
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 October 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1280970
This article is part of the Research Topic Omics Technologies and Fruit Postharvest Quality View all 7 articles
Anthocyanin is the main component of pigment in red-fleshed kiwifruit. ‘Jinhongguan’ is a new cultivar of Actinidia arguta with red peel and flesh after harvest. However, the specific types of anthocyanin in the ‘Jinhongguan’ fruit and its biosynthesis pathways remain largely unknown. Here, the total anthocyanin content in the fruit color conversion process was determined. The results showed that total anthocyanin content increased with the deepening color of the peel and flesh. To identify the genes related to anthocyanin biosynthesis and the types of anthocyanins in the ‘Jinhongguan’ fruit, a combined analysis of transcriptome and anthocyanin-targeted metabolome was carried out. A total of 5751 common differentially expressed genes (DEGs) at different stages of peel and flesh were identified, of which 2767 were common up-DEGs and 2976 were common down-DEGs. KEGG and GO enrichment analyses showed that the common up-DEGs were significantly enriched in anthocyanin synthesis-related pathways, suggesting some up-DEGs are involved in anthocyanin biosynthesis. In total, 29 metabolites were detected in the flesh by anthocyanin-targeted metabolome. Among these, nine were differential accumulation metabolites (DAMs) in comparison to red flesh vs green flesh. Six DAMs were up-regulated, with five of them were cyanidins. The content of cyanidin-3-O-galactoside was much higher than that of other DAMs, making it the main pigment in ‘Jinhongguan’. Moreover, a total of 36 anthocyanin synthesis-related structural genes, 27 MYB transcription factors (TFs), 37 bHLH TFs and 9 WDR TFs were screened from the common DEGs. Correlation analysis of transcriptome and metabolome revealed that 9 structural genes, 6 MYB TFs, 6 bHLH TFs and 1 WDR TF were significantly associated with cyanidin-3-O-galactoside. Further, qRT-PCR analysis demonstrated that structural genes (AaPAL3, Aa4CL3, AaCHS2/3/8/9/11, AaDFR1/2, AaANR1, UFGT3a and UFGT6b) and TFs (MYB108, bHLH30, bHLH94-1 and WD43) play important roles in cyanidin biosynthesis. Overall, this study identified cyanidin-3-O-galactoside as the main anthocyanin type and revealed key candidate genes of red coloration of post-harvest fruit in Actinidia arguta. These findings provided new insights into the color formation mechanism of post-harvest fruit and offered a theoretical basis for color regulation in kiwifruit.
Kiwifruit belongs to Actinidia, and it has a cultivation history of more than 110 years from wild to artificial domestication (Huang and Ferguson, 2007). At present, the main cultivated species in production include Actinidia chinensis, Actinidia deliciosa, Actinidia arguta and Actinidia eriantha (Hanley, 2018; Huang et al., 2013). Kiwifruit is highly favored by consumers due to its unique fruit flavor, high nutritional value, and notably high vitamin C content (Richardson et al., 2018). The fruit of kiwifruit has a variety of colors, mainly divided into green, yellow and red, among which red fruit is mainly due to the presence of anthocyanin (Sui et al., 2013). Anthocyanins possess antioxidant capacity and exhibit multiple health-promoting advantages, such as anti-aging, anti-cancer, and lowering of blood lipids (Bendokas et al., 2020). In Actinidia arguta kiwifruit, the accumulation of anthocyanin is most abundant, and some varieties of them capable of accumulating anthocyanin in both the peel and flesh, resulting in the whole fruit being red or even purple, which is highly appealing to consumers (Li et al., 2018a). The molecular mechanisms of kiwifruit fruit color conversion from green to red are highly complex (Valenta et al., 2018). Currently, some genes involved in the coloration of red kiwifruit have been identified, but they mainly play a role in the pre-harvest coloring process of kiwifruit flesh (Wang et al., 2022b; Wang et al., 2022c; Shu et al., 2023). The molecular mechanisms of post-harvest reddening of kiwifruit peel and flesh remain largely unclear.
Anthocyanins widely exist in various organs of plants, such as leaves, flowers and fruits, and they are the basis for plants to present various colors. The basic structure of anthocyanin is C6-C3-C6, which is composed of two aromatic rings and one oxygen-containing heterocyclic ring (Bloor and Abrahams, 2002). According to the different structures, anthocyanin can be divided into six categories, namely, peonidin, pelargonidin, cyanidin, delphinidin, petunidin and malvidin (Smeriglio et al., 2016). Currently, about 92% of the over 500 anthocyanins are formed by combining the hydroxyl groups of these six anthocyanins with different sugar groups (Sun et al., 2018). Different types of anthocyanins present slight differences in colors. For example, pelargonidin and its derivatives are red or orange; cyanidin and its derivatives are mainly brick red or mauve; delphinidin and its derivatives are blue or purple (Liu et al., 2018). The anthocyanin biosynthesis pathway is relatively conserved in higher plants and is an important branch of flavonoid metabolism. The biosynthesis of anthocyanins can be divided into three stages according to the enzyme reaction process. The first stage is the synthesis process of 4-acyl-CoA from phenylalanine through three enzymatic reactions, which is a common pathway of secondary metabolism in many plants; the second stage is the formation of the basic skeleton of anthocyanins, and the synthesis of dihydrokaempferol by three enzymatic reactions of 4-coumaroyl A; the third stage involves the anthocyanin precursor forms different substrates under the catalysis of different enzymes, followed by gradual modification into various anthocyanin glycosides (Jaakola, 2013).
The synthesis of anthocyanins is inseparable from the catalysis of structural gene-encoded enzymes. According to the time of participation in the reaction, structural genes can be categorized into early biosynthetic genes (EBG): phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxy-lase (C4H), 4-coumaroyl-CoA (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), flavonoid 3’, 5’-hydroxylase (F3’,5’H); and late biosynthetic genes (LBG): dihydro flavonol 4-reductase (DFR), anthocyanidin synthase (ANS), flavonoid 3-O-glucosyl-transferase (UFGT), glutathione S-transferase (GST) and so on (Liu et al., 2018). The function of these structural genes has been confirmed in various species. For instance, the expression down-regulation of SICHS resulted in the flavonoid content decrease and the color of the tomato peel lighter (Heredia et al., 2015). The expression up-regulation of the DFR gene in purple radish led to increased synthesis of anthocyanin in flavonoid metabolism, while the mutation of the DFR gene in green radish caused an obstacle to anthocyanin synthesis (Zhuang et al., 2019). In onion, two ANS allele variations resulted in the loss of anthocyanin (Kim et al., 2016).
In addition, anthocyanin synthesis is regulated by various TFs, among which the most important three types are MYB, bHLH and WDR (MBW). These TFs either individually or in combination with each other form the MBW complex, thus promoting or inhibiting anthocyanin synthesis by recognizing and binding to the promoters of structural genes (Espley et al., 2007; An et al., 2012; Zhu et al., 2015; Lai et al., 2016). For example, MdMYBA and MdMYB1 regulate anthocyanin synthesis in the peel, while MdMYB10 and MdMYB110a control anthocyanin synthesis in leaves and flesh (Lin-Wang et al., 2010; Hu et al., 2021). Similarly, in diploid strawberry, FveMYB10 and FveMYB10L independently regulate anthocyanin synthesis in fruit and petiole, respectively, and produce different anthocyanin components (Luo et al., 2023). Both MdbbHLH3 and MdbbHLH33 interact with MdbMYB10 in apple to promote fruit reddening (Espley et al., 2007). In litchi, LcbHLH1 and LcbHLH3 cooperate with LcMYB1 to regulate the synthesis and accumulation of anthocyanin (Lai et al., 2016). Overexpression of the WD40 transcription factor StAN11 in potato can enhance the expression of the DFR gene and deepen the color of the tuber epidermis (Zhang et al., 2017). In pepper, silencing CaMYBA and CaWD40 resulted in decreased expression of anthocyanin synthesis-related structural genes and anthocyanin content (Aguilar-Barragán and Ochoa-Alejo, 2014). In Arabidopsis, MYB family members PAP1 and PAP2 bind to bHLH family member TT8 and WD40 family member TTG1 to form a complex that activates the expression of CHS and DFR (Borevitz et al., 2000). In addition, some other TFs such as WRKY, ERF, SPL, COP1 and NAC have also been found to regulate anthocyanin synthesis (Zhou et al., 2015; Chen et al., 2019; Shu et al., 2023; Zou et al., 2023).
In kiwifruit, AcMYB75 has been found to bind to ANS promoting the significant accumulation of anthocyanin in the leaves of Actinidia chinensis (Li et al., 2017b). MYB110a, which was significantly expressed in the red petal offspring of the kiwifruit hybrid population but not in the white petal individuals, could directly activate the expression of F3GT1 (Fraser et al., 2013). Overexpression of MYB10 or MYB110 increased cyanidin and delphinidin anthocyanins (Peng et al., 2019; Shu et al., 2023). MYB110 has a stronger activation effect than MYB10, and overexpression of MYB10 lea to anthocyanin accumulation only in the endocarp (Wang et al., 2022a). AcMYB5-1/5-2/A1-1 promoted the accumulation of anthocyanin in kiwifruit flesh by activating the transcription of ANS and DFR (Li et al., 2017a). While the interaction between AcMYB123 and AcbHLH42 is necessary for activating the expression of anthocyanin biosynthetic enzymes AcANS and AcF3GT1 (Wang et al., 2019). So far, most of the genes involved in anthocyanin synthesis identified in kiwifruit are MYB TFs. The types of anthocyanins in red-fleshed kiwifruit and more key genes related to anthocyanin biosynthesis still need to be further identified.
In this study, Actinidia arguta ‘Jinhongguan’ is a post-harvest full-red (peel and flesh) kiwifruit cultivar, which is a good material for exploring the mechanism of anthocyanin biosynthesis in post-harvest kiwifruit. Transcriptome and metabolome were used to analyze the types and biosynthesis pathways of anthocyanins in ‘Jinhongguan’. Further, the correlation analysis between transcriptome, metabolome and qRT-PCR analysis was used to screen key candidate genes responsible for cyanidin-3-O-galactoside biosynthesis in ‘Jinhongguan’. This study identified the main types of anthocyanins in the post-harvest fruit of Actinidia arguta and the key candidate genes related to anthocyanin biosynthesis, which provide insight into the regulatory network of anthocyanin biosynthesis.
