
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 December 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1282661
This article is part of the Research TopicMulti-omics and Computational Biology in Horticultural Plants: From Genotype to Phenotype, Volume IIView all 25 articles
 Lei Luo1†
Lei Luo1† Jos Molthoff2†
Jos Molthoff2† Qiang Li1
Qiang Li1 Ying Liu3
Ying Liu3 Shuangxia Luo1
Shuangxia Luo1 Na Li1
Na Li1 Shuxin Xuan1
Shuxin Xuan1 Yanhua Wang1
Yanhua Wang1 Shuxing Shen1
Shuxing Shen1 Arnaud G. Bovy2*
Arnaud G. Bovy2* Jianjun Zhao1*
Jianjun Zhao1* Xueping Chen1*
Xueping Chen1*Eggplant (Solanum melongena L.) is a highly nutritious and economically important vegetable crop. However, the fruit peel of eggplant often shows poor coloration owing to low-light intensity during cultivation, especially in the winter. The less-photosensitive varieties produce anthocyanin in low light or even dark conditions, making them valuable breeding materials. Nevertheless, genes responsible for anthocyanin biosynthesis in less-photosensitive eggplant varieties are not characterized. In this study, an EMS mutant, named purple in the dark (pind), was used to identify the key genes responsible for less-photosensitive coloration. Under natural conditions, the peel color and anthocyanin content in pind fruits were similar to that of wildtype ‘14-345’. The bagged pind fruits were light purple, whereas those of ‘14-345’ were white; and the anthocyanin content in the pind fruit peel was significantly higher than that in ‘14-345’. Genetic analysis revealed that the less-photosensitive trait was controlled by a single dominant gene. The candidate gene was mapped on chromosome 10 in the region 7.72 Mb to 11.71 Mb. Thirty-five differentially expressed genes, including 12 structural genes, such as CHS, CHI, F3H, DFR, ANS, and UFGT, and three transcription factors MYB113, GL3, and TTG2, were identified in pind using RNA-seq. Four candidate genes EGP21875 (myb domain protein 113), EGP21950 (unknown protein), EGP21953 (CAAX amino-terminal protease family protein), and EGP21961 (CAAX amino-terminal protease family protein) were identified as putative genes associated with less-photosensitive anthocyanin biosynthesis in pind. These findings may clarify the molecular mechanisms underlying less-photosensitive anthocyanin biosynthesis in eggplant.
Anthocyanins are natural flavonoid pigments, which are responsible for blue, purple and red colors in plant tissues, such as flowers, fruits, seed coats, leaves, and stems. In addition to their role in coloring, anthocyanins attract pollinators and seed disperser and protect plants from biotic and abiotic stresses (Fan et al., 2016; Sivankalyani et al., 2016; Sun et al., 2018; Naing and Kim, 2021). Anthocyanins also show beneficial effects on human health, including antioxidation, antimutagenicity, cardiovascular disease prevention, liver protection, and the inhibition of tumor cell metastasis (Pojer et al., 2013; Liu et al., 2016).
Anthocyanins are produced through the phenylpropanoid pathway, which has been studied in many plants, such as petunia and Arabidopsis (Krol et al., 1990; Dooner et al., 1991; Timothy and Holton, 1995; Baudry et al., 2004). The anthocyanin biosynthesis is catalyzed stepwise by a series of enzymes, including phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate coenzyme A ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose: flavonoid-3-O-glycosyl transferase (UFGT). The expression of these structural genes was primarily regulated by three classes of transcription factors (MYB, bHLH, and WD40), which always formed a MYB-bHLH-WD40 (MBW) complex. (Gonzalez et al., 2008; Albert et al., 2014; Xu et al., 2015; Shan et al., 2019; Li et al., 2020). Among them, MYB TFs have been identified to be the major determinant regulator in the MBW complex (Gonzalez et al., 2008; Tang et al., 2020). In Arabidopsis, MYB TFs TT2 (TRANSPARENT TESTA 2), PAP1/PAP2 (the production of anthocyanin pigment 1/2), MYB113 and MYB114, bHLH TFs TT8 (transparent testa8), and GL3 (glabra3), WD40 repeat protein TTG1 (TRANSPARENT TESTA GLABRA1) are the main components of the MBW complexes that regulate anthocyanin production (Zhang et al., 2003; Baudry et al., 2004).
The biosynthesis of anthocyanins is often affected by environmental factors such as light (Sun et al., 2020; Zhao et al., 2022) and temperature (Gao-Takai et al., 2019; Ryu et al., 2020). Most plants accumulate anthocyanin in a light-induced manner (Meng and Liu, 2015; Kim et al., 2021; Ma et al., 2021a), and anthocyanin concentration tends to rise as the light intensity increase (Li et al., 2018; Bai et al., 2019; Hong et al., 2019). In light signal transduction, phytochromes and their downstream factors, such as COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1), HY5 (LONG HYPOCOTYL 5), and other TFs participate in light-induced anthocyanin biosynthesis (Maier et al., 2013; Meng and Liu, 2015; Hoai Nguyen, 2020; Bhatia et al., 2021; Ma et al., 2021b). In this biological process, expression of structural genes and regulatory genes, except WD40, is strongly dependent upon the existence of light (Takos et al., 2006; Bai et al., 2017; Hong et al., 2019; Liu et al., 2019). Interestingly, less- and non-photosensitive biosynthesis of anthocyanins has been reported in cherry, chrysanthemum, mango, turnip, grape and eggplant (Huang et al., 2016; Yang et al., 2017; Guo et al., 2018; He et al., 2019; Zha et al., 2019; Shi et al., 2021), although the underlying mechanism is still unclear.
Eggplant (Solanum melongena L.) is a globally cultivated vegetable crop with high economic benefits (Saini and Kaushik, 2019; Oladosu et al., 2021). The color of eggplant fruit is an important quality trait, with purple varieties exhibiting high levels of anthocyanins in the fruit peel (Nayanathara et al., 2016; Nino-Medina et al., 2017). However, low light during cultivation often leads to poor coloration, which reduces their visual quality and commercial value (Li et al., 2018). Therefore, eggplant genotypes with less- and non-photosensitive anthocyanin biosynthesis are valuable for the breeding of low-light tolerant varieties. The purple peel under the calyx is a good indicator of less- and non-photosensitive coloration (Xiang et al., 2015; He et al., 2019). Although some quantitative trait loci (QTL) that are responsible for purple peel under calyx have been identified in eggplant (Chen, 2015; Toppino et al., 2016; Mangino et al., 2022) and markers have been developed (Xiang et al., 2015; Zhang et al., 2021b), the underlying causal genes conferring less-photosensitive biosynthesis of anthocyanin in eggplant remain unknown.
In this study, a less-photosensitive anthocyanin biosynthesis mutant purple in the dark (pind) in eggplant was discovered. Under bagging conditions, the pind fruit color was light purple, whereas the wild type was white, indicating that anthocyanin biosynthesis in the pind mutant was less dependent on light. Phenotypic and genetic analysis, gene mapping, and transcriptomic analysis were performed to identify the causative genes conferring less-photosensitive anthocyanin biosynthesis in the fruit peel of pind. Taken together, our results provided novel insight into less-photosensitive anthocyanin accumulation in eggplant fruit peel.
The mutant was identified from M1 line in an ethyl methanesulfonate (EMS) mutagenized population of eggplant variety ‘14-345’, which is white under the calyx. After two generations of self-fertilization, we obtained a homozygous mutant and named it “pind” (Figure 1A). The pind mutant and wildtype ‘14-345’ were crossed to produce F1 plants. Subsequently, F2 and BC1 populations were generated from F1 plants via self- and backcross with ‘14-345’ or pind, respectively. They were used as segregating populations for genetic analysis and mapping the candidate gene. In addition, pind and a cultivated eggplant ‘18-305’ (Figure S1), which is white under the calyx, were crossed to obtain another F2 population (‘305F2’). The ‘305F2’ population was also used for gene mapping. All plants were grown in a plastic greenhouse in the experimental fields of Hebei Agricultural University, Baoding, China. During the cultivation, the temperature inside the greenhouse was ranged from 18 to 32°C, the air humidity ranged from 50 to 70%, and the daytime light intensity ranged from 40k to 55k lx.
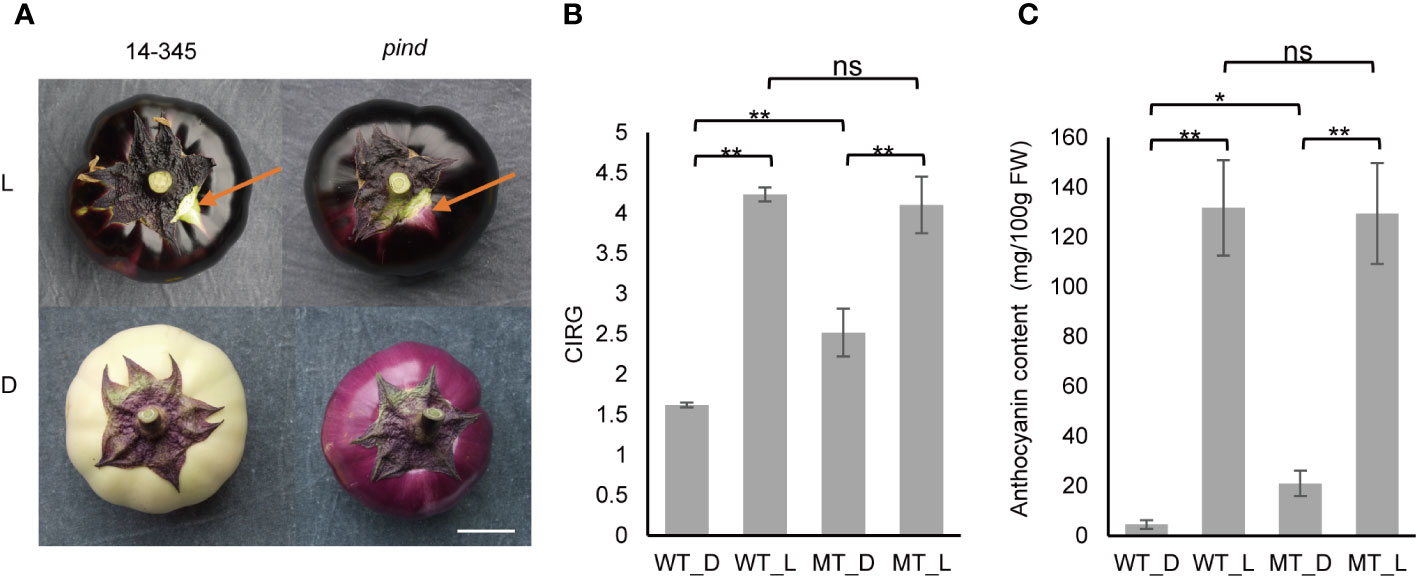
Figure 1 Effect of bagging on the peel color of ‘14-345’ (wild type) and pind (mutant) fruits. (A) Fruit color of ‘14-345’ (WT) and pind (MT) under natural light (L) and dark (D) conditions. The peel color under the calyx is indicated with arrows. White bar = 4cm. (B) CIRG (color index of red grape) value and (C) total anthocyanin content of the peel of ‘14-345’ and pind under natural and dark conditions. Data marked with *, ** and ns indicate P <0.05, P<0.01 and no significance, respectively.
