
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 08 December 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1286547
This article is part of the Research Topic Plant Biology: Told by Petunia - Model Organisms in Plant Science View all 7 articles
Salinity is a current and growing problem, affecting crops worldwide by reducing yields and product quality. Plants have different mechanisms to adapt to salinity; some crops are highly studied, and their salinity tolerance mechanisms are widely known. However, there are other crops with commercial importance that still need characterization of their molecular mechanisms. Usually, transcription factors are in charge of the regulation of complex processes such as the response to salinity. MYB-TFs are a family of transcription factors that regulate various processes in plant development, and both central and specialized metabolism. MYB-TFs have been studied extensively as mediators of specialized metabolism, and some are master regulators. The influence of MYB-TFs on highly orchestrated mechanisms, such as salinity tolerance, is an attractive research target. The versatility of petunia as a model species has allowed for advances to be made in multiple fields: metabolomic pathways, quality traits, stress resistance, and signal transduction. It has the potential to be the link between horticultural crops and lab models, making it useful in translating discoveries related to the MYB-TF pathways into other crops. We present a phylogenetic tree made with Petunia axillaris and Petunia inflata R2R3-MYB subfamily sequences, which could be used to find functional conservation between different species. This work could set the foundations to improve salinity resistance in other commercial crops in later studies.
Salinity is among the most detrimental factors affecting plant growth and crop yield. Currently, 20% of irrigated crop land is affected by salinity, and due to climate change this problem is expected to worsen (Li et al., 2023). Salt stress starts with osmotic stress, a rapid response that results in reduced water uptake by the roots, and is followed by an ionic stress, which occurs over a period of days to weeks (Munns and Tester, 2008). Osmotic stress reduces water uptake through the roots, with the first and most prominent effect being stomatal closure, which subsequently reduces the rates of CO2 assimilation and transpiration (Munns and Tester, 2008). Another consequence of osmotic stress is a reduction in the rates of cell elongation, hampering cell division and slowing down growth rates (Munns et al., 1995). Ionic stress is caused by a prolonged exposure to salinity, wherein sodium (Na+) and chloride (Cl-) accumulate in plant cells and cause, among other effects, an increased rate of leaf senescence (Rodríguez Coca et al., 2023). Due to leaf senescence and reduced leaf area expansion (caused by osmotic stress), plants may have a reduction in whole-plant light interception, and additionally leaves may show reduced photosynthetic capacity; both consequences directly impact biomass (Munns et al., 1995; Zörb et al., 2019). In most cases, yield is reduced when crops are grown under high salinity conditions (Zörb et al., 2019). In the case of fruits and vegetables, this could result in smaller final products or products that do not meet consumer quality standards (Saied et al., 2005). However, examples do exist where quality related metabolites are elevated under salinity stress, often leading to higher customer appreciation (Rouphael et al., 2018; Moles et al., 2019). Breeding for high productivity under salinity requires an understanding of different mechanisms that plants have evolved in their response to salinity.
Salt tolerance in plants involves intricate molecular mechanisms that aim to keep plants functioning. Higher Na+ ion concentrations inside plant cells act as competitive inhibitors of enzymes and other ions, such as potassium (K+), disrupt photosynthesis and cell homeostasis, and result in higher levels of Reactive Oxygen Species (ROS) (Zörb et al., 2019). Because these effects are detrimental for plant functioning, plants have developed several responses as adaptation mechanisms to soil salinity—such as osmotic adjustment, ion compartmentalization, and selective ion exclusion (Serrano and Rodriguez-Navarro, 2001; Parida and Das, 2005; Zhao et al., 2020). Several antiporters have been proposed as primary actors, to some extent, of the previously mentioned salinity stress resistance processes. These antiporters include the CHX, KEA, and NHX families, which play a critical role in the adaptation to stress of Na+ either by controlling the uptake into the membranes or by compartmentalizing into the vacuoles (Munns, 2005; Asif et al., 2011; Sze and Chanroj, 2018). To synchronize their expression in the tissues of interest, signal transduction depends on large, orchestrated changes in gene expression that can be induced by transcriptional modules (Mantri et al., 2007; Shaar-Moshe et al., 2017; Franzoni et al., 2019). In this light, we propose to investigate the involvement of a ubiquitous transcription factor (TF) family that may coordinate different physiological processes, and which in some cases have been shown to enhance salinity resistance (see below).
MYB-TFs were originally named after the avian myeloblastosis virus (MYB), and are among the most prominent TF families in plants (Klempnauer et al., 1982). They have been demonstrated to be a cornerstone of plant functioning (Allan and Espley, 2018; Wang X. et al., 2021). As an example, MYB-TFs are involved in the regulation of different central metabolism processes, such as cell fate specification in the root epidermis (Ryu et al., 2005), differentiation of cells into trichomes (Oppenheimer et al., 1991), pigmentation of flowers and tissues (Koes et al., 2005; Liu et al., 2021), and controlling stomatal aperture in guard cells (Cominelli et al., 2005; Liang et al., 2005).
