
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 November 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1286699
This article is part of the Research Topic Transcriptome & Metabolic Profiling: An Insight Into the Abiotic Stress Response Crosstalk in Plants View all 13 articles
 Hongkai Zhang1,2,3†
Hongkai Zhang1,2,3† Bowen Luo1,2,3†
Bowen Luo1,2,3† Jin Liu1,2,3
Jin Liu1,2,3 Xinwu Jin1,2,3
Xinwu Jin1,2,3 Haiying Zhang1,2,3
Haiying Zhang1,2,3 Haixu Zhong1,2,3
Haixu Zhong1,2,3 Binyang Li1,2,3
Binyang Li1,2,3 Hongmei Hu1,2,3
Hongmei Hu1,2,3 Yikai Wang1,2,3
Yikai Wang1,2,3 Asif Ali1
Asif Ali1 Asad Riaz4
Asad Riaz4 Javed Hussain Sahito2,5
Javed Hussain Sahito2,5 Muhammad Zafar Iqbal2
Muhammad Zafar Iqbal2 Xiao Zhang1,2,3
Xiao Zhang1,2,3 Dan Liu1,2,3
Dan Liu1,2,3 Ling Wu1,2,3
Ling Wu1,2,3 Duojiang Gao2
Duojiang Gao2 Shiqiang Gao2
Shiqiang Gao2 Shunzong Su3
Shunzong Su3 Shibin Gao1,2,3*
Shibin Gao1,2,3*A previous metabolomic and genome-wide association analysis of maize screened a glucose-6-phosphate 1-epimerase (ZmG6PE) gene, which responds to low-phosphorus (LP) stress and regulates yield in maize’s recombinant inbred lines (RILs). However, the relationship of ZmG6PE with phosphorus and yield remained elusive. This study aimed to elucidate the underlying response mechanism of the ZmG6PE gene to LP stress and its consequential impact on maize yield. The analysis indicated that ZmG6PE required the Aldose_epim conserved domain to maintain enzyme activity and localized in the nucleus and cell membrane. The zmg6pe mutants showed decreased biomass and sugar contents but had increased starch content in leaves under LP stress conditions. Combined transcriptome and metabolome analysis showed that LP stress activated plant immune regulation in response to the LP stress through carbon metabolism, amino acid metabolism, and fatty acid metabolism. Notably, LP stress significantly reduced the synthesis of glucose-1-phosphate, mannose-6-phosphate, and β-alanine-related metabolites and changed the expression of related genes. ZmG6PE regulates LP stress by mediating the expression of ZmSPX6 and ZmPHT1.13. Overall, this study revealed that ZmG6PE affected the number of grains per ear, ear thickness, and ear weight under LP stress, indicating that ZmG6PE participates in the phosphate signaling pathway and affects maize yield-related traits through balancing carbohydrates homeostasis.
Maize (Zea mays ssp. mays) is a globally imperative food, feed, and industrial feedstock crop (Niu et al., 2020). Growth of maize requires a substantial amount of phosphorus-based fertilizer, but it is a finite and non-renewable resource. It is concerning that phosphate rock is projected to be depleted within the next 100 years (Kochian, 2012). Roots absorb the phosphorus mainly in the form of inorganic phosphate (Pi), but microorganisms and metal cations quickly convert it into unavailable forms by precipitation and fixation (Liu et al., 2005; Chiou and Lin, 2011; Ham et al., 2018). Therefore, determining how to reduce the amount of phosphorus fertilizer while maintaining a high crop yield is an important problem deserving further research.
Plants have evolved complex and sophisticated regulatory systems to adjust to a varying soil phosphate environment to maintain growth. Phosphate starvation-induced (PSI) genes are mainly regulated by Phosphate Starvation Response-SYG1/PH081/XPRI (PHR-SPX) modules that play key roles in this network (Puga et al., 2014; Wang et al., 2018). Phosphate Starvation Response (PHR) transcription factors (a sub-family of MYB TFs) positively regulate PSI gene expressions by attaching to PHR1 Binding Sequence (P1BS) (Rubio et al., 2001; Zhou et al., 2008). The activities of PHRs are balanced post-transcriptionally by negatively regulating TFs known as SPX-domains containing proteins through recognizing soluble inositol polyphosphates (InsPs) levels in cytosol and preventing the binding of PHRs to P1BS (Wang et al., 2014; Wild et al., 2016). Phosphate Starvation Response (PHR) transcription factors induced the transcription of plasma membrane-localized phosphate transporter (PHT), which is a class of phosphorous transporters playing crucial roles in phosphate uptake/distribution/redistribution in plants and enhances the transmembrane transport of phosphate under LP stress (Gonzalez et al., 2005; Sun et al., 2012). By elucidating the mechanism of phosphorus tolerance, it is possible to effectively improve maize yield while reducing the application of phosphorous fertilizer.
Previous studies have shown that LP stress activates physiological responses in plants, leading to changes in the levels of related metabolites. Under phosphate deficiency stress, plant pile up starch and sugar, which promotes carbon allocation to the root system and facilitates further soil phosphorus excavation. Low-phosphorus (LP) stress affects starch metabolism, inhibits photosynthesis in crop plants, and triggers starch accumulation in leaves (Gibson, 2004; Morcuende et al., 2007). Sucrose upregulated the activities of D-glucuronic acid, 5-O-methylembelin, and N-acetyl-L-phenylalanine during LP conditions (Yang et al., 2020). A metabolome analysis showed that LP stress accelerated glycolysis in the roots of soybean and repressed malic acid synthesis (Li et al., 2022). Under LP conditions, LP-tolerant maize genotypes induce the transcription of genes regulating plant hormone signaling, acid phosphatase, and metabolite, which promote phosphorus absorption and utilization in maize (Jiang et al., 2017).
Metabolomic profiling was conducted on six LP tolerant and sensitive maize inbred lines, and combined with genome-wide association analysis, five genes were identified to be responsive to LP stress. Among them, ZmG6PE not only influenced phosphorus content but also responded to LP stress by affecting ear diameter and ear rows (Luo et al., 2019). Glucose‐6‐phosphate 1‐epimerase catalyzes the conversion of anomeric forms of α-D-glucose‐6‐phosphate to β-D-glucose‐6‐phosphate at the branch point of D-glucose metabolism (Wurster and Hess, 1972; Wurster and Hess, 1973; Graille et al., 2006). This enzyme is named YMR099C in Saccharomyces cerevisiae, and it has an active site with sulfate ion connected by an arginine clamp formed by a side chain of two highly conserved residues of arginine (Graille et al., 2006). A bioinformatics analysis of the budding yeast’s entire genome for finding the putative target of transcription factor “GCN4 (a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences)” has shown that GCN4 regulates YMR099C (Schuldiner et al., 1998). Trz1 (a tRNA 3' end processing endonuclease), Nuc1 (a mitochondrial nuclease), and YMR099C form highly stable heterohexamers consisting of two copies of each of the three subunits, which suggests that YMR099C and Trz1 may regulate apoptotic nucleases activity (Ma et al., 2017). Glucose-6-phosphate is an important metabolite in multiple metabolic pathways, such as glycolysis and the pentose phosphate pathway (Wu et al., 2018; He et al., 2021). Glycolysis is a metabolic precursor for producing biomass and supplies cellular energy (Tanner et al., 2018). The pentose phosphate pathway can additionally catalyze the production of nicotinamide adenine dinucleotide phosphate (NADPH) using glucose-6-phosphate dehydrogenase (G6PDH) for providing reductive power (Jiang et al., 2012; Fouquerel et al., 2014). Currently, research on glucose-6-phosphate 1-epimerase mainly focuses on microorganisms and has not been studied in plants (Wurster and Hess, 1972; Wurster and Hess, 1973; Graille et al., 2006). Therefore, the current study would be a reference for investigating the future study of glucose-6-phosphate-1 epimerase in plants.
In sum, ZmG6PE is a glucose-6-phosphate 1-epimerase that regulates carbohydrate homeostasis. ZmG6PE is an enzyme that catalyzes α-D-glucose‐6‐phosphate to β-D-glucose‐6‐phosphate and participates in glycolysis and the pentose phosphate pathway. By biochemical, omics, molecular biology, and genetics analysis, the present study found that the gene function of ZmG6PE affects maize yield and the mechanism of response to LP stress. This research provides new directions for the improvement of maize yield and the study of LP tolerance mechanisms.
Using the CRISPR/Cas9 (Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) system, the ZmG6PE gene was knocked out in the maize inbred line KN5585 by the Agrobacterium tumefaciens-mediated transformation method. The mutation site was detected using ZmG6PE-KO-F and R primers (Supplementary Table S1). Maize Research Institute of Sichuan Agricultural University provided the seeds of maize inbred lines 178 and 9782 for the current study.
Seeds were surface disinfected with 2% (V/V) sodium hypochlorite (NaClO) solution for 30 min, then rinsed with ddH2O 5-6 times to remove the NaClO solution and were germinated on filter paper in dark conditions at 28 °C for 3 days. Germinated seeds were cultivated in sand and gravel for 7 days, and the seedlings with consistent growth were selected to remove embryos and endosperms. The seedlings were then transferred to plastic containers containing 25 L of half Hoagland nutrient solution concentration for adaptation cultivation for 3 days. Subsequently, the nutrient solution was replaced with normal-phosphorus (NP) (1 mM) or LP (1 μM) solution, also containing 6 mM KNO3, 4 mM Ca (NO3)2.4H2O, 1 mM or 1 μM NH4H2PO4, 100 μM EDTA-Fe, 2 mM MgSO4.7H2O, 46 μM H3BO3, 0.146 μM MnCl2.4H2O, 0.76 μM ZnSO4.7H2O, 0.016 μM (NH4)6Mo7O24.4H2O, and 0.32 μM CuSO4.5H2O, with a pH of 5.5 (Luo et al., 2019). The nutrient solution was replaced every three days, and the ventilation pump was operated for 8 hours every day.