The fruit of the plants were bagged on the 5th day after flowering and the fruit color and anthocyanin content were investigated on the 14th day under bagging condition. Color differences were assessed by the color index of red grapes (CIRG). A CR-400 colorimeter (Konica Minolta, Chroma Meter, Osaka, Japan) was used to measure the values of L*, a*, and b*. CIRG was calculated with the equation CIRG = [(180-H)/(L* + C*)], H=arctan (b*/a*), C=(a*2 + b*2)0.5 (Carreno et al., 1995; Zhang et al., 2008).
The total anthocyanin content in the peel was quantified using the pH differential spectroscopic method (Cheng and Patrick, 1991).
One-way analysis of variance (ANOVA) was conducted on the CIRG value and anthocyanin content, and significant differences between groups were assessed by Duncan’s multiple range tests (p < 0.05) using SPSS 16.0 Statistics (SPSS Inc., Chicago, IL, USA).
In the F2 population under bagging conditions, 30 plants with purple fruit and 30 plants with white fruits were selected to construct the mixed pools ‘P’ and ‘G’, respectively. Genome DNA was extracted using the CTAB method.
Bulk segregant analysis (BSA)-based sequencing and MutMap analysis were used to map the candidate genes. The qualified DNA was randomly broken into fragments with a length of 350bp, and libraries were built using a TruSeq Library Construction Kit, followed by Illumina PE150 sequencing. BWA software was used to map the clean data to the eggplant reference genome (Li et al., 2021). The UnifiedGenotyper module of GATK3.8 software (Mckenna et al., 2010) was used to detect SNPs, and VariantFiltration was used to filter SNP detection. The SNP-index value with ‘14-345’ as the reference was calculated to identify the key genes. The candidate intervals were determined with the threshold value: SNP-index (P) > 0.67, SNP-index (G) < 0.1, and Δ(SNP-index) > 0.6.
To narrow down the candidate region, the genotype of individuals in the F2 and ‘305F2’ populations was detected using Kompetitive allele specific PCR (KASP) technology to screen recombinants. The primers used for genotype verification of the individual plants are listed in Table S1.
Total RNA was extracted using an Eastep Super Kit (Shanghai Promega, Shanghai, China). The extracted RNA was treated with DNase to remove the genome DNA and the integrity and quantity were examined using 1% agarose gels and a Nanodrop 2000c Spectrophotometer (Thermo Nanodrop Technologies, Wilmington, DE, USA). A total of 1 μg RNA per sample was used to construct RNA-seq libraries. After qualification, mRNA was isolated from total RNA using oligo-magnetic beads. The mRNA was interrupted, reverse transcribed into cDNA, and then the cDNA was purified with an AMpure XP system (Beckman Coulter, Beverly, MA, USA). Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Boston, MA, USA). Twenty-four libraries were sequenced on the Illumina Hiseq platform (Illumina, San Diego, CA, USA). The library construction and sequencing were completed by Novogene (Beijing, China).
Raw data (raw reads) in fastq format were filtered to obtain clean reads, and the Q20, Q30, and GC content of the clean data were calculated. The clean data were aligned to the eggplant reference genome (Li et al., 2021) using Hisat2 v2.0.5. software (Mortazavi et al., 2008). The featureCounts v.5.0-p3 tool in the subread software was used to count the reads mapped to each gene (Liao et al., 2014) and the expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) value was calculated based on the read count and the length of the gene. The differential expression analysis was performed using the DESeq2 R package (1.20.0), and the genes with the criteria |log2(fold change)| > 1 and padj≤ 0.05 were considered DEGs (Love et al., 2014). The relative expression level log2 (ratios) of all DEGs was clustered by the K-means method. Plant TFs were predicted by hmmscan based on iTAK software (Perez-Rodriguez et al., 2010). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment was analyzed with clusterProfile software and a padj of less than 0.05 was considered significantly enriched (Yu et al., 2012).
Fourteen genes were chosen for the validation of RNA-seq using qRT-PCR. Primer Premier 5.0 software was used to design the primers, which are listed in Table S2. A total of 1 μg RNA per sample was reverse transcribed using a PrimeScript™ reagent Kit with gDNA Eraser (TaKaRa, Beijing, China) in 20 μL of reaction mixture. The qRT-PCR was performed on a LightCycler® 96 instrument (Roche, Basel, Switzerland), using THUNDERBIRD SYBR qPCR Mix (TOYOBO, Shanghai, China) with the following reactions: 95°C 2 min; 95°C 30s, 60°C 10s, and 68°C 10s for 40 cycles. The PCR products were quantified by 2−△△CT method (Livak and Schmittgen, 2001) with normalization to the expression level of SmGAPDH (EGP1067575). Significant differences between groups were assessed by Student’s t-test (p < 0.05) using SPSS 16.0 Statistics (SPSS Inc., Chicago, IL, USA).
The pind mutant was obtained from eggplant ‘14-345’ mutagenized population with EMS. The coloration of the ‘14-345’ fruit peel is photosensitive with white-colored fruit under the calyx. Interestingly, a light purple pigmentation was observed under the calyx of pind, indicating that the coloration of pind was less photosensitive. To further characterize the less-photosensitive coloration in pind, the fruit was bagged to mimic dark conditions. Compared to the white fruit of ‘14-345’, the pind fruit was light purple (Figure 1A). Under natural conditions, the color index, CIRG value, of ‘14-345’ was similar to that of pind, whereas the CIRG value of the pind mutant was significantly higher than that of ‘14-345’ under bagging conditions (Figure 1B).
The anthocyanin contents in the peel of ‘14-345’ and pind were also quantified. The anthocyanin content in the peel of ‘14-345’ and pind was comparable under natural conditions. However, the level of anthocyanin in the bagged eggplants decreased significantly in both ‘14-345’ and pind compared with natural conditions. Anthocyanin content in the peel of bagged pind (21.1 mg/100 g FW) was significantly higher than that in the bagged ‘14-345’ (4.5 mg/100 g FW) (Figure 1C). These results indicated that light was required for the coloration of fruit peel in ‘14-345’, but less so for pind.
To investigate the inheritance of pind, segregating populations were constructed. All the F1 plants from the crosses of pind and ‘14-345’ had purple fruits under bagging conditions, which was phenocopied pind. Out of the F2 plants, 116 had purple bagged fruits, while 37 had white bagged fruits. A chi-squared test revealed that this segregation pattern agreed with the 3:1 Mendelian segregation ratio (P=0.82>0.05, χ2 = 0.05<χ2 0.05,1 = 3.84). In the W-BC1 population (the backcross progeny of F1 with ‘14-345’), 30 plants displayed white wild type fruit, whereas the remaining 23 plants produced purple fruit under bagging conditions. The segregation pattern fitted the ratio 1:1 (P=0.34>0.05, χ2 = 0.93<χ2 0.05,1 = 3.84). All M-BC1 plants, the backcross progeny of F1 with pind, exhibited purple fruits under bagging conditions (Table 1). These results suggested that the mutant gene was a single dominant allele.
To identify the genomic region responsible for less-photosensitive anthocyanin biosynthesis in the fruit peel of pind, BSA-based genome resequencing and MutMap analysis were performed. Quality control resulted in 96,777,013 and 90,196,644 high-quality reads and 98.67% and 98.64% of them were mapped on the eggplant genome (Tables S3, S4), from which 1421190 SNPs were detected in total. Based on the Δ(SNP-index), a single significant interval in a 12.2 Mb region (from 2.6 to 14.8 Mb) on chromosome 10 was identified as a candidate region of the mutation (Figure 2A). There were 2,058 SNPs, including 22 non-synonymous mutation sites.
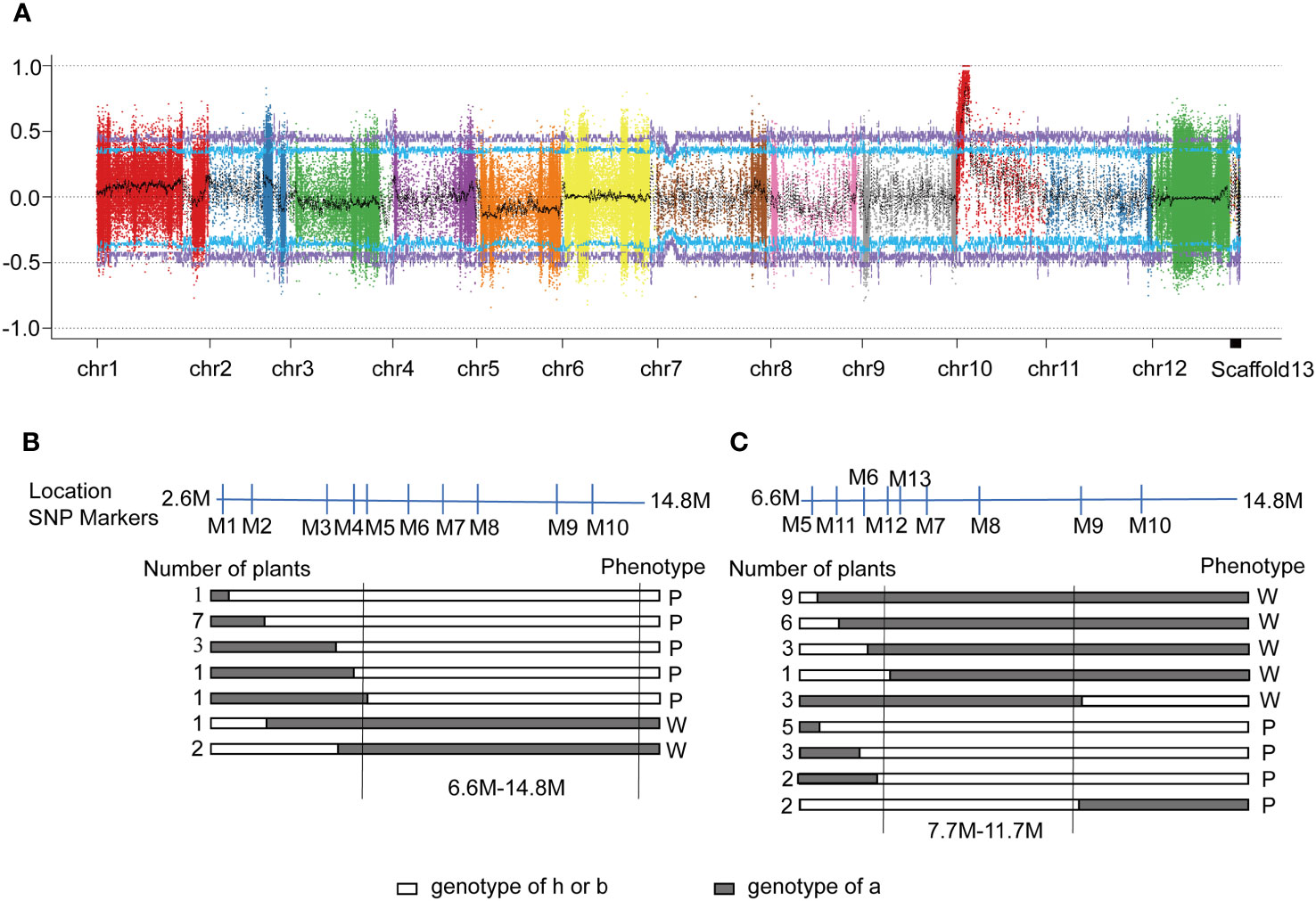
Figure 2 Mapping and identification of the candidate gene. (A) The distribution of Δ(SNP-index) on all chromosomes. X-axis: Chromosome ID; Y-axis: Δ(SNP-index). Each colored dot represents an Δ(SNP-index) value of an SNP site. Schematic representation of allelic segregation of recombinants in F2. (B) and ‘305F2’ (C). a: Genotype of wildtype ‘14-345’ (B) or ‘18-305’ (C); b: Genotype of the pind mutant; h: Heterozygote of ‘14-345’ and pind. P: Purple fruit peel under bagging conditions; W: White fruit peel under bagging conditions.