Different classifications of MYB-TFs, depending on homology, have been described (Stracke et al., 2001; Dubos et al., 2010; Allan and Espley, 2018; Pucker et al., 2020). The structure of a MYB-TF consists of a highly conserved DNA binding domain (MYB domain) with a helix-turn-helix (HTH) motif. These HTH motifs form a DNA recognition site (R), allowing the interaction with different target DNA sequences (Dubos et al., 2010). Depending on the number of R sites, the MYB-TF family is subdivided into four subfamilies: 1R-MYB, R2R3-MYB, 3R-MYB, and 4R-MYB (Dubos et al., 2010). Among the four subfamilies of MYB-TFs, the R2R3-MYB subfamily is by far the largest, the best studied (see below, Du et al., 2012). Therefore, we focus on the R2R3-MYB subfamily and its potential interaction on salinity tolerance mechanisms.
Several R2R3-MYBs have been characterized as master regulators, i.e., genes at the top of the hierarchy of a regulatory pathway (Kin Chan, 2013). MYB-TFs are an integral part of plant development, cell fate, and specialized metabolism. In particular for specialized metabolism, they have a key role as master regulators in processes related to anthocyanin biosynthesis (Quattrocchio et al., 2006; Pérez-Díaz et al., 2016; Feng et al., 2018; Zhou et al., 2019), fragrance (Verdonk et al., 2005; Yoshida et al., 2018), and lignin and secondary cell wall biosynthesis (Zhong et al., 2008). Even with the discovered functions, many other gene copies and different species remain uncharacterized, leaving many MYB-TFs with unknown roles.
The function of MYB-TFs is closely associated with different abiotic stress responses. For example, MYB-TFs were shown to be involved in drought, salt, and cold stress responses in wheat (Triticum aestivum), Arabidopsis thaliana, and rice (Oryza sativa) (Jung et al., 2008; Cai et al., 2011; Qu et al., 2022). OsMYB91 coordinates plant growth in rice and contributes to salt stress tolerance by promoting the accumulation of abscisic acid (ABA)—a phytohormone that, among its numerous roles, plays a key role in abiotic stress resistance by regulating stomatal conductance, thereby impacting rates of water loss and carbon gain (Zhu et al., 2015). Two other rice MYBs (OsMYB6 and OsMYB48-1) were shown to be involved in abiotic stress responses. Overexpression increased drought and salinity stress tolerance in rice, by inducing abiotic stress-responsive genes and promoting ABA signaling genes, respectively (Xiong et al., 2014; Tang et al., 2019).
Transcriptomic studies can be used to detect salinity responses in different crops. For example, in salinity-tolerant wild tomato (Solanum chilense) salt tolerance was shown to be dependent on differentially regulated genes related to: hormone signaling, Ca2+ signaling, ROS scavenging, and transcriptional regulation (Kashyap et al., 2020). Additionally, a comparative analysis on salt-stressed seedlings of domesticated tomato (S. lycopersicum) when cyclic guanosine monophosphate (c-GMP) was applied—a secondary messenger molecule involved in the salt stress response—showed improved plant osmotic adjustment, reduced non-stomatal water loss, and enhanced antioxidant defense pathways (Zhu et al., 2022). This improved salt tolerance coincided with the differential expression of 140 MYB-TF, suggesting that this transcription factor family could be involved in salinity responses (Zhu et al., 2022).
In rice, another connection between salinity and the MYB-TF family was observed: When growing a salinity-resistant and salinity-susceptible rice cultivar in saline conditions, transcriptomic analysis revealed two putative MYB60 transcription factors to be upregulated in the tolerant cultivar (Formentin et al., 2018). Interestingly, in Arabidopsis and grape (Vitis vinifera), these MYB60 TFs play a role in stomatal regulation (Cominelli et al., 2005; Galbiati et al., 2011). Similarly, when comparing drought-tolerant, salinity-tolerant, and susceptible rice cultivars, MYB-TFs were differentially expressed in the salinity-tolerant cultivar compared with the other two cultivars (Shankar et al., 2016). These examples demonstrate the value of transcriptomic approaches to identify important TFs. However, a targeted approach could be better suitable for known genes.
Overexpression of other MYB-TFs reduced various stresses: salt, drought, and cold stress in tomato and apple (Malus domestica) (Cao et al., 2013); salt and pathogen resistance in Arabidopsis (Shen et al., 2017); heat stress in rice (El-kereamy et al., 2012); and drought stress in cotton (Gossypium barbadense) (Chen et al., 2014). Similarly, transgenic maize (Zea mays) showed enhanced heat and drought tolerance, when OsMYB55 was overexpressed (Casaretto et al., 2016). Another example is SlMYB102, an R2R3-MYB in tomato; when it was overexpressed, ROS scavenging enzyme activities and antioxidant content increased, reducing ROS content, along with increased Na+/K+ homeostasis (Zhang et al., 2020). Whereas ectopic expression of a wheat MYB (TaMYB730) in Arabidopsis increased salinity tolerance, by improving the ionic resistance (He et al., 2012). Furthermore, GmMYB84 was characterized as a mediator of root elongation inhibition in response to drought stress in soybean (Glycine max) (Wang et al., 2017), whereas silencing AtMYB60 resulted in increased drought tolerance by constitutively reducing stomatal opening in Arabidopsis (Cominelli et al., 2005). The variety of species studied and the close relationship to specific functions opens new possibilities for understanding the diverse roles of MYB-TFs in various abiotic stress responses.