The total RNA was extracted using a Trizol reagent kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. The reverse transcription of the first strand was performed using the PrimeScript™ II 1st Strand cDNA synthesis kit by following the manual’s instructions. The RT-qPCR was carried out using the FastStart Essential DNA Green Master (Roche, Germany) on a CFX96 Real-Time PCR Detection System (Bio-Rad, USA) following the kit’s protocol. The total reaction volume consisted of 5 μl 2× FastStart Essential DNA Green Master, with 0.5 μl each of forward and reverse primers and 1 μl of the cDNA template. The amplification procedure was set as follows; initial denaturation at 95 °C for 10 min; then 40 cycles of 95 °C for 5 sec, 60 °C for 15 sec, and 72 °C for 20 sec; and finally, a melting curve process. The quantitative PCR primers were designed using Beacon Designer™ 7 software and shown in Supplementary Table S1. All experiments were conducted thrice with three technical replicates.
Approximately 0.2 g of dry tissues were digested using H2SO4-H2O2 at 420 °C to obtain the total phosphorus. Then, phosphorus in the supernatant was measured using Auto Discrete Analyzers (SmartChem 200).
A starch content kit (Code: G0507W, Grace Biotechnology Co., LTD, Suzhou, China) was used to assay the starch content of maize leaves. The soluble sugar content of maize leaves was determined by a plant soluble sugar content kit (Code: KT-1-Y, Comin Biotechnology Co., LTD, Suzhou, China). The determination of the starch and soluble sugar was carried out strictly according to the operation instructions of the kit.
The full-length coding sequence of ZmG6PE was introduced into the pCold vector and then transformed into Rosetta-competent cells. The optimal protein induction conditions were screened, and the culture broth was scaled up under the optimal conditions. The extracted protein was purified and stored at -80 °C for future use.
Kinetic reactions were carried out at 25 °C in 50 mM KCl, 50 mM imidazole, and 8 mM MgSO4 at pH 7.6. The absorbance values were measured at 340 nm using the SW18-MV Stopped Flow Spectrophotometer (Applied Photophysics, UK). Reactions were started by mixing equivalent volumes of equilibrated 2 mM nicotinamide adenine dinucleotide phosphate (NADP+) and 60 μM glucose-6-phosphate with 160 units/mL glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides (Sigma) and the varying concentrations of ZmG6PE (Graille et al., 2006).
For transcriptome analysis, the second fully expanded leaves and the roots of the mutant zmg6pe and wild type (WT) plants were harvested and frozen in liquid nitrogen immediately. The Trizol reagent (Invitrogen Life Technologies) was used to isolate total RNA from the leaves and roots following the manual’s instructions. RNA’s concentration, integrity, and quality were measured and assessed using a NanoDrop™ spectrophotometer (Thermo Scientific). RNA samples were prepared using 3 μg RNA as the initial input material. The RNA-seq and data analysis were carried out by the Shanghai Bioprofile Biotechnology Co., Ltd. (Shanghai, China). The sequencing libraries of the total RNA were constructed using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA), and the RNA libraries were sequenced by the Illumina NovaSeq 6000 platform. Each sample consists of three plants with three replicates.
For extracting the metabolites, samples were weighed, dried, and transferred into a 1.5 mL Eppendorf tube containing a 5 mm tungsten bead and grounded in a grinding mill for 1 min with 65 Hz power. Metabolite extraction was performed using an ultrasonic shaker with 1 mL precooled acetonitrile, methanol, and H2O (v:v:v, 2:2:1) at 4 °C for 1 hour. The mixture was placed at -20 °C for 1 hour and then centrifuged at 14,000 rpm for 20 min at 4 °C. The supernatants were collected and placed in a cryogenic vacuum for concentration. Subsequently, the UPLC-ESI-Q-Orbitrap-MS system (UHPLC, Shimadzu Nexera X2 LC-30AD, Shimadzu, Japan) coupled with Q-Exactive Plus (Thermo Scientific, San Jose, CA, USA) was used for Metabolomic profiling.
The MS-DIAL software was used to process the raw MS data for retention time correction, peak alignment, and peak area extraction. The accuracy mass (mass tolerance < 0.02 Da) and MS/MS data (mass tolerance < 0.02 Da) were matched with Kyoto Encyclopedia of Genes and Genomes (KEGG), MassBank, and other available databases, and a standard self-built metabolite library was used to differentiate and recognize the metabolites. Only variables greater than 50% non-zero measured values within at least 1 group were processed in extracted ion features. The samples used in the metabolite analysis were consistent with those used in the RNA-seq analysis.
To localize ZmG6PE protein in the cell, a full-length coding sequence of ZmG6PE was introduced into pCAMBIA2300-35S-eGFP using the NovoRec® plus One Step PCR Cloning Kit (NovoProtein, Shanghai, China). The constructs were mobilized into an Agrobacterium strain GV3101 by the freeze-thaw method and cultured for up to the required density. Then cells were collected and resuspended in a solution comprising 10 mM magnesium chloride and 150 mM acetosyringone (Sigma-Aldrich). Fluorescence signals of the eGFP in Nicotiana benthamiana leaves were detected with a confocal microscope (Leica).
The subcellular localization of the ZmG6PE protein was determined by introducing the combined vector pCAMBIA2300-P35S: ZmG6PE-eGFP and the nucleus localization signal (NLS) into tobacco leaves using transient transformation. The signals derived from ZmG6PE overlapped with the autofluorescence of NLS (Figure 1A). Similarly, the signals derived from ZmG6PE overlapped with the autofluorescence of the cell membrane localization signal (CMLS) (Figure 1A). These results confirmed the prediction of CELLO v.2.5, showing that the ZmG6PE protein is localized in the nucleus and cell membrane.
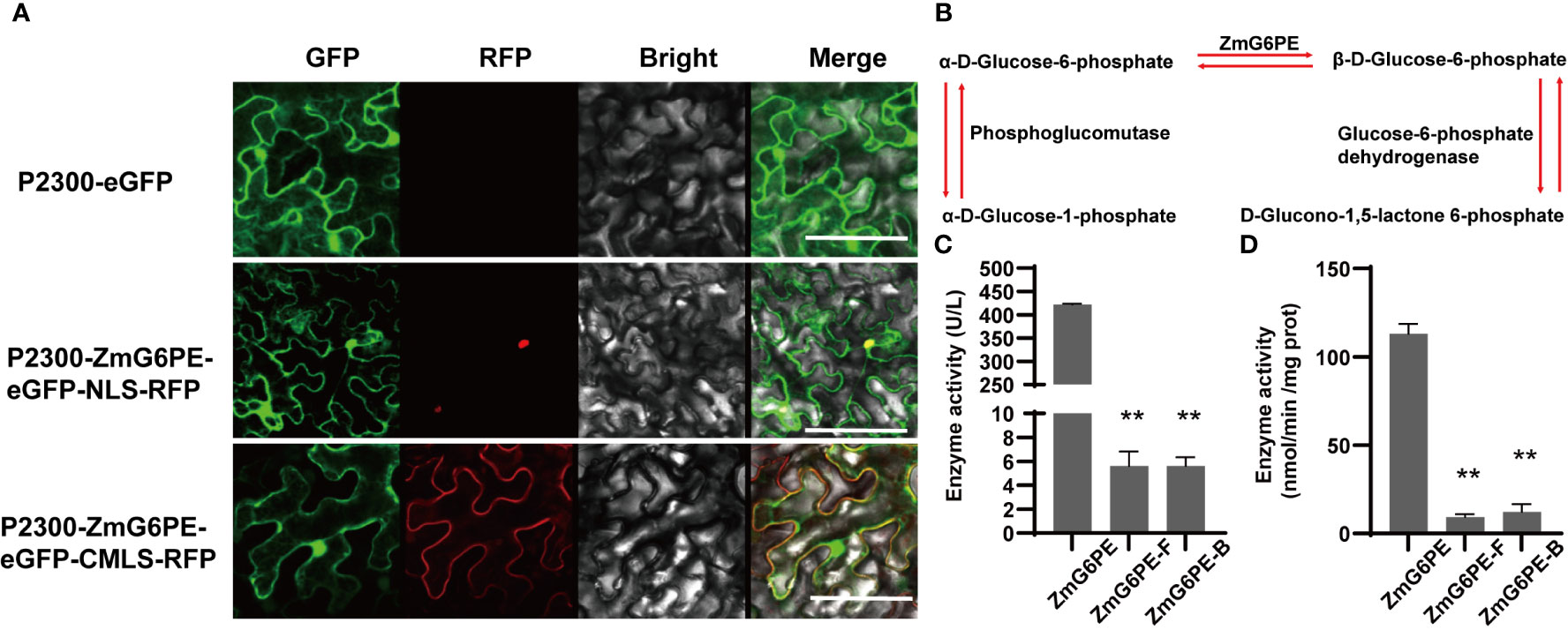
Figure 1 Subcellular localization of ZmG6PE and enzyme activity. (A) Subcellular localization of ZmG6PE in tobacco leaves, bar = 10 nm. (B) Schematic representation of enzymatic reactions involving glucose-6-phosphate anomer. (C) Determination of the enzyme activity by the enzyme kinetic method. (D) Determination of enzyme activity using an ELISA kit. “**” represents p < 0.01 by single factor ANOVA test.