To identify the causal mutation in this region, several molecular markers were designed to uniformly cover the preliminary mapping interval. Fifteen recombinant plants were identified and the candidate gene was mapped to an 8.2 Mb region (from 6.6 Mb to 14.8 Mb) between markers M5 and M10 (Figure 2B).
To further map the SNP related to less-photosensitive anthocyanin biosynthesis, another F2 population, ‘305F2’ (pind × ‘18-305’), was used to obtain recombinants. The ‘305F2’ population consisted of 408 plants, of which 305 had purple fruit and 103 had white fruit under bagging conditions (Table S5). Thirty-four recombinants were identified and the candidate gene was finally mapped to a 4.0 Mb region between the markers M12 and M9 (Figure 2C), from 7.7 Mb~11.7 Mb on chromosome 10. A total of 224 SNPs that were associated with 75 genes (Table S6), were detected in the candidate interval.
To identify the candidate gene responsible for less-photosensitive anthocyanin biosynthesis in pind, RNA-seq analysis was performed on the fruits of pind (MT) and ‘14-345’ (WT) under bagging (D) and natural (L) conditions for two stages, 2days (2d) and 5 days (5d) after bagging. For both stages, the fruits of the ‘14-345’ bagged were white, whereas the fruits of pind were light purple under bagged conditions. The fruit peel color of ‘14-345’ and pind at the two stages was almost the same under natural conditions (Figure 3A).
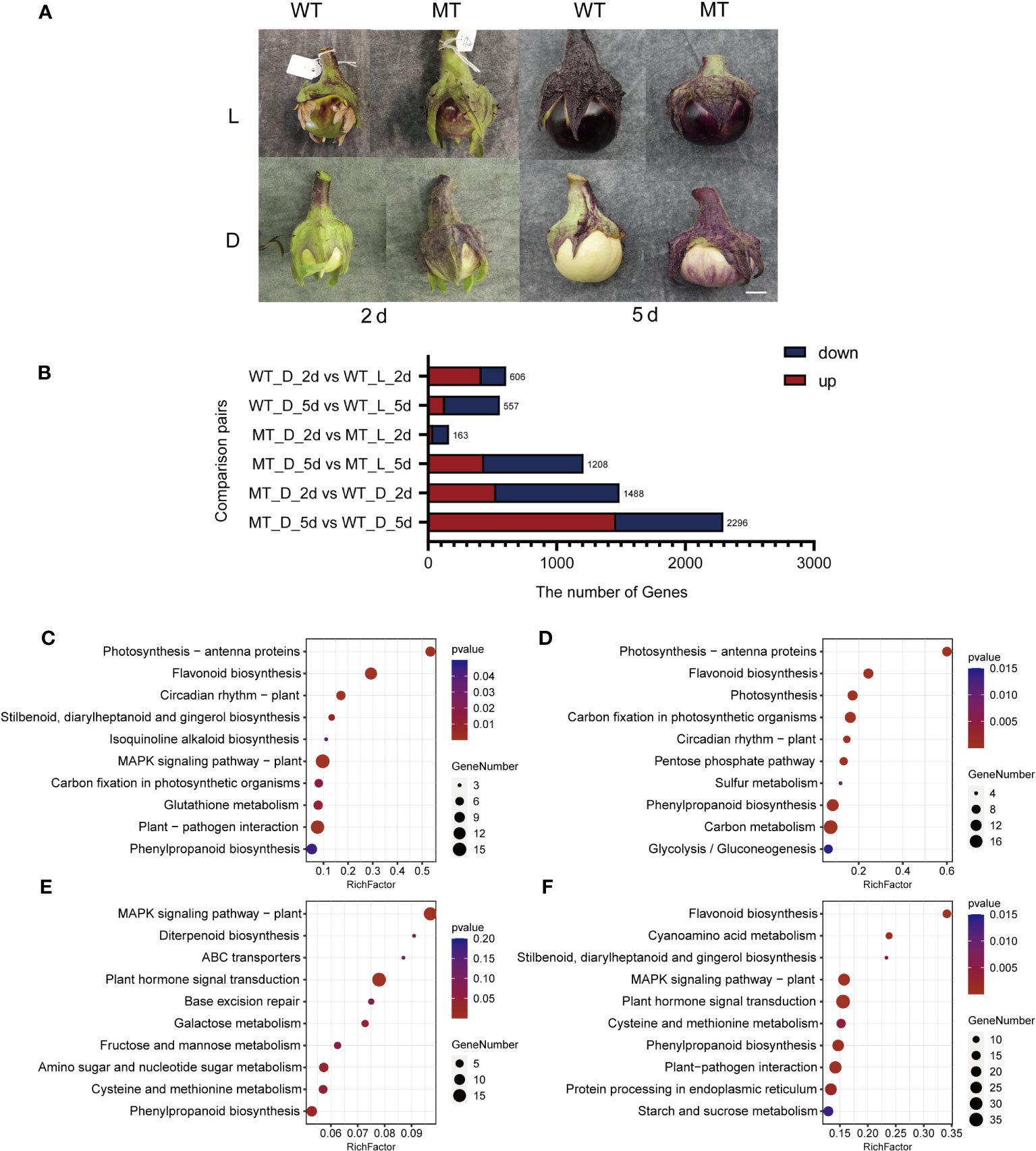
Figure 3 Overview of DEGs identified by RNA-seq analysis in the peel of ‘14-345’ and pind fruits under bagging conditions at two developmental stages. (A) The fruit color of ‘14-345’(WT) and pind (MT) under natural (L) and dark (D) conditions at 2 days (2d) and 5 days (5d) under bagging conditions. Scale bar represents 1 cm. (B) The number of DEGs between samples in each combination. (C–F) KEGG pathway enrichment of DEGs in ‘WT_D_2d vs. WT_L_2d’, ‘WT_D_5d vs.WT_L_5d’, ‘WT_D_2d vs. MT_D_2d’, and ‘WT_D_5d vs. MT_D_5d’.
To identify the key differentially expressed genes (DEGs) between the fruit peel of ‘14-345’ and pind, pairwise comparisons between bagged ‘14-345’ and pind at each stage (MT_D_2d vs. WT_D_2d, MT_D_5d vs. WT_D_5d), bagged and natural ‘14-345’ at each stage (WT_D_2d vs. WT_L_2d, WT_D_5d vs. WT_L_5d), and bagged and natural pind at each stage (MT_D_2d vs. MT_L_2d, MT_D_5d vs. MT_L_5d) were analyzed. A total of 4,338 DEGs were detected in the above-mentioned six combinations (Figure 3B).
A total of 415 and 191 genes were significantly induced and inhibited in the peel of ‘14-345’ under bagging conditions on 2d, respectively, compared with natural conditions (WT_D_2d vs. WT_L_2d). KEGG enrichment analysis showed that the pathways ‘flavonoid biosynthesis’ and ‘phenylpropanoid biosynthesis’ were significantly enriched (Figure 3C). There were 557 DEGs in the ‘WT_D_5d vs. WT_L_5d’ combination, in which the pathways ‘flavonoid biosynthesis’ and ‘phenylpropanoid biosynthesis’ were also significantly enriched (Figure 3D). The decreased expression of the eight flavonoid biosynthetic genes, including CHS (EGP24357, EGP16216), CHI (EGP24232, EGP22200), F3H (EGP30923), F3′5′H (EGP32037), DFR (EGP31016), and ANS (EGP18904) (Figures S2A, B), indicated that the anthocyanin biosynthetic pathway was repressed in ‘14-345’ under dark conditions.
A total of 1,488 DEGs were detected in the ‘MT_D_2d vs. WT_D_2d’ combination, which were significantly enriched in the ‘MAPK signal pathway’ and ‘plant signal transduction pathway’ (Figure 3E). The ‘MT_D_5d vs. WT_D_5d’ combination produced 1,463 DEGs, and the enrichment analysis revealed that among all of the pathways identified, the ‘flavonoid biosynthesis pathway’ were the most significantly enriched (Figure 3F). The expression of structural genes 4CL (EGP10904), CHS (EGP24357), CHI (EGP24232, EGP22200), F3H (EGP30923), F3′5′H (EGP32037), DFR (EGP31016), and ANS (EGP18904) (Figure S2C) involved in anthocyanin biosynthesis was significantly higher in the mutant than in the wild type at 5d under bagging conditions.
