- 1US Dairy Forage Research Center, United States Department of Agriculture-Agricultural Research Service (USDA-ARS), Madison, WI, United States
- 2James E. “Bud” Smith Plant Materials Center, United States Department of Agriculture-Natural Resources Conservation Service (USDA-NRCS), Knox City, TX, United States
- 3Corvallis Plant Materials Center, USDA-NRCS, Corvallis, OR, United States
- 4Central National Technology Support Center, USDA-NRCS, Fort Worth, TX, United States
- 5National Plant Materials Program, USDA-NRCS, Washington, DC, United States
- 6Corteva Agriscience, Connell, WA, United States
- 7Sustainable Agricultural Systems Laboratory, United States Department of Agriculture-Natural Resources Conservation Service (USDA-ARS), Beltsville, MD, United States
- 8Bayer Crop Science, Chesterfield, MO, United States
- 9School of Medicine in Kansas, University of Kansas Medical Center, Wichita, KS, United States
- 10Forage Seed and Cereal Research Unit, USDA-ARS, Corvaillis, OR, United States
Hairy vetch (Vicia villosa Roth), a winter-hardy annual legume, is a promising cover crop. To fully leverage its potential, seed production and field performance of V. villosa must be improved to facilitate producer adoption. Two classic domestication traits, seed dormancy (hard seed) and dehiscence (pod shatter), are selection targets in an ongoing breeding program. This study reports a genome-wide association study of 1,019 V. villosa individuals evaluated at two sites (Knox City, Texas and Corvallis, Oregon) for the proportion of dormant seed, visual pod dehiscence scores, and two dehiscence surrogate measures (force to dehiscence and pod spiraling score). Trait performance varied between sites, but reliability (related to heritability) across sites was strong (dormant seed proportion: 0.68; dehiscence score: 0.61; spiraling score: 0.42; force to dehiscence: 0.41). A major locus controlling seed dormancy was found (q-value: 1.29 × 10−5; chromosome 1: position: 63611165), which can be used by breeding programs to rapidly reduce dormancy in breeding populations. No significant dehiscence score QTL was found, primarily due to the high dehiscence rates in Corvallis, Oregon. Since Oregon is a potentially major V. villosa seed production region, further dehiscence resistance screening is necessary
Introduction
Cover crop adoption can improve farm productivity and reduce environmental harm (Poeplau and Don, 2015; Tiemann et al., 2015; Austin et al., 2017). Cover crops can reduce nutrient loss (Wyland, 1996; Drinkwater et al., 1998), improve soil health (Sainju and Singh, 1997), and decrease yield loss due to pests and weeds (Teasdale, 1996). A total of 6.2 million hectares of cover crops were planted in 2017 (USDA-NASS, ; Wallander et al., 2021), an increase of 50% from 2012. This indicates that 5.1% of US-harvested, non-alfalfa cropland used cover crops in 2017. Further cover crop adoption requires improved agronomic methods and breeding improved varieties in a wider suite of cover crop plant species (Kaspar and Bakker, 2015; Wallace et al., 2017; Wayman et al., 2017).
As an agricultural method, cover cropping is a broad term with goals varying by environmental constraints and producer needs. Currently, small grains (cereal rye (Secale cereale L.), wheat (Triticum aestivum L.), oats (Avena sativa L.), or barley (Hordeum vulgare L.)) collectively account for >90% of cover crop acreage (Wallander et al., 2021; note this survey did not ask about cover crop mixes). Small grains are affordable, already familiar to farmers, and established rapidly. Cereal rye is popular because it can overwinter in the northern USA and produces abundant biomass, which can provide early-season weed control (Wallace et al., 2017). However, there is concern that small grain residues break down too slowly and limit nitrogen availability for the following cash crop (Wagger et al., 1998; Wallace et al., 2017; Nevins et al., 2020; Thompson et al., 2020). The extent of this issue likely varies by location and cropping system (Snapp and Surapur, 2018; Bourgeois et al., 2022).
The inclusion of legume species (family Fabaceae) in fall-planted cover crop mixes allows atmospheric nitrogen fixation. Further, legume residue decomposes more rapidly than grass residue due to lower carbon-to-nitrogen ratios. Therefore, legumes in fall cover crop plantings will improve the nutrient availability and yield of the following cash crops (Ranells and Wagger, 1996; Clark et al., 1997; Vyn et al., 2000; Singh et al., 2020). Hairy vetch is a commonly grown legume cover crop due to its winter hardiness, high biomass yields, and nitrogen supply to the subsequent cash crop (Power and Zachariassen, 1993; Brandsæter et al., 2002; Parr et al., 2011; Mirsky et al., 2017). V. villosa is a diploid (2n = 14, Kahlaoui, 2009) allogamous species (Zhang and Mosjidis, 1995) that tolerates some self-pollination (Hamadi et al., 2012). Various bee species, including Bombus sp. and Apis mellifera, facilitate cross-pollination.
Hairy vetch has limited adoption in current agricultural systems due to its incomplete domestication (Wayman et al., 2017). V. villosa produces dormant seed (i.e., hard seed) and is prone to early shatter (pod dehiscence). Both traits are typically eliminated during crop domestication (Doebley et al., 2006; Purugganan and Fuller, 2009; Bohra et al., 2022). Due to limited historic and current investment in forage and cover crop breeding, seed dormancy and pod dehiscence persist in V. villosa cultivars (Wilke and Snapp, 2008). Like many members of the Faboideae family (Smýkal, 2014), physical seed dormancy in V. villosa is linked to a water-impermeable seed coat that prevents imbibition of water (Jones, 1928; Renzi et al., 2014). Prior reports of V. villosa seed dormancy rates range from% 0 to 100% (Jannink, 1997; Jacobsen et al., 2010; Renzi et al., 2014; Mirsky et al., 2015; Kissing Kucek et al., 2020a). Dormant seed of V. villosa persistent in the soil seed bank leads to weedy V. villosa plants, which can reduce cash crop yields (Curran, 2015; Mirsky et al., 2015). In addition, V. villosa and small grain seeds mature at the same time, therefore requiring the use of expensive and specialized threshing equipment to separate small grain seed harvests to separate volunteer V. villosa seed from small grain seed.
Pod shatter, also known as dehiscence, contributes to seed yield losses and can create weedy “volunteer” V. villosa populations (Kissing Kucek et al., 2020a). One study reported 3% to 60% shattered pods among evaluated V. villosa lines (Abd El-Moneim, 1993). For seed growers, V. villosa’s propensity for pod dehiscence, in combination with the crop’s indeterminant growth habit, makes decisions on harvest timing challenging. The result can be extensive seed yield losses. The reduced seed harvestability due to pod shattering results in expensive V. villosa seed that can become a barrier for farmers looking for an inexpensive legume cover crop seed (Renzi et al., 2023).
Using recurrent phenotypic selection, plant breeders should be able to gradually reduce seed dormancy and pod dehiscence after repeated selection cycles. Opportunities to integrate DNA markers linked to these traits could accelerate this breeding progress. Research in previously domesticated legume crops indicates that seed dormancy and pod dehiscence can be eliminated by a small number of large effect loci, and consequently, these traits are excellent candidates for marker-assisted selection. For example, seed dormancy was eliminated by a single altered gene in Lens culinaris Medic. (Ladizinsky, 1985), Lupinus angustifolius L. (Forbes and Wells, 1968), Vigna unguiculata L. (Kongjaimun et al., 2012), V. umbellata (Thunb.) (Isemura et al., 2010), and V. radiata L. (Isemura et al., 2012). In Pisum sativum L., two to three loci eliminated seed dormancy (Weeden, 2007). A single recessive allele removed physiological dormancy in Medicago truncatula Gaertn. (Chai et al., 2016). In soybean (Glycine max (L.) Merr.), a recessive single-point mutation of Gmhs1-1 created soft seed by modifying a calcium-based seed coat membrane protein (Sun et al., 2015). Alternative mutations at qHS1 in G. max also formed cracks in the seed coat that caused partial imbibition (Jang et al., 2015; Sun et al., 2015).
Similar to seed dormancy, pod dehiscence can be controlled by large-effect loci. Ladizinsky (1985) reported one to two dominant genes in Lens species. Two genes controlled dehiscence in Pisum satium L. (Weeden, 2007). Abd El-Moneim (1993) found that nonshattering in V. sativa L. could be eliminated through one large effect recessive gene. In V. radiata (L.) Wilczek), one quantitative trait loci (QTL) was associated with pod dehiscence (Isemura et al., 2012).