ZmG6PE protein sequence from 41 to 308 amino acids encodes a conserved domain of Aldose_epim (Supplementary Figure S1A). To explore the critical peptide sequences of ZmG6PE, we divided ZmG6PE into two sections, ZmG6PE-Front (ZmG6PE-F) and ZmG6PE-Behind (ZmG6PE-B). Both sections (ZmG6PE-F and ZmG6PE-B) contained the conserved Aldose_epim domains of ZmG6PE (41 to 308 aa), ZmG6PE-F (41 to 147 aa), and ZmG6PE-B (1 to 161 aa) (Supplementary Figure S1A). Unlike ZmG6PE-B and ZmG6PE-F, the tertiary structure of ZmG6PE had a putative active site pocket (Supplementary Figure S1B). To investigate the biochemical characteristics of ZmG6PE, His-tagged ZmG6PE, ZmG6PE-B, and ZmG6PE-F proteins were expressed in Escherichia coli (Supplementary Figure S1C). The enzymatic activity of protein during the interconversion of α- and β-D-glucose-6-phosphate anomers is explained by the specificity of the enzymes participating in sugar metabolism (Figure 1B). Enzyme kinetics and enzyme-linked immunosorbent assay (ELISA) were employed to evaluate the ZmG6PE activity. The results showed that ZmG6PE had glucose-6-phosphate 1-epimerase activity (Figures 1C, D); however, the ZmG6PE protein segments (ZmG6PE-F and ZmG6PE-B) exhibited negligible glucose-6-phosphate 1-epimerase activity (Figures 1C, D).
To investigate the functions of ZmG6PE in maize, two independent transgenic lines (zmg6pe-1 and zmg6pe-2) were developed using CRISPR/Cas9. The line zmg6pe-1 has a 421 bp deletion, while zmg6pe-2 has a 1 bp deletion (Supplementary Figure S2A), thus both result in a truncated protein. During the seedling stage, the fresh weight and phosphorus content in zmg6pe decreased compared to WT (Supplementary Figures S2B-D), but there was no significant difference at maturity (Supplementary Figures S2E-G). To determine whether ZmG6PE responds to LP stress, 10 days old maize seedlings were further raised in a hydroponic system and treated with 1 mM and 1 μM phosphate conditions for 15 days (Figure 2A and Supplementary Figure S2H). Under both NP and LP conditions, the leaves and roots fresh weight of the zmg6pe significantly reduced (Figures 2B, C); LP tolerance coefficients in zmg6pe leaves and roots were reduced (Supplementary Figures S2I, J). Starch contents decreased under NP conditions in leaves of zmg6pe (Figures 2D). Soluble sugar contents decreased under LP conditions in the leaves of zmg6pe (Figures 2E). The phosphorus contents decreased under NP conditions in zmg6pe leaves than in WT leaves (Figure 2F). However, the phosphorus content in roots did not significantly change in WT and zmg6pe (Figure 2G). It significantly reduced the expression levels of ZmG6PE in the leaves and roots of the zmg6pe mutant (Figures 2H, I).
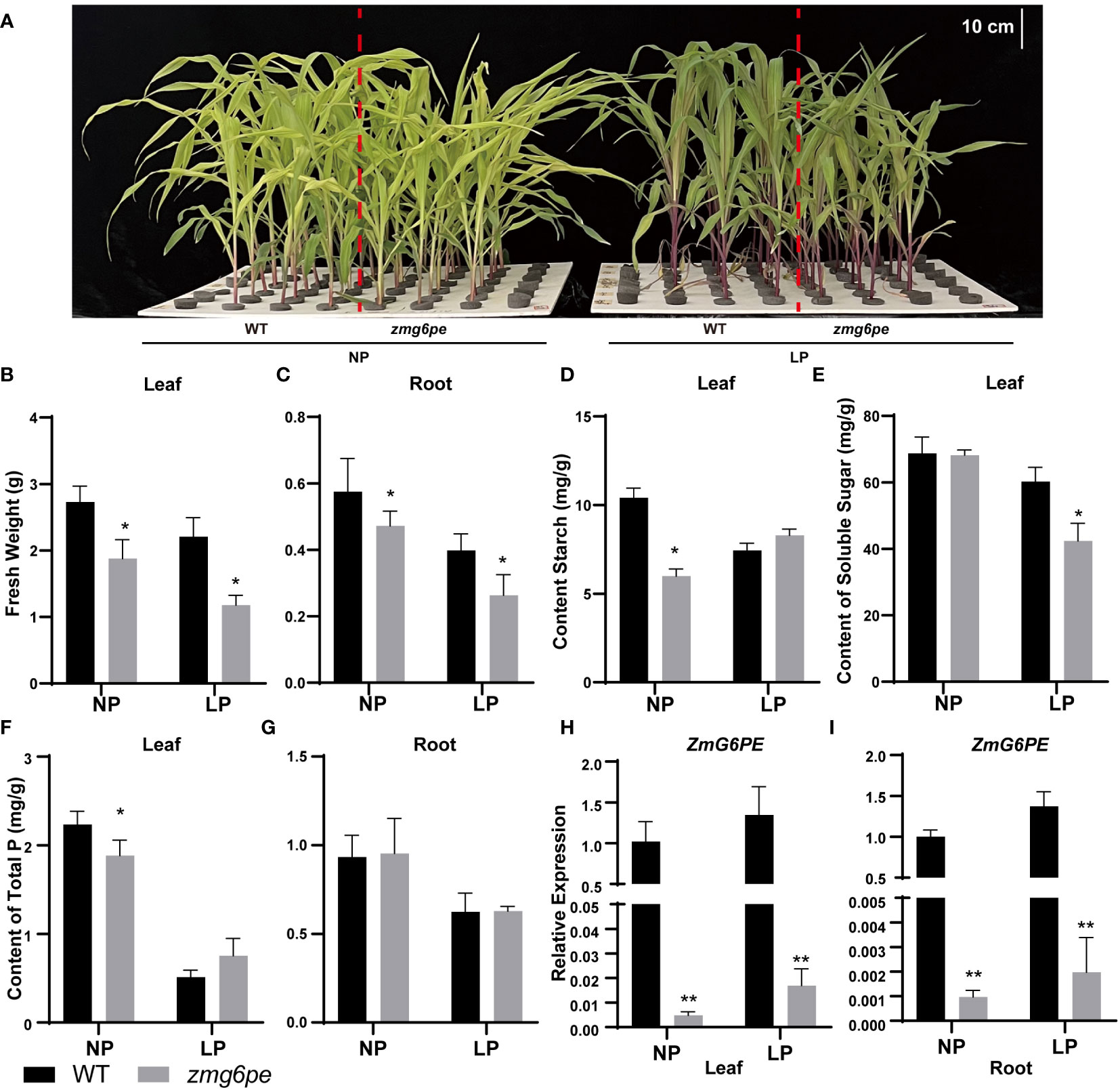
Figure 2 Responses of ZmG6PE in hydroponic culture with different phosphorous concentrations. (A) WT and zmg6pe mutants under NP and LP hydroponics. (B, C) Fresh weight. (D) Starch content. (E) Soluble sugar content. (F, G) Total phosphorus content. (H, I). The expression levels of ZmG6PE gene. “*” represents 0.01< p < 0.05, “**” represents p < 0.01, single factor ANOVA test.
The zmg6pe had reduced fresh weight under LP conditions, indicating that ZmG6PE responds to LP stress. In order to elucidate the regulatory mechanisms of ZmG6PE under LP conditions, comparative transcriptome analysis of leaves and roots was conducted between WT and zmg6pe mutant. There were 2072, 2367, and 504 overlapping differentially expressed genes (DEGs) in leaves (L), roots (R), and both in leaves and the roots (LR), respectively (Supplementary Figure S3A).
KEGG pathway enrichment analysis revealed that terms related to carbon metabolism, amino acid synthesis and metabolism, fatty acid metabolomic pathway, immune regulation, and plant hormones were highly enriched in L (Figure 3A and Supplementary Figure S3B). While terms related to carbon, amino acid and fatty acid metabolisms, amino acid degradation, secondary metabolites, and vitamins were highly enriched in the R (Figure 3B and Supplementary Figure S3C); similarly, the terms related to carbon, amino acid and fatty acid metabolisms, amino acid degradation, immune regulation, and secondary metabolites were enriched in LR (Figure 3C and Supplementary Figure S3D). As such, LP stress activated the immune response system in seedlings, and ZmG6PE regulated carbon, amino acid and fatty acid metabolisms, plant hormones, and secondary metabolites in response to LP stress.