Due to the difference in fruit color between ‘14-345’ and pind under bagged conditions (MT_D vs. WT_D) and the differences in the color of ‘14-345’ fruit between bagged and natural conditions (WT_D vs. WT_L), we screened for common DEGs in these combinations. The Venn diagram clearly illustrated the relationships of DEGs between different comparisons, and 238 DEGs (Figure 4A; Table S7) and 145 DEGs (Figure 4B; Table S8) were commonly shared by ‘MT_D vs. WT_D’ and ‘WT_D vs. WT_L’ on 2d and 5d, respectively. Moreover, 35 DEGs were commonly shared by these four combinations (Figure 4C; Table S9), which may participate in anthocyanin biosynthesis in pind under dark conditions. Therefore, these 35 DEGs, including 12 structural genes and three TFs, were used for further analysis.
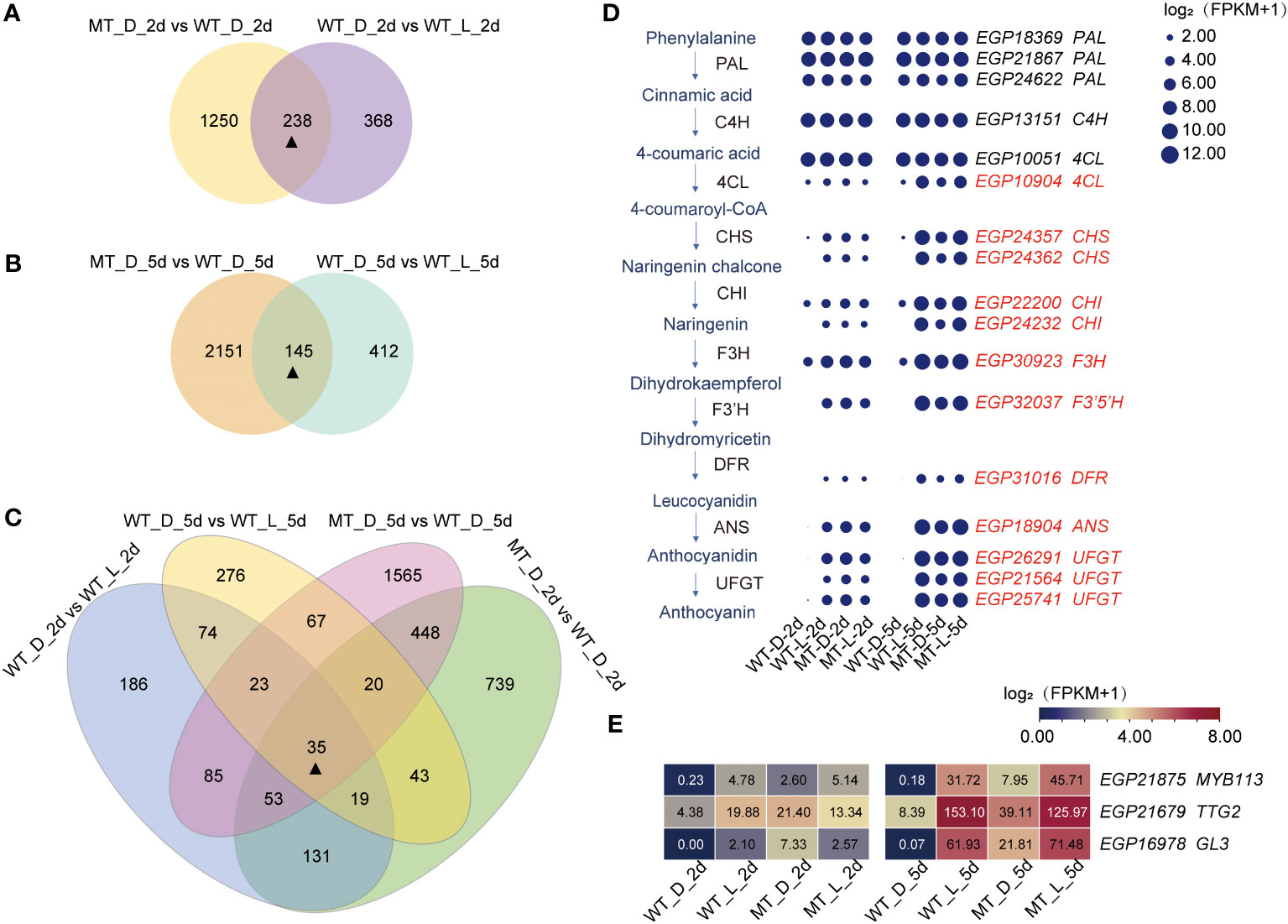
Figure 4 Identification of the DEGs related to less-photosensitive anthocyanin biosynthesis in pind. (A–C) Venn diagram of the different comparisons of DEGs. (D) Expression pattern analysis of structural genes involved in less-photosensitive anthocyanin biosynthesis in fruit peel of pind. Note: Red gene ID represents candidate DEGs related to less-photosensitive anthocyanin biosynthesis in the fruit peel of pind. The size of the circle represents log2 (FPKM+1); (E) Expression pattern analysis of TFs involved in less-photosensitive anthocyanin biosynthesis in the fruit peel of pind. The number in the box indicates the FPKM value.
Under natural conditions, there was no significant difference in the expression of structural genes between ‘14-345’ and pind fruit peel. Bagging inhibited the transcripts of most of the structural genes, including 4CL, CHS, CHI, F3H, F3′5′H, DFR, and ANS in ‘14-345’ while not in pind, as the transcriptional level of structural genes were extremely higher in pind than in ‘14-345’ (Figure 4D). These data suggest that ‘14-345’ and pind have different responses to bagging in terms of anthocyanin biosynthesis. The highly expressed structural genes in pind under dark conditions showed pind was less photosensitive.
TFs can regulate the expression of structural genes by directly binding to cis-regulatory elements in the promoter of the genes, which play important roles in plant growth and development (Chen et al., 2019; Jiang et al., 2022; Liu et al., 2023). In this study, MYB TF SmMYB113 (EGP21875), WRKY TF SmTTG2 (EGP21679), and bHLH TF SmGL3 (EGP16978) were identified from the 35 DEGs as the putative main regulators of the anthocyanin biosynthesis in a less-photosensitive manner in the fruit peel of pind. Under natural conditions, the expression of SmMYB113, SmTTG2, and SmGL3 was comparable between ‘14-345’ and pind. Under bagging conditions, SmMYB113, SmTTG2, and SmGL3 was down-regulated both in ‘14-345’ and pind compared to natural conditions (Figure 4E). Notably, the expression levels of MYB113, TTG2, and GL3 were significantly increased in the peel of pind compared to ‘14-345’ under bagged conditions (Figure 4E).
To confirm the reliability of the RNA-seq data, six structural genes (C4H, CHI, F3H, DFR, ANS, and 5GT), five TFs (MYB113, GL2, GL3, TT8, and TTG2), and three genes involved in light signaling (COP1, UVR8, and CRY3) were selected to analyze expression profiles by qRT-PCR. The transcript abundances of these genes determined using qRT-PCR agreed with those determined from transcriptome sequencing (Figure 5), indicating the reliability of the RNA-seq results.
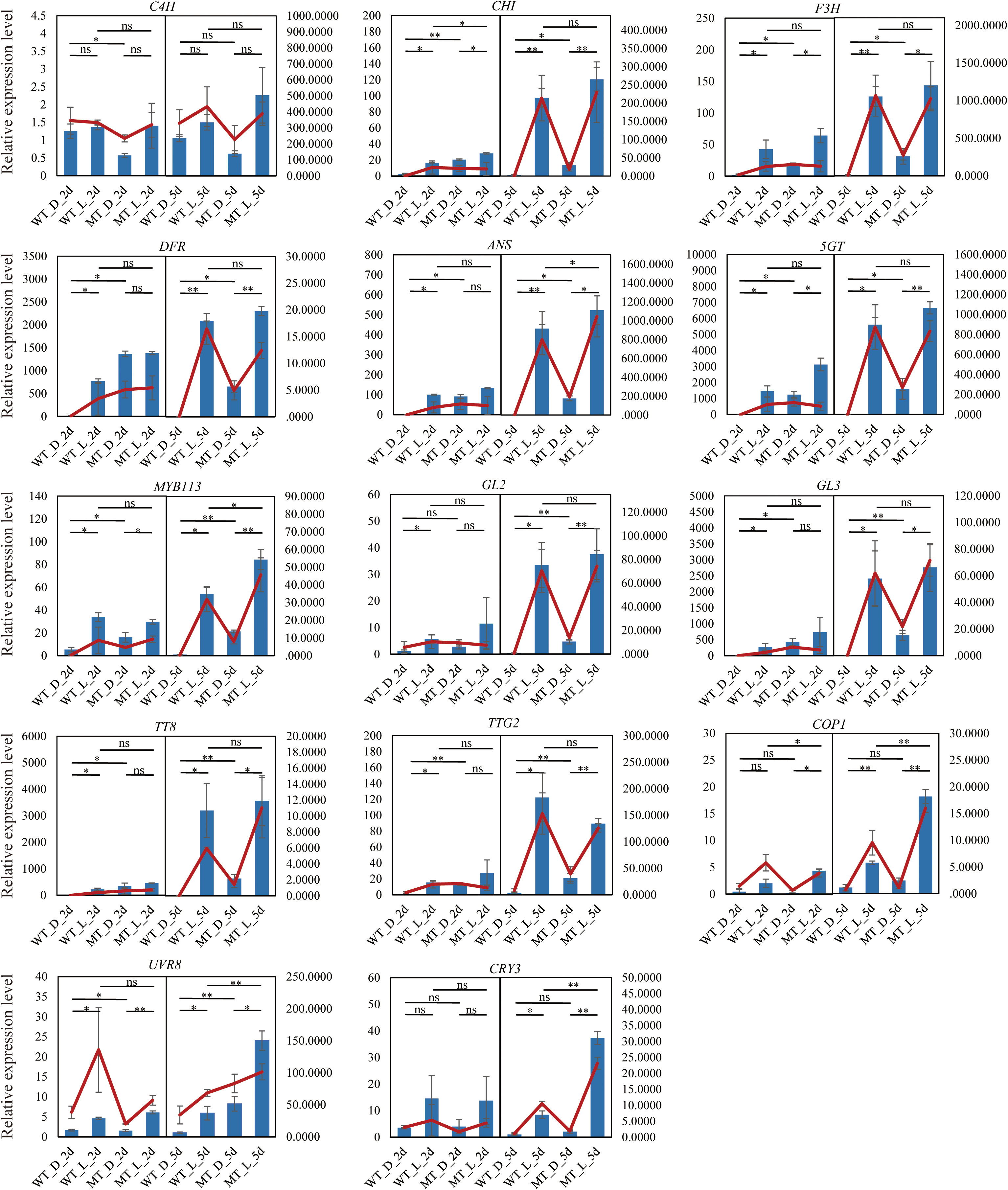
Figure 5 Validation of RNA-seq results using qRT-PCR. Y-axis: relative expression. The blue bar and red line represent the data from qRT-PCR and RNA-seq data, respectively. Asterisks indicate significant differences determined by the Student’s t-test (*P < 0.05, **P < 0.01). ns, no significance.