Potential large effect loci for domestication traits could be obscured by environmental effects such as relative humidity (Quinlivan, 1968) and temperature at seed maturity (Kissing Kucek et al., 2020a), pod maturity at harvest (Samarah, 2006), and pod handling. Postharvest seed storage can also alter dormancy in V. villosa (Renzi et al., 2014). Such mechanisms are common in winter annual species of Mediterranean origin since hot summer weather signals upcoming wet and cool fall conditions that are ideal for germination (Williams and Elliott, 1960). Within the palisade layers of the seed coat, lipids and other hydrophobic compounds degrade in high heat and allow germination (Smýkal et al., 2014).
The objective of this study is to determine trait genetic associations for pod dehiscence and seed dormancy in a diverse set of V. villosa germplasm from an existing breeding program. This population varies for both seed dormancy and pod dehiscence measurements (Kissing Kucek et al., 2020b, 2020a). These observations and the presence of large effect loci in other legumes (see above) highlight the promise of marker-assisted selection to accelerate V. villosa development. This study reports (1) the presence of significant QTL in V. villosa breeding populations and (2) estimates of the genetic reliability and phenotypic correlations among seed dormancy, dehiscence, and other surrogate traits.
Materials and methods
Germplasm and experimental design
In 2015 and 2016, researchers in the Cover Crop Breeding Network (www.covercropbreeding.com) performed a search of available local and internationally collected germplasm with the goal of finding germplasm with traits beneficial for cover crops. The team selected and acquired 27 V. villosa populations for the breeding program, including breeding material developed in Maryland, Minnesota, North Carolina, and Wisconsin. The ancestry and location of origin of these 27 populations are described in Kucek et al. (2019). In the fall of 2016, 12,500 V. villosa genotypes from the 27 populations were evaluated at six sites (Rocky Mount, North Carolina; Clayton, North Carolina; Beltsville, Maryland; Ithaca, New York; Prairie du Sac, Wisconsin; and Roseville, Minnesota). Each site assessed emergence, fall and spring vigor, winter survival, and flowering time, and two sites assessed biological nitrogen fixation (Kucek et al., 2019). The top 3% to 6% of individuals at each site were allowed to flower and contribute to crossing blocks.
The current experiment used 80 maternal half-sibling lines derived from selections from 24 of the 27 initially selected populations (Table 1). The 80 parents were selected based on divergent variations in seed dormancy and pod dehiscence traits. Individual open-pollinated progeny from these half-sibling lines were assigned to one of two sites (Oregon and Texas; 10 progeny per maternal line per site). The Oregon site was planted at the USDA-Natural Resources Conservation Service (NRCS), Corvallis Plant Materials Center in Oregon (OR) on 25 September 2018, in a Woodburn silt-loam (fine-silty, mixed, superactive, mesic Aquultic Argixerolls), and the Texas site was planted at the USDA-NRCS, James E. “Bud” Smith Plant Materials Center in Knox City (TX) on 16th October 2018 in an Altus fine sandy-loam (fine-loamy, mixed, superactive, thermic Pachic Argiustolls). Individual unreplicated progeny were randomly assigned to rows within 10 balanced blocks of half-sibling families within each location, with 1.83 m between individuals and 1.83 m between rows. Field traits were measured in 2019 at both sites. The Oregon site experienced almost complete fall predation, likely from slugs (Gastropods), and therefore transplants were established into landscape fabric in late winter. Texas relied on manual rouging to remove weeds. To control for bruchid beetle (Bruchus brachialis Fahraeus) damage, Zeta-cypermethrin (Mustang Maxx; 0.0028 kg A.I. ha−1) was applied in 2019 in Oregon on June, 2 July, 11 July, 21 and July and in Texas on 10 May, 17 May, 25 May, and 31May 31.
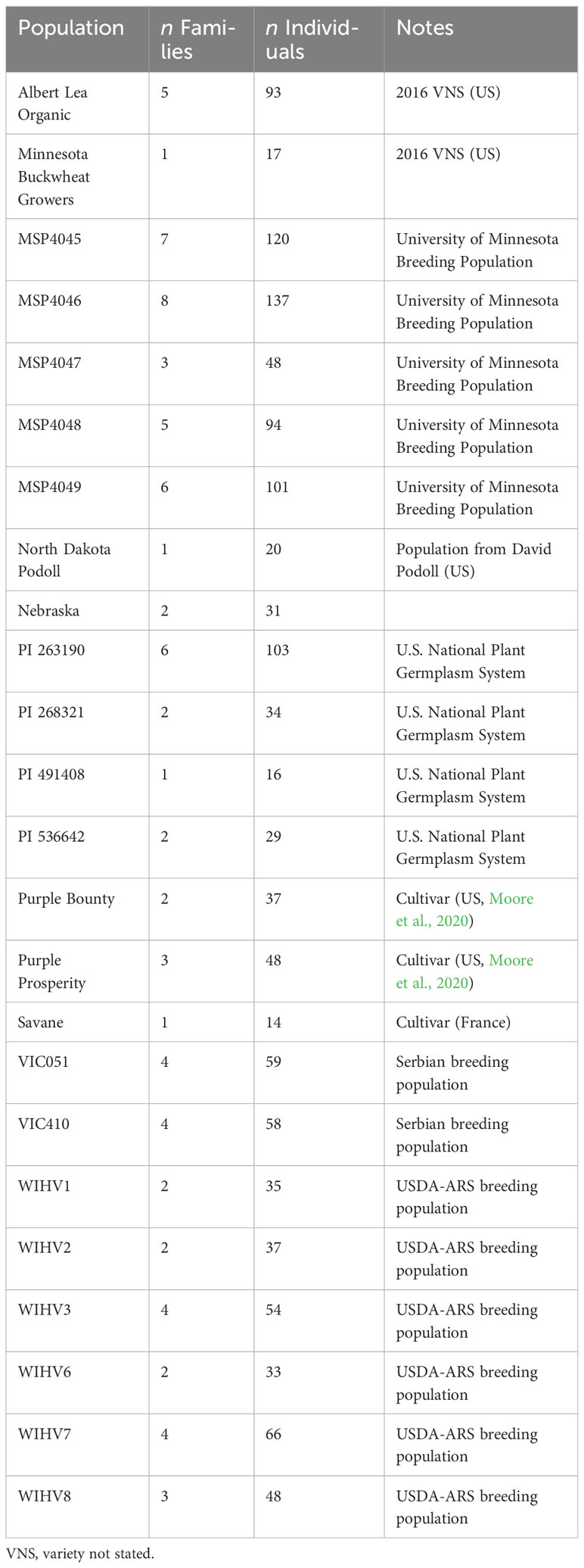
Table 1 The number (n) of families and individuals evaluated for seed dormancy and dehiscence based on initial germplasm origin.
Data collection
Maturity ratings based on Kalu and Fick (1981) were performed when 50% of individuals had flowers at the Texas location and when 50% of individuals had pods at both sites in 2019. These maturity ratings were collected due to early maturity being a potential breeding target, and plant maturity could serve as a covariate for pod dehiscence and seed dormancy.
Between early June and mid-August of 2019, pods developed a brown color, a signal of physiological maturity. Around 50 pods were collected from each individual at each site. If pods appeared to contain few seeds per pod, additional pods were collected per plant. Pods were phenotyped for traits related to pod dehiscence and seed dormancy at the USDA-ARS Dairy Forage Research Center in Madison, WI in 2020. Detailed methods of visual dehiscence ratings and seed dormancy determination are described in (Kissing Kucek et al., 2020b) and Kissing Kucek et al. (2020a), respectively. Briefly, all pods collected from an individual plant received a visual pod dehiscence score, targeting 50 pods per individual. Pods were dried at 28°C for 24 h before evaluation. Dehiscence was scored on a zero to three scale, with zero indicating a fully intact pod (no openings along sutures), one indicating one suture was partially opened (one side of the pod), two indicating two sutures were partially opened (both sides of the pod), two and a half indicating a pod was partially winged apart, and three indicating the pod had fully opened. Individuals with fewer than 10 pods rated were not included in subsequent data analysis. Individual plants with flat pods from lack of seed fill were also removed. Individual plants with >66% immature pods were removed from dehiscence measurements.
Each seed from an individual received a binomial rating for seed dormancy: “0” if the seed imbibed water and produced a radicle without scarification, or “1” if the seed did not imbibe water after 7 days and produced a radicle after scarification. Nonviable seeds or seed-imbibed water that did not produce a radicle were removed from subsequent analysis. Individuals who did not have at least 10 viable seeds observed were also removed from further analysis.