There is a close relationship between R2R3-MYB sequences and their function, even between different species (Allan and Espley, 2018). Phylogenetic trees can be used to discover new functions of different R2R3-MYBs, through homology in the aminoacidic sequences (Stracke et al., 2001; Dubos et al., 2010; Du et al., 2012; Hou et al., 2014; Pucker et al., 2020). For example, CaMYB101 represses anthocyanin biosynthesis in sweet pepper (Capsicum annuum) and is closely related to petunia PhMYB27, which has the same function (Liu et al., 2021). The functional redundancy is very well illustrated by the regulation of anthocyanin biosynthesis. In Arabidopsis, petunia, and maize, close R2R3-MYB homologs PAP1 (AtMYB75), AN2, and C1 respectively were shown to regulate the anthocyanin pathway (Koes et al., 2005). The same was shown in other species i.e., apple, litchi (Litchi chinensis), and grape (Chagné et al., 2007; Cavallini et al., 2014; Lai et al., 2019). Another example is AtMYB4, a MYB repressor of the phenylpropanoid pathway in Arabidopsis. PhMYB4 and FaMYB1 are homolog genes of AtMYB4 that fine-tune volatile production by controlling flavonoids in petunia and anthocyanins in strawberries (Fragaria × ananasa) by repressing the same gene in the phenylpropanoid pathway in both species (Jin et al., 2000; Aharoni et al., 2001; Colquhoun et al., 2011). In banana (Musa acuminata) fruits, MaMYB3 acts as a repressor in the modulation of starch degradation; when overexpressing MaMYB3 in transgenic tomatoes, this MYB prevented normal ripening of tomato fruits (Fan et al., 2018). In this context, we can take advantage of R2R3-MYBs with known functions related to abiotic stress resistance as candidates to test in other species and investigate if their function is conserved.
Petunia, together with Arabidopsis and maize, is among the first plant species where many different R2R3-MYBs with different functions were identified (Koes et al., 2005). Petunia belongs to the Solanaceae family, and is a perfect link between lab and crop models due to its close relation with common crops such as tomato, sweet pepper, potato (Solanum tuberosum), and eggplant (Solanum melongena) (Vandenbussche et al., 2016). Furthermore, petunia flowers have several known quality traits—e.g., fragrance, color, morphological patterns—that are regulated by R2R3-MYBs (Santos and Handro, 1983; Verdonk et al., 2005; Quattrocchio et al., 2006; Baumann et al., 2007; Hoballah et al., 2007). Similarly agronomic and horticultural traits—such as abiotic resistance—are expected to be regulated by MYB-TFs as well (Wang et al., 2020; Hussain et al., 2021). Nevertheless, petunia contains >100 hypothetical R2R3-MYBs that remain uncharacterized. Hence, we propose to identify homologous genes and new functions of different R2R3-MYBs, which could be used later in genome-wide association studies or functional characterization studies. Such studies should emulate the harsh conditions that field crops often grow under—such as salinity, drought, high light, or heat—and elucidate the R2R3-MYBs related to a given stress response.
We made a phylogenetic tree (Figure 1) to group the two openly available genome sequences of petunia: Petunia axillaris and Petunia inflata (Jin et al., 2017). This tree can be used to predict complete groups with potential homologous genes related to salinity resistance. R2R3-MYBs can be visualized with 27 groups of R2R3-MYBs with homologous sequences. With this strategy, we aim to have these groups as a foundation for future studies and help characterize stress tolerance in different species.