Based on the gene mapping analysis, there were 75 genes in the candidate region (Table S6), of which only nine (Figure 6) were expressed in the eggplant peel according to the transcriptome analysis. Twenty-seven SNPs were detected to be associated with these nine genes, and only one non-synonymous SNP (within EGP21953) was identified. The gene EGP21953 (Table 2) had an amino acid residue substitution at aa-262 (Ala-to-Tyr mutation). According to the annotated information, EGP21953 encodes a CAAX amino-terminal protease family protein. However, there was no significant difference in EGP21953 expression between ‘14-345’ and pind, neither in natural nor in bagged conditions.
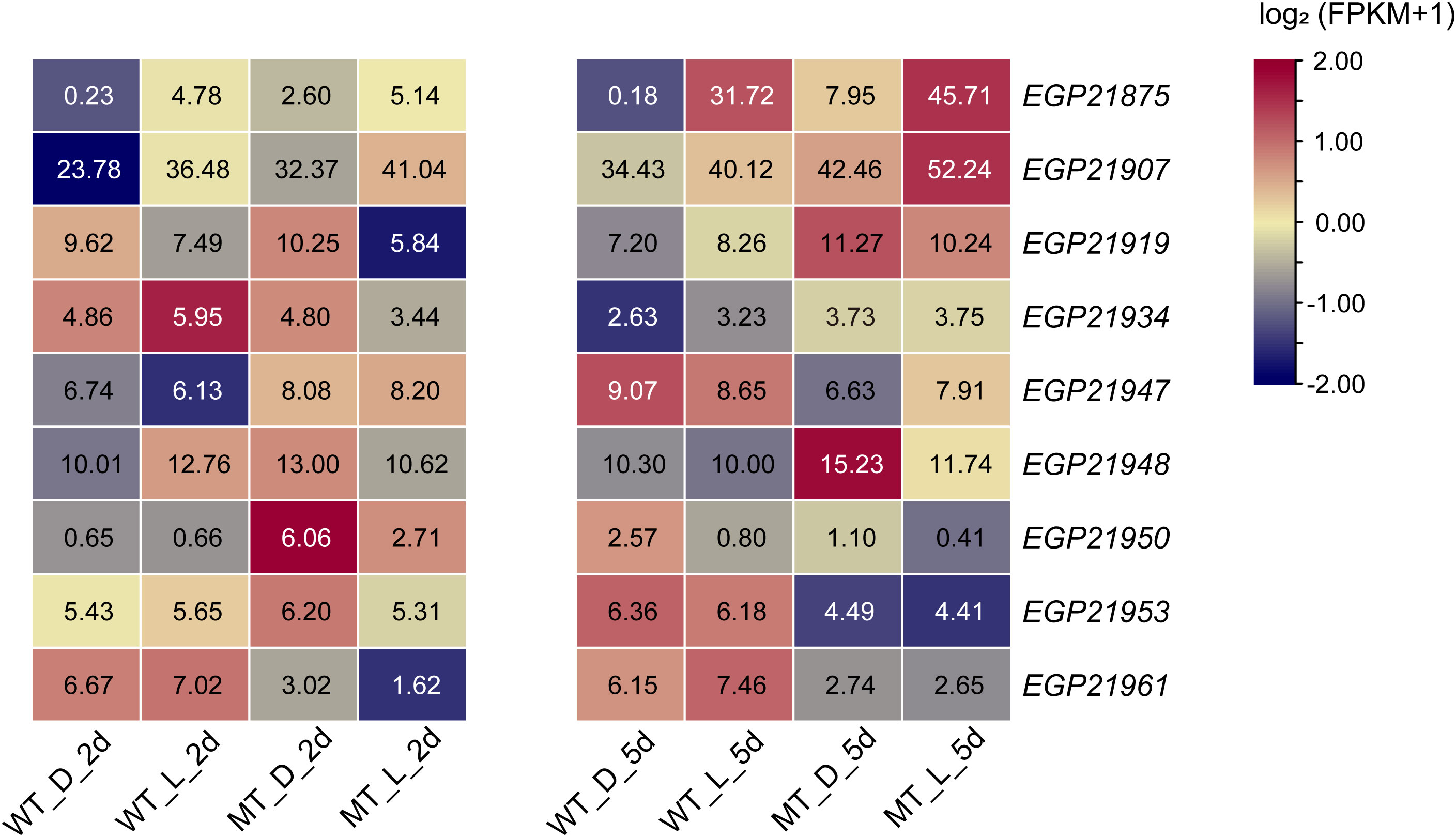
Figure 6 Expression pattern analysis of the genes in the candidate region. The number in the box indicates the FPKM value.
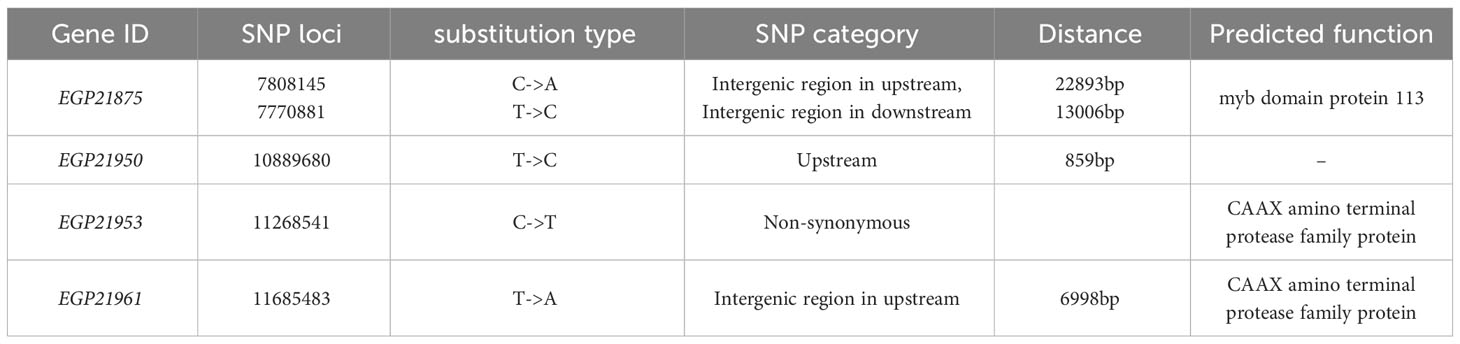
Table 2 Candidate genes related to less-photosensitive anthocyanin synthesis in the fruit peel of pind.
Among the other eight genes, whose related SNPs were detected in intergenic regions or introns, only EGP21875, EGP21950, and EGP21961, were differentially expressed between the peel of pind and ‘14-345’ (Figure 6; Table 2). EGP21875 encodes an MYB TF, “SmMYB113”, which has been shown to participate in the regulation of anthocyanin biosynthesis (Zhang et al., 2014; Jiang et al., 2016; He et al., 2019; Yang et al., 2022). The expression of SmMYB113 was inhibited in the fruit peel of ‘14-345’ under bagged conditions. Interestingly, SmMYB113 showed significantly higher expression in the peel of pind than in ‘14-345’ at both 2d and 5d stages, pinpointing the activation of SmMYB113 in pind in darkness. There was one SNP 22.9 kb upstream of the start codon and one SNP 13 kb downstream of the stop codon of SmMYB113, respectively. Because the intergenic SNPs play a potentially important role in phenotype variation in plants by altering gene expression (Schwartz et al., 2009; Ding et al., 2012; Li et al., 2012; Wang et al., 2015), these two SNPs in the intergenic region associated with SmMYB113 may affect its expression.
EGP21950 encoded an unknown functional protein, and there was one SNP 859 bp upstream of the start codon of EGP21950. The expression of EGP21950 was significantly up-regulated in the fruit peel of pind in comparison to ‘14-345’ at 2d under bagged conditions, but its expression level showed no significant difference between ‘14-345’ and pind under natural conditions and at 5d under bagged conditions. In addition to EGP21953, EGP21961 also encoded a CAAX amino-terminal protease family protein, and its associated SNP was about 7 kb upstream of the initiation codon. EGP21961 was down-regulated significantly in the peel of pind compared to ‘14-345’, in both natural and bagged conditions.
Anthocyanin is an important flavonoid type pigment in flowers and fruits, with light being an essential regulator of anthocyanin accumulation (Takos et al., 2006; Meng and Liu, 2015; Bai et al., 2017; Kim et al., 2021; Ma et al., 2021a). Proper anthocyanin accumulation in fruits or flowers can be hindered by low-light conditions (caused by weather and climate, the cultivation facility, or plant morphology) (Hong et al., 2015; Cao et al., 2016; Zhang et al., 2021a; Guo et al., 2022). Therefore, understanding the mechanism of less-and non-photosensitive anthocyanin biosynthesis and creating the less- and non-photosensitive cultivars will provide valuable resources for the breeding of low-light tolerant varieties
In this study, we discovered a less-photosensitive anthocyanin biosynthesis mutant (pind) in eggplant, and a candidate gene that was physically mapped to 7.7 ~11.7 Mb on chromosome 10 (Figure 2C). This result was consistent with previous reports, showing that the candidate genes that may correspond to loci controlling fruit peel color under calyx in eggplants were mapped to chromosome 10 (Zhang et al., 2021b; Mangino et al., 2022; Qiao et al., 2022). In addition to chromosome 10, the candidate genes controlling the fruit peel color under calyx in eggplants were also found on other chromosomes, including chromosome 1, 3, 4, and 11 (Mangino et al., 2022; Qiao et al., 2022). However, the above studies did not distinguish materials as less-photosensitive or non-photosensitive materials. When comparing the phenotypic differences between the less-photosensitive or non-photosensitive materials, it was observed that under bagging conditions, fruit coloration became significantly lighter for less-photosensitive materials (Figure 1A), whereas there was no significant difference for non-photosensitive materials (He et al., 2019; He et al., 2022). Recent studies have found that the candidate gene related to non-photosensitive anthocyanin biosynthesis in eggplant is SmFTSH10 (He et al., 2022), which was physically close to, but outside the candidate interval 7.7Mb-11.7Mb (Figure 3C) for less-photosensitive anthocyanin biosynthesis in this study, indicating less- and non-photosensitive anthocyanin biosynthesis in eggplant may be controlled by different locus.
