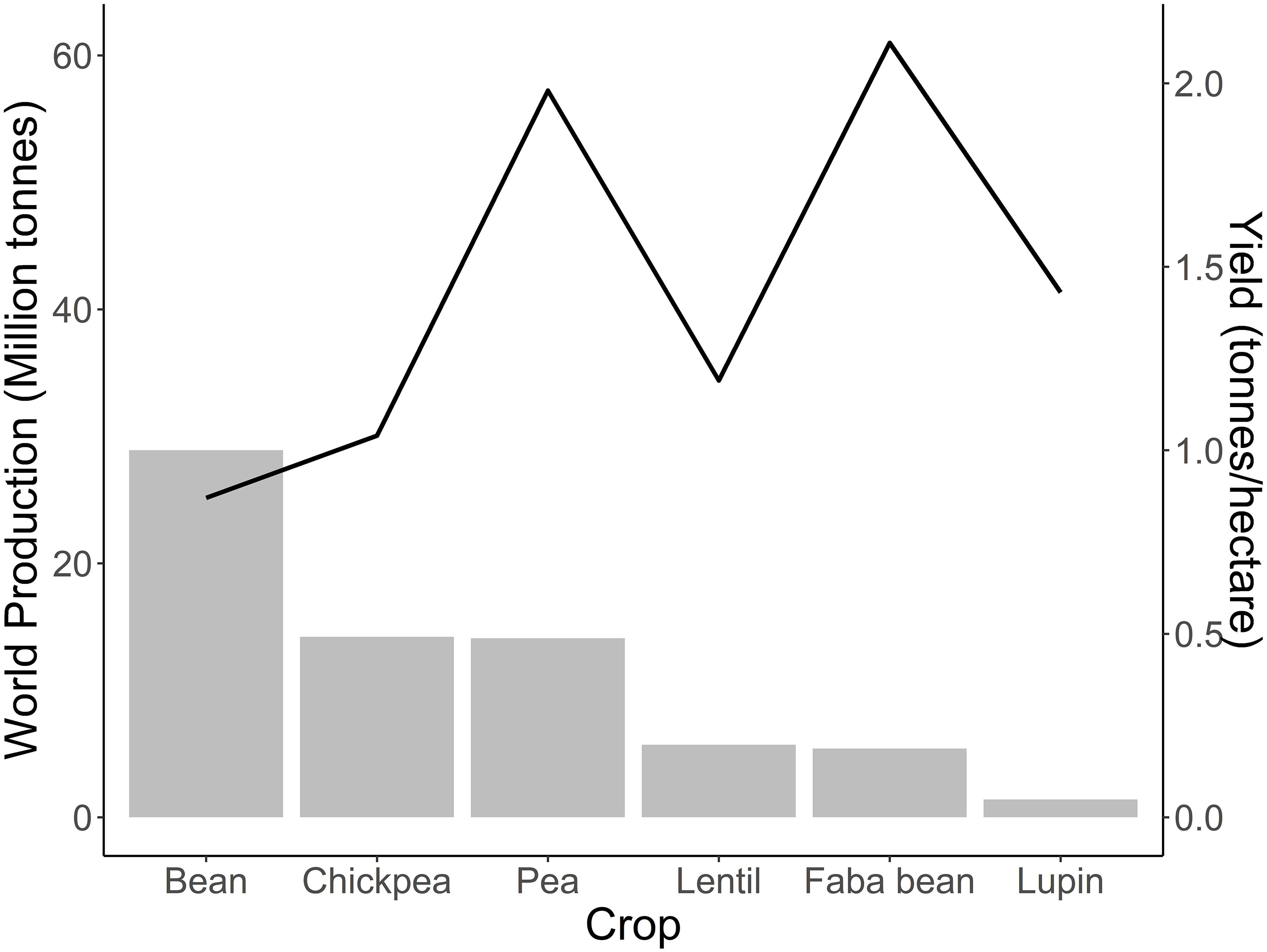
Figure 1 World production and yield of major pulses. Production in million tonnes of bean, chickpea, faba bean, lentil, lupin and pea for the year 2021. The trendline highlights the average yield of these crops as tonnes ha−1. The yield value reported was calculated by dividing world production by the total harvested area using data of 2021. Source: FAOSTAT, 2021.
Vicia faba represents one of the cheapest sources of proteins and carbohydrates. Carbohydrate content accounts for 50 to 68% (Punia et al., 2019), while protein content ranges from 20 to 40% (Dhull et al., 2022) of its seed composition, with an average of 29%. Contrarily, the oil content is low and ranges from 0.5 to 2%. Besides for high protein content, this crop is prized in the food industry because is locally grown and processed in Europe. Contrarily to imported soybean, faba bean supports “clean label” product (Asioli et al., 2017). Moreover, its seeds already have a long history of use in food worldwide (Zong et al., 2019), facilitating consumer acceptance of new products. This pulse is a healthy food source, containing potassium, calcium, magnesium, iron, and zinc, dietary fibres, phenolic compounds, 3,4-dihydroxyphenylalanine (L-DOPA), and γ-aminobutyric acid (GABA) (L’Hocine et al., 2020). Faba bean offers several agronomic and ecological advantages to growers, such as adaptability to a wide range of climates and soils, and a particularly high nitrogen-fixing ability via Rhizobium bacteria symbiosis (60-250 kg ha−1 per year) (Torres et al., 2011; Maalouf et al., 2018). Therefore, faba bean is an attractive option for both farmers and the food industry. To support the foreseen increase of its market share in the next years (Multari et al., 2015), it is necessary to develop and commercialise varieties with enhanced quality features that could facilitate the use of its protein fraction in meat or dairy analogues. Optimising the quality of the seeds for such food applications requires a synergistic approach between plant breeding and food technology research. In this way legume breeders will be guided by food technologists to the quality requirements of highly processed food including protein functionality (i.e., solubility, gelling, emulsifying or foaming property, etc.) and flavor. At the moment, the typical faba-bean-like flavor hampers consumer acceptance and is negatively perceived as “off-flavor” in meat and dairy analogues.
In this comprehensive review, we address the challenges associated with improving flavor features in faba bean, emphasising the need for specialised breeding programs tailored to ameliorate traits relevant to the modern food industry. The review will first provide a brief introduction on the cultivation, food uses, and breeding history of this crop. It will subsequently explore the molecules likely involved in flavor perception, elucidate the primary biosynthetic pathways controlling the production of these compounds, and identify the respective key regulators that can be targeted with breeding. Additionally, we will provide a perspective on breeding strategies aimed at mitigating off-flavors, discussing challenges such as high-throughput phenotyping and potential drawbacks in the process.
1.1 Traditional food uses and new trends of consumption
Faba bean is one of the earliest domesticated crops, but knowledge about its origin and pre-domestication distribution has been scarce and debatable (Duc et al., 2010). Cubero (1974) reported the Near East as the centre of origin. From this area, Vicia faba cultivation spread following human migrations to the Mediterranean basin, the Nile Valley, and Central and East Asia, all becoming regions of historical cultivation of this crop (Cubero, 1973). In area where the consumption of faba bean is a customary practice there are well known typical dishes based on faba bean such as ‘Ful Medames’ (Egypt), ‘Shahan ful’ (Ethiopia), ‘Ful bi’l-kammun’ (Tunisia), Bissara (Morocco), Bakla (Turkey), ‘Macco’, and ‘Puré di fave e cicorie’(Italy) (Pasqualone et al., 2020). In many Western nations (e.g., central-northern Europe and USA), faba bean is mainly used in animal feed. Overall, only about 15% of the total crop production targets human consumption, contributing to the perception that faba bean is an underutilised protein source. Despite being a healthy food, faba bean contains antinutritional factors, such us tannins, lectins, trypsin inhibitors that can reduce protein digestibility and negatively impact the bioavailability of minerals and polysaccharides (Kim et al., 2015; Rahate et al., 2021). Additionally, vicine and convicine cause favism, in individuals who have a hereditary loss of the enzyme glucose-6-phosphate dehydrogenase (Luzzatto and Arese, 2018).
The interest of the food industry in utilising faba bean protein fractions as food ingredients has been growing. This is witnessed by increased literature on the incorporation of faba bean flour and protein-rich fractions in formulations for bread, pasta, snack, infant formulas, plant-based meat analogues, etc. By replacing 30% of wheat flour with faba bean flour, bread formulations have shown an increase in protein content from 11.6% to 16.5% of dry matter (Coda et al., 2017). The partial substitution of wheat with faba bean ingredients in pasta has also been suggested to enhance nutritional properties for better health outcomes (Coda et al., 2017; Chan et al., 2019). Moreover, dry fractionation-derived faba bean protein concentrate has been demonstrated to be suitable for meat-like products (do Carmo et al., 2021). Fibrous textured meat analogues were successfully produced using high moisture extrusion of commercial faba bean concentrate (56% protein) or isolates prepared by aqueous fractionation (88% protein) (Ferawati et al., 2019).
The increased number of studies on incorporating faba bean protein fractions into various food products suggests that this ancient pulse is nowadays consumed in new and alternative ways. To promote the use of faba bean as reliable food ingredient, it is crucial to produce high-quality seed-derived ingredients. Plant breeding can significantly contribute to developing optimal faba bean varieties tailored for modern food applications.
1.2 Faba bean ideotypes for modern food applications are lacking in the variety portfolio
In the period 1960-1990, Europe witnessed a severe decline in the faba bean cultivation area. The primary reasons include small economic margins for farmers, economic competition with soybean, as well as low and unstable yields caused by biotic and abiotic stress (Flores et al., 2013; Karkanis et al., 2018; Mínguez and Rubiales, 2021). The lack of interest in European faba bean cultivation in the period 1960-1990 resulted in a relatively brief breeding history with limited investments, starting only in the 1980s in Europe. Seed production has been traditionally relegated to small farms, where landraces have been selected based on their agronomic performance at a local scale (Duc et al., 2010), representing the main genetic material cultivated. Consequently, there is a restricted number of varieties registered in the European catalogue in comparison to field pea and soybean (European Commission, 2023). The main varieties available in the market belong to the botanical groups Vicia faba var. major Harz. (broad bean) and Vicia faba var. minor Beck. (field bean). Broad bean and field bean mainly exhibit differences in the morphology of the seeds (i.e., seed size), with broad bean possessing larger seeds than field bean (Muratova, 1931). In Europe, broad beans are primarily utilised for food purposes, while field beans serve as animal feed or green manure (Pasqualone et al., 2020).
Following 1990, faba bean yield started to increase because of genetic selection. Breeding has been conducted to increase tolerance to winter climate conditions (frost hardiness), vernalization requirement and photoperiodic induction (Arbaoui et al., 2008). As a result, spring and winter types are available. Winter-type varieties are typically sown in autumn in cold-temperate regions, including the Mediterranean basin or some mild climates of Australia and China (Flores et al., 2012; Mínguez and Rubiales, 2021). Farmers are particularly attracted by the possibility of cultivating the winter faba bean in a wider area in Europe, even in colder climates (Neugschwandtner et al., 2019). Certain cultivars with particularly enhanced hardening responses are currently cultivated in the UK and other high-latitude regions. Spring-type varieties, on the other hand, do not require vernalization or hardening response, thus are currently the most preferred in cold climates (Flores et al., 2012; Mínguez and Rubiales, 2021).
Improving seed quality has not been a major target for breeding, mostly because there was no impulse from the food industry. The breeding efforts in seed quality have been mostly directed to contents of tannins and the alkaloids vicine and convicine (V-C) (Gutierrez and Torres, 2019; Bjornsdotter et al., 2021; Tacke et al., 2022). At the moment, the European catalogue of registered varieties includes four types with improved quality based on the reduced tannin content in the seed testa (outer protective layer of the bean) and the lower levels of V-C in the cotyledons (the part of the seed embryo that will become the first leaves of the plant upon germination). These four types refer to high or low levels of tannins, and high or low levels of V-C. The word “fevita” usually refers to the simultaneous combination of low tannins and V-C, after the first variety of this type was registered in the French catalogue under the name of FEVITA® in 2004 (Crépon et al., 2010; Zong et al., 2019). Despite the release of varieties with low antinutritional compounds on the market, varieties with high levels of tannins and V-C are still the most cultivated.
Faba bean is gaining popularity in several countries as an ingredient for plant-based products. However, faba bean ideotypes for modern food applications (i.e., plant-based meat analogues) are lacking in the market. The development of improved varieties with good techno-functional properties (i.e. solubility, emulsifying, foaming ability, etc.), together with nutritional improvement, is pivotal to expanding the use of faba bean ingredients in the food industry. Interestingly, new varieties could be bred for protein components and amino acid composition, which are intrinsic components impacting the techno-functionality of the products during processing (Rahate et al., 2021; Augustin and Cole, 2022).
2 Defining reduced off-flavor as a quality trait in faba bean breeding
One of the major obstacles to adopting a diet with reduced meat consumption is the lack of familiarity with meat-alternative products and their limited aroma, taste and texture attractiveness (Wang et al., 2022).
Flavour is a multisensory perception defined by taste and aroma, which are influenced by the food matrix and its complex physico-chemical affinity with flavor-active chemical compounds (Tournier et al., 2007; Roland et al., 2017). Aroma refers to the perception of volatile odour-active compounds via receptors in the nose (Czerny et al., 2008). The main aroma features of pulses are beany, grassy, and earthy (Wang et al., 2021). Taste is perceived via receptors on the tongue and in the oral cavity and results from non-volatile compounds present in the grains. The taste of pulses is often reported as bitter and astringent (Roland et al., 2017; Sharan et al., 2021). A specific flavor can be perceived as pleasant or unpleasant (“off-flavor”) by consumers, depending on the intended final product. For example, the typical faba-bean-like flavor is unpleasant in meat or dairy alternative analogues, as consumers expect tastes like meat, milk, and cheese rather than beans. Consequently, off-flavors hinder consumer preference and acceptance of faba bean in modern plant-based products (Wang et al., 2022). Ideal faba bean ingredients should be as bland as possible in terms of flavor.
Prior to starting breeding programs and setting breeding objectives, plant breeders need understanding about off-flavor molecules and which of them highly impact on off-flavor in the final ingredients. In this way, marker compounds for screening for unpleasant flavors can be defined. It is important to understand that not all compounds detected in a food or food ingredient contribute to its flavor, and determining the real impact of the diverse aroma and taste compounds on perception typically involves laborious methods. Furthermore, the sensory impact of compounds is not solely based on their concentration, but it also relies on their specific sensory thresholds. Some compounds are detectable by some consumers at ultra-trace levels (Ridgway, 2015). The importance of volatile compounds on flavor is usually ranked by calculating the so-called odour activity values (OAVs), which simultaneously account for their concentration and sensory threshold. Volatile compounds with high OAVs are often essential to the aroma, unless they are suppressed in the respective matrix (Grosch, 2001). However, it is worth mentioning that the sensory thresholds of compounds vary across different mediums or the matrix in which these compounds are incorporated. For example, the taste threshold of kaempferol is 20 mg/kg in 5% aqueous ethanol, but it is almost 50 mg/kg in beer (Van Gemert, 2003). Another approach to rank the importance of flavor compounds in a food matrix involves taste dilution or aroma dilution experiments. In aroma dilution experiments, gas chromatography-olfactometry (GC-O), which combines the chromatographic separation of compounds with human sensory detection, is used. The result of this analysis is expressed as a ‘flavor dilution factor’ (FD-factor) (Grosch, 2001). In taste dilution experiments, a sensory panel is involved, and the result is expressed as ‘taste dilution factor’ (TD factor) (Frank et al., 2001). Higher dilution factors imply a greater impact of the respective compound on the flavor of the food.
It is worth noting that the identification of key off-flavor compounds depends on several factors. For instance, different varieties of faba exhibit a different level of off-flavors compounds (Akkad et al., 2021), but the difference in off-flavor can also be caused by the specific processing applied to the seeds (Tuccillo et al., 2022). Currently, faba bean seeds are processed to produce flour or protein concentrates and isolates. Protein concentrate and protein isolate are two types of protein-enriched fractions (Augustin and Cole, 2022). Protein concentrates typically contain about 60-80% protein, while protein isolate, due to additional processing to remove nearly all non-protein components, results in a purer protein content of around 90-95% (Ma et al., 2022). Tuccillo et al. (2022) recently demonstrated that flour, protein concentrate and protein isolate can have different off-flavors and off-flavor intensities. This is likely linked to the different protein content of different ingredients, as protein play as a carrier of flavor compounds, but is also influenced by the distinct handling processes of the seeds (e.g., removal off the seed coats, temperatures etc.) and by the oxidative reactions occurring in the meantime.