The Actinidia arguta cultivar ‘Jinhongguan’ plants were grafted onto 5-year-old ‘RZ002’ (Actinidia arguta) rootstock and planted for more than 3 years in the National Kiwifruit Germplasm Resource Garden of China. The garden is located in the Research Institute of Fruit and Tea, Hubei Academy of Agricultural Sciences (30°29′ N, 114°16′ E), Wuhan, China. Mature fruit samples were collected in early August, with three biological duplicates, and stored in the dark at 25°C after harvest. The samples were divided into four stages based on the color of the fruit peel and flesh. Stage 1: newly harvested fruits with green peel (FP1) and flesh (FF1); Stage 2: two days after fruit harvest, the peel (FP2) began to turn red, while the flesh (FF2) was still green; Stage 3: three days after fruit harvest, the peel (FP3) became all red, and the flesh (FF3) began to turn red; Stage 4: six days after fruit harvest, both the peel (FP4) and flesh (FF4) were completely red. For the transcriptome and metabolome analyses, the fruit peel and flesh from the four stages of ‘Jinhongguan’ were separated, immediately frozen in liquid nitrogen, and then stayed at −80°C.
The microplate anthocyanin content determination kit (Grace Biotechnology, Suzhou, China) was used to measure the total anthocyanin content following the provided instructions. Briefly, the flesh samples were ground into a powder in liquid nitrogen, and then approximately 0.15 g sample powder was added to 1 mL extraction solution for oscillation extraction at 75°C for 25 minutes. The extract was centrifuged at 12,000 rpm for 10 minutes, and the supernatant was taken into a clean tube for testing. Take 100 μL of the supernatant and add 300 μL of reagent 1 or reagent 2 mix evenly, and then stand for 60 minutes at room temperature and in the dark. Afterward, 200 μL of the clear mixed liquid was transferred into a 96-well plate, and the absorbance at 530 nm and 700 nm was measured to obtain the A1530 and A1700 values for the mixture added with reagent 1, and the A2530 and A2700 values for the mixture added with reagent 2, respectively. The anthocyanin content was calculated using the equation: An (mg/kg) = [ΔA ÷ (ε × d) × V2 × 103 × Mr] ÷ (W × V1 ÷ V) × D × 1000 = 133.6 × ΔA ÷ W × D, where ΔA = (A1530 − A1700) − (A2530 − A2700),ε= 26900 L/mol/cm (extinction coefficient of cyanidin-3-glucoside); d = 0.5 cm (optical path), V2 = 4 × 10-4 L (total volume detected), Mr = 449.2 (molecular weight of cyanidin-3-glucoside), W is the sample weight, V1 = 0.1 mL (detected sample volume), V = 1 mL (sample extract volume), and D = 1 (dilution multiple). Each sample contained three biological replicates, and three measurements were taken for each biological replicate.
A total of 12 samples were collected from three biological replicates in each stage of FP1 to FP4. Similarly, 12 samples were collected from three biological replicates in each stage of FF1 to FF4. These 24 samples from the peel and flesh of ‘Jinhongguan’ were used for transcriptome sequencing. According to the standard operating procedure, total RNA extraction was carried out using the RNAprep Plant Extraction Kit (QIAGEN, Germany), and the total amount and integrity were evaluated using the RNA Nano 6000 detection reagent system. Random hexamer primers and M-MuLV reverse transcriptase were used to synthesize the first strand cDNA, and DNA polymerase I and dNTP were used to synthesize the second strand cDNA. The DNA fragments were adenylated at the 3’end and then connected with adapters containing hairpin structures for hybridization. The library fragments were purified and optimized to obtain cDNA fragments of 370-420 bp. Then the fragments were used as templates for PCR amplification, and the amplified products were purified with AMPure XP beads. Finally, the libraries were constructed and subjected to quality control to obtain qualified libraries. The detailed steps were performed as previously study described (Pan et al., 2023).
The qualified libraries were mixed according to effective concentration and target amount and then sequenced by Illumina NovaSeq 6000. The raw reads in fastq format were first filtered by an in-house Perl script to obtain clean reads. The Q20 value, Q30 value, and GC content were calculated to evaluate the quality of the clean reads, and then the clean reads were assembled in Trinity software (v2.6.6) based on the reference sequence. Gene functional annotation was performed using the following databases: NR, NT, PFAM, KOG/COG, Swiss-Prot, KO and GO (Yu et al., 2020). Differential expression analysis was conducted using the DESeq2 R package (1.20.0) with screening thresholds of |log2FC| ≥1.0 and padj < 0.05. GOseq (Young et al., 2010) and KOBAS (Bu et al., 2021) were used for GO function and KEGG pathway enrichment analysis of the DEGs. Plant transcription factors were predicted by iTAK software and identified by hmmscan based on TF family classification and definition in the database. The transcriptome data was submitted to NCBI-SRA and can be accessed through NCBI BioProject with the identifier PRJNA1018137 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1018137).
The flesh samples were vacuum freeze-dried and ground into powder. About 50 mg of the powder was dissolved in 500 μL of extract (methanol: water: hydrochloric = 500:500:1). The mixture was vortexed for 5 minutes, followed by sonication for 5 minutes, and then centrifuged for 3 minutes (12,000 r/min, 4°C). The supernatant was collected, and the above residue was re-extracted, and then the supernatant was combined twice. The supernatant was filtered with a microporous membrane (0.22 um pore size) and the filtrate was stored in an injection vial for LC-MS/MS analysis. Ultra-high performance liquid chromatography (UPLC) (ExionLCTM AD) and tandem mass spectrometry (MS/MS) (QTRAPR6500+) were used for data collection. The specific liquid phase and mass spectrometry conditions were conducted following the methods previously reported (Shao et al., 2022).
Anthocyanins were analyzed using the Scheduled Multiple Reaction Monitoring (MRM) technique and performed by Metware Biotechnology (Wuhan, China). Analyst 1.6.3 software (Sciex) was used for data collection. All metabolite quantification was performed in Multiquant 3.0.3 software (Sciex). Anthocyanin content was detected by NetWare (http://www.metware.cn/). DAMs between groups were identified by VIP≥ 0 and |log2fold change| ≥ 1.0. The metabolome data has also been uploaded to the public database MetaboLights and can be accessed through the identifier MTBLS8619 (https://www.ebi.ac.uk/metabolights/MTBLS8619).
To jointly analyze transcriptome and metabolome data, Pearson’s Correlation Coefficients (PCC) among the DEGs and DAMs were analyzed. The PCC values were calculated using the weighted gene coexpression network analysis (WGCNA) R package with default parameters (v1.4.1717). Strong correlation conditions are |PCC| ≥ 0.8 and p < 0.05. The Cytoscape software (version 3.10.0) was used to visualize the corresponding association network. The names of the 109 anthocyanins synthesis-related structural genes and TFs in Table S6 were mainly obtained from the annotation results of the NT and NR databases, such as MYB4, MYB7 and MYB13. For genes that were not annotated with specific names in the NT or NR database, they were classified based on gene category and named sequentially according to the gene ID, such as AaMYB1 to AaMYB10.
The total RNA of the fruit peel and flesh samples were extracted using the Total RNA kit (Aidlab Biotechnology, Beijing, China). The quality and concentration of the RNA were detected by agarose gel electrophoresis and Nanodrop spectrophotometer. The cDNA was synthesized by TRUEscript RT MasterMix (OneStep gDNA Removal) kit (Aidlab Biotechnology, Beijing, China). The qRT-PCR was performed according to the method described in our previous report (Ye et al., 2022). The primers used in this study were listed in Table S8.
The statistical analysis was conducted in IBM SPSS Statistics software (v26.0). One-way ANOVA and Duncan test (p < 0.05) were used to determine the significant difference. The total anthocyanin content and the gene expression level were visualized in GraphPad Prism 9 software.
‘Jinhongguan’ is a full-red Actinidia arguta cultivar with oval fruit, which ripens in early August in Wuhan, China. The average single fruit weight was 16.34 g, and the average soluble solids content was 11.2%. The most typical feature of this cultivar is that the freshly mature fruit is green, and it gradually becomes full-red after being harvested and stored at 25°C in the dark for about one week. To explore the mechanism of post-harvest fruit color conversion, we divided the color conversion process into four stages (Figures 1A, B). The peel began to turn red in FP2 and became completely red in FP3 (Figure 1A), while the flesh began to turn red in FF3 and became entirely red in FF4 (Figure 1B), indicating that the peel started turning red earlier than the flesh.
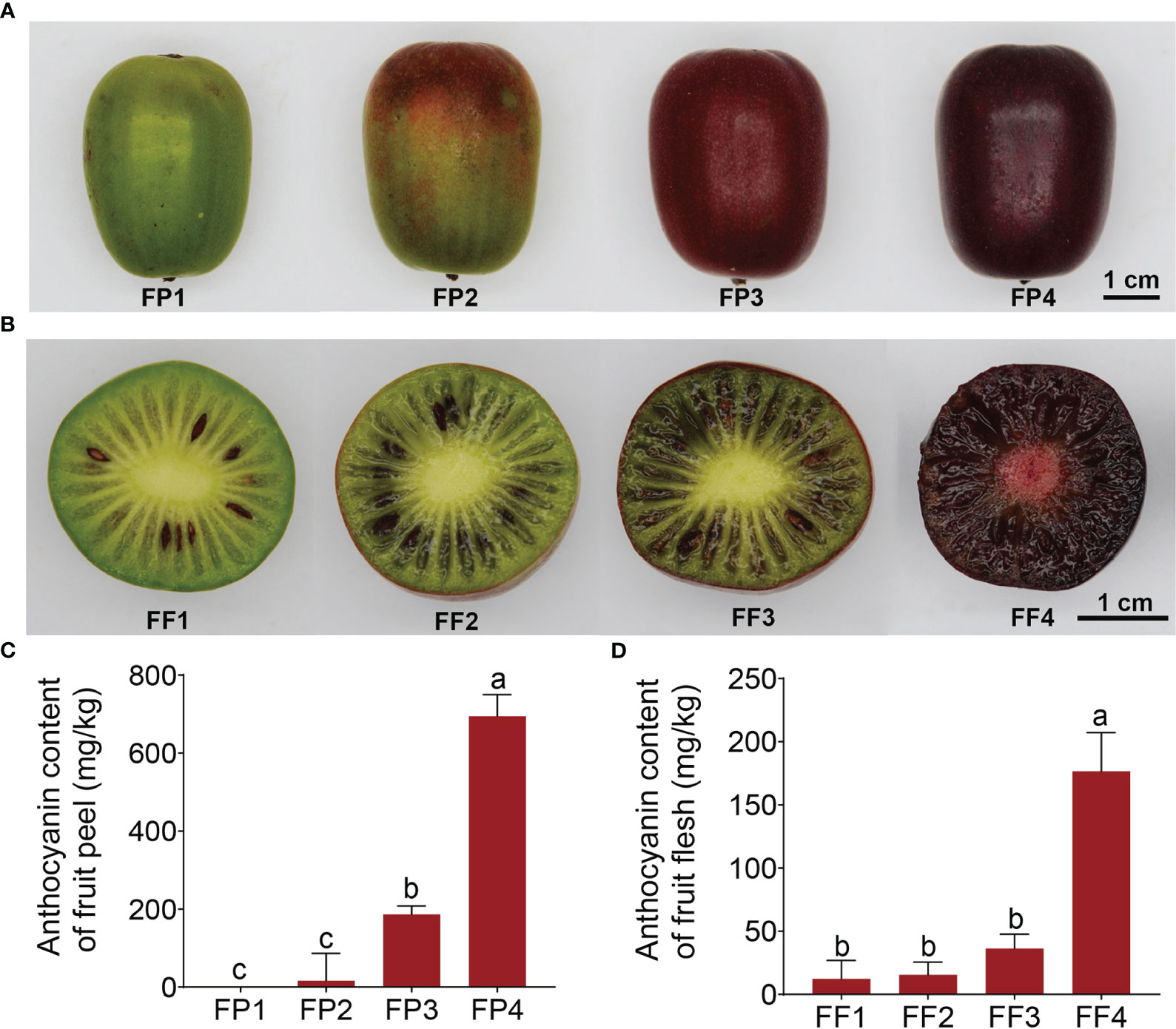
Figure 1 Coloration and anthocyanin content of the fruit peel and flesh in ‘Jinhongguan’. (A, B) The fruit peel (A) and flesh (B) color in 4 different stages after harvest. FP1-4: Stage 1 to 4 of fruit peel, FF1-4: Stage 1 to 4 in fruit flesh. (C, D) Total anthocyanin contents in fruit peel (C) and flesh (D) at different stages. Values represent the means ± SD from three biological replicates. The significantly different in total anthocyanin contents in fruit peel and flesh among different stages were evaluated by One-way ANOVA and Duncan test (p < 0.05). Bars with the same lowercase letters indicate no significant difference, while bars without shared lowercase letters indicate significant difference.