Pod samples were rated for pod spiraling on a visual scale of zero, one, and two (further details are reported in Kissing Kucek et al., 2020b). For individuals with >66% immature pods, spiraling values were set to not applicable (NA), due to a relationship between immaturity and spiraling.
Individual plants with at least three fully closed pods were screened for force to dehiscence using an MTS Insight® 1kN (MTS Systems Corporation) instrument for measuring cracking force on unopened pods. The evaluation used a 100-N load cell and TestWorks ® [v.4.11B] (MTS Systems Corporation: Eden Prairie, MN, USA) set to 96% break sensitivity, 0.1 N break threshold, and 25 Hz data acquisition rate. Individual plants with fewer than three pods tested for force to dehiscence were excluded from the analysis. Samples with immature green pods, or pods that were flat and did not fill with seeds were removed from analysis.
Hairy vetch dormancy and shatter calculation of BLUPs
Broadly, models were used to partition variance and remove covariates prior to genome-wide association studies (GWAS) analysis. The analysis did not use models with complex genotype by environment interactions between the sites since any site-specific QTL will not be useful for the breeding program. For pod dehiscence, the best linear unbiased predictors (BLUPs) of each plant were extracted using a multinomial logistic regression model implemented through ASReml-R (Butler et al., 2017), with 1,019 individuals representing all 80 maternal lines that met inclusion criteria for analysis for visual dehiscence. Visual pod dehiscence BLUPs were also generated separately by site, as the highly skewed Oregon data violated model assumptions.
Where f(k,n) indicates the cumulative probability of each pod observation in taking a value of the kth category of dehiscence, where k = 0, 1, 2, 2.5, 3, and χn is the set of explanatory variables including fixed effect maternal line i and the random effect of site j.
A total of 713 individual plants, representing all 80 maternal lines, met inclusion criteria for force-to-dehiscence analysis. Values for force to dehiscence (N) were right skewed and consequently square root transformed to create a normally distributed set of observations. BLUPs for each plant were obtained using Eq. (2):
Where Yijkl indicates the square root of force (N) required to open a closed pod from the fixed effect α of maternal line i, the random effect β of site j, the random effect δ of individual plant k, the random covariate γ of pod maturity rating, and residual error ϵ. BLUPs for each individual plant were calculated by adding random coefficients from the βj term to the fixed coefficients from the αi term.
For seed dormancy, 1,019 individual plants, representing all 80 maternal lines, met inclusion criteria for analysis. BLUPs for seed dormancy were extracted as the coefficient of individual plant j added to the coefficient of maternal line i from Eq. (3) in ASReml-R:
Where Yijklm is the log-likelihood of dormant seed (0,1) for maternal line i, individual plant j, site k, technical replicate l, and maturity covariate m; B0 is the intercept of log odds, β1 and xi1 are the partial slope and fixed variable associated with maternal line i, β2 and xj2 are the partial slope and random variable associated with individual plant j, β3 and xk3 are the partial slope and random variable associated with site k, β4 and xl4 are the partial slope and random variable associated with technical replicate l, β5 and xm5 are the partial slope and random variable associated with the pod maturity covariate m.
Spiraling data were not corrected through analysis due to only one observation per individual plant. However, it was included in the GWAS analysis after adjusting for site means.
Tissue sampling, sequencing, bioinformatics, and genome-wide association
V. villosa tissues were collected for sequencing during active vegetative growth. At the Oregon location, actively expanding penultimate leaves were sampled just before and during early bloom on three separate dates (19 May, 7 June, and 14 June), and the Texas location plants were sampled on 14 April. Each field sample was placed into a labeled coin envelope and immediately placed on ice before transport to a laboratory freezer (−20°C). The samples were subsequently frozen to −60°C and lyophilized prior to shipment. Twice. Once at the University of Wisconsin Biotechnology Center and once at Oklahoma State University on an Illumina sequencer (NovaSeq 6000). The University of Wisconsin used an NsiI-BfaI double digestion restriction enzyme digestion and Oklahoma State University Oklahoma State University used an ApeKI restriction enzyme digestion. Fragments were then ligated to barcoded adaptors prior to polymerase chain reaction amplification.
Bioinformatics sequence data processing was completed at the University of Wisconsin Biotechnology Center, which merged the two sequencing runs. Sequencing output analysis used the TASSEL analysis platform (Glaubitz et al., 2014). Barcoded sequence read outputs were collapsed into a set of unique sequence tags with counts. These tags were aligned to the reference genome (V. villosa v1.1; Fuller et al., 2023). Each tag was assigned to a position with the best unique alignment, and the occupancies of tags for each sample were observed from barcode data. Overall, 2,524,985 single-nucleotide polymorphism markers (SNPs) were present prior to filtering. A subset (n = 74) of individuals had a means<1 reads per site, relative to the mean of 4.16 reads per site (SD = 1.28) in the overall population (Figure 1). Over 90% of these samples were collected from the Texas location and were not stratified by genetic background; therefore, this was presumed to be a leaf tissue storage issue and these individuals were removed from further analysis. The 38% of markers containing<0.5 read per site and the 26% of SNPs that mapped outside the seven primary chromosomes in the assembly were removed. A random subset of 100,000 SNP markers was used to evaluate inter-relationships within the dataset using principal component analysis. This revealed a strong out-group, which was omitted from further analysis (n = 137; discussed briefly below). Finally, markers with minor allele frequencies of<0.025 were removed. In total, 1,010,403 SNP markers across 881 individuals met the filtering criteria. The imputation of missing markers (42%) was done through the Beagle program (Browning et al., 2018).
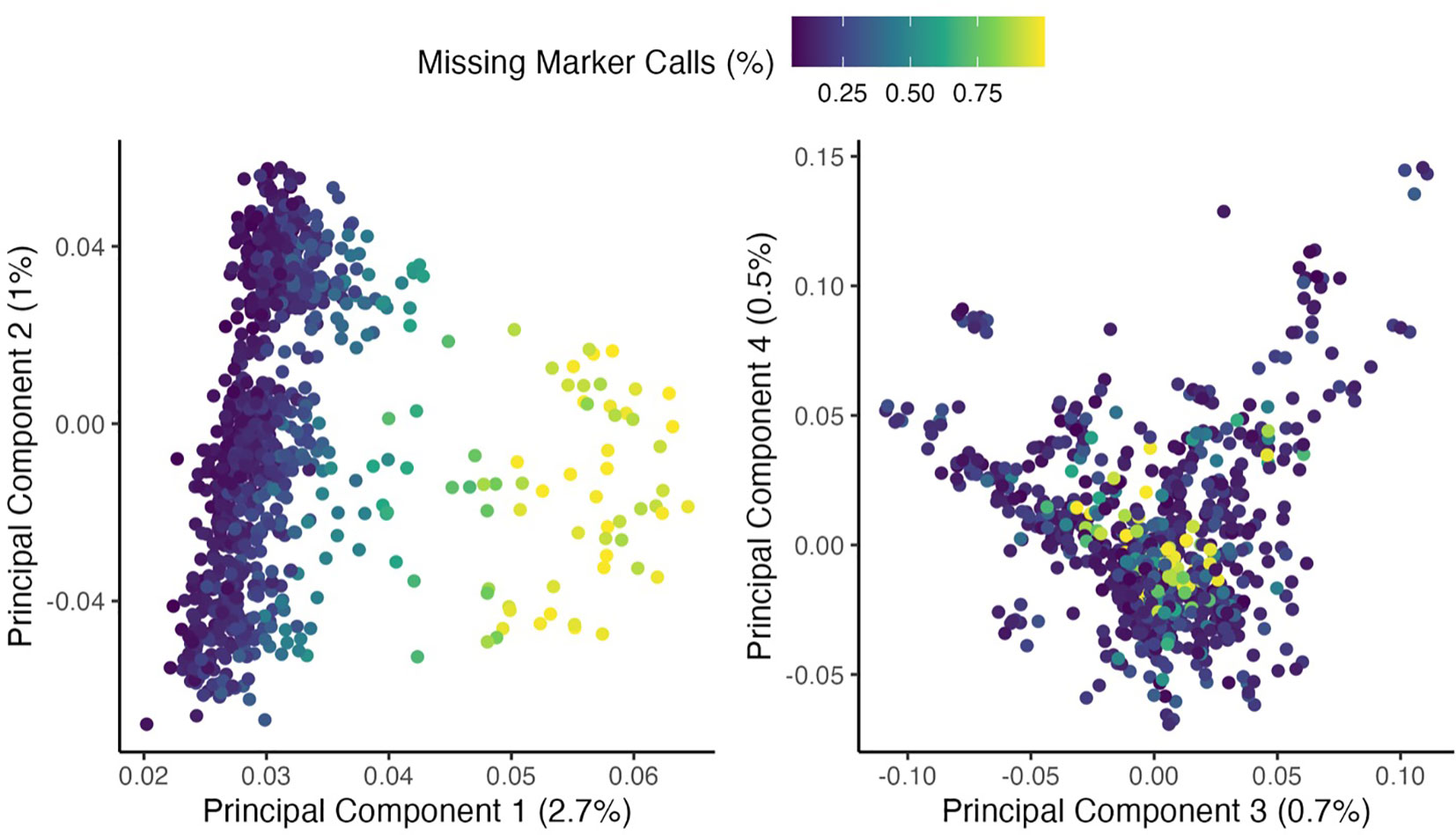
Figure 1 The first four principal components of 956 V. villosa genotypes are based on 100,000 randomly selected SNPs. Point colors indicate the proportion of missing markers in each individual prior to imputation. The percent variance of each principal component is indicated in parentheses. Individuals with<1 mean read depth per site were removed from further analysis (population mean: 4.16). This figure does not contain an out-group (out-group presented in Supplementary Figure S1).
Significant QTLs were determined using the GWAS function in the R package “sommer” v.4.3.1 (Covarrubias-Pazaran, 2016). The analysis included force-to-dehiscence BLUPs (n = 500), pod dehiscence BLUPs (n = 791), seed dormancy BLUPs (n = 853), and pod spiraling scores (n = 797). Population structure was accounted for by including the realized relationship matrix (K model), and the interactive effect of each maternal line at each site was included as a random effect. The GWAS model also allowed different error variances between sites (diagonal residual covariance structure). Multiple-testing correction was based on the false discovery rate (Benjamini and Hochberg, 1995) with a significance threshold of q = 0.01. Reliability, a measure related to heritability on an entry-difference basis, was calculated for each trait using the genotypic variance and error variance estimated within the GWAS model (Schmidt et al., 2019).
Results
Field observations and BLUP calculations
Maturity ratings at 50% flowering were not collected at the Oregon site, but maturity ratings at 50% pod development had positive (0.51) correlations between the Texas and Oregon sites. Maturity covariates did not correlate strongly with the target traits and only improved model fit for force to dehiscence during BLUP calculation (Supplementary Figure S2). Maturity covariates were not included in the models for visual shatter, spiraling, or seed dormancy. Trait correlations generally were low after accounting for differences in site means (Supplementary Figure S3). The only exception was the correlations (p< 0.001) between pod dehiscence scores and pod spiraling scores (Oregon: 0.37, Texas: 0.58), and seed dormancy and pod dehiscence scores (Oregon: 0.26, Texas: 0.32).
Mean and trait distributions of visual pod dehiscence were heavily influenced by the test site (mean Oregon: 2.70, SD: 0.53; mean Texas: 0.90, SD: 0.67; Supplementary Figure S3). The Oregon site had few observations in low pod–dehiscence classes, resulting in a strong left skew (Supplementary Figure S2). Within the Texas site, maternal line, individual plant, and error contributed similar variance to visual dehiscence (Table 2). The reliability of the pod dehiscence score was 0.61, but the correlation between mean site family performance was 0.21.
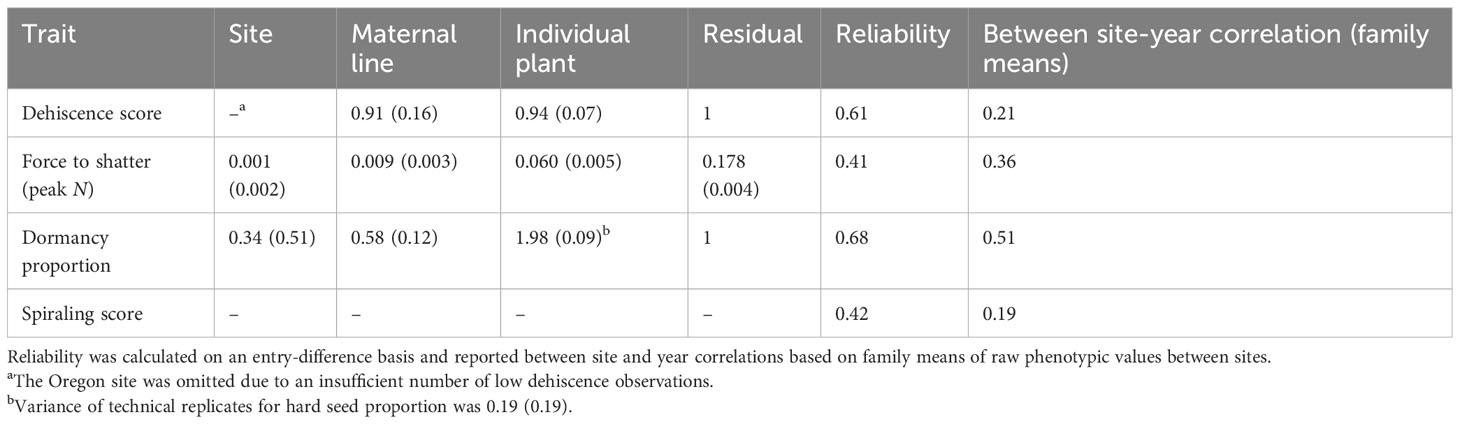
Table 2 Variance components with standard errors in parentheses for the models of pod shatter score (Eq. 1), force to shatter (Eq. 2), and hard seed (Eq. 3).
Raw force to dehiscence (N) scores had only mild right skew distributions (Supplementary Figure S2). The variance of force to dehiscence was dominated by the influence of residual error among pods within an individual plant, followed by an individual plant, followed by the maternal line, and then the site (Table 2). Force to dehiscence was only measured on the low-shatter individuals (mean visual dehiscence scores: 1.1 with force to dehiscence measurements vs. 2.8 without force-to-dehiscence measurements, p< 0.001). The site had a very small influence on force to dehiscence (mean Oregon: 2.38, SD: 1.29; mean Texas: 2.15, SD: 0.95; Table 2). Force-to-shatter reliability was 0.41, and correlation between site and mean family performance correlation was 0.36.
The mean seed dormancy was 58%, with a large number of individuals with >75% dormancy and some individuals with zero measured dormant seed (Supplementary Figure S2). Seed dormancy variance was dominated by the influence of individual plants, followed by residual error, then maternal line, site, and technical replicates (Table 2). The site had a small influence on seed dormancy means (mean Oregon: 0.66, SD: 0.28; mean Texas: 0.52, SD: 0.27; Table 2). Despite only one observation per genotype, pod spiraling had a reliability of 0.42 and between site and mean family correlation of 0.19. Similar to dehiscence, there were fewer observations of low pod spiraling rankings in the Oregon site (mean Oregon: 1.92, SD: 0.33; mean Texas: 1.03, SD: 0.82; Table 2).
Genetic relationships and GWAS analysis
Principal component analysis revealed the presence of two major subpopulations in the breeding program (Supplementary Figure S1). Germplasm in the outgroup is consistently traced back to a small number of germplasm sources (Nebraska, PI 268321, ‘Purple Bounty’, and ‘Purple Prosperity’). These individuals appear to be distantly related to the core V. villosa populations based on their poor alignment with the V. villosa genome assembly, and they also appear to not cross-pollinate with the core breeding population (Supplementary Figure S1).
Broadly, quantile–quantile plots indicated that the inclusion of the K matrix and half-sibling family effect controlled potential p-value inflation in the GWAS (Supplementary Figure S4). The GWAS panel included individuals from a relatively small number of half-sibling families (n = 80). This had the potential to introduce population structure and inflate marker effect estimates. The solution for this was to include a covariate to remove the maternal effect, which resulted in the removal of a large portion of informative genetic variance from the GWAS.
Seed dormancy BLUPs had multiple significant associations around a primary peak with the lowest q-value 1.29 × 10−5 (Figures 2, 3; Supplementary Figure S3; chromosome: 1, position: 63611165). The reference allele frequency in the population is 0.68, with an estimated additive marker effect of the alternate allele of −0.61 (TX: −0.38 and OR: −0.83). This implausibly large marker effect is due to the strong impact of heterozygotes from Oregon in the dataset (Figure 3).
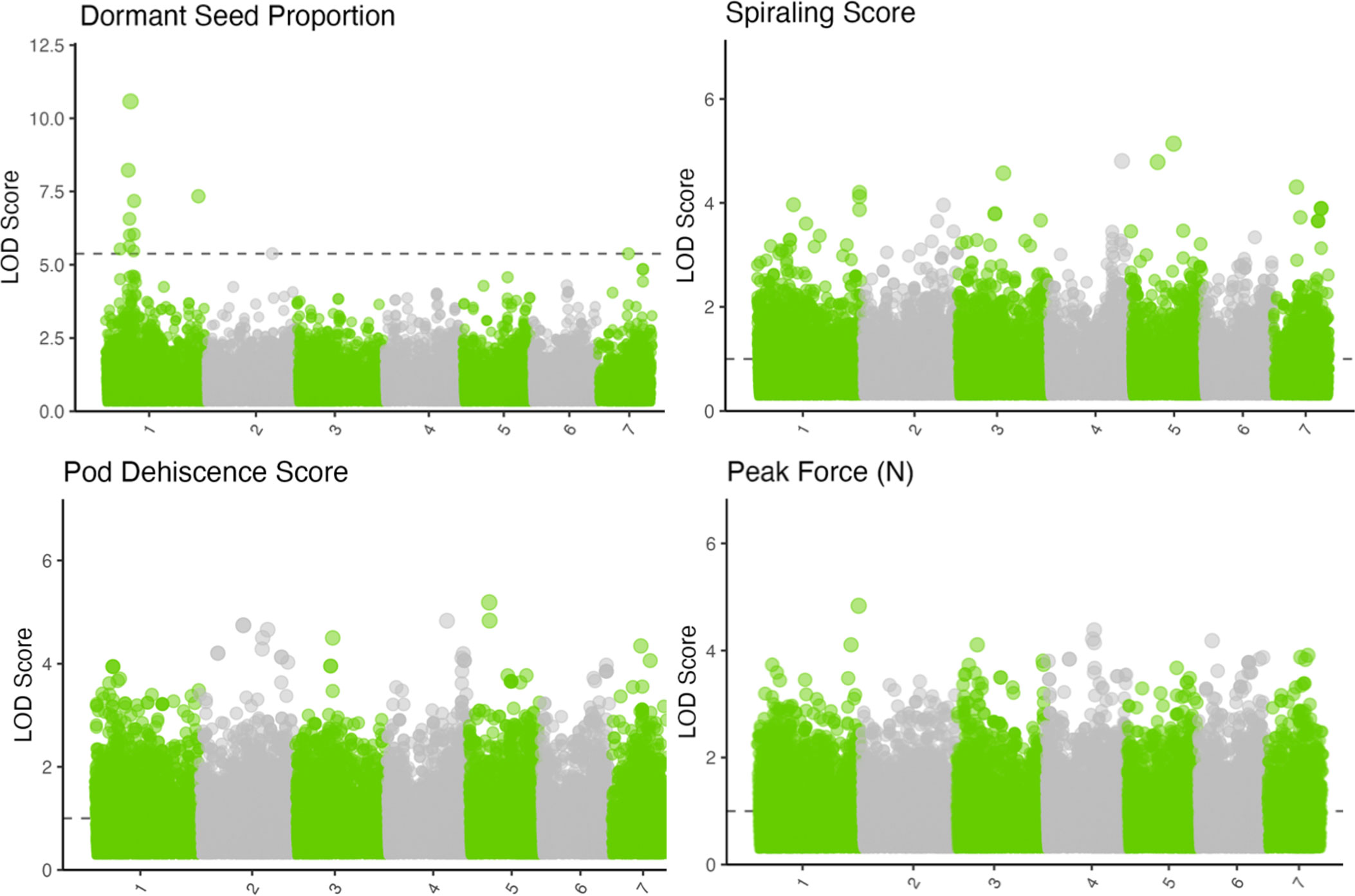
Figure 2 Manhattan plots for physical dormancy proportion, pod spiraling score, pod dehiscence score, and peak force to dehiscence. Colors alternate to differentiate between seven chromosomes. The y-axis indicates the logarithm of odds (LOD) score of the p-value for each marker model. The horizontal dotted line indicates the p-value level equivalent to a false discovery rate of 0.05. If no marker reached the false discovery rate of 0.05, a horizontal line was plotted at LOD 1. Note the unequal y-axes.
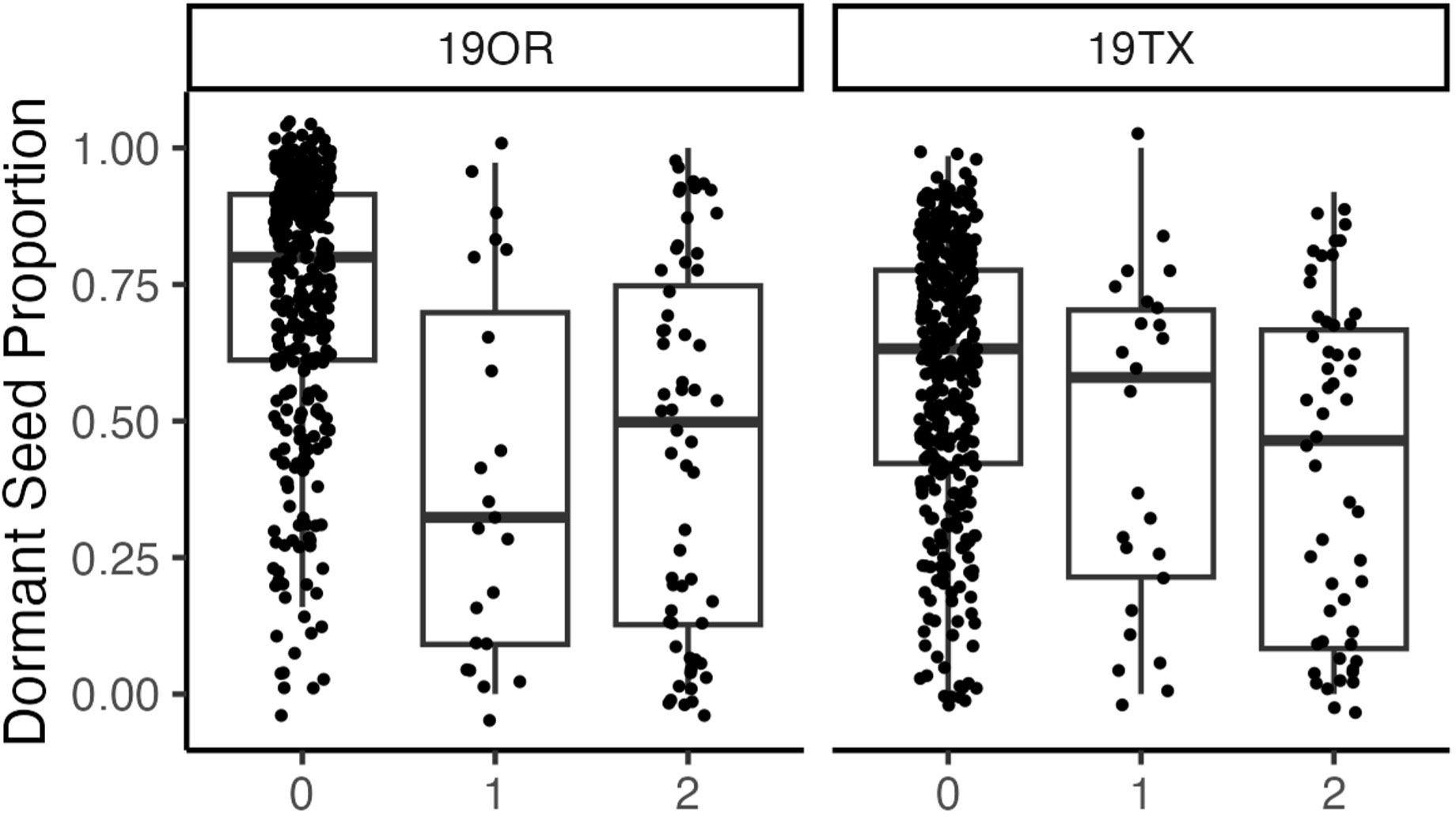
Figure 3 A boxplot indicating the relationship between allele dosage at a major quantitative trait locus (chromosome: 1, position: 63611165) and hard seed proportion at two site years (19OR: Oregon, 19TX: Texas). Each point is the unadjusted hard seed proportion for each individual with each allele dosage.
Force to dehiscence BLUPs, spiraling scores, and pod dehiscence score BLUPs had no statistically significant genome-wide associations, with the lowest q-value SNP for each trait remaining >0.20. Models run independently for Texas and Oregon sites found similarly insignificant associations within each trait. Furthermore, a multi-trait GWAS model including spiraling scores and pod dehiscence scores did not result in significant QTL despite modest correlations between the traits (Supplementary Figure S3; data not presented).
Discussion
This study found a large effect of QTL on the proportion of dormant seeds in V. villosa (Figure 3). Assays measuring seed dormancy are labor-intensive (29 min per sample) and cannot be completed prior to pollination of a crossing block. These labor-intensive phenotypic selection cycles have resulted in<10% reduction in seed dormancy per season (data not presented). Therefore, the strong QTL for seed dormancy is useful for reducing labor during breeding cycles and can allow screening prior to flowering. Eliminating seed dormancy will improve V. villosa agronomics by reducing volunteers in fields and allowing reduced seeding rates for producers (Kissing Kucek et al., 2020a). Further work is ongoing to (1) find a rapid genotyping platform for marker-assisted selection based on this QTL, (2) validate these QTL in more recent breeding material, and (3) determine potential genes responsible. Further genetic analysis will be aided by recently reported transcripts associated with seed coat formation in V. villosa (Ali et al., 2023). Prior reports of legume seed dormancy reduction during domestication are through mutations that weaken the seed coat (Smýkal et al., 2014; Sun et al., 2015; Chai et al., 2016; Sedláková et al., 2021; Soltani et al., 2021).
Given the presence of strong environmental variation, measurement uncertainties, and sequencing errors, the primary seed dormancy QTL could range from having a moderate impact to having complete control over seed dormancy (Beavis, 1998; Kissing Kucek et al., 2020a). Fortunately, there are two independent secondary QTL with q-value<0.05 both within (position: 61125989; q-value = 0.004) and outside chromosome 1 (chromosome: 7; position: 77125802; q-value = 0.046; Figure 2; Supplementary Table S1). These secondary QTLs have modest statistical support and are independent of each other and the primary QTL (r2< 0.2), indicating that they could provide additional or alternative reductions in seed dormancy.
The germplasm in this study has undergone selection for two generations in the Cover Crop Breeding Network. By tracing pedigree information, it was found that the major QTL-reducing seed dormancy was over-represented (>50% beneficial allele frequency) in progeny descended from three germplasm sources: WIHV1, WIHV2, and PI 491408. This is a greater concentration than would occur by chance based on a Chi-squared test for allele counts in WIHV1, WIHV2, and PI 491408 (all p< 0.0001). Omitting progeny from these germplasm sources, the allele frequency drops from 32% to 13%. Both WIHV1 and WIHV2 are the results of prior breeding efforts from a diversity panel in Wisconsin. PI 491408 is a collection from China (Inner Mongolia region) from 1984 and has shown generally strong winter survival in Wisconsin. Interestingly, WIHV1 is derived from PI 491408 through its maternal pedigree. These are potential genetic resources for further evaluation and introgression.
Prior studies documenting loci controlling dormant seed have focused on crosses between domesticated and wild-type populations (Forbes and Wells, 1968; Ladizinsky, 1985; Weeden, 2007; Isemura et al., 2010; Isemura et al., 2012; Kongjaimun et al., 2012; Jang et al., 2015). These studies often report that seed dormancy can be controlled by a single or small number of alleles. In contrast, this study reports the presence of one major and multiple minor QTL within the standing variation of a breeding population (Figure 2). It is likely there are multiple genetic paths that could eliminate seed dormancy, but only one largely effective QTL would be fixed in a domesticated population. This is probably because the genetic architecture required to maintain seed dormancy (or dehiscence) is polygenic, and any mutation weakening or disabling a required gene could eliminate dormancy. For example, Sun et al. (2015) reported that different G. max populations contained distinct mutations reducing seed dormancy, and each induced different seed coat alterations (increased permeability versus increased cracking). Similarly, Soltani et al. (2021) found that a QTL controlling seed water uptake occurred in only 77% of domesticated Phaseolus vulgaris L. samples, and that their reported QTL occurred at different rates in different growing environments. The exact mutation that becomes fixed in a cultivated population is likely a mixture of chance and physiological constraints. For example, Chai et al. (2016) reported a loss of seed dormancy in M. truncatula through a mutation in the KNOX II gene, but alterations in this gene influenced cutin formation across many organs. This could influence fitness. Other domestication-related mutations appear to have genetic “side-effects” that could be adaptive or maladaptive for a given environment (Weeden, 2007; Soltani et al., 2021). Therefore, semidomesticated populations or meta-populations could segregate for multiple large-effect QTL for domestication traits such as seed dormancy. However, modern cultivars are likely to have only one large effect on QTL because redundant QTL does not provide additional adaptive benefits in agronomic conditions.
No significant QTL were found for pod dehiscence scores, pod spiraling scores, or force to shatter. Interestingly, the surrogate measures for pod dehiscence scores had lower reliability estimates (force to dehiscence: 0.41; spiraling score: 0.42) than the primary phenotype of visual dehiscence scores (0.61). Kissing Kucek et al. (2020b) previously reported force to shatter is a promising trait for phenotyping to differentiate between individuals with low pod dehiscence rates, while spiraling scores are best able to differentiate among individuals with high pod dehiscence rates. In contrast to the extensive time and labor required to phenotype seed dormancy and force-to-dehiscence, pod dehiscence scores can be measured in<1 min per field replicate. Without a significant QTL for dehiscence, visual scoring of pod openness remains a fast method for evaluating dehiscence in breeding programs.
Broadly, the lack of significant pod dehiscence QTL was due to a combination of smaller sample sizes, lower trait reliability, and poor trait distributions at the Oregon site. It is also possible that major QTLs exist in the breeding program but are at a frequency below the statistical detection threshold. Oregon was a much more challenging growing environment for pod shatter traits, with >80% of pod dehiscence scores >2.5 (on a scale of 0–3). Since the force-to-shatter measurement required unshattered pods, this limited the Oregon force-to-shatter sample size to 133. The use of Oregon as a site was strategic since west-central Oregon is a major region for growing cover crop seed. The dry Oregon summers, which enable seed production, will also exacerbate pod dehiscence. Parker et al. (2020) speculated that major new alleles that reduce dehiscence occurred in P. vulgaris when introduced to more arid growing conditions. Similarly, continued selection to reduce pod dehiscence in arid seed-producing regions will be required to improve cover crop seed yields.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://datadryad.org/stash/share/cm3KNBgBAYYOmc4yQlEkMjaVV4mM6s4Eb5LjFNglgH8.
Author contributions
NT: Formal Analysis, Investigation, Visualization, Writing – original draft. LK: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – review & editing. BC: Data curation, Project administration, Supervision, Writing – review & editing. AM: Data curation, Project administration, Supervision, Writing – review & editing. JD: Data curation, Project administration, Supervision, Writing – review & editing. JE: Data curation, Project administration, Supervision, Writing – review & editing. SA: Data curation, Formal Analysis, Investigation, Supervision, Writing – review & editing. JR: Data curation, Supervision, Writing – review & editing. SB: Data curation, Formal Analysis, Writing – review & editing. SM: Data curation, Funding acquisition, Writing – review & editing. MM: Data curation, Project administration, Supervision, Writing – review & editing. SK: Data curation, Writing – review & editing. RH: Data curation, Project administration, Supervision, Writing – review & editing. MA: Data curation, Project administration, Supervision, Writing – review & editing. HR: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by USDA-ARS Projects 5090-21000-001-00D, USDA-NIFA grants 2018-67013-27570 and 2015-51300-24192.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1282187/full#supplementary-material
References
Abd El-Moneim, A. M. (1993). Selection for non-shattering common vetch, Vicia sativa L. Plant Breed 110, 168–171. doi: 10.1111/j.1439-0523.1993.tb01231.x
Ali, S., Kucek, L. K., Riday, H., Krom, N., Krogman, S., Cooper, K., et al. (2023). Transcript profiling of hairy vetch (Vicia villosa Roth) identified interesting genes for seed dormancy. Plant Genome 16, e20330. doi: 10.1002/tpg2.20330
Austin, E. E., Wickings, K., McDaniel, M. D., Robertson, G. P., Grandy, A. S. (2017). Cover crop root contributions to soil carbon in a no-till corn bioenergy cropping system. GCB Bioenergy 9, 1252–1263. doi: 10.1111/gcbb.12428
Beavis, W. D. (1998). “QTL analyses: power, precision, and accuracy,” in Molecular Dissection of Complex Traits (Boca Raton, FL, USA: CRC Press).
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bohra, A., Tiwari, A., Kaur, P., Ganie, S. A., Raza, A., Roorkiwal, M., et al. (2022). The key to the future lies in the past: insights from grain legume domestication and improvement should inform future breeding strategies. Plant Cell Physiol. 63, 1554–1572. doi: 10.1093/pcp/pcac086
Bourgeois, B., Charles, A., Van Eerd, L. L., Tremblay, N., Lynch, D., Bourgeois, G., et al. (2022). Interactive effects between cover crop management and the environment modulate benefits to cash crop yields: a meta-analysis. Can. J. Plant Sci. 102, 656–678. doi: 10.1139/cjps-2021-0177
Brandsæter, L. O., Olsmo, A., Tronsmo, A. M., Fykse, H. (2002). Freezing resistance of winter annual and biennial legumes at different developmental stages. Crop Sci. 42, 437–443. doi: 10.2135/cropsci2002.4370
Browning, B. L., Zhou, Y., Browning, S. R. (2018). A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103, 338–348. doi: 10.1016/j.ajhg.2018.07.015
Butler, D. G., Cullis, B. R., Gilmour, A. R., Gogel, B. J., Thompson, R. (2017). ASReml-R reference manual version 4 (Hemel Hempstead HP1 1ES UK: VSN Int. Ltd).
Chai, M., Zhou, C., Molina, I., Fu, C., Nakashima, J., Li, G., et al. (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. 113, 6997–7002. doi: 10.1073/pnas.1601256113
Clark, A. J., Decker, A. M., Meisinger, J. J., McIntosh, M. S. (1997). Kill date of vetch, rye, and a vetch–rye mixture: I. Cover Crop Corn Nitrogen. Agron. J. 89, 427–434. doi: 10.2134/agronj1997.00021962008900030010x
Covarrubias-Pazaran, G. (2016). Genome-assisted prediction of quantitative traits using the R package sommer. PloS One 11, e0156744. doi: 10.1371/journal.pone.0156744
Curran, W. S. (2015). Effectiveness of herbicides for control of hairy vetch (Vicia villosa) in winter wheat. Weed Technol. 29, 509–518. doi: 10.1614/WT-D-14-00139.1
Doebley, J. F., Gaut, B. S., Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Drinkwater, L. E., Wagoner, P., Sarrantonio, M. (1998). Legume-based cropping systems have reduced carbon and nitrogen losses. Nature 396, 262–265. doi: 10.1038/24376
Forbes, I., Wells, H. (1968). Hard and soft seededness in Blue Lupine, Lupinus angustifolius L.: inheritance and pheno- type classification. Crop Sci. 8, 195–197. doi: 10.2135/cropsci1968.0011183X000800020018x
Fuller, T., Koch, L. M., Kucek, L. K., Ali, S., Mangelson, H., et al. (2023). A reference assembly for the legume cover crop, hairy vetch (Vicia villosa). bioRxiv. 2023.03.28.534423.
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PloS One 9, e90346. doi: 10.1371/journal.pone.0090346
Hamadi, H., Aziza, Z., Salah, B. Y., Khouloud, C. (2012). Analysis of floral Biology of Vicia villosa Roth. Fabaceae Agric. Sci. 2, 121–127.
Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PloS One 7, e41304. doi: 10.1371/journal.pone.0041304
Isemura, T., Kaga, A., Tomooka, N., Shimizu, T., Vaughan, D. A. (2010). The genetics of domestication of rice bean, Vigna umbellata. Ann. Bot. 106, 927–944. doi: 10.1093/aob/mcq188
Jacobsen, K. L., Gallagher, R. S., Burnham, M., Bradley, B. B., Larson, Z. M., Walker, C. W., et al. (2010). Mitigation of seed germination impediments in hairy vetch. Agron. J. 102, 1346–1351. doi: 10.2134/agronj2010.0002n
Jang, S.-J., Sato, M., Sato, K., Jitsuyama, Y., Fujino, K., Mori, H., et al. (2015). A single-nucleotide polymorphism in an endo-1,4-β-glucanase gene controls seed coat permeability in soybean. PloS One 10, e0128527. doi: 10.1371/journal.pone.0128527
Jannink, J. (1997). Management and winter hardiness of hairy vetch in Maine. Maine Agric. For. Exp. Stn. Tech. Bull. 167, 1–35.
Jones, J. P. (1928). A physiological study of dormancy in vetch seed. Cornell Univ. Agric. Exp. Stn. 120, 1–50.
Kahlaoui, S. (2009). The morphology, chromosome number and nuclear DNA content of Tunisian populations of three Vicia species. Afr. J. Biotechnol. 8, 3184–3191. doi: 10.5897/AJB09.496
Kalu, B. A., Fick, G. W. (1981). Quantifying morphological development of alfalfa for studies of herbage quality. Crop Sci. 21, 267–271. doi: 10.2135/cropsci1981.0011183X002100020016x
Kaspar, T. C., Bakker, M. G. (2015). Biomass production of 12 winter cereal cover crop cultivars and their effect on subsequent no-till corn yield. J. Soil Water Conserv. 70, 353–364. doi: 10.2489/jswc.70.6.353
Kissing Kucek, L., Azevedo, M. D., Eagen, S. S., Ehlke, N. J., Hayes, R. J., Mirsky, S. B., et al. (2020a). Seed dormancy in hairy vetch (Vicia villosa Roth) is influenced by genotype and environment. Agronomy 10, 1804. doi: 10.3390/agronomy10111804
Kissing Kucek, L., Riday, H., Rufener, B. P., Burke, A. N., Eagen, S. S., Ehlke, N., et al. (2020b). Pod dehiscence in hairy vetch (Vicia villosa Roth). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00082
Kongjaimun, A., Kaga, A., Tomooka, N., Somta, P., Vaughan, D. A., Srinives, P. (2012). The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann. Bot. 109, 1185–1200. doi: 10.1093/aob/mcs048
Kucek, L. K., Riday, H., Ehlke, N., Reberg-Horton, C., Maul, J., Mirsky, S. B., et al. (2019). Environmental influences on the relationship between fall and spring vigor in hairy vetch. Crop Sci. 59, 2443–2454. doi: 10.2135/cropsci2018.09.0569
Ladizinsky, G. (1985). The genetics of hard seed coat in the genus Lens. Euphytica 34, 539–543. doi: 10.1007/BF00022952
Mirsky, S. B., Ackroyd, V. J., Cordeau, S., Curran, W. S., Hashemi, M., Reberg-Horton, S. C., et al. (2017). Hairy vetch biomass across the Eastern United States: effects of latitude, seeding rate and date, and termination timing. Agron. J. 109, 1510–1519. doi: 10.2134/agronj2016.09.0556
Mirsky, S. B., Wallace, J. M., Curran, W. S., Crockett, B. C. (2015). Hairy vetch seedbank persistence and implications for cover crop management. Agron. J. 107, 2391–2400. doi: 10.2134/agronj14.0523
Moore, V. M., Maul, J. E., Wilson, D., Curran, W. S., Brainard, D. C., Devine, T. E., et al. (2020). Registration of ‘Purple Bounty’ and ‘Purple Prosperity’ hairy vetch. J. Plant Regist. 14, 340–346. doi: 10.1002/plr2.20044
Nevins, C. J., Lacey, C., Armstrong, S. (2020). The synchrony of cover crop decomposition, enzyme activity, and nitrogen availability in a corn agroecosystem in the Midwest United States. Soil Tillage Res. 197, 104518. doi: 10.1016/j.still.2019.104518
Parker, T. A., Berny Mier y Teran, J. C., Palkovic, A., Jernstedt, J., Gepts, P. (2020). Pod indehiscence is a domestication and aridity resilience trait in common bean. New Phytol. 225, 558–570. doi: 10.1111/nph.16164
Parr, M., Grossman, J. M., Reberg-Horton, S. C., Brinton, C., Crozier, C. (2011). Nitrogen delivery from legume cover crops in no-till organic corn production. Agron. J. 103, 1578–1590. doi: 10.2134/agronj2011.0007
Poeplau, C., Don, A. (2015). Carbon sequestration in agricultural soils via cultivation of cover crops – A meta-analysis. Agric. Ecosyst. Environ. 200, 33–41. doi: 10.1016/j.agee.2014.10.024
Power, J. F., Zachariassen, J. A. (1993). Relative nitrogen utilization by legume cover crop species at three soil temperatures. Agron. J. 85, 134–140. doi: 10.2134/agronj1993.00021962008500010025x
Purugganan, M. D., Fuller, D. Q. (2009). The nature of selection during plant domestication. Nature 457, 843–848. doi: 10.1038/nature07895
Quinlivan, B. J. (1968). The softening of hard seeds of sand-plain lupin (Lupinus Varius L. Aust. J. Agric. Res. 19, 507–515. doi: 10.1071/AR9680507
Ranells, N. N., Wagger, M. G. (1996). Nitrogen release from grass and legume cover crop monocultures and bicultures. Agron. J. 88, 777–882. doi: 10.2134/agronj1996.00021962008800050015x
Renzi, J. P., Chantre, G. R., Cantamutto, M. A. (2014). Development of a thermal-time model for combinational dormancy release of hairy vetch (Vicia villosa ssp. villosa. Crop Pasture Sci. 65, 470–478. doi: 10.1071/CP13430
Renzi, J. P., Garayalde, A. F., Brus, J., Pohankova, T., Smýkal, P., Cantamulo, M. A. (2023). Environmental and agronomic determinants of hairy vetch (Vicia villosa Roth) seed yield in rainfed temperate agroecosystems. Eur. J. Agron. 147, 126822. doi: 10.1016/j.eja.2023.126822
Sainju, U. M., Singh, B. P. (1997). Winter cover crops for sustainable agricultural systems: Influence on soil properties, water quality, and crop yields. HortScience 32, 21–28. doi: 10.21273/HORTSCI.32.1.21
Samarah, N. H. (2006). Effect of air-drying immature seeds in harvested pods on seed quality of common vetch (Vicia sativa L. N. Z. J. Agric. Res. 49, 331–339. doi: 10.1080/00288233.2006.9513723
Schmidt, P., Hartung, J., Bennewitz, J., Piepho, H.-P. (2019). Heritability in plant breeding on a genotype-difference basis. Genetics 212, 991–1008. doi: 10.1534/genetics.119.302134
Sedláková, V., Hanáček, P., Grulichová, M., Zablatzká, L., Smýkal, P. (2021). Evaluation of seed dormancy, one of the key domestication traits in chickpea. Agronomy 11, 2292. doi: 10.3390/agronomy11112292
Singh, G., Dhakal, M., Yang, L., Kaur, G., Williard, K. W. J., Schoonover, J. E., et al. (2020). Decomposition and nitrogen release of cover crops in reduced- and no-tillage systems. Agron. J. 112, 3605–3618. doi: 10.1002/agj2.20268
Smýkal, P. (2014). A comparative study of ancient DNA isolated from charred pea (Pisum sativum L.) seeds from an Early Iron Age settlement in southeast Serbia: inference for pea domestication. Genet. Resour. Crop Evol. 61, 1533–1544. doi: 10.1007/s10722-014-0128-z
Smýkal, P., Vernoud, V., Blair, M. W., Soukup, A., Thompson, R. D. (2014). The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00351
Snapp, S., Surapur, S. (2018). Rye cover crop retains nitrogen and doesn’t reduce corn yields. Soil Tillage Res. 180, 107–115. doi: 10.1016/j.still.2018.02.018
Soltani, A., Walter, K. A., Wiersma, A. T., Santiago, J. P., Quiqley, M., Chitwood, D., et al. (2021). The genetics and physiology of seed dormancy, a crucial trait in common bean domestication. BMC Plant Biol. 21, 58. doi: 10.1186/s12870-021-02837-6
Sun, L., Miao, Z., Cai, C., Zhang, D., Zhao, M., Wu, Y., et al. (2015). GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat. Genet. 47, 939–943. doi: 10.1038/ng.3339
Teasdale, J. R. (1996). Contribution of cover crops to weed management in sustainable agricultural systems. J. Prod. Agric. 9, 475–479. doi: 10.2134/jpa1996.0475
Thompson, N. M., Armstrong, S. D., Roth, R. T., Ruffatti, M. D., Reeling, C. J. (2020). Short-run net returns to a cereal rye cover crop mix in a midwest corn–soybean rotation. Agron. J. 112, 1068–1083. doi: 10.1002/agj2.20132
Tiemann, L. K., Grandy, A. S., Atkinson, E. E., Marin-Spiotta, E., McDaniel, M. D. (2015). Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecol. Lett. 18, 761–771. doi: 10.1111/ele.12453
Vyn, T. J., Faber, J. G., Janovicek, K. J., Beauchamp, E. G. (2000). Cover crop effects on nitrogen availability to corn following wheat. Agron. J. 92, 915–924. doi: 10.2134/agronj2000.925915x
Wagger, M. G., Cabrera, M. L., Ranells, N. N. (1998). Nitrogen and carbon cycling in relation to cover crop residue quality. J. Soil Water Conserv. 53, 214–218.
Wallace, J. M., Williams, A., Liebert, J. A., Ackroyd, V. J., Vann, R. A., Curran, W. S., et al. (2017). Cover crop-based, organic rotational no-till corn and soybean production systems in the Mid-atlantic United States. Agriculture 7, 34. doi: 10.3390/agriculture7040034
Wallander, S., Smith, D., Bowman, M., Claassen, R. (2021). Cover Crop Trends, Programs, and Practices in the United States. EIB 222, U.S. Department of Agriculture, Economic Research Service. Available online at: https://www.ers.usda.gov/webdocs/publications/100551/eib-222.pdf (Accessed October 5, 2022).
Wayman, S., Kucek, L. K., Mirsky, S. B., Ackroyd, V., Cordeau, S., Ryan, M. R. (2017). Organic and conventional farmers differ in their perspectives on cover crop use and breeding. Renew. Agric. Food Syst. 32, 376–385. doi: 10.1017/S1742170516000338
Weeden, N. F. (2007). Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the “domestication syndrome” for legumes? Ann. Bot. 100, 1017–1025. doi: 10.1093/aob/mcm122
Wilke, B. J., Snapp, S. S. (2008). Winter cover crops for local ecosystems: linking plant traits and ecosystem function. J. Sci. Food Agric. 88, 551–557. doi: 10.1002/jsfa.3149
Williams, W. A., Elliott, J. R. (1960). Ecological significance of seed coat impermeability to moisture in crimson, subterreanean and rose clovers in a mediterranean-type climate. Ecology 41, 733–742. doi: 10.2307/1931807
Wyland, L. J. (1996). Altering surface soil dynamics with winter cover crops in a vegetable cropping system: impacts on yield, nitrate leaching, pests and management costs. Agric. Ecosyst. Environ. 59, 1–17. doi: 10.1016/0167-8809(96)01048-1
Keywords: seed dormancy, genomewide association, cover crop, legume, genomics
Citation: Tilhou N, Kucek LK, Carr B, Marion A, Douglas J, Englert J, Ali S, Raasch J, Bhamidimarri S, Mirsky SB, Monteros MJ, Krogman S, Hayes R, Azevedo M and Riday H (2023) Genome-wide association mapping in hairy vetch (Vicia villosa) discovers a large effect locus controlling seed dormancy. Front. Plant Sci. 14:1282187. doi: 10.3389/fpls.2023.1282187
Received: 23 August 2023; Accepted: 06 October 2023;
Published: 24 October 2023.
Edited by:
Petr Smýkal, Palacký University in Olomouc, CzechiaReviewed by:
Juan Pablo Renzi, Instituto Nacional de Tecnología Agropecuaria, ArgentinaOldřich Trněný, Agricultural Research Ltd., Czechia
Copyright © 2023 Tilhou, Kucek, Carr, Marion, Douglas, Englert, Ali, Raasch, Bhamidimarri, Mirsky, Monteros, Krogman, Hayes, Azevedo and Riday. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neal Tilhou, bmVhbC50aWxob3VAdXNkYS5nb3Y=
 Neal Tilhou
Neal Tilhou Lisa Kissing Kucek
Lisa Kissing Kucek Brandon Carr2
Brandon Carr2 Shahjahan Ali
Shahjahan Ali Steven Brian Mirsky
Steven Brian Mirsky Ryan Hayes
Ryan Hayes Mark Azevedo
Mark Azevedo Heathcliffe Riday
Heathcliffe Riday