The dominant flavor of faba bean has been traditionally described as dried pea flavor, bitter aftertaste and unpleasantly fruity (Seidel, 1976; Schultz et al., 1988). Specific research on the off-flavor properties of faba bean is scarce compared to soybean and pea, partially because the use of faba bean as a plant-based food ingredient is more recent. For the investigated pea matrices, key off-flavor compounds have been pointed out. Taste dilution experiments have been conducted for bitter compounds in pea protein isolates (Glaser et al., 2020). Moreover, the use of GC-O and OAV calculations using literature thresholds in water have been reported for pea milk (Zhang et al., 2020). For faba bean, Akkad et al. (2021) conducted chemical analyses of volatile compounds in flour and determined key volatile flavor compounds by calculating relative OAVs using literature thresholds in air. In another study, Tuccillo et al. (2022) combined sensory analysis with chemical analyses of volatile and non-volatile compounds, employing statistical methods to identify compounds in faba bean ingredients and extrudates that are correlated with off-flavor. The most recent study, by Karolkowski et al. (2023a), performed gas chromatography mass spectrometry (GC-MS) and GC-O following solvent assisted flavor extraction (SAFE) of various faba bean fractions, including flour, starch, and protein. However, to the best of our knowledge, no flavor dilution or taste dilution experiments have been reported yet for faba beans, and knowledge gaps regarding key off-flavor compounds (mainly non-volatile) are still relatively large. Therefore, educated guesses need to be taken to determine which compounds potentially have an impact on faba bean off-flavor, based on their presence and abundance, their sensory characteristics, and their impact known for crops like pea and soy.
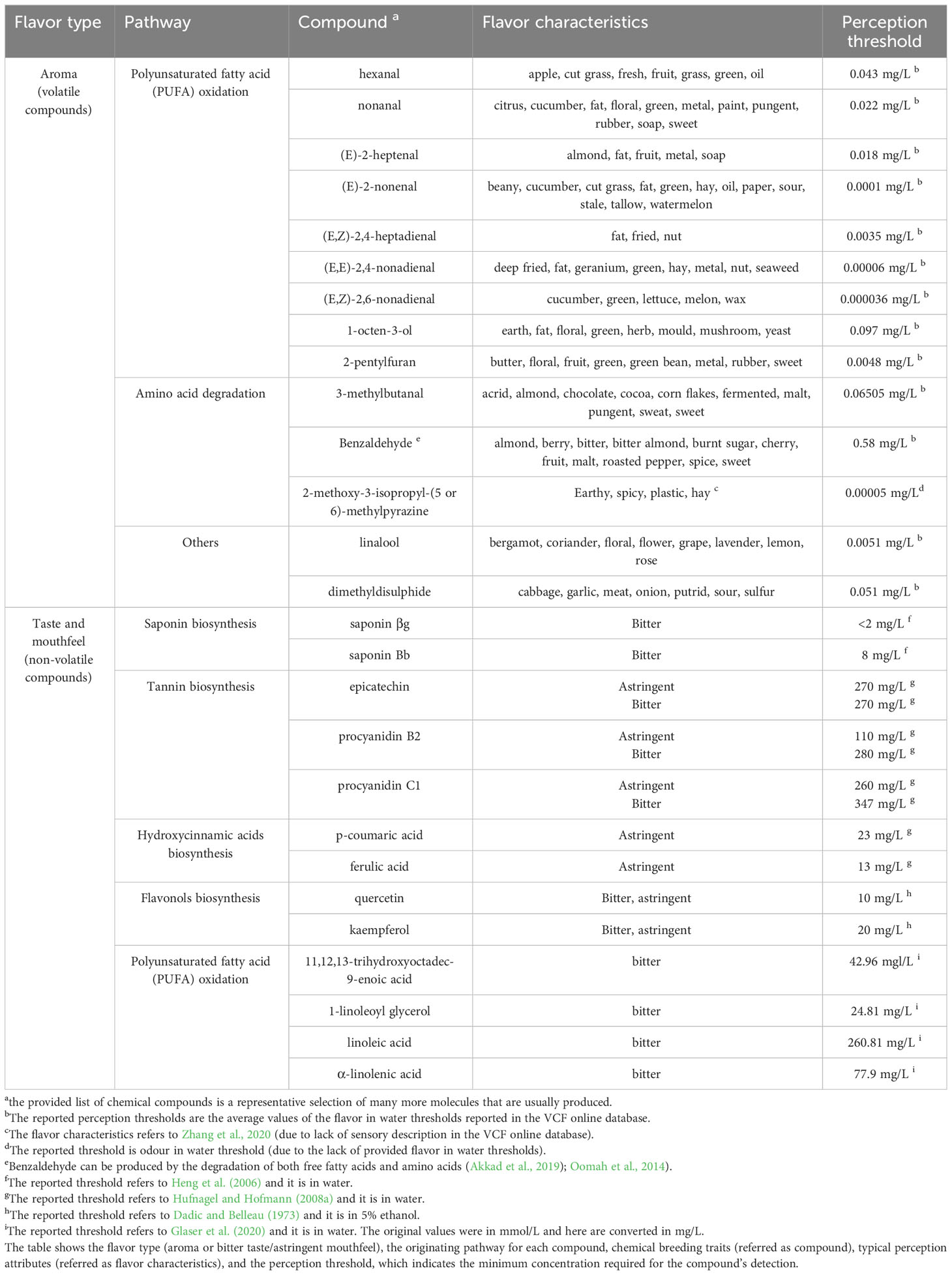
In the following subparagraphs, we discuss a summary of molecules potentially involved in off-flavor development in faba bean and their origins. Table 1 presents a shortlist of molecules that we consider suitable as marker compounds for screening faba bean ingredients for potential off-flavors. Our observations are supported by literature, such as key compounds determined by relative OAV (Akkad et al., 2021) and by GC-O (Karolkowski et al., 2023b) in faba bean, supplemented by observations in our own laboratories. A marker compound is not necessarily the one with the most significant impact on the flavor. It can also be a compound produced in large amounts and capable of indicating the occurrence of off-flavors-related chemical reactions. For this reason, the amounts of oxidation compounds formed when C18:1, C18:2, and C18:3 are oxidized (Belitz et al., 2009) were also included in the final choice of the makers for volatile compounds. It is important that the marker compounds for off-flavors are easy to detect, acting as proxy of other chemicals that might have a more substantial impact on flavor but are less detectable or whose detectability is affected by extraction techniques.
2.1 Unpleasant aroma is associated with lipids oxidation, amino acids degradation, and other chemical pathways
The unpleasant aroma of faba bean protein fractions is associated with the release of certain volatile compounds originating from non-volatile precursors like lipids, amino acids, and carbohydrates. The chemical reactions leading to the development of aroma compounds mainly occur during harvesting, postharvest processing, and storage (Roland et al., 2017). Even though faba bean is not an oil crop and has a low-fat content (0.5-2% dry matter), it is still prone to developing off-flavors as a result of lipid oxidation (Wang et al., 2014). In fact, some lipid-derived compounds can be detected by the human nose at extremely low concentrations (Akkad et al., 2021). Polyunsaturated fatty acids (PUFA) like oleic acid (C18:1), linoleic (C18:2) and linolenic (C18:3) are readily oxidised to hydroperoxides, which are subsequently degraded in volatile secondary oxidation products (i.e., aldehydes, ketones, alcohols, aromatic hydrocarbons, furans, etc.), some of which being exceptionally odorous compounds (Decker et al., 2012). The formation of beany flavor is primarily attributed to aldehydes and dienals produced through PUFA degradation catalysed by LOX. Akkad et al. (2021) reported (in order of decreasing Relative OAVs) nonanal, octanal, hexanal, 3-methylbutyric acid, decanal, 3-methylbutanal, 1-octen-3-ol, and others as key volatile compounds in faba bean flours of 4 faba bean cultivars, based on the calculation of relative odour activity values. The type of volatile compounds produced from the oxidation of PUFA mainly depend on the nature of the fatty acids involved in the reaction. The oxidation of oleic acid oxidation leads to the formation of 8-, 9-, 10-, and 11-hydroperoxides, and subsequentially to many secondary compounds such as pentanal, octanal, hexanal, and heptanal, among others (Shahidi and Hossain, 2022). The oxidation of linoleic acid results in two distinct stereoisomers of both 9- and 13-hydroperoxides. As an example, the volatile 2,4-decadienal primarily originates from 9-hydroperoxide, whereas hexanal is primarily produced from 13-hydroperoxides. Linolenic acid, on the other hand, oxidises to generate 9-, 12-, 13-, and 16-hydroperoxides (Karolkowski et al., 2021). Linoleic and linolenic-derived compounds have more impact on off-flavor compared to those derived from oleic acid.
Lipid peroxidation occurs via autoxidation or enzymatic oxidation which requires the presence of lipoxygenase. The susceptibility of fatty acids to lipid peroxidation augments proportionally with the degree of unsaturation (Belitz et al., 2009). Oleic acid results more stable than linoleic and linolenic acid. Linoleic acid is about ten times more susceptible to autooxidation and linolenic acid twenty times more (Decker et al., 2012). Moreover, lipoxygenase selectively oxidizes linoleic and linolenic acids, while oleic acid remains unaffected by this enzymatic process.
Lipid oxidation is not the only source of unpleasant aroma development in faba bean. Akkad et al. (2019) suggested that the high protein content of faba bean varieties may impact their aroma profile. Amino acid degradation can generate a myriad of volatile compounds, such as branched-chain compounds, benzene aldehydes, alcohols, acids, esters, nitrogen, and sulfur compounds (Karolkowski et al., 2021). Amino acid-derived volatiles, including 3-methylbutanal and 3-methylbutanoic acid (leucine-derived), 2-methylbutanal (isoleucine-derived), and benzacetaldehyde (phenylalanine-derived) were detected in faba bean flour (Akkad et al., 2019). In addition to the degradation pathway, amino acids generate odor-active compounds through reactions with sugars and carbonyls induced by the effect of temperature (Maillard reaction) (MacLeod and Ames, 1988).
Other compounds, such as terpenes, can also influence the aroma profile of faba bean. For example, Akkad et al. (2019) detected limonene, which typically has a citrus-like odour at high concentrations. Additionally, the presence of linalool, alpha-pinene, delta-3-carene was also reported (Akkad et al., 2019; Tuccillo et al., 2022). Naturally occurring methoxypyrazines were hypothesised to potentially play a role in off-flavor development in V. faba due to their key role in the aroma of green peas and their extremely low odour thresholds (Sharan et al., 2021). However, due to their extremely low concentrations at which they are odour-active, they are rather detected by the nose, than by GC-MS. Therefore, while GC-MS measurements of faba bean fractions failed to detect any methoxypyrazines, the GC-O measurements-which involve smelling the separated volatiles-revealed the presence of 3-isopropyl-2-methoxypyrazine and 3-isobutyl-2-methoxypyrazine (Karolkowski et al., 2023b).
2.2 Unpleasant taste is associated with polyphenols, saponins and lipid-derived compounds
The bitter and astringent unpleasant taste of legumes is associated with non-volatile compounds present in seeds. The information on the role of non-volatile compounds in the taste of faba bean is scarce, but it is widely accepted that polyphenols (such as phenolic acids, flavones, flavanones, flavonols, flavan-3-ols, and proanthocyanidins), saponins, amino acids and peptides are bitter and/or astringent compounds (Drewnowski and Gomez-Carneros, 2000; de Camargo and Schwember, 2019; Iwaniak et al., 2019), and also many non-volatile lipid oxidation compounds and oxidised phospholipids taste bitter (Sessa and Rackis, 1977; Glaser et al., 2020).
Phenolic compounds are usually either measured as total phenolic compounds with the use of the Folin-Ciocalteu assay, reporting gallic acid equivalents (GAE) measured in a spectrometric way (e.g., in Tuccillo et al., 2022), or they are measured individually by chromatographic methods (e.g., in Abu-Reidah et al., 2017). Various phenolic compounds from the classes of phenolic acids, flavanols, procyanidins, prodelphinidins, flavanones, flavonols, flavones, and others, have been detected in Vicia faba (Turco et al., 2016; Abu-Reidah et al., 2017). Many of these flavonoids activate bitter taste receptors (Roland et al., 2013), so it can be expected that they impact off-taste in this crop. For example, the flavonols quercetin and kaempferol are present in faba bean (Troszyńska et al., 2006) and are bitter (Drewnowski and Gomez-Carneros, 2000; Roland et al., 2013). The phenolic acids vanillic, caffeic, p-coumaric, and ferulic acid have been perceived as astringent (Hufnagel and Hofmann, 2008a) and have been detected in faba bean (Troszyńska et al., 2006). However, which of these phenolic compounds impacts the perception of bitterness and astringency in faba bean depends on their concentrations and the presence of other compounds in the matrix (Roland et al., 2017). In fact, free phenolics have been recently associated to the strong taste, bitterness, and mouth-drying sensation of faba bean protein contrate (Tuccillo et al., 2022), but the understanding of which specific individual compounds links to this off-taste is lacking.
An important class of polyphenols in faba bean is represented by the tannins. Tannins are mainly classified into two categories based on their reactions with hydrolytic agents: condensed tannins and hydrolysable tannins. Hydrolysable tannins have a central carbohydrate core esterified to phenolic carboxylic acids such as gallic acid and ellagic acid and are usually referred to as ellagitannins and gallotannins. Condensed tannins are mainly polymerised products of flavan-3-ols. The most occurring classes are procyanidins, formed by (epi)catechins, and prodelphinidins, formed by (epi)gallocatechins (Appeldoorn, 2009). Tannins are often determined as total tannins, using e.g. a vanillin assay and determining the Catechin Activity Equivalents (CAE) in a spectrometric way. Despite naming “tannins” in many articles, the literature about which tannins exactly are present in faba beans is scarce and few articles were published. Merghem et al. (2004) identified different types of procyanidins in faba bean, namely 2 types of procyanidin A-dimers (without being able to specify which exact A-dimers), the procyanidin dimers B1, B2, and B3, the trimer C1. A subsequent study (Abu-Reidah et al., 2014), performed metabolic profiling of faba bean seeds and tentatively identified the procyanidin dimers B1, B2, B3, and B4, and diverse prodelphinidin dimers. Tannins are bitter and/or astringent depending on their degree of polymerization. Lower-weight tannins generally have a more pronounced bitter taste, while those with a higher degree of polymerization tend to be more astringent (Peleg et al., 1999; Soares et al., 2020). According to Peleg et al., 1999, flavanol monomers (for example catechin and epicatechin) are more bitter than astringent, and the dimers and trimers (for example procyanidins) are more astringent than bitter (Peleg et al., 1999). However, a partially contradictory effects were observed by Hufnagel and Hofmann (2008a, b). In taste receptor assays, lower bitterness of the monomer epicatechin compared to the procyanidin trimer C2 was shown (Soares et al., 2013). Tannins have historically attracted the attention of faba bean breeders, as they are considered major antinutritional factors (Guevara Oquendo et al., 2022). However, condensed tannins have been shown to evoke positive effects on human cardiovascular health as well (Appeldoorn, 2009).
Saponins are known to contribute to the bitter, astringent, and metallic taste of pea and soybean (Price and Roger Fenwick, 1984; Heng et al., 2006). They are natural compounds found in many plants, especially legumes. Saponins are triterpenoid glycosides, in which an apolar sapogenol backbone is substituted with one sugar chain (at sapogenol B and sapogenol E) or two sugar chains (at sapogenol A) (Decroos et al., 2005). The length and composition of the sugar chain determines the exact type of saponin. B-type saponins are the most common in legumes. A-type saponins are reported in soy (Sundaramoorthy et al., 2019a), but not in pea and faba bean. In the intact plant tissue, B-type saponins occur as conjugates of DDMP (2,3-dihydro-2,5-dihydroxy-6-methyl-4H-pyran-4-one). They are easily degraded to non-DDMP conjugated saponins upon processing. Soyasaponin αg, βa, βg, γa, and γg- belong to DDMP-saponins, while soyasaponins Bb (in another nomenclature called saponin I), Bc (II), Bb’(III), Bc’(IV), Ba (V) etc. belong to non-DDMP conjugated saponins (Barakat et al., 2015) (Singh et al., 2017). It has been shown in sensory experiments with saponins extracted from pea that these saponins are bitter and that DDMP-saponin βg is more bitter than saponin Bb (Heng et al., 2006), but no information related to the impact of saponins on the bitter taste of faba bean is present in literature.
Alkaloid content affects bitterness in food products such as lupins, which are classified into “bitter” or “sweet” varieties based on this attribute. Faba bean contains two pyrimidine glucoside alkaloids, vicine and convicine (V-C), yet their flavor characteristics have remained unexplored (Karolkowski et al., 2023a). V-C compounds may have been overlooked in recent flavor studies due to their absence in other major legumes, especially soybean. Moreover, their potential toxicity to some consumers might hamper sensory tests. When compounds are toxic and sensory tests are unfeasible, there is an alternative to test for bitterness, namely by in vitro taste receptor assays. However, although many toxic compounds (e.g., the alkaloid strychnine) have been tested on receptor level (Meyerhof et al., 2010), vicine or convicine have not been reported to be tested yet. Tuccillo et al. (2022) reported a correlation between V-C presence, bitterness, and mouth dryness, but this correlation also accounted for free phenolics and specific amino acids (Tuccillo et al., 2022). Given that V-C are expected to be bitter, the ongoing genetic efforts aiming to entirely remove their content for nutritional reasons (Bjornsdotter et al., 2021) will also be beneficial to the taste of the seeds.