It is reported that the pigments in red fruits are mainly composed of anthocyanins (Sui et al., 2013). Considering this, the total anthocyanin content in the ‘Jinhongguan’ fruit in four stages was measured. The results showed that the anthocyanin content was increased with increasingly deepening color (Figures 1C, D). The relative content of anthocyanin in FP4 and FF4 reached 694.76 mg/kg and 176.53 mg/kg fresh weight, respectively, which was significantly higher than that in FP1 and FF1 (Figures 1C, D). These findings suggested that the major pigment component in stage 4 of the ‘Jinhongguan’ fruit peel and flesh is anthocyanin, but the specific type of anthocyanin needs to be further identified.
To explore the molecular mechanism of anthocyanin biosynthesis in ‘Jinhongguan’, high-throughput RNA-Seq analysis of 24 cDNA libraries of fruit peel and flesh was performed. The clean reads ranged from 19980145 to 23307997. The Q30 value of each library was more than 94%, and the average GC content was 47.10% (Table S1). A total of 82525 genes were identified by gene annotation in NR, NT, PFAM, KOG/COG, Swiss-Prot, KO, GO and other databases (Table S2). Based on the thresholds of |log2FC| ≥1.0 and padj < 0.05, a total of 24339 DEGs were screened with FP1 and FF1 as controls (Table S3). The distribution of DEGs in each stage was shown in Figure 2. With the red deepening of the peel and flesh, the number of identified DEGs gradually increased, and it was up to 16447 DEGs in FP4 vs FP1. Moreover, the number of DEGs in the peel was more than that in the flesh within the same stage (Figure 2A). Totally, 9204 DEGs were shared among the four stages of peel, and 8142 DEGs were shared among the four stages of flesh (Figures 2B, C). In addition, 5751 common DEGs were shared by both the peel and flesh among the four stages, including 2767 common up-DEGs and 2976 common down-DEGs (Figure 2D; Table S4). The remaining 8 common DEGs did not belong to common up-DEGs or common down-DEGs due to the inconsistent expression patterns in peel and flesh. For example, six down-DEGs in FP2 vs FP1 were up-DEGs in FF2 vs FF1 (Figure 2D; Table S4).
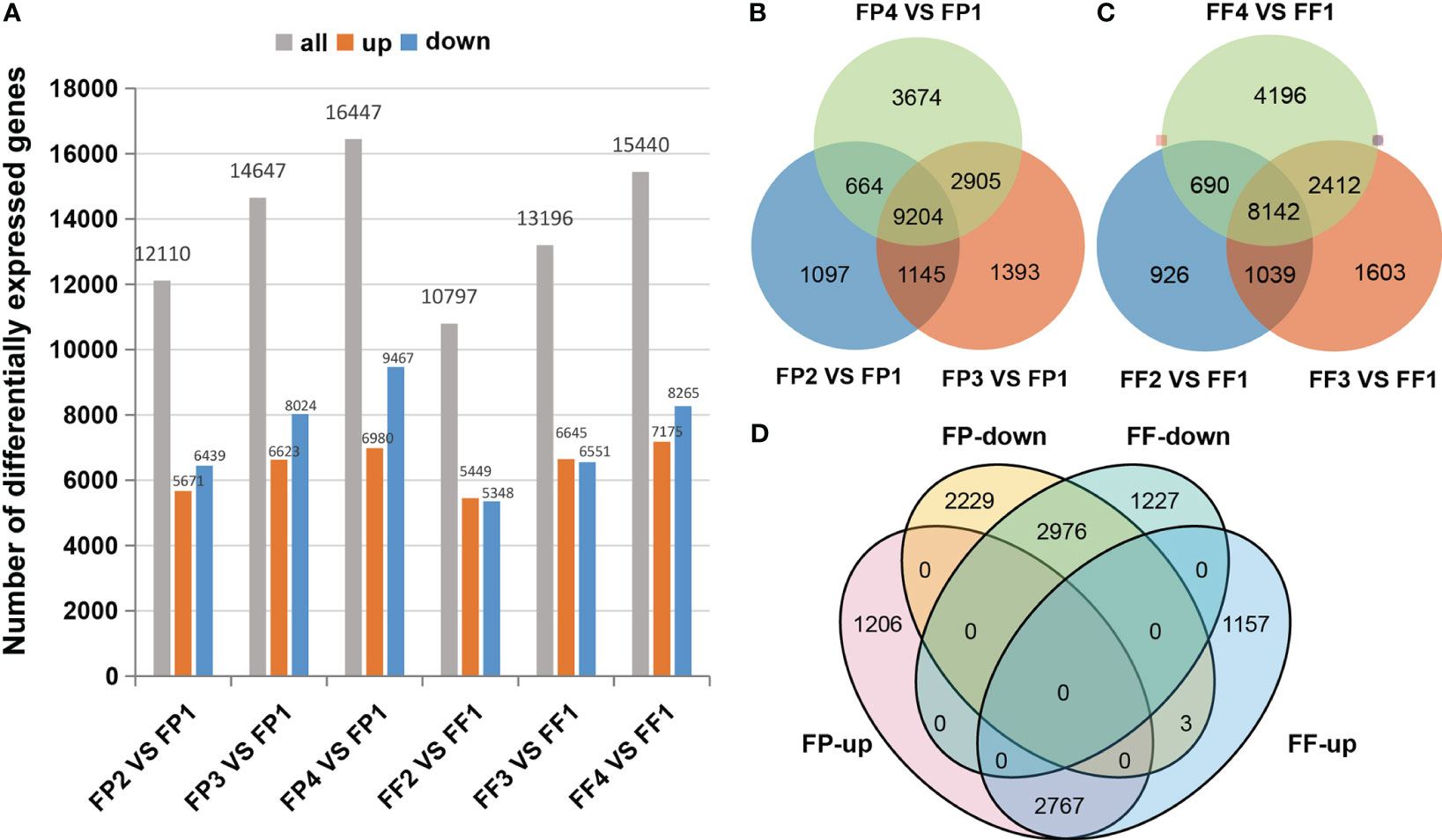
Figure 2 Differentially expressed genes in the transcriptome. (A) DEGs statistics of different stages. (B, C) Venn diagrams for DEGs between different stages of fruit peel (B) and flesh (C). (D) Venn diagrams between co-upregulated DEGs and co-downregulated DEGs of fruit peel and flesh.
Further, the expression levels of 5751 common DEGs in peel and flesh were analyzed. The heatmap showed that these common DEGs exhibited very consistent expression patterns between peel and flesh, as well as between different stages (Figure 3A). For example, a certain up-DEG tended to be up-regulated in all stages from stage 2 to stage 4, and vice versa, a down-DEG exhibited down-regulated expression in all stages (Figure 3A). This indicates that these DEGs were strongly positively or negatively correlated with the peel and flesh color conversion process. To further explore the function of these DEGs, KEGG and GO enrichment analyses of the common up-DEGs and common down-DEGs were performed. The KEGG enrichment analysis revealed that the common up-DEGs were mainly enriched in pathways such as carbohydrate metabolism, transport and catabolism, and signal transduction, while the common down-DEGs were mainly enriched in energy metabolism and cofactors and vitamins metabolism pathways (Figures 3B, C). Notably, some common up-DEGs were significantly enriched in flavonoid biosynthesis (Figure 3B). In terms of GO enrichment, our focus was mainly on the biological process pathways, and the results showed that the common up-DEGs were mainly enriched in response to chemical, abiotic stimulus and hormone pathways. Furthermore, these up-DEGs were also significantly enriched in flavonoid metabolic process, chalcone metabolic process and anthocyanin-containing compound metabolic process, all of which are relevant to anthocyanin biosynthesis (Figure 3D). On the other hand, the common down-DEGs were mainly enriched in the nucleobase-containing compound metabolic process, response to light stimulus and lipid metabolic process pathways (Figure 3E). It is worth noting that some common up-DEGs were significantly enriched in anthocyanin biosynthesis-related pathways in both KEGG and GO enrichment analysis (Figures 3B, D), and these genes might be directly involved in anthocyanin biosynthesis in the peel and flesh of ‘Jinhongguan’.