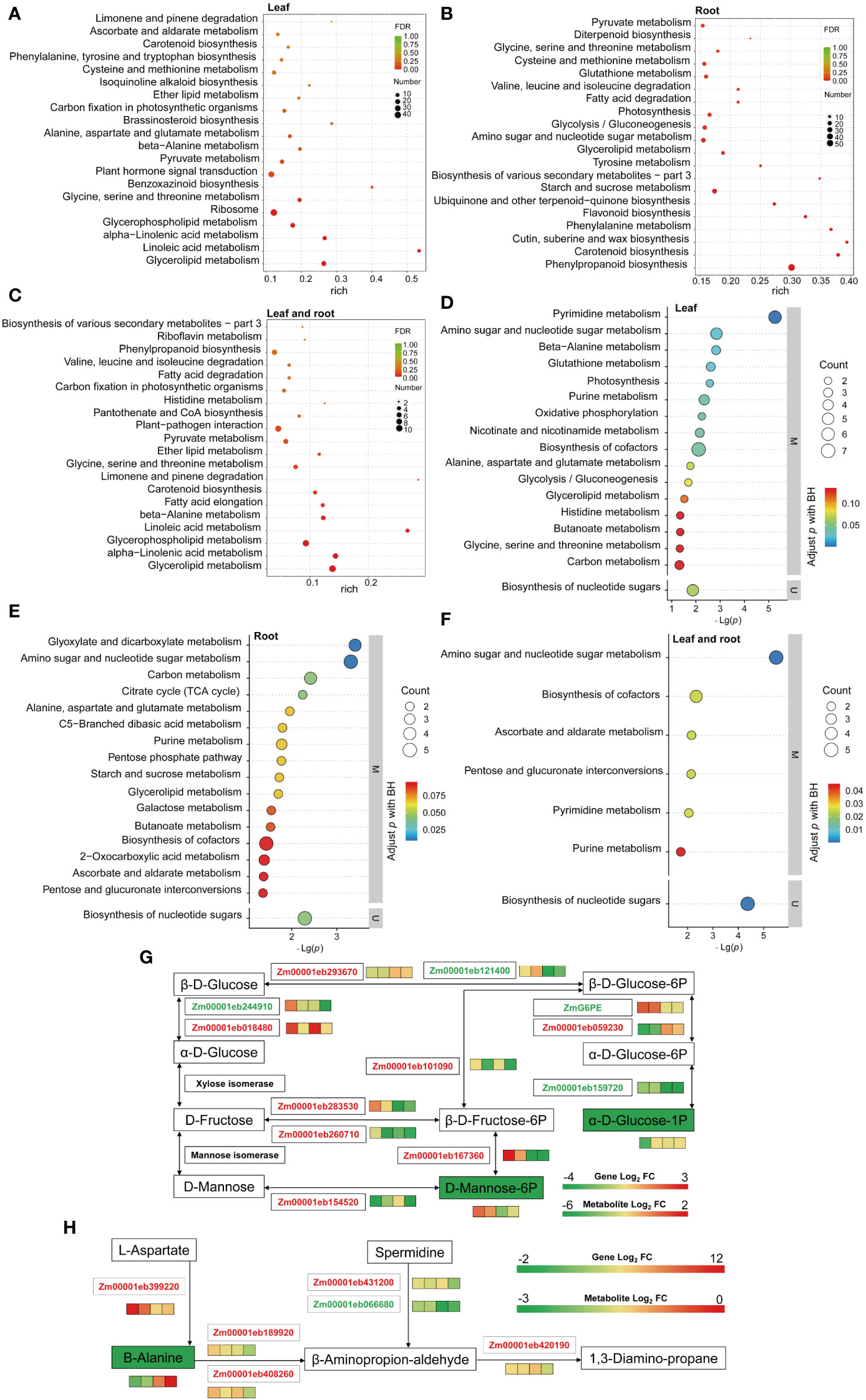
Figure 3 KEGG pathway enrichment analysis performed on the DEGs and DEMs, and channel heat map analysis. (A–C) KEGG enrichment analysis of the overlapping DEGs in leaves, roots, and shared in leaves and roots, respectively. (D–F) KEGG enrichment analysis of the overlapping DEMs in leaves, roots, and shared in leaves and roots, respectively. (G) Glycolysis/gluconeogenesis, fructose, and mannose metabolic pathways involve phosphorus transport in leaves and roots. (H) The β-alanine metabolism involves phosphorus transport in leaves and roots. Boxes are WT-NP-L vs. WT-LP-L, zmg6pe-NP-L vs. zmg6pe-LP-L, WT-NP-R vs. WT-LP-R, and zmg6pe-NP-R vs. zmg6pe-LP-R from left to right. Red and green fonts indicate upregulated and downregulated genes, respectively. Green and red solid squares indicate downregulated and upregulated metabolites, respectively.
Transcriptome KEGG enrichment analysis showed that LP treatment affected carbon and amino acid metabolisms, indicating that LP stress caused variations in the synthesis and metabolism of nitrogen- and carbon-related compounds. To further investigate the function of ZmG6PE, comparative metabolomic analyses were performed in zmg6pe and WT seedlings under LP and NP conditions. There were 164, 96, and 26 overlapping differentially expressed metabolites (DEMs) found in L, R, and LR, respectively (Supplementary Figure S4A). The metabolomic analysis of L and R samples revealed significant KEGG pathway enrichment in carbon metabolism, amino acid metabolism, genetic material synthesis, and immune regulation, with some more enrichments of fatty acid metabolism, enzymatic activity, and energy metabolism in L (Figures 3D-F and Supplementary Figures S4B-D). These results indicate that LP stress may activate the plant immune response system, whereas ZmG6PE affects LP stress by regulating the above-mentioned metabolic pathways.
A combined transcriptome and metabolome analysis was performed to examine whether there is any association between gene expression patterns and metabolite accumulation. Enrichment analysis revealed 42, 15, and 12 KEGG pathways in L, R, and LR, respectively (Supplementary Figure S5A). In the combined analysis of the KEGG pathways, the carbon metabolism, amino acid synthesis and metabolism, glycolysis/gluconeogenesis, secondary metabolite biosynthesis, and carbon fixation in photosynthesis were found enriched in L (Supplementary Figure S5B). In R, galactose metabolism, starch and sucrose metabolism, carotenoid metabolism, photosynthesis, and secondary metabolite biosynthesis were highly enriched (Supplementary Figure S5C). In LR, significant enrichment was observed in carbon, amino acid and fatty acid metabolisms, and immune regulation (Supplementary Figure S5D). These results indicate that LP stress may activate immune regulation and the ZmG6PE gene affects LP stress by regulating primary and secondary metabolism.
The combined network analysis found that α-D-glucose-1 phosphate in the glycolysis/gluconeogenesis pathway and D-mannose-6 phosphate in fructose and mannose metabolism differed significantly (Figure 3G). The pathway map showed significant reductions in α-D-glucose-1 phosphate and D-mannose-6 phosphate. Zm00001eb244910, Zm00001eb121400, Zm00001eb159720, and ZmG6PE gene expression levels decreased, while Zm00001eb293670, Zm00001eb018480, Zm00001eb059230, Zm00001eb101090, Zm00001eb283530, Zm00001eb260710, Zm00001eb154520, and Zm00001eb167360 gene expression levels increased. These results indicate that glucose-1-phosphate- and mannose-6-phosphate-related genes and metabolites respond to LP stress.
The pathway map showed significant reduction in β-alanine (Figure 3H). Zm00001eb066680 gene expression level decreased, while Zm00001eb399220, Zm00001eb431200, Zm00001eb189920, Zm00001eb408260, and Zm00001eb420190 gene expression levels increased. These results indicate that β-alanine genes and metabolites respond to LP stress.
Phosphate Starvation Response (PHR) transcriptional factors are the central regulatory factors regulating the activities of LP-responsive genes in the LP regulatory network (Nilsson et al., 2007; Zhou et al., 2008; Lu et al., 2020). The phosphate transporter (PHT) mediates the absorption and transport of Pi in the plant rhizosphere (Shin et al., 2004; Sun et al., 2012). SYG1/PH081/XPRI (SPX) is a negative regulator of PHR, and regulates phosphorus transport and homeostasis in plants (Liu et al., 2010). To further explore the pathways that regulate the phosphorus signal and expression levels of PSI genes were investigated. Transcriptome analysis of PSI genes revealed that LP treatment promoted the expression of ZmSPXs (ZmSPX1, ZmSPX2, ZmSPX3, ZmSPX4, and ZmSPX6) and ZmPHTs (ZmPHT1.2 and ZmPHT1.13), ut the expression of ZmPHRs (ZmPHR1, ZmPHR2, and ZmPHR3) did not show significant changes (Figures 4A–J). Interestingly, regardless of phosphorus status, the expression levels of ZmSPX2 and ZmPHT1.13 in zmg6pe were significantly lower than WT in both L and R (Figures 4B, J). The RT-qPCR (Figures 4K, L) confirmed the above results, suggesting that ZmG6PE is probably involved in responding to LP stress by mediating expressions of ZmSPX2 and ZmPHT1.13. Under LP conditions, the lack of significant differences in phosphorus content between WT and zmg6pe mutants might be attributed to the enhanced expression of ZmSPX2, which inhibits the expression of ZmPHRs and ZmPHT1.13.
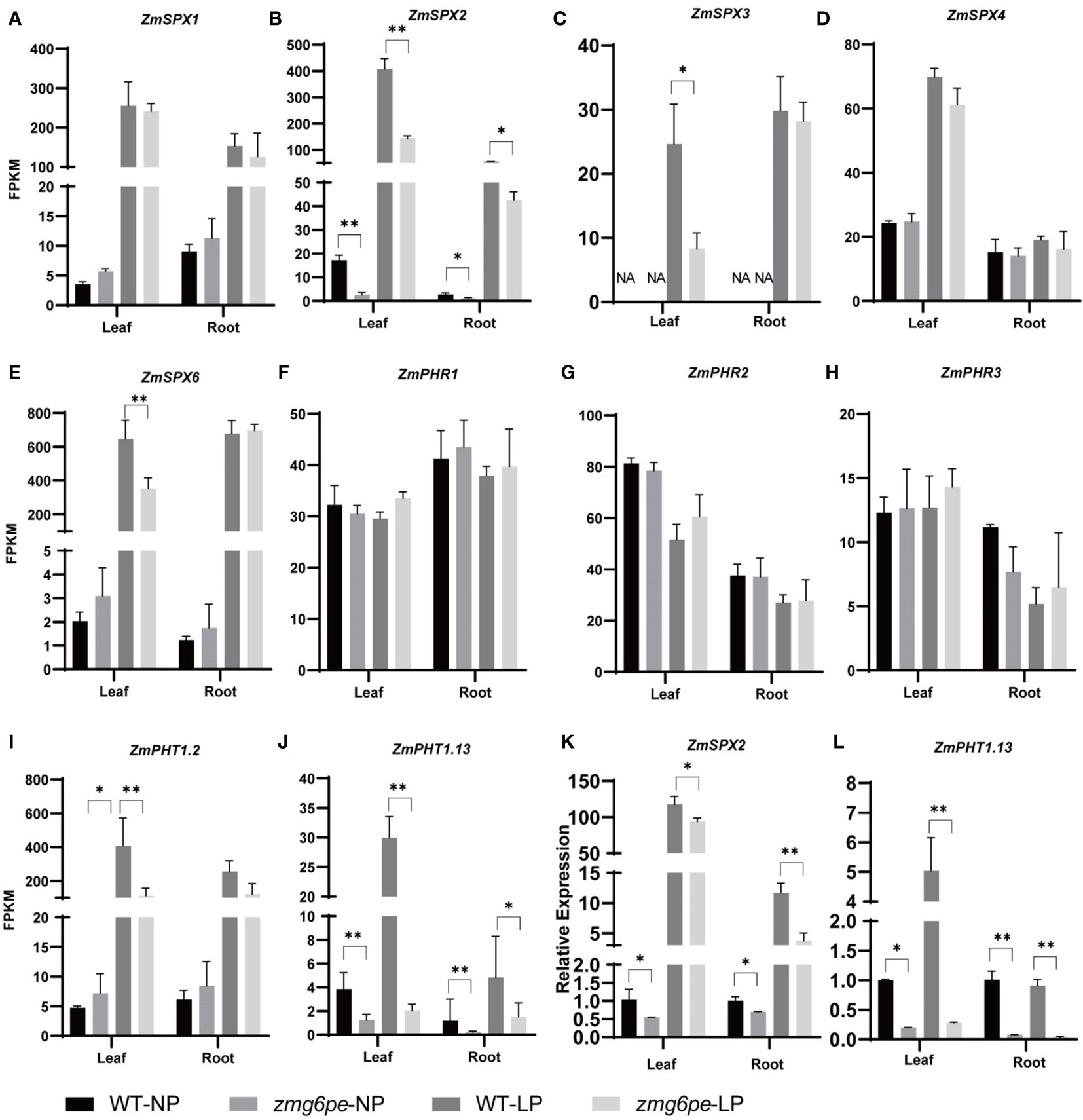
Figure 4 Analysis of PSI genes expression levels. (A–J) Expression levels (FPKM) of ZmSPX1, ZmSPX2, ZmSPX3, ZmSPX4, ZmSPX6, ZmPHR1, ZmPHR2, ZmPHR3, ZmPHT1.2, and ZmPHT1.13, respectively, in leaves and roots under NP and LP conditions. (K, L) Expression levels of ZmSPX2 and ZmPHT1.13 in leaves and roots under NP and LP, respectively, by RT-qPCR. Values are given as the mean ± SD and indicate the average FPKM values from three biological replicates of RNA-seq libraries. “*” represents p < 0.05, and ”**” represents p < 0.01 by single factor ANOVA test.