Off-taste cannot only be evoked by compounds inherently present in the seeds like described above, but it can also be evoked by non-volatile lipid oxidation compounds. In pea protein isolates, it has been shown that bitterness was mainly caused by four trihydroxy-octadecenoic acid isomers (e.g. 9,10,11-trihydroxy-octadec-12-enoic), four hydroxy-octadecadienoic acid isomers (e.g. (10E,12E)-9-hydroxyoctadeca-10,12-dienoic acid), one oxo-octadecadienoic acid, one octacosatetraen, 1-linoleoyl glycerol, 2-hydroxyoleic acid, 2-hydroxypalmitic acid, α-linolenic acid, and linoleic acid (Glaser et al., 2020). Some of these compounds showed very low sensory threshold values (e.g., 0.08 mmol/L for 9,10,13-trihydroxyoctadec-12-enoic acid). The impact of non-volatile lipid oxidation compounds on bitterness has been confirmed in other crops. In wheat, two trihydroxy octadecenoic acids (pinellic acid, 8R*,9R*,10S*-trihydroxy-octadec-6Z-enoic acid), one trihydroxy-octadecadienoic acid (9S,12S,13S-trihydroxy-octadeca-10E,15Z-dienoic acid) and 1-(octadeca-9Z,12Z-dienoyl)-sn-glycero-3-phosphocholine have been identified as bitter, and associated with a dislike for the whole wheat breads (Cong et al., 2021). Analysis of wheat mutant lines, with the lipoxygenase gene disrupted, showed a significant reduction (88–93%) in bitter compound production when compared to the unaltered, or wild type, control. This establishes that the activity of the lipoxygenase enzyme plays a crucial role in creating the bitter taste from trihydroxy fatty acids in wheat (Cong et al., 2021). For faba bean or its ingredients, such bitter non-volatile lipid oxidation compounds have not been reported in literature yet. Given the presence of PUFAs and lipoxygenases in this crop, it can be expected that these compounds can be formed in faba bean as well. However, it remains to be established for faba bean ingredients which of all the potentially bitter and astringent compounds really contribute to the bitter and astringent sensation.
3 Plant breeding as a strategy to establish Vicia faba as plant-based food ingredient
Various thermal, chemical, or enzymatic strategies have been investigated to bridge the flavor gap in plant-based proteins by eliminating, modifying, or masking the development of undesirable taste and aroma (Tangyu et al., 2019; Xu et al., 2020; Mittermeier-Klessinger et al., 2021; Sontag-Strohm et al., 2021). However, post-harvest processing is often costly and energy-intensive, and sometimes the off-flavor is persistent despite the measures taken. An alternative approach is the breeding of faba bean varieties that exhibit low levels of off-flavor precursors, off-flavors, or enzymes that contribute to off-flavor development (Roland et al., 2017). Despite the identification of molecules having an impact on the aroma and possibly on the taste and astringent mouthfeel of faba bean ingredients in previous studies (Oomah et al., 2014; Akkad et al., 2019; Karolkowski et al., 2023a), a knowledge gap persists regarding specific marker compounds that could serve as proxies for off-flavor development in breeding programs. Determining and characterising these compounds further, along with the pathways responsible for their production, is essential for devising effective plant breeding strategies aimed at preventing or minimising off-flavors in faba bean.
It is important to specify that preventing the formation of off-flavors is a breeding target for the use of faba bean as ingredient in plant-based dairy or meat analogues, but not for other applications where seeds are cooked or consumed freshly. Breeding efforts to improve seed quality in faba bean must consider the significant variation in consumer preferences influenced by cultural and personal factors (Szenderak et al., 2022). Some consumers might still prefer the traditional usage of the seeds, where the distinctive and unique flavor is actually a positive and desired feature. Breeding for quality in this context focuses on enhancing the nutritional properties of the seeds rather than removing the typical taste and aroma. Important breeding targets include the removal of antinutritional compounds such as tannins, vicine and convicine, phytic acid, lectins, trypsin inhibitors, and oligosaccharides (e.g., raffinose, stachyose), and enhance the protein content. Additionally, cooking time is increasingly recognised as an essential trait since traditional home cooking of faba bean involves extended time to achieve satisfactory softness and palatability (Dhull et al., 2022). Therefore, faba bean breeding should aim to develop varieties that cater to different product end-uses, including those for sustainable and nutritious alternatives to animal protein sources.
The following paragraphs illustrate the candidate pathways that are critical to major off-flavors development and how they can be targeted with breeding aimed at improving the quality of faba bean for plant-based meat and dairy analogues.
3.1 Targeting lipid-oxidation pathways to reduce beany flavor and lipid-derived bitterness
3.1.1 Disrupting LOX genes leads to reduced oxidation rate of fatty acids
The LOX genes are known to regulate the enzymatic oxidation rate of polyunsaturated fatty acids, such as linoleic acid (C18:2) and linolenic acid (C18:3), through the expression of the lipoxygenase enzyme (Shahidi and Oh, 2020). Consequently, disrupting LOX genes would likely result in a decrease in the oxidation rate, reducing the amount of derived volatile and non-volatile compounds causing unpleasant aroma and taste, respectively.
Lipoxygenases selectively act on these fatty acids containing a 1-cis,4-cis-pentadiene unit producing the odourless stereospecific hydroperoxides. The latter are highly unstable and are in turn decomposed into a variety of compounds, including carbonyl compounds, alcohols, ketones, furans, epoxy compounds and hydrocarbons, among others, through the action of hydroperoxide lyase (HPL) and alcohol dehydrogenase (ADH) (see Figure 2) (Baysal and Demirdoven, 2007; El Hadi et al., 2013). The stereo-specificity of LOX enzymes has significant implications on the diversity of short-chain aldehydes, alcohols, and hydrocarbons produced, and thus on the final aroma (Karolkowski et al., 2023a). Lipoxygenases found in plants predominantly demonstrate regiospecificity towards the 9 or 13 carbon positions (Belitz et al., 2009). Additionally, LOX-catalysed reactions (but also auto-oxidation) contribute to the formation of lipid-derived non-volatile compounds. The hydroperoxides that are produced during the oxidation process can be transformed into various epoxy and hydroxy fatty acids by the action of peroxygenase (POX). This represents an alternative pathway to their conversion into volatiles by hydroperoxide lyase (HPL) and alcohol dehydrogenase (ADH) (Feussner and Wasternack, 2002). Epoxy and hydroxy fatty acids were identified in pea and oat and shown to be bitter in pea (Yang et al., 2019; Glaser et al., 2020; Glaser et al., 2021).

Figure 2 Biosynthesis of lipid-derived off-flavors. The figure depicts a simplified representation of the biosynthesis of lipid-derived off-flavors. The process begins with oleic acid (C18:1) serving as precursor for the synthesis of linoleic acid (C18:2) and linolenic acid (C18:3), mediated by enzymes FAD2 and FAD3 respectively. A portion of the produced polyunsaturated fatty acids is assembled into triacylglycerides (TAGs). Free polyunsaturated fatty acids, released by lipase action on TAGs, undergo oxidation by lipoxygenase enzymes (LOX) to form hydroperoxides. Hydroperoxides are decomposed into various compounds, including aldehydes, alcohols, ketones, and hydrocarbons, through the activity of hydroperoxide lyase (HPL) and alcohol dehydrogenase (ADH). In the figure, as example, n-hexanal and (Z)-3-hexenal represent the class of aldehydes, while n-hexanol and (Z)-3-hexenol represent the class of alcohols. However, the real molecules formed, and the number of reactions happing are numerous. The figure highlights (pointing hand) FAD2, FAD3, and LOX as target genes for breeding efforts aimed at reducing the fatty acid oxidation rate. The acronym of each key enzyme is reported along the pathway: FAD2 and FAD3, fatty acid desaturase 2 and 3; LOX, lipoxygenase; HPL, hydroperoxide lyase; ADH, alcohol dehydrogenase.
LOX family genes are conserved among legumes. In fact, lipid-oxidations products, (i.e, hexanal, 1-hexanol and 2-pentylfuran, etc.) are considered markers for “beany” flavor in other legumes such us chickpea, lentil, and yellow pea (Xu et al., 2019). Additionally, aldehydes derived from lipids are well known to cause the green, earthy, fatty, leafy aroma of peas and soybean (Seo and Hummel, 2012; Akkad et al., 2019). Two lipoxygenase isoenzymes (BBL-1/BBL-2) have been isolated in faba bean, but no studies to assess whether the presence of null or mutant alleles has a downstream effect on the flavor profile have been reported (Clemente et al., 2000).
The expression of these enzymes could vary among different cultivars, potentially accounting for the differences in odour-active compounds across them. In pea, for instance, a variation in lipoxygenase activity (as much as 70%) among three distinct commercially available pea types has been reported (Haydar and Hadziyev, 1973; Trindler et al., 2021). In addition, it has been shown that LOX differential activity depends on genetic variation in soybean (Mandal et al., 2014). If such genetic variations exist in faba bean, it could potentially serve as a basis for breeding initiatives aimed at creating new cultivars with reduced beany flavor.
Dysfunctional LOX pathways lead to reduced oxidation rate and are of interest for breeding applications in pulses. Lately, the newly developed soy genotype NRC132, which features lipoxygenase2 inactivation, has shown that LOX-free variants can exhibit diminished off-flavor in soy-milk production (Kumar et al., 2021). This development presents new possibilities for incorporating soybean into a broader range of food items. Several lipoxygenase-free soybean varieties are already available for cultivation in the USA (i.e., IA1008LF, IA2053LF, IA2076LF, IA2104LF, IA3027LF, IA3045LF, IA3051LF, etc.) (Yu et al., 2016). Based on these findings, the development of lipoxygenase-free faba bean seeds is envisioned in the near future.
Although some LOX enzymes do not require the prior release of fatty acids from their storage forms, the hydrolysis of acyl lipids to free fatty acids (FFAs) is generally required for their oxidising activity in plants (Lampi et al., 2020). Lampi et al. (2020) have emphasised the need to control endogenous lipase in faba bean during food processing due to its high lipid hydrolysis activity, which can also impact exogenous lipids in food products. The investigation of lipase alleles’ functionality is important for understanding off-flavor development in view of genetic improvement of this trait. However, lipases are essential for the utilization of storage triglycerides by developing seeds (Kanai et al., 2019), thus their genetic removal may be more critical than other candidate genes hereby proposed.
3.1.2 Disrupting FAD genes causes lower unsaturation of fatty acids and increased oil stability
Ideally, faba bean cultivars to produce protein fractions for meat and dairy analogues ought to exhibit decreased levels of C18:2 and C18:3, thereby diminishing the available substrate for lipoxygenase activity. Oil stability, characterised by resistance to oxidation, is affected by the ratio between C18:1, C18:2, C18:3 in the total oil composition. Enhancing the level of C18:1 while reducing C18:2 and C18:3 is an important breeding target to reduce fatty acid oxidation.
In several species, Fatty Acid Desaturase (FAD) genes are responsible for adding double bonds to the oleic acid (C18:1) to form linoleic (C18:2) and linolenic acids (C18:3) (Zhang et al., 2008). In particular, FAD2 (omega-6 fatty acid desaturase) is the key enzyme that catalyses the desaturation of C18:1 into C18:2 in roots and developing seeds of oil crops (Traore and He, 2021), while FAD3 (omega-3 fatty acid desaturase) subsequentially converts C18:2 into C18:3 (Figure 2) (Dar et al., 2017). The faba bean research community has previously not focused on investigating these genes, as they are typically not considered pertinent in non-oil crop research. However, breeding efforts should be conducted to develop high-oleic faba bean varieties less prone to oxidation. Evidence from soybean research indicates that combining two mutant isoforms of fad2 (GmFAD2–1A and GmFAD2–1B) into the same genetic background can effectively raise oleic acid content up to 80%, by disrupting the desaturase pathway (Tang et al., 2005; Pham et al., 2010). Crispr-Cas9 successfully produced double homozygous mutant plants showing a high oleic acid phenotypes (83.9%) compared to the wild-type soybean line (20.2%) (Do et al., 2019). Likewise, mutations in the three FAD3 genes copies (fad3a, fad3b, and fad3c) have led to the development of ultra-low-linolenic soybean oil (less than 1%), positively affecting oil shelf life and reducing off-flavors (Hudson and Hudson, 2021). Additionally, combining mutations in FAD3A with mutations in FAD2–1A and FAD2–1B was proposed as a strategy to produce high oleic and low linolenic soybean oil (Demorest et al., 2016). Studies have demonstrated that developing soybean high-oleic varieties via FAD gene manipulation does not impact yield or protein production (Kim et al., 2015). This is pivotal because farmers still prioritise high-yielding and protein-rich varieties, which are the most valuable traits in the legume market.
It should be noted that both linoleic (ω6) and linolenic (ω3) acids are essential fatty acids for humans, and deficiencies in these fatty acids are linked to various health concerns (Choque et al., 2014). Hence, it could be posited that novel faba bean varieties may exhibit reduced nutritional value in comparison to conventional varieties with unmodified oil profiles. However, it is important to highlight that new varieties with reduced off-flavors should not entirely supplant traditional cultivars that have higher levels of linoleic and linolenic acids, as these varieties will only target a specific segment of the market. In addition, it is also crucial to acknowledge that faba bean is not an oil crop or a conventional source of polyunsaturated fatty acids (PUFAs). Therefore, regardless of the release of new reduced-flavor varieties on the market, it is imperative to obtain PUFAs from alternative sources to ensure a well-balanced and nutritious diet.
3.2 Targeting polyphenols and saponins to reduce bitterness and astringency
3.2.1 Condensed tannins content is regulated by the zt1 and zt2 loci, but other candidate genes can be proposed to reduce their bitter precursors
Plant breeding efforts have led to the development of several low-tannin or zero-tannin faba bean varieties that are currently sold on the market. Low tannin genotypes present a reduction in condensed tannin content from ~8% to up to 0.01% on the dry matter (Oomah et al., 2011; Subodh Kumar and Amresh, 2018). Breeding for condensed tannins reduction mainly aimed at improving the nutritional value of animal feed (Guevara Oquendo et al., 2022).
There is a pleiotropic effect between tannin content in seeds and anthocyanic pigmentation on the flowers, which has facilitated genetic selection (Filippetti and Ricciardi, 1993). Condensed tannins and anthocyanins are, in fact, both end-products of the flavonoid pathway and they share metabolic routes. Anthocyanins are coloured pigments causing distinct colours in faba bean flowers. The wild-type flower is distinguished by its white petals, which feature a pronounced black spot on each wing petal and dark striae on the standard petal. Various color variations have been noted in the faba bean flowers, including an entirely white version, yellow wing spots, solid brown, pink, diffused yellow, red, etc. (Hughes et al., 2020). The absence of condensed tannins and presence of full white flowers share a simultaneous monogenetic control of two complementary and recessive genes, zt1 and zt2 (Jaakola et al., 2002; Gutierrez and Torres, 2019). Candidate genes for zt1 and zt2, which lead to a defect in the synthesis of anthocyanins or their precursors, have been proposed. The Medicago ortholog WD40 transcription factor TTG1 (Transparent Testa Glabra 1) was identified as the causal gene for the white flower phenotype (zt-1 locus) (Webb et al., 2016), while the helix-loop-helix (bHLH) transcription factor TRANSPARENT TESTA8 (TT8) has been suggested as the candidate for zt2 locus (Gutierrez et al., 2020). Despite TTG- and bHLH-conserved domain transcription factors being known to modulate flavonoid biosynthesis (Broun, 2005), their specific role in zero-tannin genotypes has not been elucidated. However, genomic breeding tools are available to differentiate two different mutant alleles for TTG1, ttg1-a, probably caused by a mutation in the promoter region, and ttg1-b due to a deletion at the 5′end of VfTTG1. Additionally, an allele-specific diagnostic marker has been developed to distinguish zt-1 from both wild and zt-2 genotypes (Gutierrez and Torres, 2019). These tools allow for marker assisted selection (MAS) for the zero-tannin trait.
The phenylpropanoid biosynthetic pathway that leads to the synthesis of condensed tannins (depicted in Figure 3) is particularly complex and influenced by several genes and environmental factors (He et al., 2008). Various branches of this pathway yield bitter and/or astringent compounds (i.e., coumaric, caffeic, ferulic acid, myricetin, quercetin, etc.), therefore they are an interesting target for breeding for eliminating multiple compounds simultaneously. Along the pathways, L-phenylalanine, synthetised via the shikimate pathway, is converted into p-coumaroyl-Coa through a three-step reaction involving phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (CH4) and 4-coumaroyl-CoA ligase (4CL) (Figure 3). p-coumaric acids, an intermediate in subsequent reactions, can be a precursor of bitter phenolic acids such as caffeic, ferulic and sinapic acids, besides being converted to p-coumaroyl-Coa. Naringenin chalcone is synthesised from p-coumaroyl-CoA and malonyl-CoA via chalcone synthase (CHS), serving as the principal intermediate in flavonol biosynthesis. This compound undergoes catalysis by chalcone isomerase (CHI), transforming it into naringenin. Dihydroflavonols are subsequentially formed through sequential enzymatic actions involving flavanone-3-hydroxylase (F3H), flavanone-3’-hydroxylase (F3’H), and flavonoid-3’,5’-hydroxylase (F3’5’H), yielding dihydrokaempferol (DHK), dihydroquercetin (DHQ), and dihydromyricetin (DHM), respectively. Dihydroflavonols serve as the foundation for flavan-3-ols synthesis, which represents the core monomers in the complex condensed tannin structure (He et al., 2015; Rosa-Martinez et al., 2023). The enzymes anthocyanidins synthase (ANS), anthocyanidin reductase (ANR), and leucoanthocyanidin reductase (LAR) contribute to the formation of the flavan-3-ols catechin, gallocatechin, epicatechin, epigallocatechin from intermediates leucocyanidin and leucodelphinidin (Mora et al., 2022). Flavan-3-ols then undergo polymerization to create condensed tannins (proanthocyanidins), although the exact mechanism and genetic control remain still unknown. The enzymes codified by zt1 and zt2 are expected to act in the last steps of the pathway between the synthesis of dihydroflavonols and flavan-3-ols (Zanotto et al., 2020), modulating the pathway at the transcriptional level. In line with this hypothesis, Zanotto et al. (2020) found dihydroflavonols in the seeds of zt1 and zt2 genetic backgrounds, showing that zero tannins genotypes still contain the precursor molecules.