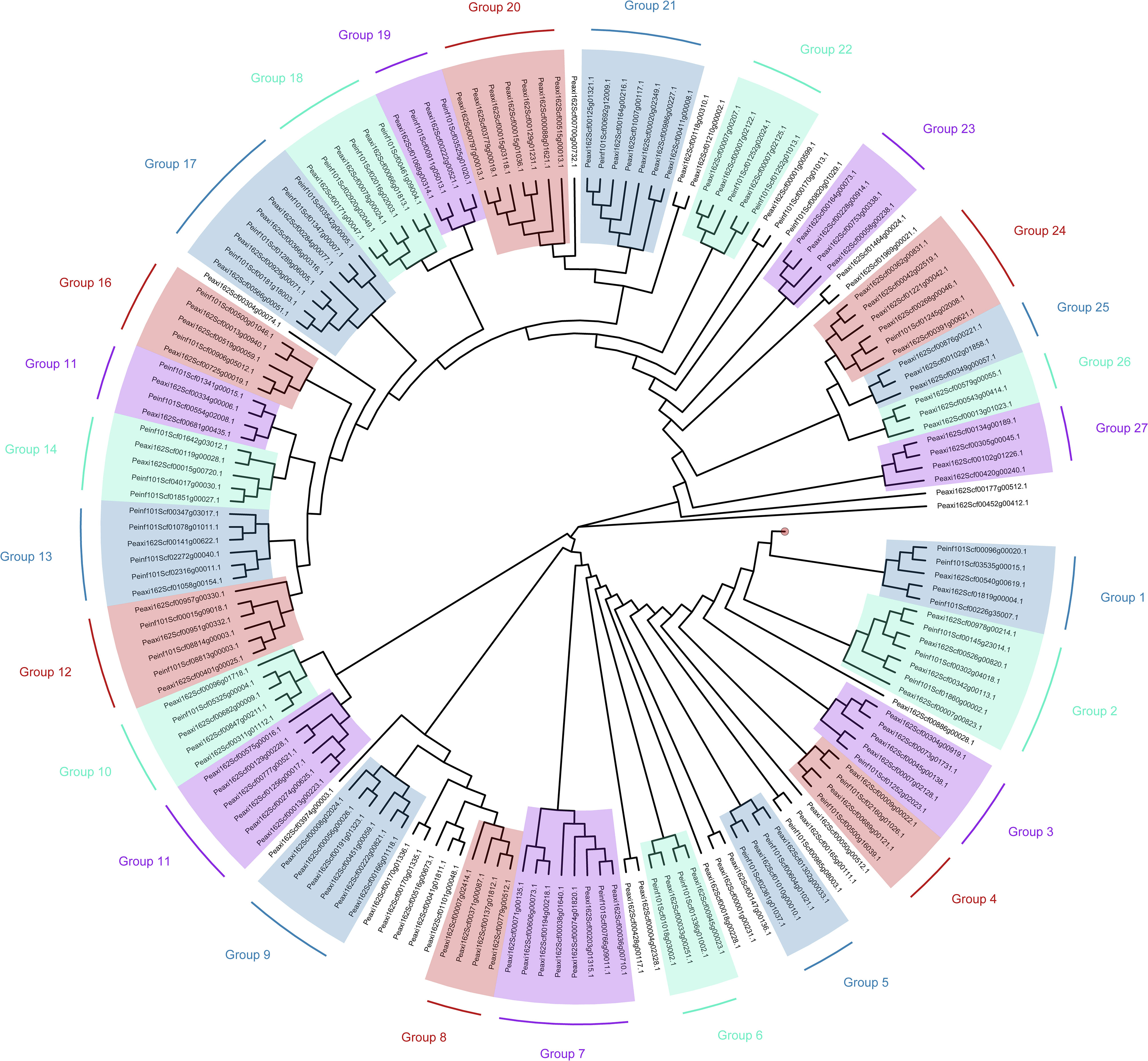
Figure 1 Phylogenetic tree with R2R3-MYBs based on aminoacidic sequences of Petunia axilallis and Petunia inflata. (1) The sequences of the MYB-TF family were retrieved from the Plant transcription factor database (Jin et al., 2017). (2) Using the Levenshtein distance method in R, the dissimilarity between sequences was measured (Pagès et al., 2023). (3) The phylogenetic tree was then built using the BioNJ algorithm (Gascuel, 1997), and (4) visually illustrating the relationships among the sequences using (Yu et al., 2017). The red circle between Group 27 and Group 1 represents 23 individual clades, each with single sequences, that were omitted from this tree.
We constructed the phylogenetic tree with information from both Petunia axilaris and Petunia inflata because of gene redundancies during their evolution (Bombarely et al., 2016). Gene redundancies are important because there are examples of paralogs and diversification in petunia: for example AN2 and AN4 are genes that code for R2R3-MYBs in charge of flower coloration. These are expressed in different locations in flower tissues and flower developmental stages, producing a clear difference in anthocyanin expression between the two wild parental lines (Bombarely et al., 2016, Supplementary note 7). To confirm the previous duplication and potential search of salinity related R2R3-MYB, we align previously mentioned MYB-TFs: OsMYB55, OsMYB91, and AtMYB60. For OsMYB55, group 10 (Peaxi162Scf00311g01112.1, Peaxi162Scf00096g01718.1, Peaxi162Scf00682g00009.1, and Peaxi162Scf00847g00211.1, including the sequence for Petunia inflata :Peinf101Scf05325g00004.1) showed the homolog sequence in the same clade. The homolog sequence, on the other hand, for OsMYB91 and AtMYB60 were part of group 11. This example suggests that groups 10 and 11 in this tree could be related to abiotic stress resistance in petunia, and it may help future studies to begin with these groups first. With this strategy, it will be possible to transfer this knowledge from petunia to other species.