To assess the effects of ZmG6PE on yield-related traits, we investigated the yield-related data for 2021-Qujing (QJ) and 2022-Chongzhou (CZ) grown materials. Results showed that the ear length, ear weight, 100-grain weight, and number of grains per ear were significantly difference in the zmg6pe compared to WT (Supplementary Table S2). Besides this, the grain size in zmg6pe was significantly smaller (Figure 5A). Moreover, the sugar contents increased, and starch contents decreased in the zmg6pe compared to the WT (Figures 5B, C). The phosphate content in the zmg6pe was significantly lower than in WT (Figure 5D). The observation of paraffin sections revealed that grain filling in WT was normal at 7 days after pollination (DAP). In contrast, zmg6pe showed abnormal grain filling (Figure 5E). At 15 DAP, the starch particles in the WT seeds were fully packed, while zmg6pe had irregular cavities. The red box area of the zmg6pe had a significantly larger cavity than WT (Figure 5E). The ZmG6PE gene expression levels significantly decreased at 7 and 15 DAP in the zmg6pe (Figure 5F). The above results demonstrate that the ZmG6PE gene affects maize yield.
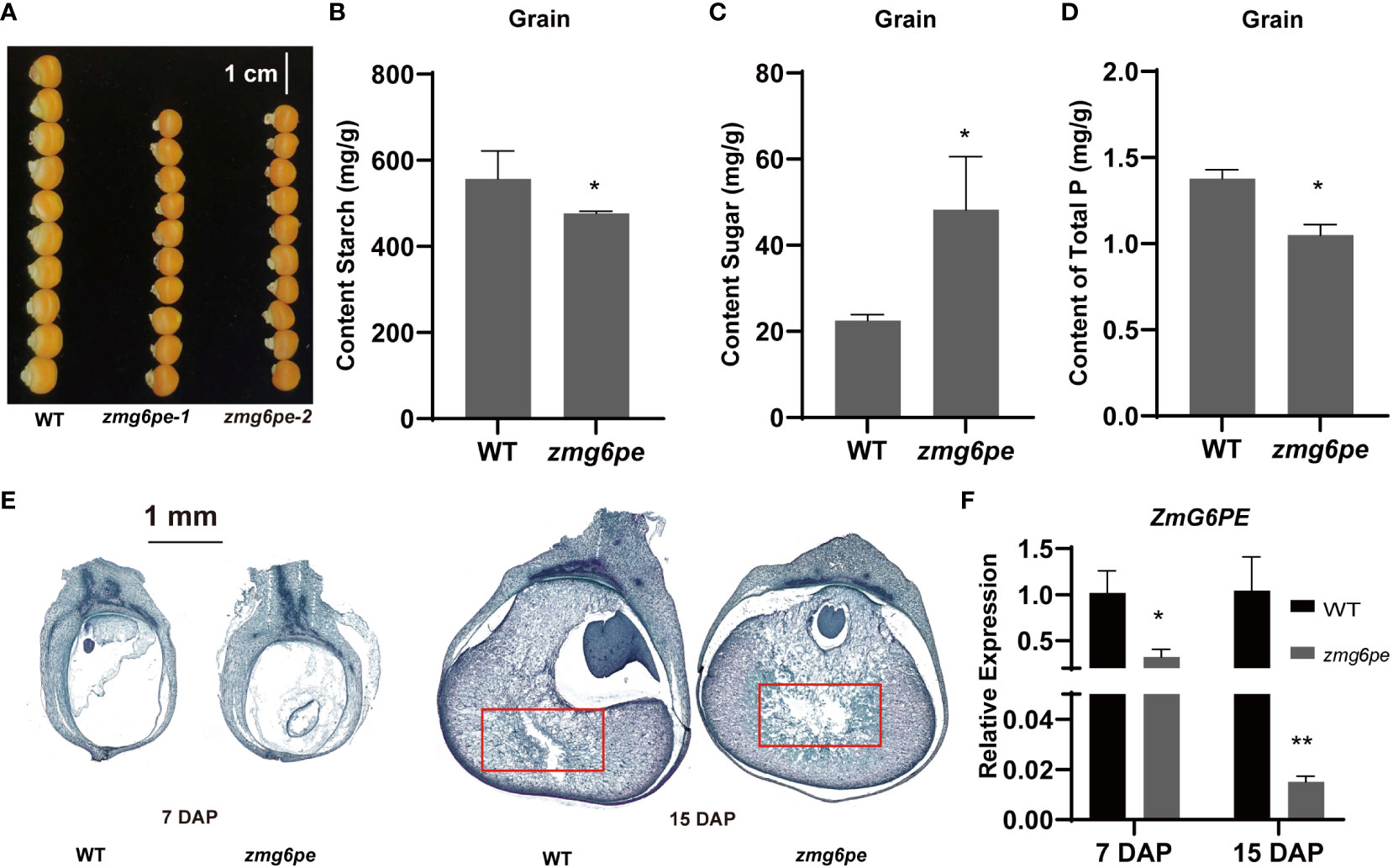
Figure 5 Analysis of maize kernel development. (A) Grain size. (B–D) Content of the grain’s starch, sugar, and total phosphorus, respectively. (E) Paraffin section of grains at 7 and 15 DAP. (F) The expression levels of ZmG6PE in grain were measured at 7 and 15 DAP. “*” represents p < 0.05, “**” represents p < 0.01 by single factor ANOVA test.
To investigate the effects of ZmG6PE on yield under LP conditions, the yield-related traits of RILs derived from inbred lines 178 and 9782 were measured. The amplification of the ZmG6PE gene coding sequences in the inbred lines 178 and 9782 revealed no differences (Supplementary Figure S6A). However, an insertion or deletion (Indel) was detected in the promoter sequence of the ZmG6PE gene in two inbred lines, with a 23 bp deletion in inbred line 178 compared to inbred line 9782 (Supplementary Figure S6B), which could be classified into two haplotypes (178 genotypes and 9782 genotypes) (Supplementary Figure S6C). Genotype statistics were performed on the RILs, and the yield-related traits were researched from the Wenjiang Farm in 2013 and the Bifengxia Farm in 2014 under LP conditions. Row number per panicle, number of grains per row, ear length, ear thickness, shaft ear thickness and ear weight of genotype 178 were significantly lower than those of genotype 9782 (Figures 6A–F). In inbred line 9782, the expression of the ZmG6PE gene varied with changes in the phosphorus concentration, while in inbred line 178, gene expression was unaffected by the phosphorus concentration (Figure 6G). These results indicate that ZmG6PE plays a role in maize yield and phosphorus content.