Through MutMap and transcriptome analysis, we obtained four candidate genes: EGP21875, EGP21950, EGP21953, and EGP21961 (Table 2). EGP21953 and EGP21961 are homologs of the gene AT1G14270.1 in Arabidopsis, which is a CAAX amino-terminal protease family member. However, at present, there is no literature or report indicating that the CAAX amino-terminal protease family proteins under investigation exert regulatory control over anthocyanin biosynthesis. EGP21950 encodes a protein with an unknown function, that contained a DUF616 (Protein of unknown function) domain (Figure S3). Additionally, proteins encoded by structural genes and regulatory genes didn’t possess CAAX prenyl endopeptidase-like domains or UDF616 domains (Figure S3), indicating their low likelihood to bind with CAAX amino-terminal protease family proteins or DUF616 proteins. Therefore, we speculate that EGP21950, EGP21953, or EGP21961 had minimal potential in contributing to the genetic basis of less-photosensitivity.
The gene EGP21875 encoded an MYB TF SmMYB113, homologous to AtMYB113 in Arabidopsis. In this study, SmMYB113 was hardly expressed in the fruit peel of wild type ‘14-345’ under bagged conditions, whereas its expression was considerably up-regulated in the pind mutant compared to ‘14-345’. It was reported that overexpression of SmMYB113 (also known as SmMYB1) resulted in the up-regulation of SmCHS, SmCHI, SmF3H, SmANS, and other genes, and a high level of anthocyanin accumulation (Zhang et al., 2014; Yang et al., 2022). Thus, anthocyanin-pigmentation in pind in the dark may be related to the mutations of SmMYB113. Collectively, we speculated that EGP21875 (SmMYB113) was the best candidate gene for the regulation of less-photosensitive anthocyanin biosynthesis in the fruit peel of pind.
In addition, the SNPs related to EGP21875 (SmMYB113) were more than 13kb from the coding region. Some studies have shown that a non-coding region far from the gene promoter may change the activity of neighboring genes. For example, the Teosinte Branched 1 (TB1) gene is mainly responsible for a major-effect QTL that controls morphological differences in plant architecture between maize (Zea mays subsp. mays) and its wild relative, teosinte (Z. mays subsp. mexicana and subsp. parviglumis) (Clark et al., 2004; Studer et al., 2011); the expression of tb1 was altered by a sequence >41kb upstream of tb1 (Clark et al., 2006). In addition, booster1 (b1) was a regulator gene responsible for the biosynthesis of flavonoid pigments in maize; a 6-kb region at about 100kb upstream of the transcription start site of gene b1 was found to be a hepta-repeat enhancer, which may increase the transcription initiation rate (Stam et al., 2002). The non-coding region located distally from the gene promoter could be considered as a putative cis-regulatory element that modulates gene expression through recruitment of transcription factors (Stadhouders et al., 2014; Visser et al., 2014; Roberts et al., 2016). Studies have shown that natural variation in cis-regulatory regions of genes played important roles in phenotypic variations by altering gene expression (Frary et al., 2000; Schwartz et al., 2009; Muños et al., 2011; Van Der Knaap et al., 2014; Chu et al., 2019). Taken together, we speculated that the position of the SNPs in the intergenic region of EGP21875 (SmMYB113) might coincide with a regulatory element that controls the SmMYB113 expression level.
Under bagging conditions, ‘14-345’ fruits exhibited white coloration while the pind mutants displayed purple coloration. Consequently, under bagging conditions, the structural genes were expressed at a minimal level in the peel of ‘14-345’ but showed higher expression in pind (Figure 4D). It is well known that the expression of structural genes is directly regulated by transcription factors, among which MYB TFs, bHLH TFs, and WD40 are key modulators (Shan et al., 2019; Li et al., 2020). In this study, expression of SmMYB113, SmGL3 (bHLH) and SmTTG2 (WRKY) was significantly higher in pind peel than ‘14-345’ in darkness (Figure 4E). Moreover, bHLH TF SmTT8 was inactive in ‘14-345’ under bagged conditions, but strongly activated in pind on 5d (Table S8; Figure 5). Therefore, under bagging conditions, the inactive transcription factors, such as SmMYB113, SmGL3, SmWRY44 and SmTT8, cannot stimulate the expression of structural genes, which is the underlying reason for the inability of ‘14-345’ to produce anthocyanins under bagging conditions (Figure 7A).
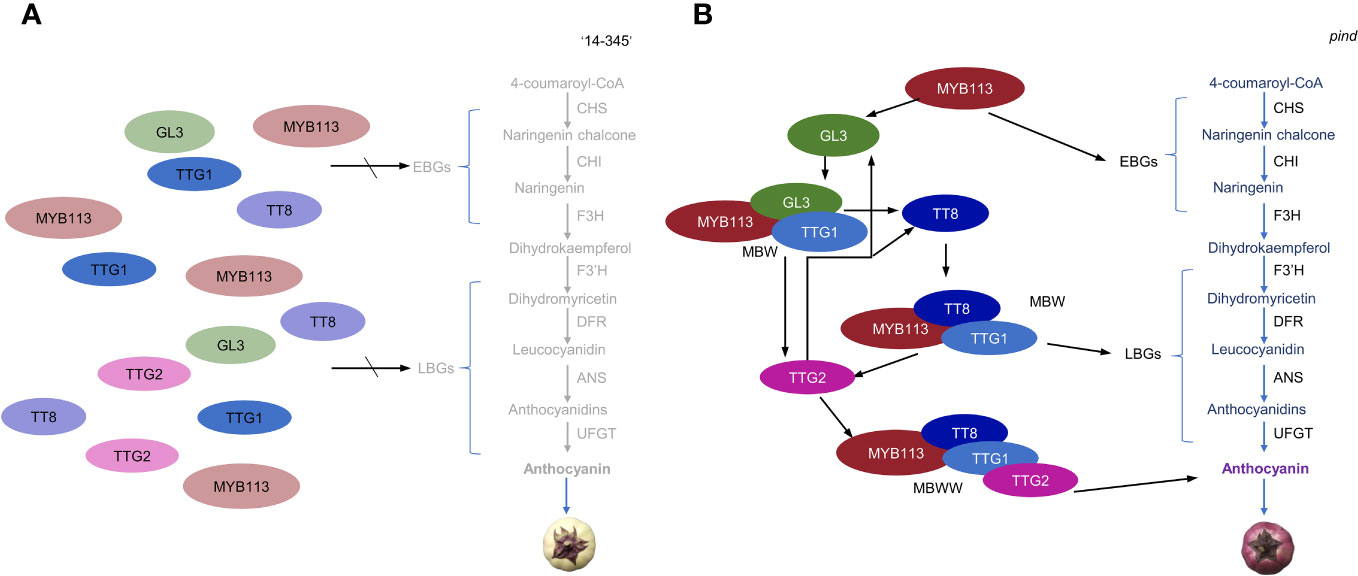
Figure 7 A hypothetical model of the mechanism for anthocyanin biosynthesis in the eggplant peel of ‘14-345’ (A) and pind (B) under dark conditions. (A) Under bagging conditions, in ‘14-345’, the inactivated state of transcription factors, SmMYB113, SmGL3, SmTTG2 and SmTT8, cannot promote the expression of structural genes, consequently resulting in the inhibition of structural gene expression and subsequently impacting the biosynthesis of anthocyanins. (B) TF MYB113 promotes the expression of EBGs and activates the expression of GL3. MYB113, and GL3 form a MBW complex with the WD40 protein TTG1 to promote the expression of TT8. Then MYB113, TT8, and TTG1 will form a new MBW complex to promote the expression of LBGs. TTG2 is activated by MBW and forms the MBWW complex, and the MBWW complex promotes the expression of genes related to anthocyanin transport. The highly expressed structural genes in pind under dark conditions led to accumulation of anthocyanin. The positive regulation of genes is marked in black arrows. The gray font represents genes with very low expression, while the lighter-colored circles indicate transcription factors with low activity.
In Arabidopsis, AtMYB113 interacts with AtTT8 or AtGL3 and AtTTG1, forming the MBW complex, to regulate the expression of late biosynthesis genes (LBGs) (Ramsay and Glover, 2005). The MYB TF is the main determinant of the MBW complex, and MYBs control the expression of early biosynthesis genes (EBGs) and LBGs (Stracke et al., 2007; Gonzalez et al., 2008; Tang et al., 2020). In tomato, after the overexpression of SlANT1 (a homolog of AtMYB113), the expression of the structural genes, and the bHLH gene SlAN1 (a homolog of AtTT8) was significantly up-regulated (Mathews et al., 2003; Kiferle et al., 2015). SlAN1 probably activates the expression of structural genes through the MYB-AN1-WD40 complex, whereas SlJAF13 (a homolog of AtGL3) regulates the transcription of SlAN1 through the MYB-SlJAF13-WD40 complex (Montefiori et al., 2015; Liu et al., 2018). Therefore, we speculated that under bagged conditions, the up-regulation of EBGs was mainly due to the activation of SmMYB113 in the peel of pind; the LBGs were directly regulated by the MBW complex composed of SmMYB113, SmTT8, and WD40 (SmTTG1), whereas the expression of SmTT8 was activated by the MBW complex SmMYB113-SmGL3-SmTTG1; and SmGL3 was activated by SmMYB113 (Figure 7B).
In addition, WRKY44 was reported to regulate anthocyanin accumulation. In tobacco, the coloration of flowers was observed to increase upon overexpression of TTG2, whereas a decrease in flower coloration was achieved through silencing TTG2 (Li et al., 2017). Anthocyanin accumulation was induced in tobacco leaves after transient overexpression of kiwifruit WRKY44 (Peng et al., 2020). WRKY44 was reported to interact with BMW to regulate the corresponding biological processes (Pesch et al., 2014; Gonzalez et al., 2016; Verweij et al., 2016; Lloyd et al., 2017). In summary, it was suggested that under bagging conditions, the expression of SmTTG2 was up-regulated by the MBW complex and SmTTG2 could bind to the MBW complex to promote the expression of structural genes in pind (Figure 7B).
Based on the expression patterns and putative function of the genes in other plants, we proposed a working model that describes the regulatory mechanism of anthocyanin biosynthesis in the peel of ‘14-345’ and pind under dark conditions (Figure 7). It is indicated that the activation of SmMYB113, SmTT8, SmTTG2 and SmTT8 is the necessary factor for anthocyanin accumulation in the dark in eggplant. Considering that SmMYB113 was the most upstream in the pathway, it is postulated that SmMYB113 was the first transcription factor to be activated and serves as the most crucial regulator. Combined with gene mapping analysis, it is speculated that the activation of SmMYB113 in pind under bagged conditions may be related to the SNP of the intergenic region of EGP21875.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1014581, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1013920.