Petunia presents a high tolerance to a number of abiotic stresses. Petunia is more salinity tolerant when compared with tomato, eggplant, or potato, which are respectively moderately salt-tolerant, moderately salt-sensitive, and sensitive to salinity (Fornes et al., 2007; Ünlükara et al., 2010; Villarino and Mattson, 2011; Charfeddine et al., 2019; Moles et al., 2019; Parkash and Singh, 2020; Altaf et al., 2023). Under stressful conditions such as high light intensity, petunia modulates vegetative anthocyanin and volatile production as a coping mechanism (Albert et al., 2009; Colquhoun et al., 2013). Nevertheless, even when petunia presents tolerance to abiotic stresses, the current trend is to study the final phenotype—physiological and metabolomics responses—but understanding transcriptomic regulation could be a more direct way to improve resistance when breeding for new varieties. Some transcriptomics studies have been conducted to identify the different pathways involved in salinity resistance mechanisms on petunia under salinity stress (Villarino et al., 2014). For example, salinity and drought resistance are partially achieved by modulating ion transport inside the vacuoles (Xu et al., 2009; Asif et al., 2011; Banjara et al., 2012). In these studies, the overexpression of AtNHX1—a vacuolar Na+/H+ antiporter gene—significantly enhanced the resistance to salinity and drought stresses. The transgenic expression of FvMYB24 in Arabidopsis enhanced the salinity tolerance of transgenic plants, and among other genes AtNHX1 was upregulated (Wang S. et al., 2021). However, the connection between those studies and intermediary regulatory pathways remains uncharacterized.
To summarize, we argue that salinity is a problem affecting commercially important crops worldwide, and that studying R2R3-MYBs in petunia could help to address this issue. Petunia, as a member of the Solanaceae family, is a suitable link between lab and crop models, and is highly salinity tolerant. We suggest the use of petunia as a model plant to study salinity resistance and to identify the R2R3-MYB regulators in response to salinity. We picked R2R3-MYB, because they are known to be related with different developmental pathways, specialized metabolisms pathways, and stress responses pathways. The most important aspect is that the function of those MYB-TFs is usually maintained between different plant species, making the extrapolation of these discoveries to other crops feasible. We proposed to functionally characterize these MYB-TFs by the use of homology and phylogeny. Here, we present an example based on Petunia axillaris and Petunia inflata, and suggest that it should be possible to extrapolate this idea to other crops with known sequences.
The original contributions presented in the study are included in the article/supplementary files, further inquiries can be directed to the corresponding author.
BZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. LM: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review and editing, Funding acquisition, Investigation. EK: Conceptualization, Writing – original draft, Writing – review and editing, Investigation, Project administration, Supervision. JV: Conceptualization, Investigation, Project administration, Supervision, Writing – original draft, Writing – review and editing, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Chilean National Agency for Research and Development (ANID) and the Chilean National Scholarship Program for Graduate Studies, DOCTORADO BECAS CHILE/2019 – 72200074.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aharoni, A., De Vos, C. H. R., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/J.1365-313X.2001.01154.X
Albert, N. W., Lewis, D. H., Zhang, H., Irving, L. J., Jameson, P. E., Davies, K. M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60, 2191–2202. doi: 10.1093/jxb/erp097
Allan, A. C., Espley, R. V. (2018). MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Sci. 23, 693–705. doi: 10.1016/j.tplants.2018.06.001
Altaf, M. A., Mandal, S., Behera, B., Mangal, V., Naz, S., Kumar, R., et al. (2023). Salinity stress tolerance in solanaceous crops: current understanding and its prospects in genome editing. J. Plant Growth Regul. 42, 4020–4036. doi: 10.1007/s00344-022-10890-0
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Mol. Biotechnol. 49, 250–256. doi: 10.1007/s12033-011-9399-1
Banjara, M., Zhu, L., Shen, G., Payton, P., Zhang, H. (2012). Expression of an Arabidopsis sodium/proton antiporter gene (AtNHX1) in peanut to improve salt tolerance. Plant Biotechnol. Rep. 6, 59–67. doi: 10.1007/s11816-011-0200-5
Baumann, K., Perez-Rodriguez, M., Bradley, D., Venail, J., Bailey, P., Jin, H., et al. (2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134, 1691–1701. doi: 10.1242/DEV.02836
Bombarely, A., Moser, M., Amrad, A., Bao, M., Bapaume, L., Barry, C. S., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2 (May), 1–9. doi: 10.1038/nplants.2016.74
Cai, H., Tian, S., Liu, C., Dong, H. (2011). Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene 485, 146–152. doi: 10.1016/j.gene.2011.06.026
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of mdoMYB121 gene confering abiotic stress tolerance in plants. PloS One 8, e69955. doi: 10.1371/journal.pone.0069955