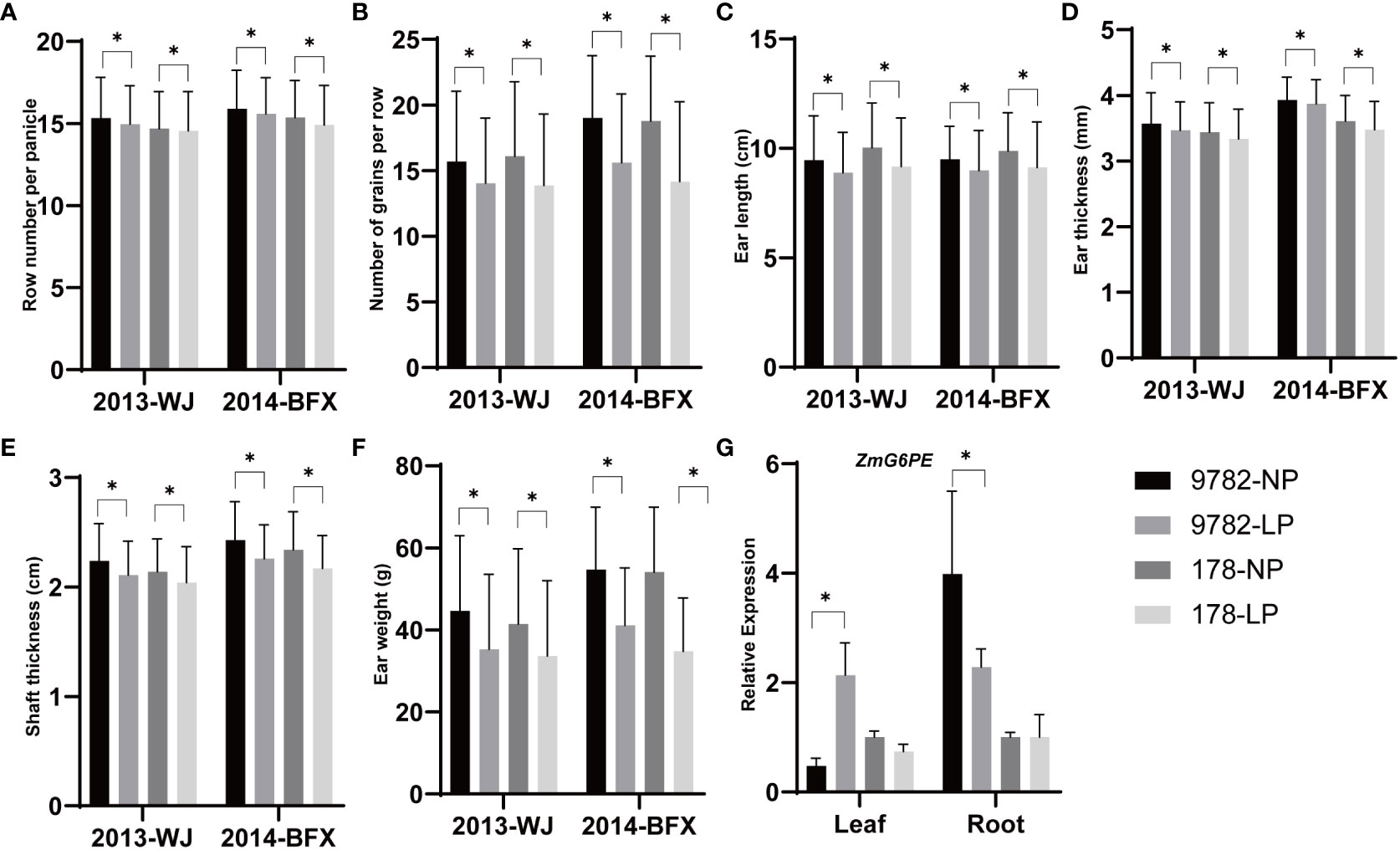
Figure 6 Yield-related analysis of the RILs. (A–F) Significantly different traits between segregating genotypes of ZmG6PE. WJ, Wenjiang Farm; BFX, Bifengxia Farm. (G) The gene expression level of ZmG6PE. “*” represents p < 0.05 by single factor ANOVA test.
Anomeric carbon is essential in glycan reactions and quickly provides metabolites for physiological processes (Gomes et al., 2015). Glucose-6-phosphate has two forms of α and β anomers (Wurster and Hess, 1972; Wurster and Hess, 1973; Graille et al., 2006). The isomerization of α-D-glucose-6-phosphate by glucose-6-phosphate isomerase produces fructose 6-phosphate (Aguilar-Pontes et al., 2018). Phosphoglucomutase catalyzes the reversible isomerization of α-D-glucose-6-phosphate to glucose-1-phosphate, which is converted into ADP-glucose (precursor of starch synthesis) by ADP-glucose-pyrophosphorylase (Geigenberger et al., 2004; Zeeman et al., 2007). Combined analysis reveals significant differences in glucose-1-phosphate levels, and starch content reduced in grains, suggesting that ZmG6PE may affect starch synthesis through the modulation of α-D-glucose-6-phosphate. β-D-glucose-6-phosphate participates in pentose phosphate metabolism to provide NADPH for life activities (Efferth et al., 2006; Mamani et al., 2016). Glycolysis and the pentose phosphate pathway occur in the cytoplasm, while the ZmG6PE protein is subcellularly localized in the nucleus and cell membrane. Omics analysis revealed significant enrichment of glycolysis/gluconeogenesis and pentose phosphate pathway-related processes. This suggests that ZmG6PE may indirectly participate in glycolysis and the pentose phosphate pathway by regulating the balance of glucose-6-phosphate. In addition, fructose-6-phosphate and mannose-6-phosphate may be potential substrates of glucose-6-phosphate-1 epimerase, indicating its broad substrate specificity (Graille et al., 2006). Significant differences were also observed in combined analysis for mannose-6-phosphate, indicating that mannose-6-phosphate may serve as a catalytic substrate for ZmG6PE, but further validation is required.
Sugars are primary metabolites and signaling molecules, and starch is the final product of photosynthesis and the dominant form of energy storage in plants. Phosphorus deficiency makes soluble sugar accumulation in leaves, while exogenously applied sugars can alleviate the phosphorus starvation response (Hermans et al., 2006; Yang et al., 2020). Similarly, LP treatment increases the starch content (Gibson, 2004; Morcuende et al., 2007). The results showed a cascade relationship between carbon metabolism and phosphorus signaling. However, the specific interaction between carbon metabolism and phosphorus remains unclear. ADP-glucosepyrophosphorylase (AGP) Large Subunit 1 (AGPL1) and AGP Small Subunit 1 (AGPS1) increases the starch and soluble sugar contents in leaves under LP, but the starch and sugar contents do not change significantly in the roots (Meng et al., 2020). The zmg6pe mutants had reduced soluble sugar content and increased starch content in the leaves under LP conditions but also had reduced starch content and increased sugar content in the grain. Sugar and starch are the main carbon sources in plants and keep a dynamic balance (Flütsch et al., 2020). Non-metabolizable sugar analogs do not affect the expression of PSI genes. Although sugar input appears to be downstream of initial Pi sensing, it is required to complete the PSI signaling pathway (Karthikeyan et al., 2007). The cellulose synthase-like family (OsCSLF6) gene mutation increases the sugar and phosphorus content, and the expression of sucrose synthase and sucrose transporter genes is also induced in rice (Jin et al., 2015). The overexpression of the sucrose transport (AtSUC2) gene promotes sucrose transport and accumulation and enhances PSI gene expression and Pi accumulation (Lei et al., 2011). Sucrose and auxin play differential roles in the developmental responses of ontogenetically distinct root traits during Pi deprivation (Jain et al., 2007). Therefore, it is crucial to investigate the potential mechanism of PSI gene expression changes mediated by glycometabolism-related genes.
KEGG enrichment analyses of the transcriptomic and metabolomic data showed that amino acids, starch and sugar synthesis, photosynthesis, plant pathogens, ABC transporters, brassinosteroids, plant hormones, and citric acid metabolism responded to LP stress. LP stress significantly increases amino acids in plants (Hernández et al., 2009; Tawaraya et al., 2014; Sung et al., 2015). Plants increase starch content and reduce photosynthesis to avoid anthocyanins accumulation in leaves in LP environments (Wissuwa et al., 2005; Plaxton and Tran, 2011; Pant et al., 2015). The current study analysis showed that LP affected carbon fixation in leaves while starch and sugar synthesis in roots. Phosphorus starvation is also associated with microbial interactions. Under LP conditions, plants establish a mutualistic symbiotic relationship with microorganisms to increase phosphorus acquisition (Smith and Smith, 2011; Oldroyd and Leyser, 2020). These microorganisms may be arbuscular mycorrhizal fungi or species of Colletotrichum. ABC transcription proteins are involved in both aluminum tolerance and the inhibition of local phosphate signaling pathways (Dong et al., 2017). Brassinosteroids and phytohormones respond to LP stress due to the regulation of local phosphorus signaling pathways by hormones interacting with different signals (Kobayashi et al., 2013). Overexpression of citrate synthase and transporter can improve the phosphorus utilization efficiency (PUE) by increasing organic acids secretion (López-Arredondo et al., 2014; Panchal et al., 2021). These pathways have all been reported in plant responses to LP stress. This evidence shows that the design of this research is reasonable and that these pathways are important in response to LP stress.
Phosphate Starvation Response (PHR) transcriptional factors are characterized by a conserved MYB domain and can participate in the transcriptional activation of PSI genes (Zhou et al., 2008; Guo et al., 2015). Phosphorus transport (PHT) regulates the uptake and transport of phosphorus (Paszkowski et al., 2002). SPX1 and SPX2 interact with PHR through their SPX domains, thereby inhibiting the binding ability of PHR to P1BS (Liu et al., 2010; Wang et al., 2014). Under LP stress, the expression of ZmSPXs increased, indicating that ZmSPXs inhibited ZmPHR and ZmPHT activities and affected the absorption and transport of phosphorus. Compared to the WT, the phosphorous content in zmg6pe mutant leaves decreased under NP conditions but remained significantly unchanged in zmg6pe mutant under LP stress conditions. Combined with the expression analysis, the expression of ZmSPX2 decreased in the zmg6pe mutant under LP stress, indicating that ZmG6PE had a stimulative effect on ZmSPX2. The regulatory effect of ZmG6PE on ZmSPX2 may be achieved by influencing carbohydrate homeostasis or by modulating the expression of relevant genes in the nucleus. ZmG6PE might play a potential role in coordinating cellular metabolism and gene expression. The mutation of AGPL1 and AGPS1 lead to the significant downregulation of OsSPX2 (Meng et al., 2020). AGPL1/AGPS1 is a starch synthesis enzyme, and ZmG6PE is an enzyme of the glycolysis pathway, suggesting that the carbon balance may affect SPX activity.