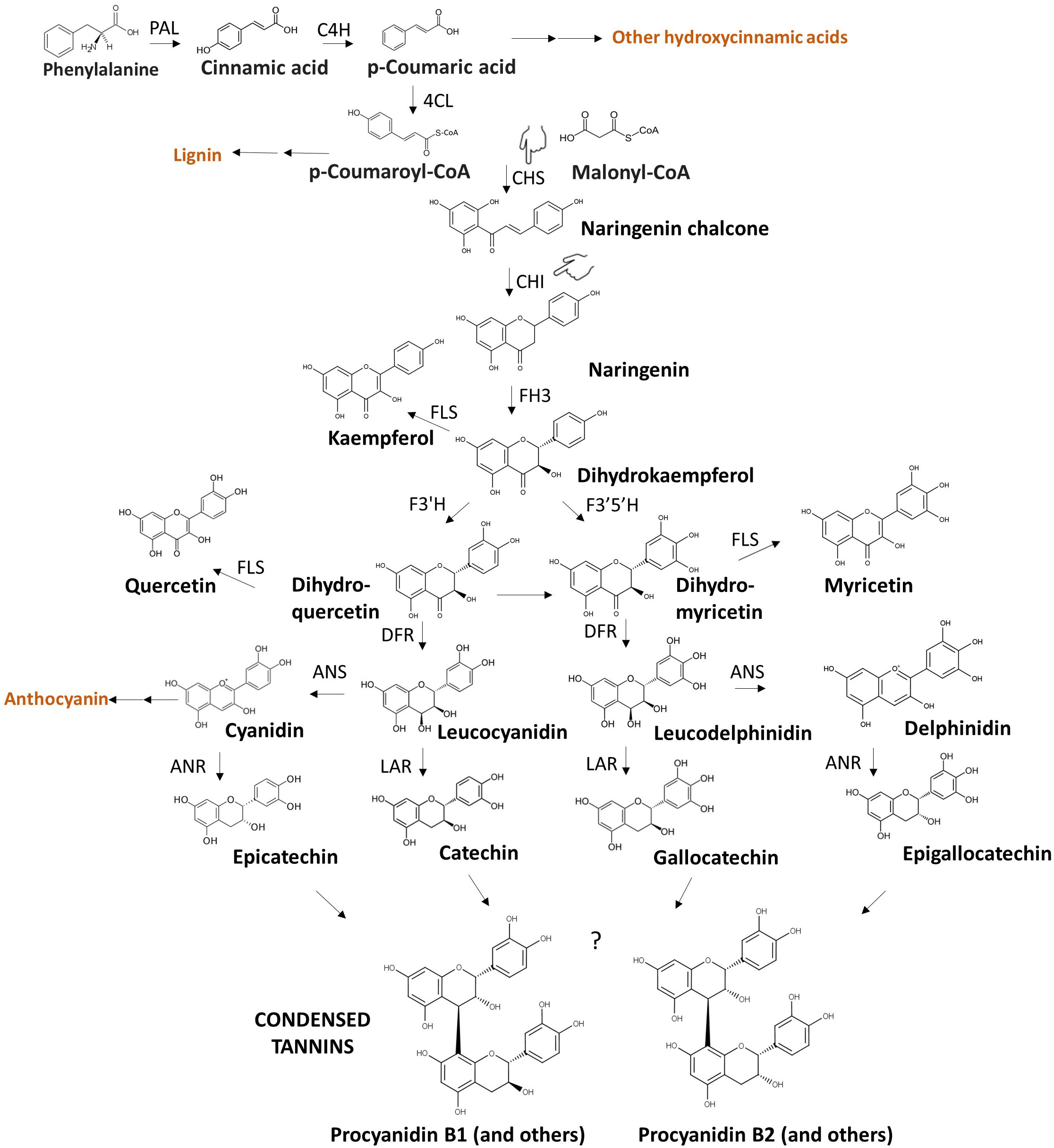
Figure 3 Condensed tannin biosynthesis. The illustration presents a streamlined version of the phenylpropanoid biosynthetic route involved in condensed tannin synthesis, resulting in various bitter and astringent polyphenols. Key intermediate compounds hereby represented belong to flavanone (naringenin), flavonol (quercetin, myricetin, kaempferol), leucoanthocyanidin (leucodelphinidin, leucocyanidin), anthocyanidin (delphinidin, cyanidin), flavan-3-ol (catechin, epicatechin, gallocatechin, epigallocatechin) classes. Compounds in orange are examples of important molecules that share precursors with condensed tannins. The acronym of each key enzyme is reported along the pathway: PAL, Phenylalanine ammonia lyase; CH4, cinnamate 4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase; F3’H, flavanone-3’-hydroxylase; F3’5’H, flavonoid-3’,5’-hydroxylase; ANS, anthocyanidins synthase; ANR, anthocyanidin reductase; LAR, leucoanthocyanidin reductase. The question mark in the last step of the pathways indicates that molecular mechanisms underlying the polymerization of flavan-3-ols into condensed tannins have not yet been elucidated. Procyanidin B1 and Procyanidin B2 are example of dimers belonging to the class of condensed tannins. The figure highlights the genes encoding CHI and CHS enzymes (pointing hand) as potential target genes for breeding applications to disrupt a portion of the phenylpropanoid biosynthetic pathway. CHI and CHS represent proposed alternative genes to zt1 and zt2. The question mark indicates that the exact genetic control of the polymerization of flavan-3-ols in condensed tannins is unknown.
Despite breeding for reduced condensed tannin content being a successful story in faba bean, additional efforts are required to develop varieties with reduced off-flavor. A more precise understanding of specific tannin compounds and associated phenylpropanoid pathway genes is crucial. Potential interventions at the initial steps in the pathway could disrupt subsequent reactions that produce several taste-active molecules. Targeting genes such as CHS and CHI (marked by two pointing hands in Figure 3) could simultaneously reduce the level of flavonols kaempferol, quercetin, myricetin, and others, likely reducing bitterness. In order crops, the involvement of chs-like genes have been speculated. Novák et al. (2006) showed that a higher and more prolonged expression of chalcone synthase-like genes was found in maturing hop cones of cultivars with high bitterness compared to those in cultivars with a lower bitter profile. However, even in successful applications, some bitterness and/or astringency is expected to remain as disrupting CHS and CHI do not affect the synthesis of phenolic acids.
3.2.2 Blocking the cyclisation of 2,3-oxydosqualene into saponins precursors as a strategy to mitigate off-taste and astringency
Amarowicz et al. (1997) isolated two distinct saponins possessing a chemical structure similar to that of soybean group B saponin in the seeds of four faba cultivars. The existence of B-type saponins (soyasaponin I/Bb) in faba bean was later verified, along with the detection of the DDMP saponin (soyasaponin βg)(Barakat et al., 2015). More recently, Stone et al. (2021) have detected and quantified the levels of the DDMP-soysaponins βa, αg and βg in faba bean seeds. Overall, the total saponin content in raw faba seeds ranges from approximately 481 to 757 µg/g, which is about half of the content detected in pea (1367 to 1701 µg/g) (Stone et al., 2021). The lower concentration of total saponins in Vicia faba compared to pea may be associated with a lower impact on taste. Recently, Tuccillo et al. (2022) corroborated the earlier findings of Heng et al. (2006) for pea, indicating that soyasaponin βg exhibits a greater degree of bitterness than soyasaponin Bb in faba bean ingredients. However, it has not been researched yet in detail whether saponins at their low content in faba bean impact the bitterness or astringency of faba bean products.
Breeding for zero saponins content has been implemented in several crops, including legumes. In soybean, for instance, the biosynthetic pathway of saponins is well-studied (Chung et al., 2020; Sundaramoorthy et al., 2019b). A simplified version of this pathway is depicted in Figure 4. The 2,3-oxidosqualene is the key compound in triterpenoid saponin biosynthesis since the molecule is the common precursor for the different classes of saponins (Xu et al., 2004). The biosynthesis steps are known to begin with the cyclisation of the 2,3-oxidosqualene by a range of oxidosqualene cyclases (OSCs) into various triterpene precursors of saponins. Among them, β-amyrin synthase (bAS) is considered a primary OSC in plants and has been molecularly characterised in soybean (Chung et al., 2007; Mugford and Osbourn, 2013). The enzyme produces the aglycone β-amyrin, which is subsequently modified through a series of oxidations and sugar chain additions to form the final saponin compounds (Chung et al., 2007; Mugford and Osbourn, 2013). Previous studies in soybean and pea have demonstrated that reducing the saponin content in seeds is a feasible strategy. Takagi et al. (2011) successfully used RNA interference (RNAi) to silence the bAS gene and suppress saponin biosynthesis in soybean. Mutant lines of peas with the homozygous mutant PsBAS1 (b-amyrin synthase1) gene accumulate virtually no saponins, paving the way for the development of peas with reduced bitterness and astringency (Vernoud et al., 2021). β-amyrin synthase genes are conserved in legumes as they have been cloned also in model legumes (Iturbe-Ormaetxe et al., 2003), suggesting that targeting bAS (indicated by the pointing hand in Figure 4) could be a viable approach for developing faba bean lines with reduced saponins and improved flavor.
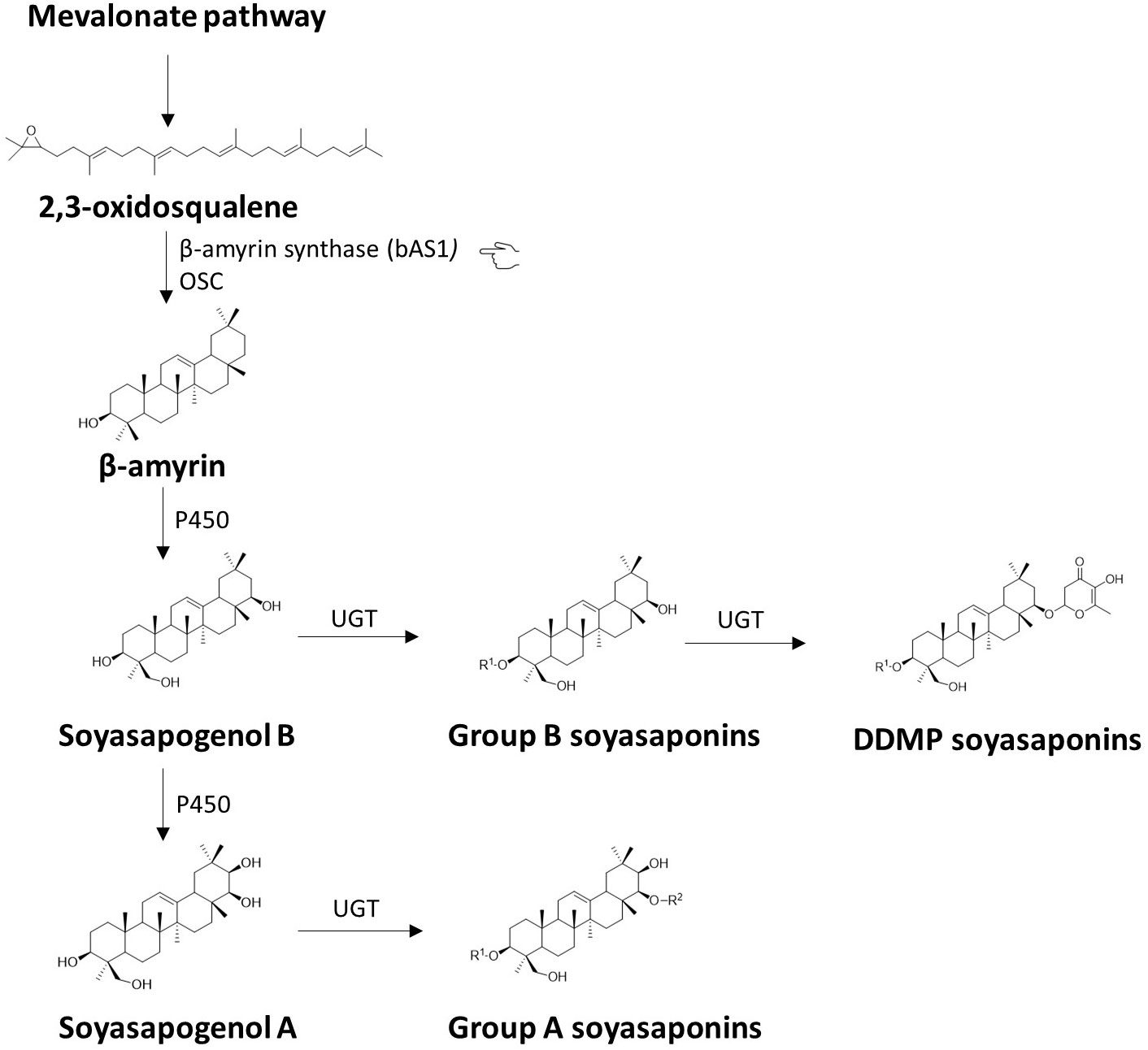
Figure 4 Biosynthesis of saponins. The figure illustrates a concise version of the proposed pathway for the synthesis of saponins in Glycine max. It is noteworthy to notice that saponin A reported in the pathway is present only in soybean, but not in faba bean. The synthesis of triterpenoid saponins begins with the cyclization of the precursor molecule 2,3-oxidosqualene into various triterpene scaffolds by the enzyme β-amyrin synthase (bAS), belonging to the family of oxidosqualene cyclases (OSCs). Among the produced scaffolds, β-amyrin is then subjected to site-specific oxidation reactions catalysed by cytochrome P450 monooxygenases (P450s), resulting in the formation of soyasapogenol B and soyasapogenol A. Subsequent glycosylation reactions catalysed by enzymes from the glycosyltransferase 1 superfamily, known as uridine diphosphate (UDP)-dependent glycosyltransferases (UGTs), produce structurally diverse triterpenoid saponins including, group B soyasaponin, DDMP-saponin and group A saponin. The highlighted β-amyrin synthase (bAS) gene (pointing hand) is a potential candidate for breeding applications because it plays a key role in the synthesis of saponins and is also located at a key point in the pathway.
4 Prospects for the development of faba bean ideotypes with neutral taste and aroma
Developing a breeding program to reduce off-flavors requires understanding the genetic basis of these traits and establishing breeding goals. Primary objectives involve reducing polyunsaturated fatty acid oxidation and polyphenol or saponin content in seeds, as schematised in Table 2. However, faba bean breeders must concurrently improve yield and stress resistance to ensure the crop’s success on the market. Faba bean is already well known to suffer yield instability, showing a large genotype by environment interaction (G × E) and lack of resilience to multiple abiotic and biotic stress conditions (Annicchiarico and Lannucci, 2008). Therefore, the inclusion of multiple traits in breeding schemes should be considered when designing breeding strategies. Selecting multiple traits will require understanding the genetic correlations between chemical attributes and agronomic performance (i.e., yield). Consequently, it will play a pivotal role to determine whether the genetic association between traits is favourable, unfavourable, or neutral and whether they are influenced by environmental components (Rana et al., 2019). Given that the biosynthesis of secondary metabolites is regulated by biotic and abiotic stress response (Thakur et al., 2019; Jan et al., 2021), it is expected that their concentration increase or decrease based on cultivation-specific conditions (Thakur et al., 2019). Oomah et al. (2014) have shown that the total extractable volatiles were highly dependent on the growing location of faba bean. Therefore, it will be crucial to assess the genotype-environment interaction for off-flavor related traits. Additionally, it is important to understand the heritability of these traits.
In plant breeding, selection strategies are largely based on the crop’s reproduction system, which typically involves either self-pollination (where the same plant acts as both the male and female parent) or cross-pollination (where one plant serves as the male parent and another as the female parent). It is important to consider that faba bean demonstrates a mixed-mating system, exhibiting partial outcrossing (cross-pollination) behaviour with an average outcrossing rate of about 30%. The co-existence of selfing and outcrossing in the flowers of the same plant of faba bean requires modifications of traditional breeding methods for self-pollinated crops while also considering that strategies for highly outcrossing species may not be entirely applicable (Gnanasambandam et al., 2012). Flavour-neutral homozygous lines can be developed through line or family selection under insect-proof enclosures. These could be directly commercialised, adhering to the rules of distinctiveness, uniformity, and stability. However, high degrees of homozygosity in faba bean may result in a significant loss of heterosis, raising concerns from a yield perspective (Adhikari et al., 2021). In the absence of true F1 hybrids with maximised heterosis, synthetic varieties offer a viable option to partially exploit hybrid vigour (Brünjes and Link, 2021) and develop faba bean cultivars with reduced off-flavors.