Casaretto, J. A., El-kereamy, A., Zeng, B., Stiegelmeyer, S. M., Chen, X., Bi, Y. M., et al. (2016). Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17, 1–15. doi: 10.1186/s12864-016-2659-5
Cavallini, E., Zenoni, S., Finezzo, L., Guzzo, F., Zamboni, A., Avesani, L., et al. (2014). Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol. 55 (3), 517–534. doi: 10.1093/pcp/pct190
Chagné, D., Carlisle, C. M., Blond, C., Volz, R. K., Whitworth, C. J., Oraguzie, N. C., et al. (2007). Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8 (1), 212. doi: 10.1186/1471-2164-8-212
Charfeddine, M., Charfeddine, S., Ghazala, I., Bouaziz, D., Bouzid, R. G. (2019). Investigation of the response to salinity of transgenic potato plants overexpressing the transcription factor StERF94. J. Biosci. 44, 1–16. doi: 10.1007/S12038-019-9959-2/FIGURES/10
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., Zhang, B. (2014). A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Colquhoun, T. A., Kim, J. Y., Wedde, A. E., Levin, L. A., Schmitt, K. C., Schuurink, R. C., et al. (2011). PhMYB4 fine-tunes the floral volatile signature of Petunia×hybrida through PhC4H. J. Exp. Bot. 62, 1133–1143. doi: 10.1093/JXB/ERQ342
Colquhoun, T. A., Schwieterman, M. L., Gilbert, J. L., Jaworski, E. A., Langer, K. M., Jones, C. R., et al. (2013). Light modulation of volatile organic compounds from petunia flowers and select fruits. Postharvest Biol. Technol. 86, 37–44. doi: 10.1016/j.postharvbio.2013.06.013
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Du, H., Feng, B.-R., Yang, S.-S., Huang, Y.-B., Tang, Y.-X. (2012). The R2R3-MYB transcription factor gene family in maize. PloS One 7, e37463. doi: 10.1371/journal.pone.0037463
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
El-kereamy, A., Bi, Y. M., Ranathunge, K., Beatty, P. H., Good, A. G., Rothstein, S. J. (2012). The rice R2R3-MYB transcription factor osMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PloS One 7, e52030. doi: 10.1371/journal.pone.0052030
Fan, Z. Q., Ba, L. J., Shan, W., Xiao, Y. Y., Lu, W. J., Kuang, J. F., et al. (2018). A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 96, 1191–1205. doi: 10.1111/tpj.14099
Feng, K., Xu, Z. S., Que, F., Liu, J. X., Wang, F., Xiong, A. S. (2018). An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica. Planta 247, 301–315. doi: 10.1007/s00425-017-2783-8
Formentin, E., Sudiro, C., Perin, G., Riccadonna, S., Barizza, E., Baldoni, E., et al. (2018). Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00204
Fornes, F., Belda, R. M., Carrión, C., Noguera, V., García-Agustín, P., Abad, M. (2007). Pre-conditioning ornamental plants to drought by means of saline water irrigation as related to salinity tolerance. Sci. Hortic. 113, 52–59. doi: 10.1016/j.scienta.2007.01.008
Franzoni, G., Cocetta, G., Trivellini, A., Ferrante, A. (2019). Transcriptional regulation in rocket leaves as affected by salinity. Plants 9 (1), 20. doi: 10.3390/plants9010020
Galbiati, M., Matus, J. T., Francia, P., Rusconi, F., Cañón, P., Medina, C., et al. (2011). The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 11 (1), 142. doi: 10.1186/1471-2229-11-142
Gascuel, O. (1997). BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695. doi: 10.1093/oxfordjournals.molbev.a025808
He, Y., Li, W., Lv, J., Jia, Y., Wang, M., Xia, G. (2012). Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 63, 1511–1522. doi: 10.1093/JXB/ERR389
Hoballah, M. E., Gübitz, T., Stuurman, J., Broger, L., Barone, M., Mandel, T., et al. (2007). Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19, 779–790. doi: 10.1105/tpc.106.048694
Hou, X. J., Li, S. B., Liu, S. R., Hu, C. G., Zhang, J. Z. (2014). Genome-wide classification and evolutionary and expression analyses of citrus MYB transcription factor families in sweet orange. PloS One 9, e112375. doi: 10.1371/journal.pone.0112375
Hussain, Q., Asim, M., Zhang, R., Khan, R., Farooq, S., Wu, J. (2021). Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules 11, 1159. doi: 10.3390/BIOM11081159
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Jung, C., Jun, S. S., Sang, W. H., Yeon, J. K., Chung, H. K., Sang, I. S., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kashyap, S. P., Prasanna, H. C., Kumari, N., Mishra, P., Singh, B. (2020). Understanding salt tolerance mechanism using transcriptome profiling and de novo assembly of wild tomato Solanum Chilense. Sci. Rep. 10 (1), 15835. doi: 10.1038/s41598-020-72474-w
Kin Chan, S. S. (2013). What is a master regulator? J. Stem Cell Res. Ther. 03, e114. doi: 10.4172/2157-7633.1000e114
Klempnauer, K.-H., Gonda, T. J., Michael Bishop, J. (1982). Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 31, 453–463. doi: 10.1016/0092-8674(82)90138-6
Koes, R., Verweij, W., Quattrocchio, F. (2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Lai, B., Du, L. N., Hu, B., Wang, D., Huang, X. M., Zhao, J. T., et al. (2019). Characterization of a novel litchi R2R3-MYB transcription factor that involves in anthocyanin biosynthesis and tissue acidification. BMC Plant Biol. 19, 62. doi: 10.1186/s12870-019-1658-5
Li, A., Wu, Y., Cao, S. (2023). Effects of land use-land cover on soil water and salinity contents. Acta Ecologica Sinica. doi: 10.1016/j.chnaes.2023.07.002. [Epub ahead of print].