Based on the above findings, we put forward a working model involving ZmG6PE in phosphorus homeostasis maintenance in maize (Figure 7). ZmG6PE is attributed with the capability to modulate the dynamic equilibrium between two carbohydrate sugar and starch. The expression of ZmSPX2 is subjected to regulation by sugar concentration, while it was found to have no discernible effect on ZmPHRs expression. Notably, despite a prominent decrease in ZmPHT1.13 expression level observed in the zmg6pe mutant compared to WT, the unaltered expression of ZmPHRs appears to compensate for the deficit by modulating the expression of other ZmPHTs. This may account for the absence of significant variations in phosphorus levels under LP conditions between the zmg6pe mutant and WT. ZmG6PE is implicated in phosphate uptake through the regulation of ZmSPX2 and ZmPHT1.13 expression. Additionally, ZmG6PE exerts an influence on starch synthesis by modulating the α-D-glucose-6-phosphate content, consequently impacting grain filling and altering grain size.
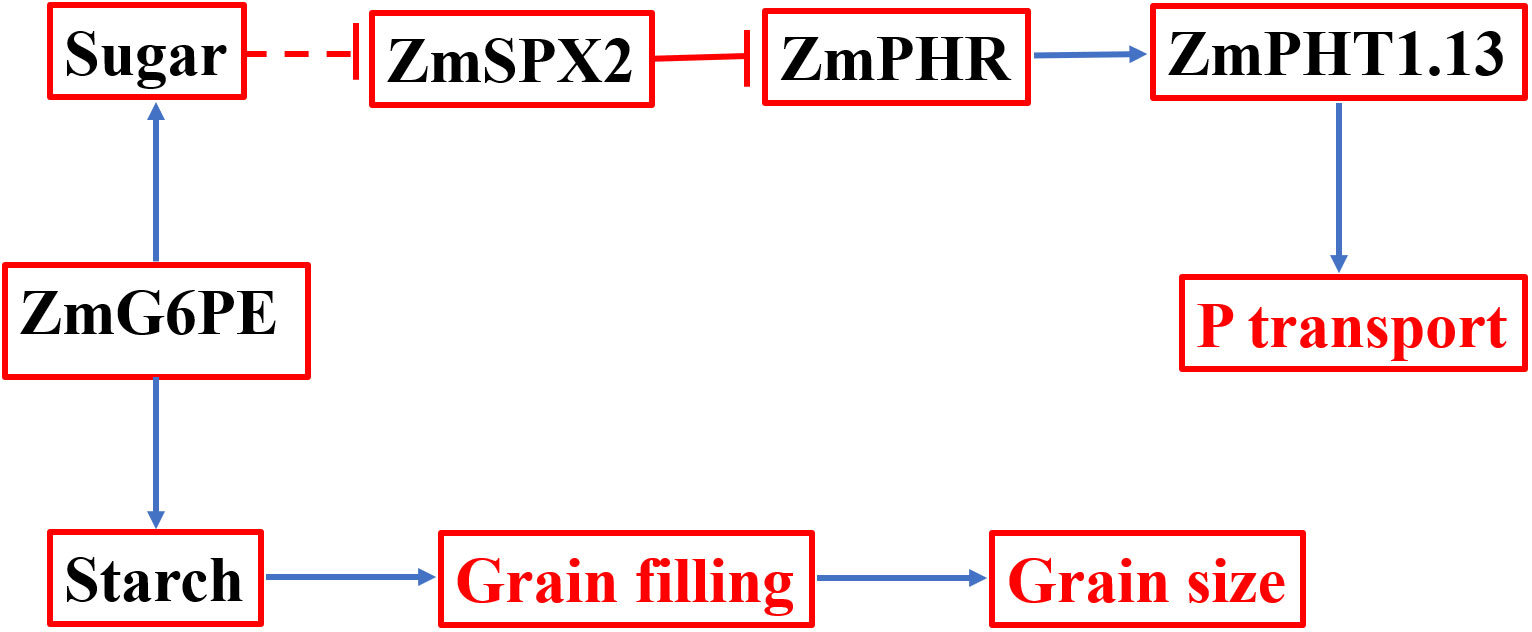
Figure 7 If working model for regulating phosphorus homeostasis by ZmG6PE. Solid lines indicate established direct correlations between different genes/proteins and between ZmG6PE, sugar, and starch. The dotted lines indicate indirect correlations between endogenous sugar level and SPX transcript abundance.
The conserved domain of ZmG6PE protein ranges from amino acid 41 to 308, which is identified as the Aldose_epimase domain. It exhibits activity as a glucose-6-phosphate-1-epimerase and may possess substrate diversity. Omics analysis reveals that the ZmG6PE gene is involved in LP stress response through pathways, such as amino acid metabolism, carbon metabolism, genetic material biosynthesis, fatty acid metabolism, and immune regulation. Glucose-1-phosphate, mannose-6-phosphate, and β-alanine are identified as marker metabolites in response to LP stress. The ZmG6PE gene regulates the expression of ZmSPX2 and ZmPHT1.13 by modulating the dynamic balance of carbohydrates, thus participating in phosphate signaling regulation. Mutant studies have demonstrated the impact of ZmG6PE on starch synthesis, grain size, and yield-related traits, while the use of RILs has revealed its influence on yield-related traits and phosphorus content. The ZmG6PE gene is a pleiotropic gene that influences maize grain size and responds to LP stress.
Transcriptomic data has been submitted to the China National Centre for Bioinformatics, Chinese Academy of Sciences, under the accession number CRA010809, and is publicly accessible at https://ngdc.cncb.ac.cn/gsa.
The manuscript presents research on animals that do not require ethical approval for their study.
HKZ: Writing – original draft, Writing – review & editing, Data curation, Validation. BWL: Writing – review & editing, Supervision. JL: Investigation, Writing – review & editing. XJ: Investigation, Writing – review & editing. HYZ: Data curation, Writing – review & editing. HXZ: Data curation, Writing – review & editing. BYL: Data curation, Writing – review & editing. HH: Data curation, Writing – review & editing. YW: Data curation, Writing – review & editing. AA: Formal Analysis, Writing – review & editing. AR: Formal Analysis, Writing – review & editing. JS: Formal Analysis, Writing – review & editing. MI: Formal Analysis, Writing – review & editing. XZ: Software, Writing – review & editing. DL: Supervision, Writing – review & editing. LW: Supervision, Writing – review & editing. DG: Resources, Writing – review & editing. SQG: Resources, Writing – review & editing. SS: Supervision, Writing – review & editing. SBG: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Key Research and Development Program of China (grant no. 2021YFF1000500 and 2021YFD1200704), the Natural Science Foundation of China (grant no. 32101655), Sichuan Science and Technology Support Project (grant no. 2021YFYZ0027, 2021YFYZ0020) and the earmarked fund for China Agriculture Research System (grant no. CARS-02-09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1286699/full#supplementary-material
Aguilar-Pontes, M. V., Brandl, J., McDonnell, E., Strasser, K., Nguyen, T. T. M., Riley, R., et al. (2018). The gold-standard genome of Aspergillus Niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi. Stud. Mycology 91, 61–78. doi: 10.1016/j.simyco.2018.10.001
Chiou, T., Lin, S. (2011). Signaling network in sensing phosphate availability in plants. Annu. Eview Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Dong, J., Piñeros, M., Li, X., Yang, H., Liu, Y., Murphy, A.S., et al. (2017). An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 10, 244–259. doi: 10.1016/j.molp.2016.11.001
Efferth, T., Schwarzl, S. M., Smith, J., Osieka, R. (2006). Role of glucose-6-phosphate dehydrogenase for oxidative stress and apoptosis. Cell Death Differentiation 13, 527–528. doi: 10.1038/sj.cdd.4401807
Flütsch, S., Wang, Y., Takemiya, A., Vialet-Chabrand, S. R. M., Klejchová, M., Nigro, A., et al. (2020). Guard cell starch degradation yields glucose for rapid stomatal opening in Arabidopsis. Plant Cell 32, 2325–2344. doi: 10.1105/tpc.18.00802
Fouquerel, E., Goellner, E. M., Yu, Z., Gagne, J. P., Barbi, D. M. M., Feinstein, T., et al. (2014). ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 8, 1819–1831. doi: 10.1016/j.celrep.2014.08.036
Geigenberger, P., Stitt, M., Fernie, A. R. (2004). Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ. 27, 655–673. doi: 10.1111/j.1365-3040.2004.01183.x
Gibson, S. I. (2004). Sugar and phytohormone response pathways: navigating a signalling network. J. Exp. Bot. 55, 253–264. doi: 10.1093/jxb/erh048
Gomes, G., Vil’, V., Terent’Ev, A., Alabugin, I. V. (2015). Stereoelectronic source of the anomalous stability of bis-peroxides. Chem. Sci. 6, 6783–6791. doi: 10.1039/c5sc02402a
Gonzalez, E., Solano, R., Rubio, V., Leyva, A., Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR 1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512. doi: 10.1105/tpc.105.036640
Graille, M., Baltaze, J. P., Leulliot, N., Liger, D., Quevillon-Cheruel, S., van Tilbeurgh, H. (2006). Structure-based functional annotation: yeast ymr099c codes for a d-hexose-6-phosphate mutarotase. J. Biol. Chem. 281, 30175–30185. doi: 10.1074/jbc.M604443200
Guo, M., Ruan, W., Li, C., Huang, F., Zeng, M., Liu, Y., et al. (2015). Integrative comparison of the role of the phosphate response1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol. 168, 1762–1776. doi: 10.1104/pp.15.00736
Ham, B. K., Chen, J., Yan, Y., Lucas, W. J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. doi: 10.1016/j.copbio.2017.07.005
He, Q., Ren, W., Xiang, J., Dabbour, M., Mintah, B. K., Li, Y., et al. (2021). Fermentation of Saccharomyces cerevisiae in a 7.5 L ultrasound-enhanced fermenter: Effect of sonication conditions on ethanol production, intracellular Ca2+ concentration and key regulating enzyme activity in glycolysis. Ultrason Sonochemistry 76, 105624. doi: 10.1016/j.ultsonch.2021.105624
Hermans, C., Hammond, J. P., White, P. J., Verbruggen, N. (2006). How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 11, 610–617. doi: 10.1016/j.tplants.2006.10.007
Hernández, G., Valdés-López, O., Ramírez, M., Goffard, N., Weiller, G., Aparicio-Fabre, R., et al. (2009). Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151, 1221–1238. doi: 10.1104/pp.109.143842
Jain, A., Poling, M. D., Karthikeyan, A. S., Blakeslee, J. J., Peer, W. A., Titapiwatanakun, B., et al. (2007). Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 144, 232–247. doi: 10.1104/pp.106.092130
Jiang, H., Zhang, J., Han, Z., Yang, J., Ge, C., Wu, Q. (2017). Revealing new insights into different phosphorus-starving responses between two maize (Zea mays) inbred lines by transcriptomic and proteomic studies. Sci. Rep. 7, 44294. doi: 10.1038/srep44294