LL: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing. JM: Conceptualization, Methodology, Software, Writing – original draft. QL: Conceptualization, Visualization, Writing – review & editing. YL: Conceptualization, Methodology, Writing – review & editing. SL: Data curation, Investigation, Writing – review & editing. NL: Visualization, Writing – review & editing. SX: Visualization, Writing – review & editing. YW: Visualization, Writing – review & editing. SS: Funding acquisition, Supervision, Writing – review & editing. AB: Conceptualization, Supervision, Writing – review & editing. JZ: Funding acquisition, Supervision, Writing – review & editing. XC: Data curation, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by S&T Program of Hebei (Grant No. 21326309D), S&T Program of Hebei (Grant No. 20592901D), and the Post-graduate’s Innovation Fund Project of Hebei (Grant No. CXZZBS2019098).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1282661/full#supplementary-material
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 26, 962–980. doi: 10.1105/tpc.113.122069
Bai, S., Sun, Y., Qian, M., Yang, F., Ni, J., Tao, R., et al. (2017). Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep-UK. 7, 1–15. doi: 10.1038/s41598-017-00069-z
Bai, S., Tao, R., Tang, Y., Yin, L., Ma, Y., Ni, J., et al. (2019). BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 17, 1985–1997. doi: 10.1111/pbi.13114
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bhatia, C., Gaddam, S. R., Pandey, A., Trivedi, P. K. (2021). COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis. Plant Sci. 303, 110760. doi: 10.1016/j.plantsci.2020.110760
Cao, L., Xu, X., Chen, S., Ma, H. (2016). Cloning and expression analysis of Ficus carica anthocyanidin synthase 1 gene. Sci. Hortic-Amsterdam. 211, 369–375. doi: 10.1016/j.scienta.2016.09.015
Carreno, J., Martinez, A., Almela, L., Fernandezlopez, J. A. (1995). Proposal of an index for the objective evaluation of the color of red table grapes. Food Res. Int. 28, 373–377. doi: 10.1016/0963-9969(95)00008-A
Chen, L., Hu, B., Qin, Y., Hu, G., Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Bioch 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Chen, X. (2015). “Study on genetic diversity and genetic linkage map construction in eggplant,” in [dissertation/doctor's thesis] (Baoding (Hebei)]: Hebei Agricultural University).
Cheng, G. W., Patrick, J. B. (1991). Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 116, 865–869. doi: 10.21273/JASHS.116.5.865
Chu, Y. H., Jang, J. C., Huang, Z., van der Knaap, E. (2019). Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct. 3, e00142. doi: 10.1002/pld3.142
Clark, R. M., Linton, E., Messing, J., Doebley, J. F. (2004). Pattern of diversity in the genomic region near the maize domestication gene tb1. P Natl. Acad. Sci. U.S.A. 101, 700–707. doi: 10.1073/pnas.2237049100
Clark, R. M., Wagler, T. N., Quijada, P., Doebley, J. (2006). A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38, 594–597. doi: 10.1038/ng1784
Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U S A. 109, 2654–2659. doi: 10.1073/pnas.1121374109
Dooner, H. K., Robbins, T. P., Jorgensen, R. A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 99. doi: 10.1146/annurev.ge.25.120191.001133
Fan, X., Fan, B., Wang, Y., Yang, W. (2016). Anthocyanin accumulation enhanced in Lc-transgenic cotton under light and increased resistance to bollworm. Plant Biotechnol. Rep. 10, 1–11. doi: 10.1007/s11816-015-0382-3
Frary, A., Nesbitt, T. C., Frary, A., Grandillo, S., Knaap, E. V. D., Cong, B., et al. (2000). fw2. 2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 289, 85–88. doi: 10.1126/science.289.5476.85
Gao-Takai, M., Katayama-Ikegami, A., Matsuda, K., Shindo, H., Uemae, S., Oyaizu, M. (2019). A low temperature promotes anthocyanin biosynthesis but does not accelerate endogenous abscisic acid accumulation in red-skinned grapes. Plant Sci. 283, 165–176. doi: 10.1016/j.plantsci.2019.01.015
Gonzalez, A., Brown, M., Hatlestad, G., Akhavan, N., Smith, T., Hembd, A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Guo, X., Shakeel, M., Wang, D., Qu, C., Yang, S., Ahmad, S., et al. (2022). Metabolome and transcriptome profiling unveil the mechanisms of light-induced anthocyanin synthesis in rabbiteye blueberry (vaccinium ashei: Reade). BMC Plant Biol. 22, 1–14. doi: 10.1186/s12870-022-03585-x
Guo, X., Wang, Y., Zhai, Z., Huang, T., Zhao, D., Peng, X., et al. (2018). Transcriptomic analysis of light-dependent anthocyanin accumulation in bicolored cherry fruits. Plant Physiol. Biochem. 130, 663–677. doi: 10.1016/j.plaphy.2018.08.016
He, Y., Chen, H., Zhou, L., Liu, Y., Chen, H. (2019). Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genomics 20, 678. doi: 10.1186/s12864-019-6023-4
He, Y., Li, S., Dong, Y., Zhang, X., Li, D., Liu, Y., et al. (2022). Fine mapping and characterization of the dominant gene SmFTSH10 conferring non-photosensitivity in eggplant (Solanum melongena L.). Theor. Appl. Genet. 135 (7), 2187–2196. doi: 10.1007/s00122-022-04078-z
Hoai Nguyen, N. (2020). HY5, an integrator of light and temperature signals in the regulation of anthocyanins biosynthesis in Arabidopsis. AIMS Mol. Science. 7, 70–81. doi: 10.3934/molsci.2020005
Hong, Y., Li, M., Dai, S. (2019). iTRAQ-based protein profiling provides insights into the mechanism of light-induced anthocyanin biosynthesis in chrysanthemum (Chrysanthemum× morifolium). Genes-Basel. 10, 1024–1042. doi: 10.3390/genes10121024
Hong, Y., Tang, X., Huang, H., Zhang, Y., Dai, S. (2015). Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genomics 16, 202. doi: 10.1186/s12864-015-1428-1
Huang, H., Li, Y., Dai, S. (2016). Investigation of germplasm in chrysanthemum cultivars with light-independent coloration. II Int. Symposium Germplasm Ornamentals 1185, 55–64. doi: 10.17660/ActaHortic.2017.1185.9
Jiang, H., Zhou, L. J., Gao, H. N., Wang, X. F., Li, Z. W., Li, Y. Y. (2022). The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 189 (4), 2044–2060. doi: 10.1093/plphys/kiac211
Jiang, M., Ren, L., Lian, H., Liu, Y., Chen, H. (2016). Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 249, 46–58. doi: 10.1016/j.plantsci.2016.04.001
Kiferle, C., Fantini, E., Bassolino, L., Povero, G., Spelt, C., Buti, S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS One 10, e0136365. doi: 10.1371/journal.pone.0136365
Kim, M. J., Kim, P., Chen, Y., Chen, B., Yang, J., Liu, X., et al. (2021). Blue and UV-B light synergistically induce anthocyanin accumulation by co-activating nitrate reductase gene expression in Anthocyanin fruit (Aft) tomato. Plant Biol. 23, 210–220. doi: 10.1111/plb.13141
Krol, A. R. V. D., Mur, L. A., Beld, M., Moi, J. N. M., Stuitje, A. R. (1990). Flavonoid genes in Petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 2, 291–299. doi: 10.1105/tpc.2.4.291
Li, S. F., Allen, P. J., Napoli, R. S., Browne, R. G., Pham, H., Parish, R. W. (2020). MYB–bHLH–TTG1 regulates Arabidopsis seed coat biosynthesis pathways directly and indirectly via multiple tiers of transcription factors. Plant Cell Physiol. 61, 1005–1018. doi: 10.1093/pcp/pcaa027
Li, P., Chen, X., Sun, F., Dong, H. (2017). Tobacco TTG2 and ARF8 function concomitantly to control flower coloring by regulating anthocyanin synthesis genes. Plant Biol. 19, 525–532. doi: 10.1111/plb.12560
Li, J., He, Y.-J., Zhou, L., Liu, Y., Jiang, M., Ren, L., et al. (2018). Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genomics 19, 1–12. doi: 10.1186/s12864-018-4587-z
Li, D., Qian, J., Li, W., Yu, N., Gan, G., Jiang, Y., et al. (2021). A high-quality genome assembly of the eggplant provides insights into the molecular basis of disease resistance and chlorogenic acid synthesis. Mol. Ecol. Resour 21 (4), 1274–1286. doi: 10.1111/1755-0998.13321
Li, X., Zhu, C., Yeh, C. T., Wu, W., Takacs, E. M., Petsch, K. A., et al. (2012). Genic and nongenic contributions to natural variation of quantitative traits in maize. Genome Res. 22, 2436–2444. doi: 10.1101/gr.140277.112
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, W., Mei, Z., Yu, L., Gu, T., Li, Z., Zou, Q., et al. (2023). The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples. Hortic. Res. 10 (5), uhad049. doi: 10.1093/hr/uhad049
Liu, C., Sun, J., Lu, Y., Bo, Y. (2016). Effects of anthocyanin on serum lipids in dyslipidemia patients: a systematic review and meta-analysis. PloS One 11, e0162089. doi: 10.1371/journal.pone.0162089
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 6, 52–68. doi: 10.3389/fchem.2018.00052
Liu, B., Wang, L., Wang, S., Li, W., Liu, D., Guo, X., et al. (2019). Transcriptomic analysis of bagging-treated ‘Pingguo’ pear shows that MYB4-like1, MYB4-like2, MYB1R1 and WDR involved in anthocyanin biosynthesis are up-regulated in fruit peels in response to light. Sci. Hortic-Amsterdam. 244, 428–434. doi: 10.1016/j.scienta.2018.09.040
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ma, Y., Ma, X., Gao, X., Wu, W., Zhou, B. (2021b). Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 22, 11116–11131. doi: 10.3390/ijms222011116
Ma, A., Wang, D., Lu, H., Wang, H., Qin, Y., Hu, G., et al. (2021a). LcCOP1 and LcHY5 control the suppression and induction of anthocyanin accumulation in bagging and debagging litchi fruit pericarp. Sci. Hortic-Amsterdam. 287, 110281–110290. doi: 10.1016/j.scienta.2021.110281
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Mangino, G., Arrones, A., Plazas, M., Pook, T., Prohens, J., Gramazio, P., et al. (2022). Newly developed MAGIC population allows identification of strong associations and candidate genes for anthocyanin pigmentation in eggplant. Front. Plant Sci. 13, 847789. doi: 10.3389/fpls.2022.847789
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 15, 1689–1703. doi: 10.1105/tpc.012963
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Meng, L. S., Liu, A. (2015). Light signaling induces anthocyanin biosynthesis via AN3 mediated COP1 expression. Plant Signal Behav. 10, e1001223. doi: 10.1080/15592324.2014.1001223
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Muños, S., Ranc, N., Botton, E., Bérard, A., Rolland, S., Duffé, P., et al. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254. doi: 10.1104/pp.111.173997
Naing, A. H., Kim, C. K. (2021). Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant 172, 1711–1723. doi: 10.1111/ppl.13373
Nayanathara, A. R., Aalolam, K. P., Reshma, J. K. (2016). Evaluation of total phenol, flavonoid and anthocyanin content in different varieties of eggplant. Emer Life Sci. Res. 2, 63–65.