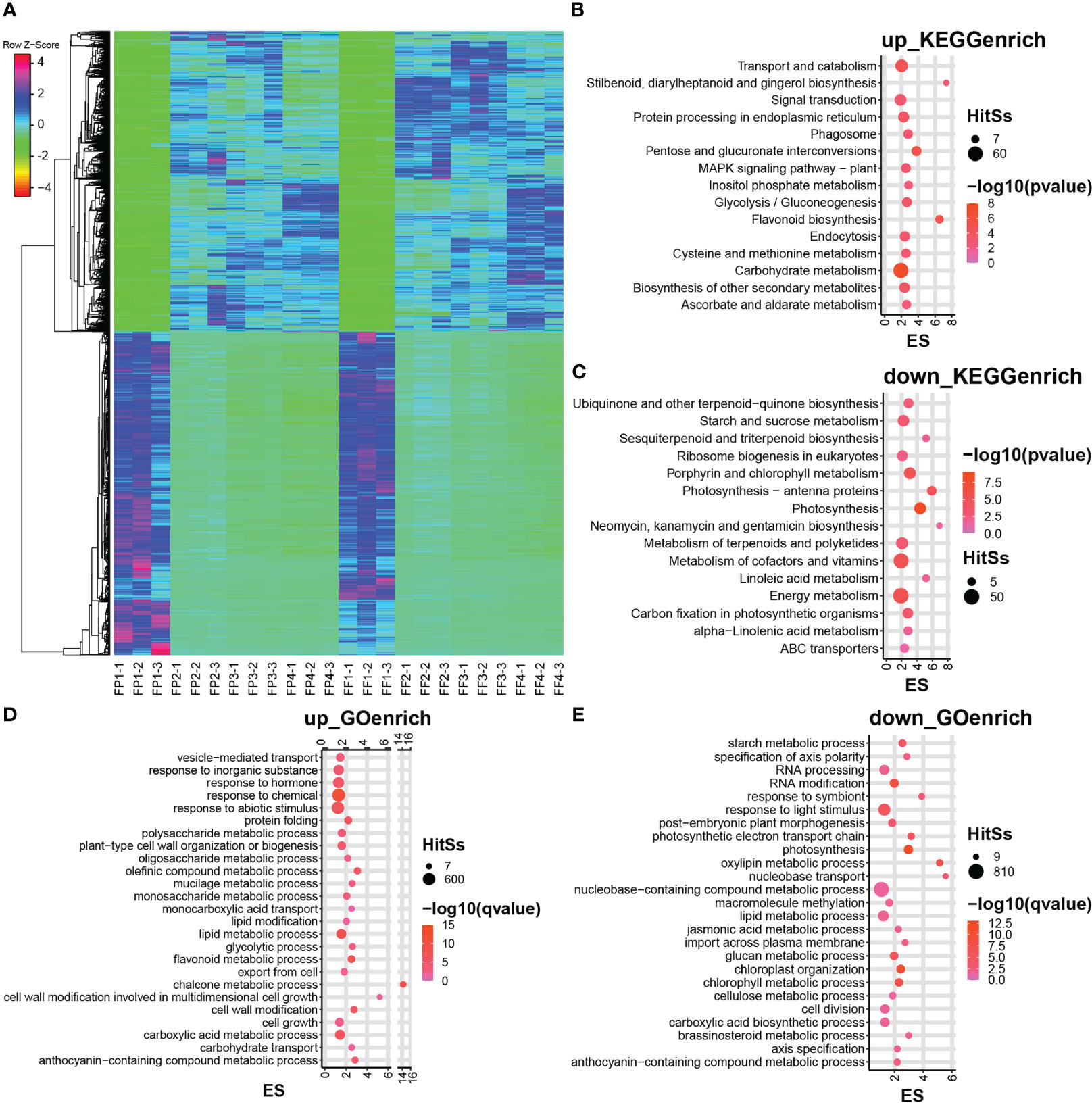
Figure 3 Heatmap and enrichment analysis of differentially expressed genes in the transcriptome. (A) Heatmap of DEGs in stage 1 to 4 of fruit peel and flesh. (B, C) The KEGG enrichment analysis of co-upregulated DEGs (B) and co-downregulated DEGs (C) of fruit peel and flesh. (D, E) The GO enrichment analysis of co-upregulated DEGs (D) and co-downregulated DEGs (E) of fruit peel and flesh.
To investigate the types of anthocyanin formed during the post-harvest color conversion of ‘Jinhongguan’ fruit, anthocyanin-targeted metabolome analysis was performed on stage 3 (FF3) and stage 4 (FF4) of the flesh (Figure 1B). The anthocyanin types in ‘Jinhongguan’ fruits were identified with 108 different anthocyanins as standard substances. A total of 29 metabolites of anthocyanin belonging to eight categories were identified, and among these categories, cyanidin was the most abundant (Figure 4A; Table S5). In the comparison between FF3 and FF4, nine DAMs were identified, of which six DAMs were up-regulated in FF4, with five being cyanidin and one being pelargonidin. Three DAMs were down-regulated, including two flavonoids and one delphinidin (Figure 4B). The difference multiple changes value log2FC of the six up-DAMs were all more than three, which was greater than that of the three down-DAMs (Figure 4C). Further, we investigated the content of the nine DAMs and found that the accumulation of cyanidin-3-O-galactoside in FF4 was significantly higher than that of other anthocyanins (Figure 4B), suggesting that cyanidin-3-O-galactoside might be the main pigment responsible for the flesh red coloration in ‘Jinhongguan’.
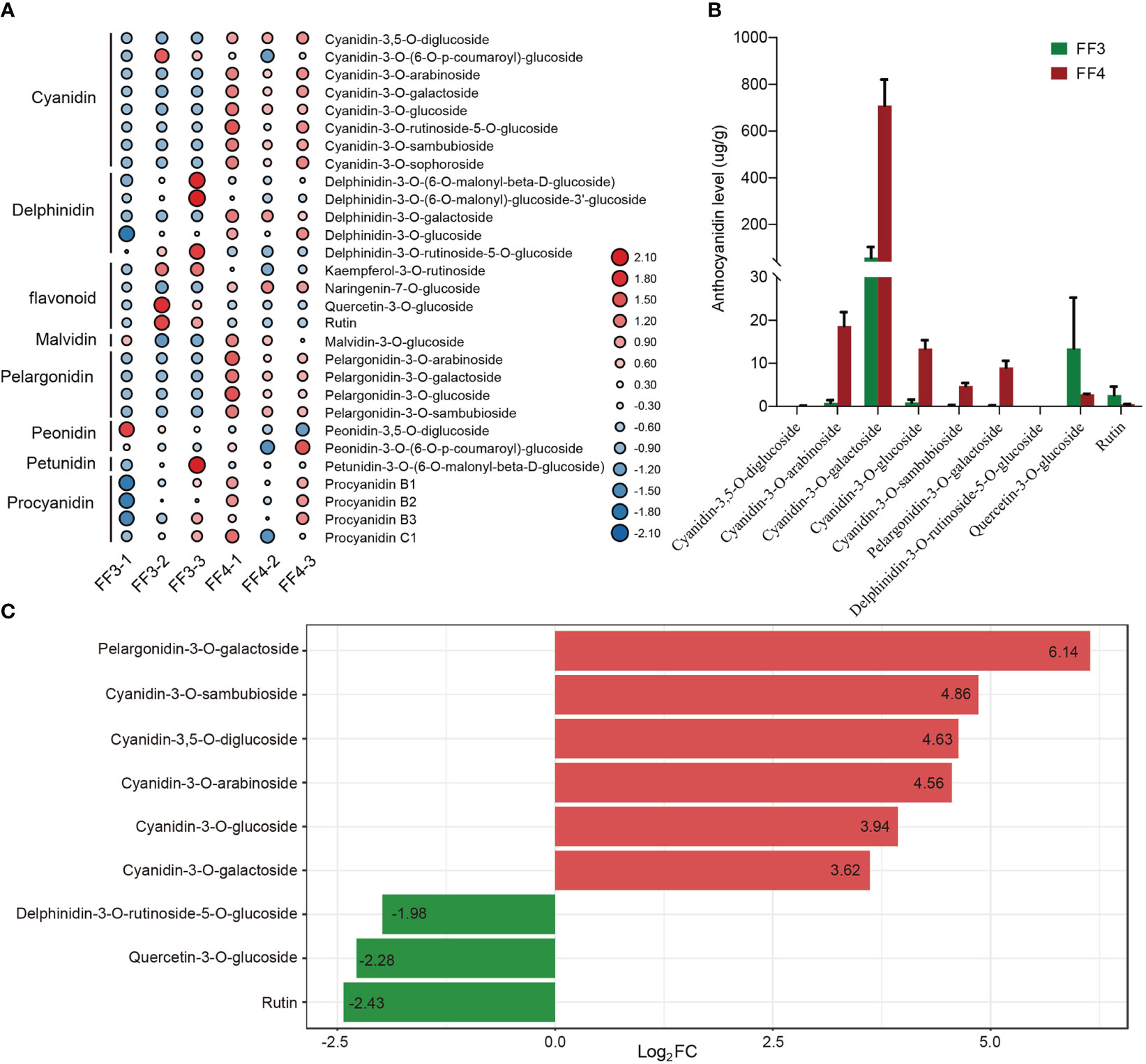
Figure 4 Statistical analysis of metabolome. (A) Heatmap of 29 anthocyanin metabolites in 8 categories identified in FF3 and FF4. (B) The content levels of 9 DAMs in FF3 and FF4. (C) Difference multiples of anthocyanins in FF3 and FF4. Red and green represent up-DAMs and down-DAMs, respectively.
To explore the biosynthesis pathway of cyanidin during the post-harvest color conversion of the ‘Jinhongguan’ fruit, we conducted a screening of the genes related to anthocyanin biosynthesis in 5751 common DEGs in the transcriptome date. A total of 109 anthocyanin biosynthesis-related genes were identified, including 36 structural genes and 73 MBW TFs (Table S6). Among the 36 structural genes, 33 were associated with cyanidin biosynthesis, and the expression heatmap of these genes was carried out. (Figure 5). Among them, 19 structural DEGs related to the early enzyme reaction were found to be significantly up-regulated, including four PAL genes, one C4H gene, two 4CL genes, ten CHS genes and two CHI genes (Figure 5; Table S6). Most of these structural genes exhibited low expression levels in stage 1 of both peel and flesh, and their expression levels increased significantly in subsequent stages. In addition, four early cyanidin biosynthesis-related genes were down-regulated, including two 4CL genes (Cluster-927.30937, Cluster-927.42872), one CHS gene (Cluster-927.31622) and one F3’H gene (Cluster-927.37247), which had the highest expression levels in stage 1 of both peel and flesh. Among the late biosynthetic genes of cyanidin, one DFR gene, four LODX genes, two UFGT genes and two GST genes were significantly up-regulated in stage 2/3/4 of both peel and flesh. DFR is a key enzyme that catalyzes the synthesis of colorless leucocyanidin from dihydro quercetin (Tanaka et al., 2008). In this study, AaDFR1 (Cluster-927.33723) was significantly up-regulated in stage 2/3/4 of both peel and flesh. Leucocyanidin is oxidized and dehydrated under the catalysis of LODX/ANS to form a colored unstable cyanidin (Jaakola, 2013). Here, four LODX genes (Cluster-927.40217, Cluster-927.40218, Cluster-927.40219 and Cluster-927.43841) were found to be highly induced in stage 2/3/4 of peel and flesh. These LODX genes are likely responsible for catalyzing the conversion of colorless anthocyanins into cyanidins. Subsequently, the unstable cyanidin requires UFGT to catalyze its glycosylation to become stable (Jaakola, 2013). Our data showed that two UFGT genes (Cluster-927.26492 and Cluster-927.18477) were induced during fruit color conversion, of which Cluster-927.26492 encodes a 3-O-beta-D-galactosyltransferase, and Cluster-927.18477 encodes a 3-O-beta-D-xylose galactosyltransferase. Metabolomics data indicated that cyanidin-3-O-galactoside was the main pigment responsible for the red color of the flesh in ‘Jinhong guan’. Based on these findings, we speculated that cyanidin might be mainly catalyzed by Cluster-927.26492 to stable cyanidin-3-O-galactose. In addition, two GST genes (Cluster-927.33115 and Cluster-927.34850) were significantly induced during the peel and flesh color conversion process, suggesting their involvement in the transport of cyanidin-3-O-galactose to vacuoles. The above results indicated that these 33 structural genes might be essential for the biosynthesis of cyanidin in ‘Jinhongguan’ fruit and participate in the process of fruit color conversion.