Jiang, S., Zhang, L. F., Zhang, H. W., Hu, S., Lu, M. H., Liang, S., et al. (2012). A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 31, 1985–1998. doi: 10.1038/emboj.2012.45
Jin, C., Fang, C., Yuan, H., Wang, S., Wu, Y., Liu, X., et al. (2015). Interaction between carbon metabolism and phosphate accumulation is revealed by a mutation of a cellulose synthase-like protein, CSLF6. J. Exp. Bot. 66, 2557–2567. doi: 10.1093/jxb/erv050
Karthikeyan, A., Varadarajan, D., Jain, A., Held, M., Carpita, N., Raghothama, K.G., et al. (2007). Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. doi: 10.1007/s00425-006-0408-8
Kobayashi, Y., Kobayashi, Y., Sugimoto, M., Lakshmanan, V., Iuchi, S., Kobayashi, M., et al. (2013). Characterization of the complex regulation of atalmt1 expression in response to phytohormones and other inducers. Plant Physiol. 162, 732–740. doi: 10.1104/pp.113.218065
Kochian, L. V. (2012). Plant nutrition: rooting for more phosphorus. Nature 488, 466–467. doi: 10.1038/488466a
Lei, M., Liu, Y., Zhang, B., Zhao, Y., Wang, X., Zhou, Y., et al. (2011). Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 156, 1116–1130. doi: 10.1104/pp.110.171736
Li, H., Xu, L., Li, J., Lyu, X., Li, S., Wang, C., et al. (2022). Multi-omics analysis of the regulatory effects of low-phosphorus stress on phosphorus transport in soybean roots. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.992036
Liu, J., Samac, D. A., Bucciarelli, B., Allan, D. L., Vance, C. P. (2005). Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J. 41, 257–268. doi: 10.1111/j.1365-313X.2004.02289.x
Liu, F., Wang, Z., Ren, H., Shen, C., Li, Y., Ling, H.Q., et al. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. doi: 10.1111/j.1365-313X.2010.04170.x
López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., López-Bucio, J., Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Lu, M., Cheng, Z., Zhang, X., Huang, P., Fan, C., Yu, G., et al. (2020). Spatial divergence of phr-pht1 modules maintains phosphorus homeostasis in soybean nodules. Plant Physiol. 184, 236–250. doi: 10.1104/pp.19.01209
Luo, B., Ma, P., Nie, Z., Zhang, X., He, X., Ding, X., et al. (2019). Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 97, 947–969. doi: 10.1111/tpj.14160
Ma, M., Lazar, N., Pellegrini, O., Lepault, J., Condon, C., et al. (2017). Trz1, the long form rnase Z from yeast, forms a stable heterohexamer with endonuclease Nuc1 and mutarotase. Biochem. J. 474, 3599–3613. doi: 10.1042/BCJ20170435
Mamani, S., Moinier, D., Denis, Y., Soulère, L., Queneau, Y., Talla, E., et al. (2016). Insights into the quorum sensing regulon of the acidophilic Acidithiobacillus ferrooxidans revealed by transcriptomic in the presence of an acyl homoserine lactone superagonist analog. Front. Microbiol. 71365. doi: 10.3389/fmicb.2016.01365
Meng, Q., Zhang, W., Hu, X., Shi, X., Chen, L., Dai, X., et al. (2020). Two ADP-glucose pyrophosphorylase subunits, OsAGPL1 and OsAGPS1, modulate phosphorus homeostasis in rice. Plant J. 104, 1269–1284. doi: 10.1111/tpj.14998
Morcuende, R., Bari, R., Gibon, Y., Zheng, W., Pant, B. D., Bläsing, O., et al. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30, 85–112. doi: 10.1111/j.1365-3040.2006.01608.x
Nilsson, L., Muller, R., Nielsen, T. H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30, 1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x
Niu, L., Hao, R., Wu, X., Wang, W., Tuberosa, R., et al. (2020). Maize mesocotyl: role in response to stress and deep-sowing tolerance. Plant Breed. 139, 466–473. doi: 10.1111/pbr.12804
Oldroyd, G., Leyser, O. (2020). A plant’s diet, surviving in a variable nutrient environment. Science 368, eaba0196. doi: 10.1126/science.aba0196
Panchal, P., Miller, A. J., Giri, J., Manavella, P. (2021). Organic acids: versatile stress-response roles in plants. J. Exp. Bot. 72, 4038–4052. doi: 10.1093/jxb/erab019
Pant, B. D., Pant, P., Alexander, E., Huhman, D., Kopka, J., Scheible, W.R.. (2015). Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor phr1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 38, 172–187. doi: 10.1111/pce.12378
Paszkowski, U., Kroken, S., Roux, C., Briggs, S. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. 99, 13324–13329. doi: 10.1073/pnas.202474599
Plaxton, W. C., Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). Spx1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Rubio, V., Linhares, F., Solano, R., Martin, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Schuldiner, O., Yanover, C., Benvenisty, N. (1998). Computer analysis of the entire budding yeast genome for putative targets of the GCN4 transcription factor. Curr. Genet. 33, 16–20. doi: 10.1007/s002940050303
Shin, H., Shin, H., Dewbre, G., Harrison, M. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Smith, S. E., Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X., Ai, P., et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159, 1571–1581. doi: 10.1104/pp.112.196345
Sung, J., Lee, S., Lee, Y., Ha, S., Song, B., Kim, T., et al. (2015). Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 241, 55–64. doi: 10.1016/j.plantsci.2015.09.027
Tanner, L. B., Goglia, A. G., Wei, M. H., Sehgal, T., Parsons, L. R., Park, J.O., et al. (2018). Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49–62. doi: 10.1016/j.cels.2018.06.003
Tawaraya, K., Horie, R., Shinano, T., Wagatsuma, T., Saito, K., Oikawa, A. (2014). Metabolite profiling of soybean root exudates under phosphorus deficiency. Soil Sci. Plant Nutr. 60, 679–694. doi: 10.1080/00380768.2014.945390
Wang, F., Deng, M., Xu, J., Zhu, X., Mao, C. (2018). Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 74, 114–122. doi: 10.1016/j.semcdb.2017.06.013
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wild, R., Gerasimaite, R., Jung, J. Y., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Wissuwa, M., Gamat, G., Ismail, A. M. (2005). Is root growth under phosphorus deficiency affected by source or sink limitations? J. Exp. Bot. 56, 1943–1950. doi: 10.1093/jxb/eri189
Wu, H., Liu, J., Chen, S., Zhao, Y., Zeng, S., Bin, P., et al. (2018). Jejunal metabolic responses to Escherichia coli infection in piglets. Front. Microbiol. 92465. doi: 10.3389/fmicb.2018.02465
Wurster, B., Hess, B. (1972). Glucose-6-phosphate-1-epimerase from baker’s yeast. A new enzyme. FEBS Lett. 23, 341–344. doi: 10.1016/0014-5793(72)80311-9
Wurster, B., Hess, B. (1973). Enzyme-catalyzed anomerization of D-glucose-6-phosphate. FEBS Lett. 38, 33–36. doi: 10.1016/0014-5793(73)80506-x
Yang, A., Kong, L., Wang, H., Yao, X., Xie, F., Wang, H., et al. (2020). Response of soybean root to phosphorus deficiency under sucrose feeding: insight from morphological and metabolome characterizations. BioMed. Res. Int. 2020, 2148011–2148032. doi: 10.1155/2020/2148032
Zeeman, S. C., Smith, S. M., Smith, A. M. (2007). The diurnal metabolism of leaf starch. Biochem. J. 401, 13–28. doi: 10.1042/BJ20061393
Keywords: maize, low-phosphorus stress, ZmG6PE, transcriptomics, metabonomics
Citation: Zhang H, Luo B, Liu J, Jin X, Zhang H, Zhong H, Li B, Hu H, Wang Y, Ali A, Riaz A, Sahito JH, Iqbal MZ, Zhang X, Liu D, Wu L, Gao D, Gao S, Su S and Gao S (2023) Functional analysis of ZmG6PE reveals its role in responses to low-phosphorus stress and regulation of grain yield in maize. Front. Plant Sci. 14:1286699. doi: 10.3389/fpls.2023.1286699
Received: 31 August 2023; Accepted: 19 October 2023;
Published: 09 November 2023.
Edited by:
Poonam Yadav, Banaras Hindu University, IndiaReviewed by:
Yiyong Zhu, Nanjing Agricultural University, ChinaCopyright © 2023 Zhang, Luo, Liu, Jin, Zhang, Zhong, Li, Hu, Wang, Ali, Riaz, Sahito, Iqbal, Zhang, Liu, Wu, Gao, Gao, Su and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shibin Gao, c2hpYmluZ2FvQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.