To be able to identify the genetic components of off-flavors and to develop breeding tools in faba bean, there is the need to explore genetic variation. In this view, zero tannin germplasm can serve as a valuable starting material in breeding schemes, as it already has reduced bitterness and astringency. In addition to the zero-tannin germplasm, further investigation of natural variation in off-flavor-related traits within the faba bean germplasm is required. It is worth mentioning that the current genetic diversity of faba bean to exploit in breeding is restricted to the cultivated gene pool (Adhikari et al., 2021), as the wild ancestors of faba bean are officially considered still missing.
To optimise the utilization of the extensively available germplasm and identify potential lines for crossbreeding, creating smaller “core collections” may be beneficial, as these would be more accessible to breeders and easier to increase seed availability from selected accessions (Malosetti and Abadie, 2001; Khazaei et al., 2013). Since there is no existing evidence to select neutral-flavor candidates from genebanks, it is strategic to begin exploring accessions with diverse genetic backgrounds, breeding history, and adaptation to various latitudes. These factors are anticipated to impact metabolic chemical profiles, potentially resulting in a range of different flavor features (Lin et al., 2014; Kusano et al., 2015). Therefore, a core collection can be established by sampling materials based on genetic diversity, assuming that this diversity is reflected in flavor variation.
4.1 High-throughput screening methods are needed to identify flavor-neutral accessions within the large faba bean germplasm
Exploring over 43,000 accessions of faba bean preserved in approximately 40 gene banks (Duc et al., 2015) to identify rare flavor-neutral traits is challenging, time-consuming and costly. To perform in-house phenotyping, breeding companies must acquire machinery and techniques appropriate for screening volatile and non-volatile chemical compounds. The high cost of acquiring such equipment may deter some investigators, but collaborative research across various sectors could make these tools more accessible (Viana and English, 2021).
For volatile flavor compounds, Gas Chromatography (GC) in combination with mass spectrometry (MS) is a widely used analysis apparatus to separate a mixture of many volatile compounds to identify (and, if wanted, to quantify) each of the compounds. Before this analysis can take place, the volatiles first need to be extracted from faba bean flour. The extraction technique chosen determines the type and quantity of compounds extracted, as not all compounds can be extracted by each extraction method. Vacuum distillation and solvent extraction are accurate, but very laborious and time-consuming (Wang et al., 2021). Faster, but still relatively laborious is StirBar Sorptive Extraction (SBSE). Headspace solid-phase microextraction (HS-SPME) has emerged as the most frequently utilised technique for the analysis of volatile compounds from protein-rich plants (Wang et al., 2022). This technique requires minimal sample preparation and can be automated. It has been used to characterise several low- and high-tannin faba bean varieties (Oomah et al., 2014; Akkad et al., 2019). Moreover, SPME-GC-MS has also been employed in large screening in the context of soybean breeding (Ning et al., 2019; Wang et al., 2020). Despite its advantages of being fast, economical, and having high extraction efficiency, it has a medium throughput. A much shorter analysis time is taken using GC- Ion Mobility Spectrometry (IMS) (a few minutes). Another type of technique is Proton Transfer Reaction-Mass Spectrometry (PTR-MS), which is a high-throughput alternative method for detecting and quantifying volatile organic compounds in real-time (Acierno et al., 2020). Its non-destructive nature allows for fast analysis (about 1 minute per sample) and has been successfully used in apple and strawberry breeding (Zini et al., 2005; Carbone et al., 2006; Acierno et al., 2020). These techniques produce very large amount of data in a very short time. A disadvantage of these fast measurements is that the time needed to analyse the measured data is much higher than the time needed to perform the measurement itself. When data analysis is automated, GC-IMS or PTR-MS, could be rapid alternative phenotyping methods for breeding faba bean for improved flavors.
For non-volatile flavor compounds, high-performance liquid chromatography (HPLC) or ultra-high-performance liquid chromatography (UPLC) coupled with mass spectrometry (MS) are commonly used. These techniques can detect and quantify saponins, lipid derivatives, tannins, phenolics, and other compounds that influence taste perception and astringent mouthfeel. Extraction steps are crucial for analysing non-volatile compounds, as solvents used to impact the solubility of compounds (Wang et al., 2022). As for volatile compounds, also for non-volatile compounds applies that what and how much is detected depends on the extraction method used. Liquid chromatography–mass spectrometry was used to characterise phenolic profiles of seed coat and flower tissue of three faba bean genotypes. In this way, Zanotto et al. (2020) have determined the contents of flavonols, tannins, hydroxybenzoic and hydroxycinnamic acids among others, which are all compounds involved in taste perception. The composition of tannins in seed coats of faba beans was investigated by Merghem et al. (2004) using HPLC-MS. In order to increase throughput, UPLC is an alternative to HPLC. It offers improved method sensitivity, resolution, and speed (Swetha et al., 2020), making it a candidate analytical methodology for screening breeding trials. However, UPLC is still considered a laborious technology with medium throughput. Similarly laborious in the extraction, but much faster in the data analysis is the use of spectrometric determinations of total phenolics and of total tannins. However, these types of methods do not identify individual compounds and are not suitable for saponins and others. Near Infrared Spectroscopy (NIRS) provides a high-throughput alternative to traditional approaches for breeding applications. It is a light-based technology that predicts the chemical composition of seeds after developing a prediction model. Model development requires reference chemical data and absorbance values obtained after scanning samples under near-infrared light (Ozaki and Morisawa, 2021). NIR has already been adopted in faba bean to quantify the presence of tannins and total polyphenols and has the potential to be applied to other chemical compounds involved in off-flavors (Johnson et al., 2020), aiding in the fast development of improved faba bean varieties. Usually, NIR can predict chemical compounds as classes, but the quantification of individual compounds is usually hampered.
4.2 The availability of a reference genome opens a new scenario facilitating molecular and quantitative breeding
The field of faba bean genomics has historically trailed behind that of other legumes, resulting in a slower pace of advancements in trait improvement and breeding programs (Torres et al., 2011). Molecular markers and isozyme polymorphisms to assist faba bean breeding have been documented, but to a lesser extent than in other major crop species (Alghamdi et al., 2012). Several breeding traits were mapped using genetic maps constructed from bi-parental or multi-parental populations as extensively reviewed by Khazaei et al. (2021). However, except for the zt1 and zt2 loci involved in the biosynthetic pathways of tannins, there has been no attempt to map quantitative trait loci (QTL) or genes controlling off-flavors development in faba bean.
The lack of a historical focus on off-flavors in faba bean breeding presents a unique opportunity for modern genomics to contribute significantly to this field. The recent release of the giant genome sequence (~13 Gb) of the faba bean cultivar Hedin/2 (Jayakodi et al., 2023) opens a multitude of new possibilities for researchers and breeders (Jayakodi et al., 2023). Furthermore, a pan-genome initiative was launched by the University of Helsinki and Luke (Natural Resources Institute Finland) (Khazaei et al., 2021). This will further facilitate efficient utilization of genetic resources, and a more comprehensive understanding of genetic diversity within faba bean.
Mapping major genes and QTL associated with off-flavor development can expedite the discovery of novel genes and the identification of natural variation. The complexity of off-flavor pathways likely involves a combined effect of multiple alleles, requiring quantitative approaches in breeding. Several examples of quantitative approaches have been reported in legume breeding literature to map QTL regulating molecules with taste and aroma properties, including saponin, flavonoid, alkaloid, fatty acid compositions, and hexenal (Phan et al., 2007; Liang et al., 2010; Li et al., 2016; Li et al., 2017; Teraishi et al., 2017; Wang et al., 2020). The current low-cost high-density genotyping platforms can accelerate the selection process for these complex traits and enhance genome-wide association studies in faba bean (GWAS) (Pavan et al., 2020). GWAS enables fine mapping, but the construction of biparental or multi-parent advanced generation inter-cross (MAGIC) populations also offers a possibility for markers development within existing breeding programs (Khazaei et al., 2018). Due to the size of the faba bean genome, whole-genome resequencing for marker-assisted selection (MAS) is still cost-prohibitive. However, reduced representation sequencing approaches such as Genotyping by Sequencing (GBS), Restriction site-associated DNA sequencing (RADseq), and Double digest restriction-site-associated sequencing (ddRAD-seq) can be utilised for high-density scans of off-flavor related traits (Davey and Blaxter, 2010; Elshire et al., 2011; Truong et al., 2012). Additionally, the newly developed Single Primer Enrichment Technology (SPET) genotyping platform (commercialised as ALLEGRO) could serve as a valuable tool for dissecting the genetic architecture of off-flavors in faba bean (IGATech, 2022). SPET assay targets and types single nucleotide polymorphism (SNP) in 90,000 genic and intergenic loci across the faba bean genome, offering more flexibility than previous SNP arrays (Scaglione et al., 2019; Jayakodi et al., 2023).
Many candidate genes involved in the production of volatile and non-volatile compounds have been proposed in this review. Their functional role in the development of off-flavor in faba bean could be validated by CRISPR/Cas9 gene editing technology, which is revolutionising the research in plant breeding (Ahmad, 2023). CRISPR/Cas9-based genome editing employs the Cas9 endonuclease to make precise cuts in DNA. These cuts are guided by specific RNA sequences, known as gRNA, ensuring targeted modifications. Typically, CRISPR-CAS9 uses a transformation system, such as Agrobacterium-mediated transformation, which involves the introgression of foreign DNA into plant cells or tissues. Editing the genomes of most legumes presents challenges due to their transformation recalcitrance. As a consequence, alternative strategies for genome editing that bypass traditional transformation methods have been proposed and are currently evaluated (Nivya and Shah, 2023). Another limitation in applying genome editing to legumes is their low regeneration capacities (Baloglu et al., 2022). This refers to the ability of a genetically transformed plant cell or tissue to regenerate into a full plant that expresses and propagates the introduced traits. Despite the mentioned difficulties in applying genome editing to legume crops, example of successful transformations include soybean (Lu and Tian, 2022), Lotus japonicus (Wang et al., 2019), Medicago truncatula (Jaudal et al., 2022), cowpea (Bridgeland et al., 2023), peanut (Yuan et al., 2019), pea (Li et al., 2023), and chickpea (Gupta et al., 2023). To date, no CRISPR/Cas9 studies have been reported for faba bean (Bhowmik et al., 2021). However, the release of the reference genome aids in designing specific gRNAs to target specific genes. A tailored system for producing transgenic faba bean through Agrobacterium-mediated transformation exists (Hanafy et al., 2005), but there is a continuous need to refine protocols to overcome transformation and regeneration obstacles, which are also genotype-dependent.
In conclusion, the combination of newly available genomic resources, the development of mapping populations tailored to off-flavor traits, and the application of advanced genetic techniques such as CRISPR/Cas9 (which is currently primarily used for research purposes) hold great promise for accelerating progress in faba bean breeding. These advancements will ultimately contribute to the development of improved faba bean varieties that are manufactured into food products that are more appealing to consumers, helping to drive the expansion of meat and dairy analogues.
4.3 Potential drawbacks and considerations for breeding strategies to reduce off-flavors
Breeding to remove off-flavor compounds from faba bean seeds has the potential to improve the taste and palatability of meat or dairy analogues. However, this approach also carries some potential drawbacks. Most of the secondary metabolites likely involved in off-flavor (e.g., lipid oxidation products, flavonoids, tannins, saponins) are required by plants for various functions, such as pigmentation, growth, reproduction, and resistance to pathogens. As such, they represent adaptive traits that have undergone natural selection during evolution (Peter Constabel et al., 2014; Ku et al., 2022). The elimination of these secondary metabolites could cause a yield decrease.
Plants augment the production of polyphenols in response to abiotic stressors as a strategy to assist adaptation to drought, heavy metal exposure, salinity, extreme temperatures, and ultraviolet radiation (Sharma et al., 2019). Tannins, for example, play a role in frost protection, acting as supercooling-promoting agents or anti-ice nucleating agents (Koyama et al., 2014). Zero-tannin faba bean cultivars resulted in higher susceptibility to cold and frost damage of seeds. Faba bean is a crop at high risk for late-season frost injury in certain environments, which results in reduced yield and marketability (Inci and Toker, 2011; Henriquez et al., 2017).
Saponin represents a chemical barrier against pathogens (Zaynab et al., 2021). Positively, the disruption of the molecular pathway controlling the saponin biosynthesis in pea by mutational breeding has not shown negative effects on the physiological (germination capacity) or nutritional quality (protein content) of the seeds under controlled greenhouse conditions (Vernoud et al., 2021). However, field-based studies are needed to evaluate the agronomical performance of these plants under the pressure of pathogens, pests and, environmental cues.
Furthermore, removing secondary metabolites might negatively impact the nutritional value of the beans. For instance, some polyphenols serve as important sources of antioxidants and are considered health-promoting compounds (Ganesan and Xu, 2017). In addition, altering the composition of fatty acids results in seeds with diminished nutritional value. Any disruption of LOX pathways needs careful evaluation anyway, as these enzymes play a pivotal role in plant defense mechanisms. LOX mediates the biosynthesis of jasmonic acid upon wounding (Bell et al., 1995), but also contributes to the production of oxylipins, enhancing plant resistance against pathogens (Wilson et al., 2001).
Considering the potential drawbacks reported, it would be preferable in breeding to specifically target genes that are expressed within the seeds only. This allows the production of secondary metabolites in other plant tissues, ensuring the production of molecules that are involved in defense mechanisms against biotic or abiotic stresses in some parts of the plant. It is also important to account that the disruption of secondary metabolism may lead to unforeseen effects elsewhere in the plant’s metabolic system, which is complex and highly interconnected. The elimination of bitter and astringent compounds could change the availability of certain nutrients, which may lead to the production of new compounds or changes in the concentration of existing ones. Therefore, careful monitoring of any changes in the bean’s composition is crucial to ensure that no harmful or unwanted compounds are produced.
Establishing strong collaborations between plant breeders and the food industry, alongside rigorous monitoring and evaluation of the breeding program outcomes, will be crucial for ensuring that the development of new faba bean varieties aligns with both consumer preferences and the long-term sustainability of the crop.
5 Conclusions
The plant-based food industry is increasingly using faba bean as a key ingredient in meat and dairy alternatives. Although technological solutions can mitigate the off-flavors present in faba bean ingredients, they are energy-intensive and costly. We firmly believe that the most sustainable solution to eliminate off-flavors involves plant breeding, with a focus on developing varieties that are tailored for these applications and that ensure minimal to no off-flavors. Previous breeding research on other crops has identified genes behind off-flavor synthesis. Moreover, as we deepen our understanding of the molecules responsible for off-flavors, and with the onset of the genomic era in faba bean research, we can uncover additional genetic pathways. Such insights can significantly benefit faba bean breeding. The success of breeding initiatives aiming to improve off-flavor profiles depend on several factors, including the availability or creation of genetic variations, the quantitative nature of the trait, its heritability, and the efficiency of high-throughput screening methods, among others. For improved faba bean varieties with reduced off-flavors to be both competitive in the market and attractive to farmers, they must also exhibit high yield and resistance to both biotic and abiotic stresses. Complementing these traits in new varieties can promote the transition toward a more sustainable and climate-resilient diets.
Author contributions
AL: Conceptualization, Formal Analysis, Visualization, Writing – original draft. WR: Formal Analysis, Investigation, Writing – review & editing. OB: Writing – review & editing. LP: Writing – review & editing, Funding acquisition, Project administration. LT: Funding acquisition, Project administration, Writing – review & editing, Conceptualization, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research is part of project ‘Pulses optimized for flavor and functionality’ co-financed by the Top Consortium for Knowledge and Innovation Agri & Food by the Dutch Ministry of Economic Affairs under contract number LWV19028.
Conflict of interest
The author(s) declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Reidah, I. M., Arraez-Roman, D., Warad, I., Fernandez-Gutierrez, A., Segura-Carretero, A. (2017). UHPLC/MS(2)-based approach for the comprehensive metabolite profiling of bean (Vicia faba L.) by-products: A promising source of bioactive constituents. Food Res. Int. 93, 87–96. doi: 10.1016/j.foodres.2017.01.014
Abu-Reidah, I. M., Del Mar Contreras, M., Arráez-Román, D., Fernández-Gutiérrez, A., Segura-Carretero, A. (2014). UHPLC-ESI-QTOF-MS-based metabolic profiling of Vicia faba L.(Fabaceae) seeds as a key strategy for characterization in foodomics. Electrophoresis 35, 1571–1581. doi: 10.1002/elps.201300646
Acierno, V., De Jonge, L., Van Ruth, S. (2020). Sniffing out cocoa bean traits that persist in chocolates by PTR-MS, ICP-MS and IR-MS. Food Res. Int. 133, 109212. doi: 10.1016/j.foodres.2020.109212
Adhikari, K. N., Khazaei, H., Ghaouti, L., Maalouf, F., Vandenberg, A., Link, W., et al. (2021). Conventional and molecular breeding tools for accelerating genetic gain in faba bean (Vicia faba L.). Front. Plant Sci. 12, 744259. doi: 10.3389/fpls.2021.744259
Ahmad, M. (2023). Plant breeding advancements with “CRISPR-Cas” genome editing technologies will assist future food security. Front. Plant Sci. 14, 1133036. doi: 10.3389/fpls.2023.1133036
Akkad, R., Buchko, A., Johnston, S. P., Han, J., House, J. D., Curtis, J. M. (2021). Sprouting improves the flavor quality of faba bean flours. Food Chem. 364, 130355. doi: 10.1016/j.foodchem.2021.130355
Akkad, R., Kharraz, E., Han, J., House, J. D., Curtis, J. M. (2019). Characterisation of the volatile flavor compounds in low and high tannin faba beans (Vicia faba var. minor) grown in Alberta, Canada. Food Res. Int. 120, 285–294. doi: 10.1016/j.foodres.2019.02.044
Alghamdi, S. S., Migdadi, H. M., Ammar, M. H., Paull, J. G., Siddique, K. H. M. (2012). Faba bean genomics: current status and future prospects. Euphytica 186, 609–624. doi: 10.1007/s10681-012-0658-4
Amarowicz, R., Yoshiki, Y., Pegg, R. B., Okubo, K. (1997). Presence of two saponins in faba bean (Vicia faba L.) seeds. Nahrung-Food 41, 352–354. doi: 10.1002/food.19970410607
Annicchiarico, P., Lannucci, A. (2008). Breeding strategy for faba bean in southern Europe based on cultivar responses across climatically contrasting environments. Crop Sci. 48, 983–991. doi: 10.2135/cropsci2007.09.0501
Appeldoorn, M. M. (2009). Dietary A-and B-type procyanidins: characterization and biofunctional potential of an abundant and diverse group of phenolics (The Netherlands: Wageningen University and Research).