Liang, Y. K., Dubos, C., Dodd, I. C., Holroyd, G. H., Hetherington, A. M., Campbell, M. M. (2005). AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 15, 1201–1206. doi: 10.1016/j.cub.2005.06.041
Liu, Y., Wu, Y., Wang, Z., Zhang, S., Liu, X., Tikunov, Y., et al. (2021). Genome-wide characterization of the R2R3-MYB transcription factors in pepper (Capsicum spp.) unveils the role of CaMYB101 as repressor in anthocyanin biosynthesis. bioRxiv. doi: 10.1101/2021.08.24.457473. [Epub ahead of print].
Mantri, N. L., Ford, R., Coram, T. E., Pang, E. C. K. (2007). Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8, 1–14. doi: 10.1186/1471-2164-8-303/TABLES/4
Moles, T. M., de Brito Francisco, R., Mariotti, L., Pompeiano, A., Lupini, A., Incrocci, L., et al. (2019). Salinity in autumn-winter season and fruit quality of tomato landraces. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01078
Munns, R. (2005). Genes and salt tolerance: Bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Munns, R., Schachtman, D. P., Condon, A. G. (1995). The significance of a two-phase growth response to salinity in wheat and barley. Funct. Plant Biol. 22, 561–569. doi: 10.1071/PP9950561
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., Marks, M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Pagès, H., Aboyoun, P., Gentleman, R., DebRoy, S. (2023) Package “Biostrings” Title Efficient manipulation of biological strings. Available at: https://bioconductor.org/packages/release/bioc/html/Biostrings.html (Accessed 06, 2023).
Parida, A. K., Das, A. B. (2005). Salt tolerance and salinity effects on plants: A review. Ecotoxicol Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Parkash, V., Singh, S. (2020). Potential of biochar application to mitigate salinity stress in eggplant. HortScience 55, 1946–1955. doi: 10.21273/HORTSCI15398-20
Pérez-Díaz, J. R., Pérez-Díaz, J., Madrid-Espinoza, J., González-Villanueva, E., Moreno, Y., Ruiz-Lara, S. (2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76. doi: 10.1007/s11103-015-0394-y
Pucker, B., Pandey, A., Weisshaar, B., Stracke, R. (2020). The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. PloS One 15, e0239275. doi: 10.1371/journal.pone.0239275
Qu, X., Zou, J., Wang, J., Yang, K., Wang, X., Le, J. (2022). A rice R2R3-type MYB transcription factor osFLP positively regulates drought stress response via osNAC. Int. J. Mol. Sci. 23, 5873. doi: 10.3390/ijms23115873
Quattrocchio, F., Verweij, W., Kroon, A., Spelt, C., Mol, J., Koes, R. (2006). PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18, 1274–1291. doi: 10.1105/tpc.105.034041
Rodríguez Coca, L. I., García González, M. T., Gil Unday, Z., Jiménez Hernández, J., Rodríguez Jáuregui, M. M., Fernández Cancio, Y. (2023). Effects of sodium salinity on rice (Oryza sativa L.) cultivation: A review. Sustainability 15, 1804. doi: 10.3390/su15031804
Rouphael, Y., Petropoulos, S. A., Cardarelli, M., Colla, G. (2018). Salinity as eustressor for enhancing quality of vegetables. Sci. Hortic. 234, 361–369. doi: 10.1016/J.SCIENTA.2018.02.048
Ryu, K. H., Kang, Y. H., Park, Y. H., Hwang, I., Schiefelbein, J., Lee, M. M. (2005). The WEREWOLF MYB protein directly regulates CAPRICEtranscription during cell fate specification in the Arabidopsis root epidermis. Development 132, 4765–4775. doi: 10.1242/DEV.02055
Saied, A. S., Keutgen, A. J., Noga, G. (2005). The influence of NaCl salinity on growth, yield and fruit quality of strawberry cvs. “Elsanta” and “Korona”. Sci. Hortic. 103, 289–303. doi: 10.1016/j.scienta.2004.06.015
Santos, R. F., Handro, W. (1983). Morphological patterns in Petunia hybrida plants regenerated from tissue cultures and differing by their ploidy. Theor. Appl. Genet. 66, 55–60. doi: 10.1007/BF00281849/METRICS
Serrano, R., Rodriguez-Navarro, A. (2001). Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13, 399–404. doi: 10.1016/S0955-0674(00)00227-1
Shaar-Moshe, L., Blumwald, E., Peleg, Z. (2017). Unique physiological and transcriptional shifts under combinations of salinity, drought, and heat. Plant Physiol. 174, 421–434. doi: 10.1104/PP.17.00030
Shankar, R., Bhattacharjee, A., Jain, M. (2016). Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 6, 23719. doi: 10.1038/srep23719