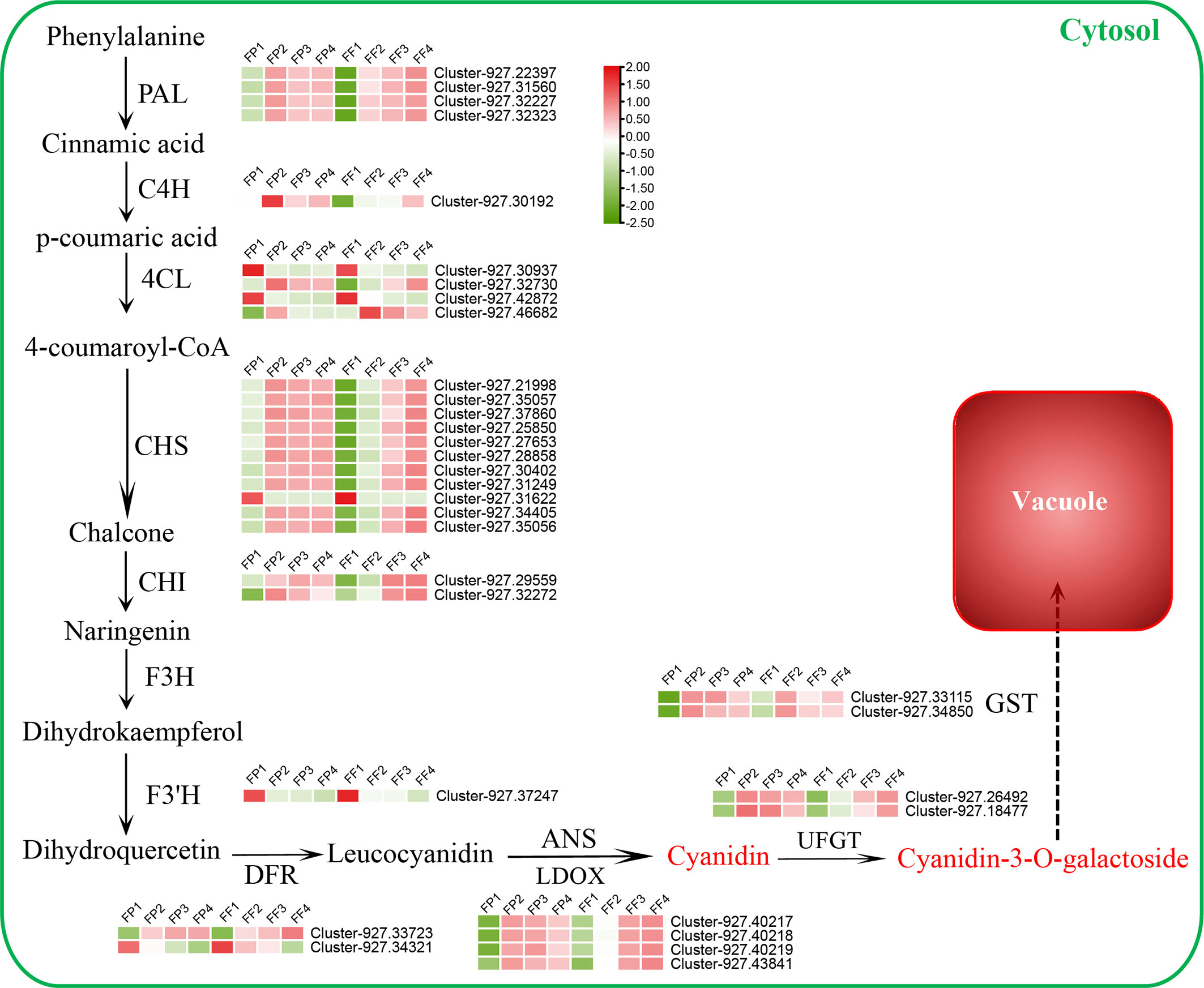
Figure 5 Differentially expressed structural genes regulatory network of cyanidin biosynthesis. The expression level from low to high is represented by green to red.
The biosynthesis of anthocyanins is regulated by various TFs, the most important of which are MYB, bHLH and WDR. In this study, a significant number of the 5751 DEGs were TFs, and 27 MYB TFs, 37 bHLH TFs and 9 WDR TFs were identified (Figure S1). The expression heatmap revealed that most of the MYB and WDR TFs were up-regulated (15 of MYB TFs, 7 of WDR TFs), while most of the bHLH TFs (27 out of 37) were down-regulated (Figure 6; Figure S1). Some TFs, such as Cluster-927.46953 (MYB), Cluster-927.31831 (bHLH), Cluster-927.43614 (WDR), exhibited down-regulated expression patterns in stage 2/3/4, while their expression levels in stage 1 (FP1 and FF1) were significantly higher (Figure 6), indicating a negative correlation with anthocyanin biosynthesis. Moreover, the expression peaks of the up-regulated TFs appeared in different stages. For example, Cluster-927.21050 (MYB) and Cluster-927.47586 (bHLH) reached their expression peak in stage 2 (FP2 and FF2), while Cluster-927.34911 (bHLH) and Cluster-927.31439 (WDR) peaked in stage3 (FP3 and FF3), Cluster-927.6100 (MYB) and Cluster-927.31199 (WDR) peaked in stage 4 (FP4 and FF4) (Figure 6). These results implied that these TFs might promote the biosynthesis of cyanidin by regulating the expression of structural genes at different stages.
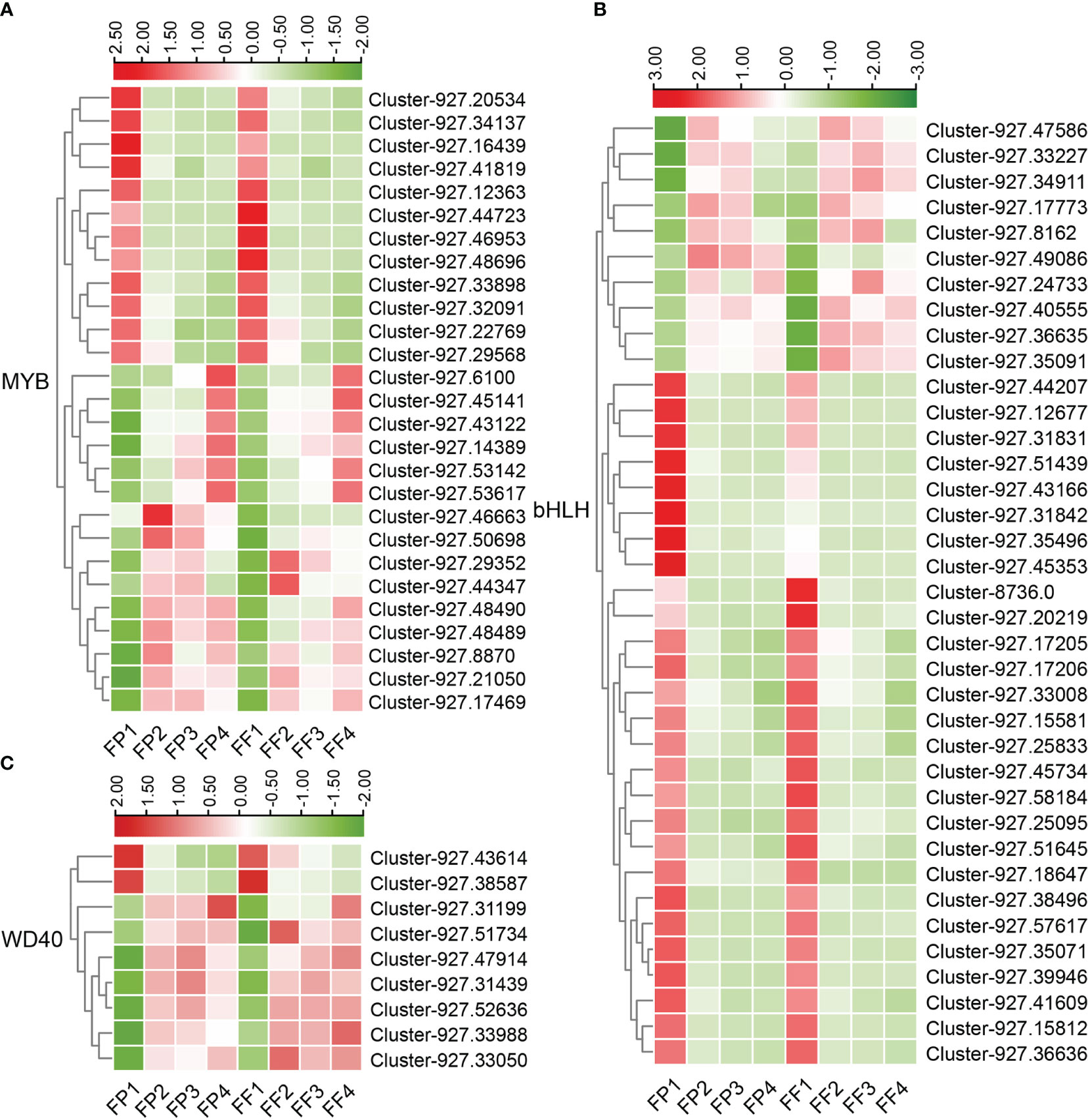
Figure 6 Heatmap of differentially expressed MBW transcription factors in the transcriptome. (A-C) Heatmap of 27 differentially expressed MYB TFs (A), 37 differentially expressed bHLH TFs (B) and 9 differentially expressed WDR TFs (C).