Arbaoui, M., Balko, C., Link, W. (2008). Study of faba bean (Vicia faba L.) winter-hardiness and development of screening methods. Field Crops Res. 106, 60–67. doi: 10.1016/j.fcr.2007.10.015
Aschemann-Witzel, J., Gantriis, R. F., Fraga, P., Perez-Cueto, F. J. A. (2021). Plant-based food and protein trend from a business perspective: markets, consumers, and the challenges and opportunities in the future. Crit. Rev. Food Sci. Nutr. 61, 3119–3128. doi: 10.1080/10408398.2020.1793730
Asioli, D., Aschemann-Witzel, J., Caputo, V., Vecchio, R., Annunziata, A., Næs, T., et al. (2017). Making sense of the “clean label” trends: A review of consumer food choice behavior and discussion of industry implications. Food Res. Int. 99, 58–71. doi: 10.1016/j.foodres.2017.07.022
Augustin, M. A., Cole, M. B. (2022). Towards a sustainable food system by design using faba bean protein as an example. Trends Food Sci. Technol. 125, 1–11. doi: 10.1016/j.tifs.2022.04.029
Baloglu, M. C., Celik Altunoglu, Y., Baloglu, P., Yildiz, A. B., Türkölmez, N., Özden Çiftçi, Y. (2022). Gene-editing technologies and applications in legumes: Progress, evolution, and future prospects. Front. Genet. 13, 859437. doi: 10.3389/fgene.2022.859437
Barakat, H., Reim, V., Rohn, S. (2015). Stability of saponins from chickpea, soy and faba beans in vegetarian, broccoli-based bars subjected to different cooking techniques. Food Res. Int. 76, 142–149. doi: 10.1016/j.foodres.2015.03.043
Baysal, T., Demirdoven, A. (2007). Lipoxygenase in fruits and vegetables: A review. Enzyme Microbial Technol. 40, 491–496. doi: 10.1016/j.enzmictec.2006.11.025
Belitz, H.-D., Grosch, W., Schieberle, P. (2009). Food chemistry (Berlin, Germany: Springer Science & Business Media), 158–245. doi: 10.1007/978-3-540-69934-7
Bell, E., Creelman, R. A., Mullet, J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92, 8675–8679. doi: 10.1073/pnas.92.19.8675
Bhowmik, P., Konkin, D., Polowick, P., Hodgins, C. L., Subedi, M., Xiang, D., et al. (2021). CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legume Sci. 3 (3). doi: 10.1002/leg3.96
Bjornsdotter, E., Nadzieja, M., Chang, W., Escobar-Herrera, L., Mancinotti, D., Angra, D., et al. (2021). VC1 catalyses a key step in the biosynthesis of vicine in faba bean. Nat. Plants 7, 923–931. doi: 10.1038/s41477-021-00950-w
Bridgeland, A., Biswas, S., Tsakirpaloglou, N., Thomson, M. J., Septiningsih, E. M. (2023). Optimization of gene editing in cowpea through protoplast transformation and agroinfiltration by targeting the phytoene desaturase gene. PloS One 18, e0283837. doi: 10.1371/journal.pone.0283837
Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279. doi: 10.1016/j.pbi.2005.03.006
Brünjes, L., Link, W. (2021). Paternal outcrossing success differs among faba bean genotypes and impacts breeding of synthetic cultivars. Theor. Appl. Genet. 134 (8), 2411–2427. doi: 10.1007/s00122-021-03832-z
Calicioglu, O., Flammini, A., Bracco, S., Bellù, L., Sims, R. (2019). The future challenges of food and agriculture: an integrated analysis of trends and solutions. Sustainability 11, 222. doi: 10.3390/su11010222
Carbone, F., Mourgues, F., Biasioli, F., Gasperi, F., Mark, T. D., Rosati, C., et al. (2006). Development of molecular and biochemical tools to investigate fruit quality traits in strawberry elite genotypes. Mol. Breed. 18, 127–142. doi: 10.1007/s11032-006-9017-2
Chan, C. K. Y., Fabek, H., Mollard, R. C., Jones, P. J. H., Tulbek, M. C., Chibbar, R. N., et al. (2019). Faba bean protein flours added to pasta reduce post-ingestion glycaemia, and increase satiety, protein content and quality. Food Funct. 10, 7476–7488. doi: 10.1039/C9FO01186B
Choque, B., Catheline, D., Rioux, V., Legrand, P. (2014). Linoleic acid: between doubts and certainties. Biochimie 96, 14–21. doi: 10.1016/j.biochi.2013.07.012
Chung, E., Cho, C. W., Kim, K. Y., Chung, J., Kim, J. I., Chung, Y. S., et al. (2007). Molecular characterization of the GmAMS1 gene encoding β-amyrin synthase in soybean plants. Russian J. Plant Physiol. 54, 518–523. doi: 10.1134/S1021443707040139
Chung, S. Y., Seki, H., Fujisawa, Y., Shimoda, Y., Hiraga, S., Nomura, Y., et al. (2020). A cellulose synthase-derived enzyme catalyses 3-o-glucuronosylation in saponin biosynthesis. Nat. Commun. 11 (1), 5664. doi: 10.1038/s41467-020-19399-0
Clemente, A., Olias, R., Olias, J. M. (2000). Purification and characterization of broad bean lipoxygenase isoenzymes. J. Agric. Food Chem. 48, 1070–1075. doi: 10.1021/jf990463s
Coda, R., Varis, J., Verni, M., Rizzello, C. G., Katina, K. (2017). Improvement of the protein quality of wheat bread through faba bean sourdough addition. Lwt-Food Sci. Technol. 82, 296–302. doi: 10.1016/j.lwt.2017.04.062
Cong, W., Schwartz, E., Tello, E., Simons, C. T., Peterson, D. G. (2021). Identification of non-volatile compounds that negatively impact whole wheat bread flavor liking. Food Chem. 364, 130362. doi: 10.1016/j.foodchem.2021.130362
Crépon, K., Marget, P., Peyronnet, C., Carrouée, B., Arese, P., Duc, G. (2010). Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crops Res. 115, 329–339. doi: 10.1016/j.fcr.2009.09.016
Cubero, J. I. (1973). Evolutionary trends in Vicia faba L. Theor. Appl. Genet. 43, 59–65. doi: 10.1007/BF00274958
Cubero, J. I. (1974). On the evolution of Vicia faba L. Theor. Appl. Genet. 45, 47–51. doi: 10.1007/BF00283475
Czerny, M., Christlbauer, M., Christlbauer, M., Fischer, A., Granvogl, M., Hammer, M., et al. (2008). Re-investigation on odour thresholds of key food aroma compounds and development of an aroma language based on odour qualities of defined aqueous odorant solutions. Eur. Food Res. Technol. 228, 265–273. doi: 10.1007/s00217-008-0931-x
Dadic, M., Belleau, G. (1973). “Polyphenols and beer flavor,” in Proceedings. Annual meeting-American Society of Brewing Chemists. (Taylor & Francis), Vol. 31. 107–114. doi: 10.1080/00960845.1973.12006011
Dar, A. A., Choudhury, A. R., Kancharla, P. K., Arumugam, N. (2017). The FAD2 gene in plants: occurrence, regulation, and role. Front. Plant Sci. 8, 1789. doi: 10.3389/fpls.2017.01789
Davey, J. W., Blaxter, M. L. (2010). RADSeq: next-generation population genetics. Brief Funct. Genomics 9, 416–423. doi: 10.1093/bfgp/elq031
de Camargo, A. C., Schwember, A. R. (2019). Phenolic-driven sensory changes in functional foods. J. Food Bioactives 5, 6–7. doi: 10.31665/JFB.2019.5173
Decker, E. A., Akoh, C. C., Wilkes, R. S. (2012). Incorporation of (n-3) fatty acids in foods: challenges and opportunities. J. Nutr. 142, 610S–613S. doi: 10.3945/jn.111.149328
Decroos, K., Vincken, J. P., Heng, L., Bakker, R., Gruppen, H., Verstraete, W. (2005). Simultaneous quantification of differently glycosylated, acetylated, and 2,3-dihydro-2,5-dihydroxy-6-methyl-4H-pyran-4-one-conjugated soyasaponins using reversed-phase high-performance liquid chromatography with evaporative light scattering detection. J. Chromatogr A 1072, 185–193. doi: 10.1016/j.chroma.2005.03.021
Demorest, Z. L., Coffman, A., Baltes, N. J., Stoddard, T. J., Clasen, B. M., Luo, S., et al. (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 16, 1–8. doi: 10.1186/s12870-016-0906-1
Dhull, S. B., Kidwai, M. K., Noor, R., Chawla, P., Rose, P. K. (2022). A review of nutritional profile and processing of faba bean (Vicia faba L.). Legume Sci. 4, e129. doi: 10.1002/leg3.129
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2–1A and GmFAD2–1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19, 1–14. doi: 10.1186/s12870-019-1906-8
do Carmo, C. S., Knutsen, S. H., Malizia, G., Dessev, T., Geny, A., Zobel, H., et al. (2021). Meat analogues from a faba bean concentrate can be generated by high moisture extrusion. Future Foods 3, 100014. doi: 10.1016/j.fufo.2021.100014
D’Odorico, P., Davis, K. F., Rosa, L., Carr, J. A., Chiarelli, D., Dell’angelo, J., et al. (2018). The global food-energy-water nexus. Rev. geophys. 56, 456–531. doi: 10.1029/2017RG000591
Drewnowski, A., Gomez-Carneros, C. (2000). Bitter taste, phytonutrients, and the consumer: a review. Am. J. Clin. Nutr. 72, 1424–1435. doi: 10.1093/ajcn/72.6.1424
Duc, G., Aleksić, J. M., Marget, P., Mikic, A., Paull, J., Redden, R. J., et al. (2015). “Faba bean,” in Grain legumes. Ed. De Ron, A. M. (New York, NY: Springer), 141–178. doi: 10.1007/978-1-4939-2797-5_5
Duc, G., Bao, S., Baum, M., Redden, B., Sadiki, M., Suso, M. J., et al. (2010). Diversity maintenance and use of Vicia faba L. Genet. resources. Field Crops Res. 115, 270–278. doi: 10.1016/j.fcr.2008.10.003
El Hadi, M. A., Zhang, F. J., Wu, F. F., Zhou, C. H., Tao, J. (2013). Advances in fruit aroma volatile research. Molecules 18, 8200–8229. doi: 10.3390/molecules18078200
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6, e19379. doi: 10.1371/journal.pone.0019379
European Commission (2023). “Plant variety database,” in Plant variety catalogues, databases & information systems. Available at: https://food.ec.europa.eu/plants/plant-reproductive-material/plant-variety-catalogues-databases-information-systems_en.
FAOSTAT (2021). Statistical database, food food and agriculture organization of the United Nations (Fao: ROME). Available at: https://www.fao.org/faostat/en/#data/QCL.
Ferawati, F., Hefni, M., Witthoft, C. (2019). Flours from Swedish pulses: Effects of treatment on functional properties and nutrient content. Food Sci. Nutr. 7, 4116–4126. doi: 10.1002/fsn3.1280
Feussner, I., Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi: 10.1146/annurev.arplant.53.100301.135248
Filippetti, A., Ricciardi, L. (1993). “Faba bean,” in Genetic improvement of vegetable crops. Eds. Kalloo, G., Bergh, B. O. (Oxford, UK: Pergamon Press), 355–385.
Flores, F., Hybl, M., Knudsen, J. C., Marget, P., Muel, F., Nadal, S., et al. (2013). Adaptation of spring faba bean types across European climates. Field Crops Res. 145, 1–9. doi: 10.1016/j.fcr.2013.01.022
Flores, F., Nadal, S., Solis, I., Winkler, J., Sass, O., Stoddard, F. L., et al. (2012). Faba bean adaptation to autumn sowing under European climates. Agron. Sustain. Dev. 32, 727–734. doi: 10.1007/s13593-012-0082-0
Frank, O., Ottinger, H., Hofmann, T. (2001). Characterization of an intense bitter-tasting 1 H, 4 H-quinolizinium-7-olate by application of the taste dilution analysis, a novel bioassay for the screening and identification of taste-active compounds in foods. J. Agric. Food Chem. 49, 231–238. doi: 10.1021/jf0010073
Fresán, U., Mejia, M. A., Craig, W. J., Jaceldo-Siegl, K., Sabaté, J. (2019). Meat analogs from different protein sources: A comparison of their sustainability and nutritional content. Sustainability 11 (11), 3231. doi: 10.3390/su11123231
Ganesan, K., Xu, B. (2017). Polyphenol-rich lentils and their health promoting effects. Int. J. Mol. Sci. 18, 2390. doi: 10.3390/ijms18112390
Glaser, P., Dawid, C., Meister, S., Bader-Mittermaier, S., Schott, M., Eisner, P., et al. (2020). Molecularization of bitter off-taste compounds in pea-protein isolates (Pisum sativum L.). J. Agric. Food Chem. 68, 10374–10387. doi: 10.1021/acs.jafc.9b06663
Glaser, P., Mittermeier-Klessinger, V. K., Spaccasassi, A., Hofmann, T., Dawid, C. (2021). Quantification and bitter taste contribution of lipids and their oxidation products in pea-protein isolates (Pisum sativum L.). J. Agric. Food Chem. 69, 8768–8776. doi: 10.1021/acs.jafc.1c02889
Gnanasambandam, A., Paull, J., Torres, A., Kaur, S., Leonforte, T., Li, H., et al. (2012). Impact of molecular technologies on faba bean (Vicia faba l.) breeding strategies. Agronomy 2 (3), 132–166. doi: 10.3390/agronomy2030132
Grosch, W. (2001). Evaluation of the key odorants of foods by dilution experiments, aroma models and omission. Chem. Senses 26, 533–545. doi: 10.1093/chemse/26.5.533
Guevara Oquendo, V. H., Rodriguez Espinosa, M. E., Yu, P. (2022). Research progress on faba bean and faba forage in food and feed types, physiochemical, nutritional, and molecular structural characteristics with molecular spectroscopy. Crit. Rev. Food Sci. Nutr. 62, 8675–8685. doi: 10.1080/10408398.2021.1931805
Gupta, S. K., Vishwakarma, N. K., Malakar, P., Vanspati, P., Sharma, N. K., Chattopadhyay, D. (2023). Development of an Agrobacterium-delivered codon-optimized CRISPR/Cas9 system for chickpea genome editing. Protoplasma 260, 1–15. doi: 10.1007/s00709-023-01856-4
Gutierrez, N., Avila, C. M., Torres, A. M. (2020). The bHLH transcription factor VfTT8 underlies zt2, the locus determining zero tannin content in faba bean (Vicia faba L.). Sci. Rep. 10, 14299. doi: 10.1038/s41598-020-71070-2
Gutierrez, N., Torres, A. M. (2019). Characterization and diagnostic marker for TTG1 regulating tannin and anthocyanin biosynthesis in faba bean. Sci. Rep. 9, 16174. doi: 10.1038/s41598-019-52575-x
Hanafy, M., Pickardt, T., Kiesecker, H., Jacobsen, H. J. (2005). Agrobacterium-mediated transformation of faba bean (Vicia faba L.) using embryo axes. Euphytica 142, 227–236. doi: 10.1007/s10681-005-1690-4
Haydar, M., Hadziyev, D. (1973). A study of lipoxidase in pea seeds and seedlings. J. Sci. Food Agric. 24, 1039–1053. doi: 10.1002/jsfa.2740240906
He, F., Pan, Q. H., Shi, Y., Duan, C. Q. (2008). Biosynthesis and genetic regulation of proanthocyanidins in plants. Molecules 13, 2674–2703. doi: 10.3390/molecules13102674
He, M., Tian, H., Luo, X., Qi, X., Chen, X. (2015). Molecular progress in research on fruit astringency. Molecules 20, 1434–1451. doi: 10.3390/molecules20011434
Heng, L., Vincken, J. P., Van Koningsveld, G., Legger, A., Gruppen, H., Van Boekel, T., et al. (2006). Bitterness of saponins and their content in dry peas. J. Sci. Food Agric. 86, 1225–1231. doi: 10.1002/jsfa.2473
Henriquez, B., Olson, M., Hoy, C., Jackson, M., Wouda, T. (2017). Frost tolerance of faba bean cultivars (Vicia faba L.) in central Alberta. Can. J. Plant Sci. 98, 509–514. doi: 10.1139/CJPS-2017-0078
Hudson, K. A., Hudson, M. E. (2021). Genetic variation for seed oil biosynthesis in soybean. Plant Mol. Biol. Rep. 39, 700–709. doi: 10.1007/s11105-020-01276-1
Hufnagel, J. C., Hofmann, T. (2008a). Orosensory-directed identification of astringent mouthfeel and bitter-tasting compounds in red wine. J. Agric. Food Chem. 56 (4), 1376–1386. doi: 10.1021/jf073031n
Hufnagel, J. C., Hofmann, T. (2008b). Quantitative reconstruction of the nonvolatile sensometabolome of a red wine. J. Agric. Food Chem. 56 (19), 9190–9199. doi: 10.1021/jf801742w
Hughes, J., Khazaei, H., Vandenberg, A. (2020). The study of genetics of flower color in faba bean reveals generous diversity to be used in the horticulture industry. Hortscience 55, 1584–1588. doi: 10.21273/HORTSCI15238-20
IGATech (2022). “Targeted Genotyping Panels (Allegro - SPET) - Fava bean – 90k loci,” in Genomics Research Services/Allegro Genotyping Panels (SPET) (Udine, Italy: IGATech). Available at: igatechnology.com.