Shen, X., Guo, X., Guo, X., Zhao, D., Zhao, W., Chen, J., et al. (2017). PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 112, 302–311. doi: 10.1016/j.plaphy.2017.01.015
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sze, H., Chanroj, S. (2018). Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 177, 875–895. doi: 10.1104/pp.18.00142
Tang, Y., Bao, X., Zhi, Y., Wu, Q., Guo, Y., Yin, X., et al. (2019). Overexpression of a myb family gene, Osmyb6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00168
Ünlükara, A., Kurunç, A., Kesmez, G. D., Yurtseven, E., Suarez, D. L. (2010). Effects of salinity on eggplant (Solanum melongena L.) growth and evapotranspiration. Irrigation Drainage 59, 203–214. doi: 10.1002/IRD.453
Vandenbussche, M., Chambrier, P., Rodrigues Bento, S., Morel, P. (2016). Petunia, your next supermodel? Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00072
Verdonk, J. C., Haring, M. A., Tunen, A.J.V., Schuurink, R. C. (2005). ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17, 1–13. doi: 10.1105/tpc.104.028837.1
Villarino, G. H., Bombarely, A., Giovannoni, J. J., Scanlon, M. J., Mattson, N. S. (2014). Transcriptomic analysis of Petunia hybrida in response to salt stress using high throughput RNA sequencing. PloS One 9, 1–13. doi: 10.1371/journal.pone.0094651
Villarino, G. H., Mattson, N. S. (2011). Assessing tolerance to sodium chloride salinity in fourteen floriculture species. Horttechnology 21, 539–545. doi: 10.21273/horttech.21.5.539
Wang, N., Zhang, W., Qin, M., Li, S., Qiao, M., Liu, Z., et al. (2017). Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 58, 1764–1776. doi: 10.1093/pcp/pcx111
Wang, S., Shi, M., Zhang, Y., Xie, X., Sun, P., Fang, C., et al. (2021). FvMYB24, a strawberry R2R3-MYB transcription factor, improved salt stress tolerance in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 569, 93–99. doi: 10.1016/j.bbrc.2021.06.085
Wang, W.-Q., Allan, A. C., Yin, X.-R. (2020). Small RNAs with a big impact on horticultural traits. CRC Crit. Rev. Plant Sci. 39, 30–43. doi: 10.1080/07352689.2020.1741923
Wang, X., Niu, Y., Zheng, Y. (2021). Multiple functions of myb transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22, 6125. doi: 10.3390/ijms22116125
Xiong, H., Li, J., Liu, P., Duan, J., Zhao, Y., Guo, X., et al. (2014). Overexpression of osMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PloS One 9, e92913. doi: 10.1371/JOURNAL.PONE.0092913
Xu, K., Hong, P., Luo, L., Xia, T. (2009). Overexpression of AtNHX1, a vacuolar Na+/H+ antiporter from arabidopsis thalina, in petunia hybrida enhances salt and drought tolerance. J. Plant Biol. 52, 453–461. doi: 10.1007/s12374-009-9058-2
Yoshida, K., Oyama-Okubo, N., Yamagishi, M. (2018). An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Mol. Breed. 38, 144. doi: 10.1007/s11032-018-0902-2
Yu, G., Smith, D. K., Zhu, H., Guan, Y., Lam, T. T. (2017). GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang, X., Chen, L., Shi, Q., Ren, Z. (2020). SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 291, 110356. doi: 10.1016/j.plantsci.2019.110356
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Zhu, N., Cheng, S., Liu, X., Du, H., Dai, M., Zhou, D. X., et al. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236, 146–156. doi: 10.1016/j.plantsci.2015.03.023
Zhu, X., Su, M., Wang, B., Wei, X. (2022). Transcriptome analysis reveals the main metabolic pathway of c-GMP induced by salt stress in tomato. Funct. Plant Biol. 49 (9), 784–798. doi: 10.1071/FP21337
Keywords: MYB, transcription factors, salinity, petunia, quality traits
Citation: Zepeda B, Marcelis LFM, Kaiser E and Verdonk JC (2023) Petunia as a model for MYB transcription factor action under salt stress. Front. Plant Sci. 14:1286547. doi: 10.3389/fpls.2023.1286547
Received: 31 August 2023; Accepted: 20 November 2023;
Published: 08 December 2023.
Edited by:
Sachin Teotia, Sharda University, IndiaReviewed by:
Rongfang Guo, Fujian Agriculture and Forestry University, ChinaCopyright © 2023 Zepeda, Marcelis, Kaiser and Verdonk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian C. Verdonk, anVsaWFuLnZlcmRvbmtAd3VyLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.