To further clarify the relationship between DEGs and DAMs, the correlation between nine anthocyanin DAMs and 36 anthocyanin synthesis structure DEGs or 73 MBW TFs was analyzed. The results showed that nine DAMs were significantly correlated with 26 structural DEGs and 29 MBW TFs. There were 266 significantly correlated pairs, of which 90 pairs were negatively correlated, and 176 pairs were positively correlated (Figure 7A; Table S7). Some DEGs were significantly correlated with 8 DAMs simultaneously, such as AaPAL3/4, Aa4CL2/4, AaCHS2/3/5/9/10, AaDFR2 and bHLH30 (Figure 7A; Table S7). Except for bHLH30, all these genes belonged to early structural genes involved in anthocyanin biosynthesis, highlighting the crucial role of early structural genes in anthocyanin biosynthesis. Furthermore, seven out of the nine DAMs showed significant correlations with more than 30 DEGs, while the remaining two down-DAMs were correlated with 14 and 23 DEGs, respectively (Table S7). Given that cyanidin-3-O-galactoside was the most abundant pigment in the fruit color conversion of ‘Jinhongguan’ (Figure 4B), the correlation between cyanidin-3-O-galactoside and structural DEGs and MBW TFs was analyzed. A total of 34 DEGs (21 structural DEGs and 13 MBW TFs) were significantly correlated with cyanidin-3-O-galactoside, most of which were positively correlated (Table S7). Ten DEGs showed negative correlation with cyanidin-3-O-galactoside, half of which belonged to the bHLH TF family. Only AabHLH6 exhibited a positive correlation with cyanidin-3-O-galactoside, while all six MYB TFs were positively correlated with it (Figure 7B). These findings suggested that bHLH TFs might mainly negatively regulate cyanidin-3-O-galactoside synthesis, while MYB TFs might positively regulate it.
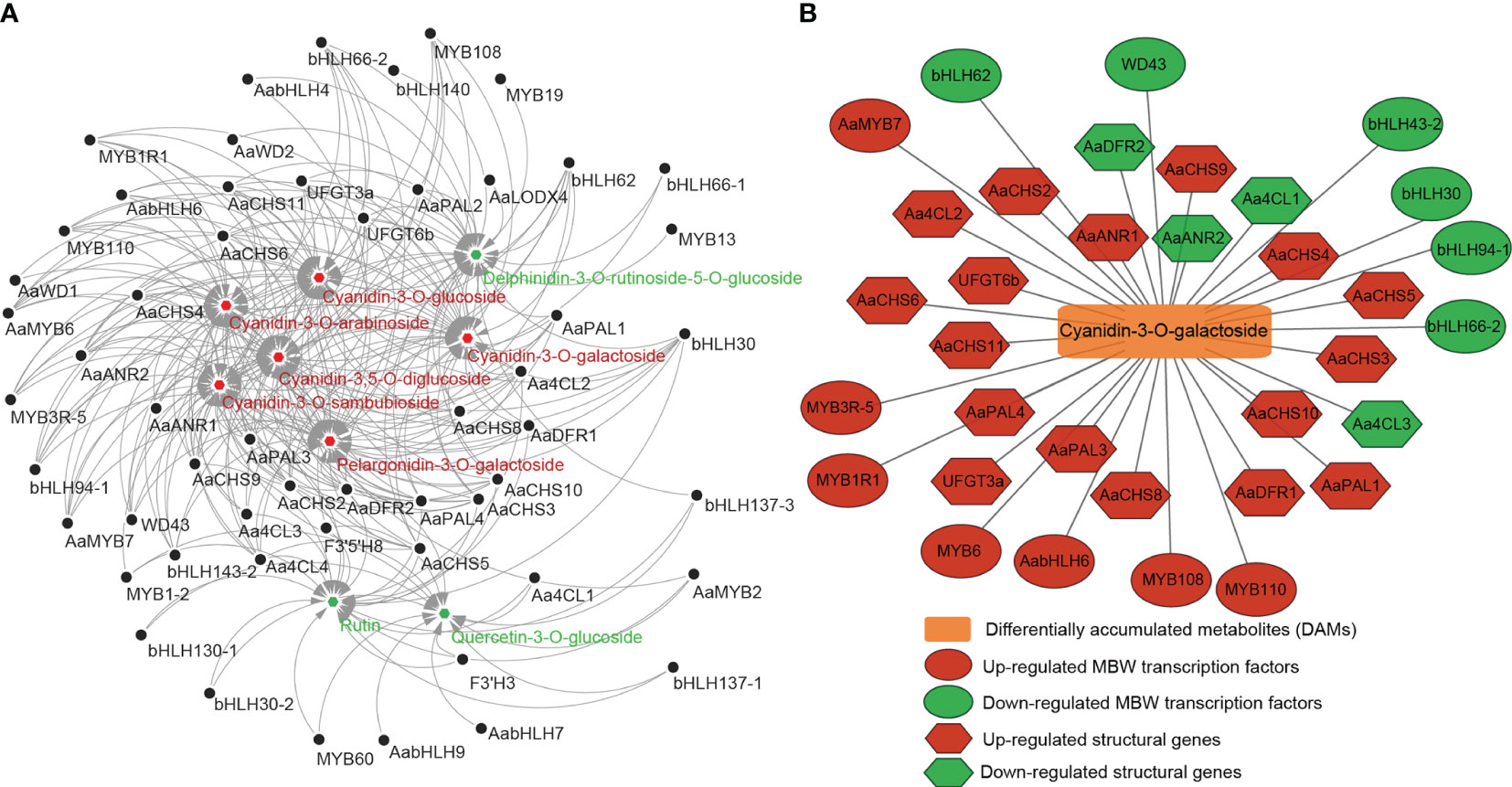
Figure 7 The correlation between differentially accumulated metabolites and differentially expressed structural genes and MBW transcription factors. (A) The correlation between 9 DAMs and 26 structural DEGs and 29 MBW TFs. Red: up-regulated DAMs, green: down-regulated DAMs. (B) The correlation between cyanidin-3-O-galactoside and 21 structural DEGs and 13 MBW TFs.
To further screen structural genes and MBW TFs, we performed qRT-PCR analysis of 24 genes based on transcriptome and metabolome data. Among the 17 structural genes, AaPAL3, AaCHS2/3/8/9, UFGT3a and UFGT6b were significantly highly expressed in peel and flesh in stage 2/3/4, reaching their peak in flesh at FF4 (Figure 8), which was consistent with the phenotype of the flesh completely turning red at FF4 (Figure 1B). This indicated that these seven structural genes were closely related to the biosynthesis of cyanidin in ‘Jinhongguan’. The expression level of AaANR1 gradually increased from FP1 to FP4 with higher expression in the peel than in the flesh. Its highest expression was observed in FP4, while AaDFR1 exhibited the highest expression in FF4 (Figure 8), indicating that the structural genes involved in the synthesis of cyanidin might differ between the peel and flesh. Regarding the MBW TFs, the expression patterns of bHLH30, bHLH94-1 and WD43 were similar to those of structural genes Aa4CL3, AaCHS11 and AaDFR2 (Figure 8), indicating that these TFs might directly bind to the promoters of these structural genes to regulate their transcription, thus promoting cyanidin biosynthesis. MYB108 showed significantly higher expression in FP4 and FF4 compared to other stages. (Figure 8), suggesting its potential role in the transport and accumulation of cyanidin. Taken together, all the above results indicated that structural genes including AaPAL3, Aa4CL3, AaCHS2/3/8/9/11, AaDFR1/2, AaANR1, UFGT3a and UFGT6b, as well as TFs including MYB108, bHLH30, bHLH94-1 and WD43 played important roles in cyanidin biosynthesis in ‘Jinhongguan’.
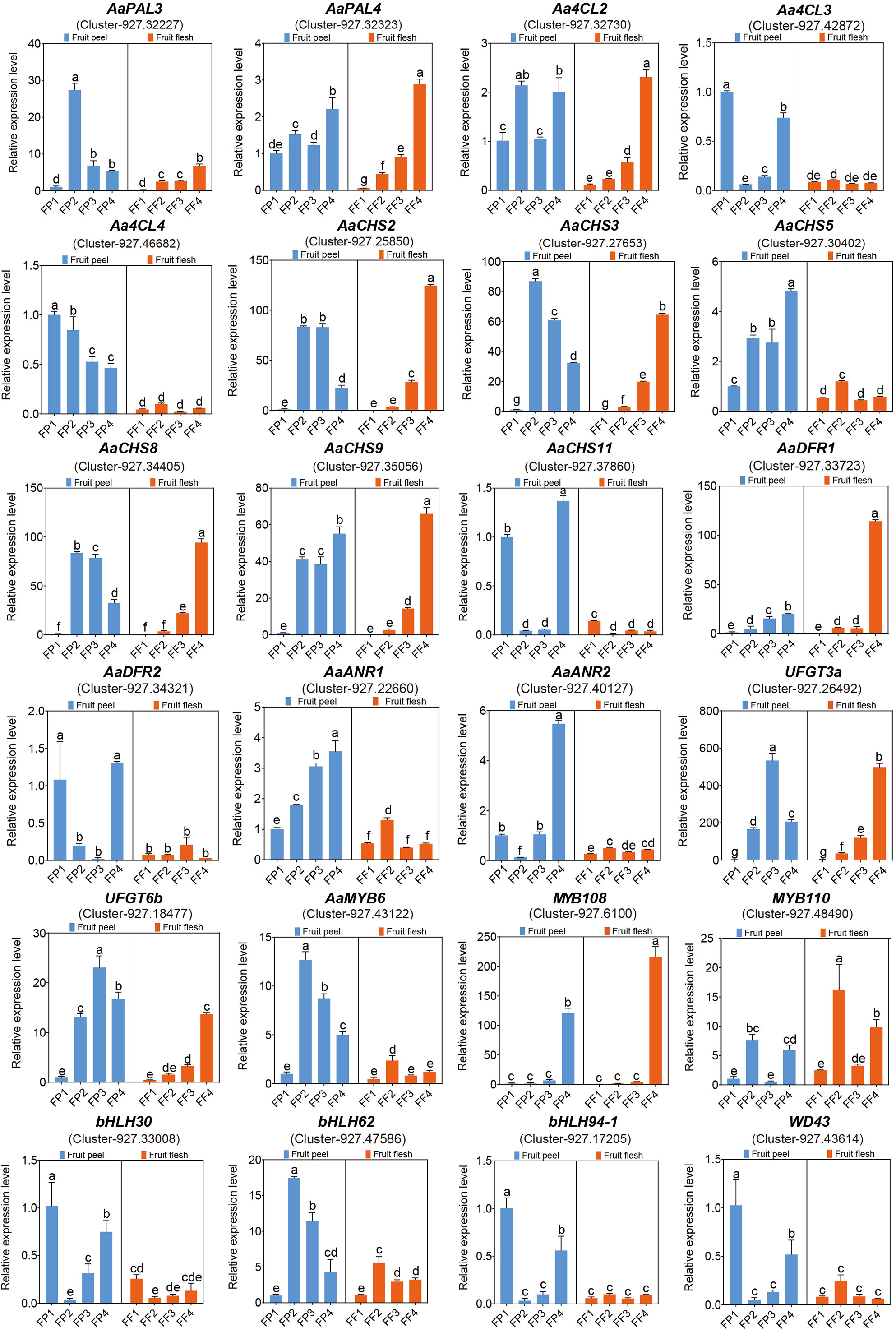
Figure 8 The qRT-PCR analysis of candidate genes related to anthocyanin biosynthesis. The significantly different expression levels of genes in fruit peel and flesh among different stages were evaluated by One-way ANOVA and Duncan test (p < 0.05). Bars with the same lowercase letters indicate no significant difference, while bars without shared lowercase letters indicate significant difference.