Inci, N. E., Toker, C. (2011). Screening and selection of faba beans (Vicia faba L.) for cold tolerance and comparison to wild relatives. Genet. Resour. Crop Evol. 58, 1169–1175. doi: 10.1007/s10722-010-9649-2
Iriti, M., Varoni, E. M. (2017). Pulses, healthy, and sustainable food sources for feeding the planet. Int. J. Mol. Sci. 18 (2), 255. doi: 10.3390/ijms18020255
Iturbe-Ormaetxe, I., Haralampidis, K., Papadopoulou, K., Osbourn, A. E. (2003). Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol. Biol. 51, 731–743. doi: 10.1023/A:1022519709298
Iwaniak, A., Hrynkiewicz, M., Bucholska, J., Minkiewicz, P., Darewicz, M. (2019). Understanding the nature of bitter-taste di- and tripeptides derived from food proteins based on chemometric analysis. J. Food Biochem. 43, e12500. doi: 10.1111/jfbc.12500
Jaakola, L., Maatta, K., Pirttila, A. M., Torronen, R., Karenlampi, S., Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/pp.006957
Jan, R., Asaf, S., Numan, M., Kim, K. M. (2021). Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy-Basel 11, 968. doi: 10.3390/agronomy11050968
Jaudal, M., Mayo-Smith, M., Poulet, A., Whibley, A., Peng, Y., Zhang, L., et al. (2022). MtING2 encodes an ING domain PHD finger protein which affects Medicago growth, flowering, global patterns of H3K4me3, and gene expression. Plant J. 112, 1029–1050. doi: 10.1111/tpj.15994
Jayakodi, M., Golicz, A. A., Kreplak, J., Fechete, L. I., Angra, D., Bednar, P., et al. (2023). The giant diploid faba genome unlocks variation in a global protein crop. Nature 615, 652–659. doi: 10.1038/s41586-023-05791-5
Johnson, J. B., Walsh, K., Naiker, M. (2020). Application of infrared spectroscopy for the prediction of nutritional content and quality assessment of faba bean (Vicia faba L.). Legume Sci. 2, e40. doi: 10.1002/leg3.40
Kanai, M., Yamada, T., Hayashi, M., Mano, S., Nishimura, M. (2019). Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9, 8924. doi: 10.1038/s41598-019-45331-8
Karkanis, A., Ntatsi, G., Lepse, L., Fernandez, J. A., Vagen, I. M., Rewald, B., et al. (2018). Faba bean cultivation - revealing novel managing practices for more sustainable and competitive European cropping systems. Front. Plant Sci. 9, 1115. doi: 10.3389/fpls.2018.01115
Karolkowski, A., Belloir, C., Briand, L., Salles, C. (2023a). Non-volatile compounds involved in bitterness and astringency of pulses: A review. Molecules 28, 3298. doi: 10.3390/molecules28083298
Karolkowski, A., Gourrat, K., Bouzidi, E., Albouy, J. F., Levavasseur, L., Briand, L., et al. (2023b). Origins of volatile compounds and identification of odour-active compounds in air-classified fractions of faba bean (Vicia faba L. minor). Food Res. Int. 163, 112260. doi: 10.1016/j.foodres.2022.112260
Karolkowski, A., Guichard, E., Briand, L., Salles, C. (2021). Volatile compounds in pulses: A review. Foods 10 (12), 3140. doi: 10.3390/foods10123140
Khazaei, H., O’sullivan, D. M., Stoddard, F. L., Adhikari, K. N., Paull, J. G., Schulman, A. H., et al. (2021). Recent advances in faba bean genetic and genomic tools for crop improvement. Legum Sci. 3, e75. doi: 10.1002/leg3.75
Khazaei, H., Stoddard, F. L., Purves, R. W., Vandenberg, A. (2018). A multi-parent faba bean (Vicia faba L.) population for future genomic studies. Plant Genet. Resources-Characterization Utilization 16, 419–423. doi: 10.1017/S1479262118000242
Khazaei, H., Street, K., Bari, A., Mackay, M., Stoddard, F. L. (2013). The FIGS (focused identification of germplasm strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PloS One 8, e63107. doi: 10.1371/journal.pone.0063107
Kim, H. J., Ha, B. K., Ha, K. S., Chae, J. H., Park, J. H., Kim, M. S., et al. (2015). Comparison of a high oleic acid soybean line to cultivated cultivars for seed yield, protein and oil concentrations. Euphytica 201, 285–292. doi: 10.1007/s10681-014-1210-5
Koyama, T., Inada, T., Kuwabara, C., Arakawa, K., Fujikawa, S. (2014). Anti-ice nucleating activity of polyphenol compounds against silver iodide. Cryobiology 69, 223–228. doi: 10.1016/j.cryobiol.2014.07.009
Ku, Y.-S., Ng, M.-S., Cheng, S.-S., Luk, C.-Y., Ludidi, N., Chung, G., et al. (2022). Soybean secondary metabolites and flavors: The art of compromise among climate, natural enemies, and human culture. Soybean Physiol. Genet. 102, 295–347. doi: 10.1016/bs.abr.2022.03.001
Kumar, V., Rani, A., Rawal, R. (2021). First Indian soybean variety free from off-flavour generating lipoxygenase-2 gene identified for release for commercial cultivation. Natl. Acad. Sci. Letters-India 44, 477–480. doi: 10.1007/s40009-021-01046-x
Kusano, M., Baxter, I., Fukushima, A., Oikawa, A., Okazaki, Y., Nakabayashi, R., et al. (2015). Assessing metabolomic and chemical diversity of a soybean lineage representing 35 years of breeding. Metabolomics 11, 261–270. doi: 10.1007/s11306-014-0702-6
Kyriakopoulou, K., Dekkers, B., van der Goot, A. J. (2019). “Plant-based meat analogues,” in Sustainable meat production and processing. (Elsevier), 103–126. doi: 10.1016/B978-0-12-814874-7.00006-7
Lampi, A. M., Yang, Z., Mustonen, O., Piironen, V. (2020). Potential of faba bean lipase and lipoxygenase to promote formation of volatile lipid oxidation products in food models. Food Chem. 311, 125982. doi: 10.1016/j.foodchem.2019.125982
L’Hocine, L., Martineau-Côté, D., Achouri, A., Wanasundara, J. P., Loku Hetti Arachchige, G. W. (2020). “Broad bean (faba bean),” in Pulses: processing and product development. (Switzerland: Springer International Publishing), 27–54.
Li, B., Fan, S., Yu, F., Chen, Y., Zhang, S., Han, F., et al. (2017). High-resolution mapping of QTL for fatty acid composition in soybean using specific-locus amplified fragment sequencing. Theor. Appl. Genet. 130, 1467–1479. doi: 10.1007/s00122-017-2902-8
Li, G., Liu, R., Xu, R., Varshney, R. K., Ding, H., Li, M., et al. (2023). Development of an Agrobacterium-mediated CRISPR/Cas9 system in pea (Pisum sativum L.). Crop J. 11, 132–139. doi: 10.1016/j.cj.2022.04.011
Li, M. W., Munoz, N. B., Wong, C. F., Wong, F. L., Wong, K. S., Wong, J. W., et al. (2016). QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front. Plant Sci. 7, 854. doi: 10.3389/fpls.2016.00854
Liang, H. Z., Yu, Y. L., Wang, S. F., Lian, Y., Wang, T. F., Wei, Y. L., et al. (2010). QTL mapping of isoflavone, oil and protein contents in soybean (Glycine max L. Merr.). Agric. Sci. China 9, 1108–1116. doi: 10.1016/S1671-2927(09)60197-8
Lin, H., Rao, J., Shi, J., Hu, C., Cheng, F., Wilson, Z. A., et al. (2014). Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J. Integr. Plant Biol. 56, 826–836. doi: 10.1111/jipb.12228
Lu, Q. S. M., Tian, L. (2022). An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 22, 7. doi: 10.1186/s12896-022-00737-7
Luzzatto, L., Arese, P. (2018). Favism and glucose-6-phosphate dehydrogenase deficiency. New Engl. J. Med. 378, 60–71. doi: 10.1056/NEJMra1708111
Ma, K. K., Greis, M., Lu, J., Nolden, A. A., Mcclements, D. J., Kinchla, A. J. (2022). Functional performance of plant proteins. Foods 11, 594. doi: 10.3390/foods11040594
Maalouf, F., Hu, J., O’sullivan, D. M., Zong, X., Hamwieh, A., Kumar, S., et al. (2018). Breeding and genomics status in faba bean (Vicia faba). Plant Breed. 138, 465–473. doi: 10.1111/pbr.12644
MacLeod, G., Ames, J. (1988). Soy flavor and its improvement. Crit. Rev. Food Sci. Nutr. 27, 219–400. doi: 10.1080/10408398809527487
Malosetti, M., Abadie, T. (2001). Sampling strategy to develop a core collection of Uruguayan maize landraces based on morphological traits. Genet. Resour. Crop Evol. 48, 381–390. doi: 10.1023/A:1012003611371
Mandal, S., Dahuja, A., Kar, A., Santha, I. M. (2014). In vitro kinetics of soybean lipoxygenase with combinatorial fatty substrates and its functional significance in off flavor development. Food Chem. 146, 394–403. doi: 10.1016/j.foodchem.2013.08.100
Merghem, R., Jay, M., Brun, N., Voirin, B. (2004). Qualitative analysis and HPLC isolation and identification of procyanidins from Vicia faba. Phytochem. Anal. 15, 95–99. doi: 10.1002/pca.731
Metayer, N. (2004). Vicia faba breeding for sustainable agriculture in Europe: Identification of regional priorities and definition of target genotypes (Paris: GIE Févérole), 5.
Meyerhof, W., Batram, C., Kuhn, C., Brockhoff, A., Chudoba, E., Bufe, B., et al. (2010). The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 35, 157–170. doi: 10.1093/chemse/bjp092
Mínguez, M. I., Rubiales, D. (2021). “Faba bean,” in Crop Physiology Case Histories For Major Crops. (Cambridge, MA, USA: Academic Press), 452–481. doi: 10.1016/B978-0-12-819194-1.00015-3
Mittermeier-Klessinger, V. K., Hofmann, T., Dawid, C. (2021). Mitigating off-flavors of plant-based proteins. J. Agric. Food Chem. 69, 9202–9207. doi: 10.1021/acs.jafc.1c03398
Mora, J., Pott, D. M., Osorio, S., Vallarino, J. G. (2022). Regulation of plant tannin synthesis in crop species. Front. Genet. 13, 870976. doi: 10.3389/fgene.2022.870976
Mugford, S. T., Osbourn, A. (2013). “Saponin synthesis and function,” in Isoprenoid synthesis in plants and microorganisms: New concepts and experimental approaches. (New York, NY, USA: Springer), 405–424. doi: 10.1007/978-1-4614-4063-5_28
Multari, S., Stewart, D., Russell, W. R. (2015). Potential of fava bean as future protein supply to partially replace meat intake in the human diet. Compr. Rev. Food Sci. Food Saf. 14, 511–522. doi: 10.1111/1541-4337.12146
Muratova, V. (1931). Common beans: (Vicia faba L.). Bull. Appl. Bot. Genet. Pl. Breed. Suppl. 50, 1–298.
Nawaz, M. A., Tan, M. V., Oiseth, S., Buckow, R. (2022). An emerging segment of functional legume-based beverages: A review. Food Rev. Int. 38, 1064–1101. doi: 10.1080/87559129.2020.1762641
Neugschwandtner, R. W., Bernhuber, A., Kammlander, S., Wagentristl, H., Klimek-Kopyra, A., Kaul, H. P. (2019). Agronomic potential of winter grain legumes for Central Europe: Development, soil coverage and yields. Field Crops Res. 241, 107576. doi: 10.1016/j.fcr.2019.107576
Ning, X., Yan, W.-B., Wang, X.-Q., Shao, Y.-P., Yang, M.-M., Wang, Z.-K., et al. (2019). Genetic dissection of hexanol content in soybean seed through genome-wide association analysis. J. Integr. Agric. 18, 1222–1229. doi: 10.1016/S2095-3119(19)62674-2
Nivya, V., Shah, J. M. (2023). Recalcitrancy to transformation, a hindrance for genome editing of legumes. Front. Genome Editing 5, 1247815. doi: 10.3389/fgeed.2023.1247815
Novák, P., Krofta, K., Matousek, J. (2006). Chalcone synthase homologues from Humulus lupulus: some enzymatic properties and expression. Biol. plantarum 50, 48–54. doi: 10.1007/s10535-005-0073-y
Nowacka, M., Trusinska, M., Chraniuk, P., Piatkowska, J., Pakulska, A., Wisniewska, K., et al. (2023). Plant-based fish analogs-A review. Appl. Sciences-Basel 13, 4509. doi: 10.3390/app13074509
Oomah, B. D., Luc, G., Leprelle, C., Drover, J. C., Harrison, J. E., Olson, M. (2011). Phenolics, phytic acid, and phytase in Canadian-grown low-tannin faba bean (Vicia faba L.) genotypes. J. Agric. Food Chem. 59, 3763–3771. doi: 10.1021/jf200338b
Oomah, B. D., Razafindrainibe, M., Drover, J. C. (2014). Headspace volatile components of Canadian grown low-tannin faba bean (Vicia faba L.) genotypes. J. Sci. Food Agric. 94, 473–481. doi: 10.1002/jsfa.6272
Ozaki, Y., Morisawa, Y. (2021). “Principles and characteristics of NIR spectroscopy,” in Near-Infrared Spectroscopy: Theory, Spectral Analysis, Instrumentation, and Applications. (Singapore: Springer), 11–35. doi: 10.1007/978-981-15-8648-4_2
Pasqualone, A., Abdallah, A., Summo, C. (2020). Symbolic meaning and use of broad beans in traditional foods of the Mediterranean Basin and the Middle East. J. Ethnic Foods 7, 1–13. doi: 10.1186/s42779-020-00073-1
Pavan, S., Delvento, C., Ricciardi, L., Lotti, C., Ciani, E., D’agostino, N. (2020). Recommendations for choosing the genotyping method and best practices for quality control in crop genome-wide association studies. Front. Genet. 11, 447. doi: 10.3389/fgene.2020.00447
Peleg, H., Gacon, K., Schlich, P., Noble, A. C. (1999). Bitterness and astringency of flavan-3-ol monomers, dimers and trimers. J. Sci. Food Agric. 79, 1123–1128. doi: 10.1002/(SICI)1097-0010(199906)79:8<1123::AID-JSFA336>3.0.CO;2-D
Peter Constabel, C., Yoshida, K., Walker, V. (2014). Diverse ecological roles of plant tannins: plant defense and beyond. Recent Adv. polyphenol Res. 4, 115–142. doi: 10.1002/9781118329634.ch5
Pham, A. T., Lee, J. D., Shannon, J. G., Bilyeu, K. D. (2010). Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 10, 195. doi: 10.1186/1471-2229-10-195
Phan, H. T., Ellwood, S. R., Adhikari, K., Nelson, M. N., Oliver, R. P. (2007). The first genetic and comparative map of white lupin (Lupinus albus L.): identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res. 14, 59–70. doi: 10.1093/dnares/dsm009
Price, K. R., Roger Fenwick, G. (1984). Soyasaponin I, a compound possessing undesirable taste characteristics isolated from the dried pea (Pisum sativum L.). J. Sci. Food Agric. 35, 887–892. doi: 10.1002/jsfa.2740350812
Punia, S., Dhull, S. B., Sandhu, K. S., Kaur, M. (2019). Faba bean (Vicia faba) starch: Structure, properties, and in vitro digestibility—A review. Legume Sci. 1, e18. doi: 10.1002/leg3.18
Rahate, K. A., Madhumita, M., Prabhakar, P. K. (2021). Nutritional composition, anti-nutritional factors, pretreatments-cum-processing impact and food formulation potential of faba bean (Vicia faba L.): A comprehensive review. Lwt-Food Sci. Technol. 138, 110796. doi: 10.1016/j.lwt.2020.110796
Rana, M., Sood, A., Hussain, W., Kaldate, R., Sharma, T. R., Gill, R., et al. (2019). “Gene pyramiding and multiple character breeding,” in Lentils. (Amsterdam, The Netherlands: Elsevier), 83–124. doi: 10.1016/B978-0-12-813522-8.00006-6
Ridgway, K. (2015). “Analysis of taints and off-flavors,” in Flavour Development, Analysis and Perception in Food and Beverages. (Sawston, UK: Woodhead Publishing), 63–81. doi: 10.1016/B978-1-78242-103-0.00004-7
Roland, W. S. U., Pouvreau, L., Curran, J., Van De Velde, F., De Kok, P. M. T. (2017). Flavor aspects of pulse ingredients. Cereal Chem. 94, 58–65. doi: 10.1094/CCHEM-06-16-0161-FI
Roland, W. S., Van Buren, L., Gruppen, H., Driesse, M., Gouka, R. J., Smit, G., et al. (2013). Bitter taste receptor activation by flavonoids and isoflavonoids: modeled structural requirements for activation of hTAS2R14 and hTAS2R39. J. Agric. Food Chem. 61, 10454–10466. doi: 10.1021/jf403387p
Rosa-Martinez, E., Bovy, A., Plazas, M., Tikunov, Y., Prohens, J., Pereira-Dias, L. (2023). Genetics and breeding of phenolic content in tomato, eggplant and pepper fruits. Front. Plant Sci. 14, 1135237. doi: 10.3389/fpls.2023.1135237
Scaglione, D., Pinosio, S., Marroni, F., Di Centa, E., Fornasiero, A., Magris, G., et al. (2019). Single primer enrichment technology as a tool for massive genotyping: a benchmark on black poplar and maize. Ann. Bot. 124, 543–552. doi: 10.1093/aob/mcz054
Schultz, M., Hoppe, K., Schmandke, H. (1988). Off-flavor reduction in vicia-faba bean protein isolate. Food Chem. 30, 129–135. doi: 10.1016/0308-8146(88)90150-1
Seidel, U. (1976). Sensorische und gaschromatographische Untersuchungen zum off-Flavour von Ackerbohnensamen und-proteinisolat: ein Beitrag zur” Subjektiven Gustometrie und Olfaktometrie” und zur Reaktionskinetik sensorischer Veränderungen. Sensory and gas chromatographic investigations on the off-flavour of field bean seeds and protein isolate: a contribution to 'Subjective Gustometry and Olfactometry. Dissertation A, Humboldt University of Berlin, Section of Food Economy and Food Technology.