Nino-Medina, G., Urias-Orona, V., Muy-Rangel, M. D., Heredia, J. B. (2017). Structure and content of phenolics in eggplant (Solanum melongena). S Afr J. Bot. 111, 161–169. doi: 10.1016/j.sajb.2017.03.016
Oladosu, Y., Rafii, M. Y., Arolu, F., Chukwu, S. C., Salisu, M. A., Olaniyan, B. A., et al. (2021). Genetic diversity and utilization of cultivated eggplant germplasm in varietal improvement. Plants (Basel). 10, 1714–1734. doi: 10.3390/plants10081714
Peng, Y., Thrimawithana, A. H., Cooney, J. M., Jensen, D. J., Espley, R. V., Allan, A. C. (2020). The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 10, 14161. doi: 10.1038/s41598-020-70977-0
Perez-Rodriguez, P., Riano-Pachon, D. M., Correa, L. G., Rensing, S. A., Kersten, B., Mueller-Roeber, B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38, D822–D827. doi: 10.1093/nar/gkp805
Pesch, M., Dartan, B., Birkenbihl, R., Somssich, I. E., Hulskamp, M. (2014). Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell. 26, 4067–4083. doi: 10.1105/tpc.114.129379
Pojer, E., Mattivi, F., Johnson, D., Stockley, C. S. (2013). The case for anthocyanin consumption to promote human health: a review. Compr. Rev. Food Sci. F. 12, 483–508. doi: 10.1111/1541-4337.12024
Qiao, J., Liu, J., Li, S., Wang, L. (2022). Prediction of fruit color genes under the calyx of eggplant based on genome-wide resequencing in an extreme mixing pool (in Chinese). Acta Hortic. Sinica. 49, 613. doi: 10.16420/j.issn.0513-353x.2021-0018
Ramsay, N. A., Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Roberts, A. R., Vecellio, M., Chen, L., Ridley, A., Cortes, A., Knight, J. C., et al. (2016). An ankylosing spondylitis-associated genetic variant in the IL23R-IL12RB2 intergenic region modulates enhancer activity and is associated with increased Th1-cell differentiation. Ann. Rheumatol. Dis. 75, 2150–2156. doi: 10.1136/annrheumdis-2015-208640
Ryu, S., Han, J. H., Cho, J. G., Jeong, J. H., Lee, S. K., Lee, H. J. (2020). High temperature at véraison inhibits anthocyanin biosynthesis in berry skins during ripening in ‘Kyoho’grapevines. Plant Physiol. Biochem. 157, 219–228. doi: 10.1016/j.plaphy.2020.10.024
Saini, D. K., Kaushik, P. (2019). Visiting eggplant from a biotechnological perspective: A review. Sci. Hortic-Amsterdam. 253, 327–340. doi: 10.1016/j.scienta.2019.04.042
Schwartz, C., Balasubramanian, S., Warthmann, N., Michael, T. P., Lempe, J., Sureshkumar, S., et al. (2009). Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics. 183 723-732, 721SI–727SI. doi: 10.1534/genetics.109.104984
Shan, X., Li, Y., Yang, S., Gao, R., Zhou, L., Bao, T., et al. (2019). A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 141, 60–72. doi: 10.1016/j.plaphy.2019.05.015
Shi, B., Wu, H. X., Zheng, B., Qian, M. J., Gao, A. P., Zhou, K. B. (2021). Analysis of light-independent anthocyanin accumulation in mango (Mangifera indica L.). Horticulturae. 7, 423–434. doi: 10.3390/horticulturae7110423
Sivankalyani, V., Feygenberg, O., Diskin, S., Wright, B., Alkan, N. (2016). Increased anthocyanin and flavonoids in mango fruit peel are associated with cold and pathogen resistance. Postharvest Biol. Tec. 111, 132–139. doi: 10.1016/j.postharvbio.2015.08.001
Stadhouders, R., Aktuna, S., Thongjuea, S., Aghajanirefah, A., Pourfarzad, F., Van Ijcken, W., et al. (2014). HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 124, 1699–1710. doi: 10.1172/JCI71520
Stam, M., Belele, C., Dorweiler, J. E., Chandler, V. L. (2002). Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918. doi: 10.1101/gad.1006702
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Studer, A., Zhao, Q., Ross-Ibarra, J., Doebley, J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163. doi: 10.1038/ng.942
Sun, L., Li, S., Tang, X., Fan, X., Zhang, Y., Jiang, J., et al. (2020). Transcriptome analysis reveal the putative genes involved in light-induced anthocyanin accumulation in grape ‘Red Globe’(V. vinifera L.). Gene 728, 144284. doi: 10.1016/j.gene.2019.144284
Sun, X. H., Zhou, T. T., Wei, C. H., Lan, W. Q., Zhao, Y., Pan, Y. J., et al. (2018). Antibacterial effect and mechanism of anthocyanin rich Chinese wild blueberry extract on various foodborne pathogens. Food Control. 94, 155–161. doi: 10.1016/j.foodcont.2018.07.012
Takos, A. M., Jaffe, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tang, B., Li, L., Hu, Z., Chen, Y., Tan, T., Jia, Y., et al. (2020). Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper. J. Agr Food Chem. 68, 12152–12163. doi: 10.1021/acs.jafc.0c02460
Timothy, A., Holton, E. C. C. (1995). genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 7, 1071–1083. doi: 10.2307/3870058
Toppino, L., Barchi, L., Lo Scalzo, R., Palazzolo, E., Francese, G., Fibiani, M., et al. (2016). Mapping quantitative trait loci affecting biochemical and morphological fruit properties in eggplant (Solanum melongena L.). Front. Plant Sci. 7, 256. doi: 10.3389/fpls.2016.00256
Van Der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5, 227. doi: 10.3389/fpls.2014.00227
Verweij, W., Spelt, C. E., Bliek, M., De Vries, M., Wit, N., Faraco, M., et al. (2016). Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis. Plant Cell. 28, 786–803. doi: 10.1105/tpc.15.00608
Visser, M., Palstra, R. J., Kayser, M. (2014). Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum. Mol. Genet. 23, 5750–5762. doi: 10.1093/hmg/ddu289
Wang, H., Niu, Q. W., Wu, H. W., Liu, J., Ye, J., Yu, N., et al. (2015). Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 84, 404–416. doi: 10.1111/tpj.13018
Xiang, C. C., Zhang, W. W., Luo, S. X., Zhao, J. J., Zhang, T. T., Shen, S. X., et al. (2015). Genetic analysis on fruit color under calyx and correlation analysis of SSR maker in eggplant (in Chinese). J. Agric. Univ. Hebei. 38, 50–55. doi: 10.13320/j.cnki.jauh.2015.0083
Xu, W., Dubos, C., Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yang, J. F., Chen, Y. Z., Kawabata, S., Li, Y. H., Wang, Y. (2017). Identification of light-independent anthocyanin biosynthesis mutants induced by Ethyl Methane Sulfonate in turnip "Tsuda" (Brassica rapa). Int. J. Mol. Sci. 18, 1288–1303. doi: 10.3390/ijms18071288
Yang, G., Li, L., Wei, M., Li, J., Yang, F. (2022). SmMYB113 is a key transcription factor responsible for compositional variation of anthocyanin and color diversity among eggplant peels. Front. Plant Sci. 13, 843996. doi: 10.3389/fpls.2022.843996
Yu, G., Wang, L.-G., Han, Y., He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16, 284–287. doi: 10.1089/omi.2011.0118
Zha, Q., Xi, X. J., He, Y., Jiang, A. L. (2019). Bagging affecting sugar and anthocyanin metabolism in the ripening period of grape berries. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 47, 1194–1205. doi: 10.15835/nbha47411577
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, X. T., He, Y. J., Liu, Y., Chen, H. Y. (2021b). Development and validation of molecular marker for photosensitivity of anthocyanin production in eggplant fruit (in Chinese). J. Nanjing Agric. University. 44, 637–645.
Zhang, Y., Hu, Z., Chu, G., Huang, C., Tian, S., Zhao, Z., et al. (2014). Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J. Agr Food Chem. 62, 2906–2912. doi: 10.1021/jf404574c
Zhang, W. S., Li, X., Zheng, J. T., Wang, G. Y., Sun, C. D., Ferguson, I. B., et al. (2008). Bioactive components and antioxidant capacity of Chinese bayberry (Myrica rubra Sieb. and Zucc.) fruit in relation to fruit maturity and postharvest storage. Eur. Food Res. Technol. 227, 1091–1097. doi: 10.1007/s00217-008-0824-z
Zhang, B., Zhu, Z. Z., Qu, D., Wang, B. C., Hao, N. N., Yang, Y. Z., et al. (2021a). MdBBX21, a B-Box protein, positively regulates light-induced anthocyanin accumulation in apple peel. Front. Plant Sci. 12, 774446. doi: 10.3389/fpls.2021.774446
Keywords: eggplant, EMS mutants, anthocyanin, less-photosensitive, MutMap
Citation: Luo L, Molthoff J, Li Q, Liu Y, Luo S, Li N, Xuan S, Wang Y, Shen S, Bovy AG, Zhao J and Chen X (2023) Identification of candidate genes associated with less-photosensitive anthocyanin phenotype using an EMS mutant (pind) in eggplant (Solanum melongena L.). Front. Plant Sci. 14:1282661. doi: 10.3389/fpls.2023.1282661
Received: 24 August 2023; Accepted: 24 November 2023;
Published: 11 December 2023.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Junxiang Zhang, Shenyang Agricultural University, ChinaCopyright © 2023 Luo, Molthoff, Li, Liu, Luo, Li, Xuan, Wang, Shen, Bovy, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnaud G. Bovy, YXJuYXVkLmJvdnlAd3VyLm5s; Jianjun Zhao, amp6MTk3MUBhbGl5dW4uY29t; Xueping Chen, Y2hlbnh1ZXBpbmdAaGViYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.