Kiwifruit flesh is rich in color, exhibiting extensive variations among different species and within species. Currently, the cultivated varieties of kiwifruit mainly consist of Actinidia deliciosa with green flesh (such as ‘Hayward’), Actinidia chinensis with yellow flesh (such as ‘Hort16A’) or red flesh core (such as ‘Hongyang’, ‘Donghong’), and Actinidia arguta with red flesh (such as ‘Hongbaoshixing’) (McGhie and Ainge, 2002; Nishiyama, 2007). The main pigments in the green, yellow and red flesh of kiwifruit are chlorophyll, carotene and anthocyanin, respectively, and pigment content, proportion and spatial distribution determine the color of kiwifruit (Nishiyama, 2007; Montefiori et al., 2009). Initially, the flesh color of yellow and green kiwifruit is green at the early stage of development. As the fruit matures, the chlorophyll in yellow kiwifruit degrades, while its original carotenoid is retained, resulting in a yellow flesh color. By contrast, the content of chlorophyll in green kiwifruit remains unchanged, maintaining a green flesh color throughout ripening (Pilkington et al., 2012). Most kiwifruit cultivars have completed the color conversion process before reaching maturity. However, in this study, the fruit peel and flesh of ‘Jinhongguan’ (a new cultivar of Actinidia arguta) gradually turned fully red during storage in the dark at 25°C for about one week, which differs from previous studies. Anthocyanin was identified as the main pigment in the ‘Jinhongguan’ fruit, and total anthocyanin content was higher in the peel than in the flesh at the same storage stage, which may be due to the higher water content in the flesh than that in the peel. Previous studies have identified cyanidin-based anthocyanins as the pigments in red-fleshed kiwifruit (Montefiori et al., 2011). Recent studies have also reported the presence of a small number of delphinidin derivatives in the flesh of Actinidia melanandra and Actinidia arguta, as well as in the peel of Actinidia chinensis ‘Hongyang’ (Montefiori et al., 2009; Liu et al., 2017). Similarly, in this study, the anthocyanin-targeted metabolome analysis revealed that the main pigment in the flesh of ‘Jinhongguan’ was cyanidin, and a small number of pelargonidins were also detected. In addition, cyanidin mainly exist in the form of cyanidin-3-O-galactoside, and pelargonidin exist as pelargonidin-3-O-galactoside. These findings have further revealed the composition and existence form of pigments in red-fleshed kiwifruit.
The genes related to anthocyanin synthesis mainly include the structural genes encoding various enzymes and the TFs regulating the structural genes (Liu et al., 2016; Wang et al., 2022b). Some structural genes and TFs have been confirmed to be involved in the synthesis of anthocyanin in kiwifruit. For example, in Actinidia arguta Sieb. Zucc, AaF3H, AaLDOX, AaUFGT, AaMYB, AabHLH and AaHB2 have been reported to regulate flavonoid biosynthesis in ‘Hongbaoshixing’ and AaLDOX controls anthocyanin biosynthesis in the flesh of ‘Tianyuanhong’ (Li et al., 2018a; Li et al., 2018b). The cMYBF110-AcbHLH1-AcWDR1 complex can bind to the promoters of structural genes including AcCHS, AcF3′H, AcANS, AcUFGT3a, AcUFGT6b and AcGST1 to activate their expression (Liu et al., 2021). In this study, transcriptome analysis of ‘Jinhongguan’ identified a total of 5751 common DEGs in the peel and flesh. Furthermore, 36 structural DEGs and 73 MBW TFs related to anthocyanin synthesis were screened by gene annotation, KEGG and GO enrichment analysis. The expression heatmap analysis showed that multiple structural genes related to the catalyzed reactions of anthocyanin biosynthesis were significantly up-regulated, such as PAL, C4H, 4CL, CHS, CHI, DFR, LDOX, UFGT and GST genes. These genes exhibited low expression levels in stage 1 of peel and flesh and were increased significantly increased in the late stages, indicating that these structural genes were functionally conserved and were closely related to anthocyanin synthesis in ‘Jinhongguan’. Among the 5751 common DEGs, 27 MYB TFs, 37 bHLH TFs, and 9 WDR TFs were identified. The expression heatmap also demonstrated that most of the MYB and WDR TFs were up-regulated, while the majority of bHLH TFs were down-regulated. These down-regulated TFs might have a negative regulatory role in anthocyanin synthesis. At present, some TFs have been reported to negatively regulate anthocyanin synthesis. For example, PtrMYB57 and LcbHLH92 are negative regulators of anthocyanin/proanthocyanidin accumulation in Poplar and Sheepgrass (Leymus chinensis), respectively (Wan et al., 2017; Zhao et al., 2018). In Chimonanthus praecox (L.), ectopic overexpression experiments in model plants have revealed that CpbHLH1 can reduce anthocyanin accumulation by inhibiting the expression of genes involved in the flavonoid biosynthesis pathway (Zhao et al., 2020).
To further screen cyanidin biosynthesis-related candidate genes, we performed a correlation analysis between the transcriptome and metabolome. A total of 21 structural DEGs were significantly correlated with cyanidin-3-O-galactoside metabolism, most of which exhibited a positive correlation. Notably, AaPAL4, Aa4CL2, AaDFR2 and AaCHS2/3/5/9/10 were significantly correlated with eight DAMs including cyanidin-3-O-galactoside. These findings suggested that these structural genes were key genes involved in the anthocyanin synthesis process in ‘Jinhongguan’. In addition, 13 MBW TFs including six MYB, six bHLH and one WDR were also significantly correlated with cyanidin-3-O-galactoside. Specifically, all the MYB TFs showed a positive correlation with cyanidin-3-O-galactoside, while five bHLH TFs and one WDR TF showed a negative correlation with this metabolite, indicating that the MYB TFs positively regulated the biosynthesis of cyanidin-3-O-galactoside, while bHLH and WDR TFs mainly negatively regulated it. The qRT-PCR results further confirmed that these structural genes and MBW TFs played important roles in cyanidin biosynthesis in ‘Jinhongguan’. However, the specific functions and regulatory mechanisms of these genes remain to be further explored.
Here, we used transcriptome and metabolome analysis of anthocyanin biosynthesis in post-harvest fruits of a new full red-type kiwifruit (Actinidia arguta) ‘Jinhongguan’. The determination of total anthocyanin content showed that anthocyanin was the main pigment accumulated during the reddening process of the ‘Jinhongguan’ fruit. Additionally, the anthocyanin-targeted metabolome analysis revealed that the main form of anthocyanin was cyanidin-3-O-galactoside. Through correlation analysis between the transcriptome, metabolome and qRT-PCR, 12 structural genes and 4 MBW TFs were identified as key candidate genes responsible for cyanidin-3-O-galactoside biosynthesis. This study provides insight into the composition of anthocyanins in Actinidia arguta and lays a foundation for revealing the molecular mechanism of anthocyanin synthesis in post-harvest fruits of Actinidia arguta.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
LY: Investigation, Funding acquisition, Writing – original draft. FB: Validation, Writing – review and editing, Data curation. LZ: Funding acquisition, Writing – review and editing, Resources. ML: Investigation, Supervision, Writing – review and editing. LG: Investigation, Methodology, Writing – review and editing. ZW: Data curation, Validation, Writing – review and editing. JP: Investigation, Writing – review and editing. QC: Conceptualization, Writing – review and editing. XL: Conceptualization, Writing – review and editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of Hubei Province (2023AFB234), the China Agriculture Research System of MOF and MARA (CARS-026), the Hubei Key R&D Project (2021BBA100), the National Key R&D Program of China (2022YFD1600700), the Youth Foundation of Hubei Academy of Agricultural Sciences (2021NKYJJ12), the National Natural Science Foundation of China (32302519) and the Hubei Provincial Agricultural Science and Technology Innovation Fund (2021-620-000-001-023).
The authors would like to thank reviewers for comments on this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1280970/full#supplementary-material
Supplementary Figure 1 | Transcription factor classification statistics of common differentially expressed genes in the transcriptome.
Aguilar-Barragán, A., Ochoa-Alejo, N. (2014). Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins in chilli pepper fruit. Biol. Plantarum 58 (3), 567–574. doi: 10.1007/s10535-014-0427-4
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169 (7), 710–717. doi: 10.1016/j.jplph.2012.01.015
Bendokas, V., Skemiene, K., Trumbeckaite, S., Stanys, V., Passamonti, S., Borutaite, V., et al. (2020). Anthocyanins: From plant pigments to health benefits at mitochondrial level. Crit. Rev. Food Sci. Nutr. 60 (19), 3352–3365. doi: 10.1080/10408398.2019.1687421
Bloor, S. J., Abrahams, S. (2002). The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59 (3), 343–346. doi: 10.1016/s0031-9422(01)00460-5
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 12 (12), 2383–2394. doi: 10.1105/tpc.12.12.2383
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 49(1) 317-325. doi: 10.1093/nar/gkab447.
Chen, L., Hu, B., Qin, Y., Hu, G., Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 (3), 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fraser, L. G., Seal, A. G., Montefiori, M., McGhie, T. K., Tsang, G. K., Datson, P. M., et al. (2013). An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genomics 14, 28. doi: 10.1186/1471-2164-14-28
Hanley, Z. (2018). Kiwifruit (Actinidia spp.) Breeding: Volume 3. Advances in Plant Breeding Strategies: Fruits.
Heredia, A., Heredia-Guerrero, J. A., Dominguez, E. (2015). CHS silencing suggests a negative cross-talk between wax and flavonoid pathways in tomato fruit cuticle. Plant Signal Behav. 10 (5), e1019979. doi: 10.1080/15592324.2015.1019979
Hu, Y., Cheng, H., Zhang, Y., Zhang, J., Niu, S., Wang, X., et al. (2021). The MdMYB16/MdMYB1-miR7125-MdCCR module regulates the homeostasis between anthocyanin and lignin biosynthesis during light induction in apple. New Phytol. 231 (3), 1105–1122. doi: 10.1111/nph.17431
Huang, H. W., Ferguson, A. R. (2007). Actinida in China: natural diversity, phylogeographical evolution, interspecific gene flow and kiwifruit cultivar improvement. Acta Hortic. 753, 31–40. doi: 10.17660/ActaHortic.2007.753.1
Huang, S., Ding, J., Deng, D., Tang, W., Sun, H., Liu, D., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4, 2640. doi: 10.1038/ncomms3640
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18 (9), 477–483. doi: 10.1016/j.tplants.2013.06.003
Kim, E. Y., Kim, C. W., Kim, S. (2016). Identification of two novel mutant ANS alleles responsible for inactivation of anthocyanidin synthase and failure of anthocyanin production in onion (Allium cepa L.). Euphytica 212 (3), 427–437. doi: 10.1007/s10681-016-1774-3
Lai, B., Du, L. N., Liu, R., Hu, B., Su, W. B., Qin, Y. H., et al. (2016). Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00166
Li, W., Ding, Z., Ruan, M., Yu, X., Peng, M., Liu, Y. (2017b). Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 7 (1), 16861. doi: 10.1038/s41598-017-16905-1
Li, Y., Fang, J., Qi, X., Lin, M., Zhong, Y., Sun, L. (2018a). A key structural gene, AaLDOX, is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit (Actinidia arguta) based on transcriptome analysis. Gene 648, 31–41. doi: 10.1016/j.gene.2018.01.022
Li, Y., Fang, J., Qi, X., Lin, M., Zhong, Y., Sun, L., et al. (2018b). Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 19 (5), 1471. doi: 10.3390/ijms19051471
Li, B., Xia, Y., Wang, Y., Qin, G., Tian, S. (2017a). Characterization of genes encoding key enzymes involved in anthocyanin metabolism of kiwifruit during storage period. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00341
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10, 50. doi: 10.1186/1471-2229-10-50
Liu, X., Luo, Y., Wu, H., Xi, W., Yu, J., Zhang, Q., et al. (2016). Systematic analysis of O-methyltransferase gene family and identification of potential members involved in the formation of O-methylated flavonoids in Citrus. Gene 575 (2 Pt 2), 458–472. doi: 10.1016/j.gene.2015.09.048
Liu, Y., Ma, K., Qi, Y., Lv, G., Ren, X., Liu, Z., et al. (2021). Transcriptional regulation of anthocyanin synthesis by MYB-bHLH-WDR complexes in kiwifruit (Actinidia chinensis). J. Agric. Food Chem. 69 (12), 3677–3691. doi: 10.1021/acs.jafc.0c07037
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front. Chem. 6. doi: 10.3389/fchem.2018.00052
Liu, Y., Zhou, B., Qi, Y., Chen, X., Liu, C., Liu, Z., et al. (2017). Expression differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front. Plant Sci. 8, 1507. doi: 10.3389/fpls.2017.01507
Luo, X., Plunkert, M., Teng, Z., Mackenzie, K., Guo, L., Luo, Y., et al (2023). Two MYB activators of anthocyanin biosynthesis exhibit specialized activities in petiole and fruit of diploid strawberry. J Exp Bot 74(5) 1517-1531. doi: 10.1093/jxb/erac507.
McGhie, T. K., Ainge, G. D. (2002). Color in fruit of the genus actinidia: carotenoid and chlorophyll compositions. J. Agric. Food Chem. 50 (1), 117–121. doi: 10.1021/jf010677l
Montefiori, M., Comeskey, D. J., Wohlers, M., McGhie, T. K. (2009). Characterization and quantification of anthocyanins in red kiwifruit ( Actinidia spp.). J. Agric. Food Chem. 57 (15), 6856–6861. doi: 10.1021/jf900800z
Montefiori, M., Espley, R. V., Stevenson, D., Cooney, J., Datson, P. M., Saiz, A., et al. (2011). Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 65 (1), 106–118. doi: 10.1111/j.1365-313X.2010.04409.x
Nishiyama, I. (2007). Fruits of the actinidia genus. Adv. Food Nutr. Res. 52, 293–324. doi: 10.1016/S1043-4526(06)52006-6
Pan, X., Ding, J.-H., Zhao, S.-Q., Shi, H.-C., Miao, W.-L., Wu, F.-A., et al. (2023). Identification and functional study of detoxification-related genes in response to tolfenpyrad stress in Glyphodes pyloalis Walker (Lepidoptera Pyralidae). Pestic. Biochem. Physiol. 194, 105503. doi: 10.1016/j.pestbp.2023.105503
Peng, Y., Lin-Wang, K., Cooney, J. M., Wang, T., Espley, R. V., Allan, A. C. (2019). Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res. 6, 3. doi: 10.1038/s41438-018-0076-4
Pilkington, S. M., Montefiori, M., Jameson, P. E., Allan, A. C. (2012). The control of chlorophyll levels in maturing kiwifruit. Planta 236 (5), 1615–1628. doi: 10.1007/s00425-012-1723-x
Richardson, D. P., Ansell, J., Drummond, L. N. (2018). The nutritional and health attributes of kiwifruit: a review. Eur. J. Nutr. 57 (8), 2659–2676. doi: 10.1007/s00394-018-1627-z
Shao, D., Liang, Q., Wang, X., Zhu, Q. H., Liu, F., Li, Y., et al. (2022). Comparative metabolome and transcriptome analysis of anthocyanin biosynthesis in white and pink petals of cotton (Gossypium hirsutum L.). Int. J. Mol. Sci. 23 (17), 10137. doi: 10.3390/ijms231710137
Shu, P., Zhang, Z., Wu, Y., Chen, Y., Li, K., Deng, H., et al. (2023). A comprehensive metabolic map reveals major quality regulations in red-flesh kiwifruit (Actinidia chinensis). New Phytol. 238 (5), 2064–2079. doi: 10.1111/nph.18840
Smeriglio, A., Barreca, D., Bellocco, E., Trombetta, D. (2016). Chemistry, pharmacology and health benefits of anthocyanins. Phytother. Res. 30 (8), 1265–1286. doi: 10.1002/ptr.5642
Sui, L., Liu, Y., Zhong, C., Huang, H. (2013). Geographical distribution and morphological diversity of red-fleshed kiwifruit germplasm (Actinidia chinensis Planchon) in China. Genet. Resour. Crop Evol. 60 (6), 1873–1883. doi: 10.1007/s10722-013-9961-8
Sun, H., Zhang, P., Zhu, Y., Lou, Q., He, S. (2018). Antioxidant and prebiotic activity of five peonidin-based anthocyanins extracted from purple sweet potato (Ipomoea batatas (L.) Lam.). Sci. Rep. 8 (1), 5018. doi: 10.1038/s41598-018-23397-0
Tanaka, Y., Sasaki, N., Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54 (4), 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Valenta, K., Kalbitzer, U., Razafimandimby, D., Omeja, P., Ayasse, M., Chapman, C. A., et al. (2018). The evolution of fruit colour: phylogeny, abiotic factors and the role of mutualists. Sci. Rep. 8 (1), 14302. doi: 10.1038/s41598-018-32604-x
Wan, S., Li, C., Ma, X., Luo, K. (2017). PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 36 (8), 1263–1276. doi: 10.1007/s00299-017-2151-y
Wang, W. Q., Moss, S. M. A., Zeng, L., Espley, R. V., Wang, T., Lin-Wang, K., et al. (2022a). The red flesh of kiwifruit is differentially controlled by specific activation-repression systems. New Phytol. 235 (2), 630–645. doi: 10.1111/nph.18122
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99 (2), 359–378. doi: 10.1111/tpj.14330
Wang, Y., Wang, Y., Sun, J., Dai, Y., Yang, F., Jiang, H., et al. (2022b). Metabolomic and transcriptomic analysis of flavonoid biosynthesis in two main cultivars of Actinidia arguta sieb.zucc. grown in Northern China. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.911203
Wang, Y., Zhang, M., Dong, K., Yin, X., Hao, C., Zhang, W., et al. (2022c). Metabolomic and transcriptomic exploration of the uric acid-reducing flavonoids biosynthetic pathways in the fruit of Actinidia arguta Sieb. Zucc. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1025317
Ye, L. X., Luo, M. M., Wang, Z., Bai, F. X., Luo, X., Gao, L., et al. (2022). Genome-wide analysis of MADS-box gene family in kiwifruit (Actinidia chinensis var. chinensis) and their potential role in floral sex differentiation. Front. Genet. 13. doi: 10.3389/fgene.2022.1043178
Young, M.D., Wakefield, M.J., Smyth, G.K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2) R14. doi: 10.1186/gb-2010-11-2-r14.
Yu, X., Tang, W., Yang, Y., Zhang, J., Lu, Y. (2020). Comparative transcriptome analysis revealing the different germination process in aryloxyphenoxypropionate-resistant and app-susceptible asia minor bluegrass (polypogon fugax). Plants (Basel) 9 (9), 1191. doi: 10.3390/plants9091191
Zhang, H., Yang, B., Liu, J., Guo, D., Hou, J., Chen, S., et al. (2017). Analysis of structural genes and key transcription factors related to anthocyanin biosynthesis in potato tubers. Sci. Hortic. 225, 310–316. doi: 10.1016/j.scienta.2017.07.018
Zhao, P., Li, X., Jia, J., Yuan, G., Chen, S., Qi, D., et al. (2018). bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy. J. Exp. Bot. 70 (1), 269–284. doi: 10.1093/jxb/ery335
Zhao, R., Song, X., Yang, N., Chen, L., Xiang, L., Liu, X.-Q., et al. (2020). Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 39 (7), 891–907. doi: 10.1007/s00299-020-02537-9
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82 (1), 105–121. doi: 10.1111/tpj.12792
Zhu, Z., Wang, H., Wang, Y., Guan, S., Wang, F., Tang, J., et al. (2015). Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 66 (13), 3775–3789. doi: 10.1093/jxb/erv173
Zhuang, H., Lou, Q., Liu, H., Han, H., Wang, Q., Tang, Z., et al. (2019). Differential regulation of anthocyanins in green and purple turnips revealed by combined de novo transcriptome and metabolome analysis. Int. J. Mol. Sci. 20 (18), 4387. doi: 10.3390/ijms20184387
Keywords: Actinidia arguta, transcriptome, metabolome, red fruit, post-harvest, anthocyanin, cyanidin
Citation: Ye L, Bai F, Zhang L, Luo M, Gao L, Wang Z, Peng J, Chen Q and Luo X (2023) Transcriptome and metabolome analyses of anthocyanin biosynthesis in post-harvest fruits of a full red-type kiwifruit (Actinidia arguta) ‘Jinhongguan’. Front. Plant Sci. 14:1280970. doi: 10.3389/fpls.2023.1280970
Received: 21 August 2023; Accepted: 22 September 2023;
Published: 09 October 2023.
Edited by:
Ana Margarida Fortes, University of Lisbon, PortugalReviewed by:
Xianbao Deng, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Ye, Bai, Zhang, Luo, Gao, Wang, Peng, Chen and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghong Chen, a2l3aWhiQDE2My5jb20=; Xuan Luo, bHVveHVhbkBoYmFhcy5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.