Seo, H.-S., Hummel, T. (2012). “Smell, taste, and flavor,” in Food Flavors–Chemical, Sensory and Technological Properties. Ed. Jelen, H. (Boca Raton, FL: CRC Press), 35–65.
Sessa, D. J., Rackis, J. J. (1977). Lipid-Derived flavors of legume protein products. J. Am. Oil Chemists’ Soc. 54, 468–473. doi: 10.1007/BF02671039
Shahidi, F., Hossain, A. (2022). Role of lipids in food flavor generation. Molecules 27, 5014. doi: 10.3390/molecules27155014
Shahidi, F., Oh, W. Y. (2020). Lipid-derived flavor and off-flavor of traditional and functional foods: an overview. J. Food Bioactives 10. doi: 10.31665/JFB.2020.10224
Sharan, S., Zanghelini, G., Zotzel, J., Bonerz, D., Aschoff, J., Saint-Eve, A., et al. (2021). Fava bean (Vicia faba L.) for food applications: From seed to ingredient processing and its effect on functional properties, antinutritional factors, flavor, and color. Compr. Rev. Food Sci. Food Saf. 20, 401–428. doi: 10.1111/1541-4337.12687
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24, 2452. doi: 10.3390/molecules24132452
Singh, B., Singh, J. P., Singh, N., Kaur, A. (2017). Saponins in pulses and their health promoting activities: A review. Food Chem. 233, 540–549. doi: 10.1016/j.foodchem.2017.04.161
Soares, S., Brandao, E., Guerreiro, C., Soares, S., Mateus, N., De Freitas, V. (2020). Tannins in food: insights into the molecular perception of astringency and bitter taste. Molecules 25 (11). doi: 10.3390/molecules25112590
Soares, S., Kohl, S., Thalmann, S., Mateus, N., Meyerhof, W., De Freitas, V. (2013). Different phenolic compounds activate distinct human bitter taste receptors. J. Agric. Food Chem. 61, 1525–1533. doi: 10.1021/jf304198k
Sontag-Strohm, T., Stoddard, F., Schauman, C. (2021) Thermal treatment of faba bean for flavor improvement. Available at: https://www.legumehub.eu/wp-content/uploads/2021/11/PN33_Thermal-treatment-of-faba-bean-for-flavor.pdf.
Stone, A. K., Waelchli, K. N., Cabuk, B., Mcintosh, T. C., Wanasundara, J., Arntfield, S. D., et al. (2021). The levels of bioactive compounds found in raw and cooked Canadian pulses. Food Sci. Technol. Int. 27, 528–538. doi: 10.1177/1082013220973804
Subodh Kumar, S., Amresh, K. (2018). Condensed tannin: a major anti-nutritional constituent of faba bean (Vicia faba L.). Horticulture Int. J. 2 (2), 31–32. doi: 10.15406/hij.2018.02.00022
Sundaramoorthy, J., Palaniswamy, S., Park, G. T., Son, H. R., Tsukamoto, C., Lee, J. D., et al. (2019a). Characterization of a new sg-5 variant with reduced biosynthesis of group a saponins in soybean (Glycine max (L.) merr.). Mol. Breed 39 (11), 144. doi: 10.1007/s11032-019-1066-4
Sundaramoorthy, J., Park, G. T., Komagamine, K., Tsukamoto, C., Chang, J. H., Lee, J. D., et al. (2019b). Biosynthesis of DDMP saponins in soybean is regulated by a distinct UDP-glycosyltransferase. New Phytol. 222 (1), 261–274. doi: 10.1111/nph.15588
Swetha, S. R., Bhavya, S. K., Mounika, C. (2020). A review on comparative study of HPLC and UPLC. Res. J. Pharm. Technol. 13, 1570–1574. doi: 10.5958/0974-360X.2020.00284.X
Szenderak, J., Frona, D., Rakos, M. (2022). Consumer acceptance of plant-based meat substitutes: A narrative review. Foods 11, 1274. doi: 10.3390/foods11091274
Tacke, R., Ecke, W., Höfer, M., Sass, O., Link, W. (2022). Fine-mapping of the major locus for vicine and convicine in faba bean (Vicia faba) and marker-assisted breeding of a novel, low vicine and convicine winter faba bean population. Plant Breed. 141, 644–657. doi: 10.1111/pbr.13039
Takagi, K., Nishizawa, K., Hirose, A., Kita, A., Ishimoto, M. (2011). Manipulation of saponin biosynthesis by RNA interference-mediated silencing of beta-amyrin synthase gene expression in soybean. Plant Cell Rep. 30, 1835–1846. doi: 10.1007/s00299-011-1091-1
Tang, G. Q., Novitzky, W. P., Carol Griffin, H., Huber, S. C., Dewey, R. E. (2005). Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J. 44, 433–446. doi: 10.1111/j.1365-313X.2005.02535.x
Tangyu, M., Muller, J., Bolten, C. J., Wittmann, C. (2019). Fermentation of plant-based milk alternatives for improved flavor and nutritional value. Appl. Microbiol. Biotechnol. 103, 9263–9275. doi: 10.1007/s00253-019-10175-9
Teraishi, M., Tojo, Y., Yamada, N., Okumoto, Y. (2017). Identification of environmentally stable QTLs controlling Saponin content in Glycine max. Breed Sci. 67, 123–128. doi: 10.1270/jsbbs.16086
Thakur, M., Bhattacharya, S., Khosla, P. K., Puri, S. (2019). Improving production of plant secondary metabolites through biotic and abiotic elicitation. J. Appl. Res. Medicinal Aromatic Plants 12, 1–12. doi: 10.1016/j.jarmap.2018.11.004
Torres, A. M., Avila, C. M., Stoddard, F. L., Cubero, J. I. (2011). “Faba bean,” in Genetics, genomics and breeding of cool season grain legumes (genetics, genomics and breeding in crop plants). Eds. Pérez de la vega, M., Torres, A. M., Cubero, J. I., Kole, C. (New Hampshire: Science Pubs), 50–97.
Tournier, C., Sulmont-Rossé, C., Guichard, E. (2007). Flavour perception: aroma, taste and texture interactions. Food 1.2, 246–257.
Traore, S. M., He, G. (2021). Soybean as a model crop to study plant oil genes: mutations in FAD2 gene family. Model. Organisms Plant Genet. 6, 87–100. doi: 10.5772/intechopen.99752
Trindler, C., Annika Kopf-Bolanz, K., Denkel, C. (2021). Aroma of peas, its constituents and reduction strategies - Effects from breeding to processing. Food Chem. 376, 131892. doi: 10.1016/j.foodchem.2021.131892
Troszyńska, A., Amarowicz, R., Lamparski, G., Wołejszo, A., Baryłko-Pikielna, N. (2006). Investigation of astringency of extracts obtained from selected tannins-rich legume seeds. Food Qual. preference 17, 31–35. doi: 10.1016/j.foodqual.2005.04.006
Truong, H. T., Ramos, A. M., Yalcin, F., De Ruiter, M., van der Poel, H. J., Huvenaars, K. H., et al. (2012). Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PloS One 7, e37565. doi: 10.1371/journal.pone.0037565
Tsolakis, N., Wood, T., Privat, I., Srai, J. S., Griffiths, H., Terrier, B., et al. (2019). Developing fava beans as a sustainable source of high-quality protein for food supply chains through optimised genetics, farming and processing. 23rd Cambridge International Manufacturing Symposium University of Cambridge, 26–27 September 2019. doi: 10.17863/CAM.45884
Tuccillo, F., Kantanen, K., Wang, Y., Martin Ramos Diaz, J., Pulkkinen, M., Edelmann, M., et al. (2022). The flavor of faba bean ingredients and extrudates: Chemical and sensory properties. Food Res. Int. 162, 112036. doi: 10.1016/j.foodres.2022.112036
Turco, I., Ferretti, G., Bacchetti, T. (2016). Review of the health benefits of Faba bean (Vicia faba L.) polyphenols. J. Food Nutr. Res. 55, 283–293.
Van Gemert, L. (2003). Compilations of flavor threshold values in water and other media (Zeist, The Netherlands: Boelens Aroma Chemical Information Service).
Vernoud, V., Lebeigle, L., Munier, J., Marais, J., Sanchez, M., Pertuit, D., et al. (2021). beta-amyrin synthase1 controls the accumulation of the major saponins present in pea (Pisum sativum). Plant Cell Physiol. 62, 784–797. doi: 10.1093/pcp/pcab049
Viana, L., English, M. (2021). The application of chromatography in the study of off-flavor compounds in pulses and pulse by-products. Lwt-Food Sci. Technol. 150, 111981. doi: 10.1016/j.lwt.2021.111981
Wang, Z. K., Bao, G. G., Yang, C., Yang, M. M., Zhao, X., Shao, Y. P., et al. (2020). A genome-wide association study of hexanal content related to soymilk off-flavors in seed of soybean (Glycine max). Crop Pasture Sci. 71, 552–561. doi: 10.1071/CP20068
Wang, J., Liu, H., Ren, G. (2014). Near-infrared spectroscopy (NIRS) evaluation and regional analysis of Chinese faba bean (Vicia faba L.). Crop J. 2, 28–37. doi: 10.1016/j.cj.2013.10.001
Wang, L., Rubio, M. C., Xin, X., Zhang, B., Fan, Q., Wang, Q., et al. (2019). CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 224, 818–832. doi: 10.1111/nph.16077
Wang, Y., Tuccillo, F., Lampi, A. M., Knaapila, A., Pulkkinen, M., Kariluoto, S., et al. (2022). Flavor challenges in extruded plant-based meat alternatives: A review. Compr. Rev. Food Sci. Food Saf. 21, 2898–2929. doi: 10.1111/1541-4337.12964
Wang, B., Zhang, Q., Zhang, N., Bak, K. H., Soladoye, O. P., Aluko, R. E., et al. (2021). Insights into formation, detection and removal of the beany flavor in soybean protein. Trends Food Sci. Technol. 112, 336–347. doi: 10.1016/j.tifs.2021.04.018
Webb, A., Cottage, A., Wood, T., Khamassi, K., Hobbs, D., Gostkiewicz, K., et al. (2016). A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.). Plant Biotechnol. J. 14, 177–185. doi: 10.1111/pbi.12371
Wilson, R. A., Gardner, H. W., Keller, N. P. (2001). Cultivar-dependent expression of a maize lipoxygenase responsive to seed infesting fungi. Mol. Plant Microbe Interact. 14, 980–987. doi: 10.1094/MPMI.2001.14.8.980
Xu, R., Fazio, G. C., Matsuda, S. P. (2004). On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291. doi: 10.1016/j.phytochem.2003.11.014
Xu, M., Jin, Z., Gu, Z., Rao, J., Chen, B. (2020). Changes in odor characteristics of pulse protein isolates from germinated chickpea, lentil, and yellow pea: Role of lipoxygenase and free radicals. Food Chem. 314, 126184. doi: 10.1016/j.foodchem.2020.126184
Xu, M., Jin, Z., Lan, Y., Rao, J., Chen, B. (2019). HS-SPME-GC-MS/olfactometry combined with chemometrics to assess the impact of germination on flavor attributes of chickpea, lentil, and yellow pea flours. Food Chem. 280, 83–95. doi: 10.1016/j.foodchem.2018.12.048
Yang, Z., Piironen, V., Lampi, A. M. (2019). Epoxy and hydroxy fatty acids as non-volatile lipid oxidation products in oat. Food Chem. 295, 82–93. doi: 10.1016/j.foodchem.2019.05.052
Yu, K. F., Woodrow, L., Poysa, V. (2016). Registration of lipoxygenase free food grade soybean Germplasm, HS-151. Can. J. Plant Sci. 96, 148–150. doi: 10.1139/cjps-2015-0167
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19, 1–7. doi: 10.1186/s12896-019-0516-8
Zanotto, S., Khazaei, H., Elessawy, F. M., Vandenberg, A., Purves, R. W. (2020). Do faba bean genotypes carrying different zero-tannin genes (zt1 and zt2) differ in phenolic profiles? J. Agric. Food Chem. 68, 7530–7540. doi: 10.1021/acs.jafc.9b07866
Zaynab, M., Sharif, Y., Abbas, S., Afzal, M. Z., Qasim, M., Khalofah, A., et al. (2021). Saponin toxicity as key player in plant defense against pathogens. Toxicon 193, 21–27. doi: 10.1016/j.toxicon.2021.01.009
Zhang, P., Burton, J. W., Upchurch, R. G., Whittle, E., Shanklin, J., Dewey, R. E. (2008). Mutations in a Δ9-stearoyl-ACP-desaturase gene are associated with enhanced stearic acid levels in soybean seeds. Crop Sci. 48, 2305–2313. doi: 10.2135/cropsci2008.02.0084
Zhang, C., Hua, Y., Li, X., Kong, X., Chen, Y. (2020). Key volatile off-flavor compounds in peas (Pisum sativum L.) and their relations with the endogenous precursors and enzymes using soybean (Glycine max) as a reference. Food Chem. 333, 127469. doi: 10.1016/j.foodchem.2020.127469
Zini, E., Biasioli, F., Gasperi, F., Mott, D., Aprea, E., Mark, T. D., et al. (2005). QTL mapping of volatile compounds in ripe apples detected by proton transfer reaction-mass spectrometry. Euphytica 145, 269–279. doi: 10.1007/s10681-005-1645-9
Keywords: plant based, off-flavors, faba bean, breeding, tannins, saponins, lypoxygenase
Citation: Lippolis A, Roland WSU, Bocova O, Pouvreau L and Trindade LM (2023) The challenge of breeding for reduced off-flavor in faba bean ingredients. Front. Plant Sci. 14:1286803. doi: 10.3389/fpls.2023.1286803
Received: 31 August 2023; Accepted: 17 October 2023;
Published: 30 October 2023.
Edited by:
Frédéric Marsolais, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Pankaj Kumar Bhowmik, National Research Council Canada (NRC), CanadaStefano Pavan, University of Bari Aldo Moro, Italy
Copyright © 2023 Lippolis, Roland, Bocova, Pouvreau and Trindade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luisa M. Trindade, bHVpc2EudHJpbmRhZGVAd3VyLm5s
 Antonio Lippolis
Antonio Lippolis Wibke S. U. Roland2
Wibke S. U. Roland2 Ornela Bocova
Ornela Bocova Luisa M. Trindade
Luisa M